
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 16 February 2023
Sec. Microbiome in Health and Disease
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1109634
 Yuanmeihui Tao1†
Yuanmeihui Tao1† Yajun Ge1,2†
Yajun Ge1,2† Jing Yang1,3
Jing Yang1,3 Weitao Song4
Weitao Song4 Dong Jin1,3
Dong Jin1,3 Hong Lin4
Hong Lin4 Han Zheng1,3
Han Zheng1,3 Shan Lu1,3
Shan Lu1,3 Wenbo Luo1
Wenbo Luo1 Yuyuan Huang1
Yuyuan Huang1 Zhenhong Zhuang4*
Zhenhong Zhuang4* Jianguo Xu1,3,5*
Jianguo Xu1,3,5*The species in the genus Erwinia are Gram-stain-negative, facultatively anaerobic, motile, and rod-shaped. Most species in the genus Erwinia are phytopathogens. Also, Erwinia persicina was involved in several human infections. Based on the reverse microbial etiology principles, it is worth analyzing the pathogenicity of species in this genus. In this study, we isolated and sequenced two species of Erwinia. Phylogenetic, phenotypic, biochemical, and chemotaxonomic analyses were performed to identify its taxonomy position. The virulence tests on plant leaves and pear fruits were used to identify the plant pathogenicity of two species of Erwinia. Bioinformatic methods predicted the possible pathogenic determinants based on the genome sequence. Meanwhile, adhesion, invasion, and cytotoxicity assays on RAW 264.7 cells were applied to identify animal pathogenicity. We isolated two Gram-stain-negative, facultatively anaerobic, motile, and rod-shaped strains from the feces of ruddy shelducks in the Tibet Plateau of China, designated J780T and J316. Distinct phylogenetic, genomic, phenotypic, biochemical, and chemotaxonomic characters of J780T and J316 identified they were novel species and belonged to the genus Erwinia, for which the name Erwinia sorbitola sp. nov. was proposed, the type strain was J780T (= CGMCC 1.17334T = GDMCC 1.1666T = JCM 33839T). Virulence tests showed blight and rot on the leaves and pear fruits confirmed Erwinia sorbitola sp. nov. was a phytopathogen. Predicted gene clusters of motility, biofilm formation, exopolysaccharides, stress survival, siderophores, and Type VI secretion system might be the causes of pathogenicity. In addition, predicted polysaccharide biosynthesis gene clusters on the genome sequence, and the high capacity for adhesion, invasion, and cytotoxicity to animal cells confirmed it has pathogenicity on animals. In conclusion, we isolated and identified a novel phytopathogen Erwinia sorbitola sp. nov. in ruddy shelducks. A predefined pathogen is beneficial for preventing from suffering potential economic losses caused by this new pathogen.
The genus Erwinia was first described by Winslow et al., with Erwinia amylovora being the type species. Cells of the genus Erwinia are mainly characterized as Gram-stain-negative, facultatively anaerobic, motile, rod-shaped bacteria with 49.8-54.1 mol% of G+C contents (Hauben et al., 1998). Their major fatty acids are C12:0, C14:0, and C16:0 (Hauben et al., 1998). Thus far, there are 20 species in the genus Erwinia with valid names (https://lpsn.dsmz.de/genus/erwinia). Most strains are plant pathogens, including E. amylovora (Hauben et al., 1998), E. aphidicola (Harada et al., 1997), E. mallotivora (Amin et al., 2010), E. papaya (Gardan et al., 2004), E. pyrifoliae (Kim et al., 1999), E. piriflorinigrans (Lopez et al., 2011), and E. uzenensis (Matsuura et al., 2012). Among these phytopathogens, E. amylovora can cause fire blight, a plant disease which threatens host species, including Maloideae (commercial apple and pear), Rosoideae (blackberry and raspberry), and Spiraeoideae. The pathogenesis of E. amylovora is that cells enter susceptible hosts through the nectarthodes of flowers and other natural openings, rapidly moving through the parenchyma and then establishing a systemic infection through the xylem vessels, finally provoking extensive lesions; in late stages, the infected plants will appear brown or black (Pique et al., 2015).
In the pathogenic mechanism of the genus Erwinia, the Type III secretion system (T3SS), the exopolysaccharide (EPS) amylovoran, biofilm formation, and motility were characterized as virulent determinants in species of Erwinia (Kharadi et al., 2021). Due to its pathogenicity to many host plants (including many fruit plants), it is necessary to conduct in-depth research into it.
Subsequent studies have also found that: E. persicina has been isolated from human urine (O'Hara et al., 1998); E. tasmaniensis was involved in a cervical lymphadenitis case (Shin et al., 2008b); and E. billingiae has been associated with dermohypodermitis (Prod'homme et al., 2017) and septic arthritis (Bonnet et al., 2019).
The Erwinia sorbitola sp. nov. was isolated from ruddy shelduck (Tadorna ferruginea), a migratory species that were involved in the highly pathogenic avian influenza (HPAI) H5N1 that broke out at Qinghai Lake (Liu et al., 2005). Due to migrating large distances between their breeding and wintering areas, several novel microorganisms have been isolated from ruddy shelduck (Marinova et al., 2015; Ge et al., 2020), which is a reminder it may be a potential vector of more novel pathogens.
Advanced pathogen isolation and identification are efficient ways to avoid spreading and properly treat infectious diseases. There have been several successful reverse microbial etiology cases reported recently. Anaplasma capra was first isolated and identified, and then the patients related to this Anaplasma were informed (Li et al., 2015b; Wei et al., 2017). Additionally, Escherichia marmotae was identified as a human pathogen when four infected cases were reported to have been caused by Escherichia marmotae after our team first isolated it in 2015 (Liu et al., 2015; Liu et al., 2019; Sivertsen et al., 2022).
Herein, we characterized a novel species in the genus Erwinia (named Erwinia sorbitola sp. nov.) from the potential microbial pathogen vector ruddy shelduck, and the pathogenicity of this pathogen on plants and animals was explored in this study.
Immediately after ruddy shelducks (T. ferruginea) took flight, droppings samples (20 samples) of these birds were freshly collected from the ground and stored in sterile tubes. The fecal samples were collected at two different geographical locations (E 91°48′22″/N27°54′23″ and E 91°58′53″/N27°59′16″) in Cona County, Tibet. Pieces of material (approximate 1-2 g) were diluted in sterile water from 10-1 to 10-6 and homogenized by shaking, then 100 μL of the 10-4 dilution was spread onto Nutrient Agar (NA) plates. After five days of aerobic incubation at 28 °C, different morphological colonies were randomly picked and separated. Strains J780T and J316 from among all the beige and opaque colony isolates (J178, J312, J316, J556, and J780T) were selected based on a comparative analysis of 16S rRNA gene sequences. The reference strains E. aphidicola DSM 19347T, E. persicina GDMCC 1.331T, and E. rhapontici ATCC 29283T were obtained from the Japan Collection Microorganisms (JCM), Guangdong Microbial Culture Collection Center (GDMCC), and China General Microbiological Culture Collection Center (CGMCC), respectively.
The genomic DNA of strains J780T and J316 was extracted using the Wizard Genomic DNA Purification Kit (Promega). The 16S rRNA genes were cloned by universal bacterial primers 27F/1492R (5’-agagtttgatcmtggctcag-3’and 5’-ggytaccttgttacgactt-3’) (Ge et al., 2020) and deposited into the GenBank nucleotide database of NCBI (length 1,507 bp, accession numbers MN203624 and MN708966). The sequence similarities to the related type strains were calculated using BLASTn. Sequences of novel and related type strains were aligned using the Clustal_W program (Larkin et al., 2007). Phylogenetic trees based on 16S rRNA gene sequences were constructed with the software package MEGA 7.0 (Kumar et al., 2016), through the algorithms of neighbor-joining (NJ) (Saitou and Nei, 1987), maximum-likelihood (ML) (Felsenstein, 1981), and maximum parsimony (MP) (Kolaczkowski and Thornton, 2004). Confidence levels of branching were determined by bootstrap analysis with 1,000 replicates (Felsenstein, 1985). Pseudomonas aeruginosa LMG 1242T was used as an outgroup.
The optimal growth conditions for strains J780T and J316 were detected in nutrient broth at different temperatures (4, 16, 28, 30, 35, 37, and 42°C), salinities [0.5–10% (w/v) NaCl at intervals of 0.5%], and pH [(3–12) at intervals of 1.0 pH unit]. Cell morphology, Gram-staining, and colony appearance were observed using isolated bacterial stains grown on NA plates at 28°C for 24 h. Cell morphology was observed by a scanning electron microscope (SU8010, Hitachi). Gram-staining was performed using a Gram-staining kit (Baso) (Austrian, 1960). With E. aphidicola DSM 19347T, E. persicina GDMCC 1.331T, and E. rhapontici ATCC 29283T as positive controls for the motility assay, cell motility was tested by growing in semi-solid agar at 22°C for 48 h (Xu et al., 2013). The catalytic activity of catalase was detected using 3% (v/v) H2O2. Enzymatic activity and acid production from various carbohydrates of the novel strains were determined by API ZYM, API 50 CHB, API 20E, and API 20NE strips (bioMérieux), and strains E. aphidicola DSM 19347T, E. persicina GDMCC1.331T, and E. rhapontici ATCC 29283T were used as the parallel references.
