- 1Department of Microbiology, Faculty of Medicine, University Hospital Pilsen, Charles University, Pilsen, Czechia
- 2Biomedical Center, Faculty of Medicine, Charles University, Pilsen, Czechia
- 3Department of Natural Sciences, School of Arts and Sciences, Lebanese American University, Byblos, Lebanon
- 4Department of Pathology and Laboratory Medicine, American University of Beirut Medical Center, Beirut, Lebanon
Resistance to ceftolozane/tazobactam (C/T) in Pseudomonas aeruginosa is a health concern. In this study, we conducted a whole-genome-based molecular characterization to correlate resistance patterns and β-lactamases with C/T resistance among multi-drug resistant P. aeruginosa clinical isolates. Resistance profiles for 25 P. aeruginosa clinical isolates were examined using disk diffusion assay. Minimal inhibitory concentrations (MIC) for C/T were determined by broth microdilution. Whole-genome sequencing was used to check for antimicrobial resistance determinants and reveal their genetic context. The clonal relatedness was evaluated using MLST, PFGE, and serotyping. All the isolates were resistant to C/T. At least two β-lactamases were detected in each with the blaOXA-4, blaOXA-10, blaOXA-50, and blaOXA-395 being the most common. blaIMP-15, blaNDM-1, or blaVIM-2, metallo-β-lactamases, were associated with C/T MIC >256 μg/mL. Eight AmpC variants were identified, and PDC-3 was the most common. We also determined the clonal relatedness of the isolates and showed that they grouped into 11 sequence types (STs) some corresponding to widespread clonal complexes (ST111, ST233, and ST357). C/T resistance was likely driven by the acquired OXA β-lactamases such as OXA-10, and OXA-50, ESBLs GES-1, GES-15, and VEB-1, and metallo- β-lactamases IMP-15, NDM-1, and VIM-2. Collectively, our results revealed C/T resistance determinants and patterns in multi-drug resistant P. aeruginosa clinical isolates. Surveillance programs should be implemented and maintained to better track and define resistance mechanisms and how they accumulate and interact.
Introduction
Antimicrobial resistance has been a soaring global health care tolling problem (Tacconelli et al., 2018; Moghnieh et al., 2019). Pseudomonas aeruginosa is one of the leading multidrug resistant (MDR) nosocomial pathogens worldwide and defined by the World Health Organization as a critical health concern with limited effective treatment options and is associated with poor clinical outcomes (Weiner et al., 2016; Tacconelli et al., 2018). MDR P. aeruginosa isolates have a broad variety of mechanisms mediating antimicrobial resistance (Gellatly and Hancock, 2013; López-Causapé et al., 2018). These include the up-regulation of efflux pumps, loss of outer membrane porins, the production of AmpC, extended-spectrum β-lactamases (ESBLs) and carbapenemases, and modification of antimicrobial target sites (Lister et al., 2009; Zavascki et al., 2010; Bassetti et al., 2019). The overproduction of AmpC β-lactamase was also linked to cephalosporins resistance and which was not reversed by β-lactamase inhibitors such as tazobactam and clavulanic acid (Sligl et al., 2015).
Two drug combinations were developed to treat infections caused by resistant Gram-negative bacteria, namely ceftazidime/avibactam and ceftolozane/tazobactam (C/T) (Sold under the brand name Zerbaxa) (Wright et al., 2017). Ceftolozane is a cephalosporin derivative of ceftazidime with an intrinsic broad activity and is not hydrolyzed by most broad-spectrum β-lactamases such as ESBLs and AmpCs (van Duin and Bonomo, 2016). Ceftolozane is particularly active against P. aeruginosa exhibiting AmpC efflux pumps overexpression (Moyá et al., 2012; Giacobbe et al., 2018), and has a heavier pyrazole substituent at the 3-position side chain instead of the lighter pyridium in ceftazidime, enhancing steric hindrance and interfering with AmpC hydrolytic activity (van Duin and Bonomo, 2016). The combination of ceftolozane and the β-lactamase inhibitor tazobactam proved to be active against many, but not all, ESBL-producing Gram-negative bacteria (Farrell et al., 2014). In particular, C/T combination was active against MDR and carbapenem-resistant P. aeruginosa (Giacobbe et al., 2018). The overall clinical success rate was reported to be 76.2% among MDR and extensively drug-resistant (XDR) P. aeruginosa (Maraolo et al., 2020). C/T drug combination was also more effective compared to polymyxins or aminoglycosides (Pogue et al., 2019). However, regional variations in C/T resistance within MDR P. aeruginosa were reported (Farrell et al., 2014; Sid Ahmed et al., 2019; O’Neall et al., 2020), and AmpC hyperproduction was a factor linked to C/T resistance in P. aeruginosa (Cabot et al., 2014). C/T was also less efficient against ESBL producing Escherichia coli and P. aeruginosa (Ortiz de la Rosa et al., 2019). The C/T activity against the Gram-negative ESBL producers, including E. coli, Klebsiella pneumoniae, and P. aeruginosa, recovered from clinical settings in Lebanon was recently tested by Araj et al. (2020), and which revealed that the C/T against MDR E. coli and K. pneumoniae isolates were much lower (Araj et al., 2020). In light of the rapid increase in the number of C/T P. aeruginosa resistant isolates we thought of studying C/T activity against MDR P. aeruginosa, determining the susceptibilities of the isolates, and conducting a genome-based molecular characterization.
Materials and Methods
Ethical Approval
Ethical approval was not required. The isolates were collected as part of routine clinical care and patient data collection. No additional isolates were collected beyond those obtained from routine clinical care, and no diagnostic or treatment decisions were affected by the outcomes of this study.
Bacterial Isolates and Identification
A total of 25 P. aeruginosa isolates were collected and identified by the Matrix-Assisted Laser Desorption/Ionization Time of Flight (MALDI-TOF) system (Bruker Daltonik, GmbH, Bremen, Germany) at the clinical microbiology laboratory of the American University of Beirut Medical Center (AUBMC), Beirut, Lebanon between December 2017 and November 2018. AUBMC is around 350-bed tertiary care major hospital in the country. The isolates were designated as ZBX-P1 to ZBX-P25.
