- 1Department of Pharmacy, Shaoxing Hospital of Traditional Chinese Medicine Affiliated to Zhejiang Chinese Medical University, Shaoxing, China
- 2Department of Pharmacy, Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 3Department of Clinical Laboratory, The Quzhou Affiliated Hospital of Wenzhou Medical University, Quzhou People’s Hospital, Quzhou, China
- 4Core Facility, The Quzhou Affiliated Hospital of Wenzhou Medical University, Quzhou People’s Hospital, Quzhou, China
Objectives: To characterize one OXA-232-producing wzi93-KL112-O1 carbapenem-resistant Klebsiella pneumoniae (CRKP) co-harboring chromosomal blaCTX-M-15 and one rmpA2-associated virulence plasmid.
Methods: Minimum inhibitory concentrations (MICs) were measured via broth microdilution method. Conjugation, chemical transformation, string test and Galleria mellonella infection model experiments were also conducted. Whole-genome sequencing (WGS) was performed on the Illumina and Nanopore platforms. Antimicrobial resistance determinants were identified using ABRicate program with ResFinder database. Insertion sequences (ISs) were identified using ISfinder. Bacterial virulence factors were identified using virulence factor database (VFDB). Wzi, capsular polysaccharide (KL) and lipoolygosaccharide (OCL) were analyzed using Kleborate with Kaptive. Phylogenetic analysis of 109 ST15 K. pneumoniae strains was performed using core genome multilocus sequence typing (cgMLST) on the Ridom SeqSphere+ server. MLST, replicons type, SNP strategies and another cgMLST analysis for 45 OXA-232-producing K. pneumoniae strains were further conducted using BacWGSTdb server.
Results: K. pneumoniae KPTCM strain belongs to ST15 with wzi93, KL112 and O1. It possessed a multidrug-resistant (MDR) profile and was resistant to carbapenems (meropenem and ertapenem), ciprofloxacin and amikacin. Virulence assays demonstrated KPTCM strain possesses a low virulence phenotype. WGS revealed it contained one circular chromosome and nine plasmids. The carbapenemase-encoding gene blaOXA-232 was located in a 6141-bp ColKP3-type non-conjugative plasmid and flanked by ΔISEcp1 and ΔlysR-ΔereA. Interestingly, blaCTX-M-15 was located in the chromosome mediated by ISEcp1-based transposon Tn2012. Importantly, it harbored a rmpA2-associated pLVPK-like virulence plasmid with iutA-iucABCD gene cluster and one IS26-mediated MDR fusion plasmid according to 8-bp (AGCTGCAC or GGCCTTTG) target site duplications (TSD). Based on the cgMLST and SNP analysis, data showed OXA-232-producing ST15 K. pneumoniae isolates were mainly isolated from China and have evolved in recent years.
Conclusions: Early detection of CRKP strains carrying chromosomal blaCTX-M-15, OXA-232 carbapenemase and pLVPK-like virulence plasmid is recommended to avoid the extensive spread of this high-risk clone.
Introduction
Klebsiella pneumoniae is an important opportunistic pathogen associated with hospital- and community-acquired infections that has gained public attention due to its capability of acquiring resistance genes and plasmids (Kong et al., 2021a; Xu et al., 2021). Specifically, the emergence of carbapenem-resistant K. pneumoniae (CRKP) has limited effective therapies and poses a tremendous challenge in clinical settings (He et al., 2022). It is well documented that K. pneumoniae usually is classified into classical K. pneumoniae (cKP) and hypervirulent K. pneumoniae (hvKP) based on different virulence levels (Tian et al., 2021). In general, CRKP belong to the cKP group. When compared with cKP, hvKP mainly causes bloodstream infections (BSIs) and liver abscesses, both of which pose huge threat to patients (Wang et al., 2018; Wanford et al., 2021). Hypervirulent phenotype is mainly relevant to rmpA/rmpA2, aerobactin (iucABCD and iutA) and salmochelin (iroBCDN). These virulence factors are usually on a pLVPK-like virulence plasmid. (Kong et al., 2021b). Of greater concern, the amount of carbapenem-resistant hypervirulent K. pneumoniae (CR-hvKP) is increasing and infections caused by CR-hvKP strains generally lead to severe outcomes and mortality (Gu et al., 2018; Chen et al., 2021a).
OXA-232, a type of OXA-48-like enzyme, differs from OXA-48 and OXA-181 by five and one amino acid substitutions, respectively (Potron et al., 2013; Lahlaoui et al., 2017). OXA-232 could hydrolyze temocillin, penicillins, cefotaxime, cefepime and carbapenems with different hydrolytic activities (Potron et al., 2013). Catalytic activities of OXA-232 for all penicillins except temocillin (ten-fold lower) were two- to six-fold higher than OXA-181 and OXA-48, respectively (Potron et al., 2013; Oueslati et al., 2015). The catalytic activity of OXA-232 for imipenem was quite lower than OXA-181 and OXA-48. Hydrolysis abilities of meropenem and ertapenem were similar among OXA-232, OXA-48 and OXA-181 (Potron et al., 2013).
OXA-232-producing K. pneumoniae strains have been reported in Zhejiang and Shanghai, China (Shen et al., 2020b; Shi et al., 2020; Jia et al., 2021), but to our knowledge, no comprehensive phylogenetic analysis for ST15 K. pneumoniae isolates collected from various countries has been performed before. Moreover, a study by Zhu et al. in Yancheng, China reported clonal dissemination of ST15 OXA-232-producing K. pneumoniae strains (Zhu et al., 2021). Based on the short-read Illumina whole-genome sequencing (WGS), blaCTX-M-15 was found in an IncFIIK-type plasmid (Zhu et al., 2021). In addition, there are no further corresponding description about rmpA2-related virulence plasmid structure and virulence assessment experiments in their ST15 OXA-232-producing K. pneumoniae strains.
Here, one OXA-232-producing sequence type (ST) 15 CRKP in human bloodstream infection was isolated in January 2022 in China and the complete genetic characteristics were further investigated. This study provides a comprehensive description of the complete genomic features of a wzi93-KL112-O1 OXA-232-producing ST15 CRKP, which harbors a rmpA2-related pLVPK-like virulence plasmid and the chromosomal blaCTX-M-15 mediated by ISEcp1-based transposon Tn2012. Moreover, a multi-drug resistance plasmid may have evolved through IS26-mediated co-integration. This information will offer help to prevent and control the extensive spread of OXA-232-producing ST15 K. pneumoniae isolates. Importantly, the combination of short-read Illumina and long-read MinION WGS was performed to provide complete insight into the genomic structure features of resistance and virulence plasmids of K. pneumoniae.
Materials and methods
Flow chart illustrating the study process
A flow chart (Figure 1) was constructed to detail all experiments and procedures associated with this study using CmapTools v6.04 (https://cmap.ihmc.us) (Behzadi and Gajdacs, 2021). Based on different contents of experiments and bioinformatic analysis, this study was divided into two parts, consisting of Wet Lab and Dry Lab.
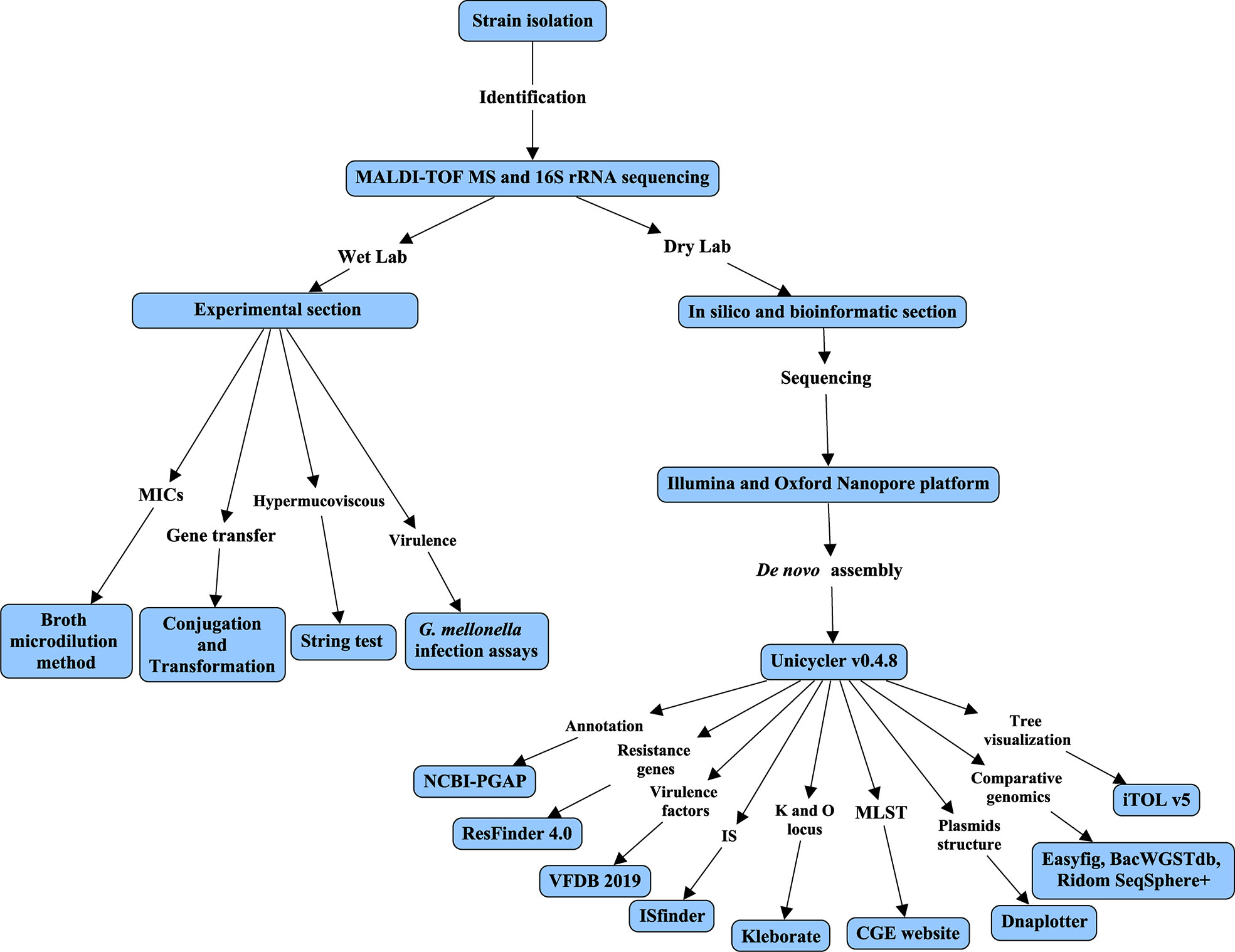
Figure 1 Flow chart of this study. The flow chart was constructed using CmapTools v6.04. The flow chart was divided into Wet and Dry labs based on experiments or bioinformatic analysis.
