
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 07 April 2022
Sec. Molecular Bacterial Pathogenesis
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.869314
This article is part of the Research Topic Antibiotic Resistance of Escherichia and Its Mechanism View all 4 articles
 Mabel Kamweli Aworh1,2*
Mabel Kamweli Aworh1,2* Eme Ekeng3
Eme Ekeng3 Pernille Nilsson4
Pernille Nilsson4 Beverly Egyir5
Beverly Egyir5 Christian Owusu-Nyantakyi5
Christian Owusu-Nyantakyi5 Rene S. Hendriksen4
Rene S. Hendriksen4Introduction: Beef cattle, one of the food-producing animals, are linked to humans through a shared environment and the food chain as a major source of animal protein. Antimicrobial drugs are readily accessible for use in food animal production in Nigeria. Beef cattle and abattoir environments harbor pathogenic bacteria such as Escherichia coli (E. coli) which have developed resistance to antimicrobial agents used for prophylaxis or treatment. This study investigated the zoonotic transmission of extended-spectrum beta-lactamase-producing E. coli (ESBL-EC) among humans, beef cattle, and abattoir environments in Abuja and Lagos, Nigeria.
Materials and Methods: We conducted a cross-sectional study among abattoir workers, beef cattle, and abattoir environments in Abuja and Lagos. Stool, cecal, and environmental samples were collected from apparently healthy workers, slaughtered cattle, and abattoir environments from May to December 2020. Data were collected electronically using open data kit app installed on a mobile phone. Antimicrobial susceptibility patterns were determined using the Kirby–Bauer disk diffusion method against a panel of 16 antimicrobial agents. Phenotypic and genotypic characterizations of the isolates were conducted. Data were analyzed with descriptive statistics.
Results: From 21.7% (n = 97) of 448 samples, ESBL-EC were isolated and further characterized. Prevalence of ESBL-EC was highest in cattle (45.4%; n = 44), abattoir workers (41.2%; n = 40), and abattoir environment (13.4%; n = 13). Whole-genome sequencing of ESBL-EC showed dissemination of blaCTX-M-15 (90.7%; n = 88); blaCTX-M-14 (5.2%; n = 5); and blaCTX-M-55 (2.1%; n = 2) genes. The blaCTX-M-15 coexisted with blaCTX-M-14 and blaTEM-1 genes in 2.1% (n = 2) and 39.2% (n = 38) of the isolates, respectively. The presence of blaCTX-M-14 and blaCTX-M-15 genes was significantly associated with isolates originating from abattoir workers when compared with beef cattle isolates (p = 0.05; p < 0.01). The most prevalent sequence types (ST) were ST10 (n = 11), ST215 (n = 7), ST4684 (n = 7), and ST2178 (n = 6). ESBL-EC strain (ST205/B1) harbored mcr-1.1 and blaCTX-M15 and was isolated from a worker at Lagos abattoir. In 91 ESBL-EC isolates, 219 mobile genetic elements (MGEs) harbored resistance genes out of which β-lactam genes were carried on 64 different MGEs. Isolates showed equal distribution of insertion sequences and miniature inverted repeats although only a few composite transposons were detected (humans n = 12; cattle n = 9; environment n = 4). Two isolates of human and cattle origin (ST46/A) harboring ESBL genes and carried by MGEs were clonally related.
Conclusions: This is the first report of blaCTX-M-55 gene in humans and cattle in Nigeria. This study demonstrates the horizontal transfer of ESBL genes possibly by MGEs and buttresses the importance of genomic surveillance. Healthcare workers should be sensitized that people working closely with cattle or in abattoir environments are a high-risk group for fecal carriage of ESBL-EC when compared with the general population.
Antimicrobial-resistant food-borne bacteria have become an emerging problem in both human and animal populations (Rahman et al., 2017; Pormohammad et al., 2019; Ramos et al., 2020). The global spread of drug-resistant Enterobacterales has been associated with blood stream and urinary tract infections in humans (Runcharoen et al., 2017). During treatment of Gram-negative bacterial infections, an important resistance mechanism observed is the reduction in the efficacy of cephalosporins and monobactams (Geser et al., 2012). This is achieved by producing enzymes capable of inactivating these antimicrobial agents by hydrolyzing their beta-lactam ring. Extended-spectrum beta-lactamase (ESBL) efficiently hydrolyze 3rd- and 4th-generation cephalosporins with negative impact on patient care resulting in increased length of hospital stay, high morbidity, mortality, and high treatment costs (Geser et al., 2012; Runcharoen et al., 2017). ESBL genes were previously detected on chromosomes; however, these genes are now harbored on plasmids with resultant derivatives of plasmid-mediated β-lactamases such as blaTEM including blaCTX-M derived from the environment (Overdevest et al., 2011; Mensah et al., 2016).
The first description of ESBL production among Enterobacterales was from hospitalized patients although in recent years there has been reports of community-acquired ESBL infections (Reist et al., 2013; Song et al., 2020). There have also been increased reports of ESBL-producing Enterobacterales in humans, food-producing animals, and the environment in many countries (Reist et al., 2013; Rahman et al., 2017; Dantas Palmeira and Ferreira, 2020; Ramos et al., 2020; Song et al., 2020) although most bacteremias observed in humans may not be associated with animal reservoirs (Day et al., 2019). This has been attributed to the growing reservoirs as well as the indiscriminate use of antimicrobials (Overdevest et al., 2011; Veenemans et al., 2014). The ESBL-EC-Tricycle project uses a model that targets monitoring the indicator bacteria across the human, animal, and environmental sectors (WHO, 2021). Our recently published work showed that ESBL-EC, a zoonotic pathogen was detected among apparently healthy poultry workers, chickens, and the poultry farm/market environment (Aworh et al., 2020).
Food-producing animals including cattle have been reported as potential reservoirs for the spread of ESBL-EC along the food chain (Overdevest et al., 2011; Nguyen et al., 2019; Falgenhauer et al., 2019). Beef cattle, one of the food-producing animals, are linked to humans through shared environment and the food chain as a major source of animal protein (Marshall and Levy, 2011). In Nigeria like many developing economies, many families depend on beef cattle production as a means of livelihood as well as a major source of animal protein, hence the increased consumption of beef (Bettencourt et al., 2015). One of the gaps identified from an antimicrobial resistance (AMR) and use situation analysis across human, animal, and environmental health was the nonavailability of studies in Nigeria on ESBL-EC using a One Health approach (Nigeria Centre for Disease Control, 2017; Aworh et al., 2019). It is therefore imperative to assess the impact of healthy cattle as a possible reservoir for ESBL-EC along the food chain and to humans.
We hypothesized that slaughtered beef cattle harboring ESBL-EC can become potential sources of transmission of resistant genes along the food chain or to abattoir workers exposed based on their occupation as well as to the abattoir environments. We investigated the prevalence of fecal carriage of ESBL-EC among abattoir workers, beef cattle, and the abattoir environment to better understand if an association exists using antimicrobial susceptibility testing and whole-genome sequencing.
This cross-sectional study was carried out in one abattoir each in two major cities—Abuja (North/dry) and Lagos (South/humid), Nigeria, from May 2020 to December 2020. Using sterile stool containers, we collected freshly passed stool from apparently healthy consenting abattoir workers who were randomly selected. Cecal contents were randomly collected from the cecum of slaughtered beef cattle using sterile universal containers. For this study, we did not have access to information on animal husbandry and antimicrobial use in the slaughtered animals. Environmental samples comprising 30 g of lairage litter, 100 ml of abattoir waste water, and meat stall swabs were randomly collected from different locations in the selected abattoirs using sterile containers. All samples were transported in cool boxes to the National Reference Laboratory, Nigeria Centre for Disease Control Gaduwa, Abuja and processed for the presence of ESBL-EC.
