- Wellcome Sanger Institute, Wellcome Genome Campus, Cambridge, United Kingdom
CRISPR editing has enabled the rapid creation of fluorescent Plasmodium transgenic lines, facilitating a deeper understanding of parasite biology. The impact of genetic perturbations such as gene disruption or the introduction of drug resistance alleles on parasite fitness is typically quantified in competitive growth assays between the query line and a wild type reference. Although fluorescent reporter lines offer a facile and frequently used method to measure relative growth, this approach is limited by the strain background of the existing reporter, which may not match the growth characteristics of the query strains, particularly if these are slower-growing field isolates. Here, we demonstrate an efficient CRISPR-based approach to generate fluorescently labelled parasite lines using mNeonGreen derived from the LanYFP protein in Branchiostoma lanceolatum, which is one of the brightest monomeric green fluorescent proteins identified. Using a positive-selection approach by insertion of an in-frame blasticidin S deaminase marker, we generated a Dd2 reporter line expressing mNeonGreen under the control of the pfpare (P. falciparum Prodrug Activation and Resistance Esterase) locus. We selected the pfpare locus as an integration site because it is highly conserved across P. falciparum strains, expressed throughout the intraerythrocytic cycle, not essential, and offers the potential for negative selection to further enrich for integrants. The mNeonGreen@pare line demonstrates strong fluorescence with a negligible fitness defect. In addition, the construct developed can serve as a tool to fluorescently tag other P. falciparum strains for in vitro experimentation.
Introduction
Transgenic parasites expressing fluorescent proteins are a powerful tool in parasitology research. The ability to identify and track whole parasites or tagged parasite proteins in vitro has been integral for gaining insights into the biology of parasites and their interactions with hosts. In the study of malaria, the generation of fluorescent Plasmodium reporter lines has been instrumental in interrogating gene function, drug activity, life cycle, and host-parasite interactions (Frischknecht et al., 2006; Talman et al., 2010; Wilson et al., 2010; Portugaliza et al., 2019; Voorberg-van der Wel et al., 2020; Thommen et al., 2022). Reporter lines have also been instrumental in facilitating scaled-up analyses and the development of new methodologies. One important use of reporter lines is to quantify parasite fitness using competitive head-to-head growth assays. A query line, typically with an engineered mutation of interest such as a drug resistance allele or gene knockout, is mixed with a fluorescent isogenic wild type parasite. The change in relative abundance over time provides a measure of the fitness impact of the mutation of interest (Baragaña et al., 2015; Gabryszewski et al., 2016a; Ross et al., 2018; Stokes et al., 2021).
The first reporter lines developed in Plasmodium parasites expressed the exogenous proteins, firefly luciferase and chloramphenicol, using episomes (Goonewardene et al., 1993; Wu et al., 1995; Horrocks and Kilbey, 1996). Shortly after, green fluorescent protein (GFP) from Aequorea victoria, was developed as a reporter and adapted into P. falciparum and became widely used as it provided stable and strong fluorescence without requiring a cofactor (Chalfie et al., 1994; VanWye and Haldar, 1997). The reporter lines enabled functional and genetic analyses, particularly the bioluminescent and fluorescent reporters which supported imaging. The application of other reporters such as mCherry and yellow fluorescent protein and the transition to integrated reporters were gradual and hindered by challenges with homologous recombination-based integration (Armstrong and Goldberg, 2007; Engelmann et al., 2009). The advent of CRISPR/Cas9 genome editing and its application in P. falciparum made the development of reporter lines rapid and straightforward, allowing for the selective integration of fluorescent markers into specific genomic sites (Mogollon et al., 2016; Kuang et al., 2017; Miyazaki et al., 2020). Favourable integration sites could be selected based on the essentiality and phenotype of the gene and its expression profile, allowing for the development of reporter lines with stage-specific or multi-stage expression (Marin-Mogollon et al., 2019; Miyazaki et al., 2020). More recently developed fluorescent proteins i.e., mNeonGreen and mGreenLantern could generate better performing Plasmodium reporter lines as these proteins have a 3 and 6-fold increase in brightness compared to mEGFP, respectively. Faster maturation, improved acid tolerance, increased photostability and thermostability are also features that are often enhanced in these fluorescent proteins compared to standard fluorescent proteins (Shaner et al., 2013; Campbell et al., 2020).
In this work, we developed an efficient CRISPR/Cas9 approach to generate mNeonGreen-expressing P. falciparum lines, with the potential to use positive and negative selection to remove untagged parasites and obviate the need for clonal isolation. We inserted mNeonGreen in the genome at the non-essential pfpare (P. falciparum Prodrug Activation and Resistance Esterase) locus, so that its integration is stable, and its expression is endogenously driven by the pfpare promoter. We demonstrate that mNeonGreen@pare exhibits strong fluorescence throughout the intraerythrocytic cycle and has robust fitness compared to existing reporter lines.
Methods
Sequence alignment
DNA sequences for pfpare locus (PF3D7_0709700) in P. falciparum 3D7, HB3, 7G8, GB4, CD01, GA01, IT and Dd2 strains were obtained from PlasmoDB and alignment figures were generated using Clustal Omega (Goujon et al., 2010; Sievers et al., 2011; Amos et al., 2021). Transcriptomic data of 3D7 pfpare expression in the intraerythrocytic life cycle was obtained from PlasmoDB, specifically RNA-sequencing data from Chappell et al. was used (Chappell et al., 2020).
Construct design and generation
The construct backbone used was pDC2-coCas9-gRNA, encoding Cas9, a gRNA expression cassette, hDHFR resistance for selection for plasmid uptake in P. falciparum culture and ampicillin resistance cassette for plasmid propagation in E. coli (Adjalley and Lee, 2022). Into this base vector, we inserted a guide RNA targeting pfpare (GGACAGTCAGAAGGATGGAA), and a donor region with flanking homology to 3D7 pfpare (255 bp and 466 bp homology regions; see Figure 1B). We synthesised mNeonGreen codon-optimised for P. falciparum and cloned this downstream of the blasticidin S deaminase (BSD) selectable marker. To facilitate the expression of unfused proteins, 2A linkers were included separating the upstream pfpare fragment, BSD, and mNeonGreen (see Figure 1B). Cloning was performed by Gibson assembly (NEBuilder DNA HiFi Assembly), and transformations were performed in XL-10 Gold Ultracompetent cells (Agilent). Plasmid sequences were confirmed by Sanger sequencing and plasmids were amplified and isolated using Midiprep plasmid extraction kits (Macherey-Nagel).