Chemotaxonomic analyses were performed after bacteria had grown on NA plates at 28°C for 24 h. The cellular fatty acids were extracted and analyzed in parallel according to the instructions of the Sherlock Microbial Identification System (MIDI) (Sasser, 1990). The respiratory quinones were analyzed by HPLC (Minnikin et al., 1984; Tamaoka, 1986).
The whole genome sequence of strain J780T was determined using PacBio RSII single-molecule real-time (SMRT) sequencing (PacBio). Through the hierarchical genome assembly process (HGAP) protocol (Version 4.0) in SMRT Analysis (Version 2.3.0), de novo assembly of sequencing reads was performed, and one circular genome without gaps was generated. With strains E. aphidicola DSM 19347T and E. rhapontici ATCC 29283T as references, strain J316 was sequenced on the Illumina X Ten platform following a paired-end protocol and assembled by the SOAP de novo (Illumina) (Luo et al., 2012; Li et al., 2015a), and the protein-coding sequences (CDSs), tRNAs, and rRNAs were predicted using GeneMarkS+4.2 software (Berlin et al., 2015). The average nucleotide identity (ANI) and digital DNA-DNA hybridization (dDDH) values were examined with the OrthoANIu tool (https://www.ezbiocloud.net/tools/ani) (Yoon et al., 2017) and the genome-to-genome distance calculation (GGDC 2.1) (Auch et al., 2010; Meier-Kolthoff et al., 2013), respectively. To further elucidate the taxonomy position of strains J780T and J316 in the phylogenetic relationship, core genes (Chen et al., 2013) were extracted from the genome sequences of the isolates and 13 species in the genus Erwinia and aligned to generate the phylogenomic tree through FastTree (Price et al., 2009). The phylogenetic tree was further edited with Dendroscope.
The pathogenicity of isolates was predicted by the Pathogen-Host Interactions database (PHI) and the Virulence Factors of Pathogenic Bacteria (VFDB). Amino acid sequences were compared in the databases by the program DIAMOND (cutoff: 40% identity). When the annotation results were over one, the best annotation was selected by ensuring the biological functions. The arrangement and similarities of gene clusters were illustrated with Easyfig (Version 2.2.2). The amino acid similarities of type VI secretion system effector genes were compared through BlastP on the NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
The virulence tests on these bacterial isolates were performed by adopting leaf-clipping and pear-fruit inoculation methods as previously described (Dow et al., 2003; Tian et al., 2017). The leaf models included arabidopsis (Arabidopsis thaliana, At), tobacco (Nicotiana benthamiana, Nb), potato (Solanum tuberosum, St), Chinese flowering crabapple (Malus halliana, Mh), and cherry blossom (Prunus campanulate, Pc), and the plant leaves were gifted by FAFU, (Fujian Agriculture and Forestry University). The pear fruits were purchased from a commercial market (Changping, Beijing). In short, the bacteria (J178, J312, J316, J556, J780T, E. aphidicola DSM 19347T, and E. persicina GDMCC 1.331T) were harvested at the logarithmic phase by centrifugation, and the bacterial precipitates were washed with PBS three times before inoculating the plants. The leaves were clipped and inoculated with an equal load of bacteria. Pear fruits were inoculated with 100 μL bacteria suspension through disposable needles (1 mL range). All the infection models were incubated in high humidity at 28°C. Symptoms were observed and recorded within the following days.
Adhesion assay was done according to the following description (Chen et al., 2017). RAW264.7 cells (mice macrophage) were cultured in DMEM (supplemented with 10% FBS), and 1×105 cells/well were plated to 24 multi-well plates one day before the experiment. 1×107 CFU/well logarithmic phase bacteria (strain: J178, J312, J316, J556, and J780T) were mixed with RAW264.7 cells washed with PBS after they were centrifuged and resuspended in DMEM (without FBS). Plates were incubated at 37°C supplied with 5% CO2 for two hours, then washed five times with PBS. The infected RAW264.7 cells were lysed for 10 min with 1% saponin (Sigma-Aldrich), and the lysates were diluted with PBS from 1:1000 to 1:10000.
Based on the above, the invasion assay was conducted through another two hours of antibiotic treatments. The infected RAW264.7 cells were also lysed for 10 min with 1% saponin (Sigma-Aldrich), and the lysates were diluted with PBS from 1:100 to 1:1000. Plate count was adopted for quantitation of the colony forming unit (CFU) of live bacteria in samples. All the assays were done three times. The cytotoxicity assay was carried out following the protocol of the CytoTox 96® Non-Radioactive Cytotoxicity Assay (Promega).
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequence of strains J780T and J316 are MN203624 and MN708966, respectively. The GenBank/EMBL/DDBJ accession numbers for the genome sequence of strains J780T, J316, E. rhapontici ATCC 29283T, and E. aphidicola DSM 19347T are CP046509–CP046510, WLZX00000000, WUUO00000000, and JACXBP000000000, respectively.
The bacterial cells of J780T grown on the NA plate were observed under the scanning electron microscope. The result showed that the J780T strain was rod-shaped (1.5-2.0 mm in length and 0.6-1.0 mm in diameter and Gram-stain-negative (stained with a Gram-staining kit (Baso) (Austrian, 1960)). The catalase activity was tested by treating the J780T strain with 3% (v/v) H2O2, and the results showed that the enzymatic activity of this strain was positive. By growing in semi-solid agar at 22°C for 48 h, the observed results showed that the J780T strain was motile. The colonies of the strain J780T grown on NA were circular, low convex, smooth, entire edge, and beige. It was different from the pink color of E. persicina GDMCC 1.331T grown on TSA at 28°C for 24 h. Through the research on serial growth conditions, it was found that the best combination conditions for growth in both strains J780T and J316 were under 28°C (the screening range: 4-40°C), at pH7.0 (pH4.0-10.0), and with 0.5% (w/v) NaCl (0.5-5.0% (w/v)).
All strains (including J780T, J316, E. persicina, and E. rhapontic) emerged in a state of diffusion growth within 24 h when inoculated into semi-solid agar at 22°C. Any one of these tests, including the tests for N-acetyl-β-glucosaminidase, potassium 5-ketogluconate, and citrate, d-Sorbitol, and d-Turanose utilization, could distinguish the strains J780T and J316 from their closely related species E. aphidicola DSM 19347T, E. persicina GDMCC 1.331T, and E. rhapontici ATCC 29283T (Table 1).
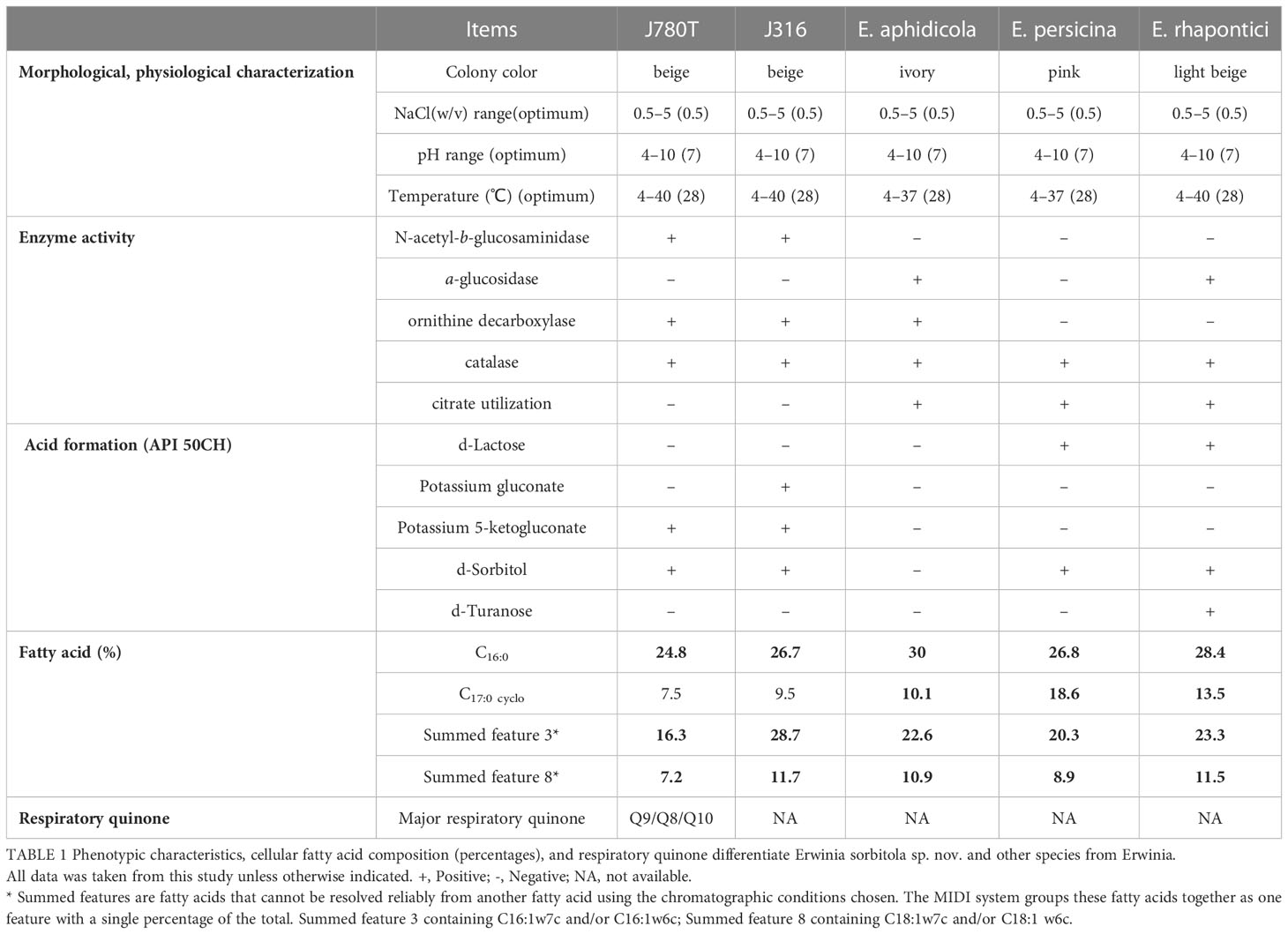
Table 1 Phenotypic characteristics, cellular fatty acid composition (percentages), and respiratory quinone differentiate Erwinia sorbitola sp. nov. and other species from Erwinia.
