
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol., 12 October 2022
Sec. Microbes and Innate Immunity
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.996778
 Wang Yangyanqiu1,2,3†
Wang Yangyanqiu1,2,3† Han Shuwen1,2,3*
Han Shuwen1,2,3*The incidence of cancer is high worldwide, and biological factors such as viruses and bacteria play an important role in the occurrence of cancer. Helicobacter pylori, human papillomavirus, hepatitis B viruses and other organisms have been identified as carcinogens. Cancer is a disease driven by the accumulation of genome changes. Viruses can directly cause cancer by changing the genetic composition of the human body, such as cervical cancer caused by human papillomavirus DNA integration and liver cancer caused by hepatitis B virus DNA integration. Recently, bacterial DNA has been found around cancers such as pancreatic cancer, breast cancer and colorectal cancer, and the idea that bacterial genes can also be integrated into the human genome has become a hot topic. In the present paper, we reviewed the latest phenomenon and specific integration mechanism of bacterial DNA into the human genome. Based on these findings, we also suggest three sources of bacterial DNA in cancers: bacterial DNA around human tissues, free bacterial DNA in bacteremia or sepsis, and endogenous bacterial DNA in the human genome. Clarifying the theory that bacterial DNA integrates into the human genome can provide a new perspective for cancer prevention and treatment.
According to the statistics of the World Health Organization, it is estimated that there will be 19.3 million new cancer cases and nearly 10 million cancer deaths in 2020 around the world (Sung et al., 2021). Assuming that the estimated global incidence rate in 2020 remains unchanged, there will be 28.4 million new cancer cases in 2040, an increase of 47% over the corresponding 19.3 million cases in 2020 (Sung et al., 2021). The consequences are not optimistic. The occurrence of cancer is caused by the interaction between genetic factors and carcinogenic factors in the external environment. Among the carcinogenic factors, there are many physical and chemical factors, such as ultraviolet rays (Rivas et al., 2020) and nitrosamines (Liu et al., 2021). Another factor is the biological factor that causes cancer, that is, infection caused by microorganisms, viruses and bacteria (De Martel et al., 2017; Watanabe et al., 2021). Using metagenome sequencing, 16S rRNA detection and other technologies, the microbial community composition of various cancers, such as colon cancer, pancreatic cancer, breast cancer, and lung cancer, was studied and identified, and the existence of bacteria, viruses and other microorganisms was found (Nejman et al., 2020). At present, the role of specific microorganisms in the pathogenesis of cancer has been widely studied. Helicobacter pylori (H. pylori) (Ford et al., 2022), human papillomavirus (HPV) (Rebolj et al., 2022), and hepatitis B viruses (HBV) (Kamiza et al., 2022) are considered the main infection sources of 1.9 million new cancer cases worldwide every year. These cancer-causing microorganisms can lead to carcinogenesis by triggering inflammation (Chung et al., 2018), releasing metabolites (Yang et al., 2022), inducing DNA damage (Wang et al., 2016) and changing the cancer immune microenvironment (Zegarra-Ruiz et al., 2021).
Cancer is a disease driven by the accumulation of genome changes. The essence of cancer is always genetic changes. The methods of gene change include gene mutation (Greathouse et al., 2018), gene integration (Sung et al., 2012), gene repair damage (Imai et al., 2021), etc. Nonlong terminal repeats, such as long interspersed elements (LINEs) (Lander et al., 2001), and mobile elements, such as Alu elements (Steely et al., 2021), can jump actively in the human genome and induce cancers by causing gene mutations. For example, LINE-1 is inserted into the adenomatous polyposis coli (APC) gene of colon cancer (Scott et al., 2016), and Alu is inserted into mixed-lineage leukemia (MLL) genes (Stumpel et al., 2013). The Whitehead Institute of Biomedicine found that even a single base insertion in some genes may change the original operon structure and then have a certain impact on the functions of related genes. For example, the formation of the LIM domain only 2 (LMO2) oncogene expression enhancer caused by insertion mutation transforms somatic cells into primary leukemia cancer cells (Abraham et al., 2017). The mismatch repair gene mutL homolog 1 (MLH1) and mismatch repair gene mutL homolog 2 (MSH2) genes are related to hereditary nonpolyposis colorectal cancer syndrome (HNPCC) (Bouvet et al., 2019). MLH1 can induce a leukemia phenotype by changing the protein structure and inactivating inherent MLL1 function (Whitman et al., 2005).
Viruses can integrate into the human genome and cause cancer-related mutations. The integration of HPV is the key event leading to HPV-related cancers. Up to 80-100% of cervical cancers have HPV16 or HPV18 integration (Corden et al., 1999; Melsheimer et al., 2004). There are more than 1500 integration sites reported in the literature, such as 3q28, 8q24.21, 13q22.1, 2q22.3, 3p14.2, 8q24.22, 14q24.1, 17p11.1, 17q23.1, and 17q23.2 (Bodelon et al., 2016). When HPV integration leads to the deletion of E1 and E2 proteins, cell proliferation increases aberrantly (Pokrývková et al., 2019; Hirose et al., 2020). The integration of HBV DNA into the host hepatocyte genome is key to hepatocellular carcinoma (HCC) (Sung et al., 2012). Telomerase reverse transcriptase (TERT) (Sze et al., 2021), MLL4 (Saigo et al., 2008) and cyclin E1 (CCNE1) (Bayard et al., 2018) are common insertion genes for HBV integration. A region from nt1100 to nt1500 containing multiple HCC risk mutation sites (OR > 1) was identified as a potential HCC-related mutational hot zone (Zheng et al., 2021). The chromosome change rate and p53 mutation inactivation rate of HBV-related cancers are high (Song et al., 2021), leading to the activation of the mTOR signaling pathway (Luo et al., 2021) or WNT/β-catenin pathway (Dang et al., 2019), thus increasing the proliferation of cancer cells.
The number of bacteria in a healthy adult is only one-tenth the number of viruses. However, bacteria should not be underestimated. In the human body, there are more bacterial cells than human cells (Sender et al., 2016). Bacteria have been neglected in cancer research because of their extremely low biomass (Salter et al., 2014; Eisenhofer et al., 2019). Recently, next-generation sequencing methods for microbiome research have become more sensitive than ever to study microbial communities, their genomes and their functions. Bacterial DNA from 1010 tumor samples and 516 normal samples from 7 cancer types (Nejman et al., 2020) was quantitatively analyzed by 16S rDNA sequencing. Some bacteria were found to accumulate in the cytoplasm, and they were called intracellular bacteria. These bacteria might be cancer specific. Different cancer tissues harbor different bacteria (Figure 1). Therefore, this article will review the latest phenomena related to bacteria and cancer and the specific integration mechanism of bacterial DNA. The integration of bacterial DNA into the human genome may act as a cis-element to affect the function of host genes, activate proto-oncogenes, silence tumor suppressor genes and regulate cancer-related pathways. The integration of bacterial DNA is an important mechanism of carcinogenesis, which can provide theoretical support for the search for new cancer diagnostic and therapeutic targets.
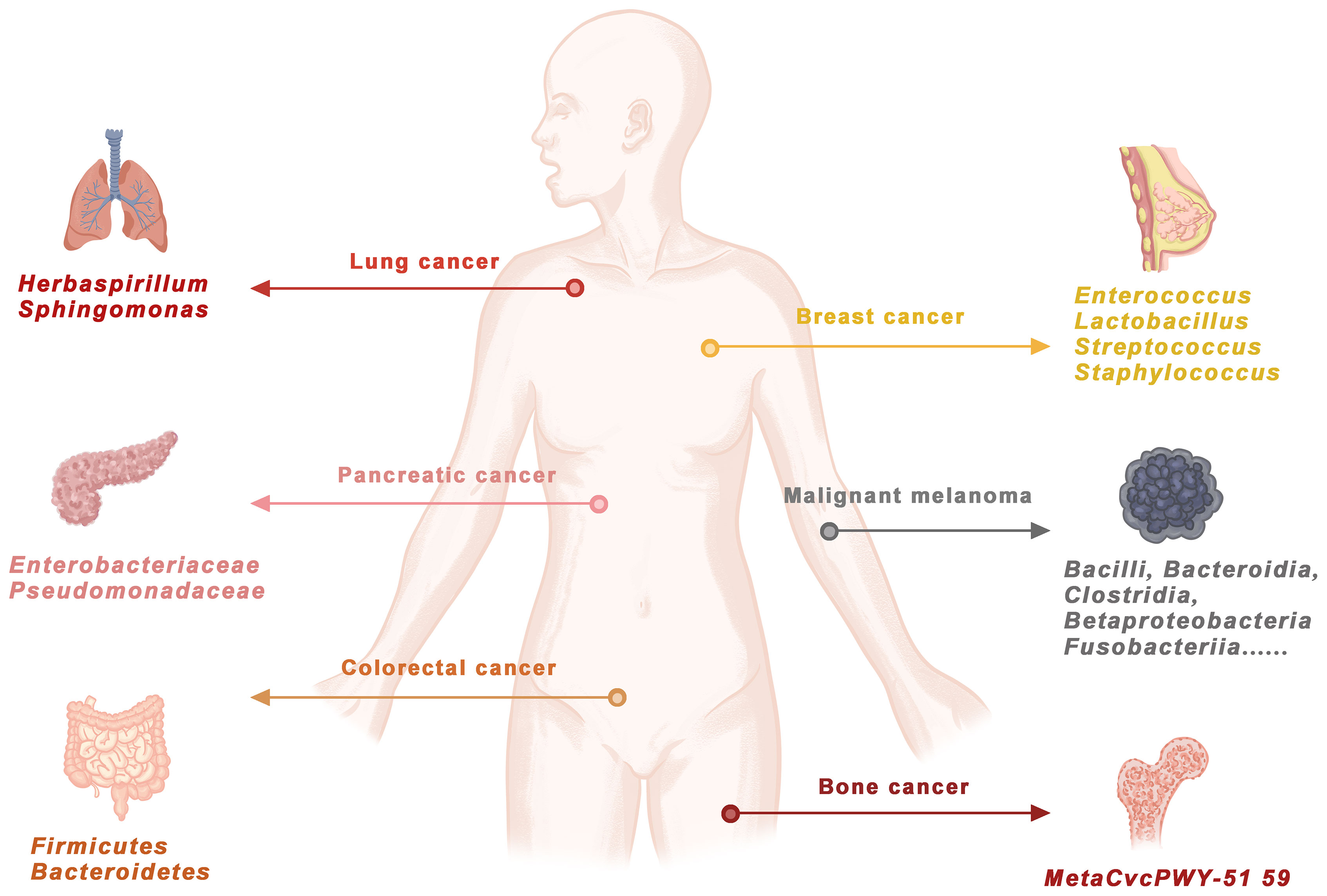
Figure 1 Bacterial DNA exists in human cancers.Through genome sequencing, PCR amplification, 16S RNA detection and KEGG/TCGA database comparison, bacterial DNA was confirmed in lung cancer, pancreatic cancer, breast cancer, malignant melanoma, colorectal cancer and bone cancer. DOI: 10.1126/science.aay9189.