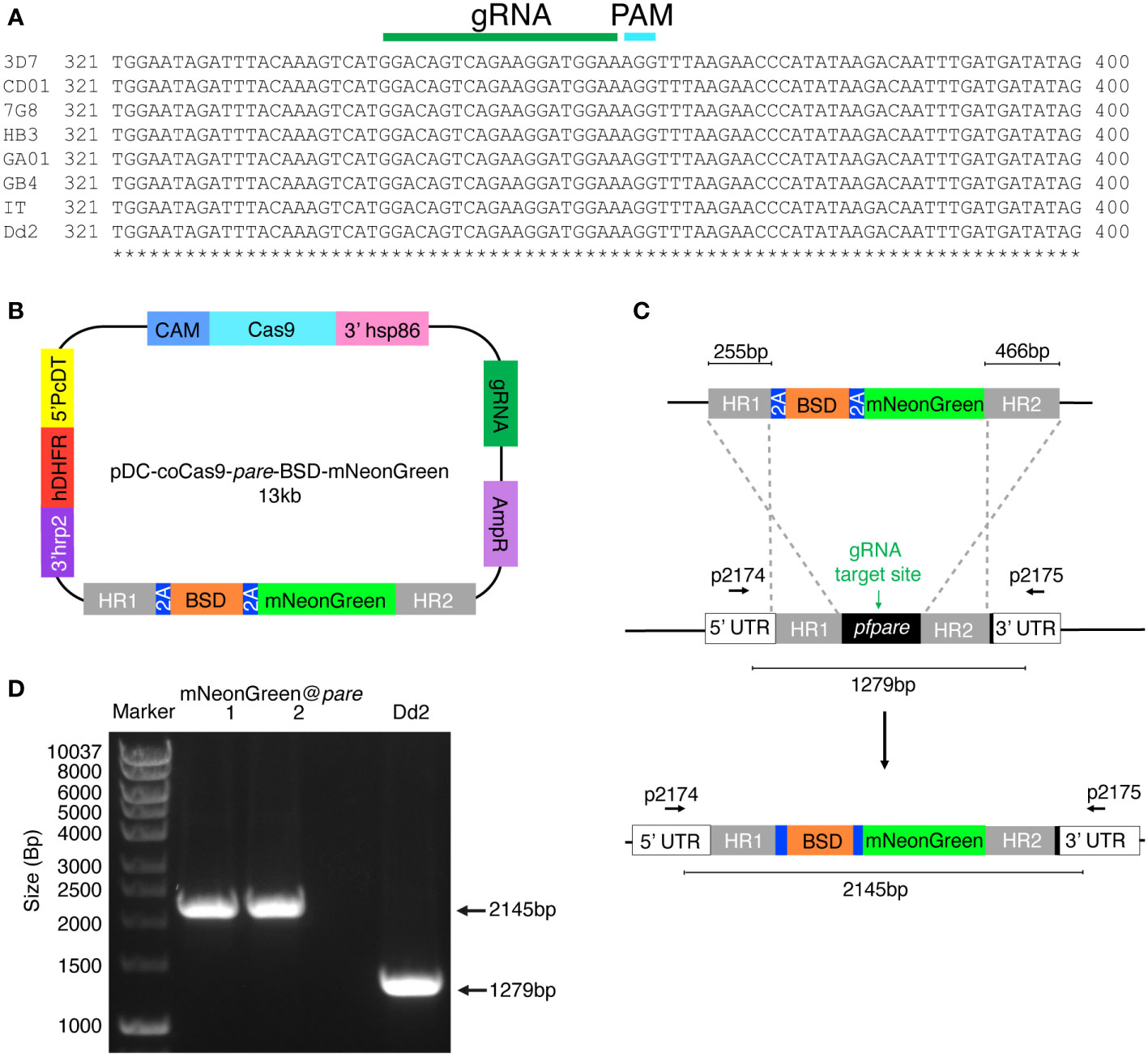
Figure 1 Generation of a P. falciparum Dd2 reporter line expressing mNeonGreen under the control of the highly conserved pfpare locus (mNeonGreen@pare). (A) DNA sequences for pfpare (PF3D7_0709700) in P. falciparum 3D7, HB3, 7G8, GB4, CD01, GA01, IT and Dd2 strains were obtained from PlasmoDB and Clustal Omega was used to create a multiple sequence alignment (Goujon et al., 2010; Sievers et al., 2011; Amos et al., 2021). (B) Schematic of the pDC2-coCas9-pare-BSD-mNeonGreen plasmid. The plasmid encodes a codon-optimised Cas9 enzyme, guide RNA cassette containing a guide targeting pfpare, an ampicillin resistance cassette for plasmid propagation in E coli and the hDHFR marker for selection for plasmid uptake in P. falciparum. The donor region contains a blasticidin S deaminase (BSD) selectable marker flanked by 2A linkers with mNeonGreen downstream. The payload is flanked by two homology regions for the pfpare locus. (C) Upon transfection of the pDC2-coCas9-pare-BSD-mNeonGreen plasmid into P. falciparum Dd2, the pfpare-targeting gRNA directs Cas9 to make a site-directed cut of pfpare. The donor region facilitates homology-directed repair and the insertion of BSD and mNeonGreen. (D) The pfpare locus of blasticidin-resistant recrudesced parasites (mNeonGreen@pare.1 and mNeonGreen@pare.2) was PCR-amplified using 5’ and 3’ UTR primers (p2174 and p2175) to check for correct insertion of mNeonGreen. Lengths of PCR products were quantified using gel electrophoresis. No detectable wild type product was observed in the transfected bulk culture.
