- 1Laboratory of Veterinary Hygiene, School/Faculty of Veterinary Medicine, Hokkaido University, Sapporo, Japan
- 2Graduate School of Infectious Diseases, Hokkaido University, Sapporo, Japan
- 3Department of Microbiology, Sapporo Medical University School of Medicine, Sapporo, Japan
- 4Department of Veterinary Internal Medicine, Tottori University, Tottori, Japan
- 5Laboratory of Food Microbiology and Food Safety, Department of Health and Environmental Sciences, School of Veterinary Medicine, Rakuno Gakuen University, Ebetsu, Japan
Transmission of colistin-resistant Enterobacterales from companion animals to humans poses a clinical risk as colistin is a last-line antimicrobial agent for treatment of multidrug-resistant Gram-negative bacteria including Enterobacterales. In this study, we investigated the colistin susceptibility of 285 Enterobacterales (including 140 Escherichia coli, 86 Klebsiella spp., and 59 Enterobacter spp.) isolated from companion animals in Japan. We further characterized colistin-resistant isolates by multilocus sequence typing (MLST), phylogenetic analysis of hsp60 sequences, and population analysis profiling, to evaluate the potential clinical risk of companion animal-derived colistin-resistant Enterobacterales to humans in line with the One Health approach. All E. coli isolates were susceptible to colistin, and only one Klebsiella spp. isolate (1.2%, 1/86 isolates) was colistin resistant. Enterobacter spp. isolates were frequently colistin resistant (20.3%, 12/59 isolates). In colistin-resistant Enterobacter spp., all except one isolate exhibited colistin heteroresistance by population analysis profiling. These colistin-heteroresistant isolates belonged to clusters I, II, IV, VIII, and XII based on hsp60 phylogeny. MLST analysis revealed that 12 colistin-resistant Enterobacter spp. belonged to the Enterobacter cloacae complex; five Enterobacter kobei (four ST591 and one ST1577), three Enterobacter asburiae (one ST562 and two ST1578), two Enterobacter roggenkampii (ST606 and ST1576), and Enterobacter hormaechei (ST1579) and E. cloacae (ST765) (each one strain). Forty-two percent of the colistin-resistant E. cloacae complex isolates (predominantly ST562 and ST591) belonged to lineages with human clinical isolates. Four E. kobei ST591 isolates were resistant to third-generation cephalosporines, aminoglycosides, and fluroquinolones but remained susceptible to carbapenems. In conclusion, our study is the first to our knowledge to report the frequent isolation of the colistin-resistant E. cloacae complex from companion animals. Furthermore, a subset of isolates belonged to human-associated lineages with resistance to multiple classes of antibiotics. These data warrant monitoring carriage of the colistin-resistant E. cloacae complex in companion animals as part of a domestic infection control procedure in line with the One Health approach.
Introduction
Colistin is a last-line antimicrobial agent used for treatment of multidrug-resistant Gram-negative bacteria, including Enterobacterales (Falagas and Kasiakou, 2005). Spread of colistin resistance therefore poses a major therapeutic problem for the treatment of human infections. The plasmid-mediated colistin resistance gene, mcr, has spread worldwide (Hussein et al., 2021). mcr encodes a phosphoethanolamine transferase and reduces colistin susceptibility by modifying the lipid A component of lipopolysaccharide with phosphoethanolamine (Hussein et al., 2021). The mcr variants from mcr-1 to mcr-10 have been known, whereas few countries have reported mcr-9 and mcr-10 (Wang et al., 2020). Recently, we identified mcr-9 and mcr-10 in Enterobacter asburiae and Enterobacter roggenkampii, respectively, from companion animals in Japan (Sato et al., 2021a; Sato et al., 2021b). These genes were encoded by plasmids with high sequence similarity to those from human clinical isolates (Sato et al., 2021a; Sato et al., 2021b). These observations suggested that horizontal transfer of the plasmid encoding mcr-9 and mcr-10 occurs between isolates from humans and companion animals, which may pose a major risk to human health. However, the overall risk of transmission of colistin-resistant Enterobacterales from companion animals to humans was unclear, partly because of an incomplete characterization of carriage of these organisms in companion animals.
In this study, we investigated the colistin susceptibility of Enterobacterales isolates derived from companion animals in Japan and characterized colistin-resistant isolates using molecular epidemiological analysis.
Materials and Methods
Bacterial Isolates
Our bacterial collection encompassed 285 Enterobacterales [140 Escherichia coli, 86 Klebsiella spp. (84 Klebsiella pneumoniae and two Klebsiella quasipneumoniae), and 59 Enterobacter spp. (51 Enterobacter cloacae complex and eight Enterobacter aerogenes)]. These isolates were derived from clinical specimens of 285 companion animals (211 dogs and 74 cats) from 2003 to 2016 at veterinary clinics throughout Japan (Harada et al., 2012; Harada et al., 2016; Harada et al., 2017). This study was carried out in accordance with the Guidelines for Proper Conduct of Animal Experiments of the Science Council of Japan (Harada et al., 2012; Harada et al., 2016; Harada et al., 2017). Bacterial strains were isolated as part of a routine veterinary examination without invasive operations on the companion animals.
Antimicrobial Susceptibility
Susceptibility to colistin was determined using the broth microdilution method according to the Clinical Laboratory Standard Institution (CLSI) guidelines (Clinical and Laboratory Standards Institute, 2020). E. coli strain ATCC 25922 was used as a reference. Isolates were classified as resistant if their colistin minimum inhibitory concentrations (MIC) was greater than 4 mg/l according to CLSI breakpoint guidelines (Clinical and Laboratory Standards Institute, 2020). To evaluate the “skip-well” phenomenon, bacterial density in each well of the microbroth dilution plate was determined by measuring their optical density at 600nm (OD600nm) using a TECAN Infinite M200 PRO microplate reader (Tecan, Kawasaki, Japan). OD600nm values of less than 0.1 were considered as no growth. Antimicrobial susceptibilities of a subset of isolates to cefotaxime, ceftazidime, meropenem, gentamicin, and ciprofloxacin were reported previously (Harada et al., 2017).
