
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 08 September 2022
Sec. Clinical Microbiology
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.988236
This article is part of the Research TopicMobile Genetic Elements as Dissemination Drivers of Multidrug-Resistant Gram-Negative BacteriaView all 5 articles
 Zhijian Yu1†
Zhijian Yu1† Zhengrong Zhang2†
Zhengrong Zhang2† Lile Shi3†
Lile Shi3† Shengni Hua4
Shengni Hua4 Ting Luan5
Ting Luan5 Qiuping Lin6
Qiuping Lin6 Zhixiong Zheng6
Zhixiong Zheng6 Xiaosan Feng7*
Xiaosan Feng7* Mubiao Liu8*
Mubiao Liu8* Xiaobin Li6*
Xiaobin Li6*Carbapenem-resistant Enterobacterales poses a global urgent antibiotic resistance threat because of its ability to transfer carbapenemase genes to other bacteria via horizontal gene transfer mediated by mobile genetic elements such as plasmids. Oxacillinase-181 (OXA-181) is one of the most common OXA-48-like carbapenemases, and OXA-181-producing Enterobacterales has been reported in many countries worldwide. However, systematic research concerning the overall picture of plasmids harboring blaOXA-181 in Enterobacterales is currently scarce. In this study, we aimed to determine the phylogeny and evolution of blaOXA-181-positive (gene encoding OXA-181) plasmids. To characterize the plasmids harboring blaOXA-181 in Enterobacterales, we identified 81 blaOXA-181-positive plasmids from 35,150 bacterial plasmids downloaded from the NCBI RefSeq database. Our results indicated that diverse plasmid types harbored blaOXA-181 but was predominantly carried by IncX3-type plasmids. We systematically compared the host strains, plasmid types, conjugative transfer regions, and genetic contexts of blaOXA-181 among the 66 blaOXA-181-positive IncX3 plasmids. We found that IncX3 plasmids harboring blaOXA-181 were mostly ColKP3-IncX3 hybrid plasmids with a length of 51 kb each and were mainly distributed in Escherichia coli and Klebsiella pneumoniae. Most of the IncX3 plasmids harboring blaOXA-181 were human origin. Almost all the blaOXA-181-positive IncX3 plasmids were found to carry genes coding for relaxases of the MOBP family and VirB-like type IV secretion system (T4SS) gene clusters, and all the 66 IncX3 plasmids were found to carry the genes encoding type IV coupling proteins (T4CPs) of the VirD4/TraG subfamily. Most IncX3 plasmids harbored both blaOXA-181 and qnrS1 in their genomes, and the two antibiotic resistance genes were found to a composite transposon bracketed by two copies of insertion sequence IS26 in the same orientation. Our findings provide important insights into the phylogeny and evolution of blaOXA-181-positive IncX3 plasmids and further address their role in acquiring and spreading blaOXA-181 genes in Enterobacterales.
The rapid increase in carbapenemase-producing Enterobacterales has become a public-health threat (Kim et al., 2021). Surveillance studies have shown that OXA-48-like carbapenemases are the most common carbapenemases in Enterobacterales in certain regions of the world (Pitout et al., 2019). Oxacillinase-181 (OXA-181) is a carbapenem-hydrolyzing class D β-lactamase, a variant of OXA-48 differing by four amino acid substitutions, possessing a higher ability to hydrolyze carbapenems (Potron et al., 2011; Oueslati et al., 2015). The OXA-181 was first reported in clinical carbapenem-resistant Klebsiella pneumoniae (K. pneumoniae) and Enterobacter cloacae strains in Indian hospitals in 2007 (Castanheira et al., 2011). Since then, OXA-181-producing Enterobacterales, mainly K. pneumoniae and Escherichia coli (E. coli), have been reported in several countries worldwide (Balm et al., 2013; Liu et al., 2015; Rojas et al., 2017; Piazza et al., 2018; Mouftah et al., 2019), indicating a trend of increasing prevalence.
Enterobacterales cause both hospital- and community-acquired infections (Rood and Li, 2017). Carbapenem-resistant Enterobacterales (CRE) has now emerged worldwide as an urgent antibiotic resistance threat because these bacteria can transfer carbapenemase genes to other bacteria via horizontal gene transfer mediated by mobile genetic elements such as plasmids (Nordmann et al., 2011). Antimicrobial resistance (AMR) in CRE isolates is frequently encoded by plasmid-borne genes, and can disseminate clonally or horizontally (Rozwandowicz et al., 2018). Four different plasmid types belonging to the ColKP3, IncX3, IncN1, and IncT replicon types have been reported to harbor OXA-181 gene (blaOXA-181) (Pitout et al., 2019).
Conjugative plasmids are important vehicles for the dissemination of antibiotic resistant genes (ARGs) (Smillie et al., 2010; Ravi et al., 2018). These plasmids typically have the conserved backbone regions and the variable accessory regions (Brown et al., 2013; Sitter et al., 2021). The former contains genes encoding plasmid-related traits, such as replication control and conjugation functions, while the latter have accessory genes, such as genes encoding antibiotic resistance, which are usually located on the transposons or integrons (Norman et al., 2009; Norberg et al., 2011). However, systematic research on the backbone and accessory regions of plasmids harboring blaOXA-181 in Enterobacterales is currently scarce.
In this study, we performed in silico typing and comparative analysis of blaOXA-181-positive plasmids from 35,150 bacterial plasmids downloaded from the NCBI RefSeq database. We analyzed and compared the host strains, plasmid replicon types, conjugative transfer regions, and genetic contexts of the blaOXA-181 gene among the blaOXA-181-positive plasmids. This study provides important insights into the phylogeny and evolution of blaOXA-181-positive plasmids and further addresses their role in the acquisition and spread of ARGs.
RefSeq database at NCBI (O’Leary et al., 2016) is a comprehensive, integrated, non-redundant, well-annotated set of reference sequences. Via the FTP release directory “Plasmid (https://ftp.ncbi.nih.gov/refseq/release/plasmid/),” we accessed and downloaded all the plasmids available in the RefSeq database. A total of 35,150 bacterial plasmid genomic sequences were downloaded on July 14, 2021 from the NCBI RefSeq database (Table S1). The genome data (FASTA DNA format) were downloaded in batches using two Bioperl modules including Bio::DB::GenBank and Bio::SeqIO.
The β-lactamase genes within the genomes of plasmids were identified in the 35,150 plasmids using the ResFinder software version 4.1 (https://cge.cbs.dtu.dk/services/ResFinder/) (Bortolaia et al., 2020), with a minimum coverage of 60% and minimum identity of 90%. The term “blaOXA-181” was used to search in the “Resistance gene” list within the ResFinder results to determine blaOXA-181-positive plasmids.
Plasmid replicon typing was performed using the PlasmidFinder software (https://cge.cbs.dtu.dk/services/PlasmidFinder/) (Carattoli and Hasman, 2020). Selecting the database “Enterobacterales”, the DNA files in FASTA format were analyzed in batches using the PlasmidFinder software, with minimum coverage of 60% and minimum identity of 95%.
