- 1Applied Microbial Processes and Environmental Health Research Group, Faculty of Life Sciences, University of Benin, Benin City, Nigeria
- 2Stellenosch Institute for Advanced Study (STIAS), Wallenberg Research Centre at Stellenbosch University, Stellenbosch, South Africa
- 3Department of Microbiology, College of Natural and Applied Sciences, Western Delta University, Oghara, Nigeria
- 4Department of Environmental Management and Toxicology, Faculty of Life Sciences, University of Benin, Benin City, Nigeria
- 5South African Medical Research Council (SAMRC) Microbial Water Quality Monitoring Centre, University of Fort Hare, Alice, Eastern Cape, South Africa
- 6Department of Microbiology, University of Medical Sciences, Ondo City, Ondo, Nigeria
- 7Department of Environmental Health Sciences, College of Health Sciences, University of Sharjah, Sharjah, United Arab Emirates
Introduction: Staphylococcus aureus causes staphylococcal food poisoning and several difficult-to-treat infections. The occurrence and dissemination of methicillin-resistance S. aureus (MRSA) in Nigeria is crucial and well documented in hospitals. However, findings on MRSA from meat in the country are yet to be adequately reported. The current study determined the prevalence, virulence profile and antibiogram characteristics of MRSA from a raw chicken product from retail outlets within Edo.
Methods: A total of 368 poultry meat samples were assessed for MRSA using a standard culture-based approach and characterized further using a molecular method. The antimicrobial susceptibility profile of the isolates was determined using the disc diffusion method. The biofilm profile of the isolates was assayed via the crystal violet microtitre-plate method. Virulence and antimicrobial resistance genes were screened using polymerase chain reaction via specific primers.
Results: Of the samples tested, 110 (29.9%) were positive for MRSA. All the isolates were positive for deoxyribonuclease (DNase), coagulase and beta-hemolysis production. Biofilm profile revealed 27 (24.55%) weak biofilm formers, 18 (16.36%) moderate biofilm formers, and 39 (35.45%) strong biofilm formers. The isolates harboured 2 and ≤17 virulence genes. Enterotoxin gene profiling revealed that 100 (90.9%) isolates harboured one or more genes. Resistance against the tested antibiotics followed the order: tetracycline 64(58.2%), ciprofloxacin 71(64.6%), trimethoprim 71(64.6%) and rifampin 103(93.6%). A total of 89 isolates were multidrug-resistant, while 3 isolates were resistant to all 22 antibiotics tested. The isolates harboured antimicrobial-resistant determinants such as methicillin-resistant gene (mecA), tetracycline resistance genes (tetK, tetL), erythromycin resistance genes (ermA, ermC), trimethoprim resistance gene (dfrK). All the staphylococcal cassette chromosome mec (SCCmec) IVa and SCCmec V positive isolates harboured the Panton-Valentine Leukocidin Gene (PVL).
Conclusion: In conclusion, S. aureus was resistant to commonly used antibiotics; a concern to public health concerning the transmission of these pathogens after consuming these highlight the significance of antimicrobial and enterotoxigenic monitoring of S. aureus in food chains.
1 Introduction
The commensal association of S. aureus with food animals promotes antimicrobial resistance (Zehra et al., 2019). Staphylococcus aureus (S. aureus) has been reported to acquire resistance to antibiotics of choice, such as vancomycin (Tegegne et al., 2021). It has been listed as a ‘priority pathogens’ threatening public health by the World Health Organization (WHO) (WHO, 2017). Methicillin-resistance S. aureus (MRSA) is a significant aetiology of massive healthcare-associated MRSA (HA-MRSA) infections leading to antibiotic resistance crises worldwide (Abolghait et al., 2020). MRSA is of concern since it can cause difficult-to-treat fatal infections. Although there has been a decline in invasive HA-MRSA infections over recent years, community-associated MRSA (CA-MRSA) infections have heightened among the general populace. The molecular mechanism for developing oxacillin/methicillin resistance is inserting and acquiring staphylococcal chromosome cassette mec (SCCmec) determinants, which house antimicrobial resistance genes. Differences exist in the SCCmec types of S aureus strains, which may increase by independently acquiring the mec gene. Furthermore, HA-MRSA, livestock-associated MRSA (LA-MRSA), and CA-MRSA can contaminate human foods, causing cases of staphylococcal food poisoning (SFP) (Sergelidis and Angelidis, 2017). S. aureus food-borne disease outbreaks have been reported recently (Le et al., 2021).
Staphylococcal food poisoning (SFP) is associated with emetic activity, sepsis-related infections, pneumonia, and toxic shock syndrome (TSS) (Fisher et al., 2018). Upon contamination of food, food triggers enterotoxins production in S. aureus, which may persist in foodstuffs after heat decimation of the bacteria, thus causing SFP (Sergelidis and Angelidis, 2017). Staphylococcal enterotoxins (SEs) comprise a superfamily of ≥23 low-molecular-weight pyrogenic exotoxins that share functional and structural similarities. Staphylococcal enterotoxins can be classified into two categories depending on their ability to invoke emesis: newly confirmed enterotoxigenic-like proteins and classical SEs (A to E). Staphylococcal enterotoxins possess potent super antigenic activity and disrupt adaptive immunity by stimulating T cells, producing inflammatory cytokines (Fisher et al., 2018). The SE-encoding elements are mainly on different mobile genetic elements (MGEs), which can extensively result in prevalence variations of SEs among S. aureus isolates. In addition, SEs are controlled by various overlapping regulatory pathways that can be multiple by various environmental factors (Fisher et al., 2018).
The ingestion of 20–100 ng performed SEs in food caused SFP (Abolghait et al., 2020; Beshiru et al., 2022). The production of SEs essentially occurred in protein-rich foods, such as meat, after the growth of enterotoxigenic S. aureus strains at high cell densities under optimal temperatures and environmental conditions (Schelin et al., 2011; Igbinosa et al., 2020). Staphylococcal enterotoxins retain activity in S. aureus-contaminated food due to their high heat stability, protein denaturation tolerance, proteolytic enzyme activity resistance, freezing, drying, and low pH conditions (Abolghait et al., 2020). Additionally, they are acid-stable and retain their activity in the digestive tract (Fisher et al., 2018). Chicken products can be a potential reservoir of MRSA infections of zoonotic origin. Chicken meat not kept at refrigeration temperature can be contaminated with SEs-producing MRSA, which may create a health hazard. Consuming or handling contaminated food could result in the spread to humans (Igbinosa et al., 2021a; Beshiru et al., 2022). Sequel to concerns from the Nigerian government regarding food safety, significant proportions of data concerning the characteristics and Prevalence of MRSA, including SEs-producing MRSA in retail foods in Nigeria, is essential.
S. aureus is biofilm-producing bacteria and can perpetuate its contamination on contact surfaces in meat processing (Igbinosa et al., 2020; Abbasi et al., 2021; Beshiru et al., 2021) and could increase bacterial resistance and spread in farm animals, favor their persistence in the environment, and improve their survival in meat products. Some S. aureus lineages have adapted to chicken (Lowdera et al., 2009; Murray et al., 2017) and harbor multiple virulence and antimicrobial resistance (AMR) genes. Biofilm attachment involves bacterial surface component sensing adhesive and matrix molecules, whereas biofilm maturation consists of the expression of intercellular adhesion gene cluster (icaABCD) operon-encoded polysaccharide adhesion molecules (Nemati et al., 2009; Bernier-Lachance et al., 2020). The importance of biofilm-forming MRSA in foods of animal origins is not adequately documented in Nigeria and other developing countries. Thus, the present study was designed to assay the occurrence and distribution of biofilm-associated determinants, virulence, and resistance elements in MRSA recovered from chicken carcasses in Nigeria.
