
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 31 August 2023
Sec. Antibiotic Resistance and New Antimicrobial drugs
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1153387
This article is part of the Research Topic Development and Evaluation of Novel Protein Vaccines against Streptococcus pneumoniae View all 5 articles
 Yongli Wu1,2,3
Yongli Wu1,2,3 Jiankang Zhao2,3
Jiankang Zhao2,3 Ziyao Li2,3,4
Ziyao Li2,3,4 Xinmeng Liu2,3,5
Xinmeng Liu2,3,5 Yanning Hu2,3,5
Yanning Hu2,3,5 Feilong Zhang2,3,5
Feilong Zhang2,3,5 Yulin Zhang2,3
Yulin Zhang2,3 Danni Pu1,2,3
Danni Pu1,2,3 Chen Li5
Chen Li5 Xianxia Zhuo2,3,6
Xianxia Zhuo2,3,6 Huihui Shi7
Huihui Shi7 Binghuai Lu1,2,3,4,5*
Binghuai Lu1,2,3,4,5*Background: Colistin, as the antibiotic of “last resort” for carbapenem-resistant Klebsiella, develop resistance during administration of this antimicrobial agent. We identified an NDM-1-producing Klebsiella quasipneumonuae subsp. similipneumoniae (KQSS) strain KQ20605 recovered from a child, which developed resistance to colistin (KQ20786) through acquiring an IS903B element between the -27th and -26th bp of mgrB promoter region after 6-day colistin usage.
Objectives: The aim of this study is to explore the source of IS903B in the disruptive mgrB gene and its underlying mechanisms.
Materials and methods: Antibiotics susceptibility testing was conducted via microbroth dilution method. The in vitro colistin-induced experiment of KQ20605 was performed to mimic the in vivo transition from colistin-sensitive to resistant. Whole-genome sequencing was used to molecular identification of colistin resistance mechanism.
Results: The IS903B element integrated into mgrB gene of KQ20786 had a 100% nucleotide identity and coverage match with one IS903B on plasmid IncR, and only 95.1% (1005/1057) identity to those on chromosome. In vitro, upon the pressure of colistin, KQ20605 could also switch its phenotype from colistin-sensitive to resistant with IS elements (e.g., IS903B and IS26) frequently inserted into mgrB gene at “hotspots”, with the insertion site of IS903B nearly identical to that of KQ20786. Furthermore, IS26 elements in this isolate were only encoded by plasmids, including IncR and conjugative plasmid IncN harboring blaNDM.
Conclusion: Mobilizable IS elements on plasmids tend to be activated and integrated into mgrB gene at “hotspots” in this KQSS, thereby causing the colistin resistance emergence and further dissemination.
Carbapenemase-producing Klebsiella spp. are key pathogens for global nosocomial dominance (Tzouvelekis et al., 2012). Colistin is currently considered the “last resort” antibiotic for carbapenemase-producing Klebsiella spp. Yet, over the years, colistin-resistant Klebsiella strains have been emerging (Biswas et al., 2012; Grundmann et al., 2017; Binsker et al., 2022), further increasing the threat to public health. The underlying mechanism of this shift remains unclear, thus posing a major concern.
Klebsiella mainly consisted of three phylogroups, K. pneumoniae (phylogroup I), K. quasipneumoniae (phylogroup II), and K. variicola (phylogroup III). K. quasipneumoniae, subclassified into K. quasipneumoniae subsp. similipneumoniae (phylogroup IIB, hereafter referred to as KQSS), and K. quasipneumoniae subsp. quasipneumoniae (phylogroup IIA), are new species that can be distinguished from K. pneumoniae (Long et al., 2017). In a Japanese study that examined 119 Klebsiella isolates collected from blood samples, 11 (9.2%) species were identified as K. quasipneumoniae using genomic analysis but misidentified as K. pneumoniae in routine microbiology (Imai et al., 2019), thus resulting in underreported infections caused by this species (Long et al., 2017). Ahmed et al. suggested that colistin resistance in Klebsiella is predominantly mediated by the decrease of negative charge in the outer membrane due to modified lipopolysaccharide (LPS), thereby impairing the binding affinity between colistin and Lipid A (El-Sayed Ahmed et al., 2020). The modifications of LPS include 4-amino-4-deoxy-l-arabinopyranose (Ara4N) and phosphoethanolamine (pEtN) to lipid A via arnBCADTEF (pmrK) or pmrC products, respectively, which are mainly regulated by two-component regulatory systems (TCS) involving pmrAB, phoPQ, and crrAB genes (El-Sayed Ahmed et al., 2020). In addition, Lippa et al. found that the product of mgrB has a negative feedback effect on phoPQ, weakening the modification of LPS (Lippa and Goulian, 2009). Moreover, Liu et al. discovered that acquiring mobile genetic elements carrying a member of the mobilized colistin resistance (mcr) gene family may also lead to lipid A modification (Liu et al., 2016).
The insertion sequence (IS), harboring one or two transposase (tnp) genes, is a common mobile element that acts through a transposase (Partridge et al., 2018). After being transposed, transposase usually generates direct repeats (DRs) at both ends (Weinert et al., 1983). A previous study found that IS elements with DRs had a lower transposition frequency than those without (Zhang et al., 2021). Today, IS elements-mediated insertional disruption and inactivation of the chromosomal mgrB gene and/or its promoter region (hereafter referred to as mgrB gene) are considered the main cause of colistin resistance (Cannatelli et al., 2013; Cannatelli et al., 2014; Poirel et al., 2015; Shankar et al., 2019). As previously documented, the types of IS elements inserted in the mgrB gene often involve ISKpn25, IS903B, ISKpn14, and ISKpn26, etc. (Cannatelli et al., 2014; Olaitan et al., 2014; Poirel et al., 2015; Esposito et al., 2018; Yang et al., 2020; Silva et al., 2021; Yan et al., 2021). It has been reported that 60% of K. pneumoniae carry IS903B elements on its plasmids (Fordham et al., 2022). The dominant Inc groups harboring IS903B elements are fusion plasmids IncHI2A + IncHI2, IncFIB(K), and IncR, 12.5% of which co-carry IS903B elements and carbapenemase genes, with blaNDM-1 dominating (Fordham et al., 2022). The possible association between IS903B elements inserted in the mgrB gene and colistin resistance has also been documented (Yang et al., 2020). However, little is known about the source of IS903B in the disruptive mgrB gene and its underlying mechanisms.
Herein, we reported a New Delhi metallo-β-lactamase (NDM-1)-producing KQSS strain KQ20605 isolated from a lung transplant of a 7-year-old boy, who developed resistance to colistin (KQ20786) after a 6-day colistin administration. We further analyzed the possible mechanism of this transition from colistin-sensitive to resistant.
Colistin-resistant KQSS strain KQ20786 and its parental colistin-susceptible strain KQ20605 were collected from bronchoalveolar lavage fluid (BALF) from a 7-year-old child admitted to China-Japan Friendship Hospital (CJFH), Beijing, China on March 2019. These two strains were not obtained at admission or within 48 hours of admission to the hospital. The KQ20605 was recovered on day 9 after admission, while KQ20786 was isolated on day 15. Identification of species was performed using matrix-assisted laser desorption–ionization time-of-flight mass spectrometry (MALDI-TOF MS, Bruker Daltonik, Bremen, Germany). Clinical data on the case were retrospectively reviewed.
Susceptibility to ticarcillin-clavulanic acid, piperacillin-tazobactam, ceftazidime, cefoperazone-sulbactam, cefepime, aztreonam, imipenem, meropenem, amikacin, tobramycin, ciprofloxacin, doxycycline, and minocycline was measured using VITEK®2 system (BioMérieux, Marcy l’Étoile, France) following the manufacturer’s instructions. The minimum inhibitory concentrations (MICs) of colistin and tigecycline were determined by broth microdilution as recommended by the Clinical and Laboratory Standards Institute (CLSI, 2022). Interpretation of antibiotic susceptibility was also in line with CLSI, 2022 criteria (CLSI, 2022), except for tigecycline, which followed the standard of USA Food and Drug Administration (FDA) breakpoints (https://www.fda.gov/drugs/development-resources/tigecycline-injection-products). Escherichia coli ATCC 25922 was used as a control.
