- 1The Second Affiliated Hospital and Yuying Children’s Hospital, Wenzhou Medical University, Wenzhou, China
- 2Key Laboratory of Medical Genetics of Zhejiang Province, Key Laboratory of Laboratory Medicine, Ministry of Education of China, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, China
- 3Institute of Biomedical Informatics, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, China
- 4Institute of Translational Medicine, Baotou Central Hospital, Baotou, China
Research on resistance against polymyxins induced by the mcr-1 gene is gaining interest. In this study, using agar dilution method, polymerase chain reaction, and comparative genomic analysis, we investigated the colistin resistance mechanism of clinical E. coli isolates. The minimum inhibitory concentration (MIC) analysis results revealed that of the 515 isolates tested, bacteria with significantly increased MIC levels against colistin were isolated in 2019. Approximately one-fifth (17.14% to 19.65%) of the isolates showed MIC values ≥1 mg/L against colistin in 2015, 2016, and 2017. However, in 2019, up to three-quarters (74.11%, 146/197) of the isolates showed MIC values ≥1 mg/L against colistin indicating an increase in colistin resistance. Six isolates (EC7518, EC4968, EC3769, EC16, EC117, EC195, 1.13%, 6/515) were found to carry the mcr-1 gene and a novel mcr-1 variant with Met2Ile mutation was identified in EC3769. All six strains showed higher MIC levels (MIC=4 mg/L) than any mcr-1-negative strains (MIC ≤ 2 mg/L). Whole-genome sequencing of the six mcr-1-positive isolates revealed that EC195 carried the highest number of resistance genes (n = 28), nearly a half more than those of the following EC117 (n = 19). Thus, EC195 showed a wider resistance spectrum and higher MIC levels against the antimicrobials tested than the other five isolates. Multi-locus sequence typing demonstrated that these mcr-1-positive strains belonged to six different sequence types. The six mcr-1 genes were located in three different incompatibility group plasmids (IncI2, IncHI2 and IncX4). The genetic context of mcr-1 was related to a sequence derived from Tn6330 (ISApl1-mcr-1-pap2-ISApl1). Investigations into the colistin resistance mechanism and characterization of the molecular background of the mcr genes may help trace the development and spread of colistin resistance in clinical settings.
Introduction
Polymyxins, a class of non-ribosomal polypeptides characterized by the presence of a lipophilic fatty acyl side chain, were produced by Bacillus polymyxa and discovered in the 1940s (Velkov et al., 2013). Due to their neurovirulence and limited renal clearance, polymyxins have been largely abandoned in the clinical treatment of bacterial infections since the 1970s (Ainsworth et al., 1947; Falagas and Kasiakou, 2005; Landman et al., 2008; Hartzell et al., 2009). However, this situation has been changed by the emergence of multidrug-resistant bacteria caused by antibiotic over-consumption (Laxminarayan et al., 2020). Polymyxins, especially polymyxin E (colistin), have been reintroduced as the last recourse against infections caused by gram-negative multidrug-resistant pathogens (Wang Q. et al., 2017). Nonetheless, there is an increasing threat against treatment with polymyxin drugs following the emergence of the plasmid-mediated mobilized colistin resistance (mcr) gene in bacteria. It was first identified and characterized in southern China in 2015, denoted as mcr-1. To date, ten slightly different variants of the mcr-1 gene (mcr-1 to mcr-10) have been identified in different bacteria isolated from animals, foods, farms, humans, and the environment (Liu et al., 2016; Sun et al., 2018; Hussein et al., 2021).
The mcr-1 gene is the most predominant type in MCR family and composed of 1,626 bp in length with a 49% G+C content. It encodes five transmembrane domains and an extracellular catalytic domain. Amino acid sequence analysis revealed that it showed 41% and 40% identities with the lipopolysaccharide export system protein EptA from Neisseria meningitidis and phosphoethanolamine (PEA) transfer EptC from Campylobacter jejuni, respectively, both of which belong to the PEA transferase family (Hu et al., 2016; Stojanoski et al., 2016). Electrospray mass spectrometry and liquid chromatography-mass spectrometry analyses showed that mcr-1 possesses biological activity similar to that of PEA transferase (Hinchliffe et al., 2017). It could modulate the lipid A residues of the lipopolysaccharides (LPS), leading to a lower binding affinity of colistin to its target site (Liu et al., 2016; Hussein et al., 2021).
In spite of that the possible mechanism underlying colistin resistance was preliminarily illuminated, the origin, acquisition, emergence, spread pathway and evolutionary lines are not yet completely understood. Epidemiological investigations have found that mcr-1 rapidly spreads in different ecological niches not only soil, water, wildlife, but also livestock, meat, vegetables, farmland and even humans (Wang et al., 2020; Yang et al., 2021). No less than 10 different bacterial species, including E. coli, Salmonella enterica, Klebsiella pneumoniae, Enterobacter aerogenes, Kluyvera ascorbata, Citrobacter freundii, and Citrobacter braakii, as well as more than 10 types of plasmids, including IncX1, IncHI2, IncFIB, IncFIA, IncFII, IncN, IncR, IncQ1, ColpVC, and IncHI1A, participate in mediating its transmission (Wang et al., 2018; Ragupathi et al., 2019). More worryingly, the “superbugs” evolving from mcr-1 coexisting with other drug resistance genes such as blaNDM-5 may cause a global challenge to the health care systems. Therefore, more effort should be devoted to putting the ax in the helve (Li et al., 2020; Ling et al., 2020; Xiaomin et al., 2020).
In this study, we investigated the colistin resistance phenotype and prevalence of the mcr-1 gene among 515 clinical E. coli isolates from two tertiary hospitals in Zhejiang Province, China. With the genome sequencing of the mcr-1-positive strains, the genetic context of mcr-1 and the structure of mcr-1-harboring plasmids were further analyzed.
Materials and Methods
Bacteria Isolation and Species Identification
A total of 515 non-duplicated clinical E. coli isolates were collected from two tertiary hospitals in Zhejiang Province, China. Among these isolates, 105, 173, 40, and 197 strains were isolated in 2015, 2016, 2017, and 2019, respectively. Based on the analytical profile index, all strains were identified as E. coli using the VITEK 2.0 system (bioMérieux, Durham NC, USA). The six mcr-1-positive E. coli strains were subsequently subjected to species identification verifying by 16S rDNA sequencing with polymerase chain reaction (PCR) primers (Forward: 5′-AGAGTTTGATCCTGGCTCAG-3′; Reverse: 5′-GGTTACCTTGTTACGACTT-3′), followed by genome sequencing.
