- 1Department of Biology, Muhlenberg College, Allentown, PA, United States
- 2Biofilm Research Laboratories, Levy Center for Oral Health, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA, United States
- 3Department of Preventive and Restorative Sciences, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA, United States
- 4School of Dental Medicine, University of Pennsylvania, Philadelphia, PA, United States
- 5Department of Orthodontics, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA, United States
- 6Center for Innovation & Precision Dentistry, School of Dental Medicine and School of Engineering & Applied Sciences, University of Pennsylvania, Philadelphia, PA, United States
- 7David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA, United States
Orofacial cleft disorders, including cleft lip and/or palate (CL/P), are one of the most frequently-occurring congenital disorders worldwide. The health issues of patients with CL/P encompass far more than just their anatomic anomaly, as patients with CL/P are prone to having a high incidence of infectious diseases. While it has been previously established that the oral microbiome of patients with CL/P differs from that of unaffected patients, the exact nature of this variance, including the relevant bacterial species, has not been fully elucidated; likewise, examination of anatomic locations besides the cleft site has been neglected. Here, we intended to provide a comprehensive review to highlight the significant microbiota differences between CL/P patients and healthy subjects in various anatomic locations, including the teeth inside and adjacent to the cleft, oral cavity, nasal cavity, pharynx, and ear, as well as bodily fluids, secretions, and excretions. A number of bacterial and fungal species that have been proven to be pathogenic were found to be prevalently and/or specifically detected in CL/P patients, which can benefit the development of CL/P-specific microbiota management strategies.
1 Introduction
Fusion of particular orofacial structures during early gestation is required for proper development of the upper lip and jaw (Hammond and Dixon, 2022). Failure of this process results in an orofacial cleft, which manifests as a gap in the tissue of the upper lip, the palate, or both (Mossey et al., 2009). Affecting around 1 out of 700 live births, cleft lip and/or palate (CL/P) is one of the most common congenital craniofacial disorders (Candotto et al., 2019).
CL/P significantly reduces patients’ quality of life (Mossey et al., 2009). Firstly, some patients have an abnormal nasal bone structure and shape (Tibesar et al., 2009), which may be accompanied by malformations of oral muscles (Marazita, 2007; Li et al., 2019). Thus, CL/P infants may not feed properly after birth due to impaired aspiration and deglutition (Duarte et al., 2016), hindering their overall growth and development (Bessell et al., 2011). In addition, oral muscle maldevelopment may lead to infection of the Eustachian tubes and subsequent deafness, contributing to communication difficulties (Vyas et al., 2020). Consequently, verbal communication is specifically challenging for CL/P patients, and their interpersonal relations can be damaged by their speech and outward presentation (Mossey et al., 2009). Moreover, people born with CL/P may continue to have a poor impression of their face and desire to alter certain aspects of their appearance even as adults (Meyer-Marcotty and Stellzig-Eisenhauer, 2009).
Unsurprisingly, various dental conditions, such as enamel hypoplasia, asymmetrical development of the dentition, and microdontia, are also associated with CL/P (Van Dyck et al., 2019; Vyas et al., 2020). Noticeably, CL/P-associated dental health issues encompass far more than the anatomic anomaly itself. For example, CL/P patients may experience delayed tooth development or eruption, particularly of the primary molars, generally by half a year compared to their non-CL/P peers (Van Dyck et al., 2019; Vyas et al., 2020). Moreover, compared to their healthy peers, children with CL/P present much more severe signs of poor oral health, develop dental caries at a higher rate, and have a higher risk of Eustachian tube infections (Bessell et al., 2011; Zhang et al., 2016; Rodrigues et al., 2019), all of which are associated with bacterial infections. Furthermore, CL/P patients harbor different oral microbiota compared to individuals with normal orofacial development (Zhang et al., 2016; Funahashi et al., 2019), which may potentially be attributed to the presence of an oronasal fistula that allows bacterial transmission from the nose to the mouth (Tuna et al., 2008). Another potential cause of the aberrant oral microbiomes seen in CL/P patients is the existence of a cleft (even after surgical correction, a residual cleft may exist) that markedly increases the difficulty of toothbrushing, possibly due to inaccessibility of the cleft region or unwillingness to brush because of a misconception that toothbrushing risks damaging the repaired area (Tuna et al., 2008; Rodrigues et al., 2019). In any case, the aberrant oral microbiomes that result from the development and surgical correction of CL/P can, in turn, have significant implications for the oral health of CL/P patients as well as the bacterial composition and condition of other anatomic sites (Tuna et al., 2008; Zhang et al., 2016; Funahashi et al., 2019). This review aims to further elucidate the different microbiota in various niches that differ between CL/P patients and healthy persons based on currently available publications, to benefit the development of CL/P-specific microbiota management strategies.
2 Original article searching and selection
To include all the available original studies on CL/P-related microbiomes, this study was conducted following the 2020 Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. The article search was carried out in August 2022. The following keywords were entered into the PubMed database: “cleft palate,” “cleft lip,” “biofilm,” “microbiology,” “microbiome,” “bacteria,” “fungal,” and “virus.” Although no publication date limits were initially set, articles published before 1980 were not included. Reviews, systematic reviews, and meta-analyses were excluded to avoid double-counting. Only publications that were written in English and examined human subjects were included. The publications were cataloged based on the locations of the microbiota reported and the different species found between the biofilms of CL/P patients and those of non-CL/P subjects.
3 Results
3.1 Bacterial species
3.1.1 Teeth inside and adjacent to the oral cleft
Given the increased prevalence of dental caries in CL/P-afflicted children (Grewcock et al., 2022), it is of particular importance to determine how the oral microbiota of those with CL/P differs from those without this condition and which bacterial species, particularly cariogenic species, may be associated with the plaque-biofilms formed on the teeth of these patients.
Prevotella is the most common genus differentially detected in the plaque-biofilms isolated from the teeth inside the cleft and teeth adjacent to the cleft site of CL/P patients when compared to those obtained from the teeth of healthy subjects (Table 1 and Supplementary Tables S1, S2). In particular, it has been reported that P. marshii, P. micans, P. nigrescens, P. pallens, and P. pleuritidis are prone to CL/P, while other two species, P. melaninogenica and P. oralis (Perdikogianni et al., 2009; Funahashi et al., 2019), have been reported to be prone to a non-CL/P control group over the CL/P group (Table 1). Notably, there were discrepancies among associations between Prevotella species and the CL/P condition across publications. For example, when assessing the bacteria on all teeth, Funahashi et al. suggested that P. loeschii was specific to non-CL/P subjects (Funahashi et al., 2019). However, when comparing the cleft-adjacent teeth of CL/P patients with the respective incisors and canines of healthy controls, Perdikogianni et al. found no significant difference in colony counts of P. loeschii between the CL/P and control groups (Perdikogianni et al., 2009). A possible explanation is that the distribution of P. loeschii is strikingly location-specified and closely related to the cleft, which needs to be validated through further investigation.
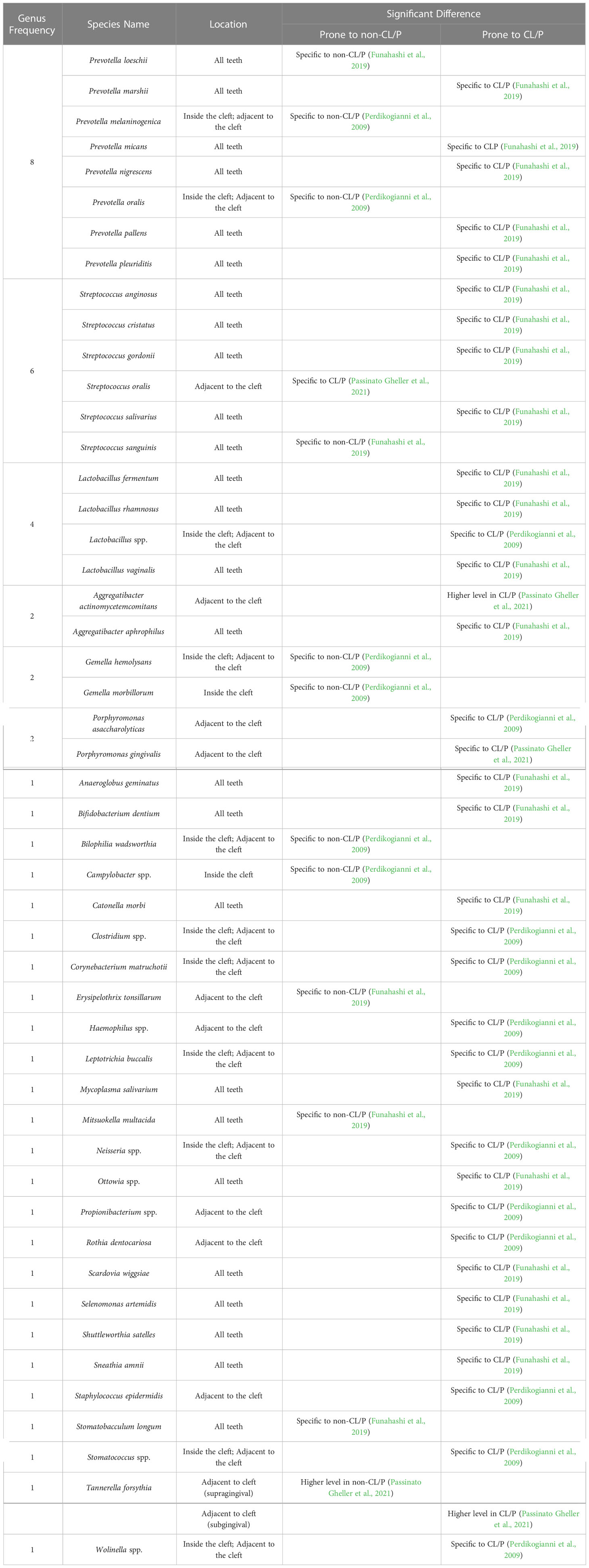
Table 1 Significant differences in bacterial species obtained from the teeth of CL/P and non-CL/P patients.
Previous studies also demonstrate that Streptococcus and Lactobacillus are the next-most common microbiota residing on the teeth that are specific to the CL/P or non-CL/P conditions (Perdikogianni et al., 2009; Funahashi et al., 2019; Passinato Gheller et al., 2021). In regard to Streptococcus, only S. anginosus, S. cristatus, S. gordonii, and S. salivarius were found to be associated with CL/P, while S. oralis and S. sanguinis were associated with non-CL/P (Funahashi et al., 2019; Passinato Gheller et al., 2021). In contrast, there was no significant difference in S. intermedius (Perdikogianni et al., 2009; Funahashi et al., 2019), S. mutans (Lucas et al., 2000), and Streptococcus spp. prevalence (Perdikogianni et al., 2009) between these two groups. The Lactobacillus species found to be more prevalent in CL/P are L. fermentum, L. rhamnosum, and L. vaginalis (Perdikogianni et al., 2009; Funahashi et al., 2019).
Other notable pathogenic microbiota in the biofilms of teeth adjacent to the cleft associated with CL/P were Aggregatibacter actinomycetemcomitans (Passinato Gheller et al., 2021), Clostridium spp. (Perdikogianni et al., 2009), Staphylococcus epidermidis (Perdikogianni et al., 2009), Tannerella forsythia (Passinato Gheller et al., 2021), and Neisseria spp. (Perdikogianni et al., 2009). Interestingly, Gheller et al. (Passinato Gheller et al., 2021) found Porphyromonas gingivalis to be present at a higher level in both the supragingival and subgingival plaque-biofilms on teeth adjacent to the cleft in CL/P patients compared to those of the non-CL/P group (Table 1). However, Perdikogianni et al. (Perdikogianni et al., 2009) found no significant difference in P. gingivalis incidence between the CL/P and control groups when sampling subgingival plaque from the teeth in and adjacent to the cleft (Table 1), again suggesting the location specificity of CL/P-related oral bacteria.