Bacterial isolation and identification
K. pneumoniae strain (KPTCM) was isolated from one patient with BSIs and liver abscess during the routine diagnostic in the hospital in Hangzhou, China in January 2022. No history of travel was reported. Unfortunately, liver abscess developed rapid and the outcome was death. According to the clinical symptoms of host, KPTCM may be a relatively hypervirulent strain and was chosen for further analysis. Isolate identification was performed by matrix-assisted laser desorption ionization-time of flight mass spectrometry ([MALDI-TOF MS] Bruker Daltonik GmbH, Bremen, Germany) and further confirmed by 16S rRNA gene-based PCR followed by sequencing (Huada, China).
Antimicrobial susceptibility testing
Minimum inhibitory concentrations (MICs) against multiple antimicrobial agents were determined using broth microdilution method and interpreted according to Clinical and Laboratory Standards Institute (CLSI) 2021 guidelines. Antibiotics, including imipenem (IPM), meropenem (MEM), ertapenem (ETP), amikacin (AMI), ciprofloxacin (CIP), colistin (COL), tigecycline (TGC), ceftazidime/avibactam (CAZ-AVI), ceftazidime (CAZ), cefepime (FEP), meropenem-vaborbactam (M/V) and cefiderocol (CFD), were investigated in this study. Escherichia coli ATCC 25922 served as the quality control strain.
Conjugation and chemical transformation experiments
To determine the transferable ability of plasmid carrying blaOXA-232, conjugation experiments using E. coli J53 (sodium azide resistant) as the recipient strain were carried out using film mating method (Yang et al., 2021b). Transconjugants were screened on Mueller–Hinton agar plates containing sodium azide (100 mg/L) and meropenem (2 mg/L). Donor or recipient bacteria alone was used as the control culture (Liu et al., 2021). The identity of putative transconjugants was confirmed via PCR and MALDI-TOF MS. Experiments were performed in triplicate independently.
Chemical transformation was further performed when conjugation failed. Plasmid harboring blaOXA-232 gene was transferred into E. coli DH5α via chemical transformation with imipenem (0.125 mg/L) for selection to yield E. coli DH5α:pOXA-232 (E. coli DH5α containing OXA-232 plasmid) (Froger and Hall, 2007). The MICs for carbapenems were measured and compared to the MICs values of E. coli DH5α.
Hypermucoviscous phenotype determination
String test assay was performed based on previous description (Shen et al., 2020a). Briefly, KPTCM strain was cultured on a sheep blood agar plate at 37 °C for an overnight culture followed by streaking an inoculation loop through a colony in the next day. Formation of a viscous string > 5 mm in length was considered as a positive phenotype.
Galleria mellonella infection model
Galleria mellonella infection model was used to assess virulence as previously described (Menard et al., 2021; Zhao et al., 2021). In brief, log-phase bacteria were centrifuged, resuspended using phosphate-buffered saline (PBS) and then diluted 10-fold to yield 1 × 108 colony forming units/mL (CFU/mL). An aliquot of 10 µL (1 × 106 CFU) was injected into G. mellonella larvae (n = 10, 0.2–0.3 g; Yuejiayin, Tianjin, China) using a Hamilton syringe (Hamilton, USA). G. mellonella larvae were incubated at 37°C and survival was recorded every 12 h and monitored for 72 h in total. K. pneumoniae NTUH-2044 was used as a positive control, and one cKP isolate (K. pneumoniae ATCC700603) served as a virulence negative control (Liao et al., 2020). Experiments were carried out in triplicate independently.
Whole genome sequencing and bioinformatics analysis
Genomic DNA was extracted from KPTCM strain using a Qiagen minikit (Qiagen, Hilden, Germany) upon the manufacturer’s recommendations with minor modifications. Whole genome was sequenced using both Illumina HiSeq platform (Illumina, San Diego, CA, USA) and Oxford Nanopore MinION platform (Nanopore, Oxford, UK). De novo assembly of the reads of Illumina and MinION was constructed using Unicycler v0.4.8 (Wick et al., 2017). Genome sequence annotation was performed using National Center for Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline (PGAP) (http://www.ncbi.nlm.nih.gov/genome/annotation_prok/) (Tatusova et al., 2016). Antimicrobial resistance determinants were identified using ABRicate program (https://github.com/tseemann/abricate) based on ResFinder database (http://genomicepidemiology.org/) (Zankari et al., 2012). Bacterial virulence factors were identified via virulence factor database (VFDB, http://www.mgc.ac.cn/VFs/) (Liu et al., 2022). Insertion sequences (ISs) were identified using ISfinder (Siguier et al., 2006). wzi, K and O antigen loci were analyzed using Kleborate with Kaptive (Lam et al., 2021; Lam et al., 2022). Plasmid structure was visualized using DNAplotter (https://www.sanger.ac.uk/tool/dnaplotter/). Plasmid comparison with p47733_OXA_181 (Genbank accession number: CP050368) (Chudejova et al., 2021) was performed using Easyfig (Sullivan et al., 2011) and visualized with Adobe Illustrator. Phylogenetic analysis of 109 ST15 K. pneumoniae strains was performed using core genome multilocus sequence typing (cgMLST) on the Ridom SeqSphere+ server (Chen et al., 2021b). MLST, replicon type, single-nucleotide polymorphism (SNP) strategies, and further cgMLST analysis for 45 OXA-232-producing K. pneumoniae strains were conducted using BacWGSTdb server (Ruan et al., 2020; Feng et al., 2021). Generation tree file was visualized using the Interactive Tree of Life (iTOL, https://itol.embl.de/) (Letunic and Bork, 2021).
Statistical analysis
G. mellonella survival rates were evaluated through Kaplan–Meier survival curves. Statistical significance was analyzed using a log-rank (Mantel–Cox) test with GraphPad Prism 8.0.2 software (GraphPad Software, San Diego California USA). P values of < 0.05 were considered statistically significant.
Results
MICs, antimicrobial resistance and virulence determinants
Antimicrobial susceptibility testing revealed KPTCM strain possessed a multidrug-resistant (MDR) profile with meropenem and ertapenem MICs of 4 mg/L and 8 mg/L, respectively. Furthermore, KPTCM strain exhibited resistance to ceftazidime (> 32 mg/L), cefepime (> 64 mg/L), ciprofloxacin (> 32 mg/L) and amikacin (> 128 mg/L). However, it was still susceptible to imipenem (1 mg/L), meropenem-vaborbactam (2 mg/L), cefiderocol (0.25 mg/L), ceftazidime/avibactam (0.5 mg/L), colistin (0.5 mg/L) and tigecycline (1 mg/L).