The files of the blaOXA-181-positive plasmids identified by ResFinder in GenBank format were downloaded in batches using the Bio::DB::GenBank and Bio::SeqIO modules. Files containing protein sequences were extracted from the files in GenBank format using the Bioperl/Bio::SeqIO module. For all blaOXA-181-positive plasmids, phylogenetic patterns based on the presence/absence of orthologous gene families were analyzed. A binary protein presence/absence matrix was created using OrthoFinder (http://www.stevekellylab.com/software/orthofinder) (Emms and Kelly, 2019) with DIAMOND for sequence similarity searches, and then a hierarchical cluster result was generated by PAST3 (Hammer et al., 2001) and eventually displayed by iTOL (https://itol.embl.de/) (Letunic and Bork, 2016).
To determine the presence/absence of oriTs, relaxase genes, T4CP genes, and gene cluster for T4SS, the files in GenBank format of the blaOXA-181-positive plasmids were analyzed in batches using the software oriTfinder (https://bioinfo-mml.sjtu.edu.cn/oriTfinder/) (Li et al., 2018) (local version). Furthermore, the types of oriTs, relaxase genes, and T4CP genes were identified based on the exhibition of oriTDB database (https://bioinfo-mml.sjtu.edu.cn/oriTDB/index.php) (Li et al., 2018). In addition, the types of gene clusters for T4SS were classified based on the SecReT4 database (https://bioinfo-mml.sjtu.edu.cn/SecReT4/) (Bi et al., 2013).
The bacterial insertion sequences within the blaOXA-181-positive plasmids in Enterobacterales were explored using ISfinder software (Siguier et al., 2006). Comparisons among the genetic contexts of blaOXA-181 were performed using BLAST Ring Image Generator (BRIG) (Alikhan et al., 2011).
Using ResFinder, 81 plasmids bearing blaOXA-181 (Table S2) were identified from 35150 bacterial plasmids downloaded from the NCBI RefSeq database. By analyzing the taxonomy of the bacterial strains containing the blaOXA-181-positive plasmids, we found that the 81 plasmids were distributed in seven different species (Figure 1A). The predominant species carrying blaOXA-181-positive plasmids was E. coli, accounting for 59.26% (48 blaOXA-181-positive plasmids), followed by K. pneumoniae, accounting for 30.10% (26 blaOXA-181-positive plasmids). Other species with blaOXA-181-positive plasmids were Enterobacter hormaechei (two plasmids), Morganella morganii (two plasmids), Citrobacter freundii (one plasmid), Klebsiella variicola (one plasmid), and Providencia rettgeri (one plasmid). Overall, all the strains harboring blaOXA-181-positive plasmids belonged to the order Enterobacterales, including family Enterobacteriaceae and family Morganellaceae.
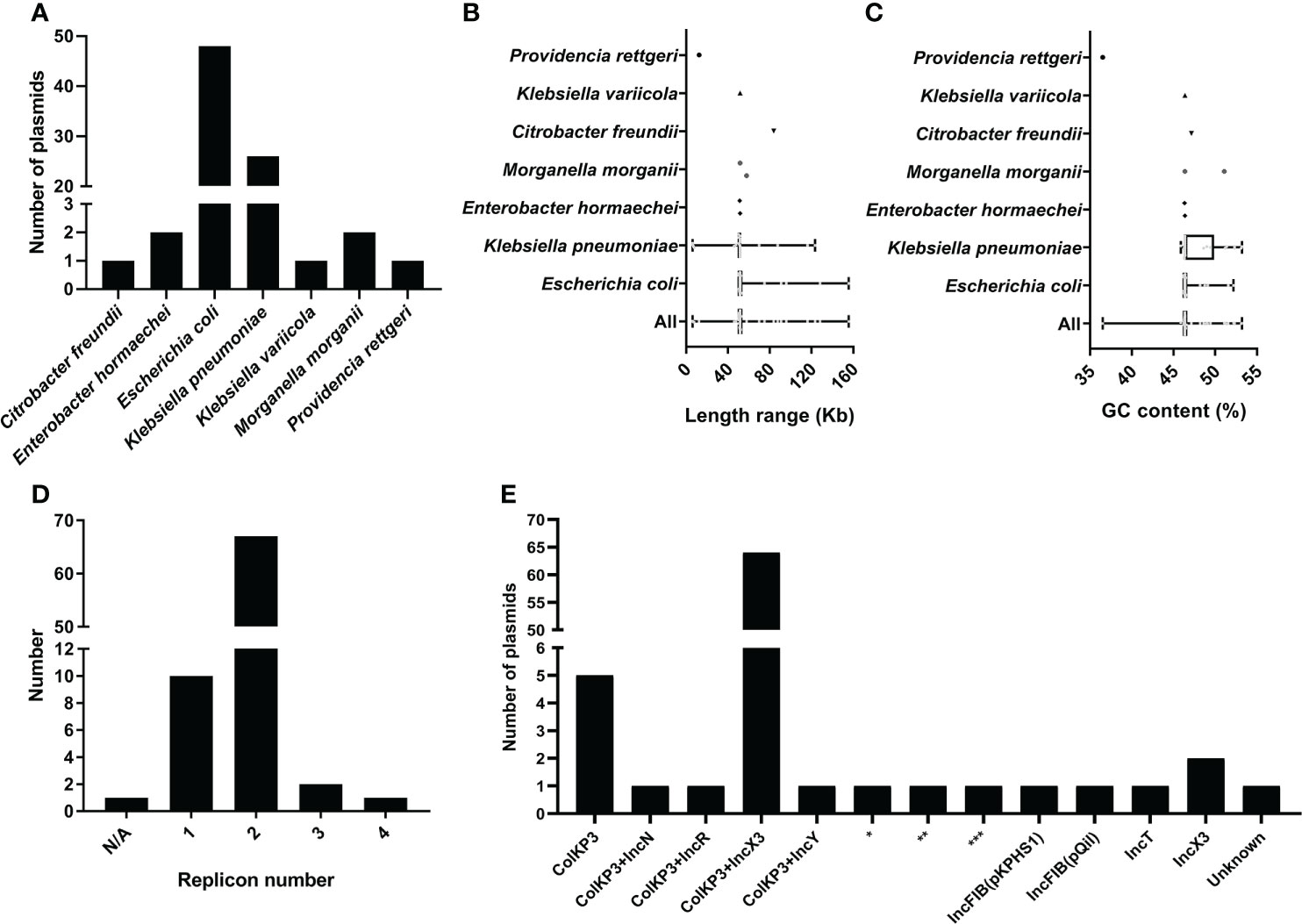
Figure 1 Characteristics of 81 blaOXA-181-positive plasmids in Enterobacterales. (A) Histogram about number of plasmids distributed in different species for the 81 blaOXA-181-positive in Enterobacterales. (B) Length distribution of the blaOXA-181-positive plasmids in different species. (C) GC content distribution of the blaOXA-181-positive plasmids in different species. (D) Histogram of number of replicons per plasmid for the 81 blaOXA-181-positive plasmids. (E) Histogram of number of combination modes of different replicons among the 81 blaOXA-181-positive plasmids. *representing ColKP3+IncFIB(pB171)+IncFII; **representing ColKP3+IncFIB(AP001918)+IncFIC(FII); ***representing ColKP3+IncFIA+IncFIB(AP001918)+IncFII(pRSB107).
