- 1Department of Laboratory Medicine, Huashan Hospital, Fudan University, Shanghai, China
- 2Department of Infectious Diseases, Huashan Hospital, Fudan University, Shanghai, China
- 3Department of Laboratory Medicine, Henan Provincial People’s Hospital & the People’s Hospital of Zhengzhou University, Zhengzhou, China
- 4Department of Clinical Laboratory, The First Affiliated Hospital of Guangxi Medical University, Nanning, China
- 5Department of Laboratory Medicine, Sixth Hospital of Shanxi Medical University, Taiyuan, China
- 6Department of Clinical Laboratory, Shandong Provincial Hospital Affiliated to Shandong University, Jinan, China
- 7Department of Laboratory Medicine, Kunming Yan’an Hospital, Kunming, China
- 8Department of Laboratory Medicine, Jinshan Hospital of Fudan University, Shanghai, China
- 9Microbiology Department. Children’s Hospital of Fudan University, Shanghai, China
- 10Zhejiang Provincial Demonstration Centre of Laboratory Medicine Experimental Teaching, Wenzhou Medical University, Wenzhou, China
- 11School of Pharmacy, Fudan University, Shanghai, China
- 12Department of Laboratory Medicine, Taizhou Municipal Hospital, Taizhou, China
Objective: To investigate the epidemiology of carbapenem-resistant hypervirulent Klebsiella pneumoniae (CR-HvKP) and hypervirulent carbapenem-resistant Klebsiella pneumoniae (Hv-CRKP).
Methods: Totally 436 K. pneumoniae strains were collected from 7 hospitals in mainland China between 2017.01 and 2018.02. Sequence types, serotypes, antimicrobial-resistance and virulence genes were analyzed. Additionally, string test, capsule stain, Periodic Acid Schiff stain, fitness analysis, quantitative real-time PCR and mouse lethality test were also performed. Molecular combinations were used to screen putative blaKPC(+)-HvKP and Hv-blaKPC(+)-KP, followed by the confirmation of mouse lethality test.
Results: Diverse detection rates were found for the virulence genes, ranging from c-rmpA (0.0%) to entB (100.0%). According to the molecular criteria, 127, 186, 9 and 26 strains were putatively denoted as HvKP, blaKPC(+)-KP, blaKPC(+)-HvKP and Hv-blaKPC(+)-KP. Mouse lethality test confirmed 2 blaKPC(+)-HvKP strains (JS184 and TZ20) and no Hv-blaKPC(+)-KP. JS184 showed K2 serotype, thin capsule, positive exopolysaccharid and string test. TZ20 presented K20 serotype, thin capsule, negative exopolysaccharide and string test. Compared with the positive control NTUH-K2044, equal galF expression and growth curves were confirmed for JS184 and TZ20.
Conclusions: Molecular determination of CR-HvKP and Hv-CRKP brings remarkable bias compared with mouse lethality test. The exact prevalence of CR-HvKP is less than 1.0%, which of Hv-CRKP is much lower.
Introduction
Klebsiella pneumoniae is a gram-negative and rod-shaped bacterium that belongs to the Enterobacteriaceae family (Adeolu et al., 2016), and was first described by Carl Friedlander in 1882. K. pneumoniae is considered a prominent nosocomial pathogen worldwide, and is a member of the “ESKAPE” (Enterococcus faecium, Staphylococcus aureus, K. pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) pathogens (Pendleton et al., 2013). General nosocomial infections caused by K. pneumoniae include pneumonia, bacteraemia, and urinary tract infections (UTIs) (Paczosa and Mecsas, 2016). The frequent use of antimicrobials has resulted in the development of carbapenem-resistant K. pneumoniae (CRKP) strains which first emerged in 1996 (Yigit et al., 2001). CRKP strains generally contain mobile genetic elements harbouring a variety of antimicrobial resistance genes, including beta-lactamase K. pneumoniae carbapenemase gene (blaKPC), New Delhi metallo-β-lactamase gene (blaNDM), and oxacillinase-48 gene (blaOXA-48) (Lee et al., 2016; Zhang et al., 2015), among which blaKPC has been found to be shared in approximately 78.6% (44/56) (Lin et al., 2020) and 89.5% (34/38) of isolates in two studies (Lin et al., 2018). CRKP strains account for over 30.0% of K. pneumoniae strains and present great challenges in clinical practice (Effah et al., 2020). CRKP is associated with mortality rates of 34.7% (17/49) for pneumonia, 37.8% (34/90) for bacteraemia, and 7.4% (9/121) for UTI (Hauck et al., 2016). Furthermore, CRKP treatment is associated with a higher medical cost than that of carbapenem-susceptible K. pneumoniae (Huang et al., 2018). CRKP constitutes a major public health issue, especially in endemic countries (Karampatakis et al., 2016). Therefore, CRKP control is considered a priority by the World Health Organization (World Health Organization, 2017). CRKP is usually denoted as classical K. pneumoniae (cKP) regarding its virulence (Russo and Marr, 2019; Zhang et al., 2020).
Hypervirulence in K. pneumoniae represents another major concern. Hypervirulent K. pneumoniae (HvKP) was first reported to cause pyogenic liver abscess (PLA) and septic endophthalmitis in seven healthy individuals (Liu et al., 1986). HvKP, which has a considerably lower median lethal dose (LD50) than that of cKP in mouse model, generally produces various virulence factors such as hypercapsules, excessive siderophores, exopolysaccharides, and fimbriae (Paczosa and Mecsas, 2016; Russo and Marr, 2019). Apart from PLA, HvKP can also cause multiple invasive infectious diseases such as endogenous endophthalmitis, necrotising fasciitis, and meningitis, and the infection can undergo metastatic spread. PLA is endemic to East Asia and associated with a morbidity rate of 15.45 per 100,000 person-years in 2011 and a mortality rate of 8.2% (Chen et al., 2016; Siu et al., 2012). It has been estimated that 60% of endogenous endophthalmitis cases are associated with PLA caused by K. pneumoniae (Wong et al., 2000). Even with intravenous and intravitreal antimicrobial treatment, 89% of endophthalmitis cases show visual acuity of light perception or worse, and over 40% of affected eyes require evisceration or enucleation (Yang et al., 2007).
Recently, a combination of hypervirulence and extreme drug resistance has been reported in K. pneumoniae, thereby exacerbating the scarcity of effective treatments and resulting in high mortality (Zhang et al., 2015; Gu et al., 2018). The prevalence of infections caused by carbapenem-resistant hypervirulent K. pneumoniae (CR-HvKP) and hypervirulent carbapenem-resistant K. pneumoniae (Hv-CRKP) presents a global concern and a great challenge in clinical practice. However, the epidemiology of CR-HvKP and Hv-CRKP has not been extensively studied. The estimated prevalence of CR-HvKP/Hv-CRKP strains ranges among 5.0–15.0% among CRKP strains in mainland China, based on molecular determination or the Galleria mellonella (greater wax moth) lethality test (Zhang et al., 2020; Zhan et al., 2017). The gold standard method for evaluating the virulence of K. pneumoniae involves the use of mouse models, rather than the G. mellonella lethality test (Russo and MacDonald, 2020). Here, we analysed 436 clinical K. pneumoniae strains using molecular techniques and mouse lethality tests to elucidate the prevalence of CR-HvKP and Hv-CRKP strains.
Materials and Methods
K. pneumoniae Strains
In this study, 436 non-duplicate and consecutive K. pneumoniae isolates were collected from seven hospitals across six provinces in China (Huashan Hospital, 180 strains; Jinshan Hospital, 28 strains; Taizhou Municipal Hospital, 84 strains; The First Affiliated Hospital of Guangxi Medical University, 20 strains; Kunming Yan’an Hospital, 34 strains; Sixth Hospital of Shanxi Medical University, 60 strains; Shandong Provincial Hospital Affiliated to Shandong University, 30 strains) from January 2017 to February 2018. The 436 isolates were collected from diverse sources: 255 isolates (58.5%) were obtained from sputum samples, 98 isolates (22.5%) from urine samples, 29 isolates (6.7%) from blood samples, and 54 isolates (12.4%) were obtained from other sources. All the isolates were cultured on sheep blood agar plates and kept at -80°C prior to use. Identification of K. pneumoniae was performed using a matrix-assisted laser desorption/ionization time-of-flight mass spectrometry system (Bruker Daltonics Inc., Fremont, CA, USA) using the standard strains P. aeruginosa ATCC 27853, K. pneumoniae ATCC 700603, and E. coli ATCC 25922 as controls.
K. pneumoniae NTUH-K2044 (Accession number: AP006725.1) obtained from the Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan, is a typical hypervirulent K. pneumoniae serotype K1 strain (Fang et al., 2004). K. pneumoniae HS11286 (Accession number: CP003200.1) isolated from the Department of Laboratory Medicine, Huashan Hospital, Fudan University, Shanghai, China, is a K. pneumoniae serotype K47 strain containing blaKPC and low virulence (Liu et al., 2012).
