- Department of Prosthodontics, Hospital of Stomatology, Guangdong Provincial Key Laboratory of Stomatology, Guanghua School of Stomatology, Sun Yat-sen University, Guangzhou, Guangdong, China
Introduction: The oral cavity and the gut tract are interconnected, and both contain abundant natural microbiota. Gut microbiota may interact with oral flora and participate in the development of periodontitis. However, the specific role of certain gut microbiota taxa for periodontitis has not been investigated. Mendelian Randomization is an ideal method to explore causal relationships avoiding reverse causality and potential confounding factors. Thus, we conducted a two-sample Mendelian Randomization study to comprehensively reveal the potential genetic causal effect of gut microbiota on periodontitis.
Methods: SNPs strongly associated with 196 gut microbiota taxa (18,340 individuals) were selected as instrument variables, and periodontitis (17,353 periodontitis cases and 28,210 controls) was used as the outcome. The causal effect was analyzed via random effect inverse variance-weighted, weighted median, and MR-Egger. The sensitivity analyses were conducted using Cochran’s Q tests, funnel plots, leave-one-out analyses, and MR-Egger intercept tests.
Results: Nine gut microbiota taxa (Prevotella 7, Lachnospiraceae UCG-008, Enterobacteriales, Pasteurellales, Enterobacteriaceae, Pasteurellaceae, Bacteroidales S24.7 group, Alistipes, and Eisenbergiella) are predicted to play a causal role in enhancing the risk of periodontitis (p< 0.05). Besides, two gut microbiota taxa (Butyricicoccus and Ruminiclostridium 6) have potentially inhibitive causal effects on the risk of periodontitis (p< 0.05). No significant estimation of heterogeneity or pleiotropy is detected.
Conclusion: Our study demonstrates the genetic causal effect of 196 gut microbiota taxa on periodontitis and provides guidance for the clinical intervention of periodontitis.
Introduction
As the leading cause of missing teeth and the most prevalent oral disease, periodontitis is a chronic, irreversible, and destructive inflammatory disease that damages the soft tissue and alveolar bone supporting the teeth (Papapanou et al., 2018). Currently, periodontitis has imposed an unbearable burden on humans. For individuals, periodontitis tends to have a tremendous impact on the patient’s masticatory function, aesthetics, and psychology, which cause a significant reduction in their life quality (Barbe et al., 2020). Periodontitis is also associated with serious systematic health problems, such as cardiovascular diseases, chronic lower respiratory disorders, and diabetes (Chapple et al., 2013; Tonetti et al., 2013; Kelly and El Karim, 2020). Globally, periodontitis is estimated to affect around 50% of adults in its mild state and is recorded as the sixth most prevalent disease in its severe state (Genco and Sanz, 2020). Furthermore, the prevalence of periodontitis increases with age and the economic impact of periodontitis accounts for a significant portion of the annual global economic burden of dental disease (Billings et al., 2018). Since periodontitis’ socioeconomic and healthcare impacts are enormous, it’s urgent to identify risk factors for periodontitis to prevent its occurrence. However, implementing public health models to prevent periodontitis is insufficient (Janakiram and Dye, 2020). Existing studies have demonstrated that many risk factors are associated with periodontitis, including smoking, alcohol consumption, poor oral hygiene, systematic health, and so on (Bartold, 2018; Baumeister et al., 2021). To further reduce the periodontitis burden, more emphasis should be laid on extra potentially modifiable risk factors.
Physiologically, the oral cavity and the gut are continuous areas connected by the gastrointestinal tract in the digestive system, and both are abundant in microorganisms. Periodontitis is triggered by dysregulated intraoral microbial communities and aberrant immune responses (Hajishengallis et al., 2020). Meanwhile, the gut microbiota is demonstrated to involve in various physiological regulations and the progression of many diseases, such as cardiovascular diseases, autoimmune diseases, and tumors (Zhou et al., 2019; Xu et al., 2022). It is of concern whether gut microbiota, as an extra-oral but oral-connected microbial community, can cause or mediate periodontitis. Traditionally, the gut microbiota was not considered to be one of the risk factors for periodontitis. Over the past few years, there has been increasing evidence of an association between gut microbiota and periodontitis. Some experimental studies point out that gut microbiota can modulate bone metabolism and play an essential role in regulating periodontal bone remodeling (Yan et al., 2016), which can affect the development of periodontitis (Jia et al., 2019). Gut microbiota may also mediate the impact of periodontitis on systemic diseases including prediabetes (Li et al., 2021). However, existing studies have not explored the causal relationship between gut microbiota and periodontitis or focused on the exact role of specific gut microbiota taxa on periodontitis. Given the above findings, we hypothesize that variations in the gut microbiota composition contribute to periodontitis. However, both gut microbiota composition and periodontitis share several risk factors including diets, smoking, alcohol consumption, and stress, which might cause spurious correlations as confounders (Bjorkhaug et al., 2019; Baumeister et al., 2021). Therefore, distinct populations studied, reverse causality, and potential confounding factors in current studies hinder the inference of the causal effect between gut microbiota and periodontitis. What’s more, the gut microbiota is a complex microorganism community containing diverse taxa, and research on the function of specific gut microbiota taxa for periodontitis is absent. Overall, confirmation of a causal relationship for this correlation and which microbiota taxa are most relevant is essential for clinical practice in the management of periodontitis.
Mendelian randomization (MR) is an alternative method to interpret observational bias, using genetic variation normally single nucleotide polymorphisms (SNPs) as instrumental variables (IV), to detect causal relationships between exposure and disease outcomes (Yavorska and Burgess, 2017). If the exposure is causally associated with the outcome, the IV related to the exposure will proportionally affect the outcome (Chen et al., 2021). The MR studies are similar to randomized controlled trials (RCT) because the probability is the same in the inheritance of either allele to an individual at random (Emdin et al., 2017). Since genetic variants are usually not associated with confounding factors, MR is more powerful in avoiding reverse causal associations and confounding factors than traditional observational studies (Hu et al., 2022). Previously, many MR studies have been applied to elucidate modest risk factors for various diseases, including cancers, cardiovascular diseases, and so on, which is effective in solving problems in epidemiology (Higbee et al., 2021). In addition, Dmitry. S reports genome-wide association studies (GWAS) for periodontitis in 17,353 participants (Shungin et al., 2019), which enables the conduction of an MR study on the potential causal links between gut microbiota and periodontitis, minimizing population differences, reverse causation, and confounding factors that interfere with the analyses.
Herein, we performed a two-sample MR study based on public large-scale GWAS data of gut microbiome and periodontitis to reveal the possible causal effects of 196 gut microbiota taxa on periodontitis. Eventually, we confirm the role of specific gut microbiota taxa in increasing or reducing the risk of periodontitis, most of which haven’t been involved in the fields of stomatology. Our findings thus not only expand the taxa of gut microbes associated with periodontitis but also reveal their specific causal relationship with periodontitis, providing a new strategy for the clinical control of periodontitis.
Materials and methods
Study design
In this study, we performed comprehensive MR analyses to reveal the causal effect between 196 gut microbiota taxa and periodontitis. The framework of our study design is presented in Figure 1. The exposure of interest was 196 gut microbiota taxa, and the outcome was periodontitis. The instrument variables for the exposure and corresponding information in the outcome were obtained and harmonized. Then MR analysis - included three approaches were performed and sensitivities analyses were conducted. A well-designed MR should follow three assumptions: (i) the genetics variant was strongly correlated with the exposure of interest (the gut microbiota); (ii) the genetics variant was not correlated with potential risk factors of the outcome (periodontitis) (p< 1×10-5); (iii) the genetic variant played role in the outcome only through exposure (Emdin et al., 2017). All these hypotheses were well-handled in our analyses. For the first hypothesis, we extracted strong instrument variables and calculated their F statistics to evaluate their strength. Since SNPs on each chromosome were randomly assigned during meiosis according to Mendel’s second law, the second hypothesis actually has been met from the study design (Veller et al., 2019). In addition, we search the PhenoScanner to remove SNPs associated with confoundings (Staley et al., 2016; Kamat et al., 2019). For the third hypothesis, we evaluated the pleiotropy using MR-Egger intercept analysis.
