- 1West African Centre for Cell Biology of Infectious Pathogens, College of Basic and Applied Sciences, University of Ghana, Accra, Ghana
- 2Department of Biochemistry Cell and Molecular Biology, College of Basic and Applied Sciences, University of Ghana, Accra, Ghana
- 3Pôle Immunophysiopathologie et Maladies Infectieuses, Institut Pasteur de Dakar, Dakar, Senegal
Ex vivo phenotyping of P. falciparum erythrocyte invasion diversity is important in the identification and down selection of potential malaria vaccine targets. However, due to the lack of appropriate laboratory facilities in remote areas of endemic countries, direct processing of P. falciparum clinical isolates is usually not feasible. Here, we investigated the combined effect of short-term cryopreservation and thawing processes on the ex vivo invasion phenotypes of P. falciparum isolates. Ex-vivo or in vitro invasion phenotyping assays were performed with P. falciparum clinical isolates prior to or following culture adaptation, respectively. All isolates were genotyped at Day 0 for parasite clonality. Subsequently, isolates that were successfully culture-adapted were genotyped again at Days 7, 15, 21, and 28-post adaptation. Invasion phenotyping assays were performed in isogenic isolates revived at different time points (3, 6, and 12 months) post-cryopreservation and the resulting data were compared to that from ex-vivo invasion data of matched isogenic parental isolates. We also show that short-term culture adaptation selects for parasite clonality and could be a driving force for variation in invasion phenotypes as compared to ex vivo data where almost all parasite clones of a given isolate are present. Interestingly, our data show little variation in the parasites’ invasion phenotype following short-term cryopreservation. Altogether, our data suggest that short-term cryopreservation of uncultured P. falciparum clinical isolates is a reliable mechanism for storing parasites for future use.
Introduction
Plasmodium falciparum uses complex mechanisms for efficiently invading human erythrocytes and evading the host immune response (Wright and Rayner, 2014; Cowman et al., 2017), therefore, a better understanding of these mechanisms is critical for developing effective vaccines. Studies with P. falciparum field isolates are important for providing a clinically relevant elucidation of the molecular mechanisms involved in host-parasite interactions and pathogenesis of malaria (Ahouidi et al., 2016; Yap et al., 2019). One of the key approaches used in investigating molecular interactions at the host-parasite interface is phenotyping parasites for their erythrocyte invasion pathways. Invasion phenotyping generates data that are more representative of the parasite’s natural biology when these assays are conducted on parasites ex vivo, or during their first replicative cycle in culture. However, direct processing of clinical isolates requires advanced laboratory equipment and technical capacities, which are not always available in the clinics in the remote areas of endemic countries. Therefore, parasites isolates collected from patients at point-of-care usually need to be processed for cryopreservation, storage and transportation to laboratories with the requisite facilities for phenotyping (Ahouidi et al., 2016). These frozen parasites are then thawed using optimized protocols that minimize erythrocyte lysis and ensure maximal parasite survival. Therefore, it is important to determine the impact of any, of the cryopreservation and thawing processes on the parasites’ biology in general and their invasion phenotypes.
In the last decade, many laboratories reported on the invasion phenotypes of P. falciparum clinical isolates. However, cross-study comparison of these data is challenged by the use of different approaches to characterize such phenotypes (Ahouidi et al., 2016). The bulk of these phenotypic data were collected from cryopreserved parasites (Okoyeh et al., 1999; Lobo et al., 2004; Nery et al., 2006; Lopez-Perez et al., 2012) while only a few were from P. falciparum clinical isolates that were directly assayed after collection (Baum et al., 2003; Jennings et al., 2007; Lantos et al., 2009). Moreover, in some cases parasites were allowed to grow in vitro for at least five cycles prior to assay set up (Okoyeh et al., 1999; Lobo et al., 2004; Lopez-Perez et al., 2012), therefore, providing room for clonal selection. Unlike laboratory strains, P. falciparum clinical isolates are usually representative of a population of different clones, presenting intrinsic characteristics that could determine their in vitro adaptability. Given that different clones of the same isolate express distinct versions of surface antigens (Cortés, 2008), it could be that different clones use distinct invasion pathways and only the dominant clones of a given isolate will be reflected during invasion phenotyping experiments.
Alongside the cryopreservation, different laboratories may use different thawing protocols prior to culturing the parasites. Therefore, the consequences of variations in cryopreservation and thawing protocols in P. falciparum in vitro culture need to be assessed, especially with respect to parasite adaptation and subsequent invasiveness during ex vivo and in vitro phenotyping assays.
Previous literature has reported similar ex vivo adaptation rates in pre-cryopreserved isolates (Bowyer et al., 2015) as compared to fresh clinical isolates (Gomez-Escobar et al., 2010). However, these conclusions were drawn from studies using isolates of different isogenic backgrounds, which were phenotypically and functionally distinct. In this study, parasites from the same isogenic backgrounds were used to investigate the effect of freeze-thaw protocols on P. falciparum ex vivo invasion phenotypes and early in vitro adaptation.
Materials and methods
Sample collection and processing
P. falciparum clinical isolates were collected from 25 symptomatic children, aged 2 to 14 years old, visiting the LEKMA Hospital, in Accra between February 2017 and January 2018. LEKMA is a sub-urban area of Accra, the capital city of Ghana, a low malaria transmission setting, with an estimated entomological inoculation rate of <50 infective mosquito bites per person/year. The study was approved by the Institutional Review Board of the Noguchi Memorial Institute for Medical Research, University of Ghana (IRB00001276) and the Ghana Health Service Ethical Review Committee (GHC-ERC: 005/12/2017). All guidelines and principles contained in the approved protocol were duly followed in the execution of the project. A written informed consent was obtained from a parent and/or legal guardian and assent obtained for older children. Venous blood samples were collected in ACD vacutainers (BD Biosciences) and transported to the laboratory for processing within two hours after collection. At the laboratory, the infected erythrocytes were separated from the leucocytes through centrifugation at 2000 rpm and washed twice with RPMI1640 medium (Sigma). About 200µL of packed erythrocytes were put straight in culture, while the remaining sample was resuspended in glycerolyte in ~500 µL vials following the standard protocol (EVIMalaR, 2013) and stored in liquid nitrogen. Frozen vials were thawed at different time intervals using two distinct sodium chloride-based protocols. Vials were thawed using either a two-step protocol (12% NaCl and 1.6% NaCl) or a three-step protocol (12% NaCl, 1.8% NaCl and 0.9% NaCl supplemented with 0.2% Glucose) as per standard procedure (EVIMalaR, 2013). To minimize the effect of possible confounders, all reagents used in this study were prepared from single batches and stored as single-use aliquots.
Plasmodium falciparum in vitro culture
P. falciparum clinical isolates were cultured as per standard protocols (EVIMalaR, 2013). In brief, isolates were maintained at 37°C in RPMI1640 medium (Sigma), supplemented with 5% Albumax (Gibco), 2 mg/ml sodium bicarbonate, 50 µg/ml gentamycin (Sigma) and 2% AB+ heat-inactivated normal human serum (PAN Biotech, UK). All cultures were adjusted to 4% hematocrit using O+ erythrocytes from a single donor and incubated in an atmosphere of 2% O2, 5% CO2 and balanced with Nitrogen. For isolates cultured upon arrival to the lab, the parasites multiplication rate (PMR), defined as the ratio of the parasitemia before and after re-invasion, was monitored for the first two in vitro cycles and fresh erythrocytes were only added after 96 hours in culture, while fresh erythrocytes were immediately added upon thawing of cryopreserved isolates. Following the addition of fresh erythrocytes, growth tests were performed to assess the PMR every 48 hours for the first three in vitro replicative cycles for the cryopreserved isolates and for up to 12 cycles for freshly culture adapted isolates. After each cycle, cultures were diluted to 0.5% parasitemia for the next cycle. Sample aliquots, taken following each replicative cycle were stained with 1 µM of Hoechst 33342 dye (Sigma Aldrich, UK) and the resulting parasitemia was assessed using flow cytometry. The growth test was considered successful only when the resulting PMR, measured after 48 hours, was greater than one (>1) (Figure 1). The median PMR of successful growth tests after twelve successive replicative cycles following the addition of fresh erythrocytes were considered as PMR of culture-adapted isolates.