Heat Shock Protein (hsp60) Phylogeny
The hsp60 phylogeny was determined as previously reported (Hoffmann and Roggenkamp, 2003). Nucleotide sequences were aligned using MAFFT (Katoh et al., 2019), and MEGA X (Kumar et al., 2018; Stecher et al., 2020) was used to generate a phylogenetic tree using the Neighbor-Joining method (Saitou and Nei, 1987). The reference strains of each cluster were used as previously reported (Hoffmann and Roggenkamp, 2003).
Genome Sequencing and Identification of STs and Antimicrobial Resistance Genes
Genomic DNA was isolated using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). Genomic libraries were prepared by Nextera DNA Flex Library Prep for Illumina sequencing according to the manufacturer’s instructions (Illumina, San Diego, CA). Genome sequencing was performed using the HiSeq 2000 instrument (Illumina). Filtered subreads (in fastq format) were de novo assembled with 300-bp paired-end reads after trimming using the CLC Genomic Workbench (Qiagen). The sequence data (in.fasta format) were used to identify E. cloacae complex species by KmerFinder 3.2 and sequence types (STs) by multilocus sequence typing (MLST) analysis. Antimicrobial resistance genes were identified using ResFinder from the Center for Genomic Epidemiology (http://www.genomicepidemiology.org).
Population Analysis Profiling
An evaluation of colistin heteroresistance by population analysis profiling (PAP) was carried out as previously described (Guerin et al., 2016). Briefly, isolates were inoculated into 1 ml of Tryptic soy broth (Becton Dickinson GmbH, Heidelberg, Germany) and incubated at 37°C overnight. One hundred microliters of each serial dilution was plated on Mueller Hinton II (MHII) agar plates (Becton Dickinson GmbH) supplemented with 0, 0.125, 0.5, or 8 mg/l of colistin and cultured for 20 h at 37°C.
After counting the number of colony-forming units (cfu), the frequency of the population displaying colistin resistance was calculated by dividing the number of cfu on colistin-containing MHII agar by the cfu on plain-MHII agar. Colistin heteroresistance was determined using a previously published method based on the following criteria (Andersson et al., 2019). 1) Bacterial growth was confirmed in >4 mg/l of colistin by microbroth dilution, and 2) the colistin concentration exhibiting the highest inhibitory effect on the growth of the resistant subpopulation (with a frequency range from 1 × 10-5 to 1 × 10-1) was at least eight-fold greater (= 8 mg/l of colistin) than the highest colistin concentration exhibiting no effect on the growth of the colistin-susceptible population (= 0.125 or 0.5 mg/l of colistin) as described in previous studies (Guerin et al., 2016; Juhasz et al., 2017).
Results
Colistin Susceptibility in Enterobacterales spp. Derived From Companion Animals
All E. coli isolates tested were susceptible to colistin, with MIC ranging from ≤0.125 to 0.5 mg/l (MIC50 and MIC90 were 0.125 and 0.25 mg/l, respectively) (Figure 1). In Klebsiella spp., only one isolate of K. pneumoniae (1.2%, 1/86 isolates) was colistin resistant with MICs ranging from ≤0.125 to >4 mg/l (MIC50 and MIC90 were both 0.25 mg/l). Instead, the colistin MICs of Enterobacter spp. ranged from 0.25 to >4 mg/l (MIC50 and MIC90, 0.25 and >4 mg/l, respectively), and the frequency of colistin resistance was significantly higher (20.3%, 12/59 isolates) than that of E. coli and Klebsiella spp. (p < 0.05). All of the 12 colistin-resistant isolates belonged to the E. cloacae complex including Enterobacter kobei (8.5%), E. asburiae (5.1%), E. roggenkampii (3.4%), Enterobacter hormaechei, and E. cloacae (each 1.7%), and no colistin-resistant E. aerogenes was observed. Six colistin-resistant Enterobacter isolates (En3, En4, En4, En30, En42, and En50 strains) exhibited a “skip-well” phenotype whereby they grew in the presence of 2 mg/l colistin, but not within the range of 0.25 to 1 mg/l colistin (Figure 2).