We modified the WHO ESBL-Tricycle project protocol for the detection of ESBL-producing E. coli for the study (WHO, 2021). Briefly, about 1 g of human stool sample, 1 g of cecal content, and 30 g of litter samples were inoculated respectively in enrichment broth (buffered peptone water) in a 1:10 sample to broth ratio and incubated at 37°C for 24 h. Subsequently, a 10-µl loop-full of overnight culture from enrichment broth was plated onto MacConkey agar supplemented with cefotaxime (1 mg/L) and incubated at 37°C for 24 h. Next, suspected pink ESBL-EC colonies were further streaked on eosin methylene blue agar and incubated at 37°C for 24 h. Colonies suggestive of ESBL-EC were confirmed biochemically using commercially available Microbact GNB 24E (Oxoid, UK) according to the manufacturer’s instructions.
We used the membrane filtration technique for the isolation of ESBL-EC from abattoir waste water samples. Each 100 ml of waste water sample was filtered using single sterile 0.45 μm pore filter disks placed in a filtration unit. Thereafter, the filter membranes were placed on MacConkey agar supplemented with cefotaxime (1 mg/L) and incubated at 37°C for 24 h. All suspected ESBL-EC isolates were further tested as previously described.
Antimicrobial susceptibility profiling of suspected ESBL-EC was conducted and interpreted according to the 2020 Clinical and Laboratory Standards Institutes (CLSI) M100 30th Edition recommendations (Clinical and Laboratory Standards Institute, 2020). The Kirby–Bauer disk diffusion method was used to determine the resistance profile of the ESBL-EC against a panel of 16 antimicrobial agents characterized by WHO to be useful in clinical case management and livestock production (WHO, 2019). Four of these were β-lactam antimicrobials comprising ampicillin, ceftazidime, cefotaxime, and cefoxitin while the remaining were non-β-lactam antimicrobials (azithromycin, chloramphenicol, ciprofloxacin, colistin, gentamicin, imipenem, meropenem, nalidixic acid, nitrofurantoin, sulphonamide, trimethoprim, and tetracycline). Briefly, one to two distinct colonies picked from the overnight culture were suspended into 5 ml of sterile saline solution with turbidity adjusted to 0.5 McFarland standard. Using a sterile cotton swab, the suspension was emulsified onto Mueller Hinton agar plate and incubated at 37°C for 18 h. Thereafter, the zone of inhibition was measured and the results were interpreted based on CLSI recommendations (Clinical and Laboratory Standards Institute, 2020). We used E. coli ATCC 25922 and K. pneumoniae strain ATCC 700603 for internal quality control. An ESBL-EC isolate observed to be resistant to three or more classes of antimicrobials was considered to be multidrug resistant (MDR).
ESBL production was confirmed phenotypically using the combination disk diffusion method as described by CLSI M100 30th Edition (Clinical and Laboratory Standards Institute, 2020). The confirmed ESBL-EC isolates had an increase in zone diameter of ≥5 mm for either cefotaxime or ceftazidime in combination with clavulanic acid when compared with either of the cephalosporin alone. We used E. coli ATCC 25922 and K. pneumoniae ATCC 700603 strains as negative and positive controls, respectively (Clinical and Laboratory Standards Institute, 2020).
Whole genome sequencing was performed at the Noguchi Memorial Institute for Medical Research, University of Ghana under the SEQAFRICA Project. Briefly, the DNA for all ESBL-EC isolates from overnight culture were extracted and purified using Qiagen Kit following the manufacturer’s instructions. This was followed by quantification of DNA concentrations using the Qubit 4.0 Fluorometer Assay Kit (Thermo Fisher Scientific, MA, USA). Thereafter, libraries were prepared using the Nextera Flex Kit according to the manufacturer’s instructions. The libraries were quantified using the 2100 Bioanalyzer System (Agilent) and Kapa Sybr Fast qPCR Kit. DNA from each isolate were pooled together and sequenced on an Illumina Miseq platform using a 2 × 300 paired-end approach (Illumina Inc., San Diego, CA, USA). Raw sequencing reads (fastq files) were quality filtered to a Phred score ≥20, filtered for a minimum read length of 50 bp, and adaptor trimmed using Trimmomatic (http://www.usadellab.org/cms/index.php?page=trimmomatic). FastQC tool was used to assess quality of reads (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The resultant high-quality reads were used for de novo assembly using the Unicycler assembler v0.4.9 (Wick et al., 2017).
The resistance genes were detected in silico using ResFinder (database version 2021-09-03) with identity threshold and minimum length set at 90% and 60%, respectively (Bortolaia et al., 2020). Genes with the highest sequence identity and coverage were retained while genes that overlapped were filtered out. Plasmid replicon types of each isolate was determined using Plasmid-Finder 2.1 (database version 2020-07-01) (Carattoli et al., 2014). The mobile genetic elements (MGEs) were predicted using MobileElementFinder (database version 2020-06-09) (Johansson et al., 2021). Multilocus sequence typing was conducted using MLSTFinder (database version 2021-10-18) with multilocus sequence typing (MLST) allele sequence and profile data from PubMLST.org which assigned sequence types (STs) based on allelic variations to seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) that matched 100% identity (Larsen et al., 2012).
Each resistance gene was classified as being carried by a MGE or having no association. The resistance gene was associated if it was located on a MGE. The integrating MGEs associated with resistance genes were grouped on the basis of MGE type.
The ESBL-EC isolates were assigned phylogenetic groups using in silico ClermonTyping 1.4.1 tool (Beghain et al., 2018). We mapped assembled genome contigs to E. coli reference genome (GenBank accession No. GCA_900636075.1) and constructed a maximum likelihood phylogenetic tree using CSI phylogeny tool from the Center for Genomic Epidemiology (CGE). Clonal relationship between ESBL-EC isolates was determined using pairwise single-nucleotide polymorphism (SNP) analysis in the E. coli core genome if two or more isolates had less than 30 different SNPs. However, isolates with fewer than 10 different SNPs were considered closely related. The SNP-based phylogenetic tree was visualized using the iTOL tool.
Electronic-structured questionnaires were administered using the open data kit (ODK) collect app which was installed on a mobile phone. Data were extracted offline onto a computer using ODK briefcase. Analysis was done by computing frequencies, proportions, and Chi-square test with a significance level of 0.05. The raw sequencing reads data for this study have been deposited in the National Center for Biotechnology Information (NCBI) under the Bio project accession number PRJNA797451.
Approval for study was obtained from the Scientific and Ethical committee of the FCT Health Research Ethics Committee (Approval Number: FHREC/2020/01/40/04-05-20). Permission was sought from the management of each abattoir. Written informed consent was obtained from each eligible abattoir workers before questionnaire administration. We assured the abattoir workers of confidentiality of information obtained. We performed all the procedures according to the ethics committee’s guidelines and requirements.
Overall, 448 samples comprising human stool (n = 118), cattle cecal content (n = 272), and abattoir environment samples (n = 58) were collected from two large abattoirs in Abuja (n = 228) and Lagos (n = 220). The sample size for each abattoir varied depending on the actual size in terms of the number of cattle slaughtered daily and the willingness of the abattoir workers to participate in the study.
The overall prevalence of ESBL-EC from all sources was 21.7% (n = 97) out of which 57.7% (n = 56) were obtained from Lagos abattoir and 42.3% (n = 41) from Abuja abattoir (Figure 1).
