
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol., 02 September 2022
Sec. Molecular Bacterial Pathogenesis
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.990312
This article is part of the Research TopicDrug-resistant Mycobacterium tuberculosisView all 21 articles
 Paolo Miotto1‡
Paolo Miotto1‡ Rita Sorrentino1‡
Rita Sorrentino1‡ Stefano De Giorgi2†
Stefano De Giorgi2† Roberta Provvedi3
Roberta Provvedi3 Daniela Maria Cirillo1
Daniela Maria Cirillo1 Riccardo Manganelli2*
Riccardo Manganelli2*Bacterial drug resistance is one of the major challenges to present and future human health, as the continuous selection of multidrug resistant bacteria poses at serious risk the possibility to treat infectious diseases in the near future. One of the infection at higher risk to become incurable is tuberculosis, due to the few drugs available in the market against Mycobacterium tuberculosis. Drug resistance in this species is usually due to point mutations in the drug target or in proteins required to activate prodrugs. However, another interesting and underexplored aspect of bacterial physiology with important impact on drug susceptibility is represented by the changes in transcriptional regulation following drug exposure. The main regulators involved in this phenomenon in M. tuberculosis are the sigma factors, and regulators belonging to the WhiB, GntR, XRE, Mar and TetR families. Better understanding the impact of these regulators in survival to drug treatment might contribute to identify new drug targets and/or to design new strategies of intervention.
One of the most fascinating aspects of bacteriology is the extremely fast and efficient responsivity of bacteria to external stimuli, which is translated in a fast and precise variation of their transcriptional profile. This capacity is founded on complex regulatory networks based on sigma factors, transcriptional repressors/activators, two component systems, small RNAs or riboswitches able to reshape bacterial physiology allowing the cells to adapt in real time to any external challenge.
Antibacterial drugs can be considered atypical stressors, both causing a direct interference with cellular physiology and inducing secondary stress due to this interference, as in the case of the oxidative stress induced by bactericidal drugs (Kohanski et al., 2007; Martínez et al., 2020). Consequently, bacteria respond to drug with profound changes in their transcriptional profile that can increase bacterial drug resistance (DR) by target overexpression, drug modification, induction of efflux systems, or simply by helping the cells to respond to the drug-induced stress.
To be noted that whereas DR is usually associated with genetic mutations (usually also referred as genetic resistance), in some cases DR can be developed without chromosomal abnormalities (defined as phenotypic resistance) (Corona and Martinez, 2013). Transcription factors can be involved in both the mechanisms of resistance.
In this paper, we review the impact of transcriptional regulation on drug susceptibility in M. tuberculosis.
Sigma factors are small interchangeable cofactors of RNA polymerase able to confer promoter specificity. The M. tuberculosis genome encodes 13 sigma factors and it is the obligate pathogen with the higher amount of sigma factor genes per megabase (Rodrigue et al., 2006). Among these sigma factors, only σA is essential. The others are dispensable and are activated in response to specific environmental signals. Following their activation, sigma factors switch RNA polymerase holoenzyme promoter-specificity, resulting in a quick change of the bacterial transcriptome leading to the adaptation to the new environment experienced by the bacterium. Mycobacterial sigma factors have been shown to be involved in the response to different conditions endangering the bacterial cells as oxidative stress, alkaline stress, surface stress, low pH, hypoxia, nutrient depletion and heat shock. Drug treatment has been shown to induce a strong stress response in bacteria and the bactericidal activity of some drugs has been primary linked to the stress they induce on the bacteria (Kohanski et al., 2007). Since sigma factors have a primary role in stress response, it is simple to imagine their implication in the establishment of the baseline resistance to drugs.
At least five M. tuberculosis sigma factors have been shown to be involved in the basal level of resistance to drugs (Table 1). σF (i) is induced upon treatment with ethambutol (EMB), rifampin (RIF), streptomycin (STR), and cycloserine (CS) (Michele et al., 1999), while a sigF null mutant in CDC1551 is more resistant to RIF (Chen et al., 2000). However, this phenotype was not confirmed in the H37Rv genetic background, suggesting a different role of σF in different mycobacterial strains (Hartkoorn et al., 2010).
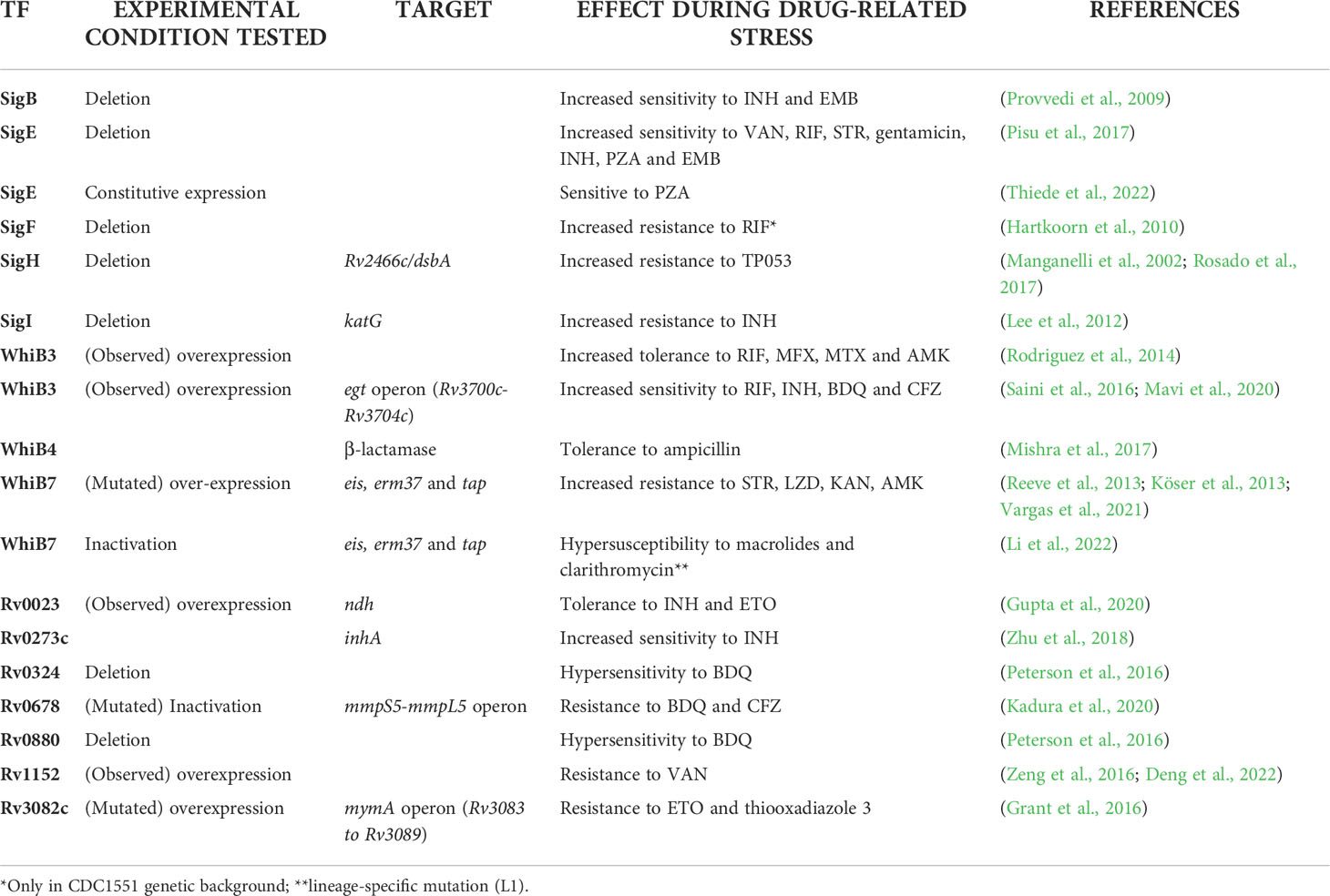
Table 1 Principal transcription factors (TF) in M. tuberculosis with established mechanisms of action involved in drug resistance (DR) (transcription factors known for their homology with other mycobacteria are described in the text but not reported in this table).
The extracytoplasmatic function (ECF) sigma factor σI (ii) directly regulates the expression of the structural gene of KatG, an enzyme required for the activation of isoniazide (INH) (Figure 1A). Consequently, a sigI null mutant was more resistant to this drug both in axenic culture and during mice infection, but surprisingly was not attenuated as predictable for a strain expressing lower levels of KatG (Lee et al., 2012).
