
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 13 April 2022
Sec. Virus and Host
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.878430
This article is part of the Research Topic The Interaction between the Host and Viruses Causing Respiratory or Gastrointestinal Tract Infections View all 8 articles
Respiratory syncytial virus (RSV) is the most common and critical viral pathogen causing acute lower respiratory tract infection in infants and young children and has a huge disease burden worldwide. At present, there are many studies on RSV transcriptomics exploring the mechanism of disease, but different studies show different gene expression patterns and results due to different sample collection platforms and data analysis strategies. A meta-analysis was performed on eight whole blood transcriptome datasets containing 436 children with acute RSV infection and 241 healthy children. A total of 319 differentially expressed genes (DEGs) (P value <0.0001) were identified in a meta-analysis using a random effect model. Functional enrichment analysis showed that several pathways related to immunity were significantly altered, including the “chemokine signaling pathway”, “natural killer cell mediated cytotoxicity” and “cytokine–cytokine receptor interaction”. Immune cell type analysis showed that the proportion of neutrophils in most RSV-infected children was higher than that in healthy children. These immune characteristics may help to provide new insights into RSV infection in children.
Human respiratory syncytial virus (RSV), a negative sense, enveloped RNA virus, belongs to the Pneumoviridae family (Amarasinghe et al., 2019). Approximately all infants become infected with RSV by the age of 2 years, making it the leading viral pathogen of acute lower respiratory tract infections (ALRIs) in children (Glezen et al., 1986; Ge et al., 2018). It is estimated that more than 3 million hospitalizations in children within 5 years of age are associated with RSV ALRI per year globally, resulting in nearly 60,000 deaths (Shi et al., 2017). Although facing a huge disease burden, the current treatment remains limited to supportive treatment, and there is no safe and effective vaccine and antiviral drugs (Behzadi and Leyva-Grado, 2019). Therefore, it is necessary to further explore the pathogenic mechanism of RSV infection and provide a new understanding for the prevention and treatment of RSV.
The microarray is used as a common tool for transcriptomics analysis of differentially expressed genes (DEGs) and pathway exploration of disease-related mechanisms (Burel and Peters, 2018). However, the microarray results are often not repeatable, and even slight disturbances are unreliable due to different sample acquisitions, platforms, and inconsistent data analysis strategies, making different microarray studies on the same disease show inconsistent gene expression patterns and results (Cahan et al., 2007). The meta-analysis of microarray data has been proposed to greatly improve the gain of information obtained from microarray experiments and to summarize and analyse gene expression data from individual research through the statistical algorithm to determine the common “higher levels” transcript spectrum (Adams et al., 2008). In particular, the addition of large samples is expected to detect genetic differences in genes that may not be discovered in individual studies and minimize false-positive discovery rates (Adams et al., 2008).
In this study, a meta-analysis was performed of eight transcriptomic studies containing genome-wide expression profiles of whole blood samples from children with acute RSV respiratory tract infection and healthy children. In addition, transcriptome changes in children with RSV respiratory tract infection were characterized, which may provide new insights into pathogenesis.
The gene expression microarray data of respiratory RSV infection in children by the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/) were searched. The keywords were “respiratory syncytial virus” and “children”. The inclusion criteria of the dataset were as follows: 1) studies containing human whole blood samples; 2) each dataset contained more than 3 samples; 3) analysis on the expression microarray platform; and 4) studies containing individual patient expression data. The details of the selected dataset are shown in Table 1.
The original expression spectra of the datasets were extracted, and the data were processed by R for statistical analysis. The results were meta-analysed using the random-effects model, and DEGs were determined according to “random P <0.0001”.
The DEGs identified in this meta-analysis were annotated by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) using the annotation, visualization, and integrated discovery database (DAVID, https://david.ncifcrf.gov/), with a filter condition of P < 0.05.
To explore the potential relationship between immune cells in children with acute respiratory infection with RSV, the mRNA expression matrix was normalized, and the CIBERSORT algorithm (https://cibersort.stanford.edu/index.php) (Newman et al., 2015) with the original CIBERSORT gene signature file LM22 and 1000 permutations was used to estimate the content of 22 human immune cells in each dataset.
A total of 8 studies containing microarray expression data from 436 children with RSV infection and 241 healthy children were included in the meta-analysis to determine DEGs. Details of the selected datasets are shown in Table 1. All samples were obtained from patients’ whole blood. Six studies used the same microarray platform, GPL10558. GSE38900 had the largest sample size, including 135 children with RSV infection and 39 healthy children. In each study, the case groups consisted of children with acute RSV respiratory infection, and the control group consisted of healthy children except GSE117827, in which the control subjects were from children having ambulatory surgery for nonacute conditions. DEGs were screened from each study with q values less than 0.001 and fold changes greater than 2 indicating upregulation; fold changes less than 0.5 indicated downregulation as criteria. There were no DEGs screened out in GSE117827 and GSE103119. The UpSet diagram shows overlapping DEGs in six studies (Figure 1), with all DEGs identified by only meta-analysis rather than in individual analysis. The overlap between GSE38900 and GSE80179 was the largest, including 254 DEGs. Few genes (only two) were identified by the five studies. It was difficult to validate certain DEGs based on low experimental repeatability according to the traditional analysis method.
