
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 28 November 2022
Sec. Molecular Bacterial Pathogenesis
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.997897
This article is part of the Research Topic Mobile Genetic Elements as Dissemination Drivers of Multidrug-Resistant Gram-Negative Bacteria View all 5 articles
A correction has been applied to this article in:
Corrigendum: Diversity of resistant determinants, virulence factors, and mobile genetic elements in Acinetobacter baumannii from India: A comprehensive in silico genome analysis
Introduction: The frequency of infections associated with multidrug resistant A. baumannii has risen substantially in India. The use of next-generation sequencing (NGS) techniques combined with comparative genomics has great potential for tracking, monitoring, and ultimately controlling the spread of this troublesome pathogen. Here, we investigated the whole genome sequences of 47 A. baumannii from India.
Methods: In brief, A. baumannii genomes were analyzed for the presence of antibiotic resistance genes (ARGs), virulence factors genes (VFGs), and mobile genetic elements (MGEs) using various in silico tools. The AbaR-type resistance islands (AbaRIs) were detected by examining the genetic environment of the chromosomal comM gene. Multilocus sequence types were determined using the Pasteur scheme. The eBURST and whole genome SNPs-based phylogenetic analysis were performed to analyze genetic diversity between A. baumannii genomes.
Results and discussion: A larger number of A. baumannii isolates belonging to the ST2 genotype was observed. The SNPs-based phylogenetic analysis showed a diversity between compared genomes. The predicted resistome showed the presence of intrinsic and acquired ARGs. The presence of plasmids, insertion sequences, and resistance islands carrying putative ARGs conferring resistance to antibiotics, quaternary ammonium compounds, and heavy metals was predicted in 43 (91%) genomes. The presence of putative VFGs related to adherence, biofilm formation and iron uptake was observed in the study. Overall, the comprehensive genome analysis in this study provides an essential insight into the resistome, virulome and mobilome of A. baumannii isolates from India.
The increasing spread of hospital-associated multidrug resistant (MDR) Acinetobacter baumannii strains is a health concern at the global level. The emergence of A. baumannii as a successful nosocomial pathogen is generally attributed to three aspects; firstly, its capacity to exhibit multidrug resistance through more than one type of mechanism. Examples include, enzymatic degradation of antibiotics, modification of target sites, upregulation of efflux pumps, and permeability alterations in the outer membrane (Asif et al., 2018; Mulani et al., 2019). The expression of multiple virulence factors (VFs) is the second attribute. Biofilm-associated proteins, lipopolysaccharides, capsular polysaccharides, porins, pili, phospholipase, outer membrane vesicle, iron, zinc acquisition system, and protein secretion systems are examples of essential VFs that assist A. baumannii in exhibiting antibiotic resistance, environmental persistence, host-pathogen interactions, and immune evasion (Sahu et al., 2012a; Sahu et al., 2012b; Gaidhani et al., 2014; Harding et al., 2018; Moubareck and Halat, 2020). The third critical aspect that makes A. baumannii a formidable pathogen is its propensity to readily acquire and spread resistance genes through different mobile genetic elements (MGEs) such as plasmids, insertion sequences, transposons, and resistance islands (Pagano et al., 2016; Cerezales et al., 2020).
In recent years, the prevalence of A. baumannii inflicted nosocomial infections in Indian health care systems is observed (Banerjee et al., 2018; Kumari et al., 2019). The reduced susceptibility of this pathogen to multiple antibiotics poses a therapeutic and economic burden (Asim et al., 2016). The reports also highlight a significant relationship between MDR A. baumannii and a higher mortality rate (Gandra et al., 2019; John et al., 2020). In the last two decades, many studies giving valuable insight into the resistance profile and prevalence of A. baumannii from different regions of India have been reported (Sengupta et al., 2001; Prashanth and Badrinath, 2006; Yavankar et al., 2007; Sivaranjani et al., 2013; Banerjee et al., 2018). The investigations exploring the epidemiology and endemicity of this nosocomial pathogen have also been carried out (Mathai et al., 2012; Dash et al., 2013). With the advancement in molecular techniques, many PCR- based oligotyping studies from India have effectively detected different types of intrinsic or acquired resistance determinants in clinically significant A. baumannii (Jones et al., 2014; Rynga et al., 2015; Pragasam et al., 2016; Kumar et al., 2019). Furthermore, several studies have used a combination of phenotypic and/or molecular techniques to establish a linkage between the presence of plasmids, insertion sequences, integron cassettes, and AbaRI in the carriage of antibiotic resistance in A. baumannii (Pardesi et al., 2007; Khajuria et al., 2014; Saranathan et al., 2014; Jose et al., 2017). Nevertheless, although most of the aforementioned studies are informative, they deal with a limited number of ARGs, VFGs or MGEs.
Contrary to these numerous individual phenotypic or genotypic characterization reports, whole genome sequencing can offer an overall insight into the architecture of A. baumannii. The prior A. baumannii genome sequences from India gave a summarized account of resistance genes present in the genomes (Singh et al., 2013; Kumar et al., 2015; Mahalingam et al., 2016). Apart from these individual genome studies, few other investigators attempted to compare multiple A. baumannii genomes from India, however the number of such studies remained limited (Balaji et al., 2015; Vijayakumar et al., 2018). Later, in 2019 a notable comparative genome analysis explained in-depth account of niche specific genome expansion by comparing complete genome of A. baumannii DS002, a soil isolate against 78 other A. baumannii publicly available genomes (Yakkala et al., 2019).
The future of pathogenic infection management in the impending post-antibiotic era depends upon the application of advanced genomics combined with functional analysis. Considering this grave scenario, the use of high-throughput sequencing techniques to investigate the outbreak, transmission, resistome, virulome, and mobilome of MDR A. baumannii isolates has become a need of the hour. This study aimed to determine diversity amongst the putative genetic determinants associated with antimicrobial resistance, virulence, and hyper genome plasticity in clinically significant A. baumannii isolates circulating in India. The present study provides a comparative genome analysis of 47 A. baumannii isolates collected from India during 2005-2020.
We have performed a genome-wide comparison of 47 A. baumannii isolates reported from India. We screened the microbial database at NCBI for the complete genome sequences of A. baumannii isolates collected from Indian hospitals. We found a total of 278 A. baumannii genomes out of which 42 were complete. These genomes belonged to isolates reported from seven different hospitals across India during 2014-2020. In addition, five A. baumannii isolates, namely, AIIMS5, AIIMS7, MMC18, SRMC9, and SRMC18, collected in 2005 and maintained as a part of our laboratory collection were also sequenced in this study. The general information regarding the collection year, specimen type, and geographical location of all A. baumannii genomes involved in this study is given in Table 1.
The genomic DNA from five A. baumannii isolates was extracted using the NaCl-CTAB method previously described (Wilson, 2001). The quality and quantity of the isolated DNA were checked on DeNovix DS-11 Spectrophotometer (DeNovix, Wilmington, DE). The QC passed DNA was used to prepare paired-end DNA libraries using the Illumina Nextera XT kit (Illumina, San Diego, CA) following the manufacturer’s guidelines. The DNA libraries were sequenced using Illumina NextSeq 500 platform with 2 x150 bp chemistry. The NGS QC toolkit v2.3.3 was used for quality check and filtering of raw reads using default parameters (PHRED quality score - 20) (Patel and Jain, 2012). Filtered high-quality reads were used to generate draft-genome using SPAdes de-novo assembler v3.11.1 (Bankevich et al., 2012). The genome coverage was calculated using BBMap v38.47 (https://sourceforge.net/projects/bbmap). The quality of assembled genome was checked using QUAST v5.0.2 (Gurevich et al., 2013). The annotations of all 47 A. baumannii genomes were carried out using Prokka v2.1.1 and comprehensive genome analysis service provided at Pathosystems Resource Integration Center (PATRIC) v3.6.5 (Seemann, 2014; Wattam et al., 2016).
The prediction of putative ARGs belonging to different resistance mechanisms was carried out using the Resistance Gene Identifier (RGI) tool provided at the Comprehensive Antibiotic Resistance Database (CARD) (Mcarthur et al., 2013). To identify ARGs, the A. baumannii genome assemblies were queried against the CARD database using perfect and strict criteria. The predicted ARGs showing sequence similarity between 97% to 100% were further classified into different resistance mechanisms (Alcock et al., 2020).
The putative VFGs present in A. baumannii genomes were predicted using an automatic pipeline called VFanalyzer provided at http://www.mgc.ac.cn/cgi-bin/VFs/v5/main.cgi (Liu et al., 2018). The VFanalyzer conducts a thorough search of the sequence similarity of VFGs with the known and predicted VFGs available in the virulence factors database for bacterial pathogens (VFDB). The identified VFGs were further classified into nine VF categories.
The prediction of plasmids in AIIMS5, AIIMS7, MMC18, SRMC9, and, SRMC18 genomes sequenced in this study was carried out using PlasmidSPAdes (Antipov et al., 2016). For analysis, the raw reads were used and based on the median chromosomal coverage the plasmid contigs were filtered. The filtered sequences were annotated using PATRIC v3.6.5. The plasmid sequences of 42 other A. baumannii genomes available at NCBI were downloaded. The presence of ARGs in all plasmid sequences was detected using CARD. The presence of insertion sequences and transposons was detected through a manual BLAST analysis of each A. baumannii genome against the ISfinder database (Siguier et al., 2006). The insertion sequences and transposons with an e-value equal to 0.0 were selected and further evaluated for the presence of ARGs. The prophage regions present within genome sequences were detected using the PHAge search tool enhanced release (PHASTER) database (Arndt et al., 2019). The putative prophage regions were further classified into bacteriophage families. The detection of AbaRIs was manually done by observing the genetic environments of the chromosomal comM gene using the interactive genome browser provided at the PATRIC server. The NCBI BLAST was used to align detected resistance island sequences against the sequences of previously known resistance islands (Altschul et al., 1990). The earlier proposed naming system for AbaRIs was used to classify the AbaRI variants predicted in this study (Bi et al., 2019). The annotations of detected AbaRIs regions obtained using PATRIC genome browser were used to construct AbaRIs gene maps in R 4.5.0.