3.1.2 Oral, nasal, pharyngeal, and ear mucosa
Next, we further summarize the microbiota composition of mucosal surfaces. The relevant anatomic regions include the mucosa in the oral region (such as the palatal cleft site, entire palate, sublingual mucosa, and dorsum of the tongue) (Supplementary Table S3), and the mucosa in non-oral regions (such as the nasal mucosa, throat, oro-nasopharynx, perineum, and ear) (Supplementary Table S4); in some studies, samples from both areas are combined (Supplementary Table S5). Although a great amount of research has been conducted on the mucosal microbiota of CL/P subjects (Mombelli et al., 1992; Machorowska-Pieniążek et al., 2017; Roode et al., 2017; Roode and Bütow, 2018; Ramdial and Madaree, 2019; Iurovschi et al., 2020), few reports compared samples from CL/P patients with those from non-CL/P subjects.
3.1.2.1 The oral mucosa
Streptococcus is by far the most common genus identified in CL/P patients’ oral mucosa samples (Supplementary Table S3). Both Rodrigues et al. and Zhang et al. found Streptococcus in approximately one third of the CL/P patients they respectively studied (Rodrigues et al., 2021; Zhang et al., 2022). A study by Machorowska-Pieniążek et al. detected Streptococcus species—specifically S. bovis biovar I, S. salivarius, and S. sanguinis—in 20-40% of the microbiota of their CL/P patients and likewise found S. mitis to be highly prevalent (Machorowska-Pieniążek et al., 2017).
Staphylococcus is the second-most-frequent genus distributed on oral mucosa based on currently available reports (Supplementary Table S3). Studies examining the cleft, dorsum of the tongue (Machorowska-Pieniążek et al., 2017), and sublingual surface (Cocco et al., 2010) agreed that S. aureus frequently appears in these mucosal regions of CL/P patients. Machorowska-Pieniążek et al. also reported that S. epidermidis occurred in around a third of neonates with CL/P and more than 80% of CL/P-afflicted infants (Machorowska-Pieniążek et al., 2017); however, other studies did not find this species to occur very frequently in patients with this condition (Cocco et al., 2010).
Several other species have also been reported as frequently occurring in CL/P subjects or occurring at high levels in oral mucosa samples from these patients (Supplementary Table S3). For example, examination of cleft samples by Iurovschi et al. revealed that P. melaninogenica, P. nigrescens, and, to a lesser degree, S. mitis and Enterobacter aerogenes, had higher mean colony counts than any other species (Iurovschi et al., 2020). Multiple studies have determined Klebsiella pneumoniae to be moderately prevalent in CL/P patients (Cocco et al., 2010; Machorowska-Pieniążek et al., 2017). Previous research has also identified Actinomyces viscous to be moderately prevalent and Gemella morbillorum and Veillonella spp. to be prevalent in the oral mucosa of CL/P patients (Mombelli et al., 1992; Machorowska-Pieniążek et al., 2017). At the phylum level, Rodrigues et al. and Zhang et al. reported Firmicutes as occurring moderately frequently, at a greater level than any other phylum, with Zhang et al. additionally identifying Proteobacteria as being less prevalent (Rodrigues et al., 2021; Zhang et al., 2022).
Interestingly, Cocco et al. collected data on CL/P patients and patients with isolated cleft lip (Cocco et al., 2010), and demonstrated that although K. pneumoniae was separately detected in the sublingual environment and oropharynx of more than half of the CL/P patients pre-operatively, only one of the ten isolated cleft lip patients tested positive for this species in these regions (Cocco et al., 2010). Likewise, methicillin-susceptible S. aureus (MSSA) was identified in nearly a quarter of the CL/P patients’ sublingual specimens but was not detected in any isolated cleft lip patients, suggesting that there were substantial differences in the microbiota composition in patients with cleft lip, cleft palate, or both (Cocco et al., 2010).
3.1.2.2 The mucosa of the ear, nose, and throat/pharynx
Little research has been done on the microbiota of CL/P patients’ ear mucosa. A relevant study by Chuo et al. reported that S. aureus occurred at a low frequency in the ear mucosa of patients with CL/P (Chuo and Timmons, 2005). In contrast, the products of ear infections, specifically otitis media secretions, have been evaluated by a greater number of studies, the results of which are discussed in Section 3.1.3.2.
Although Staphylococcus and Streptococcus are the two most-commonly detected genera occurring in the isolated nasal mucosa across the literature, only one publication (Zhang et al., 2016) has compared the prevalence of either genus in subjects with or without CL/P to date (Supplementary Table S4). This study by Zhang et al. found that while both genera were highly prevalent in CL/P patients, only Streptococcus spp. was found to be associated with the CL/P condition (Zhang et al., 2016). In comparison, other studies reported that S. aureus (Tuna et al., 2008), including MSSA (Cocco et al., 2010), is only moderately prevalent in CL/P patients’ nasal mucosa, with K. pneumoniae and S. epidermidis being less prevalent than this species (Cocco et al., 2010). In addition, Corynebacterium spp., Dolosigranulum spp., and Moraxella catarrhalis were reported as moderately prevalent, and Gemmella spp. and Neisseria spp. as less prevalent (Zhang et al., 2016). Interestingly, the genera Bacillus and Dolosigranulum were detected at a significantly lower frequency in the nasal microbiota of CL/P children than in healthy subjects (Table 2).
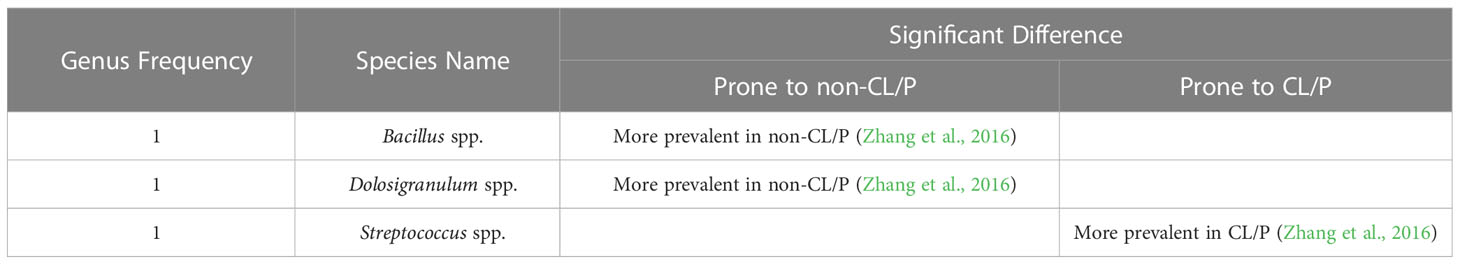
Table 2 Significant differences in bacterial species obtained from the nasal mucosa of CL/P and non-CL/P patients.
To date, only two studies have evaluated the bacterial microbiota of the isolated throat/oropharynx microbiota of CL/P patients; of these, Cocco et al. (Cocco et al., 2010) determined MSSA was prevalent and K. pneumoniae was moderately prevalent (Supplementary Table S4). Interestingly, in a study by Bos et al. examining CL/P patients who tested positive for methicillin-resistant S. aureus (MRSA), 7% of subjects were positive for this bacteria in the throat only (Bos et al., 2016). A much greater proportion of patients were found to have this bacteria in both their throat mucosa and other mucosal membranes (Bos et al., 2016), indicating the importance of evaluating the microbiota of multiple regions of the mucosa.
Many studies (Chuo and Timmons, 2005; Mÿburgh and Bütow, 2009; Narinesingh et al., 2011; Thomas et al., 2012; Bos et al., 2016; Roode et al., 2017; Roode and Bütow, 2018; Ramdial and Madaree, 2019; Roode et al., 2022) have examined the pharyngeal mucosa in combination with that of the ear, nose, and the palate or palatal cleft (Supplementary Table S5). Two publications (Narinesingh et al., 2011; Thomas et al., 2012) evaluating nasal and/or pharyngeal samples and one publication (Chuo and Timmons, 2005) evaluating ear, nose, and throat mucosa samples reported S. aureus as prevalent in CL/P subjects. In contrast, a study that solely sampled the throat did not find S. aureus to be prevalent (Rennie et al., 2009). Two studies done by Roode et al. in 2018 and 2022 evaluating the nasopharynx and the palatal cleft or cleft palate, respectively, reported a high prevalence of S. mitis and S. oralis combined (Roode and Bütow, 2018; Roode et al., 2022). Other studies sampled from part or all of the palate and the nasopharynx or oro-nasopharynx have identified H. influenzae, S. aureus, and S. viridans as being prevalent and K. pneumoniae as being moderately prevalent at these sites (Mÿburgh and Bütow, 2009; Roode et al., 2017; Roode and Bütow, 2018; Ramdial and Madaree, 2019; Roode et al., 2022). Interestingly, Narinesingh et al. noticed an association between Moraxella catarrhalis infection and oronasal fistula formation in CL/P patients; however, this species was far from prevalent in the nasal mucosa of the study’s subjects (Narinesingh et al., 2011).
3.1.3 Bodily fluids, secretions, and excretions
3.1.3.1 Saliva
Saliva has multiple constituents and physicochemical properties crucial for maintaining oral health. For example, it protects the teeth and mucosa and plays an important role in maintaining balanced microbiota (Pedersen et al., 2018). While multiple studies have evaluated S. mutans and Lactobacillus in the saliva, most (Bokhout et al., 1996; de Soet et al., 1998; van Loveren et al., 1998; Arief et al., 2005; Cheng et al., 2007; Parapanisiou et al., 2009; Antoszewska et al., 2010; Cildir et al., 2012; Ritthagol et al., 2014; Sundell et al., 2015; Sundell et al., 2018; Durhan et al., 2019; Hassani et al., 2020; Chaudhari et al., 2021) have not reported a significantly greater or lesser incidence of either species in CL/P subjects compared to healthy ones (Supplementary Table S6). However, Shashni et al. (Shashni et al., 2015) found higher S. mutans counts in CL/P children with no dental caries than in non-CL/P children without caries (Table 3). Durhan et al. (Durhan et al., 2019) found no association between CL/P and prevalence of S. mutans, but reported that Lactobacillus was more prevalent in CL/P subjects than in healthy ones. Likewise, in 2015, Sundell et al. (Sundell et al., 2015) reported that Lactobacillus was more prevalent in CL/P patients than in control subjects. Surprisingly, another study conducted in 2018 by the same group (Sundell et al., 2018) found no significant difference in the occurrence of Lactobacillus and S. mutans; however, other Streptococcus species (including S. gordonii, S. mitis, and S. salivarius), Bifidobacterium dentium, Fusobacterium nucleatum, and Veillonella parvula were reported to be significantly less prevalent in CL/P patients (Table 3). Other bacterial genera associated with the non-CL/P condition have been reported to be Bacillus and Lautropia (Zhang et al., 2016), while S. aureus has been found to be more prevalent in CL/P patients (Arief et al., 2005).
