- 1School of Biological, Environmental, and Earth Sciences, University of Southern Mississippi, Hattiesburg, MS, United States
- 2Bee Research Laboratory, Beltsville, United States Department of Agriculture, Agricultural Research Service (USDA ARS), Beltsville, MD, United States
- 3Southern Horticultural Research Unit, USDA ARS, Poplarville, MS, United States
- 4Center for Molecular and Cellular Biology, University of Southern Mississippi, Hattiesburg, Hattiesburg, MS, United States
The remarkably adaptive mite Varroa destructor is the most important honey bee ectoparasite. Varroa mites are competent vectors of deformed wing virus (DWV), and the Varroa-virus complex is a major determinant of annual honey bee colony mortality and collapse. MicroRNAs (miRNAs) are 22-24 nucleotide non-coding RNAs produced by all plants and animals and some viruses that influence biological processes through post-transcriptional regulation of gene expression. Knowledge of miRNAs and their function in mite biology remains limited. Here we constructed small RNA libraries from male and female V. destructor using Illumina’s small RNA-Seq platform. A total of 101,913,208 and 91,904,732 small RNA reads (>18 nucleotides) from male and female mites were analyzed using the miRDeep2 algorithm. A conservative approach predicted 306 miRNAs, 18 of which were upregulated and 13 downregulated in female V. destructor compared with males. Quantitative real-time PCR validated the expression of selected differentially-expressed female Varroa miRNAs. This dataset provides a list of potential miRNA targets involved in regulating vital Varroa biological processes and paves the way for developing strategies to target Varroa and their viruses.
Introduction
Varroa destructor mites are considered the most damaging ectoparasite of the European honey bee (Apis mellifera) (Hristov et al., 2020), with consequent colony losses threatening global honey production (Gallai et al., 2009; Le Conte et al., 2010; Goulson et al., 2015; Hung et al., 2018; Noël et al., 2020). Varroa mites infest honey bees, where they feed on the hemolymph and fat body of pupae and adults (Ramsey et al., 2019), depleting vital nutritional resources and suppressing the immune system in host bees (Yang and Cox-Foster, 2005; Di Prisco et al., 2016; Koleoglu et al., 2017; Koleoglu et al., 2018; Annoscia et al., 2019). Varroa are competent vectors of various viral pathogens (Gisder et al., 2009; de Miranda and Genersch, 2010; Wilfert et al., 2016), which together contribute to increased winter losses through colony collapse in many regions (Highfield et al., 2009) and requiring significant investment from beekeepers to combat infestations and replace stock. Typically, Varroa-infested hives will not survive for more than three years without treatment or cultural intervention (Martin, 1998; van Dooremalen et al., 2012). Indeed, in the United States, chemical treatment of honey bee colonies improves colony survival (Webster and Delaplane, 2001; Kulhanek et al., 2021). Although accurately costing the losses to honey bee colonies caused by Varroa infestation is technically challenging, it is safe to assume that Varroa infestation has resulted in the collapse of many thousands of honeybee colonies and billions of dollars of losses (Rosenkranz et al., 2010).
Varroa foundresses vector deformed wing virus (DWV), one of the major causes of honey bee colony collapse, to immature honey bees when they feed. There is now considerable evidence that Varroa mites are a vector of DWV (Santillán-Galicia et al., 2010; Gisder et al., 2018; Posada-Florez et al., 2019; Ryabov et al., 2019), further contributing to the negative impact of the mite on honey bees (Rosenkranz et al., 2010; Yañez et al., 2020). While DWV infection is usually commensal in honey bees and rarely pathogenic in the absence of Varroa infestation (Beaurepaire et al., 2019), inoculation of DWV during Varroa feeding significantly increases viral load in the pupal stage (Gusachenko et al., 2020). Bees symptomatic for DWV emerge with wing deformities, have reduced weight, and have a shortened lifespan or are killed prematurely by other colony members. Although bees can be asymptomatic, high viral loads adversely impact lifespan, foraging and flight capability, behavioral maturation, and immunity (Iqbal and Mueller, 2007; Natsopoulou et al., 2016; Wells et al., 2016; Benaets et al., 2017; Brettell et al., 2017; Traniello et al., 2020; Pizzorno et al., 2021). By feeding on honey bee pupae, Varroa act as an efficient vector of over a dozen viruses to their bee hosts. DWV load in the hive significantly increases upon Varroa infestation, subsequently impacting honey bee mortality (Beaurepaire et al., 2020; Gusachenko et al., 2020). These Varroa-borne viruses have several co-circulating variants that differ in virulence (Moore et al., 2011; De Miranda et al., 2013; Mordecai et al., 2016; Gisder et al., 2018; Martin and Brettell, 2019; Posada-Florez et al., 2019; Remnant et al., 2019; Ryabov et al., 2019). Although there is a clear association between DWV in Varroa and subsequent infection of honey bees with the virus, the underlying genetic mechanisms of vector competence (acquisition, maintenance, and transmission) need further clarification to develop interventions to prevent Varroa infestation and Varroa-borne virus transmission to honey bees.
The highly efficient vectoring of honey bee viruses by Varroa contributes to driving changes in virus distribution, prevalence, and virulence (Traynor et al., 2020). Varroa control and prevention are primarily based on chemical acaricides, but these contribute to Varroa resistance (Higes et al., 2020; Rinkevich, 2020), and novel, effective, and safe approaches are clearly needed to tackle this global problem. To this end, here we investigated microRNAs (miRNAs) that might regulate Varroa vector competence and therefore act as targets to disrupt pathogen transmission. MicroRNAs are single stranded non-coding RNAs 22-25 nucleotides in length derived from larger hairpin RNA precursors. MicroRNAs play significant roles in pathophysiological post-transcriptional regulation of their target genes.
MicroRNAs are now established therapeutic targets in several diseases (Bartel, 2018). In mammals, miRNAs regulate 30-60% of protein-coding genes and most cellular processes (Filipowicz et al., 2008; Yu and Pan, 2012). However, information on arthropod miRNAs and their role in arthropod physiology is limited (Barrero et al., 2011; Lucas and Raikhel, 2013; Luhur et al., 2013; Zhou et al., 2013; Asgari, 2014; Blair and Olson, 2015; He et al., 2015; Luo et al., 2015; Shao et al., 2015; Wang et al., 2015). MicroRNAs are known to play a significant role in the establishment of sexual dimorphism and in controlling sexual behavior (Lucas et al., 2015a), and the presence of male- and female-biased miRNAs and ovary- and testis-enriched miRNAs suggest a role in sex determination and gametogenesis (Fagegaltier et al., 2014). For example, dme-let-7 depletion delays germline differentiation of the early ovary, disrupts aggregation of somatic cells in the testes, and plays a significant role in sexual identity (male or female) during the late-larval to late-pupal stages and during throughout adulthood via ecdysone signaling (Fagegaltier et al., 2014). In addition, dme-miR-124 controls male sexual differentiation by targeting sex-specific splicing factor in the sex determination pathway (Weng et al., 2013). Male flies with mutant dme-miR-124 have aberrant pheromone levels, reducing mating success and receptivity by females and increasing male-male courtship. Furthermore, miRNA expression altered in different tissues post-mating following receipt of sex peptide (SP) from male flies (Fricke et al., 2014). Female flies lacking specific miRNAs (such as dme-miR-184, dme-miR-279, dme-miR-278, and dme-miR-317) demonstrated abnormal receptivity to SP from male flies (Lucas et al., 2015a).
Given the importance of Varroa as a honey bee parasite and vector of honey bee viruses, the recent availability of high-quality de novo reference genomes for V. destructor and V. jacobsoni has opened up new avenues to map non-coding small RNAs in the mite genome (Techer et al., 2019). MicroRNAs are now known to not only be critical regulators of many biological processes and pathogen infections in arthropods (Behura, 2007; Feng et al., 2018), but also play a role in replicating, harboring, and inhibiting other RNA viruses such as dengue virus (DENV), chikungunya virus (CHIKV), and other viruses and intracellular pathogens in arthropods (Sempere et al., 2003; Hussain et al., 2013; Maharaj et al., 2015). Therefore, Varroa microRNAs may also play a significant role in DWV colonization and its transmission to honey bees.
The aim of this study was to predict the repertoire of conserved and novel miRNAs in male and female Varroa to provides insights into the pathophysiological roles of Varroa miRNAs, their gene regulatory networks, and their potential biological functions. The identification of novel miRNAs in Varroa paves the way for an improved understanding of the role played by miRNAs in the transmission of Varroa-borne viruses and potential targets for the control and prevention of mites and viruses in bee colonies.
Materials and Methods
Varroa Mite Collection
Varroa mites were collected from the honey bee apiary at the University of Southern Mississippi’s Lake Thoreau Environmental Center, Hattiesburg, MS (31° 20’ 54.476” N and 89° 25’ 9.202” W) between August and October, 2019. The apiary comprises five healthy and well-established Italian Apis mellifera ligustica colonies that had successfully survived the mild winter in MS the previous year (2018) and that had not received Varroa treatment of any kind in 2019 prior to the sampling period. Capped brood and mites were collected from a single-well populated queen-right colony with two chambers hosted in a classic Langstroth hive. Adult female mites were collected from adult honey bee workers using the sugar shake method (Gregorc et al., 2018). Male Varroa were collected from sealed pupal cells by removing the wax cap and positively identifying males in infested cells by microscopy. Ten male and 20 female Varroa were collected separately in tubes containing Trizol for RNA extraction. Infection with DWV serotypes A, B, or C was determined using specific primers as described in Kevill et al. (2017).
RNA Extraction
RNA was extracted from male and female mites separately using the Trizol extraction method (Chomczynski and Mackey, 1995) with some modifications to the original protocol. Briefly, mites were collected individually and then male and female mites were pooled in separate tubes. Mites were homogenized in 500 µl Trizol using a plastic pestle. After homogenization, samples were mixed for 10 min at room temperature in a shaker followed by centrifugation at 15000 x g for 10 min at 4°C. The supernatant was transferred to a new tube and incubated for 5 min at room temperature to permit complete dissociation of the nucleoproteins. One hundred µl chilled chloroform was added to the samples, mixed, and incubated for 10 min at 4°C. Samples were centrifuged at 15000 x g for 15 mins to obtain an aqueous phase. The aqueous phase was transferred to a new tube and then 600 µl of isopropanol was added before storage overnight at -20°C. The following day, samples were centrifuged for 15 min at 4°C followed by washing the pellets in 70% ethanol. RNA pellets were dried, and RNA samples were resuspended in sterile water and quantified using a NanoDrop instrument (Thermo Fisher Scientific, Waltham, MA).
Small RNA Sequencing
The RNA concentration was 286.5 ng/ul (260/280 = 1.8) for pooled Varroa females and 185.6 ng/ul (260/280 = 1.8) for pooled Varroa males. Small RNA libraries were prepared using the Illumina TruSeq kit following the manufacturer’s instructions (Illumina, San Diego, CA). Briefly, short adapter oligonucleotides were ligated to each end of the small RNAs in the samples. Individual cDNA copies were made with reverse transcriptase, and PCR was used to add sample-specific barcodes and Illumina sequencing adapters. The final concentration of all NGS libraries was determined using a Qubit fluorometric assay. The cDNA fragment size of each library was assessed using a DNA 1000 high-sensitivity chip on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Before sequencing, samples passed the essential quality control (QC) test. After purification by polyacrylamide gel electrophoresis, the sample libraries were pooled and sequenced on an Illumina NextSeq 500 (300 cycles, single-end, 36 bases) using the TruSeq SBS kit v3 (Illumina) and protocols defined by the manufacturer. Four small RNA libraries of pooled male and female Varroa mites were sequenced via Illumina small RNA high-throughput sequencing. RNA library preparation and indexing were performed by the University of Mississippi Medical Center genomics core facility.