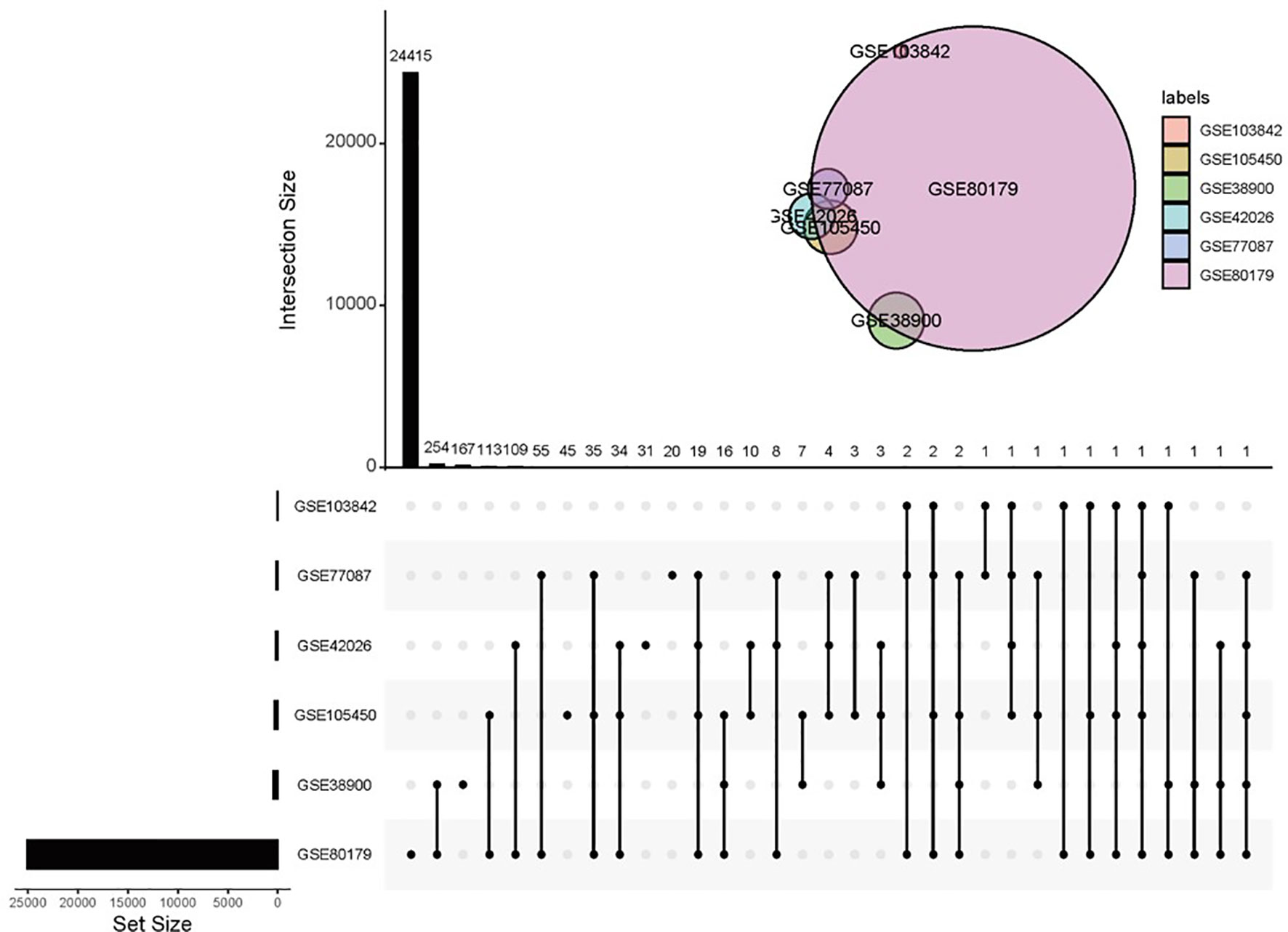
Figure 1 Overlapping DEGs in different studies. Note that there was no gene present in any of the six datasets in the UpSet diagram.
To validate the DEGs according to different studies, we performed a meta-analysis on these transcriptome datasets. The eight studies were meta-analysed using a random effect model with 105280 genes detected. A total of 319 DEGs had a P value <0.0001 (Table S1), among which the expressions of 46 genes were upregulated and the expressions of 273 genes were downregulated. Table 2 shows the top 20 most significantly upregulated and downregulated DEGs. Sialic acid binding Ig-like lectin 1 (SIGLEC1) had the largest upregulated TE value=1.42 (P value=1.29E-12), while ribosomal protein S10 pseudogene 9 (RPS10P9) had the largest downregulated TE value=-1.52 (P value=2.45E-13). The first three DEGs were TPT1 pseudogene 4 (TPT1P4), adhesion G protein-coupled receptor E4 (EMR4P) and ribosomal protein S7 pseudogene 11 (RPS7P11), with P value <5.00E-20 in each case.
Gene functional classification analysis was performed to study the function of DEGs (Figure 2). Based on molecular functions, most DEGs mapped to ribosomal structural components and immune molecules. The DEGs were mainly involved in biological processes related to signal transduction, inflammatory response and translation.