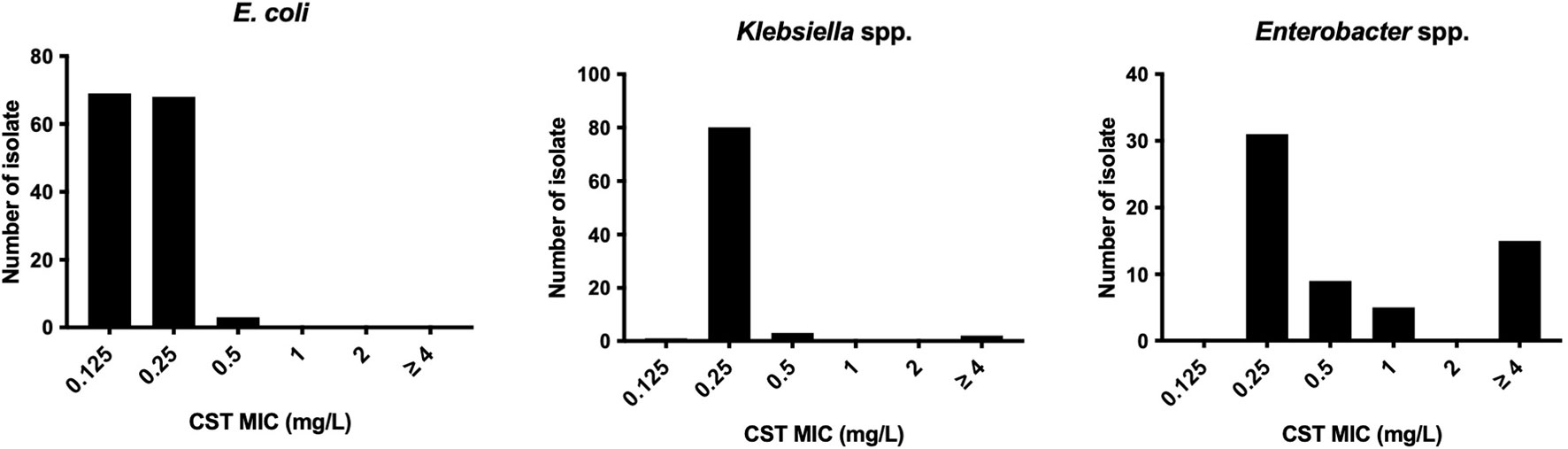
Figure 1 Colistin susceptibility in E. coli, Klebsiella spp., and Enterobacter spp. clinical isolates derived from companion animals.
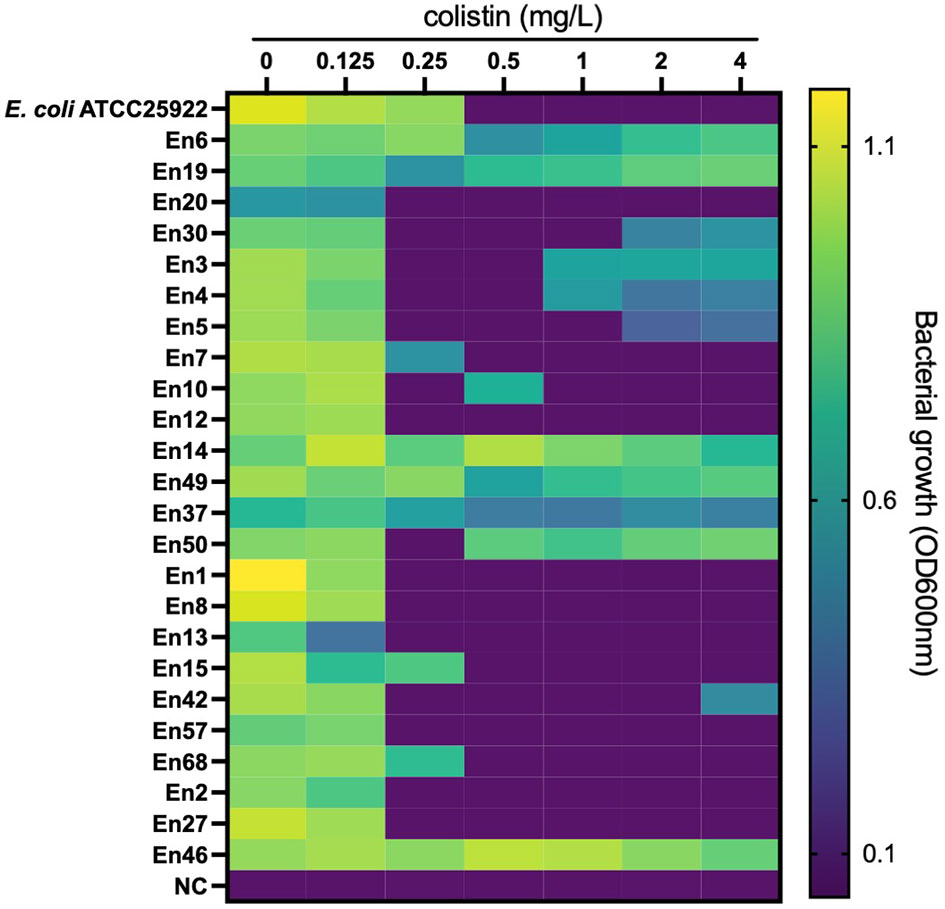
Figure 2 Bacterial growth in colistin-containing medium to evaluate the ‘skip-well’ phenomenon in Enterobacter spp. Isolates This method was identical to the broth microdilution method described in the CLSI guidelines. Bacterial growth was determined as turbidity of the medium in cation-adjusted MHII broth at the experimental endpoint (after an 18-h cultivation at 37°C in a 96-well plate). OD600nm was measured every 10 min for 18 h. An OD600nm of <0.1 was defined as no growth.
Antimicrobial Susceptibility and Detection of Antimicrobial Resistance Genes
According to clinical breakpoints, 50.0% (6 isolates) and 41.6% (n = 5 isolates) of 12 colistin-resistant E. cloacae complex isolates exhibited resistance to fluoroquinolones (ciprofloxacin) and third-generation cephalosporins (cefotaxime and ceftazidime), and aminoglycosides (gentamicin), respectively, whereas they remained susceptible to carbapenem (meropenem) (Table 1). Furthermore, 33.3% (4/12 isolates, En3, En4, En5, and En14) of the colistin-resistant isolates were co-resistant to ciprofloxacin, third-generation cephalosporins, and gentamicin.
