
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 30 May 2023
Sec. Molecular Bacterial Pathogenesis
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1175446
This article is part of the Research Topic Pathogenic mechanism and biocontrol of Xanthomonas on plants View all 6 articles
 Qi Zhou1†
Qi Zhou1† Min Tu2†
Min Tu2† Xue Fu2,3
Xue Fu2,3 Ying Chen1
Ying Chen1 Muyuan Wang1
Muyuan Wang1 Yuan Fang1
Yuan Fang1 Yichao Yan1
Yichao Yan1 Guanyun Cheng1
Guanyun Cheng1 Yikun Zhang1
Yikun Zhang1 Zhongfeng Zhu1
Zhongfeng Zhu1 Ke Yin1
Ke Yin1 Youlun Xiao4*
Youlun Xiao4* Lifang Zou1,5*
Lifang Zou1,5* Gongyou Chen1,5
Gongyou Chen1,5Bacterial leaf streak (BLS) of rice is a severe disease caused by the bacterial pathogen Xanthomonas oryzae pv. oryzicola (Xoc) that has gradually become the fourth major disease on rice in some rice-growing regions in southern China. Previously, we isolated a Bacillus velezensis strain 504 that exhibited apparent antagonistic activity against the Xoc wild-type strain RS105, and found that B. velezensis 504 was a potential biocontrol agent for BLS. However, the underlying mechanisms of antagonism and biocontrol are not completely understood. Here we mine the genomic data of B. velezensis 504, and the comparative transcriptomic data of Xoc RS105 treated by the cell-free supernatants (CFSs) of B. velezensis 504 to define differentially expressed genes (DEGs). We show that B. velezensis 504 shares over 89% conserved genes with FZB42 and SQR9, two representative model strains of B. velezensis, but 504 is more closely related to FZB42 than SQR9, as well as B. velezensis 504 possesses the secondary metabolite gene clusters encoding the essential anti-Xoc agents difficidin and bacilysin. We conclude that approximately 77% of Xoc RS105 coding sequences are differentially expressed by the CFSs of B. velezensis 504, which significantly downregulates genes involved in signal transduction, oxidative phosphorylation, transmembrane transport, cell motility, cell division, DNA translation, and five physiological metabolisms, as well as depresses an additional set of virulence-associated genes encoding the type III secretion, type II secretion system, type VI secretion system, type IV pilus, lipopolysaccharides and exopolysaccharides. We also show that B. velezensis 504 is a potential biocontrol agent for bacterial blight of rice exhibiting relative control efficiencies over 70% on two susceptible cultivars, and can efficiently antagonize against some important plant pathogenic fungi including Colletotrichum siamense and C. australisinense that are thought to be the two dominant pathogenic species causing leaf anthracnose of rubber tree in Hainan province of China. B. velezensis 504 also harbors some characteristics of plant growth-promoting rhizobacterium such as secreting protease and siderophore, and stimulating plant growth. This study reveals the potential biocontrol mechanisms of B. velezensis against BLS, and also suggests that B. velezensis 504 is a versatile plant probiotic bacterium.
Rice is an important worldwide staple crop, however, stable rice production is threatened by various diseases, including fungal and bacterial diseases. Bacterial blight (BB) of rice caused by Xanthomonas oryzae pv. oryzae (Xoo), and bacterial leaf streak (BLS) of rice caused by X. oryzae pv. oryzicola (Xoc), are important bacterial diseases in rice growing areas of Asia (Nino-Liu et al., 2006). These two diseases often result in rice yield reduction by 10-30% especially in the south of China (Li et al., 2019). Currently, the main control measures for these two bacterial diseases include the development of resistant varieties of rice and the application of chemical bactericides (Xu et al., 2015). The interaction between Xoc and rice does not conform to the gene-for-gene hypothesis (Li et al., 2019). Some rice varieties containing xa5, Xa23, Xa21 and other resistance genes showed better resistance to Xoo, but exhibited susceptibility to Xoc (Liu et al., 2021b). Many hybrid rice varieties in China are susceptible to Xoc, and some of which were highly susceptible (Li et al., 2019). At present, the control of BLS mainly depends on chemical agents, such as zinc thiazole or cupric bactericides (Yang et al., 2021). The use of these bactericides is not only easy to cause phytotoxicity and make pathogens to develop resistance, but also damages the ecological environment. Therefore, biological control measures have been strongly promoted, and the study of screening antagonistic microorganisms from agricultural ecological environment to control BB and BLS has become an important research direction (Yang et al., 2023).
Pseudomonas spp. and Bacillus spp. are used commercially as biocontrol agents against plant diseases in agricultural production. Some studies have shown that some species of Pseudomonads such as P. entomophila, P. putida group members P. soli and P. mosselii, and P. oryziphila, a novel species recently identified in our previous study, exhibited antagonistic activity against Xoo and Xoc (Li et al., 2013; Pascual et al., 2014; Yang et al., 2021). Bacillus spp. is the most commercially used biocontrol bacteria because of its good stress resistance, producing many kinds of secondary metabolites, and having the functions of disease prevention, growth promotion and environmental restoration (Baard et al., 2023; Poulaki and Tjamos, 2023). At present, B. amyloliquefaciens and B. velezensis are successful cases of Bacillus spp. used for biological control of BB and BLS. B. amyloliquefaciens Lx-11 is the first commercial biopesticide created by B. amyloliquefaciens in China (Zhang et al., 2012). It was registered for the control of BB and BLS, the control efficacy ranging from 60.2% to 70.6% (Zhang et al., 2012). B. velezensis FZB42 is a model strain of plant growth-promoting rhizobacterium (PGPR) for stimulation of induced systemic resistance, and suppression of plant pathogens such as fungi, bacteria (Xoo and Xoc), oomycetes, and nematodes (Wu et al., 2015; Fan et al., 2018; Fan et al., 2019; Han et al., 2021; Mácha et al., 2021). Previously, we identified 16 species of Bacillus including B. cereus, B. altitudinis and B. velezensis with antagonistic effects against Xoc in a biocontrol bacterial pool of 233 strains that were isolated from the 248 rhizosphere soil samples collected from 23 provinces in China (Li et al., 2019; Li et al., 2020a; Huang et al., 2021). This suggests that Bacillus is a resource reservoir to be excavated for the BLS control.
The present taxon of B. velezensis is an integrated taxon that includes all the strains previously classified as B. velezensis, B. methylotrophicus and B. amyloliquefaciens subsp. plantarum (Dunlap et al., 2015; Dunlap et al., 2016). It is predicted that about 10% of the total genome sequence of the model strain FZB42 encodes at least 13 gene clusters that devoted to synthesizing active secondary metabolites, which play effective roles in the controls of fungal, bacterial, viral and nematode diseases (Rabbee et al., 2019). These bioactive secondary metabolites have been identified in four types of cyclic lipopeptides (such as surfactin, bacillomycin-D, fengycin, and bacillibactin, an iron-siderophore), three polyketones (such as macrolactin, bacillaene and difficidin), one dipeptide antibiotic (bacilysin), two bacteriocins (plantazolicin and amylocyclicin), and volatile substances (acetoin and 2, 3-butandiol) (Chowdhury et al., 2015; Fan et al., 2018; Rabbee et al., 2019). Difficidin and bacilysin are major antibacterial agents produced by FZB42 to exert biocontrol activity of the BB and BLS caused by Xoo and Xoc, respectively (Wu et al., 2015). However, the mechanisms by which the active compounds produced by B. velezensis strains affect Xoc pathogenicity are little understood. In recent years, with the adoption of different antagonistic target pathogens, new isolates of B. velezensis, such as J17-4, NKMV-3 and HNA3, have been continuously isolated and identified (Shi et al., 2022; Vignesh et al., 2022; Zaid et al., 2022). Some new biocontrol potentials of B. velezensis strains are being explored.
Our laboratory has been working to develop biological control methods to control BLS. Previously, we isolated and identified a B. velezensis strain 504 that exhibited apparently antagonistic activity against the Xoc wild-type strain RS105 (Li et al., 2019). In this study, we completed the whole genome sequencing and conducted the comparative genomic analysis of B. velezensis 504 with its closely related B. velezensis strains. In addition, we compared the transcriptome profiling of Xoc RS105 treated by the cell-free supernatants (CFSs) of B. velezensis 504 and revealed the potential mechanisms of action of B. velezensis 504 on Xoc. We also analyzed the antagonistic spectrum of B. velezensis 504 against phytopathogenic fungi such as the pathogens of rice blast, gray mold of vegetables and leaf anthracnose of rubber tree. These studies suggest that B. velezensis 504 is a versatile plant probiotic bacterium.
B. velezensis 504 was isolated from the rhizosphere soil samples of spinach in Xiyang Village, Sanming City, Fujian province, China, cultured in Luria-Bertani (LB) medium at 28°C.
All Xanthomonas strains were cultured in nutrient agar (NA) or nutrient broth (NB) medium at 28°C. All fungal strains were cultured in Potato Dextrose Agar (PDA) medium at 28°C (Yang et al., 2023). Rice seeds Yuanfengzao, were provided by Dr. Youlun Xiao from Institute of Plant Protection, Hunan Academy of Agricultural Sciences. Rice seeds IR24 and Nipponbare used in this study were from our laboratory.
The Kirschbaum-Bauer (KB) method was used to evaluate the antagonism of B. velezensis strain 504 against Xoc RS105, Xoo PXO99A, and phytopathogenic fungi, including Fusarium graminearum causing fusarium head blight, Botrytis cinerea causing gray mold disease of vegetables, Magnaporthe oryzae causing rice blast, F. oxysporum causing root rot disease of tomato, Colletotrichum siamense and C. australisinense causing leaf anthracnose of rubber tree (Liu et al., 2018; Liu et al., 2023). All isolates were tested in triplicate, and inhibition zones against Xanthomonas were measured 2-3 days after co-cultivation at 28°C on nutrient agar (NA), and inhibition zones against fungi were measured 5-7 days after co-cultivation at 25°C. The antifungal activity of B. velezensis 504 was further studied by the following formula:
The biological functional studies of B. velezensis 504 were performed as previously reported studies (Tang et al., 2023). We detected the siderophore production from B. velezensis 504 by the assay of chrome azurol (CAS) agar plate (Milagres et al., 1999). To test the protease activity of B. velezensis 504, 1 μl bacterial solution was added to protease assay plates and incubated at 28°C for 3 days. Then, the plates were observed for the formation of transparent circles due to the decomposition of substrates. Drop 1 μl of cultured solution of B. velezensis 504 on the cellulase activity-tested medium, inorganic phosphorus culture plates, and inorganic potassium culture plates and cultured at 28°C for 4, 7, 7 days, respectively. Each inoculation of plates was conducted in three replications.
The whole genome of B. velezensis 504 was sequenced using the Pacific Biosciences platform and the Illumina Miseq platform at Personalbio (Shanghai, China). The complete genome sequences of B. velezensis 504 were deposited in GenBank under accession number, CP092439 for chromosome, and CP092440 for plasmid. After obtaining the sequence, we used various databases to add annotations of function genes. CAZy (Carbohydrate-Active enzymes) (Hmmscan, v3.1b2, February 2015) was used to predict the presence of carbohydrate active enzyme genes in the genome sequence; COG (cluster of orthologous group) (eggNOG-mapper, v4.5, November 2017) and GO (Gene Ontology) (Interpro, v66.0, November 2017) were used to annotate protein-coding genes in the whole genome; KEGG (Kyoto Encyclopedia of Genes and Genomes) (KAAS, v2.1) was used to annotate its metabolic pathway. The average nucleotide identity (ANI) values among 12 genome sequences B. velezensis 504 and other Bacillus strains, including B. velezensis FZB42, B. velezensis SQR9, B. siamensis KCTC 13613, B. amyloliquefaciens DSM 7, B. subtilis subsp. subtilis str. 168, B. halotolerans ZB201702, B. subtilis subsp. spizizenii W23, B. atrophaeus subsp. globigii BSS, B. licheniformis SCDB 14, B. mojavensis UCMB5075, and B. vallismortis Bac111 were calculated using the J Species WS Online Service (Richter et al., 2016). We obtained and compared general characteristic information for the three strains in NCBI (http://www.ncbi.nlm.nih.gov/).
Pan-genomic analysis of the B. velezensis genomes was conducted using the BPGA v1.3.0 tool (Chaudhari et al., 2016). For the extraction of the core-genome the default values of the tool were employed, i.e., USEARCH software v10.0.240 with a threshold of sequence identity equal to 0.5 or 50% (Yoon et al., 2017). The MUSCLE v3.8.31 software was used to align the concatenated amino acids of the core-genome. The pan-genomic analysis was repeated in the B. velezensis genomes, including the genomes of B. velezensis 504, B. velezensis FZB42 and B. velezensis SQR9. We also used KEGG and COG to annotate these unique, accessory and core genes. Finally, we drew a Venn diagram through TBtools software.