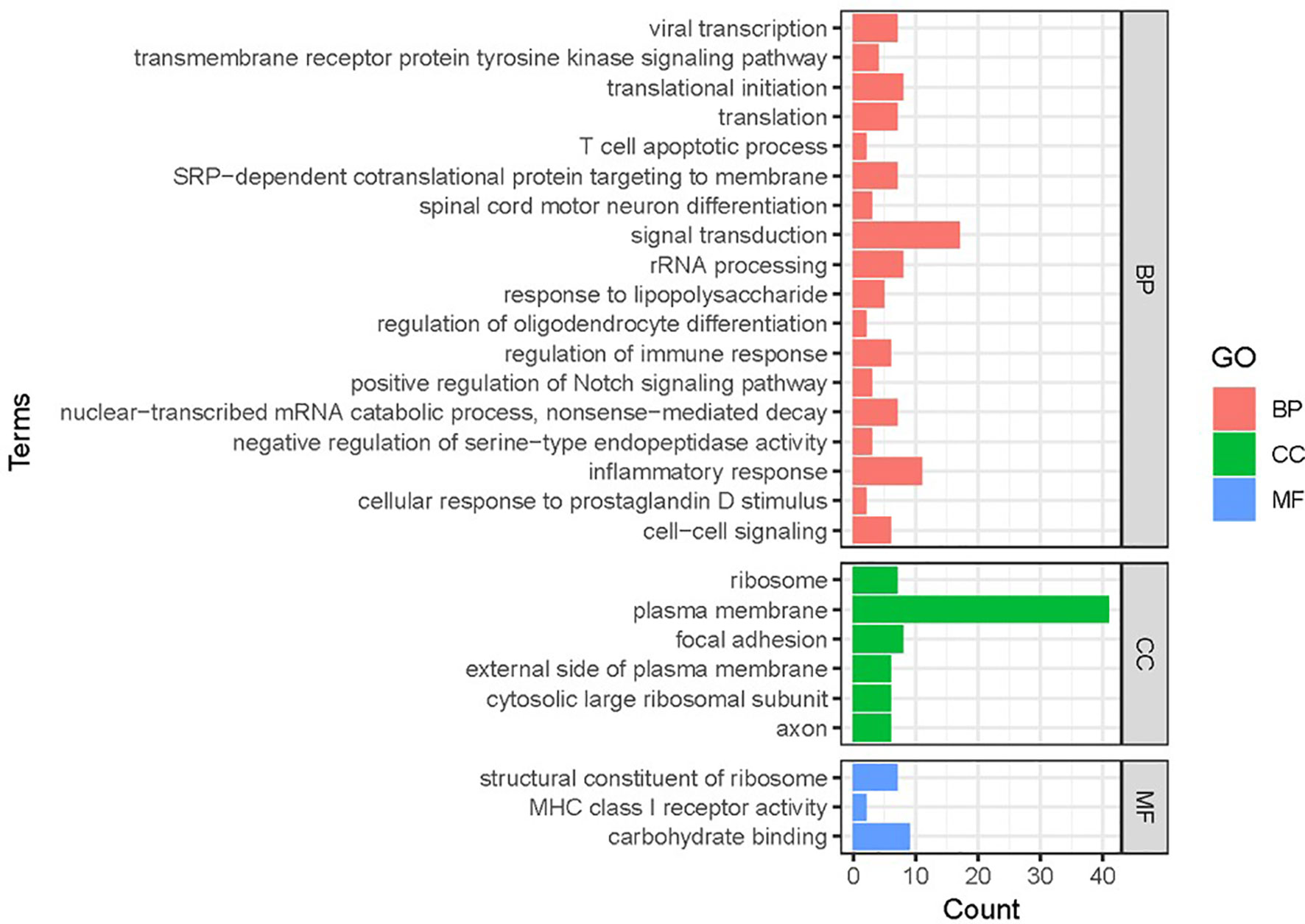
Figure 2 Gene functional classification of DEGs in RSV-infected children. BP, biological processes; CC, cellular compartment; MF, molecular function. Note that most DEGs belonged to “ribosomal processing” and “immune-related signaling”.
KEGG enrichment analysis was performed on DEGs to further investigate the related biological pathways (Figure 3). DEGs were significantly enriched in “ribosome” (P value=7.07E-07), followed by “chemokine signaling pathway” (P value=2.63E-04). Other enriched pathways included “natural killer cell mediated cytotoxicity”, “cytokine–cytokine receptor interaction”, and “cell adhesion molecules (CAMs)” (P value <1.00E-03).
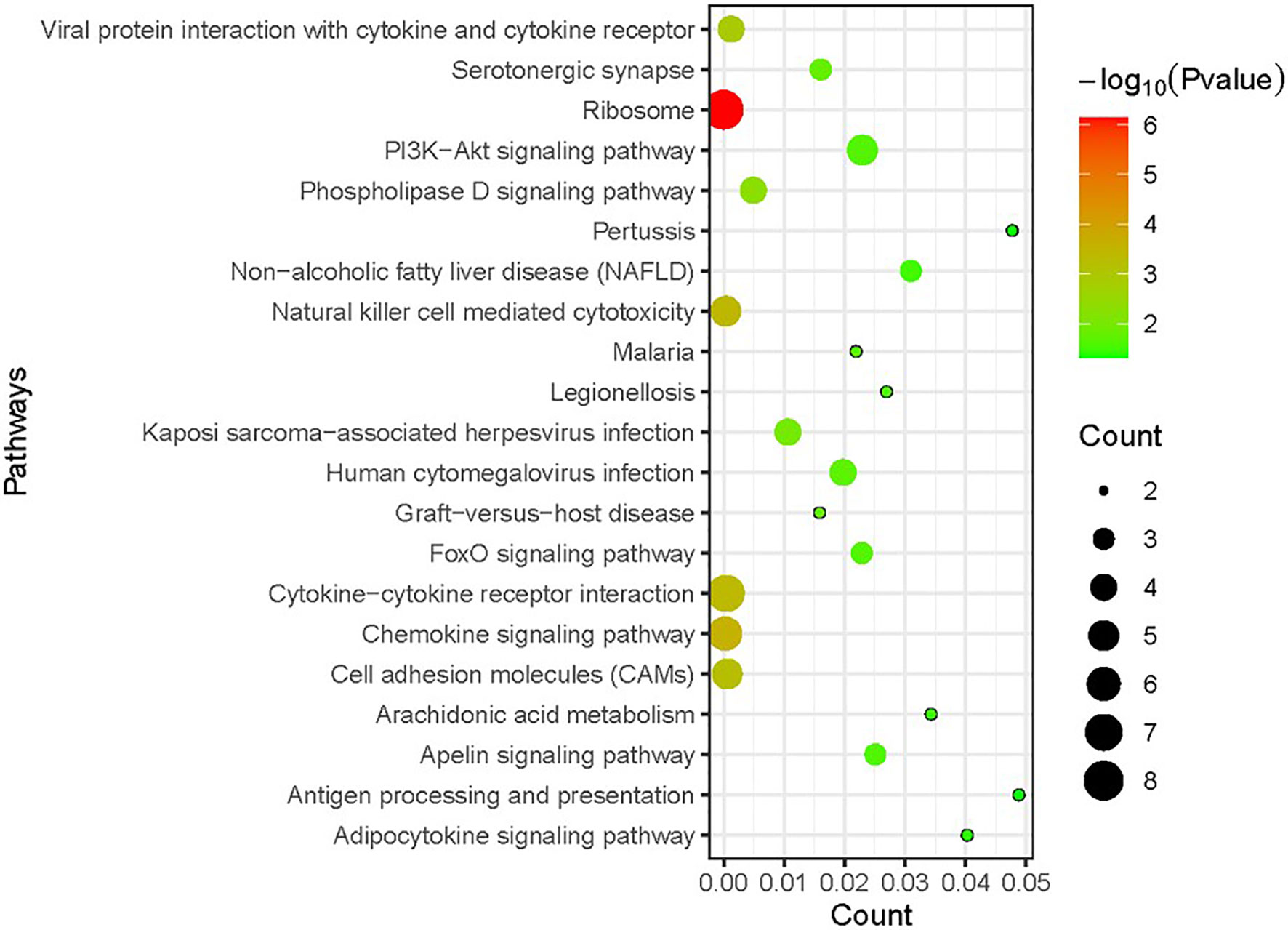
Figure 3 Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DEGs in RSV-infected children compared with healthy children. The size of the circle indicates the gene number; the colour represents the log P values. P value <0.05 and FDR < 0.01 were used as the thresholds for pathway assignment. Note that most DEGs were enriched in immune-related pathways such as “chemokine signaling pathway”, “natural killer cell-mediated cytotoxicity” and “cytokine–cytokine receptor interaction”.