Bioinformatics Analysis
The miRDeep2 version 2.0.0.8 software package (Friedländer et al., 2008; Friedländer et al., 2012) was used to process the sequencing data. The reads from all samples were combined for novel miRNA prediction. The mapper function of miRDeep2 was used to trim the adapter sequences from the reads and convert the read files from FASTQ to FASTA format. Reads shorter than 18 bases were discarded. Remaining reads were then mapped to the V. destructor reference genome (GCF_002443255.1_Vdes_3.0_genomic.fna) (Techer et al., 2019) using the default miRDeep2 mapper function parameters. Reads mapping to the genome were used to predict novel miRNAs. The Drosophila melanogaster genome was also provided as a reference genome, and mapped reads were aligned to available miRNAs of D. melanogaster in miRbase v22 and quantified. The output file included the sequence and location of the possible miRNAs, the number of reads mapping to them, and a score reflecting the likelihood that the predicted miRNA was not due to chance alone. The software aligned the reads to the reference genomes of D. melanogaster and V. destructor and looked for locations where potential miRNA reads accumulated. The regions immediately surrounding the mapped reads were examined for miRNA biogenesis features including mature miRNAs, star and precursor reads, and stem-loop folding properties. The miRDeep2 program models the miRNA biogenesis pathway using a probabilistic algorithm to score compatibility of the position and frequency of next-generation sequencing (NGS) reads with the secondary structure of the miRNA precursor. BEDtools (Quinlan and Hall, 2010) was used to determine the genomic origin of the precursors with the variant intersect.
Validation of Differentially-Expressed miRNAs by qRT-PCR
The miRprimer2 algorithm (Busk, 2014) was used to design qRT-PCR primers for the predicted miRNAs. Predicted miRNAs differentially expressed in small RNA sequencing data were validated by qRT-PCR. RNA samples from seven male and eleven female Varroa were separately pooled to extract RNA for cDNA synthesis, and the miRNA-specific qRT-PCR reaction was performed for each sample. The Mir-X miRNA qRT-PCR TB Green kit from Takara Bio (Kusatsu, Shiga, Japan; catalog # 638316) was used for cDNA synthesis and miRNA expression analysis. Conditions for qRT-PCR were: initial denaturation 95°C for 10 mins and then 40 cycles of 95°C for 5 secs and 60°C for 20 secs on a C1000 Touch Thermal cycler (Bio-Rad Laboratories, Hercules, CA; CFX96 Real-Time System). The primer sequences are shown in Supplementary Table S1.
Normalization, Differential Expression, and Statistical Analysis of miRNA Expression in Male and Female Varroa Mites
In silico differential expression (DE) analysis of predicted miRNAs was performed using the DeApp interactive web interface (Li and Andrade, 2017). DeApp is a web-based, graphical interface developed in R with the shiny package (web application framework for R). Low expression genetic features were removed after alignment if the counts per million (CPM) value was ≤1 in less than two samples. Sample normalization and a multidimensional scaling (MDS) plot are shown in Supplementary Figure S1 and include details about sample distribution after filtering out low expression genomic features. Differential expression (DE) analysis was performed using edgeR with the false discovery rate (FDR)-adjusted p-value set to 0.05 and minimum fold-change of 1.5. The interface displays a dispersion plot showing the results of overall DE analysis along with statistical significance (p-value, FDR adjusted p-value) and volcano plot corresponding to the specified parameters and cutoff values.
Prediction of the Target Genes, Proteome Re-Annotation, and Gene Ontology (GO) and KEGG Enrichment Analyses
The targeting algorithms TargetSpy (Sturm et al., 2010), MIRANDA (John et al., 2004), and PITA (Kertesz et al., 2007) were used in miRNAcons Target from sRNAtoolbox to predict the genes regulated by up- or downregulated Varroa miRNAs (Aparicio-Puerta et al., 2019). Targets common in all three programs were further considered. In silico target prediction resulted in a high number of false positives, but cross-species comparisons and combinatorial effects reduced this number (Min and Yoon, 2010). Lists of target genes were functionally characterized using the STRING webserver (Franceschini et al., 2013). The networks of target genes and the KEGG pathways significantly enriched for target genes were extracted using the STRING output (Table 3). PANNZER2 (Toronen et al., 2018) was used to functionally re-annotate the predicted proteome of up- or downregulated genes (targets of predicted miRNAs), and WEGO (Ye et al., 2006; Ye et al., 2018) was used to analyze and plot gene ontology (GO) annotations.
Results
Read Length Distribution of Small RNAs
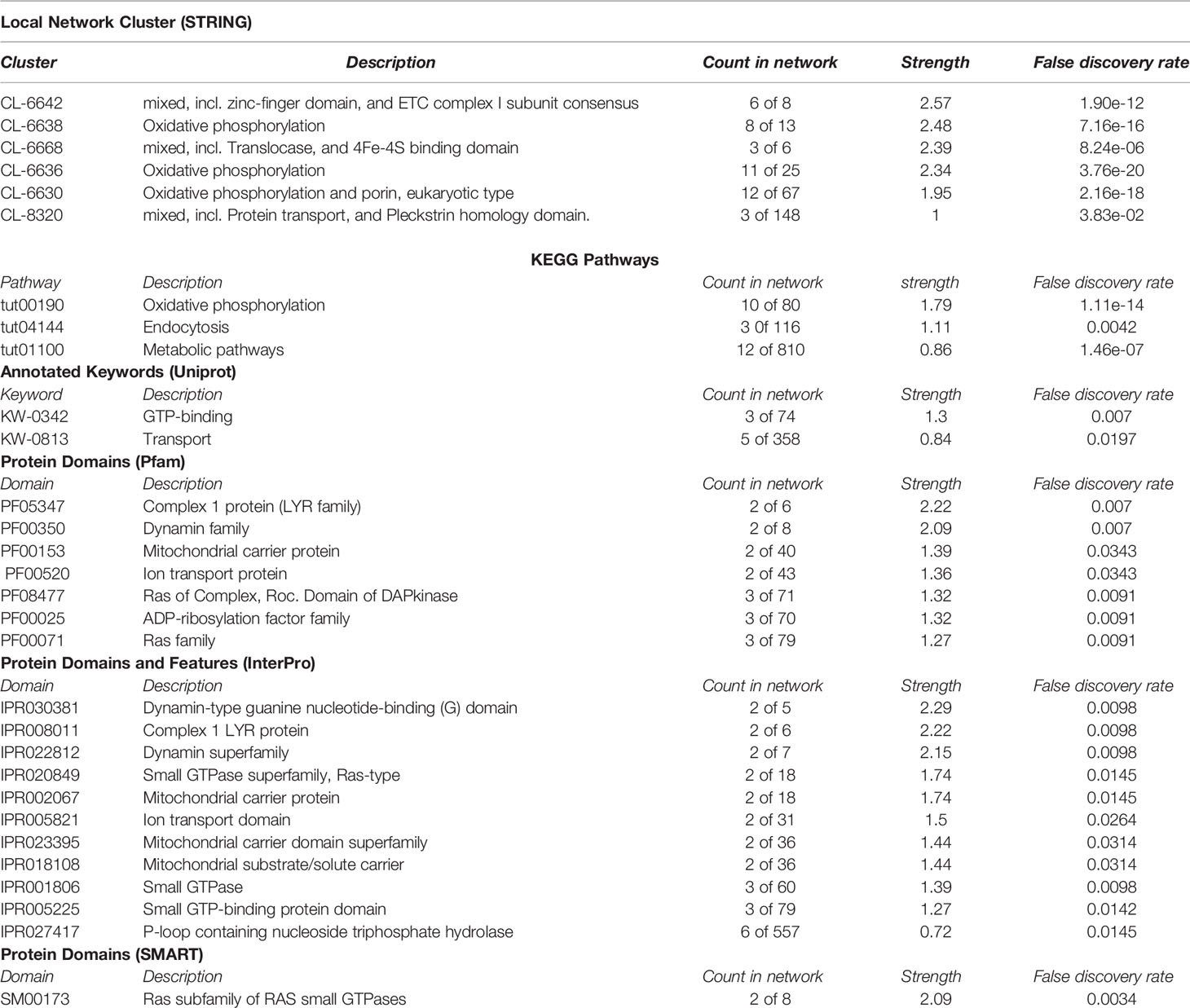
After adapter trimming and removal of short reads (≤18 nucleotides), 101,914,732 small RNA reads were available from Varroa males and 91,904,732 from females for downstream analysis. 27,204,163 male and 32,283,353 female Varroa reads matched to the Varroa genome. The read length distribution indicates the types of small RNAs present in male and female Varroa samples; Figure 1 depicts the number of short read sequences between 18 and 30 nucleotides, and three main peaks were distinguishable in both male and female samples: (i) a peak at 24 nucleotides, the highest abundance population of mature miRNAs; and (ii) relatively less abundant peaks at 22 and (iii) 23 nucleotides. There were ~2.2 x 107 and ~1.8 x 107 24 nucleotide miRNA sequences in male and female Varroa, respectively, and ~1.25 x 107 and ~1.8 x 107 22 and 23 nucleotide small RNA sequences in males and ~1.0 x107 and ~1.7 x 107 22 and 23 nucleotide small RNA sequences in females, respectively. The read length distribution in male Varroa was qualitatively identical to females.
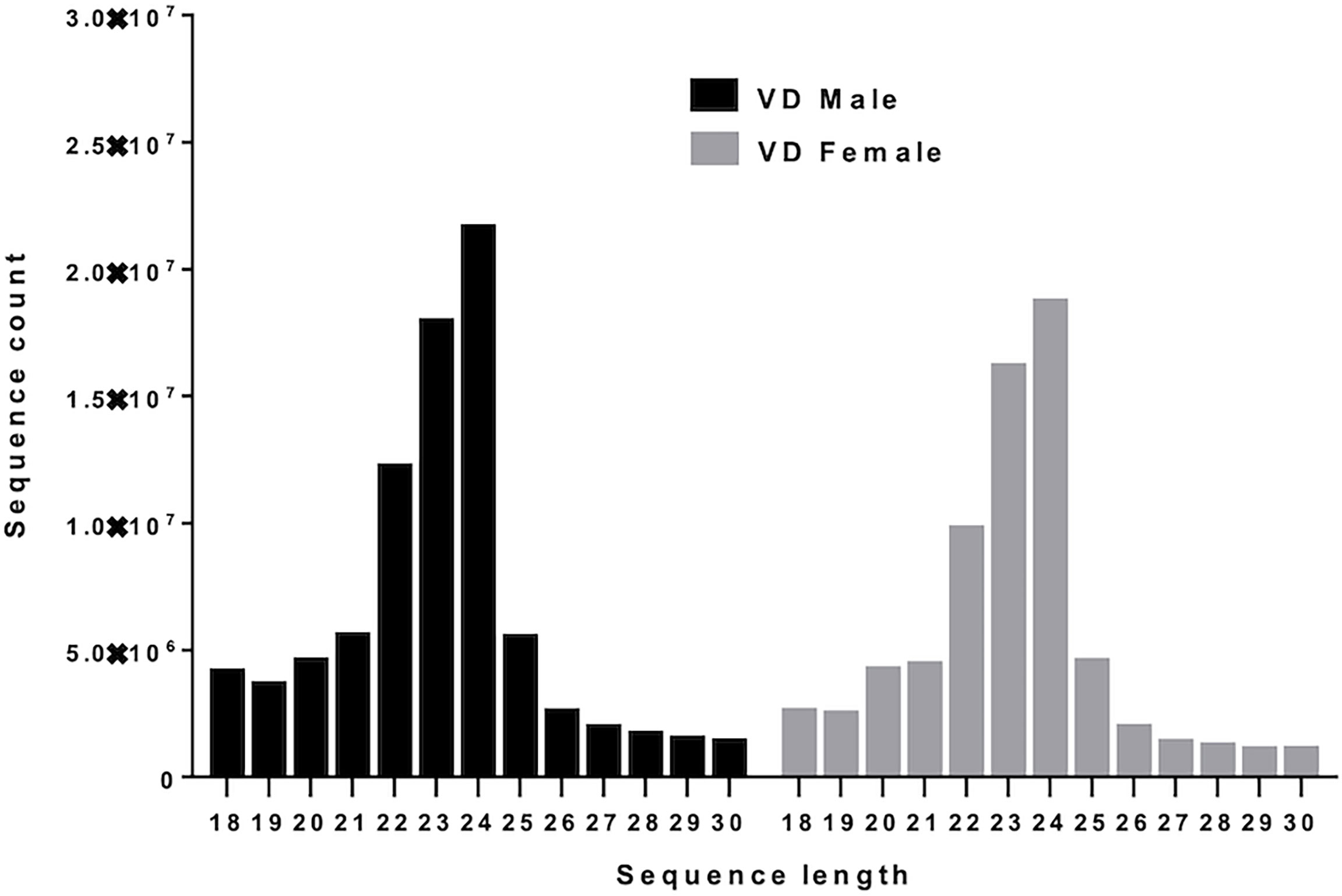
Figure 1 Small RNA sequence length distribution of microRNAs in male and female Varroa destructor (VD) mites. MicroRNAs are twenty-four (24) nucleotides in length.
