- 1Department of Microbiology and Plant Pathology, University of California, Riverside, Riverside, CA, United States
- 2Department of Biochemistry, University of California, Riverside, Riverside, CA, United States
- 3Graduate Program in Genetics, Genomics, and Bioinformatics, University of California, Riverside, Riverside, CA, United States
The commensal microbes of the gut microbiota make important contributions to host defense against gastrointestinal pathogens, including Vibrio cholerae, the etiologic agent of cholera. As interindividual microbiota variation drives individual differences in infection susceptibility, we examined both host and V. cholerae gene expression during infection of suckling mice transplanted with different model human commensal communities, including an infection-susceptible configuration representing communities damaged by recurrent diarrhea and malnutrition in cholera endemic areas and a representative infection-resistant microbiota characteristic of healthy individuals. In comparison to colonization of animals with resistant microbiota, animals bearing susceptible microbiota challenged with V. cholerae downregulate genes associated with generation of reactive oxygen/nitrogen stress, while V. cholerae in these animals upregulates biofilm-associated genes. We show that V. cholerae in susceptible microbe infection contexts are more resistant to oxidative stress and inhibitory bile metabolites generated by the action of commensal microbes and that both phenotypes are dependent on biofilm-associated genes, including vpsL. We also show that susceptible and infection-resistant microbes drive different bile acid compositions in vivo by the action of bile salt hydrolase enzymes. Taken together, these findings provide a better understanding of how the microbiota uses multiple mechanisms to modulate the infection-associated host environment encountered by V. cholerae, leading to commensal-dependent differences in infection susceptibility.
Introduction
In the transition from the aquatic reservoir into the human gastrointestinal tract, Vibrio cholerae encounters multiple host defense mechanisms, including low pH, oxidative stress, and bile acids (Louis and O’byrne, 2010). Growing evidence indicates that the resident community of gut microbes, the gut microbiota, is also an essential factor affecting host susceptibility to V. cholerae infection, using multiple mechanisms to modulate pathogen fitness and gene expression during infection (Hsiao et al., 2014; Alavi et al., 2020; Cho et al., 2021). The gut microbiota is highly complex and varies dramatically between individuals, leading to individual-specific effects on V. cholerae susceptibility driven partially by community-specific effects on V. cholerae behavior during infection (Hsiao et al., 2014; Alavi et al., 2020). In cholera-endemic areas, environmental insults such as malnutrition and diarrhea of multiple etiologies drive some individuals’ gut communities to recurring damaged or dysbiotic states that are unable to resist the colonization of V. cholerae (Hsiao et al., 2014; David et al., 2015; Alavi et al., 2020). Commensal microbes are able to modulate V. cholerae fitness and behavior during infection through numerous mechanisms including quorum sensing regulation of virulence genes (Hsiao et al., 2014) and direct competition via the type VI secretion system (T6SS) (Zhao et al., 2018). In addition to direct competition and modulating V. cholerae transcription during infection, the gut microbiota is also able to broadly modulate gut environmental conditions, such as through manipulation of oxidative conditions as well as host metabolites such as bile. Both of these factors are active during V. cholerae infection; transcriptomic studies of cholera patients reveal an increase in expression of host enzymes responsible for oxidative stress (Ellis et al., 2015; Bourque et al., 2018), and microbiota-dependent modulation of bile acid levels has been shown to affect V. cholerae virulence gene expression (Alavi et al., 2020).
A key adaptation of V. cholerae to environmental stresses is the ability to form biofilms, which in addition to protecting against acid, predation in the aquatic reservoir, and bacteriostatic molecules in the host gut such as bile also modulates V. cholerae infectivity and epidemic spread (Zhu and Mekalanos, 2003; Matz et al., 2005; Hung et al., 2006; Hay and Zhu, 2015). Here, we show using model human microbiota compositions that commensals are able to differentially affect V. cholerae fitness by modulating the expression of biofilm-associated genes in V. cholerae. This process intersects with the microbiota-specific manipulation of gut redox conditions and bile acid composition that both inhibit V. cholerae fitness in a biofilm-dependent fashion. Our findings suggest that the composition and biochemical functions of the gut microbiota are able to use reinforcing mechanisms, leading to commensal-dependent infection-permissive and restrictive conditions in the mammalian gastrointestinal tract.
Methods
Bacterial Strains and Culture Condition
All V. cholerae strains were derived from the C6706 O1 El Tor pandemic strain and grown in LB liquid media with appropriate antibiotics at 37°C with agitation. All anaerobic strains were grown in LYHBHI liquid media (BHI supplemented to 5 g/l yeast extract, 5 mg/l hemin, 1 mg/ml cellobiose, 1 mg/ml maltose, and 0.5 mg/ml cysteine–HCl) at 37°C in a Coy chamber under anaerobic conditions (5% H2, 20% CO2, balance N2) (Alavi et al., 2020). All anaerobic strains used in animal experiments were grown for 48 h, then 1:100 subcultured into fresh media and grown an additional 48 h prior to gavage. vpsL mutants were constructed using natural transformation based on the published method (Dalia et al., 2014). Approximately a 3-kb vpsL upstream (forward primer: 5′-GTTAAGAGCACCGATTGCAC-3′, reverse primer: 5′-GTCGACGGATCCCCGGAATCTTCATCACTAGACGCTCCTAAC-3′) and downstream (forward primer: 5′-GAAGCAGCTCCAGCCTACAGCGTATTAAGACAGGGCACTTG-3′, reverse primer: 5′-GCATTTTTTACCGTCAGGGTC-3′) sequence was amplified and added to ends of a trimethoprim resistance (TmR) cassette amplified from SAD530 using primers ABD123 (5′-ATTCCGGGGATCCGTCGA-3′) and ABD124 (5′-TGTAGGCTGGAGCTGCTTC-3′) by overlapping PCR using Phusion High-Fidelity PCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA). The first round of PCR consisted of an equimolar vpsL upstream fragment, a vpsL downstream fragment, and TmR cassette amplicons with a cycle condition of 98°C for 30 s, followed by 15 cycles (94°C for 10 s, 50°C for 30 s, 72°C for 4 min), and 72°C for 10 min. A vpsL upstream forward primer and a downstream reverse primer were added to the PCR reaction which was then continued using cycle conditions as described above. The final PCR product was added to 0.45 ml of instant ocean sea salt solution (7 mg/ml) with chitin-induced V. cholerae and incubated for 16–24 h at 30°C stationary, allowing natural competence. 0.5 ml LB was then added, and the mixture was incubated for 2–3 h at 30°C with agitation. The final mixture was then plated on an LB agar plate with 10 µg/ml trimethoprim for overnight incubation at 30°C to select for transformants. The final picked mutants were checked by Sanger sequencing.
Animal Experiments
Specific pathogen-free (SPF) CD-1 suckling animals were purchased from Charles River Laboratories. Upon receipt, suckling mice were fasted, and an equivalent of about 1 mg/g body weight streptomycin was gavaged with 30-gauge plastic tubing. Pups were then placed with a lactating dam for 1 day before being inoculated with communities and V. cholerae. Human gut bacterial strains were prepared by taking a total 300 µl of OD600 = 0.4 equivalent culture per animal divided evenly across constituent commensal strains. Appropriate amounts of anaerobic strain culture were pooled and pelleted by centrifugation and resuspended in 25 µl of fresh LYHBHI per animal. Mice then received 25 µl of the community mixture and 25 µl of V. cholerae (1 × 104–1 × 105 CFU) in PBS (Alavi et al., 2020). 4-day-old suckling CD-1 mice were fasted for 1.5 h prior to all gavages. 18 hours after infection, pups were sacrificed, intestinal tissue was homogenized in 5 ml of PBS, and V. cholerae was plated on a streptomycin LB-agar plate for enumeration and fitness assays. For RNA assays, 1 ml of intestinal homogenate from each infected pup was centrifuged at 14,000 rpm, the supernatant was removed, and the pellet was resuspended in 500 µl of TRIzol (Ambion, Foster City, CA, USA) at -80°C.
Quantitative PCR Analysis of Model Microbiome
DNA was extracted from an intestinal homogenate and used as template for model strain detection by quantitative PCR. 500 µl of intestinal homogenate was mixed with 210 µl of 20% SDS, 500 µl phenol:chloroform:isoamyl-alcohol (24:24:1, Fisher Scientific), and 500 µl of 0.1-mm zirconia beads (BioSpec, Bartlesville, OK, USA). The mixture was then lysed with a bead beater at 2,400 RPM for 3 min. The qPCR assay to detect model strains was performed using a universal 16S rRNA gene primer set to detect total bacterial load (forward primer: 5′-GTGSTGCAYGGYTGTCGTCA-3′, reverse primer: 5′-ACGTCRTCCMCACCTTCCTC-3′) (Horz et al., 2005). Each reaction was done in triplicates with 12.5 µl of iQ SYBR Green Supermix (BIO-RAD2), 1 µl of 10µM forward and reverse primers, 5.5 µl nuclease-free water, and 5 µl of extracted DNA (200 ng/µl).
RNA Extraction, RNA-Seq Library Prep, and Sequencing
Total RNA was extracted using TRIzol according to the manufacturer’s instructions. DNA was degraded using Baseline-ZERO DNase (Lucigen, Middleton, WI, USA), and the samples were then treated with TRIzol again for purification. A sequencing library was prepared using the Ovation Mouse RNA-Seq System with addition of V. cholerae enrichment primers (NuGEN, San Carlos, CA, USA). Final libraries were checked using an Agilent Bioanalyzer, and a 75-bp single-read sequencing was performed on Illumina NextSeq500.
RNA-Seq Analysis
All RNA-Seq-sequenced libraries were trimmed using Trimmomatic v0.39 (Bolger et al., 2014), and reads were aligned to mouse and V. cholerae genomes, using HISAT2 version 2.1.0 (Kim et al., 2015) with reference genomes GCF_000001635.26_GRCm38.p6 and GCF_000006745.1_ASM674v1. HTSeq (Anders et al., 2015) was used to calculate raw counts. After rRNA and tRNA were removed, differential expression analysis was conducted using edgeR (Robinson et al., 2009; Mccarthy et al., 2012). Statistical analysis and visualization of V. cholerae pathway profiles were performed using package clusterProfiler (Yu et al., 2012; Wu et al., 2021) in R with the selected database KEGG, and genes with FDR <0.05 and FC >1 or FC <-1 were used for further analyses.
Measurement of H2O2 Levels in Colonized Intestines
Intestinal homogenate H2O2 levels were measured using the Amplex UltraRed reagent kit (Invitrogen, Carlsbad, CA, USA). 50 µl of intestinal homogenate was collected and mixed with 50 µl of working solution (5 mM Amplex UltraRed reagent, 10 U horseradish peroxidase, HRP, 0.05 M sodium phosphate, pH 7.4) immediately after homogenization. The mixture was incubated at room temperature for 30 min, and absorbance was measured (560 nm). The H2O2 level was then determined based on the H2O2 standard curve.
V. cholerae Ex Vivo Oxidative Stress Resistance Assays
50 µl of intestinal tissue homogenates collected from V. cholerae-infected suckling mice colonized with different commensal groups was incubated with 1 mM of hydrogen peroxide for 1 h at room temperature. After incubation, 10 µl of untreated and treated homogenates was collected, serially diluted, and plated on selective agar to determine survival rates.
