
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 29 April 2022
Sec. Microbiome in Health and Disease
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.864479
This article is part of the Research Topic The Pivotal Role of Oral Microbiota Dysbiosis and Microbiota-host Interactions in Diseases - Volume II View all 11 articles
The oral cavity harbors approximately 1,000 microbial species, and both pathogenic and commensal strains are involved in the development of carcinogenesis by stimulating chronic inflammation, affecting cell proliferation, and inhibiting cell apoptosis. Moreover, some substances produced by oral bacteria can also act in a carcinogenic manner. The link between oral microbiota and chronic inflammation as well as cell proliferation has been well established. Recently, increasing evidence has indicated the association of the oral microbiota with cell migration, which is crucial in regulating devastating diseases such as cancer. For instance, increased cell migration induced the spread of highly malignant cancer cells. Due to advanced technologies, the mechanistic understanding of cell migration in carcinogenesis and cancer metastasis is undergoing rapid progress. Thus, this review addressed the complexities of cell migration in carcinogenesis and cancer metastasis. We also integrate recent findings on the molecular mechanisms by which the oral microbiota regulates cell migration, with emphasis on the effect of the oral microbiota on adhesion, polarization, and guidance. Finally, we also highlight critical techniques, such as intravital microscopy and superresolution microscopy, for studies in this field.
An eclectic and diverse assemblage of microbiota inhabits different sites within the oral cavity, such as teeth, saliva, and oral mucosal surfaces (Lamont et al., 2018). Distinct microbial communities accumulate in these sites through successive colonization events. Since van Leeuwenhoek made the first observation of oral bacteria in dental plaques using primitive microscopes in 1683, our knowledge of oral microbiology has burgeoned. Investigations of the oral microbiota in health and diseases are currently undergoing rapid progress due to the development of techniques (Willis and Gabaldon, 2020). The oral microbial community is a complex and dynamic entity, and shifts in these communities contribute to both oral diseases (e.g., dental caries, periodontitis) and systemic diseases (e.g., diabetes mellitus, cardiovascular disease) (Kleinstein et al., 2020; Hajishengallis and Lamont, 2021). Interestingly, the carcinogenic effect of the microbiome in various organs has been revealed by studies in germ-free animals (Scott et al., 2019). Recently, the microbiome has been considered an influential environmental factor modulating carcinogenesis (Scott et al., 2019; Irfan et al., 2020). Helicobacter pylori is the most known bacterial carcinogen (Alipour, 2021). There is also increasing evidence indicating a correlation of some specific species with cancer, including Porphyromonas gingivalis, Treponema denticola, Fusobacterium sp., Streptococcus sp., Peptostreptococcus sp., Prevotella sp., and Capnocytophaga (Fitzsimonds et al., 2020). In some cases, a dysbiotic community results from a shift in microbial composition rather than a specific organism associated with the carcinogenic process (Irfan et al., 2020). The current strategies to study the role of the oral microbiome in cancer have predominantly focused on detecting microbial communities present or populational shifts in specific samples and investigating host responses to specific microbial challenges, such as immunological responses, inflammatory responses, and cell proliferation. The potentially oncogenic oral bacteria and possible mechanisms of their action on the carcinogenesis of cells have been systematically reviewed (Whitmore and Lamont, 2014; Fitzsimonds et al., 2020; Irfan et al., 2020; Lamont et al., 2022). In summary, oral microbiota participate in cancer mainly via the following mechanisms: (1) stimulating chronic inflammation; (2) inducing mutagenesis, oncogene activation, and angiogenesis; (3) facilitating cell proliferation and inhibiting cellular apoptosis; and (4) producing carcinogens.
In cancer biology, the microbiome is considered an influential environmental factor modulating the carcinogenic process (Scott et al., 2019). Cell migration is an essential physiological process for the immune response, wound repair, and tissue regeneration, while abnormal cell migration is found in devastating diseases such as tumor formation and metastasis. An increasing number of studies have shown that cell migration can be induced by oral bacteria, such as P. gingivalis and Fusobacterium nucleatum. The latest study found that the tyrosine phosphatase (Ltp1) of P. gingivalis is secreted and facilitates P. gingivalis-induced proliferation, migration, and the epithelial to mesenchymal transition (EMT) of gingival epithelial cells by targeting the regulator of growth and cell cycle (RGCC). Three distinct activities occurring either simultaneously or independently are involved in cell migration: protrusion, attachment, and traction. There is a complex and discrepant scenario of these activities depending on conditions and migration types. Taking epithelial cells as an example, slow-moving epithelial cells are characterized by slipping of adhesion and retrograde actin flow, while fast-moving epithelial cells have more gripping adhesions and rapid protrusion (Jurado et al., 2005). Single-cell migration and collective cell migration are two main types of cell migration. Studies have demonstrated that epithelial cells either migrate collectively or undergo EMT, thus migrating as single mesenchymal cells (Lu and Lu, 2021). Migration and EMT are highly compatible and facilitate each other involved in development, wound healing, and cancer metastasis (Son and Moon, 2010; Stone et al., 2016). For example, cell migration induced the spread of highly malignant cancer cells (Mosier et al., 2021). Moreover, carcinoma cells migrate into adjacent tissues and invade the lymphatic system and blood vessels and then seeds in distant organs (Christiansen and Rajasekaran, 2006).
Therefore, cell migration is a potential linkage between oral microbiota and carcinogenesis. Recently, investigations in this field have attracted more attention. However, the mechanisms underlying how the oral microbiota participates in regulating cell migration remain to be elucidated. To contribute to the understanding of this issue, this review summarizes the role of cell migration in carcinogenesis and cancer metastasis and focuses on the mechanisms employed by oral microbiota for regulating cell migration. Finally, we introduced critical techniques in the field of cell migration investigation to arouse exciting science over the next decade.
EMT is a cellular biological process involved in tumor cell invasion and metastasis, which was first discovered by Elizabeth Hay during gastrulation in vertebrate embryos in 1982 (Greenburg and Hay, 1982). EMT has been traditionally defined as a process by which epithelial cells lose their vestiges of epithelial origin (e.g., cell–cell adhesion and cell polarity) while migrating and invading into mesenchymal stem cells (Stemmler et al., 2019). Research findings in recent years have found that EMT is not a binary process; there is the hybrid E/M phenotype of cells that originate from epithelial cells in the progression of cancer, known as partial, incomplete, or hybrid EMT (Bakir et al., 2020). The hybrid EMT has been demonstrated to be involved in various human primary cancers (e.g., head and neck cancer) and carcinosarcomas (e.g, esophageal carcinosarcomas) (Pastushenko and Blanpain, 2019). Theoretical and experimental efforts have provided crucial insights into the mechanisms of EMT and the coupling between EMT and other biological processes, such as cell migration, the cell cycle and apoptosis (Lovisa et al., 2015; Aiello et al., 2018; Li et al., 2019; Prakash et al., 2019). The relationship between EMT and cell migration has attracted increasing interest (Aiello et al., 2018; Li et al., 2019). In general, EMT is an important biological process for epithelial-derived cells to acquire migration and invasion during cancer development, especially during metastasis. There is mounting evidence that cancer cells exploit EMT to increase their migratory and invasive ability during the initial stage of the metastatic cascade. Furthermore, the correlation between EMT and migration is type-dependent. A highly specialized epithelial cell is relatively fixed and possesses a low migration ability due to cell–cell adhesions and the apico-basal surface. Therefore, achieving complete mesenchymal transformation is a prerequisite for the migration of a single epithelial cell. During EMT, epithelial cells gain features such as cell elongation, motility, and invasion, coordinated by reorganization of the actin cytoskeleton. In turn, cell migration occurs, which gives rise to single tumor cells capable of crossing basement membranes and invading blood vessels. This process has been utilized by many breast and colorectal cancer cell lines (Aiello et al., 2018). However, the invasion of cancer cells is usually visualized as the migration of groups of cells. The hybrid state of EMT has been associated with increased invasion and collective cell migration, in which cells retain cell–cell adhesion with each other and possess mesenchymal features, such as increased motility and loss of apical–basal polarity. For instance, Campbell et al. demonstrated that the hybrid EMT driven by Snail induced collective cell migration and seeded polyclonal metastases in Drosophila intestinal tumors (Campbell et al., 2019). Overall, cells that undergo EMT obtain increased migratory ability. Key pathways and cellular events that participate in controlling EMT are also considered to be important factors that regulate cell migration.
Three mechanisms of action have been suggested regarding EMT. The first is deconstructing cell junctions and polarity. The typical characteristics of polarized epithelial cells are tight junctions, adhesive junctions, desmosomes, and gap junctions. Over the years, numerous studies have elucidated that the junction complex plays a vital role in EMT as a medium for polarizing cell–cell contact as well as an anchor point for the actin cytoskeleton. Actin-binding proteins (ABPs) and Rho family GTPases participate in regulating the actin cytoskeleton by controlling the polymerization and disintegration of actin filaments (Tokuraku et al., 2020). Adhesive junctions can anchor capillary basal microtubule arrays and participate in collective cell migration via the interaction between E-cadherin and discoidin domain receptor 1 (DDR1) (Gadiya and Chakraborty, 2018). Microtubules and intermediate filaments are also critical for EMT by influencing cell motility, cell shape, intracellular trafficking, and forces to support protrusion (Datta et al., 2021). The second mechanism attributed to EMT is cytoskeletal changes and motility. Active remodeling of the cytoskeleton is crucial for the transformation of cells into a more motile phenotype, thereby promoting EMT (Amack, 2021; Leggett et al., 2021). In the third mechanism, mast regulators regulating gene expression that contribute to the repression of epithelial phenotype and activation of the mesenchymal phenotype drive EMT, including Snail, Slug, Twist1, Zeb1 and Zeb2 (Stemmler et al., 2019).
Since Pott noticed the association between exposure to soot in chimneys and scrotal cancer in 1775, which is considered the first research describing the cause of cancer, an explosion of research evidence on the etiology of cancer has occurred rapidly. The mechanism of carcinogenesis is a complicated process involving the regulation of various levels and pathways. Exogenous substances (e.g., chemicals) or endogenous signals (e.g., reactive intermediates generated from cellular pathways) can lead to mutations in proto-oncogenes or tumor suppressors (Patterson et al., 2018). The biological characteristics, pathways or genetic alterations during cell migration may also be related to cancer. For instance, the Abl family of tyrosine kinases can modulate actin cytoskeleton arrangement through activation of cell-surface receptors that are critical for cell motility and migration (Bradley and Koleske, 2009). They also play an important role in the progression of leukemia and solid tumors. ABL1 has been identified as an oncogene in leukemia. The upregulation and activation of ABL can also be observed in solid tumors (Greuber et al., 2013).
It has been gradually noticed that actin also exists in the nucleus (Rando et al., 2002). Actin in cytoplasm is the main provider for driving force at the front edge and responsible for the contraction of the cell body at the rear edge. Recent studies have found that increasing the level of nuclear actin monomer inhibited cell migration by regulating serum response factor (SRF) and TEA Domain (TEAD) transcription factors expression (Mcneill et al., 2020). Cell migration can be accelerated when the polymerization of nuclear membrane and actin filament is prevented (Fracchia et al., 2020). ABPs combine with actin to make it pass through the nucleus. Actin cytoskeleton dynamics are the basis of cell migration. The polymerization and degradation of actin filaments influenced by ABPs is an essential step of cell migration. ABP can regulate cell activity mediated by actin elaborately (Tokuraku et al., 2020). Additionally, ABPs induce the production of invasive structures such as filopodia (Wang et al., 2017). Meanwhile, actin and ABPs are involved in the process of carcinogenesis. Nuclear actin, as an important part of the chromatin complex, may affect DNA transcription and repair. The levels of actin and ABPs may be connected to chromatin remodeling and upregulation of oncogenes (Izdebska et al., 2020). Different kinds of ABPs in the nucleus (i.e., α-actinin-4, nerspin and cofilin) are associated with the expression of genes responsible for tumorigenesis and influence tumorigenic phenotypes (An et al., 2016; Sur-Erdem et al., 2020).