MicroRNA Profiles of Varroa
Figure 2A shows the basic hairpin loop structure of a microRNA and other parameters (dicer cut overhangs, total read count, mature read count, loop read count, total read count, randfold score, and total score) used to determine whether a hairpin loop-structured RNA is an miRNA. A total of 306 microRNAs were predicted in V. destructor, and several had homologs in D. melanogaster. The predicted miRNAs were categorized as high (n=50) and low confidence (n=80) miRNAs (Figure 2B and Table S2) based on standard criteria (Bartel, 2018). Eighteen of the predicted miRNAs were upregulated and 13 were downregulated in female Varroa compared with male Varroa (Figures 3A, B and Table 1). Two of the predicted mature microRNAs, nDS_019211455.1_10989 and nDS_019211455.1_16072, were male-specific, and three of the predicted miRNAs (nDS_019211455.1_10999, nDS_019211459.1_37116, and nDS_019211455.1_10993; Table S1, S3) were female-specific. Supplementary Table S2 includes the consensus mature, star, total read counts, and miRDeep2 scores and probabilities for every miRDeep2-predicted miRNA with an miRDeep2 score.
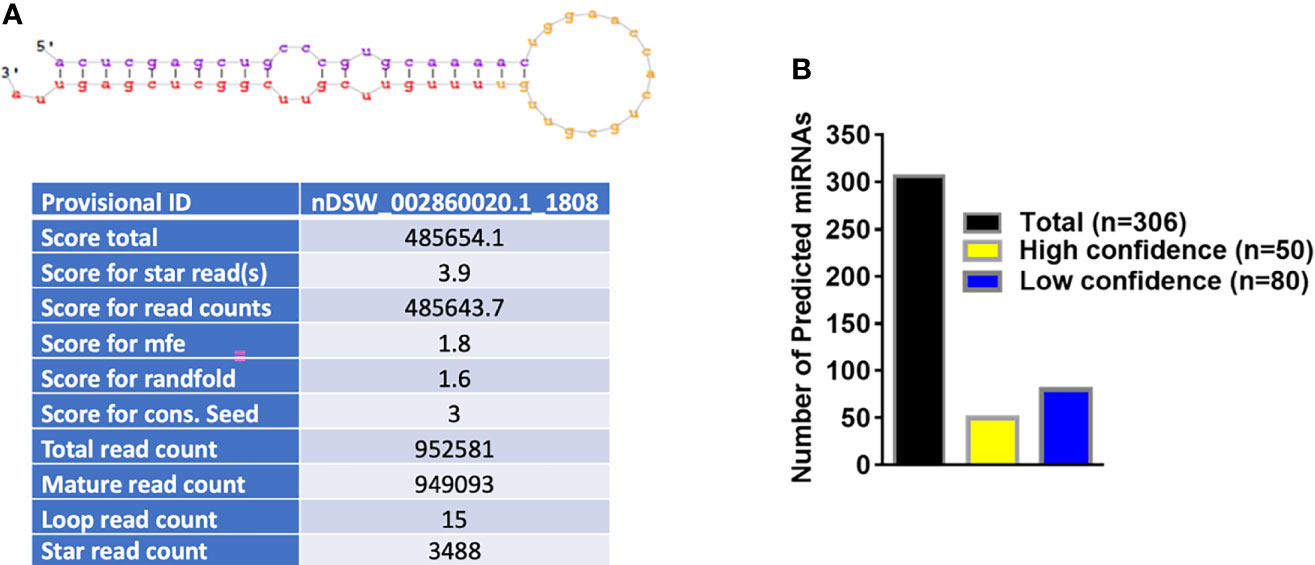
Figure 2 (A) Basic stem-loop structure of a predicted microRNA. miRDeep2 was used to identify potential miRNA precursors based on nucleotide length, star sequence, stem-loop folding, and homology to the Varroa reference genome. Shown are the predicted stem-loop structures (yellow), star (violet), and mature sequences (red). (B) Annotation of predicted Varroa destructor microRNAs. 306 microRNAs were predicted in Varroa samples. 50 were categorized as high-confidence and 80 as low-confidence miRNAs based on standard criteria.
Figure 3 (A) In silico differential expression analysis of predicted microRNAs in female Varroa destructor relative to males. EdgeR was used for differential expression analysis. 13 predicted microRNAs were downregulated, 18 upregulated, and 60 were unaffected. (B) Differential expression of individual microRNAs in Varroa females relative to males. miRNAs with a log2 fold-change expression > |1| and FDR ≤ 0.1 were considered significantly differentially expressed with respect to male miRNAs (refer to Table 1).
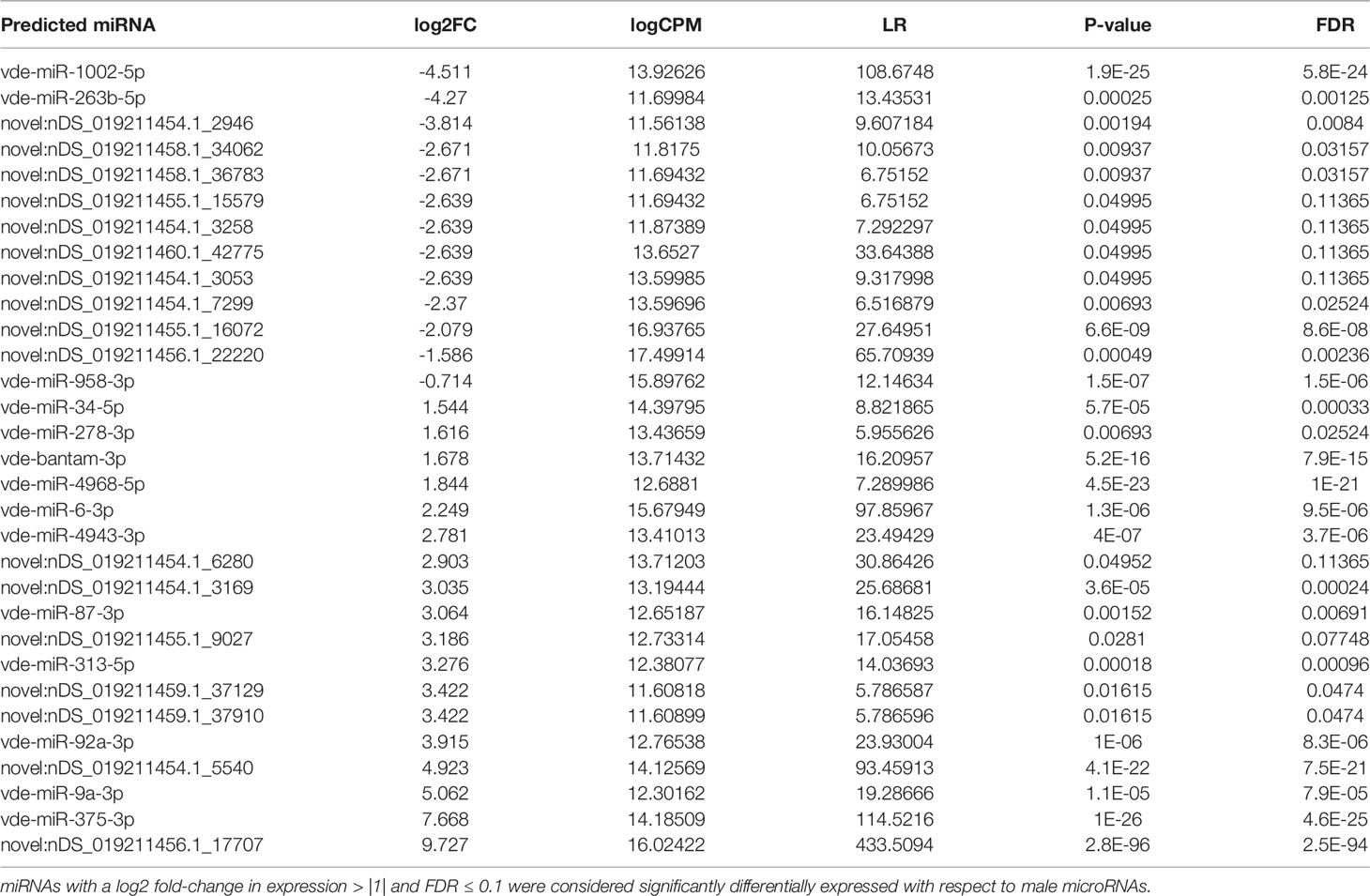
Table 1 In silico differential expression analysis of whole-body female Varroa mite microRNAs relative to male microRNAs.
In Silico Mapping of Varroa Small RNA Sequences to Apis mellifera and DWV-B (Deformed Wing Virus) Genomes
In silico mapping of Varroa small RNA sequences to the Apis mellifera genome (GCF_003254395.2_Amel_HAv3.1_genomic.fna) detected 112 miRNA orthologs (Table S4). We used the DWV-B genome (GCF_000856945.1_ViralProj15121_genomic.fna.gz; https://www.ncbi.nlm.nih.gov/assembly/GCF_000856945.1/) as the DWV reference. In silico mapping of female Varroa small RNA sequences suggested that female samples were DWV-infected, since 0.18% of female Varroa mite sequences mapped to the DWV-B genome. The few viral miRNAs predicted in the analysis had very low mIRDeep2 scores so could not be regarded as predicted viral miRNAs.
MicroRNA Differential Expression Analysis by qRT-PCR
Several of the predicted Varroa miRNAs were conserved in D. melanogaster, and previous studies revealed their conserved roles in development, replication, viral colonization, and viral inhibition in other arthropods (Figure 4A and Table 2). These high-confidence predicted miRNAs such as vde-miR-87-3p, vde-bantam-3p, vde-miR-375-3p, and vde-miR-34-5p were upregulated 100- to 20,000-fold in DWV-infected female relative to male Varroa by qRT-PCR analysis (Figure 4B).
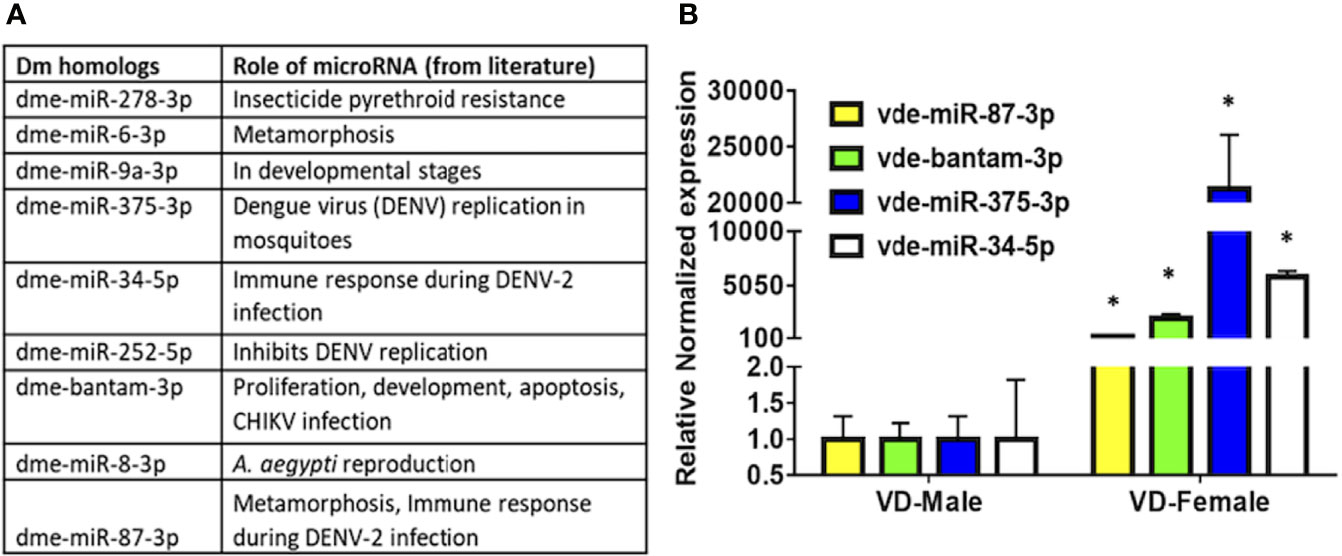
Figure 4 (A) MicroRNA candidates predicted in this study, conserved in other arthropods, and validated in the available literature play significant roles in replication, harboring, and inhibition of viral pathogens. (B) qRT-PCR expression of potential microRNAs found in the present study in DWV-B-infected male and female Varroa mites. Succinate dehydrogenase (SDHA) was used as the housekeeping gene for normalization. Expression of these microRNAs is comparatively higher in female than male Varroa. Statistical significance for qRT-PCR-based differential expression was determined using the two-tailed Student’s t-test, where * denotes p < 0.05.
