- 1Institute for Immunology and Informatics, Department of Cell and Molecular Biology, University of Rhode Island, Providence, RI, United States
- 2Viral Diseases Branch, Walter Reed Army Institute of Research, Silver Spring, MD, United States
Dengue virus (DENV) infection is the most prevalent arthropod-borne virus disease and is endemic in more than 100 countries. Several DENV proteins have been shown to target crucial human host proteins to evade innate immune responses and establish a productive infection. Here we report that the DENV NS3 protein targets RIPK1 (Receptor Interacting Protein Kinase I), a central mediator of inflammation and cell death, and decreases intracellular RIPK1 levels during DENV infection. The interaction of NS3 with RIPK1 results in the inhibition of NF-κB activation in response to TNFR or TLR3 stimulation. Also, we observed that the effects of NS3 on RIPK1 were independent of NS3 protease activity. Our data demonstrate a novel mechanism by which DENV suppresses normal cellular functions to evade host innate immune responses
Introduction
Dengue is the most prevalent arthropod-borne viral disease, with an estimated 400 million infections occurring annually (Bhatt et al., 2013). Infected individuals may develop severe symptoms leading to dengue hemorrhagic fever (DHF), dengue shock syndrome (DSS), and even death. The causative agent of dengue, dengue virus (DENV), is known to infect dendritic cells, macrophages, monocytes, B cells, T cells, hepatocytes, epithelial and endothelial cells (King et al., 1999; Martina et al., 2009; Silveira et al., 2018). Dengue disease is thought to result both from direct viral effects as well as host immune responses to infection (Mathew and Rothman, 2008; Martina et al., 2009). Effective antiviral drugs and vaccines are not available to treat dengue, which emphasizes the importance of better understanding DENV-host interactions (Low et al., 2017). DENV refers to a group of four closely related positive-sense, single-stranded RNA viruses or serotypes (DENV 1, 2, 3, 4), belonging to the Flaviviridae family. Upon entry into the host cell, the viral genome is translated into a single polypeptide, which is cleaved into its individual structural (capsid, PrM, and envelope) and non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5) by viral and host proteases (Clum et al., 1997). The DENV protease is a complex of NS2B and NS3 proteins. NS2B is a small hydrophobic protein that acts as a cofactor for NS3 proteolytic activity (Falgout et al., 1991). The N-terminal 180 acids of NS3 contain a serine protease domain (Wengler et al., 1991), while the remainder of NS3 (amino acids 180 to 618) contains an RNA helicase domain and a NTPase motif (Xu et al., 2005; Luo et al., 2008). In addition to their roles in viral RNA replication and packaging, several DENV proteins are known to interact with numerous host proteins and interfere with host antiviral immune responses. For example, the DENV protease NS2B3 interacts with and cleaves STING and downregulates RLR signaling (Aguirre et al., 2012). The DENV protease also cleaves the mitochondrial proteins MFN1 and MFN2 and thereby impairs mitochondrial dynamics as well as efficient RLR signaling (Yu et al., 2015).
RIPK1 has emerged as an important regulator that controls multiple cellular pathways involved in inflammation and cell death (Ofengeim and Yuan, 2013). RIPK1 contributes to innate immune responses to bacterial and viral infections by inducing cell death (apoptosis or necroptosis) or by inducing inflammatory signaling (Upton and Kaiser, 2017). Both kinase and scaffolding functions of RIPK1 are important for signaling. Activation of death receptors such as TNFR1 leads to the formation of a multi-protein complex including RIPK1, complex I, which facilitates the activation of NF-κB and MAPK pathways that promote cell survival by inducing anti-apoptotic gene expression and induce proinflammatory cytokine gene expression (Hsu et al., 1996). The scaffolding function of RIPK1 is critical for NF-κB activation. RIPK1 is also involved in mediating TRIF-dependent NF-κB activation in response to TLR3 and TLR4 stimulation (Meylan et al., 2004; Cusson-Hermance et al., 2005). RIPK1 can drive extrinsic apoptosis by activating caspase-8 via the formation of a distinct multi-protein complex, complex IIa (Micheau and Tschopp, 2003), and can drive necroptosis through formation of a complex with RIPK3 (complex IIb) leading to phosphorylation of the pseudokinase mixed lineage kinase domain-like (MLKL) protein (Cho et al., 2009; He et al., 2009). RIPK1 also has been shown to regulate IRF3 activation downstream of RIG-I/MDA-5 in response to RNA viruses (Rajput et al., 2011). This central role of RIPK1 makes it an ideal target for inhibition by viruses, as has been shown for HIV-1 (Wagner et al., 2015), human Rhinovirus 3C (Croft et al., 2018), and Epstein-Barr virus (Liu et al., 2018). However, no association between RIPK1 and DENV infection has been reported to date.
In the present study, we observed that RIPK1 protein levels decreased over time during DENV infection. Plasmid expression of DENV NS3, which lacks protease activity, was sufficient to decrease intracellular RIPK1 levels. Using co-immunoprecipitation and Western blot analysis, we observed that DENV NS3 physically interacted with RIPK1. DENV infection and expression of NS3 inhibited the TNFα- and poly (I:C)-stimulated activation of NF-κB. Furthermore, MLKL phosphorylation in response to TNFα was suppressed by DENV infection. We hypothesize that inhibition of RIPK1-mediated regulation of cell death may represent an additional mechanism to promote DENV replication.
Materials and methods
Cell culture and virus infection
Huh7, HEK 293T cells stably expressing TLR3 (generous gifts from Dr. Kate Fitzgerald), HEK 293T (Dharmacon, Inc.), and Vero (obtained from American Type Culture Collection (ATCC)) cells were maintained in Dulbecco’s modified minimal essential medium and HepG2 (ATCC) cells were maintained in Eagle’s Minimum Essential Medium, both supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich), 1% penicillin-streptomycin (Sigma-Aldrich), 1% non-essential amino acids (Lonza) and 1% L-Glutamine solution (Sigma-Aldrich). U937 cells stably expressing DC-SIGN (a generous gift from Dr. Anuja Mathew) were maintained in Roswell Park Memorial Institute (RPMI) medium supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich), 1% penicillin-streptomycin (Sigma-Aldrich), 1% non-essential amino acids (Lonza), and 1% L-Glutamine solution (Sigma-Aldrich). All cells were incubated in a humidified chamber at 37°C and 5% CO2. DENV2 strains DENV2 16681 and NGC were originally obtained from ATCC or the Walter Reed Army Institute of Research and were passaged in C6/36 cells (ATCC). Virus titers were determined by immunostained plaque assay on Vero cells (Liu et al., 2012; Medin et al., 2015).
Generation of MoDCs
Peripheral blood mononuclear cells (PBMCs) were isolated from buffy coat of healthy donors obtained from the Oklahoma Blood Institute (Oklahoma City, OK) by density gradient centrifugation on Ficoll-Paque Premium (GE Healthcare). Monocytes were isolated from PBMCs by positive selection using CD14+ microbeads (Miltenyi Biotec). Monocytes were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated FBS (Sigma-Aldrich), 1% penicillin- streptomycin (Sigma-Aldrich), 50 ng/mL human interleukin-4 (IL-4) and 160 ng/mL human granulocyte-macrophage colony-stimulating factor (GM-CSF) for 5 days to generate monocyte-derived dendritic cells (MoDCs). Fresh medium with cytokines was added at day 3, and cells were infected with DENV2 at day 5.
Antibodies and reagents
The following primary antibodies were used for co-immunoprecipitation and Western blot experiments: anti-Flag-HRP (sc-166355, Santa Cruz Biotechnology) anti-Flag (F1804, Sigma-Aldrich), mouse anti-RIPK3 (sc-374639, Santa Cruz Biotechnology), anti-β-actin-HRP (ab20272, Abcam), anti-V5 tag (ab27671, Abcam), anti-V5-HRP (R961-25, Invitrogen), anti-GAPDH-HRP (MA515738) anti-DENV NS3 (GTX124252, GeneTex), anti-DENV NS4B (GTX103349, GeneTex), anti-DENV NS1 (GTX103346, GeneTex), anti-DENV NS2B (GTX124246, GeneTex) anti-RIPK1 (D94C12, Cell Signaling Technology), anti-LC3B (2775S, Cell Signaling Technology), anti-p-MLKL (91689S, Cell Signaling Technology). Secondary antibodies used were goat anti-rabbit-HRP (32260, Thermo Scientific) and anti-mouse-HRP (sc516102, Santa Cruz Biotechnology). The following reagents were used for inhibitor treatment experiments: NH4Cl (A9434, Sigma-Aldrich), Chloroquine (C6628, Sigma-Aldrich), MG132 (NC9038428, Fisher Scientific), z-VAD-FMK (SC-3067, Santa Cruz Biotechnology) and DMSO (BP231-100, Fisher Scientific).