The composition of bacteria surrounding cancer tissues is different from that in normal tissues. Recently, many studies have shown that human lungs contain various microbiota closely related to the development of lung cancer. The ecological diversity in noncancerous (immediate autopsy and hospital biopsy) tissues, noncancerous adjacent tissues and cancer tissue samples was detected by 16S rRNA gene sequencing. At the phylum level, Proteobacteria (Kruskal−Wallis P = 0.0002) was increased, and Firmicutes (Kruskal−Wallis P = 0.04) was decreased in hospital lung biopsies, as well as in cancer tissues, from the National Cancer Institute-Maryland (NCI-MD) study (Huang et al., 2011). In The Cancer Genome Atlas (TCGA) study, a similar increase in Proteobacteria (Mann−Whitney P = 0.02) between noncancer lung tissues and lung cancer was also observed. This finding indicated that this was a recurring phenomenon in lung cancer (Huang et al., 2011). In the NCI-MD study, 32 significantly different genera in lung squamous cell carcinoma (SCC) (n = 47) and lung adenocarcinoma (AD) (n = 67) (Student’s t test; MW P < 0.05) were found. Nine of them, including Acidovorax, Brevundimonas, Comamonas, Tempiddimonas, Rhodoferax, Klebsiella, Leptospira, Polaromonas and anaerobes, were still significant after multiple testing correction (FDR) (Greathouse et al., 2018). These same observations were validated in the TCGA dataset (AD = 485, SCC = 489) (Mann−Whitney FDR corrected P value < 0.05). Adjusted logistic regression analysis was performed in each group, and 6/9 genera were still significantly associated with increased odds of SCC compared with AD. Research on whether these cancer-related genera would change after quitting was performed, and the results showed that Acidobacteria, Klebsiella, Tepidimonas, Rhodoferax and anaerobes remained significant (Greathouse et al., 2018).
There are thousands of microorganisms in the intestinal tract. The microbial composition of colorectal cancer (CRC) patients is significantly different from that of healthy individuals. Bacteria invading the mucous layer of the colon axis in every FAP patient were detected by 16S rRNA gene analysis and fluorescence in situ hybridization (FISH) probe labeling polyps and macroscopic normal tissues. The main biofilm members were identified as Escherichia coli (E. coli) and Bacteroides fragilis (B. fragilis). The intestinal epithelial cells of all CRC patients with bacterial biofilms in their intestines were invaded by biofilm community members. This finding was similar to those of sporadic colorectal cancer patients (Dejea et al., 2018).. Compared with healthy subjects (22% pks+E. coli and 30% B. fragilis), pks+E. coli (68%) and B. fragilis (60%) in the mucosa were increased significantly in the mucosa of familial adenomatous polyposis (FAP) patients. Pks+E. coli and B. fragilis were not associated with polyps or normal mucosa in FAP patients. The polymerase chain reaction (PCR) analysis showed that the mucosal biofilm of early FAP patients contained clbB and BFT corresponding to the carcinogenic subtypes of E. coli and B. fragilis, respectively. The proteins existed in the mucous layer, which was in direct contact with the FAP epithelium (Dejea et al., 2018). Quantitative polymerase chain reaction (qPCR) analysis showed that 9 cases (82%) of 11 quick-frozen primary cancers were positive for Fusobacterium. Fusobacterium could be isolated from 73% of cancers (n = 8/11). Fusobacterium could also be isolated from liver metastatic sites of patients with liver metastases from CRC. Fusobacterium nucleatum (F. nucleatum) was highly enriched in CRC tissues and migrated with the spread of CRC cells. Many studies have reported that F. nucleatum supports the growth and development of cancers (Yang et al., 2017; Guo et al., 2020; Hong et al., 2021). F. nucleatum increases the incidence of intestinal cancers by creating a more suitable microenvironment for cancer growth. Colon cancer cells treated with F. nucleatum had stronger division, proliferation and invasion abilities. Inhibiting the growth of F. nucleatum might inhibit the development of CRC. Based on gut microbiome sequencing, some researchers constructed CRC screening models. Twenty microbial markers for distinguishing CRC patients from healthy controls through metagenomic sequencing of fecal samples were identified (Yu et al., 2017). Similarly, an early detection model to distinguish colorectal adenoma from healthy individuals and an early detection model to distinguish adenoma from CRC were established using oral microbial markers (Zhang et al., 2020).
Pancreatic cystic lesions, including intraductal papilloma (IPMNs), are common. They are known as precursors of pancreatic cancer. The adhesion of bacteria to epithelial cells and their ability to live in epithelial cells are important factors to exert their virulence. Healthy pancreatic cells (hTERT-HPNE), early differentiated pancreatic cancer cells (Capan-2) and late differentiated pancreatic cancer cells (AsPC-1) were incubated with a single strain for two hours. This short-term coculture with pancreatic cells allowed most isolated bacteria to enter human pancreatic cells and survive (Geller et al., 2017). In all pancreatic cells, the top survivors were Enterobacter cloacae, Enterococcus faecalis, Enterococcus faecalis and Klebsiella pneumoniae. Halimi A showed that bacteria from IPMN capsule fluid could invade and survive in pancreatic cells in vitro. Cyst is a potential microbial pool that continuously provides microorganisms for healthy and cancerous pancreatic cells (Halimi et al., 2021). Bacterial DNA from 65 pancreatic cancer patients was sequenced by 16S rDNA and PCR. The results showed the presence of bacterial DNA in pancreatic cancer cells (Geller et al., 2017). Compared with the bacterial DNA in the KEGG library, the most common species in pancreatic cancer cells was γ-proteobacteria, accounting for 51.7% of the total reads. Most of them were members of Enterobacteriaceae and Pseudomonas. The presence of numerous proteobacteria in the duodenum with open pancreatic ducts suggested that retrograde migration of bacteria from the duodenum to the pancreas might be the source of bacteria associated with pancreatic duct adenocarcinoma (PDAC) (Geller et al., 2017).
Forty-one different bacteria from melanoma samples were identified by the Weizmann Institute of Science in Israel through 16S rRNA gene sequencing (Kalaora et al., 2021). Bacteria including Bacilli, Bacillus, Clostridia, Betaprotebacter, Fusobacteria, Gammaproteobacteria, Actinobacteria, and Alphabetobacteria showed high abundance in melanoma cells. The low passage cell line from the same melanoma cell was cocultured with representative intracellular bacteria, and the entry of bacteria into cells was detected by FISH. For example, 51AL melanoma cells and 55A3 melanoma cells were cocultured with Staphylococcus carinii and Actinomycetes dentatum. The presence of bacteria in melanoma cells was confirmed by immunofluorescence staining of 51AL and 55A3 cells with lipoteichoic acid. F. nucleatum and Actinomycetes were labeled by copper-free chemicals, and the labeled bacteria were cocultured with 51AL and 55A3 cells. Correlative light and electron microscopy (CLEM) analysis of cells cocultured with F. nucleatum and Actinomycetes confirmed that these bacteria entered melanoma cells (Kalaora et al., 2021). Subsequently, HLA peptidyomics was used to identify peptide libraries derived from intracellular bacteria that had been present on HLA-I and HLA-II molecules in melanoma cancers. Analysis of 17 melanoma metastases (from 9 patients) revealed 248 unique HLA-I and 35 unique HLA-II peptides from 248 bacteria. Recurrent bacterial peptides in tumors of different patients and in different tumors of the same patient were both identified (Kalaora et al., 2021).
Fresh frozen breast specimens from 221 breast cancer patients and 87 nonbreast cancer patients were analyzed. At the genus level, Pseudomonas accounted for a higher proportion of the breast microbiome in cancer tissues than in other tissues, while Proteus was the second most abundant genus in cancer tissues and was largely absent in nontumor tissues. Porphyromonas and Azomonas were also more abundant in cancer tissues than in other tissues. In particular, Porphyromonas, Lacibacter, Ezakiella, and Fusobacterium were more abundant in high-stage cancers than in low-stage cancers (Tzeng et al., 2021). Cancer tissues, adjacent tissues and lymph node tissues from paired breast cancer patients were collected, and qPCR combined with 16S sequencing was used to analyze the composition of their respective microbiota. Gram-positive bacteria were the main types, with Enterococcus, Lactobacillus, Streptococcus and Staphylococcus accounting for approximately 80% (Fu et al., 2022). Bacteria existing in the cytoplasm were observed by high-resolution electron microscopy. After clearing these bacteria with an A cell-permeable antibiotic (doxycycline), we found that the cancer tissue weight was not affected, but the lung metastasis decreased significantly. 16S RNA sequencing analysis was performed on in situ tumors, visible lung metastases, lung tissues with tiny metastases and normal lung and breast tissues. The results suggested that bacteria with early lung metastases might still bear the microbiota characteristics of cancer tissues in situ.
Circulating tumor cells carrying intracellular bacteria to distal organs were found by isolating and staining circulating cancer cells. Streptococcus, Lactobacillus, Staphylococcus and other bacteria can invade cancer cells and reshape the cytoskeleton. When metastatic cancer cells invade the circulatory system, they need to undergo fluid shear stress, which often triggers apoptosis. By placing tumor cells containing bacteria and those without bacteria into the circulation system with a peristaltic pump, which can simulate the shear stress of fluid in blood vessels, cells containing bacteria had a higher survival rate than cells without bacteria. When the living tumor cells containing bacteria and not containing bacteria were spread on the plate, the living cells with bacteria had better adhesion ability and a larger cytoskeleton. This phenotype indicated that bacteria might play a role in the actin cytoskeleton. RhoA, Rac and Cdc42 are involved in the regulation of the actin cytoskeleton in cells. Fluorescence resonance energy transfer (FRET) sensor analysis of activated RhoA and Western blot analysis of RhoA-GTP showed that the invasion of bacteria indeed suppressed RhoA and ROCK activation. The inhibition of ROCK kinase (downstream of RhoA) could also eliminate the difference in viability caused by bacterial invasion. Bacteria can help transform breast cancer cells, prevent them from being damaged in the process of metastasis, and help them transfer to distant organs in the body (Fu et al., 2022).