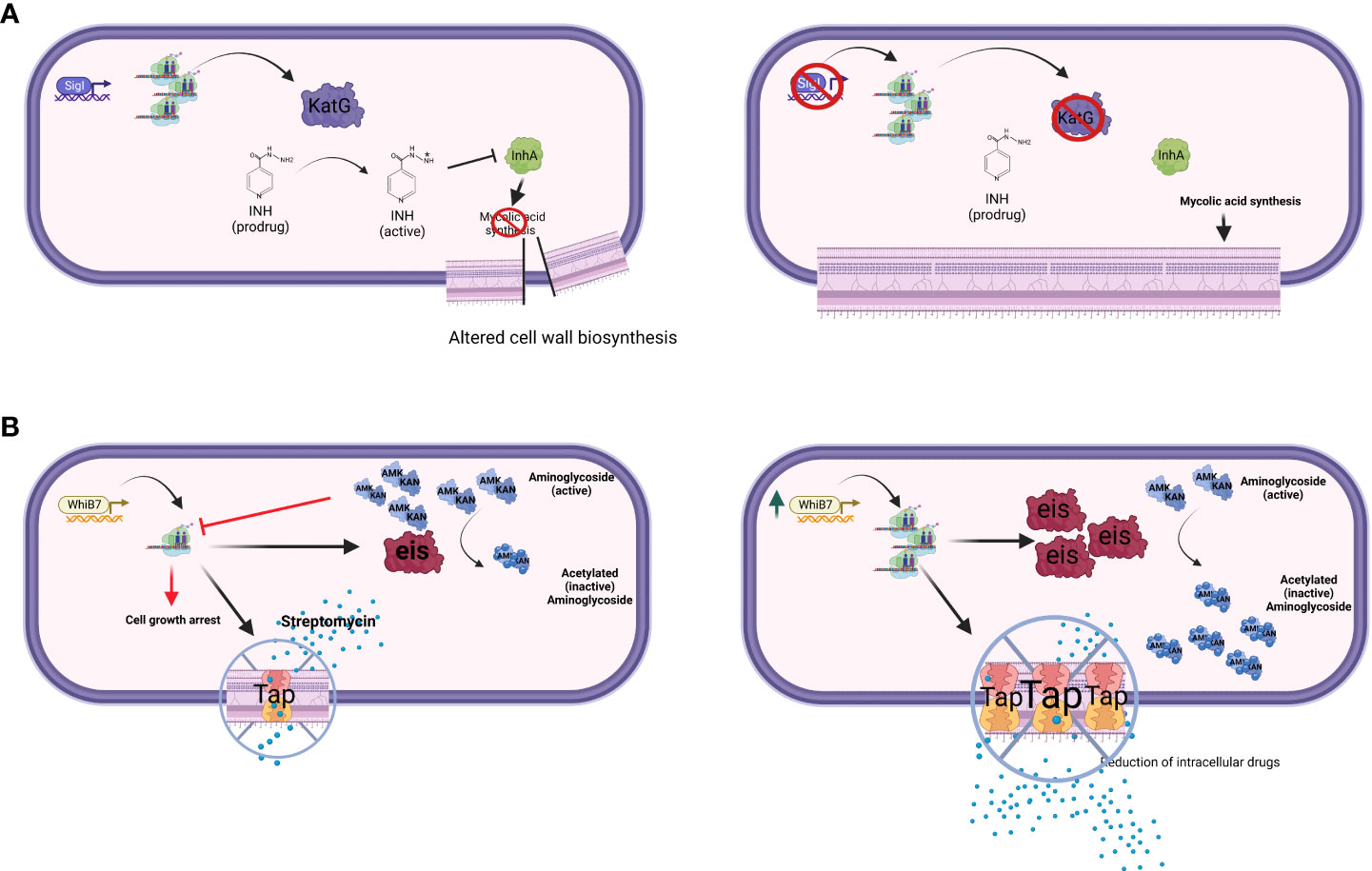
Figure 1 The figure schematizes two major drug-resistance mechanisms mediated by transcriptional factors. (A) Transcriptional factor down-regulation. The ECF sigma factor SigI controls the transcription of katG; mutant strains lacking SigI are more resistant to INH (Lee et al., 2012). (B) Transcriptional factor over-expression. WhiB7 regulon contains key genes involved in DR, such as rv2416c (eis) linked to low-level KAN and AMK resistance, and rv1258c (tap) whose overexpression is linked to low-level STR resistance (Reeves et al., 2013). Point mutations in tap, V219A and S292L, have been associated with resistance to PZA, INH, and STR (Liu et al., 2019). Created with BioRender.com.
The ECF sigma factor σH (iii) is involved in oxidative stress response. One of the genes most induced by σH in response to oxidative stress is rv2466c (Manganelli et al., 2002), encoding for the mycothiol-dependent reductase DsbA (Rosado et al., 2017). This gene is required for the activation of TP053, a promising thienopyrimidine derivative prodrug, able to kill replicating and non-replicating M. tuberculosis (Albesa-Jové et al., 2015). Consistent with these data, a sigH null mutant was shown to be resistant to TP053 (Rosado et al., 2017). Since sigH is induced upon oxidative stress, it is possible to assume that the activation of TP053 activation increases after intracellular bacteria are exposed to reactive oxygen intermediates.
Both σB (iv), a member of the primary-like sigma factors, and the ECF sigma factor σE (v) have been shown to be induced upon exposure to vancomycin (VAN) (Provvedi et al., 2009), and involved in the baseline resistance to several antitubercular drugs. In particular, Pisu and colleagues (Pisu et al., 2017) showed that a sigE null mutant was more sensitive to VAN, RIF, STR, gentamicin, INH, and EMB, while a sigB null mutant was more sensitive to INH and EMB. Moreover, Yang and colleagues reported that deletion of sigB causes increased sensitivity to para-aminosalicylic acid and sulfamethoxazole (Yang et al., 2017). Interestingly, Pisu and colleagues showed that σB and σE are also essential for the development of persistent bacteria able to survive the bactericidal activity of very high concentrations of VAN (sigE mutant), STR and INH (both mutants). These data support the hypothesis that σE represents a bistable switch involved in persistence development (Balázsi et al., 2008; Sureka et al., 2008; Manganelli and Provvedi, 2010; Tiwari et al., 2010; Zorzan et al., 2021).
Finally, a recent paper demonstrated that σE-mediated activation of surface stress response is essential for pyrazinamide (PZA) susceptibility. PZA is only active when bacteria are exposed to low pH, while its derivative pyrazinoic acid is active also at circumneutral pH. However, a mutant overexpressing sigE due to the deletion of the gene encoding its anti-sigma factor RseA (Boldrin et al., 2019) was equally sensitive to PZA at both low pH and circumneutral pH, while a sigE null mutant was resistant to both PZA and pyrazinoic acid demonstrating that the activation of the σE regulon is essential for PZA susceptibility (Thiede et al., 2022). Interestingly, both meropenem and CS, showed synergistic activity with PZA due to their activation of σE-mediated surface stress response (Thiede et al., 2022). Since it is well known that σE is activated at low pH (Bush, 2018), these data strongly suggest that the role of low pH in PZA susceptibility is the activation of the σE response.
M. tuberculosis genome encodes seven proteins belonging to the WhiB superfamily. Proteins belonging to this family are small transcriptional regulators and are exclusively present in Actinobacteria. They are characterized by four cysteine residues that bind a [4Fe–4S] cluster and by a five residues (G[V/I]WGG) motif, while their putative role is that to sense O2 and nitric oxide (Bansal et al., 2017). Two of them (WhiB1 and WhiB2) are essential for growth, while others were shown to be important for several aspects of mycobacterial physiology as redox homeostasis, DR, dormancy and reactivation (Bansal et al., 2017).
WhiB transcriptional factors are involved in stress response and their genes are often upregulated in multidrug resistant (MDR) clinical isolates. Many whiB genes showed altered expression during drug treatment: whiB6 is downregulated by CS, whiB2 is upregulated by CS, EMB and INH, whiB7 is upregulated by macrolides, fluoroquinolones and aminoglycosides (Morris et al., 2005; Geiman et al., 2006; Burian et al., 2012; Chatterjee et al., 2013).
Within the WhiB family, a key player in DR is WhiB7, a transcription factor inducing stress response and promoting low levels of resistance to several antibacterial drugs including macrolides, tetracyclines, and some aminoglycosides (Morris et al., 2005; Burian et al., 2013; Reeves et al., 2013). Recently, an elegant experiment based on CRISPR interference identified whiB7 as the unique gene responsible for increased susceptibility to ribosome-targeting drugs STR and linezolid (LZD) (Li et al., 2022). Among the genes relevant for DR to antibiotics inhibiting translation (i.e. macrolides), rv2416c (eis) encodes an aminoglycoside acetyltransferase which inactivate the drugs, whereas 23S rRNA methylation by Rv1988 (Erm37) confers resistance to macrolides. Another gene found to be associated with DR to different drugs including STR is rv1258c (tap), which encodes for an efflux pump. All these DR genes are part of the WhiB7 regulon (Figure 1B). Of particular relevance is the link between mutations affecting the expression of whiB7 observed by Reeves and colleagues leading to enhanced expression of rv2416c (eis) (low-level KAN and amikacin resistance) and of rv1258c (tap) (low-level STR resistance) (Reeves et al., 2013). Interestingly, loss-of-function mutations in the arginine biosynthesis pathway were found to up-regulate the expression of whiB7, and eis genes, thus conferring tolerance to KAN (Schrader et al., 2021). These mutations mapping in the rv2747 (argA) and rv1655 (argD) genes were associated with increased survival during RIF exposure, as well as with minimum inhibitory concentration (MIC) increase to clarithromycin, again related to the up-regulation of whiB7. Modern Beijing isolates and one phylogenetically intermediate Beijing isolate harbored a loss-of-function mutation in tap; thus whiB7 mutations would not translate into low-level STR resistance, but would lead to low-level KAN and AMK resistance only, despite this epistatic interaction has not been proven in clinical isolates yet (Köser et al., 2013; Vargas et al., 2021). Interestingly, a lineage-specific mutation present in the L1 Indo-Oceanic clade inactivates WhiB7, thus making strains belonging to this phylogenetic branch hypersusceptible to macrolides and clarithromycin (Li et al., 2022).
WhiB7 is involved also in physiological stress responses and virulence (Buriánková et al., 2004; Geiman et al., 2006; Zaunbrecher et al., 2009; Homolka et al., 2010; Adams et al., 2011; Kim et al., 2012; Larsson et al., 2012; Ramón-García et al., 2012). Interestingly, the role of WhiB7 in providing intrinsic DR is dependent on its interactions with the principal sigma factor σA, and mutations in either whiB7 or sigA preventing their interaction, have been found to cause multidrug susceptibility (Burian et al., 2013; Lilic et al., 2021). In M. smegmatis, WhiB7 was found to positively regulate ms3140, a gene homolog of rv1473 and encoding for an ABC efflux pump involved in macrolide transport (Duan et al., 2019). A similar central role for WhiB7 in DR is found in Actinobacteria, including other mycobacterial opportunistic pathogens such as Mycobacterium abscessus (Ramón-García et al., 2013; Hurst-Hess et al., 2017; Pryjma et al., 2017).
