- 1Faculty of Life Science and Technology, Kunming University of Science and Technology, Kunming, Yunnan, China
- 2Department of Infectious Diseases, The First People's Hospital of Yunnan Province, Yunnan, China
- 3School of Public Health, Nanjing Medical University, Nanjing, China
- 4Kunming Medical University, Yunnan, China
Introduction: Hepatitis C virus (HCV) infection was the primary reason causing critical hepatic Q7 diseases. Although direct-acting antiviral agents (DAA) were widely used in clinics, anti-drug mutation, the outcome of patients with different viral subtypes, and recurrence suggested that HCV pathogenic mechanism should be studied further. HCV infection, replication, and outcome were influenced by the IFNL4 and itsdownstream genes (MxA and MxB). However, whether genetic polymorphisms of these genes played necessary roles required verification in the Yunnan population.
Methods and Results: After analyzing the genotypes and allele frequencies of seven single nucleotide polymorphisms (SNP), we found the association between the genotype and allele frequencies of rs11322783 in the IFNL4 gene and HCV infection in Yunnan population. Furthermore, the genetic polymorphisms of the MxA and MxB genescould influence liver function of HCV patients. The indirect bilirubin (IBIL) and albumin (ALB) levels showed significant differences among HCV patients, who carried various genotypes. The IBIL levels were associated with genotypes of rs17000900 (P= 0.025) and rs2071430 (P= 0.037) in the MxA gene, and ALB levels were associated with genotypes of rs2838029 (P= 0.010) in the MxB gene. Similarly, the genotypes of SNPs also showed significant difference in patients infected with subtype 3a (P=0.035) and 2a (P=0.034). However, no association was identified between expression level and SNPs of the MxA and MxB genes. Furthermore, HCV subtype 3b was found to be the predominantly epidemic strain in Yunnan Province.
Conclusion: In conclusion, the association between biochemical indices/HCV subtypes and SNPs in the MxA and MxB genes was identified in Yunnan HCV population.
Introduction
Hepatitis C virus (HCV) infection could lead to critical hepatitis diseases, such as cirrhosis and hepatocellular carcinoma (HCC). Although direct-acting antivirals (DAAs) drugs are used worldwide, approximately 1.75 million new HCV infections occurred in 2015 (W.H.O, 2017). The mortality trends of infectious hepatitis disease were increasing from 2000 to 2015, differing from other popular infectious diseases, such as human immunodeficiency virus (HIV), tuberculosis (TB), and malaria. Thus, the pathogenic mechanisms of HCV infection should be studied further.
Interferons (IFNs) are the common used drugs for HCV infection therapy, and they could active Jak/STAT signaling pathway and further induce some interferon-stimulated genes (ISGs) (Heim and Thimme, 2014). The expression and function of Interferon lambda 4 (IFNL4) gene is controlled by its genetically transcriptional regulation (Zhou et al., 2020). The MyXovirus resistance (Mx) genes belongs to the ISGs family. Firstly, the MyXovirus resistance 1 (MxA or Mx1) gene was identified as one of the main protected factors for mice against influenza viruses (Lindenmann, 1964). Then, it is investigated that the MxA gene belonged to natural immune system and had wide-spectra anti-virus activity, which is induced by interferon (Haller et al., 2018). Although the antiviral activity of the MxB gene is limited compared to the MxA gene, it could interfere with HCV RNA replication by interacting with the NS5A protein (Yi et al., 2019). These reports suggested the Mxs genes were necessary natural factors for inhibiting HCV in patients.
To date, the genetic polymorphisms of the MxA gene have been reported to be associated with HCV infection or clearance of those with HCV infection (Garcia-Alvarez et al., 2017), but whether single nucleotide polymorphisms (SNPs) of the MxB gene could influence HCV infection, biochemical characteristics, or outcome of HCV patients are not clear. In this study, the SNPs of the MxA and MxB genes in HCV patients and controls from Yunnan Province were analyzed.
Materials and methods
Individuals and clinical data
We recruited 347 patients with chronic HCV infection and 448 normal controls from the First People’s Hospital of Yunnan Province from 2019 to 2022. 3 mL of whole blood was collected from each individual. The patients were diagnosed as chronic HCV infected persons by the symptoms and liver function test. All HCV-infected patients were identified to be anti-HCV positive by HCV ELISA Kit (ORTHO, USA), and all patients were without any medical treatment when we collected the samples. All patients were identified without serious liver disease, such as fibrosis, cirrhosis and hepatocellular carcinoma). The basic information and liver function data of all samples [including alanine transaminase (ALT), aspartate transaminase (AST), total bilirubin (TBIL), direct bilirubin (DBIL), indirect bilirubin (IBIL), total protein (TP), albumin (ALB), globin (GLOB)] of HCV patients were collected for further analysis. Based on the clinical phenotype and biochemical indexes, all chronic HCV infected patients were identified without Hepatitis B virus (HBV) or HIV infection by using Quantitative CLIA Kit (Autobio, China) and Anti-HIV ELISA Kit (WANTAI, China). Written informed consents of the Declaration of Helsinki were obtained from all participants prior to the study. The institutional review board of Kunming University of Science and Technology approved this study.
