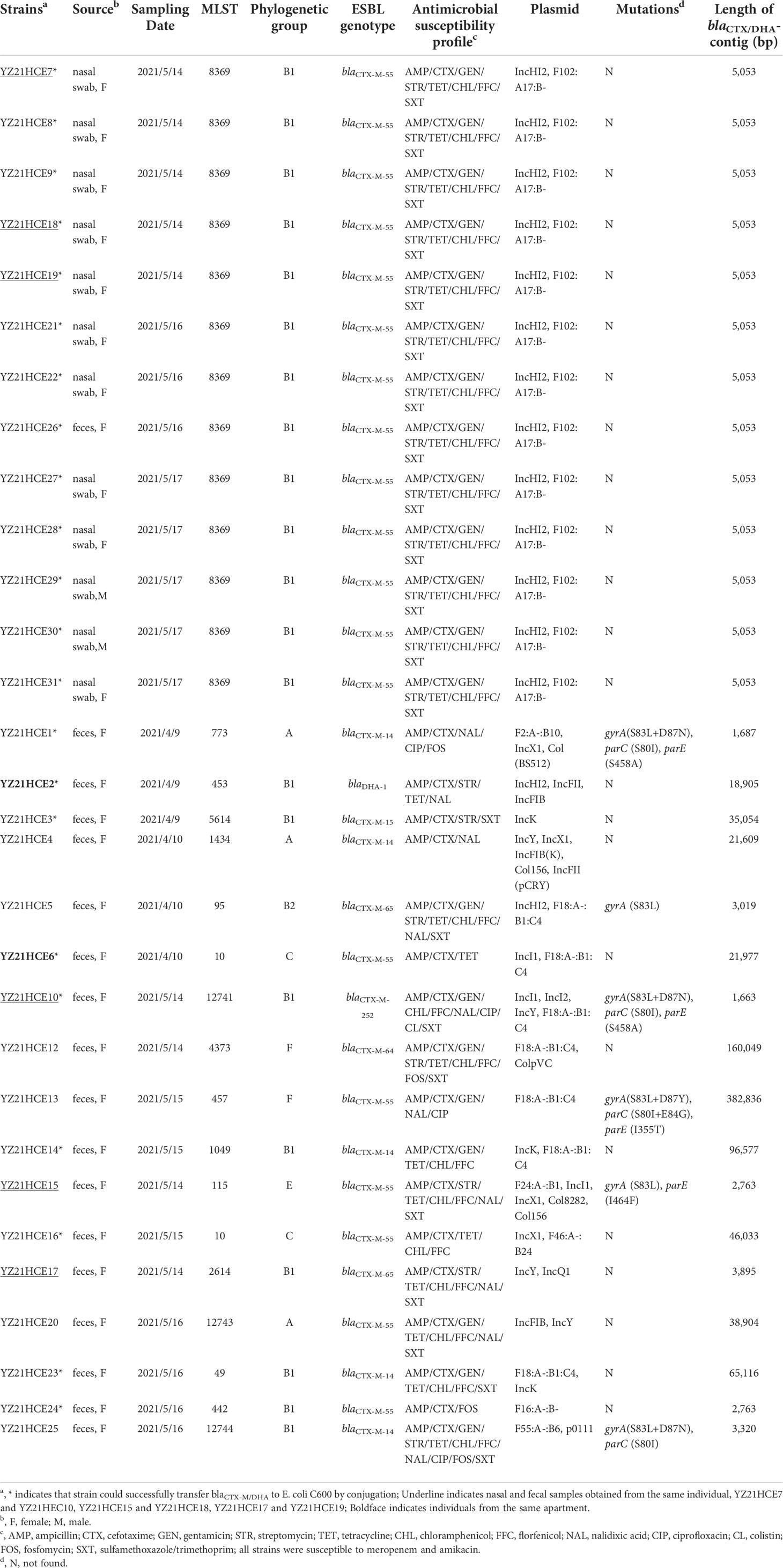
- 1Jiangsu Key Laboratory of Zoonosis, Jiangsu Co-Innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou University, Yangzhou, China
- 2Key Laboratory of Prevention and Control of Biological Hazard Factors (Animal Origin) for Agrifood Safety and Quality, Ministry of Agriculture of China, Yangzhou University, Yangzhou, China
This study aimed to investigate the prevalence and diversity of extended-spectrum β-lactamases (ESBL)-producing Escherichia coli isolates from healthy individuals in a community and to elucidate their dissemination mechanism. Cefotaxime-resistant E. coli were isolated from 95 samples of healthy persons from one community in Yangzhou, China, and were tested for minimal inhibitory concentrations of 14 antimicrobial agents. The isolates were subjected to whole genome sequencing by Illumina Hiseq or PacBio single-molecule real-time sequencing. A total of 30 cefotaxime-resistant E. coli isolates were obtained, carrying blaCTX-M (n=29) or blaDHA (n=1), of which the blaCTX-M-55 (n=19) was the most predominant genotype. One novel blaCTX-M variant blaCTX-M-252 was identified. Thirteen CTX-M-55-producing E. coli isolates belonged to ST8369 from nasal (n=12) or faecal (n=1) samples shared the identical cgMLST type, resistance profiles, resistance genes, plasmid replicons, and a 5,053-bp blaCTX-M-55 structure ΔIS26-ΔISEcp1-blaCTX-M-55-Δorf477-ΔTn2. The blaCTX-M-55 gene was located on IncHI2/ST3 plasmid in E. coli ST8369. The lengths of blaCTX-M/blaDHA-carrying contigs in the remaining 17 E. coli strains ranged from 1,663 to 382,836 bp, located on chromosome (n=4) or plasmids (n=5); the location of the other eight contigs could not be determined due to incomplete assembly. The blaCTX-M was associated with ISEcp1 as previously reported. Nasal colonization of CTX-M-55-producing ST8369 E. coli strains has occurred among healthy individuals in one community. There is a potential risk of antimicrobial resistance dissemination between humans within one community through close contact or environment via aerosols or dust. Therefore, surveillance of nasal carriage of blaCTX-M in communities is warranted to further monitor the spread of the antimicrobial resistance genes in China.
