
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 28 July 2022
Sec. Parasite and Host
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.969832
This article is part of the Research Topic Advances in Diagnosis and Therapeutic Intervention for Foodborne Parasitic Diseases, Volume III View all 5 articles
Toxoplasma gondii (T. gondii) infection can cause intestinal inflammation in rodents and significantly alters the structure of gut microbiota. However, the effects of different T. gondii genotypes on the gut microbiota of rats remain unclear. In this study, acute and chronic T. gondii infection in Fischer 344 rats was induced artificially by intraperitoneal injection of tachyzoites PYS (Chinese 1 ToxoDB#9) and PRU (Type II). Fecal 16S rRNA gene amplicon sequencing was employed to analyze the gut microbiota structure at different stages of infection, and to compare the effects of infection by two T. gondii genotypes. Our results suggested that the infection led to structural changes of gut microbiota in rats. At the acute infection stage, the microbiota diversity increased, while both diversity and abundance of beneficial bacteria decreased at the chronic infection stage. The differences of microbiota structure were caused by strains of different genotypes. However, the diversity changes were consistent. This study demonstrates that the gut microbiota plays an important role in T. gondii infection in rats. The data will improve our understanding of the association between T. gondii infection and gut microbiota in rodents.
Toxoplasma gondii (T. gondii) is an important zoonotic parasite that inhabits in the nucleated cells of various hosts (Dubey et al., 2021). Mammals and birds can be infected by T. gondii through ingesting contaminated food and water (Almeria and Dubey, 2021). T. gondii infection can destroy the epithelial barrier of host intestinal mucosa and even cause fatal intestinal inflammation (Liesenfeld et al., 1996). After the host ingests cysts or oocysts, T. gondii invades cells in the small intestine and forms a transient parasitophorous vacuole (Suss-Toby et al., 1996; Sibley, 2004). In parasitophorous vacuole, they rapidly differentiate into tachyzoites and spread rapidly through the host via the vascular or lymphatic system (Lambert et al., 2006). Visceral organs (including heart), muscles, and nervous tissue are favorite sites of T. gondii (Dubey et al., 1998). During the acute stage of T. gondii infection, the host may represent mild self-limited symptoms. In general, the individuals that have normal immunity can destroy most of T. gondii in the acute infection phase (Dunay et al., 2018), and the remaining tachyzoites differentiate into bradyzoites, which exist as cysts in muscle, brain etc. These bradyzoites will remain largely hidden from the immune system (Cerutti et al., 2020). However, it has been suggested that the immune responses to T. gondii infection is sustained throughout the chronic infection, with specific increases of IgG and IFN-γ levels (Sturge and Yarovinsky, 2014).
The population structure of T. gondii consists of three major genetic types (Types I, II and III) and other region-specific clonal lineages (Howe and Sibley, 1995). Among them, the more common genotypes are Types II, III, 12, and Chinese 1 (Wang et al., 2016). Type II mainly infects humans and is widespread in Europe, North America, and Africa (Dubey et al., 2020). Types III and 12 are often found in animals. Type 12 was recently isolated from human and wild animals (Khan et al., 2011). In China, 78% of total isolated strains belonged to Chinese 1 (ToxoDB#9) (Jiang et al., 2013; Li et al., 2014; Wang et al., 2015) that caused severe diseases in animals and humans. The virulence of a genotype is usually determined by median lethal dose (LD50) of inbred mice. In comparison of Type II (LD50 of 100-1000 tachyzoites) and III (LD50 of 105-106 tachyzoites), Type I was identified to be the most virulent, with LD100 ranging from 1-10 tachyzoites (Sibley and Boothroyd, 1992; Su et al., 2002). Chinese 1 (ToxoDB#9) was also a highly virulent genotype, and 10-100 tachyzoites could kill mice in a few days (Hou et al., 2018).
Under normal conditions, T. gondii invasion can cause the cooperation of mucosal immune system with the gut microbiota for a resistance, however, the unbalanced gut microbiota and T. gondii infection may cause inflammation (Couturier-Maillard et al., 2018). Current studies suggest that there are complex interactions among T. gondii, mucosal immune system, and gut microbiota during T. gondii infection (Burger et al., 2018; Cervantes-Barragan et al., 2019; Snyder and Denkers, 2020). T. gondii can significantly alter the gut microbiota of mice during acute and chronic infection (Shao et al., 2020). Current studies mainly focus on the infections of Type II (ME49 or PRU) strains, and the effects of some highly virulent or locally-prevalent T. gondii strains on gut microbiota are not clear. However, mice had poor resistance to highly virulent strains and could not survive at the chronic infection stage. In contrast, rats may be suitable experimental animals that can withstand infection of highly virulent strains without death (Benedetto et al., 1996; Gao et al., 2015). Therefore, using rats as an experimental animal sheds new light on the changes of gut microbiota after highly virulent T. gondii infection.
To investigate the effects of T. gondii infection on gut microbiota in rats, the infection models were constructed with two T. gondii genotypes (Chinese 1 and Type II). The structural variations of rat gut microbiota were investigated by 16S rRNA amplification sequencing after infections of two T. gondii strains. Our results showed that T. gondii infection significantly altered the gut microbiota of rats, and different T. gondii genotypes caused different patterns of microbiota change. These results suggest that gut microbiota may play an important role in host resistance to parasite invasion.
PYS (Chinese 1, ToxoDB#9) and PRU (Type II) strains of T. gondii were used in this study. The tachyzoites were subcultured in human foreskin fibroblasts (HFF) cells and purified after 5 successive passages. Female Fischer 344 rats of aged 4-6 weeks (n = 18) were randomly divided into 3 groups (n = 6 per group): PYS group, PRU group, and control group. Rats were abdominally injected with 105 tachyzoites of PYS or PRU strain suspended in 0.5 mL sterile phosphate buffered solution (PBS). The control group was inoculated with the same volume of sterile PBS without tachyzoites. All rats were kept in cages with independent ventilation system with free access to food and water. The feces of rats in all groups were collected on 0 (before infection), 7 and 40 days after infection, respectively. The feces were stored in cryopreservation tubes at -80°C until DNA extraction.
DNA was extracted from ~200 mg rat feces using E.Z.N.A ® Mag-Bind Soil DNA Kit (OMEGA, USA) according to the manufacturer’s instructions. PCR amplification of the V3-V4 region of 16S rRNA was using the primers: 341F (5’ CCTACGGGNGGCWGCAG-3’) and 806R (5’ GGACTACHVGGGTWTCTAAT-3’). The reaction of PCR was carried out in the 20 μL system that contained 4 μL 5×Taq Buffer, 2 μL dNTPs, 0.8 μL of each primer, 0.4 μL Taq DNA Polymerase, 1 μL DNA template, and 11 μL ddH2O. PCR reaction conditions are as follows: pre-denaturation at 95°C for 30 s, and followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 59°C for 30 s, and extension at 72°C for 45 s, and a final extension at 72° C for 10 min. The concentration and regional integrity of the PCR products were evaluated using the Agilent 5400 fragment analyzer system, and DNA was quantified using the Qubit3.0 DNA kit. Finally, standard procedures were used for DNA library construction, and 250 bp paired-end sequencing was performed on a NovaSeq 6000 PE250 platform.
