
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 08 September 2022
Sec. Clinical Microbiology
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.987692
This article is part of the Research Topic Interactions Between Pathogens and Host Immune System in Patients with Immunodeficiency: Estimation from High-throughput Sequencing View all 14 articles
 Lipin Liu†
Lipin Liu† Bijun Sun†
Bijun Sun† Wenjing Ying
Wenjing Ying Danru Liu
Danru Liu Ying Wang
Ying Wang Jinqiao Sun
Jinqiao Sun Wenjie Wang
Wenjie Wang Mi Yang
Mi Yang Xiaoying Hui
Xiaoying Hui Qinhua Zhou
Qinhua Zhou Jia Hou*
Jia Hou* Xiaochuan Wang*
Xiaochuan Wang*Talaromyces marneffei (T. marneffei) is an opportunistic pathogen. Patients with inborn errors of immunity (IEI) have been increasingly diagnosed with T. marneffei in recent years. The disseminated infection of T. marneffei can be life-threatening without timely and effective antifungal therapy. Rapid and accurate pathogenic microbiological diagnosis is particularly critical for these patients. A total of 505 patients with IEI were admitted to our hospital between January 2019 and June 2022, among whom T. marneffei was detected in 6 patients by metagenomic next-generation sequencing (mNGS), and their clinical and immunological characteristics were summarized. We performed a systematic literature review on T. marneffei infections with published immunodeficiency-related gene mutations. All patients in our cohort were confirmed to have genetic mutations in IL12RB1, IFNGR1, STAT1, STAT3, and CD40LG. T. marneffei was detected in both the blood and lymph nodes of P1 with IL12RB1 mutations, and the clinical manifestations were serious and included recurrent fever, weight loss, severe anemia, splenomegaly and lymphadenopathy, all requiring long-term antifungal therapy. These six patients received antifungal treatment, which relieved symptoms and improved imaging findings. Five patients survived, while one patient died of sepsis after hematopoietic stem cell transplantation. The application of mNGS methods for pathogen detection in IEI patients and comparison with traditional diagnosis methods were investigated. Traditional diagnostic methods and mNGS tests were performed simultaneously in 232 patients with IEI. Compared to the traditional methods, the sensitivity and specificity of mNGS in diagnosing T. marneffei infection were 100% and 98.7%, respectively. The reporting time for T. marneffei detection was approximately 26 hours by mNGS, 3-14 days by culture, and 6-11 days by histopathology. T. marneffei infection was first reported in IEI patients with IL12RB1 gene mutation, which expanded the IEI lineage susceptible to T. marneffei. For IEI patients with T. marneffei infection, we highlight the application of mNGS in pathogenic detection. mNGS is recommended as a front-line diagnostic test for rapidly identifying pathogens in complex and severe infections.
Talaromyces marneffi (T. marneffei), previously known as Penicillium marneffei, is an emerging pathogenic fungus causing fatal systemic mycosis in Southeast Asia, especially Thailand, northeastern India, Vietnam, southern China, Hong Kong and Taiwan (DiSalvo et al., 1973; Deng et al., 1988; Hu et al., 2013). The common manifestations of disseminated T. marneffei infection include recurrent fever, anemia, weight loss, skin lesions, hepatosplenomegaly, lymphadenopathy, and gastrointestinal and respiratory signs. When T. marneffei infection disseminates systemically, it can run a rapidly progressive course and be life-threatening without timely and effective antifungal therapy (You et al., 2021).
The vast majority of T. marneffei cases are diagnosed in HIV-positive patients (Supparatpinyo et al., 1994; Kawila et al., 2013), while cases have also been described in other immunocompromised individuals, such as patients with adult-onset acquired immunodeficiency due to autoantibodies against interferon-gamma (IFN-γ) (Guo et al., 2020), various inborn errors of immunity (IEI) (Tangye et al., 2020), hematological malignancies, diabetes mellitus and those taking corticosteroids or immunosuppressive agents (Qiu et al., 2015). In recent years, T. marneffei infection in children with IEI has been increasingly diagnosed. Genetic mutations involving STAT1, STAT3, CD40LG, CARD9 and IFNGR1 have been reported to be associated with T. marneffei infection (Ma et al., 2009; Lee et al., 2012; Du et al., 2019; Lee et al., 2019; Ba et al., 2021; You et al., 2021). However, as more cases are reported, other potential gene mutations may be discovered that predispose the host to T. marneffei infection.
Early identification and rapid diagnosis of T. marneffei infection are critical to a good prognosis. Traditional diagnostic methods of T. marneffei infection rely on culture or microscopy, which are time-consuming and have a lower positive rate. In recent years, the development of metagenomic next-generation sequencing (mNGS) has greatly improved the detection of pathogenic microorganisms (Chiu and Miller, 2019; Li et al., 2020; Qian et al., 2020; Chen et al., 2021; Su et al., 2022). mNGS can simultaneously accomplish the detection of bacteria, fungi, viruses, parasites and other pathogens with high efficiency and high sensitivity (Goldberg et al., 2015; Schlaberg et al., 2017). Currently, mNGS has matured into clinical applications in the diagnosis of complex and severe infections (Wang et al., 2020).
In this study, we reported the clinical and immunological characteristics of six patients with T. marneffei infection in a cohort of Chinese patients with IEI and emphasized the application value of mNGS in the diagnosis of T. marneffei infection. We aimed to improve the early recognition of T. marneffei infection in IEI patients. The immunodeficiency spectrum of T. marneffei infection was further broadened.
This study was approved by the Ethics Committee of the Children’s Hospital of Fudan University. Written informed consent was obtained from the parents of all patients.
Patients with IEI admitted to the Children’s Hospital of Fudan University between January 2019 and June 2022 were enrolled consecutively, and patients with T. marneffei infection were retrospectively summarized.
Children with IEI were defined as patients previously assigned a diagnosis of IEI or patients admitted for abnormal clinical manifestations, including severe/unusual infections, and then diagnosed with IEI. The clinical manifestations, immune function, genetic testing, and pathogenic microorganisms were evaluated. Routine tests of peripheral blood, urine, stool and blood culture were performed in all patients. The cultures of sputum or bronchoscopic alveolar lavage fluid (BALF) were detected in patients with pulmonary infection. Further mNGS testing was performed in patients considered to have severe or unusual pathogen infection.
Definition of terms: The age at the last follow-up was defined as the age at July 2022 or the age at death. The diagnosis of bacterial infections was based on positive culture results. The (1–3) -β-d-glucan (G) tests were defined as positive when > 100 pg/ml (Lu et al., 2011; Li et al., 2015). The galactomannan (GM) index was defined as positive when > 0.5. The GM test is highly sensitive to Aspergillus infection (Guo et al., 2010). The diagnostic criteria for Bacillus Calmette Guerin (BCG) disease were described in a previous publication (Zhou et al., 2018). Epstein-Bar virus (EBV) and cytomegalovirus (CMV) infections were based on PCR detection of DNA. Disseminated T. marneffei infection was defined as infections involving more than two organs or systems.
Routine blood counts and immunological function analyses were performed. We used nephelometry to detect immunoglobulins (Igs), including IgG, IgA, and IgM. Lymphocyte subsets were measured using flow cytometry (Becton Dickinson, Franklin Lakes, NJ, USA). The following validated antibodies were used for flow cytometry: anti-CD3 (UCHT1), anti-CD8 (RPAT8), anti-CD27 (M-T271), anti-CD45RA (HI100), anti-CD4 (RPA-T4), anti-TCRαβ (T10B9.1A-31), anti-TCRγδ (B1), anti-CD19 (HIB19), anti-CD24 (ML5), anti-CD38 (HIT2), and anti-IgD (IA6-2) (all from BD Biosciences) (Ding et al., 2018). Flow cytometry detection of CD119 and CD212 was performed.
Genomic DNA was extracted from the peripheral blood of the patients and their parents using the QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany). DNA quality was assessed using a NanoDrop ultraviolet spectrophotometer (Thermo Fisher Scientific, USA).
Next-generation sequencing was performed using a panel that included all previously reported immunodeficiency genes. Genomic DNA fragments of patients were ligated with adaptors so that two paired-end DNA libraries with insert sizes of 500 bp were formed for all samples. After enrichment, the DNA libraries were sequenced on the HiSeq 2000 platform in accordance with the manufacturer’s instructions (Illumina, San Diego, CA). The variants were annotated in ANNOVAR and VEP software and predicted with SIFT, PolyPhen-2 and MutationTaster.
T. marneffei was traditionally identified by culture or histopathology (Cao et al., 2019). In our study, T. marneffei was isolated from bacterial or fungal cultures of blood and BALF samples. This microorganism typically took approximately 3 to 14 days (Le et al., 2011; Cao et al., 2019). T. marneffei was identified by the following criteria (Segretain, 1959; Liyan et al., 2004; Cao et al., 2019) (1): Colonies of T. marneffei were verified by a MALDI-TOF mass spectrometer system (Bruker, Germany) (2). Positive fungal cultures were confirmed by Gram staining of a smear of the blood culture broth, followed by subculture onto Sabouraud dextrose agar (SDA) with incubation at 25 ˚C and 37 ˚C in room air. Yeast conversion was performed to confirm the identification of T. marneffei, and typical fungal colonies were observed (yeast phase at 37 ˚C with no red pigment production and mycelia at 25 ˚C with massive red pigment production) (3). Microscopically, the fungus had typical filamentous reproductive structures of the genus Penicillium, including the presence of conidiophore-bearing biverticillate penicilli, with each penicillus being composed of four to five metulae with smooth-walled conidia. T. marneffei. can be observed in histopathological sections, and methenamine silver or periodic acid-Schiff (PAS) staining was preferred. The presence of sausage-like cells, 2-3 μm in diameter, or elongated yeast-like organisms with clear central septum was the specific feature of T. marneffei (Liyan et al., 2004).