The role of other WhiB family members is more nuanced. WhiB4 regulates β-lactamase expression, thus inducing antibiotic tolerance in M. tuberculosis (Mishra et al., 2017). WhiB2 is part of a regulatory loop involving rv1830 (mcdR) ultimately fine-tuning mycobacterial cell division and adaptation to stress response, including increased mutation rates during antibiotic challenge (Zhou et al., 2022). Recent genome-wide association analysis (GWAS) approaches have also identified novel associations between mutations in the whiB6 region and aminoglycosides resistance (Farhat et al., 2019). Mutations in whiB2 and whiB6 have been observed within patient microevolution during antimycobacterial treatment, however their role in DR was not fully elucidated (Liu et al., 2015; Xu et al., 2018). WhiB3 was the only transcriptional regulator whose structural gene was induced in a model of adaptation to growth with lipids as the sole carbon source (Rodríguez et al., 2014). In this lipid environment, WhiB3 drove increased drug tolerance to RIF, moxifloxacin (MFX), metronidazole (MTZ), and AMK. Interestingly, similar to WhiB7, WhiB3 was found to interact with σA (Burian et al., 2013). Several antimycobacterial compounds can produce an oxidative burst as part of their antimicrobial mechanism (Gurumurthy et al., 2013; Piccaro et al., 2014; Shetty and Dick, 2018). Therefore, transcriptional factors involved in redox homeostasis are relevant in maintaining a reducing microenvironment to avoid DNA damage and macromolecules (i.e. protein and small RNA) misfolding. WhiB3 has been described to negatively regulate the egt operon (rv3700c-rv3704c) encoding for ergothioneine, which together with mycothiol, plays an important role in maintaining the oxidoreduction balance within the bacterial cell (Saini et al., 2016). Both ergothioneine and mycothiol have been proved to be triggered from a wide range of stimuli, from starvation or hypoxia to microenvironmental acidification (i.e. phagolysosome acidification post macrophage infection) (Mavi et al., 2020). Saini and colleagues tested RIF, INH, bedaquiline (BDQ) and clofazimine (CFZ) in rv3704c and rv3701c deficient strains and observed a MIC reduction for all the tested drugs. Accordingly, resistance and tolerance to antimycobacterial drugs impairing redox homeostasis have been directly linked to the intracellular accumulation of ergothioneine (Saini et al., 2016). On the other side, Xu and colleagues produced several strains with mutations in mycothiol-related enzymes demonstrating that such mutants are resistant to both INH and ethionamide (ETO) (Xu et al., 2011).
Transcription factors of the GntR family are widely shared among bacteria, and the firsts members of this family have been described as a gluconate operon repressor in B. subtilis (Vindal et al., 2007; Suvorova et al., 2015). GntR members contain a DNA-binding domain with a structural motif helix-turn-helix (HTH) at their N-terminal, conserved among all the family members, and a more variable C-terminal domain that has been used to divide the GntR factors into six subfamilies (Suvorova et al., 2015). Most of the characterized GntR family members are transcriptional repressors, although some exceptions exist. The M. tuberculosis genome encodes for a large number of GntR family transcription factors, although their role and regulation are still poorly described (Cole et al., 1998).
Among the techniques used to identify transcription factors potentially relevant for DR, over-expressing libraries for transcriptional regulators under selection on high drug concentrations have been shown to be successful tools. For example, Hu and colleagues (Hu et al., 2015) identified a hypothetical transcription factor encoded by the ms0535 gene as a potential contributor to INH resistance in M. smegmatis. Sequence analysis showed that Ms0535 belongs to the GntR family (FadR sub-family). Ms0535 acts as a transcriptional activator for the expression of its own structural gene and a major facilitator superfamily permease gene ms0534 in the same operon, thus triggering INH resistance. Interestingly, the two genes are not responsive to INH, although their over-expression increases INH resistance. Both ms0535 and ms0534 are absent in M. tuberculosis, thus they can contribute to explain the differences in INH resistance between the two species.
In M. tuberculosis there are at least eight putative GntR-like proteins: Rv0043c, Rv0165c (Mce1R), Rv0494, Rv0586 (Mce2R), Rv0792c (MoyR), Rv1152, Rv3060c, and Rv3575c (Vindal et al., 2007; Chauhan et al., 2021). Among them, Rv0494 has an ortholog in M. smegmatis (Ms2173). M. smegmatis mutants for Ms2173 showed altered INH and RIF susceptibility (its over-expression led to increased INH and RIF susceptibility), likely due to the regulatory activity of this transcription factor on membrane-associated transporter genes (Rao et al., 2012).
Rv1152 is involved in the regulation of cell wall permeability (Zeng et al., 2016; Deng et al., 2022). This transcriptional regulator is involved in acid and cell surface stress response and plays an important role in determining VAN resistance by negatively regulating genes responsive to this glycopeptide antibiotic. Indeed, M. smegmatis overexpressing M. tuberculosis Rv1152 showed an increased resistance to VAN, whereas deleting its homologous gene (ms5174) established increased sensitivity that could be restored by complementation with rv1152 (Zeng et al., 2016).
Deletion of rv0792c impaired the ability of M. tuberculosis to infect guinea pigs, however no difference was observed in survival during exposure to INH, RIF, or LEV (Chauhan et al., 2021). A role in DR for the remaining GntR family members has yet to be identified, although some of them have been found upregulated in drug resistant isolates. Interestingly, protein levels of Rv0043c were found less abundant in lineage 7 (L7), however the phenotypic outcomes of these findings remain unknown (Yimer et al., 2020).
XRE response element are among the most widespread regulatory elements in bacteria. They are characterized by a conserved HTH DNA binding domain at their N-terminus and a highly variable C-terminal region. The M. tuberculosis genome encodes for seven members of this family: Rv0023, Rv0465c (RamB), Rv0474, Rv1129 (PrpR), Rv2017, Rv2021, and Rv3849 (EspR).
Rv0023 is a transcription factor modulating nearly 900 genes, and its regulon is enriched for NAD reductases (Rustad et al., 2014). Given the link with NADH/NAD+ regulation, Gupta and coll. explored the role of Rv0023 in INH and ETO tolerance. The overexpression of Rv0023 conferred increased INH and ETO tolerance in M. smegmatis by downregulating the expression of the ndh gene, which encodes for a NADH dehydrogenase. This leads to increased NADH cellular concentration and subsequent inhibition of drug-NAD+ adducts formation, which are essential for INH activity (Gupta et al., 2020). Furthermore, the study found that Rv0023 is also a negative regulator of whiB5.
Rv0465c (RamB), and Rv1129 (PrpR) are involved in the regulation of propionate and acetate metabolisms, respectively. A link between the two metabolic pathways and conditional drug tolerance has been established; however, its biological meaning remains unclear and needs further elucidation (Hicks et al., 2018; Tang et al., 2019; Hicks et al., 2020).
rv2017 was found to be deleted or disrupted by IS6110 in several drug resistant isolates; however, the link between this gene and specific drug resistant phenotype has yet to be defined (Klopper et al., 2020; Perdigão et al., 2020; Antoine et al., 2021). Rv3849 (RspR) was predicted to regulate ponA1, a gene involved in cell wall synthesis relevant for altered fitness in M. tuberculosis during RIF treatment (Farhat et al., 2013; Kieser et al., 2015; Kieser et al., 2015). The remaining XRE transcription regulators in M. tuberculosis are uncharacterized for their role in DR.
There are at least nine genes in the genome of M. tuberculosis annotated as MarR-like proteins. One of the most studied MarR-like family transcriptional regulator is Rv0678. This is a transcriptional repressor of the mmpS5-mmpL5 operon, which encodes an efflux pump able to transport BDQ and CFZ (Milano et al., 2009; Andries et al., 2014). Mutations in rv0678 affecting its binding activity to the promoter region of the mmpS5-mmpL5 operon are relevant markers of BDQ and CFZ resistance in M. tuberculosis (Kadura et al., 2020). Peterson and coll. described BDQ tolerance mediated by Rv0880 and Rv0324, regulators belonging to the MarR and ArsR family, respectively (Peterson et al., 2016). Knockout mutant strains for rv0324 and rv0880 showed hypersensitivity to BDQ, without affecting the susceptibility to other antimycobacterial drugs with unrelated mechanisms of action. Drug tolerance is mediated by the transcriptional cascades modulated by the two transcription factors rather than being caused by genetic mutations. Interestingly, the Rv0324 regulon correlates with nutrient-limited stress condition, which has important implications since BDQ killing depends upon glycolytic pathways (Mackenzie et al., 2020).
Resistance to a novel pyrido-benzimidazole with potent mycobactericidal activity was found to be mediated by mutations in the gene encoding the MarR-like Rv2887 transcription factor (Warrier et al., 2016; Gao et al., 2017). Mutations in this gene negatively affects the ability of the transcription factor to bind its target DNA sequences, ultimately leading to the upregulation of downstream genes. Among them, rv0560c was found to encode for a benzoquinone methyltransferase able to N-methylate and thus inactivate the pyrido-benzimidazole compound (Warrier et al., 2016). Mutations in Rv2887 were also found to abrogate susceptibility to a new imidazopyridine-based drug candidate (Winglee et al., 2015). In this case, DR is mediated by the upregulation of efflux pumps yet to be further identified.
Rv2327 has been hypothesized to participate in INH antibiotic response given its role in the regulation of fbpA and fbpC (encoding antigen 85 complex A and C, respectively), which are involved in cell wall biosynthesis and over-expressed in response to INH treatment (Nguyen et al., 2005; Romero et al., 2010). However, the putative mechanism (e.g. direct interaction Rv2327-INH or other) has not been further explored. Other members of the family have not been directly linked with DR in M. tuberculosis so far.
Regulators of the TetR family usually repress transcription binding their target DNA sequence through a conserved HTH motif present at their N-terminus. Target DNA is released following a structural change of the protein caused by the binding of a specific effector molecule to a ligand-binding pocket situated at the C-terminus of the protein (Balhana et al., 2015). Among the pathways under the control of these regulators, drug efflux is probably the most studied. The TetR family regulator Rv3066 was described to repress the transcription of rv3065 (mmr), a small multidrug resistance (SMR) efflux pump (Bolla et al., 2012). Similarly, Rv1219c was reported to regulate the transcription of the ATP-binding cassette (ABC) transporter encoded by rv1217c-rv1218c (Kumar et al., 2014). Both the SMR and ABC transporters have been found overexpressed in MDR clinical isolates, however experiments with knockout strains, recently challenged their role in direct transport of relevant anti-TB drugs (Wang et al., 2013; Shahi et al., 2021; Remm et al., 2022). A recent study matching genomic mutations and increased MIC levels to several drugs identified mutations mapping in the low-affinity binding domain of Rv1219c associated with increased MIC for INH, thus proposing that the repression of the rv1217c-rv1218c is somehow linked with increased levels of resistance to this first-line drug (Consortium et al., 2021).
