- 1Department of Clinical Microbiology, Christian Medical College and Hospital, Vellore, India
- 2Cambridge Institute of Therapeutic Immunology & Infectious Disease (CITIID), Department of Medicine, University of Cambridge, Cambridge, United Kingdom
- 3Department of Pulmonary Medicine, Christian Medical College and Hospital, Vellore, India
- 4Department of Haematology, Christian Medical College and Hospital, Vellore, India
Background: Hypervirulent variants of Klebsiella pneumoniae (HvKp) were typically associated with a broadly antimicrobial susceptible clone of sequence type (ST) 23 at the time of its emergence. Concerningly, HvKp is now also emerging within multidrug-resistant (MDR) clones, including ST11, ST15, and ST147. MDR-HvKp either carry both the virulence and resistance plasmids or carry a large hybrid plasmid coding for both virulence and resistance determinants. Here, we aimed to genetically characterize a collection of MDR-HvKp ST2096 isolates haboring hybrid plasmids carrying both antimicrobial resistance (AMR) and virulence genes.
Methods: Nine K. pneumoniae ST2096 isolated over 1 year from the blood sample of hospitalized patients in southern India that were MDR and suspected to be HvKp were selected. All nine isolates were subjected to short-read whole-genome sequencing; a subset (n = 4) was additionally subjected to long-read sequencing to obtain complete genomes for characterization. Mucoviscosity assay was also performed for phenotypic assessment.
Results: Among the nine isolates, seven were carbapenem-resistant, two of which carried blaNDM-5 on an IncFII plasmid and five carried blaOXA-232 on a ColKP3 plasmid. The organisms were confirmed as HvKp, with characteristic virulence genes (rmpA2, iutA, and iucABCD) carried on a large (~320 kbp) IncFIB–IncHI1B co-integrate. This hybrid plasmid also carried the aadA2, armA, blaOXA-1, msrE, mphE, sul1, and dfrA14 AMR genes in addition to the heavy-metal resistance genes. The hybrid plasmid showed about 60% similarity to the IncHI1B virulence plasmid of K. pneumoniae SGH10 and ~70% sequence identity with the first identified IncHI1B pNDM-MAR plasmid. Notably, the hybrid plasmid carried its type IV-A3 CRISPR-Cas system which harbored spacer regions against traL of IncF plasmids, thereby preventing their acquisition.
Conclusion: The convergence of virulence and AMR is clinically concerning in K. pneumoniae. Our data highlight the role of hybrid plasmids carrying both AMR and virulence genes in K. pneumoniae ST2096, suggesting that MDR-HvKp is not confined to selected clones; we highlight the continued emergence of such genotypes across the species. The convergence is occurring globally amidst several clones and is of great concern to public health.
Introduction
Klebsiella pneumoniae (Kp) is a common cause of hospital-acquired infection (Marr and Russo, 2019). Some forms of K. pneumoniae can cause invasive diseases, affecting the liver and other internal organs, and are considered to be hypervirulent (HvKp) (Shon et al., 2013; Marr and Russo, 2019). While HvKp does not have a precise definition, it refers to isolates that carry the virulence plasmid (GenBank accession numbers CP025081, AY378100) coding for rmpA/rmpA2, iucA, iutA, and/or iroB (Marr and Russo, 2019). Although HvKp was confined to community-acquired infections, recent reports suggest that HvKp is an emerging nosocomial pathogen with the potential to cause devastating hospital outbreaks thereby establishing itself in both niches (Gu et al., 2018; Liu et al., 2020). HvKp infections are becoming prevalent globally and associated with increased mortality, and a recent study reports the gut colonization of MDR-HvKp in pregnant women (Shon et al., 2013; Marr and Russo, 2019; Huynh et al., 2020). Earlier, HvKp isolates were susceptible to the majority of clinically relevant antimicrobials as they were rarely associated with multidrug resistance plasmids (Shon et al., 2013). However, in the last decade, the organism has undergone several genomic changes and expanded its genome by acquiring multiple resistance plasmids (Lee et al., 2017; Gu et al., 2018; Liu et al., 2020).
It has been found that in the population structure of HvKp, when determined by multi-locus sequence typing (MLST) and whole-genome sequencing (WGS), most HvKp isolates belong to the clonal groups (CG) 23, 65, 86, 375, and 380 (Bialek-Davenet et al., 2014). Conversely, carbapenem-resistant K. pneumoniae (CRKp) is associated with a clonal expansion of CG258 in Europe and endemic dissemination of ST11, ST14, ST147, and ST231 clones in Asia and Europe (Qi et al., 2011; Lee et al., 2016; Navon-Venezia et al., 2017; Wyres et al., 2020). However, there have been recent reports on the the emergence of HvKp with multidrug resistance (MDR, resistant to one or more agents in ≥3 antimicrobial classes) phenotypes in divergent CGs (Lee et al., 2017; Turton et al., 2018; Lam et al., 2019), creating new strains with the ability to cause serious infection with limited treatment options (Bialek-Davenet et al., 2014; Yao et al., 2018; Yang et al., 2020). The convergence of MDR and virulence pathotypes in a single isolate occurs either by the uptake of a virulence plasmid by MDR isolate or by the uptake of plasmids carrying antimicrobial resistance genes (ARGs) by the virulent isolates (Tang et al., 2020). CRKp ST11 acquiring the pLVPK-like virulence plasmid and ST23 HvKp acquiring multiple resistance plasmids are some instances where both pathotypes have converged (Liu et al., 2020; Shankar et al., 2020).
Specifically, carbapenem-resistant hypervirulent K. pneumoniae (CR-HvKp) has arisen by the formation of mosaic plasmids and hybrid plasmids (Tang et al., 2020; Yang et al., 2021). These mosaic plasmids are typically composed of two or more different plasmid backbones and create a scenario where AMR and virulence determinants are encoded on a single large plasmid with a mosaic (medley) arrangement of resistance and virulence genes whereas the hybrid plasmids are co-integrates with two plasmid backbones (Lam et al., 2019; Turton et al., 2019). Mosaic plasmids with fragments of virulence plasmid and IncFIIK coding for resistance and virulence have been described among ST15 K. pneumoniae from Europe (Lam et al., 2019). In contrast, the hybrid plasmids with a range of replicons (IncFIB–IncHI1B, IncFIBK–IncHI1B, IncFIB–IncR) have been described in China and Europe (Lam et al., 2019; Turton et al., 2019; Li et al., 2020; Xie et al., 2020; Yang et al., 2020) and are being increasingly reported among regional MDR clones. Our understanding of these CR-HvKp hybrid plasmids is limited due to an insufficient number of complete plasmid sequences. Here, we aimed to characterize a set of MDR-HvKp belonging to ST2096, possessing hybrid plasmids that simultaneously carry both AMR and virulence genes. The complete genome sequences of four of these isolates were further generated by long-read sequencing to elucidate the detailed structure of the hybrid plasmid via comparative genomics.
Materials and Methods
Bacterial Isolates
The K. pneumoniae were isolated from patients with bacteremia admitted to the Christian Medical College, Vellore, India, in 2019. The isolates were identified using standard microbiological methods and further confirmed by VITEK MS [Database v2.0, bioMerieux, Marcy-l’Étoile, France] (Versalovic et al., 2011). The isolates were screened for hypermucoviscous phenotype using the string test (Shon et al., 2013). In addition, the mucoid phenotype-associated genes rmpA and rmpA2 were detected by PCR (Turton et al., 2010; Compain et al., 2014). The demographic and clinical details of the nine patients from whom the organisms were isolated were accessed from electronic medical records. The study was approved by the Institutional Review Board of Christian Medical College, Vellore, with minute number 9616 (01/09/2015).
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing (AST) was performed using the Kirby-Bauer disk diffusion method according to the CLSI 2019 guidelines (Clinical and Laboratory Standards Institute, 2019). The tested antimicrobials were cefotaxime (30 µg), ceftazidime (30 µg), piperacillin/tazobactam (100/10 µg), cefoperazone/sulbactam (75/30 µg), imipenem (10 µg), meropenem (10 µg), ciprofloxacin (5 µg), levofloxacin (5 µg), gentamicin (10 µg), amikacin (30 µg), and minocycline (30 µg). The minimum inhibitory concentration (MIC) of meropenem was determined by the broth microdilution (BMD) method. Escherichia coli ATCC® 25922, Enterococcus faecium ATCC® 29212, and Pseudomonas aeruginosa ATCC® 27853 were used as controls. Data were interpreted according to the 2019 CLSI guidelines (CLSI 2019).
Mucoviscosity Assay
Overnight culture of the HvKp isolates was inoculated in Luria Bertani (LB) broth (Oxoid, Hampshire, United Kingdom) and centrifuged at 1,000 rpm for 15 min as previously described (Mike et al., 2021). Briefly, the optical density (OD) of the supernatant was measured at 600-nm wavelength in the UV spectrophotometer (1st OD). 1 ml of PBS (phosphate-buffered saline) was added, and OD600 was adjusted to 1.00. This was centrifuged again at 1,000 rpm for 5 min, and the OD600 of the supernatant (3rd OD) was measured. Non-virulent isolate K. quasipneumoniae ATCC® 700603 was used as a control for mucoviscosity assay. This assay is based on the principle that hypermucoviscous isolates do not sediment easily and hence the OD600 after centrifugation will be higher than the counterparts of control and non-hypermucoviscous isolates. Hence, hypermucoviscous isolates will have a higher SAC ratio than the rest.
Sedimentation Assay Calculation (SAC) = Reading of 3rd OD600/Reading of 1st OD600.
DNA Extraction and Genome Sequencing
The isolates studied were revived from the archive of the Department of Clinical Microbiology, and a single colony was inoculated in LB broth at 37°C. Total genomic DNA was extracted from pelleted cells using the Wizard DNA Purification Kit (Promega, Madison, WI, USA). Extracted DNA was quantified using NanoDrop One spectrophotometry (Thermo Fisher Scientific, Waltham, MA, USA) and Qubit 3.0 fluorometry (Life Technologies, Carlsbad, CA, USA) and stored at -20°C until further use.
