
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell. Infect. Microbiol., 03 May 2022
Sec. Microbiome in Health and Disease
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.892232
This article is part of the Research TopicMeta-omics to investigate gut microbiome/mycobiome in cardiometabolic diseaseView all 5 articles
 Keiichi Sumida1*
Keiichi Sumida1* Zhongji Han1
Zhongji Han1 Chi-Yang Chiu2
Chi-Yang Chiu2 Tahliyah S. Mims3
Tahliyah S. Mims3 Amandeep Bajwa4
Amandeep Bajwa4 Ryan T. Demmer5,6
Ryan T. Demmer5,6 Susmita Datta7
Susmita Datta7 Csaba P. Kovesdy1,8
Csaba P. Kovesdy1,8 Joseph F. Pierre3
Joseph F. Pierre3The rapid expansion of microbiota research has significantly advanced our understanding of the complex interactions between gut microbiota and cardiovascular, metabolic, and renal system regulation. Low-grade chronic inflammation has long been implicated as one of the key mechanisms underlying cardiometabolic disease risk and progression, even before the insights provided by gut microbiota research in the past decade. Microbial translocation into the bloodstream can occur via different routes, including through the oral and/or intestinal mucosa, and may contribute to chronic inflammation in cardiometabolic disease. Among several gut-derived products identifiable in the systemic circulation, bacterial endotoxins and metabolites have been extensively studied, however recent advances in microbial DNA sequencing have further allowed us to identify highly diverse communities of microorganisms in the bloodstream from an -omics standpoint, which is termed “circulating microbiota.” While detecting microorganisms in the bloodstream was historically considered as an indication of infection, evidence on the circulating microbiota is continually accumulating in various patient populations without clinical signs of infection and even in otherwise healthy individuals. Moreover, both quantitative and compositional alterations of the circulating microbiota have recently been implicated in the pathogenesis of chronic inflammatory conditions, potentially through their immunostimulatory, atherogenic, and cardiotoxic properties. In this mini review, we aim to provide recent evidence on the characteristics and roles of circulating microbiota in several cardiometabolic diseases, such as type 2 diabetes, cardiovascular disease, and chronic kidney disease, with highlights of our emerging findings on circulating microbiota in patients with end-stage kidney disease undergoing hemodialysis.
Cardiometabolic disease, including type 2 diabetes, chronic kidney disease (CKD), and cardiovascular disease is a significant global healthcare problem with growing prevalence and substantial social and economic burden (Aron-Wisnewsky and Clément, 2016; Ralston and Nugent, 2019). The number of people with cardiovascular disease, type 2 diabetes, and CKD worldwide is reported to be 523 million (Roth et al., 2020), 422 million (Collaboration, 2016), and 847 million (Jager et al., 2019), respectively. In 2010, the global financial burden of cardiometabolic disease was estimated to be US$6.3 trillion, which is projected to double by 2030 (Arena et al., 2015).
Cardiometabolic disease stems from various factors, including genetic, behavioral and environmental factors (Ralston and Nugent, 2019). Low-grade chronic inflammation represents a key pathophysiological mechanism shared in common between the various cardiometabolic disease entities (Donath et al., 2019). A variety of mechanisms have been suggested to contribute to the perpetuation of inflammatory responses in cardiometabolic disease, including release of adipokines from obese visceral adipose tissue, renin-angiotensin-aldosterone system (RAAS) activation, cellular senescence, and accumulation of toxic metabolites (Carrero and Stenvinkel, 2010; Oishi and Manabe, 2020; Sumida et al., 2020). Over the past few decades, substantial efforts have been made to alleviate the chronic inflammation in cardiometabolic disease mainly by targeting these etiological factors, which have not been very successful; and the considerable disease burden resulting from chronic inflammation remains to be resolved. Therefore, an urgent need exists to identify novel modifiable risk factors that could help develop effective therapeutic approaches for premature morbidity and mortality in patients with cardiometabolic disease.
With recent advances in ‘-omics’ technologies, bioinformatics, and modelling approaches, a growing body of evidence suggests that microbial communities (i.e., microbiota) along the digestive tract may contribute to chronic low-grade inflammation in cardiometabolic disease (Aron-Wisnewsky and Clément, 2016; Warmbrunn et al., 2020), which in turn suggests that the microbiota could serve as a novel therapeutic target against cardiometabolic disease (Fernández-Ruiz, 2021; Sumida et al., 2021a). In a recent prospective study examining the long-term effects of a Mediterranean-style diet on the gut microbiome composition and on cardiometabolic disease risk (i.e., glucose homeostasis, lipid metabolism and inflammation), a healthy Mediterranean-style dietary pattern modified the risk of cardiometabolic disease in part through alterations of the gut microbiota (Wang et al., 2021). Given the enormous microbial load (i.e., >100 trillion individual microorganisms) in the human gastrointestinal tract and their substantial modulation of most metabolic activities (Whitman et al., 1998), it is not surprising that many diseases, including cardiometabolic disease, are related to altered gut microbiota (a.k.a. gut dysbiosis) and resultant changes in gut-derived metabolites. In contrast, the contributions of extraintestinal microbial communities circulating in the blood, which is also known as “circulating microbiota”, have been scarcely documented, let alone explored for their potential pathophysiological role in cardiometabolic disease. However, evidence that supports the roles of circulating microbiota in the onset and progression of cardiometabolic disease is steadily accumulating and receiving increasing attention. In this mini review, we summarize the current understanding of the circulating microbiota in shaping the development and progression of cardiometabolic disease with clinical and research implications from this rapidly evolving field, and highlight some of the emerging findings on the association of circulating microbiota with risk of cardiovascular disease in patients with end-stage kidney disease (ESKD) on hemodialysis.
While the colonization of microbes at specific body sites that are exposed to the external environment (e.g., the oral cavity and the gut) is both well-recognized and widely accepted (Markova, 2017), the concept of presence of microbial communities in an otherwise “sterile” milieu, such as the bloodstream, is relatively new. Traditionally, the detection of microbes in the bloodstream carried out by culturing specific microbes is interpreted as an indication of infection. However, the concept of the existence of classically “sterile” milieu in the blood of healthy humans has been challenged by mounting evidence showing the existence of blood microbes in otherwise healthy individuals (McLaughlin et al., 2002; Damgaard et al., 2015; Paisse et al., 2016). Following the seminal study by Nikkari et al. in 2001 that reported the detection of bacterial DNA in blood specimens from healthy individuals (Nikkari et al., 2001), several studies have reported the presence of blood microbes among both healthy blood donors (McLaughlin et al., 2002; Damgaard et al., 2015; Paisse et al., 2016) and various patient populations without overt infections (Rajendhran et al., 2013; Sato et al., 2014; Lelouvier et al., 2016), primarily by amplification and sequencing of the bacterial 16S ribosomal RNA (rRNA) gene. The application of archaeal 16S rRNA and fungal Internal Transcribed Spacer (ITS) rRNA sequencing and whole-genome shotgun sequencing techniques have also demonstrated the presence of archaea, fungi, and viruses in blood of healthy individuals (Dinakaran et al., 2014; Panaiotov et al., 2018; Castillo et al., 2019). It is important to note that the detection of microbial signatures in these studies is based largely on microbial DNA signatures and not viable bacteria directly, and hence do not necessarily challenge existing dogma, but rather provide deeper insights into the concept of sterility and homeostasis in the cardiovascular system.