All the K. pneumoniae strains were investigated as the flow chart in Figure 1.
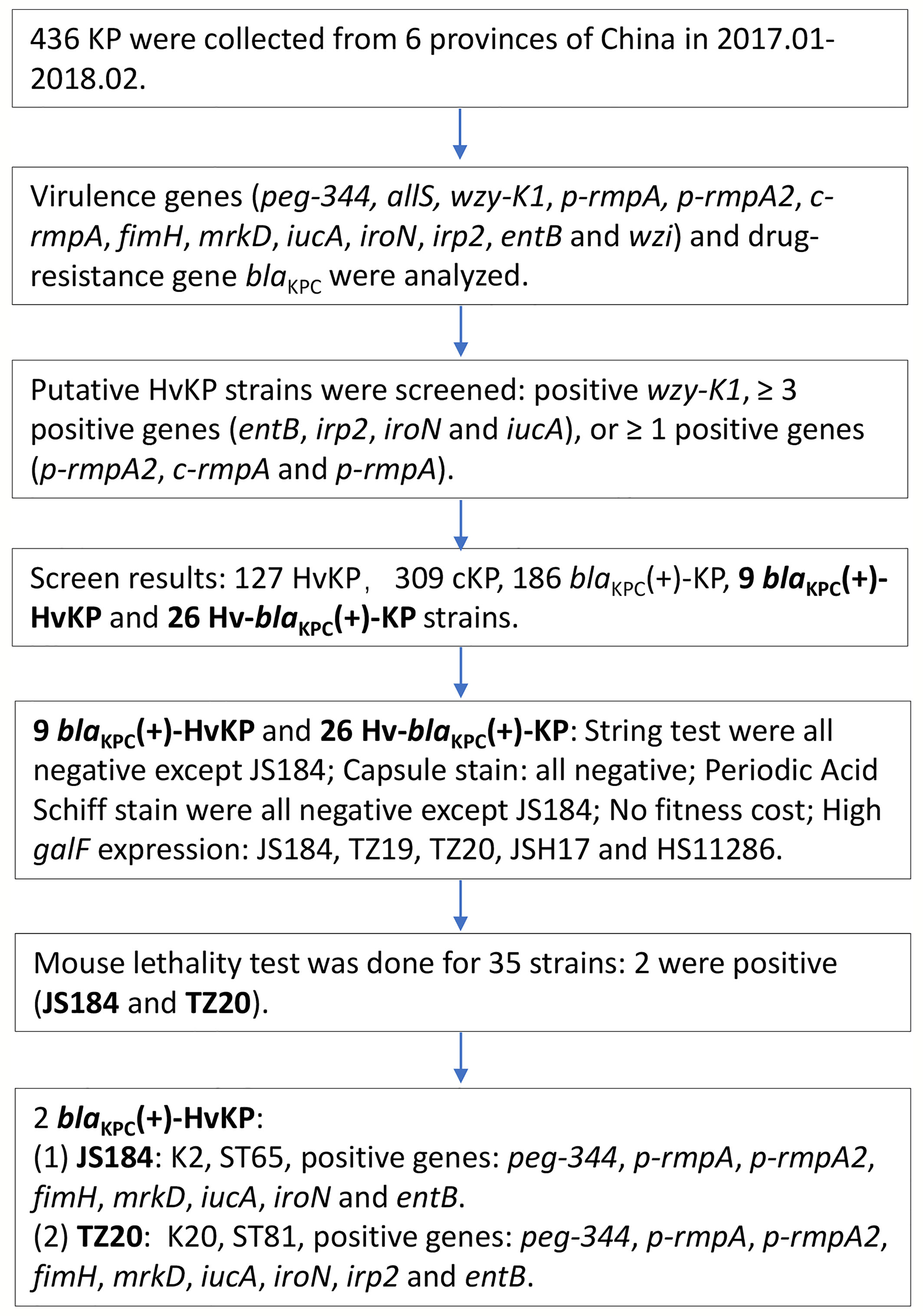
Figure 1 Experimental methodology of the study. KP, Klebsiella pneumoniae; HvKP, hypervirulent KP; blaKPC, beta-lactamase K. pneumoniae carbapenemase gene.
Multilocus Sequence Typing
DNA was extracted from the 436 K. pneumoniae strains using the QIAamp DNA mini kit (QIAGEN, Düsseldorf, Germany) according to the manufacturer’s protocol. Seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) were amplified via polymerase chain reaction (PCR) (Diancourt et al., 2005) and then sequenced using an ABI 3730XL DNA Analyser (Applied Biosystems, San Ramon, CA, USA), and then compared with sequences available on the K. pneumoniae MLST database (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html). The primers used are shown in Table S1.
Determination of Serotypes, Antimicrobial-Resistance, and Virulence Genes
The capsule type was determined via PCR amplification and sequencing of the wzi loci (Brisse et al., 2013), followed by comparison with sequences on the database of Institute Pasteur (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html).
The antimicrobial resistance gene (blaKPC) and virulence genes (wzy-K1, allS, entB, irp2, iroN, iucA, fimH, mrkD, p-rmpA2, c-rmpA, p-rmpA, peg-344, and wzi) (Compain et al., 2014; Gu et al., 2018; Russo et al., 2018) were analysed via PCR amplification, using an Applied Biosystems Veriti PCR system (Applied Biosystems). The primers used are shown in Table S1.
Determination of Putative HvKP, cKP, Hv-blaKPC(+)-KP, and blaKPC(+)-HvKP Strains
On the basis of molecular characteristics, HvKP and cKP were putatively defined as a reference (Hu et al., 2021) (Figure 1). Hv-blaKPC(+)-KP was defined as blaKPC-positive cKP which acquired key virulence genes that conferred hypervirulence. blaKPC(+)-HvKP (K1, K2, K5, K10, K20, K25, K27, and K57) (Chen et al., 2016; Siu et al., 2012) was defined as HvKP that acquired a blaKPC gene.
String Test
Overnight cultured K. pneumoniae colonies on sheep blood agar plates were stretched outward using an inoculation loop as described previously (Shon et al., 2013). The string test was considered positive when a viscous string produced was over 5 mm in length. Strain NTUH-K2044 was used as a positive control and HS11286 was used as a negative control.
Capsule Staining
Capsule staining of K. pneumoniae strains was performed according to the manufacturer’s instructions (catalog number: BA-4039; BASO, Zhuhai, China). NTUH-K2044 was used as a positive control and HS11286 was used as a negative control.
Periodic Acid-Schiff Staining
Periodic acid-Schiff staining was performed according to the manufacturer’s protocol (catalog number: BA4080A; BASO, Zhuhai, China). Strains NTUH-K2044 and HS11286 were used as positive and negative controls, respectively.
Fitness Analysis
A growth curve was generated to evaluate the fitness of K. pneumoniae strains (Liu et al., 2016). These strains were cultured overnight in Luria-Bertani broth, diluted to an optical density at 600 nm (OD600) of 0.001, and cultured at 37°C under aerobic conditions (BioTek Synergy H1, Winooski, VT, USA). The OD600 values were measured every 30 min and plotted as a curve. Strains NTUH-K2044 and HS11286 were used as positive and negative controls, respectively.
Quantitative PCR Analysis
Quantitative PCR analysis of galF mRNA together with 16S rRNA was performed using an Applied Biosystems 7500 system (Applied Biosystems, San Ramon, CA, USA). The primers used are shown in Table S1. Strains NTUH-K2044 and HS11286 were used as positive and negative controls, respectively. The analyses were performed according to the manufacturer’s protocol (catalog number: FS-Q1002; FOREVER STAR, Beijing, China).
Mouse Lethality Test
Mouse experiments were approved by the Institutional Animal Care and Use Committee of the School of Pharmacy, Fudan University (Shanghai, China) (ethical approval document NO. 201603-TY-MQ-01). Pathogen-free female BALB/c mice (age, 6 weeks), four per group, were intraperitoneally inoculated with 100 μL of K. pneumoniae strains at the mid-logarithmic growth phase (Mizuta et al., 1983). Before inoculation, K. pneumoniae strains were washed twice with normal saline and centrifuged at 10,621 × g for 4 min. A 0.6 McFarland standard equivalent to 2.0 × 108 colony forming units (CFU)/mL was prepared. The final inoculation was 102–107 CFU/mL. The mice were observed for 14 d after inoculation. LD50 was determined according to a previous study (Reed and Muench, 1938). Strains NTUH-K2044 and HS11286 were used as positive and negative controls. K. pneumoniae strains with LD50 ≤ 10 times of that of NTUH-K2044 were regarded as hypervirulent; those with LD50 > 10 times of that of NTUH-K2044 were denoted as hypovirulent.
Statistical Analysis
GraphPad Prism 8 software (GraphPad Software Inc., Sand Diego, CA, USA) was used to perform Chi-square test, one-way ANOVA, and Kruskal-Wallis test between groups; p < 0.05 was considered significant.
