- 1Department of Biology, University of Hartford, West Hartford, CT, United States
- 2Department of Computing Sciences, University of Hartford, West Hartford, CT, United States
Staphylococcus aureus is an opportunistic human pathogen that can frequently be found at various body locations, such as the upper respiratory tract, nostrils, skin, and perineum. S. aureus is responsible for causing a variety of conditions, which range from minor skin infections and food poisoning to life-threatening sepsis and endocarditis. Furthermore, S. aureus has developed resistance to numerous antimicrobial agents, which has made treatment of S. aureus infections difficult. In the present study, we examined lifestyle factors that could increase the likelihood of S. aureus carriage, the overall prevalence of S. aureus, as well as assessed the antibiotic resistance profiles of the S. aureus isolates among a population of college students. Five hundred nasal samples were collected and analyzed via selective growth media, coagulase and protein A testing, as well as polymerase chain reaction and DNA sequencing. One hundred four out of the 500 samples collected (21%) were identified as containing S. aureus. The S. aureus isolates were resistant to penicillin (74%), azithromycin (34%), cefoxitin (5%), ciprofloxacin (5%), tetracycline (4%), and trimethoprim (1%), but sensitive to gentamicin and rifampin. Lastly, we identified several lifestyle factors (i.e., pet exposure, time spent at the university recreational facility, musical instrument usage, and tobacco usage) positively correlated with S. aureus nasal colonization.
Introduction
The opportunistic pathogen, S. aureus, colonizes approximately 20% - 40% of the general population, while 20% - 60% are intermittent carriers (Kluytmans et al., 1997; Kuehnert et al., 2006; Chambers and DeLeo, 2009; Sakwinska et al., 2009; Warnke et al., 2014). Nasal carriage of S. aureus varies depending upon the population studied, method of sampling, and identification techniques utilized (Kluytmans et al., 1997). Usually, S. aureus results in minor skin infections but has the propensity to develop into life-threatening conditions. Studies have shown that individuals who have had recent surgery, experienced invasive medical device usage, or exhibit a weakened immune system, are much more susceptible to S. aureus infections (Kluytmans et al., 1997; Lowy, 1998). Hematogenous dissemination of S. aureus can result in endocarditis, osteomyelitis, septic arthritis, or epidural abscesses (Archer, 1998).
Previous reports have shown that asymptomatic individuals who carry S. aureus are at a greater risk for infection (Miles et al., 1944; Kluytmans et al., 1997; von Eiff et al., 2001) and can serve as a reservoir of transmittance via direct contact or through fomites (Lu et al., 2005; Jaradat et al., 2020). S. aureus is difficult to treat due to its high propensity to develop antibiotic resistance (Foster, 2017). In the decades following its discovery, methicillin resistant S. aureus (MRSA) (Jevons, 1961), which is resistant to β-lactam antibiotics, has been frequently implicated in healthcare-associated infections (Chambers and DeLeo, 2009; Krishnamurthy et al., 2014). However, studies have shown the emergence of MRSA transmission and infection in those without previous health-care contact, which is referred to as community-associated MRSA (CA-MRSA) (DeLeo et al., 2010; Udo, 2013). Therefore, there is a need to identify potential life-style factors that can contribute to S. aureus carriage, which could be used to mitigate transmission and subsequent infection.
The aim of the present study was to examine the prevalence of S. aureus among college-aged students and assess several life-style factors that could contribute to its nasal carriage. Among the S. aureus isolates we obtained, we assessed their antibiotic resistance to penicillin, azithromycin, ciprofloxacin, cefoxitin, tetracycline, trimethoprim, gentamicin, and rifampin.
Materials and methods
Questionnaire of potential risk factors
The participants’ risk factors were collected using a standardized paper-based questionnaire (Supplemental Material). The following potential risk factors for S. aureus colonization were assessed: public transportation usage, tobacco usage, shaving and makeup application, piercings and tattoos, number of bathroom mates, sleep, sheet changes, pet exposure, type of housing, gym usage, athletics, musical instrument usage, age, gender, race, antibiotic exposure, healthcare exposure, and previous exposure to S. aureus.
Sample collection
In accordance with the University of Hartford Institutional Review Board, nasal swabs were collected from 500 undergraduate students at the University of Hartford between September 2019 and December 2019. Samples were obtained by inserting the tip of a sterile swab (BD BBL™ CultureSwab collection and transport system) along the inner edge of the participant’s nostril. The swab was then rotated while continuously circulating along the nasal vestibule for 5 sec and then this was repeated in the second nostril using the same swab (Warnke et al., 2014). The transport swab was then labeled and sealed until it was used to inoculate enrichment media.
Staphylococci enrichment
Swabs were placed in 14 ml snap-cap tubes containing 3 ml of staph broth (BD Difco) and incubated at 35° C for 20 – 24 h, shaking at 225 rpm. The following day, cultures were streaked onto mannitol salt agar (MSA) plates (Oxoid) and grown overnight at 35° C. Plates that were negative for mannitol fermentation were discarded. From the plates that turned yellow, a single colony was selected and re-streaked for isolation onto a subsequent MSA plate (if multiple colony types appeared on a single plate, each was re-streaked) and incubated overnight. The following day, a single well-isolated colony was used to make a frozen stock of the bacterial strain, as described below.
Glycerol storage
A single isolated colony was grown overnight in approximately 3 ml of trypticase soy broth (Remel) at 35° C shaking at 225 rpm. The following day, 500 μl of the culture was added to 500 μl of 50% glycerol in a 2 ml cryovial tube, vortexed for 30 sec, and stored at -80° C.
Coagulase/protein A testing
Strains were streaked for isolation onto trypticase soy agar (TSA) plates from the frozen stocks and incubated overnight at 35° C. The Staphaurex™ Latex Agglutination Test was used according to the manufacturer’s instructions.