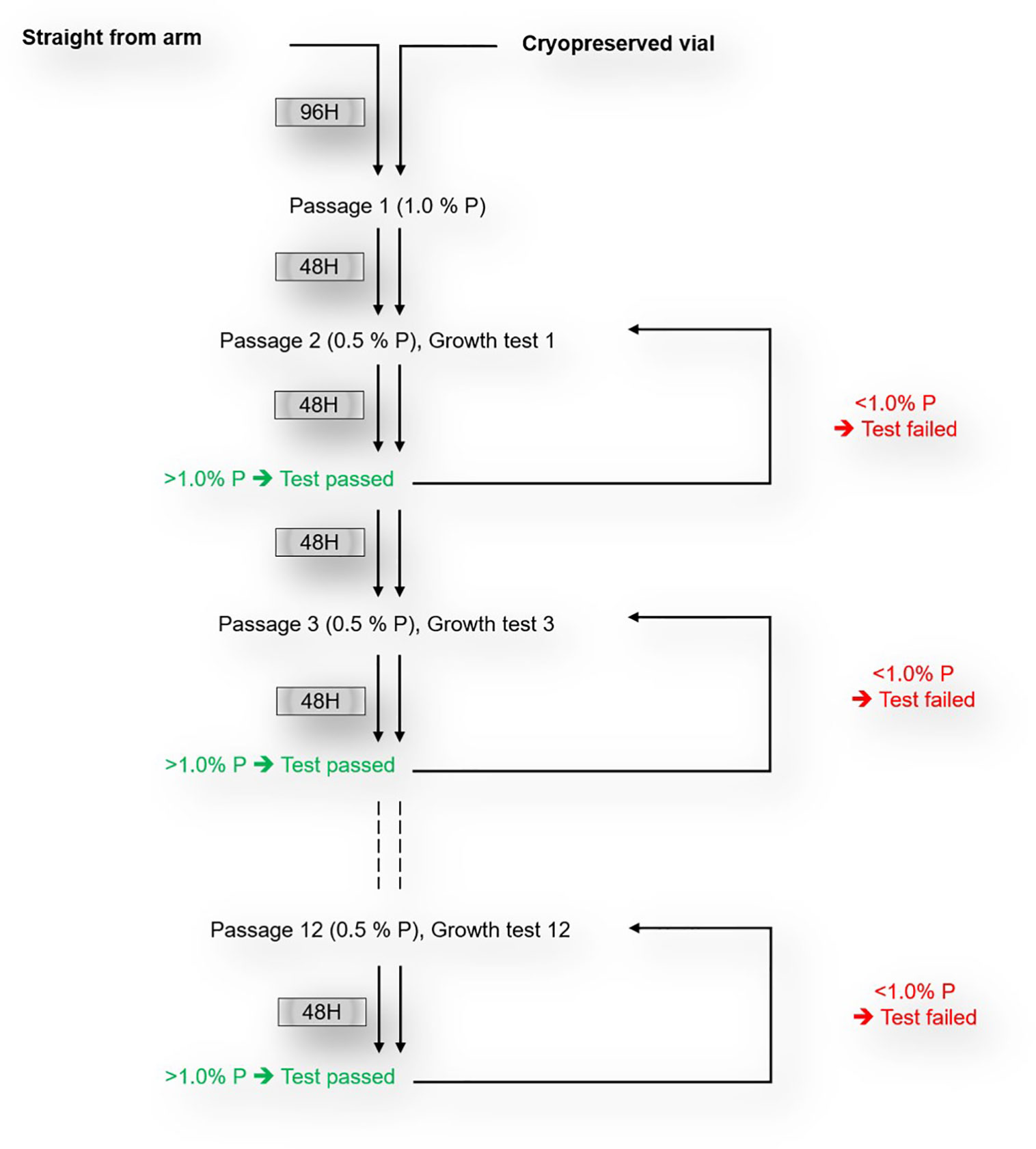
Figure 1 Schematics of in vitro culture-adaptation and growth test of freshly collected P. falciparum clinical isolates.
Plasmodium falciparum genotyping
P. falciparum genomic DNA (gDNA) was extracted from filter paper spots using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) and eluted in 30 µL elution buffer following the manufacturer’s instructions. The concentration and purity of the eluted gDNA were estimated using a NanoDrop One (Thermo Fisher Scientific, Madison, WI, USA). For each isolate, the presence of single or multiple parasite clones was assessed using a nested PCR approach as described earlier. The assays were performed using primers targeting the highly polymorphic regions of msp1 (block 2) and msp2 (block 3). All isolates that grew successfully during culture adaptation were genotyped after 7, 15, 21 and 28 days in culture and the number of clones was compared to that of the fresh isogenic isolate. The expected heterozygosity (HE) defined as the probability of being infected by at least two distinct alleles at a given locus was calculated as follows: HE = [n/(n-1)] [(1-Σpi)], with n being the number of isolates and pi the allele frequency at a given locus (Diouf et al., 2019).
Enzyme treatment and erythrocyte invasion phenotyping assays
Invasion phenotyping assays were performed using enzyme-treated erythrocytes (targets) from a single donor. Target erythrocytes were treated with either 250 mU/mL of neuraminidase, 1 mg/mL of trypsin or 1 mg/mL of chymotrypsin for 1 hour at 37° C with gentle shaking and washed thrice with RPMI1640 medium. The efficiency of enzyme treatment was assessed as previously described (Thiam et al., 2021). Treated erythrocytes were then labelled with 20 µM of carboxyfluorescein diacetate, succinimidyl ester (CFDA-SE) (Thermo Fisher Scientific) for two hours at 37° C with gentle shaking and protected from light exposure. For each isolate, schizont-infected erythrocytes were adjusted to 2% parasitemia and co-incubated with an equal volume of target erythrocytes in 96 well plates. All assays were performed in triplicates in a total volume of 100 µL at 2% haematocrit and incubated at 37° C for 24 hours. Parasitemia were adjusted to 1% for all isolates and invasion assays were considered successful only when invasion efficiency into control erythrocytes (untreated) was at least two-fold greater than the starting parasitemia. Plates were removed from the incubator and spun at 2,000 rpm for 3 minutes, after which the supernatant was discarded, replaced with a solution of 1 mM Hoechst 33342 to label the parasites’ DNA and incubated for an hour at 37° C. Plates were subsequently washed with 1X PBS and flow cytometry analyses were performed on a BD LSR Fortessa X-20 cytometer (BD Biosciences, Belgium). Invasion into target erythrocytes was determined by analysis of the proportion of Hoechst positive erythrocytes in 50,000 counted CFDA-SE positive cells. Percent invasion into enzyme-treated erythrocytes was expressed as a percentage of the invasion efficiency into labelled untreated erythrocytes.
Results
P. falciparum clinical isolates show different growth patterns during early in vitro culture adaptation
Samples used in this study were collected from Ghanaian children with uncomplicated malaria visiting the LEKMA hospital in Accra. To monitor the in vitro growth patterns of freshly collected P. falciparum clinical isolates, parasites were initially allowed to grow in the patient-derived erythrocytes for a minimum period of 96 hours after which freshly washed O+ erythrocytes (from a single donor) (Thiam et al., 2021) were added and growth tests were performed for isolates that successfully grew. Of the 25 isolates used in this study, 19 (76%) yielded detectable parasitemia 48 hours following in vitro adaptation. As measured by flow cytometry, P. falciparum clinical isolates showed different growth patterns during the first 96 hours of in vitro adaptation (Figure 2A). Monitoring of the PMR during these first two in vitro cycles revealed that most of the isolates had less than two-fold increase in parasitemia from one cycle to another. Isolates with successful growth patterns were further diluted with fresh erythrocytes and the parasitemia was used to assess the parasite multiplication rate through successive growth tests for a maximum period of twelve successive replication cycles. Overall, all isolates showed a minimum of 8 successful growth tests (66.67% success rate) with a median PMR of 1.77 (range 1.13 – 2.43) (Figure 2B).