Intestinal Homogenate Ex Vivo Treatment With Pure Culture
Intestinal homogenates were prepared as described in previously published methods (Alavi et al., 2020). Briefly, intestines were collected from 6-day-old CD-1 suckling mice fasted for 18 h and homogenized in 2.5 ml sterile water, homogenates were centrifuged, and then aqueous supernatants were collected. The supernatant was treated at 100℃ for 30 min and filter sterilized with a 0.22-µM filter. The sample was then dried with the Savant Integrated SpeedVac System (Fisher Scientific) and resuspended in 0.5 ml sterile water. 2 ml of acetonitrile (Sigma-Aldrich, St. Louis, MO, USA) was added to the sample and incubated at room temperature for 20 min to deproteinize after vortexing (Humbert et al., 2012). The aqueous layer was then filter sterilized, dried, and resuspended in 0.5 ml sterile water. 1.5 ml of OD600 = 0.4 pure culture was pelleted and resuspended in purified homogenates then incubated anaerobically for 24 h at 37°C. The culture was heat-treated at 100℃ for 30 min then centrifuged. The aqueous layer was then filter-sterilized with a 0.22-µM filter after being cooled to room temperature and sent for mass spectrometry analysis.
Quantitative PCR Analysis of VPS Gene Expression
An overnight culture of V. cholerae was 1:100 inoculated into fresh LB liquid media containing 100 µg/ml streptomycin and grown to OD600 = 0.3. 50 µM of H2O2 or an equal volume of PBS was added to the culture and incubated for 2 h at 37°C with shaking. The cultures were collected, and RNA was extracted using TRIzol according to the manufacturer’s instructions. 2 µg of total RNA was then used for reverse transcription PCR using the SuperScript™ IV One-Step RT-PCR System (Invitrogen). qPCR assays were preformed using gene-specific primer sets targeting vpsR (forward primer: 5′-TGGCACACTGCTGCTAAA-3′, reverse primer: 5′-ACATCGACTGCACGAACC-3′), vpsT (forward primer: 5′-ACCCTGATCAAAGGCATGAG-3′, reverse primer: 5′-GTGAGGTCACGACTGAGTTAC-3′), and vpsL (forward primer: 5′-CAGTATGCGAGTGATGGATAATGG-3′, reverse primer: 5′-TCGTGGATCGCCTTTGGT-3′), with recA (forward primer: 5′-ATTGAAGGCGAAATGGGCGATAG-3′, reverse primer: 5′-TACACATACAGTTGGATTGCTTGAGG-3′) as the reference gene. Each reaction was done in triplicate with 12.5 µl of iQ SYBR Green Supermix (BIO-RAD2), 1 µl of 10 µM forward and reverse primers each, 8.5 µl nuclease-free water, and 2 µl of 2-fold-diluted RT-PCR reaction product.
Measurement of Bile Acid Processing by B. obeum BSH
Overnight cultures of E. coli bshc and E. coli vector with pZE21 (Alavi et al., 2020) were 1:100 inoculated into fresh LB liquid media containing 50 µg/ml kanamycin and incubated for 24 h at 37℃ with shaking. The cultures were normalized to 1.5 ml of OD600 = 0.4 culture, pelleted, and washed with 1.5 ml of PBS. The pellet was resuspended in 1.5 ml of PBS containing 2 mM of taurodeoxycholic acid (TDCA) and transferred to glass tubes then incubated at 37°C stationary for 24 h. After the incubation, supernatants were collected by centrifuge and filter-sterilized with 0.22-µM filters. Collected supernatants were sent for mass spectrometry analysis and tested for V. cholerae growth inhibition.
V. cholerae Growth Curve Measurements
Supernatants collected as previously described were 1:1 mixed with a 1:1,000 diluted log-phase V. cholerae culture in a 96-well plate. OD600 was then taken every 30 min for 20 h with a BioTek Synergy HTX multi-mode reader under 37°C with orbital shaking at 180 cpm (6 mm) using Gen5 2.09.
Bile Acid Extraction and Preparation From In Vitro Enzymatic Assays
Bile acids were extracted from in vitro enzymatic assays for LC-MS measurements. An aliquot of 200 µl of the assay was acidified to a final concentration of 2% acetic acid. A liquid–liquid extraction was performed with 300 µl of ethyl acetate. The supernatant was collected (200 µl) and dried down under a stream of nitrogen. The tube content after drying was resuspended in 1 ml methanol and diluted 20× for LC-MS analysis.
Liquid Chromatography Mass Spectrometry Measurement of Bile Salts
Standards for TDCA and deoxycholic acid (DCA) (1 µM) were used to determine retention times and fragmentation patterns for each compound. Chromatographic separations were conducted in a Waters I-Class UPLC System using an ACQUITY UPLC BEH C18 column (2.1 × 100 mm, 1.7 µM; Waters, Milford, MA, USA). Water with 0.1% formic acid (A) and acetonitrile with 0.1% formic acid (B) were used as mobile phases. Sample separation was conducted after 1-µl injections with the column kept at 40°C, with a flow rate of 0.4 ml/min under the following gradient: 0 min, 10% B; 1 min, 10% B; 1.5 min, 25% B; 8 min, 45% B, 10 min, 90% B; 16 min, 90% B, 17 min, 45% B, 19 min, 25% B; 21 min, 10% B; 26 min, 10% B. Mass spectrometry measurements were conducted in a SYNAPT G2-Si quadrupole time-of-flight mass spectrometer (Waters). The instrument was operated in negative ion mode under MS/MS conditions with a source capillary voltage of 2.5 kV, source temperature of 150°C, desolvation temperature of 600°C, and desolvation gas at 600 l/h. Leucine enkephalin was used as lock spray standard for mass correction. The following mass transitions (m/z) produced by the indicated collision energies (trap and transfer) were used for signal identification of TDCA and DCA [TDCA, 79.95 and 124.00 (CE 40 V); DCA, 343.26 and 391.28 (CE 25 V)]. The QuanLynx software (Waters) was used for peak integration and peak area calculation and processing. A quality control sample was produced by pooling equal amounts of samples and used to monitor system stability and reproducibility.
Bile Acid Growth Inhibition of Biofilm-Associated V. cholerae
Overnight V. cholerae cultures were 1:100 inoculated into fresh LB liquid media with 100 µg/ml streptomycin and 2 ml of dilution aliquoted into 100 × 15-mm sterilized polystyrene petri dishes with 10-mg glass beads (50–100 µm, Polysciences, Warrington, PA, USA), followed by incubation for 24 h at room temperature with slow shaking (10 rpm) (Zhu and Mekalanos, 2003). Planktonic cells were collected by pipetting media, followed by pelleting via centrifugation and resuspension in LB media containing 1 mM of different bile acids (diluted from 100× stock solution in dimethyl sulfoxide, DMSO). Beads were rinsed with 1× PBS to remove residual planktonic cells, and the biofilm-associated bacteria on the beads were then treated with 5 ml LB liquid media containing 1 mM of different bile acids. For both treatments, an identical volume of DMSO as bile acid stock was added to LB liquid media as control. Both planktonic cells and biofilm on the beads were treated for 4 h, and surviving bacteria were then enumerated by serial dilution and plating.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 9 (GraphPad Software, Inc., La Jolla, CA, USA). Student’s unpaired t-test was used to analyze in vitro and ex vivo CFU and ratio group differences. The Mann–Whitney U-test was used to analyze in vivo CFU level differences between groups. Comparisons where P < 0.05 were considered statistically significant. Results are representative of independent repeated experiments.
Results
Bacterial Community-Dependent V. cholerae Colonization
Previous studies have shown that specific human gut microbiota compositions can affect V. cholerae colonization using the antibiotic-cleared infant CD-1 suckling mouse model, with dysbiotic gut microbiota being more susceptible to infection compared to prototypical microbial communities characteristic of healthy individuals (Alavi et al., 2020). As the gut microbiota is highly diverse from individual to individual, we used simplified model groups of microbes representative of “healthy” and “unhealthy” or dysbiotic states to examine the effects of microbiota structure on V. cholerae behavior during infection. The simplified infection-resistant model community (SR) consists of the species representing the major taxa found in either the healthy individuals in Bangladesh (Hsiao et al., 2014), Blautia obeum (previously classified as Ruminococcus obeum), Bacteroides vulgatus, and Clostridium scindens. The low-diversity dysbiotic state induced by recurrent diarrhea or malnutrition was represented by Streptococcus salivarius subsp. salivarius (SS). 4-day-old suckling mice were first treated with streptomycin to remove the host microbiome then coinfected with V. cholerae and the community strains after 24 h. The intestine was collected 18 h postinfection and homogenized for CFU enumeration of V. cholerae, with aliquots of homogenates also stored for host and bacterial RNA-Seq analysis. In accordance with previous findings using healthy and dysbiotic human gut microbiota (Alavi et al., 2020), we repeatedly observed that V. cholerae was able to colonize SS-containing animals at around 107 CFU; when coinfected with SR, V. cholerae colonization was around ~4-fold lower (Figure 1A). We also verified the colonization of model microbes by measuring total bacterial load using qPCR assay with universal 16S primers after antibiotic treatment and after gavage; total bacterial 16S copy number was significantly higher after bacterial gavage compared to antibiotic treated mice (Figure 1B). These findings replicate previous studies that these model microbes are capable of colonizing mice intestine (Alavi et al., 2020).