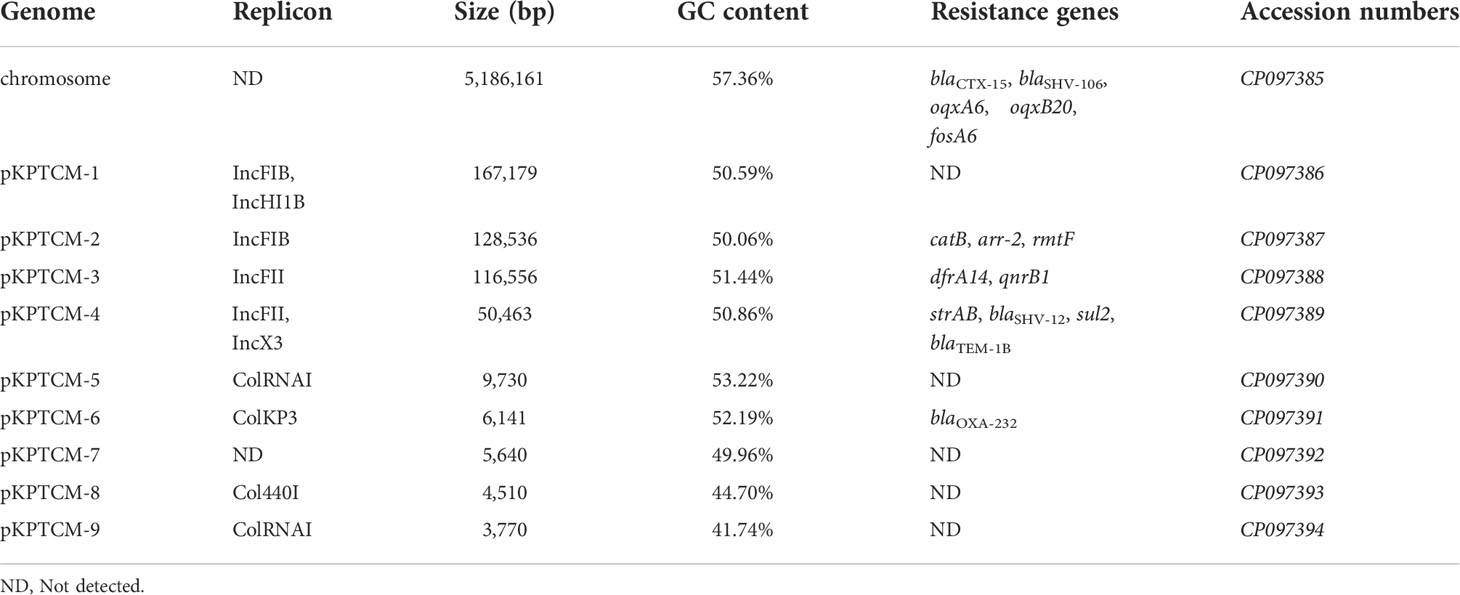
Analysis of the genome of KPTCM strain revealed that, in addition to co-harboring chromosomal blaCTX-M-15 and plasmid-mediated blaOXA-232, a series of genes conferring resistance to β-lactams (blaSHV-106, blaSHV-12 and blaTEM-1B), aminoglycosides (strAB and rmtF), sulfonamides (sul2), trimethoprim/sulfamethoxazole (dfrA14), quinolones (qnrB1), rifampicin (arr-2) and chloramphenicol (catB) were also identified (Table 1).
Many virulence factors have been observed in KPTCM strain, including rmpA2, iucABCD and iutA gene cluster, which is responsible for encoding aerobactin. Furthermore, colibactin (clbF and clbP), yersiniabactin (ybtAEPQSTUX, irp1, irp2 and fyuA), type 3 fimbriae (mrkABCDFHIJ) and genes encoding iron uptake function were further identified.
Multilocus sequence typing, wzi type, KL and OCL
Based on the MLST scheme, KPTCM strain was typed into ST15. Kaptive showed Wzi, an outer membrane protein lectin, was classified as Wzi93. Furthermore, KPTCM strain contained O locus 1 (O1), matching the 98.51% nucleotide identity. The K locus in KPTCM strain was found to be KL112, to which it matches with an overall nucleotide identity of 98.65%.
Transferability of plasmid harboring blaOXA-232
Mating assays were performed to investigate the transferability of blaOXA-232, however, blaOXA-232 failed to transfer to the recipient strain through conjugation. Concerning chemical transformation, MICs of E. coli DH5α:pOXA-232 to imipenem, meropenem and ertapenem rose to 1 mg/L, 0.25 mg/L and 2 mg/L, respectively, which was higher than that of E. coli DH5α strain (imipenem 0.06 mg/L, meropenem 0.03 mg/L and ertapenem 0.03 mg/L, respectively).
Virulence assessment of KPTCM strain
To explore the virulence level of KPTCM strain, virulence assays of string test and G. mellonella infection model were performed. As a result, negative phenotype was observed for string test experiment. In addition, G. mellonella infection model indicated virulence of KPTCM strain was significantly lower than standard hypervirulent isolate NTUH-2044 (P = 0.0005) but slightly higher than K. pneumoniae ATCC700603, used as negative control (Figure 2). However, no statistical significance was observed between KPTCM and K. pneumoniae ATCC700603 strains (P > 0.05).
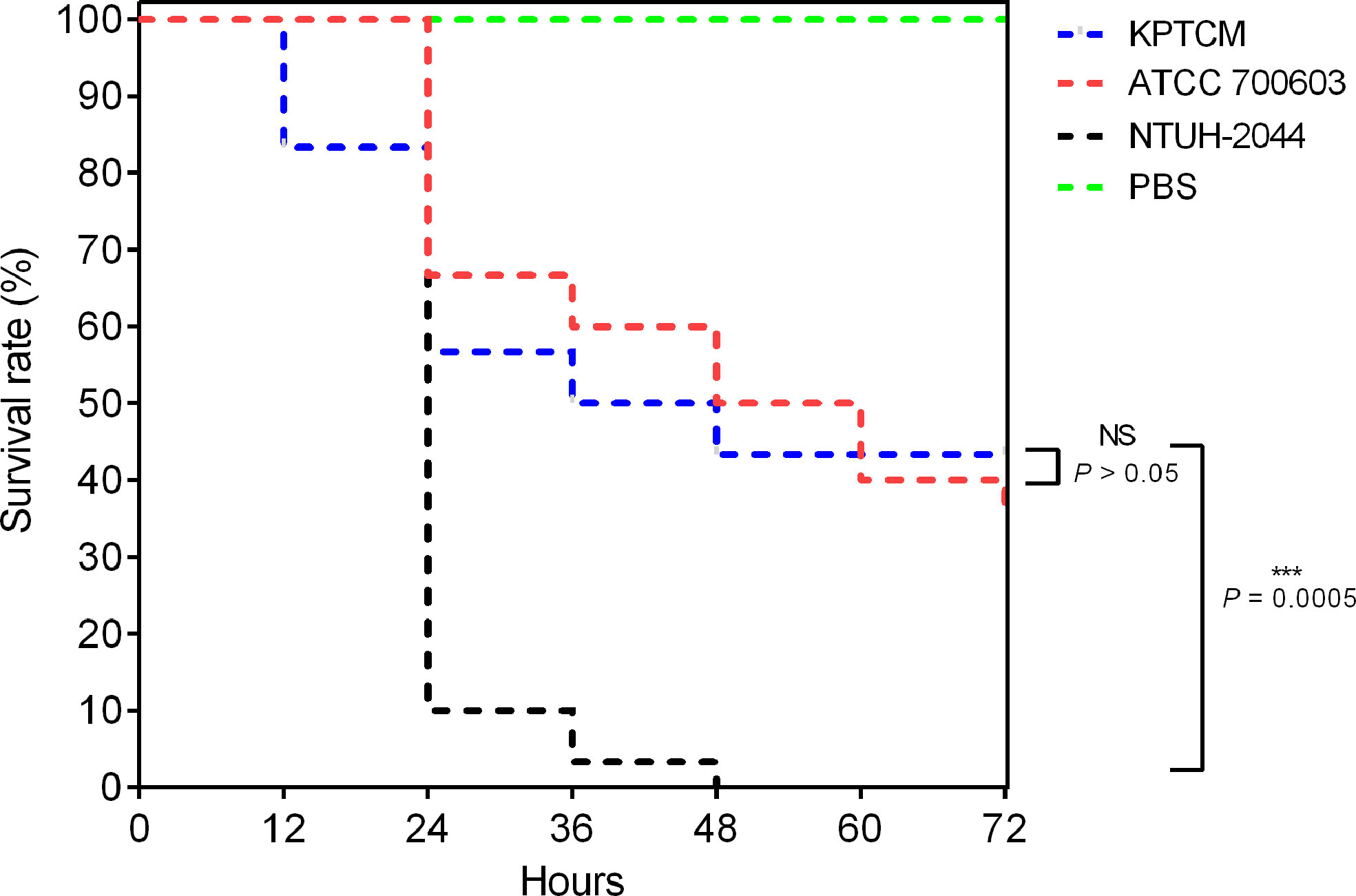
Figure 2 Survival curve in the G. mellonella infection model. G. mellonella larvae (n = 10) were inoculated with 106 colony-forming units (CFU). Survival was recorded every 12 h for 72 h. K. pneumoniae NTUH-2044 was used as a positive control, and classic K. pneumoniae ATCC700603 served as a virulence negative control. Data from three independent experiments are shown. NS: No significance, P > 0.05; ***:P = 0.0005.
Genomic characterization of the chromosome
The hybrid assembly of both Illumina and MinION reads showed KPTCM strain contains a 5,186,161-bp circular chromosome with GC content of 57.36% (Table 1). The blaCTX-M-15 gene was embedded in the chromosome via ISEcp1-based transposon Tn2012. Moreover, two amino acid mutations of gyrA (S83F) and parC (S80I) were identified, which confer resistance to fluoroquinolones.
Plasmids in KPTCM strain
Nine plasmids were solved in KPTCM clinical strain, namely pKPTCM-1 to pKPTCM-9, with sizes from 3,770-bp to 167,179-bp and GC contents ranging from 41.74% to 53.22% (Table 1). pKPTCM-1 is a rmpA2-related pLVPK-like virulence plasmid with two replicons (IncFIB and IncHI1B). pKPTCM-2, pKPTCM-3 and pKPTCM-4 plasmids were found to be MDR plasmids harboring various resistance genes. pKPTCM-6 is one ColKP3-type plasmid with one carbapenem resistance gene, blaOXA-232. However, no resistance genes were identified in pKPTCM-5, pKPTCM-7, pKPTCM-8 and pKPTCM-9.