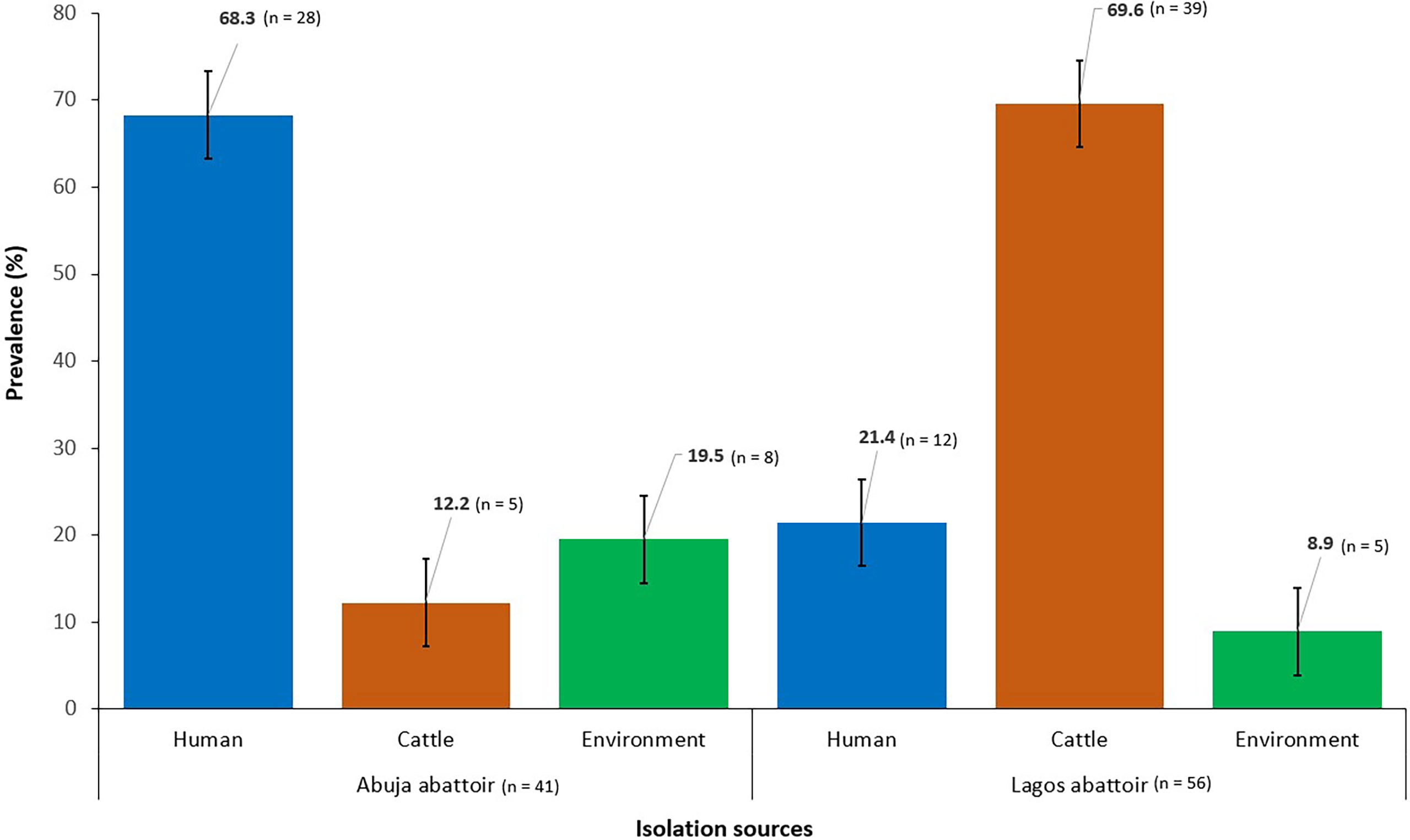
Figure 1 Prevalence of ESBL-EC isolated from humans, cattle, and abattoir environments in Abuja and Lagos Nigeria, 2021. Bars represent the proportion of ESBL-EC isolates from each isolation source with 95% confidence intervals. Error bars represent standard error of the mean prevalence. Data were obtained from two sources: Abuja and Lagos abattoirs.
Out of 97 ESBL-EC isolates, 99% (n = 96) were MDR. Out of these, 41.6% (n = 40) originated from abattoir workers, 44.8% (n = 43) were from cattle while 13.5% (n = 13) originated from the abattoir environment (Table 1). Overall, high resistance rates were observed in ESBL-EC to ceftazidime (89.7%), ciprofloxacin (95.9%), nalidixic acid (77.3%), tetracycline (82.5%), sulfonamides (78.4%), trimethoprim (76.3%), and azithromycin (52.6%), while low resistance rates were observed for gentamicin (19.6%) and chloramphenicol (18.6%). ESBL-EC isolates were however, susceptible to cefoxitin, imipenem, meropenem, and nitrofurantoin.
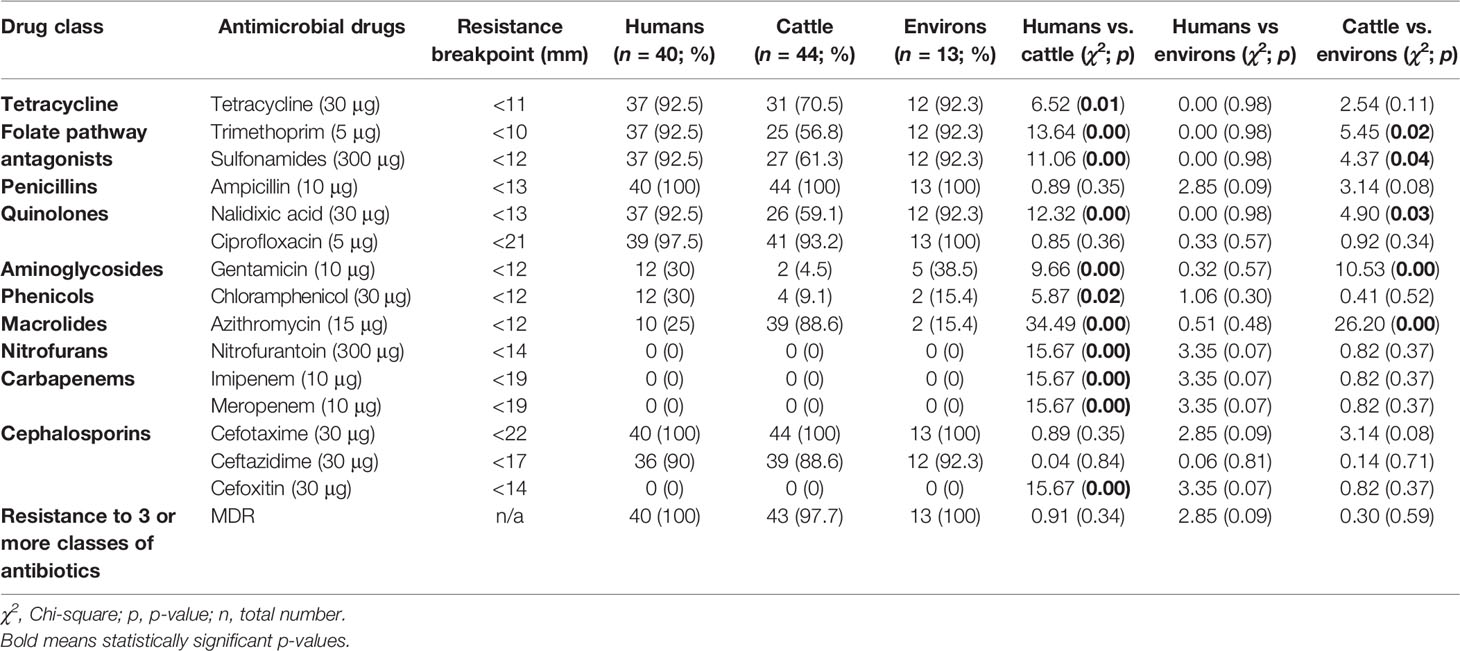
Table 1 Antimicrobial resistance profiles of ESBL-EC isolates from abattoir workers, beef cattle, and abattoir environments in Abuja and Lagos—Nigeria.
Among ESBL producers, the blaCTX-M gene was the most predominant ESBL encoding gene detected in 96.9% (n = 94) of isolates from the various sources. The blaCTX-M-15, blaCTX-M-14, and blaCTX-M-55 genes were observed in 90.7% (n = 88), 5.2% (n = 5), and 2.1% (n = 2) ESBL-EC isolates, respectively. The blaCTX-M-14 coexisted with blaCTX-M-15 gene in two isolates originating from beef cattle and abattoir environment. None of the isolates were observed to carry any carbapenemase gene. Most 84.5% (n = 82) of the isolates carried the plasmid-mediated quinolone-resistant (PMQR) gene—qnrS1. Interestingly, one isolate of human origin was observed to carry plasmid-mediated colistin-resistant (PMCR) gene—mcr 1.1. The presence of blaCTX-M-14 and blaCTX-M-15 genes were significantly associated with isolates originating from abattoir workers when compared with beef cattle isolates (p = 0.05; p < 0.01) as shown in Table 2.
