
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol., 30 May 2023
Sec. Virus and Host
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1151069
This article is part of the Research TopicEpigenetics of Virus-Host InteractionsView all 5 articles
N6-Methyladenosine (m6A) modification is the most abundant covalent modification of RNA. It is a reversible and dynamic process induced by various cellular stresses including viral infection. Many m6A methylations have been discovered, including on the genome of RNA viruses and on RNA transcripts of DNA viruses, and these methylations play a positive or negative role on the viral life cycle depending on the viral species. The m6A machinery, including the writer, eraser, and reader proteins, achieves its gene regulatory role by functioning in an orchestrated manner. Notably, data suggest that the biological effects of m6A on target mRNAs predominantly depend on the recognition and binding of different m6A readers. These readers include, but are not limited to, the YT521-B homology (YTH) domain family, heterogeneous nuclear ribonucleoproteins (HNRNPs), insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs), and many others discovered recently. Indeed, m6A readers have been recognized not only as regulators of RNA metabolism but also as participants in a variety of biological processes, although some of these reported roles are still controversial. Here, we will summarize the recent advances in the discovery, classification, and functional characterization of m6A reader proteins, particularly focusing on their roles and mechanisms of action in RNA metabolism, gene expression, and viral replication. In addition, we also briefly discuss the m6A-associated host immune responses in viral infection.
Structural and functional analyses of RNA modifications have recently become a hot area of epigenetic research. In eukaryotes, posttranslational modification of RNA is very common and over 100 types of covalent modifications have been discovered so far (Boccaletto et al., 2018). Among them, the most prevalent modification is methylation at the N6 of adenosine. The N6-methyladenosine (m6A) modification was discovered early in mRNAs. With advances in molecular detection techniques, m6A modifications have been increasingly discovered in many other types of RNAs, such as mRNAs, microRNAs, long non-coding RNAs, and circular RNAs (Chen et al., 2019; Zaccara et al., 2019). Sequence analyses have found that, in mRNAs, ~0.1%–0.4% of adenosines are N6-methylated (Wei et al., 1975; Sommer et al., 1978) and ~25% of cellular mRNAs contain multiple m6A residues (Wei and Moss, 1977; Csepany et al., 1990). Epitranscriptomic analyses have found that m6A sites are highly conserved and generally enriched in the consensus motif RRACH (R= G or A, and H = A, C, or U), which is more likely to be detected in the 3′-untranslated regions (3′UTRs), near stop codons and within internal long exons (Dominissini et al., 2012; Meyer et al., 2012). The presence of a long internal exon, which is defined as an exon that is much longer than the ~140-bp length of a typical exon, is considered to be an inducer of m6A deposition within a transcript (Dominissini et al., 2012; Ke et al., 2015; Ke et al., 2017).
The m6A level of an mRNA is dynamically maintained at a relatively balanced level via the crosstalk of several enzymes, including methyltransferases (writers) and demethylases (erasers). For example, in HIV and Enterovirus infection, viral infection can increase writer (METTL3/14 (methyltransferase 3/14)) and decrease eraser (FTO (fat mass and obesity-associated protein) or ALKBH5 (alkB homolog 5)) levels and thus promote viral replication. This dynamic and functional relevance of m6A for viral replication was further verified by silencing of the m6A writer or eraser enzymes, which decreased and increased viral replication, respectively (Lichinchi G et al., Nat Microbiol. 1: 16011, 2016; Hao H et al., Nucleic Acids Res. 47: 362, 2018). METTL3, a core component with methyltransferase activity, can combine with S-adenosylmethionine (SAM) to catalyze RNA methylation in the nucleus during transcription (Liu et al., 2014; Wang et al., 2016). However, METTL14 is an allosteric activator and functions as an RNA-binding platform to form a stable heterodimer with METTL3, strengthening the catalytic effect of METTL3 (Liu et al., 2014; Wang et al., 2016). Interestingly, WTAP (WT1-associated protein) interacts with the METTL3–METTL14 complex to ensure its localization to nuclear speckles and also modulates its recruitment to mRNA targets to promote its catalytic activity (Ping et al., 2014). Some other catalytic subunits or regulatory factors, such as vir-like m6A methyltransferase associated (VIRMA, also termed KIAA1429), RNA-binding motif protein 15/15B (RBM15/RBM15B), HAKAI (also named CBLL1, Cbl proto-oncogene-like 1), and zinc finger CCCH domain-containing protein 13 (ZC3H13), have also been described. VIRMA preferentially mediates m6A modification in the 3′UTR and near the stop codon, affecting the selection of methylation sites (Yue et al., 2018). RBM15/RBM15B plays an important role in X-inactivation and gene silencing via m6A modification of long non-coding RNA XIST (X-inactive specific transcript) (Patil et al., 2016). HAKAI, an E3 ubiquitin-protein ligase, functions as an associated component of the WTAP complex in the induction of m6A methylation (Yue et al., 2018). ZC3H13 induces the nuclear localization of the Zc3h13–WTAP–Virilizer–Hakai complex to regulate m6A modification (Wen et al., 2018). Recently, some novel methyltransferase accessory factors have been discovered. For example, methyltransferase ZCCHC4 (zinc finger CCHC-type containing 4) is responsible for the m6A methylation of 28S rRNA, while METTL5 (methyltransferase 5) can modify the N6-adenosine of 18S rRNA with the assistance of TRMT112 (TRNA methyltransferase activator subunit 11-2), an allosteric activator facilitating the binding of the catalytic complex to the RNA target (Ma et al., 2019; Ren et al., 2019; van Tran et al., 2019). Another enzyme is METTL16 (methyltransferase 16). It catalyzes the formation of a single m6A in the U6 small nuclear RNA (snRNA) involved in splicing (Pendleton et al., 2017). Additionally, METTL16 can catalyze the formation of m6A in a small number of other mRNAs and non-coding RNAs (Warda et al., 2017).
m6A modifications can be removed by RNA demethylases. Currently, only two demethylases have been identified and both are AlkB family proteins. The first one is FTO, which is located in the nucleus and abrogates m6A levels via oxidative demethylation activity (Jia et al., 2011). The second is the ALKBH5, which can remove the m6A modification on nuclear RNAs and further modulate nuclear RNA export and RNA metabolism (Zheng et al., 2013). Functional studies have found that FTO is responsible for gaining body weight when having mutations (Dina et al., 2007; Frayling et al., 2007; Do et al., 2008; Jia et al., 2011), and ALKBH5 was reported to be important for mouse fertility as mice deficient in ALKBH5 have shrunken testis (Zheng et al., 2013). More recent studies have suggested that FTO and ALKBH5 have different substrate specificities. ALKBH5 demethylates transcripts containing m6A, including viral transcripts (Zheng et al., 2013; Lichinchi et al., 2016b). However, FTO has greater substrate specificity not only for mRNAs containing m6A but also for the 5′ cap-adjacent nucleotide containing m6A, 2′O-methylation (m6Am), which occurs on about 10% of transcripts (Mauer et al., 2017).
Although the m6A writer and eraser play important roles in m6A-mediated gene regulation, the effects of m6A modification on RNA metabolism, gene regulation, and many other biological functions predominantly depend on m6A recognition by different m6A-binding proteins (also called “readers”). The known reader proteins include, but are not limited to, the YT521-B homology (YTH) domain family, heterogeneous nuclear ribonucleoproteins (HNRNPs), and insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs). Several studies have demonstrated that these proteins are not only involved in normal gene regulation but also play crucial roles in modulating the pathophysiological conditions by interacting with the target mRNAs (Figure 1). In this review, we focus on recent advances in the discovery, classification, and functional characterization of m6A reader proteins, particularly focusing on their roles and mechanisms in RNA metabolism, gene regulation, and viral replication. In addition, we also briefly discuss the m6A-associated host immune responses in viral infection.
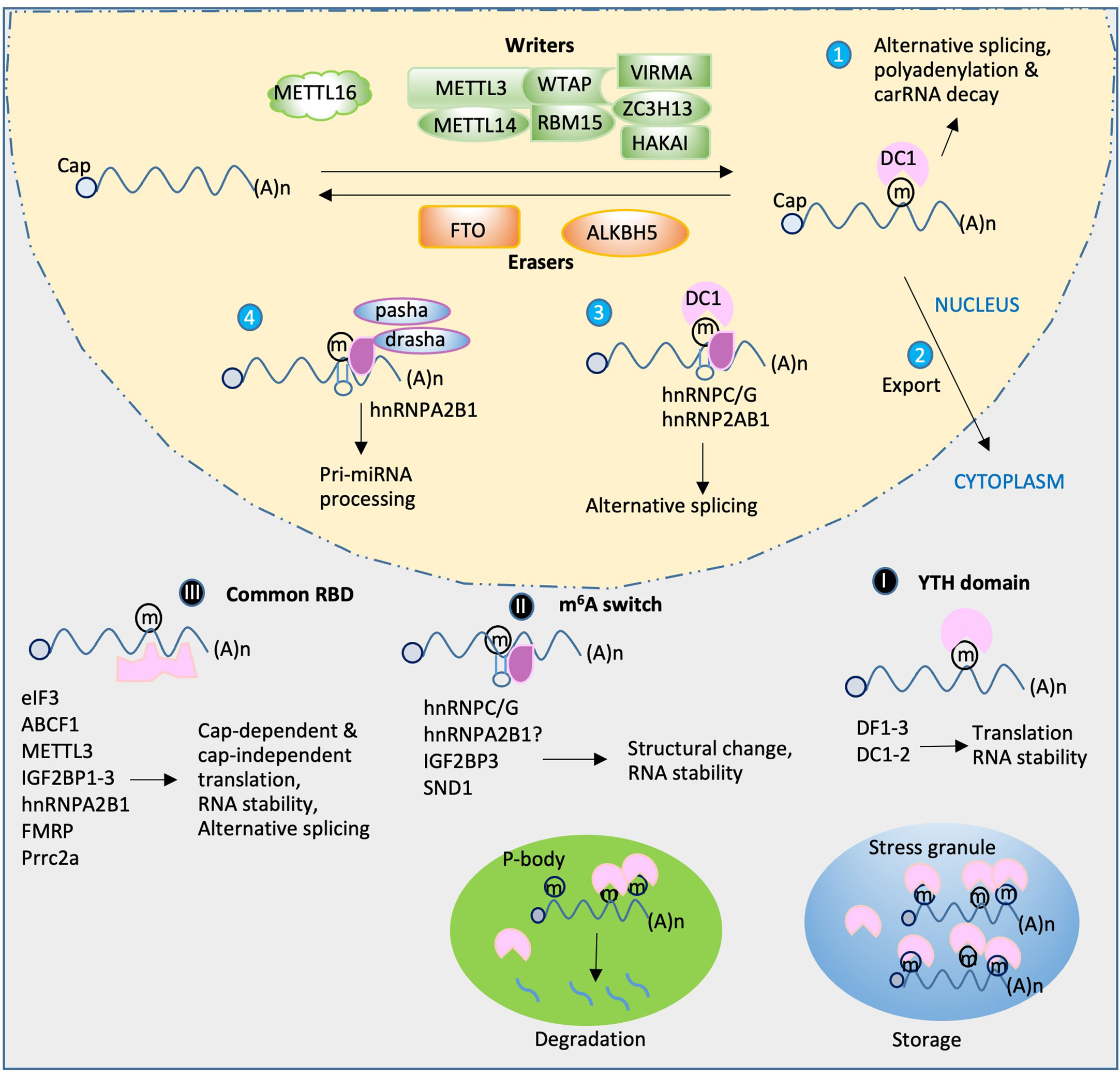
Figure 1 Functions of m6A reader proteins. RNA transcripts are m6A-methylated by a m6A writer complex composed of core subunits METTL3 and METT14 and additional accessory factors. METTL16 installs m6A on U6 snRNA and MAT2A mRNA. The m6A can be removed by demethylase FTO or ALKBH5. The reader proteins (purple) play diverse roles in RNA metabolism and gene expression. In nucleus, DC1 plays a role in alterative splicing, polyadenylation and carRNA decay (1) as well as m6A RNA export (2). The other readers hnRNPC/G or hnRNP2AB1 mediates alternative splicing (3) and pri-miRNA processing when drasha and pasha present (4). After export of the m6A RNA, three classes of readers in cytoplasm bind the m6A RNA in different mechanisms: (I) YTH readers bind m6A directly and regulate translation and RNA stability; (II) m6A switch-mediated binding. A local hairpin structure disrupted by m6A methylation favors binding events of a group of readers to regulate RNA structural changes and stability; (III) binding through tandem common RNA binding domain (RBD), such as KH, RGG and GRE domain. These readers as indicated regulate translation, splicing and RNA stability. During cellular stress, certain multiple m6A-containing RNAs and readers facilitate phase-separated compartment formation to produce stress granules for storage or transfer to P-body, leading to degradation.
m6A reader proteins, as the crucial factors in regulating m6A-modified transcripts, have been identified in many studies. The YTH domain-containing m6A readers (YTH readers) were the first discovered group of m6A readers and provided a mechanism for understanding the effects of m6A on mRNA biology (Dominissini et al., 2012). m6A methylation can destabilize the mRNA structure, which can affect the binding of diverse RNA-binding proteins (potential readers), leading to altered translation efficiency or other functional changes. With the technical advances in mapping of m6A sites and identifying m6A-binding proteins within the transcriptome, an increasing number of m6A readers have been discovered. Here, we summarize some of the major discoveries of m6A readers and their cellular functions, although controversies still exist on their functions among different reports.
The direct m6A reader group has five YTH domain-containing members, namely, three YTHDF1-3 (DF1-3) and two YTHDC1-2 (DC1-2) readers. Hereafter, we call these five proteins YTH readers. These proteins bind mRNA in an m6A-dependent manner. The YTH readers are evolutionarily conserved and independent of cell type (Wang et al., 2017). The cellular localization of these proteins differs: DF1-3 are cytosolic (Wang et al., 2014; Li et al., 2017a; Shi et al., 2017), DC1 is predominantly nuclear (Hartmann et al., 1999), and DC2 can be both nuclear and cytosolic (Wojtas et al., 2017). Structural studies have demonstrated that the selectivity of YTH readers for binding the methyl moiety of m6A is achieved mainly via a “tryptophan cage” (Luo and Tong, 2014; Xu et al., 2014; Xu et al., 2015), in which two or three tryptophans wrap around the methyl group. The three members (DF1–3) of the DF family proteins share high similarity in their amino acid (aa) sequence. In addition to the YTH domain (100–150 aa) of the protein, the remaining ~400-aa region contains several prion-like P/Q/N-rich domains (Li et al., 2014; Zhu et al., 2014) that can cause DF proteins to undergo “liquid-liquid phase separation” (LLPS). This LLPS is markedly enhanced by mRNAs that contain multiple, but not single, m6A residues. The resulting m6A-rich lipid droplets then participate in the formation of phase-separated compartments, such as stress granule and P-body (Ries et al., 2019) (Figure 1). From this point of view, PLrD may play a role in storage/protection of m6A-containing mRNA during the methylation process. Furthermore, the produced phase-separated compartments may negatively regulate certain viral replication by blocking viral particle assembly, such as that in Flavoviruses (Gokhale,N et al., Cell Host & Microbe, 20: 654, 2016). In addition, stress granule formation during viral infection is an antiviral response since stress granule is a platform of antiviral type I interferon immune signaling (Onomoto et al., 2012). However, a recent study showed that m6A modifications only play a minimal, or even no, role in mRNA partitioning into stress granules (Khong et al., 2022).