Antimicrobial Susceptibility Testing
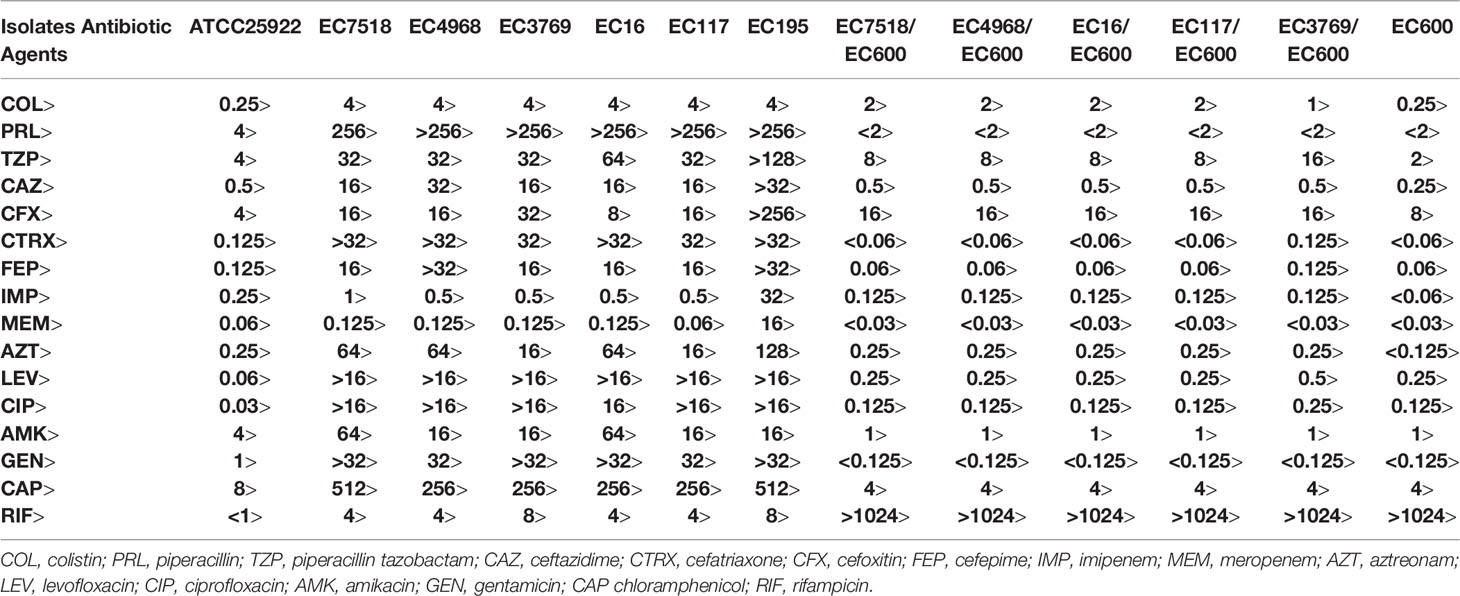
The minimum inhibitory concentrations (MICs) of clinical routine antimicrobial agents, including piperacillin (PRL), piperacillin-tazobactam (TZP), ceftazidime (CAZ), ceftriaxone (CTRX), cefoxitin (CFX), cefepime (FEP), imipenem (IMP), meropenem (MEM), aztreonam (AZT), levofloxacin (LEV), ciprofloxacin (CIP), amikacin (AMK), gentamicin (GEN), chloramphenicol (CAP) and rifampicin (RIF) were determined by the agar dilution method. The MICs of colistin were determined by the broth microdilution in cation-adjusted Mueller–Hinton broth (CAMHB) (Gajdács et al., 2020). We interpreted the results in accordance with the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2020). E. coli ATCC 25922 was used as the standard reference strain.
Conjugation Experiment
The transferability of mcr-1 was tested by conjugation experiment with mcr-1-positive E. coli as donors and rifampicin-resistant E. coli C600 as a recipient. The MacConkey agar plates containing rifampicin (512 µg/mL) and colistin (1 µg/mL) were used to select mcr-1-positive transconjugants. PCR analysis of mcr-1 and antimicrobial susceptibility testing were carried out to confirm the transconjugants.
Polymerase Chain Reaction (PCR)
All E. coli strains were screened for the presence of the mcr-1 gene by PCR using primers (Forward: 5′-CGGTCAGTCCGTTTGTTC-3′, Reverse: 5′-CTTGGTCGGTCTGTA GGG-3′) (Liu et al., 2016). The PCR products were purified and sequenced by Sanger sequencing (Generay, Shanghai, China).
Next-Generation Sequencing and Bioinformatics Analysis
Genomic DNA of six mcr-1-harboring E. coli strains was extracted using Bacterial Genomic DNA Miniprep kit (Generay, Shanghai, China), and subsequently sequenced by Illumina HiSeq 2500 (Illumina, Inc., San Diego, CA, United States). The short reads were assembled using SPAdes software (version3.14.0). EC195 showed the widest resistance spectrum and highest MIC levels against all the antimicrobials tested in this study, while the other five strains exhibited similar MIC levels, so we chose EC195 and one of the other five EC7518 as the representative isolates for whole genome sequencing. These two isolates were further sequenced by Pacific Bioscience (PacBio) RSII systems at the Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China). Then, hybrid assembly was performed using Unicycler v0.4.8, with both short and long reads. The assembled sequences were annotated using Prokka (Seemann, 2014) and then corrected by BLAST (Boratyn et al., 2013) searches against the UniProtKB/Swiss-Prot, RefSeq, ISfinder (Siguier et al., 2006), and CARD (Jia et al., 2017) databases. The multi-locus sequence typing (MLST) and plasmid replicon type (Inc groups) identification were conducted using MLST (https://pubmlst.org/) (Jolley and Maiden, 2010) and PlasmidFinder (https://cge.cbs.dtu.dk//services/PlasmidFinder/), respectively. The core genome phylogenetic tree was generated using kSNP3 (Gardner et al., 2015), and plasmid maps were generated using CGView (Grant and Stothard, 2008). Gene organization diagrams were generated using Python script and modified with Inkscape 1.0 (https://inkscape.org/).
Statistical Analyses
The differences in the colistin MIC distribution of 509 mcr-1-negative E. coli isolates among the four years were tested using the χ2 test. A P value <0.05 was considered to be statistically significant. All analyses were conducted using the SPSS software (version 22.0; SPSS Inc., Chicago, IL, USA).
Sequence Data Availability
The genome data of mcr-1-positive strains have been submitted to NCBI under the BioProject accession number PRJNA770868. The mcr-1.34 gene in this work have been submitted to GenBank under accession numbers MZ450868.
Results and Discussion
Colistin MIC of the Strains
Approximately 17.14% (18/105), 19.65% (34/173), 17.5% (7/40), and 74.11% (146/197) of the isolates showed MIC values ≥1 mg/L against colistin in 2015, 2016, 2017 and 2019, respectively. However, only six isolates showed MIC values of 4 mg/L, including three strains isolated in 2019 and one each in 2015, 2016, and 2017 (Figure 1A).
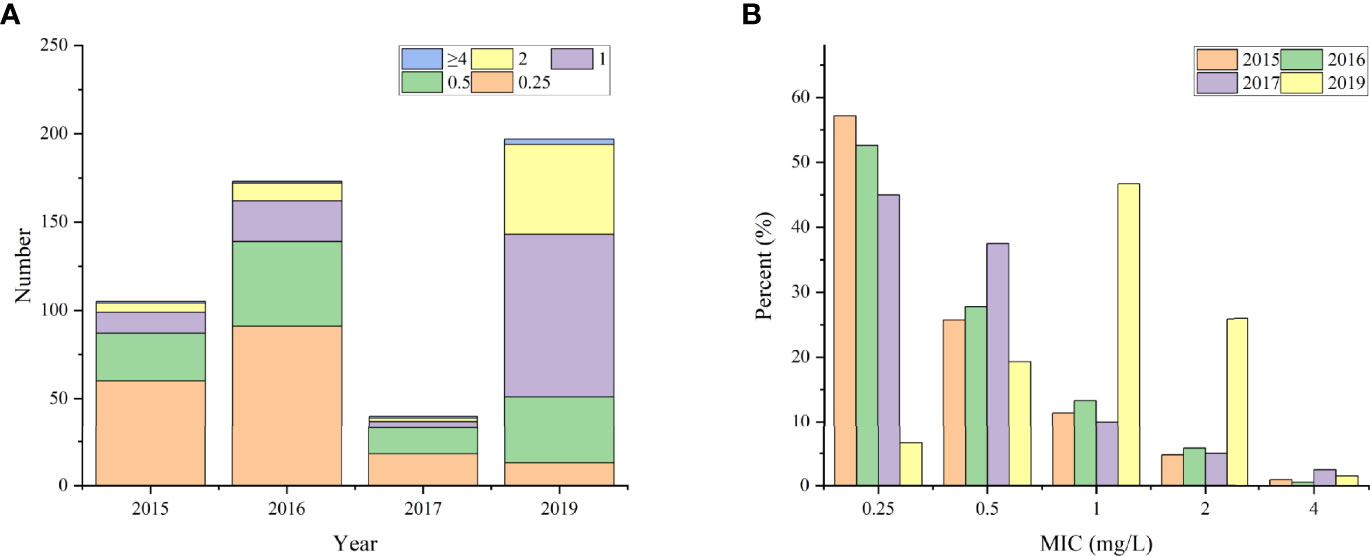
Figure 1 The distribution of clinical isolates with different MIC values. (A) the total number; (B) the percentage.