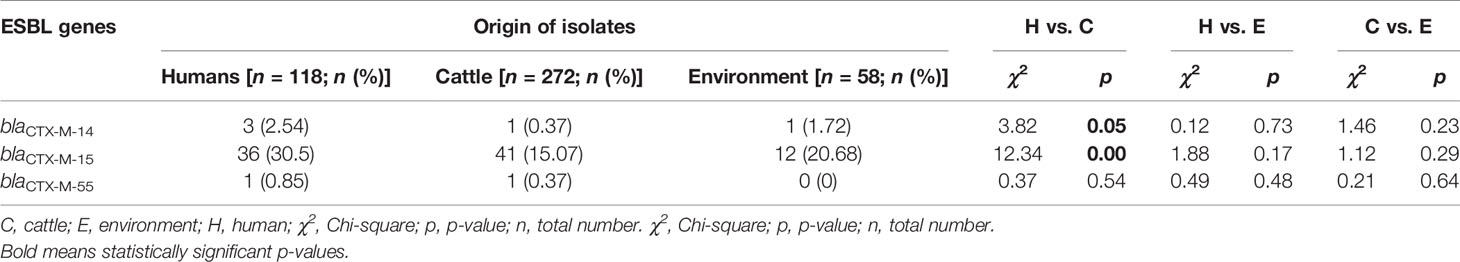
Table 2 ESBL gene variants of isolates originating from abattoir workers, beef cattle, and abattoir environments.
Forty-seven different AMR determinants were identified in 97 ESBL-EC isolates with the most prevalent displayed in Figure 2. Aminoglycosides were the most detected with 11 different variants (aadA1, aadA2, aadA5, aac(3)-Ila, aac(6)-Iaa, aac(6)-Ib3, aac(6)-Ib-cr, aph(3)-Ia, aph(3)-Ib, aph(3)-Id, and aph(6)-Id). Three ESBL-EC isolates exhibited the aac(6)-Ib-cr gene responsible for the reduction in ciprofloxacin activity. The aph(6)-Id a plasmid-encoded gene was detected in more than half of the isolates. This was followed by the detection of folate pathway antagonists with eight different variants (sul1, sul2, sul3, dfrA1, dfrA7, dfrA12, dfrA14, and dfrA17). The β-lactam genes were of five different variants (blaTEM-1, blaOXA-1, blaCTX-M-14, blaCTX-M-15, and blaCTX-M-55) out of which blaCTX-M variant was classical of ESBL-EC. Quinolone-resistant genes often associated with chromosomal mutations and categorized as critically important antimicrobials by the World Health Organization were detected in ESBL-EC isolates with six different variants (aac(6)-Ib-cr, qnrB19, qnrS1, qnrS4, qepA4, and qacE). Other AMR determinants detected in ESBL-EC isolates were polymyxin resistance (plasmid-mediated colistin resistance mcr-1.1), phenicol resistance (catA1, catA2, catB3, cmlA1, floR), fosfomycin resistance (fosA3, fosA7), macrolide resistance (erm(B), mef(B), mph(A), mdfA), and tetracycline resistance (tetA, tetB, tetM).
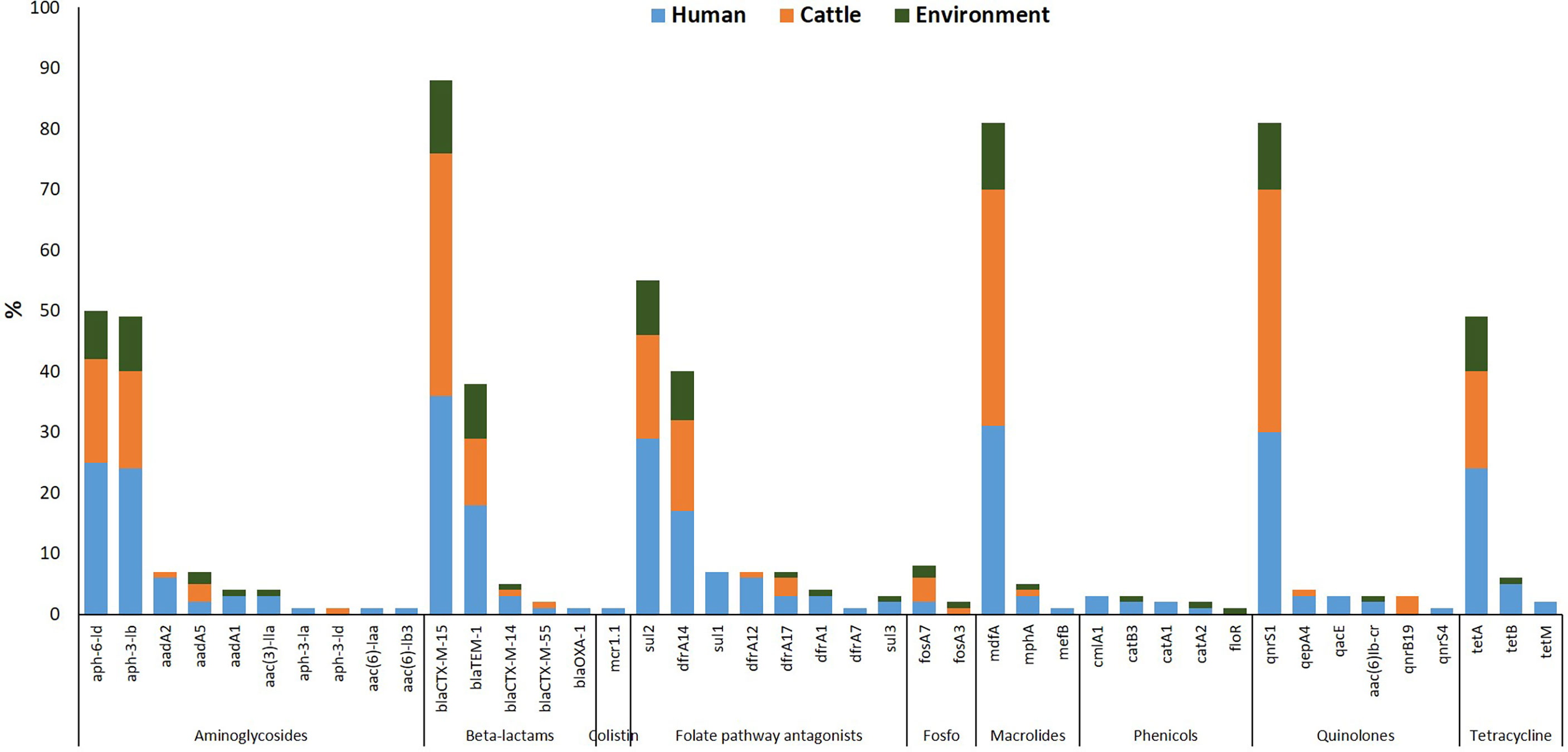
Figure 2 Resistance determinants detected in ESBL-EC isolates from humans, cattle, and abattoir environments in Abuja and Lagos—Nigeria. Each bar represents the various antimicrobial-resistant genes detected in ESBL-EC isolates obtained from abattoir workers, beef cattle, and abattoir environments.
MLST analysis showed that 97 ESBL-EC isolates were assigned to 52 known sequence types (STs). Four STs being ST 10, ST 2178, ST 4684, and ST 58 were shared by 28.9% (n = 28) of isolates from all three sources (Figure 3). Four STs were shared between abattoir workers and beef cattle (ST 46, ST 226, ST 7122, and ST 215) while ST 9529 was shared between ESBL-EC isolates from beef cattle and the abattoir environment. The most common ESBL variant detected was blaCTX-M-15 (90.7%). All blaCTX-M-15 isolates belonged to 49 different STs with eight being prevalent: ST 10 (n = 11), ST 215 (n = 7), ST 4684 (n = 7), ST 2178 (n = 6), ST 58 (n = 4), ST 226 (n = 3), ST 4429 (n = 3), and ST-7122 (n = 3).