Kirby Bauer disk diffusion susceptibility test
Samples were streaked for isolation onto TSA plates and incubated at 35° C overnight. The following morning, three to four bacterial colonies were suspended in sterile PBS (Gibco) in an autoclaved 10 mm x 100 mm glass tube. The suspension was then compared to a 0.5 McFarland standard. If needed, additional cells were added to achieve a similar turbidity. A sterile swab was then dipped into the suspension and used to inoculate a Mueller Hinton agar plate (Oxoid). The plates were allowed to dry for 5 to 15 min. The following antibiotic disks were used: (Oxoid Antimicrobial Susceptibility Test Disks) penicillin G (10 IU), cefoxitin (30 µg), tetracycline (30 µg), trimethoprim (5 µg), azithromycin (15 µg), ciprofloxacin (5 µg), gentamicin (10 µg), and rifampin (5 µg). Each disk was placed on the plate equidistant from each other using sterile forceps. Plates were then incubated at 35° C for 16 - 18 h (24 h for cefoxitin). Once incubation was complete, the diameter of the zones of inhibition were measured in mm. The values were compared to the Clinical and Laboratory Standards Institute supplement M100 (CLSI, 2021) chart to determine sensitivity or resistance. Zone breakpoints for determining resistance were as follows: penicillin (≤ 28 mm), trimethoprim (≤ 10 mm), tetracycline (≤ 14 mm), cefoxitin (≤ 24 mm), azithromycin (≤ 13 mm), gentamicin (≤ 12 mm), ciprofloxacin (≤ 15 mm), and rifampin (≤ 16 mm). The assay was performed in triplicate and the average zone of inhibition was used to initially screen for resistance.
DNA isolation
Strains were streaked for isolation onto TSA plates and incubated at 35° C overnight. One-quarter loopful (10 μl Inoculating loops) of freshly streaked bacterial colonies were suspended in a 1.5 ml microcentrifuge tube containing 200 µl of 5% Chelex (BIO-RAD) in TE buffer. The tubes were vortexed so that no clumps were present and placed in a 100° C heating block for 10 min. Samples were vortexed for 30 sec, placed on ice, then subsequently centrifuged at 15,000 rcf for 10 min. 100 µl of the supernatant were transferred into a new 1.5 ml microcentrifuge tube. DNA concentrations and purity were assessed using a Nanodrop (DeNovix DS-11FX) and samples were then subsequently stored at -20° C.
Polymerase chain reaction
Each reaction mixture contained < 250 ng of DNA, 1x GoTaq®polymerase (Promega), 0.4 µM of each primer in a final volume of 25 µl. The amplification conditions were as follows: (i) 5 min at 95°; (ii) 30 cycles of 30 s at 95° C, 30 s at X (Table S1), and 30 s for every 0.5 kb of amplicon (Table S1) at 72° C; (iii) 5 min at 72° C. The PCR products were separated on a 0.8% agarose (American Bioanalytical) gel in TBE to verify amplification.
DNA sequencing
PCR amplicons were purified using the QIAquick PCR Purification Kit (QIAGEN) according to the manufacturer’s instructions. The samples were stored at - 20° C until used for sequencing. The 27F primer was used to obtain a partial sequence of the 16S rRNA gene. Sequences of at least 550 bp, which encompass the V1 – V3 regions of the gene, were compared with the NCBI database using BLASTN (Altschul et al., 1990). Sanger sequencing reactions were completed at the W. M. KECK Biotechnology Resource Laboratory at the Yale University School of Medicine, in New Haven, CT. Reactions contained 500 ng of template, 2 μl of 4 μM primer, in a total volume of 18 μl.
Data analysis
The python scipy library was used to calculate the Fisher Exact test null-hypothesis and probability (p-value) of observing the distribution for a non-associated lifestyle factor (Virtanen et al., 2020). Here, for example, a small p-value such as p = 0.0035 means there is a 0.03% chance of observing the distribution at random and a 99.7% chance there is an association with the factor and S. aureus carriage. Since we used survey data, which possesses inherent self-reporting bias, we used a p-value cutoff of 0.15 as the other 85% of the data lacks evidence for any association. Accounting for social desirability in the survey was not possible due to validation costs related to the determination of each of the variables. Instead, we report results with the understanding that some social desirability bias is inherent in the results and that variables like “using tobacco” are under reported (Althubaiti, 2016). We also calculated and report the relative risk for the most significant associations with S. aureus. For additional details, see Supplemental Materials: Data Analysis.
Results
Staphylococci identification
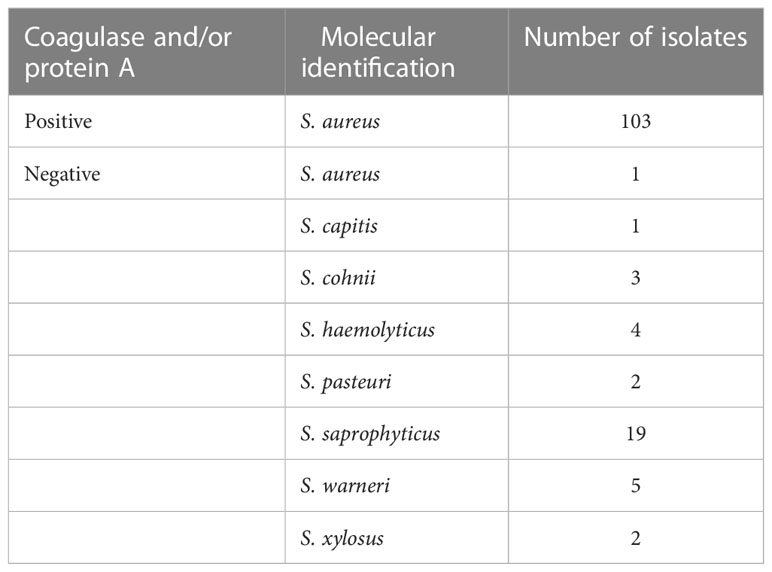
To determine the number of individuals in our study who were carrying S. aureus, after an overnight enrichment in staph broth, nasal swab samples were streaked onto mannitol salt agar plates to screen for S. aureus. Of the 500 samples collected, 140 were positive for mannitol fermentation as indicated by the change in media color from red to yellow (Table 1). We next wanted to confirm the suspected identity of these isolates as S. aureus by determining the presence of coagulase and/or protein A using a latex agglutination assay. Surprisingly, only 103 of the 140 isolates were positive for coagulase and/or protein A (Table 1). To verify the identity of the 103 isolates as S. aureus and to determine the identity of the coagulase-negative staphylococci (CoNS), we PCR amplified and partially sequenced the 16S rRNA gene for each of the 140 isolates (Table 1). Molecular identification verified the 103 coagulase and/or protein A-positive isolates as S. aureus as well as identified one of the coagulase and/or protein A-negative isolates as S. aureus (Table 1). Therefore, 104 out of the 500 individuals sampled (21%) were positive for S. aureus. The remaining 36 isolates were identified as the following CoNS: Staphylococcus capitis, Staphylococcus cohnii, Staphylococcus haemolyticus, Staphylococcus pasteuri, Staphylococcus saprophyticus, Staphylococcus warneri, and Staphylococcus xylosus isolates (Table 1).
