- 1School of Medicine, Deakin University, Geelong, VIC, Australia
- 2Department of Medical Laboratory Technology, Southern Technical University, Basra, Iraq
- 3Institute for Mental and Physical Health and Clinical Translation, Deakin University, Geelong, VIC, Australia
BCL6A is a transcriptional repressor implicated in the development and survival of B and T lymphoctyes, which is also highly expressed in many non-Hodgkin’s lymphomas, such as diffuse large B cell lymphoma and follicular lymphoma. Roles in other cell types, including macrophages and non-hematopoietic cells, have also been suggested but require further investigation. This study sought to identify and characterize zebrafish BCL6A and investigate its role in immune cell development and function, with a focus on early macrophages. Bioinformatics analysis identified a homologue for BCL6A (bcl6aa), as well as an additional fish-specific duplicate (bcl6ab) and a homologue for the closely-related BCL6B (bcl6b). The human BCL6A and zebrafish Bcl6aa proteins were highly conserved across the constituent BTB/POZ, PEST and zinc finger domains. Expression of bcl6aa during early zebrafish embryogenesis was observed in the lateral plate mesoderm, a site of early myeloid cell development, with later expression seen in the brain, eye and thymus. Homozygous bcl6aa mutants developed normally until around 14 days post fertilization (dpf), after which their subsequent growth and maturation was severely impacted along with their relative survival, with heterozygous bcl6aa mutants showing an intermediate phenotype. Analysis of immune cell development revealed significantly decreased lymphoid and macrophage cells in both homozygous and heterozygous bcl6aa mutants, being exacerbated in homozygous mutants. In contrast, the number of neutrophils was unaffected. Only the homozygous bcl6aa mutants showed decreased macrophage mobility in response to wounding and reduced ability to contain bacterial infection. Collectively, this suggests strong conservation of BCL6A across evolution, including a role in macrophage biology.
Introduction
The B cell lymphoma 6A (BCL6A) protein consists of an evolutionarily conserved domain structure, comprising an N-terminal Broad-complex, Tramtrack and Brick-a-brac/Pox virus and Zinc finger family (BTB/POZ) domain, a central PEST domain and a C-terminal zinc finger domain comprising an array of six C2H2/Krüppel-type zinc fingers (Melnick et al., 2002; Ahmad et al., 2003; Ghetu et al., 2008). It acts as a strong transcriptional repressor, with the zinc finger domain facilitating binding to specific DNA sequences (Dent et al., 1997; Liu et al., 2016) and the BTB/POZ domain enabling recruitment of corepressors, such as SMRT, NCOR, BCOR, MTA3 and CTBP1 (Basso and Dalla-Favera, 2010). BCL6-related proteins are found across a broad range of species. This includes vertebrates, which have been shown to possess distinct but highly-related BCL6A and BCL6B proteins (Okabe et al., 1998), as well as invertebrates, typified by a BCL6-related protein identified in fruit-fly that is referred to as Ken & Barbie (Ken) (Arbouzova et al., 2006).
BCL6A plays a number of critical roles in B and T cell development and function (Wang et al., 2018; Yuan et al., 2022). Bcl6a knockout mice exhibited a failure in germinal centre formation in lymph node follicles (Dent et al., 1997; Phan and Dalla-Favera, 2004; Cattoretti et al., 2005) preventing somatic hypermutation and production of high-affinity antibodies (Basso and Dalla-Favera, 2010). This was in part a result of significantly decreased numbers of follicular T helper (Tfh) cells (Nurieva et al., 2009), a lineage in which BCL6A acts as a master regulator (Choi and Crotty, 2021), but also of impaired B cell commitment to the germinal centre B cell lineage (Huang et al., 2014) as well as their subsequent survival (Basso and Dalla-Favera, 2012). Bcl6a knockout mice also displayed reduced pre-B cell self-renewal and differentiation in the bone marrow (Duy et al., 2010), with B cell responses to cytokines affected (Basso and Dalla-Favera, 2012). Other T cell subsets were also variably affected, with T helper 2 (Th2) and Th17 cells dramatically increased (Mondal et al., 2010; Choi and Crotty, 2021) and memory T cells decreased (Ichii et al., 2004). BCL6A is also considered oncogenic, being highly expressed in many B cell lymphomas such as diffuse large B cell lymphoma (DLBCL) and follicular lymphoma (FL) (Wagner et al., 2011; Green et al., 2014). Bcl6a knockout mice also had perturbed dendritic cell development (Ohtsuka et al., 2011), while their macrophages showed altered morphology and defective motility (Pixley et al., 2005) as well as enhanced expression of inflammatory cytokines and chemokines (Toney et al., 2000; Li et al., 2020). Bcl6a-deficient mice displayed significantly decreased body weight postnatally (Dent et al., 1997). They also showed poor survival, with most not surviving past 9 weeks, attributed to severe Th2-mediated inflammation of the heart, lungs, liver and spleen (Dent et al., 1997; Yoshida et al., 1999).
Zebrafish is now well established as a model for immune cell development and function. It possesses B, T and NK cells, neutrophils, macrophages, dendritic cells and other immune lineages (Gore et al., 2018). These are generated through conserved developmental processes, which extends to the multiple developmental waves (Bertrand and Traver, 2009), and the associated transcription factors (Kwan and North, 2017). Moreover, their accessibility for genetic and other manipulations, optical transparency and the availability of lineage-specific transgenic lines has enabled new insights into innate immune cell function (Linnerz and Hall, 2020; Rosowski, 2020). This study sought to use zebrafish as a model to further investigate BCL6A function, identifying and characterizing a BCL6A homologue that was ablated via genome editing to understand the impacts on overall development, growth and survival, including immune cell development and function with a focus on early macrophages.
Materials and methods
Bioinformatics
Sequence searches were performed using BLAST on relevant online genetic databases, with Genomescan (Massachusetts Institute of Technology, Cambridge, MA) used to predict protein coding sequences from genomic DNA (Yeh et al., 2001). Sequence analysis, manipulation and assembly were carried out using Sequencher version 4.10.0 (Gene Codes). ClustalX 2.1 (Jeanmougin et al., 1998) was used to generate sequence alignments, from which phylogenetic trees were generated using the Neighbor-Joining algorithm (Saitou and Nei, 1987) with replicates of 1000 and viewed with NJ plot (Perriere and Gouy, 1996) and Treeview 1.6.6 (Page, 1996). Synteny analysis was performed using Ensembl.
Zebrafish husbandry
Wild-type and Tg(mpeg1.1::GFP) (Ellett et al., 2011) zebrafish were maintained using standard husbandry practices (Lawrence, 2007). This included feeding thrice daily with a mixture of live feed (artemia and rotifers) and a dry granulated foodstuff (Otohime Hirame Japan). Embryos were obtained from spawning tanks, and in some cases were injected with either control morpholino (5’-CCTCTTACCTCAGTTACAATTTATA) or anti-sense bcl6aa morpholino targeting the intron 2/exon 3 boundary (5’-AGAGCCCACTGTGGAGAAATTATGA) at 0.5 mM. All experiments were approved by the Deakin University Animal Welfare Committee.
Genome editing
The zebrafish bcl6aa gene was targeted using genome editing with CRISPR/Cas9. Embryos were injected with guide RNA (gRNA), designed to a region of exon 3 encoding the BTB/POZ domain using the zifit protocol (Hwang et al., 2013) with the primers 5’-TAGGTCCAGACTGATGGCGTTC and 5’-AAACGAACGCCATCAGTCTGGA, along with Cas9-encoding mRNA and raised to adulthood. Founders were identified with high-resolution melt (HRM) analysis of PCR products with Precision Melt Suremix and Analysis Software (BioRad) (Garritano et al., 2009) using primers spanning the targeted region (5’-CACAGTGGGCTCTTCTACTCTATC and 5’-GGATTGCGAAACCCTCTGG). These fish were outcrossed two times to wild-type fish to remove off-target mutations before in-crossing. Sequence analysis was performed with primers 5’-GCGACCTAAAAAGTTGACTAAAATC and 5’-CCTGGACTTTATGAATCTGTGGC to identify a bcl6aa mutant allele (mdu21), which was also crossed onto the Tg(mpeg1.1::GFP) background.
Whole-mount in situ hybridization
Embryos were dechorionated and fixed in 4% (w/v) paraformaldehyde (PFA) at 4°C prior to WISH with DIG-labeled anti-sense probes, as described (Thisse and Thisse, 2008). Imaging was performed using Olympus MVX10 fluorescence microscope and DP72 camera using Cellsens Dimension 1.6 software, with ImageJ used for quantitation, as required (Abramoff et al., 2004).
Quantitative real-time reverse-transcription PCR
Total RNA was extracted from whole embryos or juvenile zebrafish using an RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol for animal tissues. This was subjected to quantitative real-time reverse-transcription PCR with immune cell gene specific primers (cd4, cd8, cd79a, ighm, mpeg1.1, mpo, nklb, nkld and tcr) (Sertori et al., 2022) along with ccr2 (5’-TGGCAACGCAAAGGCTTTCAGTGA; 5’-TCAGCTAGGGCTAGGTTGAAGAG), cxcr4b (5’-CCCATCACAAGCACCACAAG; CGATAGCATCATTTTAGACAACAG), il1b (5’-GGACTTCGCAGCACAAAATG; 5’-GTTCACTTCACGCTCTTGGATG), tgfb1 (5’-AAATAGCAGGTTTGTCCCGC; 5’-CACTTCCAGCCCAGGTCTT) and tnfa (5’-GACTGAGGAACAAGTGCTTATGAG; 5’-TGCCCAGTCTGTCTCCTTCTC). Data were normalized to β-actin (actb) and fold change calculated using the ΔΔCt method (Livak and Schmittgen, 2001).
Wounding assay
Wounding assays were performed on 3 dpf embryos (n>20 mixed progeny) by excising the end of the caudal tail fin with a scalpel after anesthesia with 0.1 mg/mL benzocaine (Hall et al., 2007) in a conservative manner as described (Meier et al., 2022), with the number of migrating cells and the number of embryos with migrating cells counted up to 8 h after wounding using fluorescence microscopy.