The major cellular fatty acids were determined by Sherlock Microbial Identification System (MIDI) (Sasser, 1990). The results showed that the major cellular fatty acids (>10%) in strain J780T and J316 were C16:0 (24.8%, 26.7%), summed feature 3 (C16:1ω7c/16:1ω6c, 16.3%, 28.7%), summed feature 2 (C14:0 3OH and/or C16:1 iso I, 15.8%, 7.1%), and summed feature 8 (C18:1ω7c and/or C18:1 ω6c 7.2%, 11.7%). The major respiratory quinones of strain J780T were Q9 (78.5%) with minor Q8 (17.9%) and Q10 (3.6%) (Table 1), which was different from the control strain E. endophytica LMG 28457T, the only known species in the genus Erwinia with only determined respiratory quinone Q8 (Ramirez-Bahena et al., 2016).
The sequencing and comparison results showed that the sequence of the 16S rRNA gene of strain J780T shared 98.2% similarity with that of E. rhapontici, 98.1% with E. persicina, and 97.9% with E. piriflorinigrans CFBP 5888T. In contrast, strain J316 displayed 98.3%, 98.0%, and 97.8% similarity with these strains, respectively. Strains J780T and J316 shared up to 99.7% 16S rRNA gene sequence similarity while sharing less than 98.7% 16S rRNA gene sequence similarity to any other strains in the genus Erwinia. In the phylogenetic tree constructed based on 16S rRNA genes (Figure 1A; Supplementary Figure 1), strains J780T and J316 clustered with E. rhapontici ATCC 29283T, E. aphidicola DSM 19347T, and E. persicina GDMCC1.331T were phylogenetically closest to E. rhapontici ATCC 29283T. Besides 16S rRNA gene-based analysis, the phylogenetic tree based on 916 core gene sequences (Figure 1B) also showed similar results of classification that strains J780T and J316 were closest with E. rhapontici ATCC 29283T and E. persicina GDMCC1.331T, which supporting the affiliation of strains J780T and J316 to the genus Erwinia.
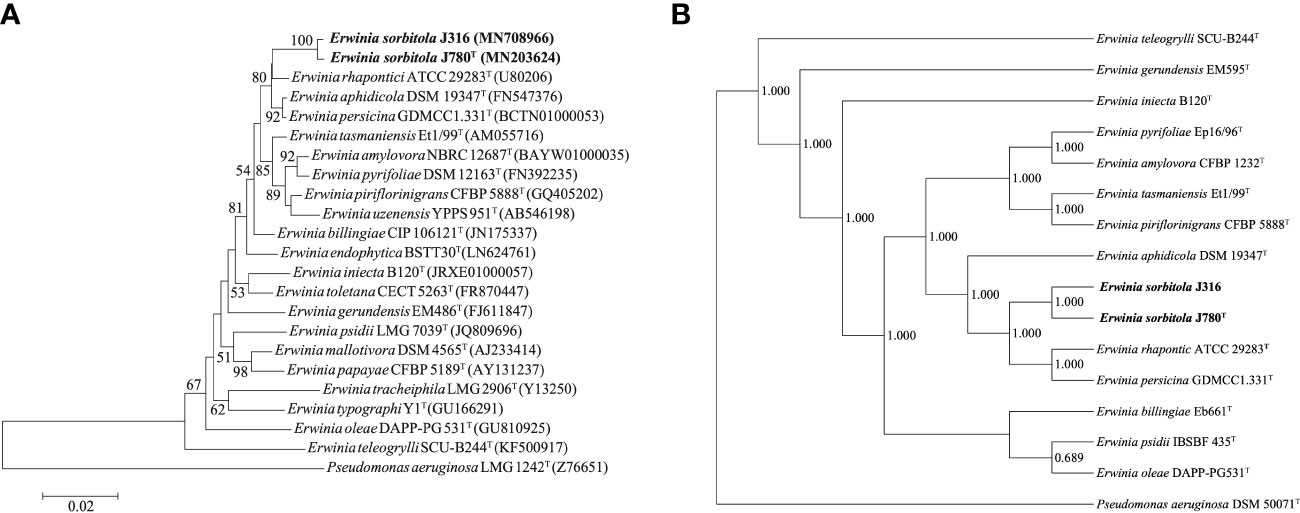
Figure 1 The phylogenetic analyses of Erwinia sorbitola sp. nov. and other species from Erwinia. (A) Neighbor-joining phylogenetic tree based on partial 16S rRNA gene sequences, showing the relationships between strains J316, J780T, and members of the genus Erwinia (Bootstrap values (> 50%) calculated for 1,000 replications are shown at branch nodes; Bar, 0.02 substitutions per nucleotide); (B) the Phylogenomic tree of strains J316 and J780T based on 916 core genes (numbers on the core gene tree indicate each split in the tree with support values from the Shimodaira-Hasegawa test calculated for 1,000 resamples). Pseudomonas aeruginosa LMG 1242T was used as the outgroup.
The ANI values of strains J780T and J316 with the closely related species E. aphidicola DSM 19347T, E. persicina GDMCC1.331T, and E. rhapontici ATCC 29283T ranged from 81.7 to 85.0%, while the dDDH values ranged from 25.1 to 29.5%. The ANI and dDDH values between strains J780T and J316 were 99.6 and 96.5%, respectively. Furthermore, the values of ANI and dDDH between each isolate and Erwinia species were lower than the proposed threshold of 95-96 and 70%, respectively, upholding the view that strains J780T and J316 were novel species of the genus Erwinia.
In conclusion, multiple in-depth analyses based on phylogenetic, physiological, and chemotaxonomic properties indicated that strains J780T and J316 belonged to the genus Erwinia. Combined with the ANI and dDDH values compared with other species in the genus, as well as differential phenotypic properties and distinguishing fatty acid profiles, this supported the proposal that these two isolates (strains J780T and J316) were a novel species, for it could utilize d-Sorbitol, which was different from its reference strains, the study named it Erwinia sorbitola sp. nov.
Most of the species in the genus Erwinia have always been known as plant pathogens. Therefore, we compared the genomes of both isolates to the PHI database using the program DIAMOND for screening the plant disease-related genes. The search showed that a few cluster genes might cause plant blight diseases (host: pear and apple) and black rot (host: radish). The detailed information is shown in Supplementary Table 1.
According to the results of the PHI database analysis, the leaves of the three plants, including Arabidopsis (A. thaliana, At), tobacco (N. benthamiana, Nb), and potato (S. tuberosum L., St), and the fruits of two Rosaceae plants, including Chinese flowering crabapple (M. halliana, Mh) and cherry blossom (P. campanulate, Pc) were constructed as infection models in this study. The result displayed no obvious infection symptoms on the leaves of both arabidopsis and tobacco compared to that of the control group, E. coli. However, all the strains (J780T, J178, J312, J316, J556, E. aphidicola DSM 19347T, and E. persicina GDMCC1.331T) caused different levels of the symptoms like the burnt by the fire on potato and cherry blossom leaves, in which strains E. persicina GDMCC1.331T, J178, and J312 induced more severe symptoms on the leaves of potato than the other strains. Moreover, a slightly burnt symptom was found on the leaves of Chinese flowering crabapple in all the experimental groups (Figure 2). All the experiments were repeated twice.