A sequencing library was prepared using the Nextera DNA Flex Library Preparation Kit (Illumina, San Diego, CA, USA). Subsequently, the paired-end library was subjected to sequencing on a HiSeq 2500 platform (Illumina, USA) generating 2 × 150-bp reads. Sequencing reads with a PHRED quality score below 20 were discarded, and adapters were trimmed using cutadapt v1.8.1 and assessed with FastQC v0.11.4 (Andrews, 2010; Martin, 2011). For a subset of four isolates, long-read sequencing was performed using an Oxford Nanopore MinION FLO-MIN106 R9 flow cell (Oxford Nanopore Technologies, UK). The long-read DNA library was prepared using the SQK-LSK108 ligation sequencing kit (v.R9) along with the ONT EXP-NBD103 Native Barcode Expansion kit (Oxford Nanopore Technologies, Oxford, UK). The library was loaded onto flow cells, run for 48 h using the standard MinKNOW software (Guppy version 3.6).
Genome Assembly and Evaluation
Draft genome sequence data generated using Illumina were assembled using SPAdes (v.3.13.0) (Bankevich et al., 2012). A hybrid de novo assembly was generated for a subset of four isolates (Vasudevan et al., 2020). The nanopore long reads were error-corrected with the standalone Canu error correction tool (v.1.7) and assembled using the Unicycler hybrid assembly pipeline (v 0.4.6) with the default settings (Koren et al., 2017; Wick et al., 2017). The genome sequences were polished using high-quality Illumina reads, as described previously using Pilon (Walker et al., 2014). The assembled genomes were subjected to quality assessment using CheckM v1.0.5 (Parks et al., 2015) and Quast v4.5 (Gurevich et al., 2013). K. pneumoniae NTUH-K2044 (GenBank accession number AP006725) was used as the reference genome since it is a well-characterized type of strain of ST23 hypervirulent K. pneumoniae.
Genome Analysis
Genome assemblies were submitted to NCBI GenBank and annotated using the NCBI Prokaryotic Genome Annotation Pipeline [PGAP v.4.1] (Tatusova et al., 2016).
The genomes described in the study are publicly available under the Bioproject ID PRJNA613369 in GenBank with accession numbers CP053765–CP053770, CP053771–CP053780, CP058798–CP058806, JAARNO010000001.1–JAARNO010000005.1, JAAQSG000000000, JAARNJ000000000, JAARMH000000000, and JAAQTC000000000. The antimicrobial resistance profile of the assembled genome sequences was identified using ResFinder v.4.1 available from CGE server (Bortolaia et al., 2020). Similarly, the presence of plasmids in the genomes was identified and characterized using PlasmidFinder (v.1.3) available at the CGE server (Carattoli et al., 2014). MLST and virulence loci (yersiniabactin, aerobactin, and other siderophore production systems) were identified using Kleborate (v.2.0.0) (Lam et al., 2021). The presence of virulence factors was confirmed using the virulence database at Pasteur Institute for K. pneumoniae (Jolley and Maiden, 2010). YbST and AbST, typing schemes based on yersiniabactin and aerobactin loci, were deduced from the database at Pasteur Institute for K. pneumoniae (https://bigsdb.pasteur.fr/cgi-bin/bigsdb/bigsdb.pl?db=pubmlst_klebsiella_seqdef&page=profiles). The K and O antigen loci were identified using Kaptive available at Kleborate (Wyres et al., 2016; Wick et al., 2018). The final assembled circular chromosomes and plasmids were visualized using CGView server v.1.0 (Grant and Stothard, 2008) and Easyfig (Sullivan et al., 2011). CRISPR regions in the genomes were identified with the CRISPRCasTyper web server (Russel et al., 2020). The genetic distance between isolates was calculated using average nucleotide identity (ANI) available at OrthoANI (Lee et al., 2016). Pairwise distance between the nine isolates was determined with BA10835 as reference using SNP-dists v 0.6.3 (Wysocka et al., 2020) from the raw reads by aligning the short reads of each isolate against the reference. An SNP-based phylogenetic tree of the complete hybrid plasmids with IncHI1B–IncFIB (pNDM-MAR) replicon types which are mentioned in Table S1 was constructed using CSI phylogeny (https://cge.cbs.dtu.dk/services/CSIPhylogeny/).
Results
Clinical Manifestations and Microbiological Characteristics of the Isolates
During the routine surveillance of HvKp, we identified isolates that were negative for the string test, positive for rmpA2 as determined by PCR, and carbapenem-resistant as determined by AST. These isolates were chosen for whole genome sequencing, and we identified nine ST2096 (a single-locus variant of ST14) K. pneumoniae associated with bacteremia in our hospital (Table 1). These nine K. pneumoniae ST2096 were resistant to all tested antimicrobials by disk diffusion assay and were initially considered to be extensively drug-resistant (XDR, non-susceptible to at least one agent in all but ≤2 classes of antimicrobials). However, upon MIC testing, two isolates were found to be susceptible to meropenem (MIC ≤0.5 µg/ml). The results of the mucoviscosity assay are mentioned in Figure S1. The isolates showed significantly higher OD600 when compared to the control strain indicating the lack of sedimentation by the hypervirulent isolates.
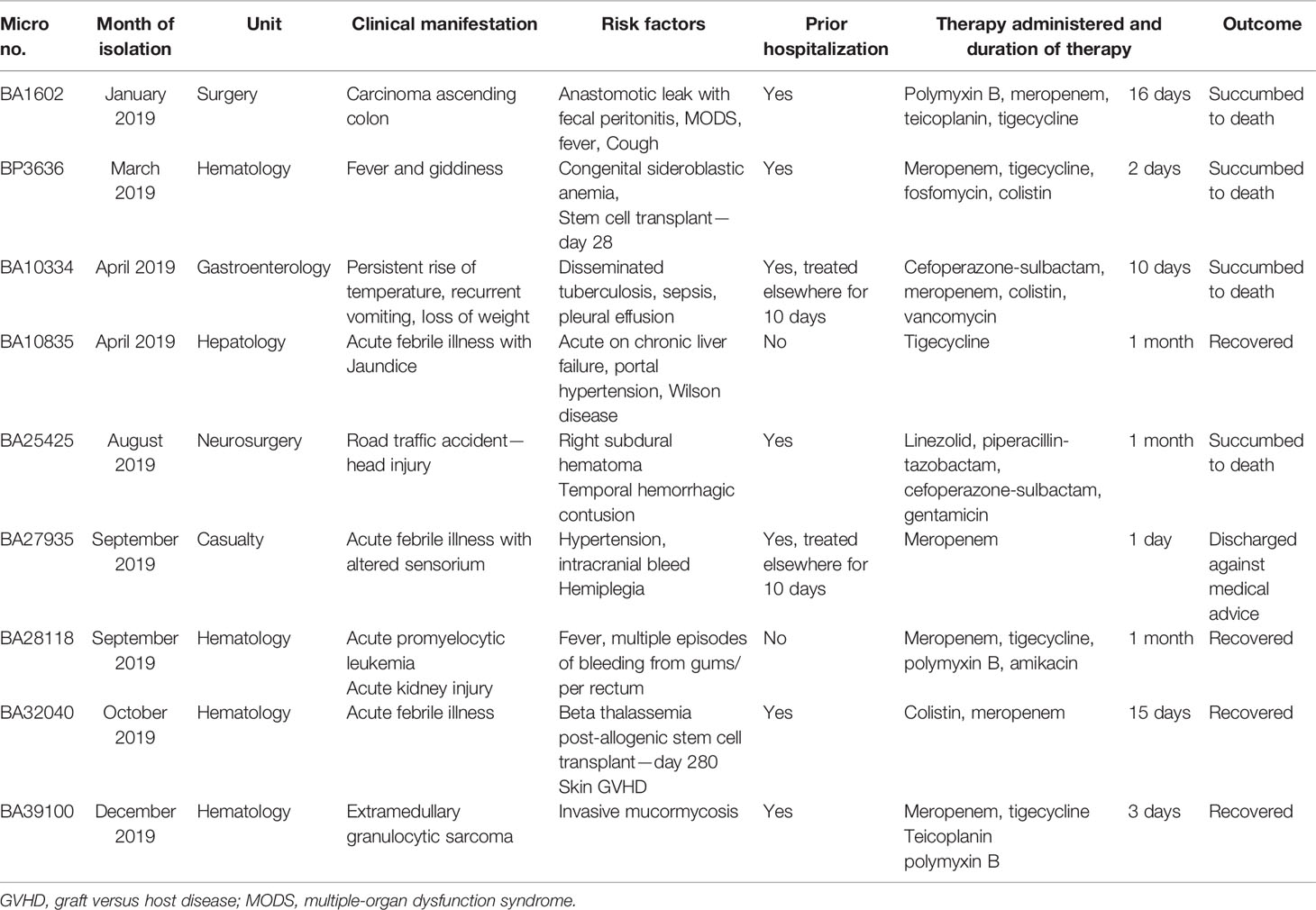
Table 1 The demographic and clinical details of the patients with bacteremia caused by hypervirulent K. pneumoniae ST2096.
From the resulting nine genome sequences, the surface capsule (K) loci were predicted to be K64 whereas the O-antigen encoding loci was determined to be O1v1 in all isolates (Tables 2 and 3). The pairwise average nucleotide identity (ANI) among the nine draft genomes was >99.8% (Figure S2A). The pairwise SNP difference among the nine isolates segregated them into two clusters, with BA10835 and BA27935 being >260 SNPs from the remaining seven sequences (Figure S2B). Within the major cluster (7 isolates), BA10334 and BA1602 were highly related (2 SNPs), and similarly, BA25425 and BP3636 (6SNPs) were related. Since this is a retrospective study, other specimen sources were not investigated to determine if there was an outbreak of ST2096 in the hospital and hence the dissemination of the plasmid cannot be explained.