Infection assay
Embryos at 4 dpf were injected with 2-5 nl ~ 5×109 CFU/mL E. coli expressing GFP (#25922GFP, ATCC) into the venous return, with bacteria visualized by fluorescence microscopy, as described (Basheer et al., 2020).
Statistics
Statistical analyses were performed using Graph Pad Prism (Version 8) software. To determine the statistical significance of various treatments, the unpaired independent student’s t test was employed, with Welch’s correction, where appropriate.
Results
Identification and characterization of BCL6-related genes in zebrafish
Bioinformatic analysis identified putative zebrafish homologues for both the BCL6A and BCL6B genes, as well as an additional related sequence, with all three genes also being present in another teleost fish, torafugu (Takifugu rubripes). One of these showed conserved synteny with human and mouse BCL6A and their adjacent genes LPP, TPRG1 and TP63 (Figure 1A), with the encoded proteins forming a clade with mammalian BCL6A (Figure 1B), and so was designated bcl6aa. The fish-specific gene showed conserved synteny across fish genomes, but not with bcl6aa or bcl6b genes (Figure 1A), but the encoded proteins formed a larger clade with the BCL6A sequences (Figure 1B), and so was named bcl6ab. The final gene showed conserved synteny with human and mouse BCL6B and their adjacent SLC16A13, ACADVL and DVL2 genes (Figure 1A), with the encoded fish and mammalian proteins divergent from the other BCL6 proteins (Figure 1B), and was designated bcl6b. Collectively, this suggests zebrafish bcl6aa and bcl6b are functional orthologues of mammalian BCL6A and BCL6B, respectively, while bcl6ab represents a fish-specific duplicate of the BCL6A gene.
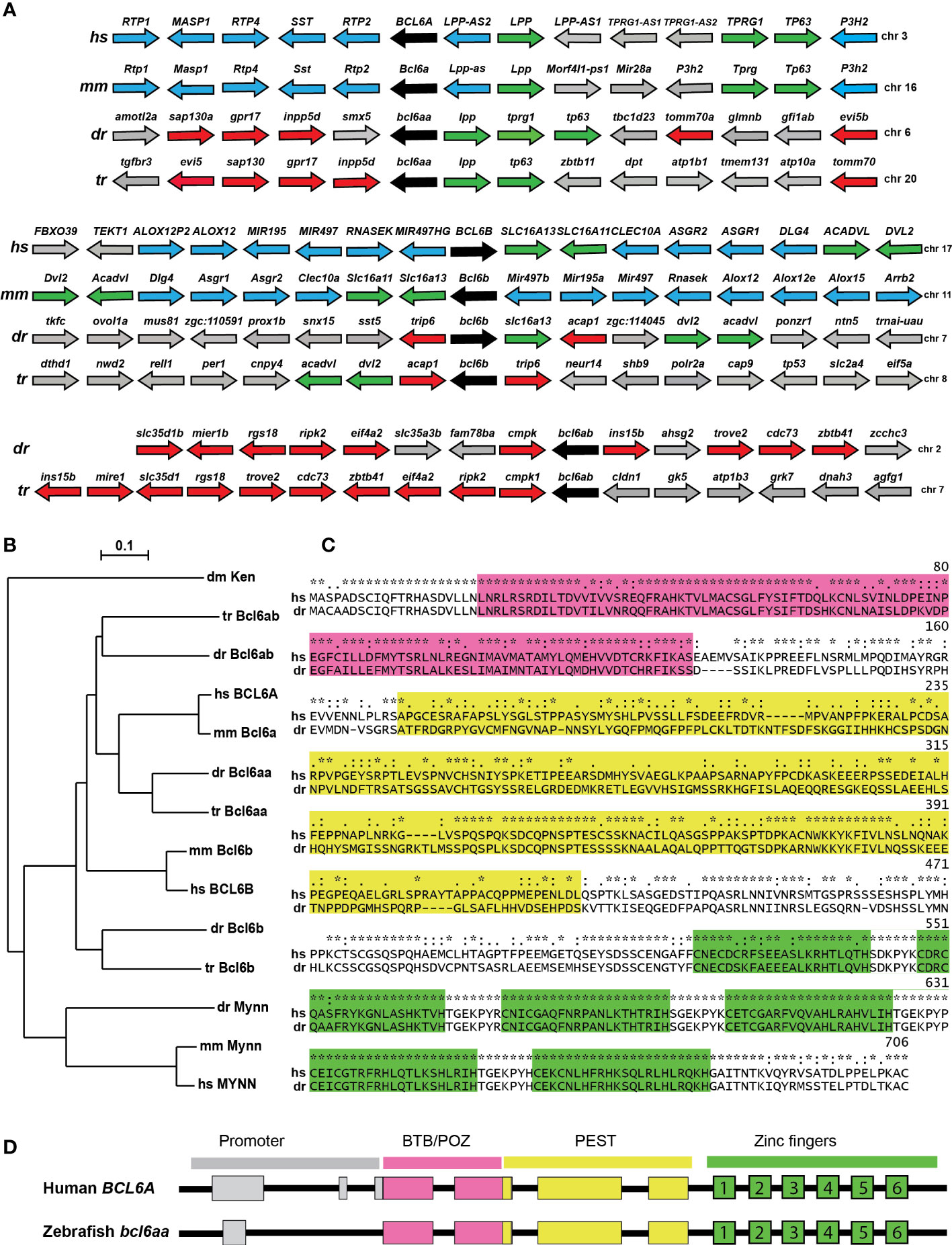
Figure 1 Conservation of BCL6A and related sequences. (A) Synteny analysis of BCL6-related genes. Arrangement of the gene neighborhood surrounding BCL6-related gene loci from human (Homo sapiens, hs), mouse (Mus musculus, mm), zebrafish (Danio rerio, dr) and torafugu (Takifugu rubripes, tr). The BCL6-related genes are in black, neighboring genes conserved between mammals and fish in green, between mammals in blue and between fish in red, with all other genes in grey. (B) Phylogenetic analysis of BCL6-related proteins. The amino acid sequences of fruit-fly Ken and Barbie (Ken) was aligned with the BCL6A and related sequences of human (hs), mouse (mm), zebrafish (dr) and torafugu (tr), and the MYNN-related sequences from human, mouse and torafugu using Clustal W. This was used to construct a phylogenetic tree using the Neighbor-Joining method with 1000 replicates, with bootstrapping values shown. (C) Conserved domains in BCL6A proteins. Human BCL6A and zebrafish Bcl6aa were aligned using Clustal X software, with specific domains highlighted (BTB/POZ in pink, PEST in yellow, zinc fingers in green). Conserved residues between the two sequences are indicated (identical *, highly similar: similar.). (D) Conserved BCL6A gene structure. Schematic diagram of human BCL6A and zebrafish bcl6aa loci, with exons shown as boxes and introns as lines. Regions corresponding to the promoter (grey) or those encoding the BTB/POZ (pink), PEST (yellow) and zinc finger (green and numbered) domains are indicated.
Alignment of the human BCL6A and zebrafish Bcl6aa proteins confirmed the presence of conserved BTB/POZ, PEST and zinc finger domains, which showed 77%, 35% and 96% identity, respectively (Figure 1C). Notably, the latter domain included a stretch of 126 identical amino acids that encompassed the last four of the six C2H2-type zinc fingers. Comparison of the genomic and mRNA (Figure 1D) revealed a strongly conserved splicing pattern between human BCL6A and zebrafish bcl6aa genes across the coding exons, with both also possessing a non-coding exon(s) in the proximal promoter region.
Expression of zebrafish bcl6aa
The embryonic expression pattern of zebrafish bcl6aa was investigated by high resolution whole-mount hybridization (WISH) on staged wild-type embryos using an anti-sense bcl6aa probe. Expression was observed from 10 hours post-fertilization (hpf) in the anterior lateral mesoderm (ALM) and the posterior lateral mesoderm (PLM), sites of early myeloid cell development (Bertrand and Traver, 2009), which continued until 24 hpf (Figures 2A, C–F). From 36 hpf, bcl6aa was expressed in the retina, cerebellum and medulla (Figures 2G, H) that continued until 7 dpf although declining after 4 dpf (Figures 2I–N). From 4 dpf bcl6aa expression was also detected in the developing thymus (Figures 2J–N), which houses T cell development (Gore et al., 2018). No staining was observed with a control sense bcl6aa probe (Figure 2B and data not shown).