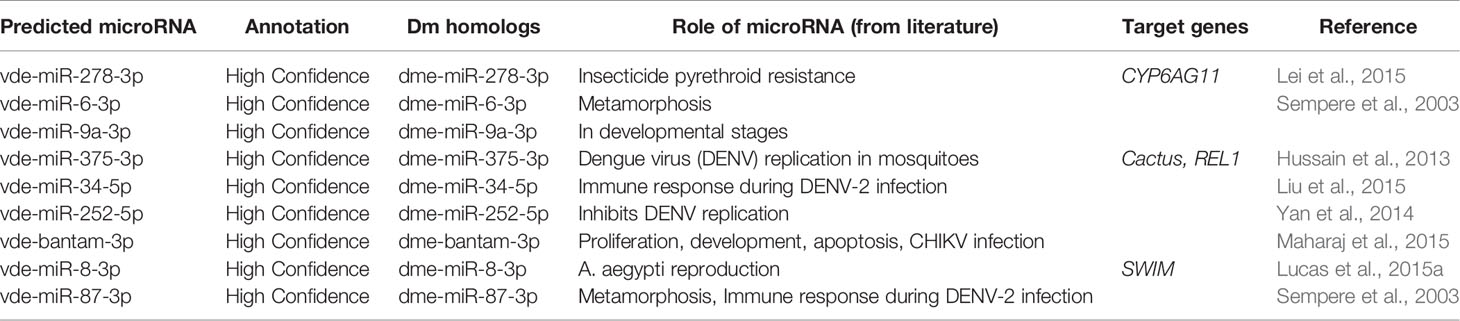
Table 2 List of potential microRNA candidates predicted in this study, conserved in Drosophila melanogaster (Dm), and with evidence in the available literature play a significant role in the development, replication, harboring, and inhibition of viral pathogens.
Other Small RNA Categories
A summary of reads from both male and female Varroa matching various small RNA categories is shown in Figure 5. Other small RNAs include signal recognition particle (SRP), protein-coding, and not annotated RNAs. Of the total small RNA reads from female Varroa, 56 x 10-4% were miRNAs, 15 x 10-2 were rRNAs, 5 x 10-1 were tRNAs,13 x 10-4 were snRNAs, 29 x 10-5 were snoRNAs, and 99.35% were others. Of the total small RNA reads from male Varroa, 25 x 10-4% were miRNAs, 15 x 10-3 were rRNAs, 94 x 10-3 were tRNAs, 4 x 10-4 were snRNAs, 2.35 x 10-5 were snoRNAs, and 99.89% were others.
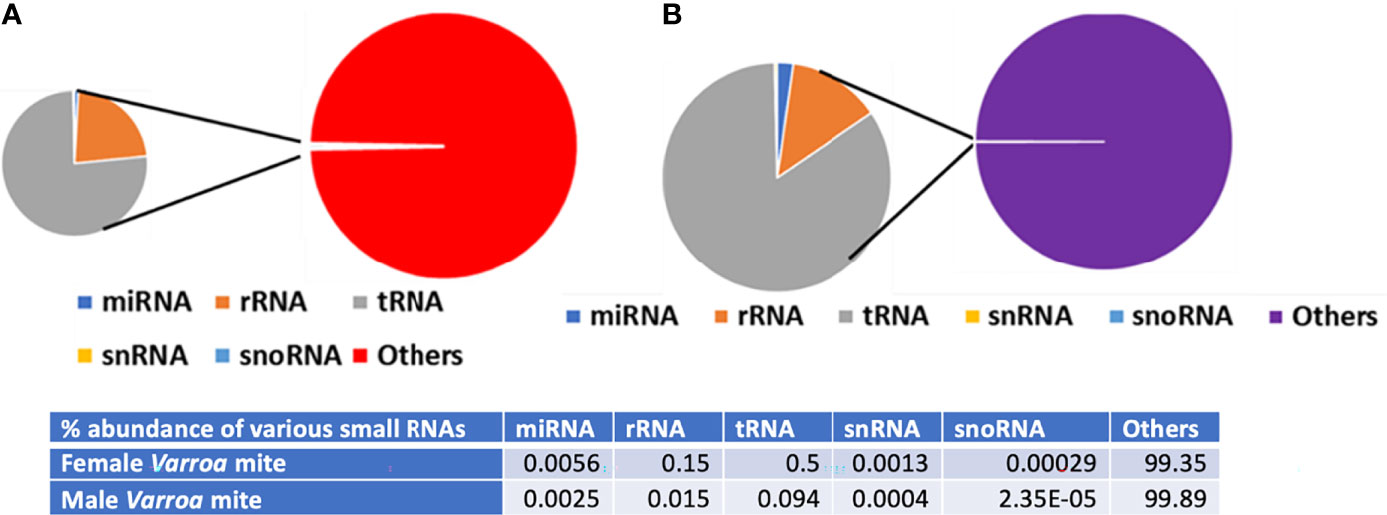
Figure 5 Summary of Varroa-derived female (A) and male (B) small RNA reads matching various small RNA categories.
Prediction of Target Genes and Gene Ontology (GO) and Functional Enrichment Analyses in the Target Network
STRING web analysis (Figure 6A) showed that target proteins for the 31 predicted miRNAs (18 upregulated and 13 downregulated) had more interactions than expected for a random set of proteins of similar size sampled from the V. destructor genome (number of nodes = 42, number of edges = 97, average node degree = 4.62, average local clustering coefficient = 0.476, expected number of edges = 25, PPI enrichment p-value < 1.0e-16). Such enrichment indicates that the proteins are at least partially biologically connected as a group. To minimize the number of false-positive targets, we opted only for targets predicted by all three miRNA target programs (TargetSpy, MIRANDA, and PITA). The predicted miRNAs were therefore of high confidence, and the main KEGG pathways involved were oxidative phosphorylation (12 out of 80), endocytosis (4 out of 116), protein processing in the endoplasmic reticulum (2 out of 109), and other metabolic pathways (14 out of 810). Many target genes were predicted for the differentially-expressed miRNAs in the small RNA-seq data (Figure 6B) using the miRNAconsTarget program from sRNAtoolbox (Aparicio-Puerta et al., 2019).
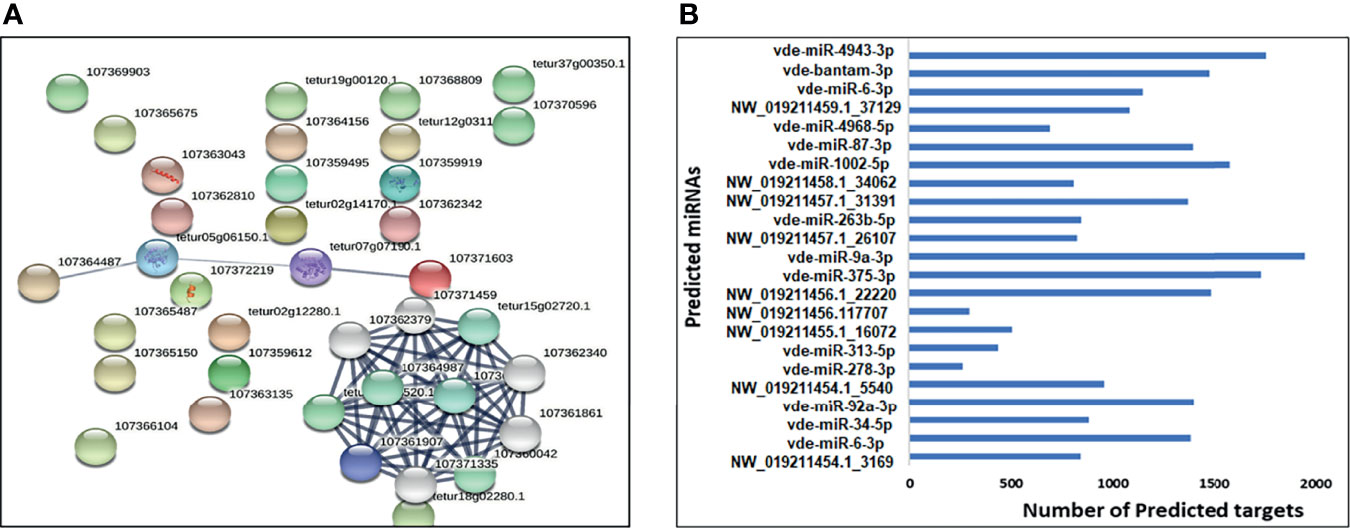
Figure 6 (A) A network built exclusively from Varroa proteins targeted by predicted upregulated/downregulated miRNAs. (B) The total number of predicted target transcripts significantly upregulated and downregulated in female Varroa miRNAs relative to male miRNAs.
Validation of Predicted miRNAs in Female Varroa by qRT-PCR
The expression of 13 predicted differentially-expressed miRNAs were validated using qRT-PCR (Figure 7). The qRT-PCR results of differentially-expressed miRNAs matched the NGS patterns for the majority of evaluated miRNAs. Inconsistencies in NGS and qRT-PCR data were found for vde-miR-4943-3p, nDS_019211456.1_17707, and vdemiR-4968-5p, which could be due to using different methodologies to quantify miRNA expression (Saldaña et al., 2017). Inconsistencies between NGS and qRT-PCR data have been reported previously (Hermance et al., 2019).
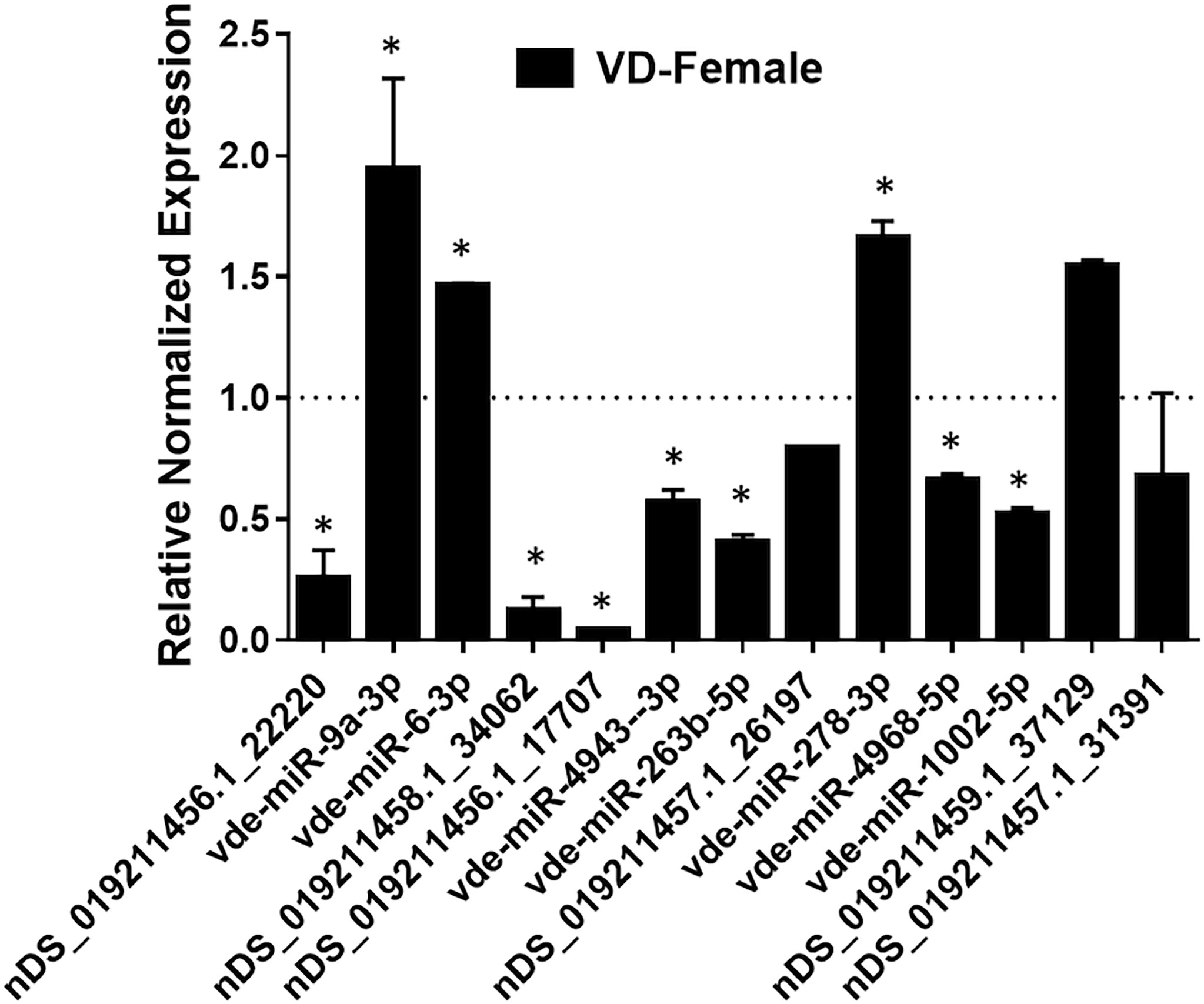
Figure 7 qPCR validation of selected mature miRNAs (high confidence) in female Varroa differentially expressed relative to male Varroa. Statistical significance for qRT-PCR-based differential expression was determined by the 2-tailed Student’s t-test, where * denotes p<0.05. miRNA expression of female Varroa was normalized to that of male Varroa (indicated as 1 on y-axis). Succinate dehydrogenase (SDHA) was used as a housekeeping control.