Parasite culturing and plasmid transfections
Blood-stage P. falciparum parasites (Dd2, Dd2bipEGFP and NF54camEGFP) were grown in RPMI media with AlbuMAX® (Gibco) and supplemented with GlutaMax® (Gibco), Gentamicin (Gibco) and HEPES (pH 7.0) with O+ human erythrocytes at 3% haematocrit. RBCs were obtained by anonymous donors from the National Health Services Blood and Transplant (NHSBT). Their use was in accordance with relevant guidelines and regulations, with approval from the NHS Cambridgeshire Research Ethics Committee and the Wellcome Sanger Institute Human Materials and Data Management Committee. Cultures were maintained in a gaseous environment of 3% CO2, 1% O2 and 96% N2 at 37°C. Parasitemia and stages were monitored using Giemsa staining and microscopy. Synchronisation was completed using sorbitol ring enrichment (Radfar et al., 2009). Gametocytes were induced using stress and media supplemented with horse serum and heparin (Fivelman et al., 2007). Transfections were completed by electroporation of parasitised red blood cells (Bio-Rad Gene Pulser Xcell) as previously described (Fidock and Wellems, 1997). Cultures containing 5% ring-stage parasites were transfected with 50μg of the plasmid. Blasticidin S drug selection (2μg/ml) was applied one-day post-electroporation and maintained continuously. Correct editing of recrudesced parasites (mNeonGreen@pare.1 and mNeonGreen@pare.2) was confirmed using primers (p2174 and p2175), which flank the 5’ and 3’ UTRs of pfpare (TGCACTTGTTTTACATTTTTATATT and TGTAACATCACTAATTAATTTATTAA). PCR amplification and gel electrophoresis were used to check insert size and Sanger sequencing to confirm the correct sequence. The Dd2bipEGFP and NF54camEGFP fluorescent lines were previously published (Baragaña et al., 2015; Gabryszewski et al., 2016b). Both lines were generated using attB × attP recombination to insert the reporter genes, however, they are driven by different constitutive promoters: Dd2bipEGFP is driven by the ER hsp70 (BiP) (PF3D7_0917900) promoter and the NF54camEGFP by the calmodulin (PF3D7_1434200) promoter.
Fluorescence microscopy
200µL of parasite culture was harvested at 5% parasitemia, spun at 3000rpm for 30 seconds in an Eppendorf tube and washed with 0.5mL of PBS. Parasites were fixed by resuspending in 4% (v/v) paraformaldehyde + 0.0075% glutaraldehyde, incubating for 30 minutes, and washed twice with 1mL of PBS. Parasite DNA was stained with 1mL of Hoechst stain (10µg/mL) for 5 minutes and resuspended in 1mL of PBS. The stained parasites were transferred to an 8-well chambered coverglass (Lab-Tek) pre-treated with 0.25mL of poly L-lysine (0.2mg/ml) for 10mins. The coverglass was transferred to an inverted fluorescence microscope (Leica DMi8). Bright-field microscopy and blue and green-filtered fluorescence microscopy were used with the 100x objective (total 1000x magnification) to image infected erythrocytes and fluorescent parasites. Image processing and analysis were completed with Leica Application Suite (LAS X).
Flow cytometric analysis of fluorescence
Parasites were transferred to a round-bottom 96-well plate, spun (3mins, 2000rpm) and the supernatant was removed. To check viability, infected erythrocytes were stained with 100nM MitoTracker Deep Red (MitoDR) solution (Thermofisher) in NaCl 0.9%/Dextrose 0.2% and incubated for 30mins in the dark at 37°C. After a 1/40 dilution in PBS, parasites were analysed on a flow cytometer (Beckman Coulter CytoFLEX S) and at least 20 000 events were recorded for each experiment. The blue 488nm laser was used to detect green fluorescence and the red 638nm laser was used to detect MitoDR. Quantification of green fluorescent (GFP and mNeonGreen) and MitoDR positive cells in the culture population was performed using FlowJo (v10).
Competition assay
Pre-assay parasitemia was measured using flow cytometry and stages were observed with microscopy. Mixed-stage lines of mNeonGreen@pare.1, mNeonGreen@pare.2 and Dd2bipEGFP were competed against Dd2 or 3D7 in triplicate biological replicates at 50:50 starting ratio with a total of 1% parasitemia. The parasitemia and fluorescence were measured using flow cytometry every 2nd or 3rd day from day 0 to day 21. Parasitemia was maintained between 0.5%-6%. The percentage of parasites expressing green fluorescence over total parasites was averaged between the triplicates and graphed over time for each line. Error bars were included representing standard deviation between triplicates. Data in Supplementary Table 1.
Results
Selection of the pfpare locus for fluorescent markers integration in P. falciparum
The pfpare locus was identified as an optimal safe-harbour site for the integration of the mNeonGreen fluorescent marker. pfpare (PF3D7_0709700) encodes a prodrug activation and resistance esterase that is not essential (Istvan et al., 2017). In addition to the dispensible nature of pfpare, loss-of-function mutations in pfpare confer resistance to the antimalarial compound MMV011438, which requires pfpare for its activation. pfpare is expressed during blood stages, therefore its promoter would facilitate the expression of a fluorescent marker throughout the intraerythrocytic life cycle. In addition, we included in the inserted sequence the blasticidin S deaminase (BSD) marker, which would only be expressed from the pfpare promoter once integrated. Thus our strategy would permit both positive and negative selection options if required for efficient isolation of a homogenous culture of mNeonGreen-tagged parasites without cloning.
To identify if pfpare is suitable as an integration site across multiple P. falciparum strains, a multiple sequence alignment of the pfpare locus for eight geographically diverse strains (3D7, HB3, 7G8, GB4, CD01, GA01, IT and Dd2) was completed. We first identified a gRNA target site that was highly conserved, with no mutations in the guide RNA or PAM sequences (Figure 1A). Examination of the flanking upstream and downstream sequences that would constitute the donor homology regions revealed no mutations in the 5’ homology region and 2 – 3 single nucleotide polymorphisms in the 3’ homology region of the donor sequence of the plasmid (Supplemental Figure 1). This level of sequence diversity would not be expected to strongly impact editing. We validated this prediction below using a 3D7-based donor sequence to edit Dd2, which has three polymorphisms relative to 3D7.
Efficient generation of endogenous mNeonGreen-expressing P. falciparum reporter lines
To design a construct to integrate mNeonGreen into pfpare, we first subcloned a codon-optimised mNeonGreen downstream of BSD, with flanking 5’ and 3’ pfpare (3D7) homology regions of 255 bp and 466 bp respectively. The resulting plasmid (pDC-coCas9-pare-BSD-mNeonGreen; Figure 1B) expresses Cas9 driven by the calmodulin promoter and transcribes a pfpare-targeting guide RNA. Plasmids were transfected into three independent P. falciparum Dd2 parasite cultures and continuous selection with blasticidin S was used to select for plasmid uptake and the insertion of the donor into pfpare by homology-directed repair (Figure 1C). Edited parasites were obtained from two of the three transfections at around three weeks post-transfection, referred to as mNeonGreen@pare.1 and mNeonGreen@pare.2. PCR-amplification of the pfpare locus demonstrated successful integration of the donor with no wild type locus detectable, indicating positive selection was sufficient to deplete any unedited parasites, obviating the need for clonal isolation (Figure 1D, primers shown in Figure 1C).