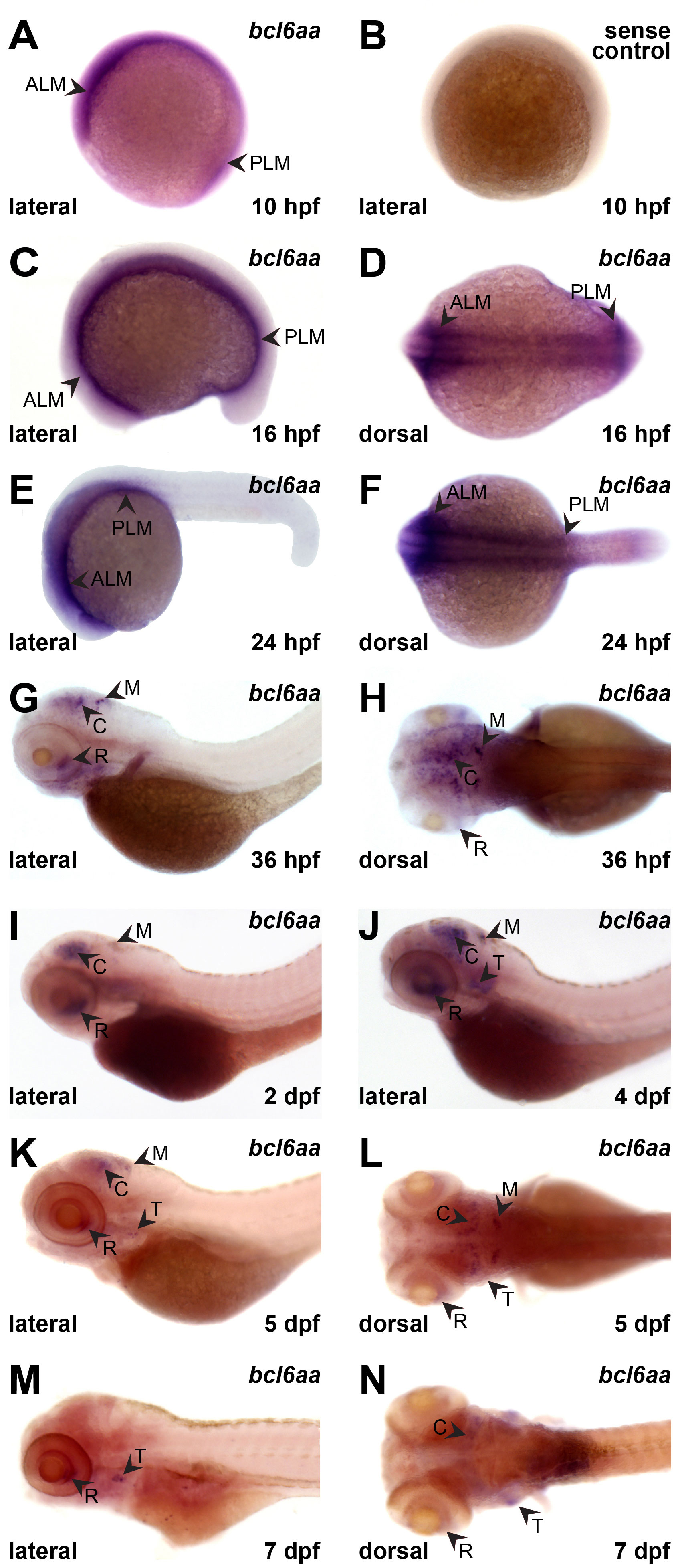
Figure 2 Expression of bcl6aa during zebrafish embryogenesis. (A–L). Representative images of wild-type embryos subjected to WISH with anti-sense (bcl6aa) and sense (sense control) bcl6aa probes as indicated at 10 hpf (A, B), 16 hpf (C, D), 24 hpf (E, F), 36 hpf (G, H), 2 dpf (I), 4 dpf (J), 5 dpf (K, L) and 7 dpf (M, N), as viewed laterally or dorsally as labelled. ALM, anterior lateral mesoderm; C, cerebellum; M, medulla; PLM, posterior lateral mesoderm; R, retina; T, thymus.
Generation and analysis of bcl6aa knockout zebrafish
The zebrafish bcl6aa gene was mutated using genome editing with CRISPR/Cas9 to target a region of exon 3 encoding the BTB/POZ domain (Supplementary Figures 1A, B). Potential founders were identified with high-resolution melt analysis of PCR products spanning the targeted region, with these outcrossed two times to wild-type fish to remove potential off-target mutations before in-crossing. Sequence analysis identified a bcl6aa allele (mdu21) that harbored a combined large deletion and insertion, predicted to encode a protein that shared just the first 70 amino acids with the wild-type protein, and then encodes 27 amino acids of unrelated sequence before a stop codon is reached (Supplementary Figure 1C). Since this represents only part of the BTB/POZ domain and none the PEST or zinc finger domains, it is anticipated that the encoded mutant protein would be non-functional.
The progeny of bcl6aawt/mdu21 in-crosses were imaged by light microscopy, with no evidence of overt developmental perturbation during embryogenesis observed in mixed groups, which should contain 25% bcl6aamdu21/mdu21 embryos (Figures 3A–C), or in individually genotyped embryos (Figure 3D and data not shown). However, this changed dramatically during the juvenile phase, such that by 21 dpf there were large and distinct differences in size that were in roughly Mendelian ratios. Genotyping of individual fish confirmed homozygote bcl6aamdu21/mdu21 mutants were the smallest, with bcl6aawt/mdu21 heterozygotes intermediate in size compared to the larger wild-type bcl6aawt/wt individuals (Figure 3E, F). Additionally, the bcl6aamdu21/mdu21 mutants showed a clearly under-developed dorsal fin, abdominal fin, tail fin, swim bladder and eye, with the bcl6aawt/mdu21 mutants again showing an intermediate phenotype.
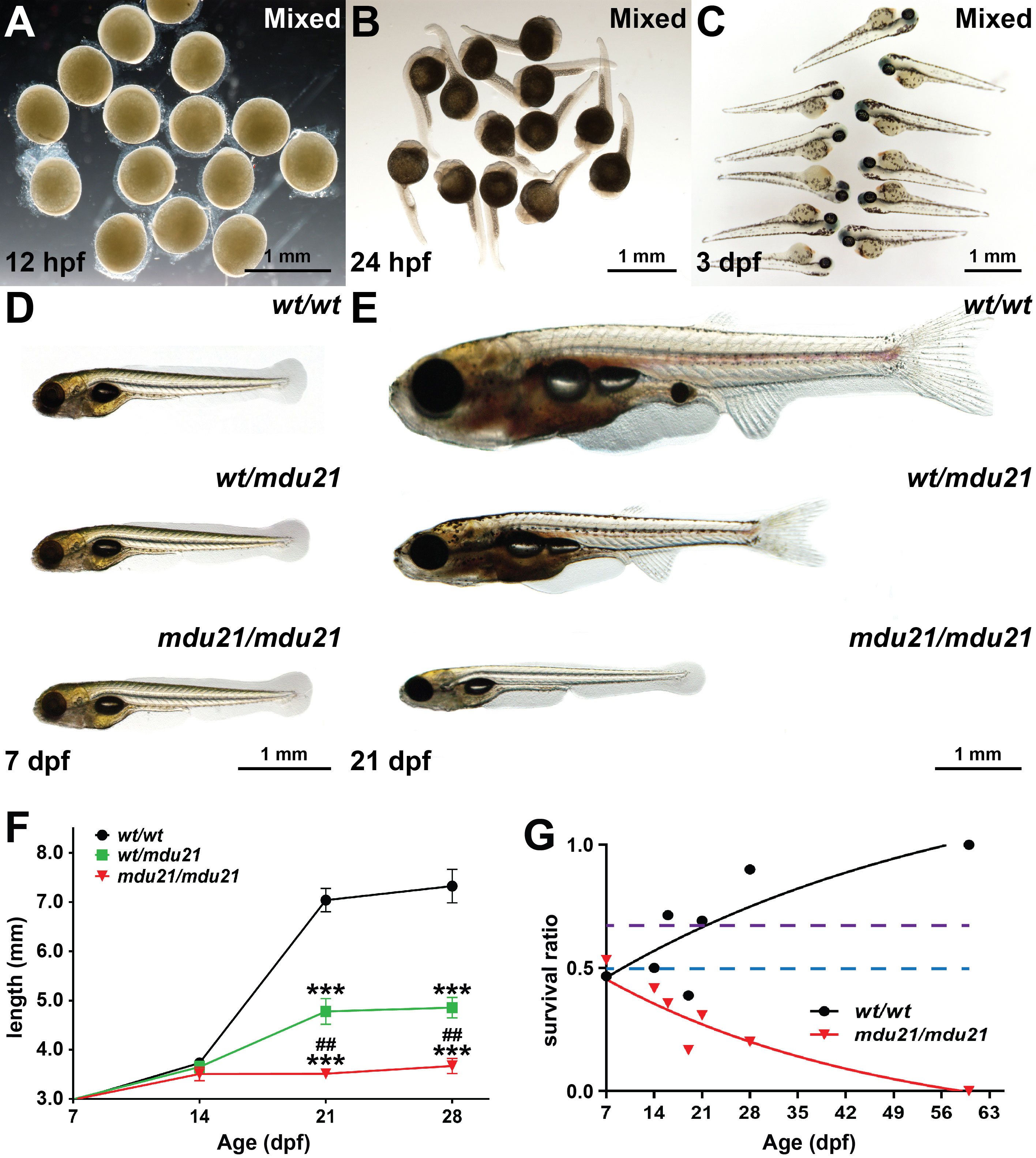
Figure 3 Impact of bcl6aa ablation on global development. (A–E). Light microscopy of representative images of mixed progeny (Mixed) derived from bcl6aawt/mdu21 in-crossing at 12 hpf (A), 24 hpf (B) and 3 dpf (C) or of individually genotyped bcl6aawt/wt (wt/wt), bcl6aawt/mdu21 (wt/mdu21) and bcl6aamdu21/mdu21 (mdu21/mdu21) embryos at 7 dpf (D) and 21 dpf (E), with 0.5 mm scale bars indicated. (F). Body length of individually genotyped bcl6aawt/wt (wt/wt), bcl6aawt/mdu21 (wt/mdu21) and bcl6aamdu21/mdu21 (mdu21/mdu21) individuals at the indicated time-points. Shown is the mean ± SEM, with statistical significance relative to wt/wt (***p<0.001 and wt/mdu21 (##p<0.01), (n>25). (G). Relative survival of bcl6aawt/wt (wt/wt) and bcl6aamdu21/mdu21 (mdu21/mdu21) individuals expressed as a ratio relative to bcl6aawt/mdu21 individuals from n>60 genotypes at each time point. The dotted lines show the expected Mendelian ratio for both wt/wt and mdu21/mdu21 individuals if all genotypes showed equivalent survival (blue) or for wt/wt individuals if they showed equivalent survival with wt/mdu21 in the absence of mdu21/mdu21 individuals (purple).
It was also noted that the proportion of smaller fish decreased over time, with none surviving to adulthood. Genotyping of adult fish confirmed that no bcl6aamdu21/mdu21 fish were present (data not shown). Close analysis of the relative proportion of bcl6aamdu21/mdu21 fish at by genotyping across multiple timepoints revealed that while present at an expected Mendelian ratio at 7 dpf, this steadily decreased, with none observed at 60 dpf (Figure 3G). The proportion of bcl6aawt/wt fish also increased to above the expected Mendelian ratio, indicating that heterozygote fish also had a milder survival defect, although a good proportion survived to adulthood and showed robust fecundity.
