
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 06 June 2022
Sec. Fungal Pathogenesis
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.879237
This article is part of the Research TopicExtracellular Vesicles in Host-Fungal InteractionsView all 3 articles
 Kamila Kulig1
Kamila Kulig1 Elzbieta Karnas2
Elzbieta Karnas2 Olga Woznicka3
Olga Woznicka3 Patryk Kuleta4
Patryk Kuleta4 Ewa Zuba-Surma2
Ewa Zuba-Surma2 Elzbieta Pyza3
Elzbieta Pyza3 Artur Osyczka4
Artur Osyczka4 Andrzej Kozik1
Andrzej Kozik1 Maria Rapala-Kozik5
Maria Rapala-Kozik5 Justyna Karkowska-Kuleta5*
Justyna Karkowska-Kuleta5*Currently, non-albicans Candida species, including C. tropicalis, C. glabrata, and C. parapsilosis, are becoming an increasing epidemiological threat, predominantly due to the distinct collection of virulence mechanisms, as well as emerging resistance to antifungal drugs typically used in the treatment of candidiasis. They can produce biofilms that release extracellular vesicles (EVs), which are nanometric spherical structures surrounded by a lipid bilayer, transporting diversified biologically active cargo, that may be involved in intercellular communication, biofilm matrix production, and interaction with the host. In this work, we characterize the size and protein composition of these structures for three species of non-albicans Candida fungi forming biofilm, indicating considerable heterogeneity of the investigated population of fungal EVs. Examination of the influence of EVs on cytokine production by the human monocytic cell line THP-1 differentiated into macrophage-like cells revealed that the tested vesicles have a stimulating effect on the secretion of tumor necrosis factor α and interleukin 8, while they reduce the production of interleukin 10. This may indicate the proinflammatory nature of the effect of EVs produced by these species on the host immune cells. Moreover, it has been indicated that vesicles may be involved in C. tropicalis biofilm resistance to fluconazole and caspofungin. This reveals the important role of EVs not only in the physiology of C. tropicalis, C. glabrata, and C. parapsilosis fungi but also in the pathogenesis of infections associated with the production of fungal biofilm.
Living in a biofilm is a natural and universal form of existence for a broad variety of microorganisms capable of infecting the human host. Bacterial and fungal pathogens that form multicellular communities might gain protection against unfavorable environmental conditions, applied antimicrobial therapies, and the host’s defense mechanisms (Nobile and Johnson, 2015). Microbial consortia also incessantly affect the host immune system and modulate its function and response, often becoming the cause of recurrent and persistent infections difficult to treat (Chandra et al., 2007; Pereira et al., 2021). One of the most widespread fungal pathogens capable of forming biofilm and causing a wide spectrum of different types of infections in humans are the opportunistic pathogenic yeasts from the Candida genus, including the most frequently identified species C. albicans (Netea et al., 2015; Tsui et al., 2016). However, other Candida species, including C. tropicalis, C. glabrata, and C. parapsilosis, are becoming an increasing epidemiological threat, predominantly due to the distinct collection of virulence mechanisms, as well as emerging resistance to antifungal drugs typically used in the treatment of candidiasis (Silva et al., 2012; Arendrup, 2013; Giacobbe et al., 2020; Zhang et al., 2020). Non-albicans Candida (NAC) species also possess the ability to produce biofilm; however, a considerable species- and strain-specific variety of biofilm production capacity for C. tropicalis, C. glabrata, and C. parapsilosis was observed, reflected in the differences in biofilm architecture, complexity, and composition of the biofilm matrix (Silva et al., 2011; Cavalheiro and Teixeira, 2018). Nevertheless, despite some interspecies diversity, generally, the ability to create a biofilm gives pathogenic Candida fungi an advantage in maintaining infection and enhancing virulence potential compared with planktonic cells (Cavalheiro and Teixeira, 2018). The adherence of microbial cells to biotic or abiotic surfaces, their coexistence in the community in high density, and embedding in a composite biofilm-supporting matrix not only might make the biofilm-forming cells different from free-floating ones but also contribute to acquiring novel virulence properties by the complex biofilm structure (Xu et al., 2022). These are mainly related to the increased difficulty in the removal of biofilm-forming pathogens from the inhabited surfaces and to the disturbances in the effective action of antimicrobial drugs (Ponde et al., 2021). In studies recently presented by Sasani et al., in 2021, C. tropicalis strains isolated from patients with candidemia and characterized by increased metabolic activity and high biofilm mass were related to higher mortality rates, and biofilms formed by these strains showed increased resistance to azole drugs (Sasani et al., 2021). Moreover, research presented previously by Tumbarello et al. (2007) demonstrated the correlation between the biofilm-forming ability of C. albicans and C. parapsilosis and the mortality of patients with candidemia caused by these species. Such reports indicate the urgent necessity to study in detail the characteristics of this sedentary form of existence of fungal pathogens, which constitute a real threat to human health and life.
In addition to various well-known mechanisms contributing to fungal virulence, the microbial cells that create biofilm produce also extracellular vesicles (EVs)—nanometric spherical structures surrounded by a lipid bilayer, carrying a diversified biologically active cargo—that are involved in intercellular communication, biofilm matrix production, and interaction with the host (Bleackley et al., 2019; Garcia-Ceron et al., 2021; Piffer et al., 2021). EVs produced by C. albicans biofilm differ from those produced by planktonic cells not only in the size and heterogeneity of the population but also in the protein composition. In studies presented by Zarnowski et al. (2018), 34% of the identified EV-related proteins were unique to the biofilm state, and several other shared proteins were also enriched in biofilm-derived EVs compared with vesicles from planktonic cells. Proteins carried by vesicles released by C. albicans biofilm might be involved in the production of the biofilm matrix through their enzymatic activity, and EVs might directly provide other matrix-building components like lipids and polysaccharides—glucan and mannan—thus contributing to the resistance of C. albicans biofilm to fluconazole by entrapping the antifungal drug within the matrix structure (Taff et al., 2012; Zarnowski et al., 2018; Zhao et al., 2021). Moreover, C. albicans biofilm-derived EVs play a role not only in biofilm matrix production and in reducing biofilm susceptibility to antifungals, but they are also involved in other stages of biofilm formation, like fungal cell adhesion and biofilm dispersion (Zarnowski et al., 2021). Hence, they contribute strongly and in multiple ways to the creation of this complex microbial community.
Candida albicans EVs have been characterized the most thoroughly so far regarding Candida fungi. For other species, the available data are incomplete. A detailed comparison has been recently performed by Zamith-Miranda et al. (2021) between EVs released by C. albicans and C. auris grown as planktonic cells, and the vesicle content was identified as significantly different for these two Candida species, including the varied abundance of 393 vesicular proteins when the two species were compared. Such interspecific variability of vesicles resulted also in diverse effects on the host cells (Zamith-Miranda et al., 2021). In our recent paper, we characterized the EVs produced by free-floating cells of three medically important NAC species—C. tropicalis, C. parapsilosis, and C. glabrata. However, given the possible dissimilarity of the EVs produced by the biofilm form, it is also particularly important to characterize the vesicles produced by the biofilm fungal community, especially that the abovementioned NAC species showing the ability to form complex sessile structures are currently the subject of growing interest and importance. Therefore, in the current research, we aimed to characterize the biofilm-derived EVs of C. tropicalis, C. parapsilosis, and C. glabrata along with the analysis of their immunoregulatory properties.
Candida glabrata (Anderson) Meyer et Yarrow strain CBS138 (ATCC® 2001™), C. tropicalis (Castellani) Berkhout strain T1 (ATCC® MYA-3404™), and C. parapsilosis (Ashford) Langeron et Talice strain CDC 317 (ATCC® MYA-4646™) were purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured at 30°C for 18 h in YPD medium (1% yeast extract, 2% soybean peptone, and 2% glucose, Sigma, St. Louis, MO, USA) on an orbital shaker MaxQ 6000 (170 rpm) (Thermo Fisher Scientific, Waltham, MA, USA). Then, cells were harvested by centrifugation (3,000g, 5 min); the cell pellets were washed twice with sterile phosphate-buffered saline (PBS), pH 7.4 (Biowest, Nuaillé, France); and 108 C. tropicalis and 109 C. glabrata or C. parapsilosis cells were inoculated into 100 ml of RPMI 1640 medium (Biowest) and cultured in roller bottles (Corning Inc., New York, NY, USA) at 37°C with roller rack speed 3 rpm for 48 h (with medium exchange after 24 h).
Medium portions collected after every 24 h of biofilm culture were pooled and EVs were then isolated using a previously described protocol (Karkowska-Kuleta et al., 2020) with minor modifications. Briefly, supernatants from cultures of C. glabrata, C. tropicalis, and C. parapsilosis were centrifuged twice at 4,000g for 15 min at 4°C, each time discarding the cell pellet, and then concentrated using an Amicon Ultra-15 Centrifugal Filter Unit with a 100-kDa cutoff (Merck, Darmstadt, Germany) with the addition of cOmplete Protease Inhibitor Cocktail (Roche, Basel, Switzerland). After centrifugation for 5 min at 5,000g and discarding the pellet, the concentrated medium was filtered using an Ultrafree-CL Centrifugal Filter with a pore size of 0.65 µm (Merck) and ultracentrifuged in an Optima™ L-90K Ultracentrifuge (Beckman Coulter, Brea, CA, USA) at 4°C for 1 h at a rotor speed of 45,000 rpm, which corresponds to a relative centrifugal field of 184,000g (k factor 85), using a fixed-angle type 50.2 Ti Rotor and polycarbonate thick wall centrifuge tubes (13 × 64 mm) with 13-mm-diameter Delrin tube adapters. After washing with PBS buffer once, the ultracentrifugation step was repeated and the obtained EVs were transferred in 400 µl of PBS to Eppendorf tubes and stored at −80°C.