Western blot analysis
Cells were lysed in RIPA buffer freshly supplemented with protease and phosphatase inhibitor cocktail (Sigma-Aldrich). Lysates were incubated on ice for 30 min with vortexing every 5 min and centrifuged at 10,000 rpm for 5 min at 4°C. Supernatants were used as total cell lysates. Protein concentrations were determined using Pierce BCA Protein Assay kit (Thermo Fisher Scientific) following manufacturer’s instructions and measured using an Envision plate reader (PerkinElmer). BOLT LDS Sample Buffer and Reducing Agent (Thermo Fisher Scientific) were added to cell lysates at a final concentration of 1x and samples were denatured at 70°C for 10 min. Proteins were separated on 4–12% or 8% BOLT Bis-Tris Plus gel (Thermo Fisher Scientific) and transferred onto nitrocellulose membranes using the Trans-Blot® TurboTM RTA Mini Nitrocellulose Transfer kit (Bio-Rad). Membranes were blocked in 5% milk in phosphate buffered saline with 0.1% Tween 20 for 1 h at room temperature (rt) followed by incubation with the appropriate primary antibody overnight at 4°C. Membranes were washed and incubated with the appropriate secondary antibody diluted in blocking buffer for 1 hour at rt. For β-actin and GAPDH analysis, membranes were stained for 1 hour with corresponding antibodies. Detection of blots was done using Amersham ECL Select Western Blotting Detection Reagent (GE Healthcare Life Sciences) following the manufacturer’s guidelines and images were captured with ChemiDocTM XRS+ System (Bio-Rad). Detection and quantification of band intensities were performed using Image Lab 5.1 (Bio-Rad).
Co-immunoprecipitation
Cells were lysed with RIPA buffer containing a cocktail of protease and phosphatase inhibitors. Protein G dynabeads (Thermo Fisher Scientific) were incubated with either mouse anti-Flag (Sigma-Aldrich), mouse anti-V5 (abcam) or mouse anti-NS3 (Genetex) for 1 hr at rt. Antibody-bead complexes were washed with antibody binding and washing buffer (Thermo Fisher Scientific). Cell lysates (100μg) were incubated with antibody-coated beads for 30 min at RT with rotation. Bead-antibody-antigen complexes were washed 4 times with antibody binding and washing buffer followed by the washing buffer provided (Thermo Fisher Scientific). The immunocomplex was then eluted under denaturing conditions using elution buffer (Thermo Fisher Scientific), dithiothreitol (Thermo Scientific) and Bolt sample buffer (Life Technologies) according to the manufacturer’s recommendations, and then examined by Western blot analysis using the indicated antibodies.
Plasmids and transfections
Plasmids pNS4B5-eGFP and pNS2B3-V5 have previously been described (Medin et al., 2015). pNS2B3-S135A was created by site-directed mutagenesis (Quik Change II site-directed mutagenesis kit, Agilent Technologies) of the pNS2B3-V5 plasmid. Plasmid pNS3-V5 was generated by PCR amplification of the NS3 coding sequence from cDNA of DENV2 strain New Guinea C obtained from BEI Resources, which was cloned into the Gateway pcDNA-DEST40 vector (ThermoFisher Scientific) following the manufacturer’s protocol. Primer sequences were: forward primer NS2B3-S135A-V5: 5’-GCT GTA TCT CTG GAC TTT TCT CCT GGA ACG GCC GGC TCT CCA ATT ATC GAC-3’; reverse primer NS2B3-S135A: 5’-GTC GAT AAT TGG AGA GCC GGC CGT TCC AGG AGA AAA GTC CAG AGA TAC AGC-3’; forward primer NS3-V5; 5’-GGG GAC AAG TTT GTA CAA AAA AGC AGG CTG CCA CCA TGG CTG GAG TAT TGT GGG ATG TC-3’; reverse primer NS3-V5; GGG GAC CAC TTT GTA CAA GAA AGC TGG GTT CTT TCT TCC AGC TGC AAA CTC-3’. The plasmid sequences were verified by DNA sequencing. Plasmid pNS2B3-pro contains a codon-optimized sequence encoding the full length of NS2B and the protease domain (residues 1-185) of NS3 derived from the S16681 isolate of DENV2 (Genbank: NC001474), which was synthesized by GenScript and cloned into in the pcDNA3.1/Zeo vector. Plasmids expressing Flag-RIPK1 and RIPK3-GFP were obtained from Addgene. The luciferase reporter constructs containing a tandem repeat of the consensus binding site for the transcription factor NF-κB, and a Renilla reniformis luciferase were obtained from Strategene. Transfection of cells with plasmids was performed using GeneJuice® Transfection Reagent (EMD Millipore), according to the manufacturer’s instructions.
RT-qPCR
Total RNA was extracted from cell pellets using the RNeasy Mini Kit (Qiagen). Quantitative RT- PCR was performed using iTaq Universal probes 1 step kit (Biorad) or iTaq Universal SYBR Green 1 step kit (Biorad) on a Bio-Rad CFX machine, and gene expression was normalized to β-actin expression level. Taqman gene expression assays (RIPK1) were purchased from Thermofisher Scientific. DENV primers were purchased from Integrated DNA Technology (IDT) and RT-qPCR was performed to detect relative DENV RNA in cell lysates (Sadon et al., 2008).
Luciferase reporter assays
HEK 293T cells were plated in 96 well plates and were co-transfected with a NF-κB luciferase reporter, a Renilla luciferase reporter, and a plasmid encoding Flag-tagged RIPK1 using GeneJuice® Transfection Reagent (EMD Millipore), according to the manufacturer’s protocol. Co-transfected cells were infected with DENV2 (NGC, MOI=3) and were cultured for 72hrs or transfected with plasmids expressing DENV NS3, NS2B3, NS2B3-S135A, or NS1 and cultured for 48hrs. Cells were then stimulated with hTNFα (50ng/ml, Peprotech) for the final 6hrs. Luciferase activity was quantified using the dual Glo luciferase assay system (Promega). HEK 293T cells stably expressing TLR3 were co-transfected and infected with DENV2 or overexpressed DENV NS3, NS2B3, NS2B3-S135A, or NS1 as described above and were stimulated with poly I:C (20µg/ml) for the final 24hrs. At the end of the treatment, luciferase activity was measured using the dual Glo luciferase assay system (Promega). The luciferase signal for each well was normalized by the Renilla luciferase signal.
Necroptosis assay
U937-DC-SIGN cells (6 x 105) were plated in a 6 well plate and infected with DENV2 (MOI=6) for 48hrs. Cells were treated with 50µM Nec-1 (Sigma Aldrich), 20µM zVAD-FMK (Santa Cruz Biotechnology), 50ng/ml TNFα (Peprotech), and 10µM Smac mimetic (Fisher Scientific) for 6 hours. Cells were harvested by centrifugation at 1600rpm for 5 minutes and lysed with RIPA buffer supplemented with protease inhibitors. Cell lysates were analyzed by Western blot.
Statistical analysis
The statistical analyses were performed using one-way analysis of variance (ANOVA) or the unpaired, two-tailed, Student’s t-test, with Shapiro-Wilk and Brown-Forsythe tests for normality and equal variance. P values were calculated using Graphpad Prism 8 program. Differences were considered statistically significant at P values less than 0.05.
Results
RIPK1 protein levels decrease with DENV infection
DENV is known to infect hepatocytes and the liver is one of the major organs affected during DHF (Samanta, 2015). We tested the effect of DENV infection on the human hepatoma cell line, Huh7. A time-course analysis of DENV2 infection revealed a reduction in intracellular RIPK1 protein concomitant with the expression of DENV proteins, beginning as early as 6h post-infection (Figure 1A). In order to see if this was a general effect, we infected cell lines originated from different human tissues with DENV2 as well as monocyte-derived dendritic cells (MoDC), which is a relevant primary cell system (Ho et al., 2001). We observed a significant decrease in endogenous RIPK1 protein levels with DENV infection in all the cells tested (Figure 1B). Overexpressed Flag-tagged RIPK1 was also decreased in a dose-response manner in DENV-infected HEK 293T cells (Figure 1C). This effect was specific for RIPK1 since endogenous or overexpressed RIPK3 levels were not affected by DENV infection (Figure 1D).