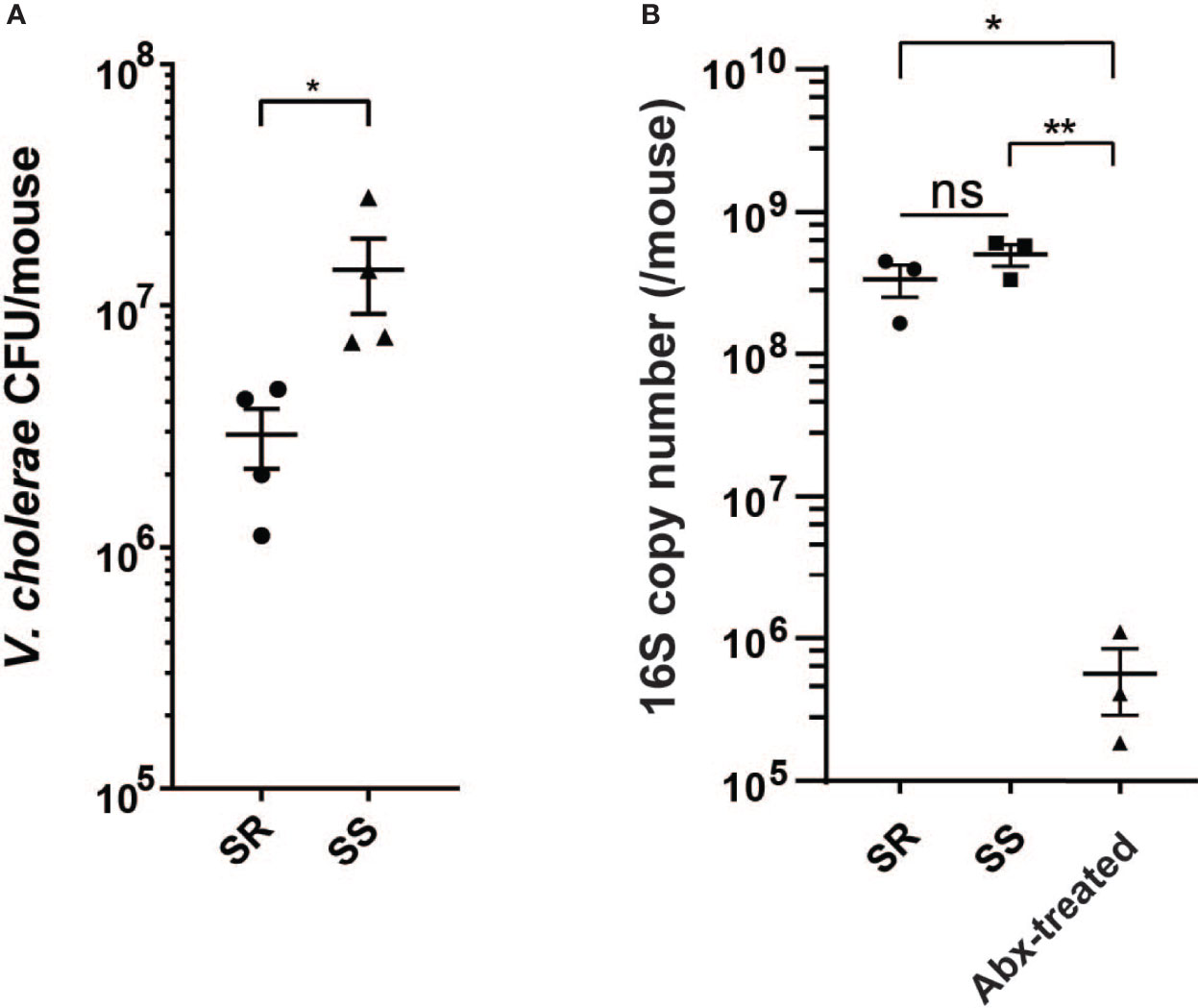
Figure 1 V. cholerae colonization and fitness varies with model commensal microbiota. (A) V. cholerae colonization of antibiotic-cleared 5-day-old suckling CD-1 mice after co-gavage with model microbial mixtures. (B) Bacterial 16S copy number in intestinal homogenates (n = 3–4, *P value < 0.05, **P value < 0.01, ns, not significant).
RNA-Seq Analysis Reveals Microbiota-Dependent Expression of Biofilm-Associated Genes
Alavi et al. determined that one contributor of microbiota-dependent variation in infection resistance was a variable level of commensal-encoded bile salt hydrolase (BSH) that de-conjugates the attached amino acid of the bile acid taurocholic acid (TCA), leading to differences in induction of virulence gene expression; B. obeum (an SR organism) was a key BSH producer, while S. salivarius lacked BSH activity (Alavi et al., 2020). To examine other potential contributors to SR/SS infection differences, we examined V. cholerae and host gene expression during infection of suckling mice. We extracted RNA from intestinal homogenates of animals colonized with both communities and infected with V. cholerae and used RNA-Seq to examine community-dependent V. cholerae gene expression patterns. Using pathway analysis with the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000; Kanehisa, 2019; Kanehisa et al., 2021), we found that V. cholerae coinfected with SS significantly upregulated biofilm formation and phosphotransferase system pathways relative to infection of SR mice; in SR mice, 16 pathways were increased in expression relative to SS, predominantly metabolic pathways including carbon metabolism, amino acid metabolism, and oxidative phosphorylation (Figure 2A). Biofilm is a population of microbes aggregated and attached to a surface in a matrix of extracellular polysaccharide (EPS), nucleic acid, and proteins. The production and manipulation of Vibrio extracellular polysaccharide (VPS) are driven by gene products encoded by two gene clusters vps-I (vpsU, vpsA-K) and vps-II (vpsL-Q) (Yildiz and Schoolnik, 1999; Fong et al., 2010). We observed that the expression of most VPS genes is upregulated 2–4-fold during infection of SS-colonized animals compared to SR-colonized animals. Among VPS genes, vpsL displayed the strongest difference in expression between microbiota groups, with 4.5-fold higher in expression in SS vs. SR. VpsL is known to be essential for forming a stable biofilm structure (Fong et al., 2010). Other than the exopolysaccharide matrix production genes, another vps-coregulated protein, Bap1 (biofilm associated protein), also displayed a 2-fold increase in expression in infection of SS animals (Figure 2B). Bap1 is an extracellular matrix protein that modulates biofilm structure and pellicle formation and mediates cell-to-surface adhesion (Fong and Yildiz, 2008; Yan et al., 2016). Biofilms protect microbes, including V. cholerae, from environmental stress including extreme environments and host defense mechanism (Roilides et al., 2015; Yin et al., 2019), and increases infectivity (Zhu and Mekalanos, 2003; Tamayo et al., 2010). Our findings suggest that infection of SS-like permissive, dysbiotic microbiota could induce V. cholerae to form a more robust biofilm structure either directly or indirectly, leading to stronger defenses against environmental stress.
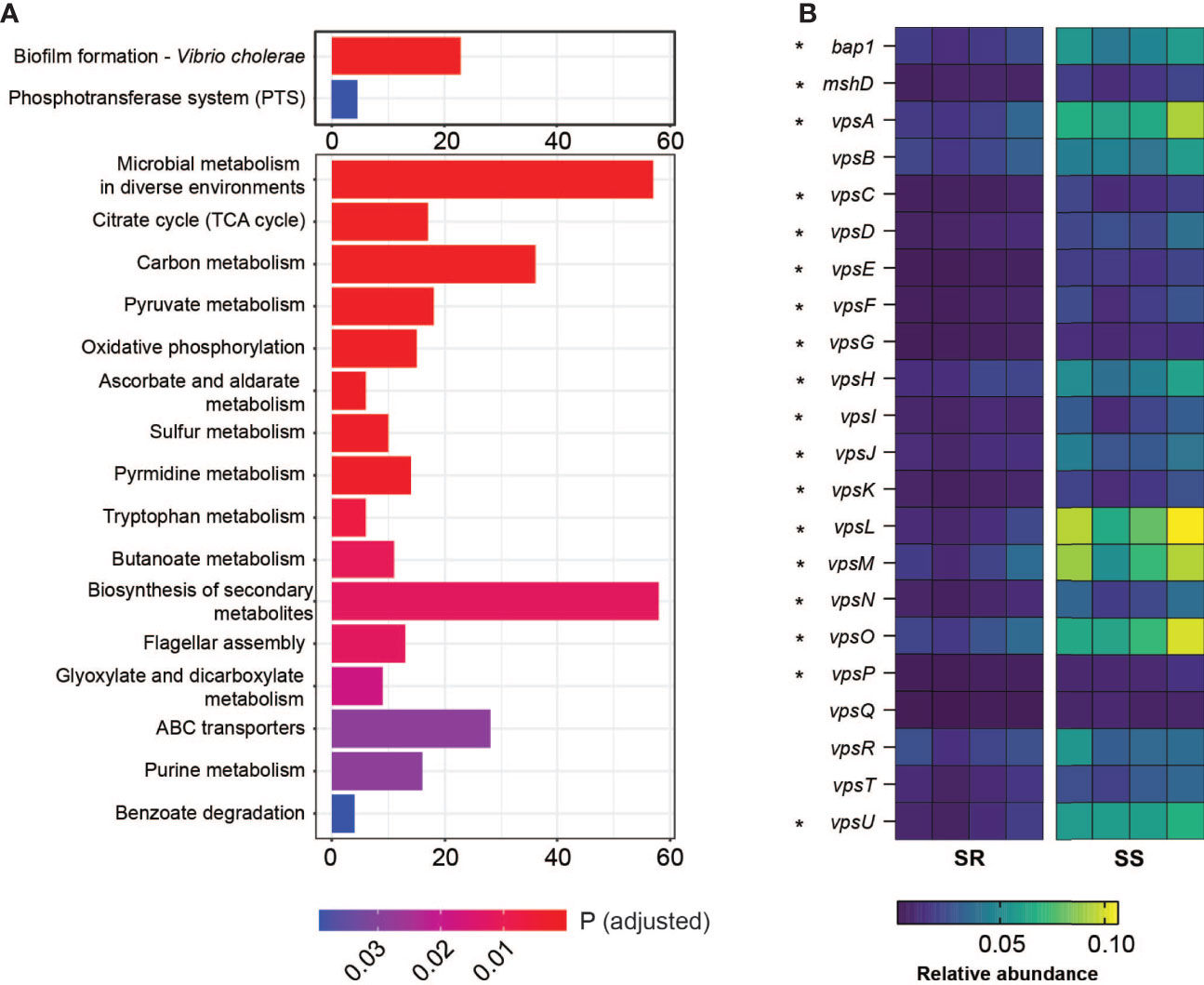
Figure 2 Differential gene regulation in V. cholerae on coinfection with different model communities. (A) Number of genes involved in indicated KEGG gene pathways relatively significantly upregulated in V. cholerae coinfected with SS vs. SR (upper panel) and SR vs. SS (bottom panel). (B) Relative abundance (%) of genes involved in Vibrio polysaccharide formation and attachment from V. cholerae coinfected with SR or SS. Starred genes (*) are significantly differentially expressed across microbiota groups (*P < 0.005).
Host iNOS and DUOX Response Toward Microbiome Structure
Given that the SS triggered the expression of the genes associated with biofilm formation in V. cholerae, and the critical role of biofilms in defense against environmental stresses, we next examined whether the host environment during infection was modulated by the microbiota through transcriptomic measures of host responses to V. cholerae infection in the presence of SR and SS model microbes. We found that mice coinfected with SR and V. cholerae exhibited a significantly higher expression of NOS1, NOS2, and DUOX2 in the total intestine (Figures 3A–C). NOS2 and DUOX2 are known to be involved in the production of compounds such as nitric oxide (NO) and extracellular hydrogen peroxide (H2O2), respectively (Geiszt et al., 2003; El Hassani et al., 2005). In response to numerous gastrointestinal infections, host enzymes are able to generate increased local reactive nitrogen stress (RNS) and reactive oxygen stress (ROS), which collectively form a key mechanism to inhibit pathogens (Paiva and Bozza, 2014). Nitric oxide synthases (NOS) catalyze the production of nitric oxide (NO), which includes three different isoforms, NOS1–3. The NO synthesized by NOS1 and NOS3 mainly acts as a neurotransmitter in the brain and peripheral nervous system and maintains a healthy cardiovascular system (Forstermann and Munzel, 2006; Knott and Bossy-Wetzel, 2009). NOS2 is involved in immune response and NO synthesis. ROS synthesis involves the expression of dual oxidase (DUOX) that catalyzes hydrogen peroxide production (Sommer and Backhed, 2015). Prior studies have demonstrated an increase in both ROS and RNS during V. cholerae infection; Ellis et al. showed that cholera patients show higher nitric oxide synthase 2 (NOS2) protein abundant during the acute phase (Ellis et al., 2015), while an examination of cholera patients’ mucosal gene expression by Bourque et al. demonstrated that dual oxidase 2 (DUOX2) and NOS2 are higher in expression during the acute phase (Bourque et al., 2018).
