- Department of Biological Sciences, Northern Illinois University, DeKalb, IL, United States
The study of the intestinal microbiome is an evolving field of research that includes comprehensive analysis of the vast array of microbes – bacterial, archaeal, fungal, and viral. Various gastrointestinal (GI) diseases, such as Crohn’s disease and ulcerative colitis, have been associated with instability of the gut microbiota. Many studies have focused on importance of bacterial communities with relation to health and disease in humans. The role of viruses, specifically bacteriophages, have recently begin to emerge and have profound impact on the host. Here, we comprehensively review the importance of viruses in GI diseases and summarize their influence in the complex intestinal environment, including their biochemical and genetic activities. We also discuss the distribution of the gut virome as it relates with treatment and immunological advantages. In conclusion, we suggest the need for further studies on this critical component of the intestinal microbiome to decipher the role of the gut virome in human health and disease.
Introduction
The human gut virome consists of the total population of viruses and their genomes that are found throughout the gastrointestinal tract. It is estimated that the human intestines harbor about 35 – 2800 active bacterial viruses in 1g of feces (Kim et al., 2011; Manrique et al., 2016). The intestinal virome includes both bacteriophages (hereafter referred to as phages) and eukaryotic viruses. Maintaining homeostasis in the complex intestinal microbial environment plays a significant role in improving the health status of the host which could otherwise contribute to the development of disease conditions (Weinbauer, 2004). Major clinical conditions such as diabetes and Crohn’s diseases have been associated with the disruption in the composition of commensal flora of the gut (dysbiosis) (Frank et al., 2007; Larsen et al., 2010; Perez-Brocal et al., 2013). Dysbiosis leads to dynamic changes in the gut community as well as phage activities (Flores et al., 2011; Ogilvie and Jones, 2015). Bacterial populations in the gut are controlled by various factors ranging from the host dietary content to the immune system and the predatory effects of phages. For example, 10-80% of total bacterial death in nature can be attributed to phage attack, hence signifying their roles in microbial community (Weinbauer and Rassoulzadegan, 2004). Further, phages are involved in the lateral transfer of genes between bacteria and serve as a determinant for genetic variability (Canchaya et al., 2003). Several recent studies have reported the role the virome, particularly phages, plays in the development of certain clinical conditions, including bowel disorders and cancer (Frank et al., 2007; Tong et al., 2013; Brooks and Watson, 2015). In this review, we provide a comprehensive compilation of the role that the virome plays in GI health and diseases.
The Virome Development
Microbiome (virome inclusive) composition varies with age, dietary intake, host immunological status, drug intake, and environmental factors. Major bacterial phyla that dominate the gut at the early stage of infancy include, Firmicutes, Bacteroidetes, Actinobacteria, Verrucomicrobia, Proteobacteria and Fusobacteria. Phage-bacteria relationship at this stage is inversely related where an increase in phage density is observed with low bacterial population and vice-versa. This relationship indicates that the prey (bacteria) distribution influences the predator’s (phage’s) diversities (reversed predator-prey dynamics) (Lim et al., 2015). The Global Virome Database indicates that 97.7% of the human gut virome are phages, 2.1% are eukaryotic viruses, and 0.1% are archaeal viruses; 88% of these phages have yet to be classified by the International Committee on Taxonomy of Viruses (Gregory et al., 2020).
The dynamism of human virome progresses from childhood to adulthood. The colonization of the human intestine by microbes starts right after delivery with usually very low populations of microorganisms consisting mainly of bacteria (Jimenez et al., 2008). Gregory et al. determined that infants (0-3 years) and adults (18-65 years) show higher viral richness, with decreases for children (3-18 years) and the elderly (65+ years). Bacteriophage richness followed this trend whereas eukaryotic virus richness was high in infants and steadily decreased throughout life (Figure 1) (Gregory et al., 2020).
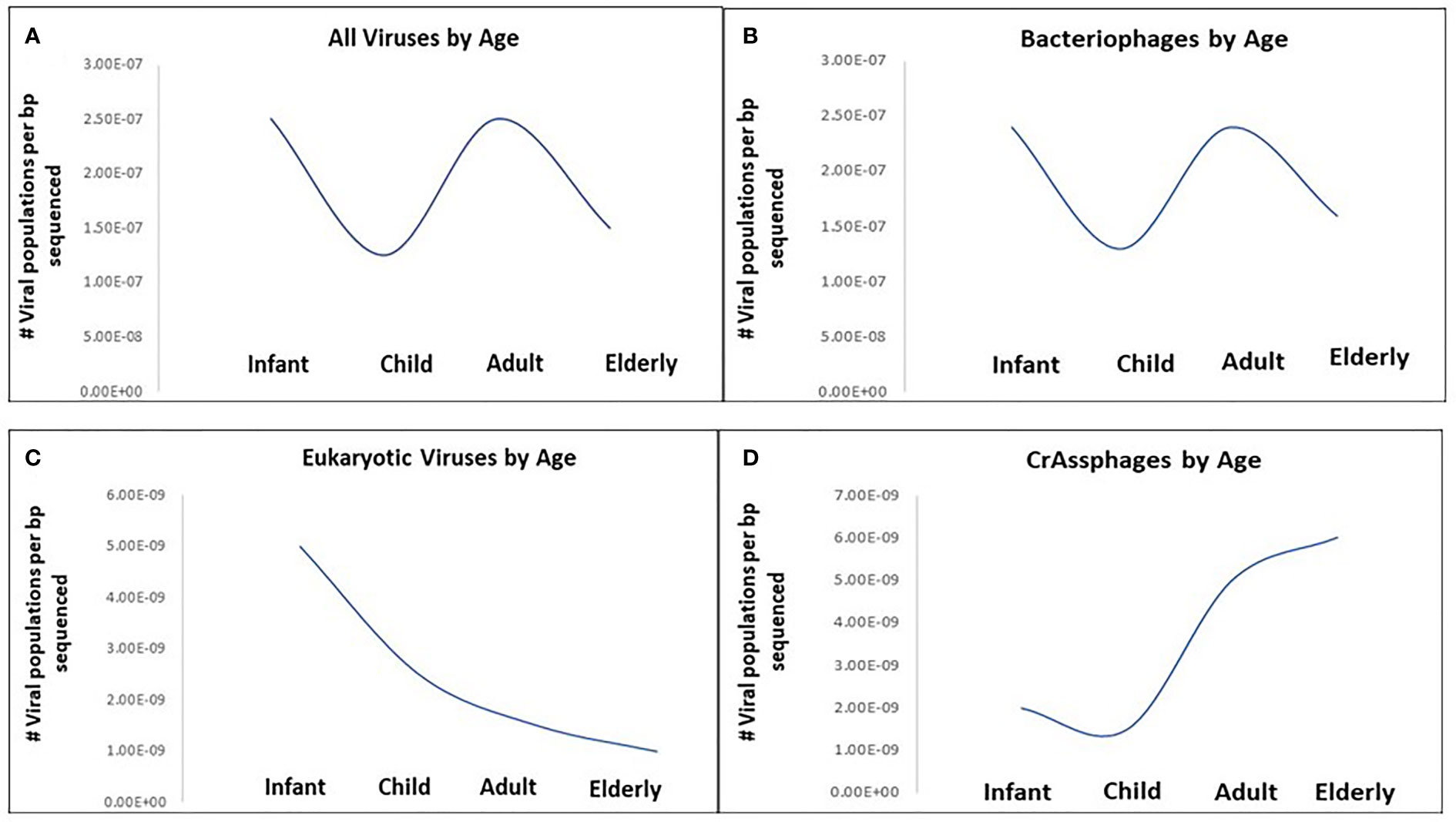
Figure 1 Viral richness changes with age in healthy humans across four age categories: Infant (0-3 years), Child (3-18 years), Adult (18-65 years) and Elderly (65+ years). (A) All viruses by age, (B) bacteriophages by age, (C) eukaryotic viruses by age, (D) CrAssphages by age. Viral richness quantified as the median number of viral populations per base-pair sequenced for each age category based on results from Gregory et al., 2020.
Gut virome in neonates initially is comprised of phages which infect the pioneer bacteria, followed by eukaryotic virus diversification associated with environmental exposures, particularly breastmilk (Liang et al., 2020). The immediate environment contributes to the diversity of the gut phage among the neonates (Huurre et al., 2008; Lim et al., 2015). Other studies in children (ages 0 to 3 years) have reported the predominance of DNA phages from the Caudiovirales order, mostly comprising of Myoviridae, Siphoviridae and Podoviridae family (Reyes et al., 2015). High phage abundance of Caudiovirales, and to a lesser extent of Microviridae, in this age group is attributed to the early colonization of the gut by Bacteroides, Proteobacteria and Actinobacteria bacteria (Reyes et al., 2010; Lim et al., 2015). Less abundant DNA eukaryotic viruses in the Anelloviridae and Herpesviridae families have also been detected in infant stool (Breitbart et al., 2008). The single-stranded (ss) DNA viruses of the Anelloviridae family (particularly Torque Teno Virus species) has been shown to be the most abundant in the few months after birth (Lim et al., 2015). Prevalence of Anelloviridae is directly associated with host immunosuppression, and pediatric febrile illness (McElvania TeKippe et al., 2012; De Vlaminck et al., 2013). This early onset of DNA viruses is attributed to immature host immune system, and its subsequent reduced load correlates to development of a fully competent immune response (Fulci et al., 2021).
