
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 02 June 2023
Sec. Molecular Bacterial Pathogenesis
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1190859
This article is part of the Research Topic The Role of Bacterial Quorum Sensing and Quorum Quenching in Pathogenic Microorganisms View all 4 articles
Introduction: Burkholderia thailandensis is a study model for Burkholderia pseudomallei, a highly virulent pathogen, known to be the causative agent of melioidosis and a potential bioterrorism agent. These two bacteria use an (acyl-homoserine lactone) AHL-mediated quorum sensing (QS) system to regulate different behaviors including biofilm formation, secondary metabolite productions, and motility.
Methods: Using an enzyme-based quorum quenching (QQ) strategy, with the lactonase SsoPox having the best activity on B. thailandensis AHLs, we evaluated the importance of QS in B. thailandensis by combining proteomic and phenotypic analyses.
Results: We demonstrated that QS disruption largely affects overall bacterial behavior including motility, proteolytic activity, and antimicrobial molecule production. We further showed that QQ treatment drastically decreases B. thailandensis bactericidal activity against two bacteria (Chromobacterium violaceum and Staphylococcus aureus), while a spectacular increase in antifungal activity was observed against fungi and yeast (Aspergillus niger, Fusarium graminearum and Saccharomyces cerevisiae).
Discussion: This study provides evidence that QS is of prime interest when it comes to understanding the virulence of Burkholderia species and developing alternative treatments.
Most bacteria use a molecular communication system, referred to as quorum sensing (QS). It relies on the production and detection of small molecules known as autoinducers. QS enables the adaptation of bacterial behaviors in a cell density-dependent manner and allows bacteria to act cooperatively and induce different physiological functions (Mion et al., 2019). Among the mechanisms activated by QS, bioluminescence was the first described in Vibrio sp. (Tobias et al., 2020). Other mechanisms were later described, such as biofilm formation, virulence factor production, and antibiotic production (Zhao et al., 2020). Many autoinducers have been identified, including oligopeptides which are mainly used by Gram-positive bacteria, acyl-homoserine lactones (AHLs) by Gram-negative bacteria, and the diester AI-2 which is used by both (Rémy et al., 2018; Tobias et al., 2020). A wide panel of AHLs has been reported. These molecules share a lactone ring but their lateral acyl chain varies in term of modifications and/or length (Billot et al., 2020). Depending on the bacterial species, one or several specific AHLs can be produced and recognized by the cells to trigger virulence and biofilm formation . Biofilm formation allows bacteria to settle and limits the effectiveness of antimicrobial treatments (Rémy et al., 2018; Bowler et al., 2020). To counteract AHL-mediated bacterial virulence, strategies to disrupt QS, referred to as quorum quenching (QQ), have been developed. Various approaches have been reported, such as the use of natural or synthetic QS inhibitors (Rémy et al., 2018), autoinducer sequesters (Grandclément et al., 2016; Rémy et al., 2018) and QQ enzymes (QQEs), which can inactivate autoinducers (Rémy et al., 2018). One major advantage of QQ is that this strategy interferes with QS to prevent noxious behaviors without killing bacteria, thus minimizing the appearance of resistance mechanisms (Rémy et al., 2018). QQEs are particularly promising in light of their capacity to act catalytically and extracellularly for efficient QS disruption (Billot et al., 2020).
Among the QQEs, the archaeal enzyme SsoPox (Hiblot et al., 2013) has been particularly investigated over the past decade considering its lactonase activity against a large panel of AHLs (Hiblot et al., 2013) and its tremendous resistance to temperature, process and long-term storage (Del Vecchio et al., 2009; Rémy et al., 2016). Enzyme engineering approaches made it possible to generate several SsoPox variants with enhanced properties (Billot et al., 2022). The QQ potential of these variants has been demonstrated against different bacterial strains, including human pathogens (such as Pseudomonas aeruginosa) and environmental strains (such as Chromobacterium violaceum). Significant phenotypic alterations upon SsoPox treatment have been reported, including biofilm disruption (Rémy et al., 2020; Mion et al., 2021), reduced virulence in in vitro (Rémy et al., 2020; Mion et al., 2021) and in vivo models (Hraiech et al., 2014), as well as modulation of antimicrobial production (Mion et al., 2021).
Here, the QQ effect of SsoPox against Burkholderia thailandensis was evaluated. Besides some rare cases of human infections (Chang et al., 2017; Gee et al., 2018), B. thailandensis is of special interest as it constitutes a relevant study model for Burkholderia pseudomallei due to their high genome relatedness (Majerczyk et al., 2014b). B. pseudomallei is the causative agent of melioidosis (Klaus et al., 2018) and a potential bioterrorism agent (Oliveira et al., 2020). B. thailandensis constitutes a safe surrogate for studying and tackling B. pseudomallei virulence. Three QS systems were identified in B. thailandensis, each system being composed of an AHL synthase and a receptor to produce and sense a specific AHL signal. These three systems are shared with B. pseudomallei (Majerczyk et al., 2014b), QS-1 (BtaI1-BtaR1) QS-2 (BtaI2-BtaR2) and QS-3 (BtaI3-BtaR3) related to the production of C8-HSL, 3-OH-C10-HSL and 3-OH-C8-HSL respectively (Majerczyk et al., 2014b). In addition, two orphan receptors were also identified (Majerczyk et al., 2014a) (BtaR4 or MalR (Gupta et al., 2017) and BtaR5). QS regulates various phenotypes in B. thailandensis such as biofilm formation (Tseng et al., 2016), motility (Ulrich et al., 2004) and antimicrobial molecules including bactobolin (Klaus, 2020), 4-hydroxy-3-methyl-2-alkenylquinoline (Majerczyk et al., 2014a; Majerczyk et al., 2014b) and malleilactone (Majerczyk et al., 2014a).
In this study, the effect of SsoPox V82I, an improved monovariant of SsoPox with enhanced lactonase activity (Billot et al., 2022), was selected and evaluated on B. thailandensis. The impact of SsoPox V82I lactonase was assessed through a proteomic approach complemented by phenotypic analysis. The results obtained show that SsoPox V82I treatment significantly disturbs B. thailandensis proteome and QS-related phenotypes, including the modulation of antimicrobial compound production.
Burkholderia thailandensis E264 (ATCC 700388), Staphylococcus aureus clinical isolate, Chromobacterium violaceum ATCC 12472 were used in this study. All bacteria were grown at 37°C in Lysogeny Broth (LB) (10 g.L-1 NaCl, 10 g.L-1 tryptone, 5 g.L-1 yeast extract, pH 7), except for B. thailandensis which was cultivated in standard succinate media (Meyer, 1978) (6 g.L-1 K2HPO4, 3 g.L-1 KH2PO4, 0.2 g.L-1 MgSO4.7H2O, 1 g.L-1 [NH4]2SO4, 4 g.L-1 succinic acid, pH adjusted at 7 with NaOH). B. thailandensis was inoculated from a single colony and pre-cultivated for 6 hours at 37°C in LB. The pre-culture was diluted 1/1,000 in SSM and 3 mL were added to a 12 well plate. Cultures were incubated for 16 hours with shaking (350 rpm). SsoPox enzyme was added at a final concentration of 0.5 mg.mL-1 in the culture. Saccharomyces cerevisiae DSM 1333 was cultivated in Yeast extract-Peptone-Dextrose (YPD) medium (Mion et al., 2021) (10 g.L-1 yeast extract, 20 g.L-1 peptone, 10 g.L-1 glucose, pH 6) at 30°C and Fusarium graminearum DSM 1096, Aspergillus niger DSM 2182 were cultivated in Potato-Dextrose broth (PDB, pH 5) (Sigma Aldrich, USA) at 25°C, respectively.
In this study, SsoPox V82I was used as lactonase enzyme. Enzyme production was performed, as described previously (Hiblot et al., 2013; Hraiech et al., 2014), using Escherichia coli BL21 (DE3)-pGro7/GroEL strain containing a SsoPox V82I plasmid. Precultures were incubated at 37°C for 16 hours in LB supplemented with ampicillin (100 mg.mL-1) and chloramphenicol (34 mg.mL-1). Bacteria were then inoculated at 1/100 in ZYP-5052 autoinducer medium supplemented with ampicillin and chloramphenicol, and incubated at 37°C while being stirred (180 rpm). When cultures reached an OD600 nm between 0.8 and 1, bacteria were induced by adding 0.2% L-arabinose for chaperon production and CoCl2 at 0.2 mM final, to stabilize the active site of SsoPox V82I enzyme, the temperature was also decreased to 23°C and cultures were grown for 20 hours at 180 rpm. Cells were harvested by centrifugation at 4,000 x g for 20 minutes at 10°C. Pellets were solubilized in lysis buffer (HEPES 50 mM pH 8, NaCl 150 mM, DNAseI 10 µg.mL-1, lysozyme 0.25 mg.mL-1 and PMSF 0.1 mM). Cells in the lysis buffer were stored at -80°C during at least 16 hours. Thawed cells were sonicated three times for 30 seconds, with an amplitude of 45% (Q700 sonicator®, QSonica, USA). Finally, cells were heated at 80°C for 30 minutes and then centrifuged to pellet the debris. The supernatant was collected and saturated with 75% of ammonium sulfate at 4°C for 16 hours to precipitate SsoPox V82I. The enzyme was pelleted down by centrifugation at 10,000 x g for 15 minutes at 10°C. The pellet was resuspended in 8 mL of SsoPox buffer (HEPES 50 mM, NaCl 150 mM and pH 8) and then filtered at 0.8 µm. Ammonium sulfate was removed via desalting (HiPrep 26/10 desalting, ÄKTA pure, GE Healthcare, USA). The sample was concentrated using 30 kDa centricon and then injected into a size-exclusion chromatography column (GF Hiload 16/600 Superdex 75pg) to purify the enzyme. Enzyme purity was then checked by SDS-PAGE and the concentration was measured using a Bradford assay.
Lactonase kinetic parameters was identified for four SsoPox enzymes: SsoPox W263F, SsoPox W263I, SsoPox V82I-A275G, and SsoPox V82I. The hydrolysis of lactones (C8-HSL, 3-OH-C8-HSL and 3-OH-C10-HSL) was performed in a lactonase buffer (2.5 mM Bicine pH 8.3, 150 mM NaCl, 0.2 mM CoCl2, 0.25 mM Cresol purple and 0.5% DMSO, pH adjusted at 8.3 with NaOH), in a range of concentrations between 0 and 2 mM. The reaction of AHL degradation was followed at 577 nm (SynergyHT, BioTek, USA). Mean values were fitted to the Michaelis-Menten equation using Graph-Pad Prism 7.04 software to obtain the catalytic parameters.
To identify AHLs production in the liquid medium, a B. thailandensis 20 mL culture, both untreated and treated with lactonase, was grown for 16 hours. Supernatants were collected by a centrifugation for 10 minutes at 10,000 × g, then 0.22 µm filtered and freeze-dried for 48 hours. The sample was solubilized in 2 mL of ultrapure water. AHLs extraction was performed by adding the same volume of ethyl acetate (v/v) and energetically vortexing the sample. The organic phase was transferred to a glass tube and dried with a nitrogen evaporator. Finally, the dried sample was resuspended in 20 µL in DMSO to concentrate the sample 1,000 times (Rémy et al., 2020). The presence of AHLs in the extract was investigated using the reporter strain E. coli MT102. This strain expresses gfp in response to AHLs detection (Andersen et al., 2001). The reporter strain was pre-cultivated overnight in LB supplemented with tetracycline 10 µg.mL-1 at 30°C, then diluted to 1/10 in fresh LB. A volume of 195 µL of culture was distributed in a 96 well-plate and supplemented with 5 µL of the B. thailandensis supernatant extract. The fluorescence was followed every 20 minutes for 16 hours using a plate reader with an excitation wavelength of 485 nm and emission detection at 528 nm.
Biofilm formation was measured by crystal violet (Sigma®) staining (Hoffman et al., 2005). Planktonic cells were carefully removed from the 12 well plates. Wells were washed with phosphate buffered saline (PBS) and completely dried at 37°C. Then 3 mL of 0.05% crystal violet was added to stain the attached biofilm. Crystal violet was gently removed, and the wells were washed with PBS. The fixed crystal violet was finally dissolved in 3 mL of absolute ethanol, 200 µL were transferred to a 96-well plate, and absorbance was quantified in a microplate reader at 595 nm.