Although all three DF (DF1–3) proteins can enhance m6A-mRNA phase separation, there is conflicting evidence about whether they each have specialized effects on m6A-modified mRNAs. Earlier reports found that each of the three DF proteins has a different effect on m6A mRNAs; for instance, DF1 promotes the translation of m6A-modified mRNAs through interaction with the translation initiation factor eIF3 (eukaryotic translation initiation factor 3) rather than with the m7G-cap structure (Wang et al., 2015), DF2 enhances m6A mRNA degradation by recruiting m6A mRNAs to decay sites (Wang et al., 2014), and DF3 not only enhances translation by interacting with DF1 but also promotes degradation by associating with DF2 (Wang et al., 2014; Shi et al., 2017; Patil et al., 2018). However, other studies demonstrated that all three DFs have similar roles in mRNA degradation by recruiting the carbon catabolite repression 4 (CCR4)—negative on the TATA-less (NOT) deadenylation complex to m6A mRNAs (Du et al., 2016; Kennedy et al., 2016). Further studies on the crosstalk of these three DF proteins found that, remarkably, DF3 can facilitate mRNA translation of DF1/3 common targets, but not DF3 unique targets. Intriguingly, DF3 depletion decreases the binding of DF1 and DF2 to their target transcripts, while DF1 or DF2 loss reduces the amount of RNA that is bound by DF3 (Shi et al., 2017). Given the high sequence similarity among the three DF proteins, it is unclear how DFs mediate different functions. Another issue is whether DF proteins can bind different or the same m6A sites in mRNAs. Some recent studies have suggested that most m6A residues only bind one of the three DF paralogues (Shi et al., 2017), whereas others have suggested that all m6A sites bind all DF paralogues in largely equivalent manners (Patil et al., 2016). Since the m6A sites are largely located in the 3′UTR, stop codon region, the 5′UTR, and sometimes the coding sequences themselves (Dominissini et al., 2012; Meyer et al., 2012), binding of the reader to different m6A sites may mediate distinct functions. Additional studies are needed to clarify mechanisms of binding of DF proteins and their functional consequences.
As mentioned earlier, the three DFs and two YTHDCs have differential cellular localization. DC1 is usually found in the nucleus (Xu et al., 2014) and can bind mRNAs shortly after they are transcribed and methylated. Upon binding, DC1 may induce alternative splicing (i.e., promoting exon inclusion) by recruiting RNA splicing factor SRSF3 (serine and arginine-rich splicing factor 3) and blocking SRSF10 (serine and arginine-rich splicing factor 10) from binding to mRNAs (Xiao et al., 2016). DC1 also interacts with SRSF3 and NXF1 (nuclear RNA export factor 1) to facilitate m6A mRNA nuclear export (Roundtree et al., 2017). Furthermore, DC1 appears to mediate the function of m6A in non-coding RNA, such as XIST, a non-coding RNA that contributes to X chromosome inactivation and silencing of genes on the X chromosome (Patil et al., 2016). Another study on the non-coding chromosome-associated regulatory RNA (carRNA) found that m6A on carRNA facilitates transcriptional downregulation of proximal genes by inducing the decay of carRNA transcripts that regulate the chromatin state of proximal loci (Liu et al., 2020). DC2 is an RNA-induced ATPase with 3-to-5′ RNA helicase activity (Wojtas et al., 2017). Unlike other YTH domain-containing proteins, which are ubiquitously expressed, DC2 is enriched in the testes (Bailey et al., 2017; Hsu et al., 2017; Wojtas et al., 2017; Liu et al., 2021b). DC2 knockout mice show defects in spermatogenesis without other obvious developmental defects (Bailey et al., 2017; Hsu et al., 2017; Wojtas et al., 2017; Liu et al., 2021b). The binding properties of DC2 are unusual. It binds to m6A mRNA weaker than the four other YTH readers. Like other YTH proteins, the DC2 domain retains the “tryptophan cage” to bind methylated adenosine. However, the DC2’s YTH domain shows sequence discrepancy in the region that is predicted to bind m6A-adjacent residues (Patil et al., 2018). Transcriptome-wide mapping of DC2-binding sites by CLIP (CAP-Gly domain-containing linker protein 1) has shown low overlap with m6A sites (Patil et al., 2016). Thus, DC2 may bind select m6A sites or affect m6A mRNA through other binding mechanisms. Additionally, DC2 contains a helicase domain, R3H domain, and ankyrin repeats (Kretschmer et al., 2018), which may increase translation efficiency of target mRNAs by facilitating the interactions between m6A RNA and small subunit ribosomes (Hsu et al., 2017; Mao et al., 2019). Indeed, translation was increased when DC2 was artificially tethered to a reporter RNA (Hsu et al., 2017). Other studies suggest that DC2 mediates mRNA degradation through recruitment of the 5′–3′ exoribonuclease Xrn1 (Wojtas et al., 2017; Kretschmer et al., 2018) and thus decreases target gene expression.
The direct m6A readers that lack an YTH domain include eIF3, ATP-binding cassette F1 protein (ABCF1), METTL3, and heterogeneous nuclear ribonucleoproteins (HNRNPs). These readers preferentially bind m6A in the 5′UTR or non-coding regions. Although they can all bind m6A directly, HNRNPs seem to require m6A-induced RNA structural changes to facilitate binding.
Since eIF3, ABCF1, and METTL3 can all bind the 5′UTR to participate in translation initiation control, these m6A readers can be discussed as a group (Figure 1). eIF3 is a multiprotein complex that functions during the initiation phase of eukaryotic translation (Aitken and Lorsch, 2012). Recent studies have demonstrated that m6A modifications in the 5′UTR, especially under cellular stress conditions (Coots et al., 2017; Perlegos et al., 2022), destabilize the local RNA secondary structure, facilitating the direct binding of reader eIF3 to initiate translation, independent of the m7G-cap and cap-binding protein eIF4E. The typical example of such is the translation of heat shock protein 70 (Hsp70) mRNA during heat shock condition (Figure 2). This pattern of translation initiation is incompatible with another cap-independent mechanism characterized by the requirement of an internal ribosome entry site (IRES) within the 5′UTR (Meyer et al., 2015).
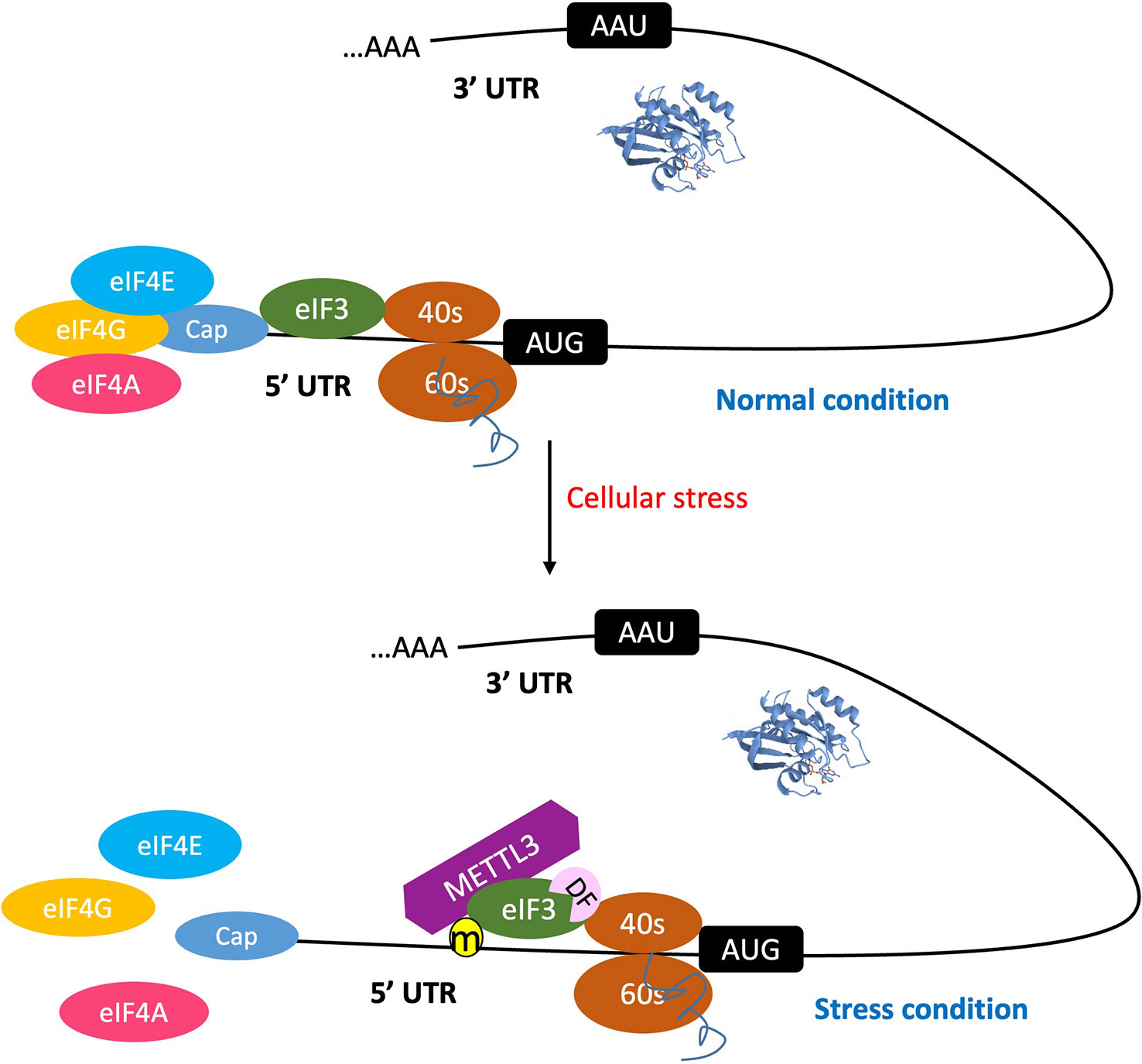
Figure 2 The 5'UTR m6A promotes cap-independent mRNA translation initiation. Under normal condition, the process of mRNA translation initiation occurs through a cap-dependent mechanism. When cells are subjected to stress, the m6A methylation machinery is activated. This leads to the destabilization of the mRNA secondary structure, thereby promoting the binding of reader protein eIF3 and/or METTL3 to the 5' UTR m6A site, and consequently initiating translation. This process is independent of the m7G-cap, as well as cap-binding proteins eIF4E and eIF4A. Furthermore, the reader DF-1 promotes translation by interacting with eIF3.
Other potential m6A reader proteins involved in the regulation of cap-independent translation initiation are ABCF1 and METTL3. Recent studies revealed that ABCF1 plays a crucial role in cap-independent translation of m6A mRNAs. This notion is supported by the observation of selective translation of heat shock-induced protein 70 (Hsp70) mRNA during cellular stress (Coots et al., 2017). A further investigation of quantitative proteomic data revealed that eIF2a, eIF5, and ABCF1 are closely associated with stress-induced Hsp70 mRNA. ABCF1-mediated promotion of translation initiation of m6A mRNA is supported by data, demonstrating that in cells lacking ABCF1, heat shock-induced Hsp70 translation was severely impaired.
METTL3 is a nuclear enzyme responsible for adding m6A on transcripts and is thus usually recognized as a writer. However, a recent study discovered that METTL3 also has a cytosolic role in translation, acting as a “reader” rather than a “writer” of methylated transcripts as its m6A catalytic (writer) activity is dispensable in METTL3-promoted translation (Lin et al., 2016). Further, Coots et al. provided evidence to verify that METTL3 directly binds to internal m6A but not to the 5′ m7G cap of mRNA and that depletion of METTL3 selectively inhibits translation of mRNAs bearing 5′UTR methylation but not 5′TOP mRNA barely having m6A (Coots et al., 2017).
Unlike eIF3 and its related factors, the other three non-YTH m6A readers, namely, HNRNPC, HNRNPG, and HNRNPA2B1, have all been found in the nucleus. These readers convey the molecular roles of m6A on pre-mRNA splicing, mRNA stability, translation, and storage control as the YTH readers do (Alarcon et al., 2015a; Liu et al., 2015; Liu et al., 2017) (Figure 1). HNRNPC and HNRNPG preferentially bind m6A-modified RNAs via a “m6A-switch” mechanism in which the m6A modifications destabilize the RNA hairpin structure, leading to the exposure of single-stranded RNA that facilitates protein binding (Liu et al., 2015; Liu et al., 2017). Conversely, HNRNPA2B1 recognizes its m6A-modified targets not via an m6A-switch mechanism but via direct binding of the methylated RGAC consensus site (Alarcon et al., 2015a). The binding of HNRNPA2B1 to m6A mRNA facilitates microRNA biogenesis via recruiting the microprocessor complex (drosha and pasha) to the pri-miRNA (Alarcon et al., 2015a; Alarcon et al., 2015b). However, a recent study indicated that HNRNPA2B1 is an indirect m6A reader because it does not specifically recognize m6A-containing RNA in vitro. This study also found that very few m6A sites exhibit proximal HNRNPA2B1 binding in vivo (Wu et al., 2018).
Very recently, a new m6A reader SND1 (Staphylococcal nuclease domain-containing protein 1) was discovered. It can stimulate the lytic reactivation of Kaposi’s sarcoma-associated herpesvirus (KSHV) through binding to ORF50/RTA (Baquero-Perez et al., 2019). Deletion of SND1 leads to inhibition of KSHV early gene expression, showing that SND1 is essential for KSHV lytic replication.
m6A mRNAs also indirectly recruit RNA-binding proteins. The known candidates include three insulin-like growth factor 2 mRNA binding proteins (IGF2BP1-3) and the fragile X messenger ribonucleoprotein (FMRP). Each of these RNA-binding proteins seems to enhance m6A mRNA stability (Edupuganti et al., 2017; Huang et al., 2018). IGF2BP1 relocalizes into mRNA aggregates in stress granules following heat stress, suggesting that it might also take part in the storage of translationally inactive mRNAs under stress conditions, thereby contributing to preventing their degradation (Huang et al., 2018). FMR1 negatively regulates the translation of a subset of m6A-modified transcripts, acting downstream of the m6A signal (Edupuganti et al., 2017).
Although both IGF2BPs and FMRP participate in the modulation of mRNA stability or translation efficiency in an m6A-dependent manner, their patterns that bind m6A mRNA directly or indirectly are not quite clear. Even though IGF2BPs may be direct m6A readers requiring an m6A structure switch (Sun et al., 2019), many other studies suggested that they are indirect m6A readers for the following reasons (Berlivet et al., 2019): first, several motif analyses of peaks by CLIP identified that FMRP and IGF2BP proteins bind a sequence motif that resembles the m6A consensus motif DRACH (Edupuganti et al., 2017; Huang et al., 2018); however, other CLIP studies suggested that IGF2BPs bind to a different consensus sequence. Second, the transcriptome-wide binding pattern determined by the distribution of their CLIP reads does not match the striking stop codon-enriched m6A distribution (Edupuganti et al., 2017; Huang et al., 2018). Third, in contrast to YTH domains, FMRP and IGF2BP proteins show weak binding affinities for m6A RNA and poor capacity to discriminate between these RNAs and non-methylated RNA (Edupuganti et al., 2017; Huang et al., 2018). Fourth, FMRP was found to directly bind reader DF2 (Youn et al., 2018; Zhang et al., 2018). Thus, FMRP may indirectly associate with m6A RNA through its interaction with DF proteins. Similarly, IGF2BP proteins interact with DF proteins in pulldown studies (Youn et al., 2018) and thus probably interact with m6A RNA indirectly. Recently, a proline-rich coiled-coil 2A (Prrc2a) protein was reported as a new reader, which specifically binds to a consensus GGm6ACU motif via a new Prrc2a domain (named GRE domain) to stabilize the critical transcript in neural cells (Wu et al., 2019). It is not clear whether Prrc2 in other organs contributes to the regulation of biological activities.
m6A readers not only play a critical role in normal and pathological conditions by interacting with cellular mRNAs but also are very important mediators that regulate viral infection by recognizing m6As on viral RNA. m6A modification of viral RNA was first discovered in nuclear viruses. These viruses are largely DNA viruses, such as simian virus 40 (SV40), adenovirus-2, and herpes simplex virus; m6A was later found in multiple nuclear RNA viruses such as retroviruses and influenza A virus (Lichinchi et al., 2016a; Courtney et al., 2017). Thus, early studies led the field to believe that the nucleus was the primary site of m6A modification of viral transcripts. Furthermore, as writer and eraser enzymes all reside in the nucleus at the steady state, it is reasonable to anticipate that any potential m6A modifications of viral RNAs would exclusively occur in the nucleus and would not apply to cytoplasmic RNA viruses. However, recent studies reported that the writer and eraser proteins can be detected in the cytoplasm of cells under stress conditions (Gokhale et al., 2016; Lichinchi et al., 2016b), indicating that these proteins can shuttle between the cytoplasm and the nucleus in response to stressors, such as viral infections. Thus, these discoveries raised the possibility that cytoplasmic viruses might also be m6A-modified. Indeed, in recent years, an increasing number of investigations have revealed that like DNA viruses, many RNA viruses, such as flaviviruses (e.g., hepatitis C virus (HCV), dengue virus and Zika virus (ZIKV)), enteroviruses, and coronaviruses, have m6A modifications on their viral genomic RNAs (Gokhale et al., 2016; Lichinchi et al., 2016b; Hao et al., 2019). These m6A methylations differentially regulate viral replication depending on the viral species (Table 1).
