- 1Department of Microbiology, Faculty of Medicine, University Hospital in Pilsen, Charles University, Pilsen, Czechia
- 2Biomedical Center, Faculty of Medicine, Charles University, Pilsen, Czechia
- 3Unit of Microbiology and Clinical Microbiology, Department of Clinical-Surgical, Diagnostic and Pediatric Sciences, University of Pavia, Pavia, Italy
Antimicrobial resistance is well-known to be a global health and development threat. Due to the decrease of effective antimicrobials, re-evaluation in clinical practice of old antibiotics, as fosfomycin (FOS), have been necessary. FOS is a phosphonic acid derivate that regained interest in clinical practice for the treatment of complicated infection by multi-drug resistant (MDR) bacteria. Globally, FOS resistant Gram-negative pathogens are raising, affecting the public health, and compromising the use of the antibiotic. In particular, the increased prevalence of FOS resistance (FOSR) profiles among Enterobacterales family is concerning. Decrease in FOS effectiveness can be caused by i) alteration of FOS influx inside bacterial cell or ii) acquiring antimicrobial resistance genes. In this review, we investigate the main components implicated in FOS flow and report specific mutations that affect FOS influx inside bacterial cell and, thus, its effectiveness. FosA enzymes were identified in 1980 from Serratia marcescens but only in recent years the scientific community has started studying their spread. We summarize the global epidemiology of FosA/C2/L1-2 enzymes among Enterobacterales family. To date, 11 different variants of FosA have been reported globally. Among acquired mechanisms, FosA3 is the most spread variant in Enterobacterales, followed by FosA7 and FosA5. Based on recently published studies, we clarify and represent the molecular and genetic composition of fosA/C2 genes enviroment, analyzing the mechanisms by which such genes are slowly transmitting in emerging and high-risk clones, such as E. coli ST69 and ST131, and K. pneumoniae ST11. FOS is indicated as first line option against uncomplicated urinary tract infections and shows remarkable qualities in combination with other antibiotics. A rapid and accurate identification of FOSR type in Enterobacterales is difficult to achieve due to the lack of commercial phenotypic susceptibility tests and of rapid systems for MIC detection.
Highlights
*Antimicrobial resistance currently represents a concern for human health and the reintroduction in clinical practice of old antibiotics as fosfomycin can provide further option in treatment of multi-drug resistant (MDR) bacterial infections.
*However, there is a global increasement of fosfomycin resistance bacteria, especially Enterobacterales, reducing its effectiveness.
*Considering this increasement, it would be crucial to understand and clarify the several mechanisms involved in fosfomycin resistance among clinically and veterinary relevant Enterobacterales.
*Moreover, knowledge on the global epidemiology of acquired fosfomycin resistance genes would provide information about the major transmission routes of such resistance profiles.
Introduction
Antimicrobial resistance (AMR) is one of the major global public health threats in 21st century that affects prevention and treatment of a wide range of bacterial infections (Prestinaci et al., 2015). In the last 20 years, several strategies have been developed and suggested to combat AMR. In 2012, World Health Organization (WHO) published The Evolving Threat of Antimicrobial Resistance – Options for Action, which presented interventions that will strength the health systems and enhance surveillance through improving the usage of antimicrobials in hospitals and communities, infection prevention, and encouraging the development of appropriate new drugs and vaccines (Prestinaci et al., 2015). In accordance with WHO report published in 2020, 43 antibiotics and combinations are currently in clinical development and, since 2017, 11 new antimicrobial drugs have been approved for clinical use. However, WHO claims that none of the 43 antibiotics sufficiently address the problem of AMR in the most clinically problematic bacteria (e.g., Escherichia coli, Klebsiella pneumoniae). As the antibiotics availability is decreasing with time, the old antibiotics retaining effectiveness against some multi-drug resistant (MDR) pathogens are re-introduced (Theuretzbacher and Paul, 2015). This temporary solution allowed the renaissance of molecules such as colistin, nitrofurantoin and fosfomycin (FOS).
Fosfomycin
FOS, originally called phosphonomycin, is a phosphonic acid derivate discovered in 1969 by the Medina Foundation (Fundación Medina, Granada, Spain) from soil Streptomyces fradiae and Pseudomonas syringae. The same year, Christensen et al. determined the FOS molecular formula (–)-(1R, 2S)-1,2-epoxy propyl phosphonic acid (Christensen et al., 1969). FOS interferes with the early stages of peptidoglycan production, inhibiting UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) enzyme. MurA enzyme catalyzes the formation of peptidoglycan precursor, N-acetylmuramic acid. The binding of FOS to MurA and, thus, the inability to proceed in peptidoglycan formation result in a bactericidal activity of the drug (Candel et al., 2019). Since both Gram-positive and -negative bacteria requires the formation of N-acetylmuramic acid for peptidoglycan, FOS presents a broad-spectrum antibiotic activity against the main genera in clinical practice, including carbapenemase- and/or extended-spectrum β-lactamase (ESβL)-producing Enterobacterales, methicillin-resistant Staphylococcus aureus (MRSA), glycopeptide-resistant enterococci and multidrug-resistant (MDR) Pseudomonas aeruginosa (Putensen et al., 2019). Chemically, FOS has a simple structure consisting in an active epoxic group bonded, through a carbon molecule, to a phosphorous (Baron et al., 1986). FOS has some unique features such as low molecular weight (138.06 g/mol) and protein binding capabilities, providing it with high tissue penetration (volume of distribution of 0.3 L/kg) (Candel et al., 2019). FOS mode of action was first described in 1974 by Kahan and colleagues, and the in vitro standardization testing was provided by Andrews et al. in 1983 (Hirschl et al., 1980; Andrews et al., 1983). Despite FOS advantages, intravenous use of FOS almost disappeared from clinical practice, partly due to its incongruency of in vitro results in early susceptibility testing (Barnett et al., 1969). FOS is available in three formulations: two orally used calcium salt form (C3H5O4PCa;194.2) and FOS tromethamione (C7H18NO7P; 259.194), and an intravenously used disodium salt (C3H5O4PNa2; 182.03) (Falagas et al., 2016). In 1996, Food and Drugs Administration (FDA) approved the clinical use of oral FOS (Monurol) in the treatment of uncomplicated lower urinary tract infections (UTIs), as acute cystitis. In the following years, FOS oral formulation was also approved in perioperative prophylaxis for transrectal prostate biopsy in adult man, post-operative treatment of UTIs, recurrent UTIs, acute uncomplicated UTIs in children and acute cystitis during pregnancy. In 2020, the European Medicine Agency (EMA) approved FOS for infusion in the treatment of a wide range of conditions (e.g. complicated urinary tract infections, bone and joint infections, bacterial meningitis) when the commonly recommended drugs are considered inappropriate (Figure 1). Some European countries such as Austria, France, Germany, Greece, and Spain allow the use of FOS intravenously with other antibiotics, such as β-lactam antibiotics or fluoroquinolones in critically ill patients suffering from carbapenem-resistant Enterobacterales infections (Michalopoulos et al., 2011). This is due to FOS’ unique mechanism of action and to the absence of side effects as nephrotoxicity, typical of aminoglycosides or colistin (Michalopoulos et al., 2010). FOS usage in veterinary settings is forbidden in China and European countries, while in Central and South America regions, such as Brazil and Argentina, is largely administered in diseased broiler chickens and pigs (Pérez et al., 2014; Wang et al., 2017). In 2016, WHO categorized phosphonic acid derivatives as critically important antibiotic in human medicine highlighting their high frequency use in human medicine and their role as available therapy to treat serious bacterial infections in people. Despite the relevance in human medicine, data concerning FOS susceptibility profiles have not been included yet in annual report on antimicrobial resistance by WHO or ECDC. Consequently, the global epidemiology of FOS resistant profiles and FOS-modifying enzymes is still incomplete and not well monitored.
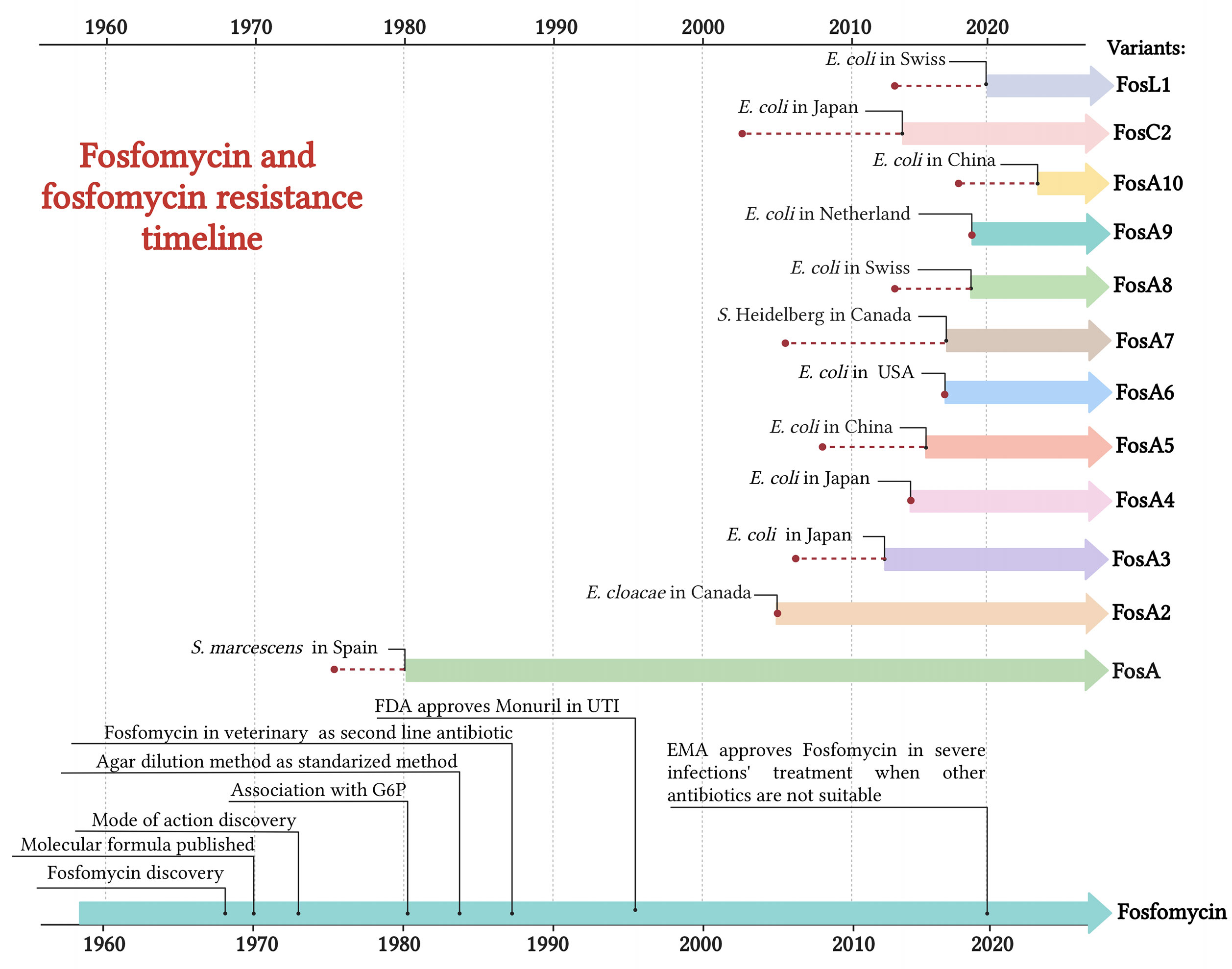
Figure 1 Timeline of FOS usage and the emergence of acquired FOS resistance determinants. Red dot = year of isolation. Created with BioRender.com.
FOS target
FOS binds and inhibits the UDP-GlcNAc enolpyruvyl transferase (MurA), acting as a phosphoenolpyruvate (PEP) analogue (Brown et al., 1995; Aghamali et al., 2019). MurA is a fundamental enzyme involved in the initial steps of peptidoglycan biosynthesis (Brown et al., 1995; Aghamali et al., 2019). FOS carries out its inhibiting activity to MurA through a covalent binding between the thyol group of a cysteine and the MurA active site, Cys115 (Figure 2). This inhibitory effect occurs in the cytoplasm and impairs an earlier stage of peptidoglycan biosynthesis when compared with that of β-lactamases or glycopeptides (Kahan et al., 1974; Eschenburg et al., 2005)..
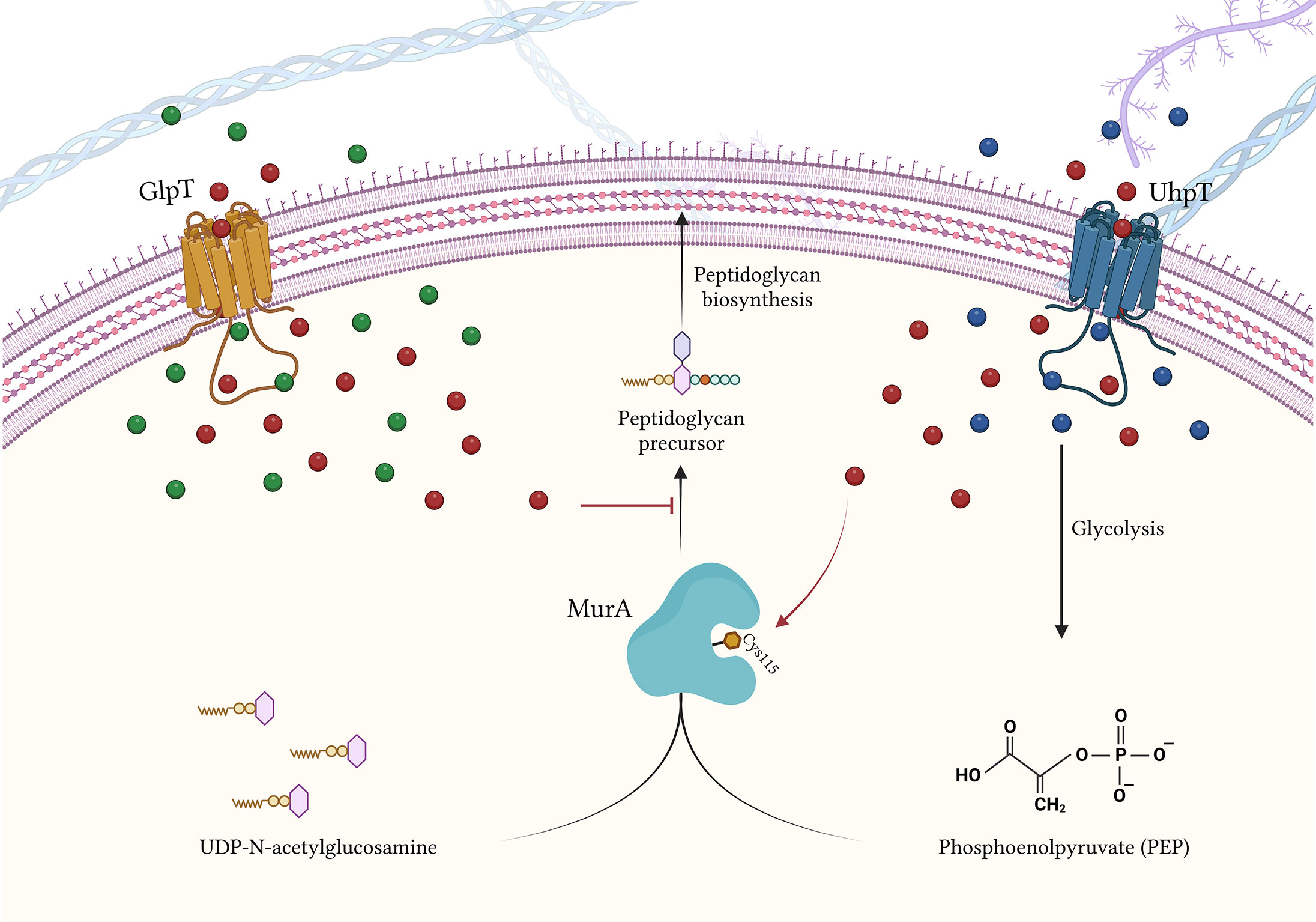
Figure 2 FOS influx inside the bacterial cell via GlpT and UhpT transportes, and FOS mode of action. Blue bubbles = G6P; green bubbles = G3P; red bubbles = FOS. Created with BioRender.com.
FOS transportation into the bacterial cell
The FOS intake has been mainly characterized in E. coli. To overcome bacterial wall, FOS takes advantage of the transportation activity of GlpT (glycerol-3-phosphate transporter) and UhpT (hexose-6-phosphate: phosphate antiporter) (Ambudkar et al., 1990; Aghamali et al., 2019) (Figure 2).
GlpT transporter
GlpT is a member of the organophosphate phosphate antiporter (OPA) family and is highly conserved in several species such as Escherichia spp., Klebsiella spp., Salmonella spp., and Citrobacter spp (Kahan et al., 1974).. GlpT is structured into two transmembrane domains, each composed of six highly conserved α-helices, that are linked by a long central loop (Lemieux et al., 2004). The glpT gene is part of the glp regulon, that controls the catabolism of G3P, glycerol and glycerophosphodiesters (Yang and Larson, 1998) (Figure 3). The extracellular G3P enters the bacterial cell through GlpT and control the expression of GlpT itself (Castañeda-García et al., 2013). In details, G3P binds to GlpR (G3P regulon repressor) that regulates the transcription of glp regulon, including glpT (Yang and Larson, 1998; Lemieux et al., 2004; Escapa et al., 2013) (Figure 3). In absence of G3P, GlpR binds to the operators of glp regulon, located in proximity of the promotor regions, and decreases the expression levels of glp regulon, including glpT (Yang and Larson, 1998) (Figure 3). When present, G3P binds to GlpR and lower GlpR-binding affinity with glp regulon, preventing the binding of GlpR to glpT promotor. The inability to bind the operator blocks glpT repression, leading to an increase of its expression levels (Cozzarelli et al., 1968; Law et al., 2009) (Figure 3).
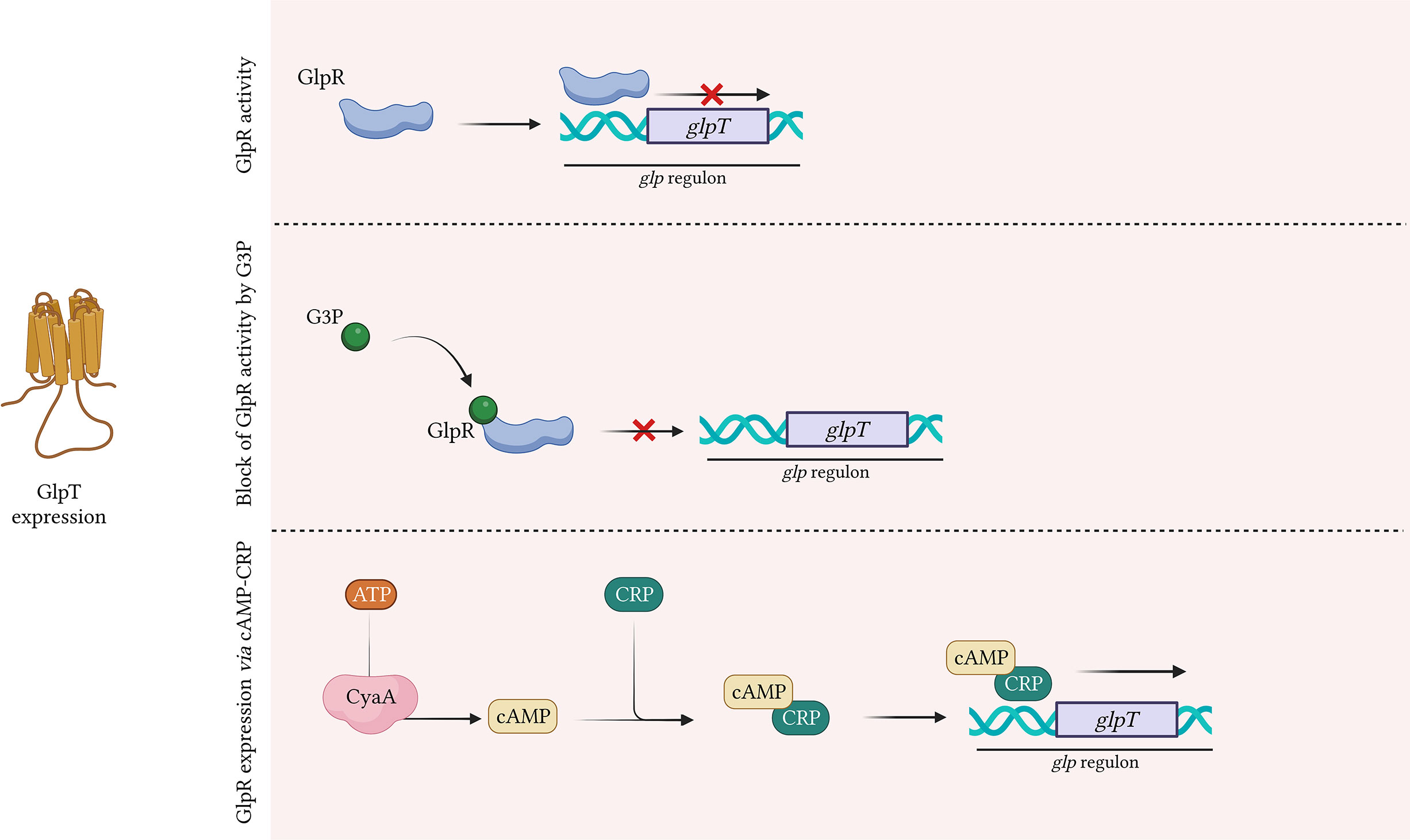
Figure 3 GlpT expression by GlpR. G3P positively regulates glpT expression. G3P binds GlpR repressor, reducing its affinity to glpT promotor. Without G3P, the repressor GlpR binds glpT promotor, derepressing glpT expression. CRP-bound cAMP binds glpT promotor and positively regulates glpT expression. Created with BioRender.com.