HCV RNA loading quantification and genotyping
HCV RNA was abstracted from 100 µL serum of patients by using TIANamp Virus RNA Kit (TIANGEN, China), and viral RNA load was quantified by using the primers and probe in our previous study (Song et al., 2019). The detection limit was set at 1,000 copies/mL. HCV RNA was further transformed as log10‐transformation. The NS5B gene of HCV was genotyped in each patient by using the method described in our previous study (Zhang et al., 2014).
SNP selection and genotyping
Genomic DNA was extracted from whole blood by using the Blood Genomic DNA Miniprep Kit (Axygen, USA). Seven tagSNPs were genotyped by using SnapShot method according to previous study (Zheng et al., 2022), including two SNPs (rs11322783 and rs117648444) in the IFNL4 gene, two SNPs (rs2071430 and rs17000900) in the MxA gene and three SNPs (rs9982944, rs408825, and rs2838029) in the MxB gene,. Then, 10% of the genotyping results were further identified by using Sanger sequencing method.
RNA expression level detection
Total RNA was extracted from 200 µL whole blood of HCV patients using the RNAsimple Total RNA Kit (TIANGEN, China). Then, the level of RNA expression of the MxA and MxB genes was detected using fluorescent quantitative real-time PCR. The primers for quantifying target genes were GTGCATTGCAGAAGGTCAGA/CTGGTGATAGGCCATCAGGT (for the MxA gene) and CAGAGGCAGCGGAATCGTAA/TGAAGCTCTAGCTCGGTGTTC (for the MxB gene). The GAPDH gene was used as the reference gene, and the primers for quantification were GGCATCCTGGGCTACACTGAG/CATACCAGGAAATGAGCTTGAC. The ChamQ SYBR® qPCR Master Mix (Vazyme, China) and Quant Gene 9600 were used to determine the RNA expression levels of the MxA and MxB genes of HCV patients.
Data analysis
The genotype and allele frequencies between HCV patients and controls are analyzed by using Chi-square text. A student t test (two tailed) was used to compare viral load between two HCV subtypes, biochemical indices among patients with different genotypes, and RNA level expression of MxA and MxB genes. The correlation analysis was used to determine the relationship between viral load of HCV and RNA level of the MxA or MxB gene (two-sided). A P-value less than 0.05 is considered statistically significant.
Results
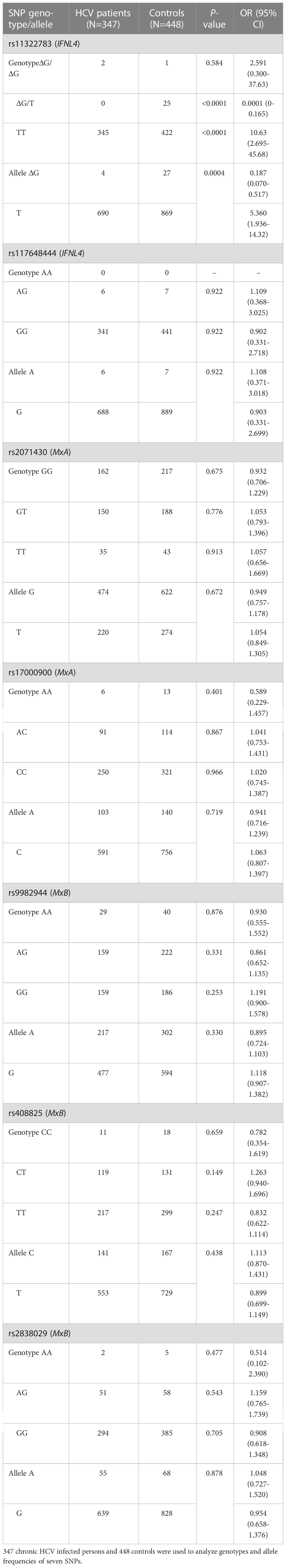
The gender ratio (male: female) was similar between HCV patient group (1.4:1) and controls (1.6:1). There were 201 males and 146 females in the HCV patients, and 275 males and 173 females in the control group. The mean age was 44.70 ± 0.89 and 40.58 ± 0.53 years in the HCV patients and controls, respectively. The genotype and allele frequencies, excluding SNP rs11322783, showed no significant difference between HCV patients and controls. Genotype ΔG/T of rs11322783 showed significantly lower frequency in HCV patients (0%) than that in controls (5.58%); however, the frequency of genotype TT was statistically higher in HCV patients (99.42%) than that in controls (94.20%) (Table 1). Similarly, allele ΔG of rs11322783 was a protective factor for HCV infection in Yunnan, with a frequency of 0.58% (4/694) and 3.01% (27/896) in patients and controls, respectively. This result suggested genotype and allele of rs11322783 could influence HCV infection in the Yunnan population.
