
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 01 May 2023
Sec. Molecular Bacterial Pathogenesis
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1177857
This article is part of the Research Topic Quorum-sensing in Gram-positive pathogens – mechanisms, role in infection, and potential as a therapeutic target View all 5 articles
 Hannah N. Agnew1
Hannah N. Agnew1 John M. Atack2,3
John M. Atack2,3 Ann R.D. Fernando1
Ann R.D. Fernando1 Sophie N. Waters1
Sophie N. Waters1 Mark van der Linden4
Mark van der Linden4 Erin Smith5
Erin Smith5 Andrew D. Abell5
Andrew D. Abell5 Erin B. Brazel1
Erin B. Brazel1 James C. Paton1*
James C. Paton1* Claudia Trappetti1*
Claudia Trappetti1*Streptococcus pneumoniae is capable of randomly switching their genomic DNA methylation pattern between six distinct bacterial subpopulations (A-F) via recombination of a type 1 restriction-modification locus, spnIII. These pneumococcal subpopulations exhibit phenotypic changes which favor carriage or invasive disease. In particular, the spnIIIB allele has been associated with increased nasopharyngeal carriage and the downregulation of the luxS gene. The LuxS/AI-2 QS system represent a universal language for bacteria and has been linked to virulence and biofilm formation in S. pneumoniae. In this work, we have explored the link between spnIII alleles, the luxS gene and virulence in two clinical pneumococcal isolates from the blood and cerebrospinal fluid (CSF) of one pediatric meningitis patient. The blood and CSF strains showed different virulence profiles in mice. Analysis of the spnIII system of these strains recovered from the murine nasopharynx showed that the system switched to different alleles commensurate with the initial source of the isolate. Of note, the blood strain showed high expression of spnIIIB allele, previously linked with less LuxS protein production. Importantly, strains with deleted luxS displayed different phenotypic profiles compared to the wildtype, but similar to the strains recovered from the nasopharynx of infected mice. This study used clinically relevant S. pneumoniae strains to demonstrate that the regulatory network between luxS and the type 1 restriction-modification system play a key role in infections and may support different adaptation to specific host niches.
Streptococcus pneumoniae (the pneumococcus) is a Gram-positive bacterium that asymptomatically colonizes the nasopharynx in up to 95% of infants and 25% of adults (Trimble et al., 2020). However, it can migrate to other niches in the body and cause localized diseases, such as otitis media and sinusitis, or it can disseminate to sterile sites to cause invasive pneumococcal diseases (IPD) such as pneumonia, bacteremia and meningitis. The broad range of both localized and invasive diseases it causes, results in more than 190 million infections with 1.2 million deaths world-wide each year, making it one of the most pathogenic bacterial species (Lavelle and Ward, 2022).
The mechanism by which S. pneumoniae transit from colonizer to pathogen is not well understood. However, pneumococci are capable of randomly switching their genomic DNA methylation pattern between six distinct bacterial subpopulations (A-F) via recombination of a type 1 restriction-modification locus, spnIII, with any given culture comprising a mixture of genotypes. These pneumococcal subpopulations exhibit phenotypic changes which favor carriage or invasive disease. Of particular interest, the spnIIIA allele was found to be associated with higher quantities of opaque phase pneumococcal colonies and had a more invasive phenotype. In contrast, the spnIIIB allele has been associated with increased nasopharyngeal carriage and the downregulation of the luxS gene (Manso et al., 2014).
LuxS is an essential component of the “universal” Quorum sensing (QS) LuxS/Autoinducer-2 (AI-2) system present in both Gram-positive and Gram-negative bacteria (Miller and Bassler, 2001; Von Bodman et al., 2008). The metabolic enzyme LuxS (an S-ribosylhomocysteine lyase) synthesizes AI-2 as a by-product of the conversion of S-ribosyl-homocysteine to homocysteine, an essential reaction of the activated methyl cycle. There are numerous QS systems, each allowing bacteria to detect stimuli, through autoinducers, and respond by inducing changes in gene expression of target cells (De Kievit and Iglewski, 2000). Although, the response to the LuxS/AI-2 system has been well characterized in Gram-negative bacteria (Vendeville et al., 2005), it is not well understood in Gram-positive bacteria, and in particular S. pneumoniae. Importantly in pneumococci, the LuxS/AI-2 QS system has been linked to virulence and biofilm formation (Stroeher et al., 2003; Trappetti et al., 2011a).
Biofilms can be defined as highly complex and dynamic surface associated structures, which provide S. pneumoniae a means of evading the host immune response and antimicrobial treatment, making biofilms indispensable for the persistence of pneumococcal disease (Costerton et al., 1999; Kolenbrander, 2000). In the laboratory strain D39, addition of extracellular AI-2 induced biofilm formation, with a deletion of the luxS gene resulting in decreased biofilm formation (Trappetti et al., 2011b) and decreased virulence in a murine model (Stroeher et al., 2003). The effect of luxS expression on biofilm formation has also been assessed in clinical isolates (Trappetti et al., 2013; Tikhomirova et al., 2022). We have previously found that serotype 3 pneumococcal isolates from the blood of 12 distinct patients had higher levels of luxS expression compared with isolates from the ears of 13 distinct patients, indicating that the role of luxS in biofilm formation may differ depending on the site of isolation (Trappetti et al., 2013). Recently we have investigated the role of LuxS in the middle ear isolate 947 serotype 14 sequence type (ST) 15 (Tikhomirova et al., 2022). We found that while biofilm formation was similar between the wildtype 947 strain and the luxS deletion mutant, the luxS mutant appeared more virulent than the parent strain in an intranasal murine infection model. These findings suggest that LuxS/AI-2 QS system in the middle ear isolate may have an opposite effect to that of the invasive strain D39 (Stroeher et al., 2003; Tikhomirova et al., 2022).
In the current study, we have characterized luxS in two S. pneumoniae clinical isolates obtained at the same time from the blood and cerebral spinal fluid (serotype 15C, ST 8711) of a single pediatric meningitis patient. Previously we have shown that these clinical isolates display different virulence phenotypes in a murine infection model (Agnew et al., 2022). Here, we aimed to investigate the role of luxS in these two clinical isolates.
The wildtype S. pneumoniae strains used in this study (Table 1) were provided to us by the German National Reference Center for Streptococci (Aachen) (Agnew et al., 2022). Pneumococci serotype 15C blood and CSF isolates (designated 60B and 60CSF, respectively) were isolated in 2015 from a child, aged 2, admitted to hospital with meningitis. The cells were routinely grown on Columbia agar supplemented with 5% (vol/vol) horse blood (BA), with or without gentamicin (5 µg/mL) or spectinomycin (200 μg/mL), at 37°C in 5% CO2 overnight. Growth assays were carried out using pneumococci grown in a chemically defined media (CDM) which is composed of SILAC RPMI 1640 Flex Media supplemented with amino acids, vitamins, choline chloride and catalase (10 U/mL) as described previously (Agnew et al., 2022), and either 0.5% (w/v) glucose or 0.5% (w/v) galactose.
The luxS gene was deleted from S. pneumoniae 60B and 60CSF strains and replaced with a spectinomycin resistance cassette by transformation. A linear DNA fragment containing the resistance cassette was constructed by overlap extension PCR (Iannelli and Pozzi, 2004), using primers listed in Table 1.
The strains were grown in flat-bottom 96-well microtiter plates (Costar) with a final volume of 200 µL as previously described (Minhas et al., 2019). Bacterial strains were inoculated at a starting optical density at 600 nm (OD600) of 0.05 in CDM supplemented with either 0.5% (w/v) glucose or 0.5% (w/v) galactose and AI-2 (refer to Supplementary data for synthesis of AI-2) at concentrations of 4 µM, 10 µM, 100 µM and 200 µM, then incubated at 37°C with 5% CO2. The OD600 was measured every 30 min for a total of 24 h in a SpectroSTAR Omega spectrophotometer (BMG Labtech). Assays were conducted in triplicate with at least two repeated independent experiments. Statistically significant differences in both final OD600 and mid-exponential phase OD600 between strains was determined using two-tailed Student’s t test; P < 0.05 were deemed statistically significant.
Strains were grown overnight on BA plates at 37°C with 5% CO2. Cells were harvested, washed and resuspended in 1 mL of CDM to a final OD600 of 0.2. Bacterial suspensions were incubated at 37°C with 5% CO2. RNA extractions were carried our using a Qiagen RNeasy Minikit as per the manufacturer’s instructions. Differences in gene expression levels were determined using one-step relative real-time qRT-PCR in a Roche LC480 real-time cycler, as described previously (Minhas et al., 2019). Primers used for luxS and gyrA (internal control) are listed in Table 1 and were used at a final concentration of 200 nM per reaction. Amplification data were analyzed using the comparative critical threshold (2-ΔΔCT) method (Livak and Schmittgen, 2001). Assays were performed in triplicate and statistical analyses were performed using two-tailed Student’s t test; P values < 0.05 were deemed as statistically significant.
Carbon phenotype microarray (PM) analysis using the PM microplate PM1 (Biolog Inc.) was performed on the strains to test for catabolism of 95 different carbon sources as previously described (Minhas et al., 2019). Each well in the microplate contained a different carbon source. The cells were inoculated to a final OD590 of 0.06 in the buffer provided, in accordance with the manufacturer’s instructions. This suspension was added in 100 µL aliquots to the wells, and the plate was incubated at 37°C, 5% CO2. The OD590 was measured every 15 min for 24 h. Catabolic activity was measured through colorimetric analysis, in which a colorless tetrazolium dye was reduced by NADH produced during catabolism. The level of metabolism for each carbon source was arbitrarily determined by comparison with the zero carbon source blank.