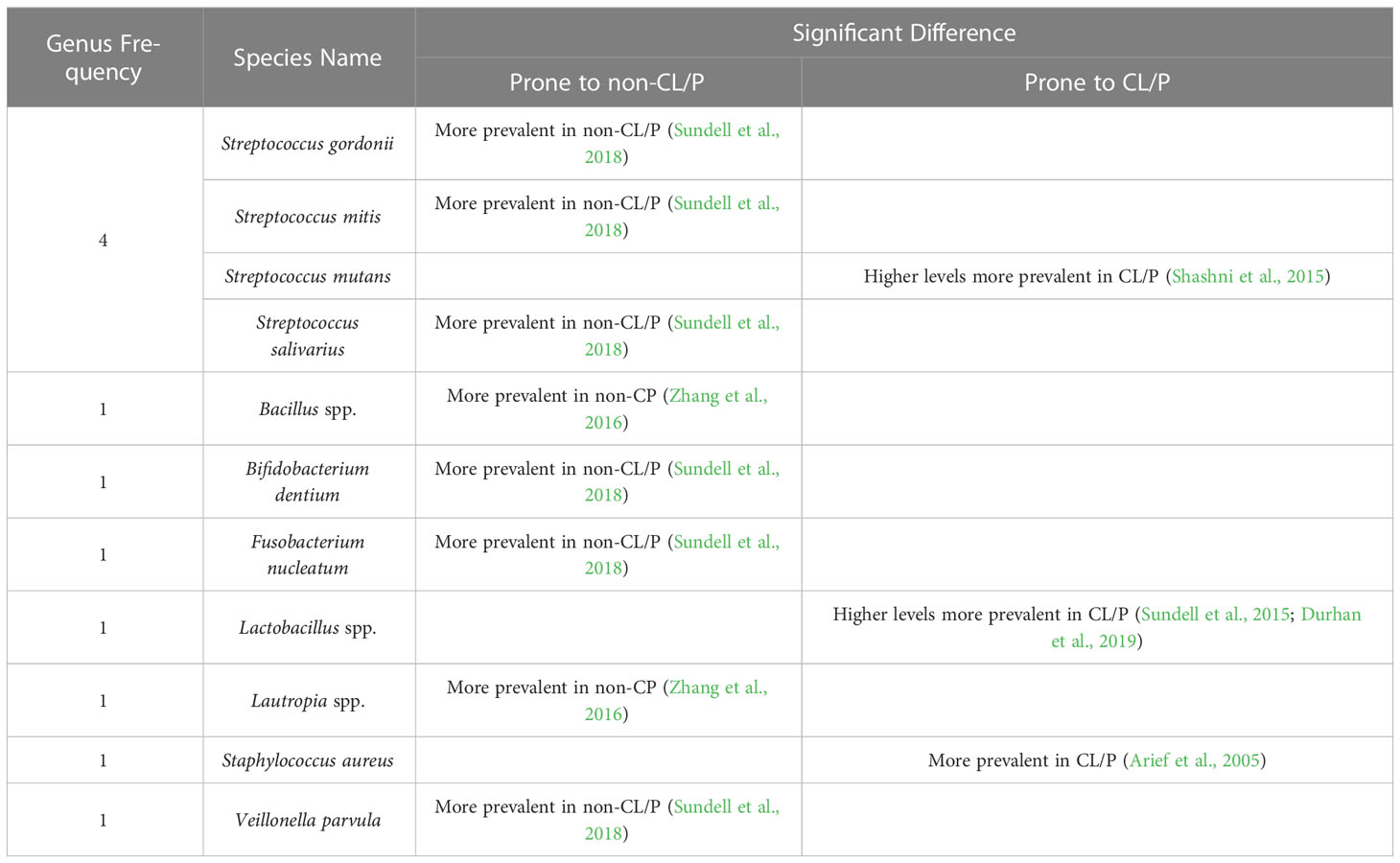
Table 3 Significant differences in bacterial species obtained from the saliva of CL/P and non-CL/P patients.
3.1.3.2 Other bodily fluids, secretions, and excretions
Little research has been done to compare the bacterial compositions of the blood, feces, and otitis media secretions between CL/P and healthy populations (Supplemental Table S6). However, studies solely examining CL/P patients have identified bacterial species as prevalent in these fluids. For example, Adeyemo et al. reported that coagulase-negative Staphylococcus was by far the most commonly-occurring bacteria in the blood of CL/P patients shortly after surgery, with over a third of the subjects testing positive for this species (Adeyemo et al., 2013). Other species, such as coagulase-positive S. aureus, E. coli, and E. cloacae, only occurred in a small percentage of these patients (Adeyemo et al., 2013). Meanwhile, Vieira et al. studied the presence of Bacteroides spp., Bifidobacterium spp., and Lactobacillus spp. in the feces of CL/P patients before and after surgical revision but did not evaluate their prevalence (Vieira et al., 2013). No doubt, further research is warranted to gain a more comprehensive understanding of the bacteria found in the blood and feces of CL/P patients compared to subjects without CL/P.
In comparison with the dearth of studies on the bacterial species in the blood and feces of CL/P patients, more research has been done on otitis media secretions (Supplementary Table S6). Weckworth et al. reported that Pseudomonas aeruginosa was somewhat prevalent in otitis media secretions, while other species, such as S. aureus, E. faecalis, and Proteus mirabilis, occurred at a lower level of frequency (Weckwerth et al., 2009). A later study by the same group found P. aeruginosa to be the most common species and moderately prevalent, and S. aureus to also be prevalent (Weckwerth et al., 2014). Interestingly, when PCR analysis was utilized, the most prevalent species appeared to be F. nucleatum, in nearly 40% of samples (Weckwerth et al., 2014). These studies are in contrast to an earlier publication by Jousimies-Somer and Rintala, in which M. catarrhalis, S. epidermidis, and Streptococcus pneumoniae (but not S. aureus) were found in most CL/P infants and adolescents (Jousimies-Somer et al., 1986). While these previous investigations do contribute to current knowledge of which bacterial genera and species are prevalent in the secretions of young CL/P patients, research directly comparing CL/P patients with their healthy counterparts must be conducted to gain a more informed understanding of how the microbiota in otitis media secretions differ between these two groups.
3.1.4 Bacterial species across all sites
More limited research has been done on the bacterial phyla in the tissues and fluids of CL/P patients (Supplementary Table S7). However, previous studies have reported Firmicutes to be moderately prevalent in the cleft site (Rodrigues et al., 2021) and either moderately prevalent (Zhang et al., 2022) or somewhat prevalent (Liu et al., 2016) in the saliva; Proteobacteria has also been found to be somewhat prevalent in the saliva (Liu et al., 2016; Zhang et al., 2022).
We also noticed that the association of certain bacterial species with the CL/P or non-CL/P condition varies among different anatomic locations. Indeed, one study may report a given species as being CL/P-specific in one location, while another might find the same species to be associated with the non-CL/P condition in another location (Table 4). As such, it is important to further evaluate these differences and better understand the relationship of these species with CL/P.
3.2 Fungi
Candida species typically form colonies in the mouth, digestive tract, skin, and vagina; excessive growth of these colonies may result in fungal infections that can lead to hospitalization or even death (Singh et al., 2020). Candida albicans is particularly pathogenic and has great capability for forming biofilms (Poulain, 2015; Ponde et al., 2021). Thus, examining the prevalence of C. albicans in the oral flora of CL/P patients is critical for understanding the nontypical nature of CL/P oral biofilms (Table 5).
Previous research sampling various oral mucosae has reported Candida spp. as being highly prevalent in CL/P patients (Rawashdeh et al., 2011; Boriollo et al., 2022; de Souza et al., 2022), although this association was disputed in a study by Durhan et al., in which the saliva was evaluated (Durhan et al., 2019). Other studies also reported this fungus as being moderately prevalent (Durhan et al., 2019), somewhat prevalent (Silva et al., 2018), and not prevalent in CL/P patients (Yilmaz et al., 2020). It is worth noting that although C. albicans is by far the most commonly-investigated fungal species in studies examining the oral environment of CL/P patients, the prevalence of C. albicans in CL/P patients varies across studies, ranging from less than 10% to as high as 70% (Mattos et al., 2009; Rawashdeh et al., 2011; Machorowska-Pieniążek et al., 2017; Yilmaz et al., 2020; Boriollo et al., 2022). Interestingly, Boriollo et al. reported that colonization by non-albicans Candida species only (namely, Candida krusei and Candida tropicalis) was more prevalent in CL/P patients than in the control (Boriollo et al., 2022). On the contrary, Rawashdeh et al. (Rawashdeh et al., 2011) reported that C. kefyr was non-significantly more prevalent in healthy subjects who tested positive for Candida species compared to their CL/P counterparts (Supplementary Table S8).
The prevalence of different Candida species in the same location also varies. Silva et al. detected Candida spp. in the cleft site in 40% of CL/P patients, while the prevalence of C. albicans in the same site was only around 15% (Silva et al., 2018). In comparison, other Candida species, such as C. krusei and C. tropicalis, were found at even lower levels in the cleft (Silva et al., 2018). Similarly, Roode and Bütow reported that nearly a third of the CL/P subjects harbored C. albicans at the soft palate margin and the nasopharyngeal mucosa, while C. tropicalis and C. krusei were much less prevalent at these locations (Roode and Bütow, 2018).
3.3 Viruses
The association between viral infection and CL/P has also been evaluated (Cerný et al., 1991; Acs et al., 2005; Métneki et al., 2005; Divya et al., 2017; Ács et al., 2020); however, there is no available data to demonstrate if any viruses participate in CL/P-specific oral or nasal biofilm formation (Supplementary Table S9).
4 Discussion
The current review summarized the significant differences between the microbiota of those with and without CL/P. Many CL/P-prone microbes, whether specific to CL/P patients or more prevalent in CL/P patients than in their healthy counterparts, have been implicated in oral health and disease (Figure 1). For example, there are a multitude of CL/P-prone microbes associated with caries, including Bifidobacterium dentium, Lactobacillus spp., a Neisseria sp., Rothia dentocariosa, Scardovia wiggsiae, Streptococcus cristatus, Streptococcus mutans, and Streptococcus salivarius (Kressirer et al., 2017; Inquimbert et al., 2019; Xiao et al., 2021), while Anaeroglobus geminatus and Shuttleworthia satelles have been found to be more prevalent in patients with caries (Wang et al., 2019). Indeed, several CL/P-prone microbes, such as P. marshii, P. micans, S. wiggsiae, Selenomonas artemidis, and S. satelles, produce acidic byproducts from sugar metabolism, contributing to the development of an acidic oral environment, and, potentially, caries (Moore et al., 1987; Downes et al., 2002; Downes et al., 2005; Downes et al., 2009; Kressirer et al., 2017). Moreover, Catonella morbi has been implicated in a more serious affliction of the teeth, primary endodontic infections, while Propionibacterium has been implicated in secondary endodontic infections and lesions (Siqueira and Rôças, 2006; Dioguardi et al., 2020).