The source of a circulating microbiota remains a topic of considerable deliberation, and it is still controversial whether the circulating microbiota is allochthonous or autochthonous (Castillo et al., 2019). Currently, the presence of circulating microbiota is thought to be largely attributable to microbial translocation from other body sites rich in microbiota (Velmurugan et al., 2020). The initial entry of microorganisms into the systemic circulation has been thought to occur prenatally by vertical transmission of maternal microbiotas in the umbilical cord, placenta, and/or amniotic fluid, although the conventional dogma suggests that the placental barrier prevents such a microbial translocation into infants during pregnancy (Funkhouser and Bordenstein, 2013). The other possible major sources of circulating microbiota include gastrointestinal tract, mouth, and skin (Whittle et al., 2018). Aside from the microbial translocation as a result of injuries, infections, and non-surgical procedures (e.g., intravenous injections and catheter placement), microbial translocation from these body sites is most likely to occur through loosening of epithelial barriers due to their structural and functional alterations. For example, microbial translocation from the oral cavities into the systemic circulation has been reported in oral diseases, such as gingivitis and periodontitis (Emery et al., 2021), and even after chewing, tooth brushing and dental flossing, all of which can lead to the oral mucosal barrier disruption (Forner et al., 2006; Lockhart et al., 2008). The identification of known members of the oral microbiota in the coronary artery tissues from patients with sudden cardiac death may also support these findings (Lehtiniemi et al., 2005), and prior work has also demonstrated the recovery of viable bacteria from atheromatous plaques (Rafferty et al., 2011).
The impairment of intestinal barrier integrity is another important mechanism that allows microbial translocation from the gut into the systemic circulation. This so-called “leaky gut” phenomenon has been well-studied and recognized in certain populations, such as those with inflammatory bowel disease (Maloy and Powrie, 2011), portal hypertension (Lutz et al., 2015), and heart failure (Verbrugge et al., 2013; Yuzefpolskaya et al., 2020). In recent years, this phenomenon has also been recognized in a variety of chronic diseases and conditions, including diabetes mellitus, CKD, and cardiovascular disease, partly due to gut dysbiosis, lifestyle and dietary factors (e.g., high-fat diet and alcohol intake), medications (e.g., non-steroidal anti-inflammatory drugs), and intestinal ischemia and/or edema (de Kort et al., 2011; Kitai and Tang, 2018; Nagpal et al., 2018; Sumida and Kovesdy, 2019). Among possible gut-derived products that can be identified in the bloodstream, bacterial lipopolysaccharide (i.e., endotoxin) and metabolites (e.g., indoxyl sulfate and trimethylamine N-oxide) have been most extensively investigated for their unique atherogenic and immunostimulatory properties (Evenepoel et al., 2009; Wiesner et al., 2010; Tang et al., 2013; Bowman et al., 2017; Donath et al., 2019); however, the translocation of gut microbiota into the systemic circulation may also occur as a result of the increased gut permeability, as supported by a few studies demonstrating a similarity in the microbial signatures between the fecal and blood samples of patients with cardiometabolic disease (Sato et al., 2014; Lelouvier et al., 2016). Importantly, both quantitative and compositional changes in the microbiota translocated into the systemic circulation have been implicated in the pathogenesis of cardiometabolic disease.
Bacterial DNA fragments can be detected by amplifying the highly conserved 16S rRNA subunit or through whole genome sequencing approaches (Jiang et al., 2009). Among various microbial components, bacterial DNA fragments are readily detectable and are easily discerned from human DNA (Szeto et al., 2018), and have thus been suggested as a better quantitative marker of the bacterial load circulating in the blood compared to measuring bacterial lipopolysaccharide (i.e., endotoxin) which is limited to detect only Gram-negative bacteria (Wiedermann et al., 1999). Furthermore, circulating bacterial DNA has been suggested to have unique pathogenic roles through recognition as pathogen-associated molecular pattern ligands (PAMPs) in host immune and cardiovascular systems.
Bacterial DNA contains unmethylated cytosine–guanine dinucleotide (CpG) flanked by two purine 5’ and two pyrimidine 3’ (Klinman et al., 1997). These DNA structures are recognized by toll-like receptors (TLRs), specifically endogenous TLR-9 (as a bacterial DNA receptor). This in turn triggers a cell signaling pathway including activation of the nuclear factor kappa B and the mitogen-activated protein kinases (Krieg, 2002). In inflammatory immune cells like polymorphonuclear neutrophils, bacterial DNA products exert profound effects on chemokine expression, regulation of adhesion molecules, cellular trafficking, and phagocyte activity. They promote the survival of mononuclear cells by inducing interleukin-6 (IL-6) (Schindler et al., 2004; Navarro et al., 2007) and rescues polymorphonuclear neutrophils from constitutive apoptosis (El Kebir et al., 2008). The activation of host immune system induced by bacterial DNA can in turn induce a plethora of other compounding pathological pathways that include metabolic dysfunction (e.g., through insulin resistance) and endothelial injury (e.g., through induction of endothelial cell apoptosis) (Merino et al., 2008; Donath et al., 2019), eventually contributing to the pathogenesis of cardiometabolic disease. In fact, elevated levels of circulating bacterial DNA have been reported in individuals with cardiometabolic disease who had no clinical evidence of systemic infection (Amar et al., 2011; Kwan et al., 2013; Dinakaran et al., 2014; Szeto et al., 2018).
In addition to its proinflammatory and atherogenic properties, it has been shown that bacterial DNA induces suppression of cardiac myocyte contraction in a dose-dependent manner (Paladugu et al., 2004). In line with this observation, higher plasma bacterial DNA levels have been demonstrated to be independently associated with subsequent risk of composite cardiovascular events (comprised mostly of hospitalization for heart failure) in peritoneal dialysis patients without overt infections (Szeto et al., 2015). Of interest, compared with bacterial endotoxin levels, the association with cardiovascular events was more pronounced for bacterial DNA levels (Szeto et al., 2015), suggesting that circulating bacterial DNA could play a more dominant role in the development and progression of cardiovascular disease.
Compositional changes in the microbiota are usually represented by alpha (α) diversity (e.g., assessed by Shannon, Simpson, Chao1 and Richness indices) and relative abundance of microbial communities. While the advent of sequencing technology has greatly advanced our understanding of the roles of compositional alterations of the gut microbiota in various diseases (Ley et al., 2006; Schwabe and Jobin, 2013; Ramezani and Raj, 2014; Wang and Kasper, 2014; Autenrieth, 2017), studies reporting the compositional changes in circulating microbiota and their association with cardiometabolic disease are still extremely limited (Table 1).
