- 1Institute for Research in Molecular Medicine (INFORMM), Universiti Sains Malaysia, Penang, Malaysia
- 2Universiti Kuala Lumpur-Royal College of Medicine Perak, Ipoh, Perak, Malaysia
- 3School of Pharmaceutical Sciences, Universiti Sains Malaysia, Penang, Malaysia
- 4Department of Zoology, Faculty of Life Sciences, Government College University, Faisalabad, Pakistan
An increase in the occurrence of viral infectious diseases is a global concern for human health. According to a WHO report, dengue virus (DENV) is one of the most common viral diseases affecting approximately 400 million people annually, with worsening symptoms in nearly 1% of cases. Both academic and industrial researchers have conducted numerous studies on viral epidemiology, virus structure and function, source and route of infection, treatment targets, vaccines, and drugs. The development of CYD-TDV or Dengvaxia® vaccine has been a major milestone in dengue treatment. However, evidence has shown that vaccines have some drawbacks and limitations. Therefore, researchers are developing dengue antivirals to curb infections. DENV NS2B/NS3 protease is a DENV enzyme essential for replication and virus assembly, making it an interesting antiviral target. For faster hit and lead recognition of DENV targets, methods to screen large number of molecules at lower costs are essential. Similarly, an integrated and multidisciplinary approach involving in silico screening and confirmation of biological activity is required. In this review, we discuss recent strategies for searching for novel DENV NS2B/NS3 protease inhibitors from the in silico and in vitro perspectives, either by applying one of the approaches or by integrating both. Therefore, we hope that our review will encourage researchers to integrate the best strategies and encourage further developments in this area.
1 Introduction
Dengue virus (DENV), an RNA virus belonging to the family Flaviviridae and genus Flavivirus, is a fatal pathogenic arthropod-borne virus (arboviruses). It is predominantly transmitted by Aedes aegypti and, to a lesser extent, Aedes albopictus. The disease is widespread in more than 110 countries, infects approximately 400 million people, and results in approximately 20,000 deaths annually (Liang Gao and Gould, 2015; World Health Organization, 2020). Over the past few years, the occurrence of dengue fever (DF), dengue hemorrhagic fever (DHF), and dengue shock syndrome (DSS) has significantly increased in major tropical regions, with alarming frequency, magnitude, and bearing dire consequences (Tiga-Loza et al., 2021). DENV has four antigenically distinct serotypes (DENV1–DENV4) with 65–70% identical genome sequences. Each DENV serotype comprises four–seven genotypes that differ by 10% at the amino acid level across the envelope protein. The four serotypes differ not only in sequence similarity but also in infection dynamics. For example, DENV-1 is the most common serotype, followed by DENV-2, which is more frequently associated with severe infections. However, the mechanisms underlying dengue infections, as well as the entire set of distinctions across serotypes, remain unknown. However, a few recent studies have investigated the differences between the serotypes (Delli Ponti and Mutwil, 2021; Katzelnick et al., 2021; Stica et al., 2022). In light of the above, we hope that the source, breadth, and impact of antigenic heterogeneity can be better understood, which will aid in the exploration of effective dengue inhibitors or vaccines.
One of the major milestones in combating dengue infection was the first licensed vaccine, CYD-TDV or Dengvaxia®. Nevertheless, owing to some drawbacks and limitations in the ongoing trials, it was found that the vaccine increased the risk of developing a severe form of dengue infection in some receivers (Redoni et al., 2020; Tully and Griffiths, 2021). This has led researchers to accentuate the development of potent inhibitors that can curb infection. Therefore, it is crucial to explore drugs directed at viral targets or critical host mechanisms that can be used as prophylaxis or treatment for the disease. Drug efficacy in the effective amelioration of the disease or the reduction of disease severity and fatalities is needed to lower the burden of dengue (Low Gatsinga et al., 2018).
Pharmacological interventions for DENV replication can be targeted for antiviral treatments. Over the years, DENV enzymes, such as NS2B/3 protease (Yusof et al., 2000; Leung et al., 2001; Li et al., 2005; Erbel et al., 2006), NS3 helicase/NTPase/RTPase (Egloff Benarroch et al., 2002; Wang et al., 2009; Basavannacharya and Vasudevan, 2014), NS5 methyltransferase (Lim et al., 2008; Lim et al., 2011; Barral et al., 2013; Lim et al., 2013), and NS5 polymerase (Nomaguchi et al., 2003; Selisko et al., 2006; Niyomrattanakit et al., 2010; Niyomrattanakit et al., 2015) have been studied comprehensively for pharmacological intervention. Among these enzymes, NS2B/3 protease, which plays multiple roles in the viral life cycle, is an attractive target for dengue antiviral drug discovery. One of the methods for faster hit and lead recognition for DENV targets is to screen a large number of chemical molecules using high-throughput screening at lower costs. An integrated and multidisciplinary approach that integrates biochemical approach and virtual simulations are frequently used in drug discovery.
This study examined 105 studies published in the Scopus citation database, MEDLINE, PubMed, and Google Scholar from 2015 to 2022. The indexed articles focused on discovering potential DENV NS2B/NS3 protease inhibitors using in silico and in vitro approaches, either by integrating both or applying one of them. Hence, search strings tailored to each database were devised for the dengue NS2B/NS3. Mendeley (Elsevier, London, England) was used to compile references for the identified articles, and duplicates were removed. All identified abstracts were examined and selected based on preset criteria. A systematic review of this paper began by tabulating significant potential inhibitors in silico and in vitro studies, followed by a Venn diagram illustrating the strategy distribution (Figure 1). This review seeks to explore a better understanding of NS2B/NS3 proteases and their therapeutic inhibitory potential and thus enlighten researchers on integrating the best strategies in this area.
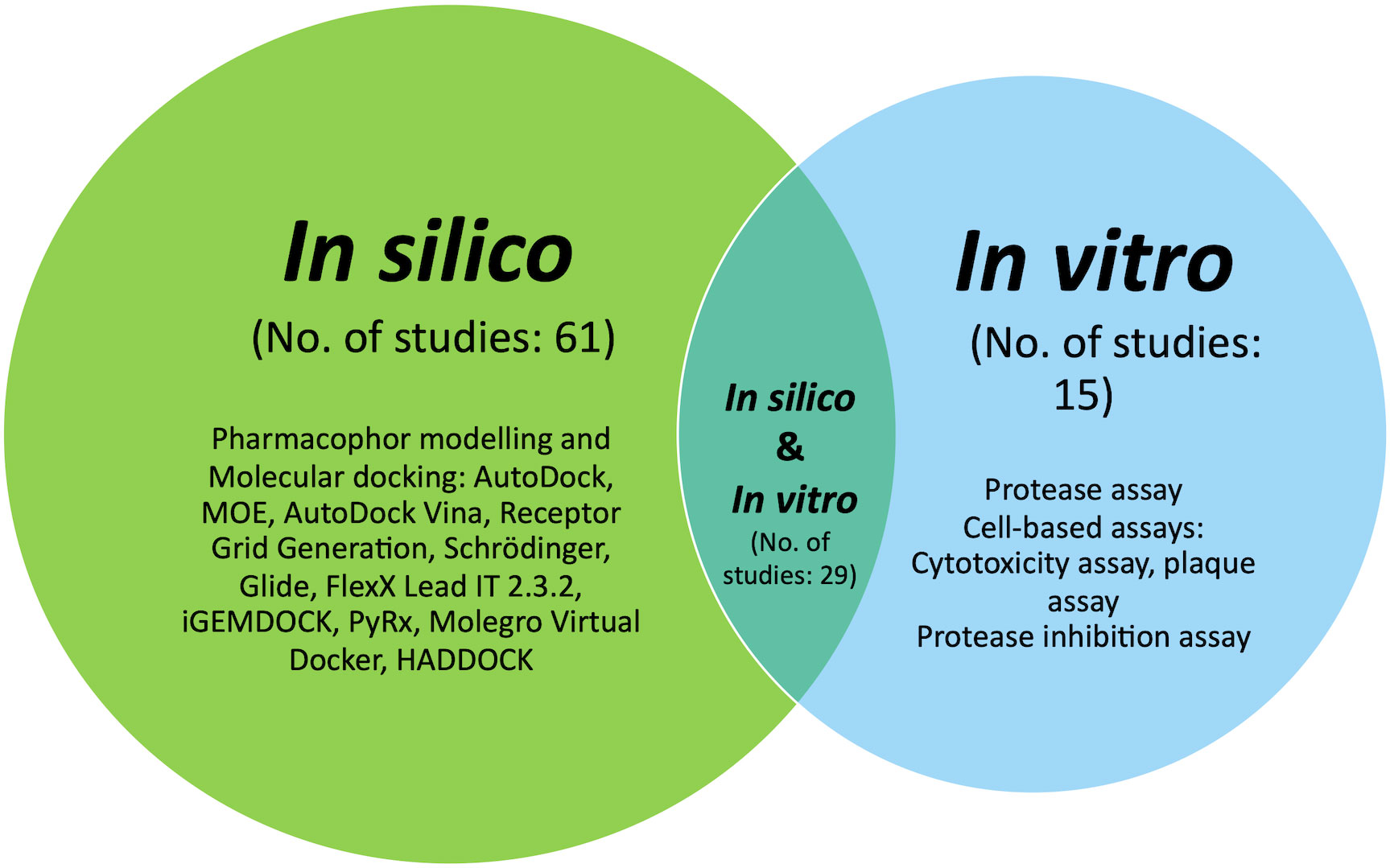
Figure 1 Venn diagram illustrating the distribution of the in silico, in vitro and approach integrating both strategies.
2 DENV polyproteins
Morphologically, dengue viruses are approximately 50 nm in diameter with an open reading frame (ORF) of over 10,000 bases (Hahn et al., 1990). Upon infection, the positive-sense single-stranded RNA genome is replicated and translated in the endoplasmic reticulum (ER), where host ribosomes translate RNA into polyproteins. These nascent proteins are further broken down by host and viral proteases into structural and nonstructural (NS) proteins (Figure 2).