Discussion
MicroRNAs are indispensable post-transcriptional regulators of gene expression in various biological pathways including cell growth, apoptosis, metamorphosis, development, and vector competence (Kloosterman and Plasterk, 2006). However, little is known about miRNA expression in parasitic mites such as Varroa destructor. Therefore, identifying and understanding miRNA expression profiles in male and female Varroa expand our fundamental knowledge of mite miRNAs and provide insight into the development and vector competence of Varroa.
A conservative in silico approach identified 306 miRNAs, among which 80 were low-confidence and 50 high-confidence miRNAs. Among these high-confidence miRNAs, 18 were up- and 13 were downregulated in female compared with male Varroa. qRT-PCR validation revealed upregulation of several predicted miRNAs including vde-miR-87-3p, vde-bantam-3p, vde-miR-375-3p, and vde-miR-34-5p in DWV-B-infected female Varroa samples, which have known roles in DENV replication, CHIKV infection, and immune responses to DENV-2 infection in mosquitoes (Sempere et al., 2003; Hussain et al., 2013; Yan et al., 2014; Liu et al., 2015; Lucas et al., 2015a; Maharaj et al., 2015). This small RNA dataset provides a new resource to characterize miRNA function in mite biology. Other predicted Varroa miRNAs have also been implicated in development, pesticide resistance, metamorphosis, immune responses, and reproduction (Figure 4A and Table 2) in other arthropods (Sempere et al., 2003; Hussain et al., 2013; Yan et al., 2014; Lei et al., 2015; Liu et al., 2015; Lucas et al., 2015a; Maharaj et al., 2015). The new and novel miRNAs predicted in this study will require functional validation using miRNA inhibitory experiments.
Among the predicted miRNAs in this study, many were novel and are likely Varroa-specific (Table S2). Several were homologous to the miRNAs already identified in other arthropods such as Apis mellifera, Drosophila melanogaster, and Ixodes scapularis (Tables S2, S4), suggesting conserved roles for these miRNAs (Marco et al., 2010; Fonseca et al., 2021). One hundred and twelve miRNAs detected in Varroa in this study have already been identified in A. mellifera. Indeed, it has been shown that siRNA molecules ingested by A. mellifera can be transferred to ectoparasitic Varroa mites and vice-versa. Small RNA exchange of this kind suggests cross-kingdom interactions and could possibly contribute to the shared miRNA profile (Garbian et al., 2012; Fonseca et al., 2021). In general, the most conserved miRNAs are involved in common cellular, biological, and development processes.
Several of the predicted miRNAs identified in this study are also conserved in D. melanogaster such as let-7-5p, bantam-3p, miR-8-3p, miR-34-5p, miR-263a-5p, miR-87-3p, miR-252-5p, miR-12-5p, miR-375-3p, miR-9a-3p, miR-306-5p, miR-133-3p, miR-6-3p, miR-276-a-3p, miR-1002-5p, and miR-304-5p, which are known to have conserved roles in other arthropods as well. bantam-3p targets the proapoptotic gene hid and is involved in several cellular processes such as proliferation, apoptosis, development, and the circadian clock (Brennecke et al., 2003; Kadener et al., 2009). In Aedes aegypti, bantam-3p is significantly upregulated during pupal developmental, and the highest expression has been reported at the mid-pupal period (Bryant et al., 2010). In another study, bantam-3p was most abundantly expressed in both pupal and adult male and female mosquitoes, indicating its functional importance (Feng et al., 2018) and a possible role in Varroa development. Another conserved miRNA, miR-8-3p, was significantly upregulated in Aedes aegypti during pupation and has the highest expression in the mid-pupal period. Bryant et al. (2010) showed upregulation of miR-8-3p in the fat body of a blood-fed female mosquito and suggested a potential regulatory role in Ae. aegypti reproduction. Different to in Ae. aegypti, miR-8-3p is abundantly expressed in Anopheles stephensi developmental stages (Feng et al., 2018) and found to be equally expressed in uninfected or infected Ae. albopictus saliva upon infection with CHIKV (Maharaj et al., 2015). In Ae. aegypti, miR-8-3p has been validated to target SWIM (secreted wingless-interacting molecule), thereby regulating reproductive events (Lucas et al., 2015a). Additionally, miR-8-3p shows cell type-specific expression and it is expressed in S2 cells (a cell line derived from Drosophila melanogaster embryos). Its temporal expression is less restricted in Drosophila and has been observed across all developmental stages, occasionally with significant variation in expression (Jin et al., 2012). Based on these studies and the conserved nature of miR-8-3p, it is also likely to play a role in Varroa development, reproduction, and virus infection and requires further investigation.
Another Varroa-predicted miRNA common to Drosophila was miR-34-5p, the expression of which is more pronounced in female midguts in A. gambiae (Winter et al., 2007). In contrast, miR-34-5p is downregulated in Drosophila during metamorphosis (pupa to adult stage transition) (Sempere et al., 2003). Interestingly, in A. gambiae, miR-34-5p expression was downregulated in the midgut upon Plasmodium falciparum infection (Dennison et al., 2015). miR-34-5p has been suggested to contribute to anti-pathogen and immune responses during DENV-2 infection in Ae. albopictus (Liu et al., 2015). Again, the conserved nature of this miRNA suggests a possible role in development regulation in Varroa, but this requires further functional validation. qRT-PCR showed ~5000-fold upregulation of miR-34-5p in DWV-B-infected Varroa females compared with DWV-B infected males, indicating a possible role for this miRNA in anti-DWV-B and immune responses during DWV-B infection, or perhaps facilitating DWV-B survival inside Varroa. Similarly, miR-87-3p was upregulated in female Varroa, and previous studies have suggested a role for this miRNA in Drosophila development (Sempere et al., 2003) and anti-viral immune responses during DENV-2 infection in Ae. albopictus (Liu et al., 2015). Our qRT-PCR analysis also demonstrated ~300-fold upregulation in DWV-B-infected Varroa females, suggesting a possible role either in anti-DWV-B immune responses or in harboring DWV-B.
miR-375-3p was another miRNA of interest in our analysis. In Ae. aegypti, miR-375-3p is expressed in blood-fed mosquitoes, and miR-375 was found to regulate DENV replication, enhancing DENV-2 infection in an Ae. aegypti cell line (Hussain et al., 2013). In Ae. Aegypti, miR-375 targets include Cactus and REL1, and the injection of an miRNA mimic into mosquitoes altered the expression of immune gene transcripts, suggesting that aae-miR-375 enhances DENV-2 infection in Ae. aegypti (Hussain et al., 2013). Our qRT-PCR data revealed ~20,000-fold upregulation in DWV-B-infected Varroa, suggesting a possible role in DWV-B infection. Interestingly, miR-263a-5p was reported as upregulated in uninfected and CHIKV-infected Ae. aegypti saliva (Maharaj et al., 2015). miR263a-5p is constitutively expressed across many developmental stages in several mosquito species (Hu et al., 2015). Another conserved miRNA, dme-miR-252-5p, was induced over three-fold after DENV-2 infection in the Ae. albopictus C6/36 cell line and inhibited DENV replication by suppressing the expression of the DENV envelope (E) protein (Yan et al., 2014). In An. gambiae, dme-miR-12-5p was found to be expressed in the thorax of males and females but predominantly in midguts and constitutively expressed in their heads (Winter et al., 2007). dme-miR-12-5p targets DNA replication licensing factor (MCM6) and monocarboxylate transporter (MCT1) genes, as validated in Ae. aegypti, through which it affects Wolbachia density in host cells (Osei-Amo et al., 2012). dme-mir-279-3p is expressed evenly and ubiquitously throughout the An. gambiae body (Winter et al., 2007). dme-miR-278-3p was also predicted in our data, and it is highly and abundantly expressed in several reported studies (Feng et al., 2018). dme-miR-278-3p was upregulated in Culex pipiens pallens upon exposure to pyrethroid, a widely and indiscriminately used insecticide, where miR-278-3p targets CYP6AG11 to regulate pyrethroid resistance (Lei et al., 2015). The pyrethroid tau-fluvalinate (Apistan®) was one of the first synthetic varroacides registered in the United States, and pyrethroid-resistant Varroa is a major challenge in controlling this mite. Pyrethroid-resistant Varroa are associated with point mutations in the voltage-gated sodium channel gene at position 925 (Millán-Leiva et al., 2021). Further dissection of these mechanisms might provide a fruitful means to overcome pyrethroid resistance in Varroa.
The miRNAs predicted through Varroa small RNA-sequencing allow us to generate a list of conserved miRNA targets that may be involved in regulating key biological processes. As discussed earlier, conserved homologous Drosophila miRNAs predicted in Varroa (Figure 4A and Table 2) include vde-miR-87-3p, vde-bantam-3p, vde-miR-375-3p, and vde-miR-34-5p. These miRNAs have been shown to play a role in inhibiting or replicating DENV and CHIKV in mosquito species (Sempere et al., 2003; Hussain et al., 2013; Maharaj et al., 2015). These miRNAs were significantly upregulated (100-20,000-fold) in DWV-B-infected Varroa females (Figure 4B), indicating a functional role for these miRNAs in viral load warranting a detailed study to characterize these miRNAs in Varroa-borne virus transmission studies. As the expression of vde-miR-87-3p, vde-bantam-3p, vde-miR-375-3p, and vde-miR-34-5p was significantly higher in females than males, it is also possible that upregulation of these miRNAs could be sex- or age-specific rather than being related to DWV-B infection, and this requires further clarification. Indeed, some of the predicted mature miRNAs such as nDS_019211455.1_10989 and nDS_019211455.1_16072 were male specific and three of the predicted miRNAs (nDS_019211455.1_10999, nDS_019211459.1_37116, and nDS_019211455.1_10993; Tables S1, S3) were female-specific. A previous study in the nematode Ascaris suum predicted the role of gender-specific miRNAs as elongation factors, heat shock proteins, and growth factors essential for organism development (Xu et al., 2013). Moreover, sperm proteins and sperm cell motility proteins were targets of male-specific miRNAs, while ovarian message proteins were targets of female-specific miRNAs (Xu et al., 2013). These male- and female-specific miRNAs in Varroa need further functional exploration.
In this study, the predominant peak of identified miRNAs corresponded to 24 nucleotides, consistent with previous genome-wide miRNA identification studies in Varroa (Fonseca et al., 2021). These miRNAs are major contributors to the total small RNA complement in male and female Varroa. It has been suggested that miRNAs might be actively involved in vector competence of Varroa-borne viruses. Using publicly-available Varroa genome data, we annotated and evaluated possible targets and functions of putative female Varroa miRNAs showing in silico upregulation or downregulation relative to males. The most conserved miRNAs were involved in several biological, cellular, and developmental processes. Genes controlled by miRNAs conserved in D. melanogaster and Varroa identified in this study regulate development, metamorphosis, proliferation, apoptosis, reproduction (Aedes aegypti), insecticide resistance, and also antiviral immune responses (Table 2). Interestingly, analysis of GO terms related to genes regulated by the shared miRNAs revealed many specific processes known to be regulated by the same miRNAs in D. melanogaster such as developmental processes, metamorphosis, and immune responses, reinforcing their conserved role (Figure 8 and Table 2). The main KEGG pathways predicted by STRING web analysis were oxidative phosphorylation, endocytosis, protein processing in the endoplasmic reticulum (ER), and other metabolic pathways (Table 3). Proteins involved in endocytosis and protein processing in the ER could provide significant clues about viral infection (Wang et al., 2013; Rosche et al., 2021) in Varroa.