UhpT transporter
An alternative route for FOS influx is via UhpT transport system. UhpT is a monomer consisting of twelve transmembrane α-helical segments, which show high amino acid sequence homology with GlpT (Ambudkar et al., 1990). UhpT is a member of the Major Facilitator Superfamily (MFS) and promotes the entry of G6P, fructose-6-phosphate and mannose-6-phosphate inside bacterial cell (Hall and Maloney, 2005). The UhpT system is exclusive to Enterobacteriaceae, except for Proteus spp. and Staphylococcus spp (Silver, 2017)..
In the presence of G6P, UhpT expression is highly induced (Yang et al, 2016), leading to an increase of FOS flow inside the cell (Xu et al., 2017).
UhpABC system
To induce the expression of UhpT, G6P interacts with the UhpABC system, composed of three proteins: the transcriptional regulatory protein UhpA, the signal transduction histidine-protein kinase/phosphatase UhpB and the membrane sensor protein UhpC (Västermark and Saier, 2014) (Figure 4). UhpC senses external G6P and interacts with UhpB, stimulating the autokinase activity of UhpB (Friedrich and Kadner, 1987). The activated UhpB transfers its phosphate to UhpA, activating it. Thus, UhpA induces the transcription of uhpT by binding specifically to the uhpT promoter gene (Dahl et al., 1997) (Figure 4). In addition, to completely activate uhpT transcription, UhpA requires the presence of cAMP-CRP complex (Escapa et al., 2013).
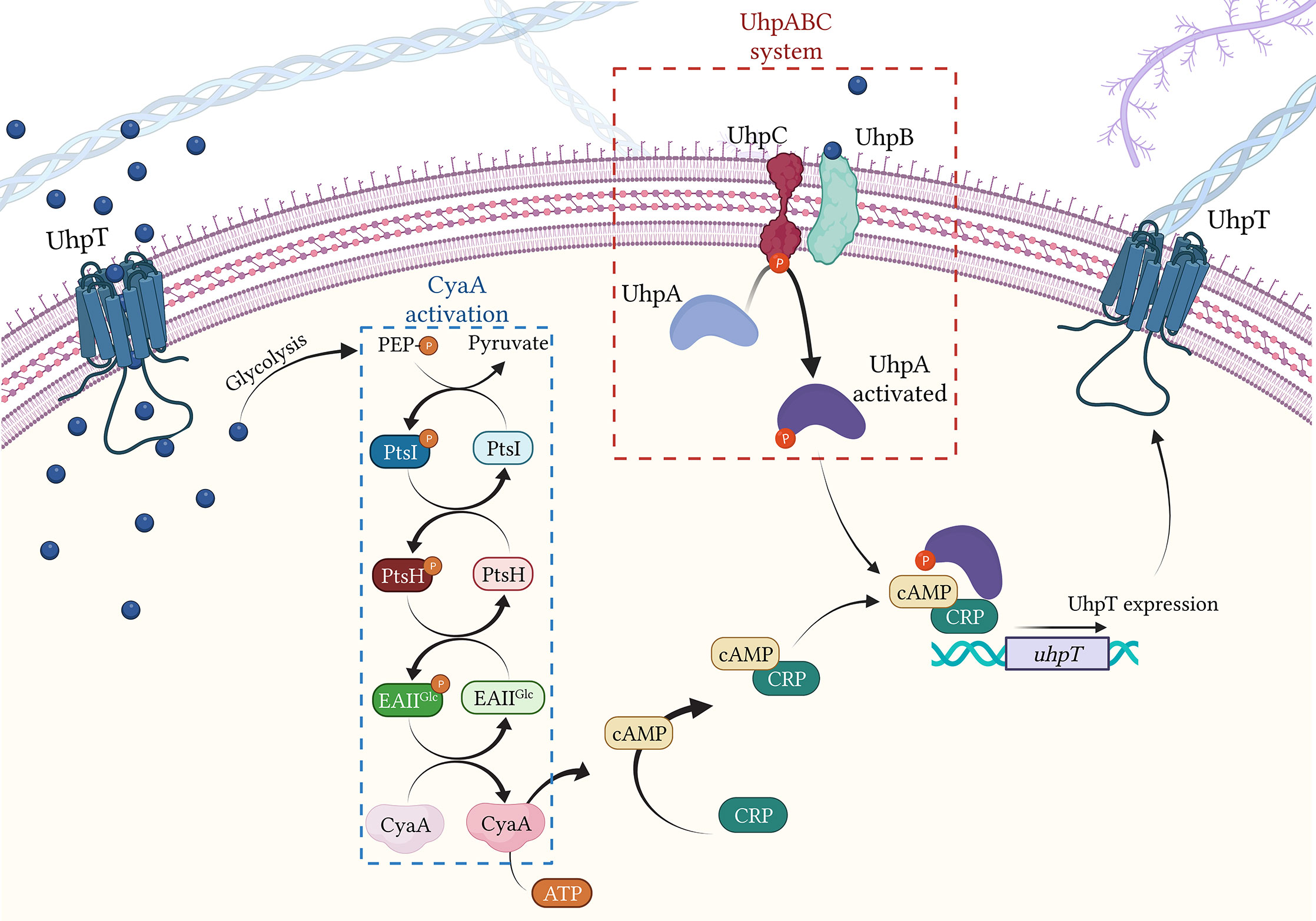
Figure 4 Regulation of UhpT expression by CyaA activation and UhpABC system. Blue bubbles = G6P; green bubbles = G3P; red bubbles = FOS. Created with BioRender.com.
cAMP and adenylate cyclase CyaA
The transcription of both glpT and uhpT is under the control of the adenylate cyclase CyaA. CyaA catalyzes the formation of cAMP (cyclic adenosine monophosphate) from ATP (Figure 4). Once produced, cAMP showed high affinity with the transcriptional regulator CRP (DNA-binding transcriptional dual regulator) and binds together leading to the formation of the cAMP-CRP complex. Concerning UhpT expression, the cAMP-CRP complex binds the activated UhpA and together attach the uhpT promotor, inducing its transcription (Castañeda-García et al., 2013). Similarly, regarding GlpT expression, the cAMP-CRP complex alone attaches to glpT promotor (Castañeda-García et al., 2013) (Figure 4).
Activation of CyaA
The activation of CyaA requires the presence of G6P and of the PTS system, the carbohydrate phosphotransferase system (Postma et al., 1993) (Figure 4). The PTS system is a sugar-phosphorylating system described in E. coli and requires three different entities: Enzyme I (PtsI), the heat-stable, histidine-phosphorylatable protein HPr (PtsH) and Enzyme II (composed by the domains EIIAGlc) (Deutscher et al., 2014). Once in the bacterial cell, G6P enters the glycolysis cycle, which leads to the production of the PEP. The formed PEP undergoes to the PTS system, transferring a P group to PtsI (Deutscher et al., 2014). Thus, PtsI activates through phosphorylation PtsH, which consequently activates EIIAGlc, transferring the P group to EIIAGlc (Saffen et al., 1987). Then, the activated EIIAGlc induces the activation of CyaA (Mazé et al., 2014) (Figure 4).
Mechanisms of fosfomycin resistance
The recent use of FOS and co-selection phenomena have contributed to the development of FOS resistance and its dissemination. FOSR mechanisms can be divided into three major groups: (a) modification of the antibiotic target MurA, (b) reduced permeability to FOS, and (c) acquisition of AMR genes. According to the recent literature, the reduction of FOS permeability is considered as the most frequent resistance mechanism (Nilsson et al., 2003; Castañeda-García et al., 2013; Silver, 2017).
Modification of the target
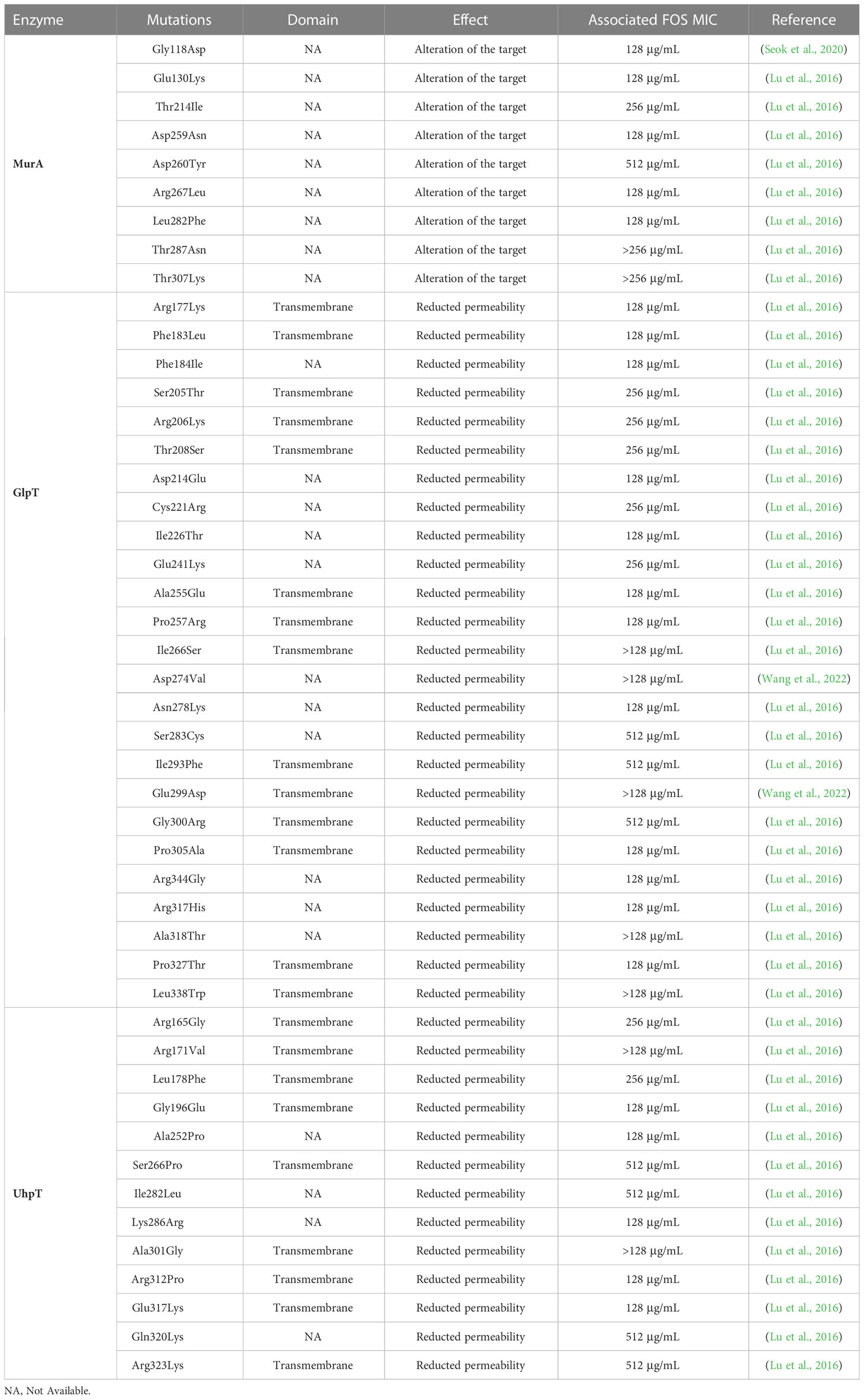
FOS inactivates MurA by binding to its active site, Cys115 (Skarzynski et al., 1996). Kim and colleagues demonstrated that Cys115 substitutions in MurA, as Cys115Asp, lead to in vitro FOSR (MIC > 512 mg/ml) in E. coli (Kim et al., 1996). However, mutations in MurA are uncommon in clinical isolates and none occurred in the catalytic site of MurA (Castañeda-García et al., 2013). Indeed, the first reports of mutations occurring in MurA from clinical E. coli isolates dated to 2010 in Japan, where the substitutions Asp369Asn and Leu370Ile were suggested to lead to development of FOSR in vivo (Takahata et al., 2010). Both mutations, occurring in two highly conserved residues, decreasing the susceptibility to FOS with MIC up to 512 mg/ml (Takahata et al., 2010). Subsequently, mutations in MurA associated to FOSR profiles have been detected from clinical E. coli isolates in China (Li et al., 2015; Bi et al., 2017), Taiwan (Tseng et al., 2015) and South Korea (Seok et al., 2020) (Table 1). Regarding clinical isolates of K. pneumoniae, Lu and coauthors reported several alterations in MurA sequence of ESβL-producing K. pneumoniae from Taiwan associated with FOSR profiles (MICs = 128 μg/mL) (Lu et al., 2016). The exposition of FOS to bacterial strains can induce covalent modifications in MurA, increasing the enzyme synthesis (Marquardt et al., 1992). Interestingly, the overexpression of murA gene in E. coli is able to confer clinical levels of FOSR (MIC=32 µg/mL) with a low fitness cost (5%) (Horii et al., 1999; Couce et al., 2012) (Table 2).
Permeability impairment
GlpT system
Impairment in GlpT activity is one of the most common mechanisms of FOSR. Strains defective in GlpT transport are not able to grow using G3P as sole carbon source (Aghamali et al., 2019). In literature, there are several reports of common mutations in GlpT associated with reduced permeability and thus increased FOS MICs (Table 1). The deletion and/or truncation in GlpT protein are associated with reduction in permeability and loss of function in E. coli strains (Li et al., 2015; Ohkoshi et al., 2017). In 2020 Sorlozano-Puerto and colleagues investigated the effect of several mutations in GlpT from E. coli clinical isolates from Spain. The biological impact of such mutations was predicted through bioinformatic tool and tested by carbon grow test. The study identified possible alterations with a deleterious effect on GlpT activity, such as Gly84Asp, Pro212Leu, Leu373Arg, and thus a direct involvement in FOSR (Sorlozano-Puerto et al., 2020) (Table 1). Differently, deletion W28del occurring in GlpT has been associate to FOS MICs >128 μg/mL in clinical ST131 E. coli from clinical setting in Czech Republic (Mattioni Marchetti et al., 2023). Another study evaluated mutations in GlpT from ESβL-producing K. pneumoniae from hospitals in Taiwan. In this study, Lu and colleagues identified several single amino acid substitutions, occurring in the transmembrane domains, such as Arg206Lys, Ile266Ser and Ile293Phe and associated with FOS resistance at high levels (FOS MICs = 256 μg/mL) (Lu et al., 2016) (Table 2).
UhpT system
Similar to GlpT, mutations in UhpT are likely to reduce G6P entry inside bacterial cell and thus FOS permeability. Indeed, the complete loss of UhpT peptide leads to the complete loss of the transport function and leads to FOSR at high levels (FOS MICs >128 μg/mL) (Takahata et al, 2010; Li et al., 2015; Ohkoshi et al., 2017; Falagas et al., 2019). Different mutations have been reported in both E. coli and K. pneumoniae clinical strains, occurring in both transmembrane and topological domain, associated with a wide MICs range of FOSR (64 μg/mL - 512 μg/mL) (Tseng et al., 2015; Seok et al., 2020; Ortiz-Padilla et al., 2022). Interestingly, Ballestero-Téllez and coauthors described the in vitro effect of premature Gln345stop in UhpT, which showed FOS MICs higher than 1,024 μg/mL in E. coli (Ballestero-Téllez et al., 2017).
UhpABC system
Impairment in the activity of UhpABC system might reduce the effectiveness of bacterial transportation systems and, consequently, reduce FOS influx into the bacterial cell (Kadner and Shattuck-Eidens, 1983). The loss of entire UhpA portion leads to different extent of FOSR (MIC > 32 μg/mL) (Ohkoshi et al., 2017; Falagas et al., 2019), while deletion of 163-188 aa or premature stop codon in UhpA contribute to high level of FOSR (MIC = 1,024 μg/mL) (Lucas et al., 2017; Ohkoshi et al., 2017). A study conducted by Cattoir et al. demonstrated the in vitro effect of mutations Gly469Arg in UhpB and Phe384Leu in UhpC. Both alterations showed a loss of function in the two regulators’ activity and an increased FOS MICs to resistance range (MIC = 128 μg/mL) (Cattoir et al., 2020). Another study conducted in 2017 highlighted the in vitro effect of mutations in UhpB (Thr27Stop, Gln262Stop, Trp181Stop, Leu255Stop, MIC = 1,024 μg/mL) and UhpC (Thr72Pro, = 1,024 μg/mL) in selected E. coli single-gene deletion mutants (ΔglpT, ΔuhpT, ΔcyaA and ΔptsI) (Ballestero-Téllez et al., 2017)(Table 1).
Regulation in cAMP levels
Despite the relevant implication of CyaA activity in GlpT and UhpT expression, investigation of mutations in CyaA and its eventual effect on FOS MICs are still not clear, with just few reports conducted in E. coli strains (Yang et al, 2016; Ohkoshi et al., 2017) (Table 1).
Acquisition of antibiotic resistance genes
FosA family
FosA group is a class of metalloenzymes able to disrupt the epoxide ring of FOS drug. It depends on manganese (II) and potassium as cofactors, and on glutathione (GSH) as nucleophilic molecule. Nowadays, 11 different and genetically related variants have been deposited in GenBank Database and 10 of these are reported in the global dissemination scenario (Figures 5–7). In accordance with Ito et al., 2017, fosA genes are chromosomally distributed in Providencia stuartii, Providencia rettgeri, K. pneumoniae, Klebsiella oxytoca, Serratia marcescens, Enterobacter aerogenes and Enterobacter cloacae genomes, while they are rarely reported in E. coli, Citrobacter freundii, Proteus mirabilis and Acinetobacter baumannii (Zurfluh et al., 2020).
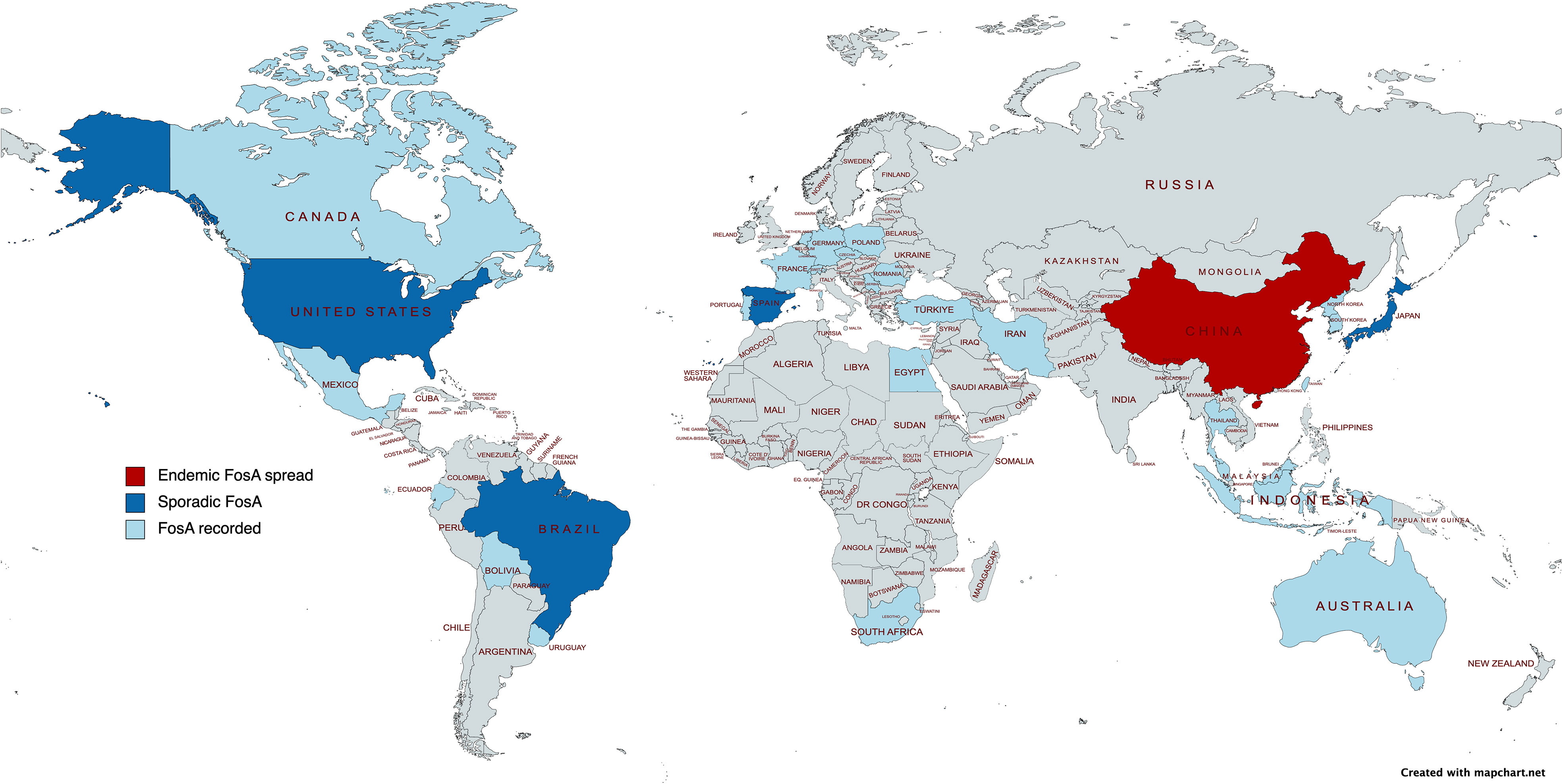
Figure 5 Epidemiological map of FosA among Enterobacterales. Created with mapchart.net.
