
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 12 May 2022
Sec. Clinical Microbiology
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.902774
This article is part of the Research Topic Klebsiella pneumoniae: Antimicrobial Resistance, Virulence and Therapeutic Strategies View all 8 articles
 Xi Li1†
Xi Li1† Weizhong Wang1†
Weizhong Wang1† Xi Jin1†
Xi Jin1† Xiaofan Zhang1
Xiaofan Zhang1 Xuehan Zou1
Xuehan Zou1 Qiang Ma2
Qiang Ma2 Qingfeng Hu1*
Qingfeng Hu1* Haijun Huang3*
Haijun Huang3* Yuexing Tu4*
Yuexing Tu4*Objectives: To characterize two plasmids co-harboring carbapenem resistance genes and tmexCD2-toprJ2 in carbapenem-resistant Klebsiella pneumoniae (CRKP) strains.
Methods: Two clinical CRKP strains were isolated and characterized by antimicrobial susceptibility testing, conjugation assays, whole-genome sequencing, and bioinformatics analysis.
Results: The two CRKP strains NB4 and NB5 were both resistant to imipenem, meropenem and tigecycline. Whole-genome sequencing revealed that two CRKP strains belonged to the ST11 type and carried multiple resistance genes. The tmexCD2-toprJ2 clusters in both strains were located on the IncFIB(Mar)-like/HI1B-like group of hybrid plasmids, which co-harbored the metallo-β-lactamase gene blaNDM-1. In addition, the co-existence of blaNDM-1 and blaKPC-2 and the presence of tmexCD2-toprJ2 in CRKP strain NB5 was observed.
Conclusions: In this study, tmexCD2-toprJ2 gene clusters were identified in two NDM-1-producing CRKP ST11 strains. These gene clusters will likely spread into clinical high-risk CRKP clones and exacerbate the antimicrobial resistance crisis. In addition, we detected the co-occurrence of blaNDM-1, blaKPC-2 and tmexCD2-toprJ2 in a single strain, which will undoubtedly accelerate the formation of a “superdrug resistant” bacteria. Hence, effective control measures should be implemented to prevent the further dissemination of such organisms in clinical settings.
Carbapenem resistance genes have been widely identified in various species of Enterobacteriaceae, posing a significant threat, especially in clinical environments. Antimicrobial options for the treatment of carbapenem-resistant Enterobacterales (CRE) infections are increasingly limited due to the extensive distribution of CRE and the emergence of mobile colistin resistance (mcr) genes (Jiang et al., 2020). Tigecycline (TGC) has been regarded as one of the last resort treatment options for infections caused by CRE. Regrettably, the increasing prevalence of CRE has inevitably resulted in increased use of TGC, accelerating the emergence of TGC-resistant isolates (Wang et al., 2018). Of note, TGC-resistant strains have been increasingly observed in clinics since the new drug was approved in 2005. Currently, TGC resistance occurs in chromosome and plasmid factors in gram-negative bacteria. The overexpression of chromosomal multidrug-resistant efflux pumps, such as resistance nodulation division (RND) pumps, AcrAB-TolC pumps, multidrug and toxic compound extrusion (MATE) pumps, and their regulator factors, or mutations, within ribosomal drug-binding sites are considered to be the most common mechanisms for increasing bacterial drug resistance (Sun et al., 2013). However, a growing concern is that the emergence of TGC resistance genes in plasmids may exacerbate transferable resistance among bacterial species. The plasmid-mediated genes tet(X3), tet(X4), tet(X5), and tet(X6), which encode enzymatic inactivation proteins against tigecycline, have been detected in animal and clinical isolates (Bai et al., 2019; Chen et al., 2019a; Chen et al., 2019b; He et al., 2019; Sun et al., 2019a; Sun et al., 2019b; Wang et al., 2019; He et al., 2020).
Recently, a novel plasmid-encoded RND efflux pump, the tmexCD1-toprJ1 gene cluster, was identified in Klebsiella pneumoniae isolates from animals, foods, and humans in China (Lv et al., 2020). Subsequently, its orthologous variants tmexCD2-toprJ2 and tmexCD3-toprJ3 were reported in Raoultella ornithinolytica and Proteus mirabilis, respectively (Wang et al., 2021a; Wang et al., 2021b). Likely originating from Pseudomonas spp., tmexCD-toprJ gene clusters appear to achieve horizontal transfer using adjacent site-specific integrases that confer multidrug resistance (including tetracycline, eravacycline, quinolones, cephalosporins, and aminoglycosides) (Lv et al., 2020). This gene cluster was mainly carried in K. pneumoniae but has also been identified in other clinical CRKP strains (Qin et al., 2021). In addition, these strains have various clone types, such as ST15 (Yang et al., 2021), ST37 (Sun et al., 2020) and ST2667 (Qin et al., 2021). Nevertheless, these gene clusters have rarely occurred in ST11-type CRKP, which is a prevalent clinical CRKP clone in China.
However, we report here two plasmids co-harbouring the tmexCD2-toprJ2 gene cluster and carbapenem resistance genes in two clinical ST11 CRKP strains.
Based on the surveillance of carbapenem resistance organisms (CRO) from clinical specimens of inpatients, all collected strains were identified by MALDI-TOF technology (bioMérieux, Marcy l’Etoile, France) as well as screened for the tmexCD-toprJ gene cluster by PCR and Sanger sequencing. Finally, two CRKP strains NB4 and NB5 showed positive for tmexCD2-toprJ2 gene cluster.
Antimicrobial susceptibility testing was performed according to the reference Clinical and Laboratory Standards Institute (CLSI) (CLSI, 2020). Broth microdilution method was used to measure MIC values for ceftazidime, cefepime, amoxicillin-clavulanic acid, amikacin, ciprofloxacin, meropenem, ertapenem, imipenem, tigecycline and colistin. The results of MICs were interpreted according to CLSI guidelines (CLSI, 2020), except tigecycline and colistin, for which were interpreted according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) criteria for Enterobacteriaceae (http://www.eucast.org/clinical_breakpoints). E. coli ATCC 25922 was used as a quality control strain.