Impact of bcl6aa ablation on immune cells
The effect of bcl6aa ablation on immune cells was investigated by WISH analysis with specific markers during embryogenesis, before any growth or survival defects were present. Homozygote bcl6aamdu21/mdu21 embryos showed a significant decrease in expression of ikzf1, a marker of T cell progenitors in the developing thymus (Willett et al., 2001), compared to bcl6aawt/wt and bcl6aawt/mdu21 siblings at both 3.5 dpf (Figures 4A, B) and 5 dpf (Figures 4C, D). Expression of rag1, a marker of mature T cells (Willett et al., 1997) was significantly decreased in both bcl6aawt/mdu21 and bcl6aamdu21/mdu21 embryos compared to bcl6aawt/wt siblings at both 3.5 dpf (Figures 4E, F) and 5 dpf (Figures 4G, H), but to a much greater extent in bcl6aamdu21/mdu21 embryos across both timepoints (Figures 4E–H). In contrast, no significant difference was observed in the number of cells expressing mpo, a marker of neutrophils (Lieschke et al., 2001), in either bcl6aawt/mdu21 or bcl6aamdu21/mdu21 compared to bcl6aawt/wt embryos at 5 dpf (Figures 4I, J). However, the number of cells expressing lcp1, a marker of leukocytes including macrophages (Bennett et al., 2001), was significantly decreased in both bcl6aawt/mdu21 and bcl6aamdu21/mdu21 compared to bcl6aawt/wt embryos, although again the quantity in bcl6aamdu21/mdu21 embryos was also significantly reduced compared to heterozygotes (Figures 4K, L). To facilitate further analysis of macrophages the bcl6aamdu21 allele was crossed onto the Tg(mpeg1.1:GFP) background, in which macrophages are marked with GFP (Wittamer et al., 2011). Tg(mpeg1.1:GFP) bcl6aawt/mdu21 fish were in-crossed and visualized by fluorescence microscopy that revealed a significant decrease in GFP+ cells at 4 dpf in bcl6aamdu21/mdu21 compared to the bcl6aawt/wt and bcl6aawt/mdu21 embryos (Figures 4M, N). To confirm the effects of bcl6aa on macrophages, embryos were injected with an anti-sense morpholino targeting the intron 2/exon 3 splice site. This also resulted in a decrease of lcp1+ cells at 22 hpf in wild-type embryos (Figures 4O, P) and in GFP+ cells in Tg(mpeg1.1:GFP) embryos at 3 dpf (Figures 4Q, S) in comparison to those injected with a control morpholino. Macrophage morphology was also altered in the bcl6aa morpholino-injected embryos with a statistically significant decrease in those with an amoeboid morphology (Figure 4R).
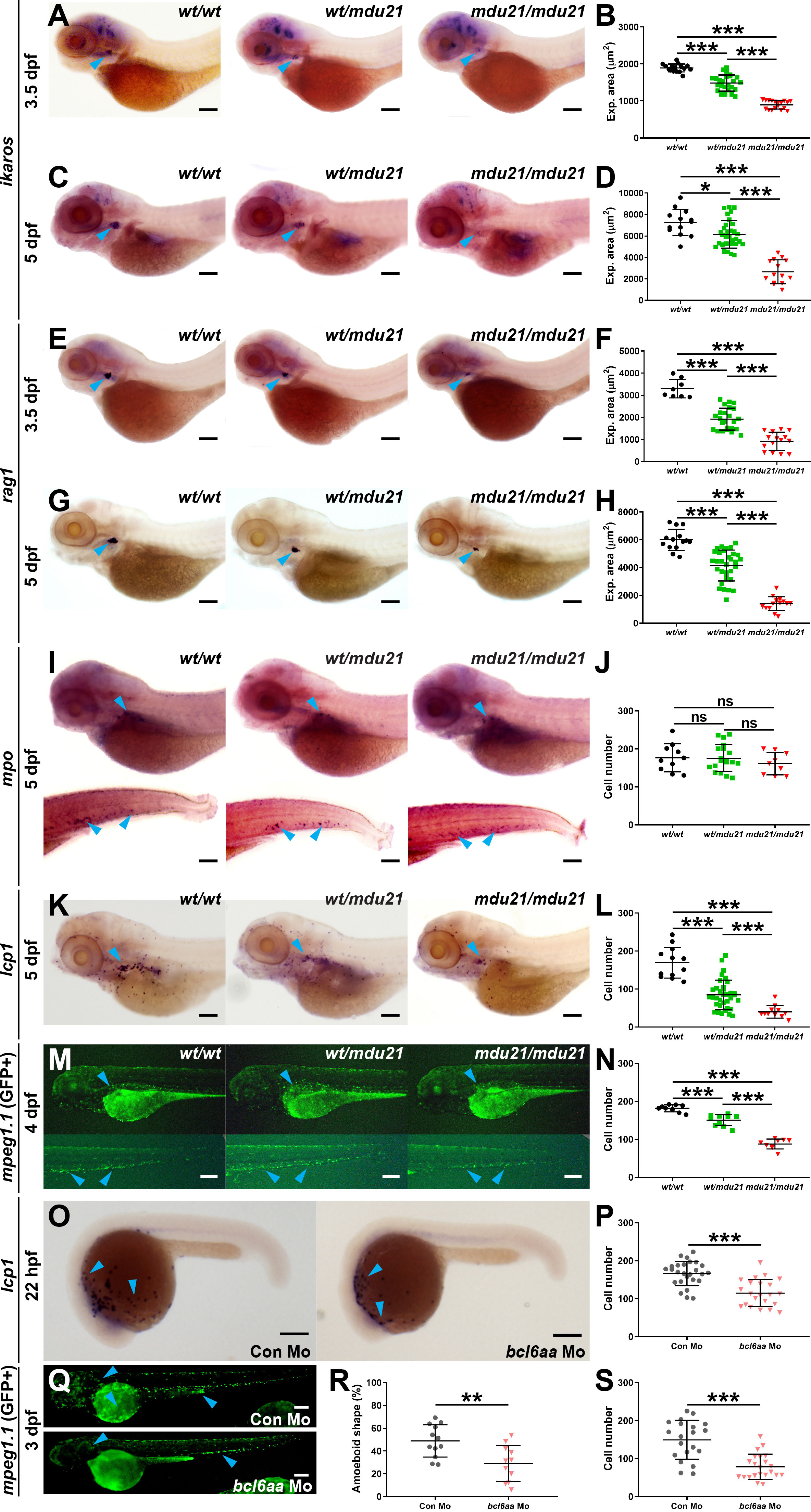
Figure 4 Analysis of lymphoid and myeloid cells in bcl6aa mutant zebrafish. (A, C, E, G, I, K). Representative bcl6aawt/wt (wt/wt), bcl6aawt/mdu21 (wt/mdu21) and bcl6aamdu21/mdu21 (mdu21/mdu21) embryos analyzed by WISH with ikzf1 at 3.5 dpf (A) and 5 dpf (C), rag1 at 3.5 dpf (E) and 5 dpf (G), mpo at 5 dpf (I) and lcp1 at 5 dpf (K). (M). Fluorescence imaging of representative bcl6aawt/wt (wt/wt), bcl6aawt/mdu21 (wt/mdu21) and bcl6aamdu21/mdu21 (mdu21/mdu21) embryos on the Tg(mpeg1.1:GFP) background at 4 dpf. (O). Representative wild-type embryos injected with control (Con) or bcl6aa morpholino (Mo) as indicated analyzed by WISH with lcp1 at 22 hpf. (Q). Representative Tg(mpeg1.1:GFP) embryos injected with control (Con) or bcl6aa morpholino (Mo) subjected to fluorescence microscopy at 3 dpf. Domains of expression are indicated with arrowheads, and scale bars represent 200 μm. (B, D, F, H, J, L, N, P, R, S). Quantification of cell markers, either using expression area (B, D, F, H), number of discrete cells (J, L, N, P, S) or the proportion with an amoeboid morphology (R), showing values for individual embryos, as well as mean ± SEM (*p<0.05; **p<0.01; ***p<0.001; ns, not significant; n>30).
Functional analysis of bcl6aa mutants
To further understand the effect of bcl6aa ablation on macrophages, a wounding assay was performed on the progeny of Tg(mpeg1.1:GFP) bcl6aawt/mdu21 fish as described (Hall et al., 2007), with individual embryos subsequently imaged at various times, after which they were genotyped. This revealed that in bcl6aawt/wt embryos GFP+ cells peaked at the wound at 4 hours post wounding (hpw) before decreasing at later time points (Figures 5A, B). In both bcl6aawt/mdu21 and bcl6aamdu21/mdu21 embryos, GFP+ cells peaked at 8 hpw, however, the numbers observed in bcl6aawt/mdu21 embryos were equivalent to or exceeded those of bcl6aawt/wt embryos, whereas they were significantly reduced in bcl6aamdu21/mdu21 embryos. When normalized to total GFP+ cells, the number of GFP+ cells remained reduced to a statistically significant level at both 4 and 24 hpw (data not shown). The experiment was repeated but examining embryos at 0.25 h intervals until the first GFP+ cell reached the wound site, after which embryos were genotyped. For almost all bcl6aawt/wt and bcl6aawt/mdu21 embryos this occurred by 1 hpw, but for bcl6aamdu21/mdu21 embryos this was around 3 hpw (Figure 5C).