As previously described (Karkowska-Kuleta et al., 2020), to observe EVs, a JEOL JEM-2100 HT transmission electron microscope (JEOL, Tokyo, Japan) and negative stain transmission electron microscopy were used with formvar-coated, 300-mesh copper grids prepared for each EV sample using 2% uranyl acetate (Chemapol, Prague, Czech Republic). The size and concentration of EVs were measured as described in a previous work (Karkowska-Kuleta et al., 2020), using the nanoparticle tracking analysis (NTA) and NanoSight NS300 system with camera type sCMOS, laser Blue488, and NTA software Version 3.4 (Malvern Instruments, Malvern, UK).
Protein concentration was measured with o-phthalaldehyde (OPA; Sigma) in four biological replicates in samples containing vesicles of each Candida species, and the fluorescence intensity was measured with excitation and emission wavelengths of 340 and 455 nm, respectively. The phospholipid concentration in the EV preparations was determined using the Phospholipid Assay Kit MAK122 (Sigma) and the carbohydrate concentration with the Total Carbohydrate Assay Kit MAK104 (Sigma) in three biological replicates for each species, strictly in accordance with the manufacturer’s instructions. Absorbance at 570 nm (MAK122) or 490 nm (MAK104) and fluorescence intensity (OPA) were measured using a Synergy H1 Microplate Reader (BioTek Instruments, Winooski, VT, USA).
For each tested Candida strain, 1 × 1010 EVs were suspended in 150 µl of 100 mM Tris–HCl buffer, pH 7.6, with 1% sodium dodecyl sulfate and sonicated in four cycles of 30 s with UP50H Compact Lab Homogenizer (amplitude 80%, cycle 0.5, 50 W, 30 kHz; Hielscher Ultrasonics, Teltow, Germany). Then, the solutions were shaken for 5 min at 95°C and centrifuged for 15 min at 12,000g, and after that, the proteins in the supernatant were precipitated by the addition of one volume of trichloroacetic acid to four volumes of the sample. After overnight incubation at −20°C, the samples were centrifuged at 10,000g for 15 min at 10°C and washed two times with ice-cold acetone. The obtained pellets were suspended in 100 µl of 10 mM HEPES buffer, pH 8.5. Further processing and analysis of the samples were according to the protocols described by Surman et al. (2019). Briefly, the samples were prepared using paramagnetic bead technology based on the Single-Pot Solid-Phase-Enhanced Sample Preparation (SP3) (Hughes et al., 2014) with the use of SpeedBeads™ GE45152105050250 and GE65152105050250 (Sigma-Aldrich) mixed in an equal ratio, and proteins were reduced with dithiothreitol, alkylated with iodoacetamide, and digested with Trypsin/Lys-C Mix (Promega, Mannheim, Germany). Peptides obtained after tryptic digestion were identified using MS/MS analysis after prior separation on a trap column (Acclaim PepMap 100 C18, 75 μm × 20 mm, 3 μm particle, Thermo Fisher Scientific) and on an analytical column (Acclaim PepMap RSLC C18, 75 µm × 500 mm, 2 µm particle, Thermo Fisher Scientific) with the use of UltiMate 3000 RSLCnano System coupled with Q-Exactive mass spectrometer (Thermo Fisher Scientific) with DPV-550 Digital PicoView nanospray source (New Objective, Woburn, MA, USA). The Q-Exactive mass spectrometer was operated in a data-dependent mode applying a top-eight method. Full-scan MS spectra were obtained with a resolution of 70,000 at m/z 200 with an automatic gain control (AGC target) of 1 × 106. The MS/MS spectra were obtained with a resolution of 35,000 at m/z 200 with an AGC target of 3 × 106. The maximum ion accumulation times were 120 and 110 ms for the full MS and MS/MS scans, respectively. Peptides were dynamically excluded from fragmentation within 30 s. The RAW files were processed by the Proteome Discoverer platform (v.1.4, Thermo Fisher Scientific) and searched using a locally installed MASCOT search engine (v.2.5.1, Matrix Science, London, UK). The NCBI database (released April 2022) was used with the following taxonomy restrictions: C. glabrata (25,798 sequences), C. parapsilosis (8,627 sequences), or Candida tropicalis (7,188 sequences). The following parameters were applied: fixed modification—cysteine carbamidomethylation; variable modifications—methionine oxidation; peptide mass tolerance—10 ppm; and fragment mass tolerance—20 mmu. Only tryptic peptides with up to one missed cleavage were considered. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Perez-Riverol et al., 2022) with the dataset identifier PXD033327 (C. glabrata), PXD033299 (C. parapsilosis), and PXD033300 (C. tropicalis).
The human monocytic cell line THP-1 (Sigma) was cultured in RPMI 1640 medium (Biowest) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific) at 37°C in an atmosphere of 5% CO2 and 95% humidity. Differentiation from monocytes to macrophage-like cells was performed by treating cells with 10 ng/ml of phorbol 12-myristate 13-acetate (PMA) added to the medium with 100 U/ml penicillin and 100 mg/ml streptomycin (both from Biowest) for 48 h (with medium exchange after 24 h). After differentiation, the medium was replaced with fresh medium without PMA for 3 h, but with 5% FBS, 100 U/ml penicillin, and 100 mg/ml streptomycin, and then the macrophage-like cells (5 × 105 cells per well of a 24-well microplate) were incubated with EVs of each investigated Candida species with a cell-to-EV ratio of 1:100,000 for 24 h, at 37°C. After THP-1 cell stimulation by EVs, the cells were discarded by centrifugation (1,000 rpm, 5 min), and the supernatants were collected for further analysis of cytokine production. For each Candida species, three independent experiments were performed.
The levels of selected cytokines [IL-8, tumor necrosis factor α (TNF-α), IL-10] produced by macrophage-like cells were measured using Human IL-8 ELISA Set, Human TNF ELISA Set, and Human IL-10 ELISA Set (BD OptEIA™) strictly in accordance with the manufacturer’s instructions.
Candida tropicalis 1 × 105 cells were placed in 100 µl of RPMI 1640 medium (Biowest) into the wells of a Corning® 96-well black/clear flat bottom polystyrene high bind microplate (Corning Inc., Corning, NY, USA) for 90 min at 37°C in an atmosphere of 5% CO2 and 95% humidity. After removing the medium with non-adherent cells and washing the wells once with 200 µl of PBS buffer, 1 × 108 C. tropicalis biofilm-derived EVs in 100 µl of RPMI 1640 medium were added to the adherent cells and further incubated for 90 min under the same conditions. After this time, an additional 100 µl of the medium was added to the wells, and fluconazole or caspofungin (both from Sigma) was introduced into some of the wells to a final concentration of 0.5 or 0.01 µg/ml, respectively. After 24 h of incubation under the same conditions, the biofilms were washed three times with 200 µl of PBS buffer, and optical density was measured at 600 nm with the nine-point area scan reading method. Then, to estimate the biofilm metabolic activity, the XTT (sodium 3′-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis (4-methoxy6-nitro) benzene sulfonic acid hydrate) (Thermo Fisher Scientific) test was carried out. For this purpose, 100 µl of RPMI 1640 medium without phenol red (Biowest) was added to the biofilm, and then a 50-µl mixture containing XTT at a final concentration of 1 mg/ml and PMS (N-methyl dibenzopyrazine methyl sulfate) at a final concentration of 5 µg/ml (Sigma) was added for 60 min. Independently, cells from each well were mechanically detached and, after thorough mixing, transferred to YPD agar plates for CFU counting after 48 h of incubation at 30°C.
The fungal culture initiated by the defined number of Candida cells was further maintained in rotating culture bottles in RPMI 1640 medium to generate a complex 48-h biofilm, and then the vesicles were isolated from the concentrated post-culture supernatants. The presence of EVs in the obtained preparations was confirmed by TEM imaging, and the achieved pictures for C. glabrata (Figure 1A), C. parapsilosis (Figure 1C), and C. tropicalis (Figure 1E) indicated the presence of spherical structures of various sizes.