Antimicrobial Susceptibility Tests
Antimicrobial susceptibility was tested against 10 antibiotics including C/T, amikacin, aztreonam, cefepime, ceftazidime, ciprofloxacin, gentamicin, imipenem, tazobactam and colistin. Data obtained were interpreted according to the CLSI guidelines (CLSI, 2022). For C/T breakpoints in P. aeruginosa, the established EUCAST (2022) (EUCAST, 2022) clinical zone diameter breakpoints were followed; S ≥ 24 mm and R < 24 mm and the MIC breakpoints of S ≤ 4 mg/L and R > 4 mg/L. For disk diffusion, the disk content was set at CXA-TAZ 30-10 µg, by both EUCAST and CLSI (CLSI, 2022; EUCAST, 2022).
Pulsed-Field Gel Electrophoresis (PFGE)
PFGE fingerprinting was performed using the SpeI restriction enzyme (ThermoScientific, Waltham, MA, USA), 1% SeaKem agarose gel, and the universal laboratory standard Salmonella enterica subsp. enterica serovar Braenderup (ATCC® BAA664™) digested with XbaI restriction enzyme according to the standard PulseNet protocol (http://www.pulsenetinternational.org). Electrophoresis was performed using the Bio-Rad laboratories CHEF DR-III system (Bio-Rad Laboratories, Bio-Rad Laboratories Inc., Hercules, CA, USA) with a run time of 16 h and switch time of 5–40 s (https://www.cdc.gov/pulsenet/). Gels were stained with ethidium bromide. PFGE profiles were analyzed with the BioNumerics software version 7.6.1 (Applied Maths, Sint-Martens-Latem, Belgium), with banding patterns showing a difference in three or more bands being placed under distinct pulsotypes (Tenover et al., 1995). Pulsotypes were clustered using the BioNumerics software version 7.6.1 (Applied Maths, Sint-Martens-Latem, Belgium) with an optimization of 0.5% and tolerance of 0.5%.
Whole-Genome Sequencing
DNA extraction was performed using the NucleoSpin Microbial DNA kit (Macherey-Nagel, Germany) according to the manufacturer’s instructions followed by long- read sequencing of the isolates (ZBX-1 to ZBX-25). PacBio long-read sequencing on the Sequel I platform (Pacific Biosciences, CA, USA) was performed. Library preparation was according to the manufacturer’s instructions for microbial isolate multiplexing. G-tubes (Covaris, USA) were used for DNA shearing, and no size selection was performed. Resulting contigs were polished with Pilon, v1.23 (Walker et al., 2014), and the overlapping ends of chromosomes were trimmed after manual inspection of reads mapped by BWA-MEM algorithm as implemented in BWA, v0.7.17 (Li and Durbin, 2010), and Bowtie, v2.3.4.2 (Langmead and Salzberg, 2012). Genome assembly was done using HGAP4 with minimum seed coverage of 30x (Chin et al., 2013).
Genome Analysis
Genomes were annotated using the RAST server (http://rast.nmpdr.org) (Aziz et al., 2012). Resfinder (Zankari et al., 2012), the Comprehensive Antibiotic Resistance Database CARD v3.1.0 (Alcock et al., 2020), MLST v1.8 (Larsen et al., 2012), ISfinder database (Siguier et al., 2006), PlasmidFinder (Carattoli et al., 2014) and BLASTn (Wheeler et al., 2003) were used to identify resistance genes, sequence types (STs), ISs, and plasmid incompatibility group identification and manual curation of annotations, respectively. The serotypes of the isolates were determined using the PAst v1.0. (Thrane et al., 2016) PAst types were split into 12 serogroups covering the 20 serotypes (Thrane et al., 2016). Mutations in AmpC, AmpR, and other target proteins were examined by aligning and comparing the corresponding amino acid sequences to P. aeruginosa PAO1 reference genome (Accession no. GCF_000006765.1).
Porin Analysis
Sequences of 27 genes encoding for porins were extracted from P. aeruginosa PAO1 reference genome (Accession no. GCF_000006765.1) available on the Pseudomonas Genome Database (https://pseudomonas.com/) (Winsor et al., 2016). These sequences were used to build a dataset in MyDbFinder 2.0 (https://cge.cbs.dtu.dk/services/MyDbFinder/). Genomes were blasted against the dataset using a 60% ID threshold and a 40% minimum length for comparison.
Data Availability
The Whole Genome Shotgun project was deposited at DDBJ/ENA/GenBank under the accession numbers presented in S1 Table.
Results
Patient Data
The mean patients’ age was 62.3 ± 18.9 years old. 64% (n=16) were males and 36% (n=9) were females and were collected from various sites of infection including: deep tracheal aspirate (DTA) (32%; n=8), urine (32%; n=8), wound (12%; n=3), sputum (12%; n=3), fluids, bone, and blood (4%; n=1).
Genome Statistics and Isolate Genotyping
The average genome size of the sequenced isolates was 6,972,377 ± 255,784 bp, and the average G+C content was 65.9 ± 0.2%. The clonal relatedness was evaluated using MLST, PFGE, and serotyping. The isolates were grouped into eleven STs (Figure 1) (one new: ST-3425). ST111 (n = 4), ST654 (n= 1), ST308 (n= 1), and ST357 (n= 4) are widespread “high-risk” clones (Woodford et al., 2011; Oliver et al., 2015; Papagiannitsis et al., 2017). The isolates were distributed within seven serotypes and 23 pulsotypes (PT1 to 23) showing ≥84% similarity based on PFGE (Figure 1).