Conjugation and electroporation experiments were performed according to our previous study (Quan et al., 2017). Briefly, conjugation experiments were performed with E. coli J53 (AziR) and C600 (RifR) as the recipient strains. 6h growth cultures of the donor strain and the recipient strains were mixed at a ratio of 1:2 in LB broth, and the mixture was then diluted and spread on a MH agar plate containing tigecycline (0.5 mg/liter) and sodium azide (300 mg/liter) or rifampicin (600 mg/liter) for selecting transconjugants. Plasmid DNA was extracted using a Qiagen plasmid midi kit (Qiagen, Germany), then was transformed into electrocompetent E. coli DH5α cells. Luria-Bertani agar plates containing tigecycline (0.5 mg/liter) were used to select the transformants, which were further confirmed by PCR targeting at tmexCD2-toprJ2 gene cluster, 16S rRNA, and antimicrobial susceptibility testing.
The genomic DNA of the K. pneumoniae NB4 and NB5 strains was obtained using a QIAamp DNA MiniKit (Qiagen, Valencia, CA, USA) following the manufacturer’s recommendations. The combination Oxford Nanopore (MinION system, Nanopore, Oxford, UK) and Illumina sequencing (NovaSeq system, Illumina Inc, San Diego, U.S.A) were used to achieve the complete chromosomes and plasmid sequences, respectively.
The Illumina reads and Nanopore reads were assembled using the hybird assembly tool Unicycler version 0.4.8 (Wick et al., 2017). Annotation of the plasmid genomes was performed using the RAST annotation website server (http://rast.nmpdr.org/rast/cgi).
Antibiotic resistance genes (ARGs), plasmid replicon types, and sequence type of the strains were obtained by the ResFinder 4.1, PlasmidFinder 1.3 and MLST 2.1 servers, which are available at the Center for Genomic Epidemiology (http://www.genomicepidemiology.org/). The virulence factors were identified using the kleborate software (https://github.com/katholt/Kleborate). BRIG and Easyfig were used to visualize the plasmid comparisons and genetic context comparisons, respectively (Alikhan et al., 2011; Sullivan et al., 2011).
The complete genome sequences of K. pneumoniae NB4 and NB5 reported in the present study were deposited in the GenBank nucleotide database under accession no. CP091986-CP091987, CP091992 and CP092653-CP092656.
The two CRKP strains NB4 and NB5 were both isolated from the urine of hospitalized patients in 2017 and displayed an almost consistent susceptibility pattern. They were resistant to amoxicillin-clavulanic acid, cefepime, ceftazidime, ertapenem, imipenem, meropenem, amikacin, ciprofloxacin and tigecycline, but susceptible to colistin (Table 1). Whole-genome sequencing analysis showed that the two K. pneumoniae strains were classified as sequence type 11 (ST11).
CRKP strain NB4 carried a 5.30-Mb chromosome and three plasmids. Among three plasmids, the blaNDM-1 gene was carried on plasmid pNB4_NDM. It carried twenty known antibiotic resistance genes (ARGs), including blaNDM-1, blaDHA-1, and fosA3. CRKP strain NB5 carried a 5.30-Mb chromosome and three plasmids, such as pNB5_NDM (355.4 Kb) and pNB5_KPC-2 (71.6 Kb). It carried twenty-eight known ARGs, including blaNDM-1, blaDHA-1, blaKPC-2 and fosA3 (Table S1). To the best of our knowledge, we are reporting the co-existence of blaNDM-1 and blaKPC-2 and the presence of tmexCD2-toprJ2 in CRKP for the first time.
BLASTn against the virulence genes database (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html) displayed that two strains carried virulence genes ipaH (invasion plasmid antigen) and acrB (acriflavine resistance protein B). Virulence plasmid-bearing virulence genes, such as iro, iuc, rmpA/rmpA2 were not present in the two strains.
To investigate the core genetic environment of tmexCD2-toprJ2 in the two K. pneumoniae strains, two complete tmexCD2-toprJ2-carrying plasmids were successfully obtained using a hybrid assembly strategy combining short-read and long-read data. The two tmexCD2-toprJ2-carrying plasmids were designated pNB4_NDM in strain NB4 and pNB5_NDM in strain NB5. Both belonged to the IncFIB(Mar)-like/HI1B-like group of multi-replicon plasmids, which were different from the first discovered tmexCD2-toprJ2-positive IncFIBK plasmid pHNNC189-2 found in R. ornithinolytica. Plasmid sequence comparison showed that pNB4_NDM and pNB5_NDM had highly conserved plasmid synteny and structure, with 100% nucleotide identities (Figure 1). Furthermore, the backbones of pNB4_NDM and pNB5_NDM were similar to those of the two tmexCD2-toprJ2-carrying plasmids pNUITM-VK4 and pNUITM-VK10 in the nr database, which were harboured by K. quasipneumoniae (Figure 2). In addition, these two plasmids could not be transferred to recipient cells by conjugation or transformation in E. coli J53 and E. coli C600 strains after three attempts.