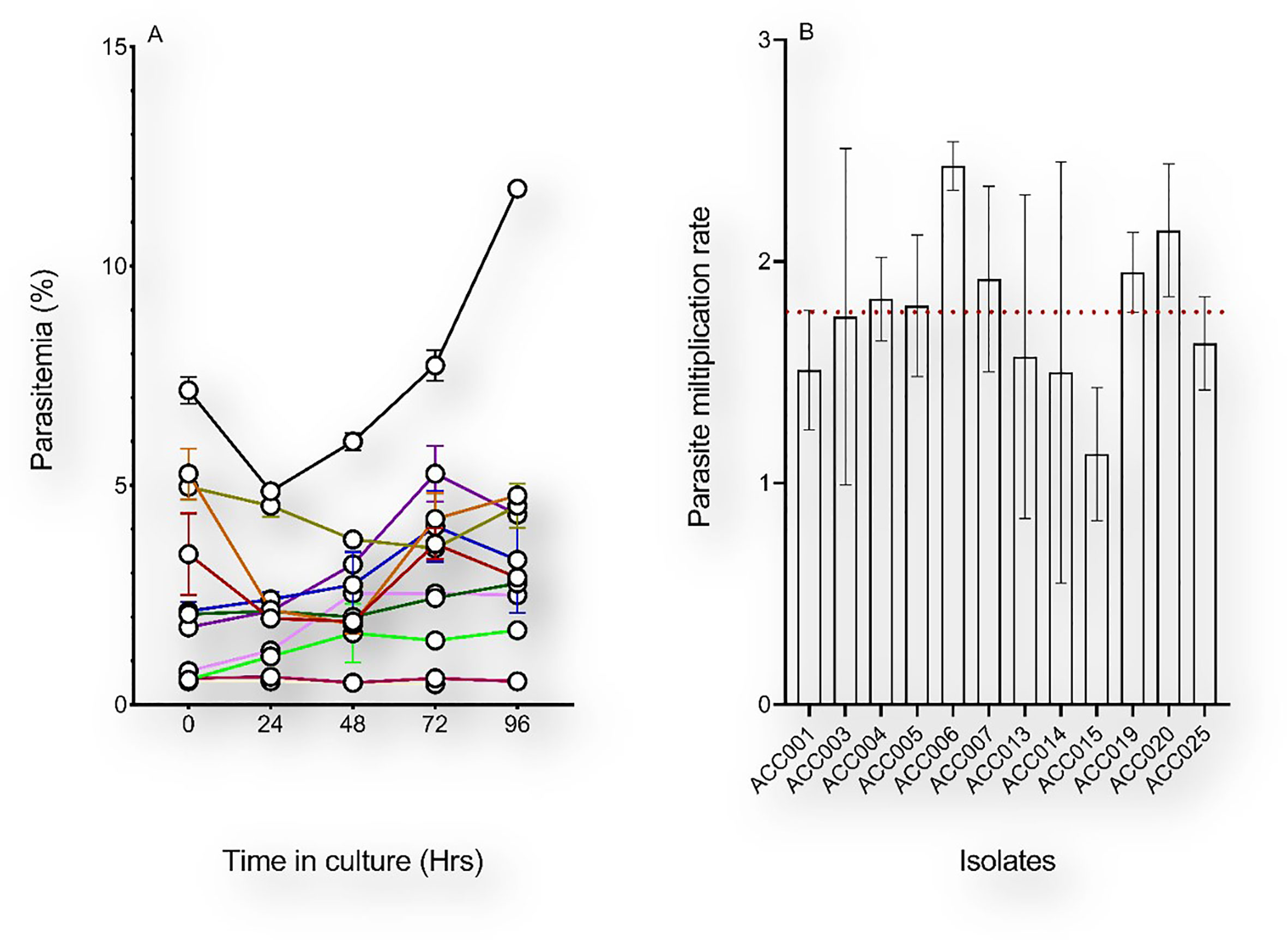
Figure 2 Early in vitro adaptation of P. falciparum clinical isolates. (A) The initial parasitemia (at H0) of each sample was recorded upon arrival from the field and the parasitemia of ex vivo-cultured isolates were monitored for 96 hours. The medium in the culture flasks was changed daily and supplemented with fresh erythrocytes only after 96 hours in vitro. (B) Parasite multiplication rates of successfully culture-adapted parasites following culture dilution with fresh erythrocytes. Depicted on the graph are the mean and standard errors of the PMRs of parasites with successful growth tests over a period of 28 days in vitro, and the red dotted line represents the median PMR (1.77).
P. falciparum clinical isolates represent genotypically diverse parasite populations
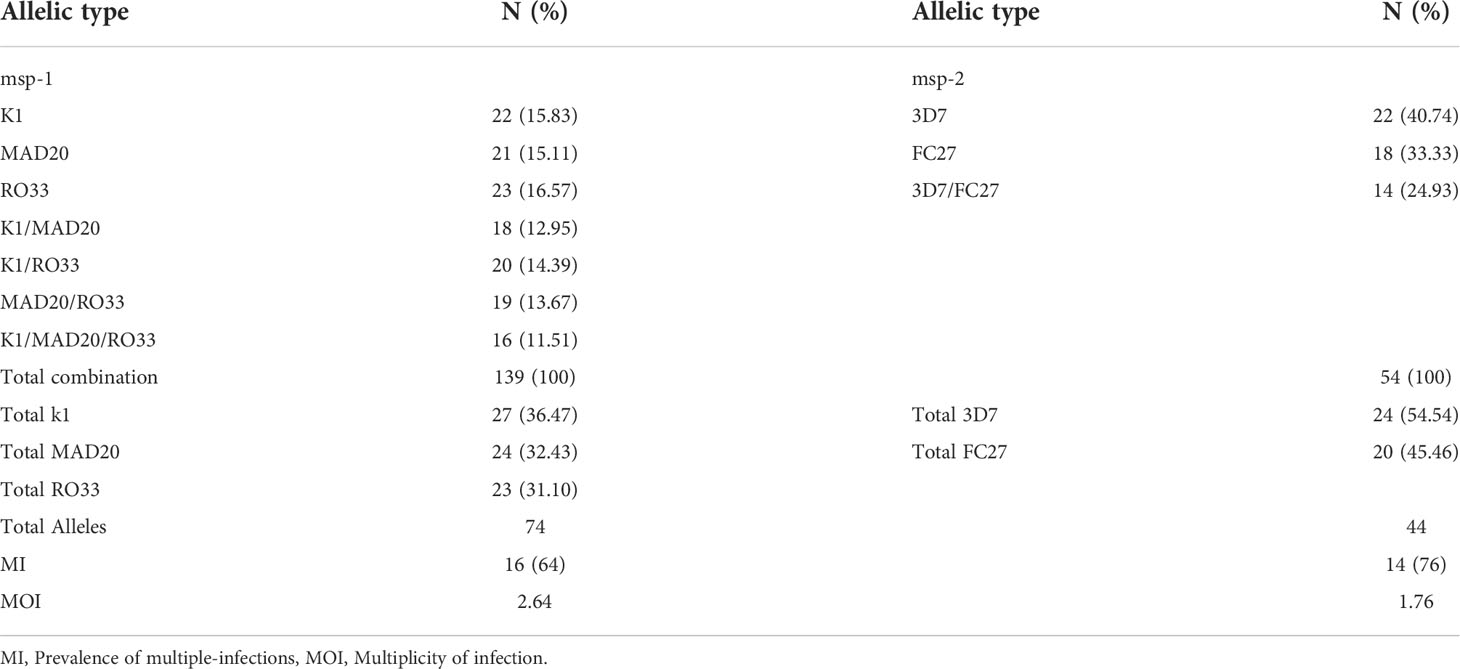
Unlike laboratory strains, P. falciparum clinical isolates frequently harbour multiple parasite clones, which could modulate the parasite’s in vitro adaptability. Here, we assessed the presence of multiple parasite clones in each of the tested isolates using the highly polymorphic regions of msp1 and msp2 genes. All 25 isolates were shown to be polyclonal and the number of alleles detected were 74 and 44 for msp1 and msp2, respectively (Table 1). For msp1, the allele frequencies were 36.47% (27/74), 32.43% (24/74) and 31.10% (23/74) for K1, MAD20 and RO33, respectively, while those for the msp2 allelic families were 54.54% (24/44) and 45.46% (20/44) for 3D7 and FC27, respectively. The prevalence of multiple infections was respectively 16 and 14 for msp1 and msp2, while the multiplicity of infections was 2.6 and 1.76, respectively for the two genes (Table 1). Besides, the HE was 0.35 and 0.52 for msp1 and msp2, respectively.
Short-term culture adapted isolates harbour lower number of parasite clones
To measure the effect of short-term culture adaptation on the parasites’ genotypes, ten of the culture-adapted isolates were genotyped at days 7, 15, 21 and 28 post-adaptation using the msp1 and 2 allelic families. Overall, there was little variation in the proportion of allelic families across the different time points (Figure 3). However, our analysis showed a reduction in the number of clones per isolate at day 28 as compared to day 0 (Table 2). The maximum number of clones per isolate was reduced from 4 to 1 for the msp1 gene, while that of msp2 was reduced from 2 to 1 (Table 2). Of all msp1 allelic families, K1 was the most predominant at both day 0 and day 28, with a percentage of 41.94% and 58.33%, respectively (Table 2). MAD20 and RO33, which were present at the same proportion at day 0 (29.03%, each), represented respectively 16.67% and 25.00% of the total number of alleles (Table 2). For msp2, there were little changes in the proportions of the respective allelic families, with 3D7 being the most predominant allele representing 63.16% and 61.55% at day 0 and 28, respectively (Table 2). Out of the eight isolates that harboured the MAD20 allelic family at day 0, only two were detected with a copy of the allele at day 28, while the RO33 allele which was initially present in nine isolates at day 0, was detected in only three isolates at day 28. K1, initially detected in all ten isolates, was still present in seven of them at day 28. Of all three msp1 allelic families, only K1 and RO33 were simultaneously detected in the same isolates at day 28, while MAD20 was only detected as single infections (Table 2).