The web-based sequence typing of the 47 A. baumannii genomes was carried out using MLST v2.0 services available at http://www.genomicepidemiology.org (Larsen et al., 2012). For the sequence typing, we used the Pasteur scheme which is designed to identify the seven house-keeping genes namely 60-kDa chaperonin (cpn60), elongation factor EF-G (fusA), citrate synthase (gltA), CTP synthase (pyrG), homologous recombination factor (recA), 50S ribosomal protein L2 (rplB), and RNA polymerase subunit B (rpoB). Furthermore, to understand the possible genetic relatedness between ST profiles we performed eBURST analysis using the goeBURST algorithm available at http://goeBURST.phyloviz.net (Francisco et al., 2009). To determine founding STs, we uploaded the numeric data of ST profiles obtained from MLST 2.0 in the flat text file format to goeBURST v1.2.1.
The phylogenetic relationship between 47 A. baumannii genomes was determined through multiple sequence alignment of genomes based on the single nucleotide polymorphisms (SNPs). The genome alignment and detection of SNPs in each genome were carried out using novel SNP procedure provided at http://cge.cbs.dtu.dk/services/CSIPhylogeny/ (Kaas et al., 2014). The novel SNP procedure filters, concatenates, aligns, and constructs a maximum likelihood phylogenetic tree with 100 bootstraps. The A. baumannii ATCC 19606 genome was used as a reference. The final version of the phylogenetic tree was visualized in iTOL v6 (Letunic and Bork, 2021).
The Welch’s t-test was performed to identify difference between the number of acquired ARGs predicted between genomes (n1 = 18, , S1 = 2.43, ) belonging to ST2 genotype and genomes (n2 = 29, , S2 = 5.21, ) belonging to other STs detected in the study. The difference was considered significant at p<0.05.
The assembled genomes of AIIMS5, AIIMS7, MMC18, SRMC9, and, SRMC18 showed an average of 38.95% GC content. The average assembly size of five sequenced genomes ranged between 3.7 to 3.8 Mb. The comprehensive genome analysis using PATRIC3.6.5 predicted a mean total of 3652 protein-coding sequences, 276 subsystems, and 62 RNAs. The Supplementary Table S1 represents the assembly statistics for five A. baumannii genomes. The Table 1 provides a summarized account of genomic features of all 47 Indian A. baumannii genomes analyzed in this study.
The web-based MLST analysis of 47 genomes using the Pasteur scheme predicted a total of 15 STs. Out of the 47 genomes analyzed, 18 (38.3%) belonged to ST2, while 10 (21.2%) belonged to ST622. We also detected presence of ST10 (n=5), ST25 (n=2), and ST1 (n=2) in the study. The remaining ten isolates belonged to the ten STs namely, ST85, ST94, ST150, ST285, ST374, ST494, ST575, ST1512, ST1545, and ST1549.
The eBURST analysis grouped STs into clusters based on single, double, and triple locus variation between ST profiles detected. A total of three clusters and five singletons were observed in the population of 15 STs (Figure 1A). The largest cluster included six STs namely, ST10, 494, 575, 1512, 1545, and 25. In this cluster, ST10 and ST575 were single locus variant while ST10 and ST1512 were double locus variants. The remaining STs in this cluster were triple variants. The second cluster included the double locus variant ST1 and ST94 while the third cluster included triple locus variants ST150 and ST374.
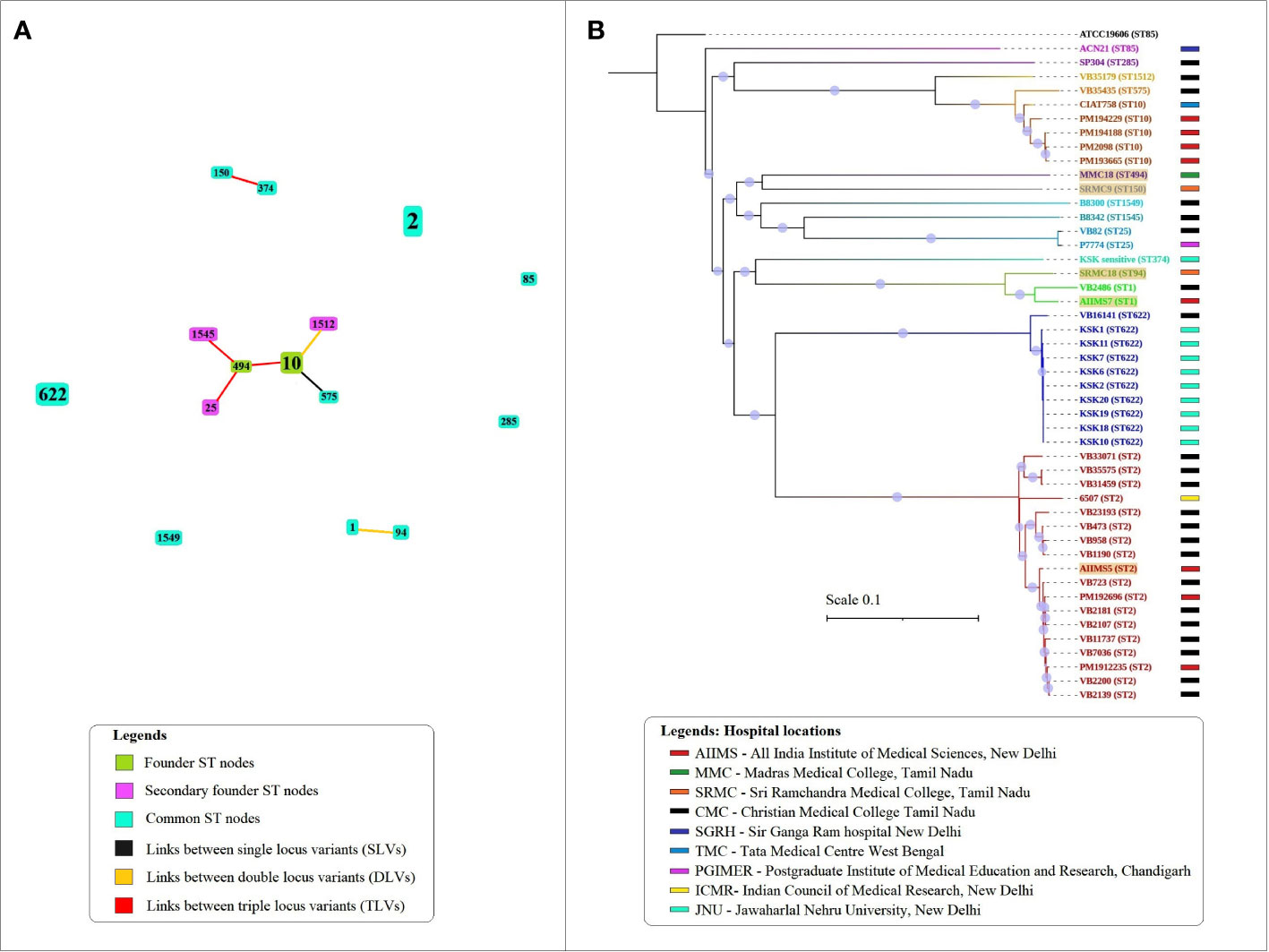
Figure 1 The sequence types and phylogenetic relationship between A. baumannii isolates analyzed in the study. (A) The eBURST analysis showing relationship between 15 STs detected in the population using the Pasteur scheme. The size of nodes is proportional to the number of isolates presenting that ST in the population under study. (B) The alignments of concatenated SNPs with 100 bootstrap replicates explain the phylogenetic relationship between 47 Indian A. baumannii genomes. The A. baumannii ATCC 19606 was used as the reference genome for alignment and SNP calling. The tree is rooted at reference genome. Different colors represent the grouping by sequence types.
The SNP-based phylogeny showed presence of two main subclades (Figure 1B). The first subclade included eight genome sequences namely SP304 (ST285), VB35179 (ST1512), VB35435 (ST575), CIAT758 (ST10), PM194229 (ST10), PM194188 (ST10), PM2098 (ST10), and PM193665 (ST10) while the second subclade included rest of A. baumannii genomes. The phylogenetic analysis also showed that the A. baumannii genomes belonging to ST2 and ST622 formed two major monophyletic clades. The results of MLST and eBURST analysis when compared with the SNP phylogeny, showed that single and double locus variants were situated under same clades while the triple locus variants such as ST150 and ST374 were separated into different clades. The SNP data matrix showed variation between similar STs such as ST2, ST10, ST25 and ST1 (Figure S1). In case of ST622, nine (KSK1, KSK2, KSK6, KSK7, KSK10, KSK11, KSK18, KSK19, and KSK20) out of 10 genomes showed very low variation, with the only exception of VB16141 which showed higher variation. Overall, the phylogenetic analysis based on the alignment of concatenated SNP showed accordance with the STs detected in this study.