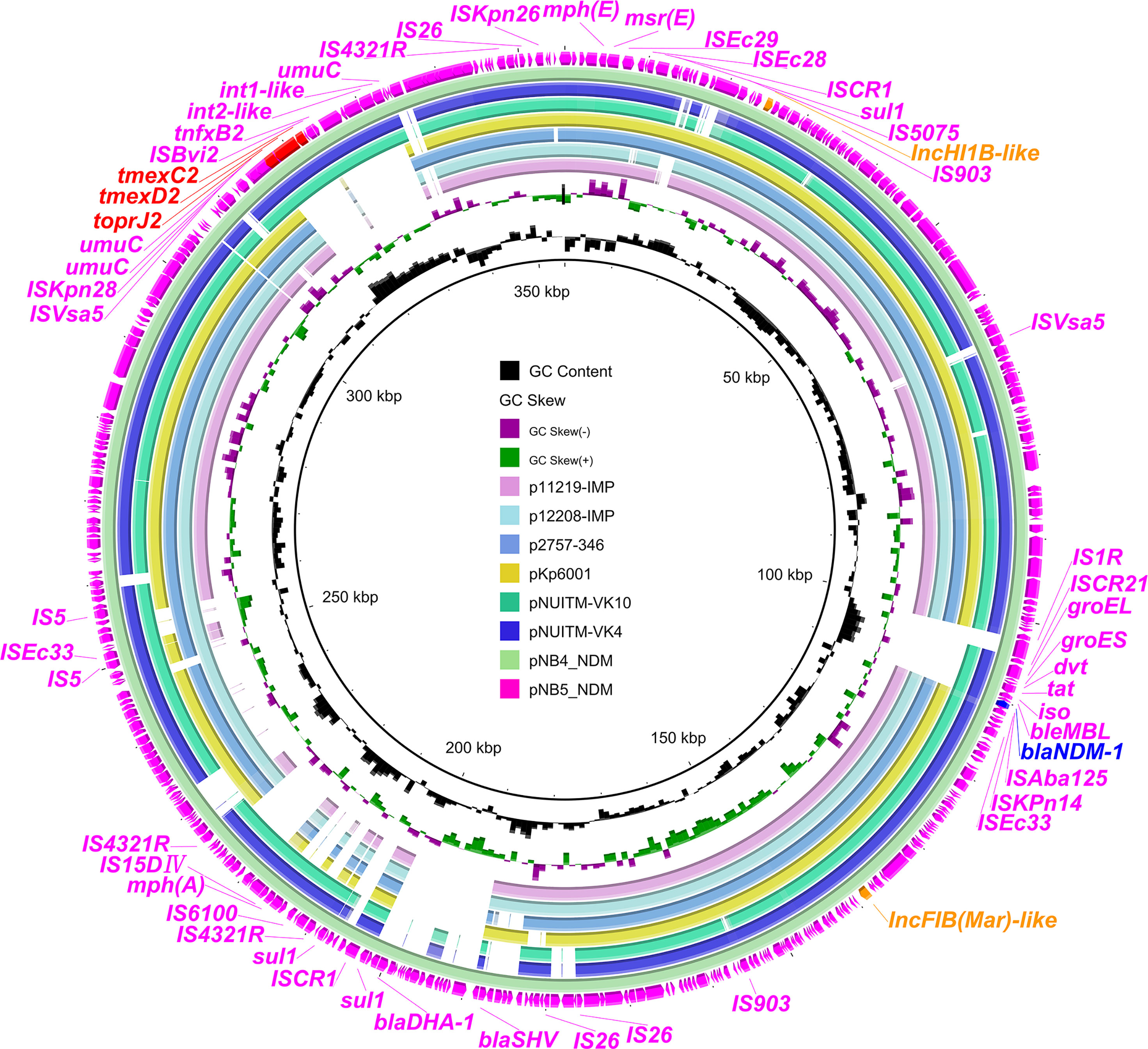
Figure 1 Comparison analysis between tmexCD2-toprJ2-bearing plasmids and other similar plasmids. The two external rings represent the structures of pNB5_NDM (pink) and pNB4_NDM (aqua). Other similar plasmids were pNUITM-VK4 (GenBank accession no. AP025165.1), pNUITM-VK10 (GenBank accession no. AP025166.1), pKp6001 (GenBank accession no. CP082291.1), p2757-346 (GenBank accession no. CP060810.1), p12208-IMP (GenBank accession no. MF344562.1), and p11219-IMP (GenBank accession no. MF344561.1). The figure was constructed using BRIG.
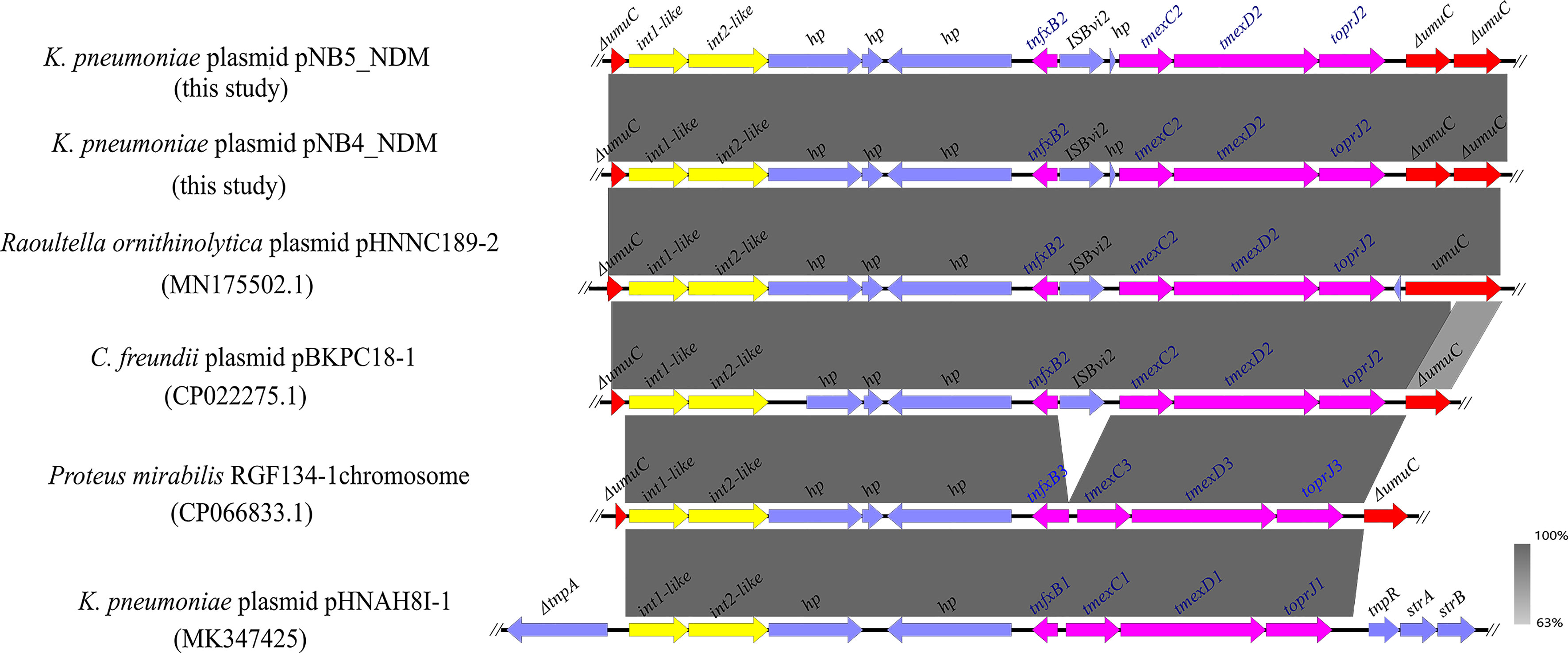
Figure 2 Comparison analysis of the genetic context of tmexCD2-toprJ2 with that of closely related sequences. The pink arrows indicate tnfxB2-tmexCD2-toprJ2 and tnfxB1-tmexCD1-toprJ1-like gene clusters. The yellow arrows indicate the int-like genes. The gene umuC is indicated by red arrows. ISBvi2 and hp are indicated by blue arrows. The symbol Δ indicates that the gene is truncated.
