- Department of Veterinary Microbiology and Pathology, Washington State University, Pullman, WA, United States
Borrelia burgdorferi, the Lyme disease pathogen, is maintained in its enzootic life cycle through complex gene regulatory pathways encoded on its uniquely fragmented genome. This genome consists of over 20 plasmids, and the regulatory mechanisms of plasmid maintenance and replication are largely unknown. The bbd21 gene, encoded on lp17 and a member of the paralogous family 32 proteins, was originally proposed to be a putative parA orthologue involved with plasmid partitioning; however, this function has not been confirmed to date. To determine the role of bbd21 in B. burgdorferi, we utilized targeted gene deletion and discovered bbd21 and bbd22 are co-transcribed. The effects of bbd21 and bbd22 deletion on plasmid copy number and mammalian infectivity were assessed. By qPCR, lp17 copy number did not differ amongst strains during mid-exponential and stationary growth phases. However, after in vitro passaging, the mutant strain demonstrated an 8-fold increase in lp17 copies, suggesting a cumulative defect in plasmid copy number regulation. Additionally, we compared lp17 copy number between in vitro and mammalian host-adapted conditions. Our findings showed 1) lp17 copy number was significantly different between these growth conditions for both the wild type and bbd21-bbd22 deletion mutant and 2) under mammalian host-adapted cultivation, the absence of bbd21-bbd22 resulted in significantly decreased copies of lp17. Murine infection studies using culture and qPCR demonstrated bbd21-bbd22 deletion resulted in a tissue colonization defect, particularly in the heart. Lastly, we showed bbd21 transcription appears to be independent of direct rpoS regulation based on similar expression levels in wild type and ΔrpoS. Altogether, our findings indicate the bbd21-bbd22 genetic region is involved with regulation of lp17 plasmid copy number. Furthermore, we propose the possibility that lp17 plasmid copy number is important for microbial pathogenesis by the Lyme disease spirochete.
Introduction
Lyme disease is the most prevalent vector-borne illness in North America with over 30,000 infections (Kugeler et al., 2021) and a health-related cost of $786M USD (Mac et al., 2019) reported each year in the United States alone. The causative agent, Borrelia burgdorferi, survives in an enzootic life cycle that requires a tick and a mammalian host. The bacterium has adapted to this unique lifestyle through the evolution of a small, complex genome composed of a single linear chromosome and up to 22 circular and linear plasmids, depending on the strain (Barbour and Garon, 1987; Barbour, 1988; Ferdows and Barbour, 1989; Stevenson et al., 1996; Casjens et al., 1997; Fraser et al., 1997; Casjens et al., 2000; Iyer et al., 2003). Generally, genes encoded on the chromosome are involved with housekeeping functions such as DNA replication, transcription, translation, solute transport, and metabolism (Casjens et al., 1997; Fraser et al., 1997), while those on plasmids encode virulence factors considered important for host adaptation (Coburn et al., 2021). Global expression of these genes is responsive to environmental cues and known to be regulated by three pathways: the BosR/Rrp2/RpoN/RpoS alternative sigma (σ) factor cascade, the Hk1/Rrp1 two-component system and c-di-GMP, and the stringent response mediated by RelBbu and DksA (Samuels et al., 2022). Linear versus circular plasmid topology has also been shown to play a factor in gene expression (Beaurepaire and Chaconas, 2007). While regulation of plasmid gene expression has been well-studied, less is known regarding regulation of plasmid maintenance and replication.
Typically, bacterial plasmid replication is tightly regulated by fixed copy numbers and permitted growth under specific conditions to achieve stable coexistence of plasmids and minimization of metabolic requirements (del Solar and Espinosa, 2000). In B. burgdorferi, plasmids are similarly reported to be maintained in low copy number of approximately one per chromosome (Hinnebusch and Barbour, 1992; Beaurepaire and Chaconas, 2007), in which there is an estimated one per cell (Morrison et al., 1999). All Borrelia plasmids have a set of related putative plasmid maintenance genes (Dunn et al., 1994; Zückert and Meyer, 1996; Fraser et al., 1997; Casjens et al., 2000). These genes are usually arranged in clusters and can confer autonomous replication (Stewart et al., 2001; Eggers et al., 2002; Stewart et al., 2003; Byram et al., 2004; Beaurepaire and Chaconas, 2005; Dulebohn et al., 2011). The genes encode five paralogous families (PFs) of proteins designated PF32, PF49, PF50, PF57, and PF62 (Fraser et al., 1997; Casjens et al., 2000). The roles of PF49 and PF50 are unknown. PF57 and PF62 are grouped together based on their limited homology and similar function as replication initiators (Stewart et al., 2001; Eggers et al., 2002; Stewart et al., 2003; Beaurepaire and Chaconas, 2005).
Except for lp5 and cp9, a PF32 member is present on all Borrelia plasmids (Casjens et al., 2000), signifying their importance to spirochete survival. The original study on these maintenance genes reported that PF32 is the only Borrelia PF with sequence homology to known DNA maintenance proteins in the ParA superfamily, including ParA involved with plasmid partitioning in Escherichia coli, Soj involved with chromosome segregation in Bacillus subtilis, and RepB involved with plasmid copy number control in Enterococcus faecalis (Zückert and Meyer, 1996). Interestingly, subsequent studies have proposed the bbd21 gene residing on the lp17 plasmid as a putative parA orthologue solely to the ParA protein of E.coli phage P1 (Casjens et al., 2000; Beaurepaire and Chaconas, 2005; Deneke and Chaconas, 2008). The bbd21 locus (NC_001849.2) is composed of a 741 bp open reading frame (ORF) that overlaps by 10 bp with bbd22, a 270 bp ORF (NC_001849.2), and the genes are oriented in the same direction. While bbd22 is predicted as a hypothetical protein with unknown function, there is evidence to support bbd21 as a putative parA orthologue, including a ~25% sequence homology with the ParA family and presence of a Walker box motif characteristic of the ParA proteins (Zückert and Meyer, 1996; Casjens et al., 2000). Additionally, bbd21 has demonstrated ATPase activity required for segregation of plasmids in other bacteria (Deneke and Chaconas, 2008).
Despite this body of evidence, confirmation that bbd21 acts as a parA orthologue with partitioning function is lacking. ParA requires ParB, a DNA binding adaptor protein, to function in plasmid partitioning (Sengupta and Austin, 2011), yet a parB orthologue has not been described in the Borrelia genome to date. This is not surprising, given that the ParB proteins are considered diverse, and sequence homology can be uncommon (Gerdes et al., 2000). Nonetheless, when bbd21 was deleted along with a large segment of lp17 in the noninfectious B31A strain, there were no reported aberrations in plasmid copy number, plasmid stability, or plasmid incompatibility (Beaurepaire and Chaconas, 2005). Previous work from our laboratory likewise found that deletion of a similar region in the infectious B31-5A4 strain did not result in changes to lp17 stability (Casselli et al., 2012). Another study reported multiple transposon insertions in bbd21, suggesting the gene is not required for lp17 replication and maintenance (Lin et al., 2012). Others have also found the PF32 member on cp26 to be dispensable (Tilly et al., 2012). One proposed alternative function to plasmid partitioning in the PF32 genes includes preventing plasmid incompatibility within the cp32 plasmids (Eggers et al., 2002). Another alternative function involves plasmid replication based on physical interaction of recombinant bbd14 and bbd21, although additional experiments in the same study did not support this proposition (Deneke and Chaconas, 2008). These aforementioned studies were limited by qualitative gel electrophoresis techniques, which lacks sensitivity to subtle quantifiable changes in plasmid copy number, replication, and incompatibility. Moreover, in the most recent article on this topic, authors of some of the previous studies casted doubt on the function of bbd21 as a parA orthologue, describing the designation as “premature and potentially inaccurate” (Chaconas and Norris, 2013). As a result, the role of bbd21 in B. burgdorferi remains unknown. Given the high retention of lp17 in B. burgdorferi clinical isolates and during cultivation (Purser and Norris, 2000; Casjens et al., 2018), and the increasing evidence that lp17 serves as a regulatory hub that modulates expression of virulence factors important for host infection (Sarkar et al., 2011; Casselli et al., 2012; Casselli et al., 2019; Medina-Pérez et al., 2020), it is critical to elucidate the mechanisms of lp17 plasmid regulation in order to understand disease pathogenesis and identify potential targets for Lyme disease prevention and therapeutic intervention.
To investigate the role of bbd21, we generated a targeted gene deletion mutant and studied the effect on plasmid copy number and mammalian infectivity. We demonstrate co-transcription with bbd22, alterations to lp17 copy number in vitro and in mammalian host-adapted spirochetes, and impaired mammalian tissue colonization. Our findings indicate the bbd21-bbd22 region is involved with regulation of lp17 plasmid copy number, and furthermore, suggest plasmid copy number is important for microbial infection and tissue colonization by the Lyme disease spirochete.
Materials and Methods
Ethics Statement
The experiments on mice and rats were carried out according to the protocols and guidelines approved by the American Association for Accreditation of Laboratory Animal Care (AAALAC) and by the Office of the Campus Veterinarian at Washington State University (Animal Welfare Assurance A3485-01 and USDA registration number 91-R-002). These guidelines are in compliance with the U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals. The animals were housed and maintained in an AAALAC-accredited facility at Washington State University, Pullman, WA. The Washington State University Institutional Animal Care and Use Committee approved the experimental procedures carried out during the current studies (Animal Safety Approval Forms # 6760 and 6701).
Bacterial Strains and Culture Conditions
Detailed descriptions of the B. burgdorferi strains used in these studies are presented in Supplementary Table 1. The B. burgdorferi isolate B31-5A4 (5A4) was kindly provided by George Chaconas and the 5A4 ΔrpoS mutant was kindly provided by Melissa Caimano. The B31 strain has been sequenced (Fraser et al., 1997; Casjens et al., 2000) and the infectivity and plasmid profiles have been previously described (Purser and Norris, 2000). B. burgdorferi were grown at 35°C and 1.5% CO2 in modified BSK-II (Pollack et al., 1993) supplemented with 6% rabbit serum (Cedarlane Laboratories). Mutant strains were grown with appropriate antibiotics, including kanamycin (200 µg/ml), gentamicin (100 µg/ml), or streptomycin (100 µg/ml). Borrelia cultures were passaged no more than three times prior to use in experiments. Cell densities and growth phase were monitored by dark-field microscopy and enumerated using a Petroff-Hausser counting chamber.
E. coli strains were grown in Luria broth (LB) or LB agar with appropriate antibiotics, including kanamycin (50µg/ml) or gentamicin (5µg/ml).