The resistome analysis of 47 A. baumannii genomes showed presence of a total of 79 types of ARGs of intrinsic as well as acquired origin belonging to six resistance mechanisms, namely antibiotic efflux (n=21), antibiotic inactivation (n=51), antibiotic target alteration (n=3), antibiotic target protection (n=1), antibiotic target replacement (n=2), and reduced permeability to antibiotics (n=1) (Figure 2). All predicted β-lactamase genes showed 100% sequence similarity with the respective β-lactamase alleles present in the CARD database. Apart from the β-lactamase genes, the remaining ARGs showed 97 to 100% sequence similarity with the ARG sequences present in the CARD database. Three types of efflux pump associated ARGs, namely the resistance-nodulation cell division (RND) family, the major facilitator superfamily (MFS), and the small multidrug resistance (SMR) family were predicted in the study. We predicted 15 alleles of OXA-51 type and 12 alleles of ADC-type β-lactamases in this study. Additionally, OXA-23 and its allele OXA-422 as well as OXA-58 and its allele OXA-420 were predicted in the study. The most dominant type of family encoding β-lactam inactivating enzymes predicted was blaOXA-23 (n=34/47), followed by blaADC-73 (n=13/47). Overall, 46 out of 47 A. baumannii genomes showed the coexistence of at least one type of the OXA-type-β-lactamases with one type of the ADC-type of cephalosporinases. The presence of putative genes for aminoglycoside acyltransferases (AAC family), aminoglycoside adenyltransferases (ANT family), and aminoglycoside phosphotransferases (APH family) was detected in 10, 46, and 38 genomes, respectively. The putative sulphonamide resistance conferring gene sul1 was predicted in 34 genomes, while sul2 was detected in 42 genomes. The presence of lpsB gene associated with reduced permeability to antibiotics was predicted in 42 genomes. However, the genomes of AIIMS5, AIIMS7, MMC18, SRMC9, and SRMC18 did not show the presence of the lpsB gene. The quinolone resistance-determining region (QRDR) associated parC and gyrA were detected in 47 and 43 genomes, respectively. However, the presence of nonmutated gyrA observed in SRM9, SP304, B8342, and B8300 indicates a potential susceptibility of these isolates to quinolones. In case of acquired ARGs no significant difference was found between acquired ARGs predicted in ST2 as compared to the acquired ARGs predicted in rest of the 14 STs detected in the study (t = 0.882, p = 0.191, critical t = 1.756). Amongst the 18 A. baumannii isolates belonging to ST2, 14 (77%) showed the presence of blaOXA-23. A variation in the number of acquired ARGs was observed in the study. The genomes of SRMC9, SP304, B8300, and B8342 showed absence of acquired ARGs. A lower number of acquired ARGs was observed in AIIMS5 (n = 4), MMC18 (n = 1), and SRMC18 (n = 1) which were collected during 2005 in comparison to the remaining genomes which showed a relatively higher number (13 ≥ n ≥ 5) of acquired ARGs (Figure 3).
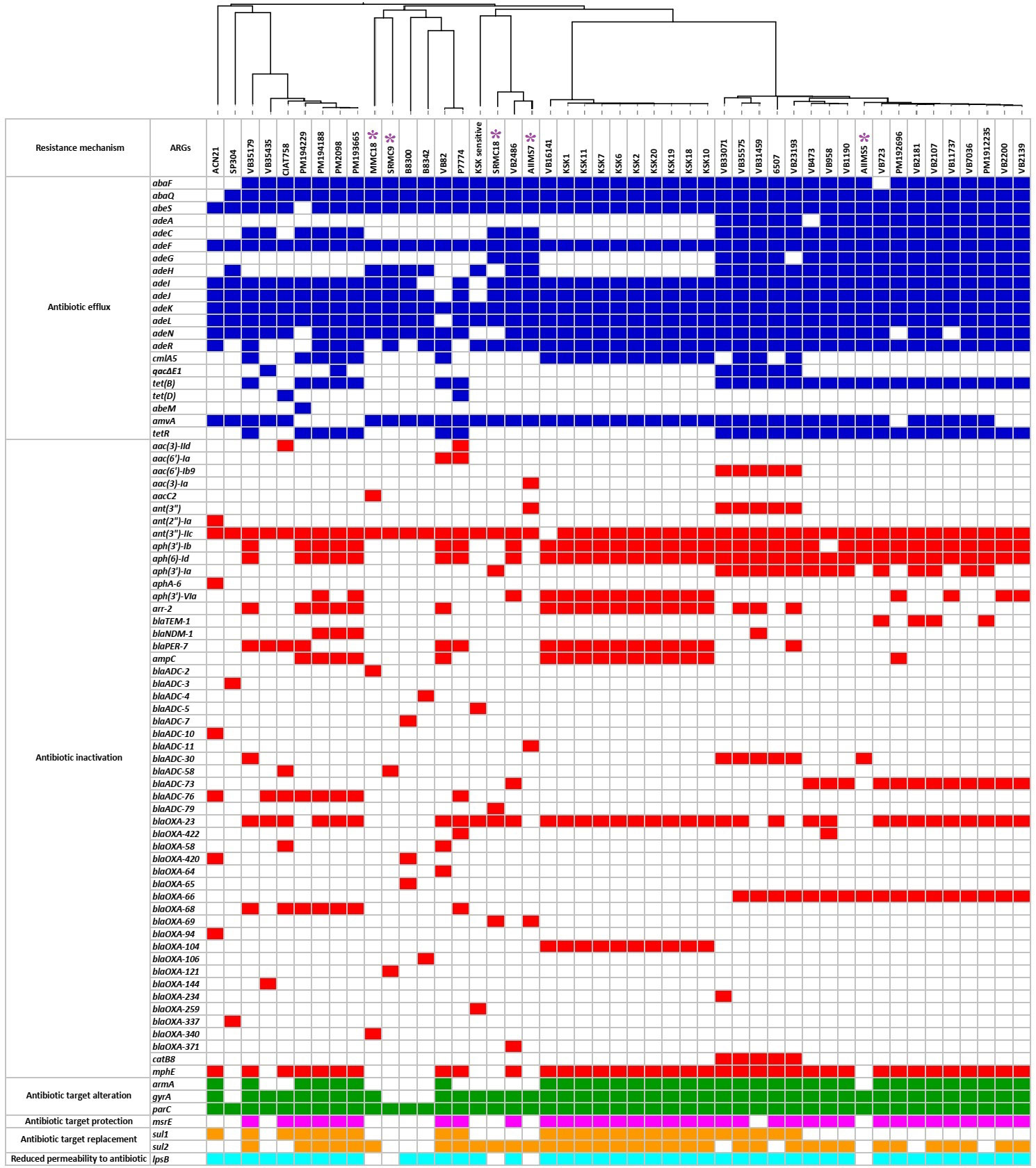
Figure 2 The predicted resistome of 47 A. baumannii genomes from India. The colored squares represent presence of ARGs while the empty or colorless squares represent absence of ARGs. The purple asterisks represent five A. baumannii isolates sequenced in this study.
A total of 56 types of putative VFGs were predicted in 47 A. baumannii genomes (Figure 4). The predicted VFGs were grouped under nine categories namely, adherence (n=4), biofilm formation (n=14), enzyme (n=2), immune evasion (n=8), iron uptake (n=19), regulation (n=4), serum resistance (n=2), antiphagocytosis (n=2), and stress adaptation (n=1). The putative VFGs predicted in all 47 genomes included outer membrane protein A (ompA) involved in adherence and invasion, efflux pump adeFGH involved in biofilm synthesis, pgaABCD locus encoding poly-β-1-6-N-acetylglucosamine required for biofilm formation, phospholipase C and D, the capsular polysaccharide biosynthesis gene cluster K, the lpx genes involved in lipid biosynthesis, the two-component signal transduction system BfmRS, and penicillin binding protein G (pbpG). Except for the basI in VB35179, all putative genes associated with acinetobactin biosynthesis and intake (barAB, basABCDFGHIJ, bauABCDEF, and entE) were detected in all genomes. The presence of putative bap gene encoding biofilm associated protein, the putative csuA/BABCDE operon associated with pili synthesis and assembly, the putative hemO encoding the heme oxygenase, and the putative abaI inducer and its receptor abaR involved in the quorum sensing were predicted in 37, 39, 38, and 40 genomes respectively. A noticeable variation was observed in the putative genes encoding virulence factors such as O-antigen polysaccharide (lpsO), type IV pilin protein (pilE), flagellar biosynthesis protein (flip), enzyme involved in biosynthesis of dTDP-l-rhamnose (rmlD), catalase (katA), and the bifunctional enzyme UDP‐N‐acetylglucosamine 2‐epimerase/ManNAc kinase associated with antiphagocytic capsular polysaccharide synthesis (wbjD/wecB). Altogether, a vast variety of VFGs were predicted in this study.
