- 1Department of Microbiology, Yongchuan Hospital of Chongqing Medical University, Chongqing, China
- 2Department of Microbiology, Affiliated Hangzhou Xixi Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 3Department of Microbiology, Yongchuan District Center for Disease Control and Prevention of Chongqing, Chongqing, China
- 4Department of Molecular biology, Jiaxing Maternal and Child Health Hospital, Jiaxing, China
Background: The aim of this study was to clarify the molecular characterization of NDM-1-producing carbapenem-resistant Enterobacter cloacae complex (CREL) at a teaching hospital in Chongqing, China.
Methods: Antimicrobial susceptibility and resistance genes were analyzed. Epidemiological relationship was analyzed by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). Conjugation experiments were performed to determine the transferability of plasmids. Whole-genome sequencing (WGS) of strains was implemented, and the genetic environment of the blaNDM-1- and mcr-9-carrying plasmids was analyzed.
Results: A total of 10 blaNDM-1-positive CREL isolates were identified. All isolates harbored multiple resistance genes. ECL68 and ECL78 co-produce blaNDM-1 and mcr-9. Among the four different sequence types (STs) detected, ST1466 was assigned as a novel ST. Six isolates exhibited highly similar PFGE patterns. Conjugation assay proved that all plasmids containing blaNDM-1 or mcr-9 could be transferred to the recipient Escherichia coli. WGS indicated that blaNDM-1 genes were carried by diverse plasmids, including IncHI2/IncN, IncX3, and one unclassified plasmid type. The backbone structure of these plasmids is involved in replication initiation (repAB), partitioning (parABM), and conjugation/type IV secretion (tra/virB). Analysis of the genetic environment showed that blaNDM-1 in three plasmids exhibited a highly similar structure to protype Tn125. Co-existence of blaNDM-1 and the colistin resistance gene mcr-9 was detected in the two isolates, ECL68 and ECL78. In ECL68, blaNDM-1 and mcr-9 were present on the same plasmid while located in two separate plasmids in ECL78. The genetic environment of mcr-9 was organized as IS26-wbuC-mcr-9-IS903-pcoS-pcoE-rcnA-rcnR, and the two-component system encoding genes qseC and qseB was not found in two plasmids, which could explain mcr-9-harboring strains’ colistin susceptibility.
Conclusions: We first report a nosocomial outbreak of NDM-1-producing E. cloacae complex ST177 in China. Conjugative plasmids contributed to the horizontal transfer of antibiotic resistance genes. The prevalence and even coexistence of blaNDM-1 and mcr-9 may further threaten public health. Our results highlight further surveillance for blaNDM-1, and mcr-9 is essential to prevent its dissemination.
Introduction
In recent years, the prevalence of carbapenem-resistant Enterobacteriaceae (CRE) has become the major reason for clinical anti-infective treatment failure and posed a serious threat to clinical management (Perez and Bonomo, 2019). Infections caused by CRE have a higher chance of having severe clinical consequences compared with other pathogens (Falagas et al., 2014). The main mechanism of carbapenem resistance includes production of carbapenemases, active efflux of bacteria, and mutation of outer membrane proteins (Majewski et al., 2016).
Enterobacter cloacae complex (ECL), a species of the Enterobacteriaceae family, is inherently resistant to first- and second-generation cephalosporin due to chromosomally mediated AmpC β-lactamase (Jacoby, 2009). The ECL bacteria are related to a series of nosocomial infections including pneumonia, urinary tract infections, and septicemia. The overuse of broad-spectrum antibiotics resulted in the emergence of multidrug-resistant and even carbapenem-resistant E. cloacae (CREL) around the world. According to CHINET surveillance data, the detected rate of CREL ranked third among all Enterobacteriaceae, just after carbapenem-resistant Klebsiella pneumoniae and Escherichia coli (Lee et al., 2017; Annavajhala et al., 2019; Tetsuka et al., 2019).
In 2009, New Delhi metallo-β-lactamase-1 (NDM-1), also known as Ambler class B metallo-β-lactamase, was first reported in a clinic K. pneumoniae in India and commonly located in conjugative plasmids (Yong et al., 2009). In the following few years, clinical strains carrying the blaNDM-1 appeared in more than 50 countries (Kumarasamy et al., 2010; Walsh, 2010; Nordmann et al., 2011). This spread and prevalence across species indicates that blaNDM-1 is seriously threatening public health in a new way (Boyd et al., 2020). However, there have been little epidemiological data on NDM-1-producing CREL in our region. Thus, our study aimed to investigate the prevalence of CREL and molecular characteristics of the blaNDM-1-carrying plasmids in Chongqing.
Materials and methods
Collection and identification of bacterial isolates
From 30 June 2018 to 30 June 2020, 10 non-duplicated NDM-1-producing CREL isolates were collected from the Yongchuan Hospital Affiliated of Chongqing Medical University. Bacterial species identification was performed by using the VITEK2 compact automated system (bioMerieux, France). The Enterobacter cloacae complex was further identified to the subspecies level by 16S rRNA as described previously (Mezzatesta et al., 2012). All isolates were preserved at −80°C until further study.
Antimicrobial susceptibility testing
Initial antibiotic susceptibility was analyzed by using the VITEK2 compact system. MICs of imipenem (IPM), meropenem (MEM), levofloxacin (LEV), amikacin (AMK), tigecycline (TGC), and polymyxin B (PB) were evaluated using the broth microdilution method. The results were interpreted according to the interpretive criteria from CLSI 2020. The tigecycline was interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines. E. coli ATCC 25922 served as the quality control strain for susceptibility testing.
Phenotype testing and detection of antibiotic-resistant genes
Phenotypic screening was based on the CLSI 2020 guidelines. The modified carbapenem inactivation method (mCIM) and the EDTA-modified carbapenem inactivation method (eCIM) were used to screen carbapenemase production. The PCR was performed to detect the presence of carbapenemase-related genes (blaKPC, blaNDM, blaVIM, blaIMP, and blaOXA−48). Moreover, ESBLs, AmpC, and resistance genes for fluoroquinolones were determined by using primers as described previously (Gong et al., 2018).
Multilocus sequence typing and pulsed-field gel electrophoresis
Multilocus sequence typing (MLST) was used to detect the sequence types. Seven housekeeping genes (dnaA, fusA, gyrB, leuS, pyrG, rplB, and rpoB) were submitted to the MLST database (https://pubmlst.org/ecloacae/). New allele and profile were approved by the MLST website. Molecular phylogenetic analyses were carried out on MEGA.X software. Moreover, pulsed-field gel electrophoresis (PFGE) was used to further determine the genetic relatedness based on the protocol (Tenover et al., 1995). The DNA patterns were analyzed by BioNumerics software v6.6 (Applied Maths, Kortrijk, Belgium). Isolates were allocated into genetic similarity clusters using an 80% cutoff value (Zhao et al., 2020).
Conjugation experiment
The transferability of resistance gene was determined using a conjugation experiment. As previously described (Wang et al., 2015; Zeng et al., 2021), the conjugation test was performed by the membrane bonding method using NDM-1-producing CREL as the donor and rifampin-resistant E. coli EC600 as the recipient. Briefly, both the donor and the recipient strains were mixed on Luria-Bertani agar at a ratio of 1:2, and the mixtures were incubated at 37°C overnight. Transconjugants were selected on Mueller-Hinton agar plates supplemented with a combination of 1 µg/ml MEM and 600 µg/ml rifampicin. The resistance gene and MIC in the tranconjugants were confirmed by PCR and antimicrobial susceptibility testing.
