- 1Department of Laboratory Medicine, The Second People’s Hospital of Lianyungang City, Affiliated to Bengbu Medical College, Lianyungang, China
- 2Department of Medical Laboratory, Bengbu Medical College, Bengbu, China
- 3School of Biotechnology, Jiangsu University of Science and Technology, Zhenjiang, China
- 4Department of Central Laboratory, Lianyungang Second People’s Hospital affiliated to Jiangsu University, Lianyungang, China
- 5Lianyungang Second People’s Hospital affiliated to Xuzhou Medical University, Lianyungang, China
Highly virulent Klebsiella pneumoniae often causes invasive infections with high morbidity and mortality rates, posing an immense clinical challenge. Rapid and accurate detection of pathogenic bacteria is of great significance for treatment and preventive control. Conventional detection by polymerase chain reaction (PCR) is limited by a dependence on laboratory equipment and professional staff. Recombinase polymerase amplification (RPA) combined with a lateral flow strip (LFS) can rapidly amplify and visualize target genes in a short period of time. The aim of this study was to develop an RPA-LFS technique for detection of the K. pneumoniae virulence gene rmpA2. Primers were designed against conserved sequences specific to the virulence gene, and primer probe design was optimized by introducing base substitution to obtain a specific and sensitive primer-probe combination for clinical detection. We tested 65 actual samples collected from clinics to evaluate the performance of the newly established RPA-LFS system in comparison with conventional PCR methods and qPCR methods. The RPA-LFS assay was performed at for 25 min a constant temperature of 37°C, and results could be observed without instrumentation. The system could specifically identify highly virulent K. pneumoniae carrying the virulence gene rmpA2 with a minimum detection limit of 10−1 ng/μL and 10 copies/μL. For the 65 clinical samples tested, The RPA-LFS assay results were in complete agreement with the qPCR results and PCR results. The RPA-LFS assay provides a rapid, accurate, and simple method for identification of highly virulent K. pneumoniae carrying rmpA2.
Introduction
Klebsiella pneumoniae is a common causative agent of hospital-acquired infections occurring in patients with multiple comorbidities in hospitals or long-term care facilities (Shon and Russo, 2012). K. pneumoniae strains can be divided into two categories based on their mucoid nature: classical K. pneumoniae (cKP)and hypervirulent K. pneumoniae (hvKP). The first case of liver abscess combined with infectious endophthalmitis due to highly virulent K. pneumoniae infection was reported in Taiwan, China, in 1986, and eventually led to blindness in most patients despite aggressive treatment with numerous antibiotics (Liu et al., 1986). hvKP mainly causes invasive infections, such as bloodstream infections and purulent liver abscesses, which mainly infect young, immunocompetent individuals. hvKP virulence factors include podoconjugate, Fimbriae, lipopolysaccharide, and iron uptake (Siu et al., 2012). These features contribute to its unique pathogenic capacity.
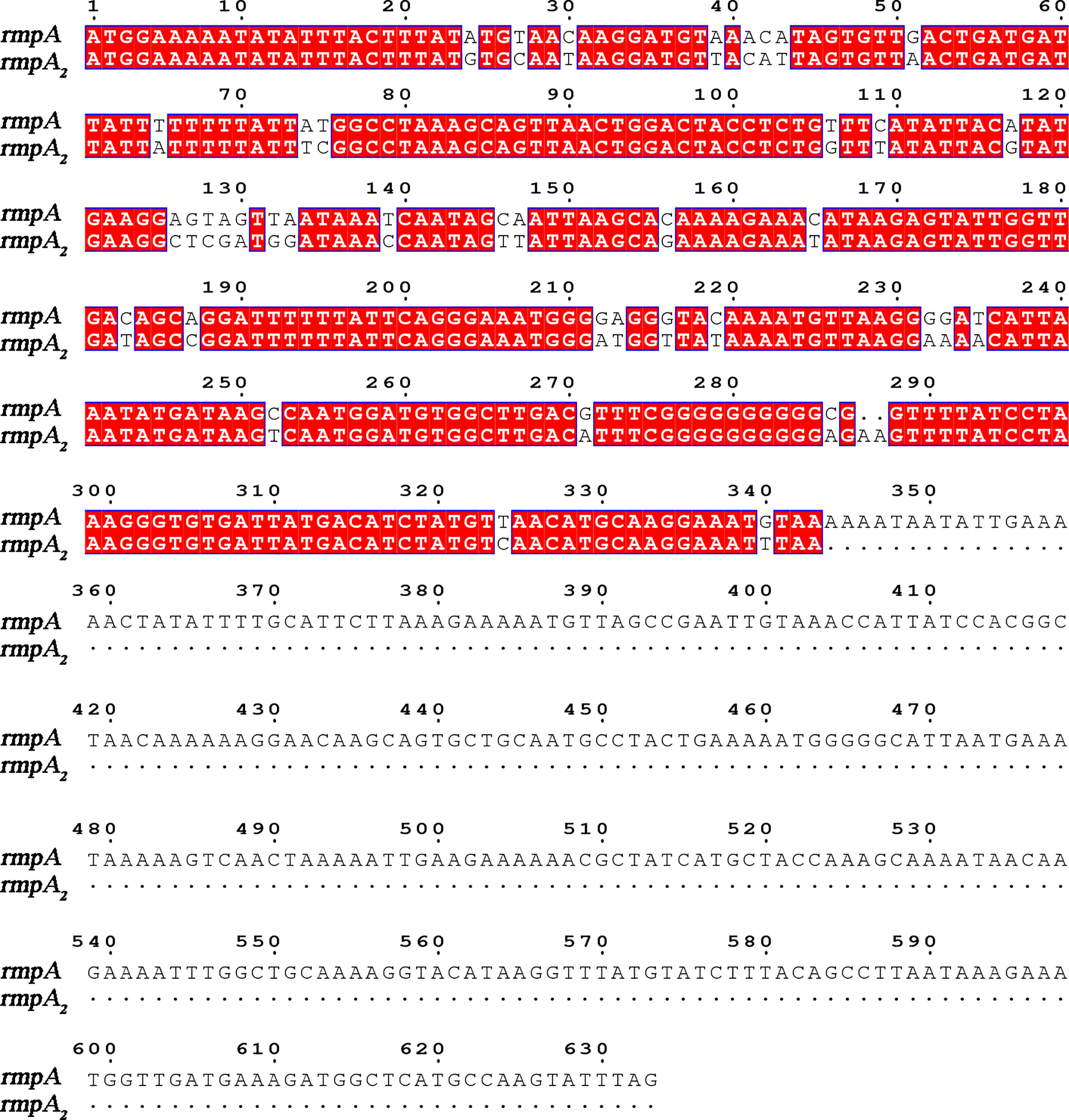
There is no accepted definition of hvKP to date. The mouse infection assay is currently the gold standard for identifying the virulence of K. pneumoniae (Yu et al., 2007). With the development of genomics, virulence plasmids such as pK2044, pLVPK, and pVir-CR-hvK4 have been identified in hvKP strains (Chen et al., 2004; Wu et al., 2009; Gu et al., 2018). Toxigenic plasmids can be spontaneously transferred between strains, and classical K. pneumoniae can be characterized as highly virulent due to the acquisition of virulence plasmids (Yang et al., 2019). The virulence factors causing the hypervirulence phenotype are encoded by genes present on these plasmids, including iuc, peg-344, iro, rmpA, and rmpA2. rmpA/rmpA2 (mucus phenotype regulatory gene A) is important for mediating the expression of the hypervirulence phenotype, regulating the production of large amounts of podocytes (Figure 1) (Liu et al., 1986; Cheng et al., 1991; Fierer et al., 2011). Its expression increases podocyte production and enhances anti-macrophage phagocytosis, thereby enhancing the virulence of K. pneumoniae (Cheng et al., 2010). K. pneumoniae strains are divided into highly virulent and non-highly virulent groups based on the results of infection experiments in mice. The sensitivity of rmpA2 assay was 88.89%, the specificity was 93.1%, the positive predictive value was 90.91%, and the negative predictive value was 91.53% (Li et al., 2020); therefore, detection of the virulence gene rmpA2 can help to predict the virulence of K. pneumoniae.
