
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 28 April 2023
Sec. Fungal Pathogenesis
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1134921
This article is part of the Research Topic Diversity and Molecular Diagnostics of Fungi and Oomycetes in Plants View all 18 articles
Heterobasidion annosum is one of the most aggressive pathogens of Pinus forests in Europe, causing considerable economic losses. To detect H. annosum for disease diagnosis and control, we developed a loop-mediated isothermal amplification (LAMP) reaction with a primer set designed from the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) DNA sequences of H. annosum. In our study, this LAMP assay was found to be capable of efficiently amplifying the target gene within 60 min at 63°C. In specificity tests, H. annosum was positively detected, and other species were negative. The detection limit of this assay was found to be 100 pg·μL-1, and the assay was also successfully tested for use with basidiospore suspensions and wood samples. This study provides a rapid method for diagnosing root and butt rot caused by H. annosum, which will be of use in port surveillance of logs imported from Europe.
Heterobasidion annosum (Fr.) Bref. sensu lato (s.l.) has been studied intensively over several decades, with interfertility studies showing that H. annosum s.l. is a species complex (Korhonen, 1978; Dai and Korhonen, 1999; Dai et al., 2003). Recently, species of Heterobasidion have been divided into three groups using multilocus phylogenetic approaches; furthermore, the pathogenic species H. annosum sensu stricto (s.s.) has been found to be a sister to H. irregulare Garbel. & Otrosina. Most taxa of H. annosum s.l. are distributed in the conifer forests of the northern hemisphere (Chen et al., 2015; Dai et al., 2021a, b; Yuan et al., 2021; Wu et al., 2022).
Heterobasidion annosum is one of the most aggressive pathogens in the destruction of pine plantations in Europe (Edmonds et al., 1989; Woodward et al., 1998; Dai and Korhonen, 1999). The root and butt rot caused by Heterobasidion s.l. species can destroy the most valuable part of the tree (Korhonen and Stenlid, 1998; Niemelä and Korhonen, 1998; Seifert, 2007), depreciating the usability of the timber (Aza et al., 2021) and lowering the tree’s resistance to strong winds (Oliva et al., 2008). Furthermore, H. annosum s.l. may remain active in residual stumps and roots for decades until the next rotation (Rishbeth, 1951; Greig and Pratt, 1976). Significantly, H. annosum s.l. grows more quickly in dead trees than in living trees (Bendz-Hellgren et al., 1999). Hence, poor thinning and logging operations may increase the incidence of annosum-related rot (Shaw et al., 1995; Morrison and Johnson, 1999). Dai et al. (2021a) proposed that the most aggressive conifer pathogens, H. abietinum, H. annosum s. s., H. irregulare, H. occidentale, and H. parviporum, should be identified as quarantine fungi, as they are not found in China. Therefore, effective detection of annosum-related rot is important.
Over the past several decades, various methods for H. annosum detection have been developed, mainly focusing on morphological characters, mating tests, and molecular strategies. Traditionally, morphological identification of H. annosum has relied on macroscopic and microscopic observations (Rishbeth, 1951; Greig and Pratt, 1976; Tokuda et al., 2009; Aberg et al., 2016). However, once the basidiomata can be observed, it is already too late to protect the trees in question from decay (Garbelotto and Gonthier, 2013). Although mating tests are a relatively reliable method to determine compatibility with known species, they take time (Korhonen, 1978; Mitchelson and Korhonen, 1998; Dai and Korhonen, 1999; Dai et al., 2002). In fact, as all the classical diagnostic methods are complicated, time-consuming, and require professional skill, researchers have been investigating molecular methods of assay. A potential polymerase chain reaction (PCR) method offers great promise for detection of pathogenic fungi because of its speed and specificity (Schulze, 1999). Multiplex real-time PCR assay, with better resolution than traditional technology, has already been conducted by several researchers (Hietala et al., 2003; Ioos et al., 2019), and qPCR technology has been used to measure the distribution of species of Heterobasidion (Oliva et al., 2017).
Although PCR technology has already been applied in detection of H. annosum due to its sensitivity and specificity, long periods of time and expensive laboratory instruments are still required for these procedures. These intrinsic disadvantages prevent this method from being used in resource-limited regions. Loop-mediated isothermal amplification (LAMP) is an alternative method that amplifies target DNA sequences with high sensitivity and specificity under isothermal conditions (Notomi et al., 2000). The technology has previously been applied in pathogen detection (Sillo et al., 2017; Kong et al., 2020; Vettraino et al., 2021). So far, LAMP technology has been widely used in the medical field (Parida et al., 2005; Parida et al., 2007; Santiago, 2021), food science (Petersen et al., 2021), and plant protection (Franco Ortega et al., 2019; Enicks et al., 2020). The North American species H. irregulare Garbel. & Otrosina was detected by LAMP using a HirrSC3 gene within cytochrome P450 monooxygenase (Sillo et al., 2017). However, methods for rapid detection of H. annosum have rarely been reported.
LAMP utilizes a Bst DNA polymerase with stand-displacement activity, along with two inner primers (FIP, BIP) and two outer primers (F3, B3) that recognize six separate regions within a target DNA sequence (Notomi et al., 2000). Correct recognition of all six regions by the primers ensures the specificity of the assay. Positive reactions can be examined in the products, as follows: turbidity of magnesium phosphate increases (Mori et al., 2001); ladder-like bands can be observed on gel electrophoresis; and color changes can be induced in the reaction system through the addition of DNA-intercalating dyes (Goto et al., 2009). The metal ion hydroxynaphthol blue (HNB) is a reliable indicator of DNA amplification because of the low risk of cross-contamination along with sensitivity equivalent to that of SYBR green, and the results can easily be judged with the naked eye (Goto et al., 2009).
In this study, we aimed to develop a simple LAMP detection method for specific identification of H. annosum and to evaluate its accuracy in detecting wood decay caused by H. annosum.