We analyzed and compared the genome sizes of the blaOXA-181-positive plasmids, and compared the sizes of the blaOXA-181-positive plasmids distributed in different species. The genome sizes of the 81 blaOXA-181-positive plasmids varied from 6.103 kb to 155.5 kb, with the 25th percentile, median, and 75th percentile being 51.47 kb, 51.48 kb, and 51.48 kb, respectively (Figure 1B). The sizes of the 48 blaOXA-181-positive plasmids in E. coli varied from 50.14 kb to 155.5 kb (25th percentile = 51.48 kb; 75th percentile = 51.48 kb), with a median size of 51.48 kb. The sizes of the 26 blaOXA-181-positive plasmids in K. pneumoniae varied from 6.103 kb to 123.3 kb (25th percentile = 49.50 kb; 75th percentile = 51.48 kb), with a median size of 51.48 kb.
We calculated the GC contents of the 81 blaOXA-181-positive plasmids. The GC contents of the 81 blaOXA-181-positive plasmids ranged from 36.51% to 53.20% (25th percentile = 46.37%; 75th percentile = 46.38%), with a median GC content of 46.37% (Figure 1C). The GC contents of the 48 blaOXA-181-positive plasmids of E. coli varied from 46.22% to 52.19% (25th percentile = 46.37%; 75th percentile = 46.38%), with a median GC content of 46.37%. The GC contents of the 26 blaOXA-181-positive plasmids of K. pneumoniae varied from 45.88% to 53.20% (25th percentile = 46.37%; 75th percentile = 49.87%), with a median GC content of 46.37%.
Among the 81 blaOXA-181-bearing plasmids, 80 were successfully identified their replicon types, including 10 single-replicon plasmids and 70 multi-replicon plasmids (67 plasmids with two replicons, two plasmids with three replicons, and one plasmid with four replicons) (Figure 1D). Of the 67 plasmids with two replicons, 64 plasmids were ColKP3-IncX3 hybrid plasmids (Figure 1E). Of the 10 single-replicon plasmids, five plasmids had ColKP3 replicon (Figure 1E). Notably, 75 of the 81 blaOXA-181-positive plasmids in our study carried ColKP3 replicon.
To obtain a comprehensive overview of blaOXA-181-positive plasmids, we constructed phylogenetic trees (Figure 2). Based on the phylogenetic patterns, replicon types, and conjugative transfer regions, the 81 blaOXA-181-positive plasmids were classified into two main clades. One clade contained 66 blaOXA-181-positive plasmids (81.48%) and represented the most common conjugative plasmid pattern carrying blaOXA-181 gene. Of the 66 blaOXA-181-positive plasmids, 65 carried genes encoding relaxases of the MOBP family. All the 66 blaOXA-181-positive plasmids harbored genes encoding type IV coupling proteins (T4CPs) of the VirD4/TraG subfamily. In addition, 65 of the 66 blaOXA-181-positive plasmids contained VirB-like type IV secretion system (T4SS) gene clusters (Figures 2, 3). Most of the VirB-like T4SS gene clusters were composed of 11 core genes (Figure 3). The current version of oriTfinder could not identify the oriT sites of the clade; however, 354-bp intergenic sequences flanking the relaxase genes were identified as oriT–like regions and had the inverted repeat (IR) sequence “TAACTA..TAGTTA” (Figure S1). Of the 66 blaOXA-181-positive conjugative IncX3 plasmids, 64 plasmids were identified with two replicons: ColKP3 and IncX3, and the other two plasmids were single-replicon IncX3 plasmids.
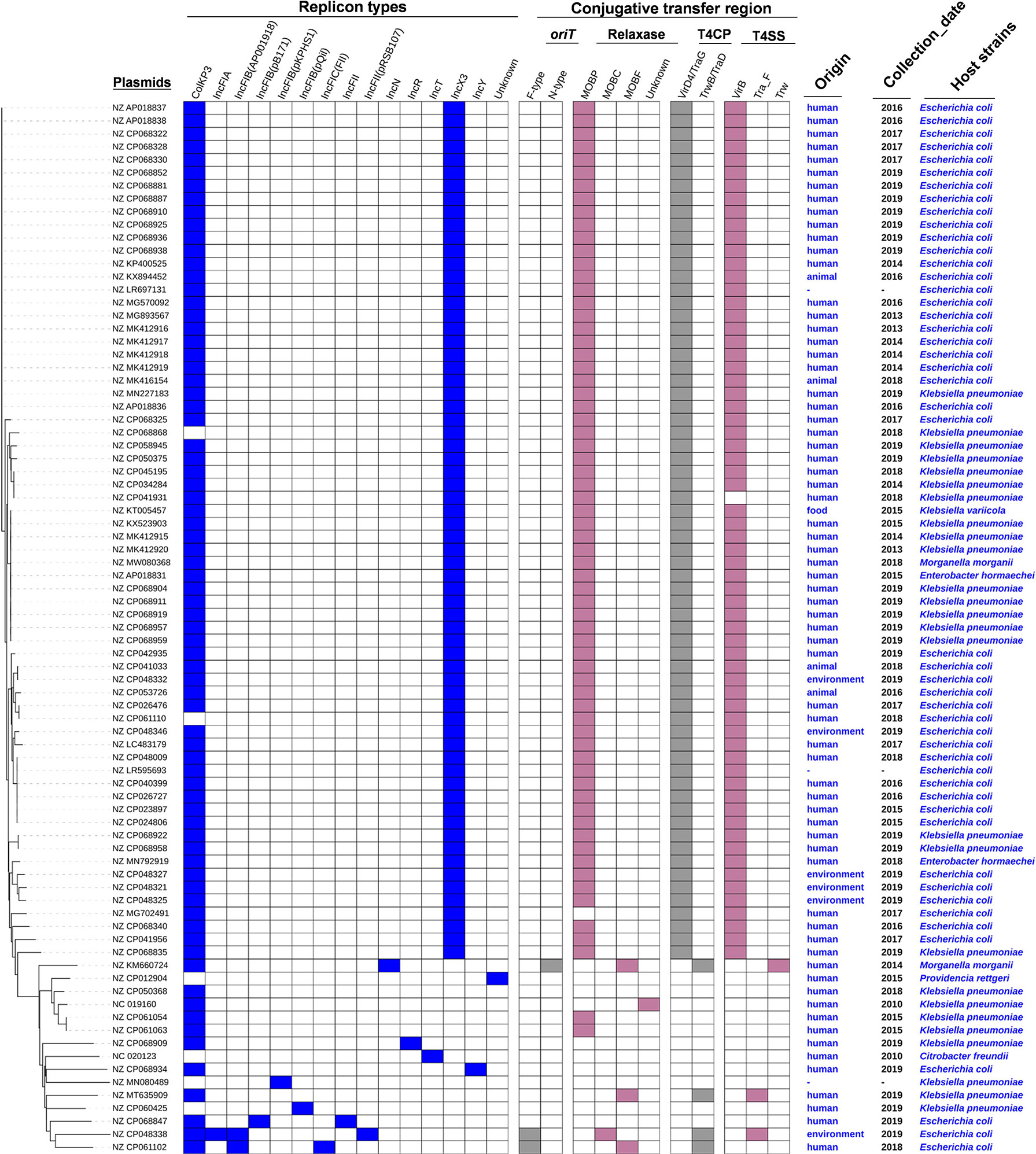
Figure 2 Details of replicon types, conjugative transfer regions, and host strains of the 81 blaOXA-181-positive plasmids in Enterobacterales. The six categories of information present in this figure include the phylogenetic tree, replicon types, conjugative transfer regions (oriT, relaxase, T4CP, and T4SS), origin, collection date, and host strains of 81 blaOXA-181-positive plasmids.