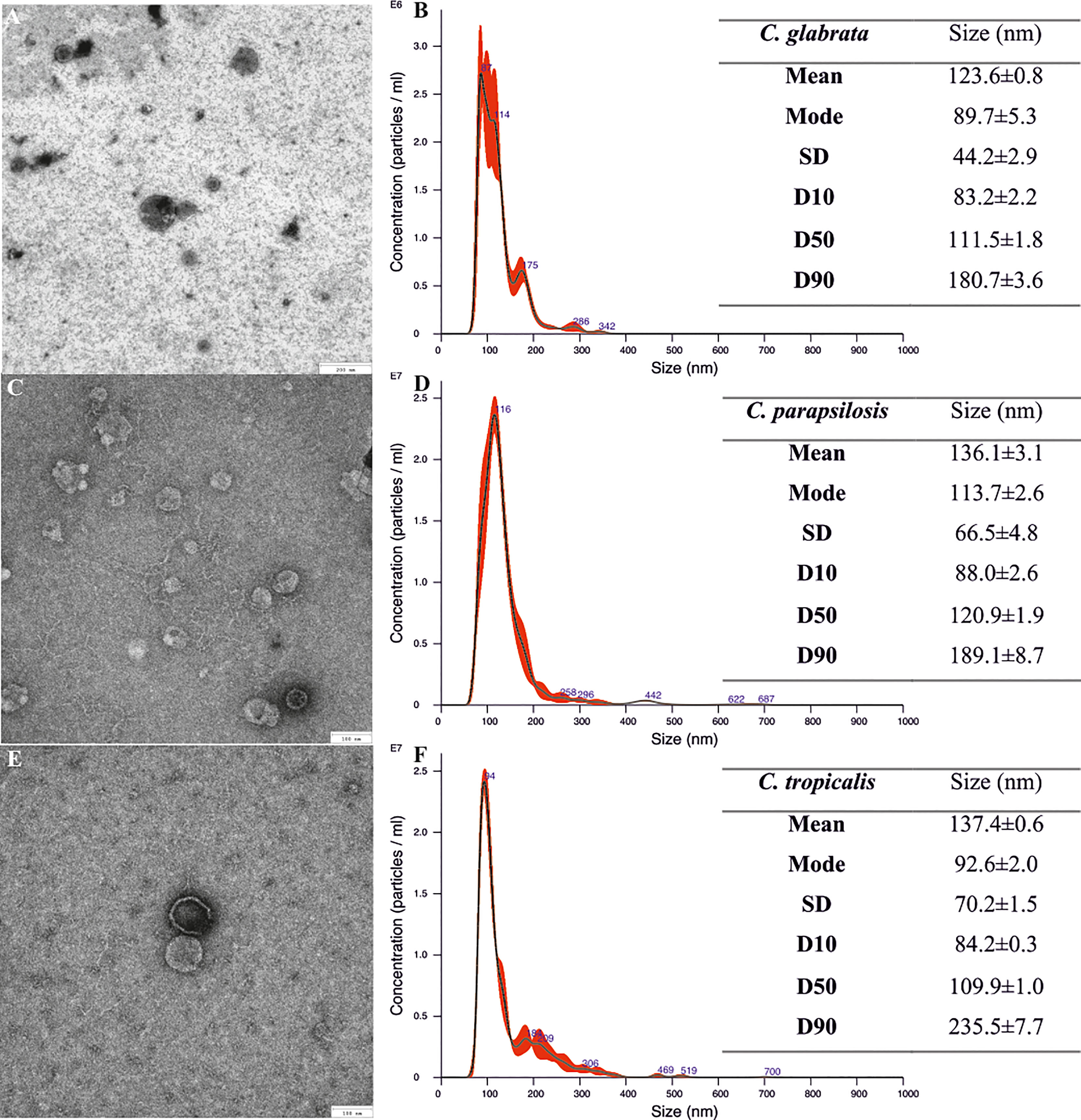
Figure 1 Size characteristics of Candida glabrata, Candida parapsilosis, and Candida tropicalis biofilm-derived extracellular vesicles (EVs). TEM images of EVs (A, C, E) and NTA particle size distribution analysis (B, D, F); representative histograms of the average size distribution from three measurements of a single sample (black line). Numbers show the maxima of particular peaks and red areas indicate the standard deviation (SD) between measurements. Tables include EVs’ size parameters measured by NTA; factors D10, D50, and D90 mean that 10%, 50%, and 90% of the EV population had a diameter of less than or equal to the given value. Data are presented as means ± SEM.
NTA that was performed to determine the number of particles in EV-containing samples showed that biofilms started by 1 × 109 C. glabrata cells produced 7.73 ± 0.07 × 109 particles (about 8 per cell) and those by 1 × 109 C. parapsilosis cells produced 2.64 ± 0.37 × 1010 particles (about 26 per cell), while biofilms initiated by 1 × 108 C. tropicalis cells produced 4.36 ± 0.1 × 109 particles (about 44 per cell). The average particle sizes were also measured using NTA and the diameters of EVs produced by C. glabrata (Figure 1B) and C. parapsilosis (Figure 1D) ranged from around 80 to 180 nm for 90% of the total particle population, while for C. tropicalis (Figure 1F), these values were almost up to 245 nm.
The measured average concentrations of proteins, phospholipids, and carbohydrates per 1 × 1010 EVs for each Candida species are listed in Table 1. The highest protein, lipid, and carbohydrate contents were observed in EVs produced by C. tropicalis biofilms. The lowest protein and lipid concentrations were identified in EVs from C. parapsilosis biofilms, and the lowest average carbohydrate content was indicated for C. glabrata biofilm-originated EVs.
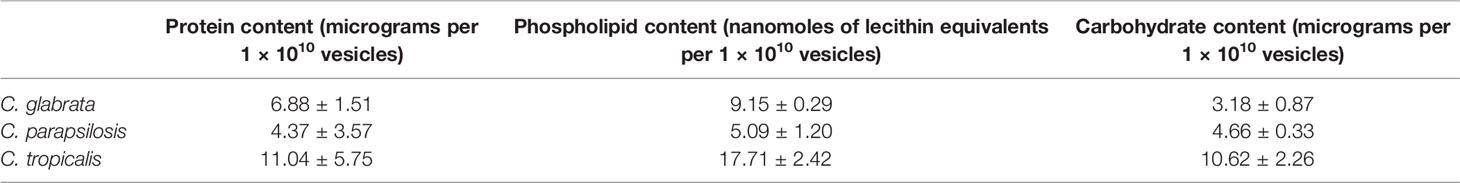
Table 1 Average protein, lipid, and carbohydrate contents in EVs released by Candida glabrata, Candida parapsilosis, and Candida tropicalis biofilms.
Further characterization of fungal EVs included vesicular protein identification using liquid chromatography-coupled tandem mass spectrometry analysis preceded by EV lysis and protein hydrolysis with trypsin. There were 288 proteins identified from C. glabrata, 86 from C. parapsilosis, and 87 from C. tropicalis biofilms (Supplementary Table 1). Among the proteins identified for C. glabrata EVs, an abundant group of proteins involved in cell wall organization might be distinguished, including among others the cell wall protein Scw11, the cell wall mannoprotein Cwp1, the 1,3-beta-glucan-linked cell wall protein Pir1, the GPI-anchored protein Ecm33, and glycoside hydrolase of the Gas/Phr family Gas1. For EVs from C. parapsilosis, this group of proteins was represented by the cell surface mannoprotein Mp65, the GPI-anchored cell wall protein Ecm33, the secreted yeast cell wall protein Ywp1, and 1,3-beta-glucosyltransferase Bgl2. Proteins involved in cell wall organization identified in C. tropicalis EVs were the cell surface mannoprotein Mp65, 1,3-beta-glucosidase Bgl2, GPI-anchored cell wall transglycosylase Crh11, and beta-1,3-glucanosyltransferase Pga4. There were some proteins identified in EVs which were shared between species (Figure 2). Five proteins were shared among all the tested species: protein similar to alpha agglutinin anchor subunit Tos1, cell wall protein Scw11, protein from the Hsp70 chaperone family, 60S ribosomal protein L12, and glyceraldehyde-3-phosphate dehydrogenase Tdh3. Five proteins—ABC transporter, the GPI-anchored cell wall protein Ecm33, mitochondrial F-ATPase beta subunit Atp2, and two proteins from the SUR7 family—were common to C. glabrata and C. parapsilosis. Moreover, five proteins—cell surface mannoproteins Mp65, Ywp1, and Bgl2; autophagy-related protein 9 Atg9; and the adhesin-like protein Sim1—were common to C. parapsilosis and C. tropicalis. Four proteins shared by C. glabrata and C. tropicalis were enolase, alcohol dehydrogenase, elongation factor 2, and ATP synthase alpha subunit Atp1.
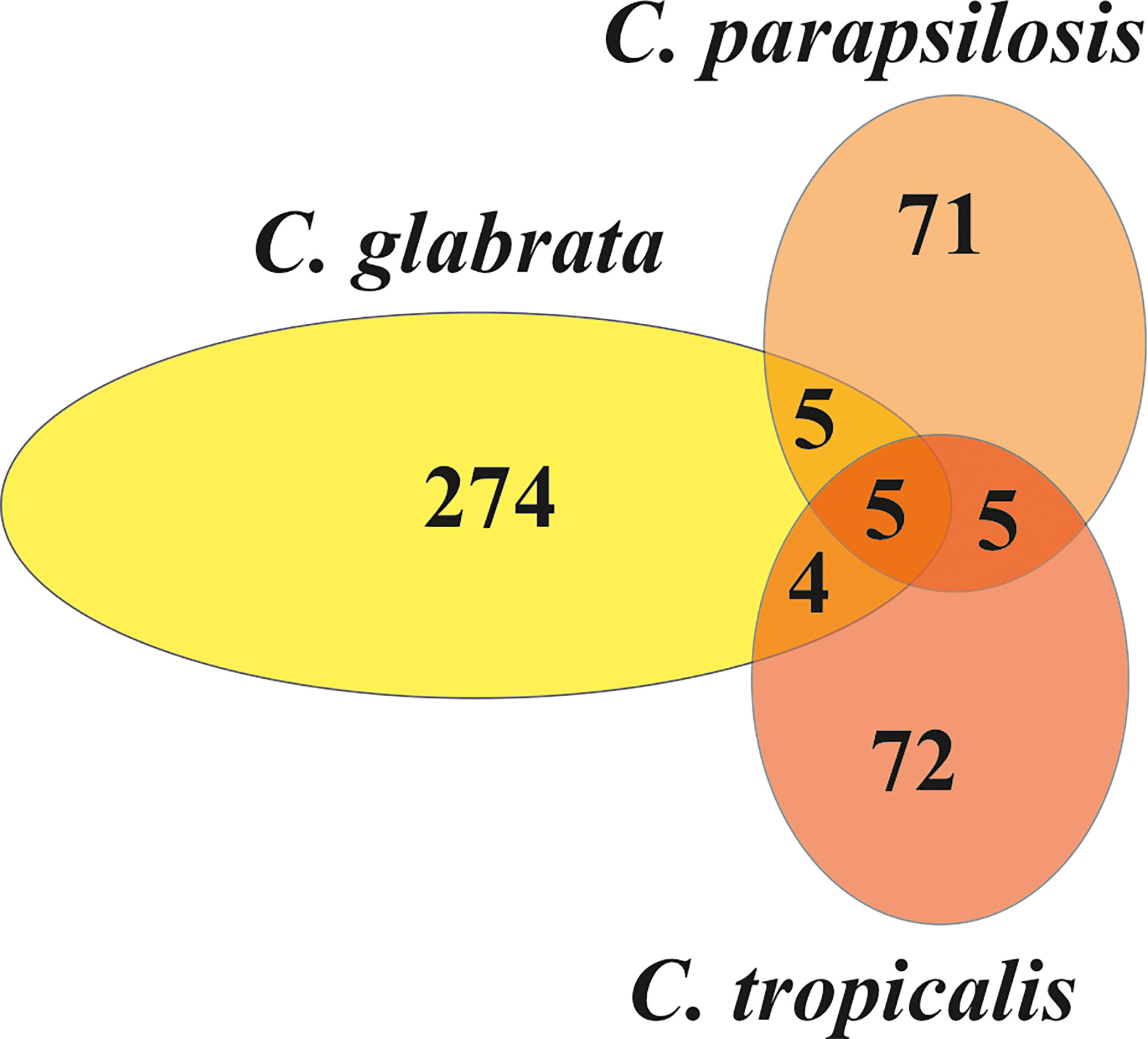
Figure 2 The Venn diagram representing the numbers of individual and common proteins among EVs from C. glabrata, C. parapsilosis, and C. tropicalis biofilms.