Blood and lymph node biopsy tissue samples were collected according to standard procedures. Plasma was prepared from blood samples, and circulating cell-free DNA (cfDNA) was isolated from plasma with the QIAamp Circulating Nucleic Acid Kit (Qiagen) according to the manufacturer’s protocols. Lymph node biopsy tissue was extracted using the QIAamp DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer’s protocols. The quantity and quality of DNA were assessed using Qubit (Thermo Fisher Scientific) and NanoDrop (Thermo Fisher Scientific), respectively.
DNA libraries were prepared using the KAPA Hyper Prep kit (KAPA Biosystems) according to the manufacturer’s protocols. An Agilent 2100 was used for quality control, and DNA libraries were 75 bp single-end sequenced on an Illumina NextSeq 550Dx (Illumina).
Raw sequencing data were split by bcl2fastq2, and high-quality sequencing data were generated using Trimmomatic by removing low-quality reads, adapter contamination, duplicates and shot (length<36 bp) reads. Human host sequences were subtracted by mapping to the human reference genome (hs37d5) using Bowtie2. Reads that could not be mapped to the human genome were retained and aligned with the microorganism genome database for microbial identification by Kraken and for species abundance estimation by Bracken. The microorganism genome database contained genomes or scaffolds of bacteria, fungi, viruses and parasites (downloaded from GenBank release 238, ftp://ftp.ncbi.nlm.nih.gov/genomes/genbank/).
We used the following criteria for positive mNGS results: For Mycobacterium, Nocardia and Legionella pneumophila, the result was considered positive if a species detected by mNGS had a species-specific read number ≥1. For bacteria (excluding Mycobacterium, Nocardia and Legionella pneumophila), fungi, viruses and parasites, the result was considered positive if a species detected by mNGS had at least 3 nonoverlapping reads. Pathogens detected in the negative ‘no-template’ control (NTC) were included only if the detected reads were ≥10-fold greater than those in the NTC.
Data were analyzed using SPSS 26.0 (IBM Corp., Armonk, NY, USA). Categorical variables were displayed as numbers and percentiles. The paired McNemar chi-square test was used to analyze the diagnostic efficiency of mNGS vs. conventional culture methods. Two-sided P values < 0.05 were considered statistically significant.
From January 2019 to June 2022, a total of 505 cases of IEI were admitted to our hospital (Table 1). One hundred and three (20.4%) patients were diagnosed with IEI before admission, while 402 (79.6%) patients were definitively diagnosed with IEI after admission. Most of the patients (425/505) were admitted with various infectious manifestations. All patients underwent routine blood cultures. Seventy-three patients underwent histopathology. Nearly half of the patients (232/505) underwent further mNGS testing. Therefore, 232 patients (45.9%) underwent both culture and mNGS detection.
T. marneffei infection was detected in six patients. The frequency of T. marneffei infection was 1.2% (6/505) in our cohort. The genetic mutations of T. marneffei-infected patients included IL12RB1, IFNGR1, STAT3, STAT1 and CD40LG (Figure 1A). Detailed clinical (Table 2) and immunological (Table 3) information of each patient is described as follows.
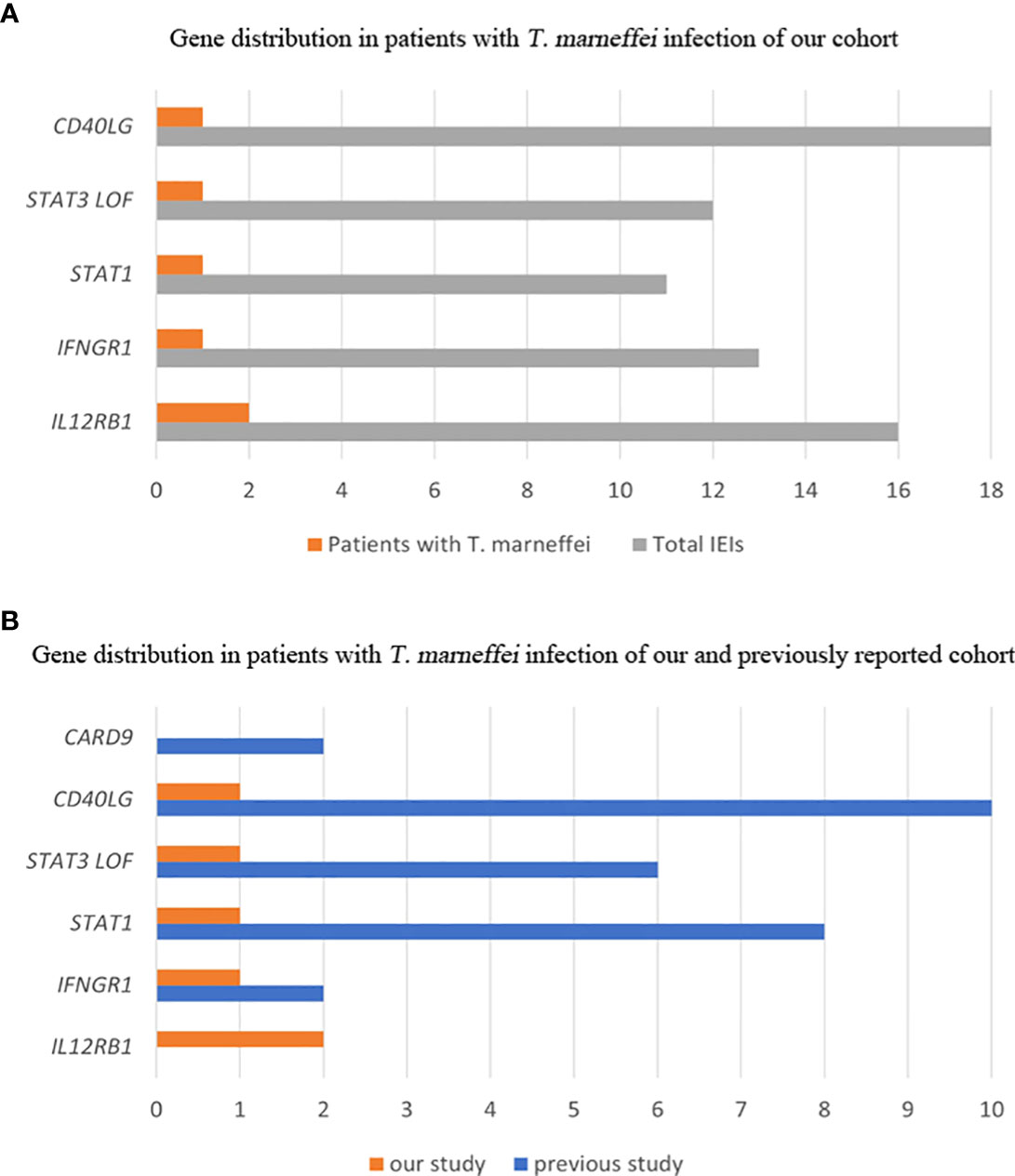
Figure 1 Gene distribution of patients with T. marneffei infection in IEIs. Gene distribution in our cohort (A). Gene distribution in previously reported cases (B).
Table 3 Hematological and immunological parameters at the time of T. marneffei infection in the six patients.
A 3-year-old girl was admitted to our hospital with the chief complaint of lymphadenopathy and intermittent fever. The girl was vaccinated with BCG after birth, and she developed left subaxillary and cervical lymph node enlargement at the age of 3 months. The local hospital performed lymph node resection and administered anti-tuberculosis treatment for 6 months. Nine months after drug withdrawal, the cervical lymph node enlargement recurred, and anti-tuberculosis therapy was administered again. IL12RB1 complex heterozygous mutations were detected: exon 14-16 deletion (maternal) and c.875-885 deletion (paternal). The expression of IL12RB1 protein was impaired (Figure 2). At the age of 3, the child developed recurrent fever, severe anemia, hepatosplenomegaly, and lymphadenopathy (Figure 3A), and the blood culture suggested T. marneffei. The serum G test (271.89 pg/ml) and GM test (6.682) were significantly increased. T. marneffei was detected in the blood (reads, 14) and lymph nodes (reads, 108380) by mNGS (Figure 3B). Therefore, isoniazid, rifampicin and ethambutol were used for antituberculosis, itraconazole for antifungal, and IFN-γ for immune regulation. The erythrocyte sedimentation rate (ESR) (120 mm/H) and G test (112.6 pg/ml) remained high after 2 months of anti-infective therapy, despite negative pathogens in blood by mNGS. Imaging examination revealed that the lymph nodes were still enlarged (Figure 4A2), and the liver and spleen lesions were smaller than before. After nearly 3 months of treatment with itraconazole, the biopsy of cervical lymph nodes suggested granulomatous inflammation (Figure 3C). PAS staining of the cervical lymph node tissues revealed fungal spores (Figure 3D), and T. marneffei was still identified by mNGS (reads, 15545702). Antifungal therapy was adjusted to amphotericin B liposomes intravenously for 2 weeks, followed by oral itraconazole. The follow-up CT scans revealed that the lymph nodes were obviously shrunken (Figure 4A3). ESR and G tests were reduced to normal, and then she was discharged.