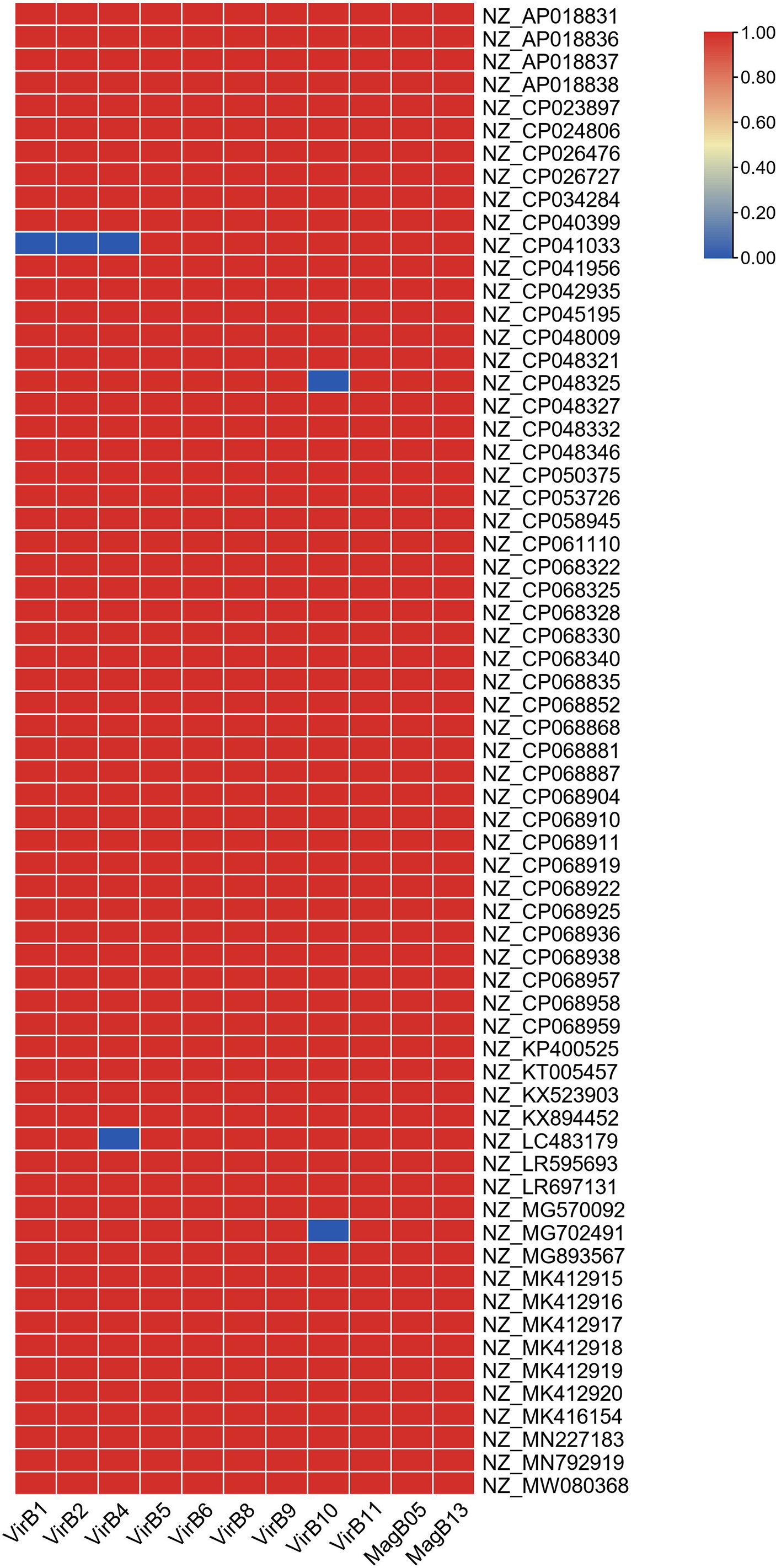
Figure 3 Details of the VirB-like T4SS gene clusters in the 66 blaOXA-181-positive IncX3 plasmids in Enterobacterales. Red/blue colors indicate gene presence/absence.
In this study, the blaOXA-181-positive conjugative IncX3 plasmids were mainly distributed in the strains of E. coli (44 plasmids) and K. pneumoniae (18 plasmids) (Figure 2 and Figure S2). Overall, all the blaOXA-181-positive conjugative IncX3 plasmids included 65 blaOXA-181-positive conjugative plasmids belonging to family Enterobacteriaceae and one plasmid belonging to family Morganellaceae (Figure 2 and Figure S2).
Of the 66 blaOXA-181-harboring IncX3 plasmids, 54 were human origin, accounting for 81.82% (Figure 2 and Table S3). In addition, four blaOXA-181-harboring IncX3 plasmids were found to be animal origin, five IncX3 plasmids harboring blaOXA-181 were animal origin, and one blaOXA-181-bearing IncX3 plasmid was food origin (Figure 2 and Table S3).
We analyzed and compared the genome sizes of all the 66 blaOXA-181-harboring conjugative plasmids. The genome sizes of 66 blaOXA-181-positive IncX3 plasmids varied from 45.90 kb to 74.95 kb (25th percentile = 51.48 kb; 75th percentile = 51.48 kb), with the median size of 51.48 kb (Figure 4). The genome sizes of the 44 blaOXA-181-positive conjugative IncX3 plasmids in E. coli ranged from 45.14 kb to 74.95 kb (25th percentile = 51.48 kb; 75th percentile = 51.48 kb), with a median size of 51.48 kb (Figure 4). The genome sizes of the 18 blaOXA-181-positive conjugative IncX3 plasmids in K. pneumoniae ranged from 45.90 kb to 69.76 kb (25th percentile = 50.98 kb; 75th percentile = 51.48 kb), with a median of 51.48 kb (Figure 4).