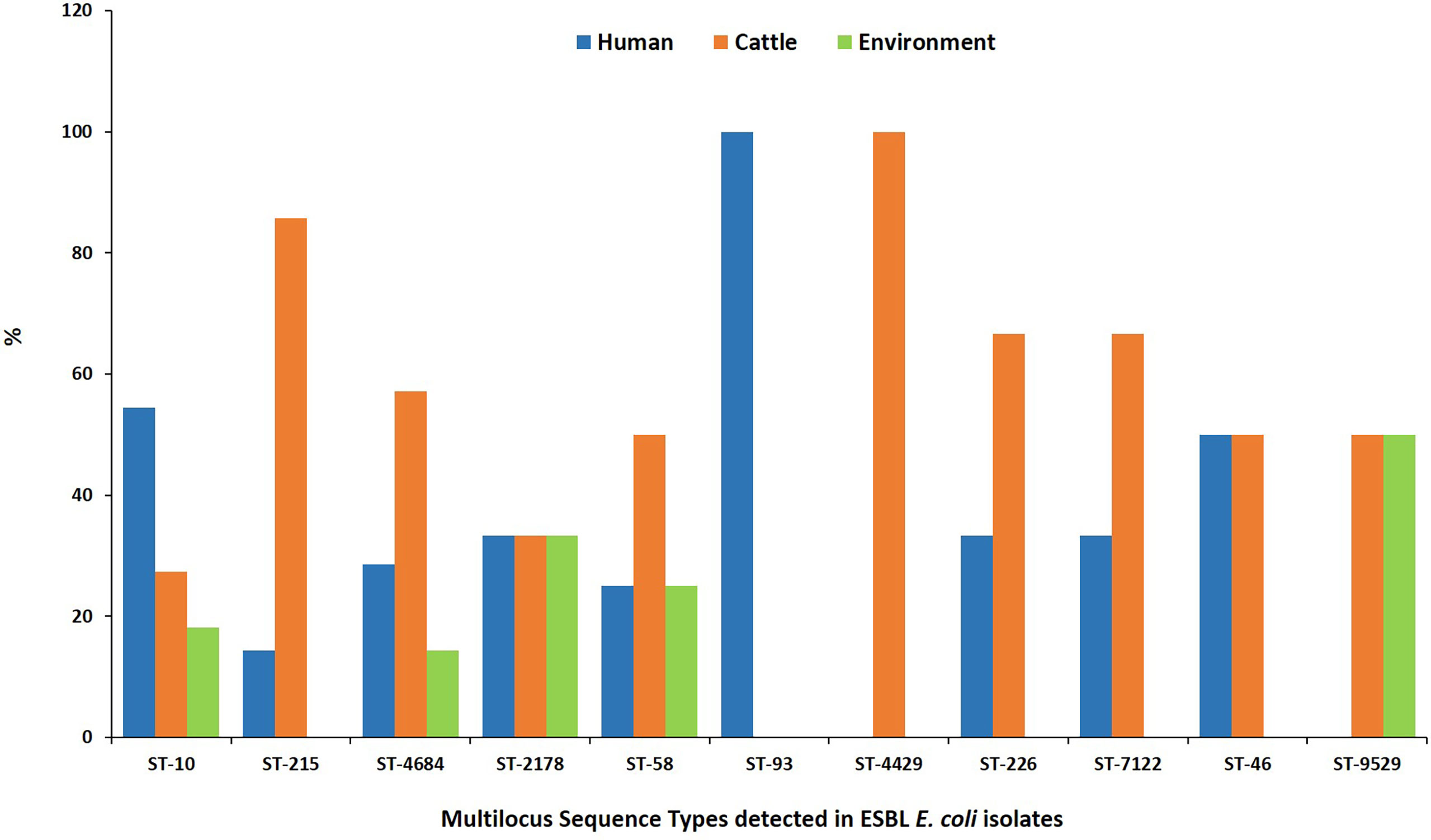
Figure 3 Multilocus sequence types for ESBL-EC isolated from humans, beef cattle, and abattoir environments in Abuja and Lagos—Nigeria. Each bar represents the various ESBL-EC sequence types for isolates obtained from abattoir workers, beef cattle, and abattoir environments.
Isolates from all sources showed equal distribution of insertion sequences (ISs) and miniature inverted repeats (MITEs). Of 97 ESBL-ECs, 25.8% (n = 25) were composite transposons (CTs) while 10.3% (n = 10) were unit transposons (UTs). Of 25 CTs, 48% (n = 12), 36% (n = 9), and 16% (n = 4) were detected in isolates from humans, cattle, and abattoir environments respectively with 52% from Lagos and 48% from Abuja abattoirs. The 10 UTs were detected in equal proportions between ESBL-EC isolates from humans and cattle. In 91 ESBL-EC isolates, 219 MGEs were observed to harbor AMR genes out of which β-lactam genes were carried on 64 different MGEs. Of these, 31 (48.4%) carried blaCTX-M-15 + qnrS1; 29 (45.3%) carried blaCTX-M-15; 2 (3.1%) carried blaCTX-M-55 + qnrS4 while 2 (3.1%) carried blaCTX-M-55 + qnrS4.The β-lactam genes were more frequently associated with MGEs in isolates from beef cattle (67.2%) than those from abattoir workers (23.4%) and the abattoir environment (9.4%). Eight different types of ISs and one MITE type were observed to harbor ESBL genes with ISEc9 (48.4%, n = 31), ISKpn19 (37.5%, n = 24), and MITTEc1 (7.8%, n = 5) being the most prevalent (Figure 4).
In the phylogenetic analysis, all isolates were assigned a phylo-group in descending order phylo-groups A (52.6%, n = 51), B1 (41.2%, n = 40), D (4.1%, n = 4), B2 (1%, n = 1), and E (1%, n = 1). Overall, 97 isolates were used to construct a SNP-based maximum likelihood phylogenetic tree (Figure 5). Only very few plasmid replicons were detected in the ESBL-EC isolates. It was evident that clonal relationship existed among isolates originating from different sources. ESBL-EC isolates were clustered into two main phylogroups and seven different STs. The pairwise distance matrix of SNPs based on core genome of 97 ESBL-EC strains showed 30 clonal relationships with a pairwise SNP difference of 30 out of which 11 (36.7%) were closely related with a pairwise SNP difference below 10. The clonally related isolates were clustered together based on the phylogenetic tree. Two isolates from the Abuja abattoir of human and cattle origin belonging to phylogroup A and sharing ST46 were observed to be closely related with a SNP difference of 2 (Table 3). In these two isolates, insertion sequences (ISKpn19 of ISKra4 family) were observed to carry the ciprofloxacin resistance gene (qnrS1). A combination of qnrS1 and blaCTX-M-15 were carried by the insertion sequence in the cattle isolate.

Figure 5 SNP-based phylogeny of ESBL E. coli isolates from humans, cattle, and abattoir environments in Abuja and Lagos. SNP-based maximum likelihood phylogeny of E. coli isolates visualized in iterative Tree of life tool (iTol). The tree was rooted in a reference isolate E. coli strain NCTC11129. Clustering of isolates was found to be following the core genome and SNP-based phylogenies. The clustering of isolates belonging to the same phylogenetic group and sequence type was consistent. Shown for each isolate is the source: humans, cattle or environment, location, phylogroup, sequence types (STs), AMR genes and plasmid replicons, and number of MGEs.
Many studies globally have reported that E. coli isolated from food animals including beef cattle are often resistant to β-lactam antimicrobials (Tadesse et al., 2018; Ramos et al., 2020; Song et al., 2020; Dantas Palmeira and Ferreira, 2020). Cattle have been reported as an important source of ESBL-EC transmission to humans and a threat to the world (Sudarwanto et al., 2016; Saleem et al., 2017; Dantas Palmeira and Ferreira, 2020). The risk for people working in close proximity with animals has been poorly elucidated.