The flaviviruses are positive single-stranded RNA (ssRNA) viruses that replicate in the cytoplasm. Two laboratories have mapped m6A methylation on the RNA genomes of some Flaviviridae family members including HCV, ZIKV, dengue virus, yellow fever virus, and West Nile virus (Gokhale et al., 2016; Lichinchi et al., 2016b). Further studies found that during HCV infection, m6A modification negatively regulates virus replication. These findings raised the question of why these viruses would retain m6A if it negatively impacts their life cycles. One possible explanation is that the m6A modifications may facilitate viral escape from host antiviral immune responses. This speculation is supported by other studies using several in vitro synthesized m6A RNAs, which suppress recognition by host pattern recognition receptors, TLR3, TLR7, TLR8, and RIG-1 (Kariko et al., 2005; Durbin et al., 2016).
In investigating the potential involvement of reader proteins in flavivirus replication, DF1–3 were silenced to identify whether altered reader protein expression affects m6A-mediated negative regulation of ZIKV and HCV RNA. In both viruses, knockdown of DF1-3 increased the levels of extracellular viral RNA (Lichinchi et al., 2016b). Furthermore, in ZIKV-infected cells, these observations were verified by DF1–3 overexpression, which reduced extracellular viral RNA levels. The studies also showed the discriminatory binding of YTH proteins to HCV and ZIKV RNA by immunoprecipitation. All these results revealed that the modulation of RNA levels in HCV and ZIKV is functionally linked to DF binding of m6A methylated viral RNA. Additionally, by site-directed mutagenesis of four potential m6A sites on the E1 gene, they found that E1-mutated HCV RNA was bound more efficiently by the HCV core protein, enhancing its packaging into nascent virions. These data demonstrated a specific mechanism by which the reader proteins facilitate m6A-mediated negative regulation of viral replication, likely through competition with core proteins to bind the region of the E1 gene to suppress packaging of viral RNA into infectious viral particles.
Enterovirus is the most-studied genus in the Picornaviridae family, which is composed of positive, ssRNA viruses that replicate in the cytoplasm. Poliovirus (PV) and enterovirus are the typical representatives of the Enterovirus genus. To date, although m6A modifications in poliovirus RNA have been reported (McIntyre et al., 2018), their specific modification sites on the genome have not been assessed. Recently, m6A modification of enterovirus 71 (EV71) RNA has been investigated. It was found that m6A sites distribute in genes encoding the VP1 capsid protein, the 3D RNA-dependent RNA polymerase, and the non-structural protein 2C (Hao et al., 2019). Mutations of the m6A sites in VP1 and 2C genes decreased EV71 replication, suggesting that m6A modifications positively regulate EV71 infection. In determining the role of m6A reader proteins in EV71 replication, knocking down reader proteins, including DF1–3 and DC1, with siRNA in AD cells enhanced viral replication. However, knockdown and overexpression of DF2 and DF3 in Vero cells resulted in decreased and increased viral replication, respectively. In addition, this study also found that EV71 infection upregulated DF1–3 and DC1 expression and led to the partial relocalization of DF1 and DF2 to the nucleus, whereas the nuclear reader DC1 relocated to the cytoplasm. Another study of PV and coxsackievirus found that three readers DF1–3 were all cleaved by viral protease 2A at a very early phase of infection. The authors further demonstrated that DF3 acts as a positive regulator of antiviral JAK/STAT signaling in response to positive ssRNA virus infection, enhancing type I interferon (T1IFN)-mediated gene regulation in infected cells. The authors proposed that EV 2A proteases cleave DF proteins to antagonize interferon-stimulated gene (ISG) induction in infected cells (Kastan et al., 2021). For the epitranscriptomic analysis of host m6A after EV infection, a recent study using clinical samples from children infected with EV identified 957 m6A-modified cellular genes. The different m6A methylations were increased in CDS regions but decreased in the 3′UTR and stop codon regions in the neurological symptom group. The authors further revealed the differences in related cellular functions and signaling pathways associated with m6A methylation patterns (Zhu et al., 2021).
The genome of coronaviruses consists of a positive-sense ssRNA molecule. Coronaviruses also produce subgenomic RNAs. The first identification of m6A in a member of the Coronaviridae family was conducted in the porcine epidemic diarrhea virus (PEDV) (Chen et al., 2020), shortly followed by SARS-CoV-2 (severe acute respiratory syndrome coronavirus-2) (Kim et al., 2020b). PEDV, a member of the alpha-coronavirus genus, causes high mortality associated with severe diarrhea and vomiting in piglets younger than 1 week of age. m6A-seq revealed that the PEDV genome contains seven m6A peaks, mostly located in ORF1b, which encodes non-structural proteins. Depletion of m6A reader DF1 or DF2 increases PEDV replication while FTO depletion decreases replication. The reader proteins, especially DF2, inhibit viral replication by reducing viral RNA stability. SARS-CoV-2 is a member of the beta-coronavirus genus (Wu et al., 2020), mainly causing acute respiratory distress syndrome (ARDS) and acute cardiac injury (Huang et al., 2020; Xu et al., 2020). A recent study using combined m6A-seq and miCLIP analyses identified eight m6A modifications on the SARS-CoV-2 genome at single-base resolution (Liu et al., 2021a), mainly located at the ORF1ab, ORF7a, N, and ORF10 regions (Li et al., 2021; Liu et al., 2021a). However, since these experiments were carried out using fragmented total RNA, the authors could not confirm whether these modifications are on the genome or subgenomic RNA or both. Of the three DF1–3 readers, only depletion of DF2 affected viral replication, which was increased compared with control cells. Therefore, as in PEDV, m6A modifications negatively regulate SARS-CoV-2 infection. However, Burgess and colleagues presented a contrasting result when utilizing human A549 lung carcinoma cells. Their findings demonstrated that the replication of SARS-CoV-2 was hindered upon the depletion of METTL3 or DF1 and DF3 (Burgess et al., 2021). Moreover, the utilization of STM2457, a novel and highly selective small-molecule inhibitor of METTL3, markedly decreased the production of infectious HCoV-OC43 virus by over 100-fold and SARS-CoV-2 virus by 300-fold. As such, STM2457 represents a promising therapeutic agent to specifically target the m6A pathway and curb the reproduction of coronavirus.
Chikungunya virus (CHIKV) is a member of the Togaviridae family. Its genome is a positive-sense ssRNA molecule (Weaver and Lecuit, 2015). The m6A modifications on CHIKV RNA were identified using a novel method called viral cross-linking and solid-phase purification (VIR–CLASP), which captures interactions between the pre-replicated viral genome and cellular proteins (Kim et al., 2020a). VIR–CLASP revealed that DF2 and DF3 interact with the CHIKV RNA. Moreover, m6A modifications were found to be abundant on the 5′ end of CHIKV genomic RNA. In addition, the effect of m6A on CHIKV replication is subject to combinatorial regulation by DF proteins. Knockdown and overexpression studies demonstrated the different effects of the three DF readers on viral RNA transcription, particularly on different (plus and minus) strands. Therefore, the authors moved on to testing whether the early effects of DF proteins on CHIKV persist until the release of new viral particles. They showed that knockdown of DF1 and DF3 increased both extracellular viral RNA levels and mature virions, while knockdown of DF2 had the opposite effect. These data suggest that DF1 and DF3 restrict CHIKV replication while DF2 promotes CHIKV replication (Kim et al., 2020a). It is worth highlighting that CHIKV expresses the viral non-structural and structural proteins from its genomic and subgenomic viral RNAs, respectively. However, whether the m6A modifications in the genomic and subgenomic RNAs are differentially regulated and carry out distinct functions during the complex viral life cycle are still unknown.
Respiratory syncytial virus (RSV) is a non‐segmented, negative‐sense ssRNA virus in the Pneumoviridae family (Afonso et al., 2016). This virus mainly causes upper and lower respiratory tract infections in infants and children (Collins and Graham, 2008). m6A-seq analysis found that RSV viral negative-sense genomic RNA (gRNA), complementary RNA (cRNA), and the multiple viral RNA transcripts were internally m6A-modified (Xue et al., 2019). The viral G transcript encoding the attachment glycoprotein present on the surface of the RSV virion was the most extensively modified of the 10 viral mRNA transcripts. m6A writers positively regulated RSV replication, while m6A erasers showed the opposite effect. Overexpression of DF1–3 readers significantly increased RSV protein expression, gRNA and mRNA synthesis, and viral particle formation. It appears that overexpression of DF1–3 enhanced the ability of the RSV polymerase to synthesize both replicate and transcript. It should also be noted that overexpression of DF1 protein in A549 cells did not significantly affect the growth or survival of the host cells. Synonymous mutations of conserved m6A motifs under the three m6A peaks in the G transcript showed reduced viral replication and pathogenesis in vitro and in the respiratory tracts of cotton rats. Since m6A function is mediated via the binding of reader proteins, the authors also verified the specific binding of DF1–3 with the RSV G mRNA (Xue et al., 2019). Together, these results demonstrate a positive role for m6A in RSV infection. These findings highlight the viral m6A machinery as a possible novel target for rational design of live attenuated vaccines.
Human metapneumonium virus (HMPV) is the first human member of the Metapneumovirus genus in the Pneumovirinae subfamily within the Paramyxoviridae family. It is an enveloped negative-sense ssRNA virus. The genome includes eight genes coding for nine different proteins (van den Hoogen et al., 2002). m6A-seq analysis found that both the viral genome and anti-genome were m6A-modified and the strongest m6A peaks were in the G gene. The m6A peak regions identified in both strands are largely overlapping, and HMPV-induced m6A modifications promote HMPV replication (Lu et al., 2020). Not only does promotion of HMPV replication occur due to the action of writer proteins but also the reader proteins play an important role. For example, transient overexpression of DF1, DF2, DF3, or DC1 increases the levels of viral G and N proteins, as well as the release of infectious virus, anti-genome, and N and G mRNAs in A549 cells. This proviral function was also observed in HeLa cells stably overexpressing DF1, DF2, or DF3 using recombinant GFP-expressing HMPV (Lu et al., 2020). Although HMPV infection has a minimal impact on the methylation of cellular mRNAs, it significantly upregulates genes that are involved in type I IFN signaling, such as retinoic acid-inducible gene-I (RIG-I). These in vitro data were further supported by studies using cotton rats (Lu et al., 2020), suggesting that HMPV may utilize m6A modification to mimic host RNA and avoid detection by the innate immune system.
Vesicular stomatitis virus (VSV) is a negative‐sense, non‐segmented (NNS) ssRNA virus in the Rhabdoviridae family. m6A-seq revealed that its anti-genome contains 18 m6A sites (Qiu et al., 2021). During VSV replication, the produced double-stranded (dsRNA) intermediate serves as a signal in activating RIG-I‐like receptors (RLRs) that recognize RNA intracellularly, triggering an antiviral mechanism. However, m6A readers can function as trans‐acting factors to compete with RLRs and inhibit RLRs from binding to m6A-modified RNAs (Lu et al., 2020). Another study found that among the three DF1–3 readers, only DF3 suppressed ISG expression under basal conditions by promoting translation of the transcription corepressor forkhead box protein O3 (FOXO3) (Zhang et al., 2019b). In this process, DF3 cooperated with two cofactors, PABP1 and eIF4G2, to promote FOXO3 translation by binding to the translation initiation region of FOXO3 mRNA. Moreover, DF3 KO mice had increased ISG levels and were resistant to several viral infections. These findings uncover the role of DF3 as a negative regulator of antiviral immunity. However, a recent study, using VSV and several other NNS RNA viruses including HMPV in the Pneumoviridae family and SeV (Sendai virus) and MeV (measles virus) in the Paramyxoviridae family, found that m6A on the genomes and anti-genomes of several families of NNS RNA viruses enabled viral RNA to escape recognition by RIG-I. In addition, the m6A reader DF2 is essential for suppression of T1IFN responses (Lu et al., 2021). These data suggest that the three families of NNS RNA viruses have evolved a common mechanism, which is to mask their genome and anti-genome with m6A, to mimic the host RNA and evade host innate immunity that is dependent on RNA sensor RIG-I.
Rotavirus (RV), a member of the family Reoviridae, is a non-enveloped virus with 11 segments of dsRNA. Children under the age of five are at high risk of rotavirus infection, which causes severe diarrhea, dehydration, and death (Crawford et al., 2017). RV infection induces global m6A modifications on host mRNA transcripts by down-regulating the m6A eraser ALKBH5. During RV infection, m6A modification of small bowel intestinal epithelial cells (IECs) has a negative effect on IFN response. Specifically, mice lacking the m6A writer enzymes METTL3 in IECs (Mettl3ΔIEC) showed increased expression of IFNs and ISGs and thus were resistant to RV infection (Wang et al., 2022). This study further found IRF7 to be one of the primary m6A modification targets during virus infection. In the absence of METTL3, IECs showed increased Irf7 mRNA stability and enhanced type I and III IFN expression. Deficiency in IRF7 attenuated the elevated expression of IFNs and ISGs and restored susceptibility to RV infection on Mettl3ΔIEC mice. However, in this m6A-IRF7-IFN signal cascade, the functional roles of reader proteins have not been elucidated.
Influenza A virus (IAV) is a member of the Orthomyxoviridae family, which is composed of negative, segmented, ssRNA viruses that replicate in the nucleus. IAV was the first virus found to express mRNA with internal m6A residues; the highest number of m6A modifications was detected on hemagglutinin (HA) mRNA segments encoding the viral envelop protein (Krug et al., 1976; Narayan et al., 1987). Recently, Courtney et al. reported that m6A modification of IAV RNA positively regulates viral gene expression and virion production as mutating m6A sites by CRISPR/Cas9 or inhibition of METTL3 with inhibitor 3-deazaadenosine (DAA) reduced the levels of both viral mRNA and proteins (Courtney et al., 2017). By using the PAR-CLIP method with three reader proteins DF1–3 as well as the PA-m6A-seq technique to map the m6A residues in IAV-infected cells, the authors found that m6A residues were present on multiple locations of the viral mRNA encoding structural proteins and at lower levels on the mRNA-encoding RNA polymerase subunits. A recent large-scale comparative m6A analysis using 70,030 complete HA sequences was conducted to investigate the conservation patterns of the DRACH motifs (Bayoumi and Munir, 2021). This bioinformatic analysis revealed the highest degree of DRACH conservation among all H1 sequences that clustered largely in the middle and in proximity of the 3′ end with at least four DRACH motifs conserved in all mRNA sequences. Interestingly, the total number and the conserved DRACH motifs in the viral RNA were found to be much lower than those observed in the mRNA. In their functional study of the readers in IAV infection, Courtney et al. reported that although DF1 and DF3 did not show an effect on viral replication, DF2 overexpression significantly promoted IAV replication and viral particle production. Taken together, this study provides evidence to suggest that m6A modifications in IAV RNA positively regulate viral infection and pathogenesis. Although the underlying mechanism is unclear thus far, this positive regulation is not likely through the downregulation of host antiviral innate immune response proteins, including RIG-1, MGA5, and interferon-β (Courtney et al., 2017).