Comparative analysis demonstrated that a similar genetic context like tmexCD2-toprJ2 was observed in the tmexCD2-toprJ2-bearing plasmids pNB4_NDM and pNB5_NDM. Meanwhile, we found that the tnfxB2-tmexCD2-toprJ2 gene clusters were inserted into the umuC gene. A similar structure was also found in the plasmids of Raoultella ornithinolytica and C. freundii in the nr database (Figure 3), which indicated that the two int-like genes may contribute to the mobilization of the tmexCD2-toprJ2 gene cluster. Moreover, the umuC gene was an integration hotspot for the two integrases.
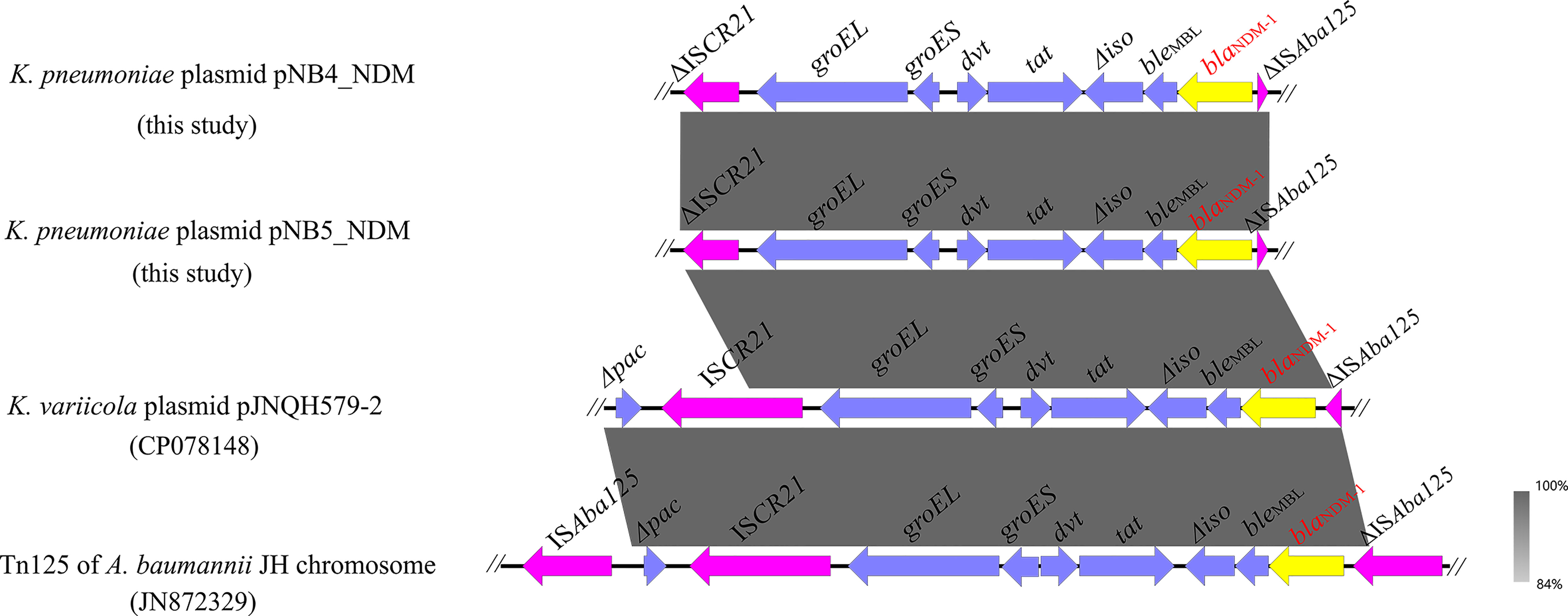
Figure 3 Genetic context of blaNDM-1 located on plasmids pNB4_NDM and pNB5_NDM compared with other similar sequences. The yellow arrows indicate the blaNDM-1 genes.
Apart from tmexCD2-toprJ2, pNB4_NDM and pNB5_NDM also contained the carbapenemase-encoding gene blaNDM-1, which was located on the same plasmids as the tmexCD2-toprJ2 gene cluster. Genetic structure analysis showed that blaNDM-1 was located in a truncated transposon Tn125 in plasmids pNB4_NDM and pNB5_NDM (Figure 4), with the structure of “ΔISAba125-blaNDM-1-bleMBL-Δiso-tat-dvt-groES- groEL-ΔISCR21”. The structure of a truncated transposon Tn125 containing blaNDM-1 was also observed in the tmexCD2-toprJ2-carrying plasmid pJNQH579-2 (Wang et al., 2021c). Furthermore, we noticed that transposon Tn125 seems to be the major vehicle for the dissemination of blaNDM-1 genes in Klebsiella spp.