This present study investigated the prevalence of ESBL-EC among abattoir workers, beef cattle at slaughter, as well as in the abattoir environments. Our findings showed that ESBL-EC are present in the abattoir environment where beef cattle are processed for food serving as a reservoir of resistant pathogens and a health hazard to people working in such environment. The prevalence of ESBL-EC in the present study was 41.2%, 45.4%, and 13.4% in humans, beef cattle, and abattoir environments, respectively. Our study prevalence of ESBL-EC in cattle was higher than what was observed in another location in Nigeria but much lower than the prevalence reported among abattoir workers (Egbule and Yusuf, 2019; Egbule and Iweriebor, 2021). Similar studies in Europe reported a much lower prevalence of ESBL-EC in cattle and farm workers (Reist et al., 2013; Dahms et al., 2015). A possible explanation for the high prevalence observed in the present study may be as a result of over-the-counter availability of antimicrobials in Nigeria for both humans and animals as opposed to Europe where these are available based on prescription (Oluwasile et al., 2014; Oloso et al., 2018; Agyare et al., 2019).
Penicillin, cephalosporin, quinolones, tetracycline, folate pathway antagonists, and aminoglycosides accounted for most of the resistance determinants detected in the present study, and this is consistent with the reports of others (Reist et al., 2013; Oloso et al., 2018; Egbule and Yusuf, 2019; Kimera et al., 2020). Critically important antimicrobial-resistant genes PMQR—qnrS1 and PMCR—mcr 1.1 were detected in the ESBL-EC isolates. Our study detected higher resistance to ciprofloxacin when compared with nalidixic acid which is rather surprising. A possible explanation for this observation may be due to the high prevalence of qnrS1. The high prevalence of AMR genes observed may be attributed to the use of these antimicrobials in livestock production in Nigeria either for prophylaxis or therapeutic purposes (Oloso et al., 2018; Kimera et al., 2020).
The only ESBL gene variant detected in the present study was blaCTX-M in isolates from all sources, and this is consistent with findings from other studies (Dantas Palmeira and Ferreira, 2020; Kimera et al., 2020). The blaCTX-M-15 genes were the most prevalent and found to coexist with blaCTX-M-14 in two isolates from beef cattle and abattoir environment. Interestingly, blaCTX-M-55 gene was also detected in isolates from humans and cattle in this study. It is important to note that blaCTX-M-55 variant has been the second most predominant ESBL gene in Asia and has been reported in Europe but rare in Africa (Lupo et al., 2018; Dantas Palmeira and Ferreira, 2020).
Our study results showed evidence of genetic diversity of ESBL-EC isolates as we detected 52 known sequence types (STs), and this is consistent with reports of others (Day et al., 2019; Falgenhauer et al., 2019). The commonly detected STs and phylogroups in isolates originating from all sources were ST10/A, ST2178/B1, ST4684/B1, and ST58/B1, suggesting that a possible transmission may have occurred between these sources. This observation further buttresses that co-colonization of resistant foodborne bacteria from a shared source is possible. Evidence shows that E. coli ST58 is an important zoonotic strain detected in food-producing animals particularly cattle implicated in the spread of ESBL-EC to humans (Pietsch et al., 2018; Irenge et al., 2019). It is worthy of note that ST10 has been detected in E. coli isolates from healthy humans, animals, and environmental samples (Müller et al., 2016; Falgenhauer et al., 2019), and this is consistent with our study results. Our observation may be as a result of a possible circulation of host-adapted lineages of ESBL-EC.
Although, some evidence exists that food-producing animals are important reservoirs in the transmission of resistant bacteria to the human population, our study clearly shows that there is a possibility. Several isolates originating from humans, cattle, and the abattoir environment demonstrated clonal relationship with 0–30 SNP differences between the isolates especially those with ST12463, ST2178, ST58, ST4684, ST215, ST10, ST93, and ST46. Our study demonstrated that one human and one cattle isolate (ST46) were closely related with a pairwise SNP difference of 2 suggesting a possibility of horizontal transfer of resistance between the two hosts and consistent with findings from a study in Ghana (Falgenhauer et al., 2019). Although the isolates in the Ghana study originated from chickens and sick children, our sources were apparently healthy cattle and abattoir workers.
In the MGE analysis, the MITEs and ISs were more predominant than the transposons in the ESBL-EC isolates in the present study and this observation is consistent in Gram-negative bacteria (Johansson et al., 2021). Majority of the resistance genes were not carried by transposons but by MITEs and ISs as reported by other studies (Partridge et al., 2018; Johansson et al., 2021). The two closely related isolates from humans and cattle originated from the same abattoir location, belonged to the same phylogroup, and had similar resistance genes carried by identical MITEs, ISs, and transposons. It is noteworthy that no ESBL genes were harbored on plasmid replicons.
To the best of our knowledge, this study is the first in Nigeria to report PMCR gene, mcr-1.1 in abattoir workers. The mcr-1.1-positive ESBL-EC strain (ST205/B1) coexisted with blaCTX-M-15 and was isolated from a worker at Lagos abattoir. Although the blaCTX-M-15 gene was carried on an insertion sequence, our results showed that the mcr-1.1 gene was not harbored by any MGE. Colistin which is considered a drug of last resort, is often used in livestock production in Nigeria for therapeutic purposes (Oluwasile et al., 2014; Aworh et al., 2019).
This study is not without limitations as we had very few people who agreed to participate in one of the abattoirs and may have been responsible for our observations.
ESBL-EC isolates were prevalent among apparently healthy abattoir workers, beef cattle, and the abattoir environment in Abuja and Lagos. Among ESBL-EC, the highest resistance was observed to ampicillin, cefotaxime, and ciprofloxacin, which are antimicrobial classes often used in livestock production for therapeutic purposes. Our study detected ESBL genes, blaCTX-M in isolates from all the sources. The blaCTX-M coexisted with qnrS1 and mcr-1.1 genes. To our knowledge, this is the first study in Nigeria to report blaCTX-M-55 gene in humans and cattle. The possibility of horizontal transfer of resistance genes from humans to cattle or vice versa may have been facilitated by MGEs. It is imperative to sensitize healthcare workers that abattoir workers or people working in such vicinities are a high-risk group for fecal carriage of ESBL-EC, hence pose a higher risk to the general population for horizontal transfer of resistance. The Government should develop and enforce regulations that prohibit the use of critically important antimicrobials for human health in food-producing animals.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by the FCT Health Research Ethics Committee (Approval Number: FHREC/2020/01/40/04-05-20). The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Scientific and Ethical Committee of the Ahmadu Bello University Committee on Animal Use and Care (Approval Number: ABUCAUC/2020/35).
MA was the principal investigator, designed data collection tools, collected data, isolated the organism, performed antibiotic sensitivity testing on the isolates, analyzed and interpreted the data, and wrote the first draft of the manuscript. RH made substantial contributions to conception and design. EE, PN, and RH supervised the laboratory aspect of the research. BE and CO generated the whole genome sequence profile for all the isolates. MA performed bioinformatics analysis. EE, PN, BE, CO, and RH revised the article critically for important intellectual content. All authors read and approved the final manuscript.