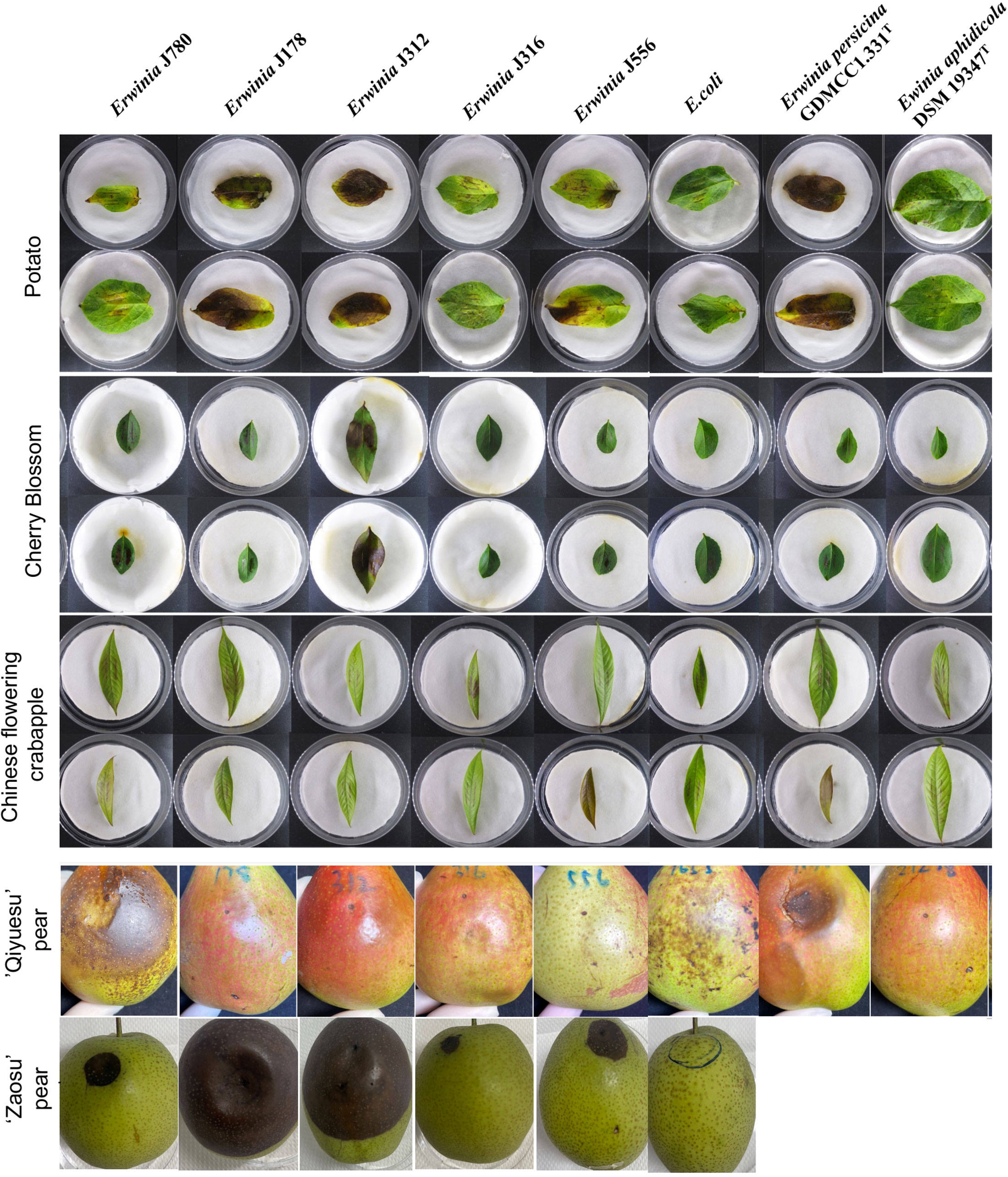
Figure 2 The virulence tests on leaves and pear fruits of E. sorbitola sp. nov. and the type strains of the reference strains (strains: J780T, J178, J312, J316, and J556; reference strains: E. persicina GDMCC 1.331T and E. aphidicola DSM 19347T). The virulence tests on leaves of E. sorbitola sp. nov. were conducted via leaf-clipping method (leaves: potato, cherry blossom, Chinese flowering crabapple); the virulence tests on fruits of E. sorbitola sp. nov. were conducted via injection into pear fruits by needle (fruits: ‘Qiyuesu’ pear, ‘Zaosu’pear). The infection dose was 1×107 CFU/mL, leaves were incubated for three days, and fruits were incubated for 14 days. E. coli was used as negative control strain. Experiments were repeated twice.
In the fruit models, pears fruits were inoculated with isolates, and E. coli was taken as the negative control. Depending on the different bacterial species infected, a discrepancy in the appearance of infected symptoms was present on the infected pear fruits. After 14 d inoculation, the pear fruits (‘Qiyuesu’ pear (P. pseudopashia, Pp), ‘Zaosu’ pear (P. bretschneideri cv. Zaosu, Pbz), and ‘Anhui Dangshan’ pear (P. bretschneideri Rend, Pbr)) infected by J780T and E. persicina GDMCC1.331T showed symptoms of rot, but no obvious appearance of infected symptom showed up on the pears infected by the other bacterial strains (Figure 2, ‘Qiyuesu’ pear). After 14 d inoculation, the pear fruits (‘Zaosu’ pear) inoculated with strains of E. sorbitola sp. nov. all presented symptoms of water-soaking and necrosis-infection, but no obvious infected symptoms were developed in the control group (Figure 2, ‘Zaosu’ pear). A similar test was carried out with the ‘Anhui Dangshan’ pear (Pyrus bretschneideri, Pb) and ‘Guoguang’ apple (Ralls Janet, Rj) fruits. The results showed that symptoms of rot were present on the pears infected by Erwinia persicina GDMCC1.331T (Supplementary Figure 2), but no symptom could be observed in the apple model (data not shown). These results proposed that, similar to other members of the genus Erwinia, the novel isolates of E. sorbitola sp. nov. were phytopathogenic to pear fruits, and E. persicina GDMCC1.331T may be a hypervirulent strain with a wide range of susceptible hosts.
Considering the phytopathogenic phenotype of these two isolates of Erwinia, their potential virulence factors were characterized by analysis through the VFDB and PHI database based on the genomes of both isolates. Moreover, the results showed that the pathogenicity of this novel bacterium might be determined by the following pathogenic characteristics, including motility (Supplementary Table 2), biofilm formation (Supplementary Table 3), and exopolysaccharides (EPS) amylovoran Figure 3D; stress survival abilities and siderophores (Supplementary Table 5); and Type VI secretion system (Supplementary Table 6). Based on the predicted results, serial follow-up experiments were carried out.
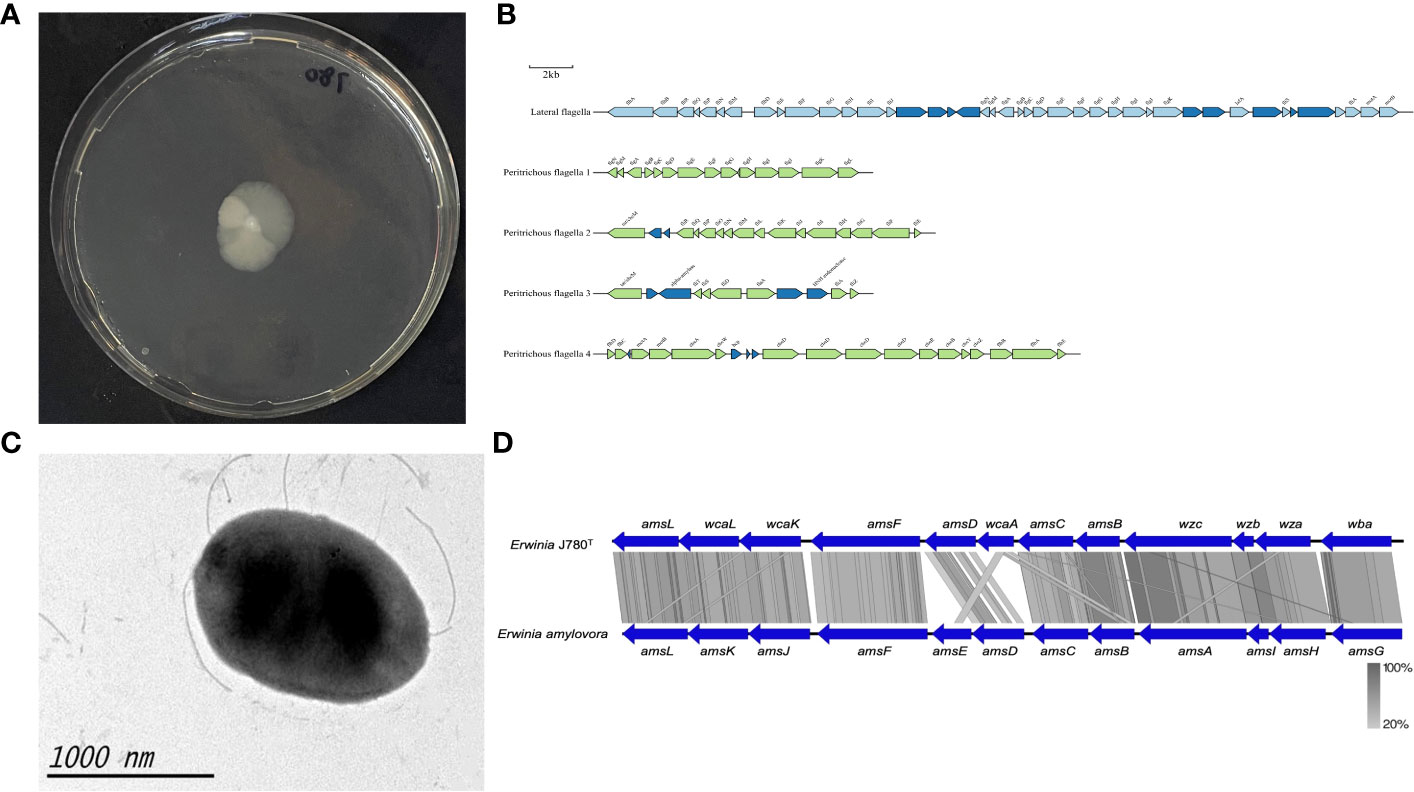
Figure 3 The motility and amylovoran biosynthesis genes of E. sorbitola sp. nov. (strain: J780T). (A) Bacterial swimming assay of the strain J780T; (B) schematic representation of flagella gene clusters of the strain J780T. Proposed gene nomenclature is shown above the corresponding ORFs (the scale bar represents 2kb); (C) transmission electron micrograph of the strain J780T (the scale bar represents 1000nm); (D) the comparison of amylovoran biosynthesis genes between the strain J780T and Erwinia amylovora. The figure was compared using BlastX and pictured with Easyfig.