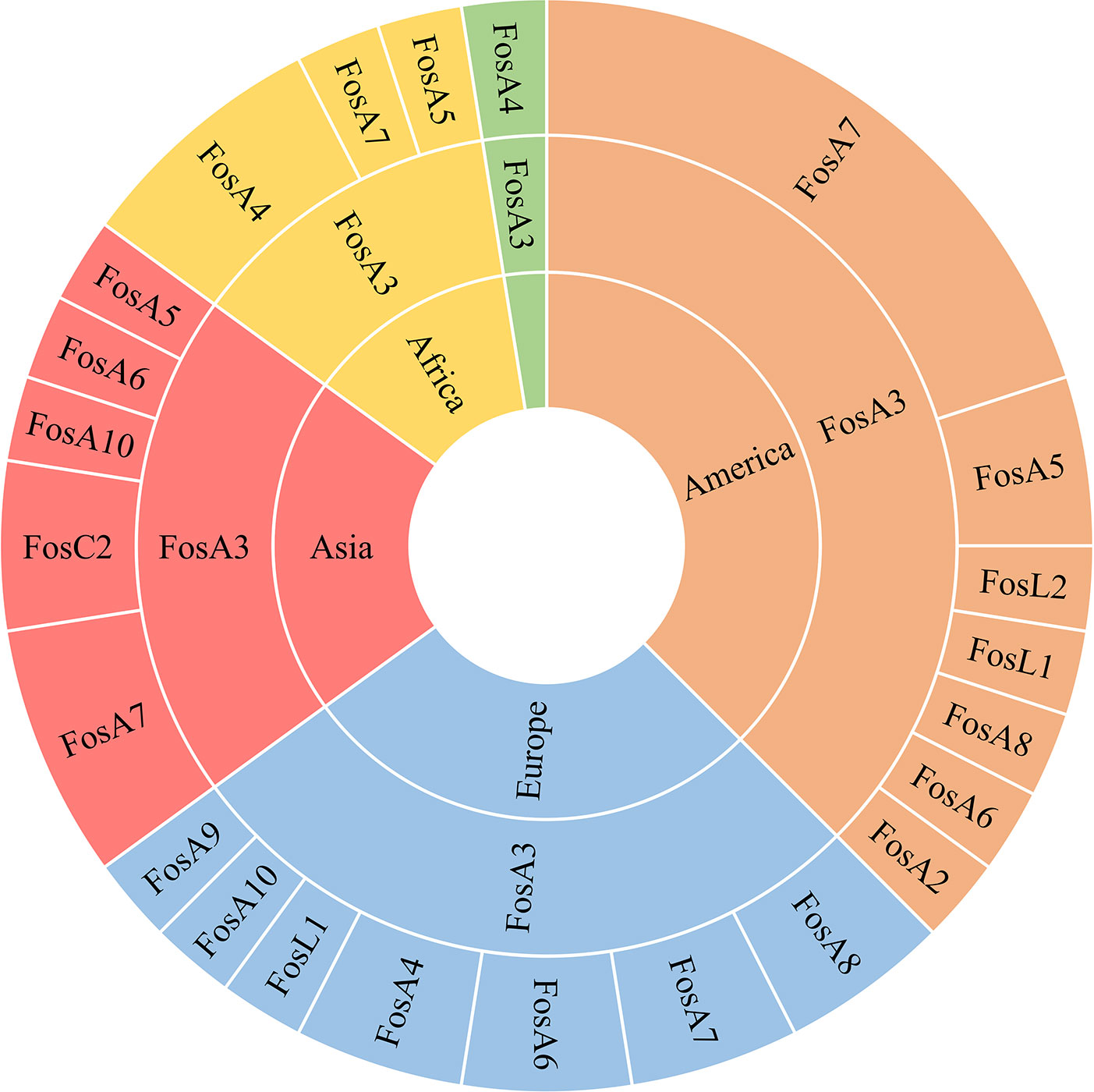
Figure 6 Hierarchy representation of FosA/C2/L1 prevalence in the continents Africa (yellow), America (orange), Asia (red), Europe (light blue) and Oceania (green).
FosA and FosA2
The first plasmid-mediated fosA was identified and isolated from a clinical sample of S. marcescens in Spain in 1980 (Mendoza et al., 1980) (Figure 1). FosA was located on a Tn2921 cassette on the plasmid pSU912 (Seoane et al., 2010) (Figure 7A). The origin of FosA is linked with the FOS-modifying enzyme FosEC, located on E. cloacae chromosome (100% identity) (García-Lobo and Ortiz, 1982; Ito et al., 2017).
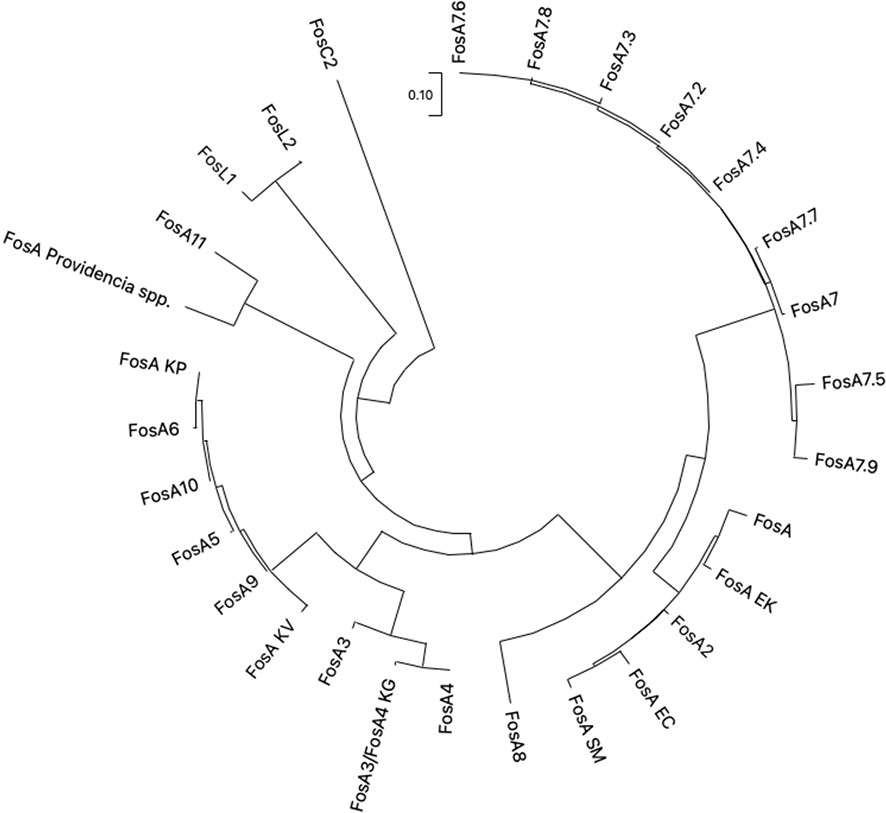
Figure 7 The evolutionary analysis and phylogenetic tree of FosA/C2/L1-2 proteins found in Enterobacterales were inferred by using the Maximum Likelihood method and ITT matrix-based model using MEGA 11. FosA KP (CDO16183.1), FosA KV (AWG41960.1), FosA EC (AWG41971.1), FosA EK (VAX69325.1), FosA P (WP_154635598.1), FosA SM (QOW96986.1), FosA2 (WP_025205684.1), FosA3 (WP_014839980.1), FosA4 (WP_034169466.1), FosA4 KG (WP_064548962.1), FosA5 (WP_012579083.1), FosA6 (WP_069174570.1), FosA7 (WP_000941934.1), FosA7.2 (WP_000941935.1), FosA7.3 (WP_023231494.1), FosA7.4 (WP_023216493.1), FosA7.5 (WP_000941933.1), FosA7.6 (WP_061377147), FosA7.7 (WP_058653118.1), FosA7.8 (WP_079820715.1), FosA7.9 (WP_071684814.1), FosA8 (WP_063277905.1), FosA9 (WP_114473955.1), FosA10 (WP_004214174.1), FosA11 (QZL11398.1), FosL1 (WP_161667239.1), FosL2 (WP_188331883.1). KP = K. pneumoniae, KV = K. variicola, KG = K. georgiana, EK = E. kobei, SM = S. marcescens, EC = E. cloacae.
FosA2 variant was first reported in 2011 (Xu et al., 2011) in E. cloacae chromosome from a water sample in Canada (Figure 1). Currently, fosA2 reports are correlated with chromosomal location only.
FosA3
FosA3 is the plasmid-acquired subtype mostly disseminated and reported (Figure 8). FosA3 shows close relation (>94% identity) to the chromosomally encoded FosAKG from Kluyvera georgiana. The first report is dated 2010 from a clinical isolates E. coli in Japan (Figure 1). Shortly after, in 2013, a fosA3 plasmid-mediated dissemination among food-chain animals in Chinese region was reported (Hou et al., 2012; Ho et al., 2013). Currently, China has the highest dissemination of plasmid-mediated fosA3 among both clinical and veterinary settings (Figures 5, 8). Concerning Chinese veterinary field, several animal species have been identified as silent reservoir, ranging from pets, as dogs and cats, to food-chain animals, as pigs and bovines, and wild animals, as pigeons. FosA3 is organized in a composite transposon, of 4 kb in size, consisting in two IS26 elements with the same orientation, that flank the cassette fosA3-orf1-orf2-Δorf3 (Wachino et al., 2010) (Figure 9A). fosA3 genes are located 316 bp downstream of IS26, while the spacer region between the 3′ end of fosA3 and IS26 can varies in size (1,758 bp, 536 bp and 370 bp) (Figure 9A). Interestingly, the 1,758 bp region shows 79% nucleotide identity with part of K. pneumoniae strain 342 chromosome (Hou et al., 2012). Based on the transposon composition, five major fosA3 environment can be classified in Enterobacterales (named type A-E): A) IS26-fosA3-orf1-orf2-Δorf3-IS26, B) IS26-fosA3-orf1-Δorf2-IS26, C) IS26-fosA3-Δorf1-IS26, D) IS26-ISEcp1-blaCTX-M-14-ΔIS903D-fosA3-orf1-Δorf2-IS26, E) IS26-ISEcp1-blaCTX-M-65-ΔIS903D-fosA3-orf1-orf2-Δorf3-IS26 (Figures 9A–E). The type A is the predominant type, and it is associated to IncF, IncI1, IncN, IncB/O and several untypable plasmids (Ho et al., 2013; Liu et al., 2022). FosA3 can be easily co-harbors on the same plasmid with other ESβLs, as blaCTX-M-3, blaCTX-M-8, -14, -55, -65 and -123 (Ho et al., 2013; Mazé et al., 2014; Xie et al., 2016; Yang et al, 2016; Dantas Palmeira et al., 2018). The first dissemination of FOSR in several E. coli strains from veterinary settings was reported by Hou et al. (2012). The study identified E. coli isolates co-harboring fosA3 and blaCTX-M-65 on IncF plasmids. The fosA3 cassette consisted of fosA3 transposon Type B (Figure 9B), with a spacer region between the 3’ end of fosA3 and IS26 of 536 bp (Hou et al., 2012).
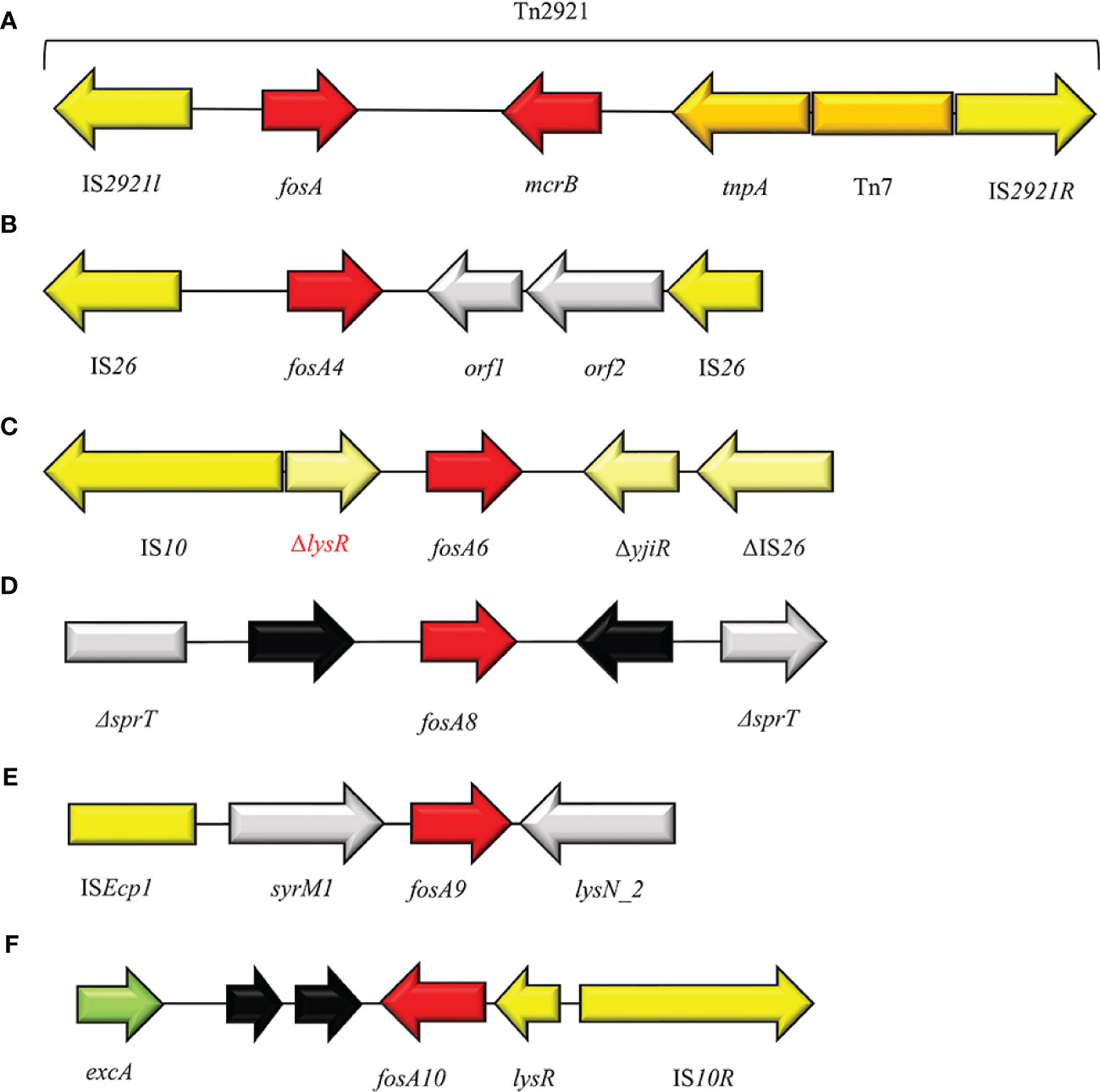
Figure 8 Structure of representative genetic environments of (A) fosA (FJ829469.1), (B) fosA4 (Loras et al., 2021), (C) fosA6 (KU254579.1), (D) fosA8 (SAMN12496803), (E) fosA9 (Wang et al., 2019), (F) fosA10 (MT074415.1). Yellow = IS, light yellow = deleted IS, red = antimicrobial resistance genes, gray = open-reading frame, black = unknown proteins, green = surface exclusion protein.
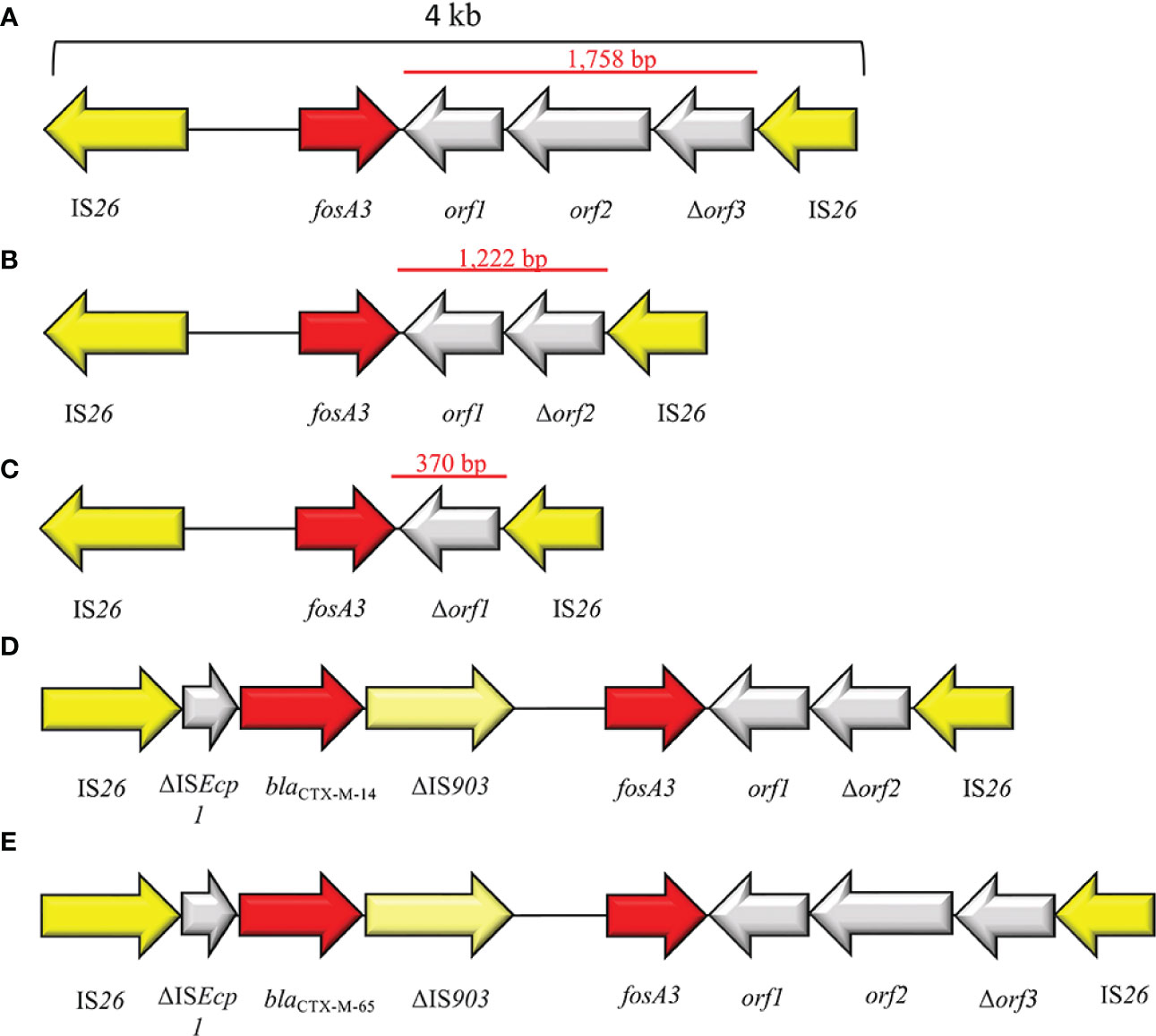
Figure 9 Representation of genetic environments of fosA3. (A) (JQ432559), (B) (JX442752), (C) (JX442751), (D) (JQ823170), (E) (JX442753). Yellow = IS, light yellow = deleted IS, red = antimicrobial resistance genes, gray = open-reading frame.