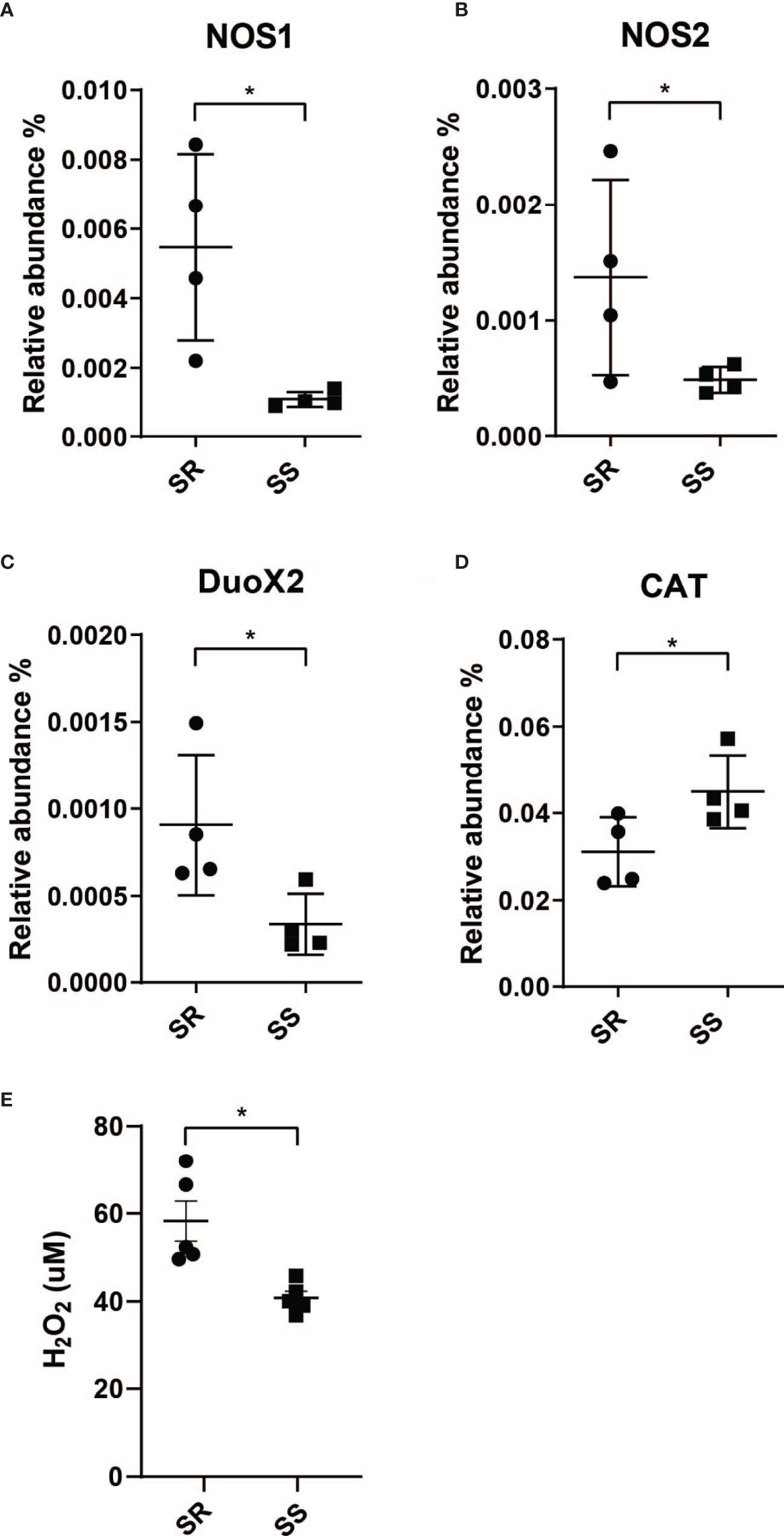
Figure 3 Community-dependent host oxidative stress responses during V. cholerae expression. Host genes involved in the synthesis of reactive nitrogen species. (A) Nitric oxide synthase 1 (NOS1). (B) Nitric oxide synthase 2 (NOS2) and reactive oxygen species. (C) Dual oxidase 2 (DUOX2). (D) Catalase (CAT). (E) H2O2 level measured from collected intestinal homogenates (n = 4, *P < 0.01).
The resulting increase in NO and H2O2 production leads to high local oxidative stress, which forms a key element of the initial host response toward pathogen infection (Kim and Lee, 2014; Król and Kepinska, 2021). These antimicrobial effects are mediated by several pathways, including conversion of NO to peroxynitrite (OONO-), and form OH· and nitrogen dioxide (·NO2) (Nathan and Ding, 2010) to cause oxidative damage on phagocytosed pathogens, and pathogen in phagosome, and H2O2 or through a catalytic reaction to form hydroxyl radicals (OH·) which leads to pathogen DNA breakage, base oxidation and deamination, and lipid peroxidation (Paiva and Bozza, 2014). This increase in ROS production has been observed in cholera patients, as measured by NO metabolite levels in serum and urine (Janoff et al., 1997). Cholera patients’ duodenum samples at the acute phase showed more abundant NOS2 and DUOX2 transcript levels compared to the convalescent phase (Ellis et al., 2015; Bourque et al., 2018). Other than higher ROS/RNS enzyme expression, host catalase (CAT) gene expression is also induced by infection in the presence of SS (Figure 3D). Since catalase is an antioxidant enzyme that promotes the conversion and detoxification of ROS and H2O2 to prevent oxidative damage to host cells, this further supports an increase in ROS stress during infection of individuals with SR-like microbiota compared to individuals with an SS-like commensal microbe. We then measured H2O2 production in the homogenates from each model microbiome and found that colonization with the SR community induced significantly higher production of H2O2 (Figure 3E). Given that a significant difference in overall H2O2 was observed from total organ samples, local differences in levels of H2O2 at epithelial microenvironments as a function of microbiome and infection may be even higher (Bhattacharyya et al., 2014; Taylor and Colgan, 2017; Burgueno et al., 2019; Imlay, 2019).
Differential Regulation of Biofilm Formation Leads to Oxidative Stress Resistance
Previous studies have shown that biofilm formation enhances V. cholerae resistance toward oxidative stresses driven by early infection (Wang et al., 2018), and our data suggest that SS can induce higher biofilm formation gene expression in V. cholerae. To test whether microbiome-dependent differences in biofilm-associated gene expression lead to an increase in oxidative stress resistance, we used an ex vivo ROS survival assay. We coinfected suckling animals with wild-type and biofilm defective vpsL mutants (Figure 4A) and our two model commensal collections. The mutant showed no significant differences in colonization between the two model microbiota (Figure 4B). We then isolated V. cholerae and associated extracellular structures by homogenizing infected small and large intestinal tissues and then treated V. cholerae-containing homogenates with H2O2. Survival rates of V. cholerae were then determined by plating. We found that wild-type V. cholerae exhibited >90% H2O2 survival after infection of the SS-co-colonized small intestine, while V. cholerae that colonized SR-bearing animals exhibited only ~55% survival. When the vpsL mutant was co-gavaged with the model microbes, this SS-dependent advantage was ablated (Figure 4C), suggesting that biofilm formation was necessary for this difference in oxidative stress resistance. We observed the same pattern from the large intestine where the wild-type V. cholerae from SS animals exhibited higher levels of ex vivo ROS resistance compared to V. cholerae from SR animals, and vpsL mutants in either microbiota context (Figure 4D), although the rate of survival in both colonization contacts was significantly reduced compared to the small intestine, which may indicate that the regulatory effects of commensal microbes are concentrated at the preferred site of colonization of V. cholerae, the distal small intestine. To determine whether biofilm biogenesis was induced by hydrogen peroxide, we treated V. cholerae culture with H2O2 in vitro and examined vpsR, vpsT, and vpsL expression. Quantitative RT-PCR analysis showed no significant differences in expression level between H2O2-treated and untreated cultures (Figure 4E), suggesting that other mechanisms, potentially microbial in nature, are responsible for observed in vivo microbiome-specific differences in V. cholerae biofilm production. Taken together, these findings suggest that i) microbiota-associated biofilm expression leads to different levels of biofilm-dependent ROS resistance in V. cholerae and ii) that this effect is much more substantial in the preferred site of colonization in the distal small intestine. In combination with higher levels of oxidative stress during infection of SR-like communities, this suggests that modulation of biofilm or biofilm formation potentially contributes to the higher V. cholerae colonization level observed during infection of SS-bearing individuals.
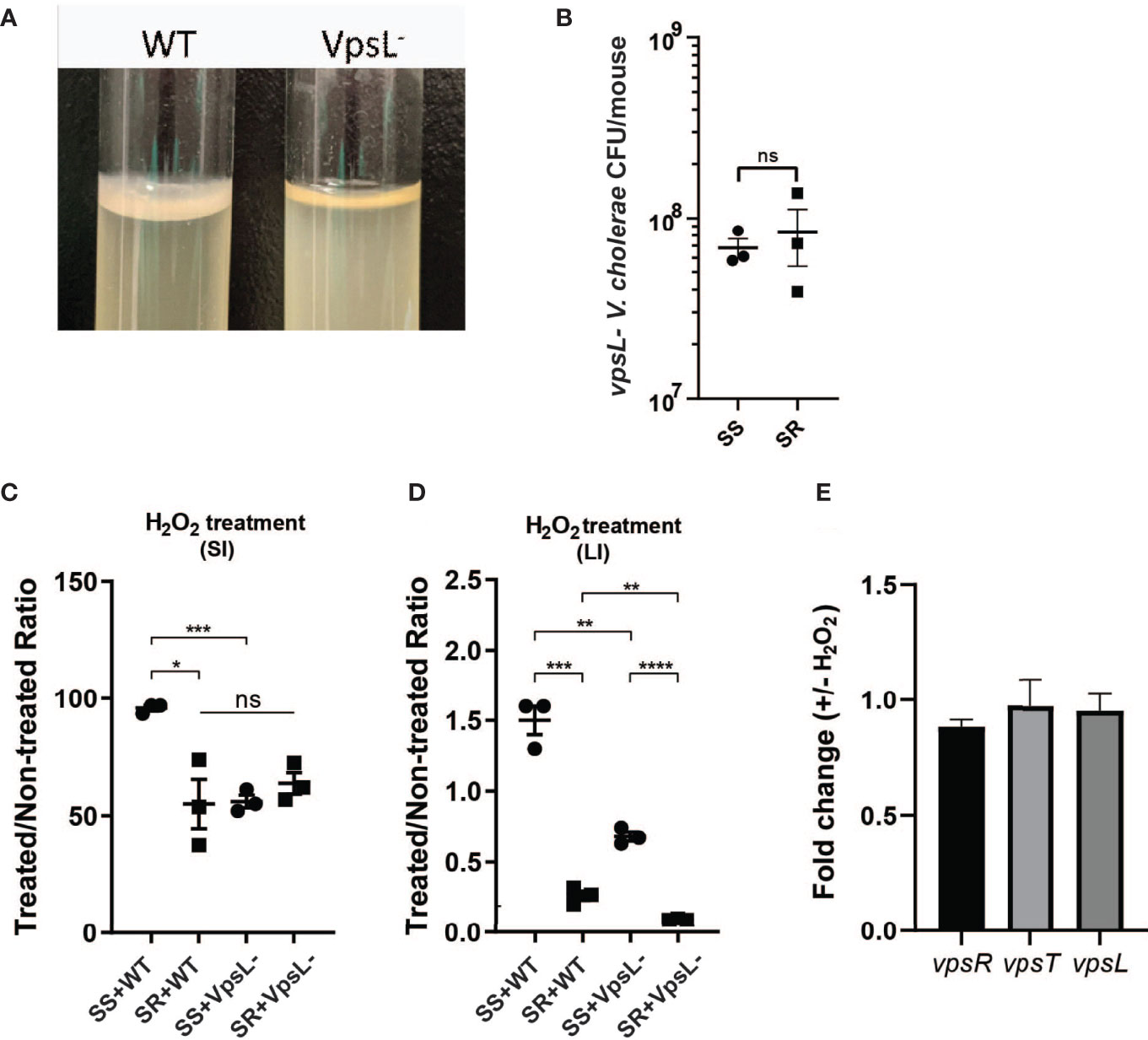
Figure 4 Presence of different commensal microbes during infection drives different biofilm-dependent V. cholerae oxidative stress resistances. (A) Confirmation of disruption of biofilm formation by vpsL gene deletion. (B) vpsL mutant V. cholerae colonization co-gavaged with indicated model microbiota. Survival of V. cholerae strains in (C) small intestine and (D) large intestine of antibiotic-cleared suckling mice co-colonized with indicated commensal communities after hydrogen peroxide treatment. (E) qPCR quantification of VPS gene expression in vitro after hydrogen peroxide treatment (n = 3, t test, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant).