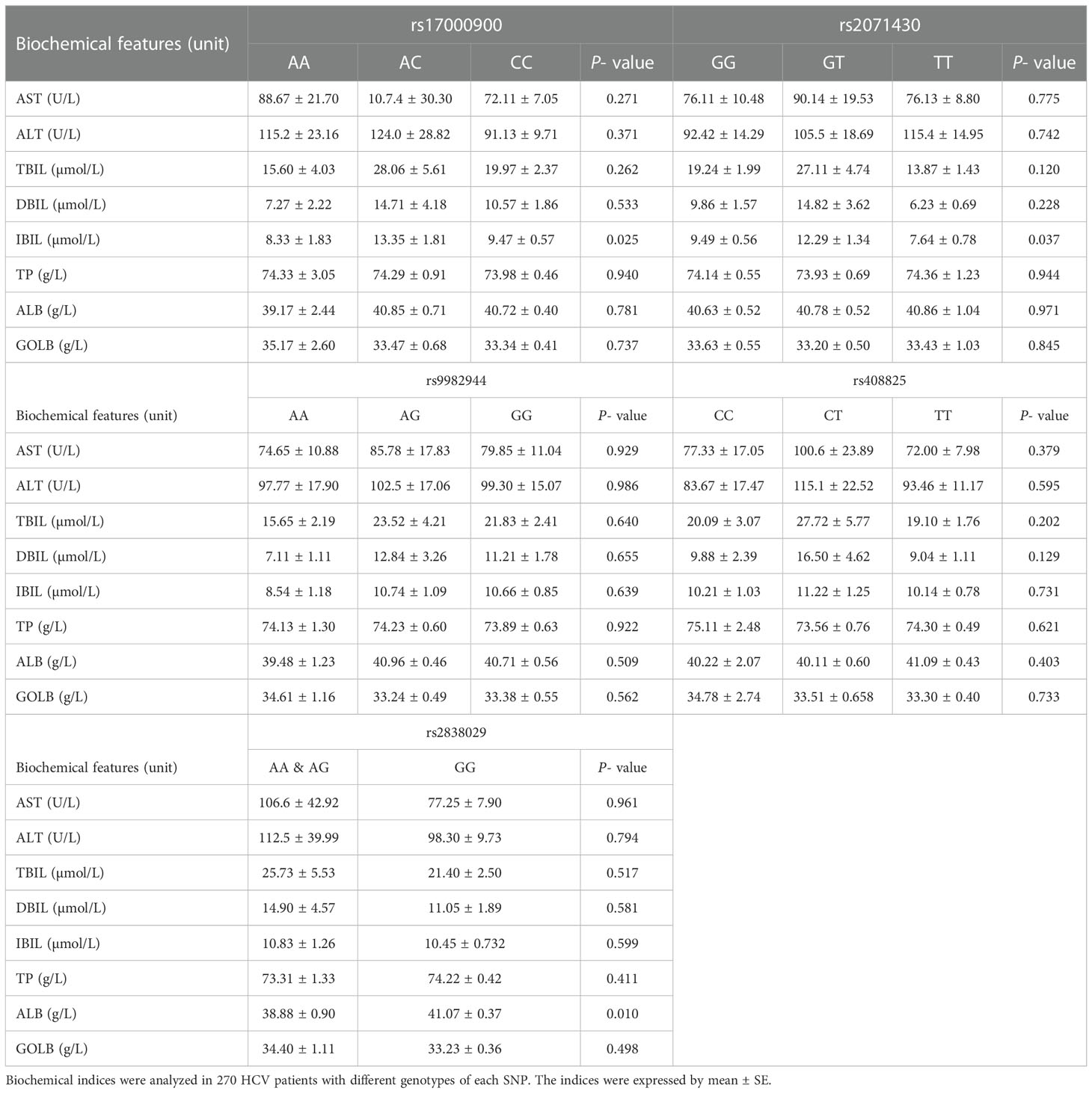
The biochemical data was analyzed among 270 HCV patients with different genotypes of each SNP (Table 2). The IBIL levels showed a significant difference among various genotypes of two SNPs in the MxA gene of HCV patients. In HCV patients with genotype AC of rs17000900 or genotype GT of rs2071430, the IBIL level were significantly higher than that in the other patients. The ALB levels were much lower in HCV patients carried genotype AA and AG of rs2838029 than other patients. These results suggested genetic polymorphisms of the MxA and MxB genes could influence the liver function of HCV patients.