The Prevalence of MRSA varies in different food, animals, and country of origin (Lim et al., 2010; Wu et al., 2019). In Nigeria, there is sparse information on the prevalence of MSRA in food and food animals, with few unrelated studies reporting MRSA infection rates (Beshiru et al., 2016; Igbinosa et al., 2016a; Igbinosa et al., 2016b; Igbinosa and Beshiru, 2019; Abolghait et al., 2020; Beshiru et al., 2021). S. aureus and MRSA have been recovered from poultry birds, frozen fish, humans, environmental samples, milk, pork, beef, ready-to-eat food, livestock, dressed chicken, pet, and stray dogs in Nigeria (Igbinosa et al., 2016a; Igbinosa et al., 2016b; Beshiru et al., 2021). However, reports on the molecular characterization of MRSA from chicken are scarce. A better understanding of the epidemiology and identifying the genetic profile of MRSA is crucial in developing preventive mechanisms against infectious of its origin. Likewise, understanding the genetics of MRSA circulating in different milieus is vital for its evolutional tracking in various niches. Hence, this study aimed to determine the antibiotic resistance, prevalence, enterotoxigenic, and other genetic profiles of MRSA in retail chicken meat sold at open markets in Nigeria.
2 Materials and methods
2.1 Sample collection
Frozen chicken carcass samples were obtained from cold rooms operating on a large scale from open markets in Edo, Nigeria. The selection of the markets was based on their strategic location in conjunction with consumer patronage. The cold rooms selected usually distribute chicken carcasses at a cheaper wholesale rate to retail traders who re-sell to direct consumers in their local markets. The sample size was determined via the sample size determination formula:
N = Number of expected samples; Z1-α/2 = Standard normal variant at 5% type I error (P< 0.05); P = prevalence expected based on previous studies [1.3% (Bernier-Lachance et al., 2020), 9.89% (Baghbaderani et al., 2020), 13.9% (Abbasi et al., 2021), 20.5 (Li et al., 2019), 29.1% (Rortana et al., 2021), 35.4% (Tegegne et al., 2021), 66.67% (Savariraj et al., 2020), 89.5% (Lika et al., 2021)]; d = Complete precision or error (which is 5%). Thus, the expected size of the sample was ≥351. The markets sampled include Ikpoba Hill (n=30) [6.3496° N, 5.6609° E], New Benin (n=32) [6.3448° N, 5.6340° E], Oba (n=32) [6.3348° N, 5.6201° E], Santana (n=30) [6.2915° N, 5.6325° E], Aduwawa (n=32) [6.3688° N, 5.6849° E], Uselu (n=31) [6.3744° N, 5.6134° E], Ileha (n=30) [6.3460° N, 5.6097° E], Ekiosa (n=31) [6.3231° N, 5.6363° E], Oka (n=30) [6.2905° N, 5.6623° E], Egor (n=30) [6.3642° N, 5.6090° E], Oregbeni (n=30) [6.3501° N, 5.6592° E], and Ugbor (n=30) [6.2629° N, 5.6063° E] markets. The sampling was conducted from June 2018 to April 2019. Seasonal durations in Nigeria include a wet season characterized by heavy rainfall with a temperature reaching 35 ± 2°C (March to September) and a dry season characterized by low to no rainfall with a temperature reaching 39 ± 2°C (November to February). A total of 368 frozen fresh chicken meat samples were collected. In all cases, 50 g of samples were collected into a plastic tube from retail outlets within Benin City and transported on ice to the laboratory (Applied Microbial Processes & Environmental Health Research Laboratory, University of Benin, Nigeria).
2.2 MRSA isolation
Twenty-five grams of the sample was inoculated into 225 ml trypticase soy broth (Merck, Darmstadt, Germany), incubated at 37 °C for 24 h, and subcultured via streaking onto MRSA selective agar plate (CHROMagar™ MRSA-ITK Diagnostics BV, Netherlands) and incubated at 37°C for 24 h. Rose to mauve colonies on MRSA selective agar plates were presumptive MRSA isolates. The isolates were identified based on cultural, morphological, and biochemical tests such as Gram-reactions, 3% potassium hydroxide (3% KOH), catalase, coagulase, β-haemolysis, DNAse activity, anaerobic utilization of glucose and mannitol (Tallent et al., 2019). One colony per plate was purified in nutrient agar (Lab M, Lancashire, United Kingdom), further incubated for 18 h at 37°C, and preserved on nutrient agar slants at 4°C. The positive control used includes S. aureus (ATCC 12600).
2.3 Phenotypic confirmation of MRSA isolates
Phenotypic detection of MRSA was performed using cefoxitin disk assay (CLSI, 2020). For individual isolates, colonies of isolated S. aureus from an overnight-grown culture were transferred into the nutrient broth. S. aureus suspensions at a 0.5 McFarland standard equivalent density in nutrient broth were spread onto Mueller–Hinton agar (Lab M, Lancashire, UK) plate in duplicate with cefoxitin (30 µg), methicillin (5 µg), cloxacillin (5 µg), and oxacillin (1 µg) disc (Mast Diagnostics, UK). The plates were incubated at 37°C for 24 h. Isolates identified with cefoxitin resistance (≤ 21 mm zone diameter) were categorized as MRSA.
2.4 PCR detection and characterization of the MRSA isolates
The genomic DNA of the MRSA isolates and positive S. aureus (S. aureus ATCC 12600) control were extracted using the DNA MiniPrep kit following the manufacturer’s instructions. The polymerase chain reaction (PCR) uses to identify Staphylococcus aureus using specific primers (Supplementary Table 1) and the PCR reaction previously described (Brakstad et al., 1992) by targeting the nuc gene. AMR genes such as methicillin resistance (mecA), trimethoprim (dfrG, dfrK, dfrD), aminoglycosides [ant(4´)-Ia, aac(6´)-Ie-aph(2´´)-Ia, aph(3´)-IIIa], chloramphenicol (cat::pC194, cat::pC223, cat::pC221), erythromycins (ermA, ermB, ermC), tetracyclines (tetO, tetM, tetL, tetK) and beta-lactamase (BlaZ); and virulence genes such as intercellular adhesion protein (icaA, icaB, icaC, icaD), toxic shock syndrome toxin 1 (tsst-1), exfoliative toxin (eta, etb), enterotoxins (sea to seu), haemolysins (hla, hlb), Panton-Valentine Leucocidin (PVL), staphylococci protein A (spa), and coagulase (coa) were amplified as described previously (Jackson and Landolo, 1986; Dale et al., 1995; Frenay et al., 1996; Monday and Bohach, 1999; Martineau et al., 2000; Fueyo et al., 2005). SCCmec I to V and subtype SCCmec (IVa to d) of the MRSA isolates was carried out using PCR as described previously (Okuma et al., 2002; Ma et al., 2005; Zhang et al., 2005) using specific primer sets in Supplementary Table 1. PCR products were performed in a 1% agarose gel electrophoresis for 45 min at 110 V, viewed after staining with ethidium bromide in a transilluminator (Vilber Lourmat, EBOX VX5, France).
2.5 Antimicrobial susceptibility testing
The antibiogram profiling of the MRSA isolates was conducted using the Kirby-Bauer disc diffusion procedure. Antibiotics used includes Penicillin G (10 units), ceftaroline (30µg), gentamicin (10µg), amikacin (30µg), kanamycin (30µg), azithromycin (15µg), clarithromycin (15µg), erythromycin (15µg), doxycycline (30µg), minocycline (30µg), tetracycline (30µg), ciprofloxacin (5µg), levofloxacin (5µg), moxifloxacin (5µg), nitrofurantoin (300µg), clindamycin (2µg), trimethoprim-sulfamethoxazole (1.25/23.75µg), trimethoprim (5µg), chloramphenicol (30µg), sulfonamides (300µg), linezolid (30µg) and rifampin (5µg). The interpretation of the zone of inhibitions of the isolates as resistance, intermediate, or susceptibility was based on the Clinical and Laboratory Standard Institute’s interpretative chart (CLSI, 2020) to determine the sensitivity, intermediate and resistance profiles of the isolates to the antibiotics used. For vancomycin (1-32 µg/mL), oritavancin (0.12-0.50 µg/mL), teicoplanin (4-64 µg/mL), daptomycin (0.5-4 µg/mL), and tedizolid (0.25-4 µg/mL) antibiotics, the minimum inhibitory concentration (MIC) procedure was adopted, and data were interpreted based on interpretive categories and MIC breakpoints, µg/mL (CLSI, 2020). Multidrug resistance and multiple antibiotic resistance index were determined as described elsewhere (Igbinosa et al., 2022).