Genetic features of pLVPK-like virulence plasmid pKPTCM-1
KPTCM clinical isolate harbored a 167,179-bp pLVPK-like virulence plasmid, namely pKPTCM-1, which was 99.65% identical to 177,790-bp pCR-HvKP1-VIR plasmid (GenBank accession number: CP040534) at 91% coverage (Figure 3A). pKPTCM-1 had an average GC content of 50.59% and consisted of four different regions, including virulence gene region, tellurium resistance region, silver/copper resistance region and mercury resistance region. rmpA2 and iucABCD-iutA gene cluster were located in the virulence gene region. In addition, frameshift mutation was observed in rmpA2 gene (Figure 3B and Figure S1).
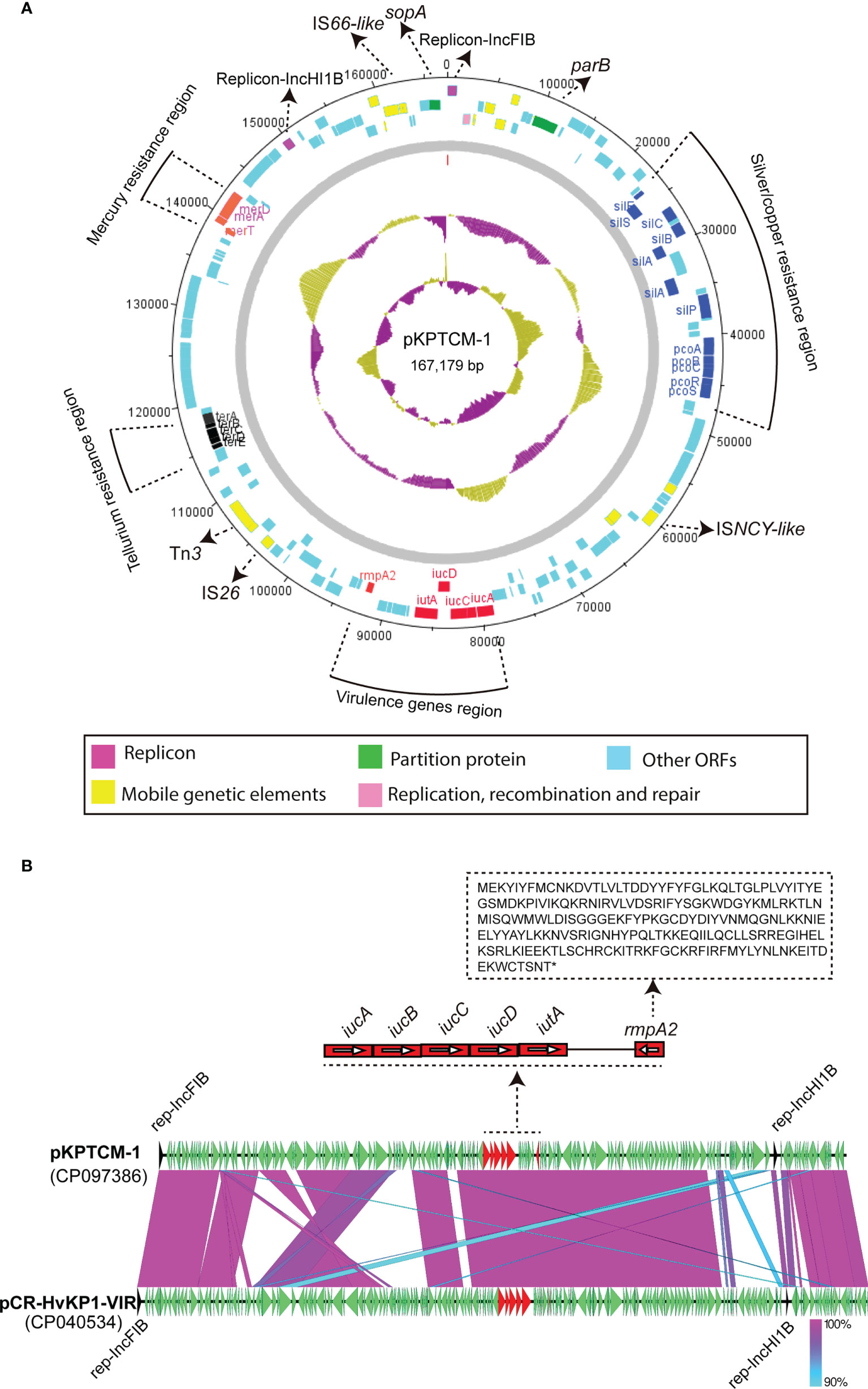
Figure 3 Circular map of pKPTCM-1 and comparison with pCR-HvKP1-VIR plasmid. (A) Circular map of pKPTCM-1 plasmid. Arrows show the direction of transcription of open reading frames (ORFs). Virulence genes region (red), tellurium resistance region (black), silver/copper resistance region (blue) and mercury resistance region (purple) are shown in different colors. ORFs of replicon, partition protein, mobile genetic elements (MGEs), replication, recombination and repair were further labeled. Light blue filled boxes represent other ORFs. (B) Structure of pKPTCM-1 compared with plasmid pCR-HvKP1-VIR (GenBank accession number: CP040534). The sequence of rmpA2 with the frameshift mutation was shown. Shades indicate regions with 90% to 100% identity.
IS26-mediated plasmids fusion of pKPTCM-4
pKPTCM-4 is an IncFII-IncX3-typed plasmid with GC content of 50.86% (Table 1). Five resistance genes and five IS26 genetic elements were identified in pKPTCM-4 plasmid (Figure 4A). In addition to IS26, IS5075, ISKpn21 and Tn3-like mobile genetic elements were also found. All resistance genes (strAB, blaSHV-12, sul2 and blaTEM-1B) were flanked by various mobile genetic elements. Moreover, 8-bp target site duplications ([TSD] AGCTGCAC) were observed upstream of all five IS26 genetic elements. The other 8-bp TSD (GGCCTTTG) were identified downstream of five IS26 in pKPTCM-4 (Figure 4B).
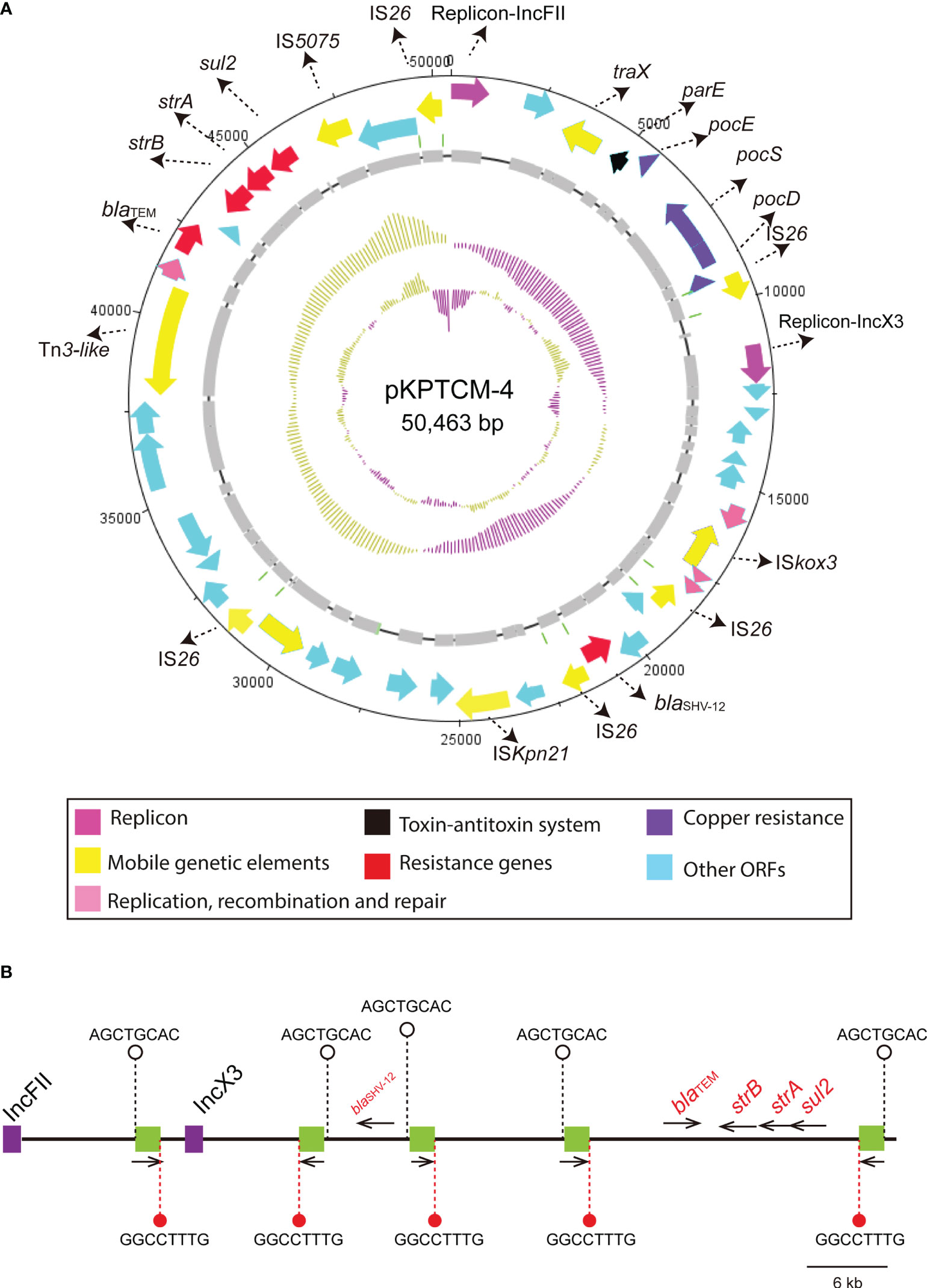
Figure 4 Circular map of pKPTCM-4 and IS26 distribution. (A) Circular map of pKPTCM-4 plasmid. Arrows show the direction of transcription of ORFs. Red arrows indicate the antimicrobial resistance genes. Pink arrows indicate the replicons. ORFs of resistance genes, MGEs, copper resistance, recombination and repair were further labeled. (B) IS26 elements are shown as green filled boxes. 8-bp target site duplications ([TSD] AGCTGCAC) were labeled on the left side, and the other 8-bp TSD (GGCCTTTG) were labeled on the right side of the IS26. Resistance genes were labeled as red with black thin arrows indicating ORF directions.