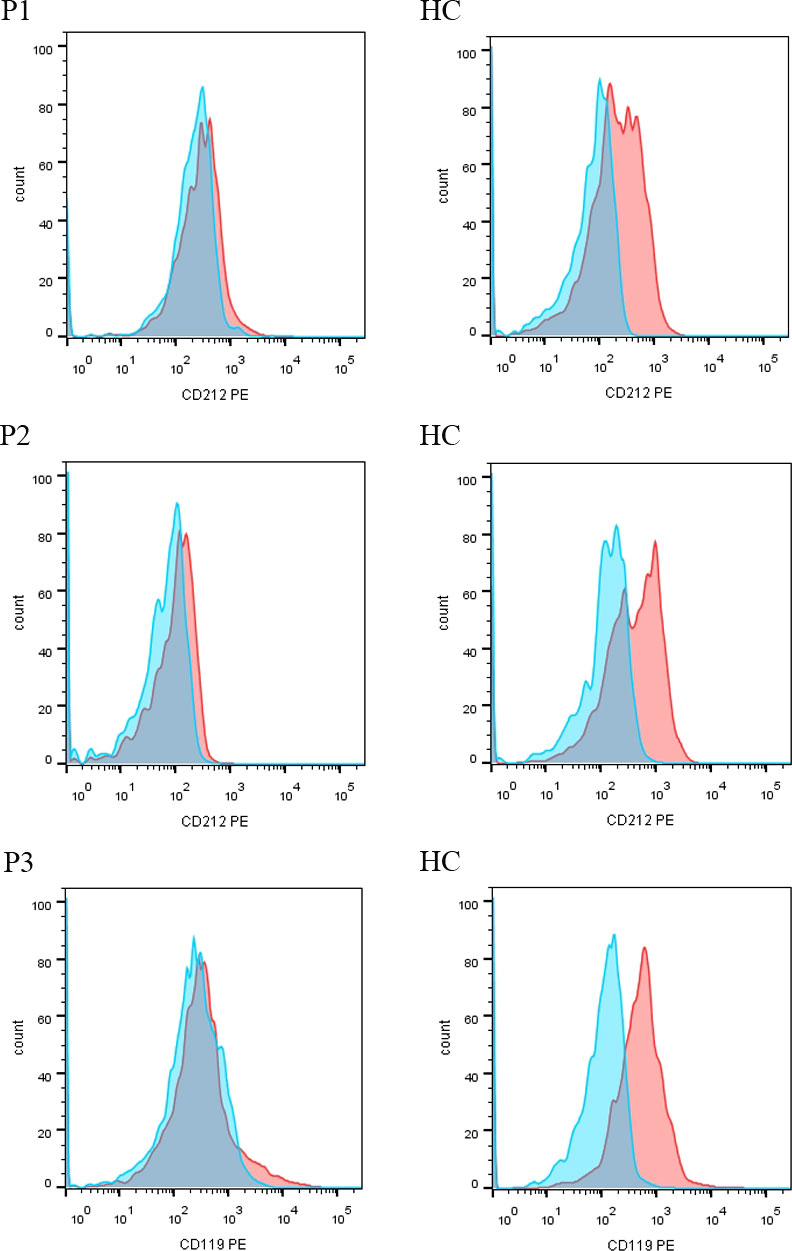
Figure 2 The expression of IL12RB1 (CD212) and IFNGR1 (CD119) protein in P1-P3. IL12RB1 protein was not expressed in P1 and P2. IFNGR1 protein was not expressed in P3.
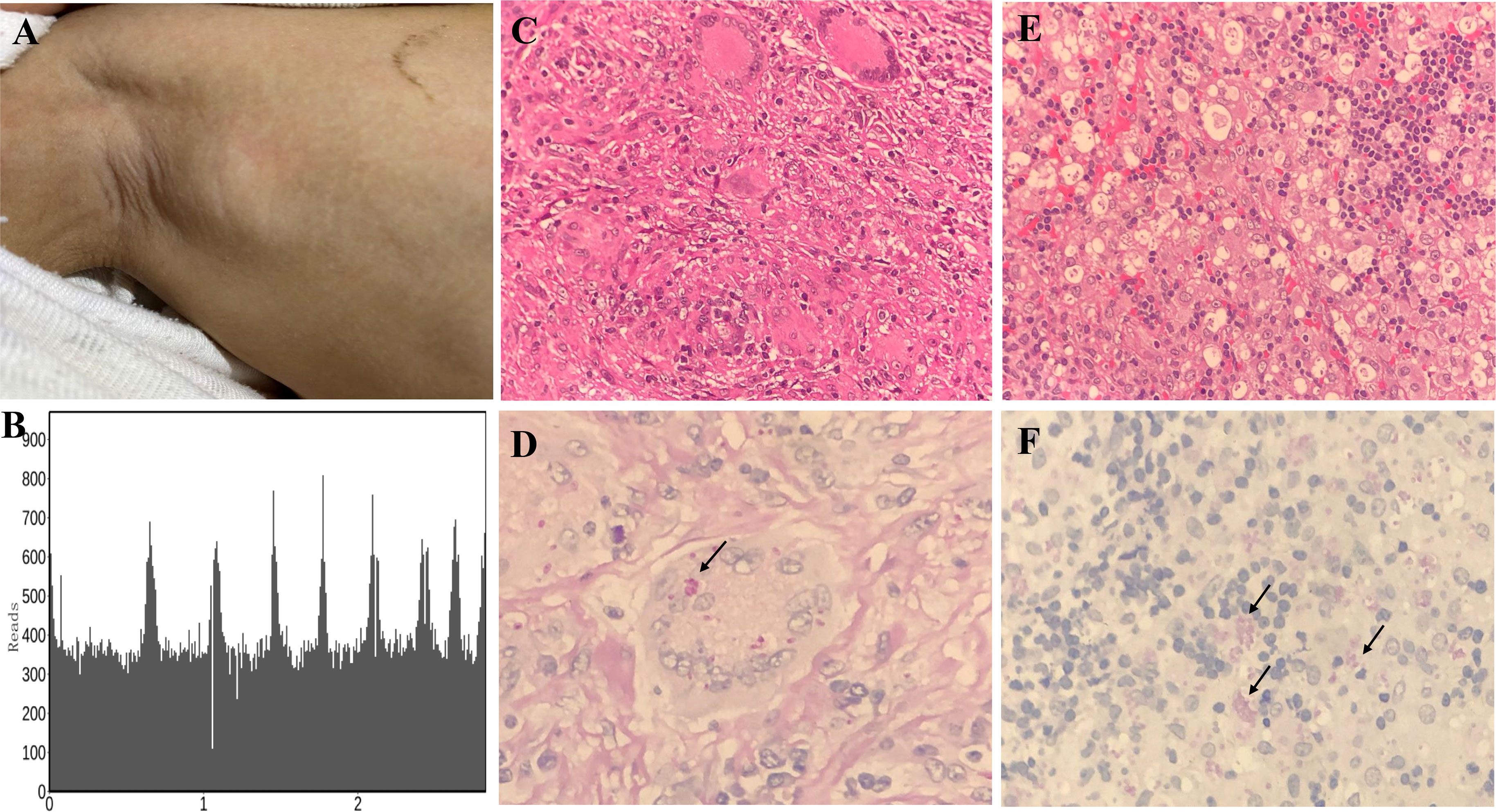
Figure 3 Histopathological staining and mNGS results in P1 and P5. Lymph node enlargement of the right axilla was seen in P1 (A). Confirmation of T. marneffei-specific amplification from lymph node tissue by mNGS showed 108,380 unique sequence reads of T. marneffei, accounting for 18.93% of the genome coverage (B). Granulomatous inflammation observed during histopathological examination of the cervical lymph node (C). PAS staining of the cervical lymph node revealed fungal spores (arrows) (magnification × 400) (D). A large number of neutrophil infiltrates were observed in the histopathological examination of abdominal lymph nodes of P5, and a patchy distribution of tissue cells was observed. Fungal spore-like substances were scattered or clustered in some tissue cells (magnification × 400) (E). PAS staining of the abdominal lymph node tissues revealed fungal spores (arrows) (magnification × 400) (F).
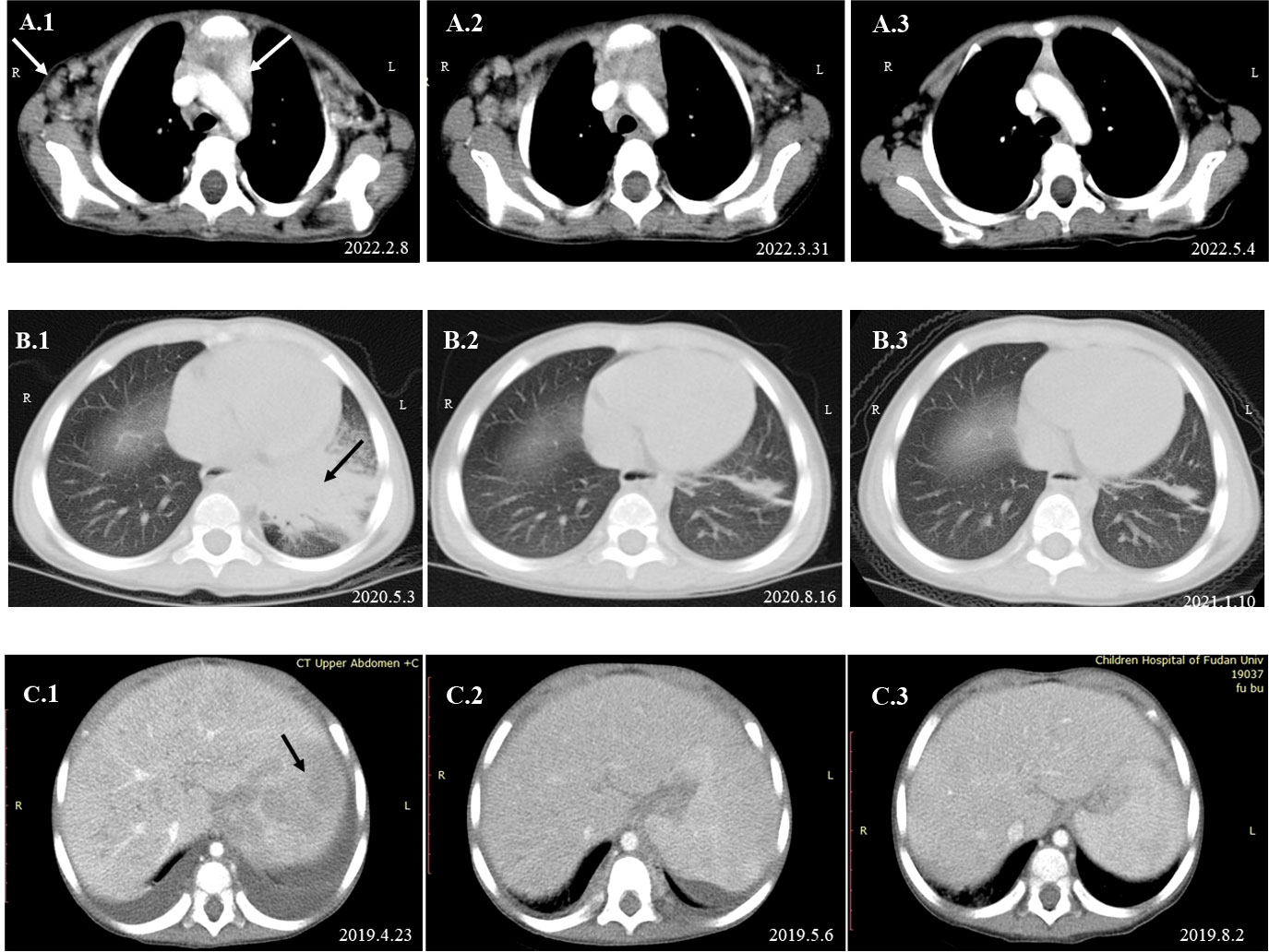
Figure 4 Dynamic changes in imaging examinations of patients during follow-up. Imaging examination of P1 revealed that the axillary and mediastinal lymph nodes were enlarged (A.1). After oral itraconazole and anti-tuberculous therapy for nearly 2 months, chest CT re-examination showed that the axillary and mediastinal lymph nodes were still enlarged (A.2). A lymph node biopsy was performed on April 19, 2022, and mNGS indicated high reads of T. marneffei. Antifungal therapy was adjusted to amphotericin B for 2 weeks, followed by oral itraconazole. One month later, the imaging examination suggested that the lymph nodes were smaller than before (A.3). Chest CT of P3 suggested pneumonia and partial consolidation of the left lung with slight pleural effusion in the acute phase (B.1), and the patient received oral itraconazole antifungal combined with anti-tuberculosis therapy. Chest CT re-examination revealed significant improvement in the lungs after 3 months (B.2) and 8 months (B.3). Abdominal CT of P5 indicated hepatosplenomegaly and multiple abnormal lesions at the beginning (C.1). He received intravenous voriconazole treatment for 25 days and then oral voriconazole. The multiple abnormal lesions improved after treatment for 2 weeks (C.2) and 3.5 months (C.3).