Further analysis demonstrated that isolates from 2015, 2016, and 2017 exhibited similar percentage of MIC to colistin, ranging from 0.25 mg/L to 2 mg/L. The percentages of different MIC levels were 53.65% (169/315), 28.57% (90/315), 12.38% (39/315), and 5.40% (17/315) at 0.25, 0.5, 1, and 2 mg/L, respectively. In 2019, nearly half (47.42%, 92/194) of the strains had a MIC value of 1 mg/L, and approximately 26.29% (51/194) of the strain had a MIC of up to 2 mg/L. Moreover, the MIC value of 0.25 mg/L was found in only 6.7% (13/194) of the bacteria isolated in 2019, which was just one-eighth (53.65%) of that from the previous years (Figure 1B). This might be associated with the increase in use of polymyxin to treat bacterial infections in humans (http://app1.nmpa.gov.cn/data_nmpa/face3/dir.html), which contributes to the emergence and increase of polymyxin-resistant bacteria.
mcr-1 Gene Detection and Sequencing
In this study, six (1.17%) mcr-1-positive E. coli were identified. Previous publications reported mcr-1 positive rates of 0.35% (12/3434), 0.69% (2/291), and 0.6% (4/700) in clinical E. coli isolates from Chinese hospitals between 2002 and 2016 (Lu et al., 2018), in 2018 (Liao et al., 2020), and from 2014 to 2015 (He et al., 2017), respectively. One study reported an mcr-1 positive rate of 0.87% (6/689) in clinical Salmonella spp. between 2009 and 2018 (Fan et al., 2020). A higher mcr-1-positive rate of 3.5% (24/688) in E. coli was reported from 2016 to 2018 in Guangzhou, China (Shen et al., 2020). These results revealed the increasing prevalence of the mcr-1 gene in clinical settings.
Of the six mcr-1-positive strains, EC3769, EC4968, and EC7518 were isolated in 2015, 2016, and 2017, respectively, whereas EC16, EC117, and EC195 were isolated in 2019. The results showed that the six mcr-1-harboring strains exhibited higher MICs against colistin than the negative strains. All six mcr-1 positive strains found in this study were resistant to colistin with a MIC value of 4 mg/L, which was in accordance with previous studies (He et al., 2017; Liao et al., 2020). Furthermore, all mcr-1-positive strains showed resistance to multiple antimicrobial agents, exhibiting similar MIC levels against 14 antimicrobials tested, except MEM and IMP (Table 1). One of the mcr-1-positive isolates, namely, EC195, showed the broadest resistance spectrum and highest MIC levels against all 14 antimicrobials tested in this study, especially for MEM and IMP. EC195 showed MIC levels >8 mg/L to MEM and IMP, approximately ≥16-fold higher than the other five strains, which showed MIC ≤ 1 mg/L to either MEM or IMP (Table 1).
Sequencing the mcr-1 gene revealed that five of the six isolates exhibited 100% identity to the primary reported mcr-1 gene (KP347127). By contrast, the mcr-1 gene of EC3769 was a novel variant (designated mcr-1.34 in this study) with a one-point mutation at nucleotide position 6 (G to A), which led to the substitution of methionine residue to isoleucine residue (Met to Ile) in comparison with the MCR-1.1 protein (AKF16168). The MCR-1 protein contains an N-terminal inner membrane-bound domain (residues 1–241) and a C-terminal soluble catalytic domain (residues 215–541) (Gao et al., 2016). The substitution of the novel variant in this study was very close to the start of the transmembrane domain.
Whole-Genome Sequencing of mcr-1-Positive Strains
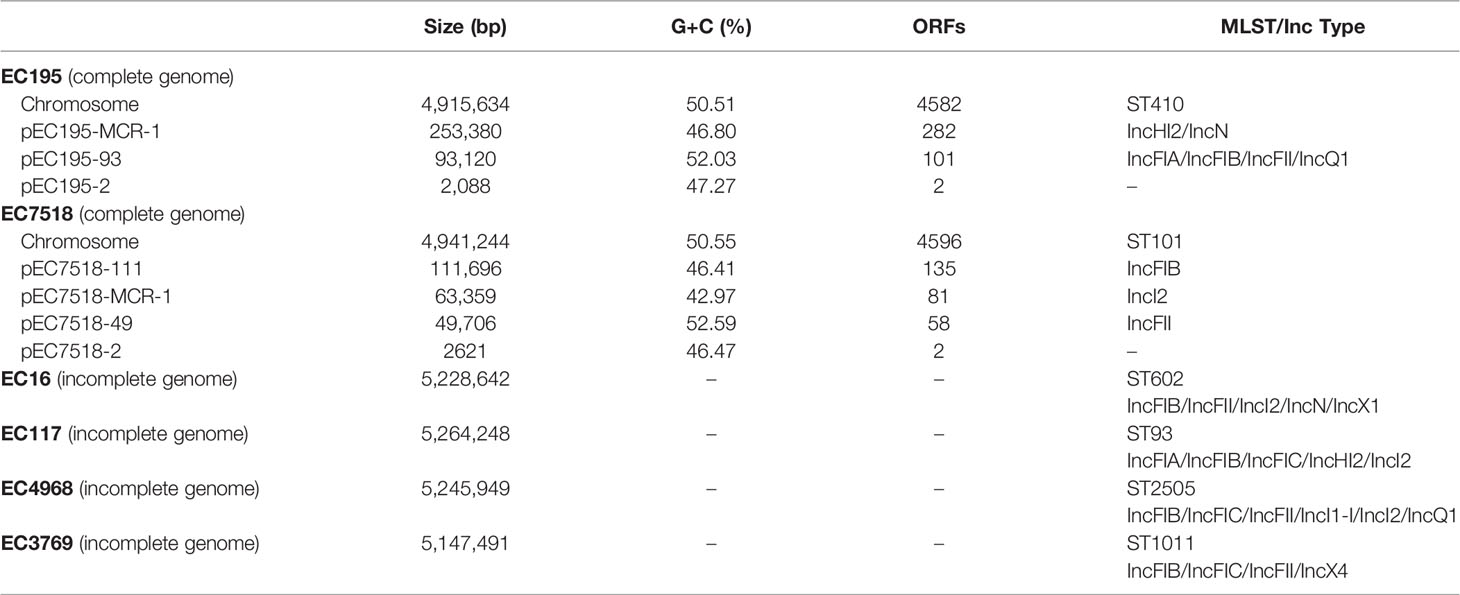
The general features of the genomes of the six mcr-positive isolates (four draft genomes of EC3769, EC117, EC16, and EC4968 and two complete genomes of EC195 and EC7518) are summarized in Table 2. In silico analysis revealed that these strains belonged to different MLST, including ST1011 (EC3769), ST93 (EC117), ST101 (EC7518), ST602 (EC16), ST410 (EC195), and ST2505 (EC4968) (Figure 2). ST1011 was proposed to be closely related to the poultry sector, identified in Lebanon and duck farms in southeast coastal China (Wang et al., 2021). However, an mcr-1-carrying E. coli ST1011 has also been isolated from a fecal sample of a 21-year-old male patient (Liang et al., 2021). Even though ST101 has been reported as one of the most prevalent sequence types (STs) among blaNDM-positive E. coli strains in poultry production, it has been found in hospital sewage water and is regarded as an infection-causing isolate (Wang Y. et al., 2017; Jin et al., 2018). In addition, ST410 has been frequently reported in animal husbandry-related epidemiology studies (Wang et al., 2020; Cheng et al., 2021).