Biofilm formation was measured in real-time with the real-time cell analyzer (RTCA) xCELLigence (Agilent Technologies Inc.) instrument as previously described (Agnew et al., 2022; Tikhomirova et al., 2022). This instrument detects variations in the impedance signal (expressed as the arbitrary cell index, CI) as bacterial cells attach and form biofilms on the gold-microelectrodes present at the bottom of the E-plates (Agilent Technologies Inc.). Bacterial strains were grown overnight on BA plates at 37°C with 5% CO2. The cells were harvested and resuspended in 200 µL of CDM + 0.5% (w/v) glucose or 0.5% (w/v) galactose to a final OD600 of 0.2. To the wells of the E-plate, 150 µL CDM + 0.5% (w/v) glucose or 0.5% (w/v) galactose ± 10µM AI-2 was added, before being placed in the cradle of the RTCA-DP system, within a 37°C incubator with 5% CO2 supplementation. An initial baseline impedance reading was taken before the E-plates were removed and 50 µL of bacterial suspension added to the wells for a 1 in 4 dilution, reducing the starting OD600 to 0.05. An additional 50 µL of CDM + 0.5% (w/v) glucose or 0.5% (w/v) galactose ± 10 µM AI-2 was added to the control wells. The E-plates were locked into the cradles of the RTCA-DP platform within the incubator and the impedance signal (CI) was recorded every 15 min for 24 h to monitor biofilm formation. Assays were conducted in duplicate with at least two repeated independent experiments. Statistical analysis was carried out using a two-tailed Student’s t test; P values < 0.05 were deemed statistically significant.
Adherence and invasion assays were carried out using the Detroit 562 human pharyngeal cell line as previously described (Trappetti et al., 2011a; Amin et al., 2015). Detroit 562 human pharyngeal cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS), 100 U/mL penicillin and 100 µg/mL streptomycin at 37°C in 5% CO2. Wells of 24-well tissue culture trays were seeded with Detroit cells in DMEM with 10% FCS as described previously (Trappetti et al., 2011a) and left to grow overnight. Cells were inoculated with 500 µL of each bacterial suspension grown overnight on BA plates and resuspended in CDM ± 10µM AI-2 at a final OD600 of 0.2. The same volume of each bacterial strain was added to empty wells as a control. Adherence assays were conducted after incubation of the bacteria with the Detroit cells for 2 h at 37°C. The wells were washed 3 times with PBS, the cells were detached from the plate by treatment with 100 µL 0.25% trypsin-0.02% EDTA and 400 µL of 0.1% Triton X-100 (Sigma), and samples were plated on BA plates to determine the number of adherent bacteria. Invasion assays were carried out essentially as described above. After the post-adherence washing step, cultures were incubated for 1 h in fresh media supplemented with 200 µg/mL gentamicin and 10 µg/mL penicillin to kill extracellular bacteria. Monolayers were again washed, lysed, serially diluted, and plated on BA, as described above. The assays were conducted in triplicate with at least two repeated independent experiments. Statistical analysis was carried out using a two-tailed Student’s t-test; P values < 0.05 were considered statistically significant. Data collected are presented as mean adherent or invasive bacteria ± standard error mean (SEM) in CFU/mL. The controls were used to monitor bacterial growth during the 2 h incubation period, ensuring strains grew at similar rates.
Animal experiments were approved by the University of Adelaide Animal Ethics Committee (approval number S-2022-029). Female outbred 4- to 6-week-old CD-1 (Swiss) mice were anaesthetized by intraperitoneal injection of ketamine (8 mg/mL) and xylazine (0.8 mg/mL), and were challenged intranasally with 50 µL of bacterial suspension containing 1 x 108 CFU in serum broth (SB) as previously described (Minhas et al., 2019). The challenge dose was retrospectively confirmed by serial dilution and plating on BA. At 24 h, groups of 8 mice were euthanized by CO2 asphyxiation before the blood, lungs, nasal tissue, ears, and brain were collected. Tissue samples were homogenized in 1 mL PBS, serially diluted and plated on BA plates containing 5 µg/mL gentamicin to enumerate pneumococci as previously described (Trappetti et al., 2011a). Statistical analyses of log-transformed CFU data were performed using two-tailed Student’s t test; P values < 0.05 were deemed statistically significant.
Female outbred 4- to 6-week-old CD-1 (Swiss) mice were anaesthetized and intranasally challenged as described above. The nasal tissue was harvested 24 h post infection, processed and plated as above. DNA extraction was performed on the luxS mutants, WT original challenge inoculum and colonies grown overnight on BA plates supplemented with 5 µg/mL gentamicin, using a Qiagen DNeasy Blood & Tissue kit as per the manufacturer’s instructions. The variant spnIII alleles were quantified as previously described (Manso et al., 2014). Statistical analyses were performed using two-way analysis of variance and a Tukey post-comparison test; P values < 0.05 were deemed statistically significant.
Female outbred 4- to 6-week-old CD-1 (Swiss) mice were anaesthetized and intranasally challenged as described above. The nasal tissue was harvested 24 h post infection, processed and plated as above. Bacterial colonies from each mouse (n = 5 for 60CSF, n = 6 for 60B) were resuspended in serum broth and stored at -80°C. Biolog assays were performed as above on bacteria from a single mouse, representative for each strain.
Previously, we have shown that while the CSF clinical S. pneumoniae isolate (60CSF) is found in the ears of infected mice 24 h post intranasal infection, the blood isolate (60B) was unable to survive in this niche (Agnew et al., 2022). To determine whether the observed different virulence phenotypes of 60B and 60CSF in mice could be mediated by rearrangements in the spnIII Type I restriction-modification (RM) system, the spnIII allele distribution was assessed from pneumococcal DNA extracted from the nasopharynx of intranasally infected animals. Mice were intranasally challenged with 108 colony forming units (CFU) of each strain and the bacteria were taken from the nose 24 h post-infection. The DNA was isolated and the spnIII alleles were quantified for the inoculum and recovered bacteria for both strains. The 60B inoculum used to challenge the mice had predominantly spnIIIB (21% spnIIIA, 39% spnIIIB, 9% spnIIIC, 7% spnIIID, 22% spnIIIE and 2% spnIIIF), while the 60CSF inoculum was skewed towards spnIIIA (33% spnIIIA, 27% spnIIIB, 7% spnIIIC, 15% spnIIID, 17% spnIIIE and 1% spnIIIF) (Figure 1). The bacteria recovered from the nasopharynx of both groups of mice, showed no change in the spnIII allele distribution. The spnIIIB allele remained the predominant allele in all samples from the nasopharynx of mice challenged with 60B (23% spnIIIA, 44% spnIIIB, 12% spnIIIC, 9% spnIIID, 10% spnIIIE and 2% spnIIIF), while mice challenged with 60CSF, showed a similar proportion of spnIIIA and spnIIIB (32% spnIIIA, 28% spnIIIB, 7% spnIIIC, 10% spnIIID, 16% spnIIIE and 7% spnIIIF). Interestingly, when comparing the bacteria recovered from the noses of infected mice, 60B had a significantly higher proportion of the spnIIIB allele compared to 60CSF (Figure 1).
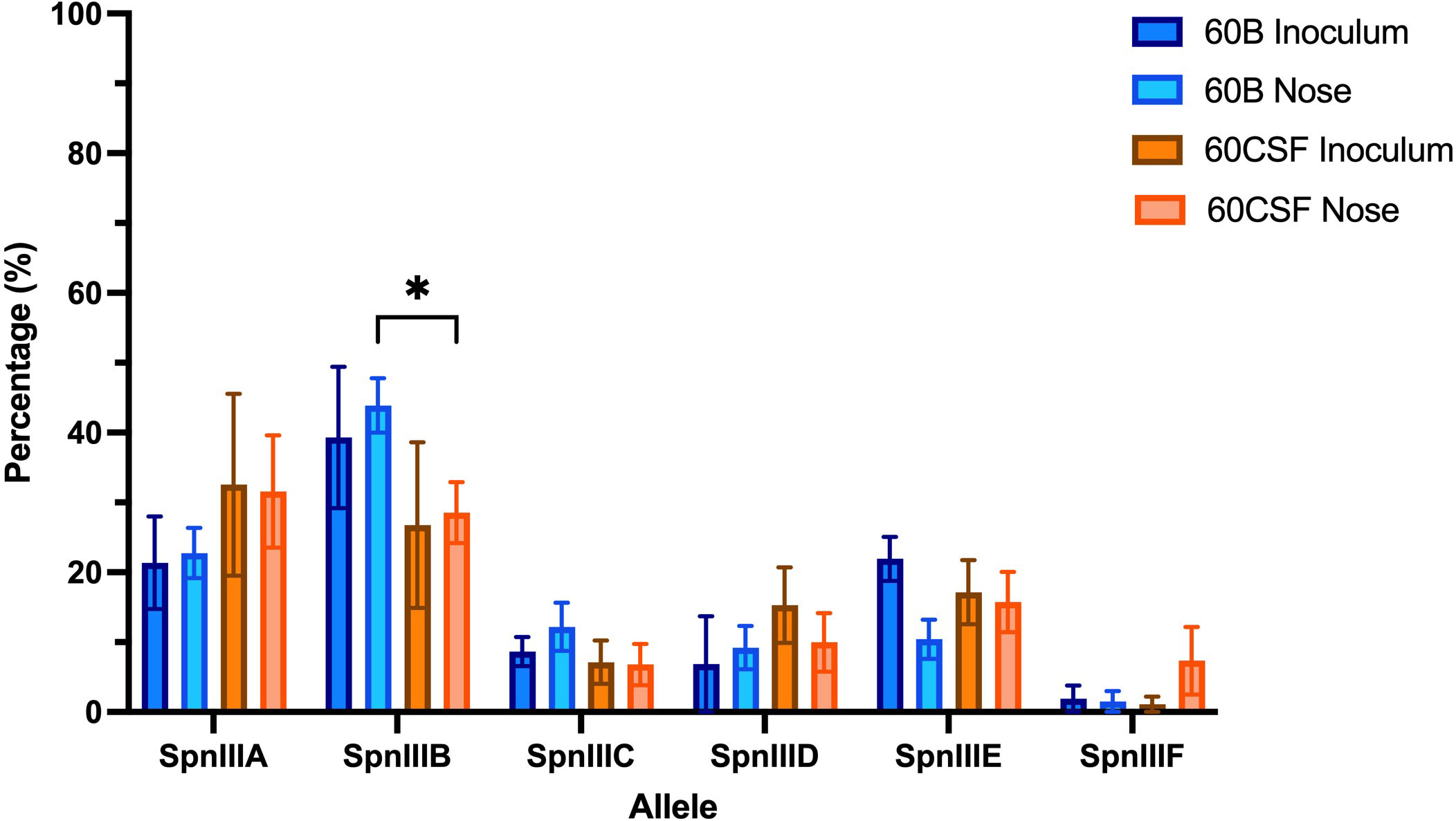
Figure 1 spnIII allele frequencies of 60B and 60CSF recovered post intranasal murine infection. Mice were intranasally challenged with 108 CFU S. pneumoniae serotype 15C ST8711 blood isolate (60B) or CSF isolate (60CSF). At 24 h, mice from each group were humanely euthanized and pneumococci in the nasal tissue were harvested. spnIII allele quantification was performed on DNA extracted from the original inoculum and colonies grown from nasopharynx samples. Allele percentages for original inoculum and bacteria recovered from noses are represented. Statistical analysis was performed using two-way analysis of variance and a Tukey post-comparison test; *P < 0.05.