Cells treated with lactonase or not were harvested, and their proteins were extracted. Four biological replicates were treated for each condition. The protein extracts were diluted with MilliQ water and NuPAGE LDS 3X sample buffer (Invitrogen, USA) supplemented with -mercaptoethanol to obtain a final concentration of proteins of 0.5 µg.µL-1 in LDS1X. The samples were heated to 99 ˚C for 5 minutes. For each sample, 20 μL (i.e. 10 µg of proteins) were subjected to a short (5 minutes) denaturing electrophoresis on a NuPAGE 4%-12% gradient gel in MES SDS running buffer (50 mM MES ([2-(N-morpholino) ethane sulfonic acid), 50 mM Tris Base, 0.1% SDS, 1 mM EDTA, pH 7.3). After electrophoresis, the gel was briefly stained with SimplyBlue SafeStain (ThermoFisher, USA) and washed extensively with milliQ water. Each proteome was extracted as a single polyacrylamide band. Each sample was processed as previously described (Hartmann et al., 2014) and then proteolyzed with trypsin Gold (Promega, USA) in the presence of ProteaseMax detergent (Promega, USA). A volume of 2 µL of the resulting peptide mixture (50 µL) was injected in a nanoscale C18 PepMap100 capillary column (3 µm, 100 Å, 75 µm id x 50 cm, LC Packings) and resolved with a 90-minutes gradient of acetonitrile (3.2%-20% for 75 minutes followed by 20%-32% for 15 minutes), 0.1% formic acid, at a flow rate of 0.2 µL.min-1. The peptides eluting from the column were identified by tandem mass spectrometry with a Q-Exactive HF mass spectrometer (ThermoFisher, USA) operated in data-dependent acquisition mode essentially, as described (Klein et al., 2016). The full scan of peptide ions was acquired at a resolution of 60,000 from m/z 350 to 1,500 and with a dynamic exclusion of 10 seconds. Each MS scan was followed by high-energy collisional dissociation and MS/MS scans at a resolution of 15,000 on the 20 most abundant precursor ions, selecting only ions with a charge of 2+ or 3+. MS/MS spectra were assigned to peptide sequences by the MASCOT Daemon 2.3.2 search engine (Matrix Science) using the B. thailandensis ATCC 700388 annotated genome, comprising 5,096 entries and totaling 1,694,428 residues. The search parameters were standard: trypsin as enzyme, a maximum of two possible miss-cleavages, peptide p-value below 0.05, oxidation of methionine as variable modification, carbamidomethylation of cysteine as fixed modification, and tolerances of 5 ppm and 0.2 Da for the MS and MS/MS signals, respectively. A protein was considered validated when at least two different peptides were detected (p-value below 0.05), resulting in a protein identification false discovery rate below 1%, as verified with a reverse decoy database search. Spectral counts (number of MS/MS spectra assigned per protein) were used as proxy for the abundance of the proteins in each condition. Protein abundances in the two conditions and all biological replicates were compared with the PatternLab Tfold procedure (Carvalho et al., 2012; PMID: 22539673). Each protein sequence with a relative abundance between two conditions of Tfold above 1.5 and p-value below 0.05 was selected as differentially detected as previously recommended (Gouveia et al., 2020; PMID: 32133743). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD040565 and 10.6019/PXD040565.
Treated and untreated condition were analyzed using Partial Least Squares discriminant analysis (PLS-DA) as previously described (Mion et al., 2021). Proteins with a fold change ≥ 1.5 and statistical p-value < 0.05 were considered as significantly impacted by the lactonase and were classified using the Burkholderia Genome Database (Winsor et al., 2008). The violin plot was constructed using ggplot2 and vioplot packages R software (R-4.1.1) (Wickham, 2016; Adler et al., 2022).
Proteolytic activity was determined by skimmed milk assay (Vial et al., 2008) on LB agar plates (10 g.L-1 NaCl, 10 g.L-1 tryptone, 5 g.L-1 yeast extract, 1.5 g.L-1 agar, pH 7) containing 2.8% of skimmed milk powder. After 16 hours of incubation, the supernatant of B. thailandensis was collected by centrifugation at 10,000 g for 10 minutes and was filtered through a 0.22 µm filter. Holes were punched in SSM agar plates and a volume of 200 µL of filtered supernatant was poured into. The plates were incubated at room temperature for at least 24 hours. Proteolytic activity was determined by measuring the clear zone surrounding the wells. Each condition was performed in triplicates.
Swarming motility was monitored on 0.7% SSM agar (pH adjusted at 7 with NAOH) by inoculating 3 µL of bacteria into the center of the plate. The SSM agar was supplemented with SsoPox V82I lactonase when indicated, at a final concentration of 0.5 mg.mL-1. Plates were incubated for at least 48 hours at 37°C under high humidity. The motility area was measured on ImageJ.
The bactericidal effect of B. thailandensis supernatants was tested against C. violaceum and S. aureus. Supernatants from 16 hours cultures of B. thailandensis were collected by centrifugation at 10,000 x g for 10 minutes at room temperature and filtered through a 0.22 µm filter. Overnight precultures from C. violaceum and S. aureus were inoculated at 1/1,000 in fresh LB. A volume of 100 µL of culture was poured into 96-well plates supplemented with 100 µL of B. thailandensis supernatant, either untreated or treated with lactonase. The culture was grown at 37°C for 24 hours while being agitated (220 rpm). Cell growth was measured by OD 600 every 30 minutes using a plate reader (Infinite M nano,Tecan, Switzerland). To test whether the bactericidal effect remained without the B. thailandensis supernatant, the cells were then serial diluted in PBS and plated on an LB-agar plate. Colony-forming units (CFU) were counted after overnight incubation at 37°C.
The antifungal activity of B. thailandensis was determined by a modified microdilution assay (Piochon et al., 2020). Spores were collected from a Potato-Dextrose Agar (PDA) plate (PDB, 1.5% agar, pH 5) and homogenized by vigorous vortexing in 5 mL sterilized water with 10 µL of tween 80 (Arendrup et al., 2015). The OD 600 nm was then adjusted to 0.09-0.1 corresponding approximately to 105 spores.mL-1 (Arendrup et al., 2015). A volume of 750 µL of 10 x serial dilutions (10-1,10-2,10-3,10-4) was poured into 24-well plates with 750 µL of B. thailandensis filtered supernatants from a 16-hour culture, either untreated or treated with enzyme. Spore formation was observed after 96 hours of incubation at 25°C.
To determine the yeasticidal activity of B. thailandensis against S. cerevisiae, cultures of B. thailandensis either untreated or treated with enzyme, were collected and centrifuged at 10,000 x g for 10 minutes at room temperature. A volume of 750 µL of filtered supernatant was mixed with an equal volume of melted YPD-agar (10 g.L-1 yeast extract, 20 g.L-1 peptone, 10 g.L-1 glucose and 1.5% agar, pH 6). A volume of 10 µL of an overnight culture of S. cerevisiae and serial dilutions (10-2,10-3,10-4) was spotted on the dry YPD-agar mixed with supernatant. The plates were incubated for at least 24 hours at 30°C (Mion et al., 2021). In addition, cell-free supernatants from B. thailandensis were mixed with an equal volume (100 µL) of serial dilutions (10-2, 10-3, 10-4) from S. cerevisiae in 96-well plates to evaluate the yeasticidal effect in liquid culture. Growth was measured at OD 600 nm every 30 minutes for 24 hours at 30°C in a plate reader. Once late stationary phase was reached, cultures were serial-diluted in PBS and 10 µL was spotted on YPD-agar to evaluate the ability of the yeast to recover growth after 24 hours in competition with the supernatant. Agar plates were incubated at 30°C for 24 hours.
B. thailandensis antimicrobial molecules were extracted as previously described for AHLs. Here, 50 mL of culture medium were freeze-dried and resuspended in 3 mL of water and extracted with an equal volume of ethyl acetate. Organic phases were evaporated under nitrogen flow and resuspended in 50 µL of HPLC-grade methanol. Sterile SSM was used as blank.
Molecules were identified using liquid chromatography coupled with mass spectrometry (LC-MS). For each sample, stored at 4°C, 1 µL was injected into a reverse phase column (Acquity BEH C18 1.7 µm 2.1 × 50 mm, Waters, USA) maintained at 35°C. Compounds were eluted at 0.5 mL.min-1 using water and methanol (LC/MS-grade) supplemented with 0.1% of formic acid, with a gradient from 30% to 95% of methanol for 2minutes, 95% of methanol for one minute, and back to the initial composition for 1 minute. Compounds were ionized in the positive mode using a Zspray electrospray ion source: capillary/cone 1.5 kV/20 V, source/desolvation 120°C/250°C. Ions were then monitored using a High Definition MS(E) data independent acquisition method, and the following settings were executed: travelling wave ion mobility survey, 50 m/z–1000 m/z, 0.1 second scan time, 6 eV low energy ion transfer, and 20–40 eV high energy for collision-induced dissociation of all ions (low/high energy alternate scans). A lockmass correction was applied to adjust mass calibration within each run (Leucin Enkephalin 556.2766 m/z). Beforehand, the Vion instrument was calibrated using a Major Mix solution (Waters, USA) to calculate collision cross section (CCS) and mass-to-charge ration (m/z). 4D peaks, corresponding to a chromatographic retention time, ion mobility drift time and parents/fragments masses, were then collected from raw data using UNIFI software (version 1.9.4, Waters, USA). Chemical structures were targeted with the following parameters: 2 ppm m/z tolerance on parent adducts (H+), isotopic profile match of chlorine atoms, and 10 mDa m/z tolerance on common bactobolin structure fragments, 383.07712, 312.04000, 294.02944 and respective [M+H]+ C14H21Cl2N2O6, C11H16Cl2NO5 and C11H14Cl2NO4 identified in MoNa-MassBank of North America (https://mona.fiehnlab.ucdavis.edu/spectra/display/CCMSLIB00004679189).
For AHLs detection, biofilm formation, proteolytic activity and motility statistical analyses were performed using Student’s t-test in GraphPad Prism 7.04. The significance level (α), or the probability of committing a type I error was set at 0.05.
B. thailandensis was shown to produce three AHLs: C8-HSL, 3-OH-C8-HSL and 3-OH-C10-HSL (Majerczyk et al., 2014b). The ability of previously reported SsoPox variants (Hiblot et al., 2013; Billot et al., 2022) to degrade these lactones was thus evaluated. Kinetic parameters were determined on synthetic AHLs using a colorimetric assay with four SsoPox variants: SsoPox W263F, SsoPox W263I, SsoPox V82I-A275G and SsoPox V82I. SsoPox W263F efficiently degraded both AHLs with the hydroxyl group, 3-OH-C8-HSL and 3-OH-C10-HSL, with kcat/KM values reaching 2.79 × 103 and 3.49 × 103 M-1.s-1 respectively but had a poor activity on C8-HSL with a kcat/KM value of 7.39 × 101 M-1.s-1. SsoPox W263I showed a better activity on C8-HSL degradation, with a kcat/KM value reaching 5.75 × 103 M-1.s-1 as compared to SsoPox W263F, but lower kcat/KM were measured against the hydroxylated AHL, 1.88 × 102 and 2.10 × 102 M-1.s-1, for 3-OH-C8-HSL and 3-OH-C10-HSL respectively. SsoPox V82I-A275G variant showed the same profile of degradation as SsoPox W263I, with an improvement of kcat/KM values on 3-OH-C10-HSL. SsoPox V82I presented a similar profile to SsoPox W263F, with an improvement in kcat/KM values for C8-HSL and 3-OH-C8-HSL. SsoPox V82I had the best degradation profile on two synthetic AHL produced by B. thailandensis, with the highest activity on C8-HSL (4.72 × 104 M-1.s-1), as compared to the three other variants (Table 1). Therefore, SsoPox V82I was selected to further investigate the impact of QQ on B. thailandensis.
