- 1Department of Microbiology, Obafemi Awolowo University, Ile-Ife, Nigeria
- 2Institute of Medical Microbiology, University Hospital Münster, Münster, Germany
- 3National Reference Centre (NRC) for Staphylococci and Enterococci, Division of Nosocomial Pathogens and Antibiotic Resistances, Department of Infectious Diseases, Robert Koch Institute, Wernigerode Branch, Wernigerode, Germany
- 4Institute for Hygiene, University Hospital Münster, Münster, Germany
We describe the identification of a methicillin-resistant, high-level mupirocin-resistant Staphylococcus argenteus. The isolate (1801221) was characterized as t6675-ST2250-SCCmecIVc, and whole-genome sequencing revealed that the isolate possessed two plasmids. One plasmid (34,870 bp), designated p1_1801221 with rep23, harboured the mupirocin resistance (mupA) gene. The second plasmid (20,644 bp), assigned as p2_1801221 with rep5a and rep16, carried the resistance determinants for penicillin (blaZ) and cadmium (cadD). Phylogenetic analysis revealed that the isolate clustered with the European ST2250 lineage. The overall high similarity of both plasmids in S. argenteus with published DNA sequences of Staphylococcus aureus plasmids strongly suggests an interspecies transfer. The pathogenic potential, community and nosocomial spread, and acquisition of antibiotic resistance gene determinants, including the mupA gene by S. argenteus, highlight its clinical significance and the need for its correct identification.
Introduction
Staphylococcus argenteus and S. schweitzeri, with S. roterodami and S. singaporensis, are recently designated species and assigned to the Staphylococcus aureus-related complex (Tong et al., 2015; Chew et al., 2021; Schutte et al., 2021). S. argenteus and S. aureus demonstrate similar reactions to key biochemical tests for phenotypic characterization with identical 16S rRNA gene sequences (Tong et al., 2015). Hence, it is difficult to distinguish these two species by routine diagnostic methods (Kaden et al., 2018; Tunsjø et al., 2018). Various tools have been developed to differentiate S. argenteus from the S. aureus-related complex (Becker et al., 2019). They include Matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Chen et al., 2018a) and PCR detection of the nonribosomal peptide synthetase (NRPS) gene (Zhang et al., 2016). S. argenteus was previously considered less virulent than S. aureus due to the lack of the carotenoid pigment, staphyloxanthin (Holt et al., 2011), which impairs oxidative stress and neutrophil killing (Liu et al., 2005). However, S. argenteus possesses similar S. aureus virulence determinants (Zhang et al., 2017), including the gene encoding Panton-Valentine leukocidin (PVL) (Chantratita et al., 2016).
There are increasing reports of S. argenteus infections worldwide (Chantratita et al., 2016; Alhussein et al., 2020; Diot et al., 2020; Hao et al., 2020; Mitsutake et al., 2020; Eshaghi et al., 2021). S. argenteus isolates are generally penicillin-resistant (blaZ-positive) (Becker et al., 2019), but in Europe, methicillin-resistant (MR)-S. argenteus (>10 isolates) have been identified in Denmark (Hansen et al., 2017), Netherlands (Bank et al., 2021) and Sweden (Hallbäck et al., 2018; Giske et al., 2019). Also, a recent study (Goswami et al., 2021) revealed that of the S. argenteus genomes deposited in the public databases, 20% were mecA-positive. Becker et al. (2019), in a position paper on the S. aureus-related complex, suggested adopting infection prevention and control measures similar to methicillin-resistant S. aureus (MRSA) guidelines on a laboratory report of MR-S. argenteus in human infections. The application of mupirocin ointment on the mucous membrane (e.g., anterior nares) is an important strategy for decolonizing patients and healthcare personnel with MRSA (Patel et al., 2009). However, the emergence of resistance is associated with unrestricted policies and antibiotic use for long periods in healthcare settings (Hetem and Bonten, 2013). Two levels of S. aureus resistance to mupirocin have been elucidated, i.e., low-level and high-level resistance (HmupR) attributed to mutation and the acquisition of plasmids, respectively (Patel et al., 2009). Whereas the prevalence of MRSA with HmupR is 5.9%, 8.0%, and 12.1% in the Americas, Europe, and Asia, respectively (Dadashi et al., 2020), it is entirely unknown in S. argenteus until now. We describe the first report of a methicillin-resistant S. argenteus that exhibited HmupR.
Materials and Methods
Identification of the Methicillin-Resistant, Mupirocin-Resistant S. argenteus
The isolate (1801221) was obtained in April 2018 from a human nasal swab and was previously identified as methicillin-resistant S. aureus (MRSA) with HmupR. For characterization, it was sent to the National Reference Center for Staphylococci and Enterococci, Robert Koch Institute, Germany. To delineate S. argenteus from S. aureus, PCR amplification of the NRPS gene (Becker et al., 2019) was performed at the Institute of Medical Microbiology, Münster. The isolate was subjected to antibiotic susceptibility testing (Vitek 2 automated system bioMérieux, Marcy l’Étoile, France). The minimum inhibitory concentration (MIC) to mupirocin was also determined using the gradient diffusion method (E-test, bioMérieux, Marcy l’Étoile, France). Methicillin and mupirocin resistance was confirmed by PCR detection of mecA (Murakami et al., 1991) and mupA (Nagant et al., 2016). We interpreted the results of the antibiotic susceptibility testing and E-test according to the EUCAST clinical breakpoints (Version 11.0).