In total, HCV subtypes were determined in 339 HCV patients (Figure 1). Subtype 3b was the main subtype in Yunnan HCV patients occurring in 42.18% of individuals (n= 143). The frequencies of HCV subtype 3a (n= 59, 17.40%), 1b (n= 51, 15.04%), 2a (n= 45, 13.27%), and 6 (n= 40, 11.80%) were similar in Yunnan population. Only one sample belonged to subtype 5b. Although HCV viral load was detected in only 137 patients, the viral load was compared among HCV patients with different subtypes (Figure 2). The results showed significantly higher viral load existed in the patients with subtype 6 than in patients with subtype 2a (P= 0.029) or 3b (P= 0.003). The viral load was statistically higher in the patients with subtype 1b than those with subtype 3b. These results suggested that the viral load of HCV patients might be affected by HCV subtypes.
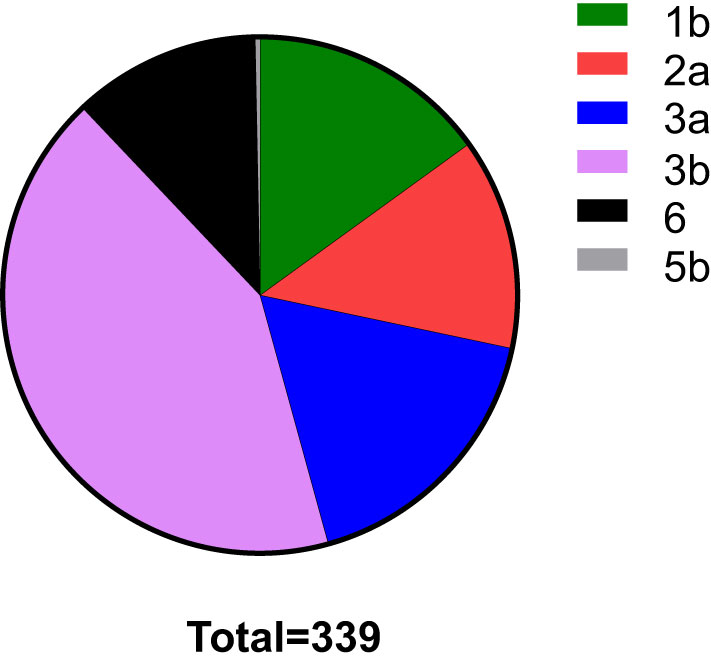
Figure 1 The distribution of HCV subtypes in this study. Totally 339 HCV patients were used to detect HCV subtypes. The subtype 3b was dominant in Yunnan Province.
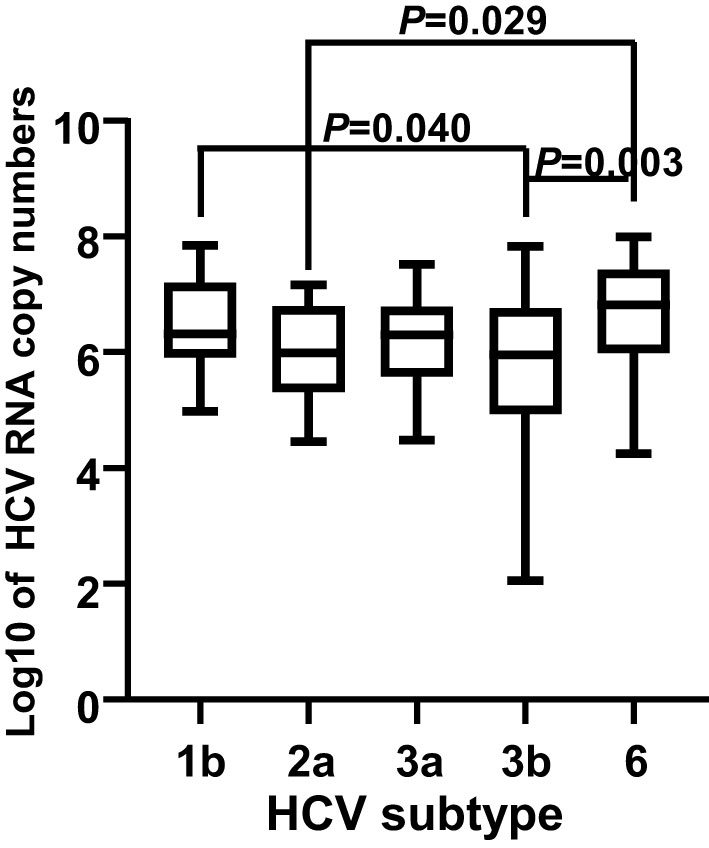
Figure 2 Comparison of viral load among 137 HCV patients with different subtypes. The viral loads were transformed into log10 of the copy numbers. A P value less than 0.05 was considered as significant difference.