Recently, highly divergent phages have been identified among healthy children, for instance, CrAssphage which are associated with Bacteroides (Dutilh et al., 2014). CrAssphages richness showed an overall upward trend with age; are abundant and persistent in the human gut virome, and functions to maintain a stable population of both resistant and sensitive bacterial hosts (Figure 1) (Gregory et al., 2020; Shkoporov et al., 2021). Metagenomic analysis of the eukaryotic viruses revealed that the diversities of the viral population are influenced by the geographical location and the health status of the host. A study on Amazonian children demonstrated the abundance of Picornaviriridae and Caliciviridae families (Siqueira et al., 2018). Other eukaryotic viruses recovered in the healthy human gut include Circoviridae, Anelloviridae, Reoviridae and Astroviridae (Altan et al., 2018). Although Breibart et al. claimed that vertical transmission of the ssDNA eukaryotic viruses, particularly Anelloviridae, occur during vaginal delivery from mother to child; factors that promote their increased abundance following first few months after delivery are unknown (Breitbart et al., 2008). Further investigations are required to identify the mechanism involved in this colonization. Furthermore, the role of internal factors such as peristalsis, immunity, drug regimen etc., on virome composition need further assessment due to their importance in gut diversity (Mackie et al., 1999).
Progressively, the microbial community in the infant gut undergoes a series of developmental shifts following the dietary change from liquid to solid (Sharon et al., 2013). These developmental shifts lead to the emergence of a balanced microbiome that is stable compositionally and functionally (Mackie et al., 1999; Minot et al., 2013). At this stage, the most commonly identified bacteria in healthy adult subjects are Bacteroidetes and Firmicutes (Mackie et al., 1999) and their dominance subsequently affects the virome composition (Minot et al., 2013). The heterogeneity in gut virome is also attributed to phages with single-stranded DNA, specifically Microviridae due to high mutations rates (Minot et al., 2013). Manrique et al. analyzed DNA phage in healthy adult stool and reported that the gut composition is made up of Podoviridae, Siphoviridae, Myoviridae, and Microviridae families. Additionally, Zhang et al., reported the abundance of Virgaviridae viruses of plant origin as the common eukaryotic RNA virus in the gut of the healthy adults suggesting dietary influence (Zhang et al., 2006). Pepper mild mottle virus (PPMV) was predominantly detected among these families whereas less common RNA viruses identified included maize chlorotic mottle virus (Tombusviridae), oat chlorotic stunt virus (Tombusviridae), grapevine asteroid mosaic virus (Tymoviridae), and panicum mosaic virus (Tombusviridae) (Zhang et al., 2006). The stability of the virome in the adult could contribute to the positive health status of the human host, however, a disturbance of the viral community may result in gut microbiome dysbiosis (Flores et al., 2011).
The Virome and the Human Host Interaction
The human body serves as a reservoir of many microorganisms including viral communities (Weinbauer and Rassoulzadegan, 2004; Pride et al., 2012; Tremaroli and Backhed, 2012). Metagenomic shotgun sequencing provides a platform to investigate the genetic potentials of indigenous viral community and aid in the identification of distinct new phages (Dinsdale et al., 2008; Perez-Brocal et al., 2013; Bibby, 2014).The most common human gut phages are grouped into three main families based on their tail structure: a complex extensive-tailed Myoviridae, long non-extensive tailed Siphoviridae, and short non-extensive tailed Podoviridae (Veesler and Cambillau, 2011). These are double-stranded DNA phages that belong to Caudovirales order and account for approximately 95% of the bacteriophages (Maniloff and Ackermann, 1998). Single-stranded DNA phage families including Microviridae and Inoviridae have also been described (Kim et al., 2011; Szekely and Breitbart, 2016; Creasy et al., 2018). However, metagenomic studies that focus on the RNA phages in the human gut are relatively few (Zhang et al., 2006; Manrique et al., 2017). Additional studies on the human gut virome have revealed that diverse genotypes of phages stably reside and play a distinct role in maintaining the host health (Minot et al., 2012; Abeles and Pride, 2014). Heterogeneity in the viral composition in other body sites such as the phages detected in skin surfaces, have also been described (Oh et al., 2016). Human microbiome compositions, including virome, are reportedly influenced by various factors such as gender, age, environmental reservoir, diet, human-to-human contact, immune status, and the health of the hosts (Breitbart et al., 2008; Arthur et al., 2009; Azad et al., 2013; Abeles et al., 2014; Hodyra-Stefaniak et al., 2015; Creasy et al., 2018). Host sex was found to be associated with changes in composition and diversity of the viral community, particularly in the oral virome, likely due to hormonal effects on the bacterial community (Abeles and Pride, 2014). Further, a comparative study involving the analysis of lactococcal virus in infant and adult stool samples shows an increased abundance of Myoviridae family probably due to change in diet (Deveau et al., 2006).
The Phage Impacts on Enteric Bacteria Virulence
Phages play a major role in bacterial evolution and exhibit viral tropism, which is influenced by bacterial rigidity, phage competence and other environmental conditions (Weinbauer, 2004; Rohwer et al., 2009; Lee et al., 2016). Phage infection may either be deleterious or beneficial to the bacteria as a result of lateral gene transfer within the bacteria community (Canchaya et al., 2003). Phages may encode toxin genes which are important in the pathogenicity of many bacteria (Gyles, 2007). The virulence effect produced by toxin-inducing prophage is best described in the role they play in Shiga toxin-producing Escherichia coli (STEC) virulence. STEC is a clinically significant foodborne pathogen which produces complications like hemolytic uremic syndrome and may be fatal in severe cases, due to the conferred effects of lambdoid phage encoded shiga-toxin (stx) genes (Riley et al., 1983; Gyles, 2007). The use of antibiotics (e.g., norfloxacin) may induce lytic cycle of stx-prophage, which can ultimately lead to fatal morbidity in the infected individuals (Zhang et al., 2000). A similar toxin-producing mechanism is observed in bacteria such as Staphylococcus aureus and Vibrio cholera with toxic shock syndrome toxin (tst) and accessory cholera exotoxin (ace), respectively (Trucksis et al., 1993; Ruzin et al., 2001). Phages can also encode for virulence factors within host bacterial cells, for instance, the temperate phage, Sopφ, encodes for sopE effector protein production which facilitates the entry of Salmonella spp. into the host intestinal epithelium (Wood et al., 1996; Hardt et al., 1998). Phages may also increase the pathogenicity of bacteria directly through the virion particle structures (e.g., hyaluronidase tightly bound to the phage) (Benchetrit et al., 1977), or by replication and transcription of phage-encoded genes as in the case of diphtheria toxin produced by Corynebacterium diphtheriae (Wagner and Waldor, 2002). Faruque et al. reported a change of a non-virulent strain of Vibrio cholera to a virulent type following CTXPhi (filamentous) phage induction (Faruque et al., 1998). Apart from horizontal transfer of virulence genes to non-pathogenic strains, several studies have demonstrated that virulence genes can also be transduced within bacterial community members. Examples include the transfer of stx genes among the Enterobacteriacae family members and an opportunistic bacteria, Acinetobacter haemolyticus, via transduction (James et al., 2001; Gamage et al., 2004; Grotiuz et al., 2006; Gorski et al., 2016). Both these bacteria cause bloody diarrhea (James et al., 2001; Grotiuz et al., 2006).
Phages can contribute to pathogenicity in bacteria by encoding genes which serve as disease factors. Phages of Staphylococcus aureus, sakϕC and sak42D, encode for the immune regulator staphylokinase (sak) which can counteract host immune responses by neutralizing antimicrobial peptides (Nguyen and Vogel, 2016) and cause host tissue death (van Wamel et al., 2006). Other phage-encoded substances, such as complement inhibitors (SCIN), protein inhibitors (CHIP) including superantigens are known to be associated with lethal outcome of S. aureus infection (de Jong et al., 2018). Phages have been reported to promote establishment of diseases by mediating biofilm formation, thus increasing adhesion and resistance to antibiotics and desiccation, of bacteria like Pseudomonas aeruginosa and E. coli in humans with cystic fibrosis (Secor et al., 2015). In mice, Waldor and Mekalanos demonstrated that filamentous phage (CTXF) is involved in the entry of V. cholera into the host’s epithelial tissue through the binding of toxin co-regulated pili (TCP) receptor (Waldor and Mekalanos, 1996).
Another mechanism by which phages increase enteric bacterial pathogenicity is through the transfer of genes encoding antibiotics resistance or genes that evade the immune system (Colomer-Lluch et al., 2011). For instance, Marta et al., reported that β-lactamase genes, blaTEM and blaCTX-M9 in the S. aureus phage can be laterally transferred into environmental isolates conferring ampicillin resistance to new S. aureus strains. Temperate phages have also been implicated as major carriers of transducing multi antibiotic-resistant genes among Salmonella Typhimurium strains (Colomer-Lluch et al., 2011). A study on the role of virome in bacterial adaptation, after antibiotic-induced stress in animal models revealed that phage genes undergo robust enrichment following antibiotic treatment. Such gene enrichment could promote the production of resistant gene against the administered drug in phages and their subsequent transfer to the bacterial community, thereby acting as a reservoir for resistant strains (Modi et al., 2013).