Results
Distribution of Key Virulence Genes
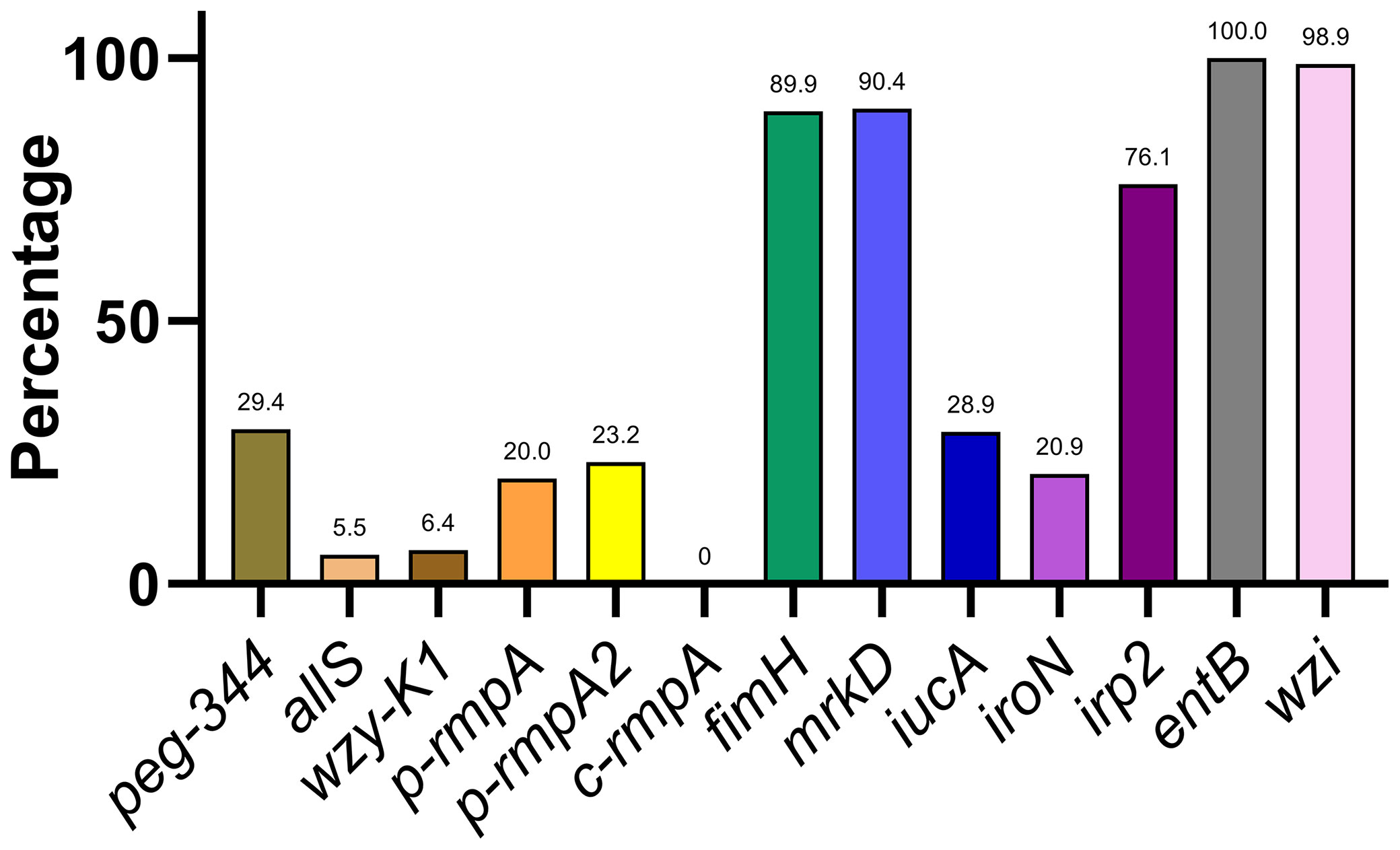
A varying distribution of virulence genes was observed, ranging from 0.0% (c-rmpA) to 100.0% (entB) (Figure 2). The 13 virulence genes could be classified into 4 categories based on rates of distribution: ≤ 10.0% (allS, wzy-K1, and c-rmpA), approximately 11.0–30.0% (peg-344, p-rmpA, p-rmpA2, iucA, and iroN), approximately 50.0–80.0% (irp2), and approximately 81.0–100.0% (fimH, mrkD, entB, and wzi).
Distribution of Predicted HvKP, blaKPC(+)-KP, blaKPC(+)-HvKP, and Hv-blaKPC(+)-KP
In total, 127 (29.1%), 186 (42.7%), 9 (2.1%), and 26 (6.0%) strains were putatively denoted as HvKP, blaKPC(+)-KP, blaKPC(+)-HvKP, and Hv-blaKPC(+)-KP strains, respectively.
Distribution of Key Virulence Genes in Putative blaKPC(+)-HvKP and Hv-blaKPC(+)-KP
The distribution of 13 key virulence genes among the putative blaKPC(+)-HvKP and Hv-blaKPC(+)-KP strains is shown in Figure 3. The 13 virulence genes were classified into four categories based on the rates of distribution: ≤ 10.0% (allS, wzy-K1, and c-rmpA), approximately 31.0–50.0% (p-rmpA and iroN), approximately 51.0–80.0% (peg-344 and p-rmpA2), and approximately 81.0–100.0% (fimH, mrkD, iucA, irp2, entB, and wzi).
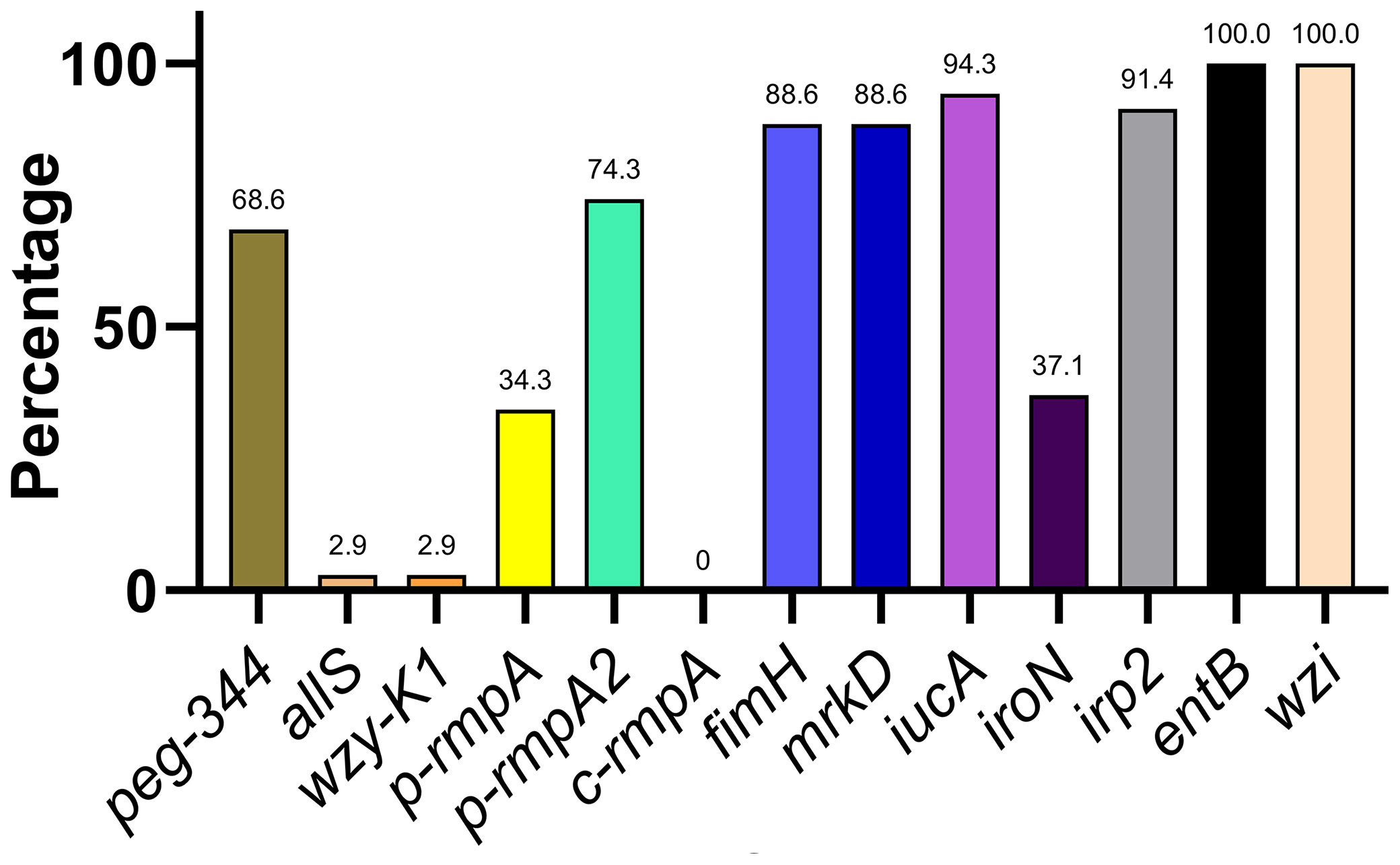
Figure 3 Distribution of key virulence genes among putative blaKPC(+)-HvKP and Hv-blaKPC(+)-KP strains. blaKPC, beta-lactamase K. pneumoniae carbapenemase gene; blaKPC(+)-HvKP, blaKPC(+) hypervirulent K. pneumoniae; Hv-blaKPC(+)-KP, hypervirulent blaKPC(+) K. pneumoniae.