Plasmid Construction
The deletion mutant construct was generated using the general protocol described in (Magunda and Bankhead, 2016). Primers are shown in Table S2. Because bbd21 and bbd22 open reading frames (ORFs) on lp17 are transcribed in the same direction and overlap by 10 bp, a targeted internal gene deletion approach of bbd21 was utilized to minimize disruption to bbd22 and its promoter region. The internal deletion was designed 260 bp upstream of the bbd22 start codon to avoid putative promoter sequences. The lp17 target region of 1195 bp including the bbd21 ORF and a 305 bp upstream and a 149 bp downstream flanking region was PCR amplified from wild-type 5A4 genomic DNA with primers P933 and P934. The amplicon was cloned into the vector pCR-Blunt-II-TOPO (Invitrogen) and maintained in One Shot Stbl3 cells (Invitrogen). The internal deletion was generated by PCR amplification with primers P996 and P997 of this plasmid to exclude a 408 bp region within the bbd21 ORF and to insert AgeI and PacI restriction sites. The gentamicin-resistance cassette (gent) driven by a flgB promoter was PCR amplified with primers P998 and P508 from pPH25 (Hove et al., 2014) with insertion of AgeI and PacI restriction sites. The amplicon was cloned into the vector pCR-Blunt-II TOPO (Invitrogen) and propagated in Top10 cells (Invitrogen). Following restriction enzyme digest with AgeI and PacI, flgB-gent was ligated into the deleted region of bbd21, generating the plasmid pJW16. The construct was maintained in EC19 cells (Hove et al., 2014) as EC276. DNA was isolated using a plasmid midikit (Sigma). Successful cloning was verified for correct size and orientation by restriction enzyme digest and sequencing before transformation into B. burgdorferi competent cells.
Mutant Generation and Confirmation
All transformations were performed using electrocompetent B. burgdorferi prepared as previously described (Bankhead and Chaconas, 2007; Samuels et al., 2018). E. coli DNA was isolated using a plasmid midikit (Sigma), and B. burgdorferi DNA was isolated using a plasmid maxikit (Sigma). The deletion mutant strain, which was found to lack expression of both bbd21 and bbd22 (Δ21-22) was generated by allelic exchange following electroporation of 50 µg of DNA isolated from EC276 and transformed into the 5A4 wild-type strain. After numerous unsuccessful attempts to complement bbd21 back into its native locus in the deletion mutant, a previously published alternative strategy using whole plasmid replacement (Grimm et al., 2004; Imai et al., 2013; Raffel et al., 2014) was utilized to generate the complement strain, lp17comp. A 5A4 lp17 mutant in which bbd01-bbd03 were replaced with a kanamycin resistance cassette (kan) by telomere resolution was utilized (Tourand et al., 2003; Casselli et al., 2019). This strain was previously reported to not affect murine infectivity or tissue colonization (Casselli et al., 2019). Approximately 18 µg of plasmid DNA was electroporated into Δ21-22 electrocompetent cells. Mutants were initially screened by PCR followed by Southern blot analyses for the respective presence or absence of the internally deleted portion of bbd21, gent, and kan (primers shown in Supplementary Table 2). Endogenous plasmid content was identified by multiplex PCR using primers previously described (Bunikis et al., 2011).
Southern Blot Hybridization
B. burgdorferi genomic DNA was isolated by phenol-chloroform extraction. Genomic DNA was separated using 0.65% agarose gel electrophoresis at 80V for 18 h with pulse field inversion and recirculating buffer followed by bidirectional transfer to two nylon membranes (Roche). Probes for bbd12-bbd13 of lp17, the internal deletion of bbd21, gent, and kan were generated from plasmid template DNA using primers indicated in Table S2 with the DIG Probe Synthesis kit (Roche), per manufacturer’s instructions. Bands were detected with the DIG Luminescent Detection kit (Roche).
In Vitro Growth Assays
Growth assays were performed as previously described (Casselli et al., 2012). Briefly, B. burgdorferi strains were grown to late log phase and subcultured to 105 cells/ml. Cell densities were determined at 24 h intervals for 9 days. Values were expressed as mean density ± standard deviation from three experiments.
In Vitro Passaging
A protocol similar to (Grimm et al., 2003) was followed for in vitro passaging of B. burgdorferi. Briefly, wild type, Δ21-22, and lp17comp were inoculated from frozen glycerol stocks and grown in parallel in 5 ml cultures of liquid BSK II medium with appropriate antibiotics to late exponential phase (5 x 107 to 1 x 108 cells/ml) and then passaged 1:500 into fresh medium 25 times. Final cultures were grown to a density of 8 x 107 to 1.0 x 108 cells/ml. All cultures were pelleted and stored at -20°C.
lp17 Plasmid Copy Number
To investigate lp17 plasmid copy number over the growth curve, genomic DNA was extracted from aliquots of 10 ml cultures of wild-type, Δ21-22, and lp17comp strains grown to mid-exponential phase (1.8 x 107 to 2.6 x 107 cells/ml) and then stationary phase (1.1 x 108 to 1.3 x 108 cells/ml). Next, the effect of multiple passages on lp17 plasmid copy number was studied. Genomic DNA was extracted from strains passaged 25 times and was compared to controls (P1), which were the same strains cultivated directly from glycerol stocks that were passaged no more than three times prior to the start of experiments. To determine the effect of different growth environments on plasmid copy number, mammalian-host adapted wild-type, Δ21-22, and lp17comp spirochetes were compared to in vitro-cultivated spirochetes. qPCR for bbd14 and flaB (primers and probes listed in Table S2) was performed in duplex with three technical replicates per sample using TaqMan probes and the QX200 Droplet Digital PCR (ddPCR) system (Bio-Rad Laboratories), including the QX200 Droplet Generator, the PX1 PCR Plate Sealer, and the QX200 Droplet Reader, all per manufacturer’s instructions. PCR was performed on the C1000 Touch Thermal Cycler (Bio-Rad Laboratories) using the following cycling conditions: enzyme activation at 95°C for 10 min, amplification for 40 cycles with a ramp rate of 2.5°C/s for denaturation at 94°C for 30 s and annealing and extension at 60°C for 1 min, enzyme deactivation at 98°C for 10 min, and an optional hold at 4°C. Values were expressed as relative copies of bbd14 per flaB and calculated as an average ± standard error mean.
Mammalian Host-Adapted Spirochetes
Mammalian host-adapted spirochetes were generated by cultivation in dialysis membrane chambers (DMCs) implanted into the peritoneal cavity of rats, as previously described (Caimano, 2018). The following modifications and specifications to the DMC protocol were applied. Sprague-Dawley rats (Envigo, Livermore, CA) were male, 7-8 weeks old, and weighed 220 – 250 g. Sterile surgical instruments included a new disposable No. 10 scalpel blade used for each animal, and an instrument pack, including 13.3 cm Mayo-Hegar needle holders, 11 cm iris scissors, 14 cm rat tooth forceps, and 14 cm Adson forceps, used aseptically on up to 3 animals. Instead of ketamine/xylazine for anesthesia, rats were maintained on isoflurane gas (1-3%; VetOne). In addition to perioperative and postoperative carprofen, rats were subcutaneously administered preoperative carprofen (5 mg/kg, VetOne) 24 h prior to surgery and perioperative buprenorphine SR (1 mg/kg, ZooPharm LLC), as well as local lidocaine (5 mg/kg, VetOne) injected as a line block along the planned incision site. Perioperative fluids (LRS or saline, 3-5% body weight) were administered to support hydration, and animal body temperature was maintained with a heating pad during surgery prep and surgery. Instead of wound clips, the skin incision was closed with a Precise Multi-Shot MS Disposable Skin Stapler (3M) and liquid skin adhesive. Each DMC contained 12 ml of diluted culture. All DMCs were explanted on day 14 post surgery.
RT-PCR/qRT-PCR
For confirmation of functional gene deletion in the Δ21-22 mutant and assessment of rpoS regulation of bbd21, three 5ml cultures of each Borrelia strain were grown to late log phase (8 x 107 to 1 x 108 cells/ml), and RNA was isolated using the RNeasy Mini Kit (Qiagen). DNA contamination was removed using the TURBO DNA-free kit (Invitrogen), per manufacturer instructions, and confirmed by PCR amplification of the flaB gene (primers P411 and P412 in Table S2). cDNA for all samples was synthesized using the iScript cDNA Synthesis kit (Bio-Rad Laboratories) and assayed in three technical replicates for transcript levels of target genes quantified using the ddPCR system described above. Primers and probes are shown in Table S2. Samples were normalized to flaB. Values were expressed as an average ± standard error mean.
Infection, Recovery, and Quantification of B. burgdorferi From Mice
Mammalian infectivity was assessed similarly to previous studies (Casselli et al., 2019). Four- to six-week-old, male immunocompetent C3H/HeJ mice (C3H; Jackson, Sacramento, CA) were infected by needle inoculation in the subcutis over the shoulder blades with 100 ul of BSK-II containing 1 x 102, 1 x 103, and 1 x 104 B. burgdorferi cells. Three mice were used per group.
For spirochete detection by culture, blood, ear, heart, urinary bladder, and tibiotarsal joint were sampled at indicated time points and cultured in BSK-II supplemented with 20μg/ml phosphomycin, 50 μg/ml rifampicin, and 2.5 μg/ml amphotericin B. The presence or absence of spirochetes were assessed by dark-field microscopy for 4 weeks. The 50% infectious dose (ID50) was calculated using the Reed-Muench method (Reed and Muench, 1938).
For bacterial burden quantification, mice were infected with 1 x 104 B. burgdorferi for 3 weeks and then humanely euthanized. Skin from the inoculation site, heart, and urinary bladder were snap-frozen in liquid nitrogen and stored at -80°C. DNA was extracted using the DNeasy Blood and Tissue kit (Qiagen) and then purified with the Genomic DNA Clean and Concentrator-10 kit (Zymo Research), both per manufacturer’s instructions. Three technical replicates were performed for each of the three biological replicates. qPCR for B. burgdorferi flaB and mouse β-actin was performed as described above with an annealing and extension temperature of 58°C. Relative copies of flaB per mouse actin were expressed as an average ± standard error mean.
Statistics
For the cell density and qPCR data over the growth curve, comparisons were performed using a paired, two-tailed Student’s t-test was performed. For all other qPCR and qRT-PCR analyses, comparisons were made using an unpaired, two-tailed Student’s t-test. P < 0.05 was considered significant unless otherwise noted.
