- 1Aix Marseille Univ Institut de Recherche pour le Développement (IRD), Assistance Publique - Hôpitaux de Marseille (APHM), Microbes, Evolution (MEPHI), Phylogénie et Infection, Marseille, France
- 2Institut Hospitalo-Universitaire (IHU)-Méditerranée Infection, Marseille, France
- 3Plateforme Génomique - Bioinformatique, Institut Pasteur d’Algérie, Rue du Petit Staouéli, Algiers, Algeria
- 4Plague Reference Laboratory, Bunia, Congo
- 5Aix Marseille Univ, Institut de Recherche pour le Développement (IRD), Assistance Publique - Hôpitaux de Marseille (APHM), Vecteurs – Infections Tropicales et Méditeranéennes (VITROME), Marseille, France
Pediculus humanus is an obligate bloodsucking parasite of humans that has two ecotypes, the head louse and the body louse, which share an intimate history of coevolution with their human host. In the present work, we obtained and analysed head and body lice collected from Mbuti pygmies living in the Orientale province of the Democratic Republic of the Congo. Cytochrome b DNA analysis was performed in order to type the six known lice clades (A, D, B, F, C and E). The results revealed the presence of two mitochondrial clades. Clade D was the most frequent (61.7% of 47), followed by clade A (38.3% of 47). Sixteen haplotypes were found in 47 samples, of which thirteen were novel haplotypes, indicating an unusually high genetic diversity that closely mirrors the diversity of their hosts. Moreover, we report for the first time the presence of the DNA of R. felis in three (6.4% of 47) head and body lice belonging to both clades A and D. Additional studies are needed to clarify whether the Pediculus lice can indeed transmit this emerging zoonotic bacterium to their human hosts.
Introduction
Human sucking lice from the genus Pediculus are obligate host-specific parasites, that co-evolved with their host (Reed et al., 2004; Amanzougaghene et al., 2020a). P. humanus includes two subspecies, each occupying a different ecological niche: hair for the head louse, P. humanus capitis, and garments for the body louse, P. h. humanus (Amanzougaghene et al., 2020a). Although morphologically almost identical, it is now possible to genetically differentiate the head louse from the body louse (Drali et al., 2013).
The body louse is the vector of three pathogenic bacteria that cause serious diseases, that killed millions of people, namely Rickettsia prowazekii (the causative agent of epidemic typhus), Bartonella quintana (trench fever) and Borrelia recurrentis (relapsing fever) (Raoult and Roux, 1999). In addition, body and head lice may harbour Yersinia pestis, the agent of plague (Houhamdi et al., 2006; Ayyadurai et al., 2010; Piarroux et al., 2013; Drali et al., 2015; Raoult, 2016). Moreover, experimental models showed that body lice have the potential to acquire, maintain and transmit R. typhi (the causative agent of endemic or murine typhus), R. rickettsii (Rocky Mountain spotted fever), R. conorii (Mediterranean spotted fever, Indian tick typhus), and Acinetobacter baumannii and A. lwoffii to rabbits (Weyer, 1952a; Houhamdi et al., 2003; Houhamdi and Raoult, 2006a; Houhamdi and Raoult, 2006b). Coxiella burnetii, the agent of Q fever, was first isolated from the body lice of individuals living in an epidemic area in Rwanda, and was subsequently found to infect body lice under experimental conditions (Giroud and Jadin, 1954; Babudieri, 1959). Recently, its DNA was found in body lice infecting homeless people in Algeria, as well as in body lice from France (Louni et al., 2018; Amanzougaghene et al., 2020b). The DNA of Anaplasma phagocytophilum was also detected in body lice from homeless people in Algeria (Louni et al., 2018). Although no reports in the field have yet demonstrated that head lice are vectors of infectious agents, several experimental and epidemiological reports indicate that they can transmit pathogens to their human host under favourable epidemiological conditions (Amanzougaghene et al., 2020a). Indeed, under laboratory conditions it has been shown that head lice retrieved from patients and rabbits infected with R. prowazekii, can be readily infected and disseminate this bacterium in their faeces, demonstrating that head lice have the potential to be a vector of this pathogen (Goldberger and Anderson, 1912; Murray and Torrey, 1975). Moreover, in a laboratory-reared head louse colony, it has been demonstrated that head lice can maintain a persistent B. quintana infection for several days following its acquisition in a bloodmeal (Previte et al., 2014). In addition, the DNA of several pathogenic bacteria have been reported in head lice, including B. quintana, C. burnetii, B. recurrentis, Y. pestis, R. aeschlimannii, Anaplasma, Ehrlichia and Acinetobacter spp. (Piarroux et al., 2013; Drali et al., 2015; Amanzougaghene et al., 2016; Amanzougaghene et al., 2017; Candy et al., 2018; Boumbanda Koyo et al., 2019; Amanzougaghene et al., 2020b; Boumbanda-Koyo et al., 2020; Hammoud et al., 2021).
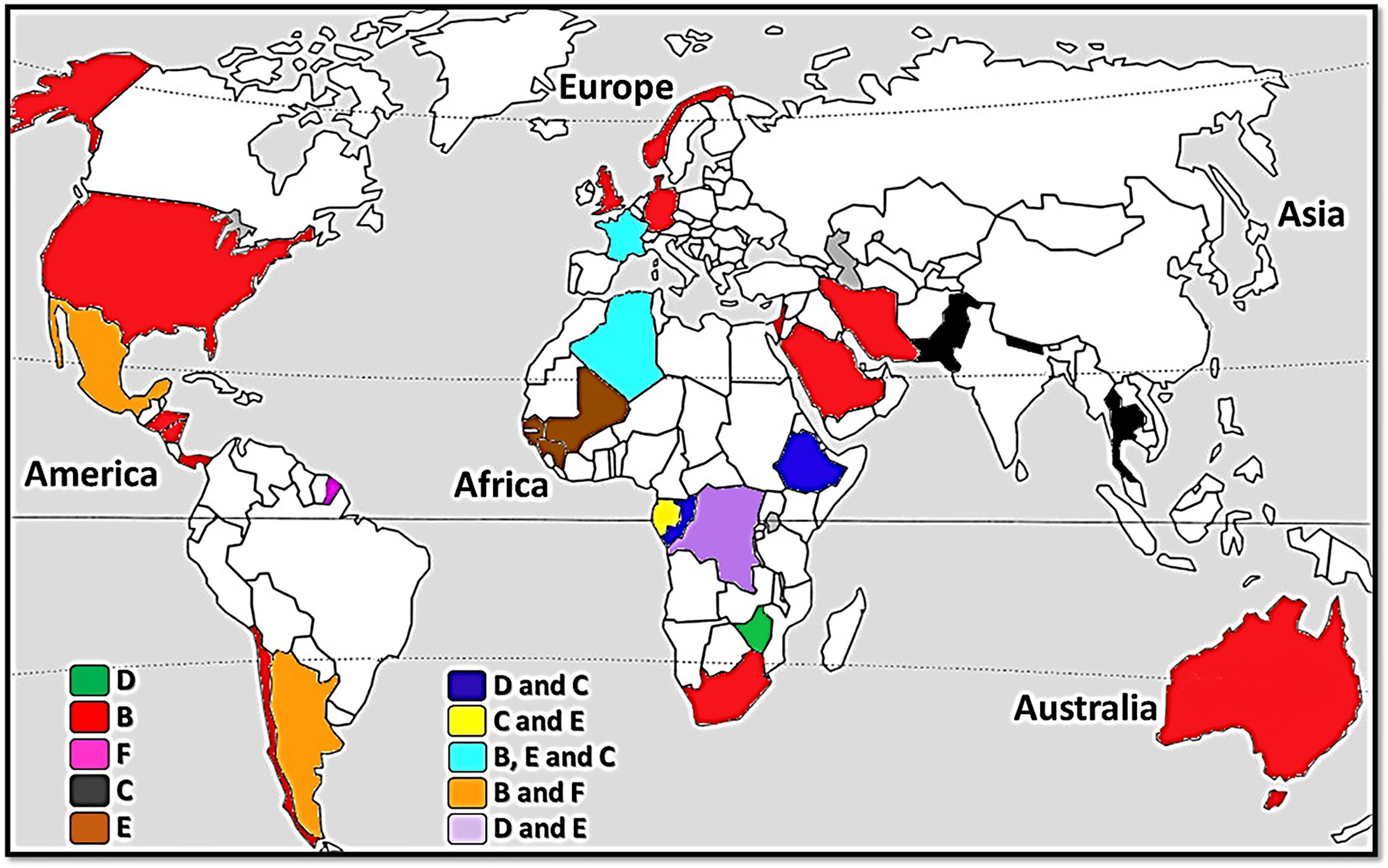
On the basis of all genetic analyses performed on human lice, only those targeting the mitochondrial genes were able to classify them into six distinctly different clades (Light et al., 2008; Ascunce et al., 2013; Ashfaq et al., 2015; Drali et al., 2015; Amanzougaghene et al., 2019). Head lice belong to all six clades (A, D, B, F, C and E), while body lice belong only to clades A and D (Amanzougaghene et al., 2019; Amanzougaghene et al., 2020a). Clade A is distributed worldwide (Ascunce et al., 2013; Ashfaq et al., 2015; Amanzougaghene et al., 2019), whereas the other clades are geographically restricted (Figure 1). Clade D is restricted to sub-Saharan African countries, including the Democratic Republic of Congo (DRC) and the Republic of Congo (Congo-Brazzaville), Ethiopia and Zimbabwe (Drali et al., 2015; Amanzougaghene et al., 2019). Clade B includes head lice found in the Americas, Western Europe, Australia, North and South Africa, Saudi Arabia and Iran (Light et al., 2008; Ascunce et al., 2013; Ashfaq et al., 2015; Boutellis et al., 2015; Al-Shahrani et al., 2017). Clade F, which is the sister group of clade B, is found in South America (Amanzougaghene et al., 2019; Amanzougaghene et al., 2020a). Clade C is, to date, limited to Africa and Asia (Ashfaq et al., 2015; Sunantaraporn et al., 2015; Amanzougaghene et al., 2016; Boumbanda-Koyo et al., 2020). Finally, clade E has mainly been found in West African countries (Senegal, Mali and Guinea) (Amanzougaghene et al., 2017; Amanzougaghene et al., 2020a; Hammoud et al., 2021).