Proteins identified from EVs produced by C. glabrata, C. parapsilosis, and C. tropicalis biofilms were grouped, as previously described (Karkowska-Kuleta et al., 2020), into seven functional classes on the basis of protein descriptions and Gene Ontology (GO) annotations included in the Candida Genome Database (CGD, http://www.candidagenome.org) (Arnaud et al., 2005), UniProtKB Database (https://www.uniprot.org) (UniProt Consortium, 2019), and KEGG (Kyoto Encyclopedia of Genes and Genomes; https://www.genome.jp/kegg/) (Kanehisa and Goto, 2000): i) fungal-type cell wall organization, ii) membrane transport and organization, iii) stress response, iv) pathogenesis, v) cell metabolism, vi) transcription and translation, and vii) proteins of unknown function. Bar charts representing the percentage contents of proteins for each functional group are shown in Figure 3. The largest group of proteins found in EVs was transcription and translation (21%) for C. glabrata and cell metabolism (22%) for C. parapsilosis and C. tropicalis (29%). The least represented functional group was pathogenesis (1%) for C. glabrata, transcription and translation (8%) for C. parapsilosis, and stress response for C. tropicalis (3%). Among the identified proteins, there were no proteins whose main function will indicate that they are involved in stress response and pathogenesis for C. parapsilosis.
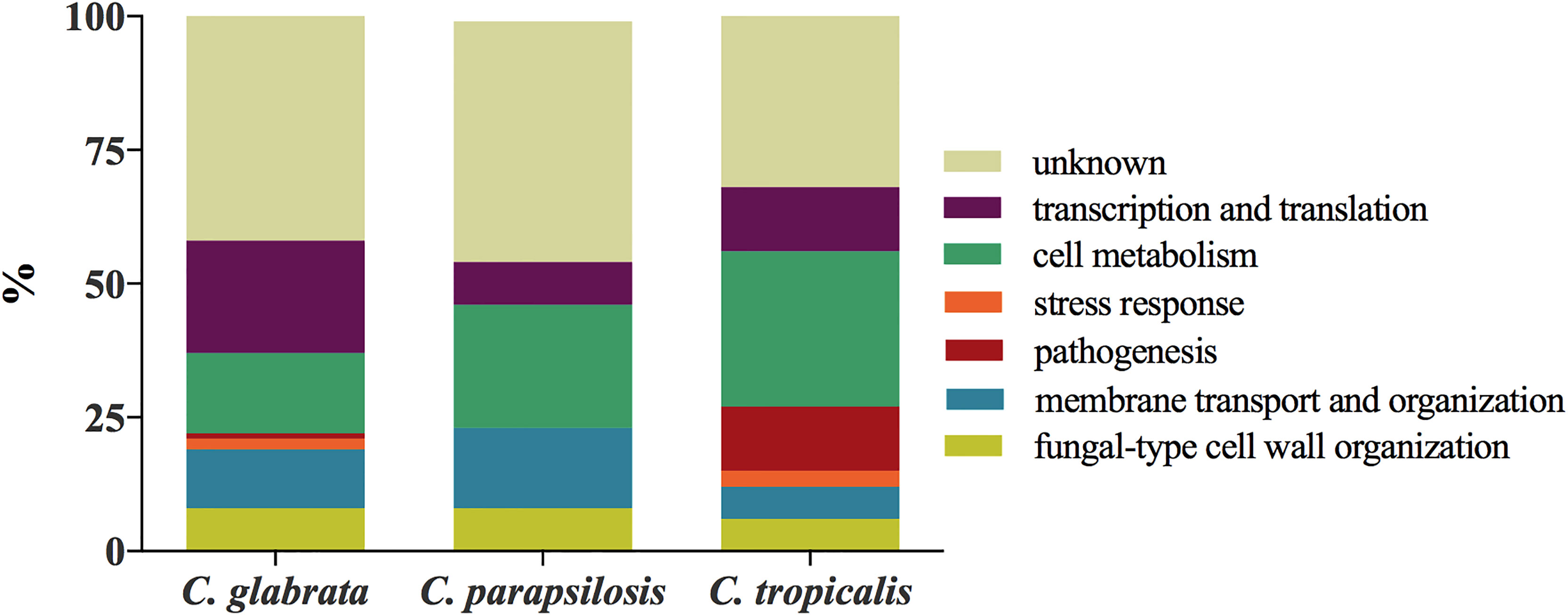
Figure 3 Functional classification of proteins identified for C. glabrata, C. parapsilosis, and C. tropicalis biofilm-originated EVs.
Previous reports showed that vesicles produced by C. albicans free-floating cells exhibited immunoregulatory properties, inducing membrane expression of co-stimulatory molecules by the host’s immune cells and influencing the production of cytokines (Vargas et al., 2015; Vargas et al., 2020; Martínez-López et al., 2022). There is still a lack of studies on the impact of candidal biofilm-originated vesicles on the host response, and the influence of EVs released by C. glabrata, C. parapsilosis, and C. tropicalis cells existing in the form of sedentary fungal communities has not yet been investigated. In the current study, we examined the effect of these particles on cytokine production by cells from the human monocytic cell line THP-1 differentiated into macrophage-like cells. Analysis of the secretion of proinflammatory and anti-inflammatory cytokines by THP-1 cells stimulated with EVs of each Candida species showed a quite comparable tendency (Figure 4). There has been a significant increase in the level of TNF-α production after contact of human cells with EVs released by the tested Candida fungi. The highest increase in the production of this cytokine, compared with the control, was observed after stimulation with EVs produced by C. glabrata biofilm, and less amount of cytokine was produced by the other two species. Similarly, the level of IL-8 secretion after incubation of human cells with candidal EVs was higher than the level for cells not stimulated with EVs, and the highest amount of cytokine was again produced under the influence of C. glabrata EVs. On the contrary, the decrease in the amount of cytokine produced was observed for IL-10 after incubation of THP-1 cells with EVs from each tested Candia species, when compared with non-stimulated cells.
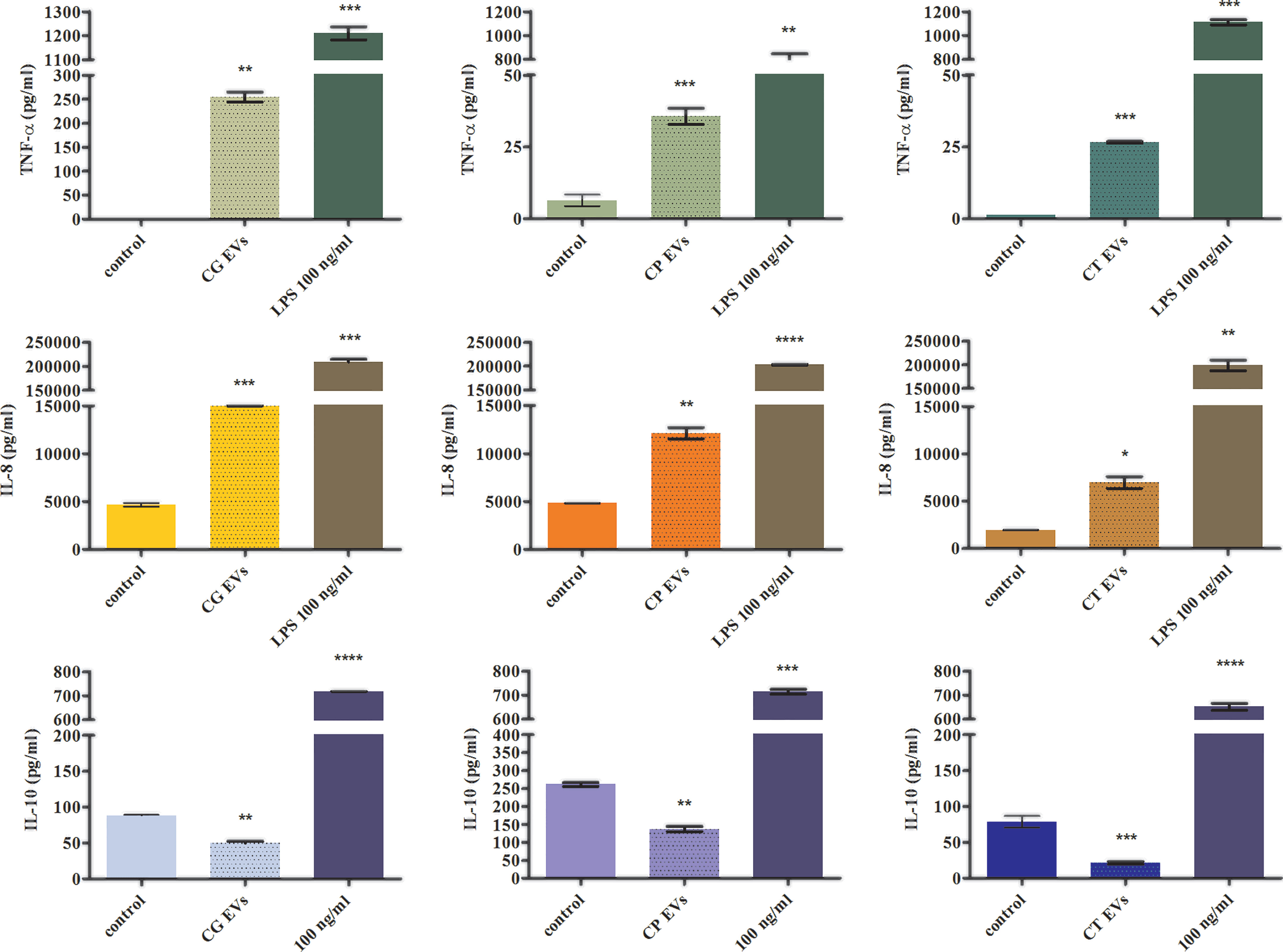
Figure 4 The analysis of cytokine production by THP-1 cells stimulated with Candida EVs. The amount of a particular cytokine was shown for stimulation with C. glabrata EVs (CG), C. parapsilosis EVs (CP), and C. tropicalis EVs (CT). A representative result of three independent experiments is presented. To analyze the statistical significance versus control, an unpaired t-test was performed with GraphPad Prism software version 7.0 (GraphPad Software, La Jolla, CA, USA). The statistical significance levels were marked with * for p < 0.05, ** for p < 0.01, *** for p < 0.001, and **** for p < 0.0001.
