
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 17 August 2022
Sec. Clinical Microbiology
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.961297
This article is part of the Research Topic Interactions Between Pathogens and Host Immune System in Patients with Immunodeficiency: Estimation from High-throughput Sequencing View all 14 articles
 Minmin Lin1,2†
Minmin Lin1,2† Kongqiu Wang1†
Kongqiu Wang1† Lidi Qiu3
Lidi Qiu3 Yingjian Liang1
Yingjian Liang1 Changli Tu1
Changli Tu1 Meizhu Chen1
Meizhu Chen1 Zhenguo Wang1
Zhenguo Wang1 Jian Wu1
Jian Wu1 Yiying Huang1
Yiying Huang1 Cuiyan Tan1
Cuiyan Tan1 Qijiu Chen1
Qijiu Chen1 Xiaobin Zheng1
Xiaobin Zheng1 Jing Liu1,2*
Jing Liu1,2*Tropheryma whipplei is the bacterium associated with Whipple’s disease (WD), a chronic systemic infectious disease primarily involving the gastrointestinal tract. T. whipplei can also be detected in different body site of healthy individuals, including saliva and feces. Traditionally, Tropheryma whipplei has a higher prevalence in bronchoalveolar lavage fluid (BALF) of immunocompromised individuals. Few studies have explored the significance of the detection of T. whipplei in BALF. Herein, we retrospectively reviewed 1725 BALF samples which detected for metagenomic next-generation sequencing (mNGS) from March 2019 to April 2022 in Zhuhai, China. Seventy BALs (70/1725, 4.0%) from 70 patients were positive for T. whipplei. Forty-four patients were male with an average age of 50 years. The main symptoms included cough (23/70), expectoration (13/70), weight loss (9/70), and/or dyspnea (8/70), but gastrointestinal symptoms were rare. Chronic liver diseases were the most common comorbidity (n=15, 21.4%), followed by diabetes mellitus (n=13, 18.6%). Only nine patients (12.9%) were immunocompromised. Twenty-four patients (34.3%) were finally diagnosed with reactivation tuberculosis and 15 patients (21.4%) were diagnosed with lung tumors, including 13 primary lung adenocarcinoma and two lung metastases. Fifteen patients (21.4%) had pneumonia. Among the 20 samples, T. whipplei was the sole agent, and Mycobacterium tuberculosis complex was the most common detected other pathogens. Among the non-tuberculosis patients, 31 (31/46, 67.4%) had ground glass nodules or solid nodules on chest CT. Our study indicates that T. whipplei should be considered as a potential contributing factor in some lung diseases. For non-immunocompromised patients, the detection of T. whipplei also needs attention. The mNGS technology improves the detection and attention of rare pathogens. In the future, the infection, colonization, and prognosis of T. whipplei in lung still need to be studied.
Tropheryma whipplei is the bacterium associated with Whipple’s disease (WD), described by George H.Whipple in 1907 (GH, 1907). Whipple’s disease is a rare, chronic, and systemic illness, which mainly involves the gastrointestinal tract. It can also lead to the involvement of joints, nervous system, and cardiovascular system. The most common symptoms of patients with classic Whipple’s disease are arthralgia, diarrhea, weight loss, lymphadenopathy, abdominal pain, and fever (Dolmans et al., 2017). Only a few studies on the prevalence and incidence rate of Whipple’s disease were performed. According to a research from the United States, the overall prevalence of Whipple’s disease in the US was 9.8 cases per 1 million people (Elchert et al., 2019). Of concern, the classic Whipple’s disease is typically found in Caucasian populations, but carriage is common in the native Asian and Africa populations (Keita et al., 2015).
T. whipplei is a rod-shaped, Gram-positive bacterium belonging to Actinomycetes. With the development of molecular biology technology, the bacterial 16S ribosomal DNA was first identified in small-bowel biopsy specimens of patients with classic Whipple’s disease by nucleotide sequencing and PCR amplification in 1991 (Relman et al., 1992). Subsequently, the T. whipplei DNA has been detected in various samples by using PCR, including stool (Schoniger-Hekele et al., 2007), saliva, urine, blood, cerebrospinal fluid, and bronchoalveolar lavage fluid (BALF) (Fenollar et al., 2012). In some studies, T. whipplei has been shown to be an etiological pathogen in pneumonia (Bousbia et al., 2010). Moreover, T. whipplei was significantly more common in BALF in HIV-positive individuals, which was considered to be a potential contributing factor with lung complications (Lozupone et al., 2013). This further indicates that T. whipplei may be involved in the occurrence of some lung diseases.
