- 1The State Key Laboratory of Agricultural Microbiology, (HZAU), Wuhan, China
- 2College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, China
- 3Department of Medicine and Infectious Diseases, Faculty of Veterinary Medicine, University of Sadat City, Sadat City, Egypt
- 4Hubei Hongshan Laboratory, Wuhan, China
- 5National Reference Laboratory of Veterinary Drug Residues (HZAU) and MAO Key Laboratory for Detection of Veterinary Drug Residues, HZAU, Wuhan, China
- 6Department of Clinical Pathology, Faculty of Veterinary Medicine, Benha University, Toukh, Egypt
- 7Hubei International Scientific and Technological Cooperation Base of Veterinary Epidemiology, Huazhong Agricultural University, Wuhan, China
- 8Infectious Diseases, Faculty of Veterinary Medicine, Benha University, Toukh, Egypt
- 9Para-Clinic Department, Faculty of Veterinary Medicine, Jalalabad, Afghanistan
Mycoplasmas as economically important and pantropic pathogens can cause similar clinical diseases in different hosts by eluding host defense and establishing their niches despite their limited metabolic capacities. Besides, enormous undiscovered virulence has a fundamental role in the pathogenesis of pathogenic mycoplasmas. On the other hand, they are host-specific pathogens with some highly pathogenic members that can colonize a vast number of habitats. Reshuffling mycoplasmas genetic information and evolving rapidly is a way to avoid their host’s immune system. However, currently, only a few control measures exist against some mycoplasmosis which are far from satisfaction. This review aimed to provide an updated insight into the state of mycoplasmas as pathogens by summarizing and analyzing the comprehensive progress, current challenge, and future perspectives of mycoplasmas. It covers clinical implications of mycoplasmas in humans and domestic and wild animals, virulence-related factors, the process of gene transfer and its crucial prospects, the current application and future perspectives of nanotechnology for diagnosing and curing mycoplasmosis, Mycoplasma vaccination, and protective immunity. Several questions remain unanswered and are recommended to pay close attention to. The findings would be helpful to develop new strategies for basic and applied research on mycoplasmas and facilitate the control of mycoplasmosis for humans and various species of animals.
Introduction
Mycoplasmas are the smallest and simplest self-replicating microorganisms. Numerous species occur as opportunistic pathogens in mammals, birds, reptiles, insects, and plants (Razin, 1992). Due to their limited metabolic capacity, which is a consequence of their small genome and lack of cell wall, they are fastidious and some of them are laborious to be cultured. As such, they can be both extracellular and intracellular pathogens whose lives depend on the largesse of their hosts (Morowitz and Tourtellotte, 1962).
Mycoplasmas are pantropic in vivo. Their favorable localizations are the mucous surface of respiratory and urogenital tracts, mammary glands, eyes, alimentary canal, and joints (Razin et al., 1998). Some Mycoplasma species (M. penetrans, M. pneumoniae, M. fermentans, M. hominis, and M. gallisepticum (MG) can adhere to and invade the targeted cells by interacting with their membranes (Baseman et al., 1995; Shibata et al., 2000; Vogl et al., 2008). When two Mycoplasma species colonize one habitat, the horizontal gene transfer (HGT) might occur leading to the virulence evolution of mycoplasmas which has a crucial impact on their pathogenesis (Bürki et al., 2015). The emergence of multidrug resistance (MDR) caused by the transfer and/or exchange of antibiotic resistance genes (ARGs) between different pathogens is a growing concern (Forsberg et al., 2012; Faucher et al., 2019). Vaccines are developed and commercially applied in the control of some Mycoplasma-related diseases but the efficacy is far from satisfaction. Therefore, novel therapeutic and preventive products are urgently needed to secure animal health, hence improving human health (Fair and Tor, 2014; Valentine-King et al., 2020).
Precisely, perceiving the comprehensive progress, current challenge, and future perspectives of mycoplasmas are helpful to settle down future plans and strategies of basic and applied research on mycoplasmas. Intriguingly, these strategies will facilitate the prevention and treatment of mycoplasmosis for various species of hosts. Therefore, this review aimed to provide an updated insight into the state of mycoplasmas focusing on them as pathogens of human and terrestrial animals. The findings would be applied to get rid of or avoid the growing threat of mycoplasmosis to the health of all affected creatures.
Clinical Implications of Mycoplasmas
Mycoplasmas are potential pathogenic organisms of humans and many animal species. Pathogenic mycoplasmas have a natural tendency to colonize certain sites in vivo called “tissue tropism”, such as respiratory, ocular and genital mucosa, and mammary glands (Razin et al., 1998)
Human Infection
Mainly, six Mycoplasma species (M. pneumoniae, M. genitalium, Ureaplasma (U) urealyticum, U. parvum, M. hominis, M. penetrans) have been demonstrated to cause human illness (Waites et al., 2005), such as acute respiratory disease (Waites et al., 2017), joint infections (Ali et al., 2021a; Mărginean et al., 2021), genital and urinary tract infections (Peter et al., 2018), and neurological disturbance (D’Alonzo et al., 2018; He et al., 2021a). On the other hand, some species mainly infecting animals like M. suis, M. ovis, and M. haemofelis have been detected in humans and regarded as zoonotic pathogens (Maggi et al., 2013b). These mycoplasmas can establish persistent infections (Yavlovich et al., 2004), alter host cell physiology, modify apoptotic pathways (Chernov et al., 2015), induce the production of inflammatory substances (Benedetti et al., 2020), and result in cellular DNA damage and cancers (Zella et al., 2018). In addition, serious consequences like chronic obstructive pulmonary disease (COPD) (Feng et al., 2021) and infertility may occur (Kusanovic et al., 2020).
Transmission of human Mycoplasma infection occurs through human-to-human contact. It mainly infects mucosal surfaces of the respiratory and urogenital tracts. Droplets containing the organism spread the infection from host to host (Waites et al., 2017). Ureaplasma species, M. genitalium, and M. hominis are genitourinary mucosal organisms and the infection can spread through direct sexual contact, Ureaplasma species mainly cause urethral and gynecological infections (Cassell et al., 1993; Lanao et al., 2022).
The clinical picture of M. pneumoniae has diversely presented from self-limiting to life-threatening disease (Saraya et al., 2014). For instance, it causes community-acquired pneumonia (CAP) in people of any age, especially in children and young adults (Li et al., 2019; Tsai et al., 2021). M. pneumoniae symptoms are variable including fever, cough, sore throat, and occasionally, acute exacerbation of asthma. In addition, severe pulmonary sickness including bronchiolitis, pleural effusion, lung abscess, and pulmonary embolism as a consequence of M. pneumoniae infection has also been reported (Meyer Sauteur et al., 2016). The culture procedure is a gold standard technique for M. pneumoniae diagnosis. On the other hand, it is recommended to use polymerase chain reaction (PCR) for diagnosing acute cases because culture methods require several days for obtaining results. Moreover, PCR is a rapid, sensitive, specific, and commercially available method, and so it is more suitable for mycoplasmas diagnosis in the clinic (Waites et al., 2017).
Mainly, macrolides, fluoroquinolones, and tetracycline are used for the treatment of M. pneumoniae infection; macrolides are the most potent antimicrobial agents for the treatment of mycoplasmosis through inhibition of the bacterial protein synthesis (Waites et al., 2017). Recently, because of the broad use of macrolides, macrolide-resistant M. pneumoniae (MRMP) has become increasingly prevalent worldwide (Yang, 2019). The macrolides resistance of M. pneumoniae has been emerging worldwide. In Taiwan (2010 to 2017), its rate was 15–30% (Yang et al., 2019a). In America and Europe (2008 to 2013), it was <30%. While in other countries and regions (China mainland, Japan, and Korea) it was about 60–90% (Waites et al., 2017). Age is also regarded as a major determinant for MRMP as the detection rate was higher for children aged ≤15 years than adults. In adults, the detection rate was higher in adolescents (16–19 years) than in older age (≥20 years) (Yamazaki and Kenri, 2016). On the other hand, fluoroquinolones and tetracyclines have more severe side effects than macrolides as tetracycline cause enamel hypoplasia and discoloration of the teeth in young children. Despite the detected hazardous effects of fluoroquinolones on joints and muscles of children, they have been successfully used for the treatment of some complicated cases of MRMP strains in young children (Jackson et al., 2016; Ahn et al., 2021).
Owing to the marked increase of M. pneumoniae antimicrobial resistance in recent years as well as the previously mentioned side effects of some antimicrobial agents, the development of protective vaccines against this pathogen is a critical requirement (Jiang et al., 2021). Recently, the designing of the next-generation vaccine approach was performed to establish an effective multi-epitope vaccine (MEV) for human protection against M. pneumoniae (Mahmood et al., 2021). To date, Mara et al. (2020) have achieved a breakthrough in explaining how the vaccine-enhanced disease (VED) occurs as a result of M. pneumoniae vaccination with lipid-associated membrane proteins (LAMPs). Intriguingly, they demonstrated that M. pneumoniae lipoproteins lipid moieties are responsible for VED occurrence. In addition, the removal of lipid molecules from LAMPs before vaccination prevents VED and reduces bacterial loads in the case of M. pneumoniae infection. Lipoproteins are the main immunogenic and antigenic constituents of the LAMPs fraction, and therefore their lipid moieties significantly reduced LAMP-stimulated TNF-α production which leads to the VED (Mara et al., 2020). These results may be widely applicable for other mycoplasmas in which vaccine-induced disease exacerbation has been described such as M. bovis (Bryson et al., 1999) and Mmm (Nicholas et al., 2004).
Nowadays, sexually transmitted antigens are of major concern. Nogueira and co-workers have conducted a recent computational study using in silico methods as “subtractive genomics and reverse vaccinology” on five strains of M. genitalium, a serious sexually transmissible pathogen. The state-of-the-art sequencing technologies with the availability of the required genomic data paved the way for conducting this work that aimed at predicting the potential vaccine targets and drug candidates. A total of 14 novel vaccine candidates and 2 novel drug targets have been obtained which need further experimental validation to ensure their efficacy for the prevention and control of M. genitalium infection (Nogueira et al., 2021). More interestingly, M. genitalium is resistant to most antibiotics and difficult to be treated and controlled. Also, it causes endometritis, premature birth, and sterility in women and urethritis in men (McGowin and Totten, 2017). Hence, Ali et al. have conducted proteome-wide vaccine targets prioritization for designing an antigenic vaccine candidate against M. genitalium infection. MEV has been constructed successfully with further determining of the different physicochemical properties of the vaccine, but this study still needs further experimental validation for the constructed MEV (Ali et al., 2021b).
More recently, M. hominis infection was reported to cause bacteremia, pneumonia, and meningitis, but its significance to cause neonatal meningitis remains elusive. Using CSF patient samples, translucent colonies were observed on chocolate agar media, and the microorganism was recognized as M. hominis with MALDI-TOF MS. The 16S rRNA gene sequencing was also carried out which showed 99% nucleotide identity to M. hominis (Kersin et al., 2020). M. hominis is characterized by a very slow growth rate that requires specific growth media and it’s resistant to many antibiotics such as β-lactams, glycopeptides, sulfonamides, and macrolides (Kersin et al., 2020; Ferreira et al., 2022).
Mycoplasma Infection and Respiratory Diseases
Traditionally, the clinical picture of Mycoplasma infections was more intimately suggestive of damage due to host immune and inflammatory responses rather than direct toxic effects induced by Mycoplasma cell components (Razin and Jacpbs, 1992). However, He et al. (2018) have shown that many direct effects, as well as indirect immune mechanisms, have been incorporated in M. pneumoniae pathogenesis. The direct effect mechanisms include adhesion damage of M. pneumoniae to targeted epithelium then membrane fusion damage via alteration in its exposed receptors (Bao et al., 2015). In addition, nutrition depletion is caused by its limited metabolic capacity (Yus et al., 2009). Invasive and toxic damages are following mycoplasmas invasion of different host cells and microbial production of H2O2 and superoxide. Besides, the produced endogenous toxic oxygen leads to an increase in the intracellular oxygen pressure in the host cells, subsequently; oxidative stress and cell death will occur (He et al., 2018) as shown in Figure 1. On the other hand, the indirect immune damage mechanisms include humoral and cell-mediated damages and inflammatory damage via an intracellular receptor protein complex (inflammasome) (Shimizu, 2016).
More recently, gene expression analysis and whole transcriptome sequencing have been performed for M. pneumoniae infected Hela cells. The results illustrated that protein-coding genes of M. pneumoniae are correlated with immune response rather than cellular processes, probably suggesting the intrinsic ability of M. pneumoniae to modulate host immune pathways (Ramos et al., 2021).
COPD is one of the foremost predisposing causes of death in the USA, killing > 130,000 individuals per year. Globally, > 3 million deaths annually of COPD (Marciniuk and Schraufnagel, 2017), meanwhile, the middle- and low-income countries are more severely affected. Moreover, the lung microbiota of COPD patients contains more M. pneumoniae (Marciniuk and Schraufnagel, 2017). Eventually, M. pneumoniae continues to significantly aggravate the onset and recurrence of asthma (Meyer Sauteur et al., 2016).
Mycoplasma Infection and Urogenital Diseases
In females, M. genitalium, a sexually transmitted pathogen, has been associated with cervicitis, pelvic inflammatory disease (PID), spontaneous abortion, preterm delivery, and infertility. In parallel, it was detected among 10% to 30% as well as 4% to 22% of women with clinical cervicitis and PID, respectively (Gaydos et al., 2009). The high susceptibility of emerging antibiotic resistance is becoming increasingly important (Peter et al., 2018). On the other hand, M. genitalium causes symptomatic and asymptomatic urethritis among men and is the etiology of approximately 15%–20% and 40% of Nongonococcal urethritis (NGU) and persistent or recurrent urethritis, respectively (Bachmann et al., 2020).
Additionally, most urinary tract infecting bacteria can be demonstrated on standard culture, but it is exceptional for Mycoplasma and Ureaplasma. In which, bacterial count in urine does not necessarily relate to the number of bacteria in the bladder wall. A significant number of these intracellular organisms may occur in the bladder wall and be absent in urine. Thereafter, unresponsiveness to antibiotics, persistent lower urinary tract infection, and pyelonephritis were previously reported (Combaz-Söhnchen and Kuhn, 2017). M. hominis and Ureaplasma infection is notably associated with women’s infertility (Latino et al., 2018; Kusanovic et al., 2020). A recent study has identified these infertility-causing pathogens using the PCR technique. A number of 2360 tissue samples have been collected by urethral and cervical canal scrapings of adult women suffering from PID. The results showed that Ureaplasma spp. and M. hominis have been identified in 543 and 179 women, respectively. In addition, 112 women had mixed infections (Piscopo et al., 2020).
Furthermore, M. hominis, U. urealyticum, and U. parvum are examples of pathogens that can invade pregnant mothers and are closely associated with neonatal pneumonia (Waites et al., 2005). In pregnant mothers, U. urealyticum is found in the lower urogenital tract flora, occasionally, it ascends and causes bacterial vaginosis, chorioamnionitis, and premature birth (Stol et al., 2021). In the fetus, it causes neonatal sepsis and meningitis (Ferreira et al., 2021). Both Ureaplasma spp. and M. hominis can produce spontaneous abortion with higher rates in the case of M. hominis (Latino et al., 2018; Kusanovic et al., 2020). Genital mycoplasmas and ureaplasmas can colonize the urogenital tract which leads to invasive infection and spread to the placenta (Huber et al., 2018). Also, congenital M. pneumoniae pneumonia may take place via invasion and hematogenous transplacental infection (Hooven and Polin, 2017).
Mycoplasma Infection and Joints, Blood, Neurological, and Bone Disorders
M. pneumoniae has been frequently involved in severe CNS diseases, such as encephalitis (Al-Zaidy et al., 2015). Besides, it is associated with acute transverse myelitis (ATM) in the form of acute bilateral lower extremity paralysis, paresthesia, and bowel and bladder dysfunction. This syndrome was observed in 15-year-old patients with a slow curing rate that paid attention to the importance of early identification of mycoplasmas infection as a causative agent of ATM and more severe neurological complications (He et al., 2021a). Several extrapulmonary lesions such as cardiovascular, digestive, musculoskeletal, and dermatological lesions during M. pneumoniae infection have been summarized in a mini-review reported by (Narita, 2016).
Clinically, arthritis associated with M. pneumoniae infection has been diagnosed in children (Azumagawa et al., 2008). In addition, septic arthritis caused by M. hominis (Ali et al., 2021a) as well as U. parvum (MacKenzie et al., 2010) has been reported in immunosuppressed patients. M. hominis has been identified as a novel periprosthetic joint infection (a rare postoperative complication) using a new tool called “metagenomic sequencing” (Wang et al., 2021a). M. hominis also might cause brain abscesses (Whitson et al., 2014). What’s more, Ureaplasma species have been reported as pathogenic agents causing CNS inflammation in premature babies and abscessation in the adults’ brains (Glaser and Speer, 2015).
Infrequently, M. orale, an organism that is generally considered non-pathogenic in humans, has been isolated from patients with immunodeficiency and, as a result, these patients suffered from multiple abscesses and destructive bone disease. For direct detection of the pathogen, surgical specimens were used to do 16S rRNA sequence analysis (Paessler et al. 2002). Recently Ketchersid et al., have described a case report of recurrent multifocal M. orale infection in an immunocompromised patient (Ketchersid et al. 2020). Eventually, hemotropic mycoplasmas (HM) are epierythrocytic pathogens that attach to red blood cells of various mammals, including humans causing severe hemolytic anemia (Maggi et al., 2013b; Ikeda et al. 2017).
Mycoplasma Infection and Cancers
In recent years, many scientists carried out in vitro studies using oral tissues (Patil et al., 2015), hepatocytes (Choi et al., 2014) cervical cells (Atallah et al., 2020), and human prostate cells (Abdul-Wahab et al., 2021). These studies concluded that mycoplasmas infection stimulates tumorigenesis by inducing cellular transformation. Yacoub et al. (2021) have demonstrated the possible relationship between mycoplasmas infection and the development of cancers. In parallel, they reported the induction of malignant transformation by mycoplasmas infection in PMNCs (Zhang et al., 2004) and in many other human cell lines such as the uterus SK-UT-1B cells (Polianskaia et al., 1998), A549 lung cells and bone tissues (Jiang et al., 2008), prostate BPH-1 cells (Namiki et al., 2009), and neuronal cell lines (Ji et al., 2019). Another study used the PCR technique to determine that M. genitalium levels in patients with prostate cancer were significantly higher than those of patients with benign prostatic hyperplasia (Namiki et al., 2009; Barykova et al., 2011). As such, the available in vitro experimental data indicate that Mycoplasma infection induces chromosomal alteration, chromosomal instability, and/or cellular transformation via genetic mutations and translocations (Paton et al., 1965; Tsai et al., 1995; Feng et al., 1999; Zhang et al., 2000). M. penetrans, M. fermentans, and M. hyorhinis have been observed to stimulate chromosomal abnormalities, which in turn alter gene expression and cause malignant cell transformation (Benedetti et al., 2020). In addition, M. hyorhinis induces hepatocellular carcinoma (HCC) cell migration via the interaction of p37 protein with an epithelial cell adhesion molecule (EpCAM). P37 protein plays a key role in facilitating metastases and invasiveness of various cancer cells (Kim et al., 2019). Benedetti et al. have described Mycoplasma chaperone DnaK protein as responsible for cellular transformation. Besides they substantiated that this chaperone protein binds to Poly-(ADP-ribose) Polymerase (PARP)-1, a protein that is involved in the repair of any possible DNA damage, and reduces its defensive action. It also binds to USP10 which acts as an essential regulator for p53 protein and minimizes the p53 anti-cancer functions (Benedetti et al., 2020). Furthermore, using the in vivo mouse model, it was stated that specific-pathogen-free (SPF) conditions reduced the possibility of tumors formation. Therefore, the diverse microbiome compositions with predominant intracellular mycoplasmas affect the association between the diverse species of Mycoplasma and human cancers (Huang et al., 2001; Pehlivan et al., 2004; Pehlivan et al., 2005). In other words, mycoplasmas have been found in many tumor types. So, it is important to identify and characterize the mycoplasmas associated with the tumors in order to determine their role in carcinogenesis (Goodman and Gardner, 2018).
The first report of empyema caused by a commensal human Mycoplasma infection was described in a case of right pleural space infection with M. salivarium that was accompanied by laryngeal cancer (Baracaldo et al., 2012). M. salivarium is commonly responsible for nonpathogenic human infections, but it causes pathogenic infections only in the case of immunosuppressed persons through invasion of the human oropharynx (Totten et al., 2021).
Animals Infection
Many species of domestic and wild animals suffer from mycoplasmosis. Of which, contagious bovine pleuropneumonia (CBPP) and contagious caprine pleuropneumonia (CCPP) are the two most serious diseases, especially in low and middle-income countries shown by pleuropneumonia accompanied by extremely painful symptoms, reduced productivity, and death. CBPP has higher morbidity compared with CCPP; however, CCPP has higher mortality rates (Bolajoko et al., 2020). Both diseases require accurate diagnostic techniques and improved vaccines which should be accessible in the affected countries (Jores et al., 2020).
Bovine Infection
CBPP is mainly a disease of cattle and water buffalo, it is caused by Mycoplasma mycoides subsp. mycoides (Mmm) and notifiable disease of cattle listed by the World Organization for Animal Health (OIE) (Grieco et al., 2001; OIE, 2021). For the time being, OIE has announced that Europe, the USA, Australia, and South Africa are free from CBPP. For Asian countries, China and India were declared to be officially free, but the disease status is currently unknown in the remaining parts of Asia (OIE, 2019). However, it is endemic in sub-Saharan Africa causing huge annual economic losses (almost 2 billion US$), high mortality (10-70%), severe fibrous bronchopneumonia in the acute cases, and pulmonary sequestra in the chronic stage (Anonymous, 2018).
Mmm infection can be summarized in several consecutive stages, firstly, inhalation of infected aerosol droplets; after that, colonization of bronchioles and alveoli, thereby; Mmm invades the blood and lymphatic vessels and causes vasculitis. Finally, Mmm passes through blood and persists in a variety of other tissues including the lung, in which, the antigen is mainly detected in lung phagocytic cells, on the alveolar and bronchiolar epithelial cells, within the wall of blood and lymphatic vessels, and inside necrotic areas (sequestra formation) as shown in Figure 3. Infected animals actively excrete the pathogen through aerosolized droplets as a potential source of infection for the closely in-contact animals (Di Teodoro et al., 2020). The attenuated CBPP vaccine can provide a moderate level of protection estimated by a reduction in lung lesions in vaccinated and challenged cattle. Annual revaccination with the live vaccine is necessary to maintain protective immunity. Additionally, this vaccine is relatively inexpensive and easy to be produced on a large scale (Jores et al., 2020). On the other hand, this vaccine has a short period of immunity with many adverse reactions because it is a live-attenuated type; therefore, its reversion to virulent form sometimes occurs. In addition, it is temperature-sensitive (Dudek et al., 2021). Often, severe inflammation at the injection site followed by skin sloughing has been reported, and so far it can lead to animal death (Jores et al., 2020).
M. californicum, M. leachii, and M. dispar are other mycoplasmas that can cause significant diseases in cattle, but the most important worldwide pathogen infecting cattle is M. bovis. It can quickly spread to all age groups. Newborn calves can get the infection from older animals that suffer from severe mastitis, arthritis, and pneumonia (Hazelton et al., 2020) that maintain the infection cycle in the herd. Following a recent survey conducted in the United Kingdom from 2006 to 2017, calves at the age of < 3 months (post-weaning) have the highest prevalence of M. bovis pneumonia Mycoplasma bovis Investigations in Cattle (2018). Mainly, M. bovis infects the upper respiratory tract of young calves during the first few weeks of life through feeding of infected milk and/or direct contact with other infected calves’ nasal secretions (Maunsell et al., 2009). Hence, to stop the infection chain, we must stop the infection spread to the new calves born after the M. bovis detection on the farm. Methods for controlling M. bovis are culling or isolating M. bovis mastitic cows, pasteurization of infected milk, raising the calves separately from older animals, and better milking hygiene and teat dipping. But unfortunately, until now the protective vaccine against this serious pathogen is commercially unavailable (Haapala et al., 2021). Moreover, it is one of the four main bacterial pathogens associated with bovine respiratory disease (BRD) with significant economic losses as a result of higher morbidity and mortality rates, reduced growth performance, and raised costs of prevention and treatment (Kudirkiene et al., 2021).
Caprine Infection
CCPP is a fatal contagious illness of goats caused by Mycoplasma capricolum subspecies capripneumoniae (Mcc) and a notifiable disease listed by OIE. It has been reported to affect wild and domestic caprines. A recent report has estimated that CCPP has different case fatality rates of 30% in goat herd (n=200) and 8% in sheep flocks (n=400) (Abd-Elrahman et al., 2020) though previous workers have found sheep to be far more resistant (Nicholas et al., 2008). In addition, It is more widely endemic in East Africa, particularly in Kenya, Tanzania, and Ethiopia (Falquet et al., 2014). The first step to establishing a successful vaccine of CCPP is to design a challenge model that can be used to perform essential immunological studies. As this microbe proved to be host and tissue-specific, a novel challenge model has been established following the recent Kenyan outbreak strain ILRI181 in 2012 (Falquet et al., 2014) rather than the old Kenyan strain F38 (MacOwan and Minette, 1976). The base of this model is using two consequent inoculations of aerosols of Mcc culture into the nasal cavity of goats than a trans-tracheal inoculation of animals. This model has a morbidity of 100% and a mortality of 50–60% which simulate the natural infection pattern (Liljander et al., 2019). The current CCPP vaccine is a bacterin with saponin adjuvant, scheduled to start vaccination at 4 months of age with revaccination every 6 months. It is expensive due to the fastidious growth requirements of the pathogen and the relatively high total protein required for one dose of the vaccine (Jores et al., 2020).
M. agalactiae is a causative agent of an OIE notefiable disease called contagious agalactia (CA) that causes mastitis in dairy goats with formidable financial losses due to arthritis and drop or complete cessation of milk secretion, cachexia, and cornea opacity that can give rise to complete blindness (Santos et al., 2015). In Brazil, the estimated prevalence of CA in goats in different Brazilian provinces such as Rio Grande do Norte, the main goat raising state, was 83.28%, São Paulo was 27.7%, (Azevedo et al., 2015) and Sergipe was 10.3% (Santos et al., 2015; Damasceno et al., 2020). More importantly, many Mediterranean countries are showing substantial losses in the goat dairy industry in France (Poumarat et al., 2016), Spain (Paterna et al., 2013), and Italy (Cillara et al., 2015). CA causes considerable economic losses in Ukraine, according to a recent serological investigation in the Artsyzk area, 168 ewes (32.4 percent) of 519 investigated animals were infected with contagious agalactia. Of which, 109 (64.9%) were in their first year of life, 52 (31.0%) in their second year, and 6 (3.6%) in the 5-6-year-old age group (Bohach et al., 2021).
Hemoplasmas are known as pleomorphic tiny bacteria; they were named because they tend to attach to the erythrocytes’ surface and may cause hemolytic anemia in a wide range of mammals as well. Two well-known hemoplasmas, M. ovis and Candidatus M. haemovis, have been proven to infect small ruminants, their severity increases in young aged and pregnant animals (Hornok et al., 2012). M. ovis is a causative agent of chronic infection in caprines. Few reports are available regarding the prevalence of M. ovis infection in goats with variable figures ranging from absence in Australia and Tunisia (Rjeibi et al., 2015) to 20% in Hungary and 94% in Malaysia (Jesse et al., 2015).
Ovine Infection
Since M. ovipneumoniae was isolated for the first time; it is widely known as “sheep atypical pneumonia” specifically infecting sheep and goats (Besser et al., 2013). Latterly, it causes a potential threat to fattening lamb flocks and the lamb industry due to lower lamb growth and decreased ewe productivity rates, it also has been reported in many worldwide epidemics (Bai et al., 2020) (Jaÿ et al., 2020).
Urie et al. had conducted a wide-scale study and estimated the overall prevalence of M. ovis infection across the USA, it was 24.3% in domestic sheep (Urie et al., 2019). In another study, M. ovis prevalence was high up to 45.8% in 504 sheep samples in China (Wang et al., 2017). Importantly, Maggi and coworkers have reported that M. ovis-like species was the most predominant hemotropic organism found in human patients; thus, M. ovis could have a zoonotic nature (Maggi et al., 2013b).
Swine Infection
M. hyopneumoniae (M. hyo) and M. hyorhinis (Stemke et al., 1992) have been recognized as the main Mycoplasma species that are responsible for various porcine respiratory disorders. Merodio et al. have applied an experimental swine infection model of M. hyorhinis, the results indicated that multiple inoculations may simulate subclinical natural infection as in the field. Besides, animals would have to be infected several times for showing a visible immune response (Merodio et al., 2021). M. hyorhinis and M. hyosynoviae, are commensal microbes of the upper respiratory tract and tonsils of swine, they cause arthritis and polyserositis in young pigs between (6-10) weeks of age. While pigs older than 3 months of age are usually suffering from mild arthritis (Neto, 2012). More frequently, M. hyosynoviae is known to cause arthritis in adult pigs, but its lesions are restricted to the joints and synovial membranes (Gomes Neto et al., 2015).
M. hyo plays a significant role in the development of the porcine respiratory disease complex (PRDC) infection via reduced animal growth performance, reduced feed efficiency, and decreased average daily gain. Mostly, an increase in mortality rate takes place with the help of complicated infections (Pasteurella multocida, Haemophilus parasuis, Streptococcus spp., and Actinomyces pyogenes) which leads to increased total fatality rates (Olaniyi et al., 2020). Multilocus variable-number tandem repeat analysis (MLVA) and multilocus sequence typing (MLST) are strain typing genetic tools that can be used for M. hyo diagnosis (You et al., 2020). A recent study concluded that the most significant histological changes recorded were thickening of alveolar septa caused by neutrophilic cellular infiltration with intraluminal cellular exudate. The majority of pulmonary lesions were chronic (75.81%) (Mucha et al., 2020). Gilts are considered the main source of pathogen inlets because they are mostly exposed to the pathogen during the lactation period (Patterson and Foxcroft, 2019). Vaccination is frequently administrated all over the world with various commercially available M. hyo vaccines for not only healthy animals but also infected herds (Maes et al., 2020). For controlling M. hyo infection, Sponheim et al., have recommended the deep tracheal catheter as a more sensitive sampling tool, used for M. hyo diagnosis, than laryngeal swabs (Sponheim et al., 2020).
Another threat to the pig industry is infectious anemia caused by three hemoplasma species, M. haemosuis, M. suis (Eperythrozoon suis), and Eperythrozoon parvum. M. suis is the main causative agent of swine hemoplasmosis, which in turn adheres to the RBCs surface and triggers their engulfing by the spleen (Petri et al., 2020), as well as causes reproductive failure mainly stillbirths as reported in Southern Brazil (Bordin et al., 2021).
More importantly, M. suis has been proved as the first member of the HM group able to invade the erythrocytes of its host. Using electron microscopy, Groebel et al., have discovered a novel M. suis invasive strain that causes severe swine anemia with a fatal illness. Such invasion enables it to escape the host’s immune response and antibiotic therapy, and the intracellular lifestyle has clarified the chronic nature of HM infections (Groebel et al., 2009). Moreover, the genus Eperythrozoon was previously transferred to the genus Mycoplasma. Now it’s classified under a new order called Mycoplasmoidales (Gupta et al., 2018).
M. haemosuis was associated with fever, anemia, and skin lesions in domestic pigs (Stadler et al., 2020). Genus Eperythrozoon has two new blood parasites species (Eperythrozoon suis and Eperythrozoon parvum), and it was associated with a severe swine disease called “anaplasmosis-like disease” (Splitter, 1950). Globally, the recently detected swine hemoplasmas, such as China (Fu et al., 2017), South Korea (Seo et al., 2019), and Germany (Stadler et al., 2020), have similar clinical signs to those were formerly concluded for M. suis infection. Porcine hemoplasmas (PHs) have been detected in the biggest three pork producers worldwide [China (Song et al., 2014), the USA (Guimaraes et al., 2011), and Brazil (Sonalio et al., 2020)], as well as Germany (Normand et al., 2020), France (Brissonnier et al., 2020), Japan (Hornok et al., 2018), and Argentina (USDA, 2020).
Avian Infection
Avian mycoplasmosis is caused by four pathogenic mycoplasmas, MG, M. synoviae (MS), M. meleagradis (MM), and M. iowae (MI). The MG and MS are OIE-listed respiratory pathogens that have been causing huge economic losses due to their dramatic drop in egg production, hatchability, weight gain, and feed conversion efficiency. On the other hand, they increase embryo mortality, carcass condemnation, and prophylaxis and treatment costs in layers, broilers, and breeders flocks (Yadav et al., 2021). MG is a major Mycoplasma affecting poultry; it causes symptomatic as well as asymptomatic infections. Clinically, it causes chronic respiratory disease in chickens with difficult breathing, sinusitis, airsacculitis, increase embryo mortality in layer parents, and reduce carcass quality in broilers. More seriously, asymptomatic infection also has a formidable impact on the birds as it can be a predisposing factor to more severe secondary bacterial infections. In addition, MG may predispose the animal to many viral contagious diseases such as Newcastle disease and infectious bronchitis (Michiels et al., 2016).
