
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 11 July 2022
Sec. Clinical Microbiology
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.922031
This article is part of the Research Topic Immune Response to Gram-negative Bacteria in the Lungs View all 7 articles
We characterized the first NDM-5 and MCR-8.2 co-harboring ST656 Klebsiella pneumoniae clinical isolate, combining with chromosomal gene-mediated resistance to colistin and tigecycline. The K. pneumoniae KP32558 was isolated from the bronchoalveolar lavage fluid from a lung transplant patient. Complete genome sequences were obtained through Illumina HiSeq sequencing and nanopore sequencing. The acquired resistance genes and mutations in chromosome-encoded genes associated with colistin and tigecycline resistance were analyzed. Comparative genomic analysis was conducted between mcr-8.2-carrying plasmids. The K. pneumoniae KP32558 was identified as a pan-drug resistant bacteria, belonging to ST656, and harbored plasmid-encoded blaNDM-5 and mcr-8.2 genes. The blaNDM-5 gene was located on an IncX3 type plasmid. The mcr-8.2 gene was located on a conjugative plasmid pKP32558-2-mcr8, which had a common ancestor with another two mcr-8.2-carrying plasmids pMCR8_020135 and pMCR8_095845. The MIC of KP32558 for colistin was 256 mg/L. The mcr-8.2 gene and mutations in the two-component system, pmrA and crrB, and the regulator mgrB, had a synergistic effect on the high-level colistin resistance. The truncation in the acrR gene, related to tigecycline resistance, was also identified. K. pneumoniae has evolved a variety of complex resistance mechanisms to the last-resort antimicrobials, close surveillance is urgently needed to monitor the prevalence of this clone.
The emergence and worldwide dissemination of carbapenem-resistant Klebsiella pneumoniae (CRKP) have posed a great threat to public health (Nordmann et al., 2009). The main resistance mechanism for CRKP is the production of carbapenemases, of which New Delhi metallo-β-lactamase (NDM) is capable of hydrolyzing all commonly-used β-lactams except monobactams. The blaNDM-5 gene was first identified from an Escherichia coli isolate in 2011 from a patient with a hospitalization history in India (Li et al., 2018). Afterward, NDM-5-producing K. pneumoniae isolates have been documented worldwide (Rojas et al., 2017; Li et al., 2018; Zhao et al., 2021). Compared to NDM-1, NDM-5 has two amino acid substitutions at Val88Leu and Met154Leu, showing increased resistance to carbapenems and broad-spectrum cephalosporins when expressed under its native promoter (Hornsey et al., 2011).
For infections caused by NDM-5-producing CRKP, tigecycline and colistin are the last treatment options (Li et al., 2021; Xu et al., 2021b). In recent years, colistin resistance in Enterobacterales isolates has been found more frequently. Several molecular mechanisms have been associated with colistin resistance, such as chromosomal mutations in the genes that encode PmrA/PmrB, PhoP/PhoQ and CrrA/CrrB two-component systems (TCS), and inactivation of the regulator MgrB (Cannatelli et al., 2013; Mcconville et al., 2020). Since the first plasmid-encoded colistin resistance gene, mcr-1, was identified in E. coli in 2015 (Liu et al., 2016), nine variants of mcr-1 (mcr-2 up to mcr-10) have also been identified in Enterobacterales (Elbediwi et al., 2021). The horizontal transferability of plasmids facilitates the rapid dissemination of colistin resistance. Furthermore, increasing consumption of tigecycline has been concurrent with increasing reports of tigecycline resistance (Osei Sekyere et al., 2016). The main mechanism is the overexpression of resistance-nodulation-cell division (RND)-type efflux pumps (acrAB and oqxAB) and mutations in efflux pump regulatory genes (ramA and acrR) and regulatory genes of the SoxS and MarAB (soxR and marR) (He et al., 2015; Osei Sekyere et al., 2016; Xu et al., 2021a).
K. pneumoniae strains, presenting with extensive resistance to carbapenems, tigecycline and colistin simultaneously, would pose a great threat to public health. In this study, we isolated a pan-drug resistant K. pneumoniae strain from the bronchoalveolar lavage fluid (BALF) specimen of a lung transplant patient. Revealing its mechanism of drug resistance will play a positive role in preventing the spread of this strain.
Two K. pneumoniae isolates (KP31166 and KP32558) were recovered from the BALF specimens of a lung transplant patient 1 and 4 months after surgery, respectively. For comparative genomic analysis, ten mcr-8.2-harboring plasmids were collected from the NCBI genome database, including plasmids pMCR8_020135 (CP037964), pMCR8_095845 (CP031883), pVNCKp115 (LC549807), pZZW20-88K (CP058962), p2019036D-mcr8-345kb (CP047337), pD120-1_83kb (CP034679), pVNCKp83 (LC549808), pSCKLB555-4 (CP043936), p2018C01-046-1_MCR8 (CP044369) and p18-29mcr-8.2 (MK262711).
In vitro susceptibility tests of amikacin, tobramycin, minocycline, doxycycline, ceftazidime, cefepime, piperacillin/tazobactam, cefoperazone/sulbactam, aztreonam, imipenem, meropenem, levofloxacin, ciprofloxacin, and sulfamethoxazole/trimethoprim were performed using the Vitek-2 system in N335 susceptibility cards (bioMérieux, France). The minimum inhibitory concentrations (MICs) of tigecycline and colistin were determined using the microdilution broth method (bio-KONT, Ltd. China). The production of carbapenemases was determined using the modified carbapenem inactivation method (mCIM) and EDTA-modified carbapenem inactivation method (eCIM) as recommended by the Clinical Laboratory Standards Institute (CLSI) (Clinical and Laboratory Standards Institute, 2019). The breakpoints of tigecycline and colistin were defined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST, version 11.0, http://www.eucast.org/). In particular, MICs > 0.5 mg/L and > 2 mg/L will be defined as resistant to tigecycline and colistin, respectively.
The string test was used to identify the hypermucoviscous phenotype of K. pneumoniae strains. Strains exhibiting a viscous string >5 mm were considered hypermucoviscous (Shon et al., 2013).
Complete genome sequencing was carried out using Illumina HiSeq 2500 platform (for both KP32558 and KP31166) as well as nanopore sequencing method on MinION flow cells (for KP32558 only). Raw reads were filtered to remove low-quality sequences and adaptors using skewer (Jiang et al., 2014) and PoreChop (https://github.com/rrwick/Porechop), respectively. De novo assembly was conducted using SPAdes Genome Assembler v3.13.1 (Prjibelski et al., 2020) and Unicycler (Wick et al., 2017). Gene prediction for 11 genomes, including one from this study and 10 from the NCBI genome database, was performed using Prokka 1.12 (Seemann, 2014). Genomic islands were predicted using IslandViewer 4 (Bertelli et al., 2017). Insertion sequences were identified using the ISfinder database (Siguier et al., 2006). The antimicrobial resistance genes, multilocus sequence types (MLST) and plasmid replicon were analyzed via the CGE server (https://cge.cbs.dtu.dk/services/). Virulence genes were identified using the BIGSdb Klebsiella genome database (http://bigsdb.Pasteur.fr/klebsiella/klebsiella.html). The single nucleotide polymorphism (SNP) was called using Snippy (https://github.com/tseemann/snippy). The heatmap was generated using an in-house R script.