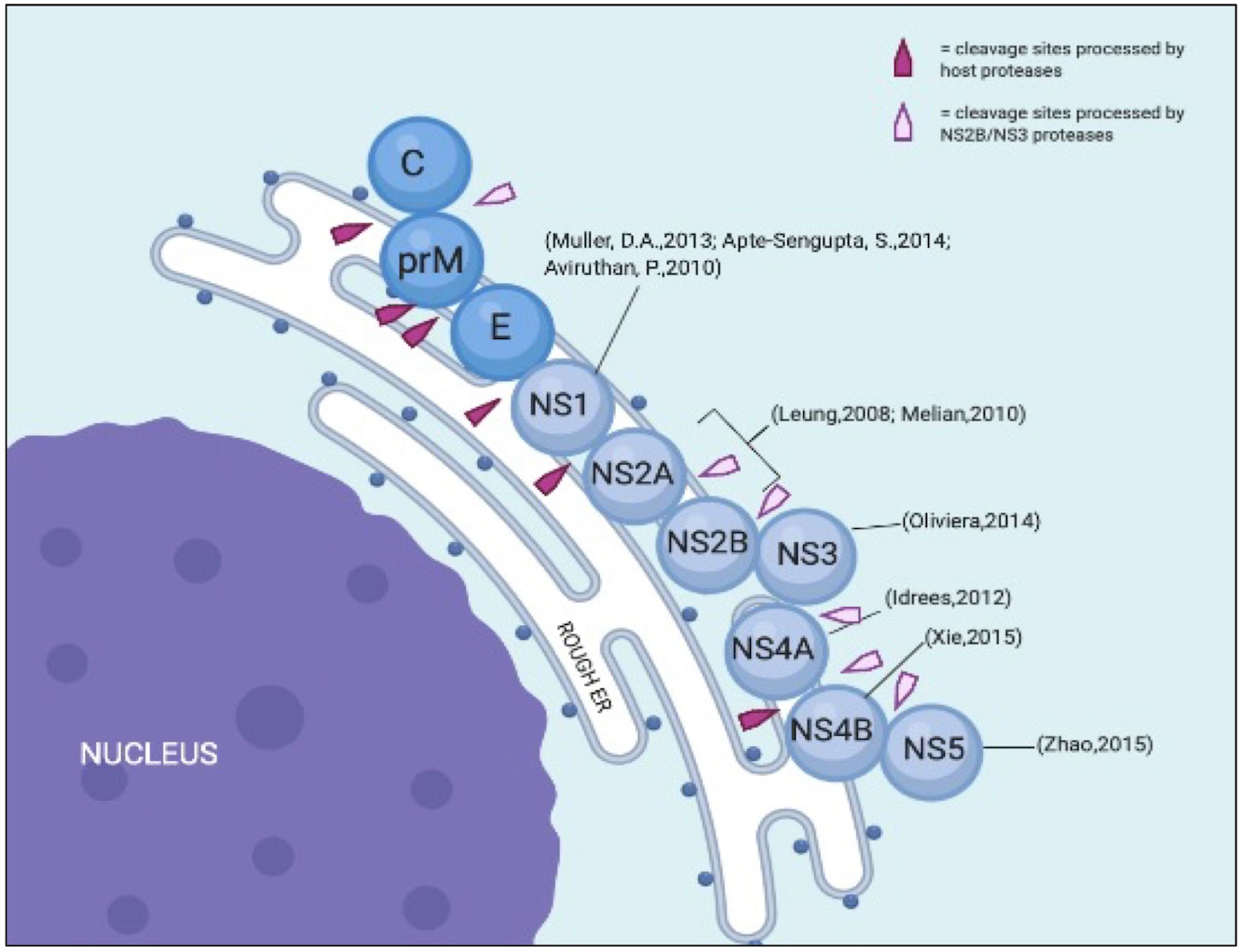
Figure 2 Illustration of DENV structural and NS proteins on the ER with its cleavage sites. Modified from Uno & Ross, 2018.
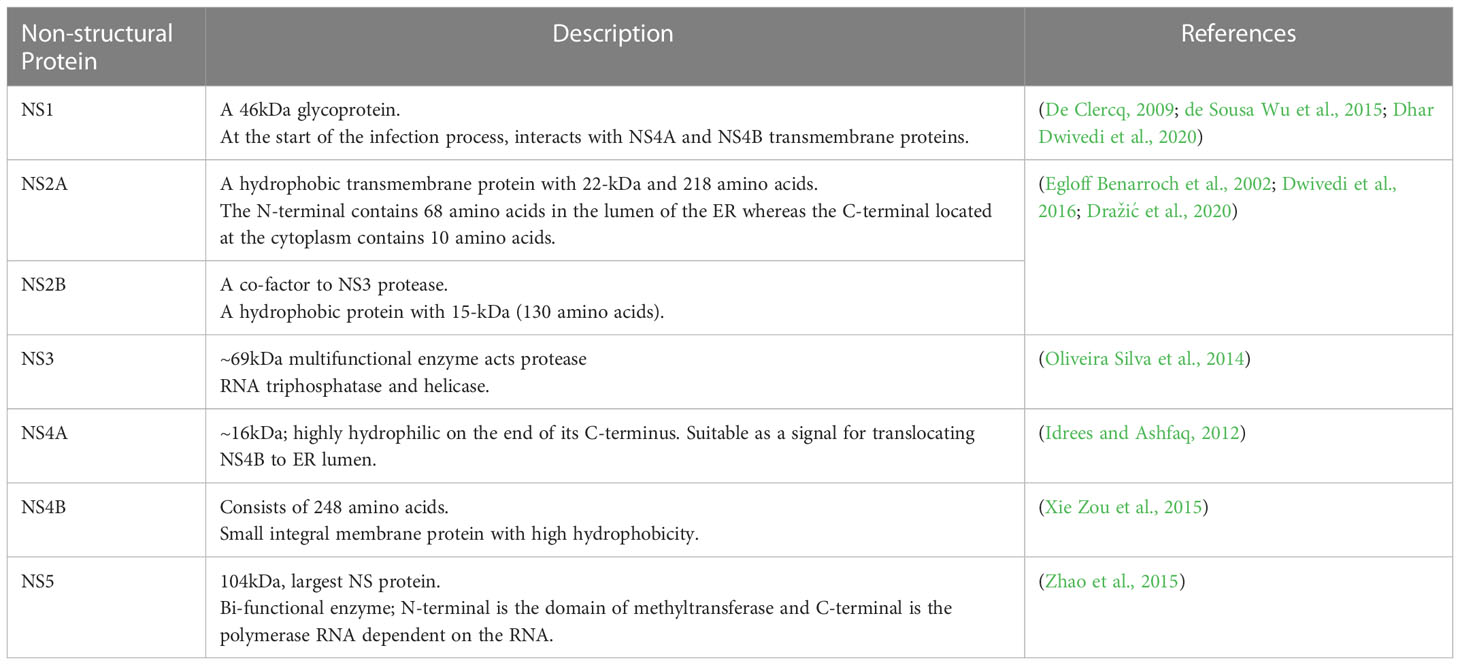
The ORF is flanked by two untranslated regions (UTRs) that contain structural and functional elements essential for viral translation and replication. The UTRs are translated into polyproteins that are processed co- and post-translationally by the host and DENV proteases to produce ten mature viral proteins. From the N-terminal region, three structural proteins are encoded in the N-terminal region: capsid protein (C, 11kDa), membrane protein (M, ~8kDa), and envelope proteins (E, 53 kDa) (Hosseini et al., 2018). NS proteins are essential for viral replication and are retained in all DENV serotypes (Ahmad and Poh, 2019). Hence, these proteins are important components of the DENV genome replication machinery. Table 1 briefly describes each protein and its relevance to viral pathogenicity.
2.1 DENV NS2B/NS3 protease as drug target
NS3 is a large multifunctional protein with serine protease (with NS2B as a cofactor), 5′-RNA triphosphatase (RTPase), nucleoside triphosphatase (NTPase), and helicase activity (Wengler and Wengler, 1991; Warrener et al., 1993; Li et al., 1999). The N-terminal 170 amino acids of NS3 have protease activity and a hydrophobic core of approximately 40 amino acids within NS2B that provides an essential cofactor function (Hahn et al., 1990; Chambers et al., 1991; Falgout et al., 1991). NS3 protease (NS3 pro) is a trypsin-like serine protease with a classic serine protease catalytic triad consisting of His51, Asp75, and Ser135 residues (Bazan and Fletterick, 1989). All four DENV serotypes have approximately 65–74% amino acid sequence homology and a common substrate preference (Li et al., 2005). The C-terminal β-hairpin of NS2B in its catalytically active form wraps around the active site of NS3 (Figure 3) (Erbel et al., 2006). Consistent with the important structural role of the C-terminal β-hairpin of NS2B, structural comparisons indicated that the amino acids within the N-terminal portion displayed similar conformations in all structures, regardless of the presence or absence of inhibitors.