This study used forty-five cultures and specimens which were maintained at the Institute of Microbiology, the Beijing Forestry University (BJFC, Beijing, P.R. China), Jiangsu Vocational College of Agriculture and Forestry (JSAFC), and the Natural Resources Institute, Finland (Luke, Helsinki, Finland) (Table 1). Fungal strains were cultured on potato dextrose agar (PDA) (Caten and Jinks, 1968; Gams et al., 1998) in 90 mm petri dishes at 25°C for 28 days. In order to obtain abundant mycelia, fungal strains were cultured on potato dextrose agar for 7 days.
Mycelia and basidiomata were ground in liquid nitrogen and subsequently collected in 1.5-mL microfuge tubes. Genomic DNA was extracted using the CTAB rapid plant genome extraction kit (Aidlab Biotechnologies Co., Ltd., Beijing, China) according to the manufacturer’s instructions, with some modifications (Chen et al., 2015). The concentration of the extracted DNA was evaluated using a Nanodrop spectrophotometer (Thermo Fisher Scientific, USA) following Kong et al. (2020); this was then diluted in 10-fold serial dilutions to produce concentrations from 10 ng·μL-1 to 10 fg·μL-1 and stored at –20°C. The cultures and specimens used were identified by morphological examination, and/or by sequencing Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Chen et al., 2015) or the internal transcribed spacer (ITS) (Table 1).
The LAMP reaction was performed according to previously described methods (Niu et al., 2012; Duan et al., 2014; Kong et al., 2020; Vettraino et al., 2021). The final LAMP reaction (26 μL volume) was performed by combining 2.5 μL 10 × ThermoPol buffer, 1.6 μmol·L-1 forward inner primer (FIP) and backward inner primer (BIP), 0.2 μmol·L-1 B3 and F3 primers, 0.8 μmol·L-1 LB and LF primers, 5 mmol·L-1 Mg2+, 0.8 mol·L-1 betaine, 1.4 mmol·L-1 dNTPs, 300 μmol·L-1 HNB, 8 U of Bst DNA polymerase, and 2 μL DNA template.
The LAMP reaction mixtures were heated at a range of reaction temperatures (viz., 61°C, 62°C, 63°C, 64°C, and 65°C) for 60 min to select the optimal temperature (Figure S1). Additionally, LAMP reactions were performed at the optimal reaction temperature (63°C) for 15 min, 30 min, 60 min, and 90 min in order to select the shortest viable reaction time (Figure S2). Runs were performed with positive controls (H. annosum), negative controls (14 Heterobasidion spp. and 24 other fungi), and controls consisting of distilled water without DNA. The assays were evaluated by observation of the HNB color change from violet to blue, which denotes positive amplification, while a negative assay remains violet. The optimum temperature and shortest viable time were identified as 63°C for 60 min. Each condition was repeated at least three times.
Basidiospore suspensions were prepared by scraping four-week-old PDA-cultured mycelium with sterile distilled water. The concentration was determined using a hemacytometer and then adjusted in sterile water to obtain the desired final concentrations, containing 104, 103, 102, 50, 10, and 0 basidiospores per 1 μL. DNA was extracted from these basidiospore suspensions in order to evaluate the effectiveness of the LAMP assay in detecting basidiospores of H. annosum.
In order to evaluate the ability of LAMP to detect H. annosum in wood, trials were conducted following Li (2014). Pinus sylvestris, a cultivar highly susceptible to Heterobasidion spp., was selected for this experiment. Six pieces of P. sylvestris almost 20 cm long and 30–35 cm in diameter were prepared for this assay. Each piece was disinfected with 75% ethanol, wiped with distilled water, and air dried. Three pieces of wood were inoculated with strains of H. annosum; the other three were sprayed only with sterile distilled water. The pieces of wood were incubated in a partially darkened room for five weeks, with the surface kept moist during this period. DNA was extracted from each piece of wood and stored at –80°C until used.
The primers were designed using the PRIMEREXPLORER V5 software program (http://primerexplorer.jp/lampv5e/index.html) based on GAPDH. Sequences were aligned using MAFFT 7 (https://mafft.cbrc.jp/alignment/server/). Regions conserved among all tested H. annosum populations but differentiating between closely related fungal species were selected for the design of LAMP primers (Figure 1). We designed a set of four primers to identify six regions of the target DNA, consisting of two inner primers (a forward inner primer FIP and a backward inner primer BIP) and two outer primers (a forward primer F3 and an outer backward primer B3). Additionally, we designed a loop forward primer (LF) and a loop backward primer (LB) to expedite the LAMP reaction. These primers were synthesized by Sangon Biotech. Nineteen sets of primers were designed for H. annosum; the set deemed suitable are listed in Table 2.