More importantly, MG has been isolated from many different bird species acting as reservoirs for commercial poultry. For example, it was identified in the tracheal swabs of racing pigeons. However, the examined birds showed unapparent symptoms, they could play a role as the potential carriers of the organism (Tsai and Lee, 2006; Michiels et al., 2016). House finches and other passerines, another free-flying avian species, are regarded as the most serious threat for uncontrollable MG infection transmission. Luttrell et al have conducted a field survey for the assessment of MG prevalence among these bird species. The testing indicated that 19.1% of 671 birds caught at farms and 11.6% of 387 birds caught at feeder sites had a positive result (Luttrell et al., 2001). MS infection sometimes remains asymptomatic, otherwise, it can show signs of lameness, synovitis, mild lower respiratory signs, and airsacculitis (Yadav et al., 2021).
MG infection can be transmitted through horizontal and vertical routes, and so prevention and control measures are mainly through biosecurity and vaccination. Live attenuated and/or recombinant live poxvirus vaccines are commercially available against MG and MS infection. Also, avirulent MG live strains (F, ts-11, and 6/85 strains) can be used safely (Yadav et al., 2021). New research proved that 3 consecutive doses of MG vaccines, one live followed by two inactivated vaccine doses, provide good protection in layers (Kiers, 2020). In terms of advantages and limitations of MG vaccines, the F strain, a field strain with moderate virulence, is preferable in places where wild-type MG is highly virulent because it can defeat this virulent MG strain. The other MG vaccines, ts-11 and 6/85, were used more safely because they were less pathogenic and transmissible toward young progeny. While, they showed a lower potency in field challenge than F strain (US Animal Health Association, 2006). The MG 6/85 vaccine strain was developed through serial passages of a field isolate originating from the United States On the other hand, MG bacterins are becoming less popular in commercial flocks, where long-term control of MG infection is critical issue. Further, bacterins are more expensive and inappropriate as they need individual vaccination of birds (Ishfaq et al., 2020).
The ts-11 strain of MG and MS-H strain of MS are temperature-sensitive strains; both of them were proved to be safe and effective for protection against challenge in both chickens and turkeys when administered by eye drop. Globally, both of them are commercially available. For instance, its administration in Australia has greatly reduced the prevalence of disease in chickens causing a tenfold reduction in the use of macrolides in poultry (Browning et al., 2011).
The strain ts-11 vaccine, a mutant induced by chemical mutagenesis, can produce long-term immunity in chickens, but the protective immunity obtained by this vaccine is dose-dependent. A strain ts-304 has been isolated from ts-11 and demonstrated to be as safe as the ts-11 strain. Surprisingly, it also has been effective but at a lower dose and protective against challenge with the MG wild-type strain. In addition, its protection lasts for at least 57 weeks after a single vaccination at 3 weeks of age (Kulappu Arachchige et al., 2021). Since live vaccines are used in many parts of the world, Sulyok and his team have developed new highly specific molecular methods to rapidly differentiate MG vaccine strains from field virulent isolates using clinical samples (Sulyok et al., 2019).
MI is primarily infecting turkeys and occasionally chickens. The natural MI infection in turkeys results in late embryo mortality, a drop in hatchability, and leg abnormalities in young chicks (Pritchard and Balish, 2015). MM is responsible for air sac disease and musculoskeletal and reproductive disorders mainly in turkeys, it also has been isolated from chickens (Béjaoui Khiari et al., 2011).
Equine Infection
Mycoplasma infection was rarely reported in horses; however, M. felis has been isolated from pleuritis and lower respiratory tract infection cases in equines (Wood et al., 1997). In Japan, using genomic DNA for nanopore sequencing, M. felis strain Myco-2 has been detected from a tracheal wash sample of a diseased horse that suffered from respiratory manifestations. This strain has 98.2% identical nucleotides to the typical reference feline strain ATCC 23391 (Kinoshita et al., 2020). In addition, M. equigenitalium (equi) is a potential cause of infertility, endometritis, and abortion in mares, besides, reduced fertility in stallions (Tortschanoff et al., 2005). It is difficult and time-consuming to identify M. equi in clinical samples, and thus, Nehra et al., have developed a species-specific PCR for M. equi diagnosis in clinical samples (Nehra et al., 2015). Two unidentified Mycoplasma strains (N3 and NI1) isolated from the equine respiratory tract were proven to have cross-reactions with strains of Mmm and M. mycoides subsp. capri (Mmc) (Lemcke et al., 1981).
M. equirhinis was isolated from 10.2% of tracheal wash samples from racehorses in Great Britain (Cardwell et al., 2013) and 16.2% from thoroughbred horses in Turkey (Mete and ÖZGÜR, 2017). More recently, using the loop-mediated isothermal amplification (LAMP) assay, M. equirhinis was isolated from 40.0% of Japanese horses (Uchida-Fujii et al., 2021).
Equine hemoplasmas were discovered for the first time in Germany in 2010, as a new species of hemoplasma (Candidatus M. haemobos- like species). After that, scientists have recorded their incidence (26.5%) using a novel real-time PCR assay (Dieckmann et al., 2012). The chronically infected animals could act as reservoirs of infection to other in-contact animals; a recent study discovered M. ovis-like species in an index horse case. What’s more amazing is that the molecular and phylogenetic analysis of the haemoplasma sequences had 100% identity with 16S rRNA of M. ovis, a hemoplasma mainly related to sheep and goats (Kalantari et al., 2020). The previous discovery means that interspecies transmission of Mycoplasma infection could occur anytime. Based on the results of R segment analysis, a species of human Mycoplasma is a group of strains that share R-segments with average nucleotide identity (ANIs) ≥97%. Moreover, R-segments are superior to 16S rRNA gene sequences and multilocus sequences for the identification and phylogenetic analysis of human Mycoplasma species and their strains (Roachford et al., 2019).
HM infection in horses seems to behave subclinically with low bacterial blood loads as represented by Dieckmann et al. (2012). In addition, the infected horses can act as a potential reservoir of infection by M. ovis-like species for both sheep and humans (Kalantari et al., 2020). Further, Manguin et al. have screened the tracheal microbial inhabitants in asthmatic horses with qPCR and determined that Mycoplasma spp. were included in the microbiome composition of tracheal mucus in horses and asthmatic children, as well (Manguin et al., 2020).
Canine Infection
More than fifteen different Mycoplasma species have been isolated from dogs. They are mostly commensal organisms with a few harmful agents. M. cynos was significantly associated with lower respiratory tract (LRT) disease in dogs. On the other hand, no significant association was detected between M. canis, M. spumans, and M. edwardii and clinical signs of canine LRT disease (Jambhekar et al., 2019). M. spumans and M. maculosum were identified by PCR and sequencing to be responsible for fertility problems in male and female dogs (Tamiozzo, 2021).
M. hemocanis (Mhc) and Candidatus M. haematoparvum (CMhp) are two hemoplasmas species that have been reported in canines. In Italy, symptomatic infection by CMhp in a dog was firstly reported by Rosanna et al., who recommended PCR as a gold standard technique for clinical diagnosis of this pathogen (Rosanna et al., 2020). In Korea, the index case of Mhc infection was reported in a dog showing clinical signs of severe hemolytic anemia (Kim et al., 2020).
M. cynos causes upper respiratory disease in dogs, and it is proved to be associated with increased severity of canine respiratory disease complex (CRDC). Clinical signs may include cough and accumulation of mucus and exudate. Potentially, this microbe often evades the immune response predisposing animals to chronic and secondary bacterial infections (Chalker, 2005).
Feline Infection
Four species of feline hemoplasmas have been characterized in domesticated cats. They include M. haemofelis (Mhf), Candidatus M. haematoparvum-like, Candidatus M. haemominutum (CMhm), and Candidatus M. turicensis (CMt). In China, the first identified feline hemoplasma in cats was Candidatus M. turicensis (CMt) (Zhang et al., 2021b). In Thailand, another study estimated that 16.1%, 24.5%, and 1.6% of the random samples collected from stray cats were infected with Mhf, CMhm, and CMt, respectively (Kamyingkird et al., 2021). In Russia, the estimated prevalence of CMhm, Mhf, and CMt was 7.6%, 5.5%, and 0.7%, respectively (Demkin and Kazakov, 2021). CMhm is the most common type of Mycoplasma species producing hemolytic anemia. Mhf causes a more severe and fatal form of hemolytic anemia in cats, whereas CMhm and CMt have lower severity, but only cause severe infection in immunocompromised cats (Willi et al., 2006). Feline infectious anemia is a disease condition of cats accompanied by severe anemia upon erythrocyte disruption. It is induced following infestation by infectious agents such as hemoplasmas (previously mentioned) and Bartonella species (intracellular vector-transmitted pathogens infecting cats) (Zhang et al., 2021b). The non-hemotropic Mycoplasma (M. felis) causes different disease lesions in cats including conjunctivitis, respiratory symptoms, and polyarthritis (Greene and Chalker, 2012).
Wild Animal Infections
For tortoises, Origgi and Jacobson stated that the most significant bacterial disease that seriously affects the endangered free-ranging and captive tortoises is mycoplasmosis (Origgi and Jacobson, 2000). Mycoplasmas cause upper respiratory tract infection in threatened species, including gopher and desert tortoises in the USA. More specifically, The M. alligatoris causes pneumonia, synovitis, and polyserositis in American alligators (Valentine-King et al., 2020). CCPP affects various species of ungulates, chiefly wildlife species, such as gazelles and some species of antelope-like gerenuks (Baziki et al., 2020). M. ovipneumoniae, another Mycoplasma species, causes pneumonia in wild caprines (Jaÿ et al., 2020). This pathogen possesses LAMPs that are considered to be the most potent stimulator of inflammatory cascades (Bai et al., 2020).
More disturbingly, many recent studies have emphasized the importance of cervids as reservoirs for mycoplasmas infection since these species are considered the essential food source for many predators (Petri et al., 2020). Boes et al. have demonstrated the first natural HM infection in white-tailed deer. Following the high identity of 16S rRNA to the previously described M. ovis organism (Messick et al., 1998), the hemoplasma detected in his study likely represents a strain variation of M. ovis, an erythrocytic parasite of ovines (Boes et al., 2012). André et al. have molecularly detected HM in wild canids for the first time in Brazil that are regarded as endangered species; therefore studies concerning their pathogenic threats to their health are critically concerned (André et al., 2011). M. ovis is a zoonotic pathogen that has already been demonstrated in reindeer (Stoffregen et al., 2006) and white-tailed deer species (Maggi et al., 2013a) in captivity in the USA as well as in free-ranging spotted deer species in Japan (Watanabe et al., 2010). In Brazil, M. ovis has been detected in free-ranging marsh deer and pampas deer species (Grazziotin et al., 2011). A recent study identified for the first time, the occurrence of M. ovis in the gray brocket deer and small red brocket deer in the Brazilian national conservation plan for endangered South American deer (André et al., 2020).
M. conjunctivae, an important contagion of wild caprinae, cause infectious keratoconjunctivitis (IKC) in the form of mild symptoms in domestic sheep and goats, while it provokes a severe inflammation of conjunctivae and cornea in wild caprinae. It was responsible for severe epidemics episodes in wild caprinae including chamois and ibex (Marco et al., 2009). In the most advanced stages of IKC, corneal ulceration and perforation, as well as 30% mortalities, have been reported. Eye blindness is a consequence of bilateral eye infection which increases the fatality rate, especially in steep rocky areas because of the falling of affected animals from cliffs (Giacometti et al., 2002).
In view of the increasing transmission of MG to house finches in the wild, and alarmingly, it was responsible for the death of over 200 million birds (Nolan et al., 1998), this marked the first epidemic of MG in the wild birds. Besides, the excessive speed at which this pathogen goes rampant among the house finch population illustrates the rapid pathogen dissemination throughout a large geographic area within a very gregarious and mobile host population (Fischer et al., 1997). Till now, MG has been expanding its host range. For instance, it was identified in many phylogenetically different birds including songbirds (Fischer et al., 1997), raptors (Wrobel et al., 2016), and wild passerines (Luttrell et al., 2001). Hence, rapid evolutionary changes of the pathogen as it expanded geographically allow it to be one of the most recognized wildlife pathogen outbreaks (Sawicka et al., 2020). A possible explanation of the aforementioned host diversity or switching might be due to a shift in CRISPR system dynamics. Also, the gradual degradation and critical functional loss of the CRISPR system in house finches MG after the host switch appears to have a great impact on the pathogen evolution (Delaney et al., 2012). More recently, Mycoplasma infections have been found in migratory wild geese, while, the question concerning the pathogens’ transmission and dispersion is still poorly understood (Sawicka-Durkalec et al., 2022)
Laboratory Animals
Laboratory Animals’ Infection
Laboratory animals are useful fundamental scientific tools; the progression of apparent or in-apparent infections with Mycoplasmas has tremendous alterations on the normal physiological responses of mice throughout experiments. When mycoplasmas are running rampant in the experimental animals’ population, the only solution will be through introducing specified pathogen-free (SPF) animals and animal facilities to avoid potential false results. M. collis, M. pulmonis, M. neurolyticum, M. muris, and M. arthritidis are the most common Mycoplasma spp. infecting mice (Masoumalinejad et al., 2018).
M. pulmonis is the most prevalent Mycoplasma pathogen in mice causing otitis media, reproductive disorders, as well as substantial respiratory consequences with a prevalence of (20-60) % (Booth et al., 2014). The most important issue regarding M. pulmonis infection in rodents is that it is probably the best model from which we have learned the most about the determinants of immunity to control human M. pneumoniae respiratory infections. Many studies have used the experimental M. pulmonis rat infection model as an ideal model in terms of the ciliary cell function and cellular kinetics (Lambert et al., 1998), neurogenic inflammation (McDonald et al., 1991), natural killer cell activity (Kamiyama et al., 1991), local and systemic immune response (Steffen and Ebersole, 1992), induction of the production of several cytokines (Faulkner et al., 1995), and polyclonal proliferation of B and T lymphocytes (Rocha Sobrinho et al., 2011). M. neurolyticum has been characterized as a mammalian brain organism responsible for nerve disorders as a result of secreted Mycoplasma toxins (Tully, 1981). Also, M. collis was isolated for the first time from the conjunctiva and nasal cavity of mice and rats (HILL, 1983). M. muris, a scarce Mycoplasma type causing a huge hazardous effect on the reproductive efficiency of female mice, has been identified in recent years (Zinatizadeh et al., 2017). M. arthritidis, another rare pathogen of mice, is regarded as the main cause of arthritis in mice with swelling of legs and fingers (Constantopoulos and McGarrity, 1987).
Animal Models for Mycoplasma Infection
The laboratory mice are the most common species used in animal experimentation in biomedical research. In addition, the experimental mouse mastitis model allows us to examine a large number of Mycoplasma strains (Dmochowski, 1967).
Saraya et al., have designed five mouse models for M. pneumoniae pneumonia to examine the pathological picture in animals with various immune statuses. Firstly, animals were immunized following different regimes (one for each animal model). Afterward, they were challenged with M. pneumoniae antigen intratracheally, only mice groups immunized with M. pneumoniae antigen and alum adjuvant or M. pneumoniae antigen with CpG adjuvant (Th2 predominant) have developed severe lymphoplasmacytic infiltration in the peri-bronchovascular areas (PBVAs). These results indicate that the adaptive host immune responses in these two models seem to be the main regulator for human M. pneumoniae pneumonia pathological features and Th2 predominant characteristics might be important to generate and simulate the typical picture of M. pneumoniae pneumonia (Saraya et al., 2014).
In vivo strategies whereby BALB/c mice were injected subcutaneously with the T-B epitope peptides resulted in strong antigen-specific serum antibody and cellular immune responses, besides decreasing the inflammatory response of the challenged mice with M. pneumoniae (Weng et al., 2017), we can take the advantage of these findings for Mycoplasma vaccine production.
Several examples of useful Mycoplasma animal models including gerbils (burrowing mouse-like rodents) inoculated intranasally with M. pneumoniae to investigate its pathogenesis of human lung infection (Rodríguez et al., 2021). A rabbit model was used in previous studies for the development of polyclonal and monoclonal antibodies against various human diseases (Kaur et al., 1998). Furthermore, hamsters were injected intratracheally with M. fermentans culture to explain the ability of the pathogen to induce pneumonia and chronic infectious diseases in humans (Yáñez et al., 2013).
Guinea pigs are regarded as the best animal model after non-human primates to study M. pneumoniae infections. In an important study, Dumke et al. have used these animals for studying the pathogen-host relationship as well as characterization and subtyping of M. pneumoniae strains isolated from human patients. The adaptation, preference, and survival of individual strains also have been investigated. They concluded that M. pneumoniae species is genetically highly homogeneous (Dumke et al., 2004). Hausner and his team have also immunized guinea pigs with a hybrid protein composed of adherence-related parts of the proteins P1 and P30 of M. pneumoniae. The results showed a dramatic decrease in its detection in pulmonary samples from vaccinated as well as subsequently infected animals (Hausner et al., 2013). Meanwhile, sera from immunized animals have been demonstrated to have crucial adherence-blocking properties. Besides, the initiation of potent stimulation of mucosal immunity was the milestone for successful vaccination with intranasal antigen as well as in combination with other biocompatible adjuvants (Zhu et al., 2012).
In vivo studies using non-human primates have also played a vital role to investigate the antigenic and immunogenic properties as well as the pathogenicity of specific mycoplasmas including M. genitalium. For instance, experimentally infected primates have been used to examine M. genitalium membrane topology, antibody accessibility, amino acid diversity, and the location of functional and antigenic epitopes for the MgpB adhesion (Iverson-Cabral et al., 2015). Another important example of using primates instead of humans for doing essential experimental work is macaque (a genus of Asian monkeys) which was used for studying the persistence, immune response, and antigenic variation of M. genitalium in an animal experimental infection model (Wood et al., 2013).
Large animal models were also established for the Mycoplasma study. The experimental infection of SPF lambs with M. ovipneumoniae resulted in the establishment of asymptomatically infected upper airways in absence of other secondary infections (Davies et al., 1981). M. bovis calf infection model was used to estimate the effectiveness of some antimicrobial agents against animal mycoplasmas (Dudek et al., 2019). These previously illustrated experimental models to study mycoplasmas are valuable in many points of view such as investigation of their infection pathogenesis and immune response, assisting in the development of therapeutic strategies and diagnostic biomarkers, and conducting the potential vaccine candidate’s trials. On the other side, some encountered limitations should be taken into account as the availability of SPF conditions to avoid false results.
Virulence-Related Factors of Mycoplasmas
The poor understanding of the pathogenesis and immune response for the genus Mycoplasma is the main restraint that hampers mycoplasmas diagnosis, prevention, and treatment. Since the shortage of effective genetic tools, late publicized genome sequences, and lack of small animal models, the discovery of virulence factors has been progressing very slowly. Generally speaking, the following virulence-related factors have been considered, including adhesion and invasion, activation of some critical molecules and pathways related to innate and acquired immunity, phenotype variation such as phase variation and antigen shift; generation of secondary metabolites such as hydrogen peroxide (H2O2), biofilm formation, etc “and so on”. Lipoproteins and secreted proteins of mycoplasmas are important components inducing these activities. The characterization of these proteins might help elucidate pathogenesis and immune response, identify novel target biomarkers, establish diagnostic methods, and make improved vaccines (Zubair et al., 2020b).
Adhesion and Host Immune Response
Adhesion is the first step of Mycoplasma infection. Because it doesn’t have a cell wall, the adhesion is mainly mediated by cellular membrane proteins. For M. pneumoniae infection, it firstly attaches to ciliated respiratory epithelial cells at the base of the cilia employing a complex terminal organelle at one end of the elongated organism. Adhesion is mediated by interactive adhesin (P1) (Razin and Jacpbs, 1992) that is translocated to the surface and localized correctly within the attachment organelle. It also maintains interactive stability with accessory structural high molecular weight proteins 1 (HMW1), 2 (HMW2), 4 (HMW4), 5 (HMW5), P90, and P65 clustered at the tip of the organelle (Widjaja et al., 2020) (Waites and Talkington, 2004). Next, M. pneumoniae produces hydrogen peroxide (H2O2) and superoxide radicals (Shimizu, 2016), which induce oxidative stress in the respiratory epithelium. It was reported that M. pneumoniae induces transforming growth factor beta-1 (TGF)-β1 in primary cultures of normal human bronchial epithelial cells and RANTES in small airway epithelial cells (Dakhama et al., 2003). Similarly, it would act in vivo by inducing TGF-β1 in large airways and RANTES in small airways together with increased IL-6 and IL-8 production on bronchial epithelial cells. On the other hand, neutrophils, the first line of body defense mechanism, secrete chemotactic signals that attract monocytes, dendritic cells (DCs), and macrophages. They produce tumor necrosis factor-alpha (TNF-α) which drives DC and macrophage differentiation and activation (Wang et al., 2018). More intriguingly, some studies have identified that M. pneumoniae surface lipoproteins can trigger Toll-like Receptor (TLR) activation, leading to the production of IL-6 pro-inflammatory cytokines. These cytokines activate the transcription factor NF-κB, which translocates to the nucleus to express pro-inflammatory genes which in turn provoke inflammation and cellular immune response (Segovia et al., 2018).
They also directly activate DCs via cell-to-cell contact through neutrophil CD11b. Afterward; neutrophils are activated to release small amounts of elastase, which induces endothelial cells to secrete molecules like CD43 allowing closer interaction and stronger binding. Then, neutrophil adhesion is facilitated by the up-regulation of pro-inflammatory cytokine (TNF-α) and endothelial cell adhesion molecules (ICAM-1&2), after that, trans-endothelial cell migration of neutrophils takes place. Yamamoto et al. found that M. pneumoniae releases a secreted protein nuclease Mpn491 that can escape neutrophil extracellular traps (NETs)-degrading the ability of neutrophils (Figure 1) (Yamamoto et al., 2017). In the absence of this enzymatic activity, NETs can be induced and the networks of extracellular molecules bind M. pneumoniae enabling neutrophils to destroy the extracellular pathogen and minimize the disturbance of their host cells (Figure 1) (Zhao et al., 2021b). Moreover, Mycoplasma lipoproteins induce TLR2 signaling that induces neutrophil NETosis. Remarkably, (NETs)-degrading ability diminishes for older ages, and thus, older patients are more vulnerable to mycoplasmas infection like M. pneumoniae (Xu et al., 2017; Christodoulides et al., 2018). Finally, inside the alveoli, if M. pneumoniae gets rid of NETs, it will attach to alveolar macrophages (AMs). Subsequently, it is recognized via TLR1, 2, and 6 on AMs which originate from blood monocytes which constitute approximately 93% of the pulmonary macrophage population and are the early effectors of innate immunity against any bacteria (Shimizu, 2016).
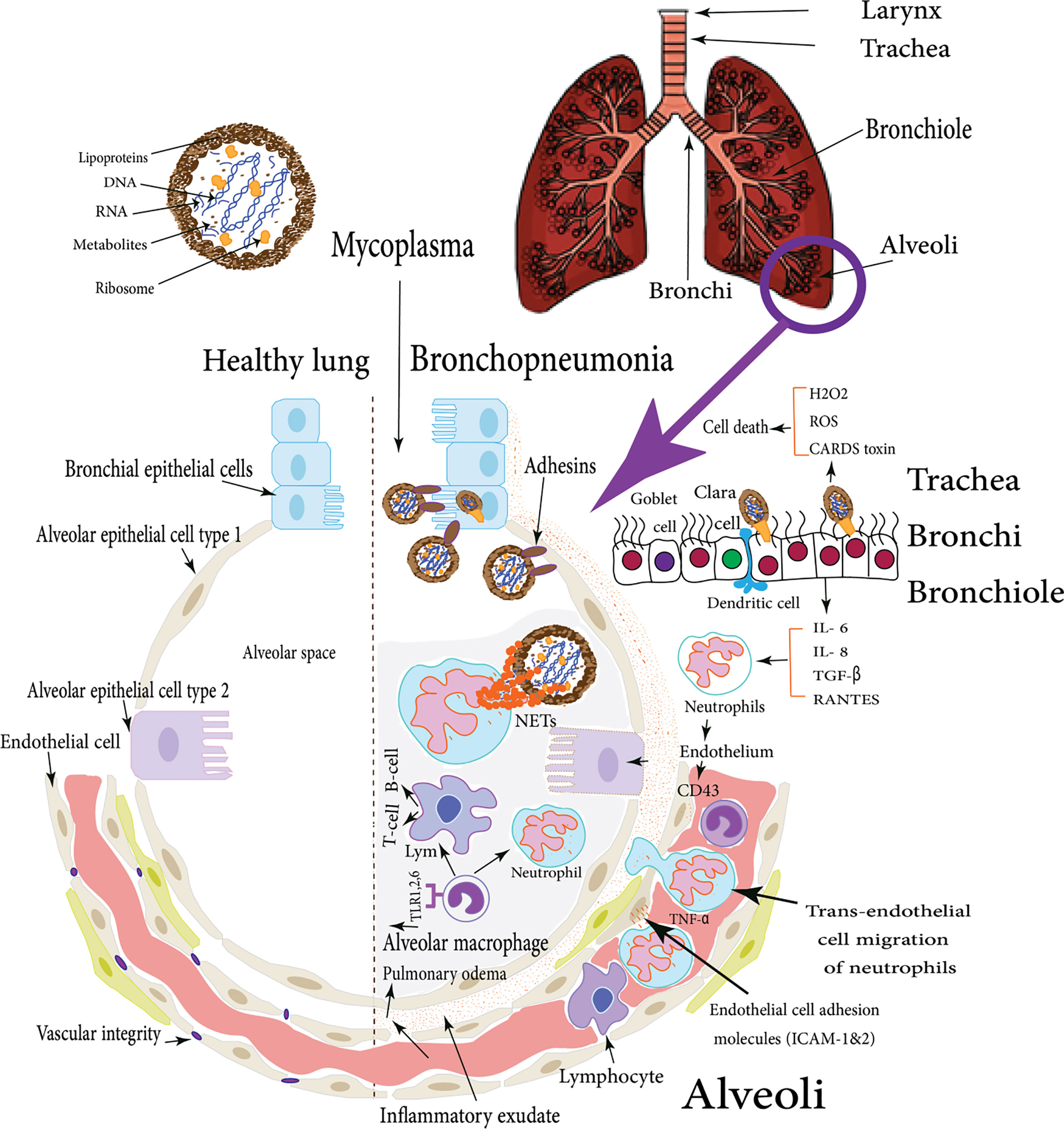
Figure 1 M. pneumoniae infection of human lung: the figure illustrates the difference between the healthy lung and Mycoplasma bronchopneumonia, the various cellular types incorporated in the respiratory defense mechanisms in case of M. pneumoniae infection of human lungs, these cells induce cytokines production which in turn stimulate both types of cellular and humoral immune responses with various virulence factors that enable Mycoplasma pathogens to adhere and colonize respiratory epithelial cells (Shimizu et al., 2008).
A cytoadhesion assay was developed to measure the interaction of Mmm with different host cells. The results indicated that Mmm cytoadherence is tissue and host-specific. In this study, the in vitro inhibitory effect of Mmm monoclonal antibodies (mAbs) against Mmm adherence to bovine lung epithelial cells (BoLEC) was investigated. Aye and coworkers demonstrated that 13 anti-Mycoplasma mycoides subspecies mycoides (AMMY) mAbs inhibited adhesion by at least 30%. More specifically, AMMY 10, a capsular polysaccharide (CPS) specific antibody, inhibited the in vitro growth of Mmm. Also, polyclonal rabbit serum against recombinant MSC_0267 blocked the adhesion of Mmm to BoLEC by 41%. Further in vivo studies are required for exploring the immune response induced by Mmm antigens recognized by these antibodies (Aye et al., 2018). A precision-cut lung slices (PCLS) infection model for Mmm has been established to study host-pathogen interactions. Using immunohistological analysis (IHA) and electron microscopy, the results of this ex-vivo infection model mimic the in vivo situation. It showed a consistent increase in the number of adherent Mmm Afadé in the bovine PCLS than caprine PCLS over time. Conversely, the adherent Mmc was not strongly affected by the type of host tissue as we observed an increase in caprine and bovine PCLS. Mmc displayed higher tropism to sub-bronchiolar tissue in caprine PCLS. Furthermore, Mmc was abundant on pulmonary endothelial cells which indicates how it causes systemic disease (Weldearegay et al., 2019).
The adhesion of animal mycoplasmas to host cells might be started by up-regulating the expression of endothelial cell P-selectin (CD62), E-selectin, vascular cell adhesion molecule-1 (VCAM-1), and intercellular adhesion molecule-1 (ICAM-1). After that, mycoplasmas attach to neutrophil L-selectin (CD62L). The second stage is presented by neutrophil activation to release elastase, which cleaves anti-adhesive molecules (CD43) from the endothelial cells, leading to stronger integrin-binding allowing the neutrophils to transiently attach to the endothelial cells as they pass along. Other well-documented adhesins were mentioned in detail (Table 1). The third stage (tethering) slows down the neutrophils, allowing them to interact more with the vascular endothelial cells (Granger and Senchenkova, 2010). M. pneumoniae cytadherence initiates inflammatory responses via an intracellular receptor protein complex called the inflammasome (Shimizu, 2016). Subsequently, extravasation and migration of neutrophils into the airways take place as a pivotal process to fight the bacterial infection. Through the endothelial cell layer and basement membrane, neutrophils attach the intercellular adhesion molecules (ICAM-1 and ICAM-2) within endothelial cell tight junctions (Chong et al., 2021). Eventually, Matrix Metalloproteases (MMPs) remodel the extracellular matrix to increase cell migration easily through tissues and move toward chemotactic agents (Figure 2) (Leick et al., 2014). Major Band Antigen (MBA), a surface-exposed lipoprotein, is a major determinant in the pathogenesis and virulence of the Ureaplasma species for causing chorioamnionitis. The potential pathogenesis for this pathogen perhaps caused by some antigenic variations of MBA leads to ureaplasmas escaping the host immune system, and colonization of the upper urogenital tract (Sweeney et al., 2017).
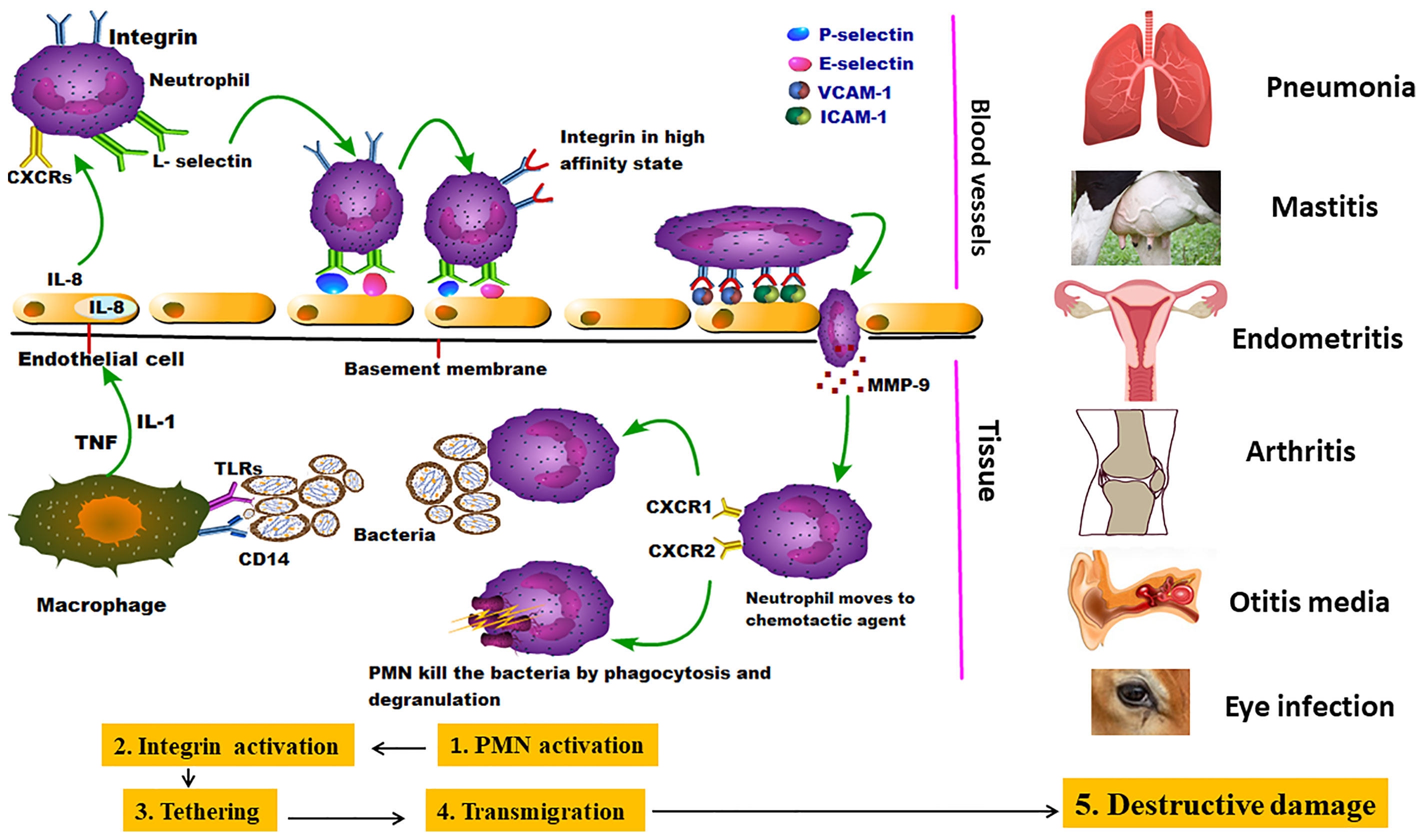
Figure 2 Invasion of mycoplasmas to target tissues and their potential interaction with immune cells: the figure showed the pantropic nature of mycoplasmas starting by stimulating the first line of immune cells (neutrophils) which in turn emit different danger signals and initialize the subsequent stages of PMNCs activation, Integrin activation, Tethering, Transmigration, and Destructive damage in many parts of the body causing different inflammatory lesions (Leick et al., 2014).