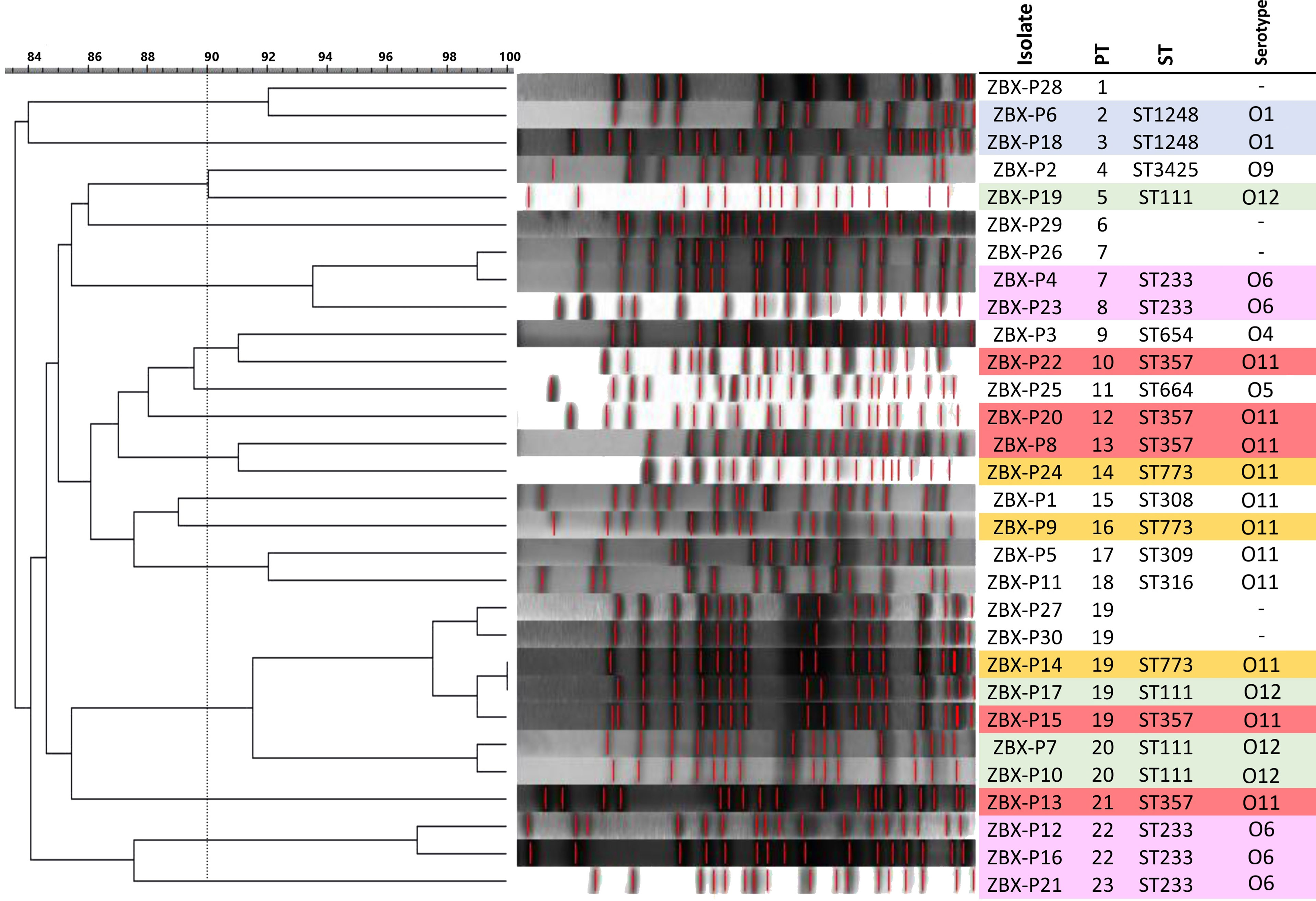
Figure 1 β-lactamases and AmpC antibiotic resistance genes identified in the isolates. Isolate’s ST is also shown.
Antibiotic Susceptibilities
All the isolates were resistant to C/T with MICs ranging between 2 to >256 μg/mL, while most also showed resistance to the other tested β-lactams including ceftazidime (88%; n=22) and imipenem (68%; n=17) (Figure 2). All the isolates were susceptible to colistin, followed by ciprofloxacin (68%; n=17), gentamicin (n = 13; 52%) and amikacin (n = 11; 44%) (Figure 2).

Figure 2 PFGE dendrogram and Serotype in the C/T P. aeruginosa isolates. Dendogram was generated by BioNumerics software version 7.6.1 showing the relationship of the isolates based on their banding patterns generated by SpeI restriction digestion. Isolates’ STs, and pulsotypes (PT) are shown.
Resistance Genomics
Whole-genome sequencing was used to check for antimicrobial resistance determinants and reveal their genetic context (Figure 3 and Table 1). Between two to four β-lactamases were detected in the sequenced genomes; all were chromosomal except for blaGES-15 (Table 1). blaIMP-15, blaNDM-1, or blaVIM-2, metallo-β-lactamases, were associated with C/T MIC >256 μg/mL. Other β-lactamases were also detected with the most common being blaOXA-395, blaOXA-50, blaOXA-4, and blaOXA-10, respectively (Figure 3). MIC distributions are shown in (Figure 2). All C/T resistant isolates with MIC > 256 were either intermediate (n =1) or resistant to ceftazidime (n = 15). Of these, nine had four β-lactamases with one being a metallo-β-lactamase.