Whole-genome sequencing and data analysis
Genomic DNA of CREL 68, 72, 78, and 112 isolates were prepared using the MagAttract HMW DNA Kit (Qiagen, Hilden, Germany) and subjected to whole-genome sequencing (WGS) using the HiSeq 2000™ platform (Illumina Inc., San Diego, CA, USA) with 2 × 100-bp paired-end reads and to long-read high-throughput sequencing (LRS) on a MinION platform (Oxford Nanopore Technologies, Oxford, UK). The long read generated by MinION was assembled using Canu v. 1.6 and polished with the short reads generated by HiSeq using Pilon v1.22 to obtain the whole genome and complete plasmid sequences. The genomic sequence was annotated using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) and Glimmer 3.02 (http://www.cbcb.umd.edu/software/glimmer/). Plasmid replicon types were identified using PlasmidFinder (https://cge.cbs.dtu.dk//services/PlasmidFinder/). Antibiotic resistance genes were identified using both the Comprehensive Antibiotic Resistance Database (CARD) and ResFinder database (https://cge.cbs.dtu.dk/services/ResFinder/). Transposon and insertion sequence (IS) elements were identified using the ISfinder database (https://www-is.biotoul.fr/). Linear comparison of sequences was performed using BLAST with the default settings (the nucleotide collection database and the megablast program), and visualized by BRIG (http://brig.sourceforge.net) or Easyfig tools (https://github.com/mjsull/Easyfig) (Zeng et al., 2021).
Nucleotide sequence accession numbers
The complete sequences of plasmids pNDM-068001, pMDR-072001, pNDM-072002, pMCR-078001, pNDM-078002, and pNDM-011201 were submitted to GenBank under accession numbers MZ156799, MZ156800, MZ156801, MZ156802, MZ156803, and MZ156804, respectively.
Results
Characteristics of collected samples
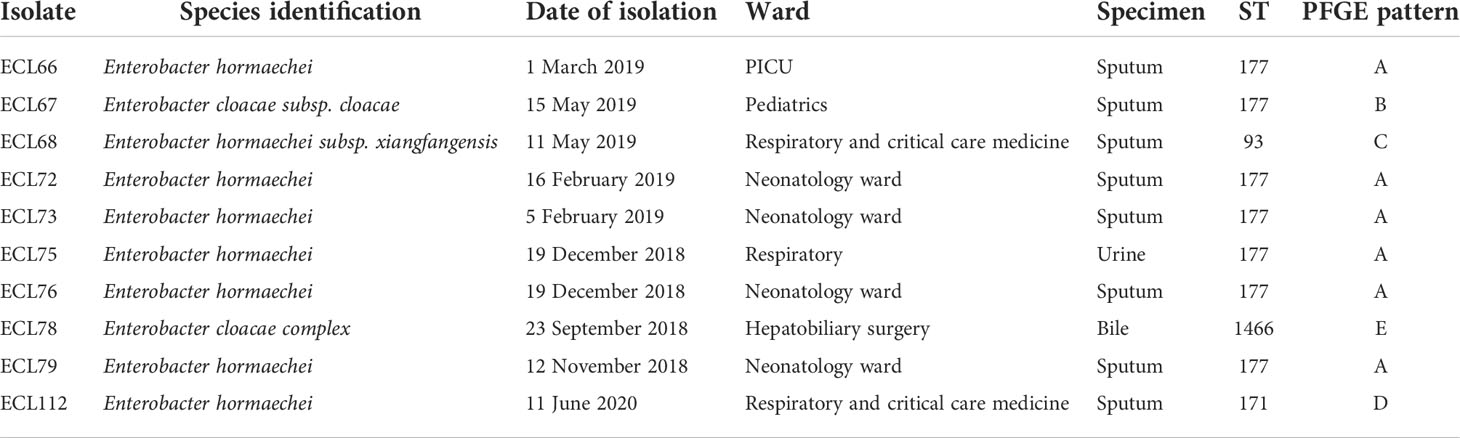
In this study, a total of 10 nonduplicate NDM-1-producing CREL isolates were investigated during the study period. The most common specimens were respiratory tract (8 cases, 80%), followed by urine (1 case, 10%) and bile (1 case, 10%). The majority of patients belonged to the neonatology ward (n = 4, 40.0%), followed by the respiratory and critical care medicine ward (n = 2, 20.0%), pediatric intensive care unit PICU (n = 1, 10.0%), pediatric ward (n = 1, 10.0%), respiratory department (n = 1, 10.0%), and hepatobiliary surgery (n = 1, 10.0%) (Table 1).
Antimicrobial susceptibility tests
All NDM-1-producing CREL isolates were defined as MDR as they were resistant to three or more classes of antimicrobial agents. Of all the antimicrobials tested, the most susceptible antimicrobial was amikacin (90%), followed by tigecycline (60%), polymyxin B (60%), ciprofloxacin (60%), and levofloxacin (40%). All strains showed high resistance rates to piperacillin-tazobactam (100%), ceftazidime (100%), cefepime (100%), and aztreonam (100%). The antibiotic susceptibility profile is shown in Table 2.
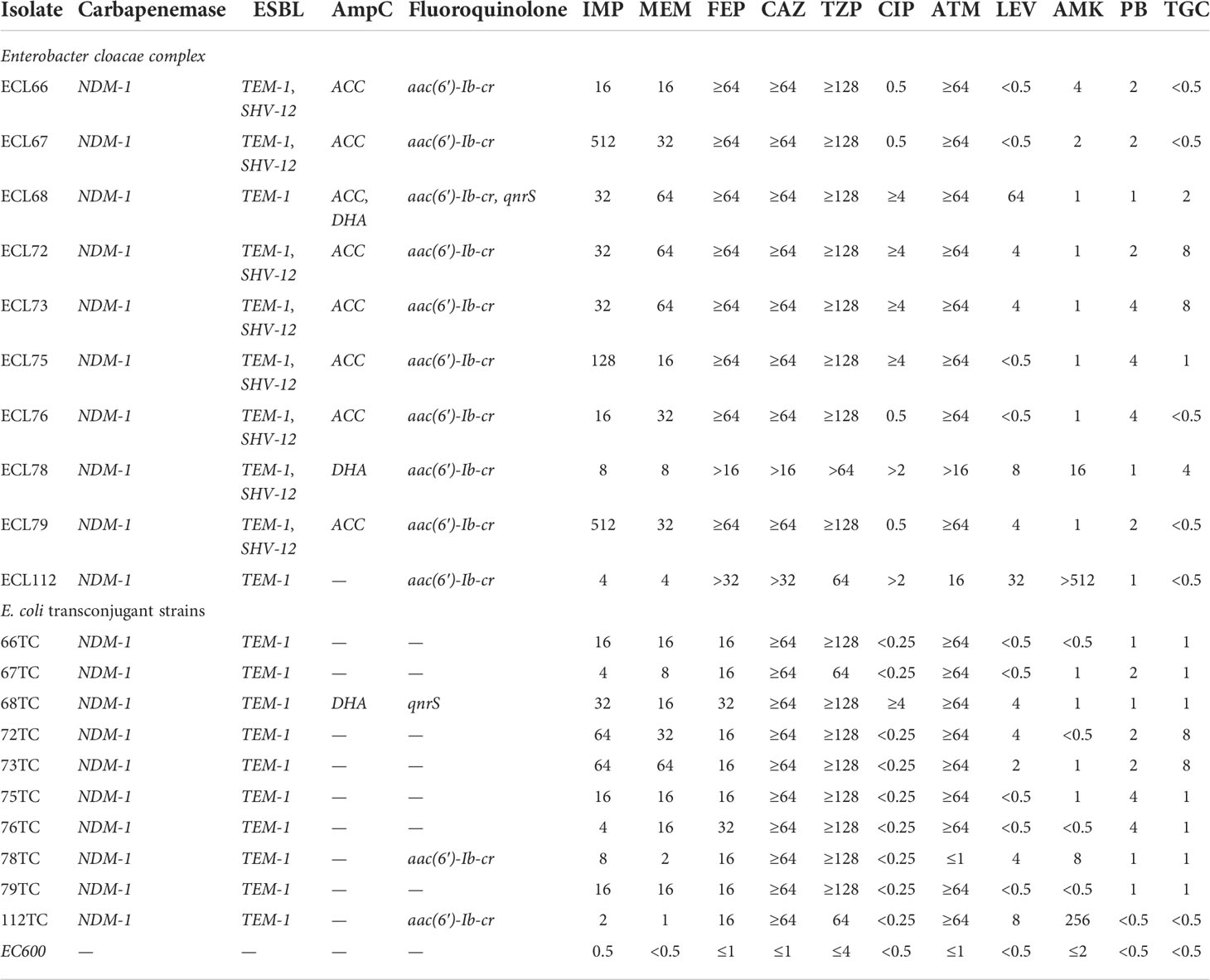
Table 2 Antibiotic susceptibilities and resistance determinants of E. cloacae isolates and their transconjugants (μg/ml).