Infection with H. pylori and a family history of gastric cancer are the main risk factors for gastric cancer (Lee et al., 2016). Approximately three thousand first-degree relatives of patients with gastric cancer were screened, and 1838 participants with H. pylori infection were randomly assigned to receive eradication therapy (lansoprazole [30 mg], amoxicillin [1000 mg], clarithromycin [500 mg], twice daily for 7 days) or placebo. During a median follow-up of 9.2 years, gastric cancer developed in 10 participants (1.2%) in the treatment group and in 23 participants (2.7%) in the placebo group (hazard ratio, 0.45; 95% confidence interval [CI], 0.21 to 0.94; log-rank test P = 0.03). Gastric cancer developed in 0.8% (5/608) of participants with eradication of H. pylori infection and developed in 2.9% (28/979) of participants with persistent infection (hazard ratio, 0.27; 95% CI, 0.10 to 0.70). It was more common in the treatment group than in the placebo group (53.0% vs. 19.1%; P < 0.001). In patients with H. pylori infection whose first-degree relatives had a family history of gastric cancer, H. pylori eradication therapy could significantly reduce the risk of gastric cancer (Choi et al., 2020).
In 2001, an important paper based on the sequencing results of the human genome proposed that some human genes may come from horizontal gene transfer of bacteria by comparing the genomes of bacteria and some animals. However, those ideas were quickly countered. Blood, liver and three types of adipose tissue samples from 40 subjects were studied recently. Bacterial genetic materials in each tissue sample were found, and the types of bacteria and the amount of bacterial DNA were different. In addition, DNA in the tissue samples was detected not only from gut bacteria but also from those commonly found in soil or water. This finding implied that viscera may be regularly exposed to exogenous genetic material (Anhê et al., 2020).
In 2020, the microbial composition of seven cancers, including breast cancer, lung cancer, ovarian cancer, pancreatic cancer, melanoma, bone cancer and brain cancer, was revealed by comparing thousands of cancer tissues with nearby normal tissues. Microbes were found in every cancer tissue from the head to the bone, and the microbe composition in every cancer tissue was different. Among them, breast cancer had the highest diversity, with 16.4 bacteria in each breast tumor sample, while other tumors had only 9 species on average (Nejman et al., 2020). Cancer tissues were found to contain bacteria in the past, but the content was very low. Some researchers think this might be the result of sample contamination. Various methods have been adopted to avoid contamination. For example, to address possible contamination of samples between storage and the laboratory, they took more than 100 sections from each paraffin block edge of paraffin embedded samples (no sample tissue) as a control group. In total, 1010 tumor samples and 516 normal tissue samples were analyzed. Bacterial DNA levels in samples from each type of cancer tissue were significantly higher than those in normal tissues and paraffin sections. Among the seven tumors, melanoma had the lowest bacterial DNA positive rate (14.3%), while breast cancer, pancreatic cancer and bone cancer had higher positive rates, all exceeding 60%. The other 400 cancer specimens were detected by immunohistochemical methods using bacterial lipopolysaccharide and lipoteichoic acid antibodies (specific components of the cell walls of Gram-negative bacteria and Gram-positive bacteria, respectively), and all types of cancers had positive results. The bacterial RNA in 400 cancer specimens was detected by FISH, and the results were also positive. Bacteria mainly exist in cancer cells and immune cells, especially in their cytoplasm. Using an optical microscope, researchers observed that most of them were located in the cytoplasm near the nuclear membrane of the nucleus, and many of them had no cell walls (Nejman et al., 2020).
Paired RNA sequencing data from patients with chronic lymphocytic leukemia and healthy donors were analyzed to look for incorporation of bacterial DNA into the human somatic cell genome (Riley et al., 2013). The Burrows−Wheeler Comparator (BWA) was applied first to the human genome and then to the bacterial genome reference. Mapping sequencing reads of the human genome, microbiome, and bacteria were distinguished, and all reads with alignment coverage less than 90% and low-complexity reads containing long poly-A/-T/-g/-C sequences were excluded. More bacterial DNA was found in chronic lymphocytic leukemia specimens, and the bacterial genera integrated in the whole human genome were Pseudomonas sp., Mesorhizobium sp., and Acinetobacter sp. (unspecified sites) (Sun and Ma, 2019). Most bacterial integration events (36%) occurred in the intronic region, 22% in the exon region, and 17% in the 3’ UTR region. Genes such as SNORD141A/B, MIR4507, MALAT1, CD74, HLA-B, LGT, and HLA-C are integration hotspots (Jima et al., 2010; Riley et al., 2013; Sun and Ma, 2019; Akimova et al., 2022). Data published by the Thousand Genomes Project were analyzed, and more than 7,000 cases of horizontal gene transfer from bacteria were found. After analyzing TCGA sequences, the researchers found 691,000 horizontal gene transfers, 99.9 percent of which came from cancer samples. One-third of the bacterial sequences in the entire human genome came from Acinetobacter and were inserted into the mitochondrial genome (Hancks and Kazazian, 2012; Riley et al., 2013). Mitochondrial genomes or nuclear mitochondria of acute myeloid leukemia samples are the most common Acinetobacter-like DNA integrations (1000 Genomes Project Consortium et al., 2010). The integration of Pseudomonas DNA in gastric adenocarcinoma samples was mainly concentrated in five genes. Among the 5 genes, 4 were upregulated proto-oncogenes, including TMSB10, CEACAM5, CEACAM6 and CD74 (Han et al., 2008; 1000 Genomes Project Consortium et al., 2010; Gold et al., 2010; Ren et al., 2020).
If bacterial integration is an important mechanism of carcinogenesis, we hope that further studies in this area will reveal similar integration from specific mobile elements and viruses, including the insertion and inactivation of coding genes and the transfer of oncogenic proteins and peptides. Is bacterial DNA integration similar in all cancers, or does each cancer have its own specific bacterial DNA integration pathway? What factors contribute to bacterial integration sites in each cancer? Which bacterial DNA integration sites are consistent? Many mechanistic issues remain to be further studied.
Natural transformation is a process by which bacteria spontaneously absorb, integrate and excrete foreign DNA (Johnston et al., 2014). This process is the source of bacterial genetic diversity and evolutionary flexibility. Many bacterial genera release their own DNA into the extracellular environment by secretion or autolysis. This behavior provides conditions for DNA uptake by other cells. DNA release from bacterial cells can be directed to selection, mainly cell division and replication (vertical gene transfer) and direct investment in the replication of genetic information in other cells (horizontal gene transfer) (Mao et al., 2014). DNA release can be mediated by secretion, for example, through the type IV secretion system and membrane capsules or by autolysis of a subset of the population (Li et al., 2005; Ibáñez de Aldecoa et al., 2017). The cagI-pathogenic island (cagPAI) of H. pylori contains 32 genes encoding the type IV secretion system (T4SS) and CagA protein (Wroblewski and Peek, 2016; Jia et al., 2018). Intraspecific variation of DNA release was found in Neisseria gonorrhoeae (mediated by both lysis and secretion) (Zaremba et al., 2014). In addition, wild-type Bacillus subtilis strains release free DNA in a lyse-independent manner (Cañas et al., 2011). The analysis of DNA release among different strains was relatively simple, and these data indicated the involvement of cells in DNA release and DNA uptake.
It is common for bacteria to transfer horizontally because the DNA of bacteria is in the cytoplasm. The probability of a piece of DNA being integrated into the bacterial genome is high if it can pass through the cell wall and cell membrane of bacteria. Bacterial cells were added to the mammalian cell line medium. After several days of cultivation, one out of every 10,000 mammalian cells was positive for bacterial plasmids. After adding an enzyme that can digest DNA outside the cell membrane and remove the cell membrane, bacterial genes could still be detected in mammalian cell lines. This confirmed that bacterial DNA could enter human cells (Waters, 2001). There is a second barrier in the genome of eukaryotic cells: the nucleus. Usually, DNA is tightly condensed in chromosomes, which restricts the splicing of foreign DNA fragments into the genome. DNA generally cannot exit the nucleus because it is a D+ protein, which is too large to pass through the nuclear pore (ignore nuclear pore identification first). Bacterial genetic material, however, is relatively small, and it is relatively easy to enter and leave the nucleus. The transmembrane transport protein complex on the nucleus is called the nuclear pore complex, which is a bifunctional and bidirectional hydrophilic nucleocytoplasmic exchange channel (Kim et al., 2018). Bacterial DNA can interact with nuclear compounds and enter the nucleus of host cells through nuclear pores. In the process of bacterial pathogenicity, the bacterial type IV secretion system (T4SS) mediates the DNA transfer of cell-to-cell binding and the cross-border protein transfer to eukaryotic host cells (Schröder et al., 2011). Bartonella henselae can transfer its cryptic plasmid into the human endothelial cell line EA.hy926 by its T4SS VirB/VirD4. DNA transfer to EA.hy926 cells were demonstrated by the insertion of reporter gene derivatives of Bartonella-specific mobile plasmids generated by the eukaryotic EGFP expression cassette. Egfp gene expression in EA.hy926 cells required cell division, suggesting that nuclear envelope rupture might facilitate passive entry of transferred ssDNA into the nucleus, a prerequisite for EGFP gene synthesis of complementary strands and transcription. During human infection with Bartonella, T4SS-dependent DNA might be naturally transferred into host cells (Schröder et al., 2011).
Many bacteria can cause single-strand breaks (SSBs) or double-strand breaks (DSBs) of human DNA (Pleguezuelos-Manzano et al., 2020; Imai et al., 2021). E. coli carries pathogenic PKS islands and encodes an enzyme that binds to Colibactin3. Colibactin3 is thought to alkylate DNA on adenine and induce DSBs in cultured cells (Imai et al., 2021). Genetically toxic pks+ E. coli were repeatedly injected into healthy human intestinal organs. Immunostaining confirmed that the pks+ E. coli-like organ model could cause double-strand cross-linking and DNA double-strand damage. Under the intervention of pks+ E. coli, the single base substitution ratio of cells increased significantly. In addition, colibactin could bind two adenines simultaneously and cross-link them. By analyzing Dutch samples from 3668 different cancer types, DNA mutation patterns caused by colibactin were found in various cancers and was more common in CRC than in other types of cancer (Pleguezuelos-Manzano et al., 2020). After, 2208 CRC samples from 100,000 genome projects in the UK were analyzed, and 4% to 5% of the patients’ mutation characteristics were consistent with the DNA mutation pattern caused by colistin. This study established a direct connection between microbes in the human body and genetic variation that drives cancer development (Pleguezuelos-Manzano et al., 2020).