Results
Generation and Characterization of Δ21-22 and lp17comp B. burgdorferi Clones
The bbd21 gene was proposed as a potential parA orthologue involved with plasmid maintenance (Zückert and Meyer, 1996; Casjens et al., 2000). However, targeted deletion of the individual bbd21 gene and detailed, quantitative analyses on its impact have not been performed to date. To investigate the function of bbd21, a deletion mutant and its complement were generated in the B31-5A4 background (Figure 1A). A detailed schematic including primer and probe descriptions is presented in Supplementary Figure 1. Briefly, the deletion mutant (Δ21-22) was made by insertion of a gentamicin resistance cassette into the bbd21 ORF through allelic exchange. After numerous unsuccessful attempts to complement bbd21 into its native locus in the deletion mutant, a previously published alternative strategy using whole plasmid replacement (Grimm et al., 2004; Imai et al., 2013; Raffel et al., 2014) was utilized to generate the complement strain, lp17comp. The lp17comp strain was generated by whole plasmid transformation of a previously published lp17 mutant in which bbd01-bbd03 were replaced by a kanamycin resistance marker, and the mutant had no reported effect on murine infectivity and tissue colonization (Tourand et al., 2003; Casselli et al., 2019). Field inversion gel electrophoresis and Southern blot analyses confirmed successful genetic manipulation of the mutant strains (Figure 1B). The field inversion gel demonstrates 1 µg of genomic DNA loaded per lane, 23 kb and 9.4 kb size molecular markers, and the location of the lp17 plasmid. The size of lp17 for wild type is 16,823 bp. For Δ21-22, the 408 bp internal deletion of bbd21 and the addition of 988 bp for the flgB promoter and gentamicin resistance cassette resulted in a size of 17,403 bp for lp17. For lp17comp, the 1395 bp deletion of bbd01-bbd03 and addition of 2608 for the flgB promoter, kanamycin resistance cassette, and part of the plasmid vector resulted in a size of 18,036 for lp17. Southern blot analyses confirmed maintenance of lp17 in all strains by detection of bbd12-bbd13 near the plasmid replication initiator; the absence of bbd21 in the deletion mutant and restoration in the complement; and the presence of the appropriate antibiotic resistance markers in the respective mutants. Additionally, PCR amplification of the region flanking the internal bbd21 deletion supported successful insertion of the antibiotic resistance gene into bbd21 and restoration of the gene in the complement (Supplementary Figure 2). RT-PCR analyses revealed that bbd21 deletion resulted in loss of bbd21 and bbd22 transcription, and that a single transcript from overlapping regions of these genes indicated co-transcription (Figure 1C). Significantly decreased transcription of in vitro bbd21 corroborated disruption of the gene in the deletion mutant (Figure 1D) and demonstrated restoration of wild-type levels in the complement clone. The endogenous plasmid profiles of both mutants were similar to the wild type except for the loss of cp9 (data not shown), a plasmid that is often lost in culture and considered nonessential for mouse infectivity (Golde and Dolan, 1995; Purser and Norris, 2000; Labandeira-Rey and Skare, 2001; McDowell et al., 2001).
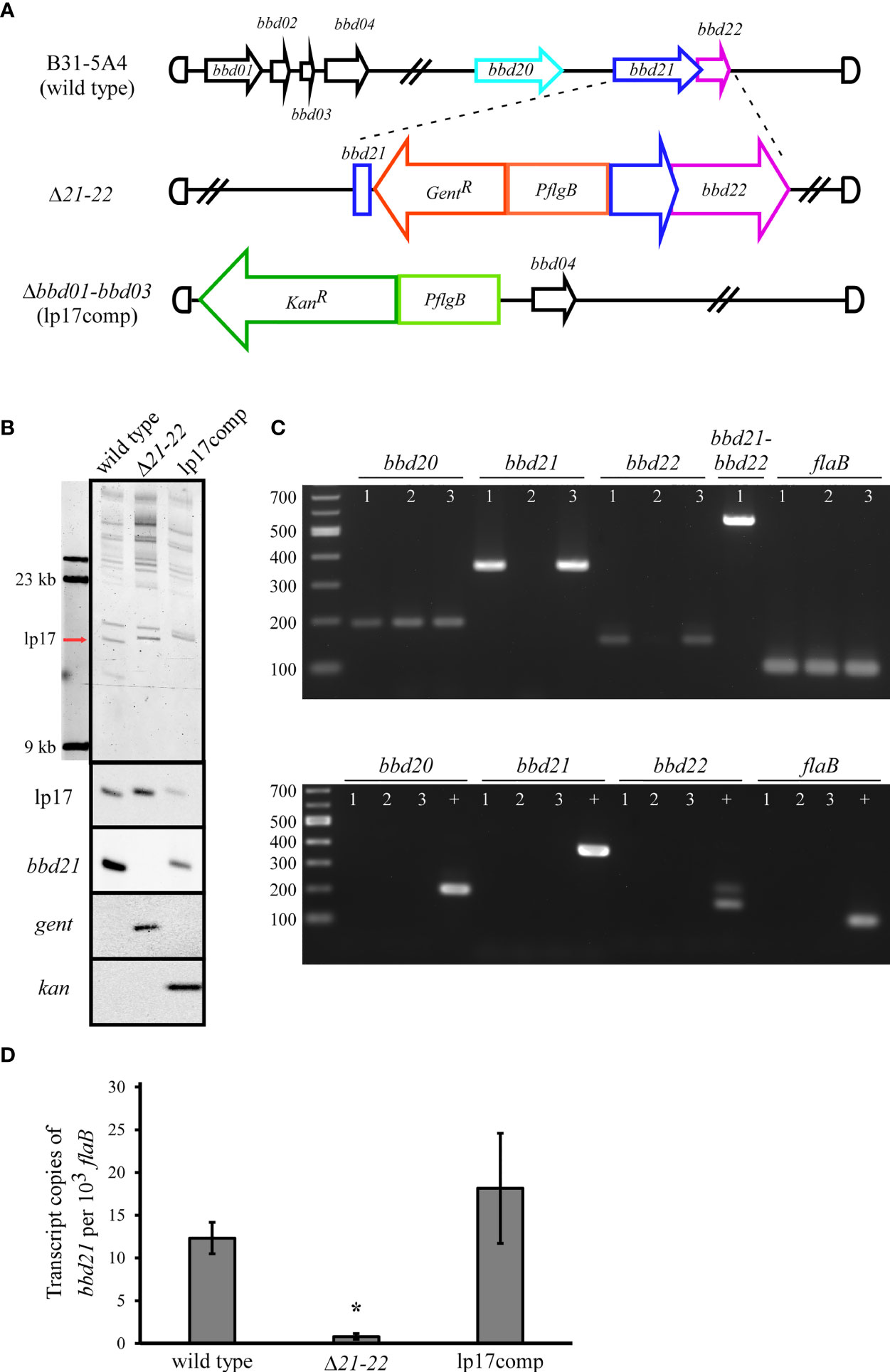
Figure 1 Characterization of the bbd21-bbd22 mutant (Δ21-22) and complement (lp17comp). (A) Schematic of the B. burgdorferi strains generated in this study. The deletion mutant was generated in the 5A4 strain by allelic exchange resulting in a targeted internal deletion within bbd21 and replacement by a gentamicin resistance cassette driven by a flgB promoter. The lp17comp clone was generated by whole plasmid transformation of a 5A4 mutant in which bbd01-bbd03 were replaced by a kanamycin resistance cassette driven by flgB. (B) The field inversion gel electrophoresis demonstrates 1 µg of genomic DNA loaded per lane, 23 kb and 9.4 kb size molecular markers, and the location of the lp17 plasmid (red arrow). The expected increased plasmid size of the deletion mutant and complement strains result from addition of antibiotic resistance markers. Southern blot analyses of wild-type, Δ21-22, and lp17comp genomic DNA probed for lp17, bbd21, kanamycin, and gentamicin. (C) The upper gel depicts reverse-transcription PCR assessment of bbd21 deletion. Total RNA was isolated, converted to complementary DNA, and used to amplify regions within bbd21, the flanking genes bbd20 and bbd22, an overlapping region of bbd21-bbd22, and flaB. 1 = wild type, 2 = Δ21-22, and 3 = lp17comp. A 194 bp region of bbd20 was amplified using P1399 and P1400, a 362 bp region within the bbd21 internal deletion was amplified using P1056 and P1057, a 157 bp region of bbd22 was amplified using P1401 and P934, a 565 bp region overlapping bbd21 and bbd22 was amplified using P1299 and P934, and a 105 bp region of flaB was amplified using P411 and P412. The lower gel depicts RNA template (no transcriptase) negative controls and wild-type genomic DNA positive controls (+). (D) qRT-PCR analyses of bbd21 transcription from in vitro-cultivated wild-type, Δ21-22, and lp17comp strains. Transcript levels for bbd21 were normalized to 103 copies of flaB using a TaqMan-assay. Values were expressed as an average of three experiments ± standard error mean. *p<0.05 (Student’s t-test).
It has been previously reported that B. burgdorferi lacking bbd16 to bbd25 had impaired growth throughout the entirety of in vitro cultivation (Casselli et al., 2012). To assess the impact of bbd21-bbd22 deletion on growth, wild-type, Δ21-22, and lp17comp strains were grown in liquid BSK-II, and averaged densities were compared every 24 h of the growth curve. The mutant clones showed altered growth compared to the wild type (Figure 2). Average bacterial densities at each time point are listed in Supplementary Table 3. The Δ21-22 strain displayed a significantly delayed early growth on days 1 and 2 as well as decreased spirochetes in the early stationary phase on day 6. Conversely, lp17comp displayed significantly increased growth density compared to wild type during exponential growth from days 2 – 7. Overall, the altered growth exhibited by these mutants indicate bbd21 and bbd22 affect spirochete replication.
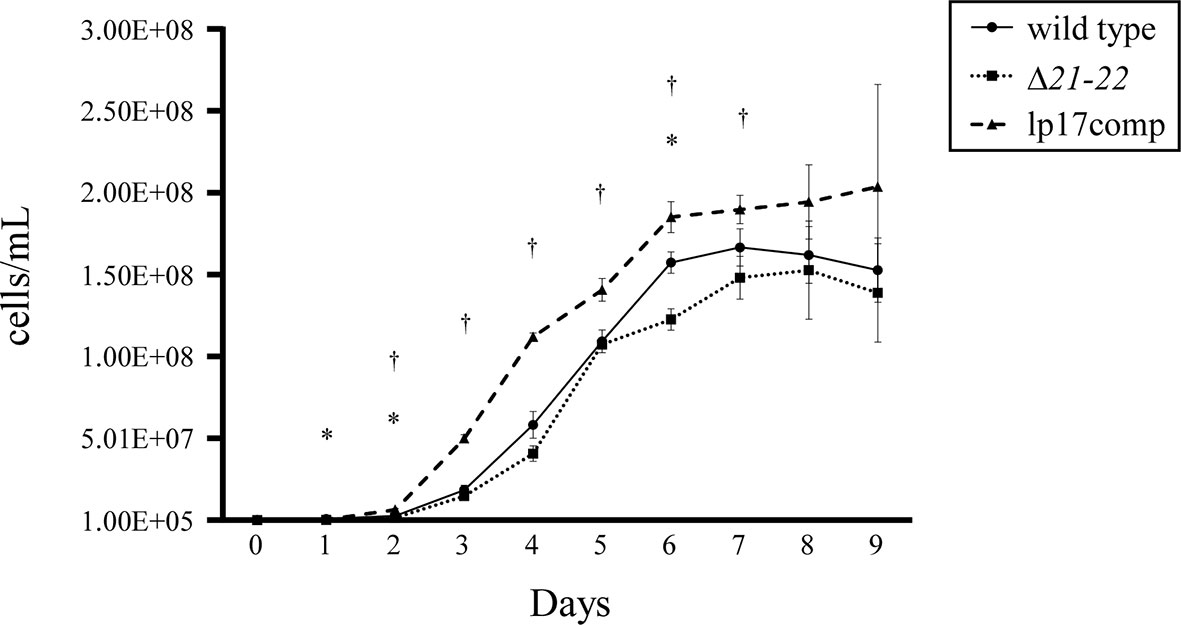
Figure 2 Deletion of bbd21-bbd22 leads to decreased growth rate in vitro. At 24-h intervals, data points represent the average of cells/ml from three experiments, and error bars represent standard deviation. *wild type vs. Δ21-22, †wild type vs. lp17comp; p<0.05 (Student’s t-test).