A total of 11 mcr-8.2-carrying plasmids were included and aligned to reference plasmid pKP32558-2-mcr8 (GenBank accession no. CP076032, this study) using Snippy. A SNP maximum likelihood (ML) tree was constructed by using FastTree 2 (Stamatakis, 2014) with a general time reversible (GTR) model of nucleotide substitution and a gamma distribution of rate heterogeneity. Phylogenetic clusters were identified using hierarchical Bayesian analysis of population structure, in which the first level of clustering was used to define clades (Cheng et al., 2013). The phylogenetic tree was annotated by using FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
The plasmid conjugation experiment was performed for K. pneumoniae strain KP32558, and azide-resistant E. coli J53 was used as the recipient strain. Transconjugants were selected on LB plates containing azide (100 mg/L) and meropenem (2 mg/L) or colistin (2 mg/L). Antimicrobial susceptibility testing, PCR amplification and S1-PFGE were performed to confirm the successful transfer. The transfer frequency was calculated as the number of transconjugants per recipient.
Total RNA was isolated from the K. pneumoniae strains using the RNeasy minikit (Qiagen), according to the manufacturer’s instructions. The expression of the acrB, acrR, crrA, crrB, crrC, phoP, phoQ, pmrA, pmrB, pmrC, pmrD, pmrK, mgrB, and rpoB genes was measured using qRT-PCR on an Applied Biosystems QuantStudio 5 real-time PCR system (ThermoFisher Scientific). The expression levels of the target genes were normalized to the rpoB gene using ΔΔCT method, and the strain K. pneumoniae ATCC 13883 was used as reference. The primers used for qRT-PCR are listed in Table S1. All tests were performed three times.
Growth curves for the recipients E. coli J53 and transconjugants were measured as follows: LB broths containing 1 × 106 CFU/mL bacteria were incubated overnight at 37°C with consecutive shaking. Meanwhile, the same broth without bacteria was set as the growth control. OD600 measurement was taken at 0, 2, 4, 6, 8, 10, and 12 h to construct a growth curve. All experiments were performed in duplicate. Statistically significant differences in growth kinetics including slope and time analysis were tested by using the R package “statmod” (Giner and Smyth, 2016). A P value of 0.05 or less was defined as significant.
A lung transplant patient was given colistin for two days on the 3rd day after the surgery for the prevention of infections. The K. pneumoniae isolates KP31166 and KP32558 were detected in his BALF specimens approximately one and four months later, respectively. In addition, the patient had normal blood routine and C-reactive protein test results and had no fever when K. pneumoniae was detected. Considering the condition of the patient and antimicrobial susceptibility results of the K. pneumoniae, no specific anti-infective treatment was given. However, K. pneumoniae was not isolated from BALF specimens since then, and the patient recovered well.
According to the antimicrobial susceptibility testing, strains KP32558 and KP31166 were all CRKP and were resistant to all tested antibiotics (Table 1). Both strains exhibited the same MICs for tigecycline (4 mg/L) and colistin (256 mg/L). ECIM and mCIM tests indicated that the two strains produced metallo-β-lactamase. Strains KP32558 and KP31166 carried the same resistance genes, and only two SNPs were found between them based on whole-genome SNP analysis (data not shown). According to a previous criterion (Schurch et al., 2018), the two strains were considered homologous and derived from the same ancestor.
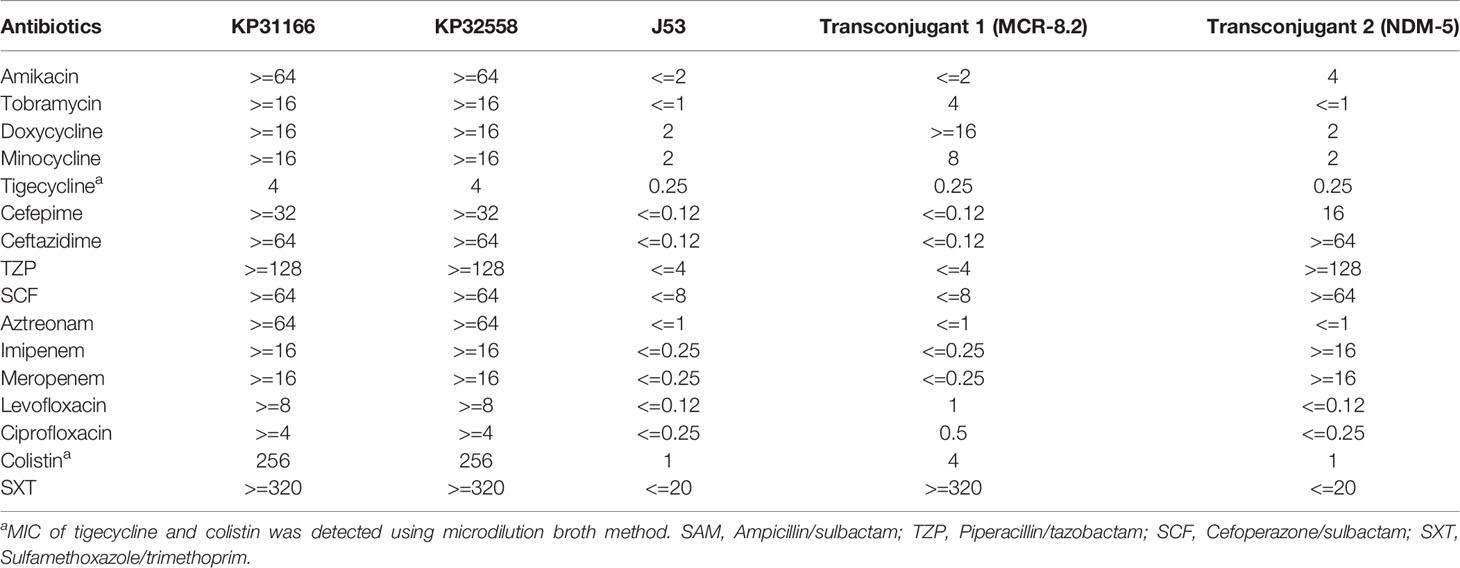
Table 1 Antibiotic resistance characteristics (MICs, mg/L) of 4 clinical K. pneumoniae strains and the transconjugant of KP32558.
Strain KP32558 was randomly selected for further analysis, which was not hypermucoviscous and assigned to ST656. Neither rmpA/rmpA2 genes nor iucABCDiutA and iroBCDN virulence gene clusters were found in this strain. Resistance gene annotation showed that about 40 resistance genes were located on its plasmids (Table S2).
The blaNDM-5 gene located in the 5th plasmid pKP32558-5-ndm5 (46161 bp in size, GenBank accession number CP076035), which belonged to the IncX3 group and did not harbor other resistance genes. Microbial nucleotide search for GenBank showed that pKP32558-5-ndm5 was nearly identical to a series of blaNDM-5 -carrying plasmids with the same size in length. Specifically, pKP32558-5-ndm5 showed 100% identity and coverage with plasmid pCREC-591_4 (CP024825, 46161 bp) from E. coli strain CREC-591, and 100% identity and 99.8% coverage with the plasmid pNDM_MGR194 (KF220657, 46253 bp), which was a typical blaNDM-5-carrying plasmid recovered from a K. pneumoniae isolate in India (Krishnaraju et al., 2015). Moreover, we found a genomic island region in pKP32558-5-ndm5, the blaNDM-5 gene and its flanking contents were also located in this region.
Similar to other blaNDM-5-carrying plasmids, several conjugal transfer genes (such as virD4, virB4, virB8 and virB9) were identified in the plasmid pKP32558-5-ndm5. Conjugation experiment showed that this plasmid could be successfully transferred to the recipient E. coli J53 as reported previously (Zhao et al., 2021) at a frequency of 10-4 (transconjugant/recipient), and the transconjugant displayed resistance to imipenem and meropenem (Table 1).