In Korea, a point prevalence study highlighted the presence of seven Enterobacteriaceae strains co-producing FosA3 and CTX-M out of 347 ESβL producers. All the seven strains harbored fosA3 + blaCTX-M-like in the same IS26-composite transposon (Sun et al., 2012). Ho et al. in 2013 evaluated the dissemination of plasmid-mediated fosA3 gene among animals and humans, highlighted 97 FosA3-producing E. coli strains out of 1,693 (Ho et al., 2013). Wei Jiang et al. screened 234 CTX-M-producing E. coli isolates collected from chickens from 2014 to 2016 in China and identified 64 fosA3+blaCTX-M-like positive E. coli located on IncFII, IncI1, IncHI2 and IncB/O. Additionally, the authors identified two novel genetic environments surrounding the fosA3 (ISEcp1-blaCTX-M-65-ΔIS903D-IS26-fosA3-orf1-orf2-Δorf3-IS26 and IS26-ISEcp1-blaCTX-M-3-orf477-blaTEM-1-IS26-fosA3-orf1-orf2-Δorf3-IS26) (Jiang et al., 2017). In E. coli, genomic studies highlighted the occurrence of FosA3 in ST10 (Seok et al., 2020), ST12 (Hameed et al., 2022), ST38 (Norizuki et al., 2018; Hameed et al., 2022), ST 46 (Hameed et al., 2022), ST57 (Hameed et al., 2022), ST69 (Hameed et al., 2022; Liu et al., 2022), ST95 (Hameed et al., 2022), ST106 (Seok et al., 2020), ST117 (Fernandes et al., 2018; Zhao et al., 2022), ST131 (Galindo-Méndez et al., 2022; Hameed et al., 2022), ST1193 (Hameed et al., 2022), ST1196 (Hameed et al., 2022), ST2736 (Wang et al., 2018), ST7584 (Hameed et al., 2022), ST10184 (Hameed et al., 2022), ST11350 (Ewbank et al., 2022), ST648 (Yang et al, 2016), ST156 (Xie et al., 2016). The occurrence of plasmid-mediated fosA-like genes turns out to be worrying in ST131 clone due to its virulence and pathogenic features (Forde et al., 2019; Galindo-Méndez et al., 2022; Hameed et al., 2022). Indeed, ST131 is a globally dominant MDR clone associated with UTI, and it is involved in the global dissemination of ESβLs as CTX-M-15 type (Forde et al., 2019; Jafari et al., 2020). However, from 2014 to 2018 Liu and colleagues evaluated the prevalence of mobile fosA3 gene in ducks from 23 Chinese farms and they highlighted the predominance of the ST69 as fosA3-harboring clones among E. coli strains (Liu et al., 2022). Similarly, Loras et al. identified an ST69 E. coli strain from urine sample in Spain and co-harboring fosA3+blaCTX-M-55 (Loras et al., 2021). Another clinical case of E. coli ST69 harboring a plasmid-mediated fosA3 (IncN) was identified in Uruguay from pediatric UTI cases (Garcia-Fulgueiras et al., 2022). ST69 is an emerging Extraintestinal Pathogenic E. coli (ExPEC) lineage detected in both humans and animal settings, that is globally involved in UTI from both the community and the hospital environment (Giacobbe et al., 2015; Hammad et al., 2019). E. coli ST69 was originally susceptible to almost all the antibiotic families, but the acquisition of β-lactams and FOSR traits could affect the use of FOS in UTI treatment (Doumith et al., 2015; Garcia-Fulgueiras et al., 2022).
First isolation of plasmid-mediated fosA3 in clinical K. pneumoniae strains was in 2012, when Lee and co-authors described the co-presence of fosA3+blaCTX-M-14 on an IncN plasmid and organized in IS26-ISEcp1-blaCTX-M-14-ΔIS903D-IS26-fosA3-orf1-orf2-Δorf3-IS26 (with a spacer sequence of 1,222 bp) (Lee et al., 2012). Lately, in 2015, Jiang Y et al. reported the characterization of 94 KPC+FosA3 co-producing K. pneumoniae collected from twelve Chinese hospitals. Additionally, the authors highlighted a clonal relation between KPC- and FosA3-producers, indicating a FOSR clonal dissemination in China (Jiang et al., 2017). In K. pneumoniae plasmid-mediated fosA3 is largely associated with isolates belonging to ST 11 (Xiang et al., 2015; Nishida et al., 2020), ST37 (Taniguchi et al., 2017), ST485 (Zhou et al., 2022). In recent years, a secondary spread of plasmid-mediated fosA3 occurred in Salmonella spp. among food-chains animals and humans in China (Wang et al., 2020; Zhang et al., 2020). Outside Chinese settings, similar cases have been recorded from pediatric patients in Spain (Vázquez et al., 2022), from clinical patients in USA (Turcotte et al., 2022), and from a wild bird in Germany (Villa et al., 2015). Noteworthy, Villa and colleagues described the first case of a Salmonella enterica Serovar Corvallis co-producing FosA3+NDM-1+CMY-16. FosA3 and blaNDM-1 were located on the same IncA/C2 plasmid and fosA3 included in a type A transposon (Villa et al., 2015). This report highlighted the bird’s migration as route for environmental diffusion of fosA3 from norther Asia to Europe (Villa et al., 2015). Among Salmonella spp. strains, transposon Type A is the most spread fosA3 environment, located on IncFII (Lin and Chen, 2015) and IncHI2 (Wong et al., 2016), followed by Type C on IncFIB (Vázquez et al., 2022) and type D on IncHI2 (Wong et al., 2016). Interestingly, a multi-replicon IncC-IncN plasmid, coharboring fosA3 Type A and blaCTX-M-14 have been already isolated from chickens in China (Zhang et al., 2020). FosA3 cases occurred in Salmonella ST32 (Vázquez et al., 2022), ST17 (Wang et al., 2022), ST34 (Wang et al., 2022), ST198 (Wang et al., 2022). Since 2017, few reports evaluate the occurrence of plasmid-mediated fosA3 in P. mirabilis from both hospitalized patient and food-chain animals (He et al., 2017; Hua et al., 2020; Lei et al., 2020). The first case focused on the chromosomal integration of blaCTX-M-14/blaCTX-M-65 and fosA3 in P. mirabilis collected in 2015 from diseased broilers in China, with the following compositions: a) IS26–ΔISEcp1–blaCTX-M-14–ΔIS903–fosA3–1,222 bp–IS26; b) IS26–ΔtraI–fip–ΔISEcp1–blaCTX-M-65–IS903D–iroN–IS26–fosA3–1758 bp–IS26. In the same study, the presence of the transposition unit “b” was detected in IncHI2 plasmid from E. coli ST117, together with the presence of minicircles that contain fosA3, blaCTX-M-65 and IS26 (He et al., 2017). Thus, the authors speculated the fosA3+blaCTX-M-65 integration into the P. mirabilis chromosome via a transposable minicircle from E. coli (He et al., 2017). Similarly, the presence of minicircles harboring IS26 and fosA3 was identified even in S. enterica from a Chinese chicken and speculations about their role in fosA3 acquisition and spread are under evaluation (Zhang et al., 2020). Similar environments containing blaCTX-M-65 + fosA3 were identified in retail meat and aquatic products from markets (Ma et al., 2022), from diseased pig (Lei et al., 2018; Song et al., 2022) and from retail chickens (Lu et al., 2021) from Chinese regions, while the co-expression CTX-M-3+FosA3 was reported from Chinese chicken (Turcotte et al., 2022). Rather worrying was the isolation of a KPC-2+CTX-M-65+FosA3 producing P. mirabilis from a Chinese 49-year-old female with a pulmonary infection (Hua et al., 2020). The blaCTX-M-65+fosA3 was located on an IncFII-33 and the authors emphasized the successful association of IS26 and IncFII-33 in spreading antimicrobial resistance features (Hua et al., 2020).
FosA3 easily fits in different plasmid environments, including single- and multi-replicons. The major vehicle of plasmid-mediated fosA3 spread is IncFII (Hou et al., 2012), followed by IncI1 (Sato et al., 2013), IncN (Liu et al., 2022), IncHI2 (Chen et al., 2021), and IncP (Hameed et al., 2022). The successful and global diffusion of fosA3 could be explain by the combination of IS26 sequences and IncFII plasmids. FosA3 genes are mainly flanked by IS26, that play a fundamental role in AMR effective transposition and in their AMR dissemination among Enterobacterales (Partridge et al., 2018; Lv et al., 2020). Moreover, as mentioned elsewhere, IS26-flanked transposons are able to form circular intermediates that could accelerate the spread of fosA3 (He et al., 2015; Harmer and Hall, 2016). The IncFII plasmids are commonly low copy number plasmids and are recognized as vehicles of ESβLs dissemination among Enterobacterales (Muthuirulandi Sethuvel et al., 2019). Moreover, researchers speculate on the role of IncFII F33:A-:B- and F2:A-:B- in fosA3 dissemination due to its high adaptation levels (Hou et al., 2012; Sun et al., 2012).
European epidemiology of fosA3 is still low, with few reports from clinical E. coli strains in Spain (Loras et al., 2021), from clinical settings in Switzerland (Mueller et al., 2019), from veterinary and enviroment in Germany (Freitag et al., 2018), from clinical and veterinary settings in France (Benzerara et al., 2017; Lupo et al., 2018) and in Portugal (Mendes et al., 2016). Although reports in literature highlight a predominant association of fosA3 with blaCTX-M-like genes, recent studies revealed an emerging co-presence with carbapenemases in E. coli (Zhao et al., 2015; Peng et al., 2019), K. pneumoniae (Xiang et al., 2015; Singkham-In et al., 2020; Hao et al., 2021), C. freundii (Feng et al., 2015), E. cloacae (Hameed et al., 2022), Cronobacter sakazakii (Liu et al., 2017) and S. enterica (Villa et al., 2015). In literature, reports highlighted the co-expression of FosA3 and carbapenemases such as FosA3+KPC (Shi et al., 2018), FosA3+NDM (Tian et al., 2020), FosA3+VIM (Tang et al., 2020), FosA3+OXA-48 (Singkham-In et al., 2020), FosA3+KPC+IMP (Tseng et al., 2017; Peng et al., 2019), FosA3+KPC+NDM (Peng et al., 2019), FosA3+NDM+OXA-48 (Singkham-In et al., 2020).
In the last six years, the co-presence of EsβL+fosA3+mcr-like genes has been already detected in both clinical and veterinary environment (Birgy et al., 2018; Hoang et al., 2022). This combination of multi-resistance strains was reported in China (Zhao et al., 2018), France (Birgy et al., 2018) and Ecuador (Hoang et al., 2022). Worryingly, the co-expression of FosA3+MCR-1 and NDM-1/KPC-2 among E. coli strains has been already identified in hospitalized patients and food-chain animals (Zhao et al., 2018; Peng et al., 2019; Tian et al., 2020). Liu and colleagues described the occurrence of MCR-1+FosA3+NDM-like in several E. coli strains collected from chicken farm in China (Liu et al., 2017). The study identified the presence of i) MCR-1+NDM-9+FosA3 coproducing ST10; ii) MCR-1+NDM-4+FosA3 co-producing ST117; iii) MCR-1+NDM-1/-9+FosA3 co-producing ST156; iv) MCR-1+NDM-4/-9+FosA3 co-producing ST297; v) MCR-1+NDM-9+FosA3 co-producing ST2973 (Liu et al., 2017). During a surveillance study in 2015 among animal farms in Shandong, two pan-drug strains of C. sakazakii were isolated from sick chickens (Liu et al., 2017). The study clarified the copresence of blaNDM-9+fosA3, located on the same conjugative IncB/O plasmid, and mcr-1 on a IncI2 plasmid (Liu et al., 2017).
FosA4
FosA4 enzyme shows 94% amino acid identity with FosA3, and speculation proposes Kluvyera georgiana as possible origin of the plasmid-mediated resistance gene fosA4 (Nakamura et al., 2014; Rodriguez et al., 2018). FosA4 epidemiology is limited and varies geographically, but it was mainly reported in E. coli isolates (Figure 8). Increasing cases of FosA4-producing E. coli have been reported among food-chain animal settings in Egypt (Soliman et al., 2021; Sadek et al., 2022) and in France (Lupo et al., 2018). Other cases, concerning clinical settings, have been described from hospitals in Madrid (Loras et al., 2021) and Australia (Mowlaboccus et al., 2020). In Southern Turkey, Cansu Önlen Güneri and co-authors described a regional diffusion of plasmid-mediated fosA4 among E. coli collected from waste-water treatment plant (Güneri et al., 2022). The fosA4 gene has been reported predominantly on IncFII plasmid type and, consequently, on IncHI2 and IncI1 (Ma et al., 2015; Loras et al., 2021; Ramadan et al., 2021). IncFII and IncI1 normally harbors additional genes responsible for resistance to other antibiotics such as penicillins, sulphonamides and aminoglocosyde (Mowlaboccus et al., 2020; Ramadan et al., 2021). FosA4-harboring plasmids often coexist with blaCTX-M- and mcr-1-harboring plasmids (Ramadan et al., 2021; Sadek et al., 2022). FosA4 is associated with a conserved cassette of 4,022 bp in size, consisting of: two IS26, fosA4, tetR/acrR family and a helix-turn-helix domain. In southern Turkey, a novel genetic enviroment was detected, replacing the upper IS26 with an IS4 (Güneri et al., 2022) (Figure 7B). MIC data for fosA4 have been reported in E. coli as >1,024 µg/ml (Güneri et al., 2022).
FosA5
In 2015, Ma Y et al. reported the first case of fosA5 from a clinical E. coli strain in an inpatient with hospital-acquired pneumonia in China (Ma et al., 2015). FosA5 enzyme shares 69% amino acid sequence similarity with FosA and 80% with FosA3. The K. pneumoniae chromosome has been identified as the origin of fosA5 variant and its spread is associated with pKP96 plasmid, as reported by Ho PL et al., 2013 (Xu et al., 2011). The genomic enviroment of fosA5 is characterized by insA and insB and an IS10 in the opposite side (Figures 10A, B). In 2019 Wang S and colleagues investigated the genomic enviroment of an IncHI2A plasmid (pIMP26) coharboring blaIMP-26, blaDHA-1 and fosA5, isolated from an E. cloacae strain involved in blood infection (Wang et al., 2019). In pIMP26, the fosA5 structure was as follow: IS4, rfaY, lysR, fosA5, rfaY, ISVsa5 (IS4-like) (Wang et al., 2019). A similar organization of the fosA5 cluster has been detected in an IncHI2/2A plasmid (pEHZJ1) from an E. hormaechei of clinical origin (Gou et al., 2020). FosA5-carrying E. coli strains were found to be highly FOSR (MIC = 512 µg/ml) (Ma et al., 2015).
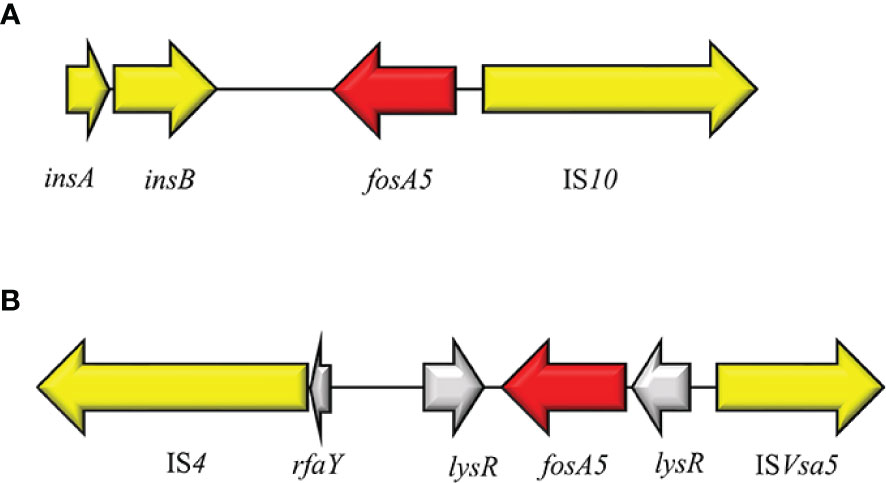
Figure 10 Structure of representative genetic environments of fosA5. (A) (KP143090), (B) (MH399264). Yellow = IS, light yellow = deleted IS, red = antimicrobial resistance genes, gray = open-reading frame.
FosA6
FosA6 was firstly described in a clinical CTX-M-2-producing E. coli ST410 from an US hospital in 2017 (Guo et al., 2016). FosA6 was carried on a self-conjugative IncFII plasmid (69 kb) and inserted in the cassette IS10R-ΔlysR-fosA6-ΔyjiR_1- ΔIS26, nearly identical to those on the chromosomes of some K. pneumoniae strains (Figure 7C). Moreover, fosA6 shared >99% sequence identity with chromosomally encoded fosA in K. pneumoniae. A point prevalence study conducted among seven Hospitals in Madrid, identified the only European case of ST354 E. coli producing FosA6 enzyme (Loras et al., 2021). FosA6-carrying E. coli had FOS MIC values of 128 to >1024 µg/ml (Guo et al., 2016).
FosA7
In 2015 Dhanani and colleagues investigated the resistome of four FOSR S. enterica serovars Heidelberg from broiler chickens among different commercial farms in Canada (Dhanani et al., 2015). As described later by Rehman et al., the 4 S. enterica strains produced a FosA-like enzyme, named FosA7, with a chromosomal location (Rehman et al., 2017).
Currently, 9 alleles of fosA7 genes are deposited in GenBank (fosA7.1-fosA7.9). All these variants have a chromosome location among different bacterial species. FosA7.5 and fosA7.9 are strictly linked with the chromosome of E. coli and C. freundii, respectively. In Salmonella spp. fosA7 is surrounded by two hypothetical proteins and located in an integrase cassette composed of Int-type II endonuclease-ATP helicase-type II methylase-RNA helicase-DNA helicase (Figure 11A). In E. coli the intercellular diffusion of fosA7.5 is due to the composite transposon flanked by ISL3 and IS3 (IS911 and ISEC52) elements (Figure 11B–E). A different composition has been highlighted for fosA7.9 in C. freundii: the fosA7.9 cassette is flanked by HNH endonuclease at both sides and organized in HNH endonuclease-fosA7.9-Fic family-type II restriction-DNA methyltransferase-AAA domain-HNH endonuclease (Figure 11F) (Mattioni Marchetti et al., 2023).