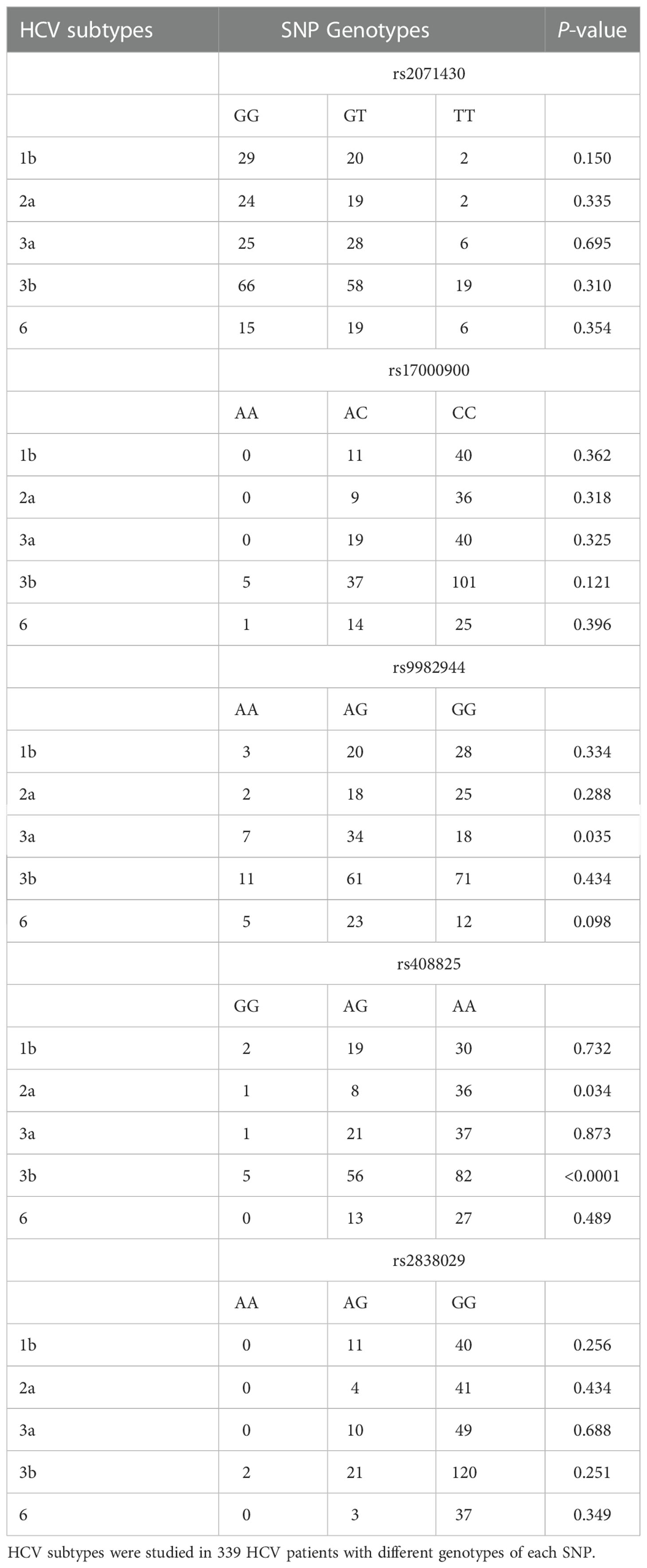
The frequencies distribution of subtypes in HCV patients with different genotypes of each SNP in the MxA and MxB genes were analyzed (Table 3). The frequency of subtype 3a was lower in patients carried genotype GG of rs9982944 (11.69%, 18/154) than in those with genotype AA (25%, 7/28) or AG (22.08%, 34/154). The frequency of subtype 2a was significantly lower in HCV patients with genotype AG of rs408825 (6.84%, 8/117) than in those with genotype AA (16.98%, 36/212) or GG (11.11%, 1/9). These results suggested the possible protective role of genotype GG of rs9982944 and genotype AG of rs408825 against HCV subtype 3b and 2a infection, respectively.
When comparing the RNA expression level of the MxA gene and MxB gene in the patients with various genotypes of each SNP of the corresponding gene (Table 4), no significant difference was identified. Similarly, no correlation was identified between the RNA expression level of the MxA gene (r= 0.083, P= 0.343) or the MxB gene (r= 0.036, P= 0.691) and the viral load of HCV patients.