PAP was conducted on the colistin-susceptible KQ20605 isolate to exclude the possibility of colistin-heteroresistance, as previously described (Mashaly and Mashaly, 2021). Briefly, serial 100-fold dilutions ranging from 108 to 102 CFU/ml were prepared from overnight culture. Then, 10μL of each dilution was spread on Mueller-Hinton agar (MHA) plates containing 0, 0.5, 1, 2, 3, 4, 5, 6 and 8 mg/L of colistin, respectively. Next, the plates were incubated for 24 h at 37°C. Isolates that grew on plates with colistin concentrations >2mg/L were considered heteroresistant (Li et al., 2006).
Complementation experiments were conducted to clarify the association between mgrB insertion and colistin resistance. Briefly, the apramycin-resistant plasmid pDK6-mgrB was used. A pDK6 derivative carrying a cloned copy of wild-type mgrB gene and some flanks were amplified by PCR using primers mgrB-F/R (F: AGCCAAGCTTGCATGCCTGCAGAGCCAGCGATGCCAGATTTA, R: ACAGGAAACAGAATTCGAGCTCCGCCAATCCATAAGATAGCCAC). The genomic DNA of strain MGH78578 was used as the template for mgrB. The plasmid pDK6 and mgrB gene products obtained by PCR were digested by the double digestion method. Then, T4 DNA Ligase (NEB, US) was selected for ligating the pDK6 and mgrB gene at 16°C overnight. The pDK6-mgrB plasmid was introduced into KQ20786 by electroporation, with pDK6 as control. Transformants were selected on Mueller-Hinton agar containing 30 mg/L apramycin.
To ascertain the within-host evolution into colistin resistance of the parental strain KQ20605, the in vitro-induced experiment was performed over 6 days, as described in a previous study (Janssen et al., 2020). We first grew the strain KQ20605 in LB broth with initial colistin concentrations of 1 mg/L. After overnight growth, 10-μl cultures were inoculated on MHA plates containing 1, 2, 4, 8, 16, 32, and 64 mg/L colistin and incubated at 37°C for 24 h. At the same time, 15 μl cultures were transferred into a 3-ml fresh LB broth supplemented with the concentration of colistin to double that of the previous day. Well-separated colonies growing on 4 mg/L colistin-containing MHA plates were selected for determining their colistin MICs and identifying the existence and completeness of the mgrB genes with PCR (primers: mgrB-F 5’-GCCAATCCATAAGATAGCCA-3’ and mgrB-R 5’-AGCCAGCGATGCCAGATT-3’) and agarose gel electrophoresis. The PCR products were purified, and Sanger sequencing was performed using ABI 3730 DNA analyzer. The sequences were compared to the database in NCBI Genbank (http://www.ncbi.nlm.nih.gov) using the BLAST algorithm. This micro-evolution of the colistin resistance experiment lasted 6 days.
To explore the transferability of the plasmid pKQ20605-IncN co-harboring IS26 and blaNDM-1, conjugation experiments were performed using KQ20605 as the donor and E. coli strain J53 as the recipient, as previously described (Zhao et al., 2022). The donor and recipient strains were mixed at a ratio of 1:3 and incubated at 37°C for 18 h. Next, the transconjugants were selected on China blue lactose agar supplemented with meropenem (2 mg/L) and sodium azide (100 mg/L). Lastly, the species identification of transconjugants was confirmed by MALDI-TOF MS, and the existence of blaNDM was confirmed by the GeneXpert system (Cepheid Inc, Sunnyvale, CA, USA).
WGS was carried out using Illumina HiSeq 2500 platform and nanopore sequencing method on MinION flow cells for KQ20605, KQ20786, KQ20605-1, KQ20605-2, and KQ20605-5. Raw reads were filtered to remove the low-quality sequences and adaptors using a skewer (Jiang et al., 2014) and PoreChop (https://github.com/rrwick/Porechop), respectively. De novo assembly was conducted via SPAdes Genome Assembler v3.13.1 (Prjibelski et al., 2020) and Unicycler (Wick et al., 2017). MLST analysis and identification of antimicrobial resistance determinants and plasmids replicon were confirmed by the Center for Genomic Epidemiology (CGE) (http://genomicepidemiology.org/services/) and Kleborate. The ISfinder database (https://www-is.biotoul.fr) was used to identify insertion sequence elements in the Sanger sequencing data and complete genome sequence. Genome annotation was performed using Prokka v4.13. Genome sequence comparison was performed using the BLAST. CGView was used to visualize the comparison results. Colistin resistance-related genes involving pmrA, pmrB, phoP, phoQ, crrA, crrB, crrC, pmrD, pmrK, pmrC, and mgrB, were defined by alignment with the Klebsiella quasipneumoniae ATCC 700603 genomes through blast+ 2.11.0 with the E value set at 1e-50. Then KQ20605 genome was used as the reference to compare the identity of the above genes among KQ20605, KQ20786, KQ20605-1, KQ20605-2, and KQ20605-5. The PROVEAN platform (http://provean.jcvi.org/index.php) was used to predict alterations in the biological functions of the above-described proteins (Li et al., 2023).
The phylogenetic tree construction was performed to determine the species of the strain KQ20605 and KQ20786. We searched the whole genome sequences with the term “Klebsiella” on NCBI, selected the subspecies of Klebsiella, including K. pneumoniae, K. variicola, and K. quasipneumoniae, and then re-determined their species by Kleborate 2.0.4 (https://github.com/katholt/Kleborate). The whole genome sequences of 12 clinical isolates worldwide were then downloaded from the GenBank database to build the phylogenetic tree, including 3 K. pneumoniae (GCF_000364385.3, GCF_000956385.1, and GCF_001022035.1), 3 K. quasipneumoniae subsp. quasipneumoniae (GCF_003285165.1, GCF_020525805.1, and GCF_024917695.1), 3 KQSS (GCF_001060825.1, GCF_001611185.1, and GCF_002248055.2) and 3 K. variicola (GCF_000941635.2, GCF_003285185.1, and GCF_003429625.1). For comparative genomic analysis, the KQSS isolate ATCC 700603 (GeneBank: GCA_001596075.2) was selected as the reference genome.
Phylogenetic analysis was performed using Snippy (https://github.com/tseemann/snippy) Gubbins and RaxML. The iTOL (https://itol.embl.de) was used for the visualization of a phylogenetic tree.
Briefly, overnight cultures were diluted to an optical density at 600 nm (OD600) of ∼0.1 and grown at 37°C with 200 rpm shaking. The culture cell density was determined every 30 min by measuring OD600 for 24h. The experiment was repeated three times independently.
GraphPad Prism 8.2.1 was used for data analyses. Significant differences were assessed using a one-way analysis of variance. P value <0.05 was considered statistically significant.
In March 2019, a 7-year-old lung-transplanted boy was admitted to National respiratory medicine center, CJFH. The KQSS strain KQ20605, producing metallo-β-lactamase, was isolated from BALF. The strain was susceptible only to colistin, tigecycline, aztreonam, trimethoprim-sulfamethoxazole and aminoglycoside, as shown in Table 1. On the 6th day of colistin administration, the patient showed no improvement, and another KQSS strain (KQ20786) was recovered, revealing to be colistin-resistant. Antibiotic treatment with aztreonam was effective, so KQSS was not found. Unfortunately, the patient died soon after due to a bloodstream infection caused by Acinetobacter baumannii. The details of the clinical data are shown in Figure 1.

Table 1 MIC values of different antibiotics to clinical KQSS strains and the induced colistin-induced resistant strains.