Antimicrobial resistance among S. aureus isolates
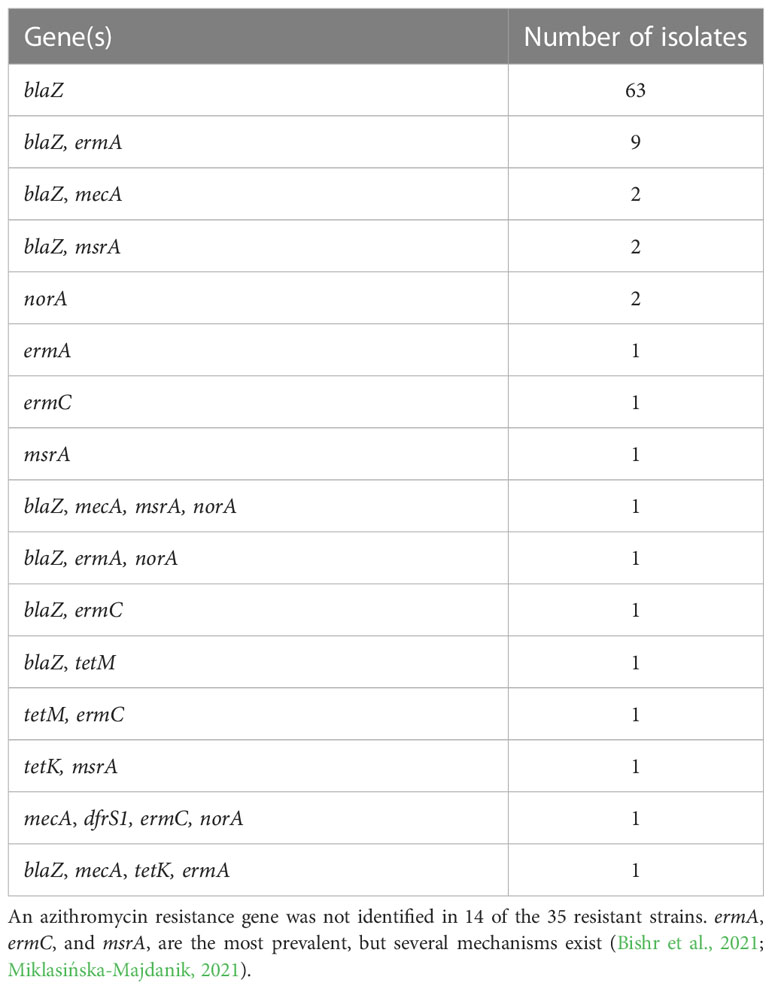
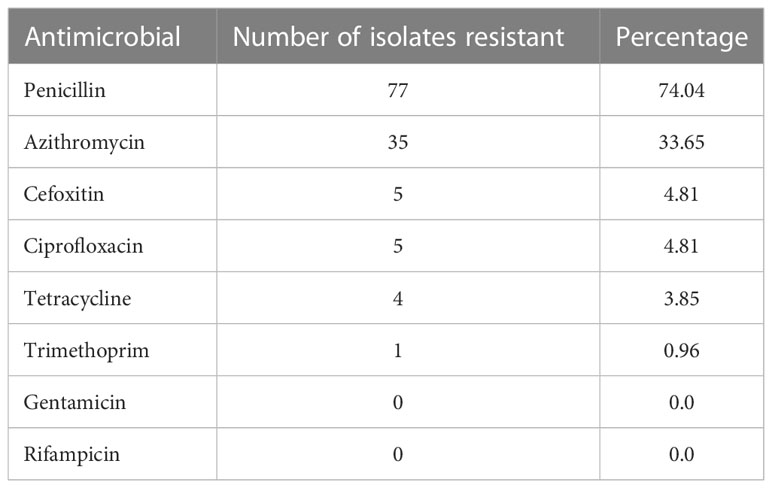
To determine the antimicrobial resistance profiles among the S. aureus isolates, we utilized the Kirby-Bauer disk diffusion susceptibility test to first screen for resistance to penicillin, cefoxitin, tetracycline, and trimethoprim. Cefoxitin is used to detect methicillin resistance as it is a better inducer of the methicillin resistance gene, mecA (CLSI, 2021). Resistance was then confirmed via PCR and sequencing of genes known to confer resistance to penicillin (blaZ), cefoxitin (mecA), tetracycline (tetM, K, O, L), trimethoprim (dfrS1), ciprofloxacin (norA), and azithromycin (ermA, ermC, msrA) (Table 2). Of the 104 S. aureus isolates, 74%, 5%, 4%, and 1% were resistant to penicillin, cefoxitin, tetracycline, and trimethoprim, respectively (Table 3). Of the five MRSA isolates, four were identified as the USA300 clone of CA-MRSA.
Lifestyle factors that contribute to S. aureus carriage
Several lifestyle factors were examined via a questionnaire to determine if there was an association between an activity and S. aureus nasal carriage. Students with pet exposure were 38% (FET, p = 0.08) more likely to carry S. aureus. Students who attended a recreational facility more than once a week for at least 30 min were 35% (FET, p = 0.06) more likely to be colonized by S. aureus. Students who play a musical instrument were 37% (FET, p = 0.08) more likely to carry S. aureus than those who do not. Lastly, tobacco product usage was the most significant factor detected in this study, revealing that smokers were 78% (FET, p = 0.005) more likely to carry S. aureus compared to non-smokers.