Collinearity of the conserved and highly orthologous genomic regions were determined and plotted among B. velezensis 504, B. velezensis FZB42 and B. velezensis SQR9 by using Mauve software v2.4.0 with default parameters (Darling et al., 2004). The colored, locally collinear blocks (LCBs) show the conserved and highly similar genomic regions. Each continuously shaded area is an LCB. The colored lines between the two genomes trace the LCBs of each homologous. We selected rhizosphere colonization-related genes among B. velezensis 504, B. velezensis FZB42 and B. velezensis SQR9 for genetic similarity comparison by using NovoPro (https://www.novopro.cn/tools/ident_sim.html).
The genomes of B. velezensis 504, B. velezensis FZB42 and B. velezensis SQR9 were analyzed by antiSMASH 5.0 with website (https://antismash.secondary-metabolites.org) to predict the putative secondary metabolite biosynthesis gene clusters (Blin et al., 2019). Detailed gene cluster information was obtained from the GenBank databases.
Biocontrol assays in rice fields were performed as previous research (Yang et al., 2023). Ten leaves from the highly susceptible rice cultivars Yuanfengzao, IR24 and Nipponbare rice at booting stage were inoculated with Xoo PXO99A (OD600 = 0.3) by spray inoculation. B. velezensis 504 cell-culture broth (CCB) preventive treatment (504-Pre) and treatment (504-Tre) indicated that spraying rice leaves with B. velezensis 504 CCB (OD600 = 1.0) 12 hours prior to and after inoculation with PXO99A, respectively. Sterile water-treated rice cultivars were used as control (CK). BB disease of rice severity under all groups was investigated after 15 days. Relative spot area was calculated by the formula: Spot area= (spot area of this leave/total area of this leave) × 100%. The inhibitory percentages (IPs) were calculated by the formula: IP = (1-spot area of treatment/spot area of control) × 100%. Three independent experiments and three technical replicates were performed. Statistical analysis of results was determined by ANOVA test with significant difference (p< 0.05), and graphs were obtained with GraphPad Prism 9 version.
To explore whether B. velezensis 504 present the ability to promoter plant growth of pak choi (B. campestris sp. chinensis L.), we measured growth parameters including dry weight of their aboveground parts. B. velezensis 504 suspension was diluted 10 and 50 times with sterile water and then irrigated to the soil prior to pakchoi sowing. Seeding with conventional method was carried out as control (CK). We randomly selected 30 plants in each treatment, and measured the dry weight of mature pakchoi cabbages after picking. Statistical analysis of results was determined by ANOVA test with significant difference (p< 0.05), and graphs were obtained with GraphPad Prism 9 version.
Cell-free supernatants (CFSs) were used to evaluate influence of B. velezensis 504 extracellular metabolites on the growth of Xoc RS105 as previous research with modifications (Jeong et al., 2017). B. velezensis 504 was shaking incubated in 250 ml NB medium at 28°C for overnight (adjusted OD600 to 2.0), centrifuged to remove bacterial cells and collected 200 ml supernatant as CFSs by 0.22um filtration. After freeze-drying, the powder of B. velezensis 504 CFSs were dissolved with sterile water (10 ml) for antagonistic activity determination. In vitro studies, 30, 40, 50, and 60 µL B. velezensis 504 CFSs (20 times concentration) was added with 2 ml RS105 inoculum (adjusted OD600 to 1.0) and incubated shaking at 28°C for overnight to determine the effective bactericidal concentration of B. velezensis 504 (Supplementary Figure 4B). The negative control consisted of sterile water and 2 ml RS105. For RNA-Seq analysis, Xoc RS105 that were centrifuged and transferred to liquid XOM3 medium (Yang et al., 2021), which is a nutrient-poor artificial medium mimicking the infection scenarios in the host rice, and can artificially induce hrp gene expression, (adjusted OD600 to 1.0) performed as negative controls. Twenty-fold concentrated B. velezensis 504 CFSs (50 µL) were added to Xoc RS105 that was re-suspended in XOM3 medium (adjusted OD600 to 1.0) and further incubated at 28°C shaking at 180 rpm for 6 h, 12 h, 24 h. Total RNAs of RS105 samples with three replicates from treatments (5046h1, 5046h2, 5046h3, 50412h1, 50412h2, 50412h3, 50424h1, 50424h2, and 50424h3) and controls (CK6h1, CK6h2, CK6h3, CK12h1, CK12h2, CK12h3, CK24h1, CK24h2, and CK24h3) grown on the tubes described at three time points were extracted for Illumina HiSeq sequencing. Finally, we sent all prepared samples above to Personalbio (Shanghai, China) for transcriptome sequencing analysis.
Based on the reference genome (https://www.ncbi.nlm.nih.gov/nuccore/CP011961.1), the company calculated the expression levels of each gene in each treatment group. The transcriptome samples were further analyzed for differential expression, enrichment, and clustering. Since we set up both control and treatment groups in the experiment, the comparison of gene expression levels between the two groups can reflect differential expression information. We used DESeq for differential analysis of gene expression, also used GO and KEGG to annotate function genes. Finally, we used Power Point software to draw a Venn diagram to show part of the results of the difference expression. Heatmaps were produced by TBtools software (Chen et al., 2020). Raw transcriptome data of the eighteen samples were deposited in the SRA database (Sequence Read Archive, NCBI) with the accession number PRJNA946931. Quantitative Real-time PCR (qRT-PCR) was employed with SYBR Green I Mix (TransGen, Beijing, China) and ABI 7500 software according to the manufacturer’s instructions. gyrB and rpoD were used as house-keeping genes for validation of fifteen differentially expressed genes in RS105. Specific primers were designed with Primer 3 online platform (Supplementary Table 1). The 2−ΔΔCT method was used for relative quantification of gene expression. Spearman correlation test was conducted to measure the correlation between qRT-PCR (log2-expression fold change data) and RNA-Seq (log2-fold change) results using SPSS Version 20.0 and determine the reliability of RNA-Seq.
Previously, we isolated the bacterial strain 504, which displayed significant antagonistic activity against Xoc, from soil samples taken from the spinach rhizosphere at Xiyang Village of Sanming City in the province of Fujian, China, on February 21, 2018 (Li et al., 2019). The BLAST analysis indicated that 504 belongs to B. velezensis by using the partial 16S rRNA gene sequences (Li et al., 2019). To further define the phylogenetic status of 504, we sequenced the complete genome of 504, and found that it has a circular chromosome of 3,905,813 base pairs with 46.60% G+C content, 3,853 protein coding sequences (CDSs), 27 rRNA genes, 86 tRNA genes and 217 other non-coding RNA genes (Figure 1A and Supplementary Tables 2, 3), as well as a plasmid of 8,564 base pairs with 39.86% G+C content (Figure 1B), which is different from B. velezensis FZB42 and SQR9, two representative model strains of B. velezensis (Supplementary Table 4). The chromosome and plasmid sequences have been deposited in GenBank under the accession numbers of CP092439 and CP092440, respectively. Further, we executed an average nucleotide identity (ANI) analysis between 504 with eleven sequenced species of Bacillus by ANI based on BLAST + (ANIb) and MUMmer (ANIm) applying the online tool JSpeciesWS, respectively. Estimated values of ANIb and ANIm between B. velezensis 504 and FZB42 were 97.60% [94.03] and 97.77% [94.77], while the ANIb and ANIm values between B. velezensis 504 and SQR9 were 97.56% [93.97] and 97.75% [89.60], respectively (Table 1). The results showed that 504 displayed an ANIb (or ANIm) value over 95% with B. velezensis FZB42 and SQR9. All ANI-values (ANIb and ANIm) between 504 and other individual species of Bacillus are below the threshold of 95% for species demarcation, indicating that 504 belongs to Bacillus velezensis, which is consistent with the result we obtained using the 16S rRNA gene sequence and alignment.
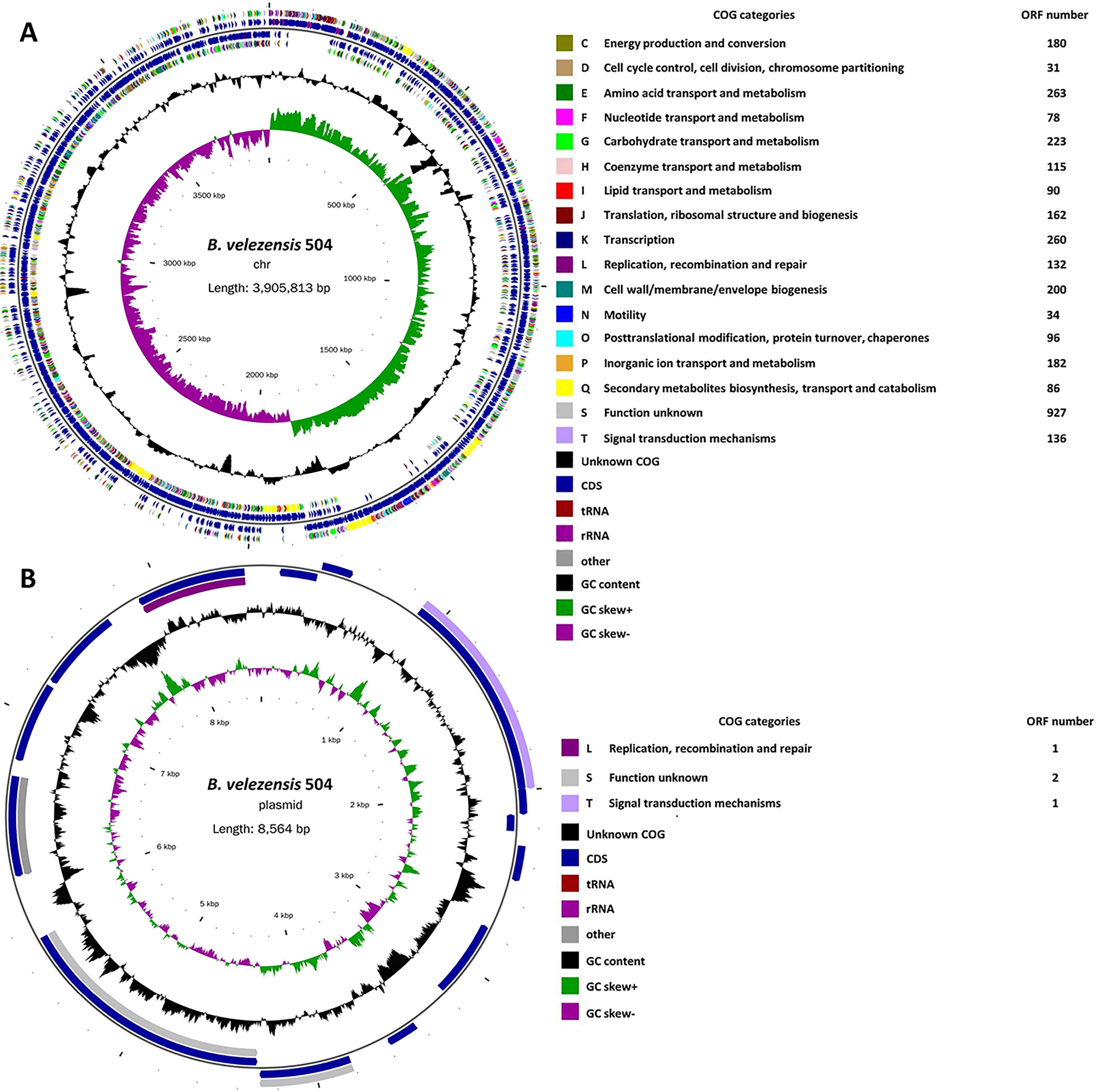
Figure 1 The complete genome map of B. velezensis 504 with its genomic features. (A) Circular map of B. velezensis 504 chromosome. The GC content of the strain 504 chromosome was 46.6% and contained 3,853 protein coding sequences (CDSs), 27 rRNA genes, 86 tRNA genes and 217 other non-coding RNA genes. (B) Circular map of B. velezensis 504 plasmid. The GC content of the 8,564 bp-plasmid was 39.86% and different from B. velezensis strains FZB42 and SQR9.
B. velezensis 504 contained complete gene clusters implicated in flagellar assembly (ko02040), bacterial motility proteins (ko02035) and chemotaxis (ko02030), including 56 genes involved in the flagellar structural components (motA-B, fliM, fliG, fliF), biosynthesis of flagella (flhA-B, flhF-G, fliP-R) and chemotactic signaling protein phospho-CheY (cheA-D, CheY-P) (Supplementary Table 5), which are required for bacterial survival by regulating the direction of flagellar rotation.