Deletion of Bbd21-bbd22 Results in Dysregulated Lp17 Copy Number Following In Vitro Passaging
Previous studies have suggested that bbd21 is involved with lp17 plasmid replication based on physical interaction of recombinant BBD21 with BBD14 (the protein product of the gene required for lp17 replication) during purification, although this was not confirmed in additional experiments by the same authors (Deneke and Chaconas, 2008). To further investigate the potential role of bbd21 on lp17 copy number, plasmid copies were evaluated over the growth curve and over multiple generations. While there were significantly increased lp17 copies for each strain in the stationary phase (~1.2 x 108 cells/ml) compared to mid-exponential phase (~2 x 107 cells/ml), there was no difference among strains at either growth phase (Figure 3A). To investigate the effect of bbd21-bbd22 deletion over multiple generations, qPCR probing for bbd14 was performed on genomic DNA to quantify the number of lp17 plasmids in each B. burgdorferi strain following 25 in vitro passages. Results showed a significant ~6-8-fold increase in lp17 copy number by qPCR in passaged Δ21-22 spirochetes compared to wild type and lp17comp (Figure 3B). These results indicate the bbd21-bbd22 region is involved with lp17 plasmid copy number after passaging.
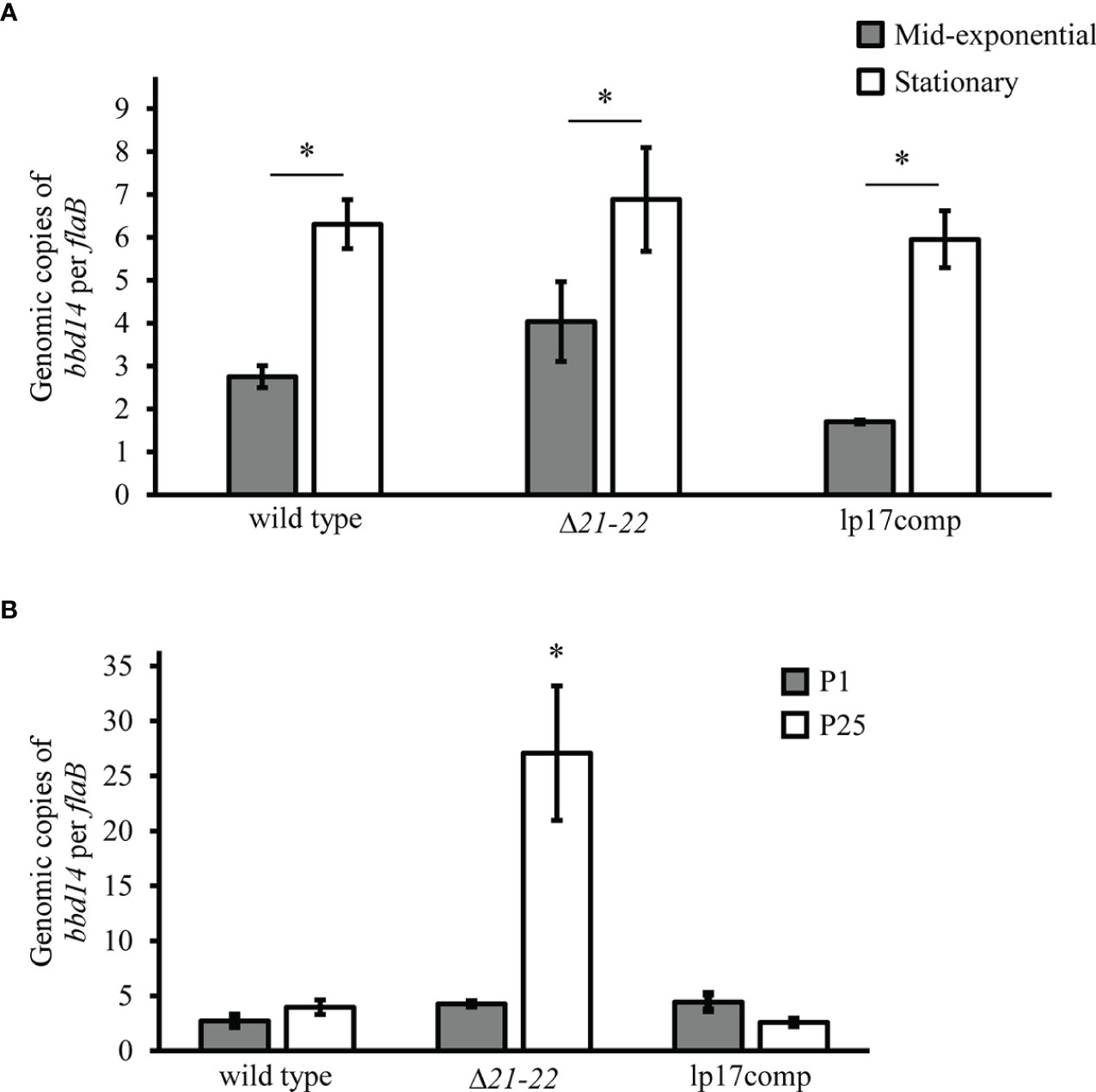
Figure 3 Deletion of bbd21-bbd22 results in changes to lp17 plasmid copy number at different stages of the growth curve and following in vitro passaging. (A) Quantitative PCR analysis of genomic DNA from wild-type, Δ21-22, and lp17comp strains grown to mid-exponential phase (~2 x 107 cells/ml; gray bars) and stationary phase (~1.2 x 108 cells/ml; white bars). Bars indicate the average relative copies of bbd14 per flaB from three experiments ± standard error mean. *p<0.05 (Student’s t-test). (B) in vitro Quantitative PCR of genomic DNA isolated from bacterial strains sepassaged 25 times in vitro. Three samples per strain were used, and values were expressed as averaged copies of bbd14 per flaB ± standard error mean. Samples passaged 25 times (P25) are displayed in white bars, and those cultivated from glycerol stocks (P1) not passaged more than three times prior to the start of experiments are displayed as gray bars. Error bars represent standard error mean. *p<0.05 (Student’s t-test).
Bbd21 Transcription and Lp17 Copy Number Differ Between In Vitro- and DMC-Cultivated Spirochetes
The B. burgdorferi 5A4 strain used in this study was isolated from a tick host (Burgdorfer et al., 1982). It is well-known that the spirochete undergoes drastic antigenic profile changes in response to host environmental signals. While differential gene expression between tick and mammalian host environments is well-studied in B. burgdorferi, differential endogenous plasmid copy number has not been investigated. To explore the role of bbd21-bbd22 in mammalian host-adapted spirochetes, bacteria were cultivated using the dialysis membrane chamber (DMC) model (Caimano, 2018). First, to establish if bbd21 is upregulated or downregulated in the mammalian host, transcript levels in wild-type spirochetes from in vitro- and DMC-cultivated spirochetes were assessed. Expression of bbd21 was significantly increased 3-fold in DMCs compared to in vitro (Figure 4A). This upregulation indicates bbd21 is induced during mammalian host adaptation.
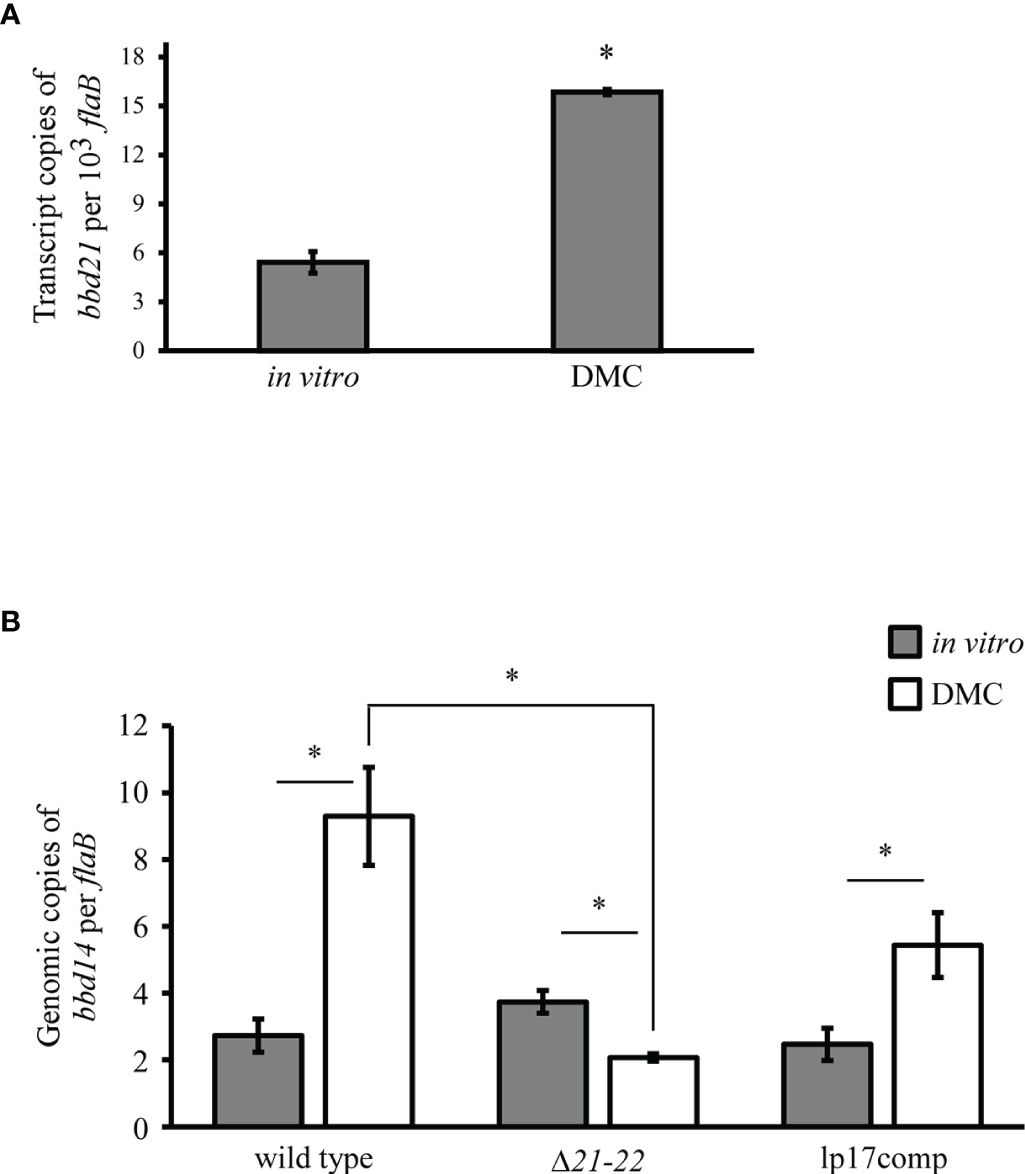
Figure 4 bbd21 transcription and lp17 copy number are expressed differently in DMC-cultivated vs. in vitro-cultivated spirochetes. Mammalian host-adapted spirochetes were generated in dialysis membrane chambers (DMCs) implanted into the peritoneal cavity of rats. (A) qRT-PCR analysis of bbd21 expression in in vitro- vs. DMC-cultivated wild type spirochetes. Bars represent the mean values of three independent experiments, and error bars represent standard error mean. *p<0.05 (Student’s t-test). (B) qPCR analyses of lp17 copy number in wild-type, Δ21-22, and lp17comp strains. In vitro conditions are depicted as gray bars, and DMC conditions are depicted as white bars. The bars represent the mean values of three independent experiments, except in the Δ21-22 DMC-cultivated spirochetes in which one animal died during surgery (n=2). The error bars represent standard error mean. *p<0.05 (Student’s t-test).