During cell migration, it is necessary for migrating cells to reduce cell rigidity and pass through narrow intercellular spaces. Genomic changes, including temporary nuclear envelope rupture, DNA damage and genomic rearrangements, can be observed in the process of immune and cancer cell migration (Denais et al., 2016; Raab et al., 2016). DNA damage is an important first step in carcinogenesis. When the replication of damaged DNA takes place before DNA repair tools come into play or the damage process occurs at a high frequency, nuclear deformation-associated genomic instability may lead to various cellular responses, ultimately leading to cancer (Basu, 2018). In the case of other cell migration, it is still necessary to discuss whether DNA will be damaged mechanically and then cause carcinogenesis.
Cancer cell migration is a critical parameter in metastatic dissemination, which allows the cells to detach from the primary tumor, migrate through the extracellular matrix (ECM), enter the lymphatic vessels or the bloodstream, spread within the tissues and then undergo metastatic growth in distant organs. The same principles of cell migration were employed by cancer cells and nonneoplastic cells (e.g., keratinocytes, fibroblasts) as mentioned above. Histopathological evidence has indicated that cancer cells spread within tissues in diverse patterns. They can disseminate using amoeboid- or mesenchymal-type cell migration or collective migration, which expand into solid cell strands, sheets, files, or clusters. Moreover, multiple forms usually exist simultaneously. Amoeboid-like cells often migrate alone or in streams. Mesenchymal-type cells tend to switch between various modes, including single-cell, in streams and collective migration (Clark and Vignjevic, 2015). For instance, oral squamous cell carcinoma (OSCC) exhibits predominantly collective cell migration when explanted in vitro. However, most solid stromal tumors disseminate via individual cells. Overall, these patterns are regulated by the molecular repertoire of cancer cells, which mainly includes integrins, matrix-degrading enzymes, cell–cell adhesion molecules and cell–cell communication. Increased contractility mediated by the Rho pathway in cancer cells migrating individually facilitates amoeboid-like migration (Sahai and Marshall, 2003). Under most conditions, the lower the differentiation stage is, the more likely the cancer cells are to spread through a single cell.
In collective migration, cell clusters retain intercellular connections and combine with ECM through integrins, cadherin, gap junctions, etc. (Shih and Yamada, 2012; Yang et al., 2019). Partial retention of epithelial characteristics and partial EMT allow cells to migrate as clusters (Shih and Yamada, 2012; Cheung and Ewald, 2016; Liao et al., 2021). Cells in migrating clusters are usually divided into two groups: leader cells and follower cells. Leader cells sense the cancer microenvironment and create a low-resistance migration path (Vilchez Mercedes et al., 2021). The traction generated by leader cells can be transmitted along with cell–cell junctions to enable the migration of follower cells (Bazellières et al., 2015). Bronsert found that cells at the metastasis front rarely expressed mesenchymal morphology or EMT markers and speculated that single-cell migration is rare or even absent in most epithelial tumors (Bronsert et al., 2014). Collective cell migration may play a key role in the process of cancer cell metastasis. Moreover, cancer seeding by collective migration has a worse clinical prognosis because of its greater metastatic and proliferative potential (Hou et al., 2012; Aceto et al., 2014).When tumor cells need to pass through the narrow channel of extracellular space in dense three-dimensional matrices, they can establish the polarized distribution of Na+/H+ pump and aquaporin, thus resulting in the inflow of water at the front of cells and the outflow of water at the back of cells, resulting in net cell displacement (Stroka et al., 2014). The method that adjusts the volume through water infiltration provides another possible cell migration mechanism that does not require actin polymerization.
The oral cavity contains one of the greatest microbiological reservoirs in the human body. The dynamic and finely balanced relationship between the oral microbiome and the host is of great importance to human health (Hajishengallis and Lamont, 2021). Bacteria are the largest contributor to the oral resident microbiota, 94% of which is composed of six major phyla (Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, Spirochaetes and Fusobacteria), and 6% contains other phyla (Zhang et al., 2018). Accumulating data support a role for oral bacterial infection in the migration of various types of cells, mainly epithelial cells, and cancer cells. The effect may vary depending on the infection method, time, and cell type (Table 1).
P. gingivalis, the key etiological agent in periodontitis, can successfully invade oral epithelial cells and live intracellularly. It adheres and internalizes to oral cells by modulating host signaling cascades such as phosphorylation/dephosphorylation of molecules and changes in the cell cytoskeleton (Andrian et al., 2006). For instance, P. gingivalis can induce gingival epithelial cells to undergo autophagy and traffic into autophagosome vacuoles to protect itself from targeted lysosome degradation mediated by selective ubiquitin (Lee et al., 2018). P. gingivalis expresses virulence and releases outer membrane vesicles (OMVs), which are involved in the communication between bacteria and the host. In general, P. gingivalis, its derivatives (e.g., heated-killed or conditioned medium from bacteria) and secreted substances (OMVs and Ltp1) can promote the migration of oral epithelial cells and OSCC cells (Inaba et al., 2014; Ha et al., 2015; Ha et al., 2016; Geng et al., 2017; Lee et al., 2017; Abdulkareem et al., 2018a; Hoppe et al., 2019; Ohshima et al., 2019; Kamarajan et al., 2020; Fitzsimonds et al., 2021; Liu et al., 2021a; Liu et al., 2021b). Comprehensive analysis of the host transcription response to P. gingivalis infection showed the mode of increased cell migration (Geng et al., 2019). The colonization of P. gingivalis has been identified as a risk factor for OSCC and is related to prognosis (Ganly et al., 2019). However, the inhibitory effect has also been observed in some studies (Laheij et al., 2013; Bhattacharya et al., 2014). This may suggest that bacterial challenge may hinder the re-epithelialization process and lead to delayed healing after the barrier function of the gingival epithelium is destroyed. We speculated that the possible reason for such a diametrically opposed situation is the relatively short infection time or different sources and locations of cells. It may also be due to differences in protocols between continuous infection and long-term culture with a short infection followed by a medium change. When serial infection experiments with P. gingivalis are performed in vitro, cells often float alive due to strong protease activity.
As a symbiotic bacterium, F. nucleatum is commonly found in the oral cavity. It expresses a variety of adhesins, such as FadA, on the surface to adhere to and invade host cells. Similarly, F. nucleatum can also release extracellular vesicles (EVs) or OMVs (Liu et al., 2019; Liu et al., 2021). Studies have shown that infection with F. nucleatum ATCC 25586 and heated-killed F. nucleatum ATCC 10953 promotes OSCC cell migration and invasion in vitro (Abdulkareem et al., 2018a; Kamarajan et al., 2020; Shao et al., 2021). The effect of its derivatives, secretions and virulence factors on cell migration still needs further study.
Butyric acid (BA), a short-chain fatty acid, is produced by periodontopathic bacteria, such as P. gingivalis and F. nucleatum. Large amounts of BA have been detected in the oral cavities of patients with periodontal disease. BA from periodontopathic bacteria does not directly affect the migration of ameloblastomas but has an indirect influence through the expression of epidermal growth factor (EGF) and transforming growth factor β1 (TGF-β1) (Ishikawa et al., 2020). Other oral bacteria are also related to cell migration. Oral streptococci, such as Streptococcus gordonii, is an important component of the oral microbiome. S. gordonii has been considered an early colonizer to host tissues. It can resist ZEB2 induction by P. gingivalis and then inhibit cell migration (Ohshima et al., 2019). The specific molecular mechanism of oral bacteria related to cell migration, interaction between different kinds of bacteria and its effect on cell migration deserve further study.
There is already evidence that oral bacteria can spread to other body sites through the bloodstream or digestive tract and participate in a variety of systemic diseases (Fiorillo et al., 2019). It may affect the pathogenesis and progression of diseases via cell migration and metastasis. P. gingivalis increases the migration of esophageal squamous cell carcinoma (Liang et al., 2020; Qi et al., 2020). F. nucleatum has been found to be enriched in colorectal cancer (CRC) tissue and acts as a pro-carcinogenic bacterium (Wang et al., 2021). It promotes CRC cancer migration in vitro and metastasis in vivo (Chen et al., 2020; Xu et al., 2021a). In addition, the metastases in mice in which breast cancer developed after infection with F. nucleatum were larger than those in the control group (Parhi et al., 2020). P. gingivalis has been found in the brains of Alzheimer’s disease patients and is involved in the disease process (Dominy et al., 2019). Gingipains, cysteine proteases secreted by P. gingivalis, induces cell migration and membrane ruffling in the human embryonic microglia clone 3 (HMC3) cell line through the protease-activated receptor 2 (PAR2)/ERK1/2 pathway (Nonaka and Nakanishi, 2020). This signaling pathway may be necessary for embryonic microglial cells to move into the infection sites.
Dental professionals are usually familiar with viruses present or cause symptoms in the oral cavity. In a broad sense, they can both be defined as oral viruses. There is not always a clear distinction between them. For example, viruses that induce symptoms affecting oral tissues can be present in the oral cavity following replication and release from other tissues or the blood circulation. Prolonged infection of certain viral genes can insert into host DNA as proto-oncogenes to turn them into oncogenes, leading to malignant transformation (Young et al., 2016; Warburton et al., 2018). There is emerging evidence that Epstein–Barr virus (EBV), as an oncogenic virus, promotes OSCC progression. P53 can promote the emergence of leader cells and then coordinate the migration of epithelial cells (Kozyrska et al., 2022). EBV decreases the stability of P53 and increases the expression of matrix metalloproteinase (MMP) through CTAR family proteins/programmed cell death protein 1 ligand, thereby promoting OSCC cell metastasis and tumorigenesis (Radaic and Kapila, 2021). Human papillomavirus (HPV) is detectable in 40-70% of oropharyngeal cancer (OPC) and 20% of non-OPC, almost the HPV16 subtype (Mehanna et al., 2013). The relationship between HPV and cervical cancer has been generally acknowledged. HPV16 can lead to enhanced migration and invasion of cervical cancer cells in vitro and in a mouse model (Hu et al., 2015; Wang et al., 2019).
Fungi are common, albeit minor, members of the oral microbiota, which are structurally and metabolically distinct from other oral microorganisms, such as bacteria and viruses. They have varied cell morphologies, including yeast, hyphae, pseudo hyphae, and chlamydospores. Although filamentous fungi can rarely be isolated from the oral cavity, yeast can be cultured from the saliva of approximately 40% of individuals, such as Candida tropicalis, Candida dubliniensis, and Candida glabrata. The most frequent oral fungal infection is Candida albicans. C. albicans significantly increased in OSCC patients compared with the control group (Hong et al., 2020). C. albicans can enhance the migratory ability of oral keratinocytes through activation of the ERK/focal adhesion kinase (FAK) pathway (Shi et al., 2009). Exposure to different concentrations of C. glabrata and Candida kefyr (MOI=1,10) was found to inhibit human buccal epithelial cell migration in vitro (Haverman et al., 2017). This inhibitory effect may play a role in the process of ulcerative mucositis.
Migration is a multistep process. First, the cells are polarized in response to the migration-promoting stimulus, and then the membrane protrusions extend in the direction of the stimulus (Lauffenburger and Horwitz, 1996). Then, the adhesion of the protrusions to the ECM produces traction, which allows the movement of the cell, disassembly of the adhesion points and retraction of the back of the cell (Ridley et al., 2003). Collective and single-cell migration share these broad mechanistic characteristics (Yang et al., 2019). Studies have found that oral microbes mainly regulate cell migration by affecting cell adhesion, polarization, guidance, etc. (Figure 1).