Morphological Characteristics
In total, 34 putative blaKPC(+)-HvKP and Hv-blaKPC(+)-KP strains, except for blaKPC(+)-HvKP strain JS184, demonstrated negative string test results. No hypercapsule was found among the 35 putative blaKPC(+)-HvKP and Hv-blaKPC(+)-KP strains (Figure 4). No exopolysaccharides were found to be produced by the putative blaKPC(+)-HvKP and Hv-blaKPC(+)-KP strains, except for JS184 (Figure 5).
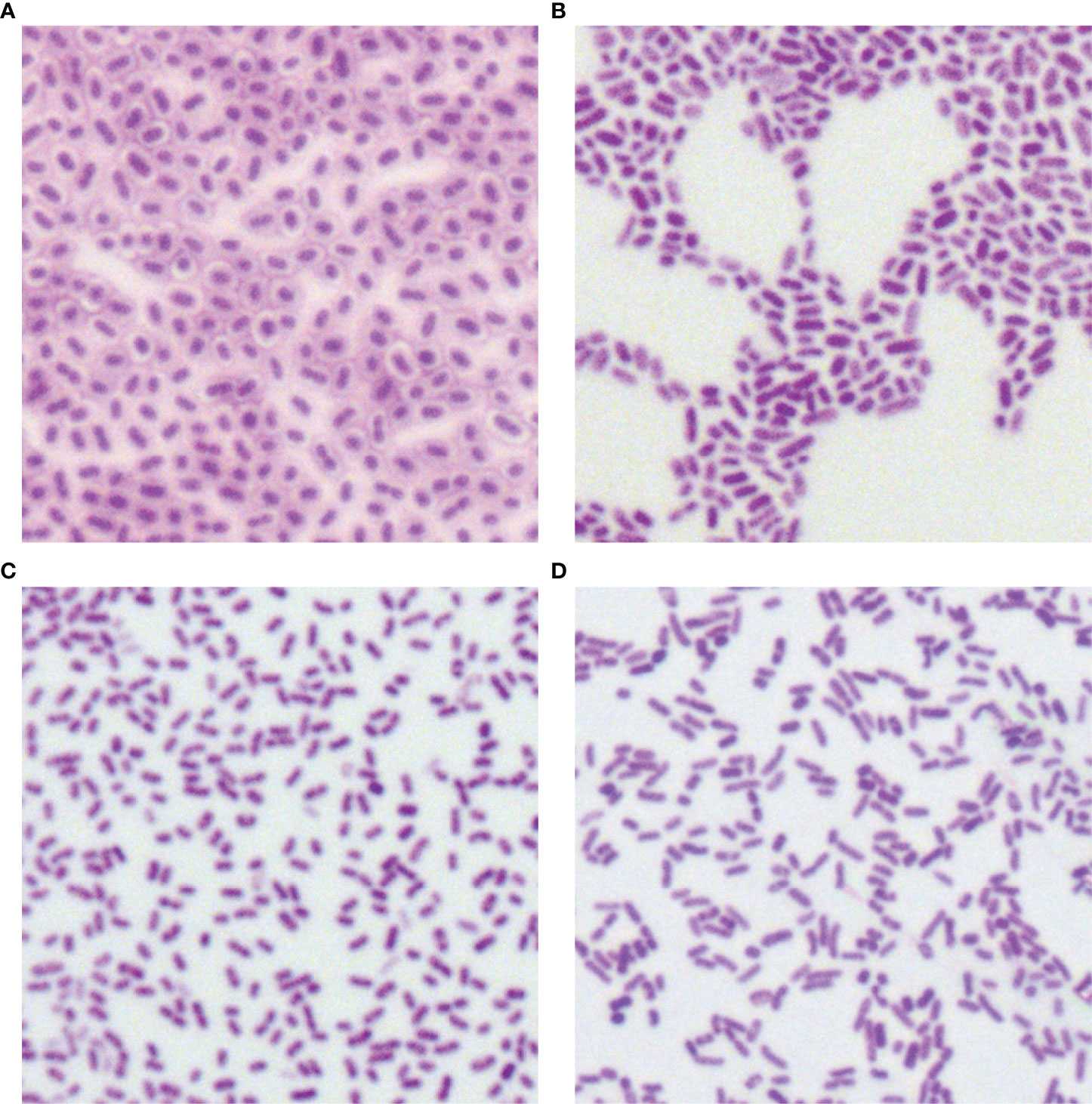
Figure 4 Capsule staining of 35 putative blaKPC(+)-HvKP and Hv-blaKPC(+)-KP strains. (A) Klebsiella pneumoniae NTUH-K2044, (B) K. pneumoniae HS11286, (C) JS184, and (D) TZ20. K. pneumoniae strains are purple and rod-shaped, and their transparent surroundings are hypercapsules (×1000). blaKPC, beta-lactamase K. pneumoniae carbapenemase gene; blaKPC(+)-HvKP, blaKPC(+) hypervirulent K. pneumoniae; Hv-blaKPC(+)-KP, hypervirulent blaKPC(+) K. pneumoniae.
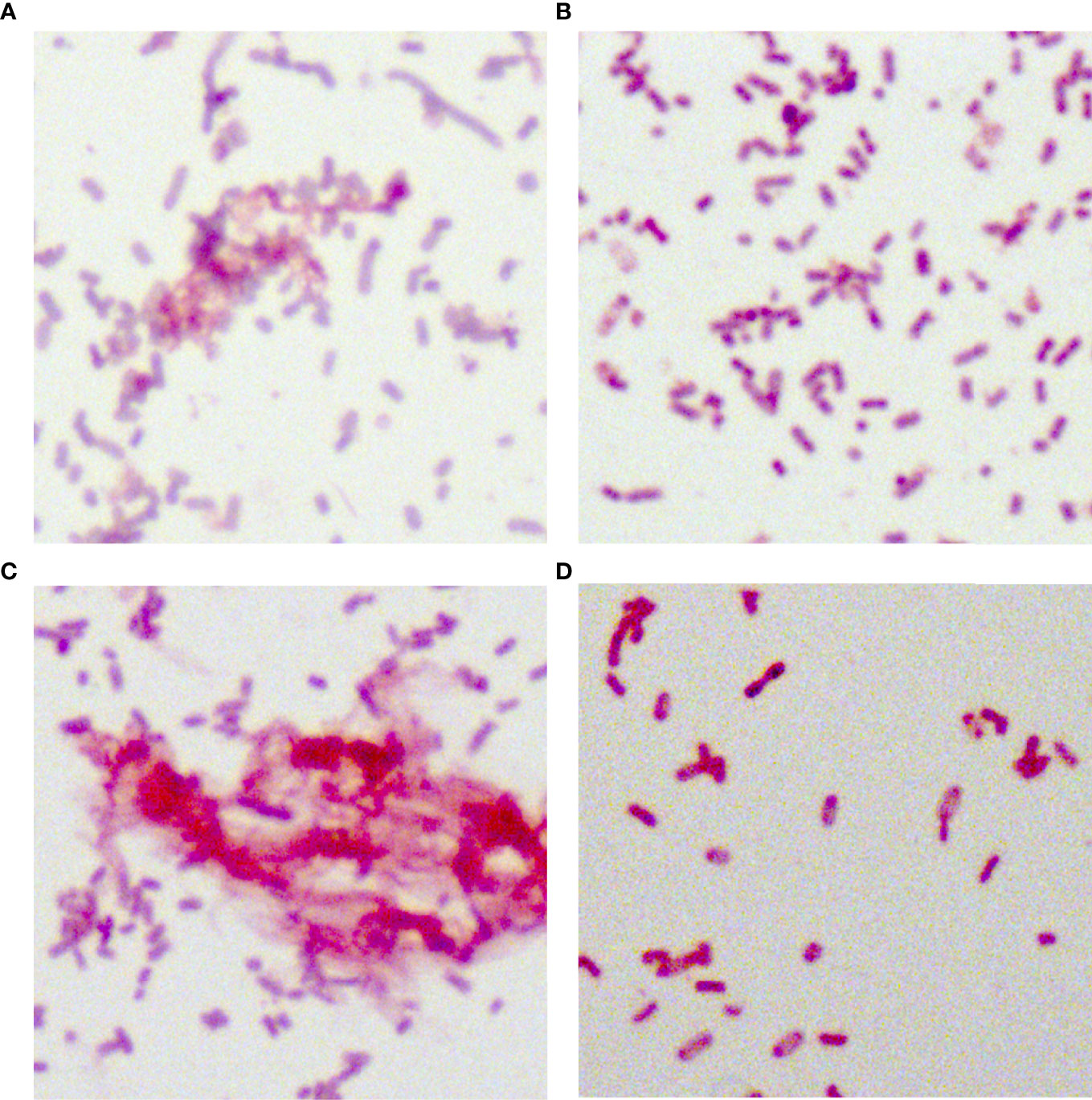
Figure 5 Periodic acid-Schiff staining of 35 putative blaKPC(+)-HvKP and Hv-blaKPC(+)-KP strains. (A) Klebsiella pneumoniae NTUH-K2044, (B) K. pneumoniae HS11286, (C) JS184, and (D) TZ20. K. pneumoniae strains were purple/red and rod-shaped; the red fluffy masses were exopolysaccharides. blaKPC, beta-lactamase K. pneumoniae carbapenemase gene; blaKPC(+)-HvKP, blaKPC(+) hypervirulent K. pneumoniae; Hv-blaKPC(+)-KP, hypervirulent blaKPC(+) K. pneumoniae.