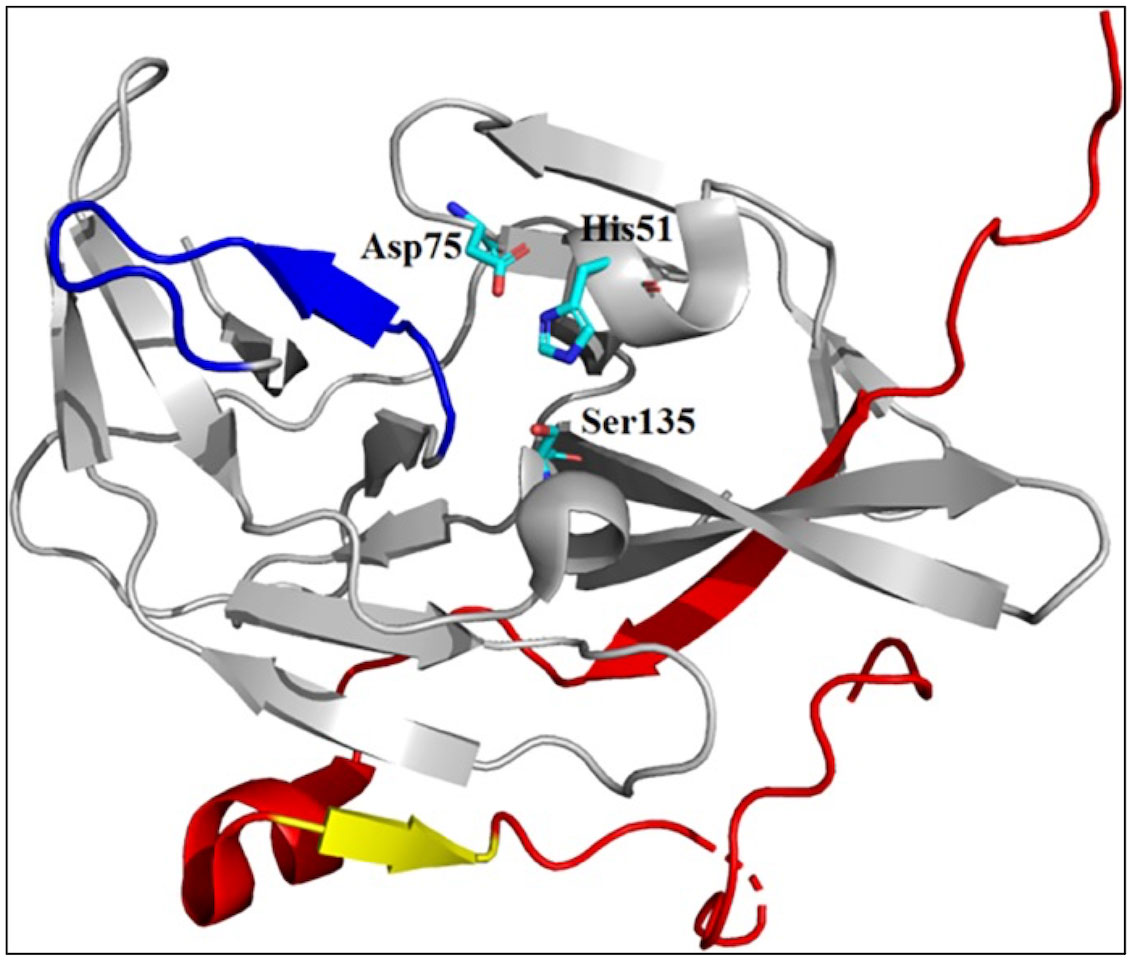
Figure 3 X-ray crystal structure of catalytically active conformation of DENV NS2B/NS3 pro (PDB code: 2FOM). Grey ribbon: NS3 structure; red ribbon: NS2B cofactor; yellow ribbon: S1-ß-hairpin; and blue ribbon: ST-loop. Figure adapted from Erbel et al. (2006).
2.2 Prospective developments
Considering the global threat of DENV and the urgent need for effective drugs, several efforts have been made to identify potential protease inhibitors. The development of NS2B/NS3pro inhibitors began with the structure-activity relationship of NS2B-NS3pro, inferred from the well-established cleavage sites of the DENV polyprotein by NS2B-NS3pro. This led to the discovery of two tetrapeptides, Bz-Nle-Lys-Arg-Arg and Bz-Nle-Lys-Thr-Arg, that have been shown to have high affinities for NS2B-NS3pro (Ki¼12.42 and 33.9 mM, respectively) (Yin et al., 2006; Othman et al., 2007; da Silva-Júnior and de Araújo-Júnior, 2019). Subsequently, efforts have been made to design peptidomimetics that have the ability to mimic the natural substrate (Gibbs et al., 2018; da Silva-Júnior and de Araújo-Júnior, 2019; Dražić et al., 2020).
The latter group of inhibitors has long been recognized as an invaluable component of medicine, and many targeted therapies have focused on these small-molecule drugs. These low molecular weight (less than 900 Da) organic compounds help to control biological targets, such as enzymes, channels, or receptors, to alter the disease cycle (Phanthanawiboon et al., 2014; Lenci and Trabocchi, 2019). At present, 90 percent of the therapeutics in the pharmaceutical market are small-molecule drugs. These include ten clinically available human immunodeficiency virus 1 (HIV-1) protease inhibitors and hepatitis C virus (HCV) protease inhibitors (De Clercq, 2009; Manns and Von Hahn, 2013). These facts also suggest that protease inhibitors of the dengue virus could be clinically effective. In the last decade, the development of small molecule NS2B/NS3pro inhibitors has involved high-throughput screening (HTS) of the natural product (Kiat et al., 2006; de Sousa Wu et al., 2015), and synthesis of rational drug design (Liu et al., 2014; Viswanathan et al., 2014; Raut et al., 2015a) with virtual screening using computer-aided drug design (CADD) being in-process (Cabarcas-Montalvo et al., 2016).
This review highlights the recent development of DENV inhibitor successors, mainly small molecules. Owing to advances in bioinformatics in drug discovery, non-peptide antiviral activity evaluation has been explored in vitro, as well as in silico and in HTS (Kanakaveti et al., 2020). Weighing the benefits of both approaches provides greater knowledge and an understanding of the anti-DENV drug development pipeline. Summarizing our findings on the methods used in developing NS2B/NS3pro inhibitors, this study highlights methods that are relevant to this co-protein only. Methods were classified into three cohorts: studies focusing on in silico methods, in vitro methods, and both. By subdividing these approaches, we hope this analysis will promote further progress in discovering potent inhibitors of fatal arbovirus infections.
3 In silico approach
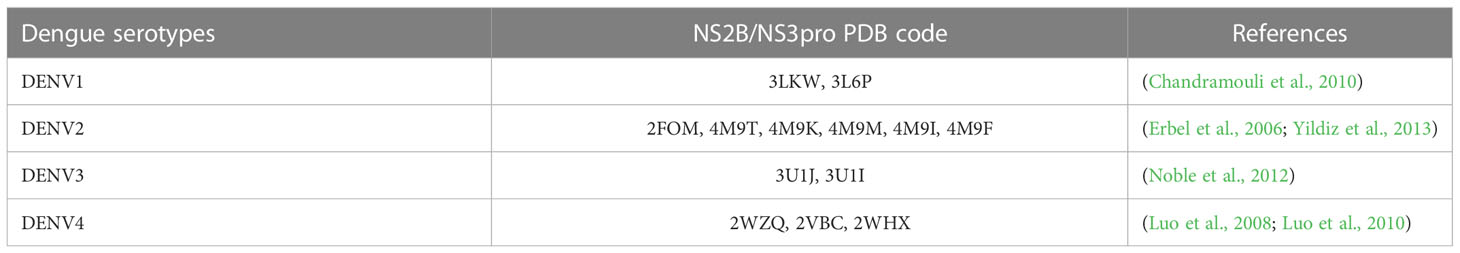
Various computational tools have been used to identify small target molecules for dengue drug discovery. Structure-based drug design (SBDD) methods, namely molecular dynamics, fragment-based drug design, pharmacophore modelling, and most importantly, molecular docking, have provided information about many molecules, including DENV protein targets, such as NS2B/NS3pro. Among the above mentioned methods, molecular docking is the most popular for searching for potential NS2B/NS3pro inhibitors. The aim of molecular docking is to determine the best ligand-binding positions in the NS2B/NS3pro binding pocket and estimate the affinity of the ligand for the protein (Jakhar et al., 2019). To date, 13 crystal structures of DENV NS2B/NS3pro with different PDB codes have been solved for all dengue serotypes (Table 2).
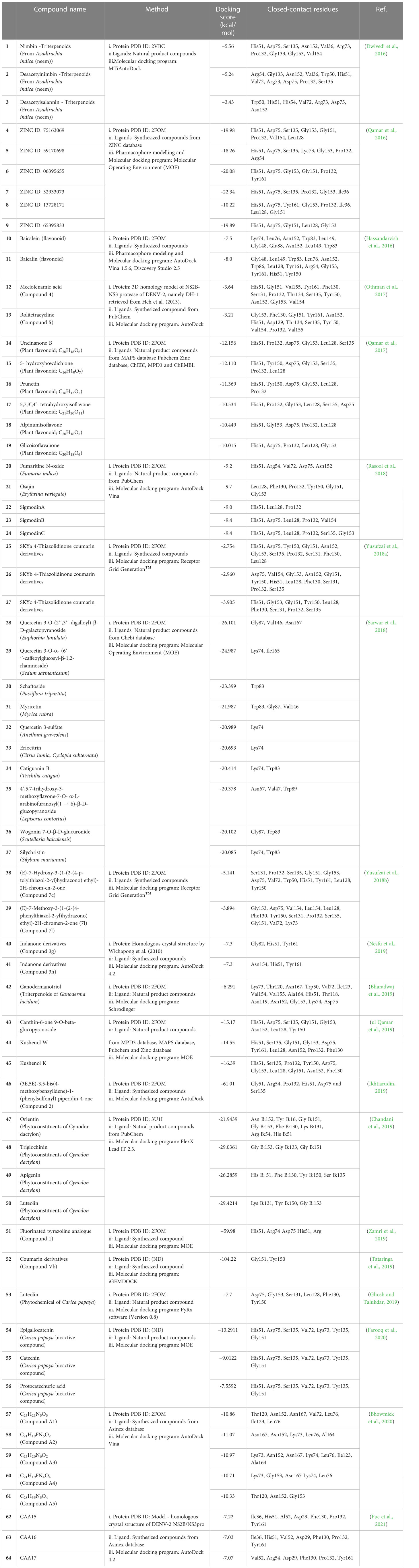
However, ligands mostly originate from virtual libraries comprising thousands to millions of compounds. The most commonly used docking software in NS2B/NS3pro studies are AutoDock, AutoDock Vina, and Molecular Operating Environment (MOE). These platforms have an algorithm for identifying the NS2B/NS3pro active site by allowing small drug-like molecules to bind to different parts of the protein. The best ligand-protein affinity and binding positions were then observed (Jakhar et al., 2019). Nevertheless, it is essential to observe the hydrogen bonding and optimize the hydrophobic interactions, as they are the key players in obtaining stable energy-favored ligands at the interface of a protein structure and help in modifying the binding affinity for the drug’s effectiveness. Studies that have applied only in silico-based approaches to explore the interaction between NS2B/NS3pro and its possible inhibitor candidates in recent years are tabulated in Table 3.