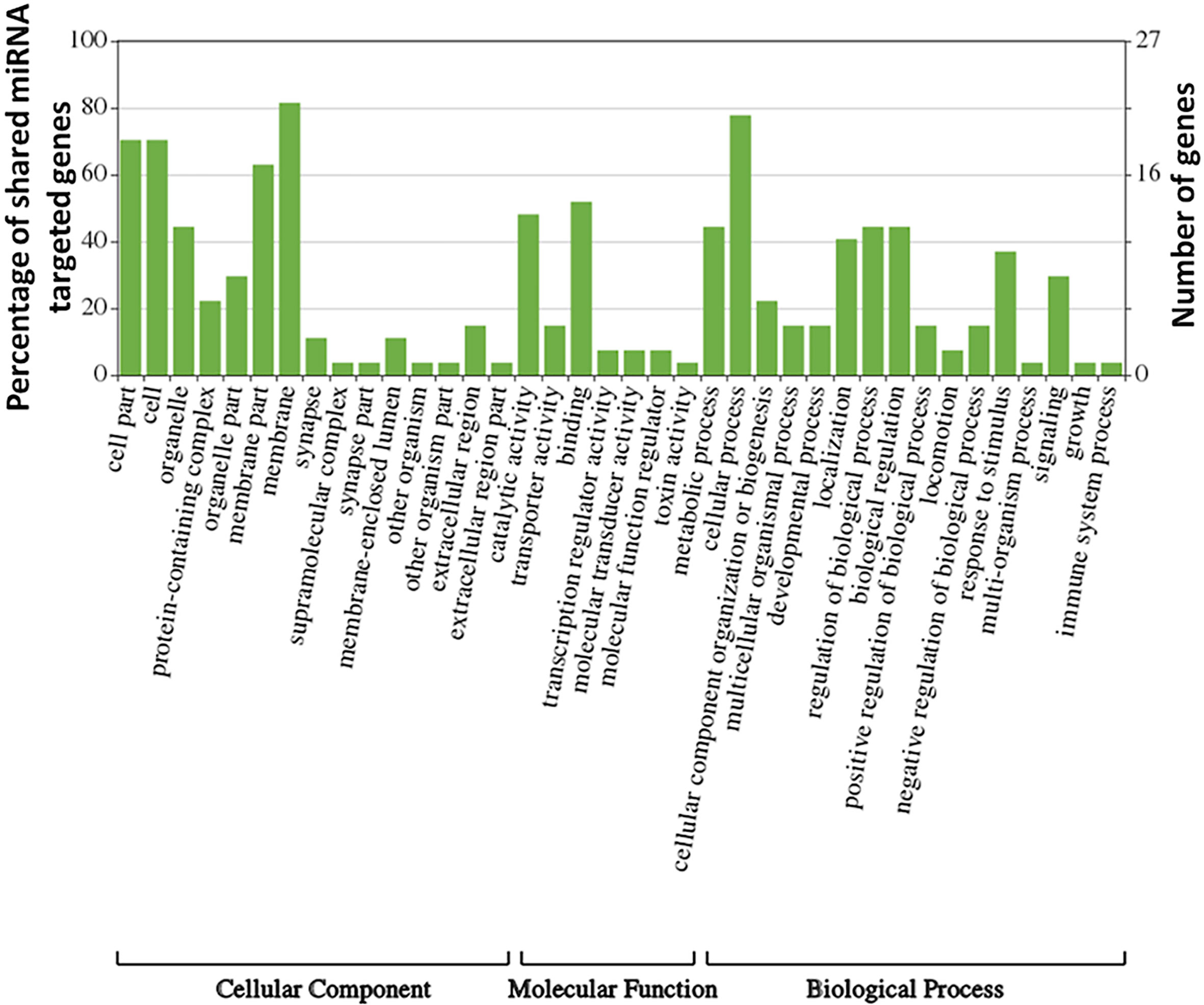
Figure 8 Gene ontology (GO)-derived biological processes related to genes targeted by upregulated/downregulated miRNAs in female V. destructor.
Conclusions
This study provides several potential miRNA targets that now require functional validation to confirm their role in the pathophysiology of Varroa. Characterization of Varroa miRNAs paves the way for a deeper understanding of Varroa biology and survival mechanisms of DWV inside Varroa so that these processes can be targeted to control and prevent both mites and the vectors they transmit.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: PRJNA794145).
Author Contributions
Conceptualization: DK and SK. Data curation: DK. Formal analysis: DK. Funding acquisition: SK and JA. Investigation: DK, MA, FT, MG, JA, and SK. Methodology: DK and SK. Project administration: SK. Resources: JA and SK. Supervision; SK. Validation: DK and SK. Visualization: DK. Writing, original draft: DK and SK. Writing, review & editing: DK, MA, MG, JA, and SK. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by a USDA ARS cooperative agreement and the Mississippi INBRE (an institutional Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under award P20GM103476).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This article reports the results of research only. Mention of a proprietary product does not constitute an endorsement or a recommendation by the USDA for its use. The USDA is an equal opportunity provider and employer.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.847000/full#supplementary-material
References
Annoscia, D., Brown, S. P., Di Prisco, G., De Paoli, E., Del Fabbro, S., Frizzera, D., et al. (2019). Haemolymph Removal by Varroa Mite Destabilizes the Dynamical Interaction Between Immune Effectors and Virus in Bees, as Predicted by Volterra's Model. Proc. Biol. Sci. 286, 20190331. doi: 10.1098/rspb.2019.0331
Aparicio-Puerta, E., Lebrón, R., Rueda, A., Gómez-Martín, C., Giannoukakos, S., Jaspez, D., et al. (2019). Srnabench and Srnatoolbox 2019: Intuitive Fast Small RNA Profiling and Differential Expression. Nucleic Acid Res. 47 (W1), W530–W535. doi: 10.1093/nar/gkz415
Asgari, S. (2014). Role of microRNAs in Arbovirus/Vector Interactions. Viruses 6, 3514–3534. doi: 10.3390/v6093514
Barrero, R. A., Keeble-Gagnère, G., Zhang, B., Moolhuijzen, P., Ikeo, K., Tateno, Y., et al. (2011). Evolutionary Conserved microRNAs Are Ubiquitously Expressed Compared to Tick-Specific miRNAs in the Cattle Tick Rhipicephalus (Boophilus) Microplus. BMC Genom. 12, 1–17. doi: 10.1186/1471-2164-12-328
Beaurepaire, A. L., Ellis, J. D., Krieger, K. J., Moritz, R. F. A. (2019). Association of Varroa Destructor Females in Multiply Infested Cells of the Honeybee Apis Mellifera. Insect Sci. 26, 128–134. doi: 10.1111/1744-7917.12529
Beaurepaire, A., Piot, N., Doublet, V., Antunez, K., Campbell, E., Chantawannakul, P., et al. (2020). Diversity and Global Distribution of Viruses of the Western Honey Bee, Apis mellifera. Insects 11 (4), 239. doi: 10.3390/insects11040239
Behura, S. K. (2007). Insect microRNAs: Structure, Function and Evolution. Insect Biochem. Mol. Biol. 37, 3–9. doi: 10.1016/j.ibmb.2006.10.006
Benaets, K., Van Geystelen, A., Cardoen, D., De Smet, L., de Graaf, D. C., Schoofs, L., et al. (2017). Covert Deformed Wing Virus Infections Have Long-Term Deleterious Effects on Honeybee Foraging and Survival. Proc. Biol. Sci. 284, 20162149. doi: 10.1098/rspb.2016.2149
Blair, C. D., Olson, K. E. (2015). The Role of RNA Interference (RNAi) in Arbovirus-Vector Interactions. Viruses 7, 820–843. doi: 10.3390/v7020820
Brennecke, J., Hipfner, D. R., Stark, A., Russell, R. B., Cohen, S. M. (2003). Bantam Encodes a Developmentally Regulated microRNA That Controls Cell Proliferation and Regulates the Proapoptotic Gene Hid in Drosophila. Cell 113, 25–36. doi: 10.1016/s0092-8674(03)00231-9
Brettell, L. E., Mordecai, G. J., Schroeder, D. C., Jones, I. M., Da Silva, J. R., Vicente-Rubiano, M., et al. (2017). A Comparison of Deformed Wing Virus in Deformed and Asymptomatic Honey Bees. Insects 8, 28. doi: 10.3390/insects8010028
Bryant, B., Macdonald, W., Raikhel, A. S. (2010). microRNA miR-275 Is Indispensable for Blood Digestion and Egg Development in the Mosquito Aedes Aegypti. Proc. Natl. Acad. Sci. U. S. A. 107, 22391–22398. doi: 10.1073/pnas.1016230107
Busk, P. K. (2014). A Tool for Design of Primers for microRNA-Specific Quantitative RT-qPCR. BMC Bioinf. 15, 29. doi: 10.1186/1471-2105-15-29
Chomczynski, P., Mackey, K. (1995). Short Technical Reports. Modification of the TRI Reagent Procedure for Isolation of RNA From Polysaccharide- and Proteoglycan-Rich Sources. Biotechniques 19 (6), 942–945.
