
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 24 August 2022
Sec. Clinical Microbiology
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.973901
This article is part of the Research Topic Klebsiella pneumoniae: Antimicrobial Resistance, Virulence and Therapeutic Strategies View all 8 articles
 Shuangshuang Li1,2†
Shuangshuang Li1,2† Xiaoyuan Jiang1†
Xiaoyuan Jiang1† Cuidan Li1†
Cuidan Li1† Yingjiao Ju1,3
Yingjiao Ju1,3 Liya Yue1
Liya Yue1 Fangzhou Chen4
Fangzhou Chen4 Lingfei Hu4
Lingfei Hu4 Jing Wang5,6,7
Jing Wang5,6,7 Xin Hu5,6
Xin Hu5,6 Bahetibieke Tuohetaerbaike5,6
Bahetibieke Tuohetaerbaike5,6 Hao Wen5,6
Hao Wen5,6 Wenbao Zhang5,6
Wenbao Zhang5,6 Dongsheng Zhou4
Dongsheng Zhou4 Zhe Yin4*
Zhe Yin4* Fei Chen1,2,3,5*
Fei Chen1,2,3,5*As a newly emerging Klebsiella pathogen, more and more Klebsiella michiganensis drug resistant strains have been reported in recent years, which posed serious threats to public health. Here we first reported a multidrug-resistant K. michiganensis strain 12084 with two blaSIM-1 and one mcr-9.2 genes isolated from the sputum specimen of a patient in the Second Affiliated Hospital of Zhejiang University School of Medicine and analyzed its genetic basis and drug-resistance phenotypes. Genetic analysis showed that this strain harbored three different incompatibility groups (IncHI2, IncHI5, and IncFIIpKPHS2:IncFIB-4.1) of plasmids (p12084-HI2, p12084-HI5, and p12084-FII). A total of 26 drug-resistance genes belonging to 12 classes of antibiotics were identified, most of which (24) were located on two plasmids (p12084-HI2 and p12084-HI5). Interestingly, two blaSIM-1 genes were identified to locate on p12084-HI2 and p12084-HI5, respectively, both of which were embedded in In630, indicating their genetic homogeny. It was noting that one blaSIM-1 gene was situated in a novel unit transposon (referred to as Tn6733) on the p12084-HI5 plasmid. We also discovered an mcr-9.2 gene on the p12084-HI2 plasmid. To the best of our knowledge, this is the first report of a blaSIM-1 and mcr-9.2 harboring K. michiganensis strain. We then investigated the population structure/classification, and antibiotic resistance for all 275 availably global K. michiganensis genomes. Population structure revealed that K. michiganensis could be divided into two main clades (Clade 1 and Clade 2); the most popular ST29 was located in Clade 1, while other common STs (such as ST50, ST27, and ST43) were located in Clade 2. Drug-resistance analysis showed 25.5% of the K. michiganensis strains (70/275) harboring at least one carbapenemase gene, indicating severe drug resistance of K. michiganensis beyond our imagination; this is a dangerous trend and should be closely monitored, especially for ST27 K. michiganensis with the most drug-resistant genes among all the STs. Overall, we reported a blaSIM-1 and mcr-9.2 harboring K. michiganensis strain, and further revealed the population structure/classification, and drug-resistance of K. michiganensis, which provided an important framework, reference, and improved understanding of K. michiganensis.
Klebsiella includes at least 27 species, such as K. pneumoniae, K. quasipneumoniae, K. variicola, K. quasivariicola, K. africana, K. terrigena, K. huaxiensis, K. michiganensis, and K. oxytoca (Merla et al., 2019; Wyres et al., 2020). K. michiganensis was originally discovered in a toothbrush holder in a Michigan household in 2012 (Saha et al., 2013). Until now, this potential emerging pathogen has been reported in clinical settings in many countries (Mediavilla et al., 2016; Chapman et al., 2020; Sands et al., 2021).
Importantly, in recent years, more and more drug resistant K. michiganensis strains, even carbapenem resistance ones, have been reported, which could lead to high morbidity and mortality (Xu et al., 2017), further causing great threats to global public health (De Oliveira et al., 2020). The prevalence of carbapenem resistance in K. michiganensis is mainly derived from carbapenemase genes on various types of drug resistance plasmids, such as blaKPC, blaNDM, blaIMP, and blaVIM genes (Tzouvelekis et al., 2012). Some K. michiganensis strains have even been reported to carry more than one carbapenemase gene, such as co-harboring of blaKPC-2, blaNDM-1, and blaNDM-5 (Zheng et al., 2018), and co-harboring of blaIMP-4 and blaNDM-1 (Li et al., 2021). The blaSIM-1 gene was first discovered in Acinetobacter pittii in a Korean hospital in 2003 (Lee et al., 2005), which could cause carbapenem resistance (Palzkill, 2013; Bean and Wareham, 2022), but it has not been reported in K. michiganensis.
Polymyxin is known to be the last line for fighting against carbapenem-resistant Gram-negative bacteria (Poirel et al., 2017). In November 2015, a plasmid-mediated polymyxin resistance gene mcr-1 was first discovered in Escherichia coli and K. pneumoniae from animal and human in China (Liu et al., 2016). In recent years, mcr-2 to mcr-10 have been reported worldwide (Wang et al., 2020). The global dissemination of mcr genes causes significant threats to the last-line of antibiotic defense. Notably, more and more multidrug-resistant (MDR) klebsiella species co-producing carbapenemases and MCR have been clinically isolated, resulting in severe clinical problems because of very limited options for the treatments of such cases (Kaye et al., 2016; Mendes et al., 2018; Wang et al., 2018; Campos-Madueno et al., 2021), but such MDR strain has not been reported in K. michiganensis.
In this study, we first reported an MDR K. michiganensis strain 12084 co-harboring carbapenemase gene blaSIM-1 and MCR gene mcr-9.2, and its genetic characteristics were comprehensively analyzed. Furthermore, we revealed the population structure/classification, and antibiotic resistance genes of K. michiganensis for the first time by analyzing all 275 available K. michiganensis genomes, which provided an important framework for K. michiganensis.
Strain 12084 was isolated in 2013 from the sputum specimen of a patient from the Second Affiliated Hospital of Zhejiang University School of Medicine in Hangzhou City, China. Strain 12084 was initially misidentified as K. oxytoca by MALDI-TOF/MS and finally identified as K. michiganensis by average nucleotide identity (ANI) analysis using Pyani-0.2.7 (Richter and Rossello-Mora, 2009).
In vitro antimicrobial susceptibility tests of ampicillin, ampicillin/sulbactam, piperacillin, piperacillin/tazobactam, cefazolin, cefuroxime, cefuroxime axetil, cefotetan, ceftazidime, ceftriaxone, aztreonam, imipenem, meropenem, amikacin, gentamicin, tobramycin, ciprofloxacin, levofloxacin, nitrofurantoin, and trimethoprim/sulfamethoxazole were performed using BioMérieux VITEK 2. The minimum inhibitory concentration (MIC) of polymyxin B was determined using the broth microdilution method following the recommendations of the Clinical and Laboratory Standards Institute (CLSI, 2020). All the above results of antimicrobial susceptibility test were interpreted according to the CLSI guidelines.
Conjugal transfer experiments were carried out with the rifampin-resistant Escherichia coli EC600 used as a recipient and the blaSIM-positive 12084 isolate as a donor. Three milliliters of overnight cultures of each donor and recipient bacteria were mixed together, harvested and resuspended in 80 μL of Brain Heart Infusion (BHI) broth (BD Biosciences). The mixture was spotted on a 1 cm2 hydrophilic nylon membrane filter with a 0.45 µm pore size (Millipore) that was placed on BHI agar (BD Biosciences) plate and then incubated for mating at 22 °C for 24 h. Bacteria were washed from filter membrane and spotted on Muller-Hinton (MH) agar (BD Biosciences) plates containing 2500 μg/mL rifampin together with 4 μg/mL meropenem for selecting an E. coli transconjugant carrying blaSIM (p12084-HI2 and p12084-HI5).