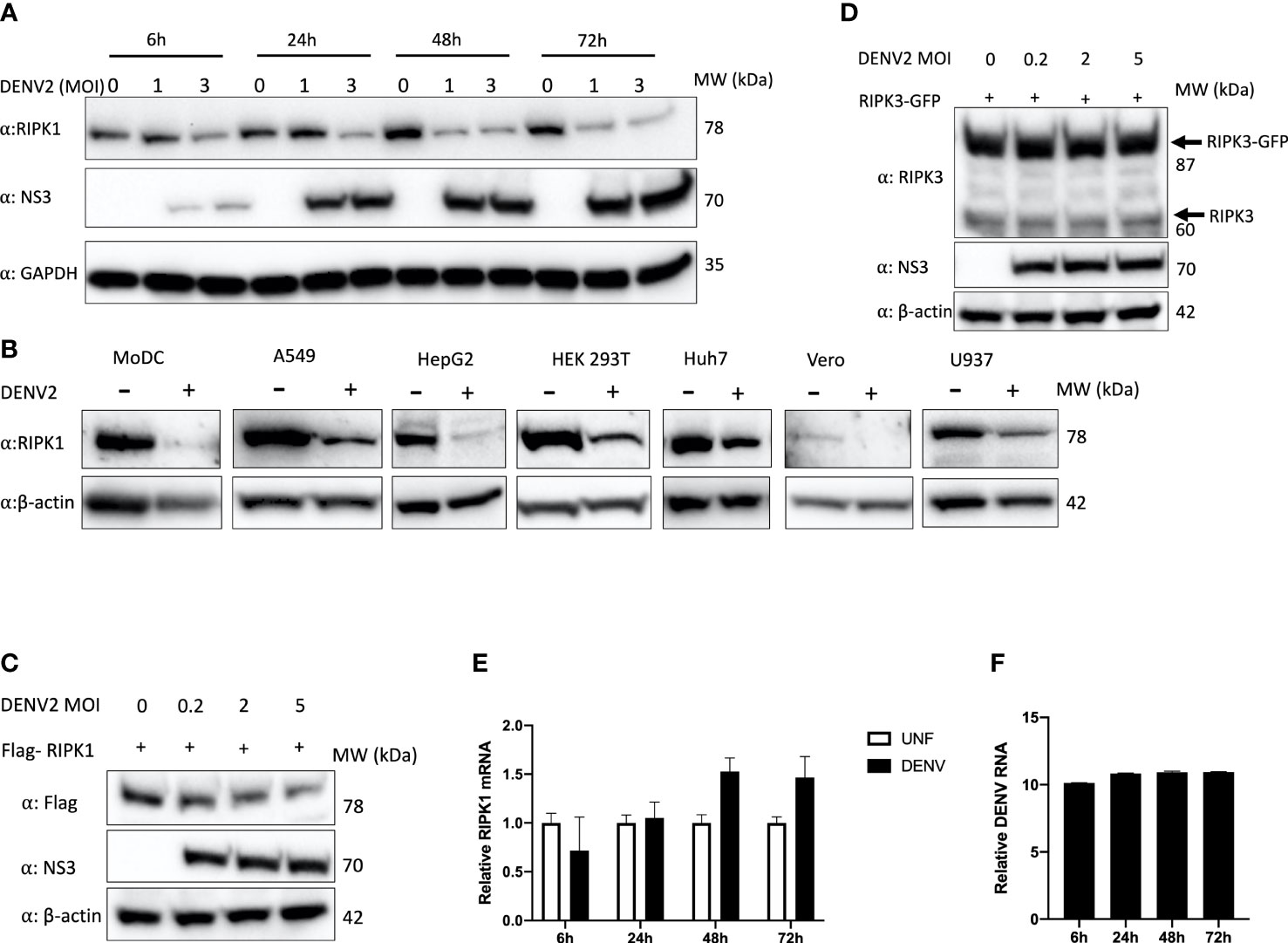
Figure 1 Endogenous and overexpressed RIPK1 decreases during DENV infection. (A) Time course study of DENV2 infection in human Huh7 cells with MOI of 1 and 3 for the indicated hours. Cell lysates were harvested for western blot analysis using the indicated antibodies. (B) Western blot analysis of RIPK1 in indicated cells infected with DENV2 (MOI=2) and cultured for 48hrs. β-actin was used as a loading control. (C) Western blot analysis of overexpressed RIPK1 and (D) RIPK3 followed by DENV2 infection in HEK 293T cells at 48h post-infection. (E) RT-qPCR analysis of total RNA isolated from Huh7 cells infected with DENV2 (MOI=2) for (E) RIPK1 or (F) intracellular DENV RNA for the indicated time points. UNF=Uninfected. Data are mean ± SD (n=3 experiments). Western blots are representative data from a minimum of 3 independent experiments (panels A–D).
Next, we investigated if DENV inhibited RIPK1 at a transcriptional level. A time-course study was done with DENV2 infection in Huh7 cells. We did not see a decrease in RIPK1 transcript levels, indicating that the downregulation of RIPK1 occurred at the post-transcriptional level (Figures 1E, F).
RIPK1 is reduced by DENV NS3 protein
RIPK1 is known to be targeted by several viral proteases to counteract its functions and enhance viral replication (Wagner et al., 2015; Croft et al., 2018). Therefore, to evaluate the mechanism of RIPK1 reduction during DENV infection, we tested whether expression of the DENV NS2B3 protease complex was sufficient to reduce RIPK1 levels. We co-transfected a Flag-tagged RIPK1 and a plasmid encoding DENV NS2B3. Previous studies with the NS2B3 plasmid shows that it efficiently self-cleaves the NS2B3-V5 product (85kDa) into NS3-V5 (70kDa) and NS2B (15kDa) (Medin et al., 2015). We observed a dose response in RIPK1 reduction with the expression of DENV NS2B3 (Figures 2A, B). However, we did not detect the accumulation of a cleaved product of RIPK1 using antibodies targeting either the C-terminal or N-terminal segment of RIPK1 (data not shown).
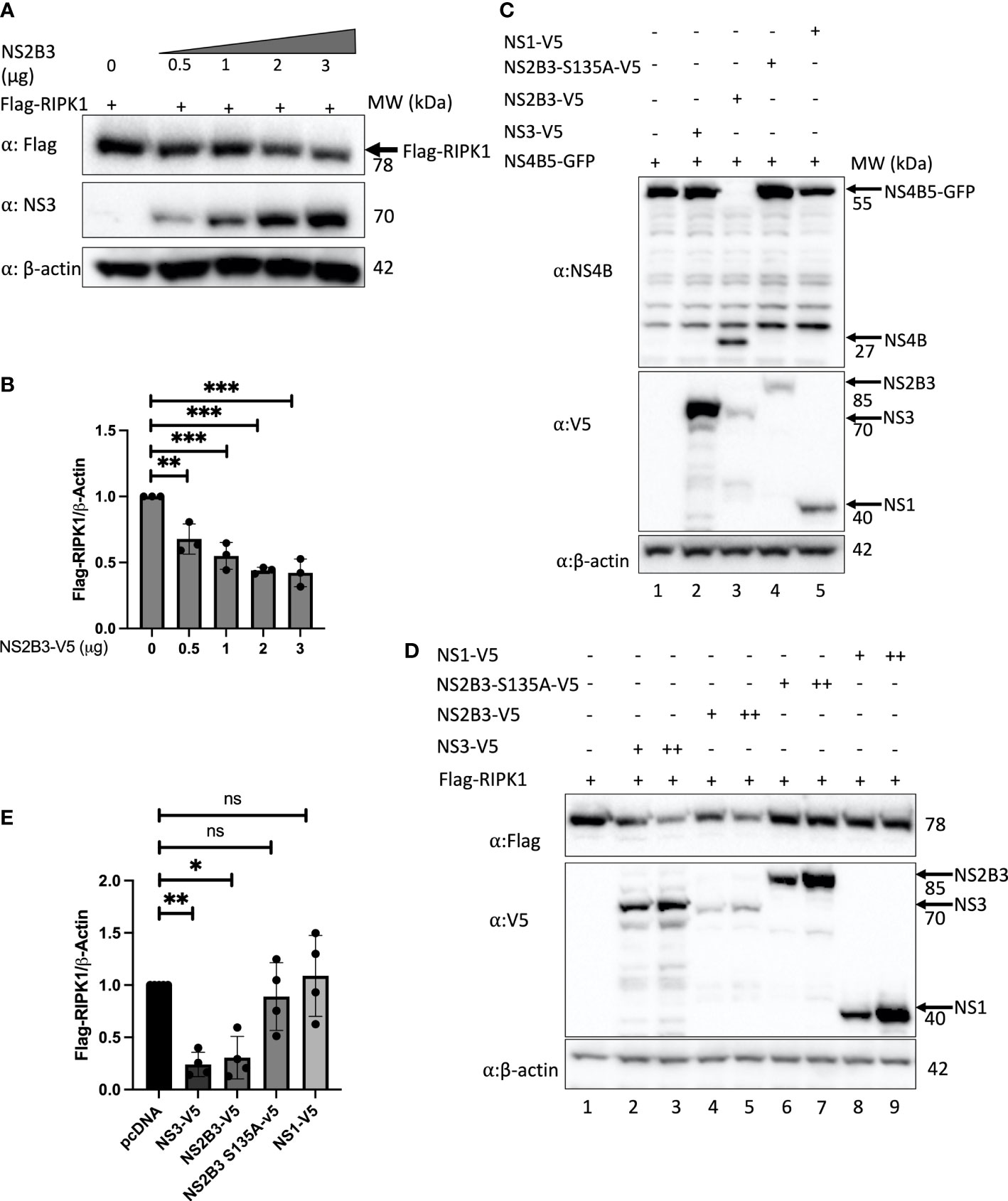
Figure 2 DENV NS3 decreases RIPK1 protein levels. (A) Western blot analysis of HEK 293T cells co-transfected with Flag-RIPK1 and increasing concentrations of a plasmid expressing the DENV NS2B3. (B) Densitometric analysis of Flag-RIPK1 band intensities normalized to β-actin levels. (C) Western blot analysis of HEK293T cells co-transfected with NS4B5-GFP and indicated plasmids at 48hrs post-transfection. (D)Western blot analysis of HEK 293T cells co-transfected with Flag-RIPK1 and 1μg (+) or 2.5μg (++) of indicated plasmids. (E) Densitometric analysis of Flag-RIPK1 band intensities normalized to β-actin levels. Results are representative of at least three independent experiments and are expressed as mean ± SD from three biological replicates. (One-way ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001). ns, not significant.