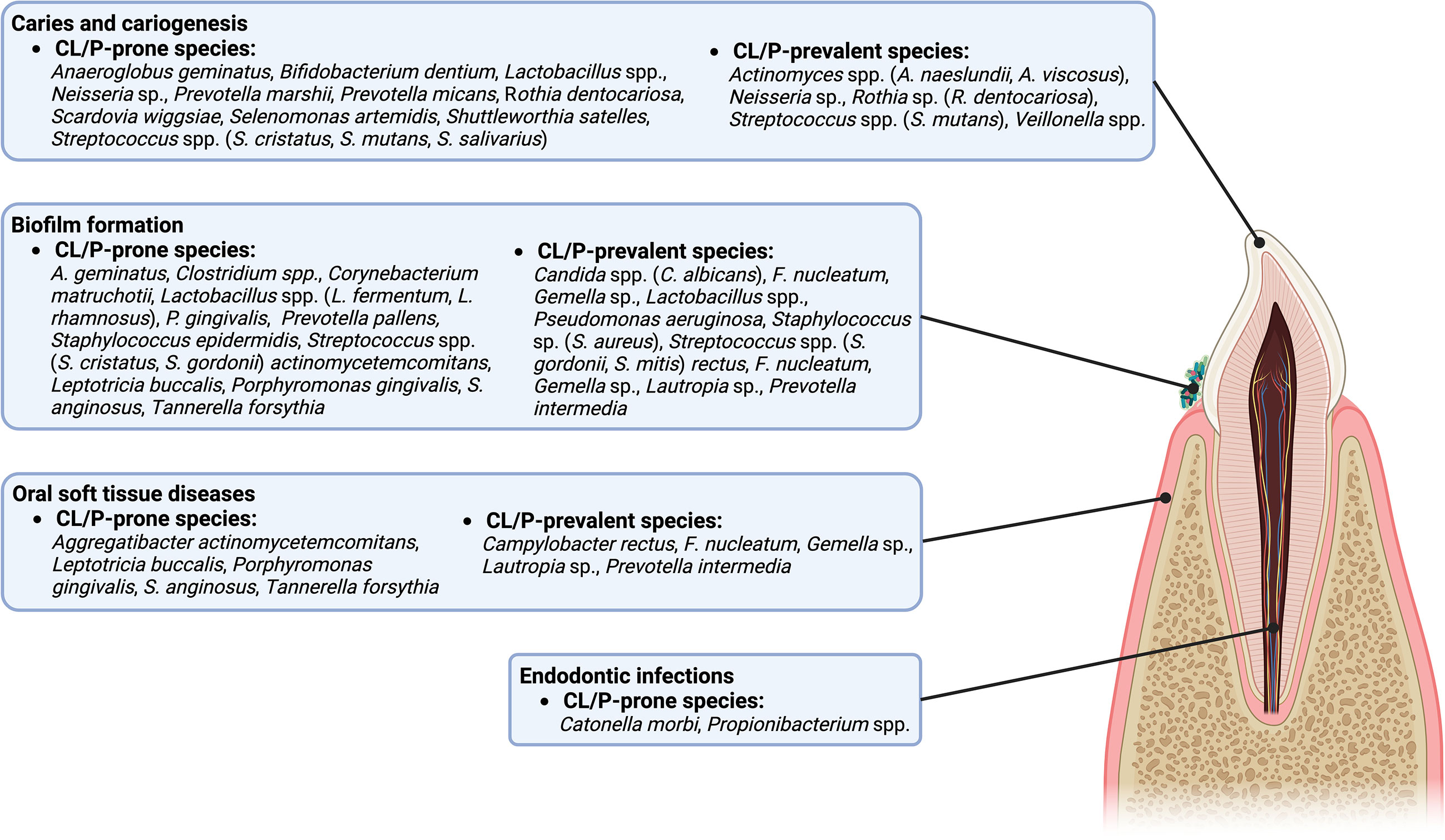
Figure 1 A diagram of the intraoral anatomic structures that are potentially influenced by CL/P-prone and CL/P-prevalent species.
CL/P-prone microbes have also been associated with oral inflammatory diseases and soft tissue destruction (Figure 1). For example, a link between gingivitis and the CL/P-prone bacteria T. forsythia (Sharma, 2010) has been reported. Meanwhile, associations between species such as A. actinomycetemcomitans, P. gingivalis, S. anginosus, and T. forsythia and periodontitis have been reported (Socransky et al., 1998; Kumagai et al., 2003; Raja et al., 2014; How et al., 2016). It is possible that L. buccalis may also be associated with one or more maladies of the oral soft tissue (Eribe and Olsen, 2017).
The CL/P-prone microbes that contribute to biofilm formation and development are particularly interesting (Figure 1). Several bacteria, such as Clostridium, Corynebacterium matruchotii, P. gingivalis, S. epidermidis, and S. gordonii are known to form biofilms or promote biofilm formation on tooth surfaces (Otto, 2009; Pantaléon et al., 2014; Gerits et al., 2017; Rath et al., 2017; Esberg et al., 2020), while P. pallens is recognized as an early colonizer (Könönen and Gursoy, 2021). Candida spp. colonization also promotes the formation of biofilms (Cavalheiro and Teixeira, 2018), potentially affecting the oral health through the development of thrush (also known as pseudomembranous candidiasis) (Akpan and Morgan, 2002). Interestingly, some microbes work in conjunction with others to enhance the formation of oral biofilms. For example, A. geminatus promotes the growth of P. intermedia (Bao et al., 2017). In contrast, some species act as antagonists, limiting the ability of other microbes to reside or accumulate on oral surfaces (Wang et al., 2009; Melo et al., 2016; Ho et al., 2017; Tahmourespour et al., 2019; He et al., 2021). Examples include CL/P-prone Lactobacillus species such as L. fermentum, which can inhibit S. aureus (Melo et al., 2016); and L. rhamnosus, which can inhibit Gardnerella (He et al., 2021) and S. mutans (Tahmourespour et al., 2019). Other antagonists include S. cristatus, which can restrict P. gingivalis colonization and virulence (Ho et al., 2017), and A. geminatus, which downregulates multiple proteins produced by S. oralis (Wang et al., 2009; Bao et al., 2017). Overall, microbiota prone to CL/P contribute to a specific microbe community which may lead to CL/P-specific infectious complications or worsen the prognosis of infections.
Besides the oral cavity, CL/P-prone microbes also affect other parts of the digestive tract (Figure 2). For example, L. fermentum and L. rhamnosus may be associated with oropharyngeal cancer (Guerrero-Preston et al., 2017); P. gingivalis and T. forsythia have been linked to esophageal cancer (Malinowski et al., 2019); Ottowia is found in higher abundance in patients with Crohn’s Disease (Sun et al., 2021); Bilophila wadsworthia can cause gastrointestinal (GI) tract inflammation and disrupt the functioning of the intestinal barrier in those who consume foods high in fat (Natividad et al., 2018) and may contribute to the initiation of colorectal cancer due to its hydrogen sulfide-releasing capabilities (Phipps et al., 2020); and P. pallens is less abundant in the saliva of those with reflux (Kawar et al., 2021), with there also being an inverse relationship between the severity of some gastritis symptoms and abundance of P. pallens (Han et al., 2019). In addition, Candida spp. can cause candidiasis in the GI tract (Sprague et al., 2022).
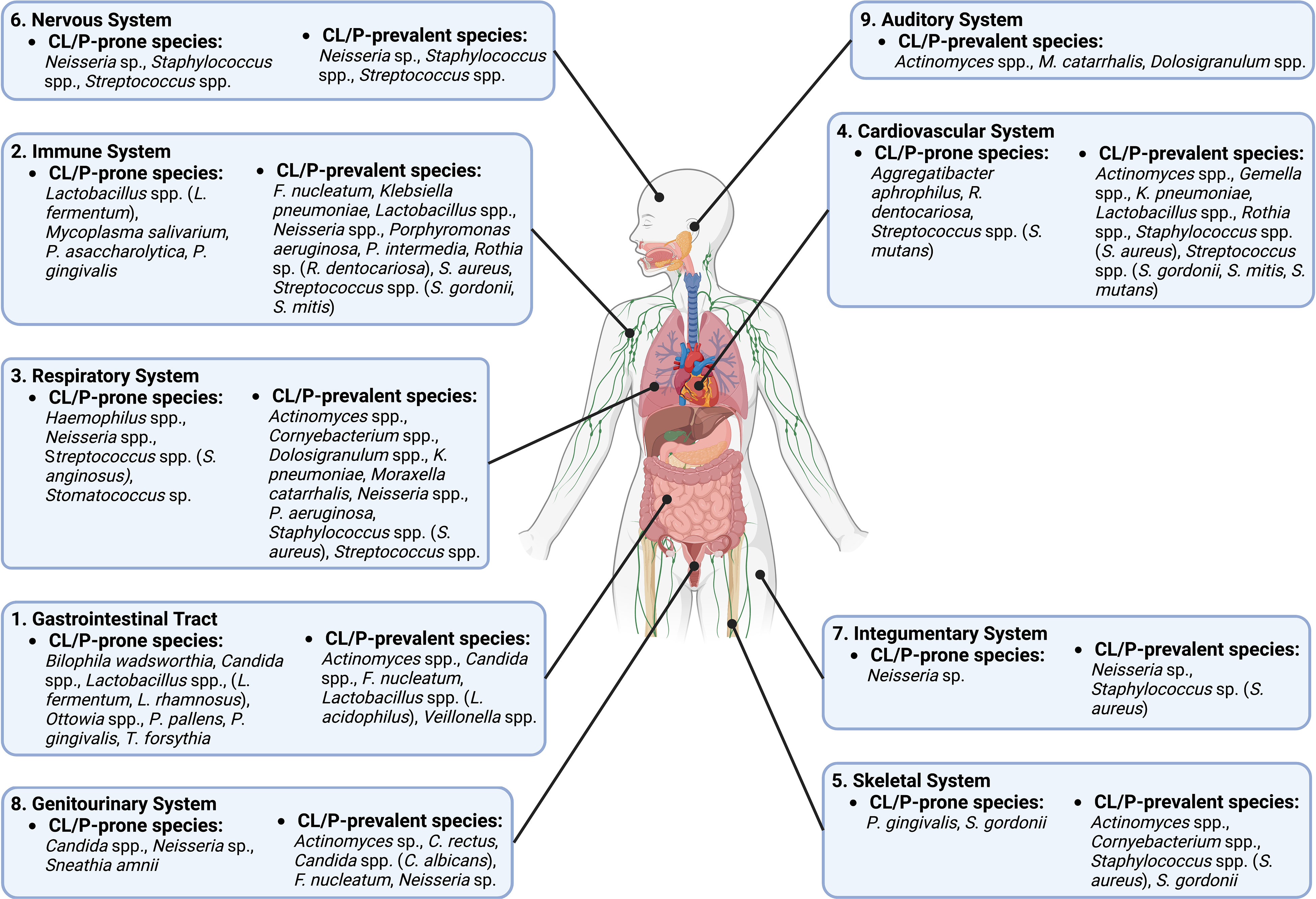
Figure 2 A diagram of different body systems that are potentially influenced by CL/P-prone and CL/P-prevalent species.
Particular CL/P-prone microbes also influence the immune system (Figure 2). For example, P. gingivalis, which has been reported to modulate inflammation in a variety of tissues (How et al., 2016; Chen et al., 2018; Bugueno et al., 2020), has an inhibitory effect on communication between Toll-like receptors and destroys cytokines produced by the host’s cells (Hajishengallis and Diaz, 2020). Mycoplasma salivarium has been shown to disrupt monocyte and macrophage activity, including phagocytosis (Nolan et al., 2016). Lactobacillus spp. likewise modulate immune cells: one particular strain of L. fermentum can upregulate specific interleukins in T-lymphocytes, while another strain has the same effect on dendritic cells (Ding et al., 2017). L. fermentum can also reduce intestinal inflammation (Aoudia et al., 2016), while P. asaccharolytica is known to boost the release of interleukins and tumor necrosis factors (Magalashvili et al., 2008).