To confirm SsoPox V82I activity on natural AHLs produced by B. thailandensis in laboratory conditions, supernatants of B. thailandensis were collected after 16 hours of growth in standard succinate medium (SSM) and AHLs were extracted. The detection of AHLs in cell-free extracts was performed using the reporter strain E. coli MT102, which produces a fluorescence signal upon the perception of AHLs (Andersen et al., 2001). The fluorescence signal induced by the control with synthetic lactones, at 100 µM, confirmed the detection of the three synthetic AHLs. Furthermore, fluorescence signals were detected for cell-free extracts confirming AHLs production by B. thailandensis. AHLs degradation by the QQE, SsoPox V82I, was then confirmed, as the enzymatic treatment completely abolished the detection of AHLs by the reporter strain (Figure 1).
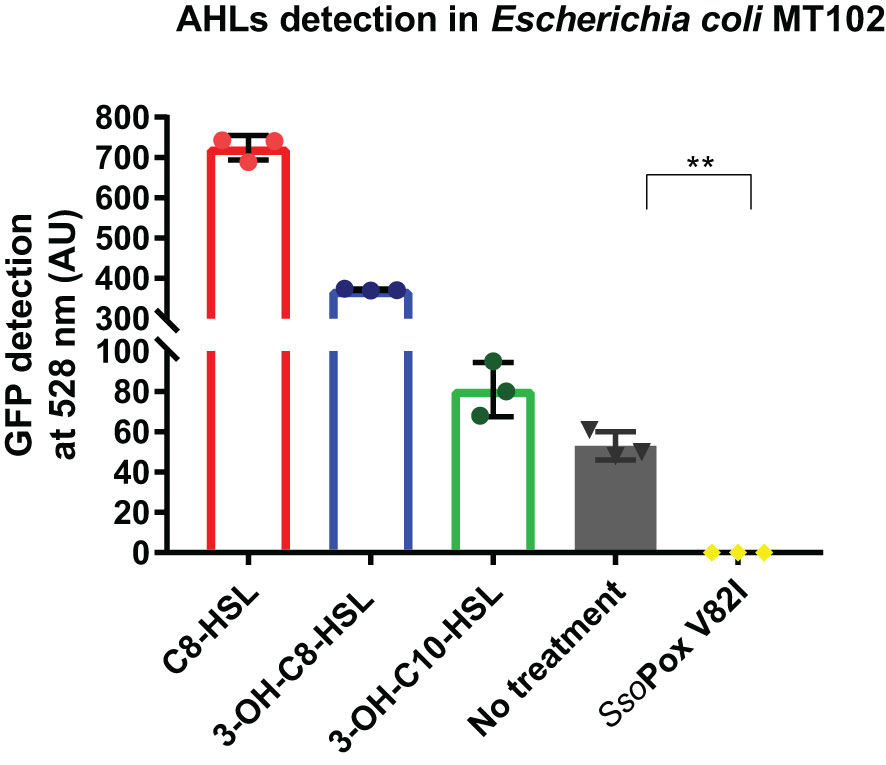
Figure 1 AHLs detection in cell-free extracts of B. thailandensis. AHL detection in B. thailandensis cell-free extracts using an E. coli MT102 reporter strain after cultivation without and with SsoPox V82I (0.5 mg.mL-1). C8-HSL, 3-OH-C8-HSL and 3-OH-C10-HSL were used as a positive control at 100 µM. Error bars represent standard deviations for n=3 biological replicates. **p-value < 0.01 according to Student’s t-test.
Finally, as biofilm formation and self-aggregation are known to be under QS regulation in B. thailandensis, these phenotypes were used to validate QQ by SsoPox V82I (Chandler et al., 2009; Tseng et al., 2016). Using crystal violet staining, a strong decrease in biofilm formation was measured when B. thailandensis was grown in the presence of SsoPox V82I at 0.5 mg.mL-1, reducing biofilm formation by more than 90% (Figure 2A). In addition, self-aggregation did not occur in cultures treated with SsoPox V82I lactonase, whereas in the untreated condition, aggregation was observed (Figure 2B). Taken as a whole, these experimental conditions were chosen to study QQ in B. thailandensis. To this end, a global approach combining proteomic and phenotypic analyses was implemented.
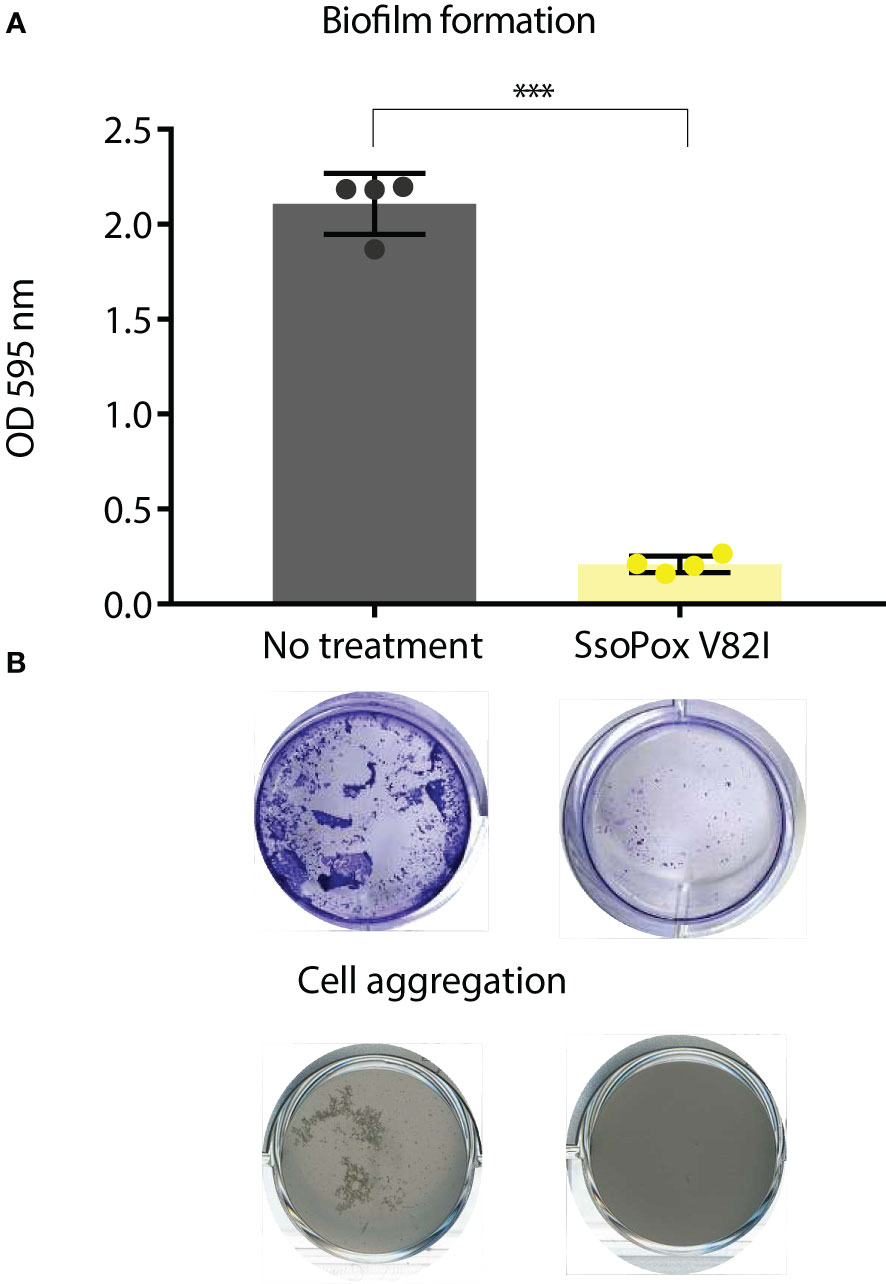
Figure 2 SsoPox V82I disrupts biofilm formation and aggregation in B. thailandensis. (A) Mean levels of biofilm formation by B. thailandensis without enzymatic treatment (gray) and treated with 0.5 mg.mL-1 SsoPox V82I (yellow). Error bars represent the standard deviations of n=4 biological replicates. ***p-value < 0.001 according to Student’s t-test. Representative pictures of crystal violet staining of biofilm formation are presented below each condition. (B) Cell aggregation in shaken culture treated with SsoPox V82I (0.5 mg.mL-1) or untreated after 16 hours culture at 37°C.
In order to get a complete overview of changes involved by QQ with SsoPox V82I on B. thailandensis, proteomic analysis was undertaken. The complete proteome of B. thailandensis comprises 5,562 proteins (Proteome ID UP000001930). In this study, 2,495 proteins (i.e. 44.9%) were detected and identified (Supplementary Data 1). Using a fold change of ≥ 1.5 and statistical p-value < 0.05 as selection criteria, 1,017 proteins (i.e. 18.3% of the entire proteome and 40.8% of the detected proteome) were identified as significantly impacted by SsoPox V82I. Statistical PLS-DA analysis revealed that the enzymatic treatment is the main separation criterion between the two conditions and accounts for 93% of the total variance (Figure 3A) indicating the huge impact that QQ and QS have upon B. thailandensis proteome.
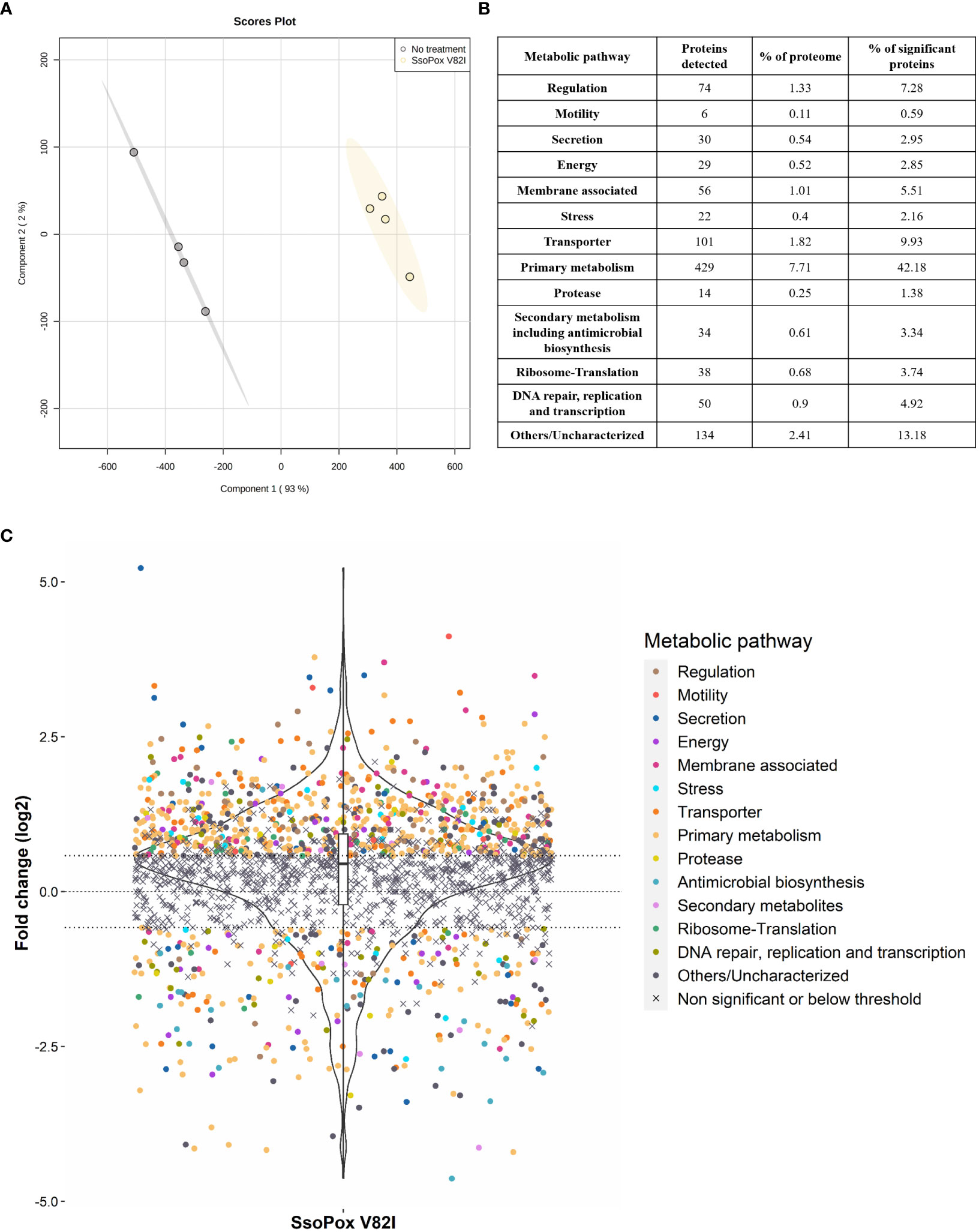
Figure 3 B. thailandensis E264 proteome changes upon SsoPox V82I treatment. (A) PLS-DA analysis of proteomic analysis of n=4 biological replicates. Untreated samples are represented in gray and samples treated with SsoPox V82I (0.5 mg.mL-1) are in yellow. (B) Table summarizing the number of proteins impacted by SsoPox V82I treatment and their respective proportion to the B. thailandensis proteome or number of significantly impacted proteins. (C) Representation of proteomic fold changes as a violin plot. Logarithmic fold changes of 2,495 proteins are plotted and proteins are classified according to their functions. The central line represents a null fold change, meaning no modification occurs with SsoPox V82I treatment. The x=0.58 and x=-0.58 lines correspond respectively to a 50% increase or decrease.