In addition to their inter clade diversity, human lice also display high intra clade diversity, as illustrated by the multiple distinct haplotypes for each clade (Ascunce et al., 2013; Ashfaq et al., 2015; Amanzougaghene et al., 2016; Drali et al., 2016; Amanzougaghene et al., 2019). The genetic diversity is also observed among global populations of Homo sapiens. It is even more accentuated among the indigenous populations of Africa, especially among pygmies. Indeed, the genetic diversity found among African pygmies is the most significant in the world. They present great diversity, both among themselves and compared to other African populations (Henn et al., 2011; Duda and Jan Zrzavý, 2016). The group is characterised by their reduced height and they survive from hunting and gathering in the rain forest in Central Africa (Jakobsson et al., 2008; Campbell and Tishkoff, 2010; Batini et al., 2011). Their number varies between 300,000 and 500,000 individuals distributed into 25 ethno-linguistic groups (Ohenjo et al., 2006; Becker et al., 2011). Molecular dating analysis based on mitochondrial DNA has revealed that they shared a common ancestor with non-pigmy populations dating to around 60 thousand years ago, whereas the separation between the West and East African pygmies dates to approximately 20 thousand years ago (Destro-Bisol et al., 2004; Batini et al., 2011; Jarvis et al., 2012). African pygmies are known to be at the root of the modern human tree (Jakobsson et al., 2008). Archaic DNA was recently characterised in the Mbuti pygmies from the Itury forest in the Republic Democratic of the Congo (RDC) (Hammer et al., 2011). Mbuti pygmies are relatively isolated from other Pygmy and neighbouring non-Pygmy populations and are directly descended from the ancestors of most Pygmy groups (Patin et al., 2009; Verdu and Destro-Bisol, 2012).
In this study, we were fortunate to obtain and analyse head and body lice collected from pygmy populations that belong to the Mbuti ethnic group living in the Orientale province of the Democratic Republic of the Congo, where the Clade D of human lice was characterised for the first time (Drali et al., 2015).
Materials and Methods
Ethics Statement
This study was approved by the by the Ethics Committee of the École de Santé Publique de l’Université de Kinshasa (N/Réf:ESP/CE/17B/2014). The head and body lice were collected from infested individuals after obtaining their verbal consent. Approval was obtained from the local authorities and representatives were present when it was performed. Moreover, the collection of lice from humans is a non-invasive procedure. This it was made according to the prescriptions of the Declaration of Helsinki of the World Medical Association.
Louse Sampling
This study was carried out in March 2014 on Mbuti Pygmies living in Shaurimoya (1°40′14″ N, 28°29′21″ E), a remote rural village, located in the Itury rain forest area near the Rethy Health District, in the Orientale province of the Democratic Republic of the Congo (Figures 2A, B). A total of 47 lice (22 body lice and 25 head lice) were collected from 22 infested volunteers aged between 4 and 30 years old. All head lice were collected exclusively from the hair, and all body lice were collected exclusively from clothing. The collected lice were then preserved in 70% ethanol before being sent to our laboratory in Marseille (France). Once received at the laboratory, the specimens were photographed on their dorsal and ventral sides using a fixed camera (Olympus DP71, Rungis, France).
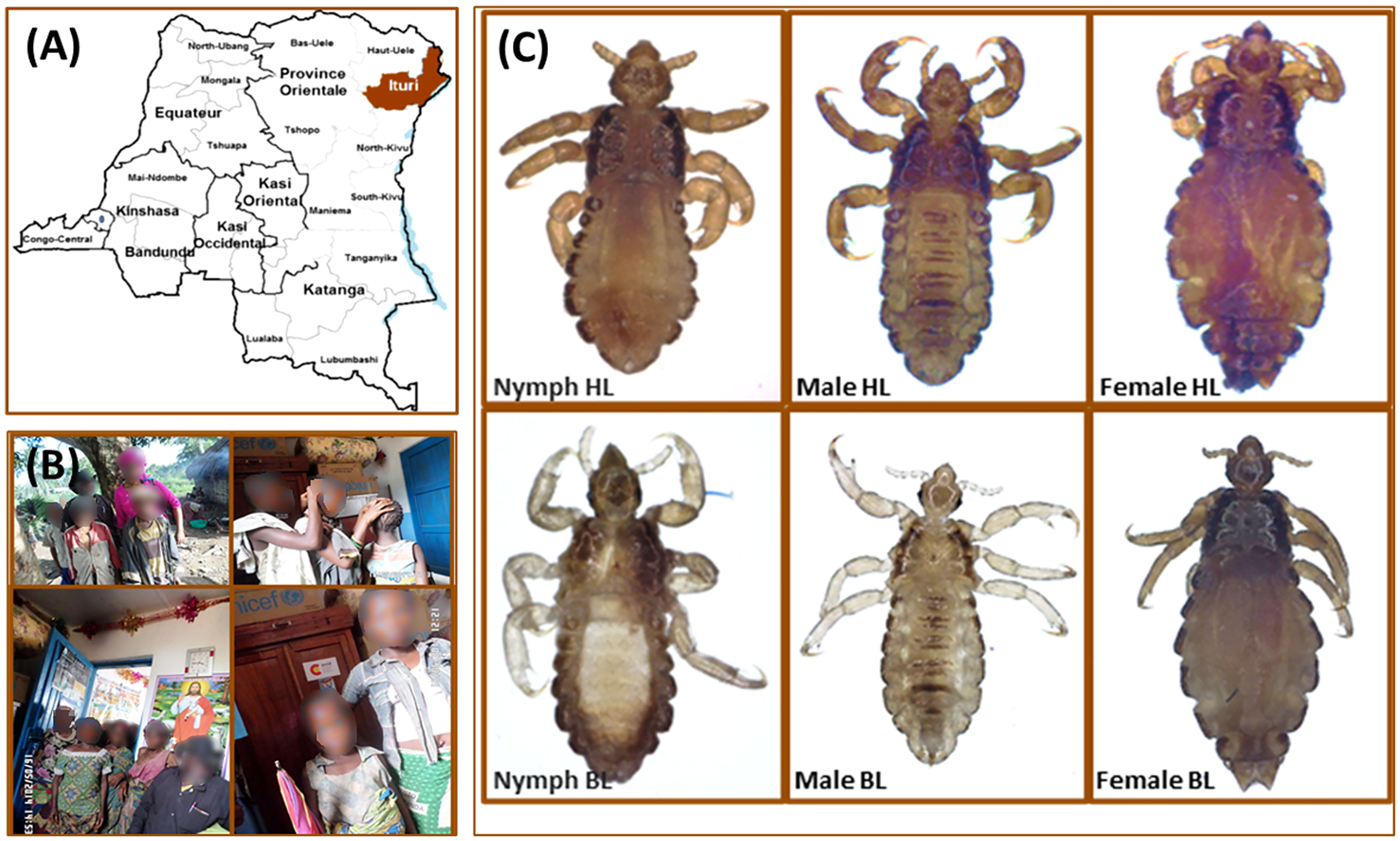
Figure 2 Head and body lice collected from Mbuti pygmies, living in the Orientale province of the Democratic Republic of the Congo. (A) Geographical location of louse sampling. (B) Mbuti pygmies infested with lice. (C) Head and body lice from Mbuti pygmies.