Currently available assays for the virulence gene rmpA2 employ polymerase chain reaction (PCR) and MALDI-TOF MS techniques (Yu et al., 2015). These techniques are time-consuming and laborious with high instrument dependence. PCR-based methods can give results in a few hours and have become the main method for pathogen detection (Salazar et al., 2015). However, the need for complex thermal cyclers and trained professionals limits their use in remote areas. By contrast, isothermal recombinant polymerase amplification (RPA) does not have these cumbersome requirements and can therefore be more widely used for rapid field assays (Daher et al., 2016; Dai et al., 2019).
In recent years, RPA has been widely used for the detection of pathogenic bacteria due to its rapidity, simplicity, and convenience. The RPA reaction amplifies a DNA target by enzymatically opening both strands of double-stranded DNA followed by strand-substitution activity under isothermal conditions. The DNA target shows exponential amplification within 20 min over a temperature range of 37–42°C (Liu et al., 2018). There are two main detection methods for RPA products: real-time fluorescence (using a conventional fluorescent quantitative PCR instrument) and the lateral flow strip (LFS; e.g., the colloidal gold immunochromatographic strip) (Wang et al., 2021). Use of LFSs as endpoint visual reads for amplified DNA targets makes the method less equipment dependent (Cordray and Richards-Kortum, 2015; Yongkiettrakul et al., 2017). Upstream and downstream primers are required in the RPA-LFS method, and the reverse primer is labeled with biotin at the 5’ end. It is also necessary to design a probe downstream of the forward primer that is modified by fluorescein isothiocyanate(FITC) at the 5’ end and closed by a C3 spacer at the 3’ end, with a base in the middle of the probe replaced by a tetrahydrofuran (THF, purine-pyrimidine-free site) (Li et al., 2019). When the product in the reaction system accumulates to a sufficient amount, the probe binds to the product. The nucleic acid endonuclease (nfo) in the reaction system then recognizes the THF site and cleaves it, exposing the 3’-OH. Strand replacement activity of Bsu polymerase ensures that the DNA strand after the THF site is replaced, and thus amplification occurs. Since the 5’ end of the reverse primer is labeled with biotin, the amplification product obtained from the RPA reaction is labeled with FITC at one end and biotin at the other (Wang et al., 2018) (Figure 2). When the diluted product is added dropwise to the sample pad, the FITC-labeled amplification product binds to anti-FITC-labeled gold nanoparticles (AuNPs) and diffuses by chromatography to the streptavidin detection line, where the biotin-labeled amplification product binds to streptavidin at one end while maintaining a positive signal through the AuNPs at the other end. Anti-FITC-labeled AuNPs that are not bound to the amplification product continue to migrate to the control line showing a red color to ensure the detection capability of the test strip (Figure 3).
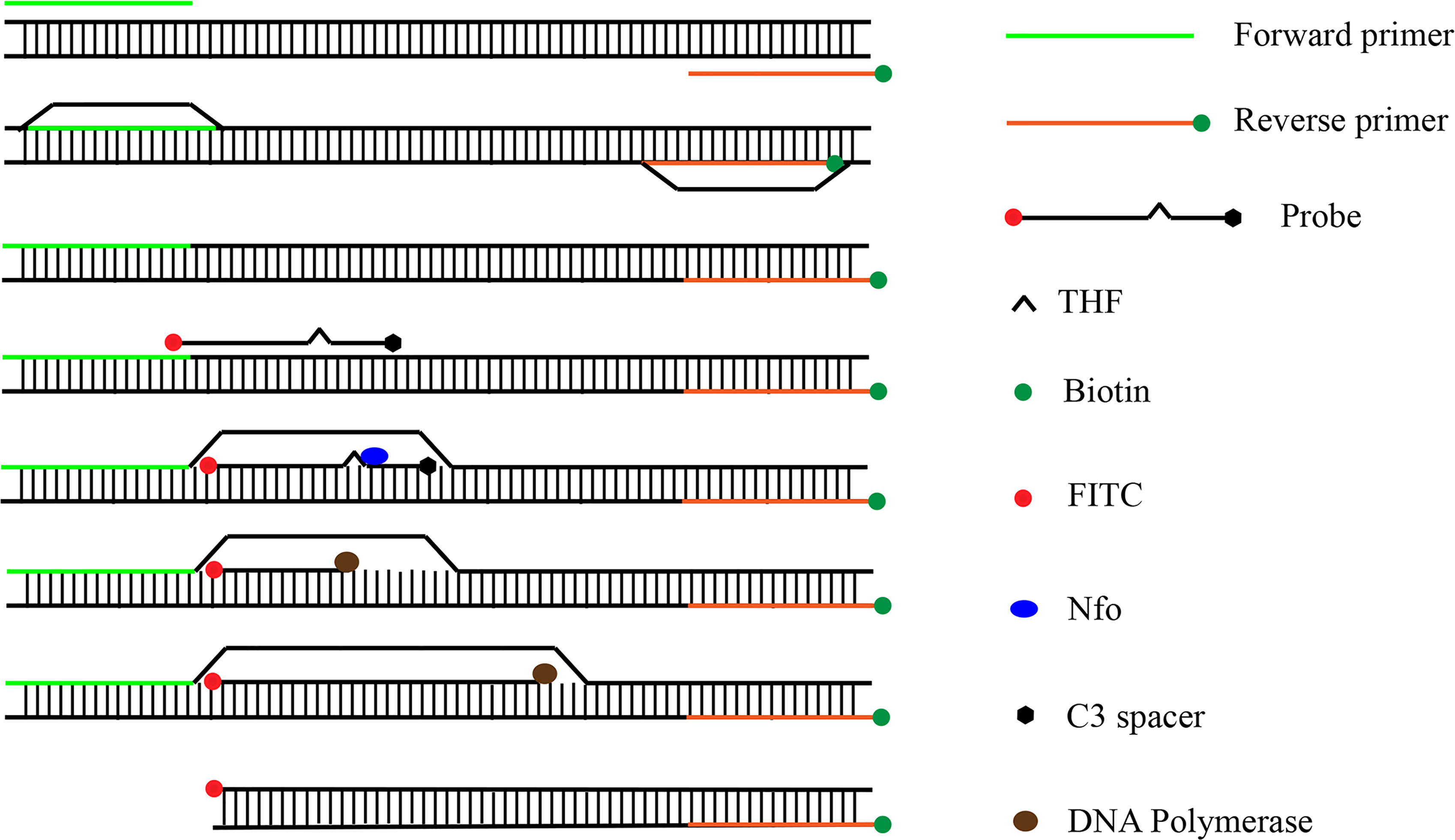
Figure 2 Amplification principle of RPA-LFS. The shapes and their representative molecules are listed on the right side of the drawing.
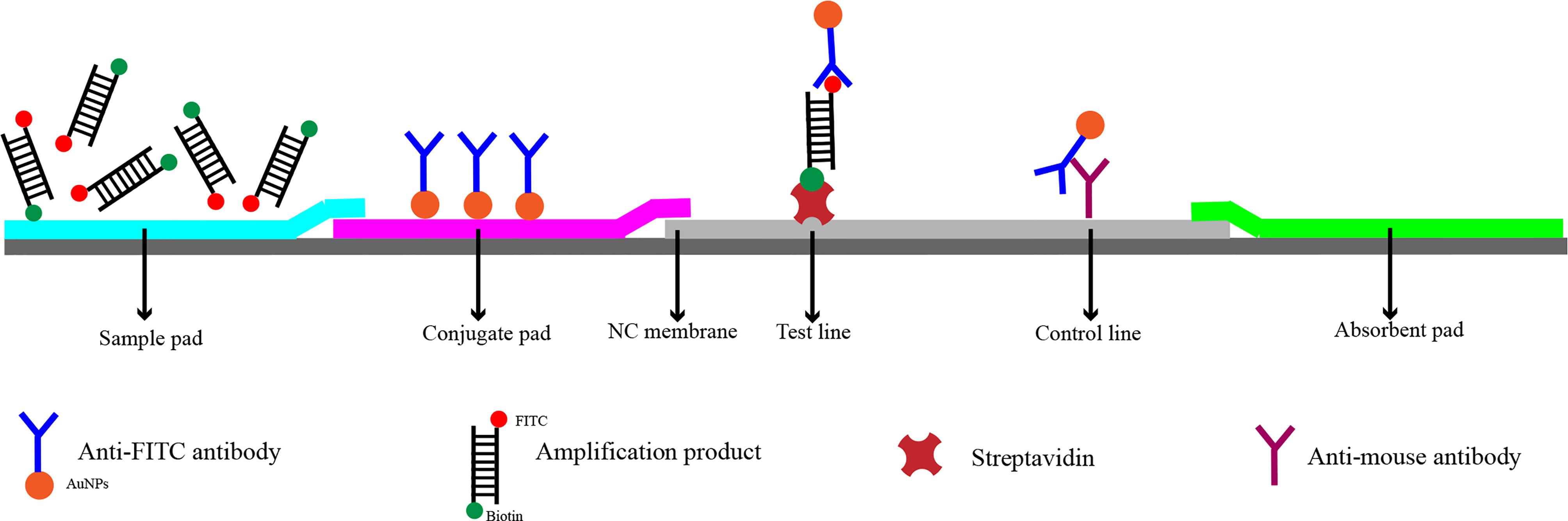
Figure 3 Detection principle of RPA-LFS. The shapes and their representing molecules are listed under the strip drawing.