Three members of these transcriptional regulators have been directly linked with DR: (i) Rv0275 (InbR) is able to directly interact with INH, and its overexpression is associated with increased resistance to this drug, whereas knockout mutants showed increased susceptibility to several anti-TB drugs (Yang et al., 2018). Genes involved in the INH pathway such as iniABC were shown to belong to its regulon; (ii) Rv3855 (EthR) is well-known for its regulatory role on ethA (rv3854c), which encodes a Baeyer-Villager monooxygenase involved in the activation of ETO (Engohang-Ndong et al., 2004). Despite the role of mutations affecting ethA in ETO resistance is evident (Ushtanit et al., 2022), mutations in its transcriptional regulator seems relatively rare in clinical isolates (da Silva et al., 2018; Mugumbate et al., 2021); (iii) Rv0273c (EtbR) is a transcriptional repressor of the inhA gene, which encodes the target of INH. EMB can bind EtbR, increasing its repressing activity on inhA transcription, thus increasing susceptibility to INH (Zhu et al., 2018).
Further relationships between TetR family transcriptional regulators and DR levels in M. tuberculosis have to be discovered yet. Of note, Rv0302, Rv1816, and Rv3249c are predicted to regulate mmpL3 and mmpL11, two relevant drug target candidates (Domenech et al., 2005; Chou et al., 2015; Delmar et al., 2015).
Among additional transcriptional regulators reported to affect DR in mycobacteria we can mention the members of the AraC/XylS and the SmtB/ArsR families. The AraC/XylS family of transcription factors includes hundreds of positive regulators (Gallegos et al., 1997; Egan, 2002). In M. tuberculosis at least nine members of the AraC/XylS family have been described: Rv0023, Rv0465c (RamB), Rv0474, Rv1129 (PrpR), Rv2017, Rv2021, Rv3082c (VirS), and Rv3849 (EspR). Mutations in rv3082c (virS) were found to mediate resistance to a new putative antimicrobial compound (defined as thiooxadiazole 3) and ETO by affecting the expression of the mymA operon, which is responsible for the activation of these molecules (Grant et al., 2016). Other members of the family are involved in several regulatory functions related to carbon metabolism, stress response, and pathogenesis.
At least 12 ArsR family homologs, including Rv0324, Rv2034 and the metal sensors Rv0827c (KmtR), Rv1994c (CmtR), Rv2358 (SmtB), and Rv3744 (NmtR), have been described in M. tuberculosis. Beside the already mentioned role of Rv0324 in BDQ tolerance (Peterson et al., 2016), no further links with DR have been found for this family of transcriptional regulators. A study showed the Rv2034, a regulator of the ArsR family, regulates whiB7 expression, but its role on the WhiB7 regulon in terms of drug tolerance/resistance has not been further explored (Gao et al., 2012).
Additional transcriptional regulators have been reported to affect DR in mycobacteria. For example, the histone-like Lsr2 protein (encoded by the rv3597c gene) is involved in several regulatory functions involving cell wall biosynthesis, transport, and responses to antibiotic treatment. Lsr2 represses INH-mediated induction of iniBAC and efpA (Colangeli et al., 2007). Interestingly, iniBAC is also under the control of another transcriptional regulator: IniR (Rv0339c) (Boot et al., 2017). Similarly, another histone-like protein, HupB (Rv2986c, also known as MDP-1), negatively regulates katG expression, thus affecting phenotypic tolerance to INH in M. tuberculosis (Niki et al., 2012). Another example is Rv1267c (EmbR), which regulates embABC transcription, well-known to be relevant for EMB resistance (Sharma et al., 2006). An additional case of interest relates to CFZ. Li and coll. described a link between the transcriptional regulator Rv1453 and CFZ resistance, where the overexpression of the gene was associated with increased MIC to the drug (Li et al., 2021).
Several transcription factors are modified post-translationally, thus adding an additional layer of control that can modify the interaction between bacteria and drugs. Among those previously cited, EmbR is positively regulated by phosphorylation, whereas HupB acetylation and methylation alter INH susceptibility in mycobacteria (Arora et al., 2021). Similarly, PknB phosphorylates the histon-like protein Lsr2, thus reducing its DNA binding affinity (Alqaseer et al., 2019). Rv3701c (part of the egt operon), is under the strict regulation of the serine/threonine-protein kinase PknD, and its phosphorylated form fails to catalyzes ergothioneine biosynthesis (Richard-Greenblatt et al., 2015). Therefore, it is plausible that these additional regulatory layers have a role in fine-tuning redox homeostasis, and thus drug tolerance. Moreover, transcription factors are not universally conserved in the M. tuberculosis complex, thus this genetic diversity has to be taken into account systematically when studying genotypic features in relationship with phenotypic drug susceptibility (Köser et al., 2012; Chiner-Oms et al., 2018; Chiner-Oms et al., 2019).
As DR has become a crucial worldwide problem, new strategies to design innovative generations of antibacterial drugs has been implemented. Targeting transcriptional regulation is one of them. An interesting example is that of EthR. The mycobacterial monoxygenase EthA is the activator of several thiocarbamide-containing drugs, including ETO. Its expression is regulated by EthR, a transcriptional repressor. Synthetic compounds selected to inhibit EthR-DNA interaction have been shown to increase ethA expression, boosting bacterial sensitivity to ETO (Willand et al., 2009; Flipo et al., 2012; Nikiforov et al., 2017). Another emerging and promising strategy is targeting riboswitches (Deigan and Ferré-D’Amaré, 2011; Lünse et al., 2014; Dar et al., 2016; Panchal and Brenk, 2021). Metabolite-binding riboswitches are non-coding RNAs that bind specifically to metabolites and regulate downstream gene expression depending on the metabolite concentration. These sequences are composed of two domains: an aptamer that binds the metabolites/ligands, and a response platform that allows the expression or repression of downstream genes. The binding of ligand leads either to transcriptional and/or translational termination of downstream gene expression (switch-OFF) or to the expression of downstream genes (switch-ON) (Bastet et al., 2018; Yadav et al., 2020). Riboswitches occur almost exclusively in prokaryotes and are involved in the metabolism of essential amino acids and metabolites. Although not yet thoroughly studied, some riboswitches are present in pathogenic bacteria, including M. tuberculosis, and play an important role in controlling essential genes. Indeed, many of the classes of riboswitches are fundamental in controlling the expression of genes involved in virulence (Blount and Breaker, 2006; Lünse et al., 2014). Being involved in the expression of essential genes and absent in eukaryotic genomes, riboswitches have become interesting targets for the design of innovative antibacterial strategies. The general idea is to target the pocket recognizing the metabolite to induce a transition of the riboswitch from the ON to the OFF configuration turning off the regulated gene. Riboswitches-based antibacterial drugs could be broad-spectrum, in the case of riboswitches present in an extended range of pathogenic species, or narrow spectrum in the case of species-specific riboswitches (Blount and Breaker, 2006; Panchal and Brenk, 2021). Using this rational some antibacterial compounds targeting riboswitches have already been identified (recently reviewed in [Panchal and Brenk, 2021)]. M. tuberculosis is predicted to have at least 16 riboswitches from the Rfam database belonging to different classes (Kalvari et al., 2018). However, only few riboswitches have been validated in this species (Schwenk and Arnvig, 2018) as the cyclic-di-AMP sensing riboswitch regulating rpfA (Arnvig and Young, 2012; Nelson et al., 2013), the riboswitch regulating rpfB (Schwenk et al., 2018), and the cobalamin-dependent riboswitch responsible for the downregulation of metE in the presence of cobalamin (Warner et al., 2007). The latter case is particularly interesting to show the potential importance of riboswitches as drug targets. M. tuberculosis has two methionine synthases: MetE, whose expression is repressed by cobalamin and MetH, which requires cobalamin for its functionality. In the clinical isolate CDC1551 MetH is not functional due to a mutation in its structural gene, so MetE is the only methionine synthase in this strain. Warner and coll. (Warner et al., 2007) showed that indeed this strain is unable to grow in the presence of cobalamin due to the repression of metE expression. Mutants resistant to cobalamin showed mutations at the level of the riboswitch confirming the role of this regulatory element in metE regulation.
Drug exposure represents a stress for the bacterium, which reacts by changing its transcriptional profile thus activating different stress regulons responsible of setting its own basal level of susceptibility to a specific drug. Sometimes, mutations in transcriptional regulators can structurally modify the bacterial transcriptional profile resulting in a constitutive change in the susceptibility to a given drug. Moreover, as outlined above, post-transcriptional regulation can further modify transcriptional regulation.
Understanding the role of transcription factors in DR pathways is critical not only to improve our knowledge in resistance mechanisms or to detect new genetic markers of resistance. Unravelling the transcriptomic networks would indeed open to new therapeutic opportunities. As an example, it has been recently reported how DR in M. tuberculosis could give rise to collateral sensitivity to β-lactam drugs, where basically anti-TB drugs induces the expression of the gene encoding the transcriptional repressor BlaI and its downstream genes atpH, and sigC, which ultimately inhibits intrinsic β-lactam resistance (Trigos et al., 2021). Finally, small non-coding RNAs and riboswitches have been described to modulate antibiotic tolerance, and resistance in several bacteria (Dar et al., 2016; Dersch et al., 2017; Zhang et al., 2020). The role (if any) in DR of transcriptional and/or post-transcriptional regulation mediated by these molecules in M. tuberculosis has not been elucidated yet and requires further investigation.
PM: Writing - original draft, conceptualization, investigation. RS: Writing - original draft, visualization, investigation. SG; Writing - review & editing, investigation. RP: Writing - review & editing. DMC: Writing - review & editing. RM: Writing - original draft, conceptualization, investigation, supervision. All authors contributed to the article and approved the submitted version.