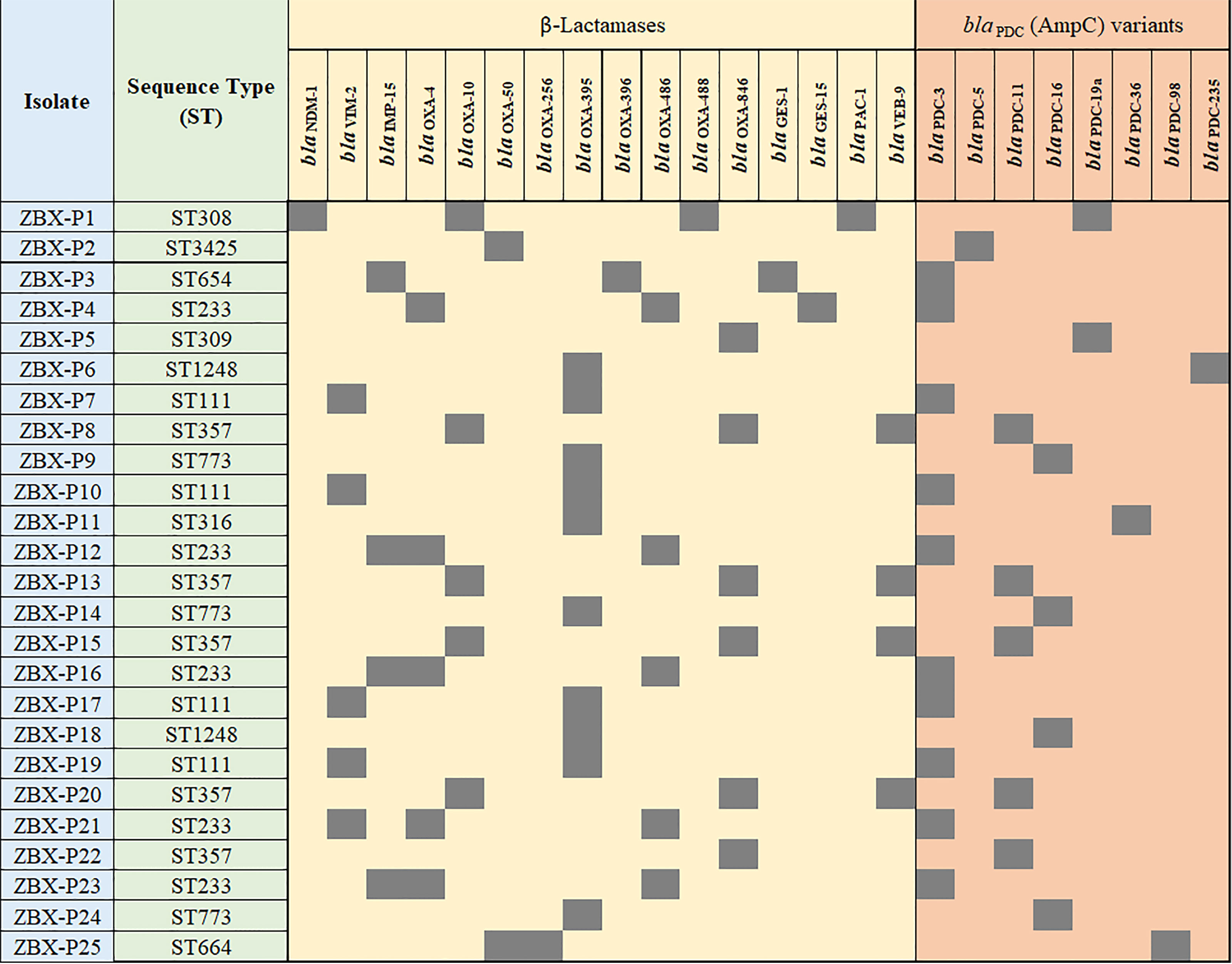
Figure 3 Detailed phenotypic and genotypic resistance information of the C/T resistant P. aeruginosa isolates. ST, Sequence type. Classes of antibiotic resistance genes are marked as follows: aminoglycoside, phenicol, macrolide, tetracycline, fosfomycin, quinolone, rifampicin, trimethoprim, and sulphonamide resistance genes. MIC, Minimum inhibitory concentration (µg/ml), DD, Diffusion diameter (mm). Black, present.
All the isolates were intrinsic AmpC producers showing eight ampC-type variants (PDC) (Figure 3). We checked for polymorphisms in the AmpR (blaPDC transcriptional regulator) aligning against PA4109 in P. aeruginosa PAO1 reference strain. Several substitutions were detected including D135N in isolates with C/T MIC < 256 μg/mL (ZBX-P9 MIC: 8 μg/mL, ZBX-P11 (MIC: 4 μg/mL, and ZBX-P24 MIC: 12 μg/mL), and another restricted to the ones typed as ST111 (E287G C/T MIC > 256 μg/mL). We also looked for OprD polymorphisms and detected the characteristic mutation (Q142X) in ZBX-P12, ZBX-P16, and ZBX-P23 (all with C/T MIC > 256 μg/mL).
Furthermore, blaNDM-1 along with floR and msr(E) were on a chromosomal 74.2 kb integrative and conjugative element (ICE) ICETn43716385. blaOXA-488 was also chromosomal, while blaVIM-2, blaOXA-4, and blaOXA-10 were all detected on chromosomal class I integrons, and blaGES-15 on IncP-6 plasmid (Table 1).
Finally, we compared 27 porin encoding genes with that of P. aeruginosa PAO1. The highest variability was observed in oprD, opdD (PA4501 and PA1025), and oprQ. Deletions and pre-mature stop codons (truncations) were detected throughout OprD, and the characteristic OprD mutation (Q142X) associated with C/T resistance (Fraile-Ribot et al., 2018) was found in ZBX-P12 and ZBX-P16, and ZBX-P23 (C/T MIC > 256 μg/mL).
Discussion
Choosing the appropriate antimicrobial agent to treat infections caused by resistant bacteria would significantly decrease infection-linked morbidity and mortality. C/T has increased activity against resistant P. aeruginosa isolates and is an important treatment option in institutions with high rates of pseudomonal infections (Humphries et al., 2017; Hirsch et al., 2020). The emergence of C/T resistance in MDR/XDR P. aeruginosa isolates, resistant to most β-lactams and having several resistance mechanisms, is very likely to happen. In this study, we used long-read whole-genome sequencing approach to study the resistance genomics and clonal relatedness of 25 C/T resistant P. aeruginosa. The isolates belonged to 11 STs and 23 pulsotypes, exhibited C/T MIC of 1.5 to > 256 μg/mL, showed resistance to the other tested β-lactams including ceftazidime (88%; n=22), ciprofloxacin (68%; n=17), and imipenem (68%; n=17). Moreover, isolates had two to four β-lactamases and 10 were positive for blaIMP-15, blaNDM-1, or blaVIM-2, and were intrinsic AmpC producers.