DEG was screened for genes associated with RSV, and the results of a meta-analysis of the four genes with the most significant expression changes are reported in Figure 4, including NOV, a member of the CCN (Cyr61, CTGF, and NOV) family, with a P value of 1.17E-16, Arachidonate 15-lipoxygenase (ALOX15, P value=5.39E-11), Charcot-Leyden Crystal Galectin (CLC, P value=4.10E-10) and Killer Cell lectin-like Receptor B1 (KLRB1, P value=7.43E-10). Based on the random effect model, the combined average difference and 95% confidence interval (CI) of the four most significant RSV-related genes were -1.28 (95% CI, -1.58 to - 0.98), -0.96 (95% CI, -1.25 to - 0.67), -1.18 (95% CI, -1.55 to -0.81), and -1.42 (95% CI, -1.87 to -0.97), respectively. Except for GSE117827, the mean fold changes of NOV and CLC in the experimental group were all downregulated by 2-fold or more than that in the control group. ALOX15 was mostly downregulated by more than 2-fold in datasets GSE105450, GSE103119, GSE103842, GSE80179, GSE77087, and GSE42026, while KLRB1 was mostly downregulated by more than 2-fold in datasets GSE105450, GSE103842, GSE80179, GSE77087, and GSE42026. Four of these genes had the largest changes in dataset GSE80179.
The ratio of different immunocytes in whole blood was analysed using CIBERSORT, as shown in Figure 5. Monocytes, neutrophils, CD8 T cells and resting NK cells were the dominant cell types in the whole blood of the healthy groups and the RSV-infected groups. In 6 of the 8 datasets, the proportion of monocytes and neutrophils in children infected with RSV was higher than that in healthy controls. The proportion of resting NK cells in the RSV-infected groups was lower than that in the healthy control groups in the 7 datasets. The proportions of CD8 T cells and primordial B cells in the healthy groups and RSV-infected groups were variable.
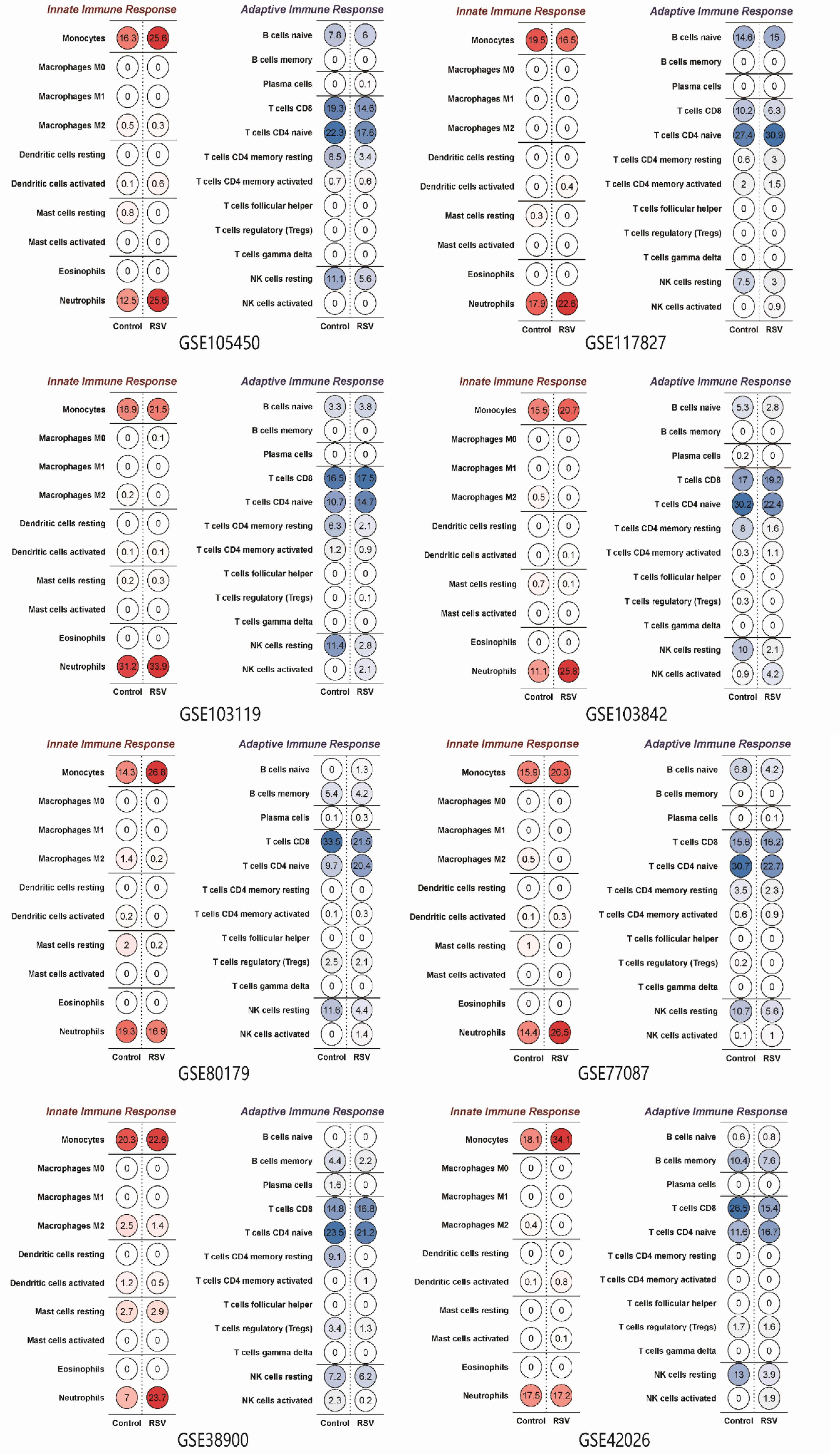
Figure 5 The proportion of immune cell types in different datasets. Note that the proportion of neutrophils in most datasets was higher in the RSV-infected groups than in the healthy groups.