With the application of metagenomic next-generation sequencing (mNGS) technology with high sensitivity in our country, the detection of T. whipplei has increased. However, at present, the epidemiology of T. whipplei in BALF in our country is still lacking. Therefore, the BALF samples which detected for mNGS in our hospital were reviewed, and the characteristics of patients with positive T. whipplei were analyzed.
We retrospectively reviewed 1725 BALF samples which detected for metagenomic next-generation sequencing from March 2019 to April 2022 at the Fifth Affiliated Hospital of Sun Yat-sen University in Zhuhai, China. The collection process of BALF was in line with clinical operation standard and followed the principle of sterility. The involved sub-segments were washed with 20 to 50ml normal saline. 3-5ml BALF samples were placed in sterile sputum containers, stored in -20°C, and then sent to BGI (Shenzhen, China) for detection.
The baseline data on patients with positive Tropheryma whipplei was collected, including demographic information, laboratory data, clinical symptoms, imaging examination results, diagnosis, pathological results, and treatment history. Immunocompromised status was defined as any of the following: (1) solid organ transplantation; (2) long-term therapy with corticosteroid (≥ 20mg prednisone or equivalent daily for ≥ 14d or a cumulative dose > 600mg of prednisone) or other immunosuppressive drugs; (3) agranulocytosis after cancer chemotherapy; (4) hematological malignancy; (5) inherited or acquired severe immunodeficiency or human immunodeficiency virus infection.
1.5mL microcentrifuge tube with 0.6mL sample and 250μL 0.5mm glass bead were attached to a horizontal platform on a vortex mixer and agitated vigorously at 2800-3200 rpm for 30 min. Then 7.2μL lysozyme was added for wall-breaking reaction. 0.3mL sample was separated into a new 1.5mL microcentrifuge tube and DNA was extracted using the TIANamp Micro DNA Kit (DP316, TIANGEN BIOTECH) according to the manufacturer’s recommendation. DNA libraries were constructed through DNA-fragmentation, end-repair, adapter-ligation and PCR amplification. Agilent 2100 was used for quality control of the DNA libraries. Quality qualified libraries were pooled, DNA Nanoball (DNB) was made and sequenced by MGISEQ-2000 platform.
High-quality sequencing data were generated by removing low-quality reads, followed by computational substraction of human host sequences mapped to the human reference genome (hg19) using Burrows-Wheeler Alignment (Li and Durbin, 2009). The remaining data by removal of low-complexity reads were classified by simultaneously aligning to Pathogens metagenomics Database (PMDB), consisting of bacteria, fungi, viruses and parasites. The classification reference databases were downloaded from NCBI (ftp://ftp.ncbi.nlm.nih.gov/genomes/). RefSeq contains 4,945 whole genome sequence of viral taxa, 6,350 bacteral genomes or scaffolds, 1064 fungi related to human infection, and 234 parasites associated with human diseases. T. whipplei was considered positive when at least 3 reads were mapped to the species level.
Categorical variables were expressed as percentages. Continuous variables subject to normal distribution were expressed by mean and SD, otherwise, median and IQR. Comparative analysis was conducted by t test, Pearson’s test, Fisher’s exact test, or Mann-Whitney U test where appropriate. Data analysis was performed with SPSS 20.0 and GraphPad Prism. P < 0.05 was considered significant.
Seventy BALs (70/1725, 4.0%) from 70 patients were positive for T. whipplei. Forty-four patients were male (62.9%). The age of the 70 patients ranged from 25 to 78 years (mean age: 50 years). All patients were hospitalized. Twenty-eight (40%) patients were hospitalized in infectious diseases department, 24 (34.3%) in thoracic surgery, 15 (21.4%) in respiratory department, two in an intensive-care unit and one in endocrine department at the sampling time of BALF. The distribution of sampling time was dispersed. The main clinical symptoms of patients included cough (23/70), expectoration (13/70), weight loss (9/70), and/or dyspnea (8/70) (Table 1). Only nine patients complained about gastrointestinal symptoms such as abdominal pain, diarrhea, vomiting, or poor appetite. One patient had joint pain, and one had neurological symptoms. Nearly half of the patients (n=32) had no clinical symptoms.