2.6 Biofilm formation profile of the MRSA isolates
Biofilm formation assay was carried out by suspending pure MRSA colonies in 4.5 mL tryptone soy broth (TSB) and incubating for 18 h at 37°C. After that, the cells were harvested at 12,000 rpm for 2 min., washed, re-suspended in phosphate-buffered solution at pH 7.2), and adjusted to 0.5 McFarland standards. A 20 mL of the suspended cell inoculant and 80 mL TSB were introduced into sterile 96-welled polystyrene microtitre plates to assess Staphylococci adherence onto a solid matrix/medium well-contained as described previously (Igbinosa et al., 2022). The negative and positive control was a well-containing TSB broth and S. aureus ATCC 12600, respectively. Each assay was done in independent biological triplicate. Biofilm-producing ability of the isolates was defined as non-producing/negative (ODi< ODc), weak/poor-producing (ODc< ODi<0.1), moderate/intermediate-producing (ODi ¼ 0.1< 0.12), or strong producer (ODi>0.12) as described somewhere else (Igbinosa et al., 2022).
3 Results
3.1 MRSA prevalence from chicken meat
From the 368 samples screened, 110(29.9%) were positive for MRSA via resistance (≤ 21 mm zone diameter) to the cefoxitin disc test. As such, representative isolates from the 110 samples were carefully screened using the S. aureus specific primer (nuc gene), where a total of 110 isolates were detected (Table 1).
3.2 Phenotypic virulence factors of the MRSA isolates
All MRSA isolates from this study were 100% positive for DNase, coagulase, and beta-hemolysis production (Table 1). The biofilm profile of the isolates revealed that 27(24.55%) were weak biofilm producers, 18(16.36%) were moderate biofilm producers, 39(35.45%) were strong biofilm producers, while 26(23.64%) were negative for biofilm formation (Table 2). An overall total of 84(76.36%) of the isolates were biofilm formers (Table 1).
3.3 Occurrence of virulence determinants from the MRSA isolates
The occurrence of virulence genes screened in this study is as follows: coa 110(100%), spa 98(89.1%), hla 110(100%), pvl 50(45.5%), hlb 23(20.95), sea 27(24.6%), seb 16(14.6%), sec 19(17.3%), sed 14(12.7%), see 13(11.8%), seg 39(35.5%), seh 13(11.8%), sei 11(10%), sej 32(29.1%), sek 4(3.6%), sel 4(3.6%), sep 20(18.2%), ser 13(11.8%), tsst 16(14.6%), etb 17(15.5%), eta 17(15.5%), icaA 45(40.9%), icaB 44(40%) (Table 1). The sem, sen, seo, seq, seu, icaD and icaC were not detected. Overall, staphylococci isolates harbored a minimum of 2 virulence genes and a maximum of 17 virulence genes (Table 1). All the isolates that possessed the adherence determinant (icaA and icaB) formed biofilms phenotypically. All the isolates that possessed the hla and hlb genes were β-hemolytic phenotypically on blood agar. All the isolates in the study were coagulase positive via biochemical process and possessed the coa determinant. Enterotoxin gene (18 sea-seu genes) profiling of the isolates revealed that 90.9% (100/110) of the isolates were enterotoxigenic, harboring either one or more genes of the 13 genes that were positive. The chain of enterotoxin gene occurrence was seg>sej>sea>sep>sec>seb>sed> see,she,ser>sei>sek,sel. All the ica gene-carrying isolates were biofilm formers (Table 1). The combined occurrence of virulence genes include: hla+hlb 23(20.9%), eta+etb 17(15.5%), icaA+IcaB 37(33.6%), pvl+hla+hlb 23(20.9%), pvl+tsst-1 16(14.5%) (Table 1).
3.4 Antimicrobial susceptibility profile and determinants of the MRSA isolates
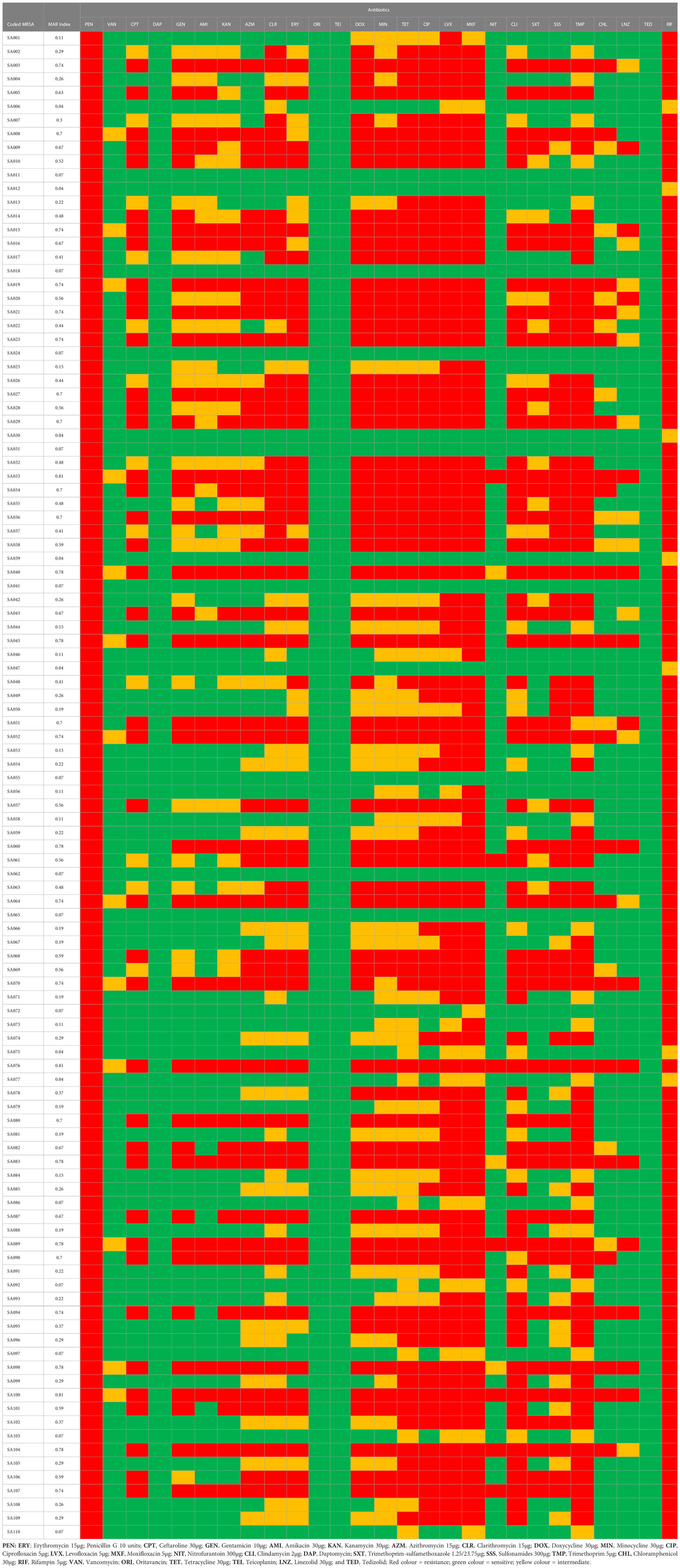
The resistance profile of the S. aureus in Table 2 to the antibiotics tested is as follows: penicillin G 110(100%), clarithromycin 53(48.2%), doxycycline 58(52.7%), minocycline 53(48.2%), tetracycline 64(58.2%), ciprofloxacin 71(64.6%), levofloxacin 84(76.4%), moxifloxacin 88(80%), clindamycin 62(56.4%), sulfonamides 53(48.2%), trimethoprim 71(64.6%), and rifampin 103(93.6%). The sensitivity profile of the isolates in Table 3 is as follows: ceftaroline 54(49.1%), gentamicin 51(46.4%), amikacin 64(58.2%), kanamycin 56(50.9%), nitrofurantoin 100(90.9%), trimethoprim-sulfamethoxazole 58(52.7%), chloramphenicol 78(70.9%), and linezolid 83(75.5%). All the isolates were vancomycin, oritavancin, teicoplanin, daptomycin, and tedizolid-sensitive (Table 2).