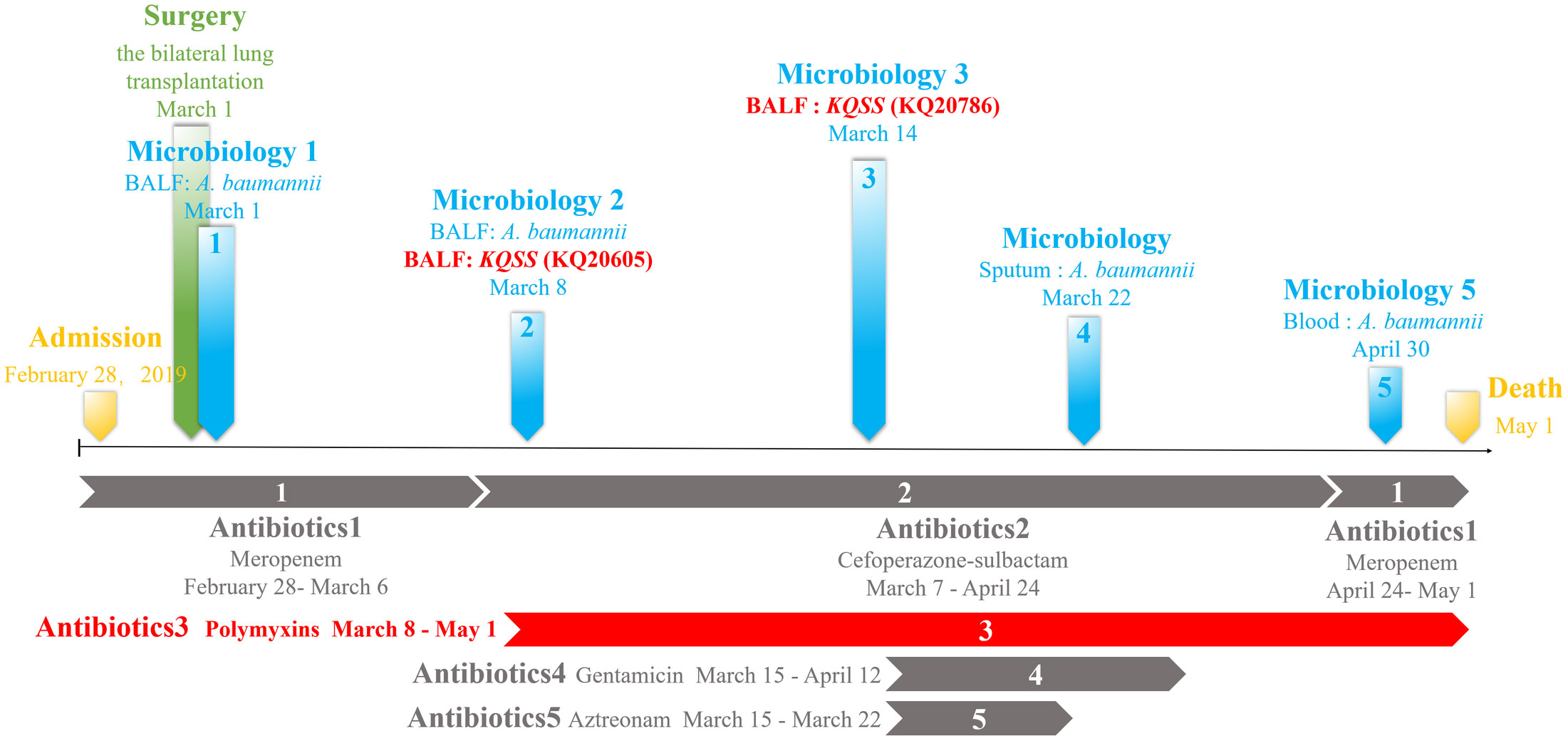
Figure 1 Clinical data of a lung transplantation recipient: time and sites of isolation of KQSS strains and other microbiological results from the patient and antimicrobial treatment. KQSS, K. quasipneumoniae subsp. similipneumoniae.
Species identification in the Kleborate database indicated that KQ20605 and KQ20786 belonged to ST2558 and KQSS, initially misidentified as K. pneumoniae by MALDI-TOF MS. Pairwise SNP analysis on both strains showed that the SNPs between them was only 21, suggesting that they are clonally related (Figure 2A). The genomic identity between KQ20605 and KQ20786 was also confirmed by Pulsed Field Gel Electrophoresis (PFGE), shown in Figure 2B. GeneXpert system showed that both isolates carried NDM. The MIC for colistin of KQ20605 was 1 mg/L, and the possibility of colistin-heteroresistance was excluded via PAP. Different from KQ20605, KQ20786 was highly resistant to colistin with a MIC of ≥ 64 mg/L. The AST results are detailed in Table 1.
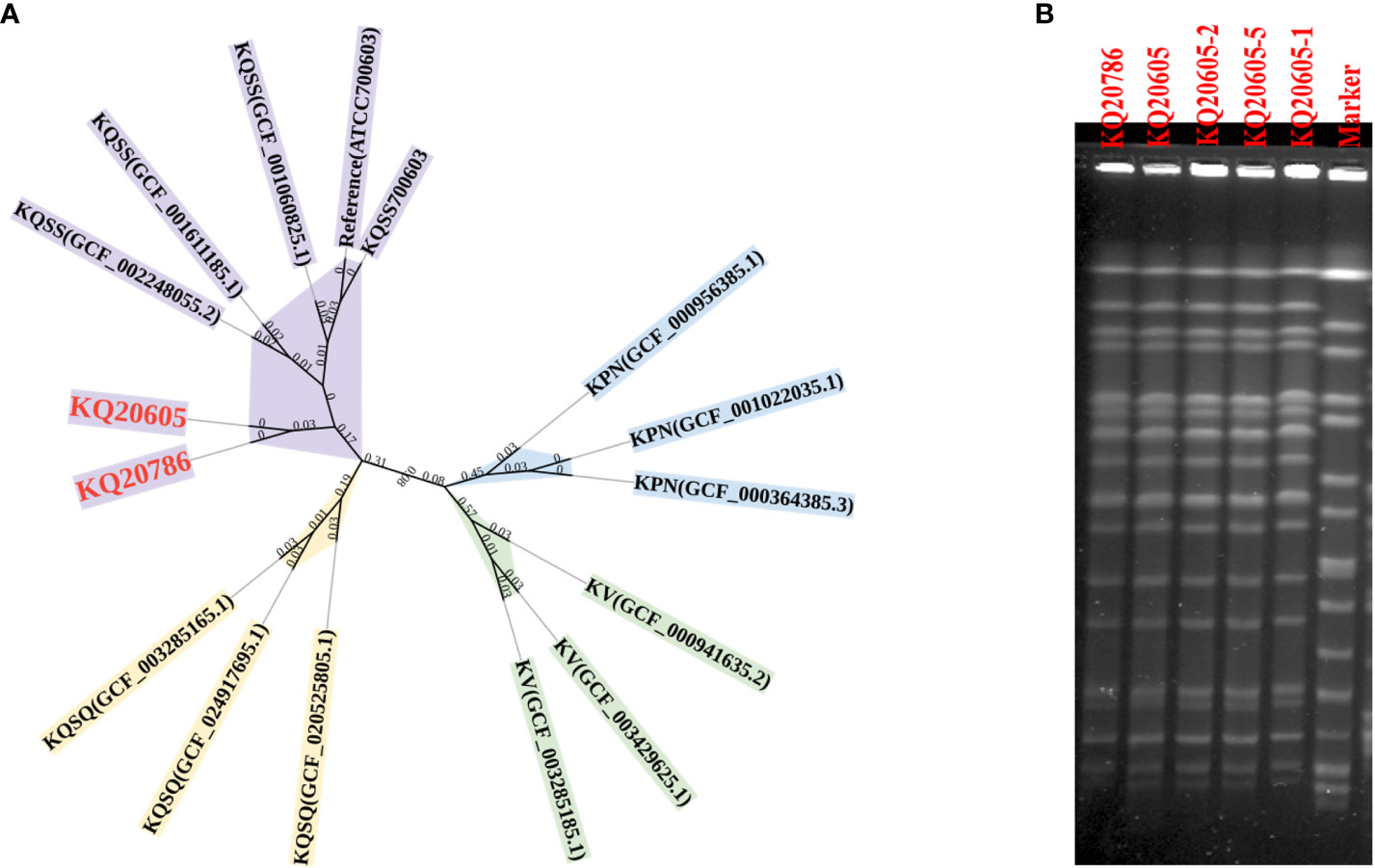
Figure 2 Species and homology analysis on strains in this study. (A) Core genome phylogenetic tree. A Core genome phylogenetic tree of 3 KPN (blue), 3 KV (green), 3 KQSS (purple), 3 KQSQ (yellow), KQ20605, and KQ20786 (red bold). ATCC 700603 was as a reference. KPN, K. pneumoniae; KV, K. variicola; KQSS, K. quasipneumoniae subsp. similipneumoniae; KQSQ, K. quasipneumoniae subsp. quasipneumoniae. (B) PFGE of 5 isolates. KQ20605 and KQ20786 were isolated from the patient. KQ20605-1, KQ20605-2, and KQ20605-5 were mgrB inactivation-induced isolates in vitro. KQ20605-1 (with IS903B insertion between -26th and -25th bp in mgrB), KQ20605-2 (with IS26 insertion between -20th and -19th bp in mgrB), KQ20605-5 (with a large segment in mgrB or mgrB-lost). PFGE, Pulsed Field Gel Electrophoresis.