Genomic DNA was isolated from 12084 isolate using a Qiagen blood & cell culture DNA maxi kit. Genome sequencing was performed with a sheared DNA library with an average size of 15 kb (ranging from 10 kb to 20 kb) on a PacBio RSII sequencer (Pacific Biosciences, CA, USA), as well as a paired-end library with an average insert size of 400 bp (ranged from 150 kb to 600 kb) on a HiSeq sequencer (Illumina, CA, USA). De novo assembly was performed using the Hierarchical Genome Assembly Process 3 (HGAP3) within SMRT Analysis v2.3.0 (https://smrt-analysis.readthedocs.io/en/latest/). Adapters and low-quality sequences were trimmed and filtered using FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/).
Open reading frames (ORFs) and pseudogenes were predicted using RAST 2.0 (Brettin et al., 2015) combined with BLASTP/BLASTN searches (Boratyn et al., 2013) against the UniProtKB/Swiss-Prot database (Boutet et al., 2016) and the RefSeq database (O’Leary et al., 2016). Annotations of resistance genes, mobile elements, and other features were carried out using the online databases including CARD (Jia et al., 2017), ResFinder (Zankari et al., 2012), ISfinder (Siguier et al., 2006), INTEGRALL (Moura et al., 2009), and Tn Number Registry (Roberts et al., 2008). Multiple and pairwise sequence comparisons were performed using MUSCLE 3.8.31 (Edgar, 2004) and BLASTN, respectively. Gene organization diagrams were drawn in Inkscape 0.48.1 (https://inkscape.org/en/). Multi locus sequence typing (MLST) analysis for K. michiganensis was performed through mlst (https://github.com/tseemann/mlst) based on the seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) from the PubMLST database (https://pubmlst.org/organisms/klebsiella-oxytoca/).
All the publicly available genomes of K. michiganensis isolates (n=306, updated on September 9, 2021) were downloaded. ANI analysis of the 306 isolates was performed against the 11 reference strains of Klebsiella spp. (K. pneumoniae HS11286, CP003200.1; K. aerogenes Ka37751, CP041925.1; K. Africana 200023, CP084874.1; K. grimontii 4928STDY7071328, LR607336.1; K. huaxiensis WCHKl090001, CP036175.1; K. oxytoca FDAARGOS_335, CP027426.1; K. pasteurii SB6415; K. quasipneumoniae KqPF26, CP065838.1; K. quasivariicola 08A119, CP084768.1; K. variicola FH-1, CP054254.1; K. michiganensis THO-011, NZ_AP022547.1). 275 K. michiganensis strains (274 publicly available strains and strain 12084) were mapped to the K. michiganensis reference strain THO-011, and single nucleotide polymorphisms (SNPs) were identified by Mummer v4 (Marçais et al., 2018). We filtered all the SNPs in the repetitive DNA regions as identified by RepeatMasker (http://www.repeatmasker.org/) and those in the mobile genetic elements including insertion sequences (ISs), transposons, integrons, and phage-related genes. Based on the final 503,605 SNPs, a maximum-likelihood clustering tree of the 275 global K. michiganensis isolates was constructed using FastTree V2.1.9 (Price et al., 2009) with a bootstrap value of 500 and visualized using iTOL v6 (https://itol.embl.de/).
The 12084-chromosome, p12084-HI2, p12084-HI5, and p12084-FII sequences were submitted to GenBank under accession numbers CP072119, MW148604, MW810613, and MZ398018, respectively.
On July 11, 2013, a 44-year-old man, who had performed a kidney transplant seven years ago, was admitted to The Second Affiliated Hospital Zhejiang University School of Medicine, due to lung infectious lesion accompanied with diarrhea, fever, cough, and acratia. After being treated with multiple antibiotics in the hospital for 27 days, the patient was not recovered and volunteered for discharge. A K. michiganensis strain named 12084 and an Acinetobacter baumannii strain were isolated from the sputum specimens during routine sampling and culturing on the 16th day of hospitalization. Since the sputum bacterial cultures of this patient were performed since the beginning of hospitalization, we had reason to infer that the two strains were both acquired through nosocomial infection. Here, the antimicrobial susceptibility tests showed that the A. baumannii strain was susceptible to carbapenems, while the K. michiganensis strain 12084 was resistant to carbapenems and selected for further analysis. The strain 12084 was initially identified as K. oxytoca through MALDI-TOF/MS and further identified as an MDR strain, displaying resistance to 13 antibiotics belonging to six categories: penicillins (ampicillin, piperacillin, ampicillin/sulbactam, and piperacillin/tazobactam), cephalosporins I (cefazolin), cephalosporins II (cefuroxime axetil, cefotetan, and cefuroxime), cephalosporins III (ceftriaxone and ceftazidime), carbapenems (meropenem and imipenem), and sulfanilamides (trimethoprim/sulfamethoxazole) (Table 1). It also showed intermediate resistance to 3 antibiotics, including furanes (nitrofurantoin), fluoroquinolones (levofloxacin), and polymyxin B. This strain was susceptible to aminoglycosides (amikacin, gentamicin, and tobramycin), fluoroquinolones (ciprofloxacin), and monobactams (aztreonam) (Table 1).
To analyze the genetic basis of strain 12084, whole-genome sequencing was performed using the long-read PacBio RSII sequencing platform (Pacific Biosciences, CA, USA) (Table S1) and short-read Illumina HiSeq sequencing platform (Illumina, CA, USA) (Table S2). The strain 12084 was further identified as K. michiganensis rather than K. oxytoca through ANI analysis among all available Klebsiella reference genomes. It showed the highest ANI value of 98.9% compared with K. michiganensis reference THO-011, while displayed lower ANI values (< 95%) with other reference genomes (Table S3; Figure S1).
Sequencing results showed that strain 12084 contained a ~6.2Mb circular chromosome and three plasmids. The three plasmids were designated as p12084-HI2, p12084-HI5, and p12084-FII, which had circularly closed DNA sequences of 237,396 bp, 273,923 bp, and 193,645 bp in length with mean G+C contents of 45.9%, 46.6%, and 51.7%, and predicted ORFs of 302 bp, 393 bp, and 268 bp, respectively (Table 2). These plasmids belonged to IncHI2, IncHI5, and IncFIIpKPHS2:IncFIB-4.1 groups, respectively, because they contained the replication genes of repHI2A + repHI2C, repHI5B + repFIB-like, and repA2-6-1-4IncFIIpKPHS2 + repIncFIB-4.1, respectively (Figure 1). Each plasmid was composed of backbone regions, together with accessory modules that were recognized with some acquired DNA regions with various mobile elements at different sites (Table 2; Figure 1).