The motility of J780T was tested via bacterial swimming assay. The result showed that it possessed high motility (Figure 3A). Prediction information from this study inferred that it might own lateral flagella and peritrichous flagella (Figure 3B). The detailed flagella related encoding genes are listed in a supplementary table (Supplementary Table 2). Moreover, other visualization under transmission electron microscopy confirmed that its flagella played an essential role in its high motility (Figure 3C). Meanwhile, our prediction information also revealed that there were four gene clusters related to pili, including type I fimbriae, PMF fimbriae, mannose-resistant fimbriae, and separated genes on its genome with the type IV pili annotation (Supplementary Table 3). The amylovoran synthesis gene cluster contains 12 structural genes (from amsA to amsL) (Bugert and Geider, 1995) in the chromosome of E. amylovora. In this study, a similar ams gene cluster associated with amylovoran biosynthesis was screened from the genomes of J780T (Figure 3D), and related detailed information is given in the supplementary materials (Supplementary Table 4). The above results displayed that the characteristics, including motility, biofilm formation, and exopolysaccharides (EPS) amylovoran, were deeply involved in the formation of virulence of this novel bacterium to host plants.
Resistance and adaptation to hostile environments are essential virulence factors for pathogens. According to VFDB prediction in this study, genes associated with the pathogens’ stress survival, including catalase and metal ion transport, were screened from the genome of E. sorbitola sp. nov. (Supplementary Table 5). In these predicted associated genes, at least three genes, including katA (J780_GM000409), katG (J780_GM000646), and ahpC (J780_GM001015), were coding as peroxiredoxin related with catalase activity, which was consistent with the positive results of catalase activity in this study. The iron uptake systems, including the direct heme uptake system (Hogle et al., 2017), ferrous iron (Fe2+) transport (Seo et al., 2014), and siderophores (Aznar et al., 2014), played an indispensable role in bacteria survival in iron-limited conditions and regulation of virulence activities. There were three siderophores predicted in the genomes of the novel isolates in this study. They were siderophores chrysobactin (Rauscher et al., 2002; Sandy and Butler, 2011), achromobactin (Berti and Thomas, 2009), and enterobactin (Gorshkov et al., 2021). Notably, in this study, the three predicted siderophores in E. sorbitola sp. nov. showed high similarities to those of Dickeya dadantii (Figures 4A–C). These results suggested that catalase and metal ion transport play an essential role in the pathogenic process of E. sorbitola sp. nov. to its host plants.

Figure 4 The comparison of siderophores genes between E. sorbitola sp. nov. (J780T) and Dickeya dadantil. (A) The comparison of achromobactin gene clusters; (B) the comparison of chrysobactin gene clusters; (C) the comparison of enterobactin gene clusters. The proposed gene nomenclature is shown above the corresponding ORFs. All the figures were compared using BlastX and pictured with Easyfig.
There were four gene clusters related to the Type VI secretion system (T6SS), including three integrated T6SSs and one auxiliary gene cluster. The T6SS constitutions of J780T are shown in Supplementary Table 6. The auxiliary gene cluster started at the locus J780_GM000434 and ended with J780_GM000441, which contained 8 ORFs. Among these ORFs, the PIN may be one of the effector candidates of the auxiliary gene cluster. The other integrated T6SSs had 21, 24, and 28 ORFs containing all 13 ORFs core genes of the T6SS (Chen et al., 2015; Crisan and Hammer, 2020). T6SS-1 was from J780_GM001628 to J780_GM001644, and T6SS-2 clusters were from J780_GM002048 to J780_GM002071. The T6SS cluster 3 (from J780_GM003723 to J780_GM003746) structure was like Citrobacter rodenthum CTS1(T6SS1) but with an additional fimbriae operon in it (Gueguen and Cascales, 2013).
The T6SS-2 and auxiliary gene clusters in the J780T genome shared homologs with E. amylovora, as predicted by the PHI database, are listed in Supplementary Table 7. Almost all the T6SS core components of E. sorbitola sp. nov. shared homologous with E. amylovora, and it was reported that the deletion of T6SS structure components and effectors in E. amylovora caused a decrease in amylovoran production and reduction in bacteria virulence (Tian et al., 2017). These findings suggested that T6SS-2 and auxiliary clusters may function as phytopathogenic determinants in E. sorbitola sp. nov. as well.
This study predicted several polysaccharide biosynthesis gene clusters, including LOS, LPS, and CPS. They were involved in immune system evasion and anti-phagocytosis (Supplementary Table 4). Type I fimbriae and mannose-resistant fimbriae also functioned as adhesive factors (Supplementary Table 3). They may promote the adherence of pathogens to bladder cells and macrophages (Mohammad Zadeh et al., 2021). These predictions inferred that E. sorbitola sp. nov. may have high adhesion and invasion abilities. To identify the adhesion, invasion abilities, and cytotoxicity of the bacteria, we counted the bacteria burdens attached to the cell surfaces and invaded cells.
As the results shown in Figure 5, the bacteria burdens attached to cells ranged from 105.4 to 106.7 CFU/mL after two hours of coculture (Figure 5A). The bacteria burdens that had invaded cells ranged from 104 to 104.7 CFU/mL after two hours of inoculation and two hours of antibiotic treatment (Figure 5B). The cytotoxicity of E. sorbitola sp. nov. on cells ranged from 20-40% at 12 hours post-infection and significantly increased at 24 hours post-infection compared with 12 hours post-infection (Figure 5C). All these results were consistent with the predicted information. These results indicated that E. sorbitola sp. nov. might have pathogenicity on mice cells.