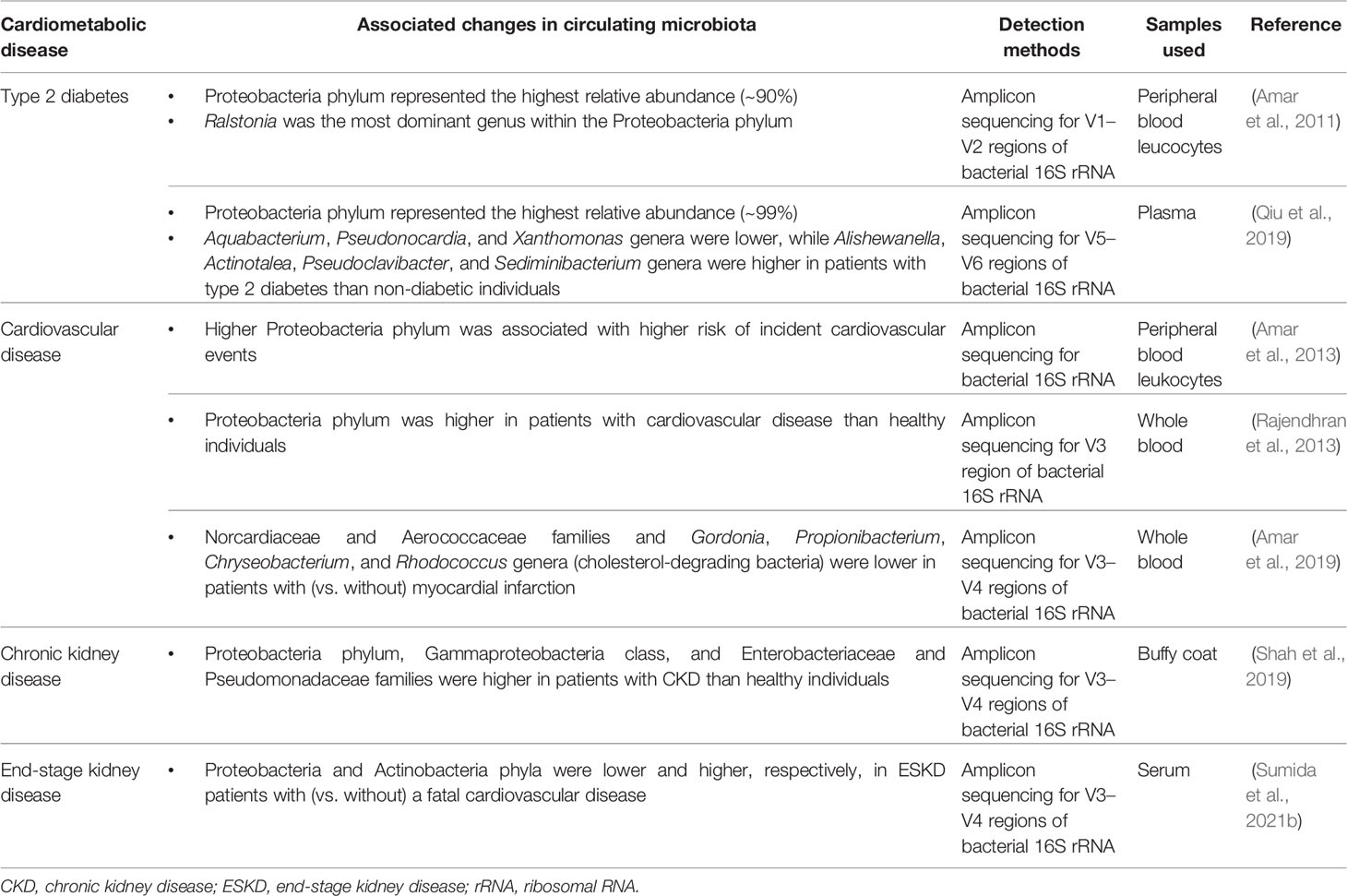
Table 1 Reported compositional changes in circulating microbiota associated with cardiometabolic disease.
Reports on the circulating microbiota in diabetes relate exclusively to type 2 diabetes, and to our knowledge no correlation studies have been reported in type 1 or gestational diabetes. In a case-control study nested from a longitudinal cohort study examining the association of blood 16S rDNA concentrations with incident diabetes and obesity in non-diabetic and non-obese adults, the pyrosequencing analysis demonstrated that Proteobacteria phylum represented the highest relative abundance (~90%) in the blood of individuals who developed diabetes, with Ralstonia being the most dominant genus within the Proteobacteria phylum (Amar et al., 2011). In a recent nested case-control study of 15 cases with incident type 2 diabetes and 100 matched controls without type 2 diabetes, the researchers investigated the composition of the circulating microbiota using bacterial 16S rRNA sequencing and found that both cases and controls had diverse bacterial communities in their pre-diagnostic blood samples, dominated largely by Proteobacteria phylum (~99%) (Qiu et al., 2019). Although there was no difference in α diversity (assessed by Simpson, Shannon, and Chao1 indices) between the two groups, the relative abundance of Aquabacterium, Pseudonocardia, and Xanthomonas genera was significantly lower among diabetic cases compared with non-diabetic controls, while that of Alishewanella, Actinotalea, Pseudoclavibacter, and Sediminibacterium genera was significantly higher among cases than controls. Of interest, the study also showed that individuals carrying the Bacteroides and Sediminibacterium genera in the blood at baseline had significantly lower and higher risk of incident type 2 diabetes, respectively, independent of potential confounders (Qiu et al., 2019), although precise mechanisms underlying these associations remain to be determined.
The profiles of blood microbes in the context of cardiovascular disease were traditionally restricted to infection-related cardiac complications, such as rheumatic heart disease and infectious peri-, myo- and endocarditis, diagnosed primarily by classical serological and/or culture-based methods (Fong, 2009). However, recent evidence regarding the association between circulating microbiota and cardiovascular disease has questioned this paradigm that links the presence of blood microbes to infectious cardiac complications.