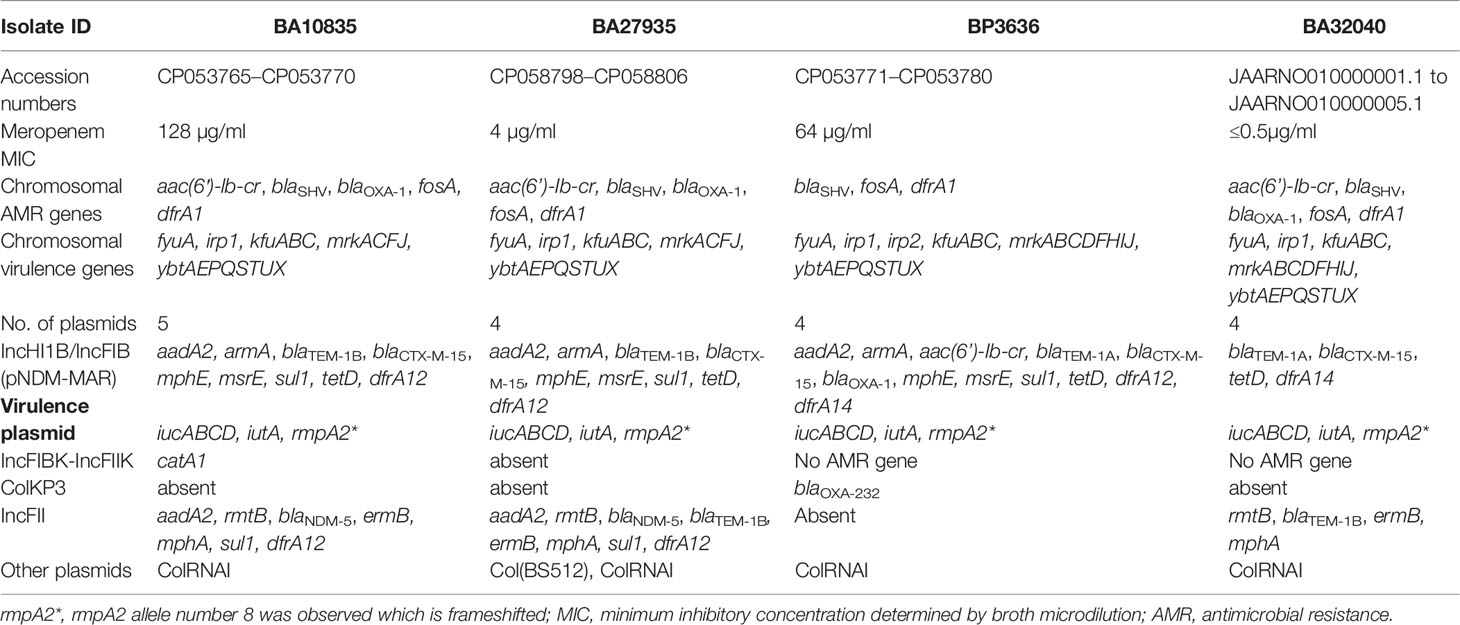
Table 2 Phenotypic and genotypic characteristics obtained using hybrid genome assembly of four Indian MDR hypervirulent K. pneumoniae ST2096.
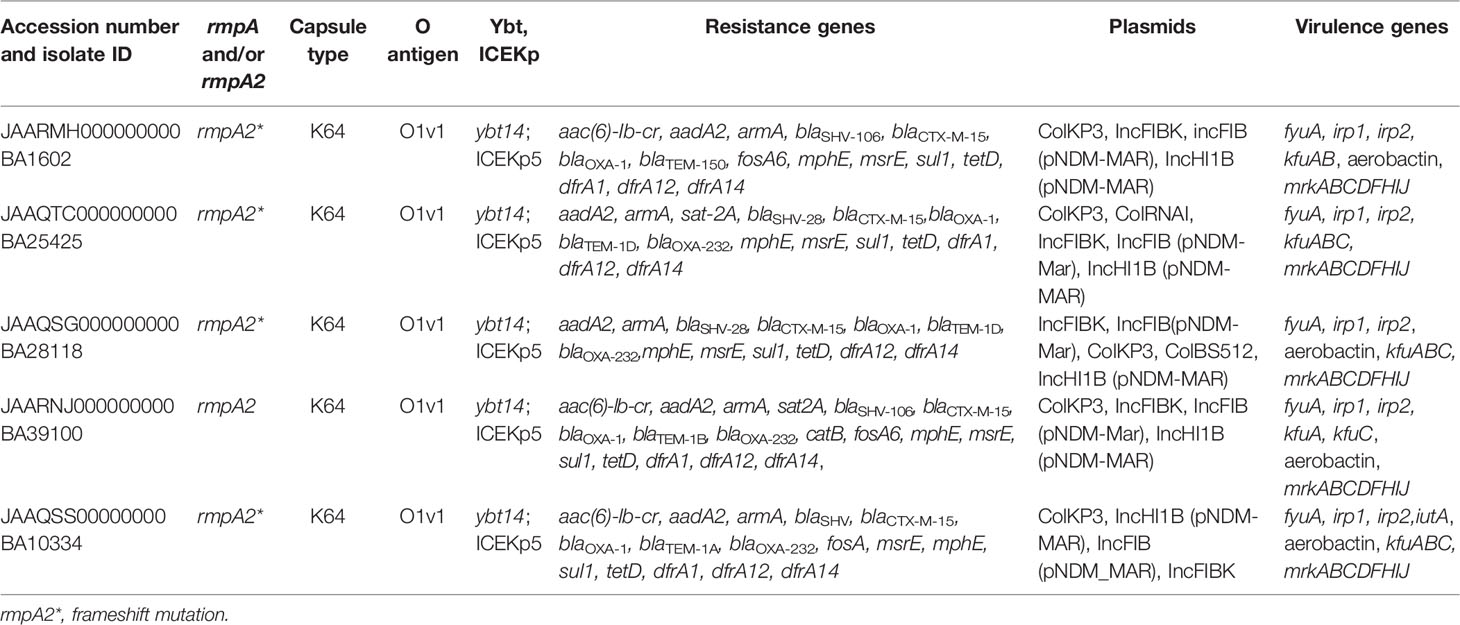
Table 3 Genotypic characteristics of multidrug-resistant hypervirulent K. pneumoniae belonging to ST2096 obtained from short read assembly.
Characterization of the HvKp ST2096 Chromosomes
Typically, K. pneumoniae chromosomes are characterized by the presence of blaSHV and fosA. In addition to these resistance genes, surprisingly, we found that three of the four MDR-HvKp isolates with complete genomes had the aac(6′)-lb-cr, blaOXA-1 and dfrA1 genes integrated into chromosome on mobile genetic elements. Specifically, aac(6′)-lb-cr and blaOXA-1 were inserted by IS26 in the central region of the chromosome at ~2.3 Mbp with a 7-bp flanking region (AGTCCGT) (Figure 1A). The dfrA1 gene was associated with ISKpn26 and a class 1 integron, intI1, at position ~5.3 Mbp. From the nine draft genome sequences, we identified a type I-E CRISPR located on the chromosome, characterized by 7–12 spacers of 32 bp and an adjacent ISKpn26 (Figure 1B).
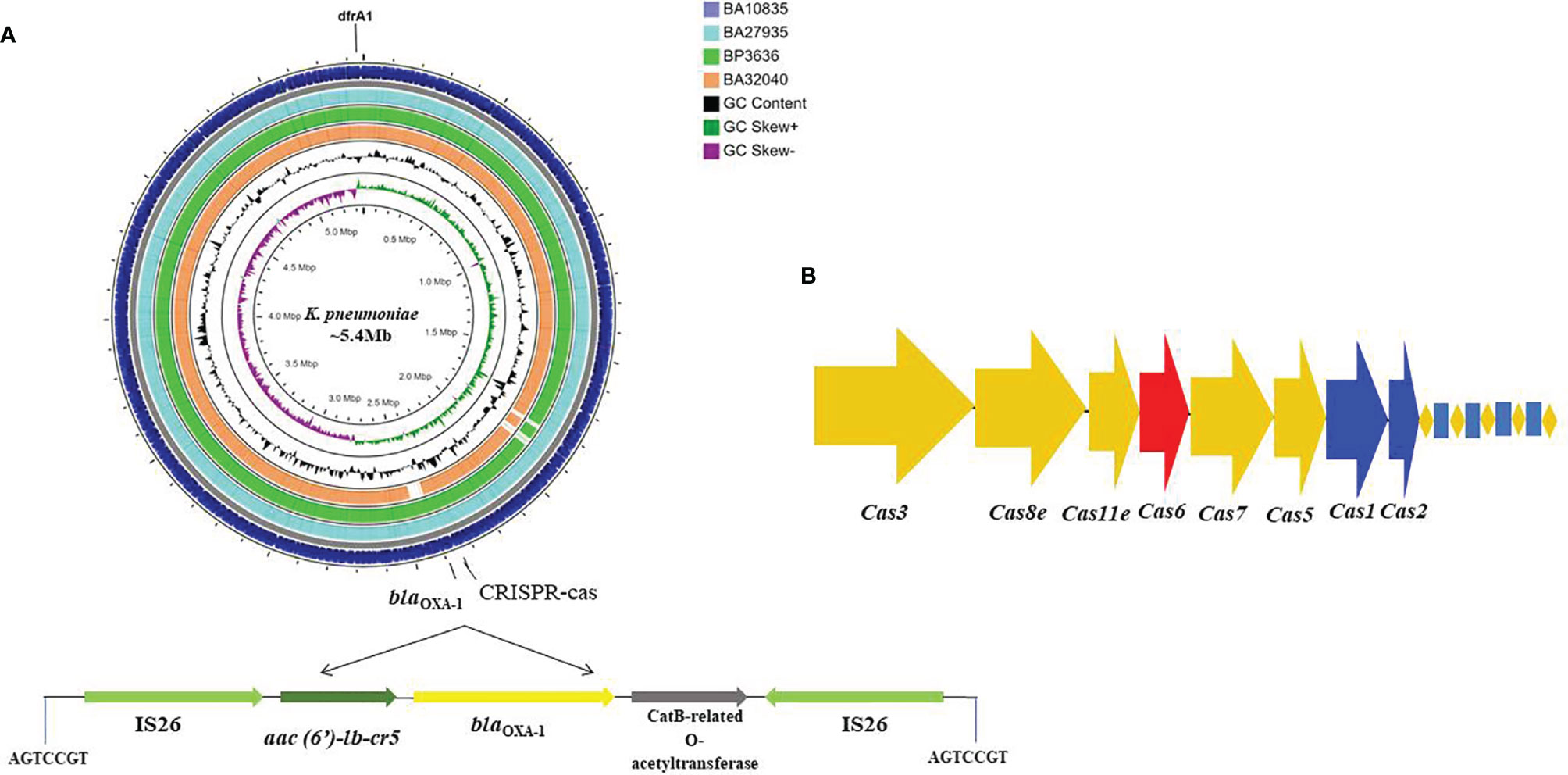
Figure 1 Circular genome map of four MDR hypervirulent K. pneumoniae ST2096 chromosomes. (A) Circles from the outside to the inside show the CDS region of BA10835 (blue), BA27935 (cyan), BP3636 (green), BA32040 (yellow), GC skew (dark green and purple), and GC content (black). Linear view of the IS26 mediated trans locatable units carrying aac(6′)-Ib-cr (fluoroquinolones and aminoglycosides), blaOXA–1 (ampicillin), and catB3 (chloramphenicol) inserted to the chromosome. A repeat region of 7 bases read as AGTCCGT was present on either ends where the insertion was observed. Map generated using CGView server (Grant and Stothard, 2008). (B) Type I-E CRISPR-Cas system identified in the chromosomes with repeat region of 28 bases.