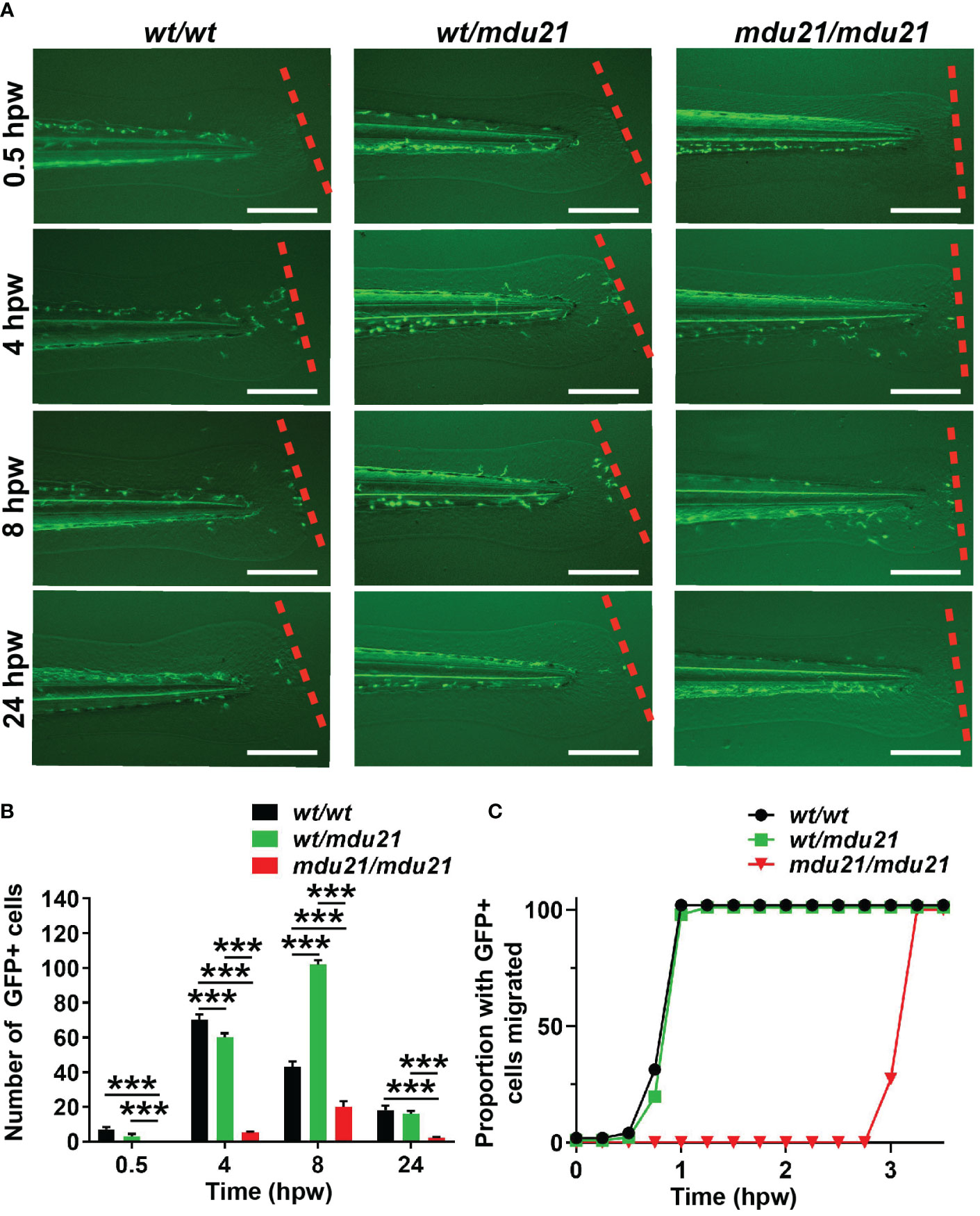
Figure 5 Analysis of macrophage migration in response to injury in bcl6aa mutant zebrafish. (A) Wounding assay on 4 dpf embryos subjected to injury via tail fin transection, showing fluorescence images of representative bcl6aawt/wt (wt/wt), bcl6aawt/mdu21 (wt/mdu21) and bcl6aamdu21/mdu21 (mdu21/mdu21) embryos on the Tg(mpeg1.1:GFP) background, as determined by retrospective genotyping, at the times indicated, with the dotted line showing the wounding site. Scale bars represent 200 μm. (B) Quantitation of the total number of GFP+ macrophage migrated at the indicated timepoint showing mean ± SEM. (***p<0.001; n>20 mixed progeny). (C) Cumulative proportion of embryos with at least 1 GFP+ cell migrated assessed at 0.25 h intervals (n>20).
The bcl6aa mutants were next investigated in a bacterial infection assay (Basheer et al., 2020). The progeny of bcl6aawt/mdu21 in-crosses were subjected to injection with GFP+ E. coli at 4 dpf. Surviving embryos were imaged by fluorescent microscopy until 24 hour post infection (hpi) and subsequently genotyped, with the fluorescence intensity used as an indicator of bacterial load. The bcl6aawt/wt, bcl6aawt/mdu21 and bcl6aamdu21/mdu21 embryos showed no difference in fluorescence at 0.5 hpi (Figures 6A, B). However, at 12 and 18 hpi the fluorescence intensity was increased in bcl6aamdu21/mdu21 compared to bcl6aawt/wt and bcl6aawt/mdu21 embryos. No difference in mortality was observed in the injected embryos at 0.5 hpi, but from 12 hpi decreased survival of bcl6aamdu21/mdu21 embryos was observed, reaching zero survival at 24 hpi, while bcl6aawt/wt and bcl6aawt/mdu21 showed similar high survival rate (Figure 6C) and fluorescence intensity (Figure 6B and data not shown). Analysis of a set of inflammatory genes indicated increased basal expression of ccr2 in bcl6aamdu21/mdu21 compared to bcl6aawt/wt embryos (Figure 6D). Infection resulted in increased expression of il1b and tnfb1b in both bcl6aawt/wt and bcl6aamdu21/mdu21 embryos (data not shown), but il1b was significantly enhanced in bcl6aamdu21/mdu21 compared to bcl6aawt/wt embryos (Figure 6D).
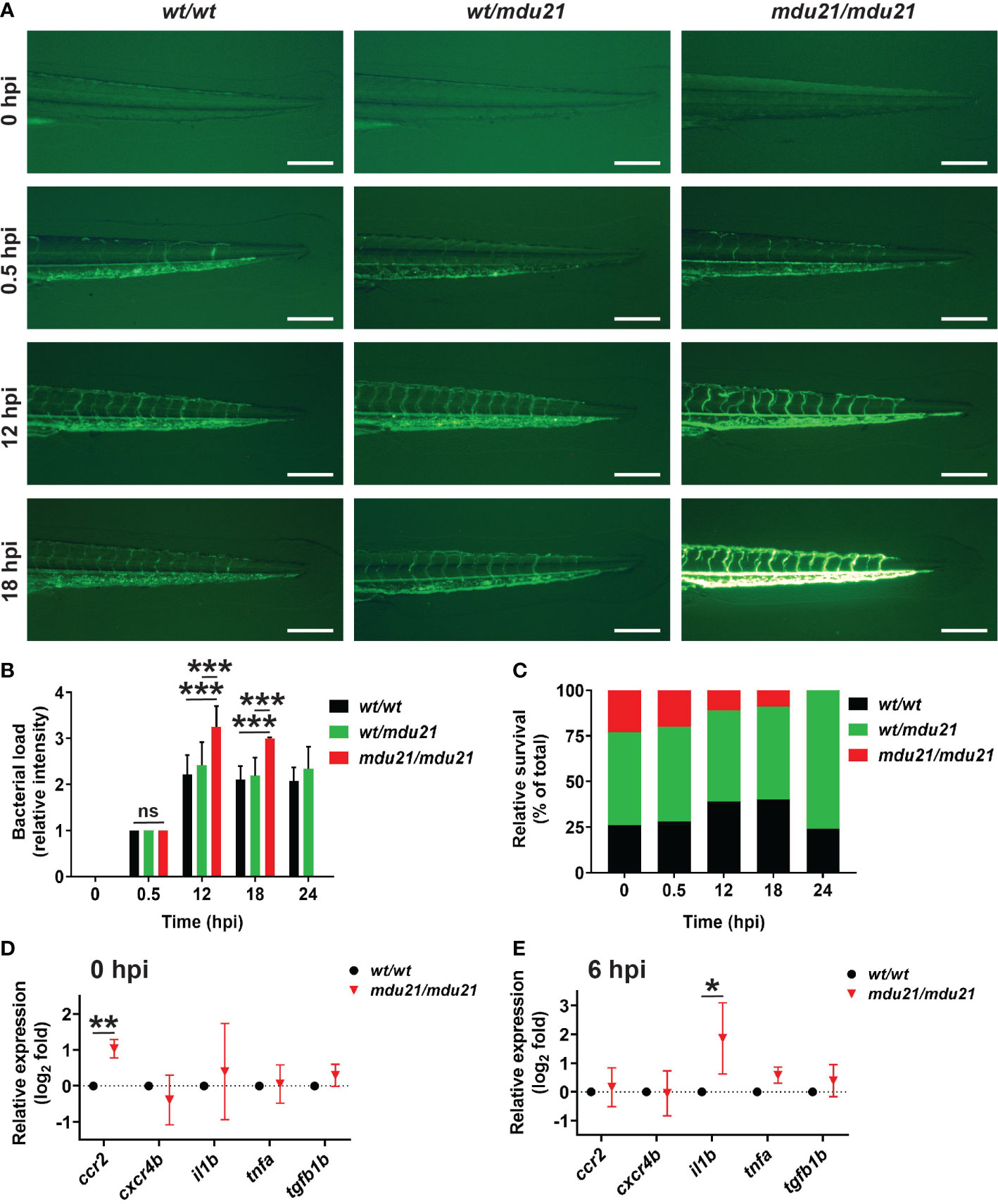
Figure 6 Analysis of bacterial infection in bcl6aa mutant zebrafish. (A) Infection of 4 dpf embryos with GFP+ E coli showing representative bcl6aawt/wt (wt/wt), bcl6aawt/mdu21 (wt/mdu21) and bcl6aamdu21/mdu21 (mdu21/mdu21) embryos at the indicated timepoints. Scale bars represent 200 μm. (B) Bacterial load intensity was assessed on a 5 point scale (0-4) for bcl6aawt/wt (wt/wt), bcl6aawt/mdu21 (wt/mdu21) and bcl6aamdu21/mdu21 (mdu21/mdu21) embryos at each timepoint relative to 0.5 hpf being 1, showing mean ± SEM (***p<0.001; ns: not significant; n≥50 total at each timepoint). (C) Relative survival of bcl6aawt/wt (wt/wt), bcl6aawt/mdu21 (wt/mdu21) and bcl6aamdu21/mdu21 (mdu21/mdu21) embryos at the indicated timepoints (n=100 total at each timepoint). (D, E) Analysis of the indicated inflammatory gene markers in homozygous bcl6aawt/wt (wt/wt) and bcl6aamdu21/mdu21 (mdu21/mdu21) individuals at 0 hpi (D) and 6 hpi (E) using qRT2-PCR with data normalized relative to actb and represented as relative fold change compared to wild-type, with mean and SD shown and statistical significance indicated (*p<0.05, **p<0.01, n=4).