Introduction
Extended-spectrum cephalosporins are widely used in human clinics and veterinary medicine to treat infections caused by multidrug-resistant Gram-negative bacteria; thus, extended-spectrum β-lactamases (ESBL), particularly CTX-M enzymes, have been increasingly reported in human clinical settings and animals worldwide (Bevan et al., 2017). Globally, incidence of CTX-M ESBLs is increasing, blaCTX-M-15 and blaCTX-M-14 are the predominant genotypes detected in many parts of the world (Woerther et al., 2013; Bevan et al., 2017). The global dissemination of blaCTX-M is mainly due to the rapid horizontal transfer mediated by conjugative plasmids; the epidemic plasmids such as IncF, IncI, and IncHI2 facilitate the global spread of blaCTX-M in Enterobacteriaceae from humans, animals and the environment, particularly in Escherichia coli (Bevan et al., 2017; Rozwandowicz et al., 2018; Partridge et al., 2018). Mobile elements such as ISEcp1, IS26, and ISCR1 have also played an essential role in the blaCTX-M transmission (Bevan et al., 2017; Partridge et al., 2018). In addition, some successful E. coli clones, such as the E. coli clone ST131 lineage diffused worldwide, are also responsible for blaCTX-M-15 global dissemination, mostly in human clinics (Bevan et al., 2017).
To date, many studies have focused on CTX-M-producing Enterobacteriaceae from clinical patients. However, the high prevalence of CTX-M-producing E. coli colonizing the intestinal tract of healthy persons in communities is of particular concern, since they could be a major reservoir of blaCTX-M (Woerther et al., 2013; Bevan et al., 2017; Chen et al., 2021). In this study, we aimed to investigate the prevalence and characterization of extended-spectrum β-lactamases (ESBL)-producing E. coli isolates from healthy individuals in a community from Yangzhou, China, to elucidate their dissemination mechanism within this small-scale community.
Materials and methods
Sample collection and antimicrobial susceptibility testing
From April 9th to May 17th 2021, 58 fecal samples and 37 nasal swabs of 72 healthy volunteers (3 male and 69 female) with no obvious disease symptoms at the age of 15-46 were collected from 37 apartments located in three buildings in one community in Yangzhou, China. Participants had been exposed to antimicrobial agents in the three months prior to sample collection were excluded from this study. This small-scale community with approximately 2000 individuals located in the urban area of Yangzhou, and included three main areas, and one building was randomly selected to sample in each area (Supplementary Figure S1). Individual written informed consent for samples was obtained from all volunteers. Samples were incubated in LB broth (OXOID, Basingstoke, UK) for 18~24 h and then cultured on the MacConkey agar (Haibo, Qingdao, China) with 2 mg/L cefotaxime. One E. coli isolate per plate was selected and identified by 16S rRNA gene sequencing using PCR and Sanger sequencing (Kim et al., 2010). The cefotaxime-resistant E. coli isolates were tested susceptibility to 14 antimicrobial agents including ampicillin, cefotaxime, meropenem, gentamicin, amikacin, streptomycin, tetracycline, chloramphenicol, florfenicol, nalidixic acid, ciprofloxacin, colistin, fosfomycin, and sulfamethoxazole/trimethoprim by using the agar dilution or broth microdilution method (limited to colistin). The results were interpreted according to Clinical Laboratory Standards Institute (CLSI) M100, 30th edition (CLSI, 2020). E. coli ATCC 25922 was used as the quality control strain.
Whole genome sequencing and analysis
All cefotaxime-resistant E. coli isolates were sequenced by Illumina Hiseq. The library was constructed using NEB NEXT Ultra DNA Library Prep Kit for Illumina (New England Biolabs, USA) and 150 bp paired-end reads were obtained. For each E. coli isolate performed WGS, at least 100-fold coverage of raw reads was collected. The 150 bp pair-end raw reads were trimmed and filtered by the NGSQC toolkit 2.3.3, then were assembled into contigs using SPAdes 3.8.2 (Bankevich et al., 2012). One representative ST8369 E. coli isolate YZ21HCE18 was sequenced using PacBio single-molecule real-time sequencing. The phylogenetic groups of E. coli were confirmed according to previously described protocol by using assembled contigs (Clermont et al., 2000). The genomes were subjected to analysis of multilocus sequence typing (MLST), core genome multilocus sequencing typing (cgMLST), resistance genes, mutations and plasmids by using the Center for Genomic Epidemiology (CGE) pipelines (http://www.genomicepidemiology/org/). The phylogenetic tree of these isolates was constructed using Parsnp (https://harvest.readthedocs.io/en/latest/content/parsnp.html) and visualized by iTOL (Letunic and Bork, 2016). The blaCTX-M/blaDHA-carrying contigs were retrieved from the draft genomes and analyzed by ISfinder (https://www-is.biotoul.fr/) and BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The blaCTX-M-bearing plasmid pYUYZ18-1 in strain YZ21HCE18 was compared with other ST8369 E. coli isolates using BRIG.