PDC variants in this study were assigned according to substitutions in AmpC described by Rodríguez-Martínez et al. (2009) and accordingly we identified eight ampC variants, PDC-3 being the most common. T79A (T105A non-processed peptide) detected in PDC-3 (Rodríguez-Martínez et al., 2009), was previously found to be prevalent in carbapenem-resistant clinical isolates. Berrazeg et al. (2015), however, overexpressed PDC-3 (T79A) in porin OprD-negative strain to test if it would potentiate the AmpC action and revealed that T79A variation doesn’t broaden the substrate spectrum of AmpC (Berrazeg et al., 2015). They concluded that PDC common polymorphisms had negligible impact on AmpC activity while confirming that mutations occurring in specific regions of the substrate-binding pocket could enhance the catalytic efficiencies and as a result increase the hydrolytic activity of P. aeruginosa AmpC on cephalosporins including ceftolozane. On the other hand, Fernández-Esgueva et al. (2020) showed that C/T resistance was associated with AmpC mutations including a novel one (PDC-388; G183V) (Fernández-Esgueva et al., 2020) and in line with this Fraile-Ribot et al. (2018) also cloned AmpC variants (PDC-221, 222, and 223) in ampC-deficient derivative of PAO1 (Fraile-Ribot et al., 2018). The cloned AmpC variants showed increased ceftolozane/tazobactam and ceftazidime/avibactam MICs compared with wild type AmpC with the associated polymorphisms being located within the Ω loop and selected in vitro upon C/T exposure (Cabot et al., 2014; Haidar et al., 2017). We detected the characteristic OprD mutation (Q142X) associated with C/T resistance (Fraile-Ribot et al., 2018) in three of the study isolates, ZBX-P12, ZBX-P16, and ZBX-P23 (C/T MIC > 256 μg/mL).
Moreover, C/T resistance emergence was previously reported in isolates producing horizontally acquired β-lactamases such as OXA-10 and OXA-2 (Fraile-Ribot et al., 2018), and ESBLs (Ortiz de la Rosa et al., 2019). In general, C/T may be ineffective against isolates carrying carbapenemases including class A and class D (OXA) β-lactamases and it’s inactive against metallo-β-lactamases (Hirsch et al., 2020; Karlowsky et al., 2021). Extended-spectrum OXAs were also noted as infrequent cause of ceftolozane resistance (Livermore et al., 2009; Juan et al., 2010). Our results mainly agreed with C/T resistance being linked to horizontally acquired β-lactamases. We detected two to four β-lactamases in the sequenced genomes. In line with this, Sid Ahmad et al. (2019) reported a strong association between MDR P. aeruginosa isolates displaying MIC256 to C/T and the presence of OXA-10, VIM-2, and OXA-488 with the highest association frequency being with class C and D β-lactamases. We also had metallo-β-lactamases including blaIMP-15, blaNDM-1, or blaVIM-2 and which were invariably (except for one isolate) linked to C/T MIC >256 μg/mL.
Infections caused by MDR P. aeruginosa could be treated with C/T, clinical studies evaluating optimal dosing and using combined therapy are recommended (Fraile-Ribot et al., 2018). Mutations occurring within the substrate-binding pocket could increase the hydrolytic activity of P. aeruginosa AmpC on cephalosporins including ceftolozane. Horizontally acquired β-lactamases, and intrinsic AmpC modifications are the main mechanisms leading to C/T resistance in MDR P. aeruginosa. Our data don’t support the accumulation of mutations leading to the overexpression or structural modification of AmpC and so it’s more likely that C/T resistance was driven by the acquired OXA β-lactamases such as OXA-10, OXA-50, ESBLs GES-1, GES-15 and VEB-1, and metallo-β-lactamases IMP-15, NDM-1, and VIM-2. Collectively, our results highlight the need to maintain active surveillance programs to better track and define resistance mechanisms and how they accumulate and interact.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
GA: Conceptualization, Validation, Writing – review and editing. ST: Conceptualization, Funding acquisition, Project administration, Supervision, Validation, Writing – review and editing. IB: Funding acquisition, Investigation, Methodology, Writing – review and editing. TS: Investigation, Methodology, Visualization, Writing – original draft, Writing – review and editing. GM: Investigation, Writing – review and editing. JH: Investigation, Methodology. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Lebanese American University Strategic Research Review Committee Grant (SRRC-R-2019-38) and by the research project grants NU20J- 05-00033 provided by Czech Health Research Council, by the Charles University Research Fund PROGRES (project number Q39), and by the project Nr. CZ.02.1.01/0.0/0.0/16_019/0000787 “Fighting Infectious Diseases” provided by the Ministry of Education Youth and Sports of the Czech Republic. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.922976/full#supplementary-material
Supplementary Table 1 | List of the isolates’ accession numbers.