The key virulence determinant carried by the chromosome of K. pneumoniae is the ybt locus, which is mobilized by ICEKp. ybt14 was carried on ICEKp5 and integrated into the chromosome in all nine sequenced isolates. The fyuA and irp1 yersiniabactin receptors were also present on the chromosome, along with a kfu gene cluster encoding for iron uptake and the mrk gene cluster, which facilities biofilm formation. YbST, typing based on yersiniabactin loci, classified all the isolates as belonging to YbST140.
The genomes of BP3636 (5,352,701 bp) and BA32040 (5,298,155 bp) were smaller than the genomes of BA10835 (5,355,460 bp) and BA27935 (5,356,693 bp) and lacked some of the iron transporters and metal transporter-encoding genes on the chromosomes. In addition, BA32040 lacked some of the genes coding for ABC transporter, MFS transporter, and LysE and LysR family transcriptional regulators when compared to the other three complete genomes (data not shown).
Characterization of the Plasmids Among HvKp ST2096
The nine HvKp isolates were found to possess an array of AMR genes associated with 4–5 plasmids per genome, including the virulence plasmid (Tables 2 and 3). Notably, blaNDM-5 was carried on the IncFII plasmid (~97 kbp) along with aadA2, rmtB, ermB, mphA, sul1, dfrA12, and blaTEM-1B (Figure 2). We also found a 293-bp segment of an IS30 family transposase, with similarity to the ISAba125, adjacent to blaNDM-5. The closest matching plasmid from the global database was from K. pneumoniae JUNP055 (GenBank accession no. LC506718), which also harbored blaNDM-5 but lacked a few IS elements (ISEc23 and IS6 family) when compared to IncFII of the ST2096 isolates (Figure 2). These pJUNP055 and IncFII plasmids from the present study shared ~80% sequence identity to those of E. coli M105 from Myanmar (GenBank accession no. AP018136), which lacked blaNDM-5. As predicted, the blaOXA-232 carbapenemase was encoded by a small 6Kb ColKP3 plasmid and was adjacent to a truncated ISEcp1 (207 bp). Notably, the two isolates (BA32040 and BA1602) that were susceptible to meropenem lacked a carbapenemase-encoding gene. Additionally, a large (~307-kbp) plasmid was present in all four of the assembled genomes and was found to be a fusion of IncFIB and IncHI1B plasmid backbones, carrying both AMR genes and virulence genes which will be referred to as p2096_hyb (Table 2). The isolates also harbored several small plasmids (<8 kbp), such as ColRNAI and Col(BS512), which did not encode either AMR or virulence genes.
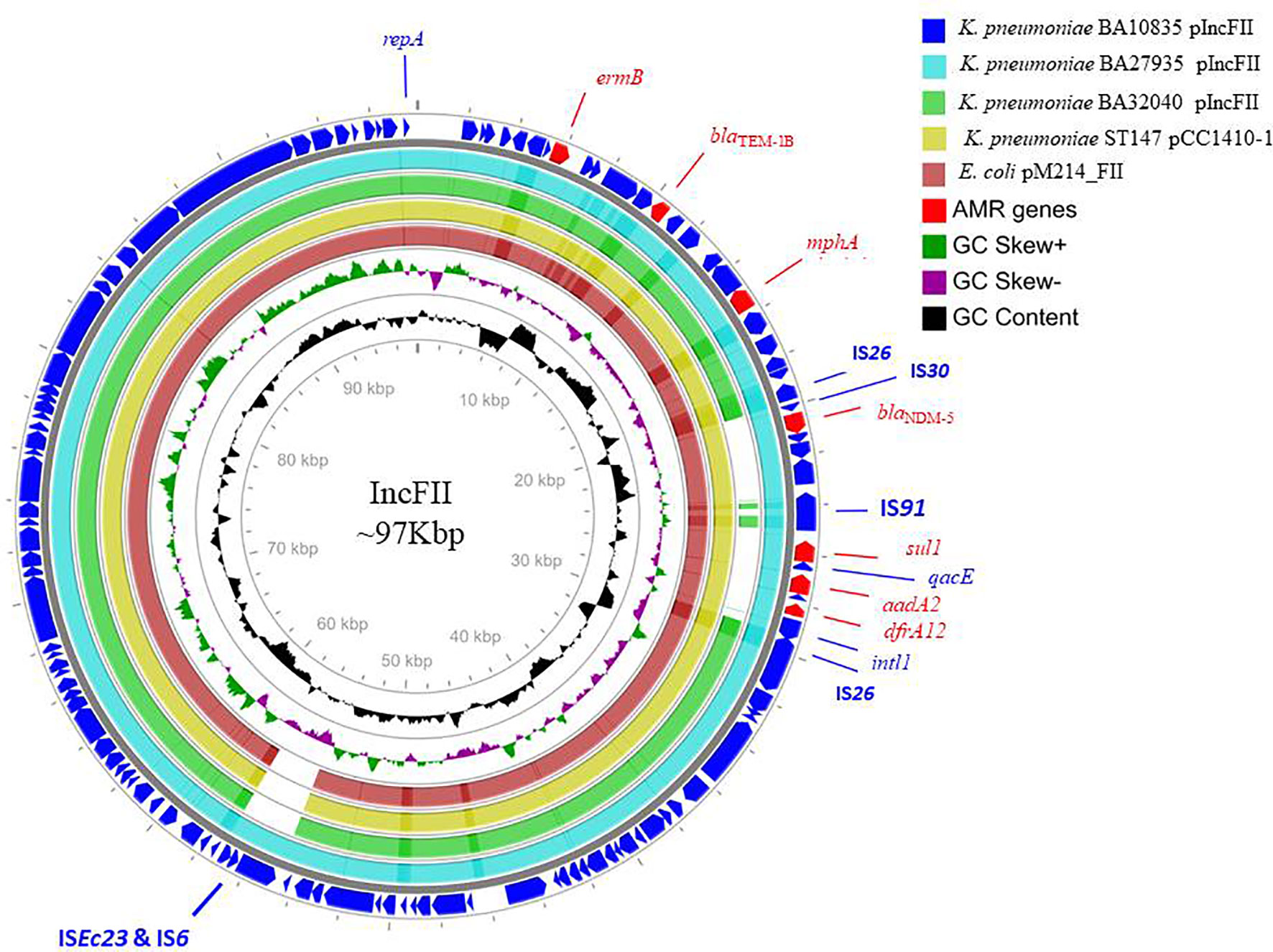
Figure 2 Alignment of IncFII plasmids of three MDR hypervirulent K. pneumoniae belonging to ST2096. Circles from the outside to the inside show the CDS region of BA10835 (blue), BA27935 (cyan), BA32040 (green), and nearest matching reference plasmids that belong to K. pneumoniae pCC1410-1 (yellow; KT725788) and E. coli pM214 (red; AP018144). GC skew (dark green and magenta) and GC content (black) of the plasmid are represented in the inner circles. Maps were generated using the CGView server.
Hybrid Plasmid Coding for Virulence and Antimicrobial Resistance
A large hybrid virulence plasmid, p2096_hyb, of ~307 kbp was the hallmark of all ST2096 isolates, and they carried a frameshifted rmpA2 and the aerobactin siderophore, encoded by iucABCD. The hybrid plasmid in the four isolates BA10835, BA27935, BP3636, and BA32040 will be referred to as p10835_hyb, p27935_hyb, p3636_hyb, and p32040_hyb, respectively. This plasmid carried both IncHI1B and IncFIB replicons on the pNDM-MAR backbone and hence was called a hybrid plasmid. This plasmid-encoded several AMR genes as listed in Table 2. The backbone of the plasmid consisted of genes related to replication, toxin–antitoxin system, conjugative transfer, DDE transposase, transcriptional regulators, and tyrosine-specific recombinases. Notably, dfrA12, aadA2, and sul1 genes were inserted into the virulence plasmid through a class 1 integron, intI1. The insertion of blaOXA-1, catB, and aac(6′)-lb-cr5 on the hybrid plasmid was through IS26, comparable to the arrangement observed in the chromosome (Figures 1A, B). A Tn3 transposon contained several AMR genes including msrE, mphE, sul1 and β-lactamases, such as blaOXA-1, blaCTX-M-15, and blaTEM-1. In addition to Tn3, blaCTX-M-15 and blaTEM-1 were associated with ISEc9, a resolvase, and IS91 insertion sequence. The hybrid plasmid in BA32040 was shorter (272 kbp) and lacked aadA2, armA, blaOXA-1, msrE, mphE, sul, dfrA14 and the CRISPR array in comparison to plasmids found in the other three genomes.
Besides the virulence genes, p2096_hyb also carried genes encoding heavy metal tolerance such as merARCTP (mercury) and terBEDWXZ (tellurium) that were possibly inserted through the Tn3 transposon, as shown in Figure 3A. Notably, a frameshift mutation was observed in rmpA2 among all the isolates, which we presumed to be associated with the negative string test results as has been previously described (Yu et al., 2015; Shankar et al., 2021b). The frameshift occurred due to the deletion of an adenine base at the 346th base in rmpA2. Non-functional rmpA2 and the absence of rmpA in these isolates contribute to the loss of a hypermucoid phenotype resulting from the decreased extracellular polysaccharide production. Aerobactin typing (AbST), a typing method using aerobactin alleles (iucA, iucB, iucC, iucD, and iutA), revealed all the study isolates that belonged to AbST-1. A type IV-A3 CRISPR-Cas system located on the hybrid plasmid of three isolates (BA10835, BA27935, BP3636) was characterized by the presence of 5–12 spacers and a 29-bp repeat region. One spacer each from the hybrid plasmid of the three isolates was comparable to traL of IncF plasmids that were found in K. pneumoniae, which may act as a potential obstacle in acquiring IncF plasmids and thereby limit the number of plasmids carried by these isolates.