In a pioneering population-based study examining the longitudinal association of circulating microbiota identified using the 16S rRNA sequencing with cardiovascular events in 3,936 adults without clinical evidence of infection, there was a significant association between relative abundance of Proteobacteria phylum (identified in peripheral blood leukocytes) and risk of incident cardiovascular events, independently of traditional cardiovascular risk factors (Amar et al., 2013). Subsequently, a cross-sectional study reported similar findings that, compared with apparently healthy individuals, patients with cardiovascular disease had a significantly higher relative abundance of Proteobacteria phylum in their whole blood (Rajendhran et al., 2013). Although these observational studies cannot conclude a causal relationship, there are several plausible explanations for the association between circulating microbiota and cardiovascular disease. In addition to the aforementioned immunostimulatory, atherogenic, and cardiotoxic properties of bacterial DNA fragments, the phylum Proteobacteria, which was found to be predominant in the blood of patients with cardiovascular disease in previous studies, has unique proinflammatory and proatherosclerotic properties (Rizzatti et al., 2017) and may thus, as a culprit pathogen, contribute to the development and progression of cardiovascular disease. Additionally, a decrease in the relative abundance of cholesterol-degrading bacteria, such as Norcardiaceae and Aerococcaceae families and Gordonia, Propionibacterium, Chryseobacterium, and Rhodococcus genera, has been implicated in the pathogenesis of atherosclerotic plaques, leading to the risk of ischemic heart disease (Amar et al., 2019).
Patients with CKD, particularly in those with ESKD undergoing dialysis therapy, are susceptible to infection due in part to altered immune responses, multiple comorbidities, use of immunosuppressants, and use of vascular access (Ishigami and Matsushita, 2019), and hence a number of studies have reported the existence of bacterial DNA in the blood and its association with inflammatory response in these patients (Navarro et al., 2007; Bossola et al., 2009; Kwan et al., 2013; Shi et al., 2014). Nevertheless, there is a paucity of data on the characteristics and roles of the circulating microbiota in patients with CKD and ESKD. In a recent pilot study comparing the composition of circulating microbiota between non-diabetic CKD patients without kidney replacement therapy and healthy individuals, patients with CKD (vs. healthy individuals) displayed a significant reduction in α diversity (Chao1 index) and had a significantly higher relative abundance of Enterobacteriaceae and Pseudomonadaceae families, Gammaproteobacteria class, and Proteobacteria phylum in their buffy coat samples (Shah et al., 2019).
More recently, using serum samples of 34 hemodialysis patients enrolled in a pilot case-control study (17 cases with a fatal cardiovascular event and 17 matched controls without such an event during a median follow-up of 2.0 years), the researchers performed a comprehensive taxonomic profile of the circulating microbiota by 16S or ITS rRNA sequencing (for bacteria, archaea, and fungi) and compared the composition of circulating microbiota between cases and controls (Sumida et al., 2021b). They found that patients who died of a cardiovascular event (i.e., cases) had significantly less Proteobacteria and greater Actinobacteria phyla compared with those who remained alive (i.e., controls) (Figure 1), although the levels of 16S rRNA and bacterial α diversity were similar between groups (Sumida et al., 2021b). Furthermore, the proportion of Proteobacteria and Actinobacteria phyla were significantly correlated with blood levels of nuclear factor erythroid 2−related factor 2 (Nrf2), a master regulator of antioxidative responses, and were marginally associated with a greater risk of cardiovascular mortality, independently of age, sex, race, dialysis vintage, and type of vascular access (Sumida et al., 2021b). These data may suggest that circulating levels of Nrf2 play a key role in the risk of premature cardiovascular mortality associated with the circulating microbiota in patients with ESKD. Of note, this study utilized serum samples to assess the circulating microbiota, in spite of the perceived concern regarding the ability to detect microbial DNA in the cell-free fraction of blood. In most previous studies, the circulating microbiota was assessed using leukocyte-containing blood fractions (Navarro et al., 2007; Paisse et al., 2016; Shah et al., 2019), the nature of which may differ from that of the circulating microbiota identified in the cell-free blood fraction. More specifically, compared with circulating microbial signatures that can be identified predominantly in the buffy coat (rich in white blood cells and platelets) presumably as a result of the microbial entrapment by white blood cells, the circulating “cell-free” microbiota located in plasma or serum fraction may exert its potential pathophysiological effects on immune cells (through their receptors [e.g., TLR-9]) and on cardiac myocytes in a more direct manner, which in turn suggests the potential of the circulating cell-free microbiota as a more clinically applicable, non-invasive diagnostic/prognostic biomarker compared with the circulating microbiota identified from other leukocyte-containing blood fractions (Sumida et al., 2021b). The observed lower proportion of Proteobacteria phylum among cardiovascular cases (vs. controls) in this study may appear contradictory to the findings of other studies. Possible explanations include differences in blood fractions used to assess the circulating microbial composition, differences in the studied patient populations, with hemodialysis patients suffering from higher comorbidity burden, and/or differences in the relative abundance of other microbial taxa (e.g., high Staphylococcus genus which belongs to the phylum Bacillota) in cardiovascular cases in this study (Sumida et al., 2021b). Uncovering the cause(s) for these differences deserves further investigation. Nonetheless, a prior study that reported lower proportion of Proteobacteria phylum in subgingival plaque to be associated with higher systemic inflammation may support the finding from hemodialysis patients, suggesting a potential influence of oral microbiota on the circulating microbiota in these patients (Demmer et al., 2017).