E. coli-induced DSBs can be identified by the suspension fragmentation labeling in situ sequencing (sBLISS) method (a DSB capture method based on sequencing), and the action of colistin directly related to AT-rich sequences can be found by DREME (motif identification method) (Dziubańska-Kusibab et al., 2020). By predicting DNA shape parameters at the central position of all 4096 possible hexanucleotides and correlating them with the sequence enrichment ratio of hexanucleotides at DSB positions in PKS+ and PKS- E. coli-infected cells, the colicin-specific six nucleotide sequences AAAATTT \ AATTTT and AAAAAATTT were detected. They had the narrowest microgroove width and the largest negative electrostatic potential. The extreme characteristics of colicin destruction motif (CDM)-related sequences were confirmed. Theoretical single base substitution (SBS) markers based on the frequency of trinucleotide sequences were deduced. The main characteristics of SBSA markers were T> C mutations at ATA, ATT and TTT trinucleotides and T> G mutations at TTT. Moreover, the prediction of the total mutational load in CRC cohort WGS by this marker is significantly correlated with the proportion of mutated CDM, indicating that the trinucleotide marker of SBSA can represent colistin-specific mutations (Dziubańska-Kusibab et al., 2020).
X (PH2A.x) is a known marker of DNA double-strand breaks and activation of the DNA damage response. Pancreatic cell damage was detected by measuring phosphorylated γH2A. In nonmalignant pancreatic cells, G. adiacens H1, E. faecalis C1 and Klebsiella oxytoca H1 strains caused strong PH2A.x (Halimi et al., 2021). In pancreatic malignant cell lines such as Capan-2 and AsPC-1, PH2A.x increased. Generally, the strongest inducer of PH2A.x is E. cloacae isolated from an intraductal papillary mucinous neoplasm (IPMN)-cancer case. Application of penicillin−streptomycin in the early stage of coculture of bacteria and cells could completely prevent DNA damage, indicating that IPMN-encapsulated bacteria could cause significant DNA damage to healthy pancreatic cells in the early and late stages of cancer (Halimi et al., 2021).
Epigenetics is a chemical marker system that annotates genetic information by affecting protein binding and chromatin structure and causing changes in gene expression. DNA methylation, histone modifications, noncoding RNAs and RNA splicing factors can change the chromatin and transcriptional program of host cells. These heritable changes, which alter gene expression but do not affect the DNA sequence, are called heterologous variations. Aberrant modifications of gene regulatory regions lead to abnormal cell development and accumulation of damaged DNA (Roadmap Epigenomics Consortium et al., 2015; Skvortsova et al., 2018). The intestinal tract has the most abundant microorganisms. Intestinal cancer microorganisms are related to the carcinogenesis of intestinal mucosal cells. By comparing DNA methylation in colonic crypt intestinal epithelial cells of germ-free mice and conventional mice through whole genome bisulfite sequencing (WGBS), global methylation levels were found to be significantly reduced in conventional mouse samples. A set of “sentinel genes” activated by demethylation in conventional mice that were responsible for normal regeneration of intestinal mucosa in the healthy gut were identified. This microbiome-dependent demethylation corresponded to the increase in the expression and activity of the DNA demethylases Tet3 and Dnmt1 in the intestinal epithelial cells of conventional mouse colon crypts, in which the deletion of specific Tet3 led to an increase in overall DNA methylation, including sensitive areas of the microbiome. In a mouse model of chemically induced sodium dextran sulfate (DSS) colitis, the phenomenon of DNA methylation reduction in intestinal mucosal cells caused by intestinal microorganisms was found. The decrease in DNA methylation was related to the activation of many genes related to intestinal inflammation and cancer, suggesting that intestinal bacteria might reprogram the DNA activity of intestinal mucosal cells (Ansari et al., 2020). Nitric oxide molecules that nitrosylated the host ALG1 protein were found by tracking the nitric oxide secreted by the model organism nematode in vivo. Excessive nitric oxide affected the normal development of nematodes, resulting in deformity. Notably, ALG1 is a highly conserved protein evolutionarily from nematodes to humans. In mammals, when commensal microorganisms produce too much nitric oxide, this protein may also be affected by the same S-nitrosylation modification, possibly causing developmental problems as well (Seth et al., 2019).
Posttranslational modification of histones is another central mechanism by which microorganisms regulate host chromatin. These types of modifications are typically not directly targeted to DNA but are covalently added to lysine residues in histone tails to affect chromatin conformation and gene expression. HDAC3, a histone deacetylase, is highly expressed in the intestinal epithelium and is sensitive to microbial signals. Cell-specific deletion of HDAC3 in intestinal mucosa increased H3K9 acetylation and gene expression, altered Paneth cell homeostasis and intestinal barrier function, and worsened the response to DSS-induced intestinal inflammation. Importantly, abnormalities in HDAC3 deletion were evident only in conventional mice but not in germ-free mice, suggesting tight communication between gut bacteria and the host genome (Alenghat et al., 2013). There are many microRNAs from small intestinal epithelial cells in mouse and human feces. These fecal microRNAs can enter bacteria, such as F. nucleatum and E. coli, and specifically regulate the transcription of bacterial genes to affect bacterial growth. If Dicer, the enzyme responsible for microRNA processing, is specifically knocked down in these two types of cells, fecal microRNA is reduced (Liu et al., 2016). Conversely, microRNAs from bacteria in feces might also enter the genome of human cells and regulate gene stability, which is worth exploring. F. nucleatum promoted the proliferation of CRC cells and the development of cancer by activating the Toll-like receptor signaling pathway in intestinal mucosal cells to nuclear factor-κB and upregulating the expression of microrNA-21. Bacteria can regulate the expression of the human host genome, thereby affecting cell functions (Yang et al., 2017).
Bacteria can destabilize the human genome, leading to events such as base substitutions and copy number variations. Staphylococcus aureus evolved into methicillin-resistant Staphylococcus aureus (MRSA) through gene mutation to resist the elimination of human immune cells and the killing effect of antibiotics. The prevalence of antibiotic-resistant staphylococcal infections is increasing, and human cells are fighting back in kind. By sequencing the whole exons of 32 patients who had persistent MRSA infections and 32 patients who were able to clear the infection from their blood, approximately 62% of patients who cleared MRSA infection were found to have had obvious genetic variation, while only 9% of patients had persistent infection. This mutation was located in the DNMT3A region of chromosome 2P (Mba Medie et al., 2019). The P53 gene was the first and most famous tumor suppressor gene discovered by human beings. The main function of the P53 protein encoded by the P53 gene is to repair DNA and prevent cell carcinogenesis (Derech-Haim et al., 2020). P53 was introduced into a mouse cancer model, and two opposite effects were observed: inhibiting the occurrence of proximal CRC but promoting the occurrence of distal CRC. Microbial abundance gradually increases from top to bottom along the intestinal tract; that is, there are fewer microorganisms in the proximal intestinal tract and more microorganisms in the distal intestinal tract. In organ-like tissues of the proximal intestine and sterile ileum, P53 could better inhibit WNT gene expression and cancer occurrence (Kadosh et al., 2020). When the intestinal microbiota in the mouse model was eliminated with antibiotics, the abnormal development of the colon and ileum was eliminated, and the activation of the WNT gene was also reduced. These results suggested that the gut microbiota could counter the p53-mediated inhibition of WNT, thereby promoting the occurrence of distal bowel cancer (Kadosh et al., 2020). Bacteria affect the stability of the human genome and affect the expression of host genes.
The DNA released by bacteria travels around human cells and then into the nucleus. Bacteria-induced changes, such as DNA strand breaks and genomic instability, provide an opportunity for bacterial DNA to integrate into the host genome. The latest research on bacterial DNA integration into the human genome is described in Part 3. Approximately one-third of healthy genomes contained bacterial DNA sequences, and even more were found in cancer cells. Bacterial genes that transfer into the human genome may initiate the transformation of healthy cells into cancer cells by stimulating protooncogenes or inhibiting oncogenes (Riley et al., 2013). Proto-oncogenes are present in normal cells but are not activated. Under the action of various environmental or genetic factors, the structure of proto-oncogenes is changed and activated into oncogenes, leading to excessive or continuous growth signals (Bolen and Veillette, 1989). Tumor suppressor genes can inhibit cell growth. They exist in normal cells and play a negative role in regulating cell proliferation. Their inactivation or deletion can lead to the survival of mutant cells and promote cancer progression (Marshall, 1991). For example, the CDKN2A, CDH1 and RUNX3 genes found in precancerous lesions of H. pylori-infected patients and gastric cancer patients could inactivate tumor suppressor genes. Gastric cancer-related mutations were identified in TP53, CDH1, ARIDIA and RHOA genes (Pandey et al., 2018; Capparelli and Iannelli, 2022). The genomes of cancer cells may be more receptive to bacterial genomes than normal cells.
The mechanism of bacterial DNA integration into the human genome and induction of cancers are shown in Figure 2.
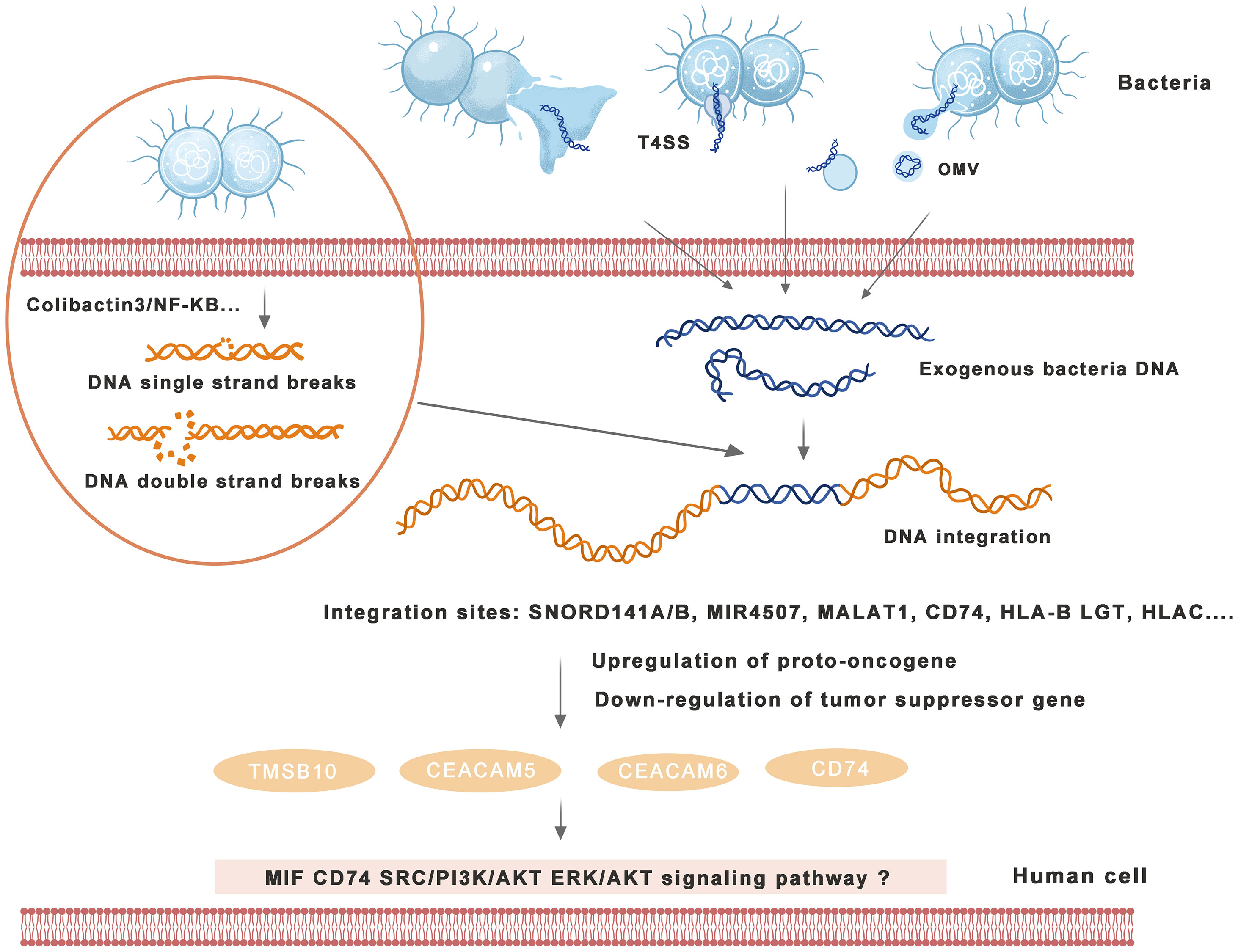
Figure 2 Mechanism of bacterial DNA integration into the human genome and induction of cancers. Bacteria excrete DNA through direct exclusion, outer membrane vesicle (OMV) excretion, type IV secretion system (T4SS), etc., and human cells receive exogenous DNA with transformation ability. Bacteria can induce human DNA single-strand breaks (SSBs) or double-strand breaks (DSBs) through a variety of signaling pathways, providing conditions for DNA integration into the human genome. Human cells integrate exogenous bacterial DNA into the exon or intron region and activate carcinogenic signaling pathways and induce tumors by enhancing proto-oncogene expression or weakening tumor suppressor gene expression in the human genome.