An mcr-8.2 gene (1698 bp) was found in the second plasmid pKP32558-2-mcr8 (CP076032) by blasting against the resfinder database. Microbial nucleotide search for GenBank suggested that pKP32558-2-mcr8 had the highest alignment score with the plasmid pKPNH54.1, which was derived from K. pneumoniae strain NH54, and did not contain the mcr gene. Comparative genomic analysis was conducted among pKP32558-2-mcr8, pKPNH54 and other 10 mcr-8.2-containing plasmids obtained from the NCBI database. The result showed that these plasmids were not identical to each other (Figure 1A), and the plasmids pKPNH54.1 (CP024917), pMCR8_020135 (CP037964) and pVNCKp115 (LC549807) could be aligned to different parts of pKP32558-2-mcr8. Despite the differences between these mcr-8.2-carrying plasmids, the commonality between them was that they had identical genetic environments around the mcr-8.2 gene, ISKpn26-orf-mcr-8.2-ISEcl1-copR-baeS-dgkA (Figure 1B). Besides, plasmid pZZW20-88k, pD120-1_83kb and p201936D-mcr8-345kb had a short fragment deletion in front of IS903B compared to other plasmids. Of note, all these genes were located on a genomic island region.
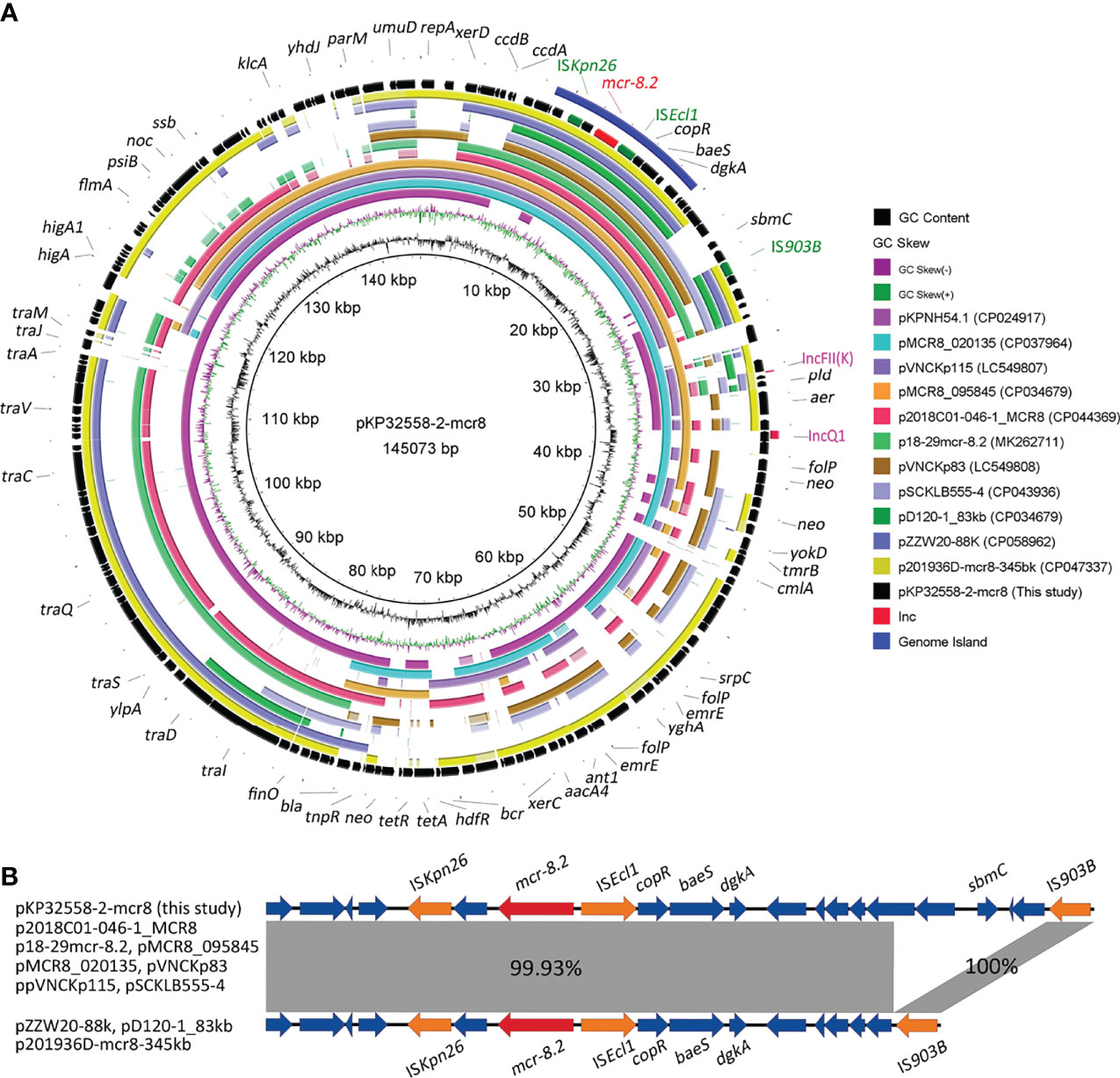
Figure 1 Genetic features of the mcr-8.2-carrying plasmid in K. pneumoniae strain KP32558. (A) Circular map of plasmid pKP32558-2-mcr8 and its reference plasmids. The outer blue circle is the genomic island region of plasmid pKP32558-2-mcr8. Red text on the plasmid map indicates mcr-8.2 gene, the green text indicates insertion sequences around mcr-8.2, and pink text represents the replicons. (B) Genetic contents of mcr-8.2 gene. Coding sequences are indicated by arrows. Sequences of shared homology between two plasmids are marked by gray shading.
Plasmid typing suggested that these mcr-8.2-carrying plasmids contained 7 different replicons, and IncFII(K) replicon was found in most plasmids (9/11) (Figure 2). In addition to IncFII(K) replicon, the plasmid pKP32558-2-mcr8 also had a truncated IncQ1 type replicon, which was also present in pMCR8_020135 and pMCR8_095845. The two plasmids were derived from different hospitals in China. The phylogenetic tree showed that these plasmids were clustered into 3 major clades. Plasmid pKP32558-2-mcr8, pMCR8_020135, pMCR8_095845 and pVNCKp115 belonged to the same clade.