In this study, we established a rapid and specific assay for the detection of highly virulent K. pneumoniae. This RPA-LFS method eliminated false-positive signals by introducing probes and base substitution. The method can be completed in 30 min at 37 °C, showing good interspecies specificity and sensitivity, and providing technical support for rapid and accurate clinical identification of highly virulent K. pneumoniae.
Materials and Methods
Ethics Statement
This study was approved by the Medical Ethics Committee of the Second People’s Hospital of Lianyungang (Permit number: 2020005). Clinical strains were collected in 2020-2021 and isolated from sputum, urine, pharyngeal swabs, drainage fluid, and other specimens. Written consent was obtained from each patient for all samples isolated.
Bacterial Strains
Six common pathogenic bacteria, Staphylococcus aureus, Streptococcus pneumoniae, Staphylococcus epidermidis, Escherichia coli, Staphylococcus cohnii, and Pseudomonas aeruginosa, were provided by the Microbiology Department of the Second People’s Hospital of Lianyungang. 65 clinical isolates of K. pneumoniae were provided by the Second People’s Hospital of Lianyungang.
DNA Extraction
Bacterial DNA isolation from clinical K. pneumoniae strains and six common pathogenic bacteria were performed using the heating and boiling method. Individual colonies were suspended in 50 μL double-distilled water (ddH2O), boiled for 10 min, and centrifuged at 12000 × g for 10 min; the supernatant was used as DNA template and stored at −20°C.
Design of RPA Primers
The rmpA2 virulence gene sequence (sequence number:MT496757) in the genome of K. pneumoniae was downloaded from National Center for Biotechnology Information(NCBI). The primers were designed using primer premier 5.0 software, and after entering the sequence for a specific target region, the parameters were set as follows: the product size was set to a minimum of 100 and a maximum of 500. The primer size was set to a minimum of 30 and a maximum of 35. The GC content was set to a minimum of 30% and a maximum of 70%, and all other parameters were set by default. Five pairs of primers were selected for testing.
RPA Procedure
Recombinant enzyme polymerase amplification reactions were performed using a DNA amplification kit according to the manufacturer’s instructions (TwistDx, Maidenhead, UK). The reaction system was added in the following order: 25 µL 2× reaction buffer, 5 µL 10× Basic e-mix, 2.5 µL 20× core mix, 2.1 µL each primer (10 µM), 9.8 µL distilled water, and 1 µL template solution; 2.5 µL magnesium acetate (280 mM) was added to initiate the reaction. After brief centrifugation, the reaction mixture was immediately incubated for 30 min at 37°C. RPA amplification products were purified using a PCR wash kit (Monad Biotech Co., Ltd, Wuhan, China) and electrophoresed on a 1.5% (w/v) agarose gel.
Design of Probes
It is known that cross-dimers formed by the probe and the reverse primer can give false positive results in RPA-LFS. Therefore, the following principles were followed when designing probes using Primer Premier 5.0 software. (1) Probe size 46–51 bp, GC content 30%–80%, and Tm 60–80°C. (2) Probe 5’ end labeled with FITC, 3’ end blocked with SpC3, and bases in the middle of the probe replaced with THF with at least 30 bp before the THF site and at least 15 bp after the THF site. (3) Use of base substitution to try to eliminate cross-dimers formed by the probe and reverse primer. (4) No modifications among the first three bases of the THF site; limited modifications to the first six bases if necessary. (5) Number of modified bases should not exceed nine bases. Primer-probe combinations were verified after base substitution for RPA false positives.
RPA-LFS Procedure
To screen probe and primer combinations, RPA-LFS experiments were performed using the TwistAmp® DNA Amplification nfo Kit (TwistDx). A total of 50 μL reaction system was prepared in the following order: 29.5 μL rehydration buffer, 2.1 μL forward primer, 2.1 μL reverse primer, 0.6 μL (10 μM) probe, 1 μL template solution, and 12.2 μL ddH2O in lyophilized powder tubes; 2.5 μL of 280 mM magnesium acetate was added to the tube cap in order to ensure the simultaneous start of all reaction systems. Immediately after transient separation, the reaction was carried out for 30 min at 37°C. RPA product (10 μL) was added to a 1.5 mL Eppendorf tube containing 190 μL ddH2O, and a LFS (Ubiquitous Biotechnology Co., Ltd., Hangzhou, China) was placed in the tube; results could be read after 3 min.
Quantitative PCR
A pair of specific primers (forward: 5’-TGATTATGACATCTAAGTCTACATGCAAGG-3’; reverse: 5’-TTTACATCTGTGACACGATAGTGTTTTCTC-3’) targeting the virulence gene rmpA2 of Klebsiella pneumoniae was used for qPCR. The qPCR reaction mixture contained 10 µl of 2 × SYBR Green qPCR Mix (Tiangen Biotech Co. Ltd), 0.4 µM of each primer, 1 µl of the template, and 8.2 µl of distilled water. The cycling program was 95°C for 30 s, followed by 40 cycles of 95°C for 10 s, then 60°C for 30 s. The melting curve analysis was set as default. Cycle threshold (Ct) values less than 32 were considered as positive.
Examination of Clinical Specimens
To evaluate the detection rate of RPA-LFS, the results were compared with those obtained by conventional PCR and qPCR, and the compliance rates of the three different assays were calculated. The PCR detection of rmpA2 followed an established method reported previously (Table 1) (Li et al., 2020). 65 clinical strains to be tested were collected and processed using the heating and boiling method. One microliter of supernatant was used as template for RPA-LFS and PCR reactions, respectively.
Results
Screening of RPA Primers and Probes
The five upstream primers designed were subjected to RPA-LFS reactions with downstream primer and probe to screen for the best upstream and downstream primer and probe combinations (Table 1). In designing the probes, we used primer premier 5.0 software to perform theoretical elimination of the cross-dimer formed by the probe and downstream primers to avoid false-positive signals (Figures 4A, B). RPA reactions were performed using genomic DNA of Klebsiella pneumoniae carrying rmpA2 as a template to verify amplification of each of the five primer pairs. As shown in the figure, all designed primers amplified the target bands as expected. The better primer pairs had brighter target bands, no non-specific amplification and had less primer dimerization (Figure 5). Therefore primer pair 5 was selected for subsequent detection. Firstly, we screened the probe and selected primer number 5 to perform RPA-LFS reaction with base substitution before (P) and base substitution after (mP), respectively. The results are shown in the figure, the test line of 5F/R/P showed color, but the test line also appeared colored in the condition without template, showing a false positive signal (Figure 4C). The primer-probe combination of 5F/R/mP showed a correct positive signal (red bands were visible in both the test and control lines), with only one red control line in the no-template control and no band in the detection line (Figure 4D). This indicates that the probe after base substitution eliminates the false positives. Next, we screened five primer-probe pairs and all five primer-probe combinations showed correct positive signals in the RPA-LFS test, with only one red control line in the no-template control and no bands in the test line (Figure 6). This indicates that none of the five primer-probe pairs showed false positives, and the primer-probe combination of 4F/R/mP had the darkest test line and therefore was the best primer-probe combination for the following RPA-LFS reactions.