DNA Extraction
Total DNA was extracted from lice using a DNA extraction kit, the QIAamp Tissue Kit (Qiagen SAS, Courtaboeuf, France) with the EZ1 apparatus following the manufacturer’s protocol. DNA from each louse was eluted in 100 μl of TE buffer and stored at -20°C under sterile conditions until the next stage of the investigation.
Lice Clade and Phylogenetic Analysis
For phylogenetic analysis, the cytochrome b (cytb) gene was chosen, as it was the most commonly used in human louse phylogenetic studies (Li et al., 2010; Ashfaq et al., 2015; Amanzougaghene et al., 2016; Amanzougaghene et al., 2019). A polymerase chain reaction (PCR) was performed to amplify a fragment of 347-bp of cytb gene using the primers described previously (Li et al., 2010), in a MiniAmp™ Plus Thermal Cycler (Thermo Fisher Scientific, Illkirch, France). PCRs consisted of 50 µl volume including 25 µl Amplitaq gold master mix (Applied Biosystems, Foster City, CA, USA), 1 µl of each primer, 5 μl of DNA template, and water. The thermal cycling profile was one incubation step at 95°C for 15 minutes, 40 cycles of one minute at 95°C, 30 seconds at 56°C and one minute at 72°C followed by a final extension for five minutes at 72°C. Negative and positive controls were included in each assay. PCR products were purified using NucleoFast 96 PCR plates (Macherey-Nagel EURL, Hoerdt, France) as per the manufacturer’s instructions. The amplicons were sequenced using the Big Dye Terminator Cycle Sequencing Kit (Perkin Elmer Applied Biosystems, Foster City, CA) with an ABI automated sequencer (Applied Biosystems). The obtained electropherograms were assembled and edited using ChromasPro software (ChromasPro 1.7, Technelysium Pty Ltd., Tewantin, Australia) and compared with those available in the GenBank database by NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). For all cytb nucleotides sequences obtained in this study, unique haplotypes were defined using DnaSPv5.10 and compared with the most recent cytb haplotype database described previously (Supplementary Table S1). Moreover, to learn more about the diversity and the genetic relatedness of lice infesting different populations of African pygmies, we recovered and analysed 160 cytb sequences from the pygmy lice of Congo Brazzaville (Amanzougaghene et al., 2016), that we compared to those included in this study. All the pygmies’ haplotypes, together with the references from all the body and head lice clades were used to construct maximum-likelihood (ML) trees and a median-joining (MJ) network. To generate the best ML tree, the Modeltest v.3.7 (Posada and Crandall, 1998) was used to examine model of nucleotide substitution and choose a best-fit model of sequence evolution. The HKY+I+G model was chosen as the best model of evolution, providing the best approximation of the data using the fewest parameters according to the Akaike Information Criterion (Huelsenbeck and Rannala, 1997; Posada and Buckley, 2004). Trees reconstructions were conducted using MEGA 6 software (Tamura et al., 2013) with ML method under HKY + I + G model with 500 bootstrap replicates. Cytb sequence from P. schaeffi (accession number: AY696067) was employed as outgroup. The median-joining (MJ) network was constructed using the Bandelt method with the NETWORK5.0 programme (www.fluxus-engineering.com/sharenet.htm) using equal weights for all mutations (Bandelt et al., 1999). For genetic diversity and haplotype analysis, population genetic indices including number of sequences (n), number of polymorphic sites (S), average number of pairwise nucleotide differences (k), nucleotide diversity (π), number of haplotypes (H) and haplotype diversity (Hd) and neutrality tests (Fu & Li’s D and Tajima’s D) were calculated for all cytb sequences, using DNASP v5.10 software (Librado and Rozas, 2009). The newly generated cytb sequences identified were submitted to the GenBank database under the accession numbers: MH117941-MH117953.
Molecular Detection of the Presence Of Pathogen DNA
Real-time quantitative PCR (qPCR) was performed to test each DNA sample for the presence of R. prowazekii, Rickettsia spp., Y. pestis, B. quintana, Borrelia spp., C. burnetii Anaplasma spp., Piroplasmida (Theileria spp. and Babesia spp.) and R. felis using previously reported specific primers and probes. All R. felis positive samples were confirmed by a second specific qPCR targeting the Orfb gene. All sequences of primers and probes used for qPCRs in this study are shown in Supplementary Table S2. All qPCRs were performed using a CFX96 Real-Time system (Bio-Rad, Marnes-la-Coquette, France) and the Roche LightCycler 480 Probes Master Mix PCR kit (Roche Applied Science, Mannheim, Germany) in accordance with the manufacturer’s instructions. We included the DNA of the target bacteria as positive controls and master mixtures as a negative control for each qPCR run.
Results
The microscopic examination of morphological criteria such as size, shape and color of the lice recovered from pygmies analyzed in this study revealed no morphological discordance in relation to what is common in human lice genus Pediculus (Figure 2C). The mitochondrial DNA analysis, of the 47 specimens (26 head lice and 21 body lice) collected from 22 mono and/or double infested individuals, showed a distribution of 38.3% for haplogroup A (12 head and 17 body lice) and 61.7% for haplogroup D (12 head and 17 body lice). Among the 22 individuals, eight (21.6%) showed dual infestation with both clades and three (8%) were simultaneously infested with both body and head lice (Table 1). An in-depth analysis of all obtained nucleotide sequences and their alignment with all publicly available haplotypes revealed the presence of 16 haplotypes in 47 samples, defined by variation of 30 nucleotide positions (18 transitions and 12 transversions) (Table 2), indicating an unusually high genetic diversity among the lice studied from Mbuti pygmies. One haplotype (12 nucleotide sequences) belonged to haplotype A5 within clade A, which is the most widely distributed haplotype in the world. Two haplotypes within clade D, haplotype D65 (seven sequences) and D67 (13 sequences), which are the most prevalent haplotypes among clade D, were also shared with pygmy lice from Congo-Brazzaville (Figures 3A, B and 4). Lastly, the remaining 13 haplotypes, A70-A71 (one sequence for each haplotype) and A72-A73 (two sequences for each haplotype), all belonged to clade A, and D77-D85 (one sequence for each haplotype) belonged to clade D, were novel and unique to lice from Mbuti pygmies (Figure 4).