Mycoplasma Invasion Processes
Cell invasion is considered one of the most beneficial processes for mycoplasmas as it hides them away from the host immune system. Besides, living intracellular may enable them to pass through different body barriers such as the mucosal epithelium, get their nutritional requirements, and avoid the harmful effect of antibiotics (Vogl et al., 2008).
Mycoplasmas and ureaplasmas are the most frequently recognized intracellular pathogens in humans (Ferreira et al., 2021). M. bovis can invade different cell types such as T and B cells, monocytes, dendritic cells, NK cells, red cells, hepatocytes, cholangiocytes, renal tubular cells, facial nerve cells, etc. This invasion is beneficial to Mycoplasma in inducing inflammatory lesions, suppressing proliferation of immune cells, moving down from upper to lower respiratory tracts, and further spreading to other tissues from the lungs (van der Merwe et al., 2010) with the help of various invasive enzymes. Several previous reports have recorded the invasion and survival of various strains of M. bovis JF4278 and L22/93 (Bürgi et al., 2018) and Mb1 and Mb304 (Maina et al., 2019) in primary bovine alveolar macrophages (Suleman et al., 2016). In addition, M. bovis survival in necrotic lung lesions for long periods was reported, even in the presence of large numbers of neutrophils and macrophages. (Khodakaram-Tafti and Lopez, 2004). Other intracellular mycoplasmas also include M. penetrans, M. pneumoniae, and M. genitalium (Baseman et al., 1995), M. suis invasive strain (Groebel et al., 2009), and MG (Vogl et al., 2008).
Invasion of the mammary gland’s epithelium by Mycoplasma pathogens is a critical determinant for inducing mastitis and is associated with an altered immune response (Sordillo and Raphael, 2013). In this concern, these microbes can invade the gut lining epithelium following enteric infection that leads to their invasion into the host via the bloodstream and lymphatic (Nakagaki et al., 2018). More importantly, when the invasion is associated with immune depression of the host, bacterial dissemination to other organs takes place, including the mammary gland (Young et al., 2015). As such, live microbes can be detected in the bloodstream of animals (Zecconi et al., 2020) and humans (Aagaard et al., 2014; Whittle et al., 2018). An in vitro infection model showed the invasion of mammary gland epithelial cells has been established using 3 bovine epithelial cell lines (Josi et al., 2018).
To date, numerous mycoplasmas have been reported to produce invasion-related enzymes, such as proteases, nucleases, sialidases, antioxidant enzymes, and hyaluronidases. Nucleases, as important factors for mycoplasmas, are essential for degrading host nucleic acids; thereby having a critical role in growth, survival, persistence, and pathogenicity (Yiwen et al., 2021). For instance, Mpn491 secreted nuclease of M. pneumoniae (Yamamoto et al., 2017) and the major membrane nuclease (MnuA) of M. bovis (Mitiku et al., 2018) can degrade NETs and evade the killing ability of neutrophils. Proteases possess immunoglobulin (Ig) degradable capacities, as [Mycoplasma immunoglobulin binding (MIB) protein- Mycoplasma immunoglobulin protease (MIP)] (MIB-MIP) system to degrade IgG antibodies. Mmc carries the MIB-MIP system that exerts serine protease activity, followed by complete cleavage of IgG, thereby contributing to the evasion of the host immune system (Nottelet et al., 2021). Sialidase and neuraminidase are pathogenic enzymes for hydrolysis of sialic acid, destruction of extracellular matrix (ECM), tissue invasion, and apoptosis (Robinson et al., 2017). MG shows tropism to ciliated respiratory epithelium, then evades the mucociliary barrier followed by cell invasion (Matyushkina et al., 2016). It undertakes the invasion through penetration. Eventually, it resides intracellular, causing chronic or latent infection (Rüger et al., 2021).
Generally, after the invasion, mycoplasmas evolve and adapt to their parasitic intracellular life, they have slow intracellular growth rates compared to other extracellular bacteria. Their slow intracellular growth rate is mainly allowing them to hide and evade the host immune system (Rüger et al., 2021).
Generation of Secondary Metabolites
Among the secondary metabolites, H2O2 is considered a critical virulent factor of Mmm (Wadher et al., 1990), but there is no direct correlation between the ability to produce H2O2 and virulence in M. bovis (Zhao et al., 2017) and M. agalactiae strains. In addition, hydrogen sulfide (H2S) is a novel potential virulence factor of M. pneumoniae (Shimizu, 2016); Nitrative stress markers are reported to be a potential virulence factor of both Mmm and M. bovis (Schott et al., 2014). A large amount of H2O2 can be produced by M. dispar, a biofilm-producing bovine respiratory pathogen with 23 identified potential virulence genes (Chen et al., 2019).
In cattle, reactive oxygen species (ROS) is released by pulmonary phagocytes in the case of Mmm, and it damages the host cells. Numerous pathways have disturbed the integrity of cellular membranes, and/or indirectly enhanced the NF-κB pathway, thereby contributing to sequestra formation as shown in Figure 3 (Karin and Delhase, 2000). Glycerol is consumed through Glycerol transporter system ATP-binding cassette (GtsABC) and the glycerol-3-phosphate oxidase (GlpO) (Pilo et al., 2007), both of them participate in glycolysis and production of ROS and H2O2. Eventually, it results in cytotoxicity and cell death. The polysaccharide capsule participates in Mmm persistence and induces cytokine production as shown in Figure 3A (Weldearegay, 2015).
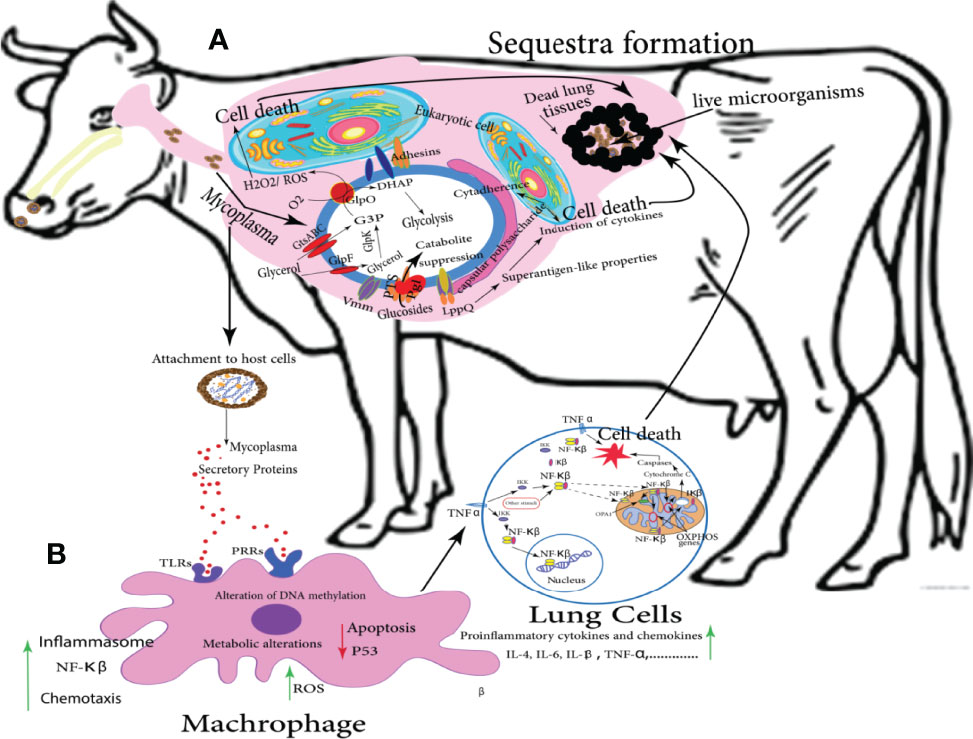
Figure 3 Potential metabolic pathways of Mmm involved in CBPP and sequestra formation: the following graph simulates the brief scenario that takes place in vivo and eventually leads to the occult problem of sequestra formation; (A) illustrates the interaction between mycoplasmas and lung eukaryotic cells following numerous pathways and release many metabolites that induce the targeted cell death. (B) showed the role of secretory proteins to stimulate the body immune cells to be directed toward the lung, activating different metabolic pathways including NF-κB, and stimulating a group of proinflammatory cytokines such as IL-4, IL-6, IL-β, TNF, etc. that is usually ended by cell death. Eventually, a collection of dead lung tissues containing live Mycoplasma pathogens called sequestra formation could be distributed in the lung (Di Teodoro et al., 2020).
The interaction of bovine lung cells with Mmm begins by attaching to mucous membranes of the respiratory epithelium followed by secretion of microbial secretory proteins that combine with specialized receptors on the surface of respiratory epithelial cells (Borchsenius et al., 2018).
Innate immune cells particularly macrophages, neutrophils, and natural killer cells are capable of recognizing pathogen-associated molecular patterns (PAMPs) of mycoplasma via toll-like receptors (Qin et al., 2019). Despite lacking a cell wall, mycoplasmas can interact with PAMPs (Demento et al., 2011). LAMPs such as MALP-2 (macrophage activating lipopeptide-2) and M161-Ag can induce TLR2 and TLR6 (Kumar et al., 2013). So far, LAMPs of M. pneumoniae can activate TLR1, TLR2, and partially TLR6 (Suzuki et al., 2003). In addition, other triacylated lipoproteins often stimulate TLR 1 and 2 but are TLR6 independent (Meylan et al., 2004). The subsequent inflammatory cascade begins with pathogen recognition by innate immune cells’ pathogen-associated molecular patterns (PAMPs) by interacting with specialized Pattern recognition receptors (PRRs) called Toll-like receptors (TLRs) (Demento et al., 2011). It also recognizes other emerged signals during tissue or cell damage that are usually known as danger-associated molecular patterns (DAMPs) (Medzhitov, 2007). This binding promotes the stimulation of macrophages which in turn induces the expression of pro-inflammatory cytokines and chemokines and the production of ROS. Additionally, the repression of p53-dependent apoptosis takes place. Eventually, it induces macrophages to produce TNF-α which acts for induction of the NF-κB inflammatory pathway (Borchsenius et al., 2020), a transcription factor that consists of a tri-subunit complex (P65, P50, and IκB) and exists in an inactive form in the cytoplasm. The activation of NF-κB only occurs when TNF-α attaches to TNF receptors. Then NF-κB activation inside mitochondria triggers cytochrome C release and the cell death occurred subsequently Figure 3B (Albensi, 2019).
Antigen Variation
To date, only a few of the surface lipoproteins from Mmm have been studied thoroughly. LppA (p72), LppB, and LppC are highly conserved lipoproteins that are present in closely related species within the M. mycoides cluster (Sacchini et al., 2011). Vmm is a small surface protein shown to have a variable expression pattern (Persson et al., 2002). LppQ is a highly antigenic lipoprotein specific to Mmm (Perez-Casal et al., 2015). Thorough characterization studies and the development of a recombinant ELISA built upon LppQ antigen showed that it is a suitable diagnostic marker. M. mycoides cluster contains many candidate proteins such as the putative ATP-binding cassette (ABC) transporter and 187 predicted surface proteins of Mmm. More antigens than just LppQ, can trigger antibody-mediated immune responses, are useful in diagnostic applications. Combinations of such antigens could thereby offer a higher specificity and sensitivity than existing methods by adding discriminative power to the current LppQ based ELISA while circumventing cross-reactivity compared to whole-cell antigen-based methods (Krasteva et al., 2014).
PARCELs (Palindromic Amphipathic Repeat Coding ELements), are a set of widely distributed and repeated protein domains or genes that were probably gained and/or exchanged through HGT. They can be disseminated by multiple gene-centric vehicles (ORFs) carrying these elements for enhancing accessory gene pools, connecting genomes of various clades, and sharing common habitats (Röske et al., 2010). A tandem repeat pattern of 25 residues was initially reported in the LppQ lipoprotein presented on the surface of Mmm. Repeats of this category show considerable sequence variation among individual copies on the surface of the Mycoplasma mycoides cluster (Röske et al., 2010). LppC is an immunodominant antigen of Mmm, its amino acid sequence and its precursor showed similarity with two Mmm lipoproteins (LppB and LppQ). The N-terminal domain of the mature LppC seems to be surface exposed, but the C-terminal domain presented an integral membrane structure (Pilo et al., 2003).
Variable surface proteins (VSPs) are major highly immunogenic lipoproteins. The expression of these proteins can be switched on or off corresponding to gene reassortment induced by environmental change. Therefore, the surface antigenic phenotypes are modified to evade the host immune response. For example, in the genome of M. bovis type strain PG45 (American strain), the cluster of the vsp gene family has 13 genes (Clampitt, 2021), however, only 2 genes are expressed each time, the remaining genes remain silent. Furthermore, the size of proteins is kinetically regulated (Lysnyansky et al., 1999). In the genome of the Chinese M. bovis HB0801 strain, the vsp gene family has only 6 genes (Qi et al., 2012). Notably, the whole parasitic intracellular life of mycoplasmas is difficult to follow, because they differ from other bacteria for their unique small size and lack of a cell wall, and so their intracellular inhabitance as silent parasites has a substantial impact on cellular metabolism and physiology and immune evasion (Benedetti et al., 2020). More interestingly, their genetic evolutions have resulted in rapid modifications in their cellular membranes due to the previously mentioned considerable variations in VSPs. Also, the membrane lipid phase variations of distinct membrane surface proteins are crucial for adhesion and intracellular colonization. For instance, sequence variations and alterations in the structural domains encode surface cytoadherence proteins (Ferreira et al., 2021).
Biofilm Formation
Biofilm formation by Mycoplasma species can increase Mycoplasma environmental persistence and survival. M. bovis has been confirmed to form a biofilm. Due to the high variation of VSPs, there is a big difference in the ability for biofilm formation among different M. bovis strains. The biofilm may induce resistance to dryness and heat indicating the enhancement of ability for environmental survival (Zbinden et al., 2015).
Mycoplasmas biofilm formation has been identified on both biotic and abiotic surfaces. Heterogeneously functional microcolonies are combined together to form one complex by bacterial polymeric substances such as polysaccharides, lipids, proteins, and extracellular DNA (Raymond et al., 2018). Mycoplasmas take the advantage of these biofilms as resistance to different environmental stressors such as antibiotics, antibodies, and host defense. Another advantage for M. bovis is to boost its environmental persistence, while inside the host leads to chronic infection (McAuliffe et al., 2006), whereas, exacerbating acute infection causes host and tissue damage after planktonic free cells are liberated from the biofilm causing host and tissue damage (Yiwen et al., 2021).
For M. pneumoniae, the more biofilms mature, the more cells encounter more morphologic changes. Additionally, H2O2, H2S, and CARDS toxin levels reach the peak at the early stage of biofilm formation but they will decrease over time indicating that the virulence of M. pneumoniae often reduces during the chronic infection stage (Feng et al., 2020). Microcolonies, the biofilm-forming unit, could also be identified in vivo in experimentally infected animals and were observed in M. suis by electron microscope on vascular endothelial cells (Sokoli et al., 2013). Raymond et al. also observed many ultrastructure molecules on the ciliated epithelium of the respiratory tracts in M. hyo infected pigs, they are essential for biofilm formation on abiotic surfaces (Raymond et al., 2018). For the first time, Awadh and coworkers have used scanning electron microscopy and confocal laser scanning microscopy to generate a 3-D image of M. fermentans biofilm architecture structure as a thin monolayer of cells to several layers thick which contain water channels allowing the diffusion of nutrients and oxygen (Awadh et al., 2021). When Mmm attaches to a solid surface, it can produce biofilms. Several extracellular binding adhesins such as pyruvate dehydrogenase were upregulated when it is included in adherent biofilm, and thus, the adherence process is essential and plays a key role in biofilm formation and commencing disease (McAuliffe et al., 2008).
Role of Mycoplasmas Secretory Proteins
Pathogenesis and Immunity
The secretory proteins usually are toxins, adhesins, and virulence determining enzymes that participate in cellular adhesion, invasion, proliferation, and inhibition of host defense. Therefore, they play a critical role in bacterial infections. The secretome is the whole proteins secreted by bacterial cells. For Mycoplasma species, the secretome research just began. By using the secretome techniques, 27 secretory proteins have been preliminarily identified for MS (Rebollo Couto et al., 2012). M. bovis was first identified to have at least 60 secreted proteins (Zubair et al., 2020a). Later using an improved proteomic technique, 178 secreted proteins were identified and 79 differential secretory proteins were determined between M.bovis virulent HB0801 (P1) and attenuated HB0801-150 (P150) strains (Zhang et al., 2021a). However, there are only a few reports relating to secretory proteins of other Mycoplasma species.
M. hominis P80 is the first known secreted protein of Mycoplasma with a type I signal peptide sequence (Hopfe et al., 2004). For Mmm, glycerol phosphate oxidase was identified in the supernatant of Mmm culture and this enzyme can cause host cell damage and induce an immune response (Pilo et al., 2005). Mycoplasma nucleases were first reported by Razin et al. (1992). Remarkably, nuclease activity was membrane-associated, for instance, M. pulmonis has substantial DNase activity exposed on the cell surface (Minion and Goguen, 1986). One secretory nuclease encoded by M. bovis MBOV_RS02825 has been identified to degrade NETs (Zhang et al., 2016). Another secreted protein MbovP280 of M. bovis can induce apoptosis of macrophages through CRYAB (Zhao et al., 2021a). For M. hyorhinis, a 200 kDa secretory protein was confirmed to inhibit the cytotoxicity of T cells and mitotic activity induced by lipopolysaccharide (Teh et al., 1988).
As it is known, although mycoplasmas have some molecules associated with secretory systems such as SecA, SecY, SecD, DnaK, P36, lepA, and SecE in M.hyo (Leal Zimmer et al., 2020), SecA, SecD, SecE, SecG, SecY and YidC in M. fermentans (Rechnitzer et al., 2011), SecA, SecG, SecE, FtsY, LspA, SecD, ffh, secY, YidC in Mcc (Chen et al., 2017) and SecA, SecD/F, SecE, SecG, VirB4 (T4SS ORF from ICE), and YidC in M.bovis, they don’t have complete Sec or Tat systems (Qi et al., 2012). Regarding the secreted mechanisms, two models have been proposed. The first is a dual secretion model shown by the secretory proteins with a dual nature as both membrane and secretory proteins. These proteins are membrane proteins when the type I signal peptide sequences of the precursor proteins are inserted into the membrane, while they become secretory proteins after the signal peptides are cleaved (Hopfe et al., 2004). The location at the membrane may help Mycoplasma adapt to environmental change; while the secretion would make them contribute more flexibly to pathogenesis and immune response. For the non-typical secreted proteins without the type I signal peptide, the extracellular vesicles (EVs) release model is proposed. Six Mycoplasma species including Mmm, Mmc, Mcc, M. agalactiae, M. fermentans, and M.bovis have been demonstrated to produce EVs under nutritional stress and the proteins in EVs include major components involved in Mycoplasma -host interaction (Gaurivaud et al., 2018). The capsular polysaccharides of M. pneumoniae (Liu et al., 2012b) and Mmm (Pilo et al., 2007) have a potent cytopathic effect that can lead to host cell death (Figure 3A).
Exotoxins previously were considered absent in the Genus Mycoplasma. Now, community-acquired respiratory distress syndrome (CARDS) toxin is a membrane-associated, ADP-ribosylating, and vacuolating substance. It was identified by Kannan and Baseman (Kannan and Baseman, 2006). In M. pneumoniae pneumonic patients, a significant seroconversion has been identified, demonstrating that CARDS toxin can be synthesized in vivo with highly immunogenic power. But, when M. pneumoniae was cultured with host cells, the toxin production raised significantly compared to using an inanimate medium for in vitro culture. These findings assure that the toxin synthesis depends on the interaction between host cells and M. pneumoniae (Waites et al., 2017). On the other hand, a few mycoplasmas can secrete hemolysins, which cause erythrocytes lysis via pores formation on the cell membrane. For example, U. parvum and U. urealyticum display hlyA and hlyC, respectively (Marques et al., 2016).
Common Proteins in Secretomes of Different Mycoplasma Species
Orthologs are genes derived from a single ancestor gene in the compared species, while paralogs are genes related through duplication in the same species. In other terms, paralogs are homologous genes that appear in single genome analysis. Since orthologs have equivalent functions, the comparative study of mycoplasmas secretomes is expected to produce a breakthrough in the exploration of common secreted proteins as novel biomarkers for mycoplasmas’ diagnosis and vaccine development (Zhang et al., 2021a). For example, M. bovis and Mmm are not only the most pathogenic cattle mycoplasmas, but also responsible for significant economic losses (Nicholas et al., 2008). They cause respiratory diseases with similar clinical and pathological symptoms. From the genome level, it was found that horizontal gene transfer (HGT) between them might occur (Citti et al., 2018). It would be probable for these events to happen when both pathogens infect the same hosts (Dudek et al., 2021). The comparative study of M. bovis and Mmm secretomes would reveal the homologous and unique secretory proteins and help understand the Mycoplasma evolution in cattle and develop common diagnostic reagents and vaccines for cattle.
The virulence-related factors identified at the protein level so far have been summarized in Table 1.
Evolution of Phylogenetically Related Mycoplasmas
A more complete view of mycoplasma evolution came from the comparative analysis of 16S rRNA oligonucleotide catalogs (Woese et al., 1980). Based on a 16S rRNA (Figure 4) sequence comparison, M. hyo and M. flocculare are known to be closely related. They are similar to the situation found in the genomes of the two closely related species M. pneumoniae and M. genitalium, whose genomes can be divided into segments with highly conserved gene organization, although the segments are arranged differently (Stemke et al., 1992), (Himmelreich et al., 1997). To identify the unique and common genes of M. flocculare, M. hyo, and M. hyorhinis and to explain the different behaviors of these species in swine respiratory tracts, Siqueira et al. took advantage of “the bidirectional best hit (BBH) approach”. Their results suggest that M. flocculare and M. hyo or M. flocculare and M. hyorhinis, share several ORF clusters (OCs) genome pairs, which can partially be attributed to HGT (Siqueira et al., 2013). Phylogenetic studies using sequence analysis of 16S rRNA genes resulted in 99.9% similarity between Mmm and Mmc, because of that they were included within a single mycoplasmas subspecies (M. mycoides subsp. capri) (Pettersson et al., 1996).
Figure 4 Evolutionary phylogenetic tree generated from 16S rRNA sequences: bars indicate distances under the corresponding tree. The pathogenic mycoplasmas of the major importance were colored and gathered in groups according to their main host, each group has a different color. The tree branches show the distance between neighboring mycoplasmas species (Chen et al., 2022).
The role of Phages and Prophages in Sustaining the Virulence in Mycoplasmas
Bacteriophages/bacteria eaters or “phages” are viruses that infect bacteria. Most of them kill their bacterial hosts. Phages have been identified for four mycoplasmas, including Mycoplasma arthritidis (phage MAV1), Mycoplasma hyorhinis (phage Hr1), Mycoplasma pulmonis (phage P1), and Mycoplasma bovirhinis (phage Br1) (Voelker and Dybvig, 1998). Of which, MAV1 was the only known virulence-associated phage. It is found in certain highly virulent strains of Mycoplasma arthritidis. In parallel, the polymorphisms within MAV1 prophage integration sites and within the prophages themselves may help to identify phylogenetic relationships among virulent M. arthritidis strains (Washburn et al., 2004).
The phage genome (prophage) is usually incorporated into the bacterial genome and transmitted vertically during replication. Prophages play a vital role in bacterial evolution, virulence determination, population shaping, and genetic transfer via horizontal gene transfer (HGT), which in turn influences bacterial traits. For example, Mycoplasma species have been proved to maintain a large complex prophage-like genomic island for the first time that carries a highly conserved gene cluster. This gene cluster is expressed in mycoplasma cells retaining resistance to three antibiotics (aminoglycosides, kanamycin, and neomycin) (Lysnyansky and Borovok, 2021).
The airway epithelium is the main place where exotic and commensal microbes interact between themselves and the host. Notably, the pulmonary surfaces (nasal and bronchial epithelium) are covered by mucus which contains mucin glycoproteins and nutrients. This environment is favorable for commensal bacteria and phage symbionts; therefore, these phages are critical for regulating bacterial populations in almost every niche (Tzani-Tzanopoulou et al., 2021). As such, it is well-known that phage communities are ampler in mucosal surfaces compared to other non-mucosal parts, and so these phages ensure a sustainable source of virulence evolution for different Mycoplasma species (Meyer, 2013)
In particular, M. hyosynoviae NPL4 strain is one of the most heavily phage-infected strains of Mycoplasma. Its genome was reported to contain two complete as well as one incomplete prophage sequences. Nevertheless, M. hyosynoviae can defend against invasion by phage as two of its strains contain a CRISPR-Cas system that keeps them resistant to infection. Interestingly, several prophage genes were discovered present within the genomes of M. hyosynoviae with significant similarity to its related species, M. arthritidis MAV1 phage (Bumgardner et al., 2015).
On the other hand, Bacteriophage-mediated immunoscreening is a promising field using an appropriate vector system. It offers a rapid and simple technique for the identification and immediate testing of putative candidate vaccines. For example, Mmm DNA vaccine, in which a whole-genome library was cloned into a bacteriophage λ ZAP, and then the phage library was plated on Escherichia coli cells (March et al., 2006).
Horizontal Gene Transfer (HGT) and Transmission of Genetic Information
Genome is dynamic in nature, and therefore epigenetic modifications have a great impact on it. To a large extent, genes may be lost, and/or the non-coding genomic regions may expand or shrink over a period of time. Furthermore, selective pressures over different genomic positions make them evolve differently. Also, epigenetic alterations in cancer include changes in DNA methylation that influence gene expression as mycoplasmas predispose their host to carcinogenesis (previously mentioned in the human infection section). Eventually, genes can be obtained via duplication within the same genome or acquisition from another organism through HGT which is considered the main regulator of microbial diversity. It is almost the final result of the infectious spread of mobile genetic elements (MGEs) in bacteria such as integrative and conjugative elements (ICEs), bacteriophages, and plasmids (Hall et al., 2017). In this concern, prevailing reports suggest that the transfer of mobile genetic elements (MGE) may represent only the tip of the iceberg (Blesa et al., 2017).
Remarkably, large chromosomal fragments can be passed across genomes. Besides, their subsequent consecutive reconstitution will be more effective, prominent, and complex than first imagined via unknown mechanisms (Husain et al., 2017). In 2014, the conjugal transfer of large chromosomal regions among ruminant Mycoplasma species has been demonstrated for the first time which had never been recognized in mycoplasmas’ research field. Intriguingly, it clearly illustrated the inter-species transmission of these pathogens between different hosts (Dordet-Frisoni et al., 2014). Mycoplasma chromosomal transfer (MCT) is a newly documented strategy that induces enormous exchanges of genomic materials. This potent mechanism has a profound impact on genetic rearrangement that reshuffled parental characteristics and created mosaics. It depends on the functional integrative conjugative element (ICE) in one partner that plays a part in the horizontal acquirement of small or large chromosomal segments as in the case of M. bovis (García-Galán et al., 2021). This has been most clearly demonstrated for M. agalactiae, numerous chromosomal DNA fragments and generated offspring comprised of a variety of genomic assortment, each proven to be unique. These gave us up to 17% of the exchanged genome. The genome has been predicted using comparative genomics that almost 18% of its genetic material has undergone HGT with mycoplasmas of the unrelated M. mycoides cluster (Dordet-Frisoni et al., 2019). A large number of ICEs copies were detected in several sequenced Mycoplasma genomes (Tardy et al., 2015), raising the likelihood that these simplest bacterial pathogens may be capable of conjugation mainly during inter-species transmission of pathogenic mycoplasmas (Table 2) (Dordet-Frisoni et al., 2019). Faucher et al. (2019) have established an advanced model to evaluate antimicrobial resistance of enrofloxacin in mycoplasmas after that; they performed a “genome-scale analysis” of major and minor determinants that lead to antimicrobial resistance. A novel protocol, for optimized conjugation in the case of M. agalactiae and M. bovis, has been adapted. It allows the horizontal transfer of ICE or chromosomal fragments carrying antibiotic resistance genes with estimating the frequency of conjugations. It can be modified also for the other Mycoplasma species (Sagné et al., 2021). Citti and Blanchard stated that the collected experimental data support their preliminary in silico predictions, and also verified that MCT has been shaping various Mycoplasma species with a mosaic-like genome (Citti and Blanchard, 2013), these mycoplasmas spp. might possess the ability to join together and support HGT strategy (Table 3).
Currently, genome sequences in databases are established for more than 60% of the known Mycoplasma species (>150) and for over 280 strains, in which almost half are available as a single circular chromosome. These figures show a fast increment, however, the already provided data guarantee a valuable source for mining total mycoplasmas genomes. Comparative genome analyses integrated with saturation transposon mutagenesis have been already established for over 20 years. The number of functional genes is likely to be closer to 450 based on synthetic genome studies. The remaining genes’ compartment was predicted to encode hypothetical proteins with little tendency to virulence factors (Citti et al., 2020). Phylogenetic analysis for the genome of many Mycoplasma species is distinct but sharing the same reservoir has contributed to the exchange of large DNA segments (Liu et al., 2012a). Bioinformatics analysis tools also predict HGT of the (MIB–MIP) system (previously mentioned in the Mycoplasma invasion processes section) between mycoplasmas infecting the same hosts, assuming that MIB–MIP is a shared system engaged in a worldwide strategy to elude the host immune system. Notably, MIP/MIB system was originally identified in Mmc (Nottelet et al., 2021). The identification of this system opens a new research route for a better understanding of the strategies exploited by minimal bacteria to escape the sophisticated immune systems of mammalian hosts (Table 2) (Arfi et al., 2016).
For the first time, an innovative technology of artificial genome was created as a minimal genome generated as a functionally competent artificial cell has been assembled by introducing a synthesized genome inside a cell envelope of a Mycoplasma designed with the help of transformation techniques (Cordova et al., 2016). Nowadays, genome transplantations have only been accomplished in atypical bacterial agents (Mollicutes). This modern technology will allow us to study comparative genomes of different mycoplasmas spp. which in role assist us in more accurate proteomics profiling and determining the potential essential common proteins (Baby et al., 2018).
Immunity, Diagnosis, and Therapy
Immune Response
The hallmark of Mycoplasma respiratory infection is the persistence of lung inflammation involving both innate and adaptive immunity. Recently, IL-17 has gained a lot of attention in respiratory Mycoplasma infection; it also has a hand in pathologic outcomes of lung infection (Luo et al., 2021b). Many recent studies, including a study, carried out in our lab, have proved that a variety of cells, particularly Th17 cells, in the lung can secrete IL-17. It contributes to respiratory Mycoplasma infection, as shown in our previous lab work using two groups of calves infected with the virulent HB0801 (P1) and attenuated HB0801 P150 strains of M. bovis (Chao et al., 2019). Peripheral blood mononuclear cells (PBMCs) also play a pivotal role to enhance innate immune response; Chao et al. have studied their transcriptome profiles. They found out 7 and 10 core differentially expressed genes (DEGs) in P1 and P150 groups, respectively. (Chao et al., 2019). Overall, the studies about immune response and pathogenesis concentrated on membrane proteins that also can be used as novel vaccine candidates (Krasteva et al., 2014).
Innate immunity plays a key role to control M. bovis infection, however, the pathogen has developed mechanisms to overrun and modulate apoptosis of bovine PBMCs and, thus, Gondaira et al. (2020) have evaluated the bovine mammary gland response following infusion of M. bovis. Somatic cells and bacterial cells counts in milk samples were increased; however, the proliferation of PBMCs and lymph node mononuclear cells (MNCs) of M. bovis-stimulated mammary glands was the same as unstimulated cells. Transcriptome analysis revealed that the mRNA levels of innate immune system-related genes in blood PBMCs, complement factor D (CFD), and tumor necrosis factor superfamily member 13 (TNFSF13) decreased. The mRNA levels of immune exhaustion-related genes, programmed cell death 1 (PD-1), programmed cell death-ligand 1 (PD-L1), lymphocyte activation gene 3 (LAG3), and cytotoxic T-lymphocyte- associated protein 4 (CTLA4)) of the milk MNCs in the infected quarter were increased as an indication of general immune suppression. While the mRNA levels of innate immune response-related genes of MNCs in the infected quarters were decreased (Gondaira et al., 2020). Much more deeply, the transcription of innate immunity-related genes in PBMCs (IL-17, IFN-γ, IL-27, and IL-36A) has been raised during M. bovis infection. These induce the triggering of T-cell subsets and cellular immune responses (Gondaira et al., 2021).