References
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: Antibiotic Resistome Surveillance With the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Araj, G. F., Berjawi, D. M., Musharrafieh, U., Beayni, N. K. E. (2020). Activity of Ceftolozane/Tazobactam Against Commonly Encountered Antimicrobial Resistant Gram-Negative Bacteria in Lebanon. J. Infect. Develop. Ctries. 14, 559–564. doi: 10.3855/jidc.12368
Aubert, D., Girlich, D., Naas, T., Nagarajan, S., Nordmann, P. (2004). Functional and Structural Characterization of the Genetic Environment of an Extended-Spectrum β-Lactamase blaVEB Gene From a Pseudomonas aeruginosa Isolate Obtained in India. Antimicrob. Agents Chemother. 48, 3284–3290. doi: 10.1128/AAC.48.9.3284-3290.2004
Aziz, R. K., Devoid, S., Disz, T., Edwards, R. A., Henry, C. S., Olsen, G. J., et al. (2012). SEED Servers: High-Performance Access to the SEED Genomes, Annotations, and Metabolic Models. PloS One 7, e48053. doi: 10.1371/journal.pone.0048053
Bassetti, M., Castaldo, N., Cattelan, A., Mussini, C., Righi, E., Tascini, C., et al. (2019). Ceftolozane/tazobactam for the Treatment of Serious Pseudomonas aeruginosa Infections: A Multicentre Nationwide Clinical Experience. Int. J. Antimicrobial. Agents 53, 408–415. doi: 10.1016/j.ijantimicag.2018.11.001
Berrazeg, M., Jeannot, K., Ntsogo Enguéné, V. Y., Broutin, I., Loeffert, S., Fournier, D., et al. (2015). Mutations in β-Lactamase AmpC Increase Resistance of Pseudomonas aeruginosa Isolates to Antipseudomonal Cephalosporins. Antimicrob. Agents Chemother. 59, 6248–6255. doi: 10.1128/AAC.00825-15
Cabot, G., Bruchmann, S., Mulet, X., Zamorano, L., Moyà, B., Juan, C., et al. (2014). Pseudomonas aeruginosa Ceftolozane-Tazobactam Resistance Development Requires Multiple Mutations Leading to Overexpression and Structural Modification of AmpC. Antimicrob. Agents Chemother. 58, 3091–3099. doi: 10.1128/AAC.02462-13
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chin, C.-S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, Finished Microbial Genome Assemblies From Long-Read SMRT Sequencing Data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
CLSI. (2022). Performance Standards for Antimicrobial Susceptibility Testing. Available at: https://clsi.org/standards/products/free-resources/access-our-free-resources/.
Ding, Y., Teo, J. W. P., Drautz-Moses, D. I., Schuster, S. C., Givskov, M., Yang, L. (2018). Acquisition of Resistance to Carbapenem and Macrolide-Mediated Quorum Sensing Inhibition by Pseudomonas aeruginosa via Icetn43716385. Commun. Biol. 1, 1–10. doi: 10.1038/s42003-018-0064-0
EUCAST. (2022). The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 12.0 (European Committee on Antimicrobial Susceptibility Testing). Available at: https://www.eucast.org/clinical_breakpoints/ (Accessed April 14, 2022).
Farrell, D. J., Sader, H. S., Flamm, R. K., Jones, R. N. (2014). Ceftolozane/tazobactam Activity Tested Against Gram-Negative Bacterial Isolates From Hospitalised Patients With Pneumonia in US and European Medical Centres, (2012). Int. J. Antimicrob. Agents 43, 533–539. doi: 10.1016/j.ijantimicag.2014.01.032
Fernández-Esgueva, M., López-Calleja, A. I., Mulet, X., Fraile-Ribot, P. A., Cabot, G., Huarte, R., et al. (2020). Characterization of AmpC β-Lactamase Mutations of Extensively Drug-Resistant Pseudomonas aeruginosa Isolates That Develop Resistance to Ceftolozane/Tazobactam During Therapy. Enferm. Infecc. Microbiol. Clin. (Engl. Ed.) 38, 474–478. doi: 10.1016/j.eimc.2020.01.017
Fraile-Ribot, P. A., Cabot, G., Mulet, X., Periañez, L., Martín-Pena, M. L., Juan, C., et al. (2018). Mechanisms Leading to In Vivo Ceftolozane/Tazobactam Resistance Development During the Treatment of Infections Caused by MDR Pseudomonas aeruginosa. J. Antimicrob. Chemother. 73, 658–663. doi: 10.1093/jac/dkx424
Garza-Ramos, J. U., Sanchez-Martinez, G., Barajas, J. M., Suarez, S., Sanchez-Perez, A., Rojas-Moreno, T., et al. (2010). Variability of the blaIMP-15-Containing Integrons, Highly Related to In95, on an Endemic Clone of Pseudomonas aeruginosa in Mexico. Microbial. Drug Resist 16, 191–195. doi: 10.1089/mdr.2010.0017
Gellatly, S. L., Hancock, R. E. W. (2013). Pseudomonas aeruginosa: New Insights Into Pathogenesis and Host Defenses. Pathog. Dis. 67, 159–173. doi: 10.1111/2049-632X.12033
Giacobbe, D. R., Bassetti, M., De Rosa, F. G., Del Bono, V., Grossi, P. A., Menichetti, F., et al. (2018). Ceftolozane/tazobactam: Place in Therapy. Expert Rev. Anti Infect. Ther. 16, 307–320. doi: 10.1080/14787210.2018.1447381
Haidar, G., Philips, N. J., Shields, R. K., Snyder, D., Cheng, S., Potoski, B. A., et al. (2017). Ceftolozane-Tazobactam for the Treatment of Multidrug-Resistant Pseudomonas aeruginosa Infections: Clinical Effectiveness and Evolution of Resistance. Clin. Infect. Dis. 65, 110. doi: 10.1093/cid/cix182
Hirsch, E. B., Brigman, H. V., Zucchi, P. C., Chen, A., Anderson, J. C., Eliopoulos, G. M., et al. (2020). Ceftolozane-Tazobactam and Ceftazidime-Avibactam Activity Against β-Lactam-Resistant Pseudomonas aeruginosa and Extended-Spectrum β-Lactamase-Producing Enterobacterales Clinical Isolates From U.S. Medical Centres. J. Global Antimicrob. Resistance 22, 689–694. doi: 10.1016/j.jgar.2020.04.017
Humphries, R. M., Hindler, J. A., Wong-Beringer, A., Miller, S. A. (2017). Activity of Ceftolozane-Tazobactam and Ceftazidime-Avibactam Against Beta-Lactam-Resistant Pseudomonas aeruginosa Isolates. Antimicrob. Agents Chemother. 61, e01858–e01817. doi: 10.1128/AAC.01858-17