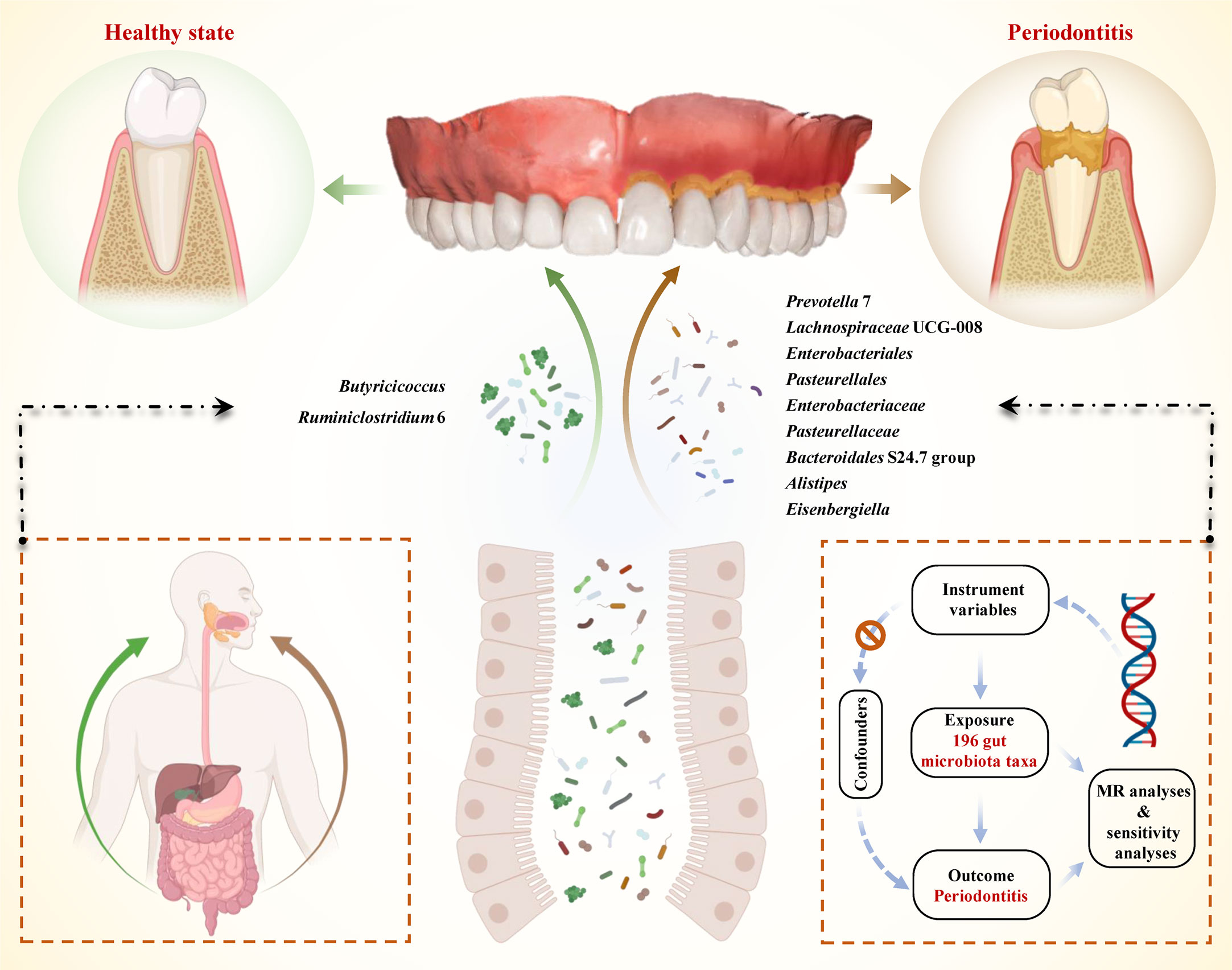
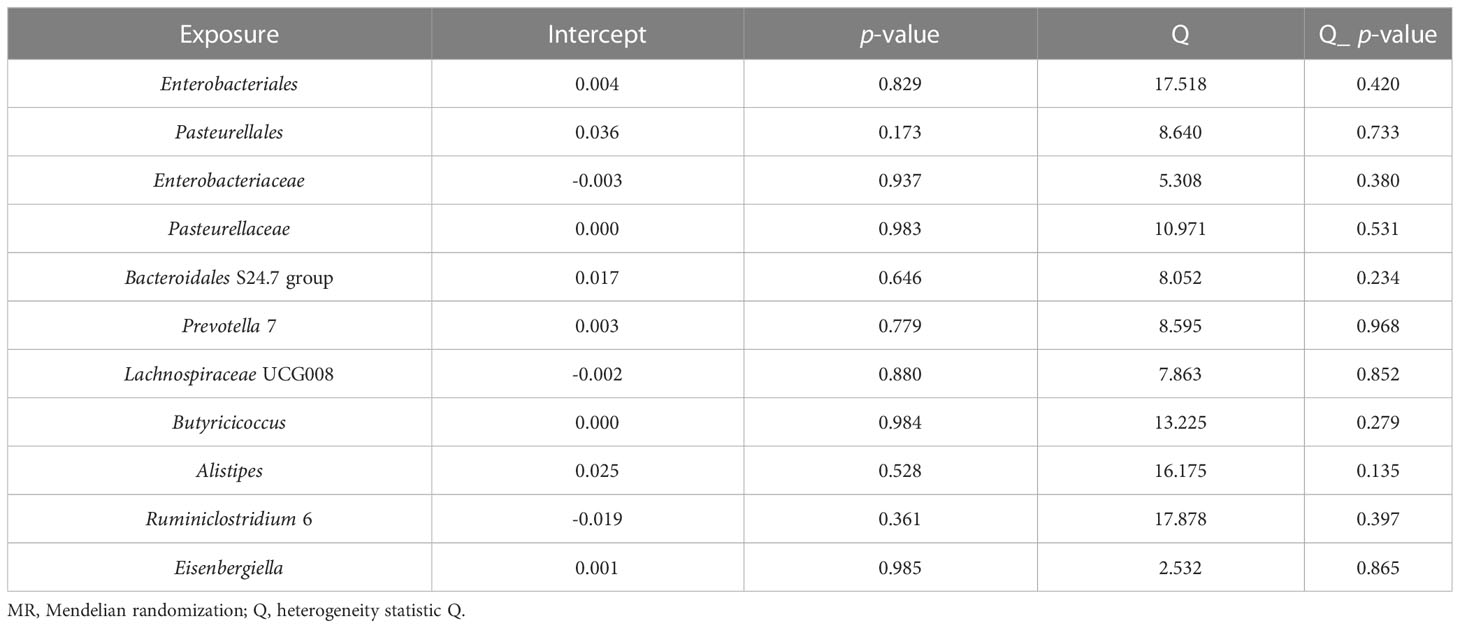
Figure 1 Schematic illustration of the causal relationship between gut microbiota and periodontitis through MR analyses. An overview of the two-sample MR study revealed the causal effect of specific gut microbiota taxa on periodontitis. There existed 9 taxa of gut microbiota that accelerated the initiation of periodontitis and 2 taxa of gut microbiota that reduced the risk of periodontitis. (MR, Mendelian randomization; SNPs, single nucleotide polymorphisms).