Next, to determine if the protease activity of NS2B3 was important for this effect, we adopted two strategies. First, we created a catalytically inactive protease mutant, NS2B3-S135A (Yu et al., 2006). Since it lacks protease activity it is unable to self-cleave and is detected as an 85kDa fusion protein on Western blot (Figure 2C). Second, we cloned the full-length NS3 coding sequence without NS2B. The NS3 plasmid yielded a stronger band with anti-V5 staining at the predicted size (70kDa) than the NS2B3 plasmid (Figure 2C). Additional fainter bands both larger and smaller than the expected size were also visible; these may reflect post-translational processing but were not studied further. Co-transfection of these constructs with a plasmid expressing a 55kDa fusion protein, NS4B5-GFP, which is a target for the DENV protease (Medin et al., 2015) confirmed the lack of protease activity for both NS2B3-S135A (Figure 2C lane 4) and NS3 alone (Figure 2C lane 2), whereas the active DENV NS2B3 plasmid cleaved the NS4B5-GFP fusion protein to yield NS4B, detected by the appearance of a new band at the expected size of 27kDa. (Other bands in Figure 2C were also seen in untransfected cell lysates stained with this rabbit polyclonal antibody.) A significant decrease in Flag-RIPK1 was detected with overexpression of NS2B3 and NS3 alone (Figure 2D lanes 2-5 & Figure 2E), whereas Flag-RIPK1 levels were not affected by the expression of DENV NS2B3-S135A or NS1 (Figure 2D lanes 6-9 & Figure 2E). Since the results with expression of NS3 alone confirm that protease activity is not required for the decrease in RIPK1, the lack of a similar effect with expression of NS2B3-S135A suggests that cleavage between NS2B and NS3 may be required for NS3 to exert this effect.
RIPK1 physically interacts with DENV NS3
To evaluate if the reduction in RIPK1 is due to a direct interaction of NS3 with RIPK1, we next performed co-immunoprecipitation experiments. HEK 293T cells were co-transfected with a plasmid expressing Flag-tagged RIPK1 and plasmids expressing either NS3 lacking the NS2B domain, wild type (WT) DENV NS2B3 or NS2B3-S135A. Plasmids expressing Flag tagged GFP or DENV NS1 and NS5 proteins served as controls. DENV NS3, WT NS2B3, NS2B3-S135A, but not NS1 or NS5, co-immunoprecipitated with RIPK1 when pulled down with anti-Flag (Figure 3A, lanes 2, 3, 4). The reciprocal immunoprecipitation using anti-V5 antibodies confirmed this interaction (Figure 3B, lanes 2, 3, 4). NS3 protease activity was not required for the interaction with RIPK1, because neither lack of the NS2B domain nor the S135A mutation prevented NS3 interacting with RIPK1. We then validated this interaction of RIPK1 and NS3 in the context of DENV infection. HEK 293T cells transiently expressing Flag-RIPK1 were infected with DENV2 for 48hrs, then RIPK1 was immunoprecipitated using an anti-Flag antibody. We detected DENV NS3 bound to RIPK1 by Western blot analysis (Figure 3C lane 2). We performed the reciprocal immunoprecipitation using an anti-DENV NS3 antibody and detected RIPK1 bound to DENV-NS3 by Western blot analysis, verifying the interaction (Figure 3C lane 2).
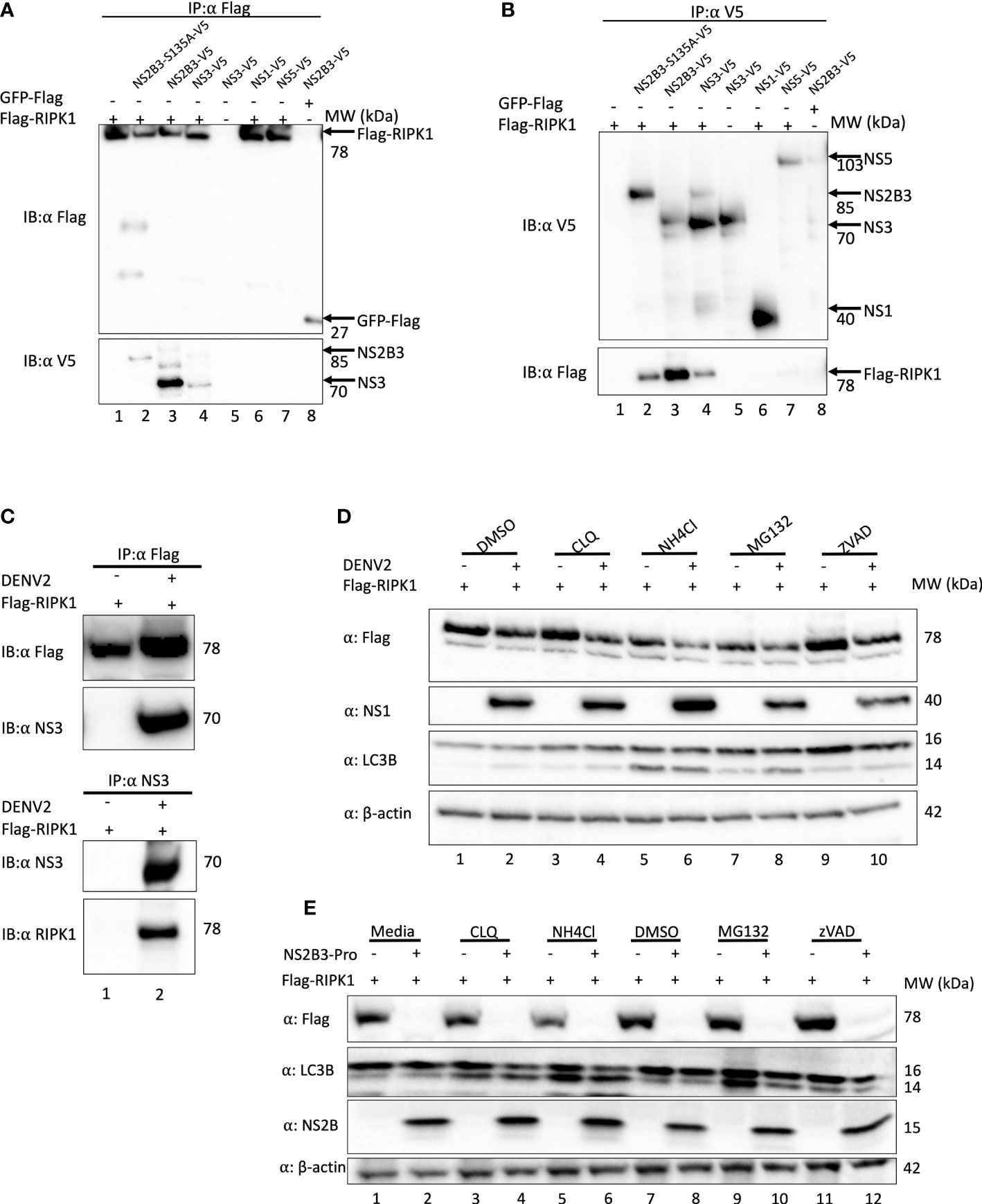
Figure 3 DENV NS3 binds to RIPK1 but does not mediate its degradation via proteasomal or lysosomal pathways. (A, B) Co-immunoprecipitation analyses of HEK 293T cells transiently overexpressing Flag-RIPK1 and indicated plasmids using α-Flag (A) or α-V5 (B) for pull down. (C) Co-immunoprecipitation analyses of HEK 293T cells transfected with Flag-RIPK1 and mock (media) or DENV2 (MOI=1) infected and cultured for 48hrs. (D) Western blot analysis of HEK293T cells infected with DENV2 for 48hrs and treated with Chloroquine (CLQ, 10μM), NH4Cl (15mM), MG132 (2.5μM), ZVAD (10μM) or DMSO for 6hrs and analyzed by indicated antibodies. β-actin is used as a loading control. (E) Co-expression of Flag-RIPK1 and DENV NS2B3-pro for 40hrs followed by treatment with indicated inhibitors for 6hrs and analyzed by western blot using indicated antibodies. Results are representative of at least three independent experiments. IP, immunoprecipitation; IB, immunoblot.
DENV has been shown to increase protein degradation via the autophagy-lysosome pathway. DENV protease co-factor NS2B interacted with and promoted autophagosome-mediated degradation of cGAS, which is a key component in the DNA sensing pathway (Aguirre et al., 2017). DENV NS2B3 interacted with and promoted lysosome mediated degradation of Nfr2, a component in the antioxidant response (Ferrari et al., 2020). Furthermore, negative regulation of RIPK1 by caspase 8 is well known. During activation of death receptors, active caspase 8 cleaves RIPK1 in the intermediate domain at Asp 324 (Lin et al., 1999).