Endogenous mNeonGreen expression generates fluorescence comparable to existing GFP lines
To determine if the integrated mNeonGreen yields fluorescent parasites, we first examined the transgenic lines by fluorescence microscopy. Infected erythrocytes were detected using Hoechst DNA stain, which does not stain uninfected erythrocytes because they are anucleate, unlike Plasmodium parasites. Erythrocytes infected with either of the mNeonGreen@pare lines showed strong green fluorescence unlike the parental Dd2 line (Figure 2A). Flow cytometry was used to quantify the level of fluorescence and distribution within the population of a mixed-stage culture. The fluorescence profiles of mNeonGreen@pare.1 and mNeonGreen@pare.2 were highly similar and are comparable to the profiles of other green-fluorescing lines used for competition assays, including Dd2bipEGFP and NF54camEGFP, which express GFP from the strong constitutive promoters of ER-Hsp70 (BiP) and calmodulin respectively (Adjalley et al., 2011; Baragaña et al., 2015). The peaks of the mNeonGreen@pare lines were modestly shifted left in comparison to the GFP lines, which means that the bulk of mNeonGreen@pare parasites in mixed culture are less fluorescent than the bulk of GFP-expressing parasites but still readily distinguishable from non-fluorescent parasites (Figure 2B). The bimodal peaks suggested that subpopulations expressed different levels of fluorescence.
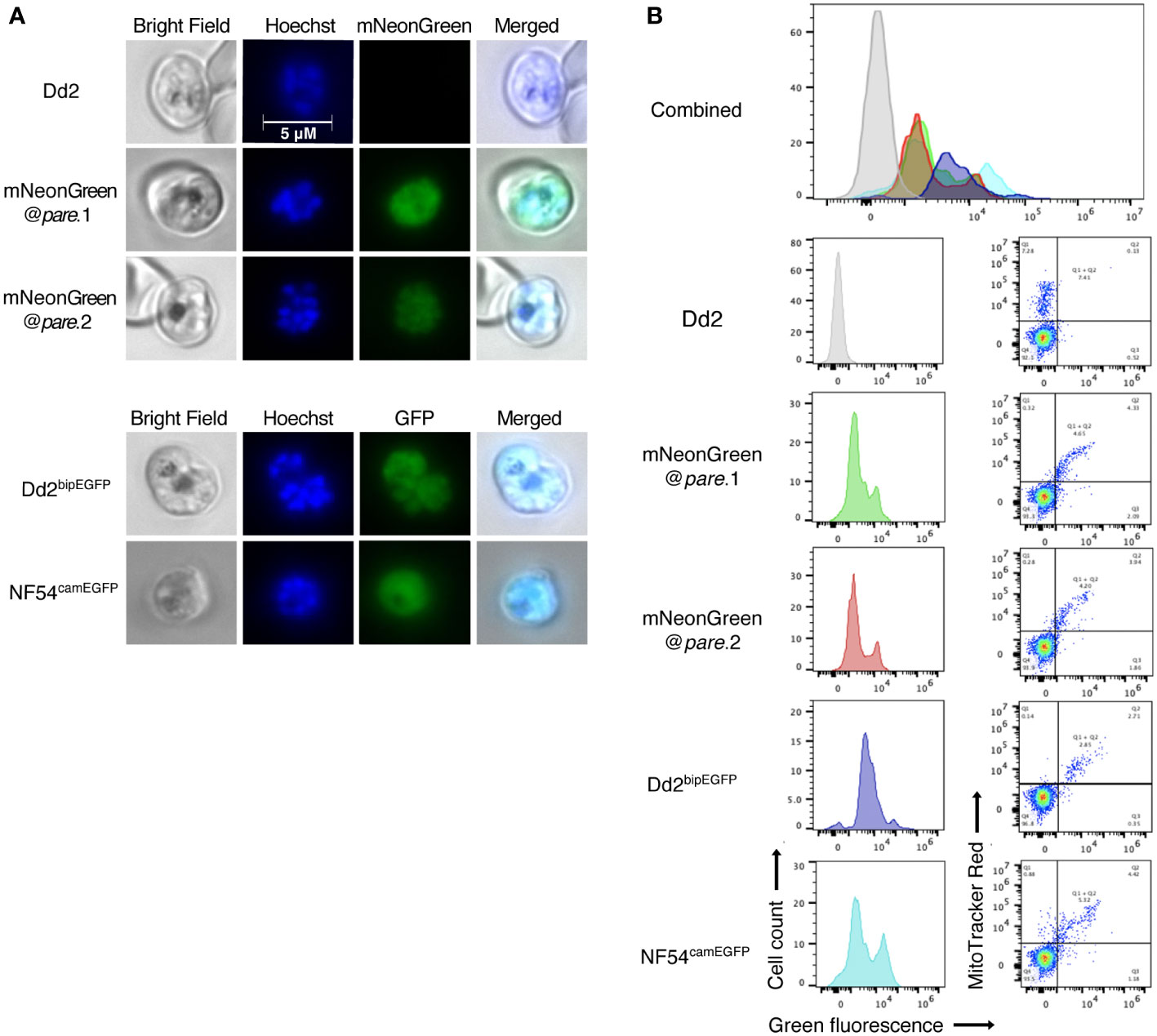
Figure 2 mNeonGreen fluorescence in P. falciparum is comparable to GFP fluorescent lines. (A) Mixed-staged parasites (Dd2, mNeonGreen@pare.1, mNeonGreen@pare.2, Dd2bipEGFP, NF54camEGFP) were fixed, stained with Hoechst DNA stain, and visualised using bright-field and fluorescence microscopy at 1000x magnification. (B) Mixed-stage parasites (Dd2, mNeonGreen@pare.1, mNeonGreen@pare.2, Dd2bipEGFP, NF54camEGFP) were stained with MitoTracker DeepRed and analysed on a flow cytometer. Quantification of green fluorescent (GFP and mNeonGreen) and MitoTracker+ cells was performed using FlowJo. All single-cell, parasitised RBCs (MitoTracker+) were gated, and histograms of green fluorescence were generated (GFP+ or mNeonGreen+).