Discussion
In the current study, we evaluated the prevalence of S. aureus for 500 college-aged students, aged 17-25 years, that attended the University of Hartford (West Hartford, CT) in fall of 2019. Of the S. aureus isolates, we determined their antibiotic resistance to azithromycin, ciprofloxacin, cefoxitin, tetracycline, trimethoprim, gentamicin, and rifampin. Finally, we assessed various lifestyle factors that we believed could increase the likelihood of S. aureus carriage.
Recent publications report high rates of S. aureus nasal carriage among healthcare workers (80.3%) (Rukan et al., 2021) and individuals with atopic dermatitis (69.8%) (Blicharz et al., 2020). These populations are either at a higher risk for S. aureus exposure or potentially have compromised immune systems, which could more easily permit S. aureus carriage. Recent studies investigating the prevalence of S. aureus in cohorts of university students report nasal carriage rates ranging from 10% (Stancu et al., 2020) to 33% (Morita et al., 2007). Our assumption is that colonization rates among populations that have increased sensitivity to S. aureus colonization or are thought to be more regularly exposed to it, most likely have higher nasal carriage rates. S. aureus nasal carriage is often reported between 20-30% for the general population (Kavanagh et al., 2018) and our data suggests that colonization rates for University of Hartford students appear on the lower end of the spectrum (21%). We contend that the cohort we studied were less sensitive to S. aureus colonization because they are younger, ostensibly healthy individuals, and were generally not exposed to healthcare facilities.
Several factors, such as geographic location, age, body site sampled, identification techniques utilized, and sample size of the study, may contribute to the large variation in S. aureus colonization rates reported among studies. For example, mannitol salt agar was originally developed in the 1940s as a rapid tool to identify S. aureus in a clinical setting (Chapman, 1945). Phenotypic tests such as this are still used in developing countries where resources are limited. However, studies have revealed that some mannitol-fermenting CoNS can be misidentified as S. aureus when solely relying on this medium (Martinez et al., 1992; Merlino et al., 1996; Kawamura et al., 1998; Zadik et al., 2001; Ayeni and Odumosu, 2016; Ayeni et al., 2017; Thakur et al., 2017). Our work reinforces the results of those previous studies, as 36 of our 140 mannitol-positive isolates were indeed CoNS. The ramifications of relying on MSA alone are twofold: studies assessing S. aureus prevalence within the population could have numerous false positives and the current clinical relevance of CoNS could be undervalued due to misidentification. Consequently, certain species of CoNS are a larger health concern than previously thought (Heilmann et al., 2019). For example, S. haemolyticus S. capitis, and S. saprophyticus, have been implicated in bacteremia, neonatal sepsis, and urinary-tract infections, respectively (Becker et al., 2014; Cameron et al., 2015; Argemi et al., 2019). While our data strengthens the notion that certain members of CoNS can be misidentified as S. aureus in the clinical setting, CoNS are most likely more prevalent within the population than what was detected by our study. One study detected S. haemolyticus, S. capitis, S. hominis, S. warneri, and S. lugdunensis in 44%, 41%, 41%, 32%, and 27% of patients sampled, respectively (Kaspar et al., 2016). The higher prevalence of CoNS found in their study is most likely due to the more vigorous enrichment procedures they performed post sampling. The importance of these members of the nasal microbiota should not be overlooked due to their capacity to serve as opportunistic pathogens (Heilmann et al., 2019), reservoirs of antibiotic resistance for more pathogenic organisms (i.e., S. aureus) (Xu et al., 2018), or conversely, some of the nonpathogenic CoNS could benefit human health by potentially serving as antagonists for S. aureus colonization (Sakr et al., 2018).
Despite the re-emergence of penicillin susceptible S. aureus, methicillin resistant S. aureus (MRSA) is still a major global health concern, as approximately 53% of S. aureus clinical isolates are resistant to methicillin in the U.S. (Lee et al., 2018). While MRSA tends to cause serious complications such as bloodstream infections, pneumonia, or surgical site infections among those who acquire it in healthcare settings, CA-MRSA is becoming a growing concern (Hiramatsu et al., 2002; DeLeo et al., 2010). For example, CA-MRSA is predominately responsible for most skin and soft tissue infections (SSTIs) presenting in the U.S. (Moran et al., 2006; Talan et al., 2011). The CDC reports that 2% of the general population carry MRSA as part of their normal flora and it has been suggested that MRSA is replacing methicillin-susceptible S. aureus (MSSA) among our natural flora (Hiramatsu et al., 2002), which has potentially led to a higher prevalence of CA-MRSA infections (Udo, 2013; Lee et al., 2018). In our study, among our sampled population, 1% (5 out of 500) possessed MRSA, which comprised approximately 5% of the S. aureus isolates, which is consistent with previous studies that detected 0.8% - 9.2% of study participants carried MRSA (Suggs et al., 1999; Creech et al., 2005). Additionally, we were able to determine that four of the five MRSA isolates in our study were the highly virulent and transmissible CA-MRSA clone, USA300 (Pan et al., 2005). Monitoring CA-MRSA and identifying transmission factors is an important step in controlling the rapid dissemination of CA-MRSA among the population. It is believed that CA-MRSA is more easily transmitted than HA-MRSA and possesses greater pathogenic capability (Hiramatsu et al., 2002; DeLeo et al., 2010).
Previous studies have demonstrated various lifestyle activities that can increase an individual’s likelihood of carrying S. aureus. Our analyses revealed that pet exposure, time spent at the recreational facility, musical instrument usage, and tobacco usage, led to an increase in S. aureus nasal carriage. While human S. aureus carriage rates are between 20-30%, rates within populations of livestock can be exceedingly higher, with up to 90% of chickens, 42% of pigs, 29% of sheep and 35% of cattle being carriers (Haag et al., 2019). Rates of S. aureus prevalence are markedly lower in domesticated animals, with 17.5% (Chai et al., 2021) to 19.17% of cats (Bierowiec et al., 2016) and between 4.9% (Sahin-Tóth et al., 2021) and 20% (Chai et al., 2021) of dogs being carriers. Evidence supports the bidirectional transfer of S. aureus from humans to pets via licking, biting and other types of human-pet interactions (Davis et al., 2012). Our survey questions did not interrogate what type(s) of pets our participants were exposed to, but our assumption was that any increased exposure to either livestock or domestic animals would predict S. aureus carriage.