There are approximately 3.8×1013 bacteria in the 70 kg standard weight reference human body, most of which exist in the human digestive tract (Sender et al., 2016). In addition, other organs communicating with the outside world, such as the lung, colorectal and bladder, may have a large number of bacteria around them (Jin et al., 2019; Mansour et al., 2020; Liu et al., 2021; Bell et al., 2022). At present, many microorganisms, including Helicobacter pylori and Escherichia coli, have been found to affect the homeostasis of the genome in different ways and even directly damage DNA groups (Tjalsma et al., 2012; Sears and Garrett, 2014; Dejea et al., 2018). Genome instability provides an opportunity for the integration of exogenous bacterial DNA. Escherichia coli, Bacillus subtilis, etc., can establish natural competence through their internal regulatory mechanisms and can “directly” ingest foreign DNA in the natural environment (Torres et al., 2019; Sun et al., 2020). With the discovery of DNA molecules and competent cells with transformation activity, the role of natural transformation in horizontal gene transfer has become a focus of research. So-called natural transformation is a type of gene transfer method in which naked DNA molecules interact with natural competent cells without any medium. Natural transformation can occur between bacteria or between bacteria and other eukaryotes (Durieux et al., 2019; Simpson et al., 2019; Torres et al., 2019; Sun et al., 2020; Bonifácio et al., 2021). In recent years, the phenomenon that bacterial cells can actively secrete DNA that has transformation activity into the environment has been observed (Durieux et al., 2019; Simpson et al., 2019). If the bacterial cells around human tissue cells can actively secrete DNA with transforming activity and human cells can actively take up the surrounding bacterial DNA, then the bacteria around human tissue may be a major source of human genes. Human cells integrate ingested exogenous DNA into the genome through mechanisms such as DNA insertion or replacement, and the resulting changes in the expression of cancer-promoting or cancer-suppressor genes may be a mechanism of bacteria-induced cancers.
Bacterial infection into the blood circulation is called bacteremia (Fontana et al., 2021). Pathogenic bacteria invade the blood circulation, grow and reproduce in the blood, and produce toxins. Acute systemic infection is called septicemia (Mirouse et al., 2020). Cell-free circulating DNA (cfDNA) refers to the DNA fragments that come from apoptosis or necrosis of cells and are free from outside the cells and widely exist in human serum and plasma. The cfDNA in the blood contains genomes from all types of tissues and cells in the entire body and theoretically also contains genomes of all types of viruses and microorganisms (Goggin et al., 2020; Xiao et al., 2021).
Stephen R. Quake and his team of Stanford University analyzed the cfDNA sequencing data of 1351 samples, extracted nonhuman sequences from them, and found that some of them could match the known bacterial genomes after assembly, indicating that there was free bacterial DNA in blood (Kowarsky et al., 2017). Based on this finding, many researchers have begun to try to detect bacteria with free bacterial DNA. Professor Samuel Yang’s team detected bacterial DNA in the blood of 350 patients with a sepsis alarm, and the results showed that 93.7% of them were consistent with blood culture, verifying that there was free bacterial DNA in the blood (Blauwkamp et al., 2019).
Human cells need a blood supply to grow continuously. At this time, bacterial DNA can reach tissue cells through the blood supply. When tissue cells come into contact with bacteria or bacterial DNA, the genes in foreign cells are likely to enter one or more tissue cells. Although there are unique enzymes in the human body that can prevent foreign genes from entering, some studies have shown that some foreign genes are similar in appearance to human cellular DNA. When genes are actively inserted into the genome of tissue cells, tissue cells will mistake foreign genes for their own genes and start reading them and using them to synthesize protein. Different bacterial DNA may have different affinities to different tissues and cells, which is related to the tumor specificity of bacterial DNA. Professor Rob Knight’s team from the University of California San Diego used the random gradient enhanced ML model to analyze the acellular microbial DNA in plasma (Poore et al., 2020) and found that acellular microbial DNA has a high degree of discrimination for various cancer types, such as prostate cancer, lung cancer and melanoma.
Therefore, we hypothesize that the free bacterial DNA in the blood may be a source of bacterial genes in human tumors, and they likely participate in the occurrence of breast cancer and other tissue tumors that are different from the external environment. The specific situation deserves further study.
Based on the first two hypotheses, human cells collect genes released from bacteria and take them for their own use. If the foreign genes are well adapted, human cells will pass on foreign genes to offspring during proliferation, so once a cell carries bacterial genes, it can be passed on to offspring, which is called vertical gene transfer. At present, the most typical example is mitochondria in cells (Ku et al., 2015a; Ku et al., 2015b).
Endosymbiosis holds that approximately 1.7 billion years ago, an aerobic bacterium called gram-negative bacteria invaded the host cell, but the host cell did not engulf the bacteria. Instead, the host cell provided a steady stream of nutrients for the bacteria, and the bacteria efficiently decomposed the nutrients into energy and provided it to the host. The cooperation between the two greatly enhanced their viability and gradually occupied a dominant position in the biological competition, and the bacteria evolved into mitochondria today after hundreds of millions of years (Dziubańska-Kusibab et al., 2020; Halimi et al., 2021). The relevant evidence is as follows: ① mitochondria have genetic material independent of the nucleus; ② mitochondria are one of a few structures with double membranes in cells, and the inner membrane of mitochondria is similar to the bacterial membrane; mitochondrial DNA is circular, similar to that of some bacteria, and the translation process of mitochondrial protein is more similar to that of some bacteria; and ④ mitochondria proliferate in a similar way to bacteria. In 2020, mitochondria were shown to originate from Proteus alpha by phylogenetic analysis based on systematic species sampling (Fan et al., 2020). Combining the two viewpoints that bacterial DNA can exist after entering human cells and that cancers have familial inheritance, we present the hypothesis that cancer-causing bacterial DNA can be inherited after being integrated into the human genome, which will lead to hereditary tumors in people with high susceptibility.
Three sources of bacterial DNA in human cancers are shown in Figure 3.

Figure 3 Three Sources of bacterial DNA in human cancers. ① Organs that are direct contact with the outside world, such as the colorectal area and lung, are full of bacteria, and tissue cells are almost bathed in bacteria. The DNA excreted by the bacteria surrounding the tissue can enter receptive human cells, resulting in gene integration. ② Breast cancer, bone cancer and other organs that are not exposed to the outside world usually cannot come into direct contact with a large number of bacteria. However, when the body is infected with bacteremia, septicemia, etc., they can come into contact with bacteria and their DNA in the blood circulation. The greater the demand for blood supply, the greater the chance of contact. Human cells come into contact with and absorb bacterial DNA, resulting in gene integration. When bacterial DNA is integrated with the human genome, it will persist in human cells without incident. Genes are vertically inherited and will be passed on to future generations. This inheritance may be the cause of endogenous bacterial DNA in the human body.
The existence of bacterial DNA in tumor tissue and its surroundings has been confirmed by a series of molecular experiments and biochemical analysis methods. We hypothesized three sources of bacterial DNA fragments: bacterial DNA transferred horizontally by bacteria near human cells, bacterial DNA remaining in human blood after bacterial infection, and bacterial DNA passed down from progenitors. For bacterial DNA to be expressed in the human body, it may undergo the following processes: it is released from bacteria, crosses the cell membrane of human cells, crosses the nucleus, and integrates into the human genome. The damage caused by bacteria to human DNA, such as inducing DNA breaks, regulating gene expression by epigenetic modifications, and causing genome instability, can facilitate the integration of bacterial DNA into the human genome. Human cells fail to remove and repair bacterial DNA in time, resulting in the persistence of bacterial DNA in the body. Changes in the host genome by bacterial DNA may activate proto-oncogenes and suppress tumor suppressor genes in human cells, causing the host cells to become cancerous.
WY: design, writing, and approval of the manuscript. HS: design, writing, and approval of the manuscript. All authors contributed to the article and approved the submitted version.