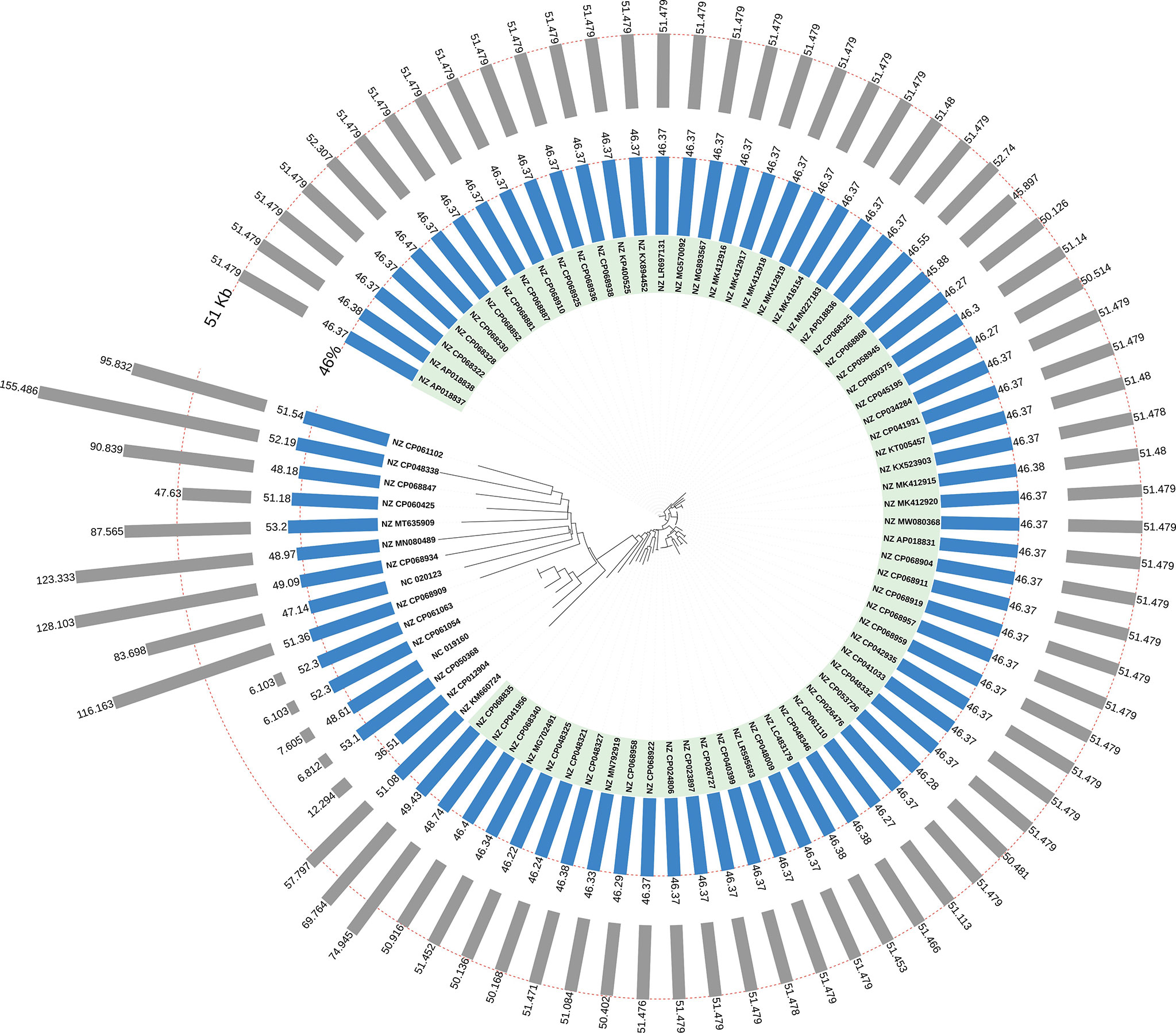
Figure 4 Distribution of lengths and GC contents of the 81 blaOXA-181-harboring plasmids in Enterobacterales. The 66 blaOXA-181-harboring IncX3 plasmids were highlighted in light green. The outermost grey bars denote the lengths of the blaOXA-181-harboring plasmids. The bars in light blue denote the GC content of the 81 blaOXA-181-harboring plasmids.
Among the 66 blaOXA-181-positive IncX3 plasmids, 63 were found to carry two acquired ARGs: blaOXA-181 and qnrS1 in their genomes; the former encoded OXA-181-type carbapenemase and the latter was responding for resistance to quinolone (Figure S3). Two blaOXA-181-positive IncX3 plasmids from K. pneumoniae (K. pneumoniae strain BA39649 plasmid pColKP3_IncX3 with GenBank accession number of NZ_CP058945 and K. pneumoniae strain RIVM_C017275 plasmid pRIVM_C017275_2 with GenBank accession number of NZ_CP068868) only contained the blaOXA-181 gene (Figure S3). In the E. coli strain EC2 IncX3 plasmid pEC2_1 (NZ_CP041956), in addition to the blaOXA-181 and qnrS1, six other acquired ARGs were also identified, including blaCTX-M-15, blaTEM-1B, blaOXA-1, aac(6’)-Ib-cr, catB3, and tet(A) (Figure S3). In the K. pneumoniae strain RIVM_C018652 IncX3 plasmid pRIVM_C018652_2 (NZ_CP068835), eight acquired ARGs [mph(A), blaTEM-1B, rmtB, blaNDM-5, sul1, aadA2, dfrA12, and blaOXA-181] were found but no qnrS1 was present in its genome (Figure S3).
In the accessory regions of the 63 IncX3 plasmids harboring both blaOXA-181 and qnrS1 in their genomes, the blaOXA-181 and qnrS1 were located in a composite transposon, which was bracketed by two copies of insertion sequence IS26 in the same orientation (Table S4 and Figure 5). Of the 63 IncX3 plasmids with both blaOXA-181 and qnrS1, 56 were found to carry almost identical genetic contexts associated with blaOXA-181 and qnrS1, the intact IS26-flanked composite transposon (Table S4). Ten blaOXA-181-positive IncX3 plasmids were found to carry the truncated IS26-flanked composite transposon (Figure 5).
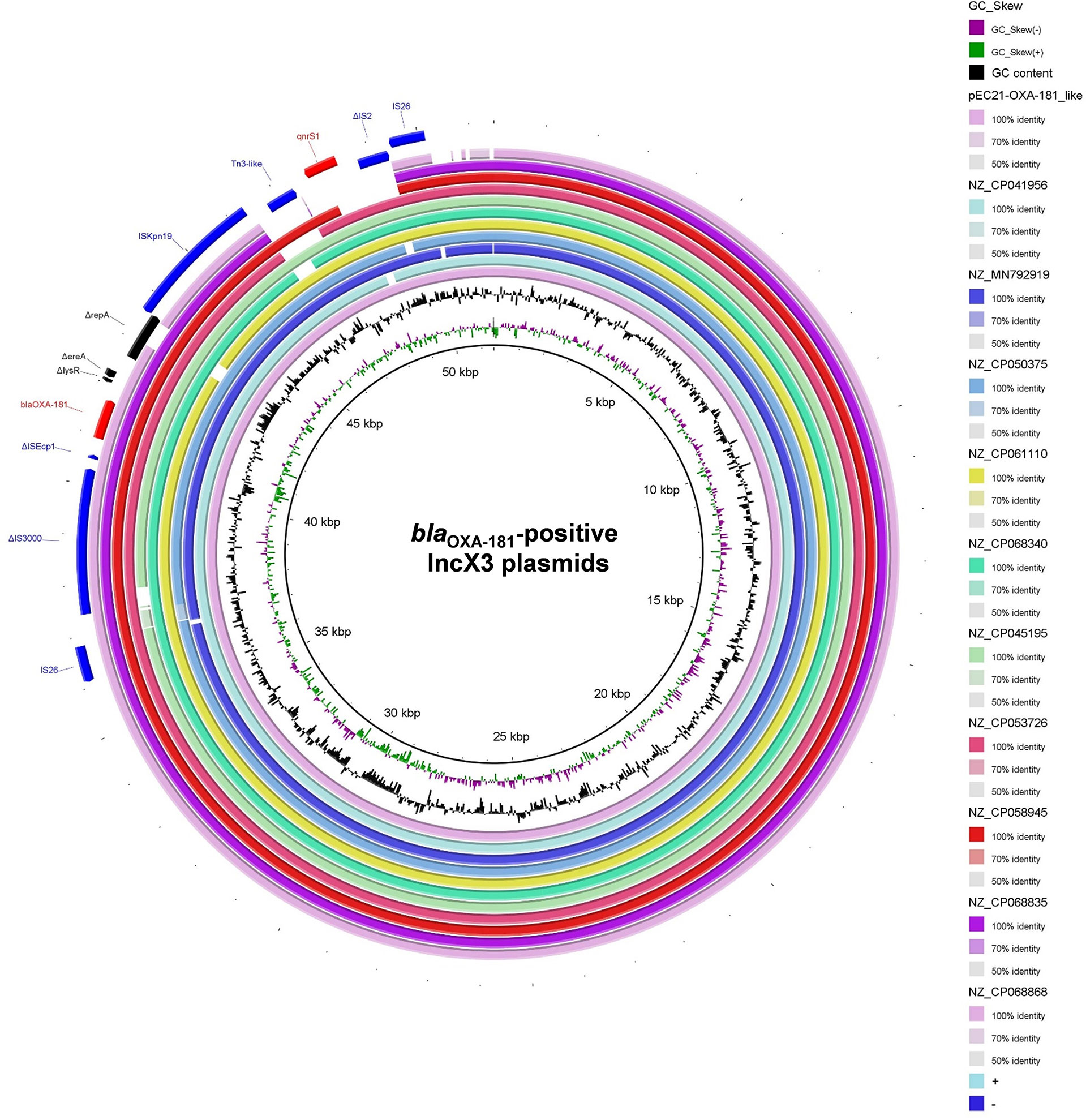
Figure 5 Schematic map of blaOXA-181-associated genetic structures identified among the 66 blaOXA-181-positive IncX3 plasmids in Enterobacterales. The pEC21-OXA-181_like represent the 56 plasmids carrying the intact IS26-flanked composite transposon.