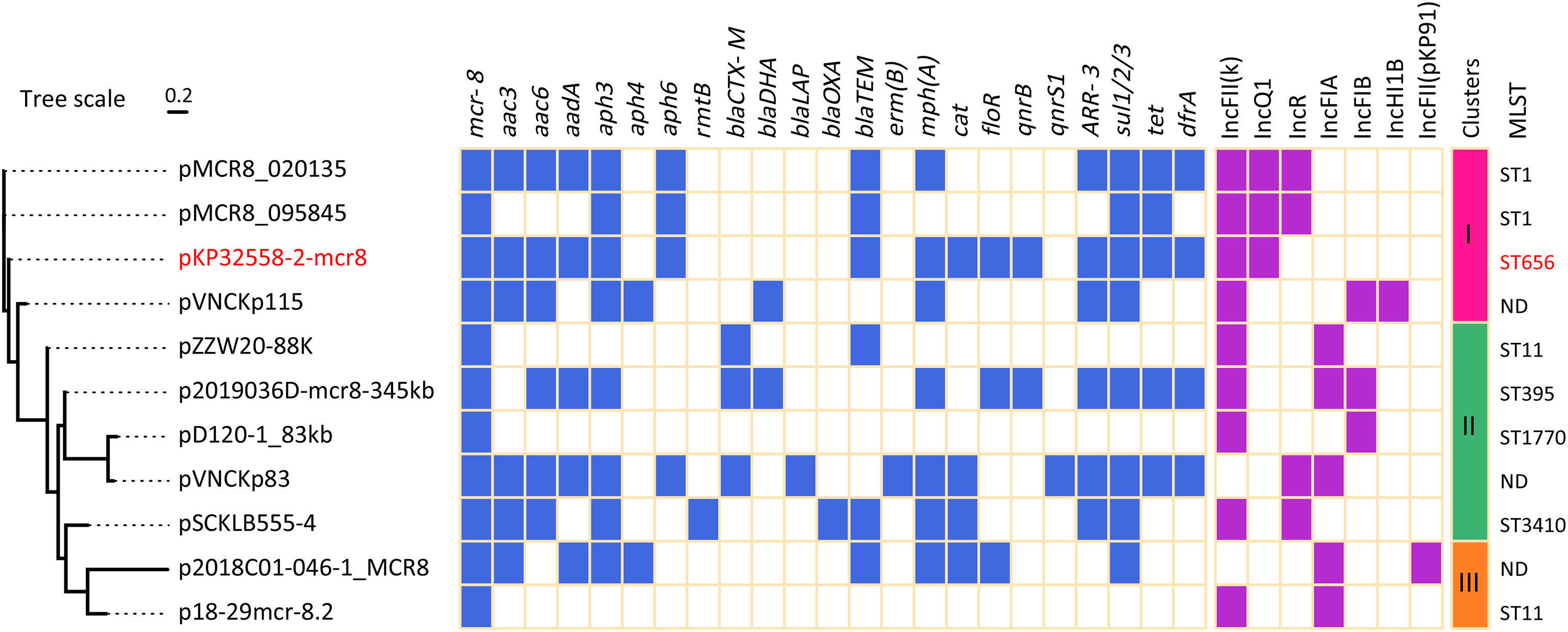
Figure 2 Resistance genes and replicons of mcr-8.2-carrying plasmids. The pKP32558-2-mcr8 (CP CP076032) was derived from K. pneumoniae KP32558 of this study. The genomic sequences of plasmids pMCR8_020135 (CP037964), pMCR8_095845 (CP031883), pVNCKp115 (LC549807), pZZW20-88K (CP058962), p2019036D-mcr8-345kb (CP047337), pD120-1_83kb (CP034679), pVNCKp83 (LC549808), pSCKLB555-4 (CP043936), p2018C01-046-1_MCR8 (CP044369) and p18-29mcr-8.2 (MK262711) were obtained from NCBI database.
Resistance genes were identified using the resfinder database. In addition to mcr-8.2, several other resistance genes were found in plasmid pKP32558-2-mcr8, including aac, aad and aph for aminoglycoside, blaTEM for β-lactam, mph(A) for macrolide, arr-3 for rifampicin, sul1/2/3/for sulphonamide, tet for tetracycline and dfrA for trimethoprim antibiotics (Figure 2). Conjugation experiments showed that the plasmid pKP32558-2-mcr8 could be transferred into E. coli J53 at a frequency of 10-7.
MLST analysis suggested that both KP31166 and KP32558 in this study belonged to ST656, and the other 10 mcr-8.2-carrying strains belonged to different ST types, including ST1, ST11, ST395, ST1770 and ST3410. Among them, ST1, ST656 and ST3410 belonged to the same clonal complex, and there was only one allele difference between them. For three isolates, their chromosomal sequences were not deposited in GenBank, therefore the MLST results could not be obtained (Figure 2).
Two-component systems PmrA/PmrB, PhoP/PhoQ and CrrA/CrrB, and the regulator mgrB gene were analyzed to identify the potential mutations involved in colistin resistance. Genes in K. pneumoniae strain MGH 78578 (CP000647) were used as controls. The results showed that strain KP32558 contained amino acid substitution in MgrB (I45F), PmrA (D86E), PmrB (T246A and G345E) and CrrB (C68S and V193G). PROVEAN was used for predicting the functional effect of the amino acid substitution, indicating that 3 mutations in MgrB, PmrA and CrrB (V193G) were deleterious (have a damaging effect on protein function), while the other 4 mutations were neutral (Table 2). These mutations might partly contribute to colistin resistance in K. pneumoniae strain KP32558. We examined the expression of the regulatory genes phoP, phoQ, pmrA, pmrB, pmrC, pmrD, pmrK, crrA, crrB, crrC and mgrB, and the result showed higher expression for all the genes except for pmrC (Figure 3).
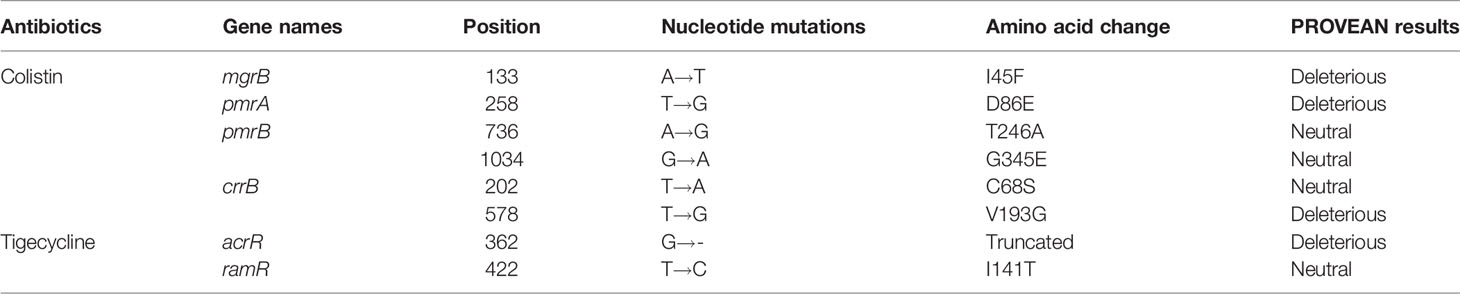
Table 2 Mutations in chromosomal genes related to colistin and tigecycline resistance for K. pneumoniae KP32558.