CL/P-prone microbes affect myriad other systems of the body (Figure 2). For example, studies indicate that S. anginosus, a Haemophilus sp., a Neisseria sp., and a Stomatococcus sp. promote infection and disease of the respiratory system (King, 2012; Deutschmann et al., 2013; Yuan et al., 2013; Noguchi et al., 2015; Li et al., 2022). The circulatory system is also affected by CL/P-prone microbes: R. dentocariosa biofilm formation in the heart can promote ineffective endocarditis (Greve et al., 2021), a condition that is also stimulated by S. mutans and A. aphrophilus (Nørskov-Lauritsen, 2014; Nomura et al., 2020); S. mutans has also been linked to the development of atherosclerotic plaque (Kesavalu et al., 2012). In addition, both P. gingivalis and S. gordonii can cause bone resorption (Kassem et al., 2015; Park et al., 2020), while Streptococcus spp. colonization may be linked to brain abscesses and meningitis (Bokhari and Mesfin, 2023; Mandziuk and Kuchar, 2023). Previously studies also showed that a Neisseria sp. can cause dermatitis (da Cruz et al., 2019) and is the cause of the sexually-transmitted disease gonorrhea, which can cause serious harm to the genitourinary system (Unemo et al., 2019). Meanwhile, another CLP-prone bacterial species, Sneathia amnii, is associated with cervical cancer and appears to be capable of binding to and having a cytotoxic effect on cervical cancer cells (Nawrot et al., 2010; Harwich et al., 2012). Moreover, S. amnii is of great concern in reproduction, as this bacterial species produces an exotoxin that permeabilizes fetal membrane cells and is associated with stillbirths and conditions such as preeclampsia that may jeopardize the lives of the mother and unborn child (Harwich et al., 2012; Vitorino et al., 2019; Gentile et al., 2020).
On the other hand, multiple microbe species that have been found to be more prevalent in non-CL/P subjects than in CL/P patients may have a positive effect on human health. For example, certain species of Dolosigranulum may be negatively correlated with the incidence of particular respiratory maladies and enhance the immune response of the respiratory epithelium (Islam et al., 2021; Nesbitt et al., 2021) and appear to provide resistance to ear infections (Pettigrew et al., 2012; Lappan et al., 2018)—a common complaint of CL/P patients (Flynn et al., 2009). Strikingly, lack of Mitsuokella multacida, another species specific to the non-CL/P condition, is associated with the early development of colon cancer (Arafat, 2020; Elkholy et al., 2020), although its presence may be linked to squamous cell lung cancer as well (Zhao et al., 2021). Undoubtedly, further research is necessary to determine if and how these microbes affect the health of normal and CL/P populations.
5 Conclusion
Significant differences in the prevalence of particular microbiota have been found in various anatomic locations of CL/P patients in comparison with those of non-CL/P subjects. Many such species have deleterious effects on specific tissues or are associated with serious diseases, making it imperative to definitively investigate the microbiota of CL/P patients to have a more complete understanding of their condition. Interestingly, characterization of a given species as CL/P-prone or non-CL/P-prone was not consistent among different tissue sampling sites, indicating that further research must be done to fully understand the non-typical bacterial and fungal species particular to CL/P patients.
Author contributions
ZZ and CL contributed to conception and design of the study. EG, BX, and MS performed the article searching and data organization. EG, YL, and BX wrote the first draft of the manuscript. EG and BX drafted the figures. HK, ZZ, and CL did critical revision of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the American Association of Orthodontists Foundation (AAOF) Orthodontic Faculty Development Fellowship Award (for CL), American Association of Orthodontists (AAO) Full-Time Faculty Fellowship Award (for CL), University of Pennsylvania School of Dental Medicine Joseph and Josephine Rabinowitz Award for Excellence in Research (for CL), and the J. Henry O’Hern Jr. Pilot Grant from the Department of Orthodontics, University of Pennsylvania School of Dental Medicine (for CL).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1159455/full#supplementary-material
References
Acs, N., Bánhidy, F., Puhó, E., Czeizel, A. E. (2005). Maternal influenza during pregnancy and risk of congenital abnormalities in offspring. Birth Defect. Res. A Clin. Mol. Teratol 73 (12), 989–996. doi: 10.1002/bdra.20195
Ács, L., Bányai, D., Nemes, B., Nagy, K., Ács, N., Bánhidy, F., et al. (2020). Maternal-related factors in the origin of isolated cleft palate-a population-based case-control study. Orthod Craniofac Res. 23 (2), 174–180. doi: 10.1111/ocr.12361
Adeyemo, W. L., Adeyemi, M. O., Ogunsola, F. T., Ogunlewe, M. O., Ladeinde, A. L., Mofikoya, B. O., et al. (2013). Prevalence and bacteriology of bacteremia associated with cleft lip and palate surgery. J. Craniofac Surg. 24 (4), 1126–1131. doi: 10.1097/SCS.0b013e31828016e8
Akpan, A., Morgan, R. (2002). Oral candidiasis. Postgrad Med. J. 78 (922), 455–459. doi: 10.1136/pmj.78.922.455
Antoszewska, J., Kawala, B., Minch, L. (2010). Selected aspects of the oral environment in cleft palate patients–a problem evidently beyond dentists’ scope. Postepy Hig. Med. Dosw (Online) 64, 659–664.
Aoudia, N., Rieu, A., Briandet, R., Deschamps, J., Chluba, J., Jego, G., et al. (2016). Biofilms of lactobacillus plantarum and lactobacillus fermentum: Effect on stress responses, antagonistic effects on pathogen growth and immunomodulatory properties. Food Microbiol. 53 (Pt A), 51–59. doi: 10.1016/j.fm.2015.04.009
Arafat, W. (2020). P-316 profile of microbiota is associated with early onset of colorectal cancer in Egyptian and Kenyan patients. Ann. Oncol. 31, (supplement 3), S192. doi: 10.1016/j.annonc.2020.04.398
Arief, E. M., Mohamed, Z., Idris, F. M. (2005). Study of viridans streptococci and staphylococcus species in cleft lip and palate patients before and after surgery. Cleft Palate Craniofac J. 42 (3), 277–279. doi: 10.1597/04-083r.1
Bao, K., Bostanci, N., Thurnheer, T., Belibasakis, G. N. (2017). Proteomic shifts in multi-species oral biofilms caused by anaeroglobus geminatus. Sci. Rep. 7 (1), 4409. doi: 10.1038/s41598-017-04594-9
Bessell, A., Hooper, L., Shaw, W. C., Reilly, S., Reid, J., Glenny, A. M. (2011). Feeding interventions for growth and development in infants with cleft lip, cleft palate or cleft lip and palate. Cochrane Database Syst. Rev. 2), CD003315. doi: 10.1002/14651858.CD003315.pub3
Bokhari, M. R., Mesfin, F. B. (2023). “Brain abscess,” in StatPearls (Treasure Island FL: StatPearls Publishing).
Bokhout, B., van Loveren, C., Hofman, F. X., Buijs, J. F., van Limbeek, J., Prahl-Andersen, B. (1996). Prevalence of streptococcus mutans and lactobacilli in 18-month-old children with cleft lip and/or palate. Cleft Palate Craniofac J. 33 (5), 424–428. doi: 10.1597/1545-1569_1996_033_0424_posmal_2.3.co_2
Boriollo, M. F. G., Oliveira, M. C., Bassinello, V., Aníbal, P. C., da Silva, T. A., da Silva, J. J., et al. (2022). Candida species biotypes and polyclonality of potentially virulent candida albicans isolated from oral cavity of patients with orofacial clefts. Clin. Oral. Investig. 26 (3), 3061–3084. doi: 10.1007/s00784-021-04290-z
Bos, M., Hopman, J., Stuiver, M. M., Voss, A. (2016). Decolonisation of meticillin-resistant staphylococcus aureus (MRSA) carriage in adopted children with cleft lip and palate. J. Glob Antimicrob. Resist. 7, 28–33. doi: 10.1016/j.jgar.2016.07.001
Bugueno, I. M., Zobairi El-Ghazouani, F., Batool, F., El Itawi, H., Anglès-Cano, E., Benkirane-Jessel, N., et al. (2020). Porphyromonas gingivalis triggers the shedding of inflammatory endothelial microvesicles that act as autocrine effectors of endothelial dysfunction. Sci. Rep. 10 (1), 1778. doi: 10.1038/s41598-020-58374-z
Candotto, V., Oberti, L., Gabrione, F., Greco, G., Rossi, D., Romano, M., et al. (2019). Current concepts on cleft lip and palate etiology. J. Biol. Regul. Homeost Agents 33 (3 Suppl. 1), 145–151.
Cavalheiro, M., Teixeira, M. C. (2018). Candida biofilms: Threats, challenges, and promising strategies. Front. Med. (Lausanne) 5. doi: 10.3389/fmed.2018.00028
Cerný, M., Fára, M., Hrivnáková, J. (1991). Aetiological, modifying and lethal factors in cleft lip and palate. Acta Chir Plast. 33 (2), 72–86.