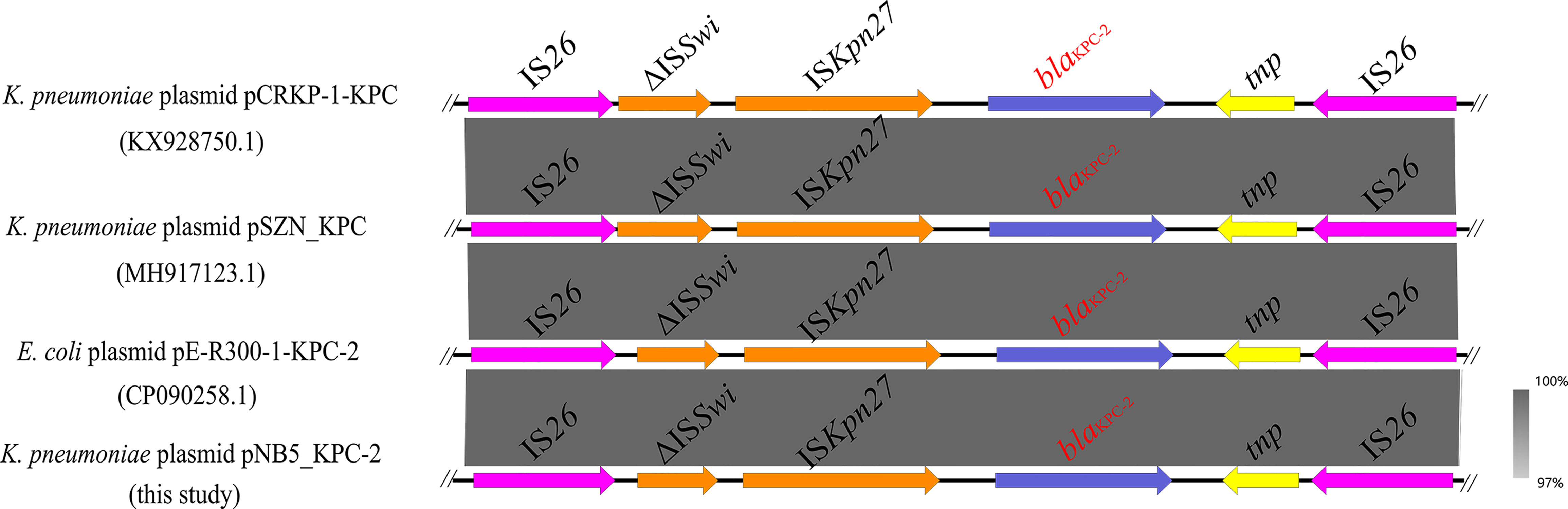
Figure 4 Genetic context of blaKPC-2 located on K. pneumoniae strain NB5 plasmid pNB5_KPC-2 compared with other similar sequences. The blue arrows indicate the blaKPC-2 genes.
Interestingly, the carbapenemase-encoding gene blaKPC-2 was also discovered in the IncN/U-type plasmid pNB5_KPC-2 (Figure 4). It is worth emphasizing that IS26-mediated transmission of the blaKPC-2 gene has been detected in many strains (Figure 4), and it is vital for the dissemination of multiple resistance genes in these bacteria to be monitored closely.
Comparative analysis of the plasmid database revealed that a total of 25 plasmids carried tmexCD-toprJ gene clusters in clinical K. pneumoniae strains (as of 06 December 2021) (Figure 5), 20 strains had tmexCD1-toprJ1, 5 strains had tmexCD2-toprJ2, and no strain carried tmexCD3-toprJ3. Of note, 20% (5/25) of the strains co-harboured carbapenem resistance genes and tmexCD-toprJ gene clusters, including 4 strains carrying the blaNDM-1 gene and 1 strain carrying the blaKPC-2 gene. In addition, 35% (7/20) of K. pneumoniae strains carrying tmexCD1-toprJ1 gene clusters were ST967, and 60% (3/5) of strains carrying tmexCD2-toprJ2 gene clusters were ST2667. Sixteen percent (4/25) of the strains carrying tmexCD-toprJ gene clusters were ST11, including 2 strains carrying tmexCD1-toprJ1 gene clusters and the 2 strains carrying tmexCD2-toprJ2 gene clusters characterized in this study.
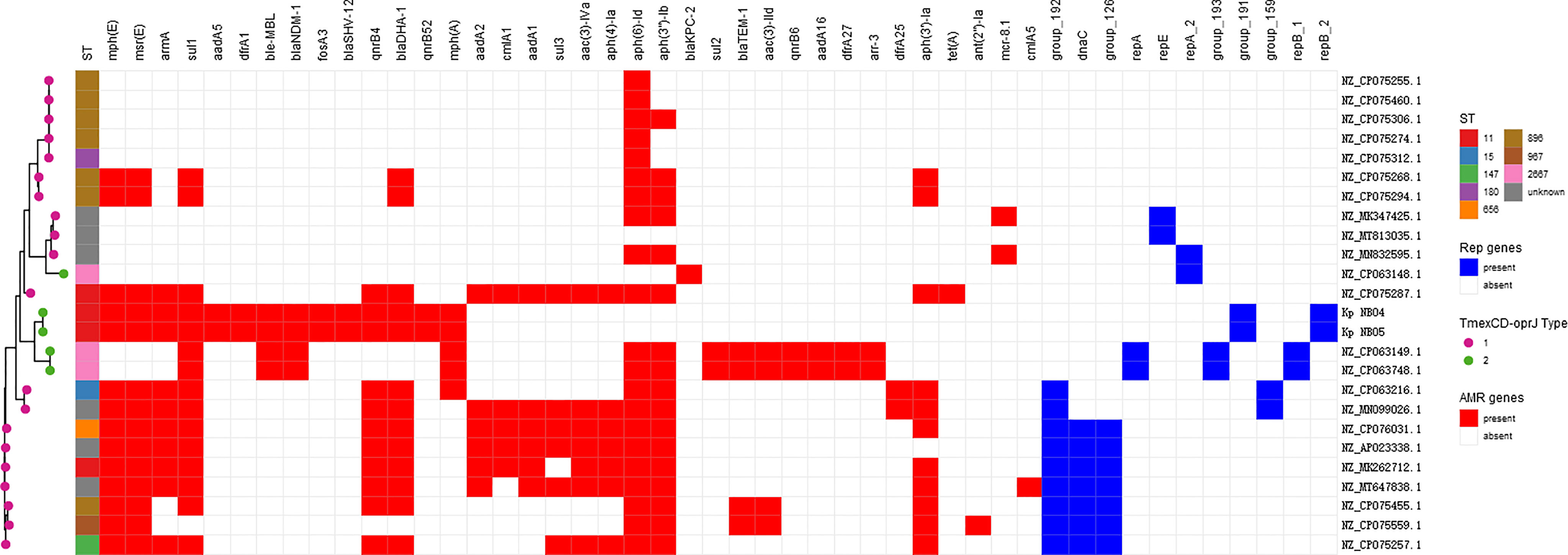
Figure 5 tmexCD-toprJ-harboring plasmids in K. pneumoniae strain. The heatmap shows the distribution of plasmid replicons (dark blue boxes) and antibiotic resistance genes (red boxes) detected within 25 tmexCD-toprJ-harboring plasmids. The variants of tmexCD-toprJ resistance genes are indicated in pink (tmexCD1-toprJ1) and green (tmexCD2-toprJ2). GenBank accession numbers and species are listed on the right-hand side.