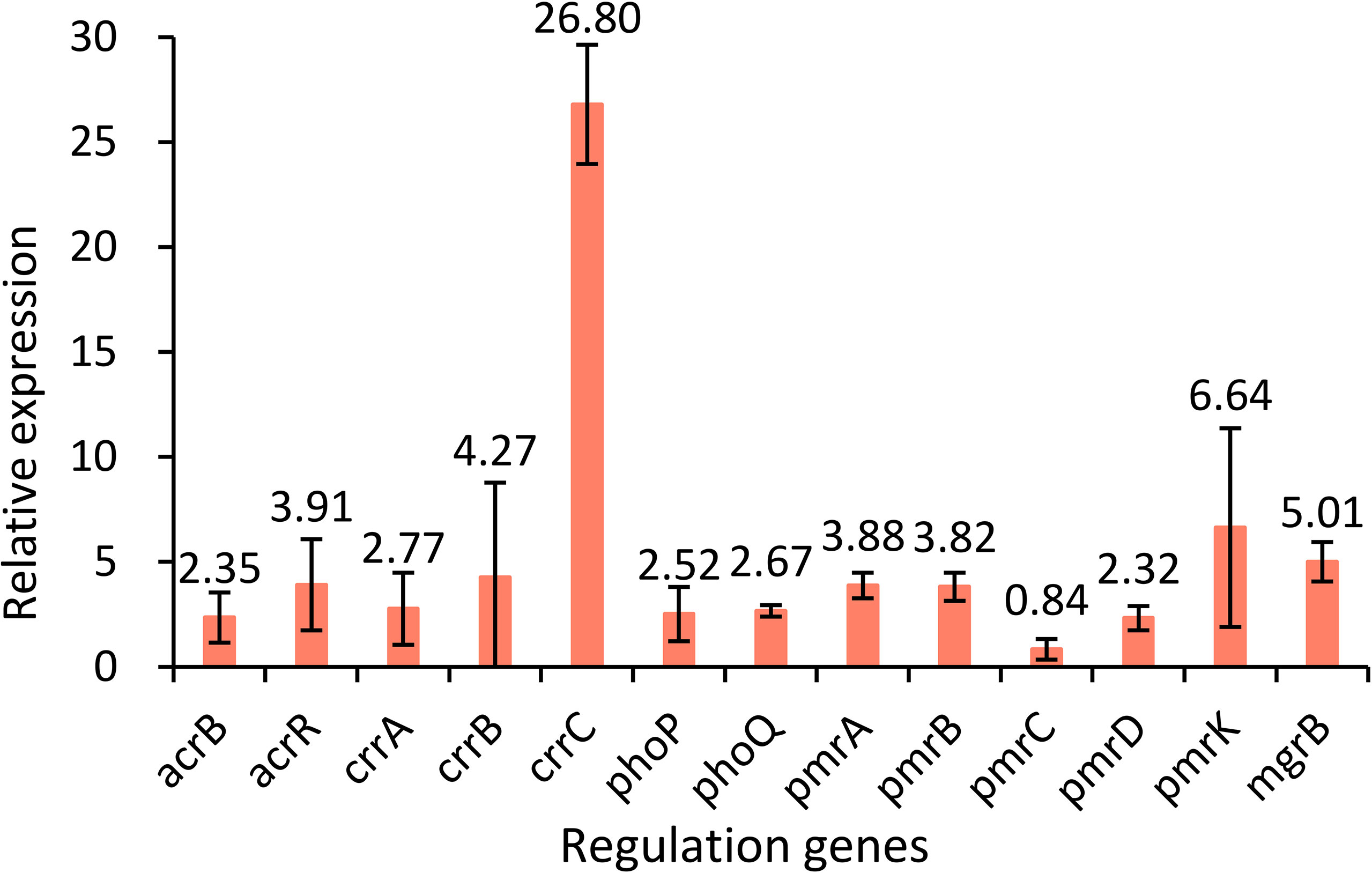
Figure 3 Relative expression levels of the acrB, acrR, crrA, crrB, crrC, phoP, phoQ, pmrA, pmrB, pmrC, pmrD, pmrK and mgrB genes in strain KP32558 compared with those in the antibiotic-susceptible K. pneumoniae strain ATCC13883. Values are the means and the standard deviations from three independent experiments.
The regulatory genes ramR, marR, soxR and acrR of KP32558 were investigated for mutation using K. pneumoniae strain MGH 78578 as a negative reference. An amino acid substitution was found in RamR (I141T), which had a neutral effect on the function. AcrR acts as a local transcriptional repressor of the AcrAB efflux pump, the full length of the acrR gene in wild-type K. pneumoniae isolate is 651 bp. A single-base deletion happened at position 362 of gene acrR, resulting in the premature termination of translation at amino acid 123, which could be associated with resistance to tigecycline (Sheng et al., 2014). We examined the expression of acrR and acrB genes, showing higher expression for both genes, which confirmed the function of AcrR (Figure 3).
Compared to a carbapenem-resistant and hypervirulent K. pneumoniae isolate KP22937 (SAMN17245924) and a standard strain K. pneumoniae ATCC 700603, KP32558 had a slower growth rate although without statistical difference. While the growth of transconjugant 1 (MCR-8.2) and transconjugant 2 (NDM-5) was almost indistinguishable from that of E. coli J53 (Figure S1).
The widespread of CRKP has aroused great attention worldwide. Once these strains are resistant to colistin and tigecycline, limited treatment options are available. In this study, we identified an NDM-5-producing K. pneumoniae isolate from a lung transplant patient, which simultaneously carried the plasmid-encoded mcr-8.2 gene and chromosomal gene-mediated resistance to colistin and tigecycline. This represents the rare report of K. pneumoniae clinical isolate with multiple resistance mechanisms to the last-resort antimicrobials.
The blaNDM gene might be located on different types of plasmids, such as IncFII, IncIB and IncX3, with IncX3 being the most frequently reported. In our study, the blaNDM gene was also identified on the IncX3 plasmid. IncX3 plasmids carrying blaNDM gene are widely found in Enterobacterales, including E. coli, K. pneumoniae, Proteus spp. and Klebsiella aerogenes (Zhang et al., 2016; Tian et al., 2020), hinting its wide transmission.
Since the mcr-8.1 gene from pig-derived K. pneumoniae strain KP91 was first reported in 2018, multiple mcr-8 variants have been identified, ranging from mcr-8.2 to mcr-8.5 (Sun et al., 2020). Currently, K. pneumoniae producing both MCR-8 and NDM has emerged, such as mcr-8.1 and blaNDM-1 (animal origin), mcr-8.2 and blaNDM-1 (human origin), mcr-8.1 and blaNDM-5 (human origin), and mcr-8.5 and blaNDM-5 (animal origin) (Wang et al., 2018; Ma et al., 2020a; Sun et al., 2020; Pei et al., 2021). In our study, we also identified the K. pneumoniae isolate co-harboring mcr-8.2 and blaNDM-5 in the lung transplantation ward, and should be alerted to its in-hospital spread.
Comparative genomic analysis was conducted to elucidate the genetic characteristics of mcr-8.2-carrying plasmid pKP32558-2-mcr8. This plasmid had two replicons, an intact IncFII(K) and a truncated IncQ1. Besides, eight of the ten mcr-8.2-carrying plasmids obtained from the GenBank database also had IncFII(K) type replicons, and the other two plasmids carried other IncF type replicons (Figure 2). This was consistent with the previous report that the IncF type plasmids are widely distributed in clinically relevant Enterobacterales isolates, and contribute to the fitness of bacterial host by providing antimicrobial resistance and virulence determinants (Johnson and Nolan, 2009). IncQ1 type plasmids are efficient antimicrobial resistance determinants among Gram-negative bacteria (Martins et al., 2020). The truncated IncQ1 replicon in the plasmid pKP32558-2-mcr8 was also observed in the other two plasmids, pMCR8_020135 and pMCR8_095845, indicating that these three plasmids may share a common ancestor. Phylogenic analysis of these mcr-8.2-carrying plasmids showed that plasmids pKP32558-2-mcr8, pMCR8_020135, pMCR8_095845 and pVNCKp115 were clustered into the same clade. At the same time, the latter three plasmids can match different parts of pKP32558-2-mcr8, indicating that there is a close genetic relationship between them.