Clustering by biological function based on the Burkholderia Genome Database (Winsor et al., 2008) was applied to significant proteins and 13 groups were identified (Figure 3B). Among the significantly impacted proteins, 72% were upregulated, while only 28% were downregulated, indicating that treatment with SsoPox V82I and QQ led to wide changes in protein regulation and activated numerous pathways (Figure 3C). Proteins involved in primary metabolism pathways including amino acid biosynthesis, fatty acid metabolism, sugar metabolism, purine metabolism and energy metabolism, were upregulated for over half of these. Several proteins associated with secondary metabolite pathways, including antimicrobial biosynthesis, were identified and were mostly downregulated, except for six proteins. QQ treatment also impacted proteins involved in secretion, transporter and membrane-associated proteins, with 50%, 73% and 84% of upregulated proteins, respectively. Proteins involved in motility and protease were observed and were mainly upregulated (84% and 57%, respectively). Proteins involved in translation, ribosome formation, DNA repair, replication, transcription and regulation were also globally upregulated (Figures 3B, C).
Interestingly, among the proteins related to the QS systems, only the BtaR3 protein (WP_009896076.1) was identified in the proteomic dataset and was downregulated (fold change of -2.53) by SsoPox V82I treatment. BtaR3 is related to QS-3 and responsible for 3-OH-C8-HSL production. Previous studies have suggested that QS-1 induces QS-3 (Majerczyk et al., 2014a). In addition, several proteins which were detected were previously described as being under QS regulation, such as proteins detected in the secondary metabolites class (Supplementary Data 1). In addition, two proteins identified as belonging to secretion system and membrane-associated classes being altered upon lactonase treatment, namely WP_009893771.1 (-7.29 fold change) and WP_009889485.1 (1.64 fold change), were also described as potentially being involved in biofilm formation (Mao et al., 2017).
SsoPox V82I lactonase treatment led to several shifts in the B. thailandensis proteome. Proteomic analyses identified proteins involved in protease production and motility as being altered upon lactonase treatment (Figure 3C).
Regarding the protease class, 13 proteins belonged to protease enzymes and one protein was described as a protease inhibitor (WP_011401901.1). Eight of these proteins were upregulated with SsoPox V82I treatment (Figure 4A). Most proteases altered by lactonase treatment belonged to the metalloprotease, aspartic protease, or serine protease classification. Two proteins were identified as belonging to the M48 family peptidase (WP_009893638.1 and WP_009889905.1), and proteins in this family are described as HtpX homologs, proteins able to degrade casein(Yoshitani et al., 2019). To support the proteomic results, in vitro assays were performed using B. thailandensis cell-free supernatants and skimmed milk agar to assess protease activity by casein degradation. Supernatants from untreated cultures showed a weak ability to degrade casein. In comparison, supernatants obtained from lactonase treated cultures harbored an increased capacity to degrade casein, with a degradation halo which was 70% larger (Figure 4B).
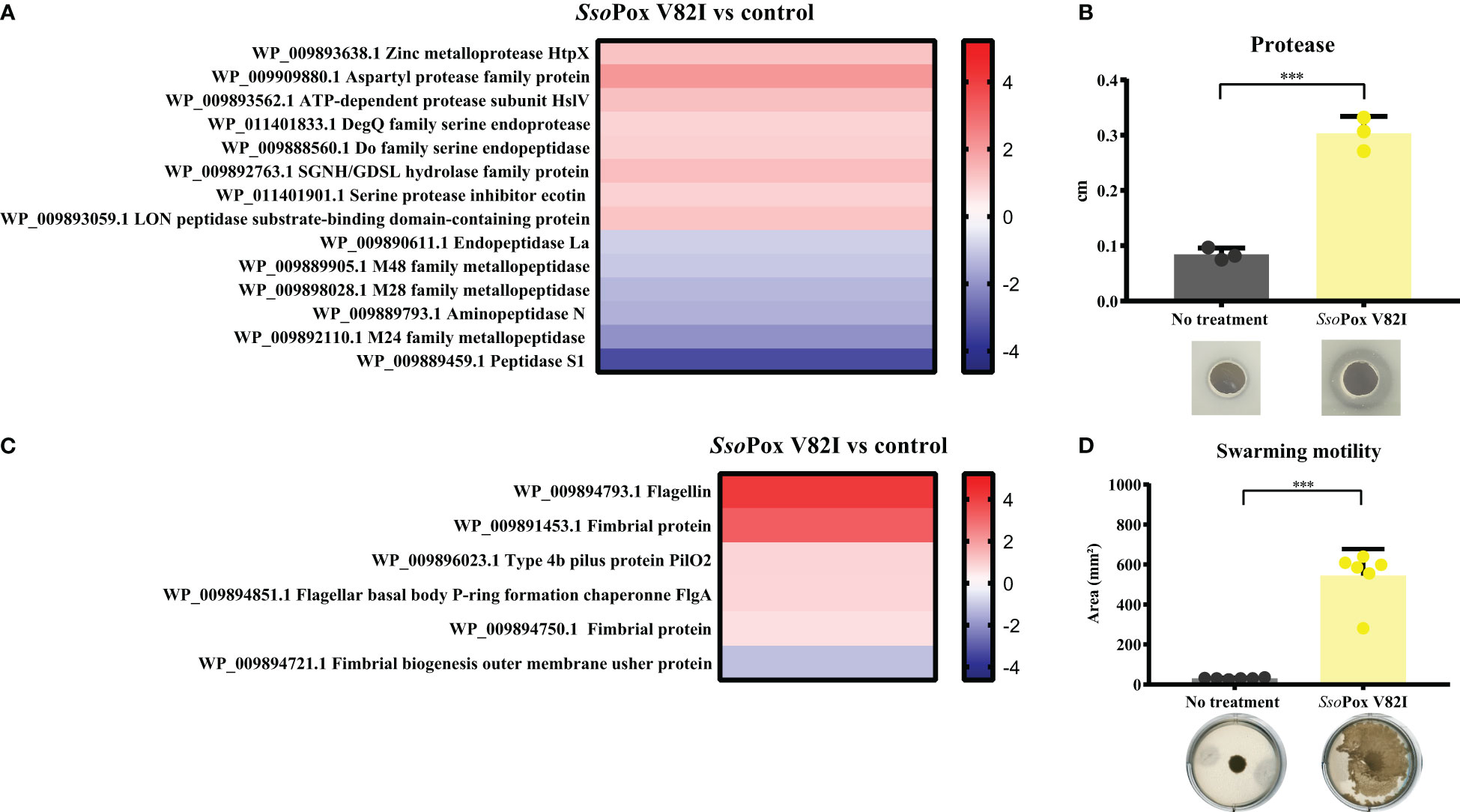
Figure 4 QS disruption in B. thailandensis increases proteolytic activity and motility. (A) Heatmap representing the fold changes of protease proteins between untreated conditions and those treated with SsoPox V82I (0.5 mg.mL-1). (B) Mean levels of B. thailandensis proteolytic activity on skimmed milk agar plates as measured with ImageJ software of treated SsoPox V82I (0.5 mg.mL-1) and untreated conditions. Error bars represent the standard deviations of n=3 biological replicates. ***p-value < 0.001 according to Student’s t-test. Pictures of proteolytic halos are presented below each condition. (C) Heatmap representing the fold changes of proteins involved in motility between untreated and treated SsoPox V82I (0.5 mg.mL-1) conditions. (D) Modulation of swarming motility in B. thailandensis after lactonase treatment with SsoPox V82I (0.5 mg.mL-1) on 0.7% agar plates. Error bars represent the standard deviations of n=6 biological replicates. ***p-value <0.001 according to Student’s t-test.
In addition, proteomic analysis identified six proteins related to motility (Figure 4C). One of these proteins was downregulated while five were upregulated. Of them, two proteins were involved in flagella structure: flagellin (WP_009894793.1) a structural protein of the flagellum that plays a role in bacterial surface adhesion (Chaban et al., 2015) and a protein related to the flagella basal body (WP_009894851.1). Flagella proteins are commonly related to swimming and swarming motility (Subramanian and Kearns, 2019). Other proteins involved in motility are related to fimbrial or pili protein (WP_009891453.1 or WP_009896023.1, WP_009894750.1 and WP_009894721.1), and are involved in cellular adhesion (Bzdyl et al., 2022) and twitching motility (Ulrich et al., 2004). To investigate a putative increase in B. thailandensis motility upon SsoPox V82I treatment, motility assays were performed. Bacteria grown with or without enzymatic treatment were spotted onto a solid surface (0.7% agar) to evaluate swarming motility. Cells from SsoPox V82I treated condition showed a high increase in swarming motility, representing an 18-fold increase of the motility area as compared to bacteria from the untreated condition (Figure 4D).
Proteomic changes upon QQ treatment were thereby supported by in vitro assays confirming that QS disruption through SsoPox V82I treatment increases proteolytic activity and motility in B. thailandensis. This confirms the validity of the large-scale molecular phenotyping carried out in this study.
To compete with other organisms, B. thailandensis produces several antimicrobial molecules (Bach et al., 2021), most of them are under QS regulation (e.g. bactobolin (Seyedsayamdost et al., 2010), thailandamide (Majerczyk et al., 2014a; Majerczyk et al., 2014b), 2-akyl-4-quinolone (Majerczyk et al., 2014a; Majerczyk et al., 2014b) as well as a type VI secretion system (T6SS) (Majerczyk et al., 2016; Lennings et al., 2019).
Here, 20 proteins related to T6SS were identified as impacted by the lactonase treatment in the proteomic analyses, most of them being downregulated (i.e. 16 proteins) (Figure 5). Similarly, 26 proteins related to the production of bacteriocin, malleilactone, bactobolin, 2-alkyl-4-quinolone, terphenyl and malleobactin were impacted by SsoPox V82I treatment. All the proteins were downregulated except the one involved in malleobactin production (Figure 6).
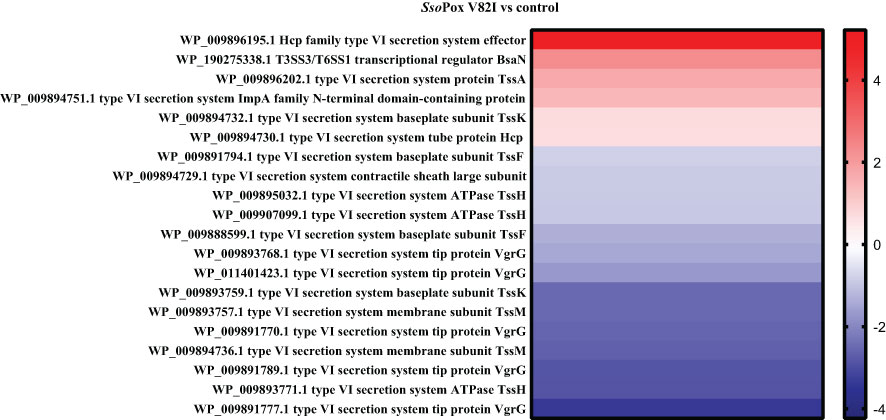
Figure 5 SsoPox V82I treatment impacts T6SS protein levels in B. thailandensis. Heatmap of fold-changes in proteins related to T6SS between cells grown in the presence of SsoPox V82I (0.5 mg.mL-1) and untreated cells.