Whole-Genome Sequencing
The S. argenteus isolate was further processed for whole-genome sequencing (WGS) on a Sequel II platform (Pacific Biosciences Inc., Menlo Park, CA, USA). Before sequencing, we constructed the sequence library using the SMRTbell Express Template Prep Kit 2.0 (Pacific Biosciences Inc.) according to the manufacturer’s recommendations. The resulting long-read sequencing data were assembled applying the “Microbial Assembly” pipeline within the SMRT Link software version 9 (Pacific Biosciences Inc.) using default parameters except for the genome size, which was adopted to 2.8 Mb. Then, we utilized the Ridom SeqSphere+ software (version 7, Ridom GmbH, Münster, Germany) to in silico predict the antimicrobial resistance and virulence genes and to extract the staphylococcal protein A (spa) type and the multilocus sequence type (ST) of the isolate. Also, we used the Plasmid Finder (version 2.1) to identify the replicon sequences (Carattoli et al., 2014). Further analysis, and annotation of the sequences, was performed using the NCBI Prokaryotic Genome Annotation Pipeline software revision 5.3 (Tatusova et al., 2016). A Neighbor-Joining (NJ) tree was constructed using sequences of a global collection of 111 S. argenteus (ST2250) isolates. Single nucleotide polymorphisms (SNPs) were extracted from 1,864 core genome genes (Leopold et al., 2014) present in all isolates. The SNPs analysis formed the basis to calculate the NJ tree with default parameters within the Ridom SeqSphere+ software version 7.
Results and Discussion
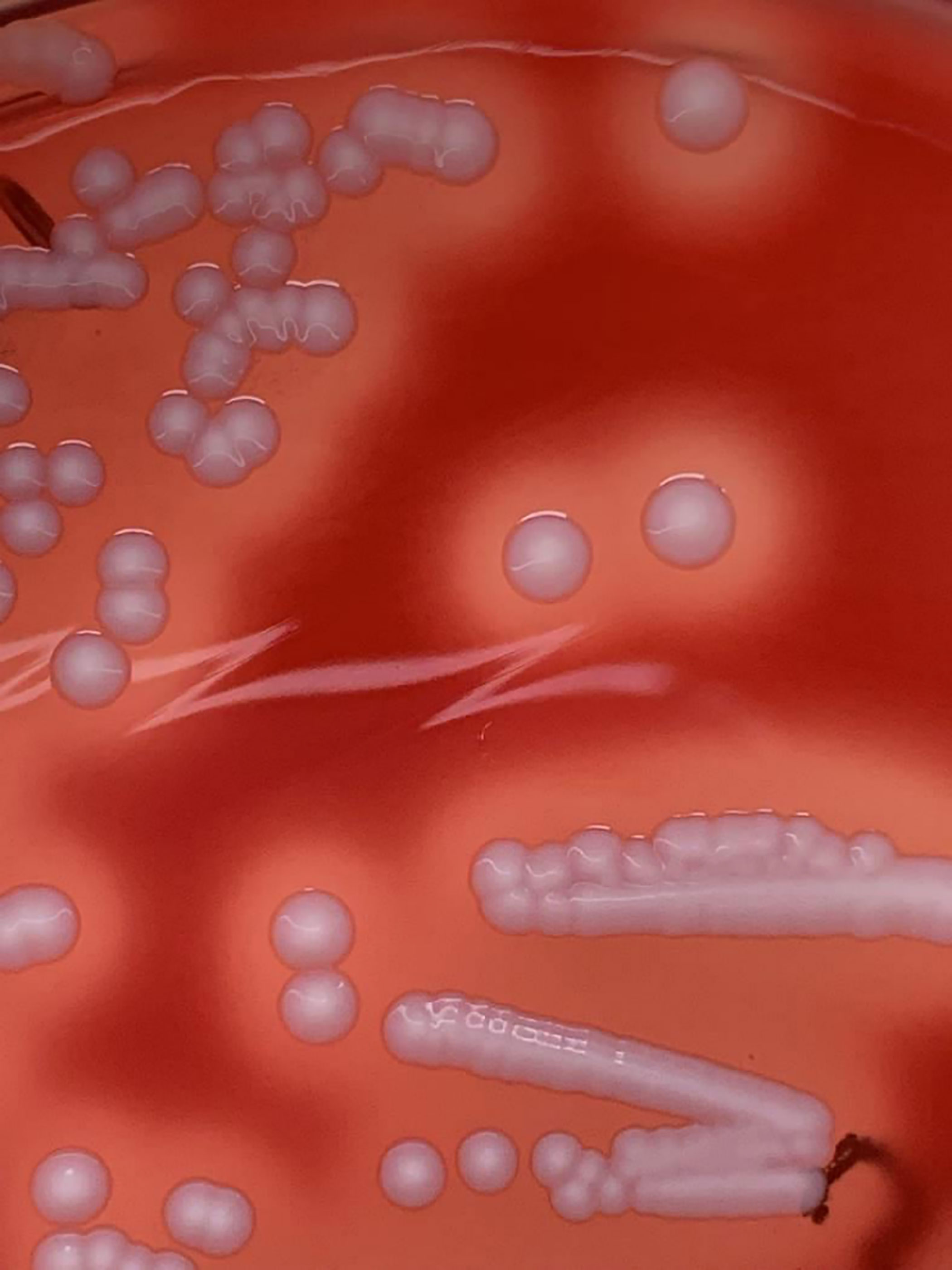
The isolate displayed creamy-white colonies with β-haemolysis on Columbia sheep blood agar (CBA, BD, Heidelberg, Germany) (Figure 1). MALDI-TOF identification using the MBT compass (Version 9) did not distinguish reliably between S. aureus (Score: 2.04) and S. argenteus (Score: 2.13). However, it was PCR-positive (360bp) for the NRPS gene, indicating that it is S. argenteus. Antibiotic susceptibility testing showed that the isolate was resistant to cefoxitin, fosfomycin, mupirocin, and trimethoprim/sulfamethoxazole. The MIC of mupirocin (≥512 μg/ml, E-test) was in agreement with the VITEK result (MIC = ≥512 μg/ml). PCR revealed that the isolate was mecA and mupA-positive. WGS confirmed the identity of the isolate as S. argenteus and its antibiotic resistance phenotype. Also, molecular typing characterized the isolate as t6675-ST2250-SCCmecIVc. It was associated with capsule type 8, positive for the immune evasion (sak, scn) gene cluster, haemolysins (hld, hlgB, hly/hla), and the intracellular adhesion (icaA, icaB, icaC, icaD, icaR) gene operon. The isolate was negative for the PVL-encoding gene.