In conclusion, this approach determines the best-fitting ligand positions in the NS2B/NS3pro binding pocket and estimates the affinity of the ligand to the protein. The in silico approach uses crystal structures of the DENV NS2B/NS3pro protein with various PDB codes as well as ligands from virtual libraries containing hundreds to millions of chemicals. The ideal ligand-protein affinity and binding location can be determined using the software. However, the crucial point is that many chemical compounds and peptides have shown significant in silico binding affinity towards viral targets, but their affinity has yet to be evaluated using in vitro methods in many cases. Hence, the mechanism underlying the inhibition of most peptides remains unknown.
4 In vitro approach
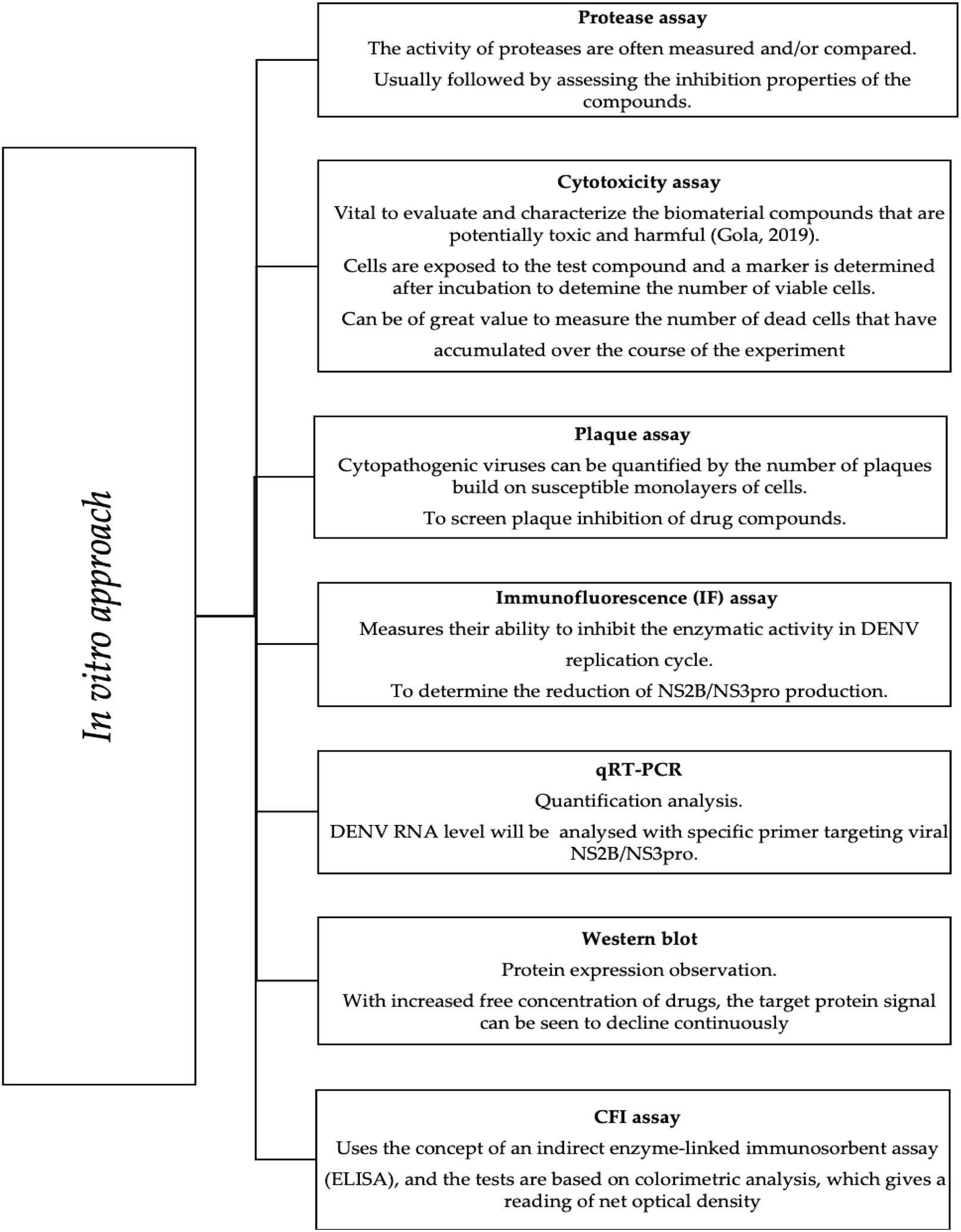
According to Lim et al., virtual hits derived from in silico docking require further validation by in vitro methods. These methods can verify on-target effects in cells (Lim, 2019). The in vitro assays are commonly performed to investigate the inhibitory properties of candidates against NS2B/NS3pro, as briefly described in Figure 4. Plaque, cytotoxicity, and immunofluorescence (IF) assays are examples of cell-based assays that provide substantial information on various cellular responses to compound exposure. Therefore, choosing the right cell type based on the target biology is critical. Among the cell types used in recent dengue inhibition studies, Vero, E6, C6/36, and BHK21 cells are effective for DENV propagation (Phanthanawiboon et al., 2014).
According to the Guidance for Industry-Antiviral Product Development by the US Food and Drug Administration (FDA), the specific antiviral activity was quantitatively measured by calculating the replication of the virus in the presence of increasing drug concentrations as opposed to replication in the absence of the drug. Therefore, to evaluate drug potency, the inhibition concentration (IC50) and effective concentration (EC50) must be measured (FDA, 2006). Nearly all recent studies have reported the IC50 of the tested compounds and their activities against the dengue enzyme. Quantification was performed using a protease inhibition assay, which measures the inhibitory activity of the drug candidates and the catalytic activity of the proteolytic enzyme.
As mentioned previously, in vitro drug potency measurements are essential for drug discovery. We reviewed recent studies that evaluated the EC50 of inhibitors through cytotoxicity tests (Chu Lee et al., 2015; Li et al., 2015; Lim et al., 2020) or plaque assays, such as the time of drug addition (Li et al., 2018; Yao et al., 2018), viral plaque reduction (Brecher et al., 2017; Li et al., 2018), and viral titer reduction assay (Brecher et al., 2017).
In addition to identifying an effective drug, it is crucial to determine the cytotoxic potential of the tested compounds in the drug discovery process (Slater, 2001). Briefly, the cytotoxic concentration (CC50) of the compounds that caused a reduction in cell viability was measured using a dilution assay. In recent studies, MTT (Raut et al., 2015b; Cabarcas-Montalvo et al., 2016; Beesetti et al., 2018; Li et al., 2018) or other cytotoxicity assays have been used to observe their effects on virus-infected host cells. The antiviral activity of the compounds was tested at different concentrations. Hence, developing potential inhibitors with lower cytotoxic concentrations is recommended (Alagarasu et al., 2022). In conclusion, many DENV NS2B/NS3 pro-inhibitor candidates have yet to be subjected to cytotoxicity investigation, making these products uncertain for further development.
Protease assays are another key pre-clinical assay in investigating protease inhibitors and their activity. For most inhibitor candidates, the target enzymatic activity was quantitatively determined to test their efficacy against the NS2B/NS3 pro-enzyme (Raut et al., 2015b; Brecher et al., 2017; Osman Idris et al., 2017; Beesetti et al., 2018; Euanorasetr et al., 2019; Hariono et al., 2019; Lim et al., 2020; Saleem et al., 2019; Sulaiman et al., 2019). The activity was determined if the tested compounds modulated the DENV NS2B/NS3 pro-enzyme function. Here, we highlight the recent five-year studies that applied only the in vitro approach (Table 4) and a combination of in vitro and in silico approaches (Table 5) to determine protease enzymatic activity.
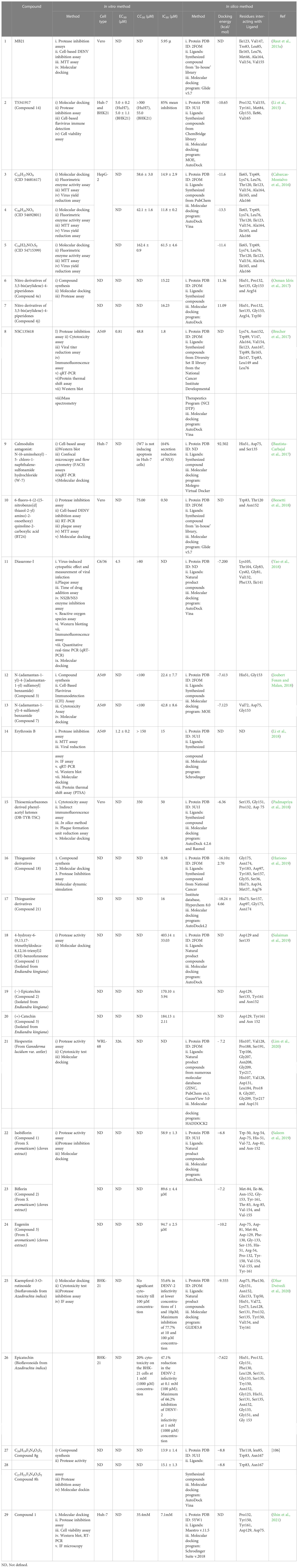
Table 5 Summary of NS2B/NS3 protease inhibitors recent development applying in vitro and in silico method.