This work is supported by key research and development project of Science and Technology Department of Zhejiang Province (No.2022C03026).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
IARC, Agency for Research on Cancer; H. pylori, Helicobacter pylori; HPV, Human papillomavirus; HBV, Hepatitis B viruses; LINE, Long interspersed element; APC, Adenomatous polyposis coli; HNPCC, Hereditary nonpolyposis colorectal cancer syndrome; MLV12, Moloney leukemia virus12; MLL, Mixed-lineage leukemia; BRCA, Breast cancer susceptibility gene; MLH, Mismatch repair gene mutL homolog; HCC, Hepatocellular carcinoma; LMO2, LIM domain only 2; NOX4, NADPH oxidase 4, TERT, Telomerase reverse transcriptase; CCNE1, Cyclin E1, NCI-MD, Cancer Institute-Maryland; SCC, Squamous cell carcinoma; AD, Adenocarcinoma; PCR, Polymerase chain reaction; PDAC, Pancreatic ductal adenocarcinoma; FISH, Fluorescence in situ hybridization; CLEM, Correlative light and electron microscopy; WGS, Whole genome sequence; TCGA, The cancer genome atlas; PRRs, Pattern recognition receptors; PAMPs, Pathogen-associated molecular patterns; LPS, Lipopolysaccharide; BWA, Burrows-Wheeler Comparator; cfDNA, Cell-free circulating DNA; T4SS, Type IV secretion system; OMV, Outer membrane vesicle; SSBs, Single strand breaks; DSBs, Double strand breaks; E.coli, Escherichia coli; B.fragilis, Bacteroides fragilis; F. nucleatum, Fusobacterium nucleatum; PDAC, Pancreatic duct adenocarcinoma; CLEM, Correlative light and electron microscopycag; PAI, Cagi-pathogenic island; sBLISS, Suspension Breaks Labeling In Situ and Sequencing; CDM, Colicin-destroying motif; IPMN, Intraductal papillary mucinous neoplasm; WGBS, Whole genome bisulfite sequencing; MRSA, Methicillin-resistant Staphylococcus aureus.
1000 Genomes Project Consortium, Abecasis, G. R., Altshuler, D., Auton, A., Brooks, L. D., Durbin, R. M., et al. (2010). A map of human genome variation from population-scale sequencing. Nature 467 (7319), 1061–1073. doi: 10.1038/nature09534
Abraham, B. J., Hnisz, D., Weintraub, A. S., Kwiatkowski, N., Li, C. H., Li, Z., et al. (2017). Small genomic insertions form enhancers that misregulate oncogenes. Nat. Commun. 8, 14385. doi: 10.1038/ncomms14385
Akimova, E., Gassner, F. J., Greil, R., Zaborsky, N., Geisberger, R. (2022). Detecting bacterial-human lateral gene transfer in chronic lymphocytic leukemia. Int. J. Mol. Sci. 23 (3), 1094. doi: 10.3390/ijms23031094
Alenghat, T., Osborne, L. C., Saenz, S. A., Kobuley, D., Ziegler, C. G., Mullican, S. E., et al. (2013). Histone deacetylase 3 coordinates commensal-bacteria-dependent intestinal homeostasis. Nature 504 (7478), 153–157. doi: 10.1038/nature12687
Anhê, F. F., Jensen, B. A. H., Varin, T. V., Servant, F., Van Blerk, S., Richard, D., et al. (2020). Type 2 diabetes influences bacterial tissue compartmentalisation in human obesity. Nat. Metab. 2 (3), 233–242. doi: 10.1038/s42255-020-0178-9
Ansari, I., Raddatz, G., Gutekunst, J., Ridnik, M., Cohen, D., Abu-Remaileh, M., et al. (2020). The microbiota programs DNA methylation to control intestinal homeostasis and inflammation. Nat. Microbiol. 5 (4), 610–619. doi: 10.1038/s41564-019-0659-3
Bayard, Q., Meunier, L., Peneau, C., Renault, V., Shinde, J., Nault, J. C., et al. (2018). Cyclin A2/E1 activation defines a hepatocellular carcinoma subclass with a rearrangement signature of replication stress. Nat. Commun. 9 (1), 5235. doi: 10.1038/s41467-018-07552-9
Bell, H. N., Rebernick, R. J., Goyert, J., Singhal, R., Kuljanin, M., Kerk, S. A., et al. (2022). Reuterin in the healthy gut microbiome suppresses colorectal cancer growth through altering redox balance. Cancer Cell. 40 (2), 185–200.e6. doi: 10.1016/j.ccell.2021.12.001
Blauwkamp, T. A., Thair, S., Rosen, M. J., Blair, L., Lindner, M. S., Vilfan, I. D., et al. (2019). Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat. Microbiol. 4 (4), 663–674. doi: 10.1038/s41564-018-0349-6
Bodelon, C., Untereiner, M. E., Machiela, M. J., Vinokurova, S., Wentzensen, N. (2016). Genomic characterization of viral integration sites in HPV-related cancers. Int. J. Cancer. 139 (9), 2001–2011. doi: 10.1002/ijc.30243
Bolen, J. B., Veillette, A. (1989). A function for the lck proto-oncogene. Trends Biochem. Sci. 14 (10), 404–407. doi: 10.1016/0968-0004(89)90288-0
Bonifácio, M., Mateus, C., Alves, A. R., Maldonado, E., Duarte, A. P., Domingues, F., et al. (2021). Natural transformation as a mechanism of horizontal gene transfer in aliarcobacter butzleri. Pathogens 10 (7), 909. doi: 10.3390/pathogens10070909
Bouvet, D., Bodo, S., Munier, A., Guillerm, E., Bertrand, R., Colas, C., et al. (2019). Methylation tolerance-based functional assay to assess variants of unknown significance in the MLH1 and MSH2 genes and identify patients with lynch syndrome. Gastroenterology 157 (2), 421–431. doi: 10.1053/j.gastro.2019.03.071
Cañas, C., Carrasco, B., García-Tirado, E., Rafferty, J. B., Alonso, J. C., Ayora, S. (2011). The stalk region of the RecU resolvase is essential for holliday junction recognition and distortion. J. Mol. Biol. 410 (1), 39–49. doi: 10.1016/j.jmb.2011.05.008
Capparelli, R., Iannelli, D. (2022). Epigenetics and helicobacter pylori. Int. J. Mol. Sci. 23 (3), 1759. doi: 10.3390/ijms23031759
Choi, I. J., Kim, C. G., Lee, J. Y., Kim, Y. I., Kook, M. C., Park, B., et al. (2020). Family history of gastric cancer and helicobacter pylori treatment. N Engl. J. Med. 382 (5), 427–436. doi: 10.1056/NEJMoa1909666
Chung, L., Thiele Orberg, E., Geis, A. L., Chan, J. L., Fu, K., DeStefano Shields, C. E., et al. (2018). Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe 23 (2), 203–214.e5. doi: 10.1016/j.chom.2018.01.007
Corden, S. A., Sant-Cassia, L. J., Easton, A. J., Morris, A. G. (1999). The integration of HPV-18 DNA in cervical carcinoma. Mol. Pathol. 52 (5), 275–282. doi: 10.1136/mp.52.5.275
Dang, S., Zhou, J., Chen, Y., Chen, P., Ji, M., Shi, B., et al. (2019). Dynamic expression of ZNF382 and its tumor-suppressor role in hepatitis b virus-related hepatocellular carcinogenesis. Oncogene 38 (24), 4804–4819. doi: 10.1038/s41388-019-0759-9
Dejea, C. M., Fathi, P., Craig, J. M., Boleij, A., Taddese, R., Geis, A. L., et al. (2018). Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359 (6375), 592–597. doi: 10.1126/science.aah3648
De Martel, C., Plummer, M., Vignat, J., Franceschi, S. (2017). Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int. J. Cancer. 141 (4), 664–670. doi: 10.1002/ijc.30716
Derech-Haim, S., Friedman, Y., Hizi, A., Bakhanashvili, M. (2020). p53 regulates its own expression by an intrinsic exoribonuclease activity through AU-rich elements. J. Mol. Med. (Berl). 98 (3), 437–449. doi: 10.1007/s00109-020-01884-0
Durieux, I., Ginevra, C., Attaiech, L., Picq, K., Juan, P. A., Jarraud, S., et al. (2019). Diverse conjugative elements silence natural transformation in legionella species. Proc. Natl. Acad. Sci. U S A. 116 (37), 18613–18618. doi: 10.1073/pnas.1909374116
Dziubańska-Kusibab, P. J., Berger, H., Battistini, F., Bouwman, B. A. M., Iftekhar, A., Katainen, R., et al. (2020). Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nat. Med. 26 (7), 1063–1069. doi: 10.1038/s41591-020-0908-2
Eisenhofer, R., Minich, J. J., Marotz, C., Cooper, A., Knight, R., Weyrich, L. S. (2019). Contamination in low microbial biomass microbiome studies: Issues and recommendations. Trends Microbiol. 27 (2), 105–117. doi: 10.1016/j.tim.2018.11.003
Fan, L., Wu, D., Goremykin, V., Xiao, J., Xu, Y., Garg, S., et al. (2020). Phylogenetic analyses with systematic taxon sampling show that mitochondria branch within alphaproteobacteria. Nat. Ecol. Evol. 4 (9), 1213–1219. doi: 10.1038/s41559-020-1239-x
Fontana, N. S., Ibrahim, K. Y., Bonazzi, P. R., Rossi, F., Almeida, S. C. G., Tengan, F. M., et al. (2021). Fluoroquinolone treatment as a protective factor for 10-day mortality in streptococcus pneumoniae bacteremia in cancer patients. Sci. Rep. 11 (1), 3699. doi: 10.1038/s41598-021-81415-0
Ford, A. C., Yuan, Y., Moayyedi, P. (2022). Long-term impact of helicobacter pylori eradication therapy on gastric cancer incidence and mortality in healthy infected individuals: A meta-analysis beyond 10 years of follow-up. Gastroenterology. 163(3), 754–756.e1. doi: 10.1053/j.gastro.2022.05.027
Fu, A., Yao, B., Dong, T., Chen, Y., Yao, J., Liu, Y., et al. (2022). Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell 185 (8), 1356–1372.e26. doi: 10.1016/j.cell.2022.02.027
Geller, L. T., Barzily-Rokni, M., Danino, T., Jonas, O. H., Shental, N., Nejman, D., et al. (2017). Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 357 (6356), 1156–1160. doi: 10.1126/science.aah5043
Goggin, K. P., Gonzalez-Pena, V., Inaba, Y., Allison, K. J., Hong, D. K., Ahmed, A. A., et al. (2020). Evaluation of plasma microbial cell-free DNA sequencing to predict bloodstream infection in pediatric patients with relapsed or refractory cancer. JAMA Oncol. 6 (4), 552–556. doi: 10.1001/jamaoncol.2019.4120
Gold, D. V., Stein, R., Burton, J., Goldenberg, D. M. (2010). Enhanced expression of CD74 in gastrointestinal cancers and benign tissues. Int. J. Clin. Exp. Pathol. 4 (1), 1–12.