Juan, C., Zamorano, L., Pérez, J. L., Ge, Y., Oliver, A. (2010). Activity of a New Antipseudomonal Cephalosporin, CXA-101 (FR264205), Against Carbapenem-Resistant and Multidrug-Resistant Pseudomonas aeruginosa Clinical Strains. Antimicrob. Agents Chemother. 54, 846–851. doi: 10.1128/AAC.00834-09
Karlowsky, J. A., Lob, S. H., Young, K., Motyl, M. R., Sahm, D. F. (2021). Activity of Ceftolozane/Tazobactam Against Gram-Negative Isolates From Patients With Lower Respiratory Tract Infections – SMART United States 2018–2019. BMC Microbiol. 21, 74. doi: 10.1186/s12866-021-02135-z
Langmead, B., Salzberg, S. L. (2012). Fast Gapped-Read Alignment With Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Li, H., Durbin, R. (2010). Fast and Accurate Long-Read Alignment With Burrows-Wheeler Transform. Bioinformatics 26, 589–595. doi: 10.1093/bioinformatics/btp698
Lister, P. D., Wolter, D. J., Hanson, N. D. (2009). Antibacterial-Resistant Pseudomonas aeruginosa: Clinical Impact and Complex Regulation of Chromosomally Encoded Resistance Mechanisms. Clin. Microbiol. Rev. 22, 582–610. doi: 10.1128/CMR.00040-09
Livermore, D. M., Mushtaq, S., Ge, Y., Warner, M. (2009). Activity of Cephalosporin CXA-101 (FR264205) Against Pseudomonas aeruginosa and Burkholderia Cepacia Group Strains and Isolates. Int. J. Antimicrob. Agents 34, 402–406. doi: 10.1016/j.ijantimicag.2009.03.021
López-Causapé, C., Cabot, G., del Barrio-Tofiño, E., Oliver, A. (2018). The Versatile Mutational Resistome of Pseudomonas aeruginosa. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00685
Maraolo, A. E., Mazzitelli, M., Trecarichi, E. M., Buonomo, A. R., Torti, C., Gentile, I. (2020). Ceftolozane/tazobactam for Difficult-to-Treat Pseudomonas aeruginosa Infections: A Systematic Review of its Efficacy and Safety for Off-Label Indications. Int. J. Antimicrob. Agents 55, 105891. doi: 10.1016/j.ijantimicag.2020.105891
Maurya, A. P., Dhar, D., Basumatary, M. K., Paul, D., Ingti, B., Choudhury, D., et al. (2017). Expansion of Highly Stable blaOXA-10 β-Lactamase Family Within Diverse Host Range Among Nosocomial Isolates of Gram-Negative Bacilli Within a Tertiary Referral Hospital of Northeast India. BMC Res. Notes 10, 145. doi: 10.1186/s13104-017-2467-2
Moghnieh, R., Araj, G. F., Awad, L., Daoud, Z., Mokhbat, J. E., Jisr, T., et al. (2019). A Compilation of Antimicrobial Susceptibility Data From a Network of 13 Lebanese Hospitals Reflecting the National Situation During 2015–2016. Antimicrobial. Resist. Infect. Control 8, 41. doi: 10.1186/s13756-019-0487-5
Moyá, B., Beceiro, A., Cabot, G., Juan, C., Zamorano, L., Alberti, S., et al. (2012). Pan-β-Lactam Resistance Development in Pseudomonas aeruginosa Clinical Strains: Molecular Mechanisms, Penicillin-Binding Protein Profiles, and Binding Affinities. Antimicrob. Agents Chemother. 56, 4771–4778. doi: 10.1128/AAC.00680-12
Oliver, A., Mulet, X., López-Causapé, C., Juan, C. (2015). The Increasing Threat of Pseudomonas aeruginosa High-Risk Clones. Drug Resist. Updates 21–22, 41–59. doi: 10.1016/j.drup.2015.08.002
O’Neall, D., Juhász, E., Tóth, Á., Urbán, E., Szabó, J., Melegh, S., et al. (2020). Ceftazidime-Avibactam and Ceftolozane-Tazobactam Susceptibility of Multidrug Resistant Pseudomonas aeruginosa Strains in Hungary. Acta Microbiol. Immunol. Hung. 67, 61–65. doi: 10.1556/030.2020.01152
Ortiz de la Rosa, J. M., Nordmann, P., Poirel, L. (2019). ESBLs and Resistance to Ceftazidime/Avibactam and Ceftolozane/Tazobactam Combinations in Escherichia Coli and Pseudomonas aeruginosa. J. Antimicrob. Chemother. 74, 1934–1939. doi: 10.1093/jac/dkz149
Papagiannitsis, C. C., Medvecky, M., Chudejova, K., Skalova, A., Rotova, V., Spanelova, P., et al. (2017). Molecular Characterization of Carbapenemase-Producing Pseudomonas aeruginosa of Czech Origin and Evidence for Clonal Spread of Extensively Resistant Sequence Type 357 Expressing IMP-7 Metallo-β-Lactamase. Antimicrob. Agents Chemother. 61, e01811–e01817. doi: 10.1128/AAC.01811-17
Pogue, J. M., Bonomo, R. A., Kaye, K. S. (2019). Ceftazidime/Avibactam, Meropenem/Vaborbactam, or Both? Clinical and Formulary Considerations. Clin. Infect. Dis. 68, 519–524. doi: 10.1093/cid/ciy576
Poirel, L., Lambert, T., Türkoglü, S., Ronco, E., Gaillard, J.-L., Nordmann, P. (2001). Characterization of Class 1 Integrons From Pseudomonas aeruginosa That Contain the blaVIM-2carbapenem-Hydrolyzing β-Lactamase Gene and of Two Novel Aminoglycoside Resistance Gene Cassettes. Antimicrob. Agents Chemother. 45, 546–552. doi: 10.1128/AAC.45.2.546-552.2001
Rodríguez-Martínez, J.-M., Poirel, L., Nordmann, P. (2009). Molecular Epidemiology and Mechanisms of Carbapenem Resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53, 4783–4788. doi: 10.1128/AAC.00574-09
Sid Ahmed, M. A., Abdel Hadi, H., Hassan, A. A. I., Abu Jarir, S., Al-Maslamani, M. A., Eltai, N. O., et al. (2019). Evaluation of In Vitro Activity of Ceftazidime/Avibactam and Ceftolozane/Tazobactam Against MDR Pseudomonas aeruginosa Isolates From Qatar. J. Antimicrob. Chemother. 74, 3497–3504. doi: 10.1093/jac/dkz379
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., Chandler, M. (2006). ISfinder: The Reference Centre for Bacterial Insertion Sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Sligl, W. I., Dragan, T., Smith, S. W. (2015). Nosocomial Gram-Negative Bacteremia in Intensive Care: Epidemiology, Antimicrobial Susceptibilities, and Outcomes. Int. J. Infect. Dis. 37, 129–134. doi: 10.1016/j.ijid.2015.06.024
Strãut, M., Dinu, S., Oprea, M., Drãgulescu, E., Lixandru, B., Surdeanu, M. (2018). Genetic Diversity of Structures Surrounding Bla Genes Identified in Pseudomonas aeruginosa Clinical Isolates From Bucharest, Romania. Bucharest, Romania. Rom. Arch. Microbiol. Imm. 77 (1), 16–27.