Data sources and instrumental variable selections
Previously, the MiBioGen consortium analyzed genome-wide genotypes and fecal microbiome data from 18,340 individuals mostly from North America, Israel, South Korea, and European (Kurilshikov et al., 2021). They performed strict quality control and those left for analysis should have a pointwise imputation quality control > 0.4. Taxa with prevalence low than 20% was discarded. The microbiome GWAS was adjusted by age, sex, technical covariates, and genetic principal components. The cutoff thresholds included: minor allele frequency > 0.05, and SNP-wise call filtering > 0.95. Alexander Kurilshikov and his colleagues reported the significant loci of host genetic variation in relation to microbial taxa (Kurilshikov et al., 2021). After excluding 15 unknown taxa, there were 196 known bacterial taxa, including 9 phyla, 16 classes, 20 orders, 32 families, and 119 genera (Table S1). The criteria for the 196 taxa included: (i) p< 1×10-5, since limited SNPs could be obtained under the genome-wide significance (p< 5×10-8) and such relaxed threshold have also been applied in many studies (Sanna et al., 2019); (ii) Linkage disequilibrium (LD) test was performed to remove linked SNPs r2< 0.1 within a window of 500 kilobase pair (Ni et al., 2022); (iii) The F-statistic was calculated for each SNP and SNPs with F< 10 were eliminated to avoid weak instruments bias (Burgess et al., 2011). The F-statistic for each SNP was calculated with a formula as
In the formula, R2 means variance of exposure explained by instrument variable; N indicates sample size. And the variance of exposure explained by the instrument variable was calculated with a formula as
In this formula, β indicated effect size for the genetic variant of interest; se indicated a standard error for β; N indicated sample size.
After strict selections, the identified IVs and corresponding F statistics were presented in Table S2.
The GWAS of periodontitis was from the GLIDE consortium which included 17,353 periodontitis cases and 28,210 controls (Shungin et al., 2019). Participants were of European ancestry. Periodontitis cases were categorized in compliance with the Community Periodontal Index (World Health Organization, 2013) or the Centers for Disease Control and Prevention/American Academy of Periodontology case definition in terms of the probing depths and the amount of deep periodontal pockets (Page and Eke, 2007). The SNP information of instrument SNPs in outcomes was obtained and harmonized with that of the exposure.
Mendelian randomization and sensitivity analyses
The MR and sensitivity analysis methods were consistent with published studies (Chen et al., 2021; Chen X. et al., 2022). Random effect inverse variance-weighted (IVW) was used as the main analysis for it was the most robust analysis and could provide a modest estimate with the presence of heterogeneity. Besides, we performed the weighted median (WM) and MR-Egger to validate the robustness of IVW estimates. MR-Egger regression could provide a test for unbalanced pleiotropy and considerable heterogeneity. When pleiotropy exists, the MR-Egger estimates were more convincing than IVW estimates (Bowden et al., 2015). And when at least half of the weighted variance provided by horizontal pleiotropy was valid, the WM estimates could provide robust estimates of effect (Bowden et al., 2016). In short, a significant estimate provided by IVW with the same direction of estimates provided by WM and MR-Egger was treated as a significant estimate.
We also conducted a series of sensitivity analyses, including Cochran’s Q tests, funnel plots, leave-one-out analyses, and MR-Egger intercept tests. To be specific, heterogeneity was detected by Cochran’s Q tests. The intercept term derived from MR-Egger regression was utilized to evaluate pleiotropy. The leave-one-out analyses were conducted to determine whether the causal estimate was driven by any single SNP.
All analyses were performed with the “Two Sample MR” (version 0.5.6) package in R software (version 4.2.1). A two-sided p< 0.05 was set as significant. All estimations were expressed as odds ratios (OR) per standard deviation (SD) increment of the corresponding exposure.
Results
An overview of IVs in taxa
Through screening the genome-wide significance threshold (p< 1×10-5), LD tests, harmonizing, and verifying F-statistics, each of the 196 bacterial taxa gets multiple SNPs as their IVs. The F-statistics of all retained SNPs are over 10, indicating sufficient correlation strength between IVs and corresponding bacterial taxon. Thus, our study has no weak instrument bias.
Associations of gut bacterial taxa with periodontitis
Preliminary results for the analyses of associations between genetically proxied gut bacterial taxa and risks of periodontitis are presented in Figure 2; Table S3. Among the 196 bacterial taxa, we find 11 gut microbiota taxa causally associated with periodontitis (Figure 3). The position of instrumental SNPs of the causal microbiota taxa and their nearest genes were listed in Table S4.
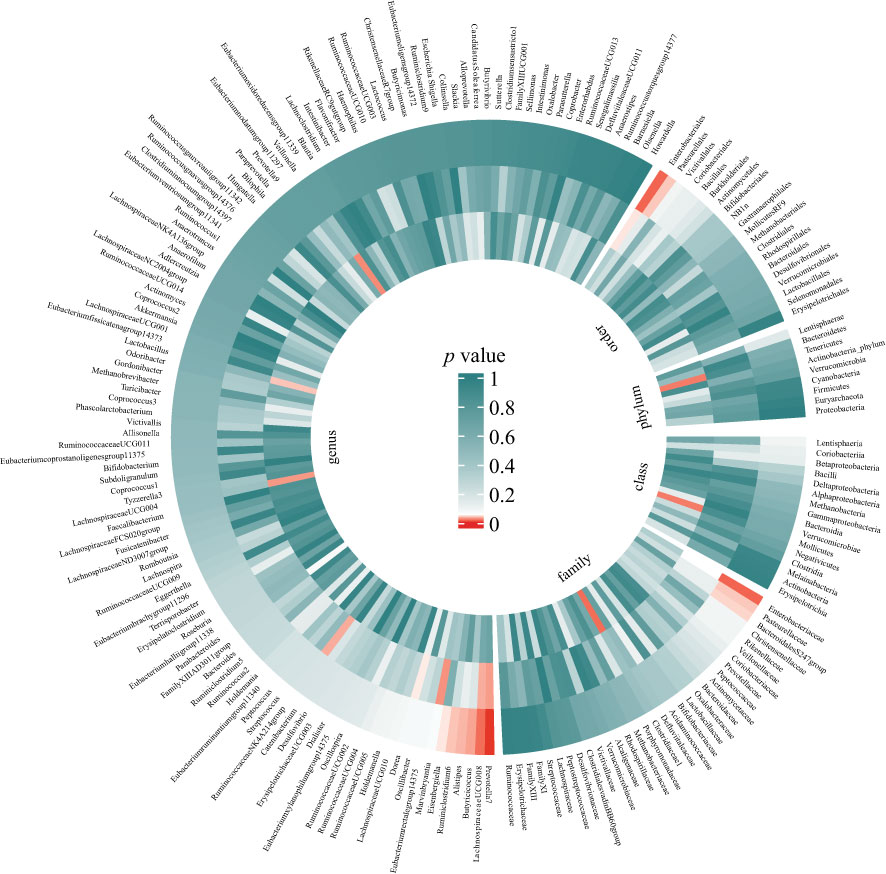
Figure 2 Preliminary MR analyses for the associations between gut microbiota and the risk of periodontitis. The circle from the outer to the inner represented the IVW, WM, and MR-Egger estimates, respectively. Gut microbiota was classified in order, phylum, class, family, and genus. The shades of color were reflections of the magnitude of the p-value as the label inside the circle. (MR, Mendelian randomization; IVW, inverse variance-weighted; WM, weighted median).