RSV is the most important viral pathogen causing acute lower respiratory tract infection in children under 5 years old worldwide, and the primary viral factor leading to hospitalization for acute respiratory infection in infants under 1 year old seriously endangers children’s health (Shi et al., 2017). In this study, we conducted a meta-analysis of peripheral blood transcriptional microarray expression data from 436 children with RSV infection and 241 healthy children. A total of 319 DEGs were identified, of which the expressions of 46 genes were upregulated and the expressions of 273 genes were downregulated. Biological information databases, such as GO and KEGG, were applied to determine important signaling pathways related to immunity, such as the “chemokine signaling pathway”, “natural killer cell-mediated cytotoxicity” and “cytokine–cytokine receptor interaction”. In addition, CIBERSORT was used to determine the proportion of different immune cells in acute RSV infection and found that the proportion of neutrophils in the peripheral blood of most children with RSV infection was higher than that of healthy subjects.
In this study, SIGLEC1 was the most upregulated gene and encodes a protein that is a lectin-like adhesion molecule that binds glycoconjugate ligands on the cell surface in a sialic acid-dependent manner (Crocker et al., 2007). Jans et al. identified that SIGLEC1 was significantly upregulated in infants infected with RSV, and SIGLEC1 was identified as the only gene that directly interacts with T cells (Jans et al., 2018). This study suggested that neonatal and adult primary T cells have low expression of the SIGLEC1 receptor CD43, thus producing IFN-γ during RSV infection (Jans et al., 2018). However, the high expression of CD43 in adult memory T cells leads to the inhibition of IFN-γ release by SIGLEC1. In addition, monocytes have an inhibitory effect on T cells through SIGLEC1 (Jans et al., 2018). This effect may also apply to other antigen-presenting cells, such as dendritic cells (DCs) and macrophages, which provides a basis for a better understanding of the antiviral immune response against RSV directly after birth.
EMR4P is associated with leukocyte adhesion (Kwakkenbos et al., 2004). In a study characterizing the expression of a novel whole-blood gene for asthma, dermatitis, and rhinitis in children and adolescents, EMR4P was consistently overexpressed in allergic diseases such as asthma, dermatitis, and rhinitis (Lemonnier et al., 2020). In addition, EMR4P has been identified as being involved in airway type 2 eosinophil inflammation (Ditz et al., 2021). In the results of this meta-analysis, EMR4P expression was lower in the RSV-infected group than in healthy children.
TPT1P4 is a tumour protein. The TPT1/TCTP gene is a tumour protein that controls translation and encodes highly structured mRNA shielded by ribosomal protein, which is very similar to virus particles, activating protein kinase R (Amson et al., 2013). The results of our study showed that TPT1P4 expression in the RSV-infected groups was lower than that in the healthy groups. The other two genes with the most significant expression changes were RPS10P9 and RPS7P11, which were both related to ribosomes (Chen et al., 2007; Beabout et al., 2015) and had low expression in children with RSV infection.
According to our meta-analysis results, we identified four DEGs with fold changes and high significance, including NOV, ALOX15, CLC and KLRB1. All of these genes play functions related to viral infection. Among them, NOV is reported to be a biomarker of acute lung injury and is associated with inflammation and apoptosis of human alveolar epithelial cells (Zhu et al., 2020). CLC and KLRB1 were identified to have downregulated expression in the whole blood of infants hospitalized with RSV (Fjaerli et al., 2006). Leonard’s study validated CLC and ALOX15 as clinical biomarkers or risk factors for RSV infection in infants (Krilov et al., 2010).
The immunization of the pulmonary tissue with RSV and other respiratory viruses begins with the recruitment of immune cells from the blood into the lungs (Nuriev and Johansson, 2019). This inflammatory process is mainly controlled by chemokines. Many chemokines are produced during the infection process, and specific cell types are recruited through several unique chemokine receptors, such as CXCL1, CXCL2, and CXCL8, which can recruit neutrophils (Culley et al., 2006; Goritzka et al., 2014; Goritzka et al., 2015; Tang et al., 2016). CX3CL1 can recruit monocytes, NK cells and T cells (Johnson et al., 2012), and CXCL10 can recruit dendritic cells and T cells (Goritzka et al., 2014). During RSV infection, almost all chemokines are positively correlated with disease severity (Russell et al., 2017). An in-depth understanding of which cell types are the main sources of chemokines and how chemokine production is regulated will contribute to understanding the occurrence and maintenance of lung inflammation.
Cytokines are small secretory molecules that play a critical role in regulating the immune response and T-cell differentiation (Shachar and Karin, 2013). Several cell types can produce and secrete cytokines, including immune cells, epithelial cells, and endothelial cells (Kasahara, 2021). Cytokines are also considered to be key factors in establishing and regulating immune and inflammatory responses. In the natural immune stage of anti-RSV infection, RSV binds to Toll-like receptors on the surface of epithelial cells, which can promote the expression of proinflammatory cytokines such as IL-6 and TNF-α, leading to an inflammatory response (Kurt-Jones et al., 2000). RSV can promote Th2 cells to produce IL-4, IL-5, IL-9, IL-10, and IL-13, resulting in a strong Th2 cytokine response, inhibiting Th1-cell and Th1/Th2 immune imbalance (Openshaw and Chiu, 2013). Determining the specific cytokine expression profile after RSV infection will therefore provide effective immune strategies for anti-RSV treatment.