Conjugation assay
The transferability of cefotaxime resistance was determined using conjugation experiments as previously described (Chen et al., 2007) and streptomycin-resistant E. coli C600 as the recipient. Transconjugants were selected using 2 mg/L cefotaxime and 3,000 mg/L streptomycin, and were confirmed by detecting blaCTX-M or blaDHA-1 using PCR and sequencing (Chen et al., 2004; Liu et al., 2007).
Results
Prevalence and genotype distribution of cefotaxime-resistant E. coli
Thirty cefotaxime-resistant E. coli isolates were obtained from 12 nasal swabs and 18 fecal samples from 27 individuals (Table 1). The blaCTX-M-55 (n=19) gene was identified as the most predominant genotype, followed by blaCTX-M-14 (n=5), blaCTX-M-65 (n=2), blaCTX-M-15 (n=1), blaCTX-M-64 (n=1), and blaDHA-1 (n=1) (Table 1). One novel blaCTX-M variant blaCTX-M-252 (GenBank accession no. OL884447) was identified, and differed from blaCTX-M-65 by a single nucleotide resulting in one amino acid change (A32V).
Characterization of cefotaxime-resistant E. coli isolates
All ESBL-producing isolates exhibited an MIC of 8 to >128 mg/L to cefotaxime, and also showed resistance to multiple antibiotics, but were susceptible to meropenem and amikacin (Table 1). Twenty-two of them successfully transfer cefotaxime resistance to E. coli C600 by conjugation (Table 1). In addition to blaCTX-M/blaDHA, they carried one to 18 resistance genes, such as blaTEM, tet(A), floR, qnrS1, fosA3, and mcr-1 (Figure 1); mutations within gyrA (S83L, D87N/Y), parC (S80I), or parE (I355T, S458A) were observed in six of them (Table 1). Phylogenetic group analysis showed that group B1 was predominant (21; 70%), which was frequently associated with commensal or intestinal pathogenic strains (Clermont et al., 2000); followed by group A (3; 10%), group C (2; 6.67%), group F (2; 6.67%) and group E (1; 3.33%) (Table 1). Only one CTX-M-65-producing isolate belonged to extraintestinal virulent group B2.
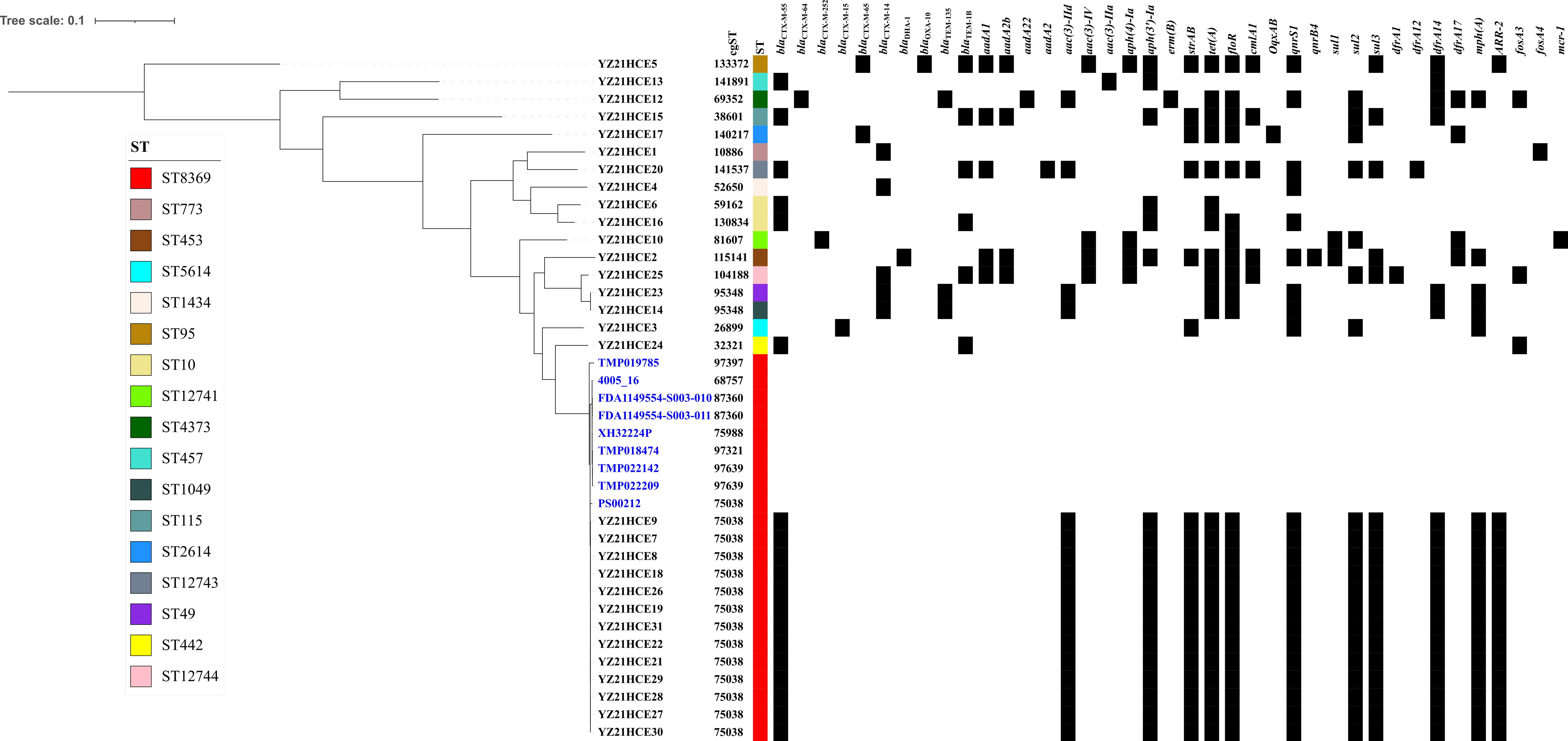
Figure 1 The maximum likelihood tree of cefotaxime-resistant E. coli isolates in this study compared with E. coli ST8369 isolates from EnteroBase (https://enterobase.warwick.ac.uk/) (in blue) based on cgSNP analysis. Antibiotic resistance genes with >90% sequence homology and coverage are shown.