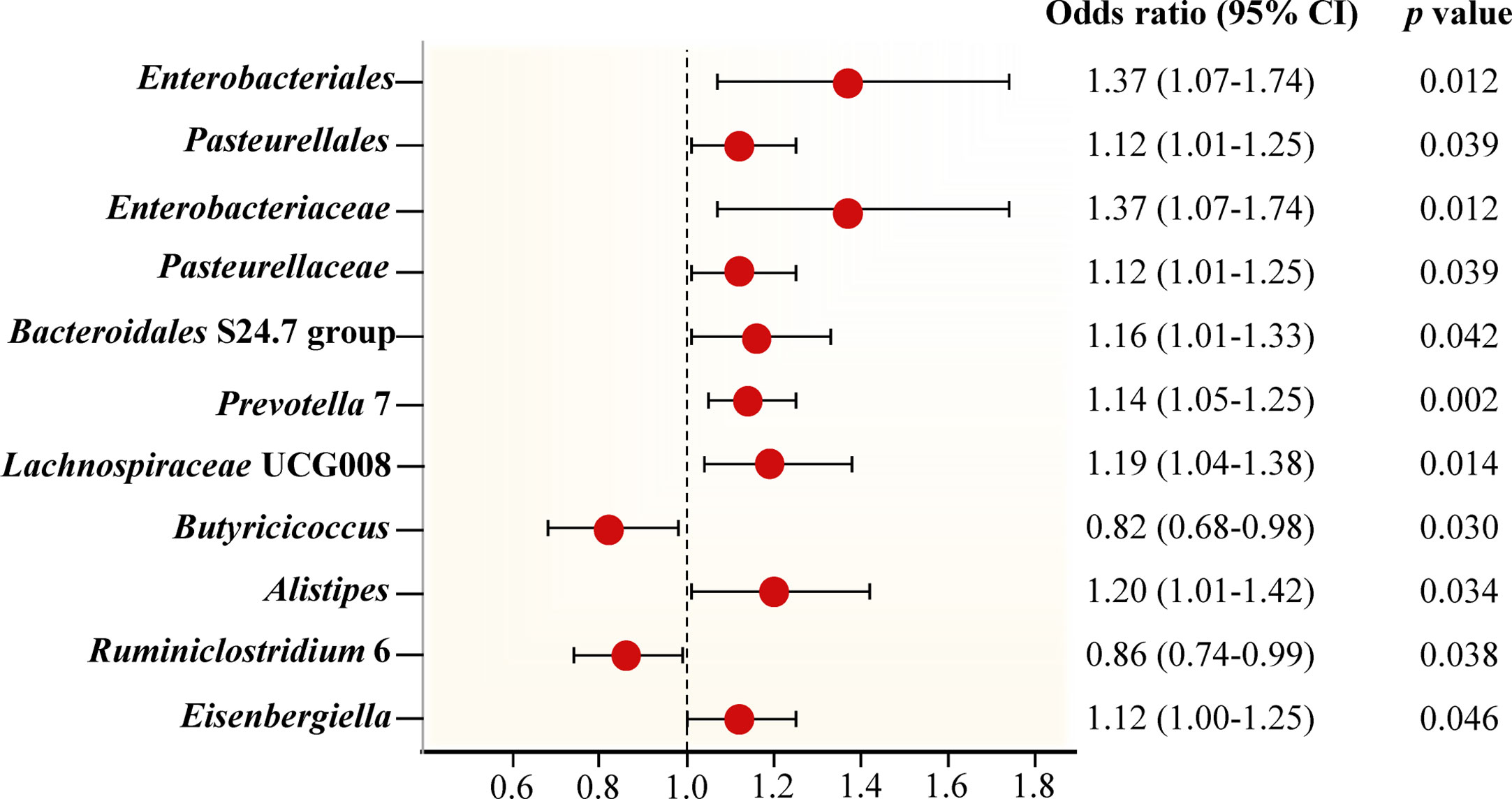
Figure 3 Forest plot of Mendelian randomization estimates between Gut microbiota and periodontitis. The figure showed the IVW estimates of significantly periodontitis-associated gut microbiota taxa. The red dots represent the IVW estimates, and the black bars represent the 95% confidence intervals of IVW estimates. The OR > 1 indicates increased risk while< 1 indicates decreased risk.
The genus Prevotella 7 is found to be positively associated with periodontitis, suggesting that genus Prevotella 7 in the human gut is causally related to an increased risk of periodontitis (IVW OR = 1.14, 95% confidence interval (CI)1.05-1.25, p = 0.002). The above result is furtherly confirmed by WM analyses (OR = 1.13, 95% CI 1.01-1.26, p = 0.028). The causal assessment from the MR-Egger analysis also supports consistent correlation but is not significant (OR = 1.12, 95% CI 0.67-1.88, p = 0.681) (Figure 4A; Table 1). The genus Lachnospiraceae UCG008 also has a progressive effect on periodontitis (IVW OR = 1.19, 95% CI 1.04-1.38, p = 0.014). The WM analysis shows similar results (OR = 1.20, 95% CI 1.02-1.42, p = 0.032). However, the MR-Egger analysis still exhibits consistent but insignificant trends (OR = 1.24, 95% CI 0.53-2.89, p = 0.633) (Figure 4B; Table 1).
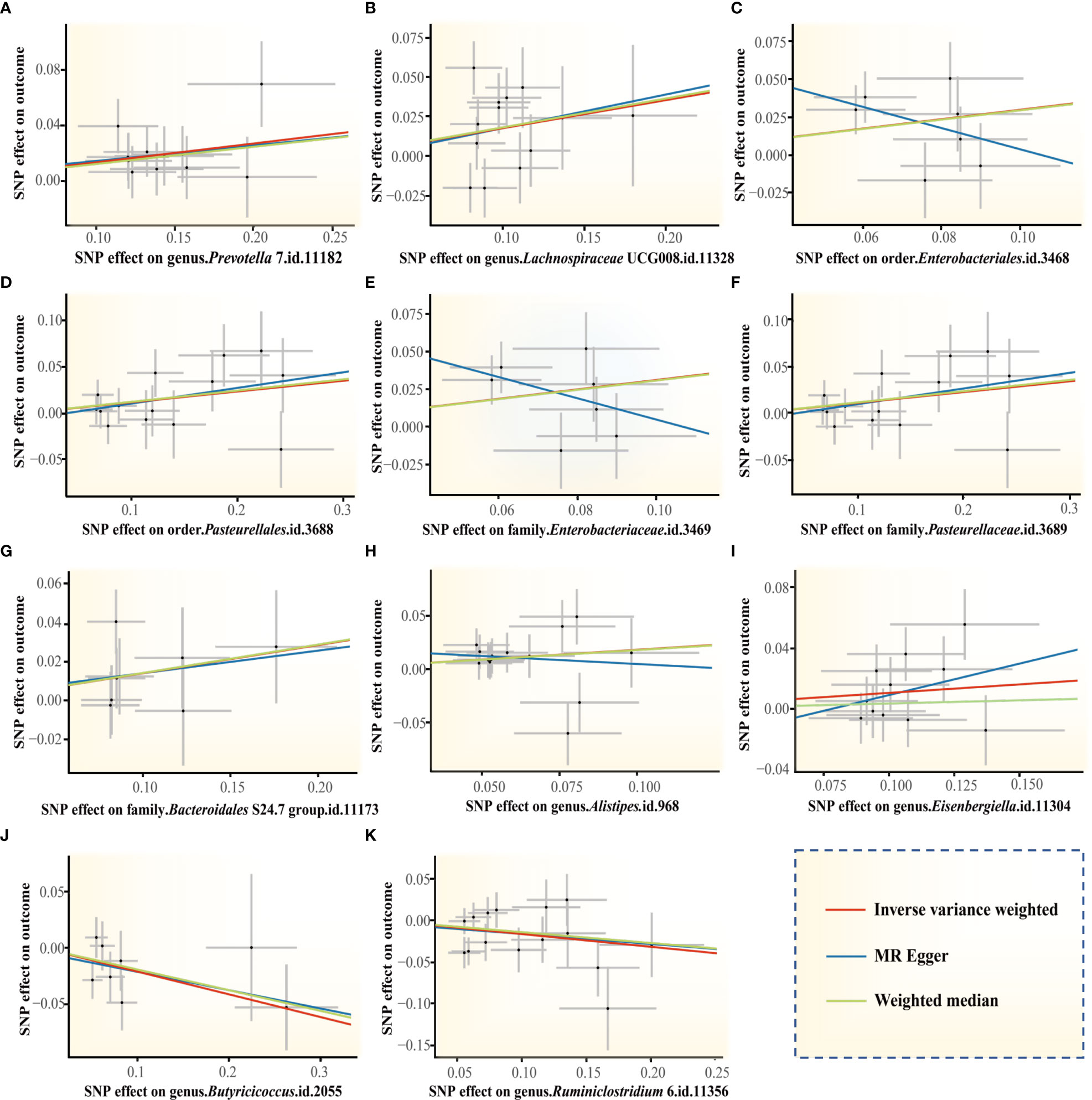
Figure 4 Scatter plots of the MR estimates for the significant causality of 11 gut microbiota taxa and the risk of periodontitis. (A) The causal effect of the genus Prevotella 7 on periodontitis; (B) The causal effect of genus Lachnospiraceae UCG008 on periodontitis; (C–K) Potential causal effect of 9 other gut microbiota taxa on periodontitis. The lines implying positive correlations moved diagonally upward from left to right, indicating a facilitative effect of gut microbiota on periodontitis. The horizontal and vertical lines indicated each correlation’s 95% confidence interval. The lines implying negative correlations move diagonally downward from left to right, indicating the inhibitory effect of gut microbiota on periodontitis. (MR, Mendelian randomization; SNPs, single nucleotide polymorphisms).