Collinearity analysis of the whole genome between B. velezensis 504, FZB42 and SQR9 showed that most genes are highly homologous with direct collinearity, but there were also gene rearrangement phenomena such as flipping and transposition (Figure 2A). Moreover, the similarity between B. velezensis 504 and FZB42 showed the higher synteny than that of SQR9, which is consistent with the comparison of the general characteristics among their genomes. Core-Pan genome analysis showed that 3306 core genes were conserved in B. velezensis 504, FZB42, and SQR9, accounting for 89.23%, 91.58%, and 87.93% of the genes in the genomes, respectively (Figure 2B). Total 280 genes unique to B. velezensis 504 don't exist in FZB42 and SQR9. 71.07% of these genes code for hypothetical proteins, 14.29% have predictive functions, 8.57% have non-predictive functions, and 5.36% code for phage portal proteins (Supplementary Table 6). 44 unique genes were predicted to encode clusters of secondary metabolites (Supplementary Table 7), of which 10 genes were predicted to encode kijanimicin (3.57%), 6 genes were predicted to encode fengycin (2.14%), and 10 genes were unknown function descriptions. (3.57%).
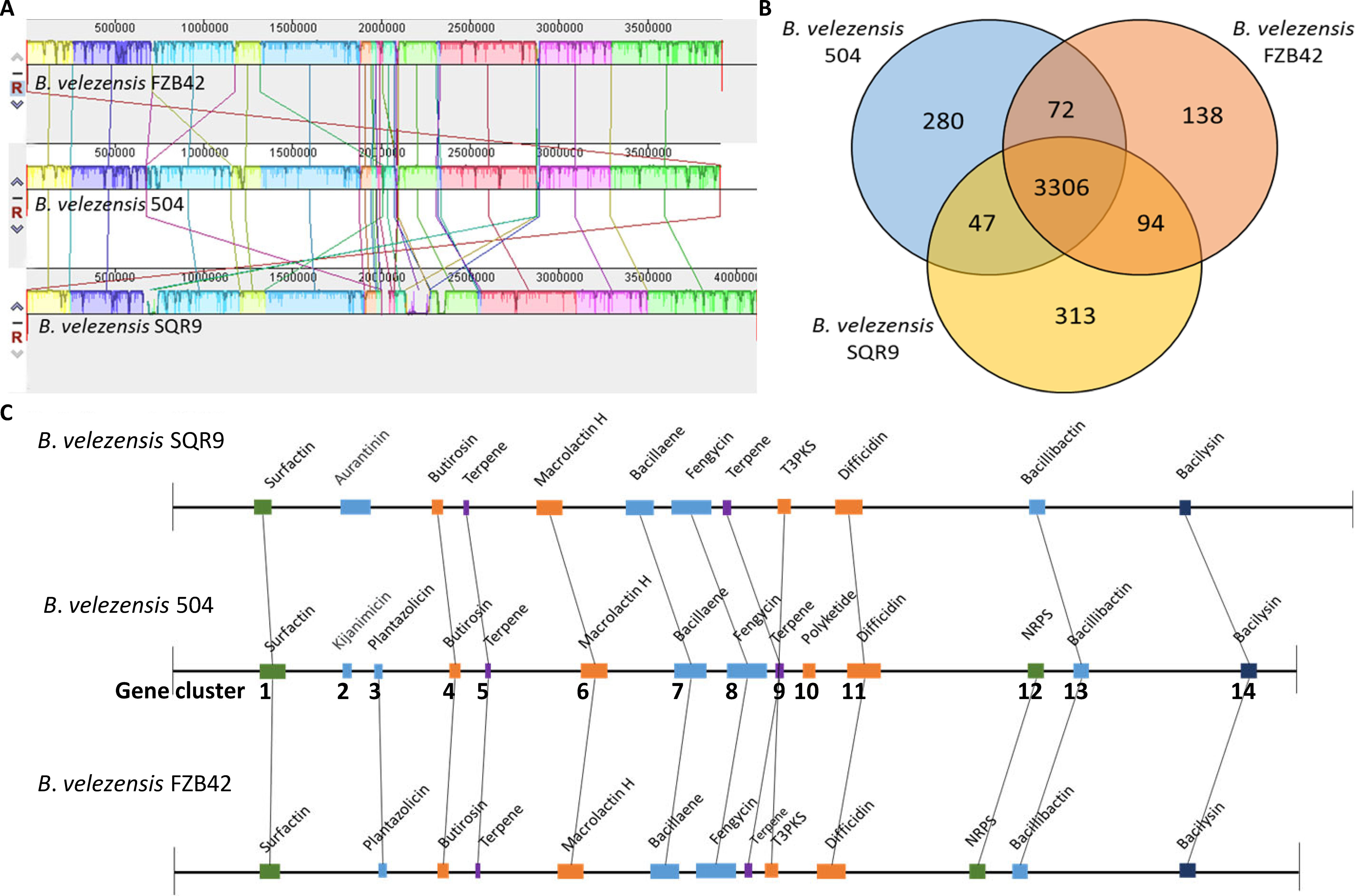
Figure 2 Comparative genomic analysis of B. velezensis 504 with other B. velezensis strains (B. velezensis FZB42 and SQR9). (A) Genome-to-genome alignment of B. velezensis 504, B. velezensis FZB42, and B. velezensis SQR9 using a progressive mauve software with B. velezensis 504 as the reference genome. Similar-colored boxes represent syntenic regions. Boxes below the horizontal line represent regions that are inverted. Rearrangements are depicted with colored lines. (B) Venn diagram illustrating the number of orthologous CDS genes that are shared and unique among three strains of B. velezensis 504, B. velezensis FZB42, and B. velezensis SQR9. (C) Gene clusters of secondary metabolite biosynthesis in B. velezensis 504, B. velezensis FZB42, and B. velezensis SQR9. The number represented the Jaccard index on the horizontal line. Connected by black lines are the colinear regions. T3PKS, type III PKS; NRPS, non-ribosomal peptide synthetase cluster.
Secondary metabolite clusters of B. velezensis 504 were compared with B. velezensis FZB42 and SQR9 using antibiotics and secondary metabolite analysis shell (anti-SMASH). The result revealed that the genome of B. velezensis 504 contains fourteen candidate gene clusters involved in nonribosomal synthesis of lipopeptides (surfactin, fengycin and bacillibactin), polyketides (difficidin, bacillaene, macrolactin H, and kijanimicin), dipeptide antibiotics (bacilysin), ribosomal synthesis of bacteriocins (plantazolicin), saccharide synthesis of butirosin, and other four unknown terpenes, polyketide and non-ribosomal peptide synthetases (Figure 2C and Supplementary Table 8). B. velezensis 504 contains several clusters that share different structures with FZB42 and SQR9, and lacks yx01, aat, ycxC, and ycxD in the Cluster 1 encoding surfactin (Supplementary Table 9). Cluster 2 involved in kijanimicin synthesis and Cluster 10 involved in polyketide synthesis exist only in B. velezensis 504, Cluster 3 and Cluster 12 encoding plantazolicin and NRPS exist in both B. velezensis 504 and FZB42, and other ten (Clusters 1, 4-9, 11, 13 and 14) conservatively present in all three strains (Figure 2C and Supplementary Table 9). Six clusters involved in macrolactin H, bacillaene, fengycin, bacillibactin, difficidin and bacilysin synthesis, share 100% homology with that of B. velezensis FZB42 (Supplementary Table 8). These bioactive molecules produced by B. velezensis FZB42 have been reported to be effective against phytopathogenic fungi and bacteria, indicating that B. velezensis 504 has the potential to control fungal and bacterial diseases (Rabbee et al., 2019). Difficidin and bacilysin have been reported to be the essential antibacterial agents produced by B. velezensis FZB42 to control BB and BLS of rice, two major rice diseases caused by Xoo and Xoc, respectively (Wu et al., 2015). Therefore, we speculated that difficidin and bacilysin are major extracellular active molecules of B. velezensis 504 to antagonize against Xoc RS105.
Our study confirmed that 50 µL 20-fold concentrated cell-free supernatants (CFSs) of B. velezensis 504 exhibited prominent antibacterial activities against Xoc RS105 (Figure 3A and Supplementary Figure 4B), suggesting that B. velezensis 504 prevented the visible growth of Xoc, most likely by synthetic metabolites that were diffusible across the 0.22 μm filter membrane. To explore the molecular mechanism underlying antibacterial effect of B. velezensis 504 on Xoc RS105, we first determined the global transcriptional response of Xoc RS105 to the CFSs of B. velezensis 504 compared to that induced by XOM3, an artificial hrp inducing medium. We isolated total RNA from Xoc RS105, at 6 hr, 12 hr, and 24 hr post induction (hpi) by the CFSs of B. velezensis 504 (20 times the concentration) dissolved in XOM3 (hereafter referred to as 504) and by XOM3 itself (hereafter called CK), respectively (Figure 3A). High-quality raw reads (Q20>98%) were mapped to the reference genome of Xoc RS105 (RefSeq assembly accession: GCF_001042875.1) in the NCBI database (Supplementary Tables 10, 11).
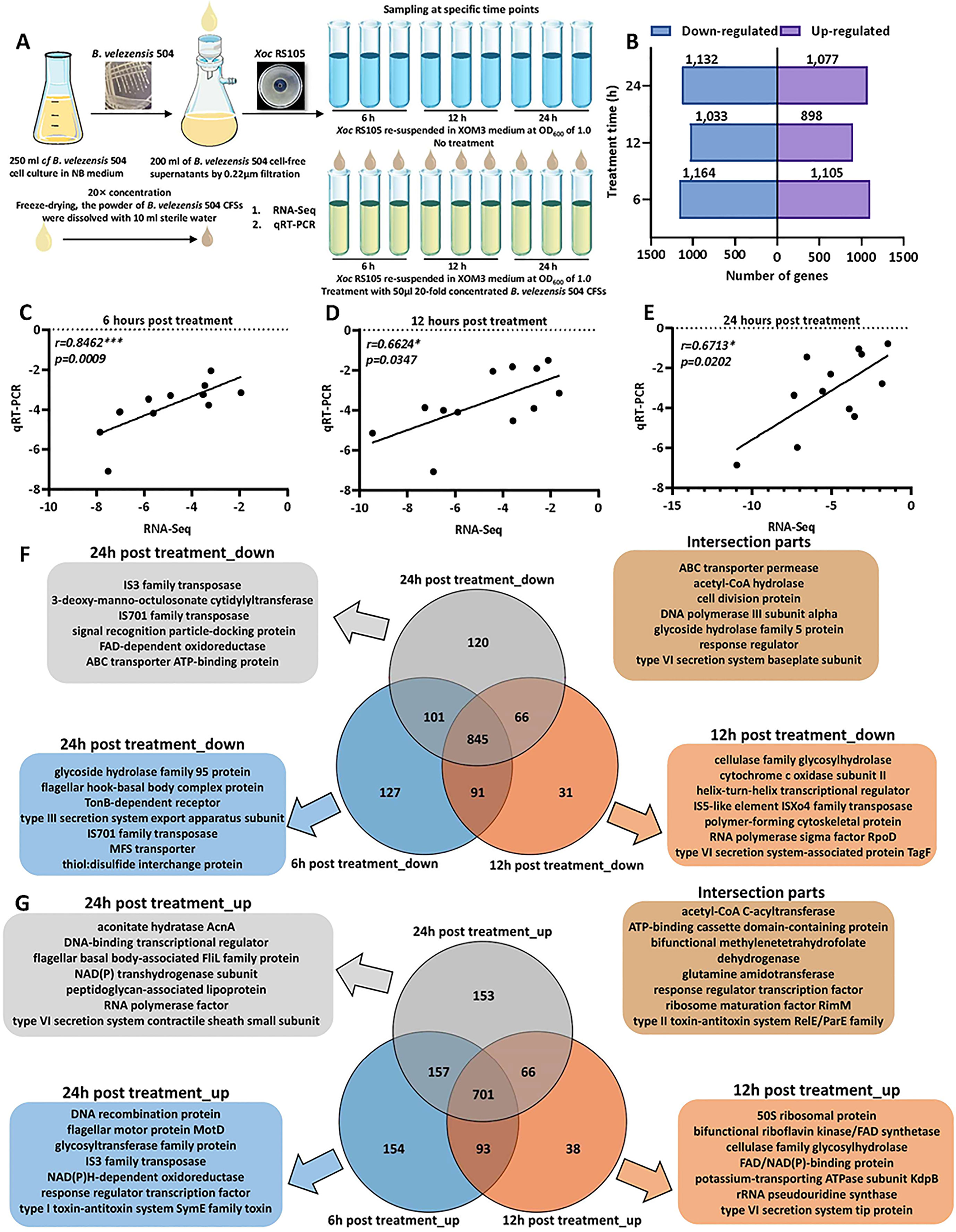
Figure 3 Overview of transcriptome of RS105 under B. velezensis 504 CFSs treatment. (A) Schematic presentation of the transcriptome assay of B. velezensis 504 CFS-treated RS105. (B) Bar chart of downregulated and upregulated DEGs distribution between RS105 and 504-treated RS105 at three distinct time points. (C–E) Gene expression patterns obtained through RNA-Seq and qRT-PCR validation have been compared. The Y-axis displays the Log2FCs of the DEGs obtained using qRT-PCR validation, whereas the X-axis represents the Log2FCs of the DEGs obtained using RNA-Seq. The equation described the correlation between X and Y. (F) Venn diagram showing the overlapping down-regulated DEGs at three distinct time points. (G) Venn diagram showing the overlapping up-regulated DEGs at three distinct time points.