Thirty ESBL-producing isolates were assigned to 14 known STs and three novel STs (ST12741, ST12743 and ST12744) (Figure 1). The most prevalent STs among them were ST8369 (n=13). So far, only nine ST8369 E. coli isolates were retrieved from EnteroBase (https://enterobase.warwick.ac.uk/) originating from humans, wild animals, and the environment (Table S1), and none of them carried resistance genes or mutations associated with quinolone resistance. To reveal the genetic differences between 22 E. coli ST8369 isolates, we analyzed their cgMLST profiles (cgSTs) based on 2513 alleles (Zhou et al., 2020). Among the identified seven cgSTs, cgST 75038 (n=14) was the dominant type shared by 13 strains in our study and one isolate PS00212 (Figure 1). Thirteen E. coli ST8369 strains from nasal (n=12) or fecal (n=1) samples of different individuals in this study shared the same cgST, resistance profiles, resistance genes, and plasmid replicons (Figure 1 and Table 1), indicating that there is a reservoir of this lineage in the community.
The genetic structures of blaCTX-M/blaDHA in 30 ESBL-producing E. coli isolates
The lengths of blaCTX-M/blaDHA-carrying contigs ranged from 1,663 to 382,836 bp, located on chromosome (n=4) or plasmids (n=5). Twenty-one contigs were short (1,663 to 5,053 bp) due to incomplete assembly and the high number of insertion elements; they did not have replicon genes or plasmid backbone; thus, it is difficult to determine their location (Table 1; Figure 2).
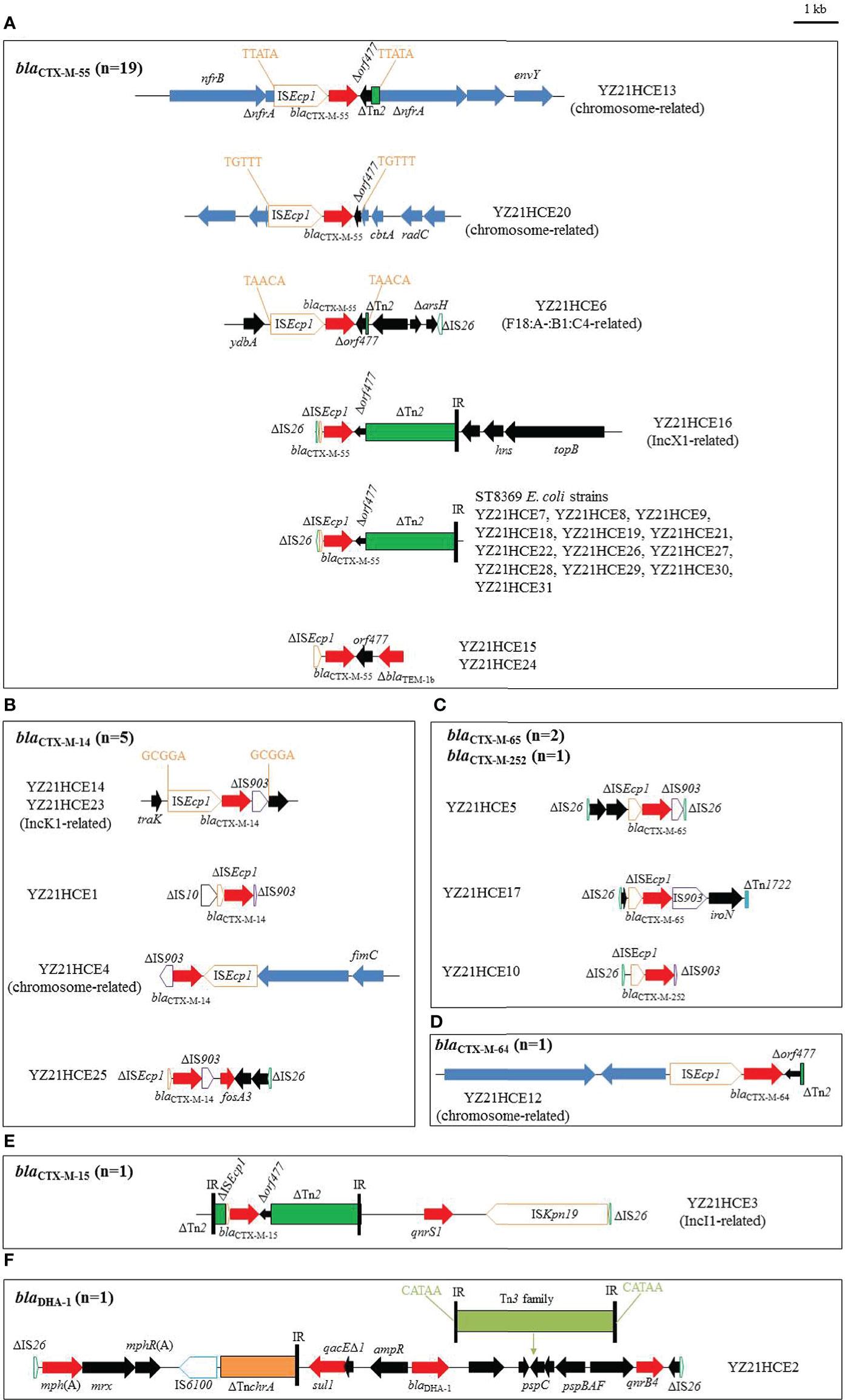
Figure 2 The genetic environments of blaCTX-M/blaDHA in 30 E. coli isolates in this study. (A) blaCTX-M-55; (B) blaCTX-M-14; (C) blaCTX-M-65/-252; (D) blaCTX-M-64; (E) blaCTX-M-15; (F) blaDHA. The extents and directions of antibiotic resistance (red arrows) and other genes (black arrows) are indicated. The blue arrows indicate chromosomal genes. ISs are shown as boxes labeled with their name. Tall bars represent the inverted repeats (IR) of transposon. Δ indicates a truncated gene or mobile element. Arrows and sequences indicate direct repeats.