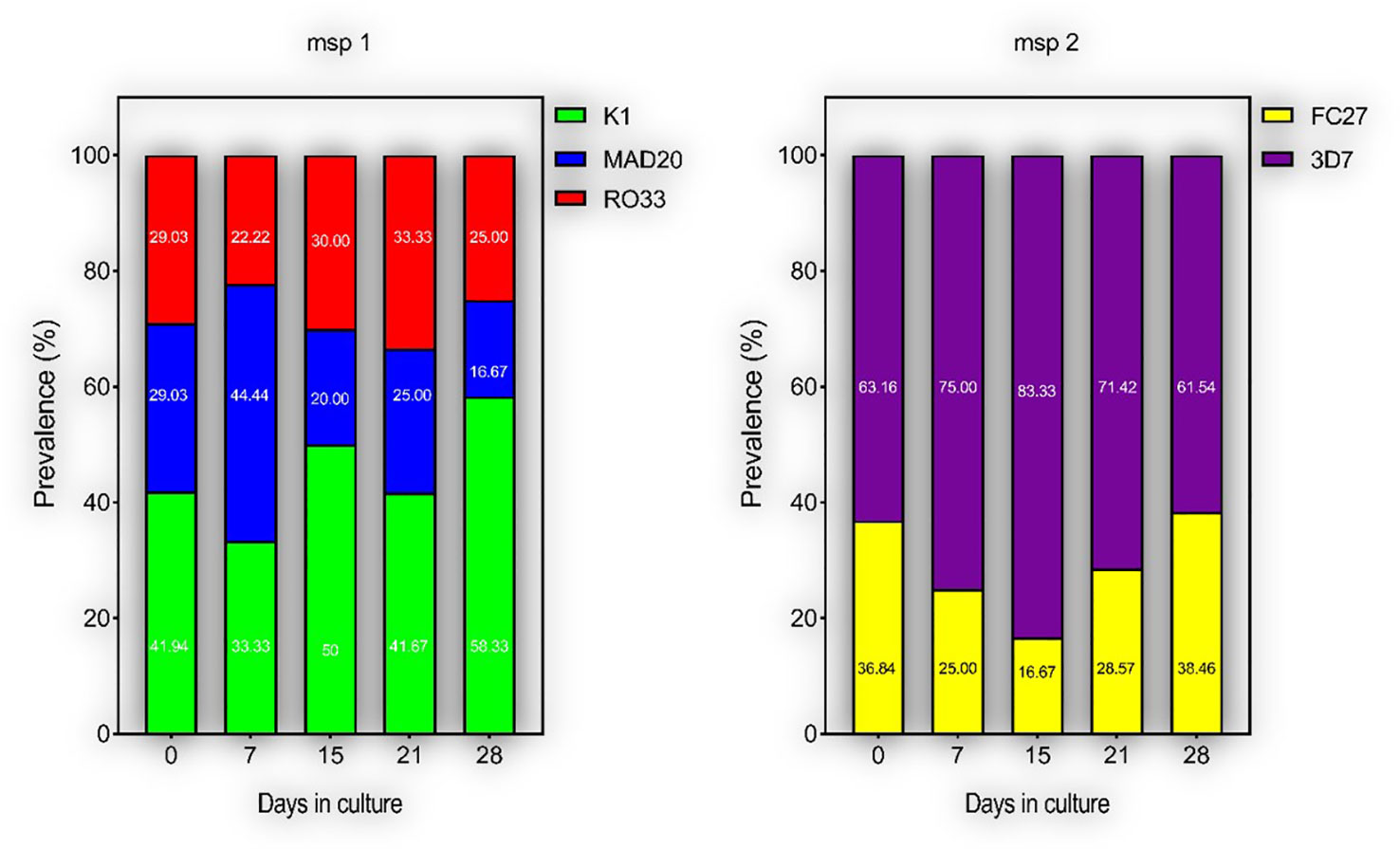
Figure 3 Proportions of different parasite clones during culture-adaptation. Parasites were all genotyped at Day 0 upon collection and successfully culture-adapted ones were further genotyped at Days 7, 15, 21 and 28-post adaptation. Represented are the proportions of individual alleles for the msp 1 and 2 genes.
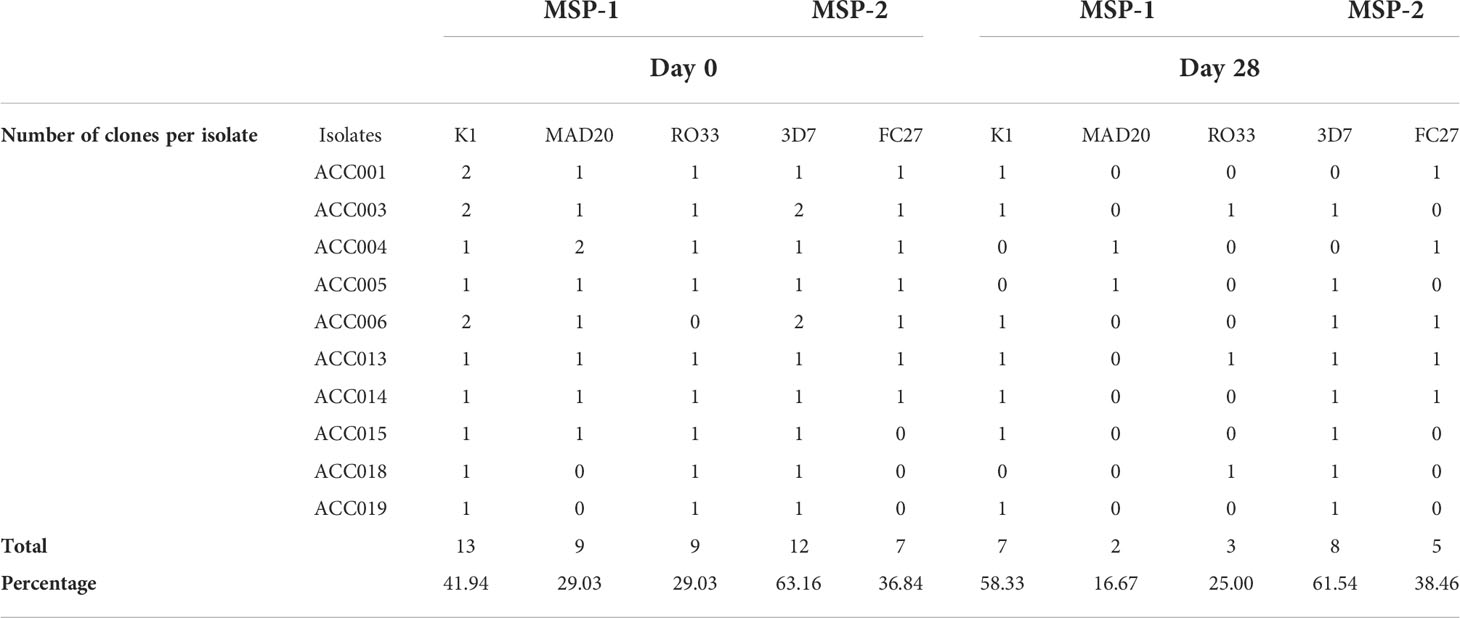
Table 2 Comparative analysis of P.falciparum genetic diversity prior and following short-term culture adaptation.
For the msp2 gene, out of the seven isolates harbouring the FC27 allelic family at day 0, only four persisted at day 28, while the 3D7 allelic family was initially present in all ten isolates at day 0 but was detected in eight of them at day 28. As for msp1, both allelic families of msp2 were present in three isolates as co-infections, while two and four isolates presented single infections of FC27 and 3D7, respectively at day 28. However, there was no specific dominant combination of msp1 and msp2 detected in our isolates at day 28. Moreover, aside those detected at day 0, there were no newly detected alleles in our isolates at day 28, therefore suggesting the absence of detectable cross-contamination during culture adaptation.