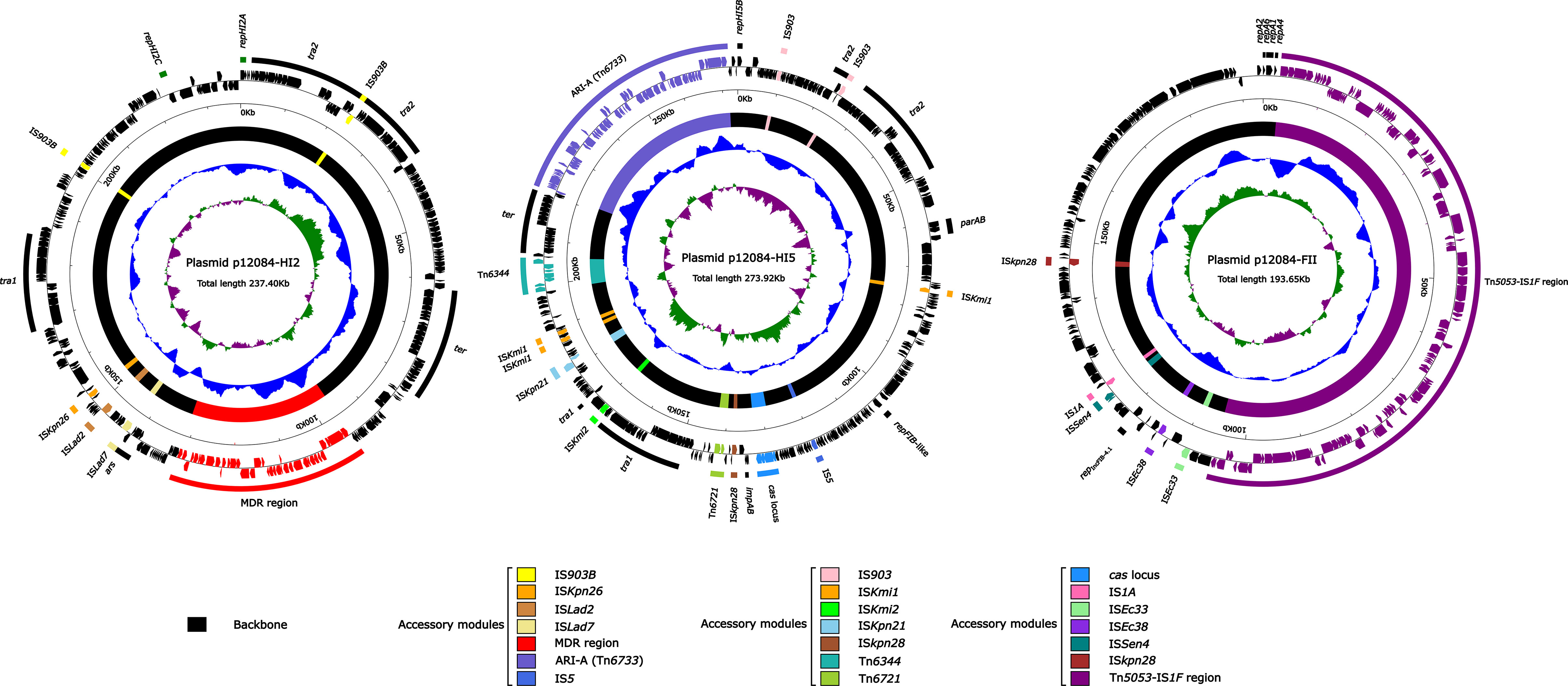
Figure 1 Schematic maps of p12084-HI2, p12084-HI5, and p12084-FII plasmids. Genes are denoted by arrows, and the backbone and accessory module regions are highlighted in black and color, respectively; The innermost circle presents GC-skew [(G-C)/(G+C)], with a window size of 500 bp and a step size of 20 bp. Inward indicates the relative content G > C, and outward indicates the relative content G < C; The next-to-innermost circle presents G and C content. Inward indicates below average G and C content, and outward indicates above average G and C content; The outermost circle shows the arrangement of genes, with genes forward and reverse in clockwise and counterclockwise directions, respectively.
Both p12084-HI2 and p12084-HI5 owned large backbone regions and small accessory modules with a Tn1696 structure. Here, the accessory modules of p12084-HI2 consisted of one MDR region and several ISs), and those of p12084-HI5 contained a new MDR region (designated as Tn6733), Tn6721, Tn6344, and several ISs (IS903/ISkmi1/ISkmi2/ISkpn21/ISkpn28) (Figure 1). P12084-FII had small backbone regions with a new large accessory module (named as Tn5053–IS1F region) (Table 2; Figure 1).
To illustrate the drug-resistance genotype of the K. michiganensis 12084, we identified drug resistance genes by BLAST against the database ResFinder (Zankari et al., 2012). A total of 26 drug resistance genes were identified, including aminoglycoside [aph(3’)-Ia, aadA15e, aadA2, and armA], β-lactam (blaCTX-M-9 and blaOXY-1), carbapenem (blaSIM-1), colistin (mcr-9.2), disinfectant (qacED1), fosfomycin (fosA3), macrolide [mph(E) and msr(E)], phenicol (catB3), quinolone (qnrA1), rifampicin (ARR-3), sulphonamide (sul1), and trimethoprim (dfrA16) (Table 3). Importantly, most drug resistance genes (92%; 24/26) were located on the accessory modules of the two MDR plasmids (p12084-HI2 and p12084-HI5) (Table 3). Here, the resistant phenotypes for all the above mentioned antibiotics of 12084 strains could be explained by the existence of corresponding drug resistant genes. In addition, although we identified some drug resistant genes related to aminoglycosides and fluoroquinolones (aph(3’)-Ia, aadA15e, aadA2, and armA for aminoglycosides; qnrA1 for fluoroquinolones), the strain 12084 was susceptible to these two drugs. Previous studies have revealed that the two drug resistant genes on the plasmids produce very low levels of resistance for aminoglycosides and fluoroquinolones. (Rodríguez-Martínez et al., 2011; Saravolatz and Stein, 2020).
It was noted that two copies of blaSIM-1 genes were observed on two plasmids (p12084-HI2 and p12084-HI5) of the strain, respectively. A colistin resistant gene mcr-9.2 was also identified on p12084-HI2 (Table 3). To our best knowledge, this is the first reported K. michiganensis strain with blaSIM-1 and mcr-9.2 genes.
Since most antibiotic resistance genes (ARGs) were located on the two MDR plasmids (p12084-HI2 and p12084-HI5), we further conducted a genetic characteristic analysis of both plasmids. The backbones of p12084-HI2 and p12084-HI5 were 195.6 kb and 197.7 kb in length, accounting for 82.4% and 72.2% of the plasmid genomes, respectively.
The p12084-HI2 backbone showed 79% BLAST query coverage and 99% nucleotide identity to the IncHI2 reference plasmid R478 (GenBank accession no. BX664015) (Gilmour et al., 2004). These two plasmids shared the core IncHI2 backbone markers, including repHI2A and repHI2C for replication initiation, the tra1 and tra2 regions for conjugal transfer, and the ter region for tellurium resistance, and the ars locus for arsenic resistance (Figure S2A). On the other hand, three major differences were identified between the two backbone sequences: IS903B was inserted between parB2 and htdA within the tra2 region of p12084-HI2; compared to R478, MDR region of p12084-HI2 was inserted at a site downstream of dcm, leading to the truncation of dcm and deletion of orf819 to orf168 region; another IS903B was inserted at a site downstream of orf450, resulting in the truncation of orf450 and loss of orf198 to orf2385 region (Figure S2A).
The p12084-HI5 backbone had 86% BLAST query coverage and 99% nucleotide identity to the IncHI5 reference plasmid pA324-IMP (GenBank accession no. CP026017) (Liang et al., 2019), with conserved IncHI5 backbone genes or gene loci (including repHI5B together with its iterons and repFIB-like for replication, parABC for partition, tra1 and tra2 for conjugal transfer, and ter for tellurium resistance) (Figure S2B). On the other hand, four different important events associated with gene acquisition/loss were identified between the backbone regions of these two plasmids. Firstly, IS903 was inserted within tivF6 (F-type type IV secretion, mating pair stabilization protein) across the tra2 region in p12084-HI5; secondly, ISKmi2 was inserted within orf3351 (superfamily I DNA helicase) across the tra1 region in p12084-HI5; Thirdly, compared to pA324-IMP (reference), p12084-HI5 had underwent the truncations of impB and umuC2 genes, and the deletion of the backbone genes of the thiF1 to umuC1 region. Importantly, two different ARI-As (Tn6733 from p12084-HI5, and Tn6382 from pA324-IMP) carried different drug-resistance profiles, although they were both inserted at a site between relB and orf342 (Figure S2B).