A total of 89(80.9%) isolates were multidrug resistant been resistant to ≥1 antibiotic in ≥3 antimicrobial classes. A total of 3 isolates were resistant to 22/27(81.5%) antibiotics used in this study. All isolates were resistant to ≥ 1 antibiotic, while 89(80.9%) isolates were resistant to ≥ 3 antibiotics (Table 2). Of the isolates, 103(93.6%) were resistant to ≥2 antibiotics. MAR index of the isolates ranged from 0.04 – 0.81. Isolates with MAR index >2 were 73(66.4%). The highest resistant phenotype was PENR+CPTR+GENR+AMIR+KANR+AZMR+CLRR+ERYR+DOXR+MINR+ TETR+CIPR+LVXR+MXFR+NITR+CLIR+SXTR+SSSR+TMPR+CHLR+LNZR+RIFR with a MAR index of 0.81. A total of 57 resistance phenotypic patterns were observed amongst all the isolates studied (Table 2).
The antibiotic-resistant genes detected includes: mecA 91/110(82.7%), blaZ 91/110(82.7%), tetK 64/110(58.2%), tetL 23/110(20.9%), ermA 55/110(50%), ermB 9/110(8.2%), ermC 21/110(19.1%), aac(6´)-Ie-aph(2´´)-Ia 37/110(33.6%), ant(4´)-Ia 28/110(25.5%), aph(3´)-IIIa 26/110(23.6%), cat::pC194 20/110(18.2%), cat::pC221 12/110(10.9%), cat::pC223 12/110(10.9%), dfrD 64/110(58.2%), dfrK 22/110(20%), dfrG 38/110(34.5%) (Table 3). Other resistant genes, such as tetM and tetO, were not detected (Table 3). A total of 91(82.7%) isolates harbored ≥1 antimicrobial resistance gene, while 19(17.3%) of the isolates didn’t harbor any antimicrobial resistance gene (Table 3). The combined occurrence of antimicrobial resistance genes includes: mecA+blaZ 91(82.7%), tetK+tetL 23(20.9%), ermA+ermB 9(8.2%), ermA+ermB+ermC 3(2.7%), aac(6´)+ ant(4´)+aph(3´) 25(22.7%), aac(6´)+ant(4´) 30(27.3%), cat::pC194+cat::pC221+ cat::pC223 7(6.4%), cat::pC194+ cat::pC221 12(10.9%), dfrD+dfrK+dfrG 17(15.5%), dfrD+dfrK 22(20%), dfrD+dfrG 37(33.6%) (Table 3).
The distribution of SCCmec is as follows: SCCmec III 18(16.4%), SCCmec IVa 33(30%) and SCCmec V 17(15.5%) (Table 3). Other SCCmec screened (SCCmec I, II, IVb, IVc, IVd, and IVh) were not detected (Table 3). The combined occurrence of SCCmec and pvl genes includes SCCmec IVa+pvl 33(30%) and SCCmec V+pvl 17(15.5%). All the SCCmec IVa and SCCmec V isolates also harbored the PVL gene, while all the SCCmec III isolates didn’t harbor the PVL gene (Table 3).
4 Discussion
Methicillin-resistance S. aureus from poultry meat poses a public health risk that can transmit to humans via the handling or consumption of contaminated poultry meat. Lower prevalence rates than those of our study (ranging from 0 – 27%) have been reported previously in North and South America, Europe, and Asia (Tang et al., 2017; Bernier-Lachance et al., 2020). Higher prevalence rates (35.4%) compared to our findings have also been documented in the Czech Republic (Tegegne et al., 2021). Differences in geographical locations, handling practices, sample size variations, seasonal variations, management practices, and methods of the experiment have been reported to cause differences in the prevalence of MRSA (Abbasi et al., 2021). These results highlight the necessity to mitigate the risk of MRSA transmission dynamics via meat products to humans.
Bernier-Lachance et al. (2020) isolates formed biofilm via the microtiter plate assay, which was higher than our study’s biofilm formers (76.36%). Biofilm occurrence meant that the MRSA isolates have adhesive potentials to the host’s extracellular matrix; these likely favor zoonotic potential, persistence, and colonization. Nigeria, where a significant proportion of the population is non-vegetarian, suggests that a large population is t risk of meat-borne hazards. Szczuka et al. (2013) reported that 76% of the biofilm-forming strains had the icaA gene (Yazdani et al., 2006), which was lower than the 100% icaA gene-carrying isolates that formed biofilm in our study. High distribution of biofilm formation genes (icaA, icaB, icaC, icaD) have been demonstrated previously (Nemati et al., 2009; Abbasi et al., 2021), which was different from our study where icaC and icaD were not detected. The icaABCD genes encode intercellular polysaccharide adhesion, which shields S. aureus in difficult environmental circumstances such as immune responses, antimicrobials, and antiseptic agents. Higher detection rates of icaABCD genes have been reported (Abbasi et al., 2021). Biofilm production ability, presence of virulence determinants, and antimicrobial capacity in the genome of MRSA constitute a severe risk to public health.
In addition to infection/colonization, MRSA strains have caused SFP outbreaks (Sergelidis and Angelidis, 2017). Most MRSA isolates by Wu et al. (2019) were MDR and harbored ≥1 SE gene, similar to our findings. Lower profiling of SEs (13 - 83%) has been documented (Kitai et al., 2005; Li et al., 2018; Savariraj et al., 2020; Tegegne et al., 2021) compared to those of our study. The chain of distribution of the enterotoxin genes from our study was seg>sej>sea>sep>sec>seb>sed>see,she,ser> sei>sek,sel. This was different from those from other studies, such as seb>seg>sei>sec>sed>sej (Savariraj et al., 2020). Kitai et al. (2005) reported the distribution of seb>sea>sec>sed. Previous studies have found that sea (Nemati, 2014), seg, and sei (Pu et al., 2011) were the prevalent toxin gene somewhat similar to our study, where seg was the most predominant, followed by sej, sea, and others. In the present study, non-classical enterotoxin genes seg and sej had higher prevalence with the exemption of classical enterotoxin sea. Previously, newly described enterotoxin genes seg, sem, sei, and sen were found among S. aureus isolates with the exemption of classical enterotoxin genes known as major etiological factors in SFP (Hwang et al., 2007). Mosaic structure in the pathogenicity islands can contribute to the shuffling and rearrangement of the enterotoxin genes in S. aureus (Banaszkiewicz et al., 2019).
Savariraj et al. (2020) reported that none of their S. aureus isolates harbored see sea, and seh either alone or in combination, which negates the findings from our study as sea and see were detected independently or in variety. Higher prevalence of seb gene in MRSA and S. aureus in retail chicken carcasses have been reported (Kitai et al., 2005; Abolghait et al., 2020). Among the SEs, seb and sea are the best elucidated. Among well-known bacterial superantigens connected with SFP, asthma, atopic dermatitis, nasal polyps, and toxic shock syndrome in humans, SEB is the most potent (Fries and Varshney, 2013). Abolghait et al. (2020) reported that the isolate positive for the sed gene did not harbor the tsst gene, which negates the finding from our study where some of the isolates carried the sed+tsst gene. No combinations of >1 of the tested SE genes (sea, sec, seb, and sed) were found by Abolghait et al. (2020), which was also different from our findings where there were multiple combinations of SEs genes that could be attributed to the larger SEs gene pool we screened (sea - seu) which aligns with other studies (Titouche et al., 2020). Most of the enterotoxigenic isolates by Abolghait et al. (2020) encoded the etb and tsst-1 genes which were similar to our findings. Wu et al. (2019) reported lower tsst-1 gene detection (3.70%) compared to ours (14.6%). None of the MRSA isolates from previous studies (Bernier-Lachance et al., 2020) harbor genes encoding eta, etb, and tsst-1, which negates our findings as these genes were detected in some of the isolates. The SEs genotypes sea–seg–sei or seg– sei, which also exists from our study, are known to be associated with SFP outbreaks (Kérouanton et al., 2007). The existence of hlb and hla genes in MRSA isolates is crucial for SFP. Ariyanti et al. (2011) showed that the hla and hlb genes were ubiquitous among S. aureus isolated from food animals, similar to our study’s findings.