NK cells kill cells infected with pathogens through a range of mechanisms, mainly through the exocrine secretion of cytosolic particles and the involvement of extracellular death receptors (Smyth et al., 2005). NK-cell lytic particles containing perforin and granules enter infected cells after the formation of immune synapses and induce apoptosis through the caspase-3-mediated signaling pathway (Moretta et al., 2006; Lanier, 2008). The death receptor Fas ligand (FASL) on the infected cells interacts with the tumour necrosis factor-related apoptosis-inducing ligand (TRAIL) on NK cells, ultimately leading to target cell apoptosis (Lieberman, 2003). NK cells promote early innate immune responses by providing an early source of IFN-γ, activating T cells, and directly killing infected cells (Culley, 2009). However, there is evidence that after RSV infection, NK cells secrete a large number of cytokines, resulting in cytotoxicity and lung immune damage (Malloy et al., 2013; van Erp et al., 2019). In addition, the activating receptor Natural Killer Group 2, Member D (NKG2D)-mediated RSV infection increases the ability of NK cells to produce excessive IFN-γ, while IFN-γ can induce NK cells to activate T cells, which may contribute to lung injury during RSV infection (Liu et al., 2018).
Neutrophils are an essential part of the innate immune system, which plays a crucial role in identifying pathogens, killing invasive pathogens, presenting antigens to T cells, recruiting other inflammatory cells and producing cytokines (Mantovani et al., 2011; Metzler et al., 2011). RSV infection causes a strongly systemic and respiratory neutrophil response. Neutrophils are the major cell type in the bronchial lavage fluid of children with severe RSV bronchiolitis and mild infection (Everard et al., 1994). Compared with the uninfected control group, neutrophil elastase (NE) levels in nasal aspirate and serum of paediatric patients with acute RSV infection increased (Emboriadou et al., 2007). RSV virus particles stimulated human neutrophils in vitro and induced ROS-dependent extracellular DNA traps named NETs through protein-arginine deiminase type 4 (PAD4) citrullination and downstream activation of the PI3K/Akt, ERK and p38 MAPK pathways (Muraro et al., 2018). The deposition of the NET product in the culture medium can capture the RSV virus in NE- and MPO-coated DNA lattices, indicating that NETs have a beneficial antiviral effect (Muraro et al., 2018). However, in calves severely infected with bovine RSV, extensive airway obstruction is caused by luminal obstruction consisting of mucin, cell debris, and NETs (Cortjens et al., 2016). In addition, the results from other experimental systems, such as allergic asthma, suggest that NETs may aggravate immunopathology. Virus-associated exacerbations of asthma were attenuated when NETs were suppressed by blocking NE or degrading NETs with DNase (Toussaint et al., 2017). These results suggest that neutrophils play a complex role in RSV infection.
This study combined the results of eight whole-blood microarray transcriptome studies associated with childhood RSV acute respiratory infection and identified 319 DEGs. Among the most significant genes, SIGLEC1 interacts with T cells, and EMR4P is related to an allergic reaction. In addition, some immune-related pathways were identified, such as the “chemokine signaling pathway”, “natural killer cell-mediated cytotoxicity” and “cytokine–cytokine receptor interaction”. The ratio of neutrophils in most RSV infections was found to be higher than that in healthy children by immunocyte infiltration analysis. The identified immune characteristics are expected to deepen the understanding of the immune response to acute RSV infection in children.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
QF performed the experiments and drafted the manuscript. SL, HL, and BY performed the data analysis. LX, XH, LH, and ZX participated in the study design and coordinated the drafting of the manuscript. All the authors read and approved the final manuscript.
This work was funded by the National Natural Science Foundation of China (82172275) and the CAMS Innovation Fund for Medical Sciences (CIFMS, 2019-I2M-5-026).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer XL declared a shared affiliation with the authors LX, QF, ZX to the handling editor at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.878430/full#supplementary-material
Adams, H. A., Southey, B. R., Robinson, G. E., Rodriguez-Zas, S. L. (2008). Meta-Analysis of Genome-Wide Expression Patterns Associated With Behavioral Maturation in Honey Bees. BMC Genomics 9, 503. doi: 10.1186/1471-2164-9-503
Amarasinghe, G. K., Ayllón, M. A., Bào, Y., Basler, C. F., Bavari, S., Blasdell, K. R., et al. (2019). Taxonomy of the Order Mononegavirales: Update 2019. Arch. Virol. 164, 1967–1980. doi: 10.1007/s00705-019-04247-4
Amson, R., Pece, S., Marine, J.-C., Fiore, P. P. D., Telerman, A. (2013). TPT1/ TCTP-Regulated Pathways in Phenotypic Reprogramming. Trends Cell Biol. 23, 37–46. doi: 10.1016/j.tcb.2012.10.002
Beabout, K., Hammerstrom, T. G., Perez, A. M., Magalhães, B. F., Prater, A. G., Clements, T. P., et al. (2015). The Ribosomal S10 Protein is a General Target for Decreased Tigecycline Susceptibility. Antimicrob. Agents Chemother. 59, 5561–5566. doi: 10.1128/aac.00547-15
Behzadi, M. A., Leyva-Grado, V. H. (2019). Overview of Current Therapeutics and Novel Candidates Against Influenza, Respiratory Syncytial Virus, and Middle East Respiratory Syndrome Coronavirus Infections. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01327
Burel, J. G., Peters, B. (2018). Discovering Transcriptional Signatures of Disease for Diagnosis Versus Mechanism. Nat. Rev. Immunol. 18, 289–290. doi: 10.1038/nri.2018.26
Cahan, P., Rovegno, F., Mooney, D., Newman, J. C., Laurent, G. S., McCaffrey, T. A. (2007). Meta-Analysis of Microarray Results: Challenges, Opportunities, and Recommendations for Standardization. Gene 401, 12–18. doi: 10.1016/j.gene.2007.06.016
Chen, D., Zhang, Z., Li, M., Wang, W., Li, Y., Rayburn, E. R., et al. (2007). Ribosomal Protein S7 as a Novel Modulator of P53-MDM2 Interaction: Binding to MDM2, Stabilization of P53 Protein, and Activation of P53 Function. Oncogene 26, 5029–5037. doi: 10.1038/sj.onc.1210327
Cortjens, B., de Boer, O. J., de Jong, R., Antonis, A. F., Piñeros, Y. S. S., Lutter, R., et al. (2016). Neutrophil Extracellular Traps Cause Airway Obstruction During Respiratory Syncytial Virus Disease. J. Pathol. 238, 401–411. doi: 10.1002/path.4660
Crocker, P. R., Paulson, J. C., Varki, A. (2007). Siglecs and Their Roles in the Immune System. Nat. Rev. Immunol. 7, 255–266. doi: 10.1038/nri2056
Culley, F. J. (2009). Natural Killer Cells in Infection and Inflammation of the Lung. Immunology 128, 151–163. doi: 10.1111/j.1365-2567.2009.03167.x
Culley, F. J., Pennycook, A. M. J., Tregoning, J. S., Hussell, T., Openshaw, P. J. M. (2006). Differential Chemokine Expression Following Respiratory Virus Infection Reflects Th1- or Th2-Biased Immunopathology. J. Virol. 80, 4521–4527. doi: 10.1128/jvi.80.9.4521-4527.2006
Ditz, B., Sarma, A., Kerstjens, H. A. M., Liesker, J. J. W., Bathoorn, E., Vonk, J. M., et al. (2021). The Sputum Transcriptome Better Predicts COPD Exacerbations After the Withdrawal of Inhaled Corticosteroids Than Sputum Eosinophils. ERJ Open Res. 7. 00097–2021. doi: 10.1183/23120541.00097-2021
Emboriadou, M., Hatzistilianou, M., Magnisali, C., Sakelaropoulou, A., Exintari, M., Conti, P., et al. (2007). Human Neutrophil Elastase in RSV Bronchiolitis. Ann. Clin. Lab. Sci. 37, 79–84.