Because bbd21 was induced during mammalian host adaptation, the impact on lp17 copy number was evaluated between in vitro and DMC cultivation. By qPCR, all strains displayed significantly different lp17 copy number between in vitro and DMC conditions. Wild-type spirochetes grown in vitro harbored 3.4-fold fewer copies of lp17 compared to spirochetes grown in DMCs (Figure 4B). The inverse was observed between Δ21-22 spirochetes grown in vitro and in DMCs. For lp17comp, lp17 copy number was 2.2-fold decreased under in vitro conditions compared to DMC. in vitro. While there was no statistical difference among strains grown under in vitro conditions, differences were observed under DMC conditions. The Δ21-22 spirochetes contained significantly fewer copies of lp17 by 4.5-fold compared to wild-type. The mutant also contained 2.6-fold fewer copies of lp17 compared to lp17comp, although this was not statistically significant. Together, these results suggest that lp17 plasmid copy number is impacted by in vitro versus DMC growth conditions and by the expression of bbd21-bbd22.
Deletion of Bbd21-Bbd22 Results in Impaired Mouse Heart Tissue Colonization
Given the difference in lp17 copy number observed in DMCs by the Δ21-22 strain combined with the recognition of lp17 as a plasmid encoding virulence factors important for mammalian adaptation (Sarkar et al., 2011; Casselli et al., 2012; Casselli et al., 2019; Medina-Pérez et al., 2020), mouse infection studies were performed to determine if the lack of bbd21-bbd22 influences the ability of B. burgdorferi to colonize mammalian tissues. C3H mice were infected, and samples of blood, ear, heart, urinary bladder, and tibiotarsal joint were collected for culture. Infection rates were dose dependent for all strains, and the ID50 for Δ21-22 was at least 10-fold higher than wild type for all tissues (Table 1).
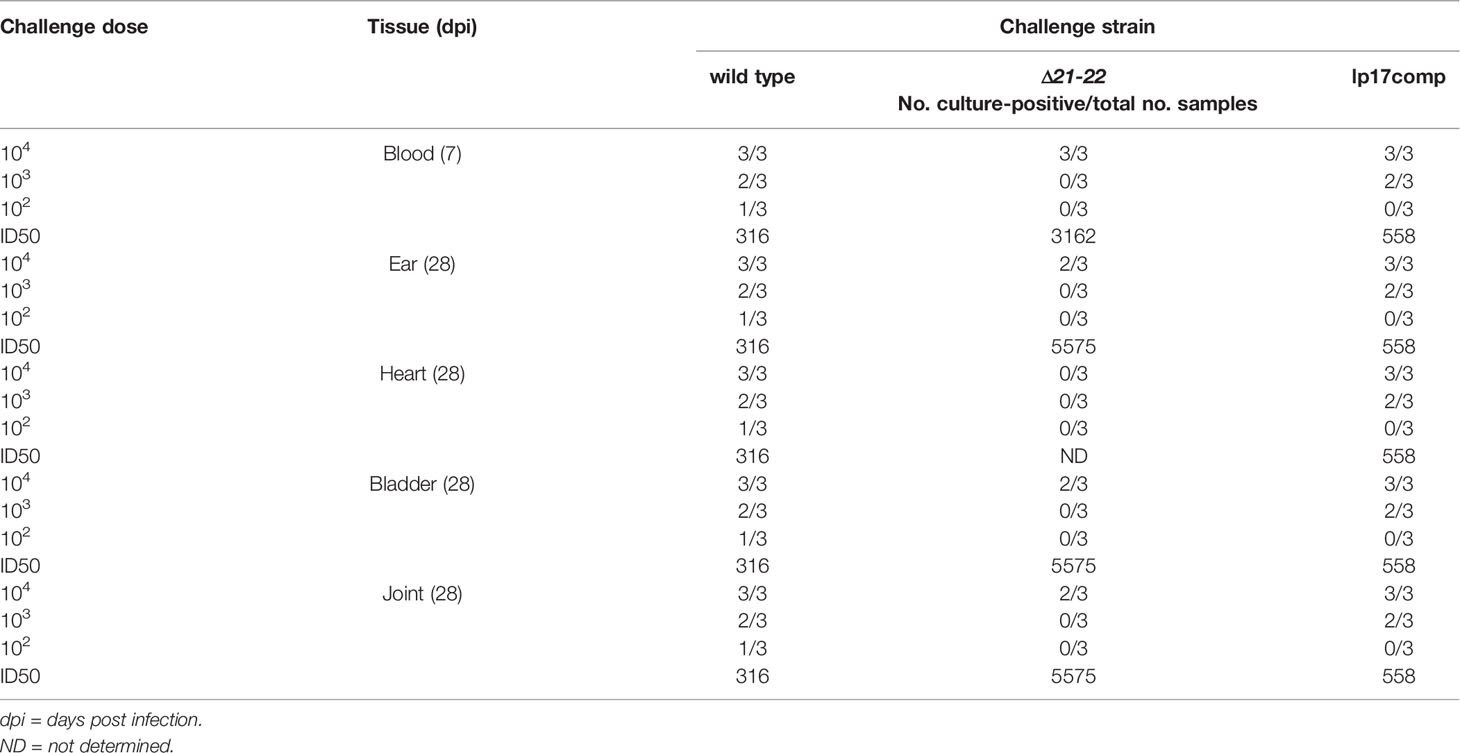
Table 1 Minimum infectious dose and tissue colonization of wild-type, Δ21-22, and lp17comp B. burgdorferi strains in immunocompetent C3H mice.
In addition to culture, qPCR was performed to quantify spirochete burden. A mouse infection study was performed identically to the infection and spirochete recovery experiments at the 104 inoculation dose. Skin at the inoculation site, heart, and bladder were harvested, and genomic DNA was analyzed by qPCR. Consistent with the culture results, spirochete burden in the heart was significantly decreased 2.4-fold for Δ21-22 compared to wild type and lp17comp (Figure 5). For the dermal inoculation site, Δ21-22 and lp17comp had mildly increased burden compared to wild type. For the bladder, lp17comp was approximately twice the burden of wild type and Δ21-22, although neither the skin nor bladder exhibited statistically significant differences. Altogether, these culture and qPCR results indicate that bbd21 is involved with mouse tissue dissemination or colonization, particularly in the heart.
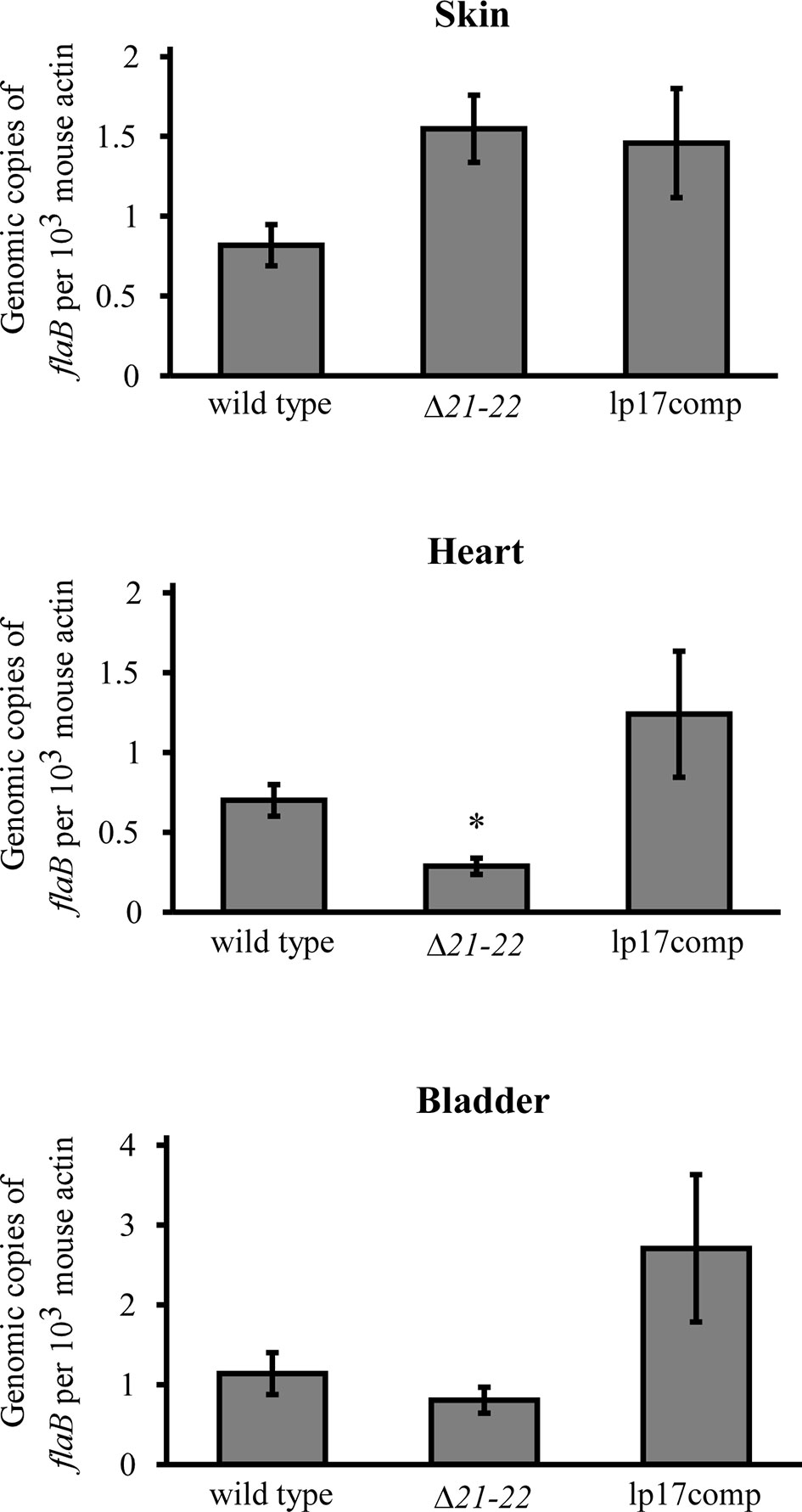
Figure 5 bbd21-bbd22 is required for efficient colonization of the heart in immunocompetent mice. Three C3H mice per group were infected in the subcutis over the dorsum with 1x104 spirochetes. Skin at the inoculation site, heart, and urinary bladder were collected at 21 days post-infection. Spirochete burden was assessed by qPCR analysis of genomic DNA isolated from these tissues. Gray bars represent the mean values of (B) burgdorferi flaB copies normalized to 103 mouse actin copies, and error bars represent standard error mean. *p<0.05 (Student’s t-test).