mNeonGreen fluorescence varies between different asexual P. falciparum blood stages
In a healthy asexual intraerythrocytic P. falciparum culture, the subpopulations include briefly free-roaming merozoites and intraerythrocytic ring, trophozoite and schizont stages. To assess the level of fluorescence in specific stages, we enriched mNeonGreen@pare cultures for different stages using synchronisation. Stage-specific levels of fluorescence were observed with schizonts producing the highest fluorescence, followed by trophozoites and lastly, ring stages (Figures 3A, C). This pattern was consistent with transcriptomic studies that demonstrate that the expression of pfpare peaks at 32-40 hours post-erythrocyte-invasion, which would suggest that the expression of mNeonGreen from the pfpare locus would also peak during late trophozoite and schizont stages (Figure 3B) (Chappell et al., 2020; Amos et al., 2021). Further, we induced gametocytogenesis and observed fluorescence in all the gametocyte stages (I-V) (Figure 3C).
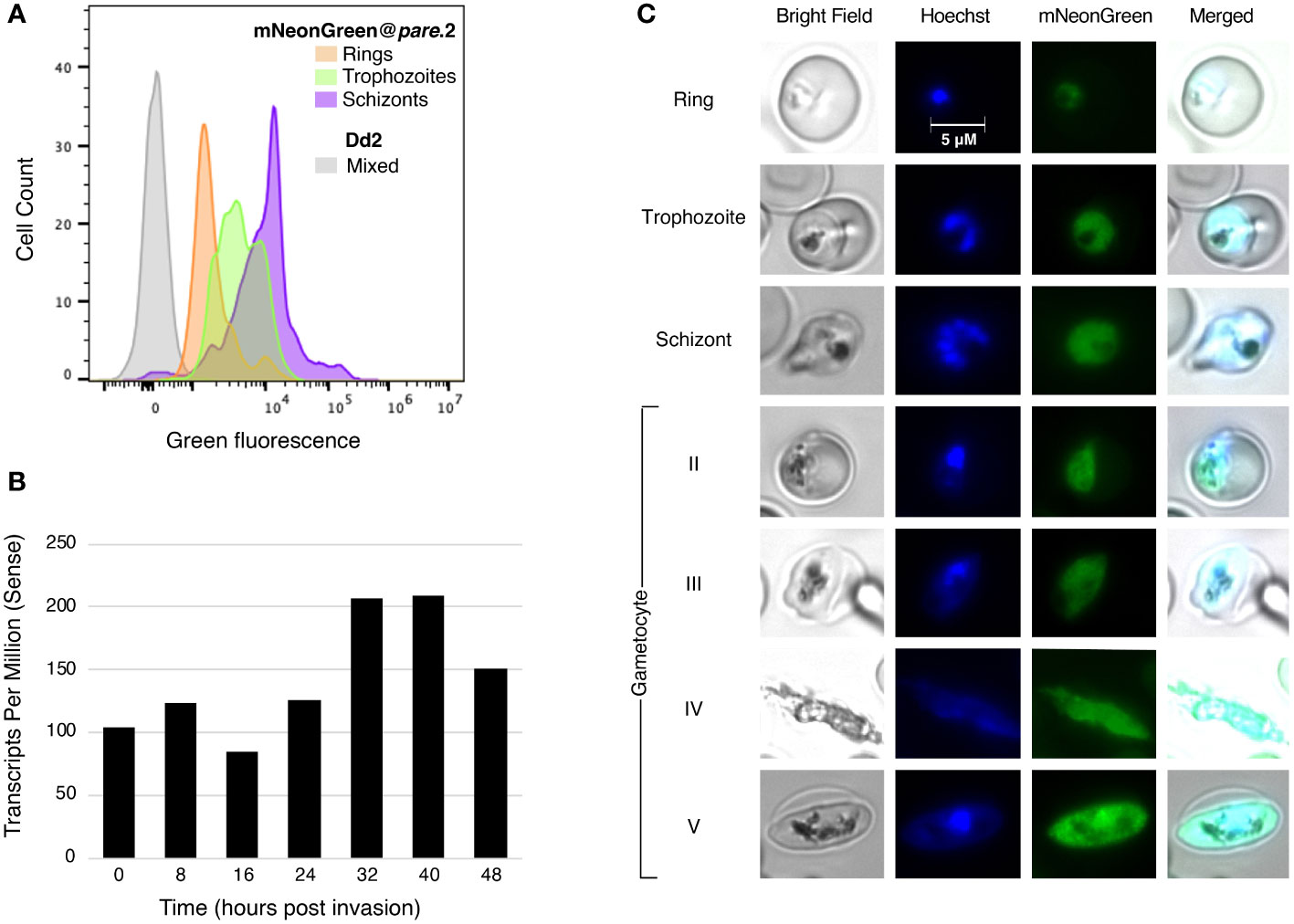
Figure 3 mNeonGreen@pare fluorescence varies between different asexual blood stages. (A) mNeonGreen@pare.2 parasites were sorbitol-synchronised and microscopy was used to confirm stages. Flow cytometry with MitoTracker DeepRed was used to enumerate parasites expressing green fluorescence in ring, trophozoite and schizont-staged cultures. FlowJo was used to gate single-cell, parasitised RBCs, quantify green fluorescence, and generate histograms. mNeonGreen@pare schizonts demonstrated the greatest fluorescence followed by trophozoites and then rings. (B) Stage-specific RNA-sequencing obtained from PlasmoDB shows that pfpare is expressed throughout the entire 48hr intraerythrocytic life cycle and expression peaks at late trophozoite and schizont stages (Chappell et al., 2020; Amos et al., 2021). (C) Fluorescence microscopy of synchronised mNeonGreen@pare parasites at asexual ring, trophozoite and schizont stages, and at gametocyte stages II-V (obtained through gametocytogenesis induction). Cultures were fixed, stained with Hoechst DNA stain, and visualised using bright-field and fluorescence microscopy at 1000x magnification.
mNeonGreen@pare lines demonstrate robust fitness
To assess the fitness of the mNeonGreen@pare lines, competition assays were performed against Dd2 and 3D7. The two populations were seeded at a 1:1 ratio of fluorescent to test line and maintained for 3 weeks. The ratio of fluorescent (mNeonGreen@pare) to non-fluorescent (Dd2 or 3D7) populations was measured every 2 - 3 days using flow cytometry to quantify the competition between the two populations. Dd2bipEGFP was also included as a control, which has been shown previously to have a slight fitness defect due to the integration of GFP (Baragaña et al., 2015). The mNeonGreen@pare lines showed nearly comparable fitness to their parent line, Dd2, over a three-week period (Figure 4A). Contrastingly when competed against 3D7, a slower-growing lab line, the mNeonGreen@pare lines outcompeted 3D7, which demonstrates the value of strain-matched competitor lines (Figure 4B).