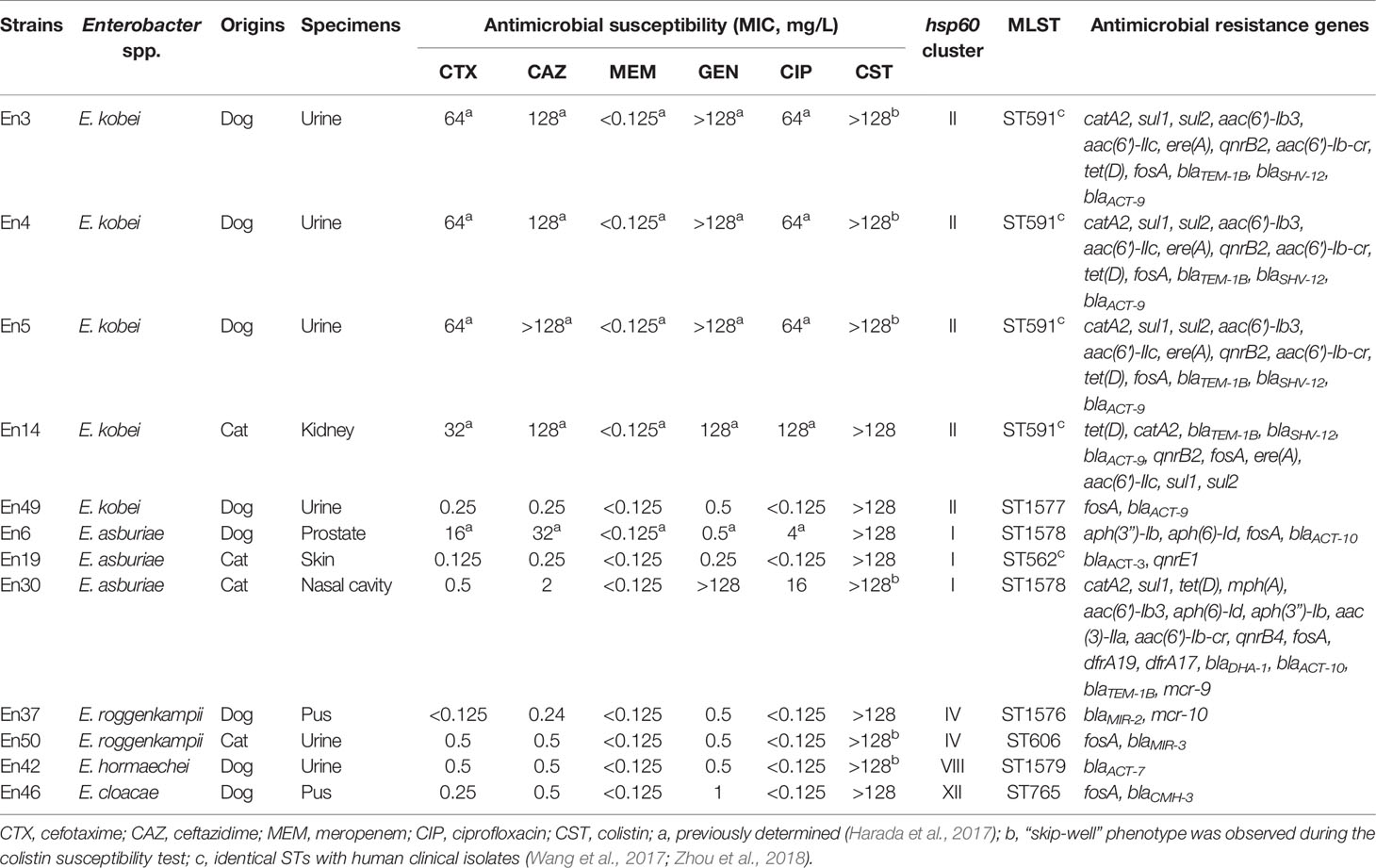
Table 1 Characteristics of colistin-resistant Enterobacter cloacae complex isolates derived from companion animals.
Using ResFinder, we found that two colistin-resistant E. cloacae complex isolates possessed mcr-9 (E. asburiae En30) (Sato et al., 2021a) and mcr-10 (E. roggenkampii En37) (Sato et al., 2021b), as previously reported. Other colistin-resistant E. cloacae complex isolates did not possess mcr genes. E. kobei En3, En4, En5, and En14 possess the extended-spectrum β-lactamase gene blaSHV-12, in addition to the class A and ampC β-lactamase genes blaTEM-1B and blaACT-9, respectively (Table 1). Additional β-lactamase genes (blaACT-3, blaACT-7, blaACT-10, blaDHA-1, blaMIR-2, blaMIR-3, blaCMH-3) were detected in other colistin-resistant isolates. The quinolone resistance gene qnrB2 was detected in five colistin-resistant E. cloacae complex isolates, and qnrE1 was also detected in the colistin-resistant strain (En19). The aminoglycoside resistance genes, aph(3″)-Ib, aph(6)-Id, aac(3)-IIa, aac(6′)-Ib3, aac(6′)-IIc, and/or aac(6′)-Ib-cr, were also detected in colistin-resistant E. cloacae complex isolates (Table 1).
hsp60 Phylogeny and MLST Analysis
Using hsp60 phylogeny, the Enterobacter spp. isolates were found to belong to 10 clusters (I, II, III, IV, V, VI, VIII, XI, XII, XIII) (Figure 3). Among them, colistin-resistant isolates belonged to five clusters (I, II, IV, VIII, and XII). Cluster II contained the majority of colistin-resistant isolates (5/12 colistin-resistant isolates) followed by clusters I (three isolates), IV (two isolates), and VIII, and XII (one strain each). Profiling by MLST analysis revealed that four and a colistin-resistant E. kobei isolate belonging to cluster II were ST591 and ST1577, respectively (Table 1). Colistin-resistant isolates of cluster I contained E. asburiae ST562 and ST1578, and those of cluster IV were E. roggenkampii ST606 and ST1576. Colistin-resistant isolates from clusters VIII and XII consisted of E. hormaechei ST1579 and E. cloacae ST765, respectively.