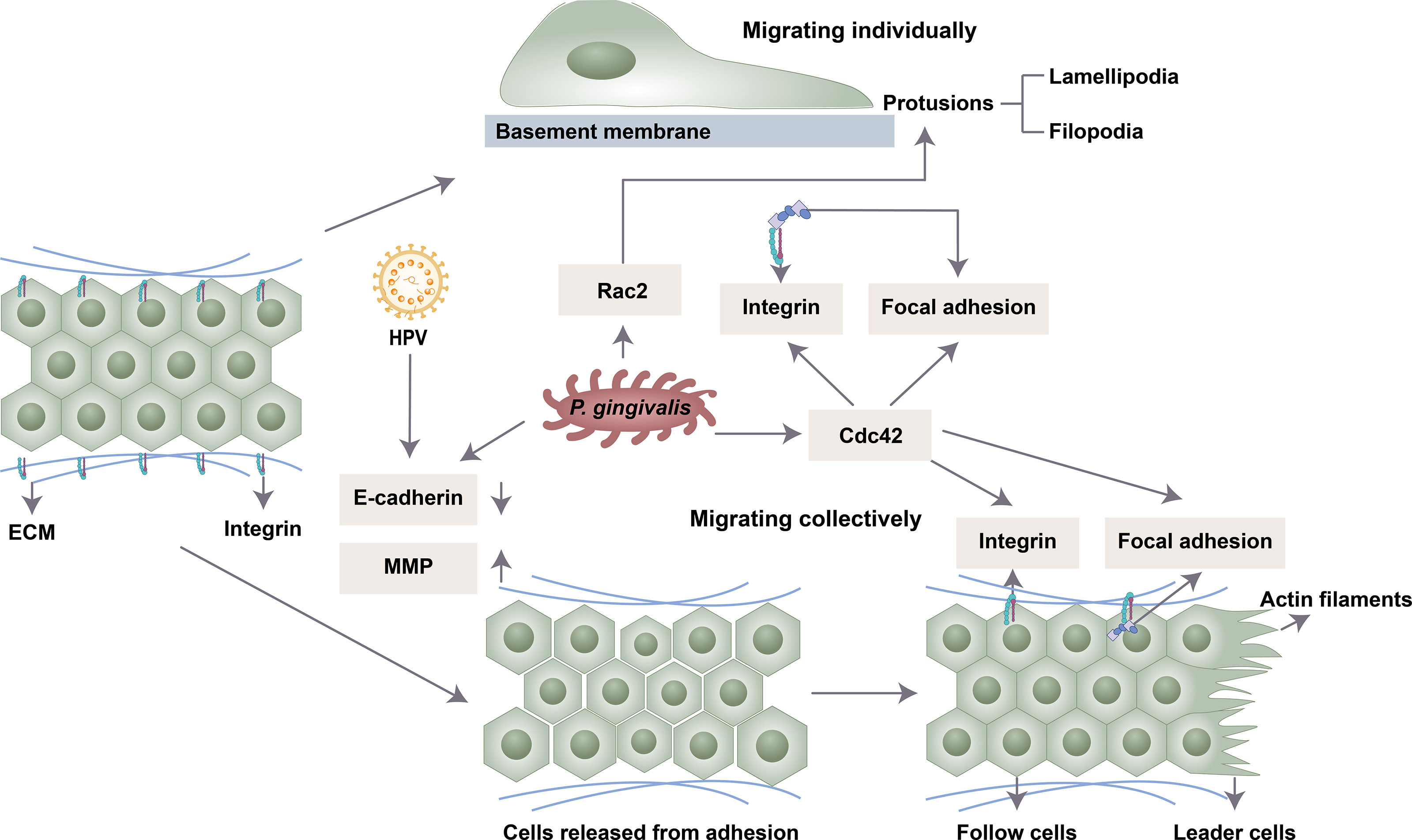
Figure 1 Possible mechanisms by which oral microbiota influence cell migration. P. gingivalis and HPV 16 release intercellular adhesions by downregulating E-cadherin and upregulating MMP. P. gingivalis enhances the expression of Cdc42 and Rac2 to induce the front and back polarity that is crucial for cell migration. P. gingivalis drives cell clusters to move orientally through the formation of integrin and focal adhesion.
Intercellular adhesion is mediated by different types of junction complexes, including tight junctions, adhesive junctions, gap junctions and desmosomes (Rusu and Georgiou, 2020). Oral microbiota can degrade epithelial cell tight junction proteins in an MMP-dependent manner or by affecting the activity of transcription factors (Ha et al., 2015). The shape of infected cells became slender, indicating absent contact inhibition, and cell junctions were weakened under a transmission electron microscope (Geng et al., 2017). MMP is involved in focal degradation of type IV collagen, breakdown of basement membrane and release of cell adhesion (Strutz et al., 2002). MMP-1, MMP-2, MMP-3, MMP-7 and MMP-9 were upregulated following exposure to P. gingivalis stimulation (Ha et al., 2016; Lee et al., 2017). One mechanism is that gingipains of P. gingivalis process the proenzyme of MMP-9 into active MMP-9 to promote cell migration and invasion (Inaba et al., 2014; Inaba et al., 2015). This process is regulated by the PAR4, p38/HSP27, ERK1/2-Ets1, and PAR2/NF-κB pathways (Inaba et al., 2014; Inaba et al., 2015). P. gingivalis could also activate the expression of MMP-2 and MMP-9 by increasing the transcription of nicotinamide N-methyltransferase (NNMT) and Gas6 (Tang et al., 2011; Geng et al., 2017).
E-cadherin, the central protein of cell–cell adherens junctions, is downregulated by a series of transcription factors (Slug, Snail, JAG1, Notch, Zeb1 and Zeb2) (Wu and Zhou, 2010). P. gingivalis can significantly enhance the activity of these transcription factors (Geng et al., 2017). Moreover, P. gingivalis infection could inhibit the expression of E-cadherin and increase the expression of Snail in oral epithelial cells by upregulating the transcription of colon cancer associated transcript 1 (CCAT1) and growth arrest specific 6 (Gas6) (Jiang et al., 2015; Guo and Hua, 2017). HPV-16 may also facilitate the migration and metastasis potential of cervical cancer through altered cadherin switching (Hu et al., 2015).
Polarity is characteristic of epithelial cells. In polarized epithelial cells, the intercellular junction complex is located asymmetrically. The spatial asymmetry of these complexes is mediated by a class of evolutionarily conserved proteins, which can be divided into three functional groups: the Crumbs2 complex, Scribble complex and Par complex. The apical domain is related to the Crumbs complex, the basolateral domain is composed of the Scribble complex, and the subapical region of the apical-basal boundary is related to the Par complex, which is composed of Par3, Par6, and atypical protein kinase C and Cdc42.
In single-cell migration, migrating epithelial cells tend to lose apical-basal polarity and rearrange their actin cytoskeleton (Gibieža and Petrikaitė, 2021). The protusions of the leading edge drive the cell to orientally migrate, and the main force of migrating is generated by the lamellipodia. Myoslin II and Rho catalyze the formation of new cell-ECM adhesion (focal adhesions) during lamellipodia extension. The stress fibers are joined to mature focal adhesions, connecting the cell to the ECM. In collective migration, cells in different positions show different expression patterns. The simple case is front/rear polarization. The leading cells with a mesenchymal phenotype guide the following cells to retain epithelial characteristics. This process is coordinated by cell-ECM and cell–cell interactions.
Rho family GTPases include RhoA, Rac and Cdc42 proteins, which participate in the formation of cytoskeletal components by inducing the accumulation of F-actin in the front of the cell (Ridley, 1999). Rho GTPases regulate cytoskeletal actin rearrangement and thus cell dynamics (Sahai and Marshall, 2003; Lawson and Ridley, 2018). Polar proteins regulated by GTPase enzymes of the Rho family induce the formation of the front and rear axes (Capuana et al., 2020). Rac protein is closely related to the formation of lamellipodia and cell migration. The Rho-dependent localization of myosin IIB in the back of the cell is necessary for the maintenance of front and back polarity and tail contraction during the process of mesenchymal migration (Vicente-Manzanares et al., 2008). Cdc42 mainly produces front and back polarity. The local activation of Cdc42 and its spatial gradient in nonpolarized cells drive the formation of the initial protruding front under uniform chemotaxis stimulation (Yang et al., 2016). When the apical junction complex is destroyed, Cdc42 and the polar protein complex relocate from the tight junction area to the front and induce the centrosome and Golgi to relocate to the front of the cell, promoting the growth of microtubules to the front of the cell and subsequent cell migration (Burute et al., 2017). Therefore, at the front end, Cdc42 and Rac promote actin polymerization, thereby promoting the formation of protrusions such as filopodia or lamelia. Increased actin reorganization and Cdc42 and Rac activity are observed in OSCC (Iwai et al., 2010). P. gingivalis enhanced the expression of Rac2 and Cdc42 in platelets and neutrophils (Börgeson et al., 2011; Senini et al., 2019). P. gingivalis fimbriae induced transendothelial migration of monocytes by activating Rac1 and PI3K (Harokopakis et al., 2006). Its effect on oral epithelial cells needs further study.
Integrin alpha V and FAK signals help mediate cell migration (Trepat et al., 2012).Cell migration depends on the binding of integrins to the ECM, which activates downstream signaling, including FAK phosphorylation and mitogen-activated protein kinase (MAPK), to recruit focal contacts. Several integrins, such as beta-6 integrin, can facilitate cell migration through actin cytoskeletal reorganization and cell polarization. P. gingivalis inhibits the induction of integrin beta-3 and -6 and the cell migratory process in oral keratinocytes (Bhattacharya et al., 2014). Periodontal pathogens (P. gingivalis, F. nucleatum, and T. denticola) promote OSCC cell migration through the activation of integrin alpha V and FAK (Kamarajan et al., 2020). The E6 protein of HPV could also promote actin cytoskeleton assembly through β1-integrin signaling (Holloway and Storey, 2014).
The other strategy employed by oral microbiota to manipulate cell migration is modulating the EMT process (Figure 2). Specifically, microbial dysbiosis results in the degradation of epithelial tight junction proteins, enhances mesenchymal characteristics, and induces at least a portion of the EMT process. For instance, P. gingivalis regulates epithelial barrier function through the degradation of E-cadherin (Sztukowska et al., 2016; Abdulkareem et al., 2018a). P. gingivalis and F. nucleatum could also enhance the expression of EMT-associated transcription factors (Zeb1, Zeb2, Slug, Snail, Jag1, Notch, Twist, OLFM4 and RGCC) in oral epithelial cells and OSCC cells through diverse pathways, including the phosphorylation of glycogen synthase kinase-3β (GSK‐3β), EGF, tumor necrosis factor-α (TNF-α) and TGF-β1 (Bhattacharya et al., 2014; Sztukowska et al., 2016; Abdulkareem et al., 2018a; Abdulkareem et al., 2018b). S. gordonii can resist ZEB2 induction by P. gingivalis by suppressing FOXO1 and activating the TAK1-NLK negative regulatory pathway (Ohshima et al., 2019). F. nucleatum can activate the signal transducer and activator of transcription 3 (STAT3) signaling pathway, which can increase the expression of EMT-associated genes (E-cadherin, Snail and Twist) (Huang et al., 2011; Wang et al., 2020). The upregulation of partial EMT genes is also observed in F. nucleatum-infected OSCC cells (Shao et al., 2021).