In the case of C. albicans biofilm, it has been shown that vesicles produced by cells in the biofilm contribute to the formation of the biofilm matrix and the occurrence of fluconazole resistance (Zarnowski et al., 2018; Zarnowski et al., 2021). Therefore, in this study we examined whether EVs might influence formation of biofilm by C. tropicalis, which of the three tested Candida species produced the most stable biofilm and was susceptible to the applied antifungal drugs ‒ fluconazole (FLU) and caspofungin (CASP) (Figure 5).
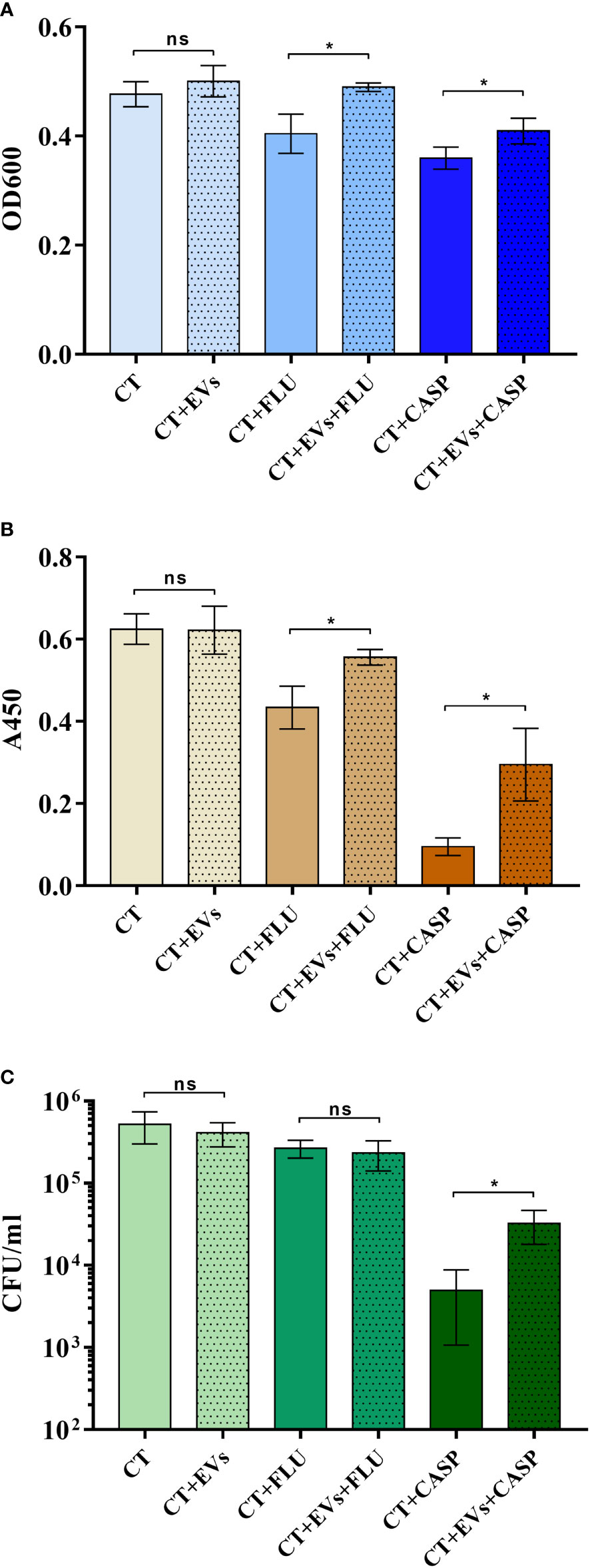
Figure 5 Candida tropicalis biofilm formation in the presence of C. tropicalis biofilm-derived EVs and the antifungal drugs fluconazole (FLU) and caspofungin (CASP). The thickness of the biofilm was investigated by measuring optical density at 600 nm in the area scan mode (A), the metabolic activity of cells in the biofilm by the XTT test (B), and the viability of the cell by counting CFUs (C). A representative result of three independent experiments is presented. To analyze the statistical significance between samples, an unpaired t-test was performed with GraphPad Prism software version 7.0. The statistical significance levels were marked with * for p < 0.05 and ns when not significant.
Antifungals were used at concentrations of 0.5 µg/ml for fluconazole and 0.01 µg/ml for caspofungin (Bizerra et al., 2008; Ferreira et al., 2009). In the first step, 105 cells were added to the wells of a microplate and incubated for 90 min to allow them to adhere to the surface. After this time, the non-adherent cells were washed away and the adherent cells were further incubated with or without vesicles produced by the 48-h C. tropicalis biofilm. Additionally, after preincubation of the adherent cells with EVs for 90 min, the antifungal drugs fluconazole or caspofungin were added to particular wells, and biofilms were further formed for 24 h, all the time in the presence of EVs where they were originally introduced. Reference samples were biofilms formed without EVs added. To investigate the impact of EVs on the thickness of biofilm formed by C. tropicalis cells in the presence of antifungals, optical density measurements in the area scan mode (OD600) were performed (Figure 5A). Biofilm metabolic activity and cell viability were analyzed with the XTT test (Figure 5B) and CFU counting (Figure 5C), respectively. It was indicated that in the presence of both antifungal drugs, the addition of EVs increased the thickness of the biofilm and the metabolic activity of the cells, as well as cell viability (only for caspofungin), when compared with samples where EVs were not added; however, in the absence of drugs, the addition of EVs did not significantly affect these parameters under the conditions tested.
Fungi from the Candida genus, including C. glabrata, C. parapsilosis, and C. tropicalis, are important commensal pathogens able to cause various types of infections in humans. Although currently these species are not as common cause of fungal diseases as C. albicans, they pose a growing threat to the immunocompromised patient population due to the everincreasing frequency of infections and emerging resistance to antifungals. During the colonization of different niches of the human organism, Candida yeasts exploit many various mechanisms of virulence, including the ability to form biofilm and the production of EVs (Yu et al., 1994; Zuza-Alves et al., 2017). EVs are nanometric structures surrounded by a lipid bilayer and filled with a plenteous and assorted cargo (Joffe et al., 2016). These structures have become more widely described in the last few decades primarily for human cells (Faure et al., 2006; Bobis-Wozowicz et al., 2017; You et al., 2020), but also for numerous bacteria (Knox et al., 1967; Lee et al., 2009; Rivera et al., 2010; Tashiro et al., 2010; Lee et al., 2013; Brown et al., 2014; Olaya-Abril et al., 2014), as well as for some fungi (Albuquerque et al., 2008; Vallejo et al., 2012; Wolf et al., 2014; Vargas et al., 2015; Joffe et al., 2016; de Toledo Martins et al., 2018; Zamith-Miranda et al., 2018). Nonetheless, fungi are the least studied so far. There are many possible functions that fungal EVs might perform, which are suggested on the basis of published but still limited research. One of the proposed functions of candidal vesicles is acting as carriers of virulence factors and signaling molecules for intercellular communication (Oliveira et al., 2010). EVs and their content might participate in the interaction between fungal pathogens and human cells during infection development, inducing also immune system response (Gil-Bona et al., 2015; de Toledo Martins et al., 2018). Moreover, Candida EVs might also play an important role in the communication between cells of the same species forming a complex community, influencing fungal cell adhesion to different surfaces, formation of biofilm structures, and further development and propagation of infection (Zarnowski et al., 2018; Zarnowski et al., 2021). In addition, the communication through EV release between Candida yeasts and other microorganisms living in the same niche might be crucial in the development of multispecies infections.
EVs might be produced by yeast cells grown in different morphological forms. Previously, we described the population of EVs released by planktonic cells and free-floating aggregates of non-albicans Candida species (Karkowska-Kuleta et al., 2020), consisting of several subpopulations of vesicles differing in size. In the current study, the heterogeneity of biofilm vesicle population has been demonstrated. Candida glabrata EVs produced by biofilm structures were rather smaller in diameter than the ones produced by planktonic cells. For C. tropicalis, the sizes were similar when comparing both forms, and in the case of C. parapsilosis, the mean sizes of biofilm EVs were larger than those determined by the NTA for the EVs released by free-floating cells (Karkowska-Kuleta et al., 2020). The dimensions of C. parapsilosis, C. glabrata, and C. tropicalis biofilm-derived vesicles were in a similar size range compared with previously described EVs released by C. albicans biofilm, constituting a population of structures of 30–200 nm in diameter (Zarnowski et al., 2018), although such smallest particles have not been measured for NAC species. The concentration of proteins and lipids was higher in EVs released by C. glabrata biofilm than in those produced by planktonic cells. In the case of C. parapsilosis EVs, the opposite was true and higher concentrations of proteins and lipids were proven for vesicles released by planktonic cells, whereas C. tropicalis biofilm vesicles contained a higher lipid content, and the protein concentration was comparable for both forms. The concentration of vesicular carbohydrates was not analyzed in a previous study; however, the presence of surface-exposed mannoproteins for EVs produced by planktonic fungal cells was demonstrated with flow cytometry (Karkowska-Kuleta et al., 2020). The current results indicate that in the case of biofilm-originated EVs, the highest concentration of polysaccharides was observed for C. tropicalis, while in the other two tested Candida species, the carbohydrate concentrations were rather within a similar range.