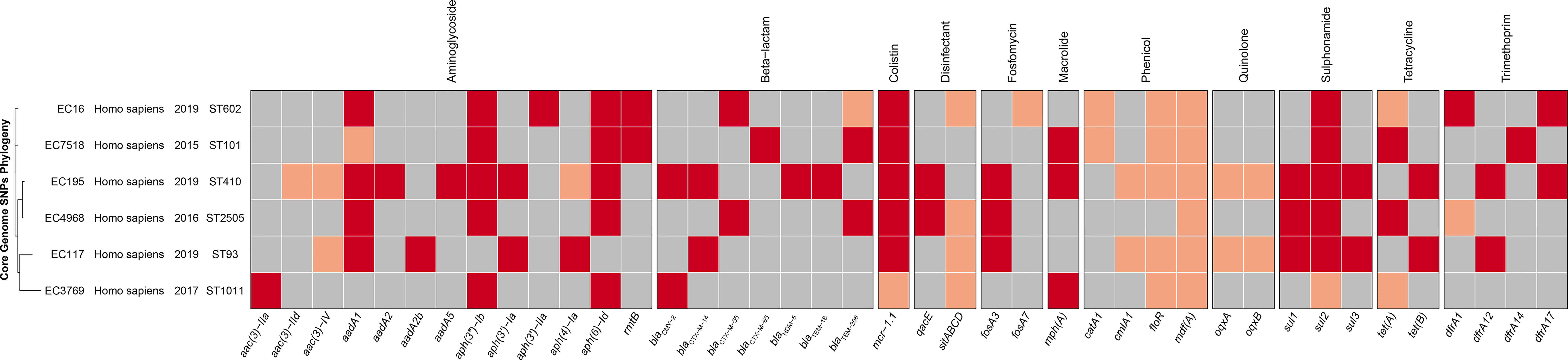
Figure 2 The phylogenetic tree and annotation of epidemiological and genomic features of six mcr-1-positive strains. The heatmap is used to display the types of acquired antimicrobial resistance genes. The presence in identity (red) or variant (pink) and absence (gray) of antimicrobial resistance genes are indicated.
ST93 was previously regarded as a similar ST to ST410; however, an isolate from the blood sample of an 84-year-old man in a Uruguay hospital with resistance to third-generation cephalosporins changed that perception (Papa-Ezdra et al., 2020). ST93 may pose a more significant threat to human health than ST410. ST602 is identified in animal lesion organs (Liu et al., 2021), whereas E. coli ST2505 is a novel ST carrying mcr-1 gene first reported in this study. However, some prevalent STs of mcr-1 carrying E. coli in previous publications such as ST10, ST131, and ST156 (He et al., 2017) were not found in this study, especially E. coli ST10, the most prevalent ST of hospital-associated E. coli (Shen et al., 2018).
Up to 41 different antibiotic resistance genes (ARGs) were identified in these mcr-1-carrying E. coli genomes, including β-lactam resistance genes, aminoglycoside, tetracycline, and sulfonamide genes (Figure 2). Moreover, aadA1, aadA2, aadA5, and the efflux genes oqxA and oqxB were also identified. This enabled them to develop multidrug resistance phenotypes. According to previous reports, blaCTX-M-65, blaTEM-206, blaCMY-2, blaNDM-5, blaCTX-M-14, blaTEM-1B, and blaCTX-M-55 belong to extended-spectrum β-lactamases, the main problem of E. coli drug resistance, which can decrease the activity of β-lactam ring and cause bacterial resistance to antimicrobials (Cullik et al., 2010; Smet et al., 2012). Genes sul1, sul2, and sul3 contribute to alleviating the damage of E. coli caused by sulfonamide, especially sul2, which is usually located in multiple resistance-determining regions of a large transferable plasmid and can thus spread along with its carrier plasmid (Enne et al., 2001). The acetyltransferase genes, including aac(3)-IIa, aac(3)-Iv, aac(3)-IId, and the phosphotransferase genes, including aph(6)-Id, aph(3′)-Ib, aph(4)-Ia, aph(3′)-Ia, and aph(3′)-IIa, are thought to increase E. coli resistance by changing the target position and decreasing medicine efficacy (Recht and Puglisi, 2001). Furthermore, since its first report in pig manure, in 2003, many studies have demonstrated that oqxA and oqxB are related to diversified drug resistance by extracellularly extruding antimicrobial poison (Sørensen et al., 2003; Hansen et al., 2007; Sato et al., 2011).
Among the six mcr-1-carrying E. coli genomes, EC195 carried the highest number (28) of resistance genes, nearly a half more than those (19) of the following EC117. Resistance genes such as aadA2, aac(3)-IIa, aadA5, and blaNDM-5 only appeared in EC195, which might contribute to its broader resistance spectrum and higher MIC levels against the antimicrobials tested than those of the other five isolates. The resistance genotype of EC195 was consistent with the phenotype. It had 12 classes of resistance genes and showed resistance to the corresponding antimicrobials (Table 1). Moreover, the specific resistance against MEM and IMP of EC195 might be related to the presence of blaNDM-5, the carbapenem resistance-related gene only identified in EC195.
Characterization and Comparative Genomic Analysis of the mcr-1-Harboring Plasmids
In the six mcr-1-positive genomes, the mcr-1 genes were all located on three types of incompatibility (Inc) group plasmids, IncHI2 (pEC195-MCR-1), IncI2 (pEC16-MCR-1, pEC117-MCR-1, pEC4968-MCR-1, and pEC7518-MCR-1), and IncX4 (pEC3769-MCR-1). Five (exclusive of pEC195-MCR-1) of the six mcr-1 carrying plasmids were transferable and the transconjugants were obtained. The transconjugants showed colistin MIC levels slightly lower than those of their corresponding original isolates (Table 1). In addition, complete plasmid sequences carrying mcr-1 were retrieved from GenBank for further analysis. IncHI2 was the most common Inc type among them, followed by IncI2 and IncX4 (Supplementary Figure S1). This result was consistent with a previous report, which showed that IncI2, IncHI2, and IncX4 accounted for more than 90% of the plasmids reported to carry mcr-1 (Berglund et al., 2018).
The IncHI2 plasmid pEC195-MCR-1 was 253,380 bp in length with a G+C content of 46.80%. It shared 99% nucleotide identity and over 95% coverage with plasmid pHNSHP45-2 (KU341381), which was the first reported IncHI2 plasmid carrying mcr-1 (Arcilla et al., 2016) (Figure 3A). The variable region of pEC195-MCR-1 consisted of a group of resistance genes including blaCTX-M-14, fosA3, aac(3)-Iv, aph(4’)-Ia, sul2, floR, aadA, cmlA6, ant3Ia, sul3, aph(4’)-Ia, sul1, oqxAB, a tellurium resistance gene cluster and mcr-1. However, additional ARGs (mrx/mphA) and mobile genetic elements (MGEs; IS1203, ISEc25, IS1A, and TnEc1) were identified in pEC195-MCR-1. The IncHI2-type plasmid has been reported to be the most diverse plasmid that contains a large MDR region composed of various ARGs and MGEs (Li et al., 2017). Co-occurrence of mcr-1 and other ARGs, especially EBSL, in plasmids can cause difficulties in clinical antibacterial treatment.