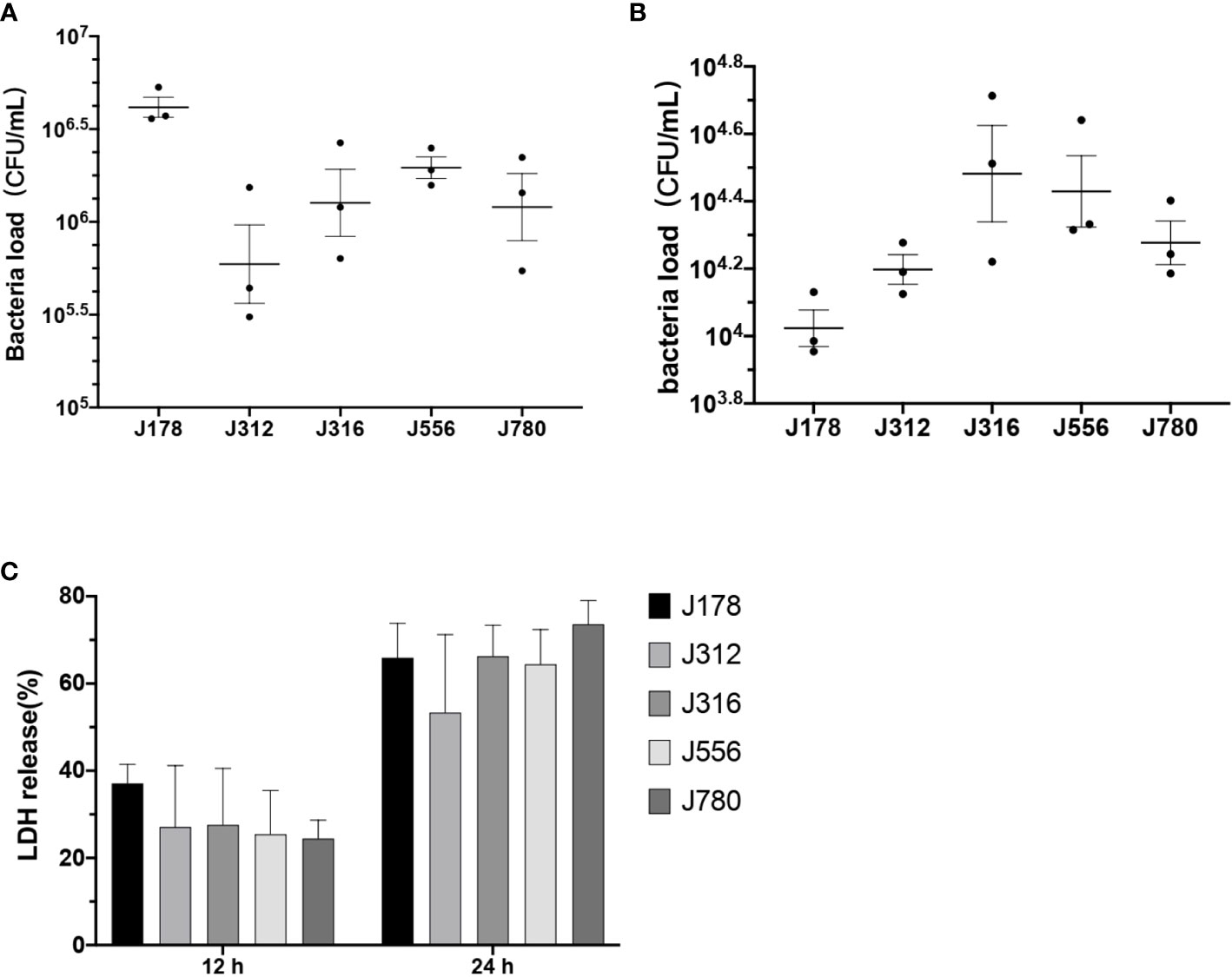
Figure 5 The adhesion, invasion, and cytotoxic ability of Erwinia sorbitola sp. nov. on RAW264.7 cells. (A) The number of bacterial loads adhered on cells; (B) the bacterial loads that invaded cells; (C) LDH released in the medium was used to assess the cytotoxicity of Erwinia sorbitola sp. nov. (Strains: J178, J312, J316, J556, and J780T. Infection dose: MOI 100. Adherence time: two hours and two hours of antibiotic treatment to kill the exocellular bacteria. All the assays were repeated three times, and each experiment was performed in triplicate. Results were presented as Mean ± SD).
Infection experiments on plant leaves and pear fruits in this study (Figure 2) identified the pathogenicity of Erwinia sorbitola sp. nov. but its mechanisms are still unclear. Using bioinformatics analysis with comparison to other closely related plant pathogens (Figures 3D, 4A–C), we preliminarily identified and analyzed several possible points related to its pathogenicity: motility, biofilm formation, and exopolysaccharides (EPS) amylovoran; stress survival and siderophores; and Type VI secretion system (Bell et al., 2004).
Firstly, Pili synthesis was predicted in E. sorbitola sp. nov. A former study determined that the defect of pili resulted in decreased virulence (Koczan et al., 2011). Also, the phytopathogenic determinant amylovoran was identified in the J780T genome and shared a high degree of homology with the plant pathogen E. amylovora (Figure 3D); exopolysaccharides (EPS) amylovoran also joined in biofilm formation and motility of microbial pathogens in the host plants, and the mutants deficient in amylovoran production were found to be not phytopathogenic (Bellemann et al., 1994; Kharadi et al., 2019). Flagella and pili together with amylovoran played vital roles in the survival of the bacteria outside the host and also for the attachment to host cell surfaces (van der Bosch et al., 1980; Connell et al., 1996; Rocha et al., 2007; Santander et al., 2014), which supported the above conclusion.
Secondly, E. sorbitola sp. nov. may significantly enhance its adaptability to adverse environments, primarily through its catalase and metal ion transport systems (Supplementary Table 5). katA and katG were found to contribute to fire blight symptoms and the progression of necrosis in immature fruits (Santander et al., 2018). Moreover, it was found in this study that siderophores in both isolate genomes shared high homology to that of D. dadantii. It was reported that the sidtheerphore in D. dadantii (previously Erwinia chrysanthemi) could cause metal homeostasis disturbances and callose deposition in host plants. Also, it contributes to a bacterial antioxidant effect by reducing ROS accumulation in bacteria (Dellagi et al., 2005; Kieu et al., 2012).
Although T3SS, a critical virulence factor of species in genus Erwinia, was not identified in strain J316 and J780T, four T6SS gene clusters were found from this new species of Erwinia (Supplementary Table 6). This study showed that the T6SS-2 and auxiliary gene clusters in the type VI secretion system of E. sorbitola sp. nov. shared a high homology to that of E. amylovora (Supplementary Table 7). It has been reported that the T6SS-2 mutant E. amylovora strain had significantly reduced virulence, possibly caused by amylovoran secretion reduction (Tian et al., 2017).
Only a part of the phytopathogenic determinants was identified and characterized in this study. There must be other determinants that still need to be discovered. Further, exploring more phytopathogenic determinants will contribute to finding the pathogenic mechanisms of E. sorbitola sp. nov.
Adherence, invasion, and cytotoxicity assays were applied in this study to confirm the capacity of this new pathogen regarding adherence, invasion, and cytotoxicity (Figure 5). Several determinants related to animal pathogenicity were identified and characterized in this study as well. Furthermore, the reference strains E. persicina (Mohamaden et al., 2019; Ceylan and Ozden, 2022), E. tasmaniensis (Shin et al., 2008a) and E. billingiae (Prod'homme et al., 2017; Bonnet et al., 2019) were related to several human infectious diseases. This study speculated that E. sorbitola sp. nov. may possess specific animal pathogenicity. However, more exploration of the animal pathogenicity of E. sorbitola sp. nov. should be done for further analysis.
In summary, this study isolated and identified a novel species (E. sorbitola sp. nov.) in the genus Erwinia from ruddy shelducks. This study characterized it as a phytopathogen through pathogenicity analysis based on genomic sequences and plant virulence assays. Moreover, a primary pathogenetic analysis based on the model of animal cells suggested it might also have a degree of animal pathogenicity.
Emerging infectious diseases are often caused by new microorganisms. Several pre-identifications of pathogens have been reported to cause human infections. Reverse microbial etiology is one way to study emerging infectious, and the methods of this study followed previous presented procedures (Xu, 2019). When emerging infectious diseases affect plants, primarily commercial crops, it can cause substantial economic losses. Our research focused on the early isolation and identification of novel pathogens to provide faster and more precise ways for the prevention and control of emerging infectious diseases, such as the new pathogen Erwinia sorbitola sp. nov. discovered in this study, might provide the early diagnosis and control for an unknown infectious disease.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by Ethical Committee of the National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention (#: ICDC-2016004).