The members of the Retroviridae family are positive ssRNA viruses that replicate in the nucleus. m6As have been identified in retroviruses, including human immunodeficiency virus-1 (HIV-1), Rous sarcoma virus, and feline leukemia virus (Gokhale and Horner, 2017; Riquelme-Barrios et al., 2018). Several years ago, three independent groups reported the mapping of m6A residues on viral RNA and further characterization of the involvement of m6A in HIV replication; however, their findings were not consistent (Kennedy et al., 2016; Lichinchi et al., 2016a; Tirumuru et al., 2016; Lu et al., 2018; Riquelme-Barrios et al., 2018). For example, all three reports showed an enrichment of m6A residues at the 3′ and/or 5′ UTR of HIV‐genomic RNA, but two of the studies discovered additional m6A residues along the viral genome (Lichinchi et al., 2016a; Tirumuru et al., 2016). This discrepancy may be due to the use of different cell lines, viral strains, and/or mapping techniques.
In functional studies of m6A, a group reported that overexpression of readers DF1–3 inhibited viral replication by blocking viral reverse transcription and promoting degradation of incoming viral RNA (Tirumuru et al., 2016). This finding was further solidified by their studies using virus-producing cells, in which they found that three DF1–3 readers preferentially bound to the m6A-modified leader sequence of viral genome RNA compared with the unmodified RNA counterpart (Lu et al., 2018). This finding on the negative role of reader proteins in HIV-1 replication is opposite to the results of Kennedy’s group. Kennedy et al. found that overexpression of DF1–3 in 293T cells increased gene expression of viral gag, ref, tat, and rev mRNAs and Gap and Nef proteins. In further studies focusing on DF2 in CEM-SS cells, they found that overexpression and knockdown of DF2 could increase and decrease HIV-1 replication, respectively (Kennedy et al., 2016). This inconsistency may be due to the cell lines or experimental conditions applied.
Recently, another study found that both DC1 and DF2 could bind to multiple distinct and overlapping sites on the HIV-1 RNA, with DC1 recruitment serving to regulate the alternative splicing of HIV-1 RNAs, while DF2 binding to m6A residues present on cellular mRNAs resulted in their destabilization, as previously reported, and DF2 binding to m6A sites on HIV-1 transcripts resulted in a marked increase in the stability of these viral RNAs. Thus, DF2 binding can exert diametrically opposite effects on RNA stability, depending on the RNA sequence context (Tsai et al., 2021). The finding of the role of DC1 in promoting viral infectivity was supported by another study (N'Da Konan et al., 2022). For reader DF3, it was reported that DF3 was incorporated into HIV particles in a nucleocapsid-dependent manner and reduced viral infectivity in the next cycle of infection. Interestingly, the authors found that HIV protease cleaves the virion-incorporated full-length DF3 protein, a process that can be blocked by FDA-approved HIV protease inhibitors (Jurczyszak et al., 2020).
In addition to the reader proteins, Lichinchi et al. also characterized the function of two m6A sites within the Rev response element (RRE) of HIV. The RRE is an important structural and functional RNA element within the HIV-1 env gene coding for an HIV-1 envelope protein. Lichinchi et al. found that m6A modification on viral RNA positively affected the interaction between HIV Rev protein and RRE RNA. This promoted the formation of Rev-RRE complexes and the nuclear export of viral transcripts, and hence viral replication (Lichinchi et al., 2016a). Although the mechanism by which m6As promote the binding of Rev to the RRE in cells is not clear, it is possible that methylation can alter the RNA structure of RRE (Liu et al., 2015; Liu et al., 2017), which facilitates Rev protein binding. An alternative explanation is that Rev is a viral m6A reader with higher affinity for methylated viral transcripts. Taken together, all these data indicate that m6A and its machinery have multiple roles in regulating HIV-1 infection depending on the stage of the viral life cycle, the position of m6A in the HIV-1 RNA, and the binding of different readers.
Endogenous retroviruses (ERVs) are abundant and heterogeneous groups of integrated retroviral sequences that affect genome regulation and cell physiology throughout their RNA-centered life cycle (Johnson, 2019). Using a genome-scale CRISPR knockout screen in mouse embryonic stem (ES) cells, Chelmicki et al. identified m6A RNA methylation as a way to restrict ERVs and that methylation of ERV mRNAs is catalyzed by the complex of METTL3/14 proteins. They also found that depletion of this writer complex, along with accessory subunits WTAP and ZC3H13, led to increased mRNA abundance of intracisternal A-particles (IAPs) and related ERVK elements. The authors further revealed that the m6A-dependent repression was proportional to the m6A content in the 5′UTR: the more m6A sites an IAPEz copy contains, the more upregulated it is in m6A-knockout cells. In addition, using controlled auxin-dependent degradation of this writer complex, they showed that IAP mRNA and protein abundance is dynamically and inversely correlated with m6A catalysis. Furthermore, using two mutant ES cell lines that exhibited loss of all three DF1–3 proteins, they found that depletion of DFs increased IAP mRNA levels, which supports the hypothesis that the m6A methylation pathway regulates IAP mRNAs in a posttranscriptional and DF-dependent manner.(Chelmicki et al., 2021) These results indicate that RNA methylation provides a protective effect in maintaining cellular integrity by clearing reactive ERV-derived RNA species, which may be especially important when transcriptional silencing is less stringent.
Simian virus 40 (SV40) is a small, double-stranded DNA (dsDNA) virus in the Polyomaviridae family. This virus replicates in the nucleus and often causes human tumors (Vilchez et al., 2003). The m6A modifications of SV40 transcripts were found over 40 years ago (Canaani et al., 1979; Finkel and Groner, 1983). However, the precise location of the m6As on SV40 viral mRNA and their functional significance in SV40 replication were unknown until recently. Tsai et al. conducted studies modifying the expression levels of the m6A machinery to identify any associated changes in viral replication (Tsai et al., 2018) and found that overexpression of DF2 elevated the expression of both the early large T antigen protein and the late structural protein VP1, leading to enhanced viral particle formation. However, a much less profound effect on viral replication was observed upon overexpression of DF3. Moreover, deletion of both DF2 and METTL3 reversed the effects observed following DF2 overexpression. These data indicate that m6A modification in SV40 RNA plays a positive role in the viral life cycle.
To further understand the underlying mechanism of these observations, the authors mapped the sites of m6As and found 13 peaks within the SV40 genome including 2 in the early transcript and 11 in the late transcript, most of which were present in the open reading frame (ORF) of structural protein VP1. They further conducted mutations of m6A on VP1 late transcript and found that mutations did not affect splicing but significantly decreased the VP1 protein levels despite the lack of significant changes in mRNA abundance. Nevertheless, comparing both cytosolic and nuclear fractions, a significant decrease in the VP1 transcripts was found in the cytosolic fraction, suggesting that m6A-mediated positive regulation of SV40 replication is due to both enhanced nuclear export and increased translation of viral late transcripts. In support of these findings, the addition of a global inhibitor of methylation, DAA, profoundly decreased SV40 late protein production (Tsai et al., 2018).
Hepatitis B virus (HBV) is a dsDNA virus belonging to the Hepadnaviridae family, which replicates in the nucleus through the reverse transcription of an RNA intermediate known as pregenomic RNA (pgRNA). A recent study found that m6A modification occurs in both HBV pgRNA and mRNA (Imam et al., 2018). In studying the effect of m6A methylation on the HBV life cycle, the authors found that knockdown of METTL3/14 increased the expression of the viral proteins, while the opposite result was obtained in cells lacking FTO or ALKBH5 expression. These results suggest that m6A negatively regulates the expression of HBV proteins. Interestingly, an increase in the expression of pgRNA was also observed for DF2 and DF3 knockdowns, suggesting that the decrease in HBV protein levels is attributed to the reduction of RNA abundance rather than decreased translation. Further m6A-seq analysis identified an m6A peak at the conserved motif situated within the epsilon stem loop, which is located in the 3′ terminus of all HBV mRNAs and both the 5′ and 3′ termini of pgRNA. Mutational analysis of the m6A site in the 5′ epsilon stem loop of pgRNA revealed that m6A at this site was required for efficient reverse transcription of pgRNA, while m6A methylation of 3′ epsilon stem loop resulted in destabilization of all HBV transcripts, suggesting that m6A has a dual functional role in the HBV life cycle.
The RIG-I signaling pathway in IFN induction is one of the host defense systems to eliminate viral infections. A recent study demonstrated that cellular m6A machinery regulates the RIG-I signaling pathway activated by virus infection (Kim et al., 2020c). Specifically, the authors revealed that DF2 inhibits RIG-I recognition of viral RNAs via interacting with m6A-modified viral RNAs, leading to a disrupted RIG-I-mediated immune response. However, another study revealed that DF2 could form a complex with IFNα-induced ISG20, a 3′–5′ exonuclease enzyme and selectively target the m6A-modified HBV transcript to perform RNA degradation (Imam et al., 2020). The functional roles of other readers were also investigated. It was reported that DC1 as well as FMRP recognized m6A-methylated HBV transcripts and facilitated their transport to the cytoplasm. In cells depleted with DC1 or FMRP, viral transcripts accumulated in the nucleus to affect the viral life cycle (Kim et al., 2021).
Adenovirus, a member of the Adenoviridae family, is a nuclear replicating dsDNA virus. While the m6A modification in viral transcripts was discovered in 1970s (Sommer et al., 1976; Chen-Kiang et al., 1979), the location of this methylation was unknown, largely due to the fact that the complex adenovirus transcriptome includes overlapping spliced units that impede accurate m6A mapping. A recent study using a combination of meRIP-seq and direct RNA long-read sequencing to profile the m6As within the transcriptome yielded both nucleotide- and transcript-resolved m6A detection (Price et al., 2020). They also found that adenovirus infection did not alter the expression of m6A machinery, such as writers (METTL3, METTL14, and WTAP) and readers (DC1, DF1, and DF2). These host proteins remained unchanged but were concentrated at sites of nascent viral RNA synthesis. Although both early and late viral transcripts contain m6As, depletion of METTL3 caused significantly more reduction of gene expression in viral late RNAs than in early RNAs. This late gene-biased effect was primarily mediated by decreased RNA splicing efficiency in the absence of METTL3 and could be extended to all of the multiply spliced adenovirus late RNAs (Price et al., 2020). Overall, these results suggest positive regulation of adenovirus by m6A modification and highlight the role of m6A in regulating the splicing and expression of a viral pathogen.
Members of the Herpesviridae family are large, dsDNA viruses. They replicate in the nucleus, causing distinct lytic and latent infections, as well as oncogenesis. While decades ago, herpes simplex virus type 1 (HSV-1) was found to contain m6A in its mRNA (Moss et al., 1977), m6A has now been mapped and functionally characterized in cells infected with human cytomegalovirus (HCMV) (Rubio et al., 2018; Winkler et al., 2019), Epstein–Barr virus (EBV) (Zheng et al., 2021), and Kaposi’s sarcoma-associated herpesvirus (KSHV) (Ye et al., 2017; Hesser et al., 2018; Tan et al., 2018).
A recent report revealed that the HSV-1 genome transcript contains 12 m6A peaks located mainly in the upstream, overlapping start, and inside the transcript region covering multiple viral transcripts. Viral infection enhances the expression of METTL3/14 and DF1–3 at an early stage and decreases their expression at the late stage; however, the expression of ALKBH5 and FTO is consistently suppressed during infection (Feng et al., 2022). Furthermore, inhibiting m6A modification by DAA or silencing writer and reader genes by siRNAs significantly decreases viral replication, whereas depleting the erasers or ectopically expressing METTL3 promotes viral replication. These data suggest that m6A modification benefits HSV-1 replication. Another study found that HSV-1 infection caused redistribution of nuclear m6A machinery including reader DC1. siRNA silencing of m6A methyltransferase reduced viral gene expression initially but less so as the infection advanced. The authors further found that this redistribution was accompanied by a wide-scale reduction in m6A addition and other RNA modifications on both host and viral mRNAs. Thus, they indicated that the m6A pathway is important for HSV-1 gene expression at the beginning of the replication cycle, becoming dispensable later (Srinivas et al., 2021).
HCMV has the largest genome among members of the Herpesviridae family and is best known for its propensity to cause disease in immunocompromised patients, especially transplant recipients, patients with advanced AIDS, and congenitally infected newborns (Boeckh and Geballe, 2011). Two recent studies in HCMV focused on how m6A impacted antiviral innate immunity via regulation of IFN production (Rubio et al., 2018; Winkler et al., 2019). One reported that in HCMV-infected or dsDNA-treated cells, deletion of METTL3 or DF2 led to an increase in the induction of ISGs. Consequently, viral propagation was suppressed in an interferon-signaling-dependent manner. Another study observed similar results by altering the expression levels of METTL14 or ALKBH5 demethylase (Rubio et al., 2018). Significantly, both studies found that mRNA of IFNβ was m6A-modified and stabilized following repression of METTL3, METTL14, or DF2. In addition, the former study showed that m6A-mediated regulation of interferon genes is conserved in mice. Mice lacking DF3 exhibited enhanced Ifna and Ifnβ induction following viral infection (Winkler et al., 2019). Together, these findings suggest that m6A serves as a negative regulator of the T1IFN response by dictating the fast turnover of IFNα and IFNβ mRNAs and consequently facilitating viral propagation.
EBV is a ubiquitous oncogenic virus that induces many types of cancers. EBV infection has three phases: the prelatent, latent, and lytic phases (Hammerschmidt, 2015). During infection, METTL14 expression is induced, and knockdown of METTL14 leads to decreased latent EBV gene expression. METTL14 is also significantly induced in EBV-positive tumors, promoting growth of EBV-transformed cells and tumors in xenograft animal models. Mechanistically, the virus-encoded latent oncoprotein EBNA3C activates transcription of METTL14 and promotes its stability by directly interacting with METTL14. This study demonstrated that EBV hijacks METTL14 to drive EBV-mediated tumorigenesis (Lang et al., 2019). Another study, examining the viral and cellular m6A epitranscriptomes, found that in the pre‐latent phase, EBV EBNA2 and BHRF1 were highly m6A‐modified upon EBV infection (Zheng et al., 2021) and that knockdown of METTL3 decreased EBNA2 expression. Further, Xia et al. demonstrated that EBV transcripts exhibited differential m6A modifications in human nasopharyngeal carcinoma biopsies, patient-derived xenograft tissues, and cells at different EBV infection stages (Xia et al., 2021). m6A-modified EBV transcripts can be recognized and destabilized by the DF1 reader, which leads to m6A-dependent suppression of EBV infection and replication. Mechanistically, DF1 accelerates viral RNA decapping and mediates RNA decay by recruiting RNA degradation complexes, including ZAP, DDX17, and DCP2, thereby posttranscriptionally downregulating the expression of EBV genes.