Previous studies have reported that both MSSA and MRSA strains have repeatedly been found on surfaces within exercise facilities with MSSA rates varying from 10% (Markley et al., 2012) to 78.3% (Bilung et al., 2018) to 90.6% (Mukherjee et al., 2016) and MRSA rates at 37.5% (Mukherjee et al., 2016). Since, recreation facilities possess numerous high-contact fomites, such as multi-user equipment, locker rooms, and water fountains, it is not surprising that S. aureus nasal carriage correlated with frequency of gym use as suggested by our data. It is likely that fomite transmission is the mechanism of transmission for students in our study at the on-campus recreation center or at an off-campus facility.
We also observed that musical instrument usage is correlated with higher S. aureus nasal carriage. Previous work has found that S. aureus can survive on clarinets for up to 5 days (Marshall and McBryde, 2014) and it has been suggested that bacteria remain viable on frequently touched regions of string instruments (Gambichler et al., 2004). Our survey asked participants to identify whether they made facial contact with their instrument. Our findings suggest that both facial and non-facial contact with musical instruments positively predicts S. aureus carriage, most likely due to some degree of fomite transmission.
Lastly, we revealed that tobacco usage was the strongest positive predictor for S. aureus nasal carriage in our study. Cigarette smoke has been found to alter buccal cavity microbiota by attenuating tobacco-sensitive bacterial growth and promoting the growth of tobacco-resistant bacteria (Grine et al., 2019). Specifically, tobacco tar-resistant S. aureus was found to be enriched in the buccal cavity of smokers and caused the increase in expression of cytokines (Fujiki et al., 2004). Other studies revealed that tobacco usage alters biofilm formation (Kumar et al., 2011) and heavily alters the composition of the oral microbiome (Pushalkar et al., 2020). Our survey asked students to identify the frequency by which they used tobacco products but did not ask if those products were single use (cigarettes/cigars) or re-usable (electronic cigarettes). Previous studies suggest that electronic cigarettes are a harbor for bacteria (Lee et al., 2019) and potentially could act as a reservoir for S. aureus transmission. Our rational as to why tobacco usage is such a strong predictor of S. aureus nasal carriage is due to two phenomena; first we entertain the possibility at least some of our tobacco using participants were using electronic cigarettes, which can act as a reservoir for S. aureus transmission and secondly due to tobacco’s effects on both the nascent nasal microbiota (Jaspers, 2014; Pfeiffer et al., 2021) and immune system of the user (Martin et al., 2016; Qiu et al., 2017). We contend that these two factors might work in a synergistic manner that leads to the disruption of the nascent nasal microbiota. This loss of competitive exclusion then allows for opportunistic pathogens, such as S. aureus, to colonize and/or increase in relative abundance. For example, previous studies have suggested that other members of the nasal microbiota may outcompete or inhibit the growth of S. aureus (Sakr et al., 2018).
Our data suggest that pet exposure, time spent at a recreational facility, musical instrument usage, and tobacco usage, can lead to a heightened likelihood of carrying S. aureus. Of these, pets and high-contact fomites found at the recreational facility may directly act as a bi-directional reservoir that allows for the transmission of S. aureus between individuals. To a lesser extent, instruments, and tobacco (i.e., cigarettes and electronic cigarettes), could also serve as a bi-directional reservoir if these fomites are shared with others. It has been reported that up to 60% of individuals are transiently colonized by S. aureus (Kluytmans et al., 1997; Fournier and Philpott, 2010). It is plausible that the latter mentioned fomites serve as a reservoir that facilitates one’s own re-acquisition of S. aureus.
S. aureus is a continued global health concern that not only impacts those in healthcare facilities but also individuals within the community due to its multitude of virulence factors and propensity to acquire antibiotic resistance. Continued investigation into potential transmission mechanisms of S. aureus could help mitigate the spread of S. aureus, while ongoing surveillance of the ever-changing antimicrobial resistance landscape of S. aureus will be essential to guide empiric antimicrobial therapy. However, the key to controlling S. aureus colonization and subsequent pathogenesis may be found in factors that shape the nasal microbiota and subsequently antagonize or potentially leave the door open for S. aureus colonization.
Data availability statement
The sequencing data is available via GenBank Accession Numbers OQ626021 – OQ626160. The datasets presented in this study can be found in the article/Supplementary Material.
Ethics statement
This study was approved by the Institutional Review Board (IRB) of the University of Hartford. The patients/participants provided their written informed consent to participate in this study.
Author contributions
SC, JG, KM, MA, DT, YA-A, AP, TB, AS contributed to the conception and design of the study. SC, JG, KM, MA, DT, YA-A, AP, AS, OR performed experiments and collected data. TB performed the statistical analysis. SC, JG, AB, AS, TB, AS wrote sections of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Faculty and Student Research Grants from the College of Arts and Sciences Dean’s Office at the University of Hartford.