Phenotype and genotype analysis
All strains were positive for mCIM and eCIM. PCR assay confirmed that all of the CREL isolates possessed blaNDM−1 while other carbapenemase genes were not detected. Besides the production of carbapenemase, all of the CREL isolates were positive for both ESBL and AmpC genes. The detection rate of blaTEM (100%) was the highest, followed by blaSHV (80%) and blaACC (80%), while the detection rate of blaDHA was only 20%. Additionally, fluoroquinolone-related genes were also detected to a certain extent, aac-(6′)-Ib-cr (100%) was present in all samples, the detection rate of qnrS was 10%, and the remaining genes were not detected. It is worth noting that two strains (ECL68 and ECL78) also detected the mcr-9 gene associated with colistin resistance (Table 2).
Molecular epidemiology of NDM-1-producing CREL isolates
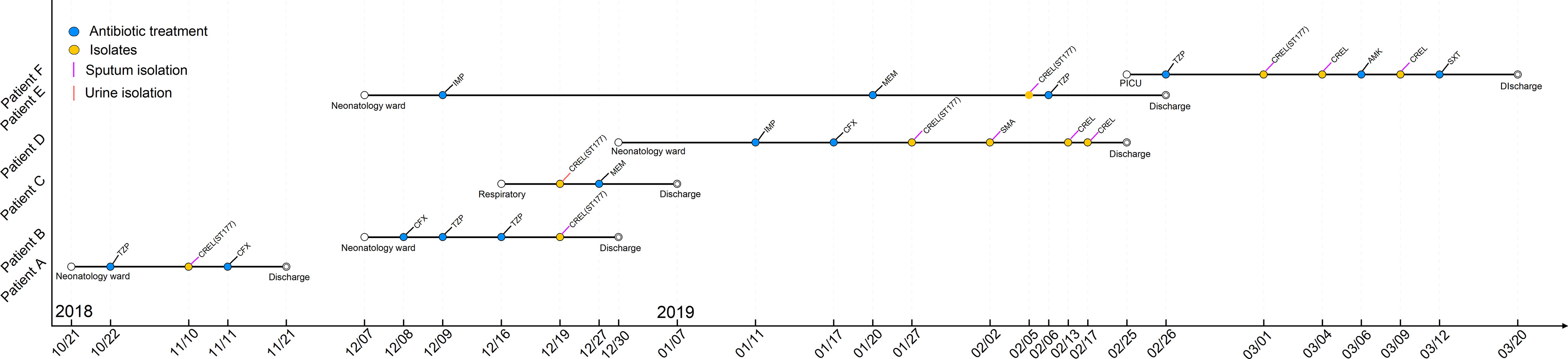
A total of four sequence types were detected in the 10 NDM-1-producing CRELs. ST177 was the most common (70%, 7/10), followed by ST93 (10%, 1/10), ST171 (10%, 1/10), and a new sequence type, ST1466 (10%, 1/10). The phylogenetic tree showed four phylogenies of these isolates (Figure 1A). It is noteworthy that ST177 was highly homologous to the high-risk clone ST93. By PFGE analyses, these isolates were grouped into A (isolate nos. 66, 72, 73, 75,76,79), B (isolate no. 67), C (isolate no. 68), D (isolate no. 112), and E (isolate no. 78) clusters. Consistent with MLST results, most strains belonging to the same ST were highly similar in their PFGE band patterns (Figure 1B). Six isolates belonged to the same clone ST177 and showed an identical PFGE pattern, indicating that outbreak of this ST has occurred in our hospital. The timeline of the outbreak case is depicted in Figure 2.
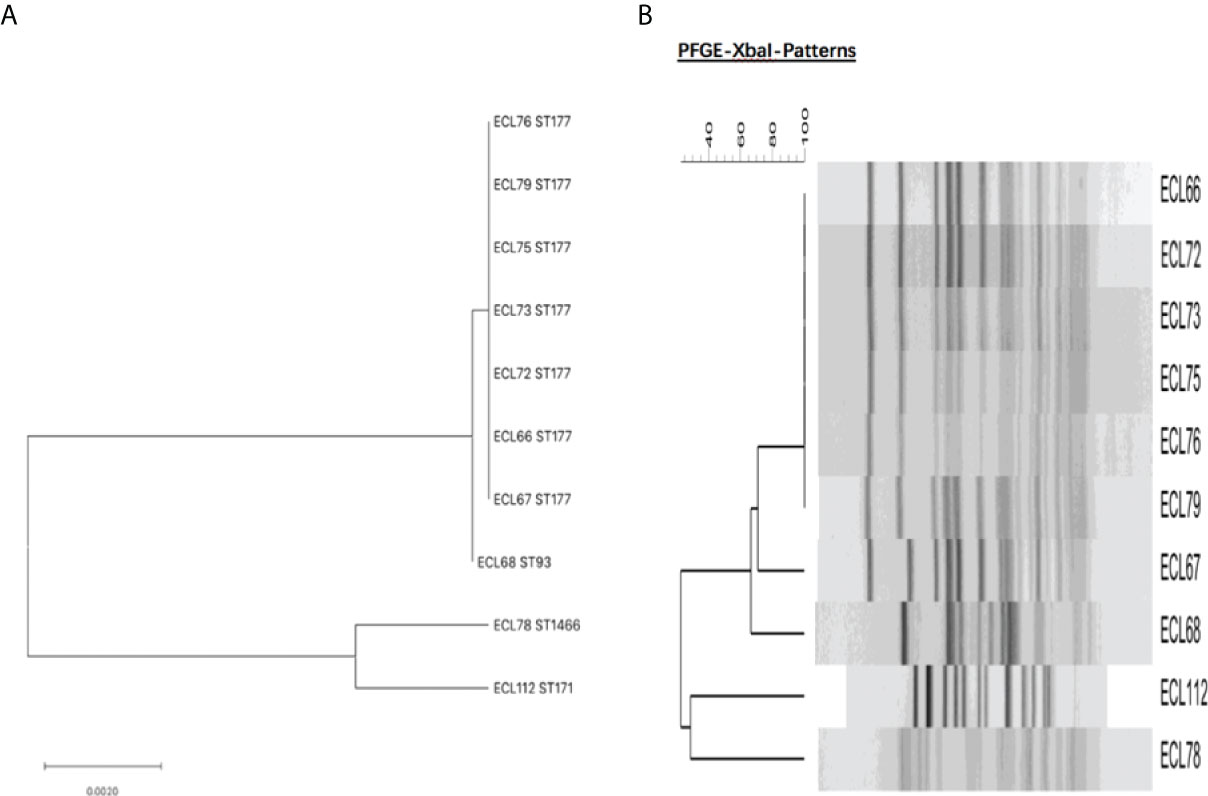
Figure 1 (A) Molecular phylogenetic analysis by MLST of NDM-1-producing CREL isolates. (B) Dendrogram of the PFGE profiles of NDM-1-producing CREL isolates.