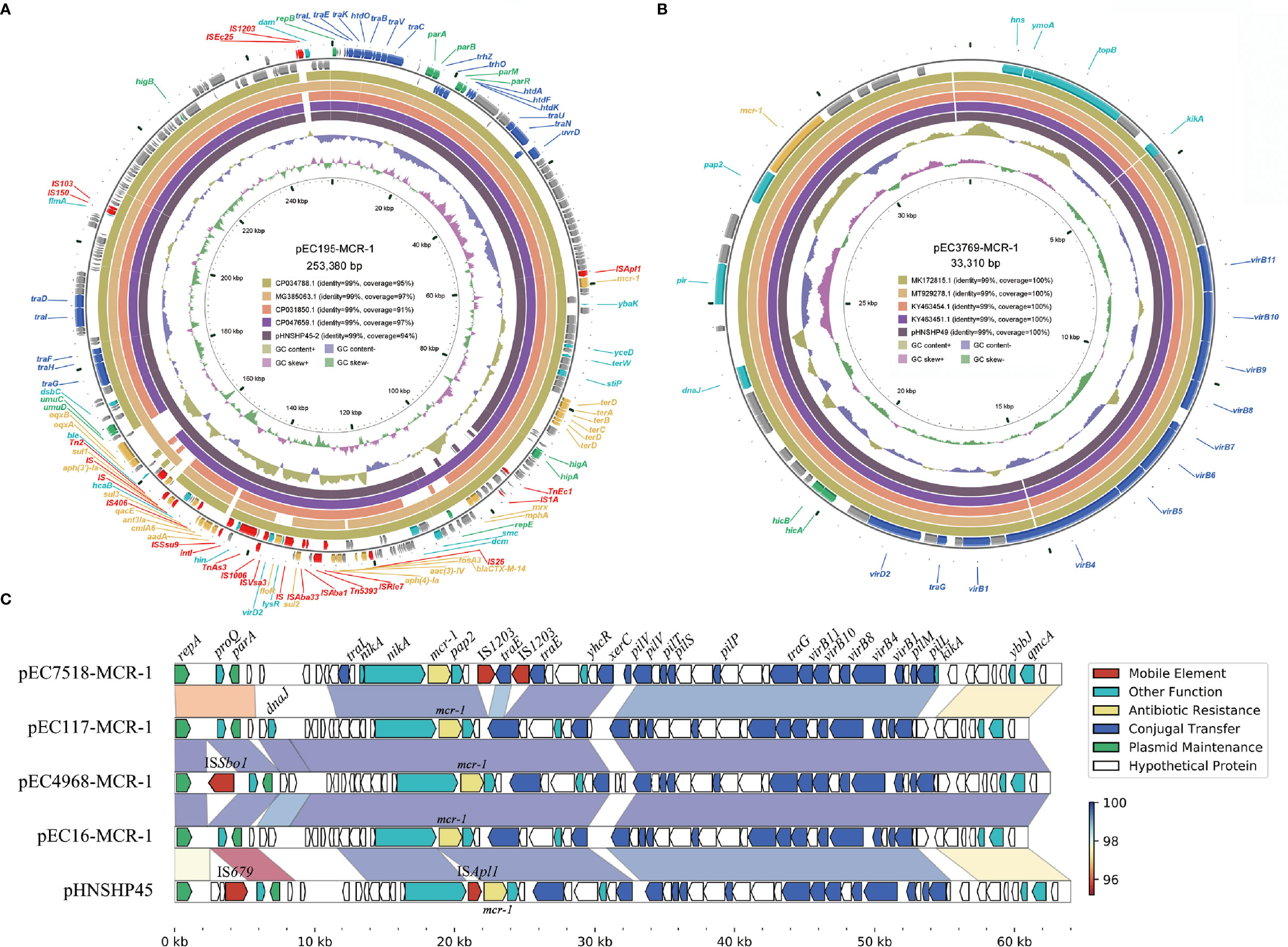
Figure 3 Diagram of the six mcr-1-harboring plasmids. (A) the plasmid map of pEC195-MCR-1; (B) the plasmid map of pEC3769-MCR-1; (C) Comparison of the four IncI2 plasmids and pHNSHP45. Genes are denoted by arrows and are colored based on gene function classification. Regions of > 95% identity are marked by gradient colors.
IncX4 plasmid pEC3769-MCR-1 was 33,310 bp in length and had an average G+C content of 41.58% (Figure 3B). The plasmid was almost identical (100% coverage, 99% identity) to other mcr-1-harboring IncX4 plasmids such as pHNSHP49 (MF774188) (Wu et al., 2018). These IncX4 plasmids have no known ARGs other than mcr-1. However, self-transmissible IncX4-type plasmids are important vehicles responsible for disseminating the mcr-1 gene among Enterobacteriaceae worldwide (Fernandes et al., 2016; Wang Q. et al., 2017).
The length of the four IncI2 plasmids ranged from 60,960 to 63,359 bp (Figure 3C). These plasmids consisted of replication genes, horizontal transfer, maintenance, and stability region and only carried one ARG mcr-1. BLASTN results revealed they were similar to the first reported mcr-1-harboring IncI2 plasmid pHNSHP45 (NZ_KP347127). These IncI2 plasmids share a highly conserved backbone and contain minor differences in the repA neighboring region. The IncI2-type plasmid was the earliest reported vector of the plasmid-mediated mcr-1 gene, which plays a significant role in rapidly mobilizing and acquiring mcr-1 (Liu et al., 2016; Petrillo et al., 2016; Zelendova et al., 2021).
Genetic Contexts of mcr-1
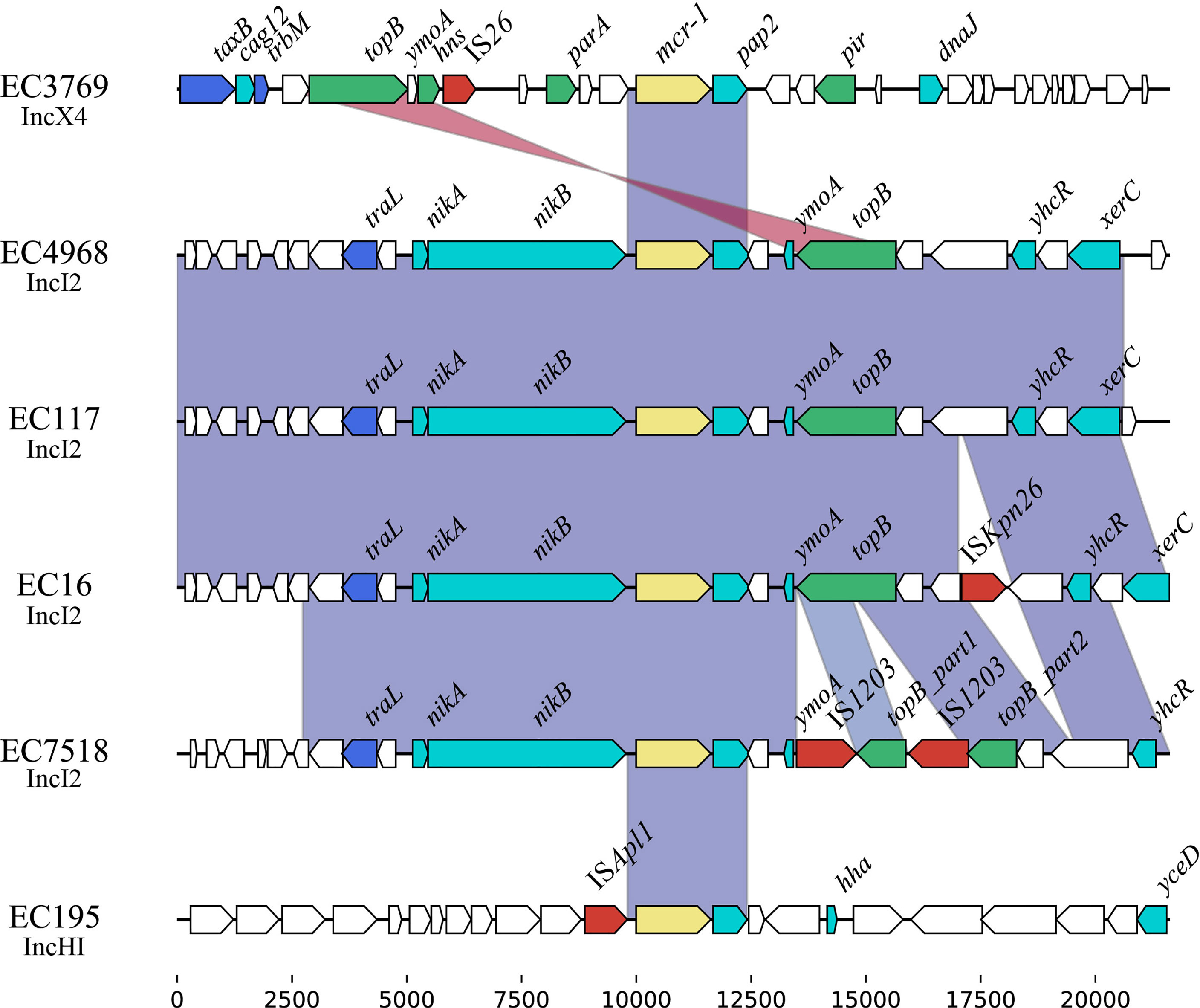
Comparative genomic analysis was performed on the ~20-kb sequences around the mcr-1 genes. The results revealed that an ~2.6-kb region encoding mcr-1-pap2 was conserved among these plasmids (Figure 4), which was in accordance with previous reports about the mcr-1 gene (Poirel et al., 2016). This 2.6-kb region is always associated with the ISApl1 element and can be mobilized in the form of Tn6330 (ISApl1-mcr-1-pap2-ISApl1) (Snesrud et al., 2018). However, among these sequenced mcr-1-bearing plasmids, only pEC195-253K carried a single copy of ISApl1 beside mcr-1, which indicated that mcr-1 was stable on these plasmids. Among these four IncI2 plasmids, the mcr-1 gene was located between tral-nikA-nikB and ymoA-topB sequences. In addition, these regions of pEC4968 and pEC117 were almost identical, but two copies of IS1203 truncated the ymoA-topB genes of pEC7518-63K. IS26 flanked by an 8-bp direct repeat was identified upstream of mcr-1 on pEC3769-MCR-1, but was not related to the mobilization of the mcr-1 context (Manageiro et al., 2019).