Genetic features of ColKP3-typed pKPTCM-6 plasmid
blaOXA-232 gene was located in a small 6,141-bp plasmid, designated pKPTCM-6 with ColKP3-typed replicon (Table 1). The plasmid was composed of repA, mob related genetic elements, ΔISEcp1, blaOXA-232, ΔlysR and ΔereA (Figure 5A). A truncated Tn2013 transposon structure was identified with 5-bp TSD (ATATA) on the right side (Figure 5B). Moreover, another ColKP3-typed plasmid p47733_OXA_181 (a blaOXA-181-carrying plasmid) showed 51% coverage and 99.9% identity to pKPTCM-6. The genetic environment between blaOXA-232 and blaOXA-181 was identical and both flanked by ΔISEcp1 and ΔlysR-ΔereA (Figure 5B).
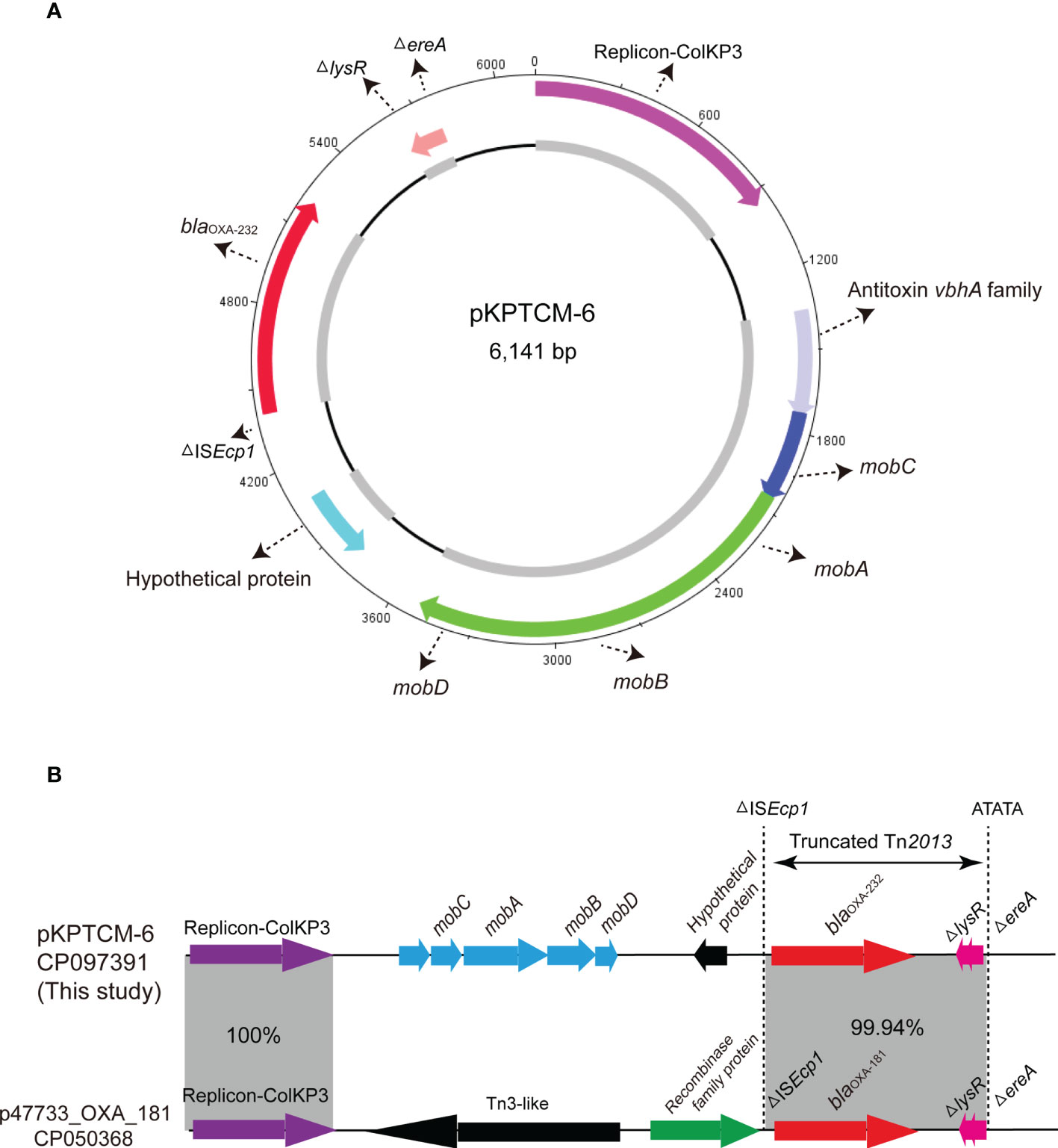
Figure 5 Circular map of pKPTCM-6. (A) Circular map of pKPTCM-6 plasmid. Arrows show the direction of ORFs. Red arrow indicate the blaOXA-232 gene. Pink arrow indicate the replicon. Light blue filled box represents hypothetical protein. Dark blue and green arrows indicate mobC, mobA, mobB and mobD. (B) Linear maps and comparison of the pKPTCM-6 and p47733_OXA_181 plasmids. Arrows show the direction of transcription of ORFs. Resistance genes are shown in red. The truncated Tn2013 was labeled with 5-bp TSD (ATATA). Homologous segments (representing ≥99.9% sequence identity) are indicated by grey shading.
Comparative genomics analysis of 109 ST15 K. pneumoniae strains and 45 OXA-232-producing K. pneumoniae strains
To analyse the characteristics of ST15 K. pneumoniae strains with close genetic relationship, genomes in public databases were searched and downloaded based on 200 SNPs threshold using BacWGSTdb server. To further study the characteristics of 109 ST15 K. pneumoniae strains from different countries, comparative genomics analysis was performed via Ridom SeqSphere+ server. All information of 109 ST15 K. pneumoniae strains is shown in Table S1. Based on the cgMLST tree, data showed ST15 K. pneumoniae isolates were mainly isolated from China (23.85%, 26/109), followed by Hungary (13.76%, 15/109), Spain (11.01%, 12/109) and United Kingdom (9.17%, 10/109) (Figure 6). ST15 K. pneumoniae strains collected from China and Hungary were shown in cluster 2 and 3, respectively. However, the number of strains isolated from other countries, such as Turkey, Nepal, Lebanon and Switzerland, was relatively small (Table S1).
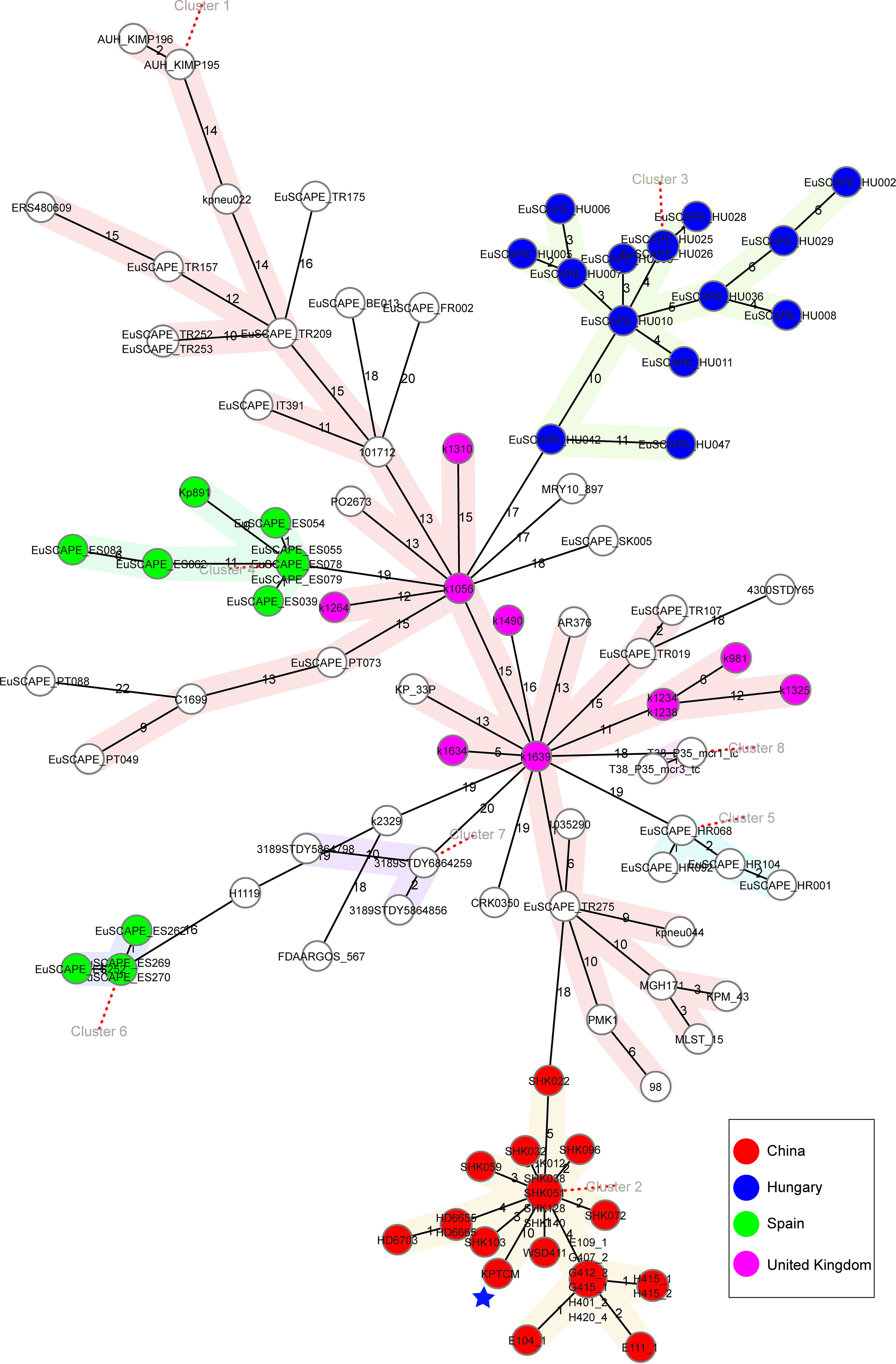
Figure 6 Phylogenetic analysis of 109 ST15 K. pneumoniae strains. cgMLST was conducted based on the 2358 alleles using Ridom SeqSphere+ server with cluster alert distance set at 15. Strains collected from China, Hungary, Spain and United Kingdom were shown as different colors. The blue pentagram indicated the position of KPTCM strain.