Fastp (v0.23.0) was used for quality control of sequencing data (Chen et al., 2018). The low-quality sequences and sequences with N-bases were removed from the original data. Demultiplexed reads were processed with QIIME2 (v2022.2.0, https://qiime2.org) (Bolyen et al., 2019) pipeline and various built-in plugins. Briefly, the raw sequence data were quality control processed and trimmed using DADA2 algorithm (Callahan et al., 2016), and the primer information and chimeric sequences were removed. DADA2 was also used to de-noise reads into amplicon sequence variants (ASVs) and obtain the profile of feature sequence. ASVs with an average abundance less than 0.001% were deleted. Taxonomy was assigned to ASVs using SILVA (138.1-SSURef-NR99) database reference sequences (Quast et al., 2012).
All statistical analyses were completed on R (v.4.1.1) platform, including diversity analysis, species composition analysis, principal component analysis, etc. Briefly, the ‘diversity’ function in the vegan package (Oksanen et al., 2020) was used to calculate Shannon index of samples. The Bray-Curtis distance between samples was calculated using the ‘vegdist’ function of vegan package; the ‘adonis’ function was used to implement permutational multivariate analysis of variance (PERMANOVA). Visualization of species composition and principal component analysis were implemented by ggplot2 package. Wilcoxon rank sum test was used to measure differences between groups, * (p < 0.05), ** (p < 0.01), and *** (p < 0.001). Lefse analysis was performed using the Galaxy online platform (Afgan et al., 2018) (https://huttenhower.sph.harvard.edu/galaxy).
A total of 54 fecal samples were divided into nine groups: PYS-0d (n = 6), PYS-7d (n = 6), PYS-40d (n = 6), PRU-0d (n = 6), PRU-7d (n = 6), PRU-40d (n = 6), Control-0d (n = 6), Control-7d (n = 6), and Control-40d (n = 6). It should be noted that it was necessary to collect samples for control group from the same period. Studies have shown that the gut microbiota of young rats is dynamically, and the structure of microbiota changes with the extension of feeding time (Flemer et al., 2017). 16S rRNA gene amplicon sequencing was performed on these samples, and a total of 580.8 Mb clean data were obtained. We used DADA2 algorithm to de-noise and cluster the data, and thus generating a total of 6,207 ASVs sequences. ASVs with an average relative abundance less than 0.001% were deleted, and 1,604 ASVs were used for the subsequent analysis.
To assess diversity changes after T. gondii infection, the Shannon index was calculated for each group. We first assessed changes in gut diversity in normal rats. Interestingly, since the beginning of the experiment, the diversity of gut microbiota of rats showed a law of first increasing and then decreasing with the extension of feeding time (Figure 1A). This was consistent with the phenomenon reported in a previous study (Flemer et al., 2017). Next, the diversity of gut microbiota in the infected rats was assessed. We found a same pattern of diversity changes in the infected and control groups, whereas a more severe diversity decline occurred during the chronic infection stage (Figures 1B, C). This indicated that the chronic infection of T. gondii was closely related to the diversity of gut microbiota.
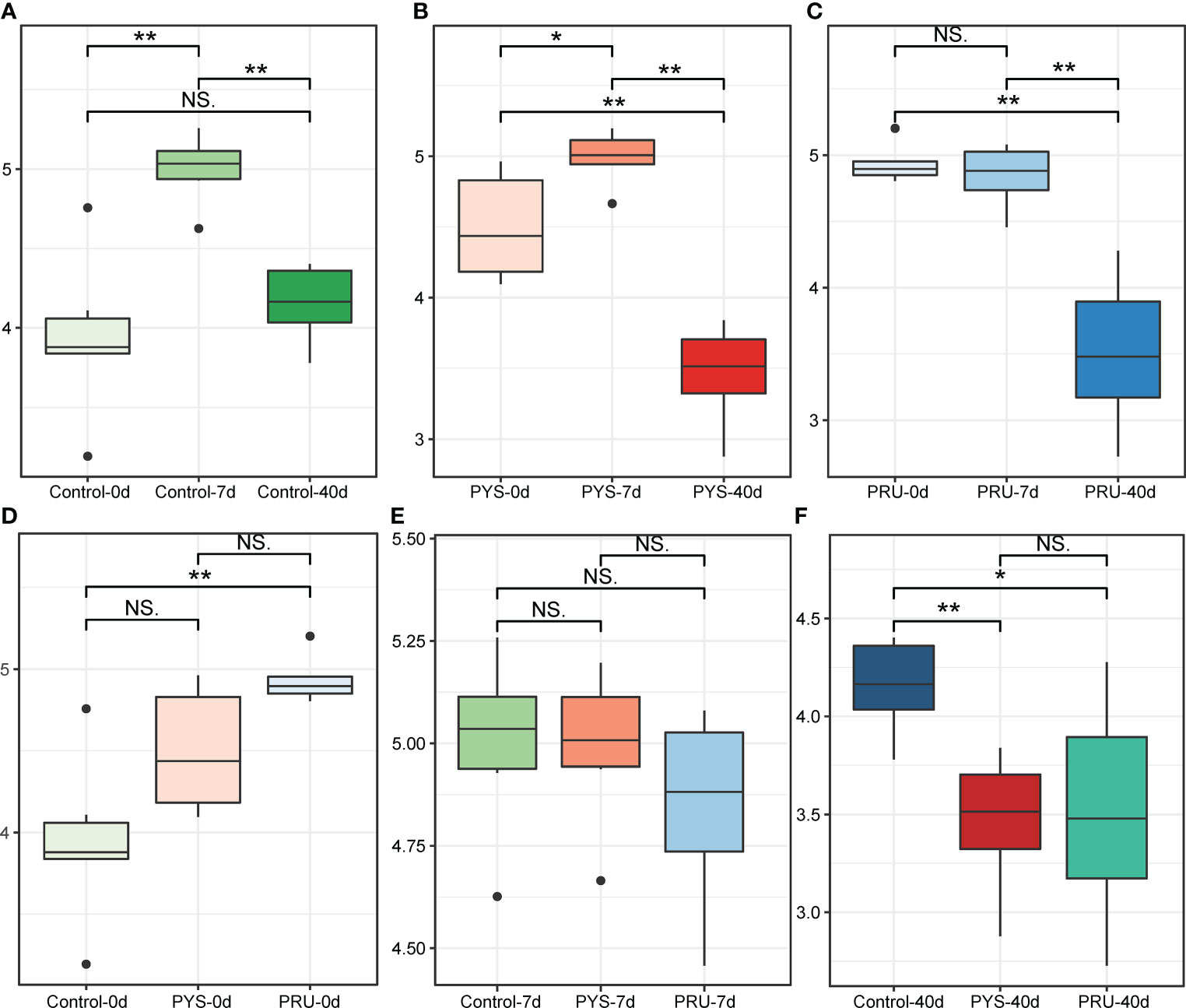
Figure 1 α diversity of gut microbiota in rats. (A–C) Shannon diversity index of control, PYS and PRU groups in different infection periods. (D–F) Shannon index of rats before infection, 7 and 40 days after infection, respectively. The significance level in the Wilcoxon rank sum test is denoted as: *p < 0.05 and **p < 0.01. For boxplot, boxes represent the interquartile range between the first and third quartiles and median (internal line); whiskers denote the lowest and highest values within 1.5 times range of the first and third quartiles, respectively; and nodes represent outliers beyond the whiskers. NS means "Not significant".