In YZ21HCE13, the 3,241-bp transposition unit (ISEcp1-blaCTX-M-55-Δorf477-ΔTn2) was inserted into the chromosome and generated 5-bp direct repeats (DRs) (Figure 2A). A similar insertion (ISEcp1-blaCTX-M-55-Δorf477) with a different location had occurred in the chromosome of YZ21HCE20. Similarly, a 3,050-bp segment (ISEcp1-blaCTX-M-55-Δorf477-ΔTn2) with DRs (5’-TAACA-3’) was inserted into the plasmid backbone in YZ21HCE6 (Figure 2A), this blaCTX-M-55-bearing contig (21,977 bp) was identical to those of F18:A-:B1:C4 plasmids, such as pTREC1 (E. coli, MN158989) (Figure S2). In YZ21HCE16, the blaCTX-M-55-carrying contig was 46,033 bp in size and similar to IncX1 plasmids such as p40EC-5 (CP070925) and pPK8277-49kb (CP080137) (Figure S2). blaCTX-M-55 was associated with the commonly observed structure ΔIS26-ΔISEcp1-blaCTX-M-55-Δorf477-ΔTn2, and three additional resistance genes aph(3’)-Ia, qnrS1 and tet(A) were co-located in this contig (Figure S2). The 2,763-bp blaCTX-M-55 region in isolates YZ21HCE15 and YZ21HCE24 were identical, including the typical transposition unit (ΔISEcp1-blaCTX-M-55-orf477) and a truncated blaTEM-1b downstream, which was commonly observed in many blaCTX-M-55-carrying plasmids, e.g., pHNMC02 (MG197489). All ST8369 E. coli isolates (n=13) had identical 5,053-bp blaCTX-M-55-positive contigs with structure ΔIS26-ΔISEcp1-blaCTX-M-55-Δorf477-ΔTn2, except strain YZ21HCE29 (one nucleotide change). The complete sequence of YZ21HCE18 as representative ST8369 E. coli was obtained (Table S2). The blaCTX-M-55 gene was located on plasmid pYUYZ18-1 with a size of 248,665 bp, belonged to IncHI2/ST3 plasmid with a similar organization to other IncHI2 plasmids. In addition to blaCTX-M-55, pYUYZ18-1 contained numerous resistance genes, including aac(3)-IId, aph(3’)-Ia, strAB, tet(A), mph(A), qnrS1, floR, sul2, sul3, dfrA14, and arr-2. Similar pYUYZ18-1 plasmids were also present in other ST8369 E. coli strains in this study (Figure S3).
Two blaCTX-M-14-positive contigs from isolates YZ21HCE14 and YZ21HCE23 displayed >99.9% similarity to IncK1 plasmids pD16EC0206-1 (E. coli, CP088610) and pJX1-2 (Klebsiella pneumoniae, CP064254) (Figure S2). The 3,060-bp blaCTX-M-14 transposition unit (ISEcp1-blaCTX-M-14-ΔIS903) with 5-bp DRs (5’-GCGGA-3’) was inserted downstream of the plasmid conjugal transfer gene traK (Figure 2B). In isolates YZ21HCE1, YZ21HCE4, and YZ21HCE25, a similar blaCTX-M-14 transposition unit was observed, differed by deletions involving ISEcp1 and/or IS903. In YZ21HCE4, the 2,996-bp fragment (ISEcp1-blaCTX-M-14-ΔIS903) was embedded in a 21,609-bp contig associated with the chromosome of E. coli. The 1,972-bp fosA3 segment (fosA3-orf1-Δorf2-ΔIS26) was present downstream of IS903 in YZ21HCE25 (Figure 2B).
As observed in Figure 2C, the typical (ISEcp1-blaCTX-M-9G-IS903-iroN) transposition unit was found in isolates YZ21HCE5 (blaCTX-M-65), YZ21HCE17 (blaCTX-M-65), and YZ21HCE10 (blaCTX-M-252), although ISEcp1 was incomplete. In YZ21HCE17, the blaCTX-M-65 unit was inserted in an incomplete Tn1722. However, IS903 was truncated by IS26 at the 3’ end in YZ21HCE5, resulting in the deletions of iroN and 701 bp of IS903. Similarly, an incomplete IS903 (80-bp) was also observed in YZ21HCE10. A 1,205-bp structure consisting of two hypothetical proteins plus ΔIS26 (76-bp) was located upstream of blaCTX-M-65 transposition unit with a 64-bp spacer in YZ21HCE5. Remnants of this structure were also identified upstream of the blaCTX-M-65 unit in YZ21HCE17 (214 bp) and YZ21HCE10 (201 bp).