Differentially expressed genes (DEGs) were defined using a P-value adjusted for the false-discovery rate (Padj<0.01), and a moderated log2-transformed fold change in gene expression (mod_lfc)>1 to define upregulated genes, and mod_lfc<-1 to define downregulated genes. We observed 2,269, 1,931, 2,209 Xoc RS105 DEGs in response to the CFSs of B. velezensis 504 at 6 hpi, 12 hpi and 24 hpi, respectively (Figure 3B). To verify the reliability of the RNA-Seq data, we selected twelve interested genes (hpa1, hrpE, hrcC, rpsQ, nuoF, nuoL, glnA, lpdA, katB, fliM, flgL, and cheY) related to ribosome (ko03010), oxidative phosphorylation (ko00190), glycolysis/gluconeogenesis (ko00010), flagellar assembly (ko02040), bacterial chemotaxis (ko02030), two-component system (ko02020) and T3SS to evaluate the abundance for validation (Supplementary Table 12). Gene expression levels that carried out by qRT-PCR showed agreement with the changes detected by RNA-Seq at 6, 12, and 24 h-post treatment (spearman correlation coefficient of 0.8462, 0.6624, and 0.6713, p value<0.05), indicating the reliability of the transcriptome results (Figures 3C–E and Supplementary Table 13). Of these genes, 1,164, 1,033 and 1,132 genes were down-regulated, while 1,105, 898 and 1,077 genes were up-regulated (Figure 3B and Supplementary Tables 14, 15). A total of 1,546 genes were found to be differentially expressed at all three time points, of which 845 were down-regulated and 701 were up-regulated (Figures 3F, G). The significantly down-regulated genes identified function associated with ABC transporter permease, cell division proteins, DNA poltmerase III subunit alpha, and type VI secretion system baseplate subunit (Figure 3F). The significantly up-regulated genes identified function associated with acetyl-CoA C-acyltransferase, ATP-binding cassette domain-containing proteins, glutamine amidotransferase and response regulator transcription factors (Figure 3G). A comparison of these gene sets identified 291 DEGs (154 up-regulated, 127 down-regulated), 69 DEGs (38 up-regulated, 31 down-regulated), and 273 DEGs (153 up-regulated, 120 down-regulated) with a 6-hr, 12-hr, and 24-hr dependent expression pattern, respectively (Figures 3F, G). We concluded that 2743 genes (1381 down-regulated, 1362 up-regulated) corresponding to 77% of Xoc RS105 coding sequences (CDSs) were differentially expressed by the CFSs of B. velezensis 504.
Further, we performed comparative RNA-seq of Xoc RS105 induced by the CFSs of B. velezensis 504 at 6 hr, 12 hr and 24 hr. This led to the identification of 435 DEGs down-regulated and 316 DEGs up-regulated with significant changes in expression during treatment by the CFSs of B. velezensis 504 (Supplementary Figure 1). A comparison of these gene sets found that 9 DEGs were consistently down-regulated over three time periods while 6 DEGs were consistently up-regulated (Supplementary Table 16).
To further investigate functions of genes regulated by the CFSs of B. velezensis 504, we performed the Gene Ontology (GO) enrichment analysis. The DEGs were enriched in the cellular process (GO:0009987), bacterial-type flagellum basal body (GO:0009425) and flagellum-dependent cell motility (GO:0001539), chemotaxis (GO:0006935), organic substance transport (GO:0071702), carbohydrate transmembrane transporter activity (GO:0015144), transcription regulator activity (GO:0140110), sigma factor activity (GO:0016987), hydrolase activity (GO:0016787) acting on carbon-nitrogen, pyrophosphate activity, nucleoside triphosphatase activity, and dTTP metabolic process (Supplementary Figure S2). In the KEGG analysis, the B. velezensis 504 CFSs-induced DEGs were significantly enriched in 88 pathways including flagellar assembly (ko02040), two-component system (ko02020) and bacterial chemotaxis (ko02030) ranking the top three rich factors, followed by ribosome (ko03010), lipopolysaccharide biosynthesis (ko00540), tyrosine metabolism (ko00350), lysine biosynthesis (ko00300), as well as glyoxylate and dicarboxylate metabolism (ko00630) (Supplementary Figure S3 and Supplementary Table 17).
To explore possible antagonistic mechanism of the B. velezensis 504 CFSs against Xoc RS105, we observed the expression of some genes associated with metabolism, cellular process, and signal transduction processing from DEGs (Supplementary Table 18). Transcript levels of 20 out of 42 genes involved in nucleotide metabolism, such as ACU12_RS19055 predicted to encode ibonucleotide-diphosphate reductase subunit beta, ACU12_RS13275 predicted to encode 5-(carboxyamino) imidazole ribonucleotide synthase, ACU12_RS02015 predicted to encode phosphomannomutase, and guaA predicted to encode glutamine-hydrolyzing GMP synthase, were down-regulated by treatment with the CFSs of B. velezensis 504 (Figure 4A). Furthermore, B. velezensis 504 CFSs affected the expression of genes involved in energy production and transformation pathway in RS105, including glyoxylate and dicarboxylate metabolism (ko00630), butanoate metabolism (ko00650), pyruvate metabolism (ko00620) and other biochemical reaction processes (Figure 4B). RNA-Seq analysis found that glnA (ACU12_RS19795), lpdA (ACU12_RS17850) and katB (ACU12_RS01600) were down-regulated at 6, 12, 24 hpi under the treatment of B. velezensis 504 CFSs, which is in agreement with the qRT-PCR analysis (Figures 5A, B, D). Transcript levels of 14 out of 21 genes involved in amino sugar and nucleotide sugar metabolism, as well as 43 out of 68 genes involved in amino acid metabolism such as phenylalanine, tyrosine and tryptophan biosynthesis, were down-regulated by the CFSs of B. velezensis 504 at three time points (Figures 4H, I). These results indicate that the genes related to the metabolisms of nucleotide, carbohydrate, amino sugar and nucleotide sugar, and amino acid are repressed by the treatment of B. velezensis 504 CFSs.

Figure 4 Defining the transcriptional response of RS105 to the CFSs of B. velezensis 504. (A) Heatmap showing the temporal pattern of the relative transcript abundance of nucleotide metabolism genes between RS105 and the CFSs of 504-treated RS105. (B) Heatmap showing the temporal pattern of the relative transcript abundance of carbohydrate metabolism genes between RS105 and the CFSs of 504-treated RS105. (C) Heatmap showing the temporal pattern of the relative transcript abundance of two-component genes between RS105 and the CFSs of 504-treated RS105. (D) Heatmap showing the temporal pattern of the relative transcript abundance of oxidative phosphorylation genes between RS105 and the CFSs of 504-treated RS105. (E) Heatmap showing the temporal pattern of the relative transcript abundance of cell motility and biofilm formation genes between RS105 and the CFSs of 504-treated RS105. (F) Heatmap showing the temporal pattern of the relative transcript abundance of translation genes between RS105 and the CFSs of 504-treated RS105. (G) Heatmap showing the temporal pattern of the relative transcript abundance of ABC transporters genes between RS105 and the CFSs of 504-treated RS105. (H) Heatmap showing the temporal pattern of the relative transcript abundance of Amino acid metabolism genes between RS105 and the CFSs of 504-treated RS105. (I) Heatmap showing the temporal pattern of the relative transcript abundance of amino sugar and nucleotide sugar metabolism genes between RS105 and the CFSs of 504-treated RS105. (J) Heatmap showing the temporal pattern of the relative transcript abundance of cell division genes between RS105 and the CFSs of 504-treated RS105.
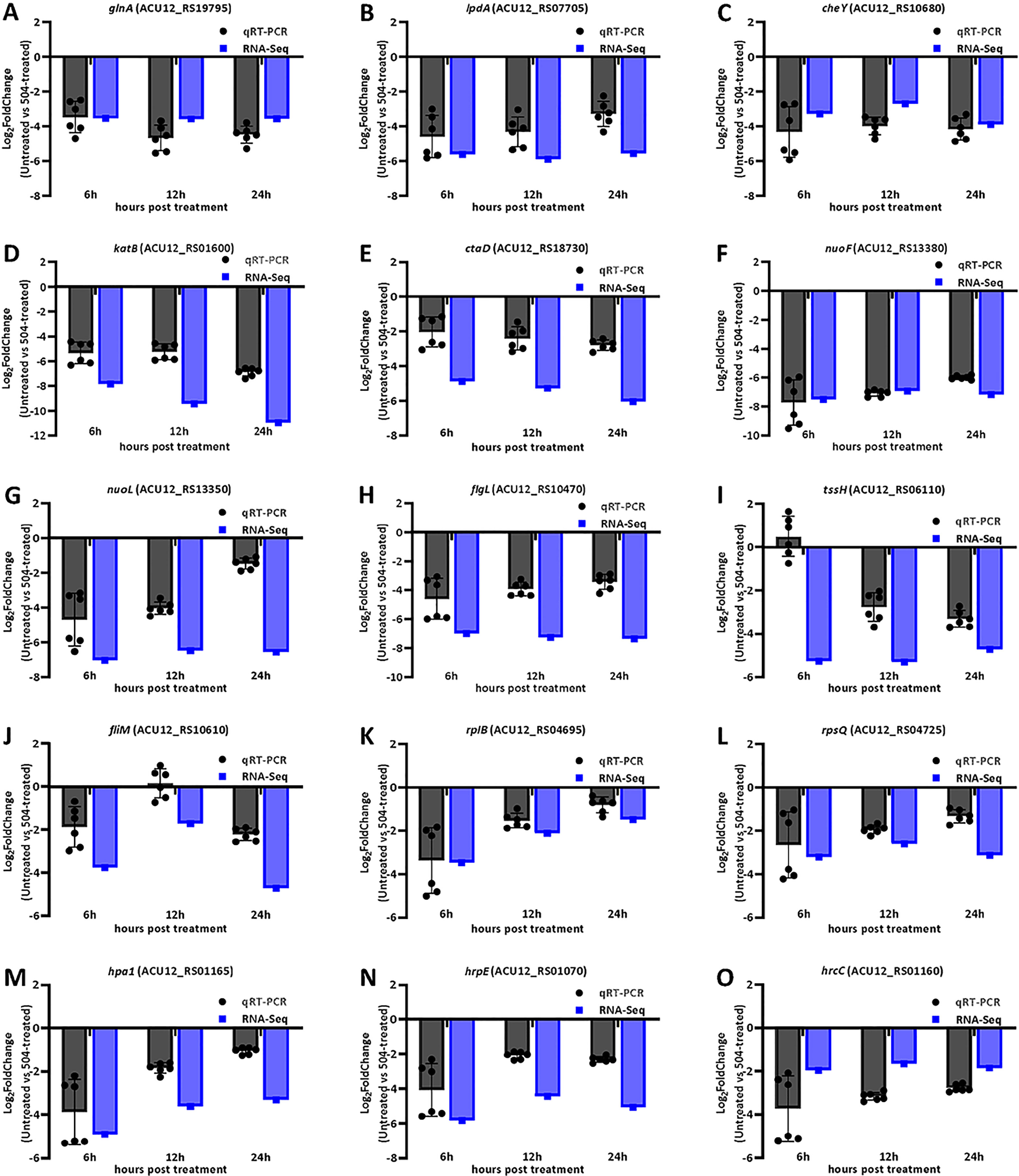
Figure 5 qRT-PCR assays for gene expression in RS105 treated with B. velezensis 504 CFSs. (A–O) The X-axis represents the 6, 12, and 24hpt. The Y-axis represents the Log2FoldChange (Untreated vs the CFSs of 504-treated) of relative expression level determined by RNA-Seq and qRT-PCR using rpoD and gyrB as an endogenous control. All experiments were repeated three times with consistent results.
Transcript levels of 35 out of 40 genes involved in oxidative phosphorylation were significantly down-regulated by the CFSs of B. velezensis 504 at 6 hpi, 12 hpi, and 24 hpi (Figure 4D). The expression pattern of ctaD (ACU12_RS18730), nuoF (ACU12_RS13380), and nuoL (ACU12_RS13350) by qRT-PCR analysis was coincident with the RNA-Seq data (Figures 5E–G). After B. velezensis 504 CFSs treatment, DEGs were mainly enriched in flagellar assembly (ko02040) and bacterial chemotaxis (ko02030), and their expression was significantly down-regulated, including flagellar and flagellin related genes (i.e., flgB-flgH, flgJ-flgL, flhA-flhB, fliE-fliG) (Figure 4E). The expression of flgL, fliM, and cheY in Xoc RS105 were downregulated by the B. velezensis 504 CFSs at 6 hpi, 12 hpi, and 24 hpi using qRT-PCR analysis. (Figures 5C, H, J), suggesting that B. velezensis 504 can inhibit flagellar assembly processing of Xoc RS105.