The S. argenteus isolate chromosome was 2,781,166 bp in size, with a GC content of 32.3%, containing 2,650 predicted coding DNA sequences (CDSs). The NJ tree based on 2,177 SNPs from a global collection of all available genomes (as of 8 December 2021) of ST2250 S. argenteus isolates (Supplementary Table) showed that it clustered with the European ST2250 clade (Figure 2). The Plasmid Finder identified sequences of two plasmids with replication (rep5a [locus tag 13590 in Supplementary Figure 1B], rep16 [locus tag 13610 in Supplementary Figure 1B], and rep23 [locus tag 13385 in Supplementary Figure 1A]) genes, respectively. The larger plasmid (34,870 bp), designated p1_1801221, with rep23 carried mupA. This gene demonstrated 100% sequence identity with the alternative isoleucyl-tRNA synthetase (ileS-2) gene conferring HmupR on a conjugative plasmid pPR9 from S. aureus (GenBank accession number GU237136). Moreover, the whole plasmid was nearly identical at sequence level with the published plasmid pPR9 (Figure 3A) using the BRIG tool (Alikhan et al., 2011). The smaller plasmid (20,644 bp), assigned as p2_1801221, with rep5a and rep16, harboured the penicillin (blaZ) and cadmium (cadD) resistance genes. Again, the genes and overall plasmid composition exhibited high homology to S. aureus resistance determinants and plasmid. Specifically, blaZ showed 99.9% sequence identity with the corresponding gene on pN315 (GenBank accession number AP003139), and the cadD gene displayed 100% homology with the resistance determinant on pSAS (GeneBank accession number BX571858). Moreover, the plasmid as a whole was nearly identical to the S. aureus plasmid p515718a of strain 515798 (GenBank accession number CP045475) (Figure 3B).