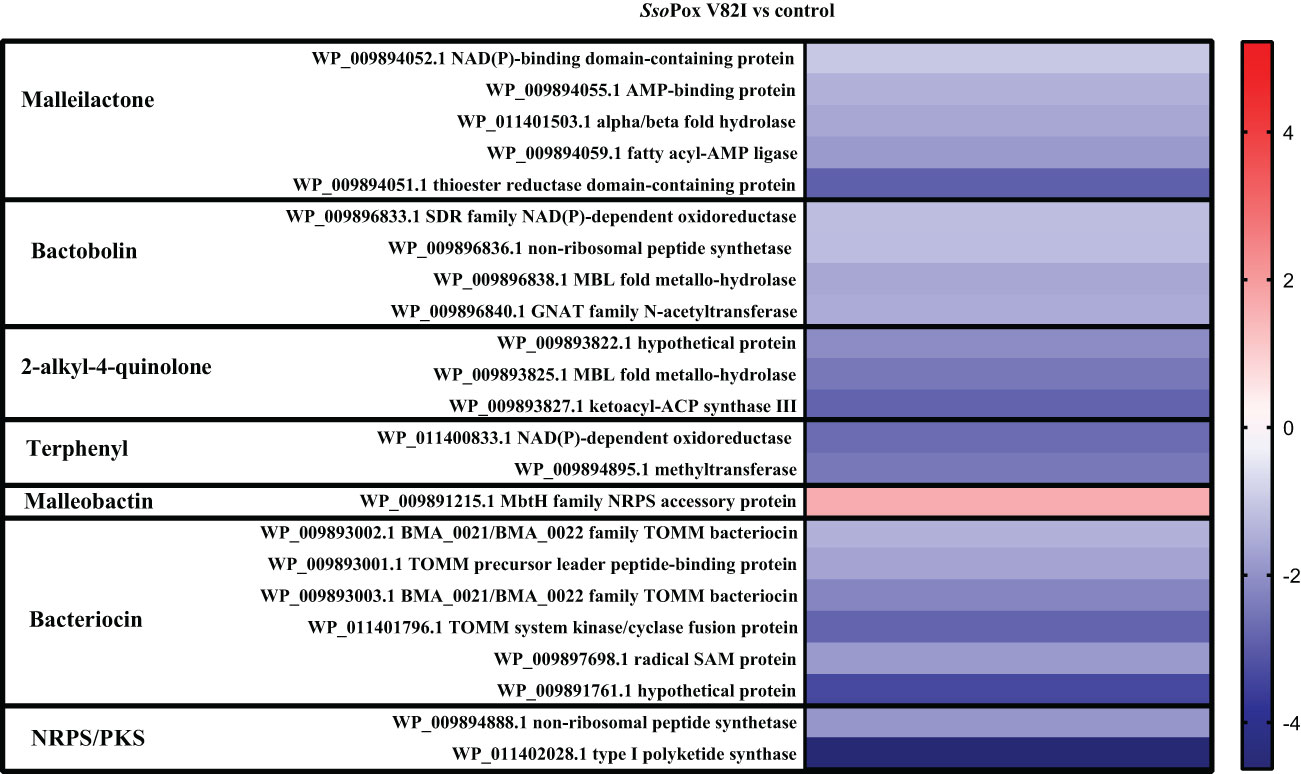
Figure 6 Impact of enzymatic treatment on antimicrobial production. Heatmap of fold-changes of proteins involved in antimicrobial molecule production between cells grown in the presence of SsoPox V82I (0.5 mg.mL-1) and untreated cells.
To investigate the impact of lactonase treatment on antimicrobial production and its consequences on B. thailandensis fitness, different competition assays were designed. A bacterial competition assay was set up using B. thailandensis cell-free supernatants, obtained from treated and non-treated cultures, and the Gram-negative bacterium C. violaceum. The supernatant obtained from the untreated cultures completely inhibited C. violaceum growth. Conversely, when the supernatant was obtained from cultures treated with SsoPox V82I, C. violaceum was able to grow (Figure 7A). The same experiment was performed with the Gram-positive bacteria S. aureus and similarly, S. aureus growth was inhibited in the presence of untreated cell-free supernatants, while cell-free supernatants obtained from cultures treated with SsoPox V82I enzyme allowed S. aureus to reach the same growth level as bacteria without supernatant (Figure 7B). Untreated supernatant inhibited the growth of both bacteria. To determine whether the effect was bacteriostatic or bactericidal, cells were further serially diluted without the addition of supernatant and bacterial survival was evaluated. The bactericidal effect of the supernatant treatment was confirmed with a stronger effect against C. violaceum (Figure S1). In conclusion, QQ decreased the production of one or several molecules involved in bacterial competition in B. thailandensis.
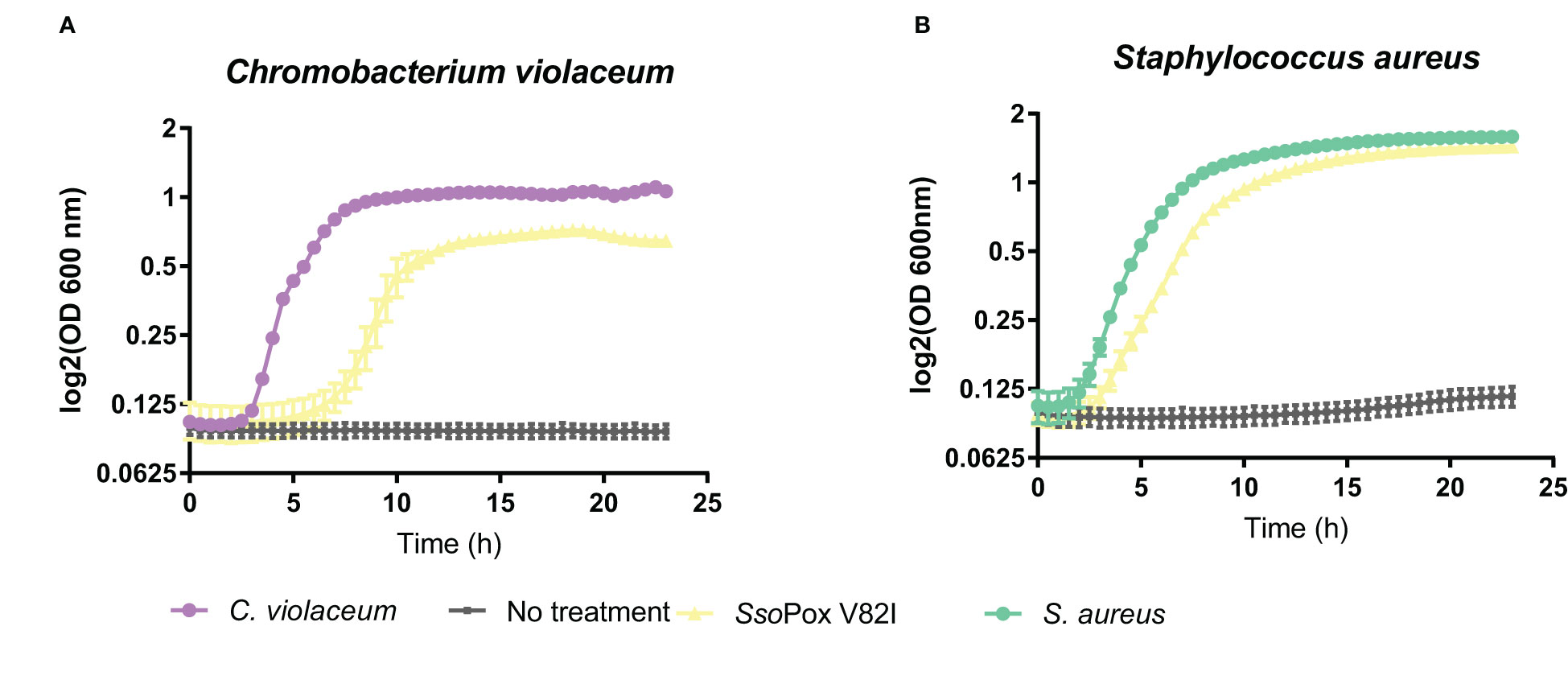
Figure 7 Decrease of bactericidal effect of B. thailandensis cell-free supernatants upon QQ. Twenty four hours of bacterial growth kinetics measured by OD 600 nm of (A) C. violaceum and (B) S. aureus in the presence of 50% of B. thailandensis supernatants obtained from cultures untreated or treated with SsoPox V82I (0.5 mg.mL-1). Error bars represent the standard deviations of n= 4 biological replicates for control growth curves without supernatant (in violet and green) and n=6 biological replicates for conditions supplemented with supernatant (in yellow and gray).
To further characterize the involvement of QS and QQ in antimicrobial compound production, other competition models with eukaryotes were developed. Competition assays were designed using two different fungi: A. niger and F. graminearum. Cell-free supernatants obtained from B. thailandensis cultures untreated or treated with SsoPox V82I were tested on both fungi, and growth was evaluated. In the presence of the untreated supernatant, both fungi were able to grow, while in the presence of the supernatant obtained from treated cultures, growth was drastically reduced or even abolished (Figure 8A), indicating that QQ increases the production of antifungal molecules. To complete these observations, the competition assay was applied to the model yeast S. cerevisiae. In the same way, colony formation was evaluated in the presence of cell-free supernatants obtained from treated and untreated cultures. S. cerevisiae was able to grow in presence of the untreated supernatant while supernatants obtained from SsoPox V82I treated cultures showed a yeasticidal activity against S. cerevisiae (Figure 8B). These results were confirmed in a microdilution broth assay in which the growth of S. cerevisiae was followed over time by measuring absorbance. S. cerevisiae growth was completely inhibited with the treated supernatant but recovered a normal growth in the presence of the untreated supernatant (Figure 8B). Moreover, this yeasticidal effect was long-lasting, and persisted for weeks. The cultures were also spotted without the addition of supernatant to evaluate any remaining effect of the supernatant. No colony formation was observed in conditions involving the pre-treated supernatant with SsoPox V82I, confirming the increase of yeasticidal activity after QQE treatment (Figure S2).
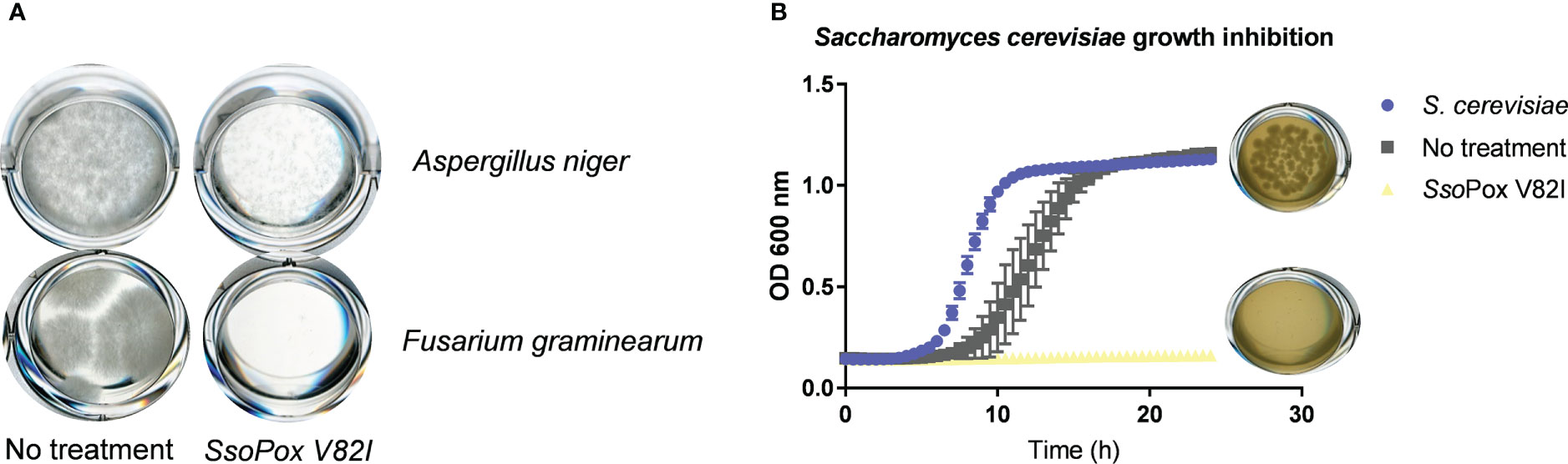
Figure 8 Lactonase treatment increases the antifungal effect of B. thailandensis supernatants (A) Representative pictures of fungal growth in the presence of B. thailandensis cell-free supernatants obtained from cultures treated with SsoPox V82I (0.5 mg.mL-1) or not on A. niger and F. graminearum. Pictures taken after 96 hours of growth at 25°C. (B) Effect of B. thailandensis cell-free supernatants obtained from cultures treated with SsoPox V82I (0.5 mg.mL-1) or untreated against S. cerevisiae. Growth was followed by measuring OD at 600 nm for 24 hours. Error bars represent the standard deviations of n= 6 biological replicates. Pictures of S. cerevisiae colonies recovered from liquid cultivation are presented next to each condition.