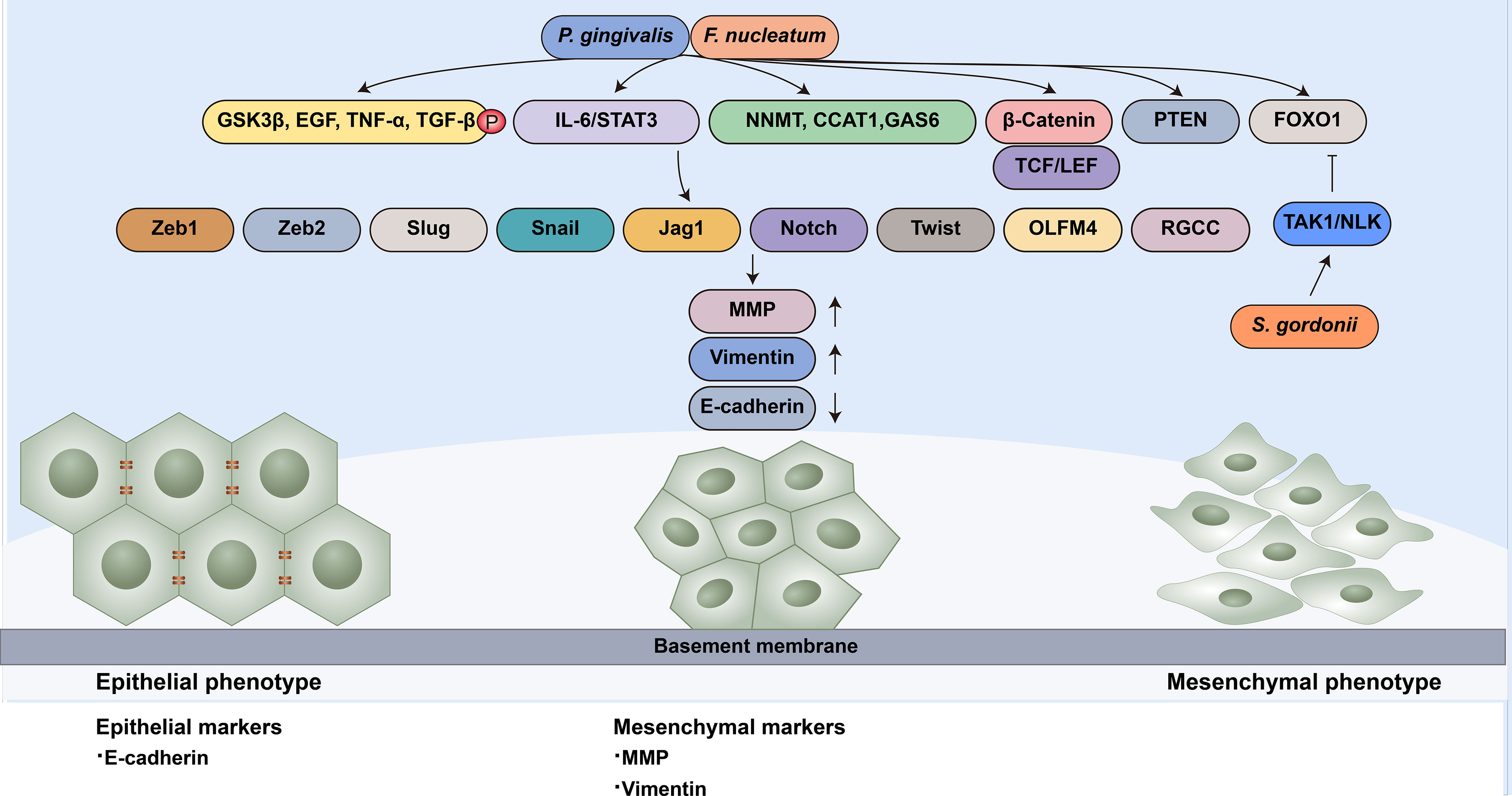
Figure 2 Oral microbiota guide epithelial-mesenchymal transition of normal epithelial cells. EMT-associated transcription factors (Zeb1, Zeb2, Slug, Snail, Jag1, Notch, Twist, OLFM4 and RGCC) can induce epithelial cells to undergo partial or complete mesenchymal transformation by downregulating epithelial markers (such as E-cadherin) and upregulating mesenchymal markers (MMP and Vimentin). P. gingivalis and F. nucleatum enhance the expression of EMT-associated transcription factors through diverse pathways: (1) the phosphorylation of GSK‐3β, EGF, TNF-α and TGF-β1; (2) the activation of the IL-6/STAT3 pathway; (3) the upregulation of NNMT, CCAT1 and GAS6;(4) the nuclear translocation of β-catenin and further activation TCF/LEF promoter elements; (5) the activation of PTEN; and (6) the upregulation of FOXO1 S. gordonii can resist EMT-associated factor induction by P. gingivalis by suppressing FOXO1 and activating the TAK1-NLK negative regulatory pathway.
Wnt/β-catenin facilitates EMT in cancer progression (Liu et al., 2017; Li et al., 2021). β-catenin in the nucleus binds to the T cell factor/lymphoid enhancer factor (TCF/LEF) transcription factor, the major end point mediators of Wnt/β-catenin signaling, to activate EMT. Vimentin is the target of Wnt/β-catenin signaling. The nuclear translocation of β-catenin and activation of vimentin can be observed in OSCC cells, which are related to poor prognosis (Chaw et al., 2012). The gingipains of P. gingivalis modulate the β-catenin pathway in gingival epithelial cells and the disassociation of the β-catenin destruction complex composed of scaffolding proteins and the kinases GSK3β and Casein Kinase 1α (CK1α). It can induce nuclear translocation of β-catenin and further activate TCF/LEF promoter elements (Zhou et al., 2015).
It has been proven that several genes are positively relevant to cell migratory and invasive ability, such as CCAT1, NNMT and Gas6. CCAT1 and Gas6 can downregulate E-cadherin and upregulate EMT-associated transcription factors, such as Snail and Twist (Jiang et al., 2015; Guo and Hua, 2017). The mRNA expression of NNMT, CCAT1 and GAS6 was increased in P. gingivalis-infected oral epithelial cells (Geng et al., 2017).
Keratin 7 (KRT7) is a type II cytokeratin and is involved in cell motile activity. It has been proven to be relevant to lymph node metastasis and poor prognosis in CRC (Bayrak et al., 2011; Hrudka et al., 2021). F. nucleatum upregulates the long noncoding RNA KRT7-antisense and stabilizes KRT7 mRNA via the NF-κB pathway (Chen et al., 2020). HPV-16 enhances actin polymerization by downregulating alpha-actinin-4 (ACTN4), leading to enhanced migration and invasion (Tentler et al., 2019; Wang et al., 2019).
With the development of new microscopy methods and fluorescent reagents specifically used for cell imaging, microscopy technology plays a central role in the research of cell biophysics. Optical microscopes allow us to view cell structures with previously unattainable spatial and temporal resolution and to image living cells in tissues and animals. Improvements in electron microscopy technology allow us to understand the molecular structure of organelles in cells in more detail. In recent years, an increasing number of technologies have emerged, such as video real-time microscopy, confocal microscopy, multiphoton microscopy, intravital microscopy, superresolution fluorescence microscopy, electrochemiluminescence microscopy, and traction force microscopy, which make the observation and analysis of cell migration more accurate and intuitive (Horwitz, 2016).
Most biological research relies on conventional experimental techniques, and static analysis is only allowed at certain time points in vitro. Visualizing cell dynamics in organisms can provide opportunities to study key biological phenomena in vivo. However, electron microscopy is usually not suitable for live or wet samples due to the need for vacuum operation conditions. Soon after the first compound microscope was invented in 1595, the intravital microscope (IVM) was used for physiological research. IVM can be combined with a variety of optical systems, such as confocal and multiphoton microscopy, to conduct deeper observations of tissues and directly observe the biological structure and dynamic behavior of objects, including single cells and living animals. With these characteristics, IVM can be used to visualize the biological morphology of various fields, such as vascular biology, immunology, stem cell biology and oncology (Choo et al., 2020). For instance, in the field of oncology, IVM can be used to observe the single-cell behavior of cancer cells and immune cells during tumor progression and metastasis (Ng et al., 2008; Pinner et al., 2009; Gabriel et al., 2018; Kuo et al., 2019). To overcome the obstacle of the traditional IVM that the limited frame rate cannot accurately observe the rapid dynamic behavior of cells, a real-time IVM capable of video image scanning (over 30 frames/sec) has been invented to visualize faster cell movement and further study cell functions and the interactions between cells (Padera et al., 2002). Moreover, the Boyden chamber assay and scratch wound assay are standard techniques for studying cell invasion and migration, but both techniques have their own limitations. The Boyden chamber assay is difficult and time-consuming, while the scratch wound assay has low repeatability. The real-time video microscope can be introduced into the incubator to generate real-time images of cell migration, which can provide accurate quantitative data for wound healing and is used to provide automatic real-time analysis of cell migration, which improves repeatability (Jain et al., 2012). This video microscope-based scratch experiment has proven to be a reliable technique for evaluating cell migration and invasion (Guy et al., 2017).
In addition, superresolution fluorescence microscopy, harmonic generation microscopy, electrochemiluminescence microscopy, and traction force microscopy are all powerful tools for studying cell migration. The5 superresolution microscope overcomes the limitations of conventional optical microscopes in resolution, dimensionality, quantification, and imaging speed. It can be used to observe the cellular processes of single cells with nanometer-level resolution, contributing to the understanding of rapid cell dynamics with a more visualized subcellular and molecular scale. Increasing methods have been developed to achieve superresolution, such as stimulated emission depletion (STED), structured illumination microscopy (SIM), photoactivation localization microscopy (PALM), stochastic optical reconstruction microscopy (STORM), and superresolution optical fluctuation imaging (SOFI) (Feng et al., 2018). These realize the nanoscale, visualization and quantitative analysis of signaling pathway molecules such as membrane proteins (Xu et al., 2021b). Second harmonic generation (SHG) is a second-order nonlinear optical process. Two photons interacting with nonlinear optical materials, such as collagen, combine to form a new photon whose frequency is twice that of the original photon, and the wavelength is halved (Wolf et al., 2009). SHG microscopy is a high-resolution nondestructive imaging method that represents an ideal method for detecting the geometry of collagen in natural and/or connective tissues. It has been used to study the morphology of fibrous collagen in a variety of tissues, which helps to understand the structure of collagen in tissues under normal or abnormal conditions. Unlike SHG microscopy, which requires specific asymmetry of the imaging structure, third harmonic generation (THG) is a combination of three photons converted into a photon with one third of the excitation wavelength and three times the energy. Therefore, compared to SHG, the application range of THG is wider. The combination of SHG or THG with fluorescence detection and intravital microscopy provides more details about tissue-tissue and cell-tissue interactions, which helps to simulate the migration of tumor cells in the tissue in a specific environment (Weigelin et al., 2012). Electrochemiluminescence microscopy can provide a clear visual contrast between the adhesion site and the noncontact domain so that the former can be selected and displayed in a label-free manner to image the cell matrix adhesion of the moving cell clusters to study the movement of cells in collective migration (Ding et al., 2020). Traction force microscopy is an experimental technique used to quantify the contractile force produced by adherent cells by placing the cells on a flexible material, such as polyethylene glycol or polyacrylamide gel with a known elastic modulus, track the displacement of the substrate caused by the cell contraction, and then convert it into a traction field (Hur et al., 2020). Traction force microscopy with integrated microfluidics can precisely control physical and chemical stimulation to detect the migration speed, traction and intercellular tension of cell clusters under different chemical gradients (Jang et al., 2019).
In the limited three-dimensional (3D) space, the situation of cell migration is more complex, which is difficult to observe by traditional methods. New engineering model systems, such as hydrogels and microchannel assays provide new insights into 3D cell migration. The synthesized hydrogel can independently modulate the hardness, composition, degradability, or other characteristics to analyze the role of a single characteristic of ECM in cell migration (Trappmann et al., 2017). Therefore, it provides a platform to see how cancer cells respond to different biochemical and mechanical signals. Other types of cells can also be introduced into these hydrogels to simultaneously observe cell-matrix and cell-cell interactions (Katz and West, 2022). Confined microchannel approach can introduce and adjust the interface geometry to explore changes in cytoskeleton, adhesion, and regulatory proteins induced by different experimental microenvironments (Mak et al., 2011). Polydimethylsiloxane (PDMS) microchannel devices are also widely used to study migration in 3D confinement. Microchannel system allows direct and real-time imaging and explores the mechanism of migration under confined conditions without shear stress. Microchannel coated with different ECM proteins can be applied to explore cells’ response to external gradients. In addition, the limited migration space in vivo can also be simulated by grooved substrates, micropatterned lines and islands, vertical confinement, patterned gels, and so on (Paul et al., 2016).