Fifteen genes (rpsQ, rplE, rpsS, rpsD, rplP, rpsA, rplN, rplW, rplB, rpsC, rpmC rplQ, rplO, rpsN, and rplV) in the ribosomal operon were down-regulated in RS105 treated with the CFSs of B. velezensis 504 (Figure 4F). rplB and rpsQ were decreased in 3.47- and 3.21-fold, 2.11- and 2.59-fold, 1.48- and 3.13-fold in RS105 at 6 hpi, 12 hpi, and 24 hpi with the addition of B. velezensis 504 CFSs, respectively (Figures 5K, L) suggesting that B. velezensis 504 can inhibit growth of Xoc RS105 by reducing the genetic information processing. Treatment of B. velezensis 504 CFSs caused significantly differential expression of 21 genes involved in cell division in Xoc RS105. (Figure 4J). Fourteen cell division (minCDE system genes, ftsA, ftsB, ftsL, ftsW, ftsY, etc.) were downregulated in RS105 treated with B. velezensis 504 CFSs at 6 h (Figure 4J). We also observed that expression levels of 21 out of 35 genes involved in ABC transporters, as well as 50 out of 81 genes involved in two-component systems were significantly down-regulated by the CFSs of B. velezensis 504 at 6 hpi, 12 hpi, and 24 hpi (Figures 4C, G). Taken together, the results indicating that B. velezensis 504 may exert antibacterial activity by affecting the entire metabolic pathway, transmembrane transport, cell motility, DNA translation and signal transduction.
The successful pathogenesis by Xoc in host rice tissues is dependent on some critical virulence factors including type III secretion system (T3SS), lipopolysaccharides (LPS), exopolysaccharides (EPS), adhesins, extracellular enzymes, toxins, cell motility, and so on (Buttner and Bonas, 2010). To further explore the biocontrol mechanisms of B. velezensis 504, we observed the expression of some major virulence related genes from DEGs (Supplementary Table 19). We observed the expression of 25 T3SS genes (hrc, hrp and hpa) encoding the T3SS secretion system of Xoc RS105, and found that 14 genes (hpa2, hpa1, hrcC, hrpB3, hrpB2, hrpB1, hrcU, hpaP, hrcR, hrpD5, hrpD6, hrpE, hpaB and hpa4) were down-regulated at 6 hpi by the CFSs of B. velezensis 504, 7 genes (hpa1, hrcC, hrcV, hpaP, hrpD5, hrpE, and hpa4) were down-regulated at 12 hpi, and 8 genes (hpa1, hrcC, hrcV, hpaP, hrpD5, hrpD6, hrpE, and hpa4) were down-regulated at 12 hpi, whereas 6 genes (hpa1, hrcC, hpaP, hrpD5, hrpE, and hpa4) were down-regulated at all three time periods (Figure 6A). This result was consistent with that by the qRT-PCR analysis of hpa1, hrpE and hrcC (Figures 5M–O). In addition, we found that genes gspD (ACU12_RS17240), gspN (ACU12_RS17250), gspG (ACU12_RS17285) encoding the critical components of type II secretion system (T2SS) of Xoc RS105 that is responsible for the secretion of extracellular enzymes were downregulated at 6 hpi, 12 hpi and 24 hpi by the CFSs of B. velezensis 504. These results indicate that the CFSs of B. velezensis 504 can weaken the functions of T3SS and T2SS of Xoc RS105.
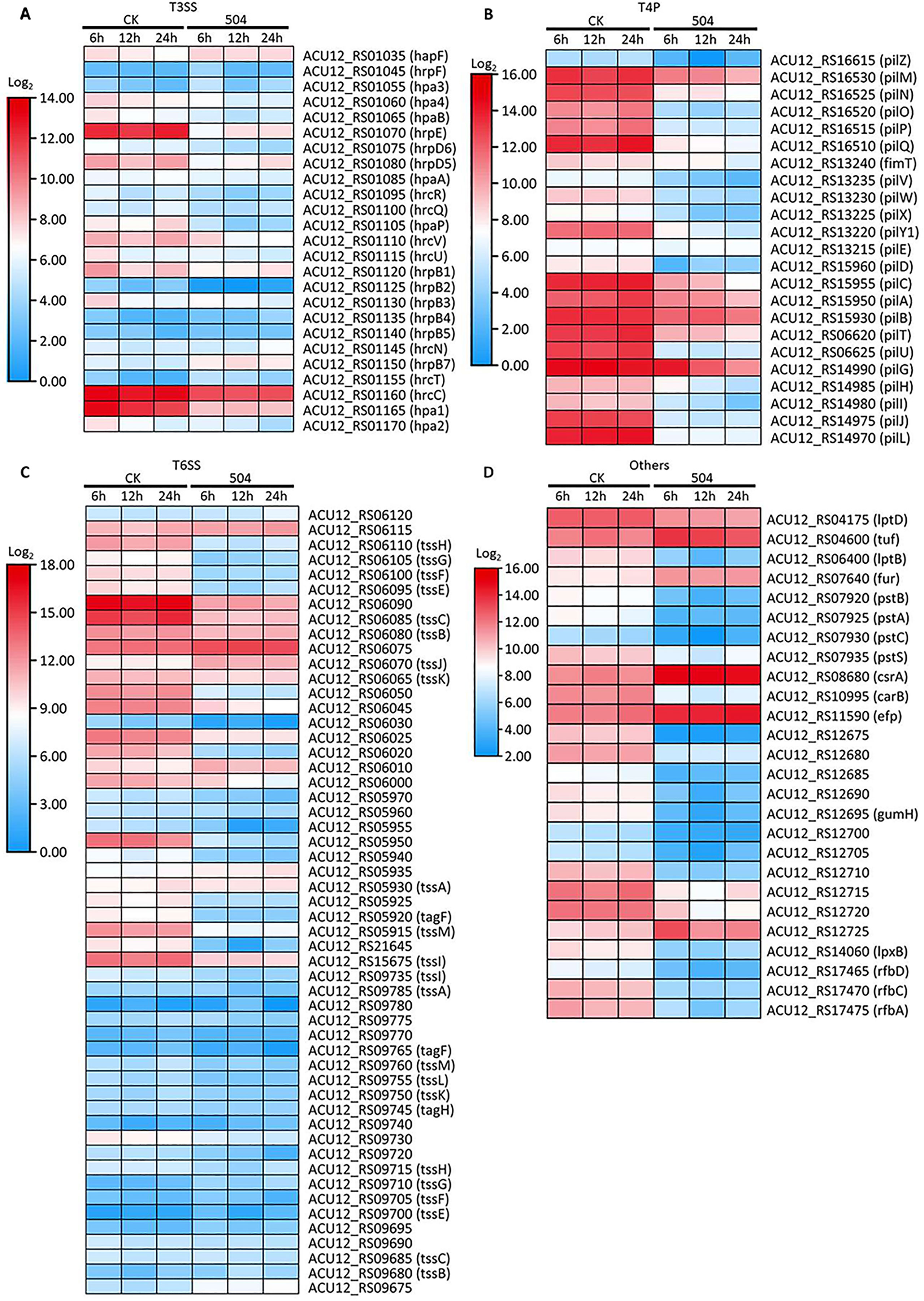
Figure 6 Defining the transcriptional response of RS105 virulence genes to the CFSs of B. velezensis 504 treatment. (A) Heatmap showing the temporal pattern of the relative transcript abundance of T3SS genes between RS105 and the CFSs of 504-treated RS105. (B) Heatmap showing the temporal pattern of the relative transcript abundance of T4P genes between RS105 and the CFSs of 504-treated RS105. (C) Heatmap showing the temporal pattern of the relative transcript abundance of T6SS genes between RS105 and the CFSs of 504-treated RS105. (D) Heatmap showing the temporal pattern of the relative transcript abundance of other genes related with virulence between RS105 and the CFSs of 504-treated RS105.
Bacterial attachment on host tissues relies on some adhesins that are classified into fimbrial and nonfimbrial adhesins (Li et al., 2020b). Xoc can use a polar flagellum and dynamic type IV pili (T4P) to contact surfaces of the host rice cells and can move across the surface using T4P-mediated twitching motility (Wei et al., 2021). We found that 23 T4P-related genes, except pilE, were down-regulated at 6 hpi, 12 hpi and 24 hpi by the CFSs of B. velezensis 504, among which the genes pilMNOP encoding the alignment complex, the genes pilVWX encoding the minor pilins, the gene pilY1 encoding the envelope protein PilY1 (adhesin), the gene pilD encoding a pre-pilin peptidase PilD, the gene pilU encoding a putative ATPase PilU were most significantly down-regulated (Figure 6B). This indicates that the CFSs of B. velezensis 504 can attenuate the function of T4P of Xoc RS105.
Type VI secretion system (T6SS) in Xanthomonas can directly inject toxins and effectors into cells of eukaryotic and prokaryotic targets by a contact apparatus, and participates in bacterial virulence and competition (Montenegro Benavides et al., 2021). We observed 53 genes associated with T6SS, among which 29 genes were significantly down-regulated, and some genes had two copies, such as tssH, tssG, tssF, tssE, tssC, tssB, tssA, tagF, tssM and tssI, etc. The expression levels of one copy of these genes such as tssH (ACU12_RS06110), tssG (ACU12_RS06105), tssF (ACU12_RS06100), tssE (ACU12_RS06095), tssM (ACU12_RS05915) were significantly reduced in RS105 treated with the CFSs of B. velezensis 504 (Figure 6C). tssM (ACU12_RS05915 and ACU12_RS09760) and tssH (ACU12_RS06110) associated with biofilm formation (ko02025), were down-regulated in 3.71-, 1.58- and 5.27-fold, 4.08-, 0.36- and 5.30-fold, and 3.73-, 2.33-, and 4.71-fold in Xoc RS105 at 6 hpi, 12 hpi, and 24 hpi by treatment of the CFSs of B. velezensis 504, respectively (Figure 6C). We chose the tssH gene for qRT–PCR analysis, the expression of it was down-regulated 2.77-fold, and 3.32-fold in Xoc RS105 at 12 hpi, and 24 hpi by the CFSs of B. velezensis 504, respectively (Figure 5I).
We found that the genes (lptD, lptB, and lpxB) predicted to encode LPS-assembly protein, LPS export ABC transporter, and lipid-A-disaccharide synthase, respectively, with the addition of the genes of gum cluster (from ACU12_RS12675 to ACU12_RS12725) associated with the EPS or LPS biosynthetic pathways were significantly down-regulated at 6 hpi, 12 hpi and 24 hpi by the CFSs of B. velezensis 504 (Figure 6D). The three genes (rfbA, rfbC and rfbD) associated with the EPS synthesis were also down-regulated at 6 hpi, 12 hpi and 24 hpi. The four genes (pstA, pstB, pstC and pstS) predicted to encode phosphate ABC transporter were depressed at 6 hpi, 12 hpi and 24 hpi by the CFSs of B. velezensis 504 (Figure 6D). Downregulation of the carB gene, a known virulence gene in Xanthomonas spp., predicted to encode carbamoyl-phosphate synthase large subunit was also observed at three time points. However, two known virulence genes (fur and csrA) predicted to encode ferric iron uptake transcriptional regulator and carbon storage regulator CsrA (or RsmA), respectively, were induced at 6 hpi, 12 hpi and 24 hpi by the CFSs of B. velezensis 504 (Figure 6D). In addition, the genes efp and tuf predicted to encode elongation factor P and Tu, respectively, were also induced at three time points (Figure 6D). Taken together, these results suggest that B. velezensis 504 may exert biocontrol effects by down-regulating key virulence genes associated with T3SS, T2SS, T4P, T6SS, EPS and LPS productions, and other virulence factors.
Our previous results showed that B. velezensis 504 displayed significant antagonistic activity against Xoo and Xoc, and demonstrated that B. velezensis 504 could effectively prevent BLS (Li et al., 2019). We found that the CFSs of B. velezensis 504 can effectively inhibit the growth of Xoo PXO99A (Supplementary Figure 4A). To further investigate the biocontrol potential of B. velezensis 504 against BB of rice caused by Xoo, we performed the field trials using three susceptible cultivars including indica rice IR24, japonica cultivar Nipponbare, and Yuanfengzao, a highly susceptible indica rice variety in Hunan province of China. The prevention (Pre) and treatment (Tre) strategies were executed as follows: Xoo PXO99A only were used as control (CK), rice leaves sprayed with B. velezensis 504 12 h before inoculation with Xoo PXO99A suspension (504-Pre), and 12 h after inoculation with Xoo PXO99A suspension (504-Tre). The BB disease severity by all treatments were investigated 15 days post inoculation. We found that B. velezensis 504 exhibited an effective biocontrol effect on IR24. Compared with the control, the 504-Pre and 504-Tre treatments dramatically reduced the severity of BB on IR24 with relative control efficiencies of 77.59% and 74.00%, respectively, in paddy fields (Figure 7A). The relative control efficiencies by the 504-Pre and 504-Tre treatments were 76.75% and 76.54% on Nipponbare (Supplementary Figures 4C, D), respectively, followed by Yuanfengzao with relative control efficiencies 78.41% and 61.03% (Figure 7B), indicating that there is no significant difference between the 504-Pre and 504-Tre treatments on Nipponbare and Yuanfengzao. Taken together, these results indicate that B. velezensis 504 is a promising biocontrol agent for BB of rice.