The genome of KQ20605 consists of a chromosome (5,131,640 bp) and three plasmids, namely, pKQ20605-IncFIB(K) (165,393 bp), pKQ20605-IncN (59,353 bp), and pKQ20605-IncR (65,658 bp). Both KQ20605 and KQ20786 harbor the resistant genes fosA, oqxA, oqxB and blaOKP-B-2 on the chromosome, coupled with qnrS1, dfrA14, and blaNDM-1 on pKQ20605-IncN (Figure 3). Chromosomal resistance mechanisms for colistin of KQ20786 were determined by investigating the nucleotide sequences and corresponding protein sequences of the genes pmrA, pmrB, phoP, phoQ, crrA, crrB, crrC, pmrD, pmrK, pmrC, and mgrB, and comparing them to sequences from its parental strain KQ20605. Genome comparison results showed that an IS903B was located between the -27th and -26th base pair of mgrB promoter in KQ20786 compared to the colistin-susceptible parental strain KQ20605 (Figure 4A). The phoPQ, pmrAB, and crrAB genes between KQ20786 and KQ20605 were 100% identical using BLASTn. No mcr gene (mcr1-10) and its variants were identified in the above two strains. Compensation experiments confirmed that the KQ20786 transformant complemented with pDK6-mgrB restored colistin susceptibility (0.5mg/L). The above investigation suggested that continual usage of colistin in the patient resulted in the within-host evolution and selection of colistin resistance through insertional inactivation of mgrB gene.
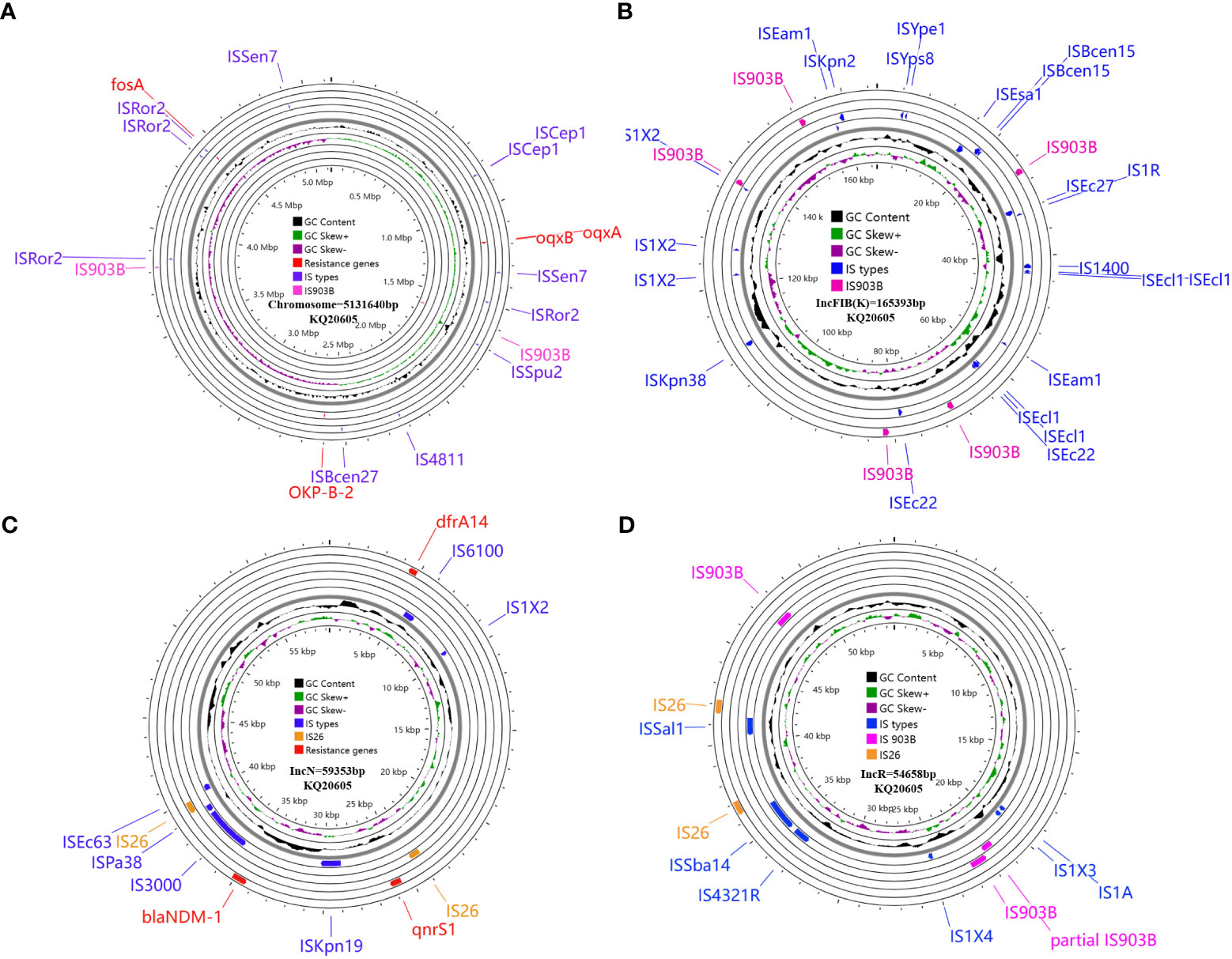
Figure 3 Circular maps and genomic analysis of all insertion sequences distribution and antibiotic resistance genes on the chromosome (A), and plasmids pKQ20605-IncFIB(K) (B), pKQ20605-IncN (C), and pKQ20605-IncR (D) of parental KQ20605 KQSS strain. KQSS, K. quasipneumoniae subsp. similipneumoniae.