Chaudhari, P. K., Kharbanda, O. P., Chaudhry, R., Pandey, R. M., Chauhan, S., Bansal, K., et al. (2021). Factors affecting high caries risk in children with and without cleft lip and/or palate: A cross-sectional study. Cleft Palate Craniofac J. 58 (9), 1150–1159. doi: 10.1177/1055665620980206
Chen, Y., Zhou, R., Yi, Z., Li, Y., Fu, Y., Zhang, Y., et al. (2018). Porphyromonas gingivalis induced inflammatory responses and promoted apoptosis in lung epithelial cells infected with H1N1 via the Bcl−2/Bax/Caspase−3 signaling pathway. Mol. Med. Rep. 18 (1), 97–104. doi: 10.3892/mmr.2018.8983
Cheng, L. L., Moor, S. L., Kravchuk, O., Meyers, I. A., Ho, C. T. (2007). Bacteria and salivary profile of adolescents with and without cleft lip and/or palate undergoing orthodontic treatment. Aust. Dent. J. 52 (4), 315–321. doi: 10.1111/j.1834-7819.2007.tb00508.x
Chuo, C. B., Timmons, M. J. (2005). The bacteriology of children before primary cleft lip and palate surgery. Cleft Palate Craniofac J. 42 (3), 272–276. doi: 10.1597/03-108.1
Cildir, S. K., Sandalli, N., Nazli, S., Alp, F., Caglar, E. (2012). A novel delivery system of probiotic drop and its effect on dental caries risk factors in cleft lip/palate children. Cleft Palate Craniofac J. 49 (3), 369–372. doi: 10.1597/10-035
Cocco, J. F., Antonetti, J. W., Burns, J. L., Heggers, J. P., Blackwell, S. J. (2010). Characterization of the nasal, sublingual, and oropharyngeal mucosa microbiota in cleft lip and palate individuals before and after surgical repair. Cleft Palate Craniofac J. 47 (2), 151–155. doi: 10.1597/08-187_1
Costa, B., Lima, J. E., Gomide, M. R., Rosa, O. P. (2003). Clinical and microbiological evaluation of the periodontal status of children with unilateral complete cleft lip and palate. Cleft Palate Craniofac J. 40 (6), 585–589. doi: 10.1597/01-083
da Cruz, M., Palma, N. Z., Ferraz, R. V., Oliveira, M., Meireles, R. (2019). Disseminated gonococcal infection: A case report of arthritis-dermatitis syndrome. J. Med. cases 10 (10), 312–314. doi: 10.14740/jmc3381
de Soet, J. J., Bokhout, B., Buijs, J. F., van Loveren, C., de Graaff, J., Prahl-Andersen, B. (1998). Transmission of mutans streptococci between mothers and children with cleft lip and/or palate. Cleft Palate Craniofac J. 35 (5), 460–464. doi: 10.1597/1545-1569_1998_035_0460_tomsbm_2.3.co_2
de Souza, P. T. D. R., Gonçalves-Wilhelmsen, N. C. V., Rosa, R. T., Correia, C. F. K. N., Pereira, T. M., Kitahara, A. B. P., et al. (2022). Oral colonization and virulence factors of candida spp. in babies with cleft palate. Cleft Palate Craniofac J. 59 (8), 1056–1063. doi: 10.1177/10556656211030437
Deutschmann, M. W., Livingstone, D., Cho, J. J., Vanderkooi, O. G., Brookes, J. T. (2013). The significance of streptococcus anginosus group in intracranial complications of pediatric rhinosinusitis. JAMA Otolaryngol Head Neck Surg. 139 (2), 157–160. doi: 10.1001/jamaoto.2013.1369
Ding, Y. H., Qian, L. Y., Pang, J., Lin, J. Y., Xu, Q., Wang, L. H., et al. (2017). The regulation of immune cells by lactobacilli: a potential therapeutic target for anti-atherosclerosis therapy. Oncotarget 8 (35), 59915–59928. doi: 10.18632/oncotarget.18346
Dioguardi, M., Alovisi, M., Crincoli, V., Aiuto, R., Malagnino, G., Quarta, C., et al. (2020). Prevalence of the genus propionibacterium in primary and persistent endodontic lesions: A systematic review. J. Clin. Med. 9 (3), 739. doi: 10.3390/jcm9030739
Divya, D. V., Prasad, M. G. S., Radhakrishna, A. N., Reddy, S. P., Pratyusha, K., Kumar, K. V. K. S., et al. (2017). The serological evidence of cytomegalovirus infection as a potent aetiological factor for cleft Lip/Palate, mental retardation and deafness. J. Clin. Diagn. Res. 11 (6), ZC51–ZC54. doi: 10.7860/JCDR/2017/25118.10067
Downes, J., Liu, M., Kononen, E., Wade, W. G. (2009). Prevotella micans sp. nov., isolated from the human oral cavity. Int. J. Syst. Evol. Microbiol. 59 (Pt 4), 771–774. doi: 10.1099/ijs.0.002337-0
Downes, J., Munson, M. A., Radford, D. R., Spratt, D. A., Wade, W. G. (2002). Shuttleworthia satelles gen. nov., sp. nov., isolated from the human oral cavity. Int. J. Syst. Evol. Microbiol. 52 (Pt 5), 1469–1475. doi: 10.1099/00207713-52-5-1469
Downes, J., Sutcliffe, I., Tanner, A. C. R., Wade, W. G. (2005). Prevotella marshii sp. nov. and prevotella baroniae sp. nov., isolated from the human oral cavity. Int. J. Syst. Evol. Microbiol. 55 (Pt 4), 1551–1555. doi: 10.1099/ijs.0.63634-0
Duarte, G. A., Ramos, R. B., Cardoso, M. C. (2016). Feeding methods for children with cleft lip and/or palate: a systematic review. Braz. J. Otorhinolaryngol 82 (5), 602–609. doi: 10.1016/j.bjorl.2015.10.020
Durhan, M. A., Topcuoglu, N., Kulekci, G., Ozgentas, E., Tanboga, I. (2019). Microbial profile and dental caries in cleft lip and palate babies between 0 and 3 years old. Cleft Palate Craniofac J. 56 (3), 349–356. doi: 10.1177/1055665618776428
Elkholy, A., Behring, M., Mohsen, M., Bajpai, P., Embaby, A., Doaa, H., et al. (2020). Abstract 6103: Absence of mitsuokella multacida is associated with early onset of colorectal cancer. Cancer Res. 80 (16_supplement), 6103. doi: 10.1158/1538-7445.AM2020-6103
Eribe, E. R. K., Olsen, I. (2017). Leptotrichia species in human infections II. J. Oral. Microbiol. 9 (1), 1368848. doi: 10.1080/20002297.2017.1368848
Esberg, A., Barone, A., Eriksson, L., Lif Holgerson, P., Teneberg, S., Johansson, I. (2020). Corynebacterium matruchotii demography and adhesion determinants in the oral cavity of healthy individuals. Microorganisms 8 (11), 1780. doi: 10.3390/microorganisms8111780
Flynn, T., Möller, C., Jönsson, R., Lohmander, A. (2009). The high prevalence of otitis media with effusion in children with cleft lip and palate as compared to children without clefts. Int. J. Pediatr. Otorhinolaryngol 73 (10), 1441–1446. doi: 10.1016/j.ijporl.2009.07.015
Funahashi, K., Shiba, T., Watanabe, T., Muramoto, K., Takeuchi, Y., Ogawa, T., et al. (2019). Functional dysbiosis within dental plaque microbiota in cleft lip and palate patients. Prog. Orthod 20 (1), 11. doi: 10.1186/s40510-019-0265-1
Gentile, G. L., Rupert, A. S., Carrasco, L. I., Garcia, E. M., Kumar, N. G., Walsh, S. W., et al. (2020). Identification of a cytopathogenic toxin from sneathia amnii. J. Bacteriol 202 (13). doi: 10.1128/JB.00162-20
Gerits, E., Verstraeten, N., Michiels, J. (2017). New approaches to combat Porphyromonas gingivalis biofilms. J. Oral. Microbiol. 9 (1), 1300366. doi: 10.1080/20002297.2017.1300366
Greve, D., Moter, A., Kleinschmidt, M. C., Pfäfflin, F., Stegemann, M. S., Kursawe, L., et al. (2021). Rothia aeria and rothia dentocariosa as biofilm builders in infective endocarditis. Int. J. Med. Microbiol. 311 (2), 151478. doi: 10.1016/j.ijmm.2021.151478
Grewcock, R. E., Innes, N. P. T., Mossey, P. A., Robertson, M. D. (2022). Caries in children with and without orofacial clefting: A systematic review and meta-analysis. Oral. Dis. 28 (5), 1400–1411. doi: 10.1111/odi.14183
Guerrero-Preston, R., White, J. R., Godoy-Vitorino, F., Rodríguez-Hilario, A., Navarro, K., González, H., et al. (2017). High-resolution microbiome profiling uncovers fusobacterium nucleatum, lactobacillus gasseri/johnsonii, and lactobacillus vaginalis associated to oral and oropharyngeal cancer in saliva from HPV positive and HPV negative patients treated with surgery and chemo-radiation. Oncotarget 8 (67), 110931–110948. doi: 10.18632/oncotarget.20677
Hajishengallis, G., Diaz, P. I. (2020). Porphyromonas gingivalis: Immune subversion activities and role in periodontal dysbiosis. Curr. Oral. Health Rep. 7 (1), 12–21. doi: 10.1007/s40496-020-00249-3
Hammond, N. L., Dixon, M. J. (2022). Revisiting the embryogenesis of lip and palate development. Oral. Dis. 28 (5), 1306–1326. doi: 10.1111/odi.14174
Han, H. S., Lee, S. Y., Oh, S. Y., Moon, H. W., Cho, H., Kim, J. H. (2019). Correlations of the gastric and duodenal microbiota with histological, endoscopic, and symptomatic gastritis. J. Clin. Med. 8 (3), 312. doi: 10.3390/jcm8030312
Harwich, M. D., Serrano, M. G., Fettweis, J. M., Alves, J. M., Reimers, M. A., Buck, G. A., et al. (2012). Genomic sequence analysis and characterization of sneathia amnii sp. nov. BMC Genomics 13 Suppl 8, S4. doi: 10.1186/1471-2164-13-S8-S4
Hassani, H., Chen, J. W., Zhang, W., Hamra, W. (2020). Comparison of microbial activity among infants with or without using presurgical nasoalveolar molding appliance. Cleft Palate Craniofac J. 57 (6), 762–769. doi: 10.1177/1055665620908150
He, Y., Na, R., Niu, X., Xiao, B., Yang, H. (2021). Lactobacillus rhamnosus and lactobacillus casei affect various stages of gardnerella species biofilm formation. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.568178
Ho, M. H., Lamont, R. J., Xie, H. (2017). Identification of streptococcus cristatus peptides that repress expression of virulence genes in porphyromonas gingivalis. Sci. Rep. 7 (1), 1413. doi: 10.1038/s41598-017-01551-4
How, K. Y., Song, K. P., Chan, K. G. (2016). Porphyromonas gingivalis: An overview of periodontopathic pathogen below the gum line. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00053
Inquimbert, C., Bourgeois, D., Bravo, M., Viennot, S., Tramini, P., Llodra, J. C., et al. (2019). The oral bacterial microbiome of interdental surfaces in adolescents according to carious risk. Microorganisms 7 (9), 319. doi: 10.3390/microorganisms7090319
Islam, M. A., Albarracin, L., Melnikov, V., Andrade, B. G. N., Cuadrat, R. R. C., Kitazawa, H., et al. (2021). Dolosigranulum pigrum modulates immunity against SARS-CoV-2 in respiratory epithelial cells. Pathogens 10 (6), 634. doi: 10.3390/pathogens10060634
Iurovschi, R., Joaquim, C. R., de Faveri, M., de Miranda, T. S., Feres, M., de Figueiredo, L. C. (2020). Evaluation of the microbiological profile of alveolar residual screws and cleft-adjacent teeth in individuals with complete unilateral fissures. Cleft Palate Craniofac J. 57 (10), 1182–1189. doi: 10.1177/1055665620945568
Jousimies-Somer, H., Grénman, R., Rintala, A. (1986). Bacteriological investigation of secretory otitis media in children with cleft palate. Scand. J. Plast. Reconstr Surg. 20 (3), 297–302. doi: 10.3109/02844318609004490
Kassem, A., Henning, P., Lundberg, P., Souza, P. P., Lindholm, C., Lerner, U. H. (2015). Porphyromonas gingivalis stimulates bone resorption by enhancing RANKL (Receptor activator of NF-κB ligand) through activation of toll-like receptor 2 in osteoblasts. J. Biol. Chem. 290 (33), 20147–20158. doi: 10.1074/jbc.M115.655787
Kawar, N., Park, S. G., Schwartz, J. L., Callahan, N., Obrez, A., Yang, B., et al. (2021). Salivary microbiome with gastroesophageal reflux disease and treatment. Sci. Rep. 11 (1), 188. doi: 10.1038/s41598-020-80170-y
Kesavalu, L., Lucas, A. R., Verma, R. K., Liu, L., Dai, E., Sampson, E., et al. (2012). Increased atherogenesis during streptococcus mutans infection in ApoE-null mice. J. Dent. Res. 91 (3), 255–260. doi: 10.1177/0022034511435101
King, P. (2012). Haemophilus influenzae and the lung (Haemophilus and the lung). Clin. Transl. Med. 1 (1), 10. doi: 10.1186/2001-1326-1-10
Könönen, E., Gursoy, U. K. (2021). Oral prevotella species and their connection to events of clinical relevance in gastrointestinal and respiratory tracts. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.798763
Kressirer, C. A., Smith, D. J., King, W. F., Dobeck, J. M., Starr, J. R., Tanner, A. C. R. (2017). Scardovia wiggsiae and its potential role as a caries pathogen. J. Oral. Biosci. 59 (3), 135–141. doi: 10.1016/j.job.2017.05.002
Kumagai, K., Sugano, N., Takane, M., Iwasaki, H., Tanaka, H., Yoshinuma, N., et al. (2003). Detection of streptococcus anginosus from saliva by real-time polymerase chain reaction. Lett. Appl. Microbiol. 37 (5), 370–373. doi: 10.1046/j.1472-765x.2003.01405.x
Lappan, R., Imbrogno, K., Sikazwe, C., Anderson, D., Mok, D., Coates, H., et al. (2018). A microbiome case-control study of recurrent acute otitis media identified potentially protective bacterial genera. BMC Microbiol. 18 (1), 13. doi: 10.1186/s12866-018-1154-3
Li, L., Mac Aogáin, M., Xu, T., Jaggi, T. K., Chan, L. L. Y., Qu, J., et al. (2022). Neisseria species as pathobionts in bronchiectasis. Cell Host Microbe 30 (9), 1311–1327.e1318. doi: 10.1016/j.chom.2022.08.005
Li, J., Rodriguez, G., Han, X., Janečková, E., Kahng, S., Song, B., et al. (2019). Regulatory mechanisms of soft palate development and malformations. J. Dent. Res. 98 (9), 959–967. doi: 10.1177/0022034519851786
Liu, L., Zhang, Q., Lin, J., Ma, L., Zhou, Z., He, X., et al. (2016). Investigating oral microbiome profiles in children with cleft lip and palate for prognosis of alveolar bone grafting. PloS One 11 (5), e0155683. doi: 10.1371/journal.pone.0155683
Lucas, V. S., Gupta, R., Ololade, O., Gelbier, M., Roberts, G. J. (2000). Dental health indices and caries associated microflora in children with unilateral cleft lip and palate. Cleft Palate Craniofac J. 37 (5), 447–452. doi: 10.1597/1545-1569_2000_037_0447_dhiaca_2.0.co_2
Machorowska-Pieniążek, A., Mertas, A., Skucha-Nowak, M., Tanasiewicz, M., Morawiec, T. (2017). A comparative study of oral microbiota in infants with complete cleft lip and palate or cleft soft palate. BioMed. Res. Int. 2017, 1460243. doi: 10.1155/2017/1460243
Magalashvili, L., Lazarovich, S., Pechatnikov, I., Wexler, H. M., Nitzan, Y. (2008). Cytokine release and expression induced by OmpA proteins from the gram-negative anaerobes, porphyromonas asaccharolytica and bacteroides fragilis. FEMS Immunol. Med. Microbiol. 53 (2), 252–259. doi: 10.1111/j.1574-695X.2008.00423.x
Malinowski, B., Węsierska, A., Zalewska, K., Sokołowska, M. M., Bursiewicz, W., Socha, M., et al. (2019). The role of tannerella forsythia and porphyromonas gingivalis in pathogenesis of esophageal cancer. Infect. Agent Cancer 14, 3. doi: 10.1186/s13027-019-0220-2
Mandziuk, J., Kuchar, E. P. (2023). “Streptococcal meningitis,” in StatPear(Treasure Island (FL: StatPearls Publishing).