If the abundance of protein categories in planktonic vesicles (Karkowska-Kuleta et al., 2020) and biofilm vesicles is compared, for C. parapsilosis, a depletion of proteins related to pathogenesis is noticeable. However, for all species, a large proportion of the proteins are not well recognized and have no assigned functions in the database; hence, the actual categorization might be slightly different. In the case of C. glabrata, the presence of hydrolytic enzymes related to the adhesive properties and virulence of this species, i.e., yapsin 1 aspartyl proteinase (Yps1) and phospholipase 1 (Plb1), is noteworthy (Kaur et al., 2007; Gómez-Molero et al., 2015). Interestingly, in the case of C. glabrata and C. parapsilosis, two proteins from the SUR7 protein family have been identified, while for C. albicans, representatives of this family have been indicated as markers for candidal EVs (Dawson et al., 2020). Despite the fact that the determined concentration of vesicular proteins was the highest for C. tropicalis, the greatest protein diversity was observed for EVs of C. glabrata, and also with regard to their functionality, where each of the distinguished categories was represented, and the most numerous was the group of proteins related to cellular metabolism and transcription and translation. Moreover, the group of proteins responsible for cell wall organization, along with proteins involved in membrane transport, was represented in each species, which certainly highlights the role of fungal vesicles in the organization of this outer compartment of the cell and, in the case of the currently studied structures, most likely also in the formation of the biofilm matrix. The role of vesicles in building the biofilm matrix was confirmed previously for C. albicans (Zarnowski et al., 2018); additionally, the involvement of particular proteins constituting the cargo of biofilm EVs in biofilm regulation has been demonstrated, including the cell wall protein Ecm33 and the cell surface mannoprotein Mp65 (Zarnowski et al., 2021). The ortholog of the Ecm33 protein in our study was identified in both C. glabrata and C. parapsilosis, while Mp65 was identified for C. parapsilosis and C. tropicalis. The latter protein in C. albicans is a well-known antigen located at the cell wall, mainly of the hyphae form, stimulating T-cell proliferation of human peripheral blood mononuclear cells and inducing dendritic cell maturation and their production of proinflammatory cytokines—TNF-α and IL-6 (Gomez et al., 1996; Nisini et al., 2001; Pietrella et al., 2006).
Vesicles as extracellularly secreted and biologically active structures might participate in the communication between fungi and human cells inducing the immune system response (Herkert et al., 2019; Munhoz da Rocha et al., 2020). For EVs released by C. albicans planktonic cells, a few studies were presented previously showing the ambiguous character of the triggered response (Vargas et al., 2015; Vargas et al., 2020; Zamith-Miranda et al., 2021). Thus far, the effect of vesicles on changes in the production of several different cytokines has been shown, including TNF-α (one of the major proinflammatory cytokines regulating immune cells), which controls apoptotic pathways and stimulates the further release of other cytokines, or IL-10 (a potent anti-inflammatory interleukin), which suppresses the secretion of proinflammatory cytokines (Zhang and An, 2007). Vargas et al. (2015) showed that both TNF-α and IL-10 were released by murine primary phagocytes and cells from cell lineages in greater amounts after stimulation with fungal EVs than in the control samples, making it challenging to have a clear definition of the type of immune response induced. These studies showed also the increased level of IL-10 produced by murine dendritic cells compared with control without EVs added (Vargas et al., 2015); however, in later studies, this tendency was observed with a lower level of IL-10 compared with the previous result, albeit it was still higher than in the control sample (Vargas et al., 2020). In contrast, in the work of Zamith-Miranda et al. (2021), the production of TNF-α and IL-10 by murine dendritic cells was not detected after their stimulation with EVs produced by C. albicans and C. auris. In the current study, the immunomodulating properties of EVs of each investigated non-albicans Candida species were demonstrated by the evaluation of the level of cytokines released by human THP-1 cells differentiated into macrophage-like cells. The obtained results revealed an increasing tendency in the production of the proinflammatory cytokine TNF-α and a decreasing release of the anti-inflammatory cytokine IL-10 compared with cells untreated with EVs. This might be a signal that C. glabrata, C. tropicalis, and C. parapsilosis biofilm-derived vesicles induce a rather proinflammatory response of the human immune cells, thereby making Candida non-albicans biofilm vesicles promising components in anti-Candida vaccines. Reducing the concentration of IL-10 at the site of infection leads to increased apoptosis and inflammation, and although it accelerates the removal of the microbial factor causing infection, it also triggers significant damage associated with the expansion of the inflammatory state (Vazquez-Torres et al., 1999; Cyktor and Turner, 2011). The observed increase in the IL-8 secretion by THP-1 cells allows us to conclude that the presence of EVs of each tested non-albicans Candida species might also stimulate response involving other immune cells, as IL-8 is a chemoattractant affecting neutrophils and stimulating their chemotaxis, degranulation, and cytotoxicity, and the enhanced production of this interleukin is a signal favoring their migration to the infectious niche. Therefore, all these observations indicate rather the stimulation of a defense response of human cells in response to Candida non-albicans vesicles at the studied contact time.
As previously demonstrated for C. albicans, EVs also contribute to the regulation of fungal biofilm (Zarnowski et al., 2018; Zarnowski et al., 2021). The analysis of the formation of biofilm by C. tropicalis cells in the presence of EVs derived from the 48-h biofilm culture, introduced at an early stage of biofilm production, corroborated the substantial role of vesicles in the process of drug resistance that occurs in this complex structure of pathogen existence. Although the introduction of vesicles into cells growing without the presence of antifungal drugs did not cause a statistically significant effect on the biofilm (it also developed without EVs), in the presence of caspofungin and fluconazole, the protective effect of the vesicles was noticeable. In the case of the two drugs tested, both the thickness of the biofilm and the metabolic activity of the cells in the biofilm were partially restored if EVs from the mature biofilm were present at the stage of biofilm initiation; similarly, in the presence of caspofungin, cell viability was higher in biofilms supported by the added EVs. In the case of C. tropicalis biofilm, this may indicate the involvement of vesicles, among other mechanisms, in resistance to antifungals. Related conclusions were presented previously for the susceptibility of C. albicans biofilm to fluconazole, but these were from studies using mutant strains impaired in vesicle secretion and with incomplete cargo (Zarnowski et al., 2018; Zarnowski et al., 2021). With regard to C. tropicalis, the mechanisms of the observed phenomenon still require further detailed studies.
The characteristic and immunobiological properties of candidal vesicles investigated in the present study reveal to some extent the complexity of functions performed by fungal biofilm-derived EVs, thus also indicating the need for further research in this area which could result in a better understanding of the mechanism of development and pathogenesis of candidal infections.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: http://www.proteomexchange.org/, PXD033327; http://www.proteomexchange.org/, PXD033299; and http://www.proteomexchange.org/,PXD033300.
KK, JK-K, EK, OW, and MR-K planned the methodology and experiments. KK, JK-K, EK, and OW executed the experiments. PK and AO provided the resources. KK and JK-K wrote the manuscript draft. MR-K, AK, EZ-S, AO, and EP contributed to the editing and preparation of the final version of the manuscript. All authors contributed to the article and approved the submitted version.
This work was financially supported by the National Science Centre of Poland (grant no. 2019/33/B/NZ6/02284 awarded to MR-K).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We acknowledge the Proteomics and Mass Spectrometry Core Facility of the Malopolska Centre of Biotechnology for protein identification.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.879237/full#supplementary-material
Albuquerque, P., Nakayasu, E., Rodrigues, M. L., Frases, S., Casadevall, A., Zancope-Oliveira, R. M., et al. (2008). Vesicular Transport in Histoplasma Capsulatum: An Effective Mechanism for Trans-Cel Wall Transfer of Proteins and Lipids in Ascomycetes. Cell. Microbiol. 10 (8), 1695–1710. doi: 10.1111/j.1462-5822.2008.01160.x
Arendrup, M. C. (2013). Candida and Candidaemia. Susceptibility and Epidemiology. Dan Med. J. 60 (11), B4698.