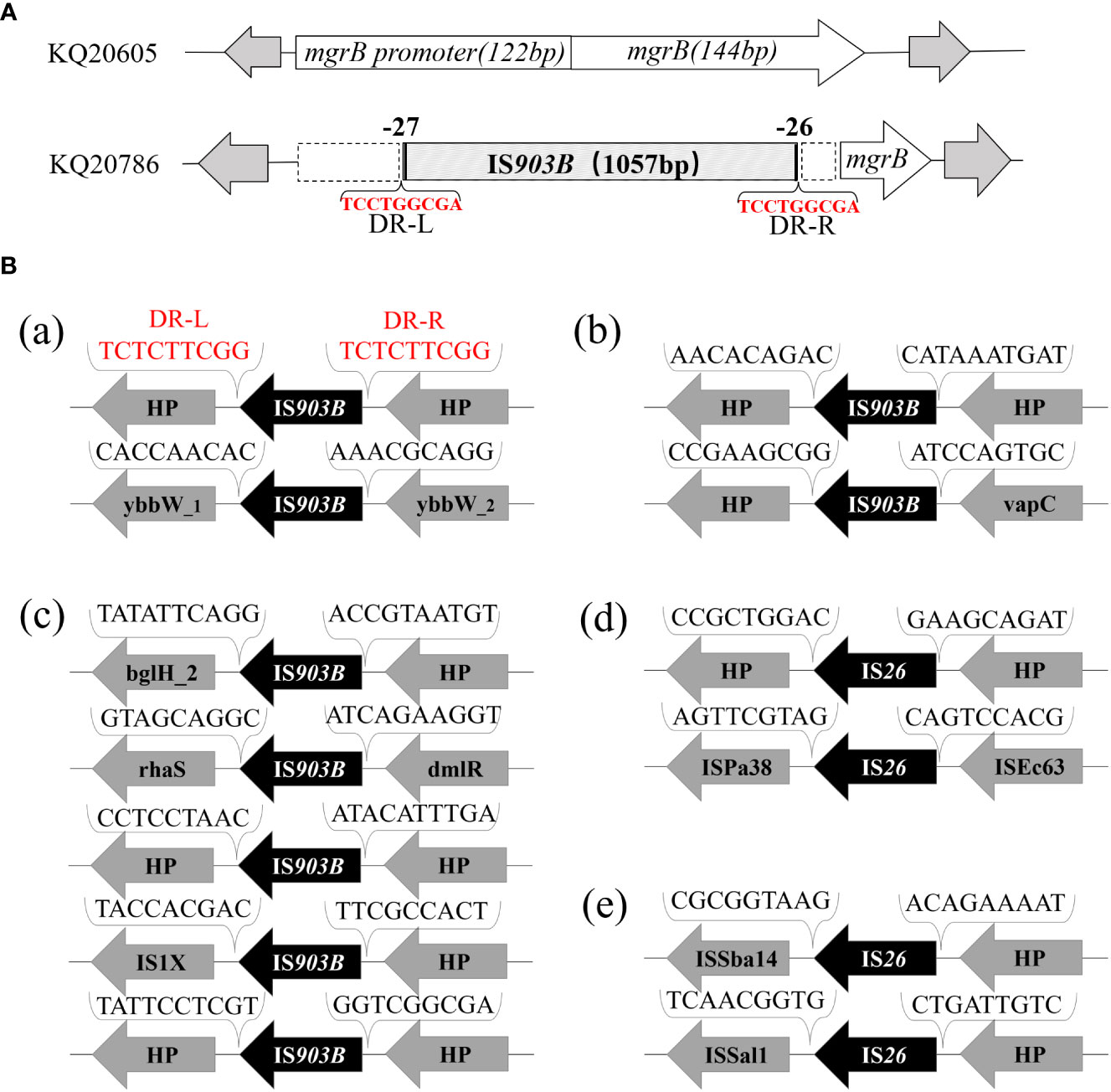
Figure 4 Analysis of insertional inactivated mgrB gene of KQ20786 and surrounding genetic environment of △mgrB-associated IS elements in KQ20605. (A) Visual figure of mgrB gene or insertional inactivation mgrB gene of KQ20605 and KQ20786. (a) Schematic diagram of mgrB genes of the parental KQSS strain (K Q20605) and the colistin-resistant variant (KQ20786) integrated with IS903B element between the -27th and -26th bp of mgrB promoter. (B) Surrounding genetic environment of IS903B or IS26 elements of KQSS strain KQ20605. (a) 2 IS903B elements located on chromosome of KQ20605: one flanked by 9-bp direct repeats (DRs) (upper) and the other not (down). (b) 2 IS903B elements on plasmid pKQ20605-IncR of KQ20605: neither flanked by DRs, with one (upper) next to a 698-bp incomplete IS903B and sharing a 100% (1057/1057) nucleotide identity and coverage match with that on insertional inactivation mgrB gene promoter of KQ20786. (c) 5 IS903B elements on plasmid pKQ20605-IncFIB(K) of KQ20605: none flanked by DRs. (d, e) 4 IS26 elements on genome of KQ20605, 2 on plasmid pKQ20605-IncN (d) and 2 on plasmid pKQ20605-IncR (e) respectively: neither flanked by DRs. KQSS, K quasipneumoniae subsp. similipneumoniae.
To trace the source of IS903B in disruptive mgrB of KQ20786, the genomes of KQ20605 were uploaded on ISFinder website and plasmidFinder database, respectively, and the results showed that it carried 9 IS903B elements in the genome, including 2 on the chromosome, 5 on pKQ20605-IncFIB(K) and 2 on pKQ20605-IncR, respectively (Figures 3, 4B). Furthermore, to clarify whether the IS903B inserted in the chromosomal mgrB promoter of KQ20786 was from chromosome or plasmid, this IS903B sequence in mgrB was aligned with all 9 IS903B elements on the whole genome. The comparison result showed that the inserted IS903B element in mgrB of KQ20786 exhibited 100% (1057/1057) and 99% (1042/1057) identity as well as 100% and 100% coverage to IS903B elements on plasmid pKQ20605-IncR of KQ20786 and KQ20605, respectively. By comparison, the identity between IS903B inserted into mgrB of KQ20786 and each IS903B element located on chromosome in KQ20605 or KQ20786 was all only 95.1% (1005/1057), indicating the role of plasmids as possible donors for IS903B elements. Also, the upstream of the IS903B element that is 100% identical to IS903B in mgrB gene of KQ20786 was close to a fragmentary IS903B (only 698 bp), as shown in Figure 3D. In addition, as shown in Figurs 4B (a), one of the two IS903B elements on the chromosome of KQ20605 was flanked by a 9-bp DRs, but no IS903B element on plasmids was flanked by DRs (Figure 4B (b) and (c)). Although nearly all IS elements could genetically exchange, the transposition frequency of IS elements with DR at both ends is lower than those without DR (Zhang et al., 2021). Therefore, we suspect that the IS903B in mutant mgrB gene of KQ20786 is more likely to come from plasmids.
To ascertain if KQ20605 might develop into colistin resistance via mgrB gene insertional inactivation and whether the other IS elements on plasmids could also be transferred and inserted into mgrB gene under the colistin pressure, the in vitro colistin-induced resistance experiment was performed on KQ20605 over 6 days. On the 3rd day, colonies were detected on MHA containing 4mg/L colistin, indicating an in vitro transit from a colistin-sensitive to a resistant phenotype occurred under the pressure of colistin. Then, the completeness of mgrB genes and promoter regions of 23 randomly selected resistant induced KQSS strains were tested using PCR and Sanger sequencing, and 43.5% (10/23) of them were inserted by IS903B elements, 8 between the -26th and -25th bp of mgrB promoter (KQ20605-1), 1 between the 69th and 70th bp (KQ20605-3), and 1 between the 44th and 45th bp (KQ20605-4) of mgrB gene, as shown in Figure 5. In addition, the insertion of another IS type (IS26 element) into the mgrB promoter between the -20th and -19th bp (KQ20605-2) was identified in 21.7% (5/23) colistin-resistant strains. No mgrB gene was identified in 30.4% (7/23) strains (KQ20605-5). The BLASTn analysis of WGS for KQ20605-5 indicated that its mgrB gene and surroundings (1038bp) were lost. Also, 4.34% (1/23) isolates contained intact mgrB gene (KQ20605’). The above strains (KQ20605-1, KQ20605-2, and KQ20605-5) were confirmed to have obvious homology to KQ20605 by AST (Table 1), PFGE (Figure 2B), species identification as well as morphological characteristics, thus excluding the possibility of contaminations. The mgrB inserted by IS elements in all colistin-resistant KQSS strains was flanked by DRs, as shown in Figures 4B, 5A. Apart from IS903B, IS26 elements were also found frequently in inactivated mgrB gene in our induced experiment. To decipher their sources, the genome of KQ20605 was uploaded to the ISFinder website and CGE website. There were 4 IS26 elements, 2 on the pKQ20605-IncN plasmid co-harboring qnrS1, dfrA14 and blaNDM-1 and 2 on pKQ20605-IncR, respectively. The IS26 elements were neither identified on chromosome nor flanked by DRs, as shown in Figures 3, 4B (d) and (e). The 100% identity and 100% coverage between IS26 inserted in mgrB of colistin-induced strains and IS26 elements on the genome of KQ20605 were identified. Through conjugation experiments, we found that pKQ20605-IncN could transfer to E.coli J53 at a frequency of 1.33x10-6, thus causing multidrug resistance in the recipient J53. The proposed colistin resistance evolution mechanism of KQ20605 mediated by insertion sequences is summarized in Figure 6. Except for mgrB and its promoter, no other colistin-resistant mechanisms were detected, such as pmrA, pmrB, phoP, phoQ, crrA, crrB, crrC, pmrD, pmrK, pmrC, among KQ20786, KQ20605-1, KQ20605-2 and KQ20605-5 using KQ20605 as reference (Table S1).