Greathouse, K. L., White, J. R., Vargas, A. J., Bliskovsky, V. V., Beck, J. A., von Muhlinen, N., et al. (2018). Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 19 (1), 123. doi: 10.1186/s13059-018-1501-6
Guo, S., Chen, J., Chen, F., Zeng, Q., Liu, W. L., Zhang, G. (2020). Exosomes derived from fusobacterium nucleatum-infected colorectal cancer cells facilitate tumour metastasis by selectively carrying miR-1246/92b-3p/27a-3p and CXCL16. Gut 71(2), e1–e3. doi: 10.1136/gutjnl-2020-321187
Halimi, A., Gabarrini, G., Sobkowiak, M. J., Ateeb, Z., Davanian, H., Gaiser, R. A., et al. (2021). Isolation of pancreatic microbiota from cystic precursors of pancreatic cancer with intracellular growth and DNA damaging properties. Gut Microbes 13 (1), 1983101. doi: 10.1080/19490976.2021.1983101
Hancks, D. C., Kazazian, H. H., Jr. (2012). Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 22 (3), 191–203. doi: 10.1016/j.gde.2012.02.006
Han, S. U., Kwak, T. H., Her, K. H., Cho, Y. H., Choi, C., Lee, H. J., et al. (2008). CEACAM5 and CEACAM6 are major target genes for Smad3-mediated TGF-beta signaling. Oncogene 27 (5), 675–683. doi: 10.1038/sj.onc.1210686
Hirose, Y., Yamaguchi-Naka, M., Onuki, M., Tenjimbayashi, Y., Tasaka, N., Satoh, T., et al. (2020). High levels of within-host variations of human papillomavirus 16 E1/E2 genes in invasive cervical cancer. Front. Microbiol. 11, 596334. doi: 10.3389/fmicb.2020.596334
Hong, J., Guo, F., Lu, S. Y., Shen, C., Ma, D., Zhang, X., et al. (2021). F. nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut 70 (11), 2123–2137. doi: 10.1136/gutjnl-2020-322780
Huang, Y. J., Nelson, C. E., Brodie, E. L., Desantis, T. Z., Baek, M. S., Liu, J., et al. (2011). Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J. Allergy Clin. Immunol. 127, 372–381. doi: 10.1016/j.jaci.2010.10.048
Ibáñez de Aldecoa, A. L., Zafra, O., González-Pastor, J. E. (2017). Mechanisms and regulation of extracellular DNA release and its biological roles in microbial communities. Front. Microbiol. 8, 1390. doi: 10.3389/fmicb.2017.01390
Imai, S., Ooki, T., Murata-Kamiya, N., Komura, D., Tahmina, K., Wu, W., et al. (2021). Helicobacter pylori CagA elicits BRCAness to induce genome instability that may underlie bacterial gastric carcinogenesis. Cell Host Microbe 29 (6), 941–958.e10. doi: 10.1016/j.chom.2021.04.006
Jia, W., Xie, G., Jia, W. (2018). Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 15, 111–128. doi: 10.1038/nrgastro.2017.119
Jima, D. D., Zhang, J., Jacobs, C., Richards, K. L., Dunphy, C. H., Choi, W. W., et al. (2010). Hematologic malignancies research consortium. deep sequencing of the small RNA transcriptome of normal and malignant human b cells identifies hundreds of novel microRNAs. Blood 116 (23), e118–e127. doi: 10.1182/blood-2010-05-285403
Jin, C., Lagoudas, G. K., Zhao, C., Bullman, S., Bhutkar, A., Hu, B., et al. (2019). Commensal microbiota promote lung cancer development via γδ T cells. Cell 176 (5), 998–1013.e16. doi: 10.1016/j.cell.2018.12.040
Johnston, C., Martin, B., Fichant, G., Polard, P., Claverys, J. P. (2014). Bacterial transformation: Distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 12 (3), 181–196. doi: 10.1038/nrmicro3199
Kadosh, E., Snir-Alkalay, I., Venkatachalam, A., May, S., Lasry, A., Elyada, E., et al. (2020). The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature 586 (7827), 133–138. doi: 10.1038/s41586-020-2541-0
Kalaora, S., Nagler, A., Nejman, D., Alon, M., Barbolin, C., Barnea, E., et al. (2021). Identification of bacteria-derived HLA-bound peptides in melanoma. Nature 592 (7852), 138–143. doi: 10.1038/s41586-021-03368-8
Kamiza, A. B., Fatumo, S., Singini, M. G., Yeh, C. C., Chikowore, T. (2022). Hepatitis b infection is causally associated with extrahepatic cancers: A mendelian randomization study. EBioMedicine 79, 104003. doi: 10.1016/j.ebiom.2022.104003
Kim, S. J., Fernandez-Martinez, J., Nudelman, I., Shi, Y., Zhang, W., Raveh, B., et al. (2018). Integrative structure and functional anatomy of a nuclear pore complex. Nature 555 (7697), 475–482. doi: 10.1038/nature26003
Kowarsky, M., Camunas-Soler, J., Kertesz, M., De Vlaminck, I., Koh, W., Pan, W., et al. (2017). Numerous uncharacterized and highly divergent microbes which colonize humans are revealed by circulating cell-free DNA. Proc. Natl. Acad. Sci. U S A. 114 (36), 9623–9628. doi: 10.1073/pnas.1707009114
Ku, C., Nelson-Sathi, S., Roettger, M., Garg, S., Hazkani-Covo, E., Martin, W. F. (2015a). Endosymbiotic gene transfer from prokaryotic pangenomes: Inherited chimerism in eukaryotes. Proc. Natl. Acad. Sci. U S A. 112 (33), 10139–10146. doi: 10.1073/pnas.1421385112
Ku, C., Nelson-Sathi, S., Roettger, M., Sousa, F. L., Lockhart, P. J., Bryant, D., et al. (2015b). Endosymbiotic origin and differential loss of eukaryotic genes. Nature 524 (7566), 427–432. doi: 10.1038/nature14963
Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., et al. (2001). International human genome sequencing consortium. initial sequencing and analysis of the human genome. Nature 409 (6822), 860–921. doi: 10.1038/35057062
Lee, Y. C., Chiang, T. H., Chou, C. K., Tu, Y. K., Liao, W. C., Wu, M. S., et al. (2016). Association between helicobacter pylori eradication and gastric cancer incidence: A systematic review and meta-analysis. Gastroenterology 150 (5), 1113–1124.e5. doi: 10.1053/j.gastro.2016.01.028
Liu, C. H., Chen, Z., Chen, K., Liao, F. T., Chung, C. E., Liu, X., et al. (2021). Lipopolysaccharide-mediated chronic inflammation promotes tobacco carcinogen-induced lung cancer and determines the efficacy of immunotherapy. Cancer Res. 81 (1), 144–157. doi: 10.1158/0008-5472.CAN-20-1994
Liu, S., da Cunha, A. P., Rezende, R. M., Cialic, R., Wei, Z., Bry, L., et al. (2016). The host shapes the gut microbiota via fecal MicroRNA. Cell Host Microbe 19 (1), 32–43. doi: 10.1016/j.chom.2015.12.005
Liu, W., Zhang, X., Xu, H., Li, S., Lau, H. C., Chen, Q., et al. (2021). Microbial community heterogeneity within colorectal neoplasia and its correlation with colorectal carcinogenesis. Gastroenterology 160 (7), 2395–2408. doi: 10.1053/j.gastro.2021.02.020
Li, J., Wolf, S. G., Elbaum, M., Tzfira, T. (2005). Exploring cargo transport mechanics in the type IV secretion systems. Trends Microbiol. 13 (7), 295–298. doi: 10.1016/j.tim.2005.05.002
Luo, Y. D., Fang, L., Yu, H. Q., Zhang, J., Lin, X. T., Liu, X. Y., et al. (2021). p53 haploinsufficiency and increased mTOR signalling define a subset of aggressive hepatocellular carcinoma. J. Hepatol. 74 (1), 96–108. doi: 10.1016/j.jhep.2020.07.036
Mansour, B., Monyók, Á, Makra, N., Gajdács, M., Vadnay, I., Ligeti, B., et al. (2020). Bladder cancer-related microbiota: examining differences in urine and tissue samples. Sci. Rep. 10 (1), 11042. doi: 10.1038/s41598-020-67443-2
Mao, D., Luo, Y., Mathieu, J., Wang, Q., Feng, L., Mu, Q., et al. (2014). Persistence of extracellular DNA in river sediment facilitates antibiotic resistance gene propagation. Environ. Sci. Technol. 48 (1), 71–78. doi: 10.1021/es404280v
Marshall, C. J. (1991). Tumor suppressor genes. Cell 64 (2), 313–326. doi: 10.1016/0092-8674(91)90641-B
Mba Medie, F., Sharma-Kuinkel, B. K., Ruffin, F., Chan, L. C., Rossetti, M., Chang, Y. L., et al. (2019). MRSA systems immunobiology group. Genetic variation of DNA methyltransferase-3A contributes to protection against persistent MRSA bacteremia in patients. Proc. Natl. Acad. Sci. U S A. 116 (40), 20087–20096. doi: 10.1073/pnas.1909849116
Melsheimer, P., Vinokurova, S., Wentzensen, N., Bastert, G., von Knebel Doeberitz, M. (2004). DNA Aneuploidy and integration of human papillomavirus type 16 e6/e7 oncogenes in intraepithelial neoplasia and invasive squamous cell carcinoma of the cervix uteri. Clin. Cancer Res. 10 (9), 3059–3063. doi: 10.1158/1078-0432.CCR-03-0565
Mirouse, A., Vigneron, C., Llitjos, J. F., Chiche, J. D., Mira, J. P., Mokart, D., et al. (2020). Sepsis and cancer: An interplay of friends and foes. Am. J. Respir. Crit. Care Med. 202 (12), 1625–1635. doi: 10.1164/rccm.202004-1116TR
Nejman, D., Livyatan, I., Fuks, G., Gavert, N., Zwang, Y., Geller, L. T., et al. (2020). The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 368 (6494), 973–980. doi: 10.1126/science.aay9189
Pandey, S., Jha, H. C., Shukla, S. K., Shirley, M. K., Robertson, E. S. (2018). Epigenetic regulation of tumor suppressors by helicobacter pylori enhances EBV-induced proliferation of gastric epithelial cells. MBio 9, e00649–e00618. doi: 10.1128/mBio.00649-18
Pleguezuelos-Manzano, C., Puschhof, J., Rosendahl Huber, A., van Hoeck, A., Wood, H. M., Nomburg, J., et al. (2020). Mutational signature in colorectal cancer caused by genotoxic pks+ e. coli. Nature 580 (7802), 269–273. doi: 10.1038/s41586-020-2080-8
Pokrývková, B., Saláková, M., Šmahelová, J., Vojtěchová, Z., Novosadová, V., Tachezy, R. (2019). Detailed characteristics of tonsillar tumors with extrachromosomal or integrated form of human papillomavirus. Viruses 12 (1), 42. doi: 10.3390/v12010042
Poore, G. D., Kopylova, E., Zhu, Q., Carpenter, C., Fraraccio, S., Wandro, S., et al. (2020). Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 579 (7800), 567–574. doi: 10.1038/s41586-020-2095-1