All MRSA isolates in our study harbored virulence determinants pivotal in toxin production, invasion, adhesion, immune modulation, tissue destruction, leucocyte, and erythrocytes lysis. Enterotoxin genes are borne on MGEs, plasmids (seb), bacteriophages (sea), or pathogenicity islands (sec) (Argudin et al., 2010), which explains their absence or presence in individual isolates either by vertical or acquisition transmission of genes respectively. The virulence genes detected in S. Aureus and MRSA in the current study have been documented in CA-MRSA isolates in humans in hospital settings (Pokhrel et al., 2016). The occurrence of these potential pathogenic MRSA isolates in chicken meat portends their role as a threat to public health. The Prevalence of SEs gene in S. aureus/MRSA varies from country to country and might likely reflect geographical differences and ecological differences in strains’ origins (Li et al., 2018; Abolghait et al., 2020).
Previous studies have reported lower MDR MRSA isolates within the range of 39.17 - 70.2% (Li et al., 2019; Zehra et al., 2019) compared to 80.9% from our study; while the MAR index was reported as 0.23% (Amoako et al., 2019) which is lower compared to most of the isolates from our study. It was reported previously that 97.1% of isolates were resistant to ≥1 antimicrobial agent (Li et al., 2019), which negates our findings as all isolates were resistant to ≥1 antimicrobial. A result from Iran revealed that 96.0% of isolates were resistant to ≥2 antimicrobials (Nemati, 2013), similar to the 93.6% reported in our study. Resistance to penicillin, ciprofloxacin, tetracycline, erythromycin and kanamycin has commonly been reported from meat samples (Li et al., 2019; Zehra et al., 2019). None of the S. aureus strains from previous studies (Tang et al., 2017; Li et al., 2019) was found to be resistant to nitrofurantoin, and less than ten strains were resistant to rifampicin, trimethoprim, chloramphenicol, teicoplanin or gentamicin. This was different from the findings from our study, where the isolates were resistant to the antibiotics mentioned with the exemption of teicoplanin, where no resistance was observed. However, from previous studies, low resistance rates varying from 2–9% were observed for clindamycin, oxacillin chloramphenicol, and ceftriaxone (Zehra et al., 2019), which negates the findings from our study. No vancomycin-resistant isolate was found in this study, similar to Okorie-Kanu et al. (2020).
Approximately 38 unique resistance phenotypic patterns were found among the chicken isolates by Zehra et al. (2019), which was lower than the 57-resistance phenotypic pattern found in our study. Antibiotics such as tetracyclines, fluoroquinolones, macrolides, and sulfonamides are important for human health and are listed by the WHO as critically essential antimicrobials (WHO, 2017). The marked resistance to such antimicrobials is perhaps not surprising since these drugs are inexpensive, orally administered, and are available from diverse sources where they are sold with or without prescription in Nigeria (Okorie-Kanu et al., 2020). The indiscriminate use of these antibiotics in food animal production is a cause for concern culminating in the upsurge of MDR S. aureus (Beshiru et al., 2016; Imanah et al., 2017; El-Ashker et al., 2020; Beshiru et al., 2021). The low levels of biosecurity practices compliance and poor husbandry practices promote indiscriminate use and overdependence of these antibiotics in water and feed as growth promoters and for prophylaxis purposes in poultry farms in Nigeria (Oviasogie et al., 2016; Igbinosa et al., 2021b; Igbinosa et al., 2023).
Similar reports of resistance to fluoroquinolones have been documented from retail meat products in South Africa, Ghana, and Bangladesh (Mkize et al., 2017). Fortunately, MRSA isolates in our study were susceptible to linezolid, vancomycin, daptomycin, tedizolid, teicoplanin, oritavancin, and nitrofurantoin in line with a previous report (Okorie-Kanu et al., 2020). These are the priority and critically essential antibiotics in human medicine (WHO, 2019). The high sensitivity observed could be because these drugs lack veterinary preparations and aren’t routinely used in a clinical setting. The efficient regulation or termination of antibiotic usage in food animals has decreased resistance to zoonotic bacteria in developed countries (Levy, 2014; El-Ashker et al., 2020). There is a need for the urgent execution of appropriate food safety strategies across all decision-makers, policy-makers, and stakeholders in environmental, animal, and human health to address the public health menace of antimicrobial resistance.
Mobile genetic elements (MGEs) carry virulence traits that encode S. aureus accessory genes (tsst-1, eta and etb) (Xia and Wolz, 2014). Therefore, upon acquiring MGEs conferring virulent traits, commensal S. aureus may become a virulent/pathogenic strain pathogen under favorable conditions (Sergelidis and Angelidis, 2017). A total of 82.7% of the cefoxitin-resistant MRSA had the mecA gene from our study. Lower mecA gene occurrence (5.5%) of examined samples has been reported previously with phenotypic MRSA-positive isolates in Egypt (Abolghait et al., 2020). Higher mecA gene occurrence (100%) from cefoxitin-resistant MRSA-positive isolates has been documented in Poland, Oklahoma, and China (Li et al., 2019). Some isolates were cefoxitin-sensitive and lacked mecA or mecC genes but were oxacillin-resistant (Shore and Coleman, 2013). There has also been a report of S. aureus isolates (5.07%) phenotypically resistant to oxacillin from India but genotypically lacking mecA gene (Zehra et al., 2019). Such a pattern may be attributed to β-lactamases’ hyperproduction of and elicitation of the variant of mecA gene and penicillin-binding protein with an altered binding affinity (Laurent et al., 2012).
A significant proportion of the β-lactamase-encoding gene (blaZ) was found among MRSA isolates in the current study in addition to mecA. Phenotypic resistance to gentamicin, tetracycline, and erythromycin was supported by detecting tetK, aphA3 and ermA/ermB/ermC. Zehra et al. (2019) isolated carried resistance genes (blaZ, mecA, aacA-aphD, ermB, ermC, tetK, tetL, and tetM) similar to the findings of our study. All the tested MRSA by Bernier-Lachance et al. (2020) harbored a much higher dfrG gene (which confers resistance to trimethoprim) compared to our research. Abbasi et al. (2021) reported that ≥1 isolate carries one of the following resistance genes blaZ, mecA, tetK, linA, tetM, ermA, ermB, and aacA-D which was much higher than the 82.7% of the isolates from our study. The presence of the tetK and tetM genes similar to our finding explained the tetracycline resistance phenotype observed by Bernier-Lachance et al. (2020).
The SCCmec typing from our study showed that few belonged to the human-associated (HA) clones’ type SCCmec III, with the majority belonging to the CA-MRSA SCCmec IVa, similar to a previous study (Li et al., 2019). All MRSA with SCCmecV also harbored the pvl gene identical to the SCCmecV+pvl variants by Zehra et al. (2019), which are molecular markers for CA-MRSA, indicating their presence in food of animal origin. Our study’s panton-valentine leukocidin (PVL) detection was slightly higher than previously detected by Okorie-Kanu et al. (2020). Kim et al. (2015) reported MRSA-SCCmecV+pvl as the commonest MRSA isolate from meat samples (Kim et al., 2015). SCCmec elements of types IV and V are the commonly found SCCmec types in CA-MRSA (Abdulgader et al., 2015). SCCmec types IVa and V have been documented in HA-MRSA in Nigeria (Ghebremedhin et al., 2009).