In addition, the changes of the diversity during the same period were further compared. It was noteworthy that the diversity of the PRU group before infection was higher than that in other groups (Figure 1D). However, this did not affect the comparison results between groups in the same period. After 7 days of infection, there was no significant difference in gut microbiota diversity between the infected and control rats (Figure 1E). After 40 days of infection, the gut microbiota diversity of the infected group was significantly lower than that of the control group (Figure 1F). In addition, there were no significant differences among different genotypes (Figure 1F).
Then, in order to further understand the differences of gut microbiota between groups, a principal coordinate analysis (PCoA) was conducted based on the Bray-Curtis distance. The results showed that the distance between different groups was far, with an obvious separation (Figure 2A). The infected and control groups in different periods were divided into three regions. The rat samples before infection were at the bottom, and samples of 7 days after infection were at the top left, and samples of 40 days after infection were at the top right. The explanatory variance of PC1 and PC2 was 30.78% and 15.84%, respectively. In addition, the PERMANOVA analysis showed significant differences among groups (R2 = 0.56, p = 0.001). PERMANOVA in the infected group showed that T. gondii infection of different genotypes also led to the variation of bacterial community structure. After 7 days of infection, there was a separation between PYS and PRU groups (Figure 2B, [R² = 0.20, p = 0.005]), whereas PYS and control groups were closer. After 40 days of infection, samples from the two infection groups were close to each other (Figure 2C, [R² = 0.098, p = 0.315]), but farther from the control group. These results suggested that the differences in infection process of T. gondii genotypes might be related to the gut microbiota.
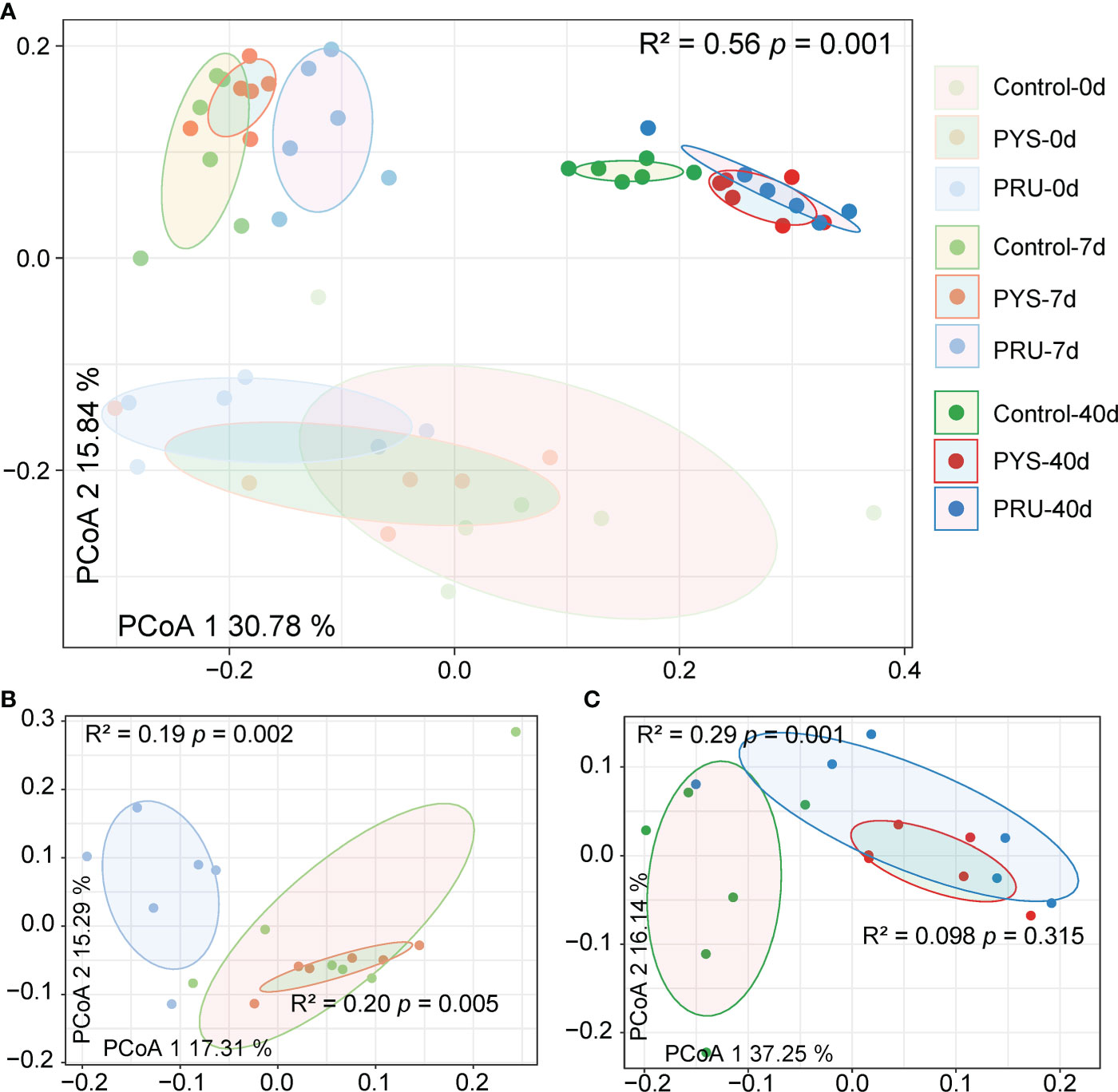
Figure 2 Differences in gut microbiota structure between groups. (A) PCoA analysis based on the Bray-Curtis distance of ASVs reveals the separation between different groups. The position of sample (represented by nodes) in the first two principal coordinates is shown below. (B) and (C) Differences between groups on day 7 and 40 post infection, respectively. R2 and p values represent the results of PERMANOVA.
To further understand the structural variation of gut microbiota in rats infected with T. gondii, the differences were assessed among groups at different taxonomic classification. At the phylum level, Firmicutes, Bacteroidetes, and Verrucomicrobia were found to be the dominant flora in the intestinal tract of rat. In the control group, the abundance of Verrucomicrobia decreased with age and then returned to earlier levels. Similarly, the abundance of Verrucomicrobia decreased and then increased in the infected group. However, after 40 days of infection, the abundance levels of Verrucomicrobia in PYS and PRU groups were significantly higher than those in the uninfected and 7-day groups, accompanied with a decrease in Bacteroides and Firmicutes (Figure 3A, Table S1). At the family level, we found that Akkermansiaceae (mainly Akkermansia) was the main differentiator. In the control group, Akkermansiaceae decreased on day 7 and recovered to day 0 level after 40 days of infection. In PYS and PRU infected groups, after 40 days of infection, the relative abundance of Akkermansiaceae did not return to the level of 0 day as the control group, but increased significantly (Figure 3B, Table S2). In addition, compared with the control group, the relative abundance of Muribaculaceae and Lachnospiraceae in the two infection groups was significantly decreased on day 40 after infection. After 7 days of infection, Akkermansiaceae of PRU group was slightly higher than that of PYS group, while after 40 days of infection, the bacterial community structures of PRU and PYS groups were almost the same. This indicated that different genotypes had different damage to intestinal tract at the early infection stage, which might be caused by the difference in virulence of T. gondii strains.