We have previously reported that cell-cell signaling via the autoinducer 2 (AI-2)/LuxS quorum-sensing system is linked to the spnIIIB variant in S. pneumoniae. In particular, the luxS gene was found to be downregulated in SpnD39IIIB locked strain compared to the other SpnD39III locked strains (Manso et al., 2014). As reported above, the 60B strain has higher proportion of spnIIIB allele, while 60CSF has comparable level of spnIIIA and spnIIIB allele (Figure 1). Thus, we analyzed luxS gene expression in the 60B and 60CSF strains. Importantly, we found downregulation of the luxS gene in the 60B strain in which the spnIIIB allele is the predominant allele (Figure 2).
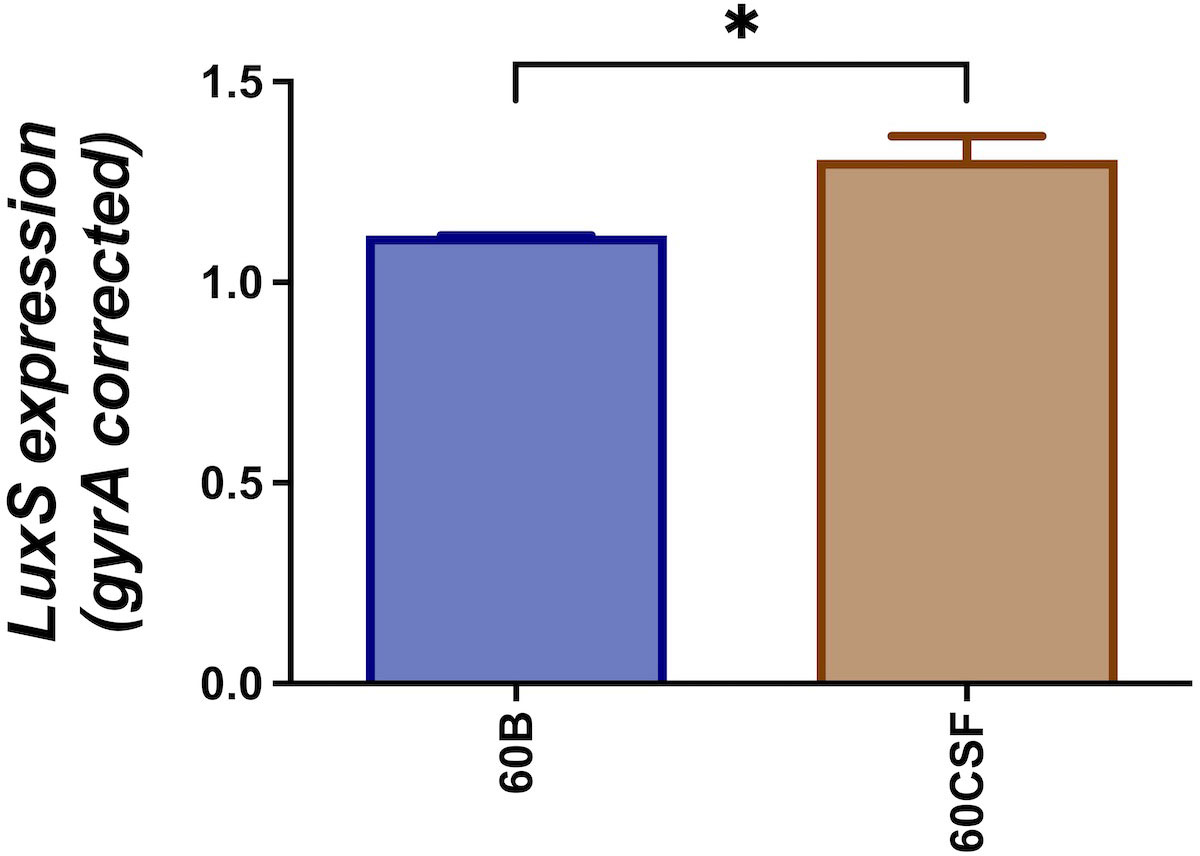
Figure 2 Expression of luxS by 60B and 60CSF isolates. Strains were resuspended in CDM at OD600 0.2 and incubated at 37°C with 5% CO2 for 30 min. RNA was extracted and luxS mRNA levels were analyzed by qRT-PCR using gyrA rRNA as an internal control (see Materials and Methods). Data are mean OD600 ± standard error mean (SEM) from three biological replicates. Significance of differences in gene expression between isolates was determined using two-tailed Student’s t test; *P < 0.05.
LuxS regulates key aspects of carbohydrate metabolism in S. pneumoniae and the capacity to utilize different carbon sources is crucial for nasopharyngeal colonization (Minhas et al., 2019; Mclean et al., 2020; Agnew et al., 2022). Thus, we employed a phenotypic microarray to compare the capacity of strains recovered from the nasopharynx of mice infected with either 60B or 60CSF to metabolize 95 different carbohydrates (see Materials and Methods). Interestingly, strains recovered from the nose of mice infected with either 60B or 60CSF showed a distinct metabolic profile to that of the inoculum strains. Importantly, the nasopharyngeal strains were now able to metabolize L-fucose, but unable to metabolize D-psicose (Supplementary Table 1).
We then created a luxS deletion mutant in both 60B and 60CSF. The metabolic profiles of these mutants were then assessed with the phenotypic microarray. Surprisingly, we found that these strains paralleled the metabolic profiles of the strains recovered from mice and not that of the original inoculating strains (Supplementary Table 1). These results indicate that the strains may undergo a change in the nose of mice that results in an increase of the spnIIIB allele to aid their colonization and resulting in reduced luxS expression, consequently leading to a similar metabolic profile to that of the luxS mutants. Thus, there appears to be an unknown link between LuxS and the spnIIIB allele, which merits further investigation.
Based on the results obtained in Section 3.3, we investigated the spnIII allele profiles of the luxS deletion mutants in 60B and 60CSF. As shown in Figure 3, it was found that 60BΔluxS possessed only spnIIIC and spnIIID (0% spnIIIA, 0% spnIIIB, 84.15% spnIIIC, 15.85% spnIIID, 0% spnIIIE and 0% spnIIIF), similar to 60CSFΔluxS (0% spnIIIA, 4.36% spnIIIB, 79.39% spnIIIC, 16.25% spnIIID, 0% spnIIIE and 0% spnIIIF). Compared to the WT 60B and 60CSF inoculums, both luxS mutants had no spnIIIA present and spnIIIB was only present in 60CSF∆luxS at a very low proportion. Interestingly, we found both mutants were now predominantly expressing spnIIIC. Similar results were seen when comparing the luxS deletion mutants with the WT strains recovered from the nose of infected mice, in which the luxS mutants had more spnIIIC, little to no spnIIIB and no spnIIIA (Supplementary Figure 1). These results support the link between LuxS and the spnIII locus, with a complete absence of LuxS resulting in a shift towards high proportions of spnIIIC and a severe decrease in spnIIIA and spnIIIB.
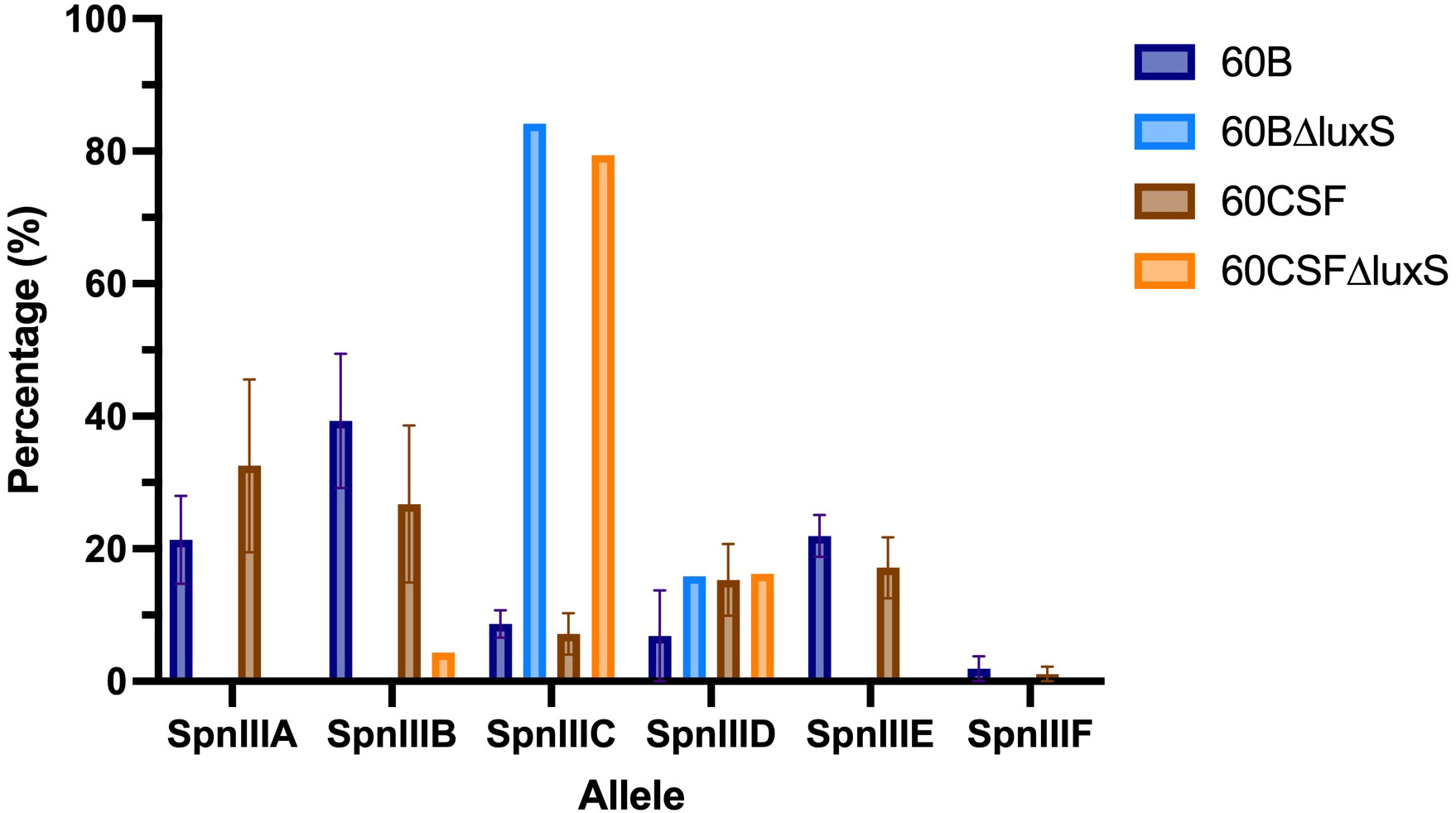
Figure 3 spnIII allele frequencies of 60B, 60CSF and luxS mutants. spnIII allele quantification was performed on DNA extracted from 60B, 60CSF, 60B∆luxS and 60CSF∆luxS colonies grown on BA plates overnight at 37°C with 5% CO2. Allele percentages are displayed for WT original inoculum (Figure 1) and luxS mutants from frozen stock.