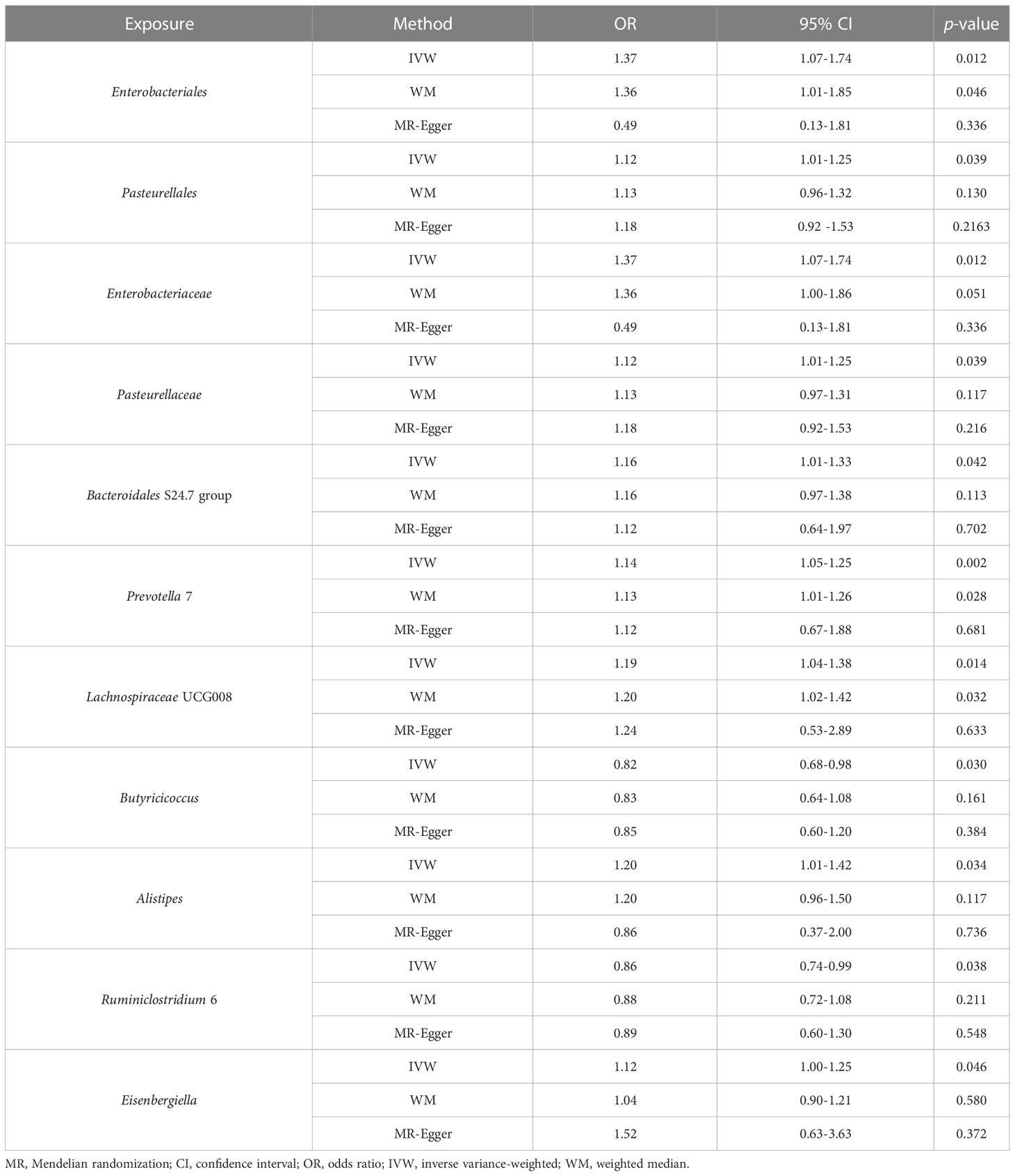
Table 1 MR estimates for the relationship between genetically instrumented gut microbiota and periodontitis.
In addition, we identify potential causal relationships between the other 9 taxa and periodontitis, as the IVW analyses results for all 9 phenotypes show significant differences (p< 0.05, Table 1). Among them, 7 taxa are determined to have a potential positive causal effect on periodontitis to increase the risk of periodontitis (Figures 4C–I), specifically containing the order Enterobacteriales (IVW OR = 1.37, 95% CI 1.07-1.74, p = 0.012), the order Pasteurellales (IVW OR = 1.12, 95% CI 1.01-1.25, p = 0.039), the family Enterobacteriaceae (IVW OR = 1.37, 95% CI 1.07-1.74, p = 0.012), the family Pasteurellaceae (IVW OR = 1.12, 95% CI 1.01-1.25, p = 0.039), the family Bacteroidales S24.7 group (IVW OR = 1.16, 95% CI 1.01-1.33, p = 0.042), the genus Alistipes (IVW OR = 1.20, 95% CI 1.01-1.42, p = 0.034), and the genus Eisenbergiella (IVW OR = 1.12, 95% CI 1.00-1.25, p = 0.046). Consequently, the above 7 taxa are confirmed to causally increase the risk of periodontitis. On the contrary, 2 taxa including the genus Butyricicoccus (IVW OR = 0.82, 95% CI 0.68-0.98, p = 0.030) and the genus Ruminiclostridium 6 (IVW OR = 0.86, 95% CI 0.74-0.99, p = 0.038) are identified as having a negative causal effect on periodontitis (Figures 4J, K) and tending to causally reduce the risk of periodontitis. Except the order Enterobacteriales, the family Enterobacteriaceae, and the genus Alistipes, the other MR analyses (WM and MR-Egger) present consistent results with the corresponding IVW analyses for the remaining 6 taxa.
For the order Enterobacteriales, the family Enterobacteriaceae, and the genus Alistipes, the causal correlations estimated by MR-Egger are the reverse of that detected by the other two MR analyses, although not significant. Therefore, the interpretation of these potential causal effects should be cautious. More investigations are needed. Nevertheless, due to the absence of horizontal pleiotropy and heterogeneity (p > 0.05, explained below), the causal relationships obtained by IVW are more accurate than the results from MR-Egger (Bowden et al., 2016; Chen et al., 2021; Chen M. et al., 2022). Consequently, it’s reasonable to recognize the IVW estimates for the order Enterobacteriales, the family Enterobacteriaceae, and the genus Alistipes, that is, they may increase the risk of periodontitis.
Sensitivity analyses and detection of pleiotropy
To avoid excessive bias effects, pleiotropic analyses are conducted. The pleiotropies are absent in the IVs of the mentioned 11 taxa causally associated with periodontitis (p > 0.05) and the conclusions are supported by the leave–one–out sensitivity (Figure S1). Funnel plots indicate that causal associations are less likely to be influenced by potential biases with SNPs symmetrically distributed (Figure S2). In view of heterogeneous results, Cochran’s Q tests demonstrate no evidence of heterogeneity among 11 taxa (p > 0.05). Besides, MR-Egger intercept tests exhibit no indication of horizontal pleiotropy within 11 taxa (Table 2). In summary, our MR analyses are verified to be reliable and robust. All these results suggest that the identified causal relationships between gut microbiota and periodontitis are likely to be mediated by the above gut bacterial taxa.