JX and HZ contributed to conception and design of the study and foundation support. JY, WS, DJ organized the database, YT, HL, SL performed the statistical analysis. YT, YG, ZZ, WL wrote the first draft of the manuscript. YH wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by grants from the National Key R&D Program of China (2019YFC1200500 and 2019YFC1200505) and Research Units of Discovery of Unknown Bacteria and Function (2018RU010).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1109634/full#supplementary-material
Amin, N. M., Bunawan, H., Redzuan, R. A., Jaganath, I. B. (2010). Erwinia mallotivora sp., a new pathogen of papaya (Carica papaya) in peninsular Malaysia. Int. J. Mol. Sci. 12 (1), 39–45. doi: 10.3390/ijms12010039
Auch, A. F., von Jan, M., Klenk, H. P., Goker, M. (2010). Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2 (1), 117–134. doi: 10.4056/sigs.531120
Austrian, R. (1960). The gram stain and the etiology of lobar pneumonia, an historical note. Bacteriol Rev. 24 (3), 261–265. doi: 10.1128/br.24.3.261-265.1960
Aznar, A., Chen, N. W., Rigault, M., Riache, N., Joseph, D., Desmaele, D., et al. (2014). Scavenging iron: a novel mechanism of plant immunity activation by microbial siderophores. Plant Physiol. 164 (4), 2167–2183. doi: 10.1104/pp.113.233585
Bellemann, P., Bereswill, S., Berger, S., Geider, K. (1994). Visualization of capsule formation by erwinia amylovora and assays to determine amylovoran synthesis. Int. J. Biol. Macromol 16 (6), 290–296. doi: 10.1016/0141-8130(94)90058-2
Bell, K. S., Sebaihia, M., Pritchard, L., Holden, M. T., Hyman, L. J., Holeva, M. C., et al. (2004). Genome sequence of the enterobacterial phytopathogen erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc. Natl. Acad. Sci. U.S.A. 101 (30), 11105–11110. doi: 10.1073/pnas.0402424101
Berlin, K., Koren, S., Chin, C. S., Drake, J. P., Landolin, J. M., Phillippy, A. M. (2015). Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 33 (6), 623–630. doi: 10.1038/nbt.3238
Berti, A. D., Thomas, M. G. (2009). Analysis of achromobactin biosynthesis by pseudomonas syringae pv. syringae B728a. J. Bacteriol 191 (14), 4594–4604. doi: 10.1128/JB.00457-09
Bonnet, I., Bozzi, B., Fourniols, E., Mitrovic, S., Soulier-Escrihuela, O., Brossier, F., et al. (2019). Erwinia billingiae as unusual cause of septic arthritis, franc. Emerg. Infect. Dis. 25 (8), 1587–1589. doi: 10.3201/eid2508.181073
Bugert, P., Geider, K. (1995). Molecular analysis of the ams operon required for exopolysaccharide synthesis of erwinia amylovora. Mol. Microbiol. 15 (5), 917–933. doi: 10.1111/j.1365-2958.1995.tb02361.x
Ceylan, A. N., Ozden, I. (2022). [NA rare bacterial pathogen in a patient with perihilar cholangiocarcinoma: Erwinia persicina; first case from Turkey]. Mikrobiyol Bul 56 (3), 574–579. doi: 10.5578/mb.20229716
Chen, W., Liu, Y., Zhang, L., Gu, X., Liu, G., Shahid, M., et al. (2017). Nocardia cyriacigeogica from bovine mastitis induced In vitro apoptosis of bovine mammary epithelial cells via activation of mitochondrial-caspase pathway. Front. Cell Infect. Microbiol. 7. doi: 10.3389/fcimb.2017.00194
Chen, C., Zhang, W., Zheng, H., Lan, R., Wang, H., Du, P., et al. (2013). Minimum core genome sequence typing of bacterial pathogens: a unified approach for clinical and public health microbiology. J. Clin. Microbiol. 51 (8), 2582–2591. doi: 10.1128/JCM.00535-13
Chen, L., Zou, Y., She, P., Wu, Y. (2015). Composition, function, and regulation of T6SS in pseudomonas aeruginosa. Microbiol. Res. 172, 19–25. doi: 10.1016/j.micres.2015.01.004
Connell, I., Agace, W., Klemm, P., Schembri, M., Marild, S., Svanborg, C. (1996). Type 1 fimbrial expression enhances escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. U.S.A. 93 (18), 9827–9832. doi: 10.1073/pnas.93.18.9827
Crisan, C. V., Hammer, B. K. (2020). The vibrio cholerae type VI secretion system: toxins, regulators and consequences. Environ. Microbiol. 22 (10), 4112–4122. doi: 10.1111/1462-2920.14976
Dellagi, A., Rigault, M., Segond, D., Roux, C., Kraepiel, Y., Cellier, F., et al. (2005). Siderophore-mediated upregulation of arabidopsis ferritin expression in response to erwinia chrysanthemi infection. Plant J. 43 (2), 262–272. doi: 10.1111/j.1365-313X.2005.02451.x
Dow, J. M., Crossman, L., Findlay, K., He, Y. Q., Feng, J. X., Tang, J. L. (2003). Biofilm dispersal in xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. U.S.A. 100 (19), 10995–11000. doi: 10.1073/pnas.1833360100
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17 (6), 368–376. doi: 10.1007/BF01734359
Felsenstein, J. (1985). Confidence-limits on phylogenies - an approach using the bootstrap. Evolution 39 (4), 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Gardan, L., Christen, R., Achouak, W., Prior, P. (2004). Erwinia papayae sp. nov., a pathogen of papaya (Carica papaya). Int. J. Syst. Evol. Microbiol. 54 (Pt 1), 107–113. doi: 10.1099/ijs.0.02718-0
Ge, Y. J., Tao, Y. M. H., Yang, J., Lai, X. H., Jin, D., Lu, S., et al. (2020). Arthrobacter yangruifuii sp. nov. and arthrobacter zhaoguopingii sp. nov., two new members of the genus arthrobacter. Int. J. Systematic Evolutionary Microbiol. 70 (10), 5287–5295. doi: 10.1099/ijsem.0.004414
Gorshkov, V., Parfirova, O., Petrova, O., Gogoleva, N., Kovtunov, E., Vorob'ev, V., et al. (2021). The knockout of enterobactin-related gene in pectobacterium atrosepticum results in reduced stress resistance and virulence towards the primed plants. Int. J. Mol. Sci. 22 (17):9594. doi: 10.3390/ijms22179594
Gueguen, E., Cascales, E. (2013). Promoter swapping unveils the role of the citrobacter rodentium CTS1 type VI secretion system in interbacterial competition. Appl. Environ. Microbiol. 79 (1), 32–38. doi: 10.1128/AEM.02504-12
Harada, H., Oyaizu, H., Kosako, Y., Ishikawa, H. (1997). Erwinia aphidicola, a new species isolated from pea aphid, acyrthosiphon pisum. J. Gen. Appl. Microbiol. 43 (6), 349–354. doi: 10.2323/jgam.43.349
Hauben, L., Moore, E. R. B., Vauterin, L., Steenackers, M., Mergaert, J., Verdonck, L., et al. (1998). Phylogenetic position of phytopathogens within the enterobacteriaceae. Systematic Appl. Microbiol. 21 (3), 384–397. doi: 10.1016/S0723-2020(98)80048-9
Hogle, S. L., Brahamsha, B., Barbeau, K. A. (2017). Direct heme uptake by phytoplankton-associated roseobacter bacteria. mSystems 2 (1):e00124-16. doi: 10.1128/mSystems.00124-16
Kharadi, R. R., Castiblanco, L. F., Waters, C. M., Sundin, G. W. (2019). Phosphodiesterase genes regulate amylovoran production, biofilm formation, and virulence in erwinia amylovora. Appl. Environ. Microbiol. 85 (1):e02233-18. doi: 10.1128/AEM.02233-18
Kharadi, R. R., Schachterle, J. K., Yuan, X., Castiblanco, L. F., Peng, J., Slack, S. M., et al. (2021). Genetic dissection of the erwinia amylovora disease cycle. Annu. Rev. Phytopathol. 59, 191–212. doi: 10.1146/annurev-phyto-020620-095540
Kieu, N. P., Aznar, A., Segond, D., Rigault, M., Simond-Cote, E., Kunz, C., et al. (2012). Iron deficiency affects plant defence responses and confers resistance to dickeya dadantii and botrytis cinerea. Mol. Plant Pathol. 13 (8), 816–827. doi: 10.1111/j.1364-3703.2012.00790.x
Kim, W. S., Gardan, L., Rhim, S. L., Geider, K. (1999). Erwinia pyrifoliae sp. nov., a novel pathogen that affects Asian pear trees (Pyrus pyrifolia nakai). Int. J. Syst. Bacteriol 49 Pt 2, 899–905. doi: 10.1099/00207713-49-2-899
Koczan, J. M., Lenneman, B. R., McGrath, M. J., Sundin, G. W. (2011). Cell surface attachment structures contribute to biofilm formation and xylem colonization by erwinia amylovora. Appl. Environ. Microbiol. 77 (19), 7031–7039. doi: 10.1128/AEM.05138-11
Kolaczkowski, B., Thornton, J. W. (2004). Performance of maximum parsimony and likelihood phylogenetics when evolution is heterogeneous. Nature 431 (7011), 980–984. doi: 10.1038/nature02917
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi: 10.1093/molbev/msw054
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23 (21), 2947–2948. doi: 10.1093/bioinformatics/btm404
Li, D., Liu, C. M., Luo, R., Sadakane, K., Lam, T. W. (2015a). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de bruijn graph. Bioinformatics 31 (10), 1674–1676. doi: 10.1093/bioinformatics/btv033