MLST analysis showed that the K. pneumoniae strain KP32558 was assigned to ST656, a rare ST type and firstly reported in China in 2012 (Wang et al., 2012). The frequently detected carbapenemase in these bacteria was KPC-2 (Liu et al., 2021) and NDM-1 (Li et al., 2020). Our study represents the first report of the emergence of NDM-5 and MCR in ST656 K. pneumoniae clinical isolate. The other 10 mcr-8.2-carrying plasmids came from bacteria with diverse ST types, including ST1, ST11, ST395, ST1770 and ST3410, suggesting the intra-species spread of mcr-8.2. It was noticeable that pMCR8_020135 and pMCR8_095845 were carried by ST1 K. pneumoniae strains, there was only one allele difference between ST656 and ST1, and they belonged to the same clonal complex. These results strengthened our hypothesis that the three plasmids share a common ancestor.
It is believed that without the antibiotic pressure, the acquisition of resistance genes in plasmids will impose fitness costs on their host (Andersson and Hughes, 2010). However, our recent study has shown that the growth of the transconjugant was almost indistinguishable from that of recipient E. coli EC600 (Zhao et al., 2021), and another study also showed that up to 75.9% (22/29) Enterobacterales strains did not produce fitness costs after obtaining the IncX3 plasmid (Ma et al., 2020b). The above results suggested that the acquisition of IncX3 plasmid might not confer a fitness cost to the host and then facilitate the rapid dissemination of the plasmid. In this study, the blaNDM-5-carrying IncX3 plasmid pKP32558-5-ndm5 could be transferred to E. coli J53 and the transconjugant had a similar growth curve compared with E. coli J53, indicating that strain KP32558 probably acquired the resistance to carbapenems by obtaining the blaNDM-5-carrying and self-transmissible plasmid.
The MIC values for colistin in mcr-8-carrying isolates are usually between 4 and 32 mg/L (Wang et al., 2018; Hadjadj et al., 2019). While in our study, the MIC of K. pneumoniae strain KP32558 and transconjugant 1 were 256 and 4 mg/L, respectively, hinting that this strain had other colistin resistance mechanisms in addition to the mcr-8.2 gene. According to a previous report, chromosomal mutations in genes encoding phoP/phoQ, pmrA/pmrB and crrA/crrB TCSs and mutations in the mgrB gene can confer resistance to colistin in K pneumoniae (Poirel et al., 2017). In isolate KP32558, a total of six amino acid substitutions were identified in MgrB, PmrA, PmrB and CrrB, among which the substitutions of I45F in MgrB, D86E in PmrA and V193G in CrrB were deleterious according to the PROVEAN tool. The D86E mutation has been described previously (Samuelsen et al., 2017), while the other two mutations are reported for the first time. PmrAB can directly regulate the pmrHFIJKLM operon and pmrC gene, MgrB negatively regulates genes phoPQ by inhibiting the phosphorylation of PhoQ, PhoPQ directly regulates pmrHFIJKLM operon and indirectly regulates pmrC through PmrD and PmrAB, mutations of crrB gene could induce the higher expression of crrC, which positively regulates the expression of pmrHFIJKLM operon through PmrAB (Cheng et al., 2018). For strain KP32558, we detected higher expression of crrC, pmrA and pmrB genes, but not pmrC, which was consistent with the above report. The higher expression of crrC gene can be the result of the mutation of crrB and the higher expression of phoP, phoQ and pmrD was related to the mgrB mutation. However, considering the deleterious mutation in pmrA and no change in the expression of pmrC, we proposed that mutation in mgrB may partly contribute to the higher colistin resistance of strain KP32558 through PhoPQ. In addition, missense mutations of crrB also lead to increased expression of H239_3064, a putative RND-type efflux pump, leading to higher resistance to colistin (Cheng et al., 2018). We cannot rule out the role of crrB mutation in strain KP32558. These results revealed that the mcr-8.2 gene and chromosomal TCS may have synergistic effects in mediating bacterial resistance to colistin, as reported in a previous study (Wu et al., 2020). Some more experiments, such as the complementation experiment are needed to verify the resistance mechanism of colistin.
High-level expression of efflux pump is the main mechanism of bacterial resistance to tigecycline (Cheng et al., 2020). Mutation in local transcriptional repressor acrR and global transcriptional activator ramA could result in the overexpression of the AcrAB efflux pump (Rosenblum et al., 2011). One previous study reported 26 tigecycline-nonsusceptible K. pneumoniae isolates, among which 23 (88.5%) had mutations in ramR, and 2 of the remaining 3 isolates contained a mutation in acrR and had a tigecycline MIC of 4 mg/L (Sheng et al., 2014). In this study, an amino acid substitution in RamR (I141T) was found in K. pneumoniae strain KP32558, and this mutation had a neutral effect on its function, as consistent with a previous report (Wang et al., 2015). In addition, we also found a single-base deletion at position 362 of acrR, which resulted in the premature termination of transcription and showed a deleterious effect on the function. Therefore, we speculated that the mutation in the acrR gene may contribute to the resistance of KP32558 to tigecycline.
Infections caused by multidrug-resistant K. pneumoniae can cause serious consequences for patients, especially those with compromised immune systems. We previously reported a K. pneumoniae isolate KP22937 with an NDM-5-producing plasmid and a hypervirulent plasmid, which caused the death of a lung transplant patient (Zhao et al., 2021). Fortunately, in this study, although pan-drug resistant K. pneumoniae isolates were isolated twice in BALF, the bacteria did not cause serious infections in this lung transplant patient and was eliminated without using specific antibiotics. Several mechanisms may contribute to the lower pathogenicity of KP32558. First, no classic hypervirulent genes were found in this strain, which limited its in-vivo dissemination. Second, the previous report showed that plasmid carriage often imposes a reduction in the fitness to the host (San Millan and Maclean, 2017). In this study, KP32558 carried 5 plasmids, growth kinetics assay showed that KP32558 had a slower growth rate compared to the standard strain K. pneumoniae ATCC 700603 and the K. pneumoniae strain KP22937 we previously reported. Although the plasmids carrying NDM-5 or MCR-8.2 did not impose fitness costs on the recipient bacteria, the other 3 plasmids might play a role.
In conclusion, we report an MCR-8.2 and NDM-5-producing pan-drug resistant ST656 K. pneumoniae isolate recovered from the BALF specimen of a lung transplant patient. The mcr-8.2 gene was located on a hybrid plasmid containing IncFII(K) and IncQ1 composition replicons, which may play synergistic effects together with chromosomal mutations in mediating colistin resistance. Efflux pump repressor mutations were also found and may involve in the tigecycline resistance of this K. pneumoniae isolate, suggesting that this bacterium has evolved and acquired a variety of complex resistance mechanisms to the last-resort antimicrobials. Therefore, close surveillance is urgently needed to monitor the prevalence of this clone.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Permission for using the information in the medical records of the patient and the K. pneumoniae isolates for research purposes was granted by the Ethics Committee of the China-Japan Friendship Hospital (2019-164-K113).
JZ, ZL, YZ, XL, and BL collected the clinical and laboratory data. JZ, BL and BC made substantial contributions to conception and design, drafted, reviewed, and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China [grant number 82102456]; Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences [grant number CIFMS 2020-I2M-2-013 and CIFMS 2021-I2M-1-030].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank all the colleagues in the Laboratory of Clinical Microbiology and Infectious Diseases for their assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.922031/full#supplementary-material
Supplementary Figure 1 | S1-PFGE profiles (A) and growth curves (B) of K. pneumoniae clinical strain, recipient bacterium E. coli J53 and transconjugants.