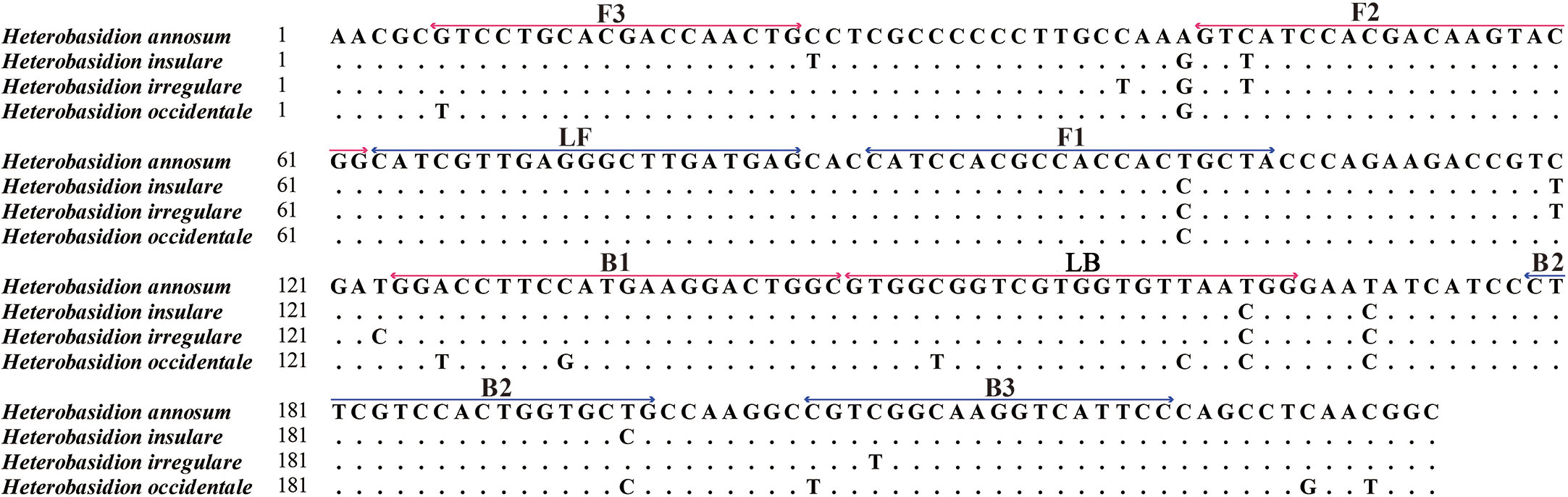
Figure 1 Genomic alignment between Heterobasidion annosum and H. insulare, H. irregulare, and H. occidentale at the locus selected for design of the LAMP primer sets. The locations of the designed primers (F3–B3) are shown: forward primer FIP includes the F1 and F2 regions; backward primer BIP includes the B1 and B2 regions; and loop primers include the LB and LF regions. (Red color represents the forward primers, and blue color represents the reverse primers).
DNA from the isolates and specimens of Heterobasidion and others, as listed in Table 1, were used to validate the specificity of the assay. The LAMP primers were found to detect the species of H. annosum accurately. A positive reaction is indicated by a color change from violet to blue in the presence of the HNB indicator (Figure 2). GAPDH primers were able to distinguish H. annosum from other Heterobasidion species, along with fungi commonly detected in wood samples. Based on visual detection using HNB, only samples of H. annosum displayed a blue color (Figure 3).
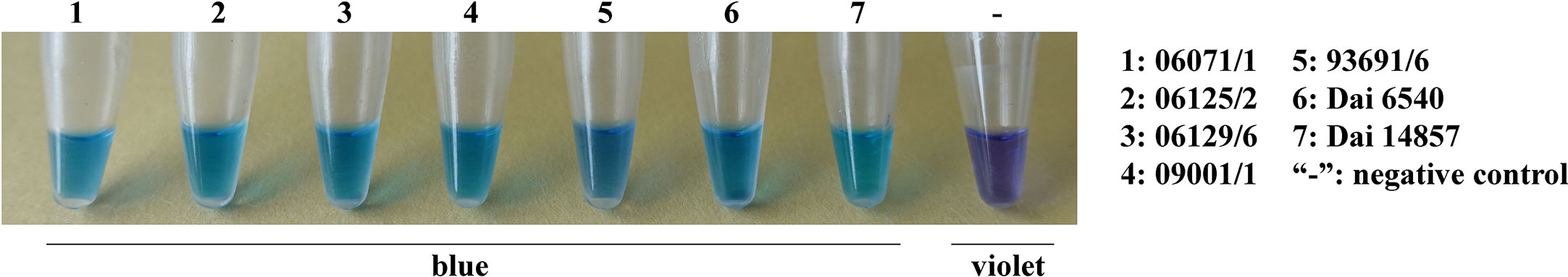
Figure 2 LAMP detection of the GAPDH gene in different isolates of Heterobasidion annosum. 1–7: H. annosum strains; “-”: negative control.
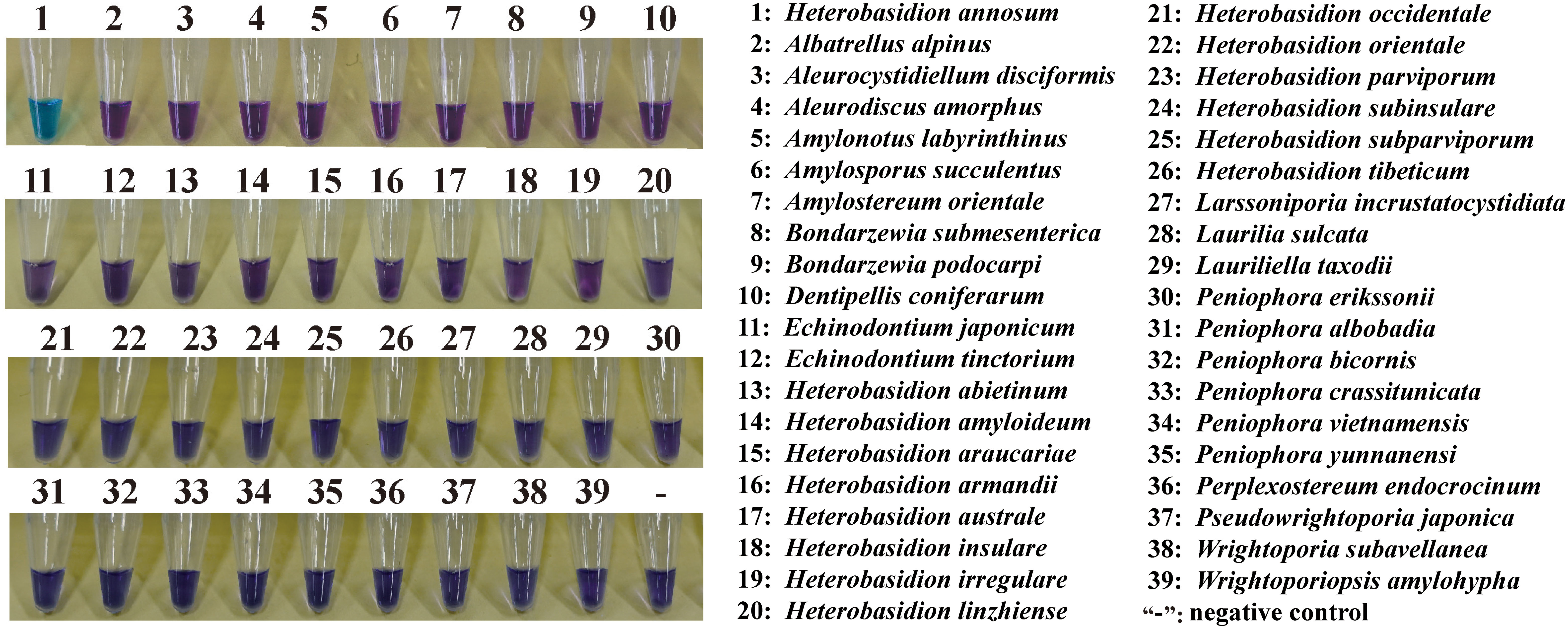
Figure 3 Ability of the LAMP assay to distinguish H. annosum from other species in Russulales. “-”: negative control.