The Virome and Enteric Diseases
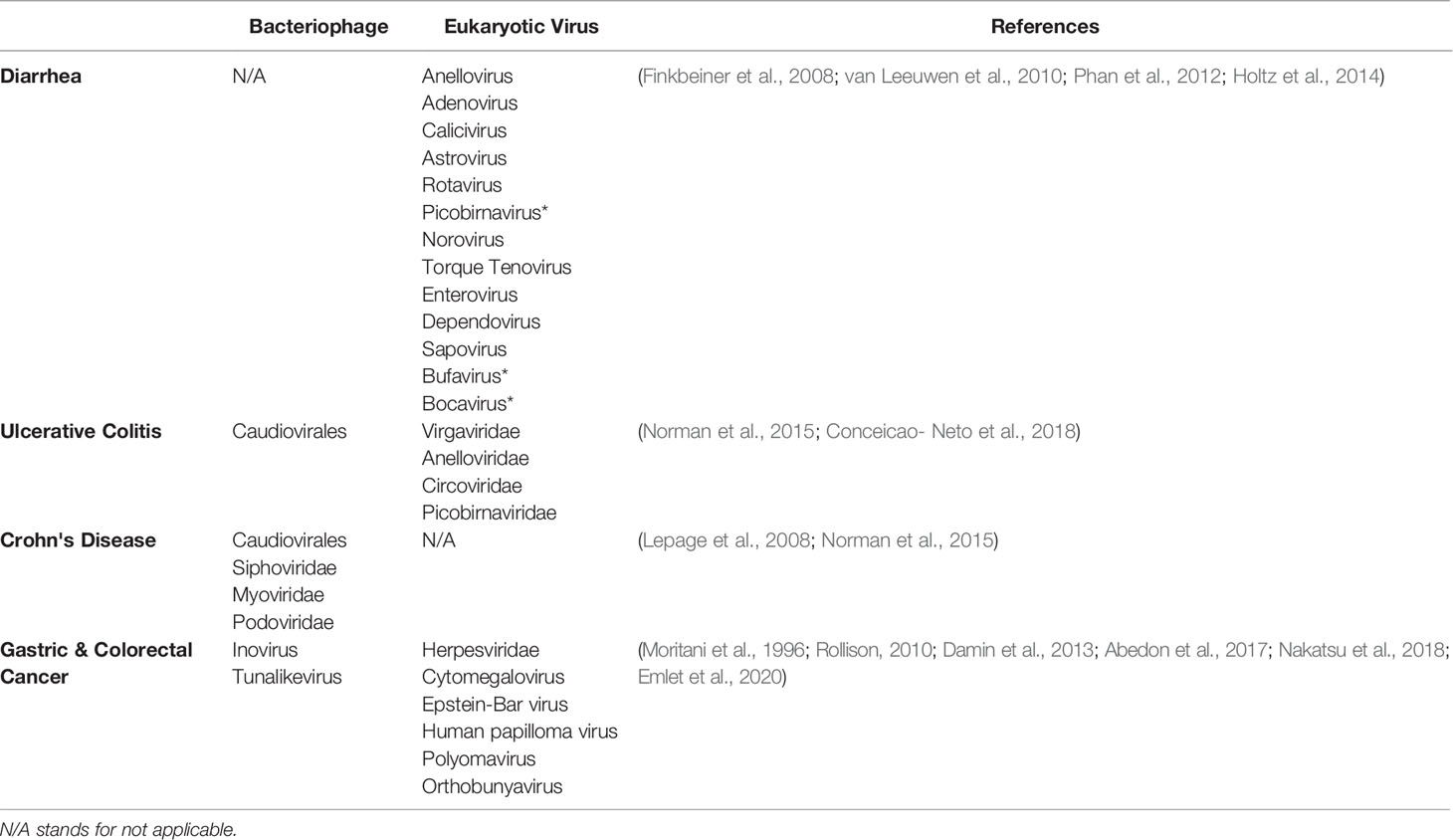
The eukaryotic viruses that are involved in enteric infections are known as enteric viruses. In infected subjects, about 106 – 108 VLPs of eukaryotic viruses are usually shed in each gram of stool (Farthing, 1989). The most frequently detected eukaryotic viruses in acute gastroenteritis are Reoviridae (rotavirus), Picornaviridae (enterovirus, echovirus etc.), Adenoviridae (adenovirus) and Caliciviridae (norovirus) (Wilhelmi et al., 2003; Finkbeiner et al., 2008a). Phages also play a significant role in the study of intestinal diseases in several ways by: (1) acting as a major determinant of virulence in enteric infections (2) associating with dysbiosis in gut during mild and chronic inflammatory bowel disease (IBD) (3) being employed as a therapeutic regimen for many human infections including intestinal disease. The human GI tract harbors abundant of diverse microbial populations that are involved in shaping human health. It is estimated that human fecal samples carry up to 105 phage per gram dry feces that specifically attack different strains of E. coli, Salmonella spp. and B. fragilis bacteria (Havelaar et al., 1990; Calci et al., 1998). The diversity of the gut virome is evolving as a major subject for host health, and its disruption has been shown to be related with disease conditions in both human and animal models (Turnbaugh et al., 2006; Tong et al., 2013; Kim and Bae, 2018). Apart from promoting virulence of the pathogens, the distribution of phages in the gut are also attributed to different disease progression, immune system functions and gut homeostasis (Gorski et al., 2016; Zhao et al., 2017; Metzger et al., 2018). The role of the virome has been implicated in intestinal conditions and even related diseases such as Crohn’s disease (Dejea et al., 2014; Gorski et al., 2016; Nakatsu et al., 2018) and ulcerative colitis (Frank et al., 2007; Brooks and Watson, 2015; Sheehan et al., 2015; Gorski et al., 2016). In this section below, we elaborate on the role of gut virome in enteric infections (Table 1).
Diarrhea
Globally, acute diarrhea is one of the health conditions with highest mortality rate, particularly among children, and viruses are identified to be one of the causative agents. In about 40% of reported diarrheal disease cases etiological agents are unidentified, however the use of metagenomics sequencing technique has enabled detection of novel viruses that may be responsible for acute infections (Finkbeiner et al., 2008b). Finkbeiner et al, studied the viral community of the fecal samples from patients with acute diarrhea and detected phages as well as common eukaryotic viruses such as, anellovirus, adenovirus, calicivirus, astrovirus and rotavirus, which belong to Anelloviridae, Adenoviridae, Caliciviridae: Astroviridae, and Reoviridae families respectively. Putative novel DNA viruses identified here, shared homology with picobirnavirus, norovirus, Torque Teno virus, and enterovirus genus, while the detected RNA virus was related to Nodaviridae family (Finkbeiner et al., 2008b). In addition to these, van Leeuwen et al., reported the identification of Picornarviridae, Retroviridae, and a novel picobirnavirus variant with distinct phylogenetic relatedness in patients with diarrhea, indicating the involvement of new emerging viruses (van Leeuwen et al., 2010). The distribution of viruses in diarrhea have been attributed to factors including population, age, etiological agent and other environmental conditions. For example, a study at two geographical locations in Australia in pediatric patients, exhibited abundance of Adenoviridae and Picornaviridae families with varying proportion (Holtz et al., 2014). In another study, Phan et al., observed that anelloviruses and dependovirus were most prevalent in West African children with acute diarrhea closely followed by sapoviruses, enteroviruses and bocaviruses (Phan et al., 2012). It is noteworthy that both anelloviruses and dependoviruses are themselves not pathogenic but have been detected by several studies and are presumably markers of infection. The mechanisms by which these viruses induce diarrhea are not well understood, however, rotavirus and adenovirus infections within enterocytes of the small intestine result in atrophy of the villi and crypt cell hyperplasia leading to fluid malabsorption (Wilhelmi et al., 2003). Novel viruses such as bufavirus and bocavirus share homology with Parvoviridae family have also been recovered in the feces of patient with diarrhea. The detection of these novel viruses depict diversity in Parvoviruses, however their possible role in this clinical condition is unknown (Arthur et al., 2009; Phan et al., 2012).
Inflammatory Bowel Disease
IBD inclusive of ulcerative colitis (UC) and Crohn’s disease (CD) is a chronic clinical condition that causes recurring inflammation of the intestine (Abraham and Cho, 2009). A common feature of CD includes a rough patch-like appearance of the inflamed tissues of the intestine. This inflammation can cause perforation of the intestinal wall and have a resultant effect on vital organs such as kidneys and uterus. Development of UC is localized in the colon where dysbiosis occurs (Nagalingam and Lynch, 2012). Intestinal microbiome and virome are crucial for human health and have been implicated to be significant factors in the IBD progression (Lepage et al., 2008). Elevation of Caudiovirales phages was found in IBD patients, particularly with CD, accompanied by a decrease in bacterial diversity, thereby demonstrating the possible influence of phages in these conditions (Lepage et al., 2008; Sheehan et al., 2015). Comparative analysis of the Viral-like particles (VLP) from the biopsy of CD patients exhibited an increase in Siphoviridae, Myoviridae and Podoviridae in patients as compared to the healthy control groups. Further, metagenomics studies revealed mild disparity in the diversity and richness in the viral composition of Caudiovirales taxa between UC and CD patients (Norman et al., 2015). This suggests that the variation in the virome may serve as a biomarker for classifying these clinical conditions. Normal et al., reported that a converse relationship exists between phage richness and bacteria diversity in CD where the rate of growth of Bacteroidaceae is greatly reduced in presence of Caudiovirales (i.e., negative phage-bacteria correlation). However, Caudiovirales population in UC showed positive correlation with the growth of Enterobacteriacae, Pastaurellacaea and Prevotellaceae (Brooks and Watson, 2015; Gorski et al., 2016).
Eukaryotic viruses, particularly Anelloviridae, are also reported to be higher in IBD patients as compared to the healthy individuals, although their role in IBD condition is not known (Norman et al., 2015). Based on the observed viral richness, diversity between UC patients undergoing fecal microbial transplant (FMT) and healthy control group, Conceicao et al., suggested that the eukaryotic viral richness could serve as a potential biomarker in diagnosis of UC (Norman et al., 2015; Conceicao-Neto et al., 2018). The four dominant viral families reported in this study include, Virgaviridae, Anelloviridae, Circoviridae, and Picobirnaviridae (Conceicao-Neto et al., 2018). While these studies have increased our knowledge about the distribution and possible involvement of the viruses in IBD, future investigations are warranted to discuss roles that diet, age, immunity plays in shaping IBD.