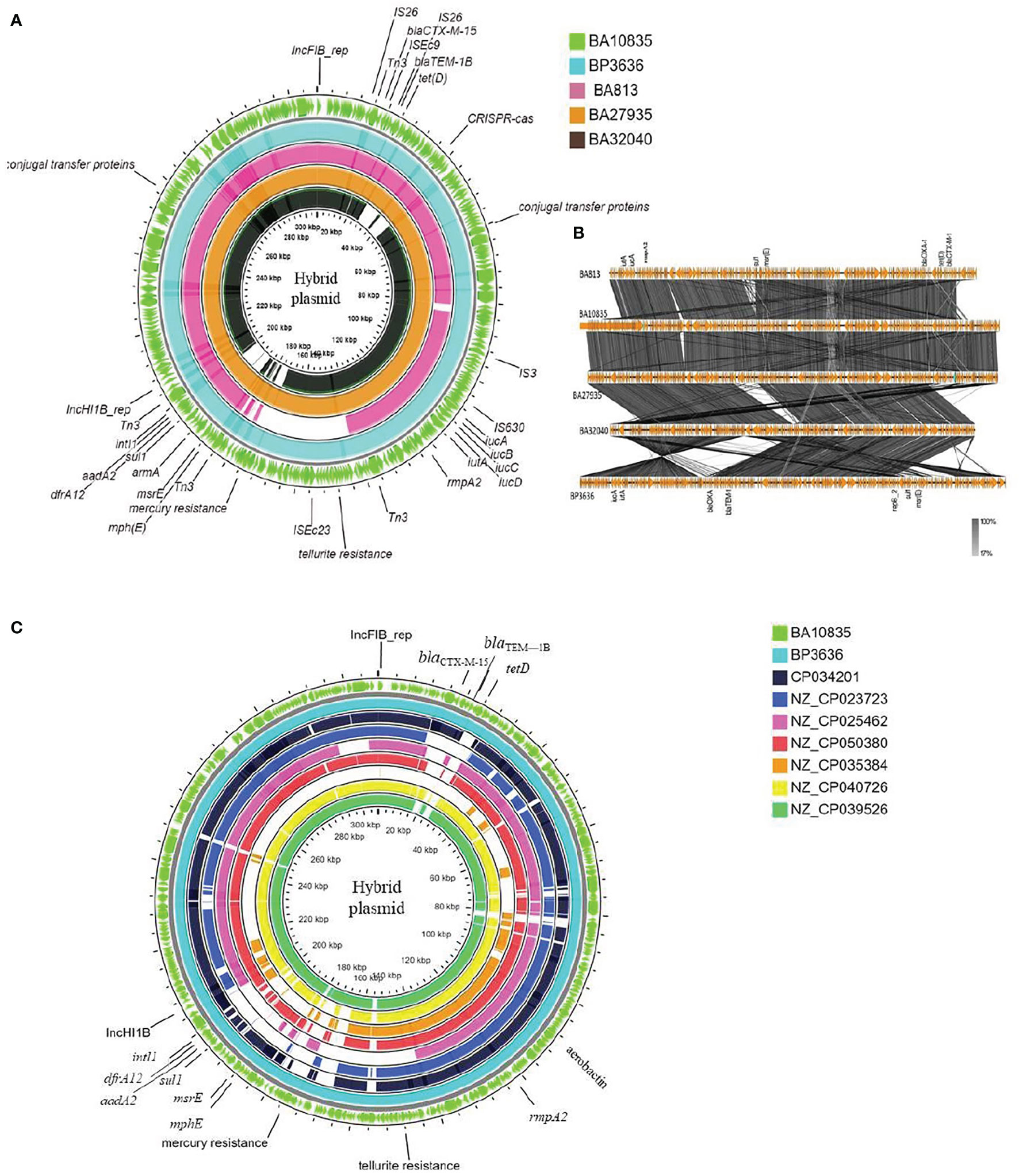
Figure 3 Maps of ST2096 mosaic plasmids in comparison to previously reported IncHI1B-IncFIB virulence plasmids. (A) Circular genome comparison map of IncFIB–IncHIB mosaic plasmids from outer to inner rings-BA10835 (light green), BP3636 (teal), BA813 (red), BA27935 (orange), and BA32040 (black). All the isolates belong to ST2096 and mosaic plasmid from BA813 (MK649825) was isolated during 2017 from the same study center. It lacks the heavy metal resistance encoding region when compared to other plasmids that were isolated during 2019. (B) Linear alignment of the mosaic plasmids obtained K. pneumoniae ST2096 using Easyfig. (C) Circular genome comparison map of IncHI1B–IncFIB mosaic plasmids from outer to inner rings-BA10835 (light green), BP3636 (teal), CP034201 (navy blue), NZ_CP023723 (indigo), NZ_CP025462 (pink), NZ_CP050380 (red), NZ_CP035384 (orange), NZ_CP040726 (yellow), and NZ_CP039526 (dark green). NZ_CP035384 shows the least similarity to the plasmids from the present study. Details of resistance and virulence genes carried by these plasmids are detailed in Supplementary Table 1.
Figures 3A, B show the BLAST comparison of the hybrid plasmids from the present study to another hybrid plasmid, MK649825, from ST2096 isolated from the same center earlier in 2017 (Wyres et al., 2020). The plasmid, MK649825, was much smaller (273 kbp) than the plasmids isolated during 2019 and lacked the genes encoding mercury and tellurite tolerance. p2096_hyb of ~307 kbp showed ≤50% sequence identity with the pLVPK (Chen et al., 2004) reference virulence plasmid having in common with the region coding for virulence genes (Figure S3). p2096_hyb showed about 60% similarity to the IncHI1B virulence plasmid of K. pneumoniae SGH10 (Lam et al., 2018) and ~70% sequence identity with the first identified IncHI1B pNDM-MAR plasmid, a 267-kbp plasmid (GenBank accession no. JN420336.1) from a K. pneumoniae ST15 (Villa et al., 2012) carrying blaNDM-1 (Figure S3; Figure 4). While the p2096_hyb among HvKp ST2096 retained the regions coding for mercury, tellurite, β-lactam (blaOXA-1 and blaCTX-M-15), chloramphenicol, and aminoglycoside resistance, it had lost the segment carrying blaNDM-1 when compared to the pNDM-MAR plasmid. The insertion of the virulence-encoding region into the pNDM-MAR plasmid is possibly through the insertion mediated by IS3 and IS66 family proteins (ISEc23) (Figure 3A). Figure 4 shows the circular comparison of two-hybrid plasmids of ST2096 to pLVPK (GenBank accession no. AY378100.1), IncHI1B (pNDM-MAR) [GenBank accession no. JN420336.1], and pittNDM (GenBank accession no NZ_CP006799.1) plasmids. The latter two plasmids lack the virulence genes but encode antimicrobial resistance genes with IncHI1B backbone.
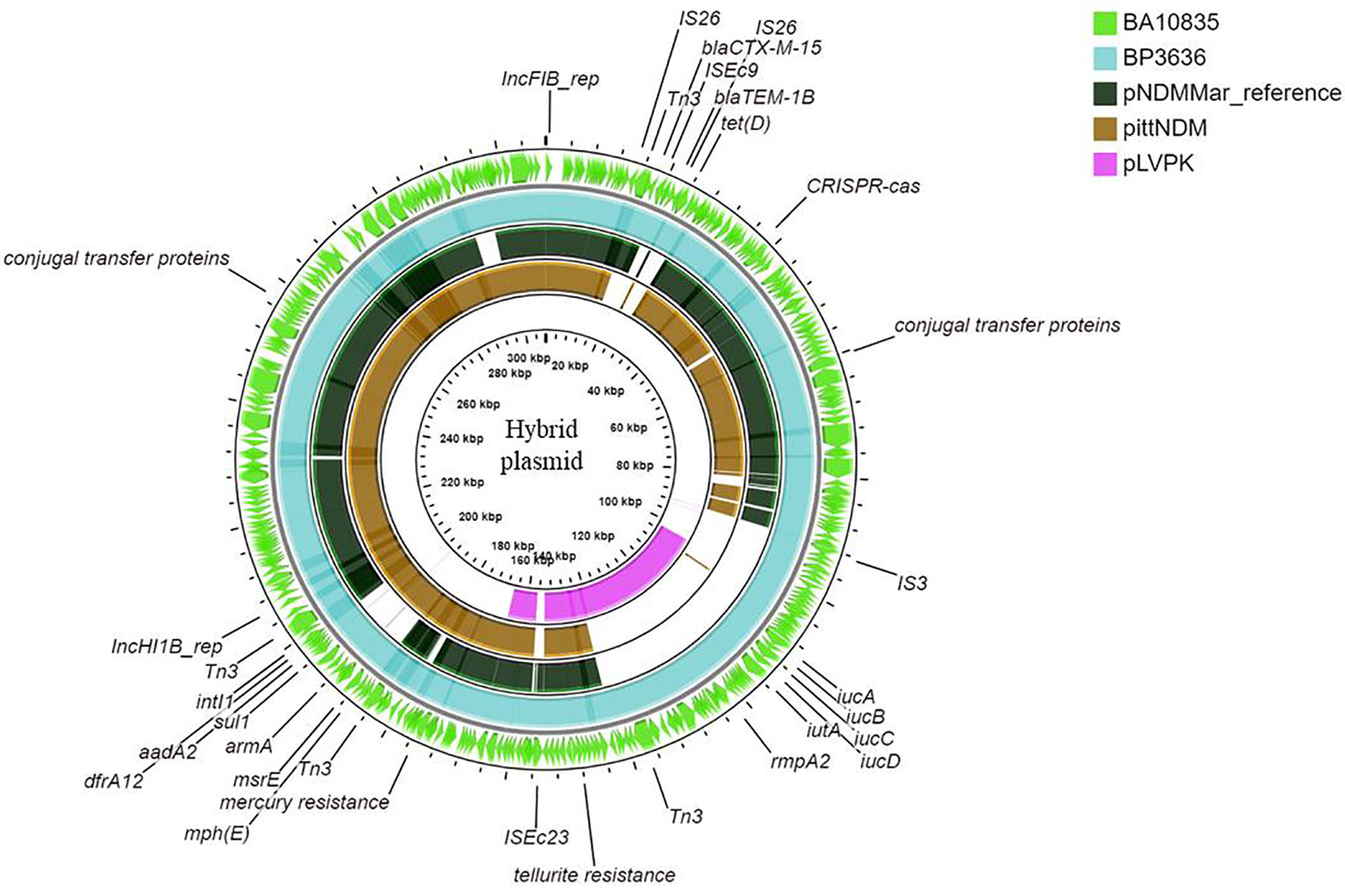
Figure 4 Maps of ST2096 mosaic plasmids in comparison to previously reported reference plasmids. Circular comparison map of plasmids from outer to inner rings—BA10836, BP3636, IncHI1B (pNDM-MAR, JN420336.1), PittNDM (NZ_CP006799.1), and pLVPK (AY378100.1) plasmids.