LAMP sensitivity was tested using 10-fold serial dilutions of target genomic DNA prepared with distilled water (10 ng·μL-1, 1 ng·μL-1, 100 pg·μL-1, 10 pg·μL-1, 1 pg·μL-1, 100 fg μL-1, 10 fg·μL-1). A Nanodrop spectrophotometer was used to measure DNA concentration. The results showed that a blue color could be detected up to the point where the DNA concentration was as low as 100 pg·μL-1. However, the color remained violet when the DNA concentration was reduced further to 10 pg·μL-1, 1 pg·μL-1, 100 fg μL-1, or 10 fg·μL-1 (Figure 4).
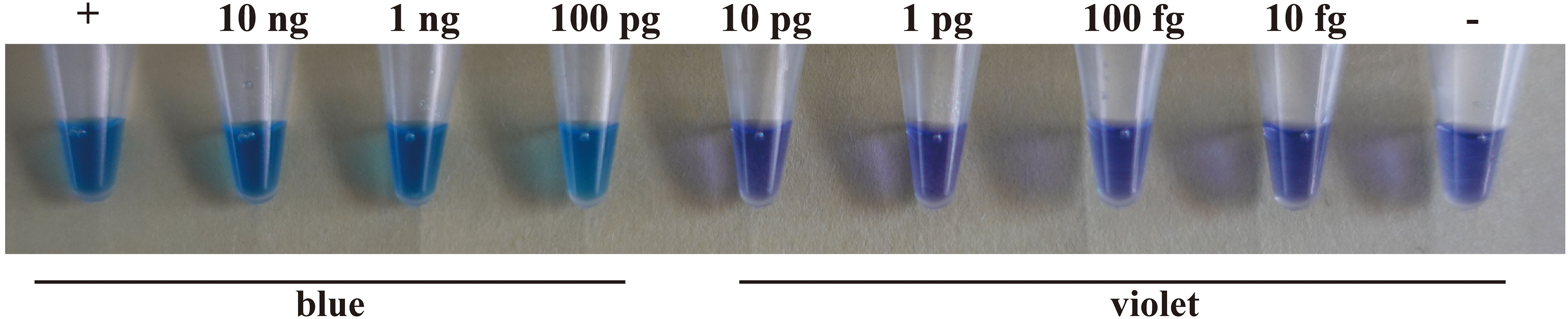
Figure 4 The sensitivity of the LAMP assay using a H. annosum s.s. 93961/6 DNA concentration gradient. “+”: positive control; “-”: negative control.
The color changed from violet to blue in the suspension of basidiospores from the positive control and other treatments containing 104, 103, 102, 50, or 10 basidiospores per 1 μL. However, in the case of the treatment without basidiospores and the negative control, the color remained violet. This pattern indicated that the LAMP assay could detect the presence of at least ten basidiospores of H. annosum per 1 μL in suspension (Figure 5).
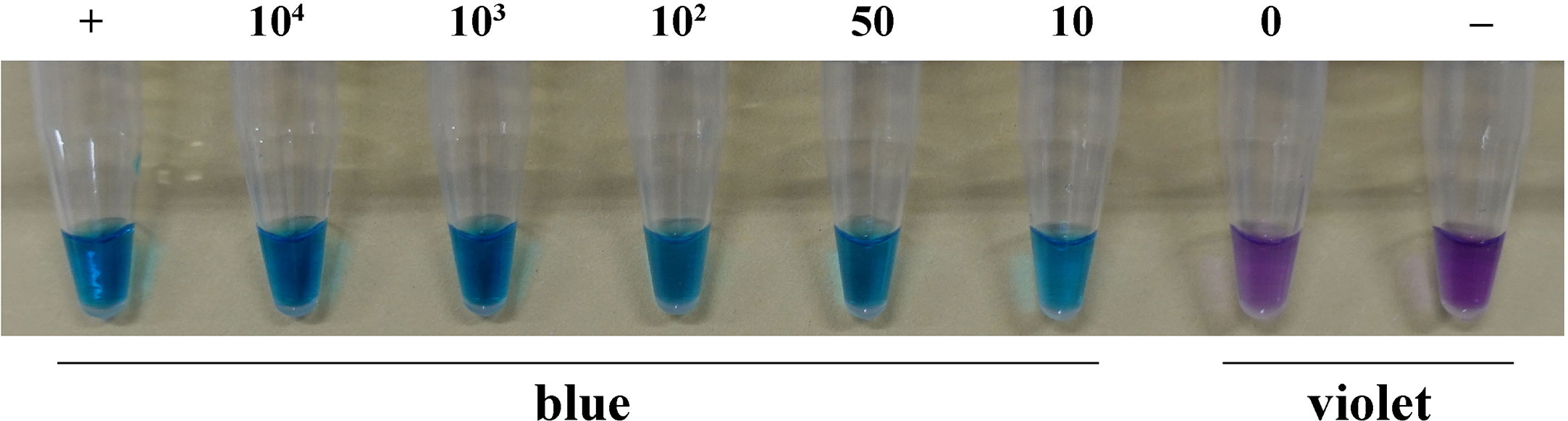
Figure 5 LAMP assay detection of H. annosum in basidiospore suspensions containing different numbers of basidiospores. “+”: positive control; “-”: negative control.