Rebolj, M., Cuschieri, K., Mathews, C. S., Pesola, F., Denton, K., Kitchener, H., et al. (2022). Extension of cervical screening intervals with primary human papillomavirus testing: observational study of English screening pilot data. BMJ 377, e068776. doi: 10.1136/bmj-2021-068776
Ren, N., Liang, B., Li, Y. (2020). Identification of prognosis-related genes in the tumor microenvironment of stomach adenocarcinoma by TCGA and GEO datasets. Biosci. Rep. 40 (10), BSR20200980. doi: 10.1042/BSR20200980
Riley, D. R., Sieber, K. B., Robinson, K. M., White, J. R., Ganesan, A., Nourbakhsh, S., et al. (2013). Bacteria-human somatic cell lateral gene transfer is enriched in cancer samples. PloS Comput. Biol. 9 (6), e1003107. doi: 10.1371/journal.pcbi.1003107
Rivas, M., Calaf, G. M., Laroze, D., Rojas, E., Mendez, J., Honeyman, J., et al. (2020). Solar ultraviolet a radiation and nonmelanoma skin cancer in arica, Chile. J. Photochem. Photobiol. B. 212, 112047. doi: 10.1016/j.jphotobiol.2020.112047
Roadmap Epigenomics Consortium, Kundaje, A., Meuleman, W., Ernst, J., Bilenky, M., Yen, A., et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature 518 (7539), 317–330. doi: 10.1038/nature14248
Saigo, K., Yoshida, K., Ikeda, R., Sakamoto, Y., Murakami, Y., Urashima, T., et al. (2008). Integration of hepatitis b virus DNA into the myeloid/lymphoid or mixed-lineage leukemia (MLL4) gene and rearrangements of MLL4 in human hepatocellular carcinoma. Hum. Mutat. 29 (5), 703–708. doi: 10.1002/humu.20701
Salter, S. J., Cox, M. J., Turek, E. M., Calus, S. T., Cookson, W. O., Moffatt, M. F., et al. (2014). Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87. doi: 10.1186/s12915-014-0087-z
Schröder, G., Schuelein, R., Quebatte, M., Dehio, C. (2011). Conjugative DNA transfer into human cells by the VirB/VirD4 type IV secretion system of the bacterial pathogen bartonella henselae. Proc. Natl. Acad. Sci. U S A. 108 (35), 14643–14648. doi: 10.1073/pnas.1019074108
Scott, E. C., Gardner, E. J., Masood, A., Chuang, N. T., Vertino, P. M., Devine, S. E. (2016). A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res. 26 (6), 745–755. doi: 10.1101/gr.201814.115
Sears, C. L., Garrett, W. S. (2014). Microbes, microbiota, and colon cancer. Cell Host Microbe 15, 317–328. doi: 10.1016/j.chom.2014.02.007
Sender, R., Fuchs, S., Milo, R. (2016). Revised estimates for the number of human and bacteria cells in the body. PloS Biol. 14 (8), e1002533. doi: 10.1371/journal.pbio.1002533
Seth, P., Hsieh, P. N., Jamal, S., Wang, L., Gygi, S. P., Jain, M. K., et al. (2019). Regulation of MicroRNA machinery and development by interspecies s-nitrosylation. Cell 176 (5), 1014–1025.e12. doi: 10.1016/j.cell.2019.01.037
Simpson, C. A., Podicheti, R., Rusch, D. B., Dalia, A. B., van Kessel, J. C. (2019). Diversity in natural transformation frequencies and regulation across vibrio species. mBio 10 (6), e02788–e02719. doi: 10.1128/mBio.02788-19
Skvortsova, K., Iovino, N., Bogdanović, O. (2018). Functions and mechanisms of epigenetic inheritance in animals. Nat. Rev. Mol. Cell Biol. 19 (12), 774–790. doi: 10.1038/s41580-018-0074-2
Song, S., Shi, Y., Wu, W., Wu, H., Chang, L., Peng, P., et al. (2021). Reticulon 3-mediated Chk2/p53 activation suppresses hepatocellular carcinogenesis and is blocked by hepatitis b virus. Gut 70 (11), 2159–2171. doi: 10.1136/gutjnl-2020-321386
Steely, C. J., Russell, K. L., Feusier, J. E., Qiao, Y., Tavtigian, S. V., Marth, G., et al. (2021). Mobile element insertions and associated structural variants in longitudinal breast cancer samples. Sci. Rep. 11 (1), 13020. doi: 10.1038/s41598-021-92444-0
Stumpel, D. J., Schneider, P., van Roon, E. H., Pieters, R., Stam, R. W. (2013). Absence of global hypomethylation in promoter hypermethylated mixed lineage leukaemia-rearranged infant acute lymphoblastic leukaemia. Eur. J. Cancer. 49 (1), 175–184. doi: 10.1016/j.ejca.2012.07.013
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. doi: 10.3322/caac.21660
Sung, W. K., Zheng, H., Li, S., Chen, R., Liu, X., Li, Y., et al. (2012). Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat. Genet. 44 (7), 765–769. doi: 10.1038/ng.2295
Sun, Y., Ma, L. (2019). New insights into long non-coding RNA MALAT1 in cancer and metastasis. Cancers 11, 216. doi: 10.3390/cancers11020216
Sun, D., Mao, X., Fei, M., Chen, Z., Zhu, T., Qiu, J. (2020). Histone-like nucleoid-structuring protein (H-NS) paralogue StpA activates the type I-e CRISPR-cas system against natural transformation in escherichia coli. Appl. Environ. Microbiol. 86 (14), e00731–e00720. doi: 10.1128/AEM.00731-20
Sze, K. M., Ho, D. W., Chiu, Y. T., Tsui, Y. M., Chan, L. K., Lee, J. M., et al. (2021). Hepatitis b virus-telomerase reverse transcriptase promoter integration harnesses host ELF4, resulting in telomerase reverse transcriptase gene transcription in hepatocellular carcinoma. Hepatology 73 (1), 23–40. doi: 10.1002/hep.31231
Tjalsma, H., Boleij, A., Marchesi, J. R., Dutilh, B. E. (2012). A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat. Rev. Microbiol. 10, 575–582. doi: 10.1038/nrmicro2819
Torres, R., Serrano, E., Alonso, J. C. (2019). Bacillus subtilis RecA interacts with and loads RadA/Sms to unwind recombination intermediates during natural chromosomal transformation. Nucleic Acids Res. 47 (17), 9198–9215. doi: 10.1093/nar/gkz647
Tzeng, A., Sangwan, N., Jia, M., Liu, C. C., Keslar, K. S., Downs-Kelly, E., et al. (2021). Human breast microbiome correlates with prognostic features and immunological signatures in breast cancer. Genome Med. 13 (1), 60. doi: 10.1186/s13073-021-00874-2
Wang, S., Ma, N., Zhao, W., Midorikawa, K., Kawanishi, S., Hiraku, Y., et al. (2016). Inflammation-related DNA damage and cancer stem cell markers in nasopharyngeal carcinoma. Mediators Inflamm. 2016, 9343460. doi: 10.1155/2016/9343460
Watanabe, Y., Oikawa, R., Kodaka, Y., Sato, Y., Ono, S., Kenmochi, T., et al. (2021). Cancer-related genetic variants of helicobacter pylori strains determined using gastric wash-based whole-genome analysis with single-molecule real-time technology. Int. J. Cancer. 148 (1), 178–192. doi: 10.1002/ijc.33257
Waters, V. L. (2001). Conjugation between bacterial and mammalian cells. Nat. Genet. 29 (4), 375–376. doi: 10.1038/ng779
Whitman, S. P., Liu, S., Vukosavljevic, T., Rush, L. J., Yu, L., Liu, C., et al. (2005). The MLL partial tandem duplication: evidence for recessive gain-of-function in acute myeloid leukemia identifies a novel patient subgroup for molecular-targeted therapy. Blood 106, 345–352. doi: 10.1182/blood-2005-01-0204
Wroblewski, L. E., Peek, R. M., Jr. (2016). Helicobacter pylori: Pathogenic enablers–toxic relationships in the stomach. Nat. Rev. Gastroenterol. Hepatol. 13, 317–318. doi: 10.1038/nrgastro.2016.68
Xiao, Q., Lu, W., Kong, X., Shao, Y. W., Hu, Y., Wang, A., et al. (2021). Alterations of circulating bacterial DNA in colorectal cancer and adenoma: A proof-of-concept study. Cancer Lett. 499, 201–208. doi: 10.1016/j.canlet.2020.11.030
Yang, J., Wei, H., Zhou, Y., Szeto, C. H., Li, C., Lin, Y., et al. (2022). High-fat diet promotes colorectal tumorigenesis through modulating gut microbiota and metabolites. Gastroenterology 162 (1), 135–149.e2. doi: 10.1053/j.gastro.2021.08.041
Yang, Y., Weng, W., Peng, J., Hong, L., Yang, L., Toiyama, Y., et al. (2017). Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-κB, and up-regulating expression of MicroRNA-21. Gastroenterology 152 (4), 851–866. doi: 10.1053/j.gastro.2016.11.018
Yu, J., Feng, Q., Wong, S. H., Zhang, D., Liang, Q. Y., Qin, Y., et al. (2017). Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 66 (1), 70–78. doi: 10.1136/gutjnl-2015-309800
Zaremba, M., Toliusis, P., Grigaitis, R., Manakova, E., Silanskas, A., Tamulaitiene, G., et al. (2014). DNA Cleavage by CgII and NgoAVII requires interaction between n- and r-proteins and extensive nucleotide hydrolysis. Nucleic Acids Res. 42 (22), 13887–13896. doi: 10.1093/nar/gku1236
Zegarra-Ruiz, D. F., Kim, D. V., Norwood, K., Kim, M., Wu, W. H., Saldana-Morales, F. B., et al. (2021). Thymic development of gut-microbiota-specific T cells. Nature 594 (7863), 413–417. doi: 10.1038/s41586-021-03531-1
Zhang, S., Kong, C., Yang, Y., Cai, S., Li, X., Cai, G., et al. (2020). Human oral microbiome dysbiosis as a novel non-invasive biomarker in detection of colorectal cancer. Theranostics 10 (25), 11595–11606. doi: 10.7150/thno.49515
Keywords: bacterial DNA, human genome, gene integration, cancerogenesis, hypothesis
Citation: Yangyanqiu W and Shuwen H (2022) Bacterial DNA involvement in carcinogenesis. Front. Cell. Infect. Microbiol. 12:996778. doi: 10.3389/fcimb.2022.996778
Received: 18 July 2022; Accepted: 27 September 2022;
Published: 12 October 2022.
Edited by:
Hui Jin, Huazhong Agricultural University, ChinaReviewed by:
Ting Wang, Guangzhou First People’s Hospital, ChinaCopyright © 2022 Yangyanqiu and Shuwen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuwen Han, c2h1d2VuaGFuOTg1QDE2My5jb20=
†ORCID: Wang Yangyanqiu, orcid.org/0000-0002-9981-1748
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.