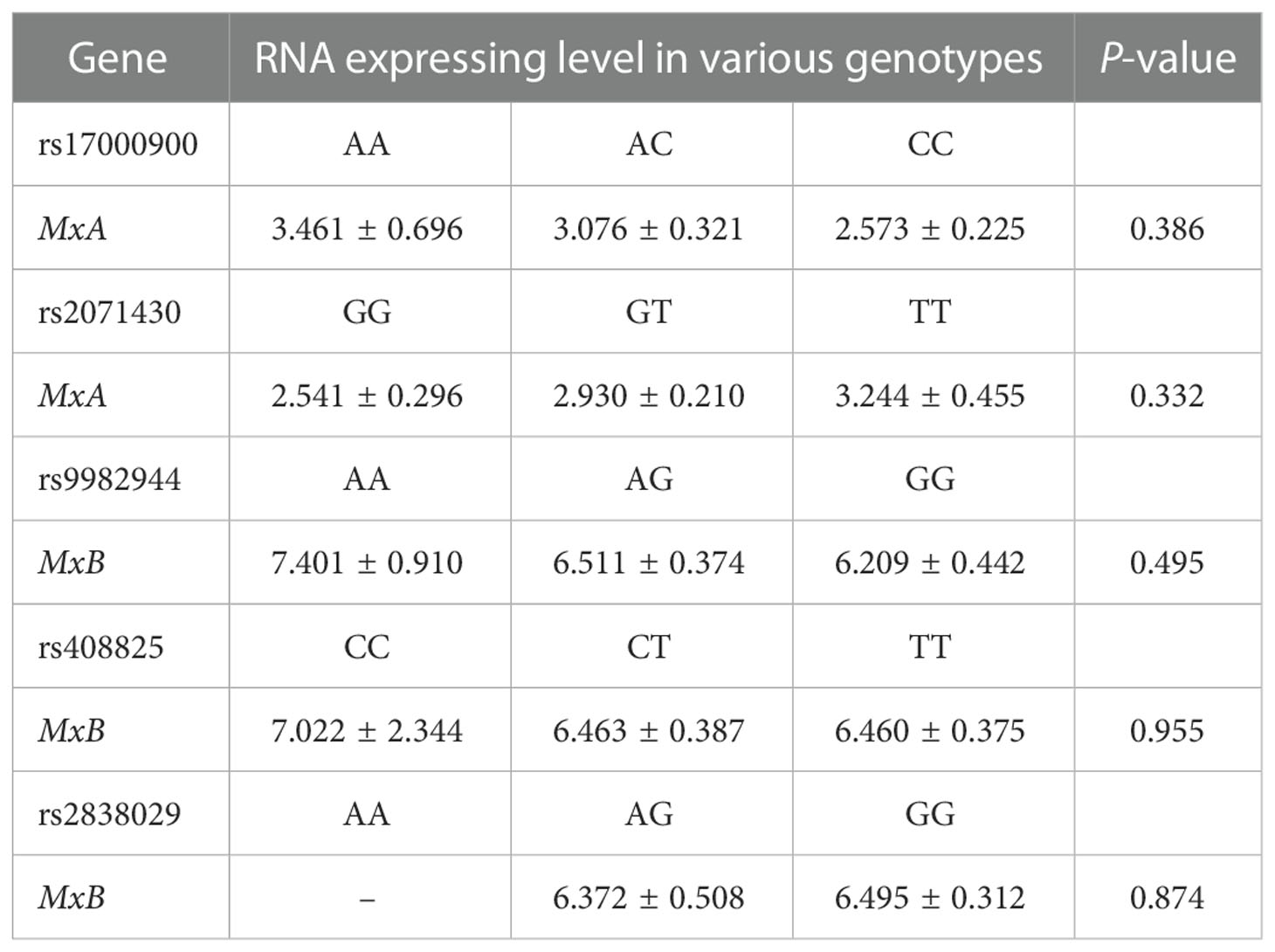
Table 4 RNA expressing level of the MxA and MxB gene in HCV patients with various genotypes of these two genes.
Discussion
Although over 90% of HCV patients could be cured by using DAA, some were still treated by using IFN-α combined Ribavirin owing to the high price of DAA and anti-drug mutation of DAA. Thus, the mechanism of IFN against HCV was necessary to further study. IFN-α could activate the Jak-STAT signaling pathway and further induce MxA production, which could inhibit HCV replication (Stevenson et al., 2011). The MxB belongs to the same family as MxA and plays wide anti-virus roles. However, reports regarding how MxB inhibited HCV replication were few. Li et al. found that MxB competitively interacted with NS5A of HCV and reduced the combination between NS5A and CypA, which provided formation of HCV infection (Li et al., 2022). These results suggested the importance of MxA and MxB in inhibiting HCV infection and replication.