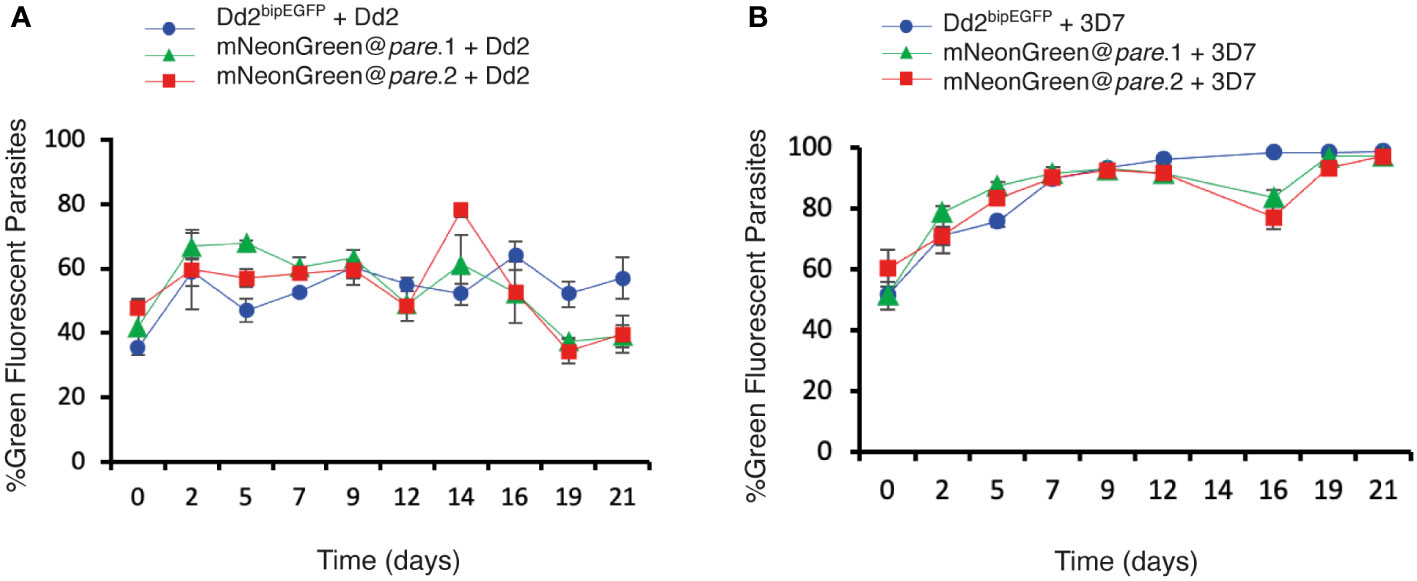
Figure 4 mNeonGreen@pare lines demonstrate comparable fitness to Dd2bipEGFP when competed against Dd2 or 3D7. Fluorescent lines (mNeonGreen@pare.1, mNeonGreen@pare.2 and Dd2bipEGFP) were competed against (A) Dd2 or (B) 3D7 in triplicate biological replicates. The two populations were seeded at a 1:1 ratio at 1% parasitemia and maintained for 3 weeks. The parasitemia and fluorescence were measured using flow cytometry every 2nd or 3rd day from day 0 to day 21. The percentage of green fluorescent parasites over total parasites was averaged between the triplicates and graphed over time for each line. The error bars represent standard deviation between triplicates. The three fluorescent lines did not outcompete Dd2 and remained close to the seeded percentages over three weeks. However, the fluorescent lines quickly outcompeted 3D7.
Discussion
Genetic engineering of fluorescent proteins has greatly improved their functionality as tools in molecular biology. As such the integration of these enhanced proteins into existing applications in malaria research should be explored. In this work, we have developed an efficient and facile approach for generating new fluorescent reporter lines in P. falciparum using CRISPR-based integration of mNeonGreen. mNeonGreen was first applied to P. falciparum to generate a tagged Exp2 protein however, here we aimed to assess its broader applicability in generating reporter lines for competitive fitness assays and visualisation (Glushakova et al., 2018). mNeonGreen has been successfully incorporated and used for these applications in other protozoans such as trypanosomes and toxoplasma (Dean et al., 2017; Markus et al., 2019). Therefore, we designed a CRISPR/Cas9 construct to integrate mNeonGreen into pfpare, a highly conserved, nonessential gene that would endogenously drive expression throughout the intraerythrocytic cycle.
We used the construct to tag the Dd2 strain and then assessed mNeonGreen@pare for features suitable in a reporter line i.e., the strength of fluorescence, localisation, stage-specificity, and impact on parasite fitness. mNeonGreen@pare demonstrated strong fluorescence that was diffused throughout the parasite. The fluorescence was expressed throughout asexual blood stage development as well as during gametocytogenesis, suggesting that these tagged lines are a valuable tool for in vitro research studying the biology of P. falciparum in these stages. Gametocyte-competent lines that express fluorescent and luciferase reporters have been useful for evaluating antimalarial activity against sexual stages, however prior to genome editing advances, these lines were laborious to construct (Adjalley et al., 2011). Our editing strategy would allow mNeonGreen and potentially other reporters to be rapidly inserted into different strains under investigation. The ability to efficiently tag Dd2 indicates that minor differences in the homology regions between the donor, based on the 3D7 sequence, and the target region are tolerated. The absence of wild type locus in bulk transfections reflects the effect of positive selection resulting from bsd expression from the endogenous pfpare promoter. Although disruption of pfpare also affords the possibility of negative selection using the commercially available compound MMV011438, this was not required in practice due to the stringency of the blasticidin S positive selection. The integration of mNeonGreen into pfpare caused a minor fitness defect leading to slightly slower growth compared to Dd2, similar to other GFP-based reporters (Baragaña et al., 2015). However, this defect was relatively minimal, as the resulting line outcompeted 3D7 in a competition assay. These findings support that the mNeonGreen@pare is a suitable reporter line and is comparable with the standard P. falciparum GFP lines that are currently used.