Differences in thawing protocols does not affect early in vitro culture adaptation of short-term cryopreserved P. falciparum clinical isolates
Culture-adaptation of P. falciparum clinical isolates has been reported as more labor-intensive than immediate ex vivo processing. Moreover, there are earlier reports of P. falciparum clonal selection during in vitro adaptation (Chen et al., 2014). To ascertain the effect of cryopreservation on P. falciparum early in vitro adaptation, we compared the PMR of short-term cryopreserved clinical isolates to that of their freshly cultured isogenic counterparts. Two vials of the same isolate were simultaneously revived using two distinct NaCl-based thawing protocols. Parasitemia were adjusted to 0.5-1% using erythrocytes from a single donor and the PMR was monitored during the first three asexual replicative cycles. Successful monitoring of the PMR of isolates prior to and following cryopreservation (one-year interval) revealed differences in PMR between cryopreserved isolates and the matched freshly culture-adapted counterparts. The median PMR was 1.62 for fresh isolates while that of cryopreserved isolates was 1.12 and 1.27 following two-step and three-step thawing, respectively (Figure 4A). However, the difference in PMR was only significant when comparing fresh isolates and cryopreserved parasites thawed using a two-step protocol (P = 0.03; Figure 4A), while no significant difference was observed between isogenic isolates thawed using distinct protocols (Figure 4A, B). Flow cytometric analysis of the DNA content of revived cryopreserved parasites revealed different fluorescence peaks following Hoechst 33342 staining suggesting the presence of different parasite stages after 48-, 96- and 144-hours post-incubation. However, no significant difference was observed in the proportions of the different parasite stages when comparing the parasites’ growth after thawing using different protocols (Figure 4C).
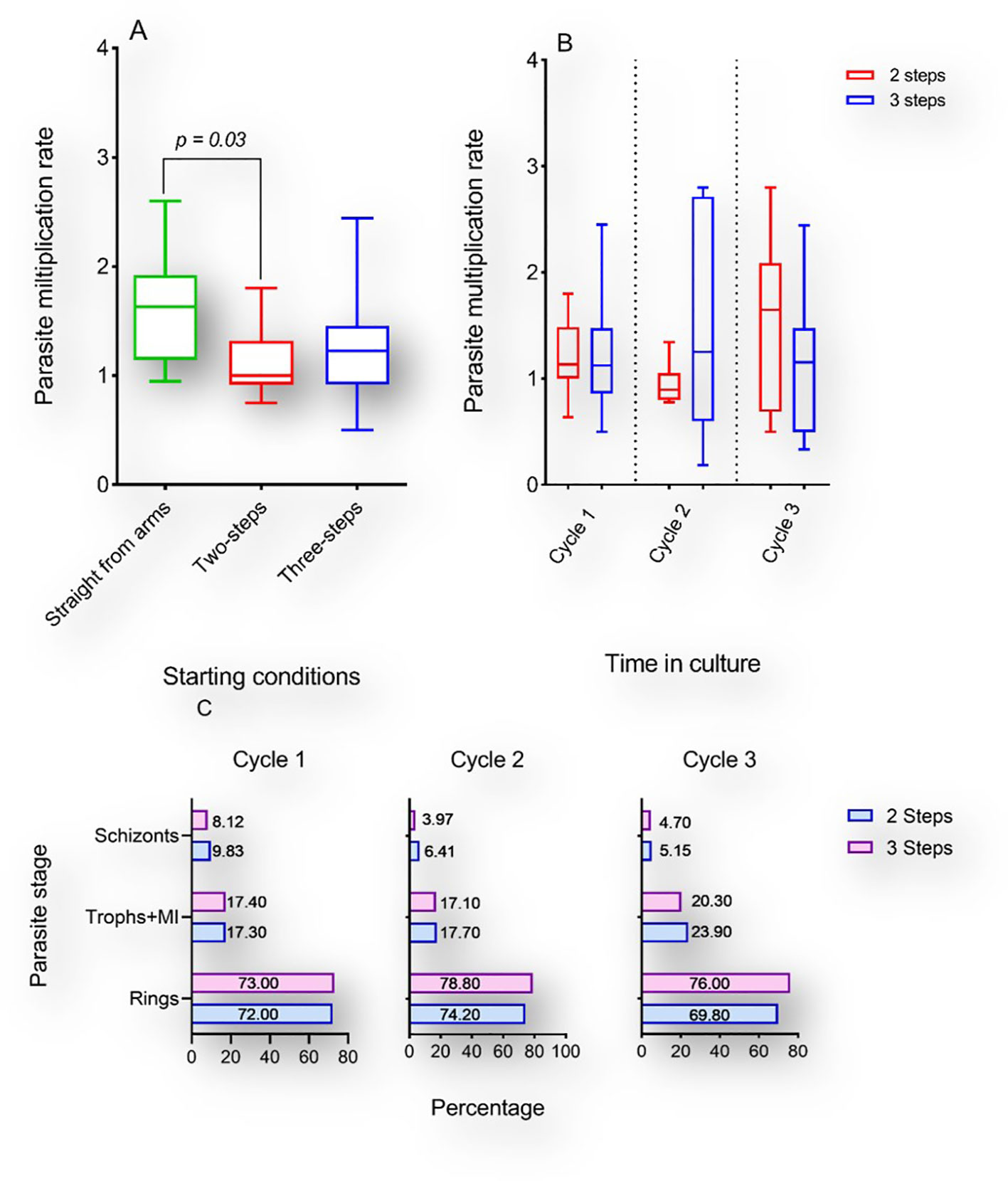
Figure 4 Multiplication rates of P. falciparum clinical isolates before and after cryopreservation. A-B: Box and whiskers plots showing the PMR of fresh cultured P. falciparum clinical isolates (green) or thawed with a two-step (red) or three-step (blue) protocol following cryopreservation. Kruskal Wallis test was conducted to compare the PMR of fresh versus cryopreserved isolates after three in vitro replicative cycles (A) or to compare the PMR of isolates thawed with different protocols after each replicative cycle (B). Represented in (C) are the proportions of different parasites stages after 1, 2 and 3 cycles following thawing with either protocol.
Short-term culture adaptation has minimal effect on P. falciparum invasion phenotype
P. falciparum invasion phenotyping has mostly been conducted following short-term culture-adaptation (Okoyeh et al., 1999; Lobo et al., 2004; Nery et al., 2006; Lopez-Perez et al., 2012). Invasion phenotypes are classified as either resistant (r) or susceptible (s) to the different enzyme treatment of the erythrocytes with invasion efficiency set at 50%. To this, we assessed the effect of short-term culture adaptation on P. falciparum invasion phenotype in the different enzyme treated erythrocytes. To allow for easy inference, we compared the ex vivo invasion phenotype of four isolates when the samples were collected to that obtained after 28 days in culture to see if there are changes. Of the four isolates tested, only one (ACC015) had a significant change in the sensitivity to enzyme treatment following culture adaptation (Figure 5), while significant changes were observed in ACC003 and ACC014 following treatment with neuraminidase and trypsin, respectively.