Expression of Bbd21 Is Independent of RpoS
The current study found that bbd21 transcription was induced in mammalian host-adapted spirochetes. Additionally, lp17 copy number was observed to be altered in the deletion mutant in this environment. This begs the question if bbd21 is regulated by the canonical alternative σ factor cascade. The BosR/Rrp2/RpoN/RpoS alternative σ factor cascade is the most well-studied global regulator of B. burgdorferi gene expression throughout the enzootic life cycle (Hubner et al., 2001; Yang et al., 2003; Caimano et al., 2007; Hyde et al., 2009; Ouyang et al., 2009; Ouyang et al., 2011; Groshong et al., 2012; Ouyang et al., 2014; Caimano et al., 2019). Despite the global regulation this network displays, neither bbd21 nor bbd22 is reported to be impacted by mutations to this pathway in microarray experiments (Caimano et al., 2007; Ouyang et al., 2009; Caimano et al., 2019). To investigate the potential role of rpoS regulation on bbd21, wild type and ΔrpoS spirochetes were cultivated in DMCs and evaluated for bbd21 transcript levels by qRT-PCR (Figure 6). The results from qRT-PCR analysis showed that there was no statistical significance between expression levels of the two strains, which further suggests that rpoS does not regulate bbd21 expression.
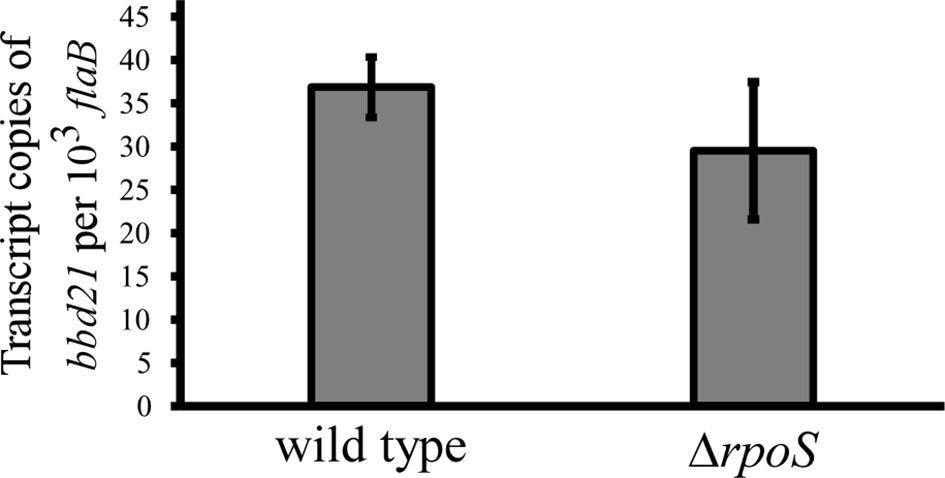
Figure 6 bbd21 transcription is independent of rpoS regulation. Mammalian host-adapted spirochetes were generated in dialysis DMCs implanted into the peritoneal cavity of rats. Transcription of bbd21 was compared by qRT-PCR analysis of wild-type and ΔrpoS spirochetes in three independent experiments. Gray bars represent mean values of bbd21 transcript copies normalized to 103 flaB, and error bars represent standard error mean. There is no statistical difference between the two strains (Student’s t-test).
Discussion
Our findings reveal that a role of the B. burgdorferi gene products of the bbd21-bbd22 genetic region is lp17 plasmid copy number regulation. We demonstrated that deletion of bbd21-bbd22 expression resulted in a significant increase of lp17 copy number following in vitro passaging, and that complementation returned plasmid copy number to wild-type levels. Copy number of lp17 was previously reported to be one copy per chromosome by DNA hybridization (Hinnebusch and Barbour, 1992), which was limited by the sensitivity of electrophoretic analysis. By qPCR, a previous study determined lp17 copy number to be ~3.1 per the chromosome end and 0.8 per the chromosome center, with the discrepancy attributed to multiple replication centers and initiation events on the chromosome (Beaurepaire and Chaconas, 2007). In the current study, wild-type and lp17comp spirochetes displayed similar lp17 copies per chromosome end ranging from 2.3 to 5.2. Additionally, an earlier study speculated that the absence of PF32 would result in loss of plasmids during in vitro cultivation (Zückert and Meyer, 1996). On the contrary, the present study revealed an 8-fold increase of lp17 copies following bbd21-bbd22 deletion following in vitro passaging. The drastic increase observed in lp17 copy number by the Δ21-22 mutant was not observed in previous reports involving bbd21 deletion (Beaurepaire and Chaconas, 2005; Casselli et al., 2012), which is likely due to conducting multiple passages coupled with quantitative analysis in the present study.
While our findings suggest that the bbd21-bbd22 genetic region is involved with plasmid copy number regulation, the precise function (i.e., plasmid replication, plasmid partitioning, or plasmid copy number control) remains obscure. The finding of increased plasmid copy number following in vitro passaging in Δ21-22 spirochetes may suggest that bbd21 functions less as a strict ParA partitioning protein and more like the RepB regulator of plasmid copy number in Enterococcus faecalis (Weaver et al., 1993). RepB is a member of the ParA superfamily of ATPases and contains sequence identity with bbd21 (Zückert and Meyer, 1996). In E. faecalis, RepB functions with RepC, the ParB orthologue, to stabilize the plasmid partitioning complex (Francia et al., 2007). However, a RepC/ParB orthologue has yet to be identified in the B. burgdorferi genome. Furthermore, the finding of significantly decreased lp17 copy number in DMC-cultivated spirochetes lacking bbd21-bbd22 expression contrasts the in vitro passaging results and may suggest bbd21-bbd22 serves a distinct function for plasmid copy number regulation during mammalian adaptation.
Several reports have speculated that PF members may act in trans to supply gene products required for plasmid maintenance, replication, or partition to another plasmid that is lacking such factors (Eggers et al., 2002; Stewart et al., 2003; Beaurepaire and Chaconas, 2005; Beaurepaire and Chaconas, 2007; Tilly et al., 2012; Chaconas and Norris, 2013). In the current study, the deletion of bbd21-bbd22 and subsequent increase in lp17 copy number following in vitro passaging and decrease in lp17 copy number in mammalian host-adapted spirochetes suggests that this PF32 member is not compensated in trans by PF32 members encoded on other plasmids in the B. burgdorferi genome. Future studies are warranted to investigate if bbd21-bbd22 may impact copy number of other endogenous plasmids as well.
In addition, we demonstrated that B. burgdorferi strains can harbor significantly different copies of lp17 under in vitro versus mammalian host-adapted conditions. This suggests that mammalian host factors not only influence transcriptome expression, but also bbd21-bbd22 and endogenous plasmid copy number. This difference in copy number raises the possibility of gene dose-associated expression. That is, with differing copy numbers of lp17 due to in vitro versus DMC cultivation or bbd21-bbd22 deletion, there may be altered expression of all genes encoded on the plasmid based on copy number variation. Given that lp17 encodes factors that regulate virulence (Sarkar et al., 2011; Casselli et al., 2012; Casselli et al., 2019; Medina-Pérez et al., 2020), as well as genes of unknown function, the impact on the transcriptome is difficult to predict. Nonetheless, a practical implication of this study is that it highlights the need for copy number analysis in mutagenesis studies, particularly if a region near a PF32 member may be affected.
In the mouse model, deletion of bbd21-bbd22 resulted in decreased infectivity based on culture results in all tissues, increased ID50, and spirochete burden in murine heart tissue. We postulate that the infectivity defect could be associated with gene dosage and dysregulated expression of virulence factors. It has been described that insufficient expression or overexpression of outer surface proteins such as OspC, which is regulated by bbd18 encoded on lp17, can lead to clearance by the host immune system (Tilly et al., 2006; Xu et al., 2006; Casselli et al., 2012). Similarly, it has been reported in other bacteria, such as Yersinia pseudotuberculosis, that upregulation of copy number for plasmids encoding key virulence factors is essential to establishing infection in a host (Wang et al., 2016). Thus, in the current study, it is possible that the attenuated murine tissue colonization may be associated with the significantly decreased lp17 copies observed in mammalian host-adapted spirochetes lacking bbd21-bbd22. Alternatively, the attenuated tissue colonization may be associated with the early growth defect which could result in partial clearance and decreased spirochete burden. Additional data is needed to support a possible link between plasmid copy number and pathogenicity in B. burgdorferi.
Expression of bbd21 does not appear to be governed by rpoS. Comparison of bbd21 transcript levels between in vitro grown wild-type and ΔrpoS spirochetes revealed no significant difference, suggesting bbd21 expression is independent of the gatekeeper pathway. This is consistent with the absence of bbd21 from transcriptomic studies involving disruption of the alternative σ factor cascade (Caimano et al., 2007; Ouyang et al., 2009; Caimano et al., 2019). One possible explanation is that several plasmid partitioning and replication proteins are known to negatively self-regulate (Funnell and Slavcev, 2014). Alternatively, bbd21 may be regulated in association with the stringent response, as expression of the gene is significantly increased in a microarray study of a DksA-deficient strain during nutrient stress (Boyle et al., 2019). Furthermore, both bbd21 and bbd22 were identified in transcriptomic experiments as upregulated genes in starvation-adapted cells starved for N-acetylglucosamine (Schneider and Rhodes, 2018). Given the increased metabolic requirement to fuel plasmid number expansion during replication, plasmid copy number dysregulation may account for the delayed growth densities observed for the Δ21-22 mutant in the current study. Current studies are underway to determine a possible link between bbd21-bbd22 plasmid copy regulation and metabolism.
Because bbd21 was previously proposed as a putative parA orthologue based on 25% sequence homology and presence of a Walker box motif (Zückert and Meyer, 1996), as well as demonstrated ATPase activity (Deneke and Chaconas, 2008), it seems plausible that the deletion of this gene is responsible for the changes in lp17 plasmid copy number. However, our findings revealed that bbd21 and bbd22 are co-transcribed. Therefore, contribution to plasmid copy number regulation from bbd22 cannot be definitively ruled out in this study. Further investigation of bbd22 is required to determine its potential role in plasmid copy number regulation, particularly as identification of a parB/repC orthologue remains elusive in the B. burgdorferi genome. Alternatively, bbd22 may not be involved with plasmid copy number and instead may serve a function not directly observed in this study.
In conclusion, our study describes a role of the bbd21-bbd22 genetic region that involves lp17 plasmid copy number regulation and raises the question of gene dosage in Lyme disease pathogenesis. Furthermore, the results described herein add to the growing body of work implicating the importance of lp17, and highlight the need for additional studies aimed at elucidating the mechanisms of bbd21-bbd22 plasmid copy control and how this putative bicistronic operon is regulated in this important pathogen.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by Washington State University Institutional Animal Care and Use Committee.