Gastric and Colorectal Cancer
In US alone, death due to cancer accounts for the second largest cause with GI cancer being associated with high morbidity and mortality rate (Siegel et al., 2015). Studies on the gut bacterial distribution and pathogenesis have shown that dysbiosis leads to development of colorectal cancer (CRC) (Baxter et al., 2016; Yu et al., 2017). Additionally, 15-20% of cancer incidence worldwide is associated with oncogenic virus infections (Chen et al., 2019). Oncogenic DNA viruses can cause cancer by interfering with cellular division or DNA repair mechanisms, while RNA viruses do so through the production of reactive oxygen species or chronic inflammation (McLaughlin-Drubin and Munger, 2008). Knowledge of the virome therefore provides useful information in the CRC screening particularly in the early stages (Zackular et al., 2014; Zeller et al., 2014; Yu et al., 2017). The virome has been associated in the development of two major GI cancer: gastric cancer and CRC (Moritani et al., 1996; Bull and Plummer, 2014; Zackular et al., 2014). The eukaryotic virus, Epstein-Bar virus (Herpesvirus), is a known etiological agent of gastric cancer (Moritani et al., 1996). Nakatsu et al. revealed that phage richness (due to the significant detection of distinctive members of Inovirus and Tunalikevirus) is common in CRC patients compared to the control group (Nakatsu et al., 2018). Inovirus species are tiny filamentous phages that are associated with regulating bacteria exopolysaccharide matrix synthesis, a precursor for biofilm formation, an underlying factor in colorectal tumor development (Dejea et al., 2014; Johnson et al., 2015; Secor et al., 2015). Enteric phages that target Bacteroides fragilis, Fusobacterium nucleatum and Escherichia coli have been associated with CRC development; the speculative mechanism of oncogenesis being phage ability to directly transfer into colonic epithelial cells as well as phage encoding for virulence genes, particularly genes regulating biofilm production (Emlet et al., 2020).
Orthobunyavirus, a eukaryotic virus,was uniquely abundant in CRC patients and may be used as a marker for CRC diagnosis (Nakatsu et al., 2018). Herpesviridae, eukaryote virus family, were more predominant in CRC, particularly Cytomegalovirus spp., which are usually implicated as an etiological agent (Dejea et al., 2014). Human papillomavirus infections were found to be associated with increased risk of CRC (Damin et al., 2013) by integrating into the host genome, but the mechanism for induction of cancer is not well understood (Emlet et al., 2020). Polyomaviruses can be oncogenic due to their encoding of T-antigen which can inactivate the p53 and pRB tumor suppressor proteins (Rollison, 2010) leading to unregulated tumor growth.
Protective Effect of the Virome
Health conditions such as cancer and neurodegenerative disorders have been treated using phage therapy with promising outcomes. For example, a study exhibited the binding potential of M13 to both β-amyloid and α-synuclein proteins, in the brain of non-human primates (Macca mulatta) suggesting phage therapeutic use in neurodegenerative disorders (Alzheimer’s and Parkinson’s diseases) (Ksendzovsky et al., 2012). In the treatment of cancer cells in mice tumor models, delivery phages have been engineered to transfer anticancer protein into the cancer cells either for malignant tumor atrophy or cancer gene therapy, thereby facilitating the cell death. In addition, phage can play a role in bolstering the immune system by preventing the reactive oxygen species (ROS) synthesis in endotoxin induced oxidative stress (Dabrowska et al., 2004). The use of phages as therapeutic agents has been extensively reviewed (Abedon et al., 2017). In this section, we focus on the role of phages in ameliorating GI tract (GIT) disorders and associated possible drawbacks with their usage.
Treatment of Intestinal Diseases
Phages have been used to treat infectious diseases of the intestines and skin in Eastern European and former Soviet Union countries since the early 1900s (Morozova et al., 2018). The long history of phage research spans many years in various countries; phages have been used with notable success to treat bacterial dysentery in France (1919), cholera in India (1927), acute colitis in Georgia (1936), and bacterial dysentery, acute colitis and salmonellosis in Russia (1968-1993) (El-Shibiny and El-Sahhar, 2017). Some prospective lysogenic phages have been used in formulating therapeutic cocktails because of their effectiveness in deleting virulence genes and adding short chain genes necessary for fatty acid metabolism in certain pathogenic bacteria (Regeimbal et al., 2016). Protective phage action was demonstrated in the evasion of Salmonella colonization in anaerobically cultivated tissue culture and had minimal or no effect on non-target bacteria (Hu et al., 2018). Phage treatment can be adapted in managing enteric diseases caused by bacterial pathogens like V. cholerae (Summers, 2001), Salmonella spp. (Goode et al., 2003); S. aureus (Mann, 2008; Oh et al., 2016), Clostridioides difficile; Listeria monocytogenes (Carlton et al., 2005; Mai et al., 2010), Campylobacter jejuni (Goode et al., 2003; Hwang et al., 2009; Gorski et al., 2016) and E. coli (Sarker et al., 2016). Therapeutic use of phage products directly in humans is not approved in most countries, including United States. However, commercially approved phage-based formulations are produced in a number of countries which can prevent food-borne intestinal infections. These phage products control bacterial pathogen contamination to increase food safety: E. coli O157:H7 (EcoShieldTM by Intralytix, USA), Listeria monocytogenes (ListShieldTM by Intralytix, USA, and LISTEXTM P100 by EBI, Netherlands) and Salmonella spp., from swine and poultry animals (BioTector by CheilJedang Corporation, Korea) (Monk et al., 2010; Endersen et al., 2014; Nannapaneni and Soni, 2015). Commercially available E. coli-targeting phages (PreforPro®) has been given experimentally to humans reporting gastrointestinal distress, showing no global disruption of the microbiota and positive outcomes, specifically, increased butyrate-producing Eubacterium and decreased bacteria closely related Clostridium perfringens (Febvre et al., 2019). The CRISPR-Cas system of E. coli is being researched with the goal of reducing drug-resistant pathogens using phages to insert the CRISPR-Cas program targeting bacterial resistance genes (Yosef et al., 2015).
On the other hand, the use of phage therapy has received many critical reviews which has limited its use both commercially and globally (Sulakvelidze et al., 2005; Kutter, 2008). Safety of the phage (Meader et al., 2013; Abedon et al., 2017), pharmacodynamics impact of phages (Carey-Smith et al., 2006), and host immunological compatibility with the formulated phages are major concerns (Hodyra-Stefaniak et al., 2015). Since, phage can infect many bacteria, the challenge of stringent specificity for target cells is hindering the use of phage regimen (Garcia et al., 2009; Gorski et al., 2016). Another setback in the use of phage is the rapid evolvement of mutant strains that are resistant to the phage treatment (Garcia et al., 2009; Kocharunchitt et al., 2009). Therefore, more studies are needed in this area before the phage therapy can be globally acceptable.
The Virome Induced Immunogenicity
The importance of viruses and phages in conferring immunity are well documented in studies involving mouse models as well as human subjects. Experimental mouse studies have suggested that enteric viruses can provide beneficial effects. For example, murine norovirus (MNV) infection was shown to reverse bacterial dysbiosis-induced GIT disease by repairing Paneth cell and crypt-villi functionality in the small intestines by upregulating the production of IFNγ and IgA (Kernbauer et al., 2014). Other investigators observed mice with latent murine gammaherpesvirus 68 or murine cytomegalovirus (mCMV) infections were protected against infection by the bacterial pathogens, Yersinia pestis and Listeria monocytogenes via persistent immune stimulation from macrophage activity and antiviral IFNγ (Barton et al., 2007). In studies involving phage effects, patients with C. difficile infections were treated with fecal filtrate showed clinical improvement and significant changes in their intestinal phage community resembling the fecal donor (Ott et al., 2017). In another study, patients with various Staphylococcus spp. infections underwent experimental phage treatment with a staphylococcal phage cocktail (MS1) resulting in increased antibody stimulation (mainly IgG and IgM) (Żaczek et al., 2016). Mucosal surfaces can be entry points for invading pathogens; increased concentrations of mucus-adherent phages have been shown to provide host immune defense against bacterial infections (Barr et al., 2013; Barr et al., 2015). Filamentous phages have experimentally shown much potential for the development of vaccines and can be engineered as therapeutic agents for managing bacterial infections and chronic diseases, such as cancer, Alzheimer’s disease, and Parkinson’s disease. These phage based vaccines have capacity for displaying various surface antigens, like bacterial LPS encoded on phage coat, to provoke various immune responses including innate immunity effectors, T cell independent antibodies and cytotoxic T lymphocytes (Henry et al., 2015).
Conclusion
The impact of phages in influencing the overall wellbeing of human hosts cannot be overemphasized due to their role in impacting virulence and potential as therapeutic agents. Therefore, phages offer a promising option in the treatment of different GI diseases, particularly in conditions like CD and UC (Abraham and Cho, 2009; Nagalingam and Lynch, 2012), where phage dysbiosis has been implicated to be a developmental factor. However, further studies are required for the phage therapy to be universally accepted in clinical practice. In addition, evolving fields such as CRISPR-Cas system (Yosef et al., 2015) of bacteria provide another platform where drug delivery to host tissue can be manipulated for cancer treatment through phage-bacteria relationship. Currently, the biological function of only 25% of viral genes have been preliminarily determined (Nayfach et al., 2021). Future studies should focus on understanding the biological relevance of the human virome which can lend to better understanding of enteric disease processes and to the development of phage therapies.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Funding
Funding provided by College of Liberal Arts and Sciences and Division of Research and Innovation Partnerships at Northern Illinois University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The graphical abstract was created with BioRender.com.