RM laboratory is founded from the Innovative Medicines Initiative 2 Joint Undertaking (JU) under grant agreement no 853989. RS was supported by the Italian Ministry of Health “Ricerca Finalizzata 2016” under grant agreement GR-2016-02364014 to PM.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adams, K. N., Takaki, K., Connolly, L. E., Wiedenhoft, H., Winglee, K., Humbert, O., et al. (2011). Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell 145, 39–53. doi: 10.1016/j.cell.2011.02.022
Albesa-Jové, D., Comino, N., Tersa, M., Mohorko, E., Urresti, S., Dainese, E., et al. (2015). The redox state regulates the conformation of Rv2466c to activate the antitubercular prodrug TP053. J. Biol. Chem. 290, 31077–31089. doi: 10.1074/jbc.M115.677039
Alqaseer, K., Turapov, O., Barthe, P., Jagatia, H., De Visch, A., Roumestand, C., et al. (2019). Protein kinase b controls Mycobacterium tuberculosis growth via phosphorylation of the transcriptional regulator Lsr2 at threonine 112. Mol. Microbiol. 112, 1847–1862. doi: 10.1111/mmi.14398
Andries, K., Villellas, C., Coeck, N., Thys, K., Gevers, T., Vranckx, L., et al. (2014). Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PloS One 9, e102135. doi: 10.1371/journal.pone.0102135
Antoine, R., Gaudin, C., Hartkoorn, R. C. (2021). Intragenic distribution of IS6110 in clinical Mycobacterium tuberculosis strains: Bioinformatic evidence for gene disruption leading to underdiagnosed antibiotic resistance. Microbiol. Spectr. 9, e0001921. doi: 10.1128/Spectrum.00019-21
Arnvig, K., Young, D. (2012). Non-coding RNA and its potential role in Mycobacterium tuberculosis pathogenesis. RNA Biol. 9, 427–436. doi: 10.4161/rna.20105
Arora, G., Bothra, A., Prosser, G., Arora, K., Sajid, A. (2021). Role of post-translational modifications in the acquisition of drug resistance in Mycobacterium tuberculosis. FEBS J. 288, 3375–3393. doi: 10.1111/febs.15582
Balázsi, G., Heath, A. P., Shi, L., Gennaro, M. L. (2008). The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 4, 225. doi: 10.1038/msb.2008.63
Balhana, R. J., Singla, A., Sikder, M. H., Withers, M., Kendall, S. L. (2015). Global analyses of TetR family transcriptional regulators in mycobacteria indicates conservation across species and diversity in regulated functions. BMC Genomics 16, 479. doi: 10.1186/s12864-015-1696-9
Bansal, R., Anil Kumar, V., Sevalkar, R. R., Singh, P. R., Sarkar, D. (2017). Mycobacterium tuberculosis virulence-regulator PhoP interacts with alternative sigma factor SigE during acid-stress response. Mol. Microbiol. 104, 400–411. doi: 10.1111/mmi.13635
Bastet, L., Turcotte, P., Wade, J. T., Lafontaine, D. A. (2018). Maestro of regulation: Riboswitches orchestrate gene expression at the levels of translation, transcription and mRNA decay. RNA Biol. 15, 679–682. doi: 10.1080/15476286.2018.1451721
Blount, K. F., Breaker, R. R. (2006). Riboswitches as antibacterial drug targets. Nat. Biotechnol. 24, 1558–1564. doi: 10.1038/nbt1268
Boldrin, F., Cioetto Mazzabò, L., Anoosheh, S., Palù, G., Gaudreau, L., Manganelli, R., et al. (2019). Assessing the role of Rv1222 (RseA) as an anti-sigma factor of the Mycobacterium tuberculosis extracytoplasmic sigma factor SigE. Sci. Rep. 9, 4513. doi: 10.1038/s41598-019-41183-4
Bolla, J. R., Do, S. V., Long, F., Dai, L., Su, C. C., Lei, H. T., et al. (2012). Structural and functional analysis of the transcriptional regulator Rv3066 of Mycobacterium tuberculosis. Nucleic Acids Res. 40, 9340–9355. doi: 10.1093/nar/gks677
Boot, M., van Winden, V. J. C., Sparrius, M., van de Weerd, R., Speer, A., Ummels, R., et al. (2017). Cell envelope stress in mycobacteria is regulated by the novel signal transduction ATPase IniR in response to trehalose. PLoS Genet. 13, e1007131. doi: 10.1371/journal.pgen.1007131
Buriánková, K., Doucet-Populaire, F., Dorson, O., Gondran, A., Ghnassia, J. C., Weiser, J., et al. (2004). Molecular basis of intrinsic macrolide resistance in the Mycobacterium tuberculosis complex. Antimicrob. Agents Chemother. 48, 143–150. doi: 10.1128/AAC.48.1.143-150.2004
Burian, J., Ramón-García, S., Howes, C. G., Thompson, C. J. (2012). WhiB7, a transcriptional activator that coordinates physiology with intrinsic drug resistance in Mycobacterium tuberculosis. Expert Rev. Anti Infect. Ther. 10, 1037–1047. doi: 10.1586/eri.12.90
Burian, J., Yim, G., Hsing, M., Axerio-Cilies, P., Cherkasov, A., Spiegelman, G. B., et al. (2013). The mycobacterial antibiotic resistance determinant WhiB7 acts as a transcriptional activator by binding the primary sigma factor SigA (RpoV). Nucleic Acids Res. 41, 10062–10076. doi: 10.1093/nar/gkt751
Bush, M. J. (2018). The actinobacterial WhiB-like (Wbl) family of transcription factors. Mol. Microbiol. 110, 663–676. doi: 10.1111/mmi.14117
Chatterjee, A., Saranath, D., Bhatter, P., Mistry, N. (2013). Global transcriptional profiling of longitudinal clinical isolates of Mycobacterium tuberculosis exhibiting rapid accumulation of drug resistance. PloS One 8, e54717. doi: 10.1371/journal.pone.0054717
Chauhan, N. K., Anand, A., Sharma, A., Dhiman, K., Gosain, T. P., Singh, P., et al. (2021). Structural and functional characterization of Rv0792c from Mycobacterium tuberculosis: identifying small molecule inhibitors against GntR protein. bioRxiv. doi: 10.1101/2021.09.17.460839
Chen, P., Ruiz, R. E., Li, Q., Silver, R. F., Bishai, W. R. (2000). Construction and characterization of a Mycobacterium tuberculosis mutant lacking the alternate sigma factor gene, sigF. Infect. Immun. 68, 5575–5580. doi: 10.1128/IAI.68.10.5575-5580.2000
Chiner-Oms, Á, Berney, M., Boinett, C., González-Candelas, F., Young, D. B., Gagneux, S., et al. (2019). Genome-wide mutational biases fuel transcriptional diversity in the Mycobacterium tuberculosis complex. Nat. Commun. 10, 3994. doi: 10.1038/s41467-019-11948-6
Chiner-Oms, Á, González-Candelas, F., Comas, I. (2018). Gene expression models based on a reference laboratory strain are poor predictors of Mycobacterium tuberculosis complex transcriptional diversity. Sci. Rep. 8, 3813. doi: 10.1038/s41598-018-22237-5
Chou, T. H., Delmar, J. A., Wright, C. C., Kumar, N., Radhakrishnan, A., Doh, J. K., et al. (2015). Crystal structure of the Mycobacterium tuberculosis transcriptional regulator Rv0302. Protein Sci. 24, 1942–1955. doi: 10.1002/pro.2802
Colangeli, R., Helb, D., Vilchèze, C., Hazbón, M. H., Lee, C. G., Safi, H., et al. (2007). Transcriptional regulation of multi-drug tolerance and antibiotic-induced responses by the histone-like protein Lsr2 in M. tuberculosis. PLoS Pathog. 3, e87. doi: 10.1371/journal.ppat.0030087
Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., et al. (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544. doi: 10.1038/31159
The CRyPTIC Consortium. , Earle, S. G., Wilson, D. J. (2022). Genome-wide association studies of global Mycobacterium tuberculosis resistance to 13 antimicrobials in 10,228 genomes identify new resistance mechanisms. PLoS Biol. 20 (8), e3001755. doi: 10.1371/journal.pbio.3001755
Corona, F., Martinez, J. L. (2013). Phenotypic resistance to antibiotics. Antibiotics (Basel) 2, 237–255. doi: 10.3390/antibiotics2020237
Dar, D., Shamir, M., Mellin, J. R., Koutero, M., Stern-Ginossar, N., Cossart, P., et al. (2016). Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science 352, aad9822. doi: 10.1126/science.aad9822
da Silva, D. A., Ferreira, N. V., Rego, A. M., Barbosa, P. C. P., Machado, R. F., Pimentel, A., et al. (2018). Integrated analysis of ethionamide resistance loci in Mycobacterium tuberculosis clinical isolates. Tuberculosis 113, 163–174. doi: 10.1016/j.tube.2018.08.010
Deigan, K. E., Ferré-D’Amaré, A. R. (2011). Riboswitches: discovery of drugs that target bacterial gene-regulatory RNAs. Acc Chem. Res. 44, 1329–1338. doi: 10.1021/ar200039b
Delmar, J. A., Chou, T. H., Wright, C. C., Licon, M. H., Doh, J. K., Radhakrishnan, A., et al. (2015). Structural basis for the regulation of the MmpL transporters of Mycobacterium tuberculosis. J. Biol. Chem. 290, 28559–28574. doi: 10.1074/jbc.M115.683797
Deng, W., Zheng, Z., Chen, Y., Yang, M., Yan, J., Li, W., et al. (2022). Deficiency of GntR family regulator MSMEG_5174 promotes Mycobacterium smegmatis resistance to aminoglycosides via manupulating purine metabolism. Front. Microbiol. 13, 919538. doi: 10.3389/fmicb.2022.919538
Dersch, P., Khan, M. A., Mühlen, S., Görke, B. (2017). Roles of regulatory RNAs for antibiotic resistance in bacteria and their potential value as novel drug targets. Front. Microbiol. 8, 803. doi: 10.3389/fmicb.2017.00803
Domenech, P., Reed, M. B., Barry, C. E. (2005). Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect. Immun. 73, 3492–3501. doi: 10.1128/IAI.73.6.3492-3501.2005