S. pneumoniae is unable to grow with fucose as the sole carbon source, therefore, none of our established assays could be used for further analysis (Higgins et al., 2014). Similarly, D-Psicose is a low energy hexoketose monosaccharide, which is a C-3 epimer of D-fructose and is rarely found in nature, thus it is unlikely to play a role in pneumococcal virulence (Matsuo et al., 2002). Therefore, to further characterize our luxS mutant we used a different approach. LuxS QS boosts the capacity of S. pneumoniae to utilize galactose as a carbon source by upregulation of the Leloir pathway, so we then assessed the capacity of the luxS mutants to grow in galactose. Glucose was used as a control sugar, as it is the major sugar present in blood (Paixão et al., 2015). In CDM + glucose or galactose, wildtype (WT) strains showed comparable growth (Supplementary Figure 2). In Figure 4A, in CDM + glucose the 60B luxS deletion mutant (60BΔluxS) had a slight reduction in the final cell density (P value = 0.0329), while in CDM + galactose the observed reduction is more prominent (P value = 0.0257). Interestingly, the 60CSF luxS deletion mutant (60CSFΔluxS) showed comparable growth to that of the WT 60CSF in CDM + glucose (Figure 4B; P value = 0.3057), but an increased generation time compared to 60CSF in CDM + galactose was observed (P value = 0.0493). These results show that luxS deletion has a different impact in the two closely related strains.
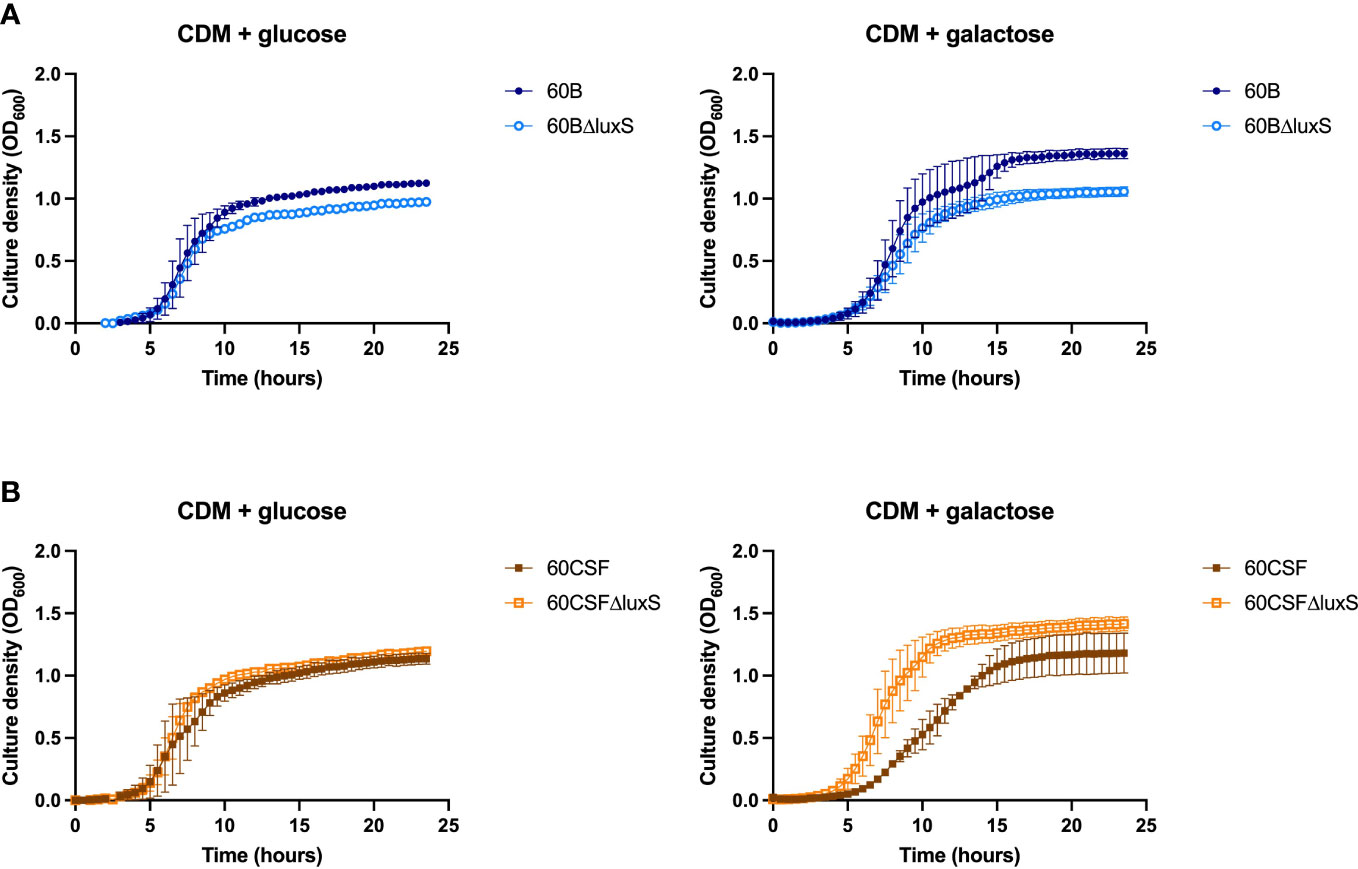
Figure 4 Growth of 60B, 60CSF and luxS mutants in CDM + glucose and CDM + galactose. 60B and its luxS mutant 60BΔluxS (A), and 60CSF and its luxS mutant 60CSFΔluxS (B) were grown in 200 µL CDM supplemented with 0.5% glucose (CDM + glucose) or 0.5% galactose (CDM + galactose). OD600 was measured every 30 min for 24 (h) Data are mean OD600 ± standard error mean (SEM) from two independent assays, each performed in triplicate.
It has previously been demonstrated that the addition of 10 µM exogenous AI-2 was able to moderately restore the defective growth of a D39 luxS deletion strain (Trappetti et al., 2017). However, it was also shown in two serotype 14 blood and ear isolates that the addition of exogenous AI-2 reduces the growth in a dose-dependent manner (Tikhomirova et al., 2022). Therefore, the effects of different concentrations of exogenous AI-2 on the growth of the blood (60B) and CSF (60 CSF) strains was assessed. In CDM + glucose, for both 60B and 60CSF strains the addition of 4 and 10 µM AI-2 had little or no effect on growth, but at the higher concentrations of 100 µM and 200 µM AI-2, reduction in growth was observed (Figure 5A). Interestingly, in CDM + galactose the addition of AI-2 caused reduced growth of 60CSF for all concentrations (4, 10, 100 and 200 µM) tested, whilst in 60B the reduced growth was only different when higher concentration of AI-2 (100 and 200 µM) were added to the media (Figure 5B).
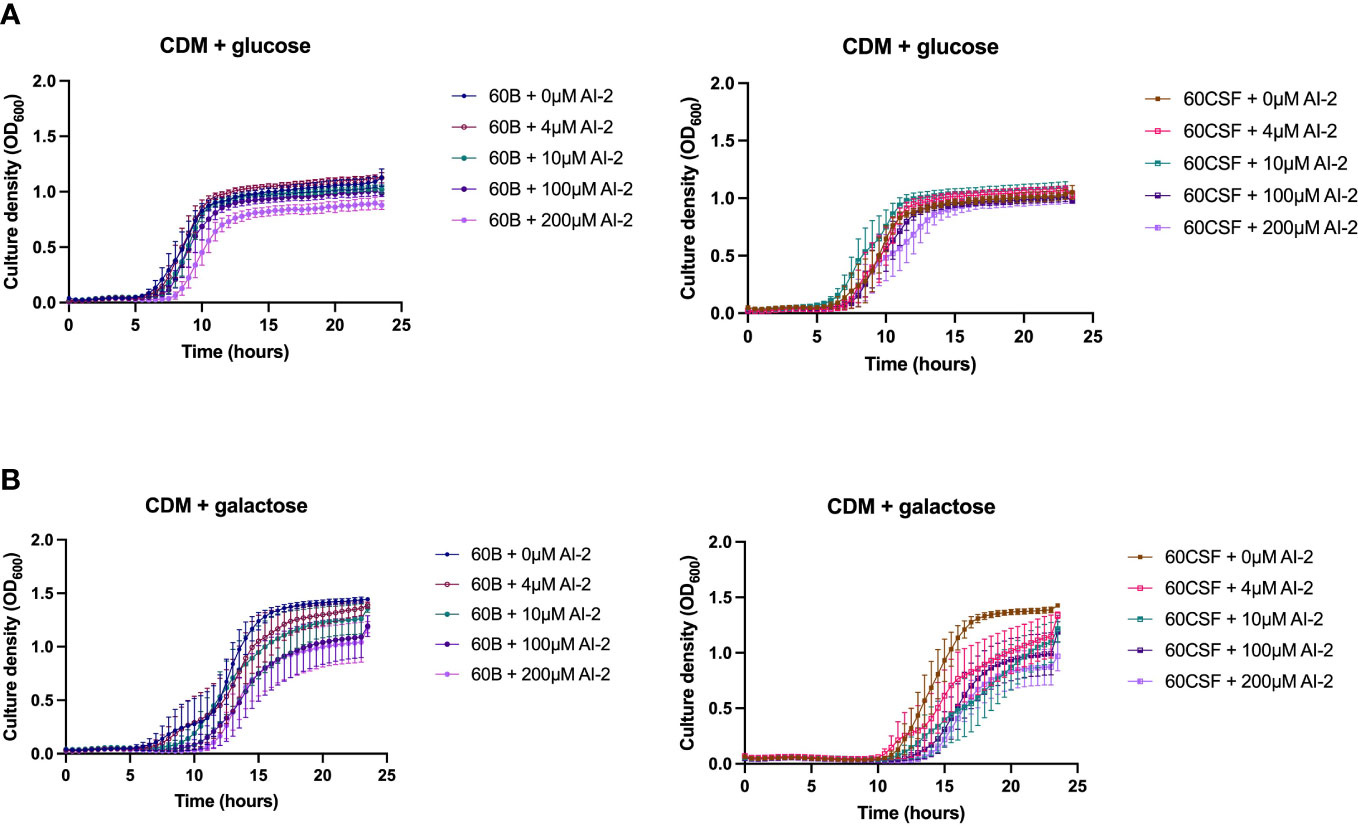
Figure 5 Growth of 60B and 60CSF with AI-2 supplementation at different concentrations. 60B and 60CSF were cultured in CDM + glucose (A) or CDM + galactose (B) supplemented with 0, 4, 10, 100 or 200 µM. OD600 was measured every 30 min for 24 (h) Data are mean OD600 ± standard error mean (SEM) from three independent assays, each performed in triplicate.