Andersson, D. I., Hughes, D. (2010). Antibiotic Resistance and its Cost: Is it Possible to Reverse Resistance? Nat. Rev. Microbiol. 8, 260–271. doi: 10.1038/nrmicro2319
Bertelli, C., Laird, M. R., Williams, K. P., Simon Fraser University Research Computing Group, Lau, B. Y., Hoad, G., et al (2017). IslandViewer 4: Expanded Prediction of Genomic Islands for Larger-Scale Datasets. Nucleic Acids Res. 45, W30–W35. doi: 10.1093/nar/gkx343
Cannatelli, A., D'andrea, M. M., Giani, T., Di Pilato, V., Arena, F., Ambretti, S., et al (2013). In Vivo Emergence of Colistin Resistance in Klebsiella Pneumoniae Producing KPC-Type Carbapenemases Mediated by Insertional Inactivation of the PhoQ/PhoP mgrB Regulator. Antimicrob. Agents Chemother. 57, 5521–5526. doi: 10.1128/AAC.01480-13
Cheng, L., Connor, T. R., Siren, J., Aanensen, D. M., Corander, J. (2013). Hierarchical and Spatially Explicit Clustering of DNA Sequences With BAPS Software. Mol. Biol. Evol. 30, 1224–1228. doi: 10.1093/molbev/mst028
Cheng, Y. H., Huang, T. W., Juan, C. H., Chou, S. H., Tseng, Y. Y., Chen, T. W., et al (2020). Tigecycline-Non-Susceptible Hypervirulent Klebsiella Pneumoniae Strains in Taiwan. J. Antimicrob. Chemother. 75, 309–317. doi: 10.1093/jac/dkz450
Cheng, Y. H., Lin, T. L., Lin, Y. T., Wang, J. T. (2018). A Putative RND-Type Efflux Pump, H239_3064, Contributes to Colistin Resistance Through CrrB in Klebsiella Pneumoniae. J. Antimicrob. Chemother. 73, 1509–1516. doi: 10.1093/jac/dky054
Clinical and Laboratory Standards Institute (2019). M100S. Performance Standards for Antimicrobial Susceptibility Testing. 29th edition (USA: Wayne, PA).
Elbediwi, M., Pan, H., Zhou, X., Rankin, S. C., Schifferli, D. M., Yue, M., et al (2021). Detection of Mcr-9-Harbouring ESBL-Producing Salmonella Newport Isolated From an Outbreak in a Large-Animal Teaching Hospital in. J. Antimicrob. Chemother. 76, 1107–1109. doi: 10.1093/jac/dkaa544
Giner, G., Smyth, G. K. (2016). Statmod: Probability Calculations for the Inverse Gaussian Distribution. R J. 8, 339–351. doi: 10.32614/RJ-2016-024
Hadjadj, L., Baron, S. A., Olaitan, A. O., Morand, S., Rolain, J. M. (2019). Co-Occurrence of Variants of Mcr-3 and Mcr-8 Genes in a Klebsiella Pneumoniae Isolate From Laos. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02720
He, F., Fu, Y., Chen, Q., Ruan, Z., Hua, X., Zhou, H., et al (2015). Tigecycline Susceptibility and the Role of Efflux Pumps in Tigecycline Resistance in KPC-Producing Klebsiella Pneumoniae. PloS One 10, e0119064. doi: 10.1371/journal.pone.0119064
Hornsey, M., Phee, L., Wareham, D. W. (2011). A Novel Variant, NDM-5, of the New Delhi Metallo-Beta-Lactamase in a Multidrug-Resistant Escherichia Coli ST648 Isolate Recovered From a Patient in the United Kingdom. Antimicrob. Agents Chemother. 55, 5952–5954. doi: 10.1128/AAC.05108-11
Jiang, H., Lei, R., Ding, S. W., Zhu, S. (2014). Skewer: A Fast and Accurate Adapter Trimmer for Next-Generation Sequencing Paired-End Reads. BMC Bioinformatics 15, 182. doi: 10.1186/1471-2105-15-182
Johnson, T. J., Nolan, L. K. (2009). Pathogenomics of the Virulence Plasmids of Escherichia Coli. Microbiol. Mol. Biol. Rev. 73, 750–774. doi: 10.1128/MMBR.00015-09
Krishnaraju, M., Kamatchi, C., Jha, A. K., Devasena, N., Vennila, R., Sumathi, G., et al (2015). Complete Sequencing of an IncX3 Plasmid Carrying blaNDM-5 Allele Reveals an Early Stage in the Dissemination of the blaNDM Gene. Indian J. Med. Microbiol. 33, 30–38. doi: 10.4103/0255-0857.148373
Li, X., Fu, Y., Shen, M., Huang, D., Du, X., Hu, Q., et al (2018). Dissemination of blaNDM-5 Gene via an IncX3-Type Plasmid Among non-Clonal Escherichia Coli in China. Antimicrob. Resist. Infect. Control 7, 59. doi: 10.1186/s13756-018-0349-6
Li, M., Guo, M., Chen, L., Zhu, C., Xiao, Y., Li, P., et al (2020). Isolation and Characterization of Novel Lytic Bacteriophages Infecting Epidemic Carbapenem-Resistant Klebsiella Pneumoniae Strains. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01554
Li, X., Song, Y., Wang, L., Kang, G., Wang, P., Yin, H., et al (2021). A Potential Combination Therapy of Berberine Hydrochloride With Antibiotics Against Multidrug-Resistant Acinetobacter Baumannii. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.660431
Liu, H., Lin, H., Sun, Z., Zhu, X., Zhang, X., Li, Q., et al (2021). Distribution of Beta-Lactamase Genes and Genetic Context of Bla KPC-2 in Clinical Carbapenemase-Producing Klebsiella Pneumoniae Isolates. Infect. Drug Resist. 14, 237–247. doi: 10.2147/IDR.S290434
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al (2016). Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Ma, K., Feng, Y., Liu, L., Yao, Z., Zong, Z. (2020a). A Cluster of Colistin- and Carbapenem-Resistant Klebsiella Pneumoniae Carrying blaNDM-1 and Mcr-8.2. J. Infect. Dis. 221, S237–S242. doi: 10.1093/infdis/jiz519
Ma, T., Fu, J., Xie, N., Ma, S., Lei, L., Zhai, W., et al (2020b). Fitness Cost of blaNDM-5-Carrying P3r-IncX3 Plasmids in Wild-Type NDM-Free Enterobacteriaceae. Microorganisms 8, 377. doi: 10.3390/microorganisms8030377
Martins, W., Nicolas, M. F., Yu, Y., Li, M., Dantas, P., Sands, K., et al (2020). Clinical and Molecular Description of a High-Copy IncQ1 KPC-2 Plasmid Harbored by the International ST15 Klebsiella Pneumoniae Clone. mSphere 5, e00756–e00720. doi: 10.1128/mSphere.00756-20
Mcconville, T. H., Annavajhala, M. K., Giddins, M. J., Macesic, N., Herrera, C. M., Rozenberg, F. D., et al (2020). CrrB Positively Regulates High-Level Polymyxin Resistance and Virulence in Klebsiella Pneumoniae. Cell Rep. 33, 108313. doi: 10.1016/j.celrep.2020.108313
Nordmann, P., Cuzon, G., Naas, T. (2009). The Real Threat of Klebsiella Pneumoniae Carbapenemase-Producing Bacteria. Lancet Infect. Dis. 9, 228–236. doi: 10.1016/S1473-3099(09)70054-4
Osei Sekyere, J., Govinden, U., Bester, L. A., Essack, S. Y. (2016). Colistin and Tigecycline Resistance in Carbapenemase-Producing Gram-Negative Bacteria: Emerging Resistance Mechanisms and Detection Methods. J. Appl. Microbiol. 121, 601–617. doi: 10.1111/jam.13169