Chronic liver diseases were the most common comorbidity (n=15, 21.4%), including hepatitis B, cirrhosis and fatty liver, followed by diabetes mellitus (n=13, 18.6%). Non-immunocompromised patients accounted for the majority. Immunodeficiency in nine patients (12.9%) was due respectively to drug including cyclosporin (n=1) or corticosteroids (n=1), and to medical conditions including HIV infection (n=5) and solid organ transplantation (n=2) (Figure 1). Four patients had extrapulmonary tumors. Almost all patients were tested for relevant inflammatory indicators after admission and before treatment. The overall inflammatory response was not strong. The average white blood cell count and haemoglobin of all 70 patients was 6.2×10^9/L (IQR 5.06-7.09×10^9/L) and 132 g/L (IQR 120-149 g/L) respectively. The median C-reactive protein was 3.5 mg/L (IQR 1-11.12 mg/L) (Table 1). The overall BMI index was normal (mean: 23.17, SD: 3.67), and patients with tuberculosis had a lower BMI (21.35 vs 24.13, p=0.002).
Twenty-one patients underwent surgery, three underwent transbronchial lung biopsy and three underwent percutaneous lung biopsy. Therefore, a total of 27 patients had lung histopathological results. The histopathological results of 15 patients were tumors, including primary lung adenocarcinoma (n=13), metastatic intestinal adenocarcinoma (n=1) and metastatic esophageal squamous cell carcinoma (n=1). Two patients showed multinucleated giant cells in histopathology, which were considered to be tuberculosis; One patient’s histopathology was positive for PAS and silver hexamine staining, which was considered as cryptococcal infection. The remaining histopathological results included nonspecific inflammatory cell infiltration (n=5), hamartoma (n=2), suspected lung cancer (n=1), and suspected multifocal micronodular alveolar epithelial hyperplasia (MMPH) (n=1).
Whipple’s disease often involves the gastrointestinal tract. Therefore, we collected the gastroscopic results. In the lamina propria of the duodenal (as well as the gastric antral region, jejunum, or ileum), foam macrophages containing large amounts of diastase-resistant PAS-positive particles was the typical histological detection. Six patients underwent gastroscopy within three months, which showed various degrees of gastritis. Other gastroscopic findings included gastric antrum ulcer (n=1), duodenal erosion and superficial ulcer (n=1), gastric mucosal intraepithelial neoplasia (n=1), gastric polyp (n=1) and esophageal cancer (n=1). Pathological biopsy of duodenal lesions showed lymphocyte and plasma cell infiltration, but PAS staining was not performed.