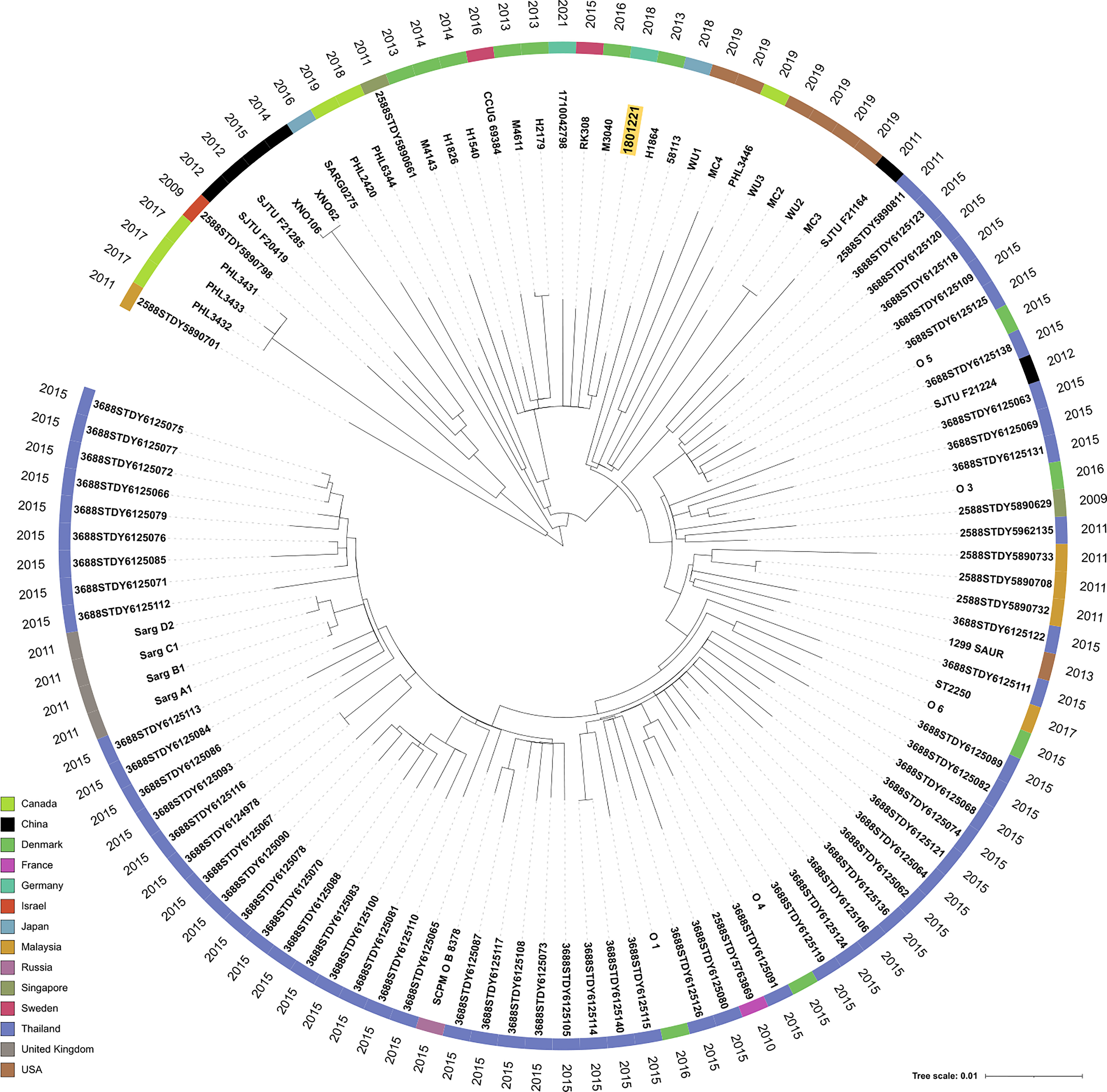
Figure 2 Neighbor-joining (NJ) tree of 111 S. argenteus ST2250 global isolates. SNPs (n = 2,177) were extracted from 1,864 core genome genes present in all isolates and formed the basis to calculate the NJ tree with default parameters within the Ridom SeqSphere+ software. We used iTOL V. 6 (Letunic and Bork, 2021) to display the tree and metadata of the strains. The leaves of the tree were annotated with the sample names. The colored circle indicates the country of isolation and the outer circle the isolation year, respectively. Isolate 1801221 is highlighted in a yellow box.
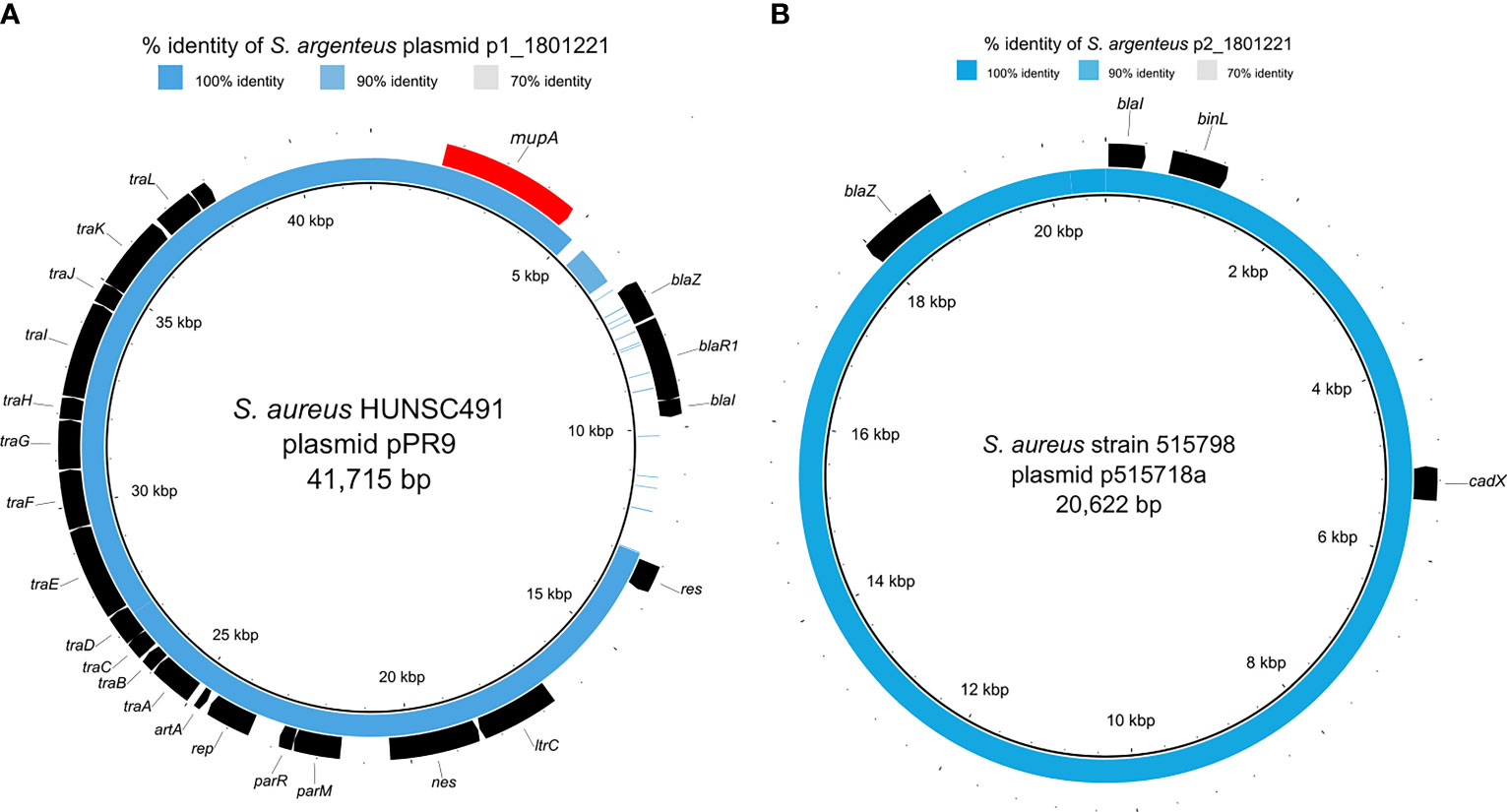
Figure 3 Comparison of S. argenteus plasmids with closely related S. aureus plasmids. The inner black ring represents the reference sequence, and the blue ring depicts the respective S. argenteus plasmid sequence. The outer black ring provides annotation information, i.e., detected ORFs, where the annotation resulted in known genes. The blue color’s intensity is related to the sequence similarity, detailed in the Supplementary Table. (A) depicts the comparison of p1_1801221 and the conjugative S. aureus plasmid pPR9 harboring the mupA gene encoding for mupirocin resistance (colored in red); (B) shows the comparison of p2_1801221 and the S. aureus plasmid p515718a harboring blaZ and cadX encoding for beta-lactam and cadmium resistance, respectively.