We further focused on the MDR regions to explore the genetic environment of antibiotic-resistant genes for the two plasmids. The MDR region (41.7 kb) from p12084-HI2 and the Tn6733 (ARI-A island, 76.2 kb) from p12084-HI5 (Figure 2) were different derivatives of Tn1696. Tn1696, a unit transposon belonging to the Tn21 subgroup of Tn3, was generated from the insertion of a class 1 integron In4 into the resolution (res) site of a primary backbone structure: IRL (inverted repeat left)–tnpA (transposase)–tnpR (resolvase)–res–mer (mercury resistance locus)–IRR (inverted repeat right) (Partridge et al., 2001).
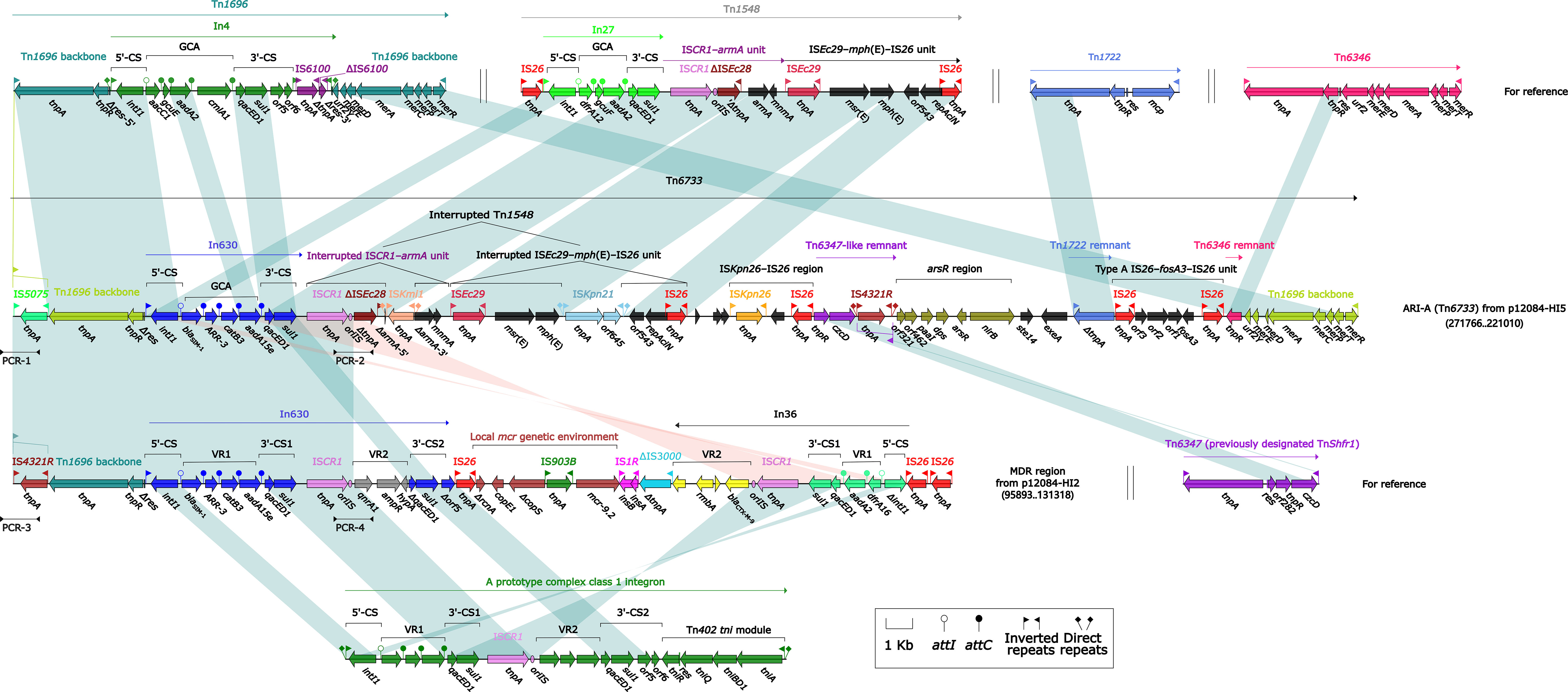
Figure 2 Organizations of the MDR regions of p12084-HI2, and p12084-HI5 compared with related reference regions. Genes are denoted by arrows. Genes, mobile elements, and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity). Numbers in brackets indicate the nucleotide positions within the corresponding plasmids. The arrowheads indicate the locations of PCR primers and the expected amplicons. The accession numbers of Tn1696, Tn1548, Tn1722, Tn6346, and Tn6347 for reference are U12338, AF550415, X61367, EU696790, and CP000447, respectively.
The MDR region from p12084-HI2 (Figure 2) contained a Tn1696-resembling backbone region with IRL-tnpA-tnpR-Δres at the 5’-end, and its IRL was interrupted by IS4321R (Partridge and Hall, 2003). But it was not an intact transposon due to its absence of IRR at the 3’-end, which was derived from the insertion of the integron-associated regions [namely In630, the mcr genetic environment (carrying mcr-9.2), In36, and five independent ISs (including IS1R, ΔIS3000, and three copies of IS26)]. Both In630 and In36 from the MDR region belonged to complex class 1 integrons (Toleman et al., 2006), which harbored two intrinsic resistance regions, namely the cassette array blaSIM-1–ARR-3–catB3–aadA15e and qnrA1 (In630), and the cassette array dfrA16–aadA2 and blaCTX-M-9 (In36).
Similar to Tn1696, the ARI-A island (Liang et al., 2019) from p12084-HI5 had paired terminal 38-bp IRL/IRR, and thus it was identified as a novel unit transposon and designated as Tn6733 (the highest coverage of 82% and identity of 99% compared to all available DNA sequences in the GenBank). ARI-A (Tn6733) differed from Tn1696 in two aspects: i) In4 of Tn1696 was replaced with some insertions at the same position within res, including a concise class 1 integron In630 (Sajjad et al., 2011) (the cassette array blaSIM-1–ARR-3–catB3–aadA15e), an mph(E) and msr(E)-carrying interrupted Tn1548, an ISKpn26-IS26 region, a Tn6347-like remnant, an arsR region (arsenic resistance), a Tn1722 remnant, a fosfomycin resistance unit type A IS26–fosA3–IS26, and a Tn6346 remnant; ii) IRL was interrupted by IS5075 that was a hunter of terminal IRs of Tn21 subgroup transposons (Figure 2) (Partridge et al., 2001).
Interestingly, a highly similar repetition region (12.8 kb in length, 100% BLAST query coverage, and 99% nucleotide identity) was observed in both p12084-HI2 and p12084-HI5, which was validated by a set of PCR amplifications targeting four key jointing fragments of the repetition region (Figure 2; Table S4). The repetition region was comprised of a Tn1696 backbone structure, with an IRL–IS5075/IS4321R–tnpA–tnpR–Δres, an In630 (the cassette array blaSIM-1–ARR-3–catB3–aadA15e) and an ISCR1, which might be resulted from the homologous recombination between the two plasmids, further contributing to extensive dissemination of drug resistance genes among various MDR plasmids.
We then explored the genetic environment of mcr-9.2 in p12084-HI2. The modular structure of the mcr genetic environment (6.2 kb in length) from p12084-HI2 (Figure 3) was organized sequentially as ΔrcnA–copE1–ΔcopS–IS903B–mcr-9.2–IS1R, which showed partial homology with the mcr genetic environment in pMCR-SCNJ07 from the strain SCNJ07 (rcnA–copE1–ΔcopS-3’–IS10R–ΔcopS-5’–IS903B–mcr-9–wbuC–qseC–qseB–IS1R) (Yuan et al., 2019).