Among 20 BALF samples, T. whipplei was the only pathogen detected. The most common detected bacterial pathogen with T. whipplei was Mycobacterium tuberculosis complex, followed by Streptococcus pneumoniae, Haemophilus influenzae, Staphylococcus aureus, Haemophilus parainfluenzae, and Actinomyces. The most common detected fungi was Candida albicans, followed by Pneumocystis jirovecii. Human gamma-herpes virus 4 (Epstein-Barr virus, EBV) was the most detected virus (Table 2). The mapped reads number of T. whipplei was normalized to reads per million (RPM). The RPM value of T. whipplei in immunodeficiency patients outnumbered those in non-immunodeficiency patients, but it did not reach statistical difference. The RPM value of T. whipplei sequences was not related to whether it was the sole agent and whether the patients had symptoms (Figure 2). Immunodeficient patients had higher detection rate of fungi and virus, but only virus reached statistical difference (66.7% vs 18.0%, p=0.005) (Supplementary Figure S1). The detection rate of Mycobacterium tuberculosis complex in patients with diabetes was significantly higher (38.5% vs 8.8%, p=0.015).
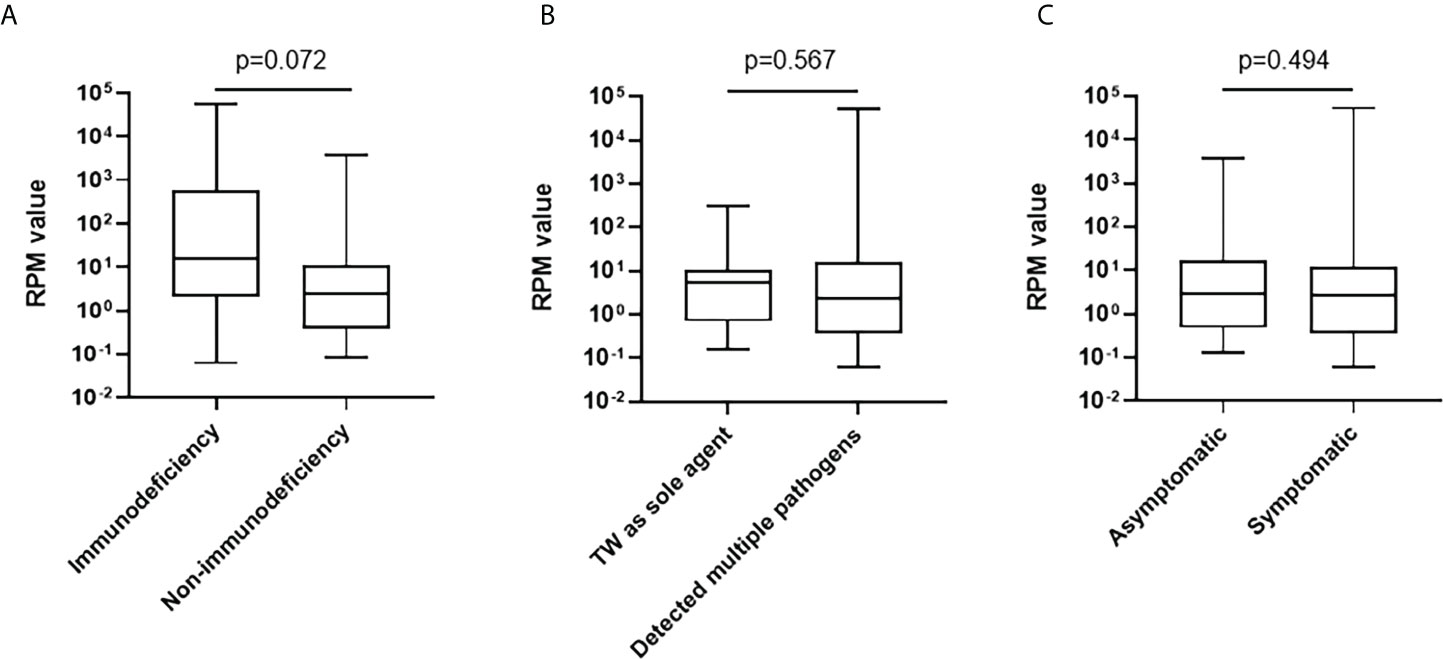
Figure 2 Comparisons of the RPM value of T. whipplei according to immune deficiency status (A), mixed detection (B), and clinical symptoms (C).
According to the classification of diseases, we simply described the chest CT findings. Among the tuberculosis (TB) patients, the most chest CT findings were common reactivation TB changes, including focal or patchy heterogeneous consolidation, poorly defined nodules or linear opacities, cavity (n=6), tuberculoma (n=1), and pleural effusion (n=1). One patient’s chest CT showed miliary tuberculosis. Among the non-TB patients, 31 (31/46, 67.4%) had ground glass nodules or solid nodules on chest CT. The remaining CT findings included patchy, mass, pleural effusion (n=6), cystic (n=2), cavity (n=2), and interstitial fibrosis (n=2). Classification of chest CT in non-tuberculosis patients, especially considering the T. whipplei as the main pathogen, it could be roughly divided into nodular type, pneumonia type and mixed type, as shown in Figure 3. In general, the chest imaging manifestations were various.
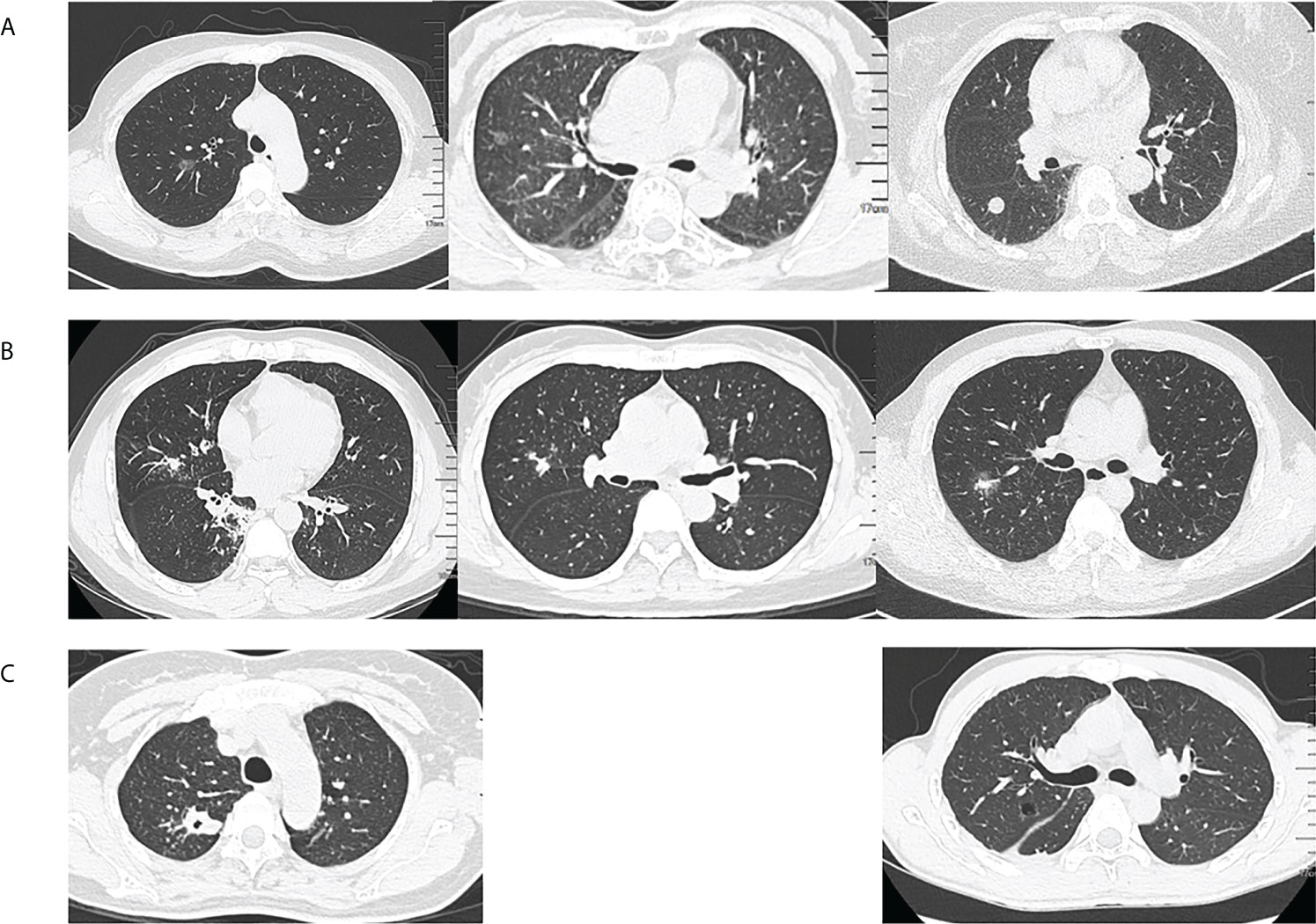
Figure 3 Classification of chest computed tomography (CT) in non-tuberculosis patients. (A) Nodular type: ground glass nodules or solid nodules, (B) Pneumonia type: focal or patchy mixed density shadow, and (C) Mixed type: other manifestations such as cavity, cystic or pleural effusion.