Microbiota Structure and Bile Salt Hydrolase Activity Modulates Bile Acid Composition and Affects V. cholerae Growth
Since different bile acids have been associated with inhibition of V. cholerae growth in vitro and regulation of biofilm production (Hay and Zhu, 2015), we examined how host bile might affect microbiota- and biofilm-dependent V. cholerae fitness during infection. Bile is a complex mixture secreted by the gallbladder into the proximal intestine in order to aid with the emulsification and absorption of dietary fats. A dominant component of bile are bile acids and their salt forms, the primary forms of which are synthesized in the liver from cholesterol, often in amino-acid conjugated forms. The cycle of bile secretion and subsequent reabsorption in the small intestine and the detergent effects of bile mean that commensals and pathogens that target this body site have evolved methods to detoxify bile components and also use them as signaling cues to time body site-specific gene expression programs. One key element of microbial bile response is the expression of bile salt hydrolase (BSH) enzymes, which catalyze the deconjugation of amino acids from primary bile, leading to a reduction in detergent activity and subsequent toxicity (Chand et al., 2017). Subsequent microbial action can also dehydroxylate the sterol backbone of bile acids, leading to a variety of different forms of bile acids that circulate within the intestine of animals housing resident microbes (De Smet et al., 1995). Taurocholic acid (TCA), an abundant amino-acid-conjugated bile species in mouse and human gut, is able to strongly activate the V. cholerae virulence regulatory cascade by promoting the formation of disulfide bonds in the upstream regulator TcpP (Yang et al., 2013). In contrast, the de-conjugated form, cholic acid (CA), and the deconjugated and dehydroxylated form deoxycholic acid (DCA) cannot strongly induce virulence. A previous study has shown that SR microbes and SS can help drive differential colonization resistance by modulating the level of TCA conjugation and virulence gene expression (Alavi et al., 2020); however, the interaction of V. cholerae with other TCA-derived bile acid metabolites has not been well described. This is especially important as DCA generated by BSH-dependent deconjugation of TCA is reabsorbed and reconjugated to taurine by the host, being secreted as taurodeoxycholic acid (TDCA) (Figure 5A). We therefore used mass spectrometry to analyze the bile salt composition in the intestinal homogenates, before and after incubation with B. obeum and S. salivarius. We found that TDCA signals in both control and SS are significantly higher than when incubated with B. obeum (Figure 5B). DCA levels in B. obeum-treated intestinal homogenates were several logs higher compared to Streptococcus-treated and control homogenates (Figure 5C). The accumulation of DCA suggests that B. obeum is able to de-conjugate TDCA to DCA in addition to de-conjugating TCA to CA. To examine whether the B. obeum BSH enzyme is able to catalyze this reaction, we examined TDCA and DCA levels in intestinal homogenates incubated with E. coli expressing B. obeum BSH (bshc) compared to isogenic vector strains lacking any endogenous BSH activity. We observed the same low TDCA and high DCA signal patterns compared to vector control (Figure 5). To confirm the conversion from TDCA to DCA via B. obeum BSH, pure solutions of TDCA were added to bshc cell pellets in PBS and incubated for 24 h. The supernatant was collected, filter sterilized, and used for mass spectrometry analysis. Consistent with our previous findings, the level of TDCA remaining was significantly higher in the vector control compared with bshc, while DCA levels were ~600-fold higher in bshc compared to vector control (Figures 6A, B). Previous studies have shown that DCA is inhibitory to V. cholerae growth (Hung et al., 2006). To confirm this, TDCA treated with bshC or vector control was added to fresh V. cholerae culture and OD600 was taken every 30 min for 20 h. We observed that V. cholerae growth was inhibited by bshc-processed supernatant compared to the vector strain(Figure 6C). Importantly, the presence of B. obeum bsh activity is able to provide inhibitory activity in the presence of Streptococcus, supporting the idea that the presence of infection-resistant microbes are able to restore colonization resistance to otherwise susceptible microbiota configurations (Alavi et al., 2020). Taken together, these results indicate that B. obeum is capable of converting TDCA to DCA via BSH expression, leading to V. cholerae growth inhibition during colonization.
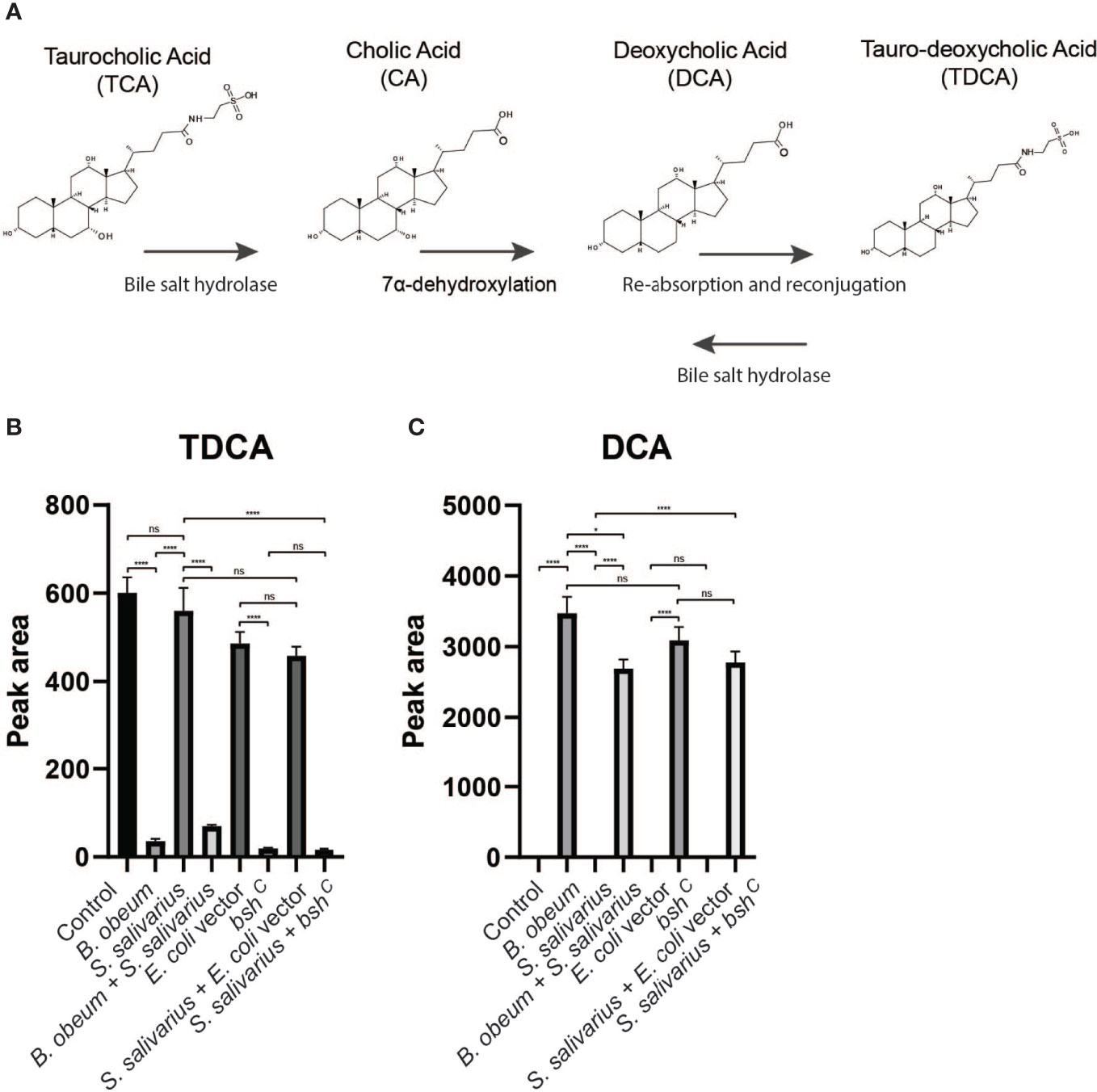
Figure 5 Bile salt hydrolase-dependent TDCA and DCA levels in infant mouse intestines. (A) Model of microbial modification of bile salt in the gut. (B) Tauro-deoxycholate (TDCA) and (C) deoxycholate (DCA) levels in intestinal homogenates after incubation with indicated strains. (n = 4, t test, *P < 0.05; ****P < 0.0001; ns, not significant).