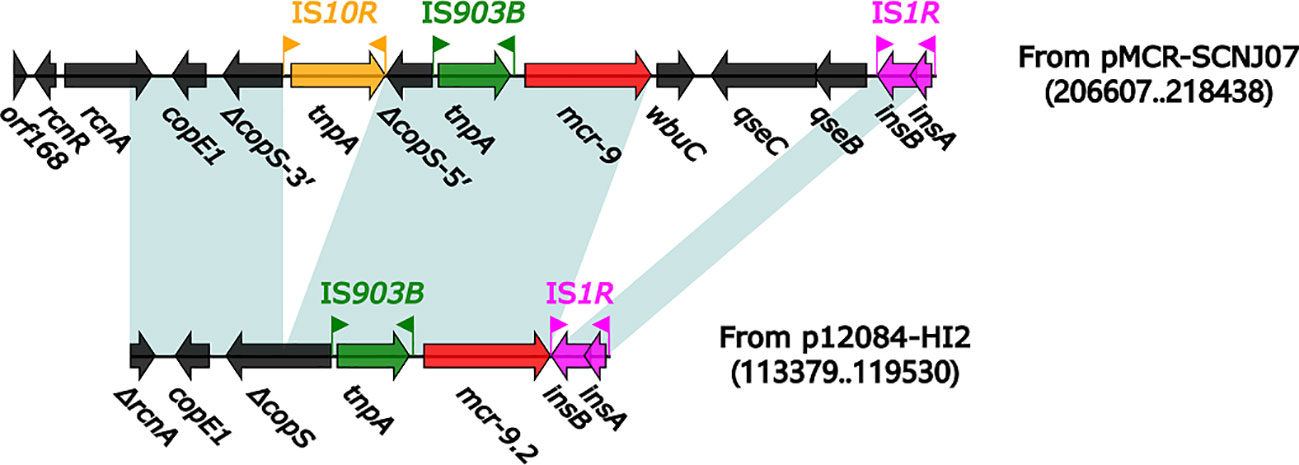
Figure 3 Organization of the mcr genetic environment from p12084-HI2 compared with related reference regions. Genes are denoted by arrows. Genes, mobile elements, and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity). Numbers in parentheses indicate the nucleotide positions within the corresponding plasmids. The accession number of the pMCR-SCNJ07 is MK933279.
Compared to pMCR-SCNJ07, the mcr genetic environment of p12084-HI2 underwent the deletions of wbuC–qseC–qseB downstream of mcr-9.2, and IS10R within copS, and the truncation of rcnA (Figure 3). The difference in mcr genetic environment between the two plasmids might result in differential mcr drug resistances between the two strains: strain 12084 showed intermediate resistance to polymyxin B (MIC=2mg/L), while the strain SCNJ07 displayed resistance to polymyxin B (MIC=8 µg/mL). The modular structures of the mcr-9 gene from ME-1 (Chavda et al., 2019), the mcr-9 gene from LM22-1 (Khalifa et al., 2020), and the mcr-9 gene from A2483 (Kananizadeh et al., 2020) all lack the qseB and qseC regulatory genes, which leads to the corresponding strains susceptible to colistin. Therefore, we speculated that strain 12084 showed intermediate resistance to polymyxin B due to the absence of qseB and qseC downstream of mcr-9.2, further indicating the importance of the qseB/qseC two-component system in mcr-9 expression.
Both p12084-HI2 and p12084-HI5 could be transferred from the strain 12084 into E. coli EC600 through conjugation, generating the transconjugants 12084-SIM-EC600 (Figure S3). These were consistent with the presence of two complete sets of tra1 and tra2 genes in p12084-HI2 and p12084-HI5, making them self-transferable.
To further investigate the population structure/classification, and antibiotic resistance of K. michiganensis, a total of 306 K. michiganensis genomes were downloaded from NCBI (Table S5). ANI analysis showed that 274 (89.5%) strains belonged to K. michiganensis based on the K. michiganensis reference strain THO-011 (ANI value ≥ 95%), including 196 human-, 16 animal-, and one environment-isolates (Table S5). The remaining 32 strains were excluded from our study due to wrong classifications, among which 19 strains belonged to K. grimontii (ANI values were more than 99% compared with the K. grimontii reference strain 4928STDY7071328), 12 strains belonged to K. pasteurii (ANI values were more than 99% compared with the K. pasteurii reference strain SB6415), and one strain (RC10) did not belong to Klebsiella (ANI values were less than 95% compared with all the Klebsiella reference strains) (Table S3; Figure S1). These results indicated more precision classification for K. michiganensis through whole genome sequences.
The phylogenetic tree was then constructed using the abovementioned 274 K. michiganensis strains and our MDR strain 12084. To the best of our knowledge, this is the first time to reveal the population structure of K. michiganensis. Here, K. michiganensis could be divided into two major clades on the tree: Clade1 and Clade2, which were also validated by ANI results (Figure 4; Figure S1). The Clade2 was further classified into three subclades: Clade2.1, Clade2.2, and Clade2.3. In addition, we performed MLST analysis on these K. michiganensis strains. The results showed that ST29 was the most common ST (12/275, 4.4%), followed by ST50 (11/275, 4%), ST27 (10/275, 3.6%), and ST43/ST84/ST85 (9/275, 3.3%) (Figure 4). The ST29 of K. michiganensis strains were located in Clade 1, while other STs (such as ST50, ST27, and ST43) were located in Clade 2. Notably, 120 isolates (43.6%) could not be assigned to known ST types. These results indicated genetic diversity and non-clonal transmission/population structure of K. michiganensis strains.
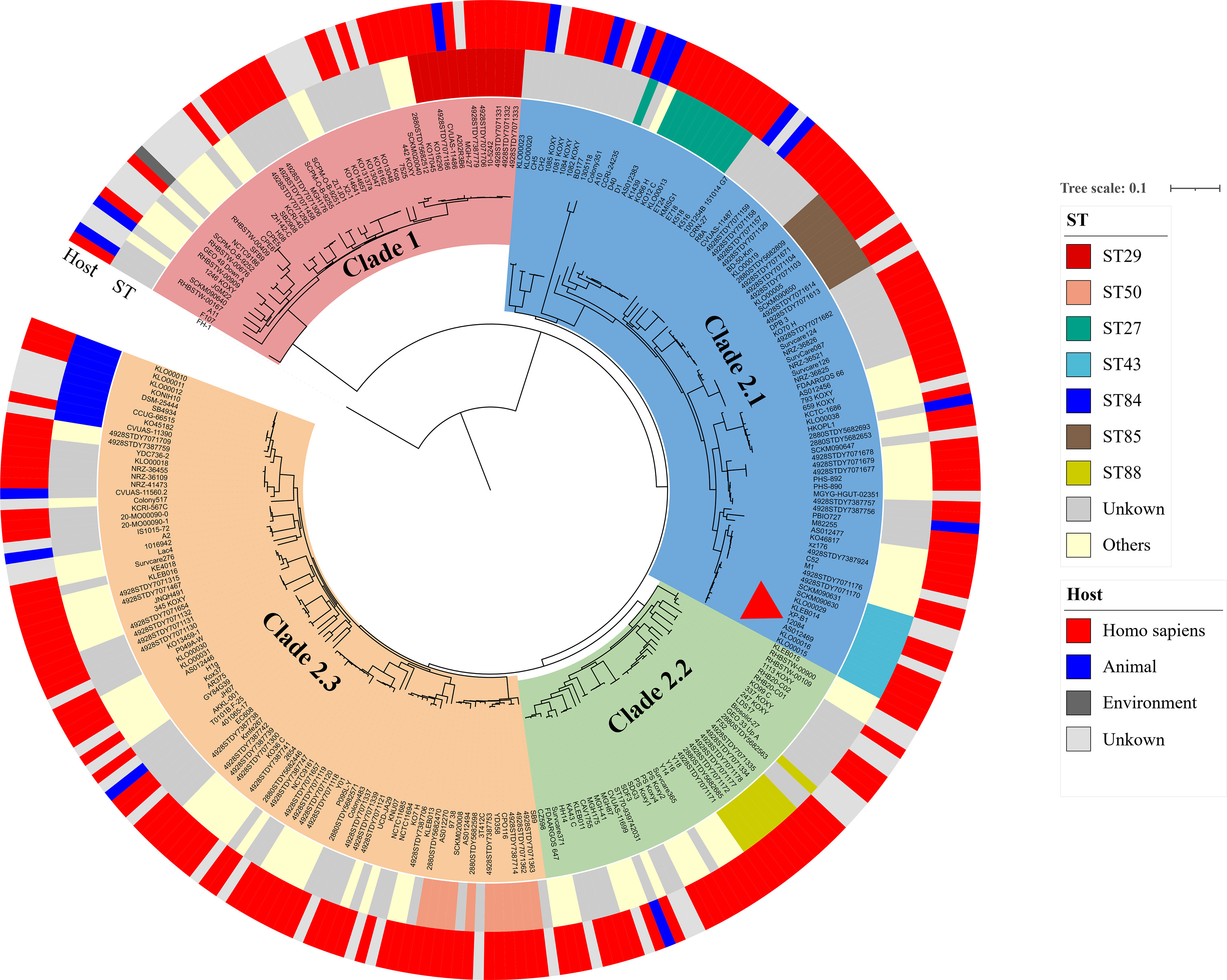
Figure 4 A maximum-likelihood clustering tree of the 275 available K. michiganensis strains. The maximum-likelihood clustering tree is constructed by 503,605 SNPs of all 275 K. michiganensis genome sequences. K. variicola FH-1 (GenBank accession no. CP054254) is used as the outgroup; The outermost circle presents the sources of the isolates; the sub-outside circle presents the STs of the isolates; the innermost circle presents different Clades of K. michiganensis; the strain 12084 is marked with a red triangle.