A 5-month-old boy (P2) was referred for enlarged axillary lymph nodes and ulceration of the BCG vaccination site. From 3 months of age, the child presented with left axillary lymph node enlargement and suppuration at the BCG vaccination site. Lymph node dissection and abscess removal were performed at 4 months of age. Acid fast bacilli were seen on pus smear, and Xpert examination of focal tissue suggested Mycobacterium tuberculosis. Mycobacterium tuberculosis (reads, 124) and Escherichia coli (reads, 952090) were detected in stool by mNGS. The child began to receive anti-tuberculosis treatment with isoniazid, rifampicin and ethambutol. At the age of 5 months, his lung CT indicated pulmonary cavities, and Mycobacterium tuberculosis was detected in BALF by mNGS. Levofloxacin was further added. In addition, the two blood GM test results were 0.589 and 1.497, respectively. Two mNGS tests of blood were performed 9 days apart, both of which were positive for T. marneffei (reads 29 and 6), and itraconazole was used for antifungal treatment. Whole-exome sequencing revealed a homozygous mutation of the IL12RB1 gene, c.632G>C, p. R211P, derived from his parents. IL12RB1 protein was not expressed (Figure 2). He received oral itraconazole for 6 months with a good response and has continued anti-tuberculosis treatment to date.
This male child was hospitalized for pneumonia during the neonatal period and was admitted with sepsis and liver dysfunction at the age of 2 months. At 3 months of age, he developed enlarged left axillary lymph nodes, redness, swelling and ulceration at the BCG vaccination site. The PPD test was strongly positive, and lymph node biopsy suggested granulomatous lesions. Isoniazid, rifampicin and ethambutol were used as anti-tuberculosis therapy, and IFN-γ was given as immunotherapy. Further genetic analysis indicated a homozygous mutation in the IFNGR1 gene: c. 655G>A, p.G219R. IFNGR1 protein was confirmed not expressed (Figure 2). His parents discontinued medication on their own. The child developed leg pain at 24 months old and mandibular swelling 2 months later. Imaging examination suggested multiple bone destruction. The anti-tuberculosis treatment was performed again. He had recurrent fever, and lung CT suggested pneumonia and partial consolidation of the left lung (Figure 4B1). T. marneffei (reads, 15) was detected in BALF by mNGS, and then itraconazole was added for antifungal treatment for 9 months. After 3 and 8 months of antifungal treatment, chest CT revealed significant improvement (Figures 4B2, 4B3).
The girl presented with recurrent skin eczema with pruritus from 1 month after birth. At the age of 9 months, the child developed low fever with a peak temperature of 38°C, accompanied by cough and expectoration, and pulmonary CT suggested bronchopneumonia. Routine blood examination revealed eosinophils of 1.58 × 109/L and IgE raised to 548 IU/ml. The BALF was positive for T. marneffei by culture and mNGS (reads, 43) and CMV DNA. The GM test of the BALF was not tested at that time. She was treated with amphotericin B for 20 days, followed by voriconazole. Fever and cough recurred, and fibrobronchoscopy was performed again at 14 months. BALF indicated an elevated result of the GM test (1.335), and Staphylococcus aureus (reads, 42) and Streptococcus pneumoniae (reads, 14) were detected by mNGS. Voriconazole and linezolid were given for anti-infective therapy, and regular intravenous immunoglobulin (IVIG) was recommended. Gene testing suggested a STAT3 gene heterozygous mutation: c.1394 C>T, p. S465F. With an NIH score of 39 (Grimbacher et al., 1999; Grimbacher et al., 1999), she was diagnosed with hyper IgE syndrome. Hematopoietic stem cell transplantation was performed at 18 months of age. She was in good condition during the six-month follow-up after transplantation.
The patient developed recurrent nausea at 27 months of age without an obvious cause. Two months later, the symptoms worsened, with lethargy and fever. The G test was elevated at 1817.7 pg/ml, and abdominal CT indicated hepatosplenomegaly and multiple abnormal lesions (Figure 4C1). Routine blood tests suggested anemia and thrombocytopenia, while elevated levels of amylase and lipase suggested pancreatitis. Multiple lymph nodes were enlarged throughout the body, and histopathological examination of the abdominal lymph node biopsy presented a large number of neutrophil infiltrates and fungal spores in the tissue cells (Figures 3E, F). T. marneffei was further detected in blood by culture and mNGS (reads, 1452), and voriconazole was used for antifungal treatment. In addition, recurrent thrush began at the age of 1 year old, and genetic testing was performed, which revealed one de novo heterozygous mutation in the STAT1 gene: c.821G>A, p.R274Q. After voriconazole treatment for 3 months, T. marneffei was negative in blood by mNGS. The multiple abnormal lesions in the liver and spleen improved (Figure 4C3). Then, he took itraconazole as a preventive antifungal treatment. Now the child has stopped antifungal drugs and is generally being well.
The male patient presented with left axillary lymph node enlargement at 2 months of age, followed by supraclavicular lymph node enlargement. The 4-month-old child developed poor appetite, and Pneumocystis jiroveci was further detected in sputum and blood. The G test results significantly increased to 767-2242 pg/ml. Isoniazid and rifampicin were given for anti-tuberculosis treatment, sulfamethoxazole (SMZ) for Pneumocystis jiroveci, linezolid for bacteria, and itraconazole for anti-fungal treatment. According to his clinical presentations, he was considered to have an IEI, and his genetic test result was hemizygous CD40LG deletion. The child developed recurrent diarrhea and fever at the age of 2 after his parents stopped the medications on their own. Lung CT suggested exudation in both lungs. Abdominal MRI suggested abnormal signals in the spleen and both kidneys, multiple small lymph nodes in the abdominal cavity and ascites. T. marneffei (reads, 31) was detected in blood, and T. marneffei (reads, 58) and Pneumocystis jiroveci (reads, 4) were detected in BALF. Gastroenteroscopy showed ulceration of rectal, colonic, and small intestinal ulcers. Cefoperazone sulbactam, metronidazole, itraconazole, and SMZ were used as anti-infection treatments, and mesalazine was used as an inhibitor of intestinal inflammation. He died of sepsis after hematopoietic stem cell transplantation.
A statistical study was performed in 232 patients who underwent both traditional standard and mNGS tests simultaneously (Table 4). We defined the patient as a positive case if T. marneffei was detected, regardless of specimen type. Traditional gold standard tests were culture and histopathology. Both mNGS and traditional methods were positive in 3 patients and negative in 226 patients. T. marneffei was detected only by mNGS in 3 patients but was negative by traditional methods. Next, we evaluated the diagnostic accuracy of mNGS in detecting T. marneffei infection. The sensitivity, specificity, positive likelihood ratio, and negative likelihood ratio of mNGS in our cohort were 100%, 98.7%, 76.9, and 0, respectively. McNemar chi-square analyses showed that there was no significant difference between the two methods (p=0.25).
The sample detection methods of these 6 patients with T. marneffei infection were analyzed in detail. T. marneffei was identified in a total of 15 samples from six patients (Figure 5). In our cohort, T. marneffei was most commonly detected by mNGS, covering 5 blood samples, 3 BALF samples, and 2 lymph node samples. Only two blood samples and one BALF sample were identified by culture. The histopathological examination of the lymph nodes in two samples indicated the presence of T. marneffei infection. Culture and mNGS were performed on 12 samples from these six patients simultaneously. T. marneffei was identified in 2 samples by the culture method (16.7%, 2/12) and 8 samples by the mNGS method (66.7%, 8/12). One biopsy sample of a cervical lymph node was tested by histopathological staining and mNGS simultaneously. T. marneffei was identified with mNGS (reads, 15545702), and the histopathological PAS staining was also positive.