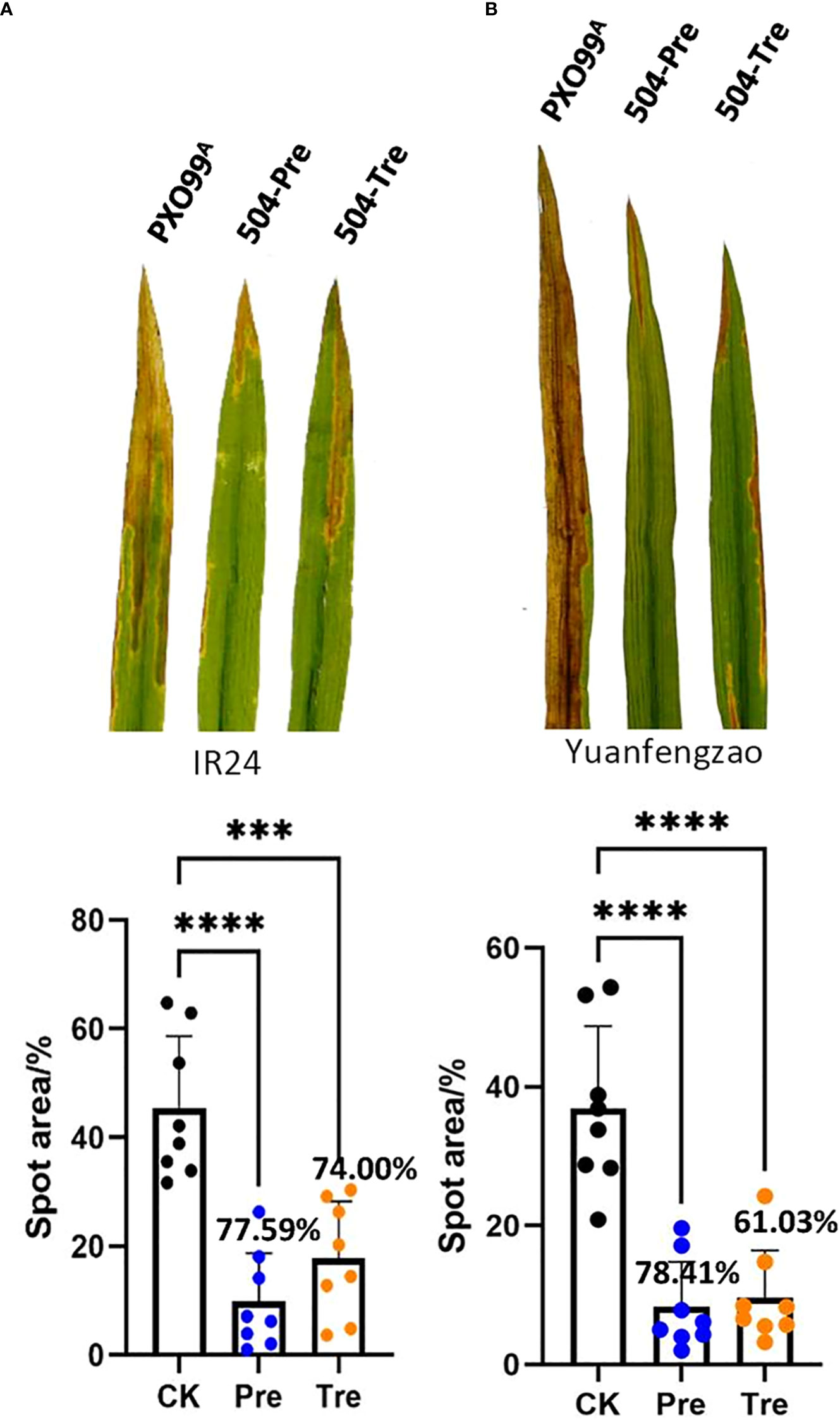
Figure 7 Efficacy of B. velezensis 504 for control of Xoo PXO99A in the field. The field trials were performed using the susceptible cultivars including indica rice IR24 (A) and japonica cultivar Yuanfengzao (B). The prevention (Pre) and treatment (Tre) strategies were executed as follows: Xoo PXO99A only (Control), rice leaves sprayed with B. velezensis 504 12 h before inoculation with Xoo PXO99A suspension (504-Pre), and 12 h after inoculation with Xoo PXO99A suspension (504-Tre). The bacterial blight disease of rice (BB) severity was investigated after 15 days. Data points represent means ± SD (n=8 independent leaves). The significant differences at ***P< 0.001 and at ****P< 0.0001.
Numerous B. velezensis strains have been reported to exhibit antagonistic activity against some important plant pathogenic fungi of the genus Rhizoctonia and Fusarium. We tested the inhibitory effect of B. velezensis 504 on four fungal pathogens including Botrytis cinerea, Magnaporthe oryzae, F. graminearum and F. oxysporum on agar plates using Oxford cups whose bottoms were filled up by bacterial suspensions or the CFSs of B. velezensis 504. The results showed that all the fungal strains spread over the entire surface of the petri dishes in the controls, whereas the growth of their hyphae was obviously suppressed in the presence of either bacterial suspensions or the CFSs of B. velezensis 504 (Figure 8A). B. cinerea, the etiological agent of gray mold, is the pathogenic fungus most influenced by B. velezensis 504 with an inhibition rate of 70.1% and 70.9% corresponding to the CFSs and bacterial suspensions treatments, respectively, followed by M. oryzae causing rice blast disease, with an inhibition rate of 52.7% and 59.2% relative to the CFSs and bacterial suspensions treatments (Figure 8A). F. graminearum and F. oxysporum were less sensitive to B. velezensis 504 and formed smaller inhibition zones, although they also were suppressed with an approximate inhibition rate of 40% (Figure 8A). In addition, we evaluated the antifungal activity of B. velezensis 504 against Colletotrichum siamense and C. australisinense that are thought to be the two dominant pathogenic species causing leaf anthracnose of rubber tree in Hainan province of China. Seven days post treatment, the inhibition rate of B. velezensis 504 against C. siamense and C. australisinense were 61.85% and 66.67% respectively (Figure 8B). These results suggest that B. velezensis 504 can efficiently inhibit the growth of B. cinerea, M. oryzae, C. siamense and C. australisinense.
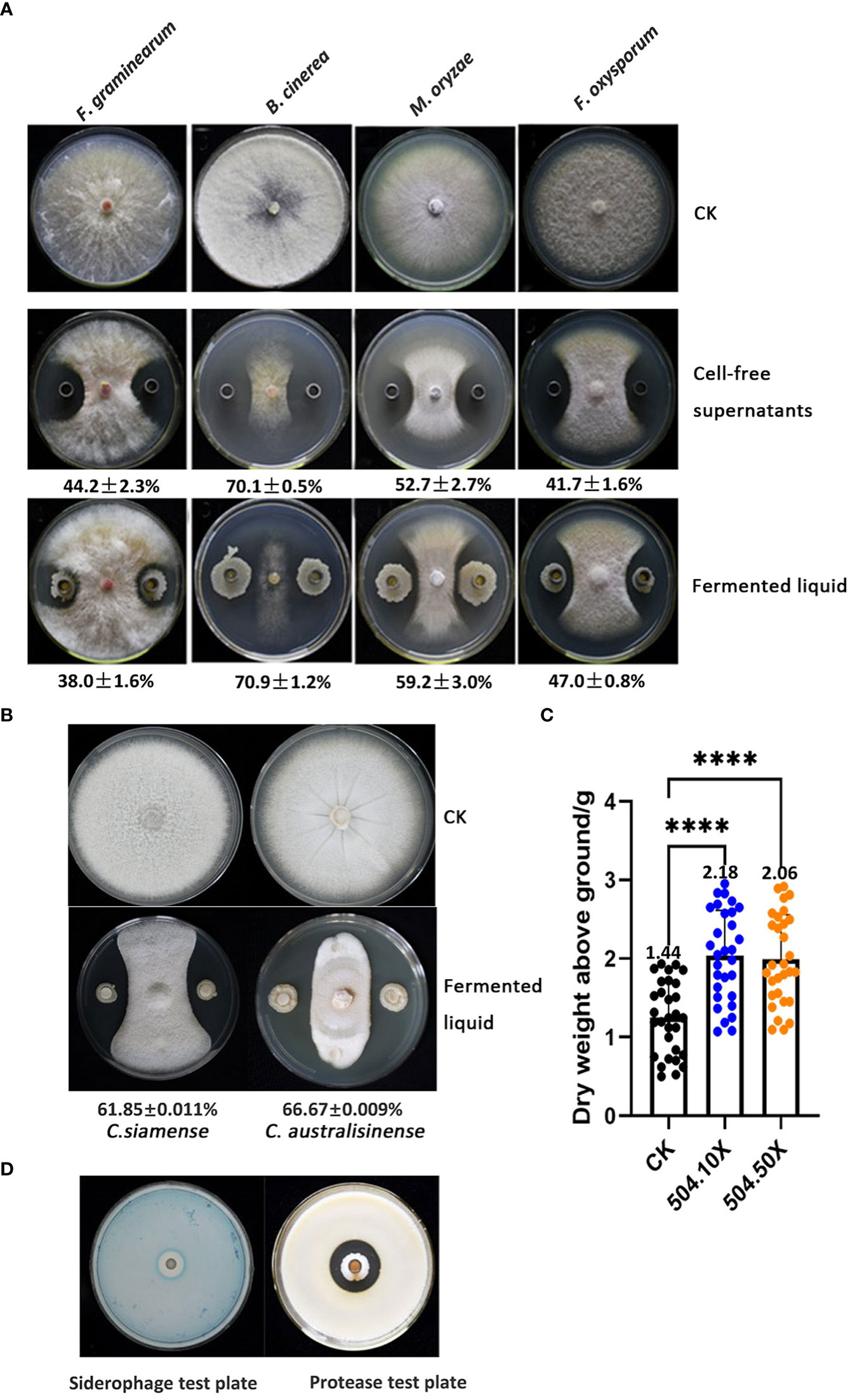
Figure 8 B. velezensis 504 can efficiently antagonize against some important plant pathogenic fungi and promote plant growth. (A) Determination of B. velezensis 504 antagonistic activity against F. graminearum, B. cinerea, M.oryzae, F. oxysporum in co-cultured assays. Sterile supernatants and fermented liquid of B. velezensis 504 were used as treatment, blank was used as control (CK). Three independent biological experiments were performed with similar results. (B) Determination of B. velezensis 504 antagonistic activities against C. siamense and C. australisinense in co-cultured assays. (C) Effect of B. velezensis 504 on growth-promotion of pak choi. (D) Siderophores and protease production of B. velezensis 504. The significant differences at ****P< 0.0001.
To determine whether B. velezensis 504 has the ability to promote plant growth, we formulated it into the commercially available bio-fertilizers in the form of liquid and powder-based, which were applied in the sowing time of pak choi (Brassica campestris sp. chinensis L.) in plastic greenhouse conditions. We found that the dry weight of pak choi was increased above 40% when the liquid bio-fertilizers of B. velezensis 504 was applied with the dilution of 10 or 50 times (Figure 8C), but no significant difference in root lengths of pak choi was observed when application of liquid and powder-based bio-fertilizers (data not shown), indicating that B. velezensis 504 is capable of stimulating plant growth. In addition, some assays relative to plant probiotic traits showed that B. velezensis 504 could secrete protease and produce siderophore (Figure 8D), but did not have cellulase activity and the ability to degrade inorganic phosphorus and potassium (Supplementary Figure S5). From these results, we conclude that B. velezensis 504 is a versatile plant probiotic bacterium.
In this study, we mined the genomic data of B. velezensis 504, and the comparative transcriptomic data of Xoc RS105 treated by the CFSs of B. velezensis 504 to define DEGs, as well as analyzed the potential of B. velezensis 504 for biological control and growth promotion. These studies revealed the potential biocontrol mechanisms of B. velezensis against BLS, and also indicated that B. velezensis 504 is a versatile plant probiotic bacterium.