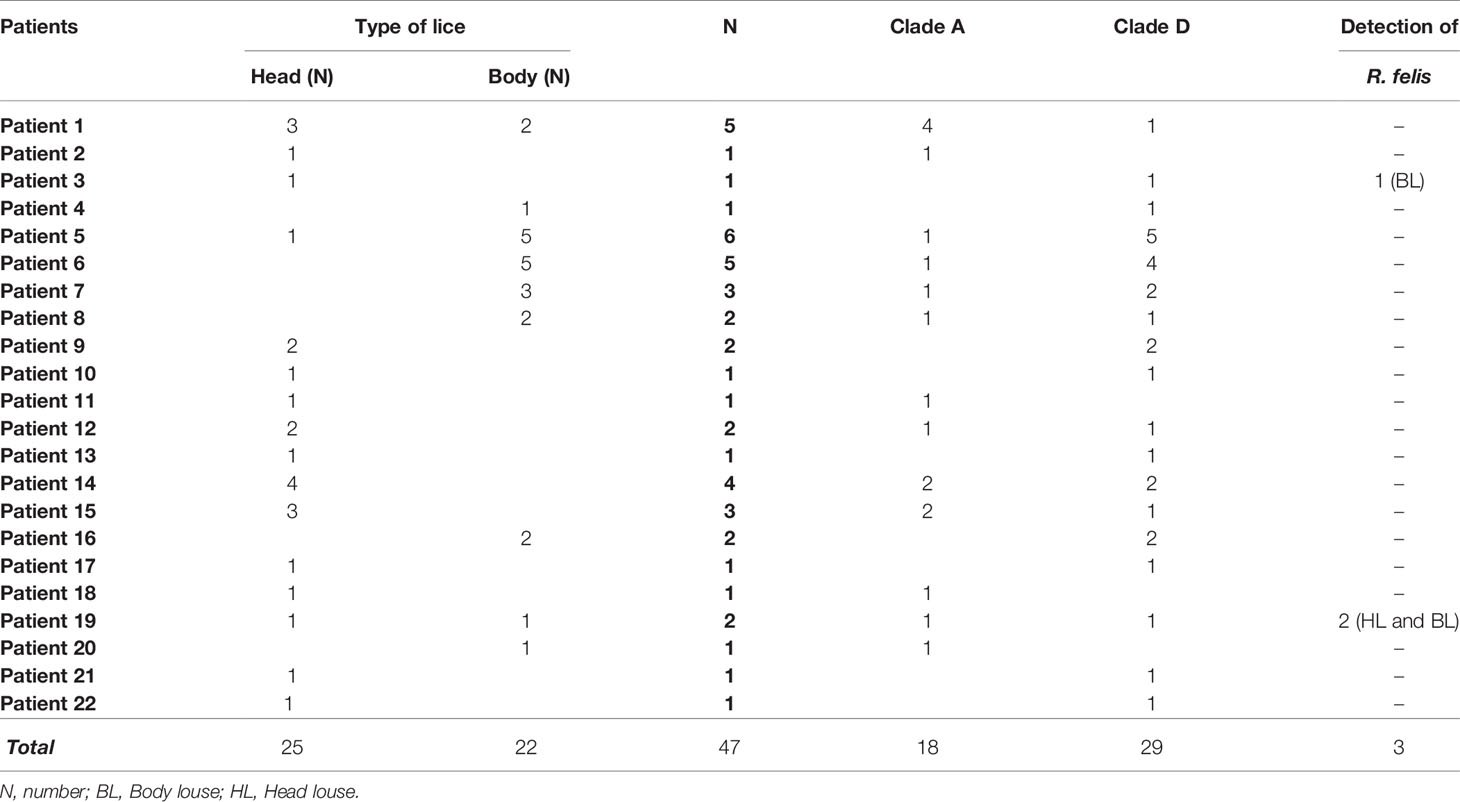
Table 1 Summary of the pathogens detected in body and head lice collected from infested Mbuti pygmies in Itury, Congo-DRC.
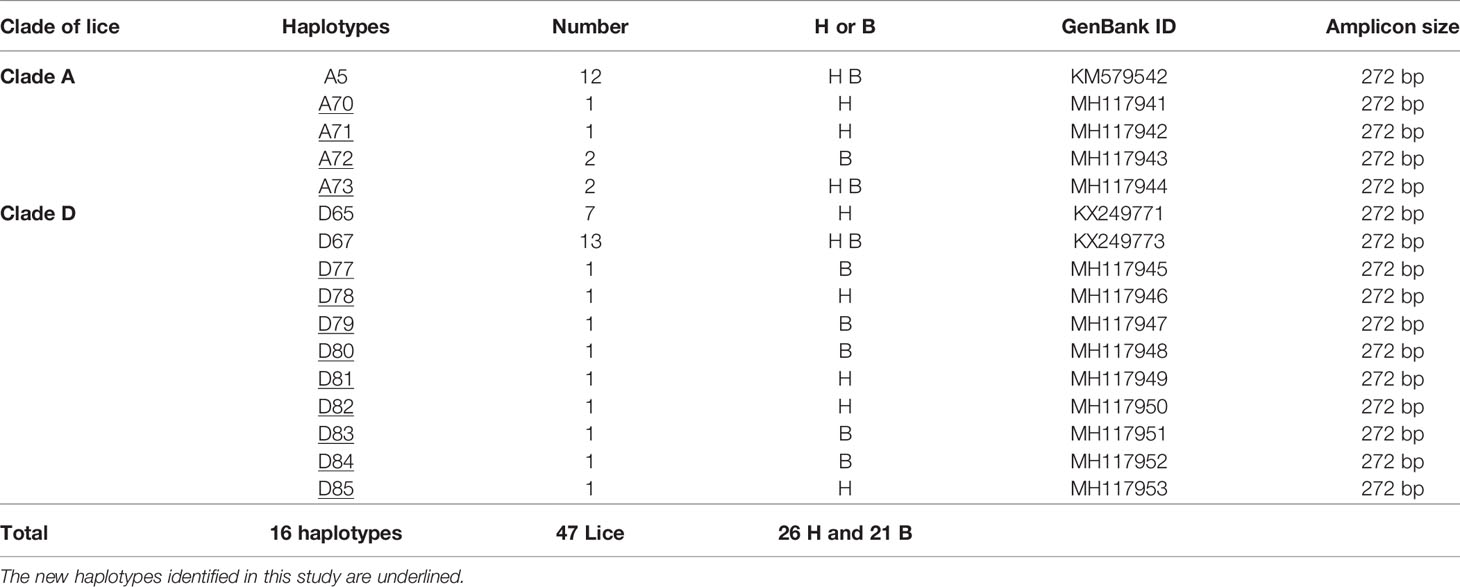
Table 2 Haplotype frequency of head and body lice identified from infested pygmy individuals in Itury, Congo-RDC.
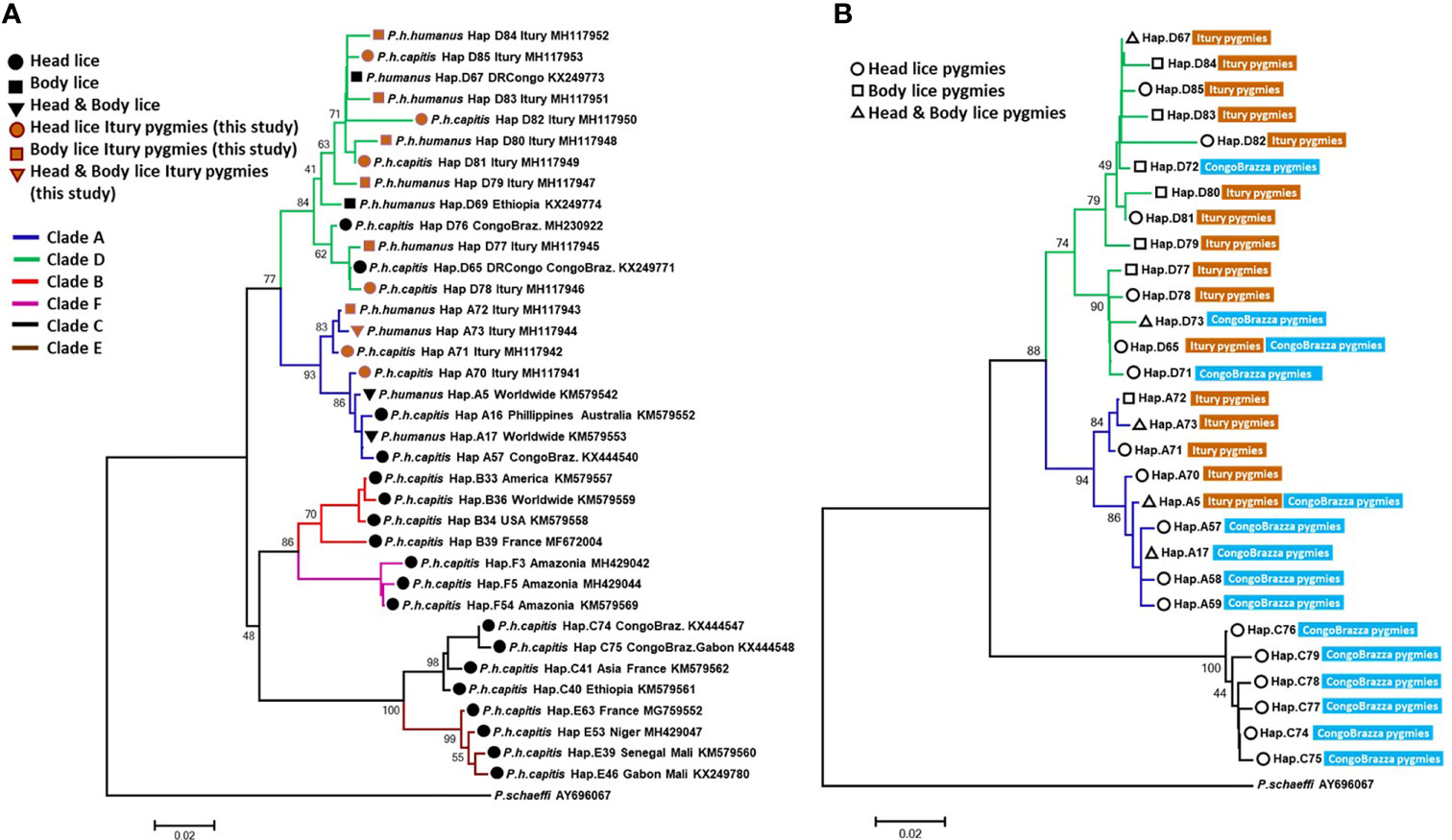
Figure 3 The phylogeny of head and body lice from Mbuti pygmies based on the cytb gene. Maximum-likelihood phylogenetic tree showing the relationship of haplotypes identified in this study (A) with the most prevalent haplotypes of P. humanus and (B) haplotypes from head lice Congo Brazzaville pygmies. Phylogenetic inference was conducted in MEGA 7 using the maximum likelihood method under HKY + I + G model with 500 bootstrap replicates. There was a total of 270 positions in the final dataset. The scale-bar represents a 2% nucleotide sequence divergence.