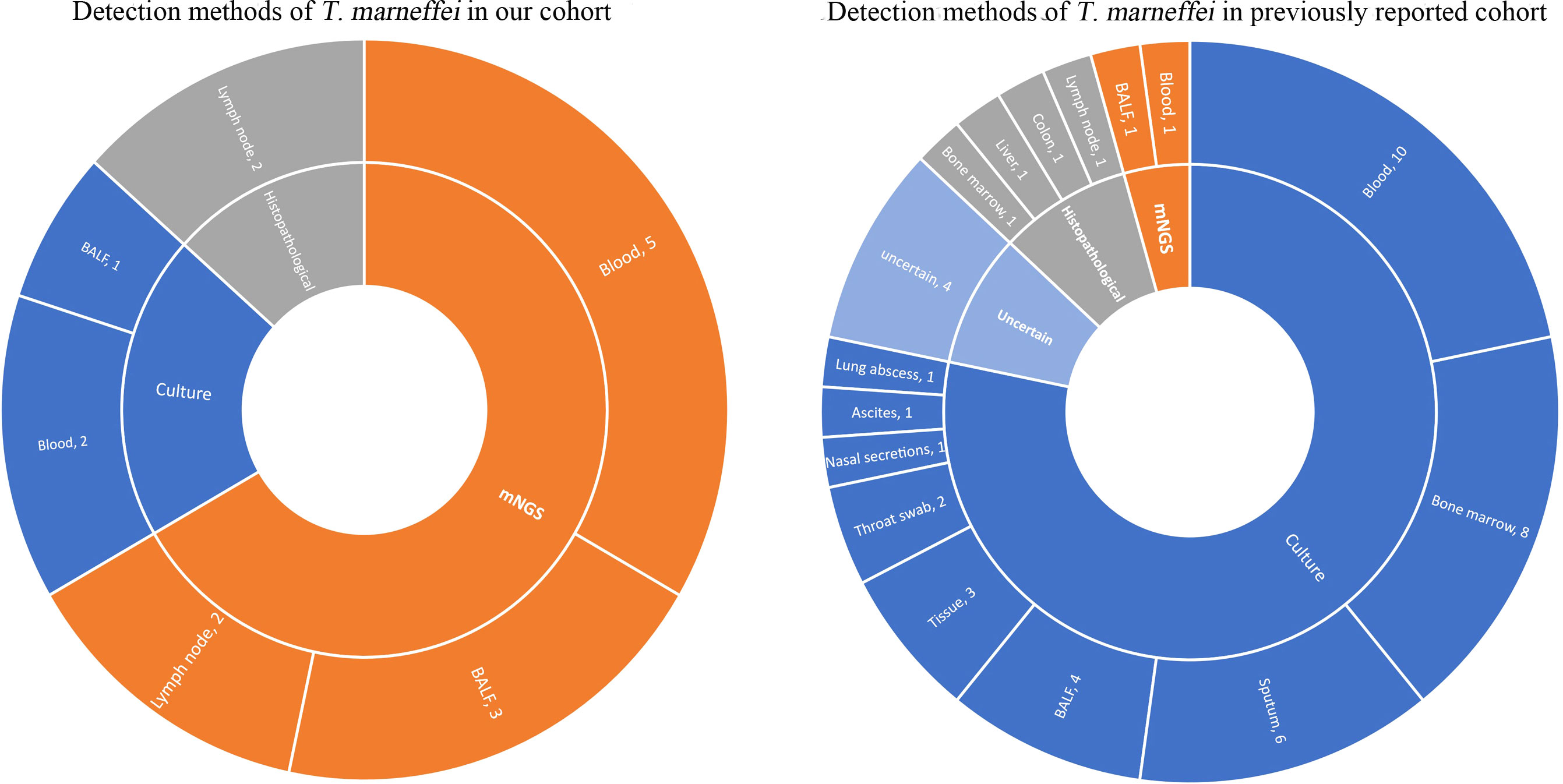
Figure 5 Distribution of sample types with T. marneffei positivity. In our cohort, T. marneffei was detected in 15 samples using different methods, including mNGS (n=10), culture (n=3), and histopathological staining (n=2). In the previous cohort, T. marneffei was mainly detected by cultures (n=36) and histopathological staining (n=4), and only two patients were detected by mNGS (n=2).
The time required for diagnosis using different methods for all six patients was further analyzed. The median reporting time for mNGS was 26 hours, with a range of 21.5-30 hours. The cultural identification periods of T.marneffei in the three positive samples were 3, 10 and 14 days. Two positive lymph node pathologic results were reported within 6 and 11 days. Therefore, the time consumption of mNGS was much less than that of culture or histopathology.
A systematic literature review was performed in PubMed on human T. marneffei infections published between 1950 and 2022 using the key words ‘‘Talaromyces marneffei’’ or ‘‘Penicillium marneffei’’ or ‘‘Penicilliosis’’ or ‘‘Talaromycosis’’. Articles reporting T. marneffei-infected patients with immune-related gene mutations were included. HIV-positive cases were excluded. As a result, a total of 28 cases with confirmed IEI from seventeen articles were analyzed. The characteristics of the patients are summarized in Table 5 (Yuen et al., 1986; Kamchaisatian et al., 2006; Ma et al., 2009; Sripa et al., 2010; Lee et al., 2012; Lee et al., 2014; Fan et al., 2018; Li et al., 2018; Du et al., 2019; Lee et al., 2019; Chen et al., 2020; Pan et al., 2020; Zhang et al., 2020; Ba et al., 2021; Chen et al., 2021; Fan et al., 2021; You et al., 2021).
The median age of patients detected with T. marneffei reported previously was 3 years, ranging from 5 months to 34 years. Most of them were Chinese (82%, 23/28), and the others were from Thailand. Twenty patients (71%) had disseminated T. marneffei disease involving blood or bone marrow, lungs, colon, skin, lymph nodes, and liver. The immunodeficiency genes included CD40LG (35%), STAT1 (29%), STAT3 (22%), CARD9 (7%) and IFNGR1 (7%). Comparisons between our cohort and previously reported gene spectra are shown in Figure 1B.
A total of 46 positive clinical samples from these 28 patients were collected (Figure 5). T. marneffei was mainly detected by cultures (78.2%) of blood, bone marrow, sputum, BALF, tissue and so on. Histopathological staining of the biopsy tissue was followed (8.7%), including lymph node, liver, colonic mucosa and bone marrow samples. Only two patients were detected to have T. marneffei by mNGS (4.3%), 566 reads in BALF (Zhang et al., 2020) and 248 reads in blood (Ba et al., 2021). The detection method of T. marneffei in the remaining four samples (8.7%), including endobronchial biopsy, lymph node biopsy and hepatic biopsy, was not available.
Nearly half of the patients (12/28) were treated with amphotericin B, followed by itraconazole or voriconazole. Other patients were treated with itraconazole (5/28), voriconazole (2/28), or amphotericin B (3/28) alone. Only one patient received intravenous amphotericin B and oral flucytosine. The treatments of five patients were unknown. Except for one patient lost to follow-up, most patients (21/28) had improved symptoms with effective antifungal therapy. Six patients (6/28) died. Two patients died of multiorgan failure, one patient died of respiratory failure due to rapid disease progression, one patient died of massive pulmonary hemorrhage at 16 years old, and the other two patients died of unknown reason.
T. marneffei is an opportunistic infectious pathogen. Inhalation of conidia is the primary route of infection, which are subsequently phagocytosed by alveolar macrophages, and then T. marneffei disseminates to the reticuloendothelial system, causing systemic infection when the host immune response is suppressed (Supparatpinyo et al., 1994). Therefore, patients susceptible to T. marneffei are usually immunosuppressed. Fungal infections in children with IEI are a growing concern.
T-cell-mediated immunity plays a central role in the immune defense mechanism against T. marneffei infection. Congenital athymic mice developed severe pulmonary and disseminated systemic Penicillium disease (Kudeken et al., 1996; Kudeken et al., 1997). A literature review showed that CD40 ligand deficiency was the most common type of immune deficiency in T. marneffei infection. CD40L mediates the interaction between T cells and different cells through ligation with its receptor CD40 (van Kooten and Banchereau, 2000; Erdos et al., 2005; Du et al., 2019). CD40 ligand-deficient patients suffered from opportunistic infections, especially Pneumocystis jiroveci and Cryptosporidium parvum infections. Recently, Pamela P. Lee discussed endemic mycoses in IEI, in which she highlighted T. marneffei infection in CD40 ligand deficiency in Southeast Asia (Lee and Lau, 2017).
Intrinsic and innate immunodeficiencies associated with T. marneffei infection have also been reported, such as STAT1 and CARD9. Caspase recruitment domain–containing protein 9 (CARD9) is an adaptor molecule in the cytosol of myeloid cells required for the induction of T-helper cells producing interleukin-17 (Th17 cells), and it can effectively integrate the recognition signals of various natural immune receptors and plays an important role in antifungal immunity (Drummond et al., 2011; Salazar and Brown, 2018; You et al., 2021). Signal transducer and activator of transcription (STAT) proteins are critical transcription factors for the appropriate regulation of cellular responses to interferons, cytokines, growth factors, and hormones. There is tight regulation of their function via several mechanisms, and STAT1 and STAT3 play a central role (Villarino et al., 2015). A consistent immunophenotype in GOF STAT1 and HIES patients is impaired development of Th17 lymphocytes, which could be the reason for their susceptibility to chronic mucocutaneous candidiasis or invasive fungal infection (Stark and Darnell, 2012; Olbrich and Freeman, 2018; Danion et al., 2020). This could explain the T. marneffei infection of P4 with STAT3 and P5 with STAT1 mutation.