As the addition of AI-2 caused a growth defect in the WT 60B and 60CSF strains (Figure 5), we examined the capacity of exogenous AI-2 to complement the growth defect of luxS deletion strain. In Figure 6, addition of exogenous AI-2 to 60BΔluxS and 60CSFΔluxS had no effect in CDM + glucose, while it enhanced the growth of the 60BΔluxS mutant in CDM + galactose. These results confirmed a dose-dependent effect previously observed in the D39 strain (Trappetti et al., 2017).
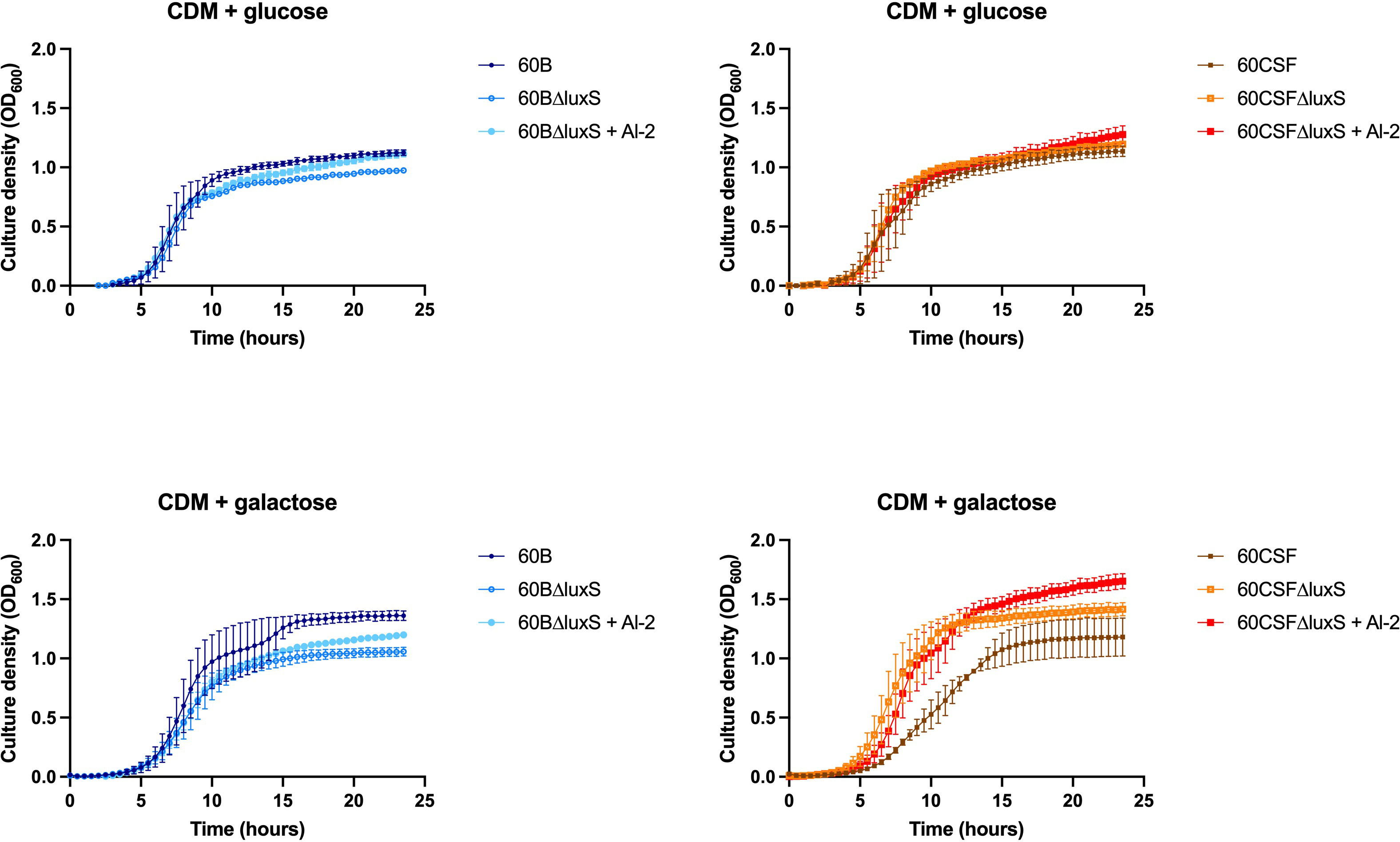
Figure 6 Impact of AI-2 on growth of luxS mutants. 60B, 60CSF, 60BΔluxS and 60CSFΔluxS were grown in 200 µL CDM supplemented with 0.5% glucose (CDM + glucose) or 0.5% galactose (CDM + galactose). OD600 was measured every 30 min for 24 h. Data are mean OD600 ± standard error mean (SEM) from two independent assays, each performed in triplicate.
Previously, luxS has been shown to play a key role in the ability of D39 to form a biofilm (Trappetti et al., 2011b). Therefore, we assessed the ability of WT 60B, 60CSF and luxS deletion mutants to form biofilm in real-time using CDM + glucose and CDM + galactose, with or without supplementation of 10 µM AI-2. In CDM + glucose and CDM + galactose both 60B and 60CSF showed the same capacity to form a biofilm (Supplementary Figure 3). The 60BΔluxS mutants formed a biofilm at a slightly increased rate compared to WT 60B in CDM + glucose (Figure 7A). Interestingly, the 60CSFΔluxS mutant had delayed biofilm formation compared to WT in CDM + glucose (Figure 7B). Addition of exogenous AI-2 greatly enhanced biofilm formation of WT strains but had no impact on luxS mutant strains (Figures 7A, B). In CDM + galactose, both luxS mutants (60BΔluxS & 60CSFΔluxS) had delayed biofilm formation compared to their WT counterparts (Figures 7C, D), with 60CSFΔluxS forming more biofilm than WT 60CSF. Similar to the earlier results in glucose, addition of AI-2 increased the biofilm formation of the WT 60B and 60CSF strains whilst appearing to have no impact in the luxS mutants.
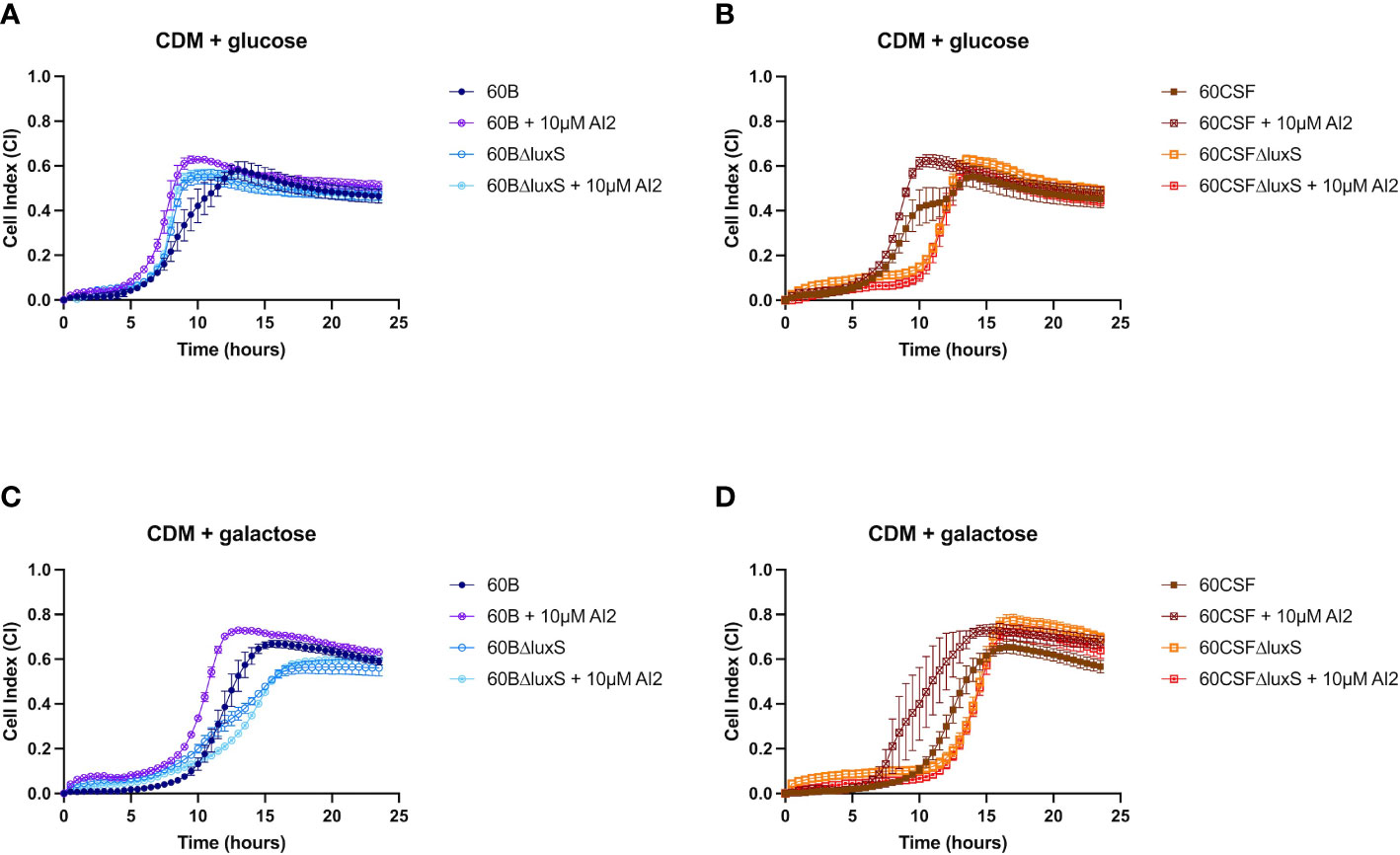
Figure 7 Biofilm formation of 60B, 60CSF, 60BΔluxS and 60CSFΔluxS strains. Bacteria were cultured overnight on BA plates before being diluted to a final OD600 of 0.05 in CDM + 0.5% glucose ± 10 µM AI-2 (A, B) or CDM + 0.5% galactose ± 10 µM AI-2 (C, D). 200 µL of each bacterial strain in each culture medium was placed into wells of a xCELLigence E-plate, with the plate being placed in the cradles of the RTCA-DP platform and incubated at 37°C with 5% CO2. Biofilm formation was determined by measuring cell index (CI) every 15 min over 24 h using real-time cell analysis (RTCA) xCELLigence technology. Data are mean CI ± standard error mean (SEM) from three independent assays, each performed in triplicate.