The blaCTX-M-64 was located on the chromosome of YZ21HCE12. The 160,049-bp blaCTX-M-64-carrying contig showed highly (>98.0%) similarity to the corresponding region of E. coli chromosome such as Z30 (CP066844) and LD67-1 (CP061185). Furthermore, a 3,045-bp region (ISEcp1-blaCTX-M-64-Δorf477-ΔTn2) in YZ21HCE12 was identical to those of chromosome of E. coli 3952 (MT773682) and plasmid pM-64-4467-1 (MT773679) from healthy humans in Hangzhou, China (Chen et al., 2021).
The blaCTX-M-15-positive contig (35,054 bp) in YZ21HCE3 was similar to IncI1 plasmid pBHBSTW-00321_3 (E. coli, CP056606) with 86% coverage and 98.36% identity (Figure S2). The blaCTX-M-15 gene was associated with genetic content ΔTn2-ΔISEcp1-blaCTX-M-15-Δorf477-ΔTn2, seen in several plasmids, such as p5908-2 (Shigella flexneri, CP045523) and p92 (E. coli, CP041521). The blaCTX-M-15 resistance module was followed by a 5,752-bp structure qnrS1-ISKpn19-ΔIS26 (Figure 2E).
The blaDHA-carrying contig (18,905 bp) of YZ21HCE2 was identical to the corresponding regions of multiple plasmids such as pM2901 (Shigella sonnei, CP061363), except for the insertion of one Tn3 family transposon within pspC flanked by 5-bp DRs in YZ21HCE2. The core structure sul1-qacEΔ1-ampR-blaDHA-1-pspDCBAF-qnrB4 is commonly observed in numerous plasmids from various species (e.g., Salmonella, Klebsiella pneumoniae, Citrobacter freundii), highlighting the co-transfer ability of blaDHA-1 and qnrB4.
Discussion
The blaCTX-M gene has been globally disseminated in different sources, with blaCTX-M-14 and blaCTX-M-15 being dominant (Bevan et al., 2017). In this study, blaCTX-M-55 is the most predominant genotype in healthy individuals, which agrees with the increasing prevalence of blaCTX-M-55 in both animals and patients in China (Bevan et al., 2017). As a variant of blaCTX-M-15, blaCTX-M-55 was first reported in clinical E. coli and K. pneumoniae isolates in Thailand in 2007 (Kiratisin et al., 2007). Recently, blaCTX-M-55 has become the predominant CTX-M genotype in E. coli and Salmonella from food animals, food products, and patients in China (Rao et al., 2014; Zhang et al., 2014; Fu et al., 2020; Huang et al., 2020; Liu et al., 2022; Zeng et al., 2022). One novel blaCTX-M variant blaCTX-M-252 was identified in this study. CTX-M-252 and CTX-M-65 differ by a single amino acid and share a similar genetic structure, suggesting a common lineage. Further investigation of blaCTX-M-252 is needed.
In communities, faecal carriage rates of blaCTX-M are increasing, particularly in developing countries (Woerther et al., 2013; Bevan et al., 2017). However, investigation of blaCTX-M in nasal samples of healthy humans is rare. Previously, one (1/77, 1.30%) CTX-M-producing E. coli isolate was obtained from the nasal sample of a healthy human working at a pig abattoir in Cameroon (Founou et al., 2018). A high nasal carriage rate (32.43%) of blaCTX-M was observed in this study, and nasal colonization of ST8369 E. coli producing CTX-M-55 among healthy persons occurred in one community in Yangzhou, China. It suggests that nasal carriage of blaCTX-M is possibly common in humans. However, the small number of samples and communities is a limitation of this study. The acquisition of blaCTX-M-55 and other resistance genes by ST8369 is mediated by the horizontal transfer of IncHI2 plasmid, followed by clonal dissemination. E. coli ST8369 is rarely described worldwide and may represent an emerging clone in humans, animals, and the environment. Nasal colonization of blaCTX-M-55-carrying ST8369 E. coli suggests a potential risk of antimicrobial resistance dissemination between humans by the spread of clonal lineages in the small-scale community through close contact or environment via aerosols or dust. Therefore, the clinical importance of nasal carriage of CTX-M-producing E. coli might be underestimated. Although horizontal transfer is the main reason for blaCTX-M dissemination, clonal spread of blaCTX-M-harbouring strains, such as E. coli ST8369 in this study, CTX-M-15-producing E. coli ST949 in water surfaces, E. coli ST2179 encoding CTX-M-65 in retail meat, and blaCTX-M-55-carrying Salmonella Typhimurium ST34 in patients (Bevan et al., 2017; Falgenhauer et al., 2021; Leão et al., 2021; Zeng et al., 2022) is another important route for blaCTX-M transmission.
Horizontal transfer mediated by plasmids and mobile elements is responsible for the global spread of blaCTX-M (Bevan et al., 2017; Partridge et al., 2018). For example, IncI, IncFII, and IncHI2 plasmids facilitate the horizontal transmission of blaCTX-M in E. coli and Salmonella from various sources (Chen et al., 2021; Guo and Zhao, 2021; Yang et al., 2014; Nadimpalli et al., 2019; Zhang et al., 2021; Zeng et al., 2021). In this study, various plasmids such as IncHI2, IncK1, IncX1, and IncI1 were associated with blaCTX-M. Although we were not able to determine the location of blaCTX-M in some E. coli isolates in this study due to incomplete assembly, sequence analysis indicates that ISEcp1 plays an important role in blaCTX-M dissemination among E. coli isolates and facilitates the horizontal transfer of blaCTX-M from plasmids to chromosomes in distinct integration sites. The chromosomal integration of blaCTX-M is increasingly reported in E. coli, K. pneumoniae, Salmonella, Proteus mirabilis and some other species of Enterobacteriaceae with the help of mobile elements (Huang et al., 2017; He et al., 2017; Zeng et al., 2022; Yoon et al., 2022). Chromosomal integration of blaCTX-M seems to be an adaptive evolution in response to antimicrobial pressure (Yoon et al., 2022).