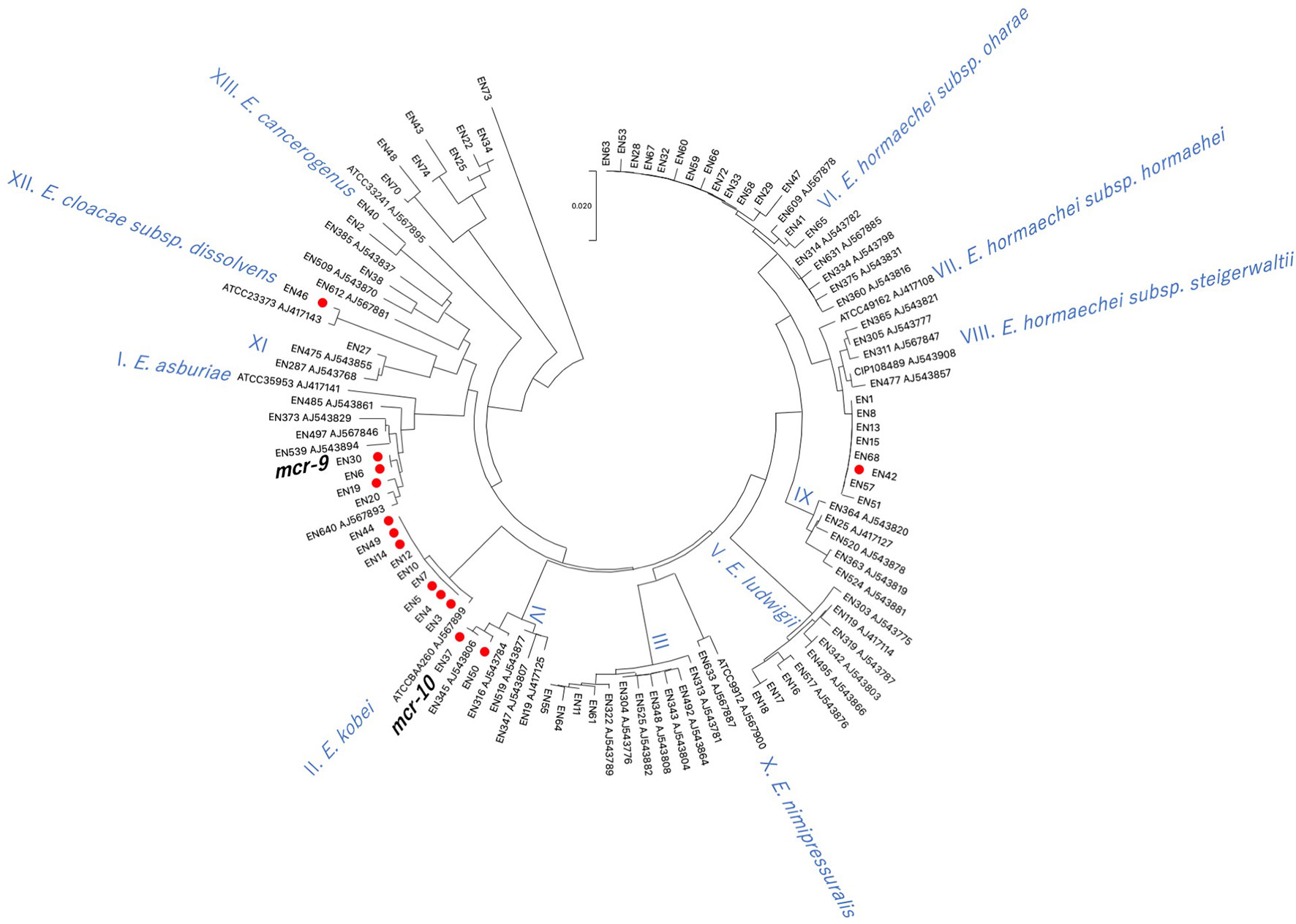
Figure 3 Phylogenetic tree based on hsp60 sequences in Enterobacter spp. isolates derived from companion animals Red circles indicate colistin-resistant isolates. Reference strains are denoted by their respective accession numbers.
PAP of the Colistin-Resistant Population
To evaluate a potential colistin heteroresistance phenotype, we calculated the frequency of colistin resistance in populations of Enterobacter spp. isolates using PAP (Figure 4). All the colistin-resistant Enterobacter spp. isolates exhibited a colistin-resistant population with a greater than 1 × 10-5 frequency. The frequency of the colistin-resistant subpopulation of 11 colistin-resistant Enterobacter spp. isolates ranged between 1.8 × 10-4 and 2.6 × 10-2. The frequency of the colistin-resistant population in the stably resistant strain En46 was 8.0 × 10-1 ± 2.0 × 10-2. Conversely, the frequency of colistin-resistant populations in colistin-susceptible isolates was lower than 1 × 10-5.