Taken together, these results show that QS modulates the production of antimicrobial compounds in B. thailandensis, repressing the expression of a bioactive compound on fungi but inducing the production of another against bacteria.
To further understand the antimicrobial effect detected in the supernatant, ethyl acetate extractions were performed to identify molecules impacted by QQ treatment.
As previously described in this study, the antibiotic activity of the supernatant was shown against S. aureus and C. violaceum and was significantly inhibited upon lactonase treatment. Proteomic analysis further revealed a significant reduction in proteins involved in antimicrobial molecule production, such as proteins involved in bactobolin biosynthesis. LC/MS analysis were then performed on B. thailandensis supernatant to identify the impact of QQ treatment on bactobolin molecules. The antibiotic activity of bactobolin was previously shown against Gram-positive and negative bacteria (Seyedsayamdost et al., 2010; Carr et al., 2011). Eight different bactobolins were reported in B. thailandensis (bactobolin A to H) varying in chlorine number (1 or 2), hydroxylation and alanine residues (Figure 9A) (Carr et al., 2011). Here, seven bactobolins were detected in the supernatant and were identified using m/z, observed retention time, common fragments (identified in bactobolin B spectrum and accessible in MoNA-MassBank of North America), and chlorine atoms. Without lactonase treatment, bactobolin E was the most abundant compound, followed by bactobolin A, bactobolin B and bactobolin F, while the abundance of remaining bactobolins was marginal. Upon lactonase treatment, a significant decrease in bactobolin E and bactobolin A was observed, with a 30- and 50-fold decrease, respectively. The signal response for bactobolin B was also half that of the treated condition. Conversely, bactobolin H was only observed in untreated samples, together with an increase in bactobolin D, but their signal response remained low as compared to bactobolins A and E (Figure 9B). Bactobolins impacted by SsoPox V82I treatments were bactobolins with a hydroxylation on C5 (Figure 9A). In addition, the protein involved in the hydroxylation reaction (i.e. BtaU) was found to be downregulated in proteomic analysis, but was not included among the proteins significantly impacted due to a fold change below the 1.5 threshold (-1.35 fold change decrease). Regarding the three bactobolins with the highest signal response, bactobolin A appears to be the most potent bactobolin, as described previously (Seyedsayamdost et al., 2010; Carr et al., 2011; Chandler et al., 2012b). A previous study also demonstrated that mutations in BtaL and in BtaP shut down the production of bactobolin (Carr et al., 2011), and both proteins were detected in proteomic analysis and were downregulated by SsoPox treatment (Figure 6).
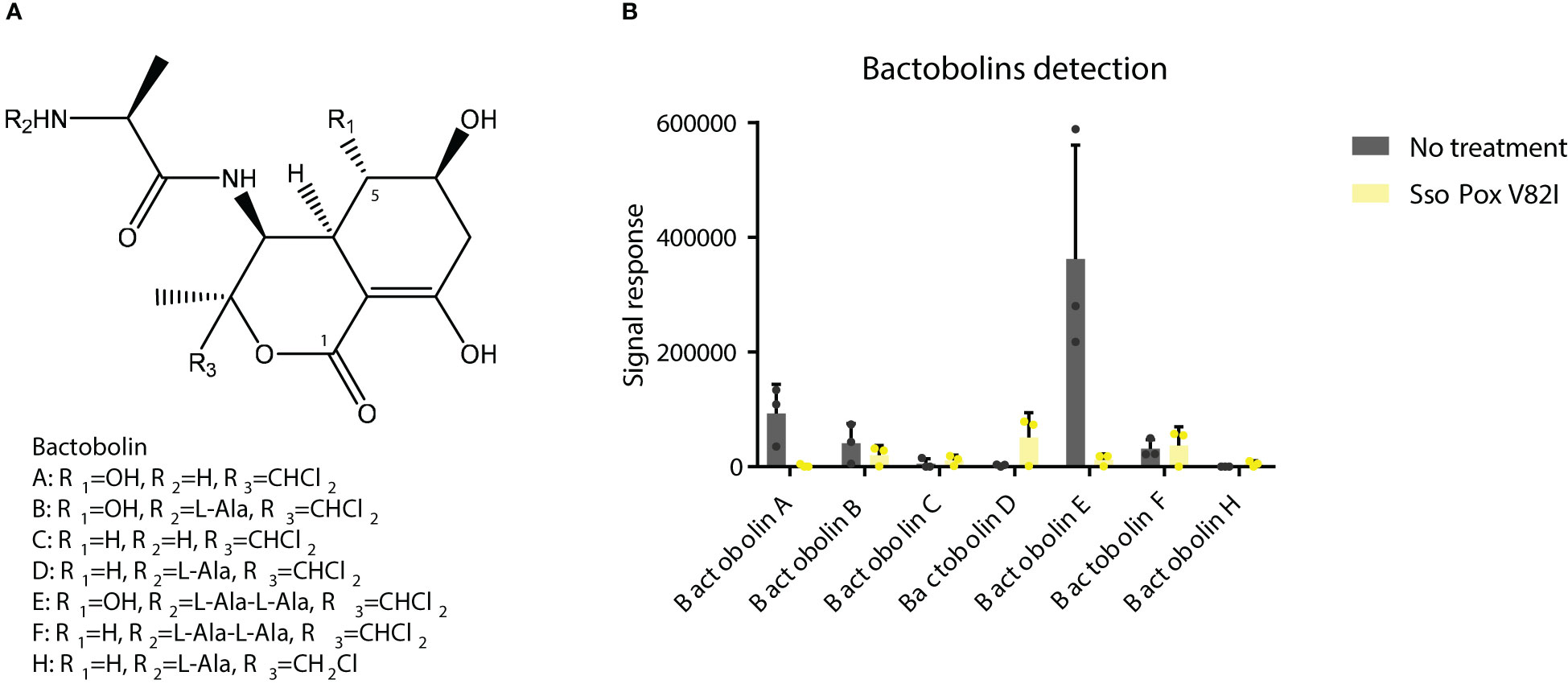
Figure 9 SsoPox treatment decreases the abundance of bactobolins in bacterial supernatants. (A) Structures of bactobolins detected in this study. (B) Signal detection in LC-MS analysis of several bactobolin molecules after extraction with ethyl acetate of B. thailandensis supernatants either untreated or treated with SsoPox V82I (0.5 mg.mL-1).
In addition, other antimicrobial molecules, such as HMAQ and its derivatives were investigated in sample extracts, as proteins involved in their synthesis were found to be altered in proteomic analysis. SsoPox V82I seemed to increase the signal detection of HAQ C9 2’, HAQNO C10 2’ and HMAQ C9, while the signal for HAQNO C8 2’, HAQNO C9 2’, HMAQNO C9 2’ seemed to be decreased (Figure S3). However, a wide variability was observed in the samples and did not enable clear conclusions to be drawn about the effect of QQ treatment on the abundance of HMAQ and related compounds.
Supernatant extractions enabled the identification of a decrease in bactobolin signals after QQ treatment, in correlation with the proteomic analysis, that could explain the decrease in bactericidal effect against S. aureus and C. violaceum in the competition model.
B. thailandensis E264 is a soil saprophyte bacteria and is considered to be a non-pathogenic homolog of B. pseudomallei. Both bacteria contain three QS-systems which regulate the expression of numerous genes as well as the production of secondary metabolites. In this study, the effect of SsoPox V82I lactonase was evaluated on B. thailandensis through proteomic and phenotypic analysis.
AHLs used by B. thailandensis were described as C8-HSL, 3-OH-C8-HSL and 3-OH-C10-HSL by LC/LC/MS analysis (Duerkop et al., 2009). Here, in vitro degradation assays towards these lactones were used to evaluate the performance of four previously described SsoPox variants, and SsoPox V82I was chosen. The QQ ability of this variant was then confirmed on B. thailandensis and a significant reduction in biofilm formation and cell aggregation, two QS-related phenotypes of B. thailandensis, was achieved (Chandler et al., 2009; Majerczyk et al., 2014a; Tseng et al., 2016). To get a broader picture of global changes involved by QS and QQ, proteomic analyses were performed.
Proteomic analyses provided evidence that QQ by SsoPox V82I treatment generates a huge shift in B. thailandensis with 18.3% of the proteome being significantly impacted. Several metabolic pathways were impacted, including primary metabolites such as sugar and amino acid metabolisms. AHLs synthesis is directly related to methionine metabolism, therefore, in addition to AHLs signal disruption, QQ might also generate an improper balance in the primary metabolism, impacting many metabolic pathways. Proteomic analysis revealed that the treatment with SsoPox V82I results in activation of numerous pathways: 72% of impacted proteins were upregulated, suggesting that QS represses several key functions in B. thailandensis. In contrast, previous studies, using other SsoPox variants on P. aeruginosa and C. violaceum did not induce such an upregulation upon lactonase treatment, suggesting that QS equally upregulated or downregulated proteins on these strains (Rémy et al., 2020; Mion et al., 2021). QS regulation is known to be complex in B. thailandensis, and could repress or activate AHL-responsive genes depending on the growth stage (Majerczyk et al., 2014a).
To confirm proteomic changes at the phenotypic level, different bioassays were performed which showed that motility and proteolytic activities were increased after SsoPox V82I treatment and that antimicrobial compound production and secretion were modulated. Indeed, competition assays using cell-free supernatants showed that the production of an antifungal molecule was induced by QQ while the production of an antibacterial molecule was repressed. Motility was already known to be negatively regulated by QS in B. thailandensis (Ulrich et al., 2004; Chandler et al., 2009; Passos da Silva et al., 2017), and the increased motility observed after QQ treatment confirmed these results. Regarding proteolytic activity, previous studies did not show any effect on protease production in a QS-mutant strain, suggesting that proteolytic activity was not related to QS (Chandler et al., 2009). Under our conditions and using a QQE, protease activity was increased, as observed on milk agar plates, suggesting that proteolytic activity is negatively regulated by QS in B. thailandensis.