In conclusion, we report nasal colonization of CTX-M-55-producing E. coli ST8369 associated with IncHI2 plasmid in healthy individuals in one community from Yangzhou, China. Therefore, continued surveillance of nasal carriage of blaCTX-M in communities is warranted.
Accession Numbers
The sequences have been deposited in the GenBank under accession number: PRJNA819533.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving human participants were reviewed and approved by Yangzhou University. The participants provided their written informed consent to participate in this study.
Author contributions
JW and Q-CL conceived and designed the experiments, Z-YW, YJ, Y-QS, H-FL, and M-JL carried out the experiments. Z-YW and JW analyzed the data and wrote the manuscript. Q-CL and XJ revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (no. 31902319), the fifth phase of the “333 project” scientific research project in Jiangsu Province (no. BRA2020002), and Postgraduate Research &Practice Innovation Program of Jiangsu Province (Yangzhou University) (no. KYCX22_3536).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.970940/full#supplementary-material
References
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bevan, E. R., Jones, A. M., Hawkey, P. M. (2017). Global epidemiology of CTX-m β-lactamases: temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72, 2145–2155. doi: 10.1093/jac/dkx146
Chen, J., Chen, S., Jiang, Y., Zhang, R., Cai, J. (2021). Fecal carriage and genetic characterization of CTX-M-1/9/1-producing escherichia coli from healthy humans in hangzhou, China. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.616687
Chen, L., Chen, Z., Liu, J., Zeng, Z., Ma, J., Jiang, H. (2007). Emergence of RmtB methylase-producing escherichia coli and enterobacter cloacae isolates from pigs in China. J. Antimicrob. Chemother. 59, 880–885. doi: 10.1093/jac/dkm065
Chen, S., Zhao, S., White, D. G., Schroeder, C. M., Lu, R., Yang, H., et al. (2004). Characterization of multiple-antimicrobial-resistant salmonella serovars isolated from retail meats. Appl. Environ. Microbiol. 70 (1), 1–7. doi: 10.1128/AEM.70.1.1-7.2004
Clermont, O., Bonacorsi, S., Bingen, E. (2000). Rapid and simple determination of the escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66, 4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000
CLSI (2020). Performance standards for antimicrobial susceptibility testing. CLSI supplement M100, 30th ed (Wayne, PA: Clinical and Laboratory Standards Institute).
Falgenhauer, L., Zur Nieden, A., Harpel, S., Falgenhauer, J., Domann, E. (2021). Clonal CTX-M-15-producing escherichia coli ST-949 are present in German surface water. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.617349
Founou, L. L., Founou, R. C., Allam, M., Ismail, A., Essack, S. Y. (2018). Draft genome sequence of an extended-spectrum β-lactamase (CTX-M-15)-producing escherichia coli ST10 isolated from a nasal sample of an abattoir worker in Cameroon. J. Glob. Antimicrob. Resist. 14, 68–69. doi: 10.1016/j.jgar.2018.06.004
Fu, Y., Xu, X., Zhang, L., Xiong, Z., Ma, Y., Wei, Y., et al. (2020). Fourth generation cephalosporin resistance among salmonella enterica serovar enteritidis isolates in shanghai, China conferred by blaCTX-M-55 harboring plasmids. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00910
Guo, L., Zhao, Y. (2021). Global spread and molecular characterization of CTX-m-producing salmonella typhimurium isolates. Antibiotics. 10 (11), 1417. doi: 10.3390/antibiotics10111417
He, D., Liu, L., Guo, B., Wu, S., Chen, X., Wang, J., et al. (2017). Chromosomal location of the fosA3 and blaCTX-m genes in Proteus mirabilis and clonal spread of escherichia coli ST117 carrying fosA3-positive IncHI2/ST3 or F2:A-:B- plasmids in a chicken farm. Int. J. Antimicrob. Agents. 49 (4), 443–448. doi: 10.1016/j.ijantimicag.2016.12.009
Huang, W., Wang, G., Sebra, R., Zhuge, J., Yin, C., Aguero-Rosenfeld, M. E., et al. (2017). Emergence and evolution of multidrug-resistant klebsiella pneumoniae with both blaKPC and blaCTX-m integrated in the chromosome. Antimicrob. Agents Chemother. 61 (7), e00076–e00017. doi: 10.1128/AAC.00076-17
Huang, Y., Zeng, L., Doi, Y., Lv, L., Liu, J. H. (2020). Extended-spectrum β-lactamase-producing escherichia coli. Lancet Infect. Dis. 20 (4), 404–405. doi: 10.1016/S1473-3099(20)30115-8
Kim, T. W., Kim, Y. H., Kim, S. E., Lee, J. H., Park, C. S., Kim, H. Y. (2010). Identification and distribution of bacillus species in doenjang by whole-cell protein patterns and 16S rRNA gene sequence analysis. J. Microbiol. Biotechnol. 20, 1210–1214. doi: 10.4014/jmb.1002.02008
Kiratisin, P., Apisarnthanarak, A., Saifon, P., Laesripa, C., Kitphati, R., Mundy, L. M. (2007). The emergence of a novel ceftazidime-resistant CTX-m extended-spectrum β-lactamase, CTX-M-55, in both community-onset and hospital-acquired infections in Thailand. Diagn. Microbiol. Infect. Dis. 58 (3), 349–355. doi: 10.1016/j.diagmicrobio.2007.02.005