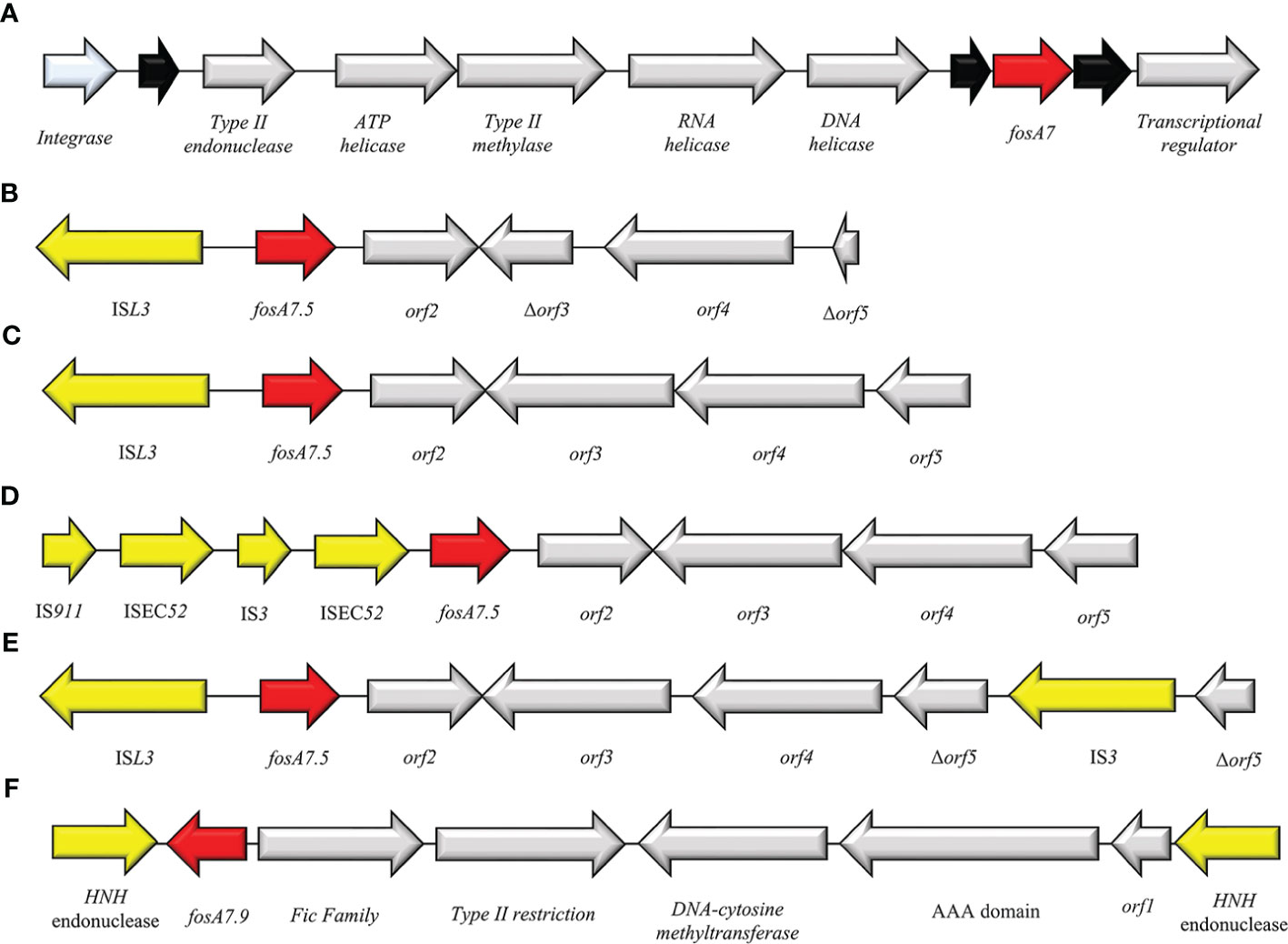
Figure 11 Structure of representative genetic environments of (A) fosA7 (GCA_000973785.1) and fosA7.5 (B) (OM355479), (C) (CP085638), (D) (CP085637), (E) (CP05525.1), (F) (CP047307). Yellow = IS, light blue = integrase, red = antimicrobial resistance genes, gray = open-reading frame, black = unknown protein.
The epidemiology of FosA7 family displays a relevant dissemination, with reports in livestock animals, clinical settings and enviroment (Balbin et al., 2020; Jovčić et al., 2020; Mosime et al., 2022). The Canadian and USA regions reported the larger diffusion of fosA7, followed by China (Pan et al., 2021). Recently, cases of FosA7 enzymes have been described in South Africa from Citrobacter koseri (Ekwanzala et al., 2020), in Czech Republic from C. freundii (Mattioni Marchetti et al., 2023) and in Poland from E. coli (Skarżyńska et al., 2021). Expression of FosA7 showed high value of FOSR MIC (>512 mg/ml) (Rehman et al., 2017).
FosA8
The newly plasmid-encoded fosA8 has been detected in clinical E. coli strains from a Swiss collection obtained from 2012 and 2013. The fosA8 gene was located on a 50 kb IncN plasmid and flanked by two copies of deleted sprT gene. FosA8 shows the highest identity with the chromosomally encoded fosA of Lecleria adecarboxylata (Poirel et al., 2019) and 96% identity with FosA7.5 from E. coli (Milner et al., 2020). Recently, Biggel et al. described a FosA8-producing K. pneumoniae, isolated from food in Switzerland, on a 65.5 kb IncN-IncR plasmid and located in the cassette IS26-ΔIS15-ardA–ccgC–ccgD–ccgEIII–ardR–ardB–mucA–mucB–ΔsprT–fosA8–orf1–ΔsprT–ardK–repA–orf2–IS26 (Biggel et al., 2021) (Figure 7D). FosA8 confers high resistance levels to FOS, with MIC > 1,024 µg/ml (Poirel et al., 2019).
FosA9
FosA9 has been reported in 2019 by Doesschate et al. from an E. coli strain causing bacteremia in Utrecht. The patient had suffered from recurrent episodes of sepsis, with blood cultures positive for K. variicola, which was identified as the source of fosA9. The fosA9 genomic environment consisted of a ISEcp1-syrM1-fosA9-lysN2 region, flanked by 5 bp DRs (AAAAA) and identical to those found in K. variicola (Wang et al., 2019) (Figure 7E). The expression of FosA9 confers FOSR at high levels, with MIC > 1,024 µg/ml (Ten Doesschate et al., 2019).
FosA10
The FosA10 enzyme has been described by Ying Huang et al. from a local broiler meat outlet in Pakistan. A 53,736 bp IncFII plasmid harbored the fosA10, inserted in a 4,328 bp variable region, flanked by two copies of IS10 element (Huang et al., 2020) (Figure 7F). Differently, the identical genomic enviroment was identified on a IncK plasmid from a clinical ST648 NDM+FosA10-producing E. coli isolated in Czech Republic (Mattioni Marchetti et al., 2023). FosA10 shares highest identity with FosA6 and FosA9 (ID = 97.84%), confirming its possible origin from K. pneumoniae species (Huang et al., 2020). In E. coli strains FosA10 induces FOSR phenotype with MIC >128 µg/ml (Huang et al., 2020).
FosC2
FosC2 is a metalloenzyme able to induce resistance profiles to FOS and shared a 56% sequence identity with FosEC of E. cloacae. It was identified for the first time in 2010 from clinical E. coli in Japan. FosC2 disseminates via plasmid but is rarely reported. Originally, fosC2 was described in integron type I structure: IS26-ΔIntI1-fosC2-dfrA17-aad5-qacEΔ1-sul1 (Wachino et al., 2010) (Figure 12). Subsequently, in 2015 Wang and colleagues reported the second clinical case of fosC2 disseminated via plasmid in a carbapenemases-producing E. cloacae strain. The plasmid (pIMP-HB623) was classified as IncL/M1 and harbored the composite cassette IS26-ΔtnpA-tnpR-tnpM-IntI1-fosC2-blaIMP34-tniR-tniQ-ΔtniA-IS26 (Wang et al., 2015). Both cases reported in literature, emphasized the co-expression of FosC2 and ESβLs. Recently, speculation on FosC2 recognizes Aliidiomarina shirensis as a possible progenitor for plasmid-mediated fosC2 (Ortiz de la Rosa et al., 2022). FosC2 expression induces broad FOSR profile (MIC value = 128 µg/ml).
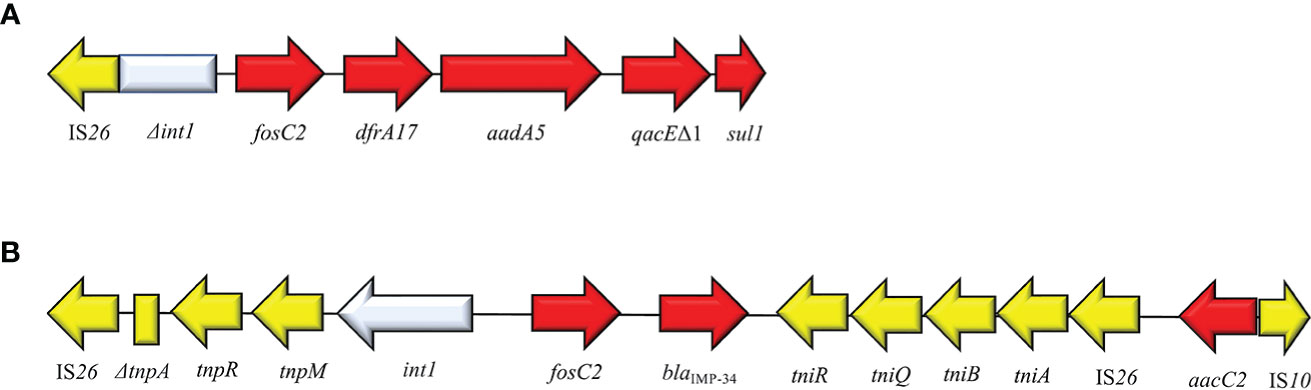
Figure 12 Structure of representative genetic environments of fosC2. (A) (AB522969) (Lucas et al., 2017), (B) (KM877517) (Guo et al., 2016). Yellow = IS, light blue = integrase, red = antimicrobial resistance genes, gray = open-reading frame, black = unknown protein.
FosL1 and FosL2
FosL1 is a novel glutathione S-transferase metalloenzyme that shared a 63% identity with FosA8. FosL1 was described on a conjugative IncX1 plasmid in a E. coli strain of a Swiss patient (Kieffer et al., 2020). The genomic enviroment surrounding fosL1 consisted of a mobile insertion cassette, flanked by ΔIS91-like at both sides. The same fosL1 cassette, was detected on an IncQ1 plasmid from a clinical S. enterica. Subsequently, an in-silico analysis of fosL1 identified a similar gene, classified as fosL2, on an IncP-like plasmid, collected from a clinical S. enterica strain. Genomic environment of fosL2 consisted of Tn7L-like-fosL1-urk-Tn7R-like and flanked by Δhyp at both sides (Kieffer et al., 2020) (Figure 13). The ancestor source for FosL1-2 remains unknown. FosL1 induces FOSR profile at high level (MIC = 1,024 µg/ml) (Kieffer et al., 2020).
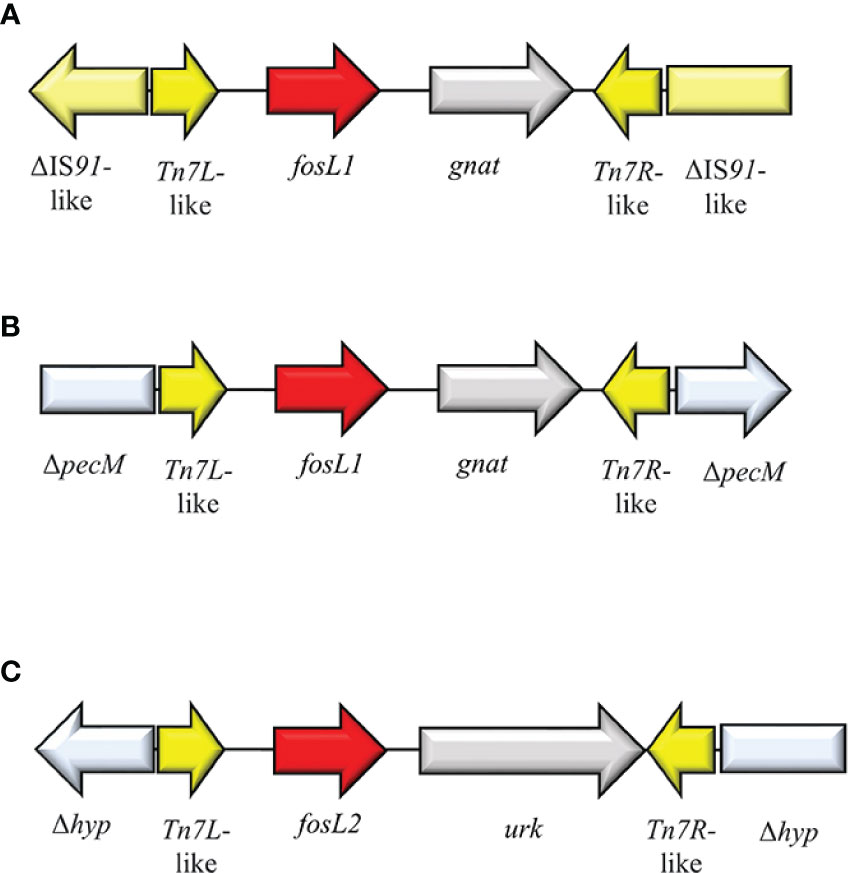
Figure 13 Structure of representative genetic environments of (A, B) fosL1 (MN464149, SAMN11620633) (Rehman et al., 2017) and (C) fosL2 (SAMN11027629) (Rehman et al., 2017). Yellow = IS, light blue = integrase, red = antimicrobial resistance genes, gray = open-reading frame.
Epidemiological breakpoints and detection strategies
According to European Committee on Antimicrobial Susceptibility Testing (EUCAST) and the Clinical and Laboratory Standards Institute (CLSI), agar dilution method (ADM) is the gold standard for FOS MIC detection in both Gram-positive and -negative bacteria but the breakpoints for FOS susceptibility have been formalized for few species and are different for CLSI and EUCAST. EUCAST breakpoints for Enterobacterales define as susceptible (S) MIC ≤ 32 mg/L and resistant (R) MIC > 32 mg/L, while CLSI breakpoints for E. coli are S ≤ 64 mg/L, I =128 mg/L, R ≥ 256 mg/L (Falagas et al., 2008).. Currently, there is a lack of fast, time-saving susceptibility tests for FOS and the limited breakpoints standardization, that highlights the difficulty in monitoring FOS profiles epidemiology and in identifying FOSR strains. In this section we describe the current available methods for the investigation of FOS susceptible profiles among Enterobacterales.
Agar dilution method (ADM)
The reference method ADM consists in the incorporation of different concentration of FOS (generally from 0.25 mg/ml up to 1,024 mg/ml) into Mueller-Hilton (MH) agar, added with 25 mg/L of G6P; Balouiri et al., 2016). Then, a 0.5 MacFarland suspension of the studied strain is prepared and diluted, to obtain the final inoculum required of 1 × 104 CFU/spot (2 μl). When replicators with 1-mm pins that deliver 0.1 to 0.2 μL are used, dilution of the initial suspension is not recommended. After inoculation, the plates are left at room temperature until the inoculation spots are completely absorbed into the agar (no more than 30 minutes). Incubate at 35 ± 2°C for 16 to 20 hours. The MIC value corresponds to the concentration in which a growth reduction of at least 80% is obtained, as compared to the control. The method should be conducted at least in duplicate. Although ADM remains the reference method for FOS MIC evaluation, it is not used routinely in diagnostic practice due to its labor-intensity and high time requirement (16-20 h) (Croughs et al., 2022). Alternative and faster methods, as gradient and disk diffusion test, or routinely used automated systems, as Vitek2, resulted unreliable due to their poor ability in detecting FOSR isolates, with high error rates (van den Bijllaardt et al., 2018; Croughs et al., 2022). According to EUCAST guidelines, the disk diffusion test is intended only in investigating FOS profiles among E. coli strains, using 200 μg FOS disk and in presence of 50 μg of G6P.
Commercial AD fosfomycin panel
A time-saving and ready-to-use alternative is represented by the commercial AD fosfomycin panel, commercialized in 2019 by Liofilchem S.r.l. (Roseto degli Abruzzi, Italy). The commercial AD fosfomycin panel allows FOS MIC evaluation and is composed of 12 wells filled with agar medium + 25 mg/L G6P and different concentrations of FOS (0.25–256 mg/L). The manufacturer’s guidelines provide for each isolates the preparation of a 0.5 McFarland bacterial suspension, consecutively diluted 1:10 in sterile saline solution. Each well is dispensed on agar surface with 2 μL (approximately 104 CFU/spot) of the diluted bacterial suspension. The incubation step requires 35 ± 2°C for 16-20 hours in ambient air. FOS MIC is recorded as the lowest concentration of FOS that completely inhibit growth.
Rapid fosfomycin/E. coli NP test
Nordmann and co-authors reported the description of a rapid test for FOS susceptibility profiles in E. coli (Nordmann et al., 2019). The rapid test is based on the microbial ability to metabolize glucose, that induce a colorimetric change of a specific pH indicator (culture medium, 2.5% MHB-CA powder, 0.005% phenol red indicator, and 1% d(+)-glucose). The test consists in preparing two solutions, named NP solutions: one solution with 25 μg/ml G6P and 40 μg/ml FOS, and one without. For bacterial suspension, a 3.0 to 3.5 McFarland solution for each tested isolate is prepared in 5 ml of sterile NaCl (0.85%). A 96-well polystyrene microtest plate is filled with both NP solutions and the bacterial suspension is directly inoculated in the presence or absence of FOS. After an incubation of 1 h 30 min at 35 ± 2°C, color changes are visually detected. FOS-resistant E. coli triggers a color change from orange to yellow, while FOS-susceptible remains orange (Nordmann et al., 2019).
This methodology showed both high rate of sensitivity (100%) and specificity (98.7%). In details, among 22 FOS-resistant E. coli isolates tested, all showed a positive result to the test (Nordmann et al., 2019). Similarly, Mueller and co-authors revealed a 100% correlation between susceptibility and resistance strains after screening 1,225 clinical ESβL-producing E. coli (Mueller et al., 2019). The rapid fosfomycin/E. coli NP test has the potential to be used as a rapid and first-step screening of FOS-resistant E. coli, thanks to its good performance and rapidity. A more recent evaluation on the accuracy of this rapid method was conducted on 149 clinical E. coli isolates, showing high rate of sensitivity and specificity (94.2% and 98.75%, respectively) and highlighting the reliability of the technique (Yunus et al., 2021). Differently, Kansak and colleagues found similar rate of sensitivity and specificity (95.9% and 100%, respectively) but a Very major Error (VME) of 22.2%, limiting the possibility to use the rapid test instead of ADM (Kansak et al., 2021). Despite the potential offered, the use of the rapid fosfomycin/E. coli NP test is still limited due to its applicability on E. coli only, the difficult in the interpretation of the results and the inability to distinguish between chromosomal and plasmid-acquired resistance mechanisms (Nordmann et al., 2019).
SuperFOS selective medium
The SuperFOS selective medium provide a first line screening for FOS resistant Enterobacterales.
The SuperFOS medium combines the differentiation features of the CHROMagar orientation medium with an optimal concentration of FOS (16 μg/ml) and G6P (25 μg/ml). To avoid any contamination by eventual Gram-positive organism and fungi, the SuperFOS medium is enriched with vancomycin (20 μg/ml) and amphotericin B (5 μg/ml).
This medium provides several advantages due to its ease in preparation, the low cost, and the excellence performance, with both sensitivity and specificity at 100%. Moreover, the medium allows a first step screening of both chromosomal and plasmid mediated FOSR mechanisms among Enterobacterales from clinical specimens (Nordmann et al., 2022).
Disk potentiation testing with PPF
The disk potentiation testing with sodium phosphonoformate (PPF) is an agar-based diffusion test requiring the presence of FOS, G6P and PPF. PPF, commercially named Foscarnet, is an anti-viral compound used primarily in the treatment of CMV infections with inhibitory properties against FosA and FosC2 enzymes (Schreiber et al., 2009; Nakamura et al., 2014). PPF is able to bind FosA/FosC2 enzymes interacting with the residue MnII(+) and Thr9 that are present in the active site of FosA/FosC2-like enzymes, leading to a inhibitory effect and, thus, restoring the FOS susceptibility (Ito et al., 2017). The test requires MH agar plates with 25 mg/L G6P, 0.5 MacFarland solution of the isolate to investigate, two disk of FOS (50 μg) and PPF (1 mg). The cutoff is set to a 5 mm enlargement in the inhibition zone of FOS+PPF disk compared with the FOS disk alone (Nakamura et al., 2014). This agar-based method shows 100% sensitivity and specificity, and successfully detects the producing of enzymes FosA/A2 (Rigsby et al., 2004), FosA3, FosA4, FosA6 (Loras et al., 2021), FosA7 (Mattioni Marchetti et al., 2023), FosA8 (Biggel et al., 2021), FosA10 (Mattioni Marchetti et al., 2023), FosC2 (Nakamura et al., 2014), and FosL1 (Kieffer et al., 2020). However, the PPF test has been validated for E. coli strains only.
Carbon source growth test
The carbon source growth test evaluates the ability of a bacterial strain to grow with G3P or G6P as the sole source of carbon. The inability to grow in presence of G3P and/or G6P is the result of a functional deficiency of the transporters GlpT and UhpT, respectively (Huang et al., 2021). This method requires the inoculation of the bacterial isolate on a M9 minimal medium agar supplemented with G3P or G6P at 0.2% (w/v) (Sorlozano-Puerto et al., 2020). After an incubation phase at 36°C for 48 h, the poor or total absence of growth is associated to an impairment in the transporter’s activity (Sorlozano-Puerto et al., 2020). The limitation of this growth test is mainly represented by the time required to perform it (72 h for results) and restricted results only on direct impairment of GlpT and UhpT activity.
Limitations
This review presents several limitations. Few studies evaluate the prevalence of amino acidic mutations in proteins involved in FOS influx and their possible effect in FOS MIC increase (Kim et al., 1996; Takahata et al, 2010; Li et al., 2015). Whereby, the knowledge on specific mutations affecting FOS influx is not clear and incomplete.
Considering plasmid-mediated mechanisms for FOSR, the update global epidemiology of fosA/fosC2/fosL1-2 gene is not completely and clearly monitored, mainly due to the lack of national surveillance plan, of fast methodology for the investigation of FOSR profiles and the lack of general interest. Moreover, the characterization of fosA-like gene variants is so far only through molecular investigations and/or WGS. These point together, explain the difficulty to draw the updated epidemiology of FosA/C2/L1-2 enzymes and to clearly specific mutation decreasing FOS MICs.