The genetic polymorphisms of the MxA gene were considered to influence HCV infection, viral clearance, and outcome in many studies (Hassany et al., 2017; Zang et al., 2018), but no association study was performed between SNPs of the MxB gene and Chinese HCV patients. In this study, although no association was identified between SNPs of the MxA and MxB genes with HCV infection in the Yunnan population, HCV subtypes and biochemical indices demonstrated significant differences among patients with various genotypes. Zang et al. reported that genotype TT of rs2071430 in the MxA gene preferred to induce patients into chronic HCV infection (Zang et al., 2018). In Egyptian and Japanese HCV patients, the promoter SNPs of the MxA gene were considered independently influenced factors in response to IFN therapy (Suzuki et al., 2004; Hassany et al., 2017).
In liver allograft patients with or without HCV infection, the expression level of the MxA gene was positively associated with AST levels (>40 U/L) in patients’ liver biopsies (Borgogna et al., 2009). SNP at nt-88 of the MxA gene was identified to be an independent determining factor for outcome of IFN therapy (Suzuki et al., 2004). Similarly, genotype TT of rs2071430 in the MxA gene influenced HCV infection chronicity of patients (Zang et al., 2018). Similarly, we found that genotype GT of rs2071430 was associated with high IBIL levels of patients. These suggested that genetic polymorphisms of the MxA gene might influence the pathogenic progress and outcome of patients. To the best of our knowledge, this was the first study to identify the association between genetic polymorphisms of the MxB gene and biochemical indices and subtypes of HCV patients.
The genotypes of rs11322783 in the IFNL4 gene were widely reported as an important host genetic factor influencing viral clearance and outcome of HCV patients (Aka et al., 2014; O'Brien et al., 2015). The allele ΔG and T of rs11322783 were found to play protective and risk roles in HCV infection. However, we could not analyze the association between genotypes of rs11322783 and biochemical indices of patients owing to the few numbers of patients with genotype ΔG/ΔG. These studies suggested that SNPs of the IFNL4 gene could influence HCV infection, disease progress, and treatment effect.
In summary, we firstly identified that genetic polymorphisms of the MxA and MxB genes were associated with biochemical indices and HCV subtypes of patients in Yunnan.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving human participants were reviewed and approved by The institutional review board of Kunming University of Science and Technology approved this study. The patients/participants provided their written informed consent to participate in this study.
Author contributions
A-MZ and XX designed this study. MH and ML performed the experiments. JG, PH and MY collected the samples. A-MZ and LL analyzed the data. A-MZ and MH prepared the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study is supported by the National Natural Science Foundation of China (32160148), Leading Reserve Talents of Academy and Science and Technology in Yunnan Province (2019HB002), and Yunnan Ten Thousand Talents Plan Young & Elite Talents Project.
Acknowledgments
We thank all the participants in this study. This study is supported by the National Natural Science Foundation of China (32160148), Leading Reserve Talents of Academy and Science and Technology in Yunnan Province (2019HB002), and Yunnan Ten Thousand Talents Plan Young & Elite Talents Project.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1119805/full#supplementary-material
References
Aka, P. V., Kuniholm, M. H., Pfeiffer, R. M., Wang, A. S., Tang, W., Chen, S., et al. (2014). Association of the IFNL4-DeltaG allele with impaired spontaneous clearance of hepatitis c virus. J. Infect. Dis. 209 (3), 350–354. doi: 10.1093/infdis/jit433
Borgogna, C., Toniutto, P., Smirne, C., Azzimonti, B., Ritta, M., Avellini, C., et al. (2009). Expression of the interferon-inducible proteins MxA and IFI16 in liver allografts. Histopathology 54 (7), 837–846. doi: 10.1111/j.1365-2559.2009.03311.x
Garcia-Alvarez, M., Berenguer, J., Jimenez-Sousa, M. A., Pineda-Tenor, D., Aldamiz-Echevarria, T., Tejerina, F., et al. (2017). Mx1, OAS1 and OAS2 polymorphisms are associated with the severity of liver disease in HIV/HCV-coinfected patients: A cross-sectional study. Sci. Rep. 7, 41516. doi: 10.1038/srep41516