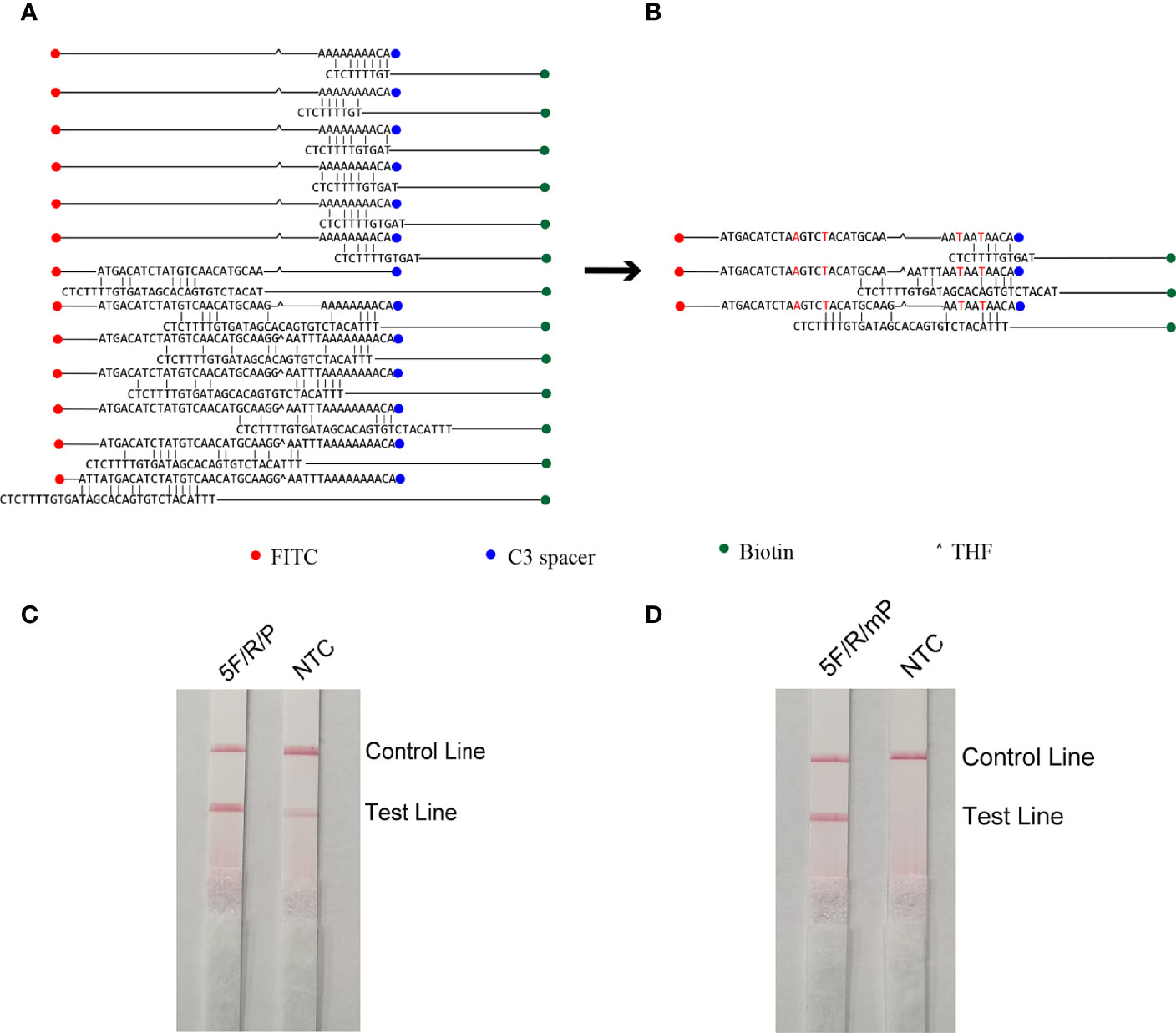
Figure 4 Probe design screening strategy. (A) Before base substitution, the probe and reverse primer form a cross-dimer. (B) After base substitution, the probe and reverse primer form a cross-dimer. (C, D) LFS results of RPA amplification products. The name of each primer-probe group is marked above the corresponding band, and the NTC lane is a template-free control for each primer-probe group.
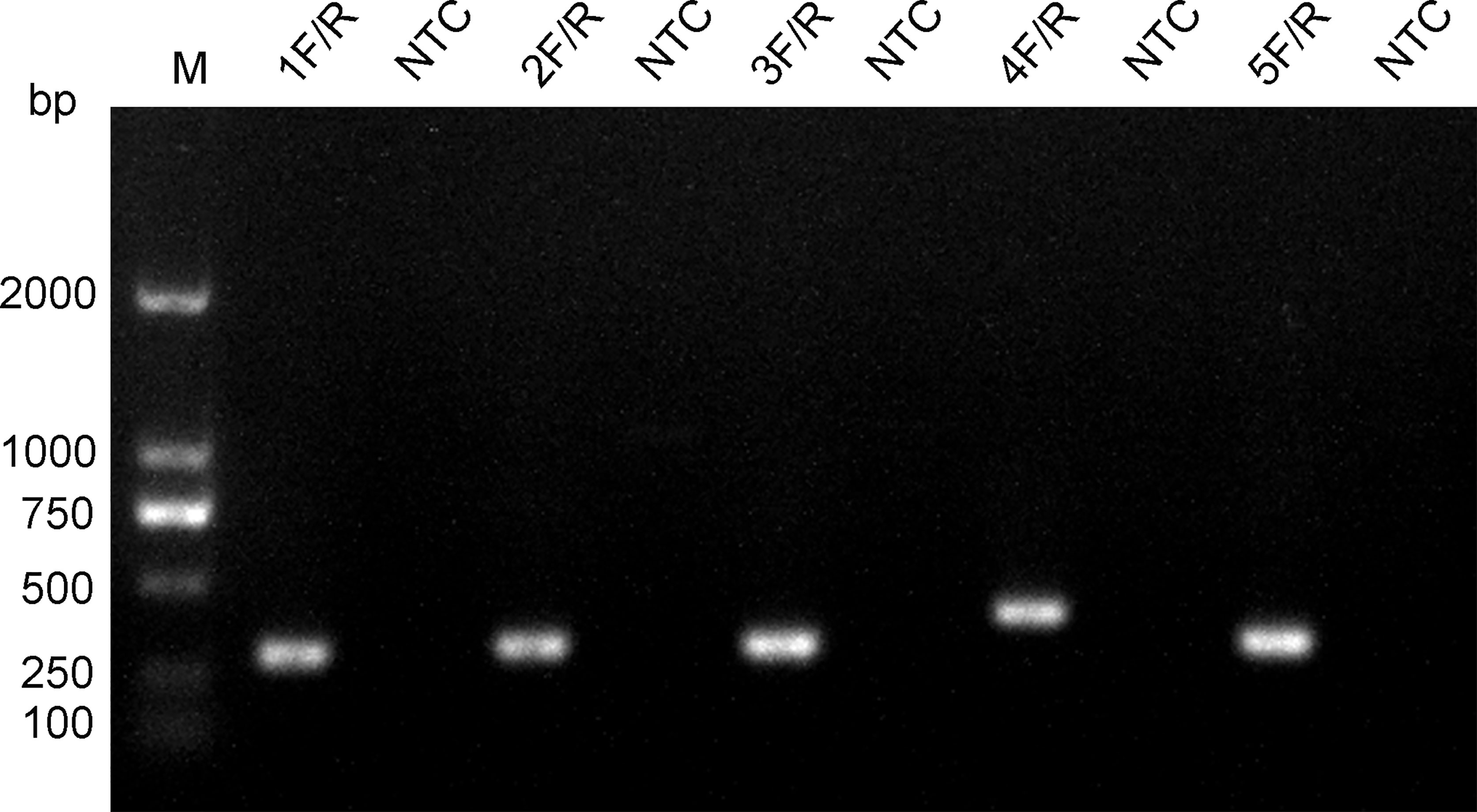
Figure 5 Screening of primers. The name of each primer pair is provided above each lane. The NTC lane immediately after is the no-template control of the respective primer pair. The band sizes of the DNA ladder are shown on the left.