Discussion
Mammalian BCL6A, and the closely-related BCL6B, are transcriptional repressors consisting of BTB/POZ, PEST and zinc finger domains (Basso and Dalla-Favera, 2010). BCL6A homologues are highly conserved, with murine BCL6A 95% identical to its human BCL6A counterpart (Fukuda et al., 1995). A zebrafish homologue (bcl6aa) was identified on the basis of phylogenetic and syntenic analysis, with the encoded protein displaying >60% identity with human BCL6A, with equivalent BTB/POZ, PEST and zinc finger domains, the latter showing 96% identity, consistent with a bcl6aa gene reported from another teleost fish (Ohtani et al., 2006). This high conservation indicates a likely conserved function across vertebrates and particular of target DNA sequences. This study also identified an additional fish-specific gene, bcl6ab, most likely a paralogue of bcl6aa, one of many teleost genes duplicated as a result of a teleost-specific whole genome duplication (WGD) event (Reams and Roth, 2015), as well as a zebrafish orthologue to the mammalian BCL6B gene (bcl6b). It will be of interest to investigate whether the bcl6ab paralogue has evolved a unique function or shares functions with bcl6aa and possibly bcl6b.
Mammalian BCL6A is strongly expressed in thymocytes from human fetal samples at 21 weeks gestation (Hyjek et al., 2001), and in the fetal mouse thymus at 17 days gestation (Bajalica-Lagercrantz et al., 1998). Zebrafish bcl6aa was similarly expressed in the developing thymus during embryogenesis, suggesting a conserved role in early T lymphocyte development across vertebrates. The bcl6aa gene was also expressed even earlier in the zebrafish ALM and PLM, which represent sites of primitive hematopoiesis in the zebrafish (Bertrand and Traver, 2009). Expression of BCL6A has also been identified in adult peripheral blood leukocytes and lymph nodes in humans (Bajalica-Lagercrantz et al., 1997), and of Bcl6a in the adult mouse thymus (Bajalica-Lagercrantz et al., 1998). The pufferfish bcl6aa homologue has been previously found to be expressed in adult thymus and kidney, the teleost bone marrow equivalent (Ohtani et al., 2006), while analysis of published single cell sequencing data (Tang et al., 2017) confirms bcl6aa is expressed in adult zebrafish T and B cells (data not shown). Collectively, this indicates potential conserved roles for BCL6A in the ongoing development of blood and immune cells in the adult.
The bcl6aa gene was also expressed in regions of the developing brain and retina, the latter confirming a previous study (Lee et al., 2013). This is consistent with expression of fruit-fly ken at the onset of gastrulation in the cephalic furrow and later in the larval eye-antenna (Arbouzova et al., 2006), with Bcl6a expression also seen in the olfactory epithelium of prenatal mice (Bajalica-Lagercrantz et al., 1998). BCL6A expression in adult human spinal cord has been described (Bajalica-Lagercrantz et al., 1997), with Bcl6a shown to be expressed in the adult mouse cerebral cortex (Bajalica-Lagercrantz et al., 1998) and bcl6aa in the adult pufferfish brain and nasal cavity (Ohtani et al., 2006). Human BCL6A was also expressed in adult skeletal muscle, thyroid, trachea, ovary and prostate (Bajalica-Lagercrantz et al., 1997), with mouse Bcl6a expressed in skeletal muscle (Albagli-Curiel et al., 1998) and pufferfish bcl6aa in skeletal muscle, intestine and ovary (Ohtani et al., 2006). Collectively, this may suggest a conserved broader role for BCL6/ken genes in non-hematopoietic tissues.
BCL6A has been shown to be a key regulator of B and T cells (Nurieva et al., 2009; Bassil et al., 2014), with specific lineages of both B and T lymphocytes impacted in Bcl6a knockout mice (Dent et al., 1997; Cattoretti et al., 2005; Nurieva et al., 2009; Mondal et al., 2010; Huang et al., 2014; Choi and Crotty, 2021). Zebrafish possess both T and B cells (Langenau and Zon, 2005; Hansen and Zapata, 1998; Trede and Zon, 1998), with zebrafish T cells precursors arising during the embryonic definitive wave of hematopoiesis and, as in mammals, generating mature T cells in the thymus (Haire et al., 2000; Bertrand et al., 2007; Seelye et al., 2016). The bcl6aamdu21/mdu21 mutants displayed a significant decrease in lymphocyte populations at this location during embryogenesis including lymphoid precursors and early T lymphocytes. This finding suggests an essential role of bcl6aa in the differentiation and/or survival of early T cells or their progenitors in zebrafish. It was more difficult to study B cells since these arise three weeks post fertilization (Danilova et al., 2000) when survival of bcl6aamdu21/mdu21 fish was already compromised. Analysis of surviving juveniles at 28 dpf showed multiple lymphocyte populations were reduced, including T, B and NK cells (Supplementary Figure 2). However, the significant developmental delay observed in bcl6aamdu21/mdu21 mutants and the reliance solely on qRT-PCR data means this result needs to be interpreted cautiously. Recent enhancements in husbandry practices have meant that bcl6aamdu21/mdu21 adults are now available, the analysis of which will provide more definitive understanding of the impacts on lymphocyte homeostasis.
BCL6A has been previously implicated in the development and function of macrophages and dendritic cells (Yamochi et al., 1997; Pantano et al., 2006; Zhang et al., 2014). A significant reduction in macrophages was observed in bcl6aamdu21/mdu21 embryos, which was also the case in juvenile fish (Supplementary Figure 2), and confirmed using morpholino-mediated gene knockdown. Significantly decreased macrophage motility was observed in response to wounding, with macrophages appearing more amoeboid following bcl6aa ablation. This is consistent with a study showing inactivation of Bcl6a in bone-marrow derived macrophages resulted in reduced macrophage motility, polarization and spreading (Pixley et al., 2005). The bcl6aamdu21/mdu21 mutants were also found to be less able to control bacterial infection, had elevated il1b and reduced survival, in agreement with data from Bcl6a knockout mice that showed increased inflammatory gene expression following LPS injection, including Il1b (Barish et al., 2010). These effects are likely a consequence of the reduced macrophage number and functionality, since T cells remain in the thymus at this stage of development, and neutrophil numbers are unchanged – although potential functional defects were not examined. Of note, fruit-fly hemocytes, which represent innate immune cells, were also found to be sensitive to the effects of Ken (Arbouzova et al., 2006). This suggests an evolutionarily conserved role for BCL6A/Ken in innate immunity.
Bcl6a knockout mice displayed severe growth retardation (Albagli-Curiel et al., 1998; Yoshida et al., 1999), which was also observed in bcl6aamdu21/mdu21 fish from 14 dpf indicating this represents another common phenotype across vertebrates. This may be mediated via a direct role on growth, since strong Bcl6a expression has been observed during skeletal muscle differentiation in mice (Bajalica-Lagercrantz et al., 1998). However, the bcl6aamdu21/mdu21 also displayed a thinner body, consistent with the reduced adipose mass seen in Bcl6a knockout mice (LaPensee et al., 2014). This might be due to a direct effect on adipose tissue, as murine Bcl6a knockouts have been shown to possess disrupted lipid metabolism (LaPensee et al., 2014). The bcl6aamdu21/mdu21 fish also displayed reduced survival. A similar phenotype was observed in Bcl6a knockout mice, which has been demonstrated to be the result of excessive inflammatory responses leading to profound myocarditis and vasculitis (Dent et al., 1997; Fukuda et al., 1997; Ye et al., 1997; Huang et al., 2014). This severe inflammation could also impact on growth and development indirectly.
Fruit-fly Ken has been found to contribute to the differentiation of photoreceptor (neuronal) cells and cone (non-neuronal) cells during eye development (Wen et al., 2000). Moreover, a previous study showed that bcl6aa knockdown in zebrafish embryos resulted in malformation of the optic cup during embryogenesis (Lee et al., 2013). We observed expression of bcl6aa in the developing zebrafish eye but no overt eye defects in bcl6aamdu21/mdu21 mutants, with none reported in Bcl6a mutant mice either. More work is required to understand the discrepancies, especially between the zebrafish studies, as well to investigate other aspects of development that are perturbed in Bcl6a knockout mice and ken mutant flies.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by Deakin University Animal Ethics Committee.
Author contributions
FA, SH, MS and CL performed experiments. FA, SH, MS, AW and CL analyzed the results and prepared figures. AW and CL designed the research. FA and AW wrote the paper, which was read and approved by all authors. All authors contributed to the article and approved the submitted version.
Funding
The authors recognize the support of funding from IMPACT at Deakin University. FA was supported by the Higher Committee for Education Development in Iraq (HCED) and both SH and MS by Deakin University International Postgraduate Research Awards.
Acknowledgments
The authors would like to thank the Deakin University Animal House staff.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.887278/full#supplementary-material
References
Abramoff, M., Magelhaes, P., Ram, S. (2004). Image processing with ImageJ. Biophoton. Int. 11, 36–42.