Both tools generated in this work, the Dd2 mNeonGreen reporter line and the construct that generates new reporter lines, will support malaria research. mNeonGreen@pare is a valuable addition to the repertoire of reporter lines in P. falciparum that facilitate experiments involving visualising, tracking and counting parasites. The pDC-coCas9-pare-BSD-mNeonGreen construct will enable the rapid generation of other fluorescently tagged P. falciparum parasites from different strains, such as field isolates, which can facilitate their study in vitro.
Data availability statement
The data supporting the findings of this study are available within the article or can be found in the Supplementary Material.
Author contributions
JH and ML conceived and designed the experiments. HJ generated the pDC2-coCas9-pare-2A-BSD construct. JH generated the pDC-coCas9-pare-BSD-mNeonGreen plasmid and performed the experiments and analysis. ML supervised the work. JH and ML wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded in whole, or in part, by Wellcome [Grant number 206194/Z/17/Z]. For the purpose of open access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.981432/full#supplementary-material
References
Adjalley, S. H., Johnston, G. L., Li, T., Eastman, R. T., Ekland, E. H., Eappen, A. G., et al. (2011). Quantitative assessment of Plasmodium falciparum sexual development reveals potent transmission-blocking activity by methylene blue. Proc. Natl. Acad. Sci. U.S.A. 108, E1214–E1223. doi: 10.1073/pnas.1112037108
Adjalley, S. H., Lee, M. C. (2022). CRISPR/Cas9 editing of the Plasmodium falciparum genome. Methods Mol. Biol. 2470, 221–239. doi: 10.1007/978-1-0716-2189-9_17
Amos, B., Aurrecoechea, C., Barba, M., Barreto, A., Basenko, E.Y., Bażant, W., et al. (2021). VEuPathDB: the eukaryotic pathogen, vector and host bioinformatics resource center. Nucleic Acids Res. 50, D898–D911. doi: 10.1093/nar/gkab929
Armstrong, C. M., Goldberg, D. E. (2007). An FKBP destabilization domain modulates protein levels in Plasmodium falciparum. Nat. Methods 4, 1007–1009. doi: 10.1038/nmeth1132
Baragaña, B., Hallyburton, I., Lee, M. C., Norcross, N. R., Grimaldi, R., Otto, T. D., et al. (2015). A novel multiple-stage antimalarial agent that inhibits protein synthesis. Nature 522, 315–320. doi: 10.1038/nature14451
Campbell, B. C., Nabel, E. M., Murdock, M. H., Lao-Peregrin, C., Tsoulfas, P., Blackmore, M. G., et al. (2020). mGreenLantern: a bright monomeric fluorescent protein with rapid expression and cell filling properties for neuronal imaging. Proc. Natl. Acad. Sci. U.S.A. 117, 30710–30721. doi: 10.1073/pnas.2000942117
Chalfie, M., Tu, Y., Euskirchen, G., Ward, W. W., Prasher, D. C. (1994). Green fluorescent protein as a marker for gene expression. Science 263, 802–805. doi: 10.1126/science.8303295
Chappell, L., Ross, P., Orchard, L., Russell, T. J., Otto, T. D., Berriman, M., et al. (2020). Refining the transcriptome of the human malaria parasite Plasmodium falciparum using amplification-free RNA-seq. BMC Genomics 21, 395. doi: 10.1186/s12864-020-06787-5
Dean, S., Sunter, J. D., Wheeler, R. J. (2017). TrypTag.org: A trypanosome genome-wide protein localisation resource. Trends Parasitol. 33, 80–82. doi: 10.1016/j.pt.2016.10.009
Engelmann, S., Silvie, O., Matuschewski, K. (2009). Disruption of Plasmodium sporozoite transmission by depletion of sporozoite invasion-associated protein 1. Eukaryot Cell 8, 640–648. doi: 10.1128/EC.00347-08
Fidock, D. A., Wellems, T. E. (1997). Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc. Natl. Acad. Sci. U.S.A. 94, 10931–10936. doi: 10.1073/pnas.94.20.10931
Fivelman, Q. L., Mcrobert, L., Sharp, S., Taylor, C. J., Saeed, M., Swales, C. A., et al. (2007). Improved synchronous production of Plasmodium falciparum gametocytes in vitro. Mol. Biochem. Parasitol. 154, 119–123. doi: 10.1016/j.molbiopara.2007.04.008
Frischknecht, F., Martin, B., Thiery, I., Bourgouin, C., Menard, R. (2006). Using green fluorescent malaria parasites to screen for permissive vector mosquitoes. Malaria J. 5, 23. doi: 10.1186/1475-2875-5-23
Gabryszewski, S. J., Dhingra, S. K., Combrinck, J. M., Lewis, I. A., Callaghan, P. S., Hassett, M. R., et al. (2016a). Evolution of fitness cost-neutral mutant PfCRT conferring P. falciparum 4-aminoquinoline drug resistance is accompanied by altered parasite metabolism and digestive vacuole physiology. PloS Pathog. 12, e1005976. doi: 10.1371/journal.ppat.1005976
Gabryszewski, S. J., Modchang, C., Musset, L., Chookajorn, T., Fidock, D. A. (2016b). Combinatorial genetic modeling of pfcrt-mediated drug resistance evolution in Plasmodium falciparum. Mol. Biol. Evol. 33, 1554–1570. doi: 10.1093/molbev/msw037
Glushakova, S., Beck, J. R., Garten, M., Busse, B. L., Nasamu, A. S., Tenkova-Heuser, T., et al. (2018). Rounding precedes rupture and breakdown of vacuolar membranes minutes before malaria parasite egress from erythrocytes. Cell Microbiol. 20, e12868. doi: 10.1111/cmi.12868
Goonewardene, R., Daily, J., Kaslow, D., Sullivan, T. J., Duffy, P., Carter, R., et al. (1993). Transfection of the malaria parasite and expression of firefly luciferase. Proc. Natl. Acad. Sci. U.S.A. 90, 5234–5236. doi: 10.1073/pnas.90.11.5234