By using the in vitro methods, the on-target effects in cells can be verified using in vitro methods. The candidate inhibitory activities against NS2B/NS3pro were evaluated using a protease inhibition assay. Moreover, plaque, cytotoxicity, and IF assays can provide valuable information on the diverse cellular responses to compound exposure. The pharmacological potency of drugs can be assessed by quantitatively measuring their specific antiviral activity. However, the cytotoxicity assessment of inhibitors during drug potency evaluation is limited, leading to uncertainty in the further development of drugs.
5 Using a combination of methods
In summary, incorporating in silico and in vitro approaches to determine the potency of dengue inhibitors can lead to the development of more potential drug candidates. Furthermore, integrating in vitro methods with in vivo assessments will reduce the number of physiologically relevant potential candidates and evaluate their characteristics simultaneously. It will also evaluate drug-drug interactions (DDI) and help comprehend the underlying mechanisms of drug candidates. Additionally, combining these approaches will help verify the relevance of in vitro results. Thus, substantiating the extrapolation of in vitro outcomes to the clinical phase of the drug development pipeline.
In addition, we would like to highlight the sources of DENV NS2B/NS3 pro candidates. In our study, the small-molecule or non-peptide candidates explored were either synthetic (25 studies) or derived from natural sources (16 studies). Currently, most medicines used in clinical practice are synthetically formulated and include chemical processes (reactions) and phytochemicals. The four anti-DENV drugs under clinical trials, celgosivir, UV4B, chloroquine, and balapiravir (Anasir et al., 2020), are small synthetic molecules developed from natural sources. As synthetic drugs have benefits such as chemical purity, a simple and cost-effective preparation process, and higher quality, more effective and safer drugs can be prepared by altering the chemical structure of the drug prototype.
Alternatively, using natural sources is a well-established method for discovering new substances with possible therapeutic effects. This class of drugs comprises new bioactive compounds that are essential for the production of modern medicines (Kumar et al., 2019). It also provides information on different classes of bioactive lead compounds for the discovery and development of novel drugs. Recent studies have focused on the bioactive compounds present in plants, such as Carica papaya (Ghosh and Talukdar, 2019; Farooq et al., 2020), Azadirachta indica (Dwivedi et al., 2016), Ganoderma lucidum (Bharadwaj et al., 2019), Ganoderma lucidum var. antler (Lim et al., 2020), Curcuma longa (Balasubramanian et al., 2019), Endiandra kingiana (Sulaiman et al., 2019), Cynodon dactylon (Chandani et al., 2019), Dryobalanops aromaticum (Salleh et al., 2019), Acorus tatarinowii Schott (Yao et al., 2018), and Syzygium aromaticum (Saleem et al., 2019). The above mentioned studies included the extraction of crude plants (or plant parts) in solvents, mainly methanol, before investigating its activity against DENV NS2B/NS3 pro. Nonetheless, from our observation, both cohorts led to potent inhibitors with promising activity against the DENV NS2B/NS3 proenzyme, which has the potential to progress to the next anti-DENV drug development phase.
6 Conclusion
The number of hits, particularly those obtained from in silico docking, should be verified using an in vitro approach. It is also important to fully characterize the hits identified from the compound libraries. Furthermore, to produce a promising DENV antiviral inhibitor, verifying its activity using in vitro methods is crucial. To further ascertain the outcome of the two approaches, incorporating in vivo assessments can be beneficial, as they can substantiate the in vitro outcomes to the clinical phase in the drug development pipeline. These combined approaches can lead to promising antiviral candidates that may curb dengue infection. Additionally, along with small drug-like molecules, the search for dengue inhibitors should focus on using peptides. As signaling molecules, this possible approach exhibits complex biological roles with high selectivity and comparatively safe criteria.
Furthermore, we emphasize that a DENV inhibitor must be effective against all four DENV serotypes, as these serotypes co-circulate in highly endemic regions [95]. Nevertheless, it is essential to remember that the plausibility of dengue serotypes, together with other factors, such as secondary infection by a heterologous serotype, age, comorbidity, poor clinical prognosis, diagnosis, virulence, and the host immune response, contribute to the development of severe dengue infection (Puc et al., 2021). Finally, considering the recent attempts to identify DENV NS2B/NS3pro inhibitors, a range of antiviral targets display antiviral intervention potential. Although small-molecule inhibitors require clinical approval, promising dengue antivirals will be possible soon.
Author contributions
LS designed the study, HN carried out the data collection, HN, KE, LH, data analysis and interpretation. LS. and HN, drafted the article. HN, RV, RA and AH edited the article. All authors read and approved the final article. Authors contributed equally for the preparation of this review.
Funding
This work was funded by the Malaysian Ministry of Education through the Higher Institution Centre of Excellence (HICoE) Grant No: 311/CIPPM/4401005 and supported by Research University Individual grant from Universiti Sains Malaysia (1001/CIPPM/8012305).
Acknowledgments
We are grateful to Emeritus Professor Satvinder Dhaliwal from Curtin University, Australia for linguistic and technical advice.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmad, Z., Poh, C. L. (2019). The conserved molecular determinants of virulence in dengue virus. Int. J. Med. Sci. 16 (3), 355–365. doi: 10.7150/ijms.29938
Alagarasu, K., Patil, P., Kaushik, M., Chowdhury, D., Joshi, R. K., Hegde, H., et al. (2022). In vitro antiviral activity of potential medicinal plant extracts against dengue and chikungunya viruses. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.866452
Anasir, M. I., Ramanathan, B., Poh, C. L. (2020). Structure-based design of antivirals against envelope glycoprotein of dengue virus. In Viruses 12 (4), 367. doi: 10.3390/v12040367
Balasubramanian, A., Pilankatta, R., Teramoto, T., Sajith, A. M., Nwulia, E., Kulkarni, A., et al. (2019). Inhibition of dengue virus by curcuminoids. Antiviral Res. 162, 71–78. doi: 10.1016/j.antiviral.2018.12.002
Barral, K., Sallamand, C., Petzold, C., Coutard, B., Collet, A., Thillier, Y., et al. (2013). Development of specific dengue virus 2’-o- and N7-methyltransferase assays for antiviral drug screening. Antiviral Res. 99 (3), 292–300. doi: 10.1016/j.antiviral.2013.06.001
Basavannacharya, C., Vasudevan, S. G. (2014). Suramin inhibits helicase activity of NS3 protein of dengue virus in a fluorescence-based high throughput assay format. Biochem. Biophys. Res. Commun. 453 (3), 539–544. doi: 10.1016/j.bbrc.2014.09.113
Bautista-Carbajal, P., Soto-Acosta, R., Angel-Ambrocio, A. H., Cervantes-Salazar, M., Loranca-Vega, C. I., Herrera-Martínez, M., et al. (2017). The calmodulin antagonist W-7 (N-(6-aminohexyl)-5-chloro-1-naphthalenesulfonamide hydrochloride) inhibits DENV infection in huh-7 cells. Virology 501, 188–198. doi: 10.1016/j.virol.2016.12.004
Bazan, J. F., Fletterick, R. J. (1989). Detection of a trypsin-like serine protease domain in flaviviruses and pestviruses. Virology 171 (2), 637–639. doi: 10.1016/0042-6822(89)90639-9
Beesetti, H., Tyagi, P., Medapi, B., Krishna, V. S., Sriram, D., Khanna, N., et al. (2018). A quinoline compound inhibits the replication of dengue virus serotypes 1–4 in vero cells. Antiviral Ther. 23, 385–394. doi: 10.3851/IMP3231
Bharadwaj, S., Lee, K. E., Dwivedi, V. D., Yadava, U., Panwar, A., Lucas, S. J., et al. (2019). Discovery of ganoderma lucidum triterpenoids as potential inhibitors against dengue virus NS2B-NS3 protease. Sci. Rep. 9 (1), 1–12. doi: 10.1038/s41598-019-55723-5
Bhowmick, S., Alissa, S. A., Wabaidur, S. M., Chikhale, R. V., Islam, M. A. (2020). Structure-guided screening of chemical database to identify NS3-NS2B inhibitors for effective therapeutic application in dengue infection. J. Mol. Recognit. 33 (7), e2838. doi: 10.1002/jmr.2838
Brecher, M., Li, Z., Liu, B., Zhang, J., Koetzner, C. A., Alifarag, A., et al. (2017). A conformational switch high-throughput screening assay and allosteric inhibition of the flavivirus NS2B-NS3 protease. PloS Pathog. 13 (5), e1006411. doi: 10.1371/journal.ppat.1006411
Cabarcas-Montalvo, M., Maldonado-Rojas, W., Montes-Grajales, D., Bertel-Sevilla, A., Wagner-Dobler, I., Sztajer, H., et al. (2016). Discovery of antiviral molecules for dengue: In silico search and biological evaluation. Eur. J. Med. Chem. 110, 87–97. doi: 10.1016/j.ejmech.2015.12.030
Chambers, T. J., Grakoui, A., Rice, C. M. (1991). Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J. Virol. 65 (11), 6042–6050. doi: 10.1128/jvi.65.11.6042-6050.1991
Chandani, S. R., Thorat, P. A., Nanda, R. K., Chitlange, S. S. (2019). Docking of phytoconstituents of cynodon dactylon on NS2B NS3 protease domain of dengue virus. Res. J. Pharm. Technol. 12 (12), 5865–5870. doi: 10.5958/0974-360X.2019.01017.5
Chandramouli, S., Joseph, J. S., Daudenarde, S., Gatchalian, J., Cornillez-Ty, C., Kuhn, P. (2010). Serotype-specific structural differences in the protease-cofactor complexes of the dengue virus family. J. Virol. 84 (6), 3059–3067. doi: 10.1128/jvi.02044-09
Chu Lee, R. C. H., Ang, M. J. Y., Wang, W. L., Lim, H. A., Wee, J. L. K., Chia, C. B., et al. (2015). Antiviral activities of 15 dengue NS2B-NS3 protease inhibitors using a human cell-based viral quantification assay. Antiviral Res. 118, 68–74. doi: 10.1016/j.antiviral.2015.03.010
da Silva-Júnior, E. F., de Araújo-Júnior, J. X. (2019). Peptide derivatives as inhibitors of NS2B-NS3 protease from dengue, West Nile, and zika flaviviruses. Bioorg. Medicinal Chem. 27 (18), 3963–3978. doi: 10.1016/j.bmc.2019.07.038
De Clercq, E. (2009). Anti-HIV drugs: 25 compounds approved within 25 years after the discovery of HIV. Int. J. Antimicrob. Agents 33 (4), 307–320. doi: 10.1016/j.ijantimicag.2008.10.010
Delli Ponti, R., Mutwil, M. (2021). Structural landscape of the complete genomes of dengue virus serotypes and other viral hemorrhagic fevers. BMC Genomics 22 (1), 1–14. doi: 10.1186/S12864-021-07638-7/FIGURES/8
de Sousa Wu, H., Nebo, L., Fernandes, J. B., Kiefer, W., Kanitz, M., Vieira, P. C., et al. (2015). Flavonoids as noncompetitive inhibitors of dengue virus NS2B-NS3 protease: Inhibition kinetics and docking studies. Bioorg. Medicinal Chem. 23 (3), 466–470. doi: 10.1016/j.bmc.2014.12.015
Dhar Dwivedi, V., Bharadwaj, S., Afroz, S., Khan, N., Ahmed Ansari, M., Yadava, U., et al. (2020). Anti-dengue infectivity evaluation of bioflavonoid from azadirachta indica by dengue virus serine protease inhibition. J. Biomol. Struct Dynamics 39 (4), 1417–1430. doi: 10.1080/07391102.2020.1734485
Dražić, T., Kopf, S., Corridan, J., Leuthold, M. M., Bertoša, B., Klein, C. D. (2020). Peptide-β-lactam inhibitors of dengue and West Nile virus NS2B-NS3 protease display two distinct binding modes. J. Medicinal Chem. 63 (1), 140–156. doi: 10.1021/acs.jmedchem.9b00759
Dwivedi, V. D., Tripathi, I. P., Mishra, S. K. (2016). In silico evaluation of inhibitory potential of triterpenoids from azadirachta indica against therapeutic target of dengue virus, NS2B-NS3 protease. J. Vector Borne Dis. 53 (2), 156.