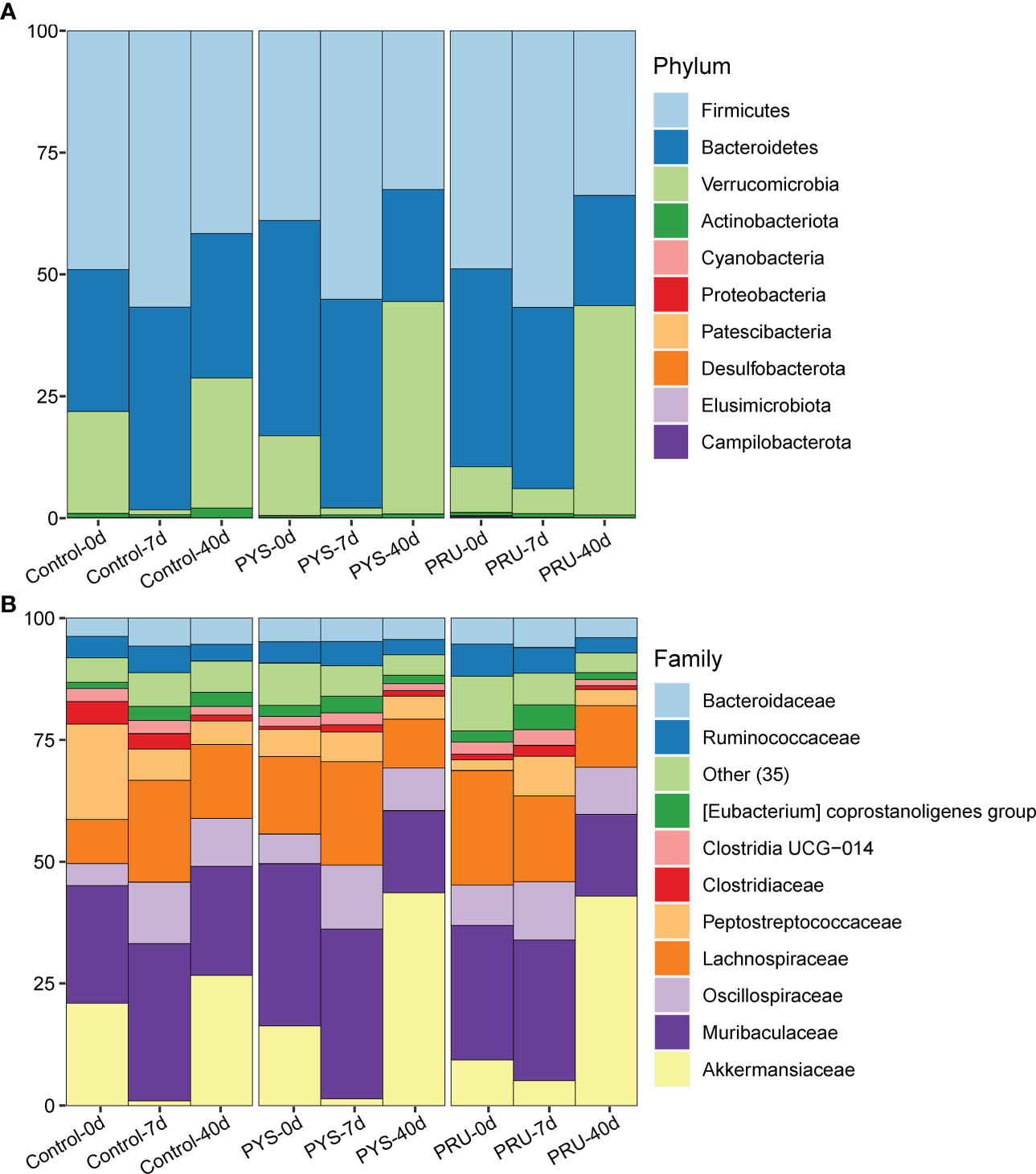
Figure 3 Gut microbiota structure of rats. (A) The gut microbiota structure at phylum level of each group. (B) The gut microbiota structure at the family level of each group, and only the top 10 families have the high abundance, while 35 low-abundance families are integrated into the “Other” category.
To look for differences between groups at the genus level, Lefse was used to identify differentially rich genera with LDA > 2.0 in each group. A total of 72 genera were significantly different between groups from day 0 to 40 of longitudinal comparison. Among them, the rats for 40 days after infection had the least number of genera, indicating that the gut microbiota in rats at chronic infection stage of T. gondii was out of order. To reduce the effects of changes in the normal gut microbiota, we focused on genera that showed no significant difference in the control group, but significant difference in the infected group. Muribaculaceae, Dorea, and Family_XIII_AD3011_group were significantly enriched in PYS-7d and PRU-7d groups, while Alistipes, Eubacterium_ventriosum_group, Negativibacillus, and Anaeroplasma were significantly absent after T. gondii infection (Figure 4A). In addition, we also found some bacteria (e.g., Prevotellaceae_NK3B31_group, Prevotellaceae_UCG-001, and Lachnospiraceae_NK4A136_group) that were opposite to the control group were enriched in Control-7d group and only few were found on days 7 and 40 in the T. gondii infected group.