Plasmid conjugation test
PCR results showed that all of the tranconjugations were positive for the blaNDM-1 gene, indicating that blaNDM-1-possessing plasmids from 10 donor strains were successfully transferred into recipient E. coli EC600. Some resistance genes were also detected in transconjugants including mcr-9, blaTEM, blaDHA, aac-(6′)-Ib-cr, and qnrS. However, blaSHV and blaACC were not detected in tranconjugations. All of the transconjugants showed multidrug resistance phenotypes, which were similar to those of the donor strain. Notably, the degree of resistance (MIC value) and the resistance rate of tranconjugations to some antibiotics were lower than those of donor strains. The results of the drug sensitivity test and resistance gene are summarized in Table 2.
Overview of WGS
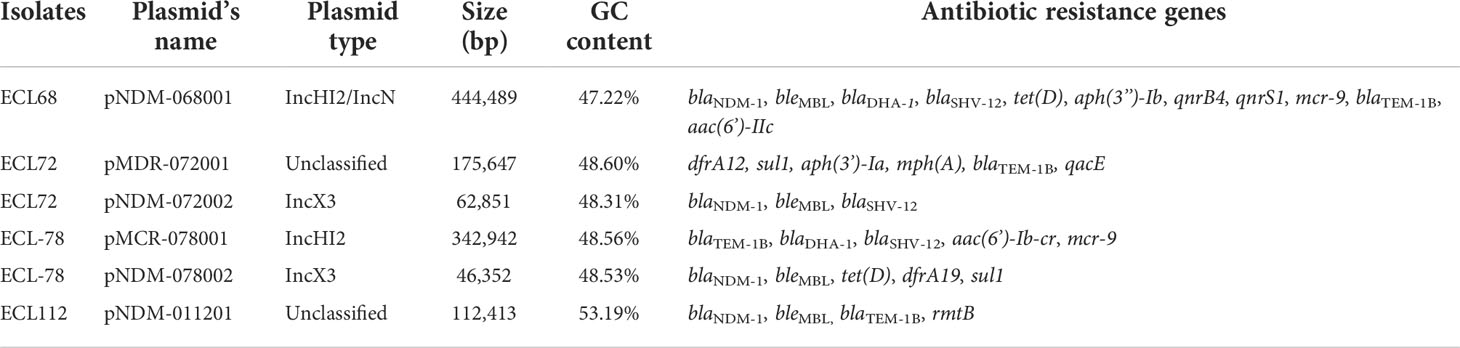
High-throughput sequencing with genomic DNA of the ECL 68, 72, 78, and 112 isolates generated six circular sequences of plasmids, of which four carried blaNDM-1 and two carried multidrug-resistant plasmids. pNDM-068001, carried by ECL68, was 444,489 bp in length, with average G+C contents of 47.22%, and contained 490 predicted coding sequences (CDSs). Strain ECL72 harbored two plasmids named pMDR-072001 (175,647 bp) and pNDM-072002 (62,851 bp) with average G+C contents of 48.6% and 48.3%, respectively. Strain ECL78 also harbored two plasmids. pMCR-078001 and pNDM-078002 were 342,942 and 46,352 bp in length, with average G+C contents of 48.56% and 48.53%, respectively. pNDM-011201, carried by ECL112, is a 112,413-bp circular plasmid with an average G+C content of 53.19% and has 120 predicted CDSs. Consistent with multidrug resistance phenotype, these strains harbored one or more plasmid carrying multiple genes mediating resistance to quinolone (qnrS1, qnrB4, and qepA), aminoglycosides [aac(6’)-IIc, aph(3’’)-Ib, aac(6’)-Ib-cr, and rmtB], β-lactams (blaNDM-1, blaTEM−1B, blaDHA-1, and blaSHV-12), bleomycin bleMBL, trimethoprims (dfrA and sul1), colistin (mcr-9), and MLS—macrolide [mph(A)] and tetracycline [tet(D)]. All six plasmids encoded plasmid replication (repAB), stability (parABM), and transfer (tra and virB) functions. The mobility of these plasmids was confirmed by conjugation assay. The major features of plasmid and schematic maps are summarized in Table 3 and Figures 3A–E.