Liu, S., Feng, J., Pu, J., Xu, X., Lu, S., Yang, J., et al. (2019). Genomic and molecular characterisation of escherichia marmotae from wild rodents in qinghai-Tibet plateau as a potential pathogen. Sci. Rep. 9 (1), 10619. doi: 10.1038/s41598-019-46831-3
Liu, S., Jin, D., Lan, R., Wang, Y., Meng, Q., Dai, H., et al. (2015). Escherichia marmotae sp. nov., isolated from faeces of marmota himalayana. Int. J. Syst. Evol. Microbiol. 65 (7), 2130–2134. doi: 10.1099/ijs.0.000228
Liu, J., Xiao, H., Lei, F., Zhu, Q., Qin, K., Zhang, X. W., et al. (2005). Highly pathogenic H5N1 influenza virus infection in migratory birds. Science 309 (5738):1206. doi: 10.1126/science.1115273
Li, H., Zheng, Y. C., Ma, L., Jia, N., Jiang, B. G., Jiang, R. R., et al. (2015b). Human infection with a novel tick-borne anaplasma species in China: a surveillance study. Lancet Infect. Dis. 15 (6), 663–670. doi: 10.1016/S1473-3099(15)70051-4
Lopez, M. M., Rosello, M., Llop, P., Ferrer, S., Christen, R., Gardan, L. (2011). Erwinia piriflorinigrans sp. nov., a novel pathogen that causes necrosis of pear blossoms. Int. J. Syst. Evol. Microbiol. 61 (Pt 3), 561–567. doi: 10.1099/ijs.0.020479-0
Luo, R., Liu, B., Xie, Y., Li, Z., Huang, W., Yuan, J., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1 (1), 18. doi: 10.1186/2047-217X-1-18
Marinova, M. H., Georgiev, B. B., Vasileva, G. P. (2015). Description of diorchis thracica n. sp. (Cestoda, hymenolepididae) from the ruddy shelduck tadorna ferruginea (Pallas) (Anseriformes, anatidae) in Bulgaria. Syst. Parasitol. 91 (3), 261–271. doi: 10.1007/s11230-015-9569-9
Matsuura, T., Mizuno, A., Tsukamoto, T., Shimizu, Y., Saito, N., Sato, S., et al. (2012). Erwinia uzenensis sp. nov., a novel pathogen that affects European pear trees (Pyrus communis l.). Int. J. Syst. Evol. Microbiol. 62 (Pt 8), 1799–1803. doi: 10.1099/ijs.0.032011-0
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., Goker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinf. 14, 60. doi: 10.1186/1471-2105-14-60
Minnikin, D. E., Odonnell, A. G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., et al. (1984). An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiological Methods 2 (5), 233–241. doi: 10.1016/0167-7012(84)90018-6
Mohamaden, W. I., Zhen-Fen, Z., Hegab, I. M., Shang-Li, S. (2019). Experimental infection in mice with erwinia persicina. Microb. Pathog. 130, 38–43. doi: 10.1016/j.micpath.2019.01.050
Mohammad Zadeh, F., Zarei, H., Honarmand Jahromy, S. (2021). Type1 and 3 fimbriae phenotype and genotype as suitable markers for uropathogenic bacterial pathogenesis via attachment, cell surface hydrophobicity, and biofilm formation in catheter-associated urinary tract infections (CAUTIs). Iran J. Basic Med. Sci. 24 (8), 1098–1106. doi: 10.22038/IJBMS.2021.53691.12079
O'Hara, C. M., Steigerwalt, A. G., Hill, B. C., Miller, J. M., Brenner, D. J. (1998). First report of a human isolate of erwinia persicinus. J. Clin. Microbiol. 36 (1), 248–250. doi: 10.1128/JCM.36.1.248-250.1998
Pique, N., Minana-Galbis, D., Merino, S., Tomas, J. M. (2015). Virulence factors of erwinia amylovora: A review. Int. J. Mol. Sci. 16 (6), 12836–12854. doi: 10.3390/ijms160612836
Price, M. N., Dehal, P. S., Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26 (7), 1641–1650. doi: 10.1093/molbev/msp077
Prod'homme, M., Micol, L. A., Weitsch, S., Gassend, J. L., Martinet, O., Bellini, C. (2017). Cutaneous infection and bactaeremia caused by erwinia billingiae: a case report. New Microbes New Infect. 19, 134–136. doi: 10.1016/j.nmni.2017.07.006
Ramirez-Bahena, M. H., Salazar, S., Cuesta, M. J., Tejedor, C., Igual, J. M., Fernandez-Pascual, M., et al. (2016). Erwinia endophytica sp. nov., isolated from potato (Solanum tuberosum l.) stems. Int. J. Syst. Evol. Microbiol. 66 (2), 975–981. doi: 10.1099/ijsem.0.000820
Rauscher, L., Expert, D., Matzanke, B. F., Trautwein, A. X. (2002). Chrysobactin-dependent iron acquisition in erwinia chrysanthemi. functional study of a homolog of the escherichia coli ferric enterobactin esterase. J. Biol. Chem. 277 (4), 2385–2395. doi: 10.1074/jbc.M107530200
Rocha, S. P., Pelayo, J. S., Elias, W. P. (2007). Fimbriae of uropathogenic Proteus mirabilis. FEMS Immunol. Med. Microbiol. 51 (1), 1–7. doi: 10.1111/j.1574-695X.2007.00284.x
Saitou, N., Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 (4), 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sandy, M., Butler, A. (2011). Chrysobactin siderophores produced by dickeya chrysanthemi EC16. J. Nat. Prod 74 (5), 1207–1212. doi: 10.1021/np200126z
Santander, R. D., Figas-Segura, A., Biosca, E. G. (2018). Erwinia amylovora catalases KatA and KatG are virulence factors and delay the starvation-induced viable but non-culturable (VBNC) response. Mol. Plant Pathol. 19 (4), 922–934. doi: 10.1111/mpp.12577
Santander, R. D., Oliver, J. D., Biosca, E. G. (2014). Cellular, physiological, and molecular adaptive responses of erwinia amylovora to starvation. FEMS Microbiol. Ecol. 88 (2), 258–271. doi: 10.1111/1574-6941.12290
Sasser, M. (1990). Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Tech. Note 101, 1–7.
Seo, S. W., Kim, D., Latif, H., O'Brien, E. J., Szubin, R., Palsson, B. O. (2014). Deciphering fur transcriptional regulatory network highlights its complex role beyond iron metabolism in escherichia coli. Nat. Commun. 5:4910. doi: 10.1038/ncomms5910
Shin, S. Y., Lee, M. Y., Song, J. H., Ko, K. S. (2008a). New erwinia-like organism causing cervical lymphadenitis. J. Clin. Microbiol. 46 (9), 3156–3158. doi: 10.1128/JCM.00716-08
Shin, S. Y., Lee, M. Y., Song, J. H., Ko, K. S. (2008b). New erwinia-like organism causing cervical lymphadenitis. J. Clin. Microbiol. 46 (9), 3156–3158. doi: 10.1128/Jcm.00716-08
Sivertsen, A., Dyrhovden, R., Tellevik, M. G., Bruvold, T. S., Nybakken, E., Skutlaberg, D. H., et al. (2022). Escherichia marmotae-a human pathogen easily misidentified as escherichia coli. Microbiol. Spectr. 10 (2), e0203521. doi: 10.1128/spectrum.02035-21
Tamaoka, J. (1986). Analysis of bacterial menaquinone mixtures by reverse-phase high-performance liquid-chromatography. Methods Enzymology 123, 251–256. doi: 10.1016/S0076-6879(86)23028-1
Tian, Y., Zhao, Y., Shi, L., Cui, Z., Hu, B., Zhao, Y. (2017). Type VI secretion systems of erwinia amylovora contribute to bacterial competition, virulence, and exopolysaccharide production. Phytopathology 107 (6), 654–661. doi: 10.1094/PHYTO-11-16-0393-R
van der Bosch, J. F., Verboom-Sohmer, U., Postma, P., de Graaff, J., MacLaren, D. M. (1980). Mannose-sensitive and mannose-resistant adherence to human uroepithelial cells and urinary virulence of escherichia coli. Infect. Immun. 29 (1), 226–233. doi: 10.1128/iai.29.1.226-233.1980
Wei, R., Liu, H. B., Jongejan, F., Jiang, B. G., Chang, Q. C., Fu, X., et al. (2017). Cultivation of anaplasma ovis in the HL-60 human promyelocytic leukemia cell line. Emerg. Microbes Infect. 6 (9), e83. doi: 10.1038/emi.2017.70
Xu, J. (2019). Reverse microbial etiology: A research field for predicting and preventing emerging infectious diseases caused by an unknown microorganism. J. Biosaf Biosecur 1 (1), 19–21. doi: 10.1016/j.jobb.2018.12.005
Xu, Y., Xu, X., Lan, R., Xiong, Y., Ye, C., Ren, Z., et al. (2013). An O island 172 encoded RNA helicase regulates the motility of escherichia coli O157:H7. PloS One 8 (6), e64211. doi: 10.1371/journal.pone.0064211
Keywords: phytopathogen, reverse microbial etiology, pathogenicity analysis, Erwinia sorbitola, ruddy shelducks
Citation: Tao Y, Ge Y, Yang J, Song W, Jin D, Lin H, Zheng H, Lu S, Luo W, Huang Y, Zhuang Z and Xu J (2023) A novel phytopathogen Erwinia sorbitola sp. nov., isolated from the feces of ruddy shelducks. Front. Cell. Infect. Microbiol. 13:1109634. doi: 10.3389/fcimb.2023.1109634
Received: 28 November 2022; Accepted: 02 February 2023;
Published: 16 February 2023.
Edited by:
Jia-Fu Jiang, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Guojian Liao, Southwest University, ChinaCopyright © 2023 Tao, Ge, Yang, Song, Jin, Lin, Zheng, Lu, Luo, Huang, Zhuang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenhong Zhuang, emhfemh1YW5nQGZhZnUuZWR1LmNu; Jianguo Xu, eHVqaWFuZ3VvQGljZGMuY24=
†These authors contributed equally to this work and should be considered co-first authors
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.