We also investigated the drug resistance profile of K. michiganensis strains. A total of 51 types of drug resistant genes were identified among these strains (Figure 5). Here, blaOXY commonly existed on the chromosome of all K. michiganensis strains (275/275), ranking first, followed by aph(3’)-Ia (152/275, top-2) and dfrA (74/275, top-3) (Figure 5). We further determined the relationship between the drug-resistant gene number and Clades/STs. Our results revealed no significant difference for the drug-resistant gene number in different clades. However, the drug-resistant gene number showed a difference among different STs: ST27 isolates possess the most drug-resistant gene number among all the STs (Figure S4A), suggesting the potential threat of ST27 K. michiganensis strains to public health. This phenomenon should be closely monitored. Incidentally, we also compared the number of ARGs between human and animal. The results showed that human-derived isolates have a greater number of ARGs than animal-derived ones (P < 0.05; Wilcoxon rank test) (Figure S4B), which was consistent with previous studies (Brisse and Duijkeren, 2005).
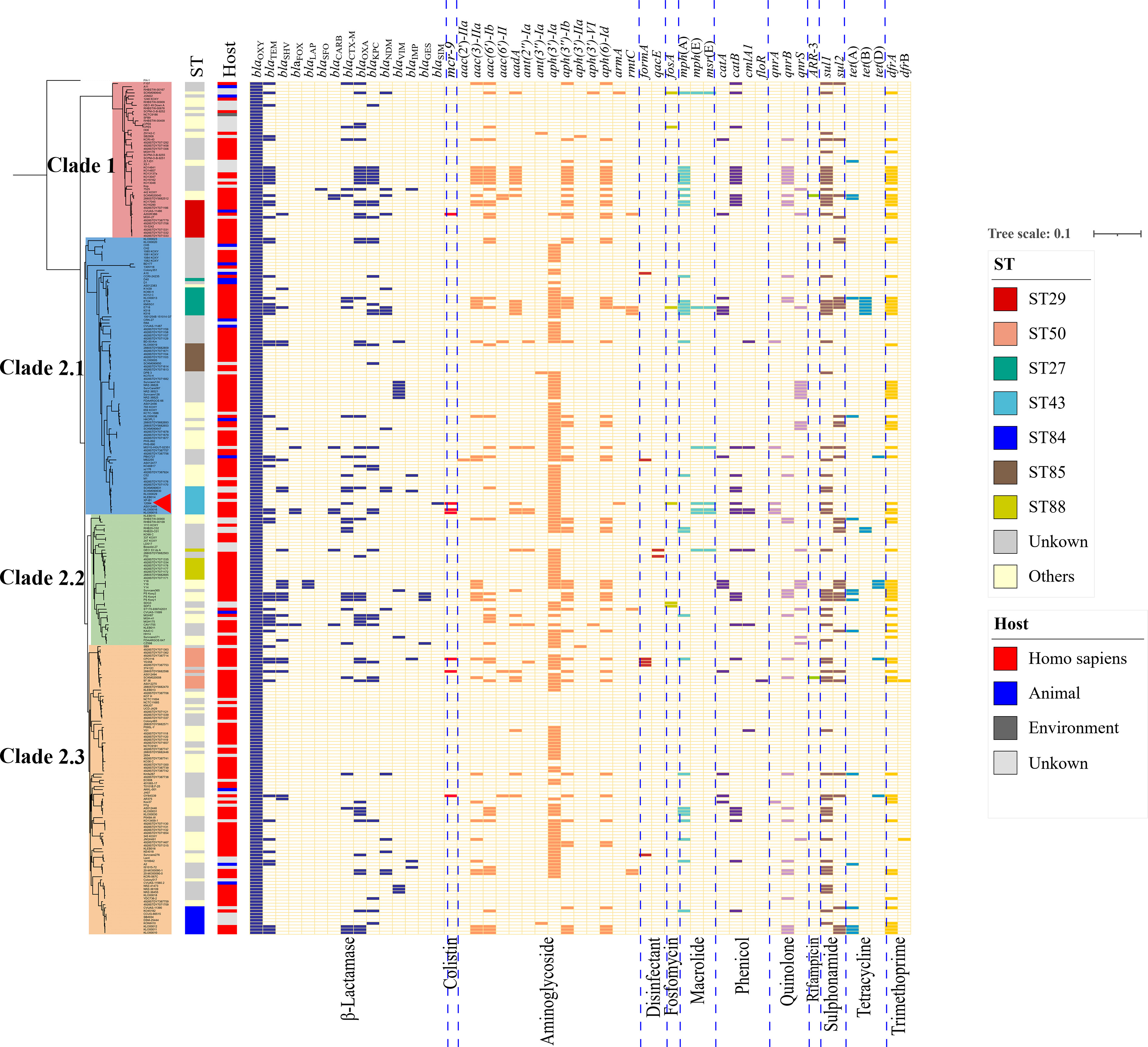
Figure 5 The maximum-likelihood tree and ARGs distributions in K. michiganensis. The matrix of ARGs is present for all K. michiganensis genomes against the dendrogram. The top of the matrix indicates the names of ARGs, while the bottom of that represents the groups of ARGs. The colored cells represent the “presence” of the ARGs, and the cells are colored based on the groups of ARGs. The strain 12084 is marked with a red triangle.
Importantly, we further focused on carbapenem-resistant genes, since carbapenem resistance was a big problem in the treatment of Klebsiella strains (Hayden and Won, 2018). Our research showed that 25.5% of K. michiganensis strains (70/275) carried at least one carbapenemase gene, indicating severe carbapenem-resistance in K. michiganensis strains throughout the world; this is a dangerous tendency and should be closely monitored. Here, the 70 carbapenem-resistant K. michiganensis strains contained 34 blaKPC genes, 17 blaNDM genes, 12 blaVIM genes, 6 blaIMP genes, 4 blaGES genes, and one blaSIM gene (Figure 5). Further analysis demonstrated that blaKPC preferred to be carried by ST27 (3, top-1), ST29 (3, top-1), and ST50 (3, top-1) strains, and blaNDM preferred to be harbored by ST27 (4, top-1) strains (Table S6). These results also indicated the potential threat of ST27 K. michiganensis strains, and therefore constant surveillance for the dissemination of ST27 strains is warranted. In addition, it was worth noting that only one blaSIM was identified in our strain 12084, indicating the emergence of the blaSIM-carrying K. michiganensis strain, which should be closely monitored.