Duan, W., Li, X., Ge, Y., Yu, Z., Li, P., Li, J., et al. (2019). Mycobacterium tuberculosis Rv1473 is a novel macrolides ABC efflux pump regulated by WhiB7. Future Microbiol. 14, 47–59. doi: 10.2217/fmb-2018-0207
Egan, S. M. (2002). Growing repertoire of AraC/XylS activators. J. Bacteriol. 184, 5529–5532. doi: 10.1128/JB.184.20.5529-5532.2002
Engohang-Ndong, J., Baillat, D., Aumercier, M., Bellefontaine, F., Besra, G. S., Locht, C., et al. (2004). EthR, a repressor of the TetR/CamR family implicated in ethionamide resistance in mycobacteria, octamerizes cooperatively on its operator. Mol. Microbiol. 51, 175–188. doi: 10.1046/j.1365-2958.2003.03809.x
Farhat, M. R., Freschi, L., Calderon, R., Ioerger, T., Snyder, M., Meehan, C. J., et al. (2019). GWAS for quantitative resistance phenotypes in Mycobacterium tuberculosis reveals resistance genes and regulatory regions. Nat. Commun. 10, 2128. doi: 10.1038/s41467-019-10110-6
Farhat, M. R., Shapiro, B. J., Kieser, K. J., Sultana, R., Jacobson, K. R., Victor, T. C., et al. (2013). Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat. Genet. 45, 1183–1189. doi: 10.1038/ng.2747
Flipo, M., Willand, N., Lecat-Guillet, N., Hounsou, C., Desroses, M., Leroux, F., et al. (2012). Discovery of novel n-phenylphenoxyacetamide derivatives as EthR inhibitors and ethionamide boosters by combining high-throughput screening and synthesis. J. Med. Chem. 55, 6391–6402. doi: 10.1021/jm300377g
Gallegos, M. T., Schleif, R., Bairoch, A., Hofmann, K., Ramos, J. L. (1997). Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61, 393–410. doi: 10.1128/mmbr.61.4.393-410.1997
Gao, Y. R., Li, D. F., Fleming, J., Zhou, Y. F., Liu, Y., Deng, J. Y., et al. (2017). Structural analysis of the regulatory mechanism of MarR protein Rv2887 in M. tuberculosis. Sci. Rep. 7, 6471. doi: 10.1038/s41598-017-01705-4
Gao, C. H., Yang, M., He, Z. G. (2012). Characterization of a novel ArsR-like regulator encoded by Rv2034 in Mycobacterium tuberculosis. PLoS One 7, e36255. doi: 10.1371/journal.pone.0036255
Geiman, D. E., Raghunand, T. R., Agarwal, N., Bishai, W. R. (2006). Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB-like genes. Antimicrob. Agents Chemother. 50, 2836–2841. doi: 10.1128/AAC.00295-06
Grant, S. S., Wellington, S., Kawate, T., Desjardins, C. A., Silvis, M. R., Wivagg, C., et al. (2016). Baeyer-villiger monooxygenases EthA and MymA are required for activation of replicating and non-replicating Mycobacterium tuberculosis inhibitors. Cell Chem. Biol. 23, 666–677. doi: 10.1016/j.chembiol.2016.05.011
Gupta, S. K., Angara, R. K., Yousuf, S., Reddy, C. G., Ranjan, A. (2020). Ectopic Expression of Rv0023 Mediates Isoniazid/Ethionamide Tolerance via Altering NADH/NAD+ in Mycobacterium smegmatis. Front. Microbiol. 11, 3. doi: 10.3389/fmicb.2020.00003
Gurumurthy, M., Rao, M., Mukherjee, T., Rao, S. P., Boshoff, H. I., Dick, T., et al. (2013). A novel F(420) -dependent anti-oxidant mechanism protects Mycobacterium tuberculosis against oxidative stress and bactericidal agents. Mol. Microbiol. 87, 744–755. doi: 10.1111/mmi.12127
Hartkoorn, R. C., Sala, C., Magnet, S. J., Chen, J. M., Pojer, F., Cole, S. T. (2010). Sigma factor f does not prevent rifampin inhibition of RNA polymerase or cause rifampin tolerance in Mycobacterium tuberculosis. J. Bacteriol 192, 5472–5479. doi: 10.1128/JB.00687-10
Hicks, N. D., Giffen, S. R., Culviner, P. H., Chao, M. C., Dulberger, C. L., Liu, Q., et al. (2020). Mutations in dnaA and a cryptic interaction site increase drug resistance in Mycobacterium tuberculosis. PloS Pathog. 16, e1009063. doi: 10.1371/journal.ppat.1009063
Hicks, N. D., Yang, J., Zhang, X., Zhao, B., Grad, Y. H., Liu, L., et al. (2018). Clinically prevalent mutations in Mycobacterium tuberculosis alter propionate metabolism and mediate multidrug tolerance. Nat. Microbiol. 3, 1032–1042. doi: 10.1038/s41564-018-0218-3
Homolka, S., Niemann, S., Russell, D. G., Rohde, K. H. (2010). Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PloS Pathog. 6, e1000988. doi: 10.1371/journal.ppat.1000988
Hurst-Hess, K., Rudra, P., Ghosh, P. (2017). Mycobacterium abscessus WhiB7 regulates a species-specific repertoire of genes to confer extreme antibiotic resistance. Antimicrob. Agents Chemother. 11, 61. doi: 10.1128/AAC.01347-17
Hu, J., Zhao, L., Yang, M. (2015). A GntR family transcription factor positively regulates mycobacterial isoniazid resistance by controlling the expression of a putative permease. BMC Microbiol. 15, 214. doi: 10.1186/s12866-015-0556-8
Kadura, S., King, N., Nakhoul, M., Zhu, H., Theron, G., Köser, C. U., et al. (2020). Systematic review of mutations associated with resistance to the new and repurposed Mycobacterium tuberculosis drugs bedaquiline, clofazimine, linezolid, delamanid and pretomanid. J. Antimicrob. Chemother. 75, 2031–2043. doi: 10.1093/jac/dkaa136
Kalvari, I., Nawrocki, E. P., Argasinska, J., Quinones-Olvera, N., Finn, R. D., Bateman, A., et al. (2018). Non-coding RNA analysis using the rfam database. Curr. Protoc. Bioinf. 62, e51. doi: 10.1002/cpbi.51
Kieser, K. J., Baranowski, C., Chao, M. C., Long, J. E., Sassetti, C. M., Waldor, M. K., et al. (2015). Peptidoglycan synthesis in Mycobacterium tuberculosis is organized into networks with varying drug susceptibility. Proc. Natl. Acad. Sci. U.S.A. 112, 13087–13092. doi: 10.1073/pnas.1514135112
Kieser, K. J., Boutte, C. C., Kester, J. C., Baer, C. E., Barczak, A. K., Meniche, X., et al. (2015). Phosphorylation of the peptidoglycan synthase PonA1 governs the rate of polar elongation in mycobacteria. PloS Pathog. 11, e1005010. doi: 10.1371/journal.ppat.1005010
Kim, K. H., An, D. R., Song, J., Yoon, J. Y., Kim, H. S., Yoon, H. J., et al. (2012). Mycobacterium tuberculosis eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7. Proc. Natl. Acad. Sci. U.S.A. 109, 7729–7734. doi: 10.1073/pnas.1120251109
Klopper, M., Heupink, T. H., Hill-Cawthorne, G., Streicher, E. M., Dippenaar, A., de Vos, M., et al. (2020). A landscape of genomic alterations at the root of a near-untreatable tuberculosis epidemic. BMC Med. 18, 24. doi: 10.1186/s12916-019-1487-2
Kohanski, M. A., Dwyer, D. J., Hayete, B., Lawrence, C. A., Collins, J. J. (2007). A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130, 797–810. doi: 10.1016/j.cell.2007.06.049
Köser, C. U., Bryant, J. M., Parkhill, J., Peacock, S. J. (2013). Consequences of whiB7 (Rv3197A) mutations in Beijing genotype isolates of the Mycobacterium tuberculosis complex. Antimicrob. Agents Chemother. 57, 3461. doi: 10.1128/AAC.00626-13
Köser, C. U., Feuerriegel, S., Summers, D. K., Archer, J. A., Niemann, S. (2012). Importance of the genetic diversity within the Mycobacterium tuberculosis complex for the development of novel antibiotics and diagnostic tests of drug resistance. Antimicrob. Agents Chemother. 56, 6080–6087. doi: 10.1128/AAC.01641-12
Kumar, N., Radhakrishnan, A., Wright, C. C., Chou, T. H., Lei, H. T., Bolla, J. R., et al. (2014). Crystal structure of the transcriptional regulator Rv1219c of Mycobacterium tuberculosis. Protein Sci. 23, 423–432. doi: 10.1002/pro.2424
Larsson, C., Luna, B., Ammerman, N. C., Maiga, M., Agarwal, N., Bishai, W. R. (2012). Gene expression of Mycobacterium tuberculosis putative transcription factors whiB1-7 in redox environments. PloS One 7, e37516. doi: 10.1371/journal.pone.0037516
Lee, J. H., Ammerman, N. C., Nolan, S., Geiman, D. E., Lun, S., Guo, H., et al. (2012). Isoniazid resistance without a loss of fitness in Mycobacterium tuberculosis. Nat. Commun. 3, 753. doi: 10.1038/ncomms1724
Li, Y., Fu, L., Zhang, W., Chen, X., Lu, Y. (2021). The transcription factor Rv1453 regulates the expression of qor and confers resistant to clofazimine in Mycobacterium tuberculosis. Infect. Drug Resist. 14, 3937–3948. doi: 10.2147/IDR.S324043
Lilic, M., Darst, S. A., Campbell, E. A. (2021). Structural basis of transcriptional activation by the Mycobacterium tuberculosis intrinsic antibiotic-resistance transcription factor WhiB7. Mol. Cell 81, 2875–2886, e2875. doi: 10.1016/j.molcel.2021.05.017
Li, S., Poulton, N. C., Chang, J. S., Azadian, Z. A., DeJesus, M. A., Ruecker, N., et al. (2022). CRISPRi chemical genetics and comparative genomics identify genes mediating drug potency in Mycobacterium tuberculosis. Nat. Microbiol. 7, 766–779. doi: 10.1038/s41564-022-01130-y
Liu, J., Shi, W., Zhang, S., Hao, X., Maslov, D. A., Shur, K. V., et al. (2019). Mutations in efflux pump Rv1258c (Tap) cause resistance to pyrazinamide, isoniazid, and streptomycin in M. tuberculosis. Front. Microbiol. 10, 216. doi: 10.3389/fmicb.2019.00216