Additionally, this review describes the FOSR mechanisms that has been investigated and reported in literature among Enterobacterales only, while does not consider other relevant FOSR sources, as S. aureus and Enterococcus faecium.
Further perspective
FOS is still a valid option against MDR Enterobacterales, but this molecule is not always monitored routinely in clinical practice or in surveillance plans and, thus, the resistance mechanisms involved are not further investigated. In a scenario of increasing FOSR, time-saving and user-friendly methods for detecting such resistance profiles turn out to be fundamental. Implementation of faster testing would allow to conduct wide surveillance studies and to monitor FOS in clinical routine.
Time-saving methodologies aforementioned are validated for E. coli only. Therefore, the validation of these methods to further species would extend the pool of strains that can be tested, providing a more in-depth knowledge about FOSR epidemiology. Moreover, a faster detection of FOS resistant bacteria and thus a further molecular characterization, could provide more information even on rarely reported FosA-like enzymes, such as FosC2, FosA4, FosA8 and FosA9, and could supply a more update epidemiology on fosA/C2/L1-2 genes spread.
Conclusion
Even though FOS is an old antimicrobial drug, it has unique and favorable features that lead in the last 20 years it to be considered as an additional resource in the treatment of MDR microorganisms’ infections (Michalopoulos et al., 2011). This review described the different mechanisms, identified so far, leading to FOS MIC increase among Enterobacterales genus. The FOS influx inside bacterial cell, that is regulated by different transporters and associated regulators, has also been described. Impairment in FOS transporters GlpT and UhpT is the most common mechanisms leading to the increase in FOS MICs, reported both in vitro and in vivo (Nilsson et al., 2003). The scientific community identified specific hotspot mutations in GlpT associated to a FOS resistance at high levels (FOS MICs > 128 μg/mL), such as W28del and Pro212Leu in E. coli, and as Arg206Lys and Ile293Phe in K. pneumoniae (Lu et al., 2016; Sorlozano-Puerto et al., 2020; Mattioni Marchetti et al., 2023). Compared with mutation frequency in GlpT and UhpT, modification in the target MurA were uncommon in vivo and no reports identified mutations in the active site (Cys115) in clinical isolates. In clinical E. coli strains the mutations Asp369Asn and Leu370Ile in MurA can likely develop FOS resistance profiles with MICs up to 512 mg/ml, while in clinical K. pneumoniae isolates the modifications Asp260Tyr and Thr307Lys has been associated to FOS MICs = 128 μg/mL (Takahata et al, 2010; Lu et al., 2016). The study of specific mutations in proteins involved in FOS influx and their eventual effect on FOS MICs is not in deep investigated and required further investigations.
Regarding acquired FOSR mechanisms, in the last twelve years there has been a global diffusion of metallo-enzymes, named FosA-like, FosC2 and FosL1-L2 (Zurfluh et al., 2020). The Chinese clinical and veterinary environments show the highest frequency of FosA/C2 enzymes but, recently, many other countries as Brazil, Japan, Spain, and USA have reported such enzymes as well (Wachino et al., 2010; Jiang et al., 2017; Loras et al., 2021; Ewbank et al., 2022; Turcotte et al., 2022). To date, 11 variants of FosA enzymes has been identified, contributing to FOS resistance at different extents. In the global scenario, fosA3 is the predominant type and it is widely reported in humans and veterinary settings. The wide and fast diffusion of fosA3 has been facilitated by the combination of IS26-mediated transposons with epidemic broad-host-range plasmids as IncFII plasmids. The versatility of these fosA3-harboring plasmids has allowed the acquisition of fosA3 genes in several clinically important ST such as E. coli ST10, E. coli ST69, E. coli ST131, K. pneumoniae ST11 and S. enterica ST32 (Xiang et al., 2015; Falagas et al., 2019; Seok et al., 2020). FosA3 is commonly co-expressed with other ESβLs, as CTX-M-65, and even with carbapenemases as KPCs, NDMs and VIMs (Villa et al., 2015; Xie et al., 2016; Jiang et al., 2017; Tang et al., 2020). Worryingly, the co-occurrence of fosA3 + mcr-type genes in carbapenemases-producing Enterobacterales has been already described in the literature (Zhao et al., 2018; Peng et al., 2019; Tian et al., 2020).
Originated from K. pneumoniae chromosome, FosA5 and FosA6 can be considered among the most frequent metallo-enzyme leading to FOSR. However, their epidemiology has not been widely investigated in strains other than K. pneumoniae and the few reported cases are confined to countries as China and Spain (Guo et al., 2016; Wang et al., 2019). The diffusion of both fosA5 and fosA6 in E. coli is linked to IS10 flaking cassettes (Xu et al., 2011; Dhanani et al., 2015).
Since the discover in 2015, FosA7 has rapidly spread among Enterobacterales, with high predominance among Salmonella spp. So far, nine alleles of fosA7 have been described and deposited in the GenBank. FosA7-like genes are strictly located on Salmonella spp. chromosome, except for fosA7.5 and fosA7.9 that are associated to E. coli and C. freundii chromosome, respectively. The current spread of fosA7-like genes includes countries as Canada and China (Dhanani et al., 2015; Pan et al., 2021).
Concurrence of impairing mutations in FOS influx and acquisition of fosA/C2/L1-2 together with ESβLs and carbapenemases genes, is worrying and could strongly affect the use of FOS in severe infections treatment.
ADM is the reference methods for FOS MICs evaluation and the few rapid methods available have been validated for E. coli only or are prone to error. The increase of surveillance plans and the implementation of new rapid approaches for the detection of FOSR Enterobacterales, would favorite a better and in-depth knowledge on the prevalence of FOSR mechanisms. Moreover, a clearer information on such mechanisms and their dissemination results of priority importance to halt eventual FOSR dissemination and to optimize therapeutic strategies.
Author contributions
VM, IB, and JH played an important role in searching the relevant literature, writing and correcting the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The study was supported by research project grants NU20J-05-00033 and NU23J-09-00067 provided by the Czech Health Research Council and by the project National Institute of Virology and Bacteriology (Program EXCELES, ID project no. LX22NPO5103), funded by the European Union–Next Generation EU.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aghamali, M., Sedighi, M., Zahedi Bialvaei, A., Mohammadzadeh, N., Abbasian, S., Ghafouri, Z., et al. (2019). Fosfomycin: mechanisms and the increasing prevalence of resistance. J. Med. Microbiol. 68 (1), 11–25. doi: 10.1099/jmm.0.000874
Ambudkar, S. V., Anantharam, V., Maloney, P. C. (1990). UhpT, the sugar phosphate antiporter of escherichia coli, functions as a monomer. J. Biol. Chem. 265 (21), 12287–12292. doi: 10.1016/S0021-9258(19)38343-7
Andrews, J. M., Baquero, F., Beltran, J. M., Canton, E., Crokaert, F., Gobernado, M., et al. (1983). International collaborative study on standardization of bacterial sensitivity to fosfomycin. J. Antimicrob. Chemother. 12 (4), 357–361. doi: 10.1093/jac/12.4.357
.Available at: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_13.0_Breakpoint_Tables.pdf.
.Available at: http://www.apteq.ch/saas/CustomUpload/374O357O340O370O356O369O350O320O367O371O356O368O320O326O/AD_Fosfomycin_0_25-256.pdf.
Balbin, M. M., Hull, D., Guest, C., Nichols, L., Dunn, R., Hull, D., et al. (2020). Antimicrobial resistance and virulence factors profile of salmonella spp. and escherichia coli isolated from different environments exposed to anthropogenic activity. J. Glob Antimicrob. Resist. , 22:578–22:583. doi: 10.1016/j.jgar.2020.05.016
Ballestero-Téllez, M., Docobo-Pérez, F., Portillo-Calderón, I., Rodríguez-Martínez, J. M., Racero, L., Ramos-Guelfo, M. S., et al. (2017). Molecular insights into fosfomycin resistance in escherichia coli. J. Antimicrob. Chemother. 72 (5), 1303–1309. doi: 10.1093/jac/dkw573
Balouiri, M., Sadiki, M., Ibnsouda, S. K. (2016). Methods for in vitro evaluating antimicrobial activity: a review. J. Pharm. Anal. 6 (2), 71–79. doi: 10.1016/j.jpha.2015.11.005
Barnett, J. A., Southern, P. M., Jr, Luby, J. P., Sanford, J. P. (1969). Efficacy of phosphonomycin in treatment of urinary-tract infections. Antimicrob. Agents Chemother. (Bethesda). 9, 349–351.
Baron, D., Desjars, P., Touze, M., et al. (1986). Clinical study on combined therapy with fosfomycin for staphylococcal infections. In Fosfomycin. Proceedings of the International Symposium. (Madrid: Instituto de Farmacologia Espaneola (CEPA)) p. 172e87.
Benzerara, Y., Gallah, S., Hommeril, B., Genel, N., Decré, D., Rottman, M., et al. (2017). Emergence of plasmid-mediated fosfomycin-resistance genes among escherichia coli isolates, France. Emerg. Infect. Dis. 23 (9), 1564–1567. doi: 10.3201/eid2309.170560
Bi, W., Li, B., Song, J., Hong, Y., Zhang, X., Liu, H., et al. (2017). Antimicrobial susceptibility and mechanisms of fosfomycin resistance in extended-spectrum β-lactamase-producing escherichia coli strains from urinary tract infections in wenzhou, China. Int. J. Antimicrob. Agents. 50 (1), 29–34. doi: 10.1016/j.ijantimicag.2017.02.010
Biggel, M., Zurfluh, K., Treier, A., Nüesch-Inderbinen, M., Stephan, R. (2021). Characteristics of fosA-carrying plasmids in e. coli and klebsiella spp. isolates originating from food and environmental samples. J. Antimicrob. Chemother. 76 (8), 2004–2011. doi: 10.1093/jac/dkab119
Birgy, A., Madhi, F., Hogan, J., Doit, C., Gaschignard, J., Caseris, M., et al. (2018). CTX-M-55-, MCR-1-, and FosA-producing multidrug-resistant escherichia coli infection in a child in France. Antimicrob. Agents Chemother. 62 (4), e00127–e00118. doi: 10.1128/AAC.00127-18
Brown, E. D., Vivas, E. I., Walsh, C. T., Kolter, R. (1995). MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in escherichia coli. J. Bacteriol. 177 (14), 4194–4197. doi: 10.1128/jb.177.14.4194-4197.1995
Candel, F. J., Matesanz David, M., Barberán, J. (2019). New perspectives for reassessing fosfomycin: applicability in current clinical practice. Rev. Esp Quimioter. 32 Suppl 1 (Suppl 1), 1–7.
Castañeda-García, A., Blázquez, J., Rodríguez-Rojas, A. (2013). Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics (Basel). 2 (2), 217–236. doi: 10.3390/antibiotics2020217
Cattoir, V., Pourbaix, A., Magnan, M., Chau, F., de Lastours, V., Felden, B., et al. (2020). Novel chromosomal mutations responsible for fosfomycin resistance in escherichia coli. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.575031
Chen, L., Ou, B., Zhang, M., Chou, C. H., Chang, S. K., Zhu, G. (2021). Coexistence of fosfomycin resistance determinant fosA and fosA3 in Enterobacter cloacae isolated from pets with urinary tract infection in Taiwan. Microb. Drug Resist. 27 (3), 415–423. doi: 10.1089/mdr.2020.0077
Christensen, B. G., Leanza, W. J., Beattie, T. R., Patchett, A. A., Arison, B. H., Ormond, R. E., et al. (1969). Phosphonomycin: structure and synthesis. Science 166, 123–125. doi: 10.1126/science.166.3901.123
Couce, A., Briales, A., Rodriguez-Rojas, A., Costas, C., Pascual, A., Blazquez, J. (2012). Genomewide overexpression screen for fosfomycin resistance in Escherichia coli: MurA confers clinical resistance at low fitness cost. Antimicrob. Agents Chemother. 56 (5), 2767–2769. doi: 10.1128/AAC.06122-11
Cozzarelli, N. R., Freedberg, W. B., Lin, E. C. (1968). Genetic control of l-alpha-glycerophosphate system in escherichia coli. J. Mol. Biol. 31 (3), 371–387. doi: 10.1016/0022-2836(68)90415-4
Croughs, P. D., Konijnendijk-de Regt, M., Yusuf, E. (2022). Fosfomycin susceptibility testing using commercial agar dilution test. Microbiol. Spectr. 10 (2), e0250421. doi: 10.1128/spectrum.02504-21
Dahl, J. L., Wei, B. Y., Kadner, R. J. (1997). Protein phosphorylation affects binding of the escherichia coli transcription activator UhpA to the uhpT promoter. J. Biol. Chem. 272 (3), 1910–1919. doi: 10.1074/jbc.272.3.1910
Dantas Palmeira, J., Ferreira, H., Madec, J. Y., Haenni, M. (2018). Pandemic escherichia coli ST648 isolate harbouring fosA3 and blaCTX-M-8 on an IncI1/ST113 plasmid: a new successful combination for the spread of fosfomycin resistance? J. Glob Antimicrob. Resist. 15, 254–255. doi: 10.1016/j.jgar.2018.10.025
Deutscher, J., Aké, F. M., Derkaoui, M., Zébré, A. C., Cao, T. N., Bouraoui, H., et al. (2014). The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol. Mol. Biol. Rev. 78 (2), 231–256. doi: 10.1128/MMBR.00001-14
Dhanani, A. S., Block, G., Dewar, K., Forgetta, V., Topp, E., Beiko, R. G., et al. (2015). Genomic comparison of non-typhoidal salmonella enterica serovars typhimurium, enteritidis, Heidelberg, hadar and Kentucky isolates from broiler chickens. PloS One 10 (6), e0128773. doi: 10.1371/journal.pone.0128773
Doumith, M., Day, M., Ciesielczuk, H., Hope, R., Underwood, A., Reynolds, R., et al. (2015). Rapid identification of major escherichia coli sequence types causing urinary tract and bloodstream infections. J. Clin. Microbiol. 53 (1), 160–166. doi: 10.1128/JCM.02562-14
Ekwanzala, M. D., Dewar, J. B., Kamika, I., Momba, M. N. B. (2020). Genome sequence of carbapenem-resistant citrobacter koseri carrying blaOXA-181 isolated from sewage sludge. J. Glob Antimicrob. Resist. 20, 94–97. doi: 10.1016/j.jgar.2019.07.011
Escapa, I. F., del Cerro, C., García, J. L., Prieto, M. A. (2013). The role of GlpR repressor in pseudomonas putida KT2440 growth and PHA production from glycerol. Environ. Microbiol. 15 (1), 93–110. doi: 10.1111/j.1462-2920.2012.02790.x
Eschenburg, S., Priestman, M., Schönbrunn, E. (2005). Evidence that the fosfomycin target Cys115 in UDP-n-acetylglucosamine enolpyruvyl transferase (MurA) is essential for product release. J. Biol. Chem. 280 (5), 3757–3763. doi: 10.1074/jbc.M411325200
Ewbank, A. C., Fuentes-Castillo, D., Sacristán, C., Cardoso, B., Esposito, F., Fuga, B., et al. (2022). Extended-spectrum β-lactamase (ESBL)-producing escherichia coli survey in wild seabirds at a pristine atoll in the southern Atlantic ocean, Brazil: first report of the O25b-ST131 clone harboring blaCTX-M-8. Sci. Total Environ. 806 (Pt 2), 150539. doi: 10.1016/j.scitotenv.2021.150539
Falagas, M. E., Athanasaki, F., Voulgaris, G. L., Triarides, N. A., Vardakas, K. Z. (2019). Resistance to fosfomycin: mechanisms, frequency and clinical consequences. Int. J. Antimicrob. Agents. 53 (1), 22–28. doi: 10.1016/j.ijantimicag.2018.09.013
Falagas, M. E., Giannopoulou, K. P., Kokolakis, G. N., Rafailidis, P. I. (2008). Fosfomycin: use beyond urinary tract and gastrointestinal infections. Clin. Infect. Dis. 46 (7), 1069–1077. doi: 10.1086/527442
Falagas, M. E., Vouloumanou, E. K., Samonis, G., Vardakas, K. Z. (2016). Fosfomycin. Clin. Microbiol. Rev. 29 (2), 321–347. doi: 10.1128/CMR.00068-15
Feng, J., Qiu, Y., Yin, Z., Chen, W., Yang, H., Yang, W., et al. (2015). Coexistence of a novel KPC-2-encoding MDR plasmid and an NDM-1-encoding pNDM-HN380-like plasmid in a clinical isolate of citrobacter freundii. J. Antimicrob. Chemother. 70 (11), 2987–2991. doi: 10.1093/jac/dkv232
Fernandes, M. R., Sellera, F. P., Moura, Q., Souza, T. A., Lincopan, N. (2018). Draft genome sequence of a CTX-M-8, CTX-M-55 and FosA3 co-producing escherichia coli ST117/B2 isolated from an asymptomatic carrier. J. Glob Antimicrob. Resist. 12, 183–184. doi: 10.1016/j.jgar.2018.01.015
Forde, B. M., Roberts, L. W., Phan, M. D., Peters, K. M., Fleming, B. A., Russell, C. W., et al. (2019). Population dynamics of an escherichia coli ST131 lineage during recurrent urinary tract infection. Nat. Commun. 10 (1), 3643. doi: 10.1038/s41467-019-11571-5
Freitag, C., Michael, G. B., Li, J., Kadlec, K., Wang, Y., Hassel, M., et al. (2018). Occurrence and characterisation of ESBL-encoding plasmids among escherichia coli isolates from fresh vegetables. Vet. Microbiol. 219, 63–69. doi: 10.1016/j.vetmic.2018.03.028
Friedrich, M. J., Kadner, R. J. (1987). Nucleotide sequence of the uhp region of escherichia coli. J. Bacteriol. 169 (8), 3556–3563. doi: 10.1128/jb.169.8.3556-3563.1987
Galindo-Méndez, M., Navarrete-Salazar, H., Baltazar-Jiménez, F., Muñoz-de la Paz, E., Sánchez-Mawcinitt, M. F., Gómez-Pardo, A., et al. (2022). Emergence of fosfomycin resistance by plasmid-mediated fos genes in uropathogenic ESBL-producing e. coli isolates in Mexico. Antibiotics (Basel). 11 (10), 1383. doi: 10.3390/antibiotics11101383
Garcia-Fulgueiras, V., Caiata, L., Bado, I., Giachetto, G., Robino, L. (2022). Antibiotic susceptibility and fosfomycin resistance characterization in a cohort of children older than 6 years of age with urinary tract infection. Rev. Argent Microbiol. 54 (2), 120–124. doi: 10.1016/j.ram.2021.04.002
García-Lobo, J. M., Ortiz, J. M. (1982). Tn292l, a transposon encoding fosfomycin resistance. J. Bacteriol. 151 (1), 477–479. doi: 10.1128/jb.151.1.477-479.1982
Giacobbe, D. R., Del Bono, V., Coppo, E., Marchese, A., Viscoli, C. (2015). Emergence of a KPC-3-Producing escherichia coli ST69 as a cause of bloodstream infections in Italy. Microb. Drug Resist. 21 (3), 342–344. doi: 10.1089/mdr.2014.0230
Gou, J. J., Liu, N., Guo, L. H., Xu, H., Lv, T., Yu, X., et al. (2020). Carbapenem-resistant Enterobacter hormaechei ST1103 with IMP-26 carbapenemase and ESBL gene blaSHV-178. Infect. Drug Resist. 13, 597–605. doi: 10.2147/IDR.S232514
Güneri, CÖ, Stingl, K., Grobbel, M., Hammerl, J. A., Kürekci, C. (2022). Different fosA genes were found on mobile genetic elements in escherichia coli from wastewaters of hospitals and municipals in Turkey. Sci. Total Environ. 824, 153928. doi: 10.1016/j.scitotenv.2022.153928