The immediate genetic environment of blaOXA-181 (IS26–ΔIS3000–ΔISEcp1–blaOXA-181–ΔlysR–Δere–ΔrepA–ISKpn19) were found to situated in all the 66 blaOXA-181-positive IncX3 plasmids. For the immediate genetic environment of qnrS1, the qnrS1 was flanked by a Tn3-like transposon (truncated by an ISKpn19) and an IS2-like insertion sequence (truncated by an IS26) (Figure 5).
OXA-181 is one of the most common OXA-48-like derivative, and the OXA-181-producing Enterobacterales has been reported in many countries worldwide (Pitout et al., 2019). To characterize the plasmids harboring blaOXA-181 in Enterobacterales, we identified and analyzed 81 blaOXA-181-positive plasmids, which were selected from 35,150 bacterial plasmids collected from all over the world. Our results show that diverse plasmid types harbor blaOXA-181 but IncX3 plasmids, especially the ColKP3-IncX3 hybrid plasmids, predominantly carry it, indicating the potential of IncX3 plasmids as vehicles in the global dissemination of OXA-181. IncX3 plasmids are narrow-host range plasmids in Enterobacterales (Johnson et al., 2012), which have been reported to carry various carbapenemase genes in CRE worldwide (Mouftah et al., 2019). To further characterize the IncX3 plasmids harboring blaOXA-181, we systematically compared the plasmid types, conjugative transfer regions, as well as the genetic features associated with blaOXA-181 in the 66 blaOXA-181-positive IncX3 plasmids.
The common species bearing the blaOXA-181-positive IncX3 plasmids were E. coli (44 plasmids) and K. pneumoniae (18 plasmids). OXA-181 was first described for K. pneumoniae (Castanheira et al., 2011; Potron et al., 2011). K. pneumoniae represents one of the most concerning pathogens known for its high frequency and diversity of AMR genes (Navon-Venezia et al., 2017; Wyres and Holt, 2018), and it has been classified as an ESKAPE organism (De Oliveira et al., 2020). Herein, we found that K. pneumoniae was the second most prevalent species harboring the blaOXA-181-positive IncX3 plasmids. In our study, E. coli was the predominant species carrying the blaOXA-181-positive IncX3 plasmids. AMR in E. coli has become an issue of concern in both human and veterinary health worldwide (Poirel et al., 2018).
Our work indicated that almost all the IncX3 plasmids harboring blaOXA-181 were ColKP3-IncX3 hybrid plasmids. ColKP3-type plasmid was also reported to harbor blaOXA-232, another gene encoding OXA-48-like carbapenemase (Shu et al., 2019). By comparing the sizes of IncX3 plasmids harboring blaOXA-181, we found that the OXA-181-encoding gene blaOXA-181 was mostly located in 51-kb IncX3-type plasmids. A previous study has demonstrated that blaOXA-181-positive IncX3 plasmid, 51-kb pOXA181_EC14828 in E. coli ST410 from China, was a self-transmissible plasmid, as it could be transferred to the recipient strain in the conjugation experiment (Liu et al., 2015).
Conjugative plasmids play a central role in facilitating horizontal genetic exchange and therefore promote the acquisition and spread of AMR genes (Partridge et al., 2018; Jiang et al., 2020). The conjugative transfer regions of plasmids typically consist of four modules: oriT region, relaxase gene, T4CP gene, and gene cluster for T4SS apparatus (de la Cruz et al., 2010). Herein, we attempted to analyze and compare the conjugative transfer regions located in the blaOXA-181-positive IncX3 plasmids in Enterobacterales using the software oriTfinder (Li et al., 2018). Almost all the IncX3 plasmids harboring blaOXA-181 contained genes coding for relaxases belonging to the MOBP family characterized by the domain “Relaxase (Pfam: PF03432)”. The TraI encoded by the IncPα plasmid RP4 (Pansegrau et al., 1993) is regarded as a representative of MOBP family. T4CPs are essential elements in conjugative T4SSs and are also key elements in many pathogenic T4SSs (Álvarez-Rodríguez et al., 2020). In this study, all the 66 blaOXA-181-positive plasmids in Enterobacterales carried the genes encoding T4CPs of the VirD4/TraG subfamily characterized by the domain “T4SS-DNA_transf (Pfam: PF02534)”. Almost all blaOXA-181-positive IncX3 plasmids contained VirB-like T4SS gene clusters, which are by far the best characterized T4SS (Guglielmini et al., 2014). Notably, we cannot identify the oriT sites in blaOXA-181-positive IncX3 plasmids using oriTfinder, but the 354-bp intergenic sequences flanking the relaxase genes were oriT–like regions.
In our study, we found that 63 of the 66 IncX3 plasmids harbored both blaOXA-181 and qnrS1 in their genomes. The two acquired ARGs were found to be embedded in a composite transposon, which is bracketed by two copies of the insertion sequence IS26 oriented in the same direction. The IS26 has been shown to play a major role in the dissemination of ARGs in gram-negative bacteria (Harmer and Hall, 2021) because IS26 can recruit ARGs into the mobile gene pool by forming transposons carrying many different resistance genes (Harmer and Hall, 2016). The blaOXA-181 harbored by all the 66 IncX3 plasmids was adjacent to ISEcp1. The ISEcp1 is a member of the IS1380 family known to mobilize adjacent DNA sequences by a so-called one-ended transposition mechanism (Poirel et al., 2005), and it has been shown to be involved in mobilization of different antibiotic resistance genes such as blaCTX-M (Poirel et al., 2003). The truncation of the ISEcp1 adjacent to blaOXA-181 in the 66 blaOXA-181-positive IncX3 plasmids suggested that the IS26-flanked composite transposon has the potential to mobilize blaOXA-181 independent of the action of ISEcp1.