Author Contributions
JW and TB designed the experiments. JW and MC performed the experiments. JW and TB analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by an Intramural grant from the College of Veterinary Medicine at Washington State University (TB), and a Poncin fellowship (JW).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank George Chaconas for the 5A4 clone and Melissa Caimano for the ΔrpoS mutant clone. The authors are grateful to Nina Woodford, Gay Lynn Clyde, John DenHerder, and Jessie McCleary for technical assistance in DMC study design for IACUC approval, animal handling and surgery training, and procuring surgical supplies. The authors appreciate Jan Luft and Julie Libby for facilitating rodent ordering, vivarium housing, and use of the surgery room. The authors thank Preeti Singh, Youki Yamasaki, and Danny Powell for critically reviewing the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.884171/full#supplementary-material
References
Bankhead, T., Chaconas, G. (2007). The Role of Vlse Antigenic Variation in the Lyme Disease Spirochete: Persistence Through a Mechanism That Differs From Other Pathogens. Mol. Microbiol. 65, 1547–1558. doi: 10.1111/j.1365-2958.2007.05895.x
Barbour, A. G. (1988). Plasmid Analysis of Borrelia Burgdorferi, the Lyme Disease Agent. J. Clin. Microbiol. 26, 475–478. doi: 10.1128/jcm.26.3.475-478.1988
Barbour, A. G., Garon, C. F. (1987). Linear Plasmids of the Bacterium Borrelia Burgdorferi Have Covalently Closed Ends. Science 237, 409–411. doi: 10.1126/science.3603026
Beaurepaire, C., Chaconas, G. (2005). Mapping of Essential Replication Functions of the Linear Plasmid Lp17 of B. Burgdorferi by Targeted Deletion Walking. Mol. Microbiol. 57, 132–142. doi: 10.1111/j.1365-2958.2005.04688.x
Beaurepaire, C., Chaconas, G. (2007). Topology-Dependent Transcription in Linear and Circular Plasmids of the Segmented Genome of Borrelia Burgdorferi. Mol. Microbiol. 63, 443–453. doi: 10.1111/j.1365-2958.2006.05533.x
Boyle, W. K., Groshong, A. M., Drecktrah, D., Boylan, J. A., Gherardini, F. C., Blevins, J. S., et al. (2019). Dksa Controls the Response of the Lyme Disease Spirochete Borrelia Burgdorferi to Starvation. J. Bacteriol. 201, e00582–e00518. doi: 10.1128/JB.00582-18
Bunikis, I., Kutschan-Bunikis, S., Bonde, M., Bergstrom, S. (2011). Multiplex PCR as a Tool for Validating Plasmid Content of Borrelia Burgdorferi. J. Microbiol. Methods 86, 243–247. doi: 10.1016/j.mimet.2011.05.004
Burgdorfer, W., Barbour, A. G., Hayes, S. F., Benach, J. L., Grunwaldt, E., Davis, J.P., et al. (1982). Lyme Disease-a Tick-Borne Spirochetosis? Science 216, 1317–1319. doi: 10.1126/science.7043737
Byram, R., Stewart, P. E., Rosa, P. (2004). The Essential Nature of the Ubiquitous 26-Kilobase Circular Replicon of Borrelia Burgdorferi. J. Bacteriol. 186, 3561–3569. doi: 10.1128/JB.186.11.3561-3569.2004
Caimano, M. J. (2018). Generation of Mammalian Host-Adapted Borrelia Burgdorferi by Cultivation in Peritoneal Dialysis Membrane Chamber Implantation in Rats. Methods Mol. Biol. (Springer New York) 1690, 35–45. doi: 10.1007/978-1-4939-7383-5_3
Caimano, M. J., Groshong, A. M., Belperron, A., Mao, J., Hawley, K. L., Luthra, A., et al. (2019). The Rpos Gatekeeper in Borrelia Burgdorferi: An Invariant Regulatory Scheme That Promotes Spirochete Persistence in Reservoir Hosts and Niche Diversity. Front. Microbiol. 10, 1923. doi: 10.3389/fmicb.2019.01923
Caimano, M. J., Iyer, R., Eggers, C. H., Gonzalez, C., Morton, E. A., Gilbert, M. A., et al. (2007). Analysis of the Rpos Regulon in Borrelia Burgdorferi in Response to Mammalian Host Signals Provides Insight Into Rpos Function During the Enzootic Cycle. Mol. Microbiol. 65, 1193–1217. doi: 10.1111/j.1365-2958.2007.05860.x
Casjens, S. R., Di, L., Akther, S., Mongodin, E. F., Luft, B. J., Schutzer, S. E., et al. (2018). Primordial Origin and Diversification of Plasmids in Lyme Disease Agent Bacteria. BMC Genomics 19, 218. doi: 10.1186/s12864-018-4597-x
Casjens, S., Palmer, N., van Vugt, R., Huang, W. M., Stevenson, B., Rosa, P., et al. (2000). A Bacterial Genome in Flux: The Twelve Linear and Nine Circular Extrachromosomal Dnas in an Infectious Isolate of the Lyme Disease Spirochete Borrelia Burgdorferi. Mol. Microbiol. 35, 490–516. doi: 10.1046/j.1365-2958.2000.01698.x
Casjens, S., van Vugt, R., Tilly, K., Rosa, P. A., Stevenson, B. (1997). Homology Throughout the Multiple 32-Kilobase Circular Plasmids Present in Lyme Disease Spirochetes. J. Bacteriol. 179, 217–227. doi: 10.1128/jb.179.1.217-227.1997
Casselli, T., Crowley, M. A., Highland, M. A., Tourand, Y., Bankhead, T. (2019). A Small Intergenic Region of Lp17 is Required for Evasion of Adaptive Immunity and Induction of Pathology by the Lyme Disease Spirochete. Cell Microbiol. 21, e13029. doi: 10.1111/cmi.13029
Casselli, T., Tourand, Y., Bankhead, T. (2012). Altered Murine Tissue Colonization by Borrelia Burgdorferi Following Targeted Deletion of Linear Plasmid 17-Carried Genes. Infect. Immun. 80, 1773–1782. doi: 10.1128/IAI.05984-11
Chaconas, G., Norris, S. J. (2013). Peaceful Coexistence Amongst Borrelia Plasmids: Getting by With a Little Help From Their Friends? Plasmid 70, 161–167. doi: 10.1016/j.plasmid.2013.05.002
Coburn, J., Garcia, B., Hu, L. T., Jewett, M. W., Kraiczy, P., Norris, S. J., et al. (2021). Lyme Disease Pathogenesis. Curr. Issues Mol. Biol. 42, 473–518. doi: 10.21775/cimb.042.473
del Solar, G., Espinosa, M. (2000). Plasmid Copy Number Control: An Ever-Growing Story. Mol. Microbiol. 37, 492–500. doi: 10.1046/j.1365-2958.2000.02005.x
Deneke, J., Chaconas, G. (2008). Purification and Properties of the Plasmid Maintenance Proteins From the Borrelia Burgdorferi Linear Plasmid Lp17. J. Bacteriol. 190, 3992–4000. doi: 10.1128/JB.00057-08
Dulebohn, D. P., Bestor, A., Rego, R. O., Stewart, P. E., Rosa, P. A. (2011). Borrelia Burgdorferi Linear Plasmid 38 is Dispensable for Completion of the Mouse-Tick Infectious Cycle. Infect. Immun. 79, 3510–3517. doi: 10.1128/IAI.05014-11
Dunn, J. J., Buchstein, S. R., Butler, L. L., Fisenne, S., Polin, D. S., Lade, B. N., et al. (1994). Complete Nucleotide Sequence of a Circular Plasmid From the Lyme Disease Spirochete, Borrelia Burgdorferi. J. Bacteriol. 176, 2706–2717. doi: 10.1128/jb.176.9.2706-2717.1994
Eggers, C. H., Caimano, M. J., Clawson, M. L., Miller, W. G., Samuels, D. S., Radolf, J. D., et al. (2002). Identification of Loci Critical for Replication and Compatibility of a Borrelia Burgdorferi Cp32 Plasmid and Use of a Cp32-Based Shuttle Vector for the Expression of Fluorescent Reporters in the Lyme Disease Spirochaete. Mol. Microbiol. 43, 281–295. doi: 10.1046/j.1365-2958.2002.02758.x
Ferdows, M. S., Barbour, A. G. (1989). Megabase-Sized Linear DNA in the Bacterium Borrelia Burgdorferi, the Lyme Disease Agent. Proc. Natl. Acad. Sci. U. S. A. 86, 5969–5973. doi: 10.1073/pnas.86.15.5969
Francia, M. V., Weaver, K. E., Goicoechea, P., Tille, P., Clewell, D. B. (2007). Characterization of an Active Partition System for the Enterococcus Faecalis Pheromone-Responding Plasmid Pad1. J. Bacteriol. 189, 8546–8555. doi: 10.1128/JB.00719-07
Fraser, C. M., Casjens, S., Huang, W. M., Sutton, G. G., Clayton, R., Lathigra, R., et al. (1997). Genomic Sequence of a Lyme Disease Spirochaete, Borrelia Burgdorferi. Nature 390, 580–586. doi: 10.1038/37551
Funnell, B. E., Slavcev, R. A. (2014). “Partition Systems of Bacterial Plasmids,” in. Plasmid. Biol. 79–103. doi: 10.1128/9781555817732.ch5
Gerdes, K., Moller-Jensen, J., Bugge Jensen, R. (2000). Plasmid and Chromosome Partitioning: Surprises From Phylogeny. Mol. Microbiol. 37, 455–466. doi: 10.1046/j.1365-2958.2000.01975.x
Golde, W. T., Dolan, M. C. (1995). Variation in Antigenicity and Infectivity of Derivatives of Borrelia Burgdorferi, Strain B31, Maintained in the Natural, Zoonotic Cycle Compared With Maintenance in Culture. Infect. Immun. 63, 4795–4801. doi: 10.1128/iai.63.12.4795-4801.1995
Grimm, D., Eggers, C. H., Caimano, M. J., Tilly, K., Stewart, P. E., Elias, A. F., et al. (2004). Experimental Assessment of the Roles of Linear Plasmids Lp25 and Lp28-1 of Borrelia Burgdorferi Throughout the Infectious Cycle. Infect. Immun. 72, 5938–5946. doi: 10.1128/IAI.72.10.5938-5946.2004
Grimm, D., Elias, A. F., Tilly, K., Rosa, P. A. (2003). Plasmid Stability During In Vitro Propagation of Borrelia Burgdorferi Assessed at a Clonal Level. Infect. Immun. 71, 3138–3145. doi: 10.1128/IAI.71.6.3138-3145.2003
Groshong, A. M., Gibbons, N. E., Yang, X. F., Blevins, J. S. (2012). Rrp2, a Prokaryotic Enhancer-Like Binding Protein, is Essential for Viability of Borrelia Burgdorferi. J. Bacteriol. 194, 3336–3342. doi: 10.1128/JB.00253-12
Hinnebusch, J., Barbour, A. G. (1992). Linear- and Circular-Plasmid Copy Numbers in Borrelia Burgdorferi. J. Bacteriol. 174, 5251–5257. doi: 10.1128/jb.174.16.5251-5257.1992