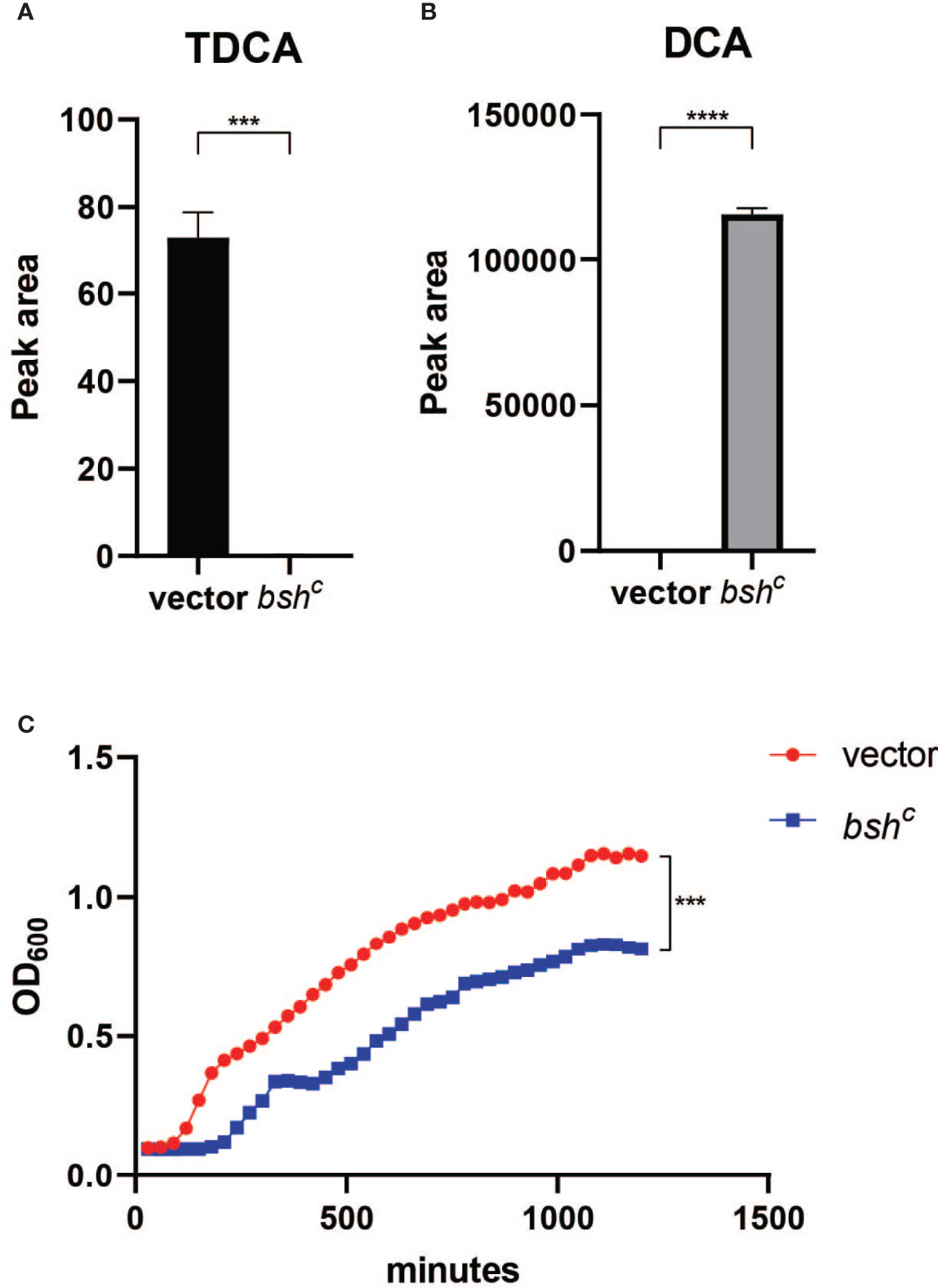
Figure 6 Blautia obeum bile salt hydrolase processes TDCA to DCA leading to inhibition of V. cholerae growth. (A) TDCA and (B) DCA levels after 24-h incubation of 2 mM TDCA with E. coli vector and bshc strains as measured by mass spectrometry. (C) Effect on V. cholerae growth in vitro of addition of filtered supernatants from TDCA incubation with the indicated strains (n = 4, t test, ***P < 0.001; ****P < 0.0001).
Biofilm-Dependent Bile Acid Resistance
Previous studies have indicated that crude bile is able to induce biofilm formation, and that biofilm-associated cells are more resistant to bile toxicity (Hung et al., 2006). To test whether biofilm will contribute to V. cholerae resistance against different microbially modified bile acids, we first generated biofilm V. cholerae populations in media with beads as a surface for microbial attachment and subsequent biofilm development. We then exposed planktonic and biofilm-formed populations with products of microbial TCA metabolism: TCA, DCA, and TDCA. We found that TCA and TDCA do not significantly affect V. cholerae planktonic cell fitness in comparison with no bile acid (Figure 7A), but that after 4 h of treatment with 1 mM of DCA, planktonic V. cholerae exhibited a significantly lower survival. In contrast, biofilm-associated V. cholerae were insensitive to all bile treatments (Figure 7B). These repeatable data suggest that the formation of biofilm provides V. cholerae resistance toward certain bile acids present in the intestinal tract.
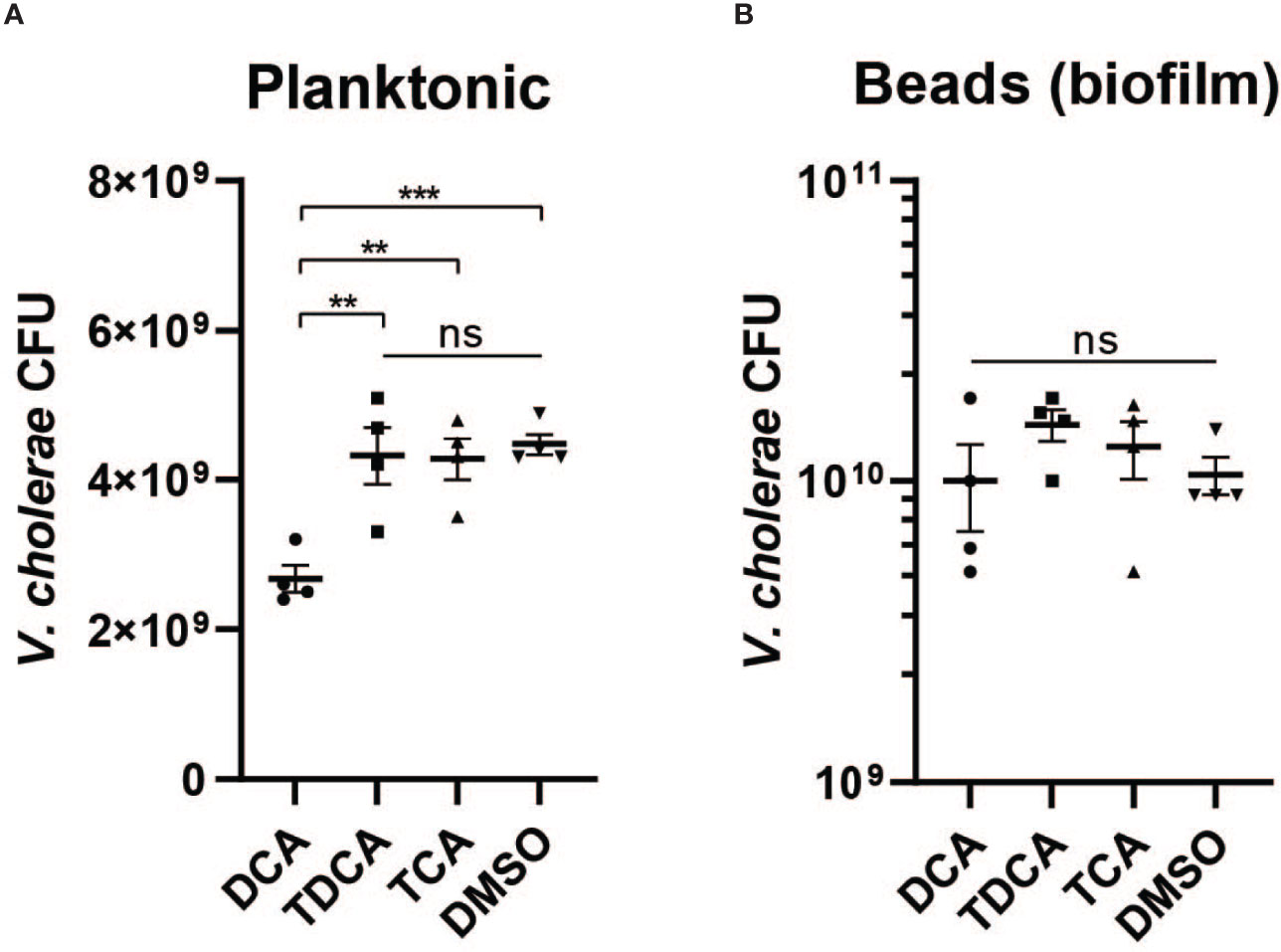
Figure 7 Biofilm provides V. cholerae resistance against secondary bile acid deoxycholate. Survival of (A) planktonic and (B) biofilm associated V. cholerae after 4 h of incubation with 1 mM of different bile acids including conjugated bile acids taurocholate (TCA) and taurodeoxycholate (TDCA), and the deconjugated secondary bile acid deoxycholate (DCA) (n = 4, T-test, **P < 0.01, ***P < 0.001; ns, not significant).
Discussion
Increasing evidence demonstrates that the composition of the gut microbiota influences host defenses against pathogens (Ribet and Cossart, 2015; Cho et al., 2021). This study demonstrates that commensal-dependent manipulation of both V. cholerae and host behavior during infection leads to differential outcomes in V. cholerae fitness. V. cholerae in the context of infection-susceptible communities expresses more biofilm-associated genes, which protects the pathogen against the inhibitory activity of host oxidative mechanisms and inhibitory bile metabolites. The commensal community present during infection in turn affects both mechanisms; SR-colonized mice during infection express significantly higher levels of ROS/RNS production enzymes and display elevated hydrogen peroxide production compared to SS-colonized animals, and we show that one bile acid-processing enzyme within the key SR community member B. obeum can accumulate deoxycholate, which is inhibitory to planktonic, but not biofilm-associated, V. cholerae. These results demonstrate the multifactorial nature of commensal-associated colonization resistance; prior studies show that the dysbiotic, high Streptococcus microbiota is more susceptible to V. cholerae infection due to the inability to ablate bile-dependent virulence gene activation compared to microbiota containing B. obeum (Alavi et al., 2020).
The exact mechanism by which commensal microbes cause differential regulation of V. cholerae is not well understood. V. cholerae biofilm production in vitro is modulated by several pathways, including quorum sensing and bile response (Hung et al., 2006; Hay and Zhu, 2015). Quorum sensing (QS) plays a key role in the regulation of multiple infection-associated pathways in V. cholerae including biofilm production. The sensing of autoinducers leads to a series of dephosphorylation events leading to the repression of the expression of the small RNA regulators Qrr 1–4 (quorum regulatory RNAs) (Lenz et al., 2004). The absence of Qrr sRNAs allows for the expression of the master QS regulator hapR, which leads to the repression of the virulence genes and biofilm formation (Miller et al., 2002; Zhu et al., 2002; Hammer and Bassler, 2003; Shao and Bassler, 2014). HapR represses biofilm formation by binding to the regulatory regions of positive regulator vpsT and polysaccharide biosynthesis glycosyltransferase vpsL, also the first gene of the vps-II cluster in V. cholerae (Waters et al., 2008). Interestingly, production of the interspecies capable autoinducer AI-2 by B. obeum has been associated with downregulation of V. cholerae virulence and biofilm formation through HapR by transcription regulator VqmA (Liu et al., 2006; Hsiao et al., 2014; Papenfort et al., 2015). While the bi-association of B. obeum and V. cholerae in adult germ-free mice revealed no specific transcriptional signatures for biofilm gene regulation in fecal samples, our results suggest significant differences in biofilm regulation in the small intestine vs. large intestine (Figures 4C, D); compartment-specific microbiota effects on transcription may thus be masked in fecal transcriptomic data. Treatment with hydrogen peroxide in vitro also caused no significant increases in VPS gene expression, further suggesting that the regulation of biofilm is a host environment-specific phenotype (Figure 4E). In vitro studies have also examined the role of bile salts/acids in biofilm regulation. Hung et al. showed that deoxycholate is able to significantly induce V. cholerae biofilm formation in culture (Hung et al., 2006). The presence of bile also has been shown to alter VqmA activity and effect on V. cholerae virulence regulation (Mashruwala and Bassler, 2020). While this suggests that the SR community, with the ability to accumulate DCA via the activity of B. obeum BSH, should promote biofilm formation, the intersection of other signaling and regulatory pathways such as quorum sensing (Mashruwala and Bassler, 2020), and host oxidative stresses, which lead to increased biofilm formation (Wang et al., 2018), may combine to yield gut compartment- and microbiota-specific effects on V. cholerae biofilm regulation.