The LAMP assay was applied to samples of wood infected with H. annosum. DNA was extracted from diseased pieces of wood under simulated field conditions; as shown in Figure 6, H. annosum was successfully detected in diseased wood samples.

Figure 6 LAMP assay detection of H. annosum in wood samples. ”+”: positive control; “-”: negative control.
Detection of wood decay based on symptoms is relatively difficult. Trees are usually asymptomatic for decades after infection by butt rot, much less for root rot. The external symptoms mostly occur after the sapwood of the tree has decayed (Greig, 1998). Omdal et al. (2004) suggested that aboveground variables can be used as reasonable indicators of root disease. However, the detection of infections caused by slow-growing wood pathogens and with less obvious outer symptoms, such as H. annosum, often requires considerable professional knowledge, especially to distinguish closely related species.
Several fragments of genes have been described for detection of species of Heterobasidion (Fabritius and Karjalainen, 1993; Kasuga and Mitchelson, 1993; Kasuga et al., 1993; Johannesson and Stenlid, 2003; Linzer et al., 2008; Chen et al., 2014, Chen, 2015; Shamoun et al., 2019; Pellicciaro et al., 2021; Yuan et al., 2021). Although the use of PCR techniques is more successful as a method of detection, it still requires specialized equipment and highly trained personnel, and it is difficult and time-consuming to implement the technique in remote areas and ports. A delay in the identification of wood pathogens causes a major threat to wood production and international trade in timber. We have developed a rapid, specific, and sensitive method of detecting wood decay caused by H. annosum, based on GAPDH sequences; furthermore we have evaluated the accuracy of this method in detecting this fungus directly on wood samples. The LAMP method is far more convenient and effective for detecting H. annosum in time- and resource- limited conditions. This fungus mostly infects pine (Pinus spp.), especially Pinus sylvestris (Chen et al., 2015), but also it can be associated with other conifer forests, such as Abies sp., Larix sp., and Picea sp. (Korhonen, 1978). Genetic evidence has confirmed the major significance of stump infection by H. annosum s.l. (Swedjemark and Stenlid, 1993) in managed forests. The fungus infects freshly cut stumps through the spores and then progresses to the roots, and is able to spread to adjacent trees through root contact (Rishbeth, 1951; Wallis, 1962; Garbelotto and Gonthier, 2013). Thus, our assay may have value during thinning periods in conifer forests.
In general, most PCR amplifications are carried out with a DNA concentration of 20 ng/μL. Conventional PCR amplifications used to detect Heterobasidion species are carried out with a DNA concentration of 20 pg/μL (Shamoun et al., 2019). However, the LAMP assay tested in our study was found to detect H. annosum with a DNA concentration of 100 pg/μL. With adjustments to the temperature and time, the sensitivity of LAMP assay for detection of H. annosum failed to increase. This point necessitates further analysis.
When wood is infected with H. annosum, the pathogen may remain active in residual stumps and roots for decades until the next rotation (Rishbeth, 1951; Greig and Pratt, 1976). Significantly, H. annosum s.l. grows more quickly in dead trees than living trees. Thus, the method presented here is applicable to the analysis of samples stored for long periods or sent over long distances.
China is one of the biggest timber importers in the world, especially in regard to logs. Conifer logs account for a large proportion of wood imports, and this proportion has climbed from 68.8% to 78.5% since 2017 (Han, 2021). Economic losses to wood decay caused by H. annosum should not be ignored, and our LAMP assay provides a quarantine tool for reducing such losses through accurate testing of wood samples.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Design of the research: ZH-M, YJ, CJ-J; performance of the research: ZH-M, LY; data analysis and interpretation: ZH-M, CJ-J, YJ; collect the materials: DY-C, YY, WC-P; writing and revising the manuscript: ZH-M, CJ-J, YJ, DY-C. All authors contributed to the article and approved the submitted version.
The research was supported by the National Natural Science Foundation of China (32161143013 & 32070022), the Science Fund of the Jiangsu Vocational College of Agriculture and Forestry (2020kj003 & 2021kj91), and the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (20KJB220003).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1134921/full#supplementary-material
SUPPLEMENTARY FIGURE 1 | The LAMP assay at different temperatures. The negative control was performed at 63°C for 60 min.
SUPPLEMENTARY FIGURE 2 | The LAMP assay at different times. The negative control was performed at 63°C for 60 min.
Aberg, A., Witzell, J., Ronnberg, J. (2016). Risk of false positives during sampling for heterobasidion annosum s.l. Plant Dis. 100 (1), 175–179. doi: 10.1094/PDIS-03-15-0269-RE
Aza, A., Kangas, A., Gobakken, T., Kallio, A. M. I. (2021). Effect of root and butt rot uncertainty on optimal harvest schedules and expected incomes at the stand level. Ann. For. Sci. 78 (3). doi: 10.1007/s13595-021-01072-1
Bendz-Hellgren, M., Brandtberg, P. O., Johansson, M. (1999). Growth rate of Heterobasidion annosum in Picea abies established on forest land and arable land. Scandinavian J. For. Res. 14 (5), 402–407. doi: 10.1080/02827589950154104
Caten, C. E., Jinks, J. L. (1968). Spontaneous variability of single isolates of Phytophthora infestans. i. cultural variation. Can. J. Bot. 46, 329–348. doi: 10.1139/b68-055
Chen, J. J. (2015). Taxonomy and phylogeny of Wrightoporia and related genera (Beijing: Beijing Forestry University).