Studies have shown that the oral microbiota can regulate the carcinogenic process of cells, and this process may involve abnormal cell migration. Partial or complete EMT is related to increased migratory ability and participates in cancer invasion and metastasis. Specific genetic changes and signaling pathway activation during cell migration may also be seen in EMT, tumorigenic phenotypes and cancer metastasis. Collective migration is the main form of cancer invasion, and it is associated with worse prognosis. Oral microbial dysbiosis is related to abnormal cell migration of oral epithelial cells. Increasing evidence has shown that P. gingivalis, F. nucleatum, Streptococci and their virulence factors regulate cell migration, whose effects vary with infection methods, time, and cell types. Some oral viruses and fungi can affect cell migration, and further study is needed. The oral microbiota also participates in systemic diseases through cell migration. The mechanism by which the oral microbiota regulates cell migration includes cell adhesion, polarization, guidance, EMT, etc. real-time microscopy, superresolution microscopy, harmonic microscopy and traction microscopy are widely used to observe and analyze the migration process. We expect that with further development of microscope imaging technology, the relevant mechanism can be observed more intuitively and accurately.
HB and JY drafted the manuscript. SM and CL edited and added valuable insights to the manuscript. All authors approved the final manuscript and agreed to be accountable for all aspects of the work.
This study was supported by the National Natural Science Foundation of China (82071108).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Porphyromonas gingivalis, P. gingivalis; Fusobacterium nucleatum, F. nucleatum; Treponema denticola, Treponema denticola; epithelial to mesenchymal transition, EMT; Actin-binding proteins, ABPs; discoidin domain receptor 1, DDR1; oral squamous cell carcinoma, OSCC; extracellular matrix, ECM; outer membrane vesicles, OMVs; extracellular vesicles, EVs; Butyric acid, BA; epidermal growth factor, EGF; transforming growth factor β1, TGFβ1; Streptococcus gordonii, S. gordonii; Epstein–Barr virus, EBV; matrix metalloproteinase, MMP; human papillomavirus, HPV; oropharyngeal cancer, OPC; nicotinamide N-methyltransferase, NNMT; growth arrest specific 6, Gas6; focal adhesion kinase, FAK; T cell factor/lymphoid enhancer factor, TCF/LEF; glycogen synthase kinase-3β, GSK‐3β; tumour necrosis factor-α, TNF-α; Keratin 7, KRT7; intravital microscope, IVM; Second harmonic generation, SHG; third harmonic generation, THG; three-dimensional, 3D; polydimethylsiloxane, PDMS.
Abdulkareem, A. A., Shelton, R. M., Landini, G., Cooper, P. R., Milward, M. R. (2018a). Periodontal Pathogens Promote Epithelial-Mesenchymal Transition in Oral Squamous Carcinoma Cells In Vitro. Cell Adh. Migr. 12, 127–137. doi: 10.1080/19336918.2017.1322253
Abdulkareem, A. A., Shelton, R. M., Landini, G., Cooper, P. R., Milward, M. R. (2018b). Potential Role of Periodontal Pathogens in Compromising Epithelial Barrier Function by Inducing Epithelial-Mesenchymal Transition. J. Periodontal Res. 53, 565–574. doi: 10.1111/jre.12546
Aceto, N., Bardia, A., Miyamoto, D. T., Donaldson, M. C., Wittner, B. S., Spencer, J. A., et al. (2014). Circulating Tumor Cell Clusters are Oligoclonal Precursors of Breast Cancer Metastasis. Cell 158, 1110–1122. doi: 10.1016/j.cell.2014.07.013
Aiello, N. M., Maddipati, R., Norgard, R. J., Balli, D., Li, J., Yuan, S., et al. (2018). EMT Subtype Influences Epithelial Plasticity and Mode of Cell Migration. Dev. Cell 45, 681–695.e684. doi: 10.1016/j.devcel.2018.05.027
Alipour, M. (2021). Molecular Mechanism of Helicobacter Pylori-Induced Gastric Cancer. J. Gastrointest. Cancer 52, 23–30. doi: 10.1007/s12029-020-00518-5
Amack, J. D. (2021). Cellular Dynamics of EMT: Lessons From Live In Vivo Imaging of Embryonic Development. Cell Commun. Signal 19, 79. doi: 10.1186/s12964-021-00761-8
Andrian, E., Grenier, D., Rouabhia, M. (2006). Porphyromonas Gingivalis-Epithelial Cell Interactions in Periodontitis. J. Dent. Res. 85, 392–403. doi: 10.1177/154405910608500502
An, H. T., Yoo, S., Ko, J. (2016). α-Actinin-4 Induces the Epithelial-to-Mesenchymal Transition and Tumorigenesis via Regulation of Snail Expression and β-Catenin Stabilization in Cervical Cancer. Oncogene 35, 5893–5904. doi: 10.1038/onc.2016.117
Bakir, B., Chiarella, A. M., Pitarresi, J. R., Rustgi, A. K. (2020). EMT, MET, Plasticity, and Tumor Metastasis. Trends Cell Biol. 30, 764–776. doi: 10.1016/j.tcb.2020.07.003
Basu, A. K. (2018). DNA Damage, Mutagenesis and Cancer. Int. J. Mol. Sci. 19, 970. doi: 10.3390/ijms19040970
Bayrak, R., Yenidünya, S., Haltas, H. (2011). Cytokeratin 7 and Cytokeratin 20 Expression in Colorectal Adenocarcinomas. Pathol. Res. Pract. 207, 156–160. doi: 10.1016/j.prp.2010.12.005
Bazellières, E., Conte, V., Elosegui-Artola, A., Serra-Picamal, X., Bintanel-Morcillo, M., Roca-Cusachs, P., et al. (2015). Control of Cell-Cell Forces and Collective Cell Dynamics by the Intercellular Adhesome. Nat. Cell Biol. 17, 409–420. doi: 10.1038/ncb3135
Bhattacharya, R., Xu, F., Dong, G., Li, S., Tian, C., Ponugoti, B., et al. (2014). Effect of Bacteria on the Wound Healing Behavior of Oral Epithelial Cells. PloS One 9, e89475. doi: 10.1371/journal.pone.0089475
Börgeson, E., Lönn, J., Bergström, I., Brodin, V. P., Ramström, S., Nayeri, F., et al. (2011). Lipoxin A₄ Inhibits Porphyromonas Gingivalis-Induced Aggregation and Reactive Oxygen Species Production by Modulating Neutrophil-Platelet Interaction and CD11b Expression. Infect. Immun. 79, 1489–1497. doi: 10.1128/IAI.00777-10
Bradley, W. D., Koleske, A. J. (2009). Regulation of Cell Migration and Morphogenesis by Abl-Family Kinases: Emerging Mechanisms and Physiological Contexts. J. Cell Sci. 122, 3441–3454. doi: 10.1242/jcs.039859
Bronsert, P., Enderle-Ammour, K., Bader, M., Timme, S., Kuehs, M., Csanadi, A., et al. (2014). Cancer Cell Invasion and EMT Marker Expression: A Three-Dimensional Study of the Human Cancer-Host Interface. J. Pathol. 234, 410–422. doi: 10.1002/path.4416
Burute, M., Prioux, M., Blin, G., Truchet, S., Letort, G., Tseng, Q., et al. (2017). Polarity Reversal by Centrosome Repositioning Primes Cell Scattering During Epithelial-To-Mesenchymal Transition. Dev. Cell 40, 168–184. doi: 10.1016/j.devcel.2016.12.004
Campbell, K., Rossi, F., Adams, J., Pitsidianaki, I., Barriga, F. M., Garcia-Gerique, L., et al. (2019). Collective Cell Migration and Metastases Induced by an Epithelial-to-Mesenchymal Transition in Drosophila Intestinal Tumors. Nat. Commun. 10, 2311. doi: 10.1038/s41467-019-10269-y
Capuana, L., Bostrom, A., Etienne-Manneville, S. (2020). Multicellular Scale Front-to-Rear Polarity in Collective Migration. Curr. Opin. Cell Biol. 62, 114–122. doi: 10.1016/j.ceb.2019.10.001
Chaw, S. Y., Abdul Majeed, A., Dalley, A. J., Chan, A., Stein, S., Farah, C. S. (2012). Epithelial to Mesenchymal Transition (EMT) Biomarkers–E-Cadherin, Beta-Catenin, APC and Vimentin–in Oral Squamous Cell Carcinogenesis and Transformation. Oral. Oncol. 48, 997–1006. doi: 10.1016/j.oraloncology.2012.05.011
Chen, S., Su, T., Zhang, Y., Lee, A., He, J., Ge, Q., et al. (2020). Fusobacterium Nucleatum Promotes Colorectal Cancer Metastasis by Modulating KRT7-As/Krt7. Gut Microbes 11, 511–525. doi: 10.1080/19490976.2019.1695494
Cheung, K. J., Ewald, A. J. (2016). A Collective Route to Metastasis: Seeding by Tumor Cell Clusters. Science 352, 167–169. doi: 10.1126/science.aaf6546
Choo, Y. W., Jeong, J., Jung, K. (2020). Recent Advances in Intravital Microscopy for Investigation of Dynamic Cellular Behavior In Vivo. BMB Rep. 53, 357–366. doi: 10.5483/BMBRep.2020.53.7.069
Christiansen, J. J., Rajasekaran, A. K. (2006). Reassessing Epithelial to Mesenchymal Transition as a Prerequisite for Carcinoma Invasion and Metastasis. Cancer Res. 66, 8319–8326. doi: 10.1158/0008-5472.CAN-06-0410
Clark, A. G., Vignjevic, D. M. (2015). Modes of Cancer Cell Invasion and the Role of the Microenvironment. Curr. Opin. Cell Biol. 36, 13–22. doi: 10.1016/j.ceb.2015.06.004
Datta, A., Deng, S., Gopal, V., Yap, K. C., Halim, C. E., Lye, M. L., et al. (2021). Cytoskeletal Dynamics in Epithelial-Mesenchymal Transition: Insights Into Therapeutic Targets for Cancer Metastasis. Cancers (Basel) 13, 1882. doi: 10.3390/cancers13081882
Denais, C. M., Gilbert, R. M., Isermann, P., Mcgregor, A. L., Te Lindert, M., Weigelin, B., et al. (2016). Nuclear Envelope Rupture and Repair During Cancer Cell Migration. Science 352, 353–358. doi: 10.1126/science.aad7297
Ding, H., Guo, W., Su, B. (2020). Imaging Cell-Matrix Adhesions and Collective Migration of Living Cells by Electrochemiluminescence Microscopy. Angew. Chem. Int. Ed. Engl. 59, 449–456. doi: 10.1002/anie.201911190
Dominy, S. S., Lynch, C., Ermini, F., Benedyk, M., Marczyk, A., Konradi, A., et al. (2019). Porphyromonas Gingivalis in Alzheimer’s Disease Brains: Evidence for Disease Causation and Treatment With Small-Molecule Inhibitors. Sci. Adv. 5, eaau3333. doi: 10.1126/sciadv.aau3333
Feng, H., Wang, X., Xu, Z., Zhang, X., Gao, Y. (2018). Super-Resolution Fluorescence Microscopy for Single Cell Imaging. Adv. Exp. Med. Biol. 1068, 59–71. doi: 10.1007/978-981-13-0502-3_6
Fiorillo, L., Cervino, G., Laino, L., D’amico, C., Mauceri, R., Tozum, T. F., et al. (2019). Porphyromonas Gingivalis, Periodontal and Systemic Implications: A Systematic Review. Dent. J. (Basel) 7, 114. doi: 10.3390/dj7040114
Fitzsimonds, Z. R., Liu, C., Stocke, K. S., Yakoumatos, L., Shumway, B., Miller, D. P., et al. (2021). Regulation of Olfactomedin 4 by Porphyromonas Gingivalis in a Community Context. Isme J. 15, 2627–2642. doi: 10.1038/s41396-021-00956-4
Fitzsimonds, Z. R., Rodriguez-Hernandez, C. J., Bagaitkar, J., Lamont, R. J. (2020). From Beyond the Pale to the Pale Riders: The Emerging Association of Bacteria With Oral Cancer. J. Dent. Res. 99, 604–612. doi: 10.1177/0022034520907341
Fracchia, A., Asraf, T., Salmon-Divon, M., Gerlitz, G. (2020). Increased Lamin B1 Levels Promote Cell Migration by Altering Perinuclear Actin Organization. Cells 9, 2161. doi: 10.3390/cells9102161
Gabriel, E. M., Fisher, D. T., Evans, S., Takabe, K., Skitzki, J. J. (2018). Intravital Microscopy in the Study of the Tumor Microenvironment: From Bench to Human Application. Oncotarget 9, 20165–20178. doi: 10.18632/oncotarget.24957
Gadiya, M., Chakraborty, G. (2018). Signaling by Discoidin Domain Receptor 1 in Cancer Metastasis. Cell Adh. Migr. 12, 315–323. doi: 10.1080/19336918.2018.1520556
Ganly, I., Yang, L., Giese, R. A., Hao, Y., Nossa, C. W., Morris, L. G. T., et al. (2019). Periodontal Pathogens are a Risk Factor of Oral Cavity Squamous Cell Carcinoma, Independent of Tobacco and Alcohol and Human Papillomavirus. Int. J. Cancer 145, 775–784. doi: 10.1002/ijc.32152
Geng, F., Liu, J., Guo, Y., Li, C., Wang, H., Wang, H., et al. (2017). Persistent Exposure to Porphyromonas Gingivalis Promotes Proliferative and Invasion Capabilities, and Tumorigenic Properties of Human Immortalized Oral Epithelial Cells. Front. Cell Infect. Microbiol. 7, 57. doi: 10.3389/fcimb.2017.00057
Geng, F., Wang, Q., Li, C., Liu, J., Zhang, D., Zhang, S., et al. (2019). Identification of Potential Candidate Genes of Oral Cancer in Response to Chronic Infection With Porphyromonas Gingivalis Using Bioinformatical Analyses. Front. Oncol. 9, 91. doi: 10.3389/fonc.2019.00091
Gibieža, P., Petrikaitė, V. (2021). The Regulation of Actin Dynamics During Cell Division and Malignancy. Am. J. Cancer Res. 11, 4050–4069.