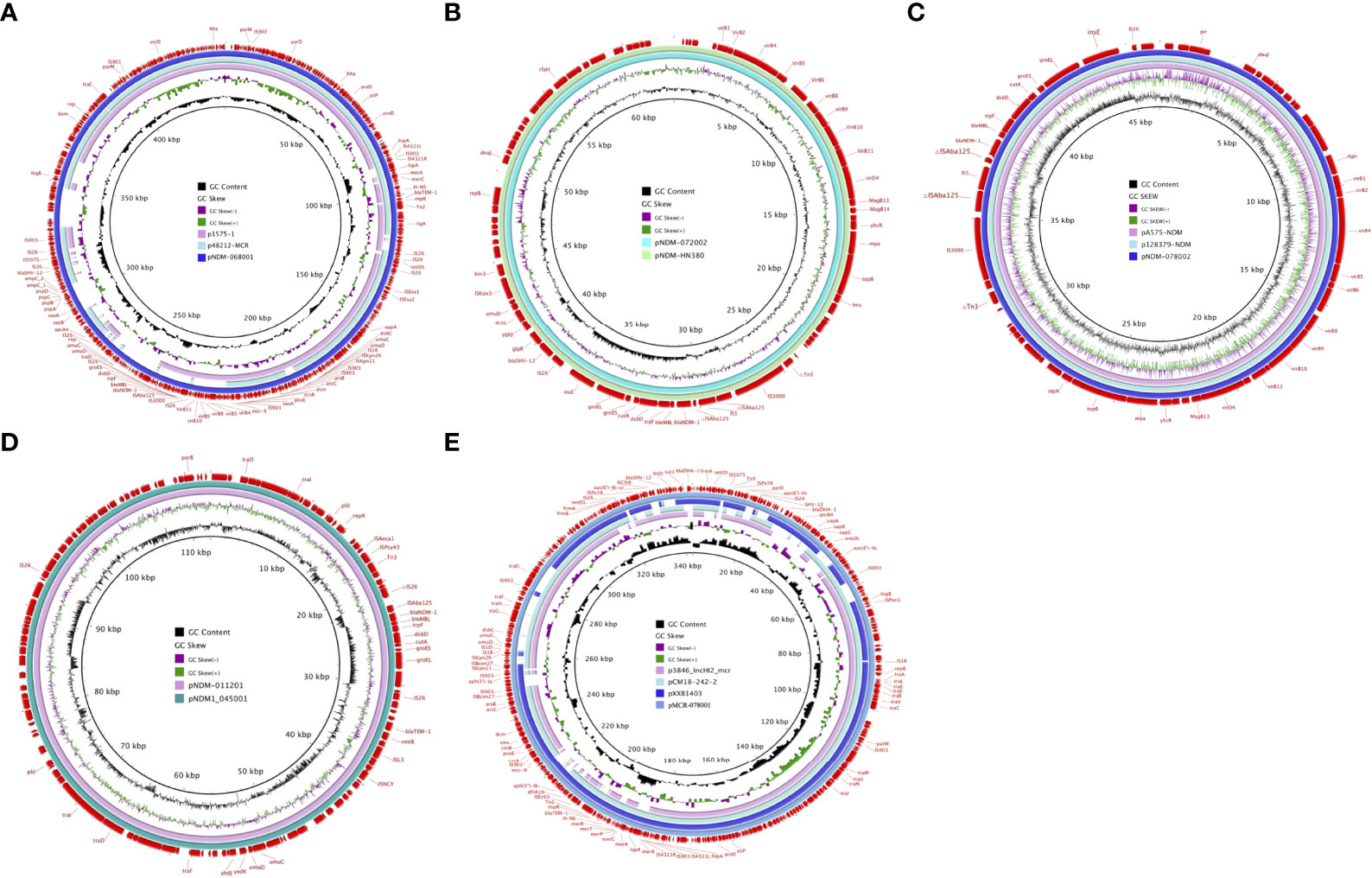
Figure 3 Schematic maps of plasmids harboring blaNDM-1 and mcr-9. (A) Circular alignments of the plasmids pNDM-068001 , p1575-1 (accession no. CP068288), and p48212-MCR (accession no. CP059413). (B) Map of the plasmids pNDM-072002 and pNDM-HN380 (accession no. NC_019162). (C) Map of the plasmids pNDM-078002, pA575-NDM (accession no. MH917283), and p128379-NDM (accession no. MF344560). (D) Map of the plasmids pNDM-011201 and pNDM1-045001 (accession no. CP043383). (E) Circular alignments of pMCR-078001 sequences with homologous mcr-carrying contigs p3846-IncHI2-mcr (accession no. CP052871), pCM18-242-2 (accession no. CP050507), and pXXB1403 (accession no. CP059887). Genes and open reading frames (ORFs) are labeled in the outermost circle. Truncated genes are indicated by Δ.
Analysis of the genetic environment of NDM-1
pNDM-072002 and pNDM-078002 are IncX3-type plasmids, which are different from the prototypical Tn125. pNDM-072002 forms a derivative of Tn125 (△Tn125). The ISAba125 located upstream has an IS5 insertion, and another ISAba125 downstream is lost and replaced by a composite transposon that contains blaSHV-12, ygbJ, and glpR and two copies of IS26 in flanks. Similarly, pNDM-078002 also shows the △Tn125; however, the “IS26-blaSHV-12 composite transposon” is missing, and only one side of IS26 is retained. Different from the insertion of IS5, pNDM-011201 belonged to the unclassified type and formed a ΔTn125 truncated by IS26. Similarly, different from the “IS26-blaSHV-12 composite transposon” structure like pNDM-072002, pNDM1-011201 forms a transposition structure dominated by the blaTEM-1 drug-resistant gene, which is specifically expressed as “IS26-Tn3-blaTEM-1-rmtB-ISL3” (Figure 4A). pNDM-068001 belongs to IncHI2/IncN type plasmid and composed of a transposon named Tn6360 rather than Tn125. The former consists of IS26, △Tn3000, ISkpn19, and △tnpA, qnrS1, IS26. The blaNDM−1 gene is located in △Tn3000 between a truncated ISAba125 and the bleMBL gene (Figure 4B).
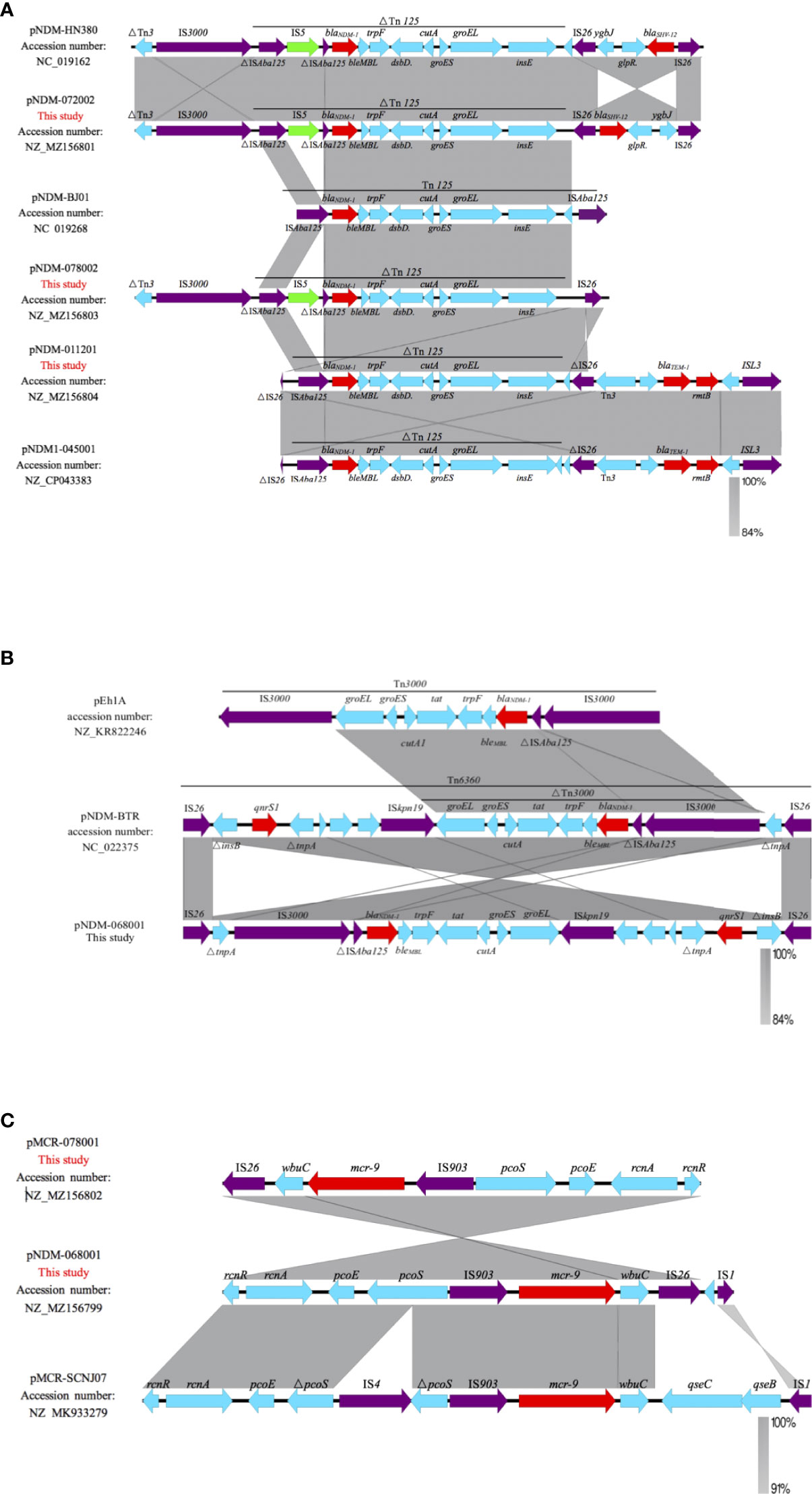
Figure 4 Linearized analyses for genetic environment of blaNDM-1 and mcr-9. (A) blaNDM-1 harbored by pNDM-HN380, pNDM-072002, pNDM-BJ01, pNDM-078002, pNDM-011201, and pNDM-045001. (B) blaNDM-1harbored by pNDM-068001, pEh1A, and pNDM-BTR. (C) mcr-9 harbored by pMCR-078001, pNDM-068001, and pMCR-SCNJ07.