Everard, M. L., Swarbrick, A., Wrightham, M., McIntyre, J., Dunkley, C., James, P. D., et al. (1994). Analysis of Cells Obtained by Bronchial Lavage of Infants With Respiratory Syncytial Virus Infection. Arch. Dis. Childhood 71, 428–432. doi: 10.1136/adc.71.5.428
Fjaerli, H.-O., Bukholm, G., Krog, A., Skjaeret, C., Holden, M., Nakstad, B. (2006). Whole Blood Gene Expression in Infants With Respiratory Syncytial Virus Bronchiolitis. BMC Infect. Dis. 6, 1–7. doi: 10.1186/1471-2334-6-175
Ge, X., Guo, Y., Chen, J., Hu, R., Feng, X. (2018). Epidemiology and Seasonality of Respiratory Viruses Detected From Children With Respiratory Tract Infections in Wuxi, East China. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 24, 1856–1862. doi: 10.12659/msm.908483
Glezen, W. P., Taber, L. H., Frank, A. L., Kasel, J. A. (1986). Risk of Primary Infection and Reinfection With Respiratory Syncytial Virus. Am. J. Dis. Children 140, 543–546. doi: 10.1001/archpedi.1986.02140200053026
Goritzka, M., Durant, L. R., Pereira, C., Salek-Ardakani, S., Openshaw, P. J. M., Johansson, C. (2014). Alpha/beta Interferon Receptor Signaling Amplifies Early Proinflammatory Cytokine Production in the Lung During Respiratory Syncytial Virus Infection. J. Virol. 88, 6128–6136. doi: 10.1128/jvi.00333-14
Goritzka, M., Makris, S., Kausar, F., Durant, L. R., Pereira, C., Kumagai, Y., et al. (2015). Alveolar Macrophage-Derived Type I Interferons Orchestrate Innate Immunity to RSV Through Recruitment of Antiviral Monocytes. J. Exp. Med. 212, 699–714. doi: 10.1084/jem.20140825
Jans, J., Unger, W. W. J., Vissers, M., Ahout, I. M. L., Schreurs, I., Wickenhagen, A., et al. (2018). Siglec-1 Inhibits RSV-Induced Interferon Gamma Production by Adult T Cells in Contrast to Newborn T Cells. Eur. J. Immunol. 48, 621–631. doi: 10.1002/eji.201747161
Johnson, C. H., Miao, C., Blanchard, E. G., Caidi, H., Radu, G. U., Harcourt, J. L., et al. (2012). Effect of Chemokine Receptor CX3CR1 Deficiency in a Murine Model of Respiratory Syncytial Virus Infection. Comp. Med. 62, 14–20.