To test if DENV NS3 also promotes RIPK1 for lysosomal or proteasomal degradation, HEK293T cells were treated with the lysosome inhibitors NH4Cl and chloroquine (CLQ), or the proteasome inhibitor MG132 following DENV2 infection and transfection of the Flag-RIPK. ZVAD-FMK, a pan caspase inhibitor, was used to test the role of caspases in the decrease in RIPK1 caused by DENV NS3. After 6 hours of inhibitor treatments, the cell lysates were analyzed by Western blotting. As previously observed, RIPK1 levels decreased with DENV2 infection (Figure 3D). However, the RIPK1 levels did not recover with the inhibition of either the proteasome or the lysosome pathways during DENV infection (Figure 3D lanes 4,6,8). Chloroquine and NH4Cl treatment clearly increased the LC3-II levels indicating efficient inhibition of lysosomal function (Figure 3D lanes 4,5,6). Caspase inhibition by ZVAD increased Flag-RIPK1 levels in uninfected cells yet the RIPK1 levels were still suppressed by DENV2 (Figure 3D lanes 9,10). Taken together these data suggest that RIPK1 is not subjected to these cellular degradation mechanisms by DENV NS3.
To confirm this result, we obtained a plasmid containing a codon-optimized sequence of the DENV2 NS2B and NS3 protease domain (see Methods). This plasmid caused a profound decrease in RIPK1 levels (Figure 3E). However, we still observed no significant lessening of this effect by treatment with proteasomal, lysosomal or caspase inhibitors (Figure 3E).
DENV impairs TNFR- and TLR3-induced NF-κB activation
RIPK1 is an adapter molecule downstream of TNFR1 and TLRs and plays an important role in downstream signaling leading to NF-κB activation (Micheau and Tschopp, 2003; Cusson-Hermance et al., 2005). Considering the observation of NS3-RIPK1 interaction and subsequent decrease in RIPK1 levels, we wanted to learn the effect of DENV in these signaling pathways. We assessed NF-κB luciferase activity in response to TNFα stimulation with DENV infection in HEK 293T cells. NF-κB luciferase activity was significantly reduced with DENV infection, and this was restored by overexpression of RIPK1 (Figure 4A). A similar pattern of inhibition of NF-κB luciferase activity in response to TNFα stimulation was detected by constructs expressing DENV NS3 or NS2B3 (Figure 4B). Expression of the protease-negative NS2B3-S135A mutant did not have a significant effect.
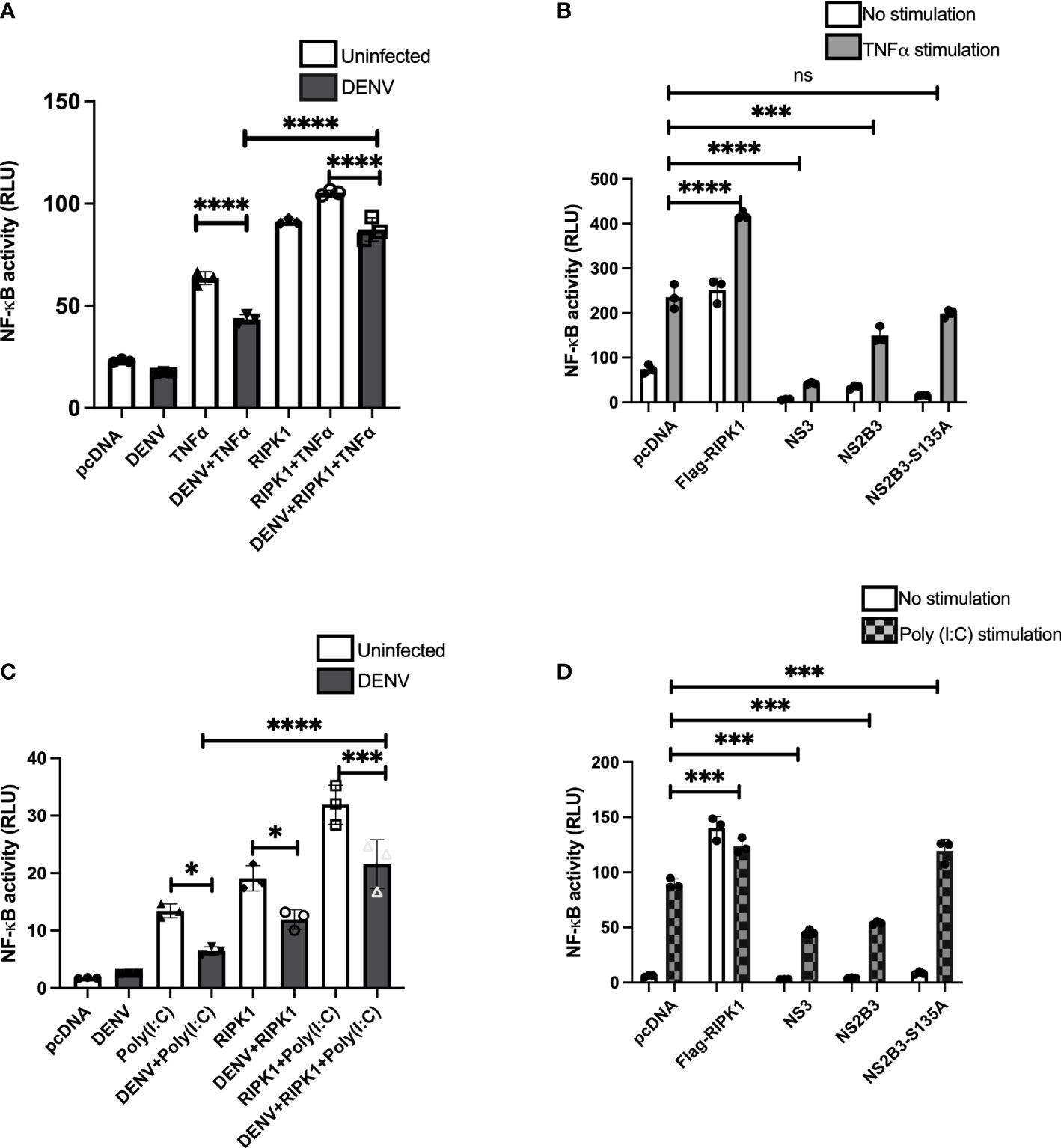
Figure 4 TNFα- and poly(I:C)-driven NF-κB activation is impaired during DENV infection and expression of DENV NS3. (A) HEK 293T cells were co-transfected with NF-κB luciferase reporter, Renilla luciferase reporter and Flag-RIPK1 followed by DENV2 infection (MOI=3). Following TNFα (50ng/ml) treatment for 6hrs, cells were lysed, and luciferase activity was measured at 72hrs post-infection. Open bars=uninfected. Closed bars=DENV infected. (B) HEK 293T cells were co-transfected with NF-κB luciferase reporter, Renilla luciferase reporter, and Flag-RIPK1 along with NS3, WT NS2B3 or NS2B3-S135A. Luciferase activity was detected at 48hrs post-transfection following 6hrs of TNFα (50ng/ml) treatment. (C) NF-κB luciferase activity was assessed in HEK 293T-TLR3 cells as in (A) and cells were stimulated with poly (I:C) 20µg/ml for 24hrs before cell lysis. Open bars=uninfected. Closed bars=DENV infected. (D) HEK 293T-TLR3 cells were co-transfected with NF-κB luciferase reporter, Renilla luciferase reporter, and Flag-RIPK1 along with NS3, WT NS2B3 or NS2B3-S135A. Luciferase activity was detected at 48hrs post-transfection following 24hrs of poly (I:C) 20µg/ml treatment. Results are representative of at least three independent experiments and are expressed as mean ± SD from three replicate wells. (One-way ANOVA, *P < 0.05, ***P < 0.001, ****P < 0.0001). ns, not significant.
We also examined the effect of DENV infection on NF-κB activation in response to TLR3 stimulation. HEK 293T cells stably expressing TLR3 were transfected with the luciferase reporter plasmids with or without the Flag-RIPK1 plasmid and then infected with DENV (MOI=3). They were further stimulated with a synthetic viral RNA mimic, poly I:C, for 24hrs and luciferase activity was measured. As with TNFR1 signaling, DENV infection impaired poly I:C-induced NF-κB activation relative to uninfected cells (Figure 4C). Also similar to the results with TNFR1 stimulation, overexpression of RIPK1 partially overcame the inhibition of poly I:C-induced NF-κB activation by DENV. NF-κB activity following stimulation with poly I:C was also inhibited with overexpression of DENV NS3 or NS2B3 (Figure 4D). Overexpression of the NS2B3-S135A mutant did not inhibit TLR3 signaling, in fact, there was a very small but reproducible increase in poly I:C induced NF-κB activation (Figure 4D). These results demonstrate that DENV infection and expression of NS3 protein impairs NF-κB activation downstream of both TNFR1 and TLR3 by decreasing RIPK1 protein levels.
DENV suppresses TSZ-induced MLKL phosphorylation
Programmed necrosis or necroptosis has been recognized as an antiviral cell death mechanism (Vandenabeele et al., 2010). Since RIPK1 is a critical component in necroptosis induction downstream of TNFR1, TLR3, TLR4 and ZBP1, viruses have evolved proteins to target and decrease RIPK1 levels or block RIPK1 signaling (Mack et al., 2008; Ofengeim and Yuan, 2013; Humphries et al., 2015; Wagner et al., 2015). Numerous cell lines have been shown to undergo necroptosis in response to TNFα, Smac mimetic and ZVAD (T/S/Z) stimulation (He et al., 2009; Cai et al., 2014), and we used U937 cells stably expressing DC-SIGN for our assays. Necroptosis can be pharmacologically inhibited by the RIPK1 inhibitor necrostatin-1 (Degterev et al., 2008). To investigate the effect of DENV on host cell necroptosis we treated DENV-infected and uninfected cells with necroptosis inducers T/S/Z and/or necrostatin-1. Since MLKL phosphorylation at T357/358 is critical for the induction of necroptosis we then performed immunoblot analysis of p-MLKL. T/S/Z treatment induced increased levels of p-MLKL in uninfected cells which was significantly suppressed by treatment with necrostatin-1 (Figures 5A lane 2, 5B). In contrast, p-MLKL was only faintly detected in DENV-infected cells following T/S/Z treatment (Figures 5A lane 5, 5B). These data indicate that DENV inhibits MLKL phosphorylation upon T/S/Z stimulation, a marker of necroptosis, potentially due to the decrease in RIPK1 levels (Figure 5C).