The immune response of mycoplasmas dictates what happens to them inside their niches. The primary Mycoplasma species that possess a prominent detrimental role worldwide is M. bovis, a major contagious pathogen in dairy and feedlot cattle that can suppress the host immune response during infection and develop a chronic inflammatory response that causes pathological immune damage in the target organs (van der Merwe et al., 2010). M. bovis develops several strategies to escape immune system elimination through inhibiting neutrophils, secreting a unique immunosuppressive peptide that inhibits the proliferation of bovine lymphocytes, and stimulating monocytes to produce anti-inflammatory factors. These factors cause apoptosis, suppress proliferation, and induce invasion of PBMCs leading to the persistence of chronic infection (Askar et al., 2021).
During M. bovis infection the host response itself contributes to the disease pathogenesis. It possesses superior strategies to elude host responses. Stimulation of both proinflammatory and anti-inflammatory cytokines takes place at the same time with skewed T-cell response accompanied by T-cell exhaustion in chronic infection with escaping immune clearance (Maunsell and Chase, 2019). Immunoglobulin-binding proteins are commonly known in many Mycoplasma species. They act to help the bacterial evasion of the host immune response (Arfi et al., 2016). To name a few, MBOVPG45_0375 (r0375) can bind to IgG and cause antibody neutralization to inhibit the antigen-antibody immune complex formation. MBOVPG45_0376, another membrane protein of M. bovis PG45 strain, is a novel IgG-cleaving protein that has a great impact on the interaction between M. bovis and host cells (Zhang et al., 2021c). P48, as an important virulence-related membrane protein of M. bovis involved in the adhesion process, its effect on EBL cells has been explored to further explain M. bovis infection mechanism. Remarkably, exogenous P48 protein inhibited EBL cells growth and induced similar apoptosis patterns as M. bovis infection, extracellularly and intracellularly (Wu et al., 2021).
Vaccination
Betlach et al. (2021) have investigated the potential impact of multiple vaccinations on reducing M. hyo transmission and infection; they found that the three-dose of commercial bacterin vaccination strategy significantly reduced bacterial load in inoculated gilts and decreased M. hyo lung lesions at 28 dpi in challenged gilts, as well. Another recent study compared M. hyo response to infection by route of exposure, concluding that intratracheal exposure produced the highest percentage of M. hyo DNA-positive pigs and higher serum antibody response which can be considered during setting a vaccination strategy (Silva et al., 2021). Another study, comparing the convenience and economic benefits of vaccinating piglets with M. hyo at 3, 7, and 14 days of age, has found that M. hyo vaccination at 3 days of age has supreme advantages over 7 or 14 days of age (Vangroenweghe, 2021). Recently, many studies concerning M. hyo vaccines evaluation have estimated the efficacy of new bivalent and trivalent vaccines containing M. hyo confirming that this vaccine provided good protection against M. hyo challenge (Yang et al., 2021a; Yang et al., 2021b).
Innate immunity regulatory factors, mainly Mannose-binding lectins (MBL) for developing new vaccines, play a key role to resist foreign pathogens invasion including Mycoplasma through selective recognizing lectins on the surface of bacteria. To date, it constitutes the first line of innate immunity against infection through activating complement, phagocytosis, and opsonization (Zhu et al., 2021a). MS bacterin was used after adding different adjuvants that can induce innate immunity. Chitosan adjuvant has enhanced lymphocyte responses and interleukins upregulation with systemic protection after subcutaneous injection (Gong et al., 2020). M. genitalium is the causative agent of several sexually transmitted infections in animals and humans. Subtractive genomics and reverse vaccinology have been applied in silico identifying potential vaccine and drug targets against five strains of M. genitalium, 14 novel vaccine candidates and 2 novel drug targets were finally predicted (Nogueira et al., 2021).
In humans, M. pneumoniae-derived lipids and membrane lipoproteins play a critical role in the inflammatory responses. Using an antibody-neutralizing assay, it was demonstrated that TLR-4 is essential for M. pneumoniae lipid-induced TNF-α and IL-1β production. NF-κB-dependent pathways also are critical for pro-inflammatory cytokines secretion (Luo et al., 2021a). Numerous types of M. pneumoniae vaccines have been designed in the form of whole-cell vaccines (inactivated or live-attenuated), subunit vaccines (involving P1, P30, P116 proteins, and CARDS toxin), and DNA vaccines (Jiang et al., 2021).
CBPP is the major threat to the cattle industry in Africa, affecting almost 25 countries. Several novel experimental vaccines have been developed over the last 2 decades to improve the T1/44 live vaccine protection ability, but mostly they have aggravated the disease (Dudek et al., 2021). The subunit vaccines formulated with a combination of recombinant proteins of Mmm showed protection against challenge with the most virulent Mmm strain (Afadé) (Nkando et al., 2016). Meanwhile, M. bovis has spread now to most cattle-raising countries. Vaccination is the basic focus for infection control because of its increasing resistance to antimicrobial therapy, but commercial effective vaccines are currently absent. In our previous work concerning M. bovis, we concluded that the protection rate of the P150 M. bovis attenuated strain was 87.7%, and thus, it is a promising candidate for a live vaccine against M. bovis infection in cattle (Zhang et al., 2014). In our previous study, we discovered 10 core DEGs in the P150 M. bovis HB0801 attenuated strain, These DEGs can be used in further studies for improving attenuated P150 strain (Chao et al., 2019). Finally, our team has determined 79 differential M. bovis secretory proteins between the virulent P1strain and attenuated P150 strain (Zhang et al., 2021a).
Nanotechnology for Diagnosis of Mycoplasmas Infection
The limitations of available options for Mycoplasma diagnosis highlighted a critical need for a new detection platform with high sensitivity and specificity. To achieve better detection efficiency, single-walled carbon nanotubes (SWCNT) coupled with colloidal gold-monoclonal antibody immunochromatographic strips (CGIC) have been used (Song et al., 2017).
As growing advanced fields, the loop-mediated isothermal amplification (LAMP) (Wang et al., 2019a) and multiple cross displacement amplification (MCDA) techniques combined with nanoparticle-based lateral flow biosensor (LFB) assay have been developed and evaluated (Wang et al., 2019b). These techniques are simple, reliable, and smart enough for the identification of M. pneumoniae. The LAMP-LFB assay specifically identified DNA templates of M. pneumoniae, and cross-reactivity with other pathogens did not occur (Wang et al., 2016; Yuan et al., 2018).
Previously, Biosensors have been designed using silver nanorod arrays (NA) for identifying M. pneumoniae in culture and throat swab samples. More interestingly, these biosensors are characterized by high specificity (95%–100%) and good sensitivity (94–100%) (Hennigan et al., 2010). Thereafter, Henderson et al. have established nanorod array-surface enhanced Raman spectroscopy (NA-SERS). It detects M. pneumoniae in true and simulated throat swabs at the qualitative endpoint of < 1 cell/μl with a sensitivity exceeding that of qPCR (Henderson et al., 2014) and high specificity and strain-typing capacity (Henderson et al., 2015). In addition, gold nanoparticles have been used for the rapid detection of M. suis in porcine plasma (Bai et al., 2018).
Today, the surface-enhanced Raman scattering (SERS) biosensor, a kind of ‘whole-organism fingerprint’, has been invented with a smart capability of identifying three Mycoplasma species M. hominis, M. genitalium, and Ureaplasma urealyticum (Berus et al., 2021). The most recent today’s technology in the field of Mycoplasma diagnosis using M. pneumoniae as a target pathogen has gained a highly sensitive DNA detection limit of 3.12 pg/μL. After 10 cycles of the PCR-coupled SERS method, it has shown enhanced detection ability. This technology is composed of a low-cost paper-based SERS substrate in the form of silver-nanowires (AgNWs) then coupled with PCR for rapid and sensitive Mycoplasma DNA determination (Lee et al., 2021).
Nanotechnology for Mycoplasmas Therapy
Generally, mycoplasmas are susceptible to antibiotics that affect proteins including tetracycline, macrolides, Lincosamides, and phenicols or nucleic acid synthesis like fluoroquinolones. However antibiotic resistance sometimes develops against these antibiotics causing a decrease in the effectiveness of certain antimicrobial agents (McDermott et al., 2016). To overcome the development of multidrug resistance (MDR) of major pathogens that threaten humans, nanotechnology-based drug delivery systems have been emerging as a talented approach. They have been used to avoid the drawbacks of traditional drugs, decrease antimicrobial resistance, and open the hypothesis for new drug formulations (Algharib et al., 2020a). Nanoparticles (NPs) are common encapsulation materials with the great advantage of increasing intracellular accumulation of the drug to overcome bacterial resistance. For example, metallic and carbon nanotubes can be used for inhibiting bacterial biofilm formation as well as solid lipid nanoparticles as nanocarriers for antimicrobial agents that cannot be administrated as free drugs (Arana et al., 2021). In our previous work to combat the growing MDR, we have designed and optimized, for the first time, chitosan nanogel to encapsulate a rifaximin. It increases bioadhesion of rifaximin, and thus targeted release in the intracellular and extracellular bacterial infection sites (Algharib et al., 2020b) as in the case of Mycoplasma infection.
Basically, the nano-drug delivery system has many advantages not achievable by conventional drugs. The drugs can be encapsulated into nanoparticles and thereby increase their solubility, enhance their absorption and uptake by cells, target them to specific organs, and release them in a controlled manner as a response to specific stimuli (Andrade et al., 2011). Additionally, some nanoparticles have great potential in medical microbiology due to their antibacterial effects with low toxicity against the hosts (Djurišić et al., 2015). Several types of nanoparticles have been designed like rifampicin/poly (lactic-co-glycolic acid) nanoparticles for delivery to the lungs by nebulization, they need further research to be tested regarding their effectiveness against mycoplasmas infection (Andrade et al., 2013). At present, silver nanoparticles have received more attention in scientific research. Yang et al., (Yang et al., 2019b) have designed and evaluated, for the first time, the anti-Mycoplasma pneumonia potential of biosynthesized herbal-based (Zingiber zerumbet) silver nanoparticles in experimental mice. The lincospectin- zinc oxide nanoparticles (ZnO-NPs) are effective against M. bovis through destructive oxidative stress to bacterial cells and disrupt their metabolic activity thereby inhibiting their growth. Nowadays, Zn is available as a food additive since it can improve the immune system, prevent biofilm formation, and has low toxicity to human cells (Fathi et al., 2019).
TLR2 agonists, effective prophylactic antigens or immunotherapeutic agents for many pathogens, are located on the cell membrane surface to recognize microbial LPS or lipopeptides. These ligands can be used as molecular adjuvants with vaccines for providing a threat signal to keep a long-lasting adaptive immune response. This idea was represented by Franzoni et al. (2021) who used a lipopeptide based on M. agalactiae surface protein (Mag-Pam2Cys) that activated antimicrobial innate immunity by polarizing porcine macrophages. More creatively, these molecules can be loaded on a compatible nanocarrier to improve their invasiveness and therapeutic power.
Discovery of Enigmatic Features of Mycoplasmas
In order to clarify the mystery regarding Mycoplasma virulence and its immune escaping ability, contemporary trends are directed toward genome transplantation and genome assembly focusing on the advancement of cutting-edge technologies (Labroussaa et al., 2019). Through using the most current technological devices, an important study on M. genitalium with a 580 kb genome, the smallest complete genome identified until now, has been conducted. They found that (55-73) % of the protein-coding genes are essential, and so they are considered the minimal set of genes essential for maintaining bacterial life (Hutchison et al., 1999). Another study on M. genitalium has identified 382 essential genes of the 482 protein-coding genes (Glass et al., 2006). The first synthetic Mycoplasma genome was created for M. genitalium with 583 kb. This technical achievement was a great step in the field of synthetic biology. More interestingly, the project of genome transplantation in bacteria has been established by changing one species into another (Lartigue et al., 2007). Afterward, a fully synthetic M. mycoides genome was transplanted into M. capricolum (Gibson et al., 2010), this genome was reduced to include the only essential genes for its life (473 genes) (Hutchison et al., 2016). These achievements will allow us to set and redesign future genomes for medical products by using computational tools (Rees-Garbutt et al., 2020). M. pneumoniae is another important bacterial model with a natural tropism to the human respiratory tract (Wodke et al., 2015). Indeed, cutting-edge technology is used to study M. pneumoniae proteome interactions with host cells using mass spectrometry which provides an accurate perception of the structural information on M. pneumoniae proteins (Kühner et al., 2009). Consequently, experimental validation of the whole metabolic network map has been conducted (Yus et al., 2009).
Other new promising directions were used to unravel the mycoplasmas panoply. To name a few, X-ray crystallography and cryo-electron microscopy tomography can explore the tridimensional structure of the various illusive virulence determinants for cytoadhesins of M. genitalium and M. pneumoniae (Vizarraga et al., 2021). It will be important to first confirm the cellular localization of these proteins using available fluorescent molecules e.g., mNeon71, or tags using electron microscopy. After that, a list of surface-exposed virulence candidates will be available that can be verified in ex vivo and in vivo biological systems (Jores et al., 2020). Also, MIB and MIP are surface proteins which present in the majority of Mycoplasma species. Cryo-electron microscopy showed how these proteins perform a “hug of death” strategy when bound to antibodies disrupting the antigen-binding sites (Nottelet et al., 2021). Acting to protect mycoplasmas from antibody-mediated agglutination, the MIB-MIP system is considered a landmark of mycoplasmas immune evasion.
Up to now, Micro RNAs (miRNAs) are key molecules that regulate gene expression in vivo with specific pathways. For instance, miR-509-5p negatively regulates the NF-κB pathway, thereby affecting the inflammatory response of M. pneumoniae in sheep (Zhu et al., 2021b). Additionally, the impact of protists/bacteria relationships has been rarely taken into account by microbiologists. Today, the understanding of protist evolution is highly dependent on prokaryotes, for example, HGT from M. hominis to symbiotic hosts allows adaptation of both species to new eukaryotes habitats (Henriquez et al., 2021). The ability of M. hominis and Trichomonas vaginalis (the most common sexually-transmitted protozoan) to establish a close symbiotic relationship opens new hypotheses on the pathogen association role in the induction of cancer. M. hominis infection dramatically upregulates the host inflammatory response to T. vaginalis. Hence, a marked chronic inflammatory state is a condition that predisposes to tumor transformation (Margarita et al., 2020).
Another novel promising trend is to create non-specific mutations into a variety of genes of Mycoplasma through DNA transformation and recombination. It was tested for creating knockout mutants ensuring that gene recombination is a successful approach for generating site-specific mutants and developing a new genetic system. This strategy will be a pioneering way to study pathogen-host interaction and pave the way to develop new genetically well-defined vaccine strains (Clampitt, 2021). Genome-scale models (GEMs) are a computational description of gene-protein reaction (GPR) associations for the whole metabolism of the target organism. Their reactions are ratio-based and mass-balanced, whereas, their formulation is based on experimentally gained gene annotation data. They help us through whole-cell analysis of an organism’s metabolic parameters. In addition, genome-scale metabolic models input external elements such as media constituents by simulating the metabolic fluxes. Then relate them to the bacterium growth, which can be described as biomass yield in the objective function of the model (Fang et al., 2020). The main computational approach that is applied to GEMs is Flux Balance Analysis (FBA) as a constraint-based model system that gives the expectation of metabolic fluxes through linear programming. It consists of a mathematical representation of the metabolic reactions (Nobile et al., 2021). More recently, Li et al. have explained the mechanisms of virulence attenuation using whole-genome sequencing and comparative genomic analysis of two M. hyo strains. The highly virulent M. hyo strain ES-2 has transformed to attenuated strain ES-2L with lower virulence after the in vitro serial passage of 200 times (Li et al., 2021).
Furthermore, proteomics techniques are frequently used. For instance, an immunoproteomics study on M. bovis has identified MbovP579 as a good diagnostic marker (Khan et al., 2016). A comparative secretome analysis on M. bovis virulent P1 and attenuated P150 strains has recognized a differential secretory protein of MbovP0145 as a potential diagnostic antigen (Zhang et al., 2021a). The in silico analysis of secretory lipoproteins revealed the apoptosis inducer MbovP280 of M. bovis via its interactive CRYAB (Zhao et al., 2021a). Above all, Multi-Omics Technology (proteomics, transcriptomics, metabolomics, etc.) by using over one omics technique would enable us to intensively and systematically understand mycoplasmas. Besides, the potential candidate targets for the development of diagnostic reagents, novel vaccines, and drugs can be discovered (Jores et al., 2020). Gaspari (Gaspari, 2021) has developed a genome-scale, constraint-based model, and metabolic modeling to determine the factors affecting the growth of M. pneumoniae. Whatsmore, she established a pioneering technique that allows the growth of M. pneumoniae on serum-free media.
Eradication of Mycoplasmosis
Eradication of CBPP in Many Parts of the World
CBPP has been completely eradicated from many parts of the world (OIE, 2019). For instance, in China the disease caused considerable economic losses to the cattle industry between the 1950s and 1970s. A potent vaccine, developed from a virulent strain of Mmm (Ben-1), was attenuated through multiple passages in rabbits. It had high immunogenicity and a remarkable protection efficacy (95-100%) in cattle for 28 months (Xin et al., 2012). Sheep were used for preparing this vaccine to increase the antigen yield and then in Tibetan sheep where it led to fewer adverse effects in domestic yaks and related species. Finally, the last CBPP case was recorded in 1989 and in 2008; OIE has announced China to be a CBPP-free country (Xin et al., 2012). More recently, this potent strain was trialled in Africa and reported to be as effective as T1/44 though the full data has not yet been available (Jores et al., 2020).
In Europe, where vaccination was prohibited for most of the 20th Century, slaughter of affected and in contact cattle has been the only method of control. This was largely successful, and it was believed that CBPP had been eradicated by the mid1960s. However, CBPP re-emerged two decades later in Portugal, Spain, France, and Italy but was finally eradicated following strict stamping out when the last case was recorded in Portugal in 1999 (Nicholas et al., 2008).
In Australia, the vaccination campaigns using attenuated vaccine strains (KH3J and T1/44) successfully reduced the number of cases. But the total eradication was achieved in 1973 only after applying strict animal movement measures and stamping out policy (Newton, 1992).
Eradication of M. bovis in New Zealand
M. bovis infection was first reported in New Zealand in 2017. The Ministry for Primary Industries (MPI) took a brave action to eradicate M. bovis from NZ despite the presented difficulties in detecting and containing the movement of infected livestock. Decisively, it was achieved via culling of infected herds, besides, the NZ Government’s decision to eradicate M. bovis was unique since no other country has attempted it previously (Boyce et al., 2021). MPI has designed a new website that contains the latest updates and M. bovis situation reports, compensation, community events, and the national surveillance concerning eradication. Subsequently, the Chair of the Technical Advisory Group to MPI for the M. bovis program on 19 August 2021 has announced that currently there are only three active properties of M. bovis. Soon afterward, the eradication may be achievable. However, long-term surveillance will be required before freedom of infection can be declared (Controlling M. bovis in NZ 2021).
M. hyo Eradication Program
More recently, Gulliksen and his co-workers have announced the eradication of M. hyo infections in the Norwegian pig population (Gulliksen et al., 2021). Their strategy was based on the implementation of numerous factors, such as well documented and effective eradication protocols, paving the way for designing accurate diagnostic tests, decreasing herd density, stopping the importation of live animals, besides, the loyalty of farmers and substantial efforts of veterinarians for rapid sampling and diagnosis. Following their path in this concern, many other mycoplasmas can be completely eradicated in the coming few years.
Conclusions
Rampant Mycoplasma pathogens have caused great concern in recent years. Up to date, few intensive studies have been performed to investigate the recent clinical implications, virulence-related factors, and the reported non-specific host infection of different mycoplasmas as host-specific and pantropic pathogens. We summarized the most recent clinical implication in human and different animals species, virulence-related factors, common proteins incorporated in Mycoplasma infection, and the pivotal influence of the gene transfer process on Mycoplasma pathogens’ evolution. Furthermore, the immune response of Mycoplasma pathogens as unique antigens with limited metabolic capacities has a greater influence than others. Future perspectives with advances in the nanotechnology field had been shown as a new ingenious field to stop the growing threat of mycoplasmosis. Therefore, the ultimate but challenging goal is referring to study and explore using of nanoparticles in the Mycoplasma field as brilliant nanocarriers. Animal models also can assist us with superior outcomes for diagnosing and treating mycoplasmosis. Subsequently, it is vital to adopt modern technological methods to verify the mechanism of mycoplasmas infection. This will lay a foundation to create a new paradigm of diagnostics and therapeutic formulations and interventions. Finally, many scenarios for achieving mycoplasmas eradication in many parts of the world have succeeded that can be followed by others.
Author Contributions
AD and AG conceived the project, reviewed the articles, and extracted the data. AD and SA wrote the manuscript. GZ, TZ, MQ, KD, ZH, MA, and IS reviewed the articles. All authors approved the submission of the manuscript.
Funding
This work was supported by National Natural Science Foundation Projects (#31772745), the Key Research and Development Program of the Ningxia Hui Autonomous Region (# 2021BEF02028), and the China Agriculture Research System (Beef/yaks) of MOF and MARA (#CARS-37).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary

References
Aagaard, K., Ma, J., Antony, K. M., Ganu, R., Petrosino, J., Versalovic, J. (2014). The Placenta Harbors a Unique Microbiome. Sci. Transl. Med. 6 (237), 237ra265. doi: 10.1126/scitranslmed.3008599
Abd-Elrahman, A. H., Khafaga, A. F., Abas, O. M. (2020). The First Identification of Contagious Caprine Pleuropneumonia (CCPP) in Sheep and Goats in Egypt: Molecular and Pathological Characterization. Trop. Anim. Health Prod 52 (3), 1179–1186. doi: 10.1007/s11250-019-02116-5
Abdul-Wahab, O. M. S., Al-Shyarba, M. H., Mardassi, B. B. A., Sassi, N., Al Fayi, M. S. S., Otifi, H., et al. (2021). Molecular Detection of Urogenital Mollicutes in Patients With Invasive Malignant Prostate Tumor. Infect. Agents Cancer 16 (1), 1–11. doi: 10.1186/s13027-021-00344-9
Ahn, J. G., Cho, H.-K., Li, D., Choi, M., Lee, J., Eun, B.-W., et al. (2021). Efficacy of Tetracyclines and Fluoroquinolones for the Treatment of Macrolide-Refractory Mycoplasma Pneumoniae Pneumonia in Children: A Systematic Review and Meta-Analysis. BMC Infect. Dis. 21 (1), 1–10. doi: 10.1186/s12879-021-06508-7
Albensi, B. (2019). What is Nuclear Factor Kappa B (Nf-κb) Doing in and to the Mitochondrion? Front. Cell Dev. Biol. 7 (7). doi: 10.3389/fcell.2019.00154
Algharib, S., Dawood, A., Xie, S. (2020a). Nanoparticles for Treatment of Bovine Staphylococcus Aureus Mastitis. Drug Deliv 27 (1), 292–308. doi: 10.1080/10717544.2020.1724209
Algharib, S., Dawood, A., Zhou, K., Chen, D., Lia, C., Meng, K., et al. (2020b). Designing, Structural Determination and Biological Effects of Rifaximin Loaded Chitosan- Carboxymethyl Chitosan Nanogel. Carbohydr. Polymers 248 (11), 67–82. doi: 10.1016/j.carbpol.2020.116782
Ali, S., Ali, S., Javed, S. O., Shoukat, S., Ahmad, S., Ali, S. S., et al. (2021b). Proteome Wide Vaccine Targets Prioritization and Designing of Antigenic Vaccine Candidate to Trigger the Host Immune Response Against the Mycoplasma Genitalium Infection. Microbial Pathogenesis 152, 104771. doi: 10.1016/j.micpath.2021.104771
Ali, G. A., Goravey, W., Hamad, A., Ibrahim, E. B., Hasan, M. R., Al Maslamani, M., et al. (2021a). An Enemy in Shadows-Mycoplasma Hominis Septic Arthritis and Iliopsoas Abscess: Case Report and Review of the Literature. IDCases 26, e01260. doi: 10.1016/j.idcr.2021.e01260
Al-Zaidy, S. A., MacGregor, D., Mahant, S., Richardson, S. E., Bitnun, A. (2015). Neurological Complications of PCR-Proven M. Pneumoniae Infections in Children: Prodromal Illness Duration may Reflect Pathogenetic Mechanism. Clin. Infect. Dis. 61 (7), 1092–1098. doi: 10.1093/cid/civ473
Andrade, F., Rafael, D., Videira, M., Ferreira, D., Sosnik, A., Sarmento, B. (2013). Nanotechnology and Pulmonary Delivery to Overcome Resistance in Infectious Diseases. Advanced Drug Deliv. Rev. 65 (13-14), 1816–1827. doi: 10.1016/j.addr.2013.07.020
Andrade, F., Videira, M., Ferreira, D., Sarmento, B. (2011). Micelle-Based Systems for Pulmonary Drug Delivery and Targeting. Drug Deliv. Lett. 1 (2), 171–185. doi: 10.2174/2210303111101020171
André, M. R., Adania, C. H., Allegretti, S. M., Machado, R. Z. (2011). Hemoplasmas in Wild Canids and Felids in Brazil. J. Zoo Wildlife Med. 42 (2), 342–347. doi: 10.1638/2010-0198.1
André, M. R., Duarte, J. M. B., Gonçalves, L. R., Sacchi, A. B. V., Jusi, M. M. G., Machado, R. Z. (2020). New Records and Genetic Diversity of Mycoplasma Ovis in Free-Ranging Deer in Brazil. Epidemiol. Infect. 148, e6, 1–7. doi: 10.1017/S0950268819002218
Anonymous (2018). “Contagious bovine pleuropneumonia (infection with Mycoplasma mycoides subsp mycoides SC). In: Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. Paris, France: OIE–World Organization for Animal Health; 2018:1097–1112.
Anonymous (2000). “Reviving Progressive Control of CBPP in Africa,” in Report of the Second Meeting of the FAO/OIE/OAU/IAEA Consultative Group on Contagious Bovine Pleuropneumonia (CBPP) (Rome, Italy: Food and Agriculture Organization of the United Nations), 1–9.
Arana, L., Gallego, L., Alkorta, I. (2021). Incorporation of Antibiotics Into Solid Lipid Nanoparticles: A Promising Approach to Reduce Antibiotic Resistance Emergence. Nanomaterials 11, 1251. doi: 10.3390/nano11051251
Arfi, Y., Minder, L., Di Primo, C., Le Roy, A., Ebel, C., Coquet, L., et al. (2016). MIB–MIP is a Mycoplasma System That Captures and Cleaves Immunoglobulin G. Proc. Natl. Acad. Sci. 113 (19), 5406–5411. doi: 10.1073/pnas.1600546113
Askar, H., Chen, S., Hao, H., Yan, X., Ma, L., Liu, Y., et al. (2021). Immune Evasion of Mycoplasma Bovis. Pathog. (Basel Switzerland) 10 (3), 297. doi: 10.3390/pathogens10030297
Atallah, S., Berçot, B., Laurence, V., Hoffmann, C. (2020). “Association of Mycoplasma Hominis and Head and Neck Cancer With Unknown Primary”, in European Annals of Otorhinolaryngology, Head and Neck Diseases. (Elsevier) vol. 137, 69–71. doi: 10.1016/j.anorl.2019.05.020
Awadh, A. A., Kelly, A. F., Forster-Wilkins, G., Wertheim, D., Giddens, R., Gould, S. W., et al. (2021). Visualisation and Biovolume Quantification in the Characterisation of Biofilm Formation in Mycoplasma Fermentans. Sci. Rep. 11(1), 1–9. doi: 10.1038/s41598-021-90455-5
Aye, R., Weldearegay, Y., Lutta, H., Chuma, F., Pich, A., Jores, J., et al. (2018). Identification of Targets of Monoclonal Antibodies That Inhibit Adhesion and Growth in Mycoplasma Mycoides Subspecies Mycoides. Vet. Immunol. Immunopathol. 204, 11–18. doi: 10.1016/j.vetimm.2018.09.002
Azevedo, E., Câmara, D., Silva, S., Guerra, M. (2015). "Agalaxia Contagiosa: Um “Novo”. problema Para Caprinos e Ovinos no Brasil." Ciec. Veterinária nos Trópicos 18 (2), 34–38.
Azumagawa, K., Kambara, Y., Murata, T., Tamai, H. (2008). Four Cases of Arthritis Associated With Mycoplasma Pneumoniae Infection. Pediatr. Int. 50 (4), 511–513. doi: 10.1111/j.1442-200X.2008.02622.x
Baby, V., Labroussaa, F., Brodeur, J., Matteau, D., Gourgues, G., Lartigue, C., et al. (2018). Cloning and Transplantation of the Mesoplasma Florum Genome. ACS Synth. Biol. 7, 209–217. doi: 10.1021/acssynbio.8b00449
Bachmann, L., Kirkcaldy, R., Geisler, W., Wiesenfeld, HC, Manhart, LE, Taylor, SN, et al. (2020). Prevalence of Mycoplasma Genitalium Infection, Antimicrobial Resistance Mutations and Symptom Resolution Following Treatment of Urethritis. Clin. Infect. Dis. 71(10):e624–e632. doi: 10.1093/cid/ciaa293
Bai, D.-P., Lin, X.-Y., Huang, Y.-F., Zhang, X.-F. (2018). Theranostics Aspects of Various Nanoparticles in Veterinary Medicine. Int. J. Mol. Sci. 19 (11), 3299. doi: 10.3390/ijms19113299
Bai, F., Wu, J., Liu, B., Wang, X., Shi, X., Lv, T., et al. (2020). Mycoplasma Ovipneumoniae-Derived Lipid-Associated Membrane Proteins Induce Cytokine Secretion in Mouse Peritoneal Macrophages Through TLR2 Signalling. Res. Veterinary Sci. 132, 474–480. doi: 10.1016/j.rvsc.2020.07.022
Bao, S., Yu, S., Guo, X., Zhang, F., Sun, Y., Tan, L., et al. (2015). Construction of a Cell-Surface Display System Based on the N-Terminal Domain of Ice Nucleation Protein and its Application in Identification of Mycoplasma Adhesion Proteins. J. Appl. Microbiol. 119 (1), 236–244. doi: 10.1111/jam.12824
Baracaldo, R., Foltzer, M., Patel, R., Bourbeau, P. (2012). Empyema Caused by Mycoplasma Salivarium. J. Clin. Microbiol. 50 (5), 1805–1806. doi: 10.1128/JCM.06839-11
Barker, E., Tasker, S. (2013). Haemoplasmas: Lessons Learnt From Cats. N. Z. Veterinary J. 61 (4), 184–192. doi: 10.1080/00480169.2013.771760
Barykova, Y. A., Logunov, D. Y., Shmarov, M. M., Vinarov, A. Z., Fiev, D. N., Vinarova, N. A., et al. (2011). Association of Mycoplasma Hominis Infection With Prostate Cancer. Oncotarget 2 (4), 289. doi: 10.18632/oncotarget.256
Baseman, J., Lange, M., Criscimagna, N., Giron, J., Thomas, C. (1995). Interplay Between Mycoplasmas and Host Target Cells. Microbial Pathogenesis 19 (2), 105–116. doi: 10.1006/mpat.1995.0050
Baziki, J., d., D., Charles, B. S., Nwankpa, N., Maina, N., Chitsungo, E., et al. (2020). Development and Evaluation of an Immuno-Capture Enzyme-Linked Immunosorbent Assay to Quantify the Mycoplasma Capricolum Subsp. Capripneumoniae (Mccp) Protein in Contagious Caprine Pleuropneumonia (CCPP) Vaccine. Veterinary Med. Int. 2020, 4236807. doi: 10.1155/2020/4236807
Béjaoui Khiari, A., Landoulsi, A., Aissa, H., Mlik, B., Amouna, F., Ejlassi, A., et al. (2011). Isolation of Mycoplasma Meleagridis From Chickens. Avian Dis. 55 (1), 8–12. doi: 10.1637/9365-041310-reg.1
Benedetti, F., Cocchi, F., Latinovic, O. S., Curreli, S., Krishnan, S., Munawwar, A., et al. (2020). Role of Mycoplasma Chaperone Dnak in Cellular Transformation. Int. J. Mol. Sci. 21 (4), 1311. doi: 10.3390/ijms21041311
Berry, I. J., Jarocki, V. M., Tacchi, J. L., Raymond, B. B. A., Widjaja, M., Padula, M. P., et al. (2017). N-Terminomics Identifies Widespread Endoproteolysis and Novel Methionine Excision in a Genome-Reduced Bacterial Pathogen. Sci. Rep. 7 (1), 11063. doi: 10.1038/s41598-017-11296-9
Berus, S., Adamczyk-Popławska, M., Młynarczyk-Bonikowska, B., Witkowska, E., Szymborski, T., Waluk, J., et al. (2021). SERS-Based Sensor for the Detection of Sexually Transmitted Pathogens in the Male Swab Specimens: A New Approach for Clinical Diagnosis. Biosensors Bioelectronics 189 (11), 33–58. doi: 10.1016/j.bios.2021.113358
Besser, T. E., Cassirer, E. F., Highland, M. A., Wolff, P., Justice-Allen, A., Mansfield, K., et al. (2013). Bighorn Sheep Pneumonia: Sorting Out the Cause of a Polymicrobial Disease. Prev. Veterinary Med. 108 (2-3), 85–93. doi: 10.1016/j.prevetmed.2012.11.018
Besser, T. E., Levy, J., Ackerman, M., Nelson, D., Manlove, K., Potter, K. A., et al. (2019). A Pilot Study of the Effects of Mycoplasma Ovipneumoniae Exposure on Domestic Lamb Growth and Performance. PloS One 14 (2), e0207420. doi: 10.1371/journal.pone.0207420
Betlach, A., Fano, E., VanderWaal, K., Pieters, M. (2021). Effect of Multiple Vaccinations on Transmission and Degree of Mycoplasma Hyopneumoniae Infection in Gilts. Vaccine 39, 767–774. doi: 10.1016/j.2020.10.096
Bischof, D. F., Janis, C., Vilei, E. M., Bertoni, G., Frey, J. (2008). Cytotoxicity of Mycoplasma Mycoides SubspMycoides Small Colony Type to Bovine Epithelial Cells. Infect. Immun. 76 (1), 263–269. doi: 10.1128/IAI.00938-07
Bischof, D. F., Vilei, E. M., Frey, J. (2009). "Functional and Antigenic Properties of Glpo From Mycoplasma Mycoides SubspMycoides SC: Characterization of a Flavin Adenine Dinucleotide-Binding Site Deletion Mutant. Veterinary Res. 40 (4), 1–12. doi: 10.1051/vetres/2009018
Blesa, A., Baquedano, I., Quintáns, N. G., Mata, C. P., Castón, J. R., Berenguer, J. (2017). The Transjugation Machinery of Thermus Thermophilus: Identification of Tdta, an Atpase Involved in DNA Donation. PloS Genet. 13 (3), e1006669. doi: 10.1371/journal.pgen.1006669
Blötz, C., Singh, N., Dumke, R., Stülke, J. (2020). Characterization of an Immunoglobulin Binding Protein (Ibpm) From Mycoplasma Pneumoniae. Front. Microbiol. 11, 685. doi: 10.3389/fmicb.2020.00685
Boes, K. M., Goncarovs, K. O., Thompson, C. A., Halik, L. A., Santos, A. P., Guimaraes, A. M., et al. (2012). Identification of a Mycoplasma Ovis-Like Organism in a Herd of Farmed White-Tailed Deer (Odocoileus Virginianus) in Rural I Ndiana. Veterinary Clin. Pathol. 41 (1), 77–83. doi: 10.1111/j.1939-165X.2011.00376.x
Bohach, D. M., Stegniy, B. T., Bohach, M. V., Pavlov, S. L., Bolotin, V. I., et al. (2021). Age and Seasonal Pattern of Contagious Agalactia in Small Ruminants in Ukraine. J. Veterinary Res. 65 (1), 67–72. doi: 10.2478/jvetres-2021-0014
Bolajoko, M. B., Van Gool, F., Peters, A. R., Martinez, J. S., Vance, C. J., Dungu, B. (2020). Field Survey of Major Infectious and Reproductive Diseases Responsible for Mortality and Productivity Losses of Ruminants Amongst Nigerian Fulani Pastoralists. Gates Open Res. 4 (162), 162. doi: 10.12688/gatesopenres.13164.1
Booth, J. L., Umstead, T. M., Hu, S., Dybvig, K. F., Cooper, T. K., Wilson, R. P., et al. (2014). Housing Conditions Modulate the Severity of Mycoplasma Pulmonis Infection in Mice Deficient in Class a Scavenger Receptor. Comp. Med. 64 (6), 424–439.