Haller, O., Arnheiter, H., Pavlovic, J., Staeheli, P. (2018). The discovery of the antiviral resistance gene mx: A story of great ideas, great failures, and some success. Annu. Rev. Virol. 5 (1), 33–51. doi: 10.1146/annurev-virology-092917-043525
Hassany, M., Gamal, A., Zaki, N., Eysa, B. (2017). Assessment of the relation between SNP in MxA gene and the responsiveness of Egyptian HCV genotype 4 patients to pegylated interferon and ribavirin treatment. Gastroenterol. Res. 10 (2), 100–105. doi: 10.14740/gr810w
Heim, M. H., Thimme, R. (2014). Innate and adaptive immune responses in HCV infections. J. Hepatol. 61 (1 Suppl), S14–S25. doi: 10.1016/j.jhep.2014.06.035
Li, Q., An, N., Yin, X., Zhang, R., Shao, H., Yi, D., et al. (2022). MxB disrupts hepatitis c virus NS5A-CypA complex: Insights from a combined theoretical and experimental approach. Front. Microbiol. 13, 849084. doi: 10.3389/fmicb.2022.849084
Lindenmann, J. (1964). Inheritance of resistance to influenza virus in mice. Proc. Soc. Exp. Biol. Med. 116, 116506–116509. doi: 10.3181/00379727-116-29292
O'Brien, T. R., Pfeiffer, R. M., Paquin, A., Lang Kuhs, K. A., Chen, S., Bonkovsky, H. L., et al. (2015). Comparison of functional variants in IFNL4 and IFNL3 for association with HCV clearance. J. Hepatol. 63 (5), 1103–1110. doi: 10.1016/j.jhep.2015.06.035
Song, Y., Yang, X., Shen, Y., Wang, Y., Xia, X., Zhang, A. M. (2019). STAT3 signaling pathway plays importantly genetic and functional roles in HCV infection. Mol. Genet. Genomic Med. 7 (8), e821. doi: 10.1002/mgg3.821
Stevenson, N. J., Murphy, A. G., Bourke, N. M., Keogh, C. A., Hegarty, J. E., O'Farrelly, C. (2011). Ribavirin enhances IFN-alpha signalling and MxA expression: a novel immune modulation mechanism during treatment of HCV. PloS One 6 (11), e27866. doi: 10.1371/journal.pone.0027866
Suzuki, F., Arase, Y., Suzuki, Y., Tsubota, A., Akuta, N., Hosaka, T., et al. (2004). Single nucleotide polymorphism of the MxA gene promoter influences the response to interferon monotherapy in patients with hepatitis c viral infection. J. Viral Hepat. 11 (3), 271–276. doi: 10.1111/j.1365-2893.2004.00509.x
W.H.O (2017). GLOBAL HEPATITIS REPORT, 2017 (World Health Organization). https://www.who.int/publications/i/item/9789241565455
Yi, D. R., An, N., Liu, Z. L., Xu, F. W., Raniga, K., Li, Q. J., et al. (2019). Human MxB inhibits the replication of hepatitis c virus. J. Virol. 93 (1), e01285–18. doi: 10.1128/JVI.01285-18
Zang, F., Yue, M., Yao, Y., Liu, M., Fan, H., Feng, Y., et al. (2018). Influence of IL28B and MxA gene polymorphisms on HCV clearance in han Chinese population. Epidemiol. Infect. 146 (3), 379–385. doi: 10.1017/S0950268817002928
Zhang, A. M., Ma, K., Song, Y., Wang, B., Feng, Y., Liu, L., et al. (2014). Genetic polymorphisms of the IFNlambda genes are associated with biochemical features in han Chinese with HCV infection from yunnan province, China. Infect. Genet. Evol. 21, 161–165. doi: 10.1016/j.meegid.2013.11.013
Zheng, K., Shen, Y., Xia, X., Song, Y., Zhang, A. M. (2022). Genetic polymorphisms in the IFNL4, MxA, and MxB genes were associated with biochemical index of chronic HBV patients from yunnan, China. PeerJ 10, e13353. doi: 10.7717/peerj.13353
Keywords: HCV infection, biochemical indices, subtype, MxA, MxB
Citation: He M, Liu M, Geng J, Liu L, Huang P, Yue M, Xia X and Zhang A-M (2023) Polymorphisms of the MxA and MxB genes are associated with biochemical indices and viral subtypes in Yunnan HCV patients. Front. Cell. Infect. Microbiol. 13:1119805. doi: 10.3389/fcimb.2023.1119805
Received: 09 December 2022; Accepted: 03 January 2023;
Published: 19 January 2023.
Edited by:
Bin Zhou, Nanjing Agricultural University, ChinaReviewed by:
Jianchao Wei, Chinese Academy of Agricultural Sciences, ChinaLimin Chen, Chinese Academy of Medical Sciences and Peking Union Medical College, China
Li Huang, Chinese Academy of Agricultural Sciences, China
Copyright © 2023 He, Liu, Geng, Liu, Huang, Yue, Xia and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: A-Mei Zhang, emFtMTk4MEB5ZWFoLm5ldA==; Xueshan Xia, b2xpdmVyeGlhMjAwMEBhbGl5dW4uY29t
 Mengzhu He1
Mengzhu He1 Peng Huang
Peng Huang Ming Yue
Ming Yue A-Mei Zhang
A-Mei Zhang