Acknowledgments
We are grateful to Dr. Preston P. Garcia of Castleton University for helpful discussion and manuscript review as well Kaelyn McFadden for technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1195758/full#supplementary-material
References
Althubaiti, A. (2016). Information bias in health research: definition, pitfalls, and adjustment methods. J. Multidiscip. Healthc. 9, 211–217. doi: 10.2147/JMDH.S104807
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 (3), 403–410. doi: 10.1016/S0022-2836(05)80360-2
Archer, G. L. (1998). Staphylococcus aureus: a well-armed pathogen. Clin. Infect. Dis. 26, 1179–1181. doi: 10.1086/520289
Argemi, X., Hansmann, Y., Prola, K., Prévost, G. (2019). Coagulase-negative staphylococci pathogenomics. Int. J. Mol. Sci. 20. doi: 10.3390/ijms20051215
Ayeni, F. A., Andersen, C., Nørskov-Lauritsen, N. (2017). Comparison of growth on mannitol salt agar, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, VITEK® 2 with partial sequencing of 16S rRNA gene for identification of coagulase-negative staphylococci. Microb. Pathog. 105, 255–259. doi: 10.1016/j.micpath.2017.02.034
Ayeni, F. A., Odumosu, B. T. (2016). False identification of other microorganisms as staphylococcus aureus in southern Nigeria. Trop. J. Pharm. Res. 15, 1941–1945. doi: 10.4314/tjpr.v15i9.19
Becker, K., Heilmann, C., Peters, G. (2014). Coagulase-negative staphylococci. Clin. Microbiol. Rev. 27, 870–926. doi: 10.1128/CMR.00109-13
Bierowiec, K., Płoneczka-Janeczko, K., Rypuła, K. (2016). Is the colonisation of staphylococcus aureus in pets associated with their close contact with owners? PloS One 11. doi: 10.1371/journal.pone.0156052
Bilung, L. M., Tahar, A. S., Kira, R., Mohd Rozali, A. A., Apun, K. (2018). High occurrence of staphylococcus aureus isolated from fitness equipment from selected gymnasiums. J. Environ. Public Health 2018. doi: 10.1155/2018/4592830
Bishr, A. S., Abdelaziz, S. M., Yahia, I. S., Yassien, M. A., Hassouna, N. A., Aboshanab, K. M. (2021). Association of macrolide resistance genotypes and synergistic antibiotic combinations for combating macrolide-resistant mrsa recovered from hospitalized patients. Biol. (Basel) 10. doi: 10.3390/biology10070624
Blicharz, L., Usarek, P., Młynarczyk, G., Skowroński, K., Rudnicka, L., Samochocki, Z. (2020). Nasal colonization by staphylococci and severity of atopic dermatitis. Dermatitis 31, 215–222. doi: 10.1097/DER.0000000000000568
Cameron, D. R., Jiang, J. H., Hassan, K. A., Elbourne, L. D. H., Tuck, K. L., Paulsen, I. T., et al. (2015). Insights on virulence from the complete genome of staphylococcus capitis. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00980
Chai, M. H., Sukiman, M. Z., Liew, Y. W., Shapawi, M. S., Roslan, F. S., Hashim, S. N., et al. (2021). Detection, molecular characterization, and antibiogram of multi-drug resistant and methicillin-resistant staphylococcus aureus (MRSA) isolated from pets and pet owners in Malaysia. Iran J. Vet. Res. 22, 277–287. doi: 10.22099/ijvr.2021.39586.5752
Chambers, H. F., DeLeo, F. R. (2009). Waves of resistance: staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7, 629–641. doi: 10.1038/nrmicro2200
Chapman, G. H. (1945). THE SIGNIFICANCE OF SODIUM CHLORIDE IN STUDIES OF STAPHYLOCOCCI. J. Bact. 50 (2), 201–203. doi: 10.1128/jb.50.2.201-203.1945
CLSI (2021). “Performance standards for antimicrobial susceptibility testing,” in CLSI supplement M100, 31st ed (Clinical and Laboratory Standards Institute).
Creech, C. B., Kernodle, D. S., Alsentzer, A., Wilson, C., Edwards, K. M. (2005). Increasing rates of nasal carriage of methicillin-resistant staphylococcus aureus in healthy children. Pediatr. Infect. Dis. J. 24, 617–621. doi: 10.1097/01.inf.0000168746.62226.a4
Davis, M. F., Iverson, S. A., Baron, P., Vasse, A., Silbergeld, E. K., Lautenbach, E., et al. (2012). Household transmission of meticillin-resistant staphylococcus aureus and other staphylococci. Lancet Infect. Dis. 12, 703–716. doi: 10.1016/S1473-3099(12)70156-1
DeLeo, F. R., Otto, M., Kreiswirth, B. N., Chambers, H. F. (2010). Community-associated meticillin-resistant staphylococcus aureus. Lancet 375, 1557–1568. doi: 10.1016/S0140-6736(09)61999-1
Foster, T. J. (2017). Antibiotic resistance in staphylococcus aureus. current status and future prospects. FEMS Microbiol. Rev. 41, 430–449. doi: 10.1093/femsre/fux007
Fournier, B., Philpott, D. (2010). Reconocimiento de carácteristicas de la degradación de pasturas en el rancho San Luis - morelia - caquetá - Colombia. Rev. Fac Cienc. Agropecu.
Fujiki, H., Takeuchi, H., Nishitani, N., Yamanaka, H., Suzuki, K., Kurusu, M., et al. (2004). Carcinogenic potential of tobacco tar-resistant staphylococcus aureus in buccal cavity. J. Cancer Res. Clin. Oncol. 130, 301–305. doi: 10.1007/s00432-004-0554-y
Gambichler, T., Boms, S., Freitag, M. (2004). Contact dermatitis and other skin conditions in instrumental musicians. BMC Dermatol. 4, 3. doi: 10.1186/1471-5945-4-3
Grine, G., Royer, A., Terrer, E., Diallo, O. O., Drancourt, M., Aboudharam, G. (2019). Tobacco smoking affects the salivary gram-positive bacterial population. Front. Public Health 7. doi: 10.3389/fpubh.2019.00196
Haag, A. F., Fitzgerald, J. R., Penadés, J. R. (2019). Staphylococcus aureus in animals. Microbiol. Spectr. 7. doi: 10.1128/microbiolspec.gpp3-0060-2019
Heilmann, C., Ziebuhr, W., Becker, K. (2019). Are coagulase-negative staphylococci virulent? Clin. Microbiol. Infect. 25, 1071–1080. doi: 10.1016/j.cmi.2018.11.012