References
Abedon, S., Mullany, P., Aminov, R., García, P. (2017). Editorial: Phage Therapy: Past, Present and Future. Front. Microbiol. 8 doi: 10.3389/fmicb.2017.00981
Abeles, S. R., Pride, D. T. (2014). Molecular Bases and Role of Viruses in the Human Microbiome. J. Mol. Biol. 426 (23), 3892–3906. doi: 10.1016/j.jmb.2014.07.002
Abeles, S. R., Robles-Sikisaka, R., Ly, M., Lum, A. G., Salzman, J., Boehm, T. K., et al. (2014). Human Oral Viruses Are Personal, Persistent and Gender-Consistent. ISME J. 8 (9), 1753–1767. doi: 10.1038/ismej.2014.31
Abraham, C., Cho, J. H. (2009). Inflammatory Bowel Disease. N. Engl. J. Med. 361 (21), 2066–2078. doi: 10.1056/NEJMra0804647
Altan, E., Aiemjoy, K., Phan, T. G., Deng, X., Aragie, S., Tadesse, Z., et al. (2018). Enteric Virome of Ethiopian Children Participating in a Clean Water Intervention Trial. PloS One 13 (8), e0202054. doi: 10.1371/journal.pone.0202054
Arthur, J. L., Higgins, G. D., Davidson, G. P., Givney, R. C., Ratcliff, R. M. (2009). A Novel Bocavirus Associated With Acute Gastroenteritis in Australian Children. PloS Pathog. 5 (4), e1000391. doi: 10.1371/journal.ppat.1000391
Azad, M., Konya, T., Maughan, H., Guttman, D., Field, C., Chari, R., et al. (2013). Gut Microbiota of Healthy Canadian Infants: Profiles by Mode of Delivery and Infant Diet at 4 Months. CMAJ 185, 385–394. doi: 10.1503/cmaj.121189
Barr, J. J., Auro, R., Furlan, M., Whiteson, K. L., Erb, M. L., Pogliano, J., et al. (2013). Bacteriophage Adhering to Mucus Provide a Non-Host-Derived Immunity. Proc. Natl. Acad. Sci. U. S. A. 110 (26), 10771–10776. doi: 10.1073/pnas.1305923110
Barr, J. J., Auro, R., Sam-Soon, N., Kassegne, S., Peters, G., Bonilla, N., et al. (2015). Subdiffusive Motion of Bacteriophage in Mucosal Surfaces Increases the Frequency of Bacterial Encounters. Proc. Natl. Acad. Sci. U. S. A. 112 (44), 13675–13680. doi: 10.1073/pnas.1508355112
Barton, E. S., White, D. W., Cathelyn, J. S., Brett-McClellan, K. A., Engle, M., Diamond, M. S., et al. (2007). Herpesvirus Latency Confers Symbiotic Protection From Bacterial Infection. Nature 447, 326–329. doi: 10.1038/nature05762
Baxter, N., Ruffin, M., Rogers, M., Schloss, P. (2016). Microbiotabased Model Improves the Sensitivity of Fecal Immunochemical Test for Detecting Colonic Lesions. Genome Med. 8, 37. doi: 10.1186/s13073-016-0290-3
Benchetrit, L. C., Gray, E. D., Wannamaker, L. W. (1977). Hyaluronidase Activity of Bacteriophages of Group A Streptococci. Infect Immun. 15 (2), 527–532. doi: 10.1128/iai.15.2.527-532.1977
Bibby, K. (2014). Improved Bacteriophage Genome Data Is Necessary for Integrating Viral and Bacterial Ecology. Microb. Ecol. 67 (2), 242–244. doi: 10.1007/s00248-013-0325-x
Breitbart, M., Haynes, M., Kelley, S., Angly, F., Edwards, R., Felts, B., et al. (2008). Viral Diversity and Dynamics in an Infant Gut. Res. Microbiol. 159, 367–373. doi: 10.1016/j.resmic.2008.04.006
Brooks, J., Watson, A. J. (2015). The Enteric Virome in Inflammatory Bowel Disease. Gastroenterology 149 (4), 1120–1121. doi: 10.1053/j.gastro.2015.08.022
Bull, M. J., Plummer, N. T. (2014). Part 1: The Human Gut Microbiome in Health and Disease. Integr. Med. (Encinitas) 13 (6), 17–22.
Calci, K., Burkhardt, W., Watkins, W., Rippey, S. (1998). Occurrence of Male-Specific Bacteriophage in Feral and Domestical Animal Wastes, Human Feces, and Human-Associated Wastewaters. Appl. Environ. Microbiol. 64, 5027–5029. doi: 10.1128/AEM.64.12.5027-5029.1998
Canchaya, C., Fournous, G., Chibani-Chenoufi, S., Dillmann, M. L., Bru¨ssow, H. (2003). Phage as Agents of Lateral Gene Transfer. Curr. Opin. Microbiol. 6, 417–424. doi: 10.1016/S1369-5274(03)00086-9
Carey-Smith, G. V., Billington, C., Cornelius, A. J., Hudson, J. A., Heinemann, J. A. (2006). Isolation and Characterization of Bacteriophages Infecting Salmonella Spp. FEMS Microbiol. Lett. 258 (2), 182–186. doi: 10.1111/j.1574-6968.2006.00217.x
Carlton, R. M., Noordman, B., Biswas, E. D., de Meester, W. H., Loessner, M. J. (2005). Bacteriophage P100 for Control of Listeria Monocytogenes in Foods Genome Sequence Bioinformatic Analyses Oral Toxicity Study and Application. Regul. Toxicol. Pharmacol. 43, 301–312. doi: 10.1016/j.yrtph.2005.08.005
Chen, X., Kost, J., Sulovari, A., Wong, N., Liang, W. S., Cao, J., et al. (2019). A Virome-Wide Clonal Integration Analysis Platform for Discovering Cancer Viral Etiology. Genome Res. 29 (5), 819–830. doi: 10.1101/gr.242529.118
Colomer-Lluch, M., Jofre, J., Muniesa, M. (2011). Antibiotic Resistance Genes in the Bacteriophage DNA Fraction of Environmental Samples. PloS One 6 (3), e17549. doi: 10.1371/journal.pone.0017549
Conceicao-Neto, N., Deboutte, W., Dierckx, T., Machiels, K., Wang, J., Yinda, K. C., et al. (2018). Low Eukaryotic Viral Richness Is Associated With Faecal Microbiota Transplantation Success in Patients With UC. Gut 67 (8), 1558–1559. doi: 10.1136/gutjnl-2017-315281
Creasy, A., Rosario, K., Leigh, B. A., Dishaw, L. J., Breitbart, M. (2018). Unprecedented Diversity of ssDNA Phages From the Family Microviridae Detected Within the Gut of a Protochordate Model Organism (Ciona Robusta). Viruses 10 (8), 404. doi: 10.3390/v10080404
Dabrowska, K., Opolski, A., Wietrzyk, J., Switala-Jelen, K., Godlewska, J., Boratynski, J., et al. (2004). Anticancer Activity of Bacteriophage T4 and Its Mutant HAP1 in Mouse Experimental Tumour Models. Anticancer Res. 24, 3991–3995.
Damin, D. C., Ziegelmann, P. K., Damin, A. P. (2013). Human Papillomavirus Infection and Colorectal Cancer Risk: A Meta-Analysis. Colorect. Dis. 15 (8), e420–e428. doi: 10.1111/codi.12257
Dejea, C. M., Wick, E. C., Hechenbleikner, E. M., White, J. R., Mark Welch, J. L., Rossetti, B. J., et al. (2014). Microbiota Organization Is a Distinct Feature of Proximal Colorectal Cancers. Proc. Natl. Acad. Sci. U. S. A. 111, 18321–18326. doi: 10.1073/pnas.1406199111
de Jong, N. W. M., Vrieling, M., Garcia, B. L., Koop, G., Brettmann, M., Aerts, P. C., et al. (2018). Identification of a Staphylococcal Complement Inhibitor With Broad Host Specificity in Equid Staphylococcus Aureus Strains. J. Biol. Chem. 293 (12), 4468–4477. doi: 10.1074/jbc.RA117.000599
Deveau, H., Labrie, S. J., Chopin, M. C., Moineau, S. (2006). Biodiversity and Classification of Lactococcal Phages. Appl. Environ. Microbiol. 72 (6), 4338–4346. doi: 10.1128/AEM.02517-05
De Vlaminck, I., Khush, K. K., Strehl, C., Kohli, B., Luikart, H., Neff, N. F., et al. (2013). Temporal Response of the Human Virome to Immunosuppression and Antiviral Therapy. Cell 155 (5), 1178–1187. doi: 10.1016/j.cell.2013.10.034
Dinsdale, E. A., Edwards, R. A., Hall, D., Angly, F., Breitbart, M., Brulc, J. M., et al. (2008). Functional Metagenomic Profiling of Nine Biomes. Nature 452 (7187), 629–632. doi: 10.1038/nature06810
Dutilh, B. E., Cassman, N., McNair, K., Sanchez, S. E., Silva, G. G., Boling, L., et al. (2014). A Highly Abundant Bacteriophage Discovered in the Unknown Sequences of Human Faecal Metagenomes. Nat. Commun. 5, 4498. doi: 10.1038/ncomms5498
El-Shibiny, A., El-Sahhar, S. (2017). Bacteriophages: The Possible Solution to Treat Infections Caused by Pathogenic Bacteria. Can. J. Microbiol. 63 (11), 865–879. doi: 10.1139/cjm-2017-0030
Emlet, C., Ruffin, M., Lamendella, R. (2020). Enteric Virome and Carcinogenesis in the Gut. Dig Dis. Sci. 65 (3), 852–864. doi: 10.1007/s10620-020-06126-4
Endersen, L., Mahony, J., Hill, C., Ross, R. P., McAuliffe, O., Coffey, A. (2014). O’ Phage Therapy in the Food Industry. Annu. Rev. Food Sci. Technol. 5, 327–349. doi: 10.1146/annurev-food-030713-092415
Farthing, M. J. G. (1989). Gut Viruses: A Role in Gastrointestinal Disease? (Welwyn Garden City, Hertfordshire, United Kingdom: Smith Kline & French, Ltd).