Guo, Q., Tomich, A. D., McElheny, C. L., Cooper, V. S., Stoesser, N., Wang, M., et al. (2016). Glutathione-s-transferase FosA6 of klebsiella pneumoniae origin conferring fosfomycin resistance in ESBL-producing escherichia coli. J. Antimicrob. Chemother. 71 (9), 2460–2465. doi: 10.1093/jac/dkw177
Hall, J. A., Maloney, P. C. (2005). Altered oxyanion selectivity in mutants of UhpT, the pi-linked sugar phosphate carrier of escherichia coli. J. Biol. Chem. 280 (5), 3376–3381. doi: 10.1074/jbc.M409965200
Hameed, M. F., Chen, Y., Bilal, H., Khan, S., Ge, H., Xiaofang, C., et al. (2022). The Co-occurrence of mcr-3 and fosA3 in IncP plasmid in ST131 escherichia coli: a novel case. J. Infect. Dev. Ctries. 16 (4), 622–629. doi: 10.3855/jidc.15943
Hammad, A. M., Hoffmann, M., Gonzalez-Escalona, N., Abbas, N. H., Yao, K., Koenig, S., et al. (2019). Genomic features of colistin resistant escherichia coli ST69 strain harboring mcr-1 on IncHI2 plasmid from raw milk cheese in Egypt. Infect. Genet. Evol. 73, 126–131. doi: 10.1016/j.meegid.2019.04.021
Hao, Y., Zhao, X., Zhang, C., Bai, Y., Song, Z., Lu, X., et al. (2021). Clonal dissemination of clinical carbapenem-resistant Klebsiella pneumoniae isolates carrying fosA3 and blaKPC-2 coharboring plasmids in Shandong, China. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.771170
Harmer, C. J., Hall, R. M. (2016). IS26-mediated formation of transposons carrying antibiotic resistance genes. mSphere. 1 (2), e00038–e00016. doi: 10.1128/mSphere.00038-16
He, S., Hickman, A. B., Varani, A. M., Siguier, P., Chandler, M., Dekker, J. P., et al. (2015). Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio. 6 (3), e00762. doi: 10.1128/mBio.00762-15
He, D., Liu, L., Guo, B., Wu, S., Chen, X., Wang, J., et al. (2017). Chromosomal location of the fosA3 and blaCTX-m genes in Proteus mirabilis and clonal spread of escherichia coli ST117 carrying fosA3-positive IncHI2/ST3 or F2:A-:B- plasmids in a chicken farm. Int. J. Antimicrob. Agents. 49 (4), 443–448. doi: 10.1016/j.ijantimicag.2016.12.009
Hirschl, A., Stanek, G., Rotter, M. (1980). Wirkungssteigerung von fosfomycin durch zusatz von glukose-6-Phosphat bei intraperitoneal infizierten mäusen [Improvement of the therapeutic efficacy of fosfomycin by addition of glucose-6-phosphate in the treatment of intraperitoneally infected mice (author's transl)]. Zentralbl Bakteriol A. 246 (4), 562–566. doi: 10.1016/S0172-5599(80)80090-3
Ho, P. L., Chan, J., Lo, W. U., Law, P. Y., Li, Z., Lai, E. L., et al. (2013). Dissemination of plasmid-mediated fosfomycin resistance fosA3 among multidrug-resistant escherichia coli from livestock and other animals. J. Appl. Microbiol. 114 (3), 695–702. doi: 10.1111/jam.12099
Hoang, H. T. T., Higashi, A., Yamaguchi, T., Kawahara, R., Calvopina, M., Bastidas-Caldés, A., et al. (2022). Fusion plasmid carrying the colistin resistance gene mcr of escherichia coli isolated from healthy residents. J. Glob Antimicrob. Resist., 30, 152–154. doi: 10.1016/j.jgar.2022.06.007
Horii, T., Kimura, T., Sato, K., Shibayama, K., Ohta, M. (1999). Emergence of fosfomycin-resistant isolates of shiga-like toxin-producing escherichia coli O26. Antimicrob. Agents Chemother. 43 (4), 789–793. doi: 10.1128/AAC.43.4.789
Hou, J., Huang, X., Deng, Y., He, L., Yang, T., Zeng, Z., et al. (2012). Dissemination of the fosfomycin resistance gene fosA3 with CTX-m β-lactamase genes and rmtB carried on IncFII plasmids among escherichia coli isolates from pets in China. Antimicrob. Agents Chemother. 56 (4), 2135–2138. doi: 10.1128/AAC.05104-11
Hua, X., Zhang, L., Moran, R. A., Xu, Q., Sun, L., van Schaik, W., et al. (2020). Cointegration as a mechanism for the evolution of a KPC-producing multidrug resistance plasmid in Proteus mirabilis. Emerg. Microbes Infect. 9 (1), 1206–1218. doi: 10.1080/22221751.2020.1773322
Huang, L., Cao, M., Hu, Y., Zhang, R., Xiao, Y., Chen, G. (2021). Prevalence and mechanisms of fosfomycin resistance among KPC-producing klebsiella pneumoniae clinical isolates in China. Int. J. Antimicrob. Agents 57 (1), 106226. doi: 10.1016/j.ijantimicag.2020.106226
Huang, Y., Lin, Q., Zhou, Q., Lv, L., Wan, M., Gao, X., et al. (2020). Identification of fosA10, a novel plasmid-mediated fosfomycin resistance gene of Klebsiella pneumoniae origin, in Escherichia coli. Infect. Drug Resist. 13, 1273–1279. doi: 10.2147/IDR.S251360
Ito, R., Mustapha, M. M., Tomich, A. D., Callaghan, J. D., McElheny, C. L., Mettus, R. T., et al. (2017). Widespread fosfomycin resistance in gram-negative bacteria attributable to the chromosomal fosA gene. mBio. 8 (4), e00749–e00717. doi: 10.1128/mBio.00749-17
Jafari, A., Falahatkar, S., Delpasand, K., Sabati, H., Sedigh Ebrahim-Saraie, H. (2020). Emergence of Escherichia coli ST131 causing urinary tract infection in Western Asia: a systematic review and meta-analysis. Microb. Drug Resist. 26 (11), 1357–1364. doi: 10.1089/mdr.2019.0312
Jiang, W., Men, S., Kong, L., Ma, S., Yang, Y., Wang, Y., et al. (2017). Prevalence of plasmid-mediated fosfomycin resistance gene fosA3 among CTX-M-Producing escherichia coli isolates from chickens in China. Foodborne Pathog. Dis. 14 (4), 210–218. doi: 10.1089/fpd.2016.2230
Jovčić, B., Novović, K., Filipić, B., Velhner, M., Todorović, D., Matović, K., et al. (2020). Genomic characteristics of colistin-resistant Salmonellaenterica subsp. enterica serovar infantis from poultry farms in the republic of Serbia. Antibiotics (Basel). 9 (12), 886. doi: 10.3390/antibiotics9120886
Kadner, R. J., Shattuck-Eidens, D. M. (1983). Genetic control of the hexose phosphate transport system of escherichia coli: mapping of deletion and insertion mutations in the uhp region. J. Bacteriol. 155 (3), 1052–1061. doi: 10.1128/jb.155.3.1052-1061.1983
Kahan, F. M., Kahan, J. S., Cassidy, P. J., Kropp, H. (1974). The mechanism of action of fosfomycin (phosphonomycin). Ann. N Y Acad. Sci. 235 (0), 364–386. doi: 10.1111/j.1749-6632.1974.tb43277.x
Kansak, N., Arıcı, N., Adaleti, R., Nakipoglu, Y., Aksaray, S. (2021). Rapid detection of fosfomycin resistance in escherichia coli and klebsiella spp. strains isolated from urinary tract infections. J. Microbiol. Methods 188, 106296. doi: 10.1016/j.mimet.2021.106296
Kieffer, N., Poirel, L., Descombes, M. C., Nordmann, P. (2020). Characterization of FosL1, a plasmid-encoded fosfomycin resistance protein identified in escherichia coli. Antimicrob. Agents Chemother. 64 (4), e02042–e02019. doi: 10.1128/AAC.02042-19
Kim, D. H., Lees, W. J., Kempsell, K. E., Lane, W. S., Duncan, K., Walsh, C. T. (1996). Characterization of a Cys115 to asp substitution in the escherichia coli cell wall biosynthetic enzyme UDP-GlcNAc enolpyruvyl transferase (MurA) that confers resistance to inactivation by the antibiotic fosfomycin. Biochemistry 35 (15), 4923–4928. doi: 10.1021/bi952937w
Law, C. J., Enkavi, G., Wang, D. N., Tajkhorshid, E. (2009). Structural basis of substrate selectivity in the glycerol-3-phosphate: phosphate antiporter GlpT. Biophys. J. 97 (5), 1346–1353. doi: 10.1016/j.bpj.2009.06.026
Lee, S. Y., Park, Y. J., Yu, J. K., Jung, S., Kim, Y., Jeong, S. H., et al. (2012). Prevalence of acquired fosfomycin resistance among extended-spectrum β-lactamase-producing escherichia coli and klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. J. Antimicrob. Chemother. 67 (12), 2843–2847. doi: 10.1093/jac/dks319
Lei, C. W., Chen, Y. P., Kang, Z. Z., Kong, L. H., Wang, H. N. (2018). Characterization of a novel SXT/R391 integrative and conjugative element carrying cfr, blaCTX-M-65, fosA3, and aac(6')-ib-cr in Proteus mirabilis. Antimicrob. Agents Chemother. 62 (9), e00849–e00818. doi: 10.1128/AAC.00849-18
Lei, C. W., Yao, T. G., Yan, J., Li, B. Y., Wang, X. C., Zhang, Y., et al. (2020). Identification of Proteus genomic island 2 variants in two clonal Proteus mirabilis isolates with coexistence of a novel genomic resistance island PmGRI1. J. Antimicrob. Chemother. 75 (9), 2503–2507. doi: 10.1093/jac/dkaa215
Lemieux, M. J., Huang, Y., Wang, D. N. (2004). The structural basis of substrate translocation by the escherichia coli glycerol-3-phosphate transporter: a member of the major facilitator superfamily. Curr. Opin. Struct. Biol. 14 (4), 405–412. doi: 10.1016/j.sbi.2004.06.003
Li, Y., Zheng, B., Li, Y., Zhu, S., Xue, F., Liu, J. (2015). Antimicrobial susceptibility and molecular mechanisms of fosfomycin resistance in clinical escherichia coli isolates in mainland China. PloS One 10 (8), e0135269. doi: 10.1371/journal.pone.0135269
Lin, D., Chen, S. (2015). First detection of conjugative plasmid-borne fosfomycin resistance gene fosA3 in salmonella isolates of food origin. Antimicrob. Agents Chemother. 59 (2), 1381–1383. doi: 10.1128/AAC.04750-14
Liu, B. T., Song, F. J., Zou, M., Zhang, Q. D., Shan, H. (2017). High incidence of escherichia coli strains coharboring mcr-1 and blaNDM from chickens. Antimicrob. Agents Chemother. 61 (3), e02347–e02316. doi: 10.1128/AAC.02347-16
Liu, F., Tian, A., Wang, J., Zhu, Y., Xie, Z., Zhang, R., et al. (2022). Occurrence and molecular epidemiology of fosA3-bearing escherichia coli from ducks in Shandong province of China. Poult Sci. 101 (3), 101620. doi: 10.1016/j.psj.2021.101620
Loras, C., González-Prieto, A., Pérez-Vázquez, M., Bautista, V., Ávila, A., Campoy, P. S., et al. (2021). Prevalence, detection and characterisation of fosfomycin-resistant escherichia coli strains carrying fosA genes in community of Madrid, Spain. J. Glob Antimicrob. Resist. 25, 137–141. doi: 10.1016/j.jgar.2021.02.032
Lu, P. L., Hsieh, Y. J., Lin, J. E., Huang, J. W., Yang, T. Y., Lin, L., et al. (2016). Characterisation of fosfomycin resistance mechanisms and molecular epidemiology in extended-spectrum β-lactamase-producing klebsiella pneumoniae isolates. Int. J. Antimicrob. Agents. 48 (5), 564–568. doi: 10.1016/j.ijantimicag.2016.08.013
Lu, J., Zhao, K., Xie, H., Li, R., Zhou, M. (2021). Identification and characterization of a novel SXT/R391 integrative and conjugative element in a Proteus mirabilis food isolate. Foodborne Pathog. Dis. 18 (10), 727–732. doi: 10.1089/fpd.2020.2886
Lucas, A. E., Ito, R., Mustapha, M. M., McElheny, C. L., Mettus, R. T., Bowler, S. L., et al. (2017). Frequency and mechanisms of spontaneous fosfomycin nonsusceptibility observed upon disk diffusion testing of escherichia coli. J. Clin. Microbiol. 56 (1), e01368–e01317. doi: 10.1128/JCM.01368-17
Lupo, A., Saras, E., Madec, J. Y., Haenni, M. (2018). Emergence of blaCTX-M-55 associated with fosA, rmtB and mcr gene variants in escherichia coli from various animal species in France. J. Antimicrob. Chemother. 73 (4), 867–872. doi: 10.1093/jac/dkx489
Lv, L., Huang, X., Wang, J., Huang, Y., Gao, X., Liu, Y., et al. (2020). Multiple plasmid vectors mediate the spread of fosA3 in extended-Spectrum-β-Lactamase-Producing Enterobacterales isolates from retail vegetables in China. mSphere. 5 (4), e00507–e00520. doi: 10.1128/mSphere.00507-20
Ma, W. Q., Han, Y. Y., Zhou, L., Peng, W. Q., Mao, L. Y., Yang, X., et al. (2022). Contamination of Proteus mirabilis harbouring various clinically important antimicrobial resistance genes in retail meat and aquatic products from food markets in China. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1086800
Ma, Y., Xu, X., Guo, Q., Wang, P., Wang, W., Wang, M. (2015). Characterization of fosA5, a new plasmid-mediated fosfomycin resistance gene in escherichia coli. Lett. Appl. Microbiol. 60 (3), 259–264. doi: 10.1111/lam.12366
Marquardt, J. L., Siegele, D. A., Kolter, R., Walsh, C. T. (1992). Cloning and sequencing of escherichia coli murZ and purification of its product, a UDP-n-acetylglucosamine enolpyruvyl transferase. J. Bacteriol. 174 (17), 5748–5752. doi: 10.1128/jb.174.17.5748-5752.1992
Mattioni Marchetti, V., Kraftova, L., Finianos, M., Sourenian, T., Hrabak, J., Bitar, I. (2023). Polyclonal spread of fosfomycin resistance among carbapenemase-producing members of the Enterobacterales in the Czech republic. Microbiol. Spectr. 26, e0009523. doi: 10.1128/spectrum.00095-23
Mazé, A., Glatter, T., Bumann, D. (2014). The central metabolism regulator EIIAGlc switches salmonella from growth arrest to acute virulence through activation of virulence factor secretion. Cell Rep. 7 (5), 1426–1433. doi: 10.1016/j.celrep.2014.04.022
Mendes, A. C., Rodrigues, C., Pires, J., Amorim, J., Ramos, M. H., Novais, Â, et al. (2016). Importation of fosfomycin resistance fosA3 gene to Europe. Emerg. Infect. Dis. 22 (2), 346–348. doi: 10.3201/eid2202.151301
Mendoza, C., Garcia, J. M., Llaneza, J., Mendez, F. J., Hardisson, C., Ortiz, J. M. (1980). Plasmid-determined resistance to fosfomycin in serratia marcescens. Antimicrob. Agents Chemother. 18 (2), 215–219. doi: 10.1128/AAC.18.2.215
Michalopoulos, A. S., Livaditis, I. G., Gougoutas, V. (2011). The revival of fosfomycin. Int. J. Infect. Dis. 15 (11), e732–e739. doi: 10.1016/j.ijid.2011.07.007
Michalopoulos, A., Virtzili, S., Rafailidis, P., Chalevelakis, G., Damala, M., Falagas, M. E. (2010). Intravenous fosfomycin for the treatment of nosocomial infections caused by carbapenem-resistant klebsiella pneumoniae in critically ill patients: a prospective evaluation. Clin. Microbiol. Infect. 16 (2), 184–186. doi: 10.1111/j.1469-0691.2009.02921.x
Milner, K. A., Bay, D. C., Alexander, D., Walkty, A., Karlowsky, J. A., Mulvey, M. R., et al. (2020). Identification and characterization of a novel FosA7 member from fosfomycin-resistant escherichia coli clinical isolates from Canadian hospitals. Antimicrob. Agents Chemother. 65 (1), e00865–e00820. doi: 10.1128/AAC.00865-20
Mosime, L. B., Newton-Foot, M., Nel, P. (2022). Fosfomycin resistance in community-acquired urinary pathogens from Western cape, south Africa. S Afr J. Infect. Dis. 37 (1), 321. doi: 10.4102/sajid.v37i1.321
Mowlaboccus, S., Daley, D., Pang, S., Gottlieb, T., Merlino, J., Nimmo, G. R., et al. (2020). Identification and characterisation of fosfomycin resistance in escherichia coli urinary tract infection isolates from Australia. Int. J. Antimicrob. Agents. 56 (4), 106121. doi: 10.1016/j.ijantimicag.2020.106121
Mueller, L., Cimen, C., Poirel, L., Descombes, M. C., Nordmann, P. (2019). Prevalence of fosfomycin resistance among ESBL-producing escherichia coli isolates in the community, Switzerland. Eur. J. Clin. Microbiol. Infect. Dis. 38 (5), 945–949. doi: 10.1007/s10096-019-03531-0
Muthuirulandi Sethuvel, D. P., Anandan, S., Devanga Ragupathi, N. K., Gajendiran, R., Kuroda, M., Shibayama, K., et al. (2019). IncFII plasmid carrying antimicrobial resistance genes in shigella flexneri: vehicle for dissemination. J. Glob Antimicrob. Resist. 16, 215–219. doi: 10.1016/j.jgar.2018.10.014
Nakamura, G., Wachino, J., Sato, N., Kimura, K., Yamada, K., Jin, W., et al. (2014). Practical agar-based disk potentiation test for detection of fosfomycin-nonsusceptible escherichia coli clinical isolates producing glutathione s-transferases. J. Clin. Microbiol. 52 (9), 3175–3179. doi: 10.1128/JCM.01094-14
Nilsson, A. I., Berg, O. G., Aspevall, O., Kahlmeter, G., Andersson, D. I. (2003). Biological costs and mechanisms of fosfomycin resistance in escherichia coli. Antimicrob. Agents Chemother. 47 (9), 2850–2858. doi: 10.1128/AAC.47.9.2850-2858.2003
Nishida, S., Matsunaga, N., Kamimura, Y., Ishigaki, S., Furukawa, T., Ono, Y. (2020). Emergence of Enterobacter cloacae complex Co-producing IMP-10 and CTX-m, and Klebsiella pneumoniae producing VIM-1 in clinical isolates in Japan. Microorganisms. 8 (11), 1816. doi: 10.3390/microorganisms8111816
Nordmann, P., Poirel, L., Mueller, L. (2019). Rapid detection of fosfomycin resistance in escherichia coli. J. Clin. Microbiol. 57 (1), e01531–e01518. doi: 10.1128/JCM.01531-18
Nordmann, P., Sadek, M., Ortiz de la Rosa, J. M., Pfister, S., Fournier, C., Poirel, L. (2022). Selective culture medium for screening of fosfomycin resistance in Enterobacterales. J. Clin. Microbiol. 60 (1), e0206321. doi: 10.1128/JCM.02063-21
Norizuki, C., Kawamura, K., Wachino, J. I., Suzuki, M., Nagano, N., Kondo, T., et al. (2018). Detection of escherichia coli producing CTX-M-1-Group extended-spectrum β-lactamases from pigs in aichi prefecture, Japan, between 2015 and 2016. Jpn J. Infect. Dis. 71 (1), 33–38. doi: 10.7883/yoken.JJID.2017.206
Ohkoshi, Y., Sato, T., Suzuki, Y., Yamamoto, S., Shiraishi, T., Ogasawara, N., et al. (2017). Mechanism of reduced susceptibility to fosfomycin in Escherichia coli clinical isolates. BioMed. Res. Int. 2017, 5470241. doi: 10.1155/2017/5470241
Ortiz de la Rosa, J. M., Nordmann, P., Zong, Z., Poirel, L. (2022). Aliidiomarina shirensis as possible source of the integron- and plasmid-mediated fosfomycin resistance gene fosC2. Antimicrob. Agents Chemother. 66 (3), e0222721. doi: 10.1128/aac.02227-21
Ortiz-Padilla, M., Portillo-Calderón, I., Velázquez-Escudero, A., Rodríguez-Baño, J., Pascual, Á, Rodríguez-Martínez, J. M., et al. (2022). Effect of glycerol on fosfomycin activity against escherichia coli. Antibiotics (Basel) 11 (11), 1612. doi: 10.3390/antibiotics11111612
Pan, Y., Hu, B., Bai, X., Yang, X., Cao, L., Liu, Q., et al. (2021). Antimicrobial resistance of non-O157 shiga toxin-producing Escherichia coli isolated from humans and domestic animals. Antibiotics (Basel). 10 (1), 74. doi: 10.3390/antibiotics10010074
Partridge, S. R., Kwong, S. M., Firth, N., Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31 (4), e00088–e00017. doi: 10.1128/CMR.00088-17
Peng, Z., Hu, Z., Li, Z., Li, X., Jia, C., Zhang, X., et al. (2019). Characteristics of a colistin-resistant Escherichia coli ST695 harboring the chromosomally-encoded mcr-1 gene. Microorganisms. 7 (11), 558. doi: 10.3390/microorganisms7110558
Pérez, D. S., Tapia, M. O., Soraci, A. L. (2014). Fosfomycin: uses and potentialities in veterinary medicine. Open Vet. J. 4 (1), 26–43.