S. argenteus was first reported in northern Australia (McDonald et al., 2006) and distinct from S. aureus based on the average nucleotide identity of 87.4% and a DNA-DNA hybridization value of 33.5% (Tong et al., 2015). About 10% of S. aureus isolates from human infections are non-pigmented (Zhang et al., 2018). Also, S. argenteus colonies on blood agar are non-pigmented (creamy-white) due to the lack of the crtOPQMN operon responsible for carotenoid pigment, staphyloxanthin (Holt et al., 2011). Hence, S. argenteus and non-pigmented S. aureus could be indistinguishable on blood agar based on colony morphology and phenotypic tests (coagulase, DNase). This scenario could be a dilemma in the clinical microbiology laboratory (Becker et al., 2019). This study provided evidence on the reliability of the PCR detection of the NRPS gene with WGS in the delineation of S. argenteus from S. aureus. ST2250 is a global S. argenteus clonal group (Eshaghi et al., 2021), and our first report of an isolate in this clone exhibiting HmupR is of public health importance. MRSA with HmupR is a serious problem as decolonization with mupirocin becomes ineffective (Patel et al., 2009). Moreover, HmupR could facilitate the spread of antibiotic resistance through the conjugative transfer of plasmid mediating HmupR with co-mobilization and co-transfer of plasmids encoding other gene determinants (Udo and Jacob, 1998; Pawa et al., 2000). Also, macrolide, gentamicin, tetracycline, and trimethoprim resistance genes have been identified on the same extra-chromosomal element with mupA (McDougal et al., 2010). In this study, the identification and high homology of both plasmids identified in S. argenteus with published DNA sequences of S. aureus plasmids suggest interspecies transfer.
S. argenteus carriage in the human population (Aung et al., 2017; Senok et al., 2020; Eshaghi et al., 2021; Jauneikaite et al., 2021) and possible person-to-person transmission (Giske et al., 2019; Eshaghi et al., 2021) have been described. Moreover, a study revealed that cases of S. argenteus bacteremia were associated with higher mortality than methicillin-susceptible S. aureus bacteremia (Chen et al., 2018b). S. argenteus with different antibiotic resistance genes have been reported (Aung et al., 2021; Eshaghi et al., 2021), including an isolate with elevated MIC (4µg/ml) to daptomycin and vancomycin in the United States (Hao et al., 2020). Recent studies from China (Chen and Wu, 2020) and Japan (Wakabayashi et al., 2021) have also identified S. argenteus from retail foods and an emerging bovine mastitis pathogen in Thailand (Pumipuntu, 2019). We could not ascertain if the study individual received mupirocin or not. Nonetheless, these increasing reports and the capacity of S. argenteus to harbor resistance gene determinants (including mupA) with its repertoire of virulence factors highlight the need for its delineation from S. aureus and correct identification. Therefore, enhanced surveillance is vital to understanding the significance of S. argenteus in clinical and non-clinical settings.
Data Availability Statement
The whole-genome sequence project for the S. argenteus; isolate (1801221) has been deposited in NCBI under the bioproject accession number PRJNA764657 with sequence accession numbers CP083805-CP083807 for the chromosome and the two plasmids.
Authors Contributions
AS, FL-N, BS, and FS designed the research. AS, M-TN, SB, and AM performed the experiments. AS, SB, and AM analyzed the data. AS wrote the initial draft of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study received support from the Deutsche Forschungsgemeinschaft (SCHA 1994/5-1, granted to AS and FS) and the Alexander von Humboldt Foundation (“Georg Forster-Forschungsstipendium” granted to AS). We acknowledge support from the Open Access Publication Fund of the University of Muenster.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledge the collaborating laboratory, MVZ Labor Limbach in Heidelberg, for sending the isolate to the National Reference Centre for Staphylococci and Enterococci, Robert Koch Institute, Germany.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.860163/full#supplementary-material
Supplementary Figures 1a and 1b | Circular illustration of the two S. argenteus plasmids and annotation of antibiotic resistance genes.