Furthermore, the 5-kb region around the mcr-1 gene of IncHI2, IncI2, and IncX4 plasmids was retrieved from GenBank of NCBI and subsequently clustered with a threshold identity >95% (Supplementary Figure S1). A complete Tn6330 transposon was identified on IncHI2 and IncI2 plasmids, demonstrating its role in the initial acquisition of mcr-1. It has been reported that the nikA-nikB-mcr-1-pap2 structure is conserved among IncI2 plasmids (Feng et al., 2019). Herein, we also found that nikB upstream of mcr-1 in all IncI2 plasmids was often truncated to different lengths. Furthermore, in Cluster 6 of IncI2 plasmids, we noticed that the pap2 gene, which encodes the PAP2-family protein to facilitate the transfer of mcr-1, was lost.
Conclusion
Although a series of mcr genes [mcr(1-10)] were detected and reported in succession, mcr-1, the first member of the MCR family discovered in 2015, is still recognized as a critical factor in polymyxin resistance. In this study, we screened mcr-1 in 515 human clinical E. coli isolates and found 1.17% (6/515) mcr-1-positive strains, and these strains showed higher resistance to colistin (with MIC levels of 4 mg/L) than the mcr-1-negative strains (all with MIC levels <4 mg/L). We identified 41 ARGs in the 6 mcr-1 positive strains, of which EC195 not only carried the highest number (28) of resistance genes but also exhibited a broader resistance spectrum and higher MIC levels. Furthermore, the unique resistance phenotype against MEM and IMP might be related to the rare identification of blaNDM-5 from the EC195 genome. MLST found that these six mcr-1-positive strains belonged to six different STs. A novel mcr-1 variant was identified in EC3769 with a one-point mutation at nucleotide position 6, causing an amino acid variation. These findings may provide a new perspective on the molecular characteristics of resistance caused by mcr-1.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
HZ, CL, and QB designed the study. HL, CF, TZ, QL, and XZ acquired data. CQ, CF, WS, LL, MG, MX, and JL performed the results analysis and interpreted data. QL, CQ, and QB wrote the first draft of the paper. KL, HZ, XL, and TX revised it critically for important intellectual content. All co-authors approved the final version.
Funding
This study was supported by the National Natural Science Foundation of China (81973382, 81960381 and 81700011), Zhejiang Provincial Natural Science Foundation of China (LQ17H010003, LY19C060002 and LQ17H190001), and the Science and Technology Project of Wenzhou City, China (N20210001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to acknowledge all study participants and individuals who contributed to this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.854534/full#supplementary-material
References
Ainsworth, G. C., Brown, A. M., Brownlee, G. (1947). Aerosporin, an Antibiotic Produced by Bacillus Aerosporus Greer. Nature 159, 263. doi: 10.1038/160263a0
Arcilla, M. S., van Hattem, J. M., Matamoros, S., Melles, D. C., Penders, J., de Jong, M. D., et al. (2016). Dissemination of the Mcr-1 Colistin Resistance Gene. Lancet Infect. Dis. 16, 147–149. doi: 10.1371/journal.ppat.1005957
Berglund, B., Chen, B., Tärnberg, M., Sun, Q., Xu, L., Welander, J., et al. (2018). Characterization of Extended-Spectrum β-Lactamase-Producing Escherichia Coli Harboring Mcr-1 and Toxin Genes From Human Fecal Samples From China. Future Microbiol. 13, 1647–1655. doi: 10.2217/fmb-2018-0242
Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., Ma, N., et al. (2013). BLAST: A More Efficient Report With Usability Improvements. Nucleic Acids Res. 41, W29–W33. doi: 10.1093/nar/gkt282
Cheng, P., Yang, Y., Cao, S., Liu, H., Li, X., Sun, J., et al. (2021). Prevalence and Characteristic of Swine-Origin Mcr-1-Positive Escherichia Coli in Northeastern China. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.712707
CLSI (2020). “Performance Standards for Antimicrobial Susceptibility Testing.” in CLSI Supplement M100, 30th ed (Wayne, PA: Clinical and Laboratory Standards Institute).