In this study, we identified 81 blaOXA-181-harboring plasmids from 35,150 bacterial plasmids downloaded from the NCBI RefSeq database. Diverse plasmid types harbored blaOXA-181 but was predominantly carried by IncX3-type plasmids. Our study mainly focused on in silico characterization of the 66 blaOXA-181-bearing IncX3 plasmids, including host strains, plasmid types, origin, conjugative transfer regions, and genetic contexts. We found that IncX3 plasmids harboring blaOXA-181 were mostly ColKP3-IncX3 hybrid plasmids with a length of 51 kb each and were mainly distributed in Escherichia coli and Klebsiella pneumoniae. Most of the IncX3 plasmids harboring blaOXA-181 were found to be human origin. For the conjugative transfer regions, almost all the blaOXA-181-positive IncX3 plasmids were found to carry genes coding for relaxases of the MOBP family and VirB-like type IV secretion system (T4SS) gene clusters, and all the 66 IncX3 plasmids were found to carry the genes encoding type IV coupling proteins (T4CPs) of the VirD4/TraG subfamily. Plasmid analysis revealed that blaOXA-181 together with the quinolone resistance gene qnrS1 were carried by an IS26-flanked composite transposon on the IncX3-type plasmids. Our findings enhance our understanding of the genetic diversity and characteristics of blaOXA-181-harboring IncX3 plasmids and further address their role in acquiring and spreading blaOXA-181 genes in Enterobacterales.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
XL, ML and XF designed the study. XL and ZY analysed all the data and drafted the manuscript. SH, TL, QL, and ZXZ contributed to the data acquisition and methodology. ZRZ and LS reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported financially by the National Natural Science Foundation of China (Grant No. 81902460 and 82002170), the Guangdong Basic and Applied Basic Research Foundation (Grant No. 2019A1515011725), the Xiangshan Talent Project of Zhuhai People’s Hospital (Grant No. 2020XSYC-02), the Cultivation Project of Zhuhai People’s Hospital (2019PY-19 and 2019PY-22).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.988236/full#supplementary-material
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., Beatson, S. A. (2011). BLAST ring image generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics 12, 402. doi: 10.1186/1471-2164-12-402
Álvarez-Rodríguez, I., Ugarte-Uribe, B., de la Arada, I., Arrondo, J. L. R., Garbisu, C., Alkorta, I. (2020). Conjugative coupling proteins and the role of their domains in conjugation, secondary structure and in vivo subcellular location. Front. Mol. Biosci. 7. doi: 10.3389/fmolb.2020.00185
Balm, M. N., La, M. V., Krishnan, P., Jureen, R., Lin, R. T., Teo, J. W. (2013). Emergence of Klebsiella pneumoniae co-producing NDM-type and OXA-181 carbapenemases. Clin. Microbiol. Infect. 19 (9), E421–E423. doi: 10.1111/1469-0691.12247
Bi, D., Liu, L., Tai, C., Deng, Z., Rajakumar, K., Ou, H. Y. (2013). SecReT4: A web-based bacterial type IV secretion system resource. Nucleic Acids Res. 41 (Database issue), D660–D665. doi: 10.1093/nar/gks1248
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75 (12), 3491–3500. doi: 10.1093/jac/dkaa345
Brown, C. J., Sen, D., Yano, H., Bauer, M. L., Rogers, L. M., van der Auwera, G. A., et al. (2013). Diverse broad-host-range plasmids from freshwater carry few accessory genes. Appl. Environ. Microbiol. 79 (24), 7684–7695. doi: 10.1128/AEM.02252-13
Carattoli, A., Hasman, H. (2020). PlasmidFinder and in silico pMLST: Identification and typing of plasmid replicons in whole-genome sequencing (WGS). Methods Mol. Biol. 2075, 285–294. doi: 10.1007/978-1-4939-9877-7_20
Castanheira, M., Deshpande, L. M., Mathai, D., Bell, J. M., Jones, R. N., Mendes, R. E. (2011). Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: Report from the SENTRY antimicrobial surveillance program 2006-2007. Antimicrob. Agents Chemother. 55 (3), 1274–1278. doi: 10.1128/aac.01497-10
de la Cruz, F., Frost, L. S., Meyer, R. J., Zechner, E. L. (2010). Conjugative DNA metabolism in gram-negative bacteria. FEMS Microbiol. Rev. 34 (1), 18–40. doi: 10.1111/j.1574-6976.2009.00195.x
De Oliveira, D. M., Forde, B. M., Kidd, T. J., Harris, P. N., Schembri, M. A., Beatson, S. A., et al. (2020). Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 33 (3), e00181–e00119. doi: 10.1128/CMR.00181-19
Emms, D. M., Kelly, S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20 (1), 238. doi: 10.1186/s13059-019-1832-y
Guglielmini, J., Néron, B., Abby, S. S., Garcillán-Barcia, M. P., de la Cruz, F., Rocha, E. P. (2014). Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res. 42 (9), 5715–5727. doi: 10.1093/nar/gku194
Hammer, Ø., Harper, D. A., Ryan, P. D. (2001). PAST: Paleontological statistics software package for education and data analysis. Palaeontol. electron. 4 (1), 9.