Egloff Benarroch, D., Selisko, B., Romette, J. L., Canard, B. ,. M. P. (2002). An RNA cap (nucleoside-2′-O-) -methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 21 (11), 2757–2768. doi: 10.1093/emboj/21.11.2757
Erbel, P., Schiering, N., D’Arcy, A., Renatus, M., Kroemer, M., Lim, S. P., et al. (2006). Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat. Struct. Mol. Biol. 13, 372–373. doi: 10.1038/nsmb1073
Euanorasetr, J., Intra, B., Thunmrongsiri, N., Limthongkul, J., Ubol, S., Anuegoonpipat, A., et al. (2019). In vitro antiviral activity of spirotetronate compounds against dengue virus serotype 2. J. Gen. Appl. Microbiol. 65 (4), 197–203. doi: 10.2323/jgam.2018.10.001
Falgout, B., Pethel, M., Zhang, Y. M., Lai, C. J. (1991). Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J. Virol. 65 (5), 2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991
Farooq, M. U., Munir, B., Naeem, S., Yameen, M., Iqbal, S. Z., Ahmad, A., et al. (2020). Exploration of carica papaya bioactive compounds as potential inhibitors of dengue NS2B, NS3 and NS5 protease. Pakistan J. Pharm. Sci. 33(1 (1 (Supplementary), 355–360. doi: 10.36721/PJPS.2020.33.1.SUP.355-360.1
FDA (2006). Guidance for industry antiviral product development–conducting and submitting virology studies to the agency (Federal Register), 32351–32352.
Ghosh, I., Talukdar, P. (2019). Molecular docking and pharmacokinetics study for selected leaf phytochemicals from carica papaya linn. against dengue virus protein, NS2B/NS3 protease. World Sci. News 124 (2), 264–278.
Gibbs, A. C., Steele, R., Liu, G., Tounge, B. A., Montelione, G. T. (2018). Inhibitor bound dengue NS2B-NS3pro reveals multiple dynamic binding modes. Biochemistry 57 (10), 1591–1602. doi: 10.1021/acs.biochem.7b01127
Hahn, C. S., Galler, R., Rice, C. M., Chambers, T. J. (1990). Flavivirus genome organization, expression, and replication. annual reviews in microbiology. Annu. Rev. Microbiol. 44 (1), 649–688. doi: 10.1146/annurev.mi.44.100190.003245
Hariono, M., Choi, S. B., Roslim, R. F., Nawi, M. S., Tan, M. L., Kamarulzaman, E. E., et al. (2019). Thioguanine-based DENV-2 NS2B/NS3 protease inhibitors: Virtual screening, synthesis, biological evaluation and molecular modelling. PloS One 14 (1), 1–21. doi: 10.1371/journal.pone.0210869
Hassandarvish, P., Rothan, H. A., Rezaei, S., Yusof, R., Abubakar, S., Zandi, K. (2016). In silico study on baicalein and baicalin as inhibitors of dengue virus replication. RSC Adv. 6 (37), 31235–31247. doi: 10.1039/C6RA00817H
Hosseini, S., Muñoz-Soto, R. B., Oliva-Ramírez, J., Vázquez-Villegas, P., Aghamohammadi, N., Rodriguez-Garcia, A., et al. (2018). Latest updates in dengue fever therapeutics: Natural, marine and synthetic drugs. Curr. Medicinal Chem. 27 (5), 719–744. doi: 10.2174/0929867325666180629124709
Idrees, S., Ashfaq, U. A. (2012). A brief review on dengue molecular virology, diagnosis, treatment and prevalence in Pakistan. Genet. Vaccines Ther. 10 (1), 6. doi: 10.1186/1479-0556-10-6
Ikhtiarudin, I. (2019). Synthesis and in silico studies of a benzenesulfonyl curcumin analogue as a new anti-dengue virus type 2 (DEN2) NS2B/NS3. Indonesian J. Pharm. 30 (2), 84–90. doi: 10.14499/indonesianjpharm30iss2pp84-90
Jakhar, R., Dangi, M., Khichi, A., Chhillar, A. K. (2019). Relevance of molecular docking studies in drug designing. Curr. Bioinf. 15 (4), 270–278. doi: 10.2174/1574893615666191219094216
Joubert, J., Foxen, E. B., Malan, S. F. (2018). Microwave optimized synthesis of n-(adamantan-1-yl)-4-[(adamantan-1-yl)-sulfamoyl] benzamide and its derivatives for anti-dengue virus activity. Molecules 23 (7), 1678. doi: 10.3390/molecules23071678
Kanakaveti, V., Shanmugam, A., Ramakrishnan, C., Anoosha, P., Sakthivel, R., Rayala, S. K., et al. (2020). Computational approaches for identifying potential inhibitors on targeting protein interactions in drug discovery. Adv. Protein Chem. Struct. Biol. 121, 25–47. doi: 10.1016/bs.apcsb.2019.11.013
Katzelnick, L. C., Escoto, A. C., Huang, A. T., Garcia-Carreras, B., Chowdhury, N., Berry, I. M., et al. (2021). Antigenic evolution of dengue viruses over 20 years. Sci. (New York N.Y.) 374 (6570), 999. doi: 10.1126/SCIENCE.ABK0058
Kiat, T. S., Pippen, R., Yusof, R., Ibrahim, H., Khalid, N., Rahman, N. A. (2006). Inhibitory activity of cyclohexenyl chalcone derivatives and flavonoids of fingerroot, boesenbergia rotunda (L.), towards dengue-2 virus NS3 protease. Bioorg. Medicinal Chem. Lett. 16 (12), 3337–3340. doi: 10.1016/j.bmcl.2005.12.075
Kumar, P., Shaunak, I., Verma, M. L. (2019). “Biotechnological application of health promising bioactive molecules,” in Biotechnological production of bioactive compounds (Elsevier), 165–189. doi: 10.1016/B978-0-444-64323-0.00006-0
Lenci, E., Trabocchi, A. (2019). “Synthetic approaches toward small molecule libraries,” in Small molecule drug discovery: Methods, molecules and applications (Elsevier), 1–34. doi: 10.1016/B978-0-12-818349-6.00001-7
Leung, D., Schroder, K., White, H., Fang, N. X., Stoermer, M. J., Abbenante, G., et al. (2001). Activity of recombinant dengue 2 virus NS3 protease in the presence of a truncated NS2B Co-factor, small peptide substrates, and inhibitors. J. Biol. Chem. 276 (49), 45762–45771. doi: 10.1074/jbc.M107360200
Liang Gao, X., Gould, E. A. G. (2015). Factors responsible for the emergence of arboviruses; strategies, challenges and limitations for their control. Emerg. Microbes Infect. 4 (3), e18. doi: 10.1038/emi.2015.18
Li, L., Basavannacharya, C., Chan, K. W. K., Shang, L., Vasudevan, S. G., Yin, Z. (2015). Structure-guided discovery of a novel non-peptide inhibitor of dengue virus NS2B-NS3 protease. Chem. Biol. Drug Design 86 (3), 255–264. doi: 10.1111/cbdd.12500
Li, H., Clum, S., You, S., Ebner, K. E., Padmanabhan, R. (1999). The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J. Virol. 73 (4), 3108–3116. doi: 10.1128/jvi.73.4.3108-3116.1999
Li, J., Lim, S. P., Beer, D., Patel, V., Wen, D., Tumanut, C., et al. (2005). Functional profiling of recombinant NS3 proteases from all four serotypes of dengue virus using tetrapeptide and octapeptide substrate libraries. J. Biol. Chem. 280 (31), 28766–28774. doi: 10.1074/jbc.M500588200
Lim, S. P. (2019). Dengue drug discovery: Progress, challenges and outlook. Antiviral Res. 163 (December), 156–178. doi: 10.1016/j.antiviral.2018.12.016
Lim, W. Z., Cheng, P. G., Abdulrahman, A. Y., Teoh, T. C. (2020). The identification of active compounds in ganoderma lucidum var. antler extract inhibiting dengue virus serine protease and its computational studies. J. Biomol. Struct Dynamics 38 (14), 4273–4288. doi: 10.1080/07391102.2019.1678523
Lim, S. P., Koh, J. H. K., Seh, C. C., Liew, C. W., Davidson, A. D., Chua, L. S., et al. (2013). A crystal structure of the dengue virus non-structural protein 5 (NS5) polymerase delineates interdomain amino acid residues that enhance its thermostability and de novo initiation activities. J. Biol. Chem. 288 (43), 31105–31114. doi: 10.1074/jbc.M113.508606