Borchsenius, S., Daks, A., Fedorova, O., Chernova, O., Barlev, N. (2018). Effects of Mycoplasma Infection on the Host Organism Response via P53/NF-κb Signaling. J. Cell Physiol. 234 (1), 171–180. doi: 10.1002/jcp.26781
Borchsenius, S., Vishnyakov, I., Chernova, O., Chernov, V., Barlev, N. (2020). Effects of Mycoplasmas on the Host Cell Signaling Pathways. Pathogens 22 (9(4), 308. doi: 10.3390/pathogens9040308
Bordin, L., Gava, D., Sonalio, K., Mechler-Dreibi, M., Zanella, J., Morés, N., et al. (2021). Investigation of Hemotropic Mycoplasmas in Fetuses and Sows With Reproductive Failure. Vet. anim. Sci. 12, 100175. doi: 10.1016/j.vas.2021.100175
Boulanger, J., Faulds, D., Eddy, E., Lingwood, C. (1995). Members of the 70 Kda Heat Shock Protein Family Specifically Recognize Sulfoglycolipids: Role in Gamete Recognition and Mycoplasma-Related Infertility. J. Cell Physiol. 165 (1), 7–17. doi: 10.1002/jcp.1041650103
Boyce, C., Jaye, C., Noller, G., Bryan, M., Doolan-Noble, F. (2021). Mycoplasma Bovis in New Zealand: A Content Analysis of Media Reporting. Kōtuitui: N. Z. J. Soc. Sci. Online, 16:2, 335–355. doi: 10.1080/1177083X.2021.1879180
Brissonnier, M., Normand, V., Lebret, A., Moalic, P., Guyomard, A., Bachy, V., et al. (2020). Frequency of Infection With Mycoplasma Suis in Gestating Sows Using Qpcr on Ten Commercial French Herds, and Impact of the Infection on Clinical, Haematological and Biochemical Parameters. Porcine Health Manage. 6 (13), 1–6. doi: 10.1186/s40813-020-00152-4
Browning, G. F., Kanci, A., Markham, P. (2011). Developing Attenuated Vaccines to Control Mycoplasmoses. Microbiol. Aust. 32 (3), 121–122. doi: 10.1071/MA11121
Brown, D., Schumacher, I., Nogueira, M., Richey, L., Zacher, L., Schoeb, T., et al. (2001). Detection of Antibodies to a Pathogenic Mycoplasma in American Alligators (Alligator Mississippiensis), Broad-Nosed Caimans (Caiman Latirostris), and Siamese Crocodiles (Crocodylus Siamensis). J. Clin. Microbiol. 39 (1), 285–292. doi: 10.1128/JCM.39.1.285-292.2001
Bryson, D. G., Ball, H. J., Brice, N., Forster, F., Pollock, D. (1999). “"Pathology of Induced Mycoplasma Bovis Calf Pneumonia in Experimentally Vaccinated Animals.",” in Mycoplasmas of Ruminants: Pathogenicity, Diagnostics, Epidemiology and Molecular Genetics (Brussels: European Commission), 128–132.
Bumgardner, E., Kittichotirat R. Bumgarner, W., Lawrence, P. (2015). Comparative Genomic Analysis of Seven Mycoplasma Hyosynoviaestrains. Microbiol. Open 4 (2), 343–359. doi: 10.1002/mbo3.242
Bürgi, N., Josi, C., Bürki, S., Schweizer, M., Pilo, P. (2018). Mycoplasma Bovis Co-Infection With Bovine Viral Diarrhea Virus in Bovine Macrophages. Veterinary Res. 49 (1), 2. doi: 10.1186/s13567-017-0499-1
Bürki, S., Frey, J., Pilo, P. (2015). Virulence, Persistence and Dissemination of Mycoplasma Bovis. Veterinary Microbiol. 179 (1-2), 15–22. doi: 10.1016/j.vetmic.2015.02.024
Cardwell, J., Smith, K., Wood, J., Newton, J. (2013). A Longitudinal Study of Respiratory Infections in British National Hunt Racehorses. Veterinary Rec. 172 (24), 637. doi: 10.1136/vr.101520
Cassell, G. H., Waits, K. B., Watson, H. L., Crouse, D. T., Harasawa, R. (1993). Ureaplasma Urealyticum: Role in Prematurity and Disease in Newborns. Clin. Microbiol. Rev. 6, 69–87. doi: 10.1128/CMR.6.1.69
Caswell, J., Bateman, K., Cai, H., Castillo-Alcala, F. (2010). "Mycoplasma Bovis in Respiratory Disease of Feedlot Cattle." Vet. Clin. North Am. Food Anim. Pract. 26, 365–379. doi: 10.1016/j.cvfa.2010.03.003
Chalker, V. J. (2005). Canine Mycoplasmas. Res. Veterinary Sci. 79 (1), 1–8. doi: 10.1016/j.rvsc.2004.10.002
Chalker, V., Stocki, T., Litt, D., Bermingham, A., Watson, J., Fleming, D., et al. (2012). Increased Detection of Mycoplasma Pneumoniae Infection in Children in England and Wales, October 2011 to January 2012. Eurosurveillance 17 (6), 20081. doi: 10.2807/ese.17.06.20081-en
Chao, J., Han, X., Liu, K., Li, Q., Peng, Q., Lu, S., et al. (2019). Calves Infected With Virulent and Attenuated Mycoplasma Bovis Strains Have Upregulated Th17 Inflammatory and Th1 Protective Responses, Respectively. Genes 10 (9), 656. doi: 10.3390/genes10090656
Chaudhry, R., Varshney, A.-K., Malhotra, P. (2007). Adhesion Proteins of Mycoplasma Pneumoniae. Front. Bioscience 12, 690–699. doi: 10.2741/2093
Chen, S., Hao, H., Zhao, P., Thiaucourt, F., He, Y., Gao, P., et al. (2017). Genome-Wide Analysis of the First Sequenced Mycoplasma Capricolum Subsp. Capripneumoniae Strain M1601. G3: Genes Genomes Genet. 7 (9), 2899–2906. doi: 10.1534/g3.117.300085
Chen, X., Huang, J., Zhu, H., Guo, Y., Khan, F. A., Menghwar, H., et al. (2018). P27 (MBOV_RS03440) is a Novel Fibronectin Binding Adhesin of Mycoplasma Bovis. Int. J. Med. Microbiol. 308 (7), 848–857. doi: 10.1016/j.ijmm.2018.07.006
Chen, R., Yu, Y., Feng, Z., Gan, R., Xie, X., Zhang, Z., et al. (2019). Featured Species-Specific Loops are Found in the Crystal Structure of Mhp Eno, a Cell Surface Adhesin From Mycoplasma Hyopneumoniae. Front. Cell. Infect. Microbiol. 9, 209. doi: 10.3389/fcimb.2019.00209
Chen, R., Zhao, L., Gan, R., Feng, Z., Cui, C., Xie, X., et al. (2022). Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases. Front. Mol. Biosci. 8, 811106. doi: 10.3389/fmolb.2021.811106
Chernov, A. V., Reyes, L., Xu, Z., Gonzalez, B., Golovko, G., Peterson, S., et al. (2015). Mycoplasma CG-and GATC-Specific DNA Methyltransferases Selectively and Efficiently Methylate the Host Genome and Alter the Epigenetic Landscape in Human Cells. Epigenetics 10 (4), 303–318. doi: 10.1080/15592294.2015.1020000
Choi, H., Lee, H., Kim, W., Kim, M., Chang, H., Lee, H., et al. (2014). Detection of Mycoplasma Infection in Circulating Tumor Cells in Patients With Hepatocellular Carcinoma. Biochem. Biophys. Res. Commun. 446, 620–625. doi: 10.1016/j.bbrc.2014.03.024
Chong, D. L. W., Rebeyrol, C., José, R. J., Williams, A. E., Brown, J. S., Scotton, C. J., et al. (2021). ICAM-1 and ICAM-2 are Differentially Expressed and Up-Regulated on Inflamed Pulmonary Epithelium, But Neither ICAM-2 Nor LFA-1: ICAM-1 are Required for Neutrophil Migration Into the Airways In Vivo. Front. Immunol. 12. doi: 10.3389/fimmu.2021.691957
Christodoulides, A., Gupta, N., Yacoubian, V., Maithel, N., Parker, J., Kelesidis, T. (2018). The Role of Lipoproteins in Mycoplasma-Mediated Immunomodulation. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01682
Cillara, G., Manca, M. G., Longheu, C., Tola, S. (2015). Discrimination Between Mycoplasma Mycoides Subsp. Capri and Mycoplasma Capricolum Subsp. Capricolum Using PCR-RFLP and PCR. Veterinary J. 205 (3), 421–423. doi: 10.1016/j.tvjl.2015.05.013
Citti, C., Baranowski, E., Dordet-Frisoni, E., Faucher, M., Nouvel, L.-X. (2020). Genomic Islands in Mycoplasmas. Genes 11 (8), 836. doi: 10.3390/genes11080836
Citti, C., Blanchard, A. (2013). Mycoplasmas and Their Host: Emerging and Re-Emerging Minimal Pathogens. Trends Microbiol. 21 (4), 196–203. doi: 10.1016/j.tim.2013.01.003
Citti, C., Dordet-Frisoni, E., Nouvel, L. X., Kuo, C. H., Baranowski, E. (2018). Horizontal Gene Transfers in Mycoplasmas. Curr. Issues Mol. Biol. 29, 3–22. doi: 10.21775/cimb.029.003
Clampitt, J. (2021). Generation of Site-Specific Mutations in Mycoplasma. A Thesis Submitted to the Graduate Faculty in Partial Fulfillment of the Requirements for the Degree of MASTER OF SCIENCE. Iowa State University. Available at: https://doi.org/10.31274/etd-20210609.
Combaz-Söhnchen, N., Kuhn, A. (2017). A Systematic Review of Mycoplasma and Ureaplasma in Urogynaecology. Geburtshilfe und Frauenheilkunde 77 (12), 1299–1303. https://doi.org/10.1055/s-0043-119687
Constantopoulos, G., McGarrity, G. (1987). Activities of Oxidative Enzymes in Mycoplasmas. J. Bacteriology 169 (5), 2012–2016. doi: 10.1128/jb.169.5.2012-2016.1987
Controlling M. bovis in NZ, f. y. o. E. R. (2021). (2021). Available at: https://www.sciencemediacentre.co.nz/2021/07/20/controlling-m-bovis-in-nz-four-years-on-expert-reaction/.
Cordova, C. M., Hoeltgebaum, D. L., Machado, L. D., SANTOS, L. D. (2016). Molecular Biology of Mycoplasmas: From the Minimum Cell Concept to the Artificial Cell. Anais da Academia Bras. Ciências 88, 599–607. doi: 10.1590/0001-3765201620150164
Dakhama, A., Kraft, M., Martin, R. J., Gelfand, E. W. (2003). Induction of Regulated Upon Activation, Normal T Cells Expressed and Secreted (RANTES) and Transforming Growth Factor-β1 in Airway Epithelial Cells by Mycoplasma Pneumoniae. Am. J. Respir. Cell Mol. Biol. 29 (3), 344–351. doi: 10.1165/rcmb.2002-0291OC
D’Alonzo, R., Mencaroni, E., Di Genova, L., Laino, D., Principi, N., Esposito, S. (2018). Pathogenesis and Treatment of Neurologic Diseases Associated With Mycoplasma Pneumoniae Infection. Front. Microbiol. 9, 2751. doi: 10.3389/fmicb.2018.02751
Damasceno, E., Pinheiro, R., Andrioli, A., Alves, F., Lima, A., Peixoto, R., et al. (2020). Seroprevalence and Associated Risk Factors of Mycoplasma Agalactiae and Investigation of Coinfection With the Caprine Lentivirus in Rio Grande do Norte, Brazil. Trop. Anim. Health Prod. 52, 2111–2117. doi: 10.1007/s11250-020-02234-5
Das, K., Garnica, O., Flores, J., Dhandayuthapani, S. (2020). Methionine Sulfoxide Reductase a (Msra) Modulates Cells and Protects Against Mycoplasma Genitalium Induced Cytotoxicity. Free Radical Biol. Med. 152, 323–335. doi: 10.1016/j.freeradbiomed.2020.03.019
Davies, D., Jones, B., Thurley, D. (1981). Infection of Specific-Pathogen-Free Lambs With Parainfluenza Virus Type 3, Pasteurella Haemolytica and Mycoplasma Ovipneumoniae. Veterinary Microbiol. 6 (4), 295–308. doi: 10.1016/0378-1135(81)90023-7
Delaney, N., Balenger, S., Bonneaud, C., Marx, C. J., Hill, G. E., Ferguson-Noel, N., et al. (2012). Ultrafast Evolution and Loss of Crisprs Following a Host Shift in a Novel Wildlife Pathogen, Mycoplasma Gallisepticum. PloS Genet. 8(2):e1002511. doi: 10.1371/journal.pgen.1002511
Demento, S. L., Siefert, A. L., Bandyopadhyay, A., Sharp, F. A., Fahmy, T. M. (2011). Pathogen-Associated Molecular Patterns on Biomaterials: A Paradigm for Engineering New Vaccines. Trends Biotechnol. 29(6):294–306. doi: 10.1016/j.tibtech.2011.02.004
Demkin, V., Kazakov, A. (2021). Prevalence of Hemotropic Mycoplasmas and Coinfection With Feline Leukemia Virus and Feline Immunodeficiency Virus in Cats in the Moscow Region, Russia. Prev. Vet. Med. 190, 105339. doi: 10.1016/j.prevetmed.2021.105339
Díaz-Sánchez, A. A., Corona-González, B., Meli, M. L., Álvarez, D. O., Cañizares, E. V., Rodríguez, O. F., et al. (2019). First Molecular Evidence of Bovine Hemoplasma Species (Mycoplasma Spp.) in Water Buffalo and Dairy Cattle Herds in Cuba. Parasites Vectors 12 (1), 1–9. doi:–10.1186/s13071-019-3325-y
Dieckmann, S. M., Hoelzle, K., Dieckmann, M. P., Straube, I., Hofmann-Lehmann, R., Hoelzle, L. E. (2012). Occurrence of Hemotrophic Mycoplasmas in Horses With Correlation to Hematological Findings. Veterinary Microbiol. 160 (1-2), 43–52. doi: 10.1016/j.vetmic.2012.05.016
Dieckmann, S. M., Winkler, M., Groebel, K., Dieckmann, M. P., Hofmann-Lehmann, R., Hoelzle, K., et al. (2010). Haemotrophic Mycoplasma Infection in Horses. Veterinary Microbiol. 145 (3-4), 351–353. doi: 10.1016/j.vetmic.2010.04.009
Di Teodoro, G., Marruchella, G., Di Provvido, A., D’Angelo, A. R., Orsini, G., Di Giuseppe, P., et al. (2020). Contagious Bovine Pleuropneumonia: A Comprehensive Overview. Veterinary Pathol. 57(4), 476–489. doi: 10.1177/0300985820921818
Djurišić, A. B., Leung, Y. H., Ng, A. M., Xu, X. Y., Lee, P. K., Degger, N., et al. (2015). Toxicity of Metal Oxide Nanoparticles: Mechanisms, Characterization, and Avoiding Experimental Artefacts. Small 11 (1), 26–44. doi:–10.1002/smll.201303947
Dmochowski, L. (1967). “"Viruses of Laboratory Rodents.",” in National Cancer Institute Monograph 20, vol. 1966 . Ed. Holdenried, R. (Washington, DC: US Government Printing Office), 180.
Dordet-Frisoni, E., Faucher, M., Sagné, E., Baranowski, E., Tardy, F., Nouvel, L., et al. (2019). Mycoplasma Chromosomal Transfer: A Distributive, Conjugative Process Creating an Infinite Variety of Mosaic Genomes. Front. Microbiol. 10, p. 2441. doi: 10.3389/fmicb.2019.02441
Dordet-Frisoni, E., Sagné, E., Baranowski, E., Breton, M., Nouvel, L. X., Blanchard, A., et al. (2014). Chromosomal Transfers in Mycoplasmas: When Minimal Genomes Go Mobile. MBio 5 (6), e01958–14. doi: 10.1128/mBio.01958-14
Dudek, K., Bednarek, D., Ayling, R. D., Kycko, A., Reichert, M., et al. (2019). Preliminary Study on the Effects of Enrofloxacin, Flunixin Meglumine and Pegbovigrastim on Mycoplasma Bovis Pneumonia. BMC Vet. Res. 15(1), 1–13. doi: 10.1186/s12917-019-2122-3
Dudek, K., Szacawa, E., Nicholas, R. (2021). Recent Developments in Vaccines for Bovine Mycoplasmoses Caused by Mycoplasma Bovis and Mycoplasma Mycoides Subsp. Mycoides. Vaccines (Basel) 24 (9(6), 549. doi: 10.3390/vaccines9060549
Dumke, R., Catrein, I., Herrmann, R., Jacobs, E. (2004). Preference, Adaptation and Survival of Mycoplasma Pneumoniae Subtypes in an Animal Model. IJMM 294 (2-3), 149–155. doi: 10.1016/j.ijmm.2004.06.020
Fair, R. J., Tor, Y. (2014). Antibiotics and Bacterial Resistance in the 21st Century. Perspect. Medicinal Chem. 6, PMC.S14459. doi: 10.4137/PMC.S14459
Falquet, L., Liljander, A., Schieck, E., Gluecks, I., Frey, J., Jores, J. (2014). Complete Genome Sequences of Virulent Mycoplasma Capricolum Subsp. Capripneumoniae Strains F38 and ILRI181. Genome Announc 2 (5), e01041–14.doi: 10.1128/genomeA.01041-14
Fang, X., Lloyd, C. J., Palsson, B. O. (2020). Reconstructing Organisms In Silico: Genome-Scale Models and Their Emerging Applications. Nat. Rev. Microbiol. 18 (12), 731–743. doi: 10.1038/s41579-020-00440-4
Fathi, A., Almuhammady, A., Sobhy, M., ElMiniawy, M., Khattab, S. (2019). Histopathological Changes After Treatment of Mycoplasma Bovis Infected Does With Zinc Oxide Nanoparticles as a New Tool. Egyptian J. Veterinary Sci. 50, 21–28. doi: 10.21608/ejvs.2019.19940.1133
Faucher, M., Nouvel, L.-X., Dordet-Frisoni, E., Sagné, E., Baranowski, E., Hygonenq, M.-C., et al. (2019). Mycoplasmas Under Experimental Antimicrobial Selection: The Unpredicted Contribution of Horizontal Chromosomal Transfer. PloS Genet. 15 (1), e1007910. doi: 10.1371/journal.pgen.1007910
Faulkner, C., Simecka, J. W., Davidson, M. K., Davis, J. K., Schoeb, T. R., Lindsey, J. R., et al. (1995). Gene Expression and Production of Tumour Necrosis Factor Alpha, Interleukin 1, Interleukin 6, and Gamma Interferon in C3H/Hen and C57BL/6N Mice in Acute Mycoplasma Pulmonis Disease. Infect. Immun. 63 (10), 4084–4090. doi: 10.1128/iai.63.10.4084-4090.1995
Felder, K. M., Hoelzle, K., Heinritzi, K., Ritzmann, M., Hoelzle, L. E. (2010). Antibodies to Actin in Autoimmune Haemolytic Anaemia. BMC Veterinary Res. 6 (1), 1–9. doi: 10.1186/1746-6148-6-18
Feng, M., Schaff, A.-C., Balish, M.-F. (2020). Mycoplasma Pneumoniae Biofilms Grown In Vitro: Traits Associated With Persistence and Cytotoxicity. Microbiology 166 (7), 629–640. doi: 10.1099/mic.0.000928
Feng, S.-H., Tsai, S., Rodriguez, J., Lo, S.-C. (1999). Mycoplasmal Infections Prevent Apoptosis and Induce Malignant Transformation of Interleukin-3-Dependent 32D Hematopoietic Cells. Mol. Cell. Biol. 19 (12), 7995–8002. doi: 10.1128/MCB.19.12.7995
Feng, C., Xu, M., Kang, J., Wen, F., Chen, Y., Zhang, J., et al. (2021). Atypical Pathogen Distribution in Chinese Hospitalized AECOPD Patients: A Multicenter Cross-Sectional Study. Int. J. Chron Obstruct Pulmon Dis. 16, 1699–1708. doi: 10.2147/copd.s300779
Ferreira, G., Blasina, F., Rey, M. R., Anesetti, G., Sapiro, R., Chavarría, L., et al. (2022). Pathophysiological and Molecular Considerations of Viral and Bacterial Infections During Maternal-Fetal and–Neonatal Interactions of SARS-Cov-2, Zika, and Mycoplasma Infectious Diseases. Biochim. Biophys. Acta (BBA)-Molecular Basis Dis. 1868 (1), 166285. doi: 10.1016/j.bbadis.2021.166285
Ferreira, G., Santander, A., Savio, F., Guirado, M., Sobrevia, L., Nicolson, G. L. (2021). SARS-Cov-2, Zika Viruses and Mycoplasma: Structure, Pathogenesis and Some Treatment Options in These Emerging Viral and Bacterial Infectious Diseases. Biochim. Biophys. Acta Mol. Basis Dis. 1867 (12), 166264. doi: 10.1016/j.bbadis.2021.166264
Findlay, G., Maccallum, F., Mackenzie, R. (1938). Rolling Disease: New Syndrome in Mice Associated With a Pleuropneumonia-Like Organism. Lancet 232 (6018), 1511–1513. doi: 10.1016/S0140-6736(00)83971-9
Fischer, J., Stallknecht P, D. E., Luttrell, P., Dhondt, A. A., Converse, K. A., et al. (1997). Mycoplasmal Conjunctivitis in Wild Songbirds: The Spread of a New Contagious Disease in a Mobile Host Population. Emerg. Infect. Dis. 3(1), 69. doi: 10.3201/eid0301.970110
Foley, J. E., Pedersen, N. C. (2001). candidatus Mycoplasma Haemominutum’, a Low-Virulence Epierythrocytic Parasite of Cats. Int. J. Systematic Evolutionary Microbiol. 51 (3), 815–817. doi: 10.1099/00207713-51-3-815
Forsberg, K., Reyes, A., Wang, B., Selleck, E., Sommer, M., OA, D. (2012). The Shared Antibiotic Resistome of Soil Bacteria and Human Pathogens. Science 337(6098), 1107–1111. doi: 10.1126/science.1220761
Foster, S., Martin, P., Braddock, J., Malik, R. (2004). A Retrospective Analysis of Feline Bronchoalveolar Lavage Cytology and Microbiology, (1995–2000). J. Feline Med. Surg. 6 (3), 189–198. doi: 10.1016/j.jfms.2003.12.001
Franzoni, G., Anfossi, A., Ciucis, D., Grazia, C., Mecocci, S., Carta, T., et al. (2021). Targeting Toll-Like Receptor 2: Polarization of Porcine Macrophages by a Mycoplasma-Derived Pam2cys Lipopeptide. Vaccines 9 (7), 692. doi: 10.3390/vaccines9070692
Fu, Y., Shi, T., Xu, L., Wei, W., Lu, F., Zhang, X., et al. (2017). Identification of a Novel Hemoplasma Species From Pigs in Zhejiang Province, China. J. Veterinary Med. Sci. 79 (5), 864–870. doi: 10.1292/jvms.16-0545
Ganter, S., Miotello, G., Manso-Silván, L., Armengaud, J., Tardy, F., Gaurivaud, P., et al. (2019). Proteases as Secreted Exoproteins in Mycoplasmas From Ruminant Lungs and Their Impact on Surface-Exposed Proteins. Appl. Environ. Microbiol. 85 (23), e01439–19. doi: 10.1128/AEM.01439-19
García-Galán, A., Baranowski, E., Hygonenq, M., Walch, M., Croville, G., Citti, C., et al. (2021). doi: 10.1101/2021.07.12.452010 Nouvel bioRxiv.
Gaspari, E. (2021). Model-Driven Design of Mycoplasma as a Vaccine Chassis (Doctoral Dissertation, Wageningen University)." Ph. D. thesis.