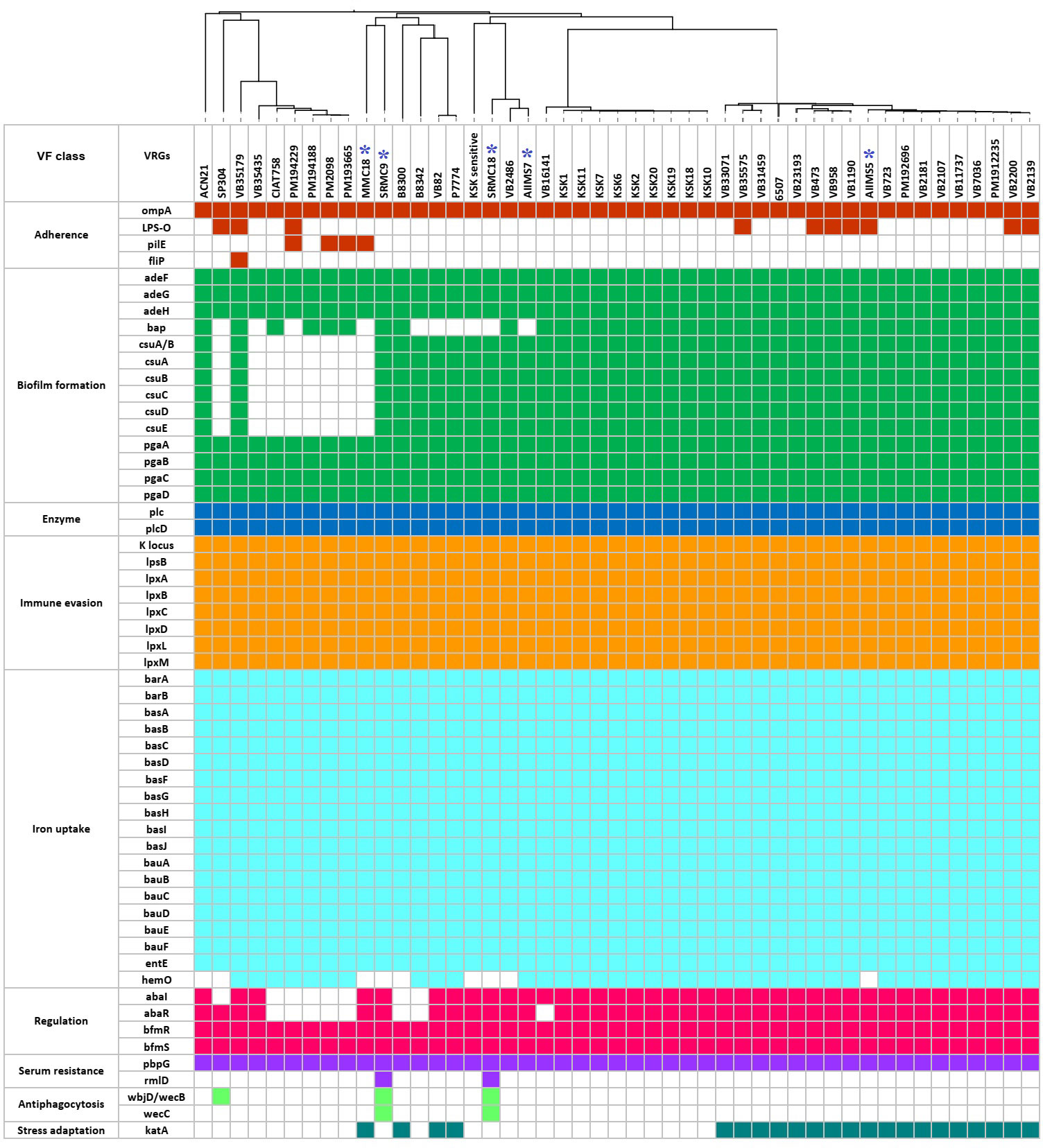
Figure 4 The predicted virulome of 47 A. baumannii genomes from India. The colored squares represent presence of VFGs while the empty or colorless squares represent absence of VFGs. The blue asterisks represent five A. baumannii sequenced in this study.
The plasmidSPAdes showed the presence of plasmid contigs in AIIMS5, AIIMS7, MMC18, and SRMC18. The CARD-based analysis of the detected plasmid contigs, as well as plasmids from other 42 A. baumannii genomes, predicted the presence of various putative ARGs associated with penems, aminoglycoside, tetracycline, macrolide, phenicol, rifamycin, sulfonamide, and heavy metal resistance (Figure 3).
Amongst the 274 prophage regions predicted, 108 were intact, 120 were incomplete and 46 were questionable. Further analysis classified predicted prophage regions into 5 families namely, siphoviridae (n=191), myoviridae (n=49), podoviridae (n=32), autographiviridae (n=1), and inoviridae (n=1) (Figure S2). The sequence showing similarity to PHAGE_Acinet_Bphi_B1251 was detected in all 47 genomes.
All 47 A. baumannii genomes analyzed in this study harbored numerous ISs of diverse bacterial origin. A total of 16 IS families, including Tn3, IS1, IS3, IS4, IS5, IS6, IS21, IS30, IS66, IS91, IS256, IS630, IS701, IS982, ISL3, and ISNCY, were detected in the study. We detected the presence of structures showing partial similarity to those of ISAba1-blaADC (n = 21/47) and ISAba1-blaOXA-23 (n = 36/47).
All Tn3-type transposons detected in this study showed presence of antibiotics and heavy metal resistance genes. A total of four types of Tn3 similar to Tn5393, TnAs3, TnAs2, and Tn3 were predicted (Figure S3). Forty out of 47 genomes studied showed the presence of at least one type of Tn3 carrying at least one resistance gene. The Tn5393 detected in 35 genomes showed the presence of ARGs associated with aminoglycoside resistance (strA/aph(3’’)-Ib, strB/aph(6)-Id). In comparison, the TnAs3 detected in 29 genomes contained putative ARGs associated with an aminoglycoside, chloramphenicol, mercury, and quaternary ammonium compound resistance. The TnAs2, a truncated form of TnAs3, only showed presence of genes associated with mercury resistance. The structure of Tn3 showed presence of putative gene encoding β-lactamase-TEM-1b.
The insertion of AbaRIs at comM gene was confirmed in 31 out of 47 genomes. The predicted AbaRIs varied in the length. The longest resistance island of size 90.4 kb was detected in VB23193 (ST2). In contrast, the shortest resistance island of size 17.7 kb was detected in AIIMS7 (ST1). Based on the sequence similarities shown with previously known AbaRIs sequences, we classify predicted AbaRIs into four types namely, ABaR3 variant, AbGRI variant, AbaR4a variant, and AbaR4b variant (Figure 5). The presence of truncated segment of Tn6022 carrying genes such as transposase (tniA), NTP-binding protein (tniB), universal stress protein (uspA), and sulfate permease (sulp) was found in all resistance islands except for the ABaR3 variant found in AIIMS7 which was a ST1 isolate (Figure 5A). The ABaR3 variant detected in ST1 (AIIMS7) possessed putative genes involved in cadmium and zinc resistance (czcD, czcR). However, it lacked the blaOXA-23 which was found in 27 out of 31 AbaRIs detected in this study. The ABaR3 variant predicted in AIIMS7 also showed presence of a class I integron gene cassette carrying putative genes conferring resistance to disinfectant (qacEΔ1), sulphonamide (sul1), and aminoglycoside (aadA, aac3). The two types of resistance islands were observed in ST2 genomes namely AbGRI variant and AbaR4a variant (Figures 5B, C). The complex AbGRI variants detected in VB23193 (ST2) and VB35575 (ST2) were longer in length as compared to the AbaR4a variant observed in the remaining ST2 genomes. Despite the complex nature and length variation in both ST2 variants (AbGRI and AbaR4) showed presence of certain common genes such as tetracycline resistance genes (tetB, tetR) as well as the truncated fragment of Tn5393 carrying strA and strB genes. Another difference we observed in AbaRI variants detected in ST2 genomes was, the presence of chloramphenicol (cmlA) and macrolides resistance (mphE, msrE) genes in AbGRI variant while their absence in AbaR4 variant. A fourth type of AbaRI variant namely AbaR4b was predicted in A. baumannii genomes belonging to ST10, ST85, ST575, ST25, ST622, and ST1512 (Figure 5D). Similar to AbGRI variant and AbaR4a variant predicted in ST2 isolates the AbaR4b showed presence of blaOXA-23. However, it lacked tetB, tetR and asrR genes. Overall, the comparative analysis of 47 A. baumannii genomes showed presence of multiple types of MGEs carrying putative resistance genes associated with antibiotics, disinfectants, and heavy metal resistance (Figure 3).