B. velezensis 504 isolated from the rhizosphere soil of spinach had a growth-promoting effect on pak choi. Some studies showed that biocontrol mechanisms of B. velezensis against pathogenic bacteria and fungi consist of the production of antimicrobial substances, induction of plant systemic resistance and promotion of plant growth (Fan et al., 2018). For examples, B. velezensis AL7 prevent wilt disease of cotton caused by Verticillium dahliae Kleb, a biocontrol function mainly contributed by fengycin (Liu et al., 2021a); X. campestris pv. campestris with B. velezensis FZB42 treatment occurred distorted cells that died by cell lysis (Mácha et al., 2021); inoculation of the B. subtilis LSBS2 that produce siderophore significantly promoted plant growth and enhanced plant biomass in sesame (Nithyapriya et al., 2021).Based on comparative analysis of the GO function annotation, we determined the presence of genes encoding hydrolytic enzyme activities such as glucanase, lipase, xylanase, and amylase in B. velezensis 504, and that hydrolytic enzyme activities are beneficial to improve the environmental adaptability and tolerance of microorganisms (Yadav et al., 2018). B. velezensis 504 produced siderophore and had extracellular protease activity, whereas genomic analysis revealed that B. velezensis 504 contained six genes (entA, entB, entC, entE, dhbF, and menF) related to the siderophore biosynthetic process, which mediate the interactions between microbial members and invasion of the eukaryotic hosts they colonized. Comparative genome analysis suggested that B. velezensis 504 was similar with SQR9 and FZB42. Genomic differences between the three strains were assumed in the numbers and sizes of hypothetical proteins, clusters encoding secondary metabolite biosynthesis, and phage portal proteins. These results implied that B. velezensis has more biocontrol potentials and probiotic mechanisms to be explored.
B. velezensis 504 showed antagonistic activity against fungal pathogens including F. graminearum, F. oxysporu, B. cinerea, M. oryzae, C. siamense and C. australisinense. Difficidin and bacilysin have been reported to be the essential antibacterial agents produced by B. velezensis FZB42 to control phytopathogenic fungi and bacteria, including Xoo and Xoc (Wu et al., 2015). Approximately ten percent of the total genome sequence of FZB42 is predicted to encode at least thirteen gene clusters devoted to the synthesis of active secondary metabolites that play an essential part in the prevention and control of fungal, bacterial, viral, and nematode diseases (Borriss et al., 2019). Based on the genome analysis by the antiSMASH, fourteen gene clusters of secondary metabolite with biological activity were predicted to be present in B. velezensis 504, including cyclic lipopeptides (e.g., surfactin, bacillomycin-D, fengycin, and bacillibactin, an iron-siderophore), polyketides (e.g., macrolactin, bacillaene and difficidin), dipeptide antibiotic (bacilysin), bacteriocins (plantazolicin, amylocyclicin), and volatiles (acetoin and 2, 3-butandiol). Among them, four clusters were demonstrated to involve in the synthesis of surfactin, plantazolicin, difficidin and bacilysin, which may essential for the antibacterial activity of secondary metabolites to inhibits phytopathogenic bacteria and fungi. Difficidin, fengycin, and bacilysin in secondary metabolites are also hypothesized to be critical in the inhibition of pathogens by B. velezensis YYC (Yan et al., 2022). Bacilysin, a compound with a special structure among the metabolites of Bacillus, is a small-molecule peptide compound, which is one of the simplest peptide antibiotics with known structure. Bacilysin is a broad-spectrum antibiotic, and has a strong inhibitory effect on fungi (Fazle Rabbee and Baek, 2020). Therefore, we speculated that diffidin, and bacilysin in B. velezensis 504 CFSs are the major secondary metabolites antagonizing Xoc and fungal pathogens. In combination with KEGG annotation analysis, it was observed that the metabolic pathways for the synthesis of 50 antibiotics such as ansamycin, penicillin, cephalosporin, and streptomycin existed in B. velezensis 504. B. velezensis 504 provides a new microbial resource for the biological control of rice, rubber tree, wheat, and grape diseases.
Based on transcriptome analysis of Xoc RS105 treated by the CFSs of B. velezensis 504, we found that 77% of Xoc RS105 CDSs were differentially expressed by the CFSs of B. velezensis 504. The most significant differences in flagellar assembly and bacterial chemotaxis of cellular processes by the KEGG pathway were identified. The KEGG pathway enrichment analysis indicated that B. velezensis 504 CFSs treatment possibly affect bacterial motility, LPS production, extracellular signal perception and carbon source metabolism in Xoc RS105. Low level expression of type I glutamate-ammonia ligase glnA inhibits glutamine biosynthesis, causing glycolysis to misdirect in the TCA cycle resulting in stunted cell growth (Baliga et al., 2002). Under aerobic conditions, large amounts of energy are generated through a series of oxidative phosphorylation processes following the TCA cycle, which is the hub for energy conversion of metabolites such as proteins, sugars, starch, and lipids (Madloo et al., 2021). B. velezensis 504 can secrete antagonistic metabolites to down-regulate the expression of five genes including rpfF, gumD, ftsZ, rrlA and glmS in Xoc RS105. rpfF is implicated in diffusible extracellular factor (DSF) formation, while gumD is responsible for extracellular polysaccharide (EPS) biosynthesis, both of which are required for bacterial virulence (Feng et al., 2023). ftsZ is associated with cell division and influences cell metabolism (Kopacz et al., 2019). glmS encodes glucosamine 6-phosphate synthetase that is important for the biosynthesis of peptidoglycan, a component of the bacterial cell wall (Klein and Ferre-D’Amare, 2006). Six genes encoding T3SS (hpa1, hrcC, hpaP, hrpD5, hrpE, and hpa4) were significantly down-regulated at all three-time points. Absence of hrcC, hrpD5 and hrpE, Xoc RS105 would lose pathogenicity on the host rice (Li et al., 2011; Guo et al., 2012), implying that B. velezensis 504 performs a biocontrol function by significantly down-regulating the expression of virulence factor T3SS.
More down-regulated genes function in flagellar assembly (ko02040), bacterial chemotaxis (ko02030) in cellular process, starch and sucrose metabolism (ko00500), and oxidative phosphorylation (ko00190) indicating that B. velezensis 504 CFSs suppress phenotypes associated with chemotaxis, and motility of Xoc RS105, both of which are related to virulence toward host colonization. Genes involved in bacterial chemotaxis (flhA, flhB, flgC, flgK, flgL, fliC genes and cheA, cheB, cheD) were downregulated to suppress bacterial flagellar synthesis and chemotactic motility, reducing bacterial pathogenicity (Cai et al., 2022). Down-regulated expression of nineteen genes in the starch and sucrose metabolism pathway could lead to inadequate intracellular energy supply to bacteria, thus affecting normal bacterial growth. Compared with the Xoc RS105 in XOM3 cultures, 20 genes of ribosome pathway in Xoc RS105 were upregulated by the CFSs of B. velezensis 504 and 16 genes were downregulated. rpmA-encoded ribosomal protein L27 interacts with tRNA substrates in the peptidyl transferase center (PTC) that located in the 50S ribosomal subunit. P. aeruginosa with decreased expression of the ribosomal protein manipulator rplU-rpmA exhibited a growth-deficient phenotype. Among them, the decreased expression of tssM (membrane subunit) affects numerous biological processes, including the biofilm formation, population growth ability, competition, and epiphytic fitness (Fei et al., 2022). Surprisingly, the expression levels of these genes were all reduced in RS105 by the CFSs of B. velezensis 504, including pstA, pstB, pstC, and pstS (related with phosphate ABC transporter), gumH (involved in EPS production), lpxB and lptB (associated to LPS synthesis and export), rfbC and rfbA (related with streptomycin synthesis). These suggested that the reduced expression of rfbAC (associated with the coding for O-antigen) in 504 CFSs-treated RS105 suppressed the biosynthesis of dTDP-rhamnose, which is the precursor for EPS, LPS and cell-wall polysaccharides.
In this study, B. velezensis 504 was demonstrated to possess antagonistic activity, specifically inhibiting the growth of Xoc, Xoo and fungal pathogens such as B. cinerea, M. oryzae, C. siamense and C. australisinense, therefore, further field experiments are needed to prove whether B. velezensis 504 is a microbial biocontrol agent with prospective applications. Several B. velezensis strains have been applied as biocontrol agents in agricultural production processes with the properties of promoting plant growth and induction of systemic resistance.
We demonstrate that B. velezensis 504 is a potential biocontrol resource for plant diseases, exhibiting strong antibacterial and antifungal activity, as well as plant growth-promoting traits. Furthermore, the transcriptome analysis shed light on mechanisms of how the biological control agent impaired the pathogen and interfered with the bacterial growth, providing a foundation for biological control in agriculture.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
LZ, QZ, and MT designed the research; YX, LZ, and MT supervised the study; QZ, XF, YC, MW, YF, YY, GuC, YZ, ZZ, and KY analyzed the data and performed part of the experiments; LZ, YY, and QZ wrote the paper; GoC, MT, and YX critically revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by Hainan Basic and Applied Basic Research (Natural Science) high-level talents (2019RC332), and Hainan Key Research and Development Program(ZDYF2021XDNY291).
We thank Dr. Ruihuan Yang (School of Agriculture and Biology, Shanghai Jiao Tong University, China) for providing technical support for biological control of rice.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1175446/full#supplementary-material
Baard, V., Bakare, O. O., Daniel, A. I., Nkomo, M., Gokul, A., Keyster, M., et al. (2023). Biocontrol potential of Bacillus subtilis and Bacillus tequilensis against four fusarium species. Pathogens 12 (2), 254. doi: 10.3390/pathogens12020254
Baliga, N. S., Pan, M., Goo, Y. A., Yi, E. C., Goodlett, D. R., Dimitrov, K., et al. (2002). Coordinate regulation of energy transduction modules in halobacterium sp. analyzed by a global systems approach. Proc. Natl. Acad. Sci. 99 (23), 14913–14918. doi: 10.1073/pnas.192558999
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., et al. (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47 (W1), W81–W87. doi: 10.1093/nar/gkz310
Borriss, R., Wu, H., Gao, X. (2019). Secondary metabolites of the plant growth promoting model rhizobacterium Bacillus velezensis FZB42 are involved in direct suppression of plant pathogens and in stimulation of plant-induced systemic resistance. Secondary. Metabol. Plant Growth Promoting. Rhizomicroorg.: Discovery Appl., 147–168. doi: 10.1007/978-981-13-5862-3_8
Buttner, D., Bonas, U. (2010). Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 34 (2), 107–133. doi: 10.1111/j.1574-6976.2009.00192.x
Cai, L., Ma, W., Zou, L., Xu, X., Xu, Z., Deng, C., et al. (2022). Xanthomonas oryzae pv. oryzicola response regulator VemR is co-opted by the sensor kinase CheA for phosphorylation of multiple pathogenicity-related targets. Front. Microbiol. 13, 928551. doi: 10.3389/fmicb.2022.928551
Chaudhari, N. M., Gupta, V. K., Dutta, C. (2016). BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6, 24373. doi: 10.1038/srep24373
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chowdhury, S. P., Hartmann, A., Gao, X., Borriss, R. (2015). Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42 - a review. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00780