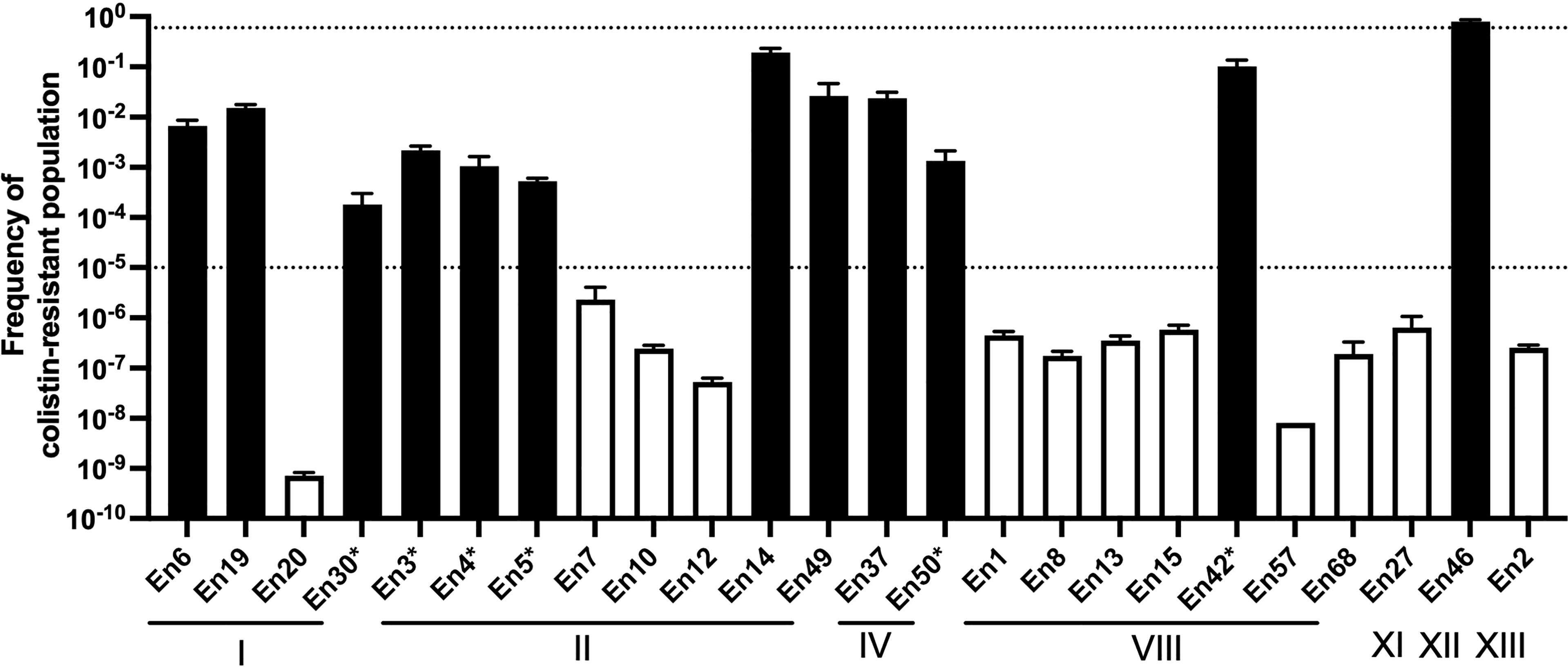
Figure 4 Population analysis profiling of colistin-resistant E. cloacae complex isolate-derived companion animals Broken lines indicate the frequency of colistin-resistant subpopulations (1 × 10-5) that should influence the colistin susceptibility test (after inoculation of 5 × 105 cfu/ml) as determined by CLSI guidelines (Clinical and Laboratory Standards Institute, 2020). Black and white bars indicate the frequencies of colistin-resistant populations in colistin-resistant and colistin-susceptible isolate cultures, respectively. *, isolates exhibited a “skip-well” phenomenon in A. Roman numerals indicate clusters by hsp60 phylogeny.
Discussion
In this study, we investigated the colistin susceptibility of Enterobacterales derived from companion animals. To the best of our knowledge, this is the first report to evaluate the colistin susceptibility of Enterobacter spp. and reveal a high frequency of colistin-resistant E. cloacae complex isolates derived from companion animals, although colistin is not used in companion animal medicine in Japan.
The E. cloacae complex comprises major nosocomial pathogens, which cause opportunistic urinary, respiratory, and bloodstream infections in human (Davin-Regli et al., 2019) as well as in veterinary clinical settings (Harada et al., 2017). The E. cloacae complex is also clinically important as one of “ESKAPE” (Rice, 2008) pathogens due to its high frequency and multidrug resistance. These observations warrant careful monitoring of the E. cloacae complex as acquisition of colistin resistance may leave very few therapeutic options in the clinic. No colistin-resistant E. cloacae complex isolates characterized in our study were resistant to carbapenems (Harada et al., 2017). However, some colistin-resistant isolates were resistant to third-generation cephalosporins (cefotaxime and ceftazidime) due to the harboring of the genes encoding ESBL (blaSHV-12) and/or AmpC (blaACT-9 and blaACT-10) β-lactamases, in addition to co-resistance to fluoroquinolone and aminoglycoside. Although it is difficult to discuss that this prevalence was high or not because antimicrobial susceptibility and the antimicrobial resistance genes have not been well elucidated by using a sufficient number of Enterobacter spp. isolates from companion animals, these colistin-resistant isolates (E. kobei En3, En4, En5, and En14) may pose a significant risk due to a lack of effective treatment options.