It is noteworthy that the IL12RB1 gene was first reported to be associated with T. marneffei infection in our cohort. T. marneffei infection has previously been reported in children with IFNGR1 gene mutations. Both the IL12RB1 and IFNGR1 genes are involved in Mendelian susceptibility to mycobacterial disease (MSMD). Susceptibility to nontuberculous mycobacteria (NTM) infection, talaromycosis, histoplasmosis, cryptococcosis, melioidosis, and nontyphoidal salmonellosis has been reported in IFN-γ knockout mice or in patients with IFN-γ signaling defects, such as MSMD (Erb et al., 1999; Tang et al., 2010; Lee et al., 2013; Chi et al., 2016). A high prevalence of anti–IFN-γ autoantibodies is the major cause of severe T. marneffei infections in HIV-negative adults in China, which suggests that IFN-γ has a role in combating this fungal infection in humans (Guo et al., 2020). Human and mouse macrophages can control T. marneffei growth and kill intracellular yeast cells when activated by T-cell-derived cytokines (Cogliati et al., 1997; Sisto et al., 2003). IFN-γ is essential in the control of intracellular pathogens, such as Mycobacterium tuberculosis and fungi (Cooper et al., 1993). The fungicidal activity of T. marneffei yeast by macrophages could be enhanced by IFN-γ via stimulation of macrophages, which involves the nitric oxide (NO)-mediated killing system (Cogliati et al., 1997; Kudeken et al., 1998). In our study, three children were confirmed to have MSMD, including IL12RB1 gene deficiency (P1, P2) and IFNGR1 deficiency (P3). They had Mycobacterium tuberculosis and T. marneffei infections simultaneously. The clinical manifestations of P1 were the most serious, with recurrent fever, weight loss, severe anemia, splenomegaly and lymphadenopathy, requiring prolonged intensive antifungal treatment, as well as concomitant anti-Mycobacterium treatment.
Recently, mNGS has been successfully applied in the diagnosis of disseminated T. marneffei infection. The traditional standard for infection diagnosis relies on culture, histopathological staining and microscopy. Studies have shown that the combination of quantitative polymerase chain reaction (qPCR) and serum GM detection can be a valuable tool for the diagnosis of T. marneffei infection (Li et al., 2020; Wei et al., 2021). Patients with fungemia often have high GM results, such as P1, P2, P5 and P6. The measurement of BALF-GM is likely to be a useful tool for diagnosing invasive aspergillosis (Guo et al., 2010); however, its role in diagnosing T. marneffei has not been well established. mNGS is a new diagnostic technology to sequence all biological genomes in various clinical samples (Voelkerding et al., 2009). Compared with culture-based methods, mNGS showed obvious advantages with respect to high detection efficiency and quick speed. The detection accuracy of microorganisms, along with the positive rate, was higher in mNGS (Tsang et al., 2021; Zhou et al., 2021), which provides a faster and more accurate diagnostic method in clinical practice. The diagnostic efficiency analysis revealed that mNGS had a high diagnostic sensitivity of 100% and specificity of 98.7%. The significantly high positive likelihood rate indicated high accuracy of mNGS in determining T. marneffei, and the significantly low negative likelihood rate predicted that negative mNGS results could exclude T. marneffei infection. However, there was no significant difference between the two methods (p=0.25). Our study was a retrospective study, and there could be a risk of bias, which may originate in the relatively few patients enrolled, data collection and incomplete clinical data.
In clinical practice, the rapid diagnosis of T. marneffei infection in patients with IEI is of great importance. Disseminated T. marneffei infection may run a rapidly progressive course and be life-threatening without timely and effective antifungal therapy. However, cultures are quite time consuming (Cao et al., 2019), with a relatively limited positive rate due to the difficulty in cultivating slow-growing and fastidious microbes (Mo et al., 2002), which results in delayed diagnosis and increased mortality. A few studies have shown that mNGS has marked advantages over conventional methods for pathogenic diagnosis, particularly opportunistic pathogens and mixed infections in patients with IEI (Parize et al., 2017; Tang et al., 2021). For novel, rare, and treatment-refractory infectious diseases and for patients with immunocompromising disease, mNGS can significantly improve the pathogen detection rate and can be used as the front-line detection method (Cao et al., 2020). Unlike previous studies, mNGS was the main methodology in our cohort. mNGS can detect T. marneffei in a variety of samples, while traditional culture methods are often negative. In addition, the detection period of T. marneffei by mNGS was approximately 26 hours. Since infection can progress rapidly in children with immunodeficiency disease, pathogens should be identified as soon as possible using mNGS, and treatment can be initiated in time. Immunodeficient children may benefit from rapid detection of pathogens by mNGS.
T. marneffei is an opportunistic pathogen, suggesting potential immune impairment in infected individuals. We reported two cases of IL12RB1 mutation in children infected with T. marneffei, extending the immunodeficiency spectrum of T. marneffei infection. For IEI patients with T. marneffei infection, we highlight the application of mNGS in the clinical diagnosis. mNGS is proposed as an important adjunctive diagnostic approach for rapidly identifying pathogens in complex and severe infections.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI repository, the accession number is PRJNA860094.
The studies involving human participants were reviewed and approved by the ethics committee of Children’s Hospital of Fudan University. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. Written informed consent was obtained from the minor(s)’ legal guardian/next of kin for the publication of any potentially identifiable images or data included in this article.
LL and BS contributed to conceptualizing and designing the study, conducting the investigation, and drafting the manuscript. JH contributed to conceptualizing and designing the study, conducting the investigation, and reviewing and revising the manuscript. XW contributed to conceptualizing the study and reviewing and revising the manuscript. JS contributed to supervising the methodology and reviewing the manuscript. WW, WY, MY, XH, and QZ contributed to collecting clinical data, conducting the investigation, carrying out the initial analyses, and reviewing the manuscript. YW and DL contributed to designing the methodology, conducting the laboratory experiments, and reviewing the manuscript. All authors approved the final manuscript as submitted and agreed to be accountable for all aspects of the work.
We gratefully acknowledge Meili Shen from Dinfectome Inc., Nanjing, China, who helped with the bioinformatics analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ba, H., Peng, H., Cheng, L., Lin, Y., Li, X., He, X., et al. (2021). Case report: Talaromyces marneffei infection in a Chinese child with a complex heterozygous CARD9 mutation. Front. Immunol. 12, 685546. doi: 10.3389/fimmu.2021.685546
Cao, Q., Chen, B., Chen, L., Chen, P., Bijie, H. (2020). Expert consensus on the clinical application of metagenomics next-generation sequencing technology for detection of infectious pathogens in China. Chin. J. Infect. Dis. 38 (11), 681–689. doi: 10.3760/cma.j.cn311365-20200731-00732
Cao, C., Xi, L., Chaturvedi, V. (2019). Talaromycosis (Penicilliosis) due to talaromyces (Penicillium) marneffei: Insights into the clinical trends of a major fungal disease 60 years after the discovery of the pathogen. MYCOPATHOL [Journal Article; Review]. 184 (6), 709–720. doi: 10.1007/s11046-019-00410-2
Chen, Z., Cheng, H., Cai, Z., Wei, Q., Li, J., Liang, J., et al. (2021). Identification of microbiome etiology associated with drug resistance in pleural empyema. Front. Cell Infect. Microbiol. [Journal Article; Res. Support Non-U.S. Gov't]. 11, 637018. doi: 10.3389/fcimb.2021.637018
Chen, K., Tan, J., Qian, S., Wu, S., Chen, Q. (2021). Case report: Disseminated talaromyces marneffei infection in a patient with chronic mucocutaneous candidiasis and a novel STAT1 gain-of-Function mutation. Front. Immunol. [Case Reports; Res. Support Non-U.S. Gov't]. 12, 682350. doi: 10.3389/fimmu.2021.682350
Chen, X., Xu, Q., Li, X., Wang, L., Yang, L., Chen, Z., et al. (2020). Molecular and phenotypic characterization of nine patients with STAT1 GOF mutations in China. J. Clin. Immunol. [Journal Article; Res. Support Non-U.S. Gov't]. 40 (1), 82–95. doi: 10.1007/s10875-019-00688-3
Chi, C. Y., Lin, C. H., Ho, M. W., Ding, J. Y., Huang, W. C., Shih, H. P., et al. (2016). Clinical manifestations, course, and outcome of patients with neutralizing anti-interferon-gamma autoantibodies and disseminated nontuberculous mycobacterial infections. Med. (Baltimore). [Journal Article]. 95 (25), e3927. doi: 10.1097/MD.0000000000003927
Chiu, C. Y., Miller, S. A. (2019). Clinical metagenomics. Nat. Rev. Genet. [Journal Article; Review]. 20 (6), 341–355. doi: 10.1038/s41576-019-0113-7
Cogliati, M., Roverselli, A., Boelaert, J. R., Taramelli, D., Lombardi, L., Viviani, M. A. (1997). Development of an in vitro macrophage system to assess penicillium marneffei growth and susceptibility to nitric oxide. Infect. Immun. 65 (1), 279–284. doi: 10.1128/iai.65.1.279-284.1997
Cooper, A. M., Dalton, D. K., Stewart, T. A., Griffin, J. P., Russell, D. G., Orme, I. M. (1993). Disseminated tuberculosis in interferon gamma gene-disrupted mice. J. Exp. Med. 178 (6), 2243–2247. doi: 10.1084/jem.178.6.2243
Danion, F., Aimanianda, V., Bayry, J., Dureault, A., Wong, S., Bougnoux, M. E., et al. (2020). Aspergillus fumigatus infection in humans with STAT3-deficiency is associated with defective interferon-gamma and Th17 responses. Front. Immunol. 11, 38. doi: 10.3389/fimmu.2020.00038
Deng, Z., Ribas, J. L., Gibson, D. W., Connor, D. H. (1988). Infections caused by penicillium marneffei in China and southeast Asia: review of eighteen published cases and report of four more Chinese cases. Rev. Infect. Dis. [Case Reports; J. Article; Res. Support Non-U.S. Gov't; Review]. 10 (3), 640–652. doi: 10.1093/clinids/10.3.640
Ding, Y., Zhou, L., Xia, Y., Wang, W., Wang, Y., Li, L., et al. (2018). Reference values for peripheral blood lymphocyte subsets of healthy children in China. J. Allergy Clin. Immunol. [Letter; Res. Support Non-U.S. Gov't]. 142 (3), 970–973. doi: 10.1016/j.jaci.2018.04.022
DiSalvo, A. F., Fickling, A. M., Ajello, L. (1973). Infection caused by penicillium marneffei: description of first natural infection in man. Am. J. Clin. Pathol. [Journal Article]. 60 (2), 259–263. doi: 10.1093/ajcp/60.2.259
Drummond, R. A., Saijo, S., Iwakura, Y., Brown, G. D. (2011). The role of Syk/CARD9 coupled c-type lectins in antifungal immunity. Eur. J. Immunol. 41 (2), 276–281. doi: 10.1002/eji.201041252
Du, X., Tang, W., Chen, X., Zeng, T., Wang, Y., Chen, Z., et al. (2019). Clinical, genetic and immunological characteristics of 40 Chinese patients with CD40 ligand deficiency. Scand. J. Immunol. 90 (4), e12798. doi: 10.1111/sji.12798
Erb, K. J., Kirman, J., Delahunt, B., Moll, H., Le Gros, G. (1999). Infection of mice with mycobacterium bovis-BCG induces both Th1 and Th2 immune responses in the absence of interferon-gamma signalling. Eur. Cytokine Netw. 10 (2), 147–154.