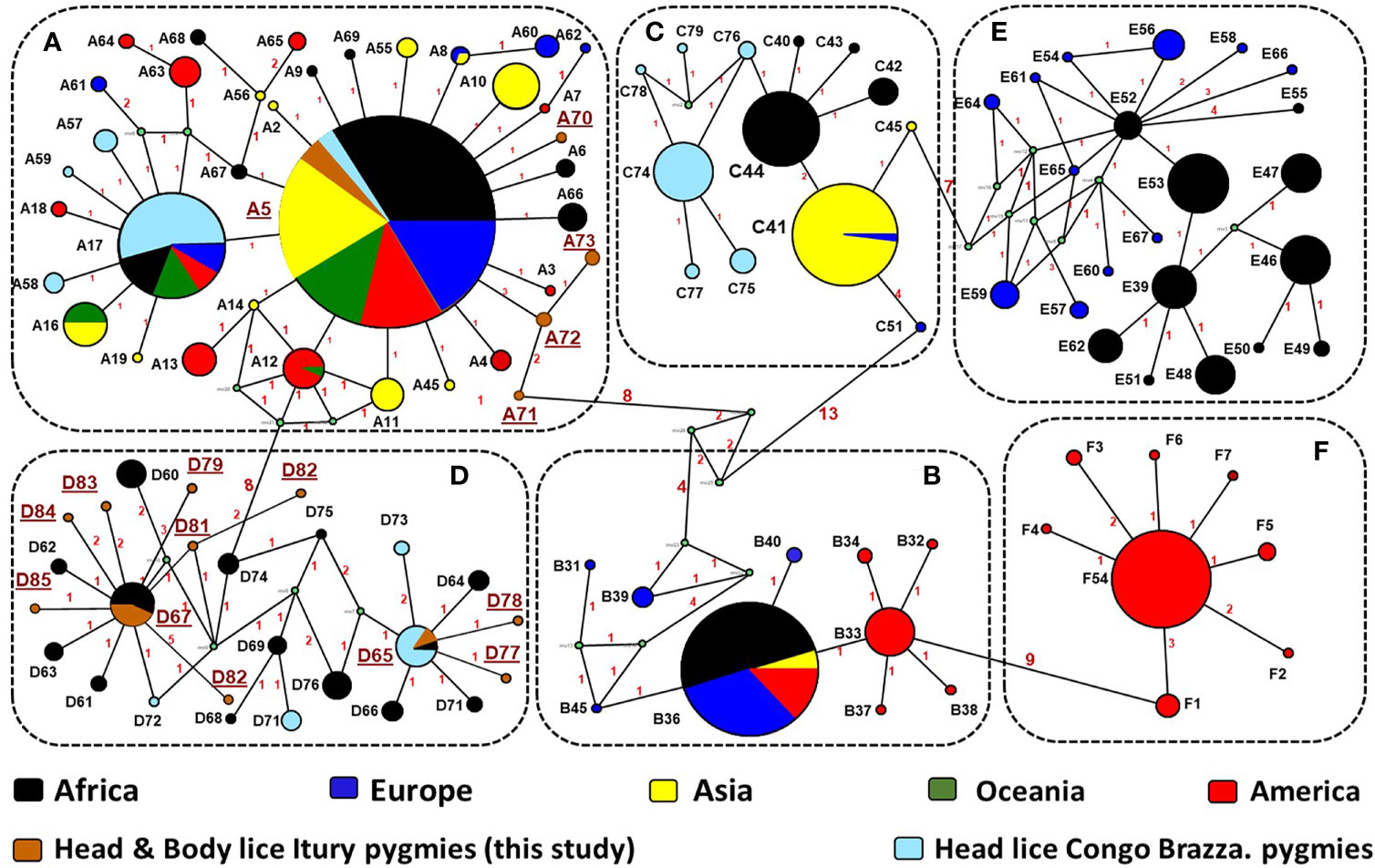
Figure 4 Cytb haplotype networks of Pediculus body and head lice. Each circle indicates a unique haplotype (272 bp) and variations in circle size are proportional to haplotype frequencies. Pie colours and sizes in circles represent the continents and the number of their sequence for a haplotype. The length of the links between nodes is proportional to the number of mutations. The types of haplotypes identified in this study are underlined.
From the 160 cytb sequences of the pygmy lice from Congo Brazzaville (Amanzougaghene et al., 2016) that we analysed, 15 haplotypes were identified (Table 3). Among them only two haplotypes (A5 and D65) were shared with those of Mbuti pygmies, as it was shown in the Figure 3B. These 15 haplotypes are well distributed across three clades, A, D and C, while those of Mbuti pygmy lice belonged only to clades A and D (Figure 3B). The MJ network analysis for all cytb haplotypes corroborated the ML phylogenetic reconstruction 3, 4), with all the cytb sequences were divided across separate clusters represented by six connected subnetworks corresponding to the six know clades A, D, B, C, E and F. Estimates of genetic diversity indices and the results of neutrality tests for all cytb sequences are presented in Table 3. The average number of nucleotide diversity (π), pairwise nucleotide differences (k) and haplotype diversity (Hd) varied among the six clades and among cytb sequences of pygmy lice from Mbuti and Congo Brazzaville. The highest haplotype diversity was found within Mbuti pygmies (Hd= 0.846) when compared to those of Congo Brazzaville pygmies (Hd= 0,773). Within the different clades, the highest haplotype diversity was found in clade D (Hd=0.831), which mainly includes the haplotypes identified in lice from Mbuti and Congo-Brazzaville pygmies (Table 3).
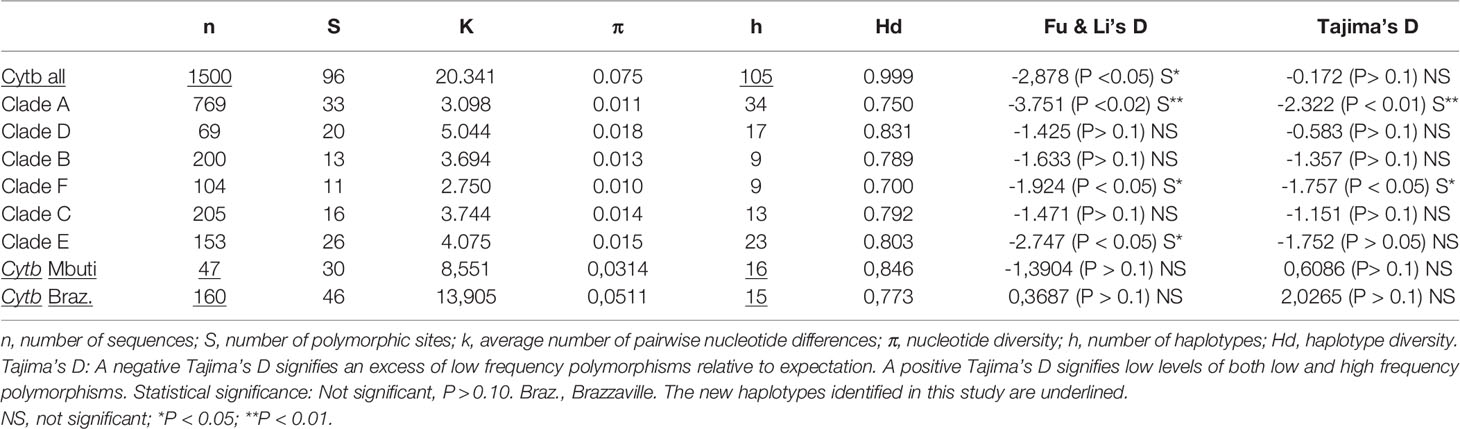
Table 3 Analysis of genetic diversity indices and neutrality tests (Fu & Li’s D and Tajima’s D) on mitochondrial cytb sequences.