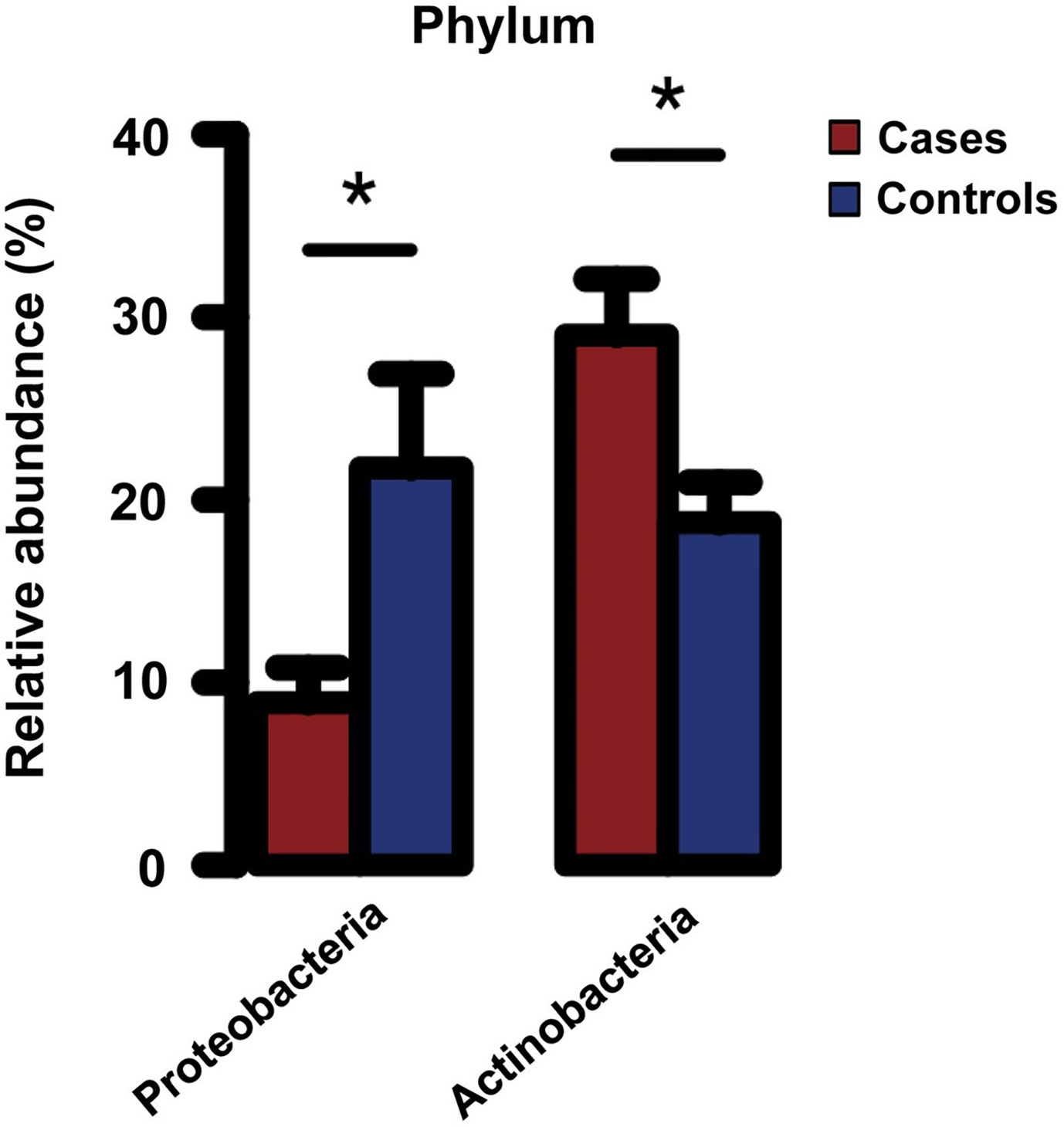
Figure 1 Relative abundance of Proteobacteria and Actinobacteria phyla in serum of hemodialysis patients with (cases) and without (controls) a fatal cardiovascular event. *P < 0.05. Error bars indicate the standard error of the mean (SEM). Reprinted with permission from Sumida et al. (2021b).
It has now become evident that highly diverse microbial communities exist in the systemic circulation of various populations and that both quantitative and compositional changes in the circulating microbiota may contribute to the development and progression of cardiometabolic disease. However, many important questions remain unanswered regarding the nature of circulating microbiota, including the sources, pathophysiological roles, localization across different blood fractions, and distinction from potential contamination, all of which need to be clarified in future in-depth basic, clinical and population-based research. In an ongoing quest to improve outcomes of patients with cardiometabolic disease, perhaps the time has come to go beyond the “gut feeling” and rigorously incorporate the potential pathophysiological insights gained from the circulating microbiota towards the development of novel biomarkers for diagnosis and prognosis, especially in personalized therapeutic approaches to premature morbidity and mortality in cardiometabolic disease where patterns are emerging.
KS and JP: conception and study design and drafting the manuscript. ZH, C-YC, TM, AB, RD, SD, and CK: revising the manuscript critically for important intellectual content, and approval of the version of the manuscript to be published. All authors contributed to the article and approved the submitted version.
This work was supported by the National Institute of Diabetes and Digestive and Kidney Disease (NIDDK) of the National Institutes of Health (NIH) under award number R01DK125586 to KS. JP is funded by NIH under award numbers R01CA253329 and R21AI163503. AB is funded by NIH under award number of R01DK117183. RD is funded by NIDDK under award number R01DK102932.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
CK is an employee of the US Department of Veterans Affairs. The interpretation and reporting of these data are the responsibility of the authors and in no way should be seen as official policy or interpretation of the Department of Veterans Affairs or the US government. The results of this paper have not been published previously in whole or part.
Amar, J., Lange, C., Payros, G., Garret, C., Chabo, C., Lantieri, O., et al. (2013). Blood Microbiota Dysbiosis Is Associated With the Onset of Cardiovascular Events in a Large General Population: The D.E.s.I.R. Study. PloS One 8 (1), e54461. doi: 10.1371/journal.pone.0054461
Amar, J., Lelouvier, B., Servant, F., Lluch, J., Burcelin, R., Bongard, V., et al. (2019). Blood Microbiota Modification After Myocardial Infarction Depends Upon Low-Density Lipoprotein Cholesterol Levels. J. Am. Heart Assoc. 8 (19), e011797. doi: 10.1161/JAHA.118.011797
Amar, J., Serino, M., Lange, C., Chabo, C., Iacovoni, J., Mondot, S., et al. (2011). Involvement of Tissue Bacteria in the Onset of Diabetes in Humans: Evidence for a Concept. Diabetologia 54 (12), 3055–3061. doi: 10.1007/s00125-011-2329-8
Arena, R., Guazzi, M., Lianov, L., Whitsel, L., Berra, K., Lavie, C. J., et al. (2015). Healthy Lifestyle Interventions to Combat Noncommunicable Disease-A Novel Nonhierarchical Connectivity Model for Key Stakeholders: A Policy Statement From the American Heart Association, European Society of Cardiology, European Association for Cardiovascular Prevention and Rehabilitation, and American College of Preventive Medicine. Eur. Heart J. 36 (31), 2097–2109. doi: 10.1093/eurheartj/ehv207
Aron-Wisnewsky, J., Clément, K. (2016). The Gut Microbiome, Diet, and Links to Cardiometabolic and Chronic Disorders. Nat. Rev. Nephrol. 12 (3), 169–181. doi: 10.1038/nrneph.2015.191
Autenrieth, I. B. (2017). The Microbiome in Health and Disease: A New Role of Microbes in Molecular Medicine. J. Mol. Med. (Berl) 95 (1), 1–3. doi: 10.1007/s00109-016-1499-8
Bossola, M., Sanguinetti, M., Scribano, D., Zuppi, C., Giungi, S., Luciani, G., et al. (2009). Circulating Bacterial-Derived DNA Fragments and Markers of Inflammation in Chronic Hemodialysis Patients. Clin. J. Am. Soc. Nephrol. 4 (2), 379–385. doi: 10.2215/CJN.03490708
Bowman, J. D., Surani, S., Horseman, M. A. (2017). Endotoxin, Toll-Like Receptor-4, and Atherosclerotic Heart Disease. Curr. Cardiol. Rev. 13 (2), 86–93. doi: 10.2174/1573403X12666160901145313
Carrero, J. J., Stenvinkel, P. (2010). Inflammation in End-Stage Renal Disease–What Have We Learned in 10 Years? Semin. Dial 23 (5), 498–509. doi: 10.1111/j.1525-139X.2010.00784.x
Castillo, D. J., Rifkin, R. F., Cowan, D. A., Potgieter, M. (2019). The Healthy Human Blood Microbiome: Fact or Fiction? Front. Cell Infect. Microbiol. 9. doi: 10.3389/fcimb.2019.00148
Collaboration, N.C.D.R.F. (2016). Worldwide Trends in Diabetes Since 1980: A Pooled Analysis of 751 Population-Based Studies With 4.4 Million Participants. Lancet 387 (10027), 1513–1530. doi: 10.1016/S0140-6736(16)00618-8
Damgaard, C., Magnussen, K., Enevold, C., Nilsson, M., Tolker-Nielsen, T., Holmstrup, P., et al. (2015). Viable Bacteria Associated With Red Blood Cells and Plasma in Freshly Drawn Blood Donations. PloS One 10 (3), e0120826. doi: 10.1371/journal.pone.0120826