Marazita, M. L. (2007). Subclinical features in non-syndromic cleft lip with or without cleft palate (CL/P): review of the evidence that subepithelial orbicularis oris muscle defects are part of an expanded phenotype for CL/P. Orthod Craniofac Res. 10 (2), 82–87. doi: 10.1111/j.1601-6343.2007.00386.x
Mattos, B. S., Sousa, A. A., Magalhães, M. H., André, M., Brito E Dias, R. (2009). Candida albicans in patients with oronasal communication and obturator prostheses. Braz. Dent. J. 20 (4), 336–340. doi: 10.1590/s0103-64402009000400013
Melo, T. A., Dos Santos, T. F., de Almeida, M. E., Junior, L. A., Andrade, E. F., Rezende, R. P., et al. (2016). Inhibition of staphylococcus aureus biofilm by lactobacillus isolated from fine cocoa. BMC Microbiol. 16 (1), 250. doi: 10.1186/s12866-016-0871-8
Métneki, J., Puhó, E., Czeizel, A. E. (2005). Maternal diseases and isolated orofacial clefts in Hungary. Birth Defect. Res. A Clin. Mol. Teratol 73 (9), 617–623. doi: 10.1002/bdra.20177
Meyer-Marcotty, P., Stellzig-Eisenhauer, A. (2009). Dentofacial self-perception and social perception of adults with unilateral cleft lip and palate. J. Orofac Orthop 70 (3), 224–236. doi: 10.1007/s00056-009-8813-9
Mombelli, A., Brägger, U., Lang, N. P. (1992). Microbiota associated with residual clefts and neighboring teeth in patients with cleft lip, alveolus, and palate. Cleft Palate Craniofac J. 29 (5), 463–469. doi: 10.1597/1545-1569_1992_029_0463_mawrca_2.3.co_2
Moore, L. V. H., Johnson, J. L., Moore, W. E. C. (1987). Selenomonas noxia sp. nov., selenomonas flueggei sp. nov., selenomonas infelix sp. nov., selenomonas dianae sp. nov., and selenomonas artemidis sp. nov., from the human gingival crevice. Int. J. Systemat. Evolutionary Microbiol. 37 (3), 271–280.
Mossey, P. A., Little, J., Munger, R. G., Dixon, M. J., Shaw, W. C. (2009). Cleft lip and palate. Lancet 374 (9703), 1773–1785. doi: 10.1016/S0140-6736(09)60695-4
Mÿburgh, H. P., Bütow, K. W. (2009). Cleft soft palate reconstruction: prospective study on infection and antibiotics. Int. J. Oral. Maxillofac. Surg. 38 (9), 928–932. doi: 10.1016/j.ijom.2009.04.022
Nørskov-Lauritsen, N. (2014). Classification, identification, and clinical significance of haemophilus and aggregatibacter species with host specificity for humans. Clin. Microbiol. Rev. 27 (2), 214–240. doi: 10.1128/CMR.00103-13
Narinesingh, S. P., Whitby, D. J., Davenport, P. J. (2011). Moraxella catarrhalis: an unrecognized pathogen of the oral cavity? Cleft Palate Craniofac J. 48 (4), 462–464. doi: 10.1597/09-054
Natividad, J. M., Lamas, B., Pham, H. P., Michel, M. L., Rainteau, D., Bridonneau, C., et al. (2018). Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat. Commun. 9 (1), 2802. doi: 10.1038/s41467-018-05249-7
Nawrot, R., Kamieniarz, K., Malinowska, M., Józefiak, A., Kedzia, W., Kwaśniewska, A., et al. (2010). The prevalence of leptotrichia amnionii in cervical swabs of HPV positive and negative women. Eur. J. Gynaecol Oncol. 31 (4), 425–428.
Nesbitt, H., Burke, C., Haghi, M. (2021). Manipulation of the upper respiratory microbiota to reduce incidence and severity of upper respiratory viral infections: A literature review. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.713703
Noguchi, S., Yatera, K., Kawanami, T., Yamasaki, K., Naito, K., Akata, K., et al. (2015). The clinical features of respiratory infections caused by the streptococcus anginosus group. BMC Pulm Med. 15, 133. doi: 10.1186/s12890-015-0128-6
Nolan, T. J., Gadsby, N. J., Hellyer, T. P., Templeton, K. E., McMullan, R., McKenna, J. P., et al. (2016). Low-pathogenicity mycoplasma spp. alter human monocyte and macrophage function and are highly prevalent among patients with ventilator-acquired pneumonia. Thorax 71 (7), 594–600. doi: 10.1136/thoraxjnl-2015-208050
Nomura, R., Matayoshi, S., Otsugu, M., Kitamura, T., Teramoto, N., Nakano, K. (2020). Contribution of severe dental caries induced by streptococcus mutans to the pathogenicity of infective endocarditis. Infect. Immun. 88 (7). doi: 10.1128/IAI.00897-19
Otto, M. (2009). Staphylococcus epidermidis – the “accidental” pathogen. Nat. Rev. Microbiol. 7 (8), 555–567. doi: 10.1038/nrmicro2182
Pantaléon, V., Bouttier, S., Soavelomandroso, A. P., Janoir, C., Candela, T. (2014). Biofilms of clostridium species. Anaerobe 30, 193–198. doi: 10.1016/j.anaerobe.2014.09.010
Parapanisiou, V., Gizani, S., Makou, M., Papagiannoulis, L. (2009). Oral health status and behaviour of Greek patients with cleft lip and palate. Eur. Arch. Paediatr. Dent. 10 (2), 85–89. doi: 10.1007/BF03321606
Park, O. J., Kwon, Y., Park, C., So, Y. J., Park, T. H., Jeong, S., et al. (2020). Streptococcus gordonii: Pathogenesis and host response to its cell wall components. Microorganisms 8 (12), 1852. doi: 10.3390/microorganisms8121852
Passinato Gheller, S. A., Porto, A. N., Borba, A. M., Veiga, K. A., Aranha, A. M. F. (2021). Periodontal findings in children and adolescents with cleft lip and/or palate: A case-control study. Pediatr. Dent. 43 (2), 133–139.