In this study we also screened all louse samples for the presence of potential louse-borne pathogens. The results showed that all qPCR investigation of 47 head and body louse samples for Borrelia spp., Rickettsia spp., Anaplasma spp., R. prowazekii, B. quintana, Y. pestis, C. burnetii and Piroplasmida (Theileria spp. and Babesia spp.) spp. produced no positive results. However, we identified R. felis in three lice specimens. The DNA of R. felis (two independent specific qPCRs targeting different genes) was detected in three of 47 (6.4%) louse specimens recovered from two individuals (9.1% of 22) (Patient 3 and Patient 19, see Table 1). Two positive lice were collected from the same patient (Patient 19): one head louse belonging to clade A and one body louse from clade D. The third positive louse also belonged to clade D and was recovered from another patient (Patient 3).
Discussion
To the best of our knowledge, this study reports for the first time molecular data for human lice, genus Pediculus, infesting Mbuti Pygmies living in the Democratic Republic of the Congo (Figure 2). The mtDNA analysis revealed that the 47 lice from Mbuti Pygmy individuals investigated in this work belong to two different cytb haplogroups, A and D, distributed through 16 haplotypes. Clades A and D have already been reported in this country and constitute the two dominant louse lineages in this area (Drali et al., 2015; Boumbanda Koyo et al., 2019). Clade A is globally distributed, while Clade D, has thus far only been found in Central African countries including DR Congo and Congo-Brazzaville, Ethiopia, and Zimbabwe (Amanzougaghene et al., 2016; Amanzougaghene et al., 2019). Clade D is the sister group of clade A from which is thought to have diverged between 0.37 and 0.54 million years ago (Ashfaq et al., 2015; Amanzougaghene et al., 2019).
Although this study includes only a small number of lice (47) and 22 humans. This is mainly due to the fact that these populations of pygmies live in small groups (the average size of a group in Mbuti varies between 15 to 60 individuals), in remote areas of the forest which are very difficult to access. However, it is possible to talk of great genetic variability among these lice. This result is supported by our finding, that, of the 47 Mbuti lice cytb sequences analyzed, sixteen haplotypes were found, of which thirteen were novel haplotypes. Moreover, we were already aware of the intimate history of the coevolution between lice and their human host and so were not surprised by the results of this study. While we did not have the opportunity to carry out genetic analyses of the carriers of these 47 lice, the literature shows that the genetic variability among pygmies is certainly the greatest in the world. They are significantly differentiated, both among themselves and compared to other African populations (Henn et al., 2011; Duda and Jan Zrzavý, 2016). This is especially true, since this study targeted Mbuti pygmies, who are known to be relatively isolated from other pygmy and neighbouring non-pygmy populations (Patin et al., 2009; Verdu and Destro-Bisol, 2012). The results obtained from the phylogenetic analysis of lice from pygmy people from the Republic of the Congo (Congo Brazzaville) have also shown a certain genetic diversity, albeit less pronounced than that observed in Mbuti lice (Amanzougaghene et al., 2016). They are well distributed across two clades, A and C, the two most well distributed clades in Africa (Amanzougaghene et al., 2016). Moreover, as is the case of lice from Itury individuals, they are absent from clades B, E and F but share clade D with them. Of the 11 haplotypes present, eight are specific to them (Figures 3B and 4) (Amanzougaghene et al., 2016). This supports the statement that pygmies in Africa have exceptional genetic diversity (Henn et al., 2011; Duda and Jan Zrzavý, 2016).
Rickettsia felis, the causative agent of flea-borne spotted fever, is a worldwide emerging zoonosis and an important cause of human febrile illness in Africa, including the DR Congo (Mediannikov et al., 2012; Angelakis et al., 2016; Legendre and Macaluso, 2017). The cat flea, Ctenocephalides felis, was considered to be the only confirmed biological vector of R. felis (Angelakis et al., 2016). However, various other arthropods including fleas, mosquitoes, ticks, mites, lice and bed bugs have been found to harbour R. felis (see Supplementary Table S3) (Mediannikov et al., 2014). Moreover, Anopheles gambiae was recently proposed as a potential vector of R. felis (Dieme et al., 2015). In the DR Congo, this bacterium was frequently detected from several arthropods, including C. felis, C. canis, Pulex irritans, Echidnophaga gallinacea, Xenopsylla brasiliensis, Tunga penetrans and Leptopsylla aethiopica (Supplementary Table S3) (Sackal et al., 2008; Mediannikov et al., 2012; Leulmi et al., 2014). In this study, we report for the first time, to the best of our knowledge, the presence of R. felis-DNA in three lice (one head and two body) recovered from two Mbuti pygmies. Although R. prowazekii is the only Rickettsia species known to be naturally associated with human lice, several experimental studies have demonstrated that the body louse may play a role, under favourable epidemiologic circumstances, in the transmission of other Rickettsia species to humans (Raoult and Roux, 1999; Amanzougaghene et al., 2020a). This is the case of R. akari (rickettsial pox) and R. typhi (endemic or murine typhus), which are typically transmitted, respectively, by acari mites and insect fleas, R. conorii (Mediterranean spotted fever, Indian tick typhus) and R. rickettsii (Rocky Mountain spotted fever), both of which are transmitted by ticks (Weyer, 1952b; Houhamdi et al., 2003; Houhamdi and Raoult, 2006b). Moreover, R. typhi has been isolated from body lice infesting sick patients during an outbreak of murine typhus that occurred in northern China and India (Kashmir State) (Liu, 1944; Kalra and Rao, 1951). Furthermore, in the epidemiological study conducted on Malian head lice, the DNA of another Rickettsia, namely R. aeschlimannii, was also detected in 2.5% of 600 head lice collected from 9.4% of 117 tested individuals (Amanzougaghene et al., 2017). Our results from the lice recovered from Mbuti pygmies, together with data from the literature, suggest that the role of human lice in the epidemiology of R. felis should be investigated further.
Conclusion
In conclusion, our finding confirms the presence of clades A and D in body and head lice infesting Mbuti Pygmies living in the Democratic Republic of the Congo. Sixteen haplotypes were found in 47 samples, of which thirteen haplotypes were novel and unique to lice from Mbuti pygmies, indicating an unusually high genetic diversity and reflecting the diversity of their pygmy hosts. Interestingly, we report for the first time the presence of R. felis-DNA from both body and head lice belonged to both clades A and D, which suggest that Pediculus lice can act as a potential vector of this Rickettsia species.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
DR, OM, and RD conceived and designed the experiments. BD and J-CS collected samples. NA conducted the experiments. NA and RD analysed the data. NA, RD, BD, OM, and DR wrote the paper. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This study was supported by the Institut Hospitalo-Universitaire (IHU) Méditerranée Infection, the National Research Agency under the “Investissements d’avenir” programme, reference ANR-10-IAHU-03, the Région Provence-Alpes-Côte d’Azur and European ERDF PRIMI funding.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to the Association pour la recherche en infectiologie (APRI) and IHU Fondation Méditerranée Infection for financial support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.834388/full#supplementary-material