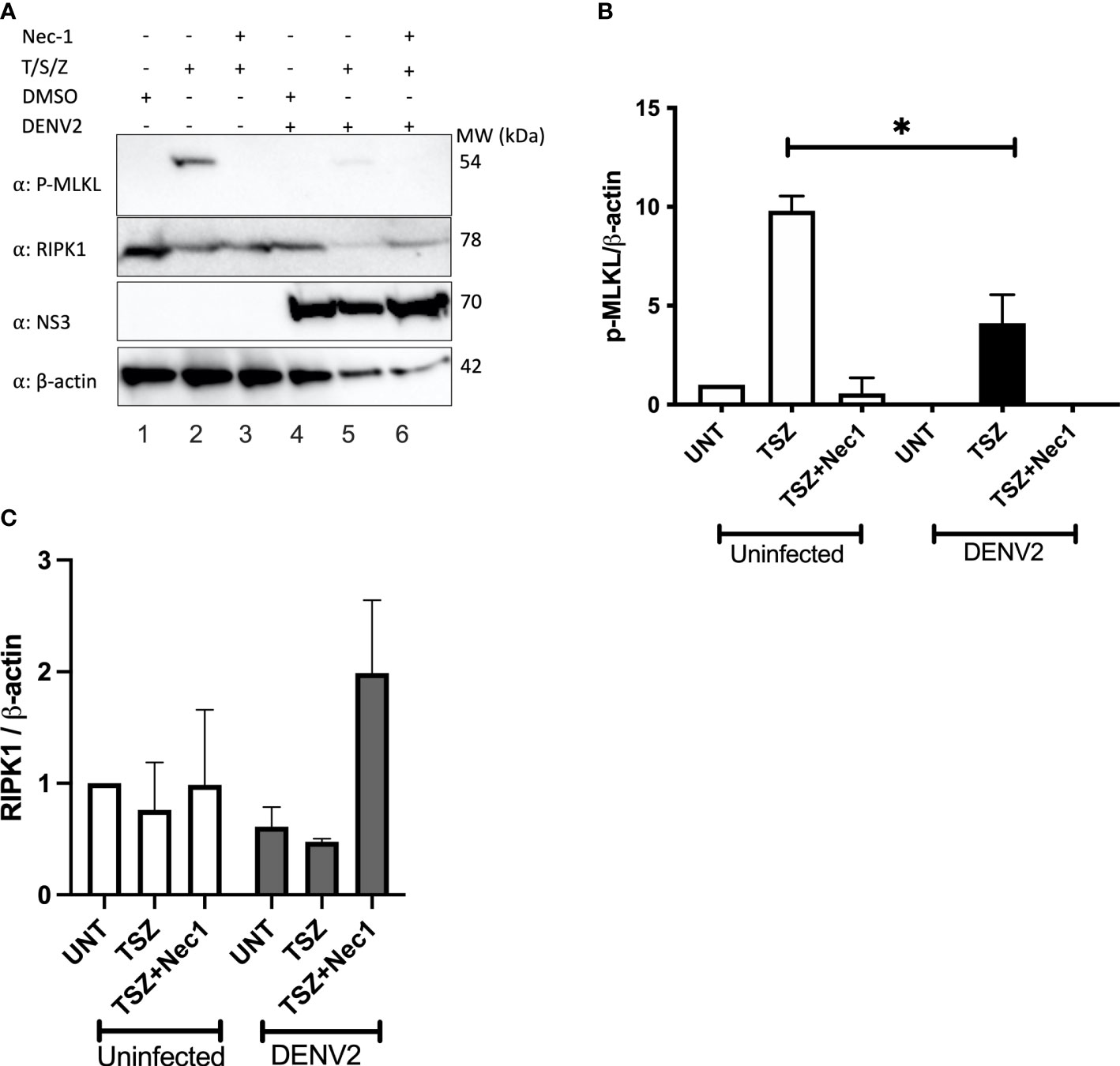
Figure 5 T/S/Z-induced MLKL phosphorylation is suppressed by DENV. (A) Western blot analysis to detect phosphorylated MLKL (p-MLKL), RIPK1, DENV NS3 and β-actin in cell lysates from DENV infected or uninfected U937-DC-SIGN cells. Cells were treated with T/S/Z, T/S/Z +Nec-1 or the vehicle control, DMSO. T-TNFα (50ng/ml), S-Smac mimetic (10μM), Z-ZVAD-FMK (20μM), Necrostatin-1 (Nec-1/50μM). (B) Densitometric analysis of phosphorylated MLKL and (C) endogenous RIPK1 band intensities normalized to β-actin levels. Results are representative of at least two independent experiments and are expressed as mean ± SD from two biological replicates. (t test, *P < 0.05).
Discussion
RIPK1 is a critical regulator in innate immune responses, hence it has emerged as an attractive target for inhibition by numerous virus families (Udawatte and Rothman, 2021). RIPK1 plays a protective role against flavivirus infections. Mice expressing kinase dead RIPK1 (RIPK1KD/KD) were more susceptible to West Nile virus infections than WT mice (Daniels et al., 2017). Furthermore, RIPK1KD/KD mice had higher viral titers than controls upon infection with Zika virus (Daniels et al., 2019). To date, a role for RIPK1 during DENV infection has not been established. In this study we show that intracellular RIPK1 levels are reduced by DENV infection in vitro, and that this reduction in intracellular RIPK1 levels is associated with inhibition of RIPK1 signaling downstream of TNFR1 (NF-κB activation and necroptosis induction) and TLR3 (NF-κB activation). We further show that these effects are caused by expression of the DENV NS3 protein. The effect was not directly dependent on NS3 protease activity or on the presence of NS2B, however, cleavage of NS2B from NS3 was necessary for the decrease in RIPK1 and for inhibition of RIPK1 signaling. These findings add RIPK1 to the list of host proteins targeted by the DENV NS3 (Yu et al., 2015; Stabell et al., 2018).
RIPK1 signaling is known to be inhibited by numerous viral proteins. HIV-1 protease (PR) has been shown to cleave RIPK1. Thus, HIV-1 PR suppress RIPK1-mediated NF-κB activation and necroptosis (Wagner et al., 2015). MCMV protein M45 is known to block RIPK1 signaling by interacting with RIPK1 and blocking RIPK1 interacting with other signaling proteins (Mack et al., 2008; Upton et al., 2008). DENV NS3 protein expression was sufficient to decrease RIPK1 levels. The protease activity did not play a role in decreasing RIPK1 levels, as shown by the significant decrease in RIPK1 with overexpression of a NS3 construct lacking the cofactor NS2B. However, a protease inactive mutant with NS2B intact interacted with RIPK1 but did not affect RIPK1 levels. Therefore, the presence of NS2B may be preventing the RIPK1 decrease by an unknown mechanism. NS2B3-S135A was also shown to interact with STING even though it did not affect STING levels (Aguirre et al., 2012). We were not able to clearly define a mechanism by which DENV NS3 promotes the decrease in RIPK1 levels. It has been reported that DENV NS3 blocks RIG-I and 14-3-3ε signaling in a proteolysis-independent manner (Chan and Gack, 2016). DENV NS2B3 was shown to promote Nrf2 for lysosomal degradation by a protease-independent mechanism (Ferrari et al., 2020). DENV NS2B was also shown to promote cGAS for lysosomal degradation. Our data suggest that neither proteasomal nor lysosomal degradation was responsible for the decrease in RIPK1 levels by DENV infection or NS3 expression (Figures 3D, E).
Increased serum TNFα levels have been reported in patients with dengue (Yadav et al., 1991). The interaction of TNFα and TNFR1 results in the formation of complex I involving RIPK1 leading to NF-κB activation (Aggarwal, 2003). RIPK1 also signals for NF-κB activation downstream of TLR3 (Meylan et al., 2004). NF-κB transcription factors regulate many genes involved in immune responses. Therefore, signaling leading to NF-κB activation serves as an anti-viral immune mechanism. Viruses inhibit NF-κB pathway activation to evade these host immune and inflammatory responses (Zhao et al., 2015). DENV infection was shown to block NF-κB activation downstream of TLR signaling resulting in downregulated cytokine production (Chang et al., 2012). Similar studies in Huh7 and monocyte-derived macrophages showed that DENV infection altered cellular responses to TNFα stimulation (Wati et al., 2011). However, the mechanism by which DENV caused these altered responses had not been defined. We demonstrated that DENV infection diminished NF-κB activation in response to TNFα or poly I:C stimulation and that this effect could be partially overcome by overexpression of RIPK1. We also observed that expression of DENV NS3 caused a similar inhibition of NF-κB activation in response to TNFα or polyI:C. These results indicate that the levels of RIPK1 are important for regulating upstream signals leading to NF-κB activation. A detailed understanding of the mechanism by which the RIPK1-NS3 interaction affects the formation of signaling complexes downstream of TNFR1 and TLR3 will require further investigation.