Fitness Analysis
Among the 9 putative blaKPC(+)-HvKP and 26 Hv-blaKPC(+)-KP strains, 8 strains were chosen to represent each serotype: JS184 (K2, ST65), JS185 (K2, ST977), JS210 (K47, ST11), TZ16 (K64, ST11), TZ19 (K20, ST81), TZ20 (K20, ST81), TZ58 (K57, ST not defined), and JSH17 (K24, ST15). One-way ANOVA analysis indicated F = 0.9081 and p = 0.5178, which demonstrated similar growth and no fitness cost for the 8 strains (Figure 6).
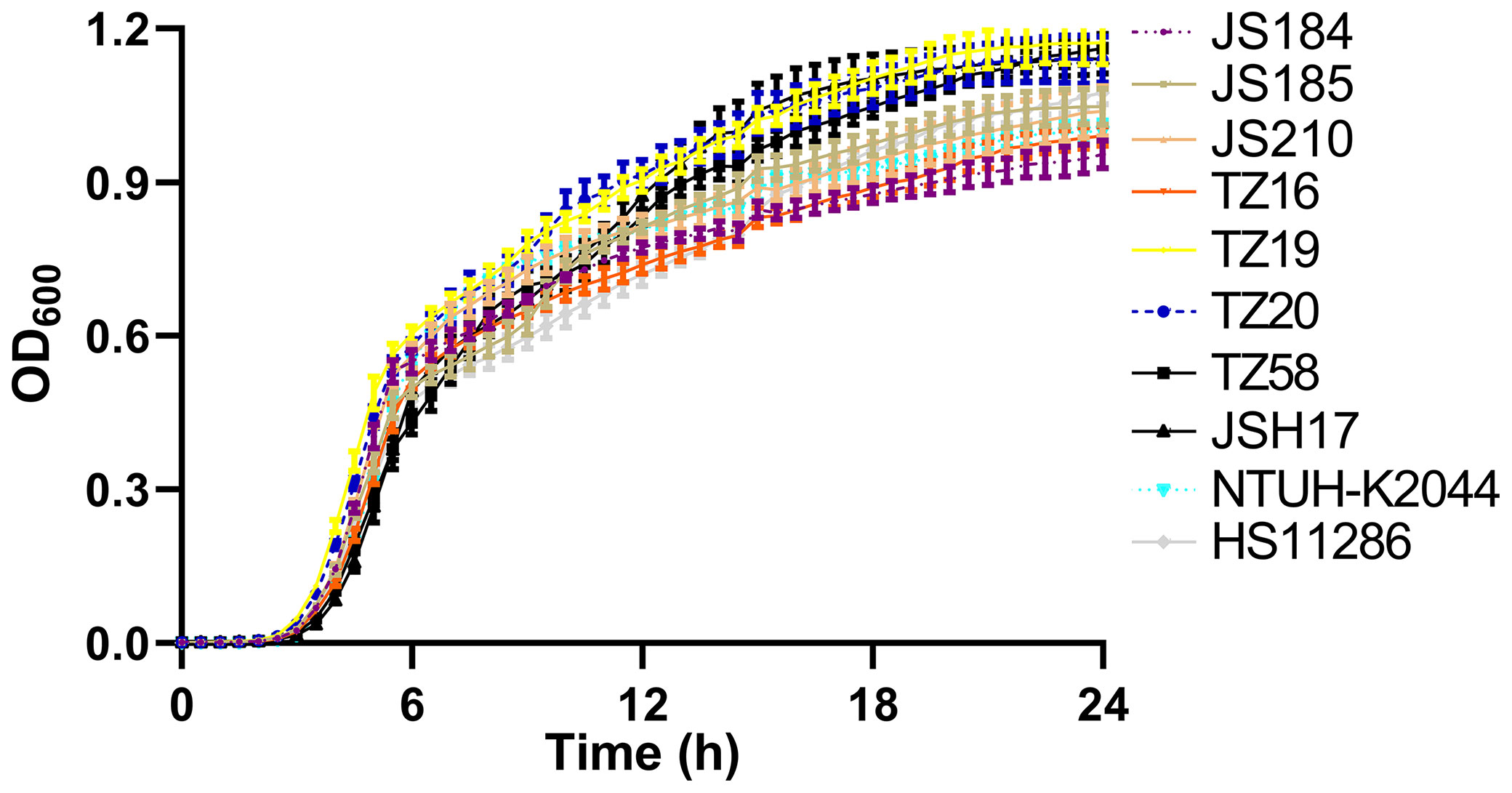
Figure 6 Growth curves of 10 putative blaKPC(+)-HvKP and Hv-blaKPC(+)-KP strains. blaKPC, beta-lactamase K. pneumoniae carbapenemase gene; blaKPC(+)-HvKP, blaKPC(+) hypervirulent K. pneumoniae; Hv-blaKPC(+)-KP, hypervirulent blaKPC(+) K. pneumoniae; OD600, optical density at 600 nm.
Expression of galF
Figure 7 shows the relative expression of galF in the putative strains compared to that in the control strains NTUH-K2044 and HS11286. JS184, TZ20, JSH17, and HS11286 showed high galF expression.
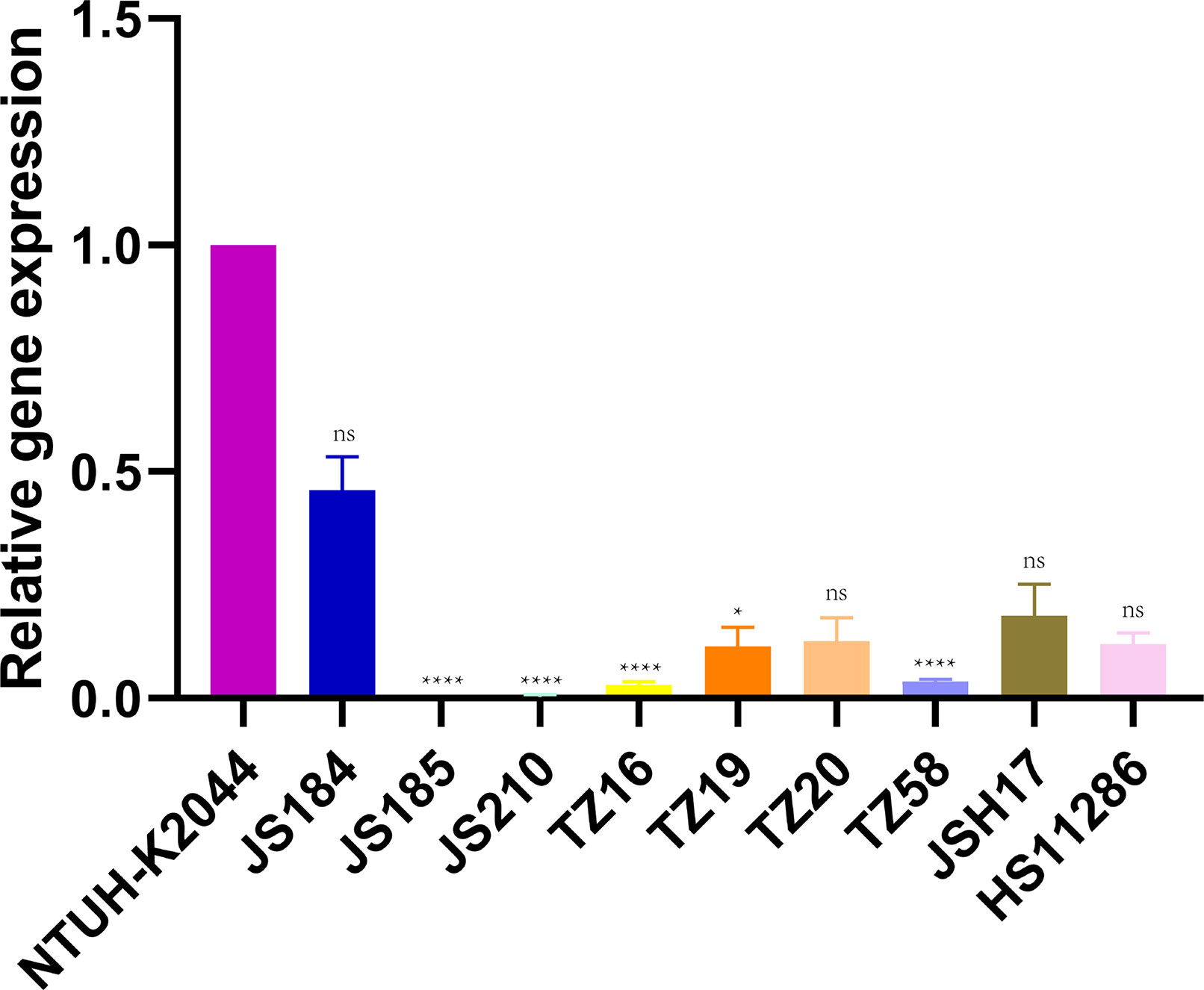
Figure 7 Expression of galF among 35 putative blaKPC(+)-HvKP and Hv-blaKPC(+)-KP strains. Klebsiella pneumoniae NTUH-K2044 was used as the standard to which others were compared. Kruskal-Wallis test was used for comparison. blaKPC, beta-lactamase K. pneumoniae carbapenemase gene; blaKPC(+)-HvKP, blaKPC(+) hypervirulent K. pneumoniae; Hv-blaKPC(+)-KP, hypervirulent blaKPC(+) K. pneumoniae; ns, not significant; ****, p < 0.0001; *, p < 0.05.