Other 19 OXA-232-producing K. pneumoniae strains were further searched based on published reports, the features of all 45 OXA-232-producing K. pneumoniae strains were analyzed using BacWGSTdb server. Interestingly, all K. pneumoniae strains carrying OXA-232 plasmid belonged to ST15 in China. However, STs in several countries, such as India (mainly ST11, ST23 and ST231), South Korea (ST14) and Italy (ST16), were quite different (Figure 7).
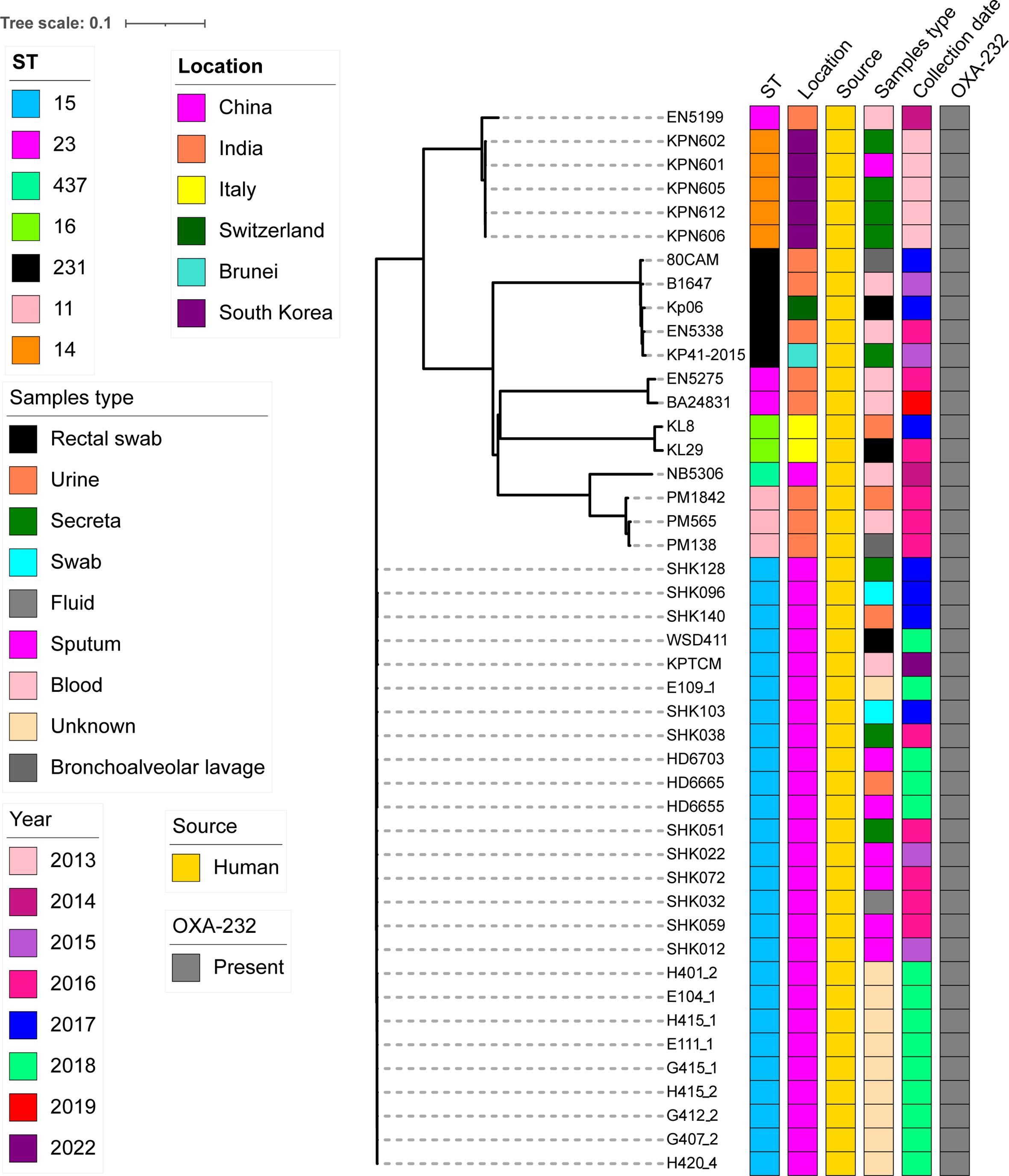
Figure 7 cgMLST analysis for 45 OXA-232-producing K. pneumoniae strains using BacWGSTdb server and visualized with iTOL v5. 26 OXA-232-producing ST15 K. pneumoniae strains from cluster 2 in Figure 6 and other 19 OXA-232-producing ST15 K. pneumoniae strains which were further searched from the published data. Isolates names, STs, locations, isolation date and collection sources are shown. The filled boxes reveal the various characteristics in different colors. The Genbank accession numbers of all 45 OXA-232-producing ST15 K. pneumoniae strains are shown in Table S2.
Of note, our data revealed that the first ST15 K. pneumoniae strain was isolated in 2015 in China. Strains were collected from several different sources, including blood, sputum, fluid, swab, urine, rectum and other sites (Figure 7). Moreover, SNP analysis was performed for the 26 OXA-232-producing ST15 strains from China. SNP strategy showed 28-222 SNPs differences among 26 ST15 K. pneumoniae isolates.
Discussion
The presence of carbapenemase-producing K. pneumoniae has become dominant in several countries, and it is being increasingly considered a quite important nosocomial pathogen (Yang et al., 2021c). More importantly, BSIs caused by CRKP present critical problems in terms of clinical therapy (Xiao et al., 2020). Hypervirulent K. pneumoniae strains could also cause serious infections among healthy individuals (Sewunet et al., 2021). These hypervirulent K. pneumoniae strains are usually resistant to antimicrobials agents and become high-risk clones in the hospital (Sewunet et al., 2021).
Mobile genetic elements, including ISs, integrons, and transposons, play a particularly important role in the resistance gene transfer among different species (Gorbunova et al., 2021). One interesting finding from this study is that blaCTX-M-15 was located in the chromosome, which was mediated by ISEcp1-based transposon Tn2012. ISEcp1 transposase gene has 14-bp inverted repeats (IRs) and is more inclined to insert into targets with the 5-bp AT-rich sites. ISEcp1 transposase gene is characterized by its capacity to transfer DNA fragments, such as resistance genes. Shu et al. reported pE109-1-CTX plasmid was found to harbor the resistance gene blaCTX-M-15 in China (Shu et al., 2019). Thus, it could be concluded that chromosomal blaCTX-M-15 in our strain may be derived from other plasmids mediated by ISEcp1-based transposon Tn2012. Moreover, the genetic environment of arr-2 gene cassette was found in a complex class 1 integron. Integrons are other mobile genetic elements, which could capture gene cassettes via attl and attC, further contributing to the global resistance crisis (Ghaly et al., 2021). More importantly, IS26 plays a key role in the spread of resistance genes in Gram-negative bacteria (Harmer et al., 2014; Hua et al., 2020). A crucial feature of IS26 is its capability to form co-integrate molecules that consists of distinct DNA segments (Hua et al., 2020). Based on the analysis of ISs and TSD, it was hypothesized that pKPTCM-4 with IncFII-IncX3 replicons was formed by five IS26-mediated co-integration events. Consequently, the joint role of diverse mobile genetic elements enables bacteria to undergo frequent genetic transposition and further leads to the MDR phenotype (Yang et al., 2021a).