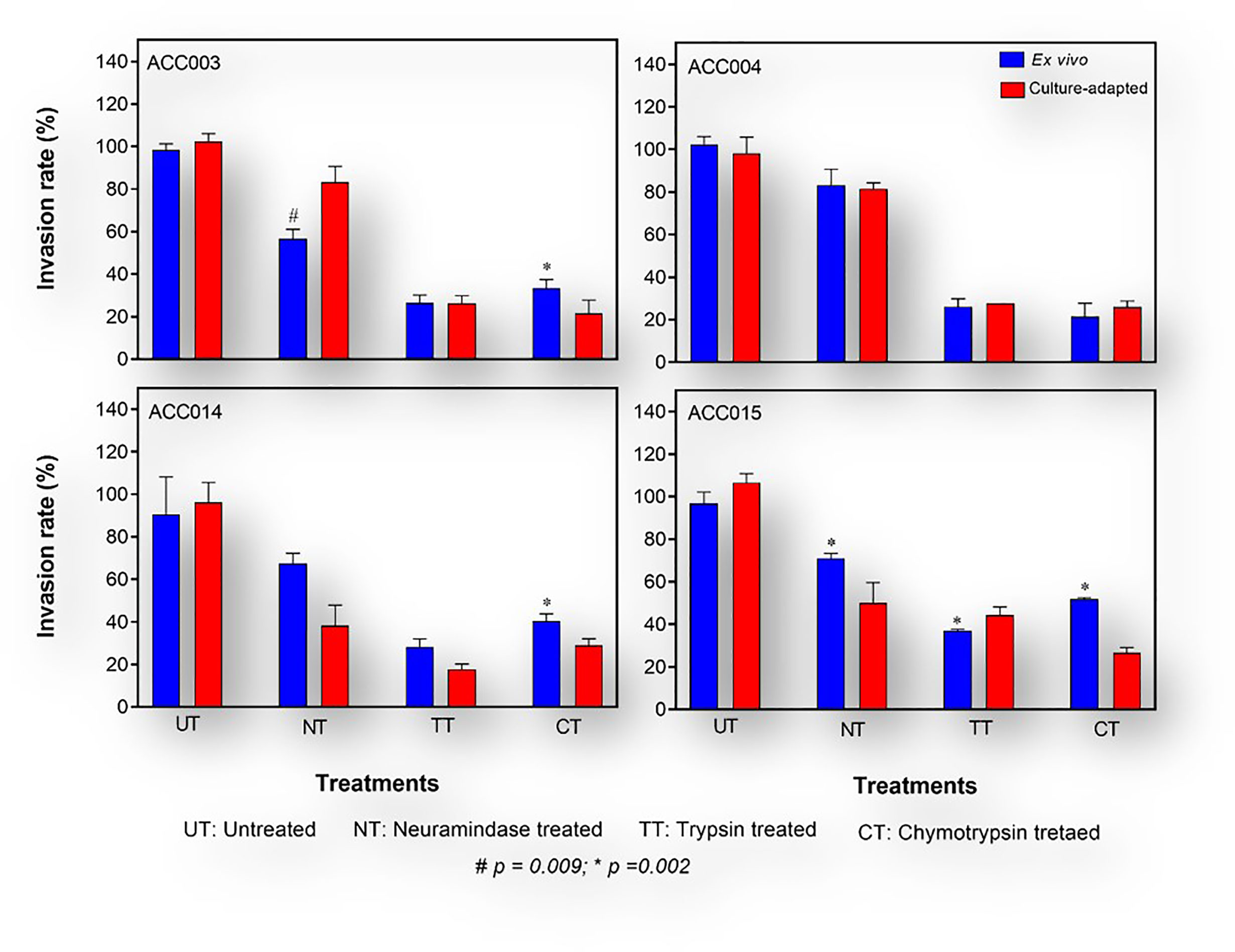
Figure 5 Invasion phenotypes of P.falciparum clinical isolates before and after short-term culture adaptation. The ex vivo phenotype of freshly collected isolates (blue bars) obtained during the first asexual replicative cycle upon arrival at the laboratory was compared to that obtained after a short-term in vitro adaptation of about 28 days (red bars). The Mann Whitney U test was used to assess the differences in invasion efficiency between the different time points.
Moreover, changes in invasion profile, defined as the combination of sensitivity to the three enzymes, was observed in two isolates (ACC014: NrTsCs → NsTsCs and ACC015: NrTsCr → NrTsCs), where N: neuraminidase, T: trypsin, C: chymotrypsin, s: sensitive and r: resistant; while → depicts the changes in profile). Interestingly, although all three msp1 alleles were initially present in these two isolates, only K1 persisted at Day 28-post adaptation, while both the 3D7 and FC27 alleles of msp2 persisted throughout the 28-day period post adaptation.
Cryopreservation has minimal effect on P. falciparum invasion phenotype
Cryopreserved isolates were thawed at different time intervals (from 3 to 12 months after cryopreservation) and assayed for their ability to invade enzyme-treated erythrocytes as compared to their fresh uncultured counterparts. Given the challenges associated with the parasites’ in vitro adaptability during the first rounds of asexual replication, and to avoid clonal selection following long-term culture adaptation, all assays were set during the first two replicative cycles. Of the 25 isolates collected in this study, seven had enough numbers of cryopreserved vials (six vials) to be thawed at all the time-points and successfully phenotyped over the course of one-year post cryopreservation. All isolates showed a sialic acid independent phenotype with the invasion of neuraminidase treated erythrocytes greater than 50% relative to that of untreated control erythrocytes (Figure 6). Overall, there was no significant change in sialic acid dependency in the parasites after cryopreservation relative to fresh uncultured isolates (Figure 6). The most common invasion profile was neuraminidase resistant, trypsin sensitive and chymotrypsin sensitive (NrTsCs). However, three out of the seven isolates showed changes in invasion profiles in trypsin and chymotrypsin treated erythrocytes following cryopreservation with the apparition of three novel phenotypes (NrTrCs, NrTsCr and NrTrCr) after six months post cryopreservation (Figure 6). Nevertheless, none of the novel phenotypes persisted after twelve months post cryopreservation, suggesting a technical or random effect associated with these changes.
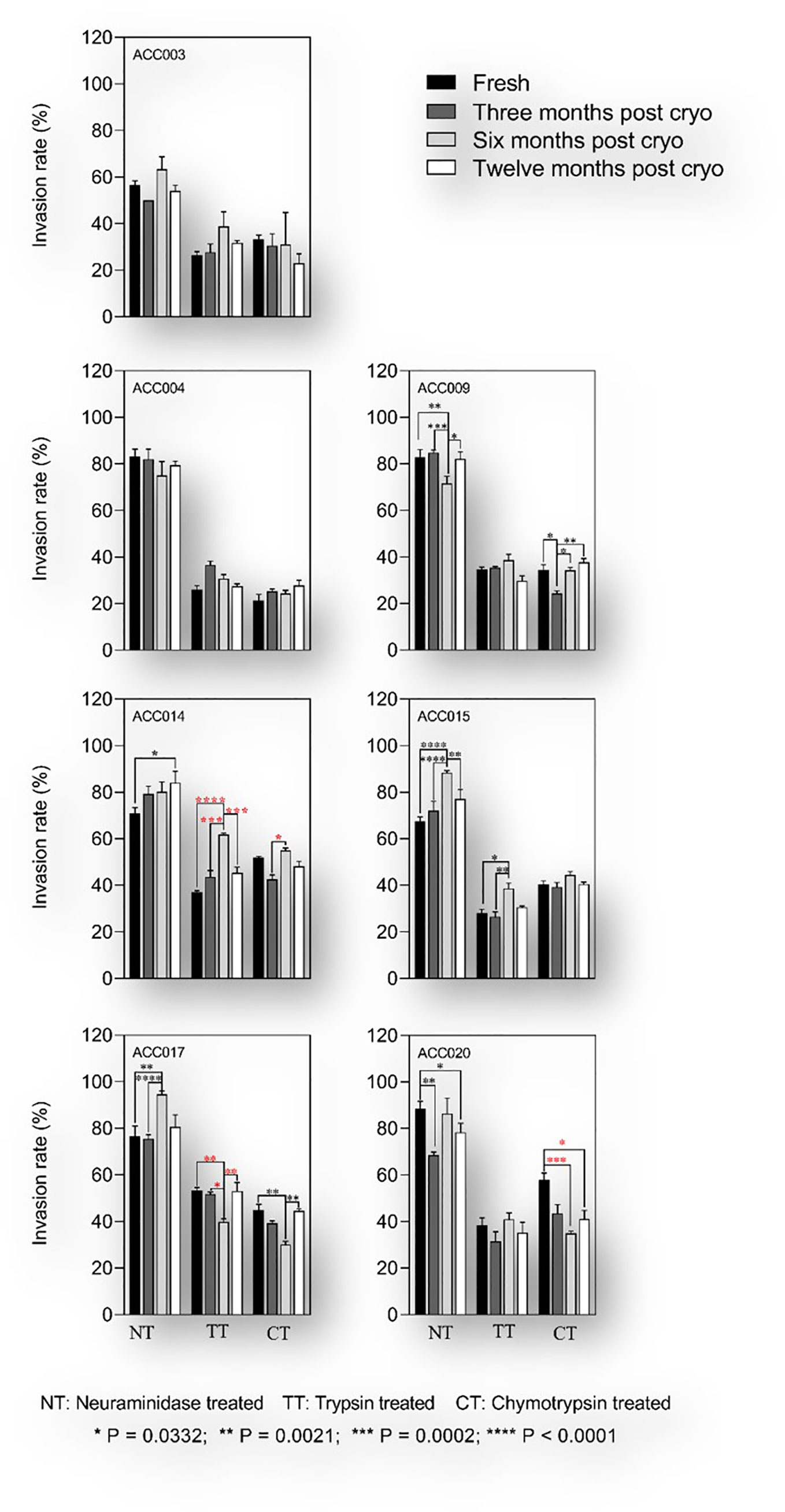
Figure 6 Invasion phenotypes of P.falciparum clinical isolates before and after short-term cryopreservation. The assays were set up between 24- and 36-hours following sample processing and the parasites were incubated for another 18 to 24 hours prior to flow cytometric analysis. For each isolate, the invasion phenotype of freshly culture adapted parasites (black bars) was compared to that obtained after three months (dark grey bars), six months (light grey bars) and twelve months (white bars) post cryopreservation. Kruskal Wallis was conducted to test for statistical differences in invasion efficiency of fresh versus cryopreserved isolates. Red stars denote significant differences associated with changes in invasion profile of a given treatment and black stars depict significant differences with no changes in the invasion profile.