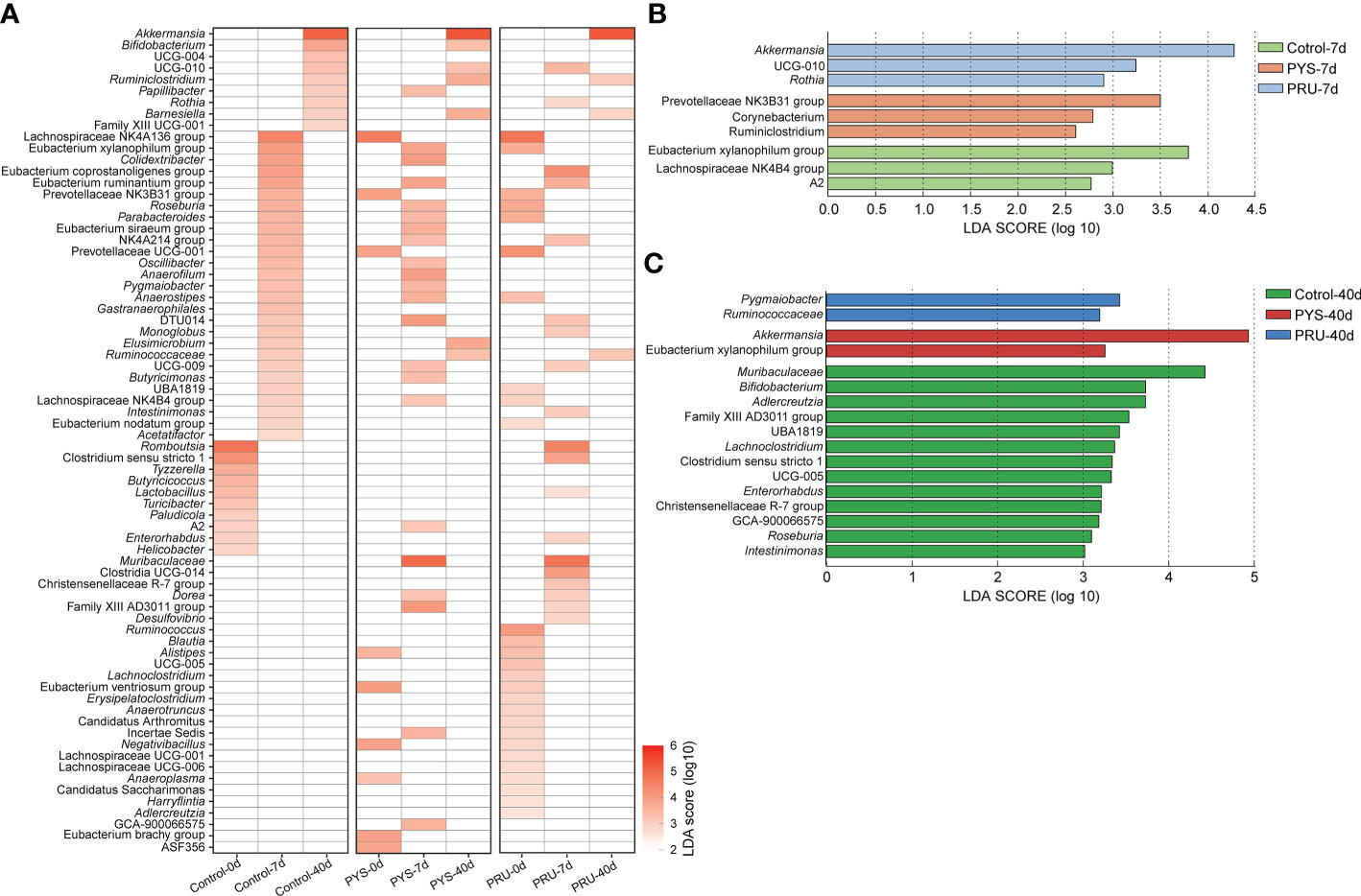
Figure 4 Lefse analysis of genus level. (A) The heat map is divided into three areas. Each area shows the different genera of control, PYS, and PRU groups in different infected periods. The red gradient represents the LDA score (log10) value. Blank means no significant difference or decline of the genus between groups. Briefly, blank means no significant difference when all three groups are blank, and enriched in one group means it is absent in the other two groups. (B) and (C) Differences at genus level between groups on day 7 and 40 post infection, respectively. The higher value of LDA Score (log10), the more significant difference is present between groups.
Then, we compared the bacterial genus differences between groups in the same infection period. After 7 days of infection, Prevotellaceae_NK3B31_group, Corynebacterium, and Ruminiclostridium were significantly enriched in PYS infected group. Akkermansia, UCG-010, and Rothia were significantly enriched in PRU infected group (Figure 4B). In contrast, Corynebacterium, Ruminiclostridium, Eubacterium Xylanophilum group, Lachnospiraceae NK4B4 group, and A2 were significantly reduced in the infected group. Akkermansia and Eubacterium_Xylanophilum_group were significantly enriched in PYS infected group after 40 days of infection. Pygmaiobacter and Ruminococcaceae were significantly enriched in PRU infected group (Figure 4C). These results suggested that differences were present in gut microbiota structure among rats infected with different T. gondii strains. Compared with acute infection period (7 days), the loss of bacterial community was more serious in chronic infection period. There were 13 genera dropped significantly in the infection group (Figure 4C), such as Muribaculaceae, Bifidobacterium, Adlercreutzia, Lachnoclostridium, Enterorhabdus, Roseburia, Intestinimonas, etc. In brief, these results suggested that during chronic T. gondii infection, the gut microbiota did not return to its healthy state, or even appeared more severe decline of bacteria.
T. gondii infection can cause inflammatory responses in intermediate hosts, and the degree is related to the virulence of different genotypes (Sanchez and Besteiro, 2021). Highly virulent genotypes cause severe inflammatory responses and even death of hosts in the acute phase. The avirulent genotype causes a persistent inflammatory response, but is not fatal. Multiple studies have shown that inflammatory responses alter the structure of microbiota in rodents’ guts (Hildebrand et al., 2013; Menghini et al., 2019). Therefore, we aimed to explore the structural variation of gut microbiota in rats during the acute and chronic stages of T. gondii infection and analyze the differences among different genotypes.
In this study, rats were infected with high virulence strain PYS and avirulent strain PRU, respectively. The difference of gut microbiota structure between acute and chronic infection stages was observed. Our results suggested that intestinal inflammation caused by T. gondii infection was closely related to changes of microbiota in guts. During acute infection period, the bacteria in rat gut showed a broad increase in α diversity (Figures 1B, C). This result is consistent with the phenomenon observed in mice previously (Shao et al., 2020). However, we did not observe an increase in the abundance of pathogenic bacteria (e.g., Proteobacteria), which might be due to the impact of T. gondii infection on rats was different from that on mice. In addition, a decline in the abundance of some beneficial bacteria, such as Alistipes, Eubacterium_ventriosum_group and Anaeroplasma, at the acute infection stage (Figure 4B). Dysregulation of these bacteria is a typical feature of inflammation in the body, especially changes in the abundance of Alistipes and Anaeroplasma are thought to be associated with a variety of diseases (Parker et al., 2020; Liu et al., 2022).
It is of interest to note that the proportions of Akkermansia, Muribaculaceae, and Dorea in rats. These bacteria are common in the guts of rats, and their increase suggests that the gut microbiota is disturbed. Akkermansia mainly refers to Akkermansia muciniphila that is a kind of probiotics widely exists in the intestinal mucus layer of mammals (Zhai et al., 2019). Its probiotic effects, including metabolic regulation, immune regulation, and intestinal health protection, have been widely studied (Cani et al., 2022). A variety of diseases, including metabolic syndrome, autoimmune, and cancer, have been reported to be associated with A. muciniphila abundance (Olson et al., 2018; Blacher et al., 2019; Derosa et al., 2022). Yan et al. found that A. muciniphila significantly increased in the guts of T. gondii infected mice after koumiss treatment (Yan et al., 2022), thus suggesting that bacteria were involved in the prevention of T. gondii infection. In addition, Liu et al. reported that A. muciniphila abundance was elevated in the guts of mice infected with Trichinella spiralis (Liu et al., 2021). A recent study showed that oral supplementation of A. muciniphila promoted T. spiralis expulsion via TLR2 in mice (Jin et al., 2022). These reports suggest that A. muciniphila plays an important role in protecting guts from parasitic invasion. However, whether the increase of these bacteria in rats infected by T. gondii infection is related to the parasite pathogenesis or the immune regulatory function remains to be further examined.