KSHV is the most studied virus in the Herpesviridae family thus far. Several groups have mapped the m6A sites in different transcripts of the virus during KSHV infection. These include KSHV ORF50/RTA mRNA, which encodes the replication and transcription activator (RTA) required for the reactivation of latent KSHV (Baquero-Perez and Whitehouse, 2015). While three of these early studies showed that m6A regulates RTA, they presented conflicting results for how m6A and its machinery control the ORF50/RTA RNA levels and/or expression, which might be due to the different cell types used. In B cells, Ye et al. identified several m6A sites crucial for RTA pre-mRNA splicing that were bound by m6A reader DC1 and its associated splicing factors, SRSF3 and SRSF10. They also found that the lytic switch protein RTA itself strongly induced m6A modification and enhanced its own pre-mRNA splicing (Ye et al., 2017). However, in the same cells, Hesser et al. found a negative effect of m6A on ORF50/RTA and KSHV lytic reactivation (Hesser et al., 2018). In epithelial cells, Tan et al. found that DF2 depletion increased the stability of many viral transcripts including ORF50/RTA during reactivation, which might or might not change its expression, indicating a potential role for m6A in KSHV lytic reactivation (Tan et al., 2018). Conversely, Hesser et al. showed that METTL3 and DF2 depletion suppressed KSHV ORF50/RTA expression, lytic reactivation, and viral particle release, suggesting that m6A promotes KSHV lytic reactivation (Hesser et al., 2018).
The m6A nuclear reader DC2 was also studied in KSHV-infected cells for its function in stabilizing RNAs, since many cellular mRNAs are degraded by KSHV endoribonuclease SOX (Macveigh-Fierro et al., 2022). This study found that the IL6 mRNA was m6A modified in its 3′UTR during KSHV lytic infection and that removal of this m6A restored susceptibility to SOX-mediated degradation. They further showed that DC2 bound to the IL6 SOX resistance element in an m6A-dependent manner and that downregulation of DC2 was sufficient to abrogate resistance to SOX. These results indicate that the m6A pathway is pivotal in the regulation of gene expression during KSHV infection, highlighting the viral-host battle for control of RNA stability. In addition to the two m6A readers mentioned above, a recent study reported a new m6A reader, SND1 (Staphylococcal nuclease domain-containing protein 1), for the lytic reactivation of KSHV through binding to ORF50/RTA. The authors found that m6A modification of the ORF50/RTA RNA was critical for SND1 binding, which in turn stabilized the ORF50 transcript. The SND1 binding was hairpin structure-dependent and preferable to the unspliced full length of ORF50/RTA. Deletion of SND1 led to inhibition of KSHV early gene expression, suggesting that SND1 is essential for KSHV lytic replication (Baquero-Perez et al., 2019).
BmNPV, a representative member of the Baculoviridae family, specifically infects the silkworm (Bombyx mori) and causes serious economic losses to the silkworm industry (Zhang et al., 2019a). BmNPV is an enveloped circular DNA virus that replicates in the nucleus. MeRIP sequence analyses found that m6A modifications are widespread in BmNPV transcripts in viral infected cells and predominantly appear in the coding sequences (CDS) and the 3′-end of the CDS. Among the viral genes related to replication and proliferation, ie-1 mRNA was found to have a higher m6A level than other viral genes. The m6A sites in the ie-1 mRNA may be negatively related to the protein expression of this mRNA. Cells overexpressing DF3 inhibited viral replication in a dose-dependent manner, and an opposite effect was found when silencing DF3 with siRNAs (Zhang et al., 2022).
Recent studies on viral m6A modifications have shifted from analyzing the m6A landscape and its effect on viral replication to investigating its regulatory mechanisms. In this regard, the T1IFN immune response is a major focus as it activates intracellular antimicrobial programs and influences the development of innate and adaptive immune responses (Shulman and Stern-Ginossar, 2020). For detailed information, readers can refer to recent reviews (Lou et al., 2021; Tang et al., 2021). The innate immune response is the first line of host antiviral response and can discriminate cellular (self) from viral (non-self) RNAs using pathogen recognition receptors (PRR). PRRs include toll-like receptor (TLR) 3, 7, and 9 in endosomes and the RIG-I like receptors (RLR) in the cytosol, of which the most important are RIG-I and the melanoma differentiation-associated protein 5 (MDA5) (Wu et al., 2014; Chow et al., 2018). Upon recognition by these RNA sensors, intracellular signaling cascades are activated, triggering the expression and secretion of IFNα and IFNβ that are recognized by the IFN receptor, activating the downstream JAK-STAT pathway. This pathway leads to transcription of a number of ISGs that establish an antiviral response (Ivashkiv and Donlin, 2014). Viruses have evolved to escape or inhibit the T1IFN response using multiple strategies, which have been very briefly mentioned above in each virus group based on availability of data; here, we would like to make a summary of the three major mechanisms by which m6A modifications modulate host immune responses.
RNA structure and posttranscriptional modifications are important molecular markers for the PRR to discriminate self from non-self RNAs. The primary ligand of RIG-I is a 5′ triphosphated or diphosphated ssRNA (Hornung et al., 2006; Pichlmair et al., 2006). Both MDA5 and RIG-I can recognize dsRNAs; MDA5 preferentially detects long dsRNA, while RIG-I detects short dsRNA (Kato et al., 2008; Schlee et al., 2009; Runge et al., 2014). Cellular mRNAs contain a unique 5′cap structure typically methylated at the guanine N7 and ribose-2′-O-position (Furuichi et al., 1977; Shuman, 2002), and this structure is not detected by RIG-I and MDA5 (Hyde and Diamond, 2015). mRNAs lacking the ribose-2′-O-methylation are recognized as the foreign RNA. Thus, certain viruses use either their own methyltransferases (e.g., WNV and VSV) (Hyde and Diamond, 2015; Johnson et al., 2018) or cellular methyltransferase FTSJ3 (e.g., HIV) to add ribose-2′-O-methylation to the RNA cap or internal regions (Ringeard et al., 2019). An additional methylation strategy is the utilization of cellular m6A methyltransferases to add m6A to viral RNA transcripts. For example, HMPV RNA uses cellular m6A machinery to add m6A and evade T1IFN detection. This notion is supported by data showing that m6A-deficient HMPV RNA induces a higher expression of RIG-I, binds efficiently to RIG-I, and facilitates the conformational change of RIG-I required to stimulate the IFN induction pathway. This finding was also verified in vivo using cotton rats infected with HMPV mutant lacking multiple m6A sites (Lu et al., 2020). Moreover, depletion of METTL3 or DF2 induces the mRNA expression of IFNβ and downstream ISGs in cells infected with HCMV, MCMV, IVA, adenovirus, or VSV (Winkler et al., 2019). Another example is RNA m6A modification of HIV, which suppresses the expression of antiviral cytokine T1IFN in differentiated human monocytic cells and primary monocyte-derived macrophages. Mechanistically, m6A of HIV-1 RNA escapes RIG-I-mediated RNA sensing and activation of the transcription factors IRF3 and IRF7 that drive IFN-I gene expression (Chen et al., 2021). Similarly, in HCV or HBV infection, depletion of METTL3/14 leads to an increase in viral RNA recognition by RIG-I, thus activating IFN production. However, overexpression of METTL3/14 reverses this outcome. The m6A modification of viral RNA renders RIG-I less effective, whereas mutation of the m6A consensus motif enhances RIG-I sensing activity. Importantly, DF2 and DF3 inhibit RIG-I-transduced signaling activated by viral RNAs by competitive binding to the m6A site to inhibit RIG-I recognition (Kim et al., 2020c). Consistently, METTL14 depletion reduces viral reproduction and stimulates HCMV- or dsDNA-induced IFNβ1 mRNA production while ALKBH5 depletion had the opposite effect (Rubio et al., 2018). This m6A-regulated innate immune response was also studied in several NNS RNA viruses including VSV, RSV, and HMPV (Lu et al., 2021; McFadden et al., 2021; Qiu et al., 2021). Qiu et al. found that m6A addition to VSV RNA decreased viral dsRNA formation, thereby reducing virus-sensing efficiency by RIG-I and MDA5 and dampening antiviral immune signaling. However, ablation of METTL3 enhanced T1IFN signaling and accelerated VSV clearance (Qiu et al., 2021). Similar results reported by Lu et al. showed that NNS RNA viruses utilized m6A as a common strategy to evade host immunity, which was supported by the fact that m6A-deficient viral RNA triggered a significantly higher level of T1IFN compared to m6A-sufficient viral RNA in a RIG-I-dependent manner. In addition, reader DF2 is essential for suppression of the T1IFN pathway (Lu et al., 2021).
Among the host molecules involved in the T1IFN signaling, IFNβ mRNA is one of the major targets. An early study found that IFNβ1 mRNA was m6A modified within both the coding sequence and the 3′UTR in HCMV-infected or dsDNA-treated cells, and these data supported the hypothesis that m6A machinery controls IFNβ production (Rubio et al., 2018). Later, it was found that loss of m6As within IFNβ1 mRNA due to METTL3 or DF2 depletion leads to the stabilization of IFNβ1 mRNA and a stronger antiviral response during HCMV infection, suggesting that m6As may serve as a negative regulator of the antiviral IFN response (Winkler et al., 2019). Other antiviral adaptor molecules, such as TRAF3, TRAF6, and MAVS in the TIIFN signaling pathway, are also the targets of m6A modification. During innate immune stimulation, the mRNAs encoding these molecules lose their m6As, catalyzed by the eraser ALKBH5 recruited to these mRNAs by RNA helicase DDX46. The loss of m6As reduces their nuclear export and translation and thus inhibits IFN production (Zheng et al., 2017). Similarly, another DDX family member, DDX3, has been found to interact with ALKBH5 during the immune response, suggesting that this group of proteins may regulate viral infection by altering the abundance and stability of these molecules (Shah et al., 2017). On the other hand, the study also showed that METTL3 promotes the splicing of the TLR signaling adaptor MYD88 and the induction of several cytokines in response to lipopolysaccharide stimulation, which indicates that m6A can also positively regulate innate immunity (Feng et al., 2018). The activation of IFN signaling induces the expression of hundreds of ISG, which is also likely regulated by m6A and its machinery. For example, DF3 was reported to negatively regulate IFN signaling and subsequent ISG expression through promotion of translation of a transcriptional repressor of ISGs named FOXO3 (Zhang et al., 2019b). Consequently, knockdown of DF3 in macrophages inhibited infection of several viruses including VSV, EMCV, and HSV-1. However, McFadden et al. showed a positive effect of m6A on antiviral activity of T1IFN signaling. They found that IFITM1, an antiviral ISG, was upregulated at the translational level by m6A and the writer proteins METTL3/14. Furthermore, reader DF1 was found to increase the expression of IFITM1 in an m6A-binding-dependent manner (McFadden et al., 2021). Another m6A reader, hnRNPA2B1, can recognize DNA virus HSV-1 in the nucleus. Upon recognition and further dimerization and demethylation, hnRNPA2B1 translocates to the cytoplasm where it activates the T1IFN signaling pathway. Additionally, hnRNPA2B1 promotes m6A modification, nucleocytoplasmic trafficking, and translation of cGAS, IFI16, and STING mRNAs to fully ensure the IFNα/β production (Zhang et al., 2020). Other m6A readers, such as FMR1 and IGFBP3, also selectively recognize m6A in viral or cellular mRNAs and modulate their function during the antiviral response (Arguello et al., 2017; Edupuganti et al., 2017).
The adaptive immune response is another arm of the immune system that specializes in the clearance of specific pathogens. It is mediated by the activation of antigen-specific T and B lymphocytes, ultimately establishing long-lasting immune memory against the given antigen. Accumulated evidence reveals that m6A exerts a vital effect on adaptive immunity through targeting certain genes regulating immune cell homeostasis. For example, early studies using conditional METTL3 knockout mice reported a novel mechanism whereby m6A functions in vivo to control T-cell differentiation and proliferation by inducible degradation of mRNAs encoding suppressor of cytokine signal (SOCS) and consequently relieves blockage of IL7 signaling and T-cell proliferation (Li et al., 2017b). This notion is based on the fact that m6A modification targets the IL-7/STAT5/SOCS signaling pathway. The IL-7/STAT5 signal axis is critical for maintaining T-cell differentiation and proliferation. SOCS-1 and -3 could act as mediators binding to the IL-7 receptor, thus preventing STAT5 activation and downstream signaling to regulate T-cell homeostasis. Thus, m6A-mediated degradation of SOCS mRNA promotes T-cell proliferation and differentiation. The same research group later found that m6A was also essential for T regulatory (Treg) cell generation and suppression. Similarly, the m6A-mediated SOCS mRNA degradation activated the IL2/STAT5 signaling pathway to sustain the suppressive function and stability of Treg cells (Tong et al., 2018).
In addition to the positive effect of m6A on T-cell homeostasis, m6A RNA modification might exert an inhibitory effect on follicular help T (Tfh) cell differentiation. A recent study revealed that E3 ligase VHL promoted Tfh development and function at the early phase upon viral infection via suppression of hypoxia-inducible factor 1a (HTF-a1)-mediated glycolysis (Zhu et al., 2019). Mechanistically, VHL deficiency results in activation of the HIF-a1-GAPDH glycolytic pathway and consequently reduction in ICOS (inducible costimulatory) expression by enhancing m6A modification on ICOS mRNA, ultimately leading to attenuated Tfh cell differentiation.
Furthermore, m6A modification also exerts essential effects on early B-cell development. Zheng et al. demonstrated that loss of writer METTL14 blocked two key transitions in B-cell development: (i) METTL14-mediated m6A modification facilitates IL-7-induced pro-B cell proliferation via its reader DF2; (ii) the large-pre-B-to-small-pre-B transition is independent of reader DF1/2. Actually, this transition is largely dependent on the METTL14-mediated proper transcriptional activation of several transcription factors (Zheng et al., 2020). Another study showed that in diffuse large B-cell lymphoma (DLBCL), METTL3 expression and m6A level were increased in both tissue and cell lines. Upregulated METTL3 promoted DLBCL cell proliferation by increasing the mRNA level of pigment epithelium-derived factor through m6A modification (Cheng et al., 2020). Similarly, WTAP was also found to be involved in the induction of DLBCL cell proliferation by enhancing m6A modification of HK2 mRNA, leading to HK2 upregulation. However, in this circumstance, WTAP was upregulated by piRNA-30743 and thus DLBCL tumorigenesis was through a piRNA/WTAP/HK2-m6A axis(Han et al., 2021). Collectively, these studies indicate that m6A modification plays an important role in early B-cell development.
With the advances in high-throughput MeRIP-seq techniques, in recent years, our understanding of the m6A landscape and its roles in regulating viral replication has greatly advanced. Study findings indicate that m6A may be proviral or antiviral depending on the viral species and which step is targeted during the viral life cycle (Zaccara et al., 2019; Baquero-Perez et al., 2021). To date, many viruses have been found to be m6A-modified in their RNA transcripts and therefore regulated by m6A machinery. Deletion or overexpression of the m6A writer, eraser, or reader proteins affects diverse facets of viral replication that are often mediated by RNA-binding protein interactions. This implies that the reader proteins of the m6A-containing transcripts play an executive role in this regulatory process. In this regard, the reader proteins may bind not only to the m6A sites of viral RNA but also to those of host transcripts. Thus, m6A-mediated regulation of viral replication is also tissue- or cell-type dependent. In brief, viral replication is regulated by m6A and their machinery, particularly the various readers, in a way that is more complex than was expected. Future experiments studying m6A in viral infection should focus on the specific modification sites of viral transcripts, their effects on the temporal dynamics of methylation, and the functions of newly identified readers.