This project was funded by the Fleming Fund Fellowship scheme through the Denmark Technical University according to grant No. 13534.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors appreciate the UK-aid Fleming Fund Fellowship scheme for providing the training and funding for this project. Special appreciation goes to the Mott MacDonald’s team especially Eileen Chappell and Elinam Segbefia for their excellent coordination of the project. The authors acknowledge the intellectual contributions of Mrs. Mba Nwando, Mr. Akinpelu Muftan, Mr. Micheal Popoola, and the bacteriology team of the National Reference Laboratory, Gaduwa, Abuja, towards the success of this research. The authors appreciate the efforts of Felicia Amoa Owusu and other members of the Bacteriology Department at Noguchi Memorial Institute for Medical Research, University of Ghana. Special appreciation goes to the mentors at the Denmark Technical University and Federal Ministry of Agriculture and Rural Development for their support towards the success of this project. The authors appreciate the support of Dr. Stephen Okeme at Karu Abattoir-Abuja, and Dr. Ayokunle Omileye at Oko-oba Agege Abattoir-Lagos.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.869314/full#supplementary-material
AMR, antimicrobial resistance; CLSI, Clinical and Laboratory Standards Institute; CTs, composite transposons; E. coli, Escherichia coli; ESBL, extended-spectrum beta-lactamase; ESBL-EC, extended-spectrum beta-lactamase-producing E. coli; ISs, insertion sequences; MDR, multidrug resistance; MIC, minimum inhibitory concentration; MITEs, miniature inverted repeats; MLST, multilocus sequence typing; MGEs, mobile genetic elements; NCBI, National Center for Biotechnology Information; ODK, open data kit; PMCR, plasmid-mediated colistin resistance; PMQR, plasmid-mediated quinolone resistance; SNP, single nucleotide polymorphisms; ST, sequence type; UT, unit transposons; WGS, whole-genome sequencing; WHO, World Health Organization.
Agyare, C., Etsiapa Boamah, V., Ngofi Zumbi, C., Boateng Osei, F. (2019). “Antibiotic Use in Poultry Production and Its Effects on Bacterial Resistance,” in Antimicrobial Resistance - A Global Threat (InTech Open Science) doi: 10.5772/intechopen.79371
Aworh, M. K., Kwaga, J., Okolocha, E., Harden, L., Hull, D., Hendriksen, R. S., et al. (2020). Extended-Spectrum ß-Lactamase-Producing Escherichia Coli Among Humans, Chickens and Poultry Environments in Abuja, Nigeria. One Heal Outlook 2 (1), 8. doi: 10.1186/s42522-020-00014-7
Aworh, M. K., Kwaga, J., Okolocha, E., Mba, N., Thakur, S. (2019). Prevalence and Risk Factors for Multi-Drug Resistant Escherichia Coli Among Poultry Workers in the Federal Capital Territory, Abuja, Nigeria. PloS One 14 (11), 1–15. doi: 10.1371/journal.pone.0225379
Beghain, J., Bridier-Nahmias, A., Nagard, H., Denamur, E., Clermont, O. (2018). Clermontyping: An Easy-to-Use and Accurate In Silico Method for Escherichia Genus Strain Phylotyping. Microb. Genomics 4 (7), 1–8. doi: 10.1099/mgen.0.000192
Bettencourt, E. M. V., Tilman, M., Narciso, V., da Silva Carvalho, M. L., de Sousa Henriques, P. D. (2015). The Livestock Roles in the Wellbeing of Rural Communities of Timor-Leste. Rev. Econ e Sociol Rural 53, S063–S080. doi: 10.1590/1234-56781806-94790053s01005
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). Resfinder 4.0 for Predictions of Phenotypes From Genotypes. J. Antimicrob. Chemother. 75 (12), 3491–3500. doi: 10.1093/jac/dkaa345
Carattoli, A., Zankari, E., Garciá-Fernández, A., Larsen, M. V., Lund, O., Villa, L., et al. (2014). In Silico Detection and Typing of Plasmids Using Plasmidfinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 58 (7), 3895–3903. doi: 10.1128/AAC.02412-14
Clinical and Laboratory Standards Institute (2020). “CLSI M100 30th Edition” in Journal of Services Marketing, 30th. USA: Clinical and Laboratory Standards Institute. ISBN 978-1-68440-066-9 (print version).
Dahms, C., Hübner, N.-O., Kossow, A., Mellmann, A., Dittmann, K., Kramer, A. (2015). Occurrence of ESBL-Producing Escherichia Coli in Livestock and Farm Workers in Mecklenburg-Western Pomerania, Germany. PloS One 10 (11), e0143326. doi: 10.1371/journal.pone.0143326
Dantas Palmeira, J., Ferreira, H. M. N. (2020). Extended-Spectrum Beta-Lactamase (ESBL)-Producing Enterobacteriaceae in Cattle Production – A Threat Around the World. Heliyon 6 (1), e03206. doi: 10.1016/j.heliyon.2020.e03206
Day, M. J., Hopkins, K. L., Wareham, D. W., Toleman, M. A., Elviss, N., Randall, L., et al. (2019). Extended-Spectrum Beta-Lactamase-Producing Escherichia Coli in Human-Derived and Foodchain-Derived Samples From England, Wales, and Scotland: An Epidemiological Surveillance and Typing Study. Lancet Infect. Dis. 19 (12), 1325–1335. doi: 10.1016/S1473-3099(19)30273-7
Egbule, O. S., Iweriebor, B. C. (2021). Beta-Lactamase-Producing Escherichia Coli Isolates Recovered From Pig Handlers in Retail Shops and Abattoirs in Selected Localities in Southern Nigeria : Implications for Public Health. Antibiotics 10 (9), 1–10. doi: 10.3390/antibiotics10010009
Egbule, O. S., Yusuf, I. (2019). Multiple Antibiotic Resistances in Escherichia Coli Isolated From Cattle and Poultry Faeces in Abraka, South-South Nigeria. Pertanika J. Trop. Agric. Sci. 42 (2), 585–594.
Falgenhauer, L., Imirzalioglu, C., Oppong, K., Akenten, C. W., Hogan, B., Krumkamp, R., et al. (2019). Detection and Characterization of ESBL-Producing Escherichia Coli From Humans and Poultry in Ghana. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.03358/full
Geser, N., Stephan, R., Hächler, H. (2012). Occurrence and Characteristics of Extended-Spectrum β-Lactamase (ESBL) Producing Enterobacteriaceae in Food Producing Animals, Minced Meat and Raw Milk. BMC Vet. Res. 8 (21), 1–9. doi: 10.1186/1746-6148-8-21
Irenge, L. M., Ambroise, J., Bearzatto, B., Durant, J. F., Chirimwami, R. B., Gala, J. L. (2019). Whole-Genome Sequences of Multidrug-Resistant Escherichia Coli in South-Kivu Province, Democratic Republic of Congo: Characterization of Phylogenomic Changes, Virulence and Resistance Genes. BMC Infect. Dis. 19 (1), 1–10. doi: 10.1186/s12879-019-3763-3
Johansson, M. H. K., Bortolaia, V., Tansirichaiya, S., Aarestrup, F. M., Roberts, A. P., Petersen, T. N. (2021). Detection of Mobile Genetic Elements Associated With Antibiotic Resistance in Salmonella Enterica Using a Newly Developed Web Tool: Mobileelementfinder. J. Antimicrob. Chemother. 76 (1), 101–109. doi: 10.1093/jac/dkaa390
Kimera, Z. I., Mshana, S. E., Rweyemamu, M. M., Mboera, L. E. G., Matee, M. I. N. (2020). Antimicrobial Use and Resistance in Food-Producing Animals and the Environment: An African Perspective. Antimicrobial Resistance Infection Control 9, 37. doi: 10.1186/s13756-020-0697-x
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 50 (4), 1355–1361. doi: 10.1128/JCM.06094-11
Lupo, A., Saras, E., Madec, J. Y., Haenni, M. (2018). Emergence of Bla CTX-M-55 Associated With Fosa, Rmtb and Mcr Gene Variants in Escherichia Coli From Various Animal Species in France. J. Antimicrob. Chemother. 73 (4), 867–872. doi: 10.1093/jac/dkx489
Marshall, B. M., Levy, S. B. (2011). Food Animals and Antimicrobials: Impacts on Human Health. Clin. Microbiol. Rev. 24 (4), 718–733. doi: 10.1128/CMR.00002-11
Mensah, D. O., Nkrumah, N. O., Bonney, E. Y., Mensah, E. O., Danso, K. T., Osei, Y. D., et al. (2016). Genetic Characterization of TEM − Type ESBL − Associated Antibacterial Resistance in Enterobacteriaceae in a Tertiary Hospital in Ghana. Ann. Clin. Microbiol. Antimicrob., 15, 29. doi: 10.1186/s12941-016-0144-2
Müller, A., Stephan, R., Nüesch-Inderbinen, M. (2016). Distribution of Virulence Factors in ESBL-Producing Escherichia Coli Isolated From the Environment, Livestock, Food and Humans. Sci. Total Environ. 541, 667–672. doi: 10.1016/j.scitotenv.2015.09.135
Nguyen, V. T., Jamrozy, D., Matamoros, S., Carrique-Mas, J. J., Ho, H. M., Thai, Q. H., et al. (2019). Limited Contribution of Non-Intensive Chicken Farming to ESBL-Producing Escherichia Coli Colonization in Humans in Vietnam: An Epidemiological and Genomic Analysis. J. Antimicrob. Chemother. 74 (3), 561–570. doi: 10.1093/jac/dky506
Nigeria Centre for Disease Control (2017) Antimicrobial Use and Resistance in Nigeria. Available at: http://www.ncdc.gov.ng/themes/common/docs/protocols/56_1510840387.pdf.