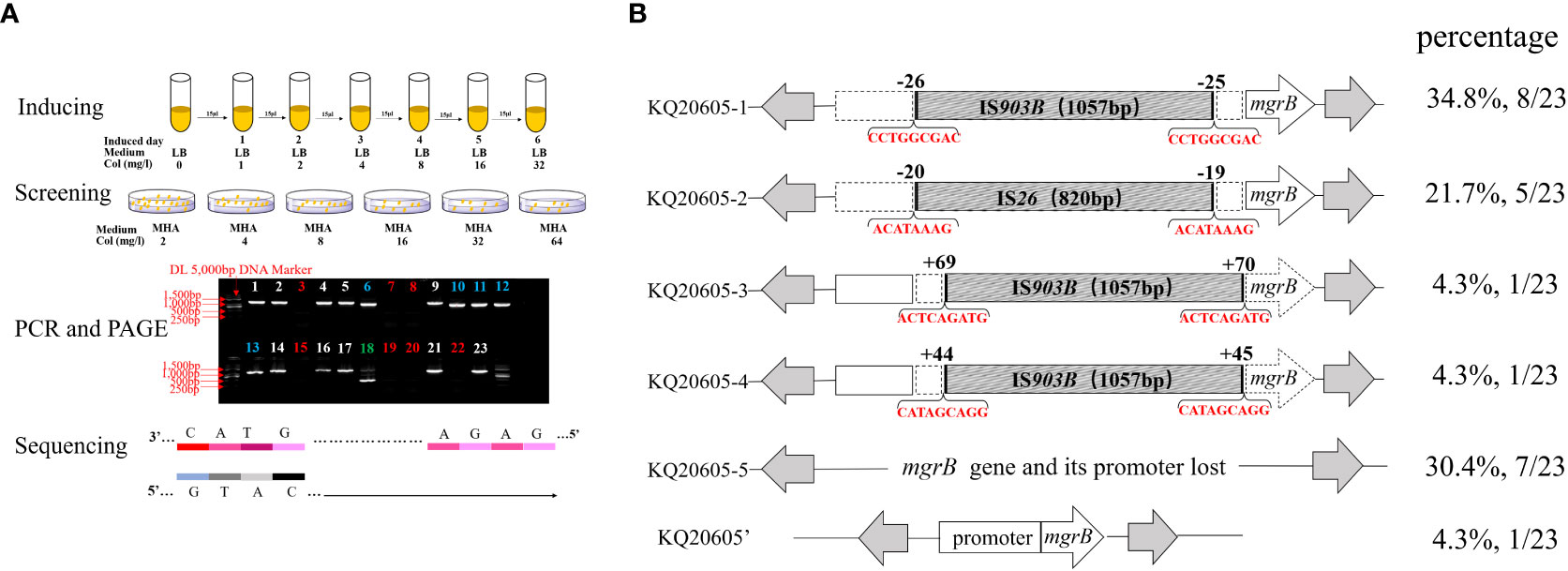
Figure 5 Experimental flow graph and analysis on insertional inactivated mgrB gene of in vitro colistin-inducted resistant variants (A) Schematic diagram of in vitro colistin-induced experiment and electropherogram results. a: In vitro colistin-induced experiment of KQ20605; b: Well-separated colonies growing on 4 mg/L colistin-containing MHA plates were selected to screen colistin-resistant isolates. c: agarose gel electrophoresis of the colistin-resistant KQSS strains performed on the 3rd day or 6th day of colistin-induced experiments. Strips 1, 2, 4, 5, 9, 14, 16, 17, 21 and 23 indicated IS903B-inserted mgrB genes, which were highlighted by white color; Strips 6, 10, 11, 12 and 13 were IS26-inserted mgrB genes, which were highlighted by blue color; Strips 3, 7, 8, 15, 19, 20 and 22 were large segment-inserted mgrB genes or mgrB-lost, which were highlighted by red color; Strips 18 was intact mgrB gene without IS insertion, which was highlighted by green color. d: All confirmed by sequencing. Col, colistin; MHA, Mueller-Hinton agar; AGE, agarose gel electrophoresis. (B) Insertional inactivation mgrB gene of KQSS strains in vitro. Schematic diagram of mgrB genes of KQ20605-1 to 5 and KQ20605’, in vitro colistin-inducted resistant variants of parental strain KQ20605. In vitro, IS26 or IS903B element inserted into mgrB gene. All IS26 elements were inserted between the -20th and -19th bp of mgrB promoter (KQ20605-2). IS903B elements shared various insertion positions including the -26th and -25th, 44th and 45th, as well as 69th and 70th bp of mgrB gene. The parental strain of these isolates is KQ20605 with an intact mgrB gene. Their proportions are listed on the right side of the figure respectively. Each IS element in the disruptive mgrB gene was flanked by DRs consisting of 8-9 bp. KQSS, K. quasipneumoniae subsp. similipneumoniae.
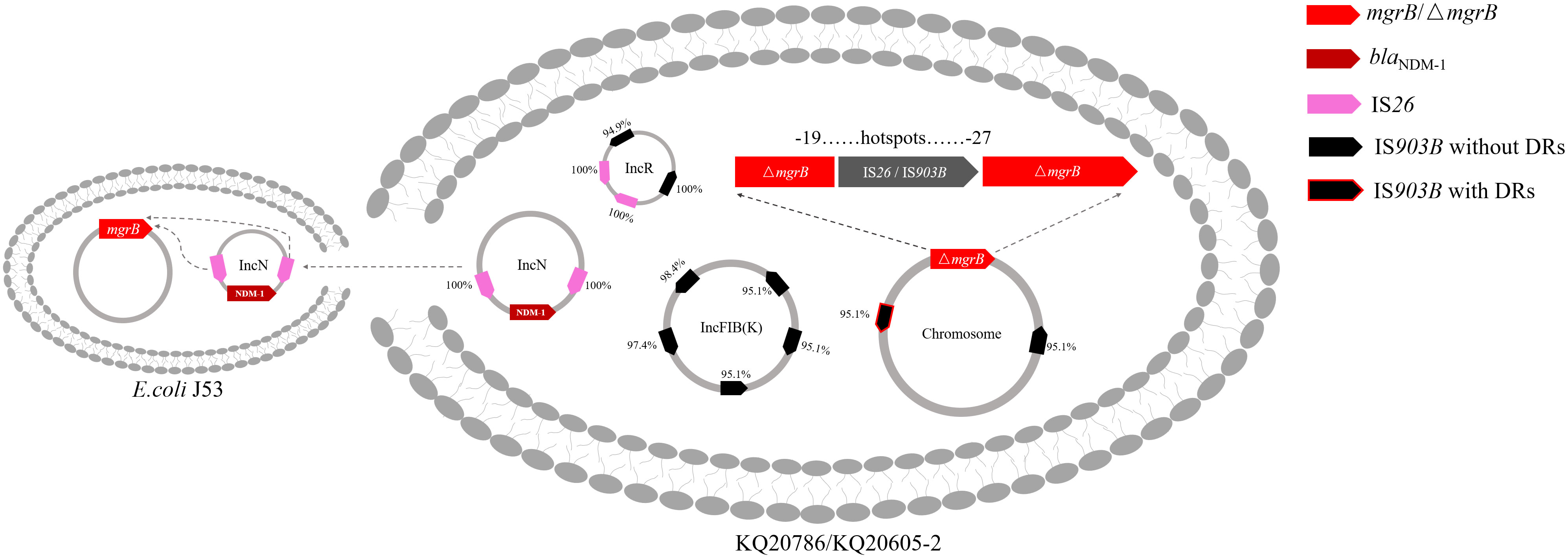
Figure 6 Proposed colistin resistance evolution mechanism of KQ20605 mediated by insertion sequences. The insertion sequence (IS) elements in mgrB possibly originated from plasmids. The mgrB gene is inserted by IS26 or IS903B at “hotspots” ranging from -27th to -19th. Black arrows: IS903B; Pink arrows: IS26. Arrows with red borders: IS elements flanked by direct repeats (DRs); Bright Red: mgrB or mutant mgrB; Dark red: blaNDM-1. Percentage next to IS elements represents the identity between IS on mgrB gene and its corresponding IS on genome.