Cullik, A., Pfeifer, Y., Prager, R., von Baum, H., Witte, W. (2010). A Novel IS26 Structure Surrounds blaCTX-M Genes in Different Plasmids From German Clinical Escherichia Coli Isolates. J. Med. Microbiol. 59, 580–587. doi: 10.1099/jmm.0.016188-0
Enne, V. I., Livermore, D. M., Stephens, P., Hall, L. M. (2001). Persistence of Sulphonamide Resistance in Escherichia Coli in the UK Despite National Prescribing Restriction. Lancet 357, 1325–1328. doi: 10.1016/s0140-6736(00)04519-0
Falagas, M. E., Kasiakou, S. K. (2005). Colistin: The Revival of Polymyxins for the Management of Multidrug-Resistant Gram-Negative Bacterial Infections. Clin. Infect. Dis. 40, 1333–1341. doi: 10.1086/429323
Fan, J., Zhang, L., He, J., Zhao, M., Loh, B., Leptihn, S., et al. (2020). Plasmid Dynamics of Mcr-1-Positive Salmonella Spp. In a General Hospital in China. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.604710
Feng, C., Wen, P., Xu, H., Chi, X., Li, S., Yu, X., et al. (2019). Emergence and Comparative Genomics Analysis of Extended-Spectrum-β-Lactamase-Producing Escherichia Coli Carrying Mcr-1 in Fennec Fox Imported From Sudan to China. mSphere 4 (6), e00732–19. doi: 10.1128/mSphere.00732-19
Fernandes, M. R., McCulloch, J. A., Vianello, M. A., Moura, Q., Pérez-Chaparro, P. J., Esposito, F., et al. (2016). First Report of the Globally Disseminated IncX4 Plasmid Carrying the Mcr-1 Gene in a Colistin-Resistant Escherichia Coli Sequence Type 101 Isolate From a Human Infection in Brazil. Antimicrob. Agents Chemother. 60, 6415–6417. doi: 10.1128/aac.01325-16
Gajdács, M., Ábrók, M., Lázár, A., Burián, K. (2020). Differential Epidemiology and Antibiotic Resistance of Lactose-Fermenting and Non-Fermenting Escherichia Coli: Is it Just a Matter of Taste? Biol. Futur. 71, 175–182. doi: 10.1007/s42977-020-00016-6
Gao, R., Hu, Y., Li, Z., Sun, J., Wang, Q., Lin, J., et al. (2016). Dissemination and Mechanism for the MCR-1 Colistin Resistance. PloS Pathog. 12, e1005957. doi: 10.1371/journal.ppat.1005957
Gardner, S. N., Slezak, T., Hall, B. G. (2015). Ksnp3.0: SNP Detection and Phylogenetic Analysis of Genomes Without Genome Alignment or Reference Genome. Bioinformatics 31, 2877–2878. doi: 10.1093/bioinformatics/btv271
Grant, J. R., Stothard, P. (2008). The CGView Server: A Comparative Genomics Tool for Circular Genomes. Nucleic Acids Res. 36, W181–W184. doi: 10.1093/nar/gkn179
Hansen, L. H., Jensen, L. B., Sørensen, H. I., Sørensen, S. J. (2007). Substrate Specificity of the OqxAB Multidrug Resistance Pump in Escherichia Coli and Selected Enteric Bacteria. J. Antimicrob. Chemother. 60, 145–147. doi: 10.1093/jac/dkm167
Hartzell, J. D., Neff, R., Ake, J., Howard, R., Olson, S., Paolino, K., et al. (2009). Nephrotoxicity Associated With Intravenous Colistin (Colistimethate Sodium) Treatment at a Tertiary Care Medical Center. Clin. Infect. Dis. 48, 1724–1728. doi: 10.1086/599225
He, Q. W., Xu, X. H., Lan, F. J., Zhao, Z. C., Wu, Z. Y., Cao, Y. P., et al. (2017). Molecular Characteristic of Mcr-1 Producing Escherichia Coli in a Chinese University Hospital. Ann. Clin. Microbiol. Antimicrob. 16, 32. doi: 10.1186/s12941-017-0207-z
Hinchliffe, P., Qiu, Y., Portal, E., Young, T., Li, H., Tooke, C. L., et al. (2017). Insights Into the Mechanistic Basis of Plasmid-Mediated Colistin Resistance From Crystal Structures of the Catalytic Domain of MCR-1. Sci. Rep. 7, 39392. doi: 10.1038/srep39392
Hu, M., Guo, J., Cheng, Q., Yang, Z., Chan, E. W. C., Chen, S., et al. (2016). Crystal Structure of Escherichia Coli Originated MCR-1, a Phosphoethanolamine Transferase for Colistin Resistance. Sci. Rep. 6, 38793. doi: 10.1038/srep38793
Hussein, N. H., Al-Kadmy, I. M. S., Taha, B. M., Hussein, J. D. (2021). Mobilized Colistin Resistance (Mcr) Genes From 1 to 10: A Comprehensive Review. Mol. Biol. Rep. 48, 2897–2907. doi: 10.1007/s11033-021-06307-y
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: Expansion and Model-Centric Curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 45, D566–d573. doi: 10.1093/nar/gkw1004
Jin, L., Wang, R., Wang, X., Wang, Q., Zhang, Y., Yin, Y., et al. (2018). Emergence of Mcr-1 and Carbapenemase Genes in Hospital Sewage Water in Beijing, China. J. Antimicrob. Chemother. 73, 84–87. doi: 10.1093/jac/dkx355
Jolley, K. A., Maiden, M. C. (2010). BIGSdb: Scalable Analysis of Bacterial Genome Variation at the Population Level. BMC Bioinf. 11, 595. doi: 10.1186/1471-2105-11-595
Landman, D., Georgescu, C., Martin, D. A., Quale, J. (2008). Polymyxins Revisited. Clin. Microbiol. Rev. 21, 449–465. doi: 10.1128/cmr.00006-08
Laxminarayan, R., Van Boeckel, T., Frost, I., Kariuki, S., Khan, E. A., Limmathurotsakul, D., et al. (2020). The Lancet Infectious Diseases Commission on Antimicrobial Resistance: 6 Years Later. Lancet Infect. Dis. 20, e51–e60. doi: 10.1016/s1473-3099(20)30003-7
Liang, G., Rao, Y., Wang, S., Chi, X., Xu, H., Shen, Y. (2021). Co-Occurrence of NDM-9 and MCR-1 in a Human Gut Colonized Escherichia Coli ST1011. Infect. Drug Resist. 14, 3011–3017. doi: 10.2147/idr.S321732
Liao, W., Lin, J., Jia, H., Zhou, C., Zhang, Y., Lin, Y., et al. (2020). Resistance and Heteroresistance to Colistin in Escherichia Coli Isolates From Wenzhou, China. Infect. Drug Resist. 13, 3551–3561. doi: 10.2147/idr.S273784
Li, L. G., Huang, Q., Yin, X., Zhang, T. (2020). Source Tracking of Antibiotic Resistance Genes in the Environment - Challenges, Progress, and Prospects. Water Res. 185, 116127. doi: 10.1016/j.watres.2020.116127
Ling, Z., Yin, W., Shen, Z., Wang, Y., Shen, J., Walsh, T. R. (2020). Epidemiology of Mobile Colistin Resistance Genes mcr-1 to mcr-9. J. Antimicrob. Chemother. 75, 3087–3095. doi: 10.1093/jac/dkaa205
Liu, Z., Liu, Y., Xi, W., Liu, S., Liu, J., Mu, H., et al. (2021). Genetic Features of Plasmid- and Chromosome-Mediated Mcr-1 in Escherichia Coli Isolates From Animal Organs With Lesions. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.707332
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/s1473-3099(15)00424-7
Li, R., Xie, M., Zhang, J., Yang, Z., Liu, L., Liu, X., et al. (2017). Genetic Characterization of Mcr-1-Bearing Plasmids to Depict Molecular Mechanisms Underlying Dissemination of the Colistin Resistance Determinant. J. Antimicrob. Chemother. 72, 393–401. doi: 10.1093/jac/dkw411
Lu, H., Wang, C., Dong, G., Xu, C., Zhang, X., Liu, H., et al. (2018). Prevalence and Molecular Characterization of Escherichia Coli Clinical Isolates Carrying Mcr-1 in a Chinese Teaching Hospital From 2002 to 2016. Antimicrob. Agents Chemother. 62, e02623-02617. doi: 10.1128/AAC.02623-17
Manageiro, V., Clemente, L., Romão, R., Silva, C., Vieira, L., Ferreira, E., et al. (2019). IncX4 Plasmid Carrying the New Mcr-1.9 Gene Variant in a CTX-M-8-Producing Escherichia Coli Isolate Recovered From Swine. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00367