Lim, S. V., Rahman, M. B. A., Tejo, B. A. (2011). Structure-based and ligand-based virtual screening of novel methyltransferase inhibitors of the dengue virus. BMC Bioinf. 12 (SUPPL. 13), 30. doi: 10.1186/1471-2105-12-S13-S24
Lim, S. P., Wen, D., Yap, T. L., Yan, C. K., Lescar, J., Vasudevan, S. G. (2008). A scintillation proximity assay for dengue virus NS5 2′-o-methyltransferase-kinetic and inhibition analyses. Antiviral Res. 80 (3), 360–369. doi: 10.1016/j.antiviral.2008.08.005
Li, Z., Sakamuru, S., Huang, R., Brecher, M., Koetzner, C. A., Zhang, J., et al. (2018). Erythrosin b is a potent and broad-spectrum orthosteric inhibitor of the flavivirus NS2B-NS3 protease. Antiviral Res. 150, 217–225. doi: 10.1016/j.antiviral.2017.12.018
Liu, H., Wu, R., Sun, Y., Ye, Y., Chen, J., Luo, X., et al. (2014). Identification of novel thiadiazoloacrylamide analogues as inhibitors of dengue-2 virus NS2B/NS3 protease. Bioorg. Medicinal Chem. 22 (22), 6344–6352. doi: 10.1016/j.bmc.2014.09.057
Low Gatsinga, R., Vasudevan, S. G., Sampath, A. ,. J. G. (2018). Dengue antiviral development: A continuing journey. in dengue and zika: Control and antiviral treatment strategies. Springer Singapore, 319–332. doi: 10.1007/978-981-10-8727-1_22
Luo, D., Wei, N., Doan, D. N., Paradkar, P. N., Chong, Y., Davidson, A. D., et al. (2010). Flexibility between the protease and helicase domains of the dengue virus NS3 protein conferred by the linker region and its functional implications. J. Biol. Chem. 285 (24), 18817–18827. doi: 10.1074/jbc.M109.090936
Luo, D., Xu, T., Hunke, C., Grüber, G., Vasudevan, S. G., Lescar, J. (2008). Crystal structure of the NS3 protease-helicase from dengue virus. J. Virol. 82 (1), 173–183. doi: 10.1128/jvi.01788-07
Manns, M. P., Von Hahn, T. (2013). Novel therapies for hepatitis c–one pill fits all? Nat. Rev. Drug Discovery 12 (8), 595. doi: 10.1038/nrd4050
Nesfu, N. Z. M., Laurain-Mattar, D., Kamarulzaman, E., Wahab, H. A., Zakaria, I. I., Hassan, M. Z., et al. (2019). The in-silico studies of benzylidene indanone derivatives towards dengue virus type-2 NS2B/NS3 protease. J. Phys. Sci. 30(Supp. 2), 191–198. doi: 10.21315/jps2019.30.s2.16
Niyomrattanakit, P., Chen, Y. L., Dong, H., Yin, Z., Qing, M., Glickman, J. F., et al. (2010). Inhibition of dengue virus polymerase by blocking of the RNA tunnel. J. Virol. 84 (11), 5678. doi: 10.1128/JVI.02451-09
Niyomrattanakit, P., Wan, K. F., Chung, K. Y., Abas, S. N., Seh, C. C., Dong, H., et al. (2015). Stabilization of dengue virus polymerase in de novo initiation assay provides advantages for compound screening. Antiviral Res. 119, 36–46. doi: 10.1016/j.antiviral.2015.04.007
Noble, C. G., Seh, C. C., Chao, A. T., Shi, P. Y. (2012). Ligand-bound structures of the dengue virus protease reveal the active conformation. J. Virol. 86 (1), 438–446. doi: 10.1128/jvi.06225-11
Nomaguchi, M., Ackermann, M., Yon, C., You, S., Padmanbhan, R. (2003). De novo synthesis of negative-strand RNA by dengue virus RNA-dependent RNA polymerase In vitro: Nucleotide, primer, and template parameters. J. Virol. 77 (16), 8831–8842. doi: 10.1128/jvi.77.16.8831-8842.2003
Oliveira Silva, M. L. D., Oliveira, A. F., Silva, C. C. D., Teixeira, R. R., De Paula, S. O. (2014). NS3 and NS5 proteins: important targets for anti-dengue drug design. J. Braz. Chem. Soc. 25 (10), 1759–1769. doi: 10.5935/0103-5053.20140057
Osman Idris, N. H., Kamarulzaman, E. E., Wahab, H. A., Hassan, M. Z. ,. H., Osman, H., Idris, N. H., et al. (2017). 3, 5-bis (arylidene)-4-piperidones as potential dengue protease inhibitors. Acta Pharm. Sin. B 7 (4), 479–484. doi: 10.1016/j.apsb.2017.04.009
Othman, R., Othman, R., Baharuddin, A., Ramakrishnan, N. R. (2017). Molecular docking studies of selected medicinal drugs as dengue virus-2 protease inhibitors. Sains Malaysiana 46 (10), 1865–1875. doi: 10.17576/jsm-2017-4610-25
Othman, R., Wahab, H. A., Yusof, R., Rahman, N. A. (2007). Analysis of secondary structure predictions of dengue virus type 2 NS2B/NS3 against crystal structure to evaluate the predictive power of the in-silico methods. In Silico Biol. 7 (2), 215–224.
Padmapriya, P., Gracy Fathima, S., Ramanathan, G., Yuvaraj, V., Khaleefathullah Sheriff, A., Kaveri, K., et al. (2018). Development of antiviral inhibitor against dengue 2 targeting Ns3 protein: In vitro and in silico significant studies. Acta Trop. 188, 1–8. doi: 10.1016/j.actatropica.2018.08.022
Phanthanawiboon, S., Atchareeya, A., Panngarm, N., Limkittikul, K., Ikuta, K., Anantapreecha, S., et al. (2014). Isolation and propagation of dengue virus in vero and BHK-21 cells expressing human DC-SIGN stably. J. Virol. Methods 209, 55–61. doi: 10.1016/j.jviromet.2014.08.023
Puc, I., Ho, T. C., Yen, K. L., Vats, A., Tsai, J. J., Chen, P. L., et al. (2021). Cytokine signature of dengue patients at different severity of the disease. Int J Mol Sci. 22 (6), 2879. doi: 10.3390/ijms22062879
Qamar, M. T., Ashfaq, U. A., Tusleem, K., Mumtaz, A., Tariq, Q., Goheer, A., et al. (2017). In-silico identification and evaluation of plant flavonoids as dengue NS2B/NS3 protease inhibitors using molecular docking and simulation approach. Pak. J. Pharm. Sci. 30 (6), 2119–2137.
Qamar, M. T. U., Kiran, S., Ashfaq, U. A., Javed, M. R., Anwar, F., Ali, M. A. (2016). Discovery of novel dengue NS2B/NS3 protease inhibitors using pharmacophore modeling and molecular docking based virtual screening of the zinc database. Int. J. Pharmacol. 12, 621–632. doi: 10.3923/ijp.2016.621.632
Rasool, N., Ashraf, A., Waseem, M., Hussain, W., Mahmood, S. (2018). Computational exploration of antiviral activity of phytochemicals against NS2B/NS3 proteases from dengue virus. Turkish J. Biochem. 44 (3), 261–277. doi: 10.1515/tjb-2018-0002
Raut, R., Beesetti, H., Tyagi, P., Khanna, I., Jain, S. K., Jeankumar, V. U., et al. (2015a). A small molecule inhibitor of dengue virus type 2 protease inhibits the replication of all four dengue virus serotypes in cell culture. Virol. J. 12 (1), 16. doi: 10.1186/s12985-015-0248-x
Raut, R., Beesetti, H., Tyagi, P., Khanna, I., Jain, S. K., Jeankumar, V. U., et al. (2015b). A small molecule inhibitor of dengue virus type 2 protease inhibits the replication of all four dengue virus serotypes in cell culture. Virol. J. 12 (1), 1–7. doi: 10.1186/S12985-015-0248-X
Redoni, M., Yacoub, S., Rivino, L., Giacobbe, D. R., Luzzati, R., di Bella, S. (2020). “Dengue: Status of current and under-development vaccines,” in Reviews in medical virology, vol. 30. (UK: John Wiley and Sons Ltd), e2101. doi: 10.1002/rmv.2101
Saleem, H. N., Batool, F., Mansoor, H. J., Shahzad-ul-Hussan, S., Saeed, M. (2019). Inhibition of dengue virus protease by eugeniin, isobiflorin, and biflorin isolated from the flower buds of syzygium aromaticum (Cloves). ACS Omega 4 (1), 1525–1533. doi: 10.1021/acsomega.8b02861
Salleh, H. M., Chong, S. L., Othman, R., Hazni, H., Ahmad, K., Mohd Yusof, M. Y. Z., et al. (2019). Dengue protease inhibition activity of selected Malaysian medicinal herbs. Trop. Biomed. 36 (2), 357–366.