Greenburg, G., Hay, E. D. (1982). Epithelia Suspended in Collagen Gels can Lose Polarity and Express Characteristics of Migrating Mesenchymal Cells. J. Cell Biol. 95, 333–339. doi: 10.1083/jcb.95.1.333
Greuber, E. K., Smith-Pearson, P., Wang, J., Pendergast, A. M. (2013). Role of ABL Family Kinases in Cancer: From Leukaemia to Solid Tumours. Nat. Rev. Cancer 13, 559–571. doi: 10.1038/nrc3563
Guo, X., Hua, Y. (2017). CCAT1: An Oncogenic Long Noncoding RNA in Human Cancers. J. Cancer Res. Clin. Oncol. 143, 555–562. doi: 10.1007/s00432-016-2268-3
Guy, J. B., Espenel, S., Vallard, A., Battiston-Montagne, P., Wozny, A. S., Ardail, D., et al. (2017). Evaluation of the Cell Invasion and Migration Process: A Comparison of the Video Microscope-Based Scratch Wound Assay and the Boyden Chamber Assay. J. Vis. Exp. 17, 56337. doi: 10.3791/56337
Hajishengallis, G., Lamont, R. J. (2021). Polymicrobial Communities in Periodontal Disease: Their Quasi-Organismal Nature and Dialogue With the Host. Periodontol. 2000 86, 210–230. doi: 10.1111/prd.12371
Ha, N. H., Park, D. G., Woo, B. H., Kim, D. J., Choi, J. I., Park, B. S., et al. (2016). Porphyromonas Gingivalis Increases the Invasiveness of Oral Cancer Cells by Upregulating IL-8 and MMPs. Cytokine 86, 64–72. doi: 10.1016/j.cyto.2016.07.013
Harokopakis, E., Albzreh, M. H., Martin, M. H., Hajishengallis, G. (2006). TLR2 Transmodulates Monocyte Adhesion and Transmigration via Rac1- and PI3K-Mediated Inside-Out Signaling in Response to Porphyromonas Gingivalis Fimbriae. J. Immunol. 176, 7645–7656. doi: 10.4049/jimmunol.176.12.7645
Haverman, T. M., Laheij, A., De Soet, J. J., De Lange, J., Rozema, F. R. (2017). Candida and Porphyromonas Gingivalis: The Effect on Wound Closure In Vitro. J. Oral. Microbiol. 9, 1328266. doi: 10.1080/20002297.2017.1328266
Ha, N. H., Woo, B. H., Kim, D. J., Ha, E. S., Choi, J. I., Kim, S. J., et al. (2015). Prolonged and Repetitive Exposure to Porphyromonas Gingivalis Increases Aggressiveness of Oral Cancer Cells by Promoting Acquisition of Cancer Stem Cell Properties. Tumour Biol. 36, 9947–9960. doi: 10.1007/s13277-015-3764-9
Holloway, A., Storey, A. (2014). A Conserved C-Terminal Sequence of High-Risk Cutaneous Beta-Human Papillomavirus E6 Proteins Alters Localization and Signalling of Beta1-Integrin to Promote Cell Migration. J. Gen. Virol. 95, 123–134. doi: 10.1099/vir.0.057695-0
Hong, B. Y., Hoare, A., Cardenas, A., Dupuy, A. K., Choquette, L., Salner, A. L., et al. (2020). The Salivary Mycobiome Contains 2 Ecologically Distinct Mycotypes. J. Dent. Res. 99, 730–738. doi: 10.1177/0022034520915879
Hoppe, T., Kraus, D., Probstmeier, R., Jepsen, S., Winter, J. (2019). Stimulation With Porphyromonas Gingivalis Enhances Malignancy and Initiates Anoikis Resistance in Immortalized Oral Keratinocytes. J. Cell Physiol. 234, 21903–21914. doi: 10.1002/jcp.28754
Hou, J. M., Krebs, M. G., Lancashire, L., Sloane, R., Backen, A., Swain, R. K., et al. (2012). Clinical Significance and Molecular Characteristics of Circulating Tumor Cells and Circulating Tumor Microemboli in Patients With Small-Cell Lung Cancer. J. Clin. Oncol. 30, 525–532. doi: 10.1200/JCO.2010.33.3716
Hrudka, J., Fišerová, H., Jelínková, K., Matěj, R., Waldauf, P. (2021). Cytokeratin 7 Expression as a Predictor of an Unfavorable Prognosis in Colorectal Carcinoma. Sci. Rep. 11, 17863. doi: 10.1038/s41598-021-97480-4
Huang, C., Yang, G., Jiang, T., Zhu, G., Li, H., Qiu, Z. (2011). The Effects and Mechanisms of Blockage of STAT3 Signaling Pathway on IL-6 Inducing EMT in Human Pancreatic Cancer Cells In Vitro. Neoplasma 58, 396–405. doi: 10.4149/neo_2011_05_396
Hur, S. S., Jeong, J. H., Ban, M. J., Park, J. H., Yoon, J. K., Hwang, Y. (2020). Traction Force Microscopy for Understanding Cellular Mechanotransduction. BMB Rep. 53, 74–81. doi: 10.5483/BMBRep.2020.53.2.308
Hu, D., Zhou, J., Wang, F., Shi, H., Li, Y., Li, B. (2015). HPV-16 E6/E7 Promotes Cell Migration and Invasion in Cervical Cancer via Regulating Cadherin Switch In Vitro and In Vivo. Arch. Gynecol. Obstet. 292, 1345–1354. doi: 10.1007/s00404-015-3787-x
Inaba, H., Amano, A., Lamont, R. J., Murakami, Y. (2015). Involvement of Protease-Activated Receptor 4 in Over-Expression of Matrix Metalloproteinase 9 Induced by Porphyromonas Gingivalis. Med. Microbiol. Immunol. 204, 605–612. doi: 10.1007/s00430-015-0389-y
Inaba, H., Sugita, H., Kuboniwa, M., Iwai, S., Hamada, M., Noda, T., et al. (2014). Porphyromonas Gingivalis Promotes Invasion of Oral Squamous Cell Carcinoma Through Induction of Prommp9 and its Activation. Cell Microbiol. 16, 131–145. doi: 10.1111/cmi.12211
Irfan, M., Delgado, R. Z. R., Frias-Lopez, J. (2020). The Oral Microbiome and Cancer. Front. Immunol. 11, 591088. doi: 10.3389/fimmu.2020.591088
Ishikawa, T., Terashima, J., Shimoyama, Y., Ohashi, Y., Mikami, T., Takeda, Y., et al. (2020). Effects of Butyric Acid, a Bacterial Metabolite, on the Migration of Ameloblastoma Mediated by Laminin 332. J. Oral. Sci. 62, 435–438. doi: 10.2334/josnusd.19-0380
Iwai, S., Yonekawa, A., Harada, C., Hamada, M., Katagiri, W., Nakazawa, M., et al. (2010). Involvement of the Wnt-Beta-Catenin Pathway in Invasion and Migration of Oral Squamous Carcinoma Cells. Int. J. Oncol. 37, 1095–1103. doi: 10.3892/ijo_00000761
Izdebska, M., Zielińska, W., Hałas-Wiśniewska, M., Grzanka, A. (2020). Involvement of Actin and Actin-Binding Proteins in Carcinogenesis. Cells 9, 2245. doi: 10.3390/cells9102245
Jain, P., Worthylake, R. A., Alahari, S. K. (2012). Quantitative Analysis of Random Migration of Cells Using Time-Lapse Video Microscopy. J. Vis. Exp., 63, e3585. doi: 10.3791/3585
Jang, H., Kim, J., Shin, J. H., Fredberg, J. J., Park, C. Y., Park, Y. (2019). Traction Microscopy With Integrated Microfluidics: Responses of the Multi-Cellular Island to Gradients of HGF. Lab. Chip 19, 1579–1588. doi: 10.1039/C9LC00173E
Jiang, T., Liu, G., Wang, L., Liu, H. (2015). Elevated Serum Gas6 Is a Novel Prognostic Biomarker in Patients With Oral Squamous Cell Carcinoma. PloS One 10, e0133940. doi: 10.1371/journal.pone.0133940
Jurado, C., Haserick, J. R., Lee, J. (2005). Slipping or Gripping? Fluorescent Speckle Microscopy in Fish Keratocytes Reveals Two Different Mechanisms for Generating a Retrograde Flow of Actin. Mol. Biol. Cell 16, 507–518. doi: 10.1091/mbc.e04-10-0860
Kamarajan, P., Ateia, I., Shin, J. M., Fenno, J. C., Le, C., Zhan, L., et al. (2020). Periodontal Pathogens Promote Cancer Aggressivity via TLR/MyD88 Triggered Activation of Integrin/FAK Signaling That is Therapeutically Reversible by a Probiotic Bacteriocin. PloS Pathog. 16, e1008881. doi: 10.1371/journal.ppat.1008881
Katz, R. R., West, J. L. (2022). Reductionist Three-Dimensional Tumor Microenvironment Models in Synthetic Hydrogels. Cancers (Basel) 14, 1225. doi: 10.3390/cancers14051225
Kleinstein, S. E., Nelson, K. E., Freire, M. (2020). Inflammatory Networks Linking Oral Microbiome With Systemic Health and Disease. J. Dent. Res. 99, 1131–1139. doi: 10.1177/0022034520926126
Kozyrska, K., Pilia, G., Vishwakarma, M., Wagstaff, L., Goschorska, M., Cirillo, S., et al. (2022). P53 Directs Leader Cell Behavior, Migration, and Clearance During Epithelial Repair. Science 375, eabl8876. doi: 10.1126/science.abl8876
Kuo, C. W., Chueh, D. Y., Chen, P. (2019). Real-Time In Vivo Imaging of Subpopulations of Circulating Tumor Cells Using Antibody Conjugated Quantum Dots. J. Nanobiotechnology 17, 26. doi: 10.1186/s12951-019-0453-7
Laheij, A. M., De Soet, J. J., Veerman, E. C., Bolscher, J. G., Van Loveren, C. (2013). The Influence of Oral Bacteria on Epithelial Cell Migration In Vitro. Mediators Inflamm. 2013, 154532. doi: 10.1155/2013/154532
Lamont, R. J., Fitzsimonds, Z. R., Wang, H., Gao, S. (2022). Role of Porphyromonas Gingivalis in Oral and Orodigestive Squamous Cell Carcinoma. Periodontol. 2000. 89, 154–165. doi: 10.1111/prd.12425
Lamont, R. J., Koo, H., Hajishengallis, G. (2018). The Oral Microbiota: Dynamic Communities and Host Interactions. Nat. Rev. Microbiol. 16, 745–759. doi: 10.1038/s41579-018-0089-x
Lauffenburger, D. A., Horwitz, A. F. (1996). Cell Migration: A Physically Integrated Molecular Process. Cell 84, 359–369. doi: 10.1016/S0092-8674(00)81280-5