Ahmad, K. F., Melnick, A., Lax, S., Bouchard, D., Liu, J., Kiang, C. L., et al. (2003). Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol. Cell 12, 1551–1564. doi: 10.1016/S1097-2765(03)00454-4
Albagli-Curiel, O., Dhordain, P., Lantoine, D., Aurade, F., Quief, S., Kerckaert, J. P., et al. (1998). Increased expression of the LAZ3 (BCL6) proto-oncogene accompanies murine skeletal myogenesis. Differentiation 64, 33–44. doi: 10.1046/j.1432-0436.1998.6410033.x
Arbouzova, N. I., Bach, E. A., Zeidler, M. P. (2006). Ken & barbie selectively regulates the expression of a subset of Jak/STAT pathway target genes. Curr. Biol. 16, 80–88. doi: 10.1016/j.cub.2005.11.033
Bajalica-Lagercrantz, S., Piehl, F., Farnebo, F., Larsson, C., Lagercrantz, J. (1998). Expression of the BCL6 gene in the pre- and postnatal mouse. Biochem. Biophys. Res. Commun. 247, 357–360. doi: 10.1006/bbrc.1998.8551
Bajalica-Lagercrantz, S., Piehl, F., Lagercrantz, J., Lindahl, J., Weber, G., Kerckeart, J. P., et al. (1997). Expression of LAZ3/BCL6 in follicular center (FC) b cells of reactive lymph nodes and FC-derived non-Hodgkin lymphomas. Leukemia 11, 594–598. doi: 10.1038/sj.leu.2400577
Barish, G. D., Yu, R. T., Karunasiri, M., Ocampo, C. B., Dixon, J., Benner, C., et al. (2010). Bcl-6 and NF-kappaB cistromes mediate opposing regulation of the innate immune response. Genes Dev. 24, 2760–2765. doi: 10.1101/gad.1998010
Basheer, F., Liongue, C., Ward, A. C. (2020). Zebrafish bacterial infection assay to study host-pathogen interactions. Bio. Protoc. 10, e3536. doi: 10.21769/BioProtoc.3536
Bassil, R., Orent, W., Olah, M., Kurdi, A. T., Frangieh, M., Buttrick, T., et al. (2014). BCL6 controls Th9 cell development by repressing Il9 transcription. J. Immunol. 193, 198–207. doi: 10.4049/jimmunol.1303184
Basso, K., Dalla-Favera, R. (2010). BCL6: master regulator of the germinal center reaction and key oncogene in b cell lymphomagenesis. Adv. Immunol. 105, 193–210. doi: 10.1016/S0065-2776(10)05007-8
Basso, K., Dalla-Favera, R. (2012). Roles of BCL6 in normal and transformed germinal center b cells. Immunol. Rev. 247, 172–183. doi: 10.1111/j.1600-065X.2012.01112.x
Bennett, C. M., Kanki, J. P., Rhodes, J., Liu, T. X., Paw, B. H., Kieran, M. W., et al. (2001). Myelopoiesis in the zebrafish, danio rerio. Blood 98, 643–651. doi: 10.1182/blood.V98.3.643
Bertrand, J. Y., Kim, A. D., Violette, E. P., Stachura, D. L., Cisson, J. L., Traver, D. (2007). Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development 134, 4147–4156. doi: 10.1242/dev.012385
Bertrand, J. Y., Traver, D. (2009). Hematopoietic cell development in the zebrafish embryo. Curr. Opin. Hematol. 16, 243–248. doi: 10.1097/MOH.0b013e32832c05e4
Cattoretti, G., Pasqualucci, L., Ballon, G., Tam, W., Nandula, S. V., Shen, Q., et al. (2005). Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large b cell lymphomas in mice. Cancer Cell 7, 445–455. doi: 10.1016/j.ccr.2005.03.037
Choi, J., Crotty, S. (2021). Bcl6-mediated transcriptional regulation of follicular helper T cells (TFH). Trends Immunol. 42, 336–349. doi: 10.1016/j.it.2021.02.002
Danilova, N., Hohman, V. S., Kim, E. H., Steiner, L. A. (2000). Immunoglobulin variable-region diversity in the zebrafish. Immunogenetics 52, 81–91. doi: 10.1007/s002510000255
Dent, A. L., Shaffer, A. L., Yu, X., Allman, D., Staudt, L. M. (1997). Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science 276, 589–592. doi: 10.1126/science.276.5312.589
Duy, C., Yu, J. J., Nahar, R., Swaminathan, S., Kweon, S. M., Polo, J. M., et al. (2010). BCL6 is critical for the development of a diverse primary b cell repertoire. J. Exp. Med. 207, 1209–1221. doi: 10.1084/jem.20091299
Ellett, F., Pase, L., Hayman, J. W., Andrianopoulos, A., Lieschke, G. J. (2011). mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117, E49–E56. doi: 10.1182/blood-2010-10-314120
Fukuda, T., Miki, T., Yoshida, T., Hatano, M., Ohashi, K., Hirosawa, S., et al. (1995). The murine BCL6 gene is induced in activated lymphocytes as an immediate early gene. Oncogene 11, 1657–1663.
Fukuda, T., Yoshida, T., Okada, S., Hatano, M., Miki, T., Ishibashi, K., et al. (1997). Disruption of the Bcl6 gene results in an impaired germinal center formation. J. Exp. Med. 186, 439–448. doi: 10.1084/jem.186.3.439
Garritano, S., Gemignani, F., Voegele, C., Nguyen-Dumont, T., Le Calvez-Kelm, F., De Silva, D., et al. (2009). Determining the effectiveness of high resolution melting analysis for SNP genotyping and mutation scanning at the TP53 locus. BMC Genet. 10, 5. doi: 10.1186/1471-2156-10-5
Ghetu, A. F., Corcoran, C. M., Cerchietti, L., Bardwell, V. J., Melnick, A., Prive, G. G. (2008). Structure of a BCOR corepressor peptide in complex with the BCL6 BTB domain dimer. Mol. Cell 29, 384–391. doi: 10.1016/j.molcel.2007.12.026
Gore, A. V., Pillay, L. M., Venero Galanternik, M., Weinstein, B. M. (2018). The zebrafish: A fintastic model for hematopoietic development and disease. Wiley Interdiscip. Rev. Dev. Biol. 7, e312. doi: 10.1002/wdev.312
Green, M. R., Vicente-Duenas, C., Romero-Camarero, I., Long Liu, C., Dai, B., Gonzalez-Herrero, I., et al. (2014). Transient expression of Bcl6 is sufficient for oncogenic function and induction of mature b-cell lymphoma. Nat. Commun. 5, 3904. doi: 10.1038/ncomms4904
Haire, R. N., Rast, J. P., Litman, R. T., Litman, G. W. (2000). Characterization of three isotypes of immunoglobulin light chains and T-cell antigen receptor alpha in zebrafish. Immunogenetics 51, 915–923. doi: 10.1007/s002510000229
Hall, C., Flores, M. V., Storm, T., Crosier, K., Crosier, P. (2007). The zebrafish lysozyme c promoter drives myeloid-specific expression in transgenic fish. BMC Dev. Biol. 7, 42. doi: 10.1186/1471-213X-7-42
Hansen, J. D., Zapata, A. G. (1998). Lymphocyte development in fish and amphibians. Immunol. Rev. 166, 199–220. doi: 10.1111/j.1600-065X.1998.tb01264.x
Huang, C., Gonzalez, D. G., Cote, C. M., Jiang, Y., Hatzi, K., Teater, M., et al. (2014). The BCL6 RD2 domain governs commitment of activated b cells to form germinal centers. Cell Rep. 8, 1497–1508. doi: 10.1016/j.celrep.2014.07.059
Hwang, W. Y., Fu, Y., Reyon, D., Maeder, M. L., Tsai, S. Q., Sander, J. D., et al. (2013). Efficient genome editing in zebrafish using a CRISPR-cas system. Nat. Biotech. 31, 227–229. doi: 10.1038/nbt.2501
Hyjek, E., Chadburn, A., Liu, Y. F., Cesarman, E., Knowles, D. M. (2001). BCL-6 protein is expressed in precursor T-cell lymphoblastic lymphoma and in prenatal and postnatal thymus. Blood 97, 270–276. doi: 10.1182/blood.V97.1.270
Ichii, H., Sakamoto, A., Kuroda, Y., Tokuhisa, T. (2004). Bcl6 acts as an amplifier for the generation and proliferative capacity of central memory CD8+ T cells. J. Immunol. 173, 883–891. doi: 10.4049/jimmunol.173.2.883
Jeanmougin, F., Thompson, J. D., Gouy, M., Higgins, D. G., Gibson, T. J. (1998). Multiple sequence alignment with clustal X. Trends Biochem. Sci. 23, 403–405. doi: 10.1016/S0968-0004(98)01285-7
Kwan, W., North, T. E. (2017). Netting novel regulators of hematopoiesis and hematologic malignancies in zebrafish. Curr. Top. Dev. Biol. 124, 125–160. doi: 10.1016/bs.ctdb.2016.11.005
Langenau, D. M., Zon, L. I. (2005). The zebrafish: a new model of T-cell and thymic development. Nat. Rev. Immunol. 5, 307–317. doi: 10.1038/nri1590
LaPensee, C. R., Lin, G., Dent, A. L., Schwartz, J. (2014). Deficiency of the transcriptional repressor b cell lymphoma 6 (Bcl6) is accompanied by dysregulated lipid metabolism. PloS One 9, e97090. doi: 10.1371/journal.pone.0097090
Lawrence, C. (2007). The husbandry of zebrafish (Danio rerio): a review. Aquaculture 269, 1–20. doi: 10.1016/j.aquaculture.2007.04.077
Lee, J., Lee, B. K., Gross, J. M. (2013). Bcl6a function is required during optic cup formation to prevent p53-dependent apoptosis and colobomata. Hum. Mol. Genet. 22, 3568–3582. doi: 10.1093/hmg/ddt211
Lieschke, G. J., Oates, A. C., Crowhurst, M. O., Ward, A. C., Layton, J. E. (2001). Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood 98, 3087–3096. doi: 10.1182/blood.V98.10.3087.h8003087_3087_3096