The growth kinetics of these isolates (KQ20605, KQ20786, KQ20605-1, KQ20605-2, KQ20605-5, and KQ20786-C) were independently investigated 3 times to determine whether the different IS types in insertional inactivation mgrB brought different fitness costs on KQSS strains. As shown in Figure S1, no significant difference was detected in the growth rates of mgrB inactivation-induced isolates and KQ20786 in LB compared to its parental isolate KQ20605. Also, no impact was detected on bacterial fitness from the complementation.
The prevalence of carbapenem-resistant Klebsiella spp. has been a major public health concern worldwide (Biswas et al., 2012; Grundmann et al., 2017; Binsker et al., 2022). Colistin is considered the “last resort” treatment for carbapenem-resistant Klebsiella spp. In this study, we reported a pair of genetically sequential and NDM-producing Klebsiella strains from a lung transplant child and explored the underlying mechanism of transition from a colistin-susceptible strain (KQ20605) to the resistant one (KQ20786) after 6-day colistin exposure.
Previous studies have suggested that the chromosomal mgrB gene might be disrupted by IS elements integration and result in colistin resistance (Cannatelli et al., 2014; Olaitan et al., 2014; Poirel et al., 2015; Esposito et al., 2018; Shankar et al., 2019; Yang et al., 2020). For example, of 32 colistin-resistant K. pneumoniae isolates from Taiwan, 21 had their mgrB genes inserted by IS elements and 1/21 by IS903B-like at +44th bp of mgrB (Yang et al., 2020). Similarly, Olaitan et al. studied mgrB insertional inactivation in 6 of 12 mgrB-disruptive and colistin-resistant clinical isolates collected in Lao PDR, Thailand, Israel, Nigeria, and France, with 3 isolates having IS903B-like insertion at nt95, nt82 and nt26 of mgrB, respectively (Olaitan et al., 2014). Several authors have attributed the rise and spread of colistin resistance in Klebsiella to insertional inactivation of the mgrB gene by IS elements from plasmids (Giordano et al., 2018; Shankar et al., 2019; Zhang et al., 2021; Fordham et al., 2022). However, clinical data and in vitro experiments that could confirm this hypothesis are lacking.
In the current study, the IS903B element in KQ20605 was mobilized and integrated into the mgrB gene promoter, resulting in colistin resistance (KQ20786) after colistin administration. Comparative genomic analysis of both strains revealed that the plasmid pKQ20605-IncR might serve as a donor for IS903B elements integrated into the chromosomal mgrB gene. The IS903B transposed into chromosomal mgrB gene has a 100% (1057/1057) nucleotide identity and coverage match with one IS903B element on plasmid pKQ20605-IncR, and only 95.1% (1005/1057) nucleotide identity to that on the chromosome. In addition, 9 IS903B elements on the genome, 2 on the chromosome, and 7 on plasmids (pKQ20605-IncFIB(K) and pKQ20605-IncR), were found. It was found that IS903B elements tended to be distributed on plasmids to mediate colistin resistance, especially on the fusion plasmid IncHI2A + IncHI2, IncFIB(K) and IncR (Fordham et al., 2022). Also, one IS903B on the chromosome was flanked by DRs, whereas none of the IS903B elements on plasmids were flanked. Previous studies confirmed that the IS element (ISKpn72) with DRs had a lower transposition frequency than without (Zhang et al., 2021). To sum up, it is reasonable to hypothesize that IS903B elements on plasmids rather than on chromosomes were more inclined to be integrated into the chromosomal mgrB gene, thereby causing colistin resistance. Thus, further experiments have been performed to prove our hypothesis.
In the in vitro colistin-induced experiment of the KQ20605 strain, the mutation types of the mgrB gene varied, involving insertional inactivation at “hotspots” sites of the mgrB gene and the loss of it, which revealed that the inactivation of the mgrB gene is an inevitable event in this strain under the colistin pressure. Specifically, 65.2% (15/23) of colistin-induced isolates were inserted by various IS elements (e.g., IS903B and IS26) in the mgrB gene. Among these 15 isolates, 8 strains were integrated IS903B element between the -26th and -25th bp of the mgrB gene, causing colistin resistance, an almost identical “hotspot” found in our patient. Moreover, 5 colistin-induced resistant isolates (KQ20605-2) were identified with the integration of IS26 element between the -20th and -19th bp of the mgrB gene. The inserted sites in the mgrB gene of this isolate were frequently focused on promoter regions ranging from -27th to -19th bp. Taken together, our data suggest the existence of insertional preferred sites, namely, “hotspots”, in the mgrB gene, both in vivo and in vitro under specific conditions, as also documented in other insertional inactivated mgrB mutants (Fordham et al., 2022).
The IS26 element, commonly located on antibiotic-resistant plasmids (Partridge et al., 2018) has been rarely reported to insert into the mgrB gene (Silva et al., 2021). A previous study confirmed that the ISKpn72-and-blaKPC harboring mobile plasmid enables the strain to develop colistin heteroresistance from susceptibility by integrating ISKpn72 into the mgrB gene from the mobile plasmid (Zhang et al., 2021). However, proof of the role of IS26 elements in the emergence and dissemination of colistin resistance was lacking. Comparative genomic analysis showed that the IS26 elements were all located on plasmids (pKQ20605-IncR and pKQ20605-IncN), and none of them were flanked by DRs in KQ20605, further demonstrating that plasmids served as donors to encode mobilizable IS elements integrating into the chromosomal mgrB gene. Moreover, the plasmid pKQ20605-IncN of the KQ20605 strain, co-harboring qnrS1, dfrA14, and blaNDM-1, could be conjugated into E. coli J53, hinting that under the colistin pressure, the IS26 elements on the carbapenem-resistant plasmid pKQ20605-IncN could be activated and mobilized into chromosomal mgrB gene, thereby causing the development, and even dissemination of colistin resistance.
In conclusion, we identified a colistin-resistant KQSS strain transited from its sensitive parental strain by acquiring an IS element in the chromosomal mgrB gene promoter in the process of using colistin. Furthermore, we provided clinical data and experimental evidence for the possibility that plasmids preferred to encode mobilizable IS elements, resulting in the insertional inactivation of chromosomal mgrB gene at “hotspots”, and subsequently causing the colistin resistance emergence and further potential dissemination.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, BioProject: PRJNA985803.
Permission for using the information in the medical records of the patient and the KQSS isolates for research purposes was granted by the Ethics Committee of the China-Japan Friendship Hospital (CJFH) (2022-KY-054).
YW, JZ, ZL, XL, FZ, YH, DP, XZ, HS, YZ, CL and BL collected the clinical and laboratory data. YW and BL made substantial contributions to conception and design, drafted, reviewed, and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the CAMS Innovation Fund for Medical Sciences (CIFMS; No.2021-I2M-1-030).
We would like to thank all our colleagues in the Laboratory of Clinical Microbiology and Infectious Diseases, CJFH, for their assistance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1153387/full#supplementary-material
Binsker, U., Käsbohrer, A., Hammerl, J. A. (2022). Global colistin use: a review of the emergence of resistant Enterobacterales and the impact on their genetic basis. FEMS Microbiol. Rev. 46, 22–26. doi: 10.1093/femsre/fuab049
Biswas, S., Brunel, J. M., Dubus, J. C., Reynaud-Gaubert, M., Rolain, J. M. (2012). Colistin: an update on the antibiotic of the 21st century’. Expert Rev. Anti Infect. Ther. 10, 917–934. doi: 10.1586/eri.12.78
Cannatelli, A., D’Andrea, M. M., Giani, T., Di Pilato, V., Arena, F., Ambretti, S., et al. (2013). In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 57, 5521–5526. doi: 10.1128/AAC.01480-13
Cannatelli, A., Giani, T., D’Andrea, M. M., Di Pilato, V., Arena, F., Conte, V., et al. (2014). MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother. 58, 5696–5703. doi: 10.1128/AAC.03110-14
CLSI (2022). Performance Standards for Antimicrobial Susceptibility Testing, 32nd Edition. CLSI supplement M100 (Wayne, PA: Clinical and Laboratory Standards Institute).