Arnaud, M. B., Costanzo, M. C., Skrzypek, M. S., Binkley, G., Lane, C., Miyasato, S. R., et al. (2005). The Genome Database (CGD), a Community Resource for Gene and Protein Information. Nucleic Acids Res. 33, D358–D363. doi: 10.1093/nar/gki003
Bizerra, F. C., Nakamura, C. V., de Poersch, C., Estivalet Svidzinski, T. I., Borsato Quesada, R. M., Goldenberg, S., et al. (2008). Characteristics of Biofilm Formation by Candida Tropicalis and Antifungal Resistance. FEMS Yeast Res. 8 (3), 442–450. doi: 10.1111/j.1567-1364.2007.00347.x
Bleackley, M. R., Dawson, C. S., Anderson, M. A. (2019). Fungal Extracellular Vesicles With a Focus on Proteomic Analysis. Proteomics 19 (8), e1800232. doi: 10.1002/pmic.201800232
Bobis-Wozowicz, S., Kmiotek, K., Kania, K., Karnas, E., Labedz-Maslowska, A., Sekula, M., et al. (2017). Diverse Impact of Xeno-Free Conditions on Biological and Regenerative Properties of hUC-MSCs and Their Extracellular Vesicles. J. Mol. Med. (Berl) 95 (2), 205–220. doi: 10.1007/s00109-016-1471-7
Brown, L., Kessler, A., Cabezas-Sanchez, P., Luque-Garcia, J. L., Casadevall, A. (2014). Extracellular Vesicles Produced by the Gram-Positive Bacterium Bacillus Subtilis are Disrupted by the Lipopeptide Surfactin. Mol. Microbiol. 93 (1), 183–198. doi: 10.1111/mmi.12650
Cavalheiro, M., Teixeira, M. C. (2018). Candida Biofilms: Threats, Challenges, and Promising Strategies. Front. Med. (Lausanne) 5, 28. doi: 10.3389/fmed.2018.00028
Chandra, J., McCormick, T. S., Imamura, Y., Mukherjee, P. K., Ghannoum, M. A. (2007). Interaction of Candida Albicans With Adherent Human Peripheral Blood Mononuclear Cells Increases C. Albicans Biofilm Formation and Results in Differential Expression of Pro- and Anti-Inflammatory Cytokines. Infect. Immun. 75 (5), 2612–2620. doi: 10.1128/IAI.01841-06
Cyktor, J. C., Turner, J. (2011). Interleukin-10 and Immunity Against Prokaryotic and Eukaryotic Intracellular Pathogens. Infect. Immun. 79 (8), 2964–2973. doi: 10.1128/IAI.00047-11
Dawson, C. S., Garcia-Ceron, D., Rajapaksha, H., Faou, P., Bleackley, M. R., Anderson, M. A. (2020). Protein Markers for Candida Albicans EVs Include Claudin-Like Sur7 Family Proteins. J. Extracell. Vesicles 9 (1), 1750810. doi: 10.1080/20013078.2020.1750810
de Toledo Martins, S., Szwarc, P., Goldenberg, S., Alves, L. R. (2018). Extracellular Vesicles in Fungi: Composition and Functions. Curr. Top. Microbiol. Immunol. 422, 45–59. doi: 10.1007/82_2018_141
Faure, J., Lachenal, G., Court, M., Hirrlinger, J., Chatellard-Causse, C., Blot, B., et al. (2006). Exosomes are Released by Cultured Cortical Neurons. Mol. Cell. Neurosci. 31 (4), 642–648. doi: 10.1016/j.mcn.2005.12.003
Ferreira, J. A., Carr, J. H., Starling, C. E., de Resende, M. A., Donlan, R. M. (2009). Biofilm Formation and Effect of Caspofungin on Biofilm Structure of Candida Species Bloodstream Isolates. Antimicrob. Agents Chemother. 53 (10), 4377–4384. doi: 10.1128/AAC.00316-09
Garcia-Ceron, D., Bleackley, M. R., Anderson, M. A. (2021). Fungal Extracellular Vesicles in Pathophysiology. Subcell. Biochem. 97, 151–177. doi: 10.1007/978-3-030-67171-6_7
Giacobbe, D. R., Maraolo, A. E., Simeon, V., Magnè, F., Pace, M. C., Gentile, I., et al. (2020). Changes in the Relative Prevalence of Candidaemia Due to non-Albicans Candida Species in Adult in-Patients: A Systematic Review, Meta-Analysis and Meta-Regression. Mycoses 63 (4), 334–342. doi: 10.1111/myc.13054
Gil-Bona, A., Llama-Palacios, A., Parra, C. M., Vivanco, F., Nombela, C., Monteoliva, L., et al. (2015). Proteomics Unravels Extracellular Vesicles as Carriers of Classical Cytoplasmic Proteins in Candida Albicans. J. Proteome Res. 14 (1), 142–153. doi: 10.1021/pr5007944
Gómez-Molero, E., de Boer, A. D., Dekker, H. L., Moreno-Martínez, A., Kraneveld, E. A., Chauhan, N., et al. (2015). Proteomic Analysis of Hyperadhesive Candida Glabrata Clinical Isolates Reveals a Core Wall Proteome and Differential Incorporation of Adhesins. FEMS Yeast Res. 15 (8), fov098. doi: 10.1093/femsyr/fov098
Gomez, M. J., Torosantucci, A., Arancia, S., Maras, B., Parisi, L., Cassone, A. (1996). Purification and Biochemical Characterization of a 65-Kilodalton Mannoprotein (MP65), a Main Target of Anti-Candida Cell-Mediated Immune Responses in Humans. Infect. Immun. 64 (7), 2577–2584. doi: 10.1128/iai.64.7.2577-2584.1996
Herkert, P. F., Amatuzzi, R. F., Alves, L. R., Rodrigues, M. L. (2019). Extracellular Vesicles as Vehicles for the Delivery of Biologically Active Fungal Molecules. Curr. Protein Pep. Sci. 20 (10), 1027–1036. doi: 10.2174/1389203720666190529124055
Hughes, C. S., Foehr, S., Garfield, D. A., Furlong, E. E., Steinmetz, L. M., Krijgsveld, J. (2014). Ultrasensitive Proteome Analysis Using Paramagnetic Bead Technology. Mol. Syst. Biol. 10, 757. doi: 10.15252/msb.20145625
Joffe, L. S., Nimrichter, L., Rodrigues, M. L., Del Poeta, M. (2016). Potential Roles of Fungal Extracellular Vesicles During Infection. mSphere 1 (4), 11–14. doi: 10.1128/mSphere.00099-16
Kanehisa, M., Goto, S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28 (1), 27–30. doi: 10.1093/nar/28.1.27
Karkowska-Kuleta, J., Kulig, K., Karnas, E., Zuba-Surma, E., Woznicka, O., Pyza, E., et al. (2020). Characteristics of Extracellular Vesicles Released by the Pathogenic Yeast-Like Fungi Candida Glabrata, Candida Parapsilosis and Candida Tropicalis. Cells 9 (7), 1722. doi: 10.3390/cells9071722
Kaur, R., Ma, B., Cormack, B. P. (2007). A Family of Glycosylphosphatidylinositol-Linked Aspartyl Proteases is Required for Virulence of Candida Glabrata. Proc. Natl. Acad. Sci. U. S. A. 104 (18), 7628–7633. doi: 10.1073/pnas.0611195104
Knox, K., Cullen, J., Work, E. (1967). An Extracellular Lipopolysaccharide-Phospholipid-Protein Complex Produced by Escherichia Coli Grown Under Lysine-Limiting Conditions. Biochem. J. 103 (1), 192–201. doi: 10.1042/bj1030192
Lee, E., Choi, D., Kim, D., Kim, J., Park, J., Kim, S., et al. (2009). Gram-Positive Bacteria Produce Membrane Vesicles: Proteomics-Based Characterization of Staphylococcus Aureus-Derived Membrane Vesicles. Proteomics 9 (24), 5425–5436. doi: 10.1002/pmic.200900338
Lee, J., Choi, C., Lee, T., Kim, S., Lee, J. C., Shin, J. H. (2013). Transcription Factor ςb Plays an Important Role in the Production of Extracellular Membrane-Derived Vesicles in Listeria Monocytogenes. PLoS One 8 (8), e73196. doi: 10.1371/journal.pone.0073196
Martínez-López, R., Hernáez, M. L., Redondo, E., Calvo, G., Radau, S., Pardo, M., et al. (2020). Candida albicans Hyphal Extracellular Vesicles Are Different From Yeast Ones, Carrying an Active Proteasome Complex and Showing a Different Role in Host Immune Response. Microbiol. Spectr., e0069822. doi: 10.1128/spectrum.00698-22
Munhoz da Rocha, I. F., Amatuzzi, R. F., Lucena, A., Faoro, H., Alves, L. R. (2020). Cross-Kingdom Extracellular Vesicles EV-RNA Communication as a Mechanism for Host-Pathogen Interaction. Front. Cell. Infect. Microbiol. 10, 593160. doi: 10.3389/fcimb.2020.593160
Netea, M. G., Joosten, L. A., van der Meer, J. W., Kullberg, B. J., van de Veerdonk, F. L. (2015). Immune Defence Against Candida Fungal Infections. Nat. Rev. Immunol. 15 (10), 630–642. doi: 10.1038/nri3897
Nisini, R., Romagnoli, G., Gomez, M. J., La Valle, R., Torosantucci, A., Mariotti, S., et al. (2001). Antigenic Properties and Processing Requirements of 65-Kilodalton Mannoprotein, a Major Antigen Target of Anti-Candida Human T-Cell Response, as Disclosed by Specific Human T-Cell Clones. Infect. Immun. 69 (6), 3728–3736. doi: 10.1128/IAI.69.6.3728-3736.2001
Nobile, C. J., Johnson, A. D. (2015). Candida Albicans Biofilms and Human Disease. Annu. Rev. Microbiol. 69, 71–92. doi: 10.1146/annurev-micro-091014-104330
Olaya-Abril, A., Prados-Rosales, R., McConnell, M., Martin-Pena, R., Gonzalez-Reyes, J. A., Jimenez-Munguia, I., et al. (2014). Characterization of Protective Extracellular Membrane-Derived Vesicles Produced by Streptococcus Pneumoniae. J. Proteomics 106, 46–60. doi: 10.1016/j.jprot.2014.04.023
Oliveira, D. L., Nakayasu, E. S., Joffe, L. S., Guimarães, A. J., Sobreira, T. J. P., Nosanchuk, J. D., et al. (2010). Characterization of Yeast Extracellular Vesicles: Evidence for the Participation of Different Pathways of Cellular Traffic in Vesicle Biogenesis. PLoS One 5 (6), e11113. doi: 10.1371/journal.pone.0011113