Harmer, C. J., Hall, R. M. (2016). IS26-mediated formation of transposons carrying antibiotic resistance genes. mSphere 1 (2), e00038-16. doi: 10.1128/mSphere.00038-16
Harmer, C. J., Hall, R. M. (2021). IS 26 cannot move alone. J. Antimicrob. Chemother. 76 (6), 1428–1432. doi: 10.1093/jac/dkab055
Jiang, Y., Wang, Y., Hua, X., Qu, Y., Peleg, A. Y., Yu, Y. (2020). Pooled plasmid sequencing reveals the relationship between mobile genetic elements and antimicrobial resistance genes in clinically isolated Klebsiella pneumoniae. Genomics Proteomics Bioinf 18 (5), 539–548. doi: 10.1016/j.gpb.2020.12.002
Johnson, T. J., Bielak, E. M., Fortini, D., Hansen, L. H., Hasman, H., Debroy, C., et al. (2012). Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68 (1), 43–50. doi: 10.1016/j.plasmid.2012.03.001
Kim, J. S., Yu, J. K., Jeon, S. J., Park, S. H., Han, S., Park, S. H., et al. (2021). Dissemination of an international high-risk clone of Escherichia coli ST410 co-producing NDM-5 and OXA-181 carbapenemases in Seoul, republic of Korea. Int. J. Antimicrob. Agents 58 (6), 106448. doi: 10.1016/j.ijantimicag.2021.106448
Letunic, I., Bork, P. (2016). Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44 (W1), W242–W245. doi: 10.1093/nar/gkw290
Liu, Y., Feng, Y., Wu, W., Xie, Y., Wang, X., Zhang, X., et al. (2015). First report of OXA-181-Producing Escherichia coli in China and characterization of the isolate using whole-genome sequencing. Antimicrob. Agents Chemother. 59 (8), 5022–5025. doi: 10.1128/aac.00442-15
Li, X., Xie, Y., Liu, M., Tai, C., Sun, J., Deng, Z., et al. (2018). oriTfinder: A web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 46 (W1), W229–w234. doi: 10.1093/nar/gky352
Mouftah, S. F., Pál, T., Darwish, D., Ghazawi, A., Villa, L., Carattoli, A., et al. (2019). Epidemic IncX3 plasmids spreading carbapenemase genes in the United Arab Emirates and worldwide. Infect. Drug Resist. 12, 1729–1742. doi: 10.2147/idr.s210554
Navon-Venezia, S., Kondratyeva, K., Carattoli, A. (2017). Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 41 (3), 252–275. doi: 10.1093/femsre/fux013
Norberg, P., Bergström, M., Jethava, V., Dubhashi, D., Hermansson, M. (2011). The IncP-1 plasmid backbone adapts to different host bacterial species and evolves through homologous recombination. Nat. Commun. 2 (1), 1–11. doi: 10.1038/ncomms1267
Nordmann, P., Naas, T., Poirel, L. (2011). Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17 (10), 1791–1798. doi: 10.3201/eid1710.110655
Norman, A., Hansen, L. H., Sørensen, S. J. (2009). Conjugative plasmids: vessels of the communal gene pool. Philos. Trans. R. Soc. B: Biol. Sci. 364 (1527), 2275–2289. doi: 10.1098/rstb.2009.0037
O’Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44 (D1), D733–D745. doi: 10.1093/nar/gkv1189
Oueslati, S., Nordmann, P., Poirel, L. (2015). Heterogeneous hydrolytic features for OXA-48-like β-lactamases. J. Antimicrob. Chemother. 70 (4), 1059–1063. doi: 10.1093/jac/dku524
Pansegrau, W., Schröder, W., Lanka, E. (1993). Relaxase (TraI) of IncP alpha plasmid RP4 catalyzes a site-specific cleaving-joining reaction of single-stranded DNA. Proc. Natl. Acad. Sci. 90 (7), 2925–2929. doi: 10.1073/pnas.90.7.2925
Partridge, S. R., Kwong, S. M., Firth, N., Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31 (4), e00088-17. doi: 10.1128/cmr.00088-17
Piazza, A., Comandatore, F., Romeri, F., Pagani, C., Floriano, A. M., Ridolfo, A., et al. (2018). First report of an ST410 OXA-181 and CTX-M-15 coproducing Escherichia coli clone in Italy: A whole-genome sequence characterization. Microb. Drug Resist. 24 (8), 1207–1209. doi: 10.1089/mdr.2017.0366
Pitout, J. D., Peirano, G., Kock, M. M., Strydom, K.-A., Matsumura, Y. (2019). The global ascendency of OXA-48-type carbapenemases. Clin. Microbiol. Rev. 33 (1), e00102–e00119. doi: 10.1128/CMR.00102-19
Poirel, L., Decousser, J.-W., Nordmann, P. (2003). Insertion sequence IS Ecp1B is involved in expression and mobilization of a bla CTX-M β-lactamase gene. Antimicrob. Agents Chemother. 47 (9), 2938–2945. doi: 10.1128/AAC.47.9.2938-2945.2003
Poirel, L., Lartigue, M.-F., Decousser, J.-W., Nordmann, P. (2005). IS Ecp1B-mediated transposition of bla CTX-M in Escherichia coli. Antimicrob. Agents chemother. 49 (1), 447–450. doi: 10.1128/AAC.49.1.447-450.2005
Poirel, L., Madec, J.-Y., Lupo, A., Schink, A.-K., Kieffer, N., Nordmann, P., et al. (2018). Antimicrobial resistance in Escherichia coli. Microbiol. Spectr. 6 (4), ARBA-0026-2017. doi: 10.1128/microbiolspec.ARBA-0026-2017
Potron, A., Nordmann, P., Lafeuille, E., Al Maskari, Z., Al Rashdi, F., Poirel, L. (2011). Characterization of OXA-181, a carbapenem-hydrolyzing class d beta-lactamase from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 55 (10), 4896–4899. doi: 10.1128/aac.00481-11
Ravi, A., Valdés-Varela, L., Gueimonde, M., Rudi, K. (2018). Transmission and persistence of IncF conjugative plasmids in the gut microbiota of full-term infants. FEMS Microbiol. Ecol. 94 (1), fix158. doi: 10.1093/femsec/fix158
Rojas, L. J., Hujer, A. M., Rudin, S. D., Wright, M. S., Domitrovic, T. N., Marshall, S. H., et al. (2017). NDM-5 and OXA-181 beta-lactamases, a significant threat continues to spread in the americas. Antimicrob. Agents Chemother. 61 (7), e00454-17. doi: 10.1128/aac.00454-17
Rood, I. G. H., Li, Q. (2017). Review: Molecular detection of extended spectrum-β-lactamase- and carbapenemase-producing Enterobacteriaceae in a clinical setting. Diagn. Microbiol. Infect. Dis. 89 (3), 245–250. doi: 10.1016/j.diagmicrobio.2017.07.013
Rozwandowicz, M., Brouwer, M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez-Zorn, B., Guerra, B., et al. (2018). Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 73 (5), 1121–1137. doi: 10.1093/jac/dkx488
Shu, L., Dong, N., Lu, J., Zheng, Z., Hu, J., Zeng, W., et al. (2019). Emergence of OXA-232 carbapenemase-producing Klebsiella pneumoniae that carries a pLVPK-like virulence plasmid among elderly patients in China. Antimicrob. Agents Chemother. 63 (3), e02246–e02218. doi: 10.1128/AAC.02246-18
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34 (Database issue), D32–D36. doi: 10.1093/nar/gkj014
Sitter, T. L., Vaughan, A. L., Schoof, M., Jackson, S. A., Glare, T. R., Cox, M. P., et al. (2021). Evolution of virulence in a novel family of transmissible mega-plasmids. Environ. Microbiol. 23 (9), 5289–5304. doi: 10.1111/1462-2920.15595
Smillie, C., Garcillán-Barcia, M. P., Francia, M. V., Rocha, E. P., de la Cruz, F. (2010). Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74 (3), 434–452. doi: 10.1128/mmbr.00020-10
Keywords: Enterobacterales, plasmid, blaOXA-181, conjugative transfer region, genetic context
Citation: Yu Z, Zhang Z, Shi L, Hua S, Luan T, Lin Q, Zheng Z, Feng X, Liu M and Li X (2022) In silico characterization of IncX3 plasmids carrying blaOXA-181 in Enterobacterales. Front. Cell. Infect. Microbiol. 12:988236. doi: 10.3389/fcimb.2022.988236
Received: 07 July 2022; Accepted: 23 August 2022;
Published: 08 September 2022.
Edited by:
Thomas Jové, INSERM U1092 Anti-Infectieux supports moléculaires des résistances et innovations thérapeutiques, FranceReviewed by:
José Alejandro Di Conza, University of Buenos Aires, ArgentinaCopyright © 2022 Yu, Zhang, Shi, Hua, Luan, Lin, Zheng, Feng, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaobin Li, bGkueGlhb2JpbjIwMDlAMTYzLmNvbQ==; Mubiao Liu, bGl1bWIxOTcyQDEyNi5jb20=; Xiaosan Feng, c2FtM0B5ZWFoLm5ldA==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.