Faruque, S. M., Asadulghani, Alim, A. R., Albert, M. J., Islam, K. M., Mekalanos, J. J. (1998). Induction of the Lysogenic Phage Encoding Cholera Toxin in Naturally Occurring Strains of Toxigenic Vibrio Cholerae O1 and O139. Infect. Immun. 66 (8), 3752–3757. doi: 10.1128/IAI.66.8.3752-3757.1998
Febvre, H. P., Rao, S., Gindin, M., Goodwin, N. D. M., Finer, E., Vivanco, J. S., et al. (2019). PHAGE Study: Effects of Supplemental Bacteriophage Intake on Inflammation and Gut Microbiota in Healthy Adults. Nutrients 11 (3), 666. doi: 10.3390/nu11030666
Finkbeiner, S. R., Allred, A. F., Tarr, P. I., Klein, E. J., Kirkwood, C. D., Wang, D. (2008a). Metagenomic Analysis of Human Diarrhea: Viral Detection and Discovery. PloS Pathog. 4 (2), e1000011. doi: 10.1371/journal.ppat.1000011
Finkbeiner, S. R., Allred, A. F., Tarr, P. I., Klein, E. J., Kirkwood, C. D., Wang, D. (2008b). Metagenomic Analysis of Human Diarrhea: Viral Detection and Discovery, PLo, P. E1000011. PloS Pathog. 4, e1000011. doi: 10.1371/journal.ppat.1000011
Flores, C. O., Meyer, J. R., Valverde, S., Farr, L., Weitz, J. S. (2011). Statistical Structure of Host-Phage Interactions. Proc. Natl. Acad. Sci. U. S. A. 108 (28), E288–E297. doi: 10.1073/pnas.1101595108
Frank, D. N., St Amand, A. L., Feldman, R. A., Boedeker, E. C., Harpaz, N., Pace, N. R. (2007). Molecular-Phylogenetic Characterization of Microbial Community Imbalances in Human Inflammatory Bowel Diseases. Proc. Natl. Acad. Sci. U. S. A. 104 (34), 13780–13785. doi: 10.1073/pnas.0706625104
Fulci, V., Stronati, L., Cucchiara, S., Laudadio, I., Carissimi, C. (2021). Emerging Roles of Gut Virome in Pediatric Diseases. Int. J. Mol. Sci. 22 (8), 4127. doi: 10.3390/ijms22084127
Gamage, S. D., Patton, A. K., Hanson, J. F., Weiss, A. A. (2004). Diversity and Host Range of Shiga Toxin-Encoding Phage. Infect. Immun. 72 (12), 7131–7139. doi: 10.1128/IAI.72.12.7131-7139.2004
Garcia, P., Madera, C., Martinez, B., Rodriguez, A., Evaristo Suarez, J. (2009). Prevalence of Bacteriophages Infecting Staphylococcus Aureus in Dairy Samples and Their Potential as Biocontrol Agents. J. Dairy Sci. 92 (7), 3019–3026. doi: 10.3168/jds.2008-1744
Goode, D., Allen, V. M., Barrow, P. A. (2003). Reduction of Experimental Salmonella and Campylobacter Contamination of Chicken Skin by Application of Lytic Bacteriophages. Appl. Environ. Microbiol. 69 (8), 5032–5036. doi: 10.1128/AEM.69.8.5032-5036.2003
Gorski, A., Miedzybrodzki, R., Weber-Dabrowska, B., Fortuna, W., Letkiewicz, S., Rogoz, P., et al. (2016). Phage Therapy: Combating Infections With Potential for Evolving From Merely a Treatment for Complications to Targeting Diseases. Front. Microbiol. 1515 (7). doi: 10.3389/fmicb.2016.01515
Gregory, A. C., Zablocki, O., Zayed, A. A., Howell, A., Bolduc, B., Sullivan, M. B. (2020). The Gut Virome Database Reveals Age-Dependent Patterns of Virome Diversity in the Human Gut. Cell Host Microbe 28 (5), 724–740 e728. doi: 10.1016/j.chom.2020.08.003
Grotiuz, G., Sirok, A., Gadea, P., Varela, G., Schelotto, F. (2006). Shiga Toxin 2-Producing Acinetobacter Haemolyticus Associated With a Case of Bloody Diarrhea. J. Clin. Microbiol. 44 (10), 3838–3841. doi: 10.1128/JCM.00407-06
Gyles, C. L. (2007). Shiga Toxin-Producing Escherichia Coli: An Overview. J. Anim. Sci. 85 (13 Suppl), E45–E62. doi: 10.2527/jas.2006-508
Hardt, W. D., Urlaub, H., Galan, J. E. (1998). A Substrate of the Centisome 63 Type III Protein Secretion System of Salmonella Typhimurium is Encoded by a Cryptic Bacteriophage. Proc. Natl. Acad. Sci. U. S. A. 95, 2574–2579. doi: 10.1073/pnas.95.5.2574
Havelaar, A. H., Pot-Hogeboom, W. M., Furuse, K., Pot, R., Hormann, M. P. (1990). F-Specific RNA Bacteriophages and Sensitive Host Strains in Faeces and Wastewater of Human and Animal Origin. J. Appl. Bacteriol 69 (1), 30–37. doi: 10.1111/j.1365-2672.1990.tb02908.x
Henry, K. A., Arbabi-Ghahroudi, M., Scott, J. K. (2015). Beyond Phage Display: Non-Traditional Applications of the Filamentous Bacteriophage as a Vaccine Carrier, Therapeutic Biologic, and Bioconjugation Scaffold. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00755
Hodyra-Stefaniak, K., Miernikiewicz, P., Drapala, J., Drab, M., Jonczyk-Matysiak, E., Lecion, D., et al. (2015). Mammalian Host-Versus-Phage Immune Response Determines Phage Fate In Vivo. Sci. Rep. 14802 (5), 14802. doi: 10.1038/srep14802
Holtz, L. R., Cao, S., Zhao, G., Bauer, I. K., Denno, D. M., Klein, E. J., et al. (2014). Geographic Variation in the Eukaryotic Virome of Human Diarrhea. Virology, 468–470, 556-564. doi: 10.1016/j.virol.2014.09.012
Hu, Y. O. O., Hugerth, L. W., Bengtsson, C., Alisjahbana, A., Seifert, M., Kamal, A., et al. (2018). Bacteriophages Synergize With the Gut Microbial Community To Combat Salmonella. mSystems 3 (5), e00119–e00118. doi: 10.1128/mSystems.00119-18
Huurre, A., Rautava, S., Rinne, M., Salminen, S., Isolauri, E. (2008). Mode of Delivery—Effects on Gut Microbiota and Humoral Immunity. Neonatology 93 (4), 236–240. doi: 10.1159/000111102
Hwang, S., Yun, J., Kim, K. P., Heu, S., Lee, S., Ryu, S. (2009). Isolation and Characterization of Bacteriophages Specific for Campylobacter Jejuni. Microbiol. Immunol. 53, 559–566. doi: 10.1111/j.1348-0421.2009.00163.x
James, C. E., Stanley, K. N., Allison, H. E., Flint, H. J., Stewart, C. S., Sharp, R. J., et al. (2001). Lytic and Lysogenic Infection of Diverse Escherichia Coli and Shigella Strains With a Verocytotoxigenic Bacteriophage. Appl. Environ. Microbiol. 67 (9), 4335–4337. doi: 10.1128/AEM.67.9.4335-4337.2001
Jimenez, E., Marin, M. L., Martin, R., Odriozola, J. M., Olivares, M., Xaus, J., et al. (2008). Is Meconium From Healthy Newborns Actually Sterile? Res. Microbiol. 159 (3), 187–193. doi: 10.1016/j.resmic.2007.12.007
Johnson, C. H., Dejea, C. M., Edler, D., Hoang, L. T., Santidrian, A. F., Felding, B. H., et al. (2015). Metabolism Links Bacterial Biofilms and Colon Carcinogenesis. Cell Metab. 21 (6), 891–897. doi: 10.1016/j.cmet.2015.04.011
Kernbauer, E., Ding, Y., Cadwell, K. (2014). An Enteric Virus Can Replace the Beneficial Function of Commensal Bacteria. Nature 516 (7529), 94–98. doi: 10.1038/nature13960
Kim, M. S., Bae, J. W. (2018). Lysogeny is Prevalent and Widely Distributed in the Murine Gut Microbiota. ISME J. 12 (4), 1127–1141. doi: 10.1038/s41396-018-0061-9
Kim, M. S., Park, E. J., Roh, S. W., Bae, J. W. (2011). Diversity and Abundance of Single-Stranded DNA Viruses in Human Feces. Appl. Environ. Microbiol. 77 (22), 8062–8070. doi: 10.1128/AEM.06331-11
Kocharunchitt, C., Ross, T., McNeil, D. L. (2009). Use of Bacteriophages as Biocontrol Agents to Control Salmonella Associated With Seed Sprouts. Int. J. Food Microbiol. 128, 453–459. doi: 10.1016/j.ijfoodmicro.2008.10.014
Ksendzovsky, A., Walbridge, S., Saunders, R. C., Asthagiri, A. R., Heiss, J. D., Lonser, R. R. (2012). Convection-Enhanced Delivery of M13bacteriophage to the Brain. J. Neurosurg. 117, 197–203. doi: 10.3171/2012.4.JNS111528
Kutter, E. (2008). Phage Therapy: Bacteriophages as Naturally Occurring Antimicrobials (Boca Raton, Fla, USA: CRC Press).