Gaurivaud, P., Ganter, S., Villard, A., Manso-Silvan, L., Chevret, D., Boulé, C., et al. (2018). Mycoplasmas are No Exception to Extracellular Vesicles Release: Revisiting Old Concepts. " PloS One 13 (11), e0208160. doi: 10.1371/journal.pone.0208160
Gaydos, C., Maldeis, N., Hardick, A., Hardick, J. (2009). Quinn Mycoplasma Genitalium as a Contributor to the Multiple Etiologies of Cervicitis in Women Attending Sexually Transmitted Disease Clinics. Sex Transm Dis. 36, 598–606. doi: 10.1097/OLQ.0b013e3181b01948
Giacometti, M., Janovsky, M., Belloy, L., Frey, J. (2002). Infectious Keratoconjunctivitis of Ibex, Chamois and Other Caprinae. Rev. Scientifique Technique (International Office Epizootics) 21 (2), 335–345. doi: 10.20506/rst.21.2.1338
Gibson, D. G., Glass, J. I., Lartigue, C., Noskov, V. N., Chuang, R. Y., Algire, M. A., et al. (2010). Creation of a Bacterial Cell Controlled by a Chemically Synthesized Genome. and Science 329 (5987), 52–56. doi: 10.1126/science.1190719
Glaser, K., Speer, C. P. (2015). Neonatal CNS Infection and Inflammation Caused by Ureaplasma Species: Rare or Relevant? Expert Rev. Anti-infective Ther. 13 (2), 233–248. doi: 10.1586/14787210.2015.999670
Glass, J. I., Assad-Garcia, N., Alperovich, N., Yooseph, S., Lewis, M. R., Maruf, M., et al. (2006). Essential Genes of a Minimal Bacterium. and Proc. Natl. Acad. Sci. U.S.A. 103 (2), 425–430. doi: 10.1073/pnas.0510013103
Gomes Neto, J. C., Bower, L., Erickson, B. Z., Wang, C., Raymond, M., Strait, E. L. (2015). Quantitative Real-Time Polymerase Chain Reaction for Detecting Mycoplasma Hyosynoviae and Mycoplasma Hyorhinis in Pen-Based Oral, Tonsillar, and Nasal Fluids. J. Vet. Sci. 16 (2), 195–201. doi: 10.4142/jvs.2015.16.2.195
Gondaira, S., Nishi, K., Iwano, H., Fujiki, J., Watanabe, R., Eguchi, A., et al. (2021). Transcriptome Analysis of Mycoplasma Bovis Stimulated Bovine Peripheral Blood Mononuclear Cells. Veterinary Immunol. Immunopathol. 232, 110–166. doi: 10.1016/j.vetimm.2020.110166
Gondaira, S., Nishi, K., Tanaka, T., Yamamoto, T., Nebu, T., Watanabe, R., et al. (2020). Immunosuppression in Cows Following Intramammary Infusion of Mycoplasma Bovis. Infect. Immun. 88, e00521–e00519. doi: 10.1128/IAI.00521-19
Gong, X., Chen, Q., Ferguson-Noel, N., Stipkovits, L., Szathmary, S., Liu, Y., et al. (2020). Evaluation of Protective Efficacy of Inactivated Mycoplasma Synoviae Vaccine With Different Adjuvants. Veterinary Immunol. Immunopathol. 220, 109995. doi: 10.1016/j.vetimm.2019.109995
Goodman, B., Gardner, H. (2018). "The Microbiome and Cancer". J. Pathol. 244, 667–676. doi: 10.1002/path.5047
Goret, J., Béven, L., Faustin, B., Contin-bordes, C., Le Roy, C., Claverol, S., et al. (2017). Interaction of Mycoplasma Hominis PG21 With Human Dendritic Cells: Interleukin- 23-Inducing Mycoplasmal Lipoproteins and Inflammasome Activation of the Cell". J. Bacteriol 199, 1–15. doi: 10.1128/JB.00213-17
Granger, D. N., Senchenkova, E. (2010). “Inflammation and the Microcirculation,” in Colloquium Series on Integrated Systems Physiology: From Molecule to Function. (Morgan & Claypool Life Sciences: Louisiana State University) Vol. 2, No. 1, pp. 1–87. doi: 10.4199/c00013ed1v01y201006is
Grazziotin, A. L., Duarte, J. M. B., Szabó, M. P. J., Santos, A. P., Guimarães, A. M. S., Mohamed, A., et al. (2011). Prevalence and Molecular Characterization of Mycoplasma Ovis in Selected Free-Ranging Brazilian Deer Populations. " ." J. Wildlife Dis. 47 (4), 1005–1011. doi: 10.7589/0090-3558-47.4.1005
Greene, C. E., Chalker, V. J. (2012). Nonhemotropic mycoplasmal, ureaplasmal, and L-form infections. In: Greene, CE (ed). Infectious diseases of the dog and cat. 4th ed. St Louis, MO: Elsevier Saunders, 319–325. doi:–10.1177/1098612X14539087
Grieco, V., Boldini, M., Luini, M., Finazzi, M., Mandelli, G., Scanziani, E. (2001). Pathological, Immunohistochemical and Bacteriological Findings in Kidneys of Cattle With Contagious Bovine Pleuropneumonia (CBPP). J. Comp. Pathol. 124 (2-3), 95–101. doi: 10.1053/jcpa.2000.0433
Groebel, K., Hoelzle, K., Wittenbrink, M. M., Ziegler, U., Hoelzle, L. E. (2009). Mycoplasma Suis Invades Porcine Erythrocytes. Infect. Immun. 77 (2), 576–584. doi: 10.1128/iai.00773-08
Gründel, A., Jacobs, E., Dumke, R. (2016). Interactions of Surface-Displayed Glycolytic Enzymes of Mycoplasma Pneumoniae With Components of the Human Extracellular Matrix. Int. J. Med. Microbiol. 306 (8), 675–685. doi: 10.1016/j.ijmm.2016.09.001
Guimaraes, A., Vieira, R., Poletto, R., Vemulapalli, R., Santos, A., De Moraes, W., et al. (2011). A Quantitative Taqman PCR Assay for the Detection of Mycoplasma Suis. J. Appl. Microbiol. 111 (2), 417–425. doi: 10.1111/j.1365-2672.2011.05053.x
Gulliksen, S. M., Baustad, B., Framstad, T., Jørgensen, A., Skomsøy, A., Kjelvik, O., et al. (2021). Successful Eradication of Mycoplasma Hyopneumoniae From the Norwegian Pig Population–10 Years Later. Porcine Health Manage. 7 (1), 1–10. doi: 10.1186/s40813-021-00216-z
Gupta, R. S., Sawnani, S., Adeolu, M., Alnajar, S., Oren, A. (2018). Phylogenetic Framework for the Phylum Tenericutes Based on Genome Sequence Data: Proposal for the Creation of a New Order Mycoplasmoidales Ord. Nov., Containing Two New Families Mycoplasmoidaceae Fam. Nov. And Metamycoplasmataceae Fam. Nov. Harbouring Eperythrozoon, Ureaplasma and Five Novel Genera. Antonie Van Leeuwenhoek 111 (9), 1583–1630. doi: 10.1007/s10482-018-1047-3
Haapala, V., Vähänikkilä, N., Kulkas, L., Tuunainen, E., Pohjanvirta, T., Autio, T., et al. (2021). Mycoplasma Bovis Infection in Dairy Herds—Risk Factors and Effect of Control Measures. J. Dairy Sci. 104 (2), 2254–2265. doi: 10.3168/jds.2020-18814
Hagemann, L., Gründel, A., Jacobs, E., Dumke, R. (2017). The Surface- Displayed Chaperones Groel and Dnak of Mycoplasma Pneumoniae Interact With Human Plasminogen and Components of the Extracellular Matrix. Pathog. Dis. 75 (3), 10.1093. doi: 10.1093/femspd/ftx017
Hall, J. P., Brockhurst, M. A., Harrison, E. (2017). Sampling the Mobile Gene Pool: Innovation via Horizontal Gene Transfer in Bacteria. Philos. Trans. R. Soc. B: Biol. Sci. 372 (1735), 20160424. doi: 10.1098/rstb.2016.0424
Hausner, M., Schamberger, A., Naumann, W., Jacobs, E., Dumke, R. (2013). Development of Protective Anti-Mycoplasma Pneumoniae Antibodies After Immunization of Guinea Pigs With the Combination of a P1-P30 Chimeric Recombinant Protein and Chitosan. Microb. .Pathog. 64, 23–32. doi: 10.1016/j.micpath.2013.07.004
Hazelton, M., Morton, J., Parker, A., Bosward, K., Sheehy, P., Dwyer, C., et al. (2020). Mycoplasma Bovis and Other Mollicutes in Replacement Dairy Heifers From Mycoplasma Bovis-Infected and Uninfected Herds: A 2-Year Longitudinal Study. J. Dairy Sci. 103 (12), 11844–11856. doi: 10.3168/jds.2020-18921
He, C. B., Lee, J. R., Kahana, M. (2021). Mycoplasma Pneumoniae Associated Acute Transverse Myelitis: An Atypical Clinical Presentation in an Adolescent Child. Cureus 13 (8), e17259. doi: 10.7759/cureus.17259
He, J., Liu, M., Ye, Z., Tan, T., Liu, X., You, X., et al. (2018). [Corrigendum] Insights Into the Pathogenesis of Mycoplasma Pneumoniae. Mol. Med. Rep. 17 (3), 4155–4155. https://doi.org/10.3892/mmr.2017.8324
Heller, M., Gicheru, N., Tjipura-Zaire, G., Muriuki, C., Yu, M., Botelho, A., et al. (2016). Development of a Novel Cocktail Enzyme-Linked Immunosorbent Assay and a Field-Applicable Lateral-Flow Rapid Test for Diagnosis of Contagious Bovine Pleuropneumonia. J. Clin. Microbiol. 54 (6), 1557–1565. doi: 10.1128/JCM.03259-15
Henderson, K. C., Benitez, A. J., Ratliff, A. E., Crabb, D. M., Sheppard, E. S., Winchell, J. M., et al. (2015). Specificity and Strain-Typing Capabilities of Nanorod Array-Surface Enhanced Raman Spectroscopy for Mycoplasma Pneumoniae Detection. PloS One 10 (6), e0131831. doi: 10.1371/journal.pone.0131831
Henderson, K. C., Sheppard, E. S., Rivera-Betancourt, O. E., Choi, J.-Y., Dluhy, R. A., Thurman, K. A., et al. (2014). The Multivariate Detection Limit for Mycoplasma Pneumoniae as Determined by Nanorod Array-Surface Enhanced Raman Spectroscopy and Comparison With Limit of Detection by Qpcr. Analyst 139 (24), 6426–6434. doi: 10.1039/C4AN01141D
Hennigan, S., Driskell, J., Dluhy, R., Zhao, Y., Tripp, R. (2010). Detection of Mycoplasma Pneumoniae in Simulated and True Clinical Throat Swab Specimens by Nanorod Array-Surface-Enhanced Raman Spectroscopy. PloS One 5 (10), e13633 doi: 10.1371/journal.pone.0013633
Henriquez, F., Mooney, R., Bandel, T., Giammarini, E., Zeroual, M., Fiori, P., et al. (2021). Paradigms of Protist/Bacteria Symbioses Affecting Human Health: Acanthamoeba Species and Trichomonas Vaginalis. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.616213
HILL, A. C. (1983). Mycoplasma Collis, a New Species Isolated From Rats and Mice. Int. J. Systematic Evolutionary Microbiol. 33 (4), 847–851. doi: 10.1099/00207713-33-4-847
Himmelreich, R., Plagens, H., Hilbert, H., Reiner, B., Herrmann, R. (1997). Comparative Analysis of the Genomes of the Bacteria Mycoplasma Pneumoniae and Mycoplasma Genitalium. Nucleic Acids Res. 25 (4), 701–712. doi: 10.1093/nar/25.4.701
Hoelzle, K., Ade, J., Hoelzle, L. E. (2020). Persistence in Livestock Mycoplasmas—A Key Role in Infection and Pathogenesis. Curr. Clin. Microbiol. Rep. 7, 81–89. doi: 10.1007/s40588-020-00149-1
Hoelzle, L. E., Zeder, M., Felder, K. M., Hoelzle, K. (2014). Pathobiology of Mycoplasma Suis. Veterinary J. 202 (1), 20–25. doi: 10.1016/j.tvjl.2014.07.023
Hooven, T. A., Polin, R. A. (2017). Pneumonia. Semin. Fetal Neonatal Med. 22 (4), 206–213. doi: 10.1016/j.siny.2017.03.002
Hopfe, M., Hoffmann, R., Henrich, B. (2004). P80, the Hint Interacting Membrane Protein, is a Secreted Antigen of Mycoplasma Hominis. BMC Microbiol. 4 (46), 1–10. doi: 10.1186/1471-2180-4-46
Hornok, S., Hajtós, I., Meli, M., Farkas, I., Gönczi, E., Meili, T., et al. (2012). First Molecular Identification of Mycoplasma Ovis and ‘Candidatus M. Haemoovis’ From Goat, With Lack of Haemoplasma PCR-Positivity in Lice. Acta Veterinaria Hungarica 60 (3), 355–360. doi: 10.1556/AVet.2012.030
Hornok, S., Sugár, L., De Mera, I. G. F., de la Fuente, J., Horváth, G., Kovács, T., et al. (2018). Tick-and Fly-Borne Bacteria in Ungulates: The Prevalence of Anaplasma Phagocytophilum, Haemoplasmas and Rickettsiae in Water Buffalo and Deer Species in Central Europe, Hungary. BMC Veterinary Res. 14 (1), 98. doi: 10.1186/s12917-018-1403-6
Howard, C. (1983). Mycoplasmas and Bovine Respiratory Disease: Studies Related to Pathogenicity and the Immune Response–a Selective Review. Yale J. Biol. Med. 56 (5-6), 789.
Huang, S., Li, J.-Y., Wu, J., Meng, L., Shou, C.-C. (2001). Mycoplasma Infections and Different Human Carcinomas. World J. Gastroenterol. 7 (2), 266. doi: 10.3748/wjg.v7.i2.266
Huang, J., Zhu, H., Wang, J., Guo, Y., Zhi, Y., Wei, H., et al. (2019). Fructose-1, 6-Bisphosphate Aldolase is Involved in Mycoplasma Bovis Colonization as a Fibronectin-Binding Adhesin. Res. Veterinary Sci. 124, 70–78. doi: 10.1016/j.rvsc.2019.02.010
Huber, B. M., Meyer Sauteur, P. M., Unger, W. W. J., Hasters, P., Eugster, M. R., Brandt, S., et al. (2018). Vertical Transmission of Mycoplasma Pneumoniae Infection. Neonatology 114 (4), 332–336. doi: 10.1159/000490610
Husain, F., Tang, K., Veeranagouda, Y., Boente, R., Patrick, S., Blakely, G., et al. (2017). Novel Large-Scale Chromosomal Transfer in Bacteroides Fragilis Contributes to its Pan-Genome and Rapid Environmental Adaptation. Microbial Genomics 3 (11), e000136. doi: 10.1099/mgen.0.000136
Hutchison, C. A., Chuang, R. Y., Noskov, V. N., Assad-Garcia, N., Deerinck, T. J., Ellisman, M. H., et al. (2016). DDesign and Synthesis of a Minimal Bacterial Genome. G. Gibson J. C. Venter Science 351 (6280), aad6253. doi: 10.1126/science.aad6253
Hutchison, C. A., Peterson, S. N., Gill, S. R., Cline, R. T., White, O., Fraser, C. M., et al. (1999). Global Transposon Mutagenesis and a Minimal Mycoplasma Genome. Science 286 (5447), 2165–2169. doi: 10.1126/science.286.5447.2165
Ikeda, P., Seki, M., Carrasco, A., Rudiak, L., Miranda, J., Goncalves, S., et al. (2017). Evidence and Molecular Characterization of Bartonella Spp. And Hemoplasmas in Neotropical Bats in Brazil. Epidemiol. Infect. 145 (10), 2038–2052. doi: 10.1017/S0950268817000966
Ishag, H. Z., Liu, M., Yang, R., Xiong, Q., Feng, Z., Shao, G. (2016). GFP as a Marker for Transient Gene Transfer and Expression in Mycoplasma Hyorhinis. SpringerPlus 5 (1), 1–4. doi: 10.1186/s40064-016-2358-3
Ishfaq, M., Hu, W., Khan et al, M. (2020). Current Status of Vaccine Research, Development, and Challenges of Vaccines for Mycoplasma Gallisepticum. Poultry Sci. 99 (9), 4195–4202. doi: 10.1016/j.psj.2020.06.014
Iverson-Cabral, S., Wood, G., Totten, P. (2015). Analysis of the Mycoplasma Genitalium Mgpb Adhesin to Predict Membrane Topology, Investigate Antibody Accessibility, Characterize Amino Acid Diversity, and Identify Functional and Immunogenic Epitopes. PloS One 10 (9), e0138244. doi: 10.1371/journal.pone.0138244
Jackson, M. A., Schutze, G. E., COMMITTEE ON INFECTIOUS DISEASES, et al. (2016). The Use of Systemic and Topical Fluoroquinolones. Pediatrics 138 (5), e20162706. doi: 10.1542/peds.2016-2706
Jambhekar, A., Robin, E., Le Boedec, K. (2019). A Systematic Review and Meta-Analyses of the Association Between 4 Mycoplasma Species and Lower Respiratory Tract Disease in Dogs. J. Veterinary Internal Med. 33 (5), 1880–1891. doi: 10.1111/jvim.15568
Jaÿ, M., Ambroset, C., Tricot, A., Colin, A., Tardy, F. (2020). Population Structure and Antimicrobial Susceptibility of Mycoplasma Ovipneumoniae Isolates in France. Vet. Microbiol. 248, 108828. doi: 10.1016/j.vetmic.2020.108828
Jesse, F., Jazid, N., Mohammed, K., Tijjani, A., Chung, E., Abba, Y., et al. (2015). Hemotropic Mycoplasma Ovis Infection in Goats With Concurrent Gastrointestinal Parasitism in MalaysiaJ. Adv. Vet. Anim. Res. 2, 464. doi: 10.5455/javar.2015.b119
Jiang, Z., Li, S., Zhu, C., Zhou, R., Leung, P. H. (2021). Mycoplasma Pneumoniae Infections: Pathogenesis and Vaccine Development. Pathogens 10 (2), 119. doi: 10.3390/pathogens10020119
Jiang, Z., Song, F., Li, Y., Xue, D., Zhao, N., Zhang, J., et al. (2017). Capsular Polysaccharide of Mycoplasma Ovipneumoniae Induces Sheep Airway Epithelial Cell Apoptosis via ROS-Dependent JNK/P38 MAPK Pathways. Oxid. Med. Cell. Longevity 2017, 6175841. doi: 10.1155/2017/6175841
Jiang, S., Zhang, S., Langenfeld, J., Lo, S. C., Rogers, M. B., et al. (2008). "Mycoplasma Infection Transforms Normal Lung Cells and Induces Bone Morphogenetic Protein 2 Expression by Post-Transcriptional Mechanisms.". J. Cell. Biochem. 104 (2), 580–594. doi: 10.1002/jcb.21647
Ji, Y., Karbaschi, M., Cooke, M. (2019). Mycoplasma Infection of Cultured Cells Induces Oxidative Stress and Attenuates Cellular Base Excision Repair Activity. Mutat. Research/Genetic Toxicol. Environ. Mutagenesis 845, 403054. doi: 10.1016/j.mrgentox.2019.05.010
Jores, J., Baldwin, C., Blanchard, A., Browning, G., Colston, A., Gerdts, V., et al. (2020). Contagious Bovine and Caprine Pleuropneumonia: A Research Community’s Recommendations for the Development of Better Vaccines. NPJ Vaccines 5(1), 66. doi: 10.1038/s41541-020-00214-2
Jores, J., Meens, J., Buettner, F. F., Linz, B., Naessens, J., Gerlach, G. F. (2009). "Analysis of the Immunoproteome of Mycoplasma Mycoides Subsp. Mycoides Small Colony Type Reveals Immunogenic Homologues to Other Known Virulence Traits in Related Mycoplasma Species. Veterinary Immunol. Immunopathol. 131 (3-4), 238–245. doi:–10.1016/j.vetimm.2009.04.016
Josi, C., Bürki, S., Stojiljkovic, A., Wellnitz, O., Stoffel, M. H., Pilo, P. (2018). Bovine Epithelial In Vitro Infection Models for Mycoplasma Bovis. Front. Cell. Infect. Microbiol. 8, 329. doi: 10.3389/fcimb.2018.00329
Kalantari, M., Sharifiyazdi, H., Ghane, M., Nazifi, S. (2020). The Occurrence of Hemotropic Mycoplasma Ovis-Like Species in Horses. Prev. Veterinary Med. 175, 104877. doi: 10.1016/j.prevetmed.2019.104877
Kamiyama, T., Saito, M., Nakagawa, M. (1991). Effects of Mycoplasma Pulmonis Infection and Exposure to Nitrogen Dioxide on Activities of Natural Killer Cells and Macrophages of the Mouse. Jikken Dobutsu 40 (10), 255–257. doi: 10.1538/EXPANIM1978.40.2_255
Kamyingkird, K., Jiyipong, T., Amavisit, P., Stich, R. W., Jittapalapong, S. (2021). Molecular Detection of Mycoplasma haemofelis, ‘Candidatus Mycoplasma haemominutum’ and ‘Candidatus Mycoplasma turicensis’ of stray cats residing in Bangkok monasteries, Thailand. Agriculture and Natural Resources 55(3), 423–430. doi: 10.34044/j.anres.2021.55.3.12
Kannan, T., Baseman, J. (2006). ADP-Ribosylating and Vacuolating Cytotoxin of Mycoplasma Pneumoniae Represents Unique Virulence Determinant Among Bacterial Pathogens. Proc. Natl. Acad. Sci. U. S. A. 103 (17), 6724–6729. doi: 10.1073/pnas.0510644103
Karin, M., Delhase, M. (2000). The Iκb Kinase (IKK) and NF-κb: Key Elements of Proinflammatory Signalling. Semin. Immunol 12(1), 85–98. doi: 10.1006/smim.2000.0210
Kaur, C., Garg, D., Mahajan, S. (1998). Experimental Mastitogenicity of Mycoplasma Capricolum Subsp. Capripneumoniae for Rabbit Mammary Glands. Indian J. Exp. Biol. 36 (4), 407–410.
Kersin, S. G., Bozan, T., Özdemir, H., Bilgen, H. S., Söyletir, G., Memişoğlu, A., et al. (2020). Clinical and Laboratory Awareness for an Under Recognized Pathogen in Newborn Meningitis: Mycoplasma Hominis: A Case Report. Turkish J. Pediatr. 62 (2), 280–283. doi: 10.24953/turkjped.2020.02.015
Ketchersid, J., Scott, J., Lew, T., Banaei, N., Kappagoda, S. (2020). Recurrent Multifocal Mycoplasma Orale Infection in an Immunocompromised Patient: A Case Report and Review. Case Rep. Infect. Dis. 2020, 8852115. doi: 10.1155/2020/8852115
Khan, F., Faisal, M., Chao, J., Liu, K., Chen, X., Zhao, G., et al. (2016). A Promising Diagnostic Biomarker for Serological Detection of Mycoplasma Bovis Infection: Immunoproteomic Identification of Mbovp579. Oncotarget 7 (26), 39376–39395. doi: 10.18632/oncotarget.9799
Khan, F.-A., Rasheed, M.-A., Faisal, M., Menghwar, H., Zubair, M., Sadique, U., et al. (2017). Proteomics Analysis and its Role in Elucidation of Functionally Significant Proteins in Mycoplasma Bovis. Microbial Pathogenesis 111, 50–59. doi: 10.1016/j.micpath.2017.08.024
Khan, F., Zhao, G., Guo, Y., Faisal, M., Chao, J., Chen, X., et al. (2018). Proteomics Identification and Characterization of Mbovp730 as a Potential DIVA Antigen of Mycoplasma Bovis. Oncotarget 9 (47), 28322–28336. doi: 10.18632/oncotarget.22265
Khodakaram-Tafti, A., Lopez, A. (2004). Immunohistopathological Findings in the Lungs of Calves Naturally Infected With Mycoplasma Bovis. J. Veterinary Med. Ser. A 51 (1), 10–14. doi: 10.1111/j.1439-0442.2004.00596.x
Kiers (2020). A Sustainable Mycoplasma Gallisepticum Control Program in Multi-Age Farms. Asian Poultry Magazine, 36–40.
Kim, J., Lee, D., Yoon, E., Bae, H., Chun, D., Kang, J.-G., et al. (2020). Clinical Case of a Transfusion-Associated Canine Mycoplasma Haemocanis Infection in the Republic of Korea: A Case Report. Korean J. Parasitology 58 (5), 565. doi: 10.3347/kjp.2020.58.5.565
Kim, M. K., Shin, S.-J., Lee, H. M., Choi, H. S., Jeong, J., Kim, H., et al. (2019). Mycoplasma Infection Promotes Tumor Progression via Interaction of the Mycoplasmal Protein P37 and Epithelial Cell Adhesion Molecule in Hepatocellular Carcinoma. Cancer Lett. 454, 44–52. doi: 10.1016/j.canlet.2019.04.007
Kinoshita, Y., Niwa, H., Uchida-Fujii, E., Nukada, T. (2020). Complete Genome Sequence of Mycoplasma Felis Strain Myco-2, Isolated From an Equine Tracheal Wash Sample in Japan. Microbiol. Resource Announcements 9 (9), e00057–20. doi: 10.1128/MRA.00057-20
Krasteva, I., Liljander, A., Fischer, A., Smith, D., Inglis, N., Scacchia, M., et al. (2014). Characterization of the In Vitro Core Surface Proteome of Mycoplasma Mycoides Subsp. Mycoides, the Causative Agent of Contagious Bovine Pleuropneumonia. Vet. Microbiol. 168 (1), 116–123. doi: 10.1016/j.vetmic.2013.10.025
Kudirkiene, E., Aagaard, A. K., Schmidt, L. M., Pansri, P., Krogh, K. M., Olsen, J. E. (2021). Occurrence of Major and Minor Pathogens in Calves Diagnosed With Bovine Respiratory Disease. Veterinary Microbiol., 259, 109135. doi: 10.1016/j.vetmic.2021.109135
Kühner, S., van Noort, V., Betts, M. J., Leo-Macias, A., Batisse, C., Rode, M., et al. (2009). Proteome Organization in a Genome-Reduced Bacterium. Science 326 (5957), 1235–1240. doi: 10.1126/science.1176343
Kulappu Arachchige, S., Young, N., Kanci Condello, A., Omotainse, O. S., Noormohammadi, A. H., Wawegama, N. K., et al. (2021). Transcriptomic Analysis of Long-Term Protective Immunity Induced by Vaccination With Mycoplasma Gallisepticum Strain Ts-304. Front. Immunol. 11, 3743. doi: 10.3389/fimmu.2020.628804
Kumar, S., Ingle, H., Prasad, D. V., Kumar, H. (2013). Recognition of Bacterial Infection by Innate Immune Sensors Crit. Rev. Microbiol., 3, 229–246. doi: 10.3109/1040841x.2012.706249
Kusanovic, J. P., Vargas, P., Ferrer, F., Díaz, F., Córdova, V., Martinovic, C., et al. (2020). Comparison of Two Identification and Susceptibility Test Kits for Ureaplasma Spp and Mycoplasma Hominis in Amniotic Fluid of Patients at High Risk for Intra-Amniotic Infection. J. Matern. Fetal Neonatal Med. 33 (20), 3409–3417. doi: 10.1080/14767058.2019.1572742
Labroussaa, F., Baby, V., Rodrigue, S., Lartigue, C. (2019). La Transplantation De Génomes. Med. Sci. (Paris) 35, 761–770. doi: 10.1051/medsci/2019154
Lambert, L., Trummell, H., Singh, A., Cassell, G. H., Bridges, R. J. (1998). Mycoplasma Pulmonis Inhibits Electrogenic Ion Transport Across Murine Tracheal Epithelial Cell Monolayers. Infect. Immun. 66 (1), 272–279. doi: 10.1128/IAI.66.1.272-279.1998
Lanao, A. E., Chakraborty, R. K., Pearson-Shaver, A. L. (2022). Mycoplasma Infections (StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing). Available at: https://www.ncbi.nlm.nih.gov/books/NBK536927/.
Lartigue, C., Glass, J. I., Alperovich, N., Pieper, R., Parmar, P. P., Hutchison, C. A., 3rd, et al. (2007). Genome Transplantation in Bacteria: Changing One Species to Another. Science 317 (5838), 632–638. doi: 10.1126/science.1144622
Latino, M. A., Botta, G., Badino, C., Maria, D., Petrozziello, A., Sensini, A., et al. (2018). Association Between Genital Mycoplasmas, Acute Chorioamnionitis and Fetal Pneumonia in Spontaneous Abortions. J. Perinat Med. 46 (5), 503–508. doi: 10.1515/jpm-2016-0305
Leal Zimmer, F. M., Paes, J. A., Zaha, A., Ferreira, H. B. (2020). Pathogenicity & Virulence of Mycoplasma Hyopneumoniae. Virulence 11 (1), 1600–1622. doi: 10.1080/21505594.2020.1842659
Lee, H., Choi, W., Yang, S., Kim, D., Park, S., Lee, M., et al. (2021). PCR-Coupled Paper-Based Surface-Enhanced Raman Scattering (SERS) Sensor for Rapid and Sensitive Detection of Respiratory Bacterial DNA. Sensors Actuators B: Chem. 326, 128802. doi: 10.1016/j.snb.2020.12
Leick, M., Azcutia, V., Newton, G., Luscinskas, F. W. (2014). Leukocyte Recruitment in Inflammation: Basic Concepts and New Mechanistic Insights Based on New Models and Microscopic Imaging Technologies. Cell Tissue Res. 355 (3), 647–656. doi: 10.1007/s00441-014-1809-9
Lemcke, R., Ernø, H., Gupta, U. (1981). The Relationship of Two Equine Mycoplasmas to Mycoplasma Mycoides. Epidemiol. Infect. 87 (1), 93–100. doi: 10.1017/S0022172400069278
Li, G., Fan, L., Wang, Y., Huang, L., Wang, M., Zhu, C., et al. (2019). High Co-Expression of TNF- α and CARDS Toxin is a Good Predictor for Refractory Mycoplasma Pneumoniae Pneumonia. Mol. Med. Rep. 25 (1), 1–10. doi: 10.1186/s10020-019-0105-2
Liljander, A., Sacchini, F., Stoffel, M. H., Schieck, E., Stokar-Regenscheit, N., Labroussaa, F., et al. (2019). Reproduction of Contagious Caprine Pleuropneumonia Reveals the Ability of Convalescent Sera to Reduce Hydrogen Peroxide Production In Vitro. Vet. Res. 50 (1), 10. doi: 10.1186/s13567-019-0628-0
Liu, W., Fang, L., Li, M., Li, S., Guo, S., Luo, R., et al. (2012a). Comparative Genomics of Mycoplasma: Analysis of Conserved Essential Genes and Diversity of the Pan-Genome. PloS One 7 (4), e35698. doi: 10.1371/journal.pone.0035698
Liu, Y.-C., Lin, I.-H., Chung, W.-J., Hu, W. S., Ng, W. V., Lu, C.-Y., et al. (2012b). Proteomics Characterization of Cytoplasmic and Lipid-Associated Membrane Proteins of Human Pathogen Mycoplasma Fermentans M64. PloS One 7 (4), e35304. doi: 10.1371/journal.pone.0035304
Liu, W., Zhou, D., Yuan, F., Liu, Z., Duan, Z., Yang, K., et al. (2019). Surface Proteins Mhp390 (P68) Contributes to Cilium Adherence and Mediates Inflammation and Apoptosis in Mycoplasma Hyopneumoniae. Microbial Pathogenesis 126, 92–100. doi: 10.1016/j.micpath.2018.10.035
Li, Z., Wang, Y., Zhang, Y., Tang, X., Wang, X., Liu, W., et al. (2021). Attenuation of Mycoplasma Hyopneumoniae Strain ES-2 and Comparative Genomic Analysis of ES-2 and its Attenuated Form ES-2l. Front. Veterinary Sci. 8, 646. doi: 10.3389/fvets.2021.696262
Luo, H., He, J., Qin, L. (2021a). Mycoplasma Pneumoniae Lipids License TLR-4 for Activation of NLRP3 Inflammasome and Autophagy to Evoke a Proinflammatory Response. Clin. Exp. Immunol. 203 (1), 66–79. doi: 10.1111/cei.13510
Luo, Y., Li, C., Zhou, Z., Gong, Z., Zhu, C., Lei, A. (2021b). Biological Functions of IL-17-Producing Cells in Mycoplasma Respiratory Infection. Immunology 164 (2), 223–230. doi: 10.1111/imm.13346
Luttrell, M. P., Stallknecht, D. E., Kleven, S. H., Kavanaugh, D. M., Corn, J. L., Fischer, J. R., et al. (2001). Mycoplasma Gallisepticum in House Finches (Carpodacus Mexicanus) and Other Wild Birds Associated With Poultry Production Facilities. Avian Dis. 45 (2), 321–329. doi: 10.2307/1592971
Lysnyansky, I., Borovok, I. (2021). A GC-Rich Prophage-Like Genomic Region of Mycoplasma Bovirhinis HAZ141_2 Carries a Gene Cluster Encoding Resistance to Kanamycin and Neomycin. Antimicrobial Agents Chemother. 65 (2), e01010–e01020. doi: 10.1128/AAC.01010-20
Lysnyansky, I., Sachse, K., Rosenbusch, R., Levisohn, S., Yogev, D. (1999). The Vsp Locus of Mycoplasma Bovis: Gene Organization and Structural Features. J. Bacteriol. 181 (18), 5734–5741. doi: 10.1128/JB.181.18.5734-5741.1999
Machado, R., André, M. (2019). Molecular Detection of Mycoplasma Suis in Captive White-Lipped Peccaries (Tayassu Pecari) and Wild Boars (Sus Scrofa) in Brazil. Comp. Immunol. Microbiol. Infect. Dis. 63, 94–96. doi: 10.1016/j.cimid.2019.01.013
MacKenzie, C. R., Nischik, N., Kram, R., Krauspe, R., Jäger, M., Henrich, B. (2010). Fatal Outcome of a Disseminated Dual Infection With Drug-Resistant Mycoplasma Hominis and Ureaplasma Parvum Originating From a Septic Arthritis in an Immunocompromised Patient. Int. J. Infect. Dis. 14 Suppl 3, e307–e309. doi: 10.1016/j.ijid.2010.02.2253
MacOwan, K. J., Minette, J. E. (1976). A Mycoplasma From Acute Contagious Caprine Pleuropneumonia in Kenya. Trop. Anim. Health Prod 8 (2), 91–95. doi: 10.1007/bf02383376
Maes, D., Boyen, F., Haesebrouck, F., Anne V. Gautier-Bouchardon, A. (2020). Antimicrobial Treatment of Mycoplasma Hyopneumoniae Infections. Vet. J. 105474, 54–74. doi: 10.1016/j.tvjl.2020.105474
Maggi, R. G., Chitwood, M. C., Kennedy-Stoskopf, S., DePerno, C. S. (2013a). Novel Hemotropic Mycoplasma Species in White-Tailed Deer (Odocoileus Virginianus). Comp. Immunol. Microbiol. Infect. Dis. 36 (6), 607–611. doi: 10.1016/j.cimid.2013.08.001
Maggi, R. G., Compton, S. M., Trull, C. L., Mascarelli, P. E., Mozayeni, B. R., Breitschwerdt, E. B. (2013b). Infection With Hemotropic Mycoplasma Species in Patients With or Without Extensive Arthropod or Animal Contact. J. Clin. Microbiol. 51 (10), 3237–3241. doi: 10.1128/JCM.01125-13
Mahmood, M., Javaid, A., Shahid, F., Ashfaq, U. A. (2021). Rational Design of Multimeric Based Subunit Vaccine Against Mycoplasma Pneumonia: Subtractive Proteomics With Immunoinformatics Framework. Infect. Genet. Evol. 91, 104795. doi: 10.1016/j.meegid.2021.104795
Maina, T., Prysliak, T., Perez-Casal, J. (2019). Mycoplasma Bovis Delay in Apoptosis of Macrophages is Accompanied by Increased Expression of Anti-Apoptotic Genes, Reduced Cytochrome C Translocation and Inhibition of DNA Fragmentation. Veterinary Immunol. Immunopathol. 208, 16–24. doi: 10.1016/j.vetimm.2018.12.004
Manguin, E., Pépin, E., Boivin, R., Leclere, M. (2020). Tracheal Microbial Populations in Horses With Moderate Asthma. J. Veterinary Internal Med. 34 (2), 986–995. doi: 10.1111/jvim.15707
Mara, A. B., Gavitt, T. D., Tulman, E. R., Geary, S. J., Szczepanek, S. M. (2020). Lipid Moieties of Mycoplasma Pneumoniae Lipoproteins are the Causative Factor of Vaccine-Enhanced Disease. NPJ Vaccines 5 (1), 1–5. doi: 10.1038/s41541-020-0181-x
March, J. B., Jepson, C. D., Clark, J. R., Totsika, M., Calcutt, M. J. (2006). Phage Library Screening for the Rapid Identification and In Vivo Testing of Candidate Genes for a DNA Vaccine Against Mycoplasma Mycoides Subsp. Mycoides Small Colony Biotype. Infect. Immun. 74 (1), 167–174. doi: 10.1128/IAI.74.1.167-174.2006
Marco, I., Mentaberre, G., Ballesteros, C., Bischof, D. F., Lavín, S., Vilei, E. M., et al. (2009). First Report of Mycoplasma Conjunctivae From Wild Caprinae With Infectious Keratoconjunctivitis in the Pyrenees (NE Spain). J. Wildlife Dis. 45 (1), 238–241. doi: 10.7589/0090-3558-45.1.238
Margarita, V., Fiori, P., Rappelli, P. (2020). Impact of Symbiosis Between Trichomonas Vaginalis and Mycoplasma Hominis on Vaginal Dysbiosis: A Mini Review. Front. Cell. Infect. Microbiol. 10, 179. doi: 10.3389/fcimb.2020.00179
Mărginean, C.,. O., Georgescu, M. A., Meliţ, E. L. (2021). Arthritis Associated With Mycoplasma Pneumoniae in a Pediatric Patient. Medicine 100 (2), e24316. doi: 10.1097/MD.0000000000024316
Marques, L., Rezende, I., Barbosa, M.-S., do Nascimento, N., Dos Santos, A., Amorim, A., et al. (2016). Ureaplasma Diversum Genome Provides New Insights About the Interaction of the Surface Molecules of This Bacterium With the Host. PloS One 11 (9), e0161926. doi: 10.1371/journal.pone.0161926
Masoumalinejad, Z., Zinatizadeh, M., Tahmasebiabdar, N. (2018). A Review of Mycoplasma in Laboratory Mice. Mod Med. Lab. J. 2 (2), 127–131. doi: 10.30699/mmlj17.2.1.15
Matyushkina, D., Pobeguts, O., Butenko, I., Vanyushkina, A., Anikanov, N., Bukato, O., et al. (2016). Phase Transition of the Bacterium Upon Invasion of a Host Cell as a Mechanism of Adaptation: A Mycoplasma Gallisepticum Model. Sci. Rep. 6, 35959. doi: 10.1038/srep35959
Maunsell, F., Chase, C. (2019). Mycoplasma Bovis: Interactions With the Immune System and Failure to Generate an Effective Immune Response. Vet. Clin. North Am. Food Anim. Pract. 35, 471–483. doi: 10.1016/j.cvfa.2019.08.003
Maunsell, F. P., Donovan, G. A., Risco, C., Brown, M. B. (2009). Field Evaluation of a Mycoplasma Bovis Bacterin in Young Dairy Calves. Vaccine 27 (21), 2781–2788. doi: 10.1016/j.vaccine.2009.02.100
McAuliffe, L., Ayling, R.-D., Ellis, R.-J., Nicholas, R. A. (2008). Biofilm-Grown Mycoplasma Mycoides Subsp. Mycoides SC Exhibit Both Phenotypic and Genotypic Variation Compared With Planktonic Cells. Vet. Microbiol. 129 (3–4), 315–324. doi: 10.1016/j.vetmic.2007.11.024
Mcauliffe, L., Ellis, R., Miles, K., Ayling, R., Nicholas, R. (2006). Biofilm Formation by Mycoplasma Species and its Role in Environmental Persistence and Survival. Microbiology 152, 913–922. doi: 10.1099/mic.0.28604-0
McDermott, A., Visentin, G., McParland, S., Berry, D., Fenelon, M., De Marchi, M. (2016). Effectiveness of Mid-Infrared Spectroscopy to Predict the Color of Bovine Milk and the Relationship Between Milk Color and Traditional Milk Quality Traits. J. Dairy Sci. 99 (5), 3267–3273. doi: 10.3168/jds.2015-10424
McDonald, D., Schoeb, T., Lindsey, J. (1991). Mycoplasma Pulmonis Infections Cause Long-Lasting Potentiation of Neurogenic Inflammation in the Respiratory Tract of the Rat. J. Clin. Invest. 87 (3), 787–799. doi: 10.1172/JCI115082
McGarrity, G., Rose, D., Kwiatkowski, V., Dion, A., Phillips, D., Tully, J. (1983). Mycoplasma Muris, a New Species From Laboratory Mice. Int. J. Systematic Evolutionary Microbiol. 33 (2), 350–355. doi: 10.1099/00207713-33-2-350
McGowin, C. L., Totten, P. A. (2017). The Unique Microbiology and Molecular Pathogenesis of Mycoplasma Genitalium. J. Infect. Dis. 216 (suppl_2), S382–S388. doi: 10.1093/infdis/jix172
Medzhitov, R. (2007). Recognition of Microorganisms and Activation of the Immune Response. Nature 449 (146), 819–826. doi: 10.1038/nature06246
Merodio, M., McDaniel, A., Poonsuk, K., Magtoto, R., Ferreyra, F., Meiroz-De-Souza-Almeida, H., et al. (2021). Evaluation of Colonization, Variable Lipoprotein-Based Serological Response, and Cellular Immune Response of Mycoplasma Hyorhinis in Experimentally Infected Swine. Vet. Microbiol. 260, 109162. doi: 10.1016/j.vetmic.2021.109162
Messick, J. B., Berent, L. M., Cooper, S. K. (1998). Development and Evaluation of a PCR-Based Assay for Detection of Haemobartonella Felis in Cats and Differentiation of H. Felis From Related Bacteria by Restriction Fragment Length Polymorphism Analysis. J. Clin. Microbiol. 36 (2), 462–466. doi: 10.1128/JCM.36.2.462-466.1998
Mete, A., ÖZGÜR, N. Y. (2017). Investigation of the Presence of Mycoplasma as an Etiologic Agent of Inflammatory Airway Diseases in Thoroughbred Racehorses in Istanbul Province. Turkish J. Veterinary Anim. Sci. 41 (3), 365–371. doi: 10.3906/vet-1606-90
Meyer, J. R. (2013). Sticky Bacteriophage Protect Animal Cells. Proc. Natl. Acad. Sci. U.S.A. 110, 10475–10476. doi: 10.1073/pnas.1307782110
Meyer Sauteur, P., Unger, W., Nadal, D., Berger, C., Vink, C., van Rossum, A. (2016). Infection With and Carriage of Mycoplasma Pneumoniae in Children. Front. Microbiol. 7, 329. doi: 10.3389/fmicb.2016.00329
Meylan, E., Burns, K., Hofmann, K., Blancheteau, V., Martinon, F., Kelliher, M., et al. (2004). RIP1 is an Essential Mediator of Toll-Like Receptor 3-Induced NF-Kappa B Activation. Nat. Immunol. 5 (5), 503–507. doi: 10.1038/ni1061
Michiels, T., Welby, S., Vanrobaeys, M., Quinet, C., Rouffaer, L., Lens, L., et al. (2016). Prevalence of Mycoplasma Gallisepticum and Mycoplasma Synoviae in Commercial Poultry, Racing Pigeons and Wild Birds in Belgium. Avian Pathol. 45 (2), 244–252. doi: 10.1080/03079457.2016.1145354
Minion, F.-C., Adams, C., Hsu, T. (2000). R1 Region of P97 Mediates Adherence of Mycoplasma Hyopneumoniae to Swine Cilia. Infect. Immun. 68 (5), 3056–3060. doi: 10.1128/IAI.68.5.3056-3060.2000
Minion, F., Goguen, J. (1986). Identification and Preliminary Characterization of External Membrane-Bound Nuclease Activities in Mycoplasma Pulmonis. Infect. Immun. 51 (1), 352–354. doi: 10.1128/iai.51.1.352-354.1986
Mitiku, F., Hartley, C. A., Sansom, F. M., Coombe, J. E., Mansell, P. D., Beggs, D. S., et al. (2018). The Major Membrane Nuclease Mnua Degrades Neutrophil Extracellular Traps Induced by Mycoplasma Bovis. Vet. Microbiol. 218, 13–19. doi: 10.1016/j.vetmic.2018.03.002
Monnerat, M.-P., Thiaucourt, F., Poveda, J. B., Nicolet, J., Frey, J. (1999). Genetic and Serological Analysis of Lipoprotein Lppa in Mycoplasma Mycoides Subsp. Mycoides LC and Mycoplasma Mycoides Subsp. Capri. Clin. Diagn. Lab. Immunol. 6 (2), 224–230. doi: 10.1128/CDLI.6.2.224-230.1999
Morowitz, H. J., Tourtellotte, M. E. (1962). The Smallest Living Cells. Sci. Am. 206, 117–126. doi: 10.1038/scientificamerican0362-117
Mucha, S. G., Ferrarini, M. G., Moraga, C., Di Genova, A., Guyon, L., Tardy, F., et al. (2020). Mycoplasma Hyopneumoniae J Elicits an Antioxidant Response and Decreases the Expression of Ciliary Genes in Infected Swine Epithelial Cells. Sci. Rep. 10 (1), 1–22. doi: 10.1038/s41598-020-70040-y
Mycoplasma bovis Investigations in Cattle . (2018) Vet. Rec. 183 (8), 256–258. doi: 10.1136/vr.k3722
Nakagaki, B. N., Vieira, A. T., Rezende, R. M., David, B. A., Menezes, G. B. (2018). Tissue Macrophages as Mediators of a Healthy Relationship With Gut Commensal Microbiota. Cell Immunol. 330, 16–26. doi: 10.1016/j.cellimm.2018.01.017
Namiki, K., Goodison, S., Porvasnik, S., Allan, R. W., Iczkowski, K. A., Urbanek, C., et al. (2009). Persistent Exposure to Mycoplasma Induces Malignant Transformation of Human Prostate Cells. PloS One 4 (9), e6872. doi: 10.1371/journal.pone.0006872
Narita, M. (2016). Classification of Extrapulmonary Manifestations Due to Mycoplasma Pneumoniae Infection on the Basis of Possible Pathogenesis. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00023
Naseem, S., Meens, J., Jores, J., Heller, M., Dübel, S., Hust, M., et al. (2010). Phage Display-Based Identification and Potential Diagnostic Application of Novel Antigens From Mycoplasma Mycoides Subsp. Mycoides Small Colony Type. Veterinary Microbiol. 142 (3-4), 285–292. doi: 10.1016/j.vetmic.2009.09.071
Nehra, K., Rana, R., Viswas, K., Arun, T., Singh, V., Singh, A. P., et al. (2015). Isolation and Molecular Identification of Mycoplasma Equigenitalium From Equine Genital Tracts in Northern India. Iranian J. Veterinary Res. 16 (2), 176. doi: 10.22099/IJVR.2015.3052
Neto, J. (2012). Diagnostic and Field Investigations in Mycoplasma Hyosynoviae and Mycoplasma Hyorhinis. Grad Theses Diss doi: 10.31274/etd-180810-253
Newton, L. (1992). Contagious Bovine Pleuropneumonia in Australia: Some Historic Highlights From Entry to Eradication. Aust. Vet. J. 69, 12, 306–317. doi: 10.1111/j.1751-0813.1992.tb09912.x
Niang, M., Rosenbusch, R., DeBey, M., Niyo, Y., Andrews, J., Kaeberle, M. (1998). Field Isolates of Mycoplasma Ovipneumoniae Exhibit Distinct Cytopathic Effects in Ovine Tracheal Organ Cultures. J. Veterinary Med. Ser. A 45 (1-10), 29–40. doi: 10.1111/j.1439-0442.1998.tb00798.x
Nicholas, R., Ayling, R., McAuliffe, L. (2008). Mycoplasma Diseases of Ruminants. 1st ed (Oxford, UK: CABI Publishing).