5 Conclusion
The MRSA recovered demonstrated MDR potential while harboring potent enterotoxin determinants and other virulence traits that could be detrimental to human health. The MRSA isolates also showed biofilm-forming capacity, which could make them more prone to antimicrobial resistance and persist on biotic and abiotic surfaces. The findings highlight the significance of surveillance studies and the need to continuously monitor the food chain for foods of animal origin for the occurrence and spread of MRSA superbugs. Our findings have revealed that raw chicken meat from Nigeria is the reservoir of MRSA. This study has also raised concerns about MRSA transmission after consuming contaminated chicken products. These findings could proactively assist industries and governments in Nigeria to improve food safety measures and enhance antimicrobial stewardship to curb the spread of critical antimicrobial-resistant pathogens.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
EI and AB, conceptualization. EI, AB, II, AGO, and TE, formal analysis. EI and AIO, funding acquisition. EI, AB, II, AGO, TE, and AIO, investigation. EI, AB, II, and AGO, methodology. EI, AB, II, and AIO, project administration. EI and AIO, resources. EI and AIO, supervision. EI, AB, and II, roles/writing - original draft. EI, AB, II, AGO, TE, and AIO, writing - review and editing. All authors have approved the final version of the manuscript for submission. All authors agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Acknowledgments
We acknowledge Alexander von Humboldt-Stiftung Foundation (AvH) Germany and the South Africa Medical Research Council (SAMRC). AB and EI thank Stellenbosch Institute for Advanced Study (STIAS), South Africa, for the short research stay and the facilities provided in preparing this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1122059/full#supplementary-material
References
Abbasi, K., Tajbakhsh, E., Momtaz, H. (2021). Antimicrobial resistance, virulence genes, and biofilm formation in Staphylococcus aureus strains isolated from meat and meat products. J. Food Saf. 41 (6), e12933. doi: 10.1111/jfs.12933
Abdulgader, S. A., Shittu, A. O., Nicol, M. B., Kabar, M. (2015). Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Africa: A systematic review. Front. Microbiol. 6, 348. doi: 10.3389/fmicb.2015.00348
Abolghait, S. K., Fathi, A. G., Youssef, F. M., Algammal, A. M. (2020). Methicillin-resistant Staphylococcus aureus (MRSA) isolated from chicken meat and giblets often produces staphylococcal enterotoxin b (SEB) in nonrefrigerated raw chicken livers. Int. J. Food Microbiol. 328, 108669. doi: 10.1016/j.ijfoodmicro.2020.108669
Amoako, D. G., Somboro, A. M., Abia, A. L. K., Molechan, C., Perrett, K., Bester, A. L., et al. (2019). Antibiotic resistance in Staphylococcus aureus from poultry and poultry products in uMgungundlovu district, south Africa, using the ‘‘Farm to fork’’ approach. Microb. Drug Res. 26 (4), 402–411. doi: 10.1089/mdr.2019.0201
Argudin, M. A., Mendoza, M. C., Rodicio, M. R. (2010). Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2, 1751–1773. doi: 10.3390/toxins2071751
Ariyanti, D., Salasia, S. I. O., Tato, S. (2011). Characterisation of haemolysin of Staphylococcus aureus isolated from food of animal origin. Ind. J. Biotechnol. 16 (1), 32–37. doi: 10.22146/ijbiotech.7834
Baghbaderani, Z. T., Shakerian, A., Rahimi, E. (2020). Phenotypic and genotypic assessment of antibiotic resistance of Staphylococcus aureus bacteria isolated from retail meat. Infect. Drug Res. 13, 1339–1349. doi: 10.2147/IDR.S241189
Banaszkiewicz, S., Calland, J. K., Mourkas, E., Sheppard, S. K., Pascoe, B., Bania, J. (2019). Genetic diversity of composite enterotoxigenic Staphylococcus epidermidis pathogenicity islands. Genome Biol. Evol. 11 (12), 3498–3509. doi: 10.1093/gbe/evz259
Bernier-Lachance, J., Arsenault, J., Usongo, V., Parent, E´, Labrie, J., Jacques, M., et al. (2020). Prevalence and characteristics of livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) isolated from chicken meat in the province of Quebec, Canada. PloS One 15 (1), e0227183. doi: 10.1371/journal.pone.0227183
Beshiru, A., Igbinosa, I. H., Igbinosa, E. O. (2016). Antimicrobial resistance of methicillin-resistant staphylococci isolated from food-producing animal POSTER presentation at the 17th international congress on infectious diseases in Hyderabad, India, from march 2 to 5, 2016. Int. J. Infect. Dis. 45S, 1–477. doi: 10.1016/j.ijid.2016.02.247
Beshiru, A., Igbinosa, I. H., Igbinosa, E. O. (2021). Characterisation of enterotoxigenic Staphylococcus aureus from ready-to-eat seafood (RTES). LWT Food Sci. Tech. 135, 110042. doi: 10.1016/j.lwt.2020.110042
Beshiru, A., Okoh, A. I., Igbinosa, E. O. (2022). Processed ready-to-eat (RTE) foods sold in yenagoa Nigeria were colonized by diarrheagenic Escherichia coli which constitute a probable hazard to human health. PloS One 17 (4), e0266059. doi: 10.1371/journal.pone.0266059
Brakstad, O. G., Aasbakk, K., Maeland, J. A. (1992). Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 30, 1654–1660. doi: 10.1128/jcm.30.7.1654-1660.1992
Clinical and Laboratory Standards Institute (CLSI) (2020). Performance standards for antimicrobial susceptibility testing; a CLSI supplement for global application (Wayne, PA 19087 USA: Clinical and Laboratory Standards Institute 950 West Valley Road, Suite 2500), 332. CLSI document M02, M07, and M11.
Dale, G. E., Langen, H., Page, M. G., Then, R. L., Stüber, D. (1995). Cloning and characterisation of a novel, plasmid-encoded trimethoprim-resistant dihydrofolate reductase from Staphylococcus haemolyticus MUR313. Antimicrob. Agents Chemother. 39, 1920–1924. doi: 10.1128/AAC.39.9.1920
El-Ashker, M., Gwida, M., Monecke, S., Ehricht, R., Elsayed, M., El-Gohary, F., et al. (2020). Microarray-based detection of resistance genes in coagulase-negative staphylococci isolated from cattle and buffalo with mastitis in Egypt. Trop. Ani Health Prod 52, 3855–3862. doi: 10.1007/s11250-020-02424-1
Fisher, E. L., Otto, M., Cheung, G. Y. C. (2018). Basis of virulence in enterotoxin-mediated staphylococcal food poisoning. Front. Microbiol. 9, 436. doi: 10.3389/fmicb.2018.00436
Frenay, H. M., Bunschote, A. E., Schouls, L. M., van Leeuwen, W. J., Vandenbroucke-Grauls, C. M., Verhoef, J., et al. (1996). Molecular typing of methicillin-resistant Staphylococcus aureus on the basis of protein a gene polymorphism. Eur. J. Clin. Microbiol. Infect. Dis. 15, 60–64. doi: 10.1007/BF01586186
Fries, B. C., Varshney, A. K. (2013). Bacterial toxins-staphylococcal enterotoxin. B. Microbiol. Spectr. 1 (2), 10–22. doi: 10.1128/microbiolspec.AID-0002-2012
Fueyo, J. M., Mendoza, M. C., Martin, M. C. (2005). Enterotoxins and toxic shock syndrome toxin in Staphylococcus aureus recovered from human nasal carriers and manually handled foods: Epidemiological and genetic findings. Microb. Infect. 7, 187–194. doi: 10.1016/j.micinf.2004.10.009
Ghebremedhin, B., Olugbosi, M. O., Raji, A. M., Layer, F., Bakare, R. A., König, B., et al. (2009). Emergence of a community-associated methicillin-resistant Staphylococcus aureus strain with a unique resistance profile in southwest Nigeria. J. Clin. Microbiol. 47, 2975–2980. doi: 10.1128/JCM.00648-09
Hwang, S. Y., Kim, S. H., Jang, E. J., Kwon, N. H., Park, Y. K., Koo, H. C., et al. (2007). Novel multiplex PCR for the detection of the Staphylococcus aureus superantigen and its application to raw meat isolates in Korea. Int. J. Food Microbiol. 117, 99–105. doi: 10.1016/j.ijfoodmicro.2007.02.013.