Hiramatsu, K., Okuma, K., Ma, X. X., Yamamoto, M., Hori, S., Kapi, M. (2002). New trends in Staphylococcus aureus infections: glycopeptide resistance in hospital and methicillin resistance in the community. Curr. Opin. Infect. Dis. 15 (4), 407–413. doi: 10.1097/00001432-200208000-00009
Jaradat, Z. W., Ababneh, Q. O., Sha’aban, S. T., Alkofahi, A. A., Assaleh, D., Al Shara, A. (2020). Methicillin resistant staphylococcus aureus and public fomites: a review. Pathog. Glob Health 114, 426–450. doi: 10.1080/20477724.2020.1824112
Jaspers, I. (2014). Cigarette smoke effects on innate immune mechanisms in the nasal mucosa: potential effects on the microbiome. Ann. Am. Thorac. Soc. 11, S38–S43. doi: 10.1513/AnnalsATS.201306-154MG
Jevons, P. M. (1961). Celbenin-resistant staphylococci. Br. Med. J 124–125. doi: 10.1136/bmj.1.5219.124-a
Kaspar, U., Kriegeskorte, A., Schubert, T., Peters, G., Rudack, C., Pieper, D. H., et al. (2016). The culturome of the human nose habitats reveals individual bacterial fingerprint patterns. Environ. Microbiol. 18, 2130–2142. doi: 10.1111/1462-2920.12891
Kavanagh, K. T., Abusalem, S., Calderon, L. E. (2018). View point: gaps in the current guidelines for the prevention of methicillin-resistant staphylococcus aureus surgical site infections 11 medical and health sciences 1103 clinical sciences. Antimicrob. Resist. Infect. Control 7. doi: 10.1186/s13756-018-0407-0
Kawamura, Y., Hou, X., Sultana, F., Hirose, K., Miyake, M., Shu, S., et al. (1998). Distribution of staphylococcus species among human clinical specimens and emended description of staphylococcus caprae. J. Clin. Microbiol. 2038–2042. doi: 10.1128/JCM.36.7.2038-2042.1998
Kluytmans, J., Van Belkum, A., Verbrugh, H. (1997). Nasal carriage of staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10 (3), 505–520. doi: 10.1128/CMR.10.3.505
Krishnamurthy, V., Saha, A., Renushri, B. V., Nagaraj, E. R. (2014). Methicillin resistant staphylococcus aureus carriage, antibiotic resistance and molecular pathogenicity among healthy individuals exposed and not exposed to hospital environment. J. Clin. Diagn. Res. 8, 4–8. doi: 10.7860/JCDR/2014/8409.4638
Kuehnert, M. J., Kruszon-Moran, D., Hill, H., McQuillan, G. (2006). Prevalence ofStaphylococcus aureusNasalColonization in the united states 2001–2002. J. Infect. Dis. 193, 172–179. doi: 10.1086/499632
Kumar, P. S., Matthews, C. R., Joshi, V., de Jager, M., Aspiras, M. (2011). Tobacco smoking affects bacterial acquisition and colonization in oral biofilms. Infect. Immun. 79, 4730–4738. doi: 10.1128/IAI.05371-11
Lee, M. S., Allen, J. G., Christiani, D. C. (2019). Endotoxin and (1→3)-β-D-glucan contamination in electronic cigarette products sold in the united states. Environ. Health Perspect. 127. doi: 10.1289/EHP3469
Lee, A. S., de Lencastre, H., Garau, J., Kluytmans, J., Malhotra-Kumar, S., Peschel, A., et al. (2018). Methicillin-resistant staphylococcus aureus. Nat. Rev. Dis. Primers 4, 18033. doi: 10.1038/nrdp.2018.33
Lowy, F. D. (1998). Staphylococcus aureus infections. N Engl. J. Med. 339, 520–531. doi: 10.1056/NEJM199808203390806
Lu, P. L., Chin, L. C., Peng, C. F., Chiang, Y. H., Chen, T. P., Ma, L., et al. (2005). Risk factors and molecular analysis of community methicillin-resistant staphylococcus aureus carriage. J. Clin. Microbiol. 43, 132–139. doi: 10.1128/JCM.43.1.132-139.2005
Markley, J. D., Edmond, M. B., Major, Y., Bearman, G., Stevens, M. P. (2012). Are gym surfaces reservoirs for staphylococcus aureus? a point prevalence survey. Am. J. Infect. Control 40, 1008–1009. doi: 10.1016/j.ajic.2012.01.015
Marshall, C., McBryde, E. (2014). The role of staphylococcus aureus carriage in the pathogenesis of bloodstream infection. BMC Res. Notes 7. doi: 10.1186/1756-0500-7-428
Martin, E. M., Clapp, P. W., Rebuli, M. E., Pawlak, E. A., Glista-Baker, E., Benowitz, N. L., et al. (2016). E-cigarette use results in suppression of immune and inflammatory-response genes in nasal epithelial cells similar to cigarette smoke. Am. J. Physiol. Lung Cell Mol. Physiol. 311, L135–L144. doi: 10.1152/ajplung.00170.2016
Martinez, O. V., Cleary, T., Baker, M., Civetta, J. (1992). Evaluation of a mannitol-Salt-Oxacillin-Tellurite medium for the isolation of methicillin-resistant staphylococcus aureus from contaminated sources. Diagn. Microbiol. Infect. Dis. 15, 207–211. doi: 10.1016/0732-8893(92)90115-A
Merlino, J., Gill, R., Robertson, G. J. (1996). Application of lipovitellin-Salt-Mannitol agar for screening, isolation, and presumptive identification of staphylococcus aureus in a teaching hospital. J. Clin. Microbiol. 1996, 3012–3015. doi: 10.1128/jcm.34.12.3012-3015.1996
Miklasińska-Majdanik, M. (2021). Mechanisms of resistance to macrolide antibiotics among staphylococcus aureus. Antibiotics 10. doi: 10.3390/antibiotics10111406
Miles, A. A., Williams, R. E. 0., Clayton-Cooper, B. (1944). The carriage of staphylococcus (Pyogenes) aureus in man and its relation to wound infection. 513–524. doi: 10.1002/path.1700560405
Moran, G. J., Krishnadasan, A., Gorwitz, R. J., Fosheim, G. E., Mcdougal, L. K., Carey, R. B., et al. (2006). Methicillin-resistant s. aureus infections among patients in the emergency department. N. Engl. J. Med. 355, 666–674. doi: 10.1056/NEJMoa055356
Morita, J. E., Fujioka, R. S., Tice, A. D., Berestecky, J., Sato, D., Seifried, S. E., et al. (2007). Survey of methicillin-resistant staphylococcus aureus (MRSA) carriage in healthy college students, hawai’i Hawaiian medicinal plants view project microbial source tracking tools view project. Hawai’i Med. J. 66, 213–215.