de Kort, S., Keszthelyi, D., Masclee, A. A. (2011). Leaky Gut and Diabetes Mellitus: What Is the Link? Obes. Rev. 12 (6), 449–458. doi: 10.1111/j.1467-789X.2010.00845.x
Demmer, R. T., Breskin, A., Rosenbaum, M., Zuk, A., LeDuc, C., Leibel, R., et al. (2017). The Subgingival Microbiome, Systemic Inflammation and Insulin Resistance: The Oral Infections, Glucose Intolerance and Insulin Resistance Study. J. Clin. Periodontol. 44 (3), 255–265. doi: 10.1111/jcpe.12664
Dinakaran, V., Rathinavel, A., Pushpanathan, M., Sivakumar, R., Gunasekaran, P., Rajendhran, J. (2014). Elevated Levels of Circulating DNA in Cardiovascular Disease Patients: Metagenomic Profiling of Microbiome in the Circulation. PloS One 9 (8), e105221. doi: 10.1371/journal.pone.0105221
Donath, M. Y., Meier, D. T., Böni-Schnetzler, M. (2019). Inflammation in the Pathophysiology and Therapy of Cardiometabolic Disease. Endocrine Rev. 40 (4), 1080–1091. doi: 10.1210/er.2019-00002
El Kebir, D., Jozsef, L., Filep, J. G. (2008). Neutrophil Recognition of Bacterial DNA and Toll-Like Receptor 9-Dependent and -Independent Regulation of Neutrophil Function. Arch. Immunol. Ther. Exp. (Warsz) 56 (1), 41–53. doi: 10.1007/s00005-008-0008-3
Emery, D. C., Cerajewska, T. L., Seong, J., Davies, M., Paterson, A., Allen-Birt, S. J., et al. (2021). Comparison of Blood Bacterial Communities in Periodontal Health and Periodontal Disease. Front. Cell. Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.577485
Evenepoel, P., Meijers, B. K., Bammens, B. R., Verbeke, K. (2009). Uremic Toxins Originating From Colonic Microbial Metabolism. Kidney Int. Suppl. 114), S12–S19. doi: 10.1038/ki.2009.402
Fernández-Ruiz, I. (2021). Modulating the Gut Microbiota With Dietary Interventions to Protect Against Cardiometabolic Disease. Nat. Rev. Cardiol. 18 (5), 305–305. doi: 10.1038/s41569-021-00537-0
Fong, I. W. (2009). New Perspectives of Infections in Cardiovascular Disease. Curr. Cardiol. Rev. 5 (2), 87–104. doi: 10.2174/157340309788166679
Forner, L., Larsen, T., Kilian, M., Holmstrup, P. (2006). Incidence of Bacteremia After Chewing, Tooth Brushing and Scaling in Individuals With Periodontal Inflammation. J. Clin. Periodontol. 33 (6), 401–407. doi: 10.1111/j.1600-051X.2006.00924.x
Funkhouser, L. J., Bordenstein, S. R. (2013). Mom Knows Best: The Universality of Maternal Microbial Transmission. PloS Biol. 11 (8), e1001631. doi: 10.1371/journal.pbio.1001631
Ishigami, J., Matsushita, K. (2019). Clinical Epidemiology of Infectious Disease Among Patients With Chronic Kidney Disease. Clin. Exp. Nephrol. 23 (4), 437–447. doi: 10.1007/s10157-018-1641-8
Jager, K. J., Kovesdy, C., Langham, R., Rosenberg, M., Jha, V., Zoccali, C. (2019). A Single Number for Advocacy and Communication-Worldwide More Than 850 Million Individuals Have Kidney Diseases. Kidney Int. 96 (5), 1048–1050. doi: 10.1016/j.kint.2019.07.012
Jiang, W., Lederman, M. M., Hunt, P., Sieg, S. F., Haley, K., Rodriguez, B., et al. (2009). Plasma Levels of Bacterial DNA Correlate With Immune Activation and the Magnitude of Immune Restoration in Persons With Antiretroviral-Treated HIV Infection. J. Infect. Dis. 199 (8), 1177–1185. doi: 10.1086/597476
Kitai, T., Tang, W. H. W. (2018). Gut Microbiota in Cardiovascular Disease and Heart Failure. Clin. Sci. (Lond) 132 (1), 85–91. doi: 10.1042/CS20171090
Klinman, D. M., Yamshchikov, G., Ishigatsubo, Y. (1997). Contribution of Cpg Motifs to the Immunogenicity of DNA Vaccines. J. Immunol. 158 (8), 3635–3639.
Krieg, A. M. (2002). Cpg Motifs in Bacterial DNA and Their Immune Effects. Annu. Rev. Immunol. 20, 709–760. doi: 10.1146/annurev.immunol.20.100301.064842
Kwan, B. C., Chow, K. M., Leung, C. B., Law, M. C., Cheng, P. M., Yu, V., et al. (2013). Circulating Bacterial-Derived DNA Fragments as a Marker of Systemic Inflammation in Peritoneal Dialysis. Nephrol. Dial Transplant. 28 (8), 2139–2145. doi: 10.1093/ndt/gft100
Lehtiniemi, J., Karhunen, P. J., Goebeler, S., Nikkari, S., Nikkari, S. T. (2005). Identification of Different Bacterial Dnas in Human Coronary Arteries. Eur. J. Clin. Invest. 35 (1), 13–16. doi: 10.1111/j.1365-2362.2005.01440.x
Lelouvier, B., Servant, F., Paisse, S., Brunet, A. C., Benyahya, S., Serino, M., et al. (2016). Changes in Blood Microbiota Profiles Associated With Liver Fibrosis in Obese Patients: A Pilot Analysis. Hepatology 64 (6), 2015–2027. doi: 10.1002/hep.28829
Ley, R. E., Turnbaugh, P. J., Klein, S., Gordon, J. I. (2006). Microbial Ecology: Human Gut Microbes Associated With Obesity. Nature 444 (7122), 1022–1023. doi: 10.1038/4441022a
Lockhart, P. B., Brennan, M. T., Sasser, H. C., Fox, P. C., Paster, B. J., Bahrani-Mougeot, F. K. (2008). Bacteremia Associated With Toothbrushing and Dental Extraction. Circulation 117 (24), 3118–3125. doi: 10.1161/CIRCULATIONAHA.107.758524
Lutz, P., Nischalke, H. D., Strassburg, C. P., Spengler, U. (2015). Spontaneous Bacterial Peritonitis: The Clinical Challenge of a Leaky Gut and a Cirrhotic Liver. World J. Hepatol. 7 (3), 304–314. doi: 10.4254/wjh.v7.i3.304
Maloy, K. J., Powrie, F. (2011). Intestinal Homeostasis and Its Breakdown in Inflammatory Bowel Disease. Nature 474 (7351), 298–306. doi: 10.1038/nature10208
Markova, N. D. (2017). L-Form Bacteria Cohabitants in Human Blood: Significance for Health and Diseases. Discov. Med. 23 (128), 305–313.