Liu, Q., Via, L. E., Luo, T., Liang, L., Liu, X., Wu, S., et al. (2015). Within patient microevolution of Mycobacterium tuberculosis correlates with heterogeneous responses to treatment. Sci. Rep. 5, 17507. doi: 10.1038/srep17507
Lünse, C. E., Schüller, A., Mayer, G. (2014). The promise of riboswitches as potential antibacterial drug targets. Int. J. Med. Microbiol. 304, 79–92. doi: 10.1016/j.ijmm.2013.09.002
Mackenzie, J. S., Lamprecht, D. A., Asmal, R., Adamson, J. H., Borah, K., Beste, D. J. V., et al. (2020). Bedaquiline reprograms central metabolism to reveal glycolytic vulnerability in Mycobacterium tuberculosis. Nat. Commun. 11, 6092. doi: 10.1038/s41467-020-19959-4
Manganelli, R., Provvedi, R. (2010). An integrated regulatory network including two positive feedback loops to modulate the activity of sigma(E) in mycobacteria. Mol. Microbiol. 75, 538–542. doi: 10.1111/j.1365-2958.2009.07009.x
Manganelli, R., Voskuil, M. I., Schoolnik, G. K., Dubnau, E., Gomez, M., Smith, I. (2002). Role of the extracytoplasmic-function sigma factor sigma(H) in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45, 365–374. doi: 10.1046/j.1365-2958.2002.03005.x
Martínez, S. R., Durantini, A. M., Becerra, M. C., Cosa, G. (2020). Real-time single-cell imaging reveals accelerating lipid peroxyl radical formation in Escherichia coli Triggered by a Fluoroquinolone Antibiotic. ACS Infect. Dis. 6, 2468–2477. doi: 10.1021/acsinfecdis.0c00317
Mavi, P. S., Singh, S., Kumar, A. (2020). Reductive stress: New insights in physiology and drug tolerance of Mycobacterium. Antioxid Redox Signal 32, 1348–1366. doi: 10.1089/ars.2019.7867
Michele, T. M., Ko, C., Bishai, W. R. (1999). Exposure to antibiotics induces expression of the Mycobacterium tuberculosis sigF gene: implications for chemotherapy against mycobacterial persistors. Antimicrob. Agents Chemother. 43, 218–225. doi: 10.1128/AAC.43.2.218
Milano, A., Pasca, M. R., Provvedi, R., Lucarelli, A. P., Manina, G., Ribeiro, A. L., et al. (2009). Azole resistance in Mycobacterium tuberculosis is mediated by the MmpS5-MmpL5 efflux system. Tuberculosis (Edinb) 89, 84–90. doi: 10.1016/j.tube.2008.08.003
Mishra, S., Shukla, P., Bhaskar, A., Anand, K., Baloni, P., Jha, R. K., et al. (2017). Efficacy of β-lactam/β-lactamase inhibitor combination is linked to WhiB4-mediated changes in redox physiology of Mycobacterium tuberculosis. Elife 26, 6. doi: 10.7554/eLife.25624.037
Morris, R. P., Nguyen, L., Gatfield, J., Visconti, K., Nguyen, K., Schnappinger, D., et al. (2005). Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 102, 12200–12205. doi: 10.1073/pnas.0505446102
Mugumbate, G., Nyathi, B., Zindoga, A., Munyuki, G. (2021). Application of computational methods in understanding mutations in Mycobacterium tuberculosis drug resistance. Front. Mol. Biosci. 8, 643849. doi: 10.3389/fmolb.2021.643849
Nelson, J. W., Sudarsan, N., Furukawa, K., Weinberg, Z., Wang, J. X., Breaker, R. R. (2013). Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat. Chem. Biol. 9, 834–839. doi: 10.1038/nchembio.1363
Nguyen, L., Chinnapapagari, S., Thompson, C. J. (2005). FbpA-dependent biosynthesis of trehalose dimycolate is required for the intrinsic multidrug resistance, cell wall structure, and colonial morphology of Mycobacterium smegmatis. J. Bacteriol. 187, 6603–6611. doi: 10.1128/JB.187.19.6603-6611.2005
Nikiforov, P. O., Blaszczyk, M., Surade, S., Boshoff, H. I., Sajid, A., Delorme, V., et al. (2017). Fragment-sized EthR inhibitors exhibit exceptionally strong ethionamide boosting effect in whole-cell Mycobacterium tuberculosis assays. ACS Chem. Biol. 12, 1390–1396. doi: 10.1021/acschembio.7b00091
Niki, M., Tateishi, Y., Ozeki, Y., Kirikae, T., Lewin, A., Inoue, Y., et al. (2012). A novel mechanism of growth phase-dependent tolerance to isoniazid in mycobacteria. J. Biol. Chem. 287, 27743–27752. doi: 10.1074/jbc.M111.333385
Panchal, V., Brenk, R. (2021). Riboswitches as Drug Targets for Antibiotics. Antibiotics. 10, 45. doi: 10.3390/antibiotics10010045
Perdigão, J., Gomes, P., Miranda, A., Maltez, F., Machado, D., Silva, C., et al. (2020). Using genomics to understand the origin and dispersion of multidrug and extensively drug resistant tuberculosis in Portugal. Sci. Rep. 10, 2600. doi: 10.1038/s41598-020-59558-3
Peterson, E. J. R., Ma, S., Sherman, D. R., Baliga, N. S. (2016). Network analysis identifies Rv0324 and Rv0880 as regulators of bedaquiline tolerance in Mycobacterium tuberculosis. Nat. Microbiol. 1, 16078. doi: 10.1038/nmicrobiol.2016.78
Piccaro, G., Pietraforte, D., Giannoni, F., Mustazzolu, A., Fattorini, L. (2014). Rifampin induces hydroxyl radical formation in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 7527–7533. doi: 10.1128/AAC.03169-14
Pisu, D., Provvedi, R., Espinosa, D. M., Payan, J. B., Boldrin, F., Palù, G., et al. (2017). The alternative sigma factors SigE and SigB are involved in tolerance and persistence to antitubercular drugs. Antimicrob. Agents Chemother. 12, 61. doi: 10.1128/AAC.01596-17
Provvedi, R., Boldrin, F., Falciani, F., Palù, G., Manganelli, R. (2009). Global transcriptional response to vancomycin in Mycobacterium tuberculosis. Microbiology 155, 1093–1102. doi: 10.1099/mic.0.024802-0
Pryjma, M., Burian, J., Kuchinski, K., Thompson, C. J. (2017). Antagonism between front-line antibiotics clarithromycin and amikacin in the treatment of Mycobacterium abscessus infections is mediated by the whiB7 gene. Antimicrob. Agents Chemother. 11, 61. doi: 10.1128/AAC.01353-17
Ramón-García, S., Mick, V., Dainese, E., Martín, C., Thompson, C. J., De Rossi, E., et al. (2012). Functional and genetic characterization of the tap efflux pump in Mycobacterium bovis BCG. Antimicrob. Agents Chemother. 56, 2074–2083. doi: 10.1128/AAC.05946-11
Ramón-García, S., Ng, C., Jensen, P. R., Dosanjh, M., Burian, J., Morris, R. P., et al. (2013). WhiB7, an Fe-S-dependent transcription factor that activates species-specific repertoires of drug resistance determinants in actinobacteria. J. Biol. Chem. 288, 34514–34528. doi: 10.1074/jbc.M113.516385
Rao, M., Liu, H., Yang, M., Zhao, C., He, Z. G. (2012). A copper-responsive global repressor regulates expression of diverse membrane-associated transporters and bacterial drug resistance in mycobacteria. J. Biol. Chem. 287, 39721–39731. doi: 10.1074/jbc.M112.383604
Reeves, A. Z., Campbell, P. J., Sultana, R., Malik, S., Murray, M., Plikaytis, B. B., et al. (2013). Aminoglycoside cross-resistance in Mycobacterium tuberculosis due to mutations in the 5' untranslated region of whiB7. Antimicrob. Agents Chemother. 57, 1857–1865. doi: 10.1128/AAC.02191-12
Remm, S., Earp, J. C., Dick, T., Dartois, V., Seeger, M. A. (2022). Critical discussion on drug efflux in Mycobacterium tuberculosis. FEMS Microbiol. Rev. 09, 46. doi: 10.1093/femsre/fuab050
Richard-Greenblatt, M., Bach, H., Adamson, J., Peña-Diaz, S., Li, W., Steyn, A. J., et al. (2015). Regulation of ergothioneine biosynthesis and its effect on Mycobacterium tuberculosis growth and infectivity. J. Biol. Chem. 290, 23064–23076. doi: 10.1074/jbc.M115.648642
Rodrigue, S., Provvedi, R., Jacques, P. E., Gaudreau, L., Manganelli, R. (2006). The sigma factors of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 30, 926–941. doi: 10.1111/j.1574-6976.2006.00040.x
Rodríguez, J. G., Hernández, A. C., Helguera-Repetto, C., Aguilar Ayala, D., Guadarrama-Medina, R., Anzóla, J. M., et al. (2014). Global adaptation to a lipid environment triggers the dormancy-related phenotype of Mycobacterium tuberculosis. mBio 5, e01125–e01114. doi: 10.1128/mBio.01125-14
Romero, I. C., Mehaffy, C., Burchmore, R. J., Dobos-Elder, K., Brennan, P., Walker, J. (2010). Identification of promoter-binding proteins of the fbp a and c genes in Mycobacterium tuberculosis. Tuberculosis 90, 25–30. doi: 10.1016/j.tube.2009.10.001
Rosado, L. A., Wahni, K., Degiacomi, G., Pedre, B., Young, D., de la Rubia, A. G., et al. (2017). The antibacterial prodrug activator Rv2466c is a mycothiol-dependent reductase in the oxidative stress response of Mycobacterium tuberculosis. J. Biol. Chem. 292, 13097–13110. doi: 10.1074/jbc.M117.797837
Rustad, T. R., Minch, K. J., Ma, S., Winkler, J. K., Hobbs, S., Hickey, M., et al. (2014). Mapping and manipulating the Mycobacterium tuberculosis transcriptome using a transcription factor overexpression-derived regulatory network. Genome Biol. 15, 502. doi: 10.1186/s13059-014-0502-3
Saini, V., Cumming, B. M., Guidry, L., Lamprecht, D. A., Adamson, J. H., Reddy, V. P., et al. (2016). Ergothioneine maintains redox and bioenergetic homeostasis essential for drug susceptibility and virulence of Mycobacterium tuberculosis. Cell Rep. 14, 572–585. doi: 10.1016/j.celrep.2015.12.056