El-Sayed Ahmed, M. A. E., Zhong, L. L., Shen, C., Yang, Y., Doi, Y., Tian, G. B. (2020). Colistin and its role in the Era of antibiotic resistance: an extended revie-2019). Emerg. Microbes Infect. 9, 868–885. doi: 10.1080/22221751.2020.1754133
Esposito, E. P., Cervoni, M., Bernardo, M., Crivaro, V., Cuccurullo, S., Imperi, F., et al. (2018). Molecular Epidemiology and Virulence Profiles of Colistin-Resistant Klebsiella pneumoniae Blood Isolates From the Hospital Agency “Ospedale dei Colli. Naples Italy’ Front. Microbiol. 9, 1463. doi: 10.3389/fmicb.2018.01463
Fordham, S. M. E., Mantzouratou, A., Sheridan, E. (2022). Prevalence of insertion sequence elements in plasmids relating to mgrB gene disruption causing colistin resistance in Klebsiella pneumoniae. Microbiologyopen 11, e1262. doi: 10.1002/mbo3.1262
Giordano, C., Barnini, S., Tsioutis, C., Chlebowicz, M. A., Scoulica, E. V., Gikas, A., et al. (2018). Expansion of KPC-producing Klebsiella pneumoniae with various mgrB mutations giving rise to colistin resistance: the role of ISL3 on plasmids. Int. J. Antimicrob. Agents 51, 260–265. doi: 10.1016/j.ijantimicag.2017.10.011
Grundmann, H., Glasner, C., Albiger, B., Aanensen, D. M., Tomlinson, C. T., Andrasević, A. T., et al. (2017). Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect. Dis. 17, 153–163. doi: 10.1016/S1473-3099(16)30257-2
Imai, K., Ishibashi, N., Kodana, M., Tarumoto, N., Sakai, J., Kawamura, T., et al. (2019). Clinical characteristics in blood stream infections caused by Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae: a comparative study, Japa-2017. BMC Infect. Dis. 19, 946. doi: 10.1186/s12879-019-4498-x
Janssen, A. B., Doorduijn, D. J., Mills, G., Rogers, M. R. C., Bonten, M. J. M., Rooijakkers, S. H. M., et al. (2020). Evolution of colistin resistance in the klebsiella pneumoniae complex follows multiple evolutionary trajectories with variable effects on fitness and virulence characteristics. Antimicrob. Agents Chemother. 65, 11. doi: 10.1128/AAC.01958-20
Jiang, H., Lei, R., Ding, S. W., Zhu, S. (2014). Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinf. 15, 182. doi: 10.1186/1471-2105-15-182
Li, Z., Liu, X., Lei, Z., Li, C., Zhang, F., Wu, Y., et al. (2023). Genetic diversity of polymyxin-resistance mechanisms in clinical isolates of carbapenem-resistant klebsiella pneumoniae: a multicenter study in China. Microbiol. Spectr. 11, e0523122. doi: 10.1128/spectrum.05231-22
Li, J., Rayner, C. R., Nation, R. L., Owen, R. J., Spelman, D., Tan, K. E., et al. (2006). Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 50, 2946–2950. doi: 10.1128/AAC.00103-06
Lippa, A. M., Goulian, M. (2009). Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PloS Genet. 5, e1000788. doi: 10.1371/journal.pgen.1000788
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Long, S. W., Linson, S. E., Ojeda Saavedra, M., Cantu, C., Davis, J. J., Brettin, T., et al. (2017). Whole-Genome Sequencing of Human Clinical Klebsiella pneumoniae Isolates Reveals Misidentification and Misunderstandings of Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae. mSphere 2, 3–8. doi: 10.1128/mSphereDirect.00290-17
Mashaly, G. E., Mashaly, M. E. (2021). Colistin-heteroresistance in carbapenemase-producing Enterobacter species causing hospital-acquired infections among Egyptian patients. J. Glob Antimicrob. Resist. 24, 108–113. doi: 10.1016/j.jgar.2020.11.019
Olaitan, A. O., Diene, S. M., Kempf, M., Berrazeg, M., Bakour, S., Gupta, S. K., et al. (2014). Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int. J. Antimicrob. Agents 44, 500–507. doi: 10.1016/j.ijantimicag.2014.07.020
Partridge, S. R., Kwong, S. M., Firth, N., Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31, 4–8. doi: 10.1128/CMR.00088-17
Poirel, L., Jayol, A., Bontron, S., Villegas, M. V., Ozdamar, M., Turkoglu, S., et al. (2015). The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J. Antimicrob. Chemother. 70, 75–80. doi: 10.1093/jac/dku323
Prjibelski, A., Antipovm, D., Meleshko, D., Lapidus, A., Korobeynikov, A. (2020). Using SPAdes de novo assembler. Curr. Protoc. Bioinf. 70 (1), e102. doi: 10.1002/cpbi.102
Shankar, C., Pragasam, A. K., Anandan, S., Veeraraghavan, B. (2019). mgrB as hotspot for insertion sequence integration: change over from multidrug-resistant to extensively drug-resistant klebsiella pneumoniae? Microb. Drug Resist. 25, 1122–1125. doi: 10.1089/mdr.2018.0415
Silva, D. M. D., Faria-Junior, C., Nery, D. R., Oliveira, P. M., Silva, L. O. R., Alves, E. G., et al. (2021). Insertion sequences disrupting mgrB in carbapenem-resistant Klebsiella pneumoniae strains in Brazil. J. Glob Antimicrob. Resist. 24, 53–57. doi: 10.1016/j.jgar.2020.11.003
Tzouvelekis, L. S., Markogiannakis, A., Psichogiou, M., Tassios, P. T., Daikos, G. L. (2012). Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25, 682–707. doi: 10.1128/CMR.05035-11
Weinert, T. A., Schaus, N. A., Grindley, N. D. (1983). ‘Insertion sequence duplication in transpositional recombination’. Science 222, 755–765. doi: 10.1126/science.6314502
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PloS Comput. Biol. 13 (6), e1005595. doi: 10.1371/journal.pcbi.1005595
Yan, W., Zhang, Q., Zhu, Y., Jing, N., Yuan, Y., Zhang, Y., et al. (2021). Molecular Mechanism of Polymyxin Resistance in Multidrug-Resistant Klebsiella pneumoniae and Escherichia coli Isolates from Henan Province, China: A Multicenter Study. Infect. Drug Resist. 14, 2657–2666. doi: 10.2147/IDR.S314490
Yang, T. Y., Wang, S. F., Lin, J. E., Griffith, B. T. S., Lian, S. H., Hong, Z. D., et al. (2020). ‘Contributions of insertion sequences conferring colistin resistance in Klebsiella pneumoniae. Int. J. Antimicrob. Agents 55, 105894. doi: 10.1016/j.ijantimicag.2020.105894
Zhang, B., Yu, B., Zhou, W., Wang, Y., Sun, Z., Wu, X., et al. (2021). Mobile plasmid mediated transition from colistin-sensitive to resistant phenotype in klebsiella pneumoniae’. Front. Microbiol. 12, 619369. doi: 10.3389/fmicb.2021.619369
Zhao, J., Li, Z., Zhang, Y., Liu, X., Lu, B., Cao, B. (2022). Convergence of MCR-8.2 and chromosome-mediated resistance to colistin and tigecycline in an NDM-5-producing ST656 klebsiella pneumoniae isolate from a lung transplant patient in China. Front. Cell Infect. Microbiol. 12, 922031. doi: 10.3389/fcimb.2022.922031
Keywords: Klebsiella quasipneumoniae subsp. similipneumoniae, colistin, mgrB gene, insertional sequence, within-host evolution
Citation: Wu Y, Zhao J, Li Z, Liu X, Hu Y, Zhang F, Zhang Y, Pu D, Li C, Zhuo X, Shi H and Lu B (2023) Within-host acquisition of colistin-resistance of an NDM-producing Klebsiella quasipneumoniae subsp. similipneumoniae strain through the insertion sequence-903B-mediated inactivation of mgrB gene in a lung transplant child in China. Front. Cell. Infect. Microbiol. 13:1153387. doi: 10.3389/fcimb.2023.1153387
Received: 07 March 2023; Accepted: 10 August 2023;
Published: 31 August 2023.
Edited by:
Maria De Lurdes Enes Dapkevicius, University of the Azores, PortugalReviewed by:
Margarida C. Gomes, University of London, United KingdomCopyright © 2023 Wu, Zhao, Li, Liu, Hu, Zhang, Zhang, Pu, Li, Zhuo, Shi and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Binghuai Lu, enMyNTA0MUAxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.