Kasahara, M. (2021). Role of Immunoproteasomes and Thymoproteasomes in Health and Disease. Pathol. Int. 71, 371–382. doi: 10.1111/pin.13088
Krilov, L. R., Palazzi, D. L., Fernandes, A. W., Klein, R. W., Mahadevia, P. J. (2010). Prevalence of Respiratory Syncytial Virus (RSV) Risk Factors and Cost Implications of Immunoprophylaxis to Infants 32 to 35 Weeks Gestation for Health Plans in the United States. Value Health 13, 77–86. doi: 10.1111/j.1524-4733.2009.00586.x
Kurt-Jones, E. A., Popova, L., Kwinn, L., Haynes, L. M., Jones, L. P., Tripp, R. A., et al. (2000). Pattern Recognition Receptors TLR4 and CD14 Mediate Response to Respiratory Syncytial Virus. Nat. Immunol. 1, 398–401. doi: 10.1038/80833
Kwakkenbos, M. J., Kop, E. N., Stacey, M., Matmati, M., Gordon, S., Lin, H.-H., et al. (2004). The EGF-TM7 Family: A Postgenomic View. Immunogenetics 55, 655–666. doi: 10.1007/s00251-003-0625-2
Lanier, L. L. (2008). Up on the Tightrope: Natural Killer Cell Activation and Inhibition. Nat. Immunol. 9, 495–502. doi: 10.1038/ni1581
Lemonnier, N., Melén, E., Jiang, Y., Joly, S., Ménard, C., Aguilar, D., et al. (2020). A Novel Whole Blood Gene Expression Signature for Asthma, Dermatitis, and Rhinitis Multimorbidity in Children and Adolescents. Allergy 75, 3248–3260. doi: 10.1111/all.14314
Lieberman, J. (2003). The ABCs of Granule-Mediated Cytotoxicity: New Weapons in the Arsenal. Nature Reviews. Immunology 3, 361–370. doi: 10.1038/nri1083
Liu, H., Osterburg, A. R., Flury, J., Huang, S., McCormack, F. X., Cormier, S. A., et al. (2018). NKG2D Regulation of Lung Pathology and Dendritic Cell Function Following Respiratory Syncytial Virus Infection. J. Infect. Dis. 218, 1822–1832. doi: 10.1093/infdis/jiy151
Malloy, A. M. W., Falsey, A. R., Ruckwardt, T. J. (2013). Consequences of Immature and Senescent Immune Responses for Infection With Respiratory Syncytial Virus. Curr. Topics Microbiol. Immunol. 372, 211–231. doi: 10.1007/978-3-642-38919-1_11
Mantovani, A., Cassatella, M. A., Costantini, C., Jaillon, S. (2011). Neutrophils in the Activation and Regulation of Innate and Adaptive Immunity. Nat. Rev. Immunol. 11, 519–531. doi: 10.1038/nri3024
Metzler, K. D., Fuchs, T. A., Nauseef, W. M., Reumaux, D., Roesler, J., Schulze, I., et al. (2011). Myeloperoxidase is Required for Neutrophil Extracellular Trap Formation: Implications for Innate Immunity. Blood 117, 953–959. doi: 10.1182/blood-2010-06-290171
Moretta, L., Bottino, C., Pende, D., Castriconi, R., Mingari, M. C., Moretta, A. (2006). Surface NK Receptors and Their Ligands on Tumor Cells. Semin. Immunol. 18, 151–158. doi: 10.1016/j.smim.2006.03.002
Muraro, S. P., Souza, G. F. D., Gallo, S. W., Silva, B. K. D., Oliveira, S. D. D., Vinolo, M. A. R., et al. (2018). Respiratory Syncytial Virus Induces the Classical ROS-Dependent NETosis Through PAD-4 and Necroptosis Pathways Activation. Sci. Rep. 8, 14166. doi: 10.1038/s41598-018-32576-y
Newman, A. M., Liu, C. L., Green, M. R., Gentles, A. J., Feng, W., Xu, Y., et al. (2015). Robust Enumeration of Cell Subsets From Tissue Expression Profiles. Nat. Methods 12, 453–457. doi: 10.1038/nmeth.3337
Nuriev, R., Johansson, C. (2019). Chemokine Regulation of Inflammation During Respiratory Syncytial Virus Infection. F1000Res. 8 (F1000 Faculty Rev), 1837 8. doi: 10.12688/f1000research.20061.1
Openshaw, P. J., Chiu, C. (2013). Protective and Dysregulated T Cell Immunity in RSV Infection. Curr. Opin. Virol. 3, 468–474. doi: 10.1016/j.coviro.2013.05.005
Russell, C. D., Unger, S. A., Walton, M., Schwarze, J. (2017). The Human Immune Response to Respiratory Syncytial Virus Infection. Clin. Microbiol. Rev. 30, 481–502. doi: 10.1128/cmr.00090-16
Shachar, I., Karin, N. (2013). The Dual Roles of Inflammatory Cytokines and Chemokines in the Regulation of Autoimmune Diseases and Their Clinical Implications. J. Leukocyte Biol. 93, 51–61. doi: 10.1189/jlb.0612293
Shi, T., McAllister, D. A., O'Brien, K. L., Simoes, E. A. F., Madhi, S. A., Gessner, B. D., et al. (2017). Global, Regional, and National Disease Burden Estimates of Acute Lower Respiratory Infections Due to Respiratory Syncytial Virus in Young Children in 2015: A Systematic Review and Modelling Study. Lancet (Lond. Engl.) 390, 946–958. doi: 10.1016/s0140-6736(17)30938-8
Smyth, M. J., Cretney, E., Kelly, J. M., Westwood, J. A., Street, S. E. A., Yagita, H., et al. (2005). Activation of NK Cell Cytotoxicity. Mol. Immunol. 42, 501–510. doi: 10.1016/j.molimm.2004.07.034
Tang, F. S. M., Ly, D. V., Spann, K., Reading, P. C., Burgess, J. K., Hartl, D., et al. (2016). Differential Neutrophil Activation in Viral Infections: Enhanced TLR-7/8-Mediated CXCL8 Release in Asthma. Respirol. (Carlton Vic.) 21, 172–179. doi: 10.1111/resp.12657
Toussaint, M., Jackson, D. J., Swieboda, D., Guedán, A., Tsourouktsoglou, T.-D., Ching, Y. M., et al. (2017). Host DNA Released by NETosis Promotes Rhinovirus-Induced Type-2 Allergic Asthma Exacerbation. Nat. Med. 23, 681–691. doi: 10.1038/nm.4332
van Erp, E. A., van Kampen, M. R., van Kasteren, P. B., de Wit, J. (2019). Viral Infection of Human Natural Killer Cells. Viruses 11, 243 doi: 10.3390/v11030243
Keywords: respiratory syncytial virus, transcriptome, microarray, meta-analysis, immune response
Citation: Feng Q, Lin S, Liu H, Yang B, Han L, Han X, Xu L and Xie Z (2022) Meta-Analysis of Whole Blood Transcriptome Datasets Characterizes the Immune Response of Respiratory Syncytial Virus Infection in Children. Front. Cell. Infect. Microbiol. 12:878430. doi: 10.3389/fcimb.2022.878430
Received: 18 February 2022; Accepted: 11 March 2022;
Published: 13 April 2022.
Edited by:
Min Zheng, Shenzhen Bay Laboratory, ChinaReviewed by:
Xiaoyan Li, Capital Medical University, ChinaCopyright © 2022 Feng, Lin, Liu, Yang, Han, Han, Xu and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lili Xu, anVzdGluZXh1bGwyNkAxNjMuY29t; Xiao Han, aGFueGlhb0BmenUuZWR1LmNu; Lifen Han, MTM2NTUwODM2MzlAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.