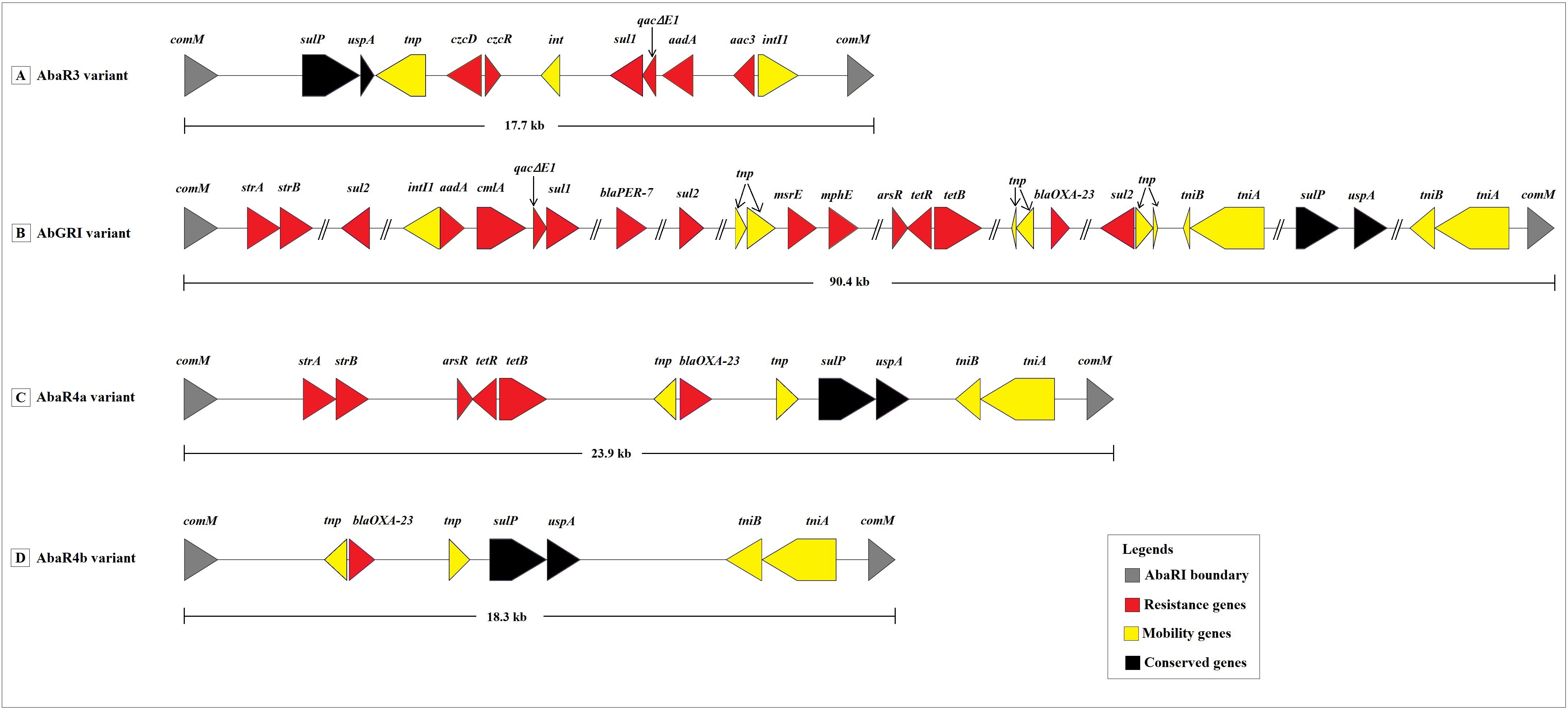
Figure 5 Schematic representation of four resistance island variants predicted in the study. (A) The AbaR3 variant predicted in AIIMS7 belonging to ST1. (B) The AbGRI variant predicted in VB23193 and VB35575 belonging to ST2. (C) The AbaR4a variant predicted in AIIMS5, VB33071, 6507, PM192696, PM1912235, VB473, VB723, VB958, VB1190, VB2107, VB2139, VB2181, VB2200, and VB7036 belonging to ST2. (D) The AbaR4b variant predicted in A. baumannii genomes belonging to ST10 (CIAT758, PM193665, PM194188), ST85 (VB82), ST575 (VB35435), ST25 (P7774), ST622 (VB16141, KSK6, KSK7, KSK10, KSK18, KSK19, KSK20), and ST1512 (VB35179). Legends: comM, ATPase; sulP, sulfate permease; uspA, universal stress protein A; tnp, Il-IS2 transposase; czcD, cobalt/zinc/cadmium resistance protein; czcR, Cd(II)/Pb(II)-responsive transcriptional regulator; int, prophage integrase; sul1 and sul2, sulfonamide resistance protein; qacΔE1, quaternary ammonium compounds efflux transporter; aadA, aminoglycoside 3’’-nucleotidyltransferase; aac3, aminoglycoside N(3)-acetyltransferase; intI1, integron integrase; tniB, NTP binding protein; tniA, transposase; strA, aminoglycoside 3’’-phosphotransferase; strB, aminoglycoside 6-phosphotransferase; arsR, transcriptional regulator; tetR, tetracycline resistance regulatory protein; tetB, tetracycline resistance efflux protein; cmlA, chloramphenicol efflux pump; blaPER-7, class A β -lactamase; msrE, erythromycin resistance; mphE, macrolide 2’ phosphotransferase; blaOXA-23, class D β-lactamase.
In India, the majority of the studies concerning A. baumannii focus on the phenotypic, genotypic or epidemiological characterization (Saranathan et al., 2014; Vijayakumar et al., 2016). The antimicrobial surveillance studies based on phenotypic characterization show that Acinetobacter spp. is the second most isolated pathogen (45%) after Pseudomonas spp. (52%). This report also emphasizes that>70% A. baumannii isolates show non-susceptibility to most antibiotics tested except colistin (Walia et al., 2019). The genotyping methods such as PCR-based oligotyping generally remain limited to molecular characterization of one or more genetic determinants associated with resistance or virulence (Kumar et al., 2010; Upadhyay et al., 2018). Despite the usefulness of routine phenotypic or molecular typing methods, the NGS provides an extraordinary insight into the genomic organization of outbreak causing pathogens. The data generated through genome sequencing can be further used to track clinically important pathogens like A. baumannii circulating through the Indian population. However, only a handful of studies based on these cutting-edge techniques exploring the dynamic genome organization of A. baumannii are reported from India (Kumar et al., 2015; Vijayakumar et al., 2019). Considering this scenario, we have performed a comprehensive comparative genome analysis of 47 clinical A. baumannii isolates collected between 2005 and 2020 from India.
The worldwide spread of carbapenem resistance via A. baumannii strains belonging to IC2 clonal lineage is a well-known factor (Gogou et al., 2011; Hamidian and Nigro, 2019). A study conducted by Kumar et al. has confirmed the presence of IC2 clones of A. baumannii carrying blaOXA-23 like carbapenamases in India (Kumar et al., 2019). Consistent with this finding, a similar trend confirming the presence of larger number (n=18, 38.3%) of A. baumannii isolates belonging to ST2 genotype (IC2 lineage) was observed in this study. The ST2 strains in our study have shown presence of significantly higher number of ARGs as compared to the other predicted STs. The presence of blaOXA-23 in almost 77% of ST2 A. baumannii genomes indicates their contribution in expansion of carbapenem resistance in Indian scenario. Apart from the ST2, the ST622 and ST10 were other major sequence types included in the current study. The occurrence of ST622, a relatively novel sequence type has been reported previously from Nepal while the ST10, a sequence type of a hypervirulent A. baumannii was reported from Iran and China (Shrestha et al., 2015; Abhari et al., 2019). Moreover, STs 622 and 10 have shown presence blaOXA-23 suggesting their potential role in spread of carbapenem resistant A. baumannii clones in India.
Gene content variation within A. baumannii genomes was reported previously (Sahl et al., 2015; Wallace et al., 2016). As per our study, a variation in the occurrence of ARGs was found between the genomes compared. The SRMC9, B8342, B8300 and SP304 genomes showed presence of a smaller number of ARGs than that predicted in the rest of the genomes included in the study. The combined action of intrinsic and acquired resistance in A. baumannii is considered as a serious barrier in the treatment of A. baumannii infections (Hood et al., 2013; Yoon et al., 2015; Juan et al., 2017; Jarlier et al., 2019). Our analysis has shown presence of ARGs of both intrinsic (AdeIJK, blaOXA-51 like β-lactamases, AmpC cephalosporinase, gyrA, parC) as well as of acquired (blaOXA-23 type, blaOXA-58 type, aac, aph, ant, cml, cat, armA, msrE, qacΔE, sul, tet) origin which signifies the clinical importance of A. baumannii. Overall, the versatile resistome predicted in this study highlights a widespread of MDR A. baumannii capable of resisting β-lactams, cephalosporins, carbapenems, aminoglycosides, and quinolones in India.
The pathogenicity illustrated by A. baumannii is a concerted action of several virulence factors together (Moubareck and Halat, 2020). The VFs that ensure survival and persistence of A. baumannii under diverse environmental conditions are mainly associated with motility, adherence, and biofilm formation (Sarshar et al., 2021). The putative VFs predicted in this study such as surface associated virulence factors such as OmpA, Bap, Csu pili, and poly-β-(1–6)-N-acetyl glucosamine (PNAG) which are known for their active role in motility, adherence and biofilm formation (Harding et al., 2018). The VFGs associated with synthesis of LPS, capsular polysaccharides, phospholipase, acinetobactin and heme oxidase predicted in our study were previously accounted for exhibition of serum resistance, immune invasion and iron acquisition capacity of A. baumannii (Sanchez-Larrayoz et al., 2017). All 47 isolates possessed an almost equal number of VFGs. In this study we did not find any linkage between VFGs and MGEs. However, considering the dynamic nature of MGEs, the possibility of acquisition of VFGs and their subsequent spread cannot be overruled. Overall, the combination of VFGs and ARGs predicted in this study provides ample evidence to suggesting the potential of A. baumannii to survive using ‘persist and resist’ strategy.
A. baumannii possesses an ‘open’ pan-genome which describes its ability to acquire new genes via various MGEs (Imperi et al., 2011) (Figure S4). The presence of numerous MGEs such as plasmids, insertion sequences, transposons, and resistance islands explain the high genome plasticity observed in this bacterium (Dziri et al., 2018; Cerezales et al., 2020). Plasmids are one of the most studied MGEs in A. baumannii. They have been shown to carry ARGs associated with carbapenems, aminoglycosides, tetracyclines, quinolones, macrolides, sulfonamides, and polymyxins resistance (Brovedan et al., 2020). In the present study, we have predicted the presence of plasmids carrying similar types of ARGs.