Previous studies largely focused on identifying the location of m6A clusters in viral RNAs. With the development of new techniques to increase the resolution of mapping at the single-nucleotide level, it is possible to study the role of specific m6A sites on viral transcripts in viral propagation by site-directed mutagenesis. In addition, since m6A modification is a dynamic and reversible process, it is very important to determine whether dynamic changes in m6A modifications correspond to altered viral replication efficiency at specific time points during infection. Furthermore, we can identify which sites are critical for these changes. As reader-binding relies on the m6A site and its surrounding conserved motif, it is very interesting to determine whether reader binding is also a dynamic process; if so, further determination of which m6A methylations are responsible for this dynamic change should be conducted. Additionally, it is known that m6A methylation affects RNA structures and that certain RNA-binding proteins require specific structures for maximum binding capacity. By identifying m6A sites at the single-nucleotide resolution, it is possible to computationally predict which modifications can significantly alter RNA secondary structure. These predictions can then be tested experimentally to determine the effect of the modifications on the binding activity of different reader proteins as well as on the viral replication efficiency.
Comparing the different groups in the m6A machinery, the reader group contains more components than the writer and eraser groups, especially when taking into account the new readers recently discovered (Williams et al., 2019; Baquero-Perez et al., 2021). However, their roles have not been well studied, especially for the “direct” readers that lack an YTH domain. For example, HNRNPA2B was previously reported to be a direct reader, but its specific binding pattern with its m6A is poorly understood. Recently, structural and biochemical data have shown that this protein may actually be an “indirect” m6A reader, as studies have shown that HNRNPA2B1 does not specifically recognize m6A-labeled RNA in vitro and in vivo (Wu et al., 2018). In addition, the roles of the three YTH domain-containing readers DF1–3 in regulating viral replication are inconsistently reported. On the one hand, this may be due to the virus and cells used; on the other hand, although previous studies have indicated different functions of these readers, their functions cannot always be directly translated to viral RNAs. In other words, these readers may not always bind to the m6A sites at the same time or with maximum binding affinity. This speculation leads to further specific questions: since all YTH-reader paralogs share a conserved m6A binding motif, do they bind to the same copy of the RNA transcript? Do they compete each other for the same sites to have an antagonistic effect or do they bind at the same location to form a complex to cause a synergistic effect? These fundamental biological questions can be answered by assays using a designed luciferase reporter construct generated by site-directed mutagenesis of the specific m6A sites.
The importance and role of the epitranscriptome in regulating viral replication and antiviral immunity open the door for novel therapeutic interventions that target either the viral or host epitranscriptome to suppress viral replication. For viruses such as HMPV, HCMV, and VSV that take advantage of m6A methylation to escape the recognition of the host innate immune response, knockdown of the methyltransferases or reader proteins with siRNA or small-molecule inhibitors is a good antiviral strategy. In addition, generation of m6A-deficient viral strains might be useful when creating attenuated live viral vaccines because m6A methylation of viral RNA negatively regulates the host innate immune response and m6A-depleted virus induces a potent T1IFN response in vivo (Lu et al., 2020). Two good examples are the RSV G-transcripts depleted of m6A and the m6A-deficient HMPV virus, which are highly attenuated yet retain high immunogenicity in cotton rats (Xue et al., 2019; Lu et al., 2020). For viruses that are suppressed by m6A modifications, similar strategies can be used for drug and vaccine development, but the approaches would be to enhance m6A methylation with specific compounds (Selberg et al., 2019) and generate mutant virus with enhanced m6A methylation. Examples include flaviviruses and SARS-CoV2 RNA, whose genomes are gradually m6A-methylated during infection in host cells; these m6A additions negatively modulate the life cycle of these viruses, while knockdown of METTL3 and/or METTL14 dramatically increases viral particle formation (Gokhale et al., 2016; Lichinchi et al., 2016b; Liu et al., 2021a). These findings suggest that strategies to enhance the m6A methylation of viral RNA may be a potential therapeutic avenue to develop vaccines or antiviral drugs for these viruses. Although the functionality of such proposed vaccines has not been evaluated, it seems plausible that infection with live attenuated vaccines based on altered m6A methylation might induce potent adaptive immune responses providing the host with long lasting immunity to the wild-type virus.
Although vast advances have been made in recent years, our understanding of how m6A modification contributes to the regulation of gene expression and viral replication is still in its infancy. In particular, viral infection is a complex process of virus–host interaction, and epitranscriptomic analysis must expand its scope to include the m6A landscape of host transcripts. Most previous studies used cell lines, which limits investigations to the host immune responses underlying the mechanisms of disease induction. Future research should involve animal models to reveal the related pathogenesis of human diseases. The recently improved m6A sequencing techniques and CRISPR/Cas9 gene editing approaches will clearly be a major help for this endeavor.
DY: writing original manuscript. GZ and HZ: revision and editing. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the Canada Institute of Health Research (PJT169202) and the Natural Sciences and Engineering Research Council (RGPIN-2017-05612).
We would like to thank Drs. Vivienne Chan and Katherine Adolphs for their critical reading and editing of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Afonso, C. L., Amarasinghe, G. K., Banyai, K., Bao, Y., Basler, C. F., Bavari, S., et al. (2016). Taxonomy of the order mononegavirales: update 2016. Arch. Virol. 161 (8), 2351–2360. doi: 10.1007/s00705-016-2880-1
Aitken, C. E., Lorsch, J. R. (2012). A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 19 (6), 568–576. doi: 10.1038/nsmb.2303
Alarcon, C. R., Goodarzi, H., Lee, H., Liu, X., Tavazoie, S., Tavazoie, S. F. (2015a). HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell 162 (6), 1299–1308. doi: 10.1016/j.cell.2015.08.011
Alarcon, C. R., Lee, H., Goodarzi, H., Halberg, N., Tavazoie, S. F. (2015b). N6-methyladenosine marks primary microRNAs for processing. Nature 519 (7544), 482–485. doi: 10.1038/nature14281
Arguello, A. E., DeLiberto, A. N., Kleiner, R. E. (2017). RNA Chemical proteomics reveals the N(6)-methyladenosine (m(6)A)-regulated protein-RNA interactome. J. Am. Chem. Soc. 139 (48), 17249–17252. doi: 10.1021/jacs.7b09213
Bailey, A. S., Batista, P. J., Gold, R. S., Chen, Y. G., de Rooij, D. G., Chang, H. Y., et al. (2017). The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. Elife 6, e26116. doi: 10.7554/eLife.26116
Baquero-Perez, B., Antanaviciute, A., Yonchev, I. D., Carr, I. M., Wilson, S. A., Whitehouse, A. (2019). The Tudor SND1 protein is an m(6)A RNA reader essential for replication of kaposi's sarcoma-associated herpesvirus. Elife 8, e47261. doi: 10.7554/eLife.47261
Baquero-Perez, B., Geers, D., Diez, J. (2021). From a to m(6)A: the emerging viral epitranscriptome. Viruses 13 (6). doi: 10.3390/v13061049
Baquero-Perez, B., Whitehouse, A. (2015). Hsp70 isoforms are essential for the formation of kaposi's sarcoma-associated herpesvirus replication and transcription compartments. PloS Pathog. 11 (11), e1005274. doi: 10.1371/journal.ppat.1005274
Bayoumi, M., Munir, M. (2021). Evolutionary conservation of the DRACH signatures of potential N6-methyladenosine m(6)A sites among influenza a viruses. Sci. Rep. 11 (1), 4548. doi: 10.1038/s41598-021-84007-0
Berlivet, S., Scutenaire, J., Deragon, J. M., Bousquet-Antonelli, C. (2019). Readers of the m(6)A epitranscriptomic code. Biochim. Biophys. Acta Gene Regul. Mech. 1862 (3), 329–342. doi: 10.1016/j.bbagrm.2018.12.008
Boccaletto, P., Machnicka, M. A., Purta, E., Piatkowski, P., Baginski, B., Wirecki, T. K., et al. (2018). MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46 (D1), D303–D307. doi: 10.1093/nar/gkx1030
Boeckh, M., Geballe, A. P. (2011). Cytomegalovirus: pathogen, paradigm, and puzzle. J. Clin. Invest. 121 (5), 1673–1680. doi: 10.1172/JCI45449
Burgess, H. M., Depledge, D. P., Thompson, L., Srinivas, K. P., Grande, R. C., Vink, E. I., et al. (2021). Targeting the m(6)A RNA modification pathway blocks SARS-CoV-2 and HCoV-OC43 replication. Genes Dev. 35 (13-14), 1005–1019. doi: 10.1101/gad.348320.121
Canaani, D., Kahana, C., Lavi, S., Groner, Y. (1979). Identification and mapping of N6-methyladenosine containing sequences in simian virus 40 RNA. Nucleic Acids Res. 6 (8), 2879–2899. doi: 10.1093/nar/6.8.2879
Chelmicki, T., Roger, E., Teissandier, A., Dura, M., Bonneville, L., Rucli, S., et al. (2021). m(6)A RNA methylation regulates the fate of endogenous retroviruses. Nature 591 (7849), 312–316. doi: 10.1038/s41586-020-03135-1
Chen, Y. G., Chen, R., Ahmad, S., Verma, R., Kasturi, S. P., Amaya, L., et al. (2019). N6-methyladenosine modification controls circular RNA immunity. Mol. Cell 76 (1), 96–109 e109. doi: 10.1016/j.molcel.2019.07.016
Chen, J., Jin, L., Wang, Z., Wang, L., Chen, Q., Cui, Y., et al. (2020). N6-methyladenosine regulates PEDV replication and host gene expression. Virology 548, 59–72. doi: 10.1016/j.virol.2020.06.008
Chen, S., Kumar, S., Espada, C. E., Tirumuru, N., Cahill, M. P., Hu, L., et al. (2021). N6-methyladenosine modification of HIV-1 RNA suppresses type-I interferon induction in differentiated monocytic cells and primary macrophages. PloS Pathog. 17 (3), e1009421. doi: 10.1371/journal.ppat.1009421
Cheng, Y., Fu, Y., Wang, Y., Wang, J. (2020). The m6A methyltransferase METTL3 is functionally implicated in DLBCL development by regulating m6A modification in PEDF. Front. Genet. 11. doi: 10.3389/fgene.2020.00955
Chen-Kiang, S., Nevins, J. R., Darnell, J. E., Jr. (1979). N-6-methyl-adenosine in adenovirus type 2 nuclear RNA is conserved in the formation of messenger RNA. J. Mol. Biol. 135 (3), 733–752. doi: 10.1016/0022-2836(79)90174-8
Chow, K. T., Gale, M., Jr., Loo, Y. M. (2018). RIG-I and other RNA sensors in antiviral immunity. Annu. Rev. Immunol. 36, 667–694. doi: 10.1146/annurev-immunol-042617-053309
Collins, P. L., Graham, B. S. (2008). Viral and host factors in human respiratory syncytial virus pathogenesis. J. Virol. 82 (5), 2040–2055. doi: 10.1128/JVI.01625-07
Coots, R. A., Liu, X. M., Mao, Y., Dong, L., Zhou, J., Wan, J., et al. (2017). m(6)A facilitates eIF4F-independent mRNA translation. Mol. Cell 68(3), 504-514 e507. doi: 10.1016/j.molcel.2017.10.002
Courtney, D. G., Kennedy, E. M., Dumm, R. E., Bogerd, H. P., Tsai, K., Heaton, N. S., et al. (2017). Epitranscriptomic enhancement of influenza a virus gene expression and replication. Cell Host Microbe 22 (3), 377–386 e375. doi: 10.1016/j.chom.2017.08.004
Crawford, S. E., Ramani, S., Tate, J. E., Parashar, U. D., Svensson, L., Hagbom, M., et al. (2017). Rotavirus infection. Nat. Rev. Dis. Primers 3, 17083. doi: 10.1038/nrdp.2017.83
Csepany, T., Lin, A., Baldick, C. J., Jr., Beemon, K. (1990). Sequence specificity of mRNA N6-adenosine methyltransferase. J. Biol. Chem. 265 (33), 20117–20122. doi: 10.1016/S0021-9258(17)30477-5
Dina, C., Meyre, D., Gallina, S., Durand, E., Korner, A., Jacobson, P., et al. (2007). Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 39 (6), 724–726. doi: 10.1038/ng2048
Do, R., Bailey, S. D., Desbiens, K., Belisle, A., Montpetit, A., Bouchard, C., et al. (2008). Genetic variants of FTO influence adiposity, insulin sensitivity, leptin levels, and resting metabolic rate in the Quebec family study. Diabetes 57 (4), 1147–1150. doi: 10.2337/db07-1267
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485 (7397), 201–206. doi: 10.1038/nature11112
Du, H., Zhao, Y., He, J., Zhang, Y., Xi, H., Liu, M., et al. (2016). YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7, 12626. doi: 10.1038/ncomms12626
Durbin, A. F., Wang, C., Marcotrigiano, J., Gehrke, L. (2016). RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. mBio 7 (5), e0083–16. doi: 10.1128/mBio.00833-16
Edupuganti, R. R., Geiger, S., Lindeboom, R. G. H., Shi, H., Hsu, P. J., Lu, Z., et al. (2017). N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24 (10), 870–878. doi: 10.1038/nsmb.3462
Feng, Z., Li, Q., Meng, R., Yi, B., Xu, Q. (2018). METTL3 regulates alternative splicing of MyD88 upon the lipopolysaccharide-induced inflammatory response in human dental pulp cells. J. Cell Mol. Med. 22 (5), 2558–2568. doi: 10.1111/jcmm.13491
Feng, Z., Zhou, F., Tan, M., Wang, T., Chen, Y., Xu, W., et al. (2022). Targeting m6A modification inhibits herpes virus 1 infection. Genes Dis. 9 (4), 1114–1128. doi: 10.1016/j.gendis.2021.02.004
Finkel, D., Groner, Y. (1983). Methylations of adenosine residues (m6A) in pre-mRNA are important for formation of late simian virus 40 mRNAs. Virology 131 (2), 409–425. doi: 10.1016/0042-6822(83)90508-1
Frayling, T. M., Timpson, N. J., Weedon, M. N., Zeggini, E., Freathy, R. M., Lindgren, C. M., et al. (2007). A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316 (5826), 889–894. doi: 10.1126/science.1141634