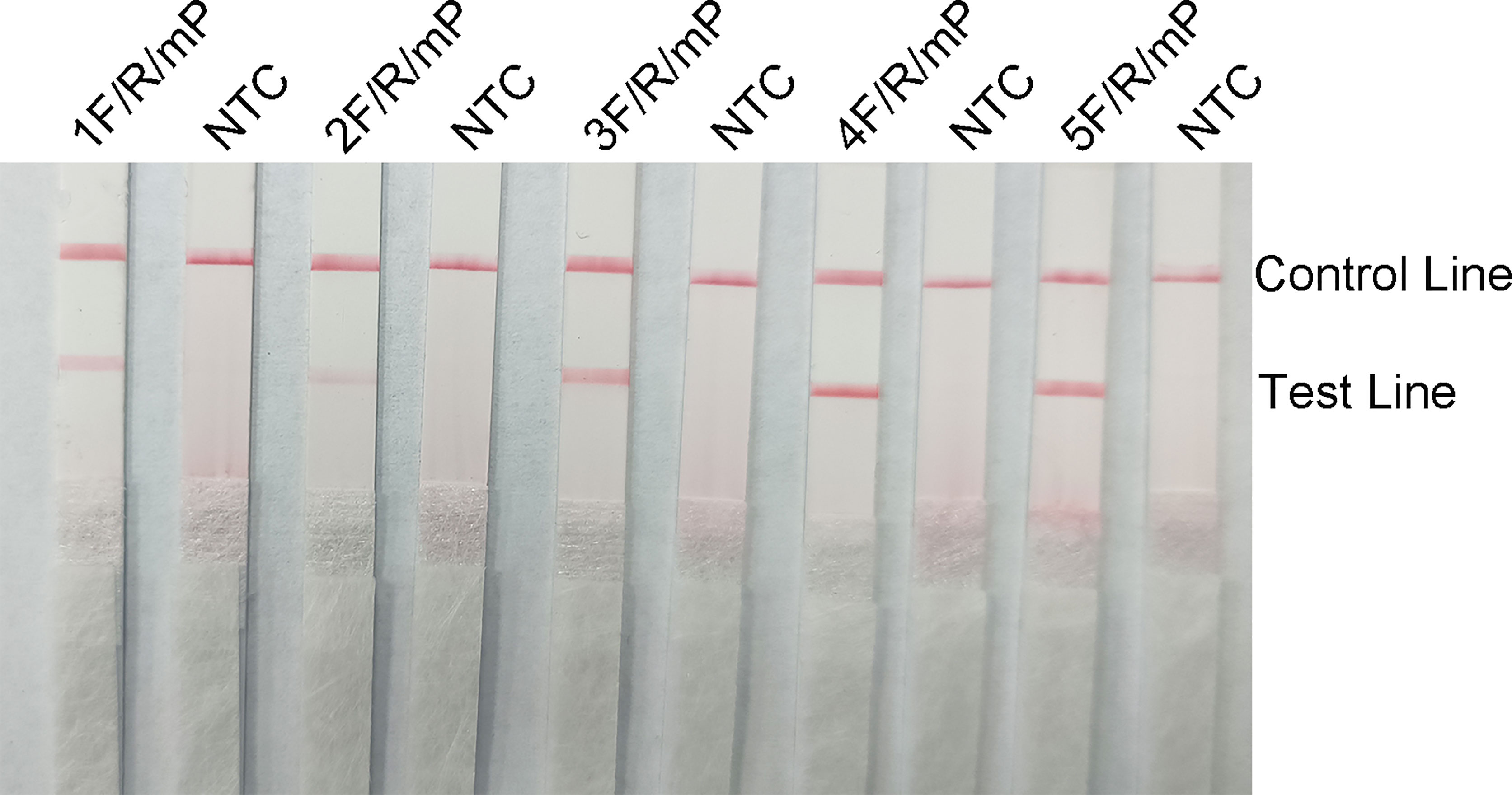
Figure 6 The performance of the primer-probe sets was tested using the RPA-LFS system. The LFS results of the RPA amplified products are shown in the figure. The name of each primer-probe set is labeled above the corresponding strip. The NTC strip is the template-free control for the corresponding RPA reaction on the left strip. The positions of the test and control lines are marked on the right side of the strip.
Optimization of Reaction Conditions
Red bands were seen on the test line at all temperatures in the RPA-LFS analysis, most precisely at 37°C (Figure 7A); therefore, we chose 37°C as the optimal reaction temperature. Red stripes appeared on the test line after 5 min of reaction time and were more obvious by 25 min. The color change was not obvious after 25 min (Figure 7B); therefore, 25 min was chosen as the optimal reaction time.
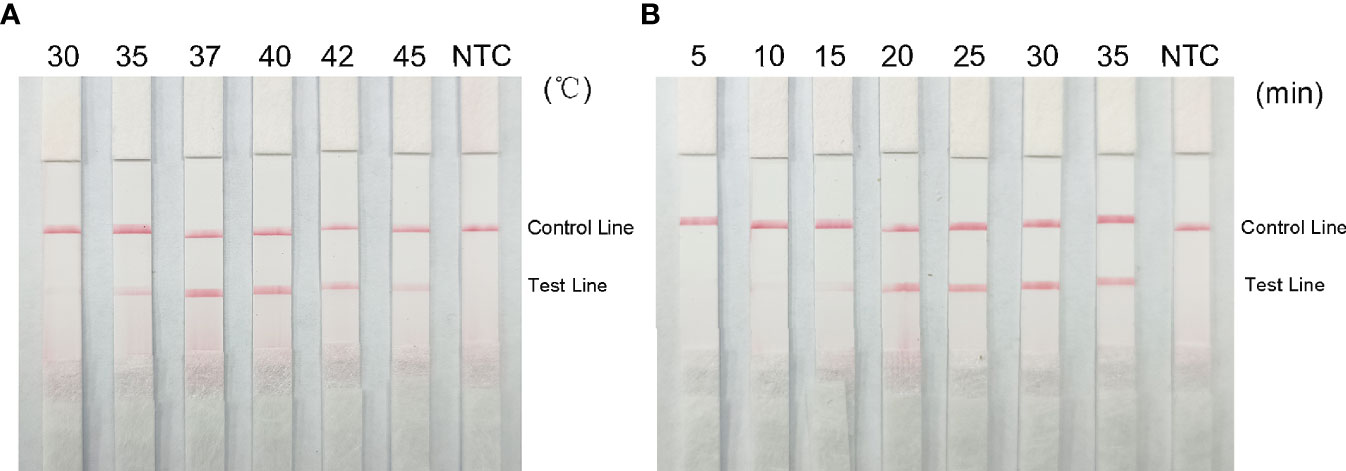
Figure 7 Optimal reaction temperature and time for the RPA-LFS system. (A) Images show the LFS results of RPA amplification at different temperatures. The temperature at which the RPA reaction was performed is shown at the top of each band. The template is the supernatant obtained from the boiling method of Klebsiella pneumoniae carrying rmpA2. The NTC strip was performed at 37°C without template control. (B) Images show the results of LFS for different lengths of RPA amplification. The length of time to perform the RPA reaction is shown at the top of each strip. The template is the supernatant obtained from the Klebsiella pneumoniae cooking method carrying rmpA2. The NTC strip was performed for 30 minutes without template control. The positions of the control and test lines are shown on the right side of the image.
Specificity of the RPA-LFS Assay
To confirm the specificity of the primer-probe set, RPA-LFS reactions were performed against six other common pathogens, namely Staphylococcus aureus, Streptococcus pneumoniae, Staphylococcus epidermidis, E. coli, Staphylococcus cloacae, and Pseudomonas aeruginosa. Using the genomes of these bacteria as templates did not produce a positive band, while using the genome of hvKP as a template produced a clear and specific amplification band (Figure 8). This suggests that the primer-probe set has good specificity for hvKP and does not cross-react with the other six pathogens and Klebsiella pneumoniae that do not carry rmpA2.