Discussion
The phenotypic diversity of P. falciparum clinical isolates has widely been reported in the last two decades (Ahouidi et al., 2016). However, conducting such assays with uncultured clinical isolates has been precluded by the lack of appropriate laboratory settings in remote areas where the highest malaria burden occurs. Consequently, the majority of the pioneering works were conducted in cryopreserved P. falciparum clinical isolates and in some cases after short-term culture adaptation following parasite thawing (Okoyeh et al., 1999; Lobo et al., 2004; Nery et al., 2006; Lopez-Perez et al., 2012), although the effect of such procedures in the parasite’s invasion phenotype has never been reported.
In this study, we investigated the effect of cryopreservation and different thawing protocols on P. falciparum in vitro adaptation and invasion phenotyping assays. We showed that P. falciparum clinical isolates show specific growth patterns during the early in vitro culture adaptation, while most of the isolates did not recover following culture dilution with fresh erythrocytes. Previous studies have reported such findings, mainly attributed to unreported antimalarial drug use prior to presentation at hospital (Baum et al., 2003).
Given that P. falciparum in vitro adaptation could also be influenced by the number of parasite clones present in a given isolate, our genotyping analysis revealed that all P. falciparum clinical isolates used in this study presented multiple parasite clones, with an overall MOI of 4.4, which is slightly higher than previously reported in Burkina Faso (Sondo et al., 2019), Ghana (Duah et al., 2016), Republique of Congo (Singana et al., 2019), Nigeria and Senegal (Oboh1 et al., 2017). However, the predominance of K1 and 3D7 allelic families of msp1 and msp2 reported here is in agreement with previous reports from Burkina Faso (Sondo et al., 2019; Sondo et al., 2020), Nigeria and Senegal (Oboh1 et al., 2017) Burkina Faso (Oboh1 et al., 2017; Sondo et al., 2019) (Kobbe et al., 2006), but contrary to studies from Uganda and Sudan which reported the predominance of RO33 and FC27, respectively for the two genes (Peyerl-Hoffmann et al., 2001; Mohammed et al., 2015).
In this study, the median PMR of short-term culture adapted isolates was 1.77, consistent with previous reports (Lantos et al., 2009), however, there was no relationship between the observed PMR and the number of parasite clones per isolate. Furthermore, of the 19 culture-adapted isolates tested in this study, 10 were successfully genotyped at day 28 post culture inoculation and our data show an apparent clonal selection following short-term culture adaptation. This is in agreement with previous data that also reported culture adaptation of P. falciparum clinical isolates as a modulator of the parasite’s susceptibility to a wide range of drugs as compared to their fresh uncultured counterparts (Chaorattanakawee et al., 2015).
Our data also show the persistence of K1 at day 28 in almost all isolates that harboured this allelic family on day 0, while MAD20 prevalence significantly decreased at day 28. Furthermore, MAD20 was outgrown by K1 in almost all isolates harbouring mixed infections (8/10 isolates), consistent with reports by Sondo and colleagues where MAD20 was outcompeted by K1 in natural malaria infections (Sondo et al., 2019).
For some of the isolates in this study, the invasion phenotype following culture adaptation was different from the parasites’ ex vivo phenotype. This, therefore, suggests a possible effect of short-term culture adaption and/or clonal selection on the parasite invasion phenotype. However, this could also result from technical variations during invasion assay set up, and given the small number of isolates tested for this experiment (only four isolates), there is a need for further confirmation with a larger number of isolates and possibly a longer culture adaptation time.
The present study also investigated the effect of cryopreservation and thawing protocols in the parasites’ early in vitro adaptation and invasion phenotype as compared to their freshly cultured isogenic counterparts. Our data show that cryopreserved P. falciparum clinical isolates used in this study have a lower multiplication rate during the first in vitro cycles as compared to their fresh isogenic counterparts. However, this difference in PMR was only significant when freshly cultured isolates were compared to those revived using a two-step NaCl protocol. This could be because of a simple artefact during culture adaptation or due to the fitness of the different parasite clones in each isolate. It is therefore possible that the parasite-induced stress during the thawing process will accentuate the low fitness of certain clones, while this effect could be minimal when parasites are thawed using a three-step protocol.
Another potential factor that could affect the early in vitro adaptation of revived cryopreserved isolates is the addition of fresh erythrocytes following parasites growth. Given the difference between the freshly added erythrocytes and the initial patient-derived erythrocytes, there is a possibility that the lower multiplication rate observed post-cryopreservation is solely due to the parasite adaptation to the new erythrocytes. However, given the observed differences in the PMR post cryopreservation, and to minimize the effect of clonal selection, invasion phenotyping assays were only performed in isolates that yielded appropriate parasitemia during the first in vitro replicative cycles.
As a result, our data show that most of the isolates assayed following cryopreservation maintained a relatively stable invasion phenotype regardless of the length of cryopreservation or the thawing protocol used for the revival of the parasites. To our knowledge, this is the first study to investigate the effect of short-term culture adaptation and cryopreservation on P. falciparum invasion phenotype using clinical isolates from the same isogenic backgrounds. Altogether, these results suggest that short-term culture adaptation could influence the invasion phenotype of P. falciparum clinical isolates due to clonal selection during in vitro culturing, but this needs further confirmatory studies with a larger number of isolates.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by Institutional Review Board of the Noguchi Memorial Institute for Medical Research, University of Ghana (IRB00001276) and the Ghana Health Service Ethical Review Committee (GHC-ERC: 005/12/2017). Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author contributions
The authors want to indicate that data presented in this manuscript forms part of thesis submitted to the University of Ghana in partial fulfillment for the award of Ph.D. to LT 28. LT, YA and GA conceived the study; LT and FA performed the experiments LT, FA, MN, YA and GA analyzed the data and drafted the manuscript; YA, GA, and MN supervised the study. All authors critically reviewed and edited the manuscript.
Funding
This work was supported by funds from a World Bank African Centres of Excellence grant (ACE02-WACCBIP: Awandare) and a DELTAS Africa grant (DEL-15-007: Awandare). LT was supported by WACCBIP-World Bank ACE PhD fellowships, respectively, FA was supported by the National Institute for Health Research (NIHR) Global Health Research program 16/136/33, using aid from the UK Government while YA was supported by a WACCBIP-DELTAS postdoctoral fellowship. The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust (107755/Z/15/Z: Awandare) and the UK government. The views expressed in this publication are those of the author(s) and not necessarily those of AAS, NEPAD Agency, Wellcome Trust or the UK government.
Acknowledgments
This work is part of the assay standardization efforts of the West African merozoite invasion network (WAMIN) consortium, and we are grateful to members for contributing ideas to this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahouidi, A. D., Amambua-Ngwa, A., Awandare, G. A., Bei, A. K., Conway, D. J., Diakite, M, et al. (2016). Malaria vaccine development: Focusing field erythrocyte invasion studies on phenotypic diversity: The West African merozoite invasion network (WAMIN). Trends Parasitol. 32, 274?–283 doi: 10.1016/j.pt.2015.11.009
Awandare, G. A., Nyarko, P. B., Aniweh, Y., Ayivor-Djanie, R., Stoute, J. A. (2018). Plasmodium falciparum strains spontaneously switch invasion phenotype in suspension culture. Sci. Rep. 8:1–10. doi: 10.1038/s41598-018-24218-0
Baum, J., Pinder, M., Conway, D. J. (2003). Erythrocyte invasion phenotypes of plasmodium falciparum in the Gambia. Infect. Immun. 71, 1856–1863. doi: 10.1128/IAI.71.4.1856-1863.2003
Bowyer, P. W., Stewart, L. B., Aspeling-Jones, H., Mensah-Brown, H. E., Ahouidi, A. D., Amambua-Ngwa, A., et al. (2015). Variation in plasmodium falciparum erythrocyte invasion phenotypes and merozoite ligand gene expression across different populations in areas of malaria endemicity. Infect. Immun. 83, 2575–2582. doi: 10.1128/IAI.03009-14
Chaorattanakawee, S., Lanteri, C. A., Sundrakes, S., Yingyuen, K., Gosi, P., Chanarat, N, et al. (2015). Attenuation of plasmodium falciparum in vitro drug resistance phenotype following culture adaptation compared to fresh clinical isolates in Cambodia. Malar J. 14, 1–7 doi: 10.1186/s12936-015-1021-8
Chen, K., Sun, L., Lin, Y., Fan, Q., Zhao, Z., Hao, M, et al. (2014). Competition between plasmodium falciparum strains in clinical infections during in vitro culture adaptation. Infect. Genet. Evol. 3, 1–10 doi: 10.1016/j.meegid.2014.03.012
Cortés, A. (2008). Switching plasmodium falciparum genes on and off for erythrocyte invasion. Trends Parasitol. 24, 517–524. doi: 10.1016/j.pt.2008.08.005
Cowman, A. F., Tonkin, C. J., Tham, W. H., Duraisingh, M. T. (2017). The molecular basis of erythrocyte invasion by malaria parasites. Cell Host Microbe. 22, 232–245 doi: 10.1016/j.chom.2017.07.003
Diouf, B., Diop, F., Dieye, Y., Loucoubar, C., Dia, I., Faye, J., et al. (2019). Association of high plasmodium falciparum parasite densities with polyclonal microscopic infections in asymptomatic children from toubacouta, Senegal. Malar J. 18, 1–8 doi: 10.1186/s12936-019-2684-3
Duah, N. O., Matrevi, S. A., Quashie, N. B., Abuaku, B., Koram, K. A., Sondo, P, et al. (2016). Genetic diversity of plasmodium falciparum isolates from uncomplicated malaria cases in Ghana over a decade. Parasites Vectors 9, 1–8. doi: 10.1186/s13071-016-1692-1
EVIMalaR (2013). “Methods in malaria research, sixth edition,”Scherf, A, Wahlgren, M, Moll, K, Kaneko, A. (Glasgow, UK: EVIMalaR: EVIMalaR; Bei Ressources). Available at: https://www.beiresources.org/portals/2/MR4/Methods_In_Malaria_Research-6th_edition.pdf.