Subedi, D., Vijay, A. K., Kohli, G. S., Rice, S. A., Willcox, M. (2018). Nucleotide Sequence Analysis of NPS-1 β-Lactamase and a Novel Integron (In1427)-Carrying Transposon in an MDR Pseudomonas aeruginosa Keratitis Strain. J. Antimicrob. Chemother. 73, 1724–1726. doi: 10.1093/jac/dky073
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3
Taiaroa, G., Samuelsen, Ø., Kristensen, T., Økstad, O. A. L., Heikal, A. (2018). Complete Genome Sequence of Pseudomonas aeruginosa K34-7, a Carbapenem-Resistant Isolate of the High-Risk Sequence Type 233. Microbiol. Resour. Announce. 7, e00886–e00818. doi: 10.1128/MRA.00886-18
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting Chromosomal DNA Restriction Patterns Produced by Pulsed-Field Gel Electrophoresis: Criteria for Bacterial Strain Typing. J. Clin. Microbiol. 33, 2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995
Thrane, S. W., Taylor, V. L., Lund, O., Lam, J. S., Jelsbak, L. (2016). Application of Whole-Genome Sequencing Data for O-Specific Antigen Analysis and In Silico Serotyping of Pseudomonas aeruginosa Isolates. J. Clin. Microbiol. 54, 1782–1788. doi: 10.1128/JCM.00349-16
van Duin, D., Bonomo, R. A. (2016). Ceftazidime/Avibactam and Ceftolozane/Tazobactam: Second-Generation β-Lactam/β-Lactamase Inhibitor Combinations. Clin. Infect. Dis. 63, 234–241. doi: 10.1093/cid/ciw243
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PloS One 9, e112963. doi: 10.1371/journal.pone.0112963
Weiner, L. M., Webb, A. K., Limbago, B., Dudeck, M. A., Patel, J., Kallen, A. J., et al. (2016). Antimicrobial-Resistant Pathogens Associated With Healthcare-Associated Infections: Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect. Control Hosp. Epidemiol. 37, 1288–1301. doi: 10.1017/ice.2016.174
Wheeler, D. L., Church, D. M., Federhen, S., Lash, A. E., Madden, T. L., Pontius, J. U., et al. (2003). Database Resources of the National Center for Biotechnology. Nucleic Acids Res. 31, 28–33. doi: 10.1093/nar/gkg033
Winsor, G. L., Griffiths, E. J., Lo, R., Dhillon, B. K., Shay, J. A., Brinkman, F. S. L. (2016). Enhanced Annotations and Features for Comparing Thousands of Pseudomonas Genomes in the Pseudomonas Genome Database. Nucleic Acids Res. 44, D646–D653. doi: 10.1093/nar/gkv1227
Woodford, N., Turton, J. F., Livermore, D. M. (2011). Multiresistant Gram-Negative Bacteria: The Role of High-Risk Clones in the Dissemination of Antibiotic Resistance. FEMS Microbiol. Rev. 35, 736–755. doi: 10.1111/j.1574-6976.2011.00268.x
Wright, H., Bonomo, R. A., Paterson, D. L. (2017). New Agents for the Treatment of Infections With Gram-Negative Bacteria: Restoring the Miracle or False Dawn? Clin. Microbiol. Infect. 23, 704–712. doi: 10.1016/j.cmi.2017.09.001
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of Acquired Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: ceftolozane/tazobactam (C/T), Pseudomonas aeruginosa, AmpC, porins, beta lactamases
Citation: Bitar I, Salloum T, Merhi G, Hrabak J, Araj GF and Tokajian S (2022) Genomic Characterization of Mutli-Drug Resistant Pseudomonas aeruginosa Clinical Isolates: Evaluation and Determination of Ceftolozane/Tazobactam Activity and Resistance Mechanisms. Front. Cell. Infect. Microbiol. 12:922976. doi: 10.3389/fcimb.2022.922976
Received: 18 April 2022; Accepted: 17 May 2022;
Published: 15 June 2022.
Edited by:
William D. Picking, University of Kansas, United StatesReviewed by:
Hamid Solgi, Isfahan University of Medical Sciences, IranVictor Meza Carmen, Michoacana University of San Nicolás de Hidalgo, Mexico
Copyright © 2022 Bitar, Salloum, Merhi, Hrabak, Araj and Tokajian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sima Tokajian, c3Rva2FqaWFuQGxhdS5lZHUubGI=
 Ibrahim Bitar
Ibrahim Bitar Tamara Salloum3
Tamara Salloum3 Georgi Merhi
Georgi Merhi Jaroslav Hrabak
Jaroslav Hrabak George F. Araj
George F. Araj Sima Tokajian
Sima Tokajian