Apart from the plasmids, the role of insertion sequences, transposons, resistance islands, and prophages in conferring antimicrobial resistance in A. baumannii has been well documented (Pagano et al., 2016; Partridge et al., 2018) The increasing adaptability and prevalence of A. baumannii strains belonging to the ST2/IC2 genotype is generally linked to its ability to acquire and spread resistance determinants through activity of MGEs (Zarrilli et al., 2013). The prevalence of A. baumannii clones containing variants of AbaRI have also been reported (Li et al., 2015). One of the characteristic feature of resistance islands is presence of transposons similar to Tn5393 and Tn2006 which are generally found associated with carriage of aminoglycoside (strA, strB) and β-lactam (blaOXA-23) resistance genes (Zhou et al., 2011; Nigro and Hall, 2015; Lee et al., 2016). Amongst the four types of AbaRIs variants predicted in this study, two variants showed presence of Tn5393 linked strA, strB while three variants showed presence of Tn2006 associated blaOXA-23 gene. Another reason for the carbapenem resistance exhibited by A. baumannii isolates is overexpression of blaOXA-23 gene due to the insertion of ISAba1 element (Viana et al., 2016). The current study predicted the presence of ISAba1 linked blaOXA-23 suggesting the possibility of A. baumannii analyzed in this study to exhibit an increased resistance to carbapenems. Similarly, the ISAba1-blaADC linkage was observed to play a significant role in cephalosporin resistance (Lopes and Amyes, 2012; Biglari et al., 2015). The ISAba1-blaADC linkage predicted in this study using in silico approach represents the potential of these isolates to confer cephalosporin resistance. The association between presence of class 1 integron cassettes and antibiotic resistance in A. baumannii was reported previously (Huang et al., 2015). The findings in this study showing presence of class 1 integron cassettes in association with resistance islands points towards the dynamic nature of A. baumannii genomes.
The role of bacterial prophages in transfer of ARGs have been reported in several studies (Chene et al., 2006; Haaber et al., 2016). In the present study, we have observed presence of multiple copies of prophages in all 47 genomes. However, no ARGs were detected within these prophage regions. Nevertheless, a further study to analyze potential of these prophage regions in transfer of ARGs is needed. In summary, the presence of plasmids, Tn3 transposons, AbaRIs associated with putative genes conferring resistance to β-lactams, aminoglycosides, chloramphenicol, quaternary ammonium compounds, and heavy metals was predicted in this study. In addition to these findings, we also observed the presence of higher number of ARGs associated with MGEs in A. baumannii isolates collected after 2014 as compared to the isolates collected in 2005 (Figure 3).
The inclusion of a fewer number of complete A. baumannii genomes from India could be considered one of the limitations of this study. Additionally, the genomes analyzed in this study only represent the north and south region of the country. The analysis does not depict a wide distribution of A. baumannii isolates throughout India and therefore provides a partial genomic diversity. Furthermore, the study limits itself to A. baumannii genomes collected from 2005 and from 2014 to 2020. The study would have been more decisive if the genomes from a wide temporal and geographical region were available.
In conclusion, this study provides a detailed account of genetic determinants associated with multidrug resistance and virulence in clinically significant A. baumannii isolates from India. Despite the diversity in sequence types, a dominance of A. baumannii isolates associated with ST2 was observed. Furthermore, the study showed a prevalence of diverse MGEs in A. baumannii genomes which highlights its ability to adapt and evolve. The study also suggests a need for the real time functional analysis of MGEs involved in dissemination of multidrug resistance. Considering this pathogen’s challenging nature, we strongly recommend the use of NGS in monitoring and thereby controlling the spread of outbreak causing A. baumannii isolates in India.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
SK conceived and designed the study. SK wrote the article, generated all tables and figures in the article. NC and DD helped with the initial quality check of the sequencing data for five A. baumannii genomes. Further, NC and DD helped with interpretation of results obtained through in silico analysis. EK helped with the statistical analysis. KP helped with manuscript reviewing and editing. All authors contributed to the article and approved the submitted version.
This work was supported by the Department of Science and Technology, India under the scheme Promotion of University Research and Scientific Excellence (DST-PURSE, GOIA670), University with Potential for Excellence Phase II program (UGC-262-A-2) implemented at Savitribai Phule Pune University, and the Director, NCCS, Pune. SK is a recipient of the CMSRF 2019 fellowship provided by SARTHI, Pune. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
We thank the bioinformatics team at the National Centre for Microbial Resource (NCMR), Pune, for their technical assistance. This work was supported by the Department of Microbiology, Savitribai Phule Pune University. The funding for bacterial genome sequencing was procured from DST-PURSE (GOI-A-670) implemented at Savitribai Phule Pune University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.997897/full#supplementary-material
Abhari, S. S., Badmasti, F., Modiri, L., Aslani, M. M., Asmar, M. (2019). Circulation of imipenem-resistant Acinetobacter baumannii ST10, ST2 and ST3 in a university teaching hospital from Tehran, Iran. J. Med. Microbiol. 68, 860–865. doi: 10.1099/JMM.0.000987/CITE/REFWORKS
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/NAR/GKZ935
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Antipov, D., Hartwick, N., Shen, M., Raiko, M., Lapidus, A., Pevzner, P. (2016). plasmidSPAdes: Assembling plasmids from whole genome sequencing data. PlasmidSPAdes Assem. Plasmids Whole Genome Seq. Data, 32 (22), 3380–3387. doi: 10.1101/048942
Arndt, D., Marcu, A., Liang, Y., Wishart, D. S., Wishart, D. (2019). PHAST, PHASTER and PHASTEST: Tools for finding prophage in bacterial genomes. Brief. Bioinform. 20, 1560–1567. doi: 10.1093/bib/bbx121
Asif, M., Alvi, I. A., Rehman, S. U. (2018). Insight into Acinetobacter baumannii: Pathogenesis, global resistance, mechanisms of resistance, treatment options, and alternative modalities. Infect. Drug Resist. 11, 1249–1260. doi: 10.2147/IDR.S166750
Asim, P., Naik, N., Muralidhar, V., Vandana, K., Varsha, A. P. (2016). Clinical and economic outcomes of Acinetobacter vis a vis non-Acinetobacter infections in an Indian teaching hospital. Perspect. Clin. Res. 7, 28. doi: 10.4103/2229-3485.173778
Balaji, V., Rajenderan, S., Anandan, S., Biswas, I. (2015). Genome sequences of two multidrug-resistant Acinetobacter baumannii clinical strains isolated from southern India. Genome Announc. 3 (5), e01010-15. doi: 10.1128/GENOMEA.01010-15
Banerjee, T., Mishra, A., Das, A., Sharma, S., Barman, H., Yadav, G. (2018). High prevalence and endemicity of multidrug resistant Acinetobacter spp. in intensive care unit of a tertiary care hospital, varanasi, India. J. Pathog. 2018, 1–8. doi: 10.1155/2018/9129083
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Biglari, S., Alfizah, H., Ramliza, R., Rahman, M. M. (2015). Molecular characterization of carbapenemase and cephalosporinase genes among clinical isolates of Acinetobacter baumannii in a tertiary medical centre in Malaysia. J. Med. Microbiol. 64, 53–58. doi: 10.1099/JMM.0.082263-0/CITE/REFWORKS
Bi, D., Xie, R., Zheng, J., Yang, H., Zhu, X., Ou, H. Y., et al. (2019). Large-Scale identification of AbaR-type genomic islands in Acinetobacter baumannii reveals diverse insertion sites and clonal lineage-specific antimicrobial resistance gene profiles. Antimicrob. Agents Chemother. 63 (4), e02526-18. doi: 10.1128/AAC.02526-18
Brovedan, M. A., Cameranesi, M. M., Limansky, A. S., Morán-Barrio, J., Marchiaro, P., Repizo, G. D. (2020). What do we know about plasmids carried by members of the Acinetobacter genus? World J. Microbiol. Biotechnol. 36, 1–15. doi: 10.1007/S11274-020-02890-7
Cerezales, M., Xanthopoulou, K., Wille, J., Krut, O., Seifert, H., Gallego, L., et al. (2020). Mobile genetic elements harboring antibiotic resistance determinants in Acinetobacter baumannii isolates from Bolivia. Front. Microbiol. 11. doi: 10.3389/FMICB.2020.00919/BIBTEX
Chene, F., Wang, K., Stewart, J., Belas, R. (2006). Induction of multiple prophages from a marine bacterium: A genomic approach. Appl. Environ. Microbiol. 72, 4995–5001. doi: 10.1128/AEM.00056-06
Dash, M., Padhi, S., Pattnaik, S., Mohanty, I., Misra, P. (2013). Frequency, risk factors, and antibiogram of Acinetobacter species isolated from various clinical samples in a tertiary care hospital in odisha, India. Avicenna J. Med. 03, 97–102. doi: 10.4103/2231-0770.120501
Dziri, O., Alonso, C. A., Dziri, R., Gharsa, H., Maraoub, A., Torres, C., et al. (2018). Metallo-β-lactamases and class d carbapenemases in south-east Tunisia: Implication of mobile genetic elements in their dissemination. Int. J. Antimicrob. Agents 52, 871–877. doi: 10.1016/J.IJANTIMICAG.2018.06.002
Francisco, A. P., Bugalho, M., Ramirez, M., Carriço, J. A. (2009). Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinforma. 10, 1–15. doi: 10.1186/1471-2105-10-152
Gaidhani, S. V., Raskar, A. V., Poddar, S., Gosavi, S., Sahu, P. K., Pardesi, K. R., et al. (2014). Time dependent enhanced resistance against antibiotics & metal salts by planktonic & biofilm form of Acinetobacter haemolyticus MMC 8 clinical isolate. Indian J. Med. Res. 140 (5), 665–71.