Erdos, M., Durandy, A., Marodi, L. (2005). Genetically acquired class-switch recombination defects: the multi-faced hyper-IgM syndrome. Immunol. Lett. 97 (1), 1–6. doi: 10.1016/j.imlet.2004.09.021
Fan, H., Huang, L., Yang, D., Lin, Y., Lu, G., Xie, Y., et al. (2018). Pediatric hyperimmunoglobulin e syndrome: A case series of 4 children in China. Med. (Baltimore). [Case Reports; J. Article]. 97 (14), e215. doi: 10.1097/MD.0000000000010215
Fan, J. H., Luo, H. Y., Yang, L. G., Wang, M. Y., Xiao, Z. H. (2021). Penicilliosis marneffei in HIV negative children: three case reports. Ann. Palliat Med. [Case Reports]. 10 (7), 8437–8447. doi: 10.21037/apm-20-2056
Goldberg, B., Sichtig, H., Geyer, C., Ledeboer, N., Weinstock, G. M. (2015). Making the leap from research laboratory to clinic: Challenges and opportunities for next-generation sequencing in infectious disease diagnostics. MBIO. [Journal Article; Review]. 6 (6), e1815–e1888. doi: 10.1128/mBio.01888-15
Grimbacher, B., Holland, S. M., Gallin, J. I., Greenberg, F., Hill, S. C., Malech, H. L., et al. (1999). Hyper-IgE syndrome with recurrent infections–an autosomal dominant multisystem disorder. N Engl. J. Med. 340 (9), 692–702. doi: 10.1056/NEJM199903043400904
Grimbacher, B., Schaffer, A. A., Holland, S. M., Davis, J., Gallin, J. I., Malech, H. L., et al. (1999). Genetic linkage of hyper-IgE syndrome to chromosome 4. Am. J. Hum. Genet. 65 (3), 735–744. doi: 10.1086/302547
Guo, Y. L., Chen, Y. Q., Wang, K., Qin, S. M., Wu, C., Kong, J. L. (2010). Accuracy of BAL galactomannan in diagnosing invasive aspergillosis: a bivariate metaanalysis and systematic review. CHEST. [Journal Article; Meta-Analysis; Systematic Review]. 138 (4), 817–824. doi: 10.1378/chest.10-0488
Guo, J., Ning, X., Ding, J., Zheng, Y., Shi, N., Wu, F., et al. (2020) Anti–IFN-γ autoantibodies underlie disseminated talaromyces marneffei infections. J. Exp. Med 217(12), e20190502. doi: 10.1084/jem.20190502
Hu, Y., Zhang, J., Li, X., Yang, Y., Zhang, Y., Ma, J., et al. (2013). Penicillium marneffei infection: an emerging disease in mainland china. MYCOPATHOLOGIA. [Journal Article; Res. Support Non-U.S. Gov't]. 175 (1-2), :57–:67. doi: 10.1007/s11046-012-9577-0
Kamchaisatian, W., Kosalaraksa, P., Benjaponpitak, S., Hongeng, S., Direkwattanachai, C., Lumbiganon, P., et al. (2006). Penicilliois in patients with X-linked hyperimmunoglobulin m syndrome (XHIGM), case reports from Thailand. J. Allergy Clin. Immunol. 117 (2), S282. doi: 10.1016/j.jaci.2005.12.1166
Kawila, R., Chaiwarith, R., Supparatpinyo, K. (2013). Clinical and laboratory characteristics of penicilliosis marneffei among patients with and without HIV infection in northern Thailand: a retrospective study. BMC Infect. Dis. [Journal Article]. 13, 464. doi: 10.1186/1471-2334-13-464
Kudeken, N., Kawakami, K., Kusano, N., Saito, A. (1996). Cell-mediated immunity in host resistance against infection caused by penicillium marneffei. J. Med. Vet. Mycol. [Journal Article]. 34 (6), 371–378. doi: 10.1080/02681219680000671
Kudeken, N., Kawakami, K., Saito, A. (1997). CD4+ T cell-mediated fatal hyperinflammatory reactions in mice infected with penicillium marneffei. Clin. Exp. Immunol. [Journal Article]. 107 (3), 468–473. doi: 10.1046/j.1365-2249.1997.d01-945.x
Kudeken, N., Kawakami, K., Saito, A. (1998). Different susceptibilities of yeasts and conidia of penicillium marneffei to nitric oxide (NO)-mediated fungicidal activity of murine macrophages. Clin. Exp. Immunol. [Journal Article; Res. Support Non-U.S. Gov't]. 112 (2), 287–293. doi: 10.1046/j.1365-2249.1998.00565.x
Lee, P. P. W., Chan, K., Lee, T., Ho, M. H., Chen, X., Li, C., et al. (2012). Penicilliosis in children without HIV infection–are they immunodeficient? Clin. Infect. Dis. 54 (2), e8–19. doi: 10.1093/cid/cir754
Lee, W., Huang, J., Wu, T., Lee, M., Chen, I. J., Yu, K., et al. (2013). Patients with inhibitory and neutralizing auto-antibodies to interferon-γ resemble the sporadic adult-onset phenotype of mendelian susceptibility to mycobacterial disease (MSMD) lacking bacille calmette–guerin (BCG)-induced diseases. IMMUNOBIOLOGY. 218 (5), 762–771. doi: 10.1016/j.imbio.2012.08.281
Lee, P. P., Lao-araya, M., Yang, J., Chan, K., Ma, H., Pei, L., et al. (2019). Application of flow cytometry in the diagnostics pipeline of primary immunodeficiencies underlying disseminated talaromyces marneffei infection in HIV-negative children. Front. Immunol. 10, 2189. doi: 10.3389/fimmu.2019.02189
Lee, P. P., Lau, Y. L. (2017). Cellular and molecular defects underlying invasive fungal infections-revelations from endemic mycoses. Front. Immunol. 8, 735. doi: 10.3389/fimmu.2017.00735
Lee, P. P., Mao, H., Yang, W., Chan, K. W., Ho, M. H., Lee, T. L., et al. (2014). Penicillium marneffei infection and impaired IFN-gamma immunity in humans with autosomal-dominant gain-of-phosphorylation STAT1 mutations. J. Allergy Clin. Immunol. [Letter; Res. Support Non-U.S. Gov't]. 133 (3), 894–896. doi: 10.1016/j.jaci.2013.08.051
Le, T., Wolbers, M., Chi, N. H., Quang, V. M., Chinh, N. T., Huong Lan, N. P., et al. (2011). Epidemiology, seasonality, and predictors of outcome of AIDS-associated penicillium marneffei infection in ho chi minh city, Viet nam. Clin. Infect. Dis. 52 (7), 945–952. doi: 10.1093/cid/cir028
Li, W. J., Guo, Y. L., Liu, T. J., Wang, K., Kong, J. L. (2015). Diagnosis of pneumocystis pneumonia using serum (1-3)-beta-D-Glucan: a bivariate meta-analysis and systematic review. J. Thorac. Dis. [Journal Article]. 7 (12), 2214–2225. doi: 10.3978/j.issn.2072-1439.2015.12.27
Li, L., Li, J., Zheng, R., Zheng, Y., Wang, W. (2018). Clinical analysis of 3 HIV negative children with penicilliosis. Jiangxi Med. J. 53 (09), 986–989. doi: 10.3969/j.issn.1006-2238.2018.9.028
Li, Y., Sun, B., Tang, X., Liu, Y. L., He, H. Y., Li, X. Y., et al. (2020). Application of metagenomic next-generation sequencing for bronchoalveolar lavage diagnostics in critically ill patients. Eur. J. Clin. Microbiol. Infect. Dis. [Journal Article]. 39 (2), 369–374. doi: 10.1007/s10096-019-03734-5
Liyan, X., Changming, L., Xianyi, Z., Luxia, W., Suisheng, X. (2004). Fifteen cases of penicilliosis in guangdong, China. MYCOPATHOLOGIA 158 (2), 151–155. doi: 10.1023/B:MYCO.0000041842.90633.86
Li, X., Zheng, Y., Wu, F., Mo, D., Liang, G., Yan, R., et al. (2020). Evaluation of quantitative real-time PCR and platelia galactomannan assays for the diagnosis of disseminated talaromyces marneffei infection. Med. Mycol. [Evaluation Study; J. Article]. 58 (2), 181–186. doi: 10.1093/mmy/myz052
Lu, Y., Chen, Y. Q., Guo, Y. L., Qin, S. M., Wu, C., Wang, K. (2011). Diagnosis of invasive fungal disease using serum (1–>3)-beta-D-glucan: a bivariate meta-analysis. Intern. Med. [Journal Article; Meta-Analysis]. 50 (22), 2783–2791. doi: 10.2169/internalmedicine.50.6175
Ma, B. H. M., Ng, C. S. H., Lam, R. K. Y., Wan, S., Wan, I. Y. P., Lee, T. W., et al. (2009). Recurrent hemoptysis with penicillium marneffei and stenotrophomonas maltophilia in job's syndrome. Can. Respir. J. 16 (4), e50–e52. doi: 10.1155/2009/586919
Mo, W., Deng, Z., Li, S. (2002). Clinical blood routine and bone marrow smear manifestations of disseminated penicilliosis marneffei. Chin. Med. J. (Engl). [Journal Article]. 115 (12), 1892–1894.