Pedersen, A. M. L., Sørensen, C. E., Proctor, G. B., Carpenter, G. H., Ekström, J. (2018). Salivary secretion in health and disease. J. Oral. Rehabil. 45 (9), 730–746. doi: 10.1111/joor.12664
Perdikogianni, H., Papaioannou, W., Nakou, M., Oulis, C., Papagiannoulis, L. (2009). Periodontal and microbiological parameters in children and adolescents with cleft lip and/or palate. Int. J. Paediatr. Dent. 19 (6), 455–467. doi: 10.1111/j.1365-263X.2009.01020.x
Pettigrew, M. M., Laufer, A. S., Gent, J. F., Kong, Y., Fennie, K. P., Metlay, J. P. (2012). Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl. Environ. Microbiol. 78 (17), 6262–6270. doi: 10.1128/AEM.01051-12
Phipps, O., Al-Hassi, H. O., Quraishi, M. N., Kumar, A., Brookes, M. J. (2020). Influence of iron on the gut microbiota in colorectal cancer. Nutrients 12 (9), 2512. doi: 10.3390/nu12092512
Ponde, N. O., Lortal, L., Ramage, G., Naglik, J. R., Richardson, J. P. (2021). Biofilms and polymicrobial interactions. Crit. Rev. Microbiol. 47 (1), 91–111. doi: 10.1080/1040841X.2020.1843400
Poulain, D. (2015). Candida albicans, plasticity and pathogenesis. Crit. Rev. Microbiol. 41 (2), 208–217. doi: 10.3109/1040841X.2013.813904
Raja, M., Ummer, F., Dhivakar, C. P. (2014). Aggregatibacter actinomycetemcomitans - a tooth killer? J. Clin. Diagn. Res. 8 (8), ZE13–ZE16. doi: 10.7860/JCDR/2014/9845.4766
Ramdial, S., Madaree, A. (2019). The spectrum of intraoral bacteria seen in patients with cleft palates in an African setting. Microbiologyopen 8 (4), e00679. doi: 10.1002/mbo3.679
Rath, H., Feng, D., Neuweiler, I., Stumpp, N. S., Nackenhorst, U., Stiesch, M. (2017). Biofilm formation by the oral pioneer colonizer streptococcus gordonii: an experimental and numerical study. FEMS Microbiol. Ecol. 93 (3). doi: 10.1093/femsec/fix010
Rawashdeh, M. A., Ayesh, J. A., Darwazeh, A. M. (2011). Oral candidal colonization in cleft patients as a function of age, gender, surgery, type of cleft, and oral health. J. Oral. Maxillofac. Surg. 69 (4), 1207–1213. doi: 10.1016/j.joms.2010.02.044
Rennie, A., Treharne, L. J., Richard, B. (2009). Throat swabs taken on the operating table prior to cleft palate repair and their relevance to outcome: a prospective study. Cleft Palate Craniofac J. 46 (3), 275–279. doi: 10.1597/08-082.1
Ritthagol, W., Saetang, C., Teanpaisan, R. (2014). Effect of probiotics containing lactobacillus paracasei SD1 on salivary mutans streptococci and lactobacilli in orthodontic cleft patients: A double-blinded, randomized, placebo-controlled study. Cleft Palate Craniofac J. 51 (3), 257–263. doi: 10.1597/12-243
Rodrigues, R., Chung, A. P., Mortensen, M. S., Fernandes, M. H., Monteiro, A. B., Furfuro, R., et al. (2021). Temporal oral microbiome changes with brushing in children with cleft lip and palate. Heliyon 7 (3), e06513. doi: 10.1016/j.heliyon.2021.e06513
Rodrigues, R., Fernandes, M. H., Bessa Monteiro, A., Furfuro, R., Carvalho Silva, C., Vardasca, R., et al. (2019). Are there any solutions for improving the cleft area hygiene in patients with cleft lip and palate? a systematic review. Int. J. Dent. Hyg 17 (2), 130–141. doi: 10.1111/idh.12385
Roode, G. J., Bütow, K. W. (2018). A descriptive study of chlorhexidine as a disinfectant in cleft palate surgery. Clin. Med. Res. 16 (1-2), 9–15. doi: 10.3121/cmr.2018.1385
Roode, G. J., Bütow, K. W., Naidoo, S. (2017). Preoperative evaluation of micro-organisms in non-operated cleft in soft palate: impact on use of antibiotics. Br. J. Oral. Maxillofac. Surg. 55 (2), 127–131. doi: 10.1016/j.bjoms.2016.09.018
Roode, G. J., Bütow, K. W., Naidoo, S. (2022). Microbial contamination profile change over a 4-year period in nonoperated cleft soft palate. J. Appl. Microbiol. 132 (1), 665–674. doi: 10.1111/jam.15193
Sharma, A. (2010). Virulence mechanisms of tannerella forsythia. Periodontol 2000 54 (1), 106–116. doi: 10.1111/j.1600-0757.2009.00332.x
Shashni, R., Goyal, A., Gauba, K., Utreja, A. K., Ray, P., Jena, A. K. (2015). Comparison of risk indicators of dental caries in children with and without cleft lip and palate deformities. Contemp Clin. Dent. 6 (1), 58–62. doi: 10.4103/0976-237X.149293
Silva, J. J. D., Silva, T. A. D., Almeida, H., Rodrigues Netto, M. F., Cerdeira, C. D., Höfling, J. F., et al. (2018). Candida species biotypes in the oral cavity of infants and children with orofacial clefts under surgical rehabilitation. Microb. Pathog. 124, 203–215. doi: 10.1016/j.micpath.2018.08.042
Singh, D. K., Tóth, R., Gácser, A. (2020). Mechanisms of pathogenic. Front. Cell Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.00094
Siqueira, J. F., Rôças, I. N. (2006). Catonella morbi and granulicatella adiacens: new species in endodontic infections. Oral. Surg. Oral. Med. Oral. Pathol. Oral. Radiol. Endod. 102 (2), 259–264. doi: 10.1016/j.tripleo.2005.09.021
Socransky, S. S., Haffajee, A. D., Cugini, M. A., Smith, C., Kent, R. L. (1998). Microbial complexes in subgingival plaque. J. Clin. Periodontol 25 (2), 134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x
Sprague, J. L., Kasper, L., Hube, B. (2022). From intestinal colonization to systemic infections: Candida albicans translocation and dissemination. Gut Microbes 14 (1), 2154548. doi: 10.1080/19490976.2022.2154548
Sun, B., Liu, B., Gao, X., Xing, K., Xie, L., Guo, T. (2021). Metagenomic analysis of saliva reveals disease-associated microbiotas in patients with periodontitis and crohn’s disease-associated periodontitis. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.719411
Sundell, A. L., Ullbro, C., Dahlén, G., Marcusson, A., Twetman, S. (2018). Salivary microbial profiles in 5-year old children with oral clefts: a comparative study. Eur. Arch. Paediatr. Dent. 19 (1), 57–60. doi: 10.1007/s40368-018-0326-z
Sundell, A. L., Ullbro, C., Marcusson, A., Twetman, S. (2015). Comparing caries risk profiles between 5- and 10- year-old children with cleft lip and/or palate and non-cleft controls. BMC Oral. Health 15, 85. doi: 10.1186/s12903-015-0067-x
Tahmourespour, A., Kasra-Kermanshahi, R., Salehi, R. (2019). Lactobacillus rhamnosus biosurfactant inhibits biofilm formation and gene expression of caries-inducing streptococcus mutans. Dent. Res. J. (Isfahan) 16 (2), 87–94. doi: 10.4103/1735-3327.250968
Thomas, G. P., Sibley, J., Goodacre, T. E., Cadier, M. M. (2012). The value of microbiological screening in cleft lip and palate surgery. Cleft Palate Craniofac J. 49 (6), 708–713. doi: 10.1597/11-063
Tibesar, R. J., Black, A., Sidman, J. D. (2009). Surgical repair of cleft lip and cleft palate. Operative Tech. Otolaryngology-Head Neck Surg. 20 (4), 245–255. doi: 10.1016/j.otot.2009.10.010
Tuna, E. B., Topçuoglu, N., Ilhan, B., Gençay, K., Kulekçi, G. (2008). Staphylococcus aureus transmission through oronasal fistula in children with cleft lip and palate. Cleft Palate Craniofac J. 45 (5), 477–480. doi: 10.1597/06-247.1
Unemo, M., Seifert, H. S., Hook, E. W., Hawkes, S., Ndowa, F., Dillon, J. R. (2019). Gonorrhoea. Nat. Rev. Dis. Primers 5 (1), 79. doi: 10.1038/s41572-019-0128-6
Van Dyck, J., Cadenas de Llano-Pérula, M., Willems, G., Verdonck, A. (2019). Dental development in cleft lip and palate patients: A systematic review. Forensic Sci. Int. 300, 63–74. doi: 10.1016/j.forsciint.2019.04.011
van Loveren, C., Buijs, J. F., Bokhout, B., Prahl-Andersen, B., Ten Cate, J. M. (1998). Incidence of mutans streptococci and lactobacilli in oral cleft children wearing acrylic plates from shortly after birth. Oral. Microbiol. Immunol. 13 (5), 286–291. doi: 10.1111/j.1399-302x.1998.tb00709.x
Vieira, N. A., Borgo, H. C., da Silva Dalben, G., Bachega, M. I., Pereira, P. C. (2013). Evaluation of fecal microorganisms of children with cleft palate before and after palatoplasty. Braz. J. Microbiol. 44 (3), 835–838. doi: 10.1590/s1517-83822013000300026
Vitorino, P., Varo, R., Castillo, P., Hurtado, J. C., Fernandes, F., Valente, A. M., et al. (2019). Sneathia amnii and maternal chorioamnionitis and stillbirth, Mozambique. Emerg. Infect. Dis. 25 (8), 1614–1616. doi: 10.3201/eid2508.190526
Vyas, T., Gupta, P., Kumar, S., Gupta, R., Gupta, T., Singh, H. P. (2020). Cleft of lip and palate: A review. J. Family Med. Prim Care 9 (6), 2621–2625. doi: 10.4103/jfmpc.jfmpc_472_20
Wang, Y., Wang, S., Wu, C., Chen, X., Duan, Z., Xu, Q., et al. (2019). Oral microbiome alterations associated with early childhood caries highlight the importance of carbohydrate metabolic activities. mSystems 4 (6). doi: 10.1128/mSystems.00450-19
Wang, B. Y., Wu, J., Lamont, R. J., Lin, X., Xie, H. (2009). Negative correlation of distributions of streptococcus cristatus and porphyromonas gingivalis in subgingival plaque. J. Clin. Microbiol. 47 (12), 3902–3906. doi: 10.1128/JCM.00072-09
Weckwerth, P. H., de Magalhães Lopes, C. A., Duarte, M. A., Weckwerth, A. C., Martins, C. H., Neto, D. L., et al. (2009). Chronic suppurative otitis media in cleft palate: microorganism etiology and susceptibilities. Cleft Palate Craniofac J. 46 (5), 461–467. doi: 10.1597/08-144.1
Weckwerth, P. H., de Mattias Franco, A. T., de Magalhães Lopes, C. A., Santos, F. D., Weckwerth, A. C., Vivan, R. R., et al. (2014). Bacterial pathogens related to chronic suppurative otitis media in individuals with cleft palate: bacteriological culture and polymerase chain reaction. Cleft Palate Craniofac J. 51 (2), 145–153. doi: 10.1597/11-325
Xiao, X., He, S., He, F., Wu, X., Zheng, Y. (2021). Metagenomic analysis reveals Neisseria bacilliformis variation in the early childhood caries plaque microbiome. Evid Based Complement Alternat Med. 2021, 2774772. doi: 10.1155/2021/2774772
Yilmaz, H. N., Hatipoglu, S., Erdem, B., Can, B., Kadir, T. (2020). Adherence frequency of CANDIDA ALBICANS on nasoalveolar molding (NAM) appliances. J. Stomatol Oral. Maxillofac. Surg. 121 (5), 473–477. doi: 10.1016/j.jormas.2020.08.005
Yuan, Z., Panchal, D., Syed, M. A., Mehta, H., Joo, M., Hadid, W., et al. (2013). Induction of cyclooxygenase-2 signaling by stomatococcus mucilaginosus highlights the pathogenic potential of an oral commensal. J. Immunol. 191 (7), 3810–3817. doi: 10.4049/jimmunol.1300883
Zhang, M., Wang, R., Liao, Y., Buijs, M. J., Li, J. (2016). Profiling of oral and nasal microbiome in children with cleft palate. Cleft Palate Craniofac J. 53 (3), 332–338. doi: 10.1597/14-162
Zhang, K., Zhou, X., Qin, J., Zhang, W., Pan, Y., Wang, H., et al. (2022). Dynamic change in oral microbiota of children with cleft lip and palate after alveolar bone grafting. Cleft Palate Craniofac J. 59 (11), 1352–1360. doi: 10.1177/10556656211044396
Keywords: cleft lip, cleft palate, biofilm, oral microbiome, bacteria, fungus, virus
Citation: Gershater E, Liu Y, Xue B, Shin MK, Koo H, Zheng Z and Li C (2023) Characterizing the microbiota of cleft lip and palate patients: a comprehensive review. Front. Cell. Infect. Microbiol. 13:1159455. doi: 10.3389/fcimb.2023.1159455
Received: 06 February 2023; Accepted: 31 March 2023;
Published: 18 April 2023.
Edited by:
Feng Chen, Peking University, ChinaReviewed by:
Tiansong Xu, Peking University Hospital of Stomatology, ChinaShuang Pan, Harbin Medical University, China
Copyright © 2023 Gershater, Liu, Xue, Shin, Koo, Zheng and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhong Zheng, bGVveno5NUBnbWFpbC5jb20=; Chenshuang Li, bGljaGVuc0B1cGVubi5lZHU=
 Elizabeth Gershater1
Elizabeth Gershater1 Yuan Liu
Yuan Liu Zhong Zheng
Zhong Zheng Chenshuang Li
Chenshuang Li