References
Al-Shahrani, S. A., Alajmi, R. A., Ayaad, T. H., Al-Shahrani, M. A., Shaurub, E.-S. H. (2017). Genetic Diversity of the Human Head Lice, Pediculus Humanus Capitis, Among Primary School Girls in Saudi Arabia, With Reference to Their Prevalence. Parasitol. Res. 116, 2637–2643. doi: 10.1007/s00436-017-5570-3
Amanzougaghene, N., Akiana, J., Mongo Ndombe, G., Davoust, B., Nsana, N. S., Parra, H.-J., et al. (2016). Head Lice of Pygmies Reveal the Presence of Relapsing Fever Borreliae in the Republic of Congo. PLoS Negl. Trop. Dis. 10, e0005142. doi: 10.1371/journal.pntd.0005142
Amanzougaghene, N., Fenollar, F., Davoust, B., Djossou, F., Ashfaq, M., Bitam, I., et al. (2019). Mitochondrial Diversity and Phylogeographic Analysis of Pediculus Humanus Reveals a New Amazonian Clade “F.” Infect. Genet. Evol. 70, 1–8. doi: 10.1016/j.meegid.2019.02.006
Amanzougaghene, N., Fenollar, F., Raoult, D., Mediannikov, O. (2020a). Where Are We With Human Lice? A Review of the Current State of Knowledge. Front. Cell. Infect. Microbiol. 9. doi: 10.3389/fcimb.2019.00474
Amanzougaghene, N., Fenollar, F., Sangaré, A. K., Sissoko, M. S., Doumbo, O. K., Raoult, D., et al. (2017). Detection of Bacterial Pathogens Including Potential New Species in Human Head Lice From Mali. PLoS One 12, e0184621. doi: 10.1371/journal.pone.0184621
Amanzougaghene, N., Mediannikov, O., Ly, T. D. A., Gautret, P., Davoust, B., Fenollar, F., et al. (2020b). Molecular Investigation and Genetic Diversity of Pediculus and Pthirus Lice in France. Parasit. Vectors 13 (1), 177. doi: 10.1186/s13071-020-04036-y
Angelakis, E., Mediannikov, O., Parola, P., Raoult, D. (2016). Rickettsia Felis: The Complex Journey of an Emergent Human Pathogen. Trends Parasitol. 32, 554–564. doi: 10.1016/j.pt.2016.04.009
Ascunce, M. S., Fane, J., Kassu, G., Toloza, A. C., Picollo, M. I., González-Oliver, A., et al. (2013). Mitochondrial Diversity in Human Head Louse Populations Across the Americas. Am. J. Phys. Anthropol. 152, 118–129. doi: 10.1002/ajpa.22336
Ashfaq, M., Prosser, S., Nasir, S., Masood, M., Ratnasingham, S., Hebert, P. D. N. (2015). High Diversity and Rapid Diversification in the Head Louse, Pediculus Humanus (Pediculidae: Phthiraptera). Sci. Rep. 5, 14188. doi: 10.1038/srep14188
Ayyadurai, S., Sebbane, F., Raoult, D., Drancourt, M. (2010). Body Lice, Yersinia Pestis Orientalis, and Black Death. Emerg. Infect. Dis. 16, 892–893. doi: 10.3201/eid1605.091280
Bandelt, H. J., Forster, P., Röhl, A. (1999). Median-Joining Networks for Inferring Intraspecific Phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Batini, C., Lopes, J., Behar, D. M., Calafell, F., Jorde, L. B., van der Veen, L., et al. (2011). Insights Into the Demographic History of African Pygmies From Complete Mitochondrial Genomes. Mol. Biol. Evol. 28, 1099–1110. doi: 10.1093/molbev/msq294
Becker, N. S. A., Verdu, P., Froment, A., Le Bomin, S., Pagezy, H., Bahuchet, S., et al. (2011). Indirect Evidence for the Genetic Determination of Short Stature in African Pygmies. Am. J. Phys. Anthropol. 145, 390–401. doi: 10.1002/ajpa.21512
Boumbanda Koyo, C. S., Amanzougaghene, N., Davoust, B., Tshilolo, L., Lekana-Douki, J. B., Raoult, D., et al. (2019). Genetic Diversity of Human Head Lice and Molecular Detection of Associated Bacterial Pathogens in Democratic Republic of Congo. Parasit. Vectors 12, 290. doi: 10.1186/s13071-019-3540-6
Boumbanda-Koyo, C. S., Mediannikov, O., Amanzougaghene, N., Oyegue-Liabagui, S. L., Imboumi-Limoukou, R. K., Raoult, D., et al. (2020). Molecular Identification of Head Lice Collected in Franceville (Gabon) and Their Associated Bacteria. Parasit. Vectors 13 (1), 410. doi: 10.1186/s13071-020-04293-x
Boutellis, A., Bitam, I., Fekir, K., Mana, N., Raoult, D. (2015). Evidence That Clade A and Clade B Head Lice Live in Sympatry and Recombine in Algeria. Med. Vet. Entomol. 29, 94–98. doi: 10.1111/mve.12058
Campbell, M. C., Tishkoff, S. A. (2010). The Evolution of Human Genetic and Phenotypic Variation in Africa. Curr. Biol. 20, R166–R173. doi: 10.1016/j.cub.2009.11.050
Candy, K., Amanzougaghene, N., Izri, A., Brun, S., Durand, R., Louni, M., et al. (2018). Molecular Survey of Head and Body Lice, Pediculus Humanus, in France. Vector. Borne. Zoonotic. Dis. 18 (5), 243–251. doi: 10.1089/vbz.2017.2206
Destro-Bisol, G., Coia, V., Boschi, I., Verginelli, F., Cagliá, A., Pascali, V., et al. (2004). The Analysis of Variation of mtDNA Hypervariable Region 1 Suggests That Eastern and Western Pygmies Diverged Before the Bantu Expansion. Am. Nat. 163, 212–226. doi: 10.1086/381405
Dieme, C., Bechah, Y., Socolovschi, C., Audoly, G., Berenger, J.-M., Faye, O., et al. (2015). Transmission Potential of Rickettsia Felis Infection by Anopheles Gambiae Mosquitoes. Proc. Natl. Acad. Sci. U. S. A. 112, 8088–8093. doi: 10.1073/pnas.1413835112
Drali, R., Abi-Rached, L., Boutellis, A., Djossou, F., Barker, S. C., Raoult, D. (2016). Host Switching of Human Lice to New World Monkeys in South America. Infect. Genet. Evol. 39, 225–231. doi: 10.1016/j.meegid.2016.02.008
Drali, R., Boutellis, A., Raoult, D., Rolain, J. M., Brouqui, P. (2013). Distinguishing Body Lice From Head Lice by Multiplex Real-Time PCR Analysis of the Phum_PHUM540560 Gene. PLoS One 8, e58088. doi: 10.1371/journal.pone.0058088
Drali, R., Shako, J.-C., Davoust, B., Diatta, G., Raoult, D. (2015). A New Clade of African Body and Head Lice Infected by Bartonella Quintana and Yersinia Pestis-Democratic Republic of the Congo. Am. J. Trop. Med. Hyg. 93, 990–993. doi: 10.4269/ajtmh.14-0686
Duda, P., Jan Zrzavý, n. (2016). Human Population History Revealed by a Supertree Approach. Sci. Rep. 6, 29890. doi: 10.1038/srep29890
Giroud, P., Jadin, J. (1954). Infection Latente Et Conservation De “Rickettsia Burnetii” Chez L’homme, Le Rôle Du Pou. Bull. Soc Pathol. Exotique. 47, 764–765.
Goldberger, J., Anderson, J. F. (1912). The Transmission of Typhus Fever, With Especial Reference to Transmission by the Head Louse (Pediculus Capitis). Public Health Rep. (1896-1970). 27, 297–307. doi: 10.2307/4567527
Hammer, M. F., Woerner, A. E., Mendez, F. L., Watkins, J. C., Wall, J. D. (2011). Genetic Evidence for Archaic Admixture in Africa. Proc. Natl. Acad. Sci. U. S. A. 108, 15123–15128. doi: 10.1073/pnas.1109300108
Hammoud, A., Louni, M., Baldé, M. C., Beavogui, A. H., Gautret, P., Raoult, D., et al. (2021). Molecular Characterization and Genetic Diversity of Haplogroup E Human Lice in Guinea, West Africa. Microorganisms 9, 257. doi: 10.3390/microorganisms9020257
Henn, B. M., Gignoux, C. R., Jobin, M., Granka, J. M., Macpherson, J. M., Kidd, J. M., et al. (2011). Hunter-Gatherer Genomic Diversity Suggests a Southern African Origin for Modern Humans. Proc. Natl. Acad. Sci. U. S. A. 108, 5154–5162. doi: 10.1073/pnas.1017511108
Houhamdi, L., Fournier, P.-E., Fang, R., Raoult, D. (2003). An Experimental Model of Human Body Louse Infection With Rickettsia Typhi. Ann. N. Y. Acad. Sci. 990, 617–627. doi: 10.1111/j.1749-6632.2003.tb07436.x
Houhamdi, L., Lepidi, H., Drancourt, M., Raoult, D. (2006). Experimental Model to Evaluate the Human Body Louse as a Vector of Plague. J. Infect. Dis. 194, 1589–1596. doi: 10.1086/508995
Houhamdi, L., Raoult, D. (2006a). Experimental Infection of Human Body Lice With Acinetobacter Baumannii. Am. J. Trop. Med. Hyg. 74, 526–531.
Houhamdi, L., Raoult, D. (2006b). Experimentally Infected Human Body Lice (Pediculus Humanus Humanus) as Vectors of Rickettsia Rickettsii and Rickettsia Conorii in a Rabbit Model. Am. J. Trop. Med. Hyg. 74, 521–525.