Furuichi, Y., LaFiandra, A., Shatkin, A. J. (1977). 5'-terminal structure and mRNA stability. Nature 266 (5599), 235–239. doi: 10.1038/266235a0
Gokhale, N. S., Horner, S. M. (2017). RNA Modifications go viral. PloS Pathog. 13 (3), e1006188. doi: 10.1371/journal.ppat.1006188
Gokhale, N. S., McIntyre, A. B. R., McFadden, M. J., Roder, A. E., Kennedy, E. M., Gandara, J. A., et al. (2016). N6-methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20 (5), 654–665. doi: 10.1016/j.chom.2016.09.015
Hammerschmidt, W. (2015). The epigenetic life cycle of Epstein-Barr virus. Curr. Top. Microbiol. Immunol. 390 (Pt 1), 103–117. doi: 10.1007/978-3-319-22822-8_6
Han, H., Fan, G., Song, S., Jiang, Y., Qian, C., Zhang, W., et al. (2021). piRNA-30473 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in DLBCL. Blood 137 (12), 1603–1614. doi: 10.1182/blood.2019003764
Hao, H., Hao, S., Chen, H., Chen, Z., Zhang, Y., Wang, J., et al. (2019). N6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Res. 47 (1), 362–374. doi: 10.1093/nar/gky1007
Hartmann, A. M., Nayler, O., Schwaiger, F. W., Obermeier, A., Stamm, S. (1999). The interaction and colocalization of Sam68 with the splicing-associated factor YT521-b in nuclear dots is regulated by the src family kinase p59(fyn). Mol. Biol. Cell 10 (11), 3909–3926. doi: 10.1091/mbc.10.11.3909
Hesser, C. R., Karijolich, J., Dominissini, D., He, C., Glaunsinger, B. A. (2018). N6-methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during kaposi's sarcoma-associated herpesvirus infection. PloS Pathog. 14 (4), e1006995. doi: 10.1371/journal.ppat.1006995
Hornung, V., Ellegast, J., Kim, S., Brzozka, K., Jung, A., Kato, H., et al. (2006). 5'-triphosphate RNA is the ligand for RIG-I. Science 314 (5801), 994–997. doi: 10.1126/science.1132505
Hsu, P. J., Zhu, Y., Ma, H., Guo, Y., Shi, X., Liu, Y., et al. (2017). Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27 (9), 1115–1127. doi: 10.1038/cr.2017.99
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., et al. (2020). Clinical features of patients infected with 2019 novel coronavirus in wuhan, China. Lancet 395 (10223), 497–506. doi: 10.1016/S0140-6736(20)30183-5
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20 (3), 285–295. doi: 10.1038/s41556-018-0045-z
Hyde, J. L., Diamond, M. S. (2015). Innate immune restriction and antagonism of viral RNA lacking 2-O methylation. Virology 479-480, 66–74. doi: 10.1016/j.virol.2015.01.019
Imam, H., Khan, M., Gokhale, N. S., McIntyre, A. B. R., Kim, G. W., Jang, J. Y., et al. (2018). N6-methyladenosine modification of hepatitis b virus RNA differentially regulates the viral life cycle. Proc. Natl. Acad. Sci. U.S.A. 115 (35), 8829–8834. doi: 10.1073/pnas.1808319115
Imam, H., Kim, G. W., Mir, S. A., Khan, M., Siddiqui, A. (2020). Interferon-stimulated gene 20 (ISG20) selectively degrades N6-methyladenosine modified hepatitis b virus transcripts. PloS Pathog. 16 (2), e1008338. doi: 10.1371/journal.ppat.1008338
Ivashkiv, L. B., Donlin, L. T. (2014). Regulation of type I interferon responses. Nat. Rev. Immunol. 14 (1), 36–49. doi: 10.1038/nri3581
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7 (12), 885–887. doi: 10.1038/nchembio.687
Johnson, W. E. (2019). Origins and evolutionary consequences of ancient endogenous retroviruses. Nat. Rev. Microbiol. 17 (6), 355–370. doi: 10.1038/s41579-019-0189-2
Johnson, B., VanBlargan, L. A., Xu, W., White, J. P., Shan, C., Shi, P. Y., et al. (2018). Human IFIT3 modulates IFIT1 RNA binding specificity and protein stability. Immunity 48(3), 487-499 e485. doi: 10.1016/j.immuni.2018.01.014
Jurczyszak, D., Zhang, W., Terry, S. N., Kehrer, T., Bermudez Gonzalez, M. C., McGregor, E., et al. (2020). HIV Protease cleaves the antiviral m6A reader protein YTHDF3 in the viral particle. PloS Pathog. 16 (2), e1008305. doi: 10.1371/journal.ppat.1008305
Kariko, K., Buckstein, M., Ni, H., Weissman, D. (2005). Suppression of RNA recognition by toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23 (2), 165–175. doi: 10.1016/j.immuni.2005.06.008
Kastan, J. P., Tremblay, M. W., Brown, M. C., Trimarco, J. D., Dobrikova, E. Y., Dobrikov, M. I., et al. (2021). Enterovirus 2A(pro) cleavage of the YTHDF m(6)A readers implicates YTHDF3 as a mediator of type I interferon-driven JAK/STAT signaling. MBio 12 (2), e00116–21. doi: 10.1128/mBio.00116-21
Kato, H., Takeuchi, O., Mikamo-Satoh, E., Hirai, R., Kawai, T., Matsushita, K., et al. (2008). Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205 (7), 1601–1610. doi: 10.1084/jem.20080091
Ke, S., Alemu, E. A., Mertens, C., Gantman, E. C., Fak, J. J., Mele, A., et al. (2015). A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation. Genes Dev. 29 (19), 2037–2053. doi: 10.1101/gad.269415.115
Ke, S., Pandya-Jones, A., Saito, Y., Fak, J. J., Vagbo, C. B., Geula, S., et al. (2017). m(6)A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31 (10), 990–1006. doi: 10.1101/gad.301036.117
Kennedy, E. M., Bogerd, H. P., Kornepati, A. V., Kang, D., Ghoshal, D., Marshall, J. B., et al. (2016). Posttranscriptional m(6)A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe 19 (5), 675–685. doi: 10.1016/j.chom.2016.04.002
Khong, A., Matheny, T., Huynh, T. N., Babl, V., Parker, R. (2022). Limited effects of m(6)A modification on mRNA partitioning into stress granules. Nat. Commun. 13 (1), 3735. doi: 10.1038/s41467-022-31358-5
Kim, B., Arcos, S., Rothamel, K., Jian, J., Rose, K. L., McDonald, W. H., et al. (2020a). Discovery of widespread host protein interactions with the pre-replicated genome of CHIKV using VIR-CLASP. Mol. Cell 78 (4), 624–640 e627. doi: 10.1016/j.molcel.2020.04.013
Kim, G. W., Imam, H., Khan, M., Siddiqui, A. (2020c). N (6)-methyladenosine modification of hepatitis b and c viral RNAs attenuates host innate immunity via RIG-I signaling. J. Biol. Chem. 295 (37), 13123–13133. doi: 10.1074/jbc.RA120.014260
Kim, G. W., Imam, H., Siddiqui, A. (2021). The RNA binding proteins YTHDC1 and FMRP regulate the nuclear export of N(6)-Methyladenosine-Modified hepatitis b virus transcripts and affect the viral life cycle. J. Virol. 95 (13), e0009721. doi: 10.1128/JVI.00097-21
Kim, D., Lee, J. Y., Yang, J. S., Kim, J. W., Kim, V. N., Chang, H. (2020b). The architecture of SARS-CoV-2 transcriptome. Cell 181 (4), 914–921 e910. doi: 10.1016/j.cell.2020.04.011
Kretschmer, J., Rao, H., Hackert, P., Sloan, K. E., Hobartner, C., Bohnsack, M. T. (2018). The m(6)A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5'-3' exoribonuclease XRN1. RNA 24 (10), 1339–1350. doi: 10.1261/rna.064238.117
Krug, R. M., Morgan, M. A., Shatkin, A. J. (1976). Influenza viral mRNA contains internal N6-methyladenosine and 5'-terminal 7-methylguanosine in cap structures. J. Virol. 20 (1), 45–53. doi: 10.1128/JVI.20.1.45-53.1976
Lang, F., Singh, R. K., Pei, Y., Zhang, S., Sun, K., Robertson, E. S. (2019). EBV epitranscriptome reprogramming by METTL14 is critical for viral-associated tumorigenesis. PloS Pathog. 15 (6), e1007796. doi: 10.1371/journal.ppat.1007796
Li, A., Chen, Y. S., Ping, X. L., Yang, X., Xiao, W., Yang, Y., et al. (2017a). Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 27 (3), 444–447. doi: 10.1038/cr.2017.10
Li, N., Hui, H., Bray, B., Gonzalez, G. M., Zeller, M., Anderson, K. G., et al. (2021). METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS-CoV-2 infection. Cell Rep. 35 (6), 109091. doi: 10.1016/j.celrep.2021.109091
Li, H. B., Tong, J., Zhu, S., Batista, P. J., Duffy, E. E., Zhao, J., et al. (2017b). m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 548 (7667), 338–342. doi: 10.1038/nature23450
Li, F., Zhao, D., Wu, J., Shi, Y. (2014). Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res. 24 (12), 1490–1492. doi: 10.1038/cr.2014.153
Lichinchi, G., Gao, S., Saletore, Y., Gonzalez, G. M., Bansal, V., Wang, Y., et al. (2016a). Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 1, 16011. doi: 10.1038/nmicrobiol.2016.11
Lichinchi, G., Zhao, B. S., Wu, Y., Lu, Z., Qin, Y., He, C., et al. (2016b). Dynamics of human and viral RNA methylation during zika virus infection. Cell Host Microbe 20 (5), 666–673. doi: 10.1016/j.chom.2016.10.002
Lin, S., Choe, J., Du, P., Triboulet, R., Gregory, R. I. (2016). The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62 (3), 335–345. doi: 10.1016/j.molcel.2016.03.021
Liu, N., Dai, Q., Zheng, G., He, C., Parisien, M., Pan, T. (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518 (7540), 560–564. doi: 10.1038/nature14234
Liu, J., Dou, X., Chen, C., Chen, C., Liu, C., Xu, M. M., et al. (2020). N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367 (6477), 580–586. doi: 10.1126/science.aay6018
Liu, R., Kasowitz, S. D., Homolka, D., Leu, N. A., Shaked, J. T., Ruthel, G., et al. (2021b). YTHDC2 is essential for pachytene progression and prevents aberrant microtubule-driven telomere clustering in male meiosis. Cell Rep. 37 (11), 110110. doi: 10.1016/j.celrep.2021.110110
Liu, J., Xu, Y. P., Li, K., Ye, Q., Zhou, H. Y., Sun, H., et al. (2021a). The m(6)A methylome of SARS-CoV-2 in host cells. Cell Res. 31 (4), 404–414. doi: 10.1038/s41422-020-00465-7
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10 (2), 93–95. doi: 10.1038/nchembio.1432
Liu, N., Zhou, K. I., Parisien, M., Dai, Q., Diatchenko, L., Pan, T. (2017). N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45 (10), 6051–6063. doi: 10.1093/nar/gkx141
Lou, X., Wang, J. J., Wei, Y. Q., Sun, J. J. (2021). Emerging role of RNA modification N6-methyladenosine in immune evasion. Cell Death Dis. 12 (4), 300. doi: 10.1038/s41419-021-03585-z
Lu, W., Tirumuru, N., St Gelais, C., Koneru, P. C., Liu, C., Kvaratskhelia, M., et al. (2018). N(6)-methyladenosine-binding proteins suppress HIV-1 infectivity and viral production. J. Biol. Chem. 293 (34), 12992–13005. doi: 10.1074/jbc.RA118.004215
Lu, M., Xue, M., Wang, H. T., Kairis, E. L., Ahmad, S., Wei, J., et al. (2021). Nonsegmented negative-sense RNA viruses utilize n (6)-methyladenosine (m(6)A) as a common strategy to evade host innate immunity. J. Virol. 95 (9), e01939–20. doi: 10.1128/JVI.01939-20
Lu, M., Zhang, Z., Xue, M., Zhao, B. S., Harder, O., Li, A., et al. (2020). N(6)-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat. Microbiol. 5 (4), 584–598. doi: 10.1038/s41564-019-0653-9
Luo, S., Tong, L. (2014). Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc. Natl. Acad. Sci. U.S.A. 111 (38), 13834–13839. doi: 10.1073/pnas.1412742111
Ma, H., Wang, X., Cai, J., Dai, Q., Natchiar, S. K., Lv, R., et al. (2019). N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 15 (1), 88–94. doi: 10.1038/s41589-018-0184-3
Macveigh-Fierro, D., Cicerchia, A., Cadorette, A., Sharma, V., Muller, M. (2022). The m(6)A reader YTHDC2 is essential for escape from KSHV SOX-induced RNA decay. Proc. Natl. Acad. Sci. U.S.A. 119 (8), e2116662119. doi: 10.1073/pnas.2116662119
Mao, Y., Dong, L., Liu, X. M., Guo, J., Ma, H., Shen, B., et al. (2019). m(6)A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 10 (1), 5332. doi: 10.1038/s41467-019-13317-9
Mauer, J., Luo, X., Blanjoie, A., Jiao, X., Grozhik, A. V., Patil, D. P., et al. (2017). Reversible methylation of m(6)Am in the 5' cap controls mRNA stability. Nature 541 (7637), 371–375. doi: 10.1038/nature21022
McFadden, M. J., McIntyre, A. B. R., Mourelatos, H., Abell, N. S., Gokhale, N. S., Ipas, H., et al. (2021). Post-transcriptional regulation of antiviral gene expression by N6-methyladenosine. Cell Rep. 34 (9), 108798. doi: 10.1016/j.celrep.2021.108798
McIntyre, W., Netzband, R., Bonenfant, G., Biegel, J. M., Miller, C., Fuchs, G., et al. (2018). Positive-sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Res. 46 (11), 5776–5791. doi: 10.1093/nar/gky029
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5' UTR m(6)A promotes cap-independent translation. Cell 163 (4), 999–1010. doi: 10.1016/j.cell.2015.10.012
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149 (7), 1635–1646. doi: 10.1016/j.cell.2012.05.003
Moss, B., Gershowitz, A., Stringer, J. R., Holland, L. E., Wagner, E. K. (1977). 5'-terminal and internal methylated nucleosides in herpes simplex virus type 1 mRNA. J. Virol. 23 (2), 234–239. doi: 10.1128/JVI.23.2.234-239.1977
N'Da Konan, S., Segeral, E., Bejjani, F., Bendoumou, M., Ait Said, M., Gallois-Montbrun, S., et al. (2022). YTHDC1 regulates distinct post-integration steps of HIV-1 replication and is important for viral infectivity. Retrovirology 19 (1), 4. doi: 10.1186/s12977-022-00589-1
Narayan, P., Ayers, D. F., Rottman, F. M., Maroney, P. A., Nilsen, T. W. (1987). Unequal distribution of N6-methyladenosine in influenza virus mRNAs. Mol. Cell Biol. 7 (4), 1572–1575. doi: 10.1128/mcb.7.4.1572-1575.1987
Onomoto, K., Jogi, M., Yoo, J. S., Narita, R., Morimoto, S., Takemura, A., et al. (2012). Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PloS One 7 (8), e43031. doi: 10.1371/journal.pone.0043031
Patil, D. P., Chen, C. K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537 (7620), 369–373. doi: 10.1038/nature19342
Patil, D. P., Pickering, B. F., Jaffrey, S. R. (2018). Reading m(6)A in the transcriptome: m(6)A-binding proteins. Trends Cell Biol. 28 (2), 113–127. doi: 10.1016/j.tcb.2017.10.001
Pendleton, K. E., Chen, B., Liu, K., Hunter, O. V., Xie, Y., Tu, B. P., et al. (2017). The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169 (5), 824–835 e814. doi: 10.1016/j.cell.2017.05.003
Perlegos, A. E., Shields, E. J., Shen, H., Liu, K. F., Bonini, N. M. (2022). Mettl3-dependent m(6)A modification attenuates the brain stress response in drosophila. Nat. Commun. 13 (1), 5387. doi: 10.1038/s41467-022-33085-3
Pichlmair, A., Schulz, O., Tan, C. P., Naslund, T. I., Liljestrom, P., Weber, F., et al. (2006). RIG-i-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science 314 (5801), 997–1001. doi: 10.1126/science.1132998
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24 (2), 177–189. doi: 10.1038/cr.2014.3