Note that KPTCM was found to carry rmpA2 and virulence-associated gene cluster (iucABCD-iutA), the virulence potential was further evaluated. Interestingly, hypermucoviscous phenotype was negative. Moreover, rmpA and rmpA2 genes have been identified to be strongly associated with the hypermucoviscous phenotype. Nevertheless, rmpA2 gene in KPTCM strain has indel mutations causing frameshifts, further rendering the gene product dysfunctional (Shen et al., 2020a). In the current study, the low virulence maybe is caused by frameshifts of rmpA2. Thus, screening for rmpA and rmpA2 genes may need to be followed by sequencing to ensure their integrity, then it could be associated with hypervirulent phenotype. Interestingly, the patient developed a liver abscess caused by KPTCM strain and the outcome was death in a short time. Based on its associated clinical disease and outcome, the KPTCM strain should be considered as a relatively hypervirulent strain. However, a huge difference was observed when compared with many phenotypic experiments. Consequently, our results manifested the complexity and difficulty with the current knowledge for the definition of hypervirulence in K. pneumoniae.
The OCL and KL gene clusters, which are responsible for the biosynthesis of the outer core of lipooligosaccharide and capsule, are potentially useful epidemiological markers and offer a key role and potential target for vaccine development (Wyres et al., 2020). In the current study, KL112 had a quite high identity but with few reports. Furthermore, analysis of the population structure of the strains with various STs showed transmission among diverse countries. Based on previous reports, various STs have been reported in OXA-232-producing K. pneumoniae strains in many countries, including UK (mainly ST14, ST147, and ST231) (Findlay et al., 2017), Italy (ST16) (Avolio et al., 2017), Switzerland (ST231) (Mancini et al., 2018) and Tunisia (ST147) (Lahlaoui et al., 2017). However, our data revealed CRKP isolates harboring OXA-232-type carbapenemase plasmid usually belong to ST15 in China. The ST15 clones in China seem to be different from those in other areas of the world in view of the allelic differences and are mainly disseminated between Zhejiang and Shanghai (two geographically adjacent regions in China) but with slight evolution based on the SNP differences.
However, some limitations in this study remain, including the use of an insect larvae model might not be the most suitable for this type of virulence assessment. Thus, a murine infection model would be more suitable. Additionally, at present, we are not able to confirm whether resistance plasmids in KPTCM strain could be transferred to other recipient bacteria via conjugation. Finally, virulence-associated studies are required to find the direct or indirect connection between the carriage of the pLVPK-like virulence plasmid and hypervirulent phenotype in K. pneumoniae.
Conclusion
This study provides a comprehensive description for the complete genome characteristics of a wzi93-KL112-O1 OXA-232-producing ST15 CRKP, which harbors a rmpA2-associated pLVPK-like virulence plasmid and chromosomal blaCTX-M-15 mediated by ISEcp1-based transposon Tn2012. Considering that ST15 CRKP strains possess the strong plasmid carrying capacity (9 plasmids in KPTCM strain), they still have the potential to become hypervirulent via acquiring another type of virulence plasmid. Therefore, early detection of K. pneumoniae strains carrying chromosomal blaCTX-M-15, OXA-232 carbapenemase and pLVPK-like virulence plasmid is recommended to avoid the extensive spread of this high-risk clone in healthcare settings.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Genbank, PRJNA838426.
Ethics statement
This study was approved by the local Ethics Committees of the Hospital with a waiver of informed consent due to this study mainly focused on bacterial genome and the retrospective nature of the study.
Author contributions
CT and MX designed the experiments, analyzed the data, and wrote the initial manuscript. CT, MX, YB, and YZ performed the majority of the experiments. MX collected the bacteria. XF, LF, and SW supervised this study and reviewed and edited the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Medical Health Science and Technology Project of Zhejiang Provincial Health Commission (2022RC278), Natural Science Foundation of Zhejiang Province (LGF20H300003, LGF20H280002, LQ19H160002), Quzhou technology projects, China (2019K36).
Acknowledgments
We thank Charlesworth Author Services for the help with revising the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.984479/full#supplementary-material
Supplementary Figure 1 | Sequence alignment of rmpA2 genes. rmpA2* indicates gene with frameshifts in pKPTCM-1 plasmid. rmpA2 is the wild type in pK2044 of K. pneumoniae NTUH-2044.
References
Avolio, M., Vignaroli, C., Crapis, M., Camporese, A. (2017). Co-Production of NDM-1 and OXA-232 by ST16 klebsiella pneumoniae, Italy 2016. Future Microbiol. 12, 1119–1122. doi: 10.2217/fmb-2017-0041
Behzadi, P., Gajdacs, M. (2021). Writing a strong scientific paper in medicine and the biomedical sciences: a checklist and recommendations for early career researchers. Biol. Futur. 72, 395–407. doi: 10.1007/s42977-021-00095-z
Chen, T., Fu, Y., Hua, X., Xu, Q., Lan, P., Jiang, Y., et al. (2021b). Acinetobacter baumannii strains isolated from cerebrospinal fluid (CSF) and bloodstream analysed by cgMLST: the dominance of clonal complex CC92 in CSF infections. Int. J. Antimicrob. Agents 58, 106404. doi: 10.1016/j.ijantimicag.2021.106404
Chen, J., Zeng, Y., Zhang, R., Cai, J. (2021a). In vivo emergence of colistin and tigecycline resistance in carbapenem-resistant hypervirulent klebsiella pneumoniae during antibiotics treatment. Front. Microbiol. 12, 702956. doi: 10.3389/fmicb.2021.702956
Chudejova, K., Kraftova, L., Mattioni Marchetti, V., Hrabak, J., Papagiannitsis, C. C., Bitar, I. (2021). Genetic plurality of OXA/NDM-encoding features characterized from enterobacterales recovered from Czech hospitals. Front. Microbiol. 12, 641415. doi: 10.3389/fmicb.2021.641415
Feng, Y., Zou, S., Chen, H., Yu, Y., Ruan, Z. (2021). BacWGSTdb 2.0: a one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic Acids Res. 49, D644–D650. doi: 10.1093/nar/gkaa821
Findlay, J., Hopkins, K. L., Loy, R., Doumith, M., Meunier, D., Hill, R., et al. (2017). OXA-48-like carbapenemases in the UK: an analysis of isolates and cases from 2007 to 2014. J. Antimicrob. Chemother. 72, 1340–1349. doi: 10.1093/jac/dkx012
Froger, A., Hall, J. E. (2007). Transformation of plasmid DNA into e. coli using the heat shock method. J. Vis. Exp. 253. doi: 10.3791/253
Ghaly, T. M., Tetu, S. G., Gillings, M. R. (2021). Predicting the taxonomic and environmental sources of integron gene cassettes using structural and sequence homology of attC sites. Commun. Biol. 4, 946. doi: 10.1038/s42003-021-02489-0
Gorbunova, V., Seluanov, A., Mita, P., Mckerrow, W., Fenyo, D., Boeke, J. D., et al. (2021). The role of retrotransposable elements in ageing and age-associated diseases. Nature 596, 43–53. doi: 10.1038/s41586-021-03542-y
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2018). A fatal outbreak of ST11 carbapenem-resistant hypervirulent klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46. doi: 10.1016/S1473-3099(17)30489-9
Harmer, C. J., Moran, R. A., Hall, R. M. (2014). Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. mBio 5, e01801–e01814. doi: 10.1128/mBio.01801-14
He, J., Du, X., Zeng, X., Moran, R. A., Van Schaik, W., Zou, Q., et al. (2022). Phenotypic and genotypic characterization of a hypervirulent carbapenem-resistant klebsiella pneumoniae ST17-KL38 clinical isolate harboring the carbapenemase IMP-4. Microbiol. Spectr. 10, e0213421. doi: 10.1128/spectrum.02134-21
Hua, X., Zhang, L., Moran, R. A., Xu, Q., Sun, L., Van Schaik, W., et al. (2020). Cointegration as a mechanism for the evolution of a KPC-producing multidrug resistance plasmid in Proteus mirabilis. Emerg. Microbes Infect. 9, 1206–1218. doi: 10.1080/22221751.2020.1773322
Jia, H., Zhang, Y., Ye, J., Xu, W., Xu, Y., Zeng, W., et al. (2021). Outbreak of multidrug-resistant OXA-232-Producing ST15 klebsiella pneumoniae in a teaching hospital in wenzhou, China. Infect. Drug Resist. 14, 4395–4407. doi: 10.2147/IDR.S329563