Discussion
In our research, a two-sample MR study successfully determines that partial gut microbiota can facilitate or prevent periodontitis. The results are examined by several analyses, namely IVW, WM, and MR-Egger analyses, almost all of which show consistent causal associations. The genus Prevotella 7, the genus Lachnospiraceae UCG-008, the order Enterobacteriales, the order Pasteurellales, the family Enterobacteriaceae, the family Pasteurellaceae, the family Bacteroidales S24.7 group, the genus Alistipes, and the genus Eisenbergiella play causal roles in promoting the initiation of the periodontitis, while the genus Butyricicoccus and the genus Ruminiclostridium 6 causally reducing the risk of periodontitis. Our findings fill the knowledge gap of whether gut microbiome can contribute to periodontitis and which taxa can accelerate or inhibit the initiation of periodontitis.
Prevotella is identified to activate key pathogenic species of periodontitis especially P. gingivalis that can result in periodontal diseases at low abundance (Hajishengallis et al., 2011). The genome of Prevotella is highly plastic and diverse, which favors its resistance to multiple exogenous factors, adaptation to variable environments, and generation of virulence (Purushe et al., 2010). This characteristic may account for the association between Prevotella and several oral infectious diseases. According to observational experiments, the genus Prevotella 7 in the gut of patients with periodontal diseases is significantly increased compared with that of populations without periodontal diseases (Abusleme et al., 2021). In our study, a higher abundance of the genus Prevotella 7 indicates a higher risk of periodontitis, suggesting the genus Prevotella 7 detected in fecal samples can be considered as a predictive biomarker and a target for efficient intervention in periodontitis.
To be noted, we found novel positively related taxa that haven’t been reported in previous literature. Among them, the genus Lachnospiraceae UCG-008 is positively correlated to the release of inflammatory factors such as IL-6, HS-CRP, and TNF-α, indicating that reducing the numbers of Lachnospiraceae UCG-008 is beneficial for controlling inflammation and can function in controlling periodontitis (Zhu et al., 2020). The pathogenic members of the order Pasteurellales and the family Pasteurellaceae are partially distributed in the mucosal of the oral cavity (Christensen et al., 2020). Our Mendelian study has shown that these two taxa distributed in the gut increase the risk of periodontitis. Combining previous studies and our findings, we assume that the order Pasteurellales and the family Pasteurellaceae may deteriorate periodontitis by regulating their abundance and pathogenicity in the oral cavity. The family Bacteroidales S24.7 group, one of the major components of gut microbiota, can trigger infections by stimulating immunity and synthesizing virulence factors (Ormerod et al., 2016). However, an observational study presents contradictory results showing the anti-inflammatory effects of metabolites of the family Bacteroidales S24.7 group. The inconsistency between studies may be ascribed to the highly individual variations in gut microbiological composition and the complicated multifactorial properties of inflammatory diseases (Eckburg et al., 2005). Our MR study can evade these issues by providing validation from a genetic perspective, obtaining clear evidence that the family Bacteroidales S24.7 group promotes the risk of periodontitis. Next, according to previous observational studies, the increase in the genus Eisenbergiella is associated with the deterioration of some chronic diseases, particularly pro-inflammatory illnesses which may include periodontitis (Bailen et al., 2020).
In addition to the above 9 taxa promoting periodontitis, we also indicate 2 negatively causative gut microbiota taxa (the genus Butyricicoccus and the genus Ruminiclostridium 6) that were first reported to be associated with periodontitis. Both the genus Butyricicoccus and the genus Ruminiclostridium 6 can degrade polysaccharides through autocrine multienzyme complexes to produce short-chain fatty acids such as butyrate, which acts as a valid anti-inflammatory mediator (Devriese et al., 2017; Wang et al., 2018). The biological function of butyrate is to maintain the intestinal epithelial barrier, balance gut microbiota, inhibit the expression of destructive cytokine, and regulate immunity and inflammation (Xiao et al., 2020), which may be the mechanisms of these two taxa to reduce risks of periodontitis. Combined with the existing results, our MR study suggests that we can possibly achieve prevention and control of periodontitis by increasing the abundance of the genus Butyricicoccus and the genus Ruminiclostridium 6 in various ways. Taken together, the correlation between mentioned gut microbiota taxa and periodontitis as verified by our MR study is reasonable.
Since the oral and gastrointestinal tracts are directly connected and both oral and gut microbiota have been demonstrated to affect the development of systemic diseases (Uchiyama et al., 2019), the interaction between the two microbiotas is meaningful to be explored. Clarification of the exact contribution of specific gut microbe taxa to periodontitis can bring new opportunities for more efficient prevention and control of periodontitis. Although efforts have already been made to elucidate the association between periodontitis and gut microbiota, no evidence for the causal effect was proposed. Furthermore, even though studies have found that periodontitis patients have a phenotype of dysbiosis of the gut microbiota, it’s the result of a multifactorial combination and the strain-specific changes of diverse microbiota taxa are inconsistent. In addition, the composition of the gut microbiota may vary due to inconsistencies in the staging of periodontitis, gender ratio, and ethnicity of the populations in different studies. The above factors hinder the inference of a specific causal effect on the risk of periodontitis and gut microbiota taxa.
As aforementioned, MR is a perfect study design to elucidate the causal effect between potential risk factors and diseases of interest. Recently, many MR studies have been applied to clarify modest risk factors for periodontitis. In these studies, the risk of periodontitis can be exaggerated by tobacco, alcohol, and genetically proxied obesity (Baumeister et al., 2021; Dong et al., 2022), while being suppressed by high micronutrients, fiber, and omega-3 fatty acids intake (Heo et al., 2022; Watson et al., 2022). Through exploring the factors that modulate the risk of periodontitis, MR studies facilitate the recommendation of public health policies and clinical interventions that effectively reduce the incidence and social burden of periodontitis. Meanwhile, there are also some factors that were linked to periodontitis in previous epidemiological observational studies but are demonstrated to have no causal association with periodontitis via MR studies, such as arthritis and psoriasis (Baurecht et al., 2022; Yin et al., 2022), which are not recommended as a target for the prevention and control of periodontitis. Compared with previous MR studies, our study is more comprehensive, revealing the causal effect of 196 gut microbiome taxa on periodontitis while previous MR studies only focus on less than 10 exposures of interest.
So far, this work is the pioneer MR study using large-scale gut microbiome and periodontitis genetic data to explore whether gut microbiota is causally related to the risk of periodontitis. The prominent advantage of our study is that the execution of the MR method robustly diminishes the interference of reverse causal associations and confounding factors. What’s more, our MR study encompasses the widest range of the population at a minimal cost, which may be more practical and convincing than conventional observational studies. However, some limitations should be noted. First, since participants in the GWAS are predominantly of European ancestry, extrapolation of our findings to other ethnic groups may be constrained. Second, given the biological plausibility and sophisticated pathobiology of periodontitis as well as the polyphasic process of statistics, the application of a strict multiple-testing correction may be so conservative that partially potential strains that are causally correlated to periodontitis are overlooked. Therefore, we didn’t implement multiple correlations. Third, since our study aimed to elucidate the risk factors for periodontitis to achieve comprehensive clinical intervention and reduce the incidence, we target the unidirectional role of 196 gut microbiota taxa on periodontitis. Fourth, the exact mechanisms by which the as-mentioned gut microbiota taxa influence the risk of periodontitis haven’t been totally investigated in this study.