Analysis of the genetic environment of mcr-9
The genetic environments of mcr-9 in the pNDM-068001 and MCR-078001 were highly similar; mcr-9 was located in an ~8-kb region surrounded by two insertion sequences IS903 and IS26. The region upstream of mcr-9 included the conserved gene structure, rcnR-pcoS-pcoE-IS903, and wbuC was located downstream of mcr-9, but lacks the two-component qseCB system. A similar structure has been reported in some IncHI2 plasmids such as p58011 (Accession no. CP049309), pN58631 (Accession no. CP049307), and pN18S2238 (Accession no. CP049312). Compared with E. hormaechei strain SCNJ07 coharboring blaNDM-1 and mcr-9, pNDM-068001 has a completed pcoS, while SCNJ07 was interrupted by IS4. The mcr-9 was surrounded by two insertion sequences, IS903 and IS1; these elements may play a crucial role in transferring mcr-9 (Figure 4C).
Discussion
Increasing antimicrobial resistance is a global emergency that is associated with adverse outcomes for infectious diseases (Uechi et al., 2019). The blaNDM-1-carrying bacteria conferred resistance to most β-lactam antibiotics, and it is a severe challenge for clinical anti-infection treatment. Previous studies have certified that CREL caused serious infections and resulted in prolonged hospital stay and increased mortality rates (da Silva et al., 2018).
Tigecycline and colistin were the last-resort antibiotics for the treatment of infections caused by multidrug-resistant bacteria including CREL (Doi, 2019). The main mechanism for reduced susceptibility to tigecycline is the upregulation of efflux pumps or mutations. In addition to this, tigecycline-resistant gene, tet(X), is another important mechanism (Li et al., 2020). Meanwhile, with the emergence of colistin-resistant genes, mcr, the treatment of CREL infection has become increasingly difficult (Lin et al., 2020). The co-production of multi-resistance genes leads to multidrug resistance, which is a serious threat to public health. Recent studies proposed that plasmid was the crucial vectors for the horizontal transfer of resistance genes (Xiang et al., 2020). In this study, blaNDM-1-possessing plasmids were successfully transferred into recipient E. coli EC600. Notably, the MIC value of most transconjugates for meropenem, imipenem, levofloxacin, and amikacin was less than donor strains, suggesting that the antibiotic resistance degree relied on multiple mechanisms such as changes in bacterial outer membrane proteins. Besides mobile genetic elements, clonal spread was another key factor associated with the prevalence of carbapenem-resistant Enterobacteriaceae.
Previous literatures have demonstrated that ST120, ST51, and ST88 NDM-1-producing CREL are widespread in China (Liu et al., 2015). In our study, ST177 was the predominant epidemic type. Interestingly, only one of the seven alleles was different between ST177 and high-risk clones, ST93, indicating that they are closely related to each other. Previous epidemiological survey of CREL in 11 cities showed that ST93 has widely spread in China (Jin et al., 2018), but it has not been reported before in our region. Therefore, epidemiological surveillance measures were necessary to control this pathogen’s further spread. Homology analysis showed that six isolates belonged to ST177 and exhibited an identical PFGE pattern, suggesting that a small-scale outbreak occurred in our hospital. Strain ECL79, retrieved from the neonatology ward, may be the index case in this event. The outbreak strains were mainly distributed in the neonatology ward. However, ECL66 and ECL75, isolated from PICU and the respiratory ward, respectively, showed highly genetic homologous strains isolated from the neonatology ward. Notably, ECL67, which belonged to ST177 and isolated from the pediatric ward, was assigned to a different cluster by PFGE analysis, indicating that this case was unrelated to outbreak. Outbreaks of NDM-1-producing E. cloacae complex ST74 and ST88 have been reported in Yunnan and Chongqing, respectively (Du et al., 2017; Jia et al., 2018). As far as we know, this is the first report of ST177 NDM-1-producing CREL outbreak in China. The outbreak of this sequence type may provide a new insight into this pathogen and should be taken seriously.
In this work, plasmid analysis revealed that pNDM-072002 and pNDM-078002 belonged to the IncX3 group, which have a broad host range and self-conjugation. pNDM-072002 is almost identical to the plasmid pNDM-HN380 (99% identity and 99.9% coverage) from K. pneumoniae from Hongkong in 2020, and there is mainly a ~3.5-kb inversion difference between the two plasmids. pNDM-078002 is highly similar to the plasmid pA575-NDM (99% identity and 99.9% coverage) from K. pneumoniae from Beijing, China, in 2020 and p128379-NDM (99% identity and 99.9% coverage) from Enterobacter hormaechei from Beijing, China, in 2020. Meanwhile, the blaNDM-1-containing plasmid IncX3 was commonly detected in Enterobacteriaceae, such as p112298-NDM (Accession no. KP987216) from Citrobacter freundii from Beijing and pNDM-SCCRK18-72 (Accession no. MN565271) from Escherichia coli from Chengdu. Our findings are consistent with previous results that show that IncX3-type plasmids are most common in Enterobacteriaceae obtained from various regions in China (Li et al., 2020). The prototype Tn125, carried by plasmid pNDM-BJ01 (Accession no. NC_019268), was isolated from Acinetobacter lwoffii and sequenced as ISAba125-blaNDM-1-bleMBL-trpF-dsbD-cutA-groES-groEL-insE-ISAba125 (Feng et al., 2015). In our study, blaNDM-1-carrying plasmids belong to the transposon Tn125 variant except for pNDM-068001. Different from prototype Tn125, the upstream ISAba125 is interrupted by IS5, and the downstream ISAba125 is replaced by IS26 in pNDM-078002 and pNDM-072002. pNDM-011201 has no IS5 insertion upstream, but ISAba125 is truncated by IS26. This variant was also found in the pNDM1-045001 (NZ_CP043383) plasmid reported in Chengdu, Sichuan Province, China. Interestingly, the MDR region in pNDM-068001 is composed of Tn6360 rather than Tn125. The former consists of △Tn3000, Tn6292, ISkpn19, △tnpA, and IS26 and was first identified in pNDM-BTR (NC_022375) (Chen et al., 2020). Thus, Tn3000 and Tn629 may be reorganized and then Tn6360 is created.