Unlike the tmexCD1-toprJ1 gene, which is primarily found in K. pneumoniae, and the tmexCD3-toprJ3 gene, which is frequently found in P. aeruginosa (Wang et al., 2021c), the tmexCD2-toprJ2 gene was identified among various bacterial species, including Raoultella ornithinolytica, Citrobacter freundii, Aeromonas hydrophila, K. quasipneumoniae, K. variicola, and K. michiganensis (Wang et al., 2021a; Wang et al., 2021c). Our study further demonstrated that tmexCD2-toprJ2 spread into CRKP strains, indicating that this gene cluster might have a wider host range than its homologous genes.
Whole-genome sequencing analysis showed that our two CRKP strains both belonged to the ST11 clone type. Currently, clonal spreading is one of the primary modes of CRKP dissemination. In China, ST11-type CRKP is a common clone lineage (Qi et al., 2011) and is frequently associated with a high fate of mortality, posing a severe challenge in clinical treatment (Giacobbe et al., 2015). In this study, we conclude that these two isolates are clonally related based on their identical STs, plasmid components, and resistomes. Our data indicate the potential clonal dissemination of tmexCD2-toprJ2-positive K. pneumoniae. As a high-risk clinical pathogen to human health, ST11 CRKP strains lack both CRISPR-Cas systems and restriction-modification (RM) systems, which usually have a special ability to acquire resistance genes with high transferability (Liao et al., 2020). We found that tmexCD2-toprJ2 and blaNDM-1 were co-encoded by the same plasmids in both the ST11 CRKP NB4 and NB5 strains. Meanwhile, many resistance genes were identified in the two strains. Furthermore, the co-existence of blaNDM-1, blaKPC-2, and tmexCD2-toprJ2 in CRKP was observed for the first time, indicating that ST11 CRKP will likely spread in the clinical environment due to its robust ability for acquiring drug resistance. In addition, comparative analysis of plasmids from the NCBI nr database revealed frequent co-occurrence of tmexCD2-toprJ2 and carbapenem resistance genes in the same plasmid harboured by CRKP. These findings suggested that such homologous plasmids were adapted by Klebsiella spp., which may be a reservoir for multiple resistance genes, such as carbapenem and tigecycline resistance genes. The co-occurrence of tmexCD2-toprJ2 and blaNDM-1 in the same plasmid should be considered seriously as a public health concern because the convergence of “mosaic” plasmids can cause both tigecycline and carbapenem resistance. Furthermore, plasmids co-harbouring the tmexCD1-toprJ1, mcr-8 and blaNDM genes have been identified (Sun et al., 2020), which will undoubtedly accelerate the formation of a “superdrug resistant”plasmid.
Of note, pNB4_NDM and pNB5_NDM share a similar plasmid backbone. Plasmid replicon analysis showed that pNB4_NDM and pNB5_NDM harboured two conserved replicon genes. In this study, we call them IncFIB(Mar)-like/IncHI1B-like, which were highly homologous to IncFIB (Mar) and IncHI1B plasmids, respectively. The IncFIB(Mar)/IncHI1B-type plasmids were mainly carried by Klebsiella spp. in food production chains according to the host range analysis and seemed to be primary vectors for the horizontal dissemination of tmexCD1-toprJ1 among Klebsiella spp. (Peng et al., 2021). Notably, IncFIB(Mar)/IncHI1B-type plasmid carrying tmexCD1-toprJ1 gene could not conjugate to E. coli J53 and E. coli C600 strains but could be transformed to E. coli DH5α strains by electroporation (Peng et al., 2021). However, the two plasmids in this study could not be transferred to recipient E. coli strains by either conjugation or transformation with E. coli strains, indicating that this IncFIB(Mar)-like/IncHI1B-like type plasmids might be more restricted by the host species. Further studies are needed to assess the contribution of two conserved replicons in the host range.
Genetic context analysis showed that tmexCD2-toprJ2 was in a conserved structure in the two plasmids. An increasing number of studies have revealed that similar structures containing the tmexCD-toprJ gene cluster were present in different bacterial species (Wang et al., 2021c). Interestingly, we found that a genetic structure containing tmexCD2-toprJ2 gene clusters and two int genes were inserted into the umuC gene in both pNB4_NDM and pNB5_NDM plasmids (Figure 2). The umuC gene appears to be a “hotspot” for tmexCD-toprJ clusters integration in chromosomes and plasmids (Peng et al., 2021). The umuCD gene was the insertion site of variable region III of SXT/R391 ICEs (Burrus et al., 2006; Carraro and Burrus, 2014), which consisted of many resistance genes such as tmexCD1-toprJ1 (Wang et al., 2021c), tet(X) (He et al., 2020; Peng et al., 2020), and blaNDM-5 (Kong et al., 2020). The prevalence of umuCD may play important role in the spreading of the tmexCD-toprJ gene cluster, while further study is needed to identify this molecular mechanism.
In addition, the blaNDM-1 carbapenem resistance gene co-existed with tmexCD2-toprJ2 in both plasmids pNB4_NDM and pNB5_NDM. blaNDM-1 is in the truncated transposon Tn125, which has been identified for years and is widely distributed on multiple plasmids in a variety of bacterial species. More attention is needed to study the genetic structure of blaNDM-1 driven from Tn125 to elucidate its possible horizontal transmission mechanisms. The structure of a truncated transposon Tn125 containing blaNDM-1 was also observed in the tmexCD2-toprJ2-carrying plasmid pJNQH579-2 (Wang et al., 2021c). Furthermore, we noticed that transposon Tn125 seems to be the major vehicle for dissemination of blaNDM-1 in Klebsiella spp.