Conclusions
In summary, this study innovatively demonstrates the causal relationship between gut microbiota and periodontitis through MR analyses and reveals the impact of specific gut microbiota taxa on the risk of periodontitis, thus providing new directions for the clinical intervention of periodontitis.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by Hospital of Stomatology, Guanghua School of Stomatology, Guangdong Provincial Key Laboratory of Stomatology, Sun Yat-sen University. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author contributions
SL, YL, and SW were responsible for the study conception and design. SL, WL, and QL participated in data analysis and interpretation. SL, MZ, and XW participated in figure and table drawing. SL and SW completed the manuscript writing and revision. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (82071158 to YL and 81900977 to SW).
Acknowledgments
We wish to acknowledge the researchers and participants of the GWAS used in our study and Dr. Yinong Huang for assisting in picture drawing. The icons in Figure 1 were referred from the Biorender with permission.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1160993/full#supplementary-material
References
Abusleme, L., Hoare, A., Hong, B. Y., Diaz, P. I. (2021). Microbial signatures of health, gingivitis, and periodontitis. Periodontol 2000. 86 (1), 57–78. doi: 10.1111/prd.12362
Bailen, M., Bressa, C., Martinez-Lopez, S., Gonzalez-Soltero, R., Lominchar, M. G. M., San Juan, C., et al. (2020). Microbiota features associated with a high-Fat/Low-Fiber diet in healthy adults. Front. Nutr. 7, 583608. doi: 10.3389/fnut.2020.583608
Barbe, A. G., Javadian, S., Rott, T., Scharfenberg, I., Deutscher, H. C. D., Noack, M. J., et al. (2020). Objective masticatory efficiency and subjective quality of masticatory function among patients with periodontal disease. J. Clin. Periodontol. 47 (11), 1344–1353. doi: 10.1111/jcpe.13364
Bartold, P. M. (2018). Lifestyle and periodontitis: the emergence of personalized periodontics. Periodontol 2000. 78 (1), 7–11. doi: 10.1111/prd.12237
Baumeister, S. E., Freuer, D., Nolde, M., Kocher, T., Baurecht, H., Khazaei, Y., et al. (2021). Testing the association between tobacco smoking, alcohol consumption, and risk of periodontitis: a mendelian randomization study. J. Clin. Periodontol. 48 (11), 1414–1420. doi: 10.1111/jcpe.13544
Baurecht, H., Freuer, D., Welker, C., Tsoi, L. C., Elder, J. T., Ehmke, B., et al. (2022). Relationship between periodontitis and psoriasis: a two-sample mendelian randomization study. J. Clin. Periodontol. 49 (6), 573–579. doi: 10.1111/jcpe.13620
Billings, M., Holtfreter, B., Papapanou, P. N., Mitnik, G. L., Kocher, T., Dye, B. A. (2018). Age-dependent distribution of periodontitis in two countries: findings from NHANES 2009 to 2014 and SHIP-TREND 2008 to 2012. J. Clin. Periodontol. 45, S130–SS48. doi: 10.1111/jcpe.12944
Bjorkhaug, S. T., Aanes, H., Neupane, S. P., Bramness, J. G., Malvik, S., Henriksen, C., et al. (2019). Characterization of gut microbiota composition and functions in patients with chronic alcohol overconsumption. Gut Microbes 10 (6), 663–675. doi: 10.1080/19490976.2019.1580097
Bowden, J., Smith, G. D., Burgess, S. (2015). Mendelian randomization with invalid instruments: effect estimation and bias detection through egger regression. Int. J. Epidemiol 44 (2), 512–525. doi: 10.1093/ije/dyv080
Bowden, J., Smith, G. D., Haycock, P. C., Burgess, S. (2016). Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol 40 (4), 304–314. doi: 10.1002/gepi.21965
Burgess, S., Thompson, S. G., Collaboration CCG (2011). Avoiding bias from weak instruments in mendelian randomization studies. Int. J. Epidemiol 40 (3), 755–764. doi: 10.1093/ije/dyr036
Chapple, I. L. C., Genco, R., AAP WGJE (2013). Diabetes and periodontal diseases: consensus report of the joint EFP/AAP workshop on periodontitis and systemic diseases. J. Clin. Periodontol. 40, S106–SS12. doi: 10.1902/jop.2013.1340011
Chen, X., Hong, X. S., Gao, W. J., Luo, S. L., Cai, J. H., Liu, G. C., et al. (2022). Causal relationship between physical activity, leisure sedentary behaviors and COVID-19 risk: a mendelian randomization study. J. Transl. Med. 20 (1), 216. doi: 10.1186/s12967-022-03407-6
Chen, X., Kong, J. Q., Pan, J. X., Huang, K., Zhou, W. H., Diao, X. Y., et al. (2021). Kidney damage causally affects the brain cortical structure: a mendelian randomization study. Ebiomedicine 72, 103592. doi: 10.1016/j.ebiom.2021.103592
Chen, M., Xie, C. R., Shi, Y. Z., Tang, T. C., Zheng, H. (2022). Gut microbiota and major depressive disorder: a bidirectional mendelian randomization. J. Affect. Disord. 316, 187–193. doi: 10.1016/j.jad.2022.08.012
Christensen, H., Bosse, J., Angen, O., Norskov-Lauritsen, N., Bisgaard, M. (2020). Immunological and molecular techniques used for determination of serotypes in pasteurellaceae. Method Microbiol. 47, 117–149. doi: 10.1016/bs.mim.2020.01.002
Devriese, S., Eeckhaut, V., Geirnaert, A., Van den Bossche, L., Hindryckx, P., Van de Wiele, T., et al. (2017). Reduced mucosa-associated butyricicoccus activity in patients with ulcerative colitis correlates with aberrant claudin-1 expression. J. Crohns Colitis 11 (2), 229–236. doi: 10.1093/ecco-jcc/jjw142
Dong, J. Y., Gong, Y. X., Chu, T. D., Wu, L. X., Li, S. S., Deng, H., et al. (2022). Mendelian randomization highlights the causal association of obesity with periodontal diseases. J. Clin. Periodontol. 49 (7), 662–671. doi: 10.1111/jcpe.13640
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308 (5728), 1635–1638. doi: 10.1126/science.1110591
Emdin, C. A., Khera, A. V., Kathiresan, S. (2017). Mendelian randomization. JAMA 318 (19), 1925–1926. doi: 10.1001/jama.2017.17219
Genco, R. J., Sanz, M. (2020). Clinical and public health implications of periodontal and systemic diseases: an overview. Periodontol 2000. 83 (1), 7–13. doi: 10.1111/prd.12344
Hajishengallis, G., Chavakis, T., Lambris, J. D. (2020). Current understanding of periodontal disease pathogenesis and targets for host-modulation therapy. Periodontol 2000. 84 (1), 14–34. doi: 10.1111/prd.12331
Hajishengallis, G., Liang, S., Payne, M. A., Hashim, A., Jotwani, R., Eskan, M. A., et al. (2011). Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10 (5), 497–506. doi: 10.1016/j.chom.2011.10.006
Heo, H., Bae, J. H., Amano, A., Park, T., Choi, Y. H. (2022). Supplemental or dietary intake of omega-3 fatty acids for the treatment of periodontitis: a meta-analysis. J. Clin. Periodontol. 49 (4), 362–377. doi: 10.1111/jcpe.13603
Higbee, D. H., Granell, R., Sanderson, E., Davey Smith, G., Dodd, J. W. (2021). Lung function and cardiovascular disease: a two-sample mendelian randomisation study. Eur. Respir. J. 58 (3), 2003196. doi: 10.1183/13993003.03196-2020