Nicholas, R., Tjipura-Zaire, G., Mbulu, R., Scacchia, M., Mettler, F., Frey, J., et al. (2004). “An Inactivated Whole Cell Vaccine and Lppq Subunit Vaccine Appear to Exacerbate the Effects of CBPP in Adult Cattle,” in Towards Sustainable CBPP Control Programmes for Africa (Rome, Italy: FAO Animal Production and Health).
Nkando, I., Perez-Casal, J., Mwirigi, M., Prysliak, T., Townsend, H., Berberov, E., et al. (2016). Recombinant Mycoplasma Mycoides Proteins Elicit Protective Immune Responses Against Contagious Bovine Pleuropneumonia. Veterinary Immunol. Immunopathol. 171, 103–114. doi: 10.1016/j.vetimm.2016.02.010
Nobile, M. S., Coelho, V., Pescini, D., Damiani, C. (2021). Accelerated Global Sensitivity Analysis of Genome-Wide Constraint-Based Metabolic Models. BMC Bioinf. 22 (2), 1–17. doi: 10.1186/s12859-021-04002-0
Nogueira, W., Jaiswal, A., Tiwari, S., Ramos, R., Ghosh, P., Barh, D., et al. (2021). Computational Identification of Putative Common Genomic Drug and Vaccine Targets in Mycoplasma Genitalium. Genomics 113 (4), 2730–2743. doi: 10.1016/j.ygeno.2021.06.011
Nolan, P., Hill, G., Stoehr, A. (1998). Sex, Size, and Plumage Redness Predict House Finch Survival in an Epidemic. Proc. R. Soc. B-Biological Sci. 265, 961. doi: 10.1098/rspb.1998.0384
Normand, V., Boulbria, G., Brissonnier, M., Bachy, V., Moalic, P.-Y., Berton, P., et al. (2020). Comparison of Qpcr and Blood Smear Microscopy for the Diagnosis of Mycoplasma Suis in a French Veterinary Practice. Porcine Health Manage. 6 (1), 1–4. doi: 10.1186/s40813-019-0143-8
Nottelet, P., Bataille, L., Gourgues, G., Anger, R., Lartigue, C., Sirand-Pugnet, P., et al. (2021). The Mycoplasma Surface Proteins MIB and MIP Promote the Dissociation of the Antibody-Antigen Interaction. Sci. Adv. 7 (10), eabf2403. doi: 10.1126/sciadv.abf2403
Oehlerking, J., Kube, M., Felder, K. M., Matter, D., Wittenbrink, M. M., Schwarzenbach, S., et al. (2011). Complete Genome Sequence of the Hemotrophic Mycoplasma Suis Strain KI3806. Am. Soc. Microbiol 2(1), e01235–13. doi: 10.1128/JB.00187-11
OIE (2019) World Animal Health Information Database. Available at: https://www.oie.int/wahis_2/public/wahid.php/Diseaseinformation/Diseasedistributionmap?disease_type_hidden¼&disease_id_hidden¼&selected_disease_name_hidden¼&disease_type¼0&disease_id_terrestrial¼6&species_t¼0&disease_id_aquatic¼999&species_a¼0&sta_method¼semesterly&selected_start_year¼2018&selected_report_period¼1&selected_start_month¼1&date_submit¼OK (Accessed December 10, 2019).
OIE (2021) Adopted by the World Assembly of Delegates of the OIE. Available at: https://www.oie.int/en/what-we-do/standards/codes-and-manuals/ (Accessed 27 May 2021).
Olaniyi, M., Ajayi, O., Alaka, O., Mustapha, O., Brown, C., Shields, J., et al. (2020). Immunohistochemical and Ultrastructural Studies of Mycoplasma Hyopneumoniae Strain in Naturally Infected Pigs in Nigeria. Folia Veterinaria 64 (1), 1–10. doi: 10.2478/fv-2020-0001
Origgi, F. C., Jacobson, E. R. (2000). Diseases of the Respiratory Tract of Chelonians. Veterinary Clinics North America: Exotic Anim. Pract. 3 (2), 537–549. doi: 10.1016/S1094-9194(17)30088-9
Paessler, M., Levinson, A., Patel, J. B., Schuster, M., Minda, M., Nachamkin, I. (2002). Disseminated Mycoplasma Orale Infection in a Patient With Common Variable Immunodeficiency Syndrome. Diagn. Microbiol. Infect. Dis. 44 (2), 201–204. doi: 10.1016/s0732-8893(02)00429-7
Paterna, A., Sánchez, A., Gómez-Martín, A., Corrales, J., de la Fe, C., Contreras, A., et al. (2013). In Vitro Antimicrobial Susceptibility of Mycoplasma Agalactiae Strains Isolated From Dairy Goats. J. dairy Sci. 96 (11), 7073–7076. doi: 10.3168/jds.2012-6492
Patil, S., Rao, R. S., Raj, A. T. (2015). Role of Mycoplasma in the Initiation and Progression of Oral Cancer. J. Int. Oral. health: JIOH 7 (7), i.
Paton, G., Jacobs, J., Perkins, F. (1965). Chromosome Changes in Human Diploid-Cell Cultures Infected With Mycoplasma. Nature 207 (4992), 43–45. doi: 10.1038/207043a0
Patterson, J., Foxcroft, G. (2019). Gilt Management for Fertility and Longevity. Animals 9 (7), 434. doi: 10.3390/ani9070434
Pehlivan, M., Itirli, G., Onay, H., Bulut, H., Koyuncuoglu, M., Pehlivan, S. (2004). Does Mycoplasma Sp. Play Role in Small Cell Lung Cancer? Lung Cancer 45 (1), 129–130. doi: 10.1016/J.LUNGCAN.2004.01.007
Pehlivan, M., Pehlivan, S., Onay, H., Koyuncuoglu, M., Kirkali, Z. (2005). Can Mycoplasma-Mediated Oncogenesis Be Responsible for Formation of Conventional Renal Cell Carcinoma? Urology 65 (2), 411–414. doi: 10.1016/j.urology.2004.10.015
Perez-Casal, J. (2020). Pathogenesis and Virulence of Mycoplasma Bovis. Veterinary Clinics North America. Food Anim. Practice. 36 (2), 269–278. doi: 10.1016/j.cvfa.2020.02.002
Perez-Casal, J., Prysliak, T., Maina, T., Wang, Y., Townsend, H., Berverov, E., et al. (2015). Analysis of Immune Responses to Recombinant Proteins From Strains of Mycoplasma Mycoides Subsp. Mycoides, the Causative Agent of Contagious Bovine Pleuropneumonia. Veterinary Immunol. Immunopathol. 168 (1-2), 103–110. doi: 10.1016/j.vetimm.2015.08.013
Persson, A., Jacobsson, K., Frykberg, L., Johansson, K.-E., Poumarat, F. (2002). Variable Surface Protein Vmm of Mycoplasma Mycoides SubspMycoides Small Colony Type. J. Bacteriol. 184 (13), 3712–3722. doi: 10.1128/JB.184.13.3712-3722.2002
Peter, C., Alec, M., Bigoni, J., Toutous-Trellu, L., Yaron, M. (2018). Update on Mycoplasma Genitalium Among Women. Rev. Medicale Suisse 14 (624), 1893–1897. doi: 10.53738/REVMED.2018.14.624.1893
Petri, F. A. M., Sonalio, K., de Souza Almeida, H. M., Ferraz, M. E. S., Storino, G. Y., de Souza, M. R., et al. (2020). Porcine Hemothropic Mycoplasmas Infection Associated With Productive Impact in Intensive Pig Production. Porcine Health Manage. 6 (1), 1–8. doi: 10.1186/s40813-020-00171-1
Pettersson, B., Leitner, T., Ronaghi, M., Bölske, G., Uhlen, M., Johansson, K.-E. (1996). Phylogeny of the Mycoplasma Mycoides Cluster as Determined by Sequence Analysis of the 16S Rrna Genes From the Two Rrna Operons. J. Bacteriol. 178 (14), 4131–4142. doi: 10.1128/jb.178.14.4131-4142.1996
Pilo, P., Frey, J., Vilei, E. M. (2007). Molecular Mechanisms of Pathogenicity of Mycoplasma Mycoides Subsp. Mycoides SC. Veterinary J. 174 (3), 513–521. doi: 10.1016/j.tvjl.2006.10.016
Pilo, P., Martig, S., Frey, J., Vilei, E. (2003). Antigenic and Genetic Characterisation of Lipoprotein Lppc From Mycoplasma Mycoides Subsp. Mycoides SC. Vet. Res. 34 (6), 761–775. doi: 10.1051/vetres:2003035
Pilo, P., Vilei, E. M., Peterhans, E., Bonvin-Klotz, L., Stoffel, M. H., Dobbelaere, D., et al. (2005). A Metabolic Enzyme as a Primary Virulence Factor of Mycoplasma Mycoides Subsp. Mycoides Small Colony. J. Bacteriol. 187 (19), 6824–6831. doi: 10.1128/JB.187.19.6824-6831
Piscopo, R., Guimarães, R., Ueno, J., Ikeda, F., Di Bella, Z., Girão, M., et al. (2020). "Increased Prevalence of Endocervical Mycoplasma and Ureaplasma Colonization in Infertile Women With Tubal Factor. J. Bras. Reprod. Assist. 24, 152–157. doi: 10.5935/1518-0557.20190078
Polianskaia, G., Efremova, T., Ender, N. A., et al. (1998). Effect of Mycoplasma Contamination of the Human Uterine Leiomyosarcoma Cell Line SK-UT-1B on Karyotype Structure. Tsitologiia 40(1), 23–30.
Poumarat, F., Gautier-Bouchardon, A., Bergonier, D., Gay, E., Tardy, F. (2016). Diversity and Variation in Antimicrobial Susceptibility Patterns Over Time in Mycoplasma Agalactiae Isolates Collected From Sheep and Goats in France. J. Appl. Microbiol. 120 (5), 1208–1218. doi: 10.1111/jam.13083
Pritchard, R. E., Balish, M. F. (2015). Mycoplasma Iowae: Relationships Among Oxygen, Virulence, and Protection From Oxidative Stress. Vet. Res. 46 (1), 36. doi: 10.1186/s13567-015-0170-7
Qi, J., Guo, A., Cui, P., Chen, Y., Mustafa, R., Ba, X., et al. (2012). Comparative Geno-Plasticity Analysis of Mycoplasma Bovis HB0801 (Chinese Isolate). PloS One 7 (5), e38239. doi: 10.1371/journal.pone.0038239
Qin, L., Chen, Y., You, X. (2019). Subversion of the Immune Response by Human Pathogenic Mycoplasmas. Front. Microbiol. 10, 1934. doi: 10.3389/fmicb.2019.01934
Ramos, E. I., Das, K., Harrison, A. L., Garcia, A., Gadad, S. S., Dhandayuthapani, S. (2021). Mycoplasma Genitalium and M. Pneumoniae Regulate a Distinct Set of Protein-Coding Genes in Epithelial Cells. Front. Immunol. 12. doi: 10.3389/fimmu.2021.738431
Raymond, B. B., Jenkins, C., Turnbull, L., Whitchurch, C. B., Djordjevic, S. P. (2018). Extracellular DNA Release From the Genome-Reduced Pathogen Mycoplasma Hyopneumoniae is Essential for Biofilm Formation on Abiotic Surfaces. Sci. Rep. 8 (1), 1–12. doi: 10.1038/s41598-018-28678-2
Razin, S. (1992). “Mycoplasma Taxonomy and Ecology,” in Mycoplasmas: Molecular Biology and Pathogenesis. Eds. Maniloff, J., McElhaney, R. N., Finch, L. R., Baseman, J. B. (Washington, D.C: American Society for Microbiology), 3–22.
Razin, S., Jacpbs, E. (1992). Mycoplasma Adhesion. Microbiology 138 (3), 407–422. doi: 10.1099/00221287-138-3-407
Razin, S., Yogev, D., Naot, Y. (1998). Molecular Biology and Pathogenicity of Mycoplasmas. Microbiol. Mol. Biol. Rev. 62 (4), 1094–1156. doi: 10.1128/MMBR.62.4.1094-1156.1998
Rebollo Couto, M. S., Klein, C. S., Voss-Rech, D., Terenzi, H. (2012). Extracellular Proteins of Mycoplasma Synoviae. ISRN Vet. Sci., 802308. doi: 10.5402/2012/802308
Rechnitzer, H., Brzuszkiewicz, E., Strittmatter, A., Liesegang, H., Lysnyansky, I., Daniel, R., et al. (2011). Genomic Features and Insights Into the Biology of Mycoplasma Fermentans. Microbiology 157 (3), 760–773. doi: 10.1099/mic.0.043208-0
Rees-Garbutt, J., Chalkley, O., Landon, S., Purcell, O., Marucci, L., Grierson, C. (2020). Designing Minimal Genomes Using Whole-Cell Models. Nat. Commun. 11 (1), 836. doi: 10.1038/s41467-020-14545-0
Rjeibi, M. R., Darghouth, M. A., Omri, H., Souidi, K., Rekik, M., Gharbi, M. (2015). First Molecular Isolation of Mycoplasma Ovis From Small Ruminants in North Africa. Onderstepoort J. Veterinary Res. 82 (1), 01–05. doi: 10.4102/ojvr.v82i1.912
Roachford, S.E.O., Nelson, K. E., Mohapatra, B. (2019). A Novel Approach for the Identification and Phylogenetic Delineation of Human Mycoplasma Species and Strains Using Genomic Segment Sequence Analysis. Infect. Genet. Evol. 68, 68–76. doi: 10.1016/j.meegid.2018.12.002
Robertson, J., Stemke, G., Davis, J., et al. (2002). Proposal of Ureaplasma Parvum Sp. Nov. And Emended Description of Ureaplasma Urealyticum. Int. J. Syst. Evol. Microbiol. 52 (Pt 2), 587–597. doi: 10.1099/00207713-52-2-587
Robinson, L.-S., Lewis, W.-G., Lewis, A.-L. (2017). The Sialate Oacetylesterase Esta From Gut Bacteroidetes Species Enables Sialidase-Mediated Cross-Species Foraging of 9- O-Acetylated Sialoglycans. J. Biol. Chem. 292 (28), 11861–11872. doi: 10.1074/jbc.M116.769232
Rocha Sobrinho, H. M., Jarach, R., et al. (2011). Mycoplasmal Lipid-Associated Membrane Proteins and Mycoplasma Arthritidis Mitogen Recognition by Serum Antibodies From Patients With Rheumatoid Arthritis. Rheumatol Int. 31 (7), 951–957. doi: 10.1007/s00296-010-1612-1
Rodríguez, F., Ramírez, A., Castro, P., Poveda, J. (2021). Pathological and Immunohistochemical Studies of Experimental Mycoplasma Pneumoniae in Gerbils (Meriones Unguiculatus). J. Comp. Pathol. 184, 37–43. doi: 10.1016/j.jcpa.2021.01.011
Rosanna, Z., Andrea, C., Isabella, B., Francesca, S., Alberto, A., Teresa, A. M., et al. (2020). Immune-Mediated Hemolytic Anemia Associated With Candidatus Mycoplasma Haematoparvum in a Splenectomized Dog in Italy. Acta Veterinaria 70 (2), 277–284. doi: 10.2478/acve-2020-0020
Röske, K., Foecking, M., Yooseph, S., Glass, J., Calcutt, M., Wise, K. (2010). A Versatile Palindromic Amphipathic Repeat Coding Sequence Horizontally Distributed Among Diverse Bacterial and Eucaryotic Microbes. BMC Genomics 11, 430. doi: 10.1186/1471-2164-11-430
Rüger, N., Sid, H., Meens, J., Szostak, M. P., Baumgärtner, W., Bexter, F., et al. (2021). New Insights Into the Host-Pathogen Interaction of Mycoplasma Gallisepticum and Avian Metapneumovirus in Tracheal Organ Cultures of Chicken. Microorganisms 9 (11). doi: 10.3390/microorganisms9112407
Sacchini, F., Naessens, J., Awino, E., Heller, M., Hlinak, A., Haider, W., et al. (2011). A Minor Role of CD4+ T Lymphocytes in the Control of a Primary Infection of Cattle With Mycoplasma Mycoides Subsp. Mycoides. Veterinary Res. 42 (1), 1–10. doi: 10.1186/1297-9716-42-77
Sagné, E., Citti, C., Dordet-Frisoni, E., Bio-protocol and, L., Bio-protocol (2021). Bacterial Conjugation Protocol for Ruminant Mycoplasmas 11, 2. doi: 10.21769/BioProtoc.3893
Santos, O. M., Campos, A. C., Santos, J. P., Santos, P. O. M., Caldas, E. L. C., Santos, A. D. F., et al. (2015). Agalaxia Contagiosa Em Ovinos E Caprinos do Estado De Sergipe: Dados Preliminares. Scientia Plena 11 (4).
Saraya, T., Kurai, D., Nakagaki, K., Sasaki, Y., Niwa, S., Tsukagoshi, H., et al. (2014). Novel Aspects on the Pathogenesis of Mycoplasma Pneumoniae Pneumonia and Therapeutic Implications. Front. Microbiol. 5, 410. doi: 10.3389/fmicb.2014.00410
Sawicka, A., Durkalec, M., Tomczyk, G., Kursa, O. (2020). Occurrence of Mycoplasma Gallisepticum in Wild Birds: A Systematic Review and Meta-Analysis. PloS One 15, e0231545. doi: 10.1371/journal.pone.0231545
Sawicka-Durkalec, A., Tomczyk, G., Kursa, O., Stenzel, T., Gyuranecz, M. (2022). Evidence of Mycoplasma Spp. Transmission by Migratory Wild Geese. Poultry Sci. 101 (1), 101526.
Schott, C., Cai, H., Parker, L., Bateman, K. G., Caswell, J. L. (2014). Hydrogen Peroxide Production and Free Radical-Mediated Cell Stress in Mycoplasma Bovis Pneumonia. J. Comp. Pathol. 150 (2-3), 127–137. doi: 10.1016/j.jcpa.2013.07.008
Segovia, J. A., Chang, T.-H., Winter, V. T., Coalson, J. J., Cagle, M. P., Pandranki, L., et al. (2018). NLRP3 is a Critical Regulator of Inflammation and Innate Immune Cell Response During Mycoplasma Pneumoniae Infection. Infect. Immun. 86 (1), e00548–e00517. doi: 10.1128/IAI.00548-17
Seo, M.-G., Kwon, O.-D., Kwak, D. (2019). Prevalence and Phylogenetic Analysis of Hemoplasma Species in Domestic Pigs in Korea. Parasites Vectors 12 (1), 378. doi: 10.1186/s13071-019-3638-x
Sharifiyazdi, H., Hasiri, M. A., Amini, A. H. (2014). “Intravascular Hemolysis Associated With Candidatus Mycoplasma Hematoparvum in a non-Splenectomized Dog in the South Region of Iran,” in Veterinary Research Forum (Urmia, Iran: Faculty of Veterinary Medicine, Urmia University).
Shaw, B.-M., Daubenspeck, J.-M., Simmons, W.-L., et al. (2013). Eps-I Polysaccharide Protects Mycoplasma Pulmonis From Phagocytosis. FEMS Microbiol. Lett. 338 (2), 155–160. doi: 10.1111/1574-6968.12048
Shibata, K.-i., Hasebe, A., Into, T., Yamada, M., Watanabe, T. (2000). The N-Terminal Lipopeptide of a 44-Kda Membrane-Bound Lipoprotein of Mycoplasma Salivarium is Responsible for the Expression of Intercellular Adhesion Molecule-1 on the Cell Surface of Normal Human Gingival Fibroblasts. J. Immunol. 165 (11), 6538–6544. doi: 10.4049/jimmunol.165.11.6538
Shimizu, T. (2016). Inflammation-Inducing Factors of Mycoplasma Pneumoniae. Front. Microbiol. 7, 414. doi: 10.3389/fmicb.2016.00414
Shimizu, T., Kida, Y., Kuwano, K. (2008). Mycoplasma Pneumoniae-Derived Lipopeptides Induce Acute Inflammatory Responses in the Lungs of Mice. Infect. Immun. 76, 270–277. doi: 10.1128/IAI.00955-07
Shirani, I., Zhang, H., Zhao, G., Lu, S., Marawan, M., Dawood, A., et al. (2020). In Silico Identification of Novel Immunogenic Secreted Proteins of Mycoplasma Bovis From Secretome Data and Experimental Verification. Pathogens 9 (9), 770. doi: 10.3390/pathogens9090770
Silva, P., Marostica, T., McDaniel, A., Arruda, B., Alonso, C., Derscheid, R., et al. (2021). Comparison of Mycoplasma Hyopneumoniae Response to Infection by Route of Exposure. Veterinary Microbiol. 258, 109118. doi: 10.1016/j.vetmic.2021.109118
Siqueira, F., de Morais, G., Higashi, S., Beier, L., Breyer, G., de Sá Godinho, C. P., et al. (2016). Mycoplasma Non-Coding RNA: Identification of Small Rnas and Targets. BMC Genomics 17, 743. doi: 10.1186/s12864-016-3061-z
Siqueira, F. M., Thompson, C. E., Virginio, V. G., Gonchoroski, T., Reolon, L., Almeida, L. G., et al. (2013). New Insights on the Biology of Swine Respiratory Tract Mycoplasmas From a Comparative Genome Analysis. BMC Genomics 14 (1), 175. doi: 10.1186/1471-2164-14-175
Sokoli, A., Groebel, K., Hoelzle, K., Amselgruber, W. M., Mateos, J. M., Schneider, M. K., et al. (2013). Mycoplasma Suis Infection Results Endothelial Cell Damage and Activation: New Insight Into the Cell Tropism and Pathogenicity of Hemotrophic Mycoplasma. Veterinary Res. 44 (1), 1–12. doi: 10.1186/1297-9716-44-6
Sonalio, K., Perles, L., Gatto, I., do Amaral, R., Almeida, H., Galdeano, J., et al. (2020). Genetic Diversity of Emerging Hemotropic Mycoplasmas in Domestic Pigs From Brazil. Transbound Emerg. Dis. doi: 10.1111/tbed.13767
Song, Q., Song, W., Zhang, W., et al. (2018). Identification of Erythrocyte Membrane Proteins Interacting With Mycoplasma Suis GAPDH and OSGEP. Res. Vet. Sci. 119, 85–90. doi: 10.1016/j.rvsc.2018.05.001
Song, M., Zhang, Y., Li, S., Zhang, C., Tao, M., Tang, Y., et al. (2017). A Sensitive and Rapid Immunoassay for Mycoplasma Pneumoniae in Children With Pneumonia Based on Single-Walled Carbon Nanotubes. Sci. Rep. 7 (1), 1–7. doi: 10.1038/s41598-017-16652-3
Song, Q., Zhang, W., Song, W., Liu, Z., Khan, M. K., He, L., et al. (2014). Seroprevalence and Risk Factors of Mycoplasma Suis Infection in Pig Farms in Central China. Prev. Veterinary Med. 117 (1), 215–221. doi: 10.1016/j.prevetmed.2014.07.006
Sordillo, L. M., Raphael, W. (2013). Significance of Metabolic Stress, Lipid Mobilization, and Inflammation on Transition Cow Disorders. Vet. Clin. North Am. Food Anim. Pract. 29 (2), 267–278. doi: 10.1016/j.cvfa.2013.03.002
Splitter, E. J. (1950). Eperythrozoon Suis N. Sp. And Eperythrozoon Parvum N. Sp., 2 New Blood Parasites of Swine. Science 111 (2889), 513–514. doi: 10.1126/science.111.2889.513
Sponheim, A., Alvarez, J., Fano, E., Schmaling, E., Dee, S., Hanson, D., et al. (2020). Comparison of the Sensitivity of Laryngeal Swabs and Deep Tracheal Catheters for Detection of Mycoplasma Hyopneumoniae in Experimentally and Naturally Infected Pigs Early and Late After Infection. Veterinary Microbiol. 241, 108500. doi: 10.1016/j.vetmic.2019.108500
Stadler, J., Ade, J., Ritzmann, M., Hoelzle, K., Hoelzle, L. E. (2020). Detection of a Novel Haemoplasma Species in Fattening Pigs With Skin Alterations, Fever and Anaemia. Veterinary Rec. doi: 10.1136/vr.105721
Steffen, M., Ebersole, J. (1992). Secretory Immune Responses to Mycoplasma Pulmonis. Infect. Immun. 60 (2), 337–344. doi: 10.1128/iai.60.2.337-344.1992
Stemke, G., Laigret, F., Grau, O., Bové, J. (1992). Phylogenetic Relationships of Three Porcine Mycoplasmas, Mycoplasma Hyopneumoniae, Mycoplasma Flocculare, and Mycoplasma Hyorhinis, and Complete 16S Rrna Sequence of M. Flocculare. Int. J. Systematic Evolutionary Microbiol. 42 (2), 220–225. doi: 10.1099/00207713-42-2-220
Stoffregen, W., Alt, D., Palmer, M., Olsen, S., Waters, W., Stasko, J. (2006). Identification of a Haemomycoplasma Species in Anemic Reindeer (Rangifer Tarandus). J. Wildlife Dis. 42 (2), 249–258. doi: 10.7589/0090-3558-42.2.249
Stol, K., Jans, J., Ott de Bruin, L., Unger, W., van Rossum, A. (2021). Perinatal Infections With Ureaplasma. Pediatr. Infect. Dis. J. 40 (5S), S26–S30. doi: 10.1097/inf.0000000000002859
Suleman, M., Prysliak, T., Clarke, K., Burrage, P., Windeyer, C., Perez-Casal, J. (2016). Mycoplasma Bovis Isolates Recovered From Cattle and Bison (Bison Bison) Show Differential In Vitro Effects on PBMC Proliferation, Alveolar Macrophage Apoptosis and Invasion of Epithelial and Immune Cells. Veterinary Microbiol. 186, 28–36. doi: 10.1016/j.vetmic.2016.02.016
Sulyok, K., Kreizinger, Z., Bekő et al, K. (2019). Development of Molecular Methods for Rapid Differentiation of Mycoplasma Gallisepticum Vaccine Strains From Field Isolates. J. Clin. Microbiol. 57 (6), e01084–e01018. doi: 10.1128/JCM.01084-18
Suzuki, N., Chen, N. J., Millar, D. G., Suzuki, S., Horacek, T., Hara, H., et al. (2003). IL-1 Receptor-Associated Kinase 4 is Essential for IL-18-Mediated NK and Th1 Cell Responses. J. Immunol. 170 (8), 4031–4035. doi: 10.4049/jimmunol.170.8.4031
Sweeney, E. L., Kallapur, S. G., Meawad, S., Gisslen, T., Stephenson, S.-A., Jobe, A. H., et al. (2017). Ureaplasma Species Multiple Banded Antigen (MBA) Variation is Associated With the Severity of Inflammation In Vivo and In Vitro in Human Placentae. Front. Cell. Infect. Microbiol. 7, 123. doi: 10.3389/fcimb.2017.00123
Tamiozzo, P. (2021). Mycoplasma Maculosum and Mycoplasma Spumans Associated With Fertility Disorders in Dogs From a Bernese Mountain Dog Kennel. Rev. Argent. Microbiología. doi: 10.1016/j.ram.2021.04.001
Tardy, F., Mick, V., Dordet-Frisoni, E., Marenda, M. S., Sirand-Pugnet, P., Blanchard, A., et al. (2015). Integrative Conjugative Elements are Widespread in Field Isolates of Mycoplasma Species Pathogenic for Ruminants. Appl. Environ. Microbiol. 81 (5), 1634–1643. doi: 10.1128/AEM.03723-14
Tasker, S., Helps, C., Day, M., Harbour, D., Shaw, S., Harrus, S., et al. (2003). Phylogenetic Analysis of Hemoplasma Species: An International Study. J. Clin. Microbiol. 41 (8), 3877–3880. doi: 10.1128/JCM.41.8.3877-3880.2003
Teh, H., Ho, M., Williams, L. D. (1988). Suppression of Cytotoxic Responses by a Supernatant Factor Derived From Mycoplasma Hyorhinis-Infected Mammalian Cell Lines. Infect. Immun. 56 (1), 197–203. doi: 10.1128/iai.56.1.197-203.1988
Thiaucourt, F., Dedieu, L., Maillard, J., Bonnet, P., Lesnoff, M., Laval, G., et al. (2003). Contagious Bovine Pleuropneumonia Vaccines, Historic Highlights, Present Situation and Hopes. Developments Biologicals 114, 147–160.