Price, A. M., Hayer, K. E., McIntyre, A. B. R., Gokhale, N. S., Abebe, J. S., Della Fera, A. N., et al. (2020). Direct RNA sequencing reveals m(6)A modifications on adenovirus RNA are necessary for efficient splicing. Nat. Commun. 11 (1), 6016. doi: 10.1038/s41467-020-19787-6
Qiu, W., Zhang, Q., Zhang, R., Lu, Y., Wang, X., Tian, H., et al. (2021). N(6)-methyladenosine RNA modification suppresses antiviral innate sensing pathways via reshaping double-stranded RNA. Nat. Commun. 12 (1), 1582. doi: 10.1038/s41467-021-21904-y
Ren, W., Lu, J., Huang, M., Gao, L., Li, D., Wang, G. G., et al. (2019). Structure and regulation of ZCCHC4 in m(6)A-methylation of 28S rRNA. Nat. Commun. 10 (1), 5042. doi: 10.1038/s41467-019-12923-x
Ries, R. J., Zaccara, S., Klein, P., Olarerin-George, A., Namkoong, S., Pickering, B. F., et al. (2019). m(6)A enhances the phase separation potential of mRNA. Nature 571 (7765), 424–428. doi: 10.1038/s41586-019-1374-1
Ringeard, M., Marchand, V., Decroly, E., Motorin, Y., Bennasser, Y. (2019). FTSJ3 is an RNA 2'-o-methyltransferase recruited by HIV to avoid innate immune sensing. Nature 565 (7740), 500–504. doi: 10.1038/s41586-018-0841-4
Riquelme-Barrios, S., Pereira-Montecinos, C., Valiente-Echeverria, F., Soto-Rifo, R. (2018). Emerging roles of N(6)-methyladenosine on HIV-1 RNA metabolism and viral replication. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00576
Roundtree, I. A., Luo, G. Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., et al. (2017). YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife 6, e31311. doi: 10.7554/eLife.31311
Rubio, R. M., Depledge, D. P., Bianco, C., Thompson, L., Mohr, I. (2018). RNA m(6) a modification enzymes shape innate responses to DNA by regulating interferon beta. Genes Dev. 32 (23-24), 1472–1484. doi: 10.1101/gad.319475.118
Runge, S., Sparrer, K. M., Lassig, C., Hembach, K., Baum, A., Garcia-Sastre, A., et al. (2014). In vivo ligands of MDA5 and RIG-I in measles virus-infected cells. PloS Pathog. 10 (4), e1004081. doi: 10.1371/journal.ppat.1004081
Schlee, M., Roth, A., Hornung, V., Hagmann, C. A., Wimmenauer, V., Barchet, W., et al. (2009). Recognition of 5' triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31 (1), 25–34. doi: 10.1016/j.immuni.2009.05.008
Selberg, S., Blokhina, D., Aatonen, M., Koivisto, P., Siltanen, A., Mervaala, E., et al. (2019). Discovery of small molecules that activate RNA methylation through cooperative binding to the METTL3-14-WTAP complex active site. Cell Rep. 26 (13), 3762–3771 e3765. doi: 10.1016/j.celrep.2019.02.100
Shah, A., Rashid, F., Awan, H. M., Hu, S., Wang, X., Chen, L., et al. (2017). The DEAD-box RNA helicase DDX3 interacts with m(6)A RNA demethylase ALKBH5. Stem Cells Int. 2017, 8596135. doi: 10.1155/2017/8596135
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27 (3), 315–328. doi: 10.1038/cr.2017.15
Shulman, Z., Stern-Ginossar, N. (2020). The RNA modification N(6)-methyladenosine as a novel regulator of the immune system. Nat. Immunol. 21 (5), 501–512. doi: 10.1038/s41590-020-0650-4
Shuman, S. (2002). What messenger RNA capping tells us about eukaryotic evolution. Nat. Rev. Mol. Cell Biol. 3 (8), 619–625. doi: 10.1038/nrm880
Sommer, S., Lavi, U., Darnell, J. E., Jr. (1978). The absolute frequency of labeled n-6-methyladenosine in HeLa cell messenger RNA decreases with label time. J. Mol. Biol. 124 (3), 487–499. doi: 10.1016/0022-2836(78)90183-3
Sommer, S., Salditt-Georgieff, M., Bachenheimer, S., Darnell, J. E., Furuichi, Y., Morgan, M., et al. (1976). The methylation of adenovirus-specific nuclear and cytoplasmic RNA. Nucleic Acids Res. 3 (3), 749–765. doi: 10.1093/nar/3.3.749
Srinivas, K. P., Depledge, D. P., Abebe, J. S., Rice, S. A., Mohr, I., Wilson, A. C. (2021). Widespread remodeling of the m6A RNA-modification landscape by a viral regulator of RNA processing and export. Proc. Natl. Acad. Sci. U.S.A. 118 (30), e2104805118. doi: 10.1073/pnas.2104805118
Sun, L., Fazal, F. M., Li, P., Broughton, J. P., Lee, B., Tang, L., et al. (2019). RNA Structure maps across mammalian cellular compartments. Nat. Struct. Mol. Biol. 26 (4), 322–330. doi: 10.1038/s41594-019-0200-7
Tan, B., Liu, H., Zhang, S., da Silva, S. R., Zhang, L., Meng, J., et al. (2018). Viral and cellular N(6)-methyladenosine and N(6),2'-o-dimethyladenosine epitranscriptomes in the KSHV life cycle. Nat. Microbiol. 3 (1), 108–120. doi: 10.1038/s41564-017-0056-8
Tang, L., Wei, X., Li, T., Chen, Y., Dai, Z., Lu, C., et al. (2021). Emerging perspectives of RNA N(6)-methyladenosine (m(6)A) modification on immunity and autoimmune diseases. Front. Immunol. 12. doi: 10.3389/fimmu.2021.630358
Tirumuru, N., Zhao, B. S., Lu, W., Lu, Z., He, C., Wu, L. (2016). N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 gag protein expression. Elife 5, e15528. doi: 10.7554/eLife.15528
Tong, J., Cao, G., Zhang, T., Sefik, E., Amezcua Vesely, M. C., Broughton, J. P., et al. (2018). m(6)A mRNA methylation sustains treg suppressive functions. Cell Res. 28 (2), 253–256. doi: 10.1038/cr.2018.7
Tsai, K., Bogerd, H. P., Kennedy, E. M., Emery, A., Swanstrom, R., Cullen, B. R. (2021). Epitranscriptomic addition of m(6)A regulates HIV-1 RNA stability and alternative splicing. Genes Dev. 35 (13-14), 992–1004. doi: 10.1101/gad.348508.121
Tsai, K., Courtney, D. G., Cullen, B. R. (2018). Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PloS Pathog. 14 (2), e1006919. doi: 10.1371/journal.ppat.1006919
van den Hoogen, B. G., Bestebroer, T. M., Osterhaus, A. D., Fouchier, R. A. (2002). Analysis of the genomic sequence of a human metapneumovirus. Virology 295 (1), 119–132. doi: 10.1006/viro.2001.1355
van Tran, N., Ernst, F. G. M., Hawley, B. R., Zorbas, C., Ulryck, N., Hackert, P., et al. (2019). The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 47 (15), 7719–7733. doi: 10.1093/nar/gkz619
Vilchez, R. A., Kozinetz, C. A., Arrington, A. S., Madden, C. R., Butel, J. S. (2003). Simian virus 40 in human cancers. Am. J. Med. 114 (8), 675–684. doi: 10.1016/s0002-9343(03)00087-1
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., et al. (2016). Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534 (7608), 575–578. doi: 10.1038/nature18298
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505 (7481), 117–120. doi: 10.1038/nature12730
Wang, S., Sun, C., Li, J., Zhang, E., Ma, Z., Xu, W., et al. (2017). Roles of RNA methylation by means of N(6)-methyladenosine (m(6)A) in human cancers. Cancer Lett. 408, 112–120. doi: 10.1016/j.canlet.2017.08.030
Wang, A., Tao, W., Tong, J., Gao, J., Wang, J., Hou, G., et al. (2022). m6A modifications regulate intestinal immunity and rotavirus infection. Elife 11, e73628. doi: 10.7554/eLife.73628
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161 (6), 1388–1399. doi: 10.1016/j.cell.2015.05.014
Warda, A. S., Kretschmer, J., Hackert, P., Lenz, C., Urlaub, H., Hobartner, C., et al. (2017). Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 18 (11), 2004–2014. doi: 10.15252/embr.201744940
Weaver, S. C., Lecuit, M. (2015). Chikungunya virus and the global spread of a mosquito-borne disease. N Engl. J. Med. 372 (13), 1231–1239. doi: 10.1056/NEJMra1406035
Wei, C. M., Gershowitz, A., Moss, B. (1975). Methylated nucleotides block 5' terminus of HeLa cell messenger RNA. Cell 4 (4), 379–386. doi: 10.1016/0092-8674(75)90158-0
Wei, C. M., Moss, B. (1977). Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 16 (8), 1672–1676. doi: 10.1021/bi00627a023
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 69 (6), 1028–1038 e1026. doi: 10.1016/j.molcel.2018.02.015
Williams, G. D., Gokhale, N. S., Horner, S. M. (2019). Regulation of viral infection by the RNA modification N6-methyladenosine. Annu. Rev. Virol. 6 (1), 235–253. doi: 10.1146/annurev-virology-092818-015559
Winkler, R., Gillis, E., Lasman, L., Safra, M., Geula, S., Soyris, C., et al. (2019). m(6)A modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 20 (2), 173–182. doi: 10.1038/s41590-018-0275-z
Wojtas, M. N., Pandey, R. R., Mendel, M., Homolka, D., Sachidanandam, R., Pillai, R. S. (2017). Regulation of m(6)A transcripts by the 3'–>5' RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol. Cell 68 (2), 374–387 e312. doi: 10.1016/j.molcel.2017.09.021
Wu, R., Li, A., Sun, B., Sun, J. G., Zhang, J., Zhang, T., et al. (2019). A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 29 (1), 23–41. doi: 10.1038/s41422-018-0113-8
Wu, B., Peisley, A., Tetrault, D., Li, Z., Egelman, E. H., Magor, K. E., et al. (2014). Molecular imprinting as a signal-activation mechanism of the viral RNA sensor RIG-I. Mol. Cell 55 (4), 511–523. doi: 10.1016/j.molcel.2014.06.010
Wu, B., Su, S., Patil, D. P., Liu, H., Gan, J., Jaffrey, S. R., et al. (2018). Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 9 (1), 420. doi: 10.1038/s41467-017-02770-z
Wu, F., Zhao, S., Yu, B., Chen, Y. M., Wang, W., Song, Z. G., et al. (2020). A new coronavirus associated with human respiratory disease in China. Nature 579 (7798), 265–269. doi: 10.1038/s41586-020-2008-3
Xia, T. L., Li, X., Wang, X., Zhu, Y. J., Zhang, H., Cheng, W., et al. (2021). N(6)-methyladenosine-binding protein YTHDF1 suppresses EBV replication and promotes EBV RNA decay. EMBO Rep. 22 (4), e50128. doi: 10.15252/embr.202050128
Xiao, W., Adhikari, S., Dahal, U., Chen, Y. S., Hao, Y. J., Sun, B. F., et al. (2016). Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61 (4), 507–519. doi: 10.1016/j.molcel.2016.01.012
Xu, C., Liu, K., Ahmed, H., Loppnau, P., Schapira, M., Min, J. (2015). Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-b homology domain family of proteins. J. Biol. Chem. 290 (41), 24902–24913. doi: 10.1074/jbc.M115.680389
Xu, Z., Shi, L., Wang, Y., Zhang, J., Huang, L., Zhang, C., et al. (2020). Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 8 (4), 420–422. doi: 10.1016/S2213-2600(20)30076-X
Xu, C., Wang, X., Liu, K., Roundtree, I. A., Tempel, W., Li, Y., et al. (2014). Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10 (11), 927–929. doi: 10.1038/nchembio.1654
Xue, M., Zhao, B. S., Zhang, Z., Lu, M., Harder, O., Chen, P., et al. (2019). Viral N(6)-methyladenosine upregulates replication and pathogenesis of human respiratory syncytial virus. Nat. Commun. 10 (1), 4595. doi: 10.1038/s41467-019-12504-y
Ye, F., Chen, E. R., Nilsen, T. W. (2017). Kaposi's sarcoma-associated herpesvirus utilizes and manipulates RNA N(6)-adenosine methylation to promote lytic replication. J. Virol. 91 (16), e00466–17. doi: 10.1128/JVI.00466-17
Youn, J. Y., Dunham, W. H., Hong, S. J., Knight, J. D. R., Bashkurov, M., Chen, G. I., et al. (2018). High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell 69 (3), 517–532 e511. doi: 10.1016/j.molcel.2017.12.020
Yue, Y., Liu, J., Cui, X., Cao, J., Luo, G., Zhang, Z., et al. (2018). VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4, 10. doi: 10.1038/s41421-018-0019-0
Zaccara, S., Ries, R. J., Jaffrey, S. R. (2019). Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20 (10), 608–624. doi: 10.1038/s41580-019-0168-5
Zhang, F., Kang, Y., Wang, M., Li, Y., Xu, T., Yang, W., et al. (2018). Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum. Mol. Genet. 27 (22), 3936–3950. doi: 10.1093/hmg/ddy292
Zhang, Y., Wang, X., Zhang, X., Wang, J., Ma, Y., Zhang, L., et al. (2019b). RNA-Binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc. Natl. Acad. Sci. U.S.A. 116 (3), 976–981. doi: 10.1073/pnas.1812536116
Zhang, F., Yuan, Y., Ma, F. (2020). Function and regulation of nuclear DNA sensors during viral infection and tumorigenesis. Front. Immunol. 11. doi: 10.3389/fimmu.2020.624556
Zhang, X., Zhang, Y., Dai, K., Liang, Z., Zhu, M., Pan, J., et al. (2019a). N (6)-methyladenosine level in silkworm Midgut/Ovary cell line is associated with bombyx mori nucleopolyhedrovirus infection. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02988
Zhang, X., Zhang, Y., Pan, J., Gong, C., Hu, X. (2022). Identification and characterization of BmNPV m6A sites and their possible roles during viral infection. Front. Immunol. 13, 869313. doi: 10.3389/fimmu.2022.869313
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49 (1), 18–29. doi: 10.1016/j.molcel.2012.10.015
Zheng, Q., Hou, J., Zhou, Y., Li, Z., Cao, X. (2017). The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nat. Immunol. 18 (10), 1094–1103. doi: 10.1038/ni.3830
Zheng, X., Wang, J., Zhang, X., Fu, Y., Peng, Q., Lu, J., et al. (2021). RNA m(6) a methylation regulates virus-host interaction and EBNA2 expression during Epstein-Barr virus infection. Immun. Inflamm. Dis. 9 (2), 351–362. doi: 10.1002/iid3.396
Zheng, Z., Zhang, L., Cui, X. L., Yu, X., Hsu, P. J., Lyu, R., et al. (2020). Control of early b cell development by the RNA N(6)-methyladenosine methylation. Cell Rep. 31 (13), 107819. doi: 10.1016/j.celrep.2020.107819
Zhu, T., Roundtree, I. A., Wang, P., Wang, X., Wang, L., Sun, C., et al. (2014). Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 24 (12), 1493–1496. doi: 10.1038/cr.2014.152
Zhu, D., Song, Y., Hu, D., Li, S., Liu, G., Li, P., et al. (2021). Characterization of enterovirus associated m6A RNA methylation in children with neurological symptoms: a prospective cohort study. Front. Neurosci. 15. doi: 10.3389/fnins.2021.791544
Keywords: N6-methyl adenosine, m6A reader, epitranscriptomics, viral replication, immune response
Citation: Yang D, Zhao G and Zhang HM (2023) m6A reader proteins: the executive factors in modulating viral replication and host immune response. Front. Cell. Infect. Microbiol. 13:1151069. doi: 10.3389/fcimb.2023.1151069
Received: 25 January 2023; Accepted: 03 May 2023;
Published: 30 May 2023.
Edited by:
Chandrabose Selvaraj, Saveetha University, IndiaReviewed by:
Balajee Ramachandran, Chobanian and Avedisian School of Medicine, Boston University, United StatesCopyright © 2023 Yang, Zhao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Decheng Yang, ZGVjaGVuZy55YW5nQGhsaS51YmMuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.