Colistin-resistant E. cloacae complex isolates derived from human clinical specimens exhibit heteroresistance (Guerin et al., 2016). Colistin-heteroresistant isolates harbor the colistin-resistant subpopulation at a certain frequency (frequency of between 1 × 10-5 and 1 × 10-1) (Andersson et al., 2019), and a skip-well phenomenon is typically observed in some cases of colistin-heteroresistant isolates derived from human clinical settings (Guerin et al., 2016; Kang et al., 2019). Consistent with this, all colistin-resistant E. cloacae complex isolates, except one strain (En46; frequency of colistin-resistant subpopulation was 8.0 × 10-1), harbored colistin-resistant subpopulations at a frequency of between 1 × 10-5 and 1 × 10-1 by PAP, and some colistin-resistant isolates exhibited the “skip-well” phenomenon in the microbroth dilution method. These suggest that like the human clinical context, E. cloacae complex isolates from companion animals also exhibit colistin heteroresistance. Most isolates that exhibited the skip-well phenomenon displayed a colistin-resistant subpopulation at a frequency of between 10-3 and 10-4. This frequency was approximately 10-fold lower than other colistin-heteroresistant isolates that grew at all tested colistin concentrations, suggesting that the skip-well phenomenon reflects the density of colistin-resistant populations.
Previous studies demonstrated that infection of mice with colistin-heteroresistant E. cloacae led to colistin treatment failure (Band et al., 2016). Thus, transmission of colistin-heteroresistant E. cloacae complex isolates from companion animals to humans poses a potential risk to human health. Previous studies demonstrated that colistin-heteroresistant E. cloacae complex isolates derived from human clinical settings belonged to distinct clusters (I, II, IV, IX, X, XI, and XII) based on hsp60 phylogeny (Guerin et al., 2016). In this study, four (I, II, IV, and XII) of the five identified clusters of colistin-resistant isolates were identical to those of human origin. In addition, the sequence types, E. kobei ST591 (En3, En4, and En5; hsp60 phylogeny cluster II) (Zhou et al., 2018) and E. asburiae ST562 (En19; hsp60 phylogeny cluster I) (Wang et al., 2017), identified in this study have been reported in the human clinical setting. Carriage of resistance to multiple classes of antimicrobial agents in E. kobei ST591 isolates (En3, En4 and En5) suggests the risk of transmission to humans where there may be few treatment options. Therefore, careful monitoring of transmission of colistin-resistant E. cloacae complex isolates between companion animals and humas is required in line with the One Health approach.
Previous studies have reported that the mechanisms underpinning colistin heteroresistance in E. cloacae complex strains are encoded on the chromosome (Kang et al., 2019). The two-component system PhoPQ causes ArnT to modify lipid A of the bacterial lipopolysaccharide with a 4-amino-4-deoxy-L-arabinose, resulting in colistin heteroresistance (Kang et al., 2019). However, the mechanism underpinning colistin heteroresistance in strains isolated from companion animals remains unclear and requires further investigation. Furthermore, the influence of mcr on colistin heteroresistance in E. cloacae requires further study, as one E. asburiae (En30) and one E. roggenkampii (En37) isolate possessed mcr-9 and mcr-10, respectively (Sato et al., 2021a; Sato et al., 2021b) but had no significant colistin-resistant subpopulation frequency compared with other non-mcr-possessing isolates from the same hsp60 phylogenetic cluster.
In conclusion, this study is the first to demonstrate the high-frequency isolation of colistin-resistant E. cloacae complex strains from companion animals in Japan. This finding provides an enhanced understanding of the spread of colistin-resistant bacteria in companion animals and highlights a strategy to prevent the spread of resistant isolates in small animal clinics and domestically. Clustering of human and companion animal-associated colistin-resistant E. cloacae complex isolates with resistance to multiple classes of antimicrobial agents by hsp60 phylogeny and STs suggests the possibility of a frequent transmission between humans and companion animals. These data highlight the need for a One Health approach in tackling the spread of drug-resistant pathogens.
Data Availability Statement
The data presented in the study are deposited in the DDBJ/ENA/GenBank repository, accession number DRX366470 - DRX366481.
Ethics Statement
The animal study was reviewed and approved by Guidelines for Proper Conduct of Animal Experiments of the Science Council of Japan. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
TS is responsible for the study design. KH and MU designed this study. KH assisted with sample collection. MU assisted in data analysis. SY and MH completed the written report. All authors contributed to the article and approved the submitted versions.
Funding
This work was supported by grants from the Japan Agency for Medical Research and Development (AMED) (JP20ak0101118h0002). This work was also partly supported by a grant from JSPS KAKENHI (JP21H03622).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Andersson, D. I., Nicoloff, H., Hjort, K. (2019). Mechanisms and Clinical Relevance of Bacterial Heteroresistance. Nat. Rev. Microbiol. 17 (8), 479–496. doi: 10.1038/s41579-019-0218-1