In summary, we report the identification of two clinical tmexCD2-toprJ2-encoding ST11 carbapenem-resistant K. pneumoniae strains. Dissemination of tmexCD-toprJ gene clusters in CRKP strains may pose a substantial threat in clinical treatment settings. TGC has become one of the few therapeutic alternatives against CRKP strains. However, the emergence of TGC resistance gene clusters in CRKP strains carrying blaNDM-1 or blaKPC-2 is a matter of major concern because colistin is presently a last-resort antibiotic, alone in its ability to treat infections caused by similar strains. It is essential to continuously monitor such resistance gene clusters in different settings to better understand their specific transmission mechanisms.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
This study was conducted in accordance with the Declaration of Helsinki and had been reviewed and approved by the Research Ethics Committee of the Zhejiang Provincial People’s Hospital (QT2022130).
Conceived and designed the experiments: QH, HH, and YT; Performed the experiments: XiZ; Analyzed the data: XuZ and QM; Wrote the manuscript: XJ and XL; All authors read and approved the final manuscript.
This study was supported by National Natural Science Foundation of China (No. 82172306), Public Technology Research Projects of Zhejiang Province, China (LGD21H190001) the Medical and Health Research Project of Zhejiang Province, China (2020KY420 and 2022KY531) and Health Science and Technology Project of Hangzhou (0020190881).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Professor Dazhi Jin (Hangzhou Medical College) for his help with revising the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.902774/full#supplementary-material
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): Simple Prokaryote Genome Comparisons. BMC Genomics 12, 402. doi: 10.1186/1471-2164-12-402
Bai, L., Du, P., Du, Y., Sun, H., Zhang, P., Wan, Y., et al. (2019). Detection of Plasmid-Mediated Tigecycline-Resistant Gene Tet(X4) in Escherichia Coli From Pork, Sichuan and Shandong Provinces, China, February 2019. Euro Surveill 24 (25), 1900340. doi: 10.2807/1560-7917
Burrus, V., Marrero, J., Waldor, M. K. (2006). The Current ICE Age: Biology and Evolution of SXT-Related Integrating Conjugative Elements. Plasmid 55, 173–183. doi: 10.1016/j.plasmid.2006.01.001
Carraro, N., Burrus, V. (2014). Biology of Three ICE Families: SXT/R391, ICEBs1, and Icest1/Icest3. Microbiol. Spectr. 2 (6). doi: 10.1128/microbiolspec.MDNA3-0008-2014
Chen, C., Cui, C. Y., Zhang, Y., He, Q., Wu, X. T., Li, G., et al. (2019a). Emergence of Mobile Tigecycline Resistance Mechanism in Escherichia Coli Strains From Migratory Birds in China. Emerg. Microbes Infect. 8, 1219–1222. doi: 10.1080/22221751.2019.1653795
Chen, C., Wu, X. T., He, Q., Chen, L., Cui, C. Y., Zhang, Y., et al. (2019b). Complete Sequence of a Tet(X4)-Harboring IncX1 Plasmid, Pyy76-1-2, in Escherichia Coli From a Cattle Sample in China. Antimicrob. Agents Chemother. 63 (12), e01528–19. doi: 10.1128/AAC.01528-19
CLSI. (2020). Performance Standards for Antimicrobial Susceptibility Testing. 30th ed Vol. CLSI Supplement M100 (Wayne, PA: Clinical and Laboratory Standards Institute).
Giacobbe, D. R., Del Bono, V., Trecarichi, E. M., De Rosa, F. G., Giannella, M., Bassetti, M., et al. (2015). Risk Factors for Bloodstream Infections Due to Colistin-Resistant KPC-Producing Klebsiella Pneumoniae: Results From a Multicenter Case-Control-Control Study. Clin. Microbiol. Infect. 21, 1106.e1101–1108. doi: 10.1016/j.cmi.2015.08.001
He, T., Wang, R., Liu, D., Walsh, T. R., Zhang, R., Lv, Y., et al. (2019). Emergence of Plasmid-Mediated High-Level Tigecycline Resistance Genes in Animals and Humans. Nat. Microbiol. 4, 1450–1456. doi: 10.1038/s41564-019-0445-2
He, D., Wang, L., Zhao, S., Liu, L., Liu, J., Hu, G., et al. (2020). A Novel Tigecycline Resistance Gene, Tet(X6), on an SXT/R391 Integrative and Conjugative Element in a Proteus Genomospecies 6 Isolate of Retail Meat Origin. J. Antimicrob. Chemother. 75, 1159–1164. doi: 10.1093/jac/dkaa012
Jiang, Y., Zhang, Y., Lu, J., Wang, Q., Cui, Y., Wang, Y., et al. (2020). Clinical Relevance and Plasmid Dynamics of Mcr-1-Positive Escherichia Coli in China: A Multicentre Case-Control and Molecular Epidemiological Study. Lancet Microbe 1, e24–e33. doi: 10.1016/S2666-5247(20)30001-X
Kong, L. H., Xiang, R., Wang, Y. L., Wu, S. K., Lei, C. W., Kang, Z. Z., et al. (2020). Integration of the blaNDM-1 Carbapenemase Gene Into a Novel SXT/R391 Integrative and Conjugative Element in Proteus Vulgaris. J. Antimicrob. Chemother. 75, 1439–1442. doi: 10.1093/jac/dkaa068
Liao, W., Liu, Y., Zhang, W. (2020). Virulence Evolution, Molecular Mechanisms of Resistance and Prevalence of ST11 Carbapenem-Resistant Klebsiella Pneumoniae in China: A Review Over the Last 10 Years. J. Glob Antimicrob. Resist. 23, 174–180. doi: 10.1016/j.jgar.2020.09.004