Hove, P. R., Haldorson, G. J., Magunda, F., Bankhead, T. (2014). Presence of Arp Specifically Contributes to Joint Tissue Edema Associated With Early-Onset Lyme Arthritis. Infect. Immun. 82, 43–51. doi: 10.1128/IAI.01061-13
Hubner, A., Yang, X., Nolen, D. M., Popova, T. G., Cabello, F. C., Norgard, M. V. (2001). Expression of Borrelia Burgdorferi Ospc and Dbpa is Controlled by a Rpon-Rpos Regulatory Pathway. Proc. Natl. Acad. Sci. 98, 12724–12729. doi: 10.1073/pnas.231442498
Hyde, J. A., Shaw, D. K., Smith Iii, R., Trzeciakowski, J. P., Skare, J. T. (2009). The Bosr Regulatory Protein of Borrelia Burgdorferi Interfaces With the Rpos Regulatory Pathway and Modulates Both the Oxidative Stress Response and Pathogenic Properties of the Lyme Disease Spirochete. Mol. Microbiol. 74, 1344–1355. doi: 10.1111/j.1365-2958.2009.06951.x
Imai, D., Holden, K., Velazquez, E. M., Feng, S., Hodzic, E., Barthold, S. W. (2013). Influence of Arthritis-Related Protein (BBF01) on Infectivity of Borrelia Burgdorferi B31. BMC Microbiol. 13, 100. doi: 10.1186/1471-2180-13-100
Iyer, R., Kalu, O., Purser, J., Norris, S., Stevenson, B., Schwartz, I. (2003). Linear and Circular Plasmid Content in Borrelia Burgdorferi Clinical Isolates. Infect. Immun. 71, 3699–3706. doi: 10.1128/IAI.71.7.3699-3706.2003
Kugeler, K. J., Schwartz, A. M., Delorey, M. J., Mead, P. S., Hinckley, A. F. (2021). Estimating the Frequency of Lyme Disease Diagnoses, United State-2018. Emerg. Infect. Dis. 27, 616–619. doi: 10.3201/eid2702.202731
Labandeira-Rey, M., Skare, J. T. (2001). Decreased Infectivity in Borrelia Burgdorferi Strain B31 is Associated With Loss of Linear Plasmid 25 or 28-1. Infect. Immun. 69, 446–455. doi: 10.1128/IAI.69.1.446-455.2001
Lin, T., Gao, L., Zhang, C., Odeh, E., Jacobs, M. B., Coutte, L., et al. (2012). Analysis of an Ordered, Comprehensive STM Mutant Library in Infectious Borrelia Burgdorferi: Insights Into the Genes Required for Mouse Infectivity. PloS One 7, e47532. doi: 10.1371/journal.pone.0047532
Mac, S., da Silva, S. R., Sander, B. (2019). The Economic Burden of Lyme Disease and the Cost-Effectiveness of Lyme Disease Interventions: A Scoping Review. PloS One 14, e0210280. doi: 10.1371/journal.pone.0210280
Magunda, P. R. H., Bankhead, T. (2016). Investigating the Potential Role of Non-Vls Genes on Linear Plasmid 28–1 in Virulence and Persistence by Borrelia Burgdorferi. BMC Microbiol. 16, 180. doi: 10.1186/s12866-016-0806-4
McDowell, J. V., Sung, S. Y., Labandeira-Rey, M., Skare, J. T., Marconi, R. T. (2001). Analysis of Mechanisms Associated With Loss of Infectivity of Clonal Populations of Borrelia Burgdorferi B31MI. Infect. Immun. 69, 3670–3677. doi: 10.1128/IAI.69.6.3670-3677.2001
Medina-Pérez, D. N., Wager, B., Troy, E., Gao, L., Norris, S. J., Lin, T., et al. (2020). The Intergenic Small Non-Coding RNA Itta is Required for Optimal Infectivity and Tissue Tropism in Borrelia Burgdorferi. PloS Pathog. 16, e1008423. doi: 10.1371/journal.ppat.1008423
Morrison, T. B., Ma, Y., Weis, J. H., Weis, J. J. (1999). Rapid and Sensitive Quantification of Borrelia Burgdorferi-Infected Mouse Tissues by Continuous Fluorescent Monitoring of PCR. J. Clin. Microbiol. 37, 987–992. doi: 10.1128/JCM.37.4.987-992.1999
Ouyang, Z., Deka, R. K., Norgard, M. V. (2011). Bosr (BB0647) Controls the Rpon-Rpos Regulatory Pathway and Virulence Expression in Borrelia Burgdorferi by a Novel DNA-Binding Mechanism. PloS Pathog. 7, e1001272. doi: 10.1371/journal.ppat.1001272
Ouyang, Z., Kumar, M., Kariu, T., Haq, S., Goldberg, M., Pal, U., et al. (2009). Bosr (BB0647) Governs Virulence Expression in Borrelia Burgdorferi. Mol. Microbiol. 74, 1331–1343. doi: 10.1111/j.1365-2958.2009.06945.x
Ouyang, Z., Zhou, J., Norgard, M. V. (2014). Synthesis of Rpos is Dependent on a Putative Enhancer Binding Protein Rrp2 in Borrelia Burgdorferi. PloS One 9, e96917. doi: 10.1371/journal.pone.0096917
Pollack, R. J., Telford, S. R., 3rd, Spielman, A. (1993). Standardization of Medium for Culturing Lyme Disease Spirochetes. J. Clin. Microbiol. 31, 1251–1255. doi: 10.1128/jcm.31.5.1251-1255.1993
Purser, J. E., Norris, S. J. (2000). Correlation Between Plasmid Content and Infectivity in Borrelia Burgdorferi. Proc. Natl. Acad. Sci. U. S. A. 97, 13865–13870. doi: 10.1073/pnas.97.25.13865
Raffel, S. J., Battisti, J. M., Fischer, R. J., Schwan, T. G. (2014). Inactivation of Genes for Antigenic Variation in the Relapsing Fever Spirochete Borrelia Hermsii Reduces Infectivity in Mice and Transmission by Ticks. PloS Pathog. 10, e1004056. doi: 10.1371/journal.ppat.1004056
Reed, L. J., Muench, H. (1938). A Simple Method of Estimating Fifty Per Cent Endpoints. Am. J. Epidemiol. 27, 493–497. doi: 10.1093/oxfordjournals.aje.a118408
Samuels, D. S., Drecktrah, D., Hall, L. S. (2018). Genetic Transformation and Complementation. Methods Mol. Biol. (Springer New York) 1690, 183–200. doi: 10.1007/978-1-4939-7383-5_15
Samuels, D. S., Lybecker, M. C., Yang, X. F., Ouyang, Z., Bourret, T. J., Boyle, W. K., et al. (2022). Gene Regulation and Transcriptomics. Curr. Issues Mol. Biol. 42, 223–266. doi: 10.21775/cimb.042.223
Sarkar, A., Hayes, B. M., Dulebohn, D. P., Rosa, P. A. (2011). Regulation of the Virulence Determinant Ospc by Bbd18 on Linear Plasmid Lp17 of Borrelia Burgdorferi. J. Bacteriol. 193, 5365–5373. doi: 10.1128/JB.01496-10
Schneider, E. M., Rhodes, R. G. (2018). N-Acetylmannosamine (Mannac) Supports the Growth of Borrelia Burgdorferi in the Absence of N-Acetylglucosamine (Glcnac). FEMS Microbiol. Lett. 365, fny243. doi: 10.1093/femsle/fny243
Sengupta, M., Austin, S. (2011). Prevalence and Significance of Plasmid Maintenance Functions in the Virulence Plasmids of Pathogenic Bacteria. Infect. Immun. 79, 2502–2509. doi: 10.1128/IAI.00127-11
Stevenson, B., Tilly, K., Rosa, P. A. (1996). A Family of Genes Located on Four Separate 32-Kilobase Circular Plasmids in Borrelia Burgdorferi B31. J. Bacteriol. 178, 3508–3516. doi: 10.1128/jb.178.12.3508-3516.1996
Stewart, P. E., Chaconas, G., Rosa, P. (2003). Conservation of Plasmid Maintenance Functions Between Linear and Circular Plasmids in Borrelia Burgdorferi. J. Bacteriol. 185, 3202–3209. doi: 10.1128/JB.185.10.3202-3209.2003
Stewart, P. E., Thalken, R., Bono, J. L., Rosa, P. (2001). Isolation of a Circular Plasmid Region Sufficient for Autonomous Replication and Transformation of Infectious Borrelia Burgdorferi. Mol. Microbiol. 39, 714–721. doi: 10.1046/j.1365-2958.2001.02256.x
Tilly, K., Checroun, C., Rosa, P. A. (2012). Requirements for Borrelia Burgdorferi Plasmid Maintenance. Plasmid 68, 1–12. doi: 10.1016/j.plasmid.2012.01.009
Tilly, K., Krum, J. G., Bestor, A., Jewett, M. W., Grimm, D., Bueschel, D., et al. (2006). Borrelia Burgdorferi Ospc Protein Required Exclusively in a Crucial Early Stage of Mammalian Infection. Infect. Immun. 74, 3554–3564. doi: 10.1128/IAI.01950-05
Tourand, Y., Kobryn, K., Chaconas, G. (2003). Sequence-Specific Recognition But Position-Dependent Cleavage of Two Distinct Telomeres by the Borrelia Burgdorferi Telomere Resolvase, Rest. Mol. Microbiol. 48, 901–911. doi: 10.1046/j.1365-2958.2003.03485.x
Wang, H., Avican, K., Fahlgren, A., Erttmann, S. F., Nuss, A. M., Dersch, P., et al. (2016). Increased Plasmid Copy Number is Essential for Yersinia T3SS Function and Virulence. Science 353, 492–495. doi: 10.1126/science.aaf7501
Weaver, K. E., Clewell, D. B., An, F. (1993). Identification, Characterization, and Nucleotide Sequence of a Region of Enterococcus Faecalis Pheromone-Responsive Plasmid Pad1 Capable of Autonomous Replication. J. Bacteriol. 175, 1900–1909. doi: 10.1128/jb.175.7.1900-1909.1993
Xu, Q., Seemanapalli, S. V., McShan, K., Liang, F. T. (2006). Constitutive Expression of Outer Surface Protein C Diminishes the Ability of Borrelia Burgdorferi to Evade Specific Humoral Immunity. Infect. Immun. 74, 5177–5184. doi: 10.1128/IAI.00713-06
Yang, X. F., Alani, S. M., Norgard, M. V. (2003). The Response Regulator Rrp2 is Essential for the Expression of Major Membrane Lipoproteins in Borrelia Burgdorferi. Proc. Natl. Acad. Sci. U. S. A. 100, 11001–11006. doi: 10.1073/pnas.1834315100
Keywords: Borrelia burgdorferi, Lyme disease, LP17, bbd21, paralogous family 32, plasmid copy number control
Citation: Wong JK, Crowley MA and Bankhead T (2022) Deletion of a Genetic Region of lp17 Affects Plasmid Copy Number in Borrelia burgdorferi. Front. Cell. Infect. Microbiol. 12:884171. doi: 10.3389/fcimb.2022.884171
Received: 25 February 2022; Accepted: 21 March 2022;
Published: 12 April 2022.
Edited by:
Tao Lin, Baylor College of Medicine, United StatesReviewed by:
Jon Blevins, University of Arkansas for Medical Sciences, United StatesJenny A. Hyde, Texas A&M Health Science Center, United States
Nikhat Parveen, Rutgers, The State University of New Jersey, United States
Copyright © 2022 Wong, Crowley and Bankhead. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Troy Bankhead, dGJhbmtoZWFkQHdzdS5lZHU=
 Jessica K. Wong
Jessica K. Wong Michael A. Crowley
Michael A. Crowley Troy Bankhead
Troy Bankhead