Chen, J. J., Cui, B. K., He, S. H., Cooper, J. A., Barrett, M. D., Chen, J. L., et al. (2016). Molecular phylogeny and global diversity of the remarkable genus Bondarzewia (Basidiomycota, russulales). Mycologia 108, 697–708. doi: 10.3852/14-216
Chen, J. J., Cui, B. K., Zhou, L. W., Dai, Y. C. (2015). Phylogeny, divergence time estimation, and biogeography of the genus Heterobasidion (Basidiomycota, russulales). Fungal Diversity 71 (1), 185–200. doi: 10.1007/s13225-014-0317-2
Chen, J. J., Korhonen, K., Li, W., Dai, Y. C. (2014). Two new species of the Heterobasidion insulare complex based on morphology and molecular data. Mycoscience 55 (4), 289–298. doi: 10.1016/j.myc.2013.11.002
Dai, Y. C., Fan, L. F., Chen, J. J., Wu, C. P., Wu, Y. D., Yuan, Y. (2021a). Species diversity of conifer pathogen Heterobasidion and related quarantine suggestions. Mycosystema 40 (8), 1958–1964. doi: 10.13346/j.mycosystema.210094
Dai, Y. C., Korhonen, K. (1999). Heterobasidion annosum Group s identified in north-eastern China. Eur. J. For. Pathol. 29 (4), 273–279. doi: 10.1046/j.1439-0329.1999.00153.x
Dai, Y. C., Vainio, E. J., Hantula, J., Niemelä, T., Korhonen, K. (2002). Sexuality and intersterility within the Heterobasidion insulare complex. Mycological Res. 106 (12), 1435–1448. doi: 10.1017/S0953756202006950
Dai, Y. C., Vainio, E. J., Hantula, J., Niemelä, T., Korhonen, K. (2003). Investigations on heterobasidion annosum s. lat. in central and eastern Asia with the aid of mating tests and DNA fingerprinting. For. Pathol. 33 (5), 269–286. doi: 10.1046/j.1439-0329.2003.00328.x
Dai, Y. C., Yang, Z. L., Cui, B. K., Wu, G., Yuan, H. S., Zhou, L. W., et al. (2021b). Diversity and systematics of the important macrofungi in Chinese forests. Mycosystema 40 (4), 770–805. doi: 10.13346/j.mycosystema.210036
Duan, Y. B., Ge, C. Y., Zhang, X. K., Wang, J. X., Zhou, M. G. (2014). A rapid detection method for the plant pathogen sclerotinia sclerotiorum based on loop-mediated isothermal amplification (LAMP). Australas. Plant Pathol. 43 (1), 61–66. doi: 10.1007/s13313-013-0239-6
Edmonds, R. L., Shaw, D. C., Hsiang, T., Driver, C. H. (1989). Impact of precommercial thinning on development of Heterobasidion annosum in Western hemlock. USDA For. Service Gen. Tech. Rep., 85–94.
Enicks, D. A., Bomberger, R. A., Amiri, A. (2020). Development of a portable LAMP assay for detection of Neofabraea perennans in commercial apple fruit. Plant Dis. 104 (9), 2346–2353. doi: 10.1094/PDIS-09-19-2036-RE
Fabritius, A. L., Karjalainen, R. (1993). Variation in Heterobasidion annosum detected by random amplified polymorphic DNAs. Eur. J. For. Pathol. 23 (4), 193–200. doi: 10.1111/j.1439-0329.1993.tb01338.x
Gams, W., Hoekstra, E. S., Aptroot, A. (1998). CBS Course on mycology (Monterey, CA, U.S: Centraalbureau voor Schimmelcultures, AG Baarn, the Netherlands).
Garbelotto, M., Gonthier, P. (2013). Biology, epidemiology, and control of Heterobasidion species worldwide. Annu. Rev. Phytopathol. 51 (1), 39–59. doi: 10.1146/annurev-phyto-082712-102225
Goto, M., Honda, E., Ogura, A., Nomoto, A., Hanaki, K.-I. (2009). Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. BioTechniques 46 (3), 167–172. doi: 10.2144/000113072
Greig, B. J. W. (1998). Field recognition and diagnosis of heterobasidion annosum. in heterobasidion annosum, biology, ecology, impact and control. Eds. Woodward, S., Stenlid, J., Karjalainen, R., Hüttermann, A. (Wallingford: CAB International), 35–41.
Greig, B. J. W., Pratt, J. E. (1976). Some observations on the longevity of Fomes annosus in conifer stumps. Eur. J. For. Pathol. 6 (4), 250–253. doi: 10.1111/j.1439-0329.1976.tb00533.x
Han, B. (2021). Detailed explanation of china's timber import trends. Construction Sci. Technol. 440 (20), 17–21. doi: 10.16116/j.cnki.jskj.2021.20.003
Hietala, A., Eikenes, M., Kvaalen, H., Solheim, H., Fossdal, C. G. (2003). Multiplex real-time PCR for monitoring Heterobasidion annosum colonization in Norway spruce clones that differ in disease resistance. Appl. Environ. Microbiol. 69 (8), 4413–4420. doi: 10.1128/AEM.69.8.4413–4420.2003
Ioos, R., Chrétien, P., Perrault, J., Jeandel, C., Dutech, C., Gonthier, P., et al. (2019). Multiplex real-time PCR assays for the detection and identification of Heterobasidion species attacking conifers in Europe. Plant Pathol. 68, 1493–1507. doi: 10.1111/ppa.13071
Johannesson, H., Stenlid, J. (2003). Molecular markers reveal genetic isolation and phylogeography of the s and f intersterility group of the wood-decay fungus Heterobasidion annosum. Mol. Phylogenet. Evol. 29 (1), 94–101. doi: 10.1016/S1055-7903(03)00087-3
Johannesson, S. H., Johannesson, K. H. P., Stenlid, J. (2000). Development of primer sets to amplify fragments of conserved genes for systematic and population studies in the genus Daldinia. Mol. Ecol. 9, 375–378. doi: 10.1046/j.1365-294x.2000.00874-6.x
Kasuga, T., Mitchelson, K. (1993). Determination of the DNA sequence of the 5.8S ribosomal gene of Heterobasidion annosum and Heterobasidion araucariae. Nucleic Acids Res. 21 (5), 1320. doi: 10.1093/nar/21.5.1320
Kasuga, T., Woods, C., Woodward, S., Mitchelson, K. (1993). Heterobasidion annosum 5.8S ribosomal DNA and internal transcribed spacer sequence: rapid identification of European intersterility groups by ribosomal DNA restriction polymorphism. Curr. Genet. 24 (5), 433–436. doi: 10.1007/BF00351853
Kong, L., Wang, H. B., Wang, S. S., Xu, P. P., Zhang, R. F., et al. (2020). Rapid detection of potato late blight using a loop-mediated isothermal amplification assay. J. Integr. Agric. 19 (5), 1274–1282. doi: 10.1016/S2095-3119(19)62816-9
Korhonen, K. (1978). Intersterility groups of Heterobasidion annosum. Communicationes Instituti Forestalis Fenniae 94, 1–25.