Igbinosa, E. O., Beshiru, A. (2019). Characterisation of antibiotic resistance and species diversity of staphylococci isolated from apparently healthy farm animals. Afr. J. Clin. Exp. Microbiol. 20 (4), 289–298. doi: 10.4314/ajcem.v20i4.4
Igbinosa, E. O., Beshiru, A., Akporehe, L. U., Ogofure, A. G. (2016a). Detection of methicillin-resistant staphylococci isolated from food producing animals: a public health implication. Vet. Sci. 3, 14. doi: 10.3390/vetsci3030014
Igbinosa, E. O., Beshiru, A., Akporehe, L. U., Oviasogie, F. E., Igbinosa, W. O. (2016b). Prevalence of methicillin-resistant Staphylococcus aureus and other Staphylococcus species in raw meat samples intended for human consumption in Benin city, Nigeria: Implications for public health. Int. J. Environ. Res. Pub. Health 13, 949–959. doi: 10.3390/ijerph13100949
Igbinosa, E. O., Beshiru, A., Igbinosa, I. H., Ogofure, A. G., Uwhuba, K. E. (2021b). Prevalence and characterization of food-borne Vibrio parahaemolyticus from African salad in southern Nigeria. Front. Microbiol. 12, 632266. doi: 10.3389/fmicb.2021.632266
Igbinosa, E. O., Beshiru, A., Igbinosa, I. H., Okoh, A. I. (2022). Antimicrobial resistance and genetic characterisation of Salmonella enterica from retail poultry meats in Benin city, Nigeria. LWT Food Sci. Technol. 169, 114049. doi: 10.1016/j.lwt.2022.114049
Igbinosa, E. O., Beshiru, A., Odjadjare, E. E. O. (2020). Diversity, antimicrobial characterization and biofilm formation of enterococci isolated from aquaculture and slaughterhouse sources in Benin city, Nigeria. Ife J. Sci. 22 (3), 51–63. doi: 10.4314/ijs.v22i3.4
Igbinosa, I. H., Amolo, C. N., Beshiru, A., Akinnibosun, O., Ogofure, A. G., El-Ashker, M., et al. (2023). Identification and characterization of MDR virulent Salmonella spp isolated from smallholder poultry production environment in edo and delta states, Nigeria. PloS One 18 (2), e0281329. doi: 10.1371/journal.pone.0281329
Igbinosa, I. H., Beshiru, A., Ikediashi, S. C., Igbinosa, E. O. (2021a). Identification and characterisation of Salmonella serovars isolated from pig farms in Benin city, edo state, Nigeria: One health perspective. Microb. Drug Res. 27 (2), 258–267. doi: 10.1089/mdr.2019.0357
Imanah, E. O., Beshiru, A., Igbinosa, E. O. (2017). Antibiogram profile of Pseudomonas aeruginosa isolated from some selected hospital environmental drains. Asian Pacific J. Trop. Dis. 7 (10), 604–609. doi: 10.12980/apjtd.7.2017D6-468
Jackson, M. P., Landolo, J. J. (1986). Sequence of the exfoliative toxin b gene of Staphylococcus aureus. J. Bacteriol. 167, 726–728. doi: 10.1128/jb.167.2.726-728.1986
Kérouanton, A., Hennekinne, J. A., Letertre, C., Petit, L., Chesneau, O., Brisabois, A., et al. (2007). Characterisation of Staphylococcus aureus strains associated with food poisoning outbreaks in France. Int. J. Food Microbiol. 115, 369–375. doi: 10.1016/j.ijfoodmicro.2006.10.050
Kim, Y., Oh, D., Song, B., Heo, E., Lim, J., Moon, J. S., et al. (2015). Molecular characterisation, antibiotic resistance, and virulence factors of methicillin-resistant Staphylococcus aureus strains isolated from imported and domestic meat in Korea. Foodborne Pathog. Dis. 12, 390–398. doi: 10.1089/fpd.2014.1885
Kitai, S., Shimizu, A., Kawano, J., Sato, E., Nakano, C., Kitagawa, H., et al. (2005). Prevalence and characterisation of Staphylococcus aureus and enterotoxigenic Staphylococcus aureus in retail raw chicken meat throughout Japan. J. Vet. Med. Sci. 67, 269–274. doi: 10.1292/jvms.67.269
Laurent, F., Chardon, H., Haenni, M., Bes, M., Reverdy, M., Madec, J., et al. (2012). MRSA harbouring mecA variant gene mecC, France. Emerg. Infect. Dis. 18, 1465–1477. doi: 10.3201/eid1809.111920
Le, H. H. T., Dalsgaard, A., Andersen, P. S., Nguyen, H. M., Ta, Y. T., Nguyen, T. T. (2021). Large-Scale Staphylococcus aureus foodborne disease poisoning outbreak among primary school children. Microbiol. Res. 12, 43–52. doi: 10.3390/microbiolres12010005
Levy, S. (2014). Reduced antibiotic use in livestock: How Denmark tackled resistance. Environ. Health Perspect. 122, 160–165. doi: 10.1289/ehp.122-A160
Li, Q., Li, Y., Tan, Y., Meng, C., Ingmer, H., Jiao, X. (2019). Prevalence and characterisation of Staphylococcus aureus and Staphylococcus argenteus in chicken from retail markets in China. Food Cont. 96, 158–164. doi: 10.1016/j.foodcont.2018.08.030
Li, S., Wang, P., Zhao, J., Zhou, L., Zhang, P., Fu, C., et al. (2018). Characterisation of toxin genes and antimicrobial susceptibility of Staphylococcus aureus from retail raw chicken meat. J. Food Prot. 81 (4), 528–533. doi: 10.4315/0362-028X.JFP-17-309
Lika, E., Puvaca, N., Jeremic, D., Stanojevic, S., Shtylla-Kika, T., Cocoli, S., et al. (2021). Antibiotic susceptibility of Staphylococcus species isolated in raw chicken meat from retail stores. Antibiotics 10, 904. doi: 10.3390/antibiotics10080904
Lim, S. K., Nam, H. M., Park, H. J., Lee, H. S., Choi, M. J., Jung, S. C., et al. (2010). Prevalence and characterisation of methicillin-resistant Staphylococcus aureus in raw meat in Korea. J. Microbiol. Biotechnol. 20, 775–788.
Lowdera, B. V., Guinanea, C. M., Zakoura, N. L. B., Weinertb, L. A., Conway-Morrisc, A., Cartwrighta, R. A., et al. (2009). Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. PNAS 106 (46), 19545–19550. doi: 10.1073/pnas.0909285106
Ma, X. X., Galiana, A., Pedreira, W., Mowszowicz, M., Christophersen, I., Machiavello, S., et al. (2005). Community-acquired methicillin-resistant Staphylococcus aureus, Uruguay. Emerg. Infect. Dis. 11, 973–976.
Martineau, F., Picard, F. J., Lansac, N., Menard, C., Roy, P. H., Ouellette, M., et al. (2000). Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. antimicrob. Agents Chemother. 44, 231–238. doi: 10.1128/AAC.44.2.231-238.2000
Mkize, N., Zishiri, O. T., Mukaratirwa, S. (2017). Genetic characterisation of antimicrobial resistance and virulence gene in Staphylococcus aureus isolated from commercial broiler chickens in the Durban metropolitan area south Africa. J. South Afr. Vet. Assoc. 88, e1–e7. doi: 10.4102/jsava.v88i0.1416
Monday, S. R., Bohach, G. A. (1999). Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J. Clin. Microbiol. 37, 3411–3414. doi: 10.1128/JCM.37.10.3411-3414.1999
Murray, S., Pascoe, B., Meric, G., Mageiros, L., Yahara, K., Hitchings, M. D., et al. (2017). Recombination-mediated host adaptation by avian Staphylococcus aureus. genome biol. Evol. 9 (4), 830–842. doi: 10.1093/gbe/evx037
Nemati, M. (2013). Antimicrobial resistance of Proteus isolates from poultry. Eur. J. Exp. Biol. 3, 499–500.