Goujon, M., Mcwilliam, H., Li, W., Valentin, F., Squizzato, S., Paern, J., et al. (2010). A new bioinformatics analysis tools framework at EMBL–EBI. Nucleic Acids Res. 38, W695–W699. doi: 10.1093/nar/gkq313
Horrocks, P., Kilbey, B. J. (1996). Physical and functional mapping of the transcriptional start sites of Plasmodium falciparum proliferating cell nuclear antigen. Mol. Biochem. Parasitol. 82, 207–215. doi: 10.1016/0166-6851(96)02737-5
Istvan, E. S., Mallari, J. P., Corey, V. C., Dharia, N. V., Marshall, G. R., Winzeler, E. A., et al. (2017). Esterase mutation is a mechanism of resistance to antimalarial compounds. Nat. Commun. 8, 14240. doi: 10.1038/ncomms14240
Kuang, D., Qiao, J., Li, Z., Wang, W., Xia, H., Jiang, L., et al. (2017). Tagging to endogenous genes of Plasmodium falciparum using CRISPR/Cas9. Parasit. Vectors 10, 595. doi: 10.1186/s13071-017-2539-0
Marin-Mogollon, C., Salman, A. M., Koolen, K. M. J., Bolscher, J. M., Van Pul, F. J. A., Miyazaki, S., et al. (2019). A P. falciparum NF54 reporter line expressing mCherry-luciferase in gametocytes, sporozoites, and liver-stages. Front. Cell Infect. Microbiol. 9, 96. doi: 10.3389/fcimb.2019.00096
Markus, B., Bell, G., Lorenzi, H., Lourido, S. (2019). Optimizing systems for Cas9 expression in Toxoplasma gondii. mSphere 43, e00386–e00319. doi: 10.1128/mSphere.00386-19
Miyazaki, S., Yang, A. S. P., Geurten, F. J. A., Marin-Mogollon, C., Miyazaki, Y., Imai, T., et al. (2020). Generation of novel Plasmodium falciparum NF135 and NF54 lines expressing fluorescent reporter proteins under the control of strong and constitutive promoters. Front. Cell Infect. Microbiol. 10, 270. doi: 10.3389/fcimb.2020.00270
Mogollon, C. M., Van Pul, F. J., Imai, T., Ramesar, J., Chevalley-Maurel, S., De Roo, G. M., et al. (2016). Rapid generation of marker-free P. falciparum fluorescent reporter lines using modified CRISPR/Cas9 constructs and selection protocol. PloS One 11, e0168362. doi: 10.1371/journal.pone.0168362
Portugaliza, H. P., Llorà-Batlle, O., Rosanas-Urgell, A., Cortés, A. (2019). Reporter lines based on the gexp02 promoter enable early quantification of sexual conversion rates in the malaria parasite Plasmodium falciparum. Sci. Rep. 9, 14595. doi: 10.1038/s41598-019-50768-y
Radfar, A., Méndez, D., Moneriz, C., Linares, M., Marín-García, P., Puyet, A., et al. (2009). Synchronous culture of Plasmodium falciparum at high parasitemia levels. Nat. Protoc. 4, 1899–1915. doi: 10.1038/nprot.2009.198
Ross, L. S., Dhingra, S. K., Mok, S., Yeo, T., Wicht, K. J., Kümpornsin, K., et al. (2018). Emerging southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat. Commun. 9, 3314. doi: 10.1038/s41467-018-05652-0
Shaner, N. C., Lambert, G. G., Chammas, A., Ni, Y., Cranfill, P. J., Baird, M. A., et al. (2013). A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10, 407–409. doi: 10.1038/nmeth.2413
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 7, 539. doi: 10.1038/msb.2011.75
Stokes, B. H., Dhingra, S. K., Rubiano, K., Mok, S., Straimer, J., Gnädig, N. F., et al. (2021). Plasmodium falciparum K13 mutations in Africa and Asia impact artemisinin resistance and parasite fitness. Elife 10, e66277. doi: 10.7554/eLife.66277.sa2
Talman, A. M., Blagborough, A. M., Sinden, R. E. (2010). A Plasmodium falciparum strain expressing GFP throughout the parasite's life-cycle. PloS One 5, e9156. doi: 10.1371/journal.pone.0009156
Thommen, B. T., Passecker, A., Buser, T., Hitz, E., Voss, T. S., Brancucci, N. M. B. (2022). Revisiting the effect of pharmaceuticals on transmission stage formation in the malaria parasite Plasmodium falciparum. Front. Cell. Infect. Microbiol. 12, 802341. doi: 10.3389/fcimb.2022.802341
VanWye, J. D., Haldar, K. (1997). Expression of green fluorescent protein in Plasmodium falciparum. Mol. Biochem. Parasitol. 87, 225–229. doi: 10.1016/S0166-6851(97)00059-5
Voorberg-van Der Wel, A. M., Zeeman, A.-M., Nieuwenhuis, I. G., van der Werff, N. M., Klooster, E. J., Klop, O., et al. (2020). A dual fluorescent Plasmodium cynomolgi reporter line reveals in vitro malaria hypnozoite reactivation. Commun. Biol. 3, 7. doi: 10.1038/s42003-019-0737-3
Wilson, D. W., Crabb, B. S., Beeson, J. G. (2010). Development of fluorescent Plasmodium falciparum for in vitro growth inhibition assays. Malaria J. 9, 152. doi: 10.1186/1475-2875-9-152
Keywords: malaria, reporter genes, fluorescence, CRISPR/Cas9, Plasmodium falciparum
Citation: Hoshizaki J, Jagoe H and Lee MCS (2022) Efficient generation of mNeonGreen Plasmodium falciparum reporter lines enables quantitative fitness analysis. Front. Cell. Infect. Microbiol. 12:981432. doi: 10.3389/fcimb.2022.981432
Received: 29 June 2022; Accepted: 30 August 2022;
Published: 16 September 2022.
Edited by:
Faiza Amber Siddiqui, University of South Florida, United StatesReviewed by:
Mariana De Niz, Universidade de Lisboa, PortugalCaroline Ng, University of Nebraska Medical Center, United States
Copyright © 2022 Hoshizaki, Jagoe and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marcus C. S. Lee, bWwzMUBzYW5nZXIuYWMudWs=
 Johanna Hoshizaki
Johanna Hoshizaki Hannah Jagoe
Hannah Jagoe Marcus C. S. Lee
Marcus C. S. Lee