Liver injury is a common finding in DENV infection and can lead to organ failure. Apoptosis of hepatocytes following DENV infection is believed to contribute to this phenomenon (Samanta, 2015). Recent findings show a protective role of RIPK1 in TNFα-mediated apoptosis in hepatocytes (Filliol et al., 2016; Farooq et al., 2019). RIPK1 knockout mice showed enhanced sensitivity to TNFα-mediated apoptosis and inflammation. Induction of NF-κB target gene transcripts was also suppressed in these mice (Filliol et al., 2016). In light of these reports, we speculate that loss of RIPK1 during DENV infection may sensitize hepatocytes to cell death and inflammation, which could contribute to liver injury in DHF.
RIPK1-mediated necroptosis has been described as a general strategy to inhibit viral replication (Huang et al., 2015; Shubina et al., 2020; Udawatte and Rothman, 2021). Several viruses are known to inhibit necroptosis by targeting RIPK1, and different mechanisms have been described (Udawatte and Rothman, 2021). To our knowledge ours is the first report that DENV infection can inhibit T/S/Z-induced phosphorylation of MLKL, an indicator of necroptosis. Modulating cell death pathways would be advantageous for non-cytolytic viruses, as inhibiting or delaying cell death could increase the number of progeny viruses, and this effect might contribute to disease pathogenesis.
This is the first description of DENV targeting RIPK1, and the first study to link inhibition of NF-κB signaling by DENV to targeting of RIPK1 by the DENV NS3 protein. Furthermore, this is the first evidence of suppression of necroptosis by DENV. These findings open new research directions on the interplay of DENV with various emerging functions of RIPK1. Further studies are required to shed light on cell- and context-dependent effects of RIPK1 reduction in vivo during DENV infection. Finally, we believe that these data expand knowledge of strategies used by DENV to evade innate immune responses.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
DU, CM, and AR contributed to the conception and design of the experiments. JC contributed critical reagents. DU and DL performed the experiments and collected the data. DU analyzed the data, prepared the final figures, and wrote the first draft of the manuscript. CM and AR obtained funding for the experiments. All authors contributed to the article and approved the submitted version.
Funding
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the (NIH) under award number P01AI034533. The funder played no role in the study. The content is solely the responsibility of the authors and does not necessarily represent the official views of the (NIH).
Acknowledgments
We would like to thank Dr. Katherine Fitzgerald for generously providing HEK 293T cells stably expressing TLR3, and Dr. Drishya Diwaker for technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aggarwal, B. B. (2003). Signalling pathways of the TNF superfamily: A double-edged sword. Nat. Rev. Immunol. 3 (9), 745–756. doi: 10.1038/nri1184
Aguirre, S., Luthra, P., Sanchez-Aparicio, M. T., Maestre, A. M., Patel, J., Lamothe, F., et al. (2017). Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat. Microbiol., 2:17037. doi: 10.1038/nmicrobiol.2017.37
Aguirre, S., Maestre, A. M., Pagni, S., Patel, J. R., Savage, T., Gutman, D., et al. (2012). DENV inhibits type I IFN production in infected cells by cleaving human STING. PloS Pathog. 8 (10), e1002934. doi: 10.1371/journal.ppat.1002934
Bhatt, S., Gething, P. W., Brady, O. J., Messina, J. P., Farlow, A. W., Moyes, C. L., et al. (2013). The global distribution and burden of dengue. Nature 496 (7446), 504–507. doi: 10.1038/nature12060
Cai, Z., Jitkaew, S., Zhao, J., Chiang, H. C., Choksi, S., Liu, J., et al. (2014). Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat. Cell Biol. 16 (1), 55–65. doi: 10.1038/ncb2883
Chan, Y. K., Gack, M. U. (2016). A phosphomimetic-based mechanism of dengue virus to antagonize innate immunity. Nat. Immunol. 17 (5), 523–530. doi: 10.1038/ni.3393
Chang, T. H., Chen, S. R., Yu, C. Y., Lin, Y. S., Chen, Y. S., Kubota, T., et al. (2012). Dengue virus serotype 2 blocks extracellular signal-regulated kinase and nuclear factor-κB activation to downregulate cytokine production. PloS One 7 (8), 4–11. doi: 10.1371/journal.pone.0041635
Cho, Y. S., Challa, S., Moquin, D., Genga, R., Ray, T. D., Guildford, M., et al. (2009). Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137 (6), 1112–1123. doi: 10.1016/j.cell.2009.05.037
Clum, S., Ebner, K. E., Padmanabhan, R. (1997). Cotranslational membrane insertion of the serine proteinase precursor NS2B-NS3(Pro) of dengue virus type 2 is required for efficient in vitro processing and is mediated through the hydrophobic regions of NS2B. J. Biol. Chem. 272 (49), 30715–30723. doi: 10.1074/jbc.272.49.30715
Croft, S. N., Walker, E. J., Ghildyal, R. (2018). Human rhinovirus 3C protease cleaves RIPK1, concurrent with caspase 8 activation. Sci. Rep. 8 (1), 1–11. doi: 10.1038/s41598-018-19839-4
Cusson-Hermance, N., Khurana, S., Lee, T. H., Fitzgerald, K. A., Kelliher, M. A. (2005). Rip1 mediates the trif-dependent toll-like receptor 3- and 4-induced NF-??B activation but does not contribute to interferon regulatory factor 3 activation. J. Biol. Chem. 280 (44), 36560–36566. doi: 10.1074/jbc.M506831200
Daniels, B. P., Kofman, S. B., Smith, J. R., Norris, G. T., Snyder, A. G., Kolb, J. P., et al. (2019). The nucleotide sensor ZBP1 and kinase RIPK3 induce the enzyme IRG1 to promote an antiviral metabolic state in neurons. Immunity 50 (1), 64–76.e4. doi: 10.1016/j.immuni.2018.11.017
Daniels, B. P., Snyder, A. G., Olsen, T. M., Orozco, S., Oguin, T. H., Tait, S. W. G., et al. (2017). RIPK3 restricts viral pathogenesis via cell death-independent neuroinflammation. Cell 169 (2), 301–313.e11. doi: 10.1016/j.cell.2017.03.011
Degterev, A., Hitomi, J., Germscheid, M., Ch’en, I. L., Korkina, O., Teng, X., et al. (2008). Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 4 (5), 313–321. doi: 10.1038/nchembio.83
Falgout, B., Pethel, M., Zhang, Y. M., Lai, C. J. (1991). Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J. Virol. 65 (5), 2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991
Farooq, M., Filliol, A., Simoes Eugénio, M., Piquet-Pellorce, C., Dion, S., Raguenes-Nicol, C., et al. (2019). Depletion of RIPK1 in hepatocytes exacerbates liver damage in fulminant viral hepatitis. Cell Death Dis. 10 (1), 12. doi: 10.1038/s41419-018-1277-3
Ferrari, M., Zevini, A., Palermo, E., Muscolini, M., Alexandridi, M., Etna, M. P., et al. (2020). Dengue virus targets Nrf2 for NS2B3-mediated degradation leading to enhanced oxidative stress and viral replication. J. Virol. 94 (24), e01551–20. doi: 10.1128/JVI.01551-20
Filliol, A., Piquet-Pellorce, C., Le Seyec, J., Farooq, M., Genet, V., Lucas-Clerc, C., et al. (2016). RIPK1 protects from TNF-α-mediated liver damage during hepatitis. Cell Death Dis. 7 (11), 1–13. doi: 10.1038/cddis.2016.362
He, S., Wang, L., Miao, L., Wang, T., Du, F., Zhao, L., et al. (2009). Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-α. Cell 137 (6), 1100–1111. doi: 10.1016/j.cell.2009.05.021
Ho, L. J., Wang, J. J., Shaio, M. F., Kao, C. L., Chang, D. M., Han, S. W., et al. (2001). Infection of human dendritic cells by dengue virus causes cell maturation and cytokine production. J. Immunol. 166 (3), 1499–1506. doi: 10.4049/jimmunol.166.3.1499
Hsu, H., Huang, J., Shu, H. B., Baichwal, V., Goeddel, D. V. (1996). TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity 4 (4), 387–396. doi: 10.1016/S1074-7613(00)80252-6
Huang, Z., Wu, S. Q., Liang, Y., Zhou, X., Chen, W., Li, L., et al. (2015). RIP1/RIP3 binding to HSV-1 ICP6 initiates necroptosis to restrict virus propagation in mice. Cell Host Microbe 17 (2), 229–242. doi: 10.1016/j.chom.2015.01.002
Humphries, F., Yang, S., Wang, B., Moynagh, P. N. (2015). RIP kinases: Key decision makers in cell death and innate immunity. Cell Death Differ 22 (2), 225–236. doi: 10.1038/cdd.2014.126
King, A. D., Nisalak, A., Kalayanrooj, S., Myint, K. S., Pattanapanyasat, K., Nimmannitya, S., et al. (1999). B cells are the principal circulating mononuclear cells infected by dengue virus. Southeast Asian J. Trop. Med. Public Health 30 (4), 718–728.