Twenty-four patients (34.3%) were finally diagnosed with reactivation tuberculosis and received anti-tuberculosis treatment. Sixteen of them improved after treatment. Fifteen patients (21.4%) were diagnosed with pneumonia, and all received antimicrobial treatment, such as β-lactams/β-lactamase inhibitor, Trimethoprim-sulfamethoxazole, doxycycline or quinolones. Six patients improved radiographically after treatment. More immunocompromised patients were diagnosed with pneumonia (p=0.001). According to histopathology, fifteen patients (21.4%) were diagnosed with lung tumors, including 13 primary lung adenocarcinoma and two lung metastases. Fourteen of them received surgery and one received chemotherapy. The remaining diagnoses include unexplained pulmonary nodules (n=4), benign pulmonary nodules (n=3), pleurisy (n=3), cryptococcal infection (n=1), and suspected lung cancer (n=1) (Figure 4). None of the patients died in hospital.
Although WD was described as early as 1907, T. whipplei was only successfully cultured in macrophages until 1997 (Schoedon et al., 1997). Due to the lack of specificity of the clinical manifestations, and the lack of understanding of WD, the disease was often misdiagnosed or undiagnosed in our country. With the development of molecular methods and application of mNGS technology, the number of positive cases detected in BALF increased. Meanwhile, there was an increasing number of cases reported about pulmonary infection caused by T. whipplei or pulmonary involvement of WD in recent years. However, the significance of T. whipplei detection in BALF still needs to be comprehensively considered. Our study found that the majority of T. whipplei positive patients in BALF were non-immunodeficient, with a large proportion of patients diagnosed with tuberculosis, lung tumors, and pneumonia. Pulmonary nodules were the main manifestation of chest CT. To our knowledge, our study has the largest sample size in the epidemiological study of T. whipplei detected in BALF in China.
In this study, mNGS was used to detect T. whipplei. In the process of sequencing and bioinformatics analysis, the technology has strictly set up positive and negative controls and carried out quality control. The distribution of departments during the patients sampling was relatively scattered, and the collection time was evenly distributed. Therefore, the possibility of nosocomial infection or outbreak could be ruled out, and the contamination in the collection process or laboratory process could also be basically ruled out. Accordingly, we consider the detected results to be credible.