Korhonen, K., Stenlid, J. (1998). “Biology of heterobasidion annosum,” in Heterobasidion annosum: biology, ecology, impact and control. Eds. Woodward, S., Stenlid, J., Karjalainen, R., Hüttermann, A. (Wallingford: CAB International), pp 43–pp 70.
Li, X. C. (2014). Biological control on heterobasidion parviporum and its decay in China (Beijing: Beijing Forestry University).
Linzer, R. E., Otrosina, W. J., Gonthier, P., Bruhn, J., Laflamme, G., Bussières, G., et al. (2008). Inferences on the phylogeography of the fungal pathogen Heterobasidion annosum, including evidence of interspecific horizontal genetic transfer and of human-mediated, long-range dispersal. Mol. Phylogenet. Evol. 46 (3), 844–862. doi: 10.1016/j.ympev.2007.12.010
Liu, S. L., Zhao, Y., Dai, Y. C., Nakasone, K., He, S. H. (2017). Phylogeny and taxonomy of Echinodontium and related genera. Mycologia 109, 1–10. doi: 10.1080/00275514.2017.1369830
Mitchelson, K., Korhonen, K. (1998). “Diagnosis and differentiation of intersterility groups,” in Heterobasidion annosum, biology, ecology, impact and control. Eds. Woodward, S., Stenlid, J., Karjalainen, R., Hüttermann, A. (Wallingford: CAB International).
Mori, Y., Nagamine, K., Tomita, N., Notomi, T. (2001). Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289 (1), 150–154. doi: 10.1006/bbrc.2001.5921
Morrison, D. J., Johnson, A. L. S. (1999). Incidence of Heterobasidion annosum in precommercial thinning stumps in coastal British Columbia. Eur. J. For. Pathol. 29 (1), 1–16. doi: 10.1046/j.1439-0329.1999.00126.x
Niemelä, T., Korhonen, K. (1998). “Taxonomy of the genus Heterobasidion,” in Heterobasidion annosum: biology, ecology, impact and control. Eds. Woodward, S., Stenlid, J., Karjalainen, R., Hüttermann, A. (Wallingford: CAB International), 27–33.
Niu, J. H., Jian, H., Guo, Q., Chen, C. L., Wang, X., Liu, Q., et al. (2012). Evaluation of loop-mediated isothermal amplification (LAMP) assays based on 5S rDNA-IGS2 regions for detecting Meloidogyne enterolobii. Plant Pathol. 61 (4), 809–819. doi: 10.1111/j.1365-3059.2011.02562.x
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28, 63–64. doi: 10.1093/nar/28.12.e63
Oliva, J., Mandy, M., Wendt, L., Elfstrand, M. (2017). Quantitative interactions between the biocontrol fungus Phlebiopsis gigantea, the forest pathogen Heterobasidion annosum and the fungal community inhabiting Norway spruce stumps. For. Ecol. Manage. 402 (10), 253–264. doi: 10.1016/j.foreco.2017.07.046
Oliva, J., Samils, N., Johansson, U., Bendz-Hellgren, M., Stenlid, J. (2008). Urea treatment reduced heterobasidion annosum s.l. root rot in Picea abies after 15 years. For. Ecol. Manage. 255 (7), 2876–2882. doi: 10.1016/j.foreco.2008.01.063
Omdal, D. W., Shaw, C. G., Jacobi, W. R. (2004). Symptom expression in conifers infected with Armillaria ostoyae and Heterobasidion annosum. Can. J. For. Res. 34 (6), 1210–1219. doi: 10.1139/X04-007
Franco Ortega, S., Bustos Lopez, M., Nari, L., Boonham, N., Gullino, M. L., Spadaro, D. (2019). Rapid detection of Monilinia fructicola and Monilinia laxa on peaches and nectarines using loop-mediated isothermal amplification. Plant Dis. 103 (9), 2305–2314. doi: 10.1094/PDIS-01-19-0035-RE
Parida, M., Horioke, K., Ishida, H., Dash, P. K., Saxena, P., Jana, A., et al. (2005). Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. J. Clin. Microbiol. 43 (6), 2895–2903. doi: 10.1128/JCM.43.6.2895–2903.2005
Parida, M. M., Santhosh, S. R., Dash, P. K., Tripathi, N. K., Lakshmi, V., Mamidi, N., et al. (2007). Rapid and real-time detection of chikungunya virus by reverse transcription loop-mediated isothermal amplification assay. J. Clin. Microbiol. 45 (2), 351–357. doi: 10.1128/JCM.01734-06
Pellicciaro, M., Lione, G., Ongaro, S., Gonthier, P. (2021). Comparative efficacy of state-of-the-art and new biological stump treatments in forests infested by the native and the alien invasive Heterobasidion species present in Europe. Pathogens 10, 1272. doi: 10.3390/pathogens10101272
Petersen, M., Ma, L. Y., Lu, X. N. (2021). Rapid determination of viable but non-culturable Campylobacter jejuni in food products by loop-mediated isothermal amplification coupling propidium monoazide treatment. Int. J. Food Microbiol. 351, 109263. doi: 10.1016/j.ijfoodmicro.2021.109263
Rishbeth, J. (1951). Observations on the biology of Fomes annosus, with particular reference to east anglian pine plantations: III. natural and experimental infection of pines, and some factors affecting severity of the disease. Ann. Bot. 15, 221–246. doi: 10.1093/oxfordjournals.aob.a083278