Mouse Lethality Tests
The survival curve for mice inoculated (106 CFU) with the two blaKPC(+)-HvKP strains, JS184 and TZ20, is shown in Figure 8. Log-rank (Mantel-Cox) test yielded values of χ2 = 11.4286, p = 0.0096 for the four groups (JS184, TZ20, HS11286, and NTUH-K2044); χ2 = 1.5521, p = 0.4602 for three groups (JS184, TZ20, and NTUH-K2044). Therefore, the virulence of JS184 and TZ20 was similar to that of NTUH-K2044, and was higher than that of HS11286. The LD50 values were 106 CFU for NTUH-K2044, 103 CFU for JS184, < 106 CFU for TZ20, > 107 CFU for HS11286 and the other 33 putative blaKPC(+)-HvKP/Hv-blaKPC(+)-KP strains.
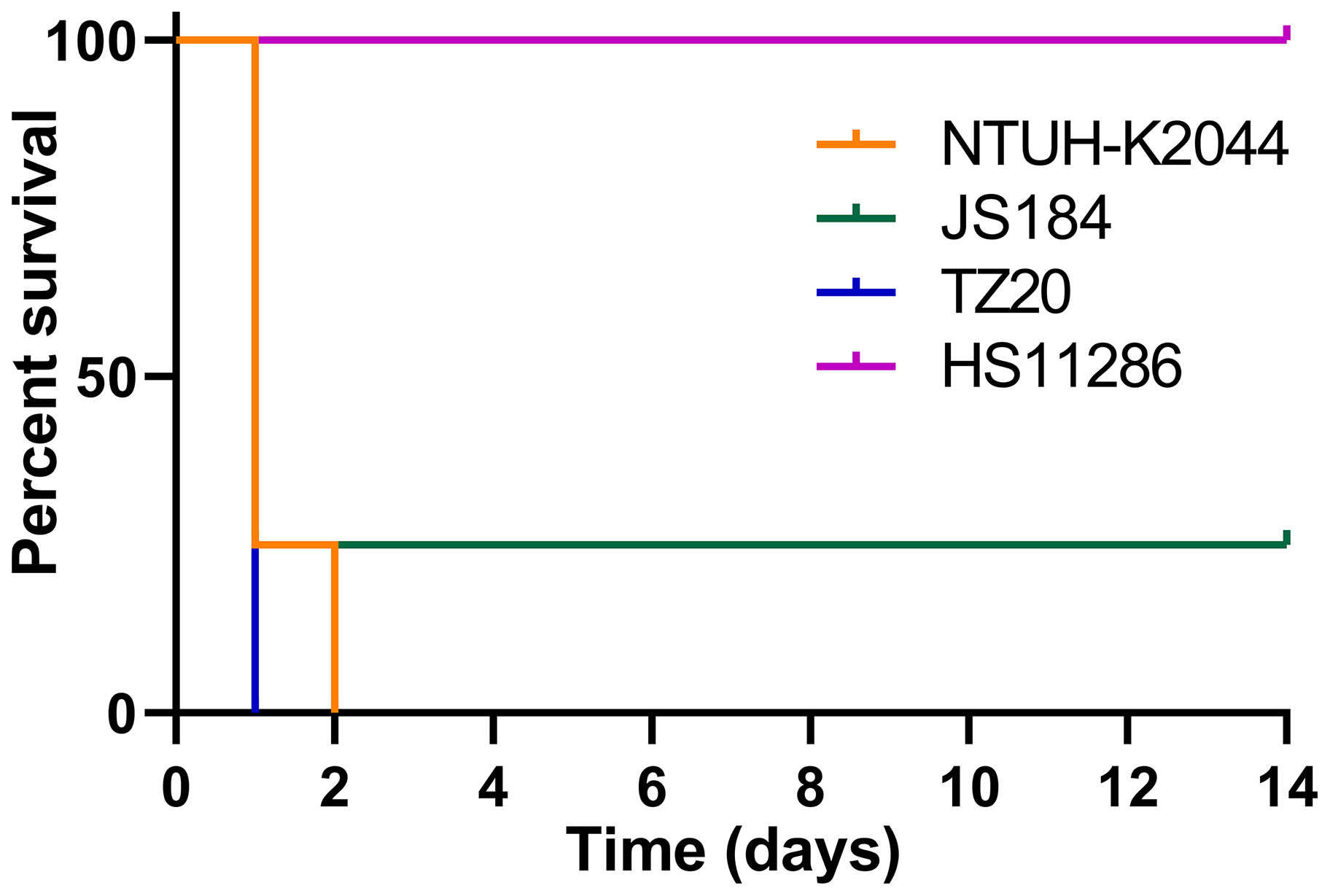
Figure 8 Survival curves of mice inoculated with two blaKPC(+)-HvKP strains. blaKPC(+)-HvKP, beta-lactamase K. pneumoniae carbapenemase gene-positive hypervirulent Klebsiella pneumoniae.
Traits of Confirmed blaKPC(+)-HvKP Strains
The two confirmed blaKPC(+)-HvKP strains, JS184 and TZ20, showed differences in ST, irp2 expression, and serotype (Table 1).
Discussion
A combination of carbapenem resistance and hypervirulence in õõõõõK. pneumoniae strains has been recently reported (Gu et al., 2018). However, the epidemiology of CR-HvKP and Hv-CRKP has not been extensively studied. To our knowledge, this is the first epidemiological surveillance study on blaKPC(+)-HvKP and Hv-blaKPC(+)-KP strains in China using a mouse lethality test to evaluate their prevalence.
In total, 13 key virulence genes in K. pneumoniae were investigated in this study; Figure 2 shows a remarkable divergence in their distribution. The extremely high detection rates of fimH, mrkD, entB, and wzi indicate ubiquitous production of type 1 fimbriae, type 2 fimbriae, enterobactins, and capsules. Another siderophore gene irp2 was present in a large proportion of the strains (76.1%), and is typically carried by ICEKp1 in the chromosome; The remaining 2 siderophore genes iucA and iroN showed detection rates of approximately 20.0–30.0%, and are usually harboured by pK2044 and pLVPK-like virulence plasmids (Struve et al., 2015). The virulence genes peg-344, p-rmpA, and p-rmpA2 are also present in pK2044- and pLVPK-like virulence plasmids (Russo and Marr, 2019), and therefore yielded similar detection rates to those of iucA and iroN (p > 0.05). The c-rmpA gene is present in the chromosome and often found in PLA specimens (Hsu et al., 2011). No such specimens were included in this study, which may explain the detection rate of 0.0%. The low detection rate of wzy-K1 (6.4%) suggests the rarity of K1 K. pneumoniae in clinical practice.