The prevalence of T. whipplei in our study was slightly lower than that reported by Lagier et al. (6.1%, 88/1438) (Lagier et al., 2016), which was close to the 3% (6/210) reported by Boubia et al. (2010). Respiratory symptoms, weight loss and fever were the main clinical manifestations of patients, while gastrointestinal symptoms were rare, which was similar with the previous case reports of pneumonia caused by T. whipplei (Urbanski et al., 2012; Guo et al., 2021). However, nearly half of the patients in our study were asymptomatic, and the overall inflammatory response was not strong. This suggests that some patients may be carriers of T. whipplei in the lungs. Our results indicated that non-immunodeficient status was predominant in T. whipplei positive patients in BALF, which was inconsistent with the previous view that T. whipplei was more prevalent among HIV-positive individuals (Lozupone et al., 2013; Garcia-Alvarez et al., 2016). The case-control study of Lagier et al. found that the prevalence of T. whipplei may not be related to immune status (Lagier et al., 2016). In addition, our results suggest that patients with diabetes and chronic liver disease may be susceptible to T. whipplei in the lungs, which has not been concerned in previous studies. The significance and causes of T. whipplei detection in non-immunocompromised patients still need to be further explored.
Our study also confirm that T. whipplei is an etiologic pathogen in acute respiratory infections (Lagier et al., 2017). T. whipplei could be co-infected with Pneumocystis jirovecii and Candida albicans, as reported in previous cases (Li et al., 2021; Yan et al., 2021), or leaded to pulmonary infection alone (Guo et al., 2021). Moreover, we also found that immunocompromised patients were more likely to be diagnosed with pneumonia, suggesting that T. whipplei should be considered more as a pathogen when detected in BALF in immunocompromised patients with clinical symptoms. Mycobacterium tuberculosis complex and T. whipplei co-infection or detection were occasionally reported in other studies (Lagier et al., 2016; Zhu et al., 2021). However, our study found that Mycobacterium tuberculosis complex was the most common pathogenic bacteria detected together, and a considerable number of patients (34.3%) were diagnosed as reactivation pulmonary tuberculosis. The reason may be that the incidence of tuberculosis in China is still high. At the same time, our hospital was a designated hospital for tuberculosis treatment, which leaded to the increase of detection rate. On the other hand, both Mycobacterium tuberculosis complex and T. whipplei belong to Actinobacteria. Mycobacterium tuberculosis complex is facultative intracellular bacterium, and T. whipplei is obligate intracellular bacterium. The infection of both bacteria have been associated with macrophages. In the pathogenesis of classic WD, macrophages in duodenal mucosa can be induced by bacteria to polarize into M2/alternatively activated macrophages (Desnues B, Lepidi et al., 2005). The impaired antigen-presenting function of macrophages and dendritic cells further weakened the response of T cells (Moos et al., 2010). In addition, the increased activity of regulatory T cells in the gut and blood further increases the immunosuppressive environment (Schinnerling et al., 2011). However, due to the lack of research on the pathogenesis of T. whipplei in lung infection, we can only speculate that the co-infection or detection of Mycobacterium tuberculosis complex and T. whipplei may be related to the impairment of pulmonary macrophage function. The interaction and causality between T. whipplei and other pathogens in the lung need to be further studied.
We performed a general classification of chest CT findings in non-pulmonary tuberculosis patients, especially those considering T. whipplei as the dominant pathogen. Therefore, our results are representative and convincing. The overall chest CT findings were diverse, with pulmonary nodules as the main manifestation, which had also been mentioned in several previous case reports (Urbanski et al., 2012; Zhang and Xu, 2021). Nevertheless, according to the histopathological results, a considerable number of pulmonary nodules were identified as tumors, most of which were early-stage lung adenocarcinoma. In the long-term follow-up survey of WD patients with a small sample size (35 patients) by Schiepatti et al., 22% (7 patients) had preneoplastic/neoplastic disorders (Schiepatti et al., 2020). At present, the risk factors for the occurrence of pulmonary nodules are not clear. We speculate that there may be a relationship between lung lesions and T. whipplei. Whether the presence of T. whipplei in the lung microbiome may promote the occurrence of pulmonary nodules or even early-stage lung cancer directly or indirectly by changing the immune microenvironment still needs to be further confirmed by prospective studies with a larger sample size.
In this study, duodenal mucosal biopsy was performed in a small number of patients, and PAS staining was not performed in suspected patients, so WD was not clearly diagnosed. As WD is a multisystem disease, untreated WD is fatal (Marth and Raoult, 2003). Therefore, we believe that duodenal mucosal biopsy, PAS staining, and immunohistochemistry can be considered for patients suspected of T. whipplei infection to further improve the diagnosis. PCR in sterile body fluids, such as blood, cerebrospinal fluid, and joint fluid, can help diagnose WD, but most of them are invasive procedures, which still need to be evaluated according to the condition. This study also had limitations. It was a single-center retrospective descriptive study without longitudinal follow-up, and partial information was not comprehensive. No control group was established, so correlation and causality could not be determined. But we believe our study is informative.