Santiago, T. D. (2021). Portable and label-free quantitative loop-mediated isothermal amplification (LF-qLamp) for reliable COVID-19 diagnostics in three minutes of reaction time: arduino-based detection system assisted by a pH microelectrode. Biosensors 11 (10), 386. doi: 10.3390/bios11100386
Schulze, S. (1999). Rapid detection of European Heterobasidion annosum intersterility groups and intergroup gene flow using taxon-specific competitive-priming PCR (TSCP-PCR). J. Phytopathol. 147 (2), 125–127. doi: 10.1046/j.1439-0434.1999.147002125.x
Seifert, T. (2007). Simulating the extent of decay caused by Heterobasidion annosum s. l. in stems of Norway spruce. For. Ecol. Manage. 248 (1–2), 95–106. doi: 10.1016/j.foreco.2007.02.036
Shamoun, S. F., Hammett, C., Sumampong, G., Li, X., Garbelotto, M. (2019). New taxon-specific Heterobasidion PCR primers detect and differentiate north American heterobasidion spp. in various substrates and led to the discovery of Heterobasidion irregulare in British Columbia, Canada. Pathogens 8 (3), 156. doi: 10.3390/pathogens8030156
Shaw, D. C., Edmonds, R. L., Littke, R. W., Browning, J. E., Russel, K. W. (1995). Incidence of wetwood and decay in precommercially thinned western hemlock stands. Can. J. For. Res. 25, 1269–1277. doi: 10.1139/x95-140
Sillo, F., Giordano, L., Gonthier, P. (2017). Fast and specific detection of the invasive forest pathogen Heterobasidion irregulare through a loop-mediated isothermal AMPlification (LAMP) assay. For. Pathol. 48 (2), e12396. doi: 10.1111/efp.12396
Swedjemark, G., Stenlid, J. (1993). Population dynamics of the root rot fungus Heterobasidion annosum following thinning of Picea abies. Oikos 66 (2), 247–254. doi: 10.2307/3544811
Tokuda, S., Hattori, T., Dai, Y. C., Ota, Y., Ota, Y. (2009). Three species of Heterobasidion (Basidiomycota, hericiales), H. parviporum, h. orientale sp. nov. and H. ecrustosum sp. nov. from East Asia. Mycoscience 50 (3), 190–202. doi: 10.1007/s10267-008-0476-7
Vettraino, A. M., Luchi, N., Rizzo, D., Pepori, A. L., Pecori, F., Santini, A. (2021). Rapid diagnostics for Gnomoniopsis smithogilvyi (syn. Gnomoniopsis castaneae) in chestnut nuts: new challenges by using LAMP and real-time PCR methods. AMB Express 11 (1), 1–11. doi: 10.1186/s13568-021-01266-w
Wallis, G. W. (1962). Survey of Fomes annosus in East anglian pine plantations. Forestry 33, 203–214. doi: 10.1093/forestry/33.2.203
White, T. J., Bruns, T., Lee, S., Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR protocols: a guide to methods and applications. Eds. Innis, M. A., Gelfand, D. H., Sninsky, J. J., White, T. J. (San Diego: Academic Press), 315–322.
Woodward, S., Stenlid, J., Karjalainen, R., Hüttermann, A. (1998). “Preface,” in Heterobasidion annosum: biology, ecology, impact and control. Eds. Woodward, S., Stenlid, J., Karjalainen, R., Huttermann, A. (Wallingford: CAB International), 1–589.
Wu, F., Man, X. W., Tohtirjap, A., Dai, Y. C. (2022). A comparison of polypore funga and species composition in forest ecosystems of China, north America, and Europe. For. Ecosyst. 4, 540–546. doi: 10.1016/j.fecs.2022.100051
Xu, Y. L., Tian, Y., He, S. H. (2023). Taxonomy and phylogeny of Peniophora sensu lato (Russulales, basidiomycota). J. Fungi 9, 93. doi: 10.3390/jof9010093
Yuan, Y., Chen, J. J., Korhonen, K., Francis, M., Dai, Y. C. (2021). An updated global species diversity and phylogeny in the forest pathogenic genus Heterobasidion (Basidiomycota, russulales). Front. Microbiol. 11. doi: 10.3389/fmicb.2020.596393
Keywords: Heterobasidion annosum, pathogens of Pinus, LAMP assay, molecular diagnosis, port quarantine
Citation: Hong-min Z, Jian Y, Ying L, Yuan Y, Cui-ping W, Yu-cheng D and Jia-jia C (2023) Rapid detection of Heterobasidion annosum using a loop-mediated isothermal amplification assay. Front. Cell. Infect. Microbiol. 13:1134921. doi: 10.3389/fcimb.2023.1134921
Received: 31 December 2022; Accepted: 11 April 2023;
Published: 28 April 2023.
Edited by:
Jianping Xu, McMaster University, CanadaReviewed by:
Banu Metin, Istanbul Sabahattin Zaim University, TürkiyeCopyright © 2023 Hong-min, Jian, Ying, Yuan, Cui-ping, Yu-cheng and Jia-jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dai Yu-cheng, eXVjaGVuZ2RAeWFob28uY29t; Chen Jia-jia, amlhamlhY2hlbkBqc2FmYy5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.