Various molecular factors were evaluated to screen for CR-HvKP and Hv-CRKP. Zhang et al. previously evaluated iucA, iroN, rmpA, and rmpA2 to check for the presence of virulence plasmids, and performed the G. mellonella lethality test to identify CR-HvKP, which yielded a rate of 5.2% (55/1052) for CR-HvKP (Zhang et al., 2020). We previously defined HvKP as: positive wzy-K1, ≥3 positive siderophore genes (entB, irp2, iroN, and iucA), or ≥1 positive capsule-regulating genes (p-rmpA2, c-rmpA/A2, and p-rmpA), and estimated a rate of 5.6% (29/521) for Hv-blaKPC(+)-KP (Shon et al., 2013), which was also applied in this study. Harada et al. defined HvKP as strains carrying virulence genes, rmpA, rmpA2, iroBCDN, iucABCD, or iutA (Harada et al., 2019). Russo et al. confirmed that the G. mellonella lethality experiment cannot accurately differentiate HvKP from cKP (Russo and MacDonald, 2020). Thus, the mouse lethality test may represent the only approach to determine the exact prevalence of CR-HvKP/Hv-CRKP. In this study, only two blaKPC(+)-HvKP strains, but no Hv-blaKPC(+)-KP strains, were eventually confirmed using a mouse lethality test, showing a rate of 0.5% (2/436) for CR-HvKP which was far lower than that reported in other studies (Zhang et al., 2020; Hu et al., 2021). Owing to the predominance of KPC-induced carbapenem resistance (Lin et al., 2018), the actual prevalence of CR-HvKP should be approximately 0.5–1.0% in mainland China in 2017. The considerable difference in prevalence rates determined between this study and other reports highlights the need to elucidate why such biomarkers are not reliable and the difference between mouse and G. mellonella lethality tests. Zhang et al., reported that only one K. pneumoniae strain has been confirmed as CR-HvKP among three strains that harbour rmpA based on a mouse lethality test (Zhang et al., 2015). The fact that rmpA genes are non-functional in cKP may be attributed to different genetic backgrounds, although rmpA-related genes, such as kvrA, kvrB, and rcsB (Palacios et al., 2018; Walker et al., 2019), were found to be widely distributed in both cKP and HvKP (data not shown).
Although Hv-CRKP and CR-HvKP strains are currently emerging worldwide (Gu et al., 2018; Karlsson et al., 2019), our study revealed that the emergence of CR-HvKP is a relatively greater concern owing to its prevalence. In this study, two confirmed blaKPC(+)-HvKP strains were found, including JS184 (K2) and TZ20 (K20), which showed no fitness cost, no hypercapsule production, and high expression of galF which is responsible for the synthesis of capsule precursor (Peng et al., 2018; Walker et al., 2019). However, JS184 showed a positive string test and exopolysaccharide production in contrast to TZ20. The reason for this is not known. In addition, JS184 and TZ20 also showed different ST and irp2 expression.
This study had a few limitations. First, only typical siderophore genes were referred to, but not their expression. Second, capsule staining is not sufficient to differentiate capsules of various thicknesses, which may impact virulence.
Taken together, our findings indicate that CR-HvKP may emerge more often than Hv-CRKP; the former accounted for less than 1.0% of the strains evaluated via mouse lethality tests among clinical K. pneumoniae strains in mainland China in 2017.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of the School of Pharmacy, Fudan University (Shanghai, China).
Author Contributions
DH, WC, and QZ contributed to conception of the study. ML, LY, ZY, YW, YH, GL, and XJ collected and identified the strains. DH, WC, QZ, PF, DT, and WW performed PCR and MLST analyses, string tests, capsular staining, periodic acid-Schiff staining, and fitness tests. PR and QM performed mouse lethality tests. DH, WC, and QZ wrote the manuscript which was revised by XJ and LY. All authors read and approved the final manuscript.
Funding
This study was supported by research grants from the National Natural Science Foundation of China (grants 81871692, 82172315, 82172315 and 81572031), Shanghai Municipal Key Clinical Specialty (Laboratory Medicine, No. shslczdzk03303), and the Shanghai Municipal Science and Technology Commission (grant number 19JC1413002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Professor Jin-Town Wang from the Department of Internal Medicine, National Taiwan University Hospital, for providing the strain NTUH-K2044.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.882210/full#supplementary-material
References
Adeolu, M., Alnajar, S., Naushad, S., Gupta, R. (2016). Genome-Based Phylogeny and Taxonomy of the 'Enterobacteriales': Proposal for Enterobacterales Ord. Nov. Divided Into the Families Enterobacteriaceae, Erwiniaceae Fam. Nov., Pectobacteriaceae Fam. Nov., Yersiniaceae Fam. Nov., Hafniaceae Fam. Nov., Morganellaceae Fam. Nov., and Budviciaceae Fam. Nov. Int. J. Syst. Evol. Microbiol. 66 (12), 5575–5599. doi: 10.1099/ijsem.0.001485
Brisse, S., Passet, V., Haugaard, A. B., Babosan, A., Kassis-Chikhani, N., Struve, C., et al. (2013). Wzi Gene Sequencing, a Rapid Method for Determination of Capsular Type for Klebsiella Strains. J. Clin. Microbiol. 51 (12), 4073–4078. doi: 10.1128/JCM.01924-13
Chen, Y. C., Lin, C. H., Chang, S. N., Shi, Z. Y. (2016). Epidemiology and Clinical Outcome of Pyogenic Liver Abscess: An Analysis From the National Health Insurance Research Database of Taiwan, 2000–2011.J. Microbiol. Immunol. Infect. 49 (5), 646–653. doi: 10.1016/j.jmii.2014.08.028
Compain, F., Babosan, A., Brisse, S., Genel, N., Audo, J., Ailloud, F., et al. (2014). Multiplex PCR for Detection of Seven Virulence Factors and K1/K2 Capsular Serotypes of Klebsiella Pneumoniae. J. Clin. Microbiol. 52 (12), 4377–4380. doi: 10.1128/JCM.02316-14
Diancourt, L., Passetet, V., Verhoef, J., Grimont, P. A., Brisse, S. (2005). Multilocus Sequence Typing of Klebsiella Pneumoniae Nosocomial Isolates. J. Clin. Microbiol. 43 (8), 4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005
Effah, C. Y., Sun, T., Liu, S., Wu, Y. (2020). Klebsiella Pneumoniae: An Increasing Threat to Public Health. Ann. Clin. Microbiol. Antimicrob. 19 (1), 1. doi: 10.1186/s12941-019-0343-8
Fang, C. T., Chuang, Y. P., Shun, C. T., Chang, S. C., Wang, J. T. (2004). A Novel Virulence Gene in Klebsiella Pneumoniae Strains Causing Primary Liver Abscess and Septic Metastatic Complications. J. Exp. Med. 199 (5), 697–705. doi: 10.1084/jem.20030857
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2018). A Fatal Outbreak of ST11 Carbapenem-Resistant Hypervirulent Klebsiella Pneumoniae in a Chinese Hospital: A Molecular Epidemiological Study. Lancet Infect. Dis. 18 (1), 37–46. doi: 10.1016/S1473-3099(17)30489-9
Harada, S., Aoki, K., Yamamoto, S., Ishii, Y., Sekiya, N., Kurai, H., et al. (2019). Clinical and Molecular Characteristics of Klebsiella Pneumoniae Isolates Causing Bloodstream Infections in Japan: Occurrence of Hypervirulent Infections in Health Care. J. Clin. Microbiol. 57 (11), e01206-19. doi: 10.1128/JCM.01206-19
Hauck, C., Cober, E., Richter, S. S., Perez, F., Salata, R. A., Kalayjian, R. C., et al. (2016). Spectrum of Excess Mortality Due to Carbapenem-Resistant Klebsiella Pneumoniae Infections. Clin. Microbiol. Infect. 22 (6), 513–519. doi: 10.1016/j.cmi.2016.01.023
Hsu, C. R., Lin, T. L., Chen, Y. C., Chou, H. C., Wang, J. T. (2011). The Role of Klebsiella Pneumoniae rmpA in Capsular Polysaccharide Synthesis and Virulence Revisited. Microbiology. 157 (12), 3446–3457. doi: 10.1099/mic.0.050336-0
Huang, W., Qiao, F., Zhang, Y., Huang, J., Deng, Y., Li, J., et al. (2018). In-Hospital Medical Costs of Infections Caused by Carbapenem-Resistant Klebsiella Pneumoniae. Clin. Infect. Dis. 67 (suppl_2), S225–S230. doi: 10.1093/cid/ciy642