The prevalence of mcr-9 is not clear because many isolates carry mcr-9 genes but still displayed colistin-susceptible phenotypes (Chavda et al., 2019). The mcr-9 gene, which may be spreading undetected in the world, was first identified in Salmonella typhimurium in 2019 (Carroll et al., 2019). Notably, we found ECL68 and ECL78 co-carrying blaNDM-1 and mcr-9. According to accessible literature, the co-existence of mcr-9 and carbapenems are not rare. Soliman et al. (2020) and Yuan et al. (2019) reported the co-existence of mcr-9 and blaVIM-4 or blaNDM-1 in the E. hormaechei isolate, respectively (Yuan et al., 2019; Soliman et al., 2020). Kananizadeh et al. (2020) revealed co-carrying of blaIMP-1 and mcr-9 in a clinic E. cloacae complex strain (Kananizadeh et al., 2020). These findings highlight that active monitoring strategies should be implemented. Tyson et al. (2020) reported sensor qseC and response regulator qseB, also known as the two-component system qseCB, to be involved in the regulation of mcr-9 (Tyson et al., 2020). In the present study, IS903 was located upstream of mcr-9, wbuC was located downstream, followed by IS26, and the two-component system qseCB was absent. Strains ECL68 and ECL78 carry mcr-9 but remain susceptible to colistin probably due to the absence of the qseC/qseB system. Considering that the prevalence of mcr-9 is not unclear, active surveillance is necessary to control the further spread.
In conclusion, we first reported a nosocomial outbreak of NDM-1-carrying E. cloacae complex ST177 in China. Prevention strategies should be implemented to prevent NDM-1-producing bacteria transmission. The potential role of mcr-9 in colistin resistance needs further investigation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MZ156799 https://www.ncbi.nlm.nih.gov/genbank/, MZ156800 https://www.ncbi.nlm.nih.gov/genbank/, MZ156801 https://www.ncbi.nlm.nih.gov/genbank/, MZ156802 https://www.ncbi.nlm.nih.gov/genbank/, MZ156803 https://www.ncbi.nlm.nih.gov/genbank/, MZ156804.
Ethics statement
Ethical review and approval was not required for the study of human participants in accordance with the local legislation and institutional requirements. Written informed consent from the patients/participants or patients/participants legal guardian/next of kin was not required to participate in this study in accordance with the national legislation and the institutional requirements.
Author contributions
All authors contributed to the article and approved the submitted version. XZ designed the study. KH and JSZ wrote this paper. JSZ, JBZ, WL, and JL conducted the experiments. KH, LZ, and JW analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the General Projects of Chongqing Natural Science Foundation (cstc2020jcyj-msxm0067), the Yongchuan Natural Science Foundation (2021yc-jckx20053), and the Talent Introduction Project of Yongchuan Hospital of Chongqing Medical University (YJYJ202005 and YJYJ202004).
Acknowledgments
We thank the curators of the Institute Pasteur MLST system (Paris, France) for approved novel alleles and profiles at http://bigsdb.pasteur.fr.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Annavajhala, M. K., Gomez-Simmonds, A., Uhlemann, A. C. (2019). Multidrug-resistant enterobacter cloacae complex emerging as a global, diversifying threat. Front. Microbiol. 10, 44. doi: 10.3389/fmicb.2019.00044
Boyd, S. E., Livermore, D. M., Hooper, D. C., Hope, W. W. (2020). Metallo-beta-Lactamases: Structure, function, epidemiology, treatment options, and the development pipeline. Antimicrob. Agents Chemother. 64 (10), e00397–20. doi: 10.1128/AAC.00397-20
Carroll, L. M., Gaballa, A., Guldimann, C., Sullivan, G., Henderson, L. O., Wiedmann, M. (2019). Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible salmonella enterica serotype typhimurium isolate. mBio 10 (3), e00853–19. doi: 10.1128/mBio.00853-19
Chavda, K. D., Westblade, L. F., Satlin, M. J., Hemmert, A. C., Castanheira, M., Jenkins, S. G., et al. (2019). First report of bla VIM-4- and mcr-9-Coharboring enterobacter species isolated from a pediatric patient. mSphere 4 (5), 00629–19. doi: 10.1128/mSphere.00629-19
Chen, Q., Lin, Y., Li, Z., Lu, L., Li, P., Wang, K., et al. (2020). Characterization of a new transposon, Tn6696, on a bla NDM- 1-carrying plasmid from multidrug-resistant enterobacter cloacae ssp. dissolvens in China. Front. Microbiol. 11, 525479. doi: 10.3389/fmicb.2020.525479
da Silva, K. E., Varella, T. R., Bet, G., Carvalhaes, C. G., Correa, M. E., Vasconcelos, N. G., et al. (2018). High mortality rate associated with KPC-producing enterobacter cloacae in a Brazilian hospital. Am. J. Infect. Control 46 (1), 108–110. doi: 10.1016/j.ajic.2017.07.011
Doi, Y. (2019). Treatment options for carbapenem-resistant gram-negative bacterial infections. Clin. Infect. Dis. 69 (Suppl 7), S565–S575. doi: 10.1093/cid/ciz830
Du, N., Liu, S., Niu, M., Duan, Y., Zhang, S., Yao, J., et al. (2017). Transmission and characterization of bla NDM-1 in enterobacter cloacae at a teaching hospital in yunnan, China. Ann. Clin. Microbiol. Antimicrob. 16 (1), 58. doi: 10.1186/s12941-017-0232-y
Falagas, M. E., Tansarli, G. S., Karageorgopoulos, D. E., Vardakas, K. Z. (2014). Deaths attributable to carbapenem-resistant enterobacteriaceae infections. Emerg. Infect. Dis. 20 (7), 1170–1175. doi: 10.3201/eid2007.121004
Feng, J., Qiu, Y., Yin, Z., Chen, W., Yang, H., Yang, W., et al. (2015). Coexistence of a novel KPC-2-encoding MDR plasmid and an NDM-1-encoding pNDM-HN380-like plasmid in a clinical isolate of citrobacter freundii. J. Antimicrob. Chemother. 70 (11), 2987–2991. doi: 10.1093/jac/dkv232
Gong, X., Zhang, J., Su, S., Fu, Y., Bao, M., Wang, Y., et al. (2018). Molecular characterization and epidemiology of carbapenem non-susceptible enterobacteriaceae isolated from the Eastern region of heilongjiang province, China. BMC Infect. Dis. 18 (1), 417. doi: 10.1186/s12879-018-3294-3
Jacoby, G. A. (2009). AmpC beta-lactamases. Clin. Microbiol. Rev. 22 (1), 161–182. doi: 10.1128/CMR.00036-08
Jia, X., Dai, W., Ma, W., Yan, J., He, J., Li, S., et al. (2018). Carbapenem-resistant e. cloacae in southwest China: Molecular analysis of resistance and risk factors for infections caused by NDM-1-Producers. Front. Microbiol. 9, 658. doi: 10.3389/fmicb.2018.02694