In summary, this study objectively analyzed the clinical characteristics of patients with positive T. whipplei detected by mNGS in BALF. Our study adds to the evidence that T. whipplei in BALF may be related to some lung diseases. T. whipplei should be considered as a potential risk factor for pneumonia when detected in immunocompromised patients. For non-immunocompromised patients, the detection of T. whipplei also needs attention because of the relationship between T. whipplei and pulmonary nodules. mNGS has improved the detection and attention of rare pathogens. Moreover, the infection, colonization, and prognosis of T. whipplei in the lung need further study.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by the Ethics Committee of the Fifth Affiliated Hospital of Sun Yat-Sen University (approve number [2022] K82-1). Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
ML participated in data collection, analysis, and manuscript drafting. KW participated in drafting the work, acquisition, and analysis. LQ and YL participated in data sorting and input. CTu, MC, ZW, JW, YH, and CTa participated in sample collection. QC and XZ participated in the conception and design of the work. JL conceived of the study, and participated in its design, and helped to draft the manuscript. All authors contributed to the article and approved the submitted version.
JL was grant from Guangdong Basic and Applied Basic Research Foundation (2020A1515011147). ML was supported by Open project of Key Laboratory of Tropical Disease Control (Sun Yat-sen University), Ministry of Education (2020kfkt04).
We are grateful to BGI for their assistance in detection methodology.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.961297/full#supplementary-material
Bousbia, S., Papazian, L., Auffray, J. P., Fenollar, F., Martin, C., Li, W., et al. (2010). Tropheryma whipplei in patients with pneumonia. Emerg. Infect. Dis. 16 (2), 258–263. doi: 10.3201/eid1602.090610
Desnues B, Lepidi, H., Raoult, D., Mege, J. L. (2005). Whipple Disease: intestinal infiltrating cells exhibit a transcriptional pattern of M2/alternatively activated macrophages. J. Infect. Dis. 192 (9), 1642–1646. doi: 10.1086/491745
Dolmans, R. A., Boel, C. H., Lacle, M. M., Kusters, J. G. (2017). Clinical manifestations, treatment, and diagnosis of tropheryma whipplei infections. Clin. Microbiol. Rev. 30 (2), 529–555. doi: 10.1128/CMR.00033-16
Elchert, J. A., Mansoor, E., Abou-Saleh, M., Cooper, G. S. (2019). Epidemiology of whipple's disease in the USA between 2012 and 2017: A population-based national study. Dig Dis. Sci. 64 (5), 1305–1311. doi: 10.1007/s10620-018-5393-9
Fenollar, F., Ponge, T., La Scola, B., Lagier, J. C., Lefebvre, M., Raoult, D. (2012). First isolation of tropheryma whipplei from bronchoalveolar fluid and clinical implications. J. Infect. 65 (3), 275–278. doi: 10.1016/j.jinf.2011.11.026
Garcia-Alvarez, L., Perez-Matute, P., Blanco, J. R., Ibarra, V., Oteo, J. A. (2016). High prevalence of asymptomatic carriers of tropheryma whipplei in different populations from the north of Spain. Enferm Infecc Microbiol. Clin. 34 (6), 340–345. doi: 10.1016/j.eimc.2015.09.006
GH, W. (1907). A hitherto undescribed disease characterized anatomically by deposits of fat and fatty acids in the intestinal and mesenteric lymphatic tissues. Bull. Johns Hopkins Hosp 18, 382–393.