The role of microbial enzymes in manipulating the host bile acid pool is complex. TCA, CA, TDCA, and DCA are all metabolic products driven by microbial action; in germ-free animals, the bile acid pool is almost exclusively amino acid-conjugated primary bile molecules such as TCA (Sayin et al., 2013). Microbial BSH activity deconjugates primary molecules such as TCA, generating CA. Further microbial enzymatic action leads to dehydroxylation of CA to DCA (Molinaro et al., 2018), which can then be reabsorbed by the host, re-conjugated to amino acids, and re-secreted as TDCA (Figure 5A). Thus, multiple commensal pathways can contribute to highly community- and individual-specific bile acid pools. Microbial communities may interact to drive bile acid compositions in vivo; both Bacteroides and C. scindens within the SR community have been shown to exhibit both BSH and dehydroxylation activity (Kawamoto et al., 1989; Kang et al., 2008). Further complexity is added by the substrate specificity of microbial BSH, which has been grouped into eight phylotypes based on amino acid sequence similarity and substrate specificity (Song et al., 2019). Previous work by Alavi et al. showed that B. obeum BSH activity leads to restriction of V. cholerae infection by processing TCA, which is a potent activator of key V. cholerae virulence genes. In this study, we show that B. obeum is also able to process TDCA to DCA; accumulation of DCA leads to inhibition of planktonic, but not biofilm-associated, V. cholerae. The reduction of the V. cholerae vps gene expression in SR communities thus reinforces the colonization-resistant nature of host intestines bearing SR microbes.
This suggests that infection-resistant commensal communities, including B. obeum, can control V. cholerae fitness during infection by multiple, reinforcing mechanisms. Production of quorum sensing autoinducers and depletion of conjugated bile acids serving as virulence regulators interrupt the finely calibrated sequence of virulence gene expression regulatory events during early infection, coincident with an increased host oxidative ROS/RNS response compared to the high Streptococcus dysbiotic and permissive microbiota. While the precise mechanism leading to lower vps gene expression in colonization-resistant communities requires additional work to elucidate, the reduced biofilm formation leads to increased susceptibility to the heightened oxidative stress encountered by V. cholerae during infection in these microbiota contexts. Furthermore, B. obeum bile salt hydrolase activity not only depletes virulence-gene activating signals such as TCA but leads to the accumulation of DCA, the growth-inhibitory effects of which are accentuated by reduced V. cholerae biofilm production.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found as follows: https://www.ncbi.nlm.nih.gov/bioproject/, 788969.
Ethics Statement
The animal studies were reviewed and approved by the Institutional Animal Care and Use Committee, University of California, Riverside.
Author Contributions
AH and JC conceived the study. JC performed the experiments. RL performed the RNA-Seq analysis. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by NIH grants GM124724 and AI157106 (to AH).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Metabolomics Core Facility at University of California, Riverside, for mass spectrometry studies.
References
Alavi, S., Mitchell, J. D., Cho, J. Y., Liu, R., Macbeth, J. C., Hsiao, A. (2020). Interpersonal Gut Microbiome Variation Drives Susceptibility and Resistance to Cholera Infection. Cell 181, 1533–1546.e1513. doi: 10.1016/j.cell.2020.05.036
Anders, S., Pyl, P. T., Huber, W. (2015). HTSeq–a Python Framework to Work With High-Throughput Sequencing Data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Bhattacharyya, A., Chattopadhyay, R., Mitra, S., Crowe, S. E. (2014). Oxidative Stress: An Essential Factor in the Pathogenesis of Gastrointestinal Mucosal Diseases. Physiol. Rev. 94, 329–354. doi: 10.1152/physrev.00040.2012
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bourque, D. L., Bhuiyan, T. R., Genereux, D. P., Rashu, R., Ellis, C. N., Chowdhury, F., et al. (2018). Analysis of the Human Mucosal Response to Cholera Reveals Sustained Activation of Innate Immune Signaling Pathways. Infect. Immun. 86. doi: 10.1128/IAI.00594-17
Burgueno, J. F., Fritsch, J., Santander, A. M., Brito, N., Fernandez, I., Pignac-Kobinger, J., et al. (2019). Intestinal Epithelial Cells Respond to Chronic Inflammation and Dysbiosis by Synthesizing H2o2. Front. Physiol. 10, 1484. doi: 10.3389/fphys.2019.01484
Chand, D., Avinash, V. S., Yadav, Y., Pundle, A. V., Suresh, C. G., Ramasamy, S. (2017). Molecular Features of Bile Salt Hydrolases and Relevance in Human Health. Biochim. Biophys. Acta Gen. Subj. 1861, 2981–2991. doi: 10.1016/j.bbagen.2016.09.024
Cho, J. Y., Liu, R., Macbeth, J. C., Hsiao, A. (2021). The Interface of Vibrio Cholerae and the Gut Microbiome. Gut Microbes 13, 1937015. doi: 10.1080/19490976.2021.1937015
Dalia, A. B., Mcdonough, E., Camilli, A. (2014). Multiplex Genome Editing by Natural Transformation. Proc. Natl. Acad. Sci. 111, 8937–8942. doi: 10.1073/pnas.1406478111
David, L. A., Weil, A., Ryan, E. T., Calderwood, S. B., Harris, J. B., Chowdhury, F., et al. (2015). Gut Microbial Succession Follows Acute Secretory Diarrhea in Humans. mBio 6, e00381–e00315. doi: 10.1128/mBio.00381-15
De Smet, I., Van Hoorde, L., Vande Woestyne, M., Christiaens, H., Verstraete, W. (1995). Significance of Bile Salt Hydrolytic Activities of Lactobacilli. J. Appl. Bacteriol. 79, 292–301. doi: 10.1111/j.1365-2672.1995.tb03140.x
El Hassani, R. A., Benfares, N., Caillou, B., Talbot, M., Sabourin, J. C., V, B., et al. (2005). Dual Oxidase2 is Expressed All Along the Digestive Tract. Am. J. Physiol. Gastrointest. Liver Physiol. 288, G933–G942. doi: 10.1152/ajpgi.00198.2004
Ellis, C. N., Larocque, R. C., Uddin, T., Krastins, B., Mayo-Smith, L. M., Sarracino, D., et al. (2015). Comparative Proteomic Analysis Reveals Activation of Mucosal Innate Immune Signaling Pathways During Cholera. Infect. Immun. 83, 1089–1103. doi: 10.1128/IAI.02765-14
Fong, J. C. N., Syed, K. A., Klose, K. E., Yildiz, F. H. (2010). Role of Vibrio Polysaccharide (Vps) Genes in VPS Production, Biofilm Formation and Vibrio Cholerae Pathogenesis. Microbiol. (Reading) 156, 2757–2769. doi: 10.1099/mic.0.040196-0
Fong, J. C., Yildiz, F. H. (2008). Interplay Between Cyclic AMP-Cyclic AMP Receptor Protein and Cyclic Di-GMP Signaling in Vibrio Cholerae Biofilm Formation. J. Bacteriol. 190, 6646–6659. doi: 10.1128/JB.00466-08
Forstermann, U., Munzel, T. (2006). Endothelial Nitric Oxide Synthase in Vascular Disease: From Marvel to Menace. Circulation 113, 1708–1714. doi: 10.1161/CIRCULATIONAHA.105.602532
Geiszt, M., Witta, J., Baffi, J., Lekstrom, K., Leto, T. L. (2003). Dual Oxidases Represent Novel Hydrogen Peroxide Sources Supporting Mucosal Surface Host Defense. FASEB J. 17, 1502–1504. doi: 10.1096/fj.02-1104fje
Hammer, B. K., Bassler, B. L. (2003). Quorum Sensing Controls Biofilm Formation in Vibrio Cholerae. Mol. Microbiol. 50, 101–114. doi: 10.1046/j.1365-2958.2003.03688.x
Hay, A. J., Zhu, J. (2015). Host Intestinal Signal-Promoted Biofilm Dispersal Induces Vibrio Cholerae Colonization. Infect. Immun. 83, 317–323. doi: 10.1128/IAI.02617-14
Horz, H. P., Vianna, M. E., Gomes, B. P., Conrads, G. (2005). Evaluation of Universal Probes and Primer Sets for Assessing Total Bacterial Load in Clinical Samples: General Implications and Practical Use in Endodontic Antimicrobial Therapy. J. Clin. Microbiol. 43, 5332–5337. doi: 10.1128/JCM.43.10.5332-5337.2005
Hsiao, A., Ahmed, A. M., Subramanian, S., Griffin, N. W., Drewry, L. L., Petri, W. A., Jr., et al. (2014). Members of the Human Gut Microbiota Involved in Recovery From Vibrio Cholerae Infection. Nature 515, 423–426. doi: 10.1038/nature13738
Humbert, L., Maubert, M. A., Wolf, C., Duboc, H., Mahe, M., Farabos, D., et al. (2012). Bile Acid Profiling in Human Biological Samples: Comparison of Extraction Procedures and Application to Normal and Cholestatic Patients. J. Chromatogr. B Analyt. Technol. BioMed. Life Sci. 899, 135–145. doi: 10.1016/j.jchromb.2012.05.015
Hung, D. T., Zhu, J., Sturtevant, D., Mekalanos, J. J. (2006). Bile Acids Stimulate Biofilm Formation in Vibrio Cholerae. Mol. Microbiol. 59, 193–201. doi: 10.1111/j.1365-2958.2005.04846.x
Imlay, J. A. (2019). Where in the World do Bacteria Experience Oxidative Stress? Environ. Microbiol. 21, 521–530. doi: 10.1111/1462-2920.14445
Janoff, E. N., Hayakawa, H., Taylor, D. N., Fasching, C. E., Kenner, J. R., Jaimes, E., et al. (1997). Nitric Oxide Production During Vibrio Cholerae Infection. Am. J. Physiol. 273, G1160–G1167. doi: 10.1152/ajpgi.1997.273.5.G1160