Jin, C., Zhang, J., Wang, Q., Chen, H., Wang, X., Zhang, Y., et al. (2018). Molecular characterization of carbapenem-resistant enterobacter cloacae in 11 Chinese cities. Front. Microbiol. 9, 1597. doi: 10.3389/fmicb.2018.01597
Kananizadeh, P., Oshiro, S., Watanabe, S., Iwata, S., Kuwahara-Arai, K., Shimojima, M., et al. (2020). Emergence of carbapenem-resistant and colistin-susceptible enterobacter cloacae complex co-harboring blaIMP-1 and mcr-9 in Japan. BMC Infect. Dis. 20 (1), 282. doi: 10.1186/s12879-020-05021-7
Kumarasamy, K. K., Toleman, M. A., Walsh, T. R., Bagaria, J., Butt, F., Balakrishnan, R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10 (9), 597–602. doi: 10.1016/S1473-3099(10)70143-2
Lee, J. H., Bae, I. K., Lee, C. H., Jeong, S. (2017). Molecular characteristics of first IMP-4-Producing enterobacter cloacae sequence type 74 and 194 in Korea. Front. Microbiol. 8, 2343. doi: 10.3389/fmicb.2017.02343
Li, P., Lin, Y., Hu, X., Liu, Y., Xue, M., Yang, L., et al. (2020). et al: Characterization of blaNDM-1- and blaSHV-12-Positive IncX3 plasmid in an enterobacter hormaechei new sequence type 1000 from China. Infect. Drug Resist. 13, 145–153. doi: 10.2147/IDR.S231366
Li, R., Lu, X., Peng, K., Liu, Z., Li, Y., Liu, Y., et al. (2020). Deciphering the structural diversity and classification of the mobile tigecycline resistance gene tet(X)-bearing plasmidome among bacteria. mSystems 5 (2), e00134–20. doi: 10.1128/mSystems.00134-20
Lin, M., Yang, Y., Yang, Y., Chen, G., He, R., Wu, Y., et al. (2020). Co-Occurrence of mcr-9 and bla NDM-1 in enterobacter cloacae isolated from a patient with bloodstream infection. Infect. Drug Resist. 13, 1397–1402. doi: 10.2147/IDR.S248342
Liu, C., Qin, S., Xu, H., Xu, L., Zhao, D., Liu, X., et al. (2015). New Delhi metallo-beta-Lactamase 1(NDM-1), the dominant carbapenemase detected in carbapenem-resistant enterobacter cloacae from henan province, China. PloS One 10 (8), e0135044. doi: 10.1145/2818302
Majewski, P., Wieczorek, P., Ojdana, D., Sienko, A., Kowalczuk, O., Sacha, P., et al. (2016). Altered outer membrane transcriptome balance with AmpC overexpression in carbapenem-resistant enterobacter cloacae. Front. Microbiol. 7, 2054. doi: 10.3389/fmicb.2016.02054
Mezzatesta, M. L., Gona, F., Stefani, S. (2012). Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol. 7 (7), 887–902. doi: 10.2217/fmb.12.61
Nordmann, P., Naas, T., Poirel, L. (2011). Global spread of carbapenemase-producing enterobacteriaceae. Emerg. Infect. Dis. 17 (10), 1791–1798. doi: 10.3201/eid1710.110655
Perez, F., Bonomo, R. A. (2019). Carbapenem-resistant enterobacteriaceae: global action required. Lancet Infect. Dis. 19 (6), 561–562. doi: 10.1016/S1473-3099(19)30210-5
Soliman, A. M., Maruyama, F., Zarad, H. O., Ota, A., Nariya, H., Shimamoto, T., et al. (2020). Emergence of a multidrug-resistant enterobacter hormaechei clinical isolate from Egypt Co-harboring mcr-9 and blaVIM-4. Microorganisms 8 (4), 595. doi: 10.3390/microorganisms8040595
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33 (9), 2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995
Tetsuka, N., Hirabayashi, A., Matsumoto, A., Oka, K., Hara, Y., Morioka, H., et al. (2019). Molecular epidemiological analysis and risk factors for acquisition of carbapenemase-producing enterobacter cloacae complex in a Japanese university hospital. Antimicrob. Resist. Infect. Control 8, 126. doi: 10.1186/s13756-019-0578-3
Tyson, G. H., Li, C., Hsu, C. H., Ayers, S., Borenstein, S., Mukherjee, S., et al. (2020). The mcr-9 gene of salmonella and escherichia coli is not associated with colistin resistance in the united states. Antimicrob. Agents Chemother. 64 (8), e00573–20. doi: 10.1128/AAC.00573-20
Uechi, K., Tada, T., Shimada, K., Nakasone, I., Kirikae, T., Fujita, J. (2019). Emergence of a carbapenem-resistant and colistin-heteroresistant enterobacter cloacae clinical isolate in Japan. J. Infect. Chemother. 25 (4), 285–288. doi: 10.1016/j.jiac.2018.09.004
Walsh, T. R. (2010). Emerging carbapenemases: a global perspective. Int. J. Antimicrob. Agents 36 Suppl 3, S8–14. doi: 10.1016/S0924-8579(10)70004-2
Wang, X., Chen, G., Wu, X., Wang, L., Cai, J., Chan, E. W., et al. (2015). Increased prevalence of carbapenem resistant enterobacteriaceae in hospital setting due to cross-species transmission of the bla NDM-1 element and clonal spread of progenitor resistant strains. Front. Microbiol. 6, 595. doi: 10.3389/fmicb.2015.00595
Xiang, T., Chen, C., Wen, J., Liu, Y., Zhang, Q., Cheng, N., et al. (2020). Resistance of klebsiella pneumoniae strains carrying bla NDM-1 gene and the genetic environment of bla NDM-1. Front. Microbiol. 11, 700. doi: 10.3389/fmicb.2020.00700
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53 (12), 5046–5054. doi: 10.1128/AAC.00774-09
Yuan, Y., Li, Y., Wang, G., Li, C., Xiang, L., She, J., et al. (2019). Coproduction of MCR-9 and NDM-1 by colistin-resistant enterobacter hormaechei isolated from bloodstream infection. Infect. Drug Resist. 12, 2979–2985. doi: 10.2147/IDR.S217168
Zeng, L., Yang, C., Zhang, J., Hu, K., Zou, J., Li, J., et al. (2021). An outbreak of carbapenem-resistant klebsiella pneumoniae in an intensive care unit of a major teaching hospital in chongqing, China. Front. Cell Infect. Microbiol. 11, 656070. doi: 10.3389/fcimb.2021.656070
Zeng, L., Zhang, J., Hu, K., Li, J., Wang, J., Yang, C., et al. (2021). Microbial characteristics and genomic analysis of an ST11 carbapenem-resistant klebsiella pneumoniae strain carrying bla KPC-2 conjugative drug-resistant plasmid. Front. Public Health 9, 809753. doi: 10.3389/fpubh.2021.809753
Keywords: carbapenem-resistant, Enterobacter cloacae complex, NDM-1, mcr-9, plasmids
Citation: Hu K, Zhang J, Zou J, Zeng L, Li J, Wang J, Long W and Zhang X (2022) Molecular characterization of NDM-1-producing carbapenem-resistant E. cloacae complex from a tertiary hospital in Chongqing, China. Front. Cell. Infect. Microbiol. 12:935165. doi: 10.3389/fcimb.2022.935165
Received: 03 May 2022; Accepted: 14 July 2022;
Published: 08 August 2022.
Edited by:
Costas C Papagiannitsis, University of Thessaly, GreeceReviewed by:
Alberto Antonelli, University of Florence, ItalyZoi Florou, University of Thessaly, Greece
Copyright © 2022 Hu, Zhang, Zou, Zeng, Li, Wang, Long and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoli Zhang, am1zenhsMTIzQDE2My5jb20=
†These authors have contributed equally to this work
 Kewang Hu1,2†
Kewang Hu1,2† Jianmin Wang
Jianmin Wang Xiaoli Zhang
Xiaoli Zhang