Biosynthesis of some antimicrobial molecules and secondary metabolites, in B. thailandensis, was also previously described to be QS-controlled (Majerczyk et al., 2014a). In our conditions, QQ treatment by SsoPox V82I led to a decrease in bactericidal effect against C. violaceum and S. aureus. Several proteins involved in antibiotic molecule synthesis (including malleilactone, terphenyl and bactobolin) were identified as being downregulated by SsoPox in proteomic analyses. Malleilactone is a cytotoxin that contributes to virulence in B. pseudomallei and B. thailandensis (Klaus et al., 2018), and its bactericidal effect against Gram positive bacteria was described (including in S. aureus and B. subtilis) (Biggins et al., 2012), and bactericidal activity against B. subtilis was reported for terphenyl (Biggins et al., 2011). Proteins related to bacteriocin biosynthesis (Russell et al., 2014; Majerczyk et al., 2016; Mao et al., 2017) were identified and downregulated by lactonase treatment in proteomic analysis. In addition, proteins involved in bactobolin production were decreased after SsoPox V82I treatment. Bactobolin is a well described antibiotic compound against different bacteria such as S. aureus (Klaus, 2020) and C. violaceum (Chandler et al., 2012a). Synthesis of bactobolin is controlled by QS-2 (3-OH-C10-HSL, BtaI2-BtaR2) (Seyedsayamdost et al., 2010; Majerczyk et al., 2014a) and has been described as the sole active antibiotic compound remaining in cell-free supernatants after 0.22 µm filtration (Klaus et al., 2020). Furthermore, LC-MS/MS analysis revealed a reduced response in bactobolin in treated samples. It is, therefore, probable that the bactericidal effect observed without SsoPox V82I treatment is due to bactobolin production. Interestingly, while decreasing antibacterial effect, QQ treatment led to a strong increase in antifungal activity against A. niger, F. graminearum and S. cerevisiae. A similar increase in antifungal activity was also reported in Burkholderia ambifaria, a member of the Burkholderia cepacia complex, when hmqG or hmqA, two genes belonging to the hmqABCDEFG operon responsible for HMAQ production and under QS regulation (Coulon et al., 2019), were mutated (Vial et al., 2008). This operon was also described in B. thailandensis and B. pseudomallei (Vial et al., 2008). In addition to an increase in antifungal activity, hmqG or hmqA mutations led to an intensification in proteolytic activity (Vial et al., 2008). Here, the HmqG protein (WP_009893822.1) was identified in the proteomic analysis as being downregulated by SsoPox V82I treatment, confirming previous results obtained in B. ambifaria. However, when extractions were performed and analyzed in LC-MS, HMAQ molecules and derivatives were identified, but a high variability between samples was observed and did not allow to draw conclusion about the impact of QQ on these molecules. Interestingly, these extractions were tested and, surprisingly, the extracted supernatants had lost their antifungal effect, suggesting a potential alteration of the bioactive compounds during the extraction step.
Previous studies have demonstrated the conservation of QS regulons and QS-regulated functions in B. pseudomallei and B. thailandensis (Majerczyk et al., 2014b) and within Burkholderia cepacia complex (Wopperer et al., 2006). In addition, previous studies investigated QS disruption using QS mutants (Ulrich et al., 2004; Chandler et al., 2009; Tseng et al., 2016) and QQ lactonase (Ulrich, 2004; Wopperer et al., 2006) in Burkholderia species have demonstrated that QS is involved in carbon metabolism and the regulation of phenotypes such as biofilm formation, motility and proteolytic activity. Here, the global proteomic approach offers an overview of the implication of QS disruption in B. thailandensis and could be extended to other Burkholderia species.
Proteomic and phenotypic analyses outline the importance of QS disruption in antimicrobial production modulation and its impact on the interbacterial or interkingdom competition model. Recent studies on several microbiota such as human gut microbiota (Coquant et al., 2021) and human oral cavity microbiota (Muras et al., 2020) have detected the presence of AHLs. In addition, their production could be related to inflammatory diseases in gut and dental plaque formation. In environmental isolates, AHLs have been detected in coral and their presence is related to coral bleaching (Zhou et al., 2020). A recent study using an SsoPox variant demonstrated biofilm disruption in a complex sample from soil and revealed that SsoPox impacted not only bacteria with an AHLs QS system but also bacteria not known to sense or produce AHLs (Schwab et al., 2019). Using a lactonase enzyme, another QQ study confirmed the role of QS on oral microbiota and the impact of a QQE on oral biofilm formation (Parga et al., 2023). These studies highlight the impact of QQ in a heterogeneous population through the modulation of antimicrobial compounds such as antibiotics, fungicides, and secretion systems. These molecules can induce dysbiosis and led to pathology genesis. Taken together, these studies open the way for further investigations of the impact of QQ on microbiota samples, to gain a better understanding of these complex microenvironments.
Using a combination of phenotypic and molecular approaches we demonstrate that lactonase-mediated quorum quenching strongly alters the behavior and the virulence of B. thailandensis, a model strain for studying the highly virulent pathogen B. pseudomallei. Biofilm formation, motility, proteolytic activity were strongly impacted upon enzymatic treatment. Interestingly we show that antimicrobial activities of B. thailandensis against other microorganisms are highly mediated by the lactonase, bactericidal effect towards both Gram positive and negative strains being significantly decreased while fungicidal activity being tremendously enhanced against both yeast and fungi. This work, combining fine and exhaustive microbiological characterizations together with proteomic analyses, will help to understand the role of QS in bacterial virulence and microbial interactions and will contribute to promote the use of quorum quenchers for controlling pathogens and modulate cooperation and competition in complex microbial populations.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD040565 and 10.6019/PXD040565.
MG, LP, ÉC, and DD designed the study; MG, LP, JA, and NA performed the experiments; MG, LP, and DD analyzed the data; MG, LP, ÉC, and DD wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was financially supported by Investissements d’Avenir programme (Méditerranée Infection 10-IAHU-03) of the French Agence Nationale de la Recherche (ANR). MG is a Ph.D. student funded by the “Emplois Jeunes Doctorants” programme of the Région Provence-Alpes-Côte d’Azur (PACA, France).
Mélodie Kielbasa (CEA) is acknowledged for her help with Q-Exactive HF mass spectrometry measurements.
MG, LP, ÉC, and DD report receiving personal fees from Gene&GreenTK during the study. ÉC, LP, and DD have filed the patents FR3093894, EP3941206, WO2020187861. ÉC, LP, and DD are shareholders in Gene&GreenTK. DD is CEO of Gene&GreenTK.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1190859/full#supplementary-material
Adler, D., Kelly, T. S., Elliot, T., Adamson, J. (2022) Vioplot: violin plot. Available at: https://github.com/TomKellyGenetics/vioplot.
Andersen, J. B., Heydorn, A., Hentzer, M., Eberl, L., Geisenberger, O., Christensen, B. B., et al. (2001). Gfp-based n-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl. Environ. Microbiol. 67, 575–585. doi: 10.1128/AEM.67.2.575-585.2001
Arendrup, M. C., Guinea, J., Cuenca-Estrella, M., Meletiadis, J., Mouton, J. W., Lagrou, K., et al. (2015). EUCAST definitive document E.Def.
Bach, E., Passaglia, L. M. P., Jiao, J., Gross, H. (2021). Burkholderia in the genomic era: from taxonomy to the discovery of new antimicrobial secondary metabolites. Crit. Rev. Microbiol. 0, 1–40. doi: 10.1080/1040841X.2021.1946009
Biggins, J. B., Liu, X., Feng, Z., Brady, S. F. (2011). Metabolites from the induced expression of cryptic single operons found in the genome of Burkholderia pseudomallei. J. Am. Chem. Soc 133, 1638–1641. doi: 10.1021/ja1087369
Biggins, J. B., Ternei, M. A., Brady, S. F. (2012). Malleilactone, a polyketide synthase-derived virulence factor encoded by the cryptic secondary metabolome of Burkholderia pseudomallei group pathogens. J. Am. Chem. Soc. 134, 13192–13195. doi: 10.1021/ja3052156
Billot, R., Plener, L., Grizard, D., Elias, M. H., Chabrière, É., Daudé, D. (2022). Applying molecular and phenotypic screening assays to identify efficient quorum quenching lactonases. Enzyme Microbial. Technol. 160, 110092. doi: 10.1016/j.enzmictec.2022.110092
Billot, R., Plener, L., Jacquet, P., Elias, M., Chabrière, E., Daudé, D. (2020). Engineering acyl-homoserine lactone-interfering enzymes toward bacterial control. J. Biol. Chem. 295, 12993–13007. doi: 10.1074/jbc.REV120.013531
Bowler, P., Murphy, C., Wolcott, R. (2020). Biofilm exacerbates antibiotic resistance: is this a current oversight in antimicrobial stewardship? Antimicrobial. Resist. Infect. Control 9, 162. doi: 10.1186/s13756-020-00830-6
Bzdyl, N. M., Moran, C. L., Bendo, J., Sarkar-Tyson, M. (2022). Pathogenicity and virulence of Burkholderia pseudomallei. Virulence 13, 1945–1965. doi: 10.1080/21505594.2022.2139063
Carr, G., Seyedsayamdost, M. R., Chandler, J. R., Greenberg, E. P., Clardy, J. (2011). Sources of diversity in bactobolin biosynthesis by Burkholderia thailandensis E264. Org. Lett. 13, 3048–3051. doi: 10.1021/ol200922s
Carvalho, P. C., Fischer, J. S. G., Xu, T., Cociorva, D., Balbuena, T. S., Valente, R. H., et al. (2012). Search engine processor: filtering and organizing peptide spectrum matches. Proteomics 12, 944–949. doi: 10.1002/pmic.201100529
Chaban, B., Hughes, H. V., Beeby, M. (2015). The flagellum in bacterial pathogens: for motility and a whole lot more. Semin. Cell Dev. Biol. 46, 91–103. doi: 10.1016/j.semcdb.2015.10.032
Chandler, J. R., Duerkop, B. A., Hinz, A., West, T. E., Herman, J. P., Churchill, M. E. A., et al. (2009). Mutational analysis of Burkholderia thailandensis quorum sensing and self-aggregation. J. Bacteriol. 191, 5901–5909. doi: 10.1128/JB.00591-09
Chandler, J. R., Heilmann, S., Mittler, J. E., Greenberg, E. P. (2012a). Acyl-homoserine lactone-dependent eavesdropping promotes competition in a laboratory co-culture model. ISME J. 6, 2219–2228. doi: 10.1038/ismej.2012.69
Chandler, J. R., Truong, T. T., Silva, P. M., Seyedsayamdost, M. R., Carr, G., Radey, M., et al. (2012b). Bactobolin resistance is conferred by mutations in the L2 ribosomal protein. mBio 3, e00499–e00412. doi: 10.1128/mBio.00499-12
Chang, K., Luo, J., Xu, H., Li, M., Zhang, F., Li, J., et al. (2017). Human infection with Burkholderia thailandensis, chin. Emerg. Infect. Dis. 23, 1416–1418. doi: 10.3201/eid2308.170048
Coquant, G., Aguanno, D., Pham, S., Grellier, N., Thenet, S., Carrière, V., et al. (2021). Gossip in the gut: quorum sensing, a new player in the host-microbiota interactions. World J. Gastroenterol. 27, 7247–7270. doi: 10.3748/wjg.v27.i42.7247