Olbrich, P., Freeman, A. F. (2018). STAT1 and STAT3 mutations: important lessons for clinical immunologists. Expert Rev. Clin. IMMU. 14 (12), 1029–1041. doi: 10.1080/1744666X.2018.1531704
Pan, M., Qiu, Y., Zeng, W., Tang, S., Wei, X., Zhang, J. (2020). Disseminated talaromyces marneffei infection presenting as multiple intestinal perforations and diffuse hepatic granulomatous inflammation in an infant with STAT3 mutation: a case report. BMC Infect. Dis. 20 (1):394. doi: 10.1186/s12879-020-05113-4
Parize, P., Muth, E., Richaud, C., Gratigny, M., Pilmis, B., Lamamy, A., et al. (2017). Untargeted next-generation sequencing-based first-line diagnosis of infection in immunocompromised adults: a multicentre, blinded, prospective study. Clin. Microbiol. Infect. [Comparative Study; Eval. Study; J. Article; Multicenter Study]. 23 (8), 571–574. doi: 10.1016/j.cmi.2017.02.006
Qian, Y. Y., Wang, H. Y., Zhou, Y., Zhang, H. C., Zhu, Y. M., Zhou, X., et al. (2020). Improving pulmonary infection diagnosis with metagenomic next generation sequencing. Front. Cell Infect. Microbiol. [Journal Article; Res. Support Non-U.S. Gov't]. 10, 567615. doi: 10.3389/fcimb.2020.567615
Qiu, Y., Liao, H., Zhang, J., Zhong, X., Tan, C., Lu, D. (2015). Differences in clinical characteristics and prognosis of penicilliosis among HIV-negative patients with or without underlying disease in southern China: a retrospective study. BMC Infect. Dis. 15 (1):525. doi: 10.1186/s12879-015-1243-y
Salazar, F., Brown, G. D. (2018). Antifungal innate immunity: A perspective from the last 10 years. J. INNATE Immun. 10 (5-6), 373–397. doi: 10.1159/000488539
Schlaberg, R., Chiu, C. Y., Miller, S., Procop, G. W., Weinstock, G. (2017). Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch. Pathol. Lab. Med. [Journal Article; Validation Study]. 141 (6), 776–786. doi: 10.5858/arpa.2016-0539-RA
Segretain, G. (1959). [Penicillium marneffei n.sp., agent of a mycosis of the reticuloendothelial system]. Mycopathol Mycol Appl. 11, 327–353. doi: 10.1007/BF02089507
Sisto, F., Miluzio, A., Leopardi, O., Mirra, M., Boelaert, J. R., Taramelli, D. (2003). Differential cytokine pattern in the spleens and livers of BALB/c mice infected with penicillium marneffei: protective role of gamma interferon. Infect. Immun. 71 (1), 465–473. doi: 10.1128/IAI.71.1.465-473.2003
Sripa, C., Mitchai, J., Thongsri, W., Sripa, B. (2010). Diagnostic cytology and morphometry of penicillium marneffei in the sputum of a hypogammaglobulinemia with hyper-IgM patient. J. Med. Assoc. Thai. [Case Reports; J. Article]. 93 Suppl 3, S69–S72.
Stark, G. R., Darnell, J. J. (2012). The JAK-STAT pathway at twenty. IMMUNITY. 36 (4), 503–514. doi: 10.1016/j.immuni.2012.03.013
Su, S. S., Chen, X. B., Zhou, L. P., Lin, P. C., Chen, J. J., Chen, C. S., et al. (2022). Diagnostic performance of the metagenomic next-generation sequencing in lung biopsy tissues in patients suspected of having a local pulmonary infection. BMC PULM Med. [Journal Article]. 22 (1), 112. doi: 10.1186/s12890-022-01912-4
Supparatpinyo, K., Khamwan, C., Baosoung, V., Nelson, K. E., Sirisanthana, T. (1994). Disseminated penicillium marneffei infection in southeast Asia. Lancet [Journal Article] 344 (8915), 110–113. doi: 10.1016/s0140-6736(94)91287-4
Tang, B. S., Chan, J. F., Chen, M., Tsang, O. T., Mok, M. Y., Lai, R. W., et al. (2010). Disseminated penicilliosis, recurrent bacteremic nontyphoidal salmonellosis, and burkholderiosis associated with acquired immunodeficiency due to autoantibody against gamma interferon. Clin. Vaccine Immunol. 17 (7), 1132–1138. doi: 10.1128/CVI.00053-10
Tangye, S. G., Al-Herz, W., Bousfiha, A., Chatila, T., Cunningham-Rundles, C., Etzioni, A., et al. (2020). Human inborn errors of immunity: 2019 update on the classification from the international union of immunological societies expert committee. J. Clin. Immunol. [Journal Article; Res. Support Non-U.S. Gov't]. 40 (1), 24–64. doi: 10.1007/s10875-019-00737-x
Tang, W., Zhang, Y., Luo, C., Zhou, L., Zhang, Z., Tang, X., et al. (2021). Clinical application of metagenomic next-generation sequencing for suspected infections in patients with primary immunodeficiency disease. Front. Immunol. [Comparative Study; J. Article; Res. Support Non-U.S. Gov't]. 12, 696403. doi: 10.3389/fimmu.2021.696403
Tsang, C. C., Teng, J., Lau, S., Woo, P. (2021). Rapid genomic diagnosis of fungal infections in the age of next-generation sequencing. J. Fungi (Basel). 7 (8), 636. doi: 10.3390/jof7080636
van Kooten, C., Banchereau, J. (2000). CD40-CD40 ligand. J. Leukoc. Biol. 67 (1), 2–17. doi: 10.1002/jlb.67.1.2
Villarino, A. V., Kanno, Y., Ferdinand, J. R., O'Shea, J. J. (2015). Mechanisms of Jak/STAT signaling in immunity and disease. J. Immunol. 194 (1), 21–27. doi: 10.4049/jimmunol.1401867
Voelkerding, K. V., Dames, S. A., Durtschi, J. D. (2009). Next-generation sequencing: from basic research to diagnostics. Clin. Chem. [Journal Article; Review]. 55 (4), 641–658. doi: 10.1373/clinchem.2008.112789
Wang, H., Lu, Z., Bao, Y., Yang, Y., de Groot, R., Dai, W., et al. (2020). Clinical diagnostic application of metagenomic next-generation sequencing in children with severe nonresponding pneumonia. PloS One [Journal Article; Res. Support Non-U.S. Gov't]. 15 (6), e232610. doi: 10.1371/journal.pone.0232610
Wei, H. Y., Liang, W. J., Li, B., Wei, L. Y., Jiang, A. Q., Chen, W. D., et al. (2021). Clinical characteristics and risk factors of talaromyces marneffei infection in human immunodeficiency virus-negative patients: A retrospective observational study. World J. Emerg. Med. [Journal Article]. 12 (4), 281–286. doi: 10.5847/wjem.j.1920-8642.2021.04.005
You, C., Hu, F., Lu, S., Pi, D., Xu, F., Liu, C., et al. (2021). Talaromyces marneffei infection in an HIV-negative child with a CARD9 mutation in China: A case report and review of the literature. MYCOPATHOLOGIA. 186 (4), 553–561. doi: 10.1007/s11046-021-00576-8
Yuen, W. C., Chan, Y. F., Loke, S. L., Seto, W. H., Poon, G. P., Wong, K. K. (1986). Chronic lymphadenopathy caused by penicillium marneffei: a condition mimicking tuberculous lymphadenopathy. Br. J. Surg. [Case Reports; J. Article]. 73 (12), 1007–1008. doi: 10.1002/bjs.1800731224
Zhang, W., Ye, J., Qiu, C., Wang, L., Jin, W., Jiang, C., et al. (2020). Rapid and precise diagnosis of t. marneffei pulmonary infection in a HIV-negative patient with autosomal-dominant STAT3 mutation: a case report. Ther. Adv. Respir. Dis. [Case Reports]. 14, 1022303031. doi: 10.1177/1753466620929225
Zhou, Q., Hui, X., Ying, W., Hou, J., Wang, W., Liu, D., et al. (2018). A cohort of 169 chronic granulomatous disease patients exposed to BCG vaccination: a retrospective study from a single center in shanghai, China (2004-2017). J. Clin. Immunol. [Journal Article; Multicenter Study; Res. Support Non-U.S. Gov't]. 38 (3), 260–272. doi: 10.1007/s10875-018-0486-y
Keywords: Talaromyces marneffei, metagenomic next-generation sequencing, inborn errors of immunity, IL12RB1 mutation, T-cell-mediated immunity, intrinsic and innate immunodeficiencies
Citation: Liu L, Sun B, Ying W, Liu D, Wang Y, Sun J, Wang W, Yang M, Hui X, Zhou Q, Hou J and Wang X (2022) Rapid diagnosis of Talaromyces marneffei infection by metagenomic next-generation sequencing technology in a Chinese cohort of inborn errors of immunity. Front. Cell. Infect. Microbiol. 12:987692. doi: 10.3389/fcimb.2022.987692
Received: 06 July 2022; Accepted: 17 August 2022;
Published: 08 September 2022.
Edited by:
Gilberto Sabino-Santos, Tulane University School of Public Health and Tropical Medicine, United StatesCopyright © 2022 Liu, Sun, Ying, Liu, Wang, Sun, Wang, Yang, Hui, Zhou, Hou and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jia Hou, ZG9jdG9yaG91amlhQGhvdG1haWwuY29t; Xiaochuan Wang, eGNod2FuZ0BzaG11LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.