De Miranda, J. R., Bailey, L., Ball, B. V., Blanchard, P., Budge, G. E., Chejanovsky, N., et al. (2013). Standard Methods for Virus Research in Apis Mellifera. J. Apicult. Res. 52, 1–56. doi: 10.3896/IBRA.1.52.4.22
de Miranda, J. R., Genersch, E. (2010). Deformed Wing Virus. J. Invertebr. Pathol. 103 Suppl 1, S48–S61. doi: 10.1016/j.jip.2009.06.01
Dennison, N. J., BenMarzouk-Hidalgo, O. J., Dimopoulos, G. (2015). MicroRNA-Regulation of Anopheles Gambiae Immunity to Plasmodium Falciparum Infection and Midgut Microbiota. Dev. Comp. Immunol. 49, 170–178. doi: 10.1016/j.dci.2014.10.016
Di Prisco, G., Annoscia, D., Margiotta, M., Ferrara, R., Varricchio, P., Zanni, V., et al. (2016). A Mutualistic Symbiosis Between a Parasitic Mite and a Pathogenic Virus Undermines Honey Bee Immunity and Health. Proc. Natl. Acad. Sci. U. S. A. 113 (12), 3203–3208. doi: 10.1073/pnas.1523515113
Fagegaltier, D., König, A., Gordon, A., Lai, E. C., Gingeras, T. R., Hannon, G. J., et al. (2014). A Genome-Wide Survey of Sexually Dimorphic Expression of Drosophila miRNAs Identifies the Steroid Hormone-Induced miRNA Let-7 as a Regulator of Sexual Identity. Genetics 198, 647–668. doi: 10.1534/genetics.114.169268
Feng, X., Zhou, S., Wang, J., Hu, W. (2018). microRNA Profiles and Functions in Mosquitoes. PLoS Neg. Trop. Dis. 12 (5), e0006463. doi: 10.1371/journal.pntd.0006463
Filipowicz, W., Bhattacharyya, S. N., Sonenberg, N. (2008). Mechanisms of Post-Transcriptional Regulation by microRNAs: Are the Answers in Sight? Nat. Rev. Genet. 9, 102–114. doi: 10.1038/nrg2290
Fonseca, P. L. C., Mucherino, J. J., Porto, J. A. M., Aemache, J. N., de Almeida, J. P. P., de Silva, F. F., et al. (2021). Genome-Wide Identification of miRNAs and Target Regulatory Newtwork in the Invasive Ectoparasitic Mite Varroa Destructor. Genomics 113, 2290–23-3. doi: 10.1016/j.ygeno.2021.05.028
Franceschini, A., Szklarczyk, D., Frankild, S., Kuhn, M., Simonovic, M., Roth, A., et al. (2013). STRING V9.1: Protein-Protein Interaction Networks, With Increased Coverage and Integration. Nucleic Acids Res. 41, D808–D815. doi: 10.1093/nar/gks1094
Fricke, C., Green, D., Smith, D., Dalmay, T., Chapman, T. (2014). MicroRNAs Influence Reproductive Responses by Females to Male Sex Peptide in Drosophila Melanogaster. Genetics 198, 1603–1619. doi: 10.1534/genetics.114.167320
Friedländer, M. R., Chen, W., Adamidi, C., Maaskola, J., Einspanier, R., Knespel, S., et al. (2008). Discovering microRNAs From Deep Sequencing Data Using miRDeep. Nat. Biotechnol. 26 (4), 407–415. doi: 10.1038/nbt1394
Friedländer, M. R., Mackowiak, S. D., Li, N., Chen, W., Rajewsky, N. (2012). Mirdeep2 Accurately Identifies Known and Hundreds of Novel microRNA Genes in Seven Animal Clades. Nucleic Acids Res. 40, 37–52. doi: 10.1093/nar/gkr688
Gallai, N., Salles, J. M., Settele, J., Vaissière, B. E. (2009). Economic Valuation of the Vulnerability of World Agriculture Confronted With Pollinator Decline. Ecol. Econ. 68, 810–821. doi: 10.1016/j.ecolecon.2008.06.014
Garbian, Y., Maori, E., Kalev, H., Shafir, S., Sela, I. (2012). Bidirectional Transfer of RNAi Between Honey Bee and Varroa Destructor: Varroa Gene Silencing Reduces Varroa Population. PLoS Pathog. 8, e1003035. doi: 10.1371/journal.ppat.1003035
Gisder, S., Aumeier, P., Genersch, E. (2009). Deformed Wing Virus: Replication and Viral Load in Mites (Varroa Destructor). J. Gen. Virol. 90, 463–467. doi: 10.1099/vir.0.005579-0
Gisder, S., Möckel, N., Eisenhardt, D., Genersch, E. (2018). In Vivo Evolution of Viral Virulence: Switching of Deformed Wing Virus Between Hosts Results in Virulence Changes and Sequence Shifts. Environ. Microbiol. 20, 4612–4628. doi: 10.1111/1462-2920.14481
Goulson, D., Nicholls, E., Botías, C., Rotheray, E. L. (2015). Bee Declines Driven by Combined Stress From Parasites, Pesticides, and Lack of Flowers. Science 347, 1255957. doi: 10.1126/science.1255957
Gregorc, A., Alburaki, M., Sampson, B., Knight, P. R., Adamczyk, J. (2018). Toxicity of Selected Acaricides to Honey Bees (Apis Mellifera) and Varroa (Varroa Destructor Anderson and Trueman) and Their Use in Controlling Varroa Within Honey Bee Colonies. Insects 9, 55. doi: 10.3390/insects9020055
Gusachenko, O. N., Woodford, L., Balbirnie-Cumming, K., Ryabov, E. V., Evans, D. J. (2020). Evidence for and Against Deformed Wing Virus Spillover From Honey Bees to Bumble Bees: A Reverse Genetic Analysis. Sci. Rep. 10, 1–10. doi: 10.1038/s41598-020-73809-3
He, Y., Lin, J., Kong, D., Huang, M., Xu, C., Kim, T. K., et al. (2015). Current State of Circulating microRNAs as Cancer Biomarkers. Clin. Chem. 61, 1138–1155. doi: 10.1373/clinchem.2015.241190
Hermance, M. E., Widen, S. G., Wood, T. G., Thangamani, S. (2019). Ixodes Scapularis Salivary Gland microRNAs Are Differentially Expressed During Powassan Virus Transmission. Sci. Rep. 9, 13110. doi: 10.1038/s41598-019-49572-5
Higes, M., Martín-Hernández, R., Hernández-Rodríguez, C. S., González-Cabrera, J. (2020). Assessing the Resistance to Acaricides in Varroa Destructor From Several Spanish Locations. Parasitol. Res. 119, 3595–3601. doi: 10.1007/s00436-020-06879-x
Highfield, A. C., El Nagar, A., Mackinder, L. C., Noël, L. M. L., Hall, M. J., Martin, S. J., et al. (2009). Deformed Wing Virus Implicated in Overwintering Honeybee Colony Losses. Appl. Environ. Microbiol. 75, 7212–7220. doi: 10.1128/AEM.02227-09
Hristov, P., Shumkova, R., Palova, N., Neov, B. (2020). Factors Associated With Honey Bee Colony Losses: A Mini-Review. Vet. Sci. 7, 166. doi: 10.3390/vetsci7040166
Hu, W., Criscione, F., Liang, S., Tu, Z. (2015). MicroRNAs of Two Medically Important Mosquito Species: Aedes Aegypti and Anopheles Stephensi. Insect Mol. Biol. 24, 240–252. doi: 10.1111/imb.12152
Hung, K. J., Kingston, J. M., Albrecht, M., Holway, D. A., Kohn, J. R. (2018). The Worldwide Importance of Honey Bees as Pollinators in Natural Habitats. Proc. Biol. Sci. 285, 20172140. doi: 10.1098/rspb.2017.2140
Hussain, M., Walker, T., O’Neill, S. L., Asgari, S. (2013). Blood Meal Induced microRNA Regulates Development and Immune Associated Genes in the Dengue Mosquito Vector, Aedes Aegypti. Insect Biochem. Mol. Biol. 43, 146–152. doi: 10.1016/j.ibmb.2012.11.005
Iqbal, J., Mueller, U. (2007). Virus Infection Causes Specific Learning Deficits in Honeybee Foragers. Proc. R. Soc. B.: Biol. Sci. 274, 1517–1521. doi: 10.1098/rspb.2007.0022
Jin, H., Kim, V. N., Hyun, S. (2012). Conserved microRNA miR-8 Controls Body Size in Response to Steroid Signaling in Drosophila. Genes Dev. 26, 1427–1432. doi: 10.1101/gad.192872.112
John, B., Enright, A. J., Aravin, A., Tuschl, T., Sander, C., Marks, D. S. (2004). Human MicroRNA Targets. PLoS Biol. 2, e363. doi: 10.1371/journal.pbio.0020363
Kadener, S., Menet, J. S., Sugino, K., Horwich, M. D., Weissbein, U., Nawathean, P., et al. (2009). A Role for microRNAs in the Drosophila Circadian Clock. Genes Dev. 23, 2179–2191. doi: 10.1101/gad.1819509
Kertesz, M., Iovino, N., Unnerstall, U., Gaul, U., Segal, E. (2007). The Role of Site Accessibility in microRNA Target Recognition. Nat. Genet. 39, 1278–1284. doi: 10.1038/ng2135
Kevill, J. L., Highfield, A., Mordecai, G. J., Martin, S. J., Schroeder, D. C. (2017). ABC Assay: Method Development and Application to Quantify the Role of Three DWV Master Variants in Overwinter Colony Losses of European Honeybees. Viruses 9, 314. doi: 10.3390/v9110314
Kloosterman, W. P., Plasterk, R. H. A. (2006). The Diverse Functions of microRNAs in Animal Development and Disease. Dev. Cell 11, 441–450. doi: 10.1016/j.devcel.2006.09.009
Koleoglu, G., Goodwin, P. H., Reyes-Quintana, M., Hamiduzzaman, M. M., Guzman-Novoa, E. (2017). Effect of Varroa Destructor, Wounding and Varroa Homogenate on Gene Expression in Brood and Adult Honey Bees. PLoS One 12, e0169669. doi: 10.1371/journal.pone.0169669
Koleoglu, G., Goodwin, P. H., Reyes-Quintana, M., Hamiduzzaman, M. M., Guzman-Novoa, E. (2018). Varroa Destructor Parasitism Reduces Hemocyte Concentrations and Prophenol Oxidase Gene Expression in Bees From Two Populations. Parasitol. Res. 117, 1175–1183. doi: 10.1007/s00436-018-5796-8
Kulhanek, K., Steinhauer, N., Wilkes, J., Wilson, M., Spivak, M., Sagili, R. R., et al. (2021). Survey-Derived Best Management Practices for Backyard Beekeepers Improve Colony Health and Reduce Mortality. PLoS One 16, e0245490. doi: 10.1371/journal.pone.0245490
Le Conte, Y., Ellis, M., Ritter, W. (2010). Varroa Mites and Honey Bee Health: Can Varroa Explain Part of the Colony Losses? Apidologie 41, 353–363. doi: 10.1051/apido/2010017
Lei, Z., Lv, Y., Wang, W., Guo, Q., Zou, F., Hu, S., et al. (2015). MiR-278-3p Regulates Pyrethroid Resistance in Culex Pipiens Pallens. Parasitol. Res. 114, 699–706. doi: 10.1007/s00436-014-4236-7
Li, Y., Andrade, J. (2017). DEApp: An Interactive Web Interface for Differential Expression Analysis of Next Generation Sequence Data. Source Code Biol. Med. 12, 2. doi: 10.1186/s13029-017-0063-4
Liu, Y., Zhou, Y., Wu, J., Zheng, P., Li, Y., Zheng, X., et al. (2015). The Expression Profile of Aedes Albopictus miRNAs Is Altered by Dengue Virus Serotype-2 Infection. Cell Biosci. 5, 1–11. doi: 10.1186/s13578-015-0009-y
Lucas, K., Raikhel, A. S. (2013). Insect microRNAs: Biogenesis, Expression Profiling and Biological Functions. Insect Biochem. Mol. Biol. 43, 24–38. doi: 10.1016/j.ibmb.2012.10.009
Lucas, K. J., Roy, S., Ha, J., Gervaise, A. L., Kokoza, V. A., Raikhel, A. S. (2015a). MicroRNA-8 Targets the Wingless Signaling Pathway in the Female Mosquito Fat Body to Regulate Reproductive Processes. Proc. Natl. Acad. Sci. U. S. A. 112, 1440–1445. doi: 10.1073/pnas.1424408112
Lucas, K. J., Zhao, B., Liu, S., Raikhel, A. S. (2015b). Regulation of Physiological Processes by microRNAs in Insects. Curr. Opin. Insect Sci. 11, 1–7. doi: 10.1016/j.cois.2015.06.004
Luhur, A., Chawla, G., Sokol, N. S. (2013). MicroRNAs as Components of Systemic Signaling Pathways in Drosophila Melanogaster. Curr. Top. Dev. Biol. 105, 97–123. doi: 10.1016/B978-0-12-396968-2.00004-X
Luo, H., Guo, W., Wang, F., You, Y., Wang, J., Chen, X., et al. (2015). miR-1291 Targets Mucin 1 Inhibiting Cell Proliferation and Invasion to Promote Cell Apoptosis in Esophageal Squamous Cell Carcinoma. Oncol. Rep. 34, 2665–2673. doi: 10.3892/or.2015.4206
Maharaj, P. D., Widen, S. G., Huang, J., Wood, T. G., Thangamani, S. (2015). Discovery of Mosquito Saliva microRNAs During CHIKV Infection. PLoS Negl. Trop. Dis. 9, e0003386. doi: 10.1371/journal.pntd.0003386
Marco, A., Hui, J. H., Ronshaugen, M., Griffiths-Jones, S. (2010). Functional Shifts in Insect microRNA Evolution. Genome Biol. Evol. 2, 686–696. doi: 10.1093/gbe/evq053