Larsen, N., Vogensen, F. K., van den Berg, F. W., Nielsen, D. S., Andreasen, A. S., Pedersen, B. K., et al. (2010). Gut Microbiota in Human Adults With Type 2 Diabetes Differs From Non-Diabetic Adults. PloS One 5 (2), e9085. doi: 10.1371/journal.pone.0009085
Lee, H., Ku, H. J., Lee, D. H., Kim, Y. T., Shin, H., Ryu, S., et al. (2016). Characterization and Genomic Study of the Novel Bacteriophage HY01 Infecting Both Escherichia Coli O157:H7 and Shigella Flexneri: Potential as a Biocontrol Agent in Food. PloS One 11 (12), e0168985. doi: 10.1371/journal.pone.0168985
Lepage, P., Colombet, J., Marteau, P., Sime-Ngando, T., Dore, J., Leclerc, M. (2008). Dysbiosis in Inflammatory Bowel Disease: A Role for Bacteriophages? Gut 57, 424–425. doi: 10.1136/gut.2007.134668
Liang, G., Zhao, C., Zhang, H., Mattei, L., Sherrill-Mix, S., Bittinger, K., et al. (2020). The Stepwise Assembly of the Neonatal Virome Is Modulated by Breastfeeding. Nature 581 (7809), 470–474. doi: 10.1038/s41586-020-2192-1
Lim, E. S., Zhou, Y., Zhao, G., Bauer, I. K., Droit, L., Ndao, I. M., et al. (2015). Early Life Dynamics of the Human Gut Virome and Bacterial Microbiome in Infants. Nat. Med. 21 (10), 1228–1234. doi: 10.1038/nm.3950
Mackie, R. I., Sghir, A., Gaskins, H. R. (1999). Developmental Microbial Ecology of the Neonatal Gastrointestinal Tract. Am. J. Clin. Nutr. 69 (5), 1035S–1045S. doi: 10.1093/ajcn/69.5.1035s
Mai, V., Ukhanova, M., Visone, L., Abuladze, T., Sulakvelidze, A. (2010). Bacteriophage Administration Reduces the Concentration of Listeria Monocytogenes in the Gastrointestinal Tract and Its Translocation to Spleen and Liver in Experimentally Infected Mice. Int. J. Microbiol. 2010, 624234. doi: 10.1155/2010/624234
Maniloff, J., Ackermann, H. W. (1998). Taxonomy of Bacterial Viruses: Establishment of Tailed Virus Genera and the Other Caudovirales. Arch. Virol. 143, 2051–2063. doi: 10.1007/s007050050442
Mann, N. H. (2008). The Potential of Phages to Prevent MRSA Infections. Res. Microbiol. 159 (5), 400–405. doi: 10.1016/j.resmic.2008.04.003
Manrique, P., Bolduc, B., Walk, S. T., van der Oost, J., de Vos, W. M., Young, M. J. (2016). Healthy Human Gut Phageome. Proc. Natl. Acad. Sci. U. S. A. 113 (37), 10400–10405. doi: 10.1073/pnas.1601060113
Manrique, P., Dills, M., Young, M. J. (2017). The Human Gut Phage Community and Its Implications for Health and Disease. Viruses 9 (6), 141. doi: 10.3390/v9060141
McElvania TeKippe, E., Wylie, K. M., Deych, E., Sodergren, E., Weinstock, G., Storch, G. A. (2012). Increased Prevalence of Anellovirus in Pediatric Patients With Fever. PloS One 7 (11), e50937. doi: 10.1371/journal.pone.0050937
McLaughlin-Drubin, M. E., Munger, K. (2008). Viruses Associated With Human Cancer. Biochim. Biophys. Acta 1782 (3), 127–150. doi: 10.1016/j.bbadis.2007.12.005
Meader, E., Mayer, M. J., Steverding, D., Carding, S. R., Narbad, A. (2013). Evaluation of Bacteriophage Therapy to Control Clostridium Difficile and Toxin Production in an In Vitro Human Colon Model System. Anaerobe 22, 25–30. doi: 10.1016/j.anaerobe.2013.05.001
Metzger, R. N., Krug, A. B., Eisenacher, K. (2018). Enteric Virome Sensing-Its Role in Intestinal Homeostasis and Immunity. Viruses 10 (4), 146. doi: 10.3390/v10040146
Minot, S., Bryson, A., Chehoud, C., Wu, G. D., Lewis, J. D., Bushman, F. D. (2013). Rapid Evolution of the Human Gut Virome. Proc. Natl. Acad. Sci. U. S. A. 110 (30), 12450–12455. doi: 10.1073/pnas.1300833110
Minot, S., Grunberg, S., Wu, G. D., Lewis, J. D., Bushman, F. D. (2012). Hypervariable Loci in the Human Gut Virome. Proc. Natl. Acad. Sci. U. S. A. 109 (10), 3962–3966. doi: 10.1073/pnas.1119061109
Modi, S. R., Lee, H. H., Spina, C. S., Collins, J. J. (2013). Antibiotic Treatment Expands the Resistance Reservoir and Ecological Network of the Phage Metagenome. Nature 499 (7457), 219–222. doi: 10.1038/nature12212
Monk, A. B., Rees, C. D., Barrow, P., Hagens, S., Harper, D. R. (2010). Bacteriophage Applications: Where Are We Now? (Letter) Appl. Microbiol. (Letter) Appl. Microbiol. 51, 363–369. doi: 10.1111/j.1472-765X.2010.02916.x
Moritani, S., Kushima, R., Sugihara, H., Hattori, T. (1996). Phenotypic Characteristics of Epstein-Barr-Virus-Associated Gastric Carcinomas. J. Cancer Res. Clin. Oncol. 122 (12), 750–756. doi: 10.1007/BF01209123
Morozova, V. V., Vlassov, V. V., Tikunova, N. V. (2018). Applications of Bacteriophages in the Treatment of Localized Infections in Humans. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01696
Nagalingam, N. A., Lynch, S. V. (2012). Role of the Microbiota in Inflammatory Bowel Diseases. Inflamm. Bowel Dis. 18 (5), 968–984. doi: 10.1002/ibd.21866
Nakatsu, G., Zhou, H., Wu, W. K. K., Wong, S. H., Coker, O. O., Dai, Z., et al. (2018). Alterations in Enteric Virome Are Associated With Colorectal Cancer and Survival Outcomes. Gastroenterology 155 (2), 529–541.e525. doi: 10.1053/j.gastro.2018.04.018
Nannapaneni, R., Soni, K. A. (2015). “Use of Bacteriophages to Remove Biofilms of Listeria Monocytogenes and Other Foodborne Bacterial Pathogens in the Food Environment,” In: Biofilms in the Food Environment, 2nd. Eds. Pometto, D.III AL (Oxford, UK: Wiley Blackwell), 131–144131–144.
Nayfach, S., Paez-Espino, D., Call, L., Low, S. J., Sberro, H., Ivanova, N. N., et al. (2021). Metagenomic Compendium of 189,680 DNA Viruses From the Human Gut Microbiome. Nat. Microbiol. 6 (7), 960–970. doi: 10.1038/s41564-021-00928-6
Nguyen, L. T., Vogel, H. J. (2016). Staphylokinase has Distinct Modes of Interaction With Antimicrobial Peptides, Modulating Its Plasminogen-Activation Properties. Sci. Rep. 6, 31817. doi: 10.1038/srep31817
Norman, J. M., Handley, S. A., Baldridge, M. T., Droit, L., Liu, C. Y., Keller, B. C., et al. (2015). Disease-Specific Alterations in the Enteric Virome in Inflammatory Bowel Disease. Cell 160 (3), 447–460. doi: 10.1016/j.cell.2015.01.002
Ogilvie, L. A., Jones, B. V. (2015). The Human Gut Virome: A Multifaceted Majority. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00918
Oh, J., Byrd, A. L., Park, M., Kong, H. H., Segre, J. A. (2016). Temporal Stability of the Human Skin Microbiome. Cell 165, 854–866. doi: 10.1016/j.cell.2016.04.008
Ott, S. J., Waetzig, G. H., Rehman, A., Moltzau-Anderson, J., Bharti, R., Grasis, J. A., et al. (2017). Efficacy of Sterile Fecal Filtrate Transfer for Treating Patients With Clostridium Difficile Infection. Gastroenterology 152, 799–811.e797. doi: 10.1053/j.gastro.2016.11.010
Perez-Brocal, V., Garcia-Lopez, R., Vazquez-Castellanos, J. F., Nos, P., Beltran, B., Latorre, A., et al. (2013). Study of the Viral and Microbial Communities Associated With Crohn's Disease: A Metagenomic Approach. Clin. Transl. Gastroenterol. 4 (6), e36. doi: 10.1038/ctg.2013.9
Phan, T. G., Vo, N. P., Bonkoungou, I. J., Kapoor, A., Barro, N., O'Ryan, M., et al. (2012). Acute Diarrhea in West African Children: Diverse Enteric Viruses and a Novel Parvovirus Genus. J. Virol. 86, 11024–11030. doi: 10.1128/jvi.01427-12
Pride, D. T., Salzman, J., Haynes, M., Rohwer, F., Davis-Long, C., White, R. A., 3rd, et al. (2012). Evidence of a Robust Resident Bacteriophage Population Revealed Through Analysis of the Human Salivary Virome. ISME J. 6, 915–926. doi: 10.1038/ismej.2011.169
Regeimbal, J. M., Jacobs, A. C., Corey, B. W., Henry, M. S., Thompson, M. G., Pavlicek, R. L., et al. (2016). Personalized Therapeutic Cocktail of Wild Environmental Phages Rescues Mice From Acinetobacter Baumannii Wound Infections. Antimicrob. Agents Chemother. 60, 5806–5816. doi: 10.1128/aac.02877-15
Reyes, A., Blanton, L. V., Cao, S., Zhao, G., Manary, M., Trehan, I., et al. (2015). Gut DNA Viromes of Malawian Twins Discordant for Severe Acute Malnutrition. Proc. Natl. Acad. Sci. U. S. A. 112 (38), 11941–11946. doi: 10.1073/pnas.1514285112
Reyes, A., Haynes, M., Hanson, N., Angly, F. E., Heath, A. C., Rohwer, F., et al. (2010). Viruses in the Faecal Microbiota of Monozygotic Twins and Their Mothers. Nature 466, 334–338. SRC - BaiduScholar. doi: 10.1038/nature09199
Riley, L. W., Remis, R. S., Helgerson, S. D. (1983). Hemorrhagic Colitis Associated With a Rare Escherichia Coli Serotype. N. Engl. J. Med. 308, 681–685. doi: 10.1056/NEJM198303243081203
Rohwer, F., Prangishvili, D., Lindell, D. (2009). Roles of Viruses in the Environment. Environ. Microbiol. 11 (11), 2771–2774. doi: 10.1111/j.1462-2920.2009.02101.x
Rollison, D. E. (2010). JC Virus Infection: A Cause of Colorectal Cancer? J. Clin. Gastroenterol. 44 (7), 466–468. doi: 10.1097/mcg.0b013e3181e0084b
Ruzin, A., Lindsay, J., Novick, R. P. (2001). Molecular Genetics of SaPI1a Mobile Pathogenicity Island in Staphylococcus Aureus. Mol. Microbiol. 41, 365–377. doi: 10.1046/j.1365-2958.2001.02488.x
Sarker, S. A., Sultana, S., Reuteler, G., Moine, D., Descombes, P., Charton, F., et al. (2016). Oral Phage Therapy of Acute Bacterial Diarrhea With Two Coliphage Preparations: Trial in Children From Bangladesh. EBioMedicine 4, 124–137. doi: 10.1016/j.ebiom.2015.12.023
Secor, P. R., Sweere, J. M., Michaels, L. A., Malkovskiy, A. V., Lazzareschi, D., Katznelson, E., et al. (2015). Filamentous Bacteriophage Promote Biofilm Assembly and Function. Cell Host Microbe 18 (5), 549–559. doi: 10.1016/j.chom.2015.10.013
Sharon, I., Morowitz, M. J., Thomas, B. C., Costello, E. K., Relman, D. A., Banfield, J. F. (2013). Time Series Community Genomics Analysis Reveals Rapid Shifts in Bacterial Species, Strains, and Phage During Infant Gut Colonization. Genome Res. 23 (1), 111–120. doi: 10.1101/gr.142315.112
Sheehan, D., Moran, C., Shanahan, F. (2015). The Microbiota in Inflammatory Bowel Disease. J. Gastroenterol. 50 (5), 495–507. doi: 10.1007/s00535-015-1064-1
Shkoporov, A. N., Khokhlova, E. V., Stephens, N., Hueston, C., Seymour, S., Hryckowian, A. J., et al. (2021). Long-Term Persistence of Crass-Like Phage Crass001 Is Associated With Phase Variation in Bacteroides Intestinalis. BMC Biol. 19 (1), 163. doi: 10.1186/s12915-021-01084-3
Siegel, R. L., Miller, K. D., Jemal, A., Jemal, A. (2015). Cancer Statistics. CA: J. Clin. 65 (1), 5–29. doi: 10.3322/caac.21254
Siqueira, J. D., Dominguez-Bello, M. G., Contreras, M., Lander, O., Caballero-Arias, H., Xutao, D., et al. (2018). Complex Virome in Feces From Amerindian Children in Isolated Amazonian Villages. Nat. Commun. 9 (1), 4270. doi: 10.1038/s41467-018-06502-9
Sulakvelidze, A., Kutter, E., Raton, F. L., C, C. R. (2005). Bacteriophage Therapy in Humans. In: Bacteriophages: Biology and Application. Kutter, E., Sulakvelidze, A. Eds. (Boca Raton, FL, USA: CRC Press) p. 381–436.