Hu, Z., Zhou, F. X., Xu, H. L. (2022). Circulating vitamin c and d concentrations and risk of dental caries and periodontitis: a mendelian randomization study. J. Clin. Periodontol. 49 (4), 335–344. doi: 10.1111/jcpe.13598
Janakiram, C., Dye, B. A. (2020). A public health approach for prevention of periodontal disease. Periodontol 2000. 84 (1), 202–214. doi: 10.1111/prd.12337
Jia, X., Jia, L., Mo, L., Yuan, S., Zheng, X., He, J., et al. (2019). Berberine ameliorates periodontal bone loss by regulating gut microbiota. J. Dent. Res. 98 (1), 107–116. doi: 10.1177/0022034518797275
Kamat, M. A., Blackshaw, J. A., Young, R., Surendran, P., Burgess, S., Danesh, J., et al. (2019). PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics 35 (22), 4851–4853. doi: 10.1093/bioinformatics/btz469
Kelly, N., El Karim, I. (2020). Periodontitis may be associated with respiratory diseases such as asthma, copd, and pneumonia. J. Evid-Based Dent. Pr. 20 (4), 101498. doi: 10.1016/j.jebdp.2020.101498
Kurilshikov, A., Medina-Gomez, C., Bacigalupe, R., Radjabzadeh, D., Wang, J., Demirkan, A., et al. (2021). Large-Scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 53 (2), 156–165. doi: 10.1038/s41588-020-00763-1
Li, L., Bao, J., Chang, Y., Wang, M., Chen, B., Yan, F. (2021). Gut microbiota may mediate the influence of periodontitis on prediabetes. J. Dent. Res. 100 (12), 1387–1396. doi: 10.1177/00220345211009449
Ni, J. J., Xu, Q., Yan, S. S., Han, B. X., Zhang, H., Wei, X. T., et al. (2022). Gut microbiota and psychiatric disorders: a two-sample mendelian randomization study. Front. Microbiol. 12, 4178. doi: 10.3389/fmicb.2021.737197
Ormerod, K. L., Wood, D. L. A., Lachner, N., Gellatly, S. L., Daly, J. N., Parsons, J. D., et al. (2016). Genomic characterization of the uncultured bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome 4, 36. doi: 10.1186/s40168-016-0181-2
Page, R. C., Eke, P. I. (2007). Case definitions for use in population - based surveillance of periodontitis. J. Periodontol. 78 (7), 1387–1399. doi: 10.1902/jop.2007.060264
Papapanou, P. N., Sanz, M., Buduneli, N., Dietrich, T., Feres, M., Fine, D. H., et al. (2018). Periodontitis: consensus report of workgroup 2 of the 2017 world workshop on the classification of periodontal and peri-implant diseases and conditions. J. Periodontol. 89, S173–SS82. doi: 10.1002/JPER.17-0721
Purushe, J., Fouts, D. E., Morrison, M., White, B. A., Mackie, R. I., Coutinho, P. M., et al. (2010). Comparative genome analysis of prevotella ruminicola and prevotella bryantii: insights into their environmental niche. Microb. Ecol. 60 (4), 721–729. doi: 10.1007/s00248-010-9692-8
Sanna, S., van Zuydam, N. R., Mahajan, A., Kurilshikov, A., Vila, A. V., Vosa, U., et al. (2019). Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 51 (4), 600–605. doi: 10.1038/s41588-019-0350-x
Shungin, D., Hawort, S., Divaris, K., Agler, C. S., Kamatani, Y., Lee, M. K., et al. (2019). Genome-wide analysis of dental caries and periodontitis combining clinical and self-reported data. Nat. Commun. 10, 2773. doi: 10.1038/s41467-019-10630-1
Staley, J. R., Blackshaw, J., Kamat, M. A., Ellis, S., Surendran, P., Sun, B. B., et al. (2016). PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics 32 (20), 3207–3209. doi: 10.1093/bioinformatics/btw373
Tonetti, M. S., Van Dyke, T. E., Working group 1 of the joint EFPAAPw (2013). Periodontitis and atherosclerotic cardiovascular disease: consensus report of the joint EFP/AAP workshop on periodontitis and systemic diseases. J. Clin. Periodontol. 40 (Suppl 14), S24–S29. doi: 10.1111/jcpe.12089
Uchiyama, K., Naito, Y., Takagi, T. (2019). Intestinal microbiome as a novel therapeutic target for local and systemic inflammation. Pharmacol. Therapeut 199, 164–172. doi: 10.1016/j.pharmthera.2019.03.006
Veller, C., Kleckner, N., Nowak, M. A. (2019). A rigorous measure of genome-wide genetic shuffling that takes into account crossover positions and mendel’s second law. Proc. Natl. Acad. Sci. U.S.A. 116 (5), 1659–1668. doi: 10.1073/pnas.1817482116
Wang, Y. Y., Sakka, M., Yagi, H., Kaneko, S., Katsuzaki, H., Kunitake, E., et al. (2018). Ruminiclostridium josui Abf62A-Axe6A: a tri-functional xylanolytic enzyme exhibiting alpha-l-arabinofuranosidase, endoxylanase, and acetylxylan esterase activities. Enzyme Microb. Tech 117, 1–8. doi: 10.1016/j.enzmictec.2018.05.016
Watson, S., Woodside, J. V., Winning, L., Wright, D. M., Srinivasan, M., McKenna, G. (2022). Associations between self-reported periodontal disease and nutrient intakes and nutrient-based dietary patterns in the UK biobank. J. Clin. Periodontol. 49 (5), 428–438. doi: 10.1111/jcpe.13604
Xiao, S. W., Liu, C., Chen, M. J., Zou, J. F., Zhang, Z. M., Cui, X., et al. (2020). Scutellariae radix and coptidis rhizoma ameliorate glycolipid metabolism of type 2 diabetic rats by modulating gut microbiota and its metabolites. Appl. Microbiol. Biot 104 (1), 303–317. doi: 10.1007/s00253-019-10174-w
Xu, Q., Ni, J. J., Han, B. X., Yan, S. S., Wei, X. T., Feng, G. J., et al. (2022). Causal relationship between gut microbiota and autoimmune diseases: a two-sample mendelian randomization study. Front. Immunol. 12. doi: 10.3389/fimmu.2021.746998
Yan, J., Herzog, J. W., Tsang, K., Brennan, C. A., Bower, M. A., Garrett, W. S., et al. (2016). Gut microbiota induce IGF-1 and promote bone formation and growth. P Natl. Acad. Sci. U.S.A. 113 (47), E7554–E7E63. doi: 10.1073/pnas.1607235113
Yavorska, O. O., Burgess, S. (2017). MendelianRandomization: an r package for performing mendelian randomization analyses using summarized data. Int. J. Epidemiol 46 (6), 1734–1739. doi: 10.1093/ije/dyx034
Yin, K. J., Huang, J. X., Wang, P., Yang, X. K., Tao, S. S., Li, H. M., et al. (2022). No genetic causal association between periodontitis and arthritis: a bidirectional two-sample mendelian randomization analysis. Front. Immunol. 13, 808832. doi: 10.3389/fimmu.2022.808832
Zhou, H., Liu, J., Zhang, Y., Fang, W., Yang, Y., Hong, S., et al. (2019). Gut microbiota and lung cancer: a mendelian randomisation study. J. Thorac. Oncol. 14 (10), S510–S5S1. doi: 10.1016/j.jtocrr.2020.100042
Keywords: periodontitis, gut microbiota, Mendelian randomization, causal effect, risk factor
Citation: Luo S, Li W, Li Q, Zhang M, Wang X, Wu S and Li Y (2023) Causal effects of gut microbiota on the risk of periodontitis: a two-sample Mendelian randomization study. Front. Cell. Infect. Microbiol. 13:1160993. doi: 10.3389/fcimb.2023.1160993
Received: 10 February 2023; Accepted: 16 May 2023;
Published: 25 May 2023.
Edited by:
Carlo Contini, University of Ferrara, ItalyReviewed by:
Hengyi Xu, The University of Texas at Austin, United StatesJiahao Cai, Guangzhou Medical University, China
Copyright © 2023 Luo, Li, Li, Zhang, Wang, Wu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuyi Wu, d3VzaHV5aUBtYWlsLnN5c3UuZWR1LmNu; Yan Li, bGl5OEBtYWlsLnN5c3UuZWR1LmNu
 Shulu Luo
Shulu Luo Yan Li
Yan Li