Martin, S. (1998). A Population Model for the Ectoparasitic Mite Varroa Jacobsoni in Honey Bee (Apis Mellifera) Colonies. Ecol. Modelling 10, 267–281. doi: 10.1016/S0304-3800(98)00059-3
Martin, S. J., Brettell, L. E. (2019). Deformed Wing Virus in Honey Bees and Other Insects. Annu. Rev. Virol. 6, 49–69. doi: 10.1146/annurev-virology-092818
Millán-Leiva, A., Marín, Ó., Christmon, K., van Engelsdorp, D., González-Cabrera, J. (2021). Mutations Associated With Pyrethroid Resistance in Varroa Mite, a Parasite of Honey Bees, Are Widespread Across the United States. Pest Manag. Sci. 77, 3241–3249. doi: 10.1002/ps.6366
Min, H., Yoon, S. (2010). Got Target? Computational Methods for microRNA Target Prediction and Their Extension. Exp. Mol. Med. 42, 233–244. doi: 10.3858/emm.2010.42.4.032
Moore, J., Jironkin, A., Chandler, D., Burroughs, N., Evans, D. J., Ryabov, E. V. (2011). Recombinants Between Deformed Wing Virus and Varroa Destructor Virus-1 may Prevail in Varroa Destructor-Infested Honeybee Colonies. J. Gen. Virol. 92, 156–161. doi: 10.1099/vir.0.025965-0
Mordecai, G. J., Brettell, L. E., Martin, S. J., Dixon, D., Jones, I. M., Schroeder, D. C. (2016). Superinfection Exclusion and the Long-Term Survival of Honey Bees in Varroa-Infested Colonies. ISME J. 10, 1182–1191. doi: 10.1038/ismej.2015.186
Natsopoulou, M. E., McMahon, D. P., Paxton, R. J. (2016). Parasites Modulate Within-Colony Activity and Accelerate the Temporal Polyethism Schedule of a Social Insect, the Honey Bee. Behav. Ecol. Sociobiol. 70, 1019–1031. doi: 10.1007/s00265-015-2019-5
Noël, A., Le Conte, Y., Mondet, F. (2020). Varroa Destructor: How Does It Harm Apis Mellifera Honey Bees and What Can Be Done About It? Emerg. Top. Life Sci. 4, 45–57. doi: 10.1042/ETLS20190125
Osei-Amo, S., Hussain, M., O’Neill, S. L., Asgari, S. (2012). Wolbachia-Induced aae-miR-12 miRNA Negatively Regulates the Expression of MCT1 and MCM6 Genes in Wolbachia-Infected Mosquito Cell Line. PLoS One 7 (11), e50049. doi: 10.1371/journal.pone.0050049
Pizzorno, M. C., Field, K., Kobokovich, A. L., Martin, P. L., Gupta, R. A., Mammone, R., et al. (2021). Transcriptomic Responses of the Honey Bee Brain to Infection With Deformed Wing Virus. Viruses 13, 287. doi: 10.3390/v13020287
Posada-Florez, F., Childers, A. K., Heerman, M. C., Egekwu, N. I., Cook, S. C., Chen, Y., et al. (2019). Deformed Wing Virus Type A, a Major Honey Bee Pathogen, Is Vectored by the Mite Varroa Destructor in a Non-Propagative Manner. Sci. Rep. 9, 1–10. doi: 10.1038/s41598-019-47447-3
Quinlan, A. R., Hall, I. M. (2010). BEDTools: A Flexible Suite of Utilities for Comparing Genomic Features. Bioinformatics 26, 841–842. doi: 10.1093/bioinformatics/btq033
Ramsey, S. D., Ochoa, R., Bauchan, G., Gulbronson, C., Mowery, J. D., Cohen, A., et al. (2019). Varroa Destructor Feeds Primarily on Honey Bee Fat Body Tissue and Not Hemolymph. Proc. Natl. Acad. Sci. U. S. A. 116, 1792–1801. doi: 10.1073/pnas.1818371116
Remnant, E. J., Mather, N., Gillard, T. L., Yagound, B., Beekman, M. (2019). Direct Transmission by Injection Affects Competition Among RNA Viruses in Honeybees. Proc. Biol. Sci. 286, 20182452. doi: 10.1098/rspb.2018.2452
Rinkevich, F. D. (2020). Detection of Amitraz Resistance and Reduced Treatment Efficacy in the Varroa Mite, Varroa Destructor, Within Commercial Beekeeping Operations. PLoS One 15, e0227264. doi: 10.1371/journal.pone.0227264
Rosche, K. L., Sidak-Loftis, L. C., Hurtado, J., Fisk, E. A. (2021). Arthropods Under Pressure: Stress Responses and Immunity at the Pathogen-Vector Interface. Front. Immunol. 11. doi: 10.3389/fimmu.2020.629777
Rosenkranz, P., Aumeier, P., Ziegelmann, B. (2010). Biology and Control of Varroa Destructor. J. Invertebr. Pathol. 103, S96–S119. doi: 10.1016/j.jip.2009.07.016
Ryabov, E. V., Childers, A. K., Lopez, D., Grubbs, K., Posada-Florez, F., Weaver, D., et al. (2019). Dynamic Evolution in the Key Honey Bee Pathogen Deformed Wing Virus: Novel Insights Into Virulence and Competition Using Reverse Genetics. PLoS Biol. 17 (10), e3000502. doi: 10.1371/journal.pbio.3000502
Saldaña, M. A., Etebari, K., Hart, C. E., Widen, S. G., Wood, T. G., Thangamani, S., et al. (2017). Zika Virus Alters the microRNA Expression Profile and Elicits an RNAi Response in Aedes Aegypti Mosquitoes. PLoS Neg. Trop. Dis. 11, e0005760. doi: 10.1371/journal.pntd.0005760
Santillán-Galicia, M. T., Ball, B. V., Clark, S. J., Alderson, P. G. (2010). Transmission of Deformed Wing Virus and Slow Paralysis Virus to Adult Bees (Apis Mellifera L.) by Varroa Destructor. J. Apic. Res. 49, 141–148. doi: 10.3896/IBRA.1.49.2.01
Sempere, L. F., Sokol, N. S., Dubrovsky, E. B., Berger, E. M., Ambros, V. (2003). Temporal Regulation of microRNA Expression in Drosophila Melanogaster Mediated by Hormonal Signals and Broad-Complex Gene Activity. Dev. Biol. 259, 9–18. doi: 10.1016/s0012-1606(03)00208-2
Shao, H., Wang, H., Tang, X. (2015). NAC Transcription Factors in Plant Multiple Abiotic Stress Responses: Progress and Prospects. Front. Plant Sci. 6, 902. doi: 10.3389/fpls.2015.00902
Sturm, M., Hackenberg, M., Langenberger, D., Frishman, D. (2010). TargetSpy: A Supervised Machine Learning Approach for microRNA Target Prediction. BMC Bioinform. 11, 292. doi: 10.1186/1471-2105-11-292
Techer, M. A., Rane, R. V., Grau, M. L., Roberts, J. M., Sullivan, S. T., Liachko, I., et al. (2019). Divergent Evolutionary Trajectories Following Speciation in Two Ectoparasitic Honey Bee Mites. Comm. Biol. 2, 1–16. doi: 10.1038/s42003-019-0606-0
Toronen, P., Medlar, A., Holm, L. (2018). PANNZER2: A Rapid Functional Annotation Web Server. Nucleic Acids Res. 46, W84–W88. doi: 10.1093/nar/gky350
Traniello, I. M., Bukhari, S. A., Kevill, J., Ahmed, A. C., Hamilton, A. R., Naeger, N. L., et al. (2020). Meta-Analysis of Honey Bee Neurogenomic Response Links Deformed Wing Virus Type A to Precocious Behavioral Maturation. Sci. Rep. 10, 1–12. doi: 10.1038/s41598-020-59808-4
Traynor, K. S., Mondet, F., de Miranda, J. R., Techer, M., Kowallik, V., Oddie, M., et al. (2020). Varroa destructor: A Complex Parasite, Crippling Honey Bees Worldwide. Trends Parasitol. 36 (7), 592–606. doi: 10.1016/j.pt.2020.04.004
van Dooremalen, C., Gerritsen, L., Cornelissen, B., van der Steen, J. J. M., van Langevelde, F., Blacquière, T. (2012). Winter Survival of Individual Honey Bees and Honey Bee Colonies Depends on Level of Varroa Destructor Infestation. PLoS One 7, e36285. doi: 10.1371/journal.pone.0036285
Wang, Y., Gosselin Grenet, A. S., Castelli, I., Cermenati, G., Ravallec, M., Fiandra, L., et al. (2013). Densovirus Crosses the Insect Midgut by Transcytosis and Disturbs the Epithelial Barrier Function. J. Virol. 87, 12380–12391. doi: 10.1128/JVI.01396-13
Wang, Y., Jiang, F., Wang, H., Song, T., Wei, Y., Yang, M., et al. (2015). Evidence for the Expression of Abundant microRNAs in the Locust Genome. Sci. Rep. 5, 1–14. doi: 10.1038/srep13608
Webster, T. C., Delaplane, K. S. (2001). Mites of the Honey Bee (Hamilton, Illinois: Dadant and Sons, Inc.).
Wells, T., Wolf, S., Nicholls, E., Groll, H., Lim, K. S., Clark, S. J., et al. (2016). Flight Performance of Actively Foraging Honey Bees Is Reduced by a Common Pathogen. Envirom. Microbiol. Rep. 8, 728–737. doi: 10.1111/1758-2229.12434
Weng, R., Chin, J. S., Yew, J. Y., Bushati, N., Cohen, S. M. (2013). miR-124 Controls Male Reproductive Success in Drosophila. eLife 2, e00640. doi: 10.7554/eLife.00640
Wilfert, L., Long, G., Leggett, H. C., Schmid-Hempel, P., Butlin, R., Martin, S. J. M., et al. (2016). Deformed Wing Virus Is a Recent Global Epidemic in Honeybees Driven by Varroa Mites. Science 351, 594–597. doi: 10.1126/science.aac9976
Winter, F., Edaye, S., Hüttenhofer, A., Brunel, C. (2007). Anopheles Gambiae miRNAs as Actors of Defence Reaction Against Plasmodium Invasion. Nucleic Acids Res. 35, 6953–6962. doi: 10.1093/nar/gkm686
Xu, M. J., Fu, J. H., Nisbet, A. J., Huang, S. Y., Zhou, D. H., Lin, R. Q., et al. (2013). Comparative Profiling of microRNAs in Male and Female Adults of Ascaris Suum. Parasitol. Res. 112, 1189–1195. doi: 10.1007/s00436-012-3250-x
Yañez, O., Piot, N., Dalmon, A., de Miranda, J. R., Chantawannakul, P., Panziera, D., et al. (2020). Bee Viruses: Routes of Infection in Hymenoptera. Front. Microbiol. 11, 943. doi: 10.3389/fmicb.2020.00943
Yang, X., Cox-Foster, D. L. (2005). Impact of an Ectoparasite on the Immunity and Pathology of an Invertebrate: Evidence for Host Immunosuppression and Viral Amplification. Proc. Natl. Acad. Sci. U. S. A. 102, 7470–7475. doi: 10.1073/pnas.0501860102
Yan, H., Zhou, Y., Liu, Y., Deng, Y., Chen, X. (2014). miR-252 of the Asian Tiger Mosquito Aedes Albopictus Regulates Dengue Virus Replication by Suppressing the Expression of the Dengue Virus Envelope Protein. J. Med. Virol. 86, 1428–1436. doi: 10.1002/jmv.23815
Ye, J., Zhang, Y., Cui, H., Liu, J., Wu, Y., Cheng, Y., et al. (2018). WEGO 2.0: A Web Tool for Analyzing and Plotting G.O. AnnotationUpdate. Nucl. Acids Res. 46 (2021), W71–W75. doi: 10.1093/nar/gky400
Ye, J., Fang, L., Zheng, H., Zhang, Y., Chen, J., Zhang, Z., et al. (2006). WEGO: A Web Tool for Plotting GO Annotations. Nucleic Acid Res. 34, W293–W297. doi: 10.1093/nar/gkl031
Yu, A. M., Pan, Y. Z. (2012). Noncoding microRNAs: Small RNAs Play a Big Role in Regulation of ADME? Acta Pharm. Sin. B. 2, 93–101. doi: 10.1016/j.apsb.2012.02.011
Keywords: Varroa destructor, microRNAs, small RNA-seq, honey bee (Apis mellifera L.), deformed wing virus
Citation: Kumar D, Alburaki M, Tahir F, Goblirsch M, Adamczyk J and Karim S (2022) An Insight Into the microRNA Profile of the Ectoparasitic Mite Varroa destructor (Acari: Varroidae), the Primary Vector of Honey Bee Deformed Wing Virus. Front. Cell. Infect. Microbiol. 12:847000. doi: 10.3389/fcimb.2022.847000
Received: 31 December 2021; Accepted: 17 February 2022;
Published: 16 March 2022.
Edited by:
Giovanni Cilia, Council for Agricultural and Economics Research (CREA), ItalyReviewed by:
Francisco José Reynaldi, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaEugene V. Ryabov, Agricultural Research Service (USDA), United States
Sofia Levin-Nikulin, University of Minnesota Twin Cities, United States
Copyright © 2022 Kumar, Alburaki, Tahir, Goblirsch, Adamczyk and Karim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shahid Karim, c2hhaGlkLmthcmltQHVzbS5lZHU=
 Deepak Kumar
Deepak Kumar Mohamed Alburaki
Mohamed Alburaki Faizan Tahir
Faizan Tahir Michael Goblirsch
Michael Goblirsch John Adamczyk3
John Adamczyk3 Shahid Karim
Shahid Karim