Huelsenbeck, J. P., Rannala, B. (1997). Phylogenetic Methods Come of Age: Testing Hypotheses in an Evolutionary Context. Science 276, 227–232. doi: 10.1126/science.276.5310.227
Jakobsson, M., Scholz, S. W., Scheet, P., Gibbs, J. R., VanLiere, J. M., Fung, H.-C., et al. (2008). Genotype, Haplotype and Copy-Number Variation in Worldwide Human Populations. Nature 451, 998–1003. doi: 10.1038/nature06742
Jarvis, J. P., Scheinfeldt, L. B., Soi, S., Lambert, C., Omberg, L., Ferwerda, B., et al. (2012). Patterns of Ancestry, Signatures of Natural Selection, and Genetic Association With Stature in Western African Pygmies. PLoS Genet. 8, e1002641. doi: 10.1371/journal.pgen.1002641
Kalra, S. L., Rao, K. N. A. (1951). Typhus Fevers In Kashmir State. Part II. Murine Typhus. Indian J. Med. Res. 39, 297–302.
Legendre, K. P., Macaluso, K. R. (2017). Rickettsia Felis: A Review of Transmission Mechanisms of an Emerging Pathogen. Trop. Med. Infect. Dis. 2 (4), 64. doi: 10.3390/tropicalmed2040064
Leulmi, H., Socolovschi, C., Laudisoit, A., Houemenou, G., Davoust, B., Bitam, I., et al. (2014). Detection of Rickettsia Felis, Rickettsia Typhi, Bartonella Species and Yersinia Pestis in Fleas (Siphonaptera) From Africa. PLoS Negl. Trop. Dis. 8 (10), e3152. doi: 10.1371/journal.pntd.0003152
Librado, P., Rozas, J. (2009). DnaSP V5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Light, J. E., Allen, J. M., Long, L. M., Carter, T. E., Barrow, L., Suren, G., et al. (2008). Geographic Distributions and Origins of Human Head Lice (Pediculus Humanus Capitis) Based on Mitochondrial Data. J. Parasitol. 94, 1275–1281. doi: 10.1645/GE-1618.1
Li, W., Ortiz, G., Fournier, P.-E., Gimenez, G., Reed, D. L., Pittendrigh, B., et al. (2010). Genotyping of Human Lice Suggests Multiple Emergencies of Body Lice From Local Head Louse Populations. PLoS Negl. Trop. Dis. 4, e641. doi: 10.1371/journal.pntd.0000641
Liu, W. T. (1944). Studies on the Murine Origin of Typhus Epidemics in North China. 3. Isolation of Murine Typhus Rickettsia From Rats, Rat-Fleas and Body-Lice of Patients During an Epidemic in a Poor House. Chin. Med. J. 62, 119–139. doi: 10.5555/cmj.0366-6999.62.02.p119.01
Louni, M., Mana, N., Bitam, I., Dahmani, M., Parola, P., Fenollar, F., et al. (2018). Body Lice of Homeless People Reveal the Presence of Several Emerging Bacterial Pathogens in Northern Algeria. PLoS Negl. Trop. Dis. 12, e0006397. doi: 10.1371/journal.pntd.0006397
Mediannikov, O., Aubadie-Ladrix, M., Raoult, D. (2014). Candidatus ‘Rickettsia Senegalensis’ in Cat Fleas in Senegal. New Microbes New Infect. 3, 24–28. doi: 10.1016/j.nmni.2014.10.005
Mediannikov, O., Davoust, B., Socolovschi, C., Tshilolo, L., Raoult, D., Parola, P. (2012). Spotted Fever Group Rickettsiae in Ticks and Fleas From the Democratic Republic of the Congo. Ticks. Tick. Borne. Dis. 3, 371–373. doi: 10.1016/j.ttbdis.2012.10.015
Murray, E. S., Torrey, S. B. (1975). Virulence of Rickettsia Prowazeki for Head Lice. Ann. N. Y. Acad. Sci. 266, 25–34. doi: 10.1111/j.1749-6632.1975.tb35086.x
Ohenjo, N., Willis, R., Jackson, D., Nettleton, C., Good, K., Mugarura, B. (2006). Health of Indigenous People in Africa. Lancet 367, 1937–1946. doi: 10.1016/S0140-6736(06)68849-1
Patin, E., Laval, G., Barreiro, L. B., Salas, A., Semino, O., Santachiara-Benerecetti, S., et al. (2009). Inferring the Demographic History of African Farmers and Pygmy Hunter-Gatherers Using a Multilocus Resequencing Data Set. PLoS Genet. 5, e1000448. doi: 10.1371/journal.pgen.1000448
Piarroux, R., Abedi, A. A., Shako, J.-C., Kebela, B., Karhemere, S., Diatta, G., et al. (2013). Plague Epidemics and Lice, Democratic Republic of the Congo. Emerging. Infect. Dis. 19, 505–506. doi: 10.3201/eid1903.121542
Posada, D., Buckley, T. R. (2004). Model Selection and Model Averaging in Phylogenetics: Advantages of Akaike Information Criterion and Bayesian Approaches Over Likelihood Ratio Tests. Syst. Biol. 53, 793–808. doi: 10.1080/10635150490522304
Posada, D., Crandall, K. A. (1998). MODELTEST: Testing the Model of DNA Substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Previte, D., Olds, B. P., Yoon, K., Sun, W., Muir, W., Paige, K. N., et al. (2014). Differential Gene Expression in Laboratory Strains of Human Head and Body Lice When Challenged With Bartonella Quintana, a Pathogenic Bacterium. Insect Mol. Biol. 23, 244–254. doi: 10.1111/imb.12077
Raoult, D. (2016). A Personal View of How Paleomicrobiology Aids Our Understanding of the Role of Lice in Plague Pandemics. Microbiol. Spectr. 4, 4. doi: 10.1128/microbiolspec.PoH-0001-2014
Raoult, D., Roux, V. (1999). The Body Louse as a Vector of Reemerging Human Diseases. Clin. Infect. Dis. 29, 888–911. doi: 10.1086/520454
Reed, D. L., Smith, V. S., Hammond, S. L., Rogers, A. R. (2004). And Clayton, DGenetic Analysis of Lice Supports Direct Contact Between Modern and Archaic Humans. H. PLoS. Biol. 2, e340. doi: 10.1371/journal.pbio.0020340
Sackal, C., Laudisoit, A., Kosoy, M., Massung, R., Eremeeva, M. E., Karpathy, S. E., et al. (2008). Bartonella Spp. And Rickettsia Felis in Fleas, Democratic Republic of Congo. Emerg. Infect. Dis. 14, 1972–1974. doi: 10.3201/eid1412.080610
Sunantaraporn, S., Sanprasert, V., Pengsakul, T., Phumee, A., Boonserm, R., Tawatsin, A., et al. (2015). Molecular Survey of the Head Louse Pediculus Humanus Capitis in Thailand and Its Potential Role for Transmitting Acinetobacter Spp. Parasit. Vectors 8, 127. doi: 10.1186/s13071-015-0742-4
Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Verdu, P., Destro-Bisol, G. (2012). African Pygmies, What’s Behind a Name? Hum. Biol. 84, 1–10. doi: 10.3378/027.084.0105
Weyer, F. (1952a). [Artificial Transmission of Rickettsia Rickettsii (Rocky Mountain Spotted Fever) on Insects, Especially on Body Lice]. Z. Hyg. Infektionskr. 135, 280–297.
Keywords: Mbuti pygmies, Pediculus humanus, genetic diversity, Rickettsia felis, Democratic Republic of Congo
Citation: Amanzougaghene N, Drali R, Shako J-C, Davoust B, Fenollar F, Raoult D and Mediannikov O (2022) High Genetic Diversity and Rickettsia felis in Pediculus humanus Lice Infesting Mbuti (pygmy people), -Democratic Republic of Congo. Front. Cell. Infect. Microbiol. 12:834388. doi: 10.3389/fcimb.2022.834388
Received: 14 December 2021; Accepted: 08 February 2022;
Published: 02 March 2022.
Edited by:
Erika Ildiko Lutter, Oklahoma State University, United StatesReviewed by:
Andrei Daniel Mihalca, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaHans-Peter Fuehrer, University of Veterinary Medicine Vienna, Austria
Copyright © 2022 Amanzougaghene, Drali, Shako, Davoust, Fenollar, Raoult and Mediannikov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oleg Mediannikov, b2xlZ3Vzc3MxQGdtYWlsLmNvbQ==
 Nadia Amanzougaghene
Nadia Amanzougaghene Rezak Drali
Rezak Drali Jean-Christophe Shako4
Jean-Christophe Shako4 Florence Fenollar
Florence Fenollar Didier Raoult
Didier Raoult Oleg Mediannikov
Oleg Mediannikov