Papa-Ezdra, R., Grill Diaz, F., Vieytes, M., García-Fulgueiras, V., Caiata, L., Ávila, P., et al. (2020). First Three Escherichia Coli Isolates Harbouring Mcr-1 in Uruguay. J. Glob Antimicrob. Resist. 20, 187–190. doi: 10.1016/j.jgar.2019.07.016
Petrillo, M., Angers-Loustau, A., Kreysa, J. (2016). Possible Genetic Events Producing Colistin Resistance Gene Mcr-1. Lancet Infect. Dis. 16, 280. doi: 10.1016/s1473-3099(16)00005-0
Poirel, L., Kieffer, N., Brink, A., Coetze, J., Jayol, A., Nordmann, P. (2016). Genetic Features of MCR-1-Producing Colistin-Resistant Escherichia Coli Isolates in South Africa. Antimicrob. Agents Chemother. 60, 4394–4397. doi: 10.1128/aac.00444-16
Ragupathi, N. K. D., Bakthavatchalam, Y. D., Mathur, P., Pragasam, A. K., Walia, K., Ohri, V. C., et al. (2019). Plasmid Profiles Among Some ESKAPE Pathogens in a Tertiary Care Centre in South India. Indian J. Med. Res. 149, 222–231. doi: 10.4103/ijmr.IJMR_2098_17
Recht, M. I., Puglisi, J. D. (2001). Aminoglycoside Resistance With Homogeneous and Heterogeneous Populations of Antibiotic-Resistant Ribosomes. Antimicrob. Agents Chemother. 45, 2414–2419. doi: 10.1128/aac.45.9.2414-2419.2001
Sørensen, A. H., Hansen, L. H., Johannesen, E., Sørensen, S. J. (2003). Conjugative Plasmid Conferring Resistance to Olaquindox. Antimicrob. Agents Chemother. 47, 798–799. doi: 10.1128/aac.47.2.798-799.2003
Sato, T., Yokota, S., Uchida, I., Okubo, T., Ishihara, K., Fujii, N., et al. (2011). A Fluoroquinolone-Resistant Escherichia Coli Clinical Isolate Without Quinolone Resistance-Determining Region Mutations Found in Japan. Antimicrob. Agents Chemother. 55, 3964–3965. doi: 10.1128/aac.00532-11
Seemann, T. (2014). Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shen, Y., Wu, Z., Wang, Y., Zhang, R., Zhou, H. W., Wang, S., et al. (2018). Heterogeneous and Flexible Transmission of Mcr-1 in Hospital-Associated Escherichia Coli. mBio 9 (4), e00943–18. doi: 10.1128/mBio.00943-18
Shen, C., Zhong, L. L., Ma, F., El-Sayed Ahmed, M. A. E., Doi, Y., Zhang, G., et al. (2020). Genomic Patterns and Characterizations of Chromosomally-Encoded Mcr-1 in Escherichia Coli Populations. Gut Pathog. 12, 55. doi: 10.1186/s13099-020-00393-2
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., Chandler, M. (2006). ISfinder: The Reference Centre for Bacterial Insertion Sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Smet, A., Boyen, F., Flahou, B., Doublet, B., Praud, K., Martens, A., et al. (2012). Emergence of CTX-M-2-Producing Escherichia Coli in Diseased Horses: Evidence of Genetic Exchanges of Bla(CTX-M-2) Linked to ISCR1. J. Antimicrob. Chemother. 67, 1289–1291. doi: 10.1093/jac/dks016
Snesrud, E., McGann, P., Chandler, M. (2018). The Birth and Demise of the ISApl1-Mcr-1-ISApl1 Composite Transposon: The Vehicle for Transferable Colistin Resistance. mBio 9 (1), e02381-17. doi: 10.1128/mBio.02381-17
Stojanoski, V., Sankaran, B., Prasad, B. V., Poirel, L., Nordmann, P., Palzkill, T. (2016). Structure of the Catalytic Domain of the Colistin Resistance Enzyme MCR-1. BMC Biol. 14, 81. doi: 10.1186/s12915-016-0303-0
Sun, J., Zhang, H., Liu, Y. H., Feng, Y. (2018). Towards Understanding MCR-Like Colistin Resistance. Trends Microbiol. 26, 794–808. doi: 10.1016/j.tim.2018.02.006
Velkov, T., Roberts, K. D., Nation, R. L., Thompson, P. E., Li, J. (2013). Pharmacology of Polymyxins: New Insights Into an 'Old' Class of Antibiotics. Future Microbiol. 8, 711–724. doi: 10.2217/fmb.13.39
Wang, Q., Sun, J., Li, J., Ding, Y., Li, X. P., Lin, J., et al. (2017). Expanding Landscapes of the Diversified Mcr-1-Bearing Plasmid Reservoirs. Microbiome 5, 70. doi: 10.1186/s40168-017-0288-0
Wang, R., van Dorp, L., Shaw, L. P., Bradley, P., Wang, Q., Wang, X., et al. (2018). The Global Distribution and Spread of the Mobilized Colistin Resistance Gene Mcr-1. Nat. Commun. 9, 1179. doi: 10.1038/s41467-018-03205-z
Wang, Y., Xu, C., Zhang, R., Chen, Y., Shen, Y., Hu, F., et al. (2020). Changes in Colistin Resistance and Mcr-1 Abundance in Escherichia Coli of Animal and Human Origins Following the Ban of Colistin-Positive Additives in China: An Epidemiological Comparative Study. Lancet Infect. Dis. 20, 1161–1171. doi: 10.1016/s1473-3099(20)30149-3
Wang, Y., Zhang, R., Li, J., Wu, Z., Yin, W., Schwarz, S., et al. (2017). Comprehensive Resistome Analysis Reveals the Prevalence of NDM and MCR-1 in Chinese Poultry Production. Nat. Microbiol. 2, 16260. doi: 10.1038/nmicrobiol.2016.260
Wang, M. G., Zhang, R. M., Wang, L. L., Sun, R. Y., Bai, S. C., Han, L., et al. (2021). Molecular Epidemiology of Carbapenemase-Producing Escherichia Coli From Duck Farms in South-East Coastal China. J. Antimicrob. Chemother. 76, 322–329. doi: 10.1093/jac/dkaa433
Wu, R., Yi, L., Yu, L., Wang, J., Liu, Y., Chen, X., et al. (2018). Fitness Advantage of Mcr-1–Bearing IncI2 and IncX4 Plasmids in Vitro. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00331
Xiaomin, S., Yiming, L., Yuying, Y., Zhangqi, S., Yongning, W., Shaolin, W. (2020). Global Impact of Mcr-1-Positive Enterobacteriaceae Bacteria on "One Health". Crit. Rev. Microbiol. 46, 565–577. doi: 10.1080/1040841x.2020.1812510
Yang, J., Wang, H. H., Lu, Y., Yi, L. X., Deng, Y., Lv, L., et al. (2021). A ProQ/FinO Family Protein Involved in Plasmid Copy Number Control Favours Fitness of Bacteria Carrying Mcr-1-Bearing IncI2 Plasmids. Nucleic Acids Res. 49, 3981–3996. doi: 10.1093/nar/gkab149
Zelendova, M., Papagiannitsis, C. C., Valcek, A., Medvecky, M., Bitar, I., Hrabak, J., et al. (2021). Characterization of the Complete Nucleotide Sequences of Mcr-1-Encoding Plasmids From Enterobacterales Isolates in Retailed Raw Meat Products From the Czech Republic. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.604067
Keywords: Escherichia coli, colistin resistance, mcr-1, plasmid, novel mcr-1 variant
Citation: Li Q, Qian C, Zhang X, Zhu T, Shi W, Gao M, Feng C, Xu M, Lin H, Lin L, Lu J, Lin X, Li K, Xu T, Bao Q, Li C and Zhang H (2022) Colistin Resistance and Molecular Characterization of the Genomes of mcr-1-Positive Escherichia coli Clinical Isolates. Front. Cell. Infect. Microbiol. 12:854534. doi: 10.3389/fcimb.2022.854534
Received: 14 January 2022; Accepted: 08 April 2022;
Published: 06 May 2022.
Edited by:
Costas C. Papagiannitsis, University of Thessaly, GreeceReviewed by:
Ibrahim Bitar, Charles University, CzechiaIva Sukkar, Central European Institute of Technology (CEITEC), Czechia
Copyright © 2022 Li, Qian, Zhang, Zhu, Shi, Gao, Feng, Xu, Lin, Lin, Lu, Lin, Li, Xu, Bao, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hailin Zhang, emhsd3o5N0Bob3RtYWlsLmNvbQ==; Changchong Li, d3psaWNoY2hAMTYzLmNvbQ==; Qiyu Bao, YmFvcXlAZ2Vub21pY3MuY24=
 Qiaoling Li
Qiaoling Li Changrui Qian
Changrui Qian Xueya Zhang1,2,3
Xueya Zhang1,2,3 Tingting Zhu
Tingting Zhu Mengdi Gao
Mengdi Gao Ming Xu
Ming Xu Hailong Lin
Hailong Lin Junwan Lu
Junwan Lu Xi Lin
Xi Lin Kewei Li
Kewei Li Teng Xu
Teng Xu Qiyu Bao
Qiyu Bao Changchong Li
Changchong Li Hailin Zhang
Hailin Zhang