Hu, D., Li, Y., Ren, P., Tian, D., Chen, W., Fu, P., et al. (2021). Molecular Epidemiology of Hypervirulent Carbapenemase-Producing Klebsiella Pneumoniae. Front. Cell. Infect. Microbiol. 11, 256. doi: 10.3389/fcimb.2021.661218
Karampatakis, T., Antachopoulos, C., Iosifidis, E. (2016). Molecular Epidemiology of Carbapenem-Resistant Klebsiella Pneumoniae in Greece. Future Microbiol. 11, 809–823. doi: 10.2217/fmb-2016-0042
Karlsson, M., Stanton, R. A., Ansari, U., McAllister, G., Chan, M. Y., Sula, E., et al. (2019). Identification of a Carbapenemase-Producing Hypervirulent Klebsiella Pneumoniae Isolate in the United States. Antimicrob. Agents Chemother. 63 (7), e00519-19. doi: 10.1128/AAC.00519-19
Lee, C. R., Lee, J. H., Park, K. S., Kim, Y. B., Jeong, B. C., Lee, S. H. (2016). Global Dissemination of Carbapenemase-Producing Klebsiella Pneumoniae: Epidemiology, Genetic Context, Treatment Options, and Detection Methods. Front. Microbiol. 7, 895. doi: 10.3389/fmicb.2016.00895
Lin, D., Chen, J., Yang, Y., Cheng, J., Sun, C. (2018). Epidemiological Study of Carbapenem-Resistant Klebsiella Pneumoniae. Open Med. 13, 460–466. doi: 10.1515/med-2018-0070
Lin, L., Xiao, X., Wang, X., Xia, M., Liu, S. (2020). In Vitro Antimicrobial Susceptibility Differences Between Carbapenem-Resistant KPC-2-Producing and NDM-1-Producing Klebsiella Pneumoniae in a Teaching Hospital in Northeast China. Microb. Drug Resist. 26 (2), 94–99. doi: 10.1089/mdr.2018.0398
Liu, Y. C., Cheng, D. L., Lin, C. L. (1986). Klebsiella Pneumoniae Liver Abscess Associated With Septic Endophthalmitis. Arch. Intern. Med. 146 (10), 1913–1916. doi: 10.1001/archinte.1986.00360220057011
Liu, P., Li, P., Jiang, X., Bi, D., Xie, Y., Tai, C., et al. (2012). Complete Genome Sequence of Klebsiella Pneumoniae Subsp. Pneumoniae HS11286, a Multidrug-Resistant Strain Isolated From Human Sputum. J. Bacteriol. 194 (7), 1841–1842. doi: 10.1128/JB.00043-12
Liu, D., Liu, Z. S., Hu, P., Cai, L., Fu, B. Q., Li, Y. S., et al. (2016). Characterization of Surface Antigen Protein 1 (SurA1) From Acinetobacter Baumannii and its Role in Virulence and Fitness. Vet. Microbiol. 186, 126–138. doi: 10.1016/j.vetmic.2016.02.018
Mizuta, K., Ohta, M., Mori, M., Hasegawa, T., Nakashima, I., Kato, N. (1983). Virulence for Mice of Klebsiella Strains Belonging to the O1 Group: Relationship to Their Capsular (K) Types. Infect. Immun. 40 (1), 56–61. doi: 10.1128/iai.40.1.56-61.1983
Paczosa, M. K., Mecsas, J. (2016). Klebsiella Pneumoniae: Going on the Offense With a Strong Defense. Microbiol. Mol. Biol. Rev. 80 (3), 629–661. doi: 10.1128/MMBR.00078-15
Palacios, M., Miner, T. A., Frederick, D. R., Sepulveda, V. E., Quinn, J. D., Walker, K. A., et al. (2018). Identification of Two Regulators of Virulence That Are Conserved in Klebsiella Pneumoniae Classical and Hypervirulent Strains. mBio. 9 (4), e01443-18. doi: 10.1128/mBio.01443-18
Pendleton, J. N., Gorman, S. P., Gilmore, B. F. (2013). Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti Infect. Ther. 11 (3), 297–308. doi: 10.1586/eri.13.12
Peng, D., Li, X., Liu, P., Zhou, X., Luo, M., Su, K., et al. (2018). Transcriptional Regulation of galF by RcsAB Affects Capsular Polysaccharide Formation in Klebsiella Pneumoniae NTUH-K2044. Microbiol. Res. 216, 70–78. doi: 10.1016/j.micres.2018.08.010
Reed, L. J., Muench, H. (1938). A Simple Method of Estimating Fifty Percent Endpoints. Am. J. Hyg. 27 (3), 493–497. doi: 10.1093/oxfordjournals.aje.a118408
Russo, T. A., MacDonald, U. (2020). The Galleria Mellonella Infection Model Does Not Accurately Differentiate Between Hypervirulent and Classical Klebsiella Pneumoniae. mSphere. 5 (1), e00850-19. doi: 10.1128/mSphere.00850-19
Russo, T. A., Marr, C. M. (2019). Hypervirulent Klebsiella Pneumoniae. Clin. Microbiol. Rev. 32 (3), e00001-19. doi: 10.1128/CMR.00001-19
Russo, T. A., Olson, R., Fang, C. T., Stoesser, N., Miller, M., MacDonald, U., et al. (2018). Identification of Biomarkers for Differentiation of Hypervirulent Klebsiella Pneumoniae From Classical K. Pneumoniae. J. Clin. Microbiol. 56 (9), e00776-18. doi: 10.1128/JCM.00776-18
Shon, A. S., Bajwa, R. P., Russo, T. A. (2013). Hypervirulent (Hypermucoviscous) Klebsiella Pneumoniae: A New and Dangerous Breed. Virulence. 4 (2), 107–118. doi: 10.4161/viru.22718
Siu, L. K., Yeh, K. M., Lin, J. C., Fung, C. P., Chang, F. Y. (2012). Klebsiella Pneumoniae Liver Abscess: A New Invasive Syndrome. Lancet Infect. Dis. 12 (11), 881–887. doi: 10.1016/S1473-3099(12)70205-0
Struve, C., Roe, C. C., Stegger, M., Stahlhut, S. G., Hansen, D. S., Engelthaler, D. M., et al. (2015). Mapping the Evolution of Hypervirulent Klebsiella Pneumoniae. mBio. 6 (4), e00630. doi: 10.1128/mBio.00630-15
Walker, K. A., Miner, T. A., Palacios, M., Trzilova, D., Frederick, D. R., Broberg, C. A., et al. (2019). A Klebsiella Pneumoniae Regulatory Mutant Has Reduced Capsule Expression But Retains Hypermucoviscosity. mBio 10 (2), e00089-19. doi: 10.1128/mBio.00089-19
Wong, J. S., Chan, T. K., Lee, H. M., Chee, S. P. (2000). Endogenous Bacterial Endophthalmitis: An East Asian Experience and a Reappraisal of a Severe Ocular Affliction. Ophthalmology 107 (8), 1483–1491. doi: 10.1016/S0161-6420(00)00216-5
World Health Organization. (2017). Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. (Geneva, Swiss).
Yang, C. S., Tsai, H. Y., Sung, C. S., Lin, K. H., Lee, F. L., Hsu, W. M. (2007). Endogenous Klebsiella Endophthalmitis Associated With Pyogenic Liver Abscess. Ophthalmology 114 (5), 876–880. doi: 10.1016/j.ophtha.2006.12.035
Yigit, H., Queenan, A. M., Anderson, G. J., Domenech-Sanchez, A., Biddle, J. W., Steward, C. D., et al. (2001). Novel Carbapenem-Hydrolyzing Beta-Lactamase, KPC-1, From a Carbapenem-Resistant Strain of Klebsiella Pneumoniae. Antimicrob. Agents Chemother. 45 (4), 1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001
Zhan, L., Wang, S., Guo, Y., Jin, Y., Duan, J., Hao, Z., et al. (2017). Outbreak by Hypermucoviscous Klebsiella Pneumoniae ST11 Isolates With Carbapenem Resistance in a Tertiary Hospital in China. Front. Cell Infect. Microbiol. 7, 182. doi: 10.3389/fcimb.2017.00182
Zhang, Y., Jin, L., Ouyang, P., Wang, Q., Wang, R., Wang, J., et al. (2020). Evolution of Hypervirulence in Carbapenem-Resistant Klebsiella Pneumoniae in China: A Multicentre, Molecular Epidemiological Analysis. J. Antimicrob. Chemother. 75 (2), 327–336. doi: 10.1093/jac/dkz446
Keywords: carbapenem-resistant hypervirulent Klebsiella pneumoniae, hypervirulent carbapenem-resistant Klebsiella pneumoniae, epidemiology, mouse lethality test, hypervirulence, carbapenemase
Citation: Hu D, Chen W, Zhang Q, Li M, Yang Z, Wang Y, Huang Y, Li G, Tian D, Fu P, Wang W, Ren P, Mu Q, Yu L and Jiang X (2022) Prevalence of Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae and Hypervirulent Carbapenem-Resistant Klebsiella pneumoniae in China Determined via Mouse Lethality Tests. Front. Cell. Infect. Microbiol. 12:882210. doi: 10.3389/fcimb.2022.882210
Received: 23 February 2022; Accepted: 20 April 2022;
Published: 01 June 2022.
Edited by:
Milena Dropa, University of São Paulo, BrazilReviewed by:
Theodoros Karampatakis, Papanikolaou General Hospital of Thessaloniki, GreeceYonghong Xiao, Zhejiang University, China
Copyright © 2022 Hu, Chen, Zhang, Li, Yang, Wang, Huang, Li, Tian, Fu, Wang, Ren, Mu, Yu and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaofei Jiang, amlhbmd4aTIxNTRAc2luYS5jb20=; Lianhua Yu, eXVsaWFuaHVhNjRAMTI2LmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Dakang Hu
Dakang Hu Wenjie Chen
Wenjie Chen Qi Zhang3†
Qi Zhang3† Yong Wang
Yong Wang Gang Li
Gang Li Dongxing Tian
Dongxing Tian Pan Fu
Pan Fu Weiwen Wang
Weiwen Wang Ping Ren
Ping Ren Qing Mu
Qing Mu Xiaofei Jiang
Xiaofei Jiang