In summary, our findings provide important references and improved understanding of K. michiganensis. We first described the complete genome of a clinical MDR K. michiganensis strain 12084 with two blaSIM-1 and one mcr-9.2 genes obtained from a Kidney transplant patient in China without a travel history. To the best of our knowledge, this is the first report of a K. michiganensis isolate with blaSIM-1 and mcr-9.2 genes. We further revealed the antibiotic resistance genes, population structure/classification, and antibiotic resistance of K. michiganensis for the first time, and provided an important framework for K. michiganensis. The emergence of resistant K. michiganensis strains, especially for those MDR strains harboring carbapenemase genes or/and mcr, poses serious threats to public health, which is a dangerous trend and should be closely monitored.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
FC, ZY, and DZ conceived the study and designed experimental procedures. XJ, WZ, FZC, and LY performed the experiments. SL, XJ, CL, YJ, and LH analyzed the data. JW, XH, BT, and HW contributed reagents and materials. FC, SL, XJ, and CL wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by Beijing Natural Science Foundation (Grant No. M21009), National Natural Science Foundation of China (NSFC) (Grant No. 31770870), Funds for International Cooperation and Exchange of the National Natural Science Foundation of China (Grant No. 32061143024).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.973901/full#supplementary-material
Supplementary Figure 1 | Pairwise average nucleotide identity (ANI) cluster map of all the downloaded genomes with Klebsiella reference strains. Shown is the hierarchical clustering and heat map of pairwise ANI values among all strains. Their final species determination (K. michiganensis, K. grimontii, and K. pasteurii) are shown as colored bars adjacent to the heat map. The strain12084 is marked with a red triangle.
Supplementary Figure 2 | Linear comparison of p12084-HI2 and p12084-HI5 with reference plasmids (A) Linear comparison of p12084-HI2 with the IncHI2 reference plasmid R478. Genes are denoted by arrows. Genes, mobile elements, and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity). (B) Linear comparison of p12084-HI5 with the IncHI5 reference pA324-IMP. Genes are denoted by arrows. Genes, mobile elements, and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity).
Supplementary Figure 3 | PCR amplifications of blaSIM, repHI2A, and repHI5B for 12084-SIM-EC600. 12084-SIM-EC600: the transconjugants. 12084 (the donor) and EC600 (the recipient) were used as the positive and negative control, respectively. repHI2A and repHI5B were the replication gene of p12084-HI2 and p12084-HI5, respectively. The 16S rDNA gene was used to identify the bacterial species of 12084-SIM-EC600.
Supplementary Figure 4 | The ARG numbers in different STs/host isolates. (A). Boxplot showed the top five ARG numbers in different STs isolates. The numbers on the y-axis represent the number of ARGs. (B). Boxplot showed the numbers of ARGs in different host isolates. The numbers on the y-axis represent the number of ARGs. The P values were obtained using the Wilcoxon rank test. **, statistically significant with P < 0.01.
Bean, D. C., Wareham, D. W. (2022). Draft genome sequence of a multidrug-resistant pseudomonas aeruginosa producing blaSIM metallo-β-lactamase: London, UK. J. Glob. Antimicrob. Resist. 29, 222–224. doi: 10.1016/j.jgar.2022.03.022
Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., Ma, N., et al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41 (Web Server issue), W29–W33. doi: 10.1093/nar/gkt282
Boutet, E., Lieberherr, D., Tognolli, M., Schneider, M., Bansal, P., Bridge, A. J., et al. (2016). UniProtKB/Swiss-prot, the manually annotated section of the uniprot knowledgebase: how to use the entry view. Methods Mol. Biol. 1374, 23–54. doi: 10.1007/978-1-4939-3167-5_2
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5, 8365. doi: 10.1038/srep08365
Brisse, S., Duijkeren, E. (2005). Identification and antimicrobial susceptibility of 100 klebsiella animal clinical isolates. Vet. Microbiol. 105 (3-4), 307–312. doi: 10.1016/j.vetmic.2004.11.010
Campos-Madueno, E. I., Moser, A. I., Risch, M., Bodmer, T., Endimiani, A. (2021). Exploring the global spread of klebsiella grimontii isolates possessing blaVIM-1 and mcr-9. Antimicrob. Agents Chemother. 65 (9), e0072421. doi: 10.1128/aac.00724-21
Chapman, P., Forde, B. M., Roberts, L. W., Bergh, H., Vesey, D., Jennison, A. V., et al. (2020). Genomic investigation reveals contaminated detergent as the source of an extended-Spectrum-β-Lactamase-Producing klebsiella michiganensis outbreak in a neonatal unit. J. Clin. Microbiol. 58 (5), e01980–19. doi: 10.1128/jcm.01980-19
Chavda, K. D., Westblade, L. F., Satlin, M. J., Hemmert, A. C., Castanheira, M., Jenkins, S. G., et al. (2019). First report of blaVIM-4- and mcr-9-Coharboring enterobacter species isolated from a pediatric patient. mSphere 4 (5), e00629–19. doi: 10.1128/mSphere.00629-19
CLSI. (2020). Performance standards for antimicrobial susceptibility testing. 30th ed. CLSI supplement M100. (Wayne, PA: Clinical and Laboratory Standards Institute.)
De Oliveira, D. M. P., Forde, B. M., Kidd, T. J., Harris, P. N. A., Schembri, M. A., Beatson, S. A., et al. (2020). Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 33 (3), e00181–19. doi: 10.1128/cmr.00181-19
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 (5), 1792–1797. doi: 10.1093/nar/gkh340
Gilmour, M. W., Thomson, N. R., Sanders, M., Parkhill, J., Taylor, D. E. (2004). The complete nucleotide sequence of the resistance plasmid R478: defining the backbone components of incompatibility group h conjugative plasmids through comparative genomics. Plasmid 52 (3), 182–202. doi: 10.1016/j.plasmid.2004.06.006
Hayden, M. K., Won, S. Y. (2018). Carbapenem-sparing therapy for extended-spectrum β-Lactamase-Producing e coli and klebsiella pneumoniae bloodstream infection: The search continues. Jama 320 (10), 979–981. doi: 10.1001/jama.2018.12565
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45 (D1), D566–D573. doi: 10.1093/nar/gkw1004
Kananizadeh, P., Oshiro, S., Watanabe, S., Iwata, S., Kuwahara-Arai, K., Shimojima, M., et al. (2020). Emergence of carbapenem-resistant and colistin-susceptible enterobacter cloacae complex co-harboring blaIMP-1 and mcr-9 in Japan. BMC Infect. Dis. 20 (1), 282. doi: 10.1186/s12879-020-05021-7
Kaye, K. S., Pogue, J. M., Tran, T. B., Nation, R. L., Li, J. (2016). Agents of last resort: Polymyxin resistance. Infect. Dis. Clin. North Am. 30 (2), 391–414. doi: 10.1016/j.idc.2016.02.005
Khalifa, H. O., Soliman, A. M., Saito, T., Kayama, S., Yu, L., Hisatsune, J., et al. (2020). First report of foodborne klebsiella pneumoniae coharboring blaVIM-1, blaNDM-1, and mcr-9. Antimicrob. Agents Chemother. 64 (9), e00882–20. doi: 10.1128/aac.00882-20