Coulon, P. M. L., Groleau, M.-C., Déziel, E. (2019). Potential of the Burkholderia cepacia complex to produce 4-Hydroxy-3-Methyl-2-Alkyquinolines. Front. Cell. Infect. Microbiol. 9. doi: 10.3389/fcimb.2019.00033
Del Vecchio, P., Elias, M., Merone, L., Graziano, G., Dupuy, J., Mandrich, L., et al. (2009). Structural determinants of the high thermal stability of SsoPox from the hyperthermophilic archaeon sulfolobus solfataricus. Extremophiles 13, 461–470. doi: 10.1007/s00792-009-0231-9
Duerkop, B. A., Varga, J., Chandler, J. R., Peterson, S. B., Herman, J. P., Churchill, M. E. A., et al. (2009). Quorum-sensing control of antibiotic synthesis in Burkholderia thailandensis. J. Bacteriol. 191, 3909–3918. doi: 10.1128/JB.00200-09
Gee, J. E., Elrod, M. G., Gulvik, C. A., Haselow, D. T., Waters, C., Liu, L., et al. (2018). Burkholderia thailandensis isolated from infected wound, Arkansas, USA. Emerg. Infect. Dis. 24, 2091–2094. doi: 10.3201/eid2411.180821
Gouveia, D., Grenga, L., Gaillard, J.-C., Gallais, F., Bellanger, L., Pible, O., et al. (2020). Shortlisting SARS-CoV-2 peptides for targeted studies from experimental data-dependent acquisition tandem mass spectrometry data. Proteomics 20, e2000107. doi: 10.1002/pmic.202000107
Grandclément, C., Tannières, M., Moréra, S., Dessaux, Y., Faure, D. (2016). Quorum quenching: role in nature and applied developments. FEMS Microbiol. Rev. 40, 86–116. doi: 10.1093/femsre/fuv038
Gupta, A., Bedre, R., Thapa, S. S., Sabrin, A., Wang, G., Dassanayake, M., et al. (2017). Global awakening of cryptic biosynthetic gene clusters in Burkholderia thailandensis. ACS Chem. Biol. 12, 3012–3021. doi: 10.1021/acschembio.7b00681
Hartmann, E. M., Allain, F., Gaillard, J.-C., Pible, O., Armengaud, J. (2014). “Taking the shortcut for high-throughput shotgun proteomic analysis of bacteria,” in Host-bacteria interactions: methods and protocols methods in molecular biology. Eds. Vergunst, A. C., O’Callaghan, D. (New York, NY: Springer), 275–285. doi: 10.1007/978-1-4939-1261-2_16
Hiblot, J., Gotthard, G., Elias, M., Chabriere, E. (2013). Differential active site loop conformations mediate promiscuous activities in the lactonase SsoPox. PloS One 8, e75272. doi: 10.1371/journal.pone.0075272
Hoffman, L. R., D’Argenio, D. A., MacCoss, M. J., Zhang, Z., Jones, R. A., Miller, S. I. (2005). Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436, 1171–1175. doi: 10.1038/nature03912
Hraiech, S., Hiblot, J., Lafleur, J., Lepidi, H., Papazian, L., Rolain, J.-M., et al. (2014). Inhaled lactonase reduces Pseudomonas aeruginosa quorum sensing and mortality in rat pneumonia. PloS One 9, e107125. doi: 10.1371/journal.pone.0107125
Klaus, J. R., Coulon, P. M. L., Koirala, P., Seyedsayamdost, M. R., Déziel, E., Chandler, J. R. (2020). Secondary metabolites from the Burkholderia pseudomallei complex: structure, ecology, and evolution. J. Ind. Microbiol. 47 (9–10), 877–887. doi: 10.1007/s10295-020-02317-0
Klaus, J. R., Deay, J., Neuenswander, B., Hursh, W., Gao, Z., Bouddhara, T., et al. (2018). Malleilactone is a Burkholderia pseudomallei virulence factor regulated by antibiotics and quorum sensing. J. Bacteriol. 200 (14), e00008-18, doi: 10.1128/JB.00008-18
Klaus, J. R., Majerczyk, C., Moon, S., Eppler, N. A., Smith, S., Tuma, E., et al. (2020). Burkholderia thailandensis methylated hydroxyalkylquinolines: biosynthesis and antimicrobial activity in cocultures. Appl. Environ. Microbiol. 86 (24), e01452-20. doi: 10.1128/AEM.01452-20
Klein, G., Mathé, C., Biola-Clier, M., Devineau, S., Drouineau, E., Hatem, E., et al. (2016). RNA-Binding proteins are a major target of silica nanoparticles in cell extracts. Nanotoxicology 10, 1555–1564. doi: 10.1080/17435390.2016.1244299
Lennings, J., West, T. E., Schwarz, S. (2019). The Burkholderia type VI secretion system 5: composition, regulation and role in virulence. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.03339
Majerczyk, C. D., Brittnacher, M. J., Jacobs, M. A., Armour, C. D., Radey, M. C., Bunt, R., et al. (2014b). Cross-species comparison of the Burkholderia pseudomallei, burkholderia thailandensis, and Burkholderia mallei quorum-sensing regulons. J. Bacteriol. 196, 3862–3871. doi: 10.1128/JB.01974-14
Majerczyk, C., Brittnacher, M., Jacobs, M., Armour, C. D., Radey, M., Schneider, E., et al. (2014a). Global analysis of the Burkholderia thailandensis quorum sensing-controlled regulon. J. Bacteriol. 196, 1412–1424. doi: 10.1128/JB.01405-13
Majerczyk, C., Schneider, E., Greenberg, E. P. (2016). Quorum sensing control of type VI secretion factors restricts the proliferation of quorum-sensing mutants. eLife 5, e14712. doi: 10.7554/eLife.14712
Mao, D., Bushin, L. B., Moon, K., Wu, Y., Seyedsayamdost, M. R. (2017). Discovery of scmR as a global regulator of secondary metabolism and virulence in Burkholderia thailandensis E264. Proc. Natl. Acad. Sci. 114, E2920–E2928. doi: 10.1073/pnas.1619529114
Meyer, J. M. (1978). The florescent pigment of Pseudomonas fluorescens biosynthesis, purification and physicalchemical properties. J. Gen. Microbiol. 107 (2), 319–328. doi: 10.1099/00221287-107-2-319
Mion, S., Carriot, N., Lopez, J., Plener, L., Ortalo-Magné, A., Chabrière, E., et al. (2021). Disrupting quorum sensing alters social interactions in Chromobacterium violaceum. NPJ Biofilms Microbiomes 7, 40. doi: 10.1038/s41522-021-00211-w
Mion, S., Rémy, B., Plener, L., Chabrière, É., Daudé, D. (2019). Quorum sensing et quorum quenching : comment bloquer la communication des bactéries pour inhiber leur virulence? médecine/sciences 35, 31–38. doi: 10.1051/medsci/2018310
Muras, A., Otero-Casal, P., Blanc, V., Otero, A. (2020). Acyl homoserine lactone-mediated quorum sensing in the oral cavity: a paradigm revisited. Sci. Rep. 10, 9800. doi: 10.1038/s41598-020-66704-4
Oliveira, M., Mason-Buck, G., Ballard, D., Branicki, W., Amorim, A. (2020). Biowarfare, bioterrorism and biocrime: a historical overview on microbial harmful applications. Forensic Sci. Int. 314, 110366. doi: 10.1016/j.forsciint.2020.110366
Parga, A., Muras, A., Otero-Casal, P., Arredondo, A., Soler-Ollé, A., Àlvarez, G., et al. (2023). The quorum quenching enzyme Aii20J modifies in vitro periodontal biofilm formation. Front. Cell. Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1118630
Passos da Silva, D., Schofield, M. C., Parsek, M. R., Tseng, B. S. (2017). An update on the sociomicrobiology of quorum sensing in gram-negative biofilm development. Pathogens 6, 51. doi: 10.3390/pathogens6040051
Piochon, M., Coulon, P. M. L., Caulet, A., Groleau, M.-C., Déziel, E., Gauthier, C. (2020). Synthesis and antimicrobial activity of Burkholderia -related 4-Hydroxy-3-methyl-2-alkenylquinolines (HMAQs) and their N -oxide counterparts. J. Nat. Prod. 83, 2145–2154. doi: 10.1021/acs.jnatprod.0c00171
Rémy, B., Mion, S., Plener, L., Elias, M., Chabrière, E., Daudé, D. (2018). Interference in bacterial quorum sensing: a biopharmaceutical perspective. Front. Pharmacol. 9. doi: 10.3389/fphar.2018.00203
Rémy, B., Plener, L., Decloquement, P., Armstrong, N., Elias, M., Daudé, D., et al. (2020). Lactonase specificity is key to quorum quenching in Pseudomonas aeruginosa. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00762
Rémy, B., Plener, L., Poirier, L., Elias, M., Daudé, D., Chabrière, E. (2016). Harnessing hyperthermostable lactonase from Sulfolobus solfataricus for biotechnological applications. Sci. Rep. 6, 37780. doi: 10.1038/srep37780
Russell, A. B., Peterson, S. B., Mougous, J. D. (2014). Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148. doi: 10.1038/nrmicro3185
Schwab, M., Bergonzi, C., Sakkos, J., Staley, C., Zhang, Q., Sadowsky, M. J., et al. (2019). Signal disruption leads to changes in bacterial community population. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00611
Seyedsayamdost, M. R., Chandler, J. R., Blodgett, J. A. V., Lima, P. S., Duerkop, B. A., Oinuma, K.-I., et al. (2010). Quorum-Sensing-Regulated bactobolin production by Burkholderia thailandensis E264. Org. Lett., 12 (4), 71–719. doi: 10.1021/ol902751x
Subramanian, S., Kearns, D. B. (2019). Functional regulators of bacterial flagella. Annu. Rev. Microbiol. 73, 225–246. doi: 10.1146/annurev-micro-020518-115725
Tobias, N. J., Brehm, J., Kresovic, D., Brameyer, S., Bode, H. B., Heermann, R. (2020). New vocabulary for bacterial communication. ChemBioChem 21, 759–768. doi: 10.1002/cbic.201900580
Tseng, B. S., Majerczyk, C. D., Passos da Silva, D., Chandler, J. R., Greenberg, E. P., Parsek, M. R. (2016). Quorum sensing influences Burkholderia thailandensis biofilm development and matrix production. J. Bacteriol. 198, 2643–2650. doi: 10.1128/JB.00047-16
Ulrich, R. L. (2004). Quorum quenching: enzymatic disruption of n-acylhomoserine lactone-mediated bacterial communication in burkholderia thailandensis. Appl. Environ. Microbiol. 70, 6173–6180. doi: 10.1128/AEM.70.10.6173-6180.2004
Ulrich, R. L., Hines, H. B., Parthasarathy, N., Jeddeloh, J. A. (2004). Mutational analysis and biochemical characterization of the burkholderia thailandensis DW503 quorum-sensing network. JB 186, 4350–4360. doi: 10.1128/JB.186.13.4350-4360.2004
Vial, L., Lépine, F., Milot, S., Groleau, M.-C., Dekimpe, V., Woods, D. E., et al. (2008). Burkholderia pseudomallei, b. thailandensis, and B. ambifaria produce 4-Hydroxy-2-Alkylquinoline analogues with a methyl group at the 3 position that is required for quorum-sensing regulation. J. Bacteriol. 190, 5339–5352. doi: 10.1128/JB.00400-08
Wickham, H. (2016). ggplot2 (Cham: Springer International Publishing). doi: 10.1007/978-3-319-24277-4
Winsor, G. L., Khaira, B., Van Rossum, T., Lo, R., Whiteside, M. D., Brinkman, F. S. L. (2008). The Burkholderia genome database: facilitating flexible queries and comparative analyses. Bioinformatics 24, 2803–2804. doi: 10.1093/bioinformatics/btn524
Wopperer, J., Cardona, S. T., Huber, B., Jacobi, C. A., Valvano, M. A., Eberl, L. (2006). A quorum-quenching approach to investigate the conservation of quorum-sensing-regulated functions within the Burkholderia cepacia complex. Appl. Environ. Microbiol. 72, 1579–1587. doi: 10.1128/AEM.72.2.1579-1587.2006
Yoshitani, K., Hizukuri, Y., Akiyama, Y. (2019). An in vivo protease activity assay for investigating the functions of the Escherichia coli membrane protease HtpX. FEBS Lett. 593, 842–851. doi: 10.1002/1873-3468.13368
Zhao, X., Yu, Z., Ding, T. (2020). Quorum-sensing regulation of antimicrobial resistance in bacteria. Microorganisms 8, 425. doi: 10.3390/microorganisms8030425
Keywords: quorum sensing, Burkholderia thailandensis, quorum quenching, lactonase, Acyl-homoserine lactone
Citation: Gonzales M, Plener L, Armengaud J, Armstrong N, Chabrière É and Daudé D (2023) Lactonase-mediated inhibition of quorum sensing largely alters phenotypes, proteome, and antimicrobial activities in Burkholderia thailandensis E264. Front. Cell. Infect. Microbiol. 13:1190859. doi: 10.3389/fcimb.2023.1190859
Received: 21 March 2023; Accepted: 15 May 2023;
Published: 02 June 2023.
Edited by:
Shaohua Chen, South China Agricultural University, ChinaReviewed by:
Izabela Podgórska-Kryszczuk, University of Life Sciences of Lublin, PolandCopyright © 2023 Gonzales, Plener, Armengaud, Armstrong, Chabrière and Daudé. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Éric Chabrière, ZXJpYy5jaGFicmllcmVAdW5pdi1hbXUuZnI=; David Daudé, ZGF2aWQuZGF1ZGVAZ2VuZS1ncmVlbnRrLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.