Figure 3C shows the BLAST comparison between the hybrid plasmid from the present study and the previously reported K. pneumoniae hybrid plasmids comprising IncHI1B (pNDM-MAR)–IncFIB (pNDM-MAR) replicon types. The complete sequences of these plasmids have been reported from 2015 till date among diverse clones of K. pneumoniae such as ST11, ST383, ST15, ST147, and ST23 and vary in size ranging from 261 to 396 kbp. The particulars of antimicrobial resistance and virulence genes carried by these plasmids are mentioned in Table S1. Interestingly, apart from the hybrid plasmid in the present study, another plasmid obtained in China, p44-1, from ST11 carried frameshifted rmpA2. Hybrid plasmids of HvKp ST2096, p10835_hyb (307 kbp), and p3636_hyb (317 kbp) showed high similarity to pKpvST383L (372 kbp) and pHB25-1-vir (396 kbp) which were isolated in England and China, respectively.
An SNP-based phylogenetic tree of the complete hybrid plasmids is shown in Figure S4. The tree was rooted at pAP855 since it was distant from the others and belonged to an ST23 isolate, lacking rmpA2 (Tian et al., 2021). All the other plasmids belonged to MDR clones such as ST11, ST15, ST147, ST383, and ST2096. The plasmids from the present study formed a separate cluster coding for mercury and tellurite tolerance, truncated rmpA2, and lacking carbapenemase. This group is closely related to two plasmids from China isolated during 2015 and 2016, one of which also carries a truncated rmpA2 (Table S1). The plasmids from England and Prague formed a separate group, and these carried rmpA and functional rmpA2 and also encoded tellurite resistance determinants (Table S1).
Discussion
MDR-Kp or CR-Kp isolates are clinically challenging, and specific carbapenemases are associated with regional/endemic clones in various regions (Lee et al., 2016; Shankar et al., 2021a). Recent reports describing the independent emergence of convergent HvKp isolates with hybrid and mosaic plasmids in multiple geographical locations make these organisms a major concern (Lam et al., 2019; Turton et al., 2019). Studies have described the acquisition of MDR plasmids by HvKp clones (ST23) and the acquisition of the pLVPK-like virulence plasmid by classical K. pneumoniae (cKp) causing invasive infections (Cejas et al., 2014; Chen and Kreiswirth, 2017; Xu et al., 2019; Shankar et al., 2020). This bidirectional convergence has resulted in the emergence of MDR-HvKp/CR-HvKp isolates within the nosocomial clones. Consequently, the circulation of nosocomial clones carrying a virulence plasmid is a matter of major public health concern (Zhan et al., 2017; Gu et al., 2018; Zhao et al., 2019; Liu et al., 2020). We report the acquisition of virulence factors among MDR clones, ST2096, crafting CR-HvKp in India.
In the present study, the ~307-kb hybrid plasmid was comparable to previously reported fusion plasmids including pKpvST147L [GenBank accession no. CM007852], pKpvST383L [GenBank accession no. CP034201], pKpvST147B [GenBank accession no. CP040726], and pBA813_1 GenBank accession no. [MK649825] (Turton et al., 2019; Wyres et al., 2020). Remarkably, these reference plasmids are associated with a diverse collection of clones, which were found to harbor blaNDM and blaOXA-48 carbapenemase genes. Moreover, the insertion of resistance cassettes carrying aadA2, armA, blaTEM-1B, blaCTX-M-15, mphE, msrE, sul1, and dfrA12 into hybrid plasmids of independent origin may accelerate the spread of MDR-HvKp. To date, the reports of hybrid plasmids are from clinical isolates indicating that the antimicrobial pressure present in this niche not only selects such plasmids but also aids in their persistence and dissemination. Here, the hybrid plasmids were a merger of IncFIB/IncHI1B (pNDM-MAR) backbones leading to convergence of AMR and virulence on a single plasmid. In India, blaNDM is endemic and widespread among several bacteria especially E. coli and K. pneumoniae and this gene is often carried on the pNDM-MAR plasmid. Under pressure and for persistence, CR-HvKp can integrate segments of virulence plasmid into the endemic plasmid types to obtain the two-fold advantage of coding antimicrobial resistance and virulence. Similar plasmids, which were the result of the fusion between IncFIB/IncHI1B and IncFIIK/IncFIBK backbones, have been reported from other regions (Lam et al., 2019; Turton et al., 2019), highlighting the susceptibility of IncF and IncH plasmids to undergo recombination to form co-integrates in K. pneumoniae. Among these HvKp with mosaic plasmids, the mosaic structures were likely formed by the integration of the virulence region from the virulence plasmid into IncFIB and IncFIIK resistance plasmids (Lam et al., 2019; Tang et al., 2020).
Furthermore, we found that the MDR-HvKp clones carrying a hybrid virulence plasmid possessed a frameshift variant of rmpA2 without rmpA. Mutations in rmpA and rmpA2 led to a lack of hypermucoviscous phenotypes (Yu et al., 2015; Shankar et al., 2021b). The effect of inactivation of rmpA2 and its stability on the hybrid plasmids carried by nosocomial clones of K. pneumoniae needs further investigation. In contrast, the hybrid plasmid in ST15 reported by Lam and colleagues carried only rmpA (Lam et al., 2019). The virulence plasmids described here also carried genes encoding heavy metal tolerance, such as tellurium and mercury. Similar hybrid plasmids, reported by Turton and colleagues, lacked the genes encoding for mercury tolerance (Turton et al., 2019). Given the community origin of HvKp, the co-occurrence of heavy-metal tolerance may provide an additional survival mechanism in harsh ecological niches found in the community and hospitals (Furlan et al., 2020).
The presence of CRISPR-Cas systems in MDR plasmids in K. pneumoniae has not been extensively studied. Recent reports of the type IV CRISPR-Cas system in K. pneumoniae mega plasmids/co-integrate plasmids suggest that these systems aid competition between plasmids (Kamruzzaman and Iredell, 2020; Newire et al., 2020). In the present study, the CRISPR-Cas system in the large co-integrate plasmid has acquired a spacer sequence identical to IncF-traL, which implies specific targeting for further gene invasion of plasmids (Kamruzzaman and Iredell, 2020). This might probably play a role in homologous recombination and integration of AMR and virulence determinants onto a single plasmid by preventing the entry of other plasmids such as IncF. The attainment of specific plasmid CRISPR spacers targeting different conjugative plasmids appears to be advantageous in K. pneumoniae to mitigate the fitness cost associated with carrying multiple AMR plasmids. Notably, the majority of the plasmids that carry plasmid-targeting spacers are co-integrate plasmids carrying IncFIB and IncHI1B replicons (Kamruzzaman and Iredell, 2020; Newire et al., 2020). This observation suggests that the plasmid-mediated CRISPR spacers not only target other plasmids but also may aid the formation of co-integrate/mega plasmids for improved stability and compatibility.
Globally, the prevalence of MDR-HvKp or CR-HvKp appears to be increasing (Siu et al., 2014; Gu et al., 2018; Liu et al., 2020). Given the large burden of MDR-HvKp and CR-HvKp infections in China, India, and Southeast Asia, these regions represent the most likely hotspot of MDR-virulence intersection and subsequent spread. Similarly, the spontaneous emergence of hybrid plasmids in these regions and their potential for clonal spread in healthcare settings represent a major focus of nosocomial outbreaks and their containment. If the incidence of the convergent clones with fusion plasmid continues, these pathotypes may replace the currently circulating CRKp clones (Liu et al., 2020). Continuous genome surveillance of MDR-HvKp would help in determining this niche shift from community to hospital. In China, ST11 was known to carry complete pLVPK with capsule-type K64 (clade1) while ST11 with K47 carried a shorter virulence plasmid with only rmpA2 and aerobactin (clade3) (Dong et al., 2018; Liu et al., 2020; Zhang et al., 2020). These MDR-HvKp infections were predominantly associated with nosocomial infections, and the sub-lineages among ST11 with different capsule types were identified only through genomic studies. Although MDR-HvKp belonged to ST11 in the study described by Dong and colleagues, there are several differences concerning the virulence profile of the two clades which comprised MDR-HvKp which can only be identified through genome surveillance (Dong et al., 2018). There can also be a selection of one of the clades of MDR-HvKp ST11 over time which has a better fitness, thereby eliminating the other two clades.
Conclusion
India has exceptionally high rates of AMR infections as stated by the annual report of the Antimicrobial Resistance Research and Surveillance Network during the year 2020 by the Indian Council of Medical Research (https://main.icmr.nic.in/sites/default/files/guidelines/AMRSN_annual_report_2020.pdf; accessed on November 15, 2021). The further generation of HvKp carrying carbapenemases on a virulence plasmid would be a potential catastrophe. The acquisition of AMR genes on the chromosome creates the further possibility of increased baseline resistance among the K. pneumoniae isolates. It is apparent that MDR-HvKp is no longer confined to selected clones and the containment of such isolates with the mosaic plasmid is very challenging. The presence of AMR and virulence among diverse Klebsiella clones present a global threat to the rapid spread of these emerging superbugs.
Data Availability Statement
The datasets presented in this study can be found in GenBank under Bioproject ID PRJNA613369. The accession numbers of the genomes are mentioned in Tables 2 and 3.
Ethics Statement
The study was approved by the Institutional Review Board of Christian Medical College, Vellore, India, with minute number 9616 (01/09/2015).
Author Contributions
CS: conceptualization, analysis, manuscript writing, and revising. KV: methodology, bioinformatics, manuscript writing. JJ: analysis, manuscript writing and revising. SB: manuscript correction and supervision. BI: resource. AN: methodology, data curation. DS: methodology. BG: resource. BV: conceptualization, manuscript revision. and supervision. All authors contributed to the article and approved the submitted version.