Sarwar, M. W., Riaz, A., Dilshad, S. M. R., Al-Qahtani, A., Nawaz-Ul-Rehman, M. S., Mubin, M. (2018). Structure activity relationship (SAR) and quantitative structure activity relationship (QSAR) studies showed plant flavonoids as potential inhibitors of dengue NS2B-NS3 protease. BMC Struct. Biol. 18 (1), 6. doi: 10.1186/s12900-018-0084-5
Selisko, B., Dutartre, H., Guillemot, J. C., Debarnot, C., Benarroch, D., Khromykh, A., et al. (2006). Comparative mechanistic studies of de novo RNA synthesis by flavivirus RNA-dependent RNA polymerases. Virology 351 (1), 145–158. doi: 10.1016/j.virol.2006.03.026
Shin, H. J., Kim, M. H., Lee, J. Y., Hwang, I., Yoon, G. Y., Kim, H. S., et al. (2021). Structure-based virtual screening: identification of a novel NS2B-NS3 protease inhibitor with potent antiviral activity against Zika and Dengue viruses. Microorganisms 9 (3), 545. doi: 10.3390/microorganisms9030545
Slater, K. (2001). Cytotoxicity tests for high-throughput drug discovery. Curr. Opin. Biotechnol. 12 (1), 70–74. doi: 10.1016/S0958-1669(00)00177-4
Stica, C. J., Barrero, R. A., Murray, R. Z., Devine, G. J., Phillips, M. J., Frentiu, F. D. (2022). Global evolutionary history and dynamics of dengue viruses inferred from whole genome sequences. Viruses 14 (4), 703. doi: 10.3390/V14040703/S1
Sulaiman, S. N., Hariono, M., Salleh, H. M., Chong, S.-L., Yee, L. S., Zahari, A., et al. (2019). Chemical constituents from endiandra kingiana (Lauraceae) as potential inhibitors for dengue type 2 NS2B/NS3 serine protease and its molecular docking. Natural Product Commun. 14 (9), 1934578X19861014. doi: 10.1177/1934578X19861014
Tataringa, G., Sathyamurthy, B., Sandu, I., Zbancioc, A. M. (2019). In silico docking study of some coumarin derivatives as potential inhibitors on different dengue viral proteins. Proteins (NS1 NS2A NS2B NS3 NS4A NS4B NS5) 10, 14. doi: 10.37358/RC.19.9.7555
Tiga-Loza, D. C., Martínez-Vega, R. A., Undurraga, E. A., Tschampl, C. A., Shepard, D. S., Ramos-Castañeda, J. (2021). Persistence of symptoms in dengue patients: A clinical cohort study. Trans. R. Soc. Trop. Med. Hyg. 114 (5), 355–364. doi: 10.1093/TRSTMH/TRAA007
Tully, D., Griffiths, C. L. (2021). Dengvaxia: the world’s first vaccine for prevention of secondary dengue. Ther. Adv. Vaccines Immunother. 9, 251513552110158. doi: 10.1177/25151355211015839
ul Qamar, M. T., Maryam, A., Muneer, I., Xing, F., Ashfaq, U. A., Khan, F. A., et al. (2019). Computational screening of medicinal plant phytochemicals to discover potent pan-serotype inhibitors against dengue virus. Sci. Rep. 9 (1), 1–16. doi: 10.1038/s41598-018-38450-1
Uno, N., Ross, T. M. (2018). Dengue virus and the host innate immune response. Emerg Microbes Infect 7 (1), 1–11.
Viswanathan, U., Tomlinson, S. M., Fonner, J. M., Mock, S. A., Watowich, S. J. (2014). Identification of a novel inhibitor of dengue virus protease through use of a virtual screening drug discovery web portal. J. Chem. Inf. Model. 54 (10), 2816–2825. doi: 10.1021/ci500531r
Wang, C. C., Huang, Z. S., Chiang, P. L., Chen, C. T., Wu, H. N. (2009). Analysis of the nucleoside triphosphatase, RNA triphosphatase, and unwinding activities of the helicase domain of dengue virus NS3 protein. FEBS Lett. 583 (4), 691–696. doi: 10.1016/j.febslet.2009.01.008
Warrener, P., Tamura, J. K., Collett, M. S. (1993). RNA-Stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J. Virol. 67 (2), 989–996. doi: 10.1128/jvi.67.2.989-996.1993
Wengler, G., Wengler, G. (1991). The carboxy-terminal part of the NS 3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology 184 (2), 707–715. doi: 10.1016/0042-6822(91)90440-M
World Health Organization (2022). Dengue and severe dengue. Available online at: http://www.searo.who.int/entity/vector_borne_tropical_diseases/data/data_factsheet/en/. (Accessed 15 April 2022).
Wu, H., Bock, S., Snitko, M., Berger, T., Weidner, T., Holloway, S., et al. (2015). Novel dengue virus NS2B/NS3 protease inhibitors. Antimicrob. Agents Chemother. 59 (2), 1100–1109. doi: 10.1128/AAC.03543-14
Xie Zou, J., Wang, Q. Y., Shi, P. Y. ,. X. (2015). Targeting dengue virus NS4B protein for drug discovery. Antiviral Res. 118, 39–45. doi: 10.1016/j.antiviral.2015.03.007
Yao, X., Ling, Y., Guo, S., He, S., Wang, J., Zhang, Q., et al. (2018). Inhibition of dengue viral infection by diasarone-I is associated with 2’O methyltransferase of NS5. Eur. J. Pharmacol. 821, 11–20. doi: 10.1016/j.ejphar.2017.12.029
Yildiz, M., Ghosh, S., Bell, J. A., Sherman, W., Hardy, J. A. (2013). Allosteric inhibition of the NS2B-NS3 protease from dengue virus. ACS Chem. Biol. 8 (12), 2744–2752. doi: 10.1021/cb400612h
Yin, Z., Wang, W.-L., Wang, G., Chan, W.-L., Ranga Rao, K. R., Alam, J., et al. (2006). Peptide inhibitors of dengue virus NS3 protease.Part 1: Warhead. Bioorg. Med. Chem. Lett. 16, 36–39. doi: 10.1016/j.bmcl.2005.09.062
Yusof, R., Clum, S., Wetzel, M., Murthy, H. M., Padmanabhan, R. (2000). Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. J. Biol. Chem. 275, 9963–9969. doi: 10.1074/jbc.275.14.9963
Yusufzai, S. K., Osman, H., Khan, M. S., Razik, B. M. A., Ezzat, M. O., Mohamad, S., et al. (2018a). 4-thiazolidinone coumarin derivatives as two-component NS2B/NS3 DENV flavivirus serine protease inhibitors: synthesis, molecular docking, biological evaluation and structure–activity relationship studies. Chem. Cent. J. 12 (1), 69. doi: 10.1186/s13065-018-0435-0
Yusufzai, S. K., Osman, H., Khan, M. S., Razik, B. M. A., Mohamad, S., Sulaiman, O., et al. (2018b). Synthesis, X-ray crystallographic study, pharmacology and docking of hydrazinyl thiazolyl coumarins as dengue virus NS2B/NS3 serine protease inhibitors. Medicinal Chem. Res. 27 (6), 1647–1665. doi: 10.1007/s00044-018-2179-8
Zamri, A., Teruna, H. Y., Wulansari, S., Herfindo, N., Frimayanti, N., Ikhtiarudin, I. (2019). 3-(3, 4-Dimethoxyphenyl)-5-(2-fluorophenyl)-1-phenyl-4, 5-dihydro-1H-pyrazole. Molbank 2019 (4), M1088. doi: 10.3390/M1088
Zhao, Y., Soh, T. S., Zheng, J., Chan, K. W. K., Phoo, W. W., Lee, C. C., et al. (2015). A crystal structure of the dengue virus NS5 protein reveals a novel inter-domain interface essential for protein flexibility and virus replication. PloS Pathog. 11 (3), e1004682. doi: 10.1371/journal.ppat.1004682
Keywords: dengue virus, NS2B/NS3pro, antiviral, drug discovery, diagnostics
Citation: Norshidah H, Leow CH, Ezleen KE, Wahab HA, Vignesh R, Rasul A and Lai NS (2023) Assessing the potential of NS2B/NS3 protease inhibitors biomarker in curbing dengue virus infections: In silico vs. In vitro approach. Front. Cell. Infect. Microbiol. 13:1061937. doi: 10.3389/fcimb.2023.1061937
Received: 05 October 2022; Accepted: 18 January 2023;
Published: 14 February 2023.
Edited by:
Vivek Dhar Dwivedi, Quanta Calculus, IndiaReviewed by:
Rahul Shukla, Central Drug Research Institute (CSIR), IndiaRashmi Rana, Sir Ganga Ram Hospital, India
Copyright © 2023 Norshidah, Leow, Ezleen, Wahab, Vignesh, Rasul and Lai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Harun Norshidah, bm9yc2hpZGFoaGFydW5AZ21haWwuY29t; Ramachandran Vignesh, dmlnbmVzaEB1bmlrbC5lZHUubXk=; Ngit Shin Lai, bGFpbmdpdHNoaW5AdXNtLm15
 Harun Norshidah
Harun Norshidah Chiuan Herng Leow
Chiuan Herng Leow Kamarulzaman Ezatul Ezleen
Kamarulzaman Ezatul Ezleen Habibah A. Wahab
Habibah A. Wahab Ramachandran Vignesh
Ramachandran Vignesh Azhar Rasul
Azhar Rasul Ngit Shin Lai
Ngit Shin Lai