McLaughlin, R. W., Vali, H., Lau, P. C., Palfree, R. G., De Ciccio, A., Sirois, M., et al. (2002). Are There Naturally Occurring Pleomorphic Bacteria in the Blood of Healthy Humans? J. Clin. Microbiol. 40 (12), 4771–4775. doi: 10.1128/jcm.40.12.4771-4775.2002
Merino, A., Nogueras, S., Garcia-Maceira, T., Rodriguez, M., Martin-Malo, A., Ramirez, R., et al. (2008). Bacterial DNA and Endothelial Damage in Haemodialysis Patients. Nephrol. Dial Transplant. 23 (11), 3635–3642. doi: 10.1093/ndt/gfn308
Nagpal, R., Mainali, R., Ahmadi, S., Wang, S., Singh, R., Kavanagh, K., et al. (2018). Gut Microbiome and Aging: Physiological and Mechanistic Insights. Nutr. Healthy Aging 4 (4), 267–285. doi: 10.3233/NHA-170030
Navarro, M. D., Carracedo, J., Ramirez, R., Madueno, J. A., Merino, A., Rodriguez, M., et al. (2007). Bacterial DNA Prolongs the Survival of Inflamed Mononuclear Cells in Haemodialysis Patients. Nephrol. Dial Transplant. 22 (12), 3580–3585. doi: 10.1093/ndt/gfm414
Nikkari, S., McLaughlin, I. J., Bi, W., Dodge, D. E., Relman, D. A. (2001). Does Blood of Healthy Subjects Contain Bacterial Ribosomal DNA? J. Clin. Microbiol. 39 (5), 1956–1959. doi: 10.1128/JCM.39.5.1956-1959.2001
Oishi, Y., Manabe, I. (2020). Organ System Crosstalk in Cardiometabolic Disease in the Age of Multimorbidity. Front. Cardiovasc. Med. 7. doi: 10.3389/fcvm.2020.00064
Paisse, S., Valle, C., Servant, F., Courtney, M., Burcelin, R., Amar, J., et al. (2016). Comprehensive Description of Blood Microbiome From Healthy Donors Assessed by 16S Targeted Metagenomic Sequencing. Transfusion 56 (5), 1138–1147. doi: 10.1111/trf.13477
Paladugu, B., Kumar, A., Parrillo, J. E., Der, S., Osman, J., Mensing, J., et al. (2004). Bacterial DNA and RNA Induce Rat Cardiac Myocyte Contraction Depression In Vitro. Shock 21 (4), 364–369. doi: 10.1097/00024382-200404000-00012
Panaiotov, S., Filevski, G., Equestre, M., Nikolova, E., Kalfin, R. (2018). Cultural Isolation and Characteristics of the Blood Microbiome of Healthy Individuals. Adv. Microbiol. 8, 406–421. doi: 10.4236/aim.2018.85027
Qiu, J., Zhou, H., Jing, Y., Dong, C. (2019). Association Between Blood Microbiome and Type 2 Diabetes Mellitus: A Nested Case-Control Study. J. Clin. Lab. Anal. 33 (4), e22842. doi: 10.1002/jcla.22842
Rafferty, B., Jonsson, D., Kalachikov, S., Demmer, R. T., Nowygrod, R., Elkind, M. S., et al. (2011). Impact of Monocytic Cells on Recovery of Uncultivable Bacteria From Atherosclerotic Lesions. J. Intern. Med. 270 (3), 273–280. doi: 10.1111/j.1365-2796.2011.02373.x
Rajendhran, J., Shankar, M., Dinakaran, V., Rathinavel, A., Gunasekaran, P. (2013). Contrasting Circulating Microbiome in Cardiovascular Disease Patients and Healthy Individuals. Int. J. Cardiol. 168 (5), 5118–5120. doi: 10.1016/j.ijcard.2013.07.232
Ralston, J., Nugent, R. (2019). Toward a Broader Response to Cardiometabolic Disease. Nat. Med. 25 (11), 1644–1646. doi: 10.1038/s41591-019-0642-9
Ramezani, A., Raj, D. S. (2014). The Gut Microbiome, Kidney Disease, and Targeted Interventions. J. Am. Soc. Nephrol. 25 (4), 657–670. doi: 10.1681/ASN.2013080905
Rizzatti, G., Lopetuso, L. R., Gibiino, G., Binda, C., Gasbarrini, A. (2017). Proteobacteria: A Common Factor in Human Diseases. BioMed. Res. Int. 2017, 9351507. doi: 10.1155/2017/9351507
Roth, G. A., Mensah, G. A., Johnson, C. O., Addolorato, G., Ammirati, E., Baddour, L. M., et al. (2020). Global Burden of Cardiovascular Diseases and Risk Factor-2019: Update From the GBD 2019 Study. J. Am. Coll. Cardiol. 76 (25), 2982–3021. doi: 10.1016/j.jacc.2020.11.010
Sato, J., Kanazawa, A., Ikeda, F., Yoshihara, T., Goto, H., Abe, H., et al. (2014). Gut Dysbiosis and Detection of “Live Gut Bacteria” in Blood of Japanese Patients With Type 2 Diabetes. Diabetes Care 37 (8), 2343–2350. doi: 10.2337/dc13-2817
Schindler, R., Beck, W., Deppisch, R., Aussieker, M., Wilde, A., Gohl, H., et al. (2004). Short Bacterial DNA Fragments: Detection in Dialysate and Induction of Cytokines. J. Am. Soc. Nephrol. 15 (12), 3207–3214. doi: 10.1097/01.ASN.0000145049.94888.26
Schwabe, R. F., Jobin, C. (2013). The Microbiome and Cancer. Nat. Rev. Cancer 13 (11), 800–812. doi: 10.1038/nrc3610
Shah, N. B., Allegretti, A. S., Nigwekar, S. U., Kalim, S., Zhao, S., Lelouvier, B., et al. (2019). Blood Microbiome Profile in CKD: A Pilot Study. Clin. J. Am. Soc. Nephrol. 14 (5), 692–701. doi: 10.2215/CJN.12161018
Shi, K., Wang, F., Jiang, H., Liu, H., Wei, M., Wang, Z., et al. (2014). Gut Bacterial Translocation may Aggravate Microinflammation in Hemodialysis Patients. Dig Dis. Sci. 59 (9), 2109–2117. doi: 10.1007/s10620-014-3202-7
Sumida, K., Han, Z., Dashputre, A. A., Potukuchi, P. K., Kovesdy, C. P. (2020). Association Between Nrf2 and CDKN2A Expression in Patients With End-Stage Renal Disease: A Pilot Study. Aging (Albany NY) 12 (16), 16357–16367. doi: 10.18632/aging.103685