Summers, W. C. (2001). Bacteriophage Therapy. Annu. Rev. Microbiol. 55, 437–451. doi: 10.1146/annurev.micro.55.1.437
Szekely, A. J., Breitbart, M. (2016). Single-Stranded DNA Phages: From Early Molecular Biology Tools to Recent Revolutions in Environmental Microbiology. FEMS Microbiol. Lett. 363 (6), fnw027. doi: 10.1093/femsle/fnw027
Tong, M., Li, X., Wegener Parfrey, L., Roth, B., Ippoliti, A., Wei, B., et al. (2013). A Modular Organization of the Human Intestinal Mucosal Microbiota and Its Association With Inflammatory Bowel Disease. PloS One 8, e80702. doi: 10.1371/journal.pone.0080702
Tremaroli, V., Backhed, F. (2012). Functional Interactions Between the Gut Microbiota and Host Metabolism. Nature 489, 242–249. doi: 10.1038/nature11552
Trucksis, M., Galen, J. E., Michalski, J., Fasano, A., Kaper, J. B. (1993). Accessory Cholera Enterotoxin (Ace), the Third Toxin of a Vibrio Cholerae Virulence Cassette. Proc. Natl. Acad. Sci. U. S. A. 90 (11), 5267–5271. doi: 10.1073/pnas.90.11.5267
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., Gordon, J. I. (2006). An Obesity-Associated Gut Microbiome With Increased Capacity for Energy Harvest. Nature 444, 1027–1031. doi: 10.1038/nature05414
van Leeuwen, M., Williams, M. M., Koraka, P., Simon, J. H., Smits, S. L., Osterhaus, A. D. (2010). Human Picobirnaviruses Identified by Molecular Screening of Diarrhea Samples. J. Clin. Microbiol. 48 (5), 1787–1794. doi: 10.1128/JCM.02452-09
van Wamel, W. J., Rooijakkers, S. H., Ruyken, M., van Kessel, K. P., Strijp, J. A. (2006). The Innate Immune Modulators Staphylococcal Complement Inhibitor and Chemotaxis Inhibitory Protein of Staphylococcus Aureus Are Located on Beta-Hemolysin-Converting Bacteriophages. J. Bacteriol 188, 1310–1315. doi: 10.1128/jb.188.4.1310-1315.2006
Veesler, D., Cambillau, C. (2011). A Common Evolutionary Origin for Tailed-Bacteriophage Functional Modules and Bacterial Machineries. Microbiol. Mol. Biol. Rev. 75 (3), 423–433. doi: 10.1128/MMBR.00014-11
Wagner, P. L., Waldor, M. K. (2002). Bacteriophage Control of Bacterial Virulence. Infect. Immun. 70 (8), 3985–3993. doi: 10.1128/IAI.70.8.3985-3993.2002
Waldor, M. K., Mekalanos, J. J. (1996). Lysogenic Conversion by a Filamentous Phage Encoding Cholera Toxin. Science 272 (5270), 1910–1914. doi: 10.1126/science.272.5270.1910
Weinbauer, M. G. (2004). Ecology of Prokaryotic Viruses. FEMS Microbiol. Rev. 28 (2), 127–181. doi: 10.1016/j.femsre.2003.08.001
Weinbauer, M. G., Rassoulzadegan, F. (2004). Are Viruses Driving Microbial Diversification and Diversity? Environ. Microbiol. 6 (1), 1–11. doi: 10.1046/j.1462-2920.2003.00539.x
Wilhelmi, I., Roman, E., Sanchez-Fauquier, A. (2003). Viruses Causing Gastroenteritis. Clin. Microbiol. Infect. 9 (4), 247–262. doi: 10.1046/j.1469-0691.2003.00560.x
Wood, M. W., Rosqvist, R., Mullan, P. B., Edwards, M. H., Galyov, E. E. (1996). SopE, a Secreted Protein of Salmonella Dublin, Is Translocated Into the Target Eukaryotic Cell via a Sip-Dependent Mechanism and Promotes Bacterial Entry. Mol. Microbiol. 22, 327–338. doi: 10.1046/j.1365-2958.1996.00116.x
Yosef, I., Manor, M., Kiro, R., Qimron, U. (2015). Temperate and Lytic Bacteriophages Programmed to Sensitize and Kill Antibiotic-Resistant Bacteria. Proc. Natl. Acad. Sci. U. S. A. 112 (23), 7267–7272. doi: 10.1073/pnas.1500107112
Yu, J., Feng, Q., Wong, S. H., Zhang, D., Liang, Q. Y., Qin, Y., et al. (2017). Metagenomic Analysis of Faecal Microbiome as a Tool Towards Targeted Non-Invasive Biomarkers for Colorectal Cancer. Gut 66, 70–78. doi: 10.1136/gutjnl-2015-309800
Zackular, J. P., Rogers, M. A., Ruffin, M. T. T., Schloss, P. D. (2014). The Human Gut Microbiome as a Screening Tool for Colorectal Cancer. Cancer Prev. Res. (Phila) 7, 1112–1121. doi: 10.1158/1940-6207.CAPR-14-0129
Żaczek, M., Łusiak-Szelachowska, M., Jończyk-Matysiak, E., Weber-Dąbrowska, B., Międzybrodzki, R., Owczarek, B., et al. (2016). Antibody Production in Response to Staphylococcal MS-1 Phage Cocktail in Patients Undergoing Phage Therapy. Front. Microbiol. 7, 1681. doi: 10.3389/fmicb.2016.01681
Zeller, G., Tap, J., Voigt, A. Y., Sunagawa, S., Kultima, J. R., Costea, P. I., et al. (2014). Potential of Fecal Microbiota for Early-Stage Detection of Colorectal Cancer. Mol. Syst. Biol. 766 (10), 766. doi: 10.15252/msb.20145645
Zhang, T., Breitbart, M., Lee, W. H., Run, J. Q., Wei, C. L., Soh, S. W., et al. (2006). RNA Viral Community in Human Feces: Prevalence of Plant Pathogenic Viruses. PloS Biol. 4 (1), e3. doi: 10.1371/journal.pbio.0040003
Zhang, X., McDaniel, A. D., Wolf, L. E., Keusch, G. T., Waldor, M. K., Acheson, D. W. (2000). Quinolone Antibiotics Induce Shiga Toxin-Encoding Bacteriophages, Toxin Production, and Death in Mice. J. Infect. Dis. 181 (2), 664–670. doi: 10.1086/315239
Keywords: virome, bacteriophage, eukaryotic virus, gastro-intestinal disease, phage therapy
Citation: Spencer L, Olawuni B and Singh P (2022) Gut Virome: Role and Distribution in Health and Gastrointestinal Diseases. Front. Cell. Infect. Microbiol. 12:836706. doi: 10.3389/fcimb.2022.836706
Received: 15 December 2021; Accepted: 10 February 2022;
Published: 10 March 2022.
Edited by:
Michael B. Sohn, University of Rochester, United StatesReviewed by:
Diane Bimczok, Montana State University, United StatesCopyright © 2022 Spencer, Olawuni and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pallavi Singh, cHNpbmdoMUBuaXUuZWR1
 Laurie Spencer
Laurie Spencer Babatunde Olawuni
Babatunde Olawuni Pallavi Singh
Pallavi Singh