Nemati, M. (2014). Prevalence of enterotoxin genes in poultry Staphylococcus aureus isolates. Bull. Environ. Pharmacol. Life Sci. 3 (2), 14–18. doi: 10.5001/omj.2015.56
Nemati, M., Hermans, K., Devriese, L. A., Maes, D., Haesebrouck, F. (2009). Screening of genes encoding adhesion factors and biofilm formation in Staphylococcus aureus isolates from poultry. Avian Pathol. 38 (6), 513–517. doi: 10.1080/03079450903349212
Okorie-Kanu, O. J., Anyanwu, M. U., Ezenduka, E. V., Mgbeahuruike, A. C., Thapaliya, D., Gerbig, G., et al. (2020). Molecular epidemiology, genetic diversity and antimicrobial resistance of Staphylococcus aureus isolated from chicken and pig carcasses, and carcass handlers. PloS One 15 (5), e0232913. doi: 10.1371/journal.pone.0232913
Okuma, K., Iwakawa, K., Turnidge, J. D., Grubb, W. B., Bell, J. M., O’Brien, F. G., et al. (2002). Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40, 4289–4294.
Oviasogie, E. F., Ogboghodo, B. I., Beshiru, A., Omoregie, O. B., Ogofure, P. (2016). The microbial burden load of eggshells from different poultry rearing systems in ekosodin village, edo state, Nigeria. J. Appl. Sci. Environ. Manag 20 (2), 227–231. doi: 10.4314/jasem.v20i2.1
Pokhrel, R. H., Aung, M. S., Thapa, B., Chaudhary, R., Mishra, S. K., Kawaguchiya, M., et al. (2016). Detection of ST772 panton-valentine leucocidin-positive methicillin-resistant Staphylococcus aureus (Bengal bay clones) and ST22 s. aureus isolates with genetic variant of elastin binding protein in Nepal. New Microbes New Infect. 11, 20–27. doi: 10.1016/j.nmni.2016.02.001
Pu, S. A. H., Wang, F., Ge, B. L. (2011). Characterisation of toxin genes and antimicrobial susceptibility of Staphylococcus aureus isolates from Louisiana retail meats. Foodbourne Pathog. Dis. 8, 299–306. doi: 10.1089/fpd.2010.0679
Rortana, C., Nguyen-Viet, H., Tum, S., Unger, F., Boqvist, S., Dang-Xuan, S., et al. (2021). Prevalence of salmonella spp. and Staphylococcus aureus in chicken meat and pork from Cambodian markets. Pathog. 10, 556. doi: 10.3390/pathogens10050556
Savariraj, W. R., Ravindran, N. B., Kannan, P., Rao, V. A. (2020). Occurrence and enterotoxin gene profiles of Staphylococcus aureus isolated from retail chicken meat. Food Sci. Technol. Int. 27 (7), 619–625. doi: 10.1177/1082013220980204
Schelin, J., Wallin-Carlquist, N., Cohn, M. T., Lindqvist, R., Barker, G. C., Rådström, P. (2011). The formation of Staphylococcus aureus enterotoxin in food environments and advances in risk assessment. Virulence 2, 580–592. doi: 10.4161/viru.2.6.18122
Sergelidis, D., Angelidis, A. S. (2017). Methicillin-resistant Staphylococcus aureus: a controversial foodborne pathogen. Lett. Appl. Microbiol. 64, 409–418. doi: 10.1111/lam.12735
Shore, A., Coleman, D. (2013). Staphylococcal cassette chromosome mec: recent advances and new insights. Int. J. Med. Microbiol. 303, 350–359. doi: 10.1016/j.ijmm.2013.02.002
Szczuka, E., Urbanska, K., Pietryka, M., Kaznowski, A. (2013). Biofilm density and detection of biofilm-producing genes in methicillin-resistant Staphylococcus aureus strains. Folia Microbiologica 58 (1), 47–52. doi: 10.1007/s12223-012-0175-9
Tallent, S., Hait, J., Bennett, R. W., Lancette, G. A. (2019). Bacteriological Analytical Manual (BAM) Main Page. BAM Chapter 12: Staphylococcus aureus. Available at: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-12-staphylococcus-aureus.
Tang, Y., Larsen, J., Kjeldgaard, J., Andersen, P. S., Skov, R., Ingmer, H. (2017). Methicillin-resistant and -susceptible Staphylococcus aureus from retail meat in Denmark. Int. J. Food Microbiol. 249, 72–76. doi: 10.1016/j.ijfoodmicro.2017.03.001
Tegegne, H. A., Kolácková, I., Florianová, M., Gelbícová, T., Madec, J., Haenni, M., et al. (2021). Detection and molecular characterisation of methicillin-resistant Staphylococcus aureus isolated from raw meat in the retail market. J. Global Antimicrob. Res. 26, 233–238. doi: 10.1016/j.jgar.2021.06.012
Titouche, Y., Houali, K., Ruiz-Ripa, L., Vingadassalon, N., Nia, Y., Fatihi, A., et al. (2020). Enterotoxin genes and antimicrobial resistance in Staphylococcus aureus isolated from food products in Algeria. J. Appl. Microbiol. 14, 665. doi: 10.1111/jam.14665
World Health Organisation (WHO) (2017) WHO list of critically important antimicrobials for human medicine (WHO CIA list) (Geneva, Switzerland). Available at: https://www.who.int/foodsafety/areas_work/antimicrobial-resistance/cia/en/ (Accessed July 18, 2018).
World Health Organization (WHO) (2019) Model lists of essential medicines (Geneva). Available at: http://www.who.int/medicines/publications/essentialmedicine/en/ (Accessed June 29, 2019).
Wu, S., Huang, J., Zhang, F., Wu, Q., Zhang, J., Pang, R., et al. (2019). Prevalence and characterisation of food-related methicillin-resistant Staphylococcus aureus (MRSA) in China. Front. Microbiol. 10, 304. doi: 10.3389/fmicb.2019.00304
Xia, G., Wolz, C. (2014). Phages of Staphylococcus aureus and their impact on host evolution. Infect. Genet. Evol. 21, 593–601. doi: 10.1016/j.meegid.2013.04.022
Yazdani, R., Oshaghi, M., Havaei, A., Pishva, E., Salehi, R., Sadeghizadeh, M., et al. (2006). Detection of icaAD gene and biofilm formation in Staphylococcus aureus isolates from wound infections. Iranian J. Public Health 35 (2), 25–28. Available at: https://ijph.tums.ac.ir/index.php/ijph/article/view/2174/2155.
Zehra, A., Gulzar, M., Singh, R., Kaur, S., Gill, J. P. S. (2019). Prevalence, multidrug resistance and molecular typing of methicillin-resistant Staphylococcus aureus (MRSA) in retail meat from punjab, India. J. Global Antimicrobial Res. 16, 152–158. doi: 10.1016/j.jgar.2018.10.005
Keywords: resistant determinants, biofilm formation, cassette chromosomes, virulence, enterotoxin
Citation: Igbinosa EO, Beshiru A, Igbinosa IH, Ogofure AG, Ekundayo TC and Okoh AI (2023) Prevalence, multiple antibiotic resistance and virulence profile of methicillin-resistant Staphylococcus aureus (MRSA) in retail poultry meat from Edo, Nigeria. Front. Cell. Infect. Microbiol. 13:1122059. doi: 10.3389/fcimb.2023.1122059
Received: 12 December 2022; Accepted: 13 February 2023;
Published: 02 March 2023.
Edited by:
Mingyu Wang, Shandong University, ChinaReviewed by:
Ben Pascoe, University of Oxford, United KingdomOlatunde Dahunsi, Bowen University, Nigeria
Ben Jesuorsemwen Enagbonma, North-West University, South Africa
Copyright © 2023 Igbinosa, Beshiru, Igbinosa, Ogofure, Ekundayo and Okoh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Etinosa O. Igbinosa, RXRpbm9zYS5JZ2Jpbm9zYUB1bmliZW4uZWR1
 Etinosa O. Igbinosa
Etinosa O. Igbinosa Abeni Beshiru
Abeni Beshiru Isoken H. Igbinosa
Isoken H. Igbinosa Abraham G. Ogofure
Abraham G. Ogofure Temitope C. Ekundayo
Temitope C. Ekundayo Anthony I. Okoh
Anthony I. Okoh