Lin, Y., Devin, A., Rodriguez, Y., Liu, Z. G. (1999). Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev. 13 (19), 2514–2526. doi: 10.1101/gad.13.19.2514
Liu, X., Li, Y., Peng, S., Yu, X., Li, W., Shi, F., et al. (2018). Epstein-Barr Virus encoded latent membrane protein 1 suppresses necroptosis through targeting RIPK1/3 ubiquitination article. Cell Death Dis. 9 (2), 53. doi: 10.1038/s41419-017-0081-9
Liu, L., Wen, K., Li, J., Hu, D., Huang, Y., Qiu, L., et al. (2012). Comparison of plaque- and enzyme-linked immunospot-based assays to measure the neutralizing activities of monoclonal antibodies specific to domain III of dengue virus envelope protein. Clin. Vaccine Immunol. 19 (1), 73–78. doi: 10.1128/CVI.05388-11
Low, J. G. H., Ooi, E. E., Vasudevan, S. G. (2017). Current status of dengue therapeutics research and development. J. Infect. Dis. 215 (Suppl 2), S96–102. doi: 10.1093/infdis/jiw423
Luo, D., Xu, T., Watson, R. P., Scherer-Becker, D., Sampath, A., Jahnke, W., et al. (2008). Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 27 (23), 3209–3219. doi: 10.1038/emboj.2008.232
Mack, C., Sickmann, A., Lembo, D., Brune, W. (2008). Inhibition of proinflammatory and innate immune signaling pathways by a cytomegalovirus RIP1-interacting protein. Proc. Natl. Acad. Sci. U.S.A. 105 (8), 3094–3099. doi: 10.1073/pnas.0800168105
Martina, B. E. E., Koraka, P., Osterhaus, A. D. M. E. (2009). Dengue virus pathogenesis: An integrated view. Clin. Microbiol. Rev. 22 (4), 564–581. doi: 10.1128/CMR.00035-09
Mathew, A., Rothman, A. L. (2008). Understanding the contribution of cellular immunity to dengue disease pathogenesis. Immunol. Rev. 225 (1), 300–313. doi: 10.1111/j.1600-065X.2008.00678.x
Medin, C. L., Valois, S., Patkar, C. G., Rothman, A. L. (2015). A plasmid-based reporter system for live cell imaging of dengue virus infected cells. J. Virol. Methods 211, 55–62. doi: 10.1016/j.jviromet.2014.10.010
Meylan, E., Burns, K., Hofmann, K., Blancheteau, V., Martinon, F., Kelliher, M., et al. (2004). RIP1 is an essential mediator of toll-like receptor 3-induced NF κB activation. Nat. Immunol. 5 (5), 503–507. doi: 10.1038/ni1061
Micheau, O., Tschopp, J. (2003). Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114 (2), 181–190. doi: 10.1016/S0092-8674(03)00521-X
Ofengeim, D., Yuan, J. (2013). Regulation of RIP1 kinase signalling at the crossroads of inflammation and cell death. Nat. Rev. Mol. Cell Biol. 14 (11), 727–736. doi: 10.1038/nrm3683
Rajput, A., Kovalenko, A., Bogdanov, K., Yang, S. H., Kang, T. B., Kim, J. C., et al. (2011). RIG-I RNA helicase activation of irf3 transcription factor is negatively regulated by caspase-8-mediated cleavage of the RIP1 protein. Immunity 34 (3), 340–351. doi: 10.1016/j.immuni.2010.12.018
Sadon, N., Delers, A., Jarman, R. G., Klungthong, C., Nisalak, A., Gibbons, R. V., et al. (2008). A new quantitative RT-PCR method for sensitive detection of dengue virus in serum samples. J. Virol. Methods 153 (1), 1–6. doi: 10.1016/j.jviromet.2008.06.023
Samanta, J. (2015). Dengue and its effects on liver. World J. Clin. cases 3 (2), 125. doi: 10.12998/wjcc.v3.i2.125
Shubina, M., Tummers, B., Boyd, D. F., Zhang, T., Yin, C., Gautam, A., et al. (2020). Necroptosis restricts influenza a virus as a stand-alone cell death mechanism. J. Exp. Med. 217 (11), e20191259. doi: 10.1084/jem.20191259
Silveira, G. F., Wowk, P. F., Cataneo, A. H. D., dos Santos, P. F., Delgobo, M., Stimamiglio, M. A., et al. (2018). Human T lymphocytes are permissive for dengue virus replication. J. Virol. 92 (10), e02181–e02117. doi: 10.1128/JVI.02181-17
Stabell, A. C., Meyerson, N. R., Gullberg, R. C., Gilchrist, A. R., Webb, K. J., Old, W. M., et al. (2018). Dengue viruses cleave STING in humans but not in nonhuman primates, their presumed natural reservoir. Elife 7, 1–25. doi: 10.7554/eLife.31919
Udawatte, D. J., Rothman, A. L. (2021). Viral suppression of RIPK1-mediated signaling. MBio. 12(4), e0172321. doi: 10.1128/mBio.01723-21
Upton, J. W., Kaiser, W. J. (2017). DAI another way: Necroptotic control of viral infection. Cell Host Microbe 21 (3), 290–293. doi: 10.1016/j.chom.2017.01.016
Upton, J. W., Kaiser, W. J., Mocarski, E. S. (2008). Cytomegalovirus M45 cell death suppression requires receptor-interacting protein (RIP) homotypic interaction motif (RHIM)-dependent interaction with RIP1. J. Biol. Chem. 283 (25), 16966–16970. doi: 10.1074/jbc.C800051200
Vandenabeele, P., Galluzzi, L., Vanden Berghe, T., Kroemer, G. (2010). Molecular mechanisms of necroptosis: An ordered cellular explosion. Nat. Rev. Mol. Cell Biol. 11 (10), 700–714. doi: 10.1038/nrm2970
Wagner, R. N., Reed, J. C., Chanda, S. K. (2015). HIV-1 protease cleaves the serine-threonine kinases RIPK1 and RIPK2. Retrovirology 12 (1), 1–16. doi: 10.1186/s12977-015-0200-6
Wati, S., Rawlinson, S. M., Ivanov, R. A., Dorstyn, L., Beard, M. R., Jans, D. A., et al. (2011). Tumour necrosis factor alpha (TNF-α) stimulation of cells with established dengue virus type 2 infection induces cell death that is accompanied by a reduced ability of TNF-α to activate nuclear factor κb and reduced sphingosine kinase-1 activity. J. Gen. Virol. 92 (4), 807–818. doi: 10.1099/vir.0.028159-0
Wengler, G., Czaya, G., Färber, P. M., Hegemann, J. H. (1991). In vitro synthesis of West Nile virus proteins indicates that the amino-terminal segment of the NS3 protein contains the active centre of the protease which cleaves the viral polyprotein after multiple basic amino acids. J. Gen. Virol. 72 (Pt 4), 851–858. doi: 10.1099/0022-1317-72-4-851
Xu, T., Sampath, A., Chao, A., Wen, D., Nanao, M., Chene, P., et al. (2005). Structure of the dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 a. J. Virol. 79 (16), 10278–10288. doi: 10.1128/JVI.79.16.10278-10288.2005
Yadav, M., Kamath, K. R., Iyngkaran, N., Sinniah, M. (1991). Dengue haemorrhagic fever and dengue shock syndrome: Are they tumour necrosis factor-mediated disorders? FEMS Microbiol. Lett. 89 (1), 45–49. doi: 10.1111/j.1574-6968.1991.tb04969.x
Yu, C.-Y., Hsu, Y.-W., Liao, C.-L., Lin, Y.-L. (2006). Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J. Virol. 80 (23), 11868–11880. doi: 10.1128/JVI.00879-06
Yu, C. Y., Liang, J. J., Li, J. K., Lee, Y. L., Chang, B. L., Su, C. I., et al. (2015). Dengue virus impairs mitochondrial fusion by cleaving mitofusins. PloS Pathog. 11 (12), 1–24. doi: 10.1371/journal.ppat.1005350
Keywords: dengue virus, NS3, ripk1, NF-κB, necroptosis
Citation: Udawatte DJ, Lang DM, Currier JR, Medin CL and Rothman AL (2022) Dengue virus downregulates TNFR1- and TLR3-stimulated NF-κB activation by targeting RIPK1. Front. Cell. Infect. Microbiol. 12:926036. doi: 10.3389/fcimb.2022.926036
Received: 22 April 2022; Accepted: 13 September 2022;
Published: 14 October 2022.
Edited by:
S. Gowri Sankar, Vector Control Research Centre (ICMR), IndiaReviewed by:
Izabela Agata Rodenhuis-Zybert, University Medical Center Groningen, NetherlandsMichelle Kelliher, University of Massachusetts Medical School, United States
Copyright © 2022 Udawatte, Lang, Currier, Medin and Rothman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alan L. Rothman, YWxhbl9yb3RobWFuQHVyaS5lZHU=
 Darshika J. Udawatte
Darshika J. Udawatte Diane M. Lang1
Diane M. Lang1 Jeffrey R. Currier
Jeffrey R. Currier Alan L. Rothman
Alan L. Rothman