luxS has been found to be crucial for adherence to epithelial cells (Tikhomirova et al., 2022). Therefore, we investigated the role of LuxS in adherence to, and invasion of, human epithelial cells, using the human nasopharyngeal Detroit 562 cell line in CDM with or without 10 µM AI-2 supplementation. No significant differences in the total number of adherent and invasive cells were observed between the WT strains and the luxS deletion mutants in CDM, with or without the addition of AI-2 (Supplementary Figure 4). These results are similar to those previously found for a D39 luxS deletion mutant in which there was no difference between WT and mutant in adherence to lung (A549) or larynx (HEp-2) derived cell lines (Stroeher et al., 2003).
The ability of S. pneumoniae to metabolise specific carbohydrates has previously been linked to pneumococci disease phenotype in a murine model (Minhas et al., 2019; Agnew et al., 2022). As luxS may play a role in carbohydrate metabolism in pneumococci, their virulence phenotypes were assessed in a murine model. It has previously been shown that luxS plays a role in the pathogenesis of S. pneumoniae D39 and ear isolate 947 (Stroeher et al., 2003; Tikhomirova et al., 2022). Specifically, in the 947 strain, deletion of luxS resulted in a significantly higher bacterial burden in the ears of infected mice (Tikhomirova et al., 2022). An earlier study of the blood and CSF strains has shown that they behave differently in a murine model, whereby 60B was unable to survive in the ear (Agnew et al., 2022). Here, we extended these studies by exploring the 60B and 60CSF luxS deletion strains in our intranasal murine infection model. CD-1 Swiss mice were intranasally challenged with 108 CFU of each strain (60B, 60CSF, 60BΔluxS & 60CSFΔluxS) and the bacterial burden in the blood, brain, ears, lungs, and nose was assessed at 24 h post challenge. At 24 h post infection, there were no significant differences observed in the bacterial burdens in the brain and nasopharynx between mice infected with WT or luxS deletion strains (Supplementary Figure 5). Consistent with previous work (Agnew et al., 2022), no bacteria were detected in the ears of mice challenged with 60B, whilst bacteria were present in the ears of 50% of the mice infected with 60CSF (Figure 8; P < 0.05). Strikingly, the bacterial burden in the ears of mice challenged with 60BΔluxS was significantly higher than the bacteria detected in both 60B (P < 0.0001) and 60CSF (Figure 8; P < 0.05). There was no significant difference between the number of 60CSFΔluxS and WT 60CSF pneumococci present in the ears of mice, with the 60CSFΔluxS strain proliferating in this niche at a slightly higher number compared to the wildtype.
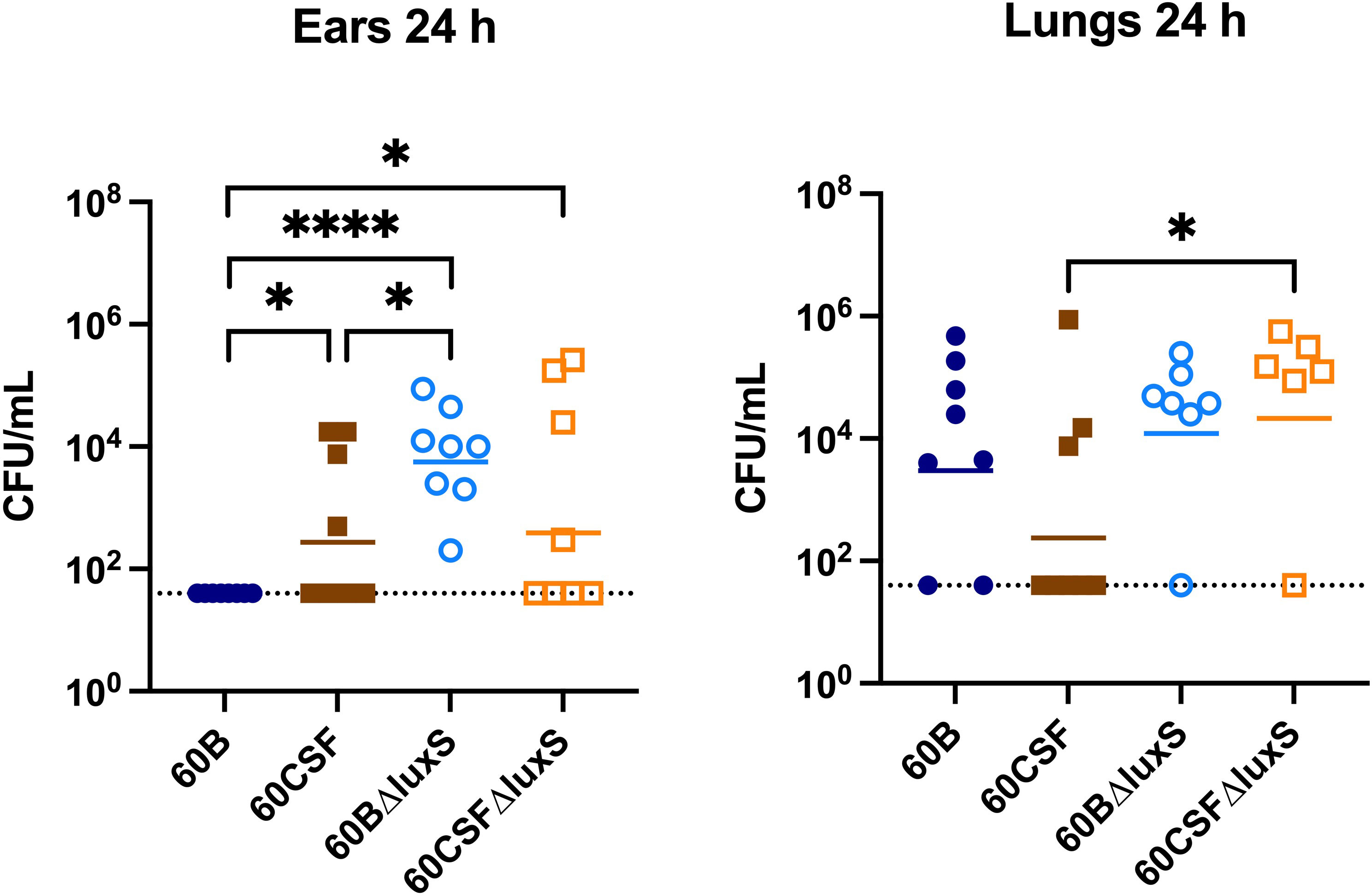
Figure 8 Virulence phenotypes of 60B, 60CSF, 60BΔluxS and 60CSFΔluxS strains. Groups of 8 mice were infected intranasally with 108 CFU of the indicated strain. At 24 h, mice from each group were humanely euthanized and pneumococci in the brain, ear, lungs and nasal tissue were enumerated. Viable bacterial counts are displayed for each mouse in the ears and lungs (brain and nose data in Supplementary Figure 5); horizontal bars indicate the geometric mean (GM) CFU for each group; the dotted line indicates the detection threshold. Significance of differences in bacterial load between groups was determined using two-tailed Student’s t test; *P < 0.05, ****P < 0.0001.
Interestingly, in the lungs of mice infected with 60CSFΔluxS, there was a significantly higher bacterial burden compared to mice challenged with 60CSF (Figure 8; P < 0.05). A similar trend was observed in the lungs of mice infected with 60BΔluxS, although the difference in bacterial numbers recovered from this niche did not reach statistical significance.
The LuxS/AI-2 QS system is dependent on the universal Autoinducer-2 (AI-2) signaling molecule, produced by LuxS. Previous studies on the pneumococcal LuxS QS system have shown that a luxS mutant strain is defective in biofilm formation and is less able to cause invasive diseases compared with wildtype pneumococci in the laboratory strain D39 (Trappetti et al., 2011b; Vidal et al., 2011). The opposite has been shown for the clinical ear isolate 947, where the luxS mutant was more able to cause invasive disease compared to the WT strain and no effect was observed on biofilm formation. In this work, we demonstrate that the different pathogenic profile previously observed in strains collected from a single pediatric patient (Agnew et al., 2022) might by linked to luxS and the restriction-modification (RM) locus spnIII.