Poirel, L., Vuillemin, X., Kieffer, N., Mueller, L., Descombes, M. C., Nordmann, P. (2019). Identification of FosA8, a plasmid-encoded fosfomycin resistance determinant from escherichia coli, and its origin in leclercia adecarboxylata. Antimicrob. Agents Chemother. 63 (11), e01403–e01419. doi: 10.1128/AAC.01403-19
Postma, P. W., Lengeler, J. W., Jacobson, G. R. (1993). Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57 (3), 543–594. doi: 10.1128/mr.57.3.543-594.1993
Prestinaci, F., Pezzotti, P., Pantosti, A. (2015). Antimicrobial resistance: a global multifaceted phenomenon. Pathog. Glob Health 109 (7), 309–318. doi: 10.1179/2047773215Y.0000000030
Putensen, C., Ellger, B., Sakka, S. G., Weyland, A., Schmidt, K., Zoller, M., et al. (2019). Current clinical use of intravenous fosfomycin in ICU patients in two European countries. Infection. 47 (5), 827–836. doi: 10.1007/s15010-019-01323-4
Ramadan, H., Soliman, A. M., Hiott, L. M., Elbediwi, M., Woodley, T. A., Chattaway, M. A., et al. (2021). Emergence of multidrug-resistant Escherichia coli producing CTX-m, MCR-1, and FosA in retail food from Egypt. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.681588
Rehman, M. A., Yin, X., Persaud-Lachhman, M. G., Diarra, M. S. (2017). First detection of a fosfomycin resistance gene, fosA7, in salmonella enterica serovar Heidelberg isolated from broiler chickens. Antimicrob. Agents Chemother. 61 (8), e00410–e00417. doi: 10.1128/AAC.00410-17
Rigsby, R. E., Rife, C. L., Fillgrove, K. L., Newcomer, M. E., Armstrong, R. N. (2004). Phosphonoformate: a minimal transition state analogue inhibitor of the fosfomycin resistance protein, FosA. Biochemistry 43 (43), 13666–13673. doi: 10.1021/bi048767h
Rodriguez, M. M., Ghiglione, B., Power, P., Naas, T., Gutkind, G. (2018). Proposing kluyvera georgiana as the origin of the plasmid-mediated resistance gene fosA4. Antimicrob. Agents Chemother. 62 (8), e00710–e00718. doi: 10.1128/AAC.00710-18
Sadek, M., Ortiz de la Rosa, J. M., Ramadan, M., Nordmann, P., Poirel, L. (2022). Molecular characterization of extended-spectrum ß-lactamase producers, carbapenemase producers, polymyxin-resistant, and fosfomycin-resistant enterobacterales among pigs from Egypt. J. Glob Antimicrob. Resist., 30, 81–87. doi: 10.1016/j.jgar.2022.05.022
Saffen, D. W., Presper, K. A., Doering, T. L., Roseman, S. (1987). Sugar transport by the bacterial phosphotransferase system. molecular cloning and structural analysis of the escherichia coli ptsH, ptsI, and crr genes. J. Biol. Chem. 262 (33), 16241–16253.
Sato, N., Kawamura, K., Nakane, K., Wachino, J., Arakawa, Y. (2013). First detection of fosfomycin resistance gene fosA3 in CTX-m-producing escherichia coli isolates from healthy individuals in Japan. Microb. Drug Resist. 19 (6), 477–482. doi: 10.1089/mdr.2013.0061
Schreiber, A., Härter, G., Schubert, A., Bunjes, D., Mertens, T., Michel, D. (2009). Antiviral treatment of cytomegalovirus infection and resistant strains. Expert Opin. Pharmacother. 10 (2), 191–209. doi: 10.1517/14656560802678138
Seoane, A., Sangari, F. J., Lobo, J. M. (2010). Complete nucleotide sequence of the fosfomycin resistance transposon Tn2921. Int. J. Antimicrob. Agents. 35 (4), 413–414. doi: 10.1016/j.ijantimicag.2009.12.006
Seok, H., Choi, J. Y., Wi, Y. M., Park, D. W., Peck, K. R., Ko, K. S. (2020). Fosfomycin resistance in Escherichia coli isolates from south Korea and in vitro activity of fosfomycin alone and in combination with other antibiotics. Antibiotics (Basel). 9 (3), 112. doi: 10.3390/antibiotics9030112
Shi, L., Feng, J., Zhan, Z., Zhao, Y., Zhou, H., Mao, H., et al. (2018). Comparative analysis of blaKPC-2- and rmtB-carrying IncFII-family pKPC-LK30/pHN7A8 hybrid plasmids from Klebsiella pneumoniae CG258 strains disseminated among multiple Chinese hospitals. Infect. Drug Resist. 11, 1783–1793. doi: 10.2147/IDR.S171953
Silver, L. L. (2017). Fosfomycin: mechanism and resistance. Cold Spring Harb. Perspect. Med. 7 (2), a025262. doi: 10.1101/cshperspect.a025262
Singkham-In, U., Muhummudaree, N., Chatsuwan, T. (2020). fosA3 overexpression with transporter mutations mediates high-level of fosfomycin resistance and silence of fosA3 in fosfomycin-susceptible klebsiella pneumoniae producing carbapenemase clinical isolates. PloS One 15 (8), e0237474. doi: 10.1371/journal.pone.0237474
Skarżyńska, M., Zaja, C. M., Bomba, A., Bocian, Ł, Kozdruń, W., Polak, M., et al. (2021). Antimicrobial resistance glides in the sky-Free-Living birds as a reservoir of resistant Escherichia coli with zoonotic potential. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.656223
Skarzynski, T., Mistry, A., Wonacott, A., Hutchinson, S. E., Kelly, V. A., Duncan, K. (1996). Structure of UDP-n-acetylglucosamine enolpyruvyl transferase, an enzyme essential for the synthesis of bacterial peptidoglycan, complexed with substrate UDP-n-acetylglucosamine and the drug fosfomycin. Structure 4 (12), 1465–1474. doi: 10.1016/S0969-2126(96)00153-0
Soliman, A. M., Ramadan, H., Zarad, H., Sugawara, Y., Yu, L., Sugai, M., et al. (2021). Coproduction of Tet(X7) conferring high-level tigecycline resistance, fosfomycin FosA4, and colistin mcr-1.1 in escherichia coli strains from chickens in Egypt. Antimicrob. Agents Chemother. 65 (6), e02084–e02020. doi: 10.1128/AAC.02084-20
Song, Z., Lei, C. W., Zuo, L., Li, C., Wang, Y. L., Tian, Y. M., et al. (2022). Whole genome sequence of Proteus mirabilis ChSC1905 strain harbouring a new SXT/R391-family ICE. J. Glob Antimicrob. Resist. 30, 279–281. doi: 10.1016/j.jgar.2022.07.004
Sorlozano-Puerto, A., Lopez-Machado, I., Albertuz-Crespo, M., Martinez-Gonzalez, L. J., Gutierrez-Fernandez, J. (2020). Characterization of fosfomycin and nitrofurantoin resistance mechanisms in Escherichia coli isolated in clinical urine samples. Antibiotics (Basel). 9 (9), 534. doi: 10.3390/antibiotics9090534
Sun, H., Li, S., Xie, Z., Yang, F., Sun, Y., Zhu, Y., et al. (2012). A novel multidrug resistance plasmid isolated from an escherichia coli strain resistant to aminoglycosides. J. Antimicrob. Chemother. 67 (7), 1635–1638. doi: 10.1093/jac/dks107
Takahata, S., Ida, T., Hiraishi, T., Sakakibara, S., Maebashi, K., Terada, S., et al. (2010). Molecular mechanisms of fosfomycin resistance in clinical isolates of escherichia coli. Int. J. Antimicrob. Agents 35 (4), 333–337. doi: 10.1016/j.ijantimicag.2009.11.011
Tang, Y., Zhou, Y., Meng, C., Huang, Y., Jiang, X. (2020). Co-Occurrence of a novel VIM-1 and FosA3-encoding multidrug-resistant plasmid and a KPC-2-encoding pKP048-like plasmid in a clinical isolate of klebsiella pneumoniae sequence type 11. Infect. Genet. Evol. 85, 104479. doi: 10.1016/j.meegid.2020.104479
Taniguchi, Y., Maeyama, Y., Ohsaki, Y., Hayashi, W., Osaka, S., Koide, S., et al. (2017). Co-Resistance to colistin and tigecycline by disrupting mgrB and ramR with IS insertions in a canine klebsiella pneumoniae ST37 isolate producing SHV-12, DHA-1 and FosA3. Int. J. Antimicrob. Agents. 50 (5), 697–698. doi: 10.1016/j.ijantimicag.2017.09.011
Ten Doesschate, T., Abbott, I. J., Willems, R. J. L., Top, J., Rogers, M. R. C., Bonten, M. M., et al. (2019). In vivo acquisition of fosfomycin resistance in escherichia coli by fosA transmission from commensal flora. J. Antimicrob. Chemother. 74 (12), 3630–3632. doi: 10.1093/jac/dkz380
Theuretzbacher, U., Paul, M. (2015). Revival of old antibiotics: structuring the re-development process to optimize usage. Clin. Microbiol. Infect. 21 (10), 878–880. doi: 10.1016/j.cmi.2015.06.019
Tian, X., Zheng, X., Sun, Y., Fang, R., Zhang, S., Zhang, X., et al. (2020). Molecular mechanisms and epidemiology of carbapenem-resistant Escherichiacoli isolated from Chinese patients during 2002-2017. Infect. Drug Resist. 13, 501–512. doi: 10.2147/IDR.S232010
Tseng, S. P., Wang, S. F., Kuo, C. Y., Huang, J. W., Hung, W. C., Ke, G. M., et al. (2015). Characterization of fosfomycin resistant extended-spectrum β-Lactamase-Producing escherichia coli isolates from human and pig in Taiwan. PloS One 10 (8), e0135864. doi: 10.1371/journal.pone.0135864
Tseng, S. P., Wang, S. F., Ma, L., Wang, T. Y., Yang, T. Y., Siu, L. K., et al. (2017). The plasmid-mediated fosfomycin resistance determinants and synergy of fosfomycin and meropenem in carbapenem-resistant klebsiella pneumoniae isolates in Taiwan. J. Microbiol. Immunol. Infect. 50 (5), 653–661. doi: 10.1016/j.jmii.2017.03.003
Turcotte, M. R., Smith, J. T., Li, J., Zhang, X., Wolfe, K. L., Gao, F., et al. (2022). Genome characteristics of clinical salmonella enterica population from a state public health laboratory, new Hampshire, USA, 2017-2020. BMC Genomics 23 (1), 537. doi: 10.1186/s12864-022-08769-1
van den Bijllaardt, W., Schijffelen, M. J., Bosboom, R. W., Cohen Stuart, J., Diederen, B., Kampinga, G., et al. (2018). Susceptibility of ESBL escherichia coli and klebsiella pneumoniae to fosfomycin in the Netherlands and comparison of several testing methods including etest, MIC test strip, Vitek2, phoenix and disc diffusion. J. Antimicrob. Chemother. 73 (9), 2380–2387. doi: 10.1093/jac/dky214
Västermark, A., Saier, M. H., Jr. (2014). The involvement of transport proteins in transcriptional and metabolic regulation. Curr. Opin. Microbiol. 18, 8–15. doi: 10.1016/j.mib.2014.01.002
Vázquez, X., Fernández, J., Rodríguez-Lozano, J., Calvo, J., Rodicio, R., Rodicio, M. R. (2022). Genomic analysis of two MDR isolates of Salmonella enterica serovar infantis from a Spanish hospital bearing the blaCTX-M-65 gene with or without fosA3 in pESI-like plasmids. Antibiotics (Basel). 11 (6), 786. doi: 10.3390/antibiotics11060786
Villa, L., Guerra, B., Schmoger, S., Fischer, J., Helmuth, R., Zong, Z., et al. (2015). IncA/C plasmid carrying bla(NDM-1), bla(CMY-16), and fosA3 in a salmonella enterica serovar Corvallis strain isolated from a migratory wild bird in Germany. Antimicrob. Agents Chemother. 59 (10), 6597–6600. doi: 10.1128/AAC.00944-15
Wachino, J., Yamane, K., Suzuki, S., Kimura, K., Arakawa, Y. (2010). Prevalence of fosfomycin resistance among CTX-m-producing escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated fosfomycin-modifying enzymes. Antimicrob. Agents Chemother. 54 (7), 3061–3064. doi: 10.1128/AAC.01834-09
Wang, Y. P., Chen, Y. H., Hung, I. C., Chu, P. H., Chang, Y. H., Lin, Y. T., et al. (2022). Transporter genes and fosA associated with fosfomycin resistance in carbapenem-resistant Klebsiella pneumoniae. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.816806
Wang, X. M., Dong, Z., Schwarz, S., Zhu, Y., Hua, X., Zhang, Y., et al. (2017). Plasmids of diverse inc groups disseminate the fosfomycin resistance gene fosA3 among escherichia coli isolates from pigs, chickens, and dairy cows in northeast China. Antimicrob. Agents Chemother. 24 61 (9), e00859–e00817. doi: 10.1128/AAC.00859-17
Wang, D., Fang, L. X., Jiang, Y. W., Wu, D. S., Jiang, Q., Sun, R. Y., et al. (2022). Comparison of the prevalence and molecular characteristics of fosA3 and fosA7 among salmonella isolates from food animals in China. J. Antimicrob. Chemother. 77 (5), 1286–1295. doi: 10.1093/jac/dkac061
Wang, Y., Lo, W. U., Lai, E. L., Chow, K. H., Ho, P. L. (2015). Complete sequence of the multidrug-resistant IncL/M plasmid pIMP-HB623 cocarrying bla IMP-34 and fosC2 in an enterobacter cloacae strain associated with medical travel to China. Antimicrob. Agents Chemother. 59 (9), 5854–5856. doi: 10.1128/AAC.00375-15
Wang, Z., Xu, H., Tang, Y., Li, Q., Jiao, X. (2020). A multidrug-resistant monophasic Salmonella typhimurium Co-harboring mcr-1, fosA3, blaCTX-M-14 in a transferable IncHI2 plasmid from a healthy catering worker in China. Infect. Drug Resist. 13, 3569–3574. doi: 10.2147/IDR.S272272
Wang, Q., Zhang, P., Zhao, D., Jiang, Y., Zhao, F., Wang, Y., et al. (2018). Emergence of tigecycline resistance in Escherichia coli co-producing MCR-1 and NDM-5 during tigecycline salvage treatment. Infect. Drug Resist. 11, 2241–2248. doi: 10.2147/IDR.S179618
Wang, S., Zhou, K., Xiao, S., Xie, L., Gu, F., Li, X., et al. (2019). A multidrug resistance plasmid pIMP26, carrying blaIMP-26, fosA5, blaDHA-1, and qnrB4 in enterobacter cloacae. Sci. Rep. 9 (1), 10212. doi: 10.1038/s41598-019-46777-6
Wong, M. H., Xie, M., Xie, L., Lin, D., Li, R., Zhou, Y., et al. (2016). Complete sequence of a F33:A-:B- conjugative plasmid carrying the oqxAB, fosA3, and blaCTX-M-55 elements from a foodborne escherichia coli strain. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01729
Xiang, D. R., Li, J. J., Sheng, Z. K., Yu, H. Y., Deng, M., Bi, S., et al. (2015). Complete sequence of a novel IncR-F33:A-:B- plasmid, pKP1034, harboring fosA3, blaKPC-2, blaCTX-M-65, blaSHV-12, and rmtB from an epidemic klebsiella pneumoniae sequence type 11 strain in China. Antimicrob. Agents Chemother. 60 (3), 1343–1348. doi: 10.1128/AAC.01488-15
Xie, M., Lin, D., Chen, K., Chan, E. W., Yao, W., Chen, S. (2016). Molecular characterization of escherichia coli strains isolated from retail meat that harbor blaCTX-m and fosA3 genes. Antimicrob. Agents Chemother. 60 (4), 2450–2455. doi: 10.1128/AAC.03101-15
Xu, S., Fu, Z., Zhou, Y., Liu, Y., Xu, X., Wang, M. (2017). Mutations of the transporter proteins GlpT and UhpT confer fosfomycin resistance in staphylococcus aureus. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00914
Xu, H., Miao, V., Kwong, W., Xia, R., Davies, J. (2011). Identification of a novel fosfomycin resistance gene (fosA2) in enterobacter cloacae from the salmon river, Canada. Lett. Appl. Microbiol. 52 (4), 427–429. doi: 10.1111/j.1472-765X.2011.03016.x
Yang, R. S., Feng, Y., Lv, X. Y., Duan, J. H., Chen, J., Fang, L. X., et al. (2016). Emergence of NDM-5- and MCR-1-Producing escherichia coli clones ST648 and ST156 from a single Muscovy duck (Cairina moschata). Antimicrob. Agents Chemother. 60 (11), 6899–6902. doi: 10.1128/AAC.01365-16
Yang, B., Larson, T. J. (1998). Multiple promoters are responsible for transcription of the glpEGR operon of escherichia coli K-12. Biochim. Biophys. Acta 1396 (1), 114–126. doi: 10.1016/s0167-4781(97)00179-6
Yang, Y., Sun, H., Liu, X., Wang, M., Xue, T., Sun, B. (2016). Regulatory mechanism of the three-component system HptRSA in glucose-6-phosphate uptake in staphylococcus aureus. Med. Microbiol. Immunol. 205 (3), 241–253. doi: 10.1007/s00430-015-0446-6
Yunus, F., Hussain, A., Mirza, I. A., Hanif, F., Hussain, W., Satti, L., et al. (2021). Diagnostic accuracy of rapid fosfomycin Np test for detection of fosfomycin resistance in escherichia coli in a tertiary care hospital in Pakistan. J. Ayub Med. Coll. Abbottabad 33 (4), 668–672.
Zhang, L. J., Gu, X. X., Zhang, J., Yang, L., Lu, Y. W., Fang, L. X., et al. (2020). Characterization of a fosA3 carrying IncC-IncN plasmid from a multidrug-resistant ST17 Salmonella Indiana isolate. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01582
Zhao, W., Li, W., Du, X. D., Yao, H. (2022). Hybrid IncFIA/FIB/FIC(FII) plasmid co-carrying blaNDM-5 and fosA3 from an escherichia coli ST117 strain of retail chicken. Int. J. Food Microbiol. 382, 109914. doi: 10.1016/j.ijfoodmicro.2022.109914
Zhao, D., Zhou, Z., Hua, X., Zhang, H., Quan, J., Li, X., et al. (2018). Coexistence of mcr-1, blaKPC-2 and two copies of fosA3 in a clinical escherichia coli strain isolated from urine. Infect. Genet. Evol. 60, 77–79. doi: 10.1016/j.meegid.2018.02.025
Zhao, J. Y., Zhu, Y. Q., Li, Y. N., Mu, X. D., You, L. P., Xu, C., et al. (2015). Coexistence of SFO-1 and NDM-1 β-lactamase genes and fosfomycin resistance gene fosA3 in an escherichia coli clinical isolate. FEMS Microbiol. Lett. 362 (1), 1–7. doi: 10.1093/femsle/fnu018
Zhou, Y., Ai, W., Cao, Y., Guo, Y., Wu, X., Wang, B., et al. (2022). The Co-occurrence of NDM-5, MCR-1, and FosA3-encoding plasmids contributed to the generation of extensively drug-resistant klebsiella pneumoniae. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.811263
Keywords: fosfomycin, Enterobacterales, fosfomycin-resistance, fosfomycin-resistant determinant, epidemiology
Citation: Mattioni Marchetti V, Hrabak J and Bitar I (2023) Fosfomycin resistance mechanisms in Enterobacterales: an increasing threat. Front. Cell. Infect. Microbiol. 13:1178547. doi: 10.3389/fcimb.2023.1178547
Received: 02 March 2023; Accepted: 31 May 2023;
Published: 04 July 2023.
Edited by:
Rachel A.F. Wozniak, University of Rochester Medical Center, United StatesReviewed by:
Balaji Veeraraghavan, Christian Medical College and Hospital, IndiaSuresh Neethirajan, Farmworx Research Institute, Netherlands
Copyright © 2023 Mattioni Marchetti, Hrabak and Bitar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vittoria Mattioni Marchetti, dml0dG9yaWEubWF0dGlvbmltYXJjaGUwMUB1bml2ZXJzaXRhZGlwYXZpYS5pdA==
 Vittoria Mattioni Marchetti
Vittoria Mattioni Marchetti Jaroslav Hrabak
Jaroslav Hrabak Ibrahim Bitar
Ibrahim Bitar