Kanehisa, M. (2019). Toward Understanding the Origin and Evolution of Cellular Organisms. Protein Sci. 28, 1947–1951. doi: 10.1002/pro.3715
Kanehisa, M., Furumichi, M., Sato, Y., Ishiguro-Watanabe, M., Tanabe, M. (2021). KEGG: Integrating Viruses and Cellular Organisms. Nucleic Acids Res. 49, D545–D551. doi: 10.1093/nar/gkaa970
Kanehisa, M., Goto, S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kang, D. J., Ridlon, J. M., Moore, D. R., 2nd, Barnes, S., Hylemon, P. B. (2008). Clostridium Scindens baiCD and baiH Genes Encode Stereo-Specific 7alpha/7beta-Hydroxy-3-Oxo-Delta4-Cholenoic Acid Oxidoreductases. Biochim. Biophys. Acta 1781, 16–25. doi: 10.1016/j.bbalip.2007.10.008
Kawamoto, K., Horibe, I., Uchida, K. (1989). Purification and Characterization of a New Hydrolase for Conjugated Bile Acids, Chenodeoxycholyltaurine Hydrolase, From Bacteroides Vulgatus. J. Biochem. 106, 1049–1053. doi: 10.1093/oxfordjournals.jbchem.a122962
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: A Fast Spliced Aligner With Low Memory Requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kim, S. H., Lee, W. J. (2014). Role of DUOX in Gut Inflammation: Lessons From Drosophila Model of Gut-Microbiota Interactions. Front. Cell Infect. Microbiol. 3, 116. doi: 10.3389/fcimb.2013.00116
Knott, A. B., Bossy-Wetzel, E. (2009). Nitric Oxide in Health and Disease of the Nervous System. Antioxid. Redox Signal 11, 541–554. doi: 10.1089/ars.2008.2234
Król, M., Kepinska, M. (2021). Human Nitric Oxide Synthase—Its Functions, Polymorphisms, and Inhibitors in the Context of Inflammation, Diabetes and Cardiovascular Diseases. Int. J. Mol. Sci. 22, 56. doi: 10.3390/ijms22010056
Lenz, D. H., Mok, K. C., Lilley, B. N., Kulkarni, R. V., Wingreen, N. S., Bassler, B. L. (2004). The Small RNA Chaperone Hfq and Multiple Small RNAs Control Quorum Sensing in Vibrio Harveyi and Vibrio Cholerae. Cell 118, 69–82. doi: 10.1016/j.cell.2004.06.009
Liu, Z., Hsiao, A., Joelsson, A., Zhu, J. (2006). The Transcriptional Regulator VqmA Increases Expression of the Quorum-Sensing Activator HapR in Vibrio Cholerae. J. Bacteriol. 188, 2446–2453. doi: 10.1128/JB.188.7.2446-2453.2006
Louis, P., O’byrne, C. P. (2010). Life in the Gut: Microbial Responses to Stress in the Gastrointestinal Tract. Sci. Prog. 93, 7–36. doi: 10.3184/003685009X12605525292307
Mashruwala, A. A., Bassler, B. L. (2020). The Vibrio Cholerae Quorum-Sensing Protein VqmA Integrates Cell Density, Environmental, and Host-Derived Cues Into the Control of Virulence. mBio 11, 1–19. doi: 10.1128/mBio.01572-20
Matz, C., Mcdougald, D., Moreno, A. M., Yung, P. Y., Yildiz, F. H., Kjelleberg, S. (2005). Biofilm Formation and Phenotypic Variation Enhance Predation-Driven Persistence of Vibrio Cholerae. Proc. Natl. Acad. Sci. U.S.A. 102, 16819–16824. doi: 10.1073/pnas.0505350102
Mccarthy, D. J., Chen, Y., Smyth, G. K. (2012). Differential Expression Analysis of Multifactor RNA-Seq Experiments With Respect to Biological Variation. Nucleic Acids Res. 40, 4288–4297. doi: 10.1093/nar/gks042
Miller, M. B., Skorupski, K., Lenz, D. H., Taylor, R. K., Bassler, B. L. (2002). Parallel Quorum Sensing Systems Converge to Regulate Virulence in Vibrio Cholerae. Cell 110, 303–314. doi: 10.1016/S0092-8674(02)00829-2
Molinaro, A., Wahlstrom, A., Marschall, H. U. (2018). Role of Bile Acids in Metabolic Control. Trends Endocrinol. Metab. 29, 31–41. doi: 10.1016/j.tem.2017.11.002
Nathan, C., Ding, A. (2010). SnapShot: Reactive Oxygen Intermediates (ROI). Cell 140951-951, e952. doi: 10.1016/j.cell.2010.03.008
Paiva, C. N., Bozza, M. T. (2014). Are Reactive Oxygen Species Always Detrimental to Pathogens? Antioxid. Redox Signal 20, 1000–1037. doi: 10.1089/ars.2013.5447
Papenfort, K., Forstner, K. U., Cong, J. P., Sharma, C. M., Bassler, B. L. (2015). Differential RNA-Seq of Vibrio Cholerae Identifies the VqmR Small RNA as a Regulator of Biofilm Formation. Proc. Natl. Acad. Sci. U.S.A. 112, E766–E775. doi: 10.1073/pnas.1500203112
Ribet, D., Cossart, P. (2015). How Bacterial Pathogens Colonize Their Hosts and Invade Deeper Tissues. Microbes Infect. 17, 173–183. doi: 10.1016/j.micinf.2015.01.004
Robinson, M. D., Mccarthy, D. J., Smyth, G. K. (2009). Edger: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Roilides, E., Simitsopoulou, M., Katragkou, A., Walsh, T. J., Ghannoum, M., Parsek, M., et al. (2015). How Biofilms Evade Host Defenses Microbiol. Spectr. 3, 3.3.22. doi: 10.1128/9781555817466.ch14
Sayin, S. I., Wahlstrom, A., Felin, J., Jantti, S., Marschall, H. U., Bamberg, K., et al. (2013). Gut Microbiota Regulates Bile Acid Metabolism by Reducing the Levels of Tauro-Beta-Muricholic Acid, A Naturally Occurring FXR Antagonist. Cell Metab. 17, 225–235. doi: 10.1016/j.cmet.2013.01.003
Shao, Y., Bassler, B. L. (2014). Quorum Regulatory Small RNAs Repress Type VI Secretion in Vibrio Cholerae. Mol. Microbiol. 92, 921–930. doi: 10.1111/mmi.12599
Sommer, F., Backhed, F. (2015). The Gut Microbiota Engages Different Signaling Pathways to Induce Duox2 Expression in the Ileum and Colon Epithelium. Mucosal Immunol. 8, 372–379. doi: 10.1038/mi.2014.74
Song, Z., Cai, Y., Lao, X., Wang, X., Lin, X., Cui, Y., et al. (2019). Taxonomic Profiling and Populational Patterns of Bacterial Bile Salt Hydrolase (BSH) Genes Based on Worldwide Human Gut Microbiome. Microbiome 7, 9–9. doi: 10.1186/s40168-019-0628-3
Tamayo, R., Patimalla, B., Camilli, A. (2010). Growth in a Biofilm Induces a Hyperinfectious Phenotype in Vibrio Cholerae. Infect. Immun. 78, 3560–3569. doi: 10.1128/IAI.00048-10
Taylor, C. T., Colgan, S. P. (2017). Regulation of Immunity and Inflammation by Hypoxia in Immunological Niches. Nat. Rev. Immunol. 17, 774–785. doi: 10.1038/nri.2017.103
Wang, H., Xing, X., Wang, J., Pang, B., Liu, M., Larios-Valencia, J., et al. (2018). Hypermutation-Induced In Vivo Oxidative Stress Resistance Enhances Vibrio Cholerae Host Adaptation. PloS Pathog. 14. doi: 10.1371/journal.ppat.1007413
Waters, C. M., Lu, W., Rabinowitz, J. D., Bassler, B. L. (2008). Quorum Sensing Controls Biofilm Formation in Vibrio Cholerae Through Modulation of Cyclic Di-GMP Levels and Repression of vpsT. J. Bacteriol. 190, 2527–2536. doi: 10.1128/JB.01756-07
Wu, T., Hu, E., Xu, S., Chen, M., Guo, P., Dai, Z., et al. (2021). Clusterprofiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation (N Y) 2, 100141. doi: 10.1016/j.xinn.2021.100141
Yang, M., Liu, Z., Hughes, C., Stern, A. M., Wang, H., Zhong, Z., et al. (2013). Bile Salt-Induced Intermolecular Disulfide Bond Formation Activates Vibrio Cholerae Virulence. Proc. Natl. Acad. Sci. U.S.A. 110, 2348–2353. doi: 10.1073/pnas.1218039110
Yan, J., Sharo, A. G., Stone, H. A., Wingreen, N. S., Bassler, B. L. (2016). Vibrio Cholerae Biofilm Growth Program and Architecture Revealed by Single-Cell Live Imaging. Proc. Natl. Acad. Sci. U.S.A. 113, E5337–E5343. doi: 10.1073/pnas.1611494113
Yildiz, F. H., Schoolnik, G. K. (1999). Vibrio Cholerae O1 El Tor: Identification of a Gene Cluster Required for the Rugose Colony Type, Exopolysaccharide Production, Chlorine Resistance, and Biofilm Formation. Proc. Natl. Acad. Sci. U.S.A. 96, 4028–4033. doi: 10.1073/pnas.96.7.4028
Yin, W., Wang, Y., Liu, L., He, J. (2019). Biofilms: The Microbial "Protective Clothing" in Extreme Environments. Int. J. Mol. Sci. 20. doi: 10.3390/ijms20143423
Yu, G. C., Wang, L. G., Han, Y. Y., He, Q. Y. (2012). Clusterprofiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omics-a J. Integr. Biol. 16, 284–287. doi: 10.1089/omi.2011.0118
Zhao, W., Caro, F., Robins, W., Mekalanos, J. J. (2018). Antagonism Toward the Intestinal Microbiota and its Effect on Vibrio Cholerae Virulence. Science 359, 210–213. doi: 10.1126/science.aap8775
Zhu, J., Mekalanos, J. J. (2003). Quorum Sensing-Dependent Biofilms Enhance Colonization in Vibrio Cholerae. Dev. Cell 5, 647–656. doi: 10.1016/S1534-5807(03)00295-8
Keywords: biofilm, Vibrio cholerae, pathogenesis, microbiota, bile acids, reactive oxygen species
Citation: Cho JY, Liu R and Hsiao A (2022) Microbiota-Associated Biofilm Regulation Leads to Vibrio cholerae Resistance Against Intestinal Environmental Stress. Front. Cell. Infect. Microbiol. 12:861677. doi: 10.3389/fcimb.2022.861677
Received: 24 January 2022; Accepted: 25 March 2022;
Published: 27 April 2022.
Edited by:
Jyl S. Matson, University of Toledo, United StatesReviewed by:
Simone Becattini, Université de Genève, SwitzerlandYang Fu, Southern University of Science and Technology, China
Copyright © 2022 Cho, Liu and Hsiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ansel Hsiao, YW5zZWwuaHNpYW9AdWNyLmVkdQ==
 Jennifer Y. Cho
Jennifer Y. Cho Rui Liu1,3
Rui Liu1,3 Ansel Hsiao
Ansel Hsiao