S. pneumoniae is able to randomly switch its genomic DNA methylation pattern between six distinct states (spnIIIA-F) via recombination at the spnIII locus. Importantly, we previously found that these pneumococcal subpopulations exhibit phenotypic changes which have a major impact on bacterial virulence (Manso et al., 2014). This system, referred to as phasevarion (for phase-variable regulon), allows bacteria to generate a range of phenotypic variants within a population to act as an extra survival strategy (Phillips et al., 2019; Seib et al., 2020). Phasevarions have been characterized in multiple bacterial species such as H. influenzae (ModA) (Srikhanta et al., 2005; Fox et al., 2007; Atack et al., 2015), M catarrhalis (ModM) (Seib et al., 2002; Blakeway et al., 2014) the pathogenic Neisseriae (ModA, ModB, ModD) (Srikhanta et al., 2009; Seib et al., 2011; Seib et al., 2015; Tan et al., 2016), Helicobacter pylori (ModH) (Srikhanta et al., 2011), Streptococcus suis (Atack et al., 2018), and Actinobacillus pleuropneumoniae (Nahar et al., 2023). Analyzing the impact of altered methylase specificity on global transcriptomic patterns is complicated by the presence of multiple spnIII alleles in any culture. However, in a previous study, analysis of global gene expression was examined in a set of D39 spnIII mutants that were locked in a single specificity (Manso et al., 2014). RNA-seq identified only two major differences between the strains; the capsular polysaccharide serotype 2 biosynthesis operon csp2 and the luxS gene, with both of these genes significantly downregulated in the SpnIIIB-locked mutant compared to all the other locked strains analyzed. Previously, we have shown that two clinical isolates 60B and 60CSF (serotype 15C, sequence type 8711) that were isolated from the blood and cerebral spinal fluid (CSF), respectively, of a single pediatric meningitis patient exhibit distinct virulence phenotypes in an intranasal murine infection model (Agnew et al., 2022). The two strains expressed similar amounts of capsular polysaccharide, but different ability to colonize the nasopharynx and the ear of mice. The first step in development of pneumococcal disease is the colonization of the human nasopharynx. S. pneumoniae is part of the commensal flora of the upper respiratory tract and shares this niche with other potentially pathogenic bacteria including Gram-negative bacteria such as Haemophilus influenzae, Neisseria meningitidis, and other Gram-positive bacteria, such as Staphylococcus aureus, and other streptococci (Faden et al., 1990). The nasopharyngeal flora is established in the first months of life (Faden et al., 1997) and it has a large turnover of colonizing species and serotypes that occupy the niche during the early years, but a balance is usually reached. In this community bacteria need to communicate to each other and thus Quorum sensing systems play a key role in this environment. In particular, the LuxS/AI-2 QS is the only QS shared between Gram-positive and Gram-negative bacteria. Apart from its role in quorum sensing, the enzyme LuxS is tightly coupled to the S-adenosylmethionine (SAM) utilization pathway. The metabolic enzyme LuxS synthesizes AI-2 as a by-product of the conversion of S-ribosyl-homocysteine to homocysteine, an integral reaction of the activated methyl cycle (AMC). Thus, the LuxS enzyme has dual roles, firstly as a critical enzyme in the AMC and secondly as an enzyme that produces the QS molecule AI-2. Homocysteine can be generated from S-adenosyl homocysteine (SAH) through two independent pathways: A) Most Gram-positive and Gram-negative bacteria use the enzyme Pfs to first produce S-ribosylhomocysteine (SRH), then LuxS catalyzes the conversion to homocysteine, simultaneously generating 4,5-dihydroxy-2,3-pentanedione (DPD), which is spontaneously converted to AI-2. B) Alternatively, bacteria such as P. aeruginosa use the enzyme SAH hydrolase (SahH) to generate homocysteine directly without generating DPD. Despite intensive research on luxS and AI-2 in the past few years, a clear separation of the possible metabolic role of AI-2 from its signaling activity, or by AMC metabolic pathway disruption, could not be achieved. In our previous paper we have shown chemical complementation of a D39ΔluxS mutant with exogenous AI-2 and identified the AI-2 receptor (Trappetti et al., 2017). Thus, unequivocally proving the role of AI-2 as a QS system in S. pneumoniae D39 strain.
In this current study, we investigated the role of luxS in the clinical isolates, 60B and 60CSF (Agnew et al., 2022). We quantified the spnIII alleles of the bacteria recovered from the nasopharynx of mice intranasally challenged with either 60B or 60CSF strains. Interestingly, bacteria recovered from mice infected with 60B had a higher proportion of the spnIIIB allele, corresponding to a low expression level of luxS, previously observed in the SpnD39IIIB locked strain (Manso et al., 2014). In contrast bacteria isolated from mice infected with 60CSF, had higher proportions of the spnIIIA and spnIIIB alleles. Importantly, when the luxS gene was deleted from 60B and 60CSF, the mutants had significantly reduced proportions of spnIIIA and spnIIIB and now had predominantly spnIIIC. These results indicate that the presence of LuxS is essential for the spnIII locus to switch to the spnIIIA and spnIIIB alleles, with the complete absence resulting in a switch to spnIIIC and spnIIID. The exact nature of this link between LuxS and the spnIII locus will require further characterization, to establish the mechanism by which LuxS influences the switching of alleles. In addition to differences in spnIII allele proportions, 60BΔluxS showed reduced ability to grow in media with only galactose. The opposite was true for 60CSF where deletion of the luxS gene improved the growth. Therefore, distinct mechanisms seem to be operating in these strains. In the case of 60B, mutation in the luxS gene decreases its ability to grow (as previously observed for the virulent strain D39 and for the ear isolate 947), while in 60CSF, deletion of luxS increased the ability of the strain to grow in galactose media. This suggested a strain-specific effect of luxS in these clinical isolates. When the ability of the strains to form a biofilm was tested, we found that both strains could form biofilm at comparable rates, but again luxS mutation was deleterious for the 60B strain, whilst improving the biofilm formation capacity of the 60CSF strain. Addition of AI-2 improved the biofilm capacity in both WT strains, while interestingly chemical complementation of the luxS mutants with exogenous AI-2 could not be achieved. Thus, LuxS seems to have more of a metabolic role rather than a role in QS in these clinical isolates, dissimilar to the findings in D39 (Trappetti et al., 2017). In a murine infection model the 60BΔluxS strain displayed an enhanced transit to the ear compared to the WT, and increased transit to the lungs for the 60CSFΔluxS strain (Figure 8). An increased transit of the 60BΔluxS strain to the ear together with the reduced capacity of the strain to form a biofilm, suggests that functional luxS may act to facilitate adherence to the nasopharynx, but may interfere with transit to other host niches, like the ear compartment. However, in the 60CSF strain, deletion of luxS increased its ability to form a biofilm and also increased the ability of the strain to persist in the lung.
These results suggested an opposite role for luxS in the infection profiles of 60B in comparison to 60CSF, with a potential role in metabolism. We then analyzed the metabolic profiles of the luxS mutants and importantly, both luxS mutants displayed the same metabolic profile, having lost the ability to metabolise psicose and having acquired the ability to metabolise fucose in comparison to the wildtype. Most importantly, strains recovered from the nasopharynx of mice intranasally infected with 60B or 60CSF displayed the same changes in metabolic profile as the luxS mutants, indicating that luxS may be downregulated in these strains during nasopharyngeal colonization. Our earlier analysis of the spnIII allele variants in 60B and 60CSF isolated from murine nasal tissue indicated both had high proportions of spnIIIB, which is associated with increased colonization ability and downregulation of luxS. Therefore, during colonization of the nasopharynx 60B and 60CSF undergo changes to increase their proportion of spnIIIB allele to aid their colonization, likely resulting in a downregulation of luxS, thereby altering their ability to metabolise fucose and psicose in a similar way as was observed for the luxS mutants. Furthermore, other factors may be contributing to the observed different phenotypes that have yet to be identified. It is important that previous and future studies on pneumococcal luxS and its involvement in virulence be interpreted in the context of the potential switching between epigenetically-different subpopulations during the course of any experimental infection study.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by University of Adelaide Animal Ethics Committee.
Conceptualization, HA, EB, JP, and CT; methodology, HA, JA, AF, SW, and CT; investigation, HA, JA, AF, MvdL, and CT; synthesis of AI-2/DPD, ES and AA; data curation, HA and CT; writing—original draft preparation, HA, EB, JP, and CT. All authors contributed to the article and approved the submitted version.
This work was supported by the National Health and Medical Research Council (NHMRC) Investigator Grant 1174876 to JP, by the Australian Research Council (ARC) Discovery Project DP190102980 to CT and JP, and by the Channel 7 Children's Research Foundation Grant 20656402 to EB.
We would like to acknowledge Peter S Zilm for access and assistance with the xCELLigence (Agilent Technologies, USA) biofilm formation system.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1177857/full#supplementary-material
Agnew, H. N., Brazel, E. B., Tikhomirova, A., van der Linden, M., Mclean, K. T., Paton, J. C., et al. (2022). Streptococcus pneumoniae strains isolated from a single pediatric patient display distinct phenotypes. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.866259
Amin, Z., Harvey, R. M., Wang, H., Hughes, C. E., Paton, A. W., Paton, J. C., et al. (2015). Isolation site influences virulence phenotype of serotype 14 streptococcus pneumoniae strains belonging to multilocus sequence type 15. Infection Immun. 83, 4781–4790. doi: 10.1128/IAI.01081-15
Atack, J. M., Srikhanta, Y. N., Fox, K. L., Jurcisek, J. A., Brockman, K. L., Clark, T. A., et al. (2015). A biphasic epigenetic switch controls immunoevasion, virulence and niche adaptation in non-typeable haemophilus influenzae. Nat. Commun. 6, 7828. doi: 10.1038/ncomms8828
Atack, J. M., Weinert, L. A., Tucker, A. W., Husna, A. U., Wileman, T. M., Hadjirin, N. F., et al. (2018). Streptococcus suis contains multiple phase-variable methyltransferases that show a discrete lineage distribution. Nucleic Acids Res. 46, 11466–11476. doi: 10.1093/nar/gky913.
Blakeway, L. V., Power, P. M., Jen, F. E., Worboys, S. R., Boitano, M., Clark, T. A., et al. (2014). ModM DNA methyltransferase methylome analysis reveals a potential role for moraxella catarrhalis phasevarions in otitis media. FASEB J. 28, 5197–5207. doi: 10.1096/fj.14-256578
Costerton, J. W., Stewart, P. S., Greenberg, E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
De Kievit, T. R., Iglewski, B. H. (2000). Bacterial quorum sensing in pathogenic relationships. Infection Immun. 68, 4839–4849. doi: 10.1128/IAI.68.9.4839-4849.2000
Faden, H., Duffy, L., Wasielewski, R., Wolf, J., Krystofik, D., Tung, Y. (1997). Relationship between nasopharyngeal colonization and the development of otitis media in children. J. Infect. Dis. 175, 1440–1445. doi: 10.1086/516477
Faden, H., Stanievich, J., Brodsky, L., Bernstein, J., Ogra, P. L. (1990). Changes in nasopharyngeal flora during otitis media of childhood. Pediatr. Infect. Dis. J. 9, 623–626.