During the chronic infection period, the bacterial diversity of rats was significantly decreased. This is probably a normal phenomenon in the growth and development of rats, similarly to a previous report (Flemer et al., 2017). However, the infected rats showed a more severe decrease in microbiota diversity than the control ones (Figures 1B, C). This indicates that the continuous inflammatory response caused by chronic infection of T. gondii may affect the normal development of gut microbiota in rats, so that the microbiota cannot be restored to the level of normal status. Compared with the acute infection period, the number of bacteria that was rich in chronic infection was significantly reduced, and the loss of bacteria was significantly increased. In addition, Akkermansia maintained a high abundance level at chronic stage. This may be due to a decrease in other bacteria, resulting in a higher percentage of Akkermansia (Figure 3B). Most of the missing bacteria have been reported as beneficial bacteria, such as Bifidobacterium, Adlercreutzia, etc.
It is well known that there were significant differences in pathogenicity between different virulence genotypes. Interestingly, although there were differences in the patterns of change in bacterial communities in PYS and PRU groups, the diversity changes in both groups were almost identical (Figures 1E, F). Thus, virulent strain PYS may not cause more severe bacterial disorder than avirulent strain PRU. This may be related to the rats’ natural resistance to T. gondii infection (Li et al., 2012). Limited studies have shown that some highly toxic strains, e.g., GUY008-ABE, can cause rats’ death and show a high parasite burden in multiple organs (Loeuillet et al., 2019). Dubey et al. reported the effects of T. gondii genotypes on parasite distribution, location, tissue cyst size, or tissue lesions (Dubey et al., 2016). However, although a considerable number of studies have assessed the pathological and immunological changes in rats infected with T. gondii (Tonin et al., 2013; Trevizan et al., 2016; Schneider et al., 2018; Machado et al., 2021), there is still a lack of comparison of intestinal damage caused by parasite genotypes in rats. Therefore, we have not yet understood the internal relationship between the structural changes of gut microbiota caused by different genotypes of T. gondii and the infection mechanism, which needs to be further studied.
Our results were in contrast with the findings by Taggart et al. (2022), who reported that the chronic infection of T. gondii did not change the gut microbiota structure of Wistar Hannover rats and concluded that chronical infection may not cause persistent inflammation in rats. The differences for these results may be due to a variety of experimental designs. For example, in the selection of rat strains, we carefully referred to the studies performed by Sergent et al. (2005) and Gao et al. (2015). Fischer 344 rats used in this study were more sensitive to T. gondii infection than Wistar and Lewis rats. This rat is closer to the clinical process of human infection and better reflects human immune resistance to T. gondii infection (Dubey and Frenkel, 1998; Zenner et al., 1999). On the other hand, in order to avoid gender influencing our judgment on the structural variation of gut microbiota, we only selected female rats for the experiments. In conclusion, our results extend the conclusions of previous studies and suggest that different strains of rats may influence the experimental results.
In conclusion, our study suggests that there is a close relationship between gut microbiota and T. gondii infection in rats. The changes in the structure of gut microbiota from the acute to chronic periods suggest a sustained inflammatory response. The comparison between PYS and PRU strains indicated that the higher virulent strain did not necessarily cause more serious imbalance of guts. However, the limitations of this study are as follows: 1) There is a lack of more assessment information on T. gondii genotypes, such as RH, which is more virulent; 2) Lack of quantitative evaluation of intestinal inflammation degree in rats; 3) Confounding factors, such as rat strain and hormone levels, may influence the final results. Therefore, it is necessary to introduce more genotypic, pathological, and immunological information in the future studies, and to evaluate the relationship between T. gondii virulence and gut microbiota through stricter variable control.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA848626/, PRJNA848626.
The animal study was reviewed and approved by Animal Ethics Committee of Qingdao Agricultural University.
HW, L-HY, HM and Q-BL, conceived of the study. Q-BL, H-YQ and Y-FQ, completed the experiment, data analysis and writing section of this study. HM, H-BN and JW, contributed to revising manuscript. All authors read and approved the final manuscript.
This work was supported by the Research Foundation for Distinguished Scholars of Qingdao Agricultural University (665-1120046).
The author would like to thank all the members of the author’s laboratory for their great help in carrying out the work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.969832/full#supplementary-material
Afgan, E., Baker, D., Batut, B., van den Beek, M., Bouvier, D., Cech, M., et al. (2018). The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46 (W1), W537–w544. doi: 10.1093/nar/gky379
Almeria, S., Dubey, J. P. (2021). Foodborne transmission of toxoplasma gondii infection in the last decade. overview. Res. vet Sci. 135, 371–385. doi: 10.1016/j.rvsc.2020.10.019
Benedetto, N., Folgore, A., Ferrara, C., Galdiero, M. (1996). Susceptibility to toxoplasmosis: correlation between macrophage function, brain cyst formation and mortality in rats. New microbiol 19 (1), 47–58.
Blacher, E., Bashiardes, S., Shapiro, H., Rothschild, D., Mor, U., Dori-Bachash, M., et al. (2019). Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature 572 (7770), 474–480. doi: 10.1038/s41586-019-1443-5;
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37 (8), 852–857. doi: 10.1038/s41587-019-0209-9
Burger, E., Araujo, A., López-Yglesias, A., Rajala, M. W., Geng, L., Levine, B., et al. (2018). Loss of paneth cell autophagy causes acute susceptibility to toxoplasma gondii-mediated inflammation. Cell Host Microbe 23 (2), 177–190.e174. doi: 10.1016/j.chom.2018.01.001
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., Holmes, S. P. (2016). DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 13 (7), 581–583. doi: 10.1038/nmeth.3869