Band, V. I., Crispell, E. K., Napier, B. A., Herrera, C. M., Tharp, G. K., Vavikolanu, K., et al. (2016). Antibiotic Failure Mediated by a Resistant Subpopulation in Enterobacter Cloacae. Nat. Microbiol. 1 (6), 16053. doi: 10.1038/nmicrobiol.2016.53
Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. CLSI supplement M100. 30th ed. Wayne, P. A. CLSI; 2020
Davin-Regli, A., Lavigne, J. P., Pages, J. M. (2019). Enterobacter Spp.: Update on Taxonomy, Clinical Aspects, and Emerging Antimicrobial Resistance. Clin. Microbiol. Rev. 32 (4), e00002–19. doi: 10.1128/CMR.00002-19
Falagas, M. E., Kasiakou, S. K. (2005). Colistin: The Revival of Polymyxins for the Management of Multidrug-Resistant Gram-Negative Bacterial Infections. Clin. Infect. Dis. 40 (9), 1333–1341. doi: 10.1086/429323
Guerin, F., Isnard, C., Sinel, C., Morand, P., Dhalluin, A., Cattoir, V., et al. (2016). Cluster-Dependent Colistin Hetero-Resistance in Enterobacter Cloacae Complex. J. Antimicrob. Chemother. 71 (11), 3058–3061. doi: 10.1093/jac/dkw260
Harada, K., Niina, A., Nakai, Y., Kataoka, Y., Takahashi, T. (2012). Prevalence of Antimicrobial Resistance in Relation to Virulence Genes and Phylogenetic Origins Among Urogenital Escherichia Coli Isolates From Dogs and Cats in Japan. Am. J. Vet. Res. 73 (3), 409–417. doi: 10.2460/ajvr.73.3.409
Harada, K., Shimizu, T., Mukai, Y., Kuwajima, K., Sato, T., Kajino, A., et al. (2017). Phenotypic and Molecular Characterization of Antimicrobial Resistance in Enterobacter Spp. Isolates From Companion Animals in Japan. PloS One 12 (3), e0174178. doi: 10.1371/journal.pone.0174178
Harada, K., Shimizu, T., Mukai, Y., Kuwajima, K., Sato, T., Usui, M., et al. (2016). Phenotypic and Molecular Characterization of Antimicrobial Resistance in Klebsiella Spp. Isolates From Companion Animals in Japan: Clonal Dissemination of Multidrug-Resistant Extended-Spectrum Beta-Lactamase-Producing Klebsiella Pneumoniae. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01021
Hoffmann, H., Roggenkamp, A. (2003). Population Genetics of the Nomenspecies Enterobacter Cloacae. Appl. Environ. Microbiol. 69 (9), 5306–5318. doi: 10.1128/AEM.69.9.5306-5318.2003
Hussein, N. H., Al-Kadmy, I. M. S., Taha, B. M., Hussein, J. D. (2021). Mobilized Colistin Resistance (Mcr) Genes From 1 to 10: A Comprehensive Review. Mol. Biol. Rep. 48 (3), 2897–2907. doi: 10.1007/s11033-021-06307-y
Juhasz, E., Ivan, M., Pinter, E., Pongracz, J., Kristof, K. (2017). Colistin Resistance Among Blood Culture Isolates at a Tertiary Care Centre in Hungary. J. Glob Antimicrob. Resist. 11, 167–170. doi: 10.1016/j.jgar.2017.08.002
Kang, K. N., Klein, D. R., Kazi, M. I., Guerin, F., Cattoir, V., Brodbelt, J. S., et al. (2019). Colistin Heteroresistance in Enterobacter Cloacae is Regulated by PhoPQ-Dependent 4-Amino-4-Deoxy-L-Arabinose Addition to Lipid A. Mol. Microbiol. 111 (6), 1604–1616. doi: 10.1111/mmi.14240
Katoh, K., Rozewicki, J., Yamada, K. D. (2019). MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief Bioinform. 20 (4), 1160–1166. doi: 10.1093/bib/bbx108
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi: 10.1093/molbev/msy096
Rice, L. B. (2008). Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 197 (8), 1079–1081. doi: 10.1086/533452
Saitou, N., Nei, M. (1987). The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 4 (4), 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sato, T., Usui, M., Harada, K., Fukushima, Y., Nakajima, C., Suzuki, Y., et al. (2021a). Complete Genome Sequence of an Mcr-9-Possessing Enterobacter Asburiae Strain Isolated From a Cat in Japan. Microbiol. Resour Announc 10 (26), e0028121. doi: 10.1128/MRA.00281-21
Sato, T., Usui, M., Harada, K., Fukushima, Y., Nakajima, C., Suzuki, Y., et al. (2021b). Complete Genome Sequence of an Mcr-10-Possessing Enterobacter Roggenkampii Strain Isolated From a Dog in Japan. Microbiol. Resour Announc 10 (30), e0042621. doi: 10.1128/MRA.00426-21
Stecher, G., Tamura, K., Kumar, S. (2020). Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 37 (4), 1237–1239. doi: 10.1093/molbev/msz312
Wang, C., Feng, Y., Liu, L., Wei, L., Kang, M., Zong, Z. (2020). Identification of Novel Mobile Colistin Resistance Gene Mcr-10. Emerg. Microbes Infect. 9 (1), 508–516. doi: 10.1080/22221751.2020.1732231
Wang, S., Xiao, S. Z., Gu, F. F., Tang, J., Guo, X. K., Ni, Y. X., et al. (2017). Antimicrobial Susceptibility and Molecular Epidemiology of Clinical Enterobacter Cloacae Bloodstream Isolates in Shanghai, China. PloS One 12 (12), e0189713. doi: 10.1371/journal.pone.0189713
Keywords: bacteria, Enterobacterales, Enterobacter spp, antimicrobial resistance, colistin
Citation: Sato T, Harada K, Usui M, Yokota SI and Horiuchi M (2022) Colistin Susceptibility in Companion Animal-Derived Escherichia coli, Klebsiella spp., and Enterobacter spp. in Japan: Frequent Isolation of Colistin-Resistant Enterobacter cloacae Complex. Front. Cell. Infect. Microbiol. 12:946841. doi: 10.3389/fcimb.2022.946841
Received: 18 May 2022; Accepted: 06 June 2022;
Published: 06 July 2022.
Edited by:
Piotr Majewski, Medical University of Bialystok, PolandReviewed by:
Mabel Kamweli Aworh, Federal Ministry of Agriculture and Rural Development (FMARD), NigeriaShigeru Kamiya, Kyorin University, Japan
Copyright © 2022 Sato, Harada, Usui, Yokota and Horiuchi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Toyotaka Sato, c2F0by50QHZldG1lZC5ob2t1ZGFpLmFjLmpw
 Toyotaka Sato
Toyotaka Sato Kazuki Harada4
Kazuki Harada4 Masaru Usui
Masaru Usui Shin-ichi Yokota
Shin-ichi Yokota