Fox, K. L., Dowideit, S. J., Erwin, A. L., Srikhanta, Y. N., Smith, A. L., Jennings, M. P. (2007). Haemophilus influenzae phasevarions have evolved from type III DNA restriction systems into epigenetic regulators of gene expression. Nucleic Acids Res. 35, 5242–5252. doi: 10.1093/nar/gkm571
Higgins, M. A., Suits, M. D., Marsters, C., Boraston, A. B. (2014). Structural and functional analysis of fucose-processing enzymes from streptococcus pneumoniae. J. Mol. Biol. 426, 1469–1482. doi: 10.1016/j.jmb.2013.12.006
Iannelli, F., Pozzi, G. (2004). Method for introducing specific and unmarked mutations into the chromosome of streptococcus pneumoniae. Mol. Biotechnol. 26, 81–86. doi: 10.1385/MB:26:1:81
Kolenbrander, P. E. (2000). Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54, 413–437. doi: 10.1146/annurev.micro.54.1.413
Lavelle, E. C., Ward, R. W. (2022). Mucosal vaccines - fortifying the frontiers. Nat. Rev. Immunol. 22, 236–250. doi: 10.1038/s41577-021-00583-2
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods (San Diego Calif.) 25, 402–408. doi: 10.1006/meth.2001.1262
Manso, A. S., Chai, M. H., Atack, J. M., Furi, L., De Ste Croix, M., Haigh, R., et al. (2014). A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat. Commun. 5, 5055–5055. doi: 10.1038/ncomms6055
Matsuo, T., Suzuki, H., Hashiguchi, M., Izumori, K. (2002). D-psicose is a rare sugar that provides no energy to growing rats. J. Nutr. Sci. Vitaminol (Tokyo) 48, 77–80. doi: 10.3177/jnsv.48.77
Mclean, K. T., Tikhomirova, A., Brazel, E. B., Legendre, S., Haasbroek, G., Minhas, V., et al. (2020). Site-specific mutations of GalR affect galactose metabolism in streptococcus pneumoniae. J. Bacteriology 203, e00180-20. doi: 10.1128/JB.00180-20
Miller, M. B., Bassler, B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199. doi: 10.1146/annurev.micro.55.1.165
Minhas, V., Harvey, R. M., Mcallister, L. J., Seemann, T., Syme, A. E., Baines, S. L., et al. (2019). Capacity to utilize raffinose dictates pneumococcal disease phenotype. mBio 10, e02596–e02518. doi: 10.1128/mBio.02596-18
Nahar, N., Tram, G., Jen, F. E.-C., Phillips, Z. N., Weinert, L. A., Bossé, J. T., et al. (2023). Actinobacillus pleuropneumoniae encodes multiple phase-variable DNA methyltransferases that control distinct phasevarions. Nucleic Acids Res 51 (7), 3240–3260. doi: 10.1093/nar/gkad091
Paixão, L., Caldas, J., Kloosterman, T. G., Kuipers, O. P., Vinga, S., Neves, A. R. (2015). Transcriptional and metabolic effects of glucose on streptococcus pneumoniae sugar metabolism. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.01041
Phillips, Z. N., Husna, A. U., Jennings, M. P., Seib, K. L., Atack, J. M. (2019). Phasevarions of bacterial pathogens - phase-variable epigenetic regulators evolving from restriction-modification systems. Microbiol. (Reading) 165, 917–928. doi: 10.1099/mic.0.000805
Seib, K. L., Jen, F. E., Tan, A., Scott, A. L., Kumar, R., Power, P. M., et al. (2015). Specificity of the ModA11, ModA12 and ModD1 epigenetic regulator N(6)-adenine DNA methyltransferases of neisseria meningitidis. Nucleic Acids Res. 43, 4150–4162. doi: 10.1093/nar/gkv219
Seib, K. L., Peak, I. R., Jennings, M. P. (2002). Phase variable restriction-modification systems in moraxella catarrhalis. FEMS Immunol. Med. Microbiol. 32, 159–165. doi: 10.1111/j.1574-695X.2002.tb00548.x
Seib, K. L., Pigozzi, E., Muzzi, A., Gawthorne, J. A., Delany, I., Jennings, M. P., et al. (2011). A novel epigenetic regulator associated with the hypervirulent neisseria meningitidis clonal complex 41/44. FASEB J. 25, 3622–3633. doi: 10.1096/fj.11-183590
Seib, K. L., Srikhanta, Y. N., Atack, J. M., Jennings, M. P. (2020). Epigenetic regulation of virulence and immunoevasion by phase-variable restriction-modification systems in bacterial pathogens. Annu. Rev. Microbiol. 74, 655–671. doi: 10.1146/annurev-micro-090817-062346
Srikhanta, Y. N., Dowideit, S. J., Edwards, J. L., Falsetta, M. L., Wu, H. J., Harrison, O. B., et al. (2009). Phasevarions mediate random switching of gene expression in pathogenic neisseria. PloS Pathog. 5, e1000400. doi: 10.1371/journal.ppat.1000400
Srikhanta, Y. N., Gorrell, R. J., Steen, J. A., Gawthorne, J. A., Kwok, T., Grimmond, S. M., et al. (2011). Phasevarion mediated epigenetic gene regulation in helicobacter pylori. PloS One 6, e27569. doi: 10.1371/journal.pone.0027569
Srikhanta, Y. N., Maguire, T. L., Stacey, K. J., Grimmond, S. M., Jennings, M. P. (2005). The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes. Proc. Natl. Acad. Sci. U.S.A. 102, 5547–5551. doi: 10.1073/pnas.0501169102
Stroeher, U. H., Paton, A. W., Ogunniyi, A. D., Paton, J. C. (2003). Mutation of luxS of streptococcus pneumoniae affects virulence in a mouse model. Infect. Immun. 71, 3206–3212. doi: 10.1128/iai.71.6.3206-3212.2003
Tan, A., Hill, D. M. C., Harrison, O. B., Srikhanta, Y. N., Jennings, M. P., Maiden, M. C. J., et al. (2016). Distribution of the type III DNA methyltransferases modA, modB and modD among neisseria meningitidis genotypes: implications for gene regulation and virulence. Sci. Rep. 6, 21015. doi: 10.1038/srep21015
Tikhomirova, A., Brazel, E. B., Mclean, K. T., Agnew, H. N., Paton, J. C., Trappetti, C. (2022). The role of luxS in the middle ear streptococcus pneumoniae isolate 947. Pathog. (Basel) 11, 216. doi: 10.3390/pathogens11020216
Trappetti, C., Mcallister, L. J., Chen, A., Wang, H., Paton, A. W., Oggioni, M. R., et al. (2017). Autoinducer 2 signaling via the phosphotransferase FruA drives galactose utilization by streptococcus pneumoniae, resulting in hypervirulence. mBio 8, e02269–e02216. doi: 10.1128/mBio.02269-16
Trappetti, C., Ogunniyi, A. D., Oggioni, M. R., Paton, J. C. (2011a). Extracellular matrix formation enhances the ability of streptococcus pneumoniae to cause invasive disease. PloS One 6, e19844–e19844. doi: 10.1371/journal.pone.0019844
Trappetti, C., Potter, A. J., Paton, A. W., Oggioni, M. R., Paton, J. C. (2011b). LuxS mediates iron-dependent biofilm formation, competence, and fratricide in streptococcus pneumoniae. Infection Immun. 79, 4550–4558. doi: 10.1128/IAI.05644-11
Trappetti, C., van der Maten, E., Amin, Z., Potter, A. J., Chen, A. Y., Van Mourik, P. M., et al. (2013). Site of isolation determines biofilm formation and virulence phenotypes of streptococcus pneumoniae serotype 3 clinical isolates. Infection Immun. 81, 505–513. doi: 10.1128/IAI.01033-12
Trimble, A., Connor, V., Robinson, R. E., Mclenaghan, D., Hancock, C. A., Wang, D., et al. (2020). Pneumococcal colonisation is an asymptomatic event in healthy adults using an experimental human colonisation model. PloS One 15. doi: 10.1371/journal.pone.0229558
Vendeville, A., Winzer, K., Heurlier, K., Tang, C. M., Hardie, K. R. (2005). Making ‘sense’ of metabolism: autoinducer-2, LUXS and pathogenic bacteria. Nat. Rev. Microbiol. 3, 383–396. doi: 10.1038/nrmicro1146
Vidal, J. E., Ludewick, H. P., Kunkel, R. M., Zähner, D., Klugman, K. P. (2011). The LuxS-dependent quorum-sensing system regulates early biofilm formation by streptococcus pneumoniae strain D39. Infect. Immun. 79, 4050–4060. doi: 10.1128/iai.05186-11
Keywords: Streptococcus pneumoniae (pneumococcus), virulence, clinical isolates bacterial, Quorum sensing, biofilm
Citation: Agnew HN, Atack JM, Fernando ARD, Waters SN, van der Linden M, Smith E, Abell AD, Brazel EB, Paton JC and Trappetti C (2023) Uncovering the link between the SpnIII restriction modification system and LuxS in Streptococcus pneumoniae meningitis isolates. Front. Cell. Infect. Microbiol. 13:1177857. doi: 10.3389/fcimb.2023.1177857
Received: 02 March 2023; Accepted: 17 April 2023;
Published: 01 May 2023.
Edited by:
Michael Otto, National Institutes of Health (NIH), United StatesReviewed by:
M. Ammar Zafar, Wake Forest University, United StatesCopyright © 2023 Agnew, Atack, Fernando, Waters, van der Linden, Smith, Abell, Brazel, Paton and Trappetti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Claudia Trappetti, Y2xhdWRpYS50cmFwcGV0dGlAYWRlbGFpZGUuZWR1LmF1; James C. Paton, amFtZXMucGF0b25AYWRlbGFpZGUuZWR1LmF1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.