Pereira, R., Dos Santos Fontenelle, R. O., de Brito, E., de Morais, S. M. (2021). Biofilm of Candida Albicans: Formation, Regulation and Resistance. J. Appl. Microbiol. 131 (1), 11–22. doi: 10.1111/jam.14949
Perez-Riverol, Y., Bai, J., Bandla, C., Hewapathirana, S., García-Seisdedos, D., Kamatchinathan, S., et al. (2022). The PRIDE Database Resources in 2022: A Hub for Mass Spectrometry-Based Proteomics Evidences. Nucleic Acids Res. 50 (D1), D543–D552. doi: 10.1093/nar/gkab1038
Pietrella, D., Bistoni, G., Corbucci, C., Perito, S., Vecchiarelli, A. (2006). Candida Albicans Mannoprotein Influences the Biological Function of Dendritic Cells. Cell. Microbiol. 8 (4), 602–612. doi: 10.1111/j.1462-5822.2005.00651.x
Piffer, A. C., Kuczera, D., Rodrigues, M. L., Nimrichter, L. (2021). The Paradoxical and Still Obscure Properties of Fungal Extracellular Vesicles. Mol. Immunol. 135, 137–146. doi: 10.1016/j.molimm.2021.04.009
Ponde, N. O., Lortal, L., Ramage, G., Naglik, J. R., Richardson, J. P. (2021). Candida Albicans Biofilms and Polymicrobial Interactions. Crit. Rev. Microbiol. 47 (1), 91–111. doi: 10.1080/1040841X.2020.1843400
Rivera, J., Cordero, R. J. B., Nakouzi, A. S., Frases, S., Nicola, A., Casadevall, A. (2010). Bacillus Anthracis Produces Membranę-Derived Vesicles Containing Biologically Active Toxins. Proc. Natl. Acad. Sci. U. S. A. 107 (44), 19002–19007. doi: 10.1073/pnas.1008843107
Sasani, E., Khodavaisy, S., Rezaie, S., Salehi, M., Yadegari, M. H. (2021). The Relationship Between Biofilm Formation and Mortality in Patients With Candida Tropicalis Candidemia. Microb. Pathog. 155, 104889. doi: 10.1016/j.micpath.2021.104889
Silva, S., Negri, M., Henriques, M., Oliveira, R., Williams, D. W., Azeredo, J. (2011). Adherence and Biofilm Formation of non-Candida Albicans Candida Species. Trends Microbiol. 19 (5), 241–247. doi: 10.1016/j.tim.2011.02.003
Silva, S., Negri, M., Henriques, M., Oliveira, R., Williams, D. W., Azeredo, J. (2012). Candida Glabrata, Candida Parapsilosis and Candida Tropicalis: Biology, Epidemiology, Pathogenicity and Antifungal Resistance. FEMS Microbiol. Rev. 36 (2), 288–305. doi: 10.1111/j.1574-6976.2011.00278.x
Surman, M., Hoja-Łukowicz, D., Szwed, S., Kędracka-Krok, S., Jankowska, U., Kurtyka, M., et al. (2019). An Insight Into the Proteome of Uveal Melanoma-Derived Ectosomes Reveals the Presence of Potentially Useful Biomarkers. Int. J. Mol. Sci. 20, 3789. doi: 10.3390/ijms20153789
Taff, H. T., Nett, J. E., Zarnowski, R., Ross, K. M., Sanchez, H., Cain, M. T., et al. (2012). A Candida Biofilm-Induced Pathway for Matrix Glucan Delivery: Implications for Drug Resistance. PLoS Path. 8 (8), e1002848. doi: 10.1371/journal.ppat.1002848
Tashiro, Y., Ichikawa, S., Shimizu, M., Toyofuku, M., Takaya, N., Nakajima-Kambe, T., et al. (2010). Variation of Physiochemical Properties and Cell Association Activity of Membrane Vesicles With Growth Phase in. Pseudomonas aeruginosa Appl. Environ. Microbiol. 76 (11), 3732–3739. doi: 10.1128/AEM.02794-09
Tsui, C., Kong, E. F., Jabra-Rizk, M. A. (2016). Pathogenesis of Candida Albicans Biofilm. Pathog. Dis. 74 (4), ftw018. doi: 10.1093/femspd/ftw018
Tumbarello, M., Posteraro, B., Trecarichi, E. M., Fiori, B., Rossi, M., Porta, R., et al. (2007). Biofilm Production by Candida Species and Inadequate Antifungal Therapy as Predictors of Mortality for Patients With Candidemia. J. Clin. Microbiol. 45 (6), 1843–1850. doi: 10.1128/JCM.00131-07
UniProt Consortium (2019). UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 47 (D1), D506–D515. doi: 10.1093/nar/gky1049
Vallejo, M. C., Nakayasu, E. S., Matsuo, A. L., Sobreira, T. J. P., Longo, L. V. G., Ganiko, L., et al. (2012). Vesicle and Vesicle-Free Extracellular Proteome of Paracoccidioides Brasiliensis: Comparative Analysis With Other Pathogenic Fungi. J. Proteome Res. 11 (3), 1676–1685. doi: 10.1021/pr200872s
Vargas, G., Honorato, L., Guimarães, A. J., Rodrigues, M. L., Reis, F., Vale, A. M., et al. (2020). Protective Effect of Fungal Extracellular Vesicles Against Murine Candidiasis. Cell. Microbiol. 22 (10), e13238. doi: 10.1111/cmi.13238
Vargas, G., Rocha, J. D., Oliveira, D. L., Albuquerque, P. C., Frases, S., Santos, S. S., et al. (2015). Compositional and Immunobiological Analyses of Extracellular Vesicles Released by Candida Albicans. Cell. Microbiol. 17 (3), 389–407. doi: 10.1111/cmi.12374
Vazquez-Torres, A., Jones-Carson, J., Wagner, R. D., Warner, T., Balish, E. (1999). Early Resistance of Interleukin-10 Knockout Mice to Acute Systemic Candidiasis. Infect. Immun. 67 (2), 670–674. doi: 10.1128/IAI.67.2.670-674.1999
Wolf, J. M., Espadas-Moreno, J., Luque-Garcia, J. L., Casadevall, A. (2014). Interaction of Cryptococcus Neoformans Extracellular Vesicles With the Cell Wall. Eukaryot. Cell 13 (12), 1484–1493. doi: 10.1128/EC.00111-14
Xu, Z., Wang, K., Min, D., Soteyome, T., Lan, H., Hong, W., et al. (2022). Regulatory Network Controls Microbial Biofilm Development, With Candida Albicans as a Representative: From Adhesion to Dispersal. Bioengineered 13 (1), 253–267. doi: 10.1080/21655979.2021.1996747
You, Y., Borgmann, K., Edara, V. V., Stacy, S., Ghorpade, A., Ikezu, T. (2020). Activated Human Astrocyte-Derived Extracellular Vesicles Modulate Neuronal Uptake, Differentiation and Firing. J. Extracell. Ves. 9 (1), 1706801. doi: 10.1080/20013078.2019.1706801
Yu, L., Lee, K. K., Ens, K., Doig, P. C., Carpenter, M. R., Staddon, W., et al. (1994). Partial Characterization of a Candida Albicans Fimbrial Adhesin. Infect. Immun. 62 (7), 2834–2842. doi: 10.1128/iai.62.7.2834-2842.1994
Zamith-Miranda, D., Heyman, H. M., Couvillion, S. P., Cordero, R., Rodrigues, M. L., Nimrichter, L., et al. (2021). Comparative Molecular and Immunoregulatory Analysis of Extracellular Vesicles From Candida Albicans and Candida Auris. mSystems 6 (4), e0082221. doi: 10.1128/mSystems.00822-21
Zamith-Miranda, D., Nimrichter, L., Rodrigues, M. L., Nosanchuk, J. D. (2018). Fungal Extracellular Vesicles: Modulating Host-Pathogen Interactions by Both the Fungus and the Host. Microbes Infect. 20 (9-10), 501–504. doi: 10.1016/j.micinf.2018.01.011
Zarnowski, R., Noll, A., Chevrette, M. G., Sanchez, H., Jones, R., Anhalt, H., et al. (2021). Coordination of Fungal Biofilm Development by Extracellular Vesicle Cargo. Nat. Commun. 12 (1), 6235. doi: 10.1038/s41467-021-26525-z
Zarnowski, R., Sanchez, H., Covelli, A. S., Dominguez, E., Jaromin, A., Bernhardt, J., et al. (2018). Candida Albicans Biofilm-Induced Vesicles Confer Drug Resistance Through Matrix Biogenesis. PLoS Biol. 16 (10), e2006872. doi: 10.1371/journal.pbio.2006872
Zhang, J. M., An, J. (2007). Cytokines, Inflammation, and Pain. Int. Anesthesiol. Clin. 45 (2), 27–37. doi: 10.1097/AIA.0b013e318034194e
Zhang, W., Song, X., Wu, H., Zheng, R. (2020). Epidemiology, Species Distribution, and Predictive Factors for Mortality of Candidemia in Adult Surgical Patients. BMC Infect. Dis. 20 (1), 506. doi: 10.1186/s12879-020-05238-6
Zhao, M., Zhang, F., Zarnowski, R., Barns, K., Jones, R., Fossen, J., et al. (2021). Turbinmicin Inhibits Candida Biofilm Growth by Disrupting Fungal Vesicle-Mediated Trafficking. J. Clin. Invest. 131 (5), e145123. doi: 10.1172/JCI145123
Keywords: extracellular vesicles, cytokines, pathogenic fungi, candidiasis, biofilm
Citation: Kulig K, Karnas E, Woznicka O, Kuleta P, Zuba-Surma E, Pyza E, Osyczka A, Kozik A, Rapala-Kozik M and Karkowska-Kuleta J (2022) Insight Into the Properties and Immunoregulatory Effect of Extracellular Vesicles Produced by Candida glabrata, Candida parapsilosis, and Candida tropicalis Biofilms. Front. Cell. Infect. Microbiol. 12:879237. doi: 10.3389/fcimb.2022.879237
Received: 19 February 2022; Accepted: 11 May 2022;
Published: 06 June 2022.
Edited by:
Roberta Peres Da Silva, University of Exeter, United KingdomReviewed by:
Ernesto Satoshi Nakayasu, Pacific Northwest National Laboratory (DOE), United StatesCopyright © 2022 Kulig, Karnas, Woznicka, Kuleta, Zuba-Surma, Pyza, Osyczka, Kozik, Rapala-Kozik and Karkowska-Kuleta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Justyna Karkowska-Kuleta, anVzdHluYS5rYXJrb3dza2FAdWouZWR1LnBs
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.