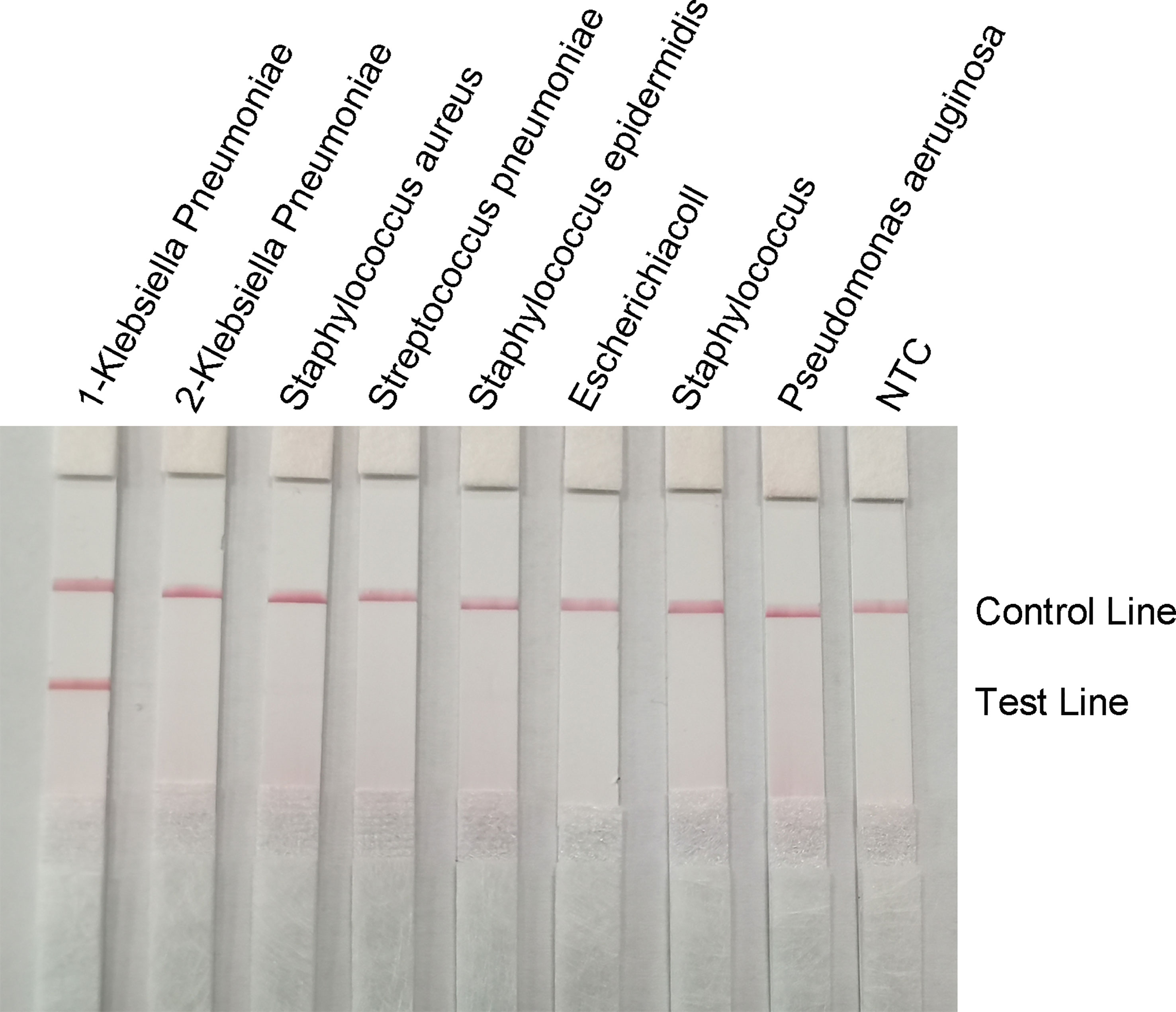
Figure 8 Specificity of the RPA-LFS system. The bacterial species is shown at the top of each strip. The NTC strip is a no-template control. The reaction was performed at 37°C for 30 minutes. The positions of the control and test lines are shown on the right side of the image. 1-Klebsiella pneumoniae means carrying rmpA2, 2-Klebsiella pneumoniae means not carrying rmpA2.
RPA-LFS Sensitivity Assay
To determine the minimum detection limit of the RPA-LFS system, we diluted genomic DNA at a gradient of 103–10-2 ng/μL. The system could detect concentrations of 10−1 ng/μL, and the red detection band gradually deepened as the template concentration increased (Figure 9A). Using genomic DNA of hvKP as a template, we amplified the rmpA2 gene using standard PCR methods. After purifying the amplification product and ligating into the pMD 19-T vector, a positive recombinant plasmid was successfully identified by PCR and sequencing. Analysis showed 100% nucleotide sequence similarity of the recombinant plasmid with the standard sequence, which further confirmed the successful construction of recombinant plasmid Ss-mb-pMD 19-T and its suitability for subsequent experiments. The construction of the mock recombinant plasmid Ss-mb-pMD 19-T was performed using SnapGene software to obtain the full base sequence of the recombinant plasmid. The conversion of ng to copy number can then be done by selecting Tools and clicking Show DNA Calculation. DNA quantity and quality of the constructed plasmids were measured using a Qubit 9000 fluorometer, copy number was calculated, and sensitivity was measured at 10-fold dilution in a 105–100 copies/μL gradient. The RPA-LFS system could detect 10 copies/μL, with the red detection band gradually deepening with increasing concentration (Figure 9B). In addition, PCR can detect concentrations up to 1 ng/μL and qPCR can detect concentrations up to 10-1 ng/μL (Figure 10). Our established RPA-LFS was more sensitive than the PCR reaction and comparable to the sensitivity of qPCR.
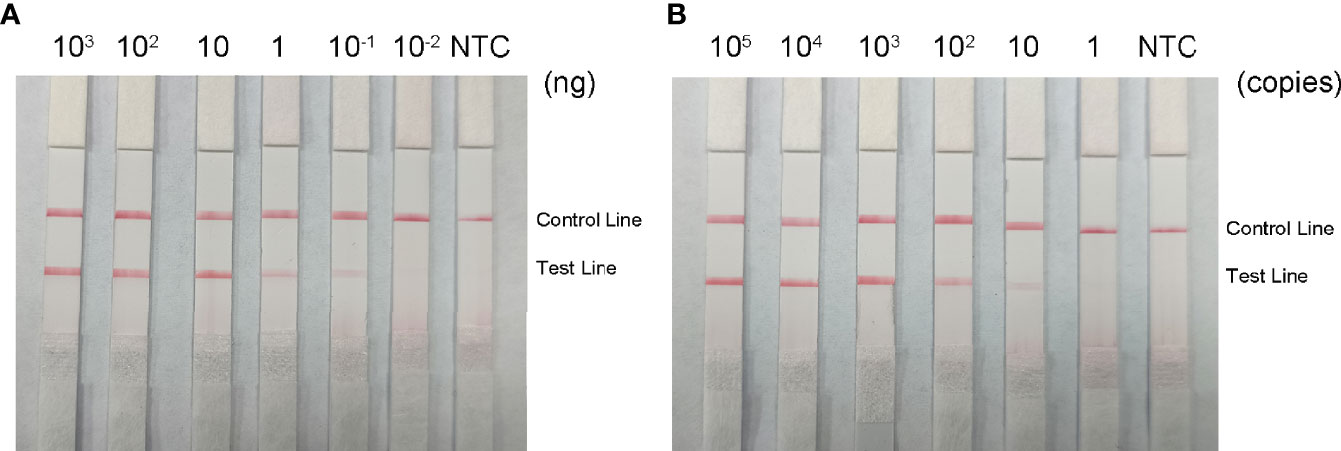
Figure 9 Sensitivity of the RPA-LFS system. (A) LFS results for different amounts of RPA carrying rmpA2 of Klebsiella pneumoniae genomic DNA. The amount added to the RPA reaction (ng/μL) is shown at the top of each band. The amplification template is the supernatant obtained from the boiling method of Klebsiella pneumoniae carrying rmpA2 (B) Images of the LFS results of RPA amplification using different amounts of recombinant plasmid Ss-mb-pMD™ 19-T. The amount added to the RPA reaction (in copies) is shown at the top of each strip. The NTC strip is the control without template. The positions of the control and test lines are shown on the right side of the image. Reactions were performed at 37°C for 30 minutes.