Funding
The study has been funded by the Indian Council of Medical Research, New Delhi, India (ref. no: AMR/Adoc/232/2020-ECD-II).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.875116/full#supplementary-material
Abbreviations
ST, sequence type; MLST, multi-locus sequence typing; MDR, multidrug-resistant; AMR, antimicrobial resistance; HvKp, hypervirulent Klebsiella pneumoniae; CRKP, carbapenem-resistant Klebsiella pneumoniae; CR-HvKp, carbapenem-resistant hypervirulent Klebsiella pneumoniae; CG, clonal group; ARG, antimicrobial resistance genes; pLVPK, large virulence plasmid of Klebsiella pneumoniae; CRISPR, clustered regularly interspaced short palindromic repeats; XDR, extensively drug-resistant; SNP, single-nucleotide polymorphism; AST, antimicrobial susceptibility testing; OD, optical density.
References
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 19 (5), pp.455–pp.477. doi: 10.1089/cmb.2012.0021
Bialek-Davenet, S., Criscuolo, A., Ailloud, F., Passet, V., Jones, L., Delannoy-Vieillard, A. S., et al. (2014). Genomic Definition of Hypervirulent and Multidrug-Resistant Klebsiella pneumoniae Clonal Groups. Emerg Infect. Dis. 20 (11), 1812. doi: 10.3201/eid2011.140206
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for Predictions of Phenotypes From Genotypes. J. Antimicrob. Chemother. 75 (12), 3491–3500. doi: 10.1093/jac/dkaa345
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother 58 (7), 3895–3903. doi: 10.1128/AAC.02412-14
Cejas, D., Fernández Canigia, L., Rincón Cruz, G., Elena, A. X., Maldonado, I., Gutkind, G. O., et al. (2014). First Isolate of KPC-2-producing Klebsiella pneumoniae Sequence Type 23 From the Americas. J. Clin. Microbiol. 52 (9), 3483–3485. doi: 10.1128/JCM.00726-14
Chen, Y. T., Chang, H. Y., Lai, Y. C., Pan, C. C., Tsai, S. F., Peng, H. L. (2004). Sequencing and Analysis of the Large Virulence Plasmid pLVPK of Klebsiella pneumoniae CG43Gene 337, 189–198.
Chen, L., Kreiswirth, B.N. (2017) Convergence of Carbapenem-Resistance and Hypervirulence in Klebsiella pneumoniae. Lancet Infect. Dis., 18 (1), 2–3. doi: 10.1016/S1473-3099(17)30517-0
Clinical and Laboratory Standards Institute. (2019). Performance Standards for Antimicrobial Susceptibility Testing; 29th Informational Supplement. M100-S29 (Wayne, PA: Clinical and Laboratory Standards Institute).
Compain, F., Babosan, A., Brisse, S., Genel, N., Audo, J., Ailloud, F., et al. (2014). Multiplex PCR for Detection of Seven Virulence Factors and K1/K2 Capsular Serotypes of Klebsiella pneumoniae. J. Clin. Microbiol. 52 (12), 4377–4380. doi: 10.1128/JCM.02316-14
Dong, N., Zhang, R., Liu, L., Li, R., Lin, D., Chan, E. W. C., et al. (2018). Genome Analysis of Clinical Multilocus Sequence Type 11 Klebsiella pneumoniae From China. Microbial. Genomics 4 (2), e000149. doi: 10.1099/mgen.0.000149
Furlan, J. P. R., Savazzi, E. A., Stehling, E. G. (2020). Genomic Insights Into Multidrug-Resistant and Hypervirulent Klebsiella pneumoniae Co-Harboring Metal Resistance Genes in Aquatic Environments. Ecotoxicol. Environ. Saf. 201, 110782. doi: 10.1016/j.ecoenv.2020.110782
Grant, J. R., Stothard, P. (2008). The CGView Server: A Comparative Genomics Tool for Circular Genomes. Nucleic Acids Res. 36 (suppl_2), W181–W184. doi: 10.1093/nar/gkn179
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2018). A Fatal Outbreak of ST11 Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae in a Chinese Hospital: A Molecular Epidemiological Study. Lancet Infect. Dis. 18 (1), 37–46. doi: 10.1016/S1473-3099(17)30489-9
Gurevich, A., Saveliev, V., Vyahhi, N., Tesler, G. (2013). QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 29 (8), 1072–1075. doi: 10.1093/bioinformatics/btt086
Huynh, B. T., Passet, V., Rakotondrasoa, A., Diallo, T., Kerleguer, A., Hennart, M., et al. (2020). Klebsiella pneumoniae Carriage in Low-Income Countries: Antimicrobial Resistance, Genomic Diversity and Risk Factors. Gut Microbes 11 (5), 1287–1299. doi: 10.1080/19490976.2020.1748257
Jolley, K. A., Maiden, M. C. (2010). BIGSdb: Scalable Analysis of Bacterial Genome Variation at the Population Level. BMC Bioinf. 11 (1), 1–11. doi: 10.1186/1471-2105-11-595
Kamruzzaman, M., Iredell, J. R. (2020). CRISPR-Cas System in Antibiotic Resistance Plasmids in Klebsiella pneumoniae. Front. Microbiol. 10, 2934. doi: 10.3389/fmicb.2019.02934
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., Phillippy, A. M. (2017). Canu: Scalable and Accurate Long-Read Assembly Via Adaptive K-Mer Weighting and Repeat Separation. Genome Res. 27 (5), 722–736. doi: 10.1101/gr.215087.116
Lam, M., Wick, R. R., Watts, S. C., Cerdeira, L. T., Wyres, K. L., Holt, K. E. (2021). A Genomic Surveillance Framework and Genotyping Tool for Klebsiella pneumoniae and Its Related Species Complex. Nat. Commun. 12 (1), 1–16. doi: 10.1038/s41467-021-24448-3
Lam, M. M., Wyres, K. L., Wick, R. R., Judd, L. M., Fostervold, A., Holt, K. E., et al. (2019). Convergence of Virulence and MDR in a Single Plasmid Vector in MDR Klebsiella pneumoniae ST15. J. Antimicrob. Chemother. 74 (5), 1218–1222. doi: 10.1093/jac/dkz028
Lee, I., Kim, Y. O., Park, S. C., Chun, J. (2016). OrthoANI: An Improved Algorithm and Software for Calculating Average Nucleotide Identity. Int. J. Syst. Evol. Microbiol. 66 (2), 1100–1103. doi: 10.1099/ijsem.0.000760
Lee, C. R., Lee, J. H., Park, K. S., Jeon, J. H., Kim, Y. B., Cha, C. J., et al. (2017). Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae: Epidemiology, Hypervirulence-Associated Determinants, and Resistance Mechanisms. Front. Cell. Infect Microbiol. 7, 483. doi: 10.3389/fcimb.2017.00483
Lee, C. R., Lee, J. H., Park, K. S., Kim, Y. B., Jeong, B. C., Lee, S. H. (2016). Global Dissemination of Carbapenemase-Producing Klebsiella pneumoniae: Epidemiology, Genetic Context, Treatment Options, and Detection Methods. Front. Microbiol. 7, 895. doi: 10.3389/fmicb.2016.00895
Li, R., Cheng, J., Dong, H., Li, L., Liu, W., Zhang, C., et al. (2020). Emergence of a Novel Conjugative Hybrid Virulence Multidrug-Resistant Plasmid in Extensively Drug-Resistant Klebsiella pneumoniae ST15. Int. J. Antimicrob. Agents 55 (6), 105952. doi: 10.1016/j.ijantimicag.2020.105952
Liu, C., Du, P., Xiao, N., Ji, F., Russo, T. A., Guo, J. (2020). Hypervirulent Klebsiella pneumoniae Is Emerging as an Increasingly Prevalent K.Pneumoniae Pathotype Responsible for Nosocomial and Healthcare-Associated Infections in Beijing, China. Virulence 11 (1), 1215–1224. doi: 10.1080/21505594.2020.1809322
Marr, C. M., Russo, T. A. (2019). Hypervirulent Klebsiella pneumoniae: A New Public Health Threat. Expert Rev. Anti-Infect. Ther. 17 (2), 71–73. doi: 10.1080/14787210.2019.1555470
Martin, M. (2011). Cutadapt Removes Adapter Sequences From High-Throughput Sequencing Reads. EMBnet J. 17 (1), 10–12. doi: 10.14806/ej.17.1.200
Mike, L. A., Stark, A. J., Forsyth, V. S., Vornhagen, J., Smith, S. N., Bachman, M. A., et al. (2021). A Systematic Analysis of Hypermucoviscosity and Capsule Reveals Distinct and Overlapping Genes That Impact Klebsiella pneumoniae Fitness. PloS Pathog. 17 (3), e1009376. doi: 10.1371/journal.ppat.1009376
Navon-Venezia, S., Kondratyeva, K., Carattoli, A. (2017). Klebsiella pneumoniae: A Major Worldwide Source and Shuttle for Antibiotic Resistance. FEMS Microbiol. Rev. 41 (3), 252–275. doi: 10.1093/femsre/fux013
Newire, E., Aydin, A., Juma, S., Enne, V. I., Roberts, A. P. (2020). Identification of a Type Iv-a CRISPR-Cas System Located Exclusively on IncHI1B/IncFIB Plasmids in Enterobacteriaceae. Front. Microbiol. 11, 1937. doi: 10.3389/fmicb.2020.01937
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., Tyson, G. W. (2015). CheckM: Assessing the Quality of Microbial Genomes Recovered From Isolates, Single Cells, and Metagenomes. Genome Res. 25 (7), 1043–1055. doi: 10.1101/gr.186072.114
Qi, Y., Wei, Z., Ji, S., Du, X., Shen, P., Yu, Y. (2011). ST11, the Dominant Clone of KPC-producing Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 66 (2), 307–312. doi: 10.1093/jac/dkq431
Russel, J., Pinilla-Redondo, R., Mayo-Muñoz, D., Shah, S. A., Sørensen, S. J. (2020). CRISPRCasTyper: Automated Identification, Annotation, and Classification of CRISPR-Cas Loci. CRISPR J. 3 (6), 462–469. doi: 10.1089/crispr.2020.0059