Sumida, K., Kovesdy, C. P. (2019). The Gut-Kidney-Heart Axis in Chronic Kidney Disease. Physiol. Int. 106 (3), 195–206. doi: 10.1556/2060.106.2019.19
Sumida, K., Lau, W. L., Kovesdy, C. P., Kalantar-Zadeh, K., Kalantar-Zadeh, K. (2021a). Microbiome Modulation as a Novel Therapeutic Approach in Chronic Kidney Disease. Curr. Opin. Nephrol. Hypertens. 30 (1), 75–84. doi: 10.1097/MNH.0000000000000661
Sumida, K., Pierre, J. F., Han, Z., Mims, T. S., Potukuchi, P. K., Yuzefpolskaya, M., et al. (2021b). Circulating Microbial Signatures and Cardiovascular Death in Patients With ESRD. Kidney Int. Rep. 6 (10), 2617–2628. doi: 10.1016/j.ekir.2021.07.023
Szeto, C. C., Kwan, B. C., Chow, K. M., Kwok, J. S., Lai, K. B., Cheng, P. M., et al. (2015). Circulating Bacterial-Derived DNA Fragment Level Is a Strong Predictor of Cardiovascular Disease in Peritoneal Dialysis Patients. PloS One 10 (5), e0125162. doi: 10.1371/journal.pone.0125162
Szeto, C. C., McIntyre, C. W., Li, P. K. (2018). Circulating Bacterial Fragments as Cardiovascular Risk Factors in CKD. J. Am. Soc. Nephrol. 29 (6), 1601–1608. doi: 10.1681/ASN.2018010068
Tang, W. H., Wang, Z., Levison, B. S., Koeth, R. A., Britt, E. B., Fu, X., et al. (2013). Intestinal Microbial Metabolism of Phosphatidylcholine and Cardiovascular Risk. N. Engl. J. Med. 368 (17), 1575–1584. doi: 10.1056/NEJMoa1109400
Velmurugan, G., Dinakaran, V., Rajendhran, J., Swaminathan, K. (2020). Blood Microbiota and Circulating Microbial Metabolites in Diabetes and Cardiovascular Disease. Trends Endocrinol. Metab. 31 (11), 835–847. doi: 10.1016/j.tem.2020.01.013
Verbrugge, F. H., Dupont, M., Steels, P., Grieten, L., Malbrain, M., Tang, W. H., et al. (2013). Abdominal Contributions to Cardiorenal Dysfunction in Congestive Heart Failure. J. Am. Coll. Cardiol. 62 (6), 485–495. doi: 10.1016/j.jacc.2013.04.070
Wang, Y., Kasper, L. H. (2014). The Role of Microbiome in Central Nervous System Disorders. Brain Behav. Immun. 38, 1–12. doi: 10.1016/j.bbi.2013.12.015
Wang, D. D., Nguyen, L. H., Li, Y., Yan, Y., Ma, W., Rinott, E., et al. (2021). The Gut Microbiome Modulates the Protective Association Between a Mediterranean Diet and Cardiometabolic Disease Risk. Nat. Med. 27 (2), 333–343. doi: 10.1038/s41591-020-01223-3
Warmbrunn, M. V., Herrema, H., Aron-Wisnewsky, J., Soeters, M. R., Van Raalte, D. H., Nieuwdorp, M. (2020). Gut Microbiota: A Promising Target Against Cardiometabolic Diseases. Expert Rev. Endocrinol. Metab. 15 (1), 13–27. doi: 10.1080/17446651.2020.1720511
Whitman, W. B., Coleman, D. C., Wiebe, W. J. (1998). Prokaryotes: The Unseen Majority. Proc. Natl. Acad. Sci. U.S.A. 95 (12), 6578–6583. doi: 10.1073/pnas.95.12.6578
Whittle, E., Leonard, M. O., Harrison, R., Gant, T. W., Tonge, D. P. (2018). Multi-Method Characterization of the Human Circulating Microbiome. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.03266
Wiedermann, C. J., Kiechl, S., Dunzendorfer, S., Schratzberger, P., Egger, G., Oberhollenzer, F., et al. (1999). Association of Endotoxemia With Carotid Atherosclerosis and Cardiovascular Disease: Prospective Results From the Bruneck Study. J. Am. Coll. Cardiol. 34 (7), 1975–1981. doi: 10.1016/S0735-1097(99)00448-9
Wiesner, P., Choi, S. H., Almazan, F., Benner, C., Huang, W., Diehl, C. J., et al. (2010). Low Doses of Lipopolysaccharide and Minimally Oxidized Low-Density Lipoprotein Cooperatively Activate Macrophages via Nuclear Factor Kappa B and Activator Protein-1: Possible Mechanism for Acceleration of Atherosclerosis by Subclinical Endotoxemia. Circ. Res. 107 (1), 56–65. doi: 10.1161/CIRCRESAHA.110.218420
Yuzefpolskaya, M., Bohn, B., Nasiri, M., Zuver, A. M., Onat, D. D., Royzman, E. A., et al. (2020). Gut Microbiota, Endotoxemia, Inflammation, and Oxidative Stress in Patients With Heart Failure, Left Ventricular Assist Device, and Transplant. J. Heart Lung Transplant. 39 (9), 880–890. doi: 10.1016/j.healun.2020.02.004
Keywords: cardiometabolic disease, cardiovascular disease, chronic kidney disease, circulating microbiota, diabetes mellitus, end-stage kidney disease, gut microbiota, inflammation
Citation: Sumida K, Han Z, Chiu C-Y, Mims TS, Bajwa A, Demmer RT, Datta S, Kovesdy CP and Pierre JF (2022) Circulating Microbiota in Cardiometabolic Disease. Front. Cell. Infect. Microbiol. 12:892232. doi: 10.3389/fcimb.2022.892232
Received: 08 March 2022; Accepted: 11 April 2022;
Published: 03 May 2022.
Edited by:
Yueqiong Ni, Leibniz Institute for Natural Product Research and Infection Biology, GermanyReviewed by:
Akihiro Yoshida, Matsumoto Dental University, JapanCopyright © 2022 Sumida, Han, Chiu, Mims, Bajwa, Demmer, Datta, Kovesdy and Pierre. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keiichi Sumida, a3N1bWlkYUB1dGhzYy5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.