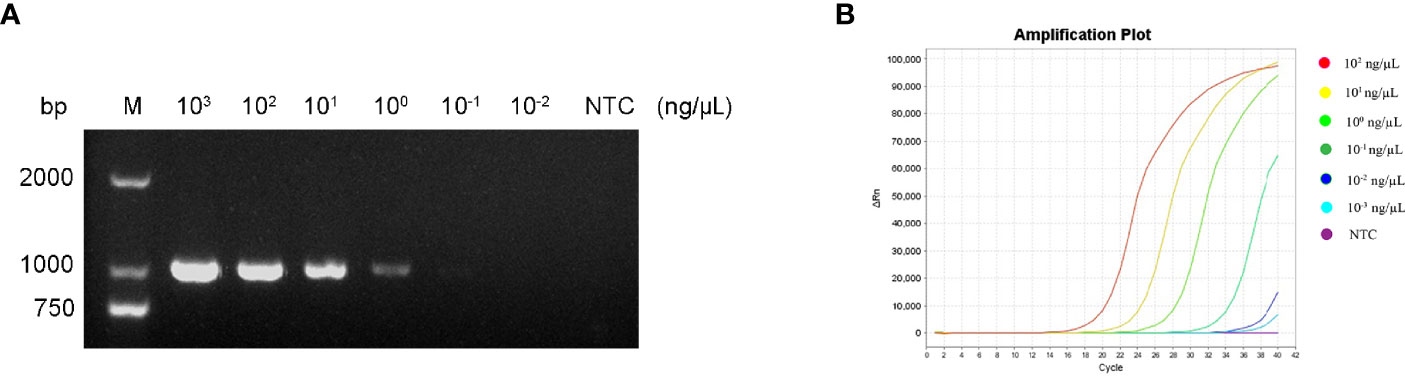
Figure 10 (A) Sensitivity of the PCR assay. (B) Sensitivity of the qPCR assay. The NTC strip is the control reaction without a template.
Detection of Clinical Samples
65 clinically confirmed strains of K. pneumoniae were examined using the RPA-LFS method 、qPCR and the conventional PCR method. 15 strains contained the virulence gene rmpA2 and 50 strains did not contain the virulence gene rmpA2, with the detection results of the three methods completely consistent (Supplementary Figure S1 and Table 2). However, the RPA-LFS method was more rapid and accurate with less instrument dependence.
Discussion
Highly virulent K. pneumoniae can cause community-invasive liver abscesses and often secondary to long-distance dissemination causing brain abscesses, lung infections, and eye infections with high mortality. Rapid and accurate detection of highly virulent K. pneumoniae is essential for clinicians to rapidly treat patients effectively. Combining the isothermal RPA reaction with the LFS endpoint reading method, with short reaction times, low isothermal conditions, simple operation, and no instrumentation, offers a promising solution (Wu et al., 2016). This RPA-LFS technique has been successfully applied in the molecular diagnosis of diseases caused by methicillin-resistant Staphylococcus aureus, Mycobacterium tuberculosis, Cryptococcus novelis, and other pathogenic bacteria with exploratory significance (Yongkiettrakul et al., 2017; Ma et al., 2019; Xu et al., 2021) However, the technique places high demands on primer design, and a very small number of primer dimers may lead to false positives.
Primer-dimer formation is influenced by buffer content, ambient temperature, mixture impurities, and other factors that are difficult to avoid. Primer-dimer formation can be reduced in PCR using a hot-start strategy; however, this approach is not applicable in the RPA-LFS technique. We found that careful screening of primer pairs and avoiding consecutive matches between primer pairs could effectively prevent primer-dimer formation. To further reduce the formation of primer dimers, we introduced probes in the RPA reaction. However, the primer-probe combinations designed in this study still showed false positives.
RPA reactions can tolerate a small number of base substitutions between template and primer or probe (Daher et al., 2015; Lillis et al., 2016). It is important to note that fewer base substitution can improve the detection performance of RPA-LFS. However, we can reduce the generation of primer dimers using appropriate base substitution. First, we designed several pairs of forward and reverse primers using Primer Premier 5.0 for the virulence gene rmpA2. From these, we selected a pair of primers with few primer dimers predicted by the software and stretched the upstream primer by about 15 bases as the probe. Base substitution of the probe and downstream primer was performed using Primer Premier 5.0 to eliminate primer dimers. Subsequently, multiple upstream primers were designed upstream of the probe, and the best primer-probe combination was selected by combining different upstream primers with downstream primers and probes. The modified primer probes showed high sensitivity while eliminating false positive signals. The modified primer probes were specific and specifically identified highly virulent K. pneumoniae. Thus, we established a primer-probe combination for the detection of highly virulent K. pneumoniae using the RPA-LFS technique.
RPA-LFS combines the advantages of the constant temperature of RPA as well as visualization using LFS, providing a rapid and accurate detection method for highly virulent K. pneumoniae. When testing the primer probe specificity with other common strains, we found that it was highly specific for highly virulent K. pneumoniae. The lower limit of detection was as low as 10−1 ng/μL and 10 copies/μL. RPA-LFS was more sensitive than the PCR reaction and comparable to the sensitivity of qPCR. In fact, the amount of bacterial DNA in clinical samples was much higher than 10 copies/μL, meeting the clinical requirements for detection.
In this study, primers and probes were designed using the target sequence of the virulence gene rmpA2, and the results were interpreted by visual inspection only at an optimal reaction temperature of 37°C after 25 min. When used for clinical specimen testing, the sample does not need to be purified; DNA can be released by boiling and used directly for testing. The results of RPA-LFS were consistent with qPCR results and conventional PCR methods, and the detection accuracy was 100%. Thus, our RPA-LFS system provides the experimental basis for rapid detection of highly virulent K. pneumoniae to help clinicians take more accurate and rapid therapeutic measures.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
This study was approved by the Medical Ethics Committee of the Second People’s Hospital of Lianyungang (Permit number: 2020005). Clinical strains were collected in 2020-2021 and isolated from sputum, urine, pharyngeal swabs, drainage fluid, and other specimens. Written consent was obtained from each patient for all samples isolated.
Author Contributions
NL, FM and WL conceived and designed the experiments and wrote the paper. LW, FW and HC performed the experiments. ST, QZ and LL analyzed the data. All authors discussed the results, provided comments and approved the final version of the manuscript.
Funding
The work was funded by the Natural Science Foundation of Jiangsu Province (BK20191210), the “Project 333” training fund of Jiangsu Province (BRA2019248), Lianyungang City “521 Project” project (LYG06521202160) and Postgraduate Research Innovation Program of Bengbu Medical College (Byycxz21007).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.877649/full#supplementary-material
Supplementary Figure 1 | (A) qPCR results of clinical samples. (B) PCR results of clinical samples.