References
Alhussein, F., Fürstenberg, J., Gaupp, R., Eisenbeis, J., Last, K., Becker, S. L., et al. (2020). Human Infections Caused by Staphylococcus Argenteus in Germany: Genetic Characterisation and Clinical Implications of Novel Species Designation. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc Clin. Microbiol. 39, 2461–2465. doi: 10.1007/s10096-020-03950-4
Alikhan, N.-F., Petty, N. K., Ben Zakour, N. L., Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): Simple Prokaryote Genome Comparisons. BMC Genomics 12, 402. doi: 10.1186/1471-2164-12-402
Aung, M. S., San, T., Aye, M. M., Mya, S., Maw, W. W., Zan, K. N., et al. (2017). Prevalence and Genetic Characteristics of Staphylococcus Aureus and Staphylococcus Argenteus Isolates Harboring Panton-Valentine Leukocidin, Enterotoxins, and TSST-1 Genes From Food Handlers in Myanmar. Toxins (Basel) 9 (241), 1–13. doi: 10.3390/toxins9080241
Aung, M. S., Urushibara, N., Kawaguchiya, M., Hirose, M., Ike, M., Ito, M., et al. (2021). Distribution of Virulence Factors and Resistance Determinants in Three Genotypes of Staphylococcus Argenteus Clinical Isolates in Japan. Pathog. (Basel Switzerland) 10, 163. doi: 10.3390/pathogens10020163
Bank, L. E. A., Bosch, T., Schouls, L. M., Weersink, A. J. L., Witteveen, S., Wolffs, P. F. G., et al. (2021). Methicillin-Resistant Staphylococcus Argenteus in the Netherlands: Not a New Arrival. Eur. J. Clin. Microbiol. Infect. Dis. 40, 1583–1585. doi: 10.1007/s10096-021-04204-7
Becker, K., Schaumburg, F., Kearns, A., Larsen, A. R., Lindsay, J. A., Skov, R. L., et al. (2019). Implications of Identifying the Recently Defined Members of the Staphylococcus Aureus Complex S. Argenteus and S. Schweitzeri: A Position Paper of Members of the ESCMID Study Group for Staphylococci and Staphylococcal Diseases (ESGS). Clin. Microbiol. Infect. 25, 1064–1070. doi: 10.1016/j.cmi.2019.02.028
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chantratita, N., Wikraiphat, C., Tandhavanant, S., Wongsuvan, G., Ariyaprasert, P., Suntornsut, P., et al. (2016). Comparison of Community-Onset Staphylococcus Argenteus and Staphylococcus Aureus Sepsis in Thailand: A Prospective Multicentre Observational Study. Clin. Microbiol. Infect. Off. Publ. Eur. Soc Clin. Microbiol. Infect. Dis. 22, 458.e11–9. doi: 10.1016/j.cmi.2016.01.008
Chen, S.-Y., Lee, H., Teng, S.-H., Wang, X.-M., Lee, T.-F., Huang, Y.-C., et al. (2018a). Accurate Differentiation of Novel Staphylococcus Argenteus From Staphylococcus Aureus Using MALDI-TOF MS. Future Microbiol. 13, 997–1006. doi: 10.2217/fmb-2018-0015
Chen, S.-Y., Lee, H., Wang, X.-M., Lee, T.-F., Liao, C.-H., Teng, L.-J., et al. (2018b). High Mortality Impact of Staphylococcus Argenteus on Patients With Community-Onset Staphylococcal Bacteraemia. Int. J. Antimicrob. Agents 52, 747–753. doi: 10.1016/j.ijantimicag.2018.08.017
Chen, C., Wu, F. (2020). Livestock-Associated Methicillin-Resistant Staphylococcus Aureus (LA-MRSA) Colonisation and Infection Among Livestock Workers and Veterinarians: A Systematic Review and Meta-Analysis. Occup. Environ. Med. 78, 530–540. doi: 10.1136/oemed-2020-106418
Chew, K. L., Octavia, S., Lai, D., Lin, R. T. P., Teo, J. W. P. (2021). Staphylococcus Singaporensis Sp. Nov., a New Member of the Staphylococcus Aureus Complex, Isolated From Human Clinical Specimens. Int. J. Syst. Evol. Microbiol. 71 (005067), 1–7. doi: 10.1099/ijsem.0.005067
Dadashi, M., Hajikhani, B., Darban-Sarokhalil, D., van Belkum, A., Goudarzi, M. (2020). Mupirocin Resistance in Staphylococcus Aureus: A Systematic Review and Meta-Analysis. J. Glob. Antimicrob. Resist. 20, 238–247. doi: 10.1016/j.jgar.2019.07.032
Diot, A., Dyon-Tafani, V., Bergot, M., Tasse, J., Martins-Simões, P., Josse, J., et al. (2020). Investigation of a Staphylococcus Argenteus Strain Involved in a Chronic Prosthetic-Joint Infection. Int. J. Mol. Sci. 21 (6245), 1–16. doi: 10.3390/ijms21176245
Eshaghi, A., Bommersbach, C., Zittermann, S., Burnham, C. A., Patel, R., Schuetz, A. N., et al. (2021). Phenotypic and Genomic Profiling of Staphylococcus argenteus in Canada and the United States and Recommendations for Clinical Result Reporting. J. Clin. Microbiol. 59, e02470-20. doi: 10.1128/JCM.02470-20