Lawson, C. D., Ridley, A. J. (2018). Rho GTPase Signaling Complexes in Cell Migration and Invasion. J. Cell Biol. 217, 447–457. doi: 10.1083/jcb.201612069
Lee, J., Roberts, J. S., Atanasova, K. R., Chowdhury, N., Han, K., Yilmaz, O. (2017). Human Primary Epithelial Cells Acquire an Epithelial-Mesenchymal-Transition Phenotype During Long-Term Infection by the Oral Opportunistic Pathogen, Porphyromonas Gingivalis. Front. Cell Infect. Microbiol. 7, 493. doi: 10.3389/fcimb.2017.00493
Lee, K., Roberts, J. S., Choi, C. H., Atanasova, K. R., Yilmaz, Ö. (2018). Porphyromonas Gingivalis Traffics Into Endoplasmic Reticulum-Rich-Autophagosomes for Successful Survival in Human Gingival Epithelial Cells. Virulence 9, 845–859. doi: 10.1080/21505594.2018.1454171
Leggett, S. E., Hruska, A. M., Guo, M., Wong, I. Y. (2021). The Epithelial-Mesenchymal Transition and the Cytoskeleton in Bioengineered Systems. Cell Commun. Signal 19, 32. doi: 10.1186/s12964-021-00713-2
Liang, G., Wang, H., Shi, H., Zhu, M., An, J., Qi, Y., et al. (2020). Porphyromonas Gingivalis Promotes the Proliferation and Migration of Esophageal Squamous Cell Carcinoma Through the miR-194/GRHL3/PTEN/Akt Axis. ACS Infect. Dis. 6, 871–881. doi: 10.1021/acsinfecdis.0c00007
Liao, C., Wang, Q., An, J., Long, Q., Wang, H., Xiang, M., et al. (2021). Partial EMT in Squamous Cell Carcinoma: A Snapshot. Int. J. Biol. Sci. 17, 3036–3047. doi: 10.7150/ijbs.61566
Li, C. F., Chen, J. Y., Ho, Y. H., Hsu, W. H., Wu, L. C., Lan, H. Y., et al. (2019). Snail-Induced Claudin-11 Prompts Collective Migration for Tumour Progression. Nat. Cell Biol. 21, 251–262. doi: 10.1038/s41556-018-0268-z
Liu, C. C., Cai, D. L., Sun, F., Wu, Z. H., Yue, B., Zhao, S. L., et al. (2017). FERMT1 Mediates Epithelial-Mesenchymal Transition to Promote Colon Cancer Metastasis via Modulation of β-Catenin Transcriptional Activity. Oncogene 36, 1779–1792. doi: 10.1038/onc.2016.339
Liu, J., Hsieh, C. L., Gelincik, O., Devolder, B., Sei, S., Zhang, S., et al. (2019). Proteomic Characterization of Outer Membrane Vesicles From Gut Mucosa-Derived Fusobacterium Nucleatum. J. Proteomics 195, 125–137. doi: 10.1016/j.jprot.2018.12.029
Liu, L., Liang, L., Yang, C., Zhou, Y., Chen, Y. (2021c). Extracellular Vesicles of Fusobacterium Nucleatum Compromise Intestinal Barrier Through Targeting RIPK1-Mediated Cell Death Pathway. Gut Microbes 13, 1–20. doi: 10.1080/19490976.2021.1902718
Liu, D., Liu, S., Liu, J., Miao, L., Zhang, S., Pan, Y. (2021b). sRNA23392 Packaged by Porphyromonas Gingivalis Outer Membrane Vesicles Promotes Oral Squamous Cell Carcinomas Migration and Invasion by Targeting Desmocollin-2. Mol. Oral. Microbiol. 36, 182–191. doi: 10.1111/omi.12334
Liu, C., Stocke, K., Fitzsimonds, Z. R., Yakoumatos, L., Miller, D. P., Lamont, R. J. (2021a). A Bacterial Tyrosine Phosphatase Modulates Cell Proliferation Through Targeting RGCC. PloS Pathog. 17, e1009598. doi: 10.1371/journal.ppat.1009598
Li, T. H., Zhao, B. B., Qin, C., Wang, Y. Y., Li, Z. R., Cao, H. T., et al. (2021). IFIT1 Modulates the Proliferation, Migration and Invasion of Pancreatic Cancer Cells via Wnt/β-Catenin Signaling. Cell Oncol. (Dordr) 44, 1425–1437. doi: 10.1007/s13402-021-00651-8
Lovisa, S., Lebleu, V. S., Tampe, B., Sugimoto, H., Vadnagara, K., Carstens, J. L., et al. (2015). Epithelial-To-Mesenchymal Transition Induces Cell Cycle Arrest and Parenchymal Damage in Renal Fibrosis. Nat. Med. 21, 998–1009. doi: 10.1038/nm.3902
Lu, P., Lu, Y. (2021). Born to Run? Diverse Modes of Epithelial Migration. Front. Cell Dev. Biol. 9, 704939. doi: 10.3389/fcell.2021.704939
Mak, M., Reinhart-King, C. A., Erickson, D. (2011). Microfabricated Physical Spatial Gradients for Investigating Cell Migration and Invasion Dynamics. PloS One 6, e20825. doi: 10.1371/journal.pone.0020825
Mcneill, M. C., Wray, J., Sala-Newby, G. B., Hindmarch, C. C. T., Smith, S. A., Ebrahimighaei, R., et al. (2020). Nuclear Actin Regulates Cell Proliferation and Migration via Inhibition of SRF and TEAD. Biochim. Biophys. Acta Mol. Cell Res. 1867, 118691. doi: 10.1016/j.bbamcr.2020.118691
Mehanna, H., Beech, T., Nicholson, T., El-Hariry, I., Mcconkey, C., Paleri, V., et al. (2013). Prevalence of Human Papillomavirus in Oropharyngeal and Nonoropharyngeal Head and Neck Cancer–Systematic Review and Meta-Analysis of Trends by Time and Region. Head Neck 35, 747–755. doi: 10.1002/hed.22015
Mosier, J. A., Schwager, S. C., Boyajian, D. A., Reinhart-King, C. A. (2021). Cancer Cell Metabolic Plasticity in Migration and Metastasis. Clin. Exp. Metastasis 38, 343–359. doi: 10.1007/s10585-021-10102-1
Ng, L. G., Mrass, P., Kinjyo, I., Reiner, S. L., Weninger, W. (2008). Two-Photon Imaging of Effector T-Cell Behavior: Lessons From a Tumor Model. Immunol. Rev. 221, 147–162. doi: 10.1111/j.1600-065X.2008.00596.x
Nonaka, S., Nakanishi, H. (2020). Secreted Gingipains From Porphyromonas Gingivalis Induce Microglia Migration Through Endosomal Signaling by Protease-Activated Receptor 2. Neurochem. Int. 140, 104840. doi: 10.1016/j.neuint.2020.104840
Ohshima, J., Wang, Q., Fitzsimonds, Z. R., Miller, D. P., Sztukowska, M. N., Jung, Y. J., et al. (2019). Streptococcus Gordonii Programs Epithelial Cells to Resist ZEB2 Induction by Porphyromonas Gingivalis. Proc. Natl. Acad. Sci. U .S. A. 116, 8544–8553. doi: 10.1073/pnas.1900101116
Padera, T. P., Stoll, B. R., So, P. T., Jain, R. K. (2002). Conventional and High-Speed Intravital Multiphoton Laser Scanning Microscopy of Microvasculature, Lymphatics, and Leukocyte-Endothelial Interactions. Mol. Imaging 1, 9–15. doi: 10.1162/153535002753395662
Parhi, L., Alon-Maimon, T., Sol, A., Nejman, D., Shhadeh, A., Fainsod-Levi, T., et al. (2020). Breast Cancer Colonization by Fusobacterium Nucleatum Accelerates Tumor Growth and Metastatic Progression. Nat. Commun. 11, 3259. doi: 10.1038/s41467-020-16967-2
Pastushenko, I., Blanpain, C. (2019). EMT Transition States During Tumor Progression and Metastasis. Trends Cell Biol. 29, 212–226. doi: 10.1016/j.tcb.2018.12.001
Patterson, A. D., Gonzalez, F. J., Perdew, G. H., Peters, J. M. (2018). Molecular Regulation of Carcinogenesis: Friend and Foe. Toxicol. Sci. 165, 277–283. doi: 10.1093/toxsci/kfy185
Paul, C. D., Hung, W. C., Wirtz, D., Konstantopoulos, K. (2016). Engineered Models of Confined Cell Migration. Annu. Rev. BioMed. Eng. 18, 159–180. doi: 10.1146/annurev-bioeng-071114-040654
Pinner, S., Jordan, P., Sharrock, K., Bazley, L., Collinson, L., Marais, R., et al. (2009). Intravital Imaging Reveals Transient Changes in Pigment Production and Brn2 Expression During Metastatic Melanoma Dissemination. Cancer Res. 69, 7969–7977. doi: 10.1158/0008-5472.CAN-09-0781
Prakash, V., Carson, B. B., Feenstra, J. M., Dass, R. A., Sekyrova, P., Hoshino, A., et al. (2019). Ribosome Biogenesis During Cell Cycle Arrest Fuels EMT in Development and Disease. Nat. Commun. 10, 2110. doi: 10.1038/s41467-019-10100-8
Qi, Y. J., Jiao, Y. L., Chen, P., Kong, J. Y., Gu, B. L., Liu, K., et al. (2020). Porphyromonas Gingivalis Promotes Progression of Esophageal Squamous Cell Cancer via TGFbeta-Dependent Smad/YAP/TAZ Signaling. PloS Biol. 18, e3000825. doi: 10.1371/journal.pbio.3000825
Raab, M., Gentili, M., De Belly, H., Thiam, H. R., Vargas, P., Jimenez, A. J., et al. (2016). ESCRT III Repairs Nuclear Envelope Ruptures During Cell Migration to Limit DNA Damage and Cell Death. Science 352, 359–362. doi: 10.1126/science.aad7611
Radaic, A., Kapila, Y. L. (2021). The Oralome and its Dysbiosis: New Insights Into Oral Microbiome-Host Interactions. Comput. Struct. Biotechnol. J. 19, 1335–1360. doi: 10.1016/j.csbj.2021.02.010
Rando, O. J., Zhao, K., Janmey, P., Crabtree, G. R. (2002). Phosphatidylinositol-Dependent Actin Filament Binding by the SWI/SNF-Like BAF Chromatin Remodeling Complex. Proc. Natl. Acad. Sci. U .S. A. 99, 2824–2829. doi: 10.1073/pnas.032662899
Ridley, A. J. (1999). Rho Family Proteins and Regulation of the Actin Cytoskeleton. Prog. Mol. Subcell. Biol. 22, 1–22. doi: 10.1007/978-3-642-58591-3_1
Ridley, A. J., Schwartz, M. A., Burridge, K., Firtel, R. A., Ginsberg, M. H., Borisy, G., et al. (2003). Cell Migration: Integrating Signals From Front to Back. Science 302, 1704–1709. doi: 10.1126/science.1092053
Rusu, A. D., Georgiou, M. (2020). The Multifarious Regulation of the Apical Junctional Complex. Open Biol. 10, 190278. doi: 10.1098/rsob.190278