Cani, P. D., Depommier, C., Derrien, M., Everard, A., de Vos, W. M. (2022). Akkermansia muciniphila: paradigm for next-generation beneficial microorganisms. Nat. Rev. Gastroenterol. Hepatol. doi: 10.1038/s41575-022-00631-9
Cerutti, A., Blanchard, N., Besteiro, S. (2020). The bradyzoite: A key developmental stage for the persistence and pathogenesis of toxoplasmosis. Pathog. (Basel Switzerland) 9 (3), 234. doi: 10.3390/pathogens9030234
Cervantes-Barragan, L., Cortez, V. S., Wang, Q., McDonald, K. G., Chai, J. N., Di Luccia, B., et al. (2019). CRTAM protects against intestinal dysbiosis during pathogenic parasitic infection by enabling Th17 maturation. Front. Immunol. 10, 1423. doi: 10.3389/fimmu.2019.01423
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinf. (Oxford England) 34 (17), i884–i890. doi: 10.1093/bioinformatics/bty560
Couturier-Maillard, A., Froux, N., Piotet-Morin, J., Michaudel, C., Brault, L., Le Bérichel, J., et al. (2018). Interleukin-22-deficiency and microbiota contribute to the exacerbation of toxoplasma gondii-induced intestinal inflammation. Mucosal Immunol. 11 (4), 1181–1190. doi: 10.1038/s41385-018-0005-8
Derosa, L., Routy, B., Thomas, A. M., Iebba, V., Zalcman, G., Friard, S., et al. (2022). Intestinal akkermansia muciniphila predicts clinical response to PD-1 blockade in patients with advanced non-small-cell lung cancer. Nat. Med. 28 (2), 315–324. doi: 10.1038/s41591-021-01655-5
Dubey, J. P., Cerqueira-Cézar, C. K., Murata, F. H. A., Kwok, O. C. H., Yang, Y. R., Su, C. (2020). All about toxoplasmosis in cats: the last decade. Vet Parasitol. 283, 109145. doi: 10.1016/j.vetpar.2020.109145
Dubey, J. P., Ferreira, L. R., Alsaad, M., Verma, S. K., Alves, D. A., Holland, G. N., et al. (2016). Experimental toxoplasmosis in rats induced orally with eleven strains of toxoplasma gondii of seven genotypes: Tissue tropism, tissue cyst size, neural lesions, tissue cyst rupture without reactivation, and ocular lesions. PloS One 11 (5), e0156255. doi: 10.1371/journal.pone.0156255
Dubey, J. P., Frenkel, J. K. (1998). Toxoplasmosis of rats: a review, with considerations of their value as an animal model and their possible role in epidemiology. Vet Parasitol. 77 (1), 1–32. doi: 10.1016/S0304-4017(97)00227-6
Dubey, J. P., Lindsay, D. S., Speer, C. A. (1998). Structures of toxoplasma gondii tachyzoites, bradyzoites, and sporozoites and biology and development of tissue cysts. Clin. Microbiol. Rev. 11 (2), 267–299. doi: 10.1128/CMR.11.2.267
Dubey, J. P., Murata, F. H. A., Cerqueira-Cézar, C. K., Kwok, O. C. H., Su, C. (2021). Epidemiological significance of toxoplasma gondii infections in wild rodents: 2009-2020. J. Parasitol. 107 (2), 182–204. doi: 10.1645/20-121
Dunay, I. R., Gajurel, K., Dhakal, R., Liesenfeld, O., Montoya, J. G. (2018). Treatment of toxoplasmosis: Historical perspective, animal models, and current clinical practice. Clin. Microbiol. Rev. 31 (4), e00057–17. doi: 10.1128/CMR.00057-17
Flemer, B., Gaci, N., Borrel, G., Sanderson, I. R., Chaudhary, P. P., Tottey, W., et al. (2017). Fecal microbiota variation across the lifespan of the healthy laboratory rat. Gut Microbes 8 (5), 428–439. doi: 10.1080/19490976.2017.1334033
Gao, J. M., Yi, S. Q., Wu, M. S., Geng, G. Q., Shen, J. L., Lu, F. L., et al. (2015). Investigation of infectivity of neonates and adults from different rat strains to toxoplasma gondii prugniaud shows both variation which correlates with iNOS and arginase-1 activity and increased susceptibility of neonates to infection. Exp. Parasitol. 149, 47–53. doi: 10.1016/j.exppara.2014.12.008
Hildebrand, F., Nguyen, T. L., Brinkman, B., Yunta, R. G., Cauwe, B., Vandenabeele, P., et al. (2013). Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 14 (1), R4. doi: 10.1186/gb-2013-14-1-r4
Hou, Z., Zhou, Y., Liu, D., Su, S., Zhao, Z., Xu, J., et al. (2018). Genotyping and virulence analysis of toxoplasma gondii isolates from a dead human fetus and dead pigs in jiangsu province, Eastern China. Acta parasitol 63 (2), 397–411. doi: 10.1515/ap-2018-0046
Howe, D. K., Sibley, L. D. (1995). Toxoplasma gondii comprises three clonal lineages: correlation of parasite genotype with human disease. J. Infect. Dis. 172 (6), 1561–1566. doi: 10.1093/infdis/172.6.1561
Jiang, H. H., Huang, S. Y., Zhou, D. H., Zhang, X. X., Su, C., Deng, S. Z., et al. (2013). Genetic characterization of toxoplasma gondii from pigs from different localities in China by PCR-RFLP. Parasites Vectors 6, 227. doi: 10.1186/1756-3305-6-227
Jin, X., Liu, Y., Wang, J., Wang, X., Tang, B., Liu, M., et al. (2022). β-glucan-triggered akkermansia muciniphila expansion facilitates the expulsion of intestinal helminth via TLR2 in mice. Carbohydr. polymers 275, 118719. doi: 10.1016/j.carbpol.2021.118719
Khan, A., Dubey, J. P., Su, C., Ajioka, J. W., Rosenthal, B. M., Sibley, L. D. (2011). Genetic analyses of atypical toxoplasma gondii strains reveal a fourth clonal lineage in north America. Int. J. Parasitol. 41 (6), 645–655. doi: 10.1016/j.ijpara.2011.01.005
Lambert, H., Hitziger, N., Dellacasa, I., Svensson, M., Barragan, A. (2006). Induction of dendritic cell migration upon toxoplasma gondii infection potentiates parasite dissemination. Cell. Microbiol. 8 (10), 1611–1623. doi: 10.1111/j.1462-5822.2006.00735.x
Liesenfeld, O., Kosek, J., Remington, J. S., Suzuki, Y. (1996). Association of CD4+ T cell-dependent, interferon-gamma-mediated necrosis of the small intestine with genetic susceptibility of mice to peroral infection with toxoplasma gondii. J. Exp. Med. 184 (2), 597–607. doi: 10.1084/jem.184.2.597
Li, M., Mo, X. W., Wang, L., Chen, H., Luo, Q. L., Wen, H. Q., et al. (2014). Phylogeny and virulence divergency analyses of toxoplasma gondii isolates from China. Parasites Vectors 7, 133. doi: 10.1186/1756-3305-7-133
Liu, X., Jin, G., Tang, Q., Huang, S., Zhang, Y., Sun, Y., et al. (2022). Life lactobacillus rhamnosus GG colonisation inhibits intestinal tumour formation. Br. J. Cancer 126 (10), 1421–1431. doi: 10.1038/s41416-021-01562-z
Liu, S., Pan, J., Meng, X., Zhu, J., Zhou, J., Zhu, X. (2021). Trichinella spiralis infection decreases the diversity of the intestinal flora in the infected mouse. J. microbiol immunol infection = Wei mian yu gan ran za zhi 54 (3), 490–500. doi: 10.1016/j.jmii.2019.09.009
Li, Z., Zhao, Z. J., Zhu, X. Q., Ren, Q. S., Nie, F. F., Gao, J. M., et al. (2012). Differences in iNOS and arginase expression and activity in the macrophages of rats are responsible for the resistance against t. gondii infection. PloS One 7 (4), e35834. doi: 10.1371/journal.pone.0035834