Lv, L., Wan, M., Wang, C., Gao, X., Yang, Q., Partridge, S. R., et al. (2020). Emergence of a Plasmid-Encoded Resistance-Nodulation-Division Efflux Pump Conferring Resistance to Multiple Drugs, Including Tigecycline, in Klebsiella Pneumoniae. mBio 11 (2), e02930–19. doi: 10.1128/mBio.02930-19
Peng, K., Li, R., He, T., Liu, Y., Wang, Z. (2020). Characterization of a Porcine Proteus Cibarius Strain Co-Harbouring Tet(X6) and Cfr. J. Antimicrob. Chemother. 75, 1652–1654. doi: 10.1093/jac/dkaa047
Peng, K., Wang, Q., Yin, Y., Li, Y., Liu, Y., Wang, M., et al. (2021). Plasmids Shape the Current Prevalence of Tmexcd1-Toprj1 Among Klebsiella Pneumoniae in Food Production Chains. mSystems 6, e0070221. doi: 10.1128/mSystems.00702-21
Qin, S., Peng, J., Deng, R., Peng, K., Yan, T., Chen, F., et al. (2021). Identification of Two Plasmids Coharboring Carbapenemase Genes and Tmexcd1-Toprj1 in Clinical Klebsiella Pneumoniae ST2667. Antimicrob. Agents Chemother. 65 (6), e00625–21. doi: 10.1128/AAC.00625-21
Qi, Y., Wei, Z., Ji, S., Du, X., Shen, P., Yu, Y. (2011). ST11, the Dominant Clone of KPC-Producing Klebsiella Pneumoniae in China. J. Antimicrob. Chemother. 66, 307–312. doi: 10.1093/jac/dkq431
Quan, J., Li, X., Chen, Y., Jiang, Y., Zhou, Z., Zhang, H., et al. (2017). Prevalence of Mcr-1 in Escherichia Coli and Klebsiella Pneumoniae Recovered From Bloodstream Infections in China: A Multicentre Longitudinal Study. Lancet Infect. Dis. 17, 400–410. doi: 10.1016/S1473-3099(16)30528-X
Sullivan, M. J., Petty, N. K., Beatson, S. A. (2011). Easyfig: A Genome Comparison Visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Sun, Y., Cai, Y., Liu, X., Bai, N., Liang, B., Wang, R. (2013). The Emergence of Clinical Resistance to Tigecycline. Int. J. Antimicrob. Agents 41, 110–116. doi: 10.1016/j.ijantimicag.2012.09.005
Sun, J., Chen, C., Cui, C. Y., Zhang, Y., Liu, X., Cui, Z. H., et al. (2019b). Plasmid-Encoded Tet(X) Genes That Confer High-Level Tigecycline Resistance in Escherichia Coli. Nat. Microbiol. 4, 1457–1464. doi: 10.1038/s41564-019-0496-4
Sun, C., Cui, M., Zhang, S., Wang, H., Song, L., Zhang, C., et al. (2019a). Plasmid-Mediated Tigecycline-Resistant Gene Tet(X4) in Escherichia Coli From Food-Producing Animals, China 2008-2018. Emerg. Microbes Infect. 8, 1524–1527. doi: 10.1080/22221751.2019.1678367
Sun, S., Gao, H., Liu, Y., Jin, L., Wang, R., Wang, X., et al. (2020). Co-Existence of a Novel Plasmid-Mediated Efflux Pump With Colistin Resistance Gene Mcr in One Plasmid Confers Transferable Multidrug Resistance in Klebsiella Pneumoniae. Emerg. Microbes Infect. 9, 1102–1113. doi: 10.1080/22221751.2020.1768805
Wang, C. Z., Gao, X., Yang, Q. W., Lv, L. C., Wan, M., Yang, J., et al. (2021a). A Novel Transferable Resistance-Nodulation-Division Pump Gene Cluster, Tmexcd2-Toprj2, Confers Tigecycline Resistance in Raoultella Ornithinolytica. Antimicrob. Agents Chemother. 65 (4), e02229–20. doi: 10.1128/AAC.02229-20
Wang, L., Liu, D., Lv, Y., Cui, L., Li, Y., Li, T., et al. (2019). Novel Plasmid-Mediated Tet(X5) Gene Conferring Resistance to Tigecycline, Eravacycline, and Omadacycline in a Clinical Acinetobacter Baumannii Isolate. Antimicrob. Agents Chemother. 64 (1), e01326–19. doi: 10.1128/AAC.01326-19
Wang, Q., Peng, K., Liu, Y., Xiao, X., Wang, Z., Li, R. (2021b). Characterization of TMexCD3-TOprJ3, an RND-Type Efflux System Conferring Resistance to Tigecycline in Proteus Mirabilis, and Its Associated Integrative Conjugative Element. Antimicrob. Agents Chemother. 65, e0271220. doi: 10.1128/AAC.02712-20
Wang, Q., Wang, X., Wang, J., Ouyang, P., Jin, C., Wang, R., et al. (2018). Phenotypic and Genotypic Characterization of Carbapenem-Resistant Enterobacteriaceae: Data From a Longitudinal Large-Scale CRE Study in China, (2012-2016). Clin. Infect. Dis. 67, S196–S205. doi: 10.1093/cid/ciy660
Wang, Y., Zhu, B., Liu, M., Dong, X., Ma, J., Li, X., et al. (2021c). Characterization of IncHI1B Plasmids Encoding Efflux Pump TmexCD2-ToprJ2 in Carbapenem-Resistant Klebsiella Variicola, Klebsiella Quasipneumoniae, and Klebsiella Michiganensis Strains. Front. Microbiol. 12, 759208. doi: 10.3389/fmicb.2021.759208
Wick, R. R., Judd, L. M., Gorrie, C. L., Holt, K. E. (2017). Unicycler: Resolving Bacterial Genome Assemblies From Short and Long Sequencing Reads. PloS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Keywords: CRKP, tmexCD2-toprJ2, blaNDM-1, plasmids, carbapenem resistance
Citation: Li X, Wang W, Jin X, Zhang X, Zou X, Ma Q, Hu Q, Huang H and Tu Y (2022) Emergence of Plasmids Co-Harboring Carbapenem Resistance Genes and tmexCD2-toprJ2 in Sequence Type 11 Carbapenem Resistant Klebsiella pneumoniae Strains. Front. Cell. Infect. Microbiol. 12:902774. doi: 10.3389/fcimb.2022.902774
Received: 23 March 2022; Accepted: 13 April 2022;
Published: 12 May 2022.
Edited by:
Ning Dong, Soochow University, ChinaReviewed by:
Yao Zhu, Harbin Veterinary Research Institute (CAAS), ChinaCopyright © 2022 Li, Wang, Jin, Zhang, Zou, Ma, Hu, Huang and Tu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingfeng Hu, bGFiX2hxZkAxMjYuY29t; Haijun Huang, aHVhbmdoYWlqdW4wODI2QDE2My5jb20=; Yuexing Tu, VHV5dWV4aW5nMTk4OEAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.