References
Chen, Y. T., Chang, H. Y., Lai, Y. C., Pan, C. C., Tsai, S. F., Peng, H. L. (2004). Sequencing and Analysis of the Large Virulence Plasmid pLVPK of Klebsiella Pneumoniae CG43. Gene 337, 189–198. doi: 10.1016/j.gene.2004.05.008
Cheng, H. Y., Chen, Y. S., Wu, C. Y., Chang, H. Y., Lai, Y. C., Peng, H. L. (2010). RmpA Regulation of Capsular Polysaccharide Biosynthesis in Klebsiella Pneumoniae CG43. J. Bacteriol. 192 (12), 3144–3158. doi: 10.1128/jb.00031-10
Cheng, D. L., Liu, Y. C., Yen, M. Y., Liu, C. Y., Wang, R. S. (1991). Septic Metastatic Lesions of Pyogenic Liver Abscess. Their Association With Klebsiella Pneumoniae Bacteremia in Diabetic Patients. Arch. Intern. Med. 151 (8), 1557–1559. doi: 10.1001/archinte.1991.00400080059010
Cordray, M. S., Richards-Kortum, R. R. (2015). A Paper and Plastic Device for the Combined Isothermal Amplification and Lateral Flow Detection of Plasmodium DNA. Malaria J. 14, 472. doi: 10.1186/s12936-015-0995-6
Daher, R. K., Stewart, G., Boissinot, M., Bergeron, M. G. (2016). Recombinase Polymerase Amplification for Diagnostic Applications. Clin. Chem. 62 (7), 947–958. doi: 10.1373/clinchem.2015.245829
Daher, R. K., Stewart, G., Boissinot, M., Boudreau, D. K., Bergeron, M. G. (2015). Influence of Sequence Mismatches on the Specificity of Recombinase Polymerase Amplification Technology. Mol. Cell. Probes 29 (2), 116–121. doi: 10.1016/j.mcp.2014.11.005
Dai, T., Yang, X., Hu, T., Jiao, B., Xu, Y., Zheng, X., et al. (2019). Comparative Evaluation of a Novel Recombinase Polymerase Amplification-Lateral Flow Dipstick (RPA-LFD) Assay, LAMP, Conventional PCR, and Leaf-Disc Baiting Methods for Detection of Phytophthora Sojae. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01884
Fierer, J., Walls, L., Chu, P. (2011). Recurring Klebsiella Pneumoniae Pyogenic Liver Abscesses in a Resident of San Diego, California, Due to a K1 Strain Carrying the Virulence Plasmid. J. Clin. Microbiol. 49 (12), 4371–4373. doi: 10.1128/jcm.05658-11
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2018). A Fatal Outbreak of ST11 Carbapenem-Resistant Hypervirulent Klebsiella Pneumoniae in a Chinese Hospital: A Molecular Epidemiological Study. Lancet Infect. Dis. 18 (1), 37–46. doi: 10.1016/s1473-3099(17)30489-9
Lillis, L., Lehman, D. A., Siverson, J. B., Weis, J., Cantera, J., Parker, M., et al. (2016). Cross-Subtype Detection of HIV-1 Using Reverse Transcription and Recombinase Polymerase Amplification. J. Virol. Methods 230, 28–35. doi: 10.1016/j.jviromet.2016.01.010
Li, J., Ma, B., Fang, J., Zhi, A., Chen, E., Xu, Y., et al. (2019). Recombinase Polymerase Amplification (RPA) Combined With Lateral Flow Immunoassay for Rapid Detection of Salmonella in Food. Foods (Basel Switzerland) 9, (1). doi: 10.3390/foods9010027
Li, G., Shi, J., Zhao, Y., Xie, Y., Tang, Y., Jiang, X., et al. (2020). Identification of Hypervirulent Klebsiella Pneumoniae Isolates Using the String Test in Combination With Galleria Mellonella Infectivity. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 39 (9), 1673–1679. doi: 10.1007/s10096-020-03890-z
Liu, Y. C., Cheng, D. L., Lin, C. L. (1986). Klebsiella Pneumoniae Liver Abscess Associated With Septic Endophthalmitis. Arch. Intern. Med. 146 (10), 1913–1916. doi: 10.1001/archinte.1986.00360220057011
Liu, L., Wang, J., Geng, Y., Wang, J., Li, R., Shi, R., et al. (2018). Equipment-Free Recombinase Polymerase Amplification Assay Using Body Heat for Visual and Rapid Point-of-Need Detection of Canine Parvovirus 2. Mol. Cell. Probes 39, 41–46. doi: 10.1016/j.mcp.2018.04.004
Ma, Q., Yao, J., Yuan, S., Liu, H., Wei, N., Zhang, J., et al. (2019). Development of a Lateral Flow Recombinase Polymerase Amplification Assay for Rapid and Visual Detection of Cryptococcus Neoformans/C. Gattii in Cerebral Spinal Fluid. BMC Infect. Dis. 19 (1), 108. doi: 10.1186/s12879-019-3744-6
Salazar, J. K., Wang, Y., Yu, S., Wang, H., Zhang, W. (2015). Polymerase Chain Reaction-Based Serotyping of Pathogenic Bacteria in Food. J. Microb. Methods 110, 18–26. doi: 10.1016/j.mimet.2015.01.009
Shon, A. S., Russo, T. A. (2012). Hypervirulent Klebsiella Pneumoniae: The Next Superbug? Future Microbiol. 7 (6), 669–671. doi: 10.2217/fmb.12.43
Siu, L. K., Yeh, K. M., Lin, J. C., Fung, C. P., Chang, F. Y. (2012). Klebsiella Pneumoniae Liver Abscess: A New Invasive Syndrome. Lancet Infect. Dis. 12 (11), 881–887. doi: 10.1016/s1473-3099(12)70205-0
Wang, F., Ge, D., Wang, L., Li, N., Chen, H., Zhang, Z., et al. (2021). Rapid and Sensitive Recombinase Polymerase Amplification Combined With Lateral Flow Strips for Detecting Candida Albicans. Anal. Biochem. 633, 114428. doi: 10.1016/j.ab.2021.114428
Wang, J., Wang, J., Li, R., Shi, R., Liu, L., Yuan, W. (2018). Evaluation of an Incubation Instrument-Free Reverse Transcription Recombinase Polymerase Amplification Assay for Rapid and Point-of-Need Detection of Canine Distemper Virus. J. Virol. Methods 260, 56–61. doi: 10.1016/j.jviromet.2018.07.007
Wu, K. M., Li, L. H., Yan, J. J., Tsao, N., Liao, T. L., Tsai, H. C., et al. (2009). Genome Sequencing and Comparative Analysis of Klebsiella Pneumoniae NTUH-K2044, a Strain Causing Liver Abscess and Meningitis. J. Bacteriol. 191 (14), 4492–4501. doi: 10.1128/jb.00315-09
Wu, Y. D., Zhou, D. H., Zhang, L. X., Zheng, W. B., Ma, J. G., Wang, M., et al. (2016). Recombinase Polymerase Amplification (RPA) Combined With Lateral Flow (LF) Strip for Equipment-Free Detection of Cryptosporidium Spp. Oocysts in Dairy Cattle Feces. Parasitol. Res. 115 (9), 3551–3555. doi: 10.1007/s00436-016-5120-4
Xu, Y., Wu, P., Zhang, H., Li, J. (2021). Rapid Detection of Mycobacterium Tuberculosis Based on Antigen 85B via Real-Time Recombinase Polymerase Amplification. Lett. Appl. Microbiol. 72 (2), 106–112. doi: 10.1111/lam.13364
Yang, X., Wai-Chi Chan, E., Zhang, R., Chen, S. (2019). A Conjugative Plasmid That Augments Virulence in Klebsiella Pneumoniae. Nat. Microbiol. 4 (12), 2039–2043. doi: 10.1038/s41564-019-0566-7
Yongkiettrakul, S., Kampeera, J., Chareanchim, W., Rattanajak, R., Pornthanakasem, W., Kiatpathomchai, W., et al. (2017). Simple Detection of Single Nucleotide Polymorphism in Plasmodium Falciparum by SNP-LAMP Assay Combined With Lateral Flow Dipstick. Parasitol. Int. 66 (1), 964–971. doi: 10.1016/j.parint.2016.10.024
Yu, V. L., Hansen, D. S., Ko, W. C., Sagnimeni, A., Klugman, K. P., von Gottberg, A., et al. (2007). Virulence Characteristics of Klebsiella and Clinical Manifestations of K. Pneumoniae Bloodstream Infections. Emerg. Infect. Dis. 13 (7), 986–993. doi: 10.3201/eid1307.070187
Keywords: highly virulent Klebsiella pneumoniae, recombinase polymerase amplification, rapid test, rmpA2, base substitution, lateral flow strip (LFS)
Citation: Li N, Wang L, Wang F, Chen H, Tao S, Zhu Q, Liu L, Liang W and Ma F (2022) Rapid Detection of Klebsiella pneumoniae Carrying Virulence Gene rmpA2 by Recombinase Polymerase Amplification Combined With Lateral Flow Strips. Front. Cell. Infect. Microbiol. 12:877649. doi: 10.3389/fcimb.2022.877649
Received: 17 February 2022; Accepted: 22 April 2022;
Published: 19 May 2022.
Edited by:
Ishwara Bhat, Indian Institute of Spices Research (ICAR), Kozhikode, IndiaReviewed by:
Bo Pang, National Institute for Communicable Disease Control and Prevention (China CDC), ChinaYingwang Ye, Hefei University of Technology, China
Copyright © 2022 Li, Wang, Wang, Chen, Tao, Zhu, Liu, Liang and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Liang, aHNsd3lzQDE2My5jb20=; Fang Ma, bWFmYW5nNjI3QHNpbmEuY29t
 Na Li
Na Li Lei Wang
Lei Wang Fang Wang
Fang Wang Huimin Chen
Huimin Chen Shuan Tao4
Shuan Tao4 Wei Liang
Wei Liang