Linnerz, T., Hall, C. J. (2020). The diverse roles of phagocytes during bacterial and fungal infections and sterile inflammation: lessons from zebrafish. Front. Immunol. 11, 1094. doi: 10.3389/fimmu.2020.01094
Liu, X., Lu, H., Chen, T., Nallaparaju, K. C., Yan, X., Tanaka, S., et al. (2016). Genome-wide analysis identifies Bcl6-controlled regulatory networks during T follicular helper cell differentiation. Cell Rep. 14, 1735–1747. doi: 10.1016/j.celrep.2016.01.038
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Li, Q., Zhou, L., Wang, L., Li, S., Xu, G., Gu, H., et al. (2020). Bcl6 modulates innate immunity by controlling macrophage activity and plays critical role in experimental autoimmune encephalomyelitis. Eur. J. Immunol. 50, 525–536. doi: 10.1002/eji.201948299
Meier, A. B., Basheer, F., Sertori, R., Laird, M., Liongue, C., Ward, A. C. (2022). Granulocyte colony-stimulating factor mediated regulation of early myeloid cells in zebrafish. Front. Biosci. (Landmark Ed.) 27, 110. doi: 10.31083/j.fbl2704110
Melnick, A., Carlile, G., Ahmad, K. F., Kiang, C. L., Corcoran, C., Bardwell, V., et al. (2002). Critical residues within the BTB domain of PLZF and bcl-6 modulate interaction with corepressors. Mol. Cell. Biol. 22, 1804–1818. doi: 10.1128/MCB.22.6.1804-1818.2002
Mondal, A., Sawant, D., Dent, A. L. (2010). Transcriptional repressor BCL6 controls Th17 responses by controlling gene expression in both T cells and macrophages. J. Immunol. 184, 4123–4132. doi: 10.4049/jimmunol.0901242
Nurieva, R. I., Chung, Y., Martinez, G. J., Yang, X. O., Tanaka, S., Matskevitch, T. D., et al. (2009). Bcl6 mediates the development of T follicular helper cells. Science 325, 1001–1005. doi: 10.1126/science.1176676
Ohtani, M., Miyadai, T., Hiroishi, S. (2006). Molecular cloning of the BCL-6 gene, a transcriptional repressor for b-cell differentiation, in torafugu (Takifugu rubripes). Mol. Immunol. 43, 1047–1053. doi: 10.1016/j.molimm.2005.06.036
Ohtsuka, H., Sakamoto, A., Pan, J., Inage, S., Horigome, S., Ichii, H., et al. (2011). Bcl6 is required for the development of mouse CD4+ and CD8alpha+ dendritic cells. J. Immunol. 186, 255–263. doi: 10.4049/jimmunol.0903714
Okabe, S., Fukuda, T., Ishibashi, K., Kojima, S., Okada, S., Hatano, M., et al. (1998). BAZF, a novel Bcl6 homolog, functions as a transcriptional repressor. Mol. Cell. Biol. 18, 4235–4244. doi: 10.1128/MCB.18.7.4235
Page, R. D. (1996). TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358. doi: 10.1093/bioinformatics/12.4.357
Pantano, S., Jarrossay, D., Saccani, S., Bosisio, D., Natoli, G. (2006). Plastic downregulation of the transcriptional repressor BCL6 during maturation of human dendritic cells. Exp. Cell. Res. 312, 1312–1322. doi: 10.1016/j.yexcr.2005.12.020
Perriere, G., Gouy, M. (1996). WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie 78, 364–369. doi: 10.1016/0300-9084(96)84768-7
Phan, R. T., Dalla-Favera, R. (2004). The BCL6 proto-oncogene suppresses p53 expression in germinal-centre b cells. Nature 432, 635–639. doi: 10.1038/nature03147
Pixley, F. J., Xiong, Y., Yu, R. Y., Sahai, E. A., Stanley, E. R., Ye, B. H. (2005). BCL6 suppresses RhoA activity to alter macrophage morphology and motility. J. Cell. Sci. 118, 1873–1883. doi: 10.1242/jcs.02314
Reams, A. B., Roth, J. R. (2015). Mechanisms of gene duplication and amplification. Cold Spring Harb. Perspect. Biol. 7, a016592. doi: 10.1101/cshperspect.a016592
Rosowski, E. E. (2020). Determining macrophage versus neutrophil contributions to innate immunity using larval zebrafish. Dis. Models Mech. 13, dmm041889 doi: 10.1242/dmm.041889
Saitou, N., Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Seelye, S. L., Chen, P. L., Deiss, T. C., Criscitiello, M. F. (2016). Genomic organization of the zebrafish (Danio rerio) T cell receptor alpha/delta locus and analysis of expressed products. Immunogenetics 68, 365–379. doi: 10.1007/s00251-016-0904-3
Sertori, R., Jones, R., Basheer, F., Rivera, L., Dawson, S., Loke, S., et al. (2022). Generation and characterization of a zebrafish IL-2Rγc SCID model. Int. J. Mol. Sci. 23, 2385. doi: 10.3390/ijms23042385
Tang, Q., Iyer, S., Lobbardi, R., Moore, J. C., Chen, H., Lareau, C., et al. (2017). Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single-cell resolution using RNA sequencing. J. Exp. Med. 214, 2875–2887. doi: 10.1084/jem.20170976
Thisse, C., Thisse, B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69. doi: 10.1038/nprot.2007.514
Toney, L. M., Cattoretti, G., Graf, J. A., Merghoub, T., Pandolfi, P. P., Dalla-Favera, R., et al. (2000). BCL-6 regulates chemokine gene transcription in macrophages. Nat. Immunol. 1, 214–220. doi: 10.1038/79749
Trede, N. S., Zon, L. I. (1998). Development of T-cells during fish embryogenesis. Dev. Comp. Immunol. 22, 253–263. doi: 10.1016/S0145-305X(98)00009-3
Wagner, S. D., Ahearne, M., Ko Ferrigno, P. (2011). The role of BCL6 in lymphomas and routes to therapy. Br. J. Haematol. 152, 3–12. doi: 10.1111/j.1365-2141.2010.08420.x
Wang, X., Pan, Y., Zhu, H., Hao, G., Huang, Y., Barnes, V., et al. (2018). An epigenome-wide study of obesity in African American youth and young adults: novel findings, replication in neutrophils, and relationship with gene expression. Clin. Epigenet. 10, 3. doi: 10.1186/s13148-017-0435-2
Wen, Y., Nguyen, D., Li, Y., Lai, Z. C. (2000). The n-terminal BTB/POZ domain and c-terminal sequences are essential for Tramtrack69 to specify cell fate in the developing drosophila eye. Genetics 156, 195–203. doi: 10.1093/genetics/156.1.195
Willett, C. E., Kawasaki, H., Amemiya, C. T., Lin, S., Steiner, L. A. (2001). Ikaros expression as a marker for lymphoid progenitors during zebrafish development. Dev. Dyn. 222, 694–698. doi: 10.1002/dvdy.1223
Willett, C. E., Zapata, A., Hopkins, N., Steiner, L. A. (1997). Expression of zebrafish rag genes during early development identifies the thymus. Dev. Biol. 182, 331–341. doi: 10.1006/dbio.1996.8446
Wittamer, V., Bertrand, J. Y., Gutschow, P. W., Traver, D. (2011). Characterization of the mononuclear phagocyte system in zebrafish. Blood 117, 7126–7135. doi: 10.1182/blood-2010-11-321448
Yamochi, T., Kitabayashi, A., Hirokawa, M., Miura, A. B., Onizuka, T., Mori, S., et al. (1997). Regulation of BCL-6 gene expression in human myeloid/monocytoid leukemic cells. Leukemia 11, 694–700. doi: 10.1038/sj.leu.2400631
Ye, B. H., Cattoretti, G., Shen, Q., Zhang, J., Hawe, N., de Waard, R., et al. (1997). The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat. Genet. 16, 161–170. doi: 10.1038/ng0697-161
Yeh, R. F., Lim, L. P., Burge, C. B. (2001). Computational inference of homologous gene structures in the human genome. Genome Res. 11, 803–816. doi: 10.1101/gr.175701
Yoshida, T., Fukuda, T., Hatano, M., Koseki, H., Okabe, S., Ishibashi, K., et al. (1999). The role of Bcl6 in mature cardiac myocytes. Cardiovasc. Res. 42, 670–679. doi: 10.1016/S0008-6363(99)00007-3
Yuan, S., Wang, X., Hou, S., Guo, T., Lan, Y., Yang, S., et al. (2022). PHF6 and JAK3 mutations cooperate to drive T-cell acute lymphoblastic leukemia progression. Leukemia 36, 370–382. doi: 10.1038/s41375-021-01392-1
Keywords: BCL6A, macrophage, immunity, zebrafish, lymphocyte
Citation: Almohaisen FLJ, Heidary S, Sobah ML, Ward AC and Liongue C (2022) B cell lymphoma 6A regulates immune development and function in zebrafish. Front. Cell. Infect. Microbiol. 12:887278. doi: 10.3389/fcimb.2022.887278
Received: 01 March 2022; Accepted: 03 October 2022;
Published: 28 October 2022.
Edited by:
Stefan Oehlers, Technology and Research (A*STAR), SingaporeReviewed by:
Yueyang Wang, Harvard Medical School, United StatesAnnemarie H. Meijer, Leiden University, Netherlands
Copyright © 2022 Almohaisen, Heidary, Sobah, Ward and Liongue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Clifford Liongue, Yy5saW9uZ3VlQGRlYWtpbi5lZHUuYXU=
†These authors have contributed equally to this work
 Farooq L. J. Almohaisen
Farooq L. J. Almohaisen Somayyeh Heidary1
Somayyeh Heidary1 Alister C. Ward
Alister C. Ward Clifford Liongue
Clifford Liongue