Mukherjee, N., Sulaiman, I. M., Banerjee, P. (2016). Characterization of methicillin-resistant staphylococcus aureus isolates from fitness centers in the Memphis metropolitan area, Tennessee. Am. J. Infect. Control 44, 1681–1683. doi: 10.1016/j.ajic.2016.06.031
Pan, E., Biep, B., Charlebois, E., Auerswald, C., Carleton, H., Sensabaugh, G., et al. (2005). Population dynamics of nasal strains of methicillin-resistant staphylococcus aureus-and their relation to community-associated disease activity. J. Infect. Dis. 192, 811–818. doi: 10.1086/432072
Pfeiffer, S., Herzmann, C., Gaede, K. I., Kovacevic, D., Krauss-Etschmann, S., Schloter, M. (2021). Different responses of the oral, nasal and lung microbiomes to cigarette smoke. Thorax 8, 191–195. doi: 10.1136/thoraxjnl-2020-216153
Pushalkar, S., Paul, B., Li, Q., Yang, J., Vasconcelos, R., Makwana, S., et al. (2020). Electronic cigarette aerosol modulates the oral microbiome and increases risk of infection. iScience 23. doi: 10.1016/j.isci.2020.100884
Qiu, F., Liang, C.-L., Liu, H., Zeng, Y.-Q., Hou, S., Huang, S., et al. (2017). Oncotarget 268 www.impactjournals.com/oncotarget impacts of cigarette smoking on immune responsiveness: up and down or upside down? Oncotarget. 8, 268–284. doi: 10.18632/oncotarget.13613
Rukan, M., Jamil, H., Bokhari, H. A., Khattak, A. A., Khan, A. N., Ullah, Z., et al. (2021). Nasal carriage of highly resistant methicillin resistant staphylococcus aureus (mrsa) strains by hospital staff in hazara region of pakistan. J. Pak Med. Assoc. 71, 47–50. doi: 10.47391/JPMA.177
Sahin-Tóth, J., Kovács, E., Tóthpál, A., Juhász, J., Forró, B., Bányai, K., et al. (2021). Whole genome sequencing of coagulase positive staphylococci from a dog-and-owner screening survey. PloS One 16, e0245351. doi: 10.1371/journal.pone.0245351
Sakr, A., Brégeon, F., Mège, J. L., Rolain, J. M., Blin, O. (2018). Staphylococcus aureus nasal colonization: an update on mechanisms, epidemiology, risk factors, and subsequent infections. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02419
Sakwinska, O., Kuhn, G., Balmelli, C., Francioli, P., Giddey, M., Perreten, V., et al. (2009). Genetic diversity and ecological success of staphylococcus aureus strains colonizing humans. Appl. Environ. Microbiol. 75, 175–183. doi: 10.1128/AEM.01860-08
Stancu, M. S., Smeding, C., Negut, A. C. (2020). The carrier status of streptococcus pneumoniae in a multi-national medical student population. Ann. Ig 32, 65–71. doi: 10.7416/ai.2020.2331
Suggs, A. H., Maranan, M. C., Boyle-Vavra, S., Daum, R. S. (1999). Methicillin-resistant and borderline methicillin-resistant asymptomatic staphylococcus aureus colonization in children without identifiable risk factors. Pediatr. Infect. Dis. J. 18, 410–414. doi: 10.1097/00006454-199905000-00003
Talan, D. A., Krishnadasan, A., Gorwitz, R. J., Fosheim, G. E., Limbago, B., Albrecht, V., et al. (2011). Comparison of staphylococcus aureus from skin and soft-tissue infections in us emergency department patients 2004 and 2008. Clin. Infect. Dis. 53, 144–149. doi: 10.1093/cid/cir308
Thakur, P., Nayyar, C., Tak, V., Saigal, K. (2017). Mannitol-fermenting and tube coagulase-negative staphylococcal isolates: unraveling the diagnostic dilemma. J. Lab. Physicians 9. doi: 10.4103/0974-2727.187926
Udo, E. E. (2013). Community-acquired methicillin-resistant staphylococcus aureus: the new face of an old foe? Med. Principles Pract. 22, 20–29. doi: 10.1159/000354201
Virtanen, P., Gommers, R., Oliphant, T. E., Haberland, M., Reddy, T., Cournapeau, D., et al. (2020). SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272. doi: 10.1038/s41592-019-0686-2
von Eiff, C., Arsten, K., Ecker, B., Onstanze, K., Achka, M., Eorg, G., et al. (2001). Nasal carriage as a source of staphylococcus aureus bacteremia. N. Engl. J. Med. 344, 11–16. doi: 10.1056/NEJM200101043440102
Warnke, P., Harnack, T., Ottl, P., Kundt, G., Podbielski, A. (2014). Nasal screening for staphylococcus aureus - daily routine with improvement potentials. PloS One 9. doi: 10.1371/journal.pone.0089667
Xu, Z., Shah, H. N., Misra, R., Chen, J., Zhang, W., Liu, Y., et al. (2018). The prevalence, antibiotic resistance and mecA characterization of coagulase negative staphylococci recovered from non-healthcare settings in London, UK. Antimicrob. Resist. Infect. Control 7. doi: 10.1186/s13756-018-0367-4
Keywords: Staphylococcus aureus, nasal carriage Stapylococcus aureus., antibiotic resistance, life-style factors, prevalence, methicillin-resistant Staphylococcus aureus (MRSA)
Citation: Congdon ST, Guaglione JA, Ricketts OMA, Murphy KV, Anderson MG, Trowbridge DA, Al-Abduladheem Y, Phillips AM, Beausoleil AM, Stanley AJ, Becker TJ and Silver AC (2023) Prevalence and antibiotic resistance of Staphylococcus aureus associated with a college-aged cohort: life-style factors that contribute to nasal carriage. Front. Cell. Infect. Microbiol. 13:1195758. doi: 10.3389/fcimb.2023.1195758
Received: 28 March 2023; Accepted: 08 June 2023;
Published: 27 June 2023.
Edited by:
Daniel Czyz, University of Florida, United StatesReviewed by:
Edet E. Udo, Kuwait University, KuwaitTimothy J. Foster, Trinity College Dublin, Ireland
Katarzyna Garbacz, Medical University of Gdansk, Poland
Copyright © 2023 Congdon, Guaglione, Ricketts, Murphy, Anderson, Trowbridge, Al-Abduladheem, Phillips, Beausoleil, Stanley, Becker and Silver. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adam C. Silver, YXNpbHZlckBoYXJ0Zm9yZC5lZHU=
 Sean T. Congdon1
Sean T. Congdon1 Timothy J. Becker
Timothy J. Becker Adam C. Silver
Adam C. Silver