Loeuillet, C., Mondon, A., Kamche, S., Curri, V., Boutonnat, J., Cavaillès, P., et al. (2019). Toxoplasma hypervirulence in the rat model parallels human infection and is modulated by the Toxo1 locus. Front. Cell. infection Microbiol. 9, 134. doi: 10.3389/fcimb.2019.00134
Machado, C. C. A., Watanabe, P. D. S., Mendes, J. D. L., Pupim, A. C. E., Ortigoza, S. M., Bergoc, H. G., et al. (2021). Toxoplasma gondii infection impairs the colonic motility of rats due to loss of myenteric neurons. Neurogastroenterol. Motil. Off. J. Eur. Gastroint Motil. Soc. 33 (1), e13967. doi: 10.1111/nmo.13967
Menghini, P., Corridoni, D., Buttó, L. F., Osme, A., Shivaswamy, S., Lam, M., et al. (2019). Neutralization of IL-1α ameliorates crohn's disease-like ileitis by functional alterations of the gut microbiome. Proc. Natl. Acad. Sci. United States America 116 (52), 26717–26726. doi: 10.1073/pnas.1915043116
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2020). Vegan community ecology package version 2.5-7 November 2020. Available at: https://CRAN.R-project.org/package=vegan
Olson, C. A., Vuong, H. E., Yano, J. M., Liang, Q. Y., Nusbaum, D. J., Hsiao, E. Y. (2018). The gut microbiota mediates the anti-seizure effects of the ketogenic diet. Cell 173 (7), 1728–1741.e1713. doi: 10.1016/j.cell.2018.04.027
Parker, B. J., Wearsch, P. A., Veloo, A. C. M., Rodriguez-Palacios, A. (2020). The genus alistipes: Gut bacteria with emerging implications to inflammation, cancer, and mental health. Front. Immunol. 11, 906. doi: 10.3389/fimmu.2020.00906
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41 (D1), D590–D596. doi: 10.1093/nar/gks1219
Sanchez, S. G., Besteiro, S. (2021). The pathogenicity and virulence of toxoplasma gondii. Virulence 12 (1), 3095–3114. doi: 10.1080/21505594.2021.2012346
Schneider, L. C. L., do Nascimento, J. C. P., Trevizan, A. R., Góis, M. B., Borges, S. C., Beraldi, E. J., et al. (2018). Toxoplasma gondii promotes changes in VIPergic submucosal neurons, mucosal intraepithelial lymphocytes, and goblet cells during acute infection in the ileum of rats. Neurogastroenterol. Motil. Off. J. Eur. Gastroint Motil. Soc. 30 (5), e13264. doi: 10.1111/nmo.13264
Sergent, V., Cautain, B., Khalife, J., Deslée, D., Bastien, P., Dao, A., et al. (2005). Innate refractoriness of the Lewis rat to toxoplasmosis is a dominant trait that is intrinsic to bone marrow-derived cells. Infection Immun. 73 (10), 6990–6997. doi: 10.1128/IAI.73.10.6990-6997.2005
Shao, D. Y., Bai, X., Tong, M. W., Zhang, Y. Y., Liu, X. L., Zhou, Y. H., et al. (2020). Changes to the gut microbiota in mice induced by infection with toxoplasma gondii. Acta tropica 203, 105301. doi: 10.1016/j.actatropica.2019.105301
Sibley, L. D. (2004). Intracellular parasite invasion strategies. Sci. (New York NY) 304 (5668), 248–253. doi: 10.1126/science.1094717
Sibley, L. D., Boothroyd, J. C. (1992). Virulent strains of toxoplasma gondii comprise a single clonal lineage. Nature 359 (6390), 82–85. doi: 10.1038/359082a0
Snyder, L. M., Denkers, E. Y. (2020). From initiators to effectors: Roadmap through the intestine during encounter of toxoplasma gondii with the mucosal immune system. Front. Cell. infection Microbiol. 10, 614701. doi: 10.3389/fcimb.2020.614701
Sturge, C. R., Yarovinsky, F. (2014). Complex immune cell interplay in the gamma interferon response during toxoplasma gondii infection. Infection Immun. 82 (8), 3090–3097. doi: 10.1128/IAI.01722-14
Su, C., Howe, D. K., Dubey, J. P., Ajioka, J. W., Sibley, L. D. (2002). Identification of quantitative trait loci controlling acute virulence in toxoplasma gondii. Proc. Natl. Acad. Sci. United States America 99 (16), 10753–10758. doi: 10.1073/pnas.172117099
Suss-Toby, E., Zimmerberg, J., Ward, G. E. (1996). Toxoplasma invasion: the parasitophorous vacuole is formed from host cell plasma membrane and pinches off via a fission pore. Proc. Natl. Acad. Sci. United States America 93 (16), 8413–8418. doi: 10.1073/pnas.93.16.8413
Taggart, P. L., Liddicoat, C., Tong, W. H., Breed, M. F., Weinstein, P., Wheeler, D., et al. (2022). Gut microbiota composition does not associate with toxoplasma infection in rats. Mol. Ecol. 31 (14), 3963–3970. doi: 10.1111/mec.16552
Tonin, A. A., Da Silva, A. S., Ruchel, J. B., Rezer, J. F., Camillo, G., Faccio, L., et al. (2013). E-NTPDase and e-ADA activities in lymphocytes associated with the immune response of rats experimentally infected with toxoplasma gondii. Exp. Parasitol. 135 (2), 325–330. doi: 10.1016/j.exppara.2013.07.014
Trevizan, A. R., Vicentino-Vieira, S. L., da Silva Watanabe, P., Góis, M. B., de Melo Gde, A., Garcia, J. L., et al. (2016). Kinetics of acute infection with toxoplasma gondii and histopathological changes in the duodenum of rats. Exp. Parasitol. 165, 22–29. doi: 10.1016/j.exppara.2016.03.015
Wang, L., He, L. Y., Meng, D. D., Chen, Z. W., Wen, H., Fang, G. S., et al. (2015). Seroprevalence and genetic characterization of toxoplasma gondii in cancer patients in anhui province, Eastern China. Parasites Vectors 8, 162. doi: 10.1186/s13071-015-0778-5
Wang, D., Liu, Y., Jiang, T., Zhang, G., Yuan, G., He, J., et al. (2016). Seroprevalence and genotypes of toxoplasma gondii isolated from pigs intended for human consumption in liaoning province, northeastern China. Parasites Vectors 9, 248. doi: 10.1186/s13071-016-1525-2
Yan, X., Han, W., Jin, X., Sun, Y., Gao, J., Yu, X., et al. (2022). Study on the effect of koumiss on the intestinal microbiota of mice infected with toxoplasma gondii. Sci. Rep. 12 (1), 1271. doi: 10.1038/s41598-022-05454-x
Zenner, L., Foulet, A., Caudrelier, Y., Darcy, F., Gosselin, B., Capron, A., et al. (1999). Infection with toxoplasma gondii RH and prugniaud strains in mice, rats and nude rats: kinetics of infection in blood and tissues related to pathology in acute and chronic infection. Pathol Res. Pract. 195 (7), 475–485. doi: 10.1016/S0344-0338(99)80051-X
Keywords: Toxoplasma gondii, fischer 344 rats, gut microbiota, 16S rRNA gene amplicon sequencing, genotype
Citation: Lv Q-B, Ma H, Wei J, Qin Y-F, Qiu H-Y, Ni H-B, Yang L-H and Cao H (2022) Changes of gut microbiota structure in rats infected with Toxoplasma gondii. Front. Cell. Infect. Microbiol. 12:969832. doi: 10.3389/fcimb.2022.969832
Received: 15 June 2022; Accepted: 07 July 2022;
Published: 28 July 2022.
Edited by:
Wei Cong, Shandong University, Weihai, ChinaReviewed by:
Jun-Jun He, Chinese Academy of Agricultural Sciences (CAAS), ChinaCopyright © 2022 Lv, Ma, Wei, Qin, Qiu, Ni, Yang and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-Hua Yang, amx5bGgxMjNAMTYzLmNvbQ==; Hongwei Cao, Y2FvaHdAeWN0dS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.