Giske, C. G., Dyrkell, F., Arnellos, D., Vestberg, N., Hermansson Panna, S., Fröding, I., et al. (2019). Transmission Events and Antimicrobial Susceptibilities of Methicillin-Resistant Staphylococcus Argenteus in Stockholm. Clin. Microbiol. Infect. 25, 1289.e5–1289.e8. doi: 10.1016/j.cmi.2019.06.003
Goswami, C., Fox, S., Holden, M., Leanord, A., Evans, T. J. (2021). Genomic Analysis of Global Staphylococcus Argenteus Strains Reveals Distinct Lineages With Differing Virulence and Antibiotic Resistance Gene Content. Front. Microbiol. 12, 795173. doi: 10.3389/fmicb.2021.795173
Hallbäck, E. T., Karami, N., Adlerberth, I., Cardew, S., Ohlén, M., Jakobsson, H. E., et al. (2018). Methicillin-Resistant Staphylococcus Argenteus Misidentified as Methicillin-Resistant Staphylococcus Aureus Emerging in Western Sweden. J. Med. Microbiol. 67, 968–971. doi: 10.1099/jmm.0.000760
Hansen, T. A., Bartels, M. D., Høgh, S. V., Dons, L. E., Pedersen, M., Jensen, T. G., et al. (2017). Whole Genome Sequencing of Danish Staphylococcus Argenteus Reveals a Genetically Diverse Collection With Clear Separation From Staphylococcus Aureus. Front. Microbiol. 8, 1512. doi: 10.3389/fmicb.2017.01512
Hao, S., Abdelghany, M., Lyden, A., Sit, R., Tan, M., Tato, C. M., et al. (2020). Genomic Profiling of Evolving Daptomycin Resistance in a Patient With Recurrent Staphylococcus Argenteus Sepsis. Antimicrob. Agents Chemother. 64, e00961–e00920. doi: 10.1128/AAC.00961-20
Hetem, D. J., Bonten, M. J. M. (2013). Clinical Relevance of Mupirocin Resistance in Staphylococcus Aureus. J. Hosp. Infect. 85, 249–256. doi: 10.1016/j.jhin.2013.09.006
Holt, D. C., Holden, M. T. G., Tong, S. Y. C., Castillo-Ramirez, S., Clarke, L., Quail, M. A., et al. (2011). A Very Early-Branching Staphylococcus Aureus Lineage Lacking the Carotenoid Pigment Staphyloxanthin. Genome Biol. Evol. 3, 881–895. doi: 10.1093/gbe/evr078
Jauneikaite, E., Pichon, B., Mosavie, M., Fallowfield, J. L., Davey, T., Thorpe, N., et al. (2021). Staphylococcus Argenteus Transmission Among Healthy Royal Marines: A Molecular Epidemiology Case-Study. J. Infect. 83, 550–553. doi: 10.1016/j.jinf.2021.08.040
Kaden, R., Engstrand, L., Rautelin, H., Johansson, C. (2018). Which Methods Are Appropriate for the Detection of Staphylococcus Argenteus and Is It Worthwhile to Distinguish S. Argenteus From S. Aureus? Infect. Drug Resist. 11, 2335–2344. doi: 10.2147/IDR.S179390
Leopold, S. R., Goering, R. V., Witten, A., Harmsen, D., Mellmann, A. (2014). Bacterial Whole-Genome Sequencing Revisited: Portable, Scalable, and Standardized Analysis for Typing and Detection of Virulence and Antibiotic Resistance Genes. J. Clin. Microbiol. 52, 2365–2370. doi: 10.1128/JCM.00262-14
Letunic, I., Bork, P. (2021). Interactive Tree Of Life (iTOL) V5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Liu, G. Y., Essex, A., Buchanan, J. T., Datta, V., Hoffman, H. M., Bastian, J. F., et al. (2005). Staphylococcus Aureus Golden Pigment Impairs Neutrophil Killing and Promotes Virulence Through Its Antioxidant Activity. J. Exp. Med. 202, 209–215. doi: 10.1084/jem.20050846
McDonald, M., Dougall, A., Holt, D., Huygens, F., Oppedisano, F., Giffard, P. M., et al. (2006). Use of a Single-Nucleotide Polymorphism Genotyping System to Demonstrate the Unique Epidemiology of Methicillin-Resistant Staphylococcus Aureus in Remote Aboriginal Communities. J. Clin. Microbiol. 44, 3720–3727. doi: 10.1128/JCM.00836-06
McDougal, L. K., Fosheim, G. E., Nicholson, A., Bulens, S. N., Limbago, B. M., Shearer, J. E. S., et al. (2010). Emergence of Resistance Among USA300 Methicillin-Resistant Staphylococcus Aureus Isolates Causing Invasive Disease in the United States. Antimicrob. Agents Chemother. 54, 3804–3811. doi: 10.1128/AAC.00351-10
Mitsutake, K., Watanabe, N., Karaushi, H., Tarumoto, N., Koyama, S., Ebihara, Y., et al. (2020). Thoracic Aortic Mycotic Aneurysm Due to Staphylococcus Argenteus: A Case Report. J. Infect. Chemother. Off. J. Jpn. Soc Chemother. 26, 1213–1215. doi: 10.1016/j.jiac.2020.05.003
Murakami, K., Minamide, W., Wada, K., Nakamura, E., Teraoka, H., Watanabe, S. (1991). Identification of Methicillin-Resistant Strains of Staphylococci by Polymerase Chain Reaction. J. Clin. Microbiol. 29, 2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991
Nagant, C., Deplano, A., Nonhoff, C., De Mendonça, R., Roisin, S., Dodémont, M., et al. (2016). Low Prevalence of Mupirocin Resistance in Belgian Staphylococcus Aureus Isolates Collected During a 10 Year Nationwide Surveillance. J. Antimicrob. Chemother. 71, 266–267. doi: 10.1093/jac/dkv286