Leão, C., Clemente, L., Moura, L., Seyfarth, A. M., Hansen, I. M., Hendriksen, R. S., et al. (2021). Emergence and clonal spread of CTX-M-65-producing escherichia coli from retail meat in Portugal. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.653595
Letunic, I., Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Liu, Z., Wang, K., Zhang, Y., Xia, L., Zhao, L., Guo, C., et al. (2022). High prevalence and diversity characteristics of blaNDM, mcr, and blaESBLs harboring multidrug-resistant escherichia coli from chicken, pig, and cattle in China. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.755545
Liu, J., Wei, S., Ma, J., Zeng, Z., Lü, D., Yang, G., et al. (2007). Detection and characterization of CTX-m and CMY-2 β-lactamases among escherichia coli isolates from farm animals in guangdong province of China. Int. J. Antimicrob. Agents. 29, 576–581. doi: 10.1016/j.ijantimicag.2006.12.015
Nadimpalli, M., Fabre, L., Yith, V., Sem, N., Gouali, M., Delarocque-Astagneau, E., et al. (2019). CTX-M-55-type ESBL-producing salmonella enterica are emerging among retail meats in Phnom Penh, Cambodia. J. Antimicrob. Chemother. 74 (2), 342–348. doi: 10.1093/jac/dky451
Partridge, S. R., Kwong, S. M., Firth, N., Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31 (4), e00088–e00017. doi: 10.1128/CMR.00088-17
Rao, L., Lv, L., Zeng, Z., Chen, S., He, D., Chen, X., et al. (2014). Increasing prevalence of extended-spectrum cephalosporin-resistant escherichia coli in food animals and the diversity of CTX-m genotypes during 2003-2012. Vet. Microbiol. 172 (3-4), 534–541. doi: 10.1016/j.vetmic.2014.06.013
Rozwandowicz, M., Brouwer, M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez-Zorn, B., Guerra, B., et al. (2018). Plasmids carrying antimicrobial resistance genes in enterobacteriaceae. J. Antimicrob. Chemother. 73 (5), 1121–1137. doi: 10.1093/jac/dkx488
Woerther, P. L., Burdet, C., Chachaty, E., Andremont, A. (2013). Trends in human fecal carriage of extended-spectrum β-lactamases in the community: toward the globalization of CTX-m. Clin. Microbiol. Rev. 26, 744–758. doi: 10.1128/CMR.00023-13
Yang, X., Liu, W., Liu, Y., Wang, J., Lv, L., Chen, X., et al. (2014). F33: A-: B-, IncHI2/ST3, and IncI1/ST71 plasmids drive the dissemination of fosA3 and blaCTX-M-55/-14/-65 in escherichia coli from chickens in China. Front. Microbiol. 5. doi: 10.3389/fmicb.2014.00688
Yoon, E. J., Choi, Y. J., Kim, D., Won, D., Choi, J. R., Jeong, S. H. (2022). Amplification of the chromosomal blaCTX-M-14 gene in escherichia coli expanding the spectrum of resistance under antimicrobial pressure. Microbiol. Spectr., 10(3):e0031933. doi: 10.1128/spectrum.00319-22
Zeng, S., Luo, J., Li, X., Zhuo, C., Wu, A., Chen, X., et al. (2021). Molecular epidemiology and characteristics of CTX-M-55 extended-spectrum β-lactamase-producing escherichia coli from guangzhou, China. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.730012
Zeng, S., Zhuo, Z., Huang, Y., Luo, J., Feng, Y., Gong, B., et al. (2022). Prevalence of chromosomally located blaCTX-M-55 in salmonella typhimurium ST34 isolates recovered from a tertiary hospital in guangzhou, China. Microbiol. Spectr., 10(3):e0277121. doi: 10.1128/spectrum.02771-21
Zhang, Z., Chang, J., Xu, X., Zhou, M., Shi, C., Liu, Y., et al. (2021). Dissemination of IncFII plasmids carrying fosA3 and blaCTX-M-55 in clinical isolates of salmonella enteritidis. Zoonoses. Public Health 68 (7), 760–768. doi: 10.1111/zph.12825
Zhang, J., Zheng, B., Zhao, L., Wei, Z., Ji, J., Li, L., et al. (2014). Nationwide high prevalence of CTX-m and an increase of CTX-M-55 in escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect. Dis. 14, 649. doi: 10.1186/s12879-014-0659-0
Keywords: blaCTX-M, chromosome, Escherichia coli, ISEcp1, plasmids, ST8369
Citation: Wang Z-Y, Jiang Y, Shao Y-Q, Lu H-F, Lu M-J, Jiao X, Li Q-C and Wang J (2022) Nasal carriage of CTX-M-55-producing Escherichia coli ST8369 in a healthy cohort in the city of Yangzhou, China. Front. Cell. Infect. Microbiol. 12:970940. doi: 10.3389/fcimb.2022.970940
Received: 16 June 2022; Accepted: 15 July 2022;
Published: 03 August 2022.
Edited by:
Yurong Wen, Xi’an Jiaotong University, ChinaReviewed by:
Stefano Leo, Université de Fribourg, SwitzerlandHanWei Jiao, Southwest University, China
Copyright © 2022 Wang, Jiang, Shao, Lu, Lu, Jiao, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiu-Chun Li, cWNsaUB5enUuZWR1LmNu; Jing Wang, d2oxMjMxQHl6dS5lZHUuY24=
†These authors have contributed equally to this article
 Zhen-Yu Wang
Zhen-Yu Wang Yue Jiang1,2†
Yue Jiang1,2† Xinan Jiao
Xinan Jiao Qiu-Chun Li
Qiu-Chun Li Jing Wang
Jing Wang