Gandra, S., Tseng, K. K., Arora, A., Bhowmik, B., Robinson, M. L., Panigrahi, B., et al. (2019). The mortality burden of multidrug-resistant pathogens in India: A retrospective, observational study. Clin. Infect. Dis. 69, 563–570. doi: 10.1093/CID/CIY955
Gogou, V., Pournaras, S., Giannouli, M., Voulgari, E., Piperaki, E.-T., Zarrilli, R., et al. (2011). Evolution of multidrug-resistant Acinetobacter baumannii clonal lineages: a 10 year study in Greece, (2000-09). J. Antimicrob. Chemother. 66, 2767–2772. doi: 10.1093/jac/dkr390
Gurevich, A., Saveliev, V., Vyahhi, N., Tesler, G. (2013). QUAST: Quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Haaber, J., Leisner, J. J., Cohn, M. T., Catalan-Moreno, A., Nielsen, J. B., Westh, H., et al. (2016). Bacterial viruses enable their host to acquire antibiotic resistance genes from neighbouring cells. Nat. Commun. 7, 1–8. doi: 10.1038/ncomms13333
Hamidian, M., Nigro, S. J. (2019). Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb. Genomics 5 (10), e000306. doi: 10.1099/MGEN.0.000306
Harding, C. M., Hennon, S. W., Feldman, M. F. (2018). Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 16, 91–102. doi: 10.1038/nrmicro.2017.148
Hood, M. I., Becker, K. W., Roux, C. M., Dunman, P. M., Skaar, E. P. (2013). Genetic determinants of intrinsic colistin tolerance in Acinetobacter baumannii. Infect. Immun. 81, 542–551. doi: 10.1128/IAI.00704-12/SUPPL_FILE/ZII999099993SO1.XLS
Huang, C., Long, Q., Qian, K., Fu, T., Zhang, Z., Liao, P., et al. (2015). Resistance and integron characterization of Acinetobacter baumannii in a teaching hospital in chongqing, China. New Microbes New Infect. 8, 103–108. doi: 10.1016/J.NMNI.2015.09.015
Imperi, F., Antunes, L. C. S., Blom, J., Villa, L., Iacono, M., Visca, P., et al. (2011). The genomics of Acinetobacter baumannii: Insights into genome plasticity, antimicrobial resistance and pathogenicity. IUBMB Life 63, 1068–1074. doi: 10.1002/iub.531
Jarlier, V., Hogberg, L. D., Heuer, O. E., Campos, J., Eckmanns, T., Giske, C. G., et al. (2019). Strong correlation between the rates of intrinsically antibiotic-resistant species and the rates of acquired resistance in gram-negative species causing bacteraemia, EU/EE. Eurosurveillance 24, 1800538. doi: 10.2807/1560-7917.ES.2019.24.33.1800538/CITE/PLAINTEXT
John, A., Paul, H., Vijayakumar, S., Anandan, S., Sudarsan, T., Abraham, O., et al. (2020). Mortality from Acinetobacter infections as compared to other infections among critically ill patients in south India: A prospective cohort study. Indian J. Med. Microbiol. 38, 24. doi: 10.4103/ijmm.IJMM_19_492
Jones, L. S., Toleman, M. A., Weeks, J. L., Howe, R. A., Walsh, T. R., Kumarasamy, K. K. (2014). Plasmid carriage of blaNDM-1in clinical Acinetobacter baumannii isolates from India. Antimicrob. Agents Chemother. 58, 4211–4213. doi: 10.1128/AAC.02500-14/ASSET/5CD2CD91-8B24-4605-A036-BD325318ED35/ASSETS/GRAPHIC/ZAC0071429870002.JPEG
Jose, J., Santhiya, K., Jayanthi, S., Ananthasubramanian, M. (2017). Insertion sequence based analysis of clinical isolates with NDM (blaNDM-1) resistance. Indian J. Biotechnol. 16, 182–188.
Juan, C., Torrens, G., González-Nicolau, M., Oliver, A. (2017). Diversity and regulation of intrinsic β-lactamases from non-fermenting and other gram-negative opportunistic pathogens. FEMS Microbiol. Rev. 41, 781–815. doi: 10.1093/FEMSRE/FUX043
Kaas, R. S., Leekitcharoenphon, P., Aarestrup, F. M., Lund, O. (2014). Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PloS One 9, e104984. doi: 10.1371/JOURNAL.PONE.0104984
Khajuria, A., Praharaj, A., Kumar, M., Grover, N. (2014). Molecular characterization of carbapenem resistant isolates of Acinetobacter baumannii in an intensive care unit of a tertiary care centre at central India. J. Clin. Diagn. Res. 8, DC38. doi: 10.7860/JCDR/2014/7749.4398
Kumari, M., Batra, P., Malhotra, R., Mathur, P. (2019). A 5-year surveillance on antimicrobial resistance of Acinetobacter isolates at a level-I trauma centre of India. J. Lab. Physicians 11, 34. doi: 10.4103/JLP.JLP_72_18
Kumar, A., Mukherjee, S., Chakraborty, R. (2010). Characterization of a novel trimethoprim resistance gene, dfra28, in class 1 integron of an oligotrophic Acinetobacter johnsonii strain, MB52, isolated from river mahananda, India. Microb. Drug Resist. 16, 29–37. doi: 10.1089/mdr.2009.0111
Kumar, S., Patil, P. P., Midha, S., Ray, P., Patil, P. B., Gautam, V. (2015). Genome sequence of Acinetobacter baumannii strain 5021_13, isolated from cerebrospinal fluid. Genome Announc. 3 (5), e01213-15. doi: 10.1128/genomeA.01213-15
Kumar, S., Patil, P. P., Singhal, L., Ray, P., Patil, P. B., Gautam, V. (2019). Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates reveals the emergence of blaOXA-23 and blaNDM-1 encoding international clones in India. Infect. Genet. Evol. 75, 103986. doi: 10.1016/J.MEEGID.2019.103986
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Lee, Y., D’Souza, R., Yong, D., Lee, K. (2016). Prediction of putative resistance islands in a carbapenem-resistant Acinetobacter baumannii global clone 2 clinical isolate. Ann. Lab. Med. 36, 320. doi: 10.3343/ALM.2016.36.4.320
Letunic, I., Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/NAR/GKAB301
Li, H., Liu, F., Zhang, Y., Wang, X., Zhao, C., Chen, H., et al. (2015). Evolution of carbapenem-resistant Acinetobacter baumannii revealed through whole-genome sequencing and comparative genomic analysis. Antimicrob. Agents Chemother. 59, 1168–1176. doi: 10.1128/AAC.04609-14/SUPPL_FILE/ZAC002153702SO1.PDF
Liu, B., Zheng, D., Jin, Q., Chen, L., Yang, J. (2018). VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 47, 687–692. doi: 10.1093/nar/gky1080
Lopes, B. S., Amyes, S. G. B. (2012). Role of ISAba1 and ISAba125 in governing the expression of bla ADC in clinically relevant Acinetobacter baumannii strains resistant to cephalosporins. J. Med. Microbiol. 61, 1103–1108. doi: 10.1099/JMM.0.044156-0/CITE/REFWORKS
Mahalingam, N., Manivannan, B., Jadhao, S., Mishra, G., Nilawe, P., Pradeep, B. (2016). Draft genome sequences of two extensively drug-resistant Acinetobacter baumannii strains isolated from pus samples. Genome Announc. 4 (2), e00161-16. doi: 10.1128/genomeA.00161-16
Mathai, A. S., Oberoi, A., Madhavan, S., Kaur, P. (2012). Acinetobacter infections in a tertiary level intensive care unit in northern India: Epidemiology, clinical profiles and outcomes. J. Infect. Public Health 5, 145–152. doi: 10.1016/J.JIPH.2011.12.002
Mcarthur, A. G., Waglechner, N., Nizam, F., Yan, A., Azad, M. A., Baylay, A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 57 (7), 3348–57. doi: 10.1128/AAC.00419-13
Moubareck, C. A., Halat, D. H. (2020). Insights into Acinetobacter baumannii: A review of microbiological, virulence, and resistance traits in a threatening nosocomial pathogen. Antibiotics 9, 119. doi: 10.3390/antibiotics9030119
Mulani, M. S., Kamble, E. E., Kumkar, S. N., Tawre, M. S., Pardesi, K. R. (2019). Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00539
Nigro, S., Hall, R. M. (2015). Distribution of the blaOXA-23-containing transposons Tn2006 and Tn2008 in Australian carbapenem-resistant Acinetobacter baumannii isolates. J. Antimicrob. Chemother. 70, 2409–2411. doi: 10.1093/JAC/DKV102
Pagano, M., Martins, A. F., Barth, A. L., Pagano, M., Martins, A. F., Barth, A. L. (2016). Mobile genetic elements related to carbapenem resistance in Acinetobacter baumannii. Braz. J. Microbiol. 47, 785–792. doi: 10.1016/j.bjm.2016.06.005
Pardesi, K., Yavankar, S., Medical, B. C.-I. J. of, and 2007, U (2007)Plasmid distribution & antimicrobial susceptibility patterns of Acinetobacter genospecies from healthy skin of a tribal population in western India (Accessed January 4, 2022).