Pei, N., Jian, Z., Liu, Y., Liang, T., Liu, W., Li, J. (2021). Draft Genome Sequence of a Polymyxin-Resistant Klebsiella Pneumoniae Clinical Strain Carrying Mcr-8.1 and Bla NDM-5. Microbiol. Resour. Announc. 10, e01224–e01220. doi: 10.1128/MRA.01224-20
Poirel, L., Jayol, A., Nordmann, P. (2017). Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/CMR.00064-16
Prjibelski, A., Antipov, D., Meleshko, D., Lapidus, A., Korobeynikov, A. (2020). Using SPAdes De Novo Assembler. Curr. Protoc. Bioinformatics 70, e102. doi: 10.1002/cpbi.102
Rojas, L. J., Hujer, A. M., Rudin, S. D., Wright, M. S., Domitrovic, T. N., Marshall, S. H., et al (2017). NDM-5 and OXA-181 Beta-Lactamases, a Significant Threat Continues To Spread in the Americas. Antimicrob. Agents Chemother. 61, e00454–e00417. doi: 10.1128/AAC.00454-17
Rosenblum, R., Khan, E., Gonzalez, G., Hasan, R., Schneiders, T. (2011). Genetic Regulation of the ramA Locus and its Expression in Clinical Isolates of Klebsiella Pneumoniae. Int. J. Antimicrob. Agents 38, 39–45. doi: 10.1016/j.ijantimicag.2011.02.012
Samuelsen, O., Overballe-Petersen, S., Bjornholt, J. V., Brisse, S., Doumith, M., Woodford, N., et al (2017). Molecular and Epidemiological Characterization of Carbapenemase-Producing Enterobacteriaceae in Norway 2007 to 2014. PloS One 12, e0187832. doi: 10.1371/journal.pone.0187832
San Millan, A., Maclean, R. C. (2017). Fitness Costs of Plasmids: A Limit to Plasmid Transmission. Microbiol. Spectr. 5, 1–12. doi: 10.1128/microbiolspec.MTBP-0016-2017
Schurch, A. C., Arredondo-Alonso, S., Willems, R. J. L., Goering, R. V. (2018). Whole Genome Sequencing Options for Bacterial Strain Typing and Epidemiologic Analysis Based on Single Nucleotide Polymorphism Versus Gene-by-Gene-Based Approaches. Clin. Microbiol. Infect. 24, 350–354. doi: 10.1016/j.cmi.2017.12.016
Seemann, T. (2014). Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sheng, Z. K., Hu, F., Wang, W., Guo, Q., Chen, Z., Xu, X., et al (2014). Mechanisms of Tigecycline Resistance Among Klebsiella Pneumoniae Clinical Isolates. Antimicrob. Agents Chemother. 58, 6982–6985. doi: 10.1128/AAC.03808-14
Shon, A. S., Bajwa, R. P., Russo, T. A. (2013). Hypervirulent (Hypermucoviscous) Klebsiella Pneumoniae: A New and Dangerous Breed. Virulence 4, 107–118. doi: 10.4161/viru.22718
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., Chandler, M. (2006). ISfinder: The Reference Centre for Bacterial Insertion Sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Stamatakis, A. (2014). RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Sun, S., Gao, H., Liu, Y., Jin, L., Wang, R., Wang, X., et al (2020). Co-Existence of a Novel Plasmid-Mediated Efflux Pump With Colistin Resistance Gene Mcr in One Plasmid Confers Transferable Multidrug Resistance in Klebsiella Pneumoniae. Emerg. Microbes Infect. 9, 1102–1113. doi: 10.1080/22221751.2020.1768805
Tian, D., Wang, B., Zhang, H., Pan, F., Wang, C., Shi, Y., et al (2020). Dissemination of the blaNDM-5 Gene via IncX3-Type Plasmid Among Enterobacteriaceae in Children. mSphere 5, e00699–e00619. doi: 10.1128/mSphere
Wang, X., Chen, J., Kang, Y., Jiang, N., An, S., Gao, Z. (2012). Prevalence and Characterization of Plasmid-Mediated blaESBL With Their Genetic Environment in Escherichia Coli and Klebsiella Pneumoniae in Patients With Pneumonia. Chin. Med. J. (Engl.) 125, 894–900. doi: 10.3760/cma.j.issn.0366-6999.2012.05.029
Wang, X., Chen, H., Zhang, Y., Wang, Q., Zhao, C., Li, H., et al (2015). Genetic Characterisation of Clinical Klebsiella Pneumoniae Isolates With Reduced Susceptibility to Tigecycline: Role of the Global Regulator RamA and its Local Repressor RamR. Int. J. Antimicrob. Agents 45, 635–640. doi: 10.1016/j.ijantimicag.2014.12.022
Wang, X., Wang, Y., Zhou, Y., Li, J., Yin, W., Wang, S., et al (2018). Emergence of a Novel Mobile Colistin Resistance Gene, Mcr-8, in NDM-Producing Klebsiella Pneumoniae. Emerg. Microbes Infect. 7, 1–9. doi: 10.1038/s41426-018-0124-z
Wick, R. R., Judd, L. M., Gorrie, C. L., Holt, K. E. (2017). Unicycler: Resolving Bacterial Genome Assemblies From Short and Long Sequencing Reads. PloS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Wu, B., Wang, Y., Ling, Z., Yu, Z., Shen, Z., Zhang, S., et al (2020). Heterogeneity and Diversity of Mcr-8 Genetic Context in Chicken-Associated Klebsiella Pneumoniae. Antimicrob. Agents Chemother. 65, e01872–e01820. doi: 10.1128/AAC.01872-20
Xu, Y., Liu, L., Zhang, H., Feng, Y. (2021b). Co-Production of Tet(X) and MCR-1, Two Resistance Enzymes by a Single Plasmid. Environ. Microbiol. 23, 7445–7464. doi: 10.1111/1462-2920.15425
Xu, Q., Sheng, Z., Hao, M., Jiang, J., Ye, M., Chen, Y., et al (2021a). RamA Upregulates Multidrug Resistance Efflux Pumps AcrAB and OqxAB in Klebsiella Pneumoniae. Int. J. Antimicrob. Agents 57, 106251. doi: 10.1016/j.ijantimicag.2020.106251
Zhang, F., Xie, L., Wang, X., Han, L., Guo, X., Ni, Y., et al (2016). Further Spread of blaNDM-5 in Enterobacteriaceae via IncX3 Plasmids in Shanghai, China. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00424
Keywords: Klebsiella pneumoniae, blaNDM-5, mcr-8.2, colistin, tigecycline
Citation: Zhao J, Li Z, Zhang Y, Liu X, Lu B and Cao B (2022) Convergence of MCR-8.2 and Chromosome-Mediated Resistance to Colistin and Tigecycline in an NDM-5-Producing ST656 Klebsiella pneumoniae Isolate From a Lung Transplant Patient in China. Front. Cell. Infect. Microbiol. 12:922031. doi: 10.3389/fcimb.2022.922031
Received: 17 April 2022; Accepted: 17 June 2022;
Published: 11 July 2022.
Edited by:
Krisztina M. Papp-Wallace, United States Department of Veterans Affairs, United StatesReviewed by:
Jiyun Li, Hunan Agricultural University, ChinaCopyright © 2022 Zhao, Li, Zhang, Liu, Lu and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Binghuai Lu, enMyNTA0MUAxMjYuY29t; Bin Cao, Y2FvYmluX2JlbkAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.