Kong, Y., Li, C., Chen, H., Zheng, W., Sun, Q., Xie, X., et al. (2021a). In vivo emergence of colistin resistance in carbapenem-resistant klebsiella pneumoniae mediated by premature termination of the mgrB gene regulator. Front. Microbiol. 12, 656610. doi: 10.3389/fmicb.2021.656610
Kong, Y., Sun, Q., Chen, H., Draz, M. S., Xie, X., Zhang, J., et al. (2021b). Transmission dynamics of carbapenem-resistant klebsiella pneumoniae sequence type 11 strains carrying capsular loci KL64 and rmpA/rmpA2 genes. Front. Microbiol. 12, 736896. doi: 10.3389/fmicb.2021.736896
Lahlaoui, H., Bonnin, R. A., Moussa, M. B., Khelifa, A. B. H., Naas, T. (2017). First report of OXA-232-producing klebsiella pneumoniae strains in Tunisia. Diagn. Microbiol. Infect. Dis. 88, 195–197. doi: 10.1016/j.diagmicrobio.2017.03.005
Lam, M. M. C., Wick, R. R., Judd, L. M., Holt, K. E., Wyres, K. L. (2022). Kaptive 2.0: updated capsule and lipopolysaccharide locus typing for the klebsiella pneumoniae species complex. Microb. Genom. 8. doi: 10.1099/mgen.0.000800
Lam, M. M. C., Wick, R. R., Watts, S. C., Cerdeira, L. T., Wyres, K. L., Holt, K. E. (2021). A genomic surveillance framework and genotyping tool for klebsiella pneumoniae and its related species complex. Nat. Commun. 124188. doi: 10.1038/s41467-021-24448-3
Letunic, I., Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Liao, W., Long, D., Huang, Q., Wei, D., Liu, X., Wan, L., et al. (2020). Rapid detection to differentiate hypervirulent klebsiella pneumoniae (hvKp) from classical k. pneumoniae by identifying peg-344 with loop-mediated isothermal amplication (LAMP). Front. Microbiol. 111189. doi: 10.3389/fmicb.2020.01189
Liu, Z., Chen, R., Xu, P., Wang, Z., Li, R. (2021). Characterization of a bla NDM-1-Bearing IncHI5-like plasmid from klebsiella pneumoniae of infant origin. Front. Cell Infect. Microbiol. 11, 738053. doi: 10.3389/fcimb.2021.738053
Liu, B., Zheng, D., Zhou, S., Chen, L., Yang, J. (2022). VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–D917. doi: 10.1093/nar/gkab1107
Mancini, S., Poirel, L., Tritten, M. L., Lienhard, R., Bassi, C., Nordmann, P. (2018). Emergence of an MDR klebsiella pneumoniae ST231 producing OXA-232 and RmtF in Switzerland. J. Antimicrob. Chemother. 73, 821–823. doi: 10.1093/jac/dkx428
Menard, G., Rouillon, A., Cattoir, V., Donnio, P. Y. (2021). Galleria mellonella as a suitable model of bacterial infection: Past, present and future. Front. Cell Infect. Microbiol. 11, 782733. doi: 10.3389/fcimb.2021.782733
Oueslati, S., Nordmann, P., Poirel, L. (2015). Heterogeneous hydrolytic features for OXA-48-like beta-lactamases. J. Antimicrob. Chemother. 70, 1059–1063. doi: 10.1093/jac/dku524
Potron, A., Rondinaud, E., Poirel, L., Belmonte, O., Boyer, S., Camiade, S., et al. (2013). Genetic and biochemical characterisation of OXA-232, a carbapenem-hydrolysing class d beta-lactamase from enterobacteriaceae. Int. J. Antimicrob. Agents 41, 325–329. doi: 10.1016/j.ijantimicag.2012.11.007
Ruan, Z., Yu, Y., Feng, Y. (2020). The global dissemination of bacterial infections necessitates the study of reverse genomic epidemiology. Brief Bioinform. 21, 741–750. doi: 10.1093/bib/bbz010
Sewunet, T., Asrat, D., Woldeamanuel, Y., Ny, S., Westerlund, F., Aseffa, A., et al. (2021). High prevalence of bla CTX-M-15 and nosocomial transmission of hypervirulent epidemic clones of klebsiella pneumoniae at a tertiary hospital in Ethiopia. JAC Antimicrob. Resist. 3, dlab001. doi: 10.1093/jacamr/dlab001
Shen, P., Berglund, B., Chen, Y., Zhou, Y., Xiao, T., Xiao, Y., et al. (2020a). Hypervirulence markers among non-ST11 strains of carbapenem- and multidrug-resistant klebsiella pneumoniae isolated from patients with bloodstream infections. Front. Microbiol. 111199. doi: 10.3389/fmicb.2020.01199
Shen, Z., Zhang, H., Gao, Q., Qin, J., Zhang, C., Zhu, J., et al. (2020b). Increased plasmid copy number contributes to the elevated carbapenem resistance in OXA-232-Producing klebsiella pneumoniae. Microb. Drug Resist. 26, 561–568. doi: 10.1089/mdr.2018.0407
Shi, Q., Han, R., Guo, Y., Zheng, Y., Yang, Y., Yin, D., et al. (2020). Emergence of ST15 klebsiella pneumoniae clinical isolates producing plasmids-mediated RmtF and OXA-232 in China. Infect. Drug Resist. 13, 3125–3129. doi: 10.2147/IDR.S257298
Shu, L., Dong, N., Lu, J., Zheng, Z., Hu, J., Zeng, W., et al. (2019). Emergence of OXA-232 carbapenemase-producing klebsiella pneumoniae that carries a pLVPK-like virulence plasmid among elderly patients in China. Antimicrob. Agents Chemother. 63. doi: 10.1128/AAC.02246-18
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Sullivan, M. J., Petty, N. K., Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Tatusova, T., Dicuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Tian, D., Wang, W., Li, M., Chen, W., Zhou, Y., Huang, Y., et al. (2021). Acquisition of the conjugative virulence plasmid from a CG23 hypervirulent klebsiella pneumoniae strain enhances bacterial virulence. Front. Cell Infect. Microbiol. 11, 752011. doi: 10.3389/fcimb.2021.752011
Wanford, J. J., Hames, R. G., Carreno, D., Jasiunaite, Z., Chung, W. Y., Arena, F., et al. (2021). Interaction of klebsiella pneumoniae with tissue macrophages in a mouse infection model and ex-vivo pig organ perfusions: an exploratory investigation. Lancet Microbe 2, e695–e703. doi: 10.1016/S2666-5247(21)00195-6
Wang, X., Xie, Y., Li, G., Liu, J., Li, X., Tian, L., et al. (2018). Whole-Genome-Sequencing characterization of bloodstream infection-causing hypervirulent klebsiella pneumoniae of capsular serotype K2 and ST374. Virulence 9, 510–521. doi: 10.1080/21505594.2017.1421894
Wick, R. R., Judd, L. M., Gorrie, C. L., Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PloS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Wyres, K. L., Cahill, S. M., Holt, K. E., Hall, R. M., Kenyon, J. J. (2020). Identification of acinetobacter baumannii loci for capsular polysaccharide (KL) and lipooligosaccharide outer core (OCL) synthesis in genome assemblies using curated reference databases compatible with kaptive. Microb. Genom. 6. doi: 10.1099/mgen.0.000339
Xiao, T., Zhu, Y., Zhang, S., Wang, Y., Shen, P., Zhou, Y., et al. (2020). A retrospective analysis of risk factors and outcomes of carbapenem-resistant klebsiella pneumoniae bacteremia in nontransplant patients. J. Infect. Dis. 221, S174–S183. doi: 10.1093/infdis/jiz559
Xu, Y., Zhang, J., Wang, M., Liu, M., Liu, G., Qu, H., et al. (2021). Mobilization of the nonconjugative virulence plasmid from hypervirulent klebsiella pneumoniae. Genome Med. 13, 119. doi: 10.1186/s13073-021-00936-5
Yang, X., Dong, N., Chan, E. W., Zhang, R., Chen, S. (2021a). Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in klebsiella pneumoniae. Trends Microbiol. 29, 65–83. doi: 10.1016/j.tim.2020.04.012
Yang, X., Dong, N., Liu, X., Yang, C., Ye, L., Chan, E. W., et al. (2021b). Co-Conjugation of virulence plasmid and KPC plasmid in a clinical klebsiella pneumoniae strain. Front. Microbiol. 12, 739461. doi: 10.3389/fmicb.2021.739461
Yang, Y., Yang, Y., Chen, G., Lin, M., Chen, Y., He, R., et al. (2021c). Molecular characterization of carbapenem-resistant and virulent plasmids in klebsiella pneumoniae from patients with bloodstream infections in China. Emerg. Microbes Infect. 10, 700–709. doi: 10.1080/22221751.2021.1906163
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zhao, D., Shi, Q., Hu, D., Fang, L., Mao, Y., Lan, P., et al. (2021). The emergence of novel sequence type strains reveals an evolutionary process of intraspecies clone shifting in ICU-spreading carbapenem-resistant klebsiella pneumoniae. Front. Microbiol. 12, 691406. doi: 10.3389/fmicb.2021.691406
Keywords: CRKP, OXA-232, chromosomal blaCTX-M-15, pLVPK-like virulence plasmid, IS26, co-integration
Citation: Tian C, Xing M, Zhao Y, Fan X, Bai Y, Fu L and Wang S (2022) Whole genome sequencing of OXA-232-producing wzi93-KL112-O1 carbapenem-resistant Klebsiella pneumoniae in human bloodstream infection co-harboring chromosomal ISEcp1-based blaCTX-M-15 and one rmpA2-associated virulence plasmid. Front. Cell. Infect. Microbiol. 12:984479. doi: 10.3389/fcimb.2022.984479
Received: 02 July 2022; Accepted: 20 September 2022;
Published: 29 September 2022.
Edited by:
Vittoria Mattioni Marchetti, Charles University, CzechiaReviewed by:
Ibrahim Bitar, Charles University, CzechiaBarbara Ghiglione, University of Buenos Aires, Argentina
Copyright © 2022 Tian, Xing, Zhao, Fan, Bai, Fu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siwei Wang, 358031289@qq.com; Liping Fu, fuliping100@163.com
†These authors have contributed equally to this work
 Chongmei Tian
Chongmei Tian