Darling, A. C., Mau, B., Blattner, F. R., Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14 (7), 1394–1403. doi: 10.1101/gr.2289704
Dunlap, C. A., Kim, S. J., Kwon, S. W., Rooney, A. P. (2015). Phylogenomic analysis shows that Bacillus amyloliquefaciens subsp. Plantarum is a later heterotypic synonym of Bacillus methylotrophicus. Int. J. Syst. Evol. Microbiol. 65 (7), 2104–2109. doi: 10.1099/ijs.0.000226
Dunlap, C. A., Kim, S. J., Kwon, S. W., Rooney, A. P. (2016). Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. Plantarum and ‘Bacillus oryzicola’ are later heterotypic synonyms of Bacillus velezensis based on phylogenomics. Int. J. Syst. Evol. Microbiol. 66 (3), 1212–1217. doi: 10.1099/ijsem.0.000858
Fan, B., Wang, C., Ding, X., Zhu, B., Song, X., Borriss, R. (2019). AmyloWiki: an integrated database for Bacillus velezensis FZB42, the model strain for plant growth-promoting bacilli. Database (Oxford). 2019, 1–7. doi: 10.1093/database/baz071
Fan, B., Wang, C., Song, X., Ding, X., Wu, L., Wu, H., et al. (2018). Bacillus velezensis FZB42 in 2018: the gram-positive model strain for plant growth promotion and biocontrol. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02491
Fazle Rabbee, M., Baek, K.-H. (2020). Antimicrobial activities of lipopeptides and polyketides of Bacillus velezensis for agricultural applications. Molecules 25 (21), 4973. doi: 10.3390/molecules25214973
Fei, N., Ji, W., Yang, L., Yu, C., Qiao, P., Yan, J., et al. (2022). Hcp of the type VI secretion system (T6SS) in Acidovorax citrulli group II strain Aac5 has a dual role as a core structural protein and an effector protein in colonization, growth ability, competition, biofilm formation, and ferric iron absorption. Int. J. Mol. Sci. 23 (17), 9632. doi: 10.3390/ijms23179632
Feng, Y.-M., Long, Z.-Q., Xiang, H.-M., Ran, J.-N., Zhou, X., Yang, S. (2023). Research on diffusible signal factor-mediated quorum sensing in Xanthomonas: a mini-review. Molecules 28 (2), 876. doi: 10.3390/molecules28020876
Guo, W., Zou, L.-f., Li, Y.-r., Cui, Y.-p., Ji, Z.-y., Cai, L.-l., et al. (2012). Fructose-bisphophate aldolase exhibits functional roles between carbon metabolism and the hrp system in rice pathogen Xanthomonas oryzae pv. oryzicola. PloS One 7 (2), e31855. doi: 10.1371/journal.pone.0031855
Han, X., Shen, D., Xiong, Q., Bao, B., Zhang, W., Dai, T., et al. (2021). The plant-beneficial rhizobacterium Bacillus velezensis FZB42 controls the soybean pathogen phytophthora sojae due to bacilysin production. Appl. Environ. Microbiol. 87 (23), e0160121. doi: 10.1128/AEM.01601-21
Huang, M., Yang, R., Yan, Y., Li, Y., Fang, Y., Zhou, Q., et al. (2021). Bacillus altitudinis 181-7, a potential biocontrol bacterium against bacterial leaf streak of rice. Acta Phytopathol. Sin. 51 (6), 962–974. doi: 10.13926/j.cnki.apps.000555
Jeong, M. H., Lee, Y. S., Cho, J. Y., Ahn, Y. S., Moon, J. H., Hyun, H. N., et al. (2017). Isolation and characterization of metabolites from Bacillus licheniformis MH48 with antifungal activity against plant pathogens. Microb. Pathog. 110, 645–653. doi: 10.1016/j.micpath.2017.07.027
Klein, D. J., Ferre-D’Amare, A. R. (2006). Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science 313 (5794), 1752–1756. doi: 10.1126/science.1129666
Kopacz, M. M., Lorenzoni, A. S., Polaquini, C. R., Regasini, L. O., Scheffers, D. J. (2019). Purification and characterization of FtsZ from the citrus canker pathogen Xanthomonas citri subsp. citri. MicrobiologyOpen 8 (5), e00706. doi: 10.1002/mbo3.706
Li, S., Chen, Y., Yang, R., Zhang, C., Liu, Z., Li, Y., et al. (2019). Isolation and identification of a Bacillus velezensis strain against plant pathogenic Xanthomonas spp. Acta Microbiol. Sin. 59 (8), 1–15. doi: 10.13343/j.cnki.wsxb.20180513
Li, S., Liu, Z., Yang, R., Chen, Y., Zhong, Y., Chen, L., et al. (2020a). Isolation and identification of a Bacillus cereus strain against plant pathogenic Xanthomonas oryzae. Jiangsu. Agric. Sci. 48 (7), 127–136. doi: 10.15889/j.issn.1002-1302.2020.07.027
Li, W., Rokni-Zadeh, H., De Vleeschouwer, M., Ghequire, M. G., Sinnaeve, D., Xie, G. L., et al. (2013). The antimicrobial compound xantholysin defines a new group of Pseudomonas cyclic lipopeptides. PloS One 8 (5), e62946. doi: 10.1371/journal.pone.0062946
Li, Y., Yan, Y., Deng, S., Zhang, C., Haq, F., Chen, T., et al. (2020b). The Xanthomonas oryzae pv. oryzae type IV pilus alignment subcomplex protein PilN contributes to regulation of bacterial surface-associated behaviours and T3SS system. Plant Pathol. 69 (4), 744–755. doi: 10.1111/ppa.13157
Li, Y. R., Zou, H. S., Che, Y. Z., Cui, Y. P., Guo, W., Zou, L. F., et al. (2011). A novel regulatory role of HrpD6 in regulating hrp-hrc-hpa genes in Xanthomonas oryzae pv. oryzicola. Mol. Plant Microbe Interact. 24 (9), 1086–1101. doi: 10.1094/MPMI-09-10-0205
Liu, X., Li, B., Cai, J., Zheng, X., Feng, Y., Huang, G. (2018). Colletotrichum species causing anthracnose of rubber trees in China. Sci. Rep. 8 (1), 10435. doi: 10.1038/s41598-018-28166-7
Liu, Y., Shi, Y. P., Zhou, D., Yang, T., Dai, L., Li, L., et al. (2023). Characterization of colletotrichum causing anthracnose on rubber trees in yunnan: two new records and two new species from China. Plant Dis. doi: 10.1094/PDIS-11-22-2685-RE
Liu, H., Zeng, Q., Yalimaimaiti, N., Wang, W., Zhang, R., Yao, J. (2021a). Comprehensive genomic analysis of Bacillus velezensis AL7 reveals its biocontrol potential against verticillium wilt of cotton. Mol. Genet. Genomics 296, 1287–1298. doi: 10.1007/s00438-021-01816-8
Liu, Z., Zhu, Y., Shi, H., Qiu, J., Ding, X., Kou, Y. (2021b). Recent progress in rice broad-spectrum disease resistance. Int. J. Mol. Sci. 22 (21), 1658. doi: 10.3390/ijms222111658
Mácha, H., Marešová, H., Juříková, T., Švecová, M., Benada, O., Škríba, A., et al. (2021). Killing effect of Bacillus velezensis FZB42 on a Xanthomonas campestris pv. campestris (Xcc) strain newly isolated from cabbage brassica oleracea convar. capitata (L.): a metabolomic study. Microorganisms 9 (7), 1410. doi: 10.3390/microorganisms9071410
Madloo, P., Lema, M., Rodríguez, V. M., Soengas, P. (2021). Transcriptomic reprograming of Xanthomonas campestris pv. campestris after treatment with hydrolytic products derived from glucosinolates. Plants 10 (8), 1656. doi: 10.3390/plants10081656
Milagres, A. M., Machuca, A., Napoleao, D. (1999). Detection of siderophore production from several fungi and bacteria by a modification of chrome azurol s (CAS) agar plate assay. J. Microbiol. Methods 37 (1), 1–6. doi: 10.1016/s0167-7012(99)00028-7
Montenegro Benavides, N. A., Alvarez, B. A., Arrieta-Ortiz, M. L., Rodriguez, R. L., Botero, D., Tabima, J. F., et al. (2021). The type VI secretion system of Xanthomonas phaseoli pv. manihotis is involved in virulence and in vitro motility. BMC Microbiol. 21 (1), 14. doi: 10.1186/s12866-020-02066-1
Nino-Liu, D. O., Ronald, P. C., Bogdanove, A. J. (2006). Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7 (5), 303–324. doi: 10.1111/j.1364-3703.2006.00344.x
Nithyapriya, S., Lalitha, S., Sayyed, R., Reddy, M., Dailin, D. J., El Enshasy, H. A., et al. (2021). Production, purification, and characterization of bacillibactin siderophore of Bacillus subtilis and its application for improvement in plant growth and oil content in sesame. Sustainability 13 (10), 5394. doi: 10.3390/su13105394
Pascual, J., Garcia-Lopez, M., Carmona, C., Sousa Tda, S., de Pedro, N., Cautain, B., et al. (2014). Pseudomonas soli sp. nov., a novel producer of xantholysin congeners. Syst. Appl. Microbiol. 37 (6), 412–416. doi: 10.1016/j.syapm.2014.07.003
Poulaki, E. G., Tjamos, S. E. (2023). Bacillus species: factories of plant protective volatile organic compounds. J. Appl. Microbiol. 134 (3), 1–10. doi: 10.1093/jambio/lxad037
Rabbee, M. F., Ali, M. S., Choi, J., Hwang, B. S., Jeong, S. C., Baek, K. H. (2019). Bacillus velezensis: a valuable member of bioactive molecules within plant microbiomes. Molecules 24 (6), 1046. doi: 10.3390/molecules24061046
Richter, M., Rossello-Mora, R., Oliver Glockner, F., Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32 (6), 929–931. doi: 10.1093/bioinformatics/btv681
Shi, Z., Hong, W., Wang, Q. (2022). Complete genome resource of Bacillus velezensis J17-4, an endophyte isolated from stem tissues of rice. Plant Dis. 106 (2), 727–729. doi: 10.1094/PDIS-05-21-0996-A
Tang, Z., Cao, X., Zhang, H. (2023). Production of iturin a by Bacillus velezensis ND and its biological control characteristics. J. Basic. Microbiol. 63 (2), 179–189. doi: 10.1002/jobm.202200473
Vignesh, M., Shankar, S. R. M., MubarakAli, D., Hari, B. N. V. (2022). A novel rhizospheric bacterium: Bacillus velezensis NKMV-3 as a biocontrol agent against alternaria leaf blight in tomato. Appl. Biochem. Biotechnol. 194 (1), 1–17. doi: 10.1007/s12010-021-03684-9
Wei, C., Wang, S., Liu, P., Cheng, S. T., Qian, G., Wang, S., et al. (2021). The PdeK-PdeR two-component system promotes unipolar localization of FimX and pilus extension in Xanthomonas oryzae pv. oryzicola. Sci. Signal 14 (700), eabi9589. doi: 10.1126/scisignal.abi9589
Wu, L., Wu, H., Chen, L., Yu, X., Borriss, R., Gao, X. (2015). Difficidin and bacilysin from Bacillus amyloliquefaciens FZB42 have antibacterial activity against Xanthomonas oryzae rice pathogens. Sci. Rep. 5, 12975. doi: 10.1038/srep12975
Xu, S., Pan, X., Luo, J., Wu, J., Zhou, Z., Liang, X., et al. (2015). Effects of phenazine-1-carboxylic acid on the biology of the plant-pathogenic bacterium Xanthomonas oryzae pv. oryzae. Pestic. Biochem. Physiol. 117, 39–46. doi: 10.1016/j.pestbp.2014.10.006
Yadav, A. N., Verma, P., Sachan, S. G., Kaushik, R., Saxena, A. K. (2018). Psychrotrophic microbiomes: molecular diversity and beneficial role in plant growth promotion and soil health. Microorg. Green Revol.: Volume. 2: Microbes Sustain. Agro-ecosystem., 197–240. doi: 10.1007/978-981-10-7146-1_11
Yan, Y., Xu, W., Hu, Y., Tian, R., Wang, Z. (2022). Bacillus velezensis YYC promotes tomato growth and induces resistance against bacterial wilt. Biol. Control. 172, 104977. doi: 10.1016/j.biocontrol.2022.104977
Yang, R., Li, S., Li, Y., Yan, Y., Fang, Y., Zou, L., et al. (2021). Bactericidal effect of Pseudomonas oryziphila sp. nov., a novel Pseudomonas species against Xanthomonas oryzae reduces disease severity of bacterial leaf streak of rice. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.759536
Yang, R., Shi, Q., Huang, T., Yan, Y., Li, S., Fang, Y., et al. (2023). The natural pyrazolotriazine pseudoiodinine from Pseudomonas mosselii 923 inhibits plant bacterial and fungal pathogens. Nat. Commun. 14 (1), 734. doi: 10.1038/s41467-023-36433-z
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie. Van. Leeuwenhoek. 110 (10), 1281–1286. doi: 10.1007/s10482-017-0844-4
Zaid, D. S., Cai, S., Hu, C., Li, Z., Li, Y. (2022). Comparative genome analysis reveals phylogenetic identity of Bacillus velezensis HNA3 and genomic insights into its plant growth promotion and biocontrol effects. Microbiol. Spectr. 10 (1), e0216921. doi: 10.1128/spectrum.02169-21
Keywords: transcriptome profiling, antagonism mechanism, biocontrol agent, Bacillus velezensis, Xanthomonas oryzae pv. oryzicola
Citation: Zhou Q, Tu M, Fu X, Chen Y, Wang M, Fang Y, Yan Y, Cheng G, Zhang Y, Zhu Z, Yin K, Xiao Y, Zou L and Chen G (2023) Antagonistic transcriptome profile reveals potential mechanisms of action on Xanthomonas oryzae pv. oryzicola by the cell-free supernatants of Bacillus velezensis 504, a versatile plant probiotic bacterium. Front. Cell. Infect. Microbiol. 13:1175446. doi: 10.3389/fcimb.2023.1175446
Received: 27 February 2023; Accepted: 27 April 2023;
Published: 30 May 2023.
Edited by:
Yangyang Zhao, Jiangsu Academy of Agricultural Sciences (JAAS), ChinaCopyright © 2023 Zhou, Tu, Fu, Chen, Wang, Fang, Yan, Cheng, Zhang, Zhu, Yin, Xiao, Zou and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lifang Zou, em91bGlmYW5nMjAyMDE4QHNqdHUuZWR1LmNu; Youlun Xiao, eGlhb3lsdW4yMDAzQGFsaXl1bi5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.