Shankar, C., Basu, S., Lal, B., Shanmugam, S., Vasudevan, K., Mathur, P., et al. (2021b). Aerobactin Seems to Be a Promising Marker Compared With Unstable RmpA2 for the Identification of Hypervirulent Carbapenem-Resistant Klebsiella Pneumoniae: In Silico and In Vitro Evidence. Front. Cell. Infect. Microbiol. 11, 709681. doi: 10.3389/fcimb.2021.709681
Shankar, C., Jacob, J. J., Sugumar, S. G., Natarajan, L., Rodrigues, C., Mathur, P., et al. (2021a). Distinctive Mobile Genetic Elements Observed in the Clonal Expansion of Carbapenem-Resistant Klebsiella pneumoniae in India. Microbial. Drug Resist. 27 (8), 1096–1104. doi: 10.1089/mdr.2020.0316
Shankar, C., Jacob, J. J., Vasudevan, K., Biswas, R., Manesh, A., Sethuvel, D. P. M., et al. (2020). Emergence of Multidrug Resistant Hypervirulent ST23 Klebsiella pneumoniae: Multidrug Resistant Plasmid Acquisition Drives Evolution. Front. Cell. Infect. Microbiol. 10, 575289. doi: 10.3389/fcimb.2020.575289
Shon, A. S., Bajwa, R. P., Russo, T. A. (2013). Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae: A New and Dangerous Breed. Virulence 4 (2), 107–118. doi: 10.4161/viru.22718
Siu, L. K. K., Huang, D. B., Chiang, T. (2014). Plasmid Transferability of KPC Into a Virulent K2 Serotype Klebsiella pneumoniae. BMC Infect. Dis. 14 (1), 1–6. doi: 10.1186/1471-2334-14-176
Sullivan, M. J., Petty, N. K., Beatson, S. A. (2011). Easyfig: A Genome Comparison Visualizer. Bioinformatics 27 (7), 1009–1010. doi: 10.1093/bioinformatics/btr039
Tang, M., Kong, X., Hao, J., Liu, J. (2020). Epidemiological Characteristics and Formation Mechanisms of Multidrug-Resistant Hypervirulent Klebsiella pneumoniae. Front. Microbiol. 11, 2774. doi: 10.3389/fmicb.2020.581543
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res. 44 (14), 6614–6624. doi: 10.1093/nar/gkw569
Tian, D., Wang, M., Zhou, Y., Hu, D., Ou, H. Y., Jiang, X. (2021). Genetic Diversity and Evolution of the Virulence Plasmids Encoding Aerobactin and Salmochelin in Klebsiella pneumoniae. Virulence 12 (1), 1323–1333. doi: 10.1080/21505594.2021.1924019
Turton, J., Davies, F., Turton, J., Perry, C., Payne, Z., Pike, R. (2019). Hybrid Resistance and Virulence Plasmids in “High-Risk”Clones of Klebsiella pneumoniae, Including Those Carrying Blandm-5. Microorganisms 7 (9), 326. doi: 10.3390/microorganisms7090326
Turton, J. F., Payne, Z., Coward, A., Hopkins, K. L., Turton, J. A., Doumith, M., et al. (2018). Virulence Genes in Isolates of Klebsiella pneumoniae From the UK During 2016, Including Among Carbapenemase Gene-Positive Hypervirulent K1-ST23 and ‘Non-Hypervirulent’types ST147, ST15 and ST383. J. Med. Microbiol. 67 (1), 118–128. doi: 10.1099/jmm.0.000653
Turton, J. F., Perry, C., Elgohari, S., Hampton, C. V. (2010). PCR Characterization and Typing of Klebsiella pneumoniae Using Capsular Type-Specific, Variable Number Tandem Repeat and Virulence Gene Targets. J. Med. Microbiol. 59 (5), 541–547. doi: 10.1099/jmm.0.015198-0
Vasudevan, K., Ragupathi, N. K. D., Jacob, J. J., Veeraraghavan, B. (2020). Highly Accurate-Single Chromosomal Complete Genomes Using IonTorrent and MinION Sequencing of Clinical Pathogens. Genomics 112 (1), 545–551. doi: 10.1016/j.ygeno.2019.04.006
Versalovic, J., Carroll, K. C., Funke, G., Jorgensen, J. H., Landry, M. L., Warnock, D. W. (2011). Manual of Clinical Microbiology (Washington, DC: ASM Press).
Villa, L., Poirel, L., Nordmann, P., Carta, C., Carattoli, A. (2012). Complete Sequencing of an IncH Plasmid Carrying the blaNDM-1, blaCTX-M-15 and qnrB1 Genes. J. Antimicrob. Chemother. 67 (7), 1645–1650. doi: 10.1093/jac/dks114
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PloS One 9 (11), e112963. doi: 10.1371/journal.pone.0112963
Wick, R. R., Heinz, E., Holt, K. E., Wyres, K. L. (2018). Kaptive Web: User-Friendly Capsule and Lipopolysaccharide Serotype Prediction for Klebsiella Genomes. J. Clin. Microbiol. 56 (6), e00197–e00118. doi: 10.1128/JCM.00197-18
Wick, R. R., Judd, L. M., Gorrie, C. L., Holt, K. E. (2017). Unicycler: Resolving Bacterial Genome Assemblies From Short and Long Sequencing Reads. PloS Comput. Biol. 13 (6), e1005595. doi: 10.1371/journal.pcbi.1005595
Wyres, K. L., Nguyen, T. N., Lam, M., Judd, L. M., van Vinh Chau, N., Dance, D. A., et al. (2020). Genomic Surveillance for Hypervirulence and Multidrug Resistance in Invasive Klebsiella pneumoniae From South and Southeast Asia. Genome Med. 12 (1), 1–16. doi: 10.1186/s13073-019-0706-y
Wyres, K. L., Wick, R. R., Gorrie, C., Jenney, A., Follador, R., Thomson, N. R., et al. (2016). Identification of Klebsiella Capsule Synthesis Loci From Whole Genome Data. Microbial. Genomics 2 (12), e000102. doi: 10.1099/mgen.0.000102
Wysocka, M., Zamudio, R., Oggioni, M. R., Gołębiewska, J., Dudziak, A., Krawczyk, B. (2020). The New Klebsiella pneumoniae ST152 Variants With Hypermucoviscous Phenotype Isolated From Renal Transplant Recipients With Asymptomatic Bacteriuria—Genetic Characteristics by WGS. Genes 11 (10), 1189. doi: 10.3390/genes11101189
Xie, M., Chen, K., Ye, L., Yang, X., Xu, Q., Yang, C., et al. (2020). Conjugation of Virulence Plasmid in Clinical Klebsiella pneumoniae Strains Through Formation of a Fusion Plasmid. Adv. Biosyst. 4 (4), 1900239. doi: 10.1002/adbi.201900239
Xu, M., Fu, Y., Fang, Y., Xu, H., Kong, H., Liu, Y., et al. (2019). High Prevalence of KPC-2-producing Hypervirulent Klebsiella pneumoniae Causing Meningitis in Eastern China. Infect. Drug Resist. 12, 641. doi: 10.2147/IDR.S191892
Yang, Q, Jia, X., Zhou, M., Zhang, H., Yang, W., Kudinha, T., Xu, Y.. (2020). Emergence of ST11-K47 and ST11-K64 Hypervirulent Carbapenem-Resistant Klebsiella pneumoniae in Bacterial Liver Abscesses from China: A Molecular, Biological and Epidemiological Study. Emerg. Microbes Infect. 9 (1), 320–331. doi: 10.1080/22221751.2020.1721334
Yang, X., Dong, N., Chan, E. W. C., Zhang, R., Chen, S. (2021). Carbapenem Resistance-Encoding and Virulence-Encoding Conjugative Plasmids in Klebsiella pneumoniae. Trends Microbiol. 29 (1), 65–83. doi: 10.1016/j.tim.2020.04.012
Yao, H., Qin, S., Chen, S., Shen, J., Du, X. D. (2018). Emergence of Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae. Lancet Infect. Dis. 18 (1), 25. doi: 10.1016/S1473-3099(17)30628-X
Yu, W. L., Lee, M. F., Tang, H. J., Chang, M. C., Chuang, Y. C. (2015). Low Prevalence of Rmpa and High Tendency of Rmpa Mutation Correspond to Low Virulence of Extended Spectrum β-Lactamase-Producing Klebsiella pneumoniae Isolates. Virulence 6 (2), 162–172. doi: 10.1080/21505594.2015.1016703
Zhang, Y., Jin, L., Ouyang, P., Wang, Q., Wang, R., Wang, J., et al. (2020). Evolution of Hypervirulence in Carbapenem-Resistant Klebsiella pneumoniae in China: A Multicentre, Molecular Epidemiological Analysis. J. Antimicrob. Chemother. 75 (2), 327–336. doi: 10.1093/jac/dkz446
Zhan, L., Wang, S., Guo, Y., Jin, Y., Duan, J., Hao, Z., et al. (2017). Outbreak by Hypermucoviscous Klebsiella pneumoniae ST11 Isolates With Carbapenem Resistance in a Tertiary Hospital in China. Front. Cell. Infect. Microbiol. 7, 182. doi: 10.3389/fcimb.2017.00182
Keywords: hypervirulent, K. pneumoniae, ST2096, hybrid plasmid, CRISPR-Cas, multidrug resistance
Citation: Shankar C, Vasudevan K, Jacob JJ, Baker S, Isaac BJ, Neeravi AR, Sethuvel DPM, George B and Veeraraghavan B (2022) Hybrid Plasmids Encoding Antimicrobial Resistance and Virulence Traits Among Hypervirulent Klebsiella pneumoniae ST2096 in India. Front. Cell. Infect. Microbiol. 12:875116. doi: 10.3389/fcimb.2022.875116
Received: 13 February 2022; Accepted: 21 March 2022;
Published: 27 April 2022.
Edited by:
Milena Dropa, University of São Paulo, BrazilReviewed by:
João Pedro Rueda Furlan, University of São Paulo, BrazilCostas C. Papagiannitsis, University of Thessaly, Greece
Copyright © 2022 Shankar, Vasudevan, Jacob, Baker, Isaac, Neeravi, Sethuvel, George and Veeraraghavan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Balaji Veeraraghavan, dmJhbGFqaUBjbWN2ZWxsb3JlLmFjLmlu
†These authors have contributed equally to this work
 Chaitra Shankar
Chaitra Shankar Karthick Vasudevan
Karthick Vasudevan Jobin John Jacob
Jobin John Jacob Stephen Baker2
Stephen Baker2 Dhiviya Prabaa Muthuirulandi Sethuvel
Dhiviya Prabaa Muthuirulandi Sethuvel Biju George
Biju George Balaji Veeraraghavan
Balaji Veeraraghavan