Lee, K., Yum, J. H., Yong, D., Lee, H. M., Kim, H. D., Docquier, J. D., et al. (2005). Novel acquired metallo-beta-lactamase gene, blaSIM-1, in a class 1 integron from acinetobacter baumannii clinical isolates from Korea. Antimicrob. Agents Chemother. 49 (11), 4485–4491. doi: 10.1128/aac.49.11.4485-4491.2005
Liang, Q., Jiang, X., Hu, L., Yin, Z., Gao, B., Zhao, Y., et al. (2019). Sequencing and genomic diversity analysis of IncHI5 plasmids. Front. Microbiol. 9 (3318). doi: 10.3389/fmicb.2018.03318
Li, X., He, J., Jiang, Y., Peng, M., Yu, Y., Fu, Y. (2021). Genetic characterization and passage instability of a hybrid plasmid Co-harboring blaIMP-4 and blaNDM-1 reveal the contribution of insertion sequences during plasmid formation and evolution. Microbiol. Spectr. 9 (3), e0157721. doi: 10.1128/Spectrum.01577-21
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16 (2), 161–168. doi: 10.1016/s1473-3099(15)00424-7
Marçais, G., Delcher, A. L., Phillippy, A. M., Coston, R., Salzberg, S. L., Zimin, A. (2018). MUMmer4: A fast and versatile genome alignment system. PLoS Comput. Biol. 14 (1), e1005944. doi: 10.1371/journal.pcbi.1005944
Mediavilla, J. R., Patrawalla, A., Chen, L., Chavda, K. D., Mathema, B., Vinnard, C., et al. (2016). Colistin- and carbapenem-resistant escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the united states. mBio 7 (4), e01191–16. doi: 10.1128/mBio.01191-16
Mendes, A. C., Novais, Â., Campos, J., Rodrigues, C., Santos, C., Antunes, P., et al. (2018) Mcr-1 in carbapenemase-producing klebsiella pneumoniae with hospitalized patients, portugal 2016-2017 Emerg. Infect. Dis. 24 (4), 762–766. doi: 10.3201/eid2404.171787
Merla, C., Rodrigues, C., Passet, V., Corbella, M., Thorpe, H. A., Kallonen, T. V. S., et al. (2019). Description of klebsiella spallanzanii sp. nov. and of klebsiella pasteurii sp. nov. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02360
Moura, A., Soares, M., Pereira, C., Leitao, N., Henriques, I., Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25 (8), 1096–1098. doi: 10.1093/bioinformatics/btp105
O’Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44 (D1), D733–D745. doi: 10.1093/nar/gkv1189
Palzkill, T. (2013). Metallo-β-lactamase structure and function. Ann. N. Y. Acad. Sci. 1277, 91–104. doi: 10.1111/j.1749-6632.2012.06796.x
Partridge, S. R., Brown, H. J., Stokes, H. W., Hall, R. M. (2001). Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45 (4), 1263–1270. doi: 10.1128/aac.45.4.1263-1270.2001
Partridge, S. R., Hall, R. M. (2003). The IS1111 family members IS4321 and IS5075 have subterminal inverted repeats and target the terminal inverted repeats of Tn21 family transposons. J. Bacteriol. 185 (21), 6371–6384. doi: 10.1128/jb.185.21.6371-6384.2003
Poirel, L., Jayol, A., Nordmann, P. (2017). Polymyxins: Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30 (2), 557–596. doi: 10.1128/cmr.00064-16
Price, M. N., Dehal, P. S., Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26 (7), 1641–1650. doi: 10.1093/molbev/msp077
Richter, M., Rossello-Mora, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106 (45), 19126–19131. doi: 10.1073/pnas.0906412106
Roberts, A. P., Chandler, M., Courvalin, P., Guedon, G., Mullany, P., Pembroke, T., et al. (2008). Revised nomenclature for transposable genetic elements. Plasmid 60 (3), 167–173. doi: 10.1016/j.plasmid.2008.08.001
Rodríguez-Martínez, J. M., Cano, M. E., Velasco, C., Martínez-Martínez, L., Pascual, A. (2011). Plasmid-mediated quinolone resistance: an update. J. Infect. Chemother. 17 (2), 149–182. doi: 10.1007/s10156-010-0120-2
Saha, R., Farrance, C. E., Verghese, B., Hong, S., Donofrio, R. S. (2013). Klebsiella michiganensis sp. nov., a new bacterium isolated from a tooth brush holder. Curr. Microbiol. 66 (1), 72–78. doi: 10.1007/s00284-012-0245-x
Sajjad, A., Holley, M. P., Labbate, M., Stokes, H. W., Gillings, M. R. (2011). Preclinical class 1 integron with a complete Tn402-like transposition module. Appl. Environ. Microbiol. 77 (1), 335–337. doi: 10.1128/aem.02142-10
Sands, K., Carvalho, M. J., Portal, E., Thomson, K., Dyer, C., Akpulu, C., et al. (2021). Characterization of antimicrobial-resistant gram-negative bacteria that cause neonatal sepsis in seven low- and middle-income countries. Nat. Microbiol. 6 (4), 512–523. doi: 10.1038/s41564-021-00870-7
Saravolatz, L. D., Stein, G. E. (2020). Plazomicin: A new aminoglycoside. Clin. Infect. Dis. 70 (4), 704–709. doi: 10.1093/cid/ciz640
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34 (Database issue), D32–D36. doi: 10.1093/nar/gkj014
Toleman, M. A., Bennett, P. M., Walsh, T. R. (2006). ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70 (2), 296–316. doi: 10.1128/mmbr.00048-05
Tzouvelekis, L. S., Markogiannakis, A., Psichogiou, M., Tassios, P. T., Daikos, G. L. (2012). Carbapenemases in klebsiella pneumoniae and other enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25 (4), 682–707. doi: 10.1128/cmr.05035-11
Wang, C., Feng, Y., Liu, L., Wei, L., Kang, M., Zong, Z. (2020). Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 9 (1), 508–516. doi: 10.1080/22221751.2020.1732231
Wang, X., Wang, Y., Zhou, Y., Li, J., Yin, W., Wang, S., et al. (2018). Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing klebsiella pneumoniae. Emerg. Microbes Infect. 7 (1), 122. doi: 10.1038/s41426-018-0124-z
Wyres, K. L., Lam, M. M. C., Holt, K. E. (2020). Population genomics of klebsiella pneumoniae. Nat. Rev. Microbiol. 18 (6), 344–359. doi: 10.1038/s41579-019-0315-1
Xu, L., Sun, X., Ma, X. (2017). Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant klebsiella pneumoniae. Ann. Clin. Microbiol. Antimicrob. 16 (1), 18. doi: 10.1186/s12941-017-0191-3
Yuan, Y., Li, Y., Wang, G., Li, C., Xiang, L., She, J., et al. (2019). Coproduction of MCR-9 and NDM-1 by colistin-resistant enterobacter hormaechei isolated from bloodstream infection. Infect. Drug Resist. 12, 2979–2985. doi: 10.2147/idr.S217168
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67 (11), 2640–2644. doi: 10.1093/jac/dks261
Keywords: Klebsiella michiganensis, drug resistance, carbapenemase, mobile colistin resistance (mcr), mobile elements, plasmid, blaSIM-1, antibiotics
Citation: Li S, Jiang X, Li C, Ju Y, Yue L, Chen F, Hu L, Wang J, Hu X, Tuohetaerbaike B, Wen H, Zhang W, Zhou D, Yin Z and Chen F (2022) A blaSIM-1 and mcr-9.2 harboring Klebsiella michiganensis strain reported and genomic characteristics of Klebsiella michiganensis. Front. Cell. Infect. Microbiol. 12:973901. doi: 10.3389/fcimb.2022.973901
Received: 20 June 2022; Accepted: 08 August 2022;
Published: 24 August 2022.
Edited by:
Ruichao Li, Yangzhou University, ChinaReviewed by:
Xuemei Yang, City University of Hong Kong, Hong Kong, SAR ChinaCopyright © 2022 Li, Jiang, Li, Ju, Yue, Chen, Hu, Wang, Hu, Tuohetaerbaike, Wen, Zhang, Zhou, Yin and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhe Yin, jerry9yin@163.com; Fei Chen, chenfei@big.ac.cn
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.