
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 26 May 2023
Sec. Extra-intestinal Microbiome
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1059339
Objective: This study assessed the impact of the cervical microbiome on reproductive outcomes in frozen embryo transfer (FET) patients.
Study design: This cross-sectional study included 120 women (aged 20–40 years) undergoing FET. A cervical sample obtained before embryo transfer was analyzed using 16S full-length assembly sequencing technology (16S-FAST), which detects full length 16S rDNA.
Results: We found that >48% of the identified Lactobacillus species were novel. The cervical microbiome was clustered into three cervical microbiome types (CMT): CMT1, dominated by L. crispatus; CMT2, dominated by L. iners; and CMT3, dominated by other bacteria. CMT1 had a significantly higher biochemical pregnancy rate (P=0.008) and clinical pregnancy rate (P=0.006) than CMT2 and CMT3. Logistic analysis showed that compared to CMT1, CMT2 and CMT3 were independent risk factors for biochemical pregnancy failure (odds ratio [OR]: 6.315, 95% confidence interval [CI]: 2.047-19.476, P=0.001; OR: 3.635, 95% CI: 1.084-12.189, P=0.037) and clinical pregnancy failure (OR: 4.883, 95% CI: 1.847-12.908, P=0.001; OR: 3.478, 95% CI: 1.221-9.911, P=0.020). A L. crispatus-dominated group as a diagnostic indicator of biochemical and clinical pregnancy positive had area under the curve (AUC) values of 0.651(P=0.008) and 0.645(P=0.007), respectively. Combining the cervical microbiome with embryonic stage optimized the diagnostic performance for biochemical and clinical pregnancy failure with AUC values of 0.743(P<0.001) and 0.702(P<0.001), respectively. Additionally, relative abundance of L. crispatus predicted biochemical pregnancy positive with AUC values of 0.679(P=0.002) and clinical pregnancy positive with AUC values of 0.659(P=0.003).
Conclusion: Cervical microbiome profiling using 16S-FAST enables stratification of the chance of becoming pregnant prior to FET. Knowledge of the cervical microbiota may enable couples to make more balanced decisions regarding the timing and continuation of FET treatment cycles.
Infertility affects 8–12% of reproductive-aged couples worldwide (Inhorn and Patrizio, 2015). Common causes of infertility include ovulatory disorders, tubal disease, inflammation, and sperm abnormalities; however, one-third of people are infertile due to unknown reasons (Bashiri et al., 2018; Carson and Kallen, 2021). In vitro fertilization and embryo transfer (IVF-ET) are currently used as effective treatments for infertility. However, the pregnancy rate remains approximately 50% (Zhang et al., 2021). Therefore, predicting the outcomes prior to the start of treatments may help to develop personalized treatment plans and reduce the psychological and financial stress for patients.
Female reproductive tract microbiota such as vaginal, cervical and uterus microbiota, may affect pregnancy outcome in IVF patients (Chen et al., 2017). A study showed that vaginal microbial community could not differ clinical pregnancy in in vitro fertilization-embryo transfer (IVF-ET) (Wang et al., 2021). While Haahr’ study showed that qPCR defined bacterial vaginosis (BV) associated abnormal vaginal microbiota was correlated to clinical pregnancy negative(Haahr et al., 2016). Microbiota of cervix which is in the transition zone between the vagina and uterus, is now getting increasing attention. A few studies have investigated the cervical bacteria and IVF pregnancy outcomes (Hao et al., 2021; Wang et al., 2021; Villani et al., 2022), but the effects of prediction were inconsistent. Although it seems the reproductive tract microbiota may have a negative impact on embryo implantation in biologically plausible, more research is needed in this field.
Lactobacillus is the most common and significant factor in the stabilization and imbalance of reproductive tract microbiome (Gajer et al., 2012; Ravel et al., 2013). The evidence of relationship between Lactobacillus and pregnancy outcome is conflicting. Lactobacillus was reported to significantly decreased in the infertile patients (Fu et al., 2020; Zhao et al., 2020; Karaer et al., 2021; Sezer et al., 2022), and women achieved biochemical pregnancy had higher level of Lactobacillus spp. (Bernabeu et al., 2019). However, another study reported that Lactobacillus showed no difference between women who got clinical pregnancy or not (Wang et al., 2021). It is supposed that the different results might be induced by insufficient identification of bacterial species levels. For instance, women with L. iners dominated microbiota were more susceptible to vaginal dysbiosis, bacterial vaginosis (BV) and sexually transmitted infections than L. crisptus (Borgdorff et al., 2014; Jespers et al., 2015; Reimers et al., 2016; van Houdt et al., 2018; Witkin et al., 2021). Thus, the effect of reproductive microbiota on pregnancy outcome might require further distinction of species level.
Until now, a few studies have used full-length of 16S rRNA sequence analysis to describe feature of microbiome. Full-length of 16S rRNA sequence analysis was reported to catalog the diversity of deeply branching phyla and identify species (Karst et al., 2018). Our previous study used 16S rDNA full-length assembly sequencing technology (16S-FAST) (Dong et al., 2021) also found improved discrimination ability on species level, because it reads the entire variable region of the 16S rRNA gene (V1-V9). In the current article, we aim to use 16S-FAST to describe the characteristics of cervical microbiota, and evaluate performance of 16S-FAST in predicting the biochemical and clinical pregnancy outcome of IVF treatment.
From September 2021 to December 2021, 230 planned FETs were performed. The inclusion criteria were as follows: female aged 20 to 40 years; good-quality cleavage-stage embryo or blastocysts transferred; endometrial thickness >8 mm; hormone replacement therapy (HRT) protocols; ethnic group is Han. The exclusion criteria were as follows: more than three previous unsuccessful FETs; uterine malformation, intrauterine adhesions, uterine fibroids, adenomyosis, or endometritis; had taken probiotics or antibiotics within 1 month; smokers or alcoholics; participation in any experimental drug study within 60 days. Finally, 120 participants were enrolled (Supplementary Figure 1). In accordance with the Declaration of Helsinki, the design of the present study was approved by the Ethics Committee of the Shengjing Hospital of China Medical University (Reference No. 2021PS016F). All participants gave written informed consents.
The endometrium was prepared with estradiol, which was administered via oral (titrated up to 2–3 mg twice per day) or transvaginal (titrated up to 1–2 mg twice per day) routes. Transvaginal ultrasound was performed to assess the endometrium after 12–14 days of estradiol administration, and progesterone was initiated once the endometrial lining was ≥8 mm and trilaminar in appearance. Progesterone (80 mg daily) was used for endometrial transformation. Cervical samples were collected before endometrium transformation according to the following steps: 1) a sterile cotton swab was placed into the patient’s cervical canal and rotated to obtain cervical samples, during this process, the operator ensured that the cotton swab did not touch the vaginal wall of the patient; 2) the samples were directly placed into the DNA storage tubes (CW2654, CwBiotech, Beijing, China).
The cervical samples were stored at room temperature and sent to a laboratory within two hours. Storage buffer contains 100 mM Tris–HCl (pH 9), 40 mM EDTA, 4 M guanidine thiocyanate (protein denaturant to inhibit bacterial growth), and 0.001% bromothymol, as previously described (Hisada et al., 2015; Nishimoto et al., 2016). Bacterial DNA was extracted using a DNA extraction kit (Qiagen Fecal DNA Extraction Kit, Qiagen, Hilden, Germany). Quantitative and qualitative analyses as well as quality control of the extracted DNA were performed. Sequencing was performed via the methods described in our previous study (Dong et al., 2021).
16S rDNA full-length sequence was generated via the methods previously described (Dong et al., 2021). Operational taxonomic unit (OTU) was clustered with a similarity threshold of 99%. Species annotations for all OTUs were performed with SILVA_132_SSURef_Nr99 database with default parameters (Quast et al., 2013). The software used is Mothur (Schloss et al., 2009), and the name of the method is wang. Muscle 3.8.31 (Edgar, 2004) was used to perform multiple sequence alignment, and FastTree 2.1.11 (Price et al., 2009; Price et al., 2010) was used to construct the evolutionary tree. Clustering of communities based on community composition and abundance was performed using complete linkage hierarchical clustering with three clusters in the R package, and R package heatmap 1.0.12 was used to draw the heatmap. The α bacterial diversity of the cervical microbiota community was estimated by QIIME1 V1.8.0 (Caporaso et al., 2010). The difference between groups was calculated by a Kruskal–Wallis sum-rank tests. Principal coordinates analysis (PCoA) was performed using the R 3.6.1 package vegan 2.5–3 analysis, and the P value was calculated by matching the Adonis method. Kruskal–Wallis sum-rank tests were used to detect significant differences between groups. To assess the impact of significant variables on pregnancy outcomes, univariate and multivariate logistic regression analyses were conducted using SPSS statistics (version 23.0; IBM, Armonk, NY, USA) to determine the odds ratio (OR) and confidence interval (CI) for the presence of pregnancy negative, then age and embryonic stage were also included into analysis as covariates considering that they are also important factors affecting pregnancy outcomes. The receiver operating characteristic (ROC) curve was modeled using GraphPad Prism 6 (GraphPad Software).
The reproductive outcomes were biochemical pregnancy rate (BPR) and clinical pregnancy rate (CPR). Biochemical pregnancy was defined as a human chorionic gonadotrophin (hCG) level ≥5 mIU/mL, and clinical pregnancy was defined as a gestational sac revealed by ultrasonography on the 21st day after a successful biochemical pregnancy.
One-hundred-and-twenty samples containing more than 2000 contigs were filtered after sequencing full-length 16S rDNA. The rarefaction curve appeared flat, indicating that the amount of sequencing data was reasonable (Supplementary Figure 2). Consistent with previous reports, Lactobacillus was the most abundant bacterium in these samples (Supplementary Figure 3). Lactobacillus identified in cervical samples was from nine lineages, which was present as 796 unique Lactobacillus 16S rDNA operational taxonomic units (OTU)s with at least a 1% genetic difference. We noticed that the majority of OTUs were derived from five lineages: L. iners, L. gasseri, L. jensenii, L. vaginalis, and L. crispatus, displaying 1100%, 43.7%, 45%, 30%, and 250% of the novel sequences, respectively; Lineage L. vaginalis was also composed of several strains previously annotated as L. iners, L. delbrueckii, L. fermentum, L. coleohominis, L. casei, and L. acidophilus. Similar phenomena were observed in the other two lineages, L. iners (comprising L. gasseri) and L. crispatus (comprising L. acidophilus and L. jensenii). Our result also showed the presence of intraspecific variations, which were dispersed in reference gene (Figure 1A). The genetic variations were most located in variable region 2 (V2) (33.3%) (Figure 1B). Then, to validate whether the novel strains identified among the OTUs were resulted from sequencing errors, we examined their mutations in the supporting reads and their distribution among patients. All 88 L. iners OTUs were observed in two or more patients, with at least five supporting full-length 16S rDNA sequences (Figure 1C). In order to identify the genetic relationships of the unique strains as well as the existing strains in the database, we draw evolutionary tree showing sequences annotated in the database and sources of the reference data (Figure 1D).
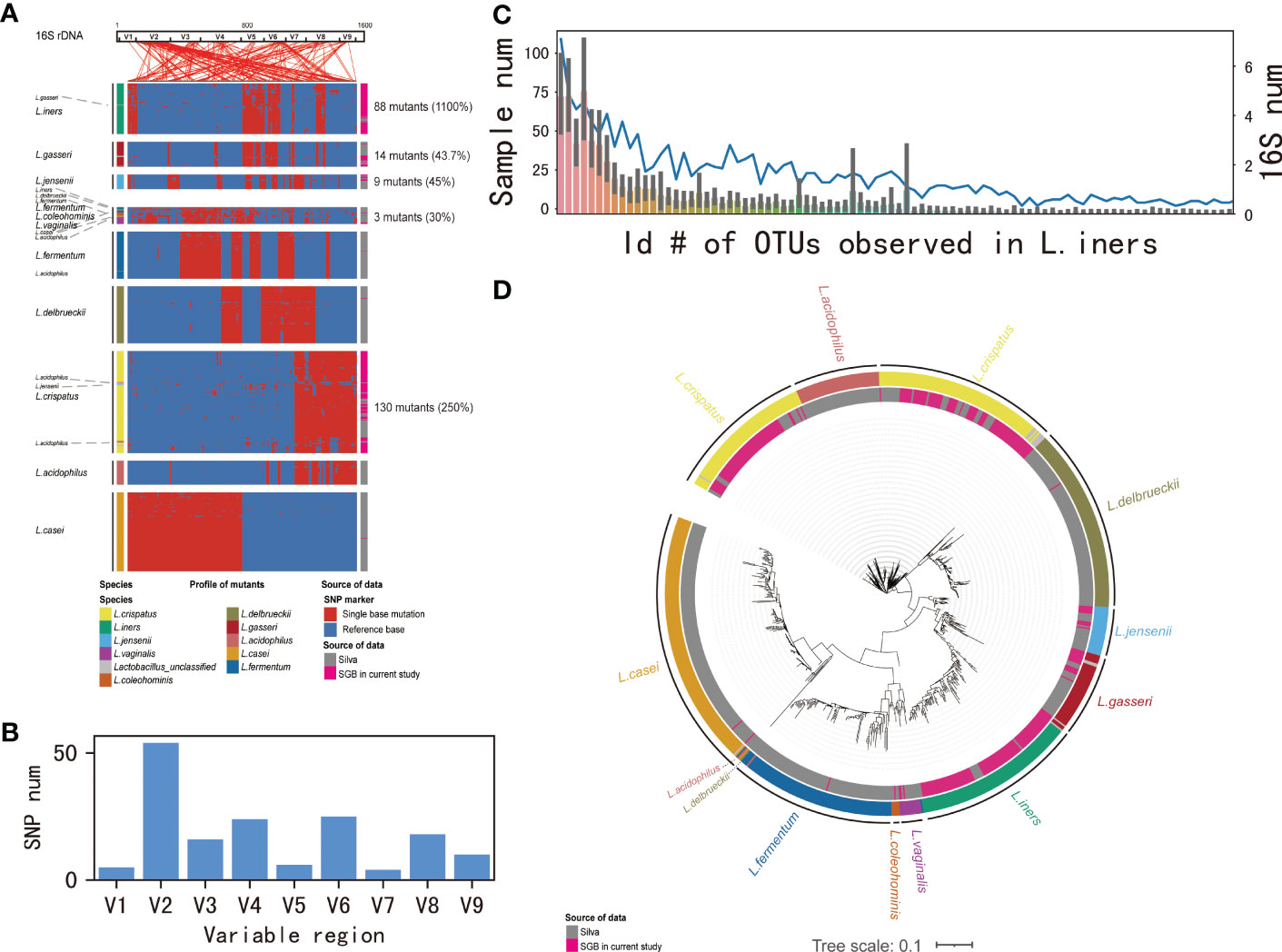
Figure 1 Identification of species in Lactobacillus by 16S-FAST. (A) Intra-species variation in Lactobacillus 16S rDNA genes. From left to right, 16S rDNA sequences annotated in the database, mutations detected in consensus sequences, and sources of reference data. Intra-species variations are clustered according to a presence/absence matrix. Coordinate positions in 16S rDNA are connected by lines above the panel. (B) SNP numbers in the variable region 1–9. (C) Numbers of samples that novel L.iners OTUs being observed. (D) Evolutionary tree showed detected 16S rDNA sequences annotated in the database and sources of reference data. Branch relations are connected by lines.
In addition, to compare the annotation differences on species levels based on 16S full-length and V3-4 region, we analyzed the top 20 strains. Compared to 16S-FAST, annotation according to V3-4 did not show L. crispatus, and OTU was extensively annotated as L. unclassified (Supplementary Figure 4). This result suggested that the 16S-FAST is indeed required for analysis in cervical microbiota.
Clustering of the cervical bacterial community was performed at the species level to assess the impact of the cervical microbiome on pregnancy outcomes. According to the 50 most abundant species, the cervical microbiome communities, which we defined as CMTs, were clustered into three major clusters dominated by L. crispatus (41.6%) (CMT1), L. iners (34.2%) (CMT2), and other bacteria (24.2%) (CMT3) such as Gardnerella vaginalis, Streptococcus gallolyticus, L. jensenii, Klebsiella pneumoniae, Atopobium vaginae, Prevotella amnii, L. gasseri, and Streptococcus agalactiae (Figure 2A). β diversity assessed by PCoA separated the three groups (Figure 2B). CMT1 had the highest α diversity, followed by CMT2 and CMT3 (Figure 2C). The abundance of the main contributors to each CMT strongly avoided each other (Figure 2D), suggesting that CMTs were groups of species that contributed to the composition of their microbiome communities.
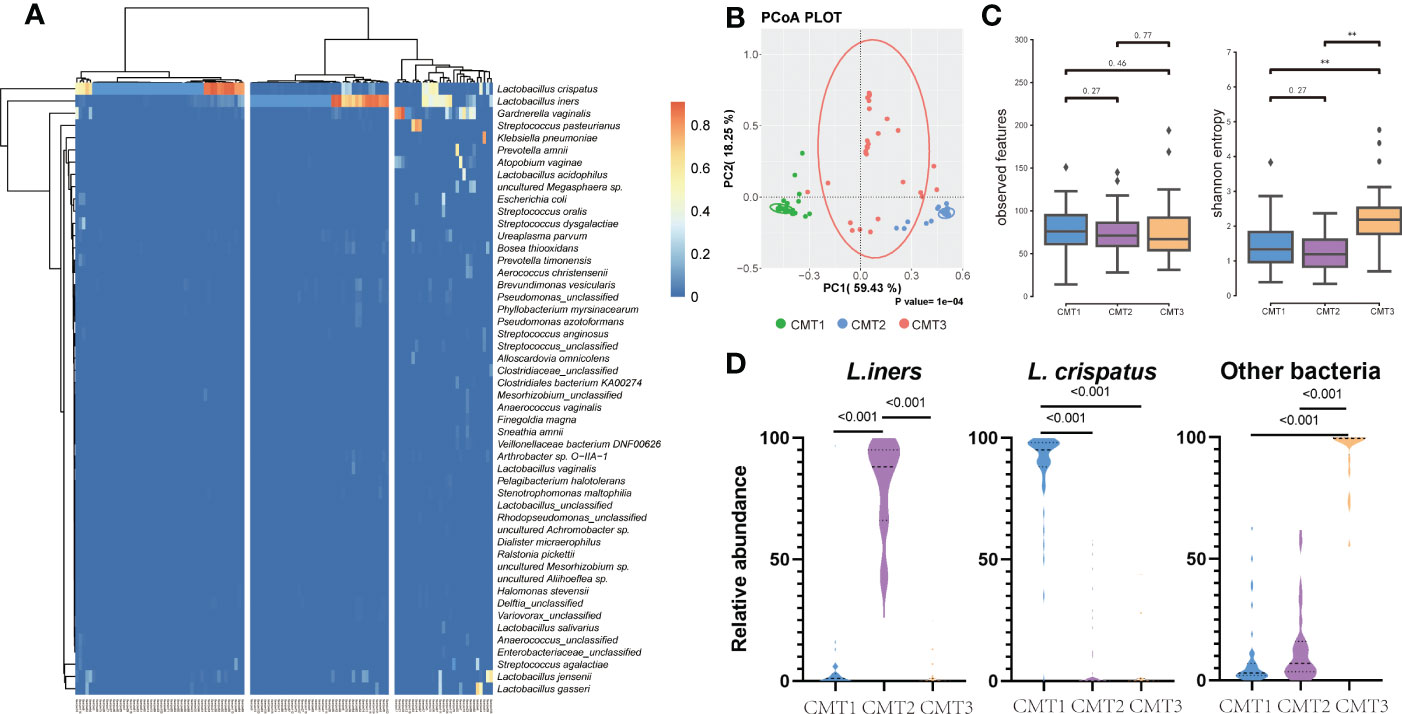
Figure 2 Clustering of cervical bacterial from 120 samples. (A) Heatmap of microbial taxa detected in the cervical bacterial communities of 120 participants. A color gradient was used to show abundance. The number of samples is shown on the bottom, and the name of species is shown on the right. Samples were clustered into three groups based on the species composition and abundance of cervical bacterial communities. (B) Principal co-ordinates analysis (PCoA) plot between three groups. (C) Observed features and Shannon entropy in three groups. (D) Relative abundance of L. crispatus, L. iners and other bacteria in three groups. “Other bacteria” refers to bacteria that is no L.crisptus and L.iners.
Data of the participants with different CMTs are listed in Table 1. Of all participants, 68.3% had a positive biochemical pregnancy, and 59.1% had a positive clinical pregnancy (Supplementary Table 1). Among these, CMT1 had significantly higher biochemical and clinical pregnancy rates (Table 1). The accompanying distribution of pregnancy outcomes is shown in Supplementary Figure 5.
To further analyze the risk factors for pregnancy failure, univariate and multivariate logistic regression analyses were conducted to determine the odds ratio (OR) and confidence interval (CI) for the presence of pregnancy negative, which showed that CMT was significantly associated with biochemical pregnancy failure. CMT2 (odds ratio [OR]: 4.109, 95% confidence interval [CI]: 1.549-10.901, P=0.05) and CMT3 (OR: 3.706, 95% CI: 1.287-10.667, P=0.015) significantly increased the risk factors for biochemical pregnancy failure compared to CMT1. After adjusting for age and embryonic stage as covariates, CMT2 and CMT3 were confirmed as independent risk factors for biochemical pregnancy failure (OR: 6.315, 95% CI: 2.047-19.476, P=0.001; OR: 3.635, 95% CI: 1.084-12.189, P=0.037). Embryonic stage day 3 was also identified as a risk factor (Table 2).
CMT was also significantly associated with clinical pregnancy failure. CMT2 (OR: 3.667, 95% CI: 1.501–8.958, P=0.004) and CMT3 (OR: 3.393, 95% CI: 1.279-9.000, P=0.014) significantly increased the risk factors for clinical pregnancy failure, and remained independent risk factors for clinical pregnancy failure after adjusted embryonic stage (OR: 4.883, 95% CI: 1.847-12.908, P=0.001; OR: 3.478, 95% CI: 1.221–9.911, P=0.020) when compared to CMT1 after adjusting for embryonic stage. Embryonic stage day 3 was also identified as a risk factor (Table 3).
We tested the diagnostic performance of CMT by dividing it into CMT1 and non-CMT1. Non-CMT1, as a diagnostic indicator of biochemical pregnancy failure and clinical pregnancy failure, had an area under the curve (AUC) of 0.651 (P=0.008) and 0.645 (P=0.007), respectively. Additionally, embryonic stage as a diagnostic indicator of biochemical pregnancy failure and clinical pregnancy failure had AUC values of 0.649 (P=0.009) and 0.614 (P=0.034), respectively. Moreover, combining CMT and embryonic stage optimized the diagnostic performance for biochemical pregnancy failure and clinical pregnancy failure with AUC values of 0.743 (P<0.001) and 0.702 (P<0.001), respectively (Figures 3A–C).
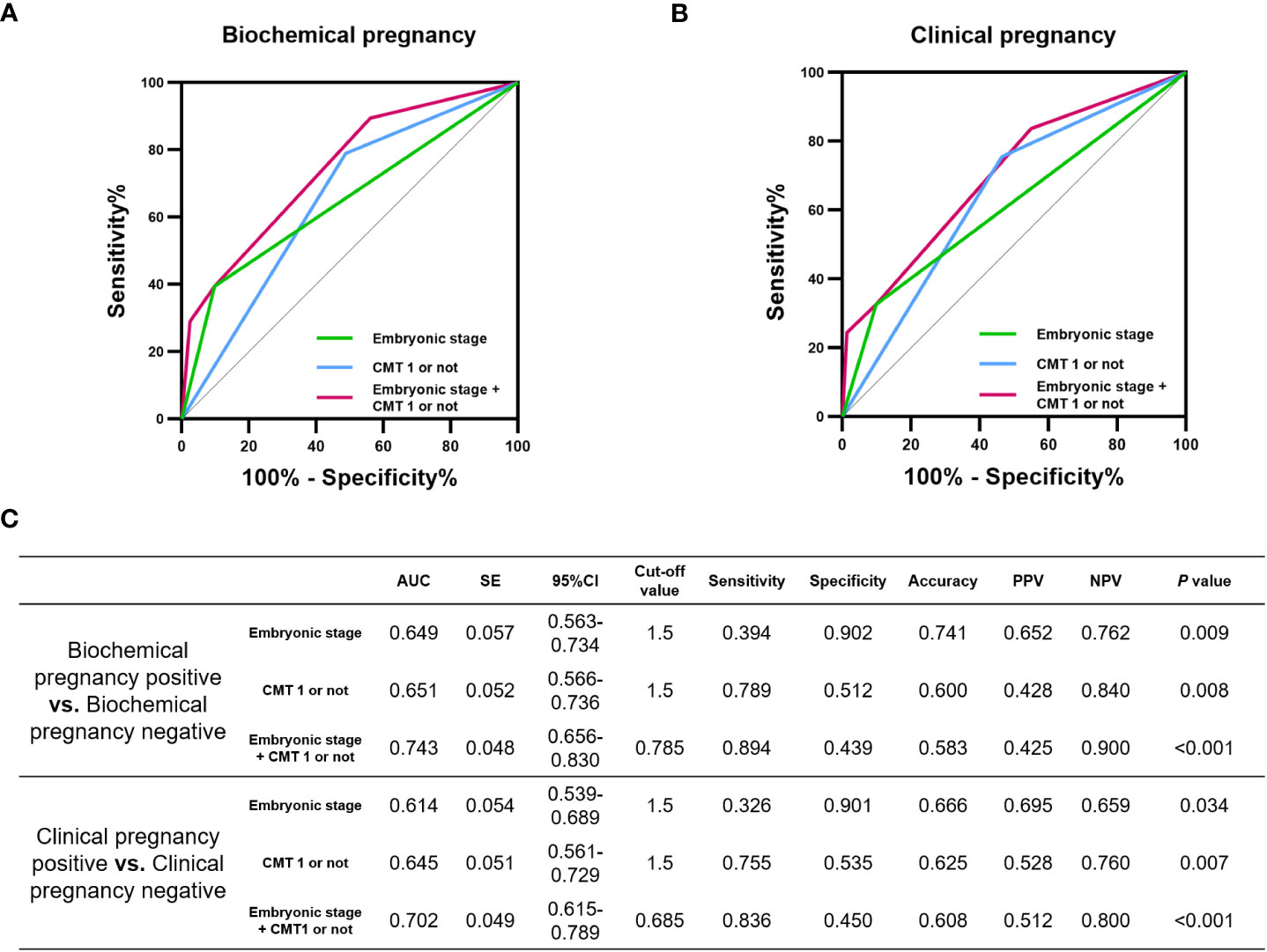
Figure 3 Diagnostic performance of CMT on the incidence of pregnancy failure. Potential of CMT in diagnosing (A) biochemical pregnancy failure and (B) clinical pregnancy failure. (C) area under the curve (AUC), standard error (SE), 95% confidence interval (CI), cut-off value, sensitivity, specificity, Accuracy, positive predictive value (PPV), negative predictive value (NPV) and P values for each ROC curve in this evaluation.
In addition, 16S-FAST can accurately identify L. crispatus, we also cared about the prediction performance of L. crispatus itself on pregnancy outcomes. The relative abundance of L. crispatus obtained by 16S-FAST had an AUC of 0.679 (P= 0.002) for biochemical pregnancy positive and an AUC of 0.659 (P= 0.003) for clinical pregnancy positive (Figure 4).
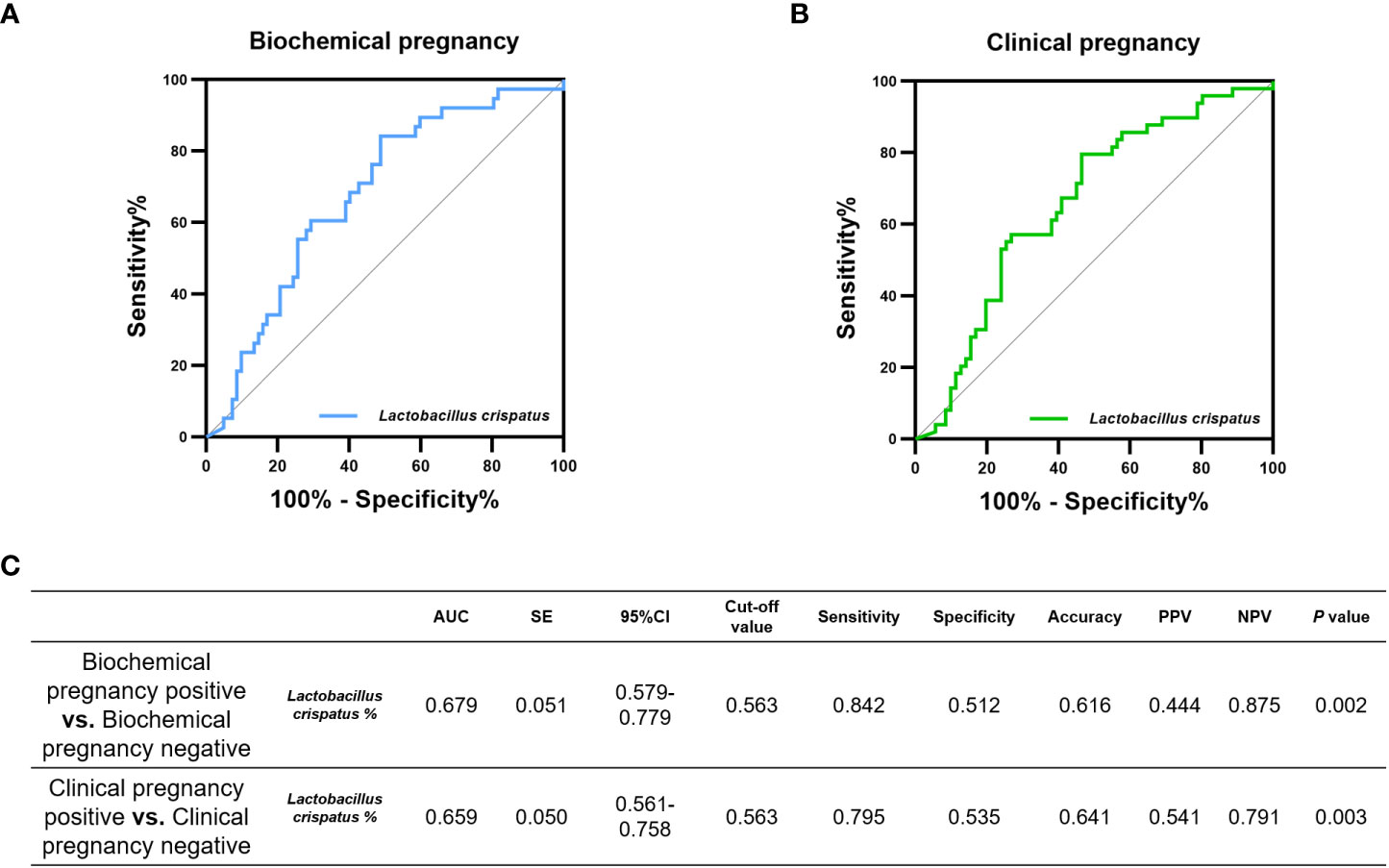
Figure 4 Diagnostic performance of relative abundance of L.crisptus measured by 16S-FAST on the incidence of pregnancy outcome. Potential of L.crisptus in diagnosing (A) biochemical pregnancy failure and (B) clinical pregnancy failure. (C) area under the curve (AUC), standard error (SE), 95% confidence interval (CI), cut-off value, sensitivity, specificity, Accuracy, positive predictive value (PPV), negative predictive value (NPV) and P values for each ROC curve in this evaluation.
In this study, we used 16S-FAST to describe the characteristics of cervical microbiome of women undergoing IVF, and we evaluated the predictive value of cervical microbiome for IVF outcomes. The cervical microbiome was clustered into three cervical microbiome types (CMT): CMT1, dominated by L. crispatus; CMT2, dominated by L. iners; and CMT3, dominated by other bacteria. CMT1 had a significantly higher biochemical and clinical pregnancy rate than CMT2 and CMT3. After reducing the bias caused by confounding factors, CMT2 and CMT3 were independent risk factors for biochemical or clinical pregnancy failure. A cervical dominance of L. crispatus was an indicator to predict good pregnancy outcomes.
We found L. crispatus-dominant microbiome was more prone to pregnancy. Previous studies had suggested that L. crispatus was a major composition of reproductive tract bacterial communities and typically had low Nugent scores (Gajer et al., 2012). It could maintain a healthy vaginal microbial ecosystem (Sehring et al., 2022), and was strongly negatively associated with dysbiosis (Borgdorff et al., 2016). Amato et al. observed that a predominance of L. crispatus of vaginal microbiome was a marker of a healthy vaginal ecosystem and was associated to intrauterine insemination (IUI) success (Amato et al., 2020). Taken together, infertile women with dominance of L. crispatus had a more chance achieving pregnancy success.
Interestingly, in our study, CMT1 and CMT2 were both Lactobacillus spp. dominant. However, CMT2 dominant by L.iners had not shown a benefit to pregnancy, which was inconsistent with previous research about pregnancy success with Lactobacillus. Karaer et al. found that the relative abundance of Lactobacillus was lower in women who failed to become pregnant using the V3-V4 regions of the 16S rRNA (Karaer et al., 2021). Bernabeu et al. investigated the vaginal microbiome influences on the IVF outcome by sequencing of V3-V4 region of 16S rRNA, reporting a greater presence of Lactobacillus spp. in pregnant women (Bernabeu et al., 2019). But it should be noticed that different species of Lactobacillus played different roles in the reproductive tract. Previous researches had shown that the L. iners-dominated reproductive tract microbiota appeared to be a risk of increased susceptibility to reproductive tract infections including Chlamydia trachomatis and BV (Macklaim et al., 2011; Santiago et al., 2012; van Houdt et al., 2018; Campisciano et al., 2020; Chen et al., 2022). Lots of researches also indicated that Chlamydia trachomatis and BV led to negative pregnancy outcomes (Wilson et al., 2002; Haahr et al., 2016; Chen et al., 2021a). In addition, traditional 16S rDNA fragment sequencing technology may not be possible to accurately distinguish the species of Lactobacillus spp. In this study, we found 16S-FAST improved discrimination ability on species level comparing with traditional sequencing of V3-V4 region of 16S rRNA. Therefore, Lactobacillus classification of species level in our study provided a better prediction of a reproductive outcome. In our results, CMT3, including Gardnerella vaginalis and Klebsiella pneumoniae, which was non-Lactobacillus dominant could also be one of risks of lower IVF pregnancy rates, which was consistent with the results of previous study (Sirota et al., 2014; Moreno et al., 2016). Moreno reported that high relative abundances of Gardnerella vaginalis and Klebsiella pneumoniae were observed in chronic endometritis in asymptomatic infertile women (Moreno et al., 2018), which indicates a negative IVF outcome (Chen et al., 2021b). In addition, non-Lactobacillus-predominant microbiota was associated with lower rates of implantation, pregnancy, ongoing pregnancy, live births, and other adverse reproductive outcomes in IVF (Wilson et al., 2002; Sirota et al., 2014; Moreno et al., 2016; Moreno and Simon, 2018).
Unsuccessful outcomes have been linked to dysregulated microbiome in endometrium (Moreno et al., 2022). However, due to the small amount of starting material and low-biomass microbiota, the endometrial microbiota is easily contaminated by exogenous bacterial DNA. Since the microbiome in the upper reproductive tract has been shown to be a continuum (Chen et al., 2017), the cervical microbiome, which is close to the uterine cavity, is convenient to obtain and contamination can be easily prevented. Currently, there are few studies that have reported differences in the cervical microbiome between women with positive assisted reproductive technology (ART) pregnancy outcomes and negative ART outcomes (Hao et al., 2021; Villani et al., 2022). However, the results are controversial and the testing is difficult to apply in clinical working, possibly because it is difficult to accurately detect bacteria at the species level. What’s more, we chose the time of sampling in the endometrial transformation stage, but not on the transfer day, so that we had enough time for testing and advised patients on whether or not to continue the transfer cycle according to the microbiome results. This could be of great benefit in achieving better pregnancy outcomes with interventions for cervical microbiome dysbiosis to postpone IVF and await a favorable profile, such as antibiotics combined with vaginal probiotic therapy (Buggio et al., 2019; Kadogami et al., 2020). A limitation of this study is that a well-defined study population was used, limiting the results to the frozen embryo transfer population. Whether these results can be translated to the general population trying to establish a pregnancy without ART cannot be determined from these data. In addition, this is a cross-sectional study and how cervical microbiome impact the uterine environment for implantation needs further studies.
In summary, we identified and analyzed the cervical microbiome of infertile patients in northern Asia and found that a microbiome dominated by L. crispatus was a protective factor for biochemical and clinical pregnancy in IVF patients. On the other hand, L. iners and other bacterial-dominated microbiomes were risk factors for pregnancy failure. The cervical community type could be an indicator of pregnancy prediction and provide a more precise treatment plan. However, the reproductive tract microbiome is rich in species and has a high diversity; its interaction with embryo implantation and the maternal host is still unclear, and further exploration and research are required.
The data presented in the study are deposited in the NCBI SRA repository, accession number PRJNA903403.
The studies involving human participants were reviewed and approved by the Ethics Committee of the Shengjing Hospital of China Medical University. The patients/participants provided their written informed consent to participate in this study.
WG: study design, execution, manuscript drafting, and critical discussion. SD: study design, execution, analysis, manuscript drafting, and critical discussion. ZW: analysis. JJ: study design, execution, analysis, manuscript drafting, and critical discussion. XW: study design and critical discussion. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (No. 81671423), National Key Research and Development Program of China (No. 2016YFC1000603), 2020 Shenyang Science and Technology Plan Program (No.20-205-4-006), Scientific and Technological Talents Applied Technology Research Program of Shenyang (No.18-014-4-56), Science and Technology Innovation Environment Creation Program of Shenyang (No.19-110-4-23), Shengjing Free Researcher Fund (No.201902), Natural Science Foundation of Zhejiang Province(No.LY21C200007, No.LQ20C200010).
We thank all the reviewers and editors for their helpful comments on this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1059339/full#supplementary-material
Amato, V., Papaleo, E., Pasciuta, R., Vigano, P., Ferrarese, R., Clementi, N., et al. (2020). Differential composition of vaginal microbiome, but not of seminal microbiome, is associated with successful intrauterine insemination in couples with idiopathic infertility: a prospective observational study. Open Forum Infect. Dis. 7 (1), ofz525. doi: 10.1093/ofid/ofz525
Bashiri, A., Halper, K. I., Orvieto, R. (2018). Recurrent implantation failure-update overview on etiology, diagnosis, treatment and future directions. Reprod. Biol. Endocrinol. 16 (1), 121. doi: 10.1186/s12958-018-0414-2
Bernabeu, A., Lledo, B., Diaz, M. C., Lozano, F. M., Ruiz, V., Fuentes, A., et al. (2019). Effect of the vaginal microbiome on the pregnancy rate in women receiving assisted reproductive treatment. J. Assist. Reprod. Genet. 36 (10), 2111–2119. doi: 10.1007/s10815-019-01564-0
Borgdorff, H., Armstrong, S. D., Tytgat, H. L., Xia, D., Ndayisaba, G. F., Wastling, J. M., et al. (2016). Unique insights in the cervicovaginal lactobacillus iners and l. crispatus proteomes and their associations with microbiota dysbiosis. PloS One 11 (3), e0150767. doi: 10.1371/journal.pone.0150767
Borgdorff, H., Tsivtsivadze, E., Verhelst, R., Marzorati, M., Jurriaans, S., Ndayisaba, G. F., et al. (2014). Lactobacillus-dominated cervicovaginal microbiota associated with reduced HIV/STI prevalence and genital HIV viral load in African women. ISME J. 8 (9), 1781–1793. doi: 10.1038/ismej.2014.26
Buggio, L., Somigliana, E., Borghi, A., Vercellini, P. (2019). Probiotics and vaginal microecology: fact or fancy? BMC Womens Health 19 (1), 25. doi: 10.1186/s12905-019-0723-4
Campisciano, G., Iebba, V., Zito, G., Luppi, S., Martinelli, M., Fischer, L., et al. (2020). Lactobacillus iners and gasseri, prevotella bivia and HPV belong to the microbiological signature negatively affecting human reproduction. Microorganisms 9 (1), 39. doi: 10.3390/microorganisms9010039
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 (5), 335–336. doi: 10.1038/nmeth.f.303
Carson, S. A., Kallen, A. N. (2021). Diagnosis and management of infertility: a review. JAMA 326 (1), 65–76. doi: 10.1001/jama.2021.4788
Chen, H., Min, S., Wang, L., Zhao, L., Luo, F., Lei, W., et al. (2022). Lactobacillus modulates chlamydia infectivity and genital tract pathology in vitro and in vivo. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.877223
Chen, C., Song, X., Wei, W., Zhong, H., Dai, J., Lan, Z., et al. (2017). The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat. Commun. 8 (1), 875. doi: 10.1038/s41467-017-00901-0
Chen, H., Wang, L., Zhao, L., Luo, L., Min, S., Wen, Y., et al. (2021a). Alterations of vaginal microbiota in women with infertility and chlamydia trachomatis infection. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.698840
Chen, W., Wei, K., He, X., Wei, J., Yang, L., Li, L., et al. (2021b). Identification of uterine microbiota in infertile women receiving in vitro fertilization with and without chronic endometritis. Front. Cell Dev. Biol. 9. doi: 10.3389/fcell.2021.693267
Dong, S., Jiao, J., Jia, S., Li, G., Zhang, W., Yang, K., et al. (2021). 16S rDNA full-length assembly sequencing technology analysis of intestinal microbiome in polycystic ovary syndrome. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.634981
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 (5), 1792–1797. doi: 10.1093/nar/gkh340
Fu, M., Zhang, X., Liang, Y., Lin, S., Qian, W., Fan, S. (2020). Alterations in vaginal microbiota and associated metabolome in women with recurrent implantation failure. mBio 11 (3), e03242–19. doi: 10.1128/mBio.03242-19
Gajer, P., Brotman, R. M., Bai, G., Sakamoto, J., Schutte, U. M., Zhong, X., et al. (2012). Temporal dynamics of the human vaginal microbiota. Sci. Transl. Med. 4 (132), 132ra152. doi: 10.1126/scitranslmed.3003605
Haahr, T., Jensen, J. S., Thomsen, L., Duus, L., Rygaard, K., Humaidan, P. (2016). Abnormal vaginal microbiota may be associated with poor reproductive outcomes: a prospective study in IVF patients. Hum. Reprod. 31 (4), 795–803. doi: 10.1093/humrep/dew026
Hao, X., Li, P., Wu, S., Tan, J. (2021). Association of the cervical microbiota with pregnancy outcome in a subfertile population undergoing In vitro fertilization: a case-control study. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.654202
Hisada, T., Endoh, K., Kuriki, K. (2015). Inter- and intra-individual variations in seasonal and daily stabilities of the human gut microbiota in Japanese. Arch. Microbiol. 197 (7), 919–934. doi: 10.1007/s00203-015-1125-0
Inhorn, M. C., Patrizio, P. (2015). Infertility around the globe: new thinking on gender, reproductive technologies and global movements in the 21st century. Hum. Reprod. Update 21 (4), 411–426. doi: 10.1093/humupd/dmv016
Jespers, V., van de Wijgert, J., Cools, P., Verhelst, R., Verstraelen, H., Delany-Moretlwe, S., et al. (2015). The significance of lactobacillus crispatus and l. vaginalis for vaginal health and the negative effect of recent sex: a cross-sectional descriptive study across groups of African women. BMC Infect. Dis. 15, 115. doi: 10.1186/s12879-015-0825-z
Kadogami, D., Nakaoka, Y., Morimoto, Y. (2020). Use of a vaginal probiotic suppository and antibiotics to influence the composition of the endometrial microbiota. Reprod. Biol. 20 (3), 307–314. doi: 10.1016/j.repbio.2020.07.001
Karaer, A., Dogan, B., Gunal, S., Tuncay, G., Arda Duz, S., Unver, T., et al. (2021). The vaginal microbiota composition of women undergoing assisted reproduction: a prospective cohort study. BJOG 128 (13), 2101–2109. doi: 10.1111/1471-0528.16782
Karst, S. M., Dueholm, M. S., McIlroy, S. J., Kirkegaard, R. H., Nielsen, P. H., Albertsen, M. (2018). Retrieval of a million high-quality, full-length microbial 16S and 18S rRNA gene sequences without primer bias. Nat. Biotechnol. 36 (2), 190–195. doi: 10.1038/nbt.4045
Macklaim, J. M., Gloor, G. B., Anukam, K. C., Cribby, S., Reid, G. (2011). At The crossroads of vaginal health and disease, the genome sequence of lactobacillus iners AB-1. Proc. Natl. Acad. Sci. U.S.A. 108 Suppl 1 (Suppl 1), 4688–4695. doi: 10.1073/pnas.1000086107
Moreno, I., Cicinelli, E., Garcia-Grau, I., Gonzalez-Monfort, M., Bau, D., Vilella, F., et al. (2018). The diagnosis of chronic endometritis in infertile asymptomatic women: a comparative study of histology, microbial cultures, hysteroscopy, and molecular microbiology. Am. J. Obstet. Gynecol. 218 (6), 602.e601–602.e616. doi: 10.1016/j.ajog.2018.02.012
Moreno, I., Codoner, F. M., Vilella, F., Valbuena, D., Martinez-Blanch, J. F., Jimenez-Almazan, J., et al. (2016). Evidence that the endometrial microbiota has an effect on implantation success or failure. Am. J. Obstet. Gynecol. 215 (6), 684–703. doi: 10.1016/j.ajog.2016.09.075
Moreno, I., Garcia-Grau, I., Perez-Villaroya, D., Gonzalez-Monfort, M., Bahceci, M., Barrionuevo, M. J., et al. (2022). Endometrial microbiota composition is associated with reproductive outcome in infertile patients. Microbiome 10 (1), 1. doi: 10.1186/s40168-021-01184-w
Moreno, I., Simon, C. (2018). Relevance of assessing the uterine microbiota in infertility. Fertil. Steril. 110 (3), 337–343. doi: 10.1016/j.fertnstert.2018.04.041
Nishimoto, Y., Mizutani, S., Nakajima, T., Hosoda, F., Watanabe, H., Saito, Y., et al. (2016). High stability of faecal microbiome composition in guanidine thiocyanate solution at room temperature and robustness during colonoscopy. Gut 65 (9), 1574–1575. doi: 10.1136/gutjnl-2016-311937
Price, M. N., Dehal, P. S., Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26 (7), 1641–1650. doi: 10.1093/molbev/msp077
Price, M. N., Dehal, P. S., Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PloS One 5 (3), e9490. doi: 10.1371/journal.pone.0009490
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41 (Database issue), D590–D596. doi: 10.1093/nar/gks1219
Ravel, J., Brotman, R. M., Gajer, P. (2013). Daily temporal dynamics of vaginal microbiota before, during and after episodes of bacterial vaginosis. Microbiome 1 (1), 29. doi: 10.1186/2049-2618-1-29
Reimers, L. L., Mehta, S. D., Massad, L. S., Burk, R. D., Xie, X., Ravel, J., et al. (2016). The cervicovaginal microbiota and its associations with human papillomavirus detection in HIV-infected and HIV-uninfected women. J. Infect. Dis. 214 (9), 1361–1369. doi: 10.1093/infdis/jiw374
Santiago, G. L., Tency, I., Verstraelen, H., Verhelst, R., Trog, M., Temmerman, M., et al. (2012). Longitudinal qPCR study of the dynamics of l. crispatus, l. iners, a. vaginae, (sialidase positive) g. vaginalis and p. bivia in the vagina. PloS One 7 (9), e45281. doi: 10.1371/journal.pone.0045281
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75 (23), 7537–7541. doi: 10.1128/AEM.01541-09
Sehring, J., Beltsos, A., Jeelani, R. (2022). Human implantation: the complex interplay between endometrial receptivity, inflammation, and the microbiome. Placenta 117, 179–186. doi: 10.1016/j.placenta.2021.12.015
Sezer, O., Soyer Caliskan, C., Celik, S., Kilic, S. S., Kuruoglu, T., Unluguzel Ustun, G., et al. (2022). Assessment of vaginal and endometrial microbiota by real-time PCR in women with unexplained infertility. J. Obstet. Gynaecol. Res. 48 (1), 129–139. doi: 10.1111/jog.15060
Sirota, I., Zarek, S. M., Segars, J. H. (2014). Potential influence of the microbiome on infertility and assisted reproductive technology. Semin. Reprod. Med. 32 (1), 35–42. doi: 10.1055/s-0033-1361821
van Houdt, R., Ma, B., Bruisten, S. M., Speksnijder, A., Ravel, J., de Vries, H. J. C. (2018). Lactobacillus iners-dominated vaginal microbiota is associated with increased susceptibility to chlamydia trachomatis infection in Dutch women: a case-control study. Sex. Transm. Infect. 94 (2), 117–123. doi: 10.1136/sextrans-2017-053133
Villani, A., Fontana, A., Barone, S., de Stefani, S., Primiterra, M., Copetti, M., et al. (2022). Identifying predictive bacterial markers from cervical swab microbiota on pregnancy outcome in woman undergoing assisted reproductive technologies. J. Clin. Med. 11 (3), 680. doi: 10.3390/jcm11030680
Wang, R., Zhou, G., Wu, L., Huang, X., Li, Y., Luo, B., et al. (2021). The microbial composition of lower genital tract may affect the outcome of in vitro fertilization-embryo transfer. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.729744
Wilson, J. D., Ralph, S. G., Rutherford, A. J. (2002). Rates of bacterial vaginosis in women undergoing in vitro fertilisation for different types of infertility. BJOG 109 (6), 714–717. doi: 10.1111/j.1471-0528.2002.01297.x
Witkin, S. S., Moron, A. F., Linhares, I. M., Forney, L. J. (2021). Influence of lactobacillus crispatus, lactobacillus iners and gardnerella vaginalis on bacterial vaginal composition in pregnant women. Arch. Gynecol. Obstet. 304 (2), 395–400. doi: 10.1007/s00404-021-05978-z
Zhang, Y., Xu, Y., Wang, Y., Xue, Q., Shang, J., Yang, X., et al. (2021). Comparison of the predictive value of progesterone-related indicators for pregnancy outcomes of women undergoing the short-acting GnRH agonist long protocol: a retrospective study. J. Ovarian Res. 14 (1), 14. doi: 10.1186/s13048-021-00768-2
Keywords: cervical microbiome, Lactobacillus crispatus, reproductive outcomes, frozen embryo transfer, 16S full-length assembly sequencing technology
Citation: Guan W, Dong S, Wang Z, Jiao J and Wang X (2023) Impact of a Lactobacillus dominant cervical microbiome, based on 16S-FAST profiling, on the reproductive outcomes of IVF patients. Front. Cell. Infect. Microbiol. 13:1059339. doi: 10.3389/fcimb.2023.1059339
Received: 08 October 2022; Accepted: 12 May 2023;
Published: 26 May 2023.
Edited by:
Liang Zhao, China Agricultural University, ChinaReviewed by:
Johanna Beth Holm, University of Maryland, Baltimore, United StatesCopyright © 2023 Guan, Dong, Wang, Jiao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiao Jiao, MTM4ODkyODQ3OTZAMTYzLmNvbQ==; Xiuxia Wang, d2FuZ3h4c2pAc2luYS5jbg==
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.