Tortschanoff, M., Aurich, C., Rosengarten, R., Spergser, J. (2005). Phase and Size Variable Surface-Exposed Proteins in Equine Genital Mycoplasmas. Veterinary Microbiol. 110, 301–306. doi: 10.1016/j.vetmic.2005.08.002
Totten, A., Xiao, L., Crabb, D., Ratliff, A., Waites, K., Hwangpo, T., et al. (2021). Septic Polyarthritis With Mycoplasma Salivarium in a Patient With Common Variable Immunodeficiency: Case Report and Review of the Literature. Access Microbiol. 3 (4). doi: 10.1099/acmi.0.000221
Tourtellotte, M., Lein, D. (1976). Infertility of Cattle Caused by Mycoplasmas. Health Lab. Sci. 13 (2), 152–158.
Tsai, H. J., Lee, C. Y. (2006). Serological Survey of Racing Pigeons for Selected Pathogens in Taiwan. Acta Veterinaria Hungarica 54, 179–189. doi: 10.1556/AVet.54.2006.2.5
Tsai, T.-A., Tsai, C.-K., Kuo, K.-C., Yu, H.-R. (2021). Rational Stepwise Approach for Mycoplasma Pneumoniae Pneumonia in Children. J. Microbiology Immunol. Infect. 54 (4), 557–565. doi: 10.1016/j.jmii.2020.10.002
Tsai, S., Wear, D. J., Shih, J., Lo, S.-C. (1995). Mycoplasmas and Oncogenesis: Persistent Infection and Multistage Malignant Transformation. Proc. Natl. Acad. Sci. 92 (22), 10197–10201. doi: 10.1073/pnas.92.22.10197
Tzani-Tzanopoulou, P., Skliros, D., Megremis P. Xepapadaki, S., Andreakos, E., Chanishvili, N., Flemetakis, E., et al. (2021). Interactions of Bacteriophages and Bacteria at the Airway Mucosa: New Insights Into the Pathophysiology of Asthma. Front. Allergy 1, 617240. doi: 10.3389/falgy.2020.617240
Uchida-Fujii, E., Kinoshita, Y., Niwa, H., Maeda, T., Nukada, T., Ueno, T. (2021). High Prevalence of Mycoplasma Equirhinis in Thoroughbred Horses With Respiratory Symptoms in Autumn 2018. J. Veterinary Med. Sci., 21–0163. doi: 10.1292/jvms.21-0163
Urie, N. J., Highland, M. A., Knowles, D. P., Branan, M. A., Herndon, D. R., Marshall, K. L. (2019). Mycoplasma Ovis Infection in Domestic Sheep (Ovis Aries) in the United States: Prevalence, Distribution, Associated Risk Factors, and Associated Outcomes. Prev. Veterinary Med. 171, 104750. doi: 10.1016/j.prevetmed.2019.104750
US Animal Health Association (2006). Report of the Committee on Transmissible Diseases of Poultry and Other Avian Species (Richmond, VA), 314–339.
USDA (2020). “United States Department of Agriculture. Livestock and Poultry,” in United States of America: World Markets and Trade; 2020.
Valentine-King, M. A., Cisneros, K., James, M. O., Huigens, R. W., Brown, M. B. (2020). Efficacy Data of Halogenated Phenazine and Quinoline Agents and an NH125 Analogue to Veterinary Mycoplasmas. BMC Veterinary Res. 16, 1–11. doi: 10.1186/s12917-020-02324-4
van der Merwe, J., Prysliak, T., Perez-Casal, J. (2010). Invasion of Bovine Peripheral Blood Mononuclear Cells and Erythrocytes by Mycoplasma Bovis. Infect. Immun. 78 (11), 4570–4578. doi: 10.1128/IAI.00707-10
Vangroenweghe, F. (2021). Convenience and Economic Benefit of Early One-Shot Mycoplasma Hyopneumoniae Vaccination at 3 Days of Age in a Commercial Sow Farm. J. Vaccines Immunol. 7 (1), 020–026. doi: 10.17352/jvi.000041
Vilei, E. M., Frey, J. (2001). Genetic and Biochemical Characterization of Glycerol Uptake in Mycoplasma Mycoides Subsp. Mycoides Sc: Its Impact on H2o2production and Virulence. Clin. Diagn. Lab. Immunol. 8 (1), 85–92.
Vizarraga, D., Torres-Puig, S., Aparicio, D., Pich, O. (2021). The Sialoglycan Binding Adhesins of Mycoplasma Genitalium and Mycoplasma Pneumoniae. Trends Microbiol. 29 (6), 477–481. doi: 10.1016/j.tim.2021.01.011
Voelker, L. L., Dybvig, K. (1998). Characterization of the Lysogenic Bacteriophage MAV1 From Mycoplasma Arthritidis. J. Bacteriology 180 (22), 5928–5931. doi: 10.1128/JB.180.22.5928-5931.1998
Vogl, G., Plaickner, A., Szathmary, S., Stipkovits, L., Rosengarten, R., Szostak, M. P. (2008). Mycoplasma Gallisepticum Invades Chicken Erythrocytes During Infection. Infect. Immun. 76 (1), 71–77. doi: 10.1128/iai.00871-07
Wadher, B., Henderson, C., Miles, R., Varsani, H. (1990). "a Mutant of Mycoplasma Mycoides Subsp. Mycoides Lacking the H2O2- Producing Enzyme L-Alpha-Glycerophosphate Oxidase. FEMS Microbiol. Lett. 60 (1-2), 127–130.
Waites, K. B., Katz, B., Schelonka, R. L. (2005). Mycoplasmas and Ureaplasmas as Neonatal Pathogens. Clin. Microbiol. Rev. 18 (4), 757–789. doi: 10.1128/cmr.18.4.757-789.2005
Waites, K. B., Talkington, D. F. (2004). Mycoplasma Pneumoniae and its Role as a Human Pathogen. Clin. Microbiol. Rev. 17 (4), 697–728. doi: 10.1128/CMR.17.4.697-728.2004
Waites, K. B., Xiao, L., Liu, Y., Balish, M. F., Atkinson, T. P. (2017). Mycoplasma Pneumoniae From the Respiratory Tract and Beyond. Clin. Microbiol. Rev. 30 (3), 747–809. doi: 10.1128/CMR.00114-16
Wang, X., Cui, Y., Zhang, Y., Shi, K., Yan, Y., Jian, F., et al. (2017). Molecular Characterization of Hemotropic Mycoplasmas (Mycoplasma Ovis and ‘Candidatus Mycoplasma Haemovis’) in Sheep and Goats in China. BMC Veterinary Res. 13 (1), 142. doi: 10.1186/s12917-017-1062-z
Wang, J., Li, Y., Pan, L., et al. (2021b). Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) Moonlights as an Adhesin in Mycoplasma Hyorhinis Adhesion to Epithelial Cells as Well as a Plasminogen Receptor Mediating Extracellular Matrix Degradation. Vet. Res. 52 (80). doi: 10.1186/s13567-021-00952-8
Wang, H., Ren, D., Li, H., Wang, S. (2021a). Periprosthetic Joint Infection Caused by Mycoplasma Hominis, Diagnosed Using Metagenomic Sequencing. Int. J. Gen. Med. 14, 7003–7006. doi: 10.2147/ijgm.s330924
Wang, Y., Wang, Y., Jiao, W., et al. (2019a). Development of Loop-Mediated Isothermal Amplification Coupled With Nanoparticle-Based Lateral Flow Biosensor Assay for Mycoplasma Pneumoniae Detection. AMB Expr 9 (196). doi: 10.1186/s13568-019-0921-3
Wang, Y., Wang, Y., Quan, S., Jiao, W., Li, J., Sun, L., et al. (2019b). Establishment and Application of a Multiple Cross Displacement Amplification Coupled With Nanoparticle-Based Lateral Flow Biosensor Assay for Detection of Mycoplasma Pneumoniae. Front. Cell. Infect. Microbiol. 9, 325. doi: 10.3389/fcimb.2019.00325
Wang, Y., Wang, Y., Zhang, L., Liu, D., Luo, L., Li, H. (2016). Multiplex, Rapid, and Sensitive Isothermal Detection of Nucleic-Acid Sequence by Endonuclease Restriction-Mediated Real-Time Multiple Cross Displacement Amplification. Front. Microbiol. 7, 753. doi: 10.3389/fmicb.2016.00753
Wang, Y., Zhang, Y., Lu, W., Wang, L. (2018). Serum Tumor Necrosis Factor-α and Interferon-γ Levels in Pediatric Mycoplasma Pneumoniae Pneumonia: A Systematic Review and Meta-Analysis. Can. Respir. J 2018.
Washburn, L., Miller, E., Mukherjee, S., Dannenbring, D. (2004). Mycoplasma Arthritidis Bacteriophage MAV1 Prophage Integration, Deletions, and Strain-Related Polymorphisms. Plasmid 52 (1), 31–47. doi: 10.1016/j.plasmid.2004.04.004
Watanabe, Y., Fujihara, M., Obara, H., Matsubara, K., Yamauchi, K., Harasawa, R. (2010). Novel Hemoplasma Species Detected in Free-Ranging Sika Deer (Cervus Nippon). J. Veterinary Med. Sci., 1007020280–1007020280. doi: 10.1292/jvms.10-0229
Weldearegay, Y. B. (2015). In Vivo and In Vitro Characterization of Mycoplasma Mycoides Subspecies Mycoides by Transcriptomic and Proteomic Analysis.
Weldearegay, Y., Müller, S., Hänske, J., Schulze, A., Kostka, A., Rüger, N., et al. (2019). Host-Pathogen Interactions of Mycoplasma Mycoides in Caprine and Bovine Precision-Cut Lung Slices (PCLS) Models. Pathogens 8 (2), 82. doi: 10.3390/pathogens8020082
Weng, J., Li, Y., Cai, L., Li, T., Peng, G., Fu, C., et al. (2017). Elimination of Mycoplasma Contamination From Infected Human Hepatocyte C3A Cells by Intraperitoneal Injection in BALB/C Mice. Front. Cell. Infect. Microbiol. 7, 440. doi: 10.3389/fcimb.2017.00440
Wesonga, H., Thiaucourt, F. (2000). Experimental Studies on Efficacy of T1 Sr and T1/44 Vaccine Strains of Mycoplasma Mycoides Subsp Mycoides (Small Colony). Rev. Elev Méd Vét Pays Trop. 53, 313–318. doi: 10.19182/remvt.9707
Whitson, W. J., Ball, P. A., Lollis, S. S., Balkman, J. D., Bauer, D. F. (2014). Postoperative Mycoplasma Hominis Infections After Neurosurgical Intervention: A Review. J. Neurosurgery: Pediatr. 14 (2), 212–218. doi: 10.3171/2014.4.PEDS13547
Whittle, E., Leonard, M. O., Harrison, R., Gant, T. W., Tonge, D. P. (2018). Multi-Method Characterization of the Human Circulating Microbiome. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.03266
Widjaja, M., Berry, I.-J., Jarocki, V.-M., et al. (2020). Cell Surface Processing of the P1 Adhesin of Mycoplasma Pneumoniae Identifies Novel Domains That Bind Host Molecules. Sci. Rep. 10 (1), 63–84. doi: 10.1038/s41598-020-63136-y
Widjaja, M., Harvey, K. L., Hagemann, L., Berry, I. J., Jarocki, V. M., Raymond, B. B. A., et al. (2017). Elongation Factor Tu is a Multifunctional and Processed Moonlighting Protein. Sci. Rep. 7 (1), 1–17. doi: 10.1038/s41598-017-10644-z
Willi, B., Boretti, F. S., Baumgartner, C., Tasker, S., Wenger, B., Cattori, V., et al. (2006). Prevalence, Risk Factor Analysis, and Follow-Up of Infections Caused by Three Feline Hemoplasma Species in Cats in Switzerland. J. Clin. Microbiol. 44 (3), 961–969. doi: 10.1128/JCM.44.3.961-969.2006
Wodke, J. A., Alibés, A., Cozzuto, L., Hermoso, A., Yus, E., Lluch-Senar, M., et al. (2015). Mympn: A Database for the Systems Biology Model Organism Mycoplasma Pneumoniae. Nucleic Acids Res. 43 (Database issue), D618–D623. doi: 10.1093/nar/gku1105
Woese, C., Maniloff, J., Zablen, L. (1980). Proc. Natl. Acad. Sci. U.S.A. 77, 494–498. doi: 10.1073/pnas.77.1.494
Wohlman, A., Yirmiya, R., Gallily, R., Weidenfeld, J. (2001). Effect of Mycoplasma Fermentans on Brain PGE2: Role of Glucocorticoids and Their Receptors. Neuroimmunomodulation 9 (3), 141–147. doi: 10.1159/000049018
Wood, J., Chanter, N., Newton, J., Burrell, M., Dugdale, D., Windsor, H., et al. (1997). An Outbreak of Respiratory Disease in Horses Associated With Mycoplasma Felis Infection. Vet. Rec. 140, 388 –3391. doi: 10.1136/vr.140.15.388
Wood, G., Iverson-Cabral, S., et al. (2013). Persistence, Immune Response, and Antigenic Variation of Mycoplasma Genitalium in an Experimentally Infected Pig-Tailed Macaque (Macaca Nemestrina). Infect. Immun. 81 (8), 2938–2951. doi: 10.1128/IAI.01322-12
Wood, G., Iverson-Cabral, S., Gillespie, C., Lowens, M., Manhart, L., Totten, P. (2020). Sequence Variation and Immunogenicity of the Mycoplasma Genitalium Mgpb and Mgpc Adherence Proteins During Persistent Infection of Men With non-Gonococcal Urethritis. PloS One 15 (10), e0240626. doi: 10.1371/journal.pone.0240626
Wrobel, E., Wilcoxen, T., Nuzzo, J. (2016). Seitz Seroprevalence of Avian Pox and Mycoplasma Gallisepticum in Raptors in Central Illinois. J. Raptor Res. 50, 289–294. J. doi: 10.3356/JRR-15-43.1
Wu, X., Zhang, S., Long, C., An, Z., Xing, X., Wen, F., et al. (2021). Mycoplasmas Bovis P48 Induces Apoptosis in EBL Cells via an Endoplasmic Reticulum Stress-Dependent Signaling Pathway. Veterinary Microbiol. 225, 109013. doi: 10.1016/j.vetmic.2021.109013
Xie, X., Hao, F., Chen, R., Wang, J., Wei, Y., Liu, J., et al. (2021). Nicotinamide Adenine Dinucleotide-Dependent Flavin Oxidoreductase of Mycoplasma Hyopneumoniae Functions as a Potential Novel Virulence Factor and Not Only as a Metabolic Enzyme. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.747421
Xin, J., Li, Y., Nicholas, R. A., Chen, C., Liu, Y., Zhang, M.-j., et al. (2012). A History of the Prevalence and Control of Contagious Bovine Pleuropneumonia in China. Veterinary J. 191 (2), 166–170. doi: 10.1016/j.tvjl.2011.02.011
Xiong, Q., Wang, J., Ji, Y., Ni, B., Zhang, B., Ma, Q., et al. (2016). The Functions of the Variable Lipoprotein Family of Mycoplasma Hyorhinis in Adherence to Host Cells. Veterinary Microbiol. 186, 82–89. doi: 10.1016/j.vetmic.2016.01.017
Xu, F., Zhang, C., Zou, Z., Fan, E. K. Y., Chen, L., Li, Y., et al. (2017). Aging-Related Atg5 Defect Impairs Neutrophil Extracellular Traps Formation. Immunology, 417–432, 151(4). doi: 10.1111/imm.12740
Yacoub, E., Saed Abdul-Wahab, O., et al. (2021). The Relationship Between Mycoplasmas and Cancer: Is it Fact or Fiction ? Narrative Review and Update on the Situation. J. Oncol. 2021, 9986550. doi: 10.1155/2021/9986550
Yadav, J., Tomar, P., Singh, Y., Khurana, S. (2021). Insights on Mycoplasma Gallisepticum and Mycoplasma Synoviae Infection in Poultry: A Systematic Review. Anim. Biotechnol. doi: 10.1080/10495398.2021.1908316
Yamamoto, T., Kida, Y., Sakamoto, Y., Kuwano, K. (2017). Mpn491, a Secreted Nuclease of Mycoplasma Pneumoniae, Plays a Critical Role in Evading Killing by Neutrophil Extracellular Traps. Cell. Microbiol. 19 (3), e12666.
Yamazaki, T., Kenri, T. (2016). Epidemiology of Mycoplasma Pneumoniae Infections in Japan and Therapeutic Strategies for Macrolide-Resistant M. Pneumoniae. Front. Microbiol. 7, 693. doi: 10.3389/fmicb.2016.00693
Yáñez, A., Martínez-Ramos, A., Calixto, T., et al. (2013). Animal Model of Mycoplasma Fermentans Respiratory Infection. BMC Res. Notes 6 (9). doi: 10.1186/1756-0500-6-9
Yang, H.-J. (2019). Benefits and Risks of Therapeutic Alternatives for Macrolide Resistant Mycoplasma Pneumoniae Pneumonia in Children. Korean J. Pediatr. 62 (6), 199. doi: 10.3345/kjp.2018.07367
Yang, S., Ahn, Y., Oh, T., Cho, H., Park, K., Chae, C. (2021a). “Field Evaluation of a Sing-Dose Bivalent Vaccine of Porcine Circovirus Type 2b and Mycoplasma Hyopneumoniae,” in Veterinary Medicine and Sci, vol. 7., 755–765. doi: 10.1002/vms3.420
Yang, T.-I., Chang, T.-H., Lu, C.-Y., Chen, J.-M., Lee, P.-I., Huang, L.-M., et al. (2019a). Mycoplasma Pneumoniae in Pediatric Patients: Do Macrolide-Resistance and/or Delayed Treatment Matter? J. Microbiol. Immunol. Infect. 52 (2), 329–335. doi: 10.1016/j.jmii.2018.09.009
Yang, S., Oh, T., Park, K., Cho, H., Suh, J., Chae, C. (2021b). Experimental Efficacy of a Trivalent Vaccine Containing Porcine Circovirus Types 2a/B (PCV2a/B) and Mycoplasma Hyopneumoniae Against PCV2d and M. Hyopneumoniae Challenges. Veterinary Microbiol. 258, 109100. doi: 10.1016/j.vetmic.2021.109100
Yang, Y., Song, W., Chen, Z., Li, Q., Liu, L. (2019b). Ameliorative Effect of Synthesized Silver Nanoparticles by Green Route Method From Zingiber Zerumbet on Mycoplasmal Pneumonia in Experimental Mice. Artif. cells nanomedicine Biotechnol. 47 (1), 2146–2154. doi: 10.1080/21691401.2019.1620757
Yavlovich, A., Tarshis, M., Rottem, S. (2004). Internalization and Intracellular Survival of Mycoplasma Pneumoniae by non-Phagocytic Cells. FEMS Microbiol. Lett. 233 (2), 241–246. doi: 10.1111/j.1574-6968.2004.tb09488.x
Yiwen, C., Yueyue, W., Lianmei, Q., Cuiming, Z., Xiaoxing, Y. (2021). Infection Strategies of Mycoplasmas: Unraveling the Panoply of Virulence Factors. Virulence 12 (1), 788–817. doi: 10.1080/21505594.2021.1889813
You, Q., Eidt, J., Bell-Rogers, P., Cai, H. Y. (2020). Diversity of Mycoplasma Hyopneumoniae Strains From Pigs Across Ontario, Canada. J. Veterinary Diagn. Invest. 32 (1), 128–131. doi: 10.1177/1040638719896283
Young, W., Hine, B. C., Wallace, O. A., Callaghan, M., Bibiloni, R. (2015). Transfer of Intestinal Bacterial Components to Mammary Secretions in the Cow. PeerJ 3, e888. doi: 10.7717/peerj.888
Yuan, X., Bai, C., Cui, Q., Zhang, H., Yuan, J., Niu, K. (2018). Rapid Detection of Mycoplasma Pneumoniae by Loop-Mediated Isothermal Amplification Assay. Med. (Baltimore) 97, 10806.
Yus, E., Maier, T., Michalodimitrakis, K., van Noort, V., Yamada, T., Chen, W.-H., et al. (2009). Impact of Genome Reduction on Bacterial Metabolism and its Regulation. science 326 (5957), 1263–1268.
Zbinden, C., Pilo, P., Frey, J., Bruckmaier, R., Wellnitz, O. (2015). The Immune Response of Bovine Mammary Epithelial Cells to Live or Heat-Inactivated Mycoplasma Bovis. Vet. Microbiol. 179 (3-4), 336–340. doi: 10.1016/j.vetmic.2015.07.007
Zecconi, A., dell’Orco, F., Rizzi, N., Vairani, D., Cipolla, M., Pozzi, P., et al. (2020). Cross-Sectional Study on the Prevalence of Contagious Pathogens in Bulk Tank Milk and Their Effects on Somatic Cell Counts and Milk Yield. Ital. J. Anim. Sci. 19, 66–74. doi: 10.1080/1828051X.2019.1693282
Zella, D., Curreli, S., Benedetti, F., Krishnan, S., Cocchi, F., Latinovic, O. S., et al. (2018). Mycoplasma Promotes Malignant Transformation In Vivo, and its Dnak, a Bacterial Chaperone Protein, has Broad Oncogenic Properties. Proc. Natl. Acad. Sci. 115 (51), E12005–E12014. doi: 10.1073/pnas.1815660115
Zhang, R., Han, X., Chen, Y., Mustafa, R., Qi, J., Chen, X., et al. (2014). Attenuated Mycoplasma Bovis Strains Provide Protection Against Virulent Infection in Calves. Vaccine 32 (25), 3107–3114. doi: 10.1016/j.vaccine.2013.12.004
Zhang, H., Hu, G., Lu, D., Zhao, G., Zhang, Y., Zubair, M., et al. (2021a). Comparative Secretome Analyses of Mycoplasma Bovis Virulent and Attenuated Strains Revealed Mbovp0145 as a Promising Diagnostic Biomarker. Front. Veterinary Sci. 8. doi: 10.3389/fvets.2021.666769
Zhang, S., Tsai, S., et al. (2004). Mycoplasma Fermentans Infection Promotes Immortalization of Human Peripheral Blood Mononuclear Cells in Culture. Blood 104 (13), 4252–4259. doi: 10.1182/blood-2004-04-1245
Zhang, S., Wear, D. J., Lo, S.-C. (2000). Mycoplasmal Infections Alter Gene Expression in Cultured Human Prostatic and Cervical Epithelial Cells. FEMS Immunol. Med. Microbiol. 27 (1), 43–50. doi: 10.1111/j.1574-695X.2000.tb01410.x
Zhang, Y., Zhang, Z., Lou, Y., Yu, Y. (2021b). Prevalence of Hemoplasmas and Bartonella Species in Client-Owned Cats in Beijing and Shanghai, China. J. Vet. Med. Sci. 83 (5), 793–797. doi: 10.1292/jvms.20-0681
Zhang, H., Zhang, Y., Wang, Z., Liu, M., Wang, P., Wu, W., et al. (2021c). MBOVPG45_0375 Encodes an Igg-Binding Protein and MBOVPG45_0376 Encodes an Igg-Cleaving Protein in Mycoplasma Bovis. Front. Vet. Sci. 8, 644224. doi: 10.3389/fvets.2021.644224
Zhang, H., Zhao, G., Guo, Y., Menghwar, H., Chen, Y., Chen, H., et al. (2016). Mycoplasma Bovis MBOV_RS02825 Encodes a Secretory Nuclease Associated With Cytotoxicity. Int. J. Mol. Sci. 17 (5), 628. doi: 10.3390/ijms17050628
Zhao, G., Zhang, H., Chen, X., Zhu, X., Guo, Y., He, C., et al. (2017). Mycoplasma Bovis NADH Oxidase Functions as Both a NADH Oxidizing and O2 Reducing Enzyme and an Adhesion. Sci. Rep. 7, 44. doi: 10.1038/s41598-017-00121-y
Zhao, Q., Zhang, T., Zhu, B., Bi, Y., Jiang, S. W., Zhu, Y., et al. (2021b). Increasing Age Affected Polymorphonuclear Neutrophils in Prognosis of Mycoplasma Pneumoniae Pneumonia. J. Inflamm. Res. 14, 3933–3943. doi: 10.2147/jir.s321656
Zhao, G., Zhu, X., Zhang, H., Chen, Y., Schieck, E., Hu, C., et al. (2021a). Novel Secreted Protein of Mycoplasma Bovis Mbovp280 Induces Macrophage Apoptosis Through CRYAB. Front. Immunol. 12, 619362. doi: 10.3389/fimmu.2021.619362
Zhou, Y., Wang, Y., Li, Y., Nick, N., Zou, X., Bai, F., et al. (2016). P19 Contributes to Mycoplasma Mycoides Subsp. Mycoides Adhesion to EBL Cells. Microbial Pathogenesis 93, 13–21. doi: 10.1016/j.micpath.2016.01.011
Zhu, X., Baranowski, E., Dong, Y., Li, X., Hao, Z., Zhao, G., et al. (2020a). An Emerging Role for Cyclic Dinucleotide Phosphodiesterase and Nanornase Activities in Mycoplasma Bovis: Securing Survival in Cell Culture. PloS Pathog. 16 (6), e1008661. doi: 10.1371/journal.ppat.1008661
Zhu, X., Dong, Y., Baranowski, E., et al. (2020b). Mbov_0503 Encodes a Novel Cytoadhesin That Facilitates Mycoplasma Bovis Interaction With Tight Junctions. Microorganisms 8(2), 164. doi: 10.3390/microorganisms8020164
Zhu, X., Dordet-Frisoni, E., Gillard, L., Ba, A., Hygonenq, M.-C., Sagné, E., et al. (2019). Extracellular DNA: A Nutritional Trigger of Mycoplasma Bovis Cytotoxicity. Front. Microbiol. 10 (27-53). doi: 10.3389/fmicb.2019.02753
Zhu, T., Liu, H., Su, L., Xiong, X., Wang, J., Xiao, Y., et al. (2021b). Microrna-18b-5p Downregulation Favors Mycobacterium Tuberculosis Clearance in Macrophages via HIF-1α by Promoting an Inflammatory Response. ACS Infect. Dis. 7 (4), 800–810. doi: 10.1021/acsinfecdis.0c00650
Zhu, M., Nan, Y., Zhai, M., Wang, M., Shao, Y., Blair, H., et al. (2021a). Comparative Profiling of the Resistance of Different Genotypes of Mannose-Binding Lectin to Mycoplasma Pneumoniae Infection in Chinese Merino Sheep Based on High-Throughput Sequencing Technology. Veterinary Immunol. Immunopathol. 223, 110183. doi: 10.1016/j.vetimm.2021.110183
Zhu, C., Wang, S., et al. (2012). Protective Efficacy of a Mycoplasma Pneumoniae P1 CDNA Vaccine Fused With the B Subunit of Escherichia Coli Heat-Labile Enterotoxin. Can. J. Microbiol. 58, 802–810. doi: 10.1139/w2012-051
Zinatizadeh, M. R., Abedini, F., Jafarpour, M., Masoumalinejad, Z. (2017). Identification of Mycoplasma Muris Isolated From Vaginal Samples of NIH Mice. Modern Med. Lab. J. 1 (3), 100–106.
Zubair, M., Khan, F., Menghwar, H., Faisal, M., Ashraf, M., Rasheed, M., et al. (2020b). Progresses on Bacterial Secretomes Enlighten Research on Mycoplasma Secretome. Microbial Pathogenesis 144, 104–160. doi: 10.1016/j.micpath.2020.104160
Keywords: mycoplasmas, pantropic pathogens, virulence factors, clinical implications, gene transfer, vaccination
Citation: Dawood A, Algharib SA, Zhao G, Zhu T, Qi M, Delai K, Hao Z, Marawan MA, Shirani I and Guo A (2022) Mycoplasmas as Host Pantropic and Specific Pathogens: Clinical Implications, Gene Transfer, Virulence Factors, and Future Perspectives. Front. Cell. Infect. Microbiol. 12:855731. doi: 10.3389/fcimb.2022.855731
Received: 15 January 2022; Accepted: 04 April 2022;
Published: 13 May 2022.
Edited by:
Erika Ildiko Lutter, Oklahoma State University, United StatesReviewed by:
Bidyut Mohapatra, The University of the West Indies, BarbadosRobin Nicholas, Consultant, Farnham, United Kingdom
Copyright © 2022 Dawood, Algharib, Zhao, Zhu, Qi, Delai, Hao, Marawan, Shirani and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aizhen Guo, YWl6aGVuQG1haWwuaHphdS5lZHUuY24=
 Ali Dawood
Ali Dawood Samah Attia Algharib
Samah Attia Algharib Gang Zhao
Gang Zhao Tingting Zhu
Tingting Zhu Mingpu Qi
Mingpu Qi Kong Delai
Kong Delai Zhiyu Hao
Zhiyu Hao Marawan A. Marawan
Marawan A. Marawan Ihsanullah Shirani
Ihsanullah Shirani Aizhen Guo
Aizhen Guo