Schrader, S. M., Botella, H., Jansen, R., Ehrt, S., Rhee, K., Nathan, C., et al. (2021). Multiform antimicrobial resistance from a metabolic mutation. Sci. Adv. 08, 7. doi: 10.1126/sciadv.abh2037
Schwenk, S., Arnvig, K. B. (2018). Regulatory RNA in Mycobacterium tuberculosis, back to basics. Pathog. Dis. 06 01, 76. doi: 10.1093/femspd/fty035
Schwenk, S., Moores, A., Nobeli, I., McHugh, T. D., Arnvig, K. B. (2018). Cell-wall synthesis and ribosome maturation are co-regulated by an RNA switch in Mycobacterium tuberculosis. Nucleic Acids Res. 46, 5837–5849. doi: 10.1093/nar/gky226
Shahi, F., Khosravi, A. D., Tabandeh, M. R., Salmanzadeh, S. (2021). Investigation of the Rv3065, Rv2942, Rv1258c, Rv1410c, and Rv2459 efflux pump genes expression among multidrug-resistant Mycobacterium tuberculosis clinical isolates. Heliyon 7, e07566. doi: 10.1016/j.heliyon.2021.e07566
Sharma, K., Gupta, M., Pathak, M., Gupta, N., Koul, A., Sarangi, S., et al. (2006). Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, EmbR, in vivo. J. Bacteriol 188, 2936–2944. doi: 10.1128/JB.188.8.2936-2944.2006
Shetty, A., Dick, T. (2018). Mycobacterial cell wall synthesis inhibitors cause lethal ATP burst. Front. Microbiol. 9, 1898. doi: 10.3389/fmicb.2018.01898
Sureka, K., Ghosh, B., Dasgupta, A., Basu, J., Kundu, M., Bose, I. (2008). Positive feedback and noise activate the stringent response regulator rel in mycobacteria. PloS One 3, e1771. doi: 10.1371/journal.pone.0001771
Suvorova, I. A., Korostelev, Y. D., Gelfand, M. S. (2015). GntR family of bacterial transcription factors and their DNA binding motifs: Structure, positioning and Co-evolution. PloS One 10, e0132618. doi: 10.1371/journal.pone.0132618
Tang, S., Hicks, N. D., Cheng, Y. S., Silva, A., Fortune, S. M., Sacchettini, J. C. (2019). Structural and functional insight into the Mycobacterium tuberculosis protein PrpR reveals a novel type of transcription factor. Nucleic Acids Res. 47, 9934–9949. doi: 10.1093/nar/gkz724
Thiede, J. M., Dillon, N. A., Howe, M. D., Aflakpui, R., Modlin, S. J., Hoffner, S. E., et al. (2022). Pyrazinamide susceptibility is driven by activation of the SigE-dependent cell envelope stress response in Mycobacterium tuberculosis. mBio 01, e0043921. doi: 10.1128/mbio.00439-21
Tiwari, A., Balázsi, G., Gennaro, M. L., Igoshin, O. A. (2010). The interplay of multiple feedback loops with post-translational kinetics results in bistability of mycobacterial stress response. Phys. Biol. 7, 036005. doi: 10.1088/1478-3975/7/3/036005
Trigos, A. S., Goudey, B. W., Bedő, J., Conway, T. C., Faux, N. G., Wyres, K. L. (2021). Collateral sensitivity to β-lactam drugs in drug-resistant tuberculosis is driven by the transcriptional wiring of BlaI operon genes. mSphere 6, e0024521. doi: 10.1128/mSphere.00245-21
Ushtanit, A., Kulagina, E., Mikhailova, Y., Makarova, M., Safonova, S., Zimenkov, D. (2022). Molecular Determinants of Ethionamide Resistance in Clinical Isolates of Mycobacterium tuberculosis. Antibiotics (Basel). 20, 11. doi: 10.3390/antibiotics11020133
Vargas, R., Freschi, L., Spitaleri, A., Tahseen, S., Barilar, I., Niemann, S., et al. (2021). Role of epistasis in amikacin, kanamycin, bedaquiline, and clofazimine resistance in Mycobacterium tuberculosis complex. Antimicrob. Agents Chemother. 65, e0116421. doi: 10.1128/AAC.01164-21
Vindal, V., Ranjan, S., Ranjan, A. (2007). In silico analysis and characterization of GntR family of regulators from Mycobacterium tuberculosis. Tuberculosis 87, 242–247. doi: 10.1016/j.tube.2006.11.002
Wang, K., Pei, H., Huang, B., Zhu, X., Zhang, J., Zhou, B., et al. (2013). The expression of ABC efflux pump, Rv1217c-Rv1218c, and its association with multidrug resistance of Mycobacterium tuberculosis in China. Curr. Microbiol. 66, 222–226. doi: 10.1007/s00284-012-0215-3
Warner, D. F., Savvi, S., Mizrahi, V., Dawes, S. S. (2007). A riboswitch regulates expression of the coenzyme B12-independent methionine synthase in Mycobacterium tuberculosis: implications for differential methionine synthase function in strains H37Rv and CDC1551. J. Bacteriol 189, 3655–3659. doi: 10.1128/JB.00040-07
Warrier, T., Kapilashrami, K., Argyrou, A., Ioerger, T. R., Little, D., Murphy, K. C., et al. (2016). N-methylation of a bactericidal compound as a resistance mechanism in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 113, E4523–E4530. doi: 10.1073/pnas.1606590113
Willand, N., Dirié, B., Carette, X., Bifani, P., Singhal, A., Desroses, M., et al. (2009). Synthetic EthR inhibitors boost antituberculous activity of ethionamide. Nat. Med. 15, 537–544. doi: 10.1038/nm.1950
Winglee, K., Lun, S., Pieroni, M., Kozikowski, A., Bishai, W. (2015). Mutation of Rv2887, a marR-like gene, confers Mycobacterium tuberculosis resistance to an imidazopyridine-based agent. Antimicrob. Agents Chemother. 59, 6873–6881. doi: 10.1128/AAC.01341-15
Xu, Y., Liu, F., Chen, S., Wu, J., Hu, Y., Zhu, B., et al. (2018). In vivo evolution of drug-resistant Mycobacterium tuberculosis in patients during long-term treatment. BMC Genomics 19, 640. doi: 10.1186/s12864-018-5010-5
Xu, X., Vilchèze, C., Av-Gay, Y., Gómez-Velasco, A., Jacobs, W. R. (2011). Precise null deletion mutations of the mycothiol synthesis genes reveal their role in isoniazid and ethionamide resistance in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 55, 3133–3139. doi: 10.1128/AAC.00020-11
Yadav, B., Sellamuthu, B., Tyagi, R. D. (2020). “12 - riboswitches and aptamers: potential future targets to control drug-resistant bacteria,” in Current developments in biotechnology and bioengineering (Amsterdam, The Netherlands:Elsevier), 421–462.
Yang, S. S., Hu, Y. B., Wang, X. D., Gao, Y. R., Li, K., Zhang, X. E., et al. (2017). Deletion of sigB causes increased sensitivity to para-aminosalicylic acid and sulfamethoxazole in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 10, 61. doi: 10.1128/AAC.00551-17
Yang, J. S., Kim, K. J., Choi, H., Lee, S. H. (2018). Delamanid, bedaquiline, and linezolid minimum inhibitory concentration distributions and resistance-related gene mutations in multidrug-resistant and extensively drug-resistant tuberculosis in Korea. Ann. Lab. Med. 38, 563–568. doi: 10.3343/alm.2018.38.6.563
Yimer, S. A., Kalayou, S., Homberset, H., Birhanu, A. G., Riaz, T., Zegeye, E. D., et al. (2020). Lineage-specific proteomic signatures in the Mycobacterium tuberculosis complex reveal differential abundance of proteins involved in virulence, DNA repair, CRISPR-cas, bioenergetics and lipid metabolism. Front. Microbiol. 11, 550760. doi: 10.3389/fmicb.2020.550760
Zaunbrecher, M. A., Sikes, R. D., Metchock, B., Shinnick, T. M., Posey, J. E. (2009). Overexpression of the chromosomally encoded aminoglycoside acetyltransferase eis confers kanamycin resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 106, 20004–20009. doi: 10.1073/pnas.0907925106
Zeng, J., Deng, W., Yang, W., Luo, H., Duan, X., Xie, L., et al. (2016). Mycobacterium tuberculosis Rv1152 is a novel GntR family transcriptional regulator involved in intrinsic vancomycin resistance and is a potential vancomycin adjuvant target. Sci. Rep. 6, 28002. doi: 10.1038/srep28002
Zhang, J., Liu, G., Zhang, X., Chang, Y., Wang, S., He, W., et al. (2020). Aminoglycoside riboswitch control of the expression of integron associated aminoglycoside resistance adenyltransferases. Virulence 11, 1432–1442. doi: 10.1080/21505594.2020.1836910
Zhou, W., Huang, S., Cumming, B. M., Zhang, Y., Tang, W., Steyn, A. J. C., et al. (2022). A feedback regulatory loop containing McdR and WhiB2 controls cell division and DNA repair in mycobacteria. mBio. Mar. 31, e0334321. doi: 10.1128/mbio.03343-21
Zhu, C., Liu, Y., Hu, L., Yang, M., He, Z. G. (2018). Molecular mechanism of the synergistic activity of ethambutol and isoniazid against. J. Biol. Chem. 293, 16741–16750. doi: 10.1074/jbc.RA118.002693
Keywords: Mycobacteria, drug resistance, transcriptional regulation, tuberculosis, sigma factors, riboswitch
Citation: Miotto P, Sorrentino R, De Giorgi S, Provvedi R, Cirillo DM and Manganelli R (2022) Transcriptional regulation and drug resistance in Mycobacterium tuberculosis. Front. Cell. Infect. Microbiol. 12:990312. doi: 10.3389/fcimb.2022.990312
Received: 09 July 2022; Accepted: 11 August 2022;
Published: 02 September 2022.
Edited by:
Ben Gold, Weill Cornell Medicine, United StatesReviewed by:
Jim Sun, University of Ottawa, CanadaCopyright © 2022 Miotto, Sorrentino, De Giorgi, Provvedi, Cirillo and Manganelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Riccardo Manganelli, cmljY2FyZG8ubWFuZ2FuZWxsaUB1bmlwZC5pdA==
†Present address: Stefano De Giorgi, Clinical Microbiology, ASST Grande Ospedale Metropolitano Niguarda, Milano, Italy
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.