Oloso, N. O., Fagbo, S., Garbati, M., Olonitola, S. O., Awosanya, E. J., Aworh, M. K., et al. (2018). Antimicrobial Resistance in Food Animals and the Environment in Nigeria: A Review. Int. J. Environ. Res. Public Health 15 (6), 1–23. doi: 10.3390/ijerph15061284
Oluwasile, B., Agbaje, M., Ojo, O., Dipeolu, M. (2014). Antibiotic Usage Pattern in Selected Poultry Farms in Ogun State. Sokoto J. Vet. Sci. 12 (1), 45. doi: 10.4314/sokjvs.v12i1.7
Overdevest, I., Willemsen, I., Rijnsburger, M., Eustace, A., Xu, L., Hawkey, P., et al. (2011). Extended-Spectrum β-Lactamase Genes of Escherichia Coli in Chicken Meat and Humans, the Netherlands. Emerg. Infect. Dis. 17 (7), 1216–1222. doi: 10.3201/eid1707.110209
Partridge, S. R., Kwong, S. M., Firth, N., Jensen, S. O. (2018). Mobile Genetic Elements Associated With Antimicrobial Resistance. Clin. Microbiol. Rev. 3131, 1–61. American Society for Microbiology. doi: 10.1128/CMR.00088-17
Pietsch, M., Irrgang, A., Roschanski, N., Brenner Michael, G., Hamprecht, A., Rieber, H., et al. (2018). Whole Genome Analyses of CMY-2-Producing Escherichia Coli Isolates From Humans, Animals and Food in Germany. BMC Genomics 19 (1), 1–17. doi: 10.1186/s12864-018-4976-3
Pormohammad, A., Nasiri, M. J., Azimi, T. (2019). Prevalence of Antibiotic Resistance in Escherichia Coli Strains Simultaneously Isolated From Humans, Animals, Food, and the Environment: A Systematic Review and Meta-Analysis. Infection Drug Resistance 12, 1181–1197. doi: 10.2147/IDR.S201324
Rahman, M. A., Rahman, A. K. M. A., Islam, M. A., Alam, M. M. (2017). Antimicrobial Resistance of Escherichia Coli Isolated From Milk, Beef and Chicken Meat in Bangladesh. Bangl. J. Vet. Med. 15 (2), 141–146.
Ramos, S., Silva, V., de Lurdes Enes Dapkevicius, M., Caniça, M., Tejedor-Junco, M. T., Igrejas, G., et al. (2020). Escherichia Coli as Commensal and Pathogenic Bacteria Among Food-Producing Animals: Health Implications of Extended Spectrum β-Lactamase (ESBL) Production. Animals 10 (12), 1–15. doi: 10.3390/ani10122239
Reist, M., Geser, N., Hächler, H., Schärrer, S., Stephan, R. (2013). ESBL-Producing Enterobacteriaceae: Occurrence, Risk Factors for Fecal Carriage and Strain Traits in the Swiss Slaughter Cattle Population Younger Than 2 Years Sampled at Abattoir Level. PloS One 8 (8), e71725. doi: 10.1371/journal.pone.0071725
Runcharoen, C., Raven, K. E., Reuter, S., Kallonen, T., Paksanont, S., Thammachote, J., et al. (2017). Whole Genome Sequencing of ESBL-Producing Escherichia Coli Isolated From Patients, Farm Waste and Canals in Thailand. Genome Med. 9 (81), 1–11. doi: 10.1186/s13073-017-0471-8
Saleem, R., Ejaz, H., Zafar, A., Younas, S., Rathore, A. W. (2017). Phenotypic Characterization of Extended-Spectrum-Beta-Lactamase Producing E. Coli From Healthy Individuals, Patients, Sewage Sludge, Cattle, Chickens and Raw Meat. Pakistan J. Med. Sci. 33 (4), 886–890. doi: 10.12669/pjms.334.12647
Song, J., Oh, S. S., Kim, J., Shin, J. (2020). Extended-Spectrum β-Lactamase-Producing Escherichia Coli Isolated From Raw Vegetables in South Korea. Sci. Rep. 10 (1), 1–7. doi: 10.1038/s41598-020-76890-w
Sudarwanto, M. B., Lukman, D. W., Latif, H., Pisestyani, H., Sukmawinata, E., Akineden, Ö, et al. (2016). CTX-M Producing Escherichia Coli Isolated From Cattle Feces in Bogor Slaughterhouse, Indonesia. Asian Pac. J. Trop. BioMed. 6 (7), 605–608. doi: 10.1016/j.apjtb.2016.05.001
Tadesse, D. A., Li, C., Mukherjee, S., Hsu, C. H., Bodeis Jones, S., Gaines, S. A., et al. (2018). Whole-Genome Sequence Analysis of CTX-M Containing Escherichia Coli Isolates From Retail Meats and Cattle in the United States. Microb. Drug Resist. 24 (7), 939–948. doi: 10.1089/mdr.2018.0206
Veenemans, J., Overdevest, I. T., Snelders, E., Willemsen, I., Hendriks, Y., Adesokan, A., et al. (2014). Next-Generation Sequencing for Typing and Detection of Resistance Genes: Performance of a New Commercial Method During an Outbreak of Extended-Spectrum-Beta-Lactamase-Producing Escherichia Coli. J. Clin. Microbiol. 52 (7), 2454–2460. doi: 10.1128/JCM.00313-14
WHO (2021). “Global Tricycle Surveillance – ESBL E.Coli - Integrated Global Surveillance on ESBL-Producing E. Coli Using a “One Health” Approach: Implementation and Opportunities,” in World Health Organization, Global Antimicrobial Resistance Surveillance System (GLASS), Surveillance Prevention and Control. Geneva: World Health Organization ISBN 978-92-4-002140-2 (electronic version). vol. 76.
Keywords: antimicrobial, resistance, abattoir, cattle, environment, Escherichia coli, Nigeria
Citation: Aworh MK, Ekeng E, Nilsson P, Egyir B, Owusu-Nyantakyi C and Hendriksen RS (2022) Extended-Spectrum ß-Lactamase-Producing Escherichia coli Among Humans, Beef Cattle, and Abattoir Environments in Nigeria. Front. Cell. Infect. Microbiol. 12:869314. doi: 10.3389/fcimb.2022.869314
Received: 04 February 2022; Accepted: 11 March 2022;
Published: 07 April 2022.
Edited by:
Xiumin Wang, Feed Research Institute (CAAS), ChinaReviewed by:
Marisa Haenni, Agence Nationale de Sécurité Sanitaire de l’Alimentation, de l’Environnement et du Travail (ANSES), FranceCopyright © 2022 Aworh, Ekeng, Nilsson, Egyir, Owusu-Nyantakyi and Hendriksen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mabel Kamweli Aworh, bWFiZWxhd29yaEB5YWhvby5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.