Patel, J. B., Gorwitz, R. J., Jernigan, J. A. (2009). Mupirocin Resistance. Clin. Infect. Dis. 49, 935–941. doi: 10.1086/605495
Pawa, A., Noble, W. C., Howell, S. A. (2000). Co-Transfer of Plasmids in Association With Conjugative Transfer of Mupirocin or Mupirocin and Penicillin Resistance in Methicillin-Resistant Staphylococcus Aureus. J. Med. Microbiol. 49, 1103–1107. doi: 10.1099/0022-1317-49-12-1103
Pumipuntu, N. (2019). Staphylococcus Argenteus: An Emerging Subclinical Bovine Mastitis Pathogen in Thailand. Vet. World 12, 1940–1944. doi: 10.14202/vetworld.2019.1940-1944
Schutte, A. H. J., Strepis, N., Zandijk, W. H. A., Bexkens, M. L., Bode, L. G. M., Klaassen, C. H. W. (2021). Characterization of Staphylococcus Roterodami Sp. Nov., a New Species Within the Staphylococcus Aureus Complex Isolated From a Human Foot Infection. Int. J. Syst. Evol. Microbiol. 71 (004996), 1–7. doi: 10.1099/ijsem.0.004996
Senok, A., Nassar, R., Kaklamanos, E. G., Belhoul, K., Abu Fanas, S., Nassar, M., et al. (2020). Molecular Characterization of Staphylococcus Aureus Isolates Associated With Nasal Colonization and Environmental Contamination in Academic Dental Clinics. Microb. Drug Resist. 26, 661–669. doi: 10.1089/mdr.2019.0318
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Tong, S. Y. C., Schaumburg, F., Ellington, M. J., Corander, J., Pichon, B., Leendertz, F., et al. (2015). Novel Staphylococcal Species That Form Part of a Staphylococcus Aureus-Related Complex: The Non-Pigmented Staphylococcus Argenteus Sp. Nov. And the Non-Human Primate-Associated Staphylococcus Schweitzeri Sp. Nov. Int. J. Syst. Evol. Microbiol. 65, 15–22. doi: 10.1099/ijs.0.062752-0
Tunsjø, H. S., Kalyanasundaram, S., Charnock, C., Leegaard, T. M., Moen, A. E. F. (2018). Challenges in the Identification of Methicillin-Resistant Staphylococcus Argenteus by Routine Diagnostics. APMIS 126, 533–537. doi: 10.1111/apm.12843
Udo, E. E., Jacob, L. E. (1998). Conjugative Transfer of High-Level Mupirocin Resistance and the Mobilization of Non-Conjugative Plasmids in Staphylococcus Aureus. Microb. Drug Resist. 4, 185–193. doi: 10.1089/mdr.1998.4.185
Wakabayashi, Y., Takemoto, K., Iwasaki, S., Yajima, T., Kido, A., Yamauchi, A., et al. (2021). Isolation and Characterization of Staphylococcus Argenteus Strains From Retail Foods and Slaughterhouses in Japan. Int. J. Food Microbiol. 363, 109503. doi: 10.1016/j.ijfoodmicro.2021.109503
Zhang, J., Suo, Y., Zhang, D., Jin, F., Zhao, H., Shi, C. (2018). Genetic and Virulent Difference Between Pigmented and Non-Pigmented Staphylococcus Aureus. Front. Microbiol. 9, 598. doi: 10.3389/fmicb.2018.00598
Zhang, D. F., Xu, X., Song, Q., Bai, Y., Zhang, Y., Song, M., et al. (2016). Identification of Staphylococcus Argenteus in Eastern China Based on a Nonribosomal Peptide Synthetase (NRPS) Gene. Future Microbiol. 11, 1113–1121. doi: 10.2217/fmb-2016-0017
Keywords: identification, methicillin-resistant Staphylococcus argenteus, high-level mupirocin resistance, plasmid, whole-genome sequencing (WGS)
Citation: Shittu AO, Layer-Nicolaou F, Strommenger B, Nguyen M-T, Bletz S, Mellmann A and Schaumburg F (2022) First Report of a Methicillin-Resistant, High-Level Mupirocin-Resistant Staphylococcus argenteus. Front. Cell. Infect. Microbiol. 12:860163. doi: 10.3389/fcimb.2022.860163
Received: 22 January 2022; Accepted: 09 February 2022;
Published: 15 March 2022.
Edited by:
Percy Schröttner, Technische Universität Dresden, GermanyReviewed by:
Florence Claude Doucet-Populaire, Université Paris-Saclay, FranceScott Wesley Long, Houston Methodist Hospital, United States
Copyright © 2022 Shittu, Layer-Nicolaou, Strommenger, Nguyen, Bletz, Mellmann and Schaumburg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adebayo Osagie Shittu, YmF5b19zaGl0dHVAeWFob28uY29t
†Present address: Adebayo Osagie Shittu, Institute of Medical Microbiology, University Hospital Münster, Münster, Germany
‡These authors have contributed equally to this work
 Adebayo Osagie Shittu
Adebayo Osagie Shittu Franziska Layer-Nicolaou
Franziska Layer-Nicolaou Birgit Strommenger
Birgit Strommenger Minh-Thu Nguyen
Minh-Thu Nguyen Stefan Bletz4
Stefan Bletz4 Alexander Mellmann
Alexander Mellmann Frieder Schaumburg
Frieder Schaumburg