Gomez-Escobar, N., Amambua-Ngwa, A., Walther, M., Okebe, J., Ebonyi, A., Conway, D. J, et al. (2010). Erythrocyte invasion and merozoite ligand gene expression in severe and mild plasmodium falciparum malaria. J. Infect. Dis. 201, 444–452 doi: 10.1086/649902
Jennings, C. V., Ahouidi, A. D., Zilversmit, M., Bei, A. K., Rayner, J., Sarr, O, et al. (2007). Molecular analysis of erythrocyte invasion in plasmodium falciparum isolates from Senegal. Infect. Immun. 75, 3531–3538. doi: 10.1128/IAI.00122-07
Kobbe, R., Neuhoff, R., Marks, F., Adjei, S., Langefeld, I., Von Reden, C, et al. (2006). Seasonal variation and high multiplicity of first plasmodium falciparum infections in children from a holoendemic area in Ghana, West Africa. Trop. Med. Int. Heal. 11, 613–619 doi: 10.1111/j.1365-3156.2006.01618.x
Lantos, P. M., Ahouidi, A. D., Bei, A. K., Jennings, C. V., Sarr, O., Ndir, O., et al. (2009). Erythrocyte invasion profiles are associated with a common invasion ligand polymorphism in Senegalese isolates of plasmodium falciparum. Parasitology. 136, 1–9 doi: 10.1017/S0031182008005167
Lobo, C. A., De Frazao, K., Rodriguez, M., Reid, M., Zalis, M., Lustigman, S, et al. (2004). Invasion profiles of Brazilian field isolates of plasmodium falciparum: Phenotypic and genotypic analyses. Infect. Immun. 72, 5886–5891. doi: 10.1128/IAI.72.10.5886-5891.2004
Lopez-Perez, M., Villasis, E., Machado, R. L.D., Póvoa, M. M., Vinetz, J. M., Blair, S, et al. (2012). Plasmodium falciparum field isolates from south America use an atypical red blood cell invasion pathway associated with invasion ligand polymorphisms. PloS One 7. doi: 10.1371/journal.pone.0047913
Mohammed, H., Mindaye, T., Belayneh, M., Kassa, M., Assefa, A., Tadesse, M, et al. (2015). Genetic diversity of plasmodium falciparum isolates based on MSP-1 and MSP-2 genes from kolla-shele area, arbaminch zuria district, southwest Ethiopia. Malar J. doi: 10.1186/s12936-015-0604-8
Nery, S., Deans, A. M., Mosobo, M., Marsh, K., Rowe, J. A., Conway, D. J., et al. (2006). Expression of plasmodium falciparum genes involved in erythrocyte invasion varies among isolates cultured directly from patients. Mol. Biochem. Parasitol. 149, 208–215. doi: 10.1016/j.molbiopara.2006.05.014
Oboh1, M. M., Ndiaye, T., Diongue, K., Ndiaye, Y. D., Sy, M., Deme, A.B., et al. (2017). Plasmodium falciparum . Emerg. Top. Life Sci. 1, 517–523. doi: 10.1042/ETLS20170099
Okoyeh, J. N., Pillai, C. R., Chitnis, C. E. (1999). Plasmodium falciparum field isolates commonly use erythrocyte invasion pathways that are independent of sialic acid residues of glycophorin a. Infect. Immun. 67, 5784–5791 doi: 10.1128/iai.67.11.5784-5791.1999
Peyerl-Hoffmann, G., Jelinek, T., Kilian, A., Kabagambe, G., Metzger, W. G., Von Sonnenburg, F, et al. (2001). Genetic diversity of plasmodium falciparum and its relationship to parasite density in an area with different malaria endemicities in West Uganda. Trop. Med. Int. Heal. 6, 607–613 doi: 10.1046/j.1365-3156.2001.00761.x
Singana, B. P., Mayengue, P. I., Niama, R. F., Ndounga, M. (2019). Genetic diversity of plasmodium falciparum infection among children with uncomplicated malaria living in pointe-noire, republic of Congo. Pan Afr. Med. J. 32, 183. doi: 10.11604/pamj.2019.32.183.15694
Sondo, P., Derra, K., Lefevre, T., Diallo-Nakanabo, S., Tarnagda, Z., Zampa, O., et al. (2019). Genetically diverse plasmodium falciparum infections, within-host competition and symptomatic malaria in humans. Sci. Rep. 9, 1–9 doi: 10.1038/s41598-018-36493-y
Sondo, P., Derra, K., Rouamba, T., Nakanabo Diallo, S., Taconet, P., Kazienga, A, et al. (2020). Determinants of plasmodium falciparum multiplicity of infection and genetic diversity in Burkina Faso. Parasites Vectors 13, 1–12. doi: 10.1186/s13071-020-04302-z
Thiam, L. G. (2019). Investigating factors influencing variation in p Falciparum Invasion Phenotyping Assays (Legon: University if Ghana). 11, pp 1–14.
Thiam, L. G., Nyarko, P. B., Kusi, K. A., Niang, M., Aniweh, Y., Awandare, G. A, et al. (2021). Blood donor variability is a modulatory factor for p. falciparum invasion phenotyping assays Sci. Rep. doi: 10.1038/s41598-021-86438-1
Wright, G. J., Rayner, J. C. (2014). Plasmodium falciparum erythrocyte invasion: Combining function with immune evasion. PloS Pathog. 3, 1–7 doi: 10.1371/journal.ppat.1003943
Keywords: Plasmodium falciparum, cryopreservation, thawing protocols, culture-adaptation, invasion phenotype
Citation: Thiam LG, Ansah F, Niang M, Awandare GA and Aniweh Y (2022) Short-term cryopreservation and thawing have minimal effects on Plasmodium falciparum ex vivo invasion profile. Front. Cell. Infect. Microbiol. 12:997418. doi: 10.3389/fcimb.2022.997418
Received: 18 July 2022; Accepted: 29 August 2022;
Published: 20 September 2022.
Edited by:
Ehsan Ahmadpour, Tabriz University of Medical Sciences, IranReviewed by:
Suchi Goel, Indian Institute of Science Education and Research, Tirupati, IndiaNirianne Querijero Palacpac, Osaka University, Japan
Copyright © 2022 Thiam, Ansah, Niang, Awandare and Aniweh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gordon A. Awandare, Z2F3YW5kYXJlQHVnLmVkdS5naA==; Yaw Aniweh, eWFuaXdlaEB1Zy5lZHUuZ2g=
†Present address: Laty G. Thiam, G4 MEGA Vaccines, Pôle Immunophysiopathologie et Maladies Infectieuses, Institut Pasteur de Dakar, Dakar, Senegal
 Laty G. Thiam
Laty G. Thiam Felix Ansah
Felix Ansah Makhtar Niang
Makhtar Niang Gordon A. Awandare
Gordon A. Awandare Yaw Aniweh
Yaw Aniweh