Sahai, E., Marshall, C. J. (2003). Differing Modes of Tumour Cell Invasion Have Distinct Requirements for Rho/ROCK Signalling and Extracellular Proteolysis. Nat. Cell Biol. 5, 711–719. doi: 10.1038/ncb1019
Scott, A. J., Alexander, J. L., Merrifield, C. A., Cunningham, D., Jobin, C., Brown, R., et al. (2019). International Cancer Microbiome Consortium Consensus Statement on the Role of the Human Microbiome in Carcinogenesis. Gut 68, 1624–1632. doi: 10.1136/gutjnl-2019-318556
Senini, V., Amara, U., Paul, M., Kim, H. (2019). Porphyromonas Gingivalis Lipopolysaccharide Activates Platelet Cdc42 and Promotes Platelet Spreading and Thrombosis. J. Periodontol. 90, 1336–1345. doi: 10.1002/JPER.18-0596
Shao, W., Fujiwara, N., Mouri, Y., Kisoda, S., Yoshida, K., Yoshida, K., et al. (2021). Conversion From Epithelial to Partial-EMT Phenotype by Fusobacterium Nucleatum Infection Promotes Invasion of Oral Cancer Cells. Sci. Rep. 11, 14943. doi: 10.1038/s41598-021-94384-1
Shih, W., Yamada, S. (2012). N-Cadherin-Mediated Cell-Cell Adhesion Promotes Cell Migration in a Three-Dimensional Matrix. J. Cell Sci. 125, 3661–3670. doi: 10.1242/jcs.103861
Shi, J., Zeng, X., Zhou, M., Chen, Q. (2009). Activation of ERK-FAK Signaling Pathway and Enhancement of Cell Migration Involved in the Early Interaction Between Oral Keratinocytes and Candida Albicans. Mycopathologia 167, 1–7. doi: 10.1007/s11046-008-9142-z
Son, H., Moon, A. (2010). Epithelial-Mesenchymal Transition and Cell Invasion. Toxicol. Res. 26, 245–252. doi: 10.5487/TR.2010.26.4.245
Stemmler, M. P., Eccles, R. L., Brabletz, S., Brabletz, T. (2019). Non-Redundant Functions of EMT Transcription Factors. Nat. Cell Biol. 21, 102–112. doi: 10.1038/s41556-018-0196-y
Stone, R. C., Pastar, I., Ojeh, N., Chen, V., Liu, S., Garzon, K. I., et al. (2016). Epithelial-Mesenchymal Transition in Tissue Repair and Fibrosis. Cell Tissue Res. 365, 495–506. doi: 10.1007/s00441-016-2464-0
Stroka, K. M., Jiang, H., Chen, S. H., Tong, Z., Wirtz, D., Sun, S. X., et al. (2014). Water Permeation Drives Tumor Cell Migration in Confined Microenvironments. Cell 157, 611–623. doi: 10.1016/j.cell.2014.02.052
Strutz, F., Zeisberg, M., Ziyadeh, F. N., Yang, C. Q., Kalluri, R., Muller, G. A., et al. (2002). Role of Basic Fibroblast Growth Factor-2 in Epithelial-Mesenchymal Transformation. Kidney Int. 61, 1714–1728. doi: 10.1046/j.1523-1755.2002.00333.x
Sur-Erdem, I., Hussain, M. S., Asif, M., Pınarbası, N., Aksu, A. C., Noegel, A. A. (2020). Nesprin-1 Impact on Tumorigenic Cell Phenotypes. Mol. Biol. Rep. 47, 921–934. doi: 10.1007/s11033-019-05184-w
Sztukowska, M. N., Ojo, A., Ahmed, S., Carenbauer, A. L., Wang, Q., Shumway, B., et al. (2016). Porphyromonas Gingivalis Initiates a Mesenchymal-Like Transition Through ZEB1 in Gingival Epithelial Cells. Cell Microbiol. 18, 844–858. doi: 10.1111/cmi.12554
Tang, S. W., Yang, T. C., Lin, W. C., Chang, W. H., Wang, C. C., Lai, M. K., et al. (2011). Nicotinamide N-Methyltransferase Induces Cellular Invasion Through Activating Matrix Metalloproteinase-2 Expression in Clear Cell Renal Cell Carcinoma Cells. Carcinogenesis 32, 138–145. doi: 10.1093/carcin/bgq225
Tentler, D., Lomert, E., Novitskaya, K., Barlev, N. A. (2019). Role of ACTN4 in Tumorigenesis, Metastasis, and EMT. Cells 8, 1427. doi: 10.3390/cells8111427
Tokuraku, K., Kuragano, M., Uyeda, T. Q. P. (2020). Long-Range and Directional Allostery of Actin Filaments Plays Important Roles in Various Cellular Activities. Int. J. Mol. Sci. 21, 3209. doi: 10.3390/ijms21093209
Trappmann, B., Baker, B. M., Polacheck, W. J., Choi, C. K., Burdick, J. A., Chen, C. S. (2017). Matrix Degradability Controls Multicellularity of 3D Cell Migration. Nat. Commun. 8, 371. doi: 10.1038/s41467-017-00418-6
Trepat, X., Chen, Z., Jacobson, K. (2012). Cell Migration. Compr. Physiol. 2, 2369–2392. doi: 10.1002/cphy.c110012
Vicente-Manzanares, M., Koach, M. A., Whitmore, L., Lamers, M. L., Horwitz, A. F. (2008). Segregation and Activation of Myosin IIB Creates a Rear in Migrating Cells. J. Cell Biol. 183, 543–554. doi: 10.1083/jcb.200806030
Vilchez Mercedes, S. A., Bocci, F., Levine, H., Onuchic, J. N., Jolly, M. K., Wong, P. K. (2021). Decoding Leader Cells in Collective Cancer Invasion. Nat. Rev. Cancer 21, 592–604. doi: 10.1038/s41568-021-00376-8
Wang, S., Liu, Y., Li, J., Zhao, L., Yan, W., Lin, B., et al. (2021). Fusobacterium Nucleatum Acts as a Pro-Carcinogenic Bacterium in Colorectal Cancer: From Association to Causality. Front. Cell Dev. Biol. 9, 710165. doi: 10.3389/fcell.2021.710165
Wang, Q., Song, R., Zhao, C., Liu, H., Yang, Y., Gu, S., et al. (2019). HPV16 E6 Promotes Cervical Cancer Cell Migration and Invasion by Downregulation of NHERF1. Int. J. Cancer 144, 1619–1632. doi: 10.1002/ijc.31876
Wang, H., Tao, L., Jin, F., Gu, H., Dai, X., Ni, T., et al. (2017). Cofilin 1 Induces the Epithelial-Mesenchymal Transition of Gastric Cancer Cells by Promoting Cytoskeletal Rearrangement. Oncotarget 8, 39131–39142. doi: 10.18632/oncotarget.16608
Wang, Q., Yu, C., Yue, C., Liu, X. (2020). Fusobacterium Nucleatum Produces Cancer Stem Cell Characteristics via EMT-Resembling Variations. Int. J. Clin. Exp. Pathol. 13, 1819–1828.
Warburton, A., Redmond, C. J., Dooley, K. E., Fu, H., Gillison, M. L., Akagi, K., et al. (2018). HPV Integration Hijacks and Multimerizes a Cellular Enhancer to Generate a Viral-Cellular Super-Enhancer That Drives High Viral Oncogene Expression. PloS Genet. 14, e1007179. doi: 10.1371/journal.pgen.1007179
Weigelin, B., Bakker, G. J., Friedl, P. (2012). Intravital Third Harmonic Generation Microscopy of Collective Melanoma Cell Invasion: Principles of Interface Guidance and Microvesicle Dynamics. Intravital 1, 32–43. doi: 10.4161/intv.21223
Whitmore, S. E., Lamont, R. J. (2014). Oral Bacteria and Cancer. PloS Pathog. 10, e1003933. doi: 10.1371/journal.ppat.1003933
Willis, J. R., Gabaldon, T. (2020). The Human Oral Microbiome in Health and Disease: From Sequences to Ecosystems. Microorganisms 8, 308. doi: 10.3390/microorganisms8020308
Wolf, K., Alexander, S., Schacht, V., Coussens, L. M., Von Andrian, U. H., Van Rheenen, J., et al. (2009). Collagen-Based Cell Migration Models In Vitro and In Vivo. Semin. Cell Dev. Biol. 20, 931–941. doi: 10.1016/j.semcdb.2009.08.005
Wu, Y., Zhou, B. P. (2010). Snail: More Than EMT. Cell Adh. Migr. 4, 199–203. doi: 10.4161/cam.4.2.10943
Xu, C., Fan, L., Lin, Y., Shen, W., Qi, Y., Zhang, Y., et al. (2021a). Fusobacterium Nucleatum Promotes Colorectal Cancer Metastasis Through miR-1322/CCL20 Axis and M2 Polarization. Gut Microbes 13, 1980347. doi: 10.1080/19490976.2021.1980347
Xu, X., Wang, Y., Choi, W. S., Sun, X., Godbout, R. (2021b). Super Resolution Microscopy Reveals DHA-Dependent Alterations in Glioblastoma Membrane Remodelling and Cell Migration. Nanoscale 13, 9706–9722. doi: 10.1039/D1NR02128A
Yang, H. W., Collins, S. R., Meyer, T. (2016). Locally Excitable Cdc42 Signals Steer Cells During Chemotaxis. Nat. Cell Biol. 18, 191–201. doi: 10.1038/ncb3292
Yang, Y., Zheng, H., Zhan, Y., Fan, S. (2019). An Emerging Tumor Invasion Mechanism About the Collective Cell Migration. Am. J. Transl. Res. 11, 5301–5312.
Young, L. S., Yap, L. F., Murray, P. G. (2016). Epstein-Barr Virus: More Than 50 Years Old and Still Providing Surprises. Nat. Rev. Cancer 16, 789–802. doi: 10.1038/nrc.2016.92
Zhang, Y., Wang, X., Li, H., Ni, C., Du, Z., Yan, F. (2018). Human Oral Microbiota and its Modulation for Oral Health. BioMed. Pharmacother. 99, 883–893. doi: 10.1016/j.biopha.2018.01.146
Keywords: oral microbiota, cell migration, EMT, carcinogenesis, metastasis
Citation: Bai H, Yang J, Meng S and Liu C (2022) Oral Microbiota-Driven Cell Migration in Carcinogenesis and Metastasis. Front. Cell. Infect. Microbiol. 12:864479. doi: 10.3389/fcimb.2022.864479
Received: 28 January 2022; Accepted: 04 April 2022;
Published: 29 April 2022.
Edited by:
Jin Xiao, University of Rochester, United StatesReviewed by:
Masae Kuboniwa, Osaka University, JapanCopyright © 2022 Bai, Yang, Meng and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shu Meng, ZHJlYW1pbmdzdWVAMTYzLmNvbQ==; Chengcheng Liu, bGl1Y2hlbmdjaGVuZzUxOUAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.