Guo, Y., Li, L., Li, Z., Sun, L., Wang, H. (2021). Tropheryma whipplei detection by nanopore sequencing in patients with interstitial lung disease. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.760696
Keita, A. K., Dubot-Peres A, Phommasone, K., Sibounheuang, B., Vongsouvath, M., Mayxay, M., et al. (2015). High prevalence of tropheryma whipplei in lao kindergarten children. PLoS Negl. Trop. Dis. 9 (2), e0003538. doi: 10.1371/journal.pntd.0003538
Lagier, J.-C., Fenollar, F., Raoult, D. (2017). Acute infections caused by tropheryma whipplei. Future Microbiol. 2017 (12), 247–254. doi: 10.2217/fmb-2017-0178
Lagier, J. C., Papazian, L., Fenollar, F., Edouard, S., Melenotte, C., Laroumagne, S., et al. (2016). Tropheryma whipplei DNA in bronchoalveolar lavage samples: A case control study. Clin. Microbiol. Infect. 22 (10), 875–879. doi: 10.1016/j.cmi.2016.07.010
Li, H., Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinf. (Oxford England) 25 (14), 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, W., Zhang, Q., Xu, Y., Zhang, X., Huang, Q., Su, Z. (2021). Severe pneumonia in adults caused by tropheryma whipplei and candida sp. infection: A 2019 case series. BMC Pulm Med. 21 (1), 29. doi: 10.1186/s12890-020-01384-4
Lozupone, C., Cota-Gomez, A., Palmer, B. E., Linderman, D. J., Charlson, E. S., Sodergren, E., et al. (2013). Widespread colonization of the lung by tropheryma whipplei in HIV infection. Am. J. Respir. Crit. Care Med. 187 (10), 1110–1117. doi: 10.1164/rccm.201211-2145OC
Marth, T., Raoult, D. (2003). Whipple's disease. Lancet (London England) 361 (9353), 239–246. doi: 10.1016/s0140-6736(03)12274-x
Moos, V., Schmidt, C., Geelhaar, A., Kunkel, D., Allers K, Schinnerling, K., et al. (2010). Impaired immune functions of monocytes and macrophages in whipple's disease. Gastroenterology 138 (1), 210–220. doi: 10.1053/j.gastro.2009.07.066
Relman, D. A., Schmidt, T. M., MacDermott, R. P., Falkow, S. (1992). Identification of the uncultured bacillus of whipple's disease. New Engl. J. Med. 327 (5), 293–301. doi: 10.1056/nejm199207303270501
Schiepatti, A., Nicolardi, M. L., Marone, P., Biagi, F. (2020). Long-term morbidity and mortality in whipple's disease a single-center experience over 20 years. Future Microbiol. 15), 847–854. doi: 10.2217/fmb-2019-0315
Schinnerling, K., Moos, V., Geelhaar, A., Allers K, Loddenkemper, C., Friebel, J., et al. (2011). Regulatory T cells in patients with whipple's disease. J. Immunol. 187 (8), 4061–4067. doi: 10.4049/jimmunol.1101349
Schoedon, G., Goldenberger, D., Forrer, R., Gunz, A., Dutly, F., Höchli, M., et al. (1997). Deactivation of macrophages with interleukin-4 is the key to the isolation of tropheryma whippelii. J. Infect. Diseases 176 (3), 672–677. doi: 10.1086/514089
Schoniger-Hekele, M., Petermann, D., Weber, B., Muller, C. (2007). Tropheryma whipplei in the environment: survey of sewage plant influxes and sewage plant workers. Appl. Environ. Microbiol. 73 (6), 2033–2035. doi: 10.1128/AEM.02335-06
Urbanski, G., Rivereau, P., Artru, L., Fenollar, F., Raoult, D., Puechal, X. (2012). Whipple Disease revealed by lung involvement: A case report and literature review. Chest 141 (6), 1595–1598. doi: 10.1378/chest.11-1812
Yan, J., Zhang, B., Zhang, Z., Shi, J., Liu, S., Qi, J., et al. (2021). Case report: Tropheryma whipplei hide in an AIDS patient with pneumocystis pneumonia. Front. Public Health 9. doi: 10.3389/fpubh.2021.663093
Zhang, W. M., Xu, L. (2021). Pulmonary parenchymal involvement caused by tropheryma whipplei. Open Med. (Wars) 16 (1), 843–846. doi: 10.1515/med-2021-0297
Keywords: Tropheryma whipplei, metagenomic next-generation sequencing, bronchoalveolar lavage fluid, Mycobacterium tuberculosis complex, pulmonary nodules
Citation: Lin M, Wang K, Qiu L, Liang Y, Tu C, Chen M, Wang Z, Wu J, Huang Y, Tan C, Chen Q, Zheng X and Liu J (2022) Tropheryma whipplei detection by metagenomic next-generation sequencing in bronchoalveolar lavage fluid: A cross-sectional study. Front. Cell. Infect. Microbiol. 12:961297. doi: 10.3389/fcimb.2022.961297
Received: 04 June 2022; Accepted: 29 July 2022;
Published: 17 August 2022.
Edited by:
Jinmin Ma, Beijing Genomics Institute (BGI), ChinaReviewed by:
Chunling Dong, Second Affiliated Hospital of Jilin University, ChinaCopyright © 2022 Lin, Wang, Qiu, Liang, Tu, Chen, Wang, Wu, Huang, Tan, Chen, Zheng and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Liu, bGl1amluZzI1QHN5c3UuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.