Partridge, S. R., Kwong, S. M., Firth, N., Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31, e00088–e00017. doi: 10.1128/CMR.00088-17
Patel, R. K., Jain, M. (2012). NGS QC toolkit: A toolkit for quality control of next generation sequencing data. PloS One 7, e30619. doi: 10.1371/journal.pone.0030619
Pragasam, A., Vijayakumar, S., Bakthavatchalam, Y., Kapil, A., Das, B., Ray, P., et al. (2016). Molecular characterisation of antimicrobial resistance in Pseudomonas aeruginosa and Acinetobacter baumannii during 2014 and 2015 collected across India. Indian J. Med. Microbiol. 34, 433–441. doi: 10.4103/0255-0857.195376
Prashanth, K., Badrinath, S. (2006). Nosocomial infections due to Acinetobacter species: Clinical findings, risk and prognostic factors. Indian J. Med. Microbiol. 24, 39–44. doi: 10.1016/S0255-0857(21)02469-5
Rynga, D., Shariff, M., Deb, M. (2015). Phenotypic and molecular characterization of clinical isolates of Acinetobacter baumannii isolated from Delhi, India. Ann. Clin. Microbiol. Antimicrob. 14, 1–8. doi: 10.1186/S12941-015-0101-5/TABLES/2
Sahl, J. W., Del Franco, M., Pournaras, S., Colman, R. E., Karah, N., Dijkshoorn, L., et al. (2015). Phylogenetic and genomic diversity in isolates from the globally distributed Acinetobacter baumannii ST25 lineage. Sci. Rep. 5, 15188. doi: 10.1038/srep15188
Sahu, P. K., Iyer, P. S., Gaikwad, M. B., Talreja, S. C., Pardesi, K. R., Chopade, et al. (2012a). An MFS transporter-like ORF from MDR Acinetobacter baumannii AIIMS 7 is associated with adherence and biofilm formation on biotic/abiotic surface. Int. J. Microbiol. 2012, 490647. doi: 10.1155/2012/490647
Sahu, P. K., Iyer, P. S., Oak, A. M., Pardesi, K. R., Chopade, B. A. (2012b). Characterization of eDNA from the clinical strain Acinetobacter baumannii AIIMS 7 and its role in biofilm formation. Sci. World J. 2012, 973436. doi: 10.1100/2012/973436
Sanchez-Larrayoz, A. F., Elhosseiny, N. M., Chevrette, M. G., Fu, Y., Giunta, P., Spallanzani, R. G., et al. (2017). Complexity of complement resistance factors expressed by Acinetobacter baumannii needed for survival in human serum. J. Immunol. 199, 2803–2814. doi: 10.4049/JIMMUNOL.1700877/-/DCSUPPLEMENTAL
Saranathan, R., Sudhakar, P., Uma Karthika, R., Singh, S. K., Shashikala, P., Kanungo, R., et al. (2014)Multiple drug resistant carbapenemases producing Acinetobacter baumannii isolates harbours multiple r-plasmids (Accessed September 15, 2022).
Sarshar, M., Behzadi, P., Scribano, D., Palamara, A. T., Ambrosi, C. (2021). Acinetobacter baumannii: An ancient commensal with weapons of a pathogen. Pathog 10, 387. doi: 10.3390/PATHOGENS10040387
Seemann, T. (2014). Genome analysis prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sengupta, S., Kumar, P., Ciraj, A. M., Shivananda, P. G. (2001). Acinetobacter baumannii — an emerging nosocomial pathogen in the burns unit manipal, India. Burns 27, 140–144. doi: 10.1016/S0305-4179(00)00094-2
Shrestha, S., Tada, T., Miyoshi-Akiyama, T., Ohara, H., Shimada, K., Satou, K., et al. (2015). Molecular epidemiology of multidrug-resistant Acinetobacter baumannii isolates in a university hospital in Nepal reveals the emergence of a novel epidemic clonal lineage. Int. J. Antimicrob. Agents 46, 526–531. doi: 10.1016/J.IJANTIMICAG.2015.07.012
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Singh, N. K., Kumar, S., Raghava, G. P. S., Mayilraj, S. (2013). Draft genome sequence of Acinetobacter baumannii strain MSP4-16. Genome Announc. 1, 137–150. doi: 10.1128/genomeA.00137-13
Sivaranjani, V., Umadevi, S., Srirangaraj, S., Kali, A., Seetha, K. S. (2013). Multi-drug resistant Acinetobacter species from various clinical samples in a tertiary care hospital from south India. Australas. Med. J. 6, 697. doi: 10.4066/AMJ.2013.1901
Upadhyay, S., Khyriem, A. B., Bhattacharya, P., Bhattacharjee, A., Joshi, S. R. (2018). High-level aminoglycoside resistance in Acinetobacter baumannii recovered from intensive care unit patients in northeastern India. Indian J. Med. Microbiol. 36, 43–48. doi: 10.4103/IJMM.IJMM_17_225
Viana, G. F., Zago, M. C. B., Moreira, R. R. B., Zarpellon, M. N., Menegucci, T. C., Cardoso, C. L., et al. (2016). ISAba1/blaOXA-23: A serious obstacle to controlling the spread and treatment of Acinetobacter baumannii strains. Am. J. Infect. Control 44, 593–595. doi: 10.1016/J.AJIC.2015.11.020
Vijayakumar, S., Anandan, S., MS, D. P., Kanthan, K., Vijayabaskar, S., Kapil, A., et al. (2019). Insertion sequences and sequence types profile of clinical isolates of carbapenem-resistant A. baumannii collected across India over four year period. J. Infect. Public Health. 13 (7), 1022–1028. doi: 10.1016/j.jiph.2019.11.018
Vijayakumar, S., Rajenderan, S., Laishram, S., Anandan, S., Balaji, V., Biswas, I. (2016). Biofilm formation and motility depend on the nature of the Acinetobacter baumannii clinical isolates. Front. Public Heal. 4. doi: 10.3389/fpubh.2016.00105
Vijayakumar, S., S, B. A., Kanthan, K., Veeraraghavan, B. (2018). Whole-genome shotgun sequences of seven colistin-resistant Acinetobacter baumannii isolates from bacteraemia. J. Glob. Antimicrob. Resist. 12, 155–156. doi: 10.1016/j.jgar.2018.01.012
Walia, K., Madhumathi, J., Veeraraghavan, B., Chakrabarti, A., Kapil, A., Ray, P., et al. (2019). Establishing antimicrobial resistance surveillance & research network in India: Journey so far. Indian J. Med. Res. 149, 164. doi: 10.4103/IJMR.IJMR_226_18
Wallace, L., Daugherty, S. C., Nagaraj, S., Johnson, J. K., Harris, A. D., Rasko, D. A. (2016). Use of comparative genomics to characterize the diversity of Acinetobacter baumannii surveillance isolates in a health care institution. Antimicrob. Agents Chemother. 60, 5933–5941. doi: 10.1128/AAC.00477-16/SUPPL_FILE/ZAC010165548SO1.PDF
Wattam, A. R., Davis, J. J., Assaf, R., Boisvert, S., Brettin, T., Bun, C., et al. (2016). Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 45, 535–542. doi: 10.1093/nar/gkw1017
Wilson, K. (2001). Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. 56, 2.4.1–2.4.5. doi: 10.1002/0471142727.mb0204s56
Yakkala, H., Samantarrai, D., Gribskov, M., Siddavattam, D. (2019). Comparative genome analysis reveals nichespecific genome expansion in Acinetobacter baumannii strains. PloS One 14, e0218204. doi: 10.1371/journal.pone.0218204
Yavankar, S., Pardesi, K., Chopade, B. (2007). Species distribution and physiological characterization of Acinetobacter genospecies from healthy human skin of tribal population in India. Indian J. Med. Microbiol. 25, 336–345. doi: 10.4103/0255-0857.37335
Yoon, E. J., Chabane, Y. N., Goussard, S., Snesrud, E., Courvalin, P., Dé, E., et al. (2015). Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. MBio 6 (2), e00309-15. doi: 10.1128/MBIO.00309-15/SUPPL_FILE/MBO002152247ST5.DOCX
Zarrilli, R., Pournaras, S., Giannouli, M., Tsakris, A. (2013). Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int. J. Antimicrob. Agents 41, 11–19. doi: 10.1016/J.IJANTIMICAG.2012.09.008
Zhou, H., Zhang, T., Yu, D., Pi, B., Yang, Q., Zhou, J., et al. (2011). Genomic analysis of the multidrug-resistant Acinetobacter baumannii strain MDR-ZJ06 widely spread in China. Antimicrob. Agents Chemother. 55, 4506–4512. doi: 10.1128/AAC.01134-10/ASSET/87E3B21D-3AF4-4F04-BCE2-F056F4725B94/ASSETS/GRAPHIC/ZAC9991002460004.JPEG
Keywords: antibiotic resistance genes (ARGs), multidrug resistance (MDR), virulence factor genes (VFGs), mobile genetic elements (MGEs), India, Acinetobacter baumannii
Citation: Kumkar SN, Kamble EE, Chavan NS, Dhotre DP and Pardesi KR (2022) Diversity of resistant determinants, virulence factors, and mobile genetic elements in Acinetobacter baumannii from India: A comprehensive in silico genome analysis. Front. Cell. Infect. Microbiol. 12:997897. doi: 10.3389/fcimb.2022.997897
Received: 19 July 2022; Accepted: 07 November 2022;
Published: 28 November 2022.
Edited by:
Carolina Silva Nodari, Institut Pasteur, FranceReviewed by:
Benjamin Andrew Evans, University of East Anglia, United KingdomCopyright © 2022 Kumkar, Kamble, Chavan, Dhotre and Pardesi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dhiraj P. Dhotre, ZGhpcmFqLmRob3RyZUBuY2NzLnJlcy